Abstract
The conditions supporting the generation of microglia-like cells in the central nervous system (CNS) after transplantation of hematopoietic stem/progenitor cells (HSPC) have been studied to advance the treatment of neurodegenerative disorders. Here, we explored the transplantation efficacy of different cell subsets and delivery routes with the goal of favoring the establishment of a stable and exclusive engraftment of HSPCs and their progeny in the CNS of female mice. In this setting, we show that the CNS environment drives the expansion, distribution and myeloid differentiation of the locally transplanted cells towards a microglia-like phenotype. Intra-CNS transplantation of HSPCs engineered to overexpress TREM2 decreased neuroinflammation, Aβ aggregation and improved memory in 5xFAD female mice. Our proof of concept study demonstrates the therapeutic potential of HSPC gene therapy for Alzheimer’s disease.
Similar content being viewed by others
Introduction
Early during development, a group of myeloid hematopoietic progenitors integrates into the central nervous system (CNS) to provide life-long support as microglia1,2. This cell population exerts crucial functions for the maintenance of brain tissue homeostasis during development as well as in adulthood, both in physiological and pathological conditions3. In fact, microglia cells actively define and reshape the neuronal network by surveying the neuronal microenvironment, pruning synaptic spines, and functionally contributing to neuroinflammation4,5. Adult microglia is a self-maintained population, but following discrete myeloablation and infusion of hematopoietic stem and progenitor cells (HSPCs), brain-resident microglia leaves space for transplant-derived myeloid cells that can, in appropriate conditions, arise and settle in the CNS, progressively acquiring morphologic, antigenic and functional features that render them similar to microglia6,7,8,9,10.
During the last decade, the contribution of HSPCs to CNS-associated myeloid cell populations upon transplantation has been deeply investigated and exploited for therapeutic purposes, demonstrating unprecedented therapeutic potential in treating monogenic neurometabolic disorders in gene therapy proof of concept preclinical studies and clinical trials11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26. In these settings, the engineered microglia-like cells progressively appearing in the CNS of transplant recipients act as vehicle for disease‐modulating therapeutics across the blood brain barrier (BBB) and contribute, with scavenging and immune surveillance functions, to the modulation of the CNS neurotoxic environment towards neuroprotection27,28,29. However, the benefit of the procedure is still limited by the slow pace and extent at which cell repopulation in different CNS regions occur. To address these limitations, the mechanisms and the conditions favoring the engraftment and generation of microglia-like cells in the CNS have been extensively studied.
Others and us demonstrated that the engraftment of transplant-derived cells in the CNS is highly favored by and strictly dependent on the recipient’s pre-transplant exposure to a myeloablative regimen based on the alkylating agent busulfan (BU) that alters the brain environment, creating space and opportunity for the local engraftment and proliferation of the transplanted cells and their progeny7,30,31. We also showed that a fraction of the HSPCs that are intravenously (IV) injected in BU-treated recipients enters the CNS shortly after transplant and proliferates locally, possibly contributing to long-term CNS hematopoietic donor chimerism7. This process is enhanced and favored when HSPCs are directly infused in the brain by intracerebroventricular injection (ICV)6.
Here, we challenge further the standard hematopoietic transplantation paradigm by exploiting and combining different HSPC subsets differentially enriched in long-term hematopoietic reconstituting potential, and by exploring different cell-infusion routes to eventually favor the establishment of a robust and exclusive donor engraftment in the CNS of recipients. In this setting, we unveil a key role exerted by the CNS environment in uniquely driving expansion, distribution and myeloid differentiation of the locally transplanted HSPCs towards a microglia-like phenotype. Importantly, we then test the therapeutic potential of engineered HSPC transplantation in the CNS in an established murine model of Alzheimer’s disease (AD) obtaining a clear example of efficacy of HSPC gene therapy in an adult onset, multifactorial neurodegenerative disease, and a proof of benefit associated with an innovative approach that determines a CNS-restricted engraftment of the genetically modified transplant progeny cells.
Results
Competitive engraftment potential of intra-CNS transplanted HSPCs
Administration of engineered hematopoietic stem and progenitor cells (HSPCs) in the lateral cerebral ventricles (ICV) of lethally myeloablated (BU-based regimen) mice can result in robust CNS-restricted myeloid engraftment and generation of microglia-like cells6. To study the mechanisms leading to the engraftment and differentiation of the CNS-transplanted HSPCs, we performed competitive transplantation experiments combining the ICV delivery of CD34+-equivalent lineage negative (Lin-) murine HSPCs with the simultaneous or delayed (after 5 days) intravenous (IV) administration of hematopoietic cell subsets that are differentially enriched in long-term hematopoietic reconstituting activity, namely total bone marrow (BM), Lin- cells and c-kit+ Sca-1+ stem cell-enriched Lin- (KSL) cells. Moreover, we explored a new intra-CNS delivery approach in the intrathecal lumbar (ITL) space as alternative to the ICV route. To discriminate the differential contribution of the transplanted and recipient cells, map their progeny and obtain relevant insights on the mechanism of HSPC engraftment and differentiation in the CNS, we took advantage of the CD45.1 (donor)/CD45.2 (recipient) allele mismatch and we differentially labeled the donor cells according to the route of administration, by using lentiviral vectors (LVs) encoding for different reporter genes (GFP for CNS-delivered cells and ∆NGFR or Cherry for IV-delivered cells). Wild type CD45.2 C57BL/6 recipients were pre-treated with BU used at myeloablative dose (25 mg/kg, from days −4 to days −1) to promote myeloid cell turnover with donor cells in the CNS7. Transplanted mice were then euthanized at different time points after the transplant and the engraftment of the donor cells in the hematopoietic tissues and CNS was assessed by flow cytometry (FC) and immunofluorescence (IF) (Fig. S1a, b). For each time point, BU-treated mice transplanted IV with ∆NGFR/Cherry Lin- cells were used as term of comparison.
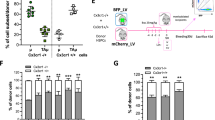
Interestingly, in all the combinations tested, we observed a high competition (in terms of extent of repopulation of the CNS compartment) between the progenies of the HSPCs transplanted through the intra-CNS versus the IV route (Fig. 1a–c and Tables S1, S2; Fig. S1c, e and f and Tables S3, S4). In particular, the contribution of the CNS-injected HSPCs exceeded that of the IV transplanted cells in all the tested conditions, although the number of HSPCs injected in CNS was substantially lower than those administered IV (0.3 × 106 vs 1 × 106, respectively) (Fig. 1a–c and Tables S1, S2; Fig. S1c, e and f and Table S1, S2). The highest competition between the IV and the intra-CNS transplanted cells was observed when Lin- cells were employed at both sites of injection, highlighting the ability of this cell population to efficiently occupy CNS niches upon both systemic and intra-CNS delivery (Fig. 1a–c and Tables S1, S2 and Fig. S1c, e and Tables S3, S4). Similar findings were observed in the spinal cord (Fig. 1a–c and Tables S1, S2; Fig. S1c, e and Tables S3, S4) with greater representation of the progeny of CNS-delivered HSPCs versus the IV-transplanted counterpart, particularly at 45 days post-transplant (post-Tx) and especially when ITL was used for intra-CNS delivery (Fig. 1a). Notably, and at odds with recent reports32, the combination of intra-CNS and IV HSPC transplantation resulted in an overall enhancement of the myeloid donor chimerism in the brain and in the spinal cord of transplant recipients, as compared to the chimerism achieved upon IV-only cell administration (Fig. 1a, b and Fig. S1e, f). This observation was confirmed by employing both the ICV and ITL routes for HSPCs delivery (Fig. 1a–c). The increased chimerism was observed as early as 45 days post-Tx and up to 90 days post-Tx, independently from the time of IV cell delivery (simultaneous vs delayed) and the cell population employed for IV (Fig. 1a, b and Fig. S1c, e and f). The difference was no longer observed at the longest tested time point, 240 days post-Tx, when the chimerism of the transplanted cell progeny reached a plateau (Fig. 1c and S1e,f). The competitive advantage of intra-CNS transplanted cells versus their IV infused counterpart, and the increased chimerism associated to combined intra-CNS and IV cell delivery were observed within both immature, transiently amplifying, CD45hiCD11b+ microglia-like cells (TAµ) and, even more significantly, within CD45lowCD11b+ cells (previously defined microglia like, µ) that according to our previously work share a large portion of the phenotypic and transcriptional features of mature microglia7 (Figs. 1d, S1d and Tables S3, S4).
Histograms showing the donor-cell chimerism within the CD45+/CD11b+ cell population in the Brain [BR], Spinal Cord [SC] and Bone marrow [BM] at 45 (a), 90 (b), and 240 (c) days post transplant (pTX). The progeny of intra-CNS transplanted cells (CNS Inj.) either ICV or ITL (as indicated), is shown in green, whereas the progeny of IV administered cells (IV inj.) are shown in blue. On the x-axis different hematopoietic subpopulations used in each combinatorial setting are shown: BM = Bone Marrow; Lin = Lineage negative cells; KSL = c-kit+, Sca-1+ Lin- cells. d Frequency of ICV (green) and IV delivered cells (blue) within the indicated brain subpopulations (μ and TAμ) at 90 days pTX. Representative dot plots showing the gating strategy used to identify μ and TAμ subpopulations are shown on the left. Histograms (a–d) show the mean ± SEM of mice at 45 days pTX [n = 8 IV Lin, n = 9 ICV Lin IV Lin, n = 7 ICV Lin IV BM, n = 9 ITL Lin IV BM mice, n = 3 experimental replicates], 90 days [n = 11 IV Lin, n = 10 ICV Lin IV Lin and ICV Lin IV BM, n = 7 ICV Lin IV KSL, n = 5 ITL Lin IV BM mice, n = 4 experimental replicates] and 240 days [n = 4 mice/group, n = 2 experimental replicates]. Statistics for a–d: 1-way ANOVA followed by Tukey post-hoc test for comparisons between the contribution of all donor-derived cells (CNS inj. + IV inj.) among the transplant settings; for additional statistical information refer to Tables S1 and S2. Source data (a–d) are provided as a Source Data file. e, f Representative laser scanning confocal microscope microphotographs of IF-stained brain sections from mice co-transplanted with Lin- both ICV and IV, at 90 days pTX [n = 5 slices from n = 3 mice]. ICV-derived cells (green signal) are stained with (e) anti-GFP antibody, IV-derived cells (red signal) with anti- ΔNGFR antibody, DAPI for nuclei (blue), and (f) Iba1 for microglia cells (arrows indicate representative co-localizations of Iba1 and GFP/ΔNGFR signals). CTX cortex, OB Olfactory Bulb, STR striatum, HPC hippocampus, CB Cerebellum. Magnification 40x. M= merge. Images were acquired at Leika SP2 confocal microscope. Scale bar (e, f) = 50 µm.
Interestingly, while both (ICV and ITL) of the intra-CNS routes of HSPC administration similarly enhanced CNS donor chimerism, they differentially contributed to marrow hematopoiesis. Indeed, while ICV injected cells did not contribute to the reconstitution of marrow hematopoiesis, as previously described6 (Figs. 1a–c and S1c), the ITL-delivered HSPCs robustly engrafted in the marrow of ITL + IV co-transplanted mice (Fig. 1a, b) with relevant translational implications.
Importantly, independently from the cell source and the setting applied, transplant-derived cells extensively colonized the brain and showed morphological and antigenic features reminiscent of endogenous microglia at confocal analysis, as documented in representative pictures at 90 days post transplant, by the presence of thin ramifications and by the expression of microglia antigens (Fig. 1e, f).
