Abstract
Transplantation of engineered hematopoietic stem/progenitor cells (HSPCs) showed curative potential in patients affected by neurometabolic diseases treated in early stage. Favoring the engraftment and maturation of the engineered HSPCs in the central nervous system (CNS) could allow enhancing further the therapeutic potential of this approach. Here we unveil that HSPCs haplo-insufficient at the Cx3cr1 (Cx3cr1−/+) locus are favored in central nervous system (CNS) engraftment and generation of microglia-like progeny cells (MLCs) as compared to wild type (Cx3cr1+/+) HSPCs upon transplantation in mice. Based on this evidence, we have developed a CRISPR-based targeted gene addition strategy at the human CX3CR1 locus resulting in an enhanced ability of the edited human HSPCs to generate mature MLCs upon transplantation in immunodeficient mice, and in lineage specific, regulated and robust transgene expression. This approach, which benefits from the modulation of pathways involved in microglia maturation and migration in haplo-insufficient cells, may broaden the application of HSPC gene therapy to a larger spectrum of neurometabolic and neurodegenerative diseases.
Similar content being viewed by others
Introduction
Transplantation of hematopoietic stem/progenitor cells (HSPCs) engineered by integrating vectors for the expression of disease-specific therapeutic transcripts has shown unprecedented curative potential in patients affected by monogenic neurometabolic disorders (NMDs) when treated in very early disease stages1,2,3,4,5,6,7. This benefit is mediated by the myeloid transplanted cell progeny in the central nervous system (CNS) that not only acts as vehicle for therapeutics, but also restores professional scavenging functions and modulates neuroinflammation, mechanisms that could be theoretically applied to a broader spectrum of neurodegenerative diseases (NDs) (Milazzo et al. in press, Nature Communications). Notably, phenotypic effects in the treated patients appeared late after transplant, likely due to the slow pace of CNS engraftment and differentiation of the engineered HSPCs into microglia-like cells (MLCs) as compared to the rapid progression of neurodegeneration. This delay thus far hampered a broader application of this approach. Moreover, unregulated gene expression driven by the currently in-use promoters could not be adequate for achieving therapeutic effects in complex neurodegenerative conditions. Further, in some cases, the requirement for a strong promoter to reach the therapeutic threshold of transgene expression could increase the risk of genotoxicity of the semi-random integrating vectors8,9,10,11. In other cases, the ectopic expression of the therapeutic gene throughout the whole hematopoiesis could cause long-term unwanted effects or toxicity12. Importantly, as neuroinflammation and microglia cell-activation play a crucial role in the pathogenesis and progression of NMDs and NDs13,14, the use of microglia-specific promoters that could function as sensors of the microenvironment to express therapeutic transcripts by HSPC transplant progeny cells in the CNS could be highly desirable.
Here, we describe a targeted gene addition approach at the CX3CR1 locus of human HSPCs (hHSPCs) that may successfully address these limitations and allow extending the application to and enhancing the therapeutic potential of HSPC gene therapy for NMDs and NDs. The CX3CR1 gene encodes for the chemokine (C-X3-C) motif receptor 1 (Cx3cr1), highly expressed in microglia. Binding to its unique ligand CX3CL1, CX3CR1 regulates several microglia functions, including their recruitment to sites of neuroinflammation, contributing to the regulation of microglia morphology and of a subset of inflammatory genes15. Several reports propose the CX3CL1/CX3CR1 axis as a potentially relevant target for therapeutic interventions in the context of NDs15,16.
By conducting conventional experiments with a Cx3cr1 haplo-insufficient microglia-reporter model, we uncovered, by serendipity, a unique phenotype of Cx3cr1−/+ HSPCs that, after transplantation in myeloablated recipients, showed a quantitative and qualitative advantage over wild type (WT) counterparts in repopulating the CNS myeloid compartment and differentiating into MLCs. Based on these findings, we designed and validated a targeted gene addition strategy at the CX3CR1 locus of hHSPCs that supported the generation of mature MLCs by CX3CR1−/+ HSPCs transplanted into immunodeficient mice and enabled a myeloid/microglia-specific transgene expression in the progeny of the engineered cells. We thus envision the preclinical and subsequent clinical development of this strategy as an HSPC-based gene therapy platform applicable to several neurodegenerative and neuroinflammatory conditions where a timely, efficient, and properly modulated transgene expression in myeloid/microglia cells could be beneficial.
