Abstract
Clinical implementation of therapeutic genome editing relies on efficient in vivo delivery and the safety of CRISPR-Cas tools. Previously, we identified PsCas9 as a Type II-B family enzyme capable of editing mouse liver genome upon adenoviral delivery without detectable off-targets and reduced chromosomal translocations. Yet, its efficacy remains insufficient with non-viral delivery, a common challenge for many Cas9 orthologues. Here, we sought to redesign PsCas9 for in vivo editing using lipid nanoparticles. We solve the PsCas9 ribonucleoprotein structure with cryo-EM and characterize it biochemically, providing a basis for its rational engineering. Screening over numerous guide RNA and protein variants lead us to develop engineered PsCas9 (ePsCas9) with up to 20-fold increased activity across various targets and preserved safety advantages. We apply the same design principles to boost the activity of FnCas9, an enzyme phylogenetically relevant to PsCas9. Remarkably, a single administration of mRNA encoding ePsCas9 and its guide formulated with lipid nanoparticles results in high levels of editing in the Pcsk9 gene in mouse liver, a clinically relevant target for hypercholesterolemia treatment. Collectively, our findings introduce ePsCas9 as a highly efficient, and precise tool for therapeutic genome editing, in addition to the engineering strategy applicable to other Cas9 orthologues.
Similar content being viewed by others
Introduction
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) systems serve as adaptive antiviral immunity mechanisms in bacteria, archaea, and large bacteriophages1,2,3. The CRISPR-associated (Cas) nucleases, repurposed for mammalian genome modification, can be programmed with single guide RNAs to target specific loci and introduce DNA double-stranded breaks (DSBs)4,5,6,7,8. These are subsequently repaired by the cellular DNA repair machinery, primarily through the non-homologous end-joining (NHEJ) pathway, leading to small insertions and deletions5,6,8,9,10,11. Alternatively, when presented with a DNA molecule homologous to the DSB locus, cells can employ homology-directed repair (HDR) to insert it into the genome12,13,14. Cas nucleases also form the basis for advanced genome editing approaches such as Base Editing15,16, Prime Editing17, and Epigenome Editing18,19,20. The wide applicability of these tools includes cell line engineering5,6,8, animal model development21,22,23, genetic screens24,25,26, cell therapies for cancer27,28,29, and curative treatments for genetic disorders30,31.
RNA-guided nucleases derived from CRISPR systems32,33, along with the recently identified OMEGA systems34,35,36, comprise a diverse toolkit for genome editing in human cells and in vivo. Streptococcus pyogenes Cas9 (SpCas9), the most studied Cas-enzyme, has proven to be a highly efficient and versatile tool for genome editing, becoming the gold standard in the field. SpCas9 was employed in clinical trials for gene therapy and in the first ex vivo genome edited cell therapy, that was recently approved by the FDA30,31,37. However, its promiscuous activity remains a concern for the wider application of genome editing in the clinic38,39,40,41,42,43,44,45,46,47,48,49,50. In response, several studies rationally engineered SpCas9 using targeted mutagenesis or applied directed evolution to eliminate its off-target activity51,52,53,54,55,56,57,58,59. Moreover, other natural and engineered Cas9 nucleases with high-fidelity properties from other prokaryotic species were introduced and suggested as additional tools for genome editing60,61,62,63,64,65,66,67,68. Our previous work characterized PsCas9 as an intrinsically high-fidelity enzyme of the Type II-B subfamily69. Viral delivery of PsCas9 to mouse liver in vivo resulted in high levels of editing with negligible off-target events and fewer chromosomal translocations, thus offering a safer alternative for therapeutic applications compared to SpCas9.
In this study, we thoroughly interrogate PsCas9 function through cellular, structural, and biochemical studies and engineer it for therapeutic genome editing applications. We found that the editing activity of PsCas9 is hampered in conditions of limited intracellular concentration, and it exhibits a relatively low affinity to DNA in vitro compared to SpCas9. We propose that the low editing activity of PsCas9 in cells could be rescued by improving its affinity to DNA. To this end, we solve a high-resolution structure of PsCas9 using cryo-electron microscopy (cryo-EM), thereby providing a foundation for the rational engineering of PsCas9. Optimization of the PsCas9 sgRNA scaffold results in a modest increase in its genome editing efficacy, while targeted enzyme mutagenesis leads to a remarkable improvement (up to 20-fold) across a wide set of genomic targets. These enhancements in gene editing activity in cells are correlated with more efficient DNA interaction in vitro. Importantly, the high-fidelity properties of the wild-type enzyme, including a favourable off-target profile and low translocation frequency, are preserved upon engineering. Employing a similar strategy, we modify the well-established member of Type II-B subfamily Francisella novicida Cas9 (FnCas9)62 and increase its activity up to 15-fold. Finally, we evaluate the performance of the engineered version of PsCas9, ePsCas9, using lipid nanoparticle (LNP) delivery to mouse liver to disrupt the Pcsk9 gene, a clinically relevant approach for hypercholesterolemia treatment70,71. We demonstrate that ePsCas9 induces a high level of editing in the liver and a concurrent decrease of Pcsk9 levels in blood plasma. Collectively, this work positions ePsCas9 as a promising tool for safe and effective in vivo genome editing applications, as well as proposes an engineering strategy to enhance the activity of other Cas9 orthologs to further expand the CRISPR toolbox for medical use.
Results
PsCas9 editing activity is limited by its intracellular concentration
We previously characterized PsCas9, a member of the Type II-B family, as a highly active and precise enzyme69. PsCas9 recognizes the same NGG PAM as SpCas9, and thus, can be conveniently benchmarked against it. In line with our previous work, PsCas9 induces genome editing in HEK293T cells with high efficiency (up to 80%) and comparable to SpCas9 at two tested targets (EMX1a and PCSK9) when delivered via plasmid vectors (Fig. 1a). This delivery approach enables sustained, high expression levels of Cas9 and its sgRNA, providing excessive amounts of functional ribonucleoproteins (RNPs), and thus, high levels of editing. To assess the activity of these enzymes in a more controlled setting, we transfected cells with plasmids overexpressing the Cas9 protein only, and then introduced synthetic sgRNA in varying quantities, thereby restricting the intracellular concentration of active Cas9 RNPs. As expected, editing efficiency increased with higher doses of delivered sgRNA for both enzymes (Fig. 1b, c). However, in stark contrast to our earlier observations, PsCas9 exhibited an order of magnitude lower efficacy than SpCas9 across all tested conditions. This finding suggests that the genome editing activity of PsCas9 is limited when the RNP abundance is constrained in the cell.
a Genome editing activity of SpCas9 and PsCas9 in HEK293T cells mediated via plasmid DNA transfection. Editing efficiency was evaluated as the percentage of reads with indels using amplicon sequencing. Data are shown as mean ± SD for n = 3 biological repeats. b, c Genome editing activity of SpCas9 and PsCsa9 in HEK293T cells mediated via plasmid DNA encoding Cas9s transfection followed by synthetic sgRNA transfection at EMX1a (b) and PCSK9 (c) sites. Editing efficiency was evaluated using amplicon sequencing. Data are shown as mean ± SD for n = 3 biological repeats. d Schematic of in vitro binding experiment of Cas9 to dsDNA “target substrate”. 38 bp substrate contains a single target site (blue) and PAM (orange). The remaining DNA (grey) was depleted of PAMs. DNA was labelled with FAM on the 3’-end of the bottom strand. e, f Sp- and PsCas9 interaction with target substrates assessed by fluorescence polarization of FAM fluorophore. The signal was fitted to a 4-parametric logistic function and binding constant Kd was extracted. Data are shown as mean ± SD for n = 3 technical replicates. g Schematic of in vitro binding experiment of Cas9 to dsDNA “no-target substrate” lacking target site. 80 bp substrate contains 17 or 19 PAMs distributed across it (orange) for EMX1a and PCSK9, respectively. DNA was labelled with FAM on the 3’-end of the bottom strand. h, i Sp- and PsCas9 interaction with no-target substrates assessed by fluorescence polarization of FAM fluorophore. Data are shown as mean ± SD for n = 3 technical replicates.
We hypothesized that the reduced editing activity of PsCas9 could be attributed to its biochemical properties. We have previously demonstrated that PsCas9 RNP cleaves DNA targets at rates similar to SpCas9 in vitro69. Therefore, we next examined the target DNA binding properties of PsCas9 using a fluorescence polarization assay. For this, we designed fluorescent double-stranded (ds) DNA substrates containing a single target site (EMX1a or PCSK9) and measured their interaction with Ps- or SpCas9 loaded with the respective sgRNAs (Fig. 1d). Concentration-dependent increases in fluorescence polarization signal were observed for both Cas9 RNPs (Fig. 1e, f). SpCas9 demonstrated strong interaction with both substrates, exhibiting an affinity of around 10 nM — a value comparable to previous reports using orthogonal binding assays72,73,74. In contrast, the binding affinity of PsCas9 to DNA was ~10- and 2-fold weaker for EMX1a and PCSK9 substrates, respectively. This finding motivated us to investigate the binding mode of PsCas9 in more detail.
