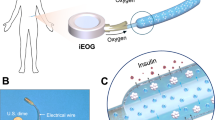
Abstract
In this study, we address the persistent challenge of providing adequate oxygen to transplanted cells by introducing a respiratoid biosystem. Central to our strategy is the chloroplast-transit-peptide (CTP), crucial for optimal oxygenation. Through conjugation of CTP with alginate, we achieve stabilization of chloroplast structure. Strategically anchored to the outer chloroplast membrane, CTP not only ensures structural integrity but also upregulates key photosynthesis-associated genes. This biosystem demonstrates exceptional efficacy in spontaneously generating oxygen, particularly under hypoxic conditions (~1% pO2). In an application, pancreatic islets encapsulated within the respiratoid biosystem and intraperitoneally implanted in diabetic mice maintain normal glucose levels effectively. Insulin secretion persists for 100 days post-xenotransplantation without the need for immunosuppressant administration, highlighting the reliance on the respiratoid biosystem’s oxygen supply and structural stability. Our study demonstrates the respiratoid biosystem as a platform in tissue engineering, offering a nature-inspired solution to the critical challenge of spontaneous oxygen supply.
Similar content being viewed by others
Introduction
Cell transplantation emerges as a prospective therapeutic strategy for addressing diseases associated with hormone or protein deficiencies. Despite notable advancements in this field, several challenges persist1. A fundamental hurdle in cell transplantation is the challenge of providing sufficient oxygen to survive encapsulated cells within 3-dimensional (3D) constructs2,3,4. Specifically, these cells are transplanted to a site with relatively low oxygen pressure and without an internal oxygen supply. In that case, they consume the available oxygen, creating a hypoxic environment inside the 3D structure. Furthermore, core regions of the 3D structure become severely hypoxic due to the limited oxygen diffusion from the surface to the core5,6. The hypoxic conditions trigger the stabilization and activation of hypoxia-inducible factors (HIFs) in encapsulated cells. In addition, hypoxia reduces the efficiency of the electron transport chain in mitochondria, leading to produce reactive oxygen species (ROS), such as superoxide radicals (O2•-), hydrogen peroxide (H2O2), and hydroxyl radicals (OH•)7,8,9. Both HIFs stabilization and ROS overproduction have the potential to damage deoxyribonucleic acid (DNA), induce mutations, and arrest the cell cycle10,11. To enhance the oxygenation of tissue constructs, various types of oxygen-releasing biomaterials (ORBs) have been developed to date. However, there are consistent challenges in the development of advanced ORBs that can address burst oxygen release and spontaneous oxygenation issues2,12,13. Also, in the oxygen decomposition process, byproducts such as ROS not only disrupt cellular pH and metabolic reactions but also induce cellular damage2.
Chloroplast, a type of membrane-bound plastid, plays a pivotal role in photosynthesis by storing solar energy in energy-storage molecules and releasing oxygen from water. Hence, they present a natural source of oxygenation. In our study, we employed a chloroplast source isolated from terrestrial plants, lowering the potential risk of biological contamination inherent in photosynthetic microorganisms14. However, the isolated chloroplasts remain structurally intact for only a few hours in a test-tube, and photosynthetic activity is typically lost within 30 min to half a day of light exposure15,16,17. The structure of the isolated chloroplasts is essential for maintaining the internal environment and enabling vital processes such as photosynthesis18. To overcome these constraints, we focused on the chloroplast proteins engaging with the translocon on the outer chloroplast membrane (TOC) and the translocon on the inner chloroplast membrane (TIC) complex19. The TOC and TIC are responsible for importing many chloroplast proteins, which contain transit peptides that signal their destination from the cytoplasm20. This TOC-TIC complex works together as a coordinated system and creates a channel or pathway across both membranes of the chloroplast envelope21. In this context, we hypothesized that the incorporation of a chloroplast-transit-peptide (CTP) derived from transmembrane domain (TMD) of TOC could potentially augment the structural stability of isolated chloroplasts through its affixation to the outer membrane.
As a proof of concept, we introduced a ‘respiratoid’ biosystem utilizing isolated chloroplasts, offering an approach to spontaneous oxygen generation (Fig. 1a). Unlike conventional alginate microcapsules (Fig. 1b), the respiratoid biosystem demonstrated the ability for continuous and self-sustained oxygen supply through activated chloroplasts with CTP, thereby improving the physiological conditions for therapeutic cells (Fig. 1c). The incorporation of CTP notably enhanced the expression of oxygen-evolving complex (OEC)-related genes and proteins within the photosystem II (PS II) complex, thereby augmenting oxygen production through strategic anchoring to the chloroplast’s outer membrane (Fig. 1d). Furthermore, the respiratoid biosystem demonstrated efficacy in sustaining oxygenation even in hypoxic conditions. Collectively, the respiratoid biosystem holds promise as a solution to address clinical challenges in tissue engineering associated with oxygen deficiency.
a The workflow for this study on the respiratoid biosystem. (1) CTP is conjugated to alginate (Al-CTP) for the matrix of respiratoid biosystem. (2) Respiratoid biosystem is implemented with Al-CTP and isolated chloroplasts. (3) CTP anchored to the outer chloroplast membrane. (4) This CTP not only augments the structural stability of isolated chloroplasts but also upregulates key photosynthesis-associated genes. b Dysfunction of encapsulated cells in alginate microcapsule due to hypoxic stress. c Maintenance of encapsulated cell’s function in the respiratoid biosystem due to sustained oxygen supply from the isolated chloroplasts activated by the CTP. The isolated chloroplasts can produce oxygen by consuming CO2, a cellular respiration by-product. d The magnified illustration depicts the CTP anchored to the isolated chloroplast’s outer membrane.
Results
Increased structural stability and oxygenation of isolated chloroplasts with CTP
To investigate whether the oxygenation efficiency of the isolated chloroplasts differs depending on the plant species, we measured their oxygen levels in hypoxic conditions (1% pO2). We selected spinach (Spinacia oleracea) as a chloroplast source due to its ability to maintain high oxygen levels for 20 h (Supplementary Fig. 1a). Moreover, the oxygen levels to the concentration of the isolated chloroplasts aimed at optimizing their functionality showed a positive correlation. We considered that excess ROS could be generated due to the Mehler reaction when chloroplasts are in excess22. Therefore, we determined that the optimal concentration to be used in vitro was 1.5 × 109 mL−1 for maximizing oxygen production while minimizing the production of cytotoxic H2O2 (Supplementary Fig. 1c)23.
First, we confirmed the effect of the CTP on isolated chloroplast. The CTP with a sequence of Met(M)-Phe(F)-Ala(A)-Phe(F)-Gln(Q)-Tyr(Y)-Leu(L)-Leu(L)-Val(V)-Met(M) was derived from the TMD of the TOC34 of Arabidopsis thaliana (At), which is known to anchor the outer membrane of the chloroplast (Fig. 2a, b)24. The purity and m/z value of CTP were 97.58% and 1261.65 Da, respectively (Supplementary Fig. 2a, b). We confirmed that CTP completely covered the surface of the isolated chloroplasts without damaging their structure for 20 h incubation, as shown by Rhodamine B staining, which confirmed the integrity of the chloroplasts (Supplementary Fig. 2c)25. The anchoring ability of CTP was maintained over 60 min as estimated by confocal microscopy. Thus, we refined the reaction time between the chloroplast and the CTP to 60 min, ensuring sufficient anchoring of chloroplast and CTP molecules (Supplementary Fig. 2d–f). The quantified release of chlorophyll notably diminishes after treatment CTP, suggesting its capacity to stabilize the structure of isolated chloroplast (Fig. 2c and Supplementary Fig. 2g)26,27. Therefore, the concentration of chloroplasts remaining after 14 days is higher following CTP treatment (Supplementary Fig. 2h).
a Schematic illustration of chloroplast’s structure and membrane. Chloroplast is constructed of a lipid bilayer membrane structure. The complexes of the translocon at the outer and inner envelope membrane of chloroplasts (TOC and TIC) mediate preprotein translocation across the chloroplast membrane. The sequence of the transmembrane domain (TMD) on TOC34, a 34-kDa integral membrane protein, is utilized as the source for chloroplast transit. Schematics is created in BioRender. Kim, H. (2024) BioRender.com/c78y188. b Chemical structure of CTP (MFAFQYLLVM). c Cumulated chlorophyll released from the destroyed chloroplasts for 14 days assessed by measuring absorbance 430 nm. Data were expressed as mean ± S.D. (n = 5 biological independent chloroplasts). Statistical significance for the comparison between chloroplast and chloroplast (+CTP) on each day was determined by unpaired, two-tailed t-test. P = 0.081 (day 0), P = 0.9240 (day 1), P = 0.0013 (day 3), P < 0.001 (day 5), P < 0.001 (day 7), P < 0.001 (day 10), P < 0.001 (day 14). d Comparison of oxygen levels of each chloroplast without (left) and with light exposure (right) at each time point under hypoxic conditions (1% pO2). Data were expressed as mean ± S.D. (n = 3 biological independent chloroplasts). a–d: statical significant differences (P < 0.05) in one-way ANOVA with two-sided Duncan’s multiple range test with adjustments for multiple comparisons. w/o light: P = 1.0 (time: 0 h), P < 0.001 (time: 5 h), P < 0.001 (time: 10 h), P < 0.001 (time: 20 h). w/ light: P = 1.0 (time: 0 h), P < 0.001 (time: 5 h), P < 0.001 (time: 10 h), P < 0.001 (time: 20 h). e Effects of CTP on the Y(II) (left) and ETR (right). Magnified Y(II) kinetics from 0 to 200 PAR in the Y(II) graph. Data were expressed as mean ± S.D. (n = 3 biological independent chloroplasts). f Schematic illustration of PS II complex structure including OEC ((Mn)4CaO5) and 3 extrinsic proteins (PsbO, PsbP, and PsbQ). g SDS-PAGE profile of each PS II complex at 14 days after isolation. Each lane and sample treatments are described in Methods. Schematics is created in BioRender. Kim, H. (2024) BioRender.com/r70l387. h Comparison of band intensity of PsbO, PsbP, and PsbQ subunit in each PS II complex according to g. Filled bar: w/o light. Empty bar: w/ light. Each protein level is expressed as relative band intensity quantified by Image J. Data were expressed as mean ± S.D. (n = 3 biological independent PS II). i Comparison of oxygen levels of each PS II at each time point under hypoxic conditions. Data were expressed as mean ± S.D. (n = 3 biological independent PS II). a–d: statical significant differences (P < 0.05) in one-way ANOVA with two-sided Duncan’s multiple range test with adjustments for multiple comparisons. P = 1.0 (time: 0 h), P < 0.001 (time: 5 h), P < 0.001 (time: 10 h), P < 0.001 (time: 20 h). Statistical significance was determined by unpaired, two-tailed t-test in c and h. Source data are provided as a Source Data file.
