Abstract
M1 macrophages induce protective immunity against infection, but also contribute to metabolic and inflammatory diseases. Here we show that the E3 ubiquitin ligase, MDM2, promotes the glycolytic and inflammatory activities of M1 macrophage by increasing the production of IL-1β, MCP-1 and nitric oxide (NO). Mechanistically, MDM2 triggers the ubiquitination and degradation of E3 ligase, SPSB2, to stabilize iNOS and increases production of NO, which s-nitrosylates and activates HIF-1α for triggering the glycolytic and pro-inflammatory programs in M1 macrophages. Myeloid-specific haplodeletion of MDM2 in mice not only blunts LPS-induced endotoxemia and NO production, but also alleviates obesity-induced adipose tissue-resident macrophage inflammation. By contrast, MDM2 haplodeletion induces higher mortality, tissue damage and bacterial burden, and also suppresses M1 macrophage response, in the cecal ligation and puncture-induced sepsis mouse model. Our findings thus identify MDM2 as an activator of glycolytic and inflammatory responses in M1 macrophages by connecting the iNOS-NO and HIF-1α pathways.
Similar content being viewed by others
Introduction
M1 macrophages protect against pathogens, at least in part, via the production of pro-inflammatory cytokines and nitric oxide (NO)1; however, its chronic activation contributes to obesity-related type 2 diabetes and sepsis2,3. M1 macrophage polarization is triggered by lipopolysaccharide (LPS) and interferon-gamma (IFNγ) stimulation and is featured by a metabolic shift from oxidative phosphorylation to glycolysis, which is similar to the Warburg effect in the tumor. This metabolic reprogramming provides a rapid supply of energy and the metabolites that allow M1 macrophage to exert their microbicidal activity, pro-inflammatory response, and adaptation to a hypoxic tissue microenvironment.
Manipulating the metabolic pathways can modulate inflammatory response in immune cells, underscoring a cross-talk between metabolism and immunity. The transcriptional factor hypoxia-inducible factor 1-alpha (HIF-1α) is a master player in connecting glycolytic and inflammatory responses in M1 macrophages4. HIF-1α induces a cluster of glycolytic genes including glucose transporter 1 (GLUT1), lactate dehydrogenase A (LDHA), monocarboxylate transporter 4 (MCT4) and the M1 pro-inflammatory factors including inducible nitric oxide (iNOS), interleukin-1beta (IL1B) and IL-6 in macrophages4,5. The increase of glycolysis is also accompanied by the tricarboxylic acid (TCA) cycle breakages, leading to an accumulation of succinate and subsequent inactivation of prolyl hydrolase (PHD), which in turn stabilizes HIF-1α and augments IL-1β production in M1 macrophages6. In addition, LPS triggers pyruvate kinase M2 (PKM2) dimerization that interacts with HIF-1α in the nucleus, thereby potentiating HIF-1α-dependent glycolysis and inflammatory response in M1 macrophages7. Recent studies indicate that accumulation of L-2-hydroxyglutarate (a metabolite derived from α-ketoglutarate in TCA cycle) or lactate (a major glycolytic product) activates HIF-1α via inhibition of PHD, thereby promoting HIF-1α-dependent glycolysis and inflammation in macrophages8,9. These findings exemplify how the metabolic and inflammatory pathways are intertwined, yet the understanding is still incomplete.
The E3 ligase MDM2 is a central negative regulator of tumor suppressor p53. The MDM2-p53 axis suppresses tumorigenesis, at least in part, via downregulation of glycolysis. Inhibition of MDM2 by the chemical compound Serdemetan activates p53, leading to down-regulation of glycolytic enzymes via a HIF-1α-dependent manner in the glioblastoma cell lines10. In addition, the HIF-1α interacts with MDM2 and prevents its suppressive effect on p5311. The MDM2-p53 axis also participates in immune response and inflammatory diseases. For instance, MDM2 is required for the production of thymic stromal lymphopoietin (TSLP), a septic responsive protein that induces an inflammatory response in macrophages12. MDM2 positively causes an immune response in multiple cell types, such as T cells, endothelial cells, and mouse embryonic fibroblasts, via the STAT5 and nuclear factor-kappa B (NF-κB) pathways, respectively13,14,15. Although MDM2 has been shown to control HIF-1α functions and inflammatory responses, whether this oncoprotein regulates M1 macrophage polarization, and if so, is p53 dependent or not, remains unclear.
Here we demonstrate that MDM2 mediates the cross-talk between NO and HIF-1α pathway during M1 macrophage polarization. Haplodeletion or siRNA-mediated silencing of Mdm2 inhibits HIF-1α-dependent glycolysis and pro-inflammatory cytokine production via targeting the inducible nitric oxide synthase (iNOS)-NO axis in M1 macrophages. Myeloid-specific inactivation of MDM2 prevents obesity-induced adipose tissue inflammation and endotoxin-induced sepsis but exacerbates polymicrobial-induced sepsis in mouse models. Our study reveals a pathway that MDM2 facilitates M1 macrophage functions through metabolic reprogramming by targeting the iNOS-NO-HIF-1α axis.
Results
MDM2 promotes M1 macrophage polarization
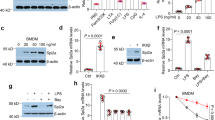
Consistent with the previous studies16,17, treatment with the M1 polarization stimuli (LPS and IFNγ) robustly induced mRNA and protein expressions of MDM2 in peritoneal macrophages and bone marrow-derived macrophages (BMDM) (Fig. 1a, e and g). To examine whether MDM2 inhibition has any impact on M1 polarization, BMDM were transfected with two pairs of siRNAs against Mdm2 (siMdm2) or scramble control (siScramble). Of note, BMDM is a well-established and idea in vitro model for understanding mechanism controlling macrophage polarization. Immunoblotting analysis showed that MDM2 level was substantially reduced in BMDM transfected with either one or two pairs of siMdm2 when compared to those transfected with siScramble (Fig. 1b). Transfection of either single pair or combination of two pairs of siMdm2 resulted in the inhibition of M1 pro-inflammatory cytokines MCP-1 and IL-1β and NO production in the culture medium (Fig. 1c, d and f). Co-transfection of two pairs of siMdm2 resulted a stronger suppressive effect on NO production (reflected by measurement of total nitrite and nitrate [stable products of NO] here and after) than transfection with the single pair of siMdm2 and thus was used in the subsequent experiments (Fig. 1f), unless otherwise specified. As expected, treatment with LPS + IFNγ upregulated the M1 genes and proteins (including Tnfa, Mcp1, inos, Il1b) in BMDM transfected with siScramble, whereas Mdm2 silencing largely abrogated these pro-inflammatory responses (Fig. 1c–g). Consistently, transcriptomic analysis indicated that the pathways involved in IFNγ, IFN-α, iNOS and TNFα were significantly downregulated by Mdm2 silencing in BMDM (Supplementary Fig. 1d). Furthermore, we reproduced the inducible action of LPS + IFNγ on MDM2 expression and inhibitory effects of MDM2 silencing on LPS + IFNγ-induced pro-inflammatory responses and NO production in human THP-1 macrophages (Supplementary Fig. 2a–d). Of note, THP-1 macrophage is a well-studied and convenient model of human macrophage polarization18. To increase the validity and generalization of our findings, we included both BMDM and human THP-1 macrophages in the subsequent experiments, unless otherwise specified.
a Immunoblotting analysis of MDM2 and GAPDH in the primary mouse peritoneal macrophages upon LPS and IFNγ or vehicle (Veh) treatment for 20 h. The lower panel is the densitometric analysis for relative abundance of MDM2 normalized with GAPDH. n = 3. (b-f) Bone marrow-derived macrophages (BMDM) were transfected with siRNA against Mdm2 (siMdm2) or scramble (siScramble) control, followed by LPS and IFNγ stimulation for 20 h. b Immunoblotting analysis of MDM2 and β-actin in the transfected BMDM. The lower panel is the densitometric analysis for relative abundance of MDM2 normalized with β-actin. n = 4. c MCP-1 and d IL-1β levels in cell culture supernatant. n = 4. e QPCR analysis of the genes related to M1 macrophage polarization in the BMDM as indicated. n = 8. f Total nitrate & nitrite in culture medium. n = 4 for siMdm2-1 and siMdm2-2. n = 8 for siScramble and siMdm2-1 + 2. g Immunoblotting analysis of iNOS, MDM2, p53 and β-actin in the BMDM. The right panel is the densitometric analysis of iNOS and p53 normalized with β-actin. n = 3. h Flow cytometry analysis of F4/80 and CD11c-double positive cells. The right panel is the mean fluorescence intensity. siScramble: n = 7, siMdm2: n = 6. Data are displayed as mean ± SEM. Statistical significance was examined using one-way ANOVA with Tukey post-hoc test. N number represents the number of biological replicates. ND Not detected.
To complement the findings in the siRNA experiments, we generated myeloid-specific MDM2 haplodeletion mice (so-called Mye-MDM2KO mice) by crossing Mdm2floxed/floxed mice with lysozyme 2 Cre knock-in mice. Noticeably, we were unable to obtain homozygous myeloid-MDM2KO mice, perhaps complete deletion of MDM2 results in over-activation of p53-dependent apoptosis and senescence and its essential role in hematopoiesis19. MDM2 protein was decreased by ~40% in BMDM from Mye-MDM2KO mice when compared to those from WT littermates (Supplementary Fig. 3a). Consistent with the findings using siRNA, haplodeletion of Mdm2 significantly inhibited the pro-inflammatory properties of M1 macrophages, as exemplified by the reduction of Tnfa, Mcp1, Il1b, and NO (Supplementary Fig. 3b–d). On the contrary, haplodeletion of Mdm2 had no apparent impact on M2 macrophage polarization (Supplementary Fig. 3e).
Inactivation of MDM2 impairs glycolysis in M1 macrophages
Metabolic reprogramming is indispensable for M1 macrophage polarization, and the MDM2-p53 axis is known to control glycolysis and mitochondrial functions20,21. We investigated whether MDM2 inactivation impacts glycolysis and mitochondrial oxidation. Seahorse XF analysis revealed that Mdm2 silencing significantly diminished glycolytic proton efflux rate (GlycoPER) in BMDM irrespective of the presence or absence of M1 stimuli during the glycolysis stress test (Fig. 2a). Biochemical assay confirmed that the M1 stimuli-induced lactate secretion was also partially blocked by Mdm2/MDM2 silencing in BMDM and THP-1 macrophages (Fig. 2b and Supplementary Fig. 2e). On the other hand, Mdm2 silencing did not exert any obvious impact on mitochondrial oxygen consumption rate in BMDM during the mitochondrial stress test (Supplementary Fig. 4a), despite of the enrichment of oxidative phosphorylation pathway in the RNAseq data (Supplementary Fig. 1). Likewise, haplodeletion of Mdm2 also reduced M1 stimuli-induced glycolysis and lactate production in the BMDM (Supplementary Fig. 3f, g).
BMDM were transfected with siMdm2 or siScramble, followed by with LPS and IFNγ for 20 h. a Glycolytic stress test was performed using Seahorse XF analyzer. Rotenone/Antimycin A (Rot/AA) and 2-deoxyglucose (2-DG) were added to inhibit mitochondrial functions and glycolysis respectively. Glycolytic proton efflux rate (GlycoPER) was measured at the basal condition and during the treatments as indicated. n = 4. P values of siScramble-LPS + IFNγ vs siMdm2-LPS + IFNγ are above the curve and highlighted in yellow while siScramble-veh vs siMdm2-veh are below the curve. b Lactate level in the conditioned medium. n = 4 biological replicates. c, d Relative abundance of metabolites in (c) glycolysis and (d) pentose phosphate pathway determined by UPLC-MS/MS. n = 5. e Immunoblotting analysis of HIF-1α, hydroxyl-HIF-1α and β-actin. n = 3. f HIF-1α transcriptional activity in the nuclear fraction of the BMDM. n = 4. g QPCR analysis of the glycolytic genes normalized with β-actin. n = 8 for Glut1, Ldha, Mct4 and Pkm2. n = 4 for Hif1a. Data are displayed as mean ± SEM. Statistical significance was examined using one-way ANOVA with Tukey post-hoc test. N number represents the number of biological replicates.