Cas9 RNP interrogation of DNA in search of PAMs is a crucial step preceding target recognition72. We used the fluorescence polarization assay to evaluate PsCas9 interaction with a DNA substrate containing PAM sequences but no target75. This method enables the assessment of Cas9 RNP interaction with DNA through transient binding to PAMs only. We designed fluorescent dsDNA EMX1a and PCSK9 substrates, containing 17 and 19 GG dinucleotide PAMs spread across the sequence, respectively; and measured their interaction with Cas9 RNPs loaded with non-matching sgRNAs (Fig. 1g). SpCas9 displayed a significant concentration-dependent increase in FP signal, indicating active DNA interrogation and PAM interaction (Fig. 1h, i). In contrast, PsCas9 showed almost no increase suggesting its PAM binding is weak.
Collectively, our findings suggest that diminished PsCas9 editing activity in conditions when RNP abundance is limited can be a result of its relatively weak binding to DNA, in particular, at the interrogation step. We hypothesize that PsCas9 activity could be boosted upon strengthening its interaction with DNA. Thus, we sought to supplement our in vitro data with structural studies of PsCas9 RNP and use them for rational engineering of the enzyme for improved activity.
Cryo-EM structure of PsCas9
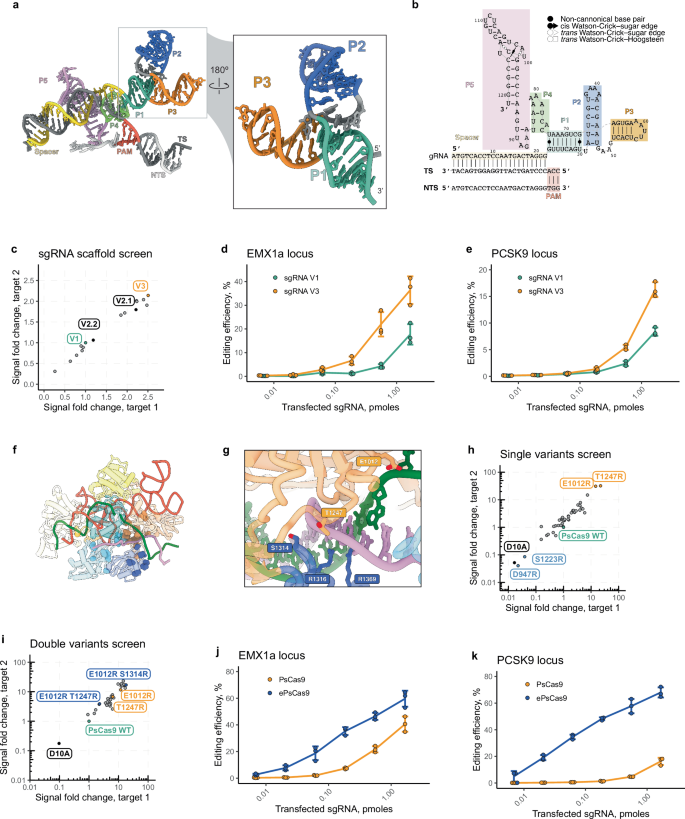
To understand the mechanisms of DNA recognition by PsCas9, we prepared a complex of PsCas9 with its cognate sgRNA and EMX1a dsDNA and acquired a cryo-EM dataset (Supplementary Data 1). During data collection, we noticed that the complex adopts a preferred orientation in ice, which we overcame by tilting the stage by −30° for the remainder of movie acquisition. After rounds of 2D and 3D classification, we obtained a cryo-EM reconstruction of PsCas9 at a global resolution of 2.9 Å (Supplementary Fig. 1). The quality of our map was sufficient for de novo modelling of the complete complex, including 1375 of the 1409 amino acids, the full 22-bp R-loop, and 121-nt of the sgRNA (Fig. 2a–c). The final 10-nt of the sgRNA (positions 122 onwards) were not resolved in the map, likely due to flexibility.
a Domain architecture of PsCas9. b 2.9 Å-resolution cryo-EM reconstruction of PsCas9 in the productive state. Both target and non-target strands (TS and NTS, respectively) have been cleaved, and the HNH domain remains positioned at the scissile phosphate of the TS. c Cartoon representation of PsCas9 model. d Structural basis of NGG PAM recognition. e Comparison of PsCas9 (green cartoon) with FnCas9 (orange cartoon, PDB ID 5B2O). The crystal structure of FnCas9 represents a dead-end, non-productive state, where the HNH domain is positioned far from the cleavage position of the TS. In the productive state, HNH is shifted by up to ~60 Å to the centre of the target strand. The REC and NUC lobes are largely consistent otherwise.
Akin to previously determined type II-A, -B, and -C CRISPR effector nucleases60,62,76,77,78, PsCas9 has a typical bilobed architecture, with the REC1, REC2 and REC3 domains constituting the REC lobe, and the Wedge, PI, RuvC and HNH domains constituting the NUC lobe (Fig. 2a, b). The sgRNA follows a tortuous path, interweaving between the flexibly tethered REC domains. Due to conformational heterogeneity, the REC2 domain is poorly resolved, as it has also been observed for SpCas958,79.
Since the PAM site of the EMX1a dsDNA target is excellently resolved in our reconstruction, we could unambiguously model the PsCas9 residues responsible for PAM recognition – R1316 and R1369. These arginine residues make a bidentate interaction with the Hoogsteen faces of the two guanosine bases in the NGG motif (Fig. 2d). While this is consistent with the mechanism of PAM readout by SpCas976,77, the two arginine residues used by SpCas9 (R1333 and R1335) are much closer together in sequence space as in comparison to PsCas9. A similar observation was documented for another Type II-B family member, FnCas962. This suggests that despite a lack of sequence homology in this region, SpCas9 and PsCas9 have evolutionarily converged on a conserved mechanism to recognise a 5’-NGG-3’ PAM motif.
Within our reconstruction, the HNH domain is docked at the target strand (TS), and the scissile phosphate of the 3rd TS position has been cleaved, indicating that our structure corresponds to the active, productive state. A previously determined structure of FnCas9 is in a non-productive state, with both DNA strands being intact62 (Fig. 2e). Comparison of our productive state structure with this non-productive state homologue reveals that the HNH domain must be repositioned by up to ~ 60 Å to successfully perform catalysis. Apart from the HNH domain, the overall structural architecture of the complexes showed a high degree of similarity between FnCas9 and PsCas9 (Fig. 2e). We observed a well-resolved density for two Mg2+ ions in the HNH and RuvC active sites and both target and non-target strands (NTS) are cleaved (Supplementary Fig. 1f). While structures of type II-A and -C Cas9 enzymes have been determined with both active sites in the productive state58,80,81, our structure represents the structure of a type II-B Cas9 enzyme in a bona fide productive state.
Type II CRISPR effector nucleases typically cleave both the target and non-target strands (TS & NTS, respectively) three nucleotides upstream of the PAM82. In our structure, we observed six NTS nucleotides between the PAM and the scissile phosphate, confirming that PsCas9 introduces double strand breaks as staggered cuts (Supplementary Fig. 1f), as suggested before69.
Collectively, we obtained a high-quality model of PsCas9 RNP, with protein-DNA interaction surfaces and sgRNA folding resolved in exceptional detail. Utilizing these structural insights, along with our biochemical studies, we sought to engineer PsCas9 for enhanced performance in cells via sgRNA scaffold optimization and targeted protein mutagenesis.
Rapid evaluation of Cas9 genome editing activity with luminescence reporter
Amplicon sequencing is a state-of-the-art approach for accurate evaluation of genome editing events. Despite the recent technological advances, NGS-based experimental readout has limited throughput and high cost. To overcome these limitations, we constructed a genome editing luminescence reporter integrated into the genome of HEK293T cells at the HBEGF locus83 (Supplementary Fig. 2a). Therein, the Nanoluciferase (Nluc) translation is interrupted by upstream stop codons, which are surrounded by nucleotide sequences with microhomologies. We anticipated that the Cas9-induced DSBs in this locus could undergo repair via the microhomology-mediated end joining (MMEJ) pathway11, resulting in the excision of the stop codon, thereby enabling translation of the Nluc gene.
To assess the reporter function and fidelity, we targeted SpCas9 and PsCas9 to the cassette locus with two independent sgRNAs to induce DSBs between the microhomology sites and then analysed the repair outcome with amplicon sequencing. We found that the programmed MMEJ deletion variant was a prevalent repair outcome for both Sp- and PsCas9 (Supplementary Fig. 2b). We further tested if reporter locus targeting with Cas9 nucleases results in Nluc expression, which we assayed using a luminescence readout. To this end, we overexpressed Sp- or PsCas9 in the reporter cells along with the delivery of corresponding sgRNAs in a fixed amount and later measured luminescence as well as analysed genomic DNA with amplicon sequencing. Background normalized luminescence signal was strongly correlated with the frequency of the MMEJ repair variant, which restores Nluc expression (Supplementary Fig. 2c). This correlation was confirmed for both Sp- and PsCas9 at a wide range of delivered sgRNA doses highlighting the reliability of our system. As such, this reporter system can be widely employed as an initial screening assay to evaluate the efficacy of compatible Cas9 nucleases without the need for amplicon sequencing analysis, providing faster results and increased throughput.
