Abstract
The formation of transcription regulatory complexes by the association of Smad4 with Smad2 and Smad3 (Smad2/3) is crucial in the canonical TGFβ pathway. Although the central requirement of Smad4 as a common mediator is emphasized in regulating TGFβ signaling, it is not obligatory for all responses. The role of Smad2/3 independently of Smad4 remains understudied. Here, we introduce a stepwise paradigm in which Smad2/3 regulate the lineage priming and differentiation of mouse embryonic stem cells (mESCs) by collaboration with different effectors. During the naïve-to-primed transition, Smad2/3 upregulate DNA methyltransferase 3b (Dnmt3b), which establishes the proper DNA methylation patterns and, in turn, enables Smad2/3 binding to the hypomethylated centers of promoters and enhancers of epiblast marker genes. Consequently, in the absence of Smad2/3, Smad4 alone cannot initiate epiblast-specific gene transcription. When primed epiblast cells begin to differentiate, Dnmt3b becomes less actively engaged in global genome methylation, and Smad4 takes over the baton in this relay race, forming a complex with Smad2/3 to support mesendoderm induction. Thus, mESCs lacking Smad4 can undergo the priming process but struggle with the downstream differentiation. This work sheds light on the intricate mechanisms underlying TGFβ signaling and its role in cellular processes.
Similar content being viewed by others
Introduction
Transforming growth factor beta (TGFβ) superfamily including TGFβ, Activin, Nodal, BMPs and many others, signals through receptor serine/threonine kinases. These cytokines are important for cell growth, differentiation, tissue regeneration and carcinogenesis. In response to activation of TGFβ, Smad2 and Smad3 (Smad2/3) are phosphorylated and form a trimer, either with or without the common mediator Smad41,2. The canonical Smad2/3-Smad4 complex imports to the nucleus and regulates transcription of target genes with other co-factors3.
Smad4 is essential for many TGFβ signaling pathways, but not obligate for all responses4,5,6,7,8. Smad2/3 still translocate to the nucleus without Smad49. In Drosophila, the disruption of oogenesis is more pronounced in mad (Smad1) mutants compared to medea (Smad4) mutants10. Specific inactivation of Smad4 gene with Cre-LoxP system in the mouse epiblast only has a minimal impact on early development, although TGFβ/BMP signaling pathways are well known to be required5. During pancreas development, Smad2/3, but not Smad4, play a key role7,8. Moreover, even though Smad4 is frequently mutated or silenced during tumor initiation and progression, TGFβ signals still activate a small group of target genes by Smad4-independent program5,11. But the underlying mechanism remains inconclusive.
DNA methylation at carbon 5 of cytosines (5-methylcytosine, 5-mC) is one of the most important epigenetic modifications, which regulate gene expression and embryogenesis. During the transition from the pre- to the post-implantation epiblast of the embryo, the de novo DNA methyltransferases, Dnmt3a and Dnmt3b (Dnmt3a/3b), are highly upregulated to establish the DNA methylation essential for cell fate specification12. Consistently, naive mouse embryonic stem cells (mESCs) from inner cell mass are characterized by low levels of Dnmt3a/3b and general hypomethylation, while primed epiblast-like cells (EpiLCs), which corresponds to the post-implantation epiblast, express high levels of Dnmt3a/3b with increased DNA methylation12,13,14. In addition, Dnmt3b-dependent methylation at the epiblast stage is essential for the subsequent determination of mesodermal and endodermal (mesendodermal) lineages, during which Dnmt3b expression was downregulated14. Mice lacking Dnmt3b exhibit embryonic lethality, whereas Dnmt3a-null mice develop to term, but most die at around 4 weeks of age15.
TGFβ has been proposed as a regulator of Dnmts in cancer. TGFβ signaling has shown differential effects on Dnmts’ activities between benign and malignant cells16,17,18,19. Early-stage embryonic bodies (EBs) formed from mESCs in leukemia inhibitory factor (LIF)-free suspension culture, recapitulate the signaling and transcriptional events of lineage priming and germ layer specification20. Smad2/3 loss in EBs results in decreased RNA levels of Dnmt3b and increased expression of Tet2 and Gadd45b that are involved in DNA demethylation21. This may be related to the ectopic extra-embryonic gene expression21. However, whether TGFβ itself or Smad4 is involved in the regulation of these genes is not explored.
The specific processes in which Smad2/3 fulfill TGFβ signaling in the absence of Smad4 and the underlying mechanisms remain unclear. To address these questions, we dissected the roles of Smad2/3 and Smad4 in mESC using knockouts of Smad2/3 (Smad2 and Smad3 double knockout, S2/3DKO) and Smad4 (Smad4 knockout, S4KO). The S2/3DKO cells exhibited challenges in DNA methylation establishment, which is important for post-implantation epiblast induction. In contrast, the S4KO cells showed comparable methylation levels as the wild-type (WT) cells, and retained the capacity to undergo epiblast formation. We found that Smad2/3 dominated this transition from the naïve mESC to the primed epiblast state by inducing Dnmt3b. The accessibility of Dnmt3b on epiblast genes was crucial for positioning Smad2/3 at the hypomethylated center of promoters and enhancers. Subsequently, Smad4, as part of the canonical Smad2/3-Smad4 complex, became more involved in the differentiation of the epiblast cells into mesendoderm cells, where Dnmt3b was downregulated. Notably, since S4KO cells remained mostly in the primed or pre-differentiated stage, the interaction between Dnmt3b and Smad2/3 was particularly evident through their shared nuclear localization and high expression levels. In all, our results demonstrate a stepwise progression of early post-implantation development, including Nodal-based Smad signaling-transduction, the naïve-to-primed transition accompanied by a shift from hypo- to hyper-methylated genomes dominated by Smad2/3-Dnmt3b, and Smad2/3-Smad4-facilitated mesendoderm induction.
Results
Distinct transcriptomes between S2/3DKO and S4KO
To investigate functional contributions of Smad2/3 and Smad4 in mESC, we generated S2/3DKO22 or S4KO using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9. Clones were selected and verified by sequencing and western blot analysis (Fig. 1A and Supplementary Fig 1A). WT, S2/3DKO and S4KO formed morphologically indistinguishable colonies with comparable alkaline phosphatase (AP) staining, indicating that pluripotency remained unchanged among all of them (Supplementary Fig. 1B). Although mESCs depleted from Smad2/3 or Smad4 showed decreased growth rate in contrast to WT cells, loss of Smad2/3 or Smad4 in mESCs didn’t significantly affect the distribution of cell cycle phases (Supplementary Fig. 1C-E).
A Western blot analysis of Smad2/3 and Smad4 proteins in WT, S2/3DKO, and S4KO cell lines. Tubulin serves as a loading control. B PCA of RNA-seq data generated in WT, S2/3DKO and S4KO in mESC (D0) and EB differentiation 4 days (D4). Samples were prepared in duplicates. C MA plot of RNA-seq datasets of D4 EBs derived from WT, S2/3DKO or S4KO. Genes upregulated (red, log2 fold-change ≥ 1) or downregulated (blue, log2 fold-change ≤ −1) in S2/3DKO or S4KO compared to WT. Two biological replicates for each condition were analyzed. D The up- and down-regulated DEGs were indicated by red and blue bars, respectively. The number of DEGs were marked on the bars. E Heatmap presentation of all DEGs in WT, S2/3DKO or S4KO at D0 or D4 EBs. Genes specifically upregulated only in S2/3DKO (C2) and genes significantly decreased in S2/3DKO (C4) were highlighted. Two biological replicates were analyzed at each condition. F The left dot-line plots showed the average expression patterns of C2 and C4 in (E). Gene Ontology (GO) analysis of genes in C2 and C4 were shown on the right. The top enriched biological processes GO terms with P values were listed. The hypergeometric test (one-side) is used to determine the statistical significance. G Lineage gene expression patterns were analyzed with GSVA of RNA-seq data from D0 to D4 in WT, S2/3DKO and S4KO. ICM, inner cell mass; PrE, primitive endoderm; PreEpi, pre-epiblast; PostEpi, post-epiblast; PS, primitive streak; End, endoderm; Mes, mesoderm; Ect, ectoderm; VE, visceral endoderm. H Heatmap showed epiblast-associated genes in mESC (D0) and D4 EBs of WT, S2/3DKO and S4KO. The color bar represents z-score normalized gene expression. I Heatmap of PS or mesendoderm genes in mESC (D0) and D4 EBs of WT, S2/3DKO and S4KO. The color bar represents z-score normalized gene expression. J qRT-PCR analysis of lineage marker genes expression in mESC (D0), EB D3 and D4 of WT, S2/3DKO and S4KO. The gene expression level is normalized to D0 WT samples. This experiment was repeated 3 times, and representative data are shown here. Data are presented as mean values +/- SD.
Next, transcriptomic profiling of WT, S2/3DKO and S4KO either in mESC condition (day 0, D0) or at day 4 (D4) EBs were carried out using RNA-seq. Principal component analysis (PCA) revealed that the transcriptome of S2/3DKO exhibited striking differences compared to WT and S4KO (Fig. 1B), reminiscent of the differential requirements for Smad2/3 and Smad4 during TGFβ-dependent patterning of early mouse embryos5. 784 genes (469 down and 315 up) at D0 and 1939 genes (913 down and 1026 up) at D4 were identified with significantly changed expression levels (Fold change > 2, P < 0.01) in S2/3DKO compared to WT ESCs. In contrast, only 339 genes (143 down and 196 up) at D0 and 254 genes (75 down and 179 up) at D4 were changed in S4KO compared to WT (Fig. 1C, D and Supplementary Fig. 1F). In D4 EBs, there was more than a 7-fold difference in differentially expressed genes (DEGs) between S2/3DKO and S4KO, although both Smad2/3 and Smad4 are assumed to be equally important in TGFβ signaling transduction.
We wondered which types of genes were differentially affected by the loss of Smad2/3 or Smad4. As shown on the heatmap, these DEGs were divided into 7 clusters (C1-C7) that were further analyzed based on their average expression patterns and by Gene Ontology (GO) analysis (Fig. 1E, F, Supplementary Fig. 1G-H). In C2, genes related to extra-embryonic development, such as placenta development, were upregulated in Smad2/3-deficient EBs compared to WT and S4KO, which was consistent with previous report21 (Fig. 1F, upper panel). As a co-Smad, Smad4 is not only required for TGFβ-Smad2/3 transduction, but also contributing to BMP signaling23. Bmp target genes Id1/2/3 were upregulated in S2/3DKO21, but not in WT and S4KO (Supplementary Fig. 1I). As a result, we postulated that the differential BMP signaling is the reason for the varying activation levels of extra-embryonic genes in S2/3DKO, WT and S4KO. Genes in C4, which are involved in embryonic patterning and development, still increased from D0 to D4 in Smad4-null EBs, albeit to a lesser extent than in WT. But they were almost completely blocked in S2/3DKO EBs (Fig. 1F, lower panel). These divergent phenotypes in C4 raised the possibility that Smad2/3 might regulate TGFβ targets genes independently of Smad4.