Next, we evaluated whether the CTP could improve the oxygen-generating capability of isolated chloroplast in a hypoxic condition with (w/) or without (w/o) light exposure. In parallel, we introduced aluminum (Al3+) to the chloroplast, which is known to inhibit oxygen evolution by damaging the subunit of photosystem II (PS II)28. CTP-treated chloroplasts maintained significantly higher oxygenation at 20 h, despite the treatment of Al3+ and/or no light (Fig. 2d and Supplementary Fig. 3a). Moreover, the CTP-treated chloroplast showed higher effective quantum efficiency of PS II (Y(II)), electron transport rate (ETR), the apparent quantum yield (α), and maximum photosynthesis rate (Pmax) values compared to those treated with Al3+ alone (Fig. 2e and Supplementary Table 1).
To investigate how CTP could improve the photosynthetic efficiency, we isolated the PS II complex comprised of three extrinsic proteins associated with oxygen-evolving (water-oxidation) activity in photosynthesis: PsbO (33 kDa), PsbP (23 kDa), and PsbQ (17 kDa) (Fig. 2f)29,30,31,32. Upon analyzing PS II by SDS-PAGE, a significant increase in the expression of PsbO, PsbP, and PsbQ subunits was maintained for 14 days after CTP treatment, compared to those treated with Al3+ alone (Fig. 2g, h, and Supplementary Fig. 3b, c). Correspondingly, the CTP-treated PS II maintained remarkably higher oxygenation for 20 h, despite the treatment of Al3+ (Fig. 2i). Based on these findings, we found that CTP could enhance the photosynthetic efficiency of chloroplast by maintaining the structural stability and increasing the content of PS II subunit in isolated chloroplasts. Additionally, we tested the effect of CTP on chloroplasts isolated from 4 different plant species to establish that CTP derived from Arbidopsis thaliana can also target chloroplasts of other plant species and improve oxygenation. The results confirmed that CTP could act universally on isolated chloroplasts regardless of the plant species (Supplementary Fig. 4).
Characterization of the molecular functionality of CTP to chloroplast with RNA-seq
To investigate what kinds of CTP-derived molecules were modulated for oxygen generation, we tried to identify the correlation between CTP and chloroplasts in molecular functions based on genetic information using RNA sequencing (RNA-seq). As a result of the analysis, a list of significantly upregulated genes was identified, and gene groups were identified using Gene Set Enrichment Analysis (GSEA)33. First, CTP treatment could significantly increase greater RNA contents (Supplementary Fig. 5). Using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) functional annotation, we compared the expression levels of the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologous groups between the without and with CTP treatment34,35. This analysis revealed that a total of 9 enriched pathways were upregulated at the gene level when chloroplast was treated with CTP. Then Gene Ontology (GO) analysis of mRNA-sequencing (mRNA-seq) quantifications was used to compare differences in gene expression (DEGs) in each pathway in chloroplast with and without CTP treatment (Fig. 3a). Much of these genes were associated with photosynthesis and were involved in the operation of thylakoid membrane proteins (Fig. 3b)36,37. In addition to the production of photosynthetic proteins, a notable upregulation of chloroplast ribosomal proteins was observed by CTP treatment (Fig. 3c). From these results, we found that the regulatory signal of CTP could overexpress the genes associated with photosynthesis and protein translation in the chloroplast. Collectively, this regulatory signal of CTP might promote the synthesis of the thylakoid membrane proteins to ultimately activate photosynthesis and oxygenation (Fig. 3d).
a Gene Ontology (GO) analysis of mRNA-sequencing (mRNA-seq) quantifications. The upregulated gene list treated with CTP (P ≤ 0.05) was subjected to DAVID functional annotation. This analysis resulted in the identification of gene clusters that corresponded to specific GO annotation terms. Extracted GO terms corresponding to biological processes (KEGG_PATHWAY). Plot depicting the significantly associated terms (P ≤ 0.05) with the gene list. The numbers of total genes for each term are represented as a percentage of the total list of KEGG genes. Additionally, the fold change and the adjusted P-value are provided for each term. Statistical significance was determined using an unpaired, two-tailed t-test. Results are expressed as -log (adjusted p-value), transforming smaller p-values into larger values to improve clarity and enable easier comparison and visualization of statistical significance. (n = 3 biological independent chloroplasts). b Functional cluster analysis of the significantly upregulated gene list (P ≤ 0.05) following CTP treatment, performed using DAVID. The analysis resulted in the identification of distinct gene clusters. The representation of the most significantly enriched cluster showcases GO terms associated with cell proliferation and movement. Filled circles represent GO terms. Schematic representation of the association between GO terms and their corresponding genes. Lines connect the GO terms to the respective genes, while the fold change for each term is indicated. c Heatmap of the relative abundance of chloroplast compositional genes profiling related to the photosynthesis, electron transport chain, and translation compared by KEGG. Z scores are calculated from Reads Per Kilobase of transcript per Million mapped reads (RPKM). d Schematic illustration depicting the regulatory signaling pathway by which CTP enhances the oxygen-releasing capacity of the chloroplast, leading to elevated rates of protein synthesis and subsequent activation of photosynthesis. e qRT-PCR results of chloroplast treated with CTP (0.2 mM) and transfected with siRNA (100 µM) of psbA, psbO, rps12, and rpl14 under hypoxic conditions (1% pO2). The relative mRNA expression level was normalized to that of ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL). Ch: Chloroplast. Sample treatments are described in Methods. Data were expressed as mean ± S.D. (n = 3 biological independent chloroplasts). f Oxygen kinetics of chloroplast treated with CTP and transfected with siRNA of psbA, psbO, rps12, and rpl14 at 37 °C for 20 h under hypoxic conditions. Data were expressed as mean ± S.D. (n = 6 biological independent chloroplasts). The shadowed area: S.D. Statistical significance was determined by unpaired, two-tailed t-test in e. Source data are provided as a Source Data file.
To further investigate chloroplast and CTP molecular profiles, we evaluated whether the treated CTP induces the gene expression related to chloroplast protein structure and photosynthesis pathway. To this end, we designed small interfering RNA (siRNA) sequences with a Turbo si-Designer algorithm for eliminating nonfunctional siRNA sites and measured the mRNA level of PS II and ribosome subunit after transfection of siRNAs that were (i) a key protein of the PS II, protein D1 genes (psbA), (ii) the largest extrinsic subunit of the OEC in PS II, protein O genes (psbO), (iii) a subunit of ribosome large units that related mRNA translation pathways, ribosomal protein S12 genes (rps12), and (iv) a subunit of ribosome small units, ribosomal protein L14 genes (rpl14). Notably, we found that CTP treatment significantly upregulated mRNA levels of psbA, psbO, rps12, and rpl14 although siRNAs were transfected into the chloroplast in hypoxic or normoxic environment (Fig. 3e and Supplementary Fig. 6). In terms of oxygenation, CTP treatment normally restored the function of siRNA-treated chloroplast regardless of siRNA transfection (Fig. 3f). Collectively, these results suggested that CTP treatment could induce a molecular reaction with oxygenation-related genes in chloroplasts.
Optimization of the respiratoid biosystem
To fabricate a respiratoid biosystem, we chemically conjugated the N-terminus of the CTP to the carboxyl group of biocompatible alginate (Al) according to the feed molar ratio by using carbodiimide chemistry (Supplementary Fig. 7a). The CTP was successfully conjugated to alginate via an amide bond to obtain Al-CTP conjugate (Supplementary Fig. 7b–k). In addition, we observed the formation of Al-CTP hydrogels which were composed of the Al-CTP conjugates via an ionotropic cross-linking reaction (Supplementary Fig. 7l). Additionally, rheological analysis was performed to verify the rigidity of the hydrogels. As a result, the storage and loss modulus of Al-CTP hydrogels were similar to those of conventional alginate hydrogel, in contrast to the 1:10 and 1:50 ratios (Supplementary Fig. 7m, n). These results demonstrated that the conjugation of CTP to alginate does not affect the formation of hydrogels. Thereby, we optimized the feed molar ratio of 1:1 for testing. The CTP conjugation efficiency was approximately 75.36%, confirming efficient conjugation to alginate at the 1:1 feed molar ratio (Supplementary Fig. 7o).
