Abstract
Inflammatory signals lead to recruitment of circulating monocytes and induce their differentiation into pro-inflammatory macrophages. Therefore, whether blocking inflammatory monocytes can mitigate disease progression is being actively evaluated. Here, we employ multiple lineage-tracing models and show that monocyte-derived macrophages (mo-mac) are the major population of immunosuppressive, liver metastasis-associated macrophages (LMAM), while the proportion of Kupffer cells (KC) as liver-resident macrophages is diminished in metastatic nodules. Paradoxically, genetic ablation of mo-macs results in only a marginal decrease in LMAMs. Using a proliferation-recording system and a KC-tracing model in a monocyte-deficient background, we find that LMAMs can be replenished either via increased local macrophage proliferation or by promoting KC infiltration. In the latter regard, KCs undergo transient proliferation and exhibit substantial phenotypic and functional alterations through epigenetic reprogramming following the vacating of macrophage niches by monocyte depletion. Our data thus suggest that a simultaneous blockade of monocyte recruitment and macrophage proliferation may effectively target immunosuppressive myelopoiesis and reprogram the microenvironment towards an immunostimulatory state.
Similar content being viewed by others
Introduction
Liver metastasis, the secondary malignancy frequently occurs in patients with gastrointestinal cancer and breast carcinoma, is an aggressive form of cancer progression with disappointing prognosis outcome and limited treatment options1. Immunotherapies have elicited durable treatment responses in some cancers but are ineffective in liver metastasis, which is enriched with various immune-suppressive cells and paralyzed effector and cytotoxic T cells2,3.
Tumour-associated hepatic myeloid cells, which consist of inflammatory monocytes and macrophages, are abundant in liver metastasis and foster a systemic immune desert microenvironment by inducing apoptosis of cytotoxic T cells4. In addition to T cell anergy, hepatic myeloid cells modulate the microenvironment of metastasis through promoting angiogenesis, inducing fibrosis, remodelling extracellular matrix5. Therapeutic approaches that precisely targeting tumour-promoting myeloid cells could offer promise6, but is hampered by a lack of knowledge about the origin and functional maintenance of liver-metastasis-associated macrophages (LMAM).
In heathy liver, the self-renewal resident macrophages, also called Kupffer cells (KC), line sinusoidal vessels and are in prime position to clear unfit red blood cells and even non-apoptotic disseminated tumour cells from portal vein7. The inflammatory milieu breaks the tissue homeostasis by disrupting KC niches and triggering an influx of blood monocytes and their differentiated macrophages8,9. However, the definitive KC tracing evidence is lacking so that it is difficult to delineate the two possibilities: 1) the replacement of KCs by monocyte-derived macrophages (mo-mac) and 2) the phenotypic plasticity of KCs.
Here we dissect the contribution of different myeloid lineages to the pool of LMAMs by mapping the fates of mo-macs and KCs. We find two distinct mechanisms—local proliferation and KC infiltration—that replenish LMAMs when the supply of circulating monocytes is blocked. After KCs infiltrate into metastatic nodules, the inflammatory cues could partially erase the ontologically epigenetic memory of KCs and reprogram KCs resemble to mo-macs. Our findings illuminate the resilience of LMAMs upon monocyte blockade and the previously underappreciated plasticity of tissue-resident macrophages, highlighting the importance of blocking monocytes and differentiated macrophages simultaneously to target tumour-promoting myelopoiesis and to switch the metastatic microenvironment from immunosuppressive to immune-stimulatory.
Results
Phenotypic and functional disparities between normal liver macrophages and LMAMs
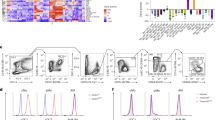
To study the differences in macrophage populations between liver metastasis tissues and normal liver tissues, we engrafted MC3810 and E077111 cells—murine colorectal cancer cells and breast cancer cells, respectively—into syngeneic immunocompetent mice via portal vein injection to establish experimental metastasis models (Fig. 1a). To extrapolate our observations to primary liver cancer, we leveraged hydrodynamic tail vein injection to generate a spontaneous cMyc overexpression and TP53 deletion hepatocellular carcinoma (HCC) model12 (Supplementary Fig. 1a). We then performed multicolour flow cytometry (FC) to identify the immune cells that were differentially enriched in normal and malignant liver tissues. Consistent with the proposed inflammatory environment in carcinoma13, the total number of CD45+ leukocytes was greater in malignant liver tissues than in nearby normal liver tissues of the same livers (Fig. 1b). Remarkably, the population of CD11blowF4/80high resident-like macrophages was diminished, whereas CD11bhighF4/80high bone marrow-derived macrophages became the dominant macrophage population in liver metastatic nodules and HCC (Fig. 1c).
a, b Schematic of the experimental liver metastasis (LvMet) models used and flow cytometry (FC) quantification of CD45+ leukocytes in LvMet and adjacent normal (adj. Normal) tissues from the same livers (n = 5 for MC38; n = 5 for E0771; n = 6 for hepatocellular carcinoma, HCC). c Multi-colour FC analysis of immune cells in LvMet and adj. Normal tissues from the same livers (n = 5 for MC38; n = 5 for E0771; n = 4 for HCC). d Schematic diagram and experimental workflow for CITE-seq. e. UMAP embedding of N-macs and LMAMs, and feature plots showing the expression of KC-identity genes and mo-mac signature genes. f Schematic design of dual-fluorescent reporter mice for tracing macrophages. g–i Representative immunofluorescence (IF) staining and FC quantification of KC markers Clec4f and Timd4 in adj. Normal and LvMet tissues from the same livers (n = 6 for E0771; scale bar, 50 μm). j Representative IF staining and quantification of Timd4 expression in liver metastasis tissue samples derived from breast cancer patients (triple negative, n = 3; HER2+, n = 3; scale bar, 50 μm). Mean ± s.e.m. shown. P values were calculated by comparing individual animals using two-tailed paired (b, g–j) Mann–Whitney U-test.
In addition to FC-based immunophenotyping, we also performed cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq)14 on fluorescence-activated cell sorting (FACS)-sorted CD45+F4/80+ macrophages from metastatic nodules and nearby normal liver tissues (Fig. 1d), which allowed us to profile the transcriptome and 119 cell surface proteins simultaneously at the single-cell level. Consistent with previously reported macrophage single-cell RNA-seq (scRNA-seq) data15,16,17, we identified 5 major clusters of LMAM populations (Spp1, Cxcl9, Ccl5, Vcan, Stmn1), revealing the heterogeneity within LMAMs. Both LMAMs and normal liver macrophages (N-mac) expressed Cd68 18, a marker of the mononuclear phagocyte system, which includes monocytes, macrophages, and dendritic cells. Inflammatory monocyte-derived macrophage (mo-mac) markers such as Ccr2 and Cx3cr119 emerged on the cell surface of LMAMs (Fig. 1e). In contrast, the transcript level of Clec4f20, a mouse KC marker, was substantially decreased in LMAMs, and this finding was confirmed by studies in dual-fluorescence Clec4f tdT-Cre;CD68EGFP mice (Fig. 1f). In this model, the expression of the nuclear-localized red fluorescent protein tdTomato (tdT) is driven by the promoter of Clec4f 21, and the expression of enhanced green fluorescent protein (EGFP) is under the control of the promoter of Cd6822. Consistent with the CITE-seq results, N-macs were positive for both tdT and EGFP, while LMAMs were negative for tdT and expressed only EGFP (Fig. 1g,h).
Moreover, Timd423,24,25, the cell membrane protein of tissue-resident macrophages that recognizes phosphoserine in the outer membrane leaflet of apoptotic cells, was not detected in LMAMs (Fig. 1e,g,i), and this finding was confirmed in liver metastases of breast cancer patients (Fig. 1j). When cocultured with apoptotic cancer cells labelled with CypHer5E26, a dye that is weakly fluorescent at neutral pH but fluoresces brightly in phagosomes, tdT+EGFP+ N-macs but not tdT–EGFP+ LMAMs began to emit a strong far-red fluorescence signal within 1 − 2 hours (Supplementary Fig. 1b-d), suggesting that the loss of Timd4 prevented LMAMs from recognizing and engulfing apoptotic cells. To determine whether KCs can also engulf metastatic cancer cells in vivo, we also engrafted E0771 cancer cells engineered to express tdT into wild-type mice via portal vein injection (Supplementary Fig. 1e). Although Clec4f+ KCs could not effectively infiltrate metastatic tissues (Supplementary Movies 1, 2), the KCs engulfed nonapoptotic tdT+ cancer cells in the periphery of metastatic nodules (Supplementary Fig. 1f). Taken together, these results suggest that the macrophages in normal liver tissues and the macrophages in metastatic nodules display profoundly distinct morphologies, transcriptional profiles, and physiological functions.
LMAMs do not originate from Kupffer cells
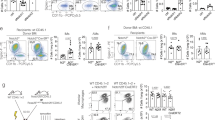
Intrigued by the phenotypic discrepancies between N-macs and LMAMs, we next sought to identify the origin of LMAMs. Accumulating evidence suggests that monocytes are recruited to inflammatory sites8 and become the dominant population of LMAMs. This hypothetical model, however, does not rule out the possibility that KCs exhibit phenotypic plasticity through loss of their lineage markers and acquisition of mo-mac markers. To investigate the origin of LMAMs in vivo, we first crossed mice expressing KC-specific Cre recombinase (Clec4f tdT-Cre mice) with the mice of the custom reporter line R26LSL-UPRT-HA-sfGFP (Fig. 2a). In the Clec4f tdT-Cre;R26LSL-UPRT-HA-sfGFP mice, KCs and their progenies express superfolder green fluorescent protein (sfGFP) and HA-tagged uracil phosphoribosyl transferase (UPRT), which can be used to label nascent RNA specifically in KCs27. In particular, even when Clec4f is silenced (e.g., in LMAMs), cells derived from KCs retain the expression of the HA tag and sfGFP and can thus be distinguished from mo-macs, which are derived from Clec4f− monocytes. According to both immunofluorescence (IF) staining and FC analysis, less than 10% of LMAMs expressed either the HA tag or sfGFP (Fig. 2b,c) in comparison with the N-macs of the same livers. To corroborate the findings in Clec4f-tracing mice, we also crossed sfGFP reporter mice with the mice expressing Cre recombinase in pre-macrophages from the yolk sac and embryo (Tnfrsf11acre)28 (Supplementary Fig. 2a). In this Tnfrsf11acre;R26LSL-UPRT-HA-sfGFP model, more than 80% of N-macs were positive for sfGFP, suggesting that foetal liver-derived KCs are maintained throughout the lifespan by self-renewal, with a minimal contribution from circulating monocytes. Similar to the findings in the KC-tracing model using Clec4fCre, only a minority of LMAMs in this Tnfrsf11acre KC-tracing model were positive for the HA tag and sfGFP (Supplementary Fig. 2b,c) compared with the N-macs of the same livers, suggesting that KCs could not effectively infiltrate into metastatic tissues and contributed to the pool of LMAMs.
