Abstract
Efficient interbacterial competitions and diverse defensive strategies employed by various bacteria play a crucial role in acquiring a hold within a dense microbial community. The gut symbiont Bacteroides fragilis secretes an antimicrobial ubiquitin homologue (BfUbb) that targets an essential periplasmic PPIase to drive intraspecies bacterial competition. However, the mechanisms by which BfUbb enters the periplasm and its potential for interspecies antagonism remain poorly understood. Here, we employ transposon mutagenesis and identify a highly conserved TonB-dependent transporter SusCD (designated as ButCD) in B. fragilis as the BfUbb transporter. As a putative protein-related nutrient utilization system, ButCD is widely distributed across diverse Bacteroides species with varying sequence similarity, resulting in distinct import efficiency of Bacteroides ubiquitin homologues (BUbb) and thereby determining the species-specific toxicity of BUbb. Cryo-EM structural and functional investigations of the BfUbb–ButCD complex uncover distinctive structural features of ButC that are crucial for its targeting by BfUbb. Animal studies further demonstrate the specific and efficient elimination of enterotoxigenic B. fragilis (ETBF) in the murine gut by BfUbb, suggesting its potential as a therapeutic against ETBF-associated inflammatory bowel disease and colorectal cancer. Our findings provide a comprehensive elucidation of the species-specific toxicity exhibited by BUbb and explore its potential applications.
Similar content being viewed by others
Introduction
As the most abundant phylum of Gram-negative bacteria in the human gut microbiota, Bacteroidota coexist with billions of other microorganisms1. Together, these microbes are crucial for proper digestion, nutrient absorption, immune system function and overall health of the host2,3,4. Although the majority of Bacteroides are commonly regarded as commensals, certain species within this genus can also exhibit pathogenicity when residing in the gut or other anatomical sites5,6. Among various pathogenic Bacteroides species, enterotoxigenic Bacteroides fragilis (ETBF) has gained increasing attention due to its association with intestinal disorders, including diarrhoea, inflammatory bowel disease (IBD), and colorectal cancer (CRC)7,8,9,10.
In the highly intricate and densely populated ecosystem of the human intestine, bacteria engage in exploitative and interference competitions, wherein they vie for limited resources and space11,12. Exploitative competition ensues when bacteria outpace their competitors in resource consumption rates, while interference competition involves the production of inhibitory compounds aimed at impeding the growth of other bacteria species13,14. These competitive interactions play a pivotal role in shaping microbial communities by exerting influence on population dynamics and keeping ecological balance15.
Exploitative competition serves as a crucial driving force for bacterial evolution and adaptation to diverse environmental conditions and niches13,15. Bacteroides are equipped with an extensive repertoire of polysaccharide utilization loci (PULs), encoding machinery for glycan degradation and uptake, enabling them to thrive in fluctuating conditions within the human gut1,16. These PULs typically harbour a diverse array of carbohydrate-active enzymes (CAZymes) responsible for breaking down complex polysaccharides17,18. Integral to each PUL systems are TonB-dependent transporters (TBDTs), such as the β-barrel forming porin SusC-like proteins, along with their closely associated cognate SusD-like lipoproteins19,20. SusD functions as a discriminating lid on the bin-like structure of SusC, facilitating selective glycan import via a “pedal-bin” transport mechanism20. Nevertheless, the function of numerous PULs in their ability to colonize and exploit a niche for Bacteroides remains unidentified.
In terms of interference competition, Bacteroides species possess a diverse array of antagonistic factors to outcompete each other. Type VI secretion systems (T6SS) facilitate contact-dependent antagonism that can lead to interspecies killing21,22,23,24, while diffusible toxins primarily result in intraspecies killing25,26,27,28,29,30,31,32. Our recent study, together with the findings of the Comstock group, demonstrated that Bacteroides fragilis secretes a ubiquitin homologue (BfUbb) to target an essential periplasmic peptidyl-prolyl isomerase (PPIase) for intraspecies antagonism25,33. Certain strains of B. fragilis can evade targeting and killing by BfUbb through a single mutation at Tyr119 in the targeted PPIase. However, the mechanism by which BfUbb gains access to the bacterial periplasm remains elusive. More importantly, it is also unclear whether BfUbb can mediate interspecies antagonism and how its antagonistic range is determined.
Here, we identified a unique yet conserved TonB-dependent transporter SusCD variant in B. fragilis (designated as ButCD for Bacteroides ubiquitin homologues (BUbb) transporter SusCD) that is exploited by BfUbb for uptake into recipient cells. Although homologues of the ButCD pair are widely distributed among other Bacteroides species, several Bacteroides species encoding BfUbb-targeted PPIase avoid BfUbb killing due to the lower sequence identity between their ButCD and ButCDBf, which prevents the transport of BfUbb into cells. The role of ButCD in determining the species-specificity of BUbb is further confirmed, as it is hijacked by BoUbb, another BUbb from B. ovatus, to antagonize several Bacteroides species that are resistant to BfUbb. The cryo-EM structure of BfUbb-ButCDBf complex reveals unique structural features of the BfUbb-targeted ButC (SusC-like) protein that facilities binding and passage of BfUbb through its porin structure. Considering its specific and potent bactericidal activity against ETBF in mice, BfUbb could potentially assist in therapy for IBD and CRC. Collectively, our study provides a comprehensive elucidation of the various factors contributing to species-specific toxicity exhibited by BUbb and explores its potential applications.
Results
BfUbb is inaccessible to certain Bacteroides species
BfUbb antagonizes certain strains of B. fragilis by targeting an essential periplasmic PPIase, which possesses Tyr119 as the determinant for sensitivity to BfUbb33. Homologues of this PPIase, containing Tyr119, are widely distributed across numerous sequenced Bacteroides species (Supplementary Fig. 1 and Fig. 1a). Consistent with sequence alignment results, the purified PPIases from different Bacteroides species interacted directly with BfUbb in vitro (Fig. 1b). However, unlike B. fragilis GS077, which is susceptible to BfUbb, B. thetaiotaomicron VPI-5482, B. xylanisolvens AM54-2NS and B. ovatus ATCC 8483 all carry the BfUbb-sensitive PPIase variant but resist BfUbb-mediated killing (Fig. 1c). Since BfUbb requires access to the periplasm of recipient cells to exert its antibacterial function, we speculated that these Bacteroides species, which would otherwise be sensitive to BfUbb killing due to carrying a BfUbb-sensitive PPIase, have an altered or decreased BfUbb uptake.
The Bacteroides species included in the analyses are B. fragilis GS077 (BfGS077), B. thetaiotaomicron VPI-5482 (Bt), B. xylanisolvens AM54-2NS (Bx), and B. ovatus ATCC 8483 (Bo). a Amino acid sequence alignment of PPIases from position 110 to 120 in the above-mentioned Bacteroides species. Conserved residues are shaded in red and similar residues in yellow. The asterisk indicates the conserved residue Tyr119 involved in BfUbb sensitivity. b Protein interaction analysis to detect the binding between purified histidine-tagged PPIases from the different Bacteroides species and untagged BfUbb. c Agar spot analysis to assess the inhibitory potential of BfUbb against the indicated Bacteroides species. Bacterial survival was determined by drop plating 10-fold serial dilutions of bacterial cultures on BHI plates supplemented with BfUbb or with Tris-buffered saline (TBS) as a control (CT). d Immunoblot detection of BfUbb in the lysates of indicated Bacteroides species pre-incubated with BfUbb before lysed to determine BfUbb accessibility. DnaK expression was included as a control, and protein size is indicated in kDa. Experiments (b–d) were conducted at least three times with consistent results. e Representative confocal microscopy images (>20 images from three independent experiments) of indicated Bacteroides species treated with Cy3-labelled BfUbb for 4 h. Over time, B. fragilis GS077 cells exhibit intense fluorescence, along with abnormal rounding and elongated shape (highlighted by light blue arrows) compared to other Bacteroides strains. Scale bar for all images is 5 μm.
To assess the capacity of B. thetaiotaomicron, B. xylanisolvens, and B. ovatus to uptake BfUbb, we investigated the accessibility of BfUbb in these species. Notably, we detected BfUbb only in cells of BfUbb-susceptible B. fragilis GS077 among the four tested Bacteroides species (Fig. 1d). Additionally, we could visually detect uptake of BfUbb, labelled with a Cy3 fluorescent probe, in the BfUbb-sensitive strain B. fragilis GS077 accompanied by an elongation and cell rounding phenotype but not in the B. thetaiotaomicron, B. xylanisolvens, and B. ovatus isolates (Fig. 1e). These results suggest that there exist cellular factors associated with BfUbb import, which contributes to toxin-sensitivity among various Bacteroides species.
A unique SusCD is required for BfUbb cytotoxicity in BfUbb-sensitive strains
To identify Bacteroides proteins involved in BfUbb uptake, we performed transposon mutagenesis in the BfUbb-sensitive strain B. fragilis GS077 to identify mutants resistant to BfUbb killing when grown in the presence of purified BfUbb under anaerobic conditions (Fig. 2a). We identified eight transposon mutants of GS077, which exhibited resistance towards BfUbb. Notably, all these mutants harboured transposon insertions within the gene GS077_4426, encoding a member of the SusC-like protein (designated as SusC*) that functions as an outer membrane TonB-dependent transporter (TBDT) (Fig. 2b).
a Overview of the transposon mutagenesis screening process in B. fragilis GS077 using purified BfUbb (blue) as a selective condition. Small yellow circles represent B. fragilis GS077 (BfGS077) mutants carrying the transposon. b Schematic representation of the transposon insertion sites (black vertical lines) in susC* gene. Coloured arrows represent genes GS077_4423 in dark blue (AAA family ATPase), GS077_4424 in light blue (VWA domain-containing protein), GS077_4425 in light purple (susD* gene encoding a SusD-like cell-surface lipoprotein), GS077_4426 in dark purple (susC* gene encoding a SusC-like TonB-dependent transporter), GS077_4427 in pink (ZnMc gene encoding a zinc-dependent metalloprotease) and GS077_4428 in brown (beta-N-acetylhexosaminidase). c Amino acid sequence alignment of SusC* from six B. fragilis strains with different BfUbb sensitivities: Bf GS077S (purple), Bf GS086S (dark blue), Bf 9343I (cyan), Bf GS075I (green), Bf GS070I (yellow), and Bf GS084I (orange). Sensitivity is indicated by superscript letters (S for BfUbb-sensitive, I for BfUbb-resistant strains). The vertical lines represent amino acid discrepancies compared to SusC* from strain Bf GS077S. On the right, percentage of sequence similarity. Above the alignment, schematic representation of the functional domains in SusC*: signal peptide (SP) in yellow, N-terminal extension (NTE) in blue, TonB Box in orange, Plug in cyan and β-barrel in purple. Numbers mark the start and end residues of each domain. d–h Assays performed with wild type B. fragilis GS077 and B. fragilis GS084, isogenic deletion mutants ΔsusC*, ΔsusD*, ΔZnMc, and complemented isogenic mutants ΔsusC*::psusCD* and ΔsusD*::psusCD* in both genomic backgrounds. Experiments were conducted at least three times with consistent results. d Agar spot assays to assess the sensitivity of the B. fragilis GS077 isogenic set to purified BfUbb, supernatant (Sup) of BfUbb-encoding strain B. fragilis NCTC 9343. e, g Western blot to determine BfUbb accessibility in the cell lysates of B. fragilis GS077 (e) and GS084 (g) isogenic sets, pre-incubated with BfUbb. f, h Representative confocal microscopy images (>20 images from three independent experiments) of B. fragilis GS077 (f) and GS084 (h) isogenic sets treated with Cy3-labelled BfUbb for 4 h. Scale bar, 5 μm.