PsCas9 sgRNA scaffold engineering
Several studies have demonstrated that the activity of various CRISPR-Cas enzymes can be enhanced through the optimization of guide RNA architecture and sequence67,84,85. Our cryo-EM structure exposed an unusual folding of PsCas9’s sgRNA (Fig. 3a, b). Most characterized Cas9 enzymes contain a sgRNA with a singular, elongated hairpin lying within the REC lobe, formed by repeat and antirepeat sequences stemming from crRNA and tracrRNA, respectively60,62,76,77,78. Intriguingly, PsCas9’s sgRNA forms a branched structure, where the P1 duplex downstream from the spacer bifurcates into two hairpins, P2 and P3. While the P1 and P3 structures interact with REC1 and Wedge domains and reside within the REC lobe, P2 appears to have minimal contact with the protein (Figs. 2c, 3a). Furthermore, the absence of the 10-nucleotide tail downstream of the terminal hairpin P5 in our cryo-EM model suggested its potential redundancy. We hypothesized that the P2 motif and the tail beyond the terminal hairpin could be trimmed without impairing the enzyme’s activity.
a, b sgRNA structure of PsCas9 and its sequence folding. c Editing activity of PsCas9 with different sgRNA scaffold variants analysed using a cell-based luminescence reporter assay. Reporter cells were transfected with plasmid DNA encoding PsCas9 followed by a single dose transfection of a synthetic sgRNA (see Supplementary Fig. 2). The luminescence signal of each sgRNA variant was normalized to the initial sgRNA, V2, for both tested targets. Data are shown as mean values for n = 3 biological repeats. d, e Genome editing activity of PsCsa9 combined with original, V1, and optimized, V3, sgRNA scaffold in HEK293T cells mediated via plasmid DNA encoding PsCas9 transfection followed by synthetic RNA transfection at EMX1a (d) and PCSK9 (e) sites. Data are shown as mean ± SD for n = 3 biological repeats. f, g Structural view of PsCas9 amino acids in the vicinity of the PAM distal DNA end. h, i Evaluation of PsCas9 mutant variants activity using cell-based luminescence reporter. Reporter cells were transfected with plasmid DNA encoding PsCas9 mutants followed by single dose transfection of synthetic sgRNA targeting the cassette. The luminescence signal was normalized to the wild-type, WT, enzyme for both target sites. The screen was performed for a set of single (h) and double (i) substitution mutant variants. Data are shown as mean values for n = 3 biological repeats. j, k Gene editing activity of PsCsa9 and its engineered E1012R S1314R mutant, ePsCas9, in HEK293T cells mediated via plasmid DNA encoding Cas9 transfection followed by synthetic RNA transfection at EMX1a (j) and PCSK9 (k) sites. Data are shown as mean ± SD for n = 3 biological repeats.
We therefore designed 15 sgRNA scaffolds with varying levels of P2 hairpin truncation and lacking the 3’- tail (Supplementary Data 2). These variants were paired with two spacers targeting the reporter locus and compared with the original sgRNA, which we denote as V1 (Fig. 3c). Complete excision of the 3’-tail from the sgRNA (V2.1) resulted in ~2-fold increase in editing activity at both target sites. Trimming of the P2 fragment up to its complete removal (V2.2) marginally affected the enzyme’s activity; however, the simultaneous removal of P2 and the 3’-tail resulted in the highest activity variant (V3) with a 2.5-fold increase over V2.
To further validate the performance of the optimized sgRNA scaffold, V3, we targeted two endogenous sites—EMX1a and PCSK9—and measured editing activity with amplicon sequencing, as described earlier (Fig. 3d, e). At both sites, PsCas9 loaded with V3 sgRNA demonstrated elevated levels of editing across conditions with up to a 2-fold increase over V2 at the highest tested dose. We next investigated whether the in vitro DNA binding of PsCas9 was altered by the optimized sgRNA scaffold. Interestingly, the binding affinity of PsCas9 to the target-containing DNA substrate was improved at both substrates (EMX1a: from ~112 nM to ~5 nM and PCSK9: from ~15 nM to 11 nM) and was in a similar range with SpCas9 (Supplementary Fig. 3a, b). Interactions with the DNA containing only PAMs but no target site were affected only marginally for both EMX1a and PCSK9 substrates, as evidenced by a negligible increase in polarization signal, suggesting that PsCas9 PAM binding remains markedly weaker than that of SpCas9 (Supplementary Fig. 3c, d).
Altogether, the engineered sgRNA scaffold of PsCas9 promotes increased genome editing likely through increased binding affinity to the DNA target sequence without affecting PAM interrogation. We hypothesize that trimming of the P2 hairpin may reduce the propensity of the sgRNA to adopt stable alternative, non-native structural conformations, thereby yielding an increased amount of active RNP complex for interaction with DNA both in vitro and inside the cell. This may also be the case for the increased activity observed for the 3’ truncated sgRNA guides.
Rational protein engineering of PsCas9
To further enhance PsCas9 genome editing properties, we designed a set of mutations in the RuvC, Wedge and PI protein domains based on the cryo-EM structure to facilitate the interaction between the PAM proximal DNA tail and the enzyme. Our primary candidates encompassed neutral and negatively charged amino acids predominantly located in flexible regions and proximate to DNA (Fig. 3f, g). We replaced these with positively charged arginine or polar amino acids to stabilize the protein interaction with the DNA backbone without imposing any sequence specificity (Supplementary Data 3).
We introduced plasmids encoding 41 single amino acid substitution mutants of PsCas9 (hereafter called variants) along with a fixed amount of sgRNAs targeting two independent sites to evaluate their relative activity in reporter cells (Fig. 3h). The control construct, which disrupts the RuvC active site (D10A), led to an almost complete loss of reporter response since Cas9-nickases typically result in minimal genome editing activity. Notably, two tested variants, D947R and S1223R, displayed nickase-like activity levels. D947, a negatively charged residue in proximity to target DNA, fits our initial candidate nomination hypothesis. However, a more careful structural analysis reveals its interaction with R921 and K933 residues in the RuvC domain (Supplementary Fig. 4a). The D947R substitution might cause strong electrostatic repulsion with these residues, disrupting RuvC folding and its function. Conversely, S1223, located in the DNA proximal surface of the Wedge domain, appears to facilitate PAM recognition through its hydroxyl group’s interaction with the amine of the guanine base at the second position of the PAM (Supplementary Fig. 4b). Substituting this serine with a bulkier arginine could impair PAM recognition, thereby substantially reducing PsCas9 activity. The majority of other variants either altered the activity marginally or increased it by more than twofold at both targets. Remarkably, two single-substitution variants (E1012R, T1247R) exhibited above 10-fold activity enhancement (Fig. 3h, orange scatters).
We then combined a subset of the activity-enhancing mutants and designed 30 double-substitution variants, which we tested in a similar fashion (Fig. 3i). As anticipated, several double mutants exhibited further activity improvements, although the effect was not strictly additive, for example with the double E1012R T1247R variant trailing the activity of each single mutant (Supplementary Data 3). Conversely, multiple other derivatives of the E1012R variant showed further improvements in editing activity using our reporter assay.
We prioritized a particularly promising variant (E1012R S1314R, named here engineered- or ePsCas9) and assessed its editing properties on endogenous genomic loci. For both tested sites, EMX1a and PCSK9, the editing activity of ePsCas9 was several folds higher compared to the wild-type PsCas9 across all tested sgRNA concentrations (Fig. 3j, k). At the lower amounts, the activity was improved most noticeably with above 10-fold increase over the wild-type. To assess if the increased genome editing activity of the engineered variant indeed stems from the improved DNA binding properties, we performed further in vitro experiments with recombinant ePsCas9 protein. The affinity of ePsCas9 loaded with sgRNA V3 scaffold was similar to the one of PsCas9 WT at target containing EMX1a and PCSK9 substrates (Supplementary Fig. 3e, f). While a noticeable increase in the FP signal was observed using EMX1a and PCSK9 substrates containing no target (Supplementary Fig. 3g, h), it was still markedly lower than that of SpCas9. This finding suggests that PAM interrogation is limiting PsCas9 activity in cells. Collectively, our structure-based rational mutagenesis campaign successfully enhanced PsCas9 genome editing activity introducing ePsCas9.
Our successful efforts in increasing the activity of PsCas9 led us to explore whether this approach could be extended to other Cas9 enzymes, particularly those of the Type II-B subfamily. The structural comparison revealed a high degree of similarity between FnCas9 and PsCas9 (Fig. 2e). Akin to PsCas9, FnCas9 has been described to have modest genome editing activity in cells86 and weak DNA interaction in vitro87, therefore making it a prime candidate for engineering. We investigated the previously published crystal structure of FnCas9 and selected 12 positions for mutagenesis, applying the same rationale used for PsCas9 (Supplementary Fig. 5a). Given that FnCas9 shows the same NGG PAM preference, we again employed our cell-based editing assay to evaluate its activity. Wild-type FnCas9 demonstrated reporter activation at both target sites, with a stronger response at target 2 (Supplementary Fig. 5b). We therefore assessed the performance of the designed FnCas9 variants at target 2. Half of the variants displayed increased activity above 2-folds over the wild-type (Supplementary Fig. 5c). Three top candidates (E1369R, N1448R and E1603R) underwent further validation across a range of sgRNA concentrations, where they exhibited significant activity enhancements (Supplementary Fig. 5d). These results indicate that our engineering approach could be applied to other Type II-B family members and enhance their genome editing efficacy.