Route-specific cell distribution in the CNS after HSPC trasplantation
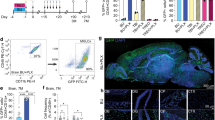
Based on the evidence that intra-CNS administration of HSPCs favors the medium to long-term CNS engraftment of their progeny, we next assessed whether the route of HSPC administration could also affect the pattern of distribution of their cell progeny in the CNS. To this goal, we first performed a short-term monitoring of cell biodistribution in the hematopoietic and CNS compartments of busulfan-treated wild type mice transplanted IV, ICV or ITL with GFP-labeled HSPCs and sacrified at 1, 5 and 14 days post injection (N = 3 mice/time point/route). Bone marrow, brain and spinal cord were collected and processed for FC and IF analysis (CNS only)(Fig. 2a). At the earliest time points ( + 1 and +5 days), the donor cells were clearly measurable only in the brain of ICV transplanted mice, while by day 14 discrete levels of CNS engraftment were observed in all the tested conditions. Based on these data, day 14 was chosen as the earliest timepoint to comparatively study the biodistribution of the CNS engrafted cells in the mice transplanted with the different routes by IF analysis. To this scope, we also analyzed animals at +30, 45 and 90 days post-transplant to follow the cell distribution over time, with the animals examined at >30 days post-transplant having received also Cherry_LV-labeled HSPCs IV, in addition to the GFP-labeled counterpart administered ICV or ITL, for assuring long-term survival after myeloablation (Fig. 2b–e). In the latter setting, the biodistribution data were thus obtained in the more challenging competitive setting.
a Short-term HSPC biodistribution in the bone marrow (BM), brain (BR) and spinal cord (SC) at 1, 5 and 14 days post-transplant (pTX) in IV (black), ICV (green) and ITL ( pink) transplanted mice as measured by flow cytometry [n = 3 mice/group across n = 2 experimental replicates]. Data are shown as mean ± SEM. 2-way ANOVA followed by Tukey post-hoc test pTX (p* = 0.0351, p**** < 0.0001). Source data (a) are provided as a Source Data file. Distribution of GFP+ donor-derived cells analyzed at 14 days (b) and 30 days pTX (c) in the brain of representative IV, ICV or ITL transplanted mice (single transplantations) [n = 4 slices from n = 3 mice/group]. Images were acquired at 25x magnification using Leica SP5 confocal microscopy after tissues clearing. Distribution of donor-derived cells, labeled with GFP (green signal) or with ΔNGFR/Cherry (blue signal), in representative mice transplanted with Lin- HSPCs via ICV + IV or ITL + IV or IV only delivery (as indicated) and analyzed at 45 (d) [n = 4 slices from n = 3 mice/group] or 90 days pTX (e) [n = 3 slices from n = 4 mice/group]. Images were acquired at Delta Vision Olympus Ix70 for brain reconstruction. The corresponding masks (b–e) were generated accordingly using Fiji software. Scale bar = 1000 µm. f Graphs showing the proportion of the total area covered by the progeny of IV injected cells, labeled with ΔNGFR or Cherry (blue bars), vs the progeny of cells injected ICV or ITL, and labeled with GFP of mice sacrified at 90 days pTX [n = 4 mice/group]. g High resolution confocal z-stacks of selected brain regions (with 5 µm step size) of IV, ICV or ITL transplanted mice at the indicated time points [n = 3 slices from n = 2 mice/group]. GFP-labeled cells are shown in green, DAPI nuclear stain is shown in blue. Images were acquired at 25x magnification and processed using Fiji open source software. Scale bar = 50 µm. CTX cortex, OB olfactory bulb, FB forebrain, CC corpus callosum, HPC hippocampus, Lv lateral ventricle, THA thalamus, HY hypothalamus, MB midbrain, PO pons, CB cerebellum, IIIv third ventricle, IVv fourth ventricle.
Interestingly, depending on the route of cell administration, we observed a differential distribution of the GFP+cells in the analyzed CNS tissues. At day 14, the progeny of the ICV-transplanted HSPCs was mostly detectable in the close proximity of the ventricular structure, in the septum and projecting rostrally and towards the basal forebrain, and a few additional clustered cells were observed in the midbrain and pons (Fig. 2b). Although the time point of observation was very precoucious, already discrete branchings of the GFP+ cells were visible suggesting an actively occurring differentiation process (Fig. 2g). At day 30, the GFP+ cells exhibited a preferential distribution across brain regions surrounding the cerebral ventricular system, exploiting a trajectory which may suggest proximity to the rostral migratory stream, and confirming the pattern of distribution observed at 14 days. Indeed, clustered cells were also observed in the proximity of the third and fourth ventricles, in the diencephalon and in the hindbrain, from which they distributed into nearby regions, colonizing the striatum, the anterior olfactory area and the brainstem (Fig. 2c). At these early time points, the distribution of the GFP+ cells in the ITL setting was substantially different. At the initial time point ( + 14 days), only a limited quantity of donor cells was documented in the brain by FC and, similarly, only a few and scattered GFP+ cells were identified by IF, with no evidence of clustering. In the spinal cord, we failed to identify a pattern, as the few detected GFP+ cells were found in different locations without any obvious clustering. By day 30, the GFP+ cells were mostly localized in the olfactory bulb and forebrain regions (Fig. 2c). A few clusters of cells could also be observed in the brainstem, consistent with its proximity to the spinal cord. Unlike the ICV route, no cells were retrieved in the subventricular zone and lateral ventricle regions. In the IV transplant setting, at day 14, very few donor/GFP+ cells were detected by FC analysis (Fig. 2a) and IF (Fig. 2g) in the brain and spinal cord of transplant recipients. These few cells showed no specific clustering and were mostly localized in the proximity of meninges and blood vessels. Unlike what was observed in ICV-transplanted mice, the GFP+ cells here displayed a round shape and no branchings, a phenotype likely consistent with immature cells (Fig. 2g). By day 30, clusters of GFP+ cells could be observed in the hippocampus, suggesting a specific recruitment of IV injected cells in this region (Fig. 2c). A few cells were also detected in the forebrain region and in the brainstem, likely infiltrating throughout the brain parenchyma from the meninges and blood vessels (Fig. 2c). The cells started to present a ramified morphology at this time point (Fig. 2g).
The analysis at later time points (i.e. 45 days post-transplant) demonstrated the presence of an abundant donor cell progeny in the brains of the transplanted animals. This suggests a robust expansion of the graft that continued up to 90 days, independently of the transplantation route applied. In the competitive settings, there was an overall relative higher frequency of the intra-CNS versus IV transplanted cell progeny in the brain (Figs. 2d, e and S2a). Interestingly, a route-specific cell distribution was confirmed also in this case. Indeed, at 45 days, we again observed that the ICV cell progeny in the brain was spreading rostrally, in a trajectory in the proximity of the rostral migratory stream, with many cells localized particularly in the forebrain and olfactory region. Notably, GFP+ cells expanded in regions that were poorly colonized at the shorter time points, as the cortex and the cerebellum, unlike IV injected Cherry+ cells whose contribution remained negligible in these areas (Figs. 2d and S2a, f). At +90 days post-transplant, the IV-transplanted cell progeny mostly localized in the thalamus, midbrain, pons and cerebellum, whereas the ICV cell progeny expanded across all the other regions, with still a preference in colonizing the cortex (as at +45 days), the hippocampus, the corpus callosum, and the olfactory bulbs (Figs. 2e and S2a). A unique distribution pattern of the IV versus intra-CNS transplanted cell progeny was observed also in the ITL + IV setting (Figs. 2c–e and S2a). While the Cherry+ progeny of the IV-transplanted cells expanded starting from the hippocampus to the cortex and the olfactory bulb, confirming the observations at the earlier time points, the ITL cell progeny preferentially colonized the hypothalamus, the striatum, the thalamus, the midbrain regions and the pons, adopting a mutually exclusive distribution compared to their IV-injected counterparts. When HSPCs were transplanted only IV, with no intra-CNS HSPC co-delivery, their progeny distributed also across regions occupied, in the co-transplanted mice, by the ICV-/ITL-transplanted cell progeny, as the forebrain, the corpus callosum, the hippocampus and the hypothalamus, indicating the physiological tendency of cells to migrate also in these regions upon transplantation (Fig. 2c–e and S2b). Notably, these findings were confirmed independently from the timing of IV cell transplant and the combination of cell source adopted, indicating that the route of cell administration is the most relevant factor affecting the regional distribution of the transplant-derived cells.
To quantify and estimate relative cell abundance and distribution, and precisely map the distribution of the intra-CNS- and IV-delivered cell progeny in different brain regions at the latest time points, we acquired and quantified the signal of each reporter gene (in terms of total area covered by the reporter positive signal divided by the total area of the corresponding CNS region analyzed) (Figs. 2f and S2c,f). The accuracy of this measurement was assessed by estimating relative abundance of the cells expressing the different reporters (intra-CNS to IV ratio) and comparing it to the data obtained by FC analysis (Fig. S2d,e), showing a good correlation between the two measures (correlation coefficient R2: 0.79). Image analysis confirmed the higher donor chimerism in mice receiving HSPCs both IV and in the CNS versus IV-only recipients, and the greater frequency of CNS-administered HSPC progeny versus IV-derived ones in the animals that received the combined transplant (IV + ICV or IV + ITL) (Figs. 2d, e and S2a, b).
Route-specific differentiation of HSPC progeny cells in the brain
We then investigated whether the route of cell administration could affect not only the engraftment, expansion and distribution of the transplanted cells and their progeny in the brain, but also their maturation and acquisition of a microglia-like phenotype. To this goal, we conducted a morphological branching analysis of the transplant-derived cells at 45 days post transplant, distinguishing between the progeny of the IV-versus intra-CNS-transplanted HSPCs in the co-transplant setting. We analyzed microglia-specific parameters (see methods for details) as per literature33 and extrapolated two criteria to provide a morphological description of the transplanted cell progeny differentiation within the complex environment of the CNS: the total length of ramifications (Sum Length) and the complexity index (CI), defined as the ratio between the total number of branches of each cell (where a branch is the segment between two nodes), and the number of primary ramifications (defined as the segments connected to the cell body) (Fig. 3a). We also performed an automated Sholl analysis34, to study the radial distribution of branches around the cell body by a quantitative method and gain information on the spatial distribution and the complexity of the cell arborization pattern (Fig. 3a). The analysis was performed on several individual cells from different brain regions based on reporter gene expression. The branching analysis revealed significant differences between the progeny of the IV versus intra-CNS transplanted HSPC (the ICV- and ITL-transplanted HSPC progeny cells shared similar features). Indeed, the progeny of the ICV- and ITL-injected cells showed a higher sum length of ramifications as compared to cells derived from IV-transplanted HSPCs in most of the brain regions analyzed (Fig. 3b and Table S5). The results of the CI analysis showed a similar tendency for the progeny of the intra-CNS transplanted cells to exceed the morphological complexity of the IV-transplanted cell progeny in the same regions (Fig. 3c and Table S5). Interestingly, the olfactory bulb, which is the region more abundantly and more rapidly reconstituted by the progeny of the transplanted cells, represents an exception as we observed an equal complexity of ICV vs IV co-injected cells (Fig. 3c and Table S5). Additional parameters were evaluated with the Sholl analysis to describe the morphology of intra-CNS and IV-transplanted progeny cells. We selected the Sum intersections and number of intersecting radii to characterize transplant-derived cells as per complexity and spatial extension of the cell arborizations, respectively. By analyzing the relationship between these two parameters, we observed that the population covering the greater surface and displaying more arborizations (and occupying the upper right—UR—quadrant in the graphs of Fig. 3d) was the progeny of the intra-CNS transplanted cells. In particular, these cells showed a structurally mature phenotype in different regions, such as the cortex and the hippocampus, as compared to the cells derived from IV-transplanted HSPCs in the co-transplanted mice (Fig. 3d, e). Overall, this analysis confirmed the results obtained with the branching analysis and suggested that HSPC administration directly in the CNS favors the maturation of the transplanted cell progeny and its differentiation towards highly ramified microglia-like cells.
a Workflow adopted for branching analysis. Confocal images after maximum intensity projection (1) are used to select individual microglia cells based on the expression of fluorescence reporter genes (dotted box). Thresholded image (2) is used to generate a skeletonized image (3). Cell body is manually removed from the image (4). Then, the following morphological parameters are extracted: sum length of all branches (highlighted in red in image (5); complexity index expressed as ratio between the total number of branches and the number of primary ramifications highlighted in blue in image (6); Sholl analysis (7). Branching analysis on microglia-like cells engrafted in the brain of co-transplanted injected mice. Total length of ramifications (Sum Length) (b) and complexity index (c) on multiple individual microglia-like cells derived from Lin- cells transplanted ICV, ITL or from HSPCs co-transplanted IV are shown within the indicated brain region. Data from n° cells/group comprised between 20 and 80 analyzed in n = 4 mice per group across n = 3 experimental replicates, are summarized as mean ± SEM. Source data (b, c) are provided as a Source Data file. Statistics for b, c: 1-way ANOVA followed by Tukey post-hoc test. For additional statistical information, refer to Table S5. d Correlation between the number intersection radii and the sum intersections for microglia-like cells derived from HSPCs co-transplanted in the CNS (ICV or ITL) and IV, and retrieved in the indicated brain regions. The vertical and horizontal dashed lines divide the graphs in four quadrants, to highlight subpopulations with different complexity, i.e.: UR = upper right quadrant, for highly complex cells characterized by high sum of intersections and high number of intersecting radii; LL = lower left quadrant for cells with low complexity; UL (upper left) and LR (lower right) quadrants for cells displaying intermediate complexity between the LL and UR quadrant. e Histograms displaying the percentage of cells retrieved in each of the four quadrants highlighted in d. Ratios are reported for ICV, ITL and IV co-injected cells in the total and indicated regions. CTX cortex, OB/FR olfactory bulb/front, STR striatum, TH thalamus, HPC/SVZ Hippocampus/subventricular zone.