To evaluate the oxygenation capacity of the isolated chloroplasts based on the bonding type between CTP and alginate, we measured the oxygen level of microcapsules made by mixing alginate with CTP and chloroplasts, as well as microcapsules made using Al-CTP conjugate and chloroplasts. We found that the oxygenation efficiency of the isolated chloroplasts is higher when CTP is conjugated to alginate than non-conjugated CTP microcapsules (Supplementary Fig. 8). To establish the comprehensive respiratoid biosystem, it is essential for the Al-CTP conjugate to interact effectively with isolated chloroplast. In addition, ensuring structural stability is crucial, especially when incorporating therapeutic cells. Consequently, we systematically assessed the feasibility of forming the respiratoid biosystem by encapsulating the Al-CTP conjugate, chloroplast, and therapeutic cells in the encapsulator (Supplementary Fig. 9). This involved optimization of conditions, considering factors such as size and spherical shape across various settings (encapsulator setting condition, like frequency, electrode, and flow rate) (Supplementary Table 2 and Supplementary Fig. 10). Furthermore, the oxygen production rate of the respiratoid biosystem was not affected although the mechanical property of the respiratoid biosystem was changed with different Ca2+ concentration (Supplementary Fig. 11), suggesting that the degradability of respiratoid biosystem could be simply controllable by different Ca2+ concentration without affecting oxygen production.
Enhanced viability and functionality of therapeutic insulin-secreting islets encapsulated in respiratoid biosystem
To investigate whether the chloroplast influenced the morphology of microcapsules, each group of microcapsules was examined by scanning electron microscopy (SEM) analysis. SEM microscopy showed that all microcapsule groups displayed a spherical shape and a compact structure (Supplementary Fig. 12). Moreover, the surface of Al-Ch and the respiratoid biosystem exhibited the existence of chloroplasts within them, as indicated by their non-homogeneous microstructure. We observed the successful encapsulation of the pancreatic islets in all microcapsule groups (Fig. 4a). To confirm the stability of the chloroplast encapsulated within the microcapsule, we measured the amount of chlorophyll released from the microcapsule (Fig. 4b and Supplementary Fig. 13a). The respiratoid biosystem showed a lower amount of chlorophyll release for 14 days, compared to Al-Ch. However, both microcapsules encapsulating chloroplasts have remained spherical shape for 14 days. (Supplementary Fig. 13b). Next, we assessed whether the respiratoid biosystem could generate oxygen under hypoxic conditions (Supplementary Fig. 14a, b). The respiratoid biosystem also remarkably increased oxygen levels and maintained them compared to other microcapsules (Fig. 4c, d). We also confirmed that the area under curve (AUC) value, an indicator of total oxygen production, of the respiratoid biosystem was significantly higher than that of both the Al-CTP and Al-Ch microcapsule (Supplementary Fig. 14c). Even though oxygen-consuming islets were encapsulated, the respiratoid biosystem still highly generated and sustained oxygen level. (Supplementary Fig. 14d–f).
a Schematic illustration and bright field images of encapsulated islets in each microcapsule. Al: alginate microcapsule. Al-CTP: microcapsule using Al-CTP conjugate, Al-Ch: microcapsule using alginate and chloroplast. Scale bar: 250 μm. Schematics is created in BioRender. Kim, H. (2024) BioRender.com/v89u207. b Cumulated chlorophyll released from the microcapsules for 14 days was assessed by measuring absorbance 430 nm. Data were expressed as mean ± S.D. (n = 4 biological independent chloroplasts). Statistical significance for the comparison between Al-Ch (w/o islet) microcapsule and respiratoid biosystem (w/o islet) on each day was determined by unpaired, two-tailed t-test in b. P = 0.094 (day 0), P = 0.1405 (day 1), P = 0.0319 (day 3), P = 0.0017 (day 5), P < 0.001 (day 7), P = 0.0010 (day 10), P = 0.0014 (day 14). Oxygen kinetics of each microcapsule under hypoxic conditions. Dissolved oxygen of microcapsules without (c) or with (600 IEQ 150 microcapsules−1) oxygen-consuming islet (d) was measured by placing the same number of microcapsules per well. Magnified oxygen kinetics from 600 to 1200 min (right). Data were expressed as mean or mean ± S.D. (n = 3 biological independent cells). a–d: statical significant differences (P < 0.05) in one-way ANOVA with two-sided Duncan’s multiple range test with adjustments for multiple comparisons. e Viability of intact islets and encapsulated islets after 20 h incubation under hypoxic conditions assessed by live/dead assay. Green fluorescence (calcein-AM): live cells. Red fluorescence (EthD-1): dead cells. Scale bar: 250 μm. (n = 4 biological independent cells). f Viability of intact islets and encapsulated islets after 20 h incubation under normoxic or hypoxic conditions assessed by quantitative CCK-8 assay. Data were expressed as mean ± S.D. (n = 4 biological independent cells). g Glucose-stimulated insulin secretion (GSIS) of intact islets and encapsulated islets after 2 h incubation with low glucose (2.8 mM) and high glucose (20.2 mM) conditions. Data were expressed as mean ± S.D. (n = 4 biological independent cells). h Stimulation indexes (SI) of intact islets and encapsulated islets assessed by GSIS assay. Data were expressed as mean ± S.D. (n = 4 biological independent cells). P < 0.001 (intact islet under normoxia versus intact islet under hypoxia), P = 0.0011 (intact islet under hypoxia versus respiratoid biosystem under hypoxia). i Representative immunofluorescence images of islets after 20 h incubation under normoxic or hypoxic conditions. Green fluorescence: the expression of insulin (INS). Red fluorescence: the expression of HIF-1α. Blue fluorescence: islet nuclei (NUC) stained with DAPI. Scale bar: 200 μm. (n = 4 biological independent cells). Statistical significance was determined by unpaired, two-tailed t-test in b, f, g, and h. Source data are provided as a Source Data file.
Next, we assessed the effect of the respiratoid biosystem on the viability of encapsulated insulin-secreting islets in hypoxic conditions. The islet enclosed within the respiratoid biosystem demonstrated sustained viability, as evidence by a reduced incidence of cell death signals compared to both intact islets and Al-CTP microcapsule (Fig. 4e). While the viability of intact islets was significantly decreased in hypoxic conditions, islets encapsulated within the respiratoid biosystem exhibited a remarkable improvement in viability (Fig. 4f). To evaluate the capacity of the respiratoid biosystem to sustain the functionality of encapsulated islets, we conducted a glucose-stimulated insulin secretion (GSIS) test. In the hypoxic environment, islets experienced impaired glucose responsiveness, leading to a malfunction in insulin secretion (Fig. 4g). Nevertheless, islets encapsulated within the respiratoid biosystem exhibited a similar insulin secretion pattern to the islet in normoxic conditions, even when subjected to hypoxic conditions. In addition, the stimulation index (SI), a marker of glucose responsiveness, for islets within the respiratoid system was fully restored despite the hypoxic environment (Fig. 4h). Immunostaining was performed to assess the expression of HIF-1α (red color) and insulin (green color) in encapsulated islets (Fig. 4i). We chose HIF-1α as a representative biomarker of hypoxia due to its high specificity for hypoxia compared to other markers38,39,40. In general, under normoxic condition (21% pO2), HIF-1α is degraded via its association with the prolyl hydroxylase domain (PHD) and von Hippel-Lndau (VHL) protein. Under hypoxic condition (1% pO2), the activity of PHD protein is inhibited due to lack of oxygen, resulting in the accumulation of HIF-1α. As result, Encapsulated islets in the respiratoid biosystem showed significantly lower HIF-1α and higher insulin intensity under hypoxic conditions compared to other microcapsule groups.
Improved therapeutic effect of insulin-secreting pancreatic islets in the respiratoid biosystem in vivo
To extend the application of respiratoid biosystem for tissue engineering, an islet xenotransplantation was implemented. We conducted subcutaneous (SQ) transplantation to assess the oxygenation of the respiratoid system. Additionally, we performed intraperitoneal (IP) transplantation to evaluate the efficacy of the islets within the respiratoid for curing type 1 diabetes (T1D) (Supplementary Fig. 15). The chloroplast concentration used in the respiratoid biosystem was optimized for in vivo experiments (Supplementary Figs. 16 and 17).
To evaluate the oxygenation capacity of the respiratoid biosystem, we measured oxygen levels in the subcutaneous region following implantation (Supplementary Fig. 18). Despite the typically low oxygen pressure in the subcutaneous region of the body, a significant increase in oxygen levels was detected after the subcutaneous implantation of the respiratoid biosystem with the oxygen-consuming islets (Fig. 5a)41,42. We found that the hydrogel microcapsules possessed sufficient water for photosynthesis even after an extended time (Supplementary Figs. 19 and 20)43. To assess glycemic correction with the respiratoid biosystem through subcutaneous transplantation, we monitored body weight and blood glucose levels of the diabetic mouse (Supplementary Fig. 21). The respiratoid biosystem exhibited superior efficacy compared to Al-CTP microcapsules in maintaining blood-glucose levels at least 14 days. Furthermore, immunostaining of the subcutaneously implanted respiratoid biosystem with islets revealed lower levels of HIF-1α (red color) and higher levels of insulin (green color) (Fig. 5b and Supplementary Fig. 22). In addition, the findings suggested that macrophages infiltration into the implanted respiratoid biosystem was not pronounced, providing enhanced protection for the islet. Moreover, the transplanted region of the respiratoid biosystem exhibited minimal fibrosis and normal histological characteristics, leading to the long-term preservation of nutrients/oxygen diffusion and insulin secretion (Fig. 5c and Supplementary Fig. 23)44,45,46.