ePsCas9 is a highly active and specific tool for genome editing
We conducted a thorough evaluation of ePsCas9’s editing properties. Firstly, we picked 18 additional endogenous target sites and compared the on-target editing capabilities of Ps-, ePs-, and SpCas9 using amplicon sequencing (Fig. 4a). Here, we used a single limiting dose of sgRNAs (0.5 pmol) to avoid editing saturation caused by Cas9 RNP excess. We observed up to 20-fold enhancement in ePsCas9 activity over the enzyme prior engineering, with a median 5-fold increase across tested targets (Fig. 4a and Supplementary Fig. 6). ePsCas9 performance was also approaching SpCas9 with comparable median efficacy over the target set (Fig. 4a). Interestingly, while SpCas9 outperformed ePsCas9 at certain loci (ATTR, B2M2, HEK3), it was outperformed by ePsCas9 at others (B2M1, PCDC1, STAT1), indicating the presence of enzyme-specific target sequence preferences (Supplementary Fig. 6). This phenomenon is well characterized in literature60,62,66,78 with high throughput screening approaches employed to decipher the enzyme-specific sequence preferences24,88,89. Yet, such data set of ~20 targets is insufficient to meaningfully determine ePsCas9’s target selection rules.
a Editing activity of SpCas9, PsCas9 and ePsCas9 across various genomic targets. Editing was induced using plasmid DNA and synthetic sgRNA transfection in HEK293T cells. Distribution of editing at 18 sites for n = 3 biological repeats is represented as combined box-violin plots. The central line shows the median, with the box edges indicating the first (Q1) and third (Q3) quartiles. Whiskers extend to 1.5 times the interquartile range (IQR) from Q1 and Q3, and individual points represent outliers. A two-way ANOVA with Tukey post-hoc test was used to evaluate statistical significance (ns p-value = 0.09, ***p-value < 0.0001; SpCas9-PsCas9 p = 2e-10, ePsCas9-PsCas9 p = 8e-6). b Specificity of SpCas9, PsCas9 and their engineered variants evaluated with CHANGE-seq in the human genome using a promiscuous sgRNA targeting HEKs4 and a specific sgRNA targeting TRAC sites. Specificity values were calculated as the number of on-target reads divided by the total number of reads accounted for by CHANGE-seq hits. Data are shown as mean ± SD for n = 3 technical replicates. c Number of targets consistently discovered across technical replicates of CHANGE-seq. d Translocation frequency induced by SpCas9, PsCas9 and their engineered variants evaluated for two independent events in HEK293T cells. Editing was induced using plasmid DNA and synthetic sgRNAs targeting two loci simultaneously. Translocation frequency was evaluated using ddPCR and normalized to the geometric mean of editing efficiencies at each site. Data are shown as mean ± SD for n = 3 biological repeats. e Schematic of in vivo study to evaluate editing capacity of SpCas9 and ePsCas9 in mice. f in vivo genome editing activity of SpCas9 and ePsCas9. Data are shown as mean ± SD for n = 3, 3 and 4 biological repeats for buffer, SpCas9 and ePsCas9, respectively. g Plasma Pcsk9 levels in mouse plasma post genome editing with SpCas9 and ePsCas9. PCKS9 abundance was evaluated with ELISA at the termination stage. The values were normalized to the mean signal of the buffer treatment group. Data are shown as mean ± SD for n = 3, 3 and 4 biological repeats for buffer, SpCas9 and ePsCas9, respectively. h Correlation between gene editing activity and plasma Pcsk9 level in mice. Pearson correlation value is indicated in the top right corner.
Secondly, we evaluated the performance of ePsCas9 in various cellular contexts. In our previous experiments we employed HEK293T cells, the primary model system for evaluation of gene editors’ performance, due to ease of DNA and sgRNA delivery. Transfection-based plasmid DNA delivery to other cell lines is often inefficient and not tolerated well. To overcome this obstacle, we synthesised ePsCas9 and SpCas9 mRNA. mRNAs with their respective synthetic sgRNAs at a single dose (0.5 pmoles) were transfected to HEK293T cells, as well as to four additional cell lines (HeLa, Huh7, DLD-1 and iPSCs) representing different tissues. We found that at 5 tested target sites editing activity of ePsCas9 remains high and comparable to SpCas9 (Supplementary Fig. 7). In some cases, editing was approaching 100% indicating high efficiency of mRNA and sgRNA delivery in cell cultures. Notably, iPSCs displayed the lowest levels of editing across our panel, potentially due to high sensitivity of these cells to transfection procedures in general.
Thirdly, the specificity of ePsCas9 was assessed by analysing its off-target DNA cleavage. Our prior study has demonstrated high fidelity of the wild-type enzyme69. However, the increased on-target activity seen in ePsCas9 could potentially suggest an increased activity overall, and thus off-target editing. Hence, we employed CHANGE-seq to examine the off-target activity of Cas9-enzymes in an unbiased manner90. We benchmarked ePsCas9 against SpCas9 and its commercially available high-fidelity variant HiFi SpCas956 with a well-characterized promiscuous sgRNA targeting the HEKs4 site43, as well as with a therapeutically-relevant specific sgRNA targeting the TRAC gene29 (Fig. 4b). For all Cas9-RNPs, the on-target sequence was readily detected amongst CHANGE-seq reads while off-target sequences exhibited considerable variation for each enzyme (Supplementary Data 4 and Supplementary Fig. 8a, b). PsCas9’s specificity was 10- and 5 times higher than that of SpCas9 for HEKs4 and TRAC targets, respectively. While SpCas9 is known to be a promiscuous enzyme, its engineered variant HiFi SpCa9 was also outperformed by PsCas9 several folds (Fig. 4b). Remarkably, ePsCas9 specificity remained high, comparable to the wild-type enzyme. We also analysed the number of off-targets consistently discovered across technical replicates (Fig. 4c). For both tested sgRNAs, ePsCas9 showed some increase in identified off-targets compared to the wild-type PsCas9, yet outperformed SpCas9 by at least an order of magnitude and its high fidelity variant HiFi SpCas9 up to 9-fold. Thus, our CHANGE-seq data suggest that the modifications enhancing PsCas9 activity have only a minimal impact on its fidelity.
Lastly, we evaluated the propensity of ePsCas9’s DSBs to promote chromosomal translocations. Large genomic rearrangements including intra- and inter-chromosomal translocations are possible consequences of Cas9 genome editing, and along with off-target editing, raise safety concerns41,42,48. We utilized our previously established translocation assays for two pairs of loci upon simultaneous DNA cuts69,91. Concurrent DSBs in HEK293T cells were induced using Cas9 nuclease overexpression and delivery of a pair of sgRNAs. The translocation events between the targeted loci were then detected using digital droplet PCR (ddPCR) and normalized to the editing efficiency measured with amplicon sequencing (Fig. 4d). While SpCas9 displayed high levels of translocations in both assays, translocations induced by PsCas9 were below the detection threshold, potentially due to its low editing activity (Fig. 4d and Supplementary Fig. 9a, b). ePsCas9 showed high levels of editing at each target and detectable levels of translocations. However, ePsCas9-induced translocations were ten and four times less frequent than that of SpCas9 in HIST1H2BC-HBEGF and PCKS9-HBEGF assays, respectively (Fig. 4d). Intriguingly, HiFi SpCas9 showed levels of translocations comparable to SpCas9. These findings reiterate that the improved editing capabilities of ePsCas9 only minimally affect its inherent editing fidelity rendering an efficient and safe editor.
In vivo editing by mRNA-encoded ePsCas9 delivered with LNPs
Lipid nanoparticles (LNPs) are recognized for their efficient and transient delivery of genome editing components to the liver and show promise in early clinical trials30,31,92,93,94. To assess the efficacy of ePsCas9 in therapeutic genome editing with LNP delivery, we focused on the PCSK9 gene, a key target for the treatment of familial hypercholesterolemia (FH)70,71. PCSK9 disruption through CRISPR-mediated genome editing offers a potentially one-time, long-term therapeutic intervention for FH. Therefore, targeting the Pcsk9 gene in mouse liver provides an ideal model to evaluate the efficacy of genome editing tools. To this end, we formulated LNPs encapsulating ePsCas9 mRNA and sgRNA targeting the mouse Pcsk9 gene using established procedure95. We also prepared LNPs containing SpCas9 mRNA and a respective sgRNA with a highly modified scaffold reported previously to facilitate genome editing activity in vivo92. These formulations were intravenously injected into C57BL/6NCrl mice, and their editing outcomes was examined in the liver after 7 days (Fig. 4e).
While SpCas9 with chemically modified sgRNA showed ~ 20% editing, the mice treated with ePsCas9 reached an average of 60% editing, indicating efficient delivery to the mouse liver in vivo (Fig. 4f). We also analysed Pcsk9 protein levels in blood plasma after LNP delivery and found a reduction in both treated groups compared to the control (Fig. 4g). As expected, a stronger decrease in Pcsk9 levels correlated with higher levels of genome editing (Fig. 4h). Furthermore, in mice treated with ePsCas9 as well as SpCas9, we observed no evident safety concerns regarding liver function and overall health condition (Supplementary Fig. 10a, b), as evaluated by body weight monitoring and liver enzymes activity. Collectively, these findings suggest that ePsCas9 is an efficient and safe editor for in vivo genome editing when delivered using LNPs.
Discussion
The advent of CRISPR editing technologies made genome modifications in vivo accessible. However, a key challenge in therapeutic genome editing lies in the efficient delivery of CRISPR components33. LNPs have recently emerged as a viable delivery modality for the liver, due to their cost-effectiveness and improved safety profile compared to viral vectors92,93,94. However, the swift metabolism and limited amount of RNA payloads present a significant obstacle, narrowing the window for efficacious genome editing.
Our previous work introduced PsCas9, a Type II-B family enzyme that demonstrated high fidelity and notable activity in vivo when delivered via viral vectors69. However, in a scenario mimicking LNP delivery within cell cultures, PsCas9 displayed limited editing activity (Fig. 1).
