Abstract
Unlocking the intricacies of protein structures and interactions within the dynamic landscape of subcellular organelles presents a significant challenge. To address this, we introduce SPACX, a method for spatially resolved protein complex profiling via biocompatible chemical cross(x)-linking with subcellular isolation, designed to monitor protein conformation, interactions, and translocation in living cells. By rapidly capturing protein complexes in their native physiological state and efficiently enriching cross-linked peptides, SPACX allows comprehensive analysis of the protein interactome within living cells. Leveraging structure refinement with cross-linking restraints, we identify subcellular-specific conformation heterogeneity of PTEN, revealing dynamic differences in its dual specificity domains between the nucleus and cytoplasm. Furthermore, by discerning conformational disparities, we identify 83 cytoplasm-exclusive and 109 nucleus-exclusive PTEN-interacting proteins, each associated with distinct biological functions. Upon induction of ubiquitin-proteasome system stress, we observe dynamic alterations in PTEN assembly and its interacting partners during translocation. These changes, including the identification of components and interaction sites, are characterized using the SPACX approach. Notably, SPACX enables identification of unique interacting proteins specific to PTEN isoforms, including PTEN and PTEN-Long, through the determination of sequence-specific cross-linking interfaces. These findings underscore the potential of SPACX to elucidate the functional diversity of proteins within distinct subcellular sociology.
Similar content being viewed by others
Introduction
Proteins engage in complex interactions and undergo conformational changes necessary for their specific functions1,2. Within the cellular environment, protein complexes experience unique factors, such as crowding, sticking, and confinement, essential for maintaining protein stability and aggregation3. Moreover, the dynamic assembly of protein complexes across different subcellular spaces forms a high-dimensional signal transduction network, facilitating exclusive structures and interactions tailored for specific functions4. Consequently, there is a pressing need to characterize the spatially dynamic conformation and interaction of protein complexes within living cells.
Various methodologies have emerged to study protein structures and interactions within cellular contexts. Cryo-electron tomography (cryo-ET) enables the visualization of large protein complexes like the nuclear pore complex5,6, which are challenging to purify intact in vitro. In-cell nuclear magnetic resonance spectroscopy (in-cell NMR) enhances the dynamic resolution of small proteins within the native crowded cellular environment7,8. Despite their merits, these approaches fall short of providing proteome-wide protein characterization.
Recently, proximity labeling technology has emerged as a powerful tool for capturing spatially adjacent protein interactions in living cells9,10,11. Leveraging proximity-dependent biotin identification technology (BioID) based mapping, comprehensive interaction networks within a human cell based on 192 subcellular markers were constructed, revealing 35,902 interactions with 4424 unique high-confidence proximity interactors12. Although proximity labeling technology surpasses affinity purification-mass spectrometry (AP-MS)13,14 in spatial resolution and capturing transient/weak interaction, it still lacks the capability to elucidate structural changes underlying interaction assembly with functional consequence.
To enhance the analysis of protein complexes, including direct interaction interfaces and structural constraints, chemical cross-linking mass spectrometry (CXMS) has emerged as a valuable complement to existing methods15,16. By inducing the formation of covalent bonds, CXMS effectively immobilizes protein conformation and interactions, enabling the comprehensive identification of intra- and inter-cross-linked site pairs within protein complexes with remarkable sensitivity. CXMS has been extensively utilized for analyzing protein complexes, ranging from purified proteins17,18 to cell lysates19,20,21, and even living cells22,23,24,25,26. However, the compatibility of chemical cross-linking with cellular viability and proteome expression remains a concern, which necessitates a thorough evaluation to ensure minimal disruption to the stability of protein complexes within the native microenvironment of living cells.
Furthermore, the inherent heterogeneity of protein complexes across different subcellular organelles often gets obscured in overall cellular analyses. To address this challenge and explore the protein interactome within specific subcellular compartments, the Heck group utilized DSSO to cross-link isolated human cell nuclei, revealing insights into chromatin-associated protein-protein interactions (PPIs)27. Similarly, the Borchers group utilized the enrichable cross-linker CBDPS to investigate PPIs within isolated yeast mitochondria, uncovering interactions involving a significant portion of the mitochondrial proteome28. Despite these advancements, existing methodologies still struggle to capture the dynamic assembly changes of protein complexes as they shuttle between different organelles within living cells, a critical aspect for deciphering the functional mechanisms of biological regulation.
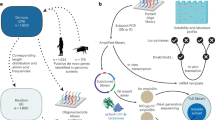
In this work, we introduce a spatially resolved protein complex profiling approach by biocompatible and deep-coverage chemical cross(x)-linking in living cells coupled with subcellular isolation (SPACX, Fig. 1). As a proof-of-concept, we successfully delineate the spatially specific assemblies of PTEN within the cytoplasm and nucleus. In addition, by using MG132 treatment, we capture the dynamic assembly of PTEN complexes during translocation from the cytoplasm to the nucleus, providing comprehensive insights into PTEN’s functional repertoire. Moreover, we identify specific interaction alterations associated with PTEN-Long (PTEN-L), an isoform of PTEN, underscoring the ability of our approach to elucidate the distinct roles of protein families in cellular functions by unraveling interactions mediated by different isoforms. Collectively, these findings show the robust performance of our developed method, SPACX, in characterizing protein dynamic assemblies in living cells during translocation.
Living cells are cross-linked, followed by separating of cytoplasm and nucleus, and cross-linked protein extraction and affinity purification. Subsequently, the extracted proteins react with azide-diazo-biotin (ADB) using bioorthogonal alkyne-azido click chemistry for biotin labeling, followed by digestion, enrichment of cross-linked peptides with streptavidin, and further release through reduction. Finally, the cross-linked peptides are subjected to LC-MS characterization for spatially specific cross-link analysis.
