Abstract
Antiviral STANDs (Avs) are bacterial anti-phage proteins evolutionarily related to immune pattern recognition receptors of the NLR family. Type 2 Avs proteins (Avs2) were suggested to recognize the phage large terminase subunit as a signature of phage infection. Here, we show that Avs2 from Klebsiella pneumoniae (KpAvs2) can recognize several different phage proteins as signature for infection. While KpAvs2 recognizes the large terminase subunit of Seuratvirus phages, we find that to protect against Dhillonvirus phages, KpAvs2 recognizes a different phage protein named KpAvs2-stimulating protein 1 (Ksap1). KpAvs2 directly binds Ksap1 to become activated, and phages mutated in Ksap1 escape KpAvs2 defense despite encoding an intact terminase. We further show that KpAvs2 protects against a third group of phages by recognizing another protein, Ksap2. Our results exemplify the evolutionary diversification of molecular pattern recognition in bacterial Avs2, and show that a single pattern recognition receptor evolved to recognize different phage-encoded proteins.
Similar content being viewed by others
Introduction
The ability to detect pathogen invasion is a hallmark of immune systems across all domains of life1,2,3,4,5,6. Pattern recognition receptors, which detect molecular signatures indicative of pathogen infection, are at the front line of the innate immune systems of many organisms1,5,6,7. These receptors can detect pathogen-associated molecular patterns (PAMPs), such as foreign nucleic acids8, conserved proteins1,7, and conserved nonproteinaceous molecules of the pathogen1.
The STAND NTPase superfamily of proteins is a large family of immune pattern recognition receptors present in animals, plants, fungi, archaea and bacteria9,10,11,12. These proteins typically have a tripartite architecture, consisting of a long C-terminal domain that senses the invader PAMP molecule, a central STAND NTPase domain of the NACHT or NB-ARC family, and an N-terminal domain that mediates cell death or an inflammatory response once an invader has been recognized1,13,14. The STAND NTPase superfamily includes animal inflammasomes and plant resistosomes, which are collectively named nucleotide-binding leucine-rich repeat-containing receptors (NLRs) because their C-terminal sensor domains typically comprise leucine-rich repeats9,15.
In bacteria, STAND NTPase proteins were broadly found to protect against phage infection7,10,12,16,17. These proteins were dubbed NLR-like proteins16,17, bacterial NACHT (bNACHT)12, antiviral ATPase/NTPase of the STAND superfamily (AVAST), or antiviral STAND (Avs)7,10. Bacterial STAND NTPases preserve the tripartite structure of proteins in this family, and their N-terminal domains usually function as direct effectors of cell death7,12. Prokaryotic STAND NTPases were detected in 4%-10% of all published bacterial genomes7,12, and are thought to be the evolutionary ancestors of eukaryotic NLRs9,12.
A recent study has characterized a large family of Avs proteins in bacteria that were documented to recognize the large subunit of the phage terminase7, a highly conserved protein essential for phage DNA packaging18. Three terminase-recognizing Avs families were identified (Avs1, Avs2 and Avs3), and a cryo-EM structure of Avs3 from Salmonella enterica complexed with the phage terminase explained the structural basis for Avs3-terminase molecular recognition7. Co-expression of phage large terminase proteins with protein representatives from the Avs1, Avs2 and Avs3 families caused cellular toxicity, indicating that recognition of the terminase by these Avs proteins activates their toxic N-terminal effectors7.
In this study, we examined a defense system from Klebsiella pneumoniae that includes an Avs2-family protein. We show that, once it recognizes phage infection, the Avs2 protein indiscriminately degrades DNA to abort phage infection. Surprisingly, our data show that during infection by phage SECphi18, the K. pneumoniae Avs2 is activated by binding a phage protein of unknown function, here called Ksap1. We demonstrate direct interaction between the C-terminus of Avs2 and Ksap1, and show that phages in which Ksap1 is deleted or mutated become resistant to the Avs2 system of K. pneumoniae. Ksap1 is a protein conserved in the Dhillonvirus phage genus, and our data show that recognition of Ksap1 explains how KpAvs2 protects against multiple phages of this family. In contrast, we found that KpAvs2 protects against phages from other families by recognizing proteins other than Ksap1. In particular, KpAvs2 recognizes the large terminase subunit of phage Bas22 (family Queuovirinae) and an unrelated protein, which we call Ksap2, in the myovirus Bas60. Our findings expand the repertoire of known phage proteins recognized by bacterial NLR-like Avs proteins and underscore how evolutionary diversification allows these proteins to adapt to a range of phage targets.
Results
An Avs2-containing operon protects E. coli against phage
A previous study identified over 2000 homologs of Avs2 in various bacterial genomes7. We became interested in a set of homologs typified by Avs2 from Klebsiella pneumoniae S_15PV (KpAvs2) that seems to be embedded in a three-gene operon, as opposed to typical Avs2 proteins that usually function as a single protein (Fig. 1A and Supplementary Data S1). This operon was frequently encoded in proximity to other known defense systems in microbial genomes, supporting the notion that its role is to defend against phages (Fig. 1B). As expected, HHpred19,20 analysis showed that KpAvs2 is divided into three distinct domains: an N-terminal endonuclease domain of the Mrr restriction endonuclease family, a central STAND NTPase domain, and a long C-terminal domain which displayed sequence homology to the C-terminal domains of previously characterized Avs2 proteins7. The two associated genes encoded a protein with a radical S-adenosylmethionine (SAM) domain, and a protein of unknown function with an HHpred19,20 hit to the pfam protein family PF19902 (domain of unknown function DUF6375) (Fig. 1A). We denote these proteins Avs-associated protein 1 (Avap1) and Avs-associated protein 2 (Avap2). About 5% of Avs2 homologs were found associated with Avap2, with the majority of these also associated with Avap1 (Supplementary Fig. S1).
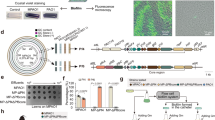
A Domain organization of KpAvs2 operon from Klebsiella pneumoniae S_15PV. Mutations used in this study are indicated below. B Gene neighborhoods of selected KpAvs2 homologs. Purple, homologs of the KpAvs2 operon; orange, known defense genes. Presented genomes are Klebsiella pneumoniae S_15PV (IMG50 scaffold identifier ID: 2701097382), Nitrospirae bacterium JdFR-85 (2728441048), Syntrophobacterales bacterium Delta_02 (2751221707), Achromobacter sp. 2789STDY5608624 (2660299158). RM: Restriction-modification, BREX: Bacteriophage Exclusion, TA: toxin-antitoxin. C Fold defense, calculated as the ratio between the efficiency of plating of phages infecting control E. coli cells and cells expressing the KpAvs2 operon at 25 °C for phages SECphi18 and SECphi6, or 37°C for λvir and T2. Bar graph represents average of 5-9 independent replicates, with individual data points overlaid. D Efficiency of plating of SECphi18 phages infecting E. coli cells expressing the KpAvs2 operon with the indicated mutations at 25 °C. Data represent plaque-forming units (PFU) per mL. Bar graph represents average of 4 independent replicates for each mutant, and 11 replicates for both negative and positive control strains, with individual data points overlaid. Negative control is a strain expressing GFP. E Liquid culture growth of E. coli cells expressing KpAvs2 operon WT, or mutated in the nuclease effector domain, or control cells expressing GFP. Cells were infected by phage SECphi18 at 25 °C. Each curve represents the average of three replicates, error bars represent standard deviation. MOI: multiplicity of infection. F Titer of SECphi18 phage propagated on E. coli cells expressing either the KpAvs2 operon or GFP as a control. SECphi18 titer, shown in PFU/mL, was measured after two or four hours from initial infection, corresponding roughly to one and two cycles of infection, divided by the original phage titer. Infection was performed at MOI = 0.1. Bar graph represents average of 4 replicates, with individual data points overlaid. G Ethidium bromide-stained agarose gel of plasmids extracted from infected cells expressing KpAvs2 operon, either WT or carrying a mutation in the nuclease domain. DNA was extracted at indicated times post-infection (min p.i.) by phage SECphi18 at MOI = 10 and 25 °C. As a control, plasmids were extracted from E. coli cultures not challenged by phages.
To test whether the KpAvs2-containing operon is able to protect against phage infection, we heterologously expressed it in Escherichia coli using an arabinose-inducible promoter. Plaque assay experiments with a panel of 12 phages showed that cells expressing this operon became resistant to multiple phages, with the strongest protection observed against SECphi18, a phage from the Dhillonvirus genus (Fig. 1C and Supplementary Fig.S2). A point mutation predicted to disrupt the active site of the N-terminal endonuclease domain of KpAvs2, K71A, abolished defense, confirming that the nuclease activity is essential for defense as expected from previous studies on Avs proteins7,12 (Fig. 1D and Supplementary Fig. S3AB). Similarly, a point mutation disrupting the STAND NTPase active site, K337A, and a deletion of the C-terminal PAMP recognition domain, also abolished defense (Fig. 1D and Supplementary Fig. S3AB).