Results
Cx3cr1 haplo-insufficient HSPC progeny display a unique phenotype
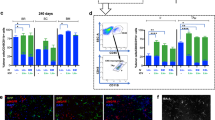
In the context of studies aimed at understanding the modalities of brain repopulation by HSPCs upon transplantation in murine models, we performed tracking experiments employing cells retrieved from CD45.2 donor mice that are haplo-insufficient for Cx3cr1 because of GFP insertion at the locus (Cx3cr1−/+) with consequent GFP signal in cells expressing Cx3cr1, such as microglia17. We FACS sorted GFP+ and GFP- cells from the lineage negative (Lin−) HSPC pool of Cx3cr1−/+ donors and independently transplanted the sorted cells in busulfan-conditioned CD45.1 recipients to evaluate their capability of engraftment in the bone marrow (BM) and brain (Supplementary Fig. 1A). At sacrifice, 45 days later, we found that only mice receiving GFP- Lin− cells showed a robust donor cell engraftment in the BM and in the brain (Fig. 1A and Supplementary Fig. 1B); no donor-derived cells were retrieved in mice receiving the GFP+ Lin- fraction, indicating that cells expressing Cx3cr1 are not capable of engraftment in transplant recipients. Unexpectedly, flow cytometry analysis revealed that the majority of the cells derived from Cx3cr1−/+ donors in the brain of transplant recipients expressed the CD45 and CD11b antigens at levels consistent with those of mature microglia (μ) cells from naïve control mice (Fig. 1B, C), rather than of more immature transiently amplifying μ (TAμ) cells that usually prevail in the transplant setting, particularly in early stages18. This was particularly evident when comparing within individual animals the cells derived from the CD45.2 Cx3cr1−/+ donors with the CD45.1 counterpart derived from the CD45.1 BM support and/or from endogenous microglia18 (Fig. 1B, C).
A Engraftment of donor cells in BM and brain of CD45.1 recipient mice that were previously transplanted with either GFP+Cx3cr1+ or GFP-Cx3cr1- HSPCs sorted from Cx3cr1+/GFP CD45.2 donor mice as detailed in Supplementary Fig. 1A, B. Mean and individual values ± SD are shown, 2 independent experiments. B, C Representative dot plots (B) and quantification (C) of the frequency of microglia (μ) and transiently amplifying microglia (Taμ), identified according to CD45 and CD11b expression levels, within donor CD45.2 cells (top) and recipient/support CD45.1 cells (bottom), in the brain of mice transplanted with GFP+Cx3cr1+ or GFP-Cx3cr1− sorted cells. Mean and individual values ± SD are shown, 2 independent experiments. D Frequency (%) of brain myeloid subsets (μ, Taμ) within donor CD45.2+ cells in the brain of mice transplanted with Cx3cr1−/+ or Cx3cr1+/+ HSPCs, sacrificed at 45 days post-transplant. Mean and individual values ± SD are shown, 3 independent experiments. E Experimental Scheme. Busulfan-conditioned CD45.1 recipients were competitively transplanted with wild type (WT, mCherry+) and Cx3cr1−/+ (BFP+) HSPCs (1:1) either intravenously (IV) or intracerebroventricularly (ICV). Mice were sacrificed at 45 days post-transplant. A dot-plot showing the engraftment of donor-derived mCherry+ (Y axis) and BFP+ (X-axis) cells in the peripheral blood of a representative transplanted mouse is also shown. F Quantification of the relative frequency (%) of Cx3cr1+/+ Cherry+ and Cx3cr1−/+ BFP+ cells within engrafted donor cells in hematopoietic organs (BM, spleen, and thymus) and brain of competitively transplanted mice (IV or, when specified, ICV), compared to the infused HSPCs (input) is shown. G Percentages of Cx3cr1+/+ (white) and Cx3cr1−/+ (gray) donor-derived cells within myeloid and lymphoid (B and T) engrafted cells in the spleen of mice competitively transplanted IV. F, G IV n = 8; ICV n = 4, 3 independent experiments. Mean values ± SEM are shown. H Frequency (%) of brain myeloid subsets (μ, Taμ) within Cx3cr1−/+ and Cx3cr1+/+ donor-derived cells in the brain of IV or ICV transplant recipients, sacrificed at 45 days post-transplant. Mean values ± SD and individual values are shown. Statistical analysis (for c, d, f–h): Mann–Whitney test, two-tailed; *p < 0.05, **p < 0.01, ***p < 0.001. Source data are provided in the Source Data file.