We further evaluated the impact of Mdm2 silencing on metabolic reprogramming by measuring metabolites derived from glycolysis, the pentose phosphate pathway (PPP) and TCA cycle. LC/MS-based metabolomics analysis showed the metabolites in glycolysis (including d-glucose-6-phosphate, dihydroxyacetone phosphate, pyruvate and lactate) and in PPP (including ribulose 5-phosphate, xylulose 5-phosphate, glyceraldehyde 3-phosphate and sedoheptulose 7-phosphate), were significantly reduced by Mdm2 silencing in the M1 BMDM (Fig. 2c, d). Despite no change in mitochondrial respiration assessed by the Seahorse XF analzyer, the TCA cycle intermediates acetyl-CoA, cis-aconitate, alpha-ketoglutarate, isocitrate and succinate were markedly reduced in BMDM with Mdm2 silencing, whereas succinyl-CoA, fumarate, malate and oxaloacetate were upregulated (Supplementary Fig. 4b). On the other hand, RNAseq and QPCR analysis showed the changes in glycolytic gene expression profiles were associated with the glycolytic metabolites in BMDM with Mdm2 silencing (Fig. 2g and Heatmap in Supplementary Fig. 1). We also found that protein and transcriptional activity of HIF-1α, the master regulator of glycolysis, was markedly suppressed in BMDM with Mdm2 silencing, despite of similar Hif1a mRNA expression (Fig. 2e, f). The reduction of glycolytic gene expression and HIF-1α activity were also observed in THP-1 macrophages with MDM2 silencing (Supplementary 2f, g) or BMDM with MDM2 haplodeletion (Supplementary Fig. 3h–i and Supplementary Fig. 8a). Taken together, these findings indicate MDM2 inactivation profoundly affects metabolic pathways, in particular glycolysis, in M1 macrophages.
The regulatory effects of MDM2 inactivation on glycolysis and inflammation in macrophage is p53-independent
MDM2 is known to restrict p53 activation22,23,24. However, inactivation of MDM2 by either siRNA or haplodeletion did not exert a noticeable effect on p53 protein expression and its dependent glycolytic (TP53-induced glycolysis and apoptosis regulator [Tigar]), apoptotic (p53 upregulated modulator of apoptosis [Puma]) and senescent (Cdkn1a; also known as p21) responses (Fig. 1g, Supplementary Fig. 5a and Supplementary Fig. 6a, b). To further confirm that the changes in M1 polarization induced by MDM2 inactivation are p53 independent, we treated the BMDM from Mye-MDM2KO mice or the siMDM2-transfected BMDM with pifithrin-α (PFT-α), a small molecule that blocks p53 transcriptional activity25. Although PFT-α reduced p53 downstream gene expression including Tigar, Cdkn1a and Puma, it was unable to reverse MDM2 inactivation-mediated suppression of pro-inflammatory cytokine expression and glycolysis in the M1 BMDM (Supplementary Fig. 5a–d and Supplementary Fig. 6b–e). These data suggest that the regulatory actions of MDM2 inactivation on inflammation and glycolysis are not due to p53 activation.
MDM2 controls M1 macrophage polarization via the iNOS-NO-HIF1α axis
As MDM2 inactivation markedly diminished HIF-1α activity and glycolysis, we hypothesized that MDM2 controls M1 macrophage functions via HIF-1α. To test this, we pre-treated the siMDM2- or siScramble-transfected BMDM or THP-1 macrophages with the HIF-1α activator CoCl2 (a chemical compound that inhibits proline hydroxylase [PHD], the upstream inhibitor of HIF-1α26), followed by induction of M1 macrophage polarization. Downregulation of HIF-1α activity by MDM2 silencing was neutralized by CoCl2 treatment in M1 macrophages (Fig. 3a and Supplementary Fig. 7a), which were accompanied by the recovery of glycolytic responses (reflected by GlycoPER, expression of the glycolytic genes and/or secreted lactate in the culture medium) (Fig. 3b–d and Supplementary Fig. 7c) and M1 pro-inflammatory cytokine expression including MCP1, TNFA and IL1B (Fig. 3e–g and Supplementary Fig. 7c). However, CoCl2 treatment could not restore NO production in BMDM or THP-1 macrophages with MDM2 silencing (Fig. 3h and Supplementary Fig. 7b). The restoration of HIF-1α activity was also observed in MDM2 haplodeficient BMDM treated with CoCl2 when compared to those treated with vehicle, which was accompanied with the restoration of pro-inflammatory cytokines including MCP-1 and TNF-α and, but not the NO production (Supplementary Fig. 8a–d).
a–f BMDM were transfected with siMdm2 or siScramble, followed by stimulation with LPS and IFNγ in the presence or absence of CoCl2 for 20 h. a Nuclear HIF-1α transcription activity. n = 4. b Glycolytic proton efflux rate (GlycoPER) measured by Seahorse XF analyzer as described in Fig. 2. n = 4. P values of siScramble-LPS + IFNγ vs siMdm2-LPS + IFNγ are under the curves while siMdm2-LPS + IFNγ vs siMdm2-CoCl2+LPS + IFNγ are above and highlighted in yellow. c QPCR analysis of the glycolytic genes normalized with β-actin. n = 4. d Lactate, e MCP-1 and f IL-1β in the cell culture supernatant. n = 4. g QPCR analysis of the pro-inflammatory genes normalized with β-actin. n = 4. h Total nitrate and nitrite in the cell culture supernatant. n = 4. Data are displayed as mean ± SEM. Statistical significance was examined using one-way ANOVA with Tukey post-hoc test. N number represents the number of biological replicates. ND Not detected, NS Not significant.
The above findings suggest that MDM2 controls the iNOS-NO axis independent of HIF-1α or the iNOS-NO axis is upstream of HIF-1α. The latter possibility is supported by the studies showing NO stabilizes HIF-1α via s-nitrosylation in the cancer cells27,28. Indeed, we found that silencing of Mdm2 or haplodeletion of Mdm2 inhibited s-nitrosylation but not hydroxylation (a post-translational modification that mediates proteasomal degradation of HIF-1α) of HIF-1α in BMDM (Fig. 2e, Fig. 4c and Supplementary Fig. 8e). We next tested whether replenishment of NO restores activity and s-nitrosylation of HIF-1α and M1 polarization in BMDM with Mdm2 silencing. siMdm2- or siScramble-transfected BMDM were pre-treated with the NO donor S-Nitroso-N-acetylpenicillamine (SNAP) or vehicle control, followed by induction of M1 macrophage polarization. Treatment with SNAP reinstated NO in the siMdm2-transfected BMDM to the level similar to that in the siScramble-transfected cells (Fig. 4a). Importantly, treatment with SNAP rescued the defective activation and s-nitrosylation of HIF-1α (Fig. 4b, c), accompanied with the restoration of the glycolytic program (Fig. 4d–f) and M1 macrophage polarization (Supplementary Fig. 9). We also replicated the restoration effects of SNAP on nitrosylation and activation of HIF-1α and inflammatory cytokine production in MDM2 haplodeficient BMDM and siMDM2-transfected THP-1 macrophages (Supplementary Fig. 7d and Supplementary Fig. 8). Given that HIF-1α activity can also be controlled by succinylation6 and MDM2 silencing led to a ~ 50% reduction in succinate level in M1 macrophages (Supplementary Fig. 4b), we examined whether replenishment of succinate is able to restore of HIF-1α activation. Unlike the SNAP treatment, treatment with cell-permeable dimethyl-succinate only partially restored HIF-1α activity (p = 0.074) and its related glycolytic gene expression and lactate production as well as IL-1β secretion (p = 0.096) in BMDM transfected with siMdm2 when compared to those treated with vehicle (Supplementary Fig. 10). Taken together, these data suggest that MDM2 controls HIF-1α-dependent glycolysis and inflammatory response mainly via the iNOS-NO axis.
Mouse BMDM were transfected with siMdm2 or siScramble for 24 h, then stimulated with LPS and IFNγ for 20 h, followed by treatment with NO donor S-Nitroso-N-acetylpenicillamine (SNAP) or DMSO for 3 h. a Total nitrate and nitrite in the cell culture medium. n = 4. b Nuclear HIF-1α transcriptional activity in the BMDM. n = 4. c The treated cells were subjected to S-nitrosylation assay, followed by immunoprecipitation (IP) of HIF-1α. Immunoblotting analysis of HIF-1α in the nuclear extract and S-nitrosylated HIF-1α (labeled by iodoTMT Reagent) was performed using an HIF-1α antibody and anti-TMT antibody respectively. Lamin B1 is a nuclear protein and used as loading control. d GlycoPER was measured using Seahorse XF analyzer. The right panel is the area under curve (AUC). n = 5. P values of siScramble-LPS + IFNγ vs siMdm2-LPS + IFNγ are under the curve while siMdm2-LPS + IFNγ vs siMdm2-SNAP + LPS + IFNγ is above and highlighted in yellow. e Lactate in the cell culture supernatant. n = 4. f Relative mRNA levels of glycolytic genes normalized with β-actin and expressed as fold change over siScramble-LPS + IFNγ. n = 4. Data are displayed as mean ± SEM. Statistical significance was examined using one-way ANOVA with Tukey post-hoc test. N number represents the number of biological replicates. Representative immunoblot images are shown from at least two independent experiments.
MDM2 enhances NO production and its promoting effect on glycolysis and M1 polarization by interacting and inhibiting SPSB2 function
Complete ablation of iNOS or pharmacological inhibition of NO has been shown to promote LPS + IFNγ-induced M1 pro-inflammatory cytokine production in BMDM29,30. We found that MDM2 inactivation leads to around 40–60% reduction of iNOS/NO in macrophages accompanied by defective M1 macrophage polarization. To address the contradictory effect of NO on M1 polarization in our study and previous studies29,30, we transfected BMDM from C57BL/6 J mice with siRNA against inos (si-inos). Expression of iNOS and NO was reduced by ~60–70% in BMDM transfected with si-inos compared to those transfected siScramble (Supplementary Fig. 11a–c). Reduction of iNOS and its derived NO resulted in a sharp reduction of M1 pro-inflammatory cytokine expression and HIF-1α activity and its downstream glycolytic program (Supplementary Fig. 11d–f), which is similar to the observation in the macrophages with MDM2 inactivation.