Expression of KpAvs2 alone conferred only weak defense against phages, suggesting that one or both of the associated genes are necessary for the full defense capacity; and neither Avap1 nor Avap2 conferred defense when expressed alone (Fig. 1D and Supplementary Fig. S3C). Expressing the three-gene operon in which avap1 was inactivated by a premature stop codon protected the culture from SECphi18 infection as efficiently as the WT three-gene operon, suggesting that Avap1 is dispensable for defense against SECphi18 (Fig. 1D and Supplementary Fig. S3AB). In contrast, inactivation of Avap1 did reduce defense against phage T2, suggesting that Avap1 is required for defense against some, but not all, phages (Supplementary Fig. S4). Attempts to delete Avap2 from the operon were unsuccessful, implying that perturbing this gene in the presence of Avap1 and KpAvs2 might lead to cellular toxicity.
Given that KpAvs2 protected against SECphi18 only in the presence of Avap2 (Fig. 1D), we investigated the possibility of an interaction between these two proteins. AlphaFold-Multimer21 analysis predicted high-confidence interactions between Avap2 and an alpha-helical bundle extending between the nuclease domain and the STAND NTPase domain of KpAvs2 (Supplementary Fig. S5AB). Co-immunoprecipitation experiments confirmed physical binding between Avap2 and KpAvs2 (Supplementary Fig. S5CD). Notably, Avap2 is not predicted to interact with the distal part of the C-terminal domain within KpAvs2 that likely comprises the phage recognition pocket (see below) (Supplementary Fig. S5AB). In agreement with the AlphaFold-Multimer prediction, Avap2 retained its ability to bind KpAvs2 even when this C-terminal domain was deleted from KpAvs2 (Supplementary Fig. S5CD). These results show that KpAvs2 binds the accessory protein Avap2, suggesting that this binding may contribute to the defensive function.
Infection experiments in liquid culture showed that the KpAvs2 operon protected the culture when cells were infected by phage SECphi18 at a low multiplicity of infection (MOI = 0.1). Infection at a high MOI resulted in growth arrest of cells expressing the KpAvs2 operon, occurring earlier than the time in which control cells lysed following infection by SECphi18 (Fig. 1E). Consistently, plaque-forming units (PFU) analysis showed that phages were unable to replicate on KpAvs2-expressing cells (Fig. 1F), confirming that KpAvs2 protects via abortive infection.
Previous studies with Avs3 and Avs4 have shown that these proteins are phage-activated DNA endonucleases that non-specifically degrade DNA upon phage recognition7. As KpAvs2 encodes an N-terminal endonuclease domain, we hypothesized that this protein, too, would degrade DNA in response to phage infection. In agreement with this hypothesis, analysis of DNA extracted from cells infected by phage SECphi18 showed a distinct DNA smear pattern indicative of non-specific DNA degradation in cells expressing KpAvs2 (Fig. 1G). Collectively, our findings identify an Avs2 variant that necessitates an accessory protein and non-specifically degrades DNA as a defense mechanism to prevent phage replication.
KpAvs2 is activated in vivo by a small phage protein
Avs2 proteins were previously shown to become toxic when co-expressed with phage large terminase subunit proteins, and it was therefore hypothesized that the large terminase is the PAMP sensed by Avs27. To test if the toxic effects of KpAvs2 can be activated by the phage terminase, we attempted to transform plasmids encoding the large terminase subunit of phage SECphi18 into cells that encode KpAvs2. We observed a substantial reduction in transformation efficiency of a plasmid encoding the SECphi18 terminase in the presence of WT KpAvs2 compared to a KpAvs2 mutant lacking the C-terminal domain (Fig. 2A). This toxicity was abolished if the nuclease domain of KpAvs2 was mutated, or if KpAvs2 was expressed alone, suggesting the full, functioning operon is necessary to observe toxicity (Fig. 2B). AlphaFold-Multimer21 predicted an interaction between the ATPase domain of the SECphi18 large terminase subunit and KpAvs2 C-terminal phage recognition domain (Fig. 2C and Supplementary Fig. S6A). This predicted interaction was verified in co-immunoprecipitation assays, showing that the SECphi18 large terminase subunit co-purifies with KpAvs2 when both proteins are mixed in vitro (Fig. 2D). The interaction between the two proteins was abolished when the C-terminus of KpAvs2 was deleted, suggesting that this domain is responsible for binding the large terminase subunit (Fig. 2D). These findings led us to initially think that the large terminase subunit may serve as the activator of KpAvs2 during infection by phage SECphi18.
A Transformation efficiency of the gene encoding the large terminase subunit of SECphi18 or SECphi6. Data represent the ratio of transformants obtained using bacteria encoding KpAvs2 operon divided by transformants obtained with the KpAvs2Δ1066-1352 deletion. Bar graph represents average of 8 replicates, with individual data points overlaid. Control indicates transformation with a plasmid encoding RFP. B Transformation efficiency of plasmids carrying either RFP or SECphi18 terminase. Data represent the ratio of terminase transformants divided by RFP transformants for the indicated strain. For each box, the central line represents the median, the edges correspond to the first (Q1) and third (Q3) quartiles, indicating the interquartile range (IQR). The whiskers extend to the smallest and largest values within 3 times the IQR from the quartiles. Data points outside this range represent potential outliers. Data represent 6 replicates, with individual datapoints overlaid. The control strain encoded GFP instead of the KpAvs2 system. C AlphaFold-Multimer21 predicted interactions between the large terminase of SECphi18 (grey) and the C-terminal domain of KpAvs2 (brown). Model confidence score: 0.83. D Co-immunoprecipitation. α-HA beads were used to immunoprecipitate HA-tagged KpAvs2 or KpAvs2Δ1066-1352, and the interacting 3xFLAG-tagged SECphi18 terminase was detected by western blot against FLAG tag. A western blot against HA tag is shown as a control for the efficiency of pulldown (lower panel). Representative of two replicates. E Schematic of the protein pulldown experiment. Cultures expressing HA-tagged KpAvs2 or KpAvs2Δ1066-1352 were infected by SECphi18 at MOI of 5. At 30 min post infection, anti-HA antibodies were used to purify KpAvs2 and its interactants from cell lysates. Mass spectrometry was used to identify proteins that were enriched when WT KpAvs2 was used as bait as compared to the mutated KpAvs2. F Mass spectrometry analysis of SECphi18 large terminase subunit and Ksap1 pulled down with KpAvs2 during infection. Data presented as the ratio between protein abundance in the WT KpAvs2 sample and the KpAvs2Δ1066-1352 sample. Protein abundance was normalized based on bait abundance for each sample. Average of 3 replicates, individual data points overlaid. A list of all identified proteins in this assay is in Supplementary Data S2.
If KpAvs2 indeed recognizes the phage large terminase subunit as a trigger for infection, one would expect KpAvs2 to bind this phage protein in vivo. To test whether this binding occurs during infection, we infected cells expressing an HA-tagged KpAvs2 with phage SECphi18, and immunoprecipitated the tagged KpAvs2 together with proteins bound to it. Surprisingly, mass spectrometry analysis of proteins that were pulled down together with KpAvs2 did not show enrichment for the phage large terminase subunit (Fig. 2E, F, Supplementary Fig. S7A and Supplementary Data S2). Rather, another phage protein, which we denote here KpAvs2-stimulating protein 1 (Ksap1), was 60-fold enriched in the KpAvs2 pulled-down sample as compared to similar samples where the KpAvs2 C-terminal recognition domain was deleted (Fig. 2F). These results implied that, counter to our original hypothesis, a phage protein other than the large terminase subunit binds KpAvs2 during infection.
Ksap1 is a 74 amino-acid long protein of unknown function that resides between a methyltransferase- and a phosphatase-encoding genes in the SECphi18 genome (Fig. 3A). Structural modeling with AlphaFold-Multimer21 showed high-scoring predicted interactions between Ksap1 and the C-terminal domain of KpAvs2, suggesting KpAvs2 can directly bind to Ksap1 (Fig. 3B and Supplementary Fig. S6B). We confirmed this prediction by co-immunoprecipitation assays, showing that a FLAG-tagged Ksap1 co-purified with an HA-tagged KpAvs2 when both proteins were mixed in vitro. In agreement with the structure predictions, the physical interactions observed in the co-immunoprecipitation assays were lost when the C-terminal phage recognition domain was deleted from KpAvs2 (Fig. 3C). These experiments demonstrated physical binding between Ksap1 and KpAvs2 WT, confirming our observations from the pulldown assay during infection (Fig. 3B).