Intrigued by these findings, we hypothesized that the haploinsufficiency of Cx3cr1 could have contributed to the observed more mature phenotype of the transplant-derived MLCs in the recipients. To test this hypothesis, we designed additional dedicated experiments. Firstly, Lin- HSPCs isolated from Cx3cr1−/+ donor mice were transplanted into CD45.1 myeloablated recipients that were then analyzed at different time points post-transplant. CD45.1 myeloablated mice transplanted with HSPCs from Cx3cr1+/+ wild-type (WT) donors were used as controls. At all the time points of analysis, WT and Cx3cr1−/+ cells engrafted equally in the BM and brain of the recipient mice (Supplementary Fig. 1C). The transplanted cells and their progeny showed Cx3cr1 expression levels consistent with their genotype, namely with low Cx3cr1 expression in bone marrow cells and MLCs of Cx3cr1−/+ HSPC transplant recipients (Supplementary Fig. 1D, E). Consistent with the previous observation, also in this setting, in the brain of mice transplanted with Cx3cr1−/+, but not with WT HSPCs, particularly at the earliest post-transplant time points, μ-like cells prevailed over TAμ cells (Fig. 1D and Supplementary Fig. 1F).
Cx3cr1 haplo-insufficient HSPCs outcompete WT cells in competitive transplantation
To further investigate the modality of Cx3cr1−/+ HSPC repopulation of the recipient brain myeloid compartment and differentiation into MLCs, we designed a competitive transplantation experiment. CD45.2+ Cx3cr1−/+ and WT HSPCs, labeled by lentiviral vector transduction with different fluorescent markers, were co-transplanted at a 1:1 ratio intravenously (IV) or intracerebroventricularly (ICV) in CD45.1 busulfan-conditioned recipients (Fig. 1E). As control, mice were transplanted with either WT or Cx3cr1−/+ HSPCs alone. Input HSPCs were comparable for the phenotype (94.53 ± 2.5% Lin− cells in Cx3cr1−/+ HSPCs; 93.9 ± 3.3% Lin- cells in WT HSPCs), marker-gene expression (99.05 ± 0.78% BFP+ in Cx3cr1−/+ HSPCs; 98.75 ± 0.78% mCherry+ in WT HSPCs, Supplementary Fig. 2A) and clonogenic potential (CFUs counts 50 ± 7.26 CFUs for Cx3cr1−/+ HSPCs; 50.25 ± 5.18 CFUs for WT HSPCs) across separate experiments. At sacrifice, 45 days later, a high and comparable engraftment of the donor cells was observed in hematopoietic organs (IV administration) and brain (IV and ICV administration) of transplant recipients in all the experimental groups (Supplementary Fig. 2B–D); no donor cells were observed in the hematopoietic organs of mice transplanted ICV, in line with previous data19,20. Unexpectedly, the proportion of Cx3cr1−/+ versus WT cells within the total of donor origin cells was uneven at sacrifice as compared to input cells, which were transplanted at a 1:1 ratio. Cx3cr1−/+ cells prevailed over the WT counterparts in hematopoietic organs (60 vs 40%) and, even more strikingly, in the brain of the recipients, where the cells derived from the Cx3cr1−/+ HSPCs constituted up to 80% of the total donor cells (Fig. 1F and Supplementary Fig. 2B). The proportion of Cx3cr1−/+ versus WT cells within hematopoietic cell lineages throughout tissues reflected the one detected on total donor cells, except than in CD3+ T lymphocytes, for which a greater unbalance between the BFP+ Cx3cr1−/+ cells and the mCherry+ WT cells was observed (almost 80 versus 20%) (Fig. 1G). Interestingly, and coherently with our initial data, also in this transplantation setting, the progeny of the Cx3cr1−/+ HSPCs at flow cytometry mostly showed a μ-like phenotype rather than TAμ cell features, at odds with the progeny of the Cx3cr1+/+ HSPCs that showed a pattern of CD45 and CD11b antigen expression consistent with standard transplants, with prevalence of TAμ cells over µ (Fig. 1H). The initial data of the single transplantation setting were also here confirmed in the controls, with the majority of the Cx3cr1−/+ cell progeny showing CD45 and CD11b expression consistent to µ cells (and of the Cx3cr1+/+ cell progeny showing features of TAµ) (Fig. 1H). Similar results were obtained by co-transplanting Cx3cr1−/− HSPCs with WT cells, even if in the absence of a dose effect (Supplementary Fig. 2E, F).
