Abstract
Autophagy, a crucial mechanism for cellular degradation, is regulated by conserved autophagy-related (ATG) core proteins across species. Impairments in autophagy result in significant developmental and reproductive aberrations in mammals. However, autophagy is thought to be functionally dispensable in Arabidopsis thaliana since most of the ATG mutants lack severe growth and reproductive defects. Here, we challenge this perception by unveiling a role for autophagy in male gametophyte development and fertility in Arabidopsis. A detailed re-assessment of atg5 and atg7 mutants found that reduced autophagy activity in germinated pollen accompanied by partial aberrations in sperm cell biogenesis and pollen tube growth, leading to compromised seed formation. Furthermore, we revealed autophagy modulates the spatial organization of actin filaments via targeted degradation of actin depolymerization factors ADF7 and Profilin2 in pollen grains and tubes through a key receptor, Neighbor of BRCA1 (NBR1). Our findings advance the understanding of the evolutionary conservation and diversification of autophagy in modulating male fertility in plants contrasting to mammals.
Similar content being viewed by others
Introduction
Autophagy is an essential and conserved catabolic pathway in eukaryotic cells that functions as a cellular scavenger by targeting of cytosol and organelles to specific vacuoles or lysosomes for degradation1,2. Macroautophagy, the most prevalent type of autophagy and referred to as autophagy hereafter, serves as a housekeeping quality control mechanism that generally takes place at a basal and low level3. When cells are challenged with biotic and/or abiotic stresses, the activity of autophagy will be significantly elevated to facilitate survival through the adverse conditions4,5. In addition, autophagy plays versatile roles in regulating cell development, growth, reproduction and senescence4,5,6,7.
The key processes of autophagy consist of de novo formation and expansion of autophagosomal membranes, sequestration of unwanted cytoplasmic elements into the autophagosome and its subsequent fusion with a vacuole or lysosome. It is evident that ATG (autophagy-related) proteins play key roles during autophagy and have been identified in various species including yeasts, animals and plants8. ATG proteins are spatiotemporally orchestrated to facilitate the biogenesis and functions of autophagosomes9. Moreover, ~18 ATG genes are found to be highly conserved across different species and therefore considered to be core ATGs for regulation of the autophagy machinery10. In particular, ATG5 and ATG7 are considered to be important because they are critically required for autophagosome formation. ATG5 mediates the formation of the autophagosome precursor by conjugation with ATG12 to form a protein complex that catalyzes the chemical binding between cytosolic ATG8 and phosphatidylethanolamine on the autophagosome membrane. ATG7 acts as an E1-like activating enzyme to facilitate the conjugation between ATG8-phosphatidylethanolamine (ATG8-PE) and ATG12 during autophagosomal membrane expansion. ATG5 and ATG7 are often considered to be the two key factors for ATG8 lipidation, which is a landmark event during autophagosome formation and autophagic flux11,12,13,14,15. ATG5- and ATG7-knockout mice died within one day after birth due to the reduced concentration of cytosolic amino acids, a deleterious condition associated with autophagy deficiency16,17. Furthermore, ATG5- and ATG7-mediated autophagy has been found to regulate spermiogenesis and male fertility in mice18,19. In plants, deletion of rice ATG7 showed severe defects in lipid metabolism and nutrient supply in anthers and caused complete sporophytic male sterility, despite the detailed underlying working mechanism remained yet to be uncovered20. Autophagy also participates in compartmental cytoplasmic deletion during tobacco pollen germination. RNA interference (RNAi) downregulation of NtATG5 and NtATG7 resulted in persistence of a layer of undegraded cytoplasm at the pollen germination aperture, which prevents extrusion of pollen tubes21. However, it is noteworthy that almost all of Arabidopsis atg mutants can survive well and accomplish their life cycle under ordinary cultivation conditions. Except for showing early senescence, Arabidopsis atg5 and atg7 mutant lines have no significant defects in growth and development and they successfully produce seeds. It is therefore believed that autophagy is not functionally required during Arabidopsis sexual reproduction10,22.
In this study, we report that ATG5- and ATG7-mediated autophagy does indeed functionally participate in the process of sexual reproduction by modulating sperm cell biogenesis and male fertility under nutrient-sufficient conditions in Arabidopsis. Despite previous study reports that Arabidopsis atg5 and atg7 mutants can produce functional seeds and present few defects in pollen tube germination in vitro, we performed systematic and detailed re-investigations on the processes of pollen germination, pollen tube growth and male fertility of atg5 and atg7 null mutants. Our results suggest that autophagy is functionally involved in: i) generation of two functional sperm cells that are derived from the mitotic division of the generative nucleus during pollen development; ii) maintenance of pollen tube growth and targeting to the ovules for fertilization; and iii) modulation of the spatial organization of actin microfilaments by specific interactions between NBR1 and actin filament depolymerization factors 7 (ADF7) and Profilin2 which the ZZ and FW domains of NBR1 are indispensable for its interaction in a ubiquitin-binding domain (UBA)-independent manner and are crucial for autophagic degradation. Taken together, our studies advance our understanding of the roles and underlying mechanisms of autophagy in modulating male gametophyte development and fertility in Arabidopsis thaliana.
Results
ATG5 and ATG7 participate in autophagy in growing pollen tubes
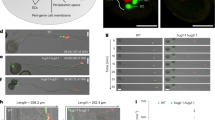
To investigate whether autophagy plays a role in sexual reproduction in Arabidopsis, we employed ATG5 and ATG7 T-DNA insertion mutants to follow and compare reproductive development with that of wild type (WT). The schematic representation of the T-DNA insertion site within atg5-1 and atg7-2 is shown in Fig. 1a. Quantitative reverse-transcription PCR (RT-qPCR) was performed to assess the gene expression levels of ATG5 and ATG7 in the atg5-1, atg7-2 and atg5-1/atg7-2 mutants respectively. ATG5 and ATG7 were barely detectable in these mutants (Fig. 1b, c). Moreover, we studied the gene expression profile of ATG5 and ATG7 in several different major cells and tissues of Arabidopsis by using the microarray data from Genevestigator (https://genevestigator.com/). Interestingly, we found that ATG5 and ATG7 are both enriched in pollens and stamens relative to other tissues in Arabidopsis (Supplementary Fig. 1). Therefore, we further performed western blotting to probe the level of autophagy activity in germinated pollens from both WT and the mutant Arabidopsis using an anti-ATG8 antibody. ATG8-PE is a reporter which indicates the recruitment of cytosolic ATG8 onto the autophagosomal membrane for the subsequent biogenesis and functions of autophagosomes9. As shown in Fig. 1d, ATG8-PE was barely detectable in atg5-1 and atg7-2 germinated pollens when compared with the WT. This result indicates that autophagy is greatly impaired in atg5-1 and atg7-2 mutant pollens.
a Schematic illustration of the T-DNA insertion sites of atg5-1 and atg7-2 of Arabidopsis thaliana. Red arrows mark the locations of primers used for RT-qPCR. b RT-qPCR analysis was performed to quantify ATG5 expression in flowers at developmental stages 11–14 of WT, atg5-1, atg5-1/atg7-2 and pATG5::GFP-ATG5/atg5-1. Statistical analysis was performed using two-tailed paired Student’s t-test, with n = 3 independent experimental replicates (error bars ± SD, ∗∗∗p < 0.001, ns, no significant). Source data are provided in Source data file. c Similarly, ATG7 expression was analyzed in flowers of WT, atg7-2, atg5-1/atg7-2 and pATG7::GFP-ATG7/atg7-2. Actin2 was used as an internal control. Statistical analysis was performed using two-tailed paired Student’s t-test, with n = 3 independent experimental replicates (error bars ± SD, ∗∗∗p < 0.001, ns, no significant). Source data are provided in Source data file. d Western blot detection of ATG8 and ATG8-PE in germinated pollen of WT, atg5-1 and atg7-2 using anti-ATG8 antibody. Three independent experimental replicates were conducted, yielding consistent results. Source data of blotting are provided in Source data file. e Co-expression of GFP-ATG5 with mCherry-ATG7, ATG6-RFP, YFP-ATG8e, GFP-ATG9 and SH3P2-RFP in growing tobacco pollen tubes respectively. The punctate dots of ATG5 and ATG7 colocalize with one another and they mainly localize in the pollen tube shank. Results of colocalization ratios are calculated by either Pearson correlation coefficients or as Spearman’s rank correlation coefficients with ImageJ. The generated r values in the range −1 to 1, where 0 indicates no discernable correlation and +1 or −1 indicate strong positive or negative correlations, respectively. Consistent results were obtained across three independent experimental replicates. Scale bar = 25 μm.