In this study, we leveraged the high-resolution cryo-EM structure of PsCas9 to guide enzyme engineering (Fig. 2). We hypothesized that PsCas9’s modest editing activity could be attributed to its limited DNA-binding ability in vitro (Fig. 1). Through a combination of protein and sgRNA modifications (Fig. 3), we achieved a boost in DNA binding and substantially improved PsCas9’s editing efficacy in cell cultures across multiple targets and cellular contexts. We also showed engineered PsCas9 applicability for therapeutically relevant in vivo genome editing (Fig. 4). At the single tested Pcsk9 target, ePsCas9 induces a higher level of editing than SpCas9 in the liver and a concurrent decrease of Pcsk9 levels in blood plasma. More in vivo experiments will be required to compare the performance of both nucleases at other loci.
Our mutagenesis efforts improved PsCas9 PAM interrogation activity modestly yet that was sufficient to enhance its editing activity up to 20-fold at some target sites. This suggests that PsCas9 initial DNA engagement is the rate limiting step for catalysing DNA cleavage in cellulo. Substantial enhancement in PsCas9 editing activity was achieved by introducing positively charged amino acids to DNA-interacting domains. It has been established that the NGG PAM recognition is a weak interaction, with an estimated dissociation constant of ~10 µM96. We propose that the E1012R E1314R and T1247R mutations contribute the enhanced DNA binding affinity of ePsCas9 through introduction of additional non-specific electrostatic contacts with the DNA phosphodiester backbone. This is reminiscent of the enhanced affinity of the PAMless variant SpRY-Cas9 which also uses non-specific contacts to enhance DNA binding affinity97. We successfully applied this approach to improve the editing efficacy of FnCas9, an enzyme phylogenetically relevant to PsCas9 albeit sharing only ~20% sequence homology. We believe this strategy could be further generalized and applied for the engineering of a broader range of RNA-guided nucleases in the Type II family.
In the future, it might be possible to add the SpRY or SpG mutations (or establish equivalent mutations that could be introduced) to ePsCas9 to broaden the DNA targeting abilities. However, one must consider that SpG and SpRY have ~25- and ~500-fold reduction in DNA cleavage rates relative to SpCas997, and given that such mutations are also likely to severely impact the on-target DNA cleavage efficiency of ePsCas9 it is worth evaluating how necessary PAM-flexible DNA targeting is for the desired genome editing application.
The therapeutic application of CRISPR-based genome editing raises multiple safety concerns38,39,40,41,42,43,44,45,46,47,48,49,50. Editing at off-target sites that share a high level of homology with the intended target is a well-documented phenomenon potentially introducing harmful unwanted mutations. Off-target editing is mitigated by the introduction of high-fidelity Cas-enzyme through rational engineering, directed evolution and natural variants discovery51,52,53,54,55,56,57,58,59. Biochemical studies74,98 proposed that differences in Cas9 RNP dissociation rates between perfect and imperfect substrates are the major determinants for the DNA cleavage, and thus, off-target editing. In our previous work, we introduced PsCas9 as an enzyme with exceptional fidelity and virtually no off-target editing in vivo, which we attributed to its strong discrimination in off- vs on-target DNA cleavage in vitro69. Here, we boosted PsCas9 activity while preserving its high biochemical fidelity and favourable off-target profile as demonstrated by only a minor alteration of the enzyme’s specificity measured by CHANGE-seq.
Large genomic rearrangements, including chromosomal translocations, have been recently identified as an on-target consequence of Cas9-based genome editing that pose an additional safety risk41,45,48,49. Moreover, multiplexed editing introduces the potential for translocations between simultaneously targeted sites. Here, we showed that engineered PsCas9 introduces fewer translocations than SpCas9, maintaining the properties of the wild-type enzyme69. We also observed that the high-fidelity SpCas9 variant, HiFi SpCas9, introduces high levels of genomic translocations, despite its higher fidelity in off-target editing56. We believe that the low translocation properties of Ps- and ePsCas9 could be attributed to their specific DNA cleavage pattern forming 5’-overhangs. Well documented observations of Cas12-family enzymes to induce fewer translocations and generate staggered DSBs further support our hypothesis.61,67,68,99. Non-matching sticky DNA ends at DSB potentially pose a bigger challenge to repair for NHEJ machinery than blunt ends, and thus, inhibit translocation formation. In this context, ePsCas9 can serve as a safer alternative to existing editing tools with high on-target and low off-target activity providing the advantage of fewer translocations, in particular for multiplexed editing.
Overall, our study presents an additional tool for the CRISPR toolbox: a high-fidelity and high-activity genome editor, which is effective for in vivo applications using LNP delivery. While SpCas9 remains the gold standard in the field due to its remarkable activity, versatility, and wealth of accumulated research, several recently introduced genome editors offer competitive activity and enhanced fidelity. Even though these emerging enzymes may perform comparably on average, specific genomic locations may be better suited to one enzyme over another. Thus, the expansion of the CRISPR toolbox with additional enzymes greatly enhances our capacity to develop efficient genome editing therapies for a wider range of targets.
Methods
Ethical statement
All mouse experiments were approved by the AstraZeneca internal committee for animal studies and the Gothenburg Ethics Committee for Experimental Animals (license numbers: 162-2015+ and 2194-2019) compliant with EU directives on the protection of animals used for scientific purpose.
Cell culture and transfection procedures
HEK293T (GenHunter Corporation, Q401), HeLa (ATCC, CCL-2) and Huh7 (Riken Cell bank, RCB1366) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% foetal bovine serum (FBS). DLD-1 (ATCC, CCL-221) cells were cultured in RPMI 1640 + 2 mM Glutamine + 10%FBS. hiPSC were generated and maintained in a feeder-free human pluripotency culturing system, Cellartis DEF-CS 500 (Takara, Japan), according to manufacturer’s instructions100. The cells were maintained at 37 °C in a humidified incubator with 5% CO2. For transfection experiments, cells were seeded at a density of 2 × 10^4 cells per well in 96-well plates, 24 h prior to transfection.
For plasmid-based sgRNA delivery, cells were transfected using a mixture of 40 ng and 80 ng of plasmids encoding Cas9 and sgRNA, respectively. The transfection was carried out using 0.3 µl of FuGene reagent (Promega) in a final volume of 5 µl.
In experiments involving synthetic sgRNA delivery, 50 ng of the Cas9-encoding plasmid was initially transfected using 0.3 µl of FuGene reagent (Promega) in a final volume of 5 µl. After a 24 h interval, varying quantities of synthetic sgRNA (Synthego or Integrated DNA technologies, IDT) were transfected into the cells using 0.5 µl of Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) in a final volume of 10 µl. Plasmid sequences are provided in Supplementary Data 5.
In experiments involving simultaneous mRNA and sgRNA transfection, 100 ng of mRNA and 0.5 pmoles of respective sgRNA were transfected using 0.3 µl of Lipofectamine MessengerMAX (Thermo Fisher Scientific) in a final volume of 10 µl.
Amplicon Sequencing and editing efficiency analysis
To assess genome editing efficacy in HEK293T cells, genomic DNA was isolated 72 h post-transfection using the QuickExtract DNA Extraction Solution (Lucigen), following the manufacturer’s instructions in 50 ul final volume.
Primary amplicons were synthesized using the Phusion Flash High-Fidelity PCR Master Mix (Thermo Fisher Scientific). Reactions were set up in a 15 µl volume, comprising 0.25 µM target-specific primers (IDT) and 1.5 µl of genomic DNA. The PCR cycling conditions were set as follows: initial denaturation at 98 °C for 1 min, followed by 32 cycles of 98 °C for 10 s, 60–65 °C for 10 s, and 72 °C for 10 s. Post amplification, PCR products were cleaned up using Ampure XP beads (Beckman Coulter) and analysed on a Fragment Analyzer (Agilent Technologies).
For the indexing step, a secondary PCR was conducted using KAPA HiFi HotStart Ready Mix (Roche). The reaction included 1 ng of the primary amplicon and 0.5 µM of indexing primers (IDT) in a 25 µl total reaction volume. The thermal cycling conditions included an initial 72 °C for 3 min, followed by 98 °C for 30 s, then 10 cycles of 98 °C for 10 s, 63 °C for 30 s, and 72 °C for 3 min; followed by a final extension at 72 °C for 5 min. Post-indexing, amplicons were again purified using Ampure XP beads and quality controled on the Fragment Analyzer.
For sequencing library quantification, the Qubit 4 Fluorometer (Thermo Fisher Scientific) was employed. High-throughput sequencing was executed on the Illumina NextSeq platform, in adherence to the manufacturer’s guidelines. All primers, amplicon reference sequences, and target sites utilized in this study are catalogued in Supplementary Data 6, 7.
NGS data was demultiplexed by using bcl2fastq software. The fastq files were analyzed by CRISPResso101 version 2.2.12 with the following parameters: -q 30 –ignore substitutions max_paired_end_reads_overlap 300 -w 15 -wc −3.
Translocations assay
Translocation frequency between two simultaneously targeted sites was evaluated using ddPCR and amplicon sequencing69,91. Briefly, balanced translocations between either HIST1H2BC-HBEGF or PCSK9-HBEGF were detected using custom FAM-labelled ddPCR assays (Bio-Rad). HEX-labelled AP3B1 assay (Bio-Rad, dHsaCP1000001) was used as reference. Sequences for the primers and probes are listed in Supplementary Data 7.