Notably, when we characterized naïve microglia from aged-matched adult Cx3cr1+/+, Cx3cr1−/+, and Cx3cr1−/− mice by flow cytometry, we did not identify significant differences among them in terms of levels of expression of the CD45 and CD11b antigens and the frequency of cells with µ versus TAµ phenotypes (Supplementary Fig. 2G, H), in agreement with previous studies21. This observation suggests that the phenotype we observed is strictly related to the post-transplant repopulation setting.
Cx3cr1 haplo-insufficient MLCs show more mature morphology and transcriptional profile as compared to WT MLCs
Based on these early findings, we performed a branching study to characterize the morphology of the WT and Cx3cr1 haplo-insufficient cell progeny in the brains of competitive-transplant recipients. The analysis was run on brain slices, identifying transplant-derived cells by fluorescent marker expression (Fig. 2A, B), once confirmed, it ensured overlapping results with the analysis of Iba-1 staining (Supplementary Fig. 3A–C). Cell morphology was described using parameters previously employed in the literature for describing microglia morphology22: (i) the total length of all cell ramifications (Sum Length); (ii) the complexity index (CI), defined as the ratio between the total number of branches of each cell (where a branch is a segment between two nodes) and the number of its primary ramifications; (iii) the covered environment area (CEA), namely the 2D total surface covered by the cell ramifications and defined as the area of the polygon formed by linking the extremities of its processes (Fig. 2B). The branching analysis revealed interesting differences between the transplanted cell progenies. Cx3cr1-/+ HSPC progeny showed a significantly higher total length of ramifications as compared to the WT HSCP progeny in both the IV and ICV competitive-transplant settings (Fig. 2C). The analysis of CI and CEA also revealed a greater complexity ramification and a larger surface area covered by the Cx3cr1−/+ MLCs versus WT MLCs (Fig. 2D, E). These results were more pronounced in the IV transplantation setting as compared to the ICV one, suggesting greater competition in the former setting. This is in line with our previous data indicating that the ICV route favors the differentiation of HSPCs toward MLCs20. We also performed an automated Sholl analysis22 to study the radial distribution of microglia branches around the cell body (Fig. 2F). The sum intersections and number of intersection radii were selected as parameters to quantitatively characterize the morphology of donor-derived cells based on the number and on the extension of cell arborizations. The analysis showed that cells derived from Cx3cr1-/+ HSPCs covered a larger surface and displayed more arborizations as compared to the WT cell progeny both in the IV and in the ICV transplantation settings (Fig. 2G–I). By plotting these two parameters (sum intersections and number of intersection radii) in a bidimensional graph, we observed that the progeny of Cx3cr1−/+ transplanted HSPCs was more abundantly distributed in the upper right—UR—quadrant that corresponds to the cells covering the greater surface and displaying more arborizations, than the Cx3cr1+/+ cell progeny (Fig. 2G–I).