To better understand the subcellular localization and spatial distribution of ATG5 and ATG7 in pollen tubes, we generated chimeric fluorescent fusion proteins of ATG5 and ATG7 and co-expressed them with several core autophagic proteins, including ATG6, ATG8e, ATG9 and SH3P2 in growing tobacco pollen tubes. ATG5 and ATG7 mainly colocalized with one another as shown in Fig. 1e and Supplementary Movie. 1. Additionally, ATG5 colocalized with ATG6, ATG8e, ATG9 and SH3P2 (Fig. 1e and Supplementary Movies. 2-5). These results suggest that ATG5 and ATG7 are involved in autophagy processes in growing pollen tube.
The seed setting rate is reduced in atg5 and atg7 Arabidopsis mutants
To further explore the modulating functions of autophagy during the processes of sexual reproduction in Arabidopsis, we began by revisiting the overall phenotypes of plant growth and development during the reproductive stage. Consistent with previous findings, no significant differences were observed for atg5-1, atg7-2, atg5-1/atg7-2 and WT that had been cultivated for 35 and 45 d (Supplementary Fig. 2a, b). Nevertheless, we found longer length of siliques produced by atg5-1, atg7-2 and atg5-1/atg7-2 compared that of the WT (Supplementary Fig. 2c). To further examine these results, we introduced atg7-3, which is another loss-of-function mutant generated by T-DNA insertion into ATG7 as previously identified15. We observed an increased length of siliques (Supplementary Fig. 2c). Statistical measurement further confirmed the observed longer length of siliques in the mutants than that in the WT (Supplementary Fig. 2d).
In addition, it is noteworthy that a portion of seed formation is abolished within the siliques of atg5-1, atg7-2, atg7-3 and atg5-1/atg7-2 as indicated by the arrows (Fig. 2a). Further statistical analysis by counting seed numbers within the siliques revealed that the seed setting rate was actually reduced by ~20-25% in atg5-1, atg7-2, atg7-3 and atg5-1/atg7-2 mutants (Fig. 2b). Meanwhile, complementation via expression of ATG5 and ATG7 under their respective native promoters successfully rescued the phenotypes observed in atg5-1 and atg7-2 mutants, restoring seed setting rate to levels comparable to those in the WT (Fig. 2a, b). Next, we conducted time-lapse microscopy imaging of in vivo pollen tube fertilization to track pollen tube germination and growth in the WT and mutants of atg5-1, atg7-2 and atg5-1/atg7-2 after 2-, 4-, 6- and 10-h pollination. The positions of maximal pollen tube growth within the styles are highlighted by the dashed lines (Fig. 2c and Supplementary Fig. 3). The pollen tubes of atg5-1, atg7-2 and atg5-1/atg7-2 were significantly shorter than that of the WT after 2-h pollination (Fig. 2c). However, no obvious differences of pollen tube growth can be observed after 4-, 6- and 10-h pollination (Fig. 2c and Supplementary Fig. 3). Statistical analysis of the maximum lengths of pollen tubes within the styles of the WT and the mutants at different time points after pollination is shown in Fig. 2d. On the other hand, a previous independent study found that both the pollen tube germination ratio and the pollen tube length of atg5-1 and atg7-2 mutants were not distinct compared with WT after 10 h germination21. We revisited this analysis with a more detailed time-lapse microscopy of in vitro pollen germination by adding more germination time points for 2, 4 and 6 h. Our results demonstrated that the pollen germination and pollen tube length of atg5-1, atg7-2 and atg5-1/atg7-2 were significantly reduced when compared with WT at 2- and 4-h germinating time points (Fig. 2e). In fact, the pollen germination ratio of atg5-1, atg7-2 and atg5-1/atg7-2 reached only ~55% and ~80% in 2 and 4 h respectively, compared to nearly 80% and 90% for WT pollen at the relative time points (Fig. 2f). Meanwhile, the length of pollen tubes of atg5-1, atg7-2 and atg5-1/atg7-2 at 2-, 4- and 6-h germinated were obviously shorter compared with WT (Fig. 2g). In summary, pollen germination was assessed both in vivo and in vitro, ATG5- and ATG7-mediated autophagy indeed modulates pollen germination and tube growth in Arabidopsis thaliana. The pollen germination ratio and tube growth are all substantially reduced at the beginning of germination in atg5-1, atg7-2 and atg5-1/atg7-2.
a Representative images of siliques harvested from one branch of inflorescence of Arabidopsis WT, atg5-1, atg7-2, atg7-3, atg5-1/atg7-2, pATG5::GFP-ATG5/atg5-1 and pATG7::GFP-ATG7/atg7-2. Scale bars = 2 mm. b Statistical calculation of seeds setting rate in WT, atg5-1, atg7-2, atg7-3 atg5-1/atg7-2, pATG5::GFP-ATG5/atg5-1 and pATG7::GFP-ATG7/atg7-2. Statistical analysis was performed using two-tailed paired Student’s t-test, with n = 50 independent experimental replicates (error bars ± SD, ∗∗∗p < 0.001, ns, no significant). Source data are provided in Source data file. c Representative time-lapse images of in vivo pollen tube germination and growth of WT, atg5-1, atg7-2 and atg5-1/atg7-2 after pollination for 2 and 10 h respectively. Dashed lines indicate the farthest position that pollen tubes could reach into the styles. Scale bars = 400 μm. d Statistical calculation of the maximum pollen tube length in the styles of WT, atg5-1, atg7-2 and atg5-1/atg7-2 after pollination for 2, 4, 6 and 10 h were performed using two-tailed paired Student’s t-test respectively, with n = 50 independent experimental replicates from each sample (error bars ± SD, ∗∗∗p < 0.001 ns, no significant). Source data are provided in Source data file. e Representative time-lapse images of in vitro Arabidopsis pollen tube germination and growth of WT, atg5-1, atg7-2 and atg5-1/atg7-2 for 2, 4 and 6 h respectively. Scale bars = 200 μm. f, g Statistical analysis of the in vitro pollen germination ratio (f) and tube length (g) of WT, atg5−1, atg7-2 and atg5−1/atg7-2 were performed using two-tailed paired Student’s t-test For the pollen germination ratio, with n = 19 independent replicates were analyzed, while for the tube length, n = 200 independent replicates were analyzed for each sample (error bars ± SD, ∗∗∗p < 0.001, ns, no significant). Source data are provided in Source data file.
Autophagy modulates spatial organization and integrity of actin filaments and maintains sperm cell biogenesis and pollen tube growth
To explore the underlying cause(s) for the reduction of the seed setting rate for the mutants of atg5-1, atg7-2 and atg5-1/atg7-2, we examined sperm cell development in mature pollen from the WT, atg5-1, atg7-2 and atg5-1/atg7-2 plants (Fig. 3a). After 4’,6-diamidino-2-phenylindole (DAPI) staining for 30 min, two well developed sperm cells can be clearly observed in the WT pollen grains by confocal 3-D imaging (Fig. 3a). In contrast, ~20% of mature pollen from atg5-1, atg7-2 and atg5-1/atg7-2 strains contained only one undivided generative nucleus as indicated by the arrows in Fig. 3a. Statistical calculations showed that less than 2% of WT pollen showed defects in sperm cell development, whereas ~20% abnormal pollens were observed in the atg mutant lines (Fig. 3b). In addition, we investigated pollen tube growth and targeting to the ovules during fertilization. Pollen tubes in the WT plants can grow generally straight and directly target the ovules for fertilization (Fig. 3c). However, portions of atg5-1, atg7-2 and atg5-1/atg7-2 mutant pollen tubes grew aberrantly twisted and accumulated outside the ovules as indicated by the arrows in Fig. 3c. To further determine whether the altered pattern of pollen tube growth observed in the mutants is caused by an abnormality in the male or female gametophyte, we performed cross-pollination between the mutants and WT. We noticed a similar irregular and twisting pattern of pollen tube growth and targeting when atg5-1 and atg7-2 pollen was applied to the WT stigma (Fig. 3c). Conversely, the WT pollen tube grew and targeted to the ovules of atg5-1 and atg7-2 without any defects (Fig. 3c). These results suggest that aberrations in the male gametophyte cause the twisting and accumulation observed in pollen tube growth and its targeting to ovules during fertilization.