a Schematic design of KC lineage tracing mice. b IF quantification of KC-derived cells (HA tag+ or sfGFP+) in adjacent normal tissues and LvMet tissues from the same livers (n = 5 for MC38; n = 5 for E0771; scale bar, 50 μm). c FC analysis of sfGFP and tdT expression in HelixNIR- CD45 + F4/80+ cells (n = 5 for MC38; n = 5 for E0771). d,e Schematic and experimental design of monocyte lineage tracing and genetic ablation (IF, immunofluorescence; bw, body weight; og, oral gavage). f-h. Representative histogram and IF image, as well as FC quantification of tdT expression in N-macs and LMAMs from the same livers (n = 7 for MC38; n = 5 for E0771; scale bar, 50 μm). i IF staining of LMAMs upon acute monocyte depletion (Cx3cr1CreERT2;R26LSL-DTA, n = 5 for MC38, n = 6 for E0771; littermate control Cx3cr1CreERT2;R26WT, n = 4 for MC38; n = 6 for E0771; scale bar, 50 μm). j FC quantification of LMAMs upon acute monocyte depletion (Cx3cr1CreERT2;R26LSL-DTA, n = 5 for MC38, n = 6 for E0771; littermate control Cx3cr1CreERT2;R26WT, n = 5 for MC38; n = 5 for E0771). k IF quantification of monocytes and LMAMs by impairing monocyte chemotaxis (Ccr2GFP/GFP: n = 6 for MC38; n = 5 for E0771; Ccr2GFP/WT littermates: n = 5 for MC38; n = 5 for E0771; scale bar, 50 μm). l Representative FC plot showing loss of Ccr2 expression in LMAMs of Ccr2GFP/GFP mice, and quantification of the percents of monocyte and LMAM in CD45+ leukocytes (Ccr2GFP/GFP: n = 6 for MC38; n = 8 for E0771; Ccr2GFP/WT littermates: n = 4 for MC38; n = 8 for E0771). Mean ± s.e.m. shown. Smaller dots are values from individual fields. Outlined circles are mean values taken over multiple fields/sections from the same mouse. P values were calculated by comparing individual animals using two-tailed paired (b, c, h) and unpaired (i-l) Mann–Whitney U-test.
Given that KCs reside in the liver and do not enter the circulation, we also performed a time-course parabiosis experiment (Supplementary Fig. 2d). After two weeks of blood circulation sharing between CD45.2+/+;Tnfrsf11acre;R26LSL-UPRT-HA-sfGFP and CD45.1+/+;CD68-EGFP parabionts, we injected cancer cells into the portal vein of CD45.1+/+ mice and waited another four weeks for the establishment of overt metastatic colonies in these mice. Indeed, very few LMAMs were positive for the HA tag (Supplementary Fig. 2e), and the majority of CD45.2+ LMAMs were negative for sfGFP (Supplementary Fig. 2f), supporting the idea that the LMAM population is composed mostly of macrophages derived from blood monocytes.
LMAMs are derived from monocytes, but can be replaced from other sources
In addition to tracing KCs, we also used an orthogonal approach of mapping the fate of inflammatory monocytes (Cx3cr1CreERT2;H11LSL-tdT for fluorescence tracing and Cx3cr1CreERT2;R26LSL-DTA for genetic ablation) (Fig. 2d, Supplementary Fig. 3a,b). KCs are established prior to birth and are independent of Cx3cr129. Conversely, mo-macs rely on Cx3cr1 for their differentiation from F4/80low monocytes to F4/80high pro-inflammatory macrophages in the context of liver injury19. Therefore, the fluorescent labelling model Cx3cr1CreERT2;H11LSL-tdT or genetic ablation model Cx3cr1CreERT2; R26LSL-DTA were useful for tracing or depleting mo-macs, respectively, in a controllable manner via the oral administration of tamoxifen (Fig. 2e). Although more than 75% of LMAMs were positive for tdT, indicating that they were derived from monocytes (Fig. 2f-h), diphtheria toxin A (DTA)-mediated depletion of inflammatory monocytes only decreased the LMAM population by 37% as determined by IF (Fig. 2i) and by 55% as determined by FC (Fig. 2j). In a complementary approach, we employed Ccr2 knockout (Ccr2GFP/GFP) mice (Supplementary Fig. 3c), in which the coding sequence of the chemokine receptor Ccr2 was replaced by that of GFP30. Notably, the protein level of Ccl2, the ligand of Ccr2, was markedly increased in the small liver metastatic lesions (Supplementary Fig. 4), indicating the potential role of the Ccl2/Ccr2 axis in monocyte recruitment. Similar to the aforementioned DTA-based ablation of monocytes, knockout of Ccr2 resulted in profound loss of monocytes (by 85% as determined by IF staining and by 90% as determined by FC) but only a marginal decrease in LMAMs (by 37% as determined by IF staining and 35% as determined by FC) (Fig. 2k,l). Collectively, these data indicate that the LMAM population is derived mostly from monocytes but may be partially replenished via other mechanisms when monocyte recruitment is blocked.
Enhanced local macrophage proliferation upon monocyte blockade
To explain how LMAMs are replenished upon monocyte depletion, we proposed two possibilities: 1) increased proliferation of local macrophages within metastatic nodules and 2) infiltration of KCs from nearby normal tissues. In fact, the proliferation of local macrophages or local monocytes to replenish vacant macrophage niches has been implied to occur in various malignant31 and normal tissues32,33,34 via colony-stimulating factor 1 (Csf1) and its receptor Csf1r. To determine whether local proliferation contributes to the replenishment of LMAMs, we established a proliferation recorder mouse model (H11DreERT2;Ki67RSR-Cre;R26LSL-UPRT-HA-sfGFP mice) (Fig. 3a), which was similar to the ProTracer system previously reported35. Unlike the method of identifying proliferating cells by IF staining for the cell proliferation marker Ki67, cells that have undergone proliferation starting from a defined time point can be marked in this proliferation recorder system. Upon treatment with one tamoxifen pulse, DreERT2 excises the rox-flanked transcriptional termination signal upstream of the Cre recombinase sequence, yielding a new Ki67-Cre genotype in which Cre recombinase can label proliferating cells at any time thereafter. Using this proliferation recording tool, we found that only approximately 30% of LMAMs were sfGFP+, whereas more than 85% of tumour-associated endothelial cells were sfGFP+ (Fig. 3b). We then leveraged clodronate liposomes (CL)36 to block the external supply of LMAMs. Although CL can effectively deplete both circulating monocytes and KCs (Fig. 3c), they cannot be engulfed by LMAMs, as revealed by the red fluorescent Dil-encaupsulated liposomes, which have similar size with CL (Fig. 3d,e); thus, macrophages within metastatic nodules are not depleted. Intravenous administration of CL increased the percentage of sfGFP+ LMAMs by two-fold compared to that in mice treated with control liposomes (Fig. 3f-h), suggesting that local proliferation of LMAMs is enhanced in response to monocyte depletion (Fig. 3i).
a Schematic design of the proliferation recorder mice (H11DreERT2;Ki67-RSR-Cre; R26LSL-UPRT-HA-sfGFP). b Representative IF staining for HA tag, macrophage marker F4/80 and endothelial marker CD31 and quantification of fraction of proliferated cells in E0771 LvMet-infiltrating macrophages and endothelial cells (TECs) (n = 5; scale bar, 50 μm). c Representative IF staining (scale bar, 50 μm) and FC quantification of Kupffer cells and mo-macs 24 h after intravenous injection of PBS control liposomes (n = 6) or clodronate liposomes (CL, n = 7). d Schematic of labelling macrophages with red fluorescent dye Dil-loaded liposomes. e Representative IF staining, and FC gating and quantification showing N-macs but not LMAMs engulfing Dil-loaded liposomes (n = 5; scale bar, 50 μm). f Schematic of experimental design. Metastasis-bearing proliferation recorder mice were pulsed with a single tamoxifen gavage, and were treated with CL or control liposomes every three days until the experimental endpoint. g Representative IF staining of HA tag+ proliferated LMAMs and Ki67+ proliferating LMAMs (indicated by white arrowheads; scale bar, 50 μm). h FC quantification of sfGFP+ proliferated LMAMs in CL-treated mice (n = 6 for MC38, n = 5 for E0771) and control mice (n = 5 for MC38; n = 4 for E0771). i Schematic showing the activation of local macrophage proliferation upon blockade of monocyte supply. Mean ± s.e.m. shown. Smaller dots are values from individual fields. Outlined circles are mean values taken over multiple fields/sections from the same mouse. P values were calculated by comparing individual animals using two-tailed paired (b, e) and unpaired (c, h) Student’s t-test.
Phenotypic plasticity of KCs
In addition to our hypothesis regarding local proliferation of LMAMs, we hypothesized that KCs could also infiltrate liver metastases when the macrophage niche was available due to monocyte deficiency. To test this hypothesis, we combined the KC tracing system with Ccr2 knockout mice (Clec4f tdT-Cre;R26LSL-UPRT-HA-sfGFP;Ccr2GFP/GFP). Because both Ccr2+ mo-macs and KC-derived LMAMs are positive for GFP, the GFP signal cannot be used to distinguish KC-derived cells from mo-macs. In addition to sfGFP and the HA tag, our Cre-inducible reporter line was also designed to incorporate a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) initially to increase mRNA transcript stability and expression; however, this element may also be used as a DNA tag for tracing Cre recombinase-derived cells in scRNA-seq analysis, as the WPRE is positioned at the 3’ end of the transcript upstream of the polyA signal, making it probable to be detected during the creation of single-cell 3’ gene expression libraries (Fig. 4a). To test whether the WPRE can be detected in sfGFP+ cells, we independently crossed an NK-specific Cre model mice37 with our reporter mice (Ncr1Cre;R26LSL-UPRT-HA-sfGFP) and sorted two NK1.1+ populations, namely, sfGFP+ NK cells and sfGFP− NKT cells, by FACS for single-cell TCR sequencing (scTCR-seq) (Supplementary Fig. 5a-c). In both normal and metastatic tissues, greater numbers of WPRE+ cells were detected in the cluster of sfGFP+ TCR− NK cells than in the cluster of sfGFP− TCR+ NKT cells (Supplementary Fig. 5d-f), suggesting that the WPRE is useful as a heritable cell tag when fluorescent protein-based tracing is not feasible.
a Bar plot showing the frequency of WPRE+ macrophages from different genetic background, and integrated genome viewer plot showing transcripts mapping to the WPRE. b UMAP embedding of N-macs and LMAMs from Ccr2GFP/GFP mice and control littermates. Feature plots showing the expression of KC-identity genes and SAM genes. c-d Re-clustering of LMAMs extracted from Ccr2GFP/GFP background and littermate control Ccr2GFP/WT background; e IF staining and quantification of HA tag+ KC-derived LMAMs (Ccr2GFP/GFP: n = 6 for MC38; n = 5 for E0771; n = 3 for HCC; Ccr2GFP/WT littermates: n = 6 for MC38; n = 5 for E0771; n = 3 for HCC). f,g Re-clustering of WPRE+ N-macs and WPRE+ LMAMs from Ccr2GFP/GFP mice (f), and pseudo time analysis revealing the dynamic gene regulatory programs along of KC trans-differentiation (g). h Heatmap showing the downregulation of KC-identity genes and upregulation of SAM genes in KCs when infiltrating into liver metastasis. i Violin plots showing the expression of selected KC-identity genes and SAM genes in normal liver KCs (WPRE+ N-macs), KC-derived LMAMs (WPRE+ LMAMs) and mo-macs (WPRE+ LMAMs). j,k IF staining and quantification of Clec4f expression (indicated by the nuclear-localized tdT) (j, n = 6 for MC38; n = 6 for E0771; n = 3 for HCC) and Timd4 expression (k, n = 5 for MC38; n = 5 for E0771; n = 3 for HCC). Mean ± s.e.m. shown. Smaller dots are values from individual fields. Outlined circles are mean values taken over multiple fields/sections from the same mouse. P values were calculated by comparing individual animals using two-tailed unpaired (e,k) and paired (j) Mann–Whitney U-test.