Gene expression profile of HSPC progeny cells
To investigate further the role of the CNS environment in driving the differentiation of transplant-derived cells, we analyzed by real time PCR the expression of microglia genes, including homeostatic genes (Tmem119, Tgfbr1, P2ry13, Mertk, and Olfml3, CX3CR1, CSF1R)35,36,37,38,39and transcriptional factors (Jun, Mafb, Mef2a)40 from isolated transplant-progeny cells (Fig. 4). To this goal, we FACS-sorted transplant-derived brain subpopulations according to CD45 and CD11b expression, distinguishing CD45highCD11blow TAμ and CD45+/lowCD11bhigh μ subsets from single IV-injected and co-transplanted (ICV/ITL + IV) mice at 45 days post-transplant. Within TAμ and μ, GFP+ (from intra-CNS delivered HSPCs) and ∆NGFR+/Cherry+ (from IV-injected HSPCs) cells were FACS-sorted separately. We selected an early post-transplant time point (45days) to increase our ability to identify early patterns of cell differentiation possibly leading to the morphometric differences described at 90 days post-transplant. Indeed, at this time point we observed the greatest difference between TAµ and µ frequency in the tested transplant conditions and the analysis of these two subsets at this time point could enhance our abilty to capture information about the process of differentiation and maturation of donor-derived myeloid cells in the CNS towards functionally equivalent microglia cells. Myeloid cells sorted from the brain of naïve adults and pups (post-natal day 1- P1) were used as reference, and values were expressed as fold change to adult microglia cells. Consistently with our previous data, the CD45+CD11b+ myeloid progeny of the transplanted cells, independently from the setting applied, displayed a microglia-enriched signature comparable to that of the adult naïve microglia (Fig. 4a and Table S6). Interestingly, we observed differences in the expression of the microglia related genes depending on the route of cell injection employed. Indeed, a higher expression of both homeostatic genes and transcriptional factors were observed in the progeny of the ICV transplanted HSPCs, as compared to the progeny of ITL and IV transplanted HSPCs (Fig. 4a, b and f and Tables S6, S8). These observations were also confirmed within the µ and TAµ fractions (Fig. 4 c, d and Table S7). Indeed, only the two GFP+ ICV-derived subpopulations expressed microglia-enriched homeostatic genes and transcriptional factors at levels overlapping those of the reference adult naïve microglia, indicating a higher maturation state reached in this transplant setting as compared to the others (IV and ITL). An intermediate level of expression of the microglia genes was observed in the ITL-progeny cells. Notably, transcriptional factors were expressed at above normal level in the ICV cell progeny, which also showed the highest expression of microglia specific genes, suggesting that the enhanced expression of the microglia associated transcriptional factors could have determined an early boosting phase driving the differentiation of transplant-derived cells (Fig. 4c, d and f and Tables S7, S8).
Expression of a pool of selected microglia genes (a) and transcription factors (TF) (b) obtained by qPCR in microglia cells FACS-sorted from the brain of IV only, co-transplanted animals sacrified at 45 days post-transplant (pTX) and pups (P1) [N = 3 mice/group]. Data are shown as mean ± SEM. Statistics for A-B: 1-way ANOVA followed by Tukey post-hoc. c-d Microglia enriched genes (c) and TF (d) shown in a, b reported as individual values in µ and TAµ subpopulations. Data are shown as fold change (calculated as 2 − DDCT) on adult control microglia. P1 mice are used as control of immature microglia. Mean values ± SEM are shown. Statistics for c, d: 1-way ANOVA followed by Tukey post-hoc test. e Heatmap of normalized expression values for microglia genes, TF and F4/80low/high signature genes from Bruttger et al. Genes (rows) are sorted according to the mean gene’s expression. RNA sequencing was performed on a pool of animals (n = 3 mice/group) analyzed at 45 days pTX upon sorting of µ and Taµ subpopulations. Endogenous microglia from untreated or busulfan-treated mice injected ICV with PBS (sham-control) were used as controls. Statistical analysis is performed in R, using DEseq2 package. f Expression of TGFBR1, Tmem119, CX3CR1 and JUN genes obtained by qPCR on GFP (ICV or ITL) or ΔNGFR (IV) labeled microglia-like cells FACS-sorted from the brain of intra-CNS and IV only transplanted mice [n = 3 mice/group]. Data are shown as fold change (calculated as 2 − DDCT) on adult control microglia. Mean values ± SEM are shown. Statistics for f: 1-way ANOVA followed by Tukey post-hoc test. Source data (a–d and f) are provided as a Source Data file and additional statistical information are shown in Tables S6–8. g, h Representative laser scanning confocal microscope microphotographs of brain sections showing the expression of selected microglia genes in the brain of intra-CNS co-transplanted mice [n = 5 slices from n = 2 mice/group] detected with RNAscope. Tissues were hybridized with probes for Tmem 119 (g) or TGFBR (h) gene (white dots) and their expression was analyzed within CNS (green) or IV co-injected cells (red). Nuclei were counterstained with DAPI (blue). Scale bar = 50 μm.
To provide a more complete characterization of the transcriptomic differences between the different myeloid brain subpopulations retrieved from the transplanted mice we performed a genome-wide expression analysis by means of an Illumina RNA sequencing (RNA-seq) platform on a pool of sorted µ and TAµ subpopulations from mice that were IV, ITL or ICV transplanted 45 days earlier with LV_GFP CD45.1 HSPCs (N = 2–3 animals/group) (Fig. 4e). Donor-derived and recipient myeloid cells were identified and sorted separately according to the expression of CD45.1 and CD45.2, respectively, within the CD11b brain compartment and characterized for the expression of microglia signature enriched genes and transcription factors commonly expressed by microglia. To characterize host microglia and newly formed microglia-like cells (µ and TAµ) in the different settings, we further extended the analysis to a published gene set that was previously used for studying microglia and engrafting myeloid cells across several models of microglia depletion41,42,43,44. Overall, we observed a very similar expression pattern between resident microglia and corresponding donor-derived µ and TAµ myeloid subpopulations retrieved from each of transplant settings. A lower expression of Sall1, commonly associated to yolk sac-derived microglia, was noticed in transplant-derived cells compared to controls, likely due to the different ontogeny42. Sall1 expression was lower in TAµ versus µ cells, further supporting the hypothesis of a differential state of maturation of the two subpopulations. As an additional control, we analyzed endogenous microglia retrieved from busulfan-treated mice injected ICV with PBS (sham control), to exclude with the reported results that the procedure per se could significantly affect the gene expression profile of resident microglia. In this setting we observed some differences that might be associated with a response to the conditioning regimen, this being the only difference when comparing endogenous microglia from untreated vs transplanted animals.
Notably, in situ RNA analysis of selected genes on cryostatic brain sections by RNAscope assay confirmed the expression of representative above-mentioned genes by the ICV/ITL derived cell progeny (Fig. 4g, h).
Overall, this data indicate that, independently from the route, we generated microglia-like cells with a signature reminiscent of microglia.
Therapeutic benefit in an Alzheimer’s disease animal model
Given the exquisite neurodegenerative nature of Alzheimer Disease (AD), we considered AD as an ideal setting to assess the therapeutic potential of our CNS-restricted HSPCs transplantation approach. In particular, we conducted a proof of concept (PoC) of efficacy study selecting the optimized protocol for ICV delivery of engineered HSPCs in a severe animal model of AD that requires timely intervention. For HSPC engineering we focused on TREM2 (Triggering receptor expressed on myeloid cells 2), a microglia cell-surface receptor required for microglial responses to neurodegeneration. TREM2 is needed to enhance signaling and to boost the metabolic capacity of microglia45 and its overexpression through BAC-mediated transgenesis showed encouraging results in enhancing the scavenging function of microglia and pathological phenotypes in AD murine models46. Importantly, elevated risk of developing AD is associated with hypomorphic variants of TREM247. We produced 3rd generation LVs expressing the human TREM2 cDNA under the control of the human phosphoglycerate kinase (hPGK) promoter, which has a ubiquitous robust expression, and confirmed vector-mediated TREM2 expression in transduced 293 T cells and BV2 murine microglia cell lines by Western blot (WB) (untrasduced cells were used as negative control) (Fig. S3a). We then proved transgene functionality by comparing the ability to engulf Aβ of mock treated versus TREM2-LV transduced BV2 microglia cells by a fluorescent imaging system. As expected, Aβ oligomer engulfment was increased in TREM2-LV transduced BV2 cells as compared to untransduced control cells (Fig. S3b). We then challenged our approach in transgenic mice with Five Familial Alzheimer’s Disease mutations (5xFAD), an established model of AD that express human APP and PSEN1 transgenes with a total of five AD-linked mutations leading to overproduction of Aβ−42 with an accelerated amyloid plaque pathology similar to that found in patients48. These mice develop early onset AD disease at approximately 6 weeks of age, with survival reduced after 10 months. Lin- HSPCs were purified from the BM of young adult 5xFAD donor mice and transduced with the TREM2-encoding LV in standardized conditions (see methods for details). The transduced Lin- cells (0.3−0.5 × 106 /mouse) were injected ICV in BU myeloablated (25 mg/kg/day for 4 consecutive days), 2 months-old 5xFAD recipient mice, which then received an unmanipulated BM support administered IV 5 days post-transplant to rescue peripheral hematopoiesis (2 × 106 total bone marrow cells from 5xFAD donors). Controls included 5xFAD mice transplanted with mock- or GFP-transduced 5xFAD Lin- cells, and wild type animals transplanted with mock- or GFP-transduced wild type Lin- cells, age matched to treated animals (Fig. 5a). We performed lentiviral Vector Copy Number (VCN) analysis on the liquid culture (LC) progeny of the pre-infused cell products and confirmed a similar LV content in the different settings. In particular, we measured VCN values of 8.45 ± 1.34 LV copies/genome in the LC progeny of WT Lin- cells transduced with the GFP_LV, and of 8.54 ± 1.09 and 10.09 ± 1.52 copies/genome in the LC progeny of 5XFAD Lin- cells transduced with the GFP_LV and hTREM2_LV, respectively. Phenotypic assessment and behavioral tests were then performed on the treated and control animals every month, starting at 4 months until they reached 12 months of age. Indeed, the 5xFAD mouse model develops an age-dependent motor phenotype in addition to working memory deficits in an alternation task, and reduced anxiety levels, as revealed in the elevated plus maze (EPM) task49,50. The EPM test, employed for the assessment of anxiety, demonstrated a clear disease progression of AD in control animals (mock/GFP-transplanted 5xFAD mice), while an ameliorated phenotype was observed in the TREM2-treated group, as shown at the 12 months-time point (Fig. 5b). For the analysis, data from mock and GFP transplanted mice were combined, to demonstrate the consistency across experimental conditions of animals that received identical treatment, consisting of four doses of Busulfan (25 mg/kg) and HSPC ICV transplantation.
a Experimental scheme of TREM2_LV HSPCs transplantation in 5xFAD mice. Performance of TREM2, mock/GFP transplanted 5xFAD mice and WT controls in the elevated plus maze (EPM) (b), analyzed monthly from 4 to 12 months and in the Morris Water Maze (MWM) test, in the hidden platform (c) and reversal platform trial (d), at 12 months (n° mice/group/time point b: N ≥ 7; c: N ≥ 12; d: N ≥ 9). Data values (b–d) are shown as MEAN ± SEM [n = 4 experimental replicates]. Statistics (b–d): 2-way ANOVA, Tukey post-hoc test [(b, p* = 0.0272 p**** < 0.0001), (c, p** = 0.0039, p**** < 0.0001), (d, p* = 0.0231, p****<0.0001)]. e VCN calculated by ddPCR on DNA from the brain of 5xFAD treated animals [GFP: n = 9, TREM2: n = 13, n = 4 experimental replicates]. Statistics: unpaired T test: not significant (P-value: 0.3881). f Correlation between VCN and TREM2 expression measured by SYBR Green qPCR on brain from transplanted mice [n = 6] and on the liquid culture progeny of the infused HSPCs (reference). Correlation analysis: Pearson r = 0.9991, R2 = 0.9981, p < 0.0001. Signal quantification for Iba1 (g—cortex, CTX and hippocampus, HPC) and GFAP (h—CTX), on brain from treated 5xFAD mice (Mock/GFP and TREM2) and controls (WT) at >12months [n° slices/mouse: g, h CTX: N > 15 slices/n = 3 WT, n = 2 Mock/GFP, n = 10 TREM2; g, HPC:N > 3 slices/n = 3 WT, n = 3 Mock/GFP, n = 6 TREM2]. Statistics (g, h): 1-way ANOVA, Tukey post-hoc test (p* = 0.416, p** = 0.0032, p**** < 0.0001). Data from mock and GFP mice are indicated by clear versus green dots, respectively. i IHC on brain slices showing the extent of neuroinflammation (Iba1 and GFAP staining) retrieved in mock vs TREM2 5xFAD transplanted mice at 12 months. j Quantification of Aβ deposits in the CTX and HPC of TREM2 transplanted animals (brain VCN > 0.5) compared to GFP/Mock animals [n > 3 slices/mouse, TREM2 n = 4, Mock/GFP n = 4]. Mean ± SEM, asterisks indicate significance at unpaired t-test (CTX p** = 0.0012; HPC p** = 0.0016). Source data (b–h, j) are provided as a Source Data file. k IHC on brain slices showing the extent of Aβ deposits in the HPC and CTX of TREM2 and mock/GFP Tx 5xFAD mice at 12 months. IHC in i and k [n > 8 slices/mouse, n = 3 mice/group].