a Representative images of mature pollen from WT, atg5-1, atg7-2 and atg5-1/atg7-2 by DAPI staining. A representative image shows that WT mature pollens contain two sperm cells (SC) and a vegetative nucleus (VN). Some of the mutant pollen grains contain only one undivided generative nucleus (GN) as the arrow indicates. Scale bars = 10 μm. b Statistical calculation of abnormal pollen that lack two sperm cells from the mature pollen of WT, atg5-1, atg7-2 and atg5-1/atg7-2 were performed using two-tailed paired Student’s t-test respectively. The number of cells examined was n = 1617 (WT), n = 1591 (atg5-1), n = 1625 (atg7-2), and n = 1624 (atg5-1/atg7-2) across three independent experiments for each sample (error bars ± SD, ∗∗∗p < 0.001). Source data are provided in Source data file. c In vivo pollen tube growth and targeting to ovules for fertilization in WT, atg5-1, atg7-2, atg5-1/atg7-2, respectively. Twisted and abnormal accumulation of pollen tubes during the ovule-targeted growth occurred as the arrows indicate. Cross-pollination between WT and atg5-1, and atg7-2 and in vivo pollen tube growth for fertilization. Arrows indicate the twisted and abnormal accumulation of pollen tubes. Three independent experimental replicates were conducted for each sample and consistently yielded similar results. Scale bars = 100 μm.
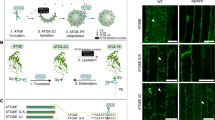
To better elucidate the mechanism(s) contributing to the undivided generative nucleus and the aberrant pollen tube morphology observed in atg5-1 and atg7-2, we performed a comprehensive investigation of actin filament organization in both mature pollen grains and tubes by phalloidin staining and confocal 3-D imaging. Actin filaments in the WT pollen grains were long cables and formed a fine network, whereas they were short and discontinuous in atg5-1 and atg7-2 pollen grains with only one generative nucleus (Fig. 4a). This pattern of actin filament organization was indicative of potential perturbations in the process of sperm cell biogenesis. To delve deeper into this phenomenon and elucidate the potential role of actin filaments in sperm cell biogenesis, we microinjected Latrunculin B (LatB) which can depolymerize actin filaments within developing flower buds as illustrated in Fig. 4b. 40 h after the microinjection, the development and blooming of LatB-injected flower buds arrested, while the control buds developed and flowered normally. Additionally, DAPI and phalloidin staining of pollen grains from LatB-treated flower buds showed that actin filaments were depolymerized and the division of the generative nucleus was halted (Fig. 4c). This provides the evidence for the functional role of actin filaments in modulating the division of the generative nucleus for sperm cell formation. Moreover, actin filaments can be assembled into elongated cables in all WT pollen tubes (Fig. 4d). However, the actin filaments were observed to be shortened and discontinuous in ~18% of pollen tubes from atg5-1 and atg7-2 (Fig. 4d). Furthermore, to investigate a potential functional link between ATG5- and ATG7-mediated autophagy and actin filament organization in other plant cell types, we examined the morphology of typical polarized cells including root hairs, epidermal leaf pavement cells and trichomes. Notably, only the length of root hairs was significantly diminished in the atg5-1 and atg7-2 mutants. Additionally, the organization of actin filaments in these root hairs was depolymerized, mirroring the defects observed in pollen and pollen tubes of these mutants (Supplementary Fig. 4). These findings suggest that ATG5- and ATG7-mediated autophagy may play important roles in cellular degradation in both pollen tubes and root hairs. Collectively, our results suggest a functional association between ATG5- and ATG7-mediated autophagy and actin filament organization in the modulation of the sperm cell biogenesis and the pollen tube growth in Arabidopsis thaliana.
a 3-D images of mature pollen grains of WT, atg5-1 and atg7-2 stained with both Alexa-488 phalloidin and DAPI. Three independent experimental replicates were conducted for each sample and consistently yielded similar results. Scale bars = 10 μm. b Schematic illustration of microinjection of 10 μL 200 nM actin polymerization inhibitor latrunculin B (LatB) into the developing flower bud (stage 10–12). After 40 h, the development and opening of the LatB-injected flower bud is strongly inhibited compared with the mock. c 3-D images of pollen grains that were released from the anthers with or without the LatB treatment and stained with both Alexa-488 phalloidin and DAPI. Three independent experimental replicates were conducted for each sample and consistently yielded similar results. Scale bars = 10 μm. d 3-D images of 6-h germinated Arabidopsis WT, atg5-1 and atg7-2 pollen tubes stained with Alexa-488 phalloidin were obtained. A total of n = 67 (WT), n = 79 (atg5-1), and n = 88 (atg7-2) pollen tubes were examined across six independent experiments for each sample. Source data are provided in Source data file. Scale bars = 10 μm.
NBR1 receptor facilitates recruitment of ADF7 and Profilin2 for degradation via autophagy
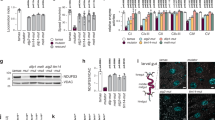
Actin Depolymerizing Factor (ADF) and Profilin are the major regulatory proteins to participate in the cleavage of actin filaments in Arabidopsis thaliana23,24,25. The ADF family comprises 11 members, with only ADF5 and ADF9 possessing actin polymerizing capabilities, while the remaining members exhibit actin depolymerizing abilities. In contrast, the Profilin family consists of five members, among which Profilin1 to Profilin3 function as actin depolymerization factors, while Profilin4 and Profilin5 are classified as actin polymerization factors (Supplementary Fig. 5). To elucidate how ATG5- and ATG7-mediated autophagy modulates the organization of actin filaments, we embarked on a series of studies. Initially, we performed a western blot analysis and found a notable elevation in ADF and Profilin protein levels in the atg5-1 and atg7-2 mutant pollen compared to the WT (Fig. 5a). Moreover, we treated mature WT Arabidopsis pollens with autophagic inhibitors 3-Methyladenine (3-MA) and wortmannin (Wort). Autophagic degradation of ADF and Profilin were strongly inhibited compared to the untreated controls by western blotting analysis with anti-ADF and anti-Profilin antibodies (Supplementary Fig. 6). Subsequently, we explored the potential recruitment and interaction between the autophagic receptor and ADF and Profilin. We selected ATG8d, an autophagy marker highly expressed in pollen, NBR1, a notable autophagic receptor, and actin filament depolymerizing regulators including actin depolymerization factors (ADFs) and Profilins for investigation. Based on the phylogenetic clustering and functional analysis of the ADF and Profilin protein families, we have identified the members specifically involved in actin depolymerization (Supplementary Fig. 5). These proteins were subsequently targeted in luciferase complementation assays to assess their interactions with NBR1. Our results showed that NBR1 can interacts with multiple ADF family members including ADF1, 2, 3, 4, 6, 7, 8, 10 and 11, with notably strongest interaction specifically with ADF7 (Supplementary Fig. 7a, b). Additionally, gene expression analysis indicated that ADF7 and ADF10 are particularly highly enriched in pollen (Supplementary Fig. 7c). Based on these findings, ADF7 was determined and selected for future study. Among the Profilin family proteins, only Profilin2 demonstrated a positive interaction with NBR1 (Supplementary Fig. 7d). Next, we conducted a yeast two-hybrid (Y2H) analysis to probe protein-protein interactions and observed that ATG8d interacted with NBR1, and NBR1 interacted with ADF7 and Profilin2 (Fig. 5b). These interactions were corroborated by luciferase complementation assay (Fig. 5c). Additionally, we have generated transgenic Arabidopsis lines expressing pADF7::GFP-ADF7 and pProfilin2::GFP-Profilin2 under the control of their native promoters. Co-immunoprecipitation (Co-IP) assays demonstrated that ADF7 and Profilin2, expressed under native promoter regulation, interact with NBR1 (Fig. 5d). Since actin organization in growing pollen tubes is closed regulated by tip-focused calcium gradient and distribution26,27, we also observed the calcium influx and gradients in pollen tube tip. We found that ~18% of the pollen tubes of atg5-1 and atg7-2 mutants exhibited disrupted tip-focused calcium gradient (Supplementary Fig. 8).