A Representative reconstruction of a brain slice from a competitively transplanted mouse where the engrafted BFP+Cx3cr1−/+ (in green) and the Cherry+Cx3cr1+/+ (in red) MLCs are visualized. Nuclei were stained with DAPI (blue signal). On top, a magnification (40X) showing the morphology of the BFP+Cx3cr1−/+ and the Cherry+Cx3cr1+/+ MLCs in the cortex. B Scheme displaying macro workflow for branching analysis. Confocal images, after maximum intensity projection, were analyzed with a standardized macro through the ImageJ software. Morphological criteria adopted to characterize donor-derived microglia cells were applied to each cell, which was thresholded, skeletonized, and deprived of the cell body. C–E Branching analysis on the BFP+Cx3cr1−/+ and the Cherry+Cx3cr1+/+ MLCs in the brain of IV or ICV competitively transplanted mice. The cumulative length of branches (C), complexity index (CI) (D), and covered environment area (CEA) (E) are shown. Mean and individual values ± SEM are shown. n > 50 cells per group, n = 3 mice/group, 3 independent experiments. An unpaired t-test with Welch’s correction, two-tailed, was performed. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. F Example of Sholl analysis on an MLC analyzed via the ImageJ software. G, H Correlation between intersection radii and sum intersection parameters obtained from the Sholl analysis on BFP+Cx3cr1−/+ and the Cherry+Cx3cr1+/+ MLCs in the brain of IV (G) and ICV (H) competitively transplanted mice. The vertical and horizontal lines divide the graphs into four quadrants, to describe the cells according to different grades of morphologic complexity, i.e., UR upper right quadrant, for very complex cells characterized by a high sum of intersections and high number of intersecting radii; LL lower left quadrant, for cells with lower complexity; UL (upper left) and LR (lower right) quadrants for cells displaying intermediate complexity between the LL and the UR quadrants. I Histograms representing the percentage of cells retrieved in each of the four quadrants displayed in Fig. G and (H) to quantify the data. Images were acquired via Leica SPE confocal in the cortex region of the brain at 40X magnification. n > 50 cells per group, n = 3 mice/group. J Gene expression of microglia enriched genes and transcription factors in MLCs sorted by fluorescent marker expression from the brain of ICV competitively transplanted mice. Data were shown as fold change (calculated as 2 − ΔΔCT) on adult control microglia cells (dotted line, as reference equal to 1). Neonatal microglia (referred to as pup) are used as examples of immature myeloid/microglia cells. n = 3 mice/group. Individual values are shown. Source data are provided as a Source Data file.
To further strengthen our observations, we analyzed the transcriptomic profile of MLCs sorted by fluorescent marker expression from the brain of competitively transplanted mice, focusing on a set of specific microglia signature genes23, comparing their expression to microglia isolated from naïve adult reference animals and neonates (Fig. 2J). Interestingly, Cx3cr1−/+ MLCs showed a pattern of expression of the microglia signature genes almost superimposable to that of the cells isolated from naïve adult control mice. On the contrary, the Cx3cr1+/+ MLC expression pattern was intermediate between the adult and neonate control cells and more variable, supporting the conclusion that Cx3cr1 haploinsufficiency could facilitate the maturation of the engrafted cells.