The domain architecture of SusC* is consistent with typical SusC proteins, featuring an N-terminal signal peptide (SP) for outer membrane translocation, an N-terminal extension (NTE) domain with unknown function, a TonB box that interacts with the periplasmic protein TonB to energize transport, a β-barrel domain that forms the channel for substrate uptake, and a plug domain that obstructs the channel (Fig. 2c). SusC-like proteins form a complex with the surface lipoprotein SusD-like proteins, and are usually encoded in adjacent genes. Downstream of susC*, a SusD-like protein (designated as SusD*) is predicted to be encoded by GS077_4425 (Fig. 2b). However, unlike the typical genetic architecture of PUL operons in Bacteroides, which contains additional genes for glycan binding and digestion, there only exists a gene encoding a predicted zinc-dependent metalloprotease (GS077_4427, ZnMc) upstream of susCD* and no additional predicted typical polysaccharide utilization-related genes around (Fig. 2b and Supplementary Fig. 2).
We further investigated if the susCD* gene locus contributed to BfUbb intoxication and uptake by deleting the susC*, susD*, or ZnMc genes in GS077. Deletion of susC* in GS077 did not alter growth when compared to wildtype in either rich medium (BHI) or minimal medium (MMF) (Supplementary Fig. 3a). However, in B. fragilis NCTC 9343, deletion of susC*Bf9343 resulted in an extended exit from lag phase in BHI medium but not in the final cell density when compared to wildtype (Supplementary Fig. 3b). Furthermore, during co-culture between B. fragilis NCTC 9343 wildtype and ΔsusC*Bf9343, the wildtype strain exhibited a growth advantage over the ΔsusC*Bf9343 mutant (Supplementary Fig. 3c). Interestingly, the SusD*Bf9343 protein is primarily detected during early exponential phase, suggesting a role for SusCD* in prioritizing nutrients present in the environment34,35 (Supplementary Fig. 3d). Consistent with the transposon screen, deletion of susC* or susD* in GS077 eliminated its sensitivity to purified BfUbb or supernatant from BfUbb-encoding strain B. fragilis NCTC 9343 (Fig. 2d). This sensitivity was restored by introducing a SusCD*-expressing plasmid into the deletion strains (Fig. 2d). However, deletion of ZnMc did not confer BfUbb tolerance to GS077 in agar spot assays (Fig. 2d).
Further, BfUbb uptake was detected in cells of wildtype, susC* complemented mutant, susD* complemented mutant and ΔZnMc mutant strains. However, BfUbb was not detected in ΔsusC* and ΔsusD* mutants (Fig. 2e). Consistently, cell-associated BfUbb-Cy3 fluorescence and BfUbb-triggered morphological changes were abolished upon deletion of susC* or susD* (Fig. 2f), indicating that both SusC* and SusD* are indispensable for the internalization and cytotoxicity of BfUbb.
SusCD* is conserved across B. fragilis
To evaluate the distribution of the SusC* homologues in B. fragilis species, we conducted a comprehensive search and observed that SusC* is highly conserved not only in BfUbb-sensitive but also in BfUbb-resistant B. fragilis strains (including BfUbb-encoding strains) (Fig. 2c and Supplementary Fig. 4). Comparative analysis of amino acid sequences between SusC* and SusD* homologues from both BfUbb-sensitive and BfUbb-resistant B. fragilis strains revealed a sequence identity of 94%–100% for SusC* homologues with respect to SusC*GS077 (Fig. 2c and Supplementary Fig. 4), while SusD* homologues were found to be 88%-100% identical in protein sequence when compared to SusD*GS077 (Supplementary Fig. 5). Consistent with the substantial sequence identity of SusCD* between BfUbb-sensitive and resistant B. fragilis strains, the western blot and confocal microscopy experiments showed that the BfUbb-resistant strain exhibits a SusCD*-dependent uptake of BfUbb, albeit without displaying elongation and cell rounding phenotypes (Fig. 2g, h). The distribution and conservation of SusCD* in B. fragilis highlight its crucial and conserved role, underscoring the significance of these proteins.
ButCD is a BfUbb transporter
Several TBDTs have been identified as crucial bacterial surface receptors or outer membrane transporters, contributing to the strains’ susceptibility to secreted bacteriocins36,37,38,39. The TonB box precedes the N-terminal plug domain of TBDTs, interacting with the C-terminal domain (CTD) of TonB to provide energy for substrate transport. Disruption of the TonB box abolishes the energy supply from TonB to TBDTs, resulting in transport failure but does not affect substrate binding20,39. In our study, we observed a complete loss of BfUbb sensitivity in B. fragilis GS077 when TonB box residues of SusC* (115DAVVV119) were deleted (ΔTonB box) or substituted with alanine (TonB box mutant, 115AAAAA119) (Fig. 3a), despite comparable levels of SusCD* expression (Supplementary Fig. 6). Subsequent immunoblotting and fluorescence microscopy analyses confirmed the absence of detectable BfUbb in both ΔtonB box and TonB box mutant cells (Fig. 3b, c). Collectively, these findings demonstrate that BfUbb utilizes SusCD* as the transporter for cellular uptake. Therefore, we rename GS077_4425-4426 (SusCD*) as ButCD for Bacteroides ubiquitin homologues (BUbb) transporter SusCD.
a–c Assays performed with wild type B. fragilis GS077 and isogenic mutants ΔTonB box (deletion of TonB box in butC) and TonB box mut (TonB box 115AAAAA119 in butC). a Agar spot assays to evaluate the inhibitory effect of BfUbb on the growth of B. fragilis GS077 isogenic sets. b Western blot to detect BfUbb accessibility in the cell lysates of B. fragilis GS077 isogenic sets, pre-incubated with BfUbb. c Confocal microscopy images to show the uptake of Cy3-labelled BfUbb (yellow) following a 4 h incubation with B. fragilis GS077 isogenic sets. Scale bar, 5 μm. d Western blot to detect BfUbb in the cell lysates of B. fragilis GS084 and isogenic mutants ΔtonB1-6, pre-incubated with BfUbb. e–h Assays performed with B. fragilis GS084 and B. fragilis GS077, isogenic deletion mutants ΔtonB3, and complemented isogenic mutant ΔtonB3::ptonB3 in both genetic backgrounds. e, g Western blot to detect BfUbb accessibility in the cell lysates of B. fragilis GS084 (e) and GS077 (g) isogenic sets, pre-incubated with BfUbb. f Confocal microscopy images to show the uptake of Cy3-labelled BfUbb (yellow) following a 4 h incubation with B. fragilis GS084 isogenic set. Scale bar, 5 μm. h Agar spot assays to evaluate the inhibitory effect of BfUbb on the growth of B. fragilis GS077 isogenic sets. i Schematic representation of the tonB3 chromosomal region in B. fragilis GS077. Functional annotation groups assigned as TonB (red), ExbB (yellow), or ExbD (blue) homologues are indicated by arrows filled with colour. PfkA (6-phosphofructokinase), LytB (putative isoprene biosynthesis related reductase), CmpK (CMP kinase), HP (conserved hypothetical protein), and TatD (DNase) are also indicated. j-k Assays performed with B. fragilis GS077, isogenic deletion mutants ΔexbB1B2, ΔexbD1D2, and complemented isogenic mutants ΔexbB1B2::pexbB1, ΔexbB1B2::pexbB2, ΔexbB1B2::pexbB1B2, ΔexbD1D2::pexbD1, ΔexbD1D2::pexbD2, ΔexbD1D2::pexbD1D2. j Western blot to detect BfUbb accessibility in the cell lysates of the B. fragilis GS077 isogenic set, pre-incubated with BfUbb. k Agar spot assays to evaluate the inhibitory effect of BfUbb on the growth of B. fragilis GS077 isogenic sets. Experiments were conducted at least three times with consistent results.
TonB3-ExbBD complex is the engine for ButCD-mediated BfUbb transportation
TonB-dependent transporters (TBDTs) actively utilize the proton motive force generated by the TonB-ExbB-ExbD (TBD) system, a nanomachine composed of three inner membrane proteins (ExbB, ExbD, and TonB), to facilitate substrate import to the periplasm40,41. Therefore, we investigated the relationship between BfUbb transport and the TBD system. We performed the genetic dissection of TBD-mediated BfUbb transport in BfUbb-resistant B. fragilis GS084 to track BfUbb uptake independent of cell death. Analysis of the B. fragilis GS084 genome revealed six tonB, four exbB and five exbD homologues (Supplementary Fig. 7). To determine which TonB protein is involved in providing energy for BfUbb transport, we constructed six individual tonB deletions in GS084. Screening all six tonB deletion mutants revealed that BfUbb was not imported in cells carrying a tonB3 deletion (Fig. 3d), but was restored when the deletion was complemented with wild-type tonB3 (Fig. 3e, f). Consistent results were also observed in the BfUbb-sensitive strain GS077 (Fig. 3g), where deletion of tonB3 conferred complete resistance to BfUbb, while BfUbb sensitivity was restored upon tonB3 complementation, further confirming that TonB3 facilitates ButCD-mediated transport of BfUbb (Fig. 3h).
Furthermore, GS077 exhibits a similar genomic architecture of the tonB3 gene locus to GS084, including two exbB and two exbD that is also observed in other Bacteroides (Fig. 3i and Supplementary Fig. 7). Deletion of either both exbB or both exbD abolished BfUbb transport. However, complementation with just one exbB or exbD is sufficient to restore BfUbb transport (Fig. 3j, k), indicating that one exbB or exbD is adequate to stimulate TonB3 for BfUbb uptake in GS077. In summary, the TonB3-ExbBD system serves as the molecular motor responsible for driving ButCD-mediated BfUbb transport.