We then FACS-sorted N-macs and LMAMs isolated from Clec4f tdT-Cre;R26LSL-UPRT-HA-sfGFP;Ccr2GFP/GFP mice and their Ccr2GFP/WT littermates and performed scRNA-seq to investigate the contribution of KCs to the pool of LMAMs. After aligning the sequencing reads to the transgenic reporter allele, we observed a significant peak over the WPRE and enrichment of WPRE+ cells in the N-mac population (Fig. 4a,b). In line with CITE-seq analysis (Fig. 1e), LMAMs can be clustered into the 5 subtypes based on the gene expression levels of Spp1, Cxcl9, Ccl5, Vcan, and Stmn1 (Fig. 4c,d). A greater percentage of WPRE+ LMAMs was detected in the Ccr2GFP/GFP mice than in the Ccr2GFP/WT littermates (Fig. 4a). This finding was confirmed by IF staining for the HA tag (Fig. 4e), suggesting that KCs may occupy inflammatory macrophage niches left vacant by monocyte depletion.
To understand the phenotypic plasticity of KCs during their infiltration into metastatic nodules, we performed pseudo-time analysis on WPRE+ cells and found that Clec4f+ KCs in the normal liver (Id3-cluster and Mrc1-cluster) could trans-differentiate into Clec4f− LMAMs (Spp1-cluster) via a status of increased proliferation (Mcm3-cluster and Mki67-cluster) (Fig. 4f,g). Interestingly, a small subset of KCs from normal liver could trans-differentiate into Ccl5+ subset when infiltrating into metastasis, which may explain why the frequency of Ccl5+ LMAMs increased in Ccr2-deficient background (Fig. 4d).
To further confirm that the KCs underwent cell division during trans-differentiation, we leveraged the HA-tagged UPRT protein in our reporter mice to profile the KC-specific transcriptome in situ27 (Supplementary Fig. 6a-e) and performed IF staining for the HA tag and the cell proliferation marker Ki67 (Supplementary Fig. 6f).
Of particular interest, these KC-derived LMAMs, similar to mo-macs, expressed high levels of scar-associated macrophage (SAM) genes (e.g., Spp1 and Trem2) that were previously identified in macrophages from fibrotic livers38 (Fig. 4h,i) and exhibited downregulation of KC marker genes, including Clec4f and Timd4, and these changes were also associated with the morphological switch from an elongated shape to a flat shape (Fig. 4j,k).
Functional alteration and epigenetic reprogramming of KCs
We therefore sought to determine whether this phenotypic plasticity of KCs can affect their functions. We generated H11DreERT2;Clec4f tdT-Cre;R26LSL-RSR-tdT-DTR mice to conditionally label or ablate KCs (Fig. 5a). In this model, the expression of CAG promoter-driven cytoplasmic tdT and DTR can be activated by the oral administration of tamoxifen, allowing the tracing of KC-derived LMAMs when Clec4f-driven nuclear tdT is repressed in liver metastasis (Fig. 5b). Consistent with the previously proposed anti-tumour effect of KCs39,40,41,42, depletion of KCs by intraperitoneal administration of diphtheria toxin (DT)43 increased the liver metastasis burden compared with that in mice treated with PBS (Fig. 5b-d). In addition to acute genetic ablation, both pharmacological depletion of KCs by CL treatment (Supplementary Fig. 7a-d) and genetic impairment of KCs by knockout of Id3 in embryonic KCs (Supplementary Fig. 7e,f) enhanced liver colonization, suggesting the potent tumour elimination effect of KCs.
a Schematic design of KC-tdT-DTR mice (H11DreERT2;Clec4f tdT-Cre;R26LSL-RSR-tdT-DTR) for tracing and ablating of KCs in normal tissues and metastatic tissues. b Acute depletion of normal liver KCs (left) and KC-derived LMAMs (right) by activation of DreERT2 using tamoxifen (TAM) and then by intraperitoneally (i.p.) administration of diphtheria toxin (DT) (scale bar, 50 μm). c Schematic diagram of pre-depletion of KCs and establishment of experimental liver metastasis models (PVi, portal vein injection). FC quantification showing DT-mediated genetic ablation of tdT+ KCs (n = 4 for PBS control group; n = 4 for DT-treated group). d Bar plots comparing liver metastasis burden of DT-treated mice (n = 5 for MC38; n = 5 for E0771) or PBS-treated control mice (n = 6 for MC38; n = 5 for E0771). The ex vivo bioluminescence value was normalised to the in vivo bioluminescence value obtained immediately after portal vein injection (day 0). e Schematic diagram of enrichment of macrophages for in vitro co-culture with Staurosporine-treated apoptotic E0771 cells labelled with CypHer5E. f FC quantification of distinct macrophage populations phagocyting apoptotic cancer cells (n = 5). g Normalised distribution of ATAC-seq or CUT&Tag-seq densities from 3 kb upstream to transcription start sites (TSS) to 3 kb downstream to transcription end sites (TES) of 118 KC-identity genes or 46 SAM genes. h,i Genome browser tracks of ATAC-seq or CUT&Tag-seq for transcriptional activator H3K27ac and transcriptional repressor H3K27me3 in the vicinities of selected SAM genes (h) and KC-identity genes (i). The enhancer regions are highlighted in yellow. j Mechanistic model showing the epigenetic reprogramming that underlies phenotypic and functional alterations of KCs in LvMet. Mean ± s.e.m. shown. P values were calculated by comparing individual animals using two-tailed unpaired (d) and paired (f) Mann–Whitney U-test.
We then sought to determine whether the decreased expression of Timd4 in KC-derived LMAMs inhibits their ability to engulf apoptotic cells. When KCs enriched from normal and metastatic tissues by anti-F4/80 magnetic beads were cocultured with CypHer5E-labelled apoptotic cancer cells (Fig. 5e), KC-derived LMAMs exhibited impaired phagocytic activity compared to KCs from normal liver tissues, yet they were still markedly different from mo-macs (Fig. 5f). Notably, despite the significant reduction in Timd4 expression, more than half of the KC-derived LMAMs remained weakly positive for Timd4 (Fig. 4k). These data suggest that both niche factors and cell ontogeny may collaboratively influence the phenotypes and functions of KCs.
To investigate the molecular mechanisms underlying the phenotypic and functional alterations, we FACS-sorted normal tdT+ KCs, metastasis-infiltrating tdT+ KCs and tdT− mo-macs from the same livers, and performed assay for transposase-accessible chromatin with sequencing (ATAC-seq) to identify open chromatin and cleavage under targets and tagmentation (CUT&Tag) to evaluate the occupancy of the macrophage-specific transcription factor PU.1 and histone modifications, including H3K27ac and H3K27me344. Surprisingly, the enhancers of most SAM genes (e.g., Spp1 and Trem2) were accessible even in KCs in the normal liver and were poised for rapid activation via the acquisition of the transcriptional activation marker H3K27ac in response to metastatic niche factors (Fig. 5g,h). Furthermore, although KC-derived LMAMs inherited a cell ontogeny-imprinted enhancer landscape by maintaining open chromatin in KC identity genes, some KC identity genes (e.g., Clec4f and Id3) could be silenced by the deposition of the transcriptional repression mark H3K27me3 (Fig. 5i). Together, these data suggest that the induction of SAM gene expression and the loss of KC marker gene expression in KC-derived LMAMs are attributed to epigenetic reprogramming within the enhancer landscapes (Fig. 5j).
Dual blockade of monocyte recruitment and macrophage proliferation reduces LMAMs more effectively
As local macrophage proliferation and KC infiltration compensated for monocyte deficiency, we proposed that dual blockade of monocyte recruitment and macrophage proliferation by knocking out Ccr2 and targeting Csf1r, respectively, can enhance the efficiency of LMAM depletion. Compared with knockout of Ccr2 alone or treatment with the Csf1r inhibitor PLX562245 alone, dual inhibition resulted in more complete reduction of metastasis-associated monocytes and macrophages (Fig. 6a-d). Importantly, we found that this combination strategy could also be applied to subcutaneous MC38 tumours and orthotopic E0771 tumours, as the population of Iba1+ tumour-associated macrophages became negligible when Ccr2 and Csf1r were simultaneously blocked (Supplementary Fig. 8). It should also be noted that the percent of Ly6G+ neutrophils, a key regulator in promoting metastasis46,47, was elevated in both CD11b+ myeloid cells and CD45+ leukocytes following the dual blockade of Ccr2 and Csf1r (Supplementary Fig. 9a,b). This increase was primarily attributed to the substantial reduction in the number of monocytes and LMAMs, as the absolute numbers of neutrophils did not show a significant increase (Supplementary Fig. 9c).
a,b Representative FC plots and quantification of LvMet infiltrating monocytes and macrophages in Ccr2GFP/GFP mice fed with Csf1r inhibitor PLX5622 diet (n = 7 for MC38; n = 5 for E0771) or control chow (n = 5 for MC38; n = 5 for E0771), and in Ccr2GFP/WT littermates fed with PLX5622 diet (n = 7 for MC38; n = 7 for E0771) or control chow (n = 5 for MC38; n = 6 for E0771). c,d Representative IF staining and quantification for LvMet infiltrating Iba1+ myeloid cells in Ccr2GFP/GFP mice fed with PLX5622 diet (n = 6 for MC38; n = 6 for E0771) or control chow (n = 6 for MC38; n = 5 for E0771; scale bar, 50 μm), and in Ccr2GFP/WT littermates fed with PLX5622 diet (n = 6 for MC38; n = 6 for E0771) or control chow (n = 5 for MC38; n = 6 for E0771). e FC quantification of immune checkpoints in CD4+ T cells and CD8+ T cells from LvMet (n = 6 for MC38; n = 4 for E0771). Representative IF staining showing CD8+ T cells expressing Pd1 (scale bar, 50 μm). f Schematic diagram showing experimental design. Bar plots comparing the effects of anti-Pd1 antibodies on the metastasis-bearing Ccr2GFP/WT mice fed with control diet (n = 6 for MC38; n = 5 for E0771) or PLX5622 (n = 5 for MC38; n = 6 for E0771) and on the metastasis-bearing Ccr2GFP/GFP littermates fed with control diet (n = 5 for MC38; n = 6 for E0771) or PLX5622 (n = 4 for MC38; n = 8 for E0771). Representative ex vivo bioluminescence imaging of liver metastases (right). Mean ± s.e.m. shown. Smaller dots are values from individual fields. Outlined circles are mean values taken over multiple fields/sections from the same mouse. P values were calculated by comparing individual animals using two-tailed unpaired Mann–Whitney U-test (b, d, f).