Cx3Cr1 haplo-insufficient MLCs display a mature microglia signature
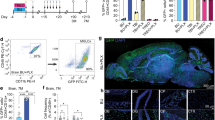
To obtain some insight into the mechanism that could be responsible for the advantage shown by Cx3cr1 haplo-insufficient HSPCs versus WT counterparts in populating the brain of myeloablated recipients with donor-derived mature MLCs, we performed a transcriptomic analysis at single-cell resolution on FACS-sorted MLCs isolated from competitively transplanted mice, using the same experimental scheme described above in Fig. 1E. At sacrifice, after extensive perfusion, we collected BM and brain samples for flow cytometry and FACS-sorted MLCs from the brain, separating WT versus haplo-insufficient cells according to fluorescent marker expression to perform a transcriptomic analysis with the 10XGenomics platform. Transplanted mice were euthanized at an early time point after transplant (15 days) to study the early dynamics of the cell reconstitution process in the brain. At this time point of analysis, flow cytometry showed that engraftment in the BM had already reached a plateau, while in the brain the donor cell frequency was still low (Fig. 3A). Consistent to previous data, the analysis confirmed a higher frequency of Cx3cr1 haplo-insufficient versus WT cells within the engrafted donor population, both in the BM and brain, also at this early time point of evaluation (Fig. 3B). The distribution of the Cx3cr1 haplo-insufficient and WT single-cell transcriptome profiles was evaluated in a two-dimensional UMAP plot (Fig. 3C and Supplementary Fig. 4A–C), on which we then identified several clusters (Fig. 3D and Supplementary Fig. 4D). To refine the characterization of each cell cluster, we analyzed the cluster-specific marker genes with a logFC >2 (Supplementary Fig. 5A) and performed cluster-specific GO analyses (Supplementary Fig. 5B). Interestingly, as far as marker-gene analysis is concerned, the two clusters more differentially enriched in Cx3Cr1 haplo-insufficient cells—namely cluster 4 containing a greater number of Cx3Cr1−/+ cells versus cluster 9 enriched in Cx3Cr1+/+ cells (Fig. 3E)—showed a mutually specular pattern. Cluster 4 was characterized by the high expression of the microglia gene ApoE, which is poorly expressed by cluster 9 cells, that instead expresses at high levels Retnlg. This gene has been recently included in a short list of genes specific to a microglia subset defined as “in transition microglia”24, suggestive of an actively occurring differentiation process. Consistently with this finding, genes highly expressed in cluster 9, which is enriched in Cx3Cr1+/+ cells, such as S100A8, S100A, LCN2, and G0S2, are known to be expressed in the early stages of myeloid differentiation and proliferation. Cluster 4, instead, showed a significant upregulation of genes associated to prototypical microglia differentiation and migration pathways (Fig. 3F). Of note, upregulation of genes related to Cdc42, a small GTPase of the Rho-subfamily, which regulates signaling pathways controlling diverse cellular functions including cell morphology, migration, endocytosis and cell cycle progression (Fig. 3F) was also observed in cluster 4. Indeed, of the 16 genes included in the gene ontology term for Cdc42 protein signal transduction, four (Nrp1, Rhob, Apoe, and Abca1) exhibited highly increased expression levels in cluster 4 as compared to the entire dataset (Fig. 3G). The heightened expression was also observed, to a lower extent, in some adjacent cell clusters enriched in migration, motility, and chemotaxis gene expression.
A Frequency of donor CD45.2+ cells in the BM and brain of competitive-transplant recipients sacrificed at 15 days post-transplant. B Frequency of Cx3cr1+/+ and Cx3cr1−/+ cells within total donor cells in the BM and brain of competitive-transplant recipients, compared to the infused HSPCs (input). For (A) and (B), n = 8, 3 independent experiments. Mean and individual values ± SEM are shown. Statistical analysis: Mann–Whitney test; *p < 0.05, **p < 0.01, ***p < 0.001. C Two-dimensional UMAP representation of the microglia single-cell transcriptome dataset. Each dot represents a cell, and colors indicate the genotypes, as indicated. D UMAP plot showing the clusters identified using the Louvain algorithm. E Bar graph showing the genotype (Cx3cr1+/+ and Cx3cr1−/+) frequency in each cluster. F Gene ontology analysis on cluster 4, enriched in Cx3cr1−/+ cells, showing a significant upregulation of Cdc42 pathway-related genes, microglia migration, and prototypical microglia differentiation pathways. G UMAP plots showing gene expression of 4 (out of 16) genes of the Cdc42 protein signal transduction pathway. H Mean fluorescent intensity (MFI) of Cdc42 expression in wild type and Cx3cr1−/+ microglia cells isolated from naïve mice. n = 10/group. Mean and individual values ± SEM are shown. Statistical analysis: Mann–Whitney test; *p < 0.05, **p < 0.01, ***p < 0.001. I Migration of Cx3cr1+/+ and Cx3cr1−/+ HSPCs isolated from naïve Cx3cr1+/+and Cx3cr1−/+ mice towards fractalkine (FLK) added at different concentrations (100 and 200 ng/mL) in a trans-well system. SDF-1 (50 nM) was employed as a positive control for HSPC migration. RAW 264.7, a murine macrophage cell line expressing high levels of Cx3cr1, was used as a positive control for migration toward FKN. The percentage of migrating cells was analyzed after 24 h. n ≥ 5 per group, 3 independent experiments. Mean v and individual values ± SD are shown. Statistical analysis: Mann–Whitney test; *p < 0.05, **p < 0.01, ***p < 0.001. Source data are provided as a Source Data file.
