Abstract
Personalized treatment for patients with advanced solid tumors critically depends on the deep characterization of tumor cells from patient biopsies. Here, we comprehensively characterize a pan-cancer cohort of 150 malignant serous effusion (MSE) samples at the cellular, molecular, and functional level. We find that MSE-derived cancer cells retain the genomic and transcriptomic profiles of their corresponding primary tumors, validating their use as a patient-relevant model system for solid tumor biology. Integrative analyses reveal that baseline gene expression patterns relate to global ex vivo drug sensitivity, while high-throughput drug-induced transcriptional changes in MSE samples are indicative of drug mode of action and acquired treatment resistance. A case study exemplifies the added value of multi-modal MSE profiling for patients who lack genetically stratified treatment options. In summary, our study provides a functional multi-omics view on a pan-cancer solid tumor cohort and underlines the feasibility and utility of MSE-based precision oncology.
Similar content being viewed by others
Introduction
Improved understanding of tumor biology and increasingly personalized treatment has led to significant improvements in the prognosis of cancer patients1,2. Tailoring drugs to specific genomic alterations enables patient stratification and informs further drug development3,4. For example, a variety of successful targeted therapies with associated companion diagnostic tests exist across various disease types, including treatments for HER2-positive breast cancers5,6,7, BRAF V600E mutated melanoma8,9,10,11, and EGFR mutated non-small cell lung cancer (NSCLC)12,13,14,15. However, several studies matching treatments to patients based on genomic alterations alone have reported limited clinical benefit16,17,18. This observation may result from two challenges: First, a significant fraction of tumors do not harbor any actionable driver mutations, and second, these studies focused on recurrent and heavily pretreated disease, where tumors often develop a complex landscape of secondary mutations leading to treatment resistance. By focusing on earlier treatment lines and applying more sophisticated treatment matching algorithms, other studies have demonstrated a clinical benefit of genomics-based precision medicine19,20,21.
Tumor biology, as well as the resulting drug resistance and sensitivity patterns, are increasingly understood to result from the interplay between different layers of regulation. These include epigenetic modifications, changes in gene expression and protein abundance, and rewiring of signaling and metabolic pathways. Multi-omics and functional approaches thus complement genomics-based precision medicine and have the potential to uncover composite biomarkers to improve patient stratification22,23,24,25,26,27,28.
However, the opportunities for comprehensive functional and molecular tumor profiling are limited to the availability of (viable) tumor material, often only accessible by invasive biopsy or surgical procedures. Malignant serous effusion (MSE) occurs in ~15% of advanced malignancies from a variety of solid tumor types, including those with the highest prevalence and mortality, i.e., lung, breast, and gastrointestinal cancers. MSE are characterized by the presence of disseminated tumor cells in fluid accumulations in serous cavities29. Since they can be accessed with minimally invasive procedures and contain metastatic cells that are adapted to growth in suspension, MSE might present an attractive alternative source of tumor material for functional and molecular profiling30. In addition, unlike rare circulating tumor cells in peripheral blood31, malignant cells in MSE are often highly abundant. Indeed, some studies reported the preliminary feasibility of using MSE in clinical decision-making based on targeted next-generation sequencing (NGS)32,33,34. In addition, MSE has been used to establish patient-derived cultures35,36,37,38,39,40,41 or organoids42,43,44 for drug response testing. However, the majority of these studies focused on demonstrating the feasibility of a small number of samples and tested drugs. Comprehensive molecular and functional characterization of primary MSE samples and integrative analyses are lacking. It, therefore, remains unclear to what extent MSE recapitulates the characteristics of the primary tumor beyond genomics and if multi-modal MSE profiling can inform precision oncology strategies.
Results
A cohort of diverse malignant serous effusions
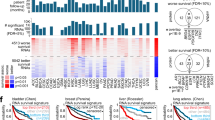
To characterize MSE samples of advanced solid tumors for precision oncology, we analyzed their molecular, cellular, and drug response landscape. We collected 261 MSE samples from 183 patients with a metastatic solid malignancy of any type. We included those 150 samples from 105 patients, which contained sufficient numbers of viable tumor cells for further profiling (see Supplementary Data 1). To capture the clinical complexity of precision oncology for metastatic solid tumors, we included MSE samples from any primary tumor, thus comprising a variety of disease types, sampling sites, treatment histories, and driver mutations (Fig. 1a, b, Supplementary Data 1). The majority of samples stemmed from patients with lung adenocarcinoma (LUAD, n = 67, 37%), followed by tubo-ovarian carcinoma (OV, n = 18, 14%) and mesothelioma (MESO, n = 14, 10%). Pleural effusion was the most common sample type and accounted for the majority of LUAD and MESO samples, while ascites were the main source of cells for OV. Treatment histories ranged from newly diagnosed patients with no prior drug exposure to recurrent disease with up to seven prior lines of systemic therapy (median: 1.0, IQR: 0.0−1.3). For 25 patients, we collected multiple samples over time, enabling a unique assessment of tumor evolution under therapy.
a Circos plot visualizing key clinical parameters. Samples taken from the same patient are connected with a gray line. For a more detailed description of the clinical parameters, see Supplementary Data 1. b Table summarizing clinical data shown in (a). Note that for the parameters “sample type” and “number of prior treatments,” the same patient can be counted in multiple categories, and thus, the patient numbers do not necessarily add up to 105. c Schematic representation of the molecular and functional profiling workflow. Cells were isolated from MSE and analyzed by pharmacoscopy (PCY), an image-based single-cell ex vivo drug response assay, which provides information on the cell type composition of each sample, as well as cell type-specific responses to a panel of 101 anticancer drugs. We further collected matched gene expression data (bulk RNA-seq, n = 131) and genomic alterations (FoundationOne CDx assay, n = 98 MSE samples and 24 patient-matched solid tissue samples), whenever samples contained a sufficient amount of tumor cells. For a subset of five LUAD samples, gene expression in response to drug perturbation (DRUG-seq) was measured. For further details, please see Supplementary Figs. 1–6 and Supplementary Data 1–10.
Single-cell ex vivo drug screening of pan-cancer MSE samples
We analyzed each sample by pharmacoscopy (PCY)26,45,46,47, measuring sample composition and high-throughput drug responses by immunofluorescence and automated confocal microscopy, after 24 hours of ex vivo culturing (Fig. 1c, Supplementary Data 2). Image analysis combined conventional single-cell image analysis with deep learning-based cell classification. Specifically, we developed sequentially nested convolutional neural networks (CNN) that recognize cell types and states based on a combination of morphology and immunofluorescence staining. This enabled the classification of 250 million imaged MSE cells into four main cell types: malignant cells, macrophages, lymphocytes/granulocytes, and ‘other’ cells. We detected apoptotic cells by nuclear and cell morphology and further stratified each main cell type into additional morphologically defined subclasses (Supplementary Figs. 1 and 2a, Supplementary Data 3). The performance of this CNN-based classification of cell type and state was validated by a strong positive correlation with fractions of explicitly IF-labeled cells in control experiments on the same MSE samples (Supplementary Fig. 2b, c), and by positive correlations between tumor cell counts measured by PCY and MSE sample-matched mutant allele frequencies measured by NGS (Supplementary Fig. 2d).
We further utilized the single-cell resolution of PCY to measure ex vivo response to a panel of 101 compounds commonly used in the clinical management of solid tumors (Supplementary Fig. 3, Supplementary Data 4, Supplementary Data 5). In our prior hemato-oncology studies, we have used a reduction in cancer fraction (RCF) to identify ‘on-target’ drug responses, which are consistently associated with improved clinical responses26,45,46,47. In the context of metastatic solid tumors, a treatment should ideally be highly tumor-specific and highly effective. However, such a treatment might not exist, and it is currently unclear whether it is better to minimize toxicity on the benign immune cells present in MSE or to maximize toxicity on the cancer cells. To this end, we quantified both the drug-induced reduction in tumor cell numbers (RCN) independent of the drug response of benign cells, as well as the reduction in tumor cell fraction (RCF), which incorporates the drug response of the benign cells (Supplementary Fig. 4a, b). Reduced tumor cell fractions were consistently associated with reduced tumor cell numbers (Supplementary Fig. 4b). Both readouts led to statistically significant hits (FDR < 0.05) across the cohort (Supplementary Fig. 4c) and consistent results across replicate wells in a plate (median Pearson correlation coefficient = 0.62 for RCF and 0.63 for RCN, respectively, Supplementary Fig. 4d). Further analysis showed that the technical replicate correlations were strongly associated with the number of significant drug effects (R2 = 0.56), and less so with the variance within DMSO wells as an orthogonal measure of technical reproducibility (R2 = 0.18), indicating that cases with low correlations of the drug response replicates stemmed predominantly from the absence of drug sensitivity rather than poor technical reproducibility (Supplementary Fig. 4e). Interestingly, the RCF readout revealed higher patient specificity (Supplementary Fig. 4d), possibly resulting from inter-patient benign cell drug response differences. In summary, PCY provided a detailed view of the sample composition both in unperturbed control conditions and in response to drug treatment.
Compositional MSE heterogeneity drives multi-omic variance
We combined this PCY-based cellular and functional MSE characterization with sample-matched transcriptomics (RNA-seq, n = 131) and targeted NGS using the FoundationOne CDx assay (324 genes, n = 98 MSE samples, n = 24 patient-matched solid tissue biopsies) (Fig. 1c, Supplementary Data 1, Supplementary Data 6). To obtain an integrated view of MSE biology and identify the main sources of variance in our cohort, we applied multi-omics factor analysis (MOFA)48,49 to the measurements we collected (Figs. 1c and 2a). Specifically, we included PCY-based cellular sample composition in unperturbed conditions (Supplementary Figs. 1 and 2a, Supplementary Data 3), both PCY-based drug response measures of tumor cells (RCN and RCF; Supplementary Fig. 3, Supplementary Data 5), expression of the top 5% most variable genes (Supplementary Fig. 5, Supplementary Data 7), and genomic alterations of genes that were mutated in at least 10 samples (Supplementary Fig. 6, Supplementary Data 8).
a Schematic depiction of the multi-omics factor analysis (MOFA) workflow. b Percentage of variance explained by each data type per factor, for the first seven factors ranked by highest overall fraction of variance explained. c Heatmap showing the top features contributing to the first seven MOFA factors for each of the 149 MSE samples. For each of the top seven MOFA factors, the values of the features that contribute most to this factor are shown. Sample type, tumor type, data type, and select feature names are annotated. d Correlation of factor 1 with either (top panel) EPCAM expression (excluding non-epithelial tumors; n = 116) or (bottom panel) expression of immune-marker PTPRC/CD45 (n = 131 samples). Linear regression line with 95% confidence bands, R squared, and corresponding P value (two-sided t test) are indicated. e Cell type composition per sample. Samples are grouped into 8 clusters based on their composition using hierarchical clustering (pearson correlation, complete linkage) and tree-cutting. f Example PCY images of MSE samples from each cluster. Images represent 8 out of 150 biological replicates, with 400 images of DMSO-treated cells per sample. Stainings and scale bars are indicated.
MOFA extracts axes of variance (factors) across multi-omic measurements in a way that each factor explains as much of the total variance as possible. Factors are associated with a set of weights specifying the contributing data types and features (Fig. 2a). Sample composition contributed strongly to the top two factors that explained the greatest variance, while gene expression features were present in all, and drug responses strongly contributed to factor 5 (Fig. 2b). In order to interpret the main sources of variability across our cohort, we visualized the highest contributing features for each of the top factors (Fig. 2c, Supplementary Fig. 7a, b). Factor 1 is related to tumor content, including the fraction of cancer cells by PCY and EPCAM gene expression, an epithelial marker commonly expressed on carcinoma cells. Consistently, factor 1 was strongly anti-correlated with expression of immune cell marker PTPRC (CD45) (Fig. 2d). Factor 2 related to immune composition, as it stratified samples by the types of immune cells present, mainly driven by the content of macrophages compared to other immune cell subsets (Supplementary Fig. 7c).