a Comparison of oxygen levels of the transplanted subcutaneous region of each group at 14 days. Data were expressed as mean ± S.D. (n = 6 biological independent animals). b Representative immunofluorescence images of the transplanted subcutaneous region at day 14. Sham group: surgery control group. Green fluorescence: the expression of insulin (INS), Red fluorescence: the expression of HIF-1α, Blue fluorescence: islet nuclei (NUC) stained with DAPI, Dashed line: epidermis border. Arrow: transplanted microcapsules. Scale bar: 100 μm. (n = 3 biological independent cells). c Hematoxylin & Eosin (HE) and masson’s trichrome (MT) staining images of subcutaneous region transplanted in each group (w/ islet) at day 14 post-transplantation. Circle: encapsulated islets in microcapsules (in HE) and fibrosis region (in MT). Scale bar: 250 µm. d Non-fasting blood glucose levels of the STZ-induced diabetic recipients after intraperitoneal xenotransplantation (1,500 IEQ per mouse) with Al-CTP microcapsules (left) and respiratoid biosystem (right). Arrow: the day of transplantation, Dashed line: normoglycemia (≤ 200 mg dL−1). e Bright field images of their abdomen after retrieving free-floating capsules by lavage, 100 days after intraperitoneal transplantation (upper) and the H&E staining images of the retrieved respiratoid biosystem from the peritoneal cavity at day 100 (bottom). Circle: the floated microcapsules in the peritoneal cavity. Scale bar: 250 µm. f Representative immunofluorescence images of encapsulated islets in Al-CTP microcapsules and respiratoid biosystem transplanted into the peritoneal cavity at day 50 and 100. Green fluorescence: the expression of insulin (INS). Red fluorescence: the expression of HIF-1α. Blue fluorescence: islet nuclei (NUC) stained with DAPI. Scale bar: 100 μm. (n = 5 biological independent animals) g Intraperitoneal glucose tolerance test (IPGTT) of mice transplanted with Al-CTP microcapsules (w/ islet) (n = 5 biological independent animals) and respiratoid biosystem (w/ islet) (n = 5 biological independent animals) at day 30, 50, and 100 post-transplantation. Healthy mice (n = 5 biological independent animals) and diabetic mice (n = 5 biological independent animals) were examined as controls. Data were expressed as mean ± S.D. The shadowed area: S.D. h Corresponding area under the curve (AUC) of blood glucose levels is calculated from IPGTT (n = 5 biological independent animals). Data were expressed as mean ± S.D. Day 30 post-transplantation: P = 0.1307 (healthy control versus respiratoid biosystem), P < 0.001 (diabetic control versus respiratoid biosystem), P = 0.0750 (Al-CTP versus respiratoid biosystem). Day 50 post-transplantation: P < 0.001 (Al-CTP versus respiratoid biosystem). Day 100 post-transplantation: P = 0.0467 (healthy control versus respiratoid biosystem), P < 0.001 (diabetic control versus respiratoid biosystem). Statistical significance was determined by unpaired, two-tailed t-test in a and h. Source data are provided as a Source Data file.
To examine the impact of fibrosis and hypoxia on the implanted respiratoid biosystem with islets, we conducted intraperitoneal implantation in diabetic mice. Mice implanted with Al-CTP microcapsule with islets experienced a return to hyperglycemia after 30 days (Fig. 5d and Supplementary Fig. 24). We retrieved some Al-CTP microcapsules at day 50 post-transplantation for analysis of their rejection (Supplementary Fig. 25a, b, and d). In contrast, mice implanted with the respiratoid biosystem with islets maintained normal glucose level for over 85 days without rejection of microcapsules. At day 100 post-transplantation, the vast majority of respiratoid biosystems were freely floating in the peritoneal cavity with minimal adhesion to the abdominal organs, while still encapsulating islets in the respiratoid biosystems (Fig. 5e). Their morphology remained clean and transparent after 100 days of implantation (Supplementary Fig. 25c).
Immunostaining of the retrieved respiratoid biosystem with islets at day 50 and 100 post-implantation showed lower HIF-1α (red color) and higher insulin (green color) expression (Fig. 5f, Supplementary Fig. 26). Intraperitoneal glucose tolerance test (IPGTT) performed at day 30, 50, and 100 post-transplantation showed that the pattern of blood glucose and serum insulin level in mice with respiratoid biosystem were similar to those in non-diabetic healthy mice (Fig. 5g and h, Supplementary Figs. 27 and 28). Collectively, our findings suggest that the respiratoid biosystem can generate oxygen to support the therapeutic effect of encapsulated cells without altering correlation between blood glucose and serum insulin.
Discussion
Oxygen-releasing biomaterials (ORBs) have shown potential for delivering oxygen to hypoxic areas but have limitations, particularly in generating harmful free radicals1. This process involves hydrolyzing solid inorganic peroxides to form H2O2, which then decomposes into water and oxygen47. Excessive H2O2 can cause oxidative stress and cell apoptosis, and it can also react with Fe2+ ions to produce toxic hydroxyl radicals48. To mitigate these effects, both enzyme and non-enzyme-based antioxidants must be incorporated into scaffold. Studies have shown that higher concentrations of calcium peroxide (CaO2) in hydrogels affect cell viability, while adding catalase enzyme49,50. Interestingly, encapsulating CaO2 in polycaprolactone (PCL) microparticles within gelatin methacryloyl (GelMA) scaffolds enhances cell viability and proliferation by providing controlled and sustained oxygen release51,52. However, CaO2-laden GelMA scaffold can maintain shortly oxygen generation. On the other hand, photosynthetic algae also have been used as oxygen-generating biomaterials (OGBs) due to their continuous oxygen production through photosynthesis, but their application is limited to light-exposed areas and requires further safety studies53,54. In this study, we first demonstrated that the respiratoid biosystem, equipped with plant chloroplasts and biocompatible alginate-CTP conjugate, can aim to provide spontaneous oxygenation without an antioxidant and light for a long time.
Upon examining the molecular dynamics, our study reveals the impact of CTP treatment on gene expression within chloroplasts, particularly in the context of photosynthesis-related proteins. Unlike conventional approaches requiring ribozyme technology, our findings suggest that CTP can induce targeted gene expression without such intervention55,56. Acting as RNA-binding proteins or regulatory elements, CTP enhances mRNA stability and translation efficiency. This translational rate regulation accelerates protein synthesis, leading to an increased production of chloroplast genome-encoded proteins associated with photosynthesis and thylakoid membrane functions. Moreover, the CTP-induced elevation of extrinsic proteins in the Oxygen-Evolving Complex (OEC) of photosystem II (PS II) suggests improved stability in water oxidation57,58. This enhancement in oxygenation, coupled with activation of photosynthesis through the electron transport chain, implies the contribution of CTP to the overall photosynthetic reaction. Additionally, the role of Mn4CaO5 cluster in scavenging reactive oxygen species (ROS) aligns with the potential of our respiratoid biosystem to generate oxygen in the absence of light, addressing a key limitation associated with other ORB59. The upregulation of extrinsic proteins within OEC, including PsbO, PsbP, and PsbQ, in response to CTP treatment draws attention. They are playing a critical role in stabilizing the active site of water oxidase (Mn4CaO5 cluster) and regulating chloride and/or calcium, suggests an improvement in oxygenation by enhancing water oxidation stability60. This intricately contributes to the activation of photosynthesis through the electron transport and proton gradient, maximizing in the synthesis of adenosine triphosphate (ATP) and nicotinamide adenine dinucleotide phosphate (NADPH) via the electron transfer chain (ETC) on the thylakoid membrane43.
To ensure the efficacy of ORBs, it is essential to conduct in vivo and clinical studies. Our study demonstrates respiratoid biosystem’s ability to provide sustained oxygenation to transplanted islets in a diabetic mice model, extending their survival for at least 85 days. This has implications for treating conditions such as T1D, where pancreatic islets exhibit a heightened oxygen demand for normal functionality61. Currently, the size of clinically implanted tissue-engineered constructs is generally limited to a few centimeters (1–2 cm) due to the diffusion constraints of oxygen and nutrients62. We anticipate that the respiratoid biosystem can overcome the size limitations by fabricating macro-scale constructs for tissue engineering, ensuring sufficient oxygenation and nutrient diffusion. To this end, we can use the respiratoid biosystem as a bio-ink to fabricate the macro-scale construct with 3-dementional bioprinting. Overall, our developed respiratoid biosystem presents an approach that holds promise for advancing the field of ORBs, particularly in the context of prolonged cellular support and potential applications in cell therapies.