Differential requirements for Smad2/3 and Smad4 during mESC differentiation
To identify the specific lineage that was differentially regulated by Smad2/3 or Smad4 during EB differentiation, we conducted a gene set variation analysis (GSVA) to compare our RNA-seq data with published single-cell transcriptome data of EBs24,25 (Fig. 1G). According to the GSVA score, S2/3DKO and S4KO exhibited the most significant differences in the determination of primitive endoderm (PrE, also referred to as hypoblast) and post-implantation epiblast (PostEpi) (Fig. 1G). In line with an earlier study21 and our findings, S2/3DKO promoted the formation of PrE, a predominantly extra-embryonic tissue responsible for giving rise to the yolk sac, which may be associated with the upregulated BMP signaling in S2/3DKO EBs (Fig. 1G and Supplementary Fig. 1I-J). Importantly, genes associated with PostEpi were significantly repressed in S2/3DKO EBs. Conversely, the expression levels of these genes were either unaffected or mildly altered in S4KO compared to WT (Fig. 1H). Arising within the epiblast at the posterior of the embryo, the primitive streak (PS) is established at the onset of gastrulation and will later give rise to the mesendoderm20. In contrast to the differential effect of Smad2/3 and Smad4 in epiblast genes, PS/mesendoderm marker genes were abrogated in both S2/3DKO and S4KO (Fig. 1I).
This observation was further verified by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 1J). Pluripotent genes like Esrrb and Oct4, decreased across all samples upon LIF removal (Fig. 1J). The expression of mesendoderm genes, such as Eomes and Wnt3, were completely inhibited in both S2/3DKO and S4KO EBs in contrast to WT (Fig. 1J). This result confirmed the requirement for both Smad2/3 and Smad4 for the formation of PS and mesendoderm, which follows the primed epiblast stage20. Remarkably, the well-known regulators of PostEpi, such as Fgf5, Brachyury (T), Dnmt3a/3b, and Sox3/426,27,28,29,30, increased upon differentiation in WT and S4KO (Fig. 1J). However, these genes were repressed in S2/3DKO EBs, indicating that Smad2/3 is crucial and sufficient for primed epiblast formation, even when Smad4 is absent.
Epiblast forms in the absence of Smad4
The transition from naïve to primed pluripotency can also be achieved by generating EpiLC from mESC31,32. Therefore, mESCs of WT, S2/3DKO and S4KO were transitioned into EpiLCs using the established system (Fig. 2A). When cultured in serum-free N2B27 media supplemented with bFGF and Activin A, WT and S4KO mESCs gradually underwent typical morphological transformation of EpiLCs, including flattening and formation of cellular protrusions. As determined by qRT-PCR, naïve pluripotent genes (Nanog and Oct4) decreased and primed epiblast genes (Fgf5, Dnmt3a, Dnmt3b and T) were induced in WT and S4KO EpiLCs (Fig. 2B). However, S2/3DKO mESC had difficulties in the transition to EpiLC, most of which kept the domed clone forms of mESC and almost didn’t grow (Fig. 2A). At D5 of the transition, very few S2/3DKO cells could emerge from EpiLC-specific bottleneck and survived, and the primed epiblast genes were silenced (Fig. 2A, B). Overexpression of Smad2/3 in S2/3DKO could fully rescue the phenotype and successfully formed EpiLCs with activation of these key epiblast genes (Fig. 2A, B and Supplementary Fig. 2A). This confirms the dominance of Smad2/3 in epiblast formation, aligning with in vivo evidence that the specific inactivation of Smad4 using the Cre-LoxP system in the epiblast has a limited impact on early mouse development5.
A The scheme showed the procedure of EpiLC transition from mESC (top). Briefly, mESCs turned into EpiLCs after mESCs were shifted from 2i+LIF medium to N2B27 medium with Activin A and bFGF for 6 days. This experiment was repeated 5 times. Representative images showing cell morphology changes during the transition in WT, S4KO, S2/3DKO, and S2/3DKO rescued with Smad2/3 (bottom) are presented here. Scale bar: 100 μm. B mRNA levels of pluripotent genes (Nanog, Oct4) and post-implantation epiblast genes (Fgf5, Dnmt3a/3b, T) during the induction of EpiLCs were analyzed by qRT-PCR. Data are presented as mean values +/- SD. n = 3 independent experiments. P values were calculated by two-way ANOVA test with Geisser-Greenhouse correction. C Venn diagram indicated the overlapping peaks among Smad4 ChIP-seq in WT, Smad2/3 ChIP-seq in WT and Smad2/3 ChIP-seq in S4KO. Genes adjacent to 7926 peaks were annotated, in which primed epiblast genes were marked below. D Heatmap of ChIP-seq tag densities for Smad2/3 and Smad4 were within −1.5/ + 1.5 kb genomic regions surrounding the centers of 11,080 high-confidence Smad2/3 binding sites in S4KO D3 EBs with AC treatment. SB, SB431542, a selective inhibitor of TGFβR; AC, Activin A, an available ligand for Nodal/Activin receptors. E Gene track view of the Fgf5, Dnmt3a, Dnmt3b and T loci. Smad2/3 and Smad4 ChIP-seq were performed in SB- or AC-treated D3 EBs and in D0 non-treated mESCs. Tag densities were normalized to reads per kilobase per million mapped reads (RPKM). The gene structures from RefSeq are schematically represented at the bottom.
Genome-wide binding profile of Smad2/3 without Smad4
Given the observation that Smad2/3 could stimulate epiblast genes even in the absence of Smad4, we sought to investigate the underlying mechanism. Previously, we conducted chromatin immunoprecipitation sequencing (ChIP-seq) of Smad2/3 and Smad4 in ESCs or D3 EBs22. Addition of Activin A (AC) or SB431542 (SB) to EBs either acutely activates or blocks the TGFβ pathway during differentiation process, respectively20,22. Using the same method, we performed Smad2/3 ChIP-seq in S4KO and compared the results with those in WT22 (Fig. 2C–E). In comparison to the Smad2/3 binding peaks observed in WT EBs (32,717 peaks), the overall peak signals of Smad2/3 in Smad4KO EBs (11,080 peaks) were reduced (Fig. 2C). Remarkably, these remaining Smad2/3 binding sites showed a high concordance in S4KO and WT. The binding of Smad2/3 to these loci increased significantly in differentiating EBs stimulated with AC compared to pluripotent ESCs or EBs treated with SB (Fig. 2D). Although the binding pattern is very similar, the overall binding peaks of Smad4 ChIP-seq (13,313 peaks) were not as pronounced as those for Smad2/3, possibly due to differential antibody affinities (Fig. 2C, D).
The disappearance of 4474 Smad2/3 peaks upon Smad4 loss, which were co-bound with Smad4 in WT EBs, strongly suggests a specific requirement for Smad4 at these loci (Fig. 2C). Intriguingly, 7926 Smad2/3 binding peaks remained in the absence of Smad4. Among these, 3787 Smad2/3 peaks were generally co-occupied by both Smad2/3 and Smad4 (Fig. 2C). Transcription factor motif analysis revealed enrichment of binding elements for Smad2, Smad3, and Smad4 in these 7926 peaks (Supplementary Fig. 2B). We annotated genes adjacent to these Smad4-independent peaks and performed GO analysis, which indicated significant enrichment of genes related to embryonic organ development and cell fate commitment. This included epiblast marker genes such as Fgf5, Dnmt3a/3b, T, Sox3/4, Pou3f1 and many others (Fig. 2C and Supplementary Fig. 2C).
The Gene track view showed that in ESCs or SB-treated EBs, there was little to no signal of Smad2/3 or Smad4 on Fgf5, Dnmt3a, Dnmt3b, and T genes. In response to the addition of AC to EBs, these typical epiblast genes were co-occupied by Smad2/3 and Smad4 at promoter and distal enhancer regions (Fig. 2D, E). If Smad4 was removed, Smad2/3 still retained its binding to these epiblast genes (Fig. 2E). Interestingly, the density of most Smad2/3 binding peaks in S4KO is nearly comparable to that in WT. This observation provides an explanation for the largely unchanged expression levels of these epiblast genes in Smad4-null EBs.
Both Smad2/3 and Smad4 are required for mesendoderm differentiation
Since Smad4 is not essential for the transition of mESCs to EpiLCs, we were curious about its importance in epiblast-derived differentiation. A two-step mesendoderm-specific in vitro differentiation model was induced from mESC-derived EpiLCs14 (Supplementary Fig. 2D). Consistent with our findings in the EB models, the loss of either Smad2/3 or Smad4 inhibits the induction of mesendoderm genes such as Wnt3, Eomes and Gsc (Supplementary Fig. 2D). At this stage of mesendoderm differentiation, we conducted cleavage under targets and tagmentation (CUT&Tag) for Smad2/3 in WT and S4KO cells. The binding of Smad2/3 to Wnt3, Eomes, and Gsc drastically decreased with the loss of Smad4 (Supplementary Fig. 2E), indicating the crucial role of the canonical Smad2/3-Smad4 complex in promoting the differentiation of PS from EpiLCs. This is also in line with the known requirement of Smad2/3 and Smad4 during gastrulation33,34. Thus, we postulate that there are two distinct waves of Smads binding and activity. The first wave underscores the key role of Smad2/3 in facilitating the transition from the naïve mESCs to the primed EpiLCs. This is followed by a secondary role of Smad4, as part of the canonical Smad2/3-Smad4 complex conducting TGFβ signaling, in inducing mesendodermal regulators.
Dnmt3b interacts with Smad2/3
To explore the potential mechanism by which Smad2/3 independently regulates epiblast genes, we conducted Smad2/3 co-immunoprecipitation with mass spectrometry (IP-MS) in D3 EBs derived from WT and S4KO, with or without SB or AC treatment. The identified proteins were compared and analyzed in three groups (S4KO vs. WT, S4KO + AC vs. S4KO + SB, S4KO vs. S4KO + SB) (Fig. 3A). Among the 16 proteins enriched in all three comparisons, Dnmt3b gained our attention for several reasons (Fig. 3A and Supplementary Fig. 3A). First, Dnmt3b was ranked on the top five most abundant proteins that was pulled down by Smad2/3 in S4KO and S4KO + AC conditions, but was hardly detectable in WT or S4KO + SB. This suggests that the interaction of Smad2/3 and Dnmt3b is Smad4-independent and TGFβ-dependent. Second, it is well known that Dnmt3b is upregulated to establish DNA methylation during epiblast formation and maturation12,13,14.