To further characterize this main source of MSE heterogeneity, we clustered the cohort based on cell composition and morphology, resulting in 8 sample groups (Fig. 2e, f): MSE samples with high tumor content were characterized by their tumor cell growth patterns: Adherent single cells (group 1), 2D multicellular colonies (group 2), 3D spheroids (group 6) or single-cell suspensions (group 7). Lower tumor content samples were further clustered based on their predominant non-malignant cell type: either macrophages (group 3), lymphocytes/granulocytes (group 5), immune cells with the intermediate phenotype (‘unassigned’, group 4), or ‘others’ possibly representing benign mesothelial cells, which are often present in MSE (group 8). Overall, the groups were not strongly associated with either tumor or sample type. Rather, we found that MSE sample composition was dynamic within individual patients, even changing between sequential samples from the same patient in the absence of any apparent change in health or treatment status (Supplementary Fig. 8). Switching between the predominant immune cell types was the most common change (9 out of 25 sequentially biopsied patients).
MSE tumor cells molecularly recapitulate their tumor origin
Beyond tumor and immune cell composition, the next MOFA factors (3, 4, 6, and 7) related to and accurately classified specific cancer types (Fig. 2c, Supplementary Fig. 7d). Factor 6 captured a series of samples from a single patient PID024 (Supplementary Fig. 7e) with an unusual carcinoma of unknown primary, further discussed in the Supplementary case studies. Factor 3 separated LUAD and OV, factor 4 defined MESO, and factor 7 breast cancer (BRCA). Motivated by this observation, we set out to analyze to what extent MSE-derived tumor cells recapitulate molecular aspects of their primary tumor. After correcting for tumor content and batch effects, gene expression profiles of samples with related tumor types were grouped together (Fig. 3a). Furthermore, analysis of genes distinguishing different tumor types retrieved known markers of the individual tumors, including those used in routine diagnostics, such as napsin A (NAPSA) for LUAD and paired box 8 (PAX8) for OV (Fig. 3b). Differential expression of selected genes was confirmed both by qPCR analysis (Supplementary Fig. 9a, b) and immunofluorescence (Supplementary Fig. 9c, d). MSE transcriptional profiles clustered with their corresponding primary solid tumors in the cancer genome atlas (TCGA; Fig. 3c, Supplementary Fig. 10a, b). Furthermore, expression profiles of matched tumor types were positively correlated (mean r = 0.31) while those of unmatched tumor types were uncorrelated (mean r = −0.04) (Supplementary Fig. 10c), and MSE tumor types could be accurately assigned (mean accuracy 93%) by a 1-nearest neighbor classifier trained on the TCGA data (Supplementary Fig. 10d). Together, this confirmed transcriptional similarity between MSE and their respective solid tumors. Lastly, we compared the genomic signatures derived from MSE to those of patient-matched solid tumor biopsies (n = 24; Fig. 3d). Even though the solid tumor biopsy and the MSE were sometimes months apart, the genomic signatures showed very high concordance, with a significantly higher median Jaccard coefficient (JC) of 0.97 compared to a JC of 0.8 for samples that were not patient-matched (Fig. 3e). Specifically, 46 out of 77 (60%) of all actionable, and 5 out of 6 (83%) druggable alterations were concordantly measured by FoundationOne CDx, with the majority of disagreements stemming from rearrangements and CNVs (Fig. 3f). For an additional 7 patients with druggable alterations, the presence of such mutations was assessed in solid tissue by tests other than FoundationOne, with concordant mutations in 6 out of 7 cases (Fig. 3f, Supplementary Data 9), leading to a total of 11 out of 13 (85%) concordant druggable mutations detected by any method. Taken together, these findings suggest that although the microenvironment differs significantly from that found in a primary tumor or solid metastasis, malignant cells derived from MSE largely retain their disease-specific gene expression and solid tumor-matched mutational profile.
a t-distributed stochastic neighbor embedding (t-SNE) based on the tumor content-corrected expression of the 5% most variable genes for 149 MSE samples. Dot color indicates tumor type. b Violin plots indicating expression of selected genes across the cohort grouped by tumor type (from left to right: LUAD, OV, MESO, BRCA, STAD). These genes were identified by differential expression (DE) analysis comparing each individual tumor type separately to all other tumor types, accounting for confounders tumor content, sequencing batch, and biological sex. c t-SNE projection of integrated transcriptomes of LUAD, OV, BRCA, MESO, and STAD samples from primary tumors in the TCGA cohort (small transparent dots, n = 2452) and this MSE cohort (n = 101, big outlined dots). The accuracy of a 1-nearest neighbor (1-NN) classifier trained on the TCGA data and evaluated for predicting the MSE tumor types is indicated (see Supplementary Fig. 10d). d Comparison of mutational profiles measured by FoundationOne CDx between patient-matched solid biopsies (n = 24 patients) and their corresponding MSE samples. e Box plots indicating the Jaccard coefficients between mutational profiles of each pair of samples, stratified by whether samples were patient-matched (left) or not (right). P value from two-sided, two-sample Wilcoxon test. Box plots indicate the median (horizontal line) and 25% and 75% ranges (box), and whiskers indicate the 1.5× interquartile range above or below the box. Outliers beyond this range are shown as individual data points. f Concordance of genomic alterations between MSE and patient-matched solid tissue biopsies evaluated for all actionable alterations measured by FoundationOne (left), only druggable alterations measured by FoundationOne (middle), and druggable alterations measured by FoundationOne on MSE and a different diagnostic test on the solid tissue (see Supplementary Data 9). Colors indicate the type of alteration (CNV copy number variation, RA rearrangement, SUB substitution), and lightness encodes concordance.
Drug-induced gene expression in MSE reflects mode-of-action
Given that MSE significantly retains molecular aspects of their solid tumor origins, we next investigated their molecular determinants of ex vivo drug sensitivity. Interestingly, the MOFA analysis highlighted overall drug sensitivity as a key source of variance, with factor 5 strongly correlating with the fraction of drugs with ‘on-target’ responses per sample (‘sensitivity score’; Fig. 4a, Supplementary Fig. 11a). To interpret the mechanisms leading to this multi-drug susceptibility, we applied gene set enrichment analysis (GSEA) on the genes associated with the sensitivity score (Fig. 4b, c). This revealed significant enrichment in genes related to DNA conformational change, chromatin remodeling, and cell division, among those positively associated with overall drug sensitivity, suggesting a key role of proliferation in governing the general ex vivo sensitivity of MSE to antineoplastic agents.
a MOFA Factor 5 plotted against the overall ex vivo ‘sensitivity score’ (fraction of drugs with RCF > 0, n = 149 samples). Linear regression lines with 95% confidence bands and corresponding P value (two-sided t test) are indicated. b Top-15 pathways resulting from gene set enrichment analysis (GSEA) for genes associated with the sensitivity score. Densities represent the t statistic of the generalized linear model (edgeR). Colors indicate −log10 FDR (Benjamini–Hochberg, BH) of the GSEA. c Volcano plot for the association between sensitivity score and gene expression. Genes belonging to the GO term “DNA conformation change” (GO:0071103) are highlighted in dark red. d Illustration of RNA-seq and drug response integration in LUAD MSE samples. For each drug, we associated RCF with baseline gene expression (top arrow; n = 59 samples). In addition, we measured drug-induced transcriptional changes in a subset of LUAD samples (bottom arrow; n = 5 samples). We then assessed whether the obtained associations or transcriptional changes were enriched in genes that are connected to the drug’s primary and secondary target(s). e Fraction of evaluable drugs for which the drug response-associated genes were significantly enriched in the drug target-proximal gene set. f BH-corrected enrichment p value (right-tailed hypergeometric test) for individual drugs in baseline and DRUG-seq analysis. Drug-target classes are annotated. g Baseline expression of CDC20 associated with ex vivo response to gemcitabine (top), and change in CDC20 expression in cells exposed to gemcitabine (bottom). P values in both panels from DE analysis using the quasi-likelihood F-test implemented in egdeR. Top: Dots represent 59 biological replicates, linear regression line with 95% confidence bands is indicated. Bottom: Box-plots as in Fig. 3e. Data from five independent samples with four technical replicates per sample. h, i Example drug target-proximal gene networks for vinorelbine (h) and palbociclib (i). Green nodes denote the drug; every other node corresponds to a gene. Colors indicate log2 fold change of this gene when comparing drug-treated to DMSO-treated cells. Select subnetworks and gene names are annotated. Enrichment statistics (right-tailed hypergeometric test, BH-adjusted) are depicted by mosaic plots (inserts).
Beyond overall sensitivity, we focused on the relationship between gene expression and the response to individual drugs. As gene expression was strongly associated with specific tumor types, we restricted this analysis to the largest subcohort (LUAD, n = 59 samples). We performed two complementary analyses (Fig. 4d): First, we correlated the baseline transcriptional signatures with their ex vivo response per drug, which yielded hardly any significant associations. Thus, while overall drug resistance was associated with the expression of cell division genes, responses to individual drugs were not explained by baseline MSE gene expression across the LUAD cohort. Second, for a subset of 5 LUAD samples selected based on their high tumor content, we measured the transcriptional response following 24 h of ex vivo treatment with 45 drugs (DRUG-seq50, Fig. 4d, Supplementary Fig. 12, Supplementary Data 10). Analysis of the resulting 932 transcriptional profiles of primary LUAD samples identified a considerable number of genes significantly differentially expressed in a drug-dependent manner across all 5 samples.
Genes whose expression relates to drug response, either at baseline or in response to treatment, may be associated with the drug mode-of-action (MoA). We, therefore, next investigated the enrichment of the ranked gene-drug associations derived from both approaches in the drug target-proximal gene sets. We obtained a drug’s direct targets from the STITCH database51,52, and extended the primary target list with reported interacting genes from the STRING database53 (Fig. 4d). Baseline RNA-seq associations with ex vivo drug response showed such enrichments for only 1 out of 73 drugs with annotated drug targets. In contrast, transcriptional changes in response to drug treatment were significantly enriched in drug target-proximal gene sets for 22 out of 35 evaluable drugs (Fig. 4e). Gemcitabine, a nucleoside analog that induces DNA damage, was enriched in both analyses (Fig. 4f). Many of the gemcitabine target-proximal genes are related to cell division. Of these, for example, the baseline expression of cell division cycle 20 (CDC20) was positively associated with the ex vivo response to gemcitabine, as well as downregulated in response to gemcitabine exposure (Fig. 4g). Among the compounds with the most drug target-enriched transcriptional signatures were vinorelbine, an inhibitor of tubulin polymerization54, and palbociclib, a selective inhibitor of cyclin-dependent kinases 4 and 6 (CDK4/6). For vinorelbine, we observed downregulation of multiple tubulin subunits as well as upregulation of genes related to apoptosis, stress response and cytokine signaling (Fig. 4h). This decrease in tubulin was further validated at the protein level in MSE cells by immunofluorescence and proteomics (Supplementary Fig. 13). For palbociclib, we found few differentially expressed genes overall (n = 61, FDR < 0.01 and absolute log fold change >0.5); however, almost all of the genes downregulated by palbociclib were related to cell cycle regulation (Fig. 4i). Taken together, our integrative analysis highlighted a link between proliferation and global drug sensitivity and revealed that, while the expression of drug-target-proximal genes did not trivially explain response to individual compounds, the transcriptional response to drug perturbation in primary MSE samples was strongly linked to, and indicative of, the drug’s mode-of-action.