Similar results were observed with the Morris Water Maze (MWM) test (Fig. 5c, d), here used to document cognitive deficits at late stage of disease (12 months). Indeed, MWM revealed a different ability of controls vs TREM2-treated animals to travel in the target quadrant (NW)(probe trial) and to reach a hidden platform over multiple trials (Figs. 5c, d and S3c). In particular, TREM2-treated animals showed a lower latency in reaching the platform and covered a significantly increased distance in the target quadrant (NW) as compared to control mock-treated 5xFAD animals, with a performance similar to wild type controls (mock/GFP transplanted wild type mice), indicating a preserved memory retention or recall as compared to 5xFAD controls (Fig. S3c). Moreover, TREM2-treated animals showed a significant improvement in the latency to find the hidden platform after 5 trials as compared to controls and a prevention of the deficit of cognitive flexibility observed in 5xFAD during reversal learning (Fig. 5c, d). After completing behavioral testing, the study was ended when treated and control mice reached 12 months of age, with euthanasia and tissue collection for molecular and histological analyses. Quantification of lentiviral VCN on the whole brain tissue documented a uniform engraftment of the engineered cell progeny in both treated and control animals (Fig. 5e). In addition, hTREM2 expression was measured on the brain of the treated animals by RT-qPCR, showing a strong correlation (R2 = 0.99) between protein expression and LV-hTREM2 content measured as VCN (Fig. 5f). To further measure disease modifying effects of the transplant, histology was performed on brain sections from the experimental animals. The extent of neuroinflammation was quantified by Iba1 and GFAP staining on brain tissue slices (Figs. 5g–i and S3d). Overall, the expression of Iba1 (Fig. 5g) and GFAP (Figs. 5h and S3d) was higher in control 5xFAD compared to wild type animals indicating the presence of both microglia and astrocyte activation. Instead, TREM2-treated animals showed a significant reduction of microgliosis (Fig. 5g), particularly in the cortex and hippocampus region. Moreover, measurement of Aβ accumulation was performed (Figs. 5j and S3d). Interestingly, the treated animals showed an overall significant reduction in Aβ deposition levels throughout the brain (Fig. S3d), that was particularly evident in the cortex and hippocampus (Fig. 5j) with dose-dependency: higher rates of Aβ reduction were observed in animals showing a VCN in the brain tissue > 0.5, average value of the study cohorts.
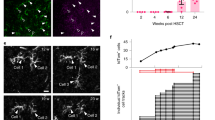
To assess whether therapeutic benefit was indeed related to an interaction of the TREM2 expressing cells with Aβ plaques, as hypothetized, we performed an in-situ hybridization (ISH) with a probe directed against the Woodchuck Hepatitis Virus (WHV) post-transcriptional regulatory element (WPRE)51, as a strategy to specifically identify the LV-transduced TREM2 expressing cells. Indeed, since WPRE sequence is not present in the mouse genome, ISH for WPRE allows to detect specifically the distribution of LV-transduced cells in the tissue52; moreover, as the WPRE sequence was added after the coding sequence o TREM2 in the LV expression cassette to stabilize the therapeutic transcript, the WPRE signal at in-situ corresponds to TREM2 expression. The ISH was combined with IF staining with an anti-Aβ antibody. Through this approach, we could detect abundant TREM2-expressing cells in the brain of the treated 5xFAD mice; these cells, which were distributed throught the brain parenchyma, were particularly abundant in the cortex and hippocampal regions, which are the brain areas where Aβ plaques were mostly detected (Fig. 6a). Interestingly, the TREM2 expressing cells were detected in the close proximity of the Aβ signal (Fig. 6b) and not in other regions as the cerebellum, where Aβ plaques were not retrieved (Fig. S3e). To elucidate the contribution of these cells to amyloid plaque phagocytosis we employed activation-associated markers such as CD68 for CNS tissue stainings on treated animals. Interestingly, the donor-derived microglia like cells identified by WPRE ISH probe positivity and Iba1 staining in the proximity of amyloid plaques showed an activated morphology (increased soma size and thickening of processes) and robust expression of CD68, allowing to hypothesize for them an active role in reducing the amyloid burden (Fig. 6c–g). On the contrary, microglia distant from plaques showed in general lower CD68 expression and showed morphological features characteristic of a more homeostatic/resting state.
a Representative epifluorescence microscope image reconstruction of a LV_TREM2 transplanted 5xFAD mouse showing the enrichment of transduced cells (identified through RNAscope with WPRE-specific probe, in red) in the cortex and hippocampus, in proximity to Aβ deposits, highlighted by IHC (in green). DAPI nuclear stain is shown in blue. Images were captured at a 20x magnification. Scale bar = 1 mm. N = 5 brain slices from n = 2 mice were analyzed. b Representative laser scanning confocal microphotographs of the cortex and hippocampal region from the brain of 5xFAD TREM2 transplanted mice (top and central figure) analyzed in a or in a WT mouse (bottom) used as negative control to demonstrate the specificity of Aβ amyloid and WPRE. Images were captured at a 40x magnification with a 1.5x digital zoom using a Leica SP5 scanning confocal microscope. CTX cortex, HPC hippocampus, THA thalamus, SVZ sub ventricular zone. Scale bar = 50 µm. c Representative laser scanning confocal microphotographs of the cortex and hippocampal region from the brain of a representative TREM2 transplanted mouse showing that reactive Iba1+ microglia/myeloid cells localized in close proximity to beta amyloid plaques are immunopositive for CD68. Images were captured at a 40x magnification with a 1.5x digital zoom using a Leica SP5 scanning confocal microscope. Scale bar = 50 µm. d Single channels of the ROI highlighted in the dashed square in c. Scale bar = 25 µm. e Representative laser scanning confocal microphotographs of the hippocampal region from the brain of 5xFAD TREM2 transplanted mice showing the presence of WPRE probe positive signal in CD68 positive cells in proximity of beta amyloid plaques. Scale bar = 50 µm. f Single channels of the ROI highlighted in the dashed squares in e. Scale bar = 25 µm. g Orthogonal view of the cell highlighted by the arrow in f. Scale bar = 25 µm. Images in c-g were analyzed in N = 4 brain slices from n = 3 mice.
Overall, these data provide clear proof of the therapeutic potential of HSPC gene therapy in a pure neurodegenerative condition as well as evidence of therapeutic benefit associated to a CNS-restricted engraftment of engineered hematopoietic cells.
Discussion
The therapeutic potential of engineered HSPC transplantation in benefiting selected monogenic neurodegenerative conditions has been extensively proven in mice and humans53. The engineered HSPCs exert therapeutic benefit through their mature progeny that represents a healthy, metabolically competent myeloid population, which acquires microglia-like features in the CNS, capable of protein delivery and restoration of the scavenging and homeostatic functions54. Therefore, enhancing the engraftment, distribution in the CNS and maturation of the transplanted cells/their progeny towards a microglia-like phenotype, as well as fine tuning the contribution of the transplant solely to the CNS could allow to further improve the efficacy of HSPC-based gene therapy protocols for CNS disorders. To this goal, we previously proposed the direct CNS-delivery of HSPCs in the lateral ventricles (ICV) as an approach for favoring the turnover of endogenous microglia with donor cells6. Confirming the interest in this approach, other groups have recently exploited the intra-CNS route of cell administration by conducting biodistribution studies on differentially transplanted animal model32,55. Intra-CNS transplantation was used to evaluate the capacity of microglia or peripheral immune cells to engraft in the brain parenchima of CSF1R-/- mice that lack microglia43,56. Interesting data were also obtained by transplanting hPSC-derived primitive macrophage progenitors via intra-CNS injection (i.e intracranial, transnasal injection)57,58.
In our work, by challenging further the hematopoietic cell transplantation paradigm we unveiled a key role exerted by the CNS environment in uniquely driving expansion, distribution and myeloid differentiation of the locally transplanted HSPCs towards a microglia-like phenotype. Importantly, we also provided a clear example of the therapeutic potential of this approach that determines a CNS-restricted engraftment of the genetically modified transplant progeny cells, in a multifactorial neurodegenerative condition, namely in a severe murine model of AD.
Firstly, we showed that intra-CNS delivery favors the transplanted HSPC engraftment over IV-delivered cells in a competitive setting. This was demonstrated by employing two different intra-CNS delivery routes, ICV and ITL. Both routes are clinically viable. Indeed, despite the ITL route, here employed for the first time for HSPC delivery, is by definition a very feasible and poorly invasive intra-CNS delivery route, protocols allowing the safe administration of drugs, proteins or cells in the cerebrospinal fluid through ICV delivery are also available in clinical practice59,60,61. Interestingly, the two intra-CNS routes equally favored CNS colonization by transplant progeny cells. Importantly, with both ICV and ITL HSPC delivery, the intra-CNS injected HSPCs exceeded the IV co-transplanted cells in contributing to the donor-cell chimerism in the brain in all the tested conditions, despite the number of cells delivered intra-CNS was substantially lower that the IV dose. This advantage was particularly evident at the earliest time points and up to 90 days post-transplant, independently from the setting employed, with strong clinical implications for diseases in which a timely and sustained therapeutic delivery to the brain is essential for an effective benefit. However, differently from the ICV injected cells that do not contribute to marrow hematopoietic reconstitution6, ITL-delivered HSPCs also robustly engrafted in the marrow of transplanted mice. This may be attributed to differences in the BBB architecture in the brain as compared to the spinal cord62. Indeed, despite the blood-spinal cord barrier (BSCB) has a similar cellular and molecular structure as the BBB, the lower number of pericytes and reduced expression of junctional proteins retrieved in the BSCB result in an increased permeability as compared to the BBB. In accordance, murine spinal cord microvascular endothelial cells in culture were shown to express reduced amounts of ZO-1, occludin, b-catenin, VE-cadherin and P-glycoprotein than brain endothelial cells63. Thus, the characteristics of the two barrier systems, i.e. the BBB and the BSCB, might be responsible for some of the differences observed between the two routes in the colonization potential of the hematopoietic organs. Notably, and in contrast to recent works32, we observed significantly higher donor engraftment when a combination of intra-CNS and IV routes were adopted compared to single IV transplantation up to 3 months post-transplant, indicating a faster early seeding of progeny cells achieved in the co-transplant setting. These results point out the clinical relevance of the combination of different HSC transplant routes, particularly for those diseases where anticipating the time of clinical benefit is crucial. Different outcomes7,30,31 could be explained by different methodologies applied, i.e. for the administration of the conditioning protocol, which is essential to favor robust CNS-engraftment of the transplant progeny cells, or for HSPC manipulation and transplantation.