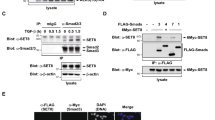
A Venn diagram of Smad2/3 co-IP and Mass spectrometry (IP-MS) analysis. Smad2/3 IP-MS were performed in D3 EBs from WT and S4KO, including non-treated, treated with SB or AC. 16 proteins were identified in all three comparisons (S4KO vs. WT, S4KO + AC vs. S4KO + SB, S4KO vs. S4KO + SB) including Dnmt3b. B Smad2/3 immunoprecipitation in WT and S4KO D3 EBs under SB or AC addition were analyzed by western blot using Dnmt3b, pSmad2/3, Smad2/3 and Smad4 antibodies. This experiment was repeated 3 times. C Western blot analysis of Dnmt3b, pSmad2/3, Smad2/3 and Smad4 for the anti-HA immunoprecipitation in Smad2/3/4TKO cells in which HA tagged Smad2/3 (HA-S2/3) or/and Smad4 (HA-S4) were overexpressed as indicated in the figure. Cells were collected after SB or AC treatment. This experiment was repeated 3 times. D Smad2/3 immunoprecipitation of chromatin-bound proteins in D3 EBs of WT and S4KO with AC treatment were analyzed by western blot using Dnmt3b, Smad2/3, Smad4 and Otx2 antibodies. This experiment was repeated 3 times. E Representative confocal images of PLA experiments on WT, S4KO and S2/3DKO D3 EBs. This experiment was repeated 3 times. Green or gray punctate, positive PLA signal; blue, DAPI. Scale bars, 20 μm. F Overexpression of HA tagged Smad2 (HA-S2) or Smad3 (HA-S3), or/and Flag tagged Dnmt3b (Flag-3b) in 293 T cells were shown in the figure. Western blot of Flag and HA were detected after anti-Flag immunoprecipitation. This experiment was repeated 3 times. G GST pull-down assays using bacterially expressed GST-Dnmt3b or GST-GFP, and 6xHistidine-tagged Smad2 (His-S2, left) or Smad3 (His-S3, right). Elution proteins were analyzed using anti-GST and anti-His antibodies. This experiment was repeated 3 times. H His pull-down assays using bacterially expressed GST-Dnmt3b or GST, and 6xHistidine-tagged Smad2 (His-S2, left) or Smad3 (His-S3, right). Elution proteins were analyzed using anti-GST and anti-His antibodies. This experiment was repeated 3 times.
Next, we repeated the co-IP experiment using WT and S4KO EBs (Fig. 3B). When AC was added to WT EBs, Smad2/3 was phosphorylated (pSmad2/3) and immunoprecipitated with Smad4, but barely with Dnmt3b. In contrast, Dnmt3b was pulled down with endogenous Smad2/3 in S4KO EBs treated with AC, and to a lower degree in S4KO EBs treated with SB. Meanwhile, co-IP was also conducted using the EBs in which HA tagged Smad2/3 (HA-S2/3) or/and Smad4 (HA-S4) were overexpressed in the triple-knockout of Smad2/3 and Smad4 (S2/3/4TKO) (Fig. 3C). When HA-S2/3 or HA-S4 alone was induced in S2/3/4TKO, Dnmt3b was significantly immunoprecipitated with pSmad2/3 when AC is added, but not with Smad4. If both HA-S2/3 and HA-S4 were expressed in S2/3/4TKO, Smad4 competed with Dnmt3b for binding Smad2/3 especially upon AC stimulation (Fig. 3B, C). An alternative explanation for this competing interaction is that EBs without Smad4 are stuck in the pre-differentiation epiblast stage compared to WT, which facilitates the interaction of highly expressed Dnmt3b and pSmad2/3 in nucleus.
To explore whether Smad2/3-Dnmt3b interaction occurs at chromatin, both WT and S4KO EBs with AC addition, were fractionated into cytosol, nucleoplasm, and chromatin (Supplementary Fig. 3B). As a control, Smad2/3 immunoprecipitated Smad4 in WT. Dnmt3b was successfully pulled down with Smad2/3 from chromatin fraction of S4KO (Fig. 3D). Otx2 is known to be highly expressed in the nuclei of epiblast or EpiSC35,36 (Supplementary Fig. 3C). In contrast, Otx2 was not immunoprecipitated with Smad2/3 (Fig. 3D). Meanwhile, the specific Otx2 antibody pulled down itself but failed to precipitate Dnmt3b, Smad2/3 or Smad4 from all the chromatin samples (Supplementary Fig. 3D). Thus, Smad2/3 specifically interacts with Dnmt3b at chromatin.
Moreover, we observed an interaction of Dnmt3b and Smad2/3 in WT, but more prominently in S4KO EBs, utilizing the in situ proximity ligation assay (PLA) (Fig. 3E and Supplementary Fig. 3E). This robust technique allows the detection of two proteins in close vicinity (distance < 40 nm)37. As a control, no positive signal was observed in S2/3DKO EBs with AC addition (Fig. 3E and Supplementary Fig. 3E).
Although Smad2 and Smad3 share a considerable amino acid sequence similarity, they could exhibit distinct functionalities in various biological contexts2,22,38,39. Notably, in mESCs and early mouse embryos, Smad2 surpasses Smad3 in abundance22,40. Consistently, both Smad2 and its phosphorylated form, pSmad2, were more effectively captured by Smad2/3 antibody than their Smad3 counterparts (Fig. 3B, upper bands vs. lower bands). To study the individual role of Smad2 and Smad3, a specific Smad2 or Smad3 antibody was used for co-IP experiment in EBs stimulated with AC (Supplementary Fig. 3F). Smad2 and Smad3 interact with each other. Smad4 was pulled down successfully by Smad2 or Smad3 in WT, but Dnmt3b exhibited a much higher efficiency of immunoprecipitation when bound to Smad2, especially in the S4KO EBs (Supplementary Fig. 3F). In addition, co-IP experiments conducted in Smad2 KO (S2KO) or Smad3 KO (S3KO) cells22 using an anti-Smad2/3 antibody showed that individual Smad2 or Smad3 could pull down Dnmt3b. Their IP efficiencies were also related to their expression levels (Supplementary Fig. 3G). Considering the variance in Smad2 and Smad3 abundance in mESC, we sought to standardize their expression levels. Therefore, we overexpressed Smad2 and Smad3 to comparable levels in S2/3/4TKO cells. Remarkably, under these conditions, both HA-S2 and HA-S3 demonstrated efficient interaction with endogenous Dnmt3b under AC stimulation (Supplementary Fig. 3H). Furthermore, human embryonic kidney 293 T (HEK293T) cells were utilized, overexpressing HA-S2, HA-S3, or/and Flag-tagged Dnmt3b (Flag-Dnmt3b). Co-IP assays revealed that Flag-Dnmt3b efficiently precipitated either HA-S2 or HA-S3 individually, as well as their co-expression (Fig. 3F). Finally, Smad2 and Smad3 were individually expressed as 6XHistidine fusion proteins (His-S2 and His-S3), while GST, GST-tagged GFP (GST-GFP) or Dnmt3b (GST-3b) were induced in E. coli. These proteins were subsequently employed in pull-down assays. GST-3b, but not GST or GST-GFP, was found to interact with His-S2 or His-S3 (Fig. 3G-H). In summary, in mESC, Smad2 is more abundant and interacts more strongly with Dnmt3b. But Smad2 and Smad3 could potentially interact with Dnmt3b at similar levels if their expression levels are similar.
Upregulation of Dnmt3b by Smad2/3 is important for epiblast formation
Next, we checked whether Dnmt3b was required for epiblast gene expression during mESC priming. Indeed, both RNA and protein levels of Dnmt3b gene were significantly upregulated during the epiblast maturation stage in WT or S4KO, encompassing D3-D4 EBs and EpiLCs (Figs. 1J, 2B, 4A, B and Supplementary Fig. 4A). Then, Dnmt3b expression sharply decreased over the course of mesendoderm differentiation from EpiLCs, indicating its reduced involvement in the subsequent differentiation process (Fig. 4B). Notably, Dnmt3b was partially downregulated in S2/3DKO EBs but was mostly blocked in S2/3/4TKO (Figs. 1J, 2B, 4A, and Supplementary Fig. 4A). This phenomenon was more pronounced in the mESC-EpiLC-mesendoderm induction models (Fig. 4B). These observations indicate that both Smad2/3 and Smad4 could regulate Dnmt3b expression, but Smad2/3 is the more dominant one. Dnmt3a and Dnmt3b are closely related in amino acid sequence, exhibiting both redundant and distinctive activities. However, the protein level of Dnmt3a remains constant from EpiLC to mesendoderm stage (Fig. 4B). In addition, Dnmt3a appears to be less affected by Smad2/3, especially in EB differentiation models (Fig. 4A-B and Supplementary Fig. 4A).
A Western blot analysis of Dnmt3a, Dnmt3b, Eomes, Smad2/3 and Smad4 protein levels during EB differentiation (D0, D3 and D4) in WT, S4KO, S2/3DKO, and S2/3/4TKO cells. Tubulin serves as a loading control. This experiment was repeated 3 times. B The two-step induction scheme of mESC-EpiLC-ME was presented (top). In brief, after changing into N2B27 medium with Activin A and bFGF for 6 days, EpiLCs were further cultured in N2B27 medium plus CHIR99021 for 24 h to form ME. Western blot showed the expression of Dnmt3a, Dnmt3b, Eomes, Smad4 and Smad2/3 in WT, S4KO and S2/3DKO. Tubulin serves as a loading control. This experiment was repeated 3 times. C qRT-PCR analysis of primed epiblast genes (Dnmt3b, Fgf5) and pluripotent gene Oct4 during EB differentiation (D0, D3) and EpiLC induction (D0, D4, D6) derived from WT, S2/3DKO, S4KO, and S4KO with Dnmt3b shRNA 1 or 2 cells. Data are presented as mean values +/- SD. n = 3 independent experiments. P values were calculated by two-way paired t-test. D Representative confocal microscope images of 5-mC (Red) and DAPI (blue) staining in WT, S4KO and S2/3DKO EBs. Scale bars, 20 μM. This experiment was repeated 3 times. E IGV view showed the WGBS signal profiles of Sox2 locus at ES and EpiLC stages from WT and Dnmt3b knockout (3bKO). The CpG island (purple) and differentially methylated regions (DMRs) (red) were labeled as well. MeDIP-qPCR analysis of DNA methylation levels on Sox2 DMR1 and DMR3 were carried out in D3 EBs derived from WT, S2/3DKO and S4KO. Data are presented as mean values +/- SD. n = 4 independent experiments. P values were calculated by two-sided Mann-Whitney test. F The mRNA level of Sox2 in WT, S2/3DKO, and S4KO cells before and after EB differentiation (D0, D3, D4) were measured by qRT-PCR. Data are presented as mean values +/- SD. n = 4 independent experiments. P values were calculated by two-way ANOVA test.
In contrast, Eomes, a key mesendoderm marker, was highly upregulated in WT during EB and EpiLC-mesendoderm differentiation processes. Its expression was blocked in S4KO and S2/3DKO, suggesting the important role of both Smad2/3 and Smad4 in regulating mesendoderm genes like Eomes (Fig. 4B).
The downregulation of Dnmt3b in S2/3DKO EBs could be one of the reasons why S2/3DKO faced challenges in the primed epiblast transition. Indeed, we observed that overexpression of pLVXI-Dnmt3b in S2/3DKO could upregulate FGF5 and T expression compared to S2/3DKO with empty vector (Supplementary Fig. 4B). Then we knocked down Dnmt3b in WT using two different shRNAs or a combination of both (WT+Dnmt3b-sh1, 2 or 1 + 2). When Dnmt3b was repressed in WT, the epiblast gene Fgf5 was mildly affected in the more heterogeneous EB models but had a greater effect in the more homogeneous EpiLCs (Supplementary Fig. 4C-D). As a control, the pluripotency gene Oct4 was not changed (Supplementary Fig. 4C-D). Interestingly, when Dnmt3b was interfered with the same shRNAs in S4KO EBs or EpiLCs, Fgf5 was significantly downregulated compared to WT or S4KO (Fig. 4C).