One important immunosuppressive role of LMAMs is to induce apoptosis in cytotoxic T cells4. Indeed, more than half of the CD4+ T cells and CD8+ T cells infiltrating liver metastatic nodules were positive for the immune checkpoint molecule programmed death-1 (Pd1) (Fig. 6e), which prompted us to investigate the effectiveness of anti-Pd1 treatment with single or dual blockade of Ccr2 and Csfr1.
Single blockade of Ccl2/Ccr2 can enhance the efficacy of immune checkpoint inhibitors (ICI) in preclinical cancer models, including bladder cancer and lung metastasis of melanoma48. Although the absolute number of CD8+ T cells remained unchanged (Supplementary Fig. 9d,e), effector CD8+ T cells (CD8+ TEFF) were more enriched in the Ccr2-deficient background than in the littermate control background (Supplementary Fig. 10a,b,e). Additionally, the exhausted T cells (CD8+ TEX) exhibited the highest clonality (Supplementary Fig. 10c), a finding consistent with observations in human breast cancer samples49. Interestingly, a subset of CD8+ TEX cells co-expressed multiple immune checkpoints and displayed a higher exhaustion score (Supplementary Fig. 10d), leading to their classification as terminally exhausted T cells (CD8+ TTEX) that are minimally responsive to ICI50,51. Despite a notable presence of CD8+ TEX, the CD8+ TTEX was less prevalent in the Ccr2GFP/GFP background compared to the Ccr2GFP/WT control background (Supplementary Fig. 10a,e). Indeed, solely blocking Ccr2 showed improvement in the efficacy of anti-Pd1 treatment (Fig. 6f). On the other hand, inhibition of Csf1r alone did not enhance the effectiveness of anti-Pd1 treatment (Fig. 6f), potentially because of the accumulation of immature monocytes (Fig. 6a) or the impairment of anti-tumour normal KCs41. Interestingly, the combined blockade of Ccr2 and Csf1r led to increased response rates and decreased liver metastasis burden (Fig. 6f) compared to a single blockade of Ccr2, indicating a promising strategy for targeting immunosuppressive myeloid cells to improve the efficacy of ICI.
Discussion
The present work reveals the heterogeneity and plasticity of macrophages in liver metastasis. Compared with other liver diseases such as metabolic dysfunction-associated steatohepatitis52,53, liver metastasis is almost devoid of KCs, and is more enriched with mo-macs, which is consistent with the niche competition model in inflammation54. The replacement of KCs with mo-macs is also observed in primary liver cancer55, a phenomenon that is driven by the Ccl2/Ccr2 signaling56,57. Counterintuitively, depletion of monocytes does not lead to a significant reduction in LMAM numbers, either due to an enhanced proliferation of macrophages within the malignancy or due to the infiltration of KCs from nearby normal liver. Our results provide direct evidence for the niche competition model on the basis of monocyte deficiency and revealed that at least a subpopulation of KCs can adapt to the inflammatory microenvironment through transient proliferation and subsequently trans-differentiate toward a cellular state similar to mo-macs.
Notably, the plasticity of tissue-resident macrophages is underappreciated, as the presence of mo-macs contaminates the resident macrophage pool58. In this study, we leveraged precise fate mapping of resident macrophage in the monocyte deficient background to uncover the plasticity of KCs in liver metastasis, which warrants further attention in other physiological and pathological contexts.
Of particular interest, both KC-derived and monocyte-derived LMAMs resemble SAMs previously identified in human and mouse liver cirrhosis38. As the maintenance of KC identity requires the niche interactions among statelet cells, sinusoidal endothelial cells and hepatocytes21,59, disruption of such niche in liver diseases drives phenotypic alterations of KCs. Mechanistically, the emergence of SAM genes and the loss of KC identity genes are attribute to the epigenetic landscape reprogramming. For example, the enhancers of SAM genes are always accessible even in KCs of normal liver and are poised for rapid activation by acquiring histone modification H3K27ac in response to metastasis niche factors. Furthermore, the KC-identity genes can be silenced by the recruitment of histone modification H3K27me3. The epigenetic silencing of Id3 could hinder the ability of KCs to engulf live cancer cells and recruit effector CD8+ T cells41, thereby impairing KCs’ anti-tumour activities in metastatic tissues.
KC-derived LMAMs exhibit partial retention of ontologically epigenetic memory, with a lower frequency of immunosuppressive Spp1+ subsets compared to mo-macs. This is associated with an increase in effector CD8+ T cells and a decrease in terminally exhausted CD8+ T cells within the tumour microenvironment. Therefore, it would be intriguing to investigate if the origins of LMAMs could serve as a predictor for the tumour microenvironment using a large cohort of clinical samples. However, since KC-derived LMAMs phenotypically resemble mo-macs, distinguishing the origins of LMAMs from transcriptome data poses a challenge. An integrated analysis of single-cell transcriptome and chromatin accessibility data will be essential to address the question.
Overall, our study points that cell ontogeny and environmental factors jointly shape the epigenetic landscape, which in turn influences phenotypic plasticity and specifies physiological functions of KCs. Given that the replenishment from fully differentiated macrophages compensates for monocyte blockade, combinatory interventions perturbing monocyte recruitment, local macrophage proliferation, and KC trans-differentiation represent a more effective strategy for targeting tumour-promoting myeloid cells, and offer promising strategies for other potential synergy.
Methods
Cell lines and cell culture
Mouse triple negative breast cancer (TNBC) E0771 cells (CH3 Biosystems, #94A001) and mouse colorectal cancer MC38 cells (iCell, #m032) were cultured in RPMI-1640 medium (Thermo Fisher, #C11875500BT) and DMEM/High Glucose medium (Thermo Fisher, #C11995500BT), respectively. Both media contained 10% FBS (Gibco, #A3160802), 100 IU ml−1 penicillin/streptomycin, 100 µg ml−1 amphotericin B (Lonza, #17-745E) and 200 nM L-Glutamine (Gibco, #25030081). The E0771 and MC38 cancer cells were engineered to express firefly luciferase (Addgene plasmid #19166, gift from Eric Campeau & Paul Kaufman) for bioluminescence imaging.
All cells were grown in a humidified incubator at 37 °C, with 5% CO2, and were tested regularly for mycoplasma contamination (Vazyme, #D101).
Animals used
All animal work was done in accordance with a protocol approved by Institutional Animal Care and Use Committee (IACUC) at the Sun Yat-sen University Cancer Center (Approval Number: L025504202108022, L025504202107004 and L025504202211002).
The C57BL/6J-Gt(ROSA)26Sorem(CAG-LSL-UPRT-T2A-NLS-sfGFP-IRES-rtTA3-WPRE-pA)1Smoc (R26LSL-UPRT-HA-sfGFP) mice in a C57BL/6 genetic background were generated by inserting CAG-LSL-UPRTHA-T2A-NLS-sfGFP-IRES-rtTA3-WPRE-pA expression cassette (Addgene #126677, a gift from Joan Massagué) into ROSA26 locus using a CRISPR–Cas9-mediated genome editing system by Shanghai Model Organisms Center (SMOC). In brief, 5 ng μl−1 Cas9 mRNA, 2 ng μl−1 gRNA targeting Rosa26 safe harbour (5’-GGGGACACACTAAGGGAGCT-3’), and 10 ng μl−1 targeting vector containing 3.3 kb at both 5’ and 3’ homologous arm of the Rosa26 target site were injected into the pronuclei of C57BL/6 J one-cell stage zygotes and transplanted into pseudopregnant female mice. The F0 chimeric offspring were then screened by PCR using two different pairs of primers: Rosa26_5p_F (5’-GCCGGGCCTCGTCGTCTG-3’) in combination with Rosa26_5p_R (5’-TGAGGGCAATCTGGGAAGGTT-3’) amplified a 5.1 kb band from the wild-type (WT) allele and a 3.4 kb band from the Rosa26-engineered allele. Additionally, Rosa26_3p_F (5’-GGGGGAGGGGAGTGTTGC-3’) paired with Rosa26_3p_R (5’-TTCTTCCTGCCTGCCTTCTGTGAC-3’) amplified a 6.5 kb band from the WT allele and a 3.6 kb band from the Rosa26-engineered allele. The F0 R26LSL-UPRT-HA-sfGFP mice were then crossed with C57BL/6 J WT mice for one generation to obtain genetically engineered F1 offspring.
C57BL/6 J (WT, #000664), Clec4ftdT-Cre (#033296)21, CD68EGFP (#026827)22, Cx3cr1CreERT2 (#021160)60, Ccr2GFP (#027619)61, CD45.1 (#033076) (all from Jackson Laboratory), H11CAG-RSR-ZsG-CAG-LSL-tdT (denoted as H11LSL-tdT, #T058308), H11-CAG-DreERT2 (denoted as H11DreERT2, #T049774), Ki67RSR-Cre (#T050101), R26LSL-DTA (#T009408) (all from GemPharmatech), Tnfrsf11aCre (#B-EM-009, from Biocytogen), Id3flox (denoted as Id3fl, #NM-CKO-2100278) and R26LSL-RSR-tdT-DTR (#NM-KI-190086, both from SMOC). Genomic DNA was prepared from mouse ear punch with Mouse Genotyping Kit (APExBio, #K1027) as per manufacturer’s protocol. The wild-type allele was used as the control to distinguish knock-in/knock-out allele. Sequences of all primers used to distinguish knock-in/knock-out allele from wild-type allele were listed in Supplementary Table 1.