Interestingly, we also showed that intra-CNS delivery of HSPC is associated to a specific pattern of distribution of progeny cells in the brain. As early as 14 days after transplant, the GFP+ progeny of the transplanted cells was located in the close proximity of the ventricular structure, projecting rostrally and towards the basal forebrain, in a trajectory which may suggest proximity to the rostral migratory stream. Interestingly, these ICV-derived cells already showed discrete branchings indicating an actively occurring differentiation process. Notably, and at odd with these findings, the progeny of the IV transplanted cells had no preferential location and was mostly identified in the proximity of meninges and blood vessels, showing a morphology consistent with immature cells, with a round shape and no branchings. While the former findings (ICV) are suggestive of anatomical proximity leading the cell distribution, the latter (IV) are in line with the emerging view of the meninges as a potential relevant route for cellular infiltration to the brain in physiological64 and pathological conditions65,66. This is possible due to the anatomical distribution of the meninges and their unique relationship with blood, lymphatic vessel and CSF. In addition, meninges consist of highly heterogeneous cell populations including endothelial cells, CNS-associated macrophages, blood borne immune cells as well as meningeal neuronal precursors capable to respond from their local niche to external chemokine-mediated stimuli and promptly react and infiltrate the brain parenchima in pathological conditions67. The analysis at the subsequent time points post transplant provided additional relevant information. Firstly, the progeny of the ICV-transplanted cells followed a path reminiscent of the rostral migratory stream and likely exploited the cerebral ventricular system, including the lateral, third and fourth ventricles, to diffuse into nearby regions such as the striatum, the anterior olfactory area and the brainstem. On the other hand, the progeny of the IV-administered HSPCs preferentially localized in the hippocampus, as a possible effect of the secretion of key chemokines as stromal cell-derived factor (SDF-1/CXCL12), and monocyte chemoattractant protein-1 (MCP-1/CCL2) from this area, as described in the literature68. The pattern of cell distribution adopted by ITL transplanted cells became also more apparent, involving the olfactory bulb and forebrain regions as well as the brainstem, consistently with the proximity to the spinal cord (injection site). Despite the differences, all three routes shared a common early colonization of the olfactory bulb/forebrain area. As neural precursor cells from the subventricular zone have been shown to migrate along the rostral migratory stream to the olfactory bulb69,70, we could hypothesize that also engrafted HSPC-progeny cells could follow the same route of distribution and rapidly reach this region upon IV and ICV injection. A different way to reach this area could be hypothesized for cells infused via ITL injection, possibly involving a specific vascular network. Interestingly, the observation at 45 and 90 days post-transplant, time points where co-transplantation (intra-CNS and IV) was performed to allow animal survival after myeloablation, allowed also to assess the relative contribution of the co-transplanted cell progeny to microgliosis. Indeed, we noticed that the areas preferentially colonized by the ICV-transplanted cell progeny were those closer to the injection site, from the lateral ventricles to hippocampus, corpus callosum, olfactory bulbs and up to the cortex, while those preferentially colonized by the ITL-transplanted cell progeny were the midbrain, pons, hypothalamus and thalamus. Interestingly, in both settings, the progeny of IV co-transplanted cells colonized alternative regions, with a mutually exclusive pattern. The finding that the transplant cell progeny could adopt a mutually exclusive distribution depending on the route of cell delivery could be interpreted in different ways. The most obvious speculation would be that cells directly injected in the CNS could have an advantage in terms of early occupancy of areas and niches that would no longer be available for brain immigrant cells incoming after intravenous injection and/or BM engraftment. According to the route of administration, cell trafficking into the CNS could be mediated by intrinsic factors or specialized tissue interfaces such as the meningeal vasculature or the choroid plexus structure71. Indeed, the choroid plexus, as critical interface between the systemic blood circulation and the CSF, exerts an important role in supporting immunosurveillance and regulation of cell activation and trafficking in response to pathological insults or specific cytokine milieu, due to the presence of specialized cell types like dendritic cells or macrophages, and specific patterns of epithelial adhesion molecule expression that could be recruited in response to conditioning and HSPC transplantation72.
Regardless of the route, we confirmed that the progeny of both IV and intra-CNS delivered cells acquires morphological features typical of homeostatic microglia. Branching and Sholl analysis demonstrated that the progeny of intra-CNS transplanted cells has a more complex morphology as compared to IV co-injected cells, with a higher number of ramifications and a more enriched arborization network, particularly in the regions in which they preferentially expanded. The observation that the progeny of the HSPCs transplanted in the CNS has a greater resemblance to endogenous microglia than the progeny of cells transplanted IV suggests that the CNS environment could have a role in favoring and driving their differentiation and maturation. This hipothesis is supported by several works showing the importance of the environment for the acquisition of microglial feature43. As described by Bohlen and colleagues, isolated microglia kept in culture in “non-neural” medium drastically reduce the expression of microglial signature genes, while reversing the loss of microglia identity when re-infused into the brain of CSF1R-/- mice56. Therefore, if the transcriptome of mature microglia could be affected by CNS signaling, it is plausible to assume that intra-CNS injected HSPCs may differentiate toward microglia faster than upon IV administration. The olfactory bulb was characterized by high but equal presence of both IV and intra-CNS cell progenies in the combinatorial transplants with no morphological differences, supporting the hypothesis that this neurogenic area may play a crucial role in the recruitment of engrafted cells independently from the route of cell injection, as demonstrated by the short term biodistribution study.
Notably, the results of a transcriptome analyses and of independently generated RNAseq data performed on microglia-like cells sorted from the brain of the transplanted mice were in line with our morphological observations, confirming the expression of microglia signature genes by newly formed cells with some differences depending on the transplantation route adopted and the brain subpopulation analyzed. Indeed, the expression of microglia signature genes35,36,73 and transcriptional factors such as mafb and jun40, required for microglia development correlates with the greater maturation stage of intra-CNS vs IV-transplanted cell origin. The analysis was performed at an early post-Tx time point (45days) to increase our ability to identify early patterns of cell differentiation possibly driving the maturation of transplant-derived cells described at 90 days post-transplant. According to our experience, this could explain the differences observed when comparing our data with those obbtained by other groups32, by which a downregulation of canonical genes associated with resident microglia, like Tmem119, was observed at later time points (12–13 months post-transplant).
Clearly, despite our transplant-derived microglia-like cells, and particularly those derived from ICV/ITL transplanted HSPCs, resemble their endogenous counterpart, their gene expression profile does not completely overlap with that of bona fide microglia isolated from adult naïve mice, as also reported by others32,74,75. This finding, which could be related to a different maturation stage between resident and newly formed microglia or to their different ontogeny, is in agreement with what reported by recent works that described how bone marrow-derived cells that colonize the brain parenchyma in response to conditioning or pathology present differences at the trascriptome level compared with adult microglia of embryonic origin, despite the fact that they resemble microglia in morphology and in the expression of many key markers42,43,44,74,76. Importantly, these differences at the transcriptome level do not necessarily result in functional impairment. On the contrary, it was demostrated that bone marrow-derived cells are even more efficient than yolk sac-derived microglia in certain conditions, mostly associated with aging, such as phagocytosis of amyloid deposits74,77.
To this regard, recent works have shown the potential of bone marrow derived microglia-like cells as cell-based disease-modyfing therapy for AD77,78. In line with these interesting findings, we here provided experimental proof of concept of the therapeutic potential of intra-CNS HSPC gene therapy for AD in a severe animal model of the disease. In this setting, we selected TREM2 as a relevant therapeutic transcript specifically intended at enhancing a disease-specific function of transplant-derived microglia-like cells for this study. Importantly, upon ICV transplantation of HSPCs from 5xFAD mutant mice transduced with a LV encoding the human TREM2 cDNA under the control of a moderate-strenght promoter, a CNS-restricted engraftment of the TREM2-expressing cells was observed up to 12 months post-transplant in busulfan myeloablated 5xFAD recipients. This resulted in substantial disease-modifying effects: a significant amelioration of the behavioral and histopathological disease manifestations was observed, with improved performance of the treated versus control mice at the Elevated plus maze and Morris Water Maze tests, as well as reduction of neuroinflammation (astrogliosis and microgliosis).
The phenotypic effects observed were accompained by a LV-dose dependent decrease of Aβ-deposits in treated mice as compared to mock treated controls. Importantly, using a WPRE ISH probe as strategy to specifically identify the LV-transduced cells in brain sections, we documented a preferential distribution of the TREM2 expressing cells in brain regions associated to β amyloid deposits such as the cortex and hippocampal area and in particular a localization of CD68+TREM2+ cells in an activated state in the close proximity of amyloid plaques.
These evidences thus support the hypothesis that TREM2 microglia could exert a key role in Aβ clearance, in agreement with previous studies that demonstrated migration of engrafting microglia to amyloid plaques in the CNS of chimeric models, followed by secretion of proteins that in turn promote the Aβ degradation and phagocytosis79. Notably, the capability to clear Aβ was, as reported, in some cases even more prominent than that of the resident microglia80.
Of note, despite the proof-of-concept nature of our findings will require proper adaptation of protocols in a clinical development plan, it is noteworty to report that, in the presence of anticonvulsant prophylaxis during conditioning administration, no short- or long-term neurotoxic effects of busulfan were noticed, at least phenotypically as per regular daily observation and at behavioral studies, nor obvious and severe signs of neural damage were observed at histopathological observation of the treated and control mice. Although specific neurotoxic effects of busulfan across epileptogenic and/or other neural circuits in this Alzheimer’s disease models remain undefined at the microscopic evaluation, such effects have not compromised treatment outcomes nor treated and control mice overall performances.
In conclusion, this work unveiled a key role exerted by the CNS environment in uniquely driving expansion, distribution and myeloid differentiation of locally transplanted HSPCs towards a microglia-like phenotype and showed the robust potential of intra-CNS HSPC gene therapy for exerting specific therapeutic effects in the CNS, paving the way for the design of ad hoc gene therapy strategies to treat severe neurodegenerative disorders.
Methods
The specific protocols covering the studies described in this paper were performed in compliance with all relevant ethical regulations and were explicitly approved by the Italian Ministry of Health, an internal ethical committee at San Raffaele Hospital (IACUC 573) and the Dana Farber Animal Facility Institutional Animal Care and Use Committee (IACUC 15-031). Experiments performed on wild type C57BL/6 mice or C57BL6/Ly5.1 mice (hereafter called CD45.2 or CD45.1 respectively) were obtained from Jackson Lab or Charles River and maintained at San Raffaele Hospital and at the Dana Farber Animal Facility. Experiments performed on 5xFAD mice (#034840-JAX) were obtained from Jackson Lab and maintained at the ARCH animal Facility at Boston Children’s hospital (approved by the Institutional Animal Care and Use Committee (IACUC 17-06-3455) and at Fondazione Istituto di Ricerca Pediatrica Città della Speranza (IRPCDS) animal facility (IACUC 46/2023-PR).
Animals used in the study were housed within temperature and humidity ranges appropriate for the species, to which they can adapt with minimal stress and physiologic alteration. Light in animal holding rooms provided for both adequate vision and neuroendocrine regulation of diurnal and circadian cycles.
Isolation, transduction and transplantation of murine hematopoietic cells
Seven/eight-week-old wild type mice were euthanized with CO2, and the bone marrow (BM) was harvested by crushing bones. After BM lysis, HSPCs were purified by Lineage- (Lin-) selection using the Miltenyi Biotec Lineage Cell Depletion Kit with Magnetic separation with the autoMACS™ Separator, following manufacturer’s instruction. For the isolation of the KSL fraction (c-kit+ sca-1+), Lin- cells were then stained with rat APC-eFluor 780 anti-mouse CD117(c-kit) (eBioscience, 1:100) and with rat Pe-cy7 anti-mouse Ly6A/E Sca-1 (Sca-1) (BD Bioscence, 1:100). At the end of the staining the cells were isolated by the cell sorter MOFLO XDP (Becton Dickinson), according to the expression of the selected markers. BM cells, isolated Lin- or KSL were transduced using different Lentiviral Vectors (LVs), for 16 h at Multiplicity of Infection (MOI) 100 81. The following LVs were used: pCCLsin.cPPT.humanPGK.GreenFluorescentProtein.Wpre for CNS injected cells (GFP-LV) pCCLsin.cPPT.humanPGK.DeletedNerveGrowthFactorReceptor.Wpre (ΔNGFR-LV) or pCCLsin.cPPT.humanPGK.mCherryProtein.Wpre (mCherry-LV) for IV injected cells. A fraction of the transduced cells was cultured for 10 days in vitro in order to assess transgene expression by cytofluorimetric analysis. Transduced cells were injected via the tail vein or directly in the CNS by means of ICV or ITL injection into seven/eight-week-old conditioned C57BL6/J female mice, 24 h after the fourth busulfan dose (25 mg/kg x four days, Merck, cod. B2635), at different concentration according to the different experimental settings. CNS injected cells (Lin- 0.3 × 106/mouse); IV injected cells (Lin-1 × 106; BM 3 × 106; KSL 0.5 × 106). 8 weeks old 5xFAD mice were transplanted ICV with 0.3 × 106 Lin- cells obtained from 5xFAD donors (6–8 weeks old) and transduced at MOI 100 with a TREM2 expressing LV (pCCLsin.cPPT.humanPGK.Trem2.Wpre) as described above. 5 days post transplantation, 2.0 × 106 BM cells from 5xFAD donors were provided IV for hematopoietic rescue. Mice were conditioned with 25 mg/kg of busulfan for 4 days prior to transplant and provided with Diazepam to avoid seizures.