20 µl ddPCR reaction mixes were prepared, each containing 1x ddPCR Supermix for Probes (no UTP) (Bio-Rad), 1x FAM-labelled custom translocation assay, 1x HEX-labelled reference assay, 1/40 HaeIII (NEB), 5 µl of 1:5 diluted QuickExtract DNA solution and ultrapure RNase- and DNase-free water (Invitrogen). An automated Droplet Generator (Bio-Rad) was used to generate droplets and C1000 Touch Thermal Cycler (Bio-Rad) for PCR amplification. Following PCR conditions were used: 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s and 61/63 °C for 1 min, followed by 98 °C for 10 min. All steps were performed with ramp rate fixed at 2 °C/s.
Droplet reading was performed with the QX 200 Droplet reader (Bio-Rad) using ddPCR Droplet Reader Oil (Bio-Rad). The QX Manager (Bio-Rad) software was used for data acquisition and analysis. The fluorescence amplitude threshold was set manually, using the midpoint between the average fluorescence amplitude of the FAM and HEX channels in positive samples and the negative control. The same threshold was applied to all wells of the ddPCR-plate using the same translocation assay. Editing efficiencies at the individual target sites were quantified using amplicon sequencing. Normalized translocation frequency was evaluated as observed frequency from ddPCR divided by geometric mean of editing efficiencies at each site.
Cell-based genome editing reporter assay
The genome editing reporter was developed employing the previously described Xential method83. In summary, HEK293T cells, at a density of 0.5 × 10 ^ 6 cells/well, were seeded onto 6-well plates. These cells were transfected with plasmid DNAs encoding Cas9, an sgRNA targeting intron 3 of the HBEGF gene, and a repair template incorporating the reporter cassette. Transfections were facilitated by the FuGene reagent (Promega) with the total transfection mixture of 150 µl, comprising 3 µg of total DNA with relative ratio of 1:1:2 (Cas9:sgRNA:repair template). A total of 9 µl of the FuGene reagent was used for each well. Seventy-two hours post-transfection, the cells underwent selection using diphtheria toxin (DT) at final concentration of 10 ng/ml for a duration of 1 week. Following this, cells were transitioned to a DT-free medium and allowed to expand.
For reporter activation, the cells were transfected with plasmid DNA encoding Cas9s and synthetic sgRNA designed to target the reporter cassette. The same transfection procedure described in “cell culture and transfection procedures” section was followed. After 72 h, the culture medium was replaced with fresh warm medium. Six to twelve hours later, the medium was harvested for luminescence assessment. 10 µl of the collected medium was mixed with 10 µl of Nano-Glo® Luciferase substrate (Promega) diluted at 1:1000 in PBS. Luminescence measurements were carried out on the PheraStar FSX plate reader (BMG Labtech). To test reporter fidelity and performance, the cassette locus was also analysed with amplicon sequencing as described above.
Modified CHANGE-seq
CHANGE-seq was performed as previously described by Lazzarotto et al.90 with additional modifications. As opposed to SpCas9, PsCas9 generates staggered ends, impeding the ligation of the sequencing adaptor to the DNA double stranded breaks (DSBs). Thus, we introduced a DNA blunting step prior to adaptor ligation, as described in our previous work69. Here, we introduced an additional end-repair step in the presence of ddNTPs prior to Cas9 cleavage to block non-specific DNA ends for adaptor ligation, and thus, reduce the background noise reads.
High Molecular weight genomic DNA (Promega) was subjected to tagmentation with a custom Tn5-transposome harbouring oCRL225/oCRL226 adaptors and the Hyperactive Tn5 Transposase (Diagenode). DNA tagmentation was performed in batches of 2 µg, utilizing 8.7 µl of the assembled transposome in a final volume of 200 µl of 1x Tagmentation Buffer (Diagenode) and incubated for 7 minutes at 55 °C. Reaction was quenched by the addition of 200 µl of SDS 0.4%, and resultant fragments were assessed on the Fragment analyzer and quantified by Qubit dsDNA BR Assay kit (Thermo Fisher Scientific). Tagmented DNA was then subjected to gap repair with Kapa Hi-Fi HotStart Uracil+ DNA Polymerase (KAPA Biosystems) and Taq DNA Ligase (NEB). Resultant gap-repaired DNA was treated with USER enzyme (NEB) and T4 polynucleotide kinase (NEB), and then circularized overnight with T4 DNA Ligase (NEB) and treated with a cocktail of exonucleases containing Plasmid-Safe ATP-dependent DNase (Lucigen), Lambda exonuclease (NEB) and Exonuclease I (NEB) to degrade residual linear DNA carryover. To avoid capturing pre-existing non-specific dsDNA breaks at the end-repair step, exonuclease-treated circles were then further subjected to 3’end blocking by incubating 160 ng circularized material with 7.5 U of T4 DNA Polymerase, 7.5 U of Klenow Fragment (3’ → 5’ exo-), in presence of 0.1 mM of ddNTP, in a final volume of 100 ul of 1x T4 DNA Ligation Buffer. The mixture was incubated at 20 degrees for 30 minutes and bead purified with AmpureXP beads at 1:1 ratio. 150 ng of circularized material were in vitro cleaved by SpCas9, HiFi SpCas9, wild-type PsCas9 and ePsCas9 RNPs in combination with HEK4 and TRAC sgRNAs (sequences details in Supplementary Data 7) in a total volume of 50 µl.
All libraries were subjected to an end-repair step using T4 DNA polymerase (NEB), so that 5‘ overhangs are filled to form blunt ends for ligation. Then, Illumina Universal Adaptor (NEB) was ligated to adenylated blunt ends, enzymatically treated with USER enzyme (NEB) and amplified with NEBNext Multiplex Oligos for Illumina for 20 amplification cycles. The quality of the amplified and bead-cleaned-up libraries was determined using a 5300 Fragment analyzer with the standard sensitivity NGS kit (Agilent). Libraries were further quantified by qPCR (Thermo Fisher Scientific), pooled and denatured according to Illumina’s recommendations and sequenced on a NextSeq550 on a PE150 configuration, to achieve a mean coverage of ~16 M reads per library. The sequenced reads were analyzed using the published version of the CHANGE-seq pipeline90 with minor modifications. The pipeline was run with the following parameters: read_threshold: 4, window_size: 3, mapq_threshold: 50, start_threshold: 1, gap_threshold: 3, mismatch_threshold: 6, search_radius: 30, merged_analysis: False, PAM = NNN. Reads with MAPQ = 0 were included in the analysis alongside those passing the MAPQ threshold defined in the parameters, to nominate putative off-targets located in non-uniquely mappable regions.
Purification of Cas9 proteins
While SpCas9 and HiFi SpCas9 proteins were procured from IDT, the PsCas9 and ePsCas9 proteins were purified using a previously established protocol69. Briefly, the E. coli BL21 λDE3 star strain was transformed with pET24a-based expression vectors. Freshly transformed colonies were cultivated overnight in LB medium, then sub-cultured into 800 ml of TB medium. This culture was maintained at 37 °C until an optical density OD600 approached ~2, under robust agitation. The growth temperature was subsequently reduced to 18 °C, and after a 1-h, isopropyl β-D-1-thiogalactopyranoside (IPTG) was introduced to a final concentration of 200 µM to induce protein expression. Post an overnight incubation, cells were harvested via centrifugation.
Cell lysis was achieved through high-pressure disintegration in a buffer comprising 20 mM HEPES (pH 7.5), 150 mM KCl, 5% glycerol, and 1 mM dithiothreitol (DTT). The resulting lysate was clarified by centrifugation and then subjected to affinity chromatography using a 5 ml HisTrap column (Cytiva). After equilibrating the column with a buffer containing 20 mM imidazole, bound proteins were eluted using a buffer with 300 mM imidazole. Relevant protein fractions were further subjected to gel filtration using a Superdex 200 10/600 column (Cytiva), pre-equilibrated with a buffer of 20 mM HEPES (pH 7.5), 300 mM NaCl, 5% glycerol, and 1 mM DTT. The purified Cas9 protein fractions were then concentrated to a final concentration of 10 mg/ml, flash frozen using liquid nitrogen, and stored at −80 °C until further use. The sequences of utilized plasmids can be found in Supplementary Data 5.
DNA binding assay using fluorescence polarization
For the DNA binding assay, we employed a fluorescence polarization technique, adapting a method that has been previously described75. FAM-labelled DNA oligonucleotides were procured from IDT. To prepare dsDNA substrates, the oligos were annealed at a concentration of 10 µM in an annealing buffer composed of 10 mM TRIS-HCl (pH 7.5) and 50 mM KCl. The solution was heated at 95 °C for 5 min and allowed to cool down gradually to room temperature. Once annealed, the dsDNA was diluted to a final concentration of 20 nM using a binding buffer containing 20 mM TRIS-HCl, 200 mM KCl, 5% Glycerol, and 10 mM CaCl2.
sgRNAs were refolded using a similar procedure, with an initial concentration of 10 µM in the annealing buffer. To form the Cas9 ribonucleoproteins (RNPs), Cas9 protein and its respective sgRNA were mixed at concentrations of 2 µM and 2.5 µM, respectively, in the binding buffer. This mixture was then incubated at room temperature (~25 °C) for 20 min. The Cas9 RNPs were subsequently serially diluted using the binding buffer. Equal volumes of the diluted RNPs and dsDNA substrates were combined, resulting in a final concentration of 10 nM DNA. The reactions were allowed to incubate at room temperature for an additional 15 minutes. Fluorescence polarization reading of FAM fluorophore was then taken using the PheraStar FSX plate reader (BMG Labtech). The sequences of utilized oligonucleotides are provided in Supplementary Data 7.