MDM2 silencing profoundly reduced protein but not mRNA expression of iNOS in BMDM and THP-1 macrophages (Fig. 1e, g and Supplementary Fig 2b), indicating that MDM2 regulates iNOS at a post-transcriptional level. Previous studies showed that LPS induces iNOS protein expression by preventing the binding of iNOS to its E3 ubiquitin ligase complex Elongin B/C-Cullin-5-SPRY domain- and SOCS box-containing protein (ECS(SPSB))31,32. SPSB2 in the ECS(SPSB) complex binds to the N-terminus of iNOS and directs its degradation by the proteasome33,34. Protein but not mRNA expression of SPSB2 was upregulated in BMDM transfected with siMDM2 compared to those transfected with siScramble (Fig. 5a, b). Treatment of the protein synthesis inhibitor cycloheximide (CHX) revealed the protein stability of SPSB2 was enhanced by Mdm2 silencing in BMDM (Fig. 5c). In THP-1 macrophages, MDM2 silencing also reduced protein but not mRNA levels of SPSB2, accompanied with increased SPSB2 ubiquitination (Fig. 5d–f), whereas overexpression of MDM2 dramatically induced SPSB2 ubiquitination and reduced its protein expression (Fig. 5g and Supplementary Fig. 12a, c). In vitro ubiquitination assay also showed that recombinant MDM2 protein directly induced SPSB2 ubiquitination in the presence of the E1 and E2 enzymes (Fig. 5h).
a–c Mouse BMDM were transfected with siMdm2 or siScramble for 24 h and stimulated with LPS + IFNγ stimulation for 20 h. a QPCR analysis of Spsb2 mRNA expression and normalized with β-actin. n = 4. b Immunoblotting of SPSB2 and β-actin and quantification of SPSB2 normalized with β-actin. n = 4. c The transfected BMDM were treated with CHX for indicated time points, followed by immunoblotting analysis of SPSB2 and β-actin. n = 3. The right panel is the densitometric analysis of SPSB2 level. d, e THP-1 macrophages were transfected with siMDM2 or siScramble for 24 h and then stimulated with LPS + IFNγ stimulation for 24 h. n = 3. d QPCR analysis of SPSB2 mRNA expression and normalized with β-ACTIN. n = 4. e Immunoblotting of iNOS, MDM2, SPSB2 and β-actin. The right panel is the quantification of SPSB2 normalized with β-actin. n = 4. f THP-1 macrophages were transfected with siMDM2 or siScramble for 48 h, followed by IP of SPSB2 and immunoblotting analysis of MDM2, SPSB2, Ub and β-actin. n = 3. g THP-1 macrophages were transfected with plasmid expressing Myc-tagged MDM2 for 48 h, followed by IP of SPSB2 and immunoblotting analysis of SPSB2, Ub and Myc. n = 3. h In vitro ubiquitination of SPSB2. The recombinant E1 and E2 enzymes, MDM2, HA‐tagged Ub proteins, and native SPSB2 protein were added to the ubiquitination reaction mix, followed by IP of SPSB2 and immunoblotting analysis as indicated. n = 4. Representative immunoblot images are shown from at least two independent experiments. Data are displayed as mean ± SEM. N number represents the number of biological replicates.
To investigate whether MDM2 controls the iNOS-NO-HIF-1α and its dependent M1 polarization via SPSB2, we co-transfected BMDM with siMdm2, siSpsb2 and/or siScramble, followed by LPS + IFNγ stimulation. We confirmed that siMdm2 and siSpsb2 transfection decreased the expression of MDM2 and SPSB2 in BMDM, respectively (Fig. 6a, b). Reduced iNOS expression and NO production in siMdm2-transfected BMDM were reversed by concomitant transfection of siSpsb2 (Fig. 6b, c), which was accompanied by the restoration of HIF-1α activity and its s-nitrosylation (Fig. 6d, e), the glycolytic program (Fig. 6f, g and Supplementary Fig. 13a) and inflammatory cytokine production (Fig. 6h, i and Supplementary Fig. 13b). To complement with the siRNA approach, we created a SPSB2 knockout THP-1 (referred to as “S-KO-THP-1”) cell line using the Crispr-Cas9 gene editing approach, as we recently described9. The protein level of SPSB2 was undetectable in S-KO-THP-1 macrophages when compared to the control THP-1 (referred to as “Ctrl-THP-1”) macrophages (Supplementary Fig. 14a). MDM2 silencing reduced HIF-1α activity, lactate secretion, IL-1β and NO production, iNOS protein expression, the glycolytic and pro-inflammatory gene expression in the Ctrl-THP-1 macrophages, but not in the S-KO-THP-1 macrophages (Supplementary Fig. 14). These findings suggest that MDM2 controls the iNOS-NO axis via targeting SPSB2, which is essential for the HIF-1α-dependent glycolytic and inflammatory response in M1 macrophages.
Mouse BMDM were transfected with siMdm2, siSpsb2 and/or siScramble as indicated for 24 h, followed by stimulation with LPS + IFNγ for 24 h. a QPCR analysis of Mdm2 and Spsb2 mRNA expression normalized with β-actin. n = 4. b Immunoblotting analysis of iNOS, MDM2 SPSB2 and nuclear HIF-1α. c Total nitrate & nitrite level in the cell culture supernatant. n = 4. d Nuclear HIF-1α transcriptional activity. n = 4. e Detection of nuclear HIF-1α and s-nitrosylated HIF-1α by immunoblotting as described in Fig. 4c. n = 3. f GlycoPER measured by Seahorse XF analyzer. n = 5. For the left panel, P values of siScramble-veh vs siMdm2-veh are under the curves while siMdm2-veh vs siMdm2+siSpsb2-veh are above and highlighted in yellow. For the right panel, P values of siScramble-LPS + IFNγ vs siMdm2-LPS + IFNγ are under the curves while siMdm2-LPS + IFNγ vs siMdm2+siSpsb2-LPS + IFNγ are above and highlighted in yellow. g Secreted lactate, h MCP-1 and i IL-1β levels in the cell culture supernatant. g, h n = 7. i: n = 4. Data are displayed as mean ± SEM. Statistical significance was examined using one-way ANOVA with Tukey post-hoc test. N number represents the number of biological replicates. ND Not detected. Representative immunoblot images are shown.
To identify which lysine residue(s) of SPSB2 is ubiquitinated by MDM2, we systematically mutated all the lysine (K) residues of SPSB2 to arginine (R) residue (a non-ubiquitinated amino acid with similar physical property with lysine). The mutants include SPSB2-K51R, SPSB2-K60R, SPSB2-K81R, SPSB2-K139R, SPSB2-K145R, SPSB2-K195R and SPSB2-K257R (Supplementary Fig. 12b). Overexpression of MDM2 promoted ubiquitination and decreased levels of wild-type SPSB2 (SPSB2-WT), SPSB2-K51R, SPSB2-K60R, SPSB2-K139R, SPSB2-K145R and SPSB2-K257R but not the SPSB2-K81R and SPSB2-K195R mutants in S-KO THP-1 macrophages (Supplementary Fig. 12c). To explore the functional relevance, we reintroduced SPSB2-WT, SPSB2-K81R or SPSB2-K195R into S-KO THP-1 macrophages (Fig. 7a). The reintroduction of SPSB2-WT restored the inhibitory effects of MDM2 silencing on NO production, HIF-1α activity and its associated glycolysis and inflammatory cytokine expression in S-KO THP-1 macrophage. However, such restorative effects of SPSB2 were lost when the ubiquitination site of SPSB2 at K81 or K195 mutated to arginine (Fig. 7b–g).
a S-KO-THP-1 and the Ctrl-THP-1 macrophages were transfected with the indicated plasmids, followed by immunoblotting analysis of SPSB2 and iNOS. n = 3. b–g S-KO-THP-1 and the Ctrl-THP-1 macrophages were co-transfected with siMDM2 or siScramble and plasmids encoding wild-type SPSB2 (WT) or the K81R or K195R mutants of SPSB2, followed by stimulation with LPS and IFNγ for 24 h. n = 3. b Total nitrate & nitrite in the conditioned medium. n = 4. c HIF-1α transcriptional activity in the nuclear fraction of the cells. n = 4. d IL-1β and e lactate in the conditioned medium. n = 4. f, g QPCR analysis of pro-inflammatory genes (f) and glycolytic genes (g) normalized with β-ACTIN. The QPCR results were displayed as fold change over Ctrl-THP-1-siScramble-GFP. n = 4. Data are displayed as mean ± SEM. Statistical significance was examined using one-way ANOVA with Tukey post-hoc test. N number represents the number of biological replicates. Representative immunoblot images are shown.
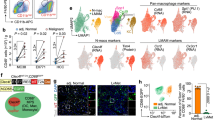
In silico analysis revealed that SPSB2 possesses two potential MDM2 binding motifs35 (Supplementary Fig. 15a), prompting us to hypothesize that the E3 ligase MDM2 binds to SPSB2 and facilitates its proteasomal degradation. Co-immunoprecipitation assay showed that LPS + IFNγ enhanced the MDM2-SPSB2 interaction in both THP-1 macrophages and BMDM in a time-dependent manner (Fig. 8a and Supplementary Fig. 15b). Of note, MDM2 did not interact with HIF-1α in the macrophages (Fig. 8a and Supplementary Fig. 15b). On the other hand, the MDM2-p53 interaction was not modulated by the presence or absence of SPSB2 in THP-1 macrophages (Supplementary Fig. 15c), indicating that MDM2-p53 and MDM2-SPSB2 complex may operate independently. The two putative MDM2-binding motifs within SPSB2 are L19/Y20/L23 and L199/Y200/V203, are found at amino acid positions 19–23 and 199–203 of SPSB2 protein, respectively (Supplementary Fig. 15a). To investigate whether MDM2 interacts with SPSB2 via these two motifs, we created FLAG-tagged human SPSB2 plasmids with the mutations: L19, Y20, and L23 were substituted with alanine residues (referred to as the Triple-1 mutant), and L199, Y200, and V203 were mutated to alanine residues (referred to as the Triple-2 mutant). To avoid interference from endogenous SPSB2 in macrophages, we employed S-KO-THP-1 macrophages for subsequent analyzes. Co-immunoprecipitation assays revealed that MDM2 interacted with SPSB2-WT and the SPSB2 Triple-2 mutant, but not with the SPSB2 Triple-1 mutant, in S-KO THP-1 macrophages (Fig. 8b). We successfully expressed SPSB2-WT and SPSB2-Triple in S-KO-THP-1 macrophages, in which SPSB levels similar to that in Ctrl-THP-1 macrophages (Fig. 8c). Furthermore, the S-KO THP-1 macrophages with MDM2 silencing were refractory to LPS-induced HIF-1α activation, NO production and inflammatory cytokine production when they were reconstituted with the SPSB2-WT but not the SPSB2 Triple-1 mutant (Fig. 8d–g and Supplementary Fig. 16). These findings suggest that the MDM2-SPSB2 interaction is crucial for the control of M1 polarization and metabolic reprogramming.
a THP-1 macrophages were treated with LPS and IFNγ for indicated time points, followed by IP using anti-MDM2 antibody and immunoblotting analysis as indicated. The chart shows the quantification of SPSB2 bound to MDM2. n = 3. b S-KO-THP-1 macrophages and Ctrl-THP-1 macrophages were transfected with plasmids expressing Myc-tagged MDM2 and FLAG-tagged SPSB2 (WT), SPSB2 (L19/Y20/L23, referred as Triple-1) or SPSB2 (L199/Y200/V203, referred as Triple-2), followed by immunoprecipitation using anti-Myc antibody and immunoblotting analysis as indicated. c S-KO-THP-1 and Ctrl-THP-1 macrophages transfected with indicated plasmids were subjected to immunoblotting analysis of SPSB2 and β-actin. d–g S-KO-THP-1 and Ctrl-THP-1 macrophages were co-transfected with indicated siRNA and plasmids for 48 h, followed by LPS + IFNγ stimulation for 24 h. n = 4. d Total nitrate & nitrite in the conditioned medium. n = 4. e IL-1β, f HIF-1α transcriptional activity and g secreted lactate. n = 4. Data are displayed as mean ± SEM. Statistical significance was examined using one-way ANOVA with Tukey post-hoc test. N number represents the number of biological replicates. Representative images were shown from at least two independent experiments.