Results
Biocompatibility of cross-linking in living cells
To achieve a comprehensive analysis of protein conformation and interactions within living cells through chemical cross-linking, it is essential to balance the amphipathy of cross-linkers, ensuring rapid membrane permeability, high reactivity with amino acids in protein complexes within a restricted distance, minimal impact on cell viability, and negligible disturbance to proteome expression. In addition, introducing an enrichable moiety enhances the sensitivity of low-abundance cross-linked peptides through specific enrichment.
Taking advantage of the high reactivity and specificity of N-hydroxysuccinimidyl (NHS) ester for lysine residues, abundant on the protein surface, we synthesized bis(succinimidyl)propargyl with nitro compound BSPNO, previously termed as CLIP29. BSPNO comprises two NHS reactive groups and one flexible enrichment group alkyne, with a maximum Cα–Cα distance restraint of 27 Å (Supplementary Figs. 1 and 2 and Supplementary Methods 2.4.1). The amphipathy and size of BSPNO were evaluated, comparing cLogP and surface area values with popular cross-linkers (Supplementary Methods 2.4.2), demonstrating better water solubility than reported enrichable cross-linkers, PIR30 and Azide-A-DSBSO31 (Fig. 2a). In addition, BSPNO is slightly larger than the linear chain cross-linkers DSS and DSSO32, but smaller than other enrichable cross-linkers. This suitable amphipathy and size afford BSPNO good solubility and membrane permeability in physiological buffer (1× PBS, pH 7.4; 1% DMSO, v:v).
a Surface area and cLogP of BSPNO and a series of widely used cross-linking reagents. b Top: Histogram of the percentages of differentially expressed proteins for BSPNO cross-linked HeLa cells under different cross-linking times. A total of 2381 proteins, identified in at least two out of the three mass spectrometry replicate experiments, were used for the analysis. The data are presented as the average ± SEM, n = 3 independent experiments (control: 3 technical replicates, experiment: 3 MS replicates). The P values were calculated using a two-sided t test with non-cross-linked samples (0 min) serving as the control. *P < 0.05, **P < 0.01, and ns indicates not significant. P (3 min) = 0.20; P (5 min) = 0.11; P (10 min) = 0.03; P (15 min) = 0.004; P (30 min) = 0.006; P (60 min) = 0.01. Bottom: Box plot of integral Log2 protein intensity distribution for the identical 2381 proteins quantified via two or three replicates at the corresponding cross-linking time points. The median, the interquartile range, the minimum, and maximum values are indicated by line, box, and whiskers (n = 2381 proteins). The P values were calculated according to a two-sided t test with non-cross-linked samples (0 min) serving as the control. No significant difference was observed: P (3 min) = 0.96; P (5 min) = 0.82; P (10 min) = 0.90; P (15 min) = 0.46; P (30 min) = 0.76; P (60 min) = 0.77. c Fluorescence imaging of BSPNO distribution across cross-linked HeLa cells reacted for 5 min. The scale bars are 10 µm. d Identified cross-link proportions among proteins, peptides and spectra from in vivo cross-linked live BEL7402 cells before and after enrichment. e Percentage distributions of BSPNO and DSS cross-linked E. coli peptides.
The influence of BSPNO cross-linking time on cell viability was investigated. As the time increased to 60 min, the size of the cross-linked cells tended to shrink with statistical significance, monitored by microscopy (Supplementary Fig. 3 and Supplementary Methods 2.3.5). Meanwhile, CCK-8 measurements indicated minimal impact on cell viability (Supplementary Fig. 4, Supplementary Methods 2.3.6), and a quantitative proteomics approach evaluating the dynamic changes in protein expression revealed negligible disturbance (Supplementary Methods 2.3.1, 2.7(3)). As depicted in Fig. 2b, within 5 min of cross-linking, no significant protein expression difference was observed. However, when the cross-linking time increased, the percentage of differential proteins significantly increased, especially with the commonly adopted chemical cross-linking time of 60 min31,33. Moreover, an artificially compacted conformation might be induced with an extended cross-linking time, as also found in a previous report termed the “Zipper effect”34.
Therefore, 5 min was determined as the optimal in vivo BSPNO cross-linking time to minimize perturbation to cellular homeostasis. Furthermore, fluorescence imaging demonstrated effective intracellular transport of BSPNO within 5 min (Fig. 2c and Supplementary Methods 2.3.7), which is crucial to capture the dynamics changes in intracellular protein complex assemblies.
Coverage of cross-linking with enrichable cross-linkers
Expanding the identification coverage of CXMS analysis is vital for unraveling the functional architecture of protein complexes. To boost the detection coverage of cross-linked products, a cleavable azide-diazo-biotin (ADB) ligand was covalently linked to the cross-linked peptides via click chemistry of alkynyl derivatization, enabling enrichment with streptavidin beads. Through this enrichment strategy, the identification of cross-linker-modified proteins, peptides, and spectra was significantly enhanced (Fig. 2d, “Methods”: “Living cell cross-linking”, Supplementary Methods 2.3.1, 2.7(1), 2.7(3)). Compared with the commonly used nonenrichable cross-linker DSS, by BSPNO, the proportion of cross-linker-modified peptides identified from E. coli lysate was increased, reducing the regular peptides from 72% to 2% (Fig. 2e, Supplementary Data 1, and Supplementary Methods 2.3.3, 2.7(4)). Moreover, the number of spectra matched for each cross-linked peptide increased (Supplementary Fig. 5), ensuring higher confidence in identification.
In vivo cross-linking of commonly used human BEL7402 cells with BSPNO resulted in the identification of 3344 cross-linked peptides by applying separate 1% false discovery rate (FDR) control for intra-protein and inter-protein results at the spectral level (“Methods”: “Living cell cross-linking”, Supplementary Methods 2.7(1), Supplementary Data 2). The abundance distribution of cross-linked proteins spanned 7 orders of magnitude, with coverage comparable to recently published data22, but with a significantly shortened cross-linking time from 1 h to 5 min, ensuring high biocompatibility in live cells (Supplementary Fig. 6). Overall, these findings demonstrate that our method, involving biocompatible in vivo cross-linking within 5 min and the selective enrichment of cross-linked peptides, enables in-depth analysis of the protein interactome.