A Genetic organization of SECphi18 genes surrounding ksap1. Mutations identified in escaper phages are shown. Δ1 and Δ2 represent deletions of part of the locus, L47R and T50P represent missense mutations at the indicated amino acids. B AlphaFold-Multimer21 predicted interactions between Ksap1 (blue) and the C-terminal domain of KpAvs2 (brown). Model confidence score: 0.84. C Co-immunoprecipitation. α-HA beads were used to immunoprecipitate HA-tagged KpAvs2 or KpAvs2Δ1066-1352, and the interacting 3xFLAG-tagged Ksap1 was detected by western blot against FLAG tag. A western blot against HA tag is shown as a control for the efficiency of pulldown (lower panel). Representative of two replicates. D Transformation efficiency of ksap1 from SECphi18 or SECphi6. Data represent the ratio of transformants obtained using bacteria expressing KpAvs2 operon divided by transformants obtained with the KpAvs2Δ1066-1352 deletion. Bar graph represents average of 4 replicates, with individual data points overlaid. E Growth curves of E. coli cells expressing either KpAvs2 operon or GFP (Control). Additionally, cells co-expressed either Ksap1 (+) or RFP as a control (-). Data represent the average of 6 replicates, error bars represent standard deviation. F Fold defense, calculated as the ratio of the efficiency of plating of SECphi18 phages on E. coli control cells that express GFP and cells expressing the KpAvs2 system. Infection was performed at 25 °C. Data presented for WT or mutated SECphi18 phages. Bar graph represents the average of 3 independent replicates, with individual data points overlaid. G SECphi18 WT and mutant (Δ2) phages were mixed to a 1:1 ratio and competed for amplification on control cells (Ctrl) or cells expressing system. Shown are the results of a PCR amplification of the ksap1 region in the phage population before competition (0) and after 1, 2 or 3 days of competition. The upper band corresponds to a DNA fragment of WT size, the lower band corresponds to Δksap1 size, as a proxy for the corresponding phage abundance. A DNA ladder in base pair (bp) is presented on the left. H Growth curves of E. coli cells co-expressing the KpAvs2 operon and Ksap1 variants. Presented data are as in panel E.
To test whether KpAvs2 becomes toxic in the presence of Ksap1, we attempted to transform plasmids carrying ksap1 into cells expressing the KpAvs2 operon. This transformation was substantially less efficient than the transformation of a control plasmid (Fig. 3D), and transformants frequently carried suppressor mutations, suggesting toxicity. Nevertheless, we managed to obtain a non-mutated transformant for further analysis. Expression of the KpAvs2 operon in the presence of Ksap1 resulted in cellular toxicity, which was abolished when KpAvs2 was mutated in its nuclease domain, confirming that Ksap1 activates KpAvs2 (Fig. 3E).
Multiple recent studies have shown that examining phage mutants that escape specific defense systems can generate valuable insights into the mechanism of defense activation, because escaper phages can evade bacterial immunity by mutations in the genes that activate the bacterial defense system22,23,24,25,26. We propagated SECphi18 phages on E. coli expressing the KpAvs2 operon, and isolated six phage mutants that were able to escape defense. Notably, all six escaper phages harbored mutations in Ksap1 (Supplementary Data S3). The mutations included partial deletion of ksap1 together with part of the upstream gene, or single point mutations altering leucine 47 or threonine 50 of Ksap1 to arginine and proline, respectively (Fig. 3A and Supplementary Data S3).
We confirmed that bacteria expressing the KpAvs2 operon failed to defend against escaper phages carrying mutations in ksap1 (Fig. 3F). Moreover, competition of a mix of SECphi18 WT and escaper phages propagated on KpAvs2 operon-expressing cells led to the disappearance of the WT phage population, confirming the importance of Ksap1 for KpAvs2-mediated defense (Fig. 3G). Co-expression of KpAvs2 with Ksap1L47R and Ksap1T50P did not cause growth arrest, suggesting that the Ksap1 variants in the mutated phages do not activate KpAvs2 and explaining why these phages escaped defense (Fig. 3H and Supplementary Fig. S8A). Notably, while one of the six escaper mutants carried a missense mutation in the gene encoding the large terminase in addition to a frameshift mutation in ksap1, the other five escaper phages were not mutated in the terminase, showing that phages could escape the KpAvs2 system defense even when expressing a wild-type terminase.
To test if KpAvs2 could bind the phage large terminase subunit during infection when Ksap1 was missing, we infected cells expressing HA-tagged KpAvs2 with a SECphi18 escaper phage deleted in ksap1, immunoprecipitated KpAvs2 during infection, and analyzed co-precipitants by mass spectrometry. We found that the terminase large subunit was mildly enriched in these pulldowns (5-fold enrichment), much lower than the 60-fold enrichment previously observed for Ksap1 (Supplementary Fig. S7B and Supplementary Data S4). These results further support the hypothesis that Ksap1, rather than the terminase, is the component sensed by KpAvs2 as a signature of SECphi18 infection.
Natural variants of Ksap1 escape KpAvs2 defense
Phage SECphi18 is taxonomically closely related to phage SECphi627. Both phages belong to the Dhillonvirus genus, and their ~45 kb genomes show 92% sequence similarity over 96% of the sequence. Despite the similarity between the two phages, the KpAvs2 operon shows no defense against phage SECphi6 while strongly defending against SECphi18 (Fig. 1C). SECphi6 harbors a homolog of Ksap1 in a locus syntenic to the Ksap1 locus in phage SECphi18, and the SECphi6 Ksap1 homolog differs from the SECphi18 Ksap1 homolog by only 14 residues (Fig. 4A). We hypothesized that the altered sequence of the SECphi6 Ksap1 protein allows the phage to escape detection by KpAvs2. In support of this hypothesis, co-expression of KpAvs2 with Ksap1 from SECphi6 did not result in cellular toxicity (Figs. 3D and 4B), suggesting that Ksap1 from this phage does not activate the KpAvs2 system.
A Amino acid sequence alignment of Ksap1 proteins from SECphi18 and SECphi6 phages. Blue color indicates the amino acids that were mutated in panel B. B Growth curves of E. coli cells co-expressing both the KpAvs2 operon and different variants of Ksap1. As controls, a plasmid expressing RFP instead of Ksap1 was used. Data represent the average of 6 replicates, error bars represent standard deviation. C Interaction predicted between indicated residues of Ksap1 from SECphi18 (in blue) and KpAvs2 (brown). Interactions were predicted by AlphaFold-Multimer21 and RING49. Shown is a close-up of the structure presented in Fig. 3B. D Transformation efficiency of the gene encoding Ksap1 from Bas16 or Bas17, or RFP as a negative control. Data represent the ratio of transformants obtained using bacteria expressing the WT KpAvs2 operon divided by transformants obtained with control cells. Bar graph represents average of 6 replicates, with individual data points overlaid. E Co-immunoprecipitation of 3xFLAG-tagged Ksap1 from Bas16, Bas17 and SECphi6 phages and the KpAvs2 system. As a negative control, we used a strain expressing RFP instead of the 3xFLAG-tagged Ksap1. α-FLAG beads were used to immunoprecipitate 3xFLAG-tagged Ksap1, and co-immunoprecipitated proteins were visualized by Coomassie staining. A ladder is presented on the left (kDa). Representative of two replicates.
We used AlphaFold-Multimer21 to predict which amino acids are in direct interaction between Ksap1 and the C-terminal domain of KpAvs2. Among the 14 residues that differed between the SECphi18 Ksap1 and its homolog in SECphi6, three were predicted to make a strong interaction with KpAvs2. These were K30, generating an ionic bond; L39, generating van der Waals interactions with three separate residues in KpAvs2; and F60, generating a π-π stacking interaction (Fig. 4C and Supplementary Data S5). These interactions were not present when we modeled the interactions of KpAvs2 with the SECphi6 version of Ksap1 (Supplementary Data S6), in which these residues are altered to valine, methionine and valine, respectively. We therefore hypothesized that the alteration of these three residues may explain why KpAvs2 does not protect from SECphi6. To examine this hypothesis, we replaced the three amino acids in the SECphi18 protein by their respective SECphi6 variants, and tested whether Ksap1K30V L39M F60V could activate the toxicity of KpAvs2. Co-expression of Ksap1K30V L39M F60V with the KpAvs2 operon was not toxic (Fig. 4B and Supplementary Fig. S8B), showing that alteration of these residues is sufficient for escape from KpAvs2 activation.
Homologs of ksap1 are found in almost all genomes of Dhillonvirus phages deposited in the NCBI database (Supplementary Data S7). To test if Ksap1 is the activator of the KpAvs2 defense system when defending against other Dhillonvirus phages, we tested whether KpAvs2 could provide defense against five other representatives of the Dhillonvirus genus from the BASEL collection28. The KpAvs2 operon provided defense against three of these phages (Bas14, Bas17 and Bas18), but not against the other two (Bas15 and Bas16) (Supplementary Fig. S9). To further examine whether the observed differences are attributed to Ksap1, we selected Ksap1 from one phage for which defense was observed, Bas17 (phage KarlBarth) and from one for which no defense was observed, Bas16 (phage GeorgBuechner) (Supplementary Fig. S10). Attempting to transform ksap1 homologs from Bas16 and Bas17 into cells expressing the KpAvs2 operon showed a substantial reduction in transformation efficiency for the Bas17 ksap1, but no reduction when using Bas16 ksap1, consistent with the defense phenotype (Fig. 4D). In agreement with these results, physical interactions were demonstrated in co-immunoprecipitation assays between Bas17 Ksap1 and KpAvs2, but no interactions were observed between Bas16 Ksap1 and KpAvs2 (Fig. 4E). A truncation in the C-terminus of the Bas16 Ksap1 can explain these observations (Supplementary Fig. S10B). The Ksap1 of SECphi6 also showed weak interactions with KpAvs2, in agreement with the absence of defense against that phage (Fig. 4E). These results provide compelling evidence that KpAvs2 protection against Dhillonvirus phages relies on the recognition of Ksap1 as the primary PAMP.