For retro-orbital injection, the mice were anaesthetized using an isoflurane-based small animal anaesthesia unit. Otherwise, ketamine (100 mg/kg)/xylazine (10 mg/kg) were used for anaesthesia.
Hydrodynamic tail-vein injection of a transposon vector overexpressing cMyc (MycOE, addgene #129775, a gift from Amaia Lujambio), a vector expressing SB13 transposase (SB13, addgene # 20207, a gift from John Ohlfest), and a CRISPR/Cas9 vector expressing a single-guide RNA targeting TP53 (Cas9-sgTP53, addgene # 59910, a gift from Tyler Jacks) was used to establish autochthonous hepatocellular carcinoma12. We dissolved 11.3 μg (2 pmol) of MycOE, 10.49 μg (2 pmol) of Cas9-sgTP53 and 2.5 μg (0.4 pmol) of SB13 in 2 ml 0.9% NaCl solution and injected the solution into the tail vein of each mouse within 5-7 seconds. The hepatocytes that receive the three plasmids will have potential to form tumour due to the stable overexpression of cMyc and inactivation of TP53.
Six to eight-week-old female and male mice of the C57BL/6 J background were used for experimental liver metastasis assays involving E0771 breast cancer cells and MC38 colorectal cancer cells, respectively, unless otherwise specified. Eight to ten-week-old female mice were used for the generation of the spontaneous HCC model. No randomization or blinding was used to allocate experimental groups. No statistical methods were used to predetermine sample size.
Mice were bred under specific pathogen free (SPF) condition, under a 12 h–12 h light-dark cycle, at around 22 °C with around 50% humidity. The experimental group mice and littermate controls were co-housed.
Liver metastases were evaluated using non-invasive bioluminescence imaging conducted weekly. Mice were euthanized approximately four weeks after portal vein injection, at which point the total photon flux ranged from 108 to 109 radiance (p/sec/cm2/sr). For subcutaneous or fat pad injections, tumour growth was monitored every other day. Mice were euthanized when the tumour diameter reached 1 cm, and the corresponding tumour weight was approximately 0.5 g, which is below the maximum allowable weight (10% of the animal’s body weight) as stipulated by the institutional animal board. The mice were euthanized by CO2.
Clinical sample collection
Human tissues were collected with informed consent from patients and were used in accordance with the approval of the Research Ethics Committee of SYSUCC (Approval Number: G2021-061-01). Six cases of liver metastasis samples of breast cancer patients (three cases HER2+ and three cases TNBC) were collected from SYSUCC. Paraffin-embedded sections of 3-μm thickness were used for haematoxylin and eosin staining and immunofluorescence staining. Before immunofluorescence staining, the sections were deparaffinized and treated with a heat-induced antigen retrieval Tris-EDTA (pH 9.0) solution (Sangon Biotech, #E673004-0100). After cooling to room temperature, the slides were first washed with running water for 5 min, then washed with 1×PBS twice for 5 min each time.
Portal vein injection
E0771 cells and MC38 cells were collected from culture with 0.25% trypsin (HyClone, #SH30042.02). The cells were washed twice with PBS, counted, then resuspended in cold PBS to a concentration of 5×106 cells ml−1 and 2×106 cells ml−1 for E0771 and MC38, respectively. Mice were anaesthetized, and two 1-inch incisions were made into the shaved abdomen and the peritoneum respectively to reveal the liver. The duodenum was gently pull out to expose the portal vein. The cancer cells were pipetted up and down several times before transferred to 31-gauge needle (BD Bioscience, #328291). 50 μl cold PBS were injected into portal vein. The incisions of the peritoneal membrane and skin were closed by absorbable sutures and would clips, respectively. The wound clips were removed on Day 7 post-injection of cancer cells.
Fat pad injection and subcutaneous injection
E0771 cells and MC38 cells were collected using 0.25% Trypsin-EDTA (Gibco). The cells were washed twice with PBS, counted, and then resuspended in 1:1 solution of PBS and growth factor reduced Matrigel (Corning, #356231). For mammary fat pad injection of E0771 cancer cells, mice were anaesthetized, and a small incision was made on the shaved abdomen to reveal mammary gland. 0.5×106 cells in 50 µl ice-cold matrigel/PBS mixture were injected directly into the fat pad. The incision was then closed using wound clips, which were removed on day 7 after injection of cancer cells. For subcutaneous injection of MC38 cancer cells, 0.5×106 cells in 50 µl ice-cold matrigel/PBS mixture were injected into loose shoulder skin on the right site of the mouse.
Bioluminescent imaging and analysis
Mice were injected retro-orbitally with 1.5 mg of D-luciferin (GoldBio, #LUCK-1) (15 mg ml−1 in PBS). Imaging was completed within 1–2 min after injection, using an IVIS Spectrum Living Imaging system and analysis software v4.2 (Perkin Elmer). For in vivo bioluminescence images, photon flux was calculated for each mouse by using a rectangular region of interest encompassing the liver of the mouse in a supine position. For ex vivo bioluminescence images, photon flux was calculated for each dissected liver by using a circular region of interest along the edge of a 35-mm cell culture dish.
Tamoxifen and diphtheria toxin treatment
Tamoxifen (MedChemExpress, #HY-13757A) was dissolved in corn oil (20 mg ml−1, Wako, #032-17016) and heated at 37 °C for 3 hours. Mice were administered with tamoxifen through oral gavage (100 mg kg−1 body weight). To activate Cx3cr1CreERT2-mediated recombinase, each mouse was fed with Tamoxifen every three days, starting from Day 12 post-intraportal injection, as indicated in Fig. 2e. For proliferation tracing experiment, DreERT2-mediated recombinase was introduced by a single dose of tamoxifen on Day 12 post-intraportal injection, as indicated in Fig. 3f. For KC specific depletion, DreERT2-mediated recombinase was introduced by 8 doses of tamoxifen gavage, as indicated in Fig. 5c. A single dose of 200 ng DT (List Laboratories, #150) was intraperitoneally introduced one week after the last Tamoxifen injection. The cancer cells were inoculated into liver through portal vein one day after DT injection.
Liposomal treatment
For fluorescent labelling or pharmacological depletion of mononuclear phagocytes, 100 μl DiI-labelled liposomes (#F70101F), clodronate liposomes (#F70101A) or PBS-control liposomes ((#F70101CA) were injected intravenously (all at the concentration established by FormuMax), as indicated in Fig. 3c-h and Supplementary Fig. 7a-d.
Csf1r inhibitor and dual ICB treatment
The liver metastasis-bearing Ccr2GFP/GFP mice or littermate control Ccr2GFP/WT mice were evaluated for metastasis burden using in vivo bioluminescence imaging. To inhibit metastasis-associated macrophages, mice were fed with 1200 ppm PLX5622 (TargetMol, # T7100, formulated in AIN-76A standard chow diet) ad libitum for 14 days before ex vivo bioluminescence imaging of liver metastasis burden. To block immune checkpoints Pd1, 100 μg anti-Pd1 antibodies (clone 29 F.1A12, BioXCell, #BE0273, Lot: 845722N1) were intraperitoneally injected into the metastasis-bearing mice starting from the day of in vivo bioluminescence imaging and every 3 days subsequently as indicated in Fig. 6f.
Tissue dissociation
The mice were euthanized by CO2 and subjected to whole-animal perfusion. In brief, an incision was immediately made in the right atrium and 10 ml PBS with 2 mM EDTA was slowly injected through the left ventricle. The change in colour of the liver should be observed for the successful perfusion. After whole body perfusion, the liver metastasis nodules and nearby normal tissues were dissected and briefly cut with scissors. The tissue pieces were transferred to a tissue digestion C-tube (Miltenyi) and incubated in digestion buffer containing 1× RPMI, Collagenase A (1 mg ml−1, Sigma, #10103586001), DNase I (0.1 mg ml−1, Sigma, #10104159001) and 1% heat-inactivated FBS, and mechanically on a Single-cell Suspension Dissociator (Miltenyi). Briefly, the “Tumor_3” program was run on the Dissociator, followed by a 15-minute incubation at 37 °C. Next, the “Tumor_3” program was run again to increase the yield of single cells, followed by a 15-min incubation at 37 °C. After a final run of the “Tumor_3” program, the digestion reaction was filtered through a 70-um cell strainer and was then transferred to 2-ml ice-cold RPMI medium to stop the reaction. Cellular debris and red blood cells were removed by the Debris Removal Solution (Miltenyi, #130-109-398) and 1× RBC Lysis Buffer (Biolegend, #420301), respectively, as per manufacturer’s instructions.
Flow cytometry
Single-cell suspensions were prepared as described in the “Tissue Dissociation” section. 0.1 − 0.5×106 cells were incubated for 15 min at 4 °C with TruStain FcX PLUS anti-mouse CD16/32 antibody (clone S17011E, #156604, 1:100, Lot: B321333) and True-Stain Monocyte Blocker (1:100, #426102, Lot: B412166) (both from Biolegend), in 100 µl FACS staining buffer (1.5% FBS and 0.5% BSA in PBS). Cells were subsequently stained with antibodies in FACS buffer at 4 °C for 25 min. The following antibodies against mouse antigens were used (1 μg ml−1): anti-CD45 (clone 30-F11, #103132, Lot: B370638), anti-CD45.2 (clone 104, #109852, Lot: B349582), anti-CD11b (clone M1/70, #109851, Lot: B316070), CD11c (clone N418, #117333, Lot: B342522), anti-F4/80 (clone BM8, #123137, Lot: B330997), anti-Ly6C (clone HK1.4, #128005, Lot: B336287), anti-Ly6G (clone 1A8, #127607, Lot: B332036), anti-Gr1 (clone RB6-8C5, #108403, Lot: B268539), anti-TCRβ (clone H57-597, #109245, Lot: B339724), anti-CD19 (clone 6D5, #115540, Lot: B336881), anti-NK1.1 (clone PK136, #108707, Lot: B320695), anti-CD4 (clone GK1.5, #100405, Lot: B335689), anti-CD8 (clone 53-6.7, #100728, Lot: B316770), anti-Timd4 (clone RMT4-54, #130007, Lot: B307560), anti-Pd1 (clone 29 F.1A12, #135209, Lot: B361178), anti-Ctla4 (clone UC10-4B9, #106309, Lot: B357050), anti-Tigit (clone 1G9, #142105, Lot: B374378), anti-Lag3 (clone C9B7W, #125209, Lot: B396078), rat IgG2aκ isotype control antibody (clone RTK2758, #400511, Lot: B382872), Armenian hamster IgG isotype control antibody (clone HTK888, #400911, Lot: B360077), mouse IgG1κ isotype control antibody (clone MOPC-21, #400119, Lot: B377860), rat IgG1κ isotype control antibody (clone RTK2071, #400411, Lot: B365119) (all from Biolegend), anti-CD45.1 (clone A20, Tonbo, #20-0453-U025, Lot: C0453011520202) and anti-F4/80 (clone CI:A3-1, BioRad, #MCA497SBV440, Lot: 100004072). 4’,6-Diamidino-2-Phenylindole Dilactate (DAPI, Biolegend, #422801) or HelixNP NIR (Biolegend, #425301) or NucFix Red (Biotium, #32010) was used to exclude dead cells. Cells were analysed or sorted using either the Beckman CytoFLEX or MoFlo Astios EQ, respectively. All data were analysed using FlowJo v10. The gating strategies were presented in Supplementary Fig. 11.