The MDM2 binding motifs can also be found in p53, and ApoB and their bindings to MDM2 have been shown to be abrogated by nutlin-3a treatment35,36. We found that treatment of nutlin-3a not only disrupted the MDM2-SPSB2 interaction but also increased SPSB2 expression in Raw264.7 macrophages (Supplementary Fig. 17a, b). These changes were accompanied by diminished iNOS expression, NO production, HIF-1α activation and its dependent glycolytic gene program and M1 polarization in BMDM (Supplementary Fig. 17c–h). siRNA-mediated knockdown of Spsb2 reversed the inhibitory effect of nutlin-3a on MCP1, NO and lactate production in the cell culture supernatant (Supplementary Fig. 17i–k), indicating that nutlin-3a controls glycolytic and pro-inflammatory responses via SPSB2.
Taken together, MDM2 interacts with SPSB2 through the L19/Y20/L23 MDM2 binding motif, leading to SPSB2 ubiquitination at K81 and K195 and subsequent proteasomal degradation. This process increases iNOS-NO expression, which promotes s-nitrosylation and stabilization of HIF1α, ultimately activating glycolytic and inflammatory program in M1 macrophages.
Myeloid MDM2 controls septic response and adipose tissue inflammation in obesity
To explore the physiological relevance of our findings, we examined the role of myeloid MDM2 in controlling sepsis and obesity-induced adipose tissue inflammation, both of which, at least in part, are due to aberrant M1 macrophage functions. The first model utilized LPS to induce an acute inflammatory and septic response in mice37. Intraperitoneal injection of a lethal dose of LPS led to significant body weight loss, hypoglycemia, hypothermia, and lethality in WT littermates (Supplementary Fig. 18a–d). However, myeloid deletion of MDM2 alleviated LPS-induced inflammation, weight loss, hypoglycemia and hypothermia but no effect on overall survival rate at 96-hour post-surgery (Supplementary Fig. 18a–d). Although there was no difference in the circulating level of MCP-1, NO, and tumor necrosis factor receptor-2 (sTNFR2, a surrogate marker of TNF-α expression) before LPS injection, these pro-inflammatory factors were significantly dampened in Mye-MDM2KO mice compared to WT littermates after LPS stimulation (Supplementary Fig. 18a–d).
Next, we explored the role of myeloid MDM2 in polymicrobial sepsis induced by cecal ligation and puncture (CLP), a commonly used model mimicking human sepsis condition38. In contrast to the endotoxin-induced sepsis model, myeloid-specific haplodeletion of MDM2 drastically exacerbated the mortality rate after CLP surgery, accompanied by worsened hypoglycemia and hypothermia (Fig. 9a–c). In addition, the circulating levels of pro-inflammatory cytokines IL-6, IL-1β, sTNFR2, and NO were also diminished in Mye-MDM2KO mice (Fig. 9d–g). Ex vivo experiments showed that the number of bacteria in the peritoneum was significantly higher in WT littermates receiving CLP than those undergoing the sham operation (Fig. 9h). Such bacterial burden was worsened by myeloid-specific haplodeletion of MDM2 (Fig. 9h). In addition, Mye-MDM2KO mice displayed more severe damage in the liver and spleen after the CLP procedure when compared to WT controls (Fig. 9i, j). BMDM or peritoneal macrophages from Mye-MDM2KO mice showed defective bacterial killing ability compared to WT controls (Fig. 9k). Such a defect could be restored by treatment with the HIF-1α activator CoCl2 (Fig. 9l). Taken together, the above findings suggest that myeloid MDM2 is required to eliminate invading pathogens by activating macrophages in the polymicrobial model but also contributes to hyperinflammation and lethality in the LPS-induced sepsis model.
a–j Cecal ligation and puncture (CLP) or sham operation were performed on Mye-MDM2KO mice and their WT controls. a Mortality of mice was monitored for 120 h after the CLP procedure. Survival rates were compared using logrank test for trend. Sham: n = 5, CLP: n = 15. b Blood glucose of the mice was measured at 24 h post-surgery. n = 4 for WT- and KO-Sham. n = 5 for WT- and KO-CLP. c Core body temperature was measured at indicated time points. n = 4 for WT- and KO-Sham. n = 10 for WT- and KO-CLP. Circulating (d) IL-6, (e) sTNFR-2, (f) IL-1β, and (g) total nitrate & nitrite in the mice. n = 410. h Peritoneal lavage fluid was collected at 24 h post-surgery, followed by bacterial culture at 37 °C overnight. Colony-forming units (CFU) of bacteria were counted. n = 4. i H&E staining of the liver and spleen collected in the CLP-operated animals. Scale bar: 50 µm. j Quantification of the necrotic area. n = 5. k, l BMDM or peritoneal macrophage (PM) isolated from Mye-MDM2KO mice or WT controls were incubated with 5 × 106 E. coli for 3 h in the presence or absence of the HIF-1α activator CoCl2, followed by bacterial culture on the agar plate overnight. CFU on the plate were counted and presented. n = 3 for PM. n = 5 for BMDM. Data are displayed as mean ± SEM. N number represents the number of biological replicates. Statistical significance was examined using one-way ANOVA with Tukey post-hoc test. ND Not detected.
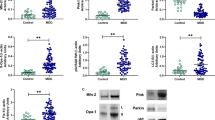
Adipose tissue-resident M1 macrophages and activation of HIF-1α in myeloid cells contribute to systemic inflammation and glucose dysregulation in obesity39,40. To investigate the role of myeloid MDM2 in adipose tissue macrophage inflammation, we fed 10-week-old male Mye-MDM2KO mice and their WT controls with standard chow (STC) or high-fat diet (HFD) to induce obesity. Mye-MDM2KO mice and their WT controls had similar body weight gain, adiposity, calorie intake, oxygen consumption, respiratory exchange ratio, and locomotor activity under STC and HFD feeding conditions (Supplementary Fig. 19). Glucose tolerance test (GTT) revealed that Mye-MDM2KO mice had a better glucose tolerance than WT controls under the HFD but not the STC feeding condition (Fig. 10a and Supplementary Fig. 20a). On the other hand, there was no difference in glucose clearance during the insulin sensitivity test (ITT) between the two groups under STC or HFD feeding conditions (Fig. 10b and Supplementary Fig. 20b). Since the improvement was only observed in Mye-MDM2KO mice fed with HFD, the animals with STC feeding were not included for subsequent analysis unless otherwise specified. Despite similar fasting glucose, Mye-MDM2KO mice had a lower level of insulin and HOMA-IR when compared to WT controls under the HFD condition (Supplementary Table 1). Myeloid-specific MDM2 haplodeletion reduced circulating levels of MCP-1 but did not alter lipid profile and adiponectin level (Fig. 10g and Supplementary Table 1). Histological staining revealed that the adipocyte size, area of the crown-like structure, and fibrosis were reduced in epididymal white adipose tissue (eWAT) of Mye-MDM2KO mice (Fig. 10c–f). MCP-1 protein was decreased in the eWAT of Mye-MDM2KO mice (Fig. 10h). Immunofluorescence staining showed that the number of F4/80 and iNOS double-positive cells were reduced in eWAT of Mye-MDM2KO mice compared to WT controls (Fig. 10i). Furthermore, expression of HIF-1α in adipose tissue-resident macrophage was also significantly decreased in Mye-MDM2KO mice (Fig. 10k). Consistently, QPCR analysis revealed that the inflammatory and pro-fibrotic genes were downregulated in eWAT of Mye-MDM2KO mice (Fig. 10j).
10-week-old male Mye-MDM2KO mice and their WT littermates were fed with high-fat diet (HFD) for 16 weeks. a Glucose tolerance test at 16-week-old. n = 6. b Insulin tolerance test at 18-week-old. n = 6. c H&E staining of epididymal white adipose tissue (eWAT). The arrows indicate immune cell infiltration. d Average adipocyte size and (e) total crown-like structure (CLS) area in the eWAT. n = 9 for (d). n = 11 for (e). f Picro Sirius Red staining of the eWAT. The right panel shows the average fibrotic area. n = 5 for WT. n = 6 for KO. g Circulating level of MCP1. n = 5. h MCP-1 level in the eWAT. n = 6. i Immunofluorescence staining of F4/80 and iNOS in the eWAT. The right panel is the quantification of F4/80+ and iNOS+ cells. n = 5. j QPCR analysis of genes related to inflammation and fibrosis in the eWAT. The gene expressions are normalized with β-actin and expressed as fold change over WT control. n = 6. k Immunofluorescence staining of F4/80 and HIF-1α in the eWAT. The right panel is the quantification of HIF-1α intensity in F4/80+ cells. Scale bar: 100 µm. Representative images were shown. n = 50. Data are displayed as mean ± SEM. Statistical significance was tested using an independent two-tailed student’s t-test. N number represents the number of biological replicates.
Discussion
Inflammatory responses and glycolysis are intertwined in M1 macrophages, but the underlying regulatory mechanism remains poorly defined. Here, we find that MDM2 induces iNOS expression and subsequent NO production via promoting SPSB2 degradation, which increases HIF-1α activity and its dependent glycolysis and inflammatory cytokine production in M1 macrophages (Supplementary Fig. 21). Our findings showed that the inflammatory axis can regulate glycolysis program in M1 macrophage, which are essential for pro-inflammatory cytokine production.
iNOS and its derived NO have been shown to reprogram metabolism and suppress inflammatory responses in M1 macrophages29,30. Completed abrogation of iNOS or treatment with iNOS inhibitor upregulates pro-inflammatory cytokines expression in macrophages, whereas the NO donor exerts the opposite effect. NO reprograms mitochondrial metabolism by inhibiting aconitase 2 (ACO2) and pyruvate dehydrogenase, leading to compensatory upregulation of glutamine-based anaplerosis. Likewise, abrogation of NO production via inhibition of the co-factor of iNOS tetrahydrobiopterin (BH4) increases IL-1β levels, modulates the TCA cycle, and increases its related metabolites including citric, succinate and itaconate41. On the contrary, Somasundaram et al. showed that intermediate to high iNOS and NO levels are required to induce M1 pro-inflammatory phenotypes42. Similarly, under obesity condition, global knockout of iNOS attenuates inflammatory cytokine expression and HIF-1α activation in adipose tissue resident-M1 macrophages and adipose tissue fibrosis43, although another study showed no effect of myeloid iNOS deletion on insulin resistance and adipose tissue function44. Herein, we found that a modest reduction of iNOS and NO by silencing iNOS or MDM2 impairs M1 macrophage polarization and glycolysis. The discrepancy in the effect of NO on M1 macrophage responses between our findings and the previous studies might be due to different NO levels in the experimental conditions ( ~60–70% reduction in our study vs. 100% reduction in the earlier studies). By replenishing NO to a level similar to that in control BMDM and THP-1 macrophages using NO donor, the inhibitory effects of MDM2 silencing on inflammatory and glycolytic responses in M1 macrophages could be completely restored.
While iNOS expression can be controlled at multiple levels45, we showed that MDM2 increases iNOS expression mainly via post-translationally by promoting ubiquitination and subsequent degradation of SPSB2. SPSB2 is a negative regulator of iNOS and NO production in macrophages33. SPSB2 interacts with iNOS via the SPRY domain and recruits E3 ubiquitin ligase complex (including Cul5) that target iNOS for proteasomal degradation33. Previous study has shown that SPSB2 mRNA levels decrease by ~60~ and 90% upon LPS + IFNγ stimulation for 2 and 4 h, respectively33. Here we show that LPS + IFNγ rapidly triggers the interaction between MDM2 and SPSB2 within 30 min. This interaction leads to the MDM2-mediated ubiquitination and degradation of SPSB2, resulting in increase in iNOS protein expression. The swift post-translational modification of SPSB2 and iNOS by MDM2 enable macrophages to instantly polarize into M1 phenotype to fight against the pathogens.