Cryo-EM data processing
PsCas9 RNP complex rapidly thawed and incubated with 4-fold excess of heat-annealed (90 °C for 5 min, and rapidly cooled to 4 °C) EMX1a dsDNA, and incubated at room temperature (~25 °C) for 30 min prior to vitrification. 2.5 µl of this complex were applied to C-flat holey carbon grids (1.2/1.3, 300 mesh), which had been plasma cleaned for 30 s in a Solarus 950 plasma cleaner (Gatan) with a 4:1 ratio of O2/H2. Grids were blotted with Vitrobot Mark IV (Thermo Fisher Scientific) for 2 s, blot force 4 at 4 °C & 100% humidty, and plunge-frozen in liquid ethane. Data were collected using a FEI Titan Krios cryo-electron microscope equipped with a K3 Summit direct electron detector (Gatan, Pleasanton, CA). Since initial data processing revealed a severe preferred orientation, the full dataset was collected with the stage tilted at −30°. Images were recorded with SerialEM102 with a pixel size of 0.81 Å. A total accumulated dose of 70 electrons/Å2during a 6 s exposure was fractionated into 80 frames, at a defocus range of −1.5 to −2.5 µm. A total of 7938 micrographs were collected, of which 6041 with CTF fits of 5 Å or better were retained. Motion correction, CTF estimation and particle picking was performed on-the-fly using cryoSPARC Live v4.0.0-privatebeta.2103. All subsequent data processing was performed in cryoSPARC v3.2104. Data processing workflow is provided in Supplementary Fig. 11.
A total of 3,932,646 particles were picked, of which 773,096 were selected after 2D classification. Multiple rounds of ab initio reconstruction and heterogeneous refinement resulted in a subset of 433,192 particles which used for a consensus reconstruction, which was resolved to 3.0 Å-resolution using non-uniform refinement. This subset of particles was then further classified using he 3D classification job within cryoSPARC (k = 10). One class of particles was well-resolved and significantly more abundant than the other classes (81,473 particles), which was then used for subsequent non-uniform refinement. After multiple rounds of CTF refinement, a 2.86 Å resolution reconstruction was determined. This map was then used for modelling.
Model building and figure preparation
An AlphaFold2 model of PsCas9 was generated, and individual domains were rigid body fitted into the unsharped reconstruction. Once all protein density had been accounted for, the individual domains were connected, and the nucleic acid chains were build de novo in Coot105. Once fully modelled, Isolde106 was used to improve the fit of the model to the map, and real-space refinement as implemented within Phenix107 was performed to optimize model geometry. All structural figures and movies were generated using ChimeraX108,109.
mRNA synthesis
mRNA is synthesised by a T7 polymerase driven in vitro transcription (IVT) reaction from a plasmid DNA template which contains a T7 polymerase promoter sequence upstream of all the elements required in the mRNA. mRNA comprises a 5’ cap structure incorporated through the inclusion of a cap analogue (CleanCap AG®) during mRNA synthesis; a 5’ untranslated region sequence; a nuclear localisation sequence genetically fused to the coding sequence of Cas9 followed by another nuclear localisation sequence; a 3’ untranslated region sequence and a defined polyA tail (80–120 bp). The mRNA is prepared with the replacement of uridine by N1-Methyl-Pseudouridine or 5-Methoxyuridine to minimize recognition of the IVT produced mRNA by the innate immune system. The DNA template is linearized downstream of the polyA tail by BspQI restriction endonuclease before IVT to ensure all mRNA molecules terminate directly after the polyA tail.
LNP Formulation for In Vivo Cas9 delivery
Lipid nanoparticles (LNPs) were synthesized in line the with previously established protocols95. Specific lipids were solubilized in ethanol at final concentration of 12.5 mM. Separately, Cas9 mRNA and corresponding sgRNAs (Axolabs) were diluted in RNase-free 50 mM citrate buffer (pH 3.0). The ethanol-based lipid solution and the aqueous mRNA/sgRNA solution were then combined at a 3:1 volume ratio utilizing the microfluidic NanoAssemblr Ignite device (Precision NanoSystems) with a set mixing flow rate of 12 mL/min. The resultant LNPs underwent an overnight dialysis using Slide-A-Lyzer G2 10 K MWCO dialysis cassettes (Thermo Fisher Scientific) in PBS, pH 7.4.
Particle size distribution and overall size were determined via dynamic light scattering (DLS) with the Zetasizer Nano-ZS instrument (Malvern Instruments). The formulated LNPs were between 70 and 78 nm in size, with a polydispersity index (PDI) in the range of 0.05 to 0.07. The LNPs were concentrated using Amicon Ultra 4 30,000 kDa MWCO centrifugal filter units, and then DLS was performed again to ascertain LNP structural integrity. To quantify the encapsulated RNA within the LNPs, the RiboGreen assay (Thermo Fisher Scientific) was utilized. The encapsulation efficiency was above 90% for all LNP samples. LNPs were then diluted to the working RNA concentration of 0.2 mg/ml.
In vivo LNP delivery
Female C57Bl/6NCrl mice were acquired from Charles River Laboratories. Animals were maintained in a controlled environment with a room temperature of 21 °C, a relative humidity ranging between 45–55%, and a 12:12 h light-dark cycle (lights on at 6:00 am, lights off at 6:00 pm). Throughout the study, mice were granted unrestricted access to a standard chow diet (R70, Lactamin AB, Stockholm, Sweden) and water. Enrichment elements, such as cardboard tubes, chew sticks, and shredded paper, were provided in the cages. Animals underwent daily health inspections and were weighed weekly.
Mice, aged between 10–12 weeks, were administered LNPs at a dose of 1 mg/kg via lateral tail vein injection. As a comparison, a control group was injected with an equivalent volume of buffer. One week following the injections, mice were euthanized, and liver tissues collected for analysis. The left median liver lobes were designated for genomic DNA extraction using the Puregene Tissue Kit (Qiagen). Subsequent analyses to determine genome editing efficiencies were executed via amplicon sequencing, as detailed in the prior section. Animal sex was not considered in the study due to the use of sex-independent genomic locus investigated.
Assessment of liver function following LNP delivery
To gauge potential hepatic impact post-LNP administration, plasma levels of alanine transaminase (ALT) and aspartate aminotransferase (AST) were assessed. Blood was collected at termination via retroorbital eye bleed. Blood samples of ~600 μL were drawn into 500 LiHep Microvette tubes. Plasma was separated from the blood within 30 min of collection using centrifugation at 1500 g for 10 minutes at 4 °C. Resultant plasma samples were preserved at −20 °C until dispatched to Charles River Laboratories (Edinburgh, UK), where the specific activities of ALT and AST were determined.
Statistics and reproducibility
No statistical method was used to predetermine sample size. Sample sizes for in vitro and in cellulo experiments were selected based on literature precedence for genome editing experiments. No data was excluded. All cell and in vitro experiment were independently repeated at least once as specified in figure legends. Mammalian cells were cultured under identical conditions, no randomization was used. Animal experiments: Three to four animals were included per group. Animals were randomized based on their weights measured prior to the experiments. The investigators were not blinded to allocation during experiments and outcome assessment. Statistical tests used described in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided as a Source Data file. The structure of PsCas9 and its associated atomic coordinates, has been deposited into the EMDB and the PDB repositories with EMDB accession number EMDB-42378 and PDB accession number 8UMF, respectively. NGS datasets generated in this study are deposited to the NIH Sequence Read Archive with BioProject accession numbers: PRJNA1154610 and PRJNA1154611. Previously reporter FnCas9 protein structure used in this work (PDB accession number 5B2O). All the sequences of sgRNAs, mRNAs, proteins and plasmids used in the study are available in the Supplementary Data file. Source data are provided with this paper. Synthetic guide RNAs and ePsCas9 are available from Synthego.
Code availability
No specific code was developed in regard to this publication.
References
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Brouns, S. J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008).
Al-Shayeb, B. et al. Clades of huge phages from across earth’s ecosystems. Nature 578, 425–431 (2020).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Cho, S. W., Kim, S., Kim, J. M. & Kim, J. S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230–232 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Jinek, M. et al. RNA-programmed genome editing in human cells. Elife 2, e00471 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Bibikova, M., Golic, M., Golic, K. G. & Carroll, D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161, 1169–1175 (2002).
Bibikova, M., Beumer, K., Trautman, J. K. & Carroll, D. Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764 (2003).
van Overbeek, M. et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 63, 633–646 (2016).
Bibikova, M. et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell Biol. 21, 289–297 (2001).
Chen, F. et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8, 753–755 (2011).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Maeder, M. L. et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979 (2013).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Swiech, L. et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33, 102–106 (2015).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379 (2013).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014).
Zhou, Y. et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509, 487–491 (2014).
Liu, X. et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res 27, 154–157 (2017).
Rupp, L. J. et al. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 7, 737 (2017).
Eyquem, J. et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017).
Frangoul, H., Ho, T. W. & Corbacioglu, S. CRISPR-Cas9 gene editing for sickle cell disease and beta-thalassemia reply. N. Engl. J. Med 384, e91 (2021).
Gillmore, J. D. et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021).
Koonin, E. V. & Makarova, K. S. Evolutionary plasticity and functional versatility of CRISPR systems. PLoS Biol. 20, e3001481 (2022).
Wang, J. Y. & Doudna, J. A. CRISPR technology: a decade of genome editing is only the beginning. Science 379, eadd8643 (2023).