KpAvs2 recognizes diverse phage proteins to activate defense
As we could not find homologs of Ksap1 in phages outside the Dhillonvirus genus, we wondered how KpAvs2 protects against other phages. To understand the extent of KpAvs2 defense, we used 48 non-Dhillonvirus phages from the BASEL phage collection28, and found that, in addition to Dhillonvirus, the KpAvs2 operon provides defense against phages from the Queuovirinae (Bas19-Bas25), Vequintavirinae (Bas48-Bas49), Stephanstirmvirinae (Bas60-62) and Studiervirinae (Bas64-68) subfamilies (Supplementary Fig. S9).
We selected one representative phage from each subfamily for which widespread defense was observed, Bas22 (Queuovirinae), Bas48 (Vequintavirinae), Bas60 (Stephanstirmvirinae) and Bas64 (Studiervirinae), and used AlphaFold-Multimer21 to predict the interactions between the C-terminal domain of KpAvs2 and all the proteins encoded by each of the selected phages (Supplementary Fig. S11). In phage Bas60, AlphaFold-Multimer21 predicted high-confidence interactions between the C-terminal domain of KpAvs2 and a small protein of unknown function, Bas60_220 (Fig. 5A and Supplementary Fig. S11A). Co-immunoprecipitation using tagged KpAvs2 as bait during infection by phage Bas60 showed that Bas60_220 was the most enriched phage protein bound to KpAvs2 (Supplementary Data S8, Supplementary Fig. S7C and Fig. 5B). Co-immunoprecipitation assays in vitro confirmed that Bas60_220 binds to KpAvs2 (Fig. 5C), and co-expression of Bas60_220 with KpAvs2 elicited KpAvs2-dependent toxicity (Fig. 5D). These results suggest that Bas60_220, which we rename KpAvs2-stimulating protein 2 (Ksap2), is the PAMP used by KpAvs2 to recognize Bas60 infection. Ksap2 is a small protein of unknown function with no sequence or structural homology to Ksap1, and HHpred19,20 and Foldseek29 searches did not find homology between Ksap2 and proteins of known function. A structural comparison of Ksap2 with the PDB database using DALI30 revealed a weak structural homology with the ATPase domain of the large terminase subunit of Geobacillus stearothermophilus bacteriophage D6E (PDB: 5OE9; DALI Z-score: 6.1). However, no significant homology was detected between Ksap2 and large terminase subunit proteins from other phages. A Blast search in the NCBI database retrieved 54 homologs, and showed that most of them are found in genomes of phages belonging to the Stephanstirmvirinae subfamily (Supplementary Data S9).
A AlphaFold-Multimer21 predicted interactions between Ksap2 (Bas60_220, in blue) and the C-terminal domain of KpAvs2 (brown). Model confidence score: 0.90. B Mass spectrometry analysis of the Bas60 large terminase subunit and Ksap2 pulled down with KpAvs2 during infection. Data presented as the ratio between protein abundance in the WT KpAvs2 sample and the KpAvs2Δ1066-1352 sample. Protein abundance was normalized based on bait abundance for each sample. Average of 3 replicates, individual data points overlaid. A list of all proteins identified by mass spectrometry in this assay is in Supplementary Data S8. C Co-immunoprecipitation of HA-tagged KpAvs2 and 3xFLAG-tagged Ksap2. α-HA beads were used to immunoprecipitate HA-tagged KpAvs2 or KpAvs2Δ1066-1352, and the interacting protein was detected by western blot against FLAG tag. A western blot against HA tag was performed on the same blot to control for efficiency of pulldown (lower panel). Representative of two replicates. D Growth curves of E. coli cells co-expressing both the KpAvs2 operon and Ksap2. Ksap2 (+) or RFP (-) control were co-expressed either the KpAvs2 operon, KpAvs2 operon mutated in the nuclease domain of KpAvs2 (KpAvs2K71A) or GFP (Control). Data represent the average of 6 replicates, error bars represent standard deviation. E AlphaFold-Multimer21 predicted interactions between Bas22 large terminase subunit (in grey) and the C-terminal domain of KpAvs2 (brown). Model confidence score: 0.72. F Mass spectrometry analysis of Bas22 large terminase subunit pulled down with KpAvs2 during infection. Data is presented as in panel B. A list of all identified proteins is in Supplementary Data S10. G Co-immunoprecipitation of HA-tagged KpAvs2 and 3xFLAG-tagged Bas22 large terminase subunit. Data is presented as in panel C. H Transformation efficiency of plasmids encoding the Bas22 large terminase subunit or RFP as control. Data represent the ratio of transformants obtained using bacteria expressing the WT KpAvs2 operon divided by transformants obtained with the KpAvs2Δ1066-1352 deletion. Bar graph represents average of 5 replicates, with individual data points overlaid.
We next examined AlphaFold-Multimer21 predictions for the other phages in the set. For Bas22, Bas48 and Bas64, the protein with the highest average scoring interactions with KpAvs2 was the large terminase subunit, with no other phage protein predicted to interact with KpAvs2 (Supplementary Fig. S11BCD and Fig. 5E). Mass spectrometry analysis of phage proteins pulled down with tagged KpAvs2 during Bas22 infection in vivo showed that the large terminase subunit was the most enriched phage protein bound to KpAvs2 (Supplementary Data S10, Supplementary Fig. S7D and Fig. 5F). We could recapitulate this physical interaction by co-immunoprecipitation in vitro (Fig. 5G), confirming that KpAvs2 is able to bind Bas22 terminase. Moreover, transformation of a plasmid carrying Bas22 terminase was extremely toxic in the presence of KpAvs2, showing that the terminase alone could activate KpAvs2 defense (Fig. 5H). Taken together, these results suggest that KpAvs2 is activated during Bas22 infection by binding to the large terminase subunit.
Our data suggest that KpAvs2 is capable of binding, and being activated by, multiple different phage proteins. AlphaFold-Multimer analyses predict that all three phage proteins interact with the same pocket in the C-terminus of KpAvs2 (Figs. 3B, 5A and E and Supplementary Fig. S12). Despite the structural unrelatedness between the three proteins, all three possess a central beta-sheet domain oriented similarly within the predicted binding pocket of KpAvs2 (Supplementary Fig. S12). Our results demonstrate a remarkable ability for a single bacterial defense protein to recognize diverse phage proteins as a signature of infection.
Discussion
In this study, we characterized KpAvs2, a type 2 Avs system, which, akin to previously described type 2 Avs proteins7, operates through the recognition of a phage protein via physical binding. Recognition of the phage protein activates an N-terminal effector in KpAvs2, which non-specifically degrades cellular DNA. A surprising discovery that emerged from our studies is that KpAvs2 is capable of recognizing multiple unrelated phage proteins as a signature for phage infection. Our data show that the ability of KpAvs2 to defend against a broad range of phages is, at least in part, a consequence of its capacity to recognize diverse phage proteins rather than its ability to recognize a conserved structure shared by many phages.
In the innate immune system of humans and animals, some pattern recognition receptors are known to respond to more than one PAMP1,31. For example, upon forming heterodimers with TLR1 and TLR6, the pattern recognition receptor TLR2 can recognize multiple chemically distinct peptidoglycans, as well as lipoproteins and lipoteichoic acid31,32,33. NLRP3, another pattern recognition receptor, has been shown to be activated by a wide variety of signals, including viral RNA, ATP or bacterial toxins, although it is unclear which of these molecules activates NLRP3 directly and which causes indirect activation34,35. Our findings suggest that recognition of multiple ligands is a trait shared between pattern recognition receptors of eukaryotes and prokaryotes. This trait can explain how a limited set of immune receptors can defend against a wider variety of pathogens.
Our data confirm that the KpAvs2 operon becomes toxic when co-expressed with the large terminase subunit of different phages, as reported7, including SECphi18. Such results were previously interpreted as if the PAMP naturally recognized by Avs2 is always the large terminase protein of the phage. However, our data surprisingly show that the KpAvs2 operon recognizes different proteins when protecting against different phages, although all these phages possess a terminase. In particular, we show that while SECphi18 large terminase subunit could bind KpAvs2 in vitro and activate toxicity in co-expression assays, a different protein was the real activator of KpAvs2 in this phage. We found that KpAvs2 binds Ksap1 during infection, and that this protein activates KpAvs2 defense. In support of these observations, the KpAvs2 system did not protect against phages mutated in Ksap1, although the large terminase subunit in these phages was intact. Moreover, KpAvs2 did not protect against SECphi6 at all, although the SECphi6 terminase seemed to be toxic in the presence of KpAvs2 based on our transformation efficiency assays (Figs. 1C and 2A). These data collectively suggest that KpAvs2 recognizes Ksap1 as its PAMP, and not the phage terminase, when protecting against SECphi18.
It is yet unclear what prevents KpAvs2 from recognizing the SECphi18 large terminase subunit during infection, considering that, at least in vitro, KpAvs2 is bound and activated by the terminase large subunit of this phage. In the absence of Ksap1, KpAvs2 was able to pull down the terminase during infection, but to a much lower levels than Ksap1, and this binding was not sufficient to achieve robust defense, suggesting that there might be a factor that prevents proper terminase recognition during infection. The SECphi18 terminase might have lower affinity for KpAvs2, or interact with a third partner during an infection such that interaction with KpAvs2 is prevented.