HIF-1α is a master regulator of glycolysis and inflammation in M1 macrophages4. The mice with myeloid-specific ablation of HIF-1α are resistant to LPS-induced cytokine and NO production, hypothermia, and lethality46,47 and dietary-induced adipose tissue macrophage inflammation and systemic insulin resisance39, but are prone to bacterial infection due to reduced bactericidal capacity and NO production48. In accordance with these studies, we observed that the haplodeletion of MDM2 in myeloid cells prevents obesity-induced adipose tissue inflammation and glucose intolerance, accompanied by reduced HIF-1α expression in adipose tissue-resident macrophages. In addition, myeloid deletion of MDM2 led to bacterial burden and higher mortality in the CLP model but alleviated LPS-induced inflammatory response, hypothermia, and lethality. The changes were associated with diminished HIF-1α activation in macrophages. However, the direct effect of MDM2 on the SPSB2-iNOS axis as well as its interplay with HIF-1α activation, in in vivo, has yet to be elucidated. Conversely, we have demonstrated that activation of HIF-1α by chemical compounds restored bactericidal ability and M1 macrophage polarization in macrophages with MDM2 inactivation. Taken in conjunction, we believe that myeloid MDM2 controls obese-induced adipose tissue inflammation and septic response, at least in part, via the HIF-1α pathway.
Apart from the transcriptional control, HIF-1α activity can be regulated at the post-translational level, including NO-dependent s-nitrosylation and PHDs-dependent hydroxylation and succinylation6,27,28,49. In our study, MDM2 silencing reduced s-nitrosylation but not hydroxylation of HIF-1α. In addition, replenishment of NO but not succinate fully restored activity and s-nitrosylation of HIF-1α in M1 macrophages with MDM2 silencing. NO s-nitrosylates HIF-1α at cysteine 533 in the oxygen-dependent degradation domain (ODD), which blocks the binding of VHL to ODD, thereby preventing degradation of HIF-1α27. NO derived from M1 macrophages stabilizes HIF-1α protein but has no effect on its mRNA expression, which leads to fibrotic in preadipocytes and adipose tissue fibrosis and inflammation in obesity condition43. MDM2 has been shown to negatively or positively control HIF-1α activity via the protein-protein interaction and post-transcriptional level in the cancer cells under hypoxia50,51. Still, we could not detect the MDM2-HIF-1α interaction in macrophages in the presence or absence of LPS + IFNγ. Although MDM2 silencing leads to a dramatic reduction of succinate in M1 macrophages, its contribution to HIF-1α activation might be minimal if compared to those effects from NO. Therefore, we propose that MDM2 indirectly enhances HIF-1α stability and activation mainly via the iNOS-NO pathway.
MDM2 interacts with p53 and mediates proteasomal degradation of p53. We and others demonstrated that MDM2 exerts its regulatory actions on metabolism, and inflammatory responses can be p53-dependent or -independent13,21,36,52. Blocking the interaction between MDM2 and p53 using the chemical compound nutlin-3a has been shown to downregulate M2 macrophage polarization and reduce pro-inflammatory cytokine secretion and iNOS mRNA expression in macrophages treated with LPS16,53. On the other hand, NO, as a stress signal, is known to stabilize and activate p53 via multiple post-translational modification including phosphorylation, ubiquitination and acetylation in different cell types54,55,56. In our study, we found that haplodeletion or siRNA-mediated silencing of MDM2 did not upregulate p53 expression and affect p53 downstream targets such as p21, PUMA, and TIGAR. The unchanged p53 protein expression in the MDM2 downregulated macrophages might be due to the residual MDM2 is sufficient to restrict p53 activation and/or the reduction of NO leads to destabilize p53. Nevertheless, we also showed that pharmacological inhibition of p53 is unable to rescue the suppressed inflammatory and glycolysis responses in MDM2 inactivated macrophages, excluding the MDM2’s effect is p53-dependent.
Consistent with our findings in BMDM, MDM2 downregulation suppresses LPS-induced IL-6 and TNF-α production in RAW264.7 cells12. Besides macrophages, MDM2 acts as a positive regulator of inflammation in multiple cell types, including endothelial cells and mouse embryonic fibroblasts13,14. Although previous studies showed that MDM2 regulates inflammatory responses mainly via the NF-κB pathway13,14, we did not observe any apparent effect of MDM2 inactivation on LPS + IFNγ-induced NF-κB activation in macrophages. The distinct effects on NF-κB activation might be due to different cell types and the percentage of MDM2 reduction.
Emerging evidence links metabolism and immune responses, and here we report that MDM2 controls glycolysis and inflammation through the modulation of the iNOS-NO axis in macrophages. This regulatory action by MDM2 is indispensable for obesity-induced adipose tissue macrophage inflammation and in the two models of sepsis, suggesting that targeting MDM2 for metabolic diseases and sepsis might be feasible. However, a cell-type specific approach is a key to success because inhibition of MDM2 in adipocytes and pancreatic β-cells are diabetogenic52,57. In addition, several MDM2 inhibitors are currently evaluated for their efficacy in cancer therapy in humans, therefore, their effects on metabolism, immunity, and infection should be closely monitored. On the other hand, the regulatory role of MDM2 on the iNOS-HIF-1α axis is not limited to sepsis and metabolic diseases, with their crucial role in a diverse range of pathologies such as cancer and autoimmune diseases.
Methods
Animal studies
All the mice are with a C57BL/6 J genetic background. Animals were housed in the barrier animal facilities of The Hong Kong Polytechnic University (PolyU) and The University of Hong Kong (HKU), with temperature control (22 ± 1 °C), humidity control (60 ± 10%), 12 h light-dark cycle and free access to food (9 kcal% fat, Catalog# 2919, Teklad or 60 kcal% fat, Catalog# D12492, Rodent Diet, Inc) and water unless specified otherwise. Myeloid-specific MDM2 haplodeficiency mice and their wild-type littermates (the so-called Mye-MDM2KO and WT mice) were generated by crossing the homozygous Mdm2flox/flox mice58 with LysMcre mice that express the Cre recombinase under the control of the lysozyme 2 promoter (Catalog# 004781, The Jackson Laboratory). For genotyping, ear biopsy was digested with DirectPCR Lysis Reagent (Catalog# 102-T, Viagen Biotech) supplemented with 0.5 mg/ml proteinase K (Catalog# P6556, Sigma–Aldrich) overnight at 55 °C, followed by an 85 °C incubation for one hour and the subsequent PCR with 2xEs Taq MasterMix (Dye) (Catalog# CW0690L, CWbiotech). Primer sequences for genotyping are indicated in Supplementary Table 4. The animals were then housed separately based on their genotype. Due to the differences in innate immunity caused by biological sex, most of the experiments were performed with male mice only, except for Supplementary Fig. 1859. For GTT and ITT, the mice were fasted for 6 h, followed by an intraperitoneal injection of D-glucose or recombinant insulin (Catalog# 91077 C, Sigma-Aldrich) at the dosage of 2 g/kg and 1 IU/kg, respectively. The blood glucose was then measured with the ACCU-Chek® Performa glucose meter (Roche). For endotoxin-induced sepsis, the mice were fasted for 6 h before intraperitoneally injected with 10 mg/kg LPS from Escherichia coli O111:B4 (Catalog# LPS25, Sigma-Aldrich). To induce polymicrobial sepsis, the cecal ligation and puncture model was adopted. Mice were first anesthetized with ketamine (400 mg/kg, Catalog# 13004, AlfaMedic) and xylazine (40 mg/kg, Catalog# 013006, AlfaMedic), placed into a biosafety cabinet, and sterilized with betadine and alcohol swabs. The peritoneal cavity was opened with a midline incision in the abdominal area, and the fecal matter in the cecum was pushed towards the cecal tip before the cecal ligation using a 4-0 nonabsorbable surgical suture. The ligated cecal tip was then punctured in a single punch-through from the mesenteric to antimesenteric direction. A small amount of fecal content was extruded from the opening to ensure its diffusion into the peritoneum. The abdominal incision was then closed with a 6-0 nonabsorbable surgical suture. For the sham controls, the cecum was taken out from the peritoneum and returned without any ligation or puncture. The core body temperature of the mice was measured with a thermometer equipped with a rectal probe for small animals (Model 4610 Precision Thermometer, Measurement Specialties). The fat mass was measured using a minispec body composition analyzer (Bruker Minispec LF90). After the completion of the experiments, animals were euthanized using an overdose of anesthesia (200 mg/kg Pentobarbitol; Catalog# 013003, AlfaMedic). All animal experimental protocols were approved by the Animal Subjects Ethics Sub-Committee at PolyU (Approval numbers: 16-17/41 and 19-20/55) and the Committee on the Use of Live Animals in Teaching and Research at HKU (Approval number: 3967-16).
Metabolic cage whole-body calorimetry
The Comprehensive Lab Animal Monitoring System (CLAMS) from Columbus Instruments was used for the measurement of whole-body metabolism in mice. The mice were housed in the CLAMS metabolic cage individually with ad libitum access to food and water for 3 days. Oxygen consumption and carbon dioxide production were measured every 10 min, while locomotor activity, food, and water consumption were measured continuously. The first day was used as an adaptive period in which the data were discarded.
Isolation, culture, and treatments of primary macrophages
L929 conditioned medium was prepared by culturing L929 mouse fibroblasts (Catalog# CCL1™, ATCC®) in DMEM (Catalog# 12800082, Gibco) supplemented with 10% fetal bovine serum (FBS, Catalog#10270106, Gibco), and 1% penicillin (100 U/ml) and streptomycin (100 μg/ml) (PS, Catalog# 15140163, Gibco) at confluence for three days. The cell culture supernatant was then passed through a 0.22 µm filter. Bone marrow cells from tibias and femurs of C57BL/6 J mice, Mye-MDM2KO mice, and their WT littermates were cultured with DMEM supplemented with 20% L929 conditioned medium, 10% FBS, and 1% PS for seven days. The differentiated BMDM were then detached using cell dissociation buffer (Catalog# 13151014, Gibco) and cultured in DMEM supplemented with 10% FBS and 1% PS (complete DMEM) for 24 h before subsequent treatments. For thioglycolate-elicited peritoneal macrophages, 3 ml of 4% thioglycolate was intraperitoneally injected into the mice. After 72 h, 5 ml DMEM was injected into the peritoneal cavity using a syringe with 25 G needle, followed by a gentle massage of the peritoneum. The content in the peritoneal cavity was then slowly withdrawn and transferred to a centrifugation tube, followed by centrifugation at 500 × g for 10 min. The cell pellet was then resuspended in DMEM with 10% FBS and seeded onto tissue culture plates. Non-adherent cells were then washed away with phosphate-buffered saline (PBS) after 3 h. For M1 macrophage polarization, macrophages were co-stimulated with 100 ng/ml LPS from Escherichia coli O111:B4 and 100 ng/ml recombinant mouse IFNγ (Catalog# 485-MI, R&D Systems) for 20–24 h. For M2 macrophage polarization, macrophage were treated with 10 ng/ml IL-4 (Catalog# 404-ML, R&D Systems) for 24 h. For replenishment of NO, macrophages were pre-treated with SNAP (250 μM; Catalog# 82250, Cayman Chemical) for 3 h before co-stimulation with LPS and IFNγ. For activation of HIF-1α pathway, 200 µM of CoCl2 (Catalog# 60818, Sigma-Aldrich) was added to the cell culture supernatant during M1 macrophage polarization by LPS and IFNγ co-treatment. Replenishment of succinate was done by pre-treating the macrophages with 1 mM cell-permeable dimethyl-succinate (Catalog# 73605, Sigma-Aldrich) for 3 h before M1 polarization. Pharmacological activation of MDM2 were achieved by treating BMDM or Raw 267.4 cells with nutlin-3a (10 µM; Catalog# HY-10029, MedChemExpress) for 3 h before LPS and IFNγ stimulation. To inhibit p53, PFTα (10 µM; Catalog# HY-15484, MedChemExpress) was incubated with the macrophages for 3 h before LPS and IFNγ treatment.