Identification of cross-links for proteome-wide profiling of protein conformation and interactions in living cells
Drawing from our comprehensive dataset of protein structural constraints and interactions in human BEL7402 cells, we identified a total of 13,215 inter-protein cross-linked sites corresponding to 3885 PPIs, involving 1567 proteins by applying separate 1% FDR control of intra-protein and inter-protein results at the spectral level. Additionally, we acquired 7768 intra-protein cross-linked sites corresponding to 2131 proteins (Supplementary Data 2, “Methods”: “Living cell cross-linking”, Supplementary Methods 2.7(1)). The constructed inter-protein interaction network presented in Supplementary Fig. 7 (Supplementary Data 3), encompasses numerous functional proteins, including histones, heat shock proteins (HSPs), transcription factors (TFs), ribosome proteins, zinc finger proteins, and calcium-related proteins. This highlights the deep-coverage cross-linking in live cells for the analysis of functional protein interactions. Analysis of the cellular component revealed the widespread distribution of PPIs throughout the cell, spanning the nucleus, cytosol, membrane, endoplasmic reticulum, and mitochondria (Fig. 3a). This demonstrates the efficacy of our method in efficiently cross-linking whole-cell protein complexes under high-activity conditions within a 5-min timeframe.
a Cellular component analysis of interacting proteins via Gene Ontology. Functional annotations were conducted whereby Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/). b Comparison of STRING score distribution between PPIs acquired by in vivo cross-linked live BEL7402 cells and from the STRING database. The cross-linked interface of BIP-PDIA6 and BIP-HYOU1 is depicted, with STRING score distributions for human PPIs in STRING (blue) and in vivo cross-linked live BEL7402 cells (yellow). c Co-IP analysis of Histone H2B and DPPA2. The experiment was repeated three times independently with similar results. d Co-IP analysis of Histone H2B and TOP2A. The experiment was repeated three times independently with similar results. e Distance distribution of all intra-protein cross-linked site pairs mapped onto available high-resolution structures from the PDB. We employed Cα–Cα Euclidean distances (ED) measured using the straight-line distance of atomic coordinates in the PDB file. f (i) Cross-linked site pairs mapped on the structure of endoplasmic reticulum chaperone BiP (PDB: 6ASY); (ii) Cross-linked site pairs mapped on the structure of voltage-dependent anion channel protein (PDB: 6TIR). (iii) Cross-linked site pairs mapped on the structure of Human mitochondrial acetoacetyl-CoA thiolase (PDB: 2F2S). Cross-linked site pairs shown as blue lines can be mapped on high-resolution structures within the distance constraint of 27 Å, whereas red lines exceeded 27 Å.
Furthermore, we compared the capability of in vivo and lysate cross-linking of BEL7402 cells in detecting PPIs (“Methods”: “Living cell cross-linking”, Supplementary Methods 2.3.4, 2.7(1), and 2.7(2)). Notably, in vivo cross-linking showed great superiority in detecting histone protein interactions, easily destroyed during cell lysis, and heat shock protein interactions, typically with weak and dynamic molecular chaperones (Supplementary Figs. 8 and 9). In addition, the relatively low-abundance transcription factor protein interactions were preferably acquired by in vivo cross-linking (Supplementary Fig. 10), likely due to the higher protein concentrations facilitated by the unique crowding and confinement of the intracellular environment.
Among the PPIs identified through in vivo cross-linking, 1258 PPIs were assigned reliability scores from the STRING database. As depicted in Fig. 3b, 57% of these interactions fell within the highest score range of 0.9–1.0, indicating their accuracy under intact cell conditions and the direct nature of the interactions determined by the cross-linked interface site. For instance, we examined the inter-cross-linked protein-protein interactions between BIP-PDIA6 and BIP-HYOU1. BIP plays a crucial role in facilitating the assembly of multimeric protein complexes within the endoplasmic reticulum, involved in protein folding and degradation of misfolded proteins. And its interacting proteins PDIA6 and HYOU1 both function as chaperones that participate in protein folding and in inhibiting the aggregation of misfolded proteins. The identification of cross-linked site pairs between these proteins provides structural insights complementary to existing functional evidence.
Furthermore, recognizing that in vivo cross-linking is adept at capturing weak or transient interactions often missed by in vitro methods like AP-MS, we conducted a comprehensive comparison of our acquired interactions with those in existing databases. Compared to STRING, BioGRID, BioPlex, and other in vivo cross-linking studies22,35,36,37, our approach identified an additional 1246 PPIs (Supplementary Fig. 11, Supplementary Data 3-PPIs database, “Methods”: “Living cell cross-linking”, Supplementary Methods 2.3.12, 2.7(1)). Molecular function analysis of proteins associated with these additionally identified PPIs highlighted enrichments in functions such as structural constituents of chromatin, RNA binding, protein heterodimerization activity, and calcium ion binding (Supplementary Fig. 12).
To validate a subset of identified interactions not annotated in existing databases, we performed co-immunoprecipitation (Co-IP) experiments (Supplementary Methods 2.3.8). Focusing on interactions enriched in chromatin-related pathways, we confirmed the interaction between Histone H2B and two other proteins: developmental pluripotency-associated protein 2 (DPPA2) and DNA topoisomerase 2-alpha (TOP2A). As illustrated in Fig. 3c, d, Co-IP results demonstrated the interaction between Histone H2B with DPPA2 and TOP2A. Drawing on the information provided in UniProt, DPPA2 plays a role in maintaining the active epigenetic state of specific genes, while TOP2A functions as a pivotal decatenating enzyme, altering DNA topology by binding to double-stranded DNA molecules. These interactions shed light on transcription and translation mechanisms, underscoring the efficacy of our method in uncovering molecular mechanisms underlying cellular processes.