To validate the single-cell RNA-seq data and investigate the possible relationship between the Cdc42 pathway and Cx3cr1, we analyzed the expression of Cdc42 by flow cytometry on microglia cells isolated from naïve WT and Cx3cr1−/+ mice. We observed a reduced expression of Cdc42 in Cx3cr1−/+ cells as compared to WT cells (Fig. 3H), confirming that Cx3cr1 deficiency could be associated with perturbations of Cdc42 expression and its pathway in microglia. Next, to assess whether these pathways could be affected in Cx3cr1 haplo-insufficient HSPCs and whether these cells could have a defective migration towards Fractalkine (FLK), Cx3cr1 ligand, we performed a trans-well migration assay using HSPCs isolated from naïve wild type and Cx3cr1−/+ mice. As control, we added in another chamber SDF-1, as HSPCs are expected to chemotactically migrate towards this chemokine25. Additionally, we performed the same assay using RAW 264.7, a murine macrophage cell line expressing high levels of Cx3cr1 and thus supposed to migrate towards an FLK gradient. As expected, increasing concentrations of FLK resulted in increased migration of RAW 264.7 cells (Fig. 3I), but no differences were observed in the migration towards SDF-1 of the wild type versus Cx3cr1−/+ HSPCs. Interestingly, despite some variability likely due to different levels of Cx3cr1 expression in the HSPC population that is heterogenous in composition, particularly during ex vivo culture, a significantly higher migration towards FLK of the Cx3cr1−/+ as compared to the Cx3cr1+/+ HSPCs was observed at both the concentrations tested (Fig. 3I).
CRISPR/Cas9 gene editing and targeted gene addition at the human CX3CR1 locus
Based on these findings, we reasoned to develop a CRISPR-Cas9-based gene editing strategy to insert an exogenous transgene expression cassette into the human CX3CR1 locus while concomitantly knocking out the expression of the edited allele. This approach could allow: (i) exploiting the positive effects of CX3CR1 haploinsufficiency in human HSPCs to enhance their ability to repopulate the brain myeloid compartment with a progeny that could efficiently differentiate towards MLCs and (ii) expressing a therapeutic transgene under the control of the endogenous CX3CR1 promoter, which is highly active in steady-state microglia and even more robustly expressed upon their activation in pathological conditions. To edit the human CX3CR1 locus, we designed and compared a targeting strategy specific for the intron 4 and one specific for the exon 5, which contains the entire gene coding sequence26,27. The exonic approach would result in gene disruption with both NHEJ-mediated INDELs and homology-directed repair (HDR)-mediated targeted integration, thus likely leading to full CX3CR1 knockout in most of the treated cells. On the contrary, the intronic approach would disrupt CX3CR1 expression mainly after HDR-integration of the transgene cassette, while most of the NHEJ-mediated INDELs should only minimally affect endogenous gene expression. Thus, this latter approach would mostly mimic an haploinsufficiency phenotype. For both approaches, we designed and tested a panel of different single guide RNAs (sgRNAs) (Fig. 4A) and identified one sgRNA for each approach that resulted in the highest cutting efficiency (sgRNA5 for the intronic region and sgRNA9 for the exon; Fig. 4B). For both the approaches, we designed a promoter-less donor DNA template that contained a splice-trapping cassette encoding for the yellow fluorescent protein (YFP) flanked by homology arms (HA) that match the nuclease cutting sites. For the exon, we also designed a similar