Methods
Material and reagents
Coupling reagents, cleavage cocktail reagents, D-sorbitol, Tris-Cl, magnesium chloride (MgCl2), sodium chloride (NaCl), EDTA, bovine serum albumin (BSA), percoll, 4-Morpholineethanesulfonic acid (MES), N-(3-Dimethylaminopropyl)-N’-ethylcarbodiimide hydrocloride (EDC·HCl), N-Hydroxysuccinimide (NHS), deuterium oxide (D2O), calcium chloride (CaCl2), HEPES, sodium cacodylate, glutaraldehyde solution, sucrose, medium 199, histopaque-1077, fetal bovine serum (FBS), D-(+)-Glucose solution, paraformaldehyde, glutaraldehyde, dimethyl sulfoxide (DMSO), and Tween-20 were purchased from Sigma-Aldrich, MO, USA. Fresh spinach from Takara species was purchased at a grocery store in Seoul, Republic of Korea. Trt-Cl resins and all Fmoc amino acids were purchased from GL Biochem (Shanghai, China). Hydroxybenzotriazole (HOBt), hexafluorophosphate benzotriazole tetramethyl uronium (HBTU), N, N-diisopropylethylamine (DIPEA), dimethylformamide (DMF), trifluoroacetic Acid (TFA), ethanedithiol (EDT), thioanisole, triisopropylsilane (TIS), ether, and trifluoroacetic acid were purchased from Daejung chemical (Korea). Sodium alginate (PRONOVA UP LVG) with a low viscosity and a G/M ratio (≥1.5) was purchased from FMC BioPolymer, Philadelphia, USA. Chloroplast-Transit-Peptide (CTP) and CTP conjugated fluorescein isothiocyanate (FITC) (CTP-FITC) were synthesized by Peptron, Daejeon, Republic of Korea. 6,000–8,000 MWCO (Molecular Weight Cut-Off) dialysis membrane, PierceTM BCA Assay Kit, Opti-MEM® Medium, Lipofectamine RNAiMAX Transfection Reagent, and Permount were purchased from Thermo Fisher Scientific, USA. RPMI-1640 medium, LIVE/DEADTM Viability/Cytotoxicity Kit, and Pico Green Quantitative DNA Assay Kit were purchased from Invitrogen, La Jolla, CA. Penicillin-streptomycin was purchased from Gibco, Carlsbad, CA. Cell counting kit-8 (CCK-8) was purchased from Dojindo Molecular Technologies, Rockville, MD, USA. Anti-HIF-1α was purchased from Abbkine, Wuhan, China, and Anti-insulin was purchased from Santa Cruz Biotechnology Inc, Texas, USA. Coomassie Protein Stain, Alexa Fluor 488 goat anti-rabbit IgG, and Alexa- Fluor 594 goat anti-mouse IgG were purchased from Abcam, MA, USA. AMPIGENE® cDNA Synthesis Kit and AMPIGENE® qPCR Green Mix Lo-ROX were purchased from Enzo Life Sciences, USA. The primer sequences used were as follows (Table 1).
Synthesis of Chloroplast-Transit-Peptide (CTP)
The Fmoc protecting group was removed by rocking in 20 % piperidine in DMF for 10 min (twice) and coupling was carried out using Fmoc amino acid (8 eq), HOBt (8 eq), HBTU (8 eq), and DIPEA (16 eq) in DMF for 2 h. In each step, the resin was washed using DMF and methanol for 2 times each. When the desired sequence was complete, the crude peptide was cleaved from the resin using a mixture of TFA/ EDT/ thioanisole/ TIS/ distilled water (D.W) (90/ 2.5/ 2.5/ 2.5/ 2.5 volume) for 2 h. The solution was precipitated with cold ether and made into pellets using a centrifuge. The solid was collected and air-dried. Crude peptide was dissolved in D.W and purified by the reverse phase HPLC (Shimadzu Prominence HPLC, Japan) using ACE 10 C18-300 column (21.2 mm × 250 mm, 10 μm, England). Elution was carried out with a water-acetonitrile linear-gradient (10~75% (v/v) of acetonitrile) containing 0.1% (v/v) trifluoroacetic acid. Collect the pure portion of CTP, which was further lyophilized.
Isolation of chloroplast from spinach
Before isolating the chloroplasts, chloroplast isolation buffer (CIB, pH 7) was prepared. To make the CIB, 0.33 M D-sorbitol, 0.1 M Tris-Cl (pH 7.8), 5 mM MgCl2, 10 mM NaCl, and 2 mM EDTA were added to deionized water. After that, fresh spinach was washed in water. 35 g of leaf fragments from spinach and 100 mL of CIB with BSA (0.1% w/v) were put together in a cold blender (BL311E; TEFAL, Republic of Korea) and ground for 30 s. The blended solution was collected in tubes using a mesh filter and centrifuged at 1295 g at 4 °C for 1 min. After centrifugation, supernatants were centrifuged at 1295 g for 7 min at 4 °C. The pellet was resuspended in CIB with BSA. The solution was layered on percoll gradients (40% and 80%) carefully and centrifuged at 3315 g at 4 °C for 10 min. Then, an intact chloroplast layer between the percoll gradients was collected and washed in CIB without BSA and stored in ice condition.
Measurement of released chlorophyll from chloroplasts
Chloroplasts (1.5 × 109 chloroplasts mL−1) diluted in a culture medium (RPMI-1640 medium with 10% FBS and 1% penicillin-streptomycin) were treated with 0.2 mM CTP and incubated at 37 °C and 5% CO2 for 14 days and the medium was changed every two days while washing to prevent contamination. The samples were centrifuged at 14,000 g for 2 min. After centrifugation, the absorbance of the supernatants was measured at 430 and 662 nm, specific for chlorophyll a, using a SpectraMax M2 microplate reader (Molecular Devices Inc., Sunnyvale, CA, USA). In the case of microcapsules, the absorbance of the culture medium was measured.
Oxygen production measurement
Chloroplast (7.5 × 108 chloroplasts mL−1) and PS II submembrane fraction (100 μg chlorophyll mL−1) samples in a culture medium and an assay medium, respectively were transferred to each 24-well plate which is attached to an oxygen sensor patch (PreSens GmbH, Regensburg, Germany). The assay medium contained 20 mM MES-NaOH (pH 6.2), 1 mM NaCl, 0.5 mM MgCl2, and 0.35 mM DCBQ (2.5-dichlorobenzoquinone) as PS II electron acceptor. In the case of chloroplast, chloroplast was treated with 0.2 mM CTP and 5 mM Al3+ for 1 h and 30 min, respectively. For analysis of CTP-treated mRNA upregulation, chloroplast was treated with 0.2 mM CTP and 100 μM siRNA for 1 h. Afterward, the dissolved oxygen concentration of samples was measured every 30 min at 37 °C for 20 h under hypoxic (1% pO2) conditions using the Sensor©DishReader system (PreSens GmbH, Regensburg, Germany) according to the manufacturer instructions. Using a fluorometric oxygen sensor, this system monitors the percentage of dissolved oxygen content (mmHg) in each medium for 20 h. The oxygen concentration was expressed as a decreasing level over time from the initial concentration after normalizing the initial concentration of each group to 100%, which means how much oxygen was maintained compared to the initial concentration.
Photosynthetic activity analysis
Before treatment, CTP and Al3+ as Al2(SO4)3 were dissolved in 1 % DMSO and 0.9% sterile saline buffer (pH 7.4), respectively. Isolated chloroplasts (7.5 × 108 chloroplasts mL−1) were treated with 0.2 mM CTP for 1 h and 5 mM Al3+ for 30 min. These samples were centrifuged at 14,000 g for 2 min and washed 2 times. Chlorophyll fluorescence of chloroplast pellets was measured using an Imaging-PAM M-series fluorometer (Heinz Walz GmbH, Germany) equipped with an IMAG-K4 CCD camera and an IMAG-MAX/L LED-array illumination unit emitting blue (450 nm) light. The samples were exposed to 4 intensities of 450 nm actinic light increasing from 0 to 1.150 × 10 μM m−2 s−1, provided by the LED ring array. Each irradiance period was 5 s to ensure the minimum (steady-state) fluorescence in actinic light (Ft) and was followed by an excitation pulse to yield the maximum fluorescence in actinic light (Fm’). Digital images of chlorophyll fluorescence were recorded immediately before and during each saturating pulse. Numerical values of the chlorophyll fluorescence parameters (Fo, Fm, Ft and Fm’) were extracted from the digital images using analytical software (IMAGING- WIN, Heinz Walz GmbH, Germany). In addition, Net photosynthetic rate data, photosynthetic capacity (Pmax), and apparent photosynthetic efficiency (α) were fitted from the equation given by Eilers and Peeters63.
Isolation of PS II submembrane fractions
In the case of light condition, chloroplasts were treated with 0.2 mM CTP and incubated in the cold room (4 °C) with 500 lux near-infrared (NIR) light (VS-PGLEDBAR; Visang, China) irradiation for 1 h. Thylakoid membranes were isolated from the chloroplast as previously described64. Then, PS II submembrane fractions were isolated from thylakoid membranes as described elsewhere with minor modifications65. Triton X-100 was added with gentle shaking for 1 min to obtain a final concentration of 0.1 mg chlorophyll mL−1. The latter solution was incubated in the dark for 20 min and centrifuged at 38,304 g for 15 min. The pellet was suspended in a buffer containing 50 mM Mes-NaOH, 15 mM NaCl (pH 6.1), 10 mM MgCl2, and 5 mM CaCl2 centrifuged at 1295 g for 2 min. Collected supernatants were centrifuged at 38,304 g for 30 min and their pellets were suspended in the same buffer. This homogenate of pellets was centrifuged at 2727 g for 10 min. Then, the pellet obtained was suspended in a buffer containing 50 mM Mes-NaOH, 15 mM NaCl (pH 6.1), 5 mM MgCl2, 5 mM CaCl2, and 400 mM sucrose and centrifuged at 24,515 g for 12 min. In the last step, the new pellet was suspended in the same buffer, and the chlorophyll content was measured at wavelengths of 645 nm and 663 nm against a blank of 80% acetone using a UV-Vis spectrophotometer. The concentrations of chlorophyll a and chlorophyll b were calculated using Porra’s equations66.
After that, the isolated PS II was stored to determine whether protein expression of it is maintained for a long term in the dark and cold room for 14 days.