In addition, to assess the reliability of our intramolecular cross-linking data in elucidating protein conformation, we mapped cross-linked sites onto high-resolution structures in the Protein Data Bank (PDB, http://www.rcsb.org) using ComMap38. In total, 2673 cross-linked sites were mapped onto their respective structures among 914 proteins, with 96% satisfaction of the maximum Cα–Cα distance restraint of 27 Å (Fig. 3e). This validated our identified cross-linked site pairs and the biocompatibility of our cross-linking conditions for protein structure stability. To assess the universality of our data, some cross-linked site pairs were mapped on representative protein structures with different cellular locations (Fig. 3f, Supplementary Fig. 13, and Supplementary Data 4). Nearly all cross-linked site pairs were mapped on the crystal structure with high structural compatibility (from 88% to 100%).
Spatially resolved profiling of protein complexes by biocompatible chemical cross-linking in living cells
Leveraging its high biocompatibility and deep-coverage capabilities for capturing protein complex assembly, coupled with high-quality subcellular isolation, our SPACX method was expanded to profile spatially specific protein conformation and interactions. Given that ~56% of all human proteins localize in multiple compartments to govern distinct functions39, it is crucial to understand the conformational and interactional dynamics in different subcellular locales and during translocation. We utilized SPACX, which specifically focuses on the subcellular compartmentalization of cytoplasm and nucleus, along with AP-MS analysis to enable subcellular-specific analysis of target protein conformation and interactions, using PTEN as an example.
Typically, studies40,41,42 have shown that wild-type PTEN, a frequently mutated gene in human cancer, is primarily expressed in the cytoplasm, with a smaller fraction localized exclusively in the nucleus. Hence, PTEN was selected as a representative case for our SPACX method (Fig. 4a). Under stress induced by the ubiquitin–proteasome system (UPS) through the proteasome inhibitor MG132, we observed a considerable increase in nuclear PTEN and a concurrent decrease in the cytoplasm, consistent with previous findings43 (Fig. 4b and Supplementary Methods 2.3.9). In addition, the malachite green phosphate assay revealed a reduction in PTEN’s phosphatase activity upon MG132 stimulation (Supplementary Fig. 14 and Supplementary Methods 2.3.10).
a Schematic outline for subcellular analysis of PTEN complexes by SPACX. Living 293APTEN-GLB cells were cross-linked, their cytoplasm and nucleus were isolated, and PTEN and its interacting proteins were affinity-purified using streptavidin beads. The purified cross-linked proteins were biotin-labeled with ADB using bioorthogonal alkyne-azido click chemistry. Following digestion into a peptide mixture, cross-linked peptides were selectively purified using streptavidin and released via a reduction reaction. These cross-linked peptides were analyzed using mass spectrometry to characterize spatially specific cross-links. b Diagram and fluorescence imaging analysis of PTEN localization and nuclear translocation. The scale bars are 10 µm. c Western blot analysis of purified cytoplasm and nucleus, using GAPDH and Lamin B as markers, alongside PTEN expression levels. d Cross-linked site pairs of PTEN from cytoplasm and nucleus before and after MG132 stimulation. (i) Cross-linked site maps. All cross-linked site pairs identified within PTEN are illustrated using the linear connectivity map. (ii) Surface distribution of cross-linked sites. The abundance distribution of cross-linked sites is mapped on an AlphaFold2 full-length structure (AF-P60484-F1-model_v2, downloaded on 2022-7-17, https://alphafold.ebi.ac.uk/entry/P60484). The red spheres represent the amino acids modified by the cross-linker. The size of each ball is proportional to the total number of spectra for the modified amino acid (including cross-links, loop-links, and mono-links), and calculated by dividing the number of spectra by 10. This size is indicative of the modification frequency. (iii) Cα–Cα distance map. Cross-linked site pairs mapped on the structure of PTEN (PDB: 1D5R). Cross-linked site pairs are shown as blue lines and could be mapped on high-resolution structure within a constraint distance of 27 Å, while red lines exceeded 27 Å. (iv) Structure ensemble refinement. Ensemble refinement for PTEN conformational changes against cross-linking restraints that satisfy all the cross-linking site pairs based on the structure of PTEN (PDB: 1D5R). The protein structural ensemble is based on the density of Cα atoms, with denser grids indicating a higher density of atoms, signifying a greater likelihood of protein conformations being localized in that specific region. The green-colored is a phosphatase tensin-type domain, and the cyan-colored is a C2 tensin-type domain.
To probe the spatial heterogeneity of PTEN’s conformation and interaction, as well as its nuclear translocation process, we expressed PTEN in a stable cell line (293APTEN-GLB) tagged with biotin and green fluorescent protein (GFP)44 (Supplementary Fig. 15 and Supplementary Methods 2.2). Subsequently, we cross-linked live 293APTEN-GLB cells, isolated cytoplasm and nucleus, and affinity-purified PTEN and its interacting proteins using streptavidin beads. We confirmed the high purity separation of cytoplasm and nucleus by Western blot (Fig. 4c). Analyzing the cross-linked peptides derived from the enriched PTEN complex, we identified 18 and 45 cross-linked site pairs of PTEN in the cytoplasm and nucleus (Supplementary Data 5). By mapping these cross-linked site pairs to the PTEN crystal structure (PDB: 1D5R), 5 and 9 cross-linked site pairs were found to exceed the maximum distance restraint of BSPNO cross-linking in the cytoplasm and nucleus, respectively. Among them, 4 and 6 cross-linked site pairs were located between the phosphatase tensin-type domain and the C2 tensin-type domain in the cytoplasm and nucleus, respectively. The ensemble structure was refined according to cross-linking restraints using the Xplor-NIH approach45,46,47 to present subcellular-specific PTEN conformations (Supplementary Methods 2.5). The representation of the protein structures is based on the density of Cα atoms, rather than a cartoon. The denser the grid, the higher the density of atoms, suggesting a higher probability of protein conformations being distributed in that particular position. In the nucleus, a broader range of dynamic conformational changes in the dual specificity domains was found than that in the cytoplasm. The differences persisted even after MG132 stimulation, indicating subcellular-specific conformational alterations that likely contribute to functional diversity in each subcellular compartment (Fig. 4d).