Tumor spheroids associate with genomic alterations and cellular states in LUAD
Given the heterogeneity in MSE-derived cancer cell morphologies and multicellular organization (Fig. 2e, f), we explored their link to clinical parameters, cellular state, and response to treatment. In the context of LUADs, the fraction of tumor cells that formed spheroids showed striking differences between patient samples in identical culture conditions. Spheroidal growth was consistent within patients and not associated with the number of prior treatments (Fig. 5a, b). Patients with higher spheroid fractions tended to respond better to the treatment following the biopsy taken for this study (Fig. 5c). This observation could, in part, be explained by an association of spheroid formation with mutational status (Fig. 5d): Spheroid growth was particularly abundant in EGFR-mutant LUAD, which clinically responds well to EGFR inhibition55 (Supplementary Discussion).
a Morphological composition of tumor cells in LUAD samples, sorted by spheroid fraction. b Example images (gray: brightfield, pink: DAPI/nuclei) of MSE samples with low and high fractions of tumor cell spheroids. Images are representative of 59 biological replicates and 400 images each. c Fraction of spheroids stratified by the response to the treatment following the biopsy, for n = 24 samples obtained prior to therapy start. d Fraction of spheroids stratified by actionable genomic alterations (BRAF V600E: 11 samples, 3 patients; EGFR: 18 samples, 9 patients; other: 34 samples, 24 patients). P values from Tukey’s honestly significant difference (HSD) test. e Example images of drug-induced spheroid dissociation (gray: brightfield, pink: DAPI/nuclei). Images are representative of 59 biological replicates with 100 images per drug treatment. f Integration of spheroid abundances from PCY and transcriptional responses from DRUG-seq (n = 5 LUAD samples and 48 drugs). TIMP1 (left) represents a gene whose expression was associated with spheroid abundance across ex vivo treatments. g Volcano plot for DRUG-seq analysis of spheroid fractions (see Methods). h Top 10 pathways (GSEA) for genes associated with spheroid fraction. Density plots and P values as in Fig. 4b. NMD nonsense-mediated decay, n-t nuclear-transcribed, PTTM protein targeting to membrane. i Pathway scores (singscore91, see Methods) for mitochondrial translation (GO:0032543) and cell adhesion (GO:0007155) associated with spheroid fraction. P value from linear regression (two-sided t test). j Spheroid dissociation in response to EGFR inhibition in 43 spheroid-containing LUAD MSE samples, stratified by the presence of any mutation in EGFR (EGFR mutant: n = 12 samples from 8 patients, EGFR wild type: n = 31 samples from 20 patients). The values correspond to the mean spheroid dissociation response across four tested EGFR inhibitors (see Supplementary Fig. 14). All box-plots as in Fig. 3e; P values in (c) and (j) from two-sided Wilcoxon rank-sum test; P values in (f) and (g) from edgeR quasi-likelihood F-test with BH correction. Lines and shaded areas in (f) and (i) correspond to a linear regression fit with 95% confidence bands.
The multicellular organization changed in response to drug treatment, where drug-induced cell death coincided with spheroid dissociation (Fig. 5e). To understand the molecular mechanisms underlying this observation, we integrated the DRUG-seq dataset with spheroid abundance across ex vivo treatments. Specifically, we associated the fraction of cells forming spheroids with the sample-matched transcriptional profiles across the 46 treatment conditions and five LUAD samples (Fig. 5f). Spheroid dissociation was associated with increased expression of genes related to protein trafficking, motility, and cell adhesion. In contrast, higher spheroid fractions were associated with mitochondrial gene expression and translation, as well as cell division and metabolism (Fig. 5g–i). Thus, MSE-based spheroidal cell patterns indicate a metabolically active and proliferative population of cell-cell contact-forming LUAD cells.
To test if EGFR-mutant LUAD spheroids were sensitive to EGFR inhibition, we developed an ex vivo response score measuring the dissociation of spheroids for samples displaying sufficient spheroidal growth (see Methods). Out of the four EGFR inhibitors tested in our assay, all induced spheroid dissociation in at least five samples (Supplementary Fig. 14). Some patient samples responded to all inhibitors, while others had more drug-specific response patterns not explained by secondary EGFR mutations conferring treatment resistance. When considering a combined readout across all inhibitors, EGFR-mutant samples, on average, responded with spheroid dissociation, while EGFR wild-type samples did not (Fig. 5j), implicating EGFR signaling in this ex vivo tumor behavior.
Multi-omics MSE profiling reveals acquired resistance mechanisms in BRAF V600E mutant LUAD
Acquired resistance to targeted therapy is one of the main challenges in the treatment of patients with advanced solid tumors. If MSE reflects tumor evolution on an individual basis, sequential sampling across a patient’s disease course may provide unique insights into the acquired resistance mechanisms. To this end, we focused on the six MSE samples from patient PID028, a 64-year-old male with a BRAF p.V600E mutant LUAD, whose samples spanned from diagnosis to death (Fig. 6a; further detailed in Supplementary case study PID028).
a Patient PID028’s clinical course (see Supplementary patient case PID028). b PCY-based cellular and morphological sample composition over time. c Genomic alterations at diagnosis (FB103), first relapse (FB209) and second relapse (FB261). d PCY-based ex vivo response to dabrafenib + trametinib in longitudinal MSE samples. Dots represent technical replicates (n = 4 for drugs, n = 16 for DMSO). e Dose-response to dabrafenib + trametinib at relapse to targeted therapy (FB215). Dots represent technical replicates (n = 3), lines and shaded areas indicate the mean and 95% confidence interval per concentration. f Fraction of phospho-ERK (pERK) positive tumor cells in response to different doses of dabrafenib + trametinib. Dots represent technical replicates (n = 3 per concentration and n = 17 for DMSO). Exact P values are provided in the Source Data. g Comparison of gene expression of FACS-purified tumor cells from diagnosis (FB103) and relapse to targeted therapy (FB215). Highlighted genes are implicated in acquired resistance to this therapy in melanoma56. h Drug-target-proximal gene network for dabrafenib and trametinib. Green dots correspond to compounds, all other nodes represent genes. Color indicates log2 fold change of dabrafenib + trametinib vs DMSO in FACS-purified tumor cells from sample FB215. Enrichment visualized by mosaic plot (insert), P value from right-tailed hypergeometric test. i Change in DUSP6 levels after exposure to dabrafenib + trametinib across five additional samples measured by DRUG-seq. Values represent average expression across four replicate wells. P value from paired two-sided Wilcoxon test. j Comparison of in situ and ex vivo transcriptional adaptation of tumor cells to dabrafenib + trametinib. X axis corresponds to log2 fold change of gene expression in tumor cells taken at diagnosis (FB103) vs relapse (FB215), and y axis to log2 fold change of gene expression of tumor cells (FB215) treated ex vivo with dabrafenib + trametinib relative to DMSO. Regression line with 95% confidence bands, Pearson’s R, and corresponding P value (two-sided t test) are indicated. P values in (d) and (f) from two-sided Student’s t test comparing treatment to control, no adjustment for multiple testing. All box plots as in Fig. 3e.
All samples had a high content of tumor cells, which adhered to the plate either as single cells or small colonies (Fig. 6b). The immune composition was dynamic over time, e.g., switching from predominant macrophages (sample FB261) to lymphocytes (sample FB263) in the span of just one week. The core mutational landscape was stable over time, with the maintenance of the majority of alterations, including BRAF p.V600E (Fig. 6c). However, compared to the sample taken at diagnosis, the relapsed samples acquired a loss of function substitution in TP53, as well as mutations possibly affecting mitogen-activated protein kinase (MAPK) signaling: A truncation in ERRFI1, which encodes a negative feedback regulator of ERBB family receptor tyrosine kinases; A substitution affecting a splice site in MKNK1, which encodes MAPK Interacting Serine/Threonine Kinase 1; And, a splice site deletion in NF1 observed in one of the two samples taken after relapse.
The ex vivo drug responses over time highlighted initial sensitivity to combined BRAF and MEK inhibition, with increasing resistance (Fig. 6d), matching the clinical course of the patient. We confirmed the dabrafenib + trametinib resistance across multiple concentrations in FB215, with neither a relative nor absolute reduction in tumor cells observed (Fig. 6e). However, measuring phospho-ERK (pERK) levels by immunofluorescence in these conditions as a readout for MAPK pathway activity revealed a strong drug-induced reduction in pERK-high cells (Fig. 6f). Thus, in this patient, the acquired resistance to MAPK inhibition was independent of the treatment-induced reduction in pERK signaling.
To understand the molecular changes underlying this acquired resistance, we compared the transcriptional signatures of FACS-purified (EpCAM positive) tumor cells from FB103 (diagnosis) and FB215 (relapse under combined TKI treatment) (Fig. 6g, Supplementary Data 12). This highlighted several key genes previously reported to be involved in acquired resistance to dabrafenib + trametinib in melanoma56. Most striking was the downregulation of dual-specificity phosphatase 6 (DUSP6), which acts as a negative feedback regulator of pERK and a tumor suppressor in LUAD57,58,59. To link this signature to transcriptional changes induced by dabrafenib + trametinib treatment, we compared gene expression between purified tumor cells from sample FB215 exposed to BRAFi + MEKi and DMSO control (Fig. 6h). Overall, the differentially expressed genes were enriched in drug target-proximal genes of dabrafenib + trametinib. Among the most strongly downregulated genes were DUSP6, EPHA, FOS, FOSL1, CCND1, and AREG, all directly involved in MAPK signaling. Interestingly, MAPKi-induced DUSP6 downregulation was independent of a BRAF p.V600E mutation, as it was observed across five additional LUAD samples with diverse mutational profiles (Fig. 6i). Finally, we globally compared the direct transcriptional effects of dabrafenib + trametinib after ex vivo treatment to the changes in gene expression observed between diagnosis and relapse in PID028. Interestingly, the strong downregulation of DUSP6, EPHA, FOS, FOSL1, and AREG were recapitulated in the downregulation of these genes in FB215 compared to FB103. Thus, the in situ transcriptional adaptation of the patient’s tumor to clinical treatment, including downregulation of DUSP6, was recapitulated ex vivo by the patient’s MSE-derived tumor cells.
Integrated analysis reveals non-genetic MET upregulation as an actionable drug target
A non-genetic actionable mechanism of acquired resistance to EGFR inhibition was identified in a 70-year-old female with lung adenocarcinoma harboring an EGFR exon 19 deletion and presenting with a malignant pleural effusion (Fig. 7a; further detailed in the supplementary case study PID038). The patient was initially treated with osimertinib15,60, and a first pleural effusion sample was profiled shortly following treatment initiation. After initial response to the EGFR inhibitor for 12 months, the patient started to show signs of progression, and two consecutive pleural effusion samples were analyzed following radiotherapy. All samples had low tumor content (Fig. 7b), with dynamic changes in the immune compartment between samples. Comparison of mutational signatures between diagnosis and relapse revealed largely consistent alterations, in particular the initial EGFR exon 19 deletion was maintained. An additional EGFR amplification and a p.L654A point mutation of unknown significance present at baseline were no longer detected at relapse. No known resistance mutations to osimertinib, such as EGFR p.C797S, MET exon 14 skipping event, or MET amplification, were detected (Fig. 7c).
a Clinical course of patient PID038. See Supplementary patient case PID038 for details. b PCY-based cellular and morphological sample composition over time. c Comparison of genomic profile between tissue at diagnosis (solid tumor biopsy) and after relapse to osimertinib (FB287) measured by FoundationOne CDx. d PCY-based ex vivo responses of the MSE sample at relapse to osimertinib (FB287) All drugs with significant on-target effects (RCF > 0, p < 0.01) are shown, as well as the previous treatment osimertinib. Exact P values (two-sided Student’s t test, no adjustment for multiple testing) per drug are provided in the Source Data. Drugs highlighted in red target MET. e Comparison of gene expression profiles in FACS-purified tumor cells between FB133 (diagnosis) and FB295 (relapse). MET expression strongly increased in the relapse sample. f Immunohistochemistry against cMET at diagnosis (top, cMET intensity 2+ in ~30% of tumor cells) and relapse (bottom, cMET intensity between 2+ and 3+ in 95% of tumor cells). Image represents 1.5% of the full scanned area, the full scans are provided in Supplementary Fig. 15. g Ex vivo response of FB295 to the combination of capmatinib and osimertinib. An on-target effect is observed across concentrations. P values in (d) and (g) from a two-sample two-sided Student’s t test comparing treatment condition to DMSO control, no adjustment for multiple testing. All box-plots as in Fig. 3e.