ButCD is distinct from canonical polysaccharide transporters
In contrast to the genomic architecture observed in classical polysaccharide utilization associated susCD pairs-containing loci, the butCD locus lacks genes predicted to be involved in polysaccharide binding and digestion except for ZnMc, suggesting this locus might confer a specialized function divergent from polysaccharide uptake. The absence of ButCD orthologues in GS077 was further confirmed by OrthoFinder42 and blastp, underscoring the potential functional uniqueness of ButCD within the SusCD superfamily in GS077.
To investigate the potential functionality of ButCD, we conducted an extensive search for annotated genes within all susCD pairs-containing loci present in strain GS077. These susCD pairs present in GS077 were further grouped into 59 putative susCD pairs-containing loci (some of which contained multiple susCD pairs), predicted their biological functions, and classified them accordingly (Supplementary Fig. 8).
By comparing annotated PULs from PULDB43 (www.cazy.org/PULDB) along with manual proofreading, 43 of 59 putative susCD pairs-containing loci were further assigned as putative PULs, potentially associated with polysaccharide metabolism and transport (Supplementary Fig. 8). These PULs can be categorized into three subtypes based on the diversity and copy number of glycoside hydrolases: Glycan transport_Sub 1 (≤2 glycoside hydrolases), Sub2 (>2 glycoside hydrolases), and Sub3 (no typical glycoside hydrolase; with genes putatively involved in polysaccharide metabolism, such as sulfatase43,44, among others) (Supplementary Fig. 8). However, a subset of susCD pairs-containing loci (12/59) lack genes with discernible functional annotations and are therefore tentatively designated as susCD pairs-containing loci with unknown functions (Supplementary Fig. 8; designated as “function unknown”). Additionally, four susCD pairs-containing loci containing putative metal-dependent peptidase domains may participate in polypeptide transport (Fig. 4a), including the GS077_4426 susCD pairs identified for BfUbb import and the GS077_3053 susCD pairs potentially associated with transport of an unidentified protein class. The peptidases within the GS077_0109 susCD pairs may be non-functional due to the presence of a stop codon truncating the immediate gene downstream of its susD. Moreover, SusC in the GS077_1440 susCD pairs lacks a complete beta-barrel domain and may not meet the requirements for transportation due to the presence of an early stop codon in its reading frame. In summary, among various types of susCD pairs, butCD is predicted to have a distinct functionality compared to other susCD pairs present in GS077, which may confer its ability to import BfUbb.
The Bacteroides species included in the experiments are B. fragilis GS077 (BfGS077), B. uniformis GS313 (Bu), B. intestinalis GS315 (Bi), B. cellulosilyticus GS316 (Bc), B. nordii (Bn), B. ovatus ATCC 8483 (Bo), B. xylanisolvens AM54-2NS (Bx), and B. thetaiotaomicron VPI-5482 (Bt). a Schematic of four susCD pair-containing loci: GS077_4426 (butCD loci, in red), GS077_3053, GS077_0109, and GS077_1440, likely involved in peptide transport. ORFs with different predicted functions are shown: SusC in dark purple, SusD in light purple, peptidase in pink and putative non-functional peptidase and truncated SusC marked by a white lightning symbol. b Unrooted phylogenetic trees based on 536 ButC homologues from Bacteroides species. Identity relative to ButC from B. fragilis GS077 is indicated for the major clades (blue and green). c Schematic representation of the ButCD loci of indicated Bacteroides species. Genes ZnMc, butC, and butD are shown as red, green, and blue arrows, respectively, with amino acid sequence identity to ButCD loci from B. fragilis GS077 inside each arrow. Percentages indicate the average sequence identity of ButCGS077, ButDGS077, and ZnMcGS077 aligned to their homologues in the specified species from the NCBI Nr database. d, i Antagonistic range analysis of BfUbb (d) and BoUbb (i) against indicated Bacteroides species. e Western blots to detect BfUbb accessibility in the cell lysates of indicated Bacteroides species expressing B. fragilis GS077 ButCD via pbutCDBf plasmid and isogenic control strains (pvector), pre-incubated with BfUbb. f Representative confocal microscopy images of indicated Bacteroides species carrying a plasmid for the expression of the ButCDBf, and treated with Cy3-BfUbb. g Agar spot assays to evaluate the inhibitory activity of BfUbb against the Bacteroides species harbouring a plasmid expressing ButCDBf (pbutCDBf). h Amino acid sequence alignment of human ubiquitin (HmUbb), BfUbb, and BoUbb, with conserved residues shaded in red and similar ones in yellow. The disulfide bond (S-S) in BfUbb and BoUbb is shown. j Representative confocal microscopy images of indicated Bacteroides species treated with Cy3-BoUbb. For (f–j) Scale bar, 5 μm. For (d-g, i, j) experiments were conducted at least three times with consistent results.
ButCD determines the species-specific toxicity of BfUbb
The presence of ButCD, the essential transporter of BfUbb, was observed across all Bacteroides species, but not in Phoecicola (except for Phoecicola coprocola), Parabacteroides or Alistipes according to the NCBI Nr database. Subsequently, we examined the similarity of ButC homologues in all Bacteroides species. A phylogenetic analysis based on the sequence alignment of ButC homologues from Bacteroides (NCBI Nr database, Bacteroides limited, Coverage > 90%) categorizes diverse Bacteroides species into two discrete clades, sharing greater than 60% and less than 60% identity with ButCGS077, respectively (Fig. 4b). However, the phylogenetic analysis of ButC homologues and the species encoding them revealed that the phylogeny of these homologues does not align with the phylogeny of their encoding species (Fig. 4b and Supplementary Fig. 9), suggesting horizontal gene transfer or rapid evolutionary diversification of the butCD genes. A similar observation was also found on the inconsistency between phylogenetic studies of T6SS E/I cassettes and species phylogenies24.
Based on this homology analysis, we found that the ButCD locus of B. uniformis exhibited the highest degree of identity to B. fragilis GS077, followed by the average sequence identity of the ButCD homologues from B. intestinalis, B. cellulosilyticus, B. nordii, B. ovatus, B. xylanisolvens, and B. thetaiotaomicron (Fig. 4c). Although all these Bacteroides species encode a BfUbb-sensitive PPIase (Supplementary Fig. 1), their sensitivity to BfUbb is correlated with the degree of sequence identity in the ButCD homologues. Among these species, B. uniformis GS313, which has a higher ButCD sequence identity, showed a certain level of sensitivity towards BfUbb, whereas those with a lower ButCD sequence identity exhibited BfUbb resistance (Fig. 4d). These findings suggest that a narrow range of moderate sequence identity in ButCD can determine the capability and efficiency of BfUbb uptake in recipients.
Furthermore, we assessed whether Bacteroides species carrying BfUbb-sensitive PPIase were resistant to BfUbb due to their ButCD homologues being unable to import BfUbb. We introduced a plasmid expressing ButCDGS077 into BfUbb-resistant Bacteroides species, such as B. ovatus, B. xylanisolvens, and B. thetaiotaomicron. Western blot analysis and florescence microscopy revealed that these strains gained the ability to transport BfUbb (Fig. 4e, f). The acquisition of BfUbb access conferred them with sensitivity to BfUbb (Fig. 4g), indicating the simultaneous attainment of both BfUbb sensitivity and import capability. Additionally, expression of either ButCGS077 or ButDGS077 alone in B. thetaiotaomicron did not confer sensitivity to this species towards BfUbb (Supplementary Fig. 10a), indicating that both ButC and ButD from B. fragilis are required for BfUbb transport. Our collective findings demonstrate that ButCD plays a pivotal role in determining the import capacity of BfUbb, thereby influencing the susceptibility of specific Bacteroides species to BfUbb and shaping its antagonistic range.
ButCD determines the species-specific toxicity of distinct Bacteroides ubiquitin homologues
During our exploration of human gut metagenome datasets, we discovered another human ubiquitin (HmUbb) homologue from B. ovatus (BoUbb), exhibiting about 61% sequence identity with both BfUbb and HmUbb. However, the prevalence of the gene encoding BoUbb in the human gut metagenomes is notably lower compared to BfUbb25, with only four strains of B. ovatus encoding BoUbb among the sequenced B. ovatus genomes in the NCBI database (~600 strains). Among the 1267 human metagenomic samples in “3 consortium gene catalogue” (3CGC) dataset44, only 1 of 499 metagenomic samples (SRR1778453) with detectable B. ovatus (~0.2%) also contained the BoUbb gene (Supplementary Table 1). Similar to BfUbb, BoUbb possesses a distinctive disulfide bond at its C-terminus (Fig. 4h), which was demonstrated to be crucial in mediating intraspecies antagonism by BfUbb33, indicating that BfUbb is likely not the sole example of an antibacterial ubiquitin homologue. However, contrary to the behaviour of BfUbb, screening results revealed that BoUbb exhibited varying levels of antagonism against strains of Bacteroides species, including B. ovatus, B. xylanisolvens, and B. thetaiotaomicron, while being ineffective against strains such as B. fragilis and B. uniformis GS313 (Fig. 4i). This observation aligns with the sequence identity of ButCD in distinct Bacteroides species (Fig. 4c).
Consistent with BfUbb, BoUbb exhibits in vitro binding to PPIases containing Tyr119 and disrupts the cell wall integrity of susceptible cells (Supplementary Fig. 10b, c). We therefore assessed the entry capability of BoUbb in four Bacteroides species and microscopy of Cy3-labelled BoUbb uptake was correlated to the species’ sensitivity to killing when exposed to BoUbb (Fig. 4j). Moreover, deletion of butC rendered B. thetaiotaomicron resistant to BoUbb, whereas complementation with a plasmid expressing ButCDBt, and not ButCDBf, reversed this outcome (Supplementary Fig. 10d), verifying the ability of the ButCDBt in recognizing and importing BoUbb. These findings indicate that alterations in strain specificity between BoUbb and BfUbb primarily arise from their accessibility into recipient cells, which is contingent upon the distinctive characteristics of ButCD.
Cryo-EM structure of BfUbb-ButCD complex
To elucidate the features of ButCD required for BfUbb recognition and uptake, we determined the structure of the BfUbb-ButCD complex using single-particle cryo-EM. We isolated BfUbb-bound ButCD complexes directly from B. fragilis GS077 (Supplementary Fig. 11a) and subjected the purified complex to cryo-EM data collection.