SDS-polyacrylamide gel electrophoresis
PS II submembrane fraction proteins were separated by polyacrylamide gel electrophoresis (SDS-PAGE) containing 12% acrylamide according to Laemmli67. Samples of PS II submembrane fractions were treated with 5 mM Al3+, incubated at room temperature in the dark for 30 min, and centrifuged at 14,000 g for 5 min. The pellets were washed twice in 20 mM Mes-NaOH (pH 6.2) centrifuged at 14,000 g for 5 min and then used for polypeptide separation in the gel. 20 mL of the different samples of PS II treated with Al3+ were loaded in each lane onto the gel. The lanes represent: Molecular weight marker (1). PS II isolated from the untreated chloroplast without (2) or with light exposure (6). PS II isolated from the untreated chloroplast and was treated with 5 mM Al3+ without (3) or with light exposure (7). PS II isolated from the chloroplast treated with 0.2 mM CTP without (4) or with light exposure (8). PS II isolated from the chloroplast treated with 0.2 mM CTP and 5 mM Al3+ without (5) or with light exposure (9). Finally, SDS-polyacrylamide gels containing separated polypeptides were stained with Coomassie brilliant blue and analyzed with the chemiluminescence reader (LuminogGraph 3 Lite) from Atto (Taito-ku, Tokyo, Japan).
mRNA-sequencing and analysis
To synthesize full-length complementary DNA (cDNA), mRNA was extracted from chloroplast samples using QIAzol Lysis Reagent and the RNeasy Mini Kit (QIAGEN, Germany). The construction of the final libraries was performed utilizing the TruSeq Stranded mRNA Sample Prep Kit (Eukaryotes). The libraries were subjected to sequencing using an Illumina NextSeq 6000 platform, performed by Theragen Bio Co., Ltd. (Gueonggi-do, South Korea). The gene list that exhibited significant upregulation upon treatment with CTP (P ≤ 0.05) was chosen for subsequent analyses due to its strong alignment with experimentally observed changes in gene expression. Functional classification and functional annotation clustering of Gene Ontology (GO) terms were conducted using the appropriate tools available within the Database for Annotation, Visualization, and Integrated Discovery (DAVID). Subsequently, pathway analysis was carried out utilizing the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways database.
siRNA transfection
To assess the efficacy of the CTP, a targeted gene knockdown experiment was conducted through transfection with small interfering RNA (siRNA). The siRNA was designed to specifically target the gene of interest. siRNA sequences used were as follows (Table 2). The Lipofectamine® RNAiMAX Reagent (Thermo Fisher Scientific, USA) was diluted in Opti-MEM® Medium (Thermo Fisher Scientific, USA) at a volume ratio of 50:3. Diluted siRNA in Opti-MEM® medium and diluted Lipofectamine® RNAiMAX reagent mixed with a 1:1 volume ratio. The siRNA-lipid complex was incubated for a duration of 5 min at room temperature. Subsequently, the siRNA-lipid complex was added to the chloroplast (7.5×108 chloroplasts mL−1).
Analysis of mRNA upregulation by qRT-PCR
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) and oxygen production measurement were conducted to evaluate CTP functionality to chloroplast. Chloroplasts (7.5×108 chloroplasts mL−1) were treated with 0.2 mM CTP and 100 μM siRNA for 1 h under the condition of light exposure. Next, RNA extraction was performed using QIAzol Lysis Reagent and RNeasy Mini Kit (QIAGEN, Germany) and reverse transcribed into AMPIGENE® cDNA Synthesis Kit (Enzo Life Sciences, USA). After preparing equal amounts of cDNA from the extracted RNA, the relative gene expression levels were quantified by RT-PCR (Applied Biosystems 7500, USA). Each sample was compared to the RbcL control and calculated using the ΔΔCt method.
Synthesis of Al-CTP conjugate
First, a solution of 10 mL of 0.1 M 4-Morpholineethanesulfonic acid (MES) buffer (pH 6) with 0.5 M NaCl was prepared. 228 mg of alginate (Al; 0.2 mM) was dissolved in 10 mL of the prepared MES buffer. When the alginate solution was sufficiently dissolved, 4 mg of EDC·HCl (2 mM) was in it. Then, 6 mg of NHS (5 mM) was added and reacted at RT for 15 min, and the pH was adjusted to 7. Synthesized CTP dissolved in 1% DMSO was added to the solution in an amount corresponding to the feed molar ratio (1:1, 1:10, and 1:50) and reacted at room temperature for 2 h. The solution was transferred to a 6000–8000 MWCO (Molecular Weight Cut-Off) dialysis membrane. Dialysis was performed in deionized water at 4 °C for 3 days. After dialysis, the solution was lyophilized for 48 h to obtain alginate-CTP (Al-CTP) conjugate powder. The lyophilized Al-CTP conjugate was stored at 4°C for further use.
Experimental animal
For this study, seven-week-old inbred BALB/C male mice and seven-week-old outbred Sprague-Dawley (SD) rats were purchased from Nara Biotech (Seoul, Republic of Korea) and Daehan-Bio Link (Chung-cheongbuk-do, Republic of Korea). All animals were housed in cages under specific pathogen-free (SPF) conditions in a temperature-controlled room with a 12-hour light/dark cycle. All animals had free access to water and food. All animal procedures were performed according to the Institutional Animal Care and Use Committee (IACUC) of Hanyang University (HY-IACUC-2022-0037A.
Rat pancreatic islets isolation and culture
Male outbred Spraque Dawley (SD) rats, weighing approximately 300 g, were used for the isolation of pancreatic islets. First, a collagenase P (Roche, Germany) solution at a concentration of 1 mg mL−1 was administered directly into the bile duct of a sacrificed rat. The pancreas was allowed to swell and then incubated in a 37 °C water bath for 15 min to facilitate digestion. The digested pancreas was rinsed twice with Medium 199, passed through a 450 μm sieve, and subsequently suspended in histopaque-1077 before being centrifuged at 1260 g for 17 min at 4 °C. The islets were then harvested from the resulting gradient and further refined through gravity sedimentation. Afterward, the purified islets were manually counted and cultured in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin-streptomycin. The islets were maintained in standard culture conditions (37 °C, 5% CO2) overnight. For experiments under hypoxic conditions, the islets were incubated at 37 °C and 1% O2 for 20 h in a customized hypoxia desiccator (84 L).
Manufacturing of microcapsules
Microcapsules were prepared via an encapsulator (B-395 Pro, BUCHI, Switzerland). To manufacture Al-CTP microcapsules, Al-CTP conjugates were dissolved in 0.9% sterile saline buffer (pH 7.4) to prepare a 1.8% (w/v) Al-CTP solution. In the case of Al-Ch or Al-CTP-Chloroplast microcapsules (respiratoid biosystem), alginate or Al-CTP conjugates were dissolved in 0.9% sterile saline buffer to prepare 3.6% (w/v) alginate or Al-CTP solution. Next, these solutions were mixed with chloroplast (1.5 × 109 chloroplasts mL−1) dissolved in 0.9% sterile saline buffer, at a volume ratio of 1:1, and transferred to a sterile syringe. To encapsulate pancreatic islets, 1 mL of Al-CTP, Al-Ch, and Al-CTP-Chloroplast solution were mixed with pancreatic islets (1200 islet equivalent (IEQ) mL−1). After loading the syringe into the encapsulator, it was operated at a frequency of 800 Hz, electrode voltage of 500 V, a flow rate of 8.0 mL min−1, and a nozzle diameter of 450 mm. The microcapsules were prepared by incubating the droplets in 100 mM CaCl2 buffer with 10 mM HEPES for 15 min. Then, the microcapsules were washed with 25 mM HEPES (pH 7.4) and culture medium for further use.
Cell viability analysis
LIVE/DEAD staining was performed to quantify the cell viability of islets using the LIVE/DEADTM Viability/Cytotoxicity Kit. Intact and encapsulated islets were treated with PBS containing 1 μM calcein-AM and 1 μM ethidium homodimer-1 (EthD-1) at 37 °C and 5% CO2 for 15 and 40 min. After that, live and dead signals were obtained using fluorescence microscopy.
Intact islets and microcapsules were treated with a culture medium containing 10% CCK-8 solution at 37°C and 5% CO2 for 2 h. The absorbance of the medium was measured at 450 nm using a SpectraMax M2 microplate reader. Then, the microcapsules were completely dissolved by treating them with 0.1 M EDTA (pH 8) solution for 15 min to retrieve the islets encapsulated in microcapsules. Next, the cell viability was normalized to the DNA content of retrieved islets using the Pico Green Quantitative DNA Assay Kit.
Glucose-stimulated insulin secretion (GSIS) assay
To investigate the responses of islets to varying glucose concentrations, the glucose-stimulated insulin secretion (GSIS) assay was performed. Islets were initially incubated in Krebs Ringer Buffered HEPES (pH 7.4) with 2.8 mM glucose for a starvation period of 30 min at 37 °C. Following this, they were treated with different concentrations of glucose (2.8 mM and 20.2 mM) at 37 °C for 2 hours. Insulin levels in the culture media were quantified using a rat insulin ELISA kit (Alpco Diagnostics, Salem, NH, USA). The insulin levels were then normalized by dividing by the DNA content. The stimulation index (SI) was calculated using the formula: (insulin secretion at high glucose, normalized to DNA) / (insulin secretion at low glucose, normalized to DNA).
Immunostaining of islets encapsulated in microcapsules
Microcapsules containing islets after normoxic and hypoxic incubation were fixed in a 4% paraformaldehyde solution at 4 °C for 1 day and then washed in PBS. Subsequently, tissue processing was carried out using the Leica TP1020 semienclosed Benchtop Tissue Processor (Leica Biosystems, Wetzlar, Hesse, Germany) in an automated manner. Following embedding in paraffin, the paraffin blocks were sectioned into slices with a thickness of 5 μm using a Leica RM 2145 Microtome (Leica Biosystems). The sectioned paraffin blocks were stained according to standard histological methods. Immunofluorescent double-staining for HIF-1α and insulin was performed using a primary antibody (anti-HIF-1α 1:100 dilution, anti-insulin 1:200 dilution) and incubated with secondary antibody (Alexa Fluor 488 goat anti-rabbit IgG 1:200 dilution, Alexa- Fluor 594 goat anti-mouse IgG 1:100 dilution) at room temperature for 1 h. The sections were also counterstained with a DAPI mounting medium. Immunostained images from the sections were obtained under fluorescence microscopy and Confocal Laser Scanning Microscope.