All the results suggest that Smad2/3 stimulates Dnmt3b expression during mESC priming process, which in turn promotes the epiblast formation. The need for Dnmt3b is particularly amplified in the context of S4KO. When Dnmt3b is downregulated, resulting in reduced involvement during epiblast-to-mesendoderm differentiation, Smad4 plays an auxiliary role alongside Smad2/3.
Catalytically active Dnmt3b is required for epiblast priming
Dnmt3b is involved in de novo DNA methylation. On one hand, DNA methylation exerts suppressive effects on gene transcription, especially in CpG islands. On the other hand, DNA methylation on gene bodies and enhancers can boost transcription and maintain correct transcription elongation41,42,43,44.To investigate how Dnmt3b mediates Smad2/3 activity in epiblast homeostasis upon Smad4 loss, S4KO+Dnmt3b-sh2 cells were infected with a lentivirus vector expressing a wild-type version of Dnmt3b (Dnmt3bWT), a catalytically inactive Dnmt3b losing DNA methylation ability (Dnmt3bMet-mut) or a PWWP domain mutation of Dnmt3b unable to bind to H3K36me2/3 (Dnmt3bPWWP-mut), respectively (Supplementary Fig. 4E). Dnmt3bWT, but not Dnmt3bMet-mut and Dnmt3bPWWP-mut, efficiently rescued the expression of Fgf5 and T in S4KO+Dnmt3b-sh2 EBs, which is comparable to their expression levels in S4KO EBs with scrambled shRNA (S4KO+Scr-sh) (Supplementary Fig. 4E). As a control, Oct4 expression pattern was not changed under these conditions (Supplementary Fig. 4E). This indicates that both DNA methylation and H3K36me2/3-association abilities of Dnmt3b are required for activating primed epiblast genes in S4KO.
The DNA methylation status of WT, S2/3DKO and S4KO were initially compared. Immunofluorescence imaging analysis via confocal microscopy showed strong 5-mC expression in D3 EBs of WT and S4KO as Dnmt3b levels increased with differentiation. In contrast, S2/3DKO failed to express a high level of 5-mC since Dnmt3b was downregulated (Figs. 4A and 4D). Previous research demonstrated that Sox2 serves as a direct target gene of Dnmt3b in EpiLCs14. Specifically, differentially methylated regions (DMR1-4) located on the Sox2 super-enhancer play a crucial role in guiding EpiLCs towards mesendoderm differentiation14. Published whole genome bisulfite sequencing (WGBS) data showed that these DMRs acquire DNA methylation at EpiLC stage, but become hypomethylated when Dnmt3b is removed (Fig. 4E, upper panel)14. Analyzing DMR1 and DMR3 of Sox2 enhancer at CpG islands during lineage priming, we performed Methylated DNA Immunoprecipitation followed by qPCR (MeDIP-qPCR) and observed a significant reduction in methylation at both loci in S2/3DKO EBs compared to WT and S4KO (Fig. 4E, lower panel). This resulted in Sox2 upregulation in S2/3DKO upon EB differentiation, whereas Sox2 expression decreased as WT and S4KO EBs exited the pluripotent ESC stage (Fig. 4F). Therefore, the timely downregulation of pluripotent factor Sox2, regulated by high level of Dnmt3b, is important for epiblast maturation in WT and S4KO EBs.
Dnmt3b mediates Smad2/3 binding to epiblast genes independently of Smad4
Next, we studied whether Dnmt3b mediates Smad2/3 activity in epiblast formation, particularly when Smad4 is lost. Integrated profiles of DNA methylation levels (WGBS14), Smad2/3 ChIP-seq and histone modifications including ChIP-seq of H3K4me1, H3K4me3 and H3K27ac in EpiLCs14, as well as H3K36me3 ChIP-seq in embryonic day (E) 6.5 epiblast45, were analyzed on epiblast key regulator Fgf5 gene (Fig. 5A). As revealed by WGBS, elevated DNA methylation levels were observed at the gene body (GB) and enhancer regions (+30, +40 and +56 kb) of Fgf5 in mESCs. As Fgf5 expression increased in EpiLCs, Fgf5 GB retained high levels of methylation, and hypomethylated regions were identified at the central area of the proximal promoter (PP) distinguished by H3K4me3 and at active enhancer centers marked by H3K27ac and H3K4me1 (Fig. 5A). It’s worth noting that H3K36me3 exhibited a high degree of enrichment within Fgf5 GB in epiblast, and to a lesser extent, it was present at Fgf5 enhancers, which were further confirmed by ChIP-qPCR (Fig. 5A and Supplementary Fig. 6B). Subsequently, Smad2/3 specifically localized to these valleys of DNA methylation at PP and enhancers, then initiated transcription of Fgf5 during the process of epiblast maturation in both WT and S4KO EBs (Fig. 5A).
A IGV view of WGBS, ChIP-seq and CUT&Tag profiles surrounding Fgf5 gene locus. WGBS as well as ChIP-seq of H3K27ac, H3K4me1 and H3K4me3 were performed at ESC or EpiLC stages14, H3K36me3 ChIP-seq was performed in E6.5 epiblast of mice45. Smad2/3 ChIP-seq were done in EBs of WT20 and S4KO (this study), Dnmt3b CUT&Tag of EBs were carried out in WT, S4KO, WT+Dnmt3b-shRNA2 (WT+3b-sh2) and S4KO+Dnmt3b-shRNA2 (S4KO + 3b-sh2), Flag CUT&Tag experiments were performed in Dnmt3a-Flag or Dnmt3b-Flag cells; Dnmt3a or Dnmt3b ChIP-seq of WT and S4KO EBs were shown at the bottom. B Heatmap of tag densities for Flag CUT&Tag in Dnmt3b-Flag, and Smad2/3 ChIP-seq in WT or S4KO, were shown within −1.5/ + 1.5 kb genomic regions surrounding the centers of 68,204 Dnmt3b binding sites in Dnmt3b-Flag EBs, 32,717 or 11,080 Smad2/3 binding sites in WT or S4KO EBs, respectively. C Gene track view of the Fgf5, Dnmt3a, and Dnmt3b loci from Smad2/3 CUT&Tag in EBs of S4KO and S4KO + 3b-sh2. D Heatmap of Smad2/3 CUT&Tag signals in S4KO and S4KO+3bsh2 surrounding 7926 Smad4-independent Smad2/3 peaks. E Peak score plot for the most significant Dnmt3b-dependent Smad2/3 binding sites (fold change > 2.0, P < 0.01). Epiblast marker genes adjacent to the peaks are indicated. Peak score plot for the most significant Dnmt3b-dependent Smad2/3 binding sites (fold change > 2.0, P < 0.01). Epiblast marker genes adjacent to the peaks are indicated. P values were generated by MACS2 callpeak strategy using Poisson test (one-sided) with Benjamini-Hochberg correction. (F)ChIP-qPCR analysis of Smad2/3 and Dnmt3b binding to Fgf5 promoter (Fgf5_PP), gene body (Fgf5_GB) and enhancers (Fgf5_+30k, Fgf5_+40k, and Fgf5_+56k) of D3 EBs treated with SB or AC in WT, S2/3KO, S4KO+ Scr-shRNA, S4KO + 3b-Sh1 or S4KO + 3b-sh2 cell lines. ChIP with anti-IgG serves as a negative control. This experiment was repeated 3 times, and representative data are shown here. Data are presented as mean values +/- SD.
To investigate the mechanistic basis for the role of Dnmt3b in facilitating Smad2/3 function, we tested the possibility that Dnmt3b interacts directly with Smad2/3 on epiblast genes. To explore this, we conducted a series of Dnmt3b CUT&Tag and ChIP-seq experiments during EB differentiation (Fig. 5A). For comparison, we also performed these experiments for Dnmt3a. Besides using a specific Dnmt3a or Dnmt3b antibody, we also employed an anti-Flag antibody to detect the Dnmt3a/3b binding pattern in Dnmt3a- or 3b-Flag cells, in which a 3XFlag epitope tag was inserted into the C-terminus of endogenous Dnmt3a (3a-Flag) or Dnmt3b (3b-Flag) genes by CRISPR/Cas9 technology, respectively (Supplementary Fig. 5A). These clones were sequenced and validated by both western blotting and immunofluorescence (Supplementary Fig. 5A-C). Furthermore, the cells formed EBs correctly and showed upregulation of primed and differentiation genes upon LIF removal, as demonstrated by qRT-PCR (Supplementary Fig. 5D). They also exhibited high levels of 5-mC, comparable to WT EBs (Fig. 4D and Supplementary Fig. 5E).
We first compared Dnmt3b binding profiles across the four assays: CUT&Tag-1, CUT&Tag-2, and ChIP-seq, all were performed with an anti-Dnmt3b antibody, and CUT&Tag-3 used an anti-Flag antibody in Dnmt3b-Flag cells. Notably, CUT&Tag-2 included an E. coli genome spike-in for calibration, which minimized errors related to low input and sequencing biases. In the three CUT&Tag assays, we identified 37,845, 57,886, and 68,204 Dnmt3b peaks (fold change ≥ 4, Q ≤ 0.01), with 57%–67% of annotated peaks overlapping, demonstrating high reproducibility among them (Supplementary Fig. 5F-G). In contrast, the ChIP-seq assay, due to its lower peak-calling performance, identified 41,702 Dnmt3b peaks even with a more lenient fold change cutoff of 3 and a P-value of 0.01 (Supplementary Fig. 5F). Additionally, only about 20% of annotated peaks overlapped between the ChIP-seq and CUT&Tag assays (Supplementary Fig. 5G).
Furthermore, we divided the genome into 1 kb bins and calculated the Pearson correlation (R) of signal intensity between each pair of assays. The R values among the three CUT&Tag datasets ranged from 0.89 to 0.93 (Supplementary Fig. 5H), consistent with a published report46. In contrast, the R values between Dnmt3b CUT&Tag and ChIP-seq assays were slightly lower, ranging from 0.75 to 0.80 (Supplementary Fig. 5H). To exclude potential ATAC-seq-like artifacts from the Dnmt3b CUT&Tag assays, we generated corresponding ATAC-seq data in WT EBs and identified 74,410 peaks using the same criteria as in CUT&Tag (fold change ≥ 4, Q ≤ 0.01, Supplementary Fig. 5F). The correlations between CUT&Tag and ATAC-seq signals, with R values ranging from 0.57 to 0.65, were lower than those observed among Dnmt3b CUT&Tag biological replicates (R = 0.89–0.93) and between CUT&Tag and ChIP-seq (R = 0.75–0.8) (Supplementary Fig. 5H and 5I). Additionally, we analyzed signal distributions across 14,461, 16,684, and 17,170 target genes from three Dnmt3b CUT&Tag assays, 14,751 target genes from the Dnmt3b ChIP-seq assay, and 18,120 target genes from the ATAC-seq assay (Supplementary Fig. 5J). In contrast to the ATAC-seq assay, all Dnmt3b binding profiles of CUT&Tag or ChIP-seq showed greater enrichment in gene body regions, consistent with published data47.