Despite the absence of MET alterations in the genomic signature, PCY analysis at relapse (sample FB287) revealed sensitivity to two out of three tested MET inhibitors (Fig. 7d), while osimertinib scored off-target. LUAD patients with EGFR driver mutations and selected MET alterations are eligible for treatment with a combination of osimertinib and a MET inhibitor (capmatinib or tepotinib61,62,63,64,65,66). To investigate the observed MET inhibitor sensitivity, we purified tumor cells from diagnosis (FB133) and relapse (FB295) by FACS and compared their transcriptional signatures (Fig. 7e, Supplementary Data 12). This revealed striking upregulation of MET mRNA expression at relapse. This transcriptional upregulation was confirmed at the protein level by immunohistochemistry against cMET (Fig. 7f, Supplementary Fig. 15), and coincided with ex vivo sensitivity to osimertinib + capmatinib combination treatment (Fig. 7g). Based on this combined functional and molecular evidence, the patient started treatment with osimertinib + capmatinib and achieved a subtotal partial response that lasted for six months, comparable to previously observed responses of patients with sensitizing MET alterations as an acquired resistance mechanism to EGFR-TKIs67.
Taken together, these results indicate that integrative analysis of multi-omics and functional data of MSE samples can identify clinically actionable changes in tumor biology that occur as a result of prior treatment, changes that are not detectable by assessing genomic alterations alone.
Discussion
We present a deep molecular, cellular, and functional characterization of 150 MSE of advanced solid tumors from diverse cancer types, showcasing the value of MSE-derived measurements for precision oncology and solid tumor research.
Our unbiased multi-omic integration identified cell type composition as a key element of MSE heterogeneity. While the composition of malignant effusions has been extensively studied in the context of cytology and diagnostics29, and more recently by scRNAseq68,69,70,71,72,73, we find that particularly the non-malignant cellular MSE composition is highly dynamic, with a switch in the predominant immune cell type (macrophage versus lymphocyte/granulocyte) observed in 9 out of 25 sequentially analyzed patients, warranting caution in the interpretation of one-time MSE snapshots. Nonetheless, patient-specific transcriptional signatures and drug responses of cancer cells were preserved. Furthermore, consistent with previous studies reporting concordant genomic profiles between MSE and solid tumors32,33,34, we show that malignant tumor cells derived from MSE faithfully recapitulate their corresponding primary solid tumors. Going beyond genomic concordance, integration of MSE transcriptomes with TCGA data reveals strong concordance of gene expression profiles between primary solid tumors and MSE. Expression of transcription factors indicative of the tumor origin could further be confirmed by immunofluorescence and imaging across the cohort. Lastly, we observed good feasibility for our multi-modal MSE profiling, with 150 out of 261 (57%) MSE samples successfully profiled. The main dropout reason was too low abundance of viable tumor cells (in 100 ml of MSE fluid analyzed), which could be overcome in future trials by analyzing larger MSE volumes or enriching tumor cells. Together, our data show that MSE samples are an easily accessible source of representative tumor cells, enabling in-depth analysis of cellular state and function for precision oncology.
Integrating drug sensitivity and baseline MSE gene expression revealed a pan-cancer proliferative signature associated with increased global drug responsiveness. Although there is generally no clear association between proliferative index and response to antineoplastic agents across tumor types74, the observed link between proliferation and drug response may reflect a drug-resistant persister cell state, as previously described in NSCLC75. We furthermore adapted multiplexed high-throughput RNA sequencing (DRUG-seq) previously developed on cell lines50,76,77 to be compatible with the high-throughput transcriptional profiling of drug responses in primary patient material. This enabled us to measure a total of 932 transcriptional profiles, covering the response of around 7600 genes to 45 drugs across five primary LUAD samples. Strikingly, the transcriptomic changes observed after drug treatment were highly enriched in genes indicative of the drug mode-of-action. And, in a case study of patient diagnosed with BRAF p.V600E mutant LUAD that was sequentially profiled, we observed that the ex vivo transcriptional response to targeted BRAF and MEK inhibition recapitulated the in situ transcriptional adaptation of the patient’s tumor to clinical treatment over time. Our results thus show feasibility and applicability of primary MSE samples to investigate drug mode of action ex vivo, and uncover recurring transcriptional adaptations to targeted therapy.
To date, individualized treatment decisions are largely driven by genomic tumor features. However, out of the 98 genomically profiled samples, the majority (84 samples) originated from patients for whom this approach is not applicable, either because their tumors lacked druggable alterations (n = 72) or because they had acquired resistance mechanisms not susceptible to targeted therapies (n = 12). This highlights the clinical need and potential utility of the multi-modal profiling that we present here, especially for the majority of patients who lack targeted therapy options. For one such patient (PID035), who had relapsed after EGFR inhibitor treatment, we identified an apparent non-genetic MET upregulation responding ex vivo and clinically to MET inhibition. While MET expression alone is not considered sufficient to predict response to capmatinib78, we find that the combination of functional and transcriptional profiling may help identify patients who do benefit from this therapy also in the absence of a genetic biomarker. In conclusion, we confirmed the feasibility of performing systematic functional and molecular measurements on MSE, revealing personal and actionable insights into the biology of advanced solid tumors. We provide a unique resource of composition, drug response, gene expression, and mutational data across a clinically annotated pan-cancer cohort of effusion samples. Moreover, we highlight the utility of our platform in uncovering determinants of ex vivo drug response, shedding light on mechanisms of acquired treatment resistance, and identifying clinically actionable drug targets.
Methods
Study design and participants
This study was conducted as a prospective, non-randomized observational clinical study with feasibility as the primary outcome. Treatment decisions during the study were solely based on current clinical guidelines and the decisions made by the treating physician and the patient. Any patient with a metastatic solid malignancy from whom a fluid sample (ascites, pericardial or pleural effusion, washing or any other fluids containing malignant cells) was collected as part of routine diagnostic or therapeutic procedures at the University Hospital Zurich (USZ), Spital Uster or Kantonsspital Winterthur was eligible for the study. Thus, there is no pre-selection bias. Patients were included if they were older than 18 years and provided written informed consent either through the USZ general consent (GC) or a study-specific informed consent form. Patients matching the inclusion criteria were identified by the cytology team at USZ, who performed diagnostics on fluid samples and consented by the treating physician. Over the course of the project (May 2019–January 2023), 261 samples from 184 patients were included in this study. Follow-up data (see Supplementary Data 1) were collected as part of clinical routine during the same time period. The research project was carried out in accordance with the research plan and with principles enunciated in the current version of the Declaration of Helsinki (DoH), the Principles of Good Clinical Practice (GCP), the Swiss Law and Swiss regulatory authority’s requirements as applicable. Ethical approval was granted by the Ethics Committee of Kanton Zurich (CEC Zurich, BASEC-Nr: 2019-01700).
Visualization of cohort statistics
Cohort statistics in Fig. 1b were visualized using the circlize R package (v.0.4.13)79.
Collection of cells from malignant effusions
100 ml of fluid was centrifuged at 200 × g for 5 min. The supernatant was discarded, and the pellet was treated with red blood cell (RBC) lysis buffer (BioLegend #420302). This step was repeated If the pellet was still red after RBC lysis. The pellet was then resuspended in growth media (Gibco RPMI1640 + GlutaMax supplemented with 10% human type AB serum (PanBiotech PANP30-2502)) and optionally passed through a 70 μm cell strainer to remove large aggregates. Cell number and viability was determined using a Countess cell counter (Invitrogen). Samples that did not contain at least 2 millions of viable cells were excluded from the study and not further processed.
Purification of cell subsets by FACS
For selected validation samples (FB103 and FB215 in Fig. 6; FB133 and FB295 in Fig. 7), cell subsets were purified from cryopreserved cells isolated from MSE by FACS. After thawing, cells were resuspended in a blocking buffer (5% (v/v) FBS in PBS) and passed through a 70 μm cell strainer. Cells were then blocked on ice for 10 min, spun down and resuspended in 100 μl of FACS buffer (1% (w/v) BSA in PBS) and stained using the following antibodies (5 μl of antibody per 5 million cells, see Supplementary Data 2 for details on the antibodies used): CD3 (Alexa Fluor 488) for T-cells, EpCAM (Alexa Fluor 555) for epithelial/adenocarcinoma cells, and CD14 (Alexa Fluor 647) for monocytes/macrophages/dendritic cells. Cells were incubated with antibodies on ice for 30 min. Then, cells were washed twice with 10 ml of cold FACS buffer, and finally resuspended in 0.5−2 ml FACS buffer to reach a concentration of ~5 million cells/ml. 1 μM SyTOX blue was added right before sorting to exclude dead cells. Cells were sorted on a BD FACS Aria Fusion using a 100 μm nozzle. We set a very inclusive gate on forward and side scatter, and excluded dead cells based on SyTOX blue intensity only. Individual cell populations were then sorted out based on single positivity for the corresponding markers.
Pharmacoscopy
The term PCY refers to short-term ex vivo culture and drug treatment of primary patient samples followed by immunohistochemistry, automated microscopy, and single-cell image analysis26,46,47. The technology thus encompasses both experimental procedures and computational analysis. PCY results in quantification of cellular and morphological sample composition, as well as cell type-resolved drug responses.
Short-term culture and ex vivo drug treatment of MSE-derived cells
The cellular component of the MSE sample was diluted to 0.1–0.2 million cells/ml in growth media (Gibco RPMI1640 + GlutaMax supplemented with 10% human type AB serum (PanBiotech PANP30-2502)), and 50 μl/well were seeded in CellCarrier 384 Ultra, clear-bottom, tissue-culture-treated plates (PerkinElmer). Cells were incubated with 10 μM of compound (see Supplementary Data 4) or matching control (DMSO for small molecules and isotype control for antibodies) for 24 h at 37॰C, 5% CO2. This drug concentration was chosen based on prior experience in hemato-oncology26,46,47. Afterwards, the media was aspirated using a microplate washer (Tecan), and cells were fixed with 20 μl/well of a periodate, lysine, formaldehyde fixative (75 mM lysine [Sigma Aldrich L5626-100G], 2.5 mg/ml sodium periodate [Sigma Aldrich 30323-100 G] and 1.25% (v/v) formalin [37% formaldehyde solution, Sigma Aldrich F8775-500ML] in PBS) for 15 min at room temperature. The fixative was removed, and 70 μl of PBS/well was added. Plates were then stored in the fridge for up to 2 weeks prior to staining.