Culture and differentiation of THP-1 macrophages
THP-1 human monocyte-like cell line (Catalog# TIB202™, ATCC®) were cultured in complete RPMI 1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin. For differentiation into macrophages, the cells were incubated with 10 ng/ml phorbol 12-myristate 13-acetate (PMA, Catalog# 10008014, Cayman Chemical) for 48 h. The cells were then washed with PBS and incubated in complete RPMI 1640 medium for another 24 h before further treatments. For polarization into M1 macrophages, THP-1 macrophages were treated with 100 ng/ml LPS and 100 ng/ml IFNγ for 24 h.
Generation of SPSB2 knockout THP-1 cells by CRISPR
THP-1 monocytes were transfected with the all-in-one pSpCas9 BB-2A-GFP (PX458) plasmid expressing Cas9, GFP and the guide RNA (gRNA) targeting human SPSB2 (guide sequence 1: 5’ GCTGCAGACTGACCACTACG 3’ and guide sequence-2: 5’ AGTCTTTGGGGTTCCAACCG 3’) using the Neon™ Transfection system (ThermoFisher). The transfected cells were then maintained in a DMEM supplemented with 10% FBS for 24 h, followed by sorting and collection of green fluorescence (GFP) positive cells using the BD FACSAria™ Cell Sorter (ULS, PolyU). Following expansion, the successful knockout of SPSB2 was confirmed through immunoblotting and DNA sequencing. Control THP-1 monocytes were transfected with the SpCas9 BB-2A-GFP (PX458) plasmid without gRNA, followed by the same sorting and culturing procedure as above.
Gene silencing in BMDM and THP-1 macrophages by siRNA
siRNA against MDM2, iNOS, SPSB2, or scramble control (GenePharma Inc.) were transfected into BMDM and THP-1 macrophages using DharmaFECT 3 transfection reagent (Catalog# T-2003-03, Dharmacon) according to the manufacturer’s instruction at a final concentration of 100 nM and described in our previous study60. After a 24-hour incubation with the siRNA complex, the cells were stimulated with the M1 polarizing agents as indicated above. The sequences of the siRNA are listed in Supplementary Table 2.
Generation of plasmids expressing SPSB2 mutants
The pcDNA3.1-SPSB2-3xFLAG-C plasmid with the wild-type sequence of SPSB2 was used. Lysine residues of potential ubiquitination sites on SPSB2 including K51, K60, K81, K139, K145, K195, and K267 were mutated and replaced by arginine residues to generate plasmids expressing the SPSB2-K51R, -K60R, -K81R, -K139R, -K145R, -K195R and -K267R mutants. Similarly, putative MDM2 binding motifs on SPSB2 (L19/Y20/L23) and (L199/Y200/V203) were mutated and replaced by alanine residues to generate plasmids expressing the SPSB2 (L19A/Y20A/L23A) and (L199A/Y200A/V203A) mutants (termed Triple-1 and Triple-2, respectively). All the above vectors were subjected to sequencing to confirm the mutation sites. The vector cloning and mutagenesis was done by GuangZhou DaHong Biosciences.
Metabolomic analysis of bone marrow-derived macrophages
Mouse BMDM transfected with siMdm2 or siScramble were stimulated with 100 ng/ml LPS and IFNγ for 20 h, followed by washing with cold PBS and snap frozen. The samples were thawed on ice, and resuspended with 100 µl of ultrapure water. 50 µl of the cell suspension was then mixed with 200 µl of cold methanol by vortexing at 2500 rpm for 2 min. The mixtures were then immediately frozen in liquid nitrogen for 5 min, followed by thawing again on ice for 5 min and 2 min of mixing with vortex. After freezing and thawing for three times, the samples were centrifuged at 13800 × g for 10 min at 4 °C. 200 µl of the supernatant was then transferred to the new tube, cooled to −20 °C for 30 min, and centrifuged again at 4 °C at 13800 × g for 10 min. 180 µl of the supernatant was passed through a protein precipitation plate and stored at −20 °C before further analysis. The remaining 50 µl of cell suspension was frozen in liquid nitrogen and thawed for 3 times, centrifuged at 13800 × g for 10 min to collect the supernatant for protein quantification with bicinchoninic acid protein assay.
For the Ultra Performance Liquid Chromatography and Tandem Mass Spectrometry (UPLC-MS/MS), Waters ACQUITY H-ClassD and QTRAP® 6500+ were used. An ACQUITY UPLC BEH Amide column (1.7 µm, 100 mm×2.1 mmi.d.) was used for the UPLC and the separation was performed using a mobile phase consisting of ultrapure water with 10 mM ammonium acetate and 0.3% ammonium hydroxide as mobile phase A and 90 acetonitrile: 10 water as mobile phase B. The flow rate was set to 0.40 mL/min with column temperature at 40 °C, a sample volume of 2 μl and a total run time of 15 min per sample. The mobile phase gradient started with 5% solvent A until 1.2 min, then increased gradually to 30% solvent A until 8 min. The gradient then further increased to 50% solvent A at 9 min and was maintained for 2 min, then back to 5% solvent A for 4 min. For MS/MS, the electrospray ionization (ESI) temperature was 550 °C, mass spectrometry voltages were 5500 V and −4500 V in positive and negative ionization mode respectively and curtain gas at 35 psi. In Q-Trap6500 + , each ion pair is scanned and detected according to the optimized Declustering Potential (DP) and Collision Energy (CE). Metabolomics analysis was performed by LC Bio (Hang Zhou, China).
Real-time quantitative PCR
Total RNA was extracted from the cells or the tissues using RNAiso Plus (Catalog# 9109, Takara) and quantified with NanoDrop™ 2000 Spectrophotometers. One µg of the total RNA was reverse transcribed into cDNA using the GoScript™ Reverse Transcription Mix (Catalog# A2801, Promega). Real-time quantitative PCR with ViiA 7 Real-Time PCR System (Applied Biosystems) using QuantiNova SYBR® Green (Catalog# 208056, QIAGEN) with the primers targeting the genes of interest. Details of the primer sequences are listed in Supplementary Table 4.
RNA-sequencing analysis
Total RNA was extracted using the RNeasy mini kit (Catalog# 74104, Qiagen) following manufacturer’s instructions. After passing quality control, the libraries were generated and sequenced with the NovaSeq PE150 according to the protocol from the company Novogene. Briefly, messenger RNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first strand cDNA was synthesized using random hexamer primers followed by the second strand cDNA synthesis. The library was ready after end repair, A-tailing, adapter ligation, size selection, amplification, and purification. The library was checked with Qubit and real-time PCR for quantification and bioanalyzer for size distribution detection. Quantified libraries were pooled and sequenced on Illumina platforms.
RNA-seq reads were preceded by removing adapters using cutadapt (v4.5) followed by quality assessment with FastQC (v0.12.1). Raw reads were mapped against the GRCm39 mouse genome assembly using STAR aligner (v2.7.10b)61 with the default options. The number of reads mapped to each of genes was counted by using featureCounts of subread package (v 2.0.3)62, with the option: -s 0 -p --countReadPairs -t exon -g gene_id. To visualize expression levels by principal component analysis (PCA), count data were first normalized using VST method from DESeq2 R package (v1.36.0)63. Differential expression analysis was done using DESeq function from DESeq2 with default settings. The significance threshold was set to FDR < 5%, |log2FC | <2 and base mean at least 20. Pathway enrichment was performed using QIAGEN IPA (QIAGEN Inc., https://digitalinsights.qiagen.com/IPA). The RNA-sequencing data is publicly available at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE276736.
Immunoblotting and immunoassay
Tissues and cells were lysed and homogenized in a RIPA lysis buffer (150 mM NaCl (Catalog#, S5886, Sigma Aldrich), 50 mM Tris HCl [pH 7.4] (Catalog# 5941, Sigma-Aldrich), 2 mM EDTA (Catalog# 03677, Sigma-Aldrich), 0.1% SDS (Catalog# 71725, Sigma-Aldrich), 1% Nonidet P-40 (Catalog# 56741, Sigma-Aldrich)) supplemented with protease inhibitors (Catalog# HY-K0010, MedChemExpress) and phosphatase inhibitors cocktail (Catalog# B15001, Bimake). Proteins were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membrane (Catalog# 1620177, BIO-RAD). The membrane was then blocked with 10% nonfat milk and probed with the primary antibodies overnight at 4 °C and the corresponding secondary antibodies conjugated with horseradish peroxidase (Catalog #7074 and #7076, Cell Signaling Technology) for one hour. The protein bands were visualized on an X-ray film by enhanced chemiluminescence reagents (Catalog #1705061 BIO-RAD) and quantified using ImageJ 1.54 f. Details of the antibodies used in this study are listed in the Supplementary Table 3.
Biochemical and immunoassays
Total nitrate and nitrite in the culture medium and serum are measured with Nitrate/Nitrite Colorimetric Assay Kit (Catalog# 780001, Cayman Chemical). Insulin, adiponectin, MCP-1, TNF-α, IL-1β, IL-6 and sTNFR-2 in cell culture supernatant and serum were quantified using High-sensitive Mouse Insulin ELISA Kit (Catalog# 32270, ImmunoDiagnostics), Mouse Adiponectin ELISA Kit (Catalog# 32010, ImmunoDiagnostics), Mouse CCL2/JE/MCP-1 DuoSet ELISA (Catalog# DY479, R&D Systems), Mouse TNF-alpha DuoSet ELISA (Catalog# DY410, R&D Systems), Mouse IL-1 beta/IL-1F2 DuoSet ELISA (Catalog# DY401, R&D Systems), Mouse IL-6 Quantikine ELISA Kit (Catalog# M6000B, R&D Systems) and Mouse sTNF RII/TNFRSF1B DuoSet ELISA (Catalog# DY426, R&D Systems), respectively. Free fatty acids, triglycerides and cholesterol in mouse serum were quantified using Free Fatty Acids, Half Micro Test (Catalog# 11383175001, Roche), Triglyceride LiquiColor® (Mono), (Catalog# 2200-430, EKF Diagnostics) and Cholesterol, LDL, Direct LiquiColor® (Catalog# 0710-080, EKF Diagnostics). All the assays were performed according to the manufacturer’s instructions.
Tissue processing and histological analysis
Tissues were fixed in 10% neutral buffered formalin (Catalog# 59201, Thermo Scientific) overnight and processed with Excelsior™ AS Tissue Processor (Thermo Scientific). The tissues were then sectioned into 5 µm sections using a microtome, followed by staining with hematoxylin and eosin (H&E) and analyzed with ImageJ. Tissue sections for immunofluorescence staining were subjected to antigen retrieval by boiling in sodium citric buffer (0.1 mol/L sodium citrate (Catalog#, S4641, Sigma-Aldrich), 0.1% Tween 20 (Catalog#, P9416, Sigma-Aldrich), pH 6.0) for 20 min and blocked by 5% FBS in PBS for one hour at room temperature. The slides were probed with primary antibodies overnight at four °C, followed by a one-hour incubation with fluorescent dye-conjugated anti-rabbit, anti-mouse, or anti-rat IgG secondary antibodies (Catalog# A-11008, A-11001 and A-11007, ThermoFisher) in the dark and mounted with ProLong™ Glass Antifade Mountant (Catalog# P36980, Invitrogen) sequentially. Images were captured with a fluorescent microscope and analyzed with ImageJ for the fluorescence intensity of iNOS and HIF-1α using fifty to sixty F4/80+ cells as the regions of interest. Sirius Red staining was performed using the Picro Sirius Red Stain Kit (Catalog# ab150681, Abcam). In brief, the adipose tissue sections were deparaffinized in xylene and rehydrated with distilled water, followed by staining with Picro Sirius Red solution for 1 h. The slides were then washed with acetic acid and ethanol, followed by dehydration and mounting. Adipocyte size was quantified using Adiposoft (https://imagej.net/plugins/adiposoft)64. Quantification of crown-like structures, fibrotic area, and sepsis-induced lesion were performed manually by adjusting the color threshold in ImageJ. Details of the antibodies used are listed in Supplementary Table 2.