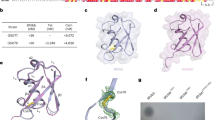
After initial two-dimensional classification, the ButCD complex exists as a dimer of ButCD in solution (designated as ButC2D2), which aligns with previous findings that SusCD-like systems exist as a dimerized complex (Supplementary Fig. 11b)19,20. Further ab-initio reconstruction and hetero-refinement revealed that, contrary to prior reports indicating the presence of three distinct conformations of open and closed dimeric SusC2D2 transporters (open-open, closed-open and closed-closed) within one dataset20, only the open-open conformation of ButC2D2 was observed (Supplementary Fig. 11c). We also found that ~30% of the dataset exhibited additional density exclusively within one ButC barrel of the ButC2D2 dimer, which was well fitted by the BfUbb protein structure. No particle populations of a ButC2D2 dimer with both barrels exhibiting a BfUbb were observed (Supplementary Fig. 11c). Consequently, we determined two ButC2D2 dimeric structures in distinct states: apo-ButC2D2 dimer (designated as the AA state; 2.97 Å) and BfUbb-ButC2D2 complex (designated as the CA state; 3.05 Å) including one apo-ButCD (Fig. 5a, Supplementary Fig. 11d–g and Supplementary Table 2).
a Cryo-EM structures of the ButC2D2 dimer alone (left) and with BfUbb (right). Top views show the outer membrane (OM) plane, and bottom views show the extracellular space. The plug domain is in dark blue, with loops L7, L8 of ButC and BfUbb marked by black arrows. b Surface views of superposition of apo-ButC, RagA, and BT1763 (light grey) within BfUbb-ButCD, RagAB (PDB: 6smq), and BT1762-1763 (PDB: 6zm1) complexes in the open state. ButD (pink), RagB (black), BT1762 (light green). c Protein structures of the open states of apo-ButCD (blue and pink), RagAB (brown and black), and BT1762-1763 (light green and forest green) complexes over blurred surface representations. Opening angles for apo-ButCD (ButD-Y71 and ButC-W352, F728), RagAB (RagB-G78 and RagA-W321, Q670), and BT1762-1763 (BT1762-G64 and BT1763-W295, Y636) are marked with black lines and labelled values. d, e Surface representations of BfUbb-bound ButC (light grey) aligned with RagA (light grey) in the RagAB (d) or BT1763 (light grey) in the BT1762-1763 (e). ButD (purple), BfUbb (orange), RagB (black), and BT1762 (light green). f, g Structural superposition of ButCs within the apo_ButCD, BfUbb-ButCD, and RagA in the RagAB (f) or BT1763 in the BT1762-1763 (g). Conformational changes among extracellular loops of ButC (cyan), apo-ButC (blue), RagA (brown), and BT1763 (forest green) are highlighted by arrows. h Close-up views of the depicted loops in (f) and (g) protein structures to illustrate the residues involved in BfUbb (orange) binding to ButC (cyan). i Western blots on B. fragilis GS077 cell lysates treated with BfUbb or indicated BfUbb mutants, and probed with antibodies against BfUbb. The BfUbb4M protein variant encompasses all four amino acid substitutions. j Overlay assays showing the susceptibility of B. fragilis GS077 to BfUbb and indicated BfUbb mutants. k Agar spot assays to evaluate the inhibitory activity of purified BfUbb or supernatant (Sup) from the BfUbb-encoding strain against B. fragilis GS077 ΔbutC deletion mutant complemented with its own butC (ButC), or with butC harbouring indicated mutations. For (i–k) experiments were conducted at least three times with consistent results.
We further explored the structural and biochemical characteristics of the ButCD complex in substrate recognition. Given the similar structure of apo-ButCD in both AA and CA states (Supplementary Fig. 12a), we used the structures of the BfUbb-ButCD complex and apo-ButCD derived from the same ButC2D2 dimer (CA) for the following structural analysis. In both structures, ButC, lacking the N-terminal extension (NTE) domain, consists of an N-terminal TonB box and plug domain that is inserted into a 22-strand β-barrel with multiple extracellular loops (Fig. 5a). Hinge loops L7 and L8 located close to ButD are responsible for binding ButD (Fig. 5a). Consistent with the previously elucidated conformation of SusCD in its open state, ButD exhibits an upward tilt and primarily interacts with hinge loops L7 and L8 of ButC, thereby exposing the substrate binding site and plug domain within the interior of the barrel structure (Fig. 5a).
In the BfUbb-ButCD complex, BfUbb is centrally positioned at the extracellular entrance of ButC, where it predominantly interacts with the extracellular loop regions of ButC while exhibiting no interactions with ButD or the plug region of ButC (Fig. 5a). Structural alignment of BfUbb-bound and apo-ButCD shows no significant conformational changes in ButCD upon binding by BfUbb (Supplementary Fig. 12b-e). These observations suggest that the BfUbb-bound ButCD complex represents a transport intermediate state where BfUbb is transferred to the entrance of ButC.
To better understand the selection of ButCD as a transporter for BfUbb, we superimposed and compared our ButCD structure with previously determined structures of SusCD-like complexes. Structural comparisons between ButCD, RagAB from Porphyromonas gingivalis, and BT1762-1763 from B. thetaiotaomicron in their open states show that the SusD-like (lid) proteins (ButD, BT1762, RagB) are angled at different degrees compared to their cognate SusC-like (barrel) proteins (ButC, BT1763, RagA, respectively) (Fig. 5b and Supplementary Fig. 12f, g). We found that the aperture angle between the SusD lid and SusC barrel is 84.98° in ButCD, versus 70.25° for RagAB and 47.17° for BT1762-1763. This difference is primarily driven by the differences observed in the hinge loops L7 and L8 of the SusC-like proteins. (Fig. 5c and Supplementary Fig. 12h). Additional comparisons of the BfUbb-ButCD complex structure reveal that the position of the SusD-like proteins, RagB or BT1762, when aligned with its cognate SusC-like proteins, RagA and BT1763, respectively, could sterically prevent access to a larger substrate such as BfUbb (Fig. 5d, e). Moreover, when compared to RagA and BT1763, extracellular loops L3, L4, L5, L9 and L10 of ButC remain in close proximity, restricting the size of the entrance to ButC (Fig. 5f, g). This structural arrangement of ButC potentially allows for establishing extensive polar interactions required for BfUbb capture (Fig. 5h).
To validate the observations above, we engineered specific amino acid mutants in both proteins to assess their impact on BfUbb transport. Alanine substitutions of BfUbb residues interacting with ButC in cryo-EM (both K6 and T14, or D32, N60, or all four residues (designated as 4M) of BfUbb) decreased BfUbb import in recipient cells at varying degrees, which resulted in corresponding reductions in cellular toxicity of sensitive cells (Fig. 5i, j). Alanine substitution of specific ButC amino acids (Y434, R628, N972 or Y1010/Q1014/K1020) involved in the interaction with BfUbb resulted in reduced import efficiency (Supplementary Fig. 12i), leading to altered sensitivity at varying degrees (Fig. 5k). Altogether, ButCD exhibits appropriate structural and biochemical characteristics that facilitate the recognition and import of BfUbb.
Additionally, we substituted ButCBf amino acids R628 and N972 with the corresponding amino acids from the ButCBt protein and found that corresponding mutations, R628Q, N972D, and R628Q/N972D, in B. fragilis strain GS077 disrupted BfUbb transport and conferred resistance to toxicity, supporting a role for residues R628 and N972 of ButCBf in substrate specificity (Supplementary Fig. 12j, k and Supplementary Fig. 13). These findings suggest the existence of a sophisticated mechanism through which BfUbb selectively recognizes ButCD, thereby contributing to the species-specific toxicity of BfUbb.
BfUbb eliminates enterotoxigenic Bacteroides fragilis in the animal gut
Enterotoxigenic Bacteroides fragilis (ETBF) is strongly associated with the development of inflammatory bowel disease (IBD) and colorectal cancer (CRC)7,8,9. Sequence examination of several characterized disease-associated ETBF strains7,8,9,45,46 reveals that they are likely sensitive to BfUbb due to the presence of the highly conserved ButCD homologues (>99.7% sequence identity) and the inclusion of the BfUbb-binding residue, Tyr119, within the targeted PPIase (Fig. 6a). Therefore, we examined whether BfUbb could decrease the abundance of these ETBF in the mammalian gut, which may serve as a potential therapeutic intervention for human diseases associated with ETBF.
a Amino acid sequence alignment of PPIase (AA 103-120) from disease-associated ETBF strains (86-5443-2-2, ATCC 43858, ATCC 43859, and ATCC 43860), B. fragilis GS077, and BfUbb-encoding B. fragilis NCTC 9343. Conserved residues are shaded in red, similar residues in yellow. The asterisk indicates the Tyr119 involved in BfUbb sensitivity. b MIC50 assays of ETBF ATCC 43860 strain with BfUbb, tetracycline, and chloramphenicol. The table summarizes molar concentrations (nM) or concentration range (in brackets) leading to 50% bacterial growth inhibition. c Agar spot assays show BfUbb inhibition against B. fragilis GS084, B. fragilis GS077, ETBF ATCC 43860, and ETBF-PPIGS084 strains. In ETBF-PPIGS084, the amino acids 110-120 of PPIaseETBF were replaced with the corresponding amino acids of PPIaseGS084. Experiments were conducted at least three times with consistent results. d Study design overview. The timeline shows C57BL/6 J mice conditions before and after gavage of strains (red triangle). Mice (n = 5/group) were colonized with ETBF (pink rods) or ETBF-PPIGS084 (blue). Group 1 and 2 received L100-55 polymer or polymer-encapsulated BfUbb, and Group 3 received water as a control. e ETBF colonization in mice. Graphs depict levels of ETBF (red) and ETBF-PPIGS084 (green) in terms of log [c.f.u. /g faeces] (y-axis) over 28 days (x-axis) in the three groups. Each group represents n = 5 mice and each dot represents an individual mouse. An unpaired two-tailed t-tests was used to analyze the statistical significance. NS, P > 0.05, not significant. ***P < 0.001. ****P < 0.0001. Exact P values are indicated in the figure. f Schematic of the postulated BfUbb functional mechanism. BfUbb enters the periplasm via ButCD and targets PPIase, eradicating susceptible Bacteroides strains such as some strains of B. fragilis (green panel). BfUbb-resistant strains employ two distinct strategies for defence: (i) producing diverse ButCD homologues to impede BfUbb entry in different Bacteroides species (blue panel) or (ii) generating off-target PPIases that fail to interact with BfUbb in resistant B. fragilis strains, thereby evading its lethal effects (yellow panel). The inner membrane (IM), peptidoglycan (P) and outer membrane (OM) of recipient bacteria are shown.