Furthermore, Dnmt3b CUT&Tag and ChIP-seq analyzes showed that Dnmt3b bound to the Fgf5 PP and enhancers, and the binding pattern was highly concordant with Smad2/3 binding genome-wide (Fig. 5A-B and Supplementary Fig. 6A). These binding peaks of Dnmt3b, especially at enhancer and GB regions, were strongly disrupted by Dnmt3b-shRNA (Fig. 5A and Supplementary Fig. 6A). In addition, Dnmt3b binding at the GB region, marked by H3K36me3, of Fgf5 was also confirmed by CUT&Tag and ChIP-qPCR (Fig. 5A and Supplementary Fig. 6B). Although Dnmt3b binding to enhancer and GB is consistent with previous reports44,47, its binding to PP was unexpected (Fig. 5A and Supplementary Fig. 5J), since earlier studies confirmed the absence of Dnmt3b around transcription start sites (TSS)47. At present, we cannot rule out the possibility that this could be potentially caused by biased binding towards nucleosome-depleted euchromatin generated by the CUT&Tag46. In comparison to the one using anti-immunoglobulin G (IgG) as a negative control, ChIP-qPCR showed that the binding of Dnmt3b to Fgf5 was slightly reduced in S2/3DKO compared to its presence in WT and S4KO EBs, probably related to the reduced expression of Dnmt3b in S2/3DKO (Fig. 5F). Addition of SB or AC didn’t affect the binding of Dnmt3b to Fgf5 in all the samples. Remarkably, the bindings of Smad2/3 to Fgf5 depended on AC stimulation in WT and S4KO EBs (Fig. 5F). Moreover, Smad2/3 exhibited a high concordance with Dnmt3b on the PP and enhancer regions of Fgf5, but not in GB (Fig. 5F). As a control, no Smad2/3 signal was detected if ChIP-qPCR was done with anti-IgG in WT or with anti-Smad2/3 in S2/3DKO EBs.
Although both Dnmt3a and Dnmt3b colocalized on Fgf5, the binding signals in GB disclosed with anti-Flag in Dnmt3b-Flag cells became less detectable in Dnmt3a-Flag cells (Fig. 5A). This discrepancy in binding preference was also evident in the ChIP-seq data for Dnmt3b and Dnmt3a (Fig. 5A). Furthermore, as shown on the heatmap, all the binding sites of Dnmt3a-Flag and Dnmt3b-Flag were analyzed and divided into four clusters (C1-C4) (Supplementary Fig. 6C). Epiblast genes were highly enriched in C1 and C2, while in C3 and C4, Dnmt3a and Dnmt3b showed different binding abilities (Supplementary Fig. 6C). Collectively, these findings indicate the overlapping yet distinct functions of Dnmt3a and Dnmt3b.
Since we found that Dnmt3b and Smad2/3 converge on cis-regulatory elements of epiblast specification genes, we wondered whether Dnmt3b enables Smad2/3 binding to these genes. Therefore, Smad2/3 CUT&Tag was performed in S4KO EBs with either Scr-sh or 3b-sh2 (Fig. 5C). The bindings of Smad2/3 to sites in Fgf5, Dnmt3b, and Dnmt3a in S4KO EBs were consistent with the observations in Smad2/3 ChIP-seq (Figs. 2E and 5C). However, Dnmt3b depletion inhibited Smad2/3 binding to a subset of these sites in the epiblast genes (Fig. 5C). Moreover, 7926 Smad2/3 binding peaks were strongly diminished in S4KO + 3b-sh2 compared to those in S4KO+Scr-sh (Figs. 2C and 5D). These Dnmt3b-dependent Smad2/3 binding peaks were analyzed, revealing a high enrichment of epiblast specification genes (Fig. 5E). Consistently, ChIP-qPCR confirmed that the binding of Dnmt3b to Fgf5 was reduced in S4KO + 3b-sh1 or 2 compared to S4KO+Scr-sh EBs, which subsequently led to fewer Smad2/3 molecules being able to access the Fgf5 gene (Fig. 5F). As a control, Dnmt3b shRNA had a mild effect on Smad2/3 binding to Fgf5 in WT EBs (Supplementary Fig. 6D). Taken together, Dnmt3b potentially facilitates Smad2/3 binding to epiblast genes including Fgf5, and this requirement is particularly amplified in the context of S4KO.
Discussion
Our research establishes the role and relevance of Smad2/3, Smad4 and Dnmt3b in the stepwise progression of early post-implantation development (Fig. 6). First, Smad2/3-Dnmt3b plays a pivotal role in orchestrating the transition from naïve pluripotency to the primed epiblast stage. Then, the canonical Smad2/3-Smad4 complex governs the differentiation of the primed epiblast into mesendoderm. We find that Smad2/3 can upregulate Dnmt3b during the naïve-to-primed transition. Dnmt3b, in turn, establishes DNA methylation patterns and ensures the timely downregulation of pluripotent factors like Sox2 by methylating DNA at DMRs. Additionally, Dnmt3b interacts with Smad2/3 to facilitate their binding to the hypomethylated centers of PP and enhancers of epiblast genes, likely relying on H3K36me2/3. Therefore, in the context of lineage priming in S2/3DKO where Dnmt3b is downregulated, Smad4 alone cannot initiate the transcription of epiblast-specific genes. Subsequently, the primed EpiLCs differentiate into mesendoderm lineages, a process that requires both Smad2/3 and Smad4. During the differentiation, Dnmt3b is downregulated and becomes less actively involved. Thus, S4KO cells are able to undergo the priming process but face difficulties in downstream differentiation. Smad4 loss intensified the interaction between Dnmt3b and Smad2/3 in the expanded population of EpiLCs or pre-differentiation cells, particularly considering the competitive binding of Dnmt3b and Smad4 to Smad2/3. Our two-step model is also consistent with the in vivo data5,33,34. Conditional knockout of Smad4 in the epiblast using the Cre-LoxP system does not prevent the development from the epiblast to the gastrulation initiation but shows focal defects in the primitive streak5. As gastrulation proceeds, Smad4-deficient embryos fail to form derivatives of the anterior primitive streak, including definitive endoderm, sharing many phenotypic similarities with the downregulation of the Nodal/Smad2/3/Foxh1 pathway5,33,34. Thus, Smad4 has a limited role during the priming process but is required for primitive streak/mesendoderm differentiation. These findings highlight the distinct roles of Smad2/3 and Smad4 in the process of lineage priming and differentiation, providing a fresh viewpoint on the complex mechanisms underlying TGFβ signaling. In addition, Dnmt3b is identified as a newly discovered partner of receptor-phosporylated Smad2/3, thereby shedding light on their collaborative regulatory mechanism underlying the priming process.
In the canonical TGFβ signaling pathway, TGFβ ligands bind to TGFβ receptors. Type II receptors phosphorylate and activate Type I receptors, which then phosphorylate Smad2/3. Phosphorylated Smad2/3 form a trimer either with or without Smad4, and transfer into nucleus. First, Smad2/3 play a dominant role in orchestrating the transition from the naïve mESC to the primed epiblast stage by inducing Dnmt3b. On one hand, Dnmt3b establishes DNA methylation patterns and ensures the timely downregulation of pluripotent factors like Sox2 by methylating its DNA at DMRs. On the other hand, Dnmt3b interacts with Smad2/3 to facilitate their binding to the hypomethylated centers of promoters and enhancers of epiblast genes, likely relying on H3K36me2/3. Second, Smad2/3-Smad4 complex plays a key role during the mesendoderm differentiation from the primed EpiLCs, in which Dnmt3b is downregulated and becomes less actively involved. The Smad2/3-Smad4 complex binds to the promoters and enhancers of mesendoderm genes, with the prebound pioneer factor Foxh162.
Of the eight Smad family members in vertebrate, Smad2 and Smad3 are substrates of TGFβ, Nodal and Activin receptors, whereas Smad1, Smad5 and Smad8 are activated by BMP and other ligands. The C-terminal phosphorylation of these receptor-regulated Smads (R-Smads) by receptor kinases generates a docking site for Smad4, enabling the formation of signaling complexes48. Smad4 has been recognized as the key effector in the canonical TGFβ pathway. However, much evidence indicates that R-Smads may substitute for or bypass the requirement for Smad4 to stimulate some TGFβ downstream signals4,5,6,7,10,34,49. Under these conditions, Smad4 appears to potentiate TGFβ signaling, but are unnecessary for it per se. In addition, TGFβ also signals via the mitogen-activated protein kinases (MAPKs), phosphatidylinositide 3-kinase (PI3K), Protein phosphatase 2 A (PP2A) and Rho family members and so on. Some of these pathways are entirely independent of Smad proteins, while others involve crosstalk with Smad signaling6.
Our understanding of signaling through R-Smads independently of Smad4 has remained limited for many decades. Trim33 (TIF1γ) has been reported as a partner of activated Smads. Smad2/3-Trim33 and Smad2/3-Smad4 function as complementary branches in the control of hematopoietic cell fate and pluripotent stem cell differentiation50,51. Despite Smad4 and Trim33 competing for binding with pSmad2/3, both complexes are essential and collaborate to fulfill their functions. But the mechanisms by which Smad2/3 alone transmit TGFβ signaling, especially in the absence of Smad4, are not yet understood. We addressed this question by using S2/3DKO and S4KO mESCs. Through a comprehensive analysis of high-throughput sequencing data and IP-MS experiments, we provide evidence suggesting that Dnmt3b potentially acts as a substitute for Smad4 in facilitating Smad2/3 binding and activating epiblast-specific genes. The formation of Smad2/3-Dnmt3b complex relies on TGFβ stimulation. In S4KO EBs, it seems that Dnmt3b can bind to the basal-state Smad2/3 under SB treatment when compared to WT, though much less robust than its binding to activated Smad2/3. Interestingly, Smad2/3-Dnmt3b is barely detectable when Smad2/3-Smad4 complex is present in EBs. We conclude that EBs lacking Smad4 couldn’t differentiate properly compared to WT. Therefore, those S4KO EBs stay in the epiblast stage and maintain a high level of Dnmt3b, which facilitates the interaction of Smad2/3 and Dnmt3b in the nucleus. When Smad2/3 or Dnmt3b were overexpressed either in 293 T cells for co-IP or in E. coli for pull-down assays, their interactions were further confirmed. Furthermore, the genome-wide binding patterns of Dnmt3b and Smad2/3 show a high concordance, especially at epiblast genes.
The effects of DNA methylation on gene expression are multifaceted and context-dependent. DNA methylation can suppress gene transcription, especially in CpG islands of promoters or in enhancers with repressive histone modifications. In contrast, Dnmt3b interacts with H3K36me2/3 by its PWWP domain to promote DNA methylation at GB and enhancer body, which correlates to active transcription of both regions41,42,43,44. Our study links Dnmt3b and Smad2/3 on the regulation of epiblast genes. This includes two layers of regulation: 1) DNA methylation is well established by Dnmt3b during the lineage priming process of WT and S4KO cells; 2) The presence of Dnmt3b on epiblast genes enables Smad2/3 access without the help of Smad4.