Staining with fluorescence-labeled antibodies
For staining, PBS was aspirated, and cells were blocked, permeabilized and stained for DNA using 20 μl of PBS supplemented with 5% fetal bovine serum (FBS, Gibco/ThermoFisher 10270106), 4′,6-diamidino-2-phenylindole (DAPI, BioLegend 422801) and 0.1% (v/v) Triton-X100 (Sigma Aldrich T8787) for 30 min at room temperature. Blocking solution was aspirated, and cells were stained with fluorescently labeled antibodies (see Supplementary Data 2) diluted in PBS + 10 mg/ml bovine serum albumin (BSA, Sigma Aldrich A7906) for 1 h at RT or overnight at 4 °C. Antibody-containing solution was then aspirated, and PBS was added on top of the cells. Every plate was first stained with a panel of antibodies only in 8 wells (Supplementary Data 2) containing no drugs. If a sample did not contain >2% malignant cells, it was excluded from analysis and not further processed. Otherwise, the whole plate was stained with an antibody panel tailored to the respective sample’s tumor marker expression (Supplementary Data 2).
Automated confocal microscopy
All samples were imaged on an automated spinning-disk confocal microscope (PerkinElmer Opera Phenix), using ×20 magnification and 25 images per well to cover the entire well area. We used five channels with non-overlapping excitation/emission filters to image the following features: Channel 1 (transmission/650–760 nm) for brightfield to capture general cell shape and texture, channel 2 (405 nm/435–480 nm) for DAPI/nuclei, channel 3 (488 nm/500–550 nm) for tumor stain, channel 4 (561 nm/570–630 nm) for a second tumor stain or additional markers, and channel 5 (640 nm/650–760 nm) for immune cells (CD45) channels.
Image analysis by CellProfiler
Raw images were first analyzed using CellProfiler v.2.2.080. Individual cells were detected based on maximum correlation thresholding of the DAPI signal. The exact parameters of the pipeline were adjusted per sample to account for differences in nucleus intensity, size and the presence of large spheroids. Staining intensities were extracted for the nucleus, and a region of 12 pixels around the nucleus that was used as a proxy for cytoplasmic intensity. For downstream analysis, intensities were log10 transformed and corrected for variation in the local background as described in ref. 81.
Filtering of segmentation artifacts and removal of outlier wells
Cells with very low DAPI intensities or abnormally high or low nucleus areas likely represent segmentation artifacts and were therefore removed from the analysis by manual gating. In addition, outlier wells (very low or high total cell numbers, or aberrant staining patterns) were removed if the observed patterns could be attributed to pipetting mistakes or the presence of large cell clumps by visual inspection.
Training of convolutional neural networks
Five different CNNs were trained (Supplementary Fig. 1). All of them are based on a ResNet18 architecture and were trained as previously described47,82. CNN 1 was trained to identify apoptotic cells using brightfield and nuclei (DAPI) images alone. Here, the training dataset was generated from test stains (2 wells without any drug treatment per sample), where we defined apoptotic cells based on the staining intensity of cleaved caspase 3 (clCASP3) and then generated a total of 95,666 2-channel single-cell crops (150 × 150 pixels corresponding to 45 × 45 μm) of apoptotic (n = 44,212) and non-apoptotic (n = 51,454) cells. CNN 2 was trained to recognize different cell types based on a combination of marker intensities and cellular morphology using all available channels. Here, the training data was generated by cropping 150 × 150 pixel images and then manually assigning them to either lymphocyte/granulocyte, macrophage, cancer, or other class. A total of 125,959 images across all samples were curated. CNN 3 classifies tumor cells into single adherent, single non-adherent, spheroid or adherent colony based on all channels and was trained on 36007 manually curated 150 × 150 pixel images. CNN 4 is used to distinguish round and elongated macrophages using the brightfield, DAPI, and 640 nm (CD45) channels, and was trained on a manually curated set of 12,846 150 × 150 pixel images.
For all CNNs, 75% of the labeled images were used for training and 25% for evaluation of classification performance.
CNN 1–4 were trained using the deep learning toolbox in Matlab R2021a and the adaptive learning rate optimization method ‘ADAM’. Except for CNN 2, equal numbers of images were used per class. Parameters were set as follows for the different CNNs: For CNN 1 and 2, the initial learning rate was set to 0.001 and kept constant. A mini-batch size of 200 and L2 regularization of 0.01 was used, and CNNs were trained for a total of 5 epochs. CNN 4 was trained the same way, except that the L2 regularization was increased to 0.1, the network was trained for 10 epochs, and the learning rate was decreased by a factor of 0.01 after the first 5 epochs. For CNN 3, the initial learning rate was set to 0.0001 and dropped by a factor of 0.01 every 5 epochs. L2 regularization was set to 0.05 and the network was trained for a total of 20 epochs with a mini batch size of 150. To strengthen the generalization of the networks, images were augmented with random rotations and reflections.
CNN 5 recognizes lymphocyte/granulocyte morphology using the brightfield, DAPI and 640 nm (CD45) channels and was obtained by transfer learning of an existing network as described in refs. 47,82. For this network, 48 × 48 pixel (14.4 × 14.4 μm) images were used to account for the smaller size of lymphocytes and granulocytes, and 4452 images from this dataset were curated for transfer learning.
Classification of cell types
The trained CNNs were used to classify every single cell in the dataset in a hierarchical manner (Supplementary Fig. 2a). First, cells were classified as apoptotic or not, and cells classified as apoptotic with a confidence of at least 0.6 were removed from the analysis. Next, cells were assigned to one of four main cell types (Lymphocyte/granulocyte, macrophage, malignant or other). If a cell could not be confidently assigned to any of these classes (maximum confidence <0.6), it was classified as ‘unassigned’ instead. Finally, each cell was assigned a morphological subtype (polarized or conventional for lymphocytes/granulocytes, round or elongated for macrophages and single adherent, single non-adherent, spheroid or adherent colony for cancer cells). No confidence cutoff was set for morphological subtypes. Cell compositions per sample that were used in the MOFA analysis described below were calculated as the mean fraction of each cell type in DMSO control wells, with the abundance of the morphological subclasses normalized to the abundance of their corresponding parent cell type.
Calculation of drug responses
We calculated two different cancer-specific drug response readouts, RCN and RCF, relative to control. These are calculated as follows:
RCN = 1−(number of cancer cells in drug-containing well/median (number of cancer cells in control wells))
RCF = 1−(fraction of cancer cells in drug-containing well/median (fraction of cancer cells in control wells))
In both cases, a positive score indicates a wanted outcome, i.e. a reduction in tumor cells. However, the RCN readout only considers what is happening to the tumor cells and thus assigns positive values to any cytotoxic treatment, regardless of its specificity to malignant cells. On the other hand, RCF only assigns a positive score to compounds that result in a reduction of malignant cells which exceeds the reduction in non-malignant cells, and thus prioritizes compounds that specifically act on malignant cells (see also Supplementary Fig. 4a). Statistical significance of the ex vivo response was assessed by a two-sided two-sample t test, comparing each drug to the matching control (DMSO for small molecules and isotype control for antibodies). In general, all small molecules were used at 10 μM, and antibodies at 10 μg/ml. The only exception to this was the drug panel used in the first 15 samples (plate barcode starting with “STP”, see Supplementary Data 2), where we additionally screened select compounds at 1 μM. An overview of all compounds used in this study is provided in Supplementary Data 4.
For downstream analysis, the drug responses were averaged across replicate wells and scaled to −1 and 1 per sample. To this end, any negative value was divided by the absolute value of the most negative value, and any positive value was divided by the strongest positive value.
Calculation of spheroid dissociation drug response
Morphological drug response was calculated on the cancer cells only, thus “fraction” refers to “the fraction of cells among cancer cells”. To avoid artifacts arising from calculating fractions from very low cell numbers, we only included samples in this analysis that had at least 5% spheroids as a fraction of tumor cells, as well as 2% spheroidal tumor cells as a fraction of all cells. We then defined spheroid dissociation as
1−(fraction of cancer cells forming spheroids in drug-containing well/median(fraction of cancer cells forming spheroids in control wells))
Thus, a positive value indicates spheroid dissociation, whereas zero indicates no change and negative values indicate increased fractions of cells in spheroids.
RNA sequencing
RNA extraction for baseline sequencing
A pellet of around 1 million cells was collected, resuspended in 350 μl of TriZol reagent (Ambion, sold by ThermoFisher 15596026) and stored at −80 °C until extraction. RNA was extracted using a Zymo DirectZol RNA miniprep kit (Zymo R2052) or a DirectZol-96 (Zymo R2056) kit for larger batches according to the manufacturer’s instructions. RNA concentration and integrity was quantified using a TapeStation with RNA reagents (Agilent Technologies).
RNA extraction of drug-treated cells
To profile transcriptomes of drug-treated cells, around 100,000 cells/well were seeded into a PerkinElmer cell carrier ultra 96-well plate and incubated with different small molecules (10 μM each) for 24 h. To avoid losing non-adherent cells, the media was afterward not completely aspirated (~50 μl left per well), and cells were lysed in 150 μl/well TriZol LS reagent (Ambion, sold by ThermoFisher 10296028). Lysates in TriZol were transferred to a PCR plate and stored at −80 °C until extraction. RNA extraction was performed using a DirectZol-96 (Zymo R2056) kit according to the manufacturer’s instructions.
Preparation and sequencing of RNA-seq libraries
All RNA-seq libraries were prepared using a customized version of the DRUG-seq protocol50. Briefly, this protocol consists of reverse transcription (RT) with barcoded poly-T primers combined with second-strand synthesis by template-switching, PCR amplification of the pooled cDNA, and sequencing library construction by tagmentation (Illumina Nextera). We made the following key modifications to the original protocol: (1) we changed the reverse transcription (RT) primer sequences to make them compatible with standard Illumina sequencing primers (see Supplementary Data 11); (2) we start from purified RNA rather than a cell lysate, in our hands this resulted in less genomic DNA contamination of the sequencing libraries and simplified sample storage; (3) we optimized the protocol for ultra-low input samples by using a Zymo IC column to purify cDNA after RT and eluting in only 10 μl of hot (80 °C) water, and by using the Nextera XT kit (Illumina FC-131-1096) for library preparation; (4) we optimized the unique mapping rates of our libraries by performing a stringent size selection with one 0.5× cleanup to remove large fragments followed by two times 0.6× cleanup using KAPA pure beads (KAPA KK8000) resulting in fragment sizes between 500–800 bp. Library size distributions were measured using a TapeStation D5000 HS kit, and library concentration was quantified using a Qubit. Libraries were sequenced at a concentration of 850 pM on a NextSeq2000 system using a mid-output 100 cycle kit (Illumina 20046811), with the following read configuration: 80 bp for read 1 to identify the transcript, 20 bp for read 2 to get the well barcode and unique molecular identifier (UMI), 6 bp index reads. We used ~4 million reads/sample for baseline RNA-seq samples and 1 million reads/well for drug-treated samples.
Processing of raw reads for RNA-seq
Raw sequencing reads were processed using a customized version of the Dropseq toolbox (https://github.com/broadinstitute/Drop-seq), which is available from GitHub83 (https://github.com/RebekkaWegmann/drugseq_toolbox). Briefly, UMIs and well barcodes are extracted from read 2, and well barcodes are removed if they do not match the set of barcodes used in the experiment (up to 2 mismatches allowed). Reads are then mapped to a reference genome (GRCh38 v93) using STAR v.2.4.2, and the number of UMIs per well barcode and gene are counted. The raw sequencing data are available from GEO under the series accession number GSE240953.
Baseline RNA-seq data analysis
QC criteria
Samples were retained if they had >104 UMIs, >5000 genes, and a RIN score of at least 3. Genes were kept if they were protein-coding or long noncoding RNAs, and detected with at least two counts in at least five samples. After applying these filtering criteria, the final dataset contained 17046 genes × 145 samples (131 unique samples), with some samples sequenced across multiple batches to control for batch effects.
Normalization
Raw counts were normalized using the method implemented in the scran R package (v.1.20.1)84. For downstream analysis and visualization, a pseudocount of 1 was added to the normalized counts before they were log2 transformed.