Both Ksap1 and Ksap2, the SECphi18 and Bas60-encoded activators of KpAvs2, respectively, are small proteins of unknown function that are restricted to a narrow phage taxon. Ksap1 homologs were identified only in phages from the genus Dhillonvirus, where this protein is conserved in almost all sequenced phages of this genus (Supplementary Fig. S13A and Supplementary Data S7). Similarly, most Ksap2 homologs were identified in genomes of Stephanstirmvirinae phages (Supplementary Data S9), although not all Stephanstirmvirinae encoded a homolog of ksap2. Sequence-based homology searches did not find significant homology of Ksap1 and Ksap2 to any protein of known function, and structure-based searches using Foldseek29 did not retrieve hits to any protein with an experimentally determined structure. Foldseek search against a database of AlphaFold236-generated protein structures (AFDB5037) did retrieve several marginal hits for Ksap1 to proteins with a predicted response regulator domain, a protein domain that can be activated by phosphorylation and is typical to two-component signaling systems in bacteria38 (Supplementary Fig. S13BC). However, a conserved aspartate residue that is essential for phosphorylation of response regulator domains is missing from Ksap1, suggesting that Ksap1 may not function as a response regulator (Supplementary Fig. S13BC)38. The presence of Ksap1 in multiple Dhillonvirus phages suggests that it has a role in the biology of these phages, but since the phages could tolerate large deletions in the ksap1 gene, we were not able to discern its functional role.
Our results show that the large terminase subunit of Bas22 is the protein likely recognized by KpAvs2 as a signature of infection. Based on AlphaFold-Multimer predictions (Supplementary Fig. S11) it is likely that KpAvs2 is also triggered by the large terminase subunit of phages Bas48 and Bas64, although experimental validation of this hypothesis awaits future studies. The large terminase subunit proteins of Bas22, Bas48 and Bas64 share no detectible sequence similarity, but they do share substantial structural homology, suggesting that KpAvs2, as previously described for other Avs2 proteins7, acts as a pattern recognition receptor that recognizes specific structural patterns.
Contrasting with previously characterized type 2 Avs, for which a single protein is sufficient to provide defense, KpAvs2 necessitates at least one additional accessory protein. We found that the accessory protein Avap2 is essential for defense against SECphi18, and that this protein physically interacts with KpAvs2. Analysis of our mass spectrometry data showed that Avap2 was pulled down with KpAvs2 both before and after phage infection (Supplementary Data S2), suggesting that Avap2 binding to KpAvs2 happens in vivo regardless of the presence of the target phage protein. The functional role of Avap2 in KpAvs2 defense is still unclear.
We also found that the other accessory protein, Avap1, is necessary for defense at least against some phages. Avap1 has a predicted radical SAM domain, similar to prokaryotic viperins (pVips) that generate antiviral compounds as a mode of defense27. We were unable to detect the modified nucleotides typical of pVip activity in cells expressing the full KpAvs2 operon during infection, and AlphaFold236 prediction of Avap1 structure does not support the presence of a catalytic site that could accommodate an NTP, the substrate of pVips. Further studies will be necessary to decipher the roles of Avap1 and Avap2 in KpAvs2 defense.
Collectively, our findings underscore the evolutionary plasticity of PAMP recognition in defense systems. Our data show that KpAvs2 can recognize several different PAMPs, and that recognition of multiple PAMPs allows KpAvs2 to provide broad defense against multiple phage families. Moreover, we show that recognition data obtained based on co-expression experiments alone can, sometimes, be insufficient to discern PAMP identity during infection. This work extends our understanding of the diversification of molecular pattern recognition in a large family of immune receptors conserved across the tree of life.
Methods
Strains and growth conditions
E. coli K-12 MG1655 was grown in MMB media (lysogeny broth (LB) supplemented with 0.1 mM MnCl2 and 5 mM MgCl2) at 37°C or 25 °C with 200 rpm shaking or on solid LB 1.5% agar plates. Ampicillin 100 μg/mL or kanamycin 50 μg/mL were added when necessary for plasmid maintenance. Induction was performed with 0.2% arabinose or 25-100 ng/mL anhydrotetracycline (aTc). Glucose 1% was used to repress the arabinose-inducible promoter when needed. All chemicals were obtained from Sigma Aldrich unless stated otherwise.
To propagate phages, wild type E. coli K-12 MG1655 was grown to OD600 ~ 0.4-0.6 and infected with a sample from a single plaque of the phage then incubated until total culture lysis at 37 °C, 200 rpm shaking. The lysate was then propagated again on a bacterial culture in the same way to increase phage titer. The lysate was then centrifuged 15 min at 3900 g and filtered through 0.2 μm filter and kept at 4°C until use. When necessary, phages were diluted in either MMB or phage buffer (50 mM Tris pH 7.4, 100 mM MgCl2, 10 mM NaCl).
A table of all plasmids, strains and phages used in this study can be found in Supplementary Data S11.
Plasmid and strain construction
Target DNA was amplified using KAPA HiFi HotStart ReadyMix (Roche) according to manufacturer instruction or synthesized by Twist Bioscience (Supplementary Data S11). Supplementary Data S12 lists all primers used in this study. All primers were obtained from Sigma Aldrich.
Some plasmids were synthesized and cloned by GenScript Corporation as indicated in Supplementary Data S11. Others were cloned as described below.
For one-fragment DNA cloning, a linear plasmid obtained by PCR was ligated using KLD enzyme mix (NEB) for 5 min at room temperature before transformation into 5-alpha Competent E. coli High Efficiency (NEB) through heat shock. Briefly, competent cells were incubated with DNA for 30 min on ice then 30 s at 42°C before resuspension in MMB media. Cells were left to recover for 1 h at 37°C, and were then plated on selective media.
For assembly of more than one fragment, PCR products were treated with FastDigest DpnI (Thermo Fisher Scientific) restriction enzyme for 30 min at 37°C to remove all remaining circular template plasmid and the enzyme was inactivated for 20 min at 80°C. The fragments were then assembled using NEBuilder HiFi DNA Assembly Master Mix (NEB) at 50°C for 30 min then transformed in 5-alpha Competent E. coli High Efficiency (NEB), as described above.
Colonies were checked by PCR using DreamTaq Green PCR Master Mix (Thermo Fisher Scientific) then Sanger sequenced through the DNA Sequencing unit of the Life Sciences core facilities of Weizmann Institute of Science; alternatively, plasmids were prepared using QIAprep Spin Miniprep Kit (Qiagen) according to manufacturer instructions and sent for whole plasmid sequencing at Plasmidsaurus (www.plasmidsaurus.com). Sequence-verified colonies were then transformed into electrocompetent E. coli MG1655 cells or into the relevant background and kept in 20% glycerol at -80°C.
Plaque assays
Phage infectivity was assessed using the small drops plaque assay as described previously39. Briefly, 300 μL of overnight culture was mixed with 30 mL melted 0.5% agar MMB media containing the relevant inducers and poured into a 10 cm square petri dish, then left to recover for ~1 h at room temperature to allow expression of the induced proteins. 10 μL drops of 10-fold serial dilutions of phages were then dropped over the lawn of bacteria, allowed to dry, and incubated overnight at either 37°C or 25 °C as needed. The following day, the number of plaque-forming units (PFU) were counted to measure phage titer and assess infectivity through the efficiency of plating (EOP). When individual plaques could not be discerned, a faint lysis zone across the drop area was considered to be 10 plaques. Pictures were acquired using BioRad gel doc XR+ and the associated Image Lab 6.1 software.
Liquid culture infection assays
Overnight cultures were diluted 1:100 in MMB with inducer and grown in tubes to OD600 0.3 at 37°C. 180 μL of culture were transferred to Nunc™ MicroWell™ 96-Well, Nunclon Delta-Treated, Flat-Bottom Microplate (Thermo Fisher Scientific) and 20 μL of phages diluted in MMB were added to reach the appropriate MOI. The 96-well plate was then incubated at 37°C or 25 °C with orbital shaking in an Infinite M200 plate reader (TECAN) for 6 h and using the i-control 2.0 software. OD600 was measured every 10 min.
Nuclease activity assays
Overnight cultures were diluted 1:100 in MMB with inducer and grown in tubes to OD600 0.3 at 37°C. Cultures were split into two halves. In one half, phages were added to an MOI of 10. No phages were added to the other half as a control. At 0, 15 and 30 min post-infection at 25 °C, samples were taken and plasmids and short DNA fragments were extracted using QIAprep Spin Miniprep Kit (Qiagen) according to manufacturer instructions. Extracted DNA was concentrated using a Concentrator plus (Eppendorf) with program V-AQ, 60°C, 15 min. The entire DNA extraction was then run on a 0.9% agarose gel containing ethidium bromide in Tris-acetate-EDTA buffer at 140 V for 25 min and imaged with Gel Doc XR+ Imaging System (Bio-Rad) to observe DNA degradation.