The upregulation of Dnmt3b during epiblast maturation is partially regulated by Smad2/3. Proper methylation levels are important for the transition from naïve pluripotency to the primed epiblast status. For example, the downregulation of pluripotent factor Sox2 during the exit from pluripotency is regulated by Dnmt3b-dependent methylation14. Meanwhile, Dnmt3b and Nodal-activated Smad2/3 cooperate in biding to common cis-regulatory elements in epiblast genes, such as Fgf5, a widely used marker for the epiblast in post-implantation embryos and epiblast stem cells28,52,53, driving this crucial developmental transition of post-implantation. Interestingly, when Dnmt3b is ablated, activated Smad2/3 lose access to the PP and enhancer regions of Fgf5, thereby blocking Fgf5 transcription. The present findings provide an expanded view of the synergistic properties of the signaling pathway and the epigenetic regulatory network, highlighting a previously unidentified level of interdependency between Smad2/3 and Dnmt3b factors.
It is worth noting that Dnmt3b exhibited a broad binding pattern in ChIP-seq data but displayed a more punctate pattern in CUT&Tag data. This discrepancy makes it difficult to ascertain whether the difference arises from the improved resolution of the CUT&Tag method or if it indicates that CUT&Tag may not be the most suitable assay for studying regulators, especially those with global functions. While Dnmt3b binding to enhancers and GB aligns with previous reports44,47, its binding to PP regions contradicts its known absence from TSS of actively transcribed genes and is inconsistent with its poor correlation with promoter-enriched H3K4me347. Currently, we cannot rule out the possibility that these observations are influenced by Tn5-directed bias toward binding in open chromatin regions during the CUT&Tag assay. Future studies may require improved technologies or methods. Given the challenging kinetics and cellular heterogeneity inherent to stem cell-based differentiation assays, as well as the rapid and potentially overlapping transitions from naïve to primed pluripotency and subsequent differentiation, clarifying how Dnmt3b precisely interacts with and facilitates the Nodal signaling pathway remains difficult and requires further investigation.
Although Smad2/3 are rarely mutated, Smad4 is frequently inactivated in various cancers54,55,56. This is often considered an indication of the silencing or dysregulation of TGFβ effects in those cancers. Recently, several reports highlight the role of Smad2/3 independently of Smad4 during tumorigenesis. Smad2/3 could behave as oncogenic factors of TGFβ in a Smad4-null human pancreatic ductal adenocarcinoma (PDAC) cancer cells11. Smad4 is necessary for epithelial-to-mesenchymal transition (EMT) but dispensable for Sox4 induction by TGFβ-Smad2/3 in PDAC4. It is of high importance to investigate whether the collaborative function of Smad2/3 and Dnmt3b that we discovered in mESCs also at play in tumors and in various contexts.
Methods
Cell culture and differentiation
E14Tg2a.IV mouse embryonic stem cells (ATCC, CRL-1821) were maintained in LIF-supplemented medium, adhering to the 0.1% gelatin coated plates as reported before20. For embryoid body (EB) formation and differentiation, mESCs were cultured on low attachment plates for 3–4 days in LIF-free medium20.
For EpiLC experiment, mESCs were first cultured in 2i/LIF medium (PD0325901, 1μM, Cayman, 13034; CHIR99021, 3μM, Cayman, 13122; LIF, 103U/ml, Millipore, ESG1107) and seeded as a density of 2 × 104 per cm2 on fibronectin-coated (Fibronectin, Millipore, FC010) plates for at least 3 days. Then the medium was changed into N2B27 supplemented with Activin A (20 ng/ml, R&D, 338-AC) and bFGF (12 ng/ml, R&D, 3139-FB) for 6 days. Medium was changed every day, and cells were passaged with Accutase on every other day at a ratio of 1:4. Subsequently, the medium was replaced with N2B27 plus 3μM CHIR99021 (Cayman, 13122) for 24 h to induce the formation of mesendoderm lineage.
HEK293T cells (ATCC, CRL-3216) were maintained in Dulbecco’s modified eagle medium (DMEM), supplemented with 10% FBS. Plasmids were introduced into 293T cells by transfection with Polyethyleneimine. Then cells were cultured in medium contained Hygromycin B or Puromycin for 48 h. Flag immunoprecipitation of whole-cell lysates were performed after treatment with Activin A for 2 h.
Immunoprecipitation and Mass spectrometry
EBs were collected at D3 either not treated or treated with Activin A (AC, 50 ng/ml, R&D, 338-AC) for 2 h or SB431542 (SB, 10 μM, Tocris, 1614) for 4-6 h. Whole-cell lysates were released using the buffer with 20 mM HEPES pH 7.9, 0.1% NP-40, 10% glycerol, 100 mM KCl, 2.5 mM MgCl2 supplemented with protease inhibitor and phosphatase inhibitor. For mass spectrometry, hypotonic buffer (10 mM HEPES pH7.5, 10 mM KCl, 1.5 mM MgCl2, 0.5% NP-40 supplemented with 1 mM DTT) was used to separate cytoplasm. Then nuclear extraction buffer (20 mM HEPES pH7.5, 300 mM NaCl, 2.5 mM MgCl2, 0.2% TritonX-100, 10% glycerol, 1 mM DTT) and RNase-Free DNase I (Lucigen, D9905K) was used. Chromatin fraction was further extracted from nuclear samples without DNase I treatment by E3 buffer (50 mM Tris-HCl pH 7.5, 20 mM NaCl, 1 mM MgCl2, 1% NP-40) plus benzonase (Millipore, 70746)57.
Cell lysate was further incubated with anti-Smad2/3 (Cell signaling technology, 8685S), anti-Smad2 (Cell signaling technology, 5339S), anti-Smad3 (Cell signaling technology, 9523S), anti-Otx2 (Proteintech, 13497-1-AP), anti-Flag M2 Affinity Gel (sigma, A2220) or anti-HA magnetic beads (Pierce, 88837) overnight at 4 °C. The next day Dynabeads Protein G (Invitrogen, 10009D) was added for 2 h (h) before washing and elution. Peptide/protein identification was analyzed and processed using Proteome Discoverer 2.5 software. For mass spectrometry analysis, we initially identified proteins enriched in S4KO + SB, S4KO, and S4KO + AC with normalized abundances > 50 and a fold change > 3 compared to WT. Subsequently, we identified proteins enriched in S4KO + AC and S4KO with a fold change > 1.5 compared to S4KO + SB. The logical relationships between all these comparisons were illustrated using a Venn diagram.
Methylated DNA Immunoprecipitation
Genomic DNA (gDNA) was extracted from D3 EBs using genomic DNA extraction kit (TIANGEN) (Cat # DP304). 4 μg gDNA was cut with a methylation-insensitive restriction enzyme BfaI (NEB, R0568) at 37 °C overnight. 500 ng of purified BfaI cut products was heat denatured at 99 °C for 10 min and quickly put on ice to obtain ssDNA. 10% was taken as input. Samples were incubated with 1 μg 5-mC antibody (Active motif, 39649) for 3 h at 4 °C, subsequently added magnetic beads for another 3 h at 4 °C. 5-mC enriched DNA fragments were eluted from magnetic beads by buffer containing 0.28 mg/ml proteinase K, 50 mM Tris-HCl, 10 mM EDTA, 0.5% SDS for 30 min at 55 °C, and purified by QIAquick PCR Purification Kit (Qiagen, 28106). Obtained DNA was analyzed by qRT-PCR, and the 5-mC levels were presented as percentages of input. Primers used in MeDIP-qPCR were listed below: 1) mSox2_DMR1: CTTCTTGGGCGTCCTGGG and GACTCCTTCGCCGACAATTC; 2) mSox2_DMR3: GTCACTTCTCTGCCCACTCT and ATGTGCCCGACTCCTGTTAA.
Immunofluorescence
mESCs were seeded on 0.1% gelatin coated microscopical cover glasses for 2D EBs culturing in LIF-free medium for 3-4 days. Cells were fixed with 4% PFA for 15 min at room temperature. After membrane permeabilization by 0.5% TritonX-100, slices were blocked with 2.5% BSA for 1 h at room temperature and incubated with Dnmt3a antibody (CST, 49768S), Dnmt3b antibody (CST, 48488S), Flag antibody (Sigma, F3165) overnight at 4 °C. For 5-mC antibody (Active motif, 39649) staining, fixed cells were incubated with 4 M HCl for 30 min at room temperature, following neutralization by 100 mM Tric-HCl (pH 7.5–8) at room temperature before blocking. The secondary antibody Alexa Fluor 555 against mouse (Invitrogen, A31570) or 488 against rabbit (Invitrogen, A21206) was added for 1 h at room temperature. Nuclear DNA was labeled by DAPI for 20 min. Images were acquired with Nikon confocal microscopy.
Plasmids
Lentiviral infections and plasmid transient transfection have already been described before51. CRISPR/Cas9 system was used to generate knockout and knock-in cell lines. sgRNA target sequence for Smad4 knockout was AGTTTGATGTGTCATAGACA. The sgRNA target sequences including TGACCGGCACGCTCCACGAT and TCTGATGACCGGTACACTCC were used for inserting a 3xFlag-tag at the C terminus of Dnmt3a or Dnmt3b, respectively. sgRNA oligos were cloned into pSpCas9(BB)−2A-puro (PX459) vector58. Donor plasmid contained about 800 bp homologous arms on each side. These plasmids were transiently delivered into cells by Lipofectamine 2000 (Invitrogen, 11668019) or Mouse ES Cell Nucleofector® Kit (Lonza, VPH-1001). S2/3DKO has been published before 22and S2/3/4TKO cells were constructed based on S2/3DKO.
Lentivirus-mediated shRNA system was an efficient mean of silencing gene expression. Two different shRNA target sequences for mouse Dnmt3b were GCTGTTCAGCCAGCACTTTAA and GCCTCAAACCCAACAAGAAGC. Designed shRNAs were cloned into LKO.1 vector (Sigma-Aldrich) by restrictive endonuclease AgeI/EcoRI. Scramble shRNA was used as a negative control. 2nd generation lentiviral system (psPAX2 and pMD2.G) (Addgene) was used to carry out the infection. The efficiency of shRNA system was evaluated by qRT-PCR.
To generate Smads protein expression plasmid, the ORFs of Smad2 and Smad4 were cloned into pLVX-EF1α-IRES-mCherry and pLVX-EF1α-IRES-zsGreen vector (Clontech), respectively. Selection marker on pLVX-Tight-Puro vector was substituted with Hygro. And then Smad3 OFRs were cloned into pLVX-Tight-Hygro. The HA-tag has been added at the N-terminal of Smads proteins. Furthermore, the CMV promoter in pLVX-Tight-Puro has been replaced by pGK promoter to avoid silencing in mouse embryonic stem cells20. The ORFs of Dnmt3b amplified from mESCs cDNA with N-terminal Flag tag was cloned into pLVX-EF1α-IRES-zsGreen vector. To prevent silencing by Dnmt3b-shRNA, synonymous mutations were introduced into Dnmt3bWT plasmid. Dnmt3bMet-mut and Dnmt3bPWWP-mut were constructed based on Dnmt3bWT expression plasmid. Point mutations of Dnmt3b were inserted using Mut Express MultiS Fast Mutagenesis Kit V2 (Vazyme, C215) and primers were listed below.