In addition, owing to these conformational disparities, distinct binding sites were exposed to interact with other proteins for functional execution in both the nucleus and cytoplasm, even under MG132 stimulation (Fig. 5a, Supplementary Fig. 16, and Supplementary Data 6). As PTEN interactions formed, specific assemblies like NTH were exclusively found in the nucleus, while PPP5, MK14, PP2AB, PROF1, and PSME2 were localized solely in the cytoplasm. Other proteins, IF5, CCND1, CSK21, and E2AK2, were identified in both cellular compartments. These localizations were further validated through immunofluorescence experiments (Supplementary Fig. 17). Furthermore, gene ontology analysis based on these subcellular-resolved PTEN interactions revealed that proteins exclusively identified in the cytoplasm were highly enriched in biological processes such as translation, apoptosis, ubiquitin–proteasome, and phosphorylation (Supplementary Fig. 18a). Conversely, proteins exclusively identified in the nucleus were primarily associated with transcription, nucleosome assembly, cell proliferation, and telomere organization (Supplementary Fig. 18b).
a Map of cross-linking site number between PTEN and interacting proteins in the cytoplasm without MG132 stimulation and nucleus after MG132 stimulation. b An alluvial diagram representing PTEN-interacting proteins transiting between cytoplasm and nucleus after MG132 stimulation. The number of proteins translocated to and from each subcellular compartment is indicated next to the labeled strata. c Gene Ontology Biological process (GOBP) analysis of PTEN-interacting proteins newly identified after MG132 stimulation. Functional annotations were conducted whereby DAVID. d Western blot analysis of PTEN-interacting proteins MK14, PP2AB, PPP5, and PROF1 via PTEN pull-down. Uncropped blots are provided in the source data file. e Immunofluorescence imaging analysis of PTEN colocalization (green, GFP) and PTEN nuclear translocation interacting proteins MK14, PP2AB, PPP5, and PROF1 (red, Alexa Fluor 647-conjugated antibody). The merged image includes a demarcated nucleus (blue, DAPI). The scale bars are 10 µm. f Localization of PTEN in respective cells before and after MG132 stimulation. NC indicates the negative control, while siMK14, siPP2AB, siPPP5, and siPROF1 indicate that the corresponding protein has been knocked out from 293APTEN-GLB cells. PTEN-GFP is depicted in green, while DAPI staining is in blue. The scale bars are 10 µm.
Upon MG132 treatment, the interaction proteins of PTEN in the nucleus and cytoplasm underwent significant changes, corresponding to PTEN translocation into the nucleus. Notably, 152 previously identified PTEN-interacting proteins were no longer observed after MG132 stimulation, while 343 additional interacting proteins were detected, primarily found in the nucleus (Fig. 5b). These proteins were highly enriched in biological processes such as transcription, phosphorylation, apoptosis, cell cycle, and ubiquitin–proteasome (Fig. 5c). Furthermore, 25 interacting proteins previously identified exclusively in the cytoplasm were found solely in the nucleus after translocation. Among them, we selected proteins with different abundance levels in the cell and considered antibody accessibility for the validation experiment. The interactions between PTEN and MK14, PP2AB, PPP5, and PROF1 were confirmed through IP–western blot analysis (Fig. 5d), demonstrating translocation from the cytoplasm to the nucleus via immunofluorescence (Fig. 5e). The biological processes of these interaction proteins were closely associated with translation, phosphorylation, DNA repair, and ubiquitin–proteasome (Supplementary Fig. 18c).
To ascertain that the identified interacting proteins were not randomly captured through cross-linking, we employed small-interfering RNA (siRNA) to knock down MK14, PP2AB, PPP5, and PROF1, as depicted in Supplementary Fig. 19 (Supplementary Methods 2.3.11). This allowed us to further assess the biological functions of these interacting proteins. Interestingly, knockdown of these genes significantly impacted the nuclear translocation of PTEN following MG132 stimulation (Fig. 5f and Supplementary Fig. 20), thereby confirming the reliability of our captured interaction proteins with PTEN during translocation. Together, these results underscored the superiority of SPACX for identifying suborganelle-shuttle protein structural and interactional dynamics in vivo, enabling comprehensive function exploration and mechanism revelation.
Furthermore, PTEN-L, known to share structural homology with PTEN, exhibits distinct functionalities44. Leveraging the site-specific resolution capabilities of CXMS, we aimed to investigate interactions involving the unique N-terminal region of PTEN-L within cells using SPACX. PTEN-L is an N-terminally extended isoform produced by alternative initiation at a CTG start codon in PTEN, adding 173 N-terminal disordered amino acids followed by the classical 403 amino acids of PTEN (Fig. 6a). Identifying specific interacting proteins associated with the unique isoform sequence is essential for discriminating their functions. In our investigations, PTEN was expressed in the stable cell line 293APTEN-GLB with biotin and GFP tags, whereas PTEN-L was expressed lacking tags (Supplementary Figs. 21 and 22). Initially, we detected conformational changes of PTEN/PTEN-L between the unique and shared sequences concerning the subcellular locations of the cytoplasm and nucleus, as well as under the states with or without MG132 treatment. However, it was unclear whether these changes were intramolecular or intermolecular (Fig. 6b and Supplementary Data 7). By identifying cross-linked sites within the interacting interface, we obtained interactions corresponding to the unique and shared sequences of PTEN and PTEN-L (Fig. 6c). This is necessary for understanding the independently formed interactional assembly, particularly for deciphering the function of the unique sequence. Before stimulation, the unique N-terminal region of PTEN-L predominantly interacted with proteins implicated in DNA-binding and chromatin regulation. However, following MG132 stimulation, this specific segment within PTEN-L displayed heightened activity in the nucleus, showing a notable affinity for binding to proteases and various enzymes, as depicted in Fig. 6d. This heightened interaction could be attributed to the distinct composition of PTEN-L, characterized by a disordered region enriched with basic residues, linear binding motifs, and numerous post-translational modification sites48. These features suggest a crucial role for PTEN-L in bolstering PTEN’s tumor-suppressive capabilities and its involvement in intracellular transport, as previously observed49. The uncovered interaction between PTEN-L and nuclear enzymes introduces a pathway that impacts fundamental cellular functions, including chromosome organization, protein synthesis, and degradation. Unveiling this mechanism is imperative for regulating cell growth and provides valuable insights into cancer biology, with potential implications for the development of targeted therapies. Therefore, our findings offer a preliminary understanding of PTEN isoform interactions in the regulation of transcription and protein homeostasis, further underscoring the capacity of SPACX to delineate the functional specificity and dynamics of protein isoforms in vivo.