a Western blotting detection of ADF and Profilin levels in Arabidopsis WT, atg5-1 and atg7-2 pollen grains by using anti-ADF and anti-Profilin antibodies. Western blotting was performed in three independent experimental replicates, yielding consistent results. Source data of blotting are provided in Source data file. b Yeast two-hybrid analysis of protein interactions between NBR1 and ATG8d, ADF7, Profilin2. Positive interactions were determined by the appearance of yeast colonies on quadruple dropout agar medium SD/–Leu/–Trp/–His/–Ade (right column). A total of three independent experimental replicates were conducted for each sample, consistently yielding similar results. c Luciferase complementation assay to investigate protein interactions between NBR1 and ATG8d, ADF7, Profilin2. Positive interactions were determined by the appearance of fluorescent colors. Three independent experimental replicates were conducted for each sample, and similar results were consistently obtained. d Immunoprecipitation assay for detection of protein interactions between NBR1 and ADF7, and Profilin2. Total proteins were extracted from transgenic Arabidopsis expressing pUBQ::GFP, pADF7::GFP-ADF7 and pProfilin2::GFP-Profilin2. Immunoprecipitation was conducted by using GFP-trap beads and determined by western blotting with indicated specific antibodies. Immunoprecipitation assays were performed in three independent experimental replicates, yielding consistent results. Source data of blotting are provided in Source data file. e Schematic illustration of different domains of the full-length Arabidopsis NBR1 and its six different types of truncations. They were subsequently used to identify the functional domain(s) for its interaction with ADF7 and Profilin2. f Yeast two hybrid analysis of the interaction between NBR1∆UBA, NBR1∆PB1∆ZZ∆FW, NBR1∆ZZ∆FW∆UBA, NBR1∆ZZ, NBR1∆FW and NBR1∆ZZ∆FW with ADF7 and Profilin2, respectively. Yeast cells transformed with various plasmids were cultured on synthetic complete medium to exam the protein interactions. Positive interactions were determined by the appearance of yeast colonies on quadruple dropout agar medium SD/–Leu/–Trp/–His/–Ade (right column). A total of three independent experimental replicates were conducted for each sample, consistently yielding similar results. g Luciferase complementation assays for determination of the interaction between NBR1∆UBA, NBR1∆PB1∆ZZ∆FW, NBR1∆ZZ∆FW∆UBA, NBR1∆ZZ, NBR1∆FW and NBR1∆ZZ∆FW with ADF7 and Profilin2, respectively. Three independent experimental replicates were conducted for each sample, and similar results were consistently obtained. Positive interactions were determined by the detection of fluorescent colors.
NBR1 could interact with cytosolic ubiquitinated proteins via its C-terminal ubiquitin-associated (UBA1 and UBA2, collectively referred to as UBA) domains for autophagic clearance in mammals28,29. To further determine whether the interaction between NBR1 and ADF7 and/or Profilin2 is through the UBA domains in Arabidopsis, we generated various truncated variants of NBR1, including NBR1∆UBA, NBR1∆PB1∆ZZ∆FW, NBR1∆ZZ∆FW∆UBA, NBR1∆ZZ, NBR1∆FW and NBR1∆ZZ∆FW for the examination (Fig. 5e). We performed Y2H assays and luciferase complementation assays with these NBR1 truncation constructs. The Y2H assay showed that NBR1∆UBA was able to interact with ADF7 and Profilin2 (Fig. 5f), implying that the two UBA domains of NBR1 are not mandatory for interaction with ADF7 and Profilin2. In contrast, activation domain (AD)-NBR1∆PB1∆ZZ∆FW could not interact with a binding domain (BD)-ADF7 and BD-Profilin2 (Fig. 5f), suggesting that the UBA domains of NBR1 cannot bind with ADF7 and Profilin2. Furthermore, a luciferase complementation assay reaffirmed that NBR1, even without the UBA domains, was still able to interact with ADF7 and Profilin2 (Fig. 5g). To further elucidate the functional domains involved in NBR1-mediated cargo recognition and interaction, we conducted Y2H and luciferase complementation assays with various NBR1 truncations. The results showed that NBR1 retained its ability to bind to ADF7 and Profilin2 when either the ZZ or FW domain was individually truncated. However, simultaneous truncation of both ZZ and FW domains abolished the interaction of NBR1 with either ADF7 or Profilin2 (Fig. 5f, g). These findings underscore the indispensable role of both the ZZ and FW domains in facilitating ADF7 and Profilin2 recognition by NBR1 during non-ubiquitinated autophagy.
On the other hand, we further generated GFP-ADF7 and GFP-Profilin2 transgenic Arabidopsis overexpression (OX) lines. We also found that ~15% of mature pollen grains from the GFP-ADF7 and GFP-Profilin2 OX lines contain one undivided generative nucleus (Supplementary Fig. 10a, b). Moreover, polar pollen tubes growth exhibited a twisted accumulation pattern during fertilization in the OX expression lines of ADF7 and Profilin2 (Supplementary Fig. 10c). Similar to the observations in the atg 5 and atg 7 mutants, the number of seeds within the siliques of the OX lines was reduced as the arrows indicated when compared with the WT (Supplementary Fig. 10d). Statistical calculation of the seed setting rate showed about ~15% reduction was observed in the OX lines (Supplementary Fig. 10e). Meanwhile, we employed a reverse genetics approach to investigate whether reducing the levels of ADF7 and/or Profilin2 could suppress the defects of seed setting rate associated with the atg5-1 and atg7-2.
ADF and Profilin consist of multiple family members with similar functions, suggesting potential functional redundancy in regulating cytoskeleton organzaition30,31. Employing a genetic approach to knock out all members of the ADF and Profilin families is time-intensive and unfeasible due to their critical roles in cytoskeleton regulation, which is likely to lead to plant lethality upon complete depletion. RNAi provides an alternative approach that enables the simultaneous knockdown of multiple ADF and Profilin family members by targeting their highly conserved sequences, thus avoiding potential lethality. Consequently, we employed RNAi to reduce the expression of multiple ADF and/or Profilin family members in the atg5-1, atg7-2 and atg5-1/atg7-2 genetic backgrounds. This strategy allowed us to effectively investigate the phenotypic defects associated with impaired autophagy. We found that the seed setting rates of ADF-RNAi/atg5-1, Profilin-RNAi/atg5-1, ADF-RNAi/atg7-2, Profilin-RNAi/atg7-2, ADF-RNAi/atg5-1/atg7-2, Profilin-RNAi/atg5-1/atg7-2, and ADF-RNAi/Profilin-RNAi/atg5-1/atg7-2 were partially restored when compared to atg5-1, atg7-2 and atg5-1/atg7-2 mutants. Nevertheless, it is noteworthy that their seed setting rates were not fully recovered to the WT levels (Supplementary Fig. 11a-d). Furthermore, we observed and systematically compared the sperm cell biogenesis and 3-D spatial organization of actin filaments in pollen grains and pollen tubes between the WT and various mutants. Although all the mutants showed exhibited varying degrees of defects in both sperm cell biogenesis and actin organization, the atg5-1, atg7-2 and atg5-1/atg7-2 mutants showed relatively higher defective ratios compared to ADF-RNAi/atg5-1, Profilin-RNAi/atg5-1, ADF-RNAi/atg7-2, Profilin-RNAi/atg7-2, ADF-RNAi/atg5-1/atg7-2, Profilin-RNAi/atg5-1/atg7-2, and ADF-RNAi/Profilin-RNAi/atg5-1/atg7-2 (Supplementary Fig. 11e, f). It suggests that down-regulation of the expression of ADF and Profilin can partially restore the defective phenotypes of atg5 and/or atg7 mutants. These results confirmed that ADF7 and Profilin2 mediated actin spatial organization is functionally essential for the sperm cell biogenesis and male fertility in Arabidopsis.
In addition, we examined the phenotypes of nbr1-c2 mutant in comparison with the WT. Notably, we observed no significant differences in ADF and Profilin protein levels, actin organization in pollen grains and pollen tubes, sperm cell formation and seed setting rate in the nbr1-2c mutant compared to the WT (Supplementary Fig. 12). This lack of phenotypic defects is consistent with the understanding that NBR1 is not the sole receptor involved in autophagy32,33. Our findings imply that the degradation of actin depolymerizing factors may not be exclusively reliant on NBR1. To further investigate the involvement of potential alternative autophagic receptor(s), we performed a screening and analysis of the interactions between various other cytoplasmic autophagic receptors, including ATI1, ATI2, ATI3, RPN10, Pex6, and Pex10, with ADF7 and Profilin2 proteins by luciferase complementation assays, respectively34,35,36,37. Our results indicate that the autophagic receptor family ATI can interact with both ADF7 and Profilin2 (Supplementary Fig. 13). These findings suggest that the autophagic degradation of ADF7 and Profilin2 proteins likely depends on both NBR1 and ATI, involving a functional coordination mechanism between NBR1 and ATI during autophagy in Arabidopsis male gametophyte development and pollen tube growth during fertilization.