Dnmt3bMet-mut: GGAAGCGGTACCAATGATCTCTCTAACGTC and GATCATTGGTACCGCTTCCACCAATCACCAA;
Dnmt3bPWWP-mut: TCCTGGTCGACCGCCATGGTGGTGTCCTGGAAA and CATGGCGGTCGACCAGGAGAAGCCCTTGATC.
Immunoblotting
Cell pellets were lysed by RIPA buffer. The protein concentration was measured using BCA Protein Assay Kit (Pierce, 23225). Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked in 5% BSA and immunoblotted with primary bodies against Smad2/3 (Cell signaling technology, 8685S), Smad4 (Santa Cruz, sc-7966X), pSmad2/3 (Cell signaling technology, 8828S), Dnmt3a (abcam, ab13888), Dnmt3b (abcam, ab13604), HA (3F10, Roche, 11867423001), Eomes (abcam, ab23345), H3 (abcam, ab1791), Otx2 (Proteintech, 13497-1-AP), Flag (sigma, F3165), 6xHis tag (abcam, ab18184), GST tag (Proteintech, 10000-0-AP) and α-Tubulin (Sigma-Aldrich, T6074) overnight. On the second day, after washing, the membranes were washed and probed with IRDye 680RD or 800CW Goat anti-Rabbit/Mouse/Rat secondary body (LI-COR) and imaged using the Odyssey CLX system.
Alkaline phosphatase staining
The mESCs were fixed with 4% PFA for 10 min at room temperature. Subsequently, they were immersed in an alkaline phosphatase substrate solution (Vector Laboratories, SK-5300) for a few minutes. Degrees of blue show the activity of alkaline phosphatase on mESCs surface.
Cell growth curves
mESCs were seeded in 48-well plates at a density of 2 × 103 per well and cultured for 7 days. Cell numbers were recorded every 24 h and each data point represents the average of three replicates.
Cell cycle analysis
2 × 106 cells were used for cell cycle analysis. Washed with cold PBS twice, cell pellets were resuspended in 70% ethanol for fixing overnight at 4 °C. The second day cells were washed with cold PBS twice to remove ethanol, following 100 μg/ml RNAse A (final concentration) treatment for 30 min at 37 °C to eliminate RNA interference with DNA. Finally, cells were stained with 50 μg/ml propidium iodide (PI) in dark for 15 min at room temperature. Then samples were assayed by flow cytometry, and the data were analyzed by ModFit LT software to obtain fitting for cell cycle fraction data.
RNA extraction and RNAseq analysis
RNA was extracted with NucleoZOL (MACHEREY-NAGEL, 740404) following the manufacturer’s instructions. 1 μg total RNA from each sample was used for cDNA synthesis with All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (Transgen, AT341). Quantitative PCR was performed with ChamQ SYBR qPCR Master Mix (Vazyme, Q311) on a Bio-Rad CFX384 RT-PCR System. GAPDH served as an internal control for calculating relative expression. Sequences of synthesized primers used for qRT-PCR assays could be found in the previous report20 or list below: 1) Dnmt3a: GAGGGAACTGAGACCCCAC and CTGGAAGGTGAGTCTTGGCA; 2) Dnmt3b: AGCGGGTATGAGGAGTGCAT and GGGAGCATCCTTCGTGTCTG; 3) Esrrb: GCACCTGGGCTCTAGTTGC and TACAGTCCTCGTAGCTCTTGC; 4) Fgf5: AAAACCTGGTGCACCCTAGA and CATCACATTCCCGAATTAAGC; 5) Oct4: CTGAGGGCCAGGCAGGAGCACGAG and CTGTAGGGAGGGCTTCGGGCACTT; 6) Sox2: GCGGAGTGGAAACTTTTGTCC and CGGGAAGCGTGTACTTATCCTT; 7) Sox3: GCCGACTGGAAACTGCTGA and CGTAGCGGTGCATCTGAGG; 8) Sox4: GACAGCGACAAGATTCCGTTC and GTTGCCCGACTTCACCTTC; 9) Gapdh: CTCCACTCACGGCAAATTCA and CGCTCCTGGAAGATGGTGAT.
For RNAseq, multiplexed sequencing libraries were run on a Hiseq2500 platform. Sequencing reads from each sample were mapped to the mouse reference genome (mouse genome mm10) by using the HISAT2 v2.2.1 tool (https://github.com/DaehwanKimLab/hisat2). Stringtie v2.2.1 software (https://ccb.jhu.edu/software/stringtie/) was applied to generate the gene counts matrix for downstream analysis of DEGs and quantitate the gene expression as fragments per kilobase per million mapped reads (FPKM). Then, we used standard DEseq2 v1.26.0 workflow to identify DEGs and the P-values are attained by the Wald test and corrected (padj) for multiple testing using the Benjamini and Hochberg method by default. In any given comparison, only genes with P-value < 0.01 and the |log2 fold change (LFC) | > 1 were considered as DEGs and visualized by heatmap based on log-transformed and scaled gene expression. The built-in R function ‘prcomp’ was used to perform principal component analysis (PCA) for multiple datasets, and the first two PCs were visualized by scatter-plot. We combined all the DEGs for downstream Weighted Correlation Network Analysis (WGCNA) to dissect different gene expression module.
We used the R package WGCNA v1.70-359 to construct the genes co-expression network. In brief, the gene expression matrix was used to calculate Pearson’s correlation matrices representing co-expression similarity of genes. Then, the weighted adjacency matrix was created by the Pearson correlation coefficient test and transformed into a topological overlap measure (TOM) matrix, which minimize the effects of noise and spurious associations. Based on the average linkage hierarchical clustering of the TOM matrix, a clustering dendrogram of genes was constructed. The minimal gene cluster/module size was set to 30 and the threshold to merge similar modules was set to 0.25. Finally, we defined 7 gene clusters with different gene expression patterns among multiple samples, which were visualized by heatmap with log-transformed and scaled gene expression.
Functional annotation of genes
We used R package clusterProfiler v3.14.360 to perform Gene Ontology (GO) analysis for DEGs or genes of interest. The results were annotated along ontology of biological processes with the following parameters: pvalueCutoff = 0.01, pAdjustMethod = “BH” (Benjamani and Hochberg) and qvalueCutoff = 0.05. Gene set variation analysis (GSVA) is a particular type of gene set enrichment method that estimates the variation of pathway activity over a sample population in an unsupervised manner. Then, we used R package GSVA v1.34.061 to calculate the enrichment score of gene sets of interest from the published paper24,25.
Chromatin immunoprecipitation
ChIP was performed as previously described20. Cells were crosslinked with 1% formaldehyde (Sigma-Aldrich, F8775) for 10 min at 37 °C, then quenched with 125 mM Glycine for 5 min at room temperature. Cells were sonicated on Branson SFX550 Sonifier to disrupt genomic DNA. 1-2% pre-cleared chromatin prior to primary antibody addition was kept as input DNA. Then samples were incubated with 3-5 μg antibody bound to Dynabeads protein G (Life Technology) overnight at 4 °C. Cross-linked ChIP DNA was reversed and purified by QIAquick PCR Purification Kit (Qiagen, 28106). Immunoprecipitated DNA was analyzed by qRT-PCR, and the content was presented as percentage of input. Primers for ChIP-qPCR were listed below. 1) Fgf5_PP: CGGCCGTGAGTACACAAA and GCTGGCACCGGGATATTTA; 2) Fgf5_GB: GCTGTGTCTCAGGGGATTGTAG and CCGGTGGAGCACTTTTCAACA; 3) Fgf5_+31k: CACCTCCCACTGTTGGTCTT and CACGGGGTAAGGTGCTTCTA; 4) Fgf5_+40k: AAGCAAAAACCCATCGGACA and GCACCCAGTAGGAACGCAAT; 5) Fgf5_+56k: GCTGGAATCTCTCTCCTGATTAC and AAGTTGGCTTGCTCAGTAGG.
For ChIP-seq, 10-50 ng of immunoprecipitated DNA were used to library preparation by VAHTS® Universal Pro DNA Library Prep Kit for Illumina (Vazyme, ND608). Quality control for the raw sequencing data was performed by fastqc v0.11.9 and cutadapt v3.4 to remove adapter sequences and low-quality sequences. Then, the clean reads were mapped to the mouse genome (mm10) using bowtie2 v2.2.4 with default parameters, and only uniquely and properly paired mapping reads were retained. The resultant BAM files were converted into bigWig files using bamCoverage tool from deepTools version 3.5.1 with default parameters. Peaks were called using macs2 v2.2.7 with the sinificance cut-off P-value ≤ 0.05 and retained when the fold change ≥ 3. The bedtools intersect command within bedtools v2.30.0 was used to report the overlap peaks between two samples. The annotatePeaks.pl function in homer v4.11.1 was used to annotate peaks with default parameters. The programs “computeMatrix” and “plotHeatmap” of deepTools v3.5.1 were used to compute the ChIP-seq signals over the region of interest and make visualization.
CUT&Tag
CUT&Tag was performed using a commercially available kit (Hyperactive Universal CUT&Tag Assay Kit for Illumina Pro, Vazyme, TD904). Briefly, 1 × 105 cells were harvested and crosslinked with 0.1% formaldehyde (Sigma-Aldrich, F8775) for 2 min at room temperature if necessary. E. coli DNA spike-in was added as 1 pg per 1 × 105 cells if needed. Sequencing was conducted on Illumina Novaseq Xplus with 150 bp paired-end reads. The analysis of CUT & Tag data parallels that of ChIP-seq data. After quality controlling, clean reads were aligned to the mouse genome (mm10). Peaks were identified using macs2 v2.2.7 with a significance cut-off of Q-value ≤ 0.01, and those with a fold change ≥ 4 were retained. The “computeMatrix” and “plotHeatmap” programs from deepTools v3.5.1 were employed for computing Cut & Tag signals across regions of interest and for visualization.
ATAC-seq
8 × 104 single-celled EBs were treated with NP-40 (10%), Tween-20 (10%), and digitonin (1%), and the reaction was stopped once the cell membrane ruptured. The Tn5 transposase (Illumina #FC-121-1030) was then incubated with the nuclei and mixed at 1000 RPM at 37 °C for 30 min. DNA fragments were subsequently purified using the DNA Clean and Concentrator kit (Zymo #D4014). DNA fragments were amplified using NEBNext® High-Fidelity 2X PCR Master Mix (NEB #M0541L) and index adapters, followed by purification to obtain the final library. The library quality was assessed using Agilent Bioanalyzer technology by Personalbio. Sequencing was performed on the Illumina NovaSeq XPlus platform. Peaks were identified using macs2 v2.2.7 with a significance cut-off of Q-value ≤ 0.01, and those with a fold change ≥ 4 were retained.