Diazepam solution (available at 5 mg/mL) was provided daily in the drinking water starting for a total of 10 days with progressive decreases in drug dose (0.25 mg/kg used for the first 8 days, then dose decreased in the two following days to 0.125 mg/kg −0.063 mg/kg and removed).
5xFAD mice transplanted with 0.3 × 106 GFP-transduced Lin-cells ICV were used as control.
Intracerebroventricular injection (ICV)
ICV transplantation was performed by surgery, upon anesthesia (ketamine (100 mg/kg) and xylazine (10 mg/kg). The head of the mouse, shaved and disinfected, was fixed with ear bars in a stereotactic frame and the skin was disclosed longitudinally. Bregma was visualized and coordinates were recorded. From bregma, injection coordinates (1 mm lateral, −0.5 mm anterior) were adjusted before the cranial bone was enclosed under visual control with a drill head of 0.7 mm diameter. Five µl of the cell suspension (0.3 × 106 cells) were injected through a 10 µl Hamilton syringe upon insertion into the brain 2.5 mm distal from the cranial bone. Following wound closure, animals received a single dose of atipamezole (1 µl/g) and were maintained in sterile conditions.
Intrathecal lumbar injection (ITL)
ITL transplantation was performed under anaesthesia with isoflurane (4% induction, 2.5% maintenance). A 10 µl Hamilton syringe connected to a 30-gauge needle containing 5 µl of cell suspension was introduced into the intervertebral foramen between L5 and L6 through the intact skin. Slow administration of 0.3 × 106 cells/mouse was performed once the proper position of the needle was confirmed by tail flick reflex. At the end of the procedure, animals received a single i.p. injection of atipamezole (1 μl/g) as analgesic and returned to the cage. Prophylactic Antibiotic (Gentamycin Sulfate, 80 mg/250 mL) was administered via the drinking water for 2 weeks following conditioning and transplantation.
Mouse tissue collection and processing for flow cytometry and histology
Mice were euthanized under deep anesthesia by extensive intra-cardiac perfusion with cold PBS for at least 10–15 min/mouse after clumping the femur. The procedure was stopped only when complete liver blanching was observed to avoid contamination with circulating blood cells. Organs were then collected and differentially processed. BM cells were collected from the clumped femur as described. Brain was removed and the two hemispheres were differently processed. For immunofluorescence analysis, one hemisphere was fixed for 24 h in 4% PFA, embedded in OCT compound and stored at −80 °C, after equilibration in sucrose gradients (from 10 to 30%). For flow cytometry analysis, cells from the other hemisphere were mechanically disaggregated to obtain a single cell suspension in 20 ml of GKN/BSA buffer (8 g/L NaCl, 0.4 g/L KCl, 1.42 g/L NaH2P04, 0.93 g/L Na2HP04, 2 g/L D+ Glucose, pH 7.4 + 0.002% BSA). The same protocol was used to process the spinal cord. Cells from BM and CNS tissues (brain and spinal cord) were analyzed by FC upon re-suspension in blocking solution (PBS 5%FBS, 1%BSA) and labeling at 4 °C for 15 min with the following specific antibodies: rat PE anti-mouse CD45 (BD Pharmingen) 1:100; rat APC anti-mouse CD45 (BD Biosciences) 1:150; rat Brilliant Violet 510 anti-mouse CD45, (BioLegend) 1:150; rat Pacific Blue antimouse CD45.2 (Bio Legend) 1:100; mouse PE anti-Mouse CD45.1 (BD bioscience) 1:100; rat APC anti-mouse CD11b (eBioscience) 1:100; mouse Alexa Fluor 647 anti-Human CD271 (NGF receptor) (BD Pharmingen) 1:30; rat APC 780 anti-mouse CD11b (eBioscience) 1:100; rat APC 780 anti-mouse CD117(c-kit) (eBioscience) 1:100; rat PE-Cy7 anti- mouse Ly-6A/E Sca-1 (Sca-1) (BD Bioscence) 1:150. 7-AAD (1 mg/ml) (Sigma-Aldrich) or DAPI was added to the cells prior to analysis for dead cell exclusion. Samples were analyzed by LSR Fortessa (Beckton Dickinson).
Immunofluorescence analysis
Brains were serially cut in the sagittal planes on a cryostat in 15 µm sections. Tissue slides were washed twice with PBS, air dried and blocked with 0.3% Triton, 2% BSA, 10% NGS (Vector Laboratories) for 2 h. Then sections were incubated over night with primary antibodies diluted in PBS, 0.1% Triton, 2% BSA, 10% NGS at 4 °C as follows; rabbit anti Iba1 (Wako) 1:100; chicken anti-GFP (Abcam) 1:250; rabbit anti GFP (Invitrogen) 1:100; mouse PE anti-human CD271 (NGF Receptor) (BD Pharmingen) 1:50; rabbit anti-cherry (Abcam) 1:100. The secondary antibodies goat IgG anti-Chicken Alexa Fluor 488, goat IgG anti-Rabbit Alexa Fluor 488, 546 or 633, goat IgG anti-Rat Alexa Fluor 546 or 633, goat IgG anti-Mouse Alexa Fluor 546 (Molecular Probes, Invitrogen) were diluted 1:500 in the same blocking solution used for primary antibodies staining and incubated with sections for 2 h at room temperature. Nuclei were stained with Topro III (Molecular Probes, Invitrogen) 1:1000 in PBS or by DAPI (Roche) 1:30 in PBS. Slices were washed in PBS, air dried and mounted with Fluorsafe Reagent (Calbiochem). Not transplanted mice were used as negative controls for the reporter transgene staining. Incubation with secondary antibody alone was performed in order to exclude the background signal. Samples were analyzed with a confocal microscope (Zeiss and Leica TCS SP2; Leica Microsystems Radiance 2100; Bio-Rad) (λexcitation = 488, 586, 660). Fluorescent signal was processed by Lasersharp 2000 software. Images were imported into Adobe Photoshop CS 8.0 software and processed by using automated level correction. For the reconstruction of brain sections, a fluorescence microscope Delta Vision Olympus Ix70 was used for the acquisition of the images, which were then processed by the Soft Work 3.5.0 software. Images were then imported into the Adobe Photoshop CS 8.0 software and reconstructed.
Branching analysis
The branching analysis was performed with ImageJ software using an automatic Macro, developed in the lab for standardizing the procedure, on multiple individual cells (comprised between 20 and 80) from different brain region of transplanted mice. Cells were identified according to the expression of different fluorescent markers (GFP/Cherry/∆NGFR) and validated reproducing the same analysis on Iba-1+ stained cells. Automated Sholl analysis was performed by applying the same strategy, to study the radial distribution of microglia branches around the cell body. The sum intersections and number of intersection radii were selected as parameters to characterize donor derived cells, in terms of complexity and spatial extension of the cell arborizations, respectively.
Area signal quantification
For area signal characterization, a mask was created from splitted channel images by using Fiji program. The positive signal was then converted in colored spotted signal to differentially mark CNS vs IV transplanted cells. For signal quantification, a deeper analysis was conducted on splitted channel images upon removal of background noise and identification of region.
Tissue clearing procedure, image acquisition and processing
Tissue clearing procedure82 was applied on 2mm-thick sagittal brain section for the characterization of the distribution pattern of short-term immigrant cells in the CNS of transplanted mice on a whole-mount 2mm-thick sagittal brain section (Tissue Clearing kit,Binaree Inc., Republic of Korea).
In particular tissues were a) soaked in Starting Solution overnight at 4 °C; b) transferred to Solution A at 37 °C for 24 h, followed by three 20-minute washes in distilled water at 4 °C; c) immersed in Solution B at 37 °C for 24 h, followed by four 30-minute washes in distilled water at 4 °C; d) stained with Nuclei staining solution (DAPI 10 µg/ml in distilled water) for 2 h at 4 °C, followed by three 10-minute washes in distilled water at 4 °C; e) placed in Mounting Solution at 37 °C for 24 h and then stored at room temperature for imaging.
A z-stack of 400–500 μm thickness (with 20 μm intervals) was obtained in both the UV and FITC channels to visualize the DAPI-stained nuclei and GFP+ cells, respectively. A composite image was generated through stitching. Four partially overlapping acquisitions were performed, focused on four different brain subregions (forebrain, hippocampus/midbrain, cerebellum and brainstem). Full brain was reconstructed by manually merging the four acquisitions (with GIMP v.2.10.8 open-source software.
Selected brain regions of interest were acquired at high-resolution on the z-axis by capturing a z-stack image of 400–500 μm thickness (with 5 μm intervals) at 25x magnification with a Leica SP5 scanning confocal microscope.
In-situ hybridization and immunohistochemistry in combinatorial transplant settings
For the analysis of microglia enriched genes, cryostatic brain sections were processed and stained using the RNAscope 2.0 kit (cat#310095, Advanced Cell Diagnostics, Hayward, CA), following the Fixed-frozen tissue sample preparation and pre-treatment protocol according to manufacturer instructions. Perfusion and post-fixation in paraformaldehyde were performed as described above. After equilibration in 30% phosphate buffered sucrose solution, 14 µm thick brain sections were mounted onto Superfrost Plus glass slides using a cryostat. Detection of Tmem119 and Tgfbr1‐mRNA was performed using the RNAscope® Fluorescent Multiplex Assay with a probe set targeting base pairs 2–1106 of mouse Tmem119‐mRNA (Cat No. 472901-C2, Advanced Cell Diagnostics, Inc., Newark, CA) or base pairs 2257–3252 of mouse Tgfbr1-mRNA (Cat No 406201, Advanced Cell Diagnostics, Inc., Newark, CA). Briefly, the sections were air dried for 20 min, washed in PBS and subsequently immersed in boiling 1x Target Retrieval solution for 5 min. After washing in distilled water and 100% ethanol for 2 min each, the sections were incubated with Protease IV solution for 30 min at 40 °C in a humidified tray. After washing in distilled water for 2 min, the sections were then hybridized with target probes for 2 hr at 40 °C followed by amplification solutions 1 (30 min), 2 (15 min), 3 (30 min), and 4 (15 min) at 40 °C. Between each amplification step, the sections were rinsed in 1x Wash Buffer for 2 min at room temperature. Finally, the sections were counterstained with 4’,6‐Diamidine‐2’‐phenylindole dihydrochloride (DAPI) for 30 s and coverslipped in PVA‐DABCO. Images were acquired at Delta Vision Olympus 10x.
In-situ hybridization and immunohistochemistry in 5xFAD mouse model
Brain samples from 5xFAD mice were processed for immunohistochemistry by Neuroscience Associates, Inc for Iba1, GFAP and β amyloid detection. After equilibration in sucrose gradients (from 10 to 30%) and 24 h treatment with 4% PFA, samples were embedded in a gelatin matrix using MultiBrain®Technology. The blocks were rapidly frozen, after curing by immersion in 2-Methylbutane chilled with crushed dry ice and mounted on a freezing stage of an AO 860 sliding microtome. The MultiBrain® blocks were sectioned coronally with desired micrometer (μ) setting on the microtome. All sections were cut through the entire length of the specimen segment and collected sequentially.
For immunohistochemistry, free floating sections were processedd for IF as per internal protocols. All incubation solutions from the blocking serum onward use Tris buffered saline (TBS) with Triton X-100 as the vehicle; all rinses are with TBS. After a hydrogen peroxide treatment and blocking serum, the sections were immunostained with the primary antibodies (i.e anti-Iba1, anti-GFAP), and incubated overnight at room temperature. Vehicle solutions contained Triton X-100 for permeabilization. Following rinses, a biotinylated secondary antibody was applied. After further rinses Vector Lab’s ABC solution (avidin-biotin-HRP complex) was applied. The sections were again rinsed, then treated with diaminobenzidine tetrahydrochloride (DAB) and hydrogen peroxide to create a visible reaction product. Following further rinses, the sections were mounted on gelatin coated glass slides, air dried. The slides were dehydrated in alcohols, cleared in xylene and coverslipped.