Collectively, a schematic hypothetical working model is proposed (Fig. 6). ATG8-NBR1-mediated autophagy can recruit and interact with actin depolymerization factors of the ADF family and Profilin2 for subsequent degradation to control their levels to sustain the dynamic organization of actin filaments in sperm cell biogenesis and pollen tube targeted growth during fertilization (Fig. 6). In atg5-1 and atg7-2, the autophagy pathway is abolished resulting in over-accumulation of ADF7 and Profilin2 in pollen grains and pollen tubes, leading to depolymerization of actin filaments. As a result, it leads to the disruption of sperm cells biogenesis and a noticeable deviation in the directional pollen tube growth and targeting to ovules during fertilization. Hence, autophagy acts as a modulator of male gametophyte development and fertility and of actin organization in Arabidopsis reproductive development.
In WT Arabidopsis, autophagy modulates sperm cell biogenesis and the directional growth of pollen tubes during fertilization. This is achieved by maintaining appropriate levels of actin depolymerization factors ADF7 and Profilin2 at appropriate levels through NBR1-mediated autophagic degradation. In the context of atg5 or atg7 mutants, compromised autophagy leads to an excessive accumulation of ADF7 and Profilin2 in pollen grains and tubes. This accumulation disrupts the spatiotemporal arrangement of actin filaments. As a result, it incites significant aberrations in spermatogenesis and directional pollen tube growth during the fertilization.
Discussion
Recent evidence has shown that autophagy is involved in multiple steps of sexual reproduction, depending on plant species, including male gametophyte development and maturation, self-incompatible pollen rejection and pollen germination20,21,22,38,39,40. Nevertheless, the role(s) of canonical autophagy during sexual reproduction in Arabidopsis thaliana is still an enigma since almost all of the autophagy-defective Arabidopsis mutants such as atg2, atg5 and atg7 exhibit normal life cycles without any deleterious phenotypes during fertilization, embryogenesis and seed formation. Plants with these mutations are nearly comparable to WT Arabidopsis with regards to progression and completion of their life cycle9,22. On the other hand, Arabidopsis plants with the ATG6 loss-of-function mutation show substantial defects in pollen germination38,41. However, ATG6 is a key component of the phosphoinositide 3-kinase (PtdIns3K) complex and therefore involved in many intracellular processes including endocytosis and endomembrane trafficking1,9. It is currently not possible to rule out these various activities as they affect pollen germination. The exact roles of ATG6 in regulating pollen germination via autophagy remain to be further studied and characterized. Due to the lack of substantial, definable phenotypes of these ATG mutants, autophagy is widely considered to be functionally irrelevant during reproduction in Arabidopsis thaliana. Despite these observations, it is curious and noteworthy that several core ATGs, including ATG5, ATG7 and ATG8h, are highly enriched in anthers or pollens compared with other tissues based on expression profile analyzes (Supplementary Fig. 1)22. Additionally, western blot analysis of ATG8-PE in germinated Arabidopsis pollens showed substantially reduced autophagic activity in atg5-1 and atg7-2 mutants compared with WT (Fig. 1). Moreover, to further elucidate the functions of ATG5 and ATG7 in autophagy during pollen tube growth, we examined their subcellular colocalization with various autophagic marker proteins involved in different developmental stages of autophagosome formation. We observed a pronounced colocalization of ATG5 and ATG7 with ATG8 and SH3P2, which is likely reflective of their critical roles in the formation and maturation of autophagosomes (Fig. 1)9. In contrast, colocalization of ATG5 with ATG6 and ATG9 was relatively lower, consistent with the primary functions of ATG6 and ATG9 in the early stages of autophagosome formation and fusion (Fig. 1)9. Collectively, these results are likely to suggest a role for autophagy in modulating functions of male gametophytes in Arabidopsis.
Emerging studies show that ATG5- and ATG7-mediated autophagy participates in the clearance of cytoplasm in the process of sperm cell differentiation in lower plants that generate motile sperm cells for fertilization42. In addition, the loss-of-function mutants of OsATG7 and OsATG9 cause male sterility in rice. Far fewer autophagosome-like structures were observed in the tapetum of Osatg7 rice20. Even though the underlying cellular mechanisms in the tapetum require further investigation, this observation indicates that autophagy is required for the development of male gametes in rice. Consistently, our results demonstrate that ATG5- and ATG7-mediated autophagy functionally participates in modulating pollen tube growth guidance and male fertility under standard cultivation conditions in Arabidopsis thaliana. In fact, it has been shown that canonical ATG5- and ATG7-mediated autophagy is required for normal sperm formation and male fertility in animals. For example, ATG5 and ATG7 were shown to induce autophagy by integrating multiple signals to maintain normal developmental processes of male fertility in mice18,19,43. ATG5-mediated autophagy controls various aspects of spermiogenesis including spermatid development, sperm individualization and sperm fertility18. Furthermore, the depletion of ATG7 disrupted autophagic flux in germ cells and resulted in irregular or nearly round-headed spermatozoa19.
In this study, we report that ATG5- and ATG7-mediated autophagy indeed participates in modulating the reproduction of Arabidopsis thaliana and controls sperm cell biogenesis and pollen tube growth. This conclusion is supported by several lines of evidence: i) detailed investigation on the seed formation in siliques of atg5-1, atg7-2, atg7-3 and atg5-1/atg7-2 revealed that ~15–20% reduction of seed set compared with WT (Fig. 2). This result could be overlooked since the seed formation defect in atg5-1, atg7-2, atg7-3 and atg5-1/atg7-2 is not severe; ii) the pollen germination ratio and tube growth of atg5-1, atg7-2 were reduced, especially at the beginning of germination, as demonstrated by time-lapse tracking of pollen tube germination (Fig. 2). Previous studies allowed the pollen from atg5-1 and atg7-2 strains to germinate for 10 h or more, which could lead to the overgrowth of pollen tubes and, therefore, obscure the phenotype1,22. In the absence of ATG5- and ATG7-mediated autophagy, the initial stages of pollen germination appeared to be inhibited, leading to reduced germination rates (Fig. 2). This could be attributed to the incapability of pollen to efficiently degrade stored materials via autophagy, thereby restricting essential metabolites availability and energy supply. Over time, however, pollens may activate alternative autophagic pathways or employ other parallel degradation mechanisms to facilitate the stored material turnover, ultimately restoring normal pollen tube growth rates during later stages of germination21,44,45; iii) more detailed studies of pollen tube growth and targeting to ovules showed that a portion of atg5-1 and atg7-2 pollen tubes were deformed. They became twisted and accumulated outside of the ovules rather than growing straight to the ovules for fertilization as observed for WT pollen tubes (Fig. 3). Although it remains to be determined whether these abnormal pollen tubes could eventually achieve fertilization, it is evident that autophagy is functionally involved in pollen tube growth and targeting during fertilization in Arabidopsis thaliana; and iv) ~20% of the mature pollens of atg5-1 and atg7-2 contain only an undivided generative nucleus. This result likely accounts for the aberrant seed formation within the siliques of the mutants (Fig. 2).
Actin filaments play crucial roles in modulating spermatogenesis in animals and mediate sperm nuclear migration during fertilization in flowering plants46,47,48,49. Moreover, abundant evidence suggests that actin filaments serve as a key regulator in controlling pollen tube cell morphogenesis and polar growth25,50,51,52. Therefore, it is highly speculated that actin filaments are regulated by canonical autophagy for sperm cell formation during pollen development and pollen tube growth during fertilization in Arabidopsis. Indeed, we observed significantly depolymerized and fragmented actin filaments in both mature pollen grains and pollen tubes from atg5-1 and atg7-2 (Fig. 4). Although the detailed mechanism of actin filaments in mediating mitosis of the generative nucleus in pollen remains ambiguous, it is evident that depolymerization of actin filaments during male gametophyte development can abolish sperm cell formation in Arabidopsis. Autophagy actually is involved in modulating the spatial organization of actin filaments in pollen grains and tubes. Similarly, it has been demonstrated that autophagy-deficient mice showed depolymerization of actin filament organization during spermatid differentiation, and thereby lead to abnormal spermatogenesis and male sterility53. Thus, autophagy is functionally conserved in modulating actin filaments to sustain sperm cell formation during male gametophyte development in both animals and plants.