Consistent with the finding above, both purified and secreted BfUbb efficiently inhibit cell viability in a disease-related ETBF ATCC 43860 strain in vitro (Supplementary Fig. 14a, b). The MIC50 of BfUbb against this ETBF strain is 100-fold lower than that of tetracycline and chloramphenicol (Fig. 6b and Supplementary Fig. 14c). The replacement of ETBF’s PPIase110-120aa with that from BfUbb-resistant B. fragilis strains reversed the sensitivity of ETBF to BfUbb, confirming that the BfUbb-sensitivity of ETBF is also determined by the PPIase (Fig. 6c and Supplementary Fig. 14b).
To investigate whether BfUbb could decrease ETBF abundance in the mammalian gut, we co-colonized three groups of antibiotic-treated mice with both BfUbb-sensitive ETBF and an equal amount of BfUbb-resistant ETBF-PPIGS084 (Fig. 6d). One day post BfUbb gavage (L100-55 polymer encapsulated BfUbb), the abundance of the BfUbb-sensitive ETBF decreased ~100-fold compared to vehicle alone (Fig. 6e, Group 1 and 2). After 7 days of daily BfUbb administration, the population of the BfUbb-sensitive ETBF dropped ~ 1000-fold, while the BfUbb-resistant ETBF-PPIGS084 remained colonization (Fig. 6e, Group 2). In contrast, ETBF in both the BfUbb-untreated and control groups showed stable colonization within the mouse intestine and maintained high abundance (Fig. 6e, Groups 1 and 3). Altogether, these results indicate that BfUbb administration can specifically and effectively eliminate certain disease-associated ETBFs in vitro and in vivo, suggesting its potential as a supporting therapeutic against ETBF-involved disorders.
Discussion
In the present study, we demonstrate that ButCD serves as a distinct and conserved TonB-dependent outer-membrane transporter in B. fragilis, facilitating the transport of BfUbb and governing antagonistic range of BfUbb across different Bacteroides species.
Here, we further identify molecular strategies employed by other resistant Bacteroides strains to evade BfUbb-mediated killing. Previously, we identified a single mutation of Tyr119 on the BfUbb-targeted PPIase, leading to evasion of BfUbb targeting by certain strains of B. fragilis and conferring resistance against intraspecies competition (Fig. 6f). Additionally, in order to evade interspecies antagonism, those Bacteroides strains harbouring susceptible PPIase successfully avoid killing from BfUbb by virtue of their ButCD homologues which lack the ability to import BfUbb (Fig. 6f). Our findings collectively elucidate the diverse and nuanced strategies employed by Bacteroides strains to mount a defence against a BfUbb threat.
Consistent with findings above, the strain-level analysis on human gut metagenome samples (3 consortium gene catalogue (3CGC44) dataset) revealed that although all other Bacteroides species encoded BfUbb-sensitive PPIase (Supplementary Fig. 1), there was no significant disparity in the relative abundance of other Bacteroides species between samples with detectable BfUbb-encoding B. fragilis strains and those without (Supplementary Fig. 15a and Supplementary Table 3). However, a significant decrease in the relative abundance of sensitive PPIase-encoding B. fragilis strains was observed in the samples where BfUbb-encoding strains were detected compared to those where they were undetected (Supplementary Fig. 15b and Supplementary Table 3). These findings further support the intraspecies antagonistic specificity of BfUbb and suggest that alterations in the abundance of sensitive PPIase-encoding B. fragilis strains may be associated with variations in the presence of BfUbb-encoding strains. However, the abundance of non-B. fragilis Bacteroides species is not correlated with the presence of BfUbb in the community.
ButCD in B. ovatus, B. xylanisolvens, and B. thetaiotaomicron exhibits a high degree of amino acid sequence conservation (Fig. 4c and Supplementary Fig. 13), allowing BoUbb to effectively exploit the ButCD of these diverse Bacteroides species and confer a broader range of antagonism (Fig. 4i). However, due to less amino acid sequence conservation of ButCD from B. fragilis compared to other Bacteroides species (Fig. 4c and Supplementary Fig. 13), BfUbb is unable to enter the periplasm of non-B. fragilis species efficiently, resulting in a narrow target in species antagonism (Fig. 4d). Therefore, the differences in strain specificity between BoUbb and BfUbb primarily stem from their ability to enter recipient cells, which is determined by the unique characteristics of ButCD within recipient cells. These findings offer a comprehensive elucidation of the species-specific toxicity exhibited by BfUbb and the broader range of antagonism demonstrated by BoUbb.
SusCD complexes, which serve as transporters of glycans, are abundant and widely distributed in Bacteroides. While several TBDTs associated with oligopeptide or bacteriocin import have been identified, no analogous cases have been reported in Bacteroides. ButCD, responsible for BfUbb import, exhibits distinct characteristics compared to other SusCD proteins in terms of gene loci and evolutionary classification, strongly suggesting that the native substrate(s) of ButCD are different from glycans. Considering the presence of a zinc-dependent metalloprotease upstream of the ButC gene locus, it is plausible that this ButCD plays a crucial role in recognizing and importing protein substrates as a source of amino acids for their incorporation into newly synthesized proteins. Therefore, BfUbb is internalized through a potential protein-specific nutrient uptake system, as hypothesized by Chatzidaki-Livanis et al.25. However, further investigations are required to elucidate exact function and biological substrates of ButCD.
It has been proposed that the archetypal SusCD-like systems generally have a total substrate size limit of ~5 kDa20. Our western blot analyses for detecting BfUbb transport did not show any degradation of BfUbb (~8.8 kDa), raising an intriguing question regarding whether BfUbb requires unfolding and refolding during import or whether ButCD can directly transport larger substrates such as BfUbb. Further structural and biochemical investigations are necessary to elucidate how ButCD recognizes and transports BfUbb.
Although the TonB-ExbB-ExbD system has been extensively investigated in Gram-negative bacteria and plays a pivotal role in bacterial survival under nutrient-limited conditions40,41, the inclusion of two ExbBs and two ExbDs within one TBD system has not been widely reported. Considering the lack of significant similarity between 2 ExbBs or 2 ExbDs, and that complementation with either exbB or exbD is sufficient to restore normal BfUbb transport, it remains unknown whether all these ExbBs and ExbDs are expressed and form the TBD complex, as well as what proportions they adopt to constitute this complex.
BfUbb shows the potential to eliminate disease-associated ETBF. With its precise targeting, BfUbb is anticipated to minimize disturbance to the gut microbiota compared to conventional antibiotic treatments. Considering significant involvement of ETBF in the onset of IBD and CRC, BfUbb holds potential as a viable therapeutic option against ETBF-associated diseases, particularly in early-stage intervention. Given that the bft gene, which encodes the B. fragilis toxin, is located on a transposable element that has mobilized across the B. fragilis species47, including those carrying non-susceptible PPIase, it is highly plausible that certain ETBF strains exhibit resistance to BfUbb. Therefore, a limitation arises due to the insensitivity of certain ETBF strains to BfUbb exposure, restricting its applicability in managing all ETBF-associated diseases.
Methods
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Supplementary Table 4. All Bacteroides strains were cultured in liquid brain heart infusion medium with L-cysteine (1 g/L), hemin (5 mg/L), and vitamin K1 (0.25 mg/L) (BHI), or on BHI agar plates at 37 °C in an anaerobic chamber48. E. coli strains were grown aerobically in lysogeny broth (LB) medium at 37 °C. Antibiotics were added to the medium as follows when required: kanamycin 100 mg/mL, ampicillin 50 mg/mL, gentamicin 200 mg/mL, erythromycin 25 mg/mL, chloramphenicol 10 mg/mL, and tetracycline 10 mg/mL. Anhydrotetracycline (aTC) was dissolved in 100% ethanol at 2 mg/mL as stock solution and diluted 10,000 × when used for counter selection.
Plasmid construction
All plasmids used in this study are presented in Supplementary Table 5. For recombinant protein expression in E. coli, BfUbb or BoUbb lacking its SPI signal was cloned into the pET28a vector, introducing an N-terminal 6 × His-SUMO (small ubiquitin-like motif) tag. PPIase genes from various Bacteroides strains were cloned into a pET15b vector including an N-terminal signal peptide pelB for periplasmic expression and linked with a C-terminal 6 × His tag. For in situ genetic manipulation in Bacteroides, ~ 900 bp overlap upstream and downstream of the target region were cloned into pSIE-Bfe1-CmR vector containing Bfe1 as counter selective marker49. A 6 × His tag was introduced into C-terminus of ButD in situ via pSIE-Bfe1-CmR to obtain ButCD complex. For expression or complementation in Bacteroides, full length tonB3, exbB1, exbB2, exbB1B2, exbD1, exbD2, exbD1D2, and their ~ 250 bp upstream were cloned into pNBU2_CmR vector, respectively. Genes like butC, butD, butCD, butCBtD, and butCDBt were cloned into pNBU2_CmR vector, introducing a tonB3 promotor to enhance expression level50. Plasmids expressing BfUbb mutants or ButC mutants were obtained using quick change strategy and other constructs were generated using the Gibson assembly strategy. All plasmids were verified by DNA sequencing.
Protein expression and purification
E. coli BL21 (DE3) carrying a plasmid expressing 6 × His-SUMO-BfUbb construct were grown in 2 L LB medium to an optical density at 600 nm (OD600) of 0.8, then induced by addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and cultures were further incubated for 12 h at 18 °C. Bacterial cells were collected by centrifugation and the pellets were resuspended in 15 mL TBS buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl) each liter culture. Bacterial cells were lysed using a high-pressure cell crusher (Union-Biotech), the supernatants were collected, run through Ni-NTA agarose resin (Qiagen), washed with 20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 20 mM imidazole. The SUMO tag was removed with homemade 6 × His-tagged ULP1 protease at 25 °C for 3 h and proteins were further purified using Superdex 75 gel-filtration chromatography51 (GE Healthcare Life Sciences). As for BfUbb mutants and BoUbb, the same purification strategy was applied.
For purification of PPIases from different Bacteroides, plasmids encoding PPIases with a C-terminal hexahistidine were transformed into E. coli BL21 (DE3) and purified by Ni affinity chromatography first (Qiagen), further purified through an anion-exchange column (Hitrap Q, GE Healthcare) and Superdex 200 Increase gel-filtration chromatography (GE Healthcare) pre-equilibrated with TBS buffer.