Quantification of necrotic area in liver and spleen
H&E-stained tissue sections were used for the quantification of necrotic area in the liver and spleen of CLP-induced septic mouse model. Three levels of the tissue which are at least 50 µm apart were used in each quantification and the average was used. The acellular necrotic region (which appeared pale pink) which featured pale cytoplasm, nuclear dissolution, cell/nuclear swelling and adjacent focus of inflammation were identified and manually traced in ImageJ, according to the guideline in previous study65. 3-4 layers of the same tissue that are at least 50 µm apart from each other were quantified. The researchers for data acquisition and analysis were blinded to the grouping of animal.
Seahorse real-time cell metabolic analysis
Real-time cell metabolic analysis for BMDM was performed using the Seahorse XFe24 analyzer (Agilent). In brief, 5 × 104 BMDM were seeded into each well of the XF24-well plate, followed by M1 activation for 24 h. The Seahorse sensor cartridge was placed into the calibrant and incubated overnight in a non-CO2 incubator at 37 °C. The cells were washed and cultured in unbuffered DMEM containing 25 mM glucose, 2 mM L-glutamine and 1 mM sodium pyruvate right before the experiment. The following compounds, including 1 µM oligomycin (Catalog# 11341, Cayman Chemical), 1.5 µM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP, Catalog# 15218, Cayman Chemical), 1 µM rotenone (Catalog# 13995, Cayman Chemical), 1 µM antimycin A (Catalog# A8674, Sigma-Aldrich) and 50 mM 2-deoxyglucose (Catalog# 14325, Cayman Chemical) were added to the cells sequentially depending on the experiments. The extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were then measured. Mitochondrial proton efflux rate (MitoPER) was determined by measuring OCR before and after rotenone/antimycin A injection and subtracted from the total proton efflux rate to generate the glycolytic proton efflux rate (GlycoPER).
Bactericidal ability assay
Macrophages isolated from mice were seeded onto 6-well plates in DMEM supplemented with 10% FBS at the density of 1 x 106 cells per well. 1 x 106 E.coli DH5α was added to each well, followed by 1-minute centrifugation at 100 × g and allowed to incubate for 3 h at 37 °C. The cells and cell culture medium were then transferred to a sterile centrifugation tube for 10-minute centrifugation at 18000 × g. The pellet was lysed with ultrapure water and the lysate was serially diluted (1:10 to 1:10000). 100 µl of the diluted lysate was then cultured on LB agar plates (Catalog# L1500, USBiological) without antibiotics overnight, and the colony-forming units were counted the next day.
Measurement of HIF-1α transcriptional activity
HIF-1α transcriptional activity was measured using the HIF-1α Transcription Factor Assay Kit (Catalog# ab133104. Abcam). The nuclear fraction was extracted according to the manufacturer’s instructions first by incubating the cells with a hypotonic solution (20 mM HEPES, pH 7.5, containing 5 mM NaF, 100 μM Na2MoO4, and 1 mM EDTA (Catalog# 54457, 201154, 243655. 331058 and 03677, Sigma-Aldrich) followed by cell rupture with Nonidet P-40 (1%, Catalog# 56741, Sigma-Aldrich). The samples were then centrifuged to remove the cytosolic fraction in the supernatant. The pellet was then resuspended in the nuclear extraction buffer and briefly vortexed and gently rocked at 4 °C. After centrifugation at 14000 × g at 4 °C for 10 min, the nuclear fraction in the supernatant was then transferred to a new tube and stored at −80 °C until use.
S-nitrosylation of HIF-1α
Detection of s-nitrosylation was performed using the commercially available Pierce™ S-Nitrosylation Western Blot Kit (Catalog# 90105, Thermo Scientific™) following the manufacturer’s instructions. Briefly, the nuclear fraction from BMDM was first lysed with HENS buffer supplied with the kit. Protein concentration was determined with Pierce™ BCA Protein Assay Kit (Catalog# 23225, Thermo Scientific™), and the amount of protein was adjusted to 100 µg per reaction. The unmodified cysteine residues on proteins were then blocked using 20 mM methyl methanethiosulfonate (MMTS), followed by protein precipitation with acetone at −20 °C to remove excess MMTS. After resuspending the precipitation protein with HENS buffer, s-nitrosylated cysteine was reduced with sodium ascorbate. The reduced cysteine residues were then irreversibly labeled with the iodoTMTzero reagent for 2 h. TMT-labeled proteins were immunoprecipitated using anti-HIF-1α antibody at 4 °C overnight, followed by incubation with protein A/G agarose (Catalog# sc-2001 and sc-2002, Santa Cruz) at 4 °C for 60 min. The immunocomplex was washed with cold lysis buffer for three times and eluted with SDS sample loading dye by heating at 99 °C for 5 min before the immunoblotting using anti-TMT antibody provided with the S-Nitrosylation Western Blot kit.
Ubiquitination assays
HEK293T (Catalog# CRL-3216™, ATCC®) cells were transfected with plasmids encoding Mye-tagged human MDM2, FLAG-tagged human SPSB2 and HA-tagged human ubiquitin using polyethylenimine. Plasmid expressing the green fluorescent protein (GFP) was used as a control. Proteasome inhibitor MG132 (25 μM; Catalog# S2619, Selleckchem) or DMSO as vehicle control were added to the cells one hour before cell harvest. After 48 h, the transfected cells were lysed with a lysis buffer containing 150 mM NaCl, 50 mM Tris-HCl, 2 mM EDTA, 1% Nonidet P-40, 1% SDS, and protease inhibitor cocktail with pH 7.5. The cells were then transferred to a 1.5 ml tube and boiled on a hotplate for 10 min. The boiled cell lysate was diluted with a buffer containing 150 mM NaCl, 50 mM Tris-HCl, 2 mM EDTA, 1% Triton X-100 (Catalog# T8787, Sigma-Aldrich), and protease inhibitor (Catalog# HY-K0010, MedChemExpress) at pH 7.5. The diluted samples were centrifuged at 20,000 x g for 30 min and the supernatant was transferred to a new tube. Cell lysate was incubated with an anti-SPSB2 antibody at 4 °C overnight on a tube rotator, and incubated with protein A/G agarose at 4 °C for 60 min. The immunocomplex was washed with cold lysis buffer for three times and eluted with SDS sample loading dye by heating at 99 °C for 5 min, followed by immunoblotting analysis.
In vitro SPSB2 ubiquitination assays
Recombinant human HA-Ub protein (Catalog# U-110-01M, R&D Systems), recombinant human GST-MDM2 protein (Catalog# E3-202, R&D Systems), recombinant human SPSB2 protein (Catalog# ab134592, Abcam) were used for in vitro SPSB2 ubiquitination assay in combination with E2-Ubiquitin Conjugation Kit (Catalog# ab139472, Abcam) containing a range of E2 enzymes and E1 following the manufacturer’s instructions. Briefly, the assay buffer, recombinant E1 and E2 from the E2-Ubiquitin Conjugation Kit were mixed with recombinant human HA-Ub protein (2.5 µM), recombinant human GST-MDM2 (100 nM) and recombinant human SPSB2 (1 µM) as the substrate, E3 ligase of interest and the target protein for ubiquitination, respectively. The mixture was then incubated at 37 °C for 60 min and the reaction was quenched using the 2X non-reducing gel loading buffer as provided. The reaction mixture was then boiled at 100 °C for 5 min, followed by immunoprecipitation of SPSB2 using anti‐SPSB2 antibody at 4 °C. The immunocomplex was subjected to immunoblotting analysis using anti-SPSB2 antibody and anti‐HA antibody.
Flow cytometry analysis of M1 macrophages
BMDM from C57BL/6 mice were subjected to siRNA-mediated silencing of Mdm2, followed by M1 macrophage polarization with LPS and IFNγ for 24 h. The cells were then collected and stained with a PE anti-F4/80 antibody (Catalog# 123110, BioLegend) and PerCP-Cy5.5 anti-CD11c antibody (Catalog# 560584, BD Biosciences) for 30 min on ice in the dark. The negative controls are the respective isotype controls (Catalog# 400508 and 560554, BioLegend and BD Biosciences). The cells were then washed and subjected to flow cytometry analysis on a BD FACSCelesta™ cell analyzer. The live cells were gated based on their forward scatter and side scatter, followed by the F4/80 and CD11c expression.
Statistical analysis
Data were analyzed with GraphPad Prism 6.0 or SPSS. Data were presented as mean ± SEM. Statistical significance between two group comparisons was determined using an unpaired Student’s t-test or nonparametric Mann-Whitney U test. Statistical significance between multiple group comparisons was determined using one-way ANOVA with Tukey post-hoc test or nonparametric Kruskal–Wallis test with Dunn’s test. Equal variance and normality were determined by Levene’s test and D’Agostino-Pearson omnibus normality test, respectively. A p-value less than 0.05 is considered statistically significant. In vitro experiments were repeated two or three times independently with similar results. Animal experiments were performed one time with each animal as a biologically independent sample. The survival rate between groups was compared using the log-rank test for trend.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The RNA-sequencing data is publicly available at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE276736. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.
References
Lian, Q. et al. Differential effects of macrophage subtypes on SARS-CoV-2 infection in a human pluripotent stem cell-derived model. Nat. Commun. 13, 2028 (2022).
Benoit, M., Desnues, B. & Mege, J. L. Macrophage polarization in bacterial infections. J. Immunol. 181, 3733–3739 (2008).
Shapouri-Moghaddam, A. et al. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 233, 6425–6440 (2018).
McGettrick, A. F. & O’Neill, L. A. J. The role of HIF in immunity and inflammation. Cell Metab. 32, 524–536 (2020).
Wang, T. et al. HIF1alpha-induced glycolysis metabolism is essential to the activation of inflammatory macrophages. Mediators Inflamm. 2017, 9029327 (2017).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496, 238–242 (2013).
Palsson-McDermott, E. M. et al. Pyruvate kinase M2 regulates Hif-1alpha activity and IL-1beta induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 21, 65–80 (2015).
Williams, N. C. et al. Signaling metabolite L-2-hydroxyglutarate activates the transcription factor HIF-1alpha in lipopolysaccharide-activated macrophages. J. Biol. Chem. 298, 101501 (2022).
Feng, T. et al. Adipocyte-derived lactate is a signalling metabolite that potentiates adipose macrophage inflammation via targeting PHD2. Nat. Commun. 13, 5208 (2022).
Lehman, J. A. et al. Serdemetan antagonizes the Mdm2-HIF1alpha axis leading to decreased levels of glycolytic enzymes. PloS one 8, e74741 (2013).
Chen, D., Li, M., Luo, J. & Gu, W. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J. Biol. Chem. 278, 13595–13598 (2003).
Han, N. R., Moon, P. D., Kim, H. M. & Jeong, H. J. TSLP exacerbates septic inflammation via murine double minute 2 (MDM2) signaling pathway. J. Clin. Med. 8, 1350 (2019).