There remains one key question to be answered: how exactly does autophagy modulate the organization of actin filaments. Shang et al. found that autophagy mediates the degradation of PDLIM1 (PDZ and LIM domain 1 [elfin]) which is a negative regulator of cytoskeleton organization. This degradation is instrumental in facilitating the morphological transitions of spermatids, particularly during acrosome biogenesis53. In the context of atg7-null mice, autophagy cannot be initiated and causes accumulation of PDLIM1 in the cytosol of the spermatids. The consequent PDLIM1 accumulation disrupts cytoskeletal organization during spermatid differentiation, culminating in a marked diminution of spermatozoa motility53. Drawing parallels with the plant kingdom, ADF and Profilin are the principal molecules mediating the depolymerization of actin filaments in various species31,54,55. In atg5-1 and atg7-2 mutants, increased accumulation of ADF and Profilin were detected and their degradation requires specific interaction with the ATG8-NBR1-medaited canonical autophagy pathway (Fig. 5). However, we observed that nbr1-c2 does not exhibit the same phenotypes as atg5-1 and atg7-2, such as increased levels of ADF and Profilin proteins and defects in the actin cytoskeleton organization related to impaired autophagy (Supplementary Fig. 11 and 12). These observations imply that the degradation of ADF and Profilin proteins may not be exclusively dependent on NBR1. Furthermore, we identified that the ATI family of autophagic receptors is also implicated in the autophagic degradation of ADF and Profilin (Supplementary Fig. 13). This finding indicates that both NBR1 and ATI may both participate in the degradation of ADF7 and Profilin2. Further investigations are required to elucidate the precise molecular mechanisms underlying the interaction and coordination between ATI and NBR1 in the autophagic degradation pathway of ADF7 and Profilin2. Understanding how these receptors cooperate and potentially compensate for each other could aid in understanding the regulatory networks governing actin cytoskeleton dynamics and autophagy. Additionally, exploring the specific conditions under which ATI or NBR1 predominates in the degradation process may provide a deeper comprehension of their roles and contributions in various physiological and stress contexts (Fig. 5 and Supplementary Fig. 13).
In addition, we identified the protein interactions between NBR1 and ARP2/3 complex that regulates the initiation of actin polymerization and the organization of filaments into y-branched networks56,57. ARP2/3 complex degradation requires ATG8-NBR1-medaited canonical autophagy pathway as previously reported by Schwarzerová et al.58. Despite of it, the gene expression level of ARP2/3 in Arabidopsis pollen is markedly low (Supplementary Fig. 9), suggesting that ARP2/3 may not play a pivotal role in actin organization within pollens. In the absence of ATG5 and ATG7, autophagy cannot be initiated, thereby it leads to excessive accumulation of ADF and Profilin proteins in pollen grains and tubes. This accumulation induces the accelerated depolymerization of actin filaments, which in turn disrupts sperm cell biogenesis and leads to the deformation of pollen tubes during fertilization in Arabidopsis. On the other hand, the partial restoration of seed setting deficiencies, the sperm cells biogenesis and actin organization in pollens and pollen tubes observed in atg5, atg7 and atg5/atg7 mutants was accomplished through the down-regulation of ADFs and/or Profilins via RNAi (Supplementary Fig. 11). These findings underscore the indispensable role of autophagy in modulating cytoskeletal organization through the degradation of key regulatory proteins in both animals and plants. In addition, it is noteworthy that atg5 and atg7 Arabidopsis mutants still managed to achieve a great portion of successful fertilization and seed production without the conventional autophagy, which is significantly distinct from mammals. They are able to generate well developed pollens containing two functional sperm cells and morphologically normal growing pollen tubes during fertilization (Fig. 2 and Supplementary Fig. 2). Hence, it will be of great importance to uncover whether and what alternative autophagic/degradative pathway(s) could be employed and its underlying mechanism(s) to partially sustain the male fertility in Arabidopsis in the future. Our study advances the understanding of the evolutionary conservation and diversification of autophagy in the modulation of male fertility in Arabidopsis compared with that in mammals.
Methods
Plant materials and growth conditions
Transgenic Arabidopsis lines expressing GFP-ADF7, GFP-Profilin2 and GCaMP5 were generated by transforming Arabidopsis thaliana Col-0 with constructs pUBQ::GFP, pUBQ::GFP-ADF7, pADF7::GFP-ADF7, pUBQ::GFP-Profilin2, pProfilin2::GFP-Profilin2 and pUBQ::GCaMP5, respectively. Complementation lines were established by expressing pATG5::GFP-ATG5 in atg5-1 mutant and pATG7::GFP-ATG7 in atg7-2 mutant background. ADF-RNAi/atg5-1, Profilin-RNAi/atg5-1, ADF-RNAi/atg7-2, Profilin-RNAi/atg7-2, ADF-RNAi/atg5-1/atg7-2, Profilin-RNAi/atg5-1/atg7-2, and ADF-RNAi/Profilin-RNAi/atg5-1/atg7-2 transgenic Arabidopsis were generated by transforming ADF-RNAi and Profilin-RNAi in atg5-1, atg7-2 and atg5-1/atg7-2 backgrounds. The primers used for the construction of these binary plasmid vectors are detailed in Supplementary Table 1. Double mutant atg5-1/atg7-2 was obtained through genetic crossing between atg5-1 and atg7-2. Primers used for the genotyping are listed in Supplementary Table 2. Arabidopsis seeds were surface sterilized and germinated on Murashige and Skoog (MS) medium containing 0.8% (w/v) agar and 3% (w/v) sucrose, pH 5.7. Seedlings were cultured in a plant growth chamber at 22 °C under a 16-h light and 8-h dark cycle prior to being transferred into soil in a plant growth room at 22 °C under a 16-h light and 8-h dark cycle. The light intensity was ~150 μmol/m2/s.
Transient expression of chimeric fluorescent fusion proteins in tobacco pollen tubes by particle bombardment
In brief, anthers were harvested freshly from ~20 tobacco flowers and transferred to 20 ml tobacco pollen germination medium. Pollens were released into the medium from anthers by vigorous agitation with a vortex for 2 min. Briefly, 15−20 Nicotiana tabacum flowers were collected and transferred into 20 ml of tobacco pollen-specific germination medium. After vortexing to release the pollen grains into the medium, 20 ml of the pollen suspension was vacuum-filtered onto pre-wetted filter paper. The filter paper, covered with pollen grains, was immediately transferred, surface-up, onto 2% agar in an 85-mm diameter Petri dish. For transient expression of reporter proteins in pollen tubes, the pollen grains were bombarded with gold particles59,60. Germinate the pollen grains for 2 hours and then observe the pollen tubes using confocal laser scanning microscopy.
Confocal laser scanning microscopy and image analysis
In general, confocal fluorescent images were collected by using a Leica TCS SP8 system with the following settings: 63x water immersion objective, 2x zoom, 750 gain, 0 background, 0.168 µm pixel size and photomultiplier tubes (PMTs) detector. A confocal dish with a cover slide at the bottom was used for live-cell imaging of pollen tubes to minimize potential mechanical damages caused by the conventional sandwich method of a sample between glass and cover slides. Colocalization analysis between two fluorescent proteins was calculated using ImageJ software with the Pearson-Spearman correlation (PSC) colocalization plug-in61. Results were presented either as Pearson correlation coefficients or as Spearman’s rank correlation coefficients, both of which produce r values in the range (−1 to 1), where 0 indicates no discernable correlation and +1 or −1 indicate strong positive or negative correlations, respectively.
Protein extraction and western blotting
Total proteins were extracted from freshly opened Arabidopsis flowers following a previously optimized method for better detection of ATG8-lipid adduct11. Equal amounts of proteins were loaded onto a 15% SDS-PAGE gel with 6 M urea. Thereafter, proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane. ATG8-PE and free ATG8 were immunodetected by anti-ATG8 antibody (Abcam, ab77003, v/v, 1:4000). ADF and Profilin were immunodetected respectively by anti-ADF antibody and anti-Profilin antibody which are self-generated and used in the previous study23.
RT-qPCR
Total RNA was isolated from freshly opened Arabidopsis flowers (S14) (Vazyme). The first strand for each cDNA was synthesized by HiScript II reverse transcriptase (Vazyme). Real-time fluorescence RT-qPCR was performed on Bio-Rad CFX Connect Real-Time PCR Detection System. At least three independent replicates were performed. Actin2 was used as an internal control. The primers used are listed in Supplementary Table 2.