Regressing out covariates for visualization
The log2-transformed normalized expression values were corrected for unwanted covariates (batch, tumor content) using a linear regression approach as implemented in the removeBatchEffect function implemented in the limma R package (v.3.48)85. To preserve biological signals that may be correlated with these covariates, tumor type, and sample type were included in the design parameter.
Visualization of expression profiles using t-SNE
Batch- and tumor content-corrected expression profiles were visualized using t-SNE. The embedding was calculated using the R package Rtsne (v.0.15)86,87,88 with all parameters set to their default values except perplexity = 5 and initial_dims = 10.
Identification of disease-specific genes
To identify genes that were specifically expressed in samples from patients with LUAD, OV, MESO, BRCA, or STAD, a differential expression (DE) analysis was run comparing one tumor type against all others. DE analysis was performed using a negative binomial generalized linear model (GLM) and a quasi-likelihood F-test implemented in edgeR (v.3.34.0)89,90. Batch, biological sex and tumor content were included as covariates in the model. Genes that were highly specific were selected for visualization in Fig. 4b.
Integration of drug responses and RNAseq data
Drug responses (scaled RCF) were integrated with baseline RNA-seq data using a negative binomial GLM and a quasi-likelihood F-test implemented in edgeR (v.3.34.0)89,90. The model included batch and tumor content as unwanted covariates and the scaled drug response as a predictor. Only genes that were detected in at least 70% of samples were included in this analysis.
Calculation of pathway scores
All pathway activation scores were calculated using singscore (v.1.12.0)91 with default parameters. For the baseline RNA-seq data, the batch- and tumor content-corrected expression values were used as input for singscore. For DRUG-seq data, normalized counts were used as input, and pathway scores were then z-scored across conditions per sample. For visualization of GSEA enrichments, only genes contributing to the enrichment (“core enrichment” gene set in clusterProfiler) were used to calculate pathway scores.
Selection of most variable genes for input to MOFA
To select the most informative genes for input to MOFA, a global mean-variance trend was fitted to the covariate-corrected expression values using a loess estimator. Genes with residual variance exceeding the 0.95 quantile of all residual variances were selected for downstream analysis. For input into MOFA, a variance stabilizing transformation (DESeq2 v.1.32.092 varianceStabilizingTransformation) was applied to the selected gene count,s and the transformed values were then z scored across samples.
RNA-seq data analysis for drug-treated samples
For RNA-seq analysis of the drug-treated samples, QC criteria were adjusted to the lower sequencing depth. Wells were kept if they had more than 104 UMIs and >3000 genes, and genes were kept if they were protein-coding or long noncoding RNAs and detected at a count of 2 in at least 2 wells per sample. Genes that were not detected across all samples were discarded. Applying these criteria, the final dimension of the dataset was 8359 genes × 932 wells, corresponding to 47 drugs measured with 3–4 replicates across five samples.
Drug-induced transcriptomic changes were assessed using a negative binomial GLM and a quasi-likelihood F-test implemented in edgeR (v.3.34.0)89,90. We included the SampleID as a covariate in the model and compared each drug to its matching control (DMSO for small compounds, isotype control for antibodies). Only genes detected in at least 70% of wells were included in the DE analysis.
Enrichment analysis for drug-target-proximal gene sets
Direct drug targets were obtained from the SITCH database51,52 (http://stitch.embl.de/, data downloaded on 24 May 2022). Data was filtered to only include interactions reported for human genes with a combined score >500. Drug-target-proximal genes were defined as any gene that functionally interacts with a direct drug target based on the STRING database. For STRING queries, the STRINGdb R package (v. 2.4.2)53 was used with the string database version 11.5. Interactions were filtered for species - human and a score threshold of 500 was used.
To calculate enrichments of DE genes in drug target-proximal gene sets, genes were ordered by the edgeR t-statistic (calculated as t.stat = sign(qlf$table$logFC) * sqrt(qlf$table$F), z = limma::zscoreT(t.stat, df = qlf$df.total), where qlf is the output of edgeR’s glmQLFTest). P values were then calculated using a one-sided hypergeometric test for enrichment of the top and bottom 100 genes among the drug target-proximal genes.
Drug-target networks were visualized using the R package igraph (v.1.2.6)93.
Comparison of expression profiles to TCGA data
The results shown in Fig. 3c and Supplementary Fig. 10 are based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. Gene counts for the different tumor types (LUAD, MESO, OV, BRCA, STAD) were obtained from the GDC data portal. Any sample that was not a primary tumor was excluded, resulting in a total of 2452 samples. Counts were then z-scored per sample, and the genes were subset to the top 5% most variable genes in the MSE cohort. For the tSNE (Fig. 3c) and sample-level clustergram (Supplementary Fig. 10b), the TCGA dataset was combined with the z-scored batch- and tumor content-corrected expression values of the corresponding tumor types in the MSE cohort. To assess the accuracy of MSE tumor-type prediction from the TCGA data, we first reduced the combined gene expression matrix to 30 dimensions using PCA. We then trained a 1-nearest neighbor classifier (knn function from the R package class) on the principal components of the TCGA samples only, and evaluated its performance on the MSE samples. To create the tumor-type level comparisons shown in Supplementary Fig. 10a,c, gene expression was first averaged per tumor type and cohort. After averaging, gene expression values were z-scored per tumor type.
Mutational profiling by FoundationOne CDx
Mutational profiling was performed using the FoundationOne®CDx assay94,95, which includes 324 cancer-related genes for the detection of base exchanges, insertions, deletions, and copy number changes. In addition, specimens are simultaneously profiled for loss of heterozygosity (LOH), tumor mutational burden (TMB) and microsatellite instability (MSI). Detailed information is available at
https://www.foundationmedicine.com/genomic-testing/foundation-one-cdx.
DNA was extracted from formalin-fixed paraffin embedded tissue cell blocks of MSE samples with at least 10% tumor content with the Maxwell 16 FFPE Plus LEV DNA Purification Kit (AS1135) according to the recommendations of the manufacturer. The DNA stock concentration was between 50 and 100 ng/µL for further analysis. Samples were assayed by adaptor ligation hybrid capture, performed for all coding exons of 309 cancer-related genes plus select introns from 34 genes. Sequencing was performed using the Illumina HiSeq instrument to a median exon coverage ≥500×, and data were analyzed for all classes of genomic alterations. The computational pipeline used to analyze sequence patterns used Bayesian algorithms to identify base substitution mutations, local assembly to identify short insertions and deletions, comparisons with process-matched normal controls to determine gene amplifications and homozygous deletions and the analysis of chimeric read pairs to identify gene rearrangements and gene fusions94. Using 0.8 to 1.1 Mb of sequenced DNA for each case, the TMB was determined using the number of somatic base substitution or indel alterations per Mb after filtering to remove germline and pathogenic mutations96.
In this manuscript, the attribution of the definition of “actionable” to any genomic alteration is based on prior knowledge as reported in ref. 97.
Visualization of mutational profiles
Mutational profiles were visualized using the oncoPrint function in the R package ComplexHeatmap (v.2.8.0)98. Alterations were colored by type and somatic impact (known versus likely, uncertain or unknown) as provided by the FoundationOne CDx report, which is based on the COSMIC database (https://cancer.sanger.ac.uk/cosmic).
Calculation of pairwise similarity between mutational profiles
Pairwise similarity between mutational profiles (Fig. 3d) was calculated using a JC as follows:
JCi,j = number of matching alterations between sample i and sample j/number of all alterations in sample i or sample j
Here, “alteration” refers to the type of change occurring in a single gene and sample, which can be wild type, substitution, rearrangement, amplification, deletion or a combination of several types. “Wild type” was also considered an alteration for calculating the JC, and only genes with at least four non-wild type samples were included in this calculation.
Proteomics analysis of vinorelbine-treated MSE cells
Sample preparation for mass spectrometry (MS) analysis
To profile the proteomes of MSE cells following 24 h of Vinorelbine treatment, frozen cells from FB002 (BRCA ascites) were thawed. Directly after thawing, 20 million cells per condition were incubated at a density of 1 million cells/ml either with DMSO or 10 μM vinorelbine for 24 h. DMSO-treated cells were used as control. For the protein extraction following the treatment, cells were split into three replicates for the vinorelbine treatment and four replicates for the DMSO controls. Cells were transferred to 50-ml tubes, centrifuged at 500 × g for 4 min at 4 °C, and washed with 10 ml of cold PBS. After two additional washing steps with cold PBS, the samples were transferred to 1.5-ml tubes and flash-frozen in liquid nitrogen after removal of PBS. Frozen cell pellets were resuspended in 300 µl cold lysis buffer (LB: 1 mM MgCl2, 150 mM KCl, 100 mM HEPES, pH 7.5), supplemented with 1× complete EDTA free protease inhibitor (Roche), transferred to bead-beater tubes and mixed with the same volume of ceramic beads (OMNI International). Subsequently, cells were lysed at 4 °C using a FastPrep‐24TM 5 G bead-beating grinder (MP Biomedicals) at a speed of 5.5 m/sec for 40 seconds. Samples were then transferred to new tubes using a gel-loader tip and spun down at 800 × g for 5 min at 4 °C to remove cell debris. Supernatants were transferred to new pre-cooled tubes, and protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Protein concentrations were adjusted to 1 µg/µl in LB buffer, and 50 µl of lysate per sample were transferred to PCR tube strips. The strip was incubated at 99 °C for 5 minutes. After incubation, the samples were cooled down to 4 °C and sodium deoxycholate (Sigma Aldrich) was added to a final concentration of 5%. Cysteine residues were reduced with 5 mM tris(2-carboxyethyl)phosphine hydrochloride (Pierce) at 37 °C for 40 minutes with shaking at 600 rpm. The reduced cysteines were alkylated by addition of 40 mM iodoacetamide (Sigma Aldrich) at room temperature for 20 min. Samples were diluted with 4 volumes of 100 mM ammonium bicarbonate (Sigma Aldrich). Proteins were digested with lysyl-endopeptidase (FUJIFILM Wako Pure Chemical Corporation) and modified trypsin (Promega) at a 1:100 enzyme to substrate ratio (wt/wt) at 37 °C on a thermoshaker overnight. Protease digestion was quenched by adding formic acid (FA, Sigma Aldrich) to a final concentration of 4%. Precipitated sodium deoxycholate was removed using the AcroPrep Advance 96-well plate with 0.2 µm wwPTFE membrane (Pall Life Sciences). Samples were desalted using a 96-well C18-Spin plate with 7–70 µg capacity (The Nest Group). Peptides were eluted with 50% acetonitrile and 0.1% FA. Eluates were dried using vacuum centrifugation and resuspended in 25 µl buffer A with iRT peptides (1:30) and transferred to 1.5 ml Eppendorf tubes. Samples were vortexed and sonicated for 10 minutes and centrifuged at maximum speed for 20 minutes at 4 °C. 10 µl of the supernatant was transferred to MS vials. A second centrifugation was carried out at maximum speed for 5 min at 4 °C. The sample acquisition volume is 2 µl.
LC-MS/MS data acquisition
Peptide digests were analyzed in randomized order on an Orbitrap Fusion Lumos Tribrid MS (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source and coupled to an Easy-nLC 1200 system (Thermo Fisher). 2 µg of peptides were separated at ambient temperature on a 25 cm × 75 µm i.d. analytical column packed with 2.0 µm C18 beads (Acclaim PepMap C18 from Thermo Fisher) using a linear gradient from 5 to 40% buffer B (B:80% Acetonitrile and 0.1% formic acid; A: 2% Acetonitrile and 0.1% formic acid) over 120 minutes at a flow rate of 300 nl/min. Full MS1 scans were acquired at a resolution of 120,000 between 350 and 1500 m/z. The automatic gain control (AGC) target of 8 × 105 and a maximum injection time of 100 ms were used. 41 variable-width windows (Supplementary Data 13) were utilized to measure fragmented precursor ions. DIA-MS2 spectra were acquired at a resolution of 30,000 an AGC target of 5 × 105, and an injection time of 54 ms. The normalized collision energy was set to 30.