Proximity ligation assay
Proximity ligation assay (PLA) was carried out according to the manufacturer’s protocol (Duolink® PLA Fluorescence Protocol, Sigma). D3 EB cells cultured on microscopical cover glasses were collected for PLA. Primary antibodies were Smad2/3 (Cell signaling technology, 8685S) and Dnmt3b (abcam, 13604). Probe anti-Rabbit PLUS, anti-Mouse MINUS, and detection reagents green were used. Confocal images were taken by Nikon confocal microscopy.
In vitro protein-protein interactions
GST, GST-GFP, GST-fused full length of Dnmt3b and 6xHis-tagged Smad2/3 were expressed using pGEX-4T1 and pET28a vectors in BL21 cells, respectively. These proteins were incubated with Ni-NTA (20503ES10, Yeasen) or Glutathione Agarose Resin (20507ES10, Yeasen), as indicated in the text.
Statistical analysis
Quantitative data are expressed as mean ± SD. Statistical significance was determined using a Mann-Whitney test, a two-way ANOVA test or a paired-t test using Prism 9 software as indicated in the text.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All RNA-Seq, ChIP-Seq, CUT&Tag and ATAC-seq data were deposited in the Gene Expression Omnibus database under accession number GSE240787. Published ChIP-seq data for Smad2/3 and Smad4 in E14 under accession code GSE7048622, H3K4me1, H3K4me3 and H3K27ac in EpiLCs under accession code GSE16841114, H3K36me3 in E6.5 epiblast under accession code GSE12421645 were reanalyzed here. WGBS in mESC and EpiLC used GSE168415 data [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE168415]14. The raw and processed mass spectrometry-based proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD057083. Uncropped western blots were provided in Supplemental information files. Correspondence and requests for materials should be addressed to Q.W. (wangqiong@shsmu.edu.cn), Y.D. (yhdu@shsmu.edu.cn), and Y.Z. (yongchun_zhang@sjtu.edu.cn). Source data are provided with this paper.
References
Wu, J. W. et al. Crystal structure of a phosphorylated Smad2. recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol. Cell 8, 1277–1289 (2001).
Jayaraman, L. & Massague, J. Distinct oligomeric states of SMAD proteins in the transforming growth factor-beta pathway. J. Biol. Chem. 275, 40710–40717 (2000).
David, C. J. & Massague, J. Contextual determinants of TGFbeta action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19, 419–435 (2018).
David, C. J. et al. TGF-beta tumor suppression through a lethal EMT. Cell 164, 1015–1030 (2016).
Chu, G. C., Dunn, N. R., Anderson, D. C., Oxburgh, L. & Robertson, E. J. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development 131, 3501–3512 (2004).
Derynck, R. & Zhang, Y. E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425, 577–584 (2003).
Bardeesy, N. et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 20, 3130–3146 (2006).
Wiater, E. & Vale, W. Roles of activin family in pancreatic development and homeostasis. Mol. Cell. Endocrinol. 359, 23–29 (2012).
Fink, S. P., Mikkola, D., Willson, J. K. & Markowitz, S. TGF-beta-induced nuclear localization of Smad2 and Smad3 in Smad4 null cancer cell lines. Oncogene 22, 1317–1323 (2003).
Wisotzkey, R. G. et al. Medea is a Drosophila Smad4 homolog that is differentially required to potentiate DPP responses. Development 125, 1433–1445 (1998).
Bertrand-Chapel, A. et al. SMAD2/3 mediate oncogenic effects of TGF-beta in the absence of SMAD4. Commun. Biol. 5, 1068 (2022).
Auclair, G., Guibert, S., Bender, A. & Weber, M. Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse. Genome Biol. 15, 545 (2014).
Leitch, H. G. et al. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311–316 (2013).
Lauria, A. et al. DNMT3B supports meso-endoderm differentiation from mouse embryonic stem cells. Nat. Commun. 14, 367 (2023).
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Zhang, Q. et al. TGF-beta regulates DNA methyltransferase expression in prostate cancer, correlates with aggressive capabilities, and predicts disease recurrence. PLoS One 6, e25168 (2011).
Koh, H. B., Scruggs, A. M. & Huang, S. K. Transforming growth factor-beta1 increases DNA Methyltransferase 1 and 3a expression through distinct post-transcriptional mechanisms in lung fibroblasts. J. Biol. Chem. 291, 19287–19298 (2016).
Luo, X. et al. Cutting edge: TGF-beta-induced expression of Foxp3 in T cells is mediated through inactivation of ERK. J. Immunol. 180, 2757–2761 (2008).
You, H., Ding, W. & Rountree, C. B. Epigenetic regulation of cancer stem cell marker CD133 by transforming growth factor-beta. Hepatology 51, 1635–1644 (2010).
Wang, Q. et al. The p53 family coordinates wnt and nodal inputs in mesendodermal differentiation of embryonic stem cells. cell stem cell 20, 70–86 (2017).
Senft, A. D. et al. Combinatorial Smad2/3 activities downstream of nodal signaling maintain embryonic/extra-embryonic cell identities during lineage priming. Cell Rep. 24, 1977–1985 e1977 (2018).
Aragon, E. et al. Structural basis for distinct roles of SMAD2 and SMAD3 in FOXH1 pioneer-directed TGF-beta signaling. Genes Dev. 33, 1506–1524 (2019).
Massague, J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 (2012).
Spangler, A., Su, E. Y., Craft, A. M. & Cahan, P. A single cell transcriptional portrait of embryoid body differentiation and comparison to progenitors of the developing embryo. Stem Cell Res. 31, 201–215 (2018).
Kim, I. S. et al. Parallel single-cell RNA-Seq and genetic recording reveals lineage decisions in developing embryoid bodies. Cell Rep. 33, 108222 (2020).
Wang, X. et al. Formative pluripotent stem cells show features of epiblast cells poised for gastrulation. Cell Res. 31, 526–541 (2021).
Brons, I. G. et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 (2007).
Nichols, J. & Smith, A. Naive and primed pluripotent states. cell stem cell 4, 487–492 (2009).
Kalkan, T. et al. Tracking the embryonic stem cell transition from ground state pluripotency. Development 144, 1221–1234 (2017).
Boroviak, T. et al. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Developmental Cell 35, 366–382 (2015).
Buecker, C. et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. cell stem cell 14, 838–853 (2014).
Hayashi, K., Ohta, H., Kurimoto, K., Aramaki, S. & Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532 (2011).
Dunn, N. R., Vincent, S. D., Oxburgh, L., Robertson, E. J. & Bikoff, E. K. Combinatorial activities of Smad2 and Smad3 regulate mesoderm formation and patterning in the mouse embryo. Development 131, 1717–1728 (2004).
Sirard, C. et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 12, 107–119 (1998).
Iwafuchi-Doi, M. et al. Transcriptional regulatory networks in epiblast cells and during anterior neural plate development as modeled in epiblast stem cells. Development 139, 3926–3937 (2012).
Acampora, D., Di Giovannantonio, L. G. & Simeone, A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development 140, 43–55 (2013).
Soderberg, O. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006).
Nomura, M. & Li, E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature 393, 786–790 (1998).
Zhu, Y., Richardson, J. A., Parada, L. F. & Graff, J. M. Smad3 mutant mice develop metastatic colorectal cancer. Cell 94, 703–714 (1998).
Peng, G. et al. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell 36, 681–697 (2016).
Nowialis, P. et al. Catalytically inactive Dnmt3b rescues mouse embryonic development by accessory and repressive functions. Nat. Commun. 10, 4374 (2019).
Yin, Y. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 (2017).
Neri, F. et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature 543, 72–77 (2017).
Rinaldi, L. et al. Dnmt3a and Dnmt3b Associate with enhancers to regulate human epidermal stem cell homeostasis. cell stem cell 19, 491–501 (2016).
Hanna, C. W. et al. Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues. Genome Biol. 20, 225 (2019).
Kaya-Okur, H. S., Janssens, D. H., Henikoff, J. G., Ahmad, K. & Henikoff, S. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 15, 3264–3283 (2020).
Baubec, T. et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247 (2015).
Shi, Y. & Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685–700 (2003).
Sirard, C. et al. Targeted disruption in murine cells reveals variable requirement for Smad4 in transforming growth factor beta-related signaling. J. Biol. Chem. 275, 2063–2070 (2000).
He, W. et al. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell 125, 929–941 (2006).
Xi, Q. et al. A poised chromatin platform for TGF-beta access to master regulators. Cell 147, 1511–1524 (2011).
Khoa le, T. P. et al. Visualization of the epiblast and visceral endodermal cells using fgf5-p2a-venus bac transgenic mice and epiblast stem cells. PLoS One 11, e0159246 (2016).
Han, D. W. et al. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell 143, 617–627 (2010).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Miyaki, M. et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene 18, 3098–3103 (1999).
Zhao, M., Mishra, L. & Deng, C. X. The role of TGF-beta/SMAD4 signaling in cancer. Int. J. Biol. Sci. 14, 111–123 (2018).
Gillotin, S. Isolation of chromatin-bound proteins from subcellular fractions for biochemical analysis. Bio Protoc. 8, e3035 (2018).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559 (2008).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 14, 7 (2013).
Charney, R. M. et al. Foxh1 occupies cis-regulatory modules prior to dynamic transcription factor interactions controlling the mesendoderm gene program. Developmental Cell 40, 595–607 e594 (2017).
Acknowledgements
We thank all the members of the Q.W. lab for insightful discussions, L. Li, Y.Ye, B. Li from SHSMU, F. Chen (Fudan U.) and X. Liu (CAS) for valuable suggestions and technical advice, J. Massagué (MSKCC) for providing plasmids and cell lines. The proteomics and mass spectrometry work were performed at the Proteomics Platform of Core Facility of Shanghai Jiao Tong University. This work was supported by National Key R&D Program of China (2021YFA1100400 to Q.W. and 2022YFA0912600 to Y.Z.), National Natural Science Foundation of China (31771512 and 32070865 to Q.W.; 32170831 to Y.Z.), Shanghai Natural Science Foundation (20ZR1430400 to Q.W.; 23ZR1455300 to Y.D.), the Fundamental Research Funds for the Central Universities, and Startup Fund of SJTU School of Medicine to Q.W.
Author information
Authors and Affiliations
Contributions
Q.W., Y.D., and Y.Z. conceived and designed the project and wrote the manuscript. B.Z., X.Y., S.M., and S.L. performed the experimental work. Y.Z., H.S., S.X., and S.L. assisted with the experiments. Y.D., J.S., Y.Y., S.X., and S.L. performed the bioinformatic analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Aristidis Moustakas and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, B., Yu, X., Shi, J. et al. A stepwise mode of TGFβ-SMAD signaling and DNA methylation regulates naïve-to-primed pluripotency and differentiation. Nat Commun 15, 10123 (2024). https://doi.org/10.1038/s41467-024-54433-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54433-5
This article is cited by
-
Systematic evaluation of porcine platelet lysate as a fetal bovine serum substitute: enhancing cell proliferation, maintaining differentiation potential, and ensuring immunophenotypic stability
Stem Cell Research & Therapy (2025)
-
Advances in the regulation of macrophage polarization by the tumor microenvironment
Discover Oncology (2025)