In vitro and in vivo Arabidopsis pollen germination
For in vitro pollen germination: mature pollens at the specific developmental stage were collected as previously described62. Mature Arabidopsis pollen grains were transferred from anthers by spreading them on Arabidopsis solid pollen germination medium (GM) containing 0.01% boric acid, 1 mM Ca (NO3)2, 1 mM MgSO4, 5 mM CaCl2, 18% (w/v) sucrose and 0.9% agarose at pH 7.0 on a standard glass microscopy slide. The slide was then incubated in a wet chamber box at 28 °C and germinated for 2, 4 and 6 h prior to imaging.
For in vivo pollen germination: Arabidopsis flowers (stage 12) were castrated and used as female recipients. Pollen grains were selected from stage 15 flowers and gently spread onto the surface of stigma of the female recipients for in vivo pollen tube germination. After 2-, 4-, 6-, 10- and 16-h pollination, the female tissues were fixed with acetic acid/ethanol 1:3 (v/v) solution for 2 h at RT. Then, the samples were rehydrated by a series of incubations in 70%, 50% and 30% ethanol for 10 min each, and rinsed with ddH2O for 10 min at RT. They were subsequently immersed in 8 M NaOH overnight at RT and then washed for 3 times with ddH2O. Finally, the samples were stained by aniline blue solution containing 0.2% (w/v) aniline blue and 0.1 M K3PO4 for 2–3 h at RT63. Confocal laser scanning microscopy imaging was then immediately performed using a TCS SP8 confocal microscope (Leica).
DAPI staining
Mature pollen grains were released from freshly opened flowers by vortexing them in Arabidopsis pollen germination medium containing 0.01% boric acid (w/v), 1 mM Ca (NO3)2, 1 mM MgSO4, 5 mM CaCl2 and 18% (w/v) sucrose at pH 7.0. The flower petals and anthers were removed with forceps. Arabidopsis pollens were concentrated by centrifugation at 800 × g for 1 min at RT. After discarding the supernatant, the pellet of pollens was gently and fully resuspended with 200 μl DAPI staining solution containing 0.25 M sodium phosphate (pH 7.0), 0.25 mM EDTA, 0.025% Triton X-100 (v/v) and 0.1 µg/ml DAPI and incubated in the dark for 30–45 min. Confocal laser scanning microscopy imaging was then immediately performed using a TCS SP8 confocal microscope (Leica).
Phalloidin staining of actin filaments in pollen grains and tubes
Actin filaments in pollen grains and tubes were stained with Alexa-488 phalloidin as previously described23. Briefly, pollen grains or germinated tubes were fixed for 1 h with 300 µM 3-maleimidobenzoic acid N-hydroxysuccinimide ester (MBS) in liquid pollen GM. Then, the samples were washed three times with TBSS (200 mM NaCl, 400 mM sucrose and 0.05% NP-40 (v/v) in 50 mM Tris-HCl, pH 7.5). Pollen grains or tubes were stained with 200 nM Alexa-488 phalloidin in TBSS buffer overnight at 4 °C and observed under a laser scanning confocal microscope (Leica SP8) equipped with a 100x oil objective. The sample was excited with the 488-nm wavelength of an argon laser, the emission was set to a range of 505–525 nm, and the Z-series images were collected with the Z-step set at 0.7 µm. 3-D images of pollen grains and tubes were generated from the maximum intensity projection of Z-serious images.
Yeast two-hybrid assay
Y2H assays were conducted using the MatchMaker GAL4 Two-Hybrid System 3 (Clontech) in accordance with the manufacturer’s instructions. A pair of pGBKT7 and pGADT7 vectors (Clontech) were used to carry target cDNAs and then transformed into the Saccharomyces cerevisiae strain AH109. After selection on SD/–Leu/–Trp medium, single transformed colonies were screened for growth on SD/–Leu/–Trp/–His/–Ade medium to determine protein interactions. The primers used are listed in Supplementary Table 3.
Luciferase complementation assay
The full-length cDNA sequence of NBR1, ATI1, ATI2, ATI3, RPN10, Pex6 and Pex10 were amplified and inserted into the pCAMBIA-35S-nLUC vector. Meanwhile, ATG8d, ADF1, ADF2, ADF3, ADF4, ADF6, ADF7, ADF8, ADF10, ADF11, Profilin1, Profilin2 and Profilin3, ARP2 and ARP3 were cloned and inserted into the pCAMBIA-35S-cLUC vector. These constructs were transformed into A. tumefaciens stain GV3101. Transformed bacteria were cultured, harvested and re-suspended in the buffer (10 mM MgCl2, 200 μM acetosyringone, 10 mM MES, pH 5.7) to a final concentration of OD600 = 0.8–1.0. The suspension was infiltrated into Nicotiana benthamiana leaves using a syringe. The tobacco plants were placed in darkness for 24 h and then incubated in a 16 h/8 h light/dark cycle for 24 h. Thereafter, the leaves were sprayed with 1 mM luciferin solution, then kept in darkness for 5 min to quench the fluorescence. A deep cooling CCD imaging apparatus (NightShade LB985; Berthold Technologies, Bad Wildbad, Germany) was used to capture fluorescence images. The primers used for the plasmid construct are listed in Supplementary Table 4.
Co-immunoprecipitation assay
Protein extraction and Co-IP were performed as described64 with minor modifications. 1 g of 7-day-old transgenic Arabidopsis seedlings expressing pUBQ::GFP, pADF7::GFP-ADF7 and pProfilin2::GFP-Profilin2 were grounded into powder with liquid nitrogen and homogenized in 2 ml IP buffer containing 50 mM of Tris-HCl at pH 7.4, 150 mM of NaCl, 1 mM of EDTA, 0.5% NP-40 (v/v), 10% glycerol (v/v) and 1x protease inhibitor cocktail. Lysates were clarified through a 0.2 μm filter (Whatman) after centrifugation at 16,000 × g. for 15 min at 4 °C, and incubated with GFP-Trap agarose beads at 4 °C for 4 h in a top-to-end rotator. The beads were then washed five times with ice-cold washing buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 10% glycerol (v/v)], harvested by centrifugation at 16,000 × g at 4 °C for 1 min and eluted by boiling in SDS sample buffer. Samples were separated appropriate antibodies. GFP and NBR1 were immunodetected by anti-GFP antibody (Invitrogen, A11122, v/v, 1:4000) and anti-NBR1 antibody (Agrisera, AS142805, v/v, 1:8000). The chemiluminescence was imaged using an image analyzer (Tanon, 5200).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided in this paper.
References
Liu, Y. & Bassham, D. C. Autophagy: pathways for self-eating in plant cells. Annu Rev. Plant Biol. 63, 215–237 (2012).
Xie, Z. & Klionsky, D. J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 (2007).
Morishita, H. & Mizushima, N. Diverse cellular roles of autophagy. Annu Rev. Cell Dev. Biol. 35, 453–475 (2019).
Bassham, D. C. et al. Autophagy in development and stress responses of plants. Autophagy 2, 2–11 (2006).
Janse van Rensburg, H. C., Van den Ende, W. & Signorelli, S. Autophagy in plants: both a puppet and a puppet master of sugars. Front Plant Sci. 10, 14 (2019).
Yan, H. et al. Autophagy and its mediated mitochondrial quality control maintain pollen tube growth and male fertility in Arabidopsis. Autophagy 19, 768–783 (2023).
Üstün, S., Hafrén, A. & Hofius, D. Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 40, 122–130 (2017).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 17, 1–382 (2021).
Marshall, R. S. & Vierstra, R. D. Autophagy: the master of bulk and selective recycling. Annu Rev. Plant Biol. 69, 173–208 (2018).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu Rev. Cell Dev. Biol. 27, 107–132 (2011).
Chung, T., Phillips, A. R. & Vierstra, R. D. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 62, 483–493 (2010).
Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R. & Vierstra, R. D. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114 (2002).
Le Bars, R., Marion, J., Le Borgne, R., Satiat-Jeunemaitre, B. & Bianchi, M. W. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 5, 4121 (2014).
Phillips, A. R., Suttangkakul, A. & Vierstra, R. D. The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–1353 (2008).
Thompson, A. R., Doelling, J. H., Suttangkakul, A. & Vierstra, R. D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110 (2005).
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005).
Kuma, A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004).
Huang, Q. et al. Autophagy core protein ATG5 is required for elongating spermatid development, sperm individualization and normal fertility in male mice. Autophagy 17, 1753–1767 (2021).
Wang, H. et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 24, 852–869 (2014).