In vitro phagocytosis assay
To trigger apoptosis, E0771 cancer cells were treated with 1 μM Staurosporine (Sigma, #S6942) for 12 hours. Dead Cell Apoptosis Kit (Thermo Fisher, #A35122) was used to find optimal time of Staurosporine treatment. The apoptotic cells were stained with CypHer5 (GE Healthcare, #PA15401) and seeded together with normal and metastasis-associated macrophages isolated from Clec4f tdT-Cre;CD68EGFP mice or H11DreERT2;Clec4f tdT-Cre;R26LSL-RSR-tdT-DTR;Ccr2GFP/GFP mice in a non-tissue culture-treated plate at 1:10 phagocyte to cancer cell ratio for 1 hour, 4 hours and 8 hours. Cells were dissociated from plate with trypsin and the phagocytes were assessed by flow cytometry analysis.
Immunofluorescence staining
The perfused livers were fixed with 4% paraformaldehyde (PFA, Leagene, #DF0131) overnight at 4 °C. After being washed with PBS three times, the fixed tissues were incubated in 30% sucrose in PBS (w/v) overnight, and frozen in optimum cutting temperature (OCT) gel (Sakura, #4583). Sections of 6-μm in thickness were cut on Leica CM1850 cryotome.
Slides were blocked in PBS with 10% normal donkey serum (Jackson ImmunoResearch, #017-000-121), 1% BSA (Jackson ImmunoResearch, #001-000-162), and 0.3% Triton X-100 (Sangon Biotech, #A110694) for 1 h at 20 °C. Primary antibodies were incubated overnight at 4 °C in the staining solution (5% normal donkey serum, 0.5% BSA and 0.3% Triton X-100). Secondary antibodies were added into the staining solution and incubated for 1 h at 20 °C. Excess antibodies were removed by washing for 5 min in PBS with 0.5% Tween-20. Nuclei were counter-stained with 1 μg ml−1 DAPI (Biolegend, #422801) for 3 min at room temperature. To reduce the background, TrueVIEW Autofluorescence Quenching Kit (Vector Labs, #SP-8400-15) was used as per manufacturer’s protocol. Primary antibodies included: anti-Clec4f (Thermo Fisher, #PA5-47396, 1:500, Lot: WF3292908), anti-Clec4f (clone 3E3F9, Biolegend, #156804, 1:100, Lot: B355441), anti-F4/80 (clone CI:A3-1, abcam, #ab6640, 1:200, Lot: GR3349811-1), anti-Glutamine Synthetase (abcam, #73593, 1:1000, Lot: GR3374144-3), anti-HA Tag (Bethyl, #A190-138A, 1:500, Lot: 1), anti-HA Tag (abcam, #ab9110, 1:500, Lot: GR3425636-1), anti-CD31 (SinoBiological, #50408-T16, 1:2000, Lot: HB07FE0711-B), anti-Iba1 (abcam, #ab178847, 1:3000, Lot: GR3229566-13), anti-TIMD4 (R&D, #AF2929, 1:100), anti-Timd4 (R&D, #AF2826, 1:100), anti-tdT (abcam, #ab167453, 1:500, Lot: GR3383449-1), anti-tdT (arigo, #ARG55724, 1:500, Lot: 11534), anti-VE cadherin (R&D, #AF1002, 1:100, Lot: FQI0119111), anti-Ki67 (clone D3B5, CTS, #12202, Lot: 6), anti-hHB-EGF (DTR, R&D, #AF259, 1:100, Lot: PX1023041), anti-Pd1 (R&D, #AF1021, 1:100, Lot: GQA0422081), anti-CD8α (clone 53-6.7, R&D, #MAB116, 1:100, Lot: FSG0523022). Secondary antibodies included: Alexa-488 donkey anti-rabbit (#711-545-152, 1:800, Lot: 152657), Cy3 donkey anti-rabbit (#711-165-152, 1:800, Lot: 153388), Cy3 donkey anti-goat (#705-165-147, 1:800, Lot: 151429), Cy3 donkey anti-rat (#712-165-153, 1:800, Lot: 151924), Alexa-647 donkey anti-rabbit (#711-605-152, 1:800, Lot: 153705), Alexa-647 donkey anti-goat (#705-605-147, 1:800, Lot: 153320, all from Jackson ImmunoResearch), Alexa-405 donkey anti rabbit (abcam, #ab175649, 1:500, Lot: GR3361021-4), Alexa-647 donkey anti rat (abcam, #ab150155, 1:500, Lot: GR3285492-3), and Alexa-488 donkey anti-rat (Bethyl, #A110-337D2,1:500, Lot: 14). Stained sections were visualized by a Zeiss LSM 980 confocal microscope or scanned with Zeiss Axioscan.Z1. ImageJ was used to manually count the number of macrophages and automatic quantify the number of nuclear foci. The isotype controls used to evaluate immunofluorescence signals from antibodies were presented in Supplementary Fig. 12.
Proinflammatory chemokines detection
Proinflammatory chemokines of normal liver and liver micro-metastasis were detected using LEGENDplex™ Mouse Proinflammatory Chemokine Panel (Biolegend, #740451). Briefly, small liver metastatic lesions (1–2 mm size) and adjacent normal liver tissues were harvested 14 days post portal vein injection and were kept in PBS containing 25 mM EDTA and protease inhibitor (cOmplete tablets, Roche, #4693159001). The tissues were then lysed with a tissue homogenizer, and centrifuged at 12,000 rpm for 30 minutes at 4 °C. The supernatant was collected and mixed with Assay buffer in a v-bottom plate. The mixture was subsequently incubated with APC-conjugated capture beads, biotinylated capture antibodies, and PE-conjugated streptavidin according to the manufacturer’s instructions. Fluorescence intensity was recorded by a CytoFLEX flow cytometer (Beckman Coulter). Data was further analysed using LEGENDplex™ Data Analysis Software Suite, version 2023-02-15.
Liver clearing, immunolabeling and three-dimensional imaging
Tissue clearing was performed with Tissue Clearing Kit (Nuohai Life Science, #NH210701-FS). Briefly, after whole-animal perfusion, livers with metastasis nodules were fixed in ice-cold 4% PFA overnight at 4 °C. After 3 times of washes in PBS for 2 hours at RT, livers were immersed in 1/2-diluted Solution 1 (S1) at 37 °C for 6 hours, and subsequently in 100% S1 for 3 days at 37 °C for rapid de-lipidation and decolorization. Then, after 3 times of washes in PBS for 2 hours at RT, livers were immersed in 1/2-diluted Solution 2 (S2) at 37 °C for 6 hours, and subsequently in 100% S2 for 3 days at 37 °C for further de-lipidation and decolorization. After de-lipidation, the livers were permeabilized in PBS with 0.3% Triton X-100 at 37 °C for 1 hour and were blocked in PBS with 10% normal donkey serum, 1% BSA, and 0.3% Triton X-100 at 37 °C for 1 day. Primary antibodies were incubated at 37 °C in a staining solution (5% normal donkey serum, 0.5% BSA and 0.3% Triton X-100) for 2 days. The livers were washed in PBS with 0.3% Triton X-100 at RT for 2 hours and were incubated with second antibody in staining solution at 37 °C for 2 days. After 3 times of washes in PBS for 2 hours at RT, livers were immersed in 1/2-diluted Solution 3 (S3) at 37 °C for 6 hours, and subsequently in 100% S3 at 37 °C for refractive index (RI) matching, until the tissues become transparent. The cleared tissue was imaged by LiTone XL Light-Sheet Microscope (Light Innovation Technology) for 3D imaging. The magnification is 10×; the numerical aperture of the detection lens is 0.6; the spatial resolution is 0.58 μm × 0.58 μm; the field view is 6.2 mm × 3.3 mm. The laser channels are 561 nm and 647 nm for the detection of red fluorescent protein tdT and Alexa Fluor 647 conjugated anti-Clec4f antibody, respectively. The original data was processed by LitScan 3.2.0 (Light Innovation Technology).
In situ RNA profiling
The in situ RNA profiling with SLAM-ITseq was conducted as previously described27. Briefly, 4-Thiouracil (4tU, Sigma, #440736) was dissolved in DMSO at 200 mg ml−1 and was then diluted in corn oil at a 1:4 ratio (50 mg ml−1). The solution was vigorously shaken and intraperitoneally injected into Clec4f tdT-Cre;R26LSL-UPRT-HA-sfGFP;Ccr2GFP/GFP mice bearing liver metastasis. Liver metastasis tissues and nearby normal tissues were harvested 4 hours after the injection of 4tU without whole animal perfusion. The collected tissues were briefly washed with PBS and immediately transferred to the lysis buffer for RNA extraction (Vazyme, #RC112). RNA was dissolved in H2O with 1 mM DTT (Sigma, #646563) to prevent oxidation of thiol group in 4tU. 1 μg RNA was reacted with Iodoacetamide (IAA, Sigma, #I1149) buffer (10 mM IAA and 500 mM NaH2PO4) at 50 °C for 15 min to alkylate the thiol group. The RNA from the alkylation reaction was then purified using RNA Clean & Concentrator (Zymo, #R1015), and 0.5 μg RNA was used as input for strand-specific library Prep Kit (Vazyme, #NR604). RNA-seq library preparation was conducted following manufacturer’s instructions with the 14 PCR cycles for library amplification. The multiplexed libraries were sequenced to generate a minimum of 100 million reads using the NovaSeq 6000 platform (Illumina) with pair-end 150 cycles.