Subcutaneous (SQ) transplantation of microcapsules
We utilized immuno-deficiency Balb/C nude and Balb/C mice as recipients for pancreatic islet transplantation. Before transplantation, diabetes was induced in the mice by administering a single intraperitoneal injection of streptozotocin (150 mg kg−1; Sigma-Aldrich, MO, USA) in citrate buffer (pH 4.5). Diabetic status was determined in mice based on non-fasting blood glucose levels exceeding 300 mg dL−1 for three consecutive days. SQ transplantation involves the transplantation of tissue or cells into the subcutaneous tissue layer, located just beneath the skin. Anesthesia induction was performed in the mice before transplantation by administering a concentration of 2–2.5 % isoflurane in oxygen. To gain access to the SQ cavity, a surgical incision was performed in the abdominal epidermis region of the mice. The microcapsules exposed to light stimulation (1 h) were transplanted into the SQ tissue. The closure of the incision was performed using sutures (Ethicon, New Jersey, USA). After transplantation, the diabetic recipient’s non-fasting blood glucose levels were measured by a portal glucometer (Accu-Chek®; Roche Diabetes Care Inc., Indianapolis, IN, USA). In the case of healthy recipients, continuous monitoring of the mouse’s oxygen level at the SQ site was performed both before and after transplantation to determine the efficacy of using a pO2 needle sensor (OXF-NX-NP/O/E, Bio-Rad, California, USA) for assessment. Data were expressed as the oxygen levels of post-transplantation at day 14 after normalizing the oxygen concentration before transplantation of each group to 100%, which means how much oxygen was maintained compared to the before transplantation.
Intraperitoneal (IP) transplantation of microcapsules
After anesthesia as above mentioned, an incision was made in the abdominal region of the mice to provide access to the IP cavity. Abdominal organs were gently moved aside to create sufficient space for the placement of the microcapsules. The microcapsules exposed to light stimulation (1 h) were carefully inserted into the peritoneal cavity. The incision was closed using sutures. Continuous monitoring of the mouse’s blood glucose level was conducted post-transplantation to ascertain the efficacy of the microcapsules in treating diabetic mellitus.
Intraperitoneal Glucose Tolerance Test (IPGTT)
IPGTT assessed glucose metabolism in mice. The mice were subjected to a period of fasting lasting roughly 16 h. Overnight fasting (16 h) mice received an IP injection of 20% glucose solution at a dose of 2 g glucose per kg body weight (2 g/kg BW). The blood glucose from tail was collected (~60 μL) and assessed at specific intervals (0, 15, 30, 45, 60, 90, and 120 min) after glucose injection using a glucometer. The samples were then centrifuged, and the serum was rapidly frozen for subsequent measurement of serum insulin using an ELISA kit.
Microcapsules retrieval from the IP space
At 100 days post-transplantation, mice were anesthetized using isoflurane and placed in a supine position. With a small incision at the time points of implantation, mice were euthanized using CO2 administration. An incision was performed using forceps and scissors along the abdominal region, encompassing the skin and peritoneal wall. To remove all capsules from the abdomen, a solution of Krebs buffer was employed as a washing agent. The capsules underwent multiple washes using Krebs buffer until the buffer was observed to be free of any remaining blood and tissue.
Histological analysis
The retrieved tissues were fixed in a 4% paraformaldehyde solution for 4 days and washed in PBS. Then processed through a graded series of ethanol for dehydration. Samples were subsequently cleared in xylene and embedded in paraffin. Paraffin blocks were sectioned into 7 µm thick slices using a microtome and mounted on glass slides. Sections were rehydrated using a graded ethanol series (100 %, 90%, 80%, and 70%) for two minutes each after being deparaffinized in xylene twice for 5 minutes each. Rinsing in distilled water was the next step. Sections were stained for 5 minutes with Harris hematoxylin for hematoxylin and eosin (HE) staining and then washed for 5 minutes under running tap water. Sections were cleaned with xylene, dehydrated using graded ethanol, counterstained for a minute with eosin Y, and mounted with a coverslip using a synthetic resin mounting medium. Weigert’s iron hematoxylin was used to stain sections for 10 minutes in order to stain the nuclei for Masson’s Trichrome staining (MT staining). After 10 minutes of washing under running tap water, the sections were dried. After that, they were dyed for 5 minutes with Biebrich scarlet-acid fuchsin to highlight the cytoplasm and muscle fibers. Sections were differentiated in phosphomolybdic-phosphotungstic acid solution for 10 minutes after being rinsed in distilled water. To emphasize collagen fibers, aniline blue staining was applied for 5 minutes. Sections were immediately dehydrated using 95% and 100% ethanol, cleaned in xylene, and mounted using a synthetic glue mounting solution after being briefly washed in 1% acetic acid for 2 minutes dehydrated quickly through 95% and 100% ethanol, cleared in xylene, and mounted with a synthetic resin mounting medium.
Statistical analysis
Data were expressed as mean ± standard deviation (S.D.). Statistical differences were analyzed using a two-tailed unpaired t-test to compare the differences of groups. In the statistical analysis for comparison between multiple data groups, one-way analysis of variance (ANOVA) followed by Duncan’s multiple comparison tests. Graft survival was compared using the log-rank (Mantel-Cox) method. The data were analyzed using the GraphPad Prism 8 program (La Jolla, CA, USA).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that all data supporting the finding of this study are available within the paper and its supplementary information files. The remaining data are available upon reasonable request from the corresponding author D.Y.L. or within the Source Data file. Source data is available for Figs. 2–5, Supplementary Figs. 1–8, 10, 12, 13, 15, 16, 18, 20, 21, 23, and 25–27, and Supplementary Tables 1 and 2 in the associated source data file. The RNA-sequencing data generated in this study have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences database under accession code GSA: CRA018274. Source data are provided with this paper.
Change history
09 December 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41467-024-55167-0
References
Yildirimer L., Seifalian A. Tissue engineering. In: Plastic and reconstructive surgery). Wiley online library (2015).
Gholipourmalekabadi, M., Zhao, S., Harrison, B. S., Mozafari, M. & Seifalian, A. M. Oxygen-generating biomaterials: a new, viable paradigm for tissue engineering? Trends Biotechnol. 34, 1010–1021 (2016).
Magliaro, C. et al. Oxygen consumption characteristics in 3D constructs depend on cell density. Front Bioeng. Biotechnol. 7, 251 (2019).
Kyun, M. L. et al. Development of an in vitro model for inflammation mediated renal toxicity using 3D renal tubules and co-cultured human immune cells. Tissue Eng. Regen. Med. 20, 1173–1190 (2023).
Kim, J. W., Ho, W. J. & Wu, B. M. The role of the 3D environment in hypoxia-induced drug and apoptosis resistance. Anticancer Res. 31, 3237 (2011).
Liverani, C. et al. A biomimetic 3D model of hypoxia-driven cancer progression. Sci. Rep. 9, 12263 (2019).
Clanton, T. L. Hypoxia-induced reactive oxygen species formation in skeletal muscle. J. Appl. Physiol. 102, 2379–2388 (2007).
Azimi, I., Petersen, R. M., Thompson, E. W., Roberts-Thomson, S. J. & Monteith, G. R. Hypoxia-induced reactive oxygen species mediate N-cadherin and SERPINE1 expression, EGFR signalling and motility in MDA-MB-468 breast cancer cells. Sci. Rep. 7, 15140 (2017).
Hamanaka, R. B. & Chandel, N. S. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr. Opin. Cell Biol. 21, 894–899 (2009).
Oh, S. H., Ward, C. L., Atala, A., Yoo, J. J. & Harrison, B. S. Oxygen generating scaffolds for enhancing engineered tissue survival. Biomaterials 30, 757–762 (2009).
Harrison, B. S., Eberli, D., Lee, S. J., Atala, A. & Yoo, J. J. Oxygen producing biomaterials for tissue regeneration. Biomaterials 28, 4628–4634 (2007).
Alemdar, N. et al. Oxygen-generating photo-cross-linkable hydrogels support cardiac progenitor cell survival by reducing hypoxia-induced necrosis. ACS Biomater. Sci. Eng. 3, 1964–1971 (2017).
Newland, B., Baeger, M., Eigel, D., Newland, H. & Werner, C. Oxygen-producing gellan gum hydrogels for dual delivery of either oxygen or peroxide with Doxorubicin. ACS Biomater. Sci. Eng. 3, 787–792 (2017).
Wang, H., Zhang, W., Chen, L., Wang, J. & Liu, T. The contamination and control of biological pollutants in mass cultivation of microalgae. Bioresour. Technol. 128, 745–750 (2013).
Leegood, R. C. & Walker, D. A. The role of transmembrane solute flux in regulation of CO2 fixation in chloroplasts. Biochem Soc. Trans. 11, 74–76 (1983).
Seftor, R. E. & Jensen, R. G. Causes for the disappearance of photosynthetic CO(2) fixation with isolated spinach chloroplasts. Plant Physiol. 81, 81–85 (1986).
Polanská, L., Vičánková, A., Dobrev, P. I., Macháv cková, I. & Vaňková, R. Viability, ultrastructure and cytokinin metabolism of free and immobilized tobacco chloroplasts. Biotechnol. Lett. 26, 1549–1555 (2004).
Kim, J., Na, Y. J., Park, S. J., Baek, S.-H. & Kim, D. H. Biogenesis of chloroplast outer envelope membrane proteins. Plant Cell Rep. 38, 783–792 (2019).
Soll, J. & Schleiff, E. Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 5, 198–208 (2004).