Quantification and statistical analysis
The DIA raw files were searched using the default directDIA+ pipeline of Spectronaut (Biognosys AG, version 18.7.240325.55695) against the human UniProt FASTA (downloaded on 17.07.2020), including the sequence of proteinase K. Search criteria included carbamidomethylation of cysteine as a fixed modification, as well as oxidation of methionine and acetylation (protein N-terminus) as variable modifications. Up to two missed cleavages were allowed. The dynamic mass tolerance strategy was applied to calculate the ideal mass tolerances for data extraction, and no correction factor was applied (correction factor = 1). The local (non-linear) regression method was used for iRT calibration using the iRT kit peptides. The mutated decoy method was used to generate label-free decoys. The false discovery rate (FDR) was estimated with the mProphet approach99 and set to 1% on precursor and protein level. Protein inference was performed using the implemented IDPicker algorithm to define protein groups100. Protein quantification was performed on the MS2 level, and the intensities of protein groups were calculated as the mean of the intensities of the top 3 most abundant peptides. Differential protein abundance was assessed using limma-trend85. The analysis was performed on 5458 proteins that were detected across conditions, had an abundance of at least 3000 in all replicates of at least one experimental condition, and did not contain any strong outliers (more than 10-fold difference in abundance within replicates of one condition).
RT-qPCR of tumor-type-specific genes
Previously extracted RNA was reverse transcribed using the iScript cDNA synthesis kit (BioRad #1708890) according to the manufacturer’s instructions. qPCR was performed using a custom TaqMan array (ThermoFisher #4413262) containing probes for the following 11 genes of interest: NAPSA (Hs00362192_m1), SFTA2 (Hs01588704_g1), NKX2-1 (Hs00968940_m1), CDH6 (Hs00191832_m1), EMX2 (Hs00244574_m1), PAX8 (Hs00247586_m1), VTN (Hs00940758_g1), CLDN15 (Hs00204982_m1), HEG1 (Hs00393516_m1), GATA3 (Hs00231122_m1), CREB3L4 (Hs00370116_m1). Additionally, the array contained four endogenous controls: 18 s rRNA (Hs99999901_s1), GAPDH (Hs99999905_m1), HPRT (Hs99999909_m1), and GUSB (Hs99999908_m1). Each gene was assayed in triplicate per sample, using 10 ng of cDNA as a template in each reaction.
Gene expression (delta cycle threshold, dCT) was quantified by subtracting the mean CT values of the four endogenous controls from the CT values of the genes of interest.
Multi-omics factor analysis
MOFA was run as implemented in the R package MOFA2 (v.1.2.2) with default parameters. The input into MOFA was 4 data modalities (Supplementary Data 6): 1) sample composition (12 variables × 149 samples, Supplementary Data 3); (2) scaled drug responses (101 variables × 2 readouts × 149 samples, Supplementary Data 5); 3) gene expression of the 10% most variable genes (853 variables × 131 samples, Supplementary Data 7); (4) binarized mutational profile (Supplementary Data 8). Here, only genes with any alteration in at least 10 samples were included (22 genes, see Supplementary Fig. 6). Mutations were then further split into the type of alteration (short variant (SV), amplification (AMP), deletion (DEL), rearrangement (RA) and whether they have a known somatic impact, resulting in a total of 176 variables measured across 98 samples. For the mutations, the MOFA likelihood parameter was set to “bernoulli”, for all others, it was left at the default (gaussian). The top-15 factors were considered for downstream analysis. One sample (FB209) for which we only had RNA-seq and mutation data was excluded from MOFA analysis.
Selection of top contributing features per factor for visualization
For visualization, we selected the top 10 features with the highest weight per factor. Only features with absolute weights of at least 0.05 were included. To avoid showing only highly correlated features from the same data type, we selected at most five features per data type.
Identifying disease-specific factors
To identify disease-specific factors, we compared the values of each factor between every single tumor type and all others. Significance was assessed using a two-sided two-sample Wilcoxon test. In addition, for each factor and tumor type, we calculated an area under the receiver operating characteristic curve (AUROC) using the factor value as a binary classifier distinguishing a given tumor type from all others. Factors that specifically separate a certain tumor type from all others will thus have high AUROC values for this tumor type.
Calculation of patient specificity per factor
Patient specificity per factor was calculated using a linear regression model. We ran one model per patient and factor, using binarized patient ID as a predictor and factor value as the response variable, and then defined patent specificity as the maximum R-squared obtained per factor. This corresponds to the maximum amount of variance that can be attributed to a single patient per factor.
Immunohistochemistry against c-MET
Immunohistochemical evaluation was performed using anti c-MET (SP44) rabbit monoclonal primary antibody (catalog # ab227637, Ventana Medical Systems, Tucson, AZ), dilution 1:50. The staining was carried out according to the manufacturer’s protocol on the BenchMark XT platform from Ventana utilizing the ultraView detection kit.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw transcriptomics data generated in this study have been deposited in Gene Expression Omnibus (GEO) under accession code GSE240953. The raw MS proteomics data generated in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under accession code PXD052582. Publicly available databases: TCGA https://www.cancer.gov/tcga/https://portal.gdc.cancer.gov/; STRING https://string-db.org/ version 11.5; STITCH http://stitch.embl.de/ version 5.0 All derived data generated in this study are provided as Supplementary Data tables. Source data are provided with this paper.
Code availability
All computational analyses were performed using Matlab R2021a or R version 4.1.0 with Bioconductor version 3.13101. The DRUG-seq processing pipeline to demultiplex RNA-seq reads is available on GitHub83: https://github.com/RebekkaWegmann/drugseq_toolbox.
References
Quaresma, M., Coleman, M. P. & Rachet, B. 40-year trends in an index of survival for all cancers combined and survival adjusted for age and sex for each cancer in England and Wales, 1971–2011: a population-based study. Lancet 385, 1206–1218 (2015).
Arnold, M. et al. Progress in cancer survival, mortality, and incidence in seven high-income countries 1995–2014 (ICBP SURVMARK-2): a population-based study. Lancet Oncol. 20, 1493–1505 (2019).
Honkala, A., Malhotra, S. V., Kummar, S. & Junttila, M. R. Harnessing the predictive power of preclinical models for oncology drug development. Nat. Rev. Drug Discov. 21, 99–114 (2021).
Attwood, M. M., Fabbro, D., Sokolov, A. V., Knapp, S. & Schiöth, H. B. Trends in kinase drug discovery: targets, indications and inhibitor design. Nat. Rev. Drug Discov. 20, 839–861 (2021).
Slamon, D. J. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 (1987).
von Minckwitz, G. et al. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer. N. Engl. J. Med. 377, 122–131 (2017).
Tripathy, D. et al. De novo versus recurrent HER2-positive metastatic breast cancer: patient characteristics, treatment, and survival from the SystHERs registry. Oncologist 25, e214–e222 (2020).
Flaherty, K. T. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363, 809–819 (2010).
Flaherty, K. T. et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 367, 1694–1703 (2012).
Robert, C. et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 372, 30–39 (2015).
Dummer, R. et al. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 19, 1315–1327 (2018).
Douillard, J.-Y. et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br. J. Cancer 110, 55–62 (2013).
Khozin, S. et al. U.S. Food and Drug Administration approval summary: erlotinib for the first-line treatment of metastatic non-small cell lung cancer with epidermal growth factor receptor exon 19 deletions or exon 21 (L858R) substitution mutations. Oncologist 19, 774–779 (2014).
Kazandjian, D. et al. FDA approval of gefitinib for the treatment of patients with metastatic EGFR mutation–positive non–small cell lung cancer. Clin. Cancer Res. 22, 1307–1312 (2016).
Soria, J.-C. et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 378, 113–125 (2018).
Le Tourneau, C. et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 16, 1324–1334 (2015).
Tsimberidou, A.-M. et al. Initiative for molecular profiling and advanced cancer therapy (IMPACT): an MD Anderson Precision Medicine Study. JCO Precis. Oncol. 2017, 1–18 (2017).
Chen, A. P. et al. Molecular profiling-based assignment of cancer therapy (NCI-MPACT): a randomized multicenter phase II trial. JCO Precis. Oncol. 5, 133–144 (2021).
Sicklick, J. K. et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat. Med. 25, 744–750 (2019).
Sicklick, J. K. et al. Molecular profiling of advanced malignancies guides first-line N-of-1 treatments in the I-PREDICT treatment-naive study. Genome Med. 13, 155 (2021).
Hoes, L. R. et al. Patients with rare cancers in the drug rediscovery protocol (DRUP) benefit from genomics-guided treatment. Clin. Cancer Res. 28, 1402–1411 (2022).
Rodon, J. et al. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat. Med. 25, 751–758 (2019).
Horak, P. et al. Comprehensive genomic and transcriptomic analysis for guiding therapeutic decisions in patients with rare cancers. Cancer Discov. 11, 2780–2795 (2021).
Kato, S. et al. Real-world data from a molecular tumor board demonstrates improved outcomes with a precision N-of-one strategy. Nat. Commun. 11, 1–9 (2020).
Irmisch, A. et al. The tumor profiler study: integrated, multi-omic, functional tumor profiling for clinical decision support. Cancer Cell 39, 288–293 (2021).
Kornauth, C. et al. Functional precision medicine provides clinical benefit in advanced aggressive hematologic cancers and identifies exceptional responders. Cancer Discov. 12, 372–387 (2022).
Malani, D. et al. Implementing a functional precision medicine tumor board for acute myeloid leukemia. Cancer Discov. 12, 388–401 (2022).
Tyner, J. W. et al. Functional genomic landscape of acute myeloid leukaemia. Nature 19, 28 (2018).
Davidson, B., Firat, P. & Michael, C. W. Serous effusions: etiology, diagnosis, prognosis and therapy. 115–175 (Springer London, 2011).
Davidson, B. Molecular testing on serous effusions. Diagn. Cytopathol. 49, 640–646 (2020).
Alix-Panabières, C. & Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 14, 623–631 (2014).
Shah, R. H. et al. Comprehensive mutation profiling by next-generation sequencing of effusion fluids from patients with high-grade serous ovarian carcinoma. Cancer Cytopathol. 123, 289–297 (2015).
Leichsenring, J. et al. Targeted deep sequencing of effusion cytology samples is feasible, informs spatiotemporal tumor evolution, and has clinical and diagnostic utility. Genes Chromosomes Cancer 57, 70–79 (2018).
Liu, L. et al. Next generation sequencing-based molecular profiling of lung adenocarcinoma using pleural effusion specimens. J. Thorac. Dis. 10, 2631–2637 (2018).
Szulkin, A. et al. Characterization and drug sensitivity profiling of primary malignant mesothelioma cells from pleural effusions. BMC Cancer 14, 709 (2014).
Lee, J. Y. et al. Patient-derived cell models as preclinical tools for genome-directed targeted therapy. Oncotarget 6, 25619–25630 (2015).
Hillerdal, C. O. et al. Ex vivo evaluation of tumor cell specific drug responses in malignant pleural effusions. Oncotarget 8, 82885–82896 (2017).
Vinayanuwattikun, C. et al. Feasibility technique of low-passage in vitro drug sensitivity testing of malignant pleural effusion from advanced-stage non-small cell lung cancer for prediction of clinical outcome. Anticancer Res. 39, 6981–6988 (2019).
Papp, E. et al. Feasibility study of in vitro drug sensitivity assay of advanced non-small cell lung adenocarcinomas. BMJ Open Respir. Res. 7, e000505 (2020).