a Sequences of PTEN and PTEN-L. b Cross-linked site pairs within PTEN-L are indicated using linear connectivity maps. The blue-colored region indicated the unique sequence of PTEN-L, and the gray-colored region denoted the shared sequence with PTEN. The blue dotted line indicating cross-linked site pairs cannot be distinguished as intra PTEN-L cross-linked site pairs or inter-cross-linked site pairs between PTEN and PTEN-L. c Venn diagram of interacting proteins between shared sequence of PTEN isoforms and PTEN-L unique sequence. d Protein functional analysis of unique PTEN-L-specific sequence interacting proteins.
Discussion
The comprehensive analysis of subcellular-specific protein conformation and interaction dynamics holds immense significance in deciphering the functional mechanisms of proteins involved in different biological processes. In this work, we introduced an approach called SPACX, which leverages chemical bioorthogonal cross-linking with deep coverage in living cells, coupled with subcellular isolation, to elucidate the dynamic changes in protein conformation and interaction within living cells, including during the translocation between different organelles.
By employing the cross-linker BSPNO, characterized by its rapid membrane permeability, high reactivity, and balanced amphipathy, we achieved in vivo cross-linking with minimal disruption to living cells within a short timeframe of 5 min. In addition, the selective enrichment of cross-linked peptides by introducing a biotin tag through alkyne-azido click chemistry ensured extensive coverage, facilitating the study of dynamic protein complex assembly within living cells.
Our study identified 13,215 inter-protein cross-linked sites in live human BEL7402 cells, mapping to 3885 PPIs involving 1567 proteins and 7768 intra-protein cross-linked sites among 2131 protein structural constraints. These interactions spanned diverse functions across nearly all cellular compartments. Encompassing highly dynamic and weak binding interactions, as well as low-abundance protein complexes, like histones, HSPs, and TFs, which are often challenging to detect using in vitro methods. This achievement was primarily facilitated by the rapid capture of interaction sites of protein complexes in the authentic microenvironment of live cells, ensuring the mapping of flexible structural regions crucial for protein recruitment and complex formation.
Furthermore, by integrating highly efficient subcellular separation techniques, we enabled the spatially resolved characterization of protein complex conformation and interactions within specific subcellular locations under varying cellular conditions. Structural refinement against cross-linking restraints unveiled a broader range of dynamic changes in PTEN conformation within the nucleus compared to the cytoplasm. Our approach also unveiled diverse interacting networks, validated through western blot and fluorescence colocalization analysis. Through biological function analysis, we delineated distinct roles of PTEN-interacting proteins in the cytoplasm and nucleus, shedding light on the spatially specific structure, interaction, and function of protein assemblies, as well as their dynamic alterations during subcellular translocation. Moreover, we demonstrated the feasibility of SPACX in studying individual interactions of PTEN-L isoform, revealing potential associations of its 173 N-terminal intrinsically disordered amino acids with the regulation of transcription and protein homeostasis.
In summary, our findings suggest that SPACX holds promise as a versatile toolkit for spatially resolved profiling of dynamic protein conformation and interactome in living cells.
Methods
Living cell cross-linking
Cells were harvested in centrifuge tubes and washed three times with 1× PBS prior to cross-linking. The cell pellet of 5 × 106 cells was resuspended and cross-linked using 300 µL of 1× PBS (1% DMSO, v/v) with 2 mM BSPNO at room temperature. The cross-linked cells were collected by centrifugation, and the cell pellet was resuspended in 300 µL of 50 mM NH4HCO3 at room temperature for 5 min to stop the cross-linking reaction. The cell pellet obtained by centrifugation was directly used for protein extraction. In this work, all cell types cross-linked at the living cell level followed the same procedure.
Cytoplasm and nucleus analysis of PTEN cross-linking
A total of 8.8 × 108 293APTEN-GLB cells were cross-linked in vivo. We then employed an SC-003 extraction kit (Invent Biotechnologies MinuteTM) for cytoplasmic and nuclear isolation. Pull-down of PTEN was performed by incubating the cytoplasmic and nuclear fractions with 1.76 mL streptavidin agarose beads, respectively. Following incubation overnight at 4 °C, streptavidin agarose beads were washed three times with three volumes of 0.2% SDS (1× PBS). The proteins on the beads were incubated with 400 µL 1% SDS (1× PBS) at 95 °C for 10 min for follow-up click chemistry cross-linked peptides enrichment.
MS sample preparation
Cross-linked BEL7402 cell samples and PTEN nucleus fraction samples were desalted and fractionated into six fractions using homemade Durashell C18 Tips. PTEN cytoplasm fraction samples were desalted and fractionated into three fractions using the same Tips. Details about the desalting and fractionation process can be found in Supplementary Methods 2.6.
Liquid chromatography-mass spectrometry (LC-MS) analysis
The living cell cross-linked samples were analyzed using an Easy-nLC 1200 system coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Samples were automatically loaded onto a C18 RP trap column (150 µm i.d. × 3 cm) and separated using a C18 capillary column (150 µm i.d. × 15 cm), packed in-house with ReproSil-Pur C18-AQ resin (1.9 µm, 120 Å).