Homologous recombination was used to add a 6 × His-tag to the C terminus of genomic ButD in B. fragilis GS077 strain52. To obtain ButCD-BfUbb complex, overnight cultures of the B. fragilis GS077 strain was grown about 12 h in BHI medium under anaerobic condition, and subcultured for 4 h next morning in fresh BHI to an OD600 ~ 0.8, then further cultured for 3 h treated with purified BfUbb at a final concentration of 10 μg/mL. Cells were harvested from 14 L BHI cultures through centrifugation, and the resulting pellets were resuspended in TBS buffer. Subsequently, high-pressure cell crusher (Union-Biotech) was employed for lysing the cells, followed by centrifugation at 17,000 g for 20 min to eliminate cellular debris. After ultra-centrifugation at 150,000 g for 60 min, the pellet was extracted with 100 mL 1.5% LDAO (n-Dodecyl-N,N-Dimethylamine-N-Oxide) in TBS buffer for 12 h by gentle stirring at 4 °C before ultracentrifugation for 30 min at 150,000 g. The supernatant was loaded onto a Ni-NTA column and after washing with TBS buffer containing 0.2% LDAO and 20 mM imidazole, protein was eluted with TBS buffer containing 0.2% LDAO and 300 mM imidazole. Protein was further purified by gel filtration using a Superdex 200 column in TBS buffer containing 0.1% LDAO. The protein from peak fraction was concentrated to 17 mg/mL for the cryo-EM experiments.
Protein interaction analysis experiment
Purified BfUbb (50 μg) and PPIase (80 μg) proteins were combined in a 3:1 ratio to test the binding of BfUbb to PPIases from other Bacteroides strains in vitro. 20 μL of Ni-agarose resin (GE Healthcare Life Sciences) was added after 1 h of incubation at 4 °C, and incubation continued for an additional 30 min. With TBS buffer containing 0.05% Triton X-100, the resins were washed three times. Using 300 mM imidazole in the TBS buffer, protein bonded to the resin was eluted. SDS-PAGE was used to evaluate the protein samples. Following the methods described above, PPIase from various Bacteroides strains was also tested for its ability to bind to BoUbb in vitro.
Overlay assay
Unless otherwise indicated, 50 ng of purified BfUbb and its variants were spotted onto standard 9 cm diameter BHI petri dishes and air-dried prior to overlaying with 4 mL of 0.8% BHI soft agar containing 100 μL of exponential phase (OD600 ~ 0.6) strains under investigation. Following anaerobic overnight incubation at 37 °C, the inhibitory zone was analyzed.
Agar spot assay
In brief, strains were cultivated anaerobically to exponential phase in 1 mL of BHI medium at 37 °C, and the density was adjusted to an OD600 ~ 0.8. In total, 200 μg pure toxin protein or 100 μL filtered supernatant concentrate was spread onto BHI plates and dried, unless otherwise indicated in the figure legend. After a 10-fold gradient dilution, 2.5 μL of each strain was spotted on plates containing pure toxin protein or filtered supernatant concentrate. After anaerobic overnight incubation at 37 °C, the inhibitory effect was assessed. When testing the toxicity of donor’s supernatant, 10 mL culture (OD600 ~ 0.8) was centrifuged at 9000 g for 10 min, filtered with a 0.22 μm filter, concentrated to ~ 500 μL.
Transposon mutagenesis
Random mutagenesis of B. fragilis GS077 was conducted using the transposon containing plasmid pMUT2D_TetR and individual mutants were screened using the agar spot assay for those that were no longer inhibited by BfUbb. To create pMUT2D_TetR, pSAM_BfN30 was modified by replacing the erythromycin resistance gene ermG with the tetracycline resistance gene TetR. The construct was verified by sequencing and transformed into E. coli S17-1 ʎ pir. This strain was used for conjugation with B. fragilis GS077 as described elsewhere53. Clones with transposon insertions were selected on 15 cm BHI agar with gentamicin, tetracycline, and 1 mg BfUbb. Transposon mutants gained BfUbb resistance were further confirmed on a BfUbb containing BHI plate then genomic DNA was purified for arbitrarily-primed PCR. The insertion sites were identified by genome walking and Sanger sequencing followed a described protocol54. Primers used for genome walking are listed in Supplementary Table 6.
Deletion mutation and complementation
Plasmids were transferred into Bacteroides strains by mating with E. coli S17-1 ʎ pir. Overnight cultures of E. coli S17-1 donor strains were diluted 100-fold in 2 mL LB medium containing ampicillin and Bacteroides recipients diluted 100-fold in 10 mL BHI medium. When the recipient strain reached an OD600 of 0.1–0.2 and the donor strain reached an OD600 of 0.2–0.6, donor and recipient strains were mixed at a 1:10 donor: recipient culture volume ratio, centrifuged at 9000 g for 10 min, resuspended in 100 μL of BHI liquid medium and spotted on 6 cm non-selective BHI agar plates for 20 h at 37 °C under aerobic condition to allow for conjugation. Mating lawns were resuspended in 1 mL LB, then 100 μL of suspension or 10-fold dilution was plated on BHI agar plates containing gentamicin and erythromycin or chloramphenicol. After cointegrates were verified by PCR, each strain was grown overnight in 1 mL BHI, then 100 μL of 10−3 dilution was plated onto BHI plate containing 200 ng/mL aTC. After 36 – 48 h, single colonies were restreaked and analyzed by PCR and DNA sequencing to distinguish wild type and mutant and confirm the loss of the selection marker. As for complementation, plasmids containing corresponding genes were inserted into attN site of each deletion mutant by conjugation and verified by PCR.
Western immunoblot analysis
For BfUbb access evaluation, strains to be tested were inoculated into 1 mL of BHI medium overnight, diluted into 1 mL fresh BHI medium at a ratio of 1:5, incubated for 1 h with 10 μg of purified BfUbb or BfUbb mutants, otherwise indicated in figure legends, then washed three times with PBS, lysed with lysis buffer (Sangon) and boiled in 6× sample buffer. Equivalent amounts of bacterial lysates were separated by electrophoresis using 15% Tricine-SDS-PAGE or 10% Glycine-SDS-PAGE gels. The contents of the gels were transferred onto polyvinylidene difluoride membranes (Millipore), which were blocked with 5% skim milk before being probed with the primary antibodies (Rabbit anti-BfUbb, this study, 1:500; Rabbit anti-DnaK, Cusabio #CSB-PA633459HA01EGW, 1:2500; Mouse anti-His, Abclonal #AE003, 1:5000) as indicated and horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit, MBL #458, 1:5000; goat anti-mouse, MBL #330, 1:5000).
Fluorescence microscopy
Strains were anaerobically grown overnight in BHI and then transferred to 1 mL of BHI with 5 μg/mL of BoUbb at 1:100 for continued culture for 4 h to investigate the morphology of BoUbb-treated strains. The medium was then supplemented with 5 μg/mL of FM 1-43 dye, and incubated for 1 h. The bacteria were examined using a Zeiss LSM900 laser scanning confocal microscope. To assess the internalization of BfUbb into Bacteroides, BfUbb was first fluorescently labelled using a Cy3-SE fluorescent dye (Solarbio, Beijing, China) and purified using gel filtration chromatography as previously described55. Strains were anaerobically cultivated overnight in BHI before being sub-cultured into 1 mL of BHI containing 40 μg of fluorescent dye Cy3 labelled BfUbb at 1:25 for continued incubation for 4 h. The samples were then washed at least three times with BHI medium and suspended in 100 μL of BHI medium before observation. The BfUbb-Cy3 treated bacteria were examined using a Zeiss LSM900 laser scanning confocal microscope.
Growth curve measurement
Bacteria single colony was picked from a fresh plate and inoculated into pre-reduced 5 mL BHI medium for overnight culture. Then, strains to be tested were diluted 1:100 in 10 mL fresh BHI medium or 1:50 in 10 mL fresh minimum media, respectively. Bacteria samples were collected every 1.5 h and OD600 readings were recorded using microplate spectrophotometer (Tecon). Prism version 9.3.0 (GraphPad Software, San Diego, CA) was used to calculate the data, which are shown as the mean of biological triplicates with the SEM depicted as error bars.
Co-culture
To simplify screening on BHI plates, the donor and recipient strains were rendered erythromycin- or chloramphenicol-resistant by harbouring a pNBU2-ermG or pNBU2-CmR plasmid. Donor and recipient strains were cultivated overnight in 5 mL of BHI medium as start culture, then sub-cultured together into 10 mL fresh BHI medium at 1:100. The co-culture mixture was collected after 8 h incubation and separately plated on selective BHI plates containing erythromycin or chloramphenicol to count the donor and recipient cell number.
Cryo-EM sample preparation and data collection
Aliquots of 4 µL of the BfUbb-ButCDBf complex at a concentration of ~17 mg/mL were applied onto glow-discharged holey carbon-coated grids (Quantifoil Au R1.2/1.3, 200 mesh, Beijing Zhongjingkeyi Technology, Beijing, China). Following a 5 s incubation on the grids under 100% humidity, the grids were blotted for 3 s at 8 °C using a blot force of 0, and then plunge-frozen into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific, Waltham, MA). The grids were transferred to a Titan Krios (Thermo Fisher Scientific) operating at 300 kV for data acquisition. EPU software (Thermo Fisher Scientific) was used for automated data collection on a Falcon 4i counting camera with a defocus range of −0.5 – −2.0 μm and at a nominal magnification of 105,000x, resulting in a calibrated pixel size of 1.18 Å. The accumulated dose was set to 60 electrons per Å2 and a total of 31 frames per movie. Data acquisition parameters can be found in Supplementary Table 2.
Image processing
A total of 5468 multi-frame movies were collected and subsequently processed using cryoSPARC56. Drift correction and dose-weighting were carried out using Patch Motion Corr. Contrast transfer function (CTF) estimation of motion-corrected micrographs was conducted using Patch CTF in cryoSPARC. Approximately 208,926 particles were automatically picked from 500 micrographs, extracted, and classified in 2D to provide templates for template-based picking. Out of 5468 micrographs, a total of 2,571,527 particles were extracted with a box size of 300 pixels for two rounds of 2D classification, after which 995,386 particles were used for 3D classification. Two rounds of ab-initio reconstruction and heterogeneous refinement were used to discard the remaining bad particles, resulting in two class particles. The two class particles were then subjected to non-uniform refinement and local refinement, with C1 and C2 symmetry, respectively, resulting in two maps at 2.97 Å and 3.05 Å. A flowchart showing the data processing is shown in Supplementary Fig. 11.