Phage burst size measurements
Overnight cultures were diluted 1:100 in MMB with inducer and grown in tubes to OD600 0.3 at 37°C, shaking 200 rpm. SECphi18 phages were added to an MOI of 0.1 and infection proceeded at 25 °C. As a control of initial phage titer, the same volume of phage was added to sterile MMB media and used as the titer of time 0 of infection. After 2 h and 4 h, corresponding to roughly one and two cycles of infection at 25°C for SECphi18, 1 mL of culture was collected, centrifuged for 3 min at 5000 g and filtered through a 0.2 μm filter. The phage titer was calculated as described above by doing a plaque assay infecting wild-type E. coli MG1655.
Growth curve experiments
Bacteria were streaked on LB 1.5% agar plates with the relevant antibiotics. One colony was resuspended in 50 μL MMB, then 5 μL of the resuspended culture were added to 195 μL MMB with antibiotic and appropriate inducer in Nunc™ MicroWell™ 96-Well, Nunclon Delta-Treated, Flat-Bottom Microplate (Thermo Fisher Scientific). Plates were then incubated at 37 °C with orbital shaking in an Infinite M200 plate reader (TECAN) for 8 h and OD600 was measured every 10 min.
Co-immunoprecipitation assays
Proteins were tagged with HA (YPYDVPDYA), 3xFLAG (DYKDHDGDYKDHDIDYKDDDDK) or TwinStrep (WSHPQFEKGGGSGGGSGGSAWSHPQFEK) tags as indicated in Supplementary Data S11, with a short linker between the tag and the protein of interest (SA for TwinStrep tag, SSG for HA and 3xFLAG). When co-expression of both proteins of interest was not possible due to toxicity, each protein was expressed in a separate strain and proteins were then mixed on beads, as described below.
Overnight cultures were diluted 1:100 in 50 mL MMB with antibiotics and grown for ~2 h to OD600 ~ 0.8 at 37°C, shaking 200 rpm. Then, the appropriate inducer was added and cells were incubated at 37°C for an additional hour. Cells were then pelleted and resuspended in 750 μL 1X Tween-Tris-buffered saline (TTBS, Biolab) with cOmplete™ ULTRA Tablets, Mini, EDTA-free, EASYpack Protease Inhibitor Cocktail (Roche) and kept on ice for the rest of the procedure. Cells were transferred to a Lysing Matrix B tube (MP biomedical) and broken down in a FastPrep-24™ Classic bead beating grinder (MP Biomedical) in two rounds of 40 s, 6 m/s shaking. Lysates were then centrifuged for 10 min at 12,000 g and 4°C and the supernatant was added to either 25 μL Pierce™ Anti-HA Magnetic Beads (Thermo Fisher Scientific) or 25 μL Pierce™ Anti-DYKDDDDK Magnetic Agarose (Thermo Fisher Scientific) washed in 1X TTBS buffer. Beads were incubated with the lysate for 1 h at 4°C with overhead shaking. Samples were placed on a magnet and the supernatant was removed. Beads were washed three times in 500 μL 1X cold TTBS buffer.
When necessary, a second lysate containing the putative partner protein was added to the beads. FLAG-tagged Ksap1 was added after the first 1 h incubation step, incubated again for 1 h at 4°C and then washed again three times in 500 μL 1X cold TTBS. Alternatively, FLAG-tagged SECphi18 terminase, Bas22 terminase and Bas60 Ksap2 lysates were mixed with the lysate containing the system on beads in one step and incubated only 1 h in total at 4°C with overhead shaking. Proteins were then eluted by adding 25 μL 4X Bolt™ LDS Sample Buffer (Thermo Fisher Scientific) and heating for 5 min at 70°C to destroy the antibody. 5-10 μL of the supernatant were then used for Coomassie staining or western blotting.
When applicable, Coomassie staining was performed by incubating the protein gel in SimplyBlue™ SafeStain (Thermo Fisher Scientific) for 15 min at room temperature with gentle shaking, before rinsing the excess dye in water and taking a picture.
Western blotting
10 μL of eluted proteins were run on Bolt 4-12% Bis-Tris Plus Gels (Thermo Fisher Scientific) for 24 min at 200 V in 1X Bolt™ MES SDS Running Buffer (Thermo Fisher Scientific). A PVDF membrane (Thermo Fisher Scientific) was activated in isopropanol for 90 s and then washed briefly in water. The proteins were transferred to the PVDF membrane in 1X Bolt™ transfer buffer (Thermo Fisher Scientific) for 1 h at 20 V in a Mini Blot module (Invitrogen). The membrane was then blocked for 30 min at room temperature or overnight at 4°C in 5% skim milk or 3% bovine serum albumin (specifically when detecting the TwinStrep tag) in 1X TTBS buffer. To detect the relevant proteins, the membrane was incubated with the appropriate primary antibody or Strep-Tactin® HRP conjugate (IBA) diluted in 1X TTBS with 3% BSA for 1 h at room temperature or overnight at 4°C. The membrane was then incubated with the secondary antibody prepared to appropriate dilution in 1X TTBS for 45 min at room temperature. No secondary antibody was used in the case of the HRP-coupled Strep-Tactin. Bands were stained using Luminata Forte Western HRP substrate ECL (Millipore) solution and imaged with ImageQuant™ LAS 4000 biomolecular imager (GE Healthcare). We used Spectra™ Multicolor Broad Range Protein Ladder (Thermo Fisher Scientific) as protein ladder. When necessary, the membranes were stripped using Restore PLUS Western Blot Stripping Buffer (Thermo Fisher Scientific) and probed with another set of primary/secondary antibodies, as detailed above. A list of all antibodies used in this study can be found in Supplementary Data S13. For presentation of full scan blots, see either in the Source data file (for main figures) or the Supplementary information file (for Supplementary Figs.).
Isolation of phage escaper mutants
Phage mutants escaping the KpAvs2 defense system were isolated as described previously23. Briefly, 100 μL of bacterial cells expressing the KpAvs2 operon were grown in MMB supplemented with 0.2% arabinose to OD600 of 0.3 and then mixed with 100 μL SECphi18 phage lysate. After 10 minutes at room temperature, 5 mL pre-melted 0.5% MMB agar with 0.2% arabinose was added and the mixture was poured onto MMB 1.1% agar plates. The double-layer plates were incubated overnight at room temperature and single plaques were picked into 90 μL phage buffer (50 mM Tris pH 7.4, 100 mM MgCl2, 10 mM NaCl). The collected phages were tested for their ability to infect both a control strain and a KpAvs2 operon-carrying strain and compared to parental SECphi18 phages by plaque assay, as described above. Phages, which propagated better on the KpAvs2 operon-carrying strain than the parental phages were further amplified from a single plaque formed on the KpAvs2 operon-carrying strain as detailed below. Escaper 1372 was propagated in a liquid culture of bacteria expressing the KpAvs2 operon grown in 1 mL MMB with 0.2% arabinose to an OD600 of 0.3. The phages were incubated with the bacteria at 37°C 200 rpm shaking for 3 h, and then an additional 9 mL of bacterial culture grown to OD600 0.3 in MMB with 0.2% arabinose was added, and incubated for another 3 hours (37°C 200 rpm). The lysate was then centrifuged at 3200 g for 10 min and the supernatant was filtered through a 0.2 μM filter to get rid of remaining bacteria. The other phages did not propagate as well in liquid culture, and thus the double-layer plaque assay method was used for propagation. For this, the single plaque formed on the defense strain in the small drop plaque assay was picked into 100 μL phage buffer and mixed with 100 μL of cells grown in MMB with 0.2% arabinose to OD600 of 0.3. For escaper 1369, KpAvs2-operon-expressing cells were used, and for escapers 1370, 1371, 1373, 1374, control cells expressing GFP were used. After incubation of the phages with the bacterial cells for 10 min at room temperature, 5 mL pre-melted 0.5% MMB agar with 0.2% arabinose was added and the mixture was poured onto MMB 1.1% agar plates. The double-layer plates were incubated overnight at room temperature and 103 -105 PFUs were scraped into 5 mL of phage buffer. After 1 hour at room temperature, the phages were centrifuged at 3200 g for 10 min, and the supernatant was filtered through a 0.2 μM filter.
The obtained lysate was then used for DNA extraction and whole-genome sequencing. DNA was extracted from 500 μL of a high titer phage lysate (>107 PFU/mL). The phage lysate was treated with DNase-I (Merck cat #11284932001) added to a final concentration of 20 μg/mL and incubated at 37°C for 1 hour to remove bacterial DNA. DNA was then extracted using the DNeasy blood and tissue kit (Qiagen, cat #69504), starting from the Proteinase-K treatment step to lyse the phages. Libraries were prepared for Illumina sequencing using a modified Nextera protocol40. Reads were aligned to SECphi18 reference genome (NCBI accession NC_073071.1) and mutations compared to the reference genome were identified using breseq (version 0.29.0) with default parameters41. Only mutations that occurred in the isolated mutants, but not in the ancestor phage, were considered. A list of escaper phages obtained in this assay and the relevant mutations they carried can be found in Supplementary Data S3.