Ganesan, I., Shi, L.-X., Labs, M. & Theg, S. M. Evaluating the Functional Pore Size of Chloroplast TOC and TIC Protein Translocons: Import of Folded Proteins. Plant Cell 30, 2161–2173 (2018).
Jin, Z. et al. Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes. Cell 185, 4788–4800.e4713 (2022).
Kozuleva, M. A., Ivanov, B. N., Vetoshkina, D. V. & Borisova-Mubarakshina, M. M. Minimizing an electron flow to molecular oxygen in photosynthetic electron transfer chain: an evolutionary view. Front. Plant Sci. 11, 211 (2020).
Neal, A. et al. Real-time imaging of intracellular hydrogen peroxide in pancreatic islets. Biochem. J. 473, 4443–4456 (2016).
Yoshizumi, T., Oikawa, K., Chuah, J.-A., Kodama, Y. & Numata, K. Selective gene delivery for integrating exogenous DNA into plastid and mitochondrial genomes using Peptide–DNA complexes. Biomacromolecules 19, 1582–1591 (2018).
An, J. et al. Visualizing the integrity of chloroplast envelope by rhodamine and Nile red staining. Front. Plant Sci. 12, 668414 (2021).
Springer, A. et al. Programmed chloroplast destruction during leaf senescence involves 13-lipoxygenase (13-LOX). Proc. Natl Acad. Sci. 113, 3383–3388 (2016).
Nugroho, E. et al. Determination of green and red spinach microgreen chlorophyll content using visible spectroscopy and wavelength selection. IOP Conf. Ser.: Earth Environ. Sci. 1183, 012049 (2023).
Hasni, I., Yaakoubi, H., Hamdani, S., Tajmir-Riahi, H. A. & Carpentier, R. Mechanism of interaction of Al3+ with the proteins composition of photosystem II. PLoS One 10, e0120876 (2015).
Shen, J. R. & Inoue, Y. Binding and functional properties of two new extrinsic components, cytochrome c-550 and a 12-kDa protein, in cyanobacterial photosystem II. Biochemistry 32, 1825–1832 (1993).
Seidler, A. The extrinsic polypeptides of Photosystem II. Biochim Biophys. Acta 1277, 35–60 (1996).
Govindjee Photosystem II: The light-driven water: Plastoquinone Oxidoreductase, edited by Thomas J. Wydrzynski and Kimiyuki Satoh, Volume 22, Advances in Photosynthesis and Respiration, Springer, Dordrecht, The Netherlands. Photosynth. Res. 87, 331–335 (2006).
Roose, J. L., Wegener, K. M. & Pakrasi, H. B. The extrinsic proteins of Photosystem II. Photosynth Res. 92, 369–387 (2007).
Reimand, J. et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 14, 482–517 (2019).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Huang, D. W. et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids Res. 35, W169–W175 (2007).
Packer, M. Autophagy-dependent and -independent modulation of oxidative and organellar stress in the diabetic heart by glucose-lowering drugs. Cardiovasc. Diabetol. 19, 62 (2020).
Dulmovits, B. M. et al. HMGB1-mediated restriction of EPO signaling contributes to anemia of inflammation. Blood 139, 3181–3193 (2022).
Batsaikhan, T., Choi, J. S., Ha, S. M., Ahn, Y. & Seo, Y. J. D-Galactose and hypoxia induce the early onset of age-related hearing loss deterioration in a mouse model. Tissue Eng. Regen. Med. 20, 779–787 (2023).
Evans, S. M., Schrlau, A. E., Chalian, A. A., Zhang, P. & Koch, C. J. Oxygen levels in normal and previously irradiated human skin as assessed by EF5 binding. J. Investig. Dermatol. 126, 2596–2606 (2006).
Chao, X. et al. Comparative Study of Two Common In Vitro Models for the Pancreatic Islet with MIN6. Tissue Eng. Regen. Med. 20, 127–141 (2023).
Lawrence, J. M. et al. Rewiring photosynthetic electron transport chains for solar energy conversion. Nat. Rev. Bioeng. 1, 887–905 (2023).
Franz, S., Rammelt, S., Scharnweber, D. & Simon, J. C. Immune responses to implants – A review of the implications for the design of immunomodulatory biomaterials. Biomaterials 32, 6692–6709 (2011).
Samsonchi, Z. et al. Transplantation of Islet-Containing microcapsules modified with constitutional isomers of sulfated alginate in diabetic mice to mitigate fibrosis for Long-term glycemic control. Chem. Eng. J. 432, 134298 (2022).
Lu, T., Cong, L., Jiang, T., Dong, X. & Song, L. Neuropeptide Y promotes the treatment of adipose stem cells on Type 2 diabetic wounds. Tissue Eng. Regen. Med. 20, 683–694 (2023).
Ashammakhi, N. et al. Advances in controlled oxygen generating biomaterials for tissue engineering and regenerative therapy. Biomacromolecules 21, 56–72 (2020).
Ighodaro, O. M. & Akinloye, O. A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 54, 287–293 (2018).
Park, S. & Park, K. M. Hyperbaric oxygen-generating hydrogels. Biomaterials 182, 234–244 (2018).
Ye, Y. et al. Reactive oxygen species scavenging hydrogel regulates stem cell behavior and promotes bone healing in Osteoporosis. Tissue Eng. Regen. Med. 20, 981–992 (2023).
Suvarnapathaki, S. et al. Oxygen generating scaffolds regenerate critical size bone defects. Bioact. Mater. 13, 64–81 (2022).
Shim, K. S. et al. Breast tissue reconstruction using polycaprolactone ball scaffolds in a partial mastectomy pig model. Tissue Eng. Regen. Med. 20, 607–619 (2023).
Centeno-Cerdas, C. et al. Development of photosynthetic sutures for the local delivery of oxygen and recombinant growth factors in wounds. Acta Biomater. 81, 184–194 (2018).
Evron, Y. et al. Oxygen supply by photosynthesis to an implantable islet cell device. Horm. Metab. Res. 47, 24–30 (2015).
Serganov, A. & Patel, D. J. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 8, 776–790 (2007).
Day, A. & Goldschmidt‐Clermont, M. The chloroplast transformation toolbox: selectable markers and marker removal. Plant Biotechnol. J. 9, 540–553 (2011).
Ifuku, K., Ishihara, S. & Sato, F. Molecular functions of oxygen‐evolving complex family proteins in photosynthetic electron flow. J. Integr. Plant Biol. 52, 723–734 (2010).
McEvoy, J. P., Gascon, J. A., Batista, V. S. & Brudvig, G. W. The mechanism of photosynthetic water splitting. Photochem. Photobiol. Sci. 4, 940–949 (2005).
Foyer, C. H. & Hanke, G. ROS production and signalling in chloroplasts: cornerstones and evolving concepts. Plant J. 111, 642–661 (2022).
Bricker, T. M., Roose, J. L., Fagerlund, R. D., Frankel, L. K. & Eaton-Rye, J. J. The extrinsic proteins of Photosystem II. Biochim. Biophys. Acta (BBA) - Bioenerg. 1817, 121–142 (2012).
Bonner-Weir, S. The microvasculature of the pancreas, with emphasis on that of the islets of Langerhans. In The Pancreas: Biology, Pathobiology, and Disease, (Eds Go, V. L. W. et al.) Ch. 39, pp. 759–768, (Raven Press, New York, 1993).
Langridge, B., Griffin, M. & Butler, P. E. Regenerative medicine for skeletal muscle loss: a review of current tissue engineering approaches. J. Mater. Sci.: Mater. Med. 32, 15 (2021).
Eilers, P. H. C. & Peeters, J. C. H. A model for the relationship between light intensity and the rate of photosynthesis in phytoplankton. Ecol. Model. 42, 199–215 (1988).
Bonente, G. et al. The Occurrence of the psbS Gene Product in Chlamydomonas reinhardtii and in other photosynthetic organisms and its correlation with energy quenching. Photochem. Photobiol. 84, 1359–1370 (2008).
Berthold, D. A., Babcock, G. T. & Yocum, C. F. A highly resolved, oxygen-evolving photosystem II preparation from spinach thylakoid membranes: EPR and electron-transport properties. FEBS Lett. 134, 231–234 (1981).
Porra, R. J., Thompson, W. A. & Kriedemann, P. E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta (BBA) - Bioenerg. 975, 384–394 (1989).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Acknowledgements
Authors S.M.J., C.R.Y., and D.Y.L. acknowledge the support from the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2020R1A2C3005834, NRF-2022R1A4A1030421) and by the Korean Fund for Regenerative Medicine (KFRM) grant funded by the Korean government (grant number: KFRM24A0105L1). We would like to especially thank Prof. EonSeon Jin and researcher Junhwan Jang of the Dept. of Life Science, Hanyang University for the experimental support for the measurement of photosynthetic efficiency.
Author information
Authors and Affiliations
Contributions
S.M.J., C.R.Y., and D.Y.L. conceived the research design. S.M.J., C.R.Y., and J.Y.K. performed the experiments. S.M.J., C.R.Y., and D.Y.L. conducted data processing. S.M.J., C.R.Y., and D.Y.L. contributed to the analysis and interpretation of data. S.M.J., C.R.Y., H.S.K., and D.Y.L. grafted the manuscript. S.M.J., C.R.Y., H.S.K., and D.Y.L. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
Dong Yun Lee reports a relationship with Elixir Pharmatech Inc. that includes: board membership and equity or stocks. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Libing Liu and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jang, S., Yoo, C., Kim, H.S. et al. Oxygenating respiratoid biosystem for therapeutic cell transplantation. Nat Commun 15, 9151 (2024). https://doi.org/10.1038/s41467-024-53246-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53246-w