The identification of LV-transduced cells in the tissues of transplanted 5xFAD mice was performed through in-situ hybridization for WPRE on 20 μm brain slices. The protocol was optimized to allow the co-detection of WPRE as well as amyloid beta, CD68 and Iba1 (stained with immunohistochemistry) on the same tissue slice. Firstly, a standard immunohistochemistry protocol was performed. Briefly, brain sections were blocked for 1 h at RT in blocking buffer (NGS 10% or BSA 3%, 0.3% Triton X-100 in PBS pH 7.4). The primary antibodies used were Mouse anti-B-Amyloid (1:500, BioLegend, SIG-39320), Rat anti-CD68 (1:100, BioRad, clone FA-11, MCA1957T) and Rabbit anti-Iba1 (1:500, Fujifilm Wako, 019–19741). After three washes with PBS, the sections were incubated with Biotinylated Goat anti-Mouse IgG (H + L) (1:200, Vector Labs, BA-9200), Biotinylated Goat anti-Rat IgG (H + L) (1:200, Vector Labs) or Biotinylated Donkey anti-Rabbit IgG (H + L) (1:200, Biotium, 20191), respectively, followed by 30’ incubation at RT with HRP-streptavidin conjugate (1:100, AAT Bioquest, 16920) and Cy3 tyramide (1:1000, AAT Bioquest, 11065) or Cy2 tyramide (1:1000, AAT Bioquest, 45100). Before starting the in-situ hybridization protocol, Avidin/Biotin blocking kit (ABCAM, AB64212) was used following manufacturer’s instructions to saturate any available biotin-binding surface in the tissue. Slices were fixed with PFA 4% and left to dry O/N at RT. In-situ hybridization was performed using the RNAscope Probe WPRE-04 (Biotechne, 540341), initially blocking peroxidases through the RNAscope H2O2 followed by protease treatment according to the manufacturer instructions (ACD, Biotechne, 322381) and subsequently amplifying the target signal with the RNAscope Multiplex Fluorescent Detection Kit V2 (ACD, Biotechne, 323110) and associating it with the Cy5 tyramide fluorophore (1:1000, AAT Bioquest,11066). Images were captured at a 40x magnification with a 1.5x digital zoom using a Leica SP5 laser scanning confocal microscope.
Gene expression analysis in sorted population from transplanted mice
Total RNA was extracted with the RNeasy Plus Micro kit (Qiagen) from sorted myeloid brain populations, RNA quantity was determined using QuantiFluor® RNA system and QuantusTM Fluorometer (Promega). cDNA was generated starting from 1 ng up to 100 ng of purified mRNA by using SuperScript VILO Master mix (Thermo Fisher Scientific). cDNA was then pre-amplified using Custom Taqman® PreAmp Pools (Thermo Fisher Scientific). Thermal cycling for cDNA generation and preamplification was performed on T100 Thermal cycler (BIO-RAD) following manufacturer’s instructions. Gene expression analysis was performed using a custom design TaqMan-based microfluidic card gene expression assay (Applied Biosystems). Real time PCR was run in standard mode on Applied Biosystems® ViiATM 7 Real-Time PCR System, using the following thermal cycling conditions: one cycle at 50 °C for 2 min, one cycle at 95 °C for 10 min, 40 cycles at 95 °C for 15 s and 60 °C for 1 min (Applied Biosystems). The ViiATM 7 Software v1.2.2 was used to extract raw data. The difference (dCT) between the threshold cycle (CT) of each gene and that of the reference gene (mean of HPRT and r18S CTs) was used to determine gene expression. Fold change expression of selected microglia genes was calculated by 2−ddCT method.
The RNA-seq gene expression analysis was performed on additional samples, corresponding to different combinations of route of administration (IV, ICV, ITL and CTRL), brain subpopulation (µ, Taµ, PBS) and markers (CD45.1, CD45.2). The complete design matrix reporting the characteristics of each sample is available as Source data in a Source data file.
Raw sequencing files were investigated for QC and aligned to the reference mouse genome mm39 (Genome assembly GRCm39) using STAR workflow83, obtaining, as a result, the gene-based count matrices. Poorly-detected genes with a cumulative reads count smaller than 10 or measured (reads count>0) in less than two samples were filtered out.
Statistical analysis has been performed in R84, using DEseq2 package85. Gene counts have been modeled using a negative binomial model. Normalized data are used for the heatmap representations and are available as Source Data in a Source data file.
Sybr green quantitative qPCR
hTREM2 expression was detected on brain tissue extracts by qPCR using a SYBR Green Kit with primers and probe from Thermofisher (Catalog number: 4448892, Hs01003898_m1, Product size: 91 bp, Dye: FAM). Primers for hTREM2 were preliminary validated on human THP1 cells, a monocytic cell line that constitutively expresses hTREM2, and on BV2 murine cells transduced with the LV_hTREM2, both used as positive controls for hTREM2 expression. All values were normalized to the housekeeping gene GAPDH. hTREM2 expression was then measured on the brain of animals transplanted ICV with LV-hTREM2 HSCs at sacrifice at 10 months after the transplant.
Droplet Digital™ PCR (ddPCR) for VCN determination
For VCN determination, ddPCR was performed on genomic DNA extracted respectively from the liquid culture grown in vitro for 14 days post transduction and from the brain tissues by using the DNeasy Blood & Tissue kit (Qiagen). ddPCR was ran in duplicate using 20 ng of gDNA as template, ddPCR Supermix for Probes (Bio-Rad Laboratories Inc) and primers specific for HIV Psi region LENTI (FAM fluorochrome, forward primer: 5’-TGAAAGCGAAAGGGAAACCA-3’; Reverse primer 5’-CCGTGCGCGCTTCAG-3’; probe: AGCTCTCTCGACGCAGGACTCGGC) and for the murine RPP30-(HEX fluorochrome, forward primer: 5’-CCAGCTCCGTTTGTGATAGT-3’, Reverse primer 5’-CAAGGCAGAGATGCCCATAA-3’), located on chromosome 19 of murine genome.
Poisson statistical analysis of the numbers of positive and negative droplets yields absolute quantification of the target sequence.
Behavioral testing in Alzheimer’s disease mouse model
For phenotypic assessment, as behavioral tests, the elevated-plus maze (EPM) test was performed on the transplanted 5xFAD animals every month from age 4 months until the age of 12 months and the Morris Water Maze (MWM) test at study termination (12 months of mice age).
The elevated-plus maze (EPM)
The EPM was performed according to previously described procedures50,86,87 using a mouse EPM (model ENV-560A) purchased from Med Associates (St. Albans, VT). All four arms are elevated 1 meter from the floor, with the drop-off detectable only in the open arms. The EPM contained two open arms (35.5 cm×6 cm) and two closed arms (35.5 cm × 6 cm) radiating from a central area (6 cm × 6 cm). A 0.5 cm high lip surrounded the edges of the open arms, whereas the closed arms were surrounded by 20 cm high walls. The EPM was cleaned with Peroxigard before the beginning of the first test session and after each subject mouse was tested, with sufficient time for the ethanol odor to dissipate before the start of the next test session. Room illumination was ~30 lux. To begin the test, the mouse was placed in the central area facing the open arm. The mouse was allowed to freely explore for 5 min. Trials were videotaped and scored with EthoVision XT video tracking software (version 12.0, Noldus Information Technologies, Leesburg, VA).
Morris Water Maze (MWM)
A white, opaque, circular tub (60 cm depth x 83 cm diameter) was filled to 29 cm deep with water that was approximately 25 °C. Four visible, distinct shapes were places in each of the four quadrants inner walls of the tub to form distinct quadrants. Trials were videotaped and scored with EthoVision XT video tracking software (version 12.0, Noldus Information Technologies, Leesburg, VA). Acquisition training (day 1) consisted of 8 trials per mouse with a white platform (10 cm diameter) 0.5 cm above the surface of the water and marked with the red reflector. Each training trial began by lowering the mouse into the water close to the pool edge. The start location for each trial was alternated in a semirandom order for each mouse. Each mouse started in each of the quadrants twice. Hidden training (day 2–3) consisted of 20 trials per mouse, 12 on day 2 and 8 on day 3. The platform was placed in a new quadrant, 1 cm below the water. Each mouse started in each of the quadrants five times. For probe testing (day 4) the platform was removed and mice started opposite the position of where the hidden platform had been located. Mice were allowed to swim for 90 s. Reversal training (day 5 consisted of 1 trials per mouse, 3 from each starting quadrant. The platform was placed in a new quadrant, 1 cm below the water. Mice were allowed a maximum of 90 s to reach the platform. During visible, hidden and reversal trials, a mouse that failed to reach the platform in 60 s was guided to the platform by the experimenter. Mice were left on the platform for 5 s before being removed. After each trial, the subject was placed in a cage lined with absorbent paper towels and allowed to rest. Time spent in each quadrant and number of crossings over the trained platform location was used to analyze probe trial performance.
Statistics and reproducibility
For the comparison of two groups, we used Unpaired T-test with Welch’s correction.
For comparisons with more than two groups, one-way ANOVA followed by Tukey post-hoc test was used; for multiple variable comparisons, two-way ANOVA followed by Sidak post-hoc test was applied. Details on the exact statistical test used as well as the number of subjects and samples are included in the respective figure legends. P-values are reported in the respective figure legends whenever possible or are available as Supplementary information.
No statistical method was used to predetermine sample size. A biological triplicate (independent experiment) was guaranteed in both in vitro and in vivo experiments, except where specifically indicated.
Blinding analysis was adopted for behavioral testing in AD mice.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or are reported as Supplementary information. Raw data will be provided upon request to the corresponding authors. The raw RNA-seq data are deposited in GEO under the accession code GSE275808, and the RNA-seq and samples’ annotation data are available in the Zenodo database under accession code 12793606 [https://doi.org/10.5281/zenodo.12793606]. Source data are provided with this paper.
References
Gomez Perdiguero, E., Schulz, C. & Geissmann, F. Development and homeostasis of “resident” myeloid cells: the case of the microglia. Glia 61, 112–120 (2013).
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Wolf, S. A., Boddeke, H. W. & Kettenmann, H. Microglia in Physiology and Disease. Annu Rev. Physiol. 79, 619–643 (2017).
Wu, Y., Dissing-Olesen, L., MacVicar, B. A. & Stevens, B. Microglia: Dynamic Mediators of Synapse Development and Plasticity. Trends Immunol. 36, 605–613 (2015).
Paolicelli, R. C. et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011).
Capotondo, A. et al. Intracerebroventricular delivery of hematopoietic progenitors results in rapid and robust engraftment of microglia-like cells. Sci. Adv. 3, e1701211 (2017).
Capotondo, A. et al. Brain conditioning is instrumental for successful microglia reconstitution following hematopoietic stem cell transplantation. Proc. Natl Acad. Sci. USA 109, 15018–15023 (2012).
Corti, S. et al. Wild-type bone marrow cells ameliorate the phenotype of SOD1-G93A ALS mice and contribute to CNS, heart and skeletal muscle tissues. Brain 127, 2518–2532 (2004).
Beam, D. et al. Outcomes of unrelated umbilical cord blood transplantation for X-linked adrenoleukodystrophy. Biol. Blood Marrow Transpl. 13, 665–674 (2007).
Mildner, A. et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat. Neurosci. 10, 1544–1553 (2007).
Peterson, C. D. et al. AAV-Mediated Gene Delivery to the Spinal Cord by Intrathecal Injection. Methods Mol. Biol. 1950, 199–207 (2019).
Eichler, F. et al. Hematopoietic Stem-Cell Gene Therapy for Cerebral Adrenoleukodystrophy. N. Engl. J. Med 377, 1630–1638 (2017).
Sessa, M. et al. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: an ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet 388, 476–487 (2016).
Ungari, S. et al. Design of a regulated lentiviral vector for hematopoietic stem cell gene therapy of globoid cell leukodystrophy. Mol. Ther. Methods Clin. Dev. 2, 15038 (2015).
Biffi, A. et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341, 1233158 (2013).
Gentner, B. et al. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci. Transl. Med 2, 58ra84 (2010).
Capotondo, A. et al. Safety of arylsulfatase A overexpression for gene therapy of metachromatic leukodystrophy. Hum. Gene Ther. 18, 821–836 (2007).
Cartier, N. et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 326, 818–823 (2009).
Biffi, A. et al. Gene therapy of metachromatic leukodystrophy reverses neurological damage and deficits in mice. J. Clin. Invest 116, 3070–3082 (2006).
Rocca, C. J. et al. CRISPR-Cas9 Gene Editing of Hematopoietic Stem Cells from Patients with Friedreich’s Ataxia. Mol. Ther. Methods Clin. Dev. 17, 1026–1036 (2020).
Ferrari, G., Thrasher, A. J. & Aiuti, A. Gene therapy using haematopoietic stem and progenitor cells. Nat. Rev. Genet 22, 216–234 (2021).
Morgan, R. A., Gray, D., Lomova, A. & Kohn, D. B. Hematopoietic Stem Cell Gene Therapy: Progress and Lessons Learned. Cell Stem Cell 21, 574–590 (2017).
Ferrua, F. et al. Lentiviral haemopoietic stem/progenitor cell gene therapy for treatment of Wiskott-Aldrich syndrome: interim results of a non-randomised, open-label, phase 1/2 clinical study. Lancet Haematol. 6, e239–e253 (2019).