Model building into cryo-EM maps
The initial model of ButCD from AlphaFold2 and BfUbb crystal structure (PDB ID:8HM1) were docked into electron microscopy density map using UCSF Chimera57, and manually adjusted in Coot58, followed by refinement using Phenix59 in real space with secondary structure and geometry restraints to prevent structure overfitting. Statistics of 3D reconstruction and model refinement are summarized in Supplementary Table 2.
Quantification of lid opening in diverse SusCD-like systems
To measure the degree of lid opening in diverse SusCD-like systems, we selected one amino acid residue (ButC_F728, RagA_Q670, and BT1762_Y636) located in the hinge loop L7 of the SusC-like protein as the original point, which is stable during the conformational changes20. Subsequently, we chose an amino acid residue (ButC_W352, RagA_W321, and BT1762_W295) from the SusC-like protein to form a reference line approximately aligned with the outer membrane (OM) plane, and another amino acid (ButD_Y71, RagB_G78, and BT1762_G64) from SusD-like proteins facing the cavity of barrel as another reference line to establish a comparable angle. The Cα atoms of these selected amino acid residues align closed during structural comparisons, respectively.
Sequence alignment and phylogenetic analysis
B. fragilis ButC (GS077_4426) and PPIase (GS077_2615) was used in a tblastn (BLAST + , v.2.12.0 + ) query (e-value≤ 1e − 10) against an in-house database composed of whole genome sequences of BfUbb sensitive strains and resistant strains. For BfUbb homologues in bacteria, the NCBI Nr database was searched (tblastn, e-value ≤ 1e − 10, bacteria limited and B. fragilis excluded) using B. fragilis BfUbb (BF9343_3779) and the top 100 hits were saved, combined, and sorted by bitscore. For ButC homologues in Bacteroides, Phoecicola, Parabacteroides, or Alistipes, the NCBI Nr database was searched (tblastn, e-value ≤ 1e − 10, Bacteroides limited) using B. fragilis ButC and the top 5000 hits were saved, combined, and sorted by bitscore. Only sequences with query coverage > 90% were included in subsequent analysis. For SusCD superfamily protein in B. fragilis GS077, we identified all possible surface lipoprotein SusD in GS077 through homology to IPR012944 (RagB/SusD domain). All possible TonB-dependent transporters (TBDTs) in GS077 were identified through homology with IPR000531 (TonB-dependent receptor-like, beta-barrel). Only those TBDTs with surface lipoprotein SusD present downstream were identified as candidate SusCD pairs in GS077.
The sorted list was parsed such that a taxonomically broad selection of top hits was retained, and the associated proteins were aligned together with the query sequence using MAFFT (v.7.487). The sequence alignment of the SusCD protein superfamily was also conducted by MAFFT, retaining the blocks in the alignment results. Maximum-likelihood phylogenetic trees were constructed using IQTREE (v.2.1.4_beta) after automatic model selection with nodal support tested via 1000 ultrafast phylogenetic bootstraps.
susCD pairs-containing loci classification of B. fragilis GS077
We first located and queried these candidate susCD gene clusters, and predicted and classified the possible biological functions of these candidate susCD pairs-containing loci. By comparing the gene functions in these 59 susCD pairs-containing loci, these susCD pairs-containing loci were initially classified. By comparing the susCD pairs-containing loci annotated in PULDB and combining with manual annotation and proofreading, the putative PULs (43 of 59 susCD pairs-containing loci) in GS077 were determined.
The classification of PUL is based on the diversity and copy number of glycoside hydrolases. Specifically, if a PUL has more types of glycoside hydrolases, then the PUL may be involved in the utilization and transport of complex polysaccharides. If the susCD pairs-containing loci only encodes peptidases and related transporters but not polysaccharide utilization-related genes, then the susCD pairs-containing loci may only be involved in peptide transport.
Elimination of enterotoxigenic Bacteroides fragilis in mice
All animal experiments were supervised and approved by the Animal Research Ethical Inspection Form of Shandong University School of Life Sciences (SYDWLL-2021-16). Seven-week-old C57BL/6 J female mice were purchased from GemPharmatech Co., Ltd. (Jiangsu, China). The mice were housed in laboratory cages under controlled conditions (25 ± 2 °C, 45 ± 5% humidity, 12 h light-dark cycle) with free access to autoclaved water and irradiated food. All the mice were given a week to acclimatize and were healthy prior to our studies described below.
Antibiotic cocktails (10 mg each of vancomycin, metronidazole, neomycin, and ampicillin per mice) were administered by oral gavage daily for 5 days. Subsequently, all antibiotic-treated mice had free access to autoclaved water supplemented with antibiotic cocktails (0.5 g/L vancomycin, 1.0 g/L metronidazole, 1.0 g/L neomycin, and 1.0 g/L ampicillin) and irradiated feed for 7 days. After antibiotic treatment, faecal pellets were collected and tested for bacterial growth on selective BHI agar (200 μg/mL gentamycin and 10 μg/mL chloramphenicol or 10 μg/mL erythromycin). Only mice without detectable bacterial growth on either medium were included in the study. The antibiotic-treated mice were randomly divided into three groups (five mice per group). All antibiotic-treated mice were inoculated with an equal mixture comprising 2.5 × 109 c.f.u. of both the BfUbb-sensitve ETBF strains and 2.5 × 109 c.f.u. of the BfUbb-insensitive ETBF-PPIGS084 strains. The faecal pellets were collected at the indicated time points. Colonization was monitored in fresh faecal samples that were weighed, mashed, and vortexed in 1 ml PBS buffer and diluted to count c.f.u. The diluted faecal samples were separately plated on selective BHI agar (200 μg/mL gentamycin and 10 μg/mL chloramphenicol or 10 μg/mL erythromycin).
To avoid protein inactivation during the oral route of BfUbb administration, L100-55 polymers were used to deliver BfUbb, as described previously60. Briefly, a total of 50 mg of BfUbb was dispersed into 5 mL of CaCl2 (0.5 M) solution first. Under gentle stirring conditions, use an injection pump to inject 5 mL of L100-55 polymer solution (10 mg/mL) into the mixture at a flow rate of 20 mL/h for 15 min. After the injection of L100-55 polymer solution, gentle stirring is still required for about 30–60 min. Then, HCl solution (pH = 1) was added to the mixture to maintain pH = 4. After centrifugation (200 g, 1 min), microspheres were collected and then stored at 4 °C before use. BfUbb was orally administered daily for 7 days, and 100 μL microspheres per mice per day. The control group was administered an equal amount of L100-55 polymer solution.
MIC test
The strains to be examined (OD600 ~ 0.6) were diluted 200-fold into 200 μL BHI medium with gradient doses of BfUbb, tetracycline, or chloramphenicol. Cell culture plates were incubated anaerobically for 12 h at 37 °C, and the OD600 was measured. For every gradient, three repeats were performed.
Strain-level relative abundance estimation in metagenomic samples
MetaPhlAn 461 and StrainPhlAn 4.162 was used to estimate the relative abundance of the indicated Bacteroides species or B. fragilis with a specific genotype in the metagenomics sample. To estimate the relative abundance of the indicated Bacteroides species, we utilized the species-specific marker genes obtained from MetaPhlAn’s reference species-specific marker genes database (updated to March 2024). Since the marker genes of B. fragilis (t__SGB1853, t__SGB1855, t__SGB104919 partial marker genes) do not encompass PPIase and BfUbb gene, we developed a customized database by incorporating the coding sequences of BfUbb and PPIase orthologues into the species-specific marker genes database of MetaPhlAn 4 to assess the relative abundance of B. fragilis with a specific genotype, following the guidelines provided in the MetaPhlAn 4 tutorial (refer to section “Customizing the database”). The PPIase subtypes in B. fragilis can be categorized as BfUbb-sensitive PPIase (B. fragilis GS077_2615 Sensitive PPIase: 100% nucleotide sequence identity) and BfUbb-insensitive PPIase (B. fragilis 9343_3784 type Insensitive PPIase: 98% nucleotide sequence identity; B. fragilis GS084_0204 Insensitive PPIase: 90% nucleotide sequence identity), based on their sensitivity to BfUbb. To ensure accurate classification of distinct subtypes of PPIase, we have selected a hypervariable region within the full-length PPIase gene as a marker, considering the high degree of nucleotide sequence identity of full-length PPIase gene. Specifically, we aimed to maintain <90% nucleotide sequence identity between each marker gene (BF9343_3784: 300-490 nt; GS077_2615: 300-490 nt; GS084_3784 500-948 nt). The full-length BfUbb gene serves as a marker gene for identifying BfUbb-encoding B. fragilis strains. The newly added marker genes were validated for correct indexing using the ‘extract_markers.py’ script in StrainPhlAn. Subsequently, the trimmed metagenomics input was mapped to the custom reference database of MetaPhlAn to determine the relative abundance of specific Bacteroides species or B. fragilis genotypes (using MetaPhlAn output parameters “-t rel_ab”). The resulting data from MetaPhlAn was subsequently utilized as input in StrainPhlan4.1 with default settings. Since the sample sizes in BfUbb-detected group and BfUbb-undetected group are unequal, unpaired two-tailed Welch’s t-test were employed to analyze the statistical significance.
It is noteworthy that in the majority of samples, we consistently observed a lower abundance of PPIase-encoding B. fragilis strains compared to the abundance of B. fragilis (determined using the default MetaPhlAn 4 markers) (Supplementary Table 3). Given that PPIase is an essential gene of B. fragilis, it would be expected that the abundance of PPIase-encoding B. fragilis strains should be close to that of B. fragilis. Therefore, a limitation in our strain-level analysis lies in the potential underestimation of the abundance of PPIase-encoding B. fragilis strains.
Statistics and reproducibility
At least three separate experiments were conducted independently, yielding consistent results. Unless otherwise stated, data are presented as the arithmetic mean ± s.d. GraphPad Prism v.9.3.0. (GraphPad) was used for all statistical analyses. Unpaired two-tailed Student’s t-tests were employed to analyze the statistical significance between two groups, unless otherwise stated. The following is an annotation of the significance of mean comparison: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant. Statistics were evaluated significant when P < 0.05. No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates and cryo-EM maps generated in this study have beendeposited in the Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB) under the accession code: 8YPT, 8YPU and EMD-39493, EMD-39494, respectively. The raw metagenomics data used in this study are available in the NCBI database under accession number SRR1778453 or listed in the ‘Metagenomics Sample-SRA ID’ column of Supplementary Table 3. Source data are provided with this paper.
References
Wexler, A. G. & Goodman, A. L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2, 17026 (2017).
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Chen, J. et al. New insights into the mechanisms of high-fat diet mediated gut microbiota in chronic diseases. iMeta 2, e69 (2023).