Kurusu, T. et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10, 878–888 (2014).
Zhao, P., Zhou, X. M., Zhao, L. L., Cheung, A. Y. & Sun, M. X. Autophagy-mediated compartmental cytoplasmic deletion is essential for tobacco pollen germination and male fertility. Autophagy 16, 2180–2192 (2020).
Zhou, X., Zhao, P. & Sun, M. X. Autophagy in sexual plant reproduction: new insights. J. Exp. Bot. 72, 7658–7667 (2021).
Jiang, Y., Chang, M., Lan, Y. & Huang, S. Mechanism of CAP1-mediated apical actin polymerization in pollen tubes. Proc. Natl Acad. Sci. USA 116, 12084–12093 (2019).
Vidali, L., McKenna, S. T. & Hepler, P. K. Actin polymerization is essential for pollen tube growth. Mol. Biol. Cell 12, 2534–2545 (2001).
Wang, Q. et al. Activation of actin-depolymerizing factor by CDPK16-mediated phosphorylation promotes actin turnover in Arabidopsis pollen tubes. PLoS Biol. 21, e3002073 (2023).
Zhao, Y. et al. The actin-related Protein2/3 complex regulates mitochondrial-associated calcium signaling during salt stress in Arabidopsis. Plant Cell 25, 4544–4559 (2013).
Wang, H. J., Wan, A. R. & Jauh, G. Y. An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 147, 1619–1636 (2008).
Kirkin, V., Lamark, T., Johansen, T. & Dikic, I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 5, 732–733 (2009).
Svenning, S., Lamark, T., Krause, K. & Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993–1010 (2011).
Zhang, J. et al. Proteomic analysis and candidate allergenic proteins in Populus deltoides CL. “2KEN8” mature pollen. Front Plant Sci. 6, 548 (2015).
Maciver, S. K. & Hussey, P. J. The ADF/cofilin family: actin-remodeling proteins. Genome Biol. 3, reviews3007 (2002).
Zhang, Y. & Chen, Z. Broad and complex roles of NBR1-mediated selective autophagy in plant stress responses. Cells 9, 2562 (2020).
Luo, S. et al. Cargo recognition and function of selective autophagy receptors in plants. Int J. Mol. Sci. 22, 1013 (2021).
Zhou, J. et al. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy 14, 487–504 (2018).
Xie, Q. et al. hfAIM: A reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy 12, 876–887 (2016).
Marshall, R. S., Li, F., Gemperline, D. C., Book, A. J. & Vierstra, R. D. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol. Cell 58, 1053–1066 (2015).
Wu, J. et al. ATI1 (ATG8-interacting protein 1) and ATI2 define a plant starvation-induced reticulophagy pathway and serve as MSBP1/MAPR5 cargo receptors. Autophagy 17, 3375–3388 (2021).
Harrison-Lowe, N. J. & Olsen, L. J. Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 4, 339–348 (2008).
Li, S., Yan, H., Mei, W. M., Tse, Y. C. & Wang, H. Boosting autophagy in sexual reproduction: a plant perspective. N. Phytol. 226, 679–689 (2020).
Norizuki, T., Minamino, N. & Ueda, T. Role of autophagy in male reproductive processes in land plants. Front Plant Sci. 11, 756 (2020).
Qin, G. et al. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 17, 249–263 (2007).
Sanchez-Vera, V. et al. Autophagy is required for gamete differentiation in the moss Physcomitrella patens. Autophagy 13, 1939–1951 (2017).
Liu, C. et al. Autophagy is required for ectoplasmic specialization assembly in Sertoli cells. Autophagy 12, 814–832 (2016).
Wang, Y. et al. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148, 1201–1211 (2008).
Obermeyer, G., Fragner, L., Lang, V. & Weckwerth, W. Dynamic adaption of metabolic pathways during germination and growth of lily pollen tubes after inhibition of the electron transport chain. Plant Physiol. 162, 1822–1833 (2013).
Bari, A. et al. Differential attentional control mechanisms by two distinct noradrenergic coeruleo-frontal cortical pathways. Proc. Natl Acad. Sci. USA 117, 29080–29089 (2020).
Crapster, J. A. et al. HIPK4 is essential for murine spermiogenesis. Elife 9, e50209 (2020).
Li, N. et al. Actin-bundling protein plastin 3 is a regulator of ectoplasmic specialization dynamics during spermatogenesis in the rat testis. Faseb j. 29, 3788–3805 (2015).
Ohnishi, Y. & Okamoto, T. Nuclear migration during karyogamy in rice zygotes is mediated by continuous convergence of actin meshwork toward the egg nucleus. J. Plant Res 130, 339–348 (2017).
Fu, Y. The cytoskeleton in the pollen tube. Curr. Opin. Plant Biol. 28, 111–119 (2015).
Gu, Y., Vernoud, V., Fu, Y. & Yang, Z. ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J. Exp. Bot. 54, 93–101 (2003).
Guan, Y., Guo, J., Li, H. & Yang, Z. Signaling in pollen tube growth: crosstalk, feedback, and missing links. Mol. Plant 6, 1053–1064 (2013).
Shang, Y. et al. Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy 12, 1575–1592 (2016).
Flynn, K. C. et al. ADF/cofilin-mediated actin retrograde flow directs neurite formation in the developing brain. Neuron 76, 1091–1107 (2012).
Fu, Y. et al. TaADF7, an actin-depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. Plant J. 78, 16–30 (2014).
Goley, E. D. & Welch, M. D. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726 (2006).
Mullins, R. D., Heuser, J. A. & Pollard, T. D. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl Acad. Sci. USA 95, 6181–6186 (1998).
Martinek, J. et al. ARP2/3 complex associates with peroxisomes to participate in pexophagy in plants. Nat. Plants 9, 1874–1889 (2023).
Wang, H. & Jiang, L. Transient expression and analysis of fluorescent reporter proteins in plant pollen tubes. Nat. Protoc. 6, 419–426 (2011).
Zhong, G., Liu, R., Zhuang, M. & Wang, H. Transient expression of chimeric fluorescent reporter proteins in pollen tubes to study protein polar secretion and dynamics. Methods Mol. Biol. 1662, 115–124 (2017).
French, A. P., Mills, S., Swarup, R., Bennett, M. J. & Pridmore, T. P. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3, 619–628 (2008).
Mori, T., Kuroiwa, H., Higashiyama, T. & Kuroiwa, T. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8, 64–71 (2006).
Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. Early flower development in Arabidopsis. Plant Cell 2, 755–767 (1990).
Thirumalaikumar, V. P. et al. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90.1 and ROF1. Autophagy 17, 2184–2199 (2021).
Acknowledgements
We apologize to those whose work could not be cited because of space restrictions. We would like to thank the members of Wang laboratory for stimulating discussions. We also thank Dr. Andrew Loria for help with manuscript editing, Prof. Shanjin Huang (Tsinghua University) for providing anti-ADF and anti-Profilin antibodies, Prof. Faqiang Li (South China Agricultural University) for sharing atg5-1, atg7-2 and atg7-3 mutant seeds (A. thaliana Col-0) and Prof. Liwen Jiang (The Chinese University of Hong Kong) for sharing nbr1-2c (A. thaliana Col-0). This work is supported by grants from the National Natural Science Foundation of China (92354302, 32270358, 91954110 and 31770196), the Natural Science Foundation of Guangdong Province (2021A1515012066) and Double First-class Discipline Promotion Project (2023B10564004) to H.W. H.Y. is supported by grants from the National Natural Science Foundation of China (32300289) and the China Postdoctoral Science Foundation (2022M721204).
Author information
Authors and Affiliations
Contributions
H.Y., Z.L., X.W., X.D. and Hao Wang conceived of the study; H.Y., Z.L., X.D., Z.Y., N.L., Z.N., X.L. and M.Y. performed the experiments; Hong Wu, L.Z. provided technical and equipment support; H.Y., X.W., Z.L. and Hao Wang analyzed the data and prepared the Figures; Hao Wang supervised the project and wrote the paper. All authors approved the manuscript submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yan, H., Lu, Z., Du, X. et al. Autophagy modulates Arabidopsis male gametophyte fertility and controls actin organization. Nat Commun 15, 10071 (2024). https://doi.org/10.1038/s41467-024-54468-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54468-8
This article is cited by
-
A single-base mutation on the 5’ UTR of BrATG5 confers the premature leaf senescence phenotype in Chinese cabbage
Theoretical and Applied Genetics (2025)