Phage competition
Bacteria expressing either the 3-gene operon or GFP as control were grown to OD ~ 0.3 at 37°C in MMB media with arabinose 0.2% and ampicillin. The WT SECphi18 phage and the Δ2 mutant phage were mixed to 1:1 PFU ratio, and the mix was used to infect the bacterial cultures with an MOI of 0.1. The infection proceeded at 25 °C for five hours, and then the cultures were centrifugated 15 min at 3900 g and filtered to recover the phages. This corresponds to “day 1” sample. For the next two days, a similar infection was performed using the phages from the previous day to infect a new bacterial culture at low MOI. After three passages of the mixed phages, a PCR amplification of the region encoding ksap1 gene on each phage population was performed and visualized on an agarose gel. The intensity of the DNA band corresponding to either the WT ksap1 or Δksap1 mutant was used as a proxy for phage abundance.
Transformation efficiency assays
Plasmids carrying phage genes as indicated, or RFP as a control, were extracted from overnight cultures using QIAprep Spin Miniprep Kit (Qiagen) according to manufacturer instructions. DNA concentration was measured using Qubit® dsDNA HS Assay (Thermo Fisher Scientific).
Bacteria were grown to OD600 ~ 0.3 in 4 mL MMB with ampicillin at 37°C, 200 rpm shaking. The samples were centrifuged 7 min at 2900 g and resuspended in 125 μL cold sterile Transformation and Storage Solution (TSS, LB broth with 10% PEG 3350 or 8000, 5% DMSO, and 40 mM MgCl2 at a final pH of 6.5) then split into 25 μL aliquots. 8 ng of each plasmid was added per tube and incubated 5 min on ice, then 5 min at room temperature and then again 5 min on ice42. Cells were allowed to recover in 1 mL MMB for 1 h at 37 °C and 200 rpm shaking. Cells were then centrifuged for 4 min at 5000 g, resuspended in 200 μL media and serially diluted by 10-fold. 10 μL drops of each dilution were plated on LB + 1.5% agar with ampicillin, kanamycin and glucose and incubated overnight at 37°C. The following day, colony-forming units (CFU) were counted as a proxy for transformation efficiency.
Protein pulldown and LC-MS
150 mL of bacteria expressing an HA-tagged KpAvs2 protein, either WT or lacking the C-terminal domain, were grown to OD600 of 0.3 in the presence of 0.2 % arabinose at 37°C. Bacteria were then infected with either WT SECphi18 phage or the Δ2 escaper mutant at 25 °C, or Bas22 or Bas60 at 37°C, in all cases at MOI = 5. For SECphi18, 50 mL aliquots were collected before infection and 30 min post infection. For the other phages, only samples after infection were collected, after 30 min of infection for SECphi18 Δ2 or after 40 min infection for Bas22 and Bas60. The samples were immunoprecipitated using anti-HA antibodies as described above, except that the samples were eluted in 5% SDS in 100 mM Tris-HCl, pH 7.4 for 10 min at room temperature with occasional shaking rather than boiled in denaturing sample buffer. The eluted samples were then subjected to tryptic digestion using an S-Trap43. The resulting peptides were analyzed using nanoflow liquid chromatography (nanoAcquity) coupled to high resolution, high mass accuracy mass spectrometry (Q Exactive Plus). Each sample was analyzed on the instrument separately in a random order in discovery mode. Raw data was processed with MetaMorpheus v1.0.2. The data was searched against the E. coli K12 UniProt proteome database, the corresponding phage proteome, and additional proteins expressed on our plasmids and common lab protein contaminants, as well as default modifications. For KpAvs2, only the common sequence between the WT and deletion mutant was considered to avoid bias. Quantification was performed using the embedded FlasLFQ44 and protein inference45 algorithms. The LFQ (Label-Free Quantification) intensities were calculated and used for further calculations using Perseus v1.6.2.3. Decoy hits and contaminants were filtered out. The LFQ intensities were log transformed and only proteins that had at least two valid values in at least one experimental group were kept. The remaining missing values were imputed by a random low-range normal distribution. A two-sided student’s t-test was performed to identify differentially represented proteins. The mass spectrometry proteomics data was deposited to the ProteomeXchange Consortium46 via the PRIDE47 partner repository with the dataset identifiers PXD048766, PXD054784 and PXD054782 respectively for SECphi18, SECphi18 Δ2 and the Basel phages.
Structural analyses and remote homology detection
Structural predictions of proteins and complexes were performed using AlphaFold236 and AlphaFold-Multimer21, respectively, version 2.3.1 with default parameters, or AlphaFold348 for the prediction of the whole KpAvs2 protein with its activators. When interrogating the whole proteome of phages against KpAvs2 C-terminal domain, the model confidence score of all predictions was analyzed and plotted. For each phage, we predicted five models. For Bas60, we computed one prediction for each of the five models, for Bas22 and Bas64 three and for Bas48 two, depending on the size of the proteome of each phage. When presenting or further analyzing predicted models, the best scoring model was used. Structures were visualized using the PyMOL Molecular Graphics System, Version 2.5.2 (Schrödinger, LLC). When applicable, proteins were structurally aligned using the cealign function of PyMOL or DALI30 server (http://ekhidna2.biocenter.helsinki.fi/dali/). To predict the list of residues interacting between two protein chains, the Residue Interaction Network Generator (https://ring.biocomputingup.it/submit)49 was used.
HHpred19,20 was used to annotate genes for which automatic sequence-based annotation did not retrieve annotations (toolkit.tuebingen.mpg.de/tools/hhpred).
Identification of KpAvs2, Ksap1 and Ksap2 homologs
Homologs of KpAvs2 were identified from both isolates and metagenomes samples from the IMG50 database in April 2020 as described previously51. The genomic neighborhood of each member of the KpAvs2 cluster was examined, and only homologs that were found in a conserved three-gene operon were retained. The list can be found in Supplementary Data S1.
Homologs of Ksap1 and Ksap2 were identified by blast search on the National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov/Blast.cgi)52,53 website. Both blastP and tblastN were used with default parameters. Ksap1 homologs that were not annotated in their genome of origin were identified manually by examining the intergenic region in the syntenic loci in all genomes of Dhillonviruses present in the NCBI Taxonomy browser that failed to show a hit in either search for Ksap1, or showed a hit only to a part of Ksap1. The genome of SECphi6 (NCBI accession CADCZA000000000.2) was similarly searched. A list of all identified homologs is found in Supplementary Data S7 and S9. All identified Ksap1 homologs were manually curated, aligned with Clustal Omega54 and a web logo was created using the online web logo tool, version 2.8.2 (weblogo.berkeley.edu/logo.cgi)55,56. An AlphaFold2-predicted protein structure of Ksap1 and Ksap2 were used to search for structural homologs using the Foldseek search29 website (search.foldseek.com/search), using default databases and 3Di/AA mode.
Avs2 phylogenetic tree
The IMG50 database downloaded in October 2017 was searched for homologs of Avs2 proteins defined in7. For this, the MMseqs2 search tool57 was used with default parameters, with a cutoff of 90% sequence similarity and 90% coverage, allowing a maximum of 1000 hits per query and no MSA prefilter. Two genes on each side of each Avs2 homolog were collected, and the PFAM domains of retrieved sequences were predicted using the HHsearch function from the HH-suite package58 with default parameters and a maximum number of HMM columns of 32000. Hits to PF19902 (DUF6375) and PF04055 (radical SAM) with a cutoff of at least 0.8 probability were used to define Avap2 and Avap1 homologs, respectively. All Avs2 used in the search were trimmed to exclude the effector domain and aligned using MAFFT59 v7.490 with default parameters. We used IQ-TREE60 multicore version 2.2.0 to build the maximum likelihood phylogenetic tree of all trimmed Avs2, and iTOL61 v6 (https://itol.embl.de/) to visualize the tree and add the annotations of the presence of Avap1 and Avap2 in the neighborhood.
Data analysis
Data was analyzed using Excel Spreadsheets (Microsoft 365) or R version 4.3.1 and Rstudio 2023.06.1 + 524, using ggplot2 package V3.5.1.
Statistics and reproducibility
No statistical method was used to predetermine sample size. We performed a minimum of three replicates for all quantitative data. Each blot was repeated at least twice, and we present a representative picture of both replicates. Likewise, agarose gel pictures in Figs. 1G and 3G are representative of two independent replicates. No relevant data points were excluded from the analysis, and all figures depict individual data points where applicable.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated in this study was made available with the manuscript in the Source data file. Protein mass spectrometry raw data was deposited to the ProteomeXchange Consortium46 via the PRIDE47 partner repository with the dataset identifiers: PXD048766 [https://www.ebi.ac.uk/pride/archive/projects/PXD048766], PXD054784, and PXD054782. We obtained the proteome sequences of phages for mass spectrometry analysis and AlphaFold-Multimer screening from GenBank repository: SECphi18: NC_073071.1, [https://www.ncbi.nlm.nih.gov/nuccore/NC_073071.1]. Bas22: MZ501091.1, [https://www.ncbi.nlm.nih.gov/nuccore/MZ501091.1/]. Bas48: MZ501054.1, [https://www.ncbi.nlm.nih.gov/nuccore/MZ501054.1/]. Bas60: MZ501100.1, [https://www.ncbi.nlm.nih.gov/nuccore/MZ501100.1/]. Bas64: MZ501081.1, [https://www.ncbi.nlm.nih.gov/nuccore/MZ501081.1/]. We obtained the scaffolds of interest of genomes of bacterial isolates for Fig. 1B from IMG repository: Klebsiella pneumoniae S_15PV: 2701097382. Nitrospirae bacterium JdFR-85: 2728441048. Syntrophobacterales bacterium Delta_02: 2751221707. Achromobacter sp. 2789STDY5608624: 2660299158 Source data are provided with this paper.