Zafar, H. & Saier, M. H., Jr. Gut Bacteroides species in health and disease. Gut Microbe. 13, 1–20 (2021).
Wexler, H. M. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621 (2007).
Chung, L. et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial Cells. Cell Host Microbe. 23, 203–214, (2018).
Wu, S. et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022 (2009).
Dejea, C. M. et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018).
El Tekle, G. & Garrett, W. S. Bacteria in cancer initiation, promotion and progression. Nat. Rev. Cancer 23, 600–618 (2023).
Cornforth, D. M. & Foster, K. R. Competition sensing: the social side of bacterial stress responses. Nat. Rev. Microbiol. 11, 285–293 (2013).
Peterson, S. B., Bertolli, S. K. & Mougous, J. D. The central role of interbacterial antagonism in bacterial life. Curr. Biol. 30, R1203–R1214 (2020).
Svanback, R. & Bolnick, D. I. Intraspecific competition drives increased resource use diversity within a natural population. Proc. Biol. Sci. 274, 839–844 (2007).
Garcia-Bayona, L. & Comstock, L. E. Bacterial antagonism in host-associated microbial communities. Science 361, eaat2456 (2018).
Ghoul, M. & Mitri, S. The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845 (2016).
Thomas, F., Hehemann, J.-H., Rebuffet, E., Czjzek, M. & Michel, G. Environmental and gut Bacteroides: the food connection. Front. Microbiol. 2, 93 (2011).
Wardman, J. F., Bains, R. K., Rahfeld, P. & Withers, S. G. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 20, 542–556 (2022).
Brown, H. A. & Koropatkin, N. M. Host glycan utilization within the Bacteroidetes Sus-like paradigm. Glycobiology 31, 697–706 (2021).
Glenwright, A. J. et al. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541, 407–411 (2017).
Gray, D. A. et al. Insights into SusCD-mediated glycan import by a prominent gut symbiont. Nat. Commun. 12, 44 (2021).
Robitaille, S. et al. Community composition and the environment modulate the population dynamics of type VI secretion in human gut bacteria. Nat. Ecol. Evol. 7, 2092–2107 (2023).
Coyne, M. J. & Comstock, L. E. Type VI secretion systems and the gut microbiota. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.PSIB-0009-2018 (2019).
Chatzidaki-Livanis, M., Geva-Zatorsky, N. & Comstock, L. E. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl Acad. Sci. USA 113, 3627–3632 (2016).
Wexler, A. G. et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl Acad. Sci. USA 113, 3639–3644 (2016).
Chatzidaki-Livanis, M. et al. Gut symbiont Bacteroides fragilis secretes a Eukaryotic-like ubiquitin protein that mediates intraspecies antagonism. mBio. 8, e01902–17 (2017).
Shumaker, A. M., McEneany, V. L., Coyne, M. J., Silver, P. A. & Comstock, L. E. Identification of a fifth antibacterial toxin produced by a single Bacteroides fragilis strain. J. Bacteriol. 201, e00577–18 (2019).
McEneany, V. L., Coyne, M. J., Chatzidaki-Livanis, M. & Comstock, L. E. Acquisition of MACPF domain-encoding genes is the main contributor to LPS glycan diversity in gut Bacteroides species. Isme J. 12, 2919–2928 (2018).
Roelofs, K. G., Coyne, M. J., Gentyala, R. R., Chatzidaki-Livanis, M. & Comstock, L. E. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition In Vivo. mBio. 7, e01055–16 (2016).
Chatzidaki-Livanis, M., Coyne, M. J. & Comstock, L. E. An antimicrobial protein of the gut symbiont Bacteroides fragilis with a MACPF domain of host immune proteins. Mol. Microbiol. 94, 1361–1374 (2014).
Bao, Y. et al. A common pathway for activation of host-targeting and bacteria-targeting toxins in human intestinal bacteria. mBio. 12, e0065621 (2021).
Evans, J. C. et al. A proteolytically activated antimicrobial toxin encoded on a mobile plasmid of Bacteroidales induces a protective response. Nat. Commun. 13, 4258 (2022).
Matano, L. M., Coyne, M. J., Garcia-Bayona, L. & Comstock, L. E. Bacteroidetocins target the essential outer membrane protein BamA of Bacteroidales symbionts and pathogens. mBio. 12, e0228521 (2021).
Jiang, K. et al. Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome. Nat. Microbiol. 9, 70–84 (2024).
Schwaim, N. D. III, Townsend, G. E. II & Groisman, E. A. Prioritization of polysaccharide utilization and control of regulator activation in Bacteroides thetaiotaomicron. Mol. Microbiol. 104, 32–45 (2017).
Pudlo, N. A. et al. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio. 6, e01282–15 (2015).
White, P. et al. Exploitation of an iron transporter for bacterial protein antibiotic import. Proc. Natl Acad. Sci. USA 114, 12051–12056 (2017).
Gomez-Santos, N., Glatter, T., Koebnik, R., Swiatek-Polatynska, M. A. & Sogaard-Andersen, L. A TonB-dependent transporter is required for secretion of protease PopC across the bacterial outer membrane. Nat. Commun. 10, 1360 (2019).
Kleanthous, C. Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nat. Rev. Microbiol. 8, 843–848 (2010).
Silale, A. & van den Berg, B. TonB-dependent transport across the bacterial outer membrane. Annu. Rev. Microbiol. 77, 67–88 (2023).
Noinaj, N. et al. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 (2010).
Braun, V., Ratliff, A. C., Celia, H. & Buchanan, S. K. Energization of outer membrane transport by the ExbB ExbD molecular motor. J. Bacteriol. 205, e0003523 (2023).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Terrapon, N. et al. PULDB: the expanded database of polysaccharide utilization loci. Nucleic Acids Res. 46, D677–D683 (2018).
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Liu, Q.-Q. et al. Enterotoxigenic Bacteroides fragilis induces the stemness in colorectal cancer via upregulating histone demethylase JMJD2B. Gut. Microbe. 12, 1788900 (2020).
Hecht, A. L., Casterline, B. W., Choi, V. M. & Wardenburg, J. B. A two-component system regulates Bacteroides fragilis toxin to maintain intestinal homeostasis and prevent lethal disease. Cell Host Microbe. 22, 443–448 (2017).
Franco, A. A. et al. Molecular evolution of the pathogenicity island of enterotoxigenic Bacteroides fragilis strains. J. Bacteriol. 181, 6623–6633 (1999).
Liu, C. et al. Enlightening the taxonomy darkness of human gut microbiomes with a cultured biobank. Microbiome 9, 119 (2021).
Garcia-Bayona, L. & Comstock, L. E. Streamlined genetic manipulation of diverse Bacteroides and Parabacteroides isolates from the human gut microbiota. mBio. 10, e01762–19 (2019).
Parker, A. C., Seals, N. L., Baccanale, C. L. & Rocha, E. R. Analysis of six tonB gene homologs in Bacteroides fragilis revealed that tonB3 is essential for survival in experimental intestinal colonization and intra-abdominal infection. Infect. Immun. 90, e0046921 (2022).
Jiang, K. et al. A strategy to enhance the insecticidal potency of Vip3Aa by introducing additional cleavage sites to increase its proteolytic activation efficiency. Eng. Microbiol. 3, 100083 (2023).
Li, R., Li, A., Zhang, Y. & Fu, J. The emerging role of recombineering in microbiology. Eng. Microbiol. 3, 100097 (2023).
Goodman, A. L. et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 6, 279–289 (2009).
Andrew, F. J., Hicks, L. D. & Minnick, M. F. A system for transposon mutagenesis of Bartonella bacilliformis. J. Microbiol. Methods 203, 106623 (2022).
Jiang, K. et al. Functional characterization of Vip3Aa from Bacillus thuringiensis reveals the contributions of specific domains to its insecticidal activity. J. Biol. Chem. 299, 103000 (2023).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF chimera − a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. Struct. Biol. 60, 2126–2132 (2004).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D. Struct. Biol. 75, 861–877 (2019).
Wang, K. et al. Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target. Science 381, eadd5787 (2023).
Blanco-Míguez, A. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. 41, 1633–1644 (2023).
Truong, D. T., Tett, A., Pasolli, E., Huttenhower, C. & Segata, N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 27, 626–638 (2017).
Acknowledgements
We thank Dr. Andrew L. Goodman (Yale University) for technical supports and Drs. Chun-Jun Guo (Cornell University) and Yongjian Huang (Vanderbilt University) for discussion and careful review of the manuscript. The cryo-EM data were collected at Biomedical Research Center for Structural Analysis, Shandong University, and we thank Drs. Lei Qi and Dongfang He for their help of data collection. We are grateful to Dianli Zhao and Kang Li (Cryo-EM facility for Marine Biology, Laoshan laboratory) for their help of cryo-EM sample screening. We also thank Yuyu Guo, Sen Wang, Jingyao Qu, Xiaoju Li, Haiyan Sui and Chengjia Zhang from the core facilities for life and environmental sciences, SKLMT of Shandong University for their assistance in laser-scanning confocal microscopy and TEM experiments. The present study was supported by the National Key R&D Program of China (grant no. 2022YFA1304200 to X.G.), the National Natural Science Foundation of China (grant nos. 32122007 to X.G. and 323B2003 to W.X.L.), the Shandong Provincial Natural Science Foundation (grant nos. ZR2021JQ09 and ZR2023ZD58 to X.G.) and the Taishan Young Scholars Program (no. tsqn202408046 to K.J.).
Author information
Authors and Affiliations
Contributions
Conceptualization, X.G.; methodology, X.G., M.T., J.H.X., W.X.L., K.J., Y.Y., Z.C., X.Y.J, X.F.M., M.Y.W., J.H., H.A.L., S.J.L., and B.L.; investigation, M.T., J.H.X., W.X.L., K.J., Y.Y., Z.C., and X.Y.J.; validation, M.T., J.H.X., W.X.L., K.J., and Z.C.; writing—original draft, X.G., M.T., J.H.X., W.X.L., K.J., X.Y.J, and B.L.; writing—review and editing, X.G., W.X.L., M.T., K.J., J.H.X., and B.L.; resources, X.G., X.F.M., M.Y.W., J.H., B.L., and S.J.L.; funding acquisition, X.G., W.X.L., and K.J.; supervision, X.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tong, M., Xu, J., Li, W. et al. A highly conserved SusCD transporter determines the import and species-specific antagonism of Bacteroides ubiquitin homologues. Nat Commun 15, 8794 (2024). https://doi.org/10.1038/s41467-024-53149-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53149-w