A customized version of SLAM-seq data processing pipeline was used for data processing. Briefly, the raw RNA reads were trimmed with Trimmomatic (v0.39) with the parameters ‘SLIDINGWINDOW:5:20 MINLEN:36’. The reverse complemented version of the trimmed R2 reads was further generated by Seqtk (v1.3). Given that Cre+ cells incorporated 4tU into nascent RNA and resulted in many T > C conversions after alkylation reaction, the mutation-tolerant aligner NextGenMap (v0.5.5) was used to align the trimmed R1 and reverse complemented R2 reads to mouse reference genome GRCm38 with options ‘-t 8 -n 1 --strata --bam --slam-seq 2’. Samblaster (v0.1.26), Samtools (v1.9), and Slamdunk (v0.4.3; options: filter -mq 2 -mi 0.8 -nm −1) were used to sort, index, and filter the resulting bam files, respectively. The bam file was further converted to mpileup file by Samtools for downstream analysis with options ‘-B -A --output-QNAME’. Varscan (v2.3.9) was used to call variants into a vcf file with options ‘mpileup2snp --strand-filter 1 --min-var-feq 0.2 --min-coverage 10 --variants 1’. The mpileup file was screened along with the vcf file to identify reads with at least two T > C base conversion events. Such base-converted reads were extracted from the raw read sets by Seqtk to run the mapping process again with NextGenMap for downstream visualization in IGV (v2.16.0) and further analysis. After transcript quantification with RSEM (v1.3.3), R package DESeq2 (v1.34.0) was used to normalise the count matrix and to identify differentially expressed genes. Enrichment of pathways pertinent to normal or metastasis-associated KCs was identified by R package clusterProfiler (v4.2.2).
Single-cell RNA sequencing
Single cell suspensions (2 ×106 cells ml−1 in ice-cold PBS) were loaded onto microwell chip using the Singeron Matrix Single Cell Processing System. Barcoding Beads were subsequently collected from the microwell chip, followed by reverse transcription of the mRNA captured by the Barcoding Beads and to obtain cDNA for PCR amplification. The amplified cDNA was then fragmented and indexed with sequencing adapters. The single-cell RNA sequencing (scRNA-seq) libraries were constructed according to the protocol of the GEXSCOPE scRNA Library Kits (Singleron Biotechnologies). Individual libraries were diluted to 4 nM, pooled, and then sequenced on Novaseq 6000 (Illumina) with 150 bp paired-end reads.
Singeron scRNA-seq analysis pipeline CeleScope (v1.9.0) was used to pre-process the raw reads and to generate gene expression matrix with default parameters. Briefly, barcodes and unique molecular identifiers (UMIs) were extracted from R1 reads and corrected. Adapter sequences and poly A tails were trimmed from R2 reads and the trimmed R2 reads were aligned against the GRCm38 (mm10) transcriptome using STAR (v2.6.1a). Uniquely mapped reads were then assigned to exons with FeatureCounts (v2.0.1). Successfully assigned reads with the same cell barcode, UMI and gene were grouped together to generate the gene expression matrix for further analysis. Seurat (v4.3)62 was used for quality control, dimensionality reduction and clustering. Cells with top 2% UMI countings were filtered out. Cells with gene count less than 200 or with top 2% gene count and cells with more than 10% mitochondrial contents were excluded for downstream analysis. Filtered data were then log normalized and scaled using ‘NormalizeData’ and ‘ScaleData’, respectively. Cell–cell variation due to UMI counts and percent mitochondrial reads regressed out. Top 2000 variable genes were selected by ‘FindVariableFeautres’ for principal component analysis (PCA). The top 30 principal components, determined by JackStraw algorithm, were used for clustering and dimensional reduction at a resolution of 1.5. For CITE-seq, the antibody-derived tag (ADT) libraries were normalized using Seurat. The cell type identity of each cluster was determined with the expression of canonical markers based on the annotation of SynEcoSys database (https://singleron.bio/product/detail-25.html). Uniform Manifold Approximation and Projection (UMAP) algorithm was applied to visualize cells in a two-dimensional space. Heatmaps, dot plots, violin plots displaying the expression of markers used to identify each cell type were generated by ‘DoHeatmap’, ‘DotPlot’, ‘FeaturePlot’, ‘Vlnplot’, respectively.
To understand the LMAM subset changes in Ccr2 knockout mice, we extracted LMAMs from Ccr2GFP/GFP background and littermate control Ccr2GFP/WT background and re-clustered for more detailed analysis following the same procedures described above and by setting the clustering resolution as 1.0 for a higher resolution map of LMAMs.
Pseudo-time trajectory analysis was performed with Monocle363 using default parameters. In brief, the Seurat object was first converted to Monocle CellDataSet. The raw count expression matrix was then pre-processed to normalise data. And top 50 principal components were used to capture variated genes across all the cells in the data set. After dimension reduction with UMAP method, the cells were put in order by how much progress they’ve made by ‘learn_graph’ function. Finally, the cells were ordered in pseudotime along a trajectory and visualized by ‘plot_cell’ function.
For cell indexing and epitopes profiling, single cell suspension was incubated with the cell hashtag antibodies (unique DNA barcodes listed in Supplementary Table 2) and the cocktail of 119 unique cell surface antibodies (Biolegend, #199901), as per manufacturer’s protocol.
To trace KC-derived LMAMs, an exogenous gene sequence, IRES-WPRE, was appended to the ends of the mouse reference genome FASTA file, accompanied by a GTF file detailing gene annotations. The reference index of this custom genome was constructed using the Celescope ‘mkref’ command for subsequent scRNA-seq analysis.
The KC gene signature and the scar-associated macrophage (SAM) gene signature38 were used to illustrate the phenotypic plasticity of different macrophage populations in liver metastasis. The KC genes were enriched in the macrophage cluster of normal mouse liver, while SAM genes were highly expressed in macrophages from chronic carbon tetrachloride (CCl4)-induced fibrotic livers.
Single-cell TCR analysis
The Cell Ranger (v4.0.0) vdj pipeline was used to assign TCR clonotype with GRCm38 (mm10) as the reference. The process involved calculating a TCR diversity metric that included clonotype frequency and barcode information. Quality control, dimensionality reduction and clustering were performed using Seurat. Cells were filtered out based on having UMI countings in the top 2% or total UMI countings less than 1000. Additionally, cells with gene count below 500 or in the top 2% gene count, as well as cells containing over 10% mitochondrial content, were excluded from downstream analysis.
Following the selection of Cd8a+ cells, the single-cell TCR sequencing data was then analysed with scRepertoire (v2.0.3) using default parameters. Only cells with one productive TCR α-chain (TRA) and one productive TCR β-chain (TRB) were retained for further analysis. Each unique TRA(s)-TRB(s) pair was considered a clonotype. Cells containing a clonotype present in at least two cells were classified as clonal, and the number of cells with such pairs indicated the level of clonality of the clonotype. As shown in Supplementary Fig. 10c, clones were categorized based on the number of cells expressing individual clonotype sequences: single (0-1 cell), small (2-5 cells), medium (6-20 cells), large (21-100 cells), hyper-expanded (101-592 cells).
The ‘combineExpression’ function in scRepertoire package64 was used to incorporate clone information into the Seurat object of scRNA-seq, enabling the calculation of clonal frequency. Additionally, the ‘AddModuleScore’ function in Seurat package was applied to selected genes (Pdcd1, Lag3, Havcr2, Ctla4, Vsir, Cd160) to compute an exhaustion score.
The cell hashtag antibodies used to distinguish CD8+ T cells from backgrounds of Ccr2GFP/WT or Ccr2GFP/GFP were detailed in Supplementary Table 3.
Epigenetic profiling
The liver metastasis and nearby normal tissues of H11DreERT2;Clec4f tdT-Cre;R26LSL-RSR-tdT-DTR;Ccr2GFP/GFP mice were dissociated as described in the “Tissue Dissociation” section. The CD45 Microbeads (RWD Life Science, #K1304) were used to enriched immune cells from single cell suspension through magnetic cell separation. 10,000 live cells (HelixNIR−) expressing F4/80 and tdT were purified by FACS sorting for ATAC-seq and CUT&Tag-seq. After trimming low-quality reads, removing duplicates and reads within blacklist regions, 5-11 million pair-end reads were retained for ATAC-seq; 9-17 million pair-end reads were retained for CUT&Tag-seq of H3K27ac and H3K27me3; 1-4 million pair-end reads were retained for CUT&Tag-seq of PU.1.
Transposase accessibility and transcription factor binding were profiled using an ATAC-seq kit (#N248) and a CUT-Tag kit (#N259, both from NovoProtein), respectively, as according to manufacturer’s instruction. The following antibodies were used for CUT&Tag assay: anti-Histone H3 acetyl K27 antibody (H3K27ac, abcam, #ab177178, 1:200, Lot: GR3202987-25), anti-Histon H3 tri-methyl K27 (H3K27me3, CTS, #9733, 1:100, Lot: 19), anti-PU.1 (abcam, #ab227835, 1:200, Lot: 1007922-11), and goat anti-rabbit IgG secondary antibodies (abcam, #ab7085, 1:200, Lot: GR3404559-6). The PCR cycles was optimized for different factors (ATAC-seq, 10 cycles; PU.1, 16 cycles; H3K27me3, 14 cycles; H3K27Ac: 14 cycles). The multiplexed libraries were sequenced using Novaseq 6000 (Illumina) for pair-end 150 cycles.
Low-quality reads were filtered with Trimmomatic (v0.39) with the parameters ‘SLIDINGWINDOW:5:20 MINLEN:36’, and were aligned to the mouse genome GRCm38 with bowtie2 (v2.4.5) with the parameters ‘--local --very-sensitive --no-unal --no-mixed --no-discordant --phred33 -I 10 -X 2000’. Picard (v2.19.0) and bedtools (v2.30.0) were used to remove duplicated reads and blacklist-region reads, respectively. The clean BAM files were converted to bigwig files with deeptools (v3.5.3) bamCoverage using the parameters ‘--effectiveGenomeSize 2652783500 --normalizeUsing RPKM --extendReads 200 --binSize 50 --outFileFormat bigwig’. The bigwig files were uploaded to local tracks of WashU Epigenome Browser (https://epigenomegateway.wustl.edu/) to visualize different histone modification markers and transcription factors simultaneously at specific genome loci.
In addition to genome browser tracks of individual genes, the bigwig files were also used to calculate scores per genome regions with deeptools computeMatrix using parameters ‘scale-regions -S --upstream 3000 --regionBodyLength 5000 --downstream 3000 --skipZeros --outFileName genes_scaled.mat.gz’. The mitochondria genes in KC signature genes were excluded to eliminate background noises. This intermediate file was used to generate heatmap and profile plot with ‘plotHeatmp’ and ‘plotProfile’, respectively.
Peak calling was performed using macs3 (3.0.0b3) with default parameters. ‘macs3 -q 0.05’ was used for ATAC-seq and CUT&Tag-seq of histone modification antibodies, and ‘macs3 -q 0.25’ was used for CUT&Tag-seq of the transcription factors. A comparison of the peak calling results from this study with published data was presented in Supplementary Fig. 13.