Saito, M. et al. Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature 620, 660–668 (2023).
Karvelis, T. et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696 (2021).
Altae-Tran, H. et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65 (2021).
FDA. CASGEVY. https://www.fda.gov/vaccines-blood-biologics/casgevy (2022).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 (2013).
Ghezraoui, H. et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell 55, 829–842 (2014).
Frock, R. L. et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33, 179–186 (2015).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Shin, H. Y. et al. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 8, 15464 (2017).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Cullot, G. et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 10, 1136 (2019).
Alanis-Lobato, G. et al. Frequent loss of heterozygosity in CRISPR-Cas9-edited early human embryos. Proc. Natl Acad. Sci. USA 118, e2004832117 (2021).
Leibowitz, M. L. et al. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat. Genet 53, 895–905 (2021).
Papathanasiou, S. et al. Whole chromosome loss and genomic instability in mouse embryos after CRISPR-Cas9 genome editing. Nat. Commun. 12, 5855 (2021).
Hoijer, I. et al. CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat. Commun. 13, 627 (2022).
Kleinstiver, B. P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Slaymaker, I. M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Chen, J. S. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017).
Casini, A. et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 36, 265–271 (2018).
Lee, J. K. et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 9, 3048 (2018).
Vakulskas, C. A. et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med 24, 1216–1224 (2018).
Schmid-Burgk, J. L. et al. Highly parallel profiling of Cas9 variant specificity. Mol. Cell 78, 794–800.e798 (2020).
Bravo, J. P. K. et al. Structural basis for mismatch surveillance by CRISPR-Cas9. Nature 603, 343–347 (2022).
Kim, Y. H. et al. Sniper2L is a high-fidelity Cas9 variant with high activity. Nat. Chem. Biol. 19, 972–980 (2023).
Nishimasu, H. et al. Crystal structure of staphylococcus aureus Cas9. Cell 162, 1113–1126 (2015).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Hirano, H. et al. Structure and engineering of francisella novicida Cas9. Cell 164, 950–961 (2016).
Kim, E. et al. In vivo genome editing with a small Cas9 orthologue derived from campylobacter jejuni. Nat. Commun. 8, 14500 (2017).
Edraki, A. et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell 73, 714–726 e714 (2019).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282 (2019).
Schmidt, M. J. et al. Improved CRISPR genome editing using small highly active and specific engineered RNA-guided nucleases. Nat. Commun. 12, 4219 (2021).
Kim, D. Y. et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 40, 94–102 (2022).
Hino, T. et al. An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis. Cell 186, 4920–4935.e23 (2023).
Bestas, B. et al. A Type II-B Cas9 nuclease with minimized off-targets and reduced chromosomal translocations in vivo. Nat. Commun. 14, 5474 (2023).
Katzmann, J. L., Cupido, A. J. & Laufs, U. Gene therapy targeting PCSK9. Metabolites 12, 70 (2022).
Ding, Q. et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res 115, 488–492 (2014).
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014).
Dagdas, Y. S., Chen, J. S., Sternberg, S. H., Doudna, J. A. & Yildiz, A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 3, eaao0027 (2017).
Gong, S., Yu, H. H., Johnson, K. A. & Taylor, D. W. DNA unwinding is the primary determinant of CRISPR-Cas9 activity. Cell Rep. 22, 359–371 (2018).
Maji, B. et al. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell 177, 1067–1079 e1019 (2019).
Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014).
Jinek, M. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997 (2014).
Yamada, M. et al. Crystal structure of the minimal Cas9 from campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems. Mol. Cell 65, 1109–1121.e1103 (2017).
Zhu, X. et al. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat. Struct. Mol. Biol. 26, 679–685 (2019).
Das, A. et al. Coupled catalytic states and the role of metal coordination in Cas9. Nat. Catal. 6, 969–977 (2023).
Sun, W. et al. Structures of neisseria meningitidis Cas9 complexes in catalytically poised and anti-CRISPR-inhibited states. Mol. Cell 76, 938–952.e935 (2019).
Gasiunas, G. et al. A catalogue of biochemically diverse CRISPR-Cas9 orthologs. Nat. Commun. 11, 5512 (2020).
Li, S. et al. Universal toxin-based selection for precise genome engineering in human cells. Nat. Commun. 12, 497 (2021).
Dang, Y. et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 16, 280 (2015).
Riesenberg, S., Helmbrecht, N., Kanis, P., Maricic, T. & Paabo, S. Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage. Nat. Commun. 13, 489 (2022).
Chen, F. et al. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat. Commun. 8, 14958 (2017).
Acharya, S. et al. Francisella novicida Cas9 interrogates genomic DNA with very high specificity and can be used for mammalian genome editing. Proc. Natl Acad. Sci. USA 116, 20959–20968 (2019).
Kim, N. et al. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat. Biotechnol. 38, 1328–1336 (2020).
Kim, H. K. et al. High-throughput analysis of the activities of xCas9, SpCas9-NG and SpCas9 at matched and mismatched target sequences in human cells. Nat. Biomed. Eng. 4, 111–124 (2020).
Lazzarotto, C. R. et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR-Cas9 genome-wide activity. Nat. Biotechnol. 38, 1317–1327 (2020).
Wimberger, S. et al. Simultaneous inhibition of DNA-PK and Polϴ improves integration efficiency and precision of genome editing. Nat. Commun. 14, 4761 (2023).
Finn, J. D. et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22, 2227–2235 (2018).
Jiang, C. et al. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 27, 440–443 (2017).
Miller, J. B. et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle Co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int Ed. Engl. 56, 1059–1063 (2017).
Lundin, A. et al. Development of an ObLiGaRe doxycycline inducible Cas9 system for pre-clinical cancer drug discovery. Nat. Commun. 11, 4903 (2020).
Cofsky, J. C., Soczek, K. M., Knott, G. J., Nogales, E. & Doudna, J. A. CRISPR-Cas9 bends and twists DNA to read its sequence. Nat. Struct. Mol. Biol. 29, 395–402 (2022).
Hibshman, G. N. et al. Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9. Nat. Commun. 15, 3663 (2024).
Liu, M. S. et al. Engineered CRISPR/Cas9 enzymes improve discrimination by slowing DNA cleavage to allow release of off-target DNA. Nat. Commun. 11, 3576 (2020).
Wang, Y. et al. Guide RNA engineering enables efficient CRISPR editing with a miniature Syntrophomonas palmitatica Cas12f1 nuclease. Cell Rep. 40, 111418 (2022).
Sjogren, A. K. et al. Critical differences in toxicity mechanisms in induced pluripotent stem cell-derived hepatocytes, hepatic cell lines and primary hepatocytes. Arch. Toxicol. 88, 1427–1437 (2014).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Punjani, A. Real-time cryo-EM structure determination. Microsc. Microanal. 27, 1156–1157 (2021).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 74, 519–530 (2018).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544 (2018).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
We thank Steve Rees for supporting this work. We thank the members of the Genome Engineering Department and Joanna Rejman for critically reading the manuscript and useful suggestions; Euan Gordon, Veronika Saez Jimenez, and Protein Science team for purification of recombinant PsCas9; Anders Gunnarsson for the help in establishing in vitro binding assays; George Thom and Salman Mustfa for the help with mRNA production; Kristina Friis and Kai Liu for formulating LNPs for in vivo study; Andrea Ahnmark, Annika Stenberg, Marie Johansson and Steven Oag for the help with in vivo experiments; Maryam Clausen and NGS team for supporting amplicon sequencing; Mike Firth for the help with processing NGS data; Kevin Holden for the sgRNA reagents. This work was supported by the AZ Postdoctoral Fellowship to I.W. and the National Institutes of Health grant R35GM138348 to D.W.T. and and Welch Foundation Research Grant F-1938 to D.W.T.
Author information
Authors and Affiliations
Contributions
D.D., M.M. and G.S. initiated the project. D.D. and J.B. performed most of the experimental work with the help from A.E., A.Z., O.K.C., J.L.T., N.S., I.W., M.F., P.A., M.P., M.M., D.T., G.S. provided technical input and guidance. A.E. designed and performed in vivo experimental work including animal handling, sampling and data processing. D.D. and J.B. prepared the manuscript with input from all authors. D.D., D.T., M.M. and G.S. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
D.D., A.E., A.Z., O.K.C., J.L.T., N.S., I.W., M.F., P.A., M.P., M.M. and G.S. are employees and shareholders of AstraZeneca. G.S. and M.M. are listed as co-inventors in a patent application filed by AstraZeneca Ab (application number: WO2022248645A1; status: published) related to this work covering aspects of protein engineering. J.B. and D.T. declare no competing interests. This work was supported by the National Institutes of Health grant R35GM138348 to D.T. and Welch Foundation Research Grant F-1938 to D.T; by the AstraZeneca Postdoctoral Fellowship to I.W.
Peer review
Peer review information
Nature Communications thanks Pranam Chatterjee, Osamu Nureki, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Degtev, D., Bravo, J., Emmanouilidi, A. et al. Engineered PsCas9 enables therapeutic genome editing in mouse liver with lipid nanoparticles. Nat Commun 15, 9173 (2024). https://doi.org/10.1038/s41467-024-53418-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53418-8
This article is cited by
-
Structural visualization of the molecular evolution of CRISPR–Cas9
Nature Structural & Molecular Biology (2026)
-
Visualization of a multi-turnover Cas9 after product release
Nature Communications (2025)