Mulay, S. R., Thomasova, D., Ryu, M. & Anders, H. J. MDM2 (murine double minute-2) links inflammation and tubular cell healing during acute kidney injury in mice. Kidney Int 81, 1199–1211 (2012).
Zeng, Y., Xu, J., Hua, Y. Q., Peng, Y. & Xu, X. L. MDM2 contributes to oxidized low-density lipoprotein-induced inflammation through modulation of mitochondrial damage in endothelial cells. Atherosclerosis 305, 1–9 (2020).
Zhou, J. et al. The ubiquitin ligase MDM2 sustains STAT5 stability to control T cell-mediated antitumor immunity. Nat. Immunol. 22, 460–470 (2021).
Li, L. et al. A unique role for p53 in the regulation of M2 macrophage polarization. Cell Death Differ. 22, 1081–1093 (2015).
Odkhuu, E. et al. Lipopolysaccharide downregulates the expression of p53 through activation of MDM2 and enhances activation of nuclear factor-kappa B. Immunobiology 220, 136–141 (2015).
Genin, M., Clement, F., Fattaccioli, A., Raes, M. & Michiels, C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 15, 577 (2015).
Abbas, H. A. et al. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell 7, 606–617 (2010).
Lacroix, M., Riscal, R., Arena, G., Linares, L. K. & Le Cam, L. Metabolic functions of the tumor suppressor p53: Implications in normal physiology, metabolic disorders, and cancer. Mol. Metab. 33, 2–22 (2020).
Arena, G. et al. Mitochondrial MDM2 regulates respiratory complex I activity independently of p53. Mol. Cell 69, 594–609.e598 (2018).
Gu, L., Findley, H. W. & Zhou, M. MDM2 induces NF-kappaB/p65 expression transcriptionally through Sp1-binding sites: a novel, p53-independent role of MDM2 in doxorubicin resistance in acute lymphoblastic leukemia. Blood 99, 3367–3375 (2002).
Thomasova, D., Mulay, S. R., Bruns, H. & Anders, H. J. p53-independent roles of MDM2 in NF-kappaB signaling: implications for cancer therapy, wound healing, and autoimmune diseases. Neoplasia 14, 1097–1101 (2012).
Haupt, Y., Maya, R., Kazaz, A. & Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 (1997).
Komarov, P. G. et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285, 1733–1737 (1999).
Piret, J. P., Mottet, D., Raes, M. & Michiels, C. CoCl2, a chemical inducer of hypoxia-inducible factor-1, and hypoxia reduce apoptotic cell death in hepatoma cell line HepG2. Ann. N. Y Acad. Sci. 973, 443–447 (2002).
Li, F. et al. Regulation of HIF-1alpha stability through S-nitrosylation. Mol. Cell 26, 63–74 (2007).
Sanhueza, C. et al. Caveolin-1-mediated tumor suppression is linked to reduced HIF1α S-nitrosylation and transcriptional activity in hypoxia. Cancers (Basel) 12, 2349 (2020).
Lu, G. et al. Myeloid cell-derived inducible nitric oxide synthase suppresses M1 macrophage polarization. Nat. Commun. 6, 6676 (2015).
Palmieri, E. M. et al. Nitric oxide orchestrates metabolic rewiring in M1 macrophages by targeting aconitase 2 and pyruvate dehydrogenase. Nat. Commun. 11, 698 (2020).
Matsumoto, K. et al. The ECS(SPSB) E3 ubiquitin ligase is the master regulator of the lifetime of inducible nitric-oxide synthase. Biochem Biophys. Res Commun. 409, 46–51 (2011).
Chen, L. et al. CHIP facilitates ubiquitination of inducible nitric oxide synthase and promotes its proteasomal degradation. Cell Immunol. 258, 38–43 (2009).
Kuang, Z. et al. The SPRY domain-containing SOCS box protein SPSB2 targets iNOS for proteasomal degradation. J. Cell Biol. 190, 129–141 (2010).
Nishiya, T. et al. Regulation of inducible nitric-oxide synthase by the SPRY domain- and SOCS box-containing proteins. J. Biol. Chem. 286, 9009–9019 (2011).
Nicholson, J. et al. A systems-wide mass spectrometric-based linear motif screen to identify dominant in-vivo interacting proteins for the ubiquitin ligase MDM2. Cell Signal 26, 1243–1257 (2014).
Lin, H. et al. Hepatic MDM2 causes metabolic-associated fatty liver disease by blocking triglyceride-VLDL secretion via ApoB degradation. Adv. Sci. (Weinh.) 9, e2200742 (2022).
Tisoncik, J. R. et al. Into the eye of the cytokine storm. Microbiol Mol. Biol. Rev. 76, 16–32 (2012).
Dejager, L., Pinheiro, I., Dejonckheere, E. & Libert, C. Cecal ligation and puncture: the gold standard model for polymicrobial sepsis? Trends Microbiol 19, 198–208 (2011).
Takikawa, A. et al. HIF-1alpha in myeloid cells promotes adipose tissue remodeling toward insulin resistance. Diabetes 65, 3649–3659 (2016).
Russo, L. & Lumeng, C. N. Properties and functions of adipose tissue macrophages in obesity. Immunology 155, 407–417 (2018).
Bailey, J. D. et al. Nitric oxide modulates metabolic remodeling in inflammatory macrophages through TCA cycle regulation and itaconate accumulation. Cell Rep. 28, 218–230 e217 (2019).
Somasundaram, V. et al. Inducible nitric oxide synthase-derived extracellular nitric oxide flux regulates proinflammatory responses at the single cell level. Redox Biol. 28, 101354 (2020).
Jang, J. E. et al. Nitric oxide produced by macrophages inhibits adipocyte differentiation and promotes profibrogenic responses in preadipocytes to induce adipose tissue fibrosis. Diabetes 65, 2516–2528 (2016).
Lu, M. et al. Inducible nitric oxide synthase deficiency in myeloid cells does not prevent diet-induced insulin resistance. Mol. Endocrinol. 24, 1413–1422 (2010).
Pautz, A. et al. Regulation of the expression of inducible nitric oxide synthase. Nitric Oxide 23, 75–93 (2010).
Peyssonnaux, C. et al. Cutting edge: essential role of hypoxia inducible factor-1alpha in development of lipopolysaccharide-induced sepsis. J. Immunol. 178, 7516–7519 (2007).
Takeda, N. et al. Differential activation and antagonistic function of HIF-{alpha} isoforms in macrophages are essential for NO homeostasis. Genes Dev. 24, 491–501 (2010).
Peyssonnaux, C. et al. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J. Clin. Invest 115, 1806–1815 (2005).
Watts, E. R. & Walmsley, S. R. Inflammation and Hypoxia: HIF and PHD Isoform Selectivity. Trends Mol. Med. 25, 33–46 (2019).
Joshi, S., Singh, A. R. & Durden, D. L. MDM2 regulates hypoxic hypoxia-inducible factor 1alpha stability in an E3 ligase, proteasome, and PTEN-phosphatidylinositol 3-kinase-AKT-dependent manner. J. Biol. Chem. 289, 22785–22797 (2014).
Nieminen, A. L., Qanungo, S., Schneider, E. A., Jiang, B. H. & Agani, F. H. Mdm2 and HIF-1alpha interaction in tumor cells during hypoxia. J. Cell Physiol. 204, 364–369 (2005).
Li, X. et al. The MDM2-p53-pyruvate carboxylase signalling axis couples mitochondrial metabolism to glucose-stimulated insulin secretion in pancreatic beta-cells. Nat. Commun. 7, 11740 (2016).
Liu, G., Park, Y. J., Tsuruta, Y., Lorne, E. & Abraham, E. p53 attenuates lipopolysaccharide-induced NF-kappaB activation and acute lung injury. J. Immunol. 182, 5063–5071 (2009).
Hofseth, L. J. et al. Nitric oxide-induced cellular stress and p53 activation in chronic inflammation. Proc. Natl Acad. Sci. USA 100, 143–148 (2003).
Wang, X., Michael, D., de Murcia, G. & Oren, M. p53 activation by nitric oxide involves down-regulation of Mdm2. J. Biol. Chem. 277, 15697–15702 (2002).
Schneiderhan, N., Budde, A., Zhang, Y. & Brune, B. Nitric oxide induces phosphorylation of p53 and impairs nuclear export. Oncogene 22, 2857–2868 (2003).
Liu, Z. et al. The dysfunctional MDM2-p53 axis in adipocytes contributes to aging-related metabolic complications by induction of lipodystrophy. Diabetes 67, 2397–2409 (2018).
Grier, J. D., Yan, W. & Lozano, G. Conditional allele of mdm2 which encodes a p53 inhibitor. Genesis 32, 145–147 (2002).
Jaillon, S., Berthenet, K. & Garlanda, C. Sexual dimorphism in innate immunity. Clin. Rev. Allergy Immunol. 56, 308–321 (2019).
Wu, K. K. L. et al. The APPL1-Rab5 axis restricts NLRP3 inflammasome activation through early endosomal-dependent mitophagy in macrophages. Nat. Commun. 12, 6637 (2021).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Galarraga, M. et al. Adiposoft: automated software for the analysis of white adipose tissue cellularity in histological sections. J. Lipid Res. 53, 2791–2796 (2012).
Elmore, S. A. et al. Recommendations from the INHAND apoptosis/necrosis working group. Toxicol. Pathol. 44, 173–188 (2016).
Acknowledgements
The authors thank Prof. Guillermina Lozano (The University of Texas M.D. Anderson Cancer Center) for providing Mdm2floxed/floxed mice. This study was mainly supported by Hong Kong Research Grant Council (RGC) General Research Fund (17100717), partially by National Natural Science Foundation of China (92357305 and 81970675) and RGC-Area of Excellence (AoE/M/707-18) and PolyU internal funding (P0040979 and P0036848), and Shenzhen Municipal Science and Technology Innovation Commission Basic Research General Programme (JCYJ20210324130202006).
Author information
Authors and Affiliations
Contributions
K.K.L.W. performed most of the experiments and drafted the manuscript. X.X., M.W., K.L., and T.Z., generated some of the data. M.H. and G.H.Y.L. conducted the analysis of RNAseq data. X.L, Q.L. H.X.H., and H.P. advised the study design and experimental procedures and provided important tools. A.X. provided myeloid-MDM2KO mice and their WT littermates and reagents. K.K.Y.C. initiated and supervised the study, provided the resources, acquired funding and wrote and finalized the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Elisabetta Liverani, Matthew Woolard and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, K.Kl., Xu, X., Wu, M. et al. MDM2 induces pro-inflammatory and glycolytic responses in M1 macrophages by integrating iNOS-nitric oxide and HIF-1α pathways in mice. Nat Commun 15, 8624 (2024). https://doi.org/10.1038/s41467-024-53006-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53006-w
This article is cited by
-
Exercise mitigates IL-27-induced muscle atrophy following myocardial infarction through anti-inflammatory mechanisms
Journal of Translational Medicine (2026)
-
The USP1-WDR48 deubiquitinase complex functions as a molecular switch regulating tumor-associated macrophage activation and anti-tumor response
Cell Death & Differentiation (2026)
-
Neuroglia and immune cells play different roles in neuroinflammation and neuroimmune response in post-stroke neural injury and repair
Acta Pharmacologica Sinica (2026)
-
Mitochondrial-targeted ROS scavenger delivery using selenium-doped layered double hydroxide nanocomposites for enhanced osteoarthritis therapy via inflammation suppression and macrophage metabolism reprogramming
Advanced Composites and Hybrid Materials (2026)
-
The role of immune cell crosstalk and signaling pathways in the pathogenesis of pulmonary fibrosis
Molecular Biology Reports (2026)