Statistics
In most in vivo experiments, group sizes were determined based on the results of preliminary experiments and no statistical method was used to predetermine sample size. Experiments were repeated at least twice independently, and the data are combined and presented. For the dot plots of IF staining results, smaller dots without an outline are values from individual fields ( ~ 0.4 mm2 fields), and circles that are outlined represent mean values taken over 4−5 fields from the same mouse, unless otherwise noted. The statistical tests were performed by comparing the individual animals. All in vitro experiments were repeated independently for three times (batches). The P values in these in vitro experiments were determined based on biological replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw sequencing reads for epigenetic profiling (Fig. 5g) and SLAM-ITseq (Supplementary Fig. 6a-e) have been deposited at the China National Center for Bioinformation (CNCB) under accession number PRJCA024187. The processed Seurat objects of scRNA-seq assays (Fig. 1e and Fig. 4b) are available at Zendo (#10809097, https://doi.org/10.1101/2024.04.23.590829). The processed Seurat objects of scTCR-seq assays (Supplementary Fig. 10) are available at Zendo (#12794202, https://doi.org/10.5281/zenodo.12794202). All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.
Code availability
The analysis code related to SLAM-ITseq, scRNA-seq, CUT-Tag and ATAC-seq have been uploaded to GitHub (https://github.com/lintian0616/KC_plasticity, https://doi.org/10.5281/zenodo.13894909).
References
Brodt, P. Role of the Microenvironment in Liver Metastasis: From Pre- to Prometastatic Niches. Clin. Cancer Res. 22, 5971–5982 (2016).
Kennecke, H. et al. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. J. Am. Soc. Clin. Oncol. 28, 3271–3277 (2010).
Tsilimigras, D. I. et al. Liver metastases. Nat. Rev. Dis. Prim. 7, 27 (2021).
Yu, J. et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat. Med. 27, 152–164 (2021).
Keirsse, J. et al. The role of hepatic macrophages in liver metastasis. Cell. Immunol. 330, 202–215 (2018).
Mantovani, A., Allavena, P., Marchesi, F. & Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 21, 799–820 (2022).
Lazarov, T., Juarez-Carreño, S., Cox, N. & Geissmann, F. Physiology and diseases of tissue-resident macrophages. Nature 618, 698–707 (2023).
Park, M. D., Silvin, A., Ginhoux, F. & Merad, M. Macrophages in health and disease. Cell 185, 4259–4279 (2022).
Guilliams, M. & Scott, C. L. Liver macrophages in health and disease. Immunity 55, 1515–1529 (2022).
Germano, G. et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 552, 116–120. (2017).
Yang, Y. et al. Immunocompetent mouse allograft models for development of therapies to target breast cancer metastasis. Oncotarget https://doi.org/10.18632/oncotarget.15695 (2017).
Ruiz de Galarreta, M. et al. β-Catenin Activation Promotes Immune Escape and Resistance to Anti-PD-1 Therapy in Hepatocellular Carcinoma. Cancer Discov. 9, 1124–1141 (2019).
Shiao, S. L., Ganesan, A. P., Rugo, H. S. & Coussens, L. M. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev. 25, 2559–2572 (2011).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Qi, J. et al. Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat. Commun. 13, 1742 (2022).
Wu, Y. et al. Spatiotemporal Immune Landscape of Colorectal Cancer Liver Metastasis at Single-Cell Level. Cancer Discov. 12, 134–153 (2022).
Bill, R. et al. CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science 381, 515–524 (2023).
Holness, C. L. & Simmons, D. L. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood 81, 1607–1613 (1993).
Lee, Y.-S. et al. CX(3)CR1 differentiates F4/80(low) monocytes into pro-inflammatory F4/80(high) macrophages in the liver. Sci. Rep. 8, 15076 (2018).
Yang, C.-Y. et al. CLEC4F is an inducible C-type lectin in F4/80-positive cells and is involved in alpha-galactosylceramide presentation in liver. PLoS One 8, e65070 (2013).
Sakai, M. et al. Liver-Derived Signals Sequentially Reprogram Myeloid Enhancers to Initiate and Maintain Kupffer Cell Identity. Immunity 51, 655–670.e8 (2019).
Iqbal, A. J. et al. Human CD68 promoter GFP transgenic mice allow analysis of monocyte to macrophage differentiation in vivo. Blood 124, e33–e44 (2014).
Miyanishi, M. et al. Identification of Tim4 as a phosphatidylserine receptor. Nature 450, 435–439 (2007).
Dick, S. A. et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 20, 29–39 (2019).
Dick, S. A. et al. Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci. Immunol. 7, eabf7777 (2022).
Morioka, S. et al. Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature 563, 714–718 (2018).
Matsushima, W. et al. SLAM-ITseq: sequencing cell type-specific transcriptomes without cell sorting. Development 145, dev164640 (2018).
Mass, E. et al. Specification of tissue-resident macrophages during organogenesis. Science 353, aaf4238 (2016).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Boring, L. et al. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J. Clin. Invest. 100, 2552–2561 (1997).
Ries, C. H. et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 25, 846–859 (2014).
Hashimoto, D. et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013).
Davies, L. C. et al. Distinct bone marrow-derived and tissue-resident macrophage lineages proliferate at key stages during inflammation. Nat. Commun. 4, 1886 (2013).
Vanneste, D. et al. MafB-restricted local monocyte proliferation precedes lung interstitial macrophage differentiation. Nat. Immunol. 24, 827–840 (2023).
He, L. et al. Proliferation tracing reveals regional hepatocyte generation in liver homeostasis and repair. Science 371, eabc4346 (2021).
van Rooijen, N., Sanders, A. & van den Berg, T. K. Apoptosis of macrophages induced by liposome-mediated intracellular delivery of clodronate and propamidine. J. Immunol. Methods 193, 93–99 (1996).
Walzer, T. et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc. Natl Acad. Sci. USA. 104, 3384–3389 (2007).
Ramachandran, P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019).
van der Bij, G. J., Oosterling, S. J., Meijer, S., Beelen, R. H. J. & van Egmond, M. Therapeutic potential of Kupffer cells in prevention of liver metastases outgrowth. Immunobiology 210, 259–265 (2005).
Li, J. et al. The ligation between ERMAP, galectin-9 and dectin-2 promotes Kupffer cell phagocytosis and antitumor immunity. Nat. Immunol. 24, 1813–1824 (2023).
Deng, Z. et al. The nuclear factor ID3 endows macrophages with a potent anti-tumour activity. Nature 626, 864–873 (2024).
Thomas, S. K. et al. Kupffer cells prevent pancreatic ductal adenocarcinoma metastasis to the liver in mice. Nat. Commun. 14, 6330 (2023).
Saito, M. et al. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat. Biotechnol. 19, 746–750 (2001).
Troutman, T. D., Kofman, E. & Glass, C. K. Exploiting dynamic enhancer landscapes to decode macrophage and microglia phenotypes in health and disease. Mol. Cell 81, 3888–3903 (2021).
Spangenberg, E. et al. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun. 10, 3758 (2019).
Yang, L. et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature 583, 133–138 (2020).
Xia, X. et al. Neutrophil extracellular traps promote metastasis in gastric cancer patients with postoperative abdominal infectious complications. Nat. Commun. 13, 1017 (2022).
Tu, M. M. et al. Inhibition of the CCL2 receptor, CCR2, enhances tumor response to immune checkpoint therapy. Commun. Biol. 3, 720 (2020).
Bassez, A. et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 27, 820–832 (2021).
Giles, J. R., Globig, A.-M., Kaech, S. M. & Wherry, E. J. CD8(+) T cells in the cancer-immunity cycle. Immunity 56, 2231–2253 (2023).
Oliveira, G. & Wu, C. J. Dynamics and specificities of T cells in cancer immunotherapy. Nat. Rev. Cancer 23, 295–316 (2023).
Daemen, S. et al. Dynamic Shifts in the Composition of Resident and Recruited Macrophages Influence Tissue Remodeling in NASH. Cell Rep. 34, 108626 (2021).
Tran, S. et al. Impaired Kupffer Cell Self-Renewal Alters the Liver Response to Lipid Overload during Non-alcoholic Steatohepatitis. Immunity 53, 627–640.e5 (2020).
Guilliams, M. & Scott, C. L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Rev. Immunol. 17, 451–460 (2017).
Lefere, S., Degroote, H., Van Vlierberghe, H. & Devisscher, L. Unveiling the depletion of Kupffer cells in experimental hepatocarcinogenesis through liver macrophage subtype-specific markers. J. Hepatol. 71, 631–633 (2019).
Wen, Y., Lambrecht, J., Ju, C. & Tacke, F. Hepatic macrophages in liver homeostasis and diseases-diversity, plasticity and therapeutic opportunities. Cell. Mol. Immunol. 18, 45–56 (2021).
Li, X. et al. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat. Rev. Cancer 21, 541–557 (2021).
Guilliams, M. & Svedberg, F. R. Does tissue imprinting restrict macrophage plasticity? Nat. Immunol. 22, 118–127 (2021).
Bonnardel, J. et al. Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 51, 638–654.e9 (2019).
Sahasrabuddhe, V. & Ghosh, H. S. Cx3Cr1-Cre induction leads to microglial activation and IFN-1 signaling caused by DNA damage in early postnatal brain. Cell Rep. 38, 110252 (2022).
Satpathy, A. T. et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14, 937–948 (2013).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Borcherding, N., Bormann, N. L. & Kraus, G. scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Research 9, 47 (2020).
Acknowledgements
We thank S-C.Zhang for critically reading the manuscript. L.T. is supported by the National Natural Science Foundation (NSFC) of China (Grant No. 82173278), the Science and Technology Projects in Guangzhou (Grant No. 202201011364) and Young Talents Program of SYSUCC (Grant No. YTP-SYSUCC-0045); Y.S. and L.T. are supported by National Key Research and Development Program of China (Grant No. 2021YFE0206300); H.J. and L.T. are supported by the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2023B1515040030); H.H. is supported by Fostering Program for NSFC Young Applicants (Tulip Talent Training Program) of SYSUCC (Grant No. TTP-SYSUCC-202406).
Author information
Authors and Affiliations
Contributions
H.H. and L.T. conceived the idea. H.H., Y.C., and L.T. performed most experiments and wrote the manuscript. X.Z and L.T performed epigenetic profiling. C.Z., K.Y., and L.T. performed bioinformatic analyses. J.Y. built a custom pipeline to process pair-end fastq data for SLAM-ITseq assay. Y.S. collected liver metastasis tissue samples from breast cancer patients. H.J. provided reagents and expertise and participated in data analysis. All co-authors have seen and approved the manuscripts.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Qiming Zhang and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, HY., Chen, YZ., Zhao, C. et al. Alternations in inflammatory macrophage niche drive phenotypic and functional plasticity of Kupffer cells. Nat Commun 15, 9337 (2024). https://doi.org/10.1038/s41467-024-53659-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53659-7
This article is cited by
-
Modeling tumor relapse using proliferation tracing and ablation transgenic mouse
npj Breast Cancer (2025)
-
Advances and emerging trends in the tumor microenvironment of colorectal cancer liver metastasis: a bibliometric analysis
Discover Oncology (2025)