References
Kawai, T. & Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 21, 317–337 (2009).
Chaplin, D. D. Overview of the immune response. J. Allergy Clin. Immunol. 125, S3–S23 (2010).
Jones, J. D. G., Vance, R. E. & Dangl, J. L. Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395 (2016).
Huiting, E. & Bondy-Denomy, J. Defining the expanding mechanisms of phage-mediated activation of bacterial immunity. Curr. Opin. Microbiol. 74, 102325 (2023).
Vance, R. E., Isberg, R. R. & Portnoy, D. A. Patterns of pathogenesis: discrimination of pathogenic and non-pathogenic microbes by the innate immune system. Cell Host Microbe 6, 10–21 (2009).
Gust, A. A., Pruitt, R. & Nürnberger, T. Sensing Danger: Key to Activating Plant Immunity. Trends Plant Sci. 22, 779–791 (2017).
Gao, L. A. et al. Prokaryotic innate immunity through pattern recognition of conserved viral proteins. Science 377, eabm4096 (2022).
Okude, H., Ori, D. & Kawai, T. Signaling Through Nucleic Acid Sensors and Their Roles in Inflammatory Diseases. Front. Immunol. 11, 625833 (2020).
Leipe, D. D., Koonin, E. V. & Aravind, L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343, 1–28 (2004).
Gao, L. et al. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369, 1077–1084 (2020).
Daskalov, A., Dyrka, W. & Saupe, S. J. NLR Function in Fungi as Revealed by the Study of Self/Non-self Recognition Systems. in Genetics and Biotechnology (eds. Benz, J. P. & Schipper, K.) 123–141 (Springer International Publishing, Cham, 2020). https://doi.org/10.1007/978-3-030-49924-2_6.
Kibby, E. M. et al. Bacterial NLR-related proteins protect against phage. Cell 186, 2410–2424.e18 (2023).
Meunier, E. & Broz, P. Evolutionary Convergence and Divergence in NLR Function and Structure. Trends Immunol. 38, 744–757 (2017).
Chou, W.-C., Jha, S., Linhoff, M. W. & Ting, J. P.-Y. The NLR gene family: from discovery to present day. Nat. Rev. Immunol. 23, 635–654 (2023).
Ting, J. P.-Y. et al. The NLR Gene Family: A Standard Nomenclature. Immunity 28, 285–287 (2008).
Rousset, F. et al. Phages and their satellites encode hotspots of antiviral systems. Cell Host Microbe 30, 740–753.e5 (2022).
Wein, T. et al. CARD-like domains mediate anti-phage defense in bacterial gasdermin systems. Preprint at https://doi.org/10.1101/2021.10.04.463034 (2022).
Rao, V. B. & Feiss, M. The Bacteriophage DNA Packaging Motor. Annu. Rev. Genet. 42, 647–681 (2008).
Gabler, F. et al. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinforma. 72, e108 (2020).
Söding, J. Protein homology detection by HMM-HMM comparison. Bioinforma. Oxf. Engl. 21, 951–960 (2005).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. 2021.10.04.463034 Preprint at https://doi.org/10.1101/2021.10.04.463034 (2022).
Bari, S. M. N. et al. A unique mode of nucleic acid immunity performed by a multifunctional bacterial enzyme. Cell Host Microbe 30, 570–582.e7 (2022).
Stokar-Avihail, A. et al. Discovery of phage determinants that confer sensitivity to bacterial immune systems. Cell 186, 1863–1876.e16 (2023).
Tal, N. et al. Bacteria deplete deoxynucleotides to defend against bacteriophage infection. Nat. Microbiol. 7, 1200–1209 (2022).
Depardieu, F. et al. A Eukaryotic-like Serine/Threonine Kinase Protects Staphylococci against Phages. Cell Host Microbe 20, 471–481 (2016).
Millman, A. et al. Bacterial Retrons Function In Anti-Phage Defense. Cell 183, 1551–1561.e12 (2020).
Bernheim, A. et al. Prokaryotic viperins produce diverse antiviral molecules. Nature 589, 120–124 (2021).
Maffei, E. et al. Systematic exploration of Escherichia coli phage–host interactions with the BASEL phage collection. PLOS Biol. 19, e3001424 (2021).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 42, 243–246 (2024).
Holm, L., Laiho, A., Törönen, P. & Salgado, M. DALI shines a light on remote homologs: One hundred discoveries. Protein Sci. 32, e4519 (2023).
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
de Oliviera Nascimento, L., Massari, P. & Wetzler, L. M. The Role of TLR2 in Infection and Immunity. Front. Immunol. 3, (2012).
Kang, J. Y. et al. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31, 873–884 (2009).
Jo, E.-K., Kim, J. K., Shin, D.-M. & Sasakawa, C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol. 13, 148–159 (2016).
Paik, S., Kim, J. K., Silwal, P., Sasakawa, C. & Jo, E.-K. An update on the regulatory mechanisms of NLRP3 inflammasome activation. Cell. Mol. Immunol. 18, 1141–1160 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
West, A. H. & Stock, A. M. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26, 369–376 (2001).
Mazzocco, A., Waddell, T. E., Lingohr, E. & Johnson, R. P. Enumeration of bacteriophages using the small drop plaque assay system. Methods Mol. Biol. Clifton NJ 501, 81–85 (2009).
Baym, M. et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PloS One 10, e0128036 (2015).
Deatherage, D. E. & Barrick, J. E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. Clifton NJ 1151, 165–188 (2014).
Chung, C. T. & Miller, R. H. Preparation and storage of competent Escherichia coli cells. Methods Enzymol. 218, 621–627 (1993).
Elinger, D., Gabashvili, A. & Levin, Y. Suspension Trapping (S-Trap) Is Compatible with Typical Protein Extraction Buffers and Detergents for Bottom-Up Proteomics. J. Proteome Res. 18, 1441–1445 (2019).
Millikin, R. J., Solntsev, S. K., Shortreed, M. R. & Smith, L. M. Ultrafast Peptide Label-Free Quantification with FlashLFQ. J. Proteome Res. 17, 386–391 (2018).
Miller, R. M. et al. Improved Protein Inference from Multiple Protease Bottom-Up Mass Spectrometry Data. J. Proteome Res. 18, 3429–3438 (2019).
Deutsch, E. W. et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 51, D1539–D1548 (2023).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Clementel, D. et al. RING 3.0: fast generation of probabilistic residue interaction networks from structural ensembles. Nucleic Acids Res. 50, W651–W656 (2022).
Chen, I.-M. A. et al. The IMG/M data management and analysis system v.7: content updates and new features. Nucleic Acids Res. 51, D723–D732 (2023).
Johnson, A. G. et al. Bacterial gasdermins reveal an ancient mechanism of cell death. Science 375, 221–225 (2022).
Sayers, E. W. et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 50, D20–D26 (2022).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Madeira, F. et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50, W276–W279 (2022).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Schneider, T. D. & Stephens, R. M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Steinegger, M. et al. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinforma. 20, 473 (2019).
Katoh, K. & Standley, D. M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30, 772–780 (2013).
Minh, B. Q. et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82 (2024).
Acknowledgements
We thank members of the Sorek laboratory for comments on earlier versions of this manuscript and in particular Jens Hör for helpful discussions on pulldown experiments design. We also thank the members of the Crown Genomics institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine for help with Sanger DNA sequencing. R.S. was supported, in part, by the European Research Council (grant no. ERC-AdG GA 101018520), Israel Science Foundation (MAPATS Grant 2720/22), the Deutsche Forschungsgemeinschaft (SPP 2330, Grant 464312965), the Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine, Dr. Barry Sherman Institute for Medicinal Chemistry, Miel de Botton, the Andre Deloro Prize, the Estate of Marjorie Plesset, and the Knell Family Center for Microbiology. N. B. was supported by a postdoctoral grant from the Azrieli foundation and a fellowship from the Deans of the Faculty of Weizmann Institute of Science.
Author information
Authors and Affiliations
Contributions
N.B., N.T. and R.S. conceptualized the work. N.B. designed and performed experiments and analyzed the corresponding data. N.T. performed the initial identification and characterization of the studied system. A.S.-A. isolated the phage mutants. A.S. and M.K. performed the protein mass spectrometry. S.M. and G.A. designed the point mutations tested in this study. R. S. supervised the study. N.B. and R.S. wrote the manuscript. All authors contributed to editing the manuscript and support the conclusions.
Corresponding author
Ethics declarations
Competing interests
R.S. is a scientific cofounder and advisor of BiomX and Ecophage. The other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Giuseppina Mariano and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Béchon, N., Tal, N., Stokar-Avihail, A. et al. Diversification of molecular pattern recognition in bacterial NLR-like proteins. Nat Commun 15, 9860 (2024). https://doi.org/10.1038/s41467-024-54214-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54214-0
This article is cited by
-
Activation of the bacterial defense-associated sirtuin system
Communications Biology (2025)
-
Expression level of anti-phage defence systems controls a trade-off between protection range and autoimmunity
Nature Microbiology (2025)