Identification of cross-links
pLink250 software was employed for cross-link identification with precursor mass accuracy at 20 ppm, and fragment ion mass accuracy at 20 ppm. The results were filtered by applying separate 1% FDR control of intra-protein and inter-protein results at the spectral level. The search parameters used: instrument, HCD; the peptide length was set to 5–60, Carbamidomethyl[C] as fixed modification, Acetyl[Protein N-term] and Oxidation[M] as variable modification. The cross-linker was set as BSPNO (cross-linking sites K and protein N terminus, cross-link mass-shift 456.176, mono-link mass-shift 474.186) and DSS (cross-linking sites K and protein N terminus, cross-link mass-shift 138.068, mono-link mass-shift 156.079). Trypsin was the protease with a maximum of three missed cleavages allowed.
Details about LC-MS analysis and data processing are in Supplementary Methods 2.7.
Ensemble refinement of PTEN against cross-linking restraints
Ensemble refinement against the cross-linking restraints was conducted using Xplor-NIH45,46,47. For each conformer, the N-terminal domain (residues 14–184) was fixed, and the C-terminal domain (residues 193–327) was moved as one unit. The linker residues between the N-terminal domain and the C-terminal domain (residues 185–192) possessed full torsion angle freedom. The conformers in the ensemble shared all cross-linking restraints and were allowed to overlap. If the two-conformer ensemble could not satisfy all cross-linking restraints, three or more conformers were supplemented to the ensemble until all the restraints were satisfied. We conducted the calculation 960 times for PTEN using different random seeds. Structures with no violation of cross-linking restraints and no clashing residues were employed for further structural analysis. Detailed procedures can be found in the Supplementary Methods 2.5.
Western blot and immunofluorescence analysis
Pull-down of PTEN and interacting proteins was conducted by incubating the cytoplasmic and nuclear fractions with streptavidin agarose beads. Following overnight incubation at 4 °C, streptavidin agarose beads were washed with 0.2% SDS (1× PBS) three times. The proteins on beads were incubated with 1% SDS (1× PBS) at 95 °C for 10 min. Equal amounts of the boiled proteins were loaded for Western blot analysis, and specific proteins were characterized using corresponding antibodies. Detailed antibody information is provided in Supplementary Methods 2.1. All western blot images were obtained using a BIO-RAD Molecular Imager with Image Lab Software.
For immunofluorescence analysis, 293APTEN-GLB cells were placed on glass coverslips with DMEM supplemented with 10% (v/v) FBS. The stimulated cells were treated with 10 µM MG132 for 24 h, then washed twice with 1× PBS, and fixed with 4% (v/v) paraformaldehyde (PFA) for 15 min at room temperature (all subsequent steps were performed at room temperature unless otherwise noted), and permeabilized using 0.25% Triton X-100 in 1× PBS for 10 min. After rinsing three times, the cells were further blocked using blocking buffer for 30 min, labeled with primary antibodies overnight at 4 °C, and incubated with secondary antibody conjugated with Alexa Fluor Plus 647 (1:500) for 1 h. After washing with 1× PBS, the cells were stained with DAPI (5 µg/mL). The antibodies used in this study are outlined in Supplementary Methods 2.1. Protein localization was characterized via GFP, DAPI, and Alexa Fluor Plus 647 using an inverted fluorescence microscope and an Andor live cell confocal imaging platform combined with a CSU-W1 Spinning Disk Field Scanning Confocal System, iXon Ultra EMCCD, with three laser lines (405 nm, 488 nm and 640 nm) using a 100× objective.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry raw data analysis has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD043485. Source data are provided with this paper.
References
Alberts, B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92, 291–294 (1998).
Scott, J. D. & Pawson, T. Cell signaling in space and time: where proteins come together and when they’re apart. Science 326, 1220–1224 (2009).
Titeca, K., Lemmens, I., Tavernier, J. & Eyckerman, S. Discovering cellular protein-protein interactions: technological strategies and opportunities. Mass Spectrom. Rev. 38, 79–111 (2019).
Lundberg, E. & Borner, G. H. H. Spatial proteomics: a powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol. 20, 285–302 (2019).
Zhang, Y. et al. Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex. Cell Res. 30, 1–9 (2020).
Kim, S. J. et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 (2018).
Breindel, L., Burz, D. S. & Shekhtman, A. Interaction proteomics by using in-cell NMR spectroscopy. J. Proteom. 191, 202–211 (2019).
Ikeya, T., Güntert, P. & Ito, Y. Protein structure determination in living cells. Int. J. Mol. Sci. 20, 2442 (2019).
Lobingier, B. T. et al. An approach to spatiotemporally resolve protein interaction networks in living cells. Cell 169, 350–360 (2017).
Ke, M. et al. Spatiotemporal profiling of cytosolic signaling complexes in living cells by selective proximity proteomics. Nat. Commun. 12, 71 (2021).
Li, J. et al. Cell-surface proteomic profiling in the fly brain uncovers wiring regulators. Cell 180, 373–386.e315 (2020).
Go, C. D. et al. A proximity-dependent biotinylation map of a human cell. Nature 595, 120–124 (2021).
Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017).
Huttlin, E. L. et al. The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 (2015).
Yu, C. & Huang, L. Cross-linking mass spectrometry: an emerging technology for interactomics and structural biology. Anal. Chem. 90, 144–165 (2018).
Iacobucci, C. et al. First community-wide, comparative cross-linking mass spectrometry study. Anal. Chem. 91, 6953–6961 (2019).
Gutierrez, C. et al. Structural dynamics of the human COP9 signalosome revealed by cross-linking mass spectrometry and integrative modeling. Proc. Natl. Acad. Sci. USA 117, 4088–4098 (2020).
Armony, G. et al. Cross-linking reveals laminin coiled-coil architecture. Proc. Natl. Acad. Sci. USA 113, 13384–13389 (2016).
Ryl, P. S. J. et al. In situ structural restraints from cross-linking mass spectrometry in human mitochondria. J. Proteome Res. 19, 327–336 (2019).
Liu, F., Rijkers, D. T., Post, H. & Heck, A. J. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 12, 1179–1184 (2015).
Tan, D. et al. Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. eLife 5, e12509 (2016).