Sheta, R. et al. Development of a 3D functional assay and identification of biomarkers, predictive for response of high-grade serous ovarian cancer (HGSOC) patients to poly-ADP ribose polymerase inhibitors (PARPis): targeted therapy. J. Transl. Med. 18, 439 (2020).
Seo, H.-Y. et al. Culture and multiomic analysis of lung cancer patient-derived pleural effusions revealed distinct druggable molecular types. Sci. Rep. 12, 6345 (2022).
Li, J. et al. Malignant ascites-derived organoid (MADO) cultures for gastric cancer in vitro modelling and drug screening. J. Cancer Res. Clin. Oncol. 145, 2637–2647 (2019).
Chen, H. et al. Short-term organoid culture for drug sensitivity testing of high-grade serous carcinoma. Gynecol. Oncol. 157, 783–792 (2020).
Wang, H.-M. et al. Using patient-derived organoids to predict locally advanced or metastatic lung cancer tumor response: a real-world study. Cell Rep. Med. 4, 100911 (2023).
Heinemann, T. et al. Deep morphology learning enhances ex vivo drug profiling-based precision medicine. Blood Cancer Discov. 3, 502–515 (2022).
Snijder, B. et al. Image-based ex-vivo drug screening for patients with aggressive haematological malignancies: interim results from a single-arm, open-label, pilot study. Lancet Haematol. 4, e595–e606 (2017).
Kropivsek, K. et al. Ex vivo drug response heterogeneity reveals personalized therapeutic strategies for patients with multiple myeloma. Nat. Cancer 4, 734–753 (2023).
Argelaguet, R. et al. MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. 21, 1–17 (2020).
Argelaguet, R. et al. Multi-omics factor analysis—a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 14, e8124 (2018).
Ye, C. et al. DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nat. Commun. 9, 4307 (2018).
Kuhn, M., von Mering, C., Campillos, M., Jensen, L. J. & Bork, P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 36, D684 (2008).
Szklarczyk, D. et al. STITCH 5: augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 44, D380 (2016).
Szklarczyk, D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021).
Jordan, M. A. & Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 4, 253–265 (2004).
Paliogiannis, P. et al. Global prognostic impact of driver genetic alterations in patients with lung adenocarcinoma: a real-life study. BMC Pulm. Med. 22, 32 (2022).
Proietti, I. et al. Mechanisms of acquired BRAF inhibitor resistance in melanoma: a systematic review. Cancers 12, 2801 (2020).
Ingram, K. et al. NKX2-1 controls lung cancer progression by inducing DUSP6 to dampen ERK activity. Oncogene 41, 293–300 (2021).
Moncho-Amor, V. et al. Role of Dusp6 phosphatase as a tumor suppressor in non-small cell lung cancer. Int. J. Mol. Sci. 20, 2036 (2019).
Ahmad, M. K., Abdollah, N. A., Shafie, N. H., Yusof, N. M. & Razak, S. R. A. Dual-specificity phosphatase 6 (DUSP6): a review of its molecular characteristics and clinical relevance in cancer. Cancer Biol. Med. 15, 14 (2018).
Ramalingam, S. S. et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med. 382, 41–50 (2020).
Friese-Hamim, M., Bladt, F., Locatelli, G., Stammberger, U. & Blaukat, A. The selective c-Met inhibitor tepotinib can overcome epidermal growth factor receptor inhibitor resistance mediated by aberrant c-Met activation in NSCLC models. Am. J. Cancer Res. 7, 962–972 (2017).
Wu, Y.-L. et al. Phase Ib/II study of capmatinib (INC280) plus gefitinib after failure of epidermal growth factor receptor (EGFR) inhibitor therapy in patients with EGFR-mutated, MET factor-dysregulated non-small-cell lung cancer. J. Clin. Oncol. 36, 3101–3109 (2018).
Gautschi, O. & Diebold, J. Intracranial activity of osimertinib plus capmatinib in a patient with EGFR and MET-driven lung cancer: case report. JTO Clin. Res. Rep. 2, 100162 (2021).
Gautschi, O., Menon, R., Bertrand, M., Murer, C. & Diebold, J. Capmatinib and osimertinib combination therapy for EGFR-mutant lung adenocarcinoma. J. Thorac. Oncol. 15, e13–e15 (2020).
Wu, Y.-L. et al. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (INSIGHT study): an open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir. Med. 8, 1132–1143 (2020).
Wu, Y.-L. et al. Capmatinib plus osimertinib versus platinum-pemetrexed doublet chemotherapy as second-line therapy in patients with stage IIIb/IIIc or IV EGFR-mutant, T790M-negative NSCLC harboring MET amplification. J. Clin. Orthod. 40, TPS9153–TPS9153 (2022).
Wu, Y.-L., Smit, E. F. & Bauer, T. M. Capmatinib for patients with non-small cell lung cancer with MET exon 14 skipping mutations: A review of preclinical and clinical studies. Cancer Treat. Rev. 95, 102173 (2021).
Huang, Z.-Y. et al. Single-cell analysis of diverse immune phenotypes in malignant pleural effusion. Nat. Commun. 12, 1–12 (2021).
Huang, X.-Z. et al. Single-cell sequencing of ascites fluid illustrates heterogeneity and therapy-induced evolution during gastric cancer peritoneal metastasis. Nat. Commun. 14, 1–22 (2023).
Carvalho, R. F. et al. Single-cell and bulk RNA sequencing reveal ligands and receptors associated with worse overall survival in serous ovarian cancer. Cell Commun. Signal. 20, 176 (2022).
Kim, N. et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 11, 2285–2285 (2020).
Eum, H. H. et al. Tumor-promoting macrophages prevail in malignant ascites of advanced gastric cancer. Exp. Mol. Med. 52, 1976–1988 (2020).
Jeong, H.-O. et al. Cellular plasticity and immune microenvironment of malignant pleural effusion are associated with EGFR-TKI resistance in non-small-cell lung carcinoma. iScience 25, 105358 (2022).
Mitchison, T. J. The proliferation rate paradox in antimitotic chemotherapy. Mol. Biol. Cell 23, 1–6 (2012).
Maynard, A. et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing, https://doi.org/10.1016/j.cell.2020.07.017. (2020).
Subramanian, A. et al. A next generation connectivity map: L1000 platform and the first 1,000,000. Profiles Cell 171, 1437–1452.e17 (2017).
Li, J. et al. DRUG-seq provides unbiased biological activity readouts for neuroscience drug discovery. ACS Chem. Biol. 17, 1401–1414 (2022).
Schuler, M. et al. Molecular correlates of response to capmatinib in advanced non-small-cell lung cancer: clinical and biomarker results from a phase I trial. Ann. Oncol. 31, 789–797 (2020).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize Implements and enhances circular visualization in. R. Bioinform. 30, 2811–2812 (2014).
Carpenter, A. E. et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Vladimer, G. I. et al. Global survey of the immunomodulatory potential of common drugs. Nat. Chem. Biol. 13, 681–690 (2017).
Shilts, J. et al. A physical wiring diagram for the human immune system. Nature 608, 397–404 (2022).
Wegmann, R. Molecular and functional landscape of malignant serous effusions for precision oncology: DRUG-seq toolbox. https://doi.org/10.5281/zenodo.13310707 (2024).
Lun, A. T. L., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with bioconductor. F1000Res 5, 2122 (2016).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
van der Maaten, L. & Hinton, G. E. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
van der Maaten, L. Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 15, 3221–3245 (2014).
Krijthe, J. H. & Van der Maaten, L. Rtsne: T-distributed stochastic neighbor embedding using Barnes-Hut implementation. https://github.com/jkrijthe/Rtsne (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Foroutan, M. et al. Single sample scoring of molecular phenotypes. BMC Bioinform. 19, 404 (2018).
Love, M. I., Anders, S. & Huber, W. Differential analysis of count data - DESeq2 Package. https://www.marcjoiret.be/pdfs/DESeq2package.pdf15, 550 (2014).
Csardi, G. & Nepusz, T. The igraph software package for complex network research. InterJournal vol. Complex Systems 1695. https://igraph.org (2006).
Frampton, G. M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1031 (2013).
He, J. et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 127, 3004–3014 (2016).
Chalmers, Z. R. et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 9, 34 (2017).
Sánchez, N. S. et al. Identification of actionable genomic alterations using circulating cell-free DNA. JCO Precis. Oncol. 3, PO.19.00017 (2019).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Reiter, L. et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 (2011).
Zhang, B., Chambers, M. C. & Tabb, D. L. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 (2007).
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
Acknowledgements
We thank the patients and their families for their trust in taking part in this study. We thank the technical staff at the cytology lab at USZ for collecting the samples and coordinating the shipment to the research lab, and Markus G. Manz for his help in the planning and setup of this study. We gratefully acknowledge the staff of the ETH Flow Cytometry Core Facility and the Functional Genomics Center Zurich for the use of and support with their instruments, and the ETH Informatikdienste team for computational infrastructure support. We thank Julien Mena for his support in designing and printing the drug assay plates. We thank all members of the Snijder lab for their comments on this manuscript and fruitful discussions throughout the course of this study. The results presented in this study are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. This project was funded by the European Research Council under the European Union’s Horizon 2020 Research and Innovation Program (SCIPER; 803063) to B.S., with additional support from the Swiss National Science Foundation (project numbers PP00P3_163961 and PP00P3_194809), the ETH Zurich, and the Institute of Molecular Systems Biology.
Author information
Authors and Affiliations
Contributions
R.W. and L.B. conceptualized and designed the study under the supervision of B.S. and C.B.; R.W., L.B., Y.F., and K.L. performed patient sample processing and PCY under the supervision of B.S; R.W. and L.B analyzed and interpreted PCY data under the supervision of B.S. and C.B.; R.W. performed integrative (MOFA & TCGA) analysis and data visualization under the supervision of B.S.; R.W. and S.L. performed qPCR validation experiments and analysis; R.W., S.L., and K.L. optimized the DRUG-seq workflow for primary patient samples and performed DRUG-seq experiments under the supervision of B.S.; R.W. analyzed DRUG-seq data under the supervision of B.S.; V.C. and A.F. performed the proteomics experiments and related analysis under the supervision of P.P.; F.A. acquired and analyzed FoundationOne data under the supervision of M.Z. and H.M.; L.B., C.B., K.J.D, D.F., D.L., and P.K.B. were responsible for patient recruitment and collection of samples and clinical data; D.L. and P.K.B. performed cytopathological assessment of samples and organized sample distribution. L.B. curated clinical data. The original draft of the manuscript was conceptualized and written by R.W., L.B., and B.S. All authors reviewed, edited, and approved the final version. H.M., C.B., and B.S. acquired funding and jointly supervised the project.
Corresponding author
Ethics declarations
Competing interests
B.S. was a scientific co-founder of Allcyte, which has been acquired by Exscientia. B.S. is a shareholder of Exscientia and a co-inventor on US patent application 15/514,045 relevant to the study. C.B. reports consulting or advisory role for AstraZeneca, Pfizer, Roche, Takeda, Janssen-Cilag, Boehringer-Ingelheim, Merck KGaA, Sanofi; research funding from Bayer; and travel, accommodation, and expenses from AstraZeneca, Takeda, Amgen. All of those are outside the presented work. H.M. is on advisory boards for Astra Zeneca, Stemline Therapeutics, Bayer, Amgen, Astella, MSD, Roche, and Merck. M.Z. receives research funds from Roche. P.P. is a scientific advisor for the company Biognosys AG (Zurich, Switzerland). All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Amit Dutt, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wegmann, R., Bankel, L., Festl, Y. et al. Molecular and functional landscape of malignant serous effusions for precision oncology. Nat Commun 15, 8544 (2024). https://doi.org/10.1038/s41467-024-52694-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52694-8