Taylor, M. et al. Hematopoietic Stem Cell Transplantation for Mucopolysaccharidoses: Past, Present, and Future. Biol. Blood Marrow Transpl. 25, e226–e246 (2019).
Stok, M. et al. Lentiviral Hematopoietic Stem Cell Gene Therapy Corrects Murine Pompe Disease. Mol. Ther. Methods Clin. Dev. 17, 1014–1025 (2020).
Chen, C. et al. Non-toxic HSC Transplantation-Based Macrophage/Microglia-Mediated GDNF Delivery for Parkinson’s Disease. Mol. Ther. Methods Clin. Dev. 17, 83–98 (2020).
Bosch, M. E. & Kielian, T. Neuroinflammatory paradigms in lysosomal storage diseases. Front Neurosci. 9, 417 (2015).
Cavalca, E. et al. Metallothioneins are neuroprotective agents in lysosomal storage disorders. Ann. Neurol. 83, 418–432 (2018).
Holley, R. J. et al. Macrophage enzyme and reduced inflammation drive brain correction of mucopolysaccharidosis IIIB by stem cell gene therapy. Brain 141, 99–116 (2018).
Sailor, K. A. et al. Hematopoietic stem cell transplantation chemotherapy causes microglia senescence and peripheral macrophage engraftment in the brain. Nat. Med 28, 517–527 (2022).
Wilkinson, F. L. et al. Busulfan conditioning enhances engraftment of hematopoietic donor-derived cells in the brain compared with irradiation. Mol. Ther. 21, 868–876 (2013).
Plasschaert, R. N. et al. High-throughput analysis of hematopoietic stem cell engraftment after intravenous and intracerebroventricular dosing. Mol. Ther. https://doi.org/10.1016/j.ymthe.2022.05.022 (2022).
Verdonk, F. et al. Phenotypic clustering: a novel method for microglial morphology analysis. J. Neuroinflammation 13, 153 (2016).
Langhammer, C. G. et al. Automated Sholl analysis of digitized neuronal morphology at multiple scales: Whole cell Sholl analysis versus Sholl analysis of arbor subregions. Cytom. A 77, 1160–1168 (2010).
Bennett, M. L. et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl Acad. Sci. USA 113, E1738–E1746 (2016).
Butovsky, O. et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014).
Chiu, I. M. et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 4, 385–401 (2013).
Grommes, C. et al. Regulation of microglial phagocytosis and inflammatory gene expression by Gas6 acting on the Axl/Mer family of tyrosine kinases. J. Neuroimmune Pharm. 3, 130–140 (2008).
Hickman, S. E. et al. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16, 1896–1905 (2013).
Matcovitch-Natan, O. et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670 (2016).
Lund, H. et al. Competitive repopulation of an empty microglial niche yields functionally distinct subsets of microglia-like cells. Nat. Commun. 9, 4845 (2018).
Cronk, J. C. et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J. Exp. Med. 215, 1627–1647 (2018).
Bennett, F. C. et al. A Combination of Ontogeny and CNS Environment Establishes Microglial Identity. Neuron 98, 1170–1183 e1178 (2018).
Bruttger, J. et al. Genetic Cell Ablation Reveals Clusters of Local Self-Renewing Microglia in the Mammalian Central Nervous System. Immunity 43, 92–106 (2015).
Ulland, T. K. & Colonna, M. TREM2 - a key player in microglial biology and Alzheimer disease. Nat. Rev. Neurol. 14, 667–675 (2018).
Lee, C. Y. D. et al. Elevated TREM2 Gene Dosage Reprograms Microglia Responsivity and Ameliorates Pathological Phenotypes in Alzheimer’s Disease Models. Neuron 97, 1032–1048 e1035 (2018).
Benitez, B. A. et al. Missense variant in TREML2 protects against Alzheimer’s disease. Neurobiol. Aging 35, 1510 e1519–1510 e1526 (2014).
Oakley, H. et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Jawhar, S., Trawicka, A., Jenneckens, C., Bayer, T. A. & Wirths, O. Motor deficits, neuron loss, and reduced anxiety coinciding with axonal degeneration and intraneuronal Abeta aggregation in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging 33, 196 e129–140 (2012).
Hogg, S. A review of the validity and variability of the elevated plus-maze as an animal model of anxiety. Pharm. Biochem Behav. 54, 21–30 (1996).
Schambach, A. et al. Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression. Gene Ther. 13, 641–645 (2006).
Tiesjema, B. et al. Viral mediated neuropeptide Y expression in the rat paraventricular nucleus results in obesity. Obes. (Silver Spring) 15, 2424–2435 (2007).
Sagoo, P. & Gaspar, H. B. The transformative potential of HSC gene therapy as a genetic medicine. Gene Ther. https://doi.org/10.1038/s41434-021-00261-x (2021).
Cartier, N., Lewis, C. A., Zhang, R. & Rossi, F. M. The role of microglia in human disease: therapeutic tool or target? Acta Neuropathol. 128, 363–380 (2014).
Xu, Z. et al. Efficient Strategies for Microglia Replacement in the Central Nervous System. Cell Rep. 32, 108041 (2020).
Bohlen, C. J. et al. Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron 94, 759–773 e758 (2017).
Xu, R. et al. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat. Commun. 11, 1577 (2020).
Parajuli, B. et al. Transnasal transplantation of human induced pluripotent stem cell-derived microglia to the brain of immunocompetent mice. Glia 69, 2332–2348 (2021).
Cohen-Pfeffer, J. L. et al. Intracerebroventricular Delivery as a Safe, Long-Term Route of Drug Administration. Pediatr. Neurol. 67, 23–35 (2017).
Slavc, I. et al. Best practices for the use of intracerebroventricular drug delivery devices. Mol. Genet Metab. 124, 184–188 (2018).
Duma, C. et al. Human intracerebroventricular (ICV) injection of autologous, non-engineered, adipose-derived stromal vascular fraction (ADSVF) for neurodegenerative disorders: results of a 3-year phase 1 study of 113 injections in 31 patients. Mol. Biol. Rep. 46, 5257–5272 (2019).
Villabona-Rueda, A., Erice, C., Pardo, C. A. & Stins, M. F. The Evolving Concept of the Blood Brain Barrier (BBB): From a Single Static Barrier to a Heterogeneous and Dynamic Relay Center. Front. Cell Neurosci. 13, 405 (2019).
Ge, S. & Pachter, J. S. Isolation and culture of microvascular endothelial cells from murine spinal cord. J. Neuroimmunol. 177, 209–214 (2006).
Bifari, F. et al. Neurogenic Radial Glia-like Cells in Meninges Migrate and Differentiate into Functionally Integrated Neurons in the Neonatal Cortex. Cell Stem Cell 20, 360–373 e367 (2017).
Benakis, C., Llovera, G. & Liesz, A. The meningeal and choroidal infiltration routes for leukocytes in stroke. Ther. Adv. Neurol. Disord. 11, 1756286418783708 (2018).
Schlager, C. et al. Effector T-cell trafficking between the leptomeninges and the cerebrospinal fluid. Nature 530, 349–353 (2016).
Decimo, I. et al. Meninges: A Widespread Niche of Neural Progenitors for the Brain. Neuroscientist 27, 506–528 (2021).
Banisadr, G. et al. Highly regionalized neuronal expression of monocyte chemoattractant protein-1 (MCP-1/CCL2) in rat brain: evidence for its colocalization with neurotransmitters and neuropeptides. J. Comp. Neurol. 489, 275–292 (2005).
Luskin, M. B. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron 11, 173–189 (1993).
Carlen, M. et al. Functional integration of adult-born neurons. Curr. Biol. 12, 606–608 (2002).
Meeker, R. B., Williams, K., Killebrew, D. A. & Hudson, L. C. Cell trafficking through the choroid plexus. Cell Adh Migr. 6, 390–396 (2012).
Kunis, G. et al. IFN-gamma-dependent activation of the brain’s choroid plexus for CNS immune surveillance and repair. Brain 136, 3427–3440 (2013).
Gosselin, D. et al. An environment-dependent transcriptional network specifies human microglia identity. Science 356, https://doi.org/10.1126/science.aal3222 (2017).
Shemer, A. et al. Engrafted parenchymal brain macrophages differ from microglia in transcriptome, chromatin landscape and response to challenge. Nat. Commun. 9, 5206 (2018).
Hohsfield, L. A. et al. Effects of long-term and brain-wide colonization of peripheral bone marrow-derived myeloid cells in the CNS. J. Neuroinflammation 17, 279 (2020).
Cuadros, M. A., Sepulveda, M. R., Martin-Oliva, D., Marin-Teva, J. L. & Neubrand, V. E. Microglia and Microglia-Like Cells: Similar but Different. Front Cell Neurosci. 16, 816439 (2022).
Li, C., Chen, Y. H. & Zhang, K. Neuroprotective Properties and Therapeutic Potential of Bone Marrow-Derived Microglia in Alzheimer’s Disease. Am. J. Alzheimers Dis. Other Demen 35, 1533317520927169 (2020).
Mishra, P. et al. Rescue of Alzheimer’s disease phenotype in a mouse model by transplantation of wild-type hematopoietic stem and progenitor cells. Cell Rep. 42, 112956 (2023).
Simard, A. R., Soulet, D., Gowing, G., Julien, J. P. & Rivest, S. Bone marrow-derived microglia play a critical role in restricting senile plaque formation in Alzheimer’s disease. Neuron 49, 489–502 (2006).
Simard, A. R. & Rivest, S. Neuroprotective properties of the innate immune system and bone marrow stem cells in Alzheimer’s disease. Mol. Psychiatry 11, 327–335 (2006).
Visigalli, I. et al. Gene therapy augments the efficacy of hematopoietic cell transplantation and fully corrects mucopolysaccharidosis type I phenotype in the mouse model. Blood 116, 5130–5139 (2010).
Peviani, M. et al. Lipophilic dye-compatible brain clearing technique allowing correlative magnetic resonance/high-resolution fluorescence imaging in rat models of glioblastoma. Sci. Rep. 10, 17974 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Team, R. C. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/ (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Bailey, K. R. & Crawley, J. N. In Methods of Behavior Analysis in Neuroscience Frontiers in Neuroscience (eds nd & J. J. Buccafusco) (2009).
Silverman, J. L. et al. Sociability and motor functions in Shank1 mutant mice. Brain Res 1380, 120–137 (2011).
Acknowledgements
We would like to thank Hind Bouzid, Guan Wang, Francisco J. Molina Estevez, Valentina Cipolla and Valeria Bortolotto for experimental support on the Alzheimer’s disease project and for supporting the optimization of RNAscope protocol and related analysis. We wish to thank Mauro Giuseppe Spatafora (University of Pavia) for the help with confocal microscope acquisitions and Claudia Balducci (“Mario Negri” Institute for Pharmacological Research) for kindly providing the anti-Amyloid beta antibody. This work was funded by ERC Consolidator Grant #617162, ERC Proof of Concept 2022 Grant TREM2MICROENGINES and sponsored research agreement funds to AB.
Author information
Authors and Affiliations
Contributions
R.M. undertook conceptualization, investigation, methodology and writing of the original draft, review and editing. A.M. carried out conceptualization, investigation, methodology, writing of the original draft, review and editing. R.K., F.F. and E.C. carried out conceptualization, investigation and methodology. S.D., P.R., P.C., Y.C. and D.P. performed methodology. A.C. carried out investigation and methodology. M.P. undertook conceptualization, investigation, methodology, review and editing. A.B. undertook supervision, conceptualization, investigation, review and editing.
Corresponding author
Ethics declarations
Competing interests
A.B., R.M., A.C. and M.P. are authors on a patent application related to this work (intracerebral transplantation of engineered HSCs) filed by Boston Children’s Hospital, Dana Farber Cancer Institute, the San Raffaele Hospital and Politecnico of Milano (application no. PCT/US17/56774); A.B. and M.P. are authors of a provisional patent application related to this work (TREM2 HSC gene therapy for AD) filed by Boston Children’s Hospital and Dana Farber Cancer Institute application no. PCT/US2020/053824). All other authors declare that they have no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Milazzo, R., Montepeloso, A., Kumar, R. et al. Therapeutic efficacy of intracerebral hematopoietic stem cell gene therapy in an Alzheimer’s disease mouse model. Nat Commun 15, 8024 (2024). https://doi.org/10.1038/s41467-024-52301-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52301-w
This article is cited by
-
Roles of TREM2 in Alzheimer’s disease
Translational Neurodegeneration (2025)
-
Advancements in Stem Cell Research for Effective Therapies Against Alzheimer’s Disease: Current Investigation and Future Insight
Molecular Neurobiology (2025)