Wheat, A. et al. Protein interaction landscapes revealed by advanced in vivo cross-linking-mass spectrometry. Proc. Natl. Acad. Sci. USA 118, e2023360118 (2021).
Wu, X. et al. In vivo protein interaction network analysis reveals porin-localized antibiotic inactivation in Acinetobacter baumannii strain AB5075. Nat. Commun. 7, 13414 (2016).
Gotze, M., Iacobucci, C., Ihling, C. H. & Sinz, A. A simple cross-linking/mass spectrometry workflow for studying system-wide protein interactions. Anal. Chem. 91, 10236–10244 (2019).
Jiang, P. L. et al. A membrane-permeable and immobilized metal affinity chromatography (IMAC) enrichable cross-linking reagent to advance in vivo cross-linking mass spectrometry. Angew. Chem. Int. Ed. Engl. 61, e202113937 (2022).
O’Reilly, F. J. et al. In-cell architecture of an actively transcribing-translating expressome. Science 369, 554–557 (2020).
Fasci, D., Ingen, H. V., Scheltema, R. A. & Heck, A. J. R. Histone interaction landscapes visualized by crosslinking mass spectrometry in intact cell nuclei. Mol. Cell Proteom. 17, 2018–2033 (2018).
Makepeace, K. A. T. et al. Improving identification of in-organello protein-protein interactions using an affinity-enrichable, isotopically coded, and mass spectrometry-cleavable chemical crosslinker. Mol. Cell Proteom. 19, 624–639 (2020).
Chowdhury, S. M. et al. Identification of cross-linked peptides after click-based enrichment using sequential collision-induced dissociation and electron transfer dissociation tandem mass spectrometry. Anal. Chem. 81, 5524–5532 (2009).
Tang, X., Munske, G. R., Siems, W. F. & Bruce, J. E. Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal. Chem. 77, 311–318 (2005).
Kaake, R. M. et al. A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol. Cell Proteom. 13, 3533–3543 (2014).
Kao, A. et al. Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell Proteom. 10, 1–17 (2011).
Chavez, J. D. et al. Cellular interactome dynamics during paclitaxel treatment article cellular interactome dynamics during paclitaxel treatment. Cell Rep. 29, 2371–2383 (2019).
Graziadei, A. & Rappsilber, J. Leveraging crosslinking mass spectrometry in structural and cell biology. Structure 30, 37–54 (2022).
Zhao, L. et al. Enhanced protein–protein interaction network construction promoted by in vivo cross-linking with acid-cleavable click-chemistry enrichment. Front. Chem. 10, 994572 (2022).
Gao, H. et al. In-depth in vivo crosslinking in minutes by a compact, membrane-permeable, and alkynyl-enrichable crosslinker. Anal. Chem. 94, 7551–7558 (2022).
Chen, J. et al. A glycosidic‐bond‐based mass‐spectrometry‐cleavable cross‐linker enables in vivo cross‐linking for protein complex analysis. Angew. Chem. Int. Ed. Engl. 62, e202212860 (2023).
Zhang, W. et al. ComMap: a software to perform large-scale structure-based mapping for cross-linking mass spectrometry. Bioinformatics 39, btad077 (2023).
Yang, L., Lv, Y., Li, T., Zuo, Y. & Jiang, W. Human proteins characterization with subcellular localizations. J. Theor. Biol. 358, 61–73 (2014).
Chen, J. H. et al. ATM-mediated PTEN phosphorylation promotes PTEN nuclear translocation and autophagy in response to DNA-damaging agents in cancer cells. Autophagy 11, 239–252 (2015).
Bononi, A. & Pinton, P. Study of PTEN subcellular localization. Methods 77-78, 92–103 (2015).
Liu, T., Wang, Y., Wang, Y. & Chan, A. M. Multifaceted regulation of PTEN subcellular distributions and biological functions. Cancers 11, 1247 (2019).
Ge, M. K. et al. FBXO22 degrades nuclear PTEN to promote tumorigenesis. Nat. Commun. 11, 1720 (2020).
Liang, H. et al. PTENalpha, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 19, 836–848 (2014).
Schwieters, C. D. et al. The Xplor-NIH NMR molecular structure determination package. J. Magn. Res. 160, 66–74 (2003).
Schwieters, C. D., Kuszewski, J. J. & Clore, G. M. Using Xplor-NIH for NMR molecular structure determination. Progr. NMR Spectrosc. 48, 47–62 (2006).
Gong, Z. et al. Visualizing the ensemble structures of protein complexes using chemical cross-linking coupled with mass spectrometry. Biophys. Rep. 1, 127–138 (2015).
Malaney, P., Uversky, V. N. & Davé, V. The PTEN long N-tail is intrinsically disordered: increased viability for PTEN therapy. Mol. BioSyst. 9, 2877 (2013).
Hopkins, B. D. et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science 341, 399–402 (2013).
Chen, Z. L. et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 10, 3404 (2019).
Acknowledgements
This work was supported by the National Natural Science Foundation (22322411 to Q.Z., 22074139 to Q.Z., 32088101 to Y.Z., 21991083 to Y.Z.), National Key Research and Development Program of China (2021YFA1301501 to Q.Z.), and the Youth Innovation Promotion Association, CAS (2020184 to Q.Z.).
Author information
Authors and Affiliations
Contributions
L. Zhao, Q.Z., and L. Zhang designed research; L. Zhao, Y.A., N.Z., H.G., and X.L. performed research; H.G., W.Z., and Z.G. contributed reagents/analytic tools; L. Zhao and Y.A. analyzed data; L. Zhao, Q.Z., and L. Zhang wrote the paper; Q.Z., B.Z., Z.L., C.T., L. Zhang, and Y.Z. conceived and directed the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Iris Smith, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, L., An, Y., Zhao, N. et al. Spatially resolved profiling of protein conformation and interactions by biocompatible chemical cross-linking in living cells. Nat Commun 15, 8331 (2024). https://doi.org/10.1038/s41467-024-52558-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52558-1