donor template where the expression of the reporter was driven by the constitutive spleen focus forming virus (SFFV) promoter (SFFV-exon) (Fig. 4C). Donor templates were cloned and produced as adeno-associated virus type 6 (AAV6) vectors to obtain efficient delivery in primary HSPCs. As control, we used previously validated sgRNA/AAV6 reagents28 that mediate the insertion of a constitutive PGK.GFP cassette into the AAV site 1 (AAVS1), a paradigmatic safe harbor for targeted transgene insertion29. To validate the editing strategies, we edited the RPMI 8226 and the K562 cell lines, which do or do not express the CX3CR1 gene, respectively. In K562 control cells, as expected, transgene expression was observed only in the SFFV-exon and AAVS1 conditions, where the expression of the fluorescent reporter is driven by the constitutive promoters encoded by the integrated cassette (Fig. 4D, E). On the contrary, edited RPMI 8226 cells showed expression of the fluorescent reporter in all the conditions, reaching up to 80% of transgene-positive cells (Fig. 4D, E). Digital droplet PCR (ddPCR) with primers designed at the vector-to-genome junction for the different constructs tested (Supplementary Fig. 6A) confirmed a comparable and high targeted integration efficiency (average of 40% of the targeted alleles) for all the tested pairs in both cell lines, molecularly verified also in cells sorted according to the expression of the fluorescent reporter (Fig. 4F, G). These data confirm that the promoter-less cassette allowed specific transgene expression by the CX3CR1 promoter, which is active in RPMI 8226 cells and not in the K562 cell line. Importantly, targeted integration at both the exonic and intronic sites resulted in transcriptional downregulation of CX3CR1 in RPMI 8226 cells (Fig. 4H).
A Schematic representation of the human CX3CR1 locus, with a zoom into intron 4 and exon 5, containing the coding sequence. Target sites of the tested sgRNAs are shown. B Cutting efficiency was measured as a percentage of INDELs of different sgRNAs delivered as RNP complexes with CRISPR/Cas9 in RPMI 8226 and K562 cells. The TIDE software was used to analyze the spectrum and frequency of INDELs. Mean and individual values ± SD are shown, 3 independent experiments. C Schematic representation of the targeted integrations with AAV6 donor templates carrying homology arms matching the chosen sgRNA cutting sites in the CX3CR1 locus. SFFV.YFP – CX exon, promoterless.YFP_SA – CX exon and promoterless.YFP_SA – CX intron donor templates were respectively designed for sgRNA9 (CX exon) and sgRNA5 (CX intron). D Representative FACS plots of RPMI-8226 and K562 cells nucleofected with CRISPR/Cas9 RNPs with the chosen sgRNAs and transduced with the generated AAV6. GFP expression is indicative of the activity of the promoter (promoterless constructs) or efficiency of targeted integration (SFFV-exon and AAVS1 safe harbor control) E Percentage of GFP assessed by FACS in RPMI-8226 and K562 cells edited with CRISPR/Cas9 + AAV6 vectors in the tested and control conditions. F, G Percentage of targeted alleles assessed by ddPCR on bulk, GFP+ and GFP- sorted RPMI-8226 (F) and K562 (G) edited cells. H CX3CR1 gene expression was evaluated with ddPCR on bulk, GFP+ and GFP− sorted RPMI-8226 edited cells. Data shown as the fold on mock. E–H Individual replicates and mean values ± SD are shown, 3 indepentent experiments. Source data are provided as a Source Data file.
We then used an established editing protocol based on CRISPR-Cas9/gRNA electroporation and AAV6 transduction to edit and obtain targeted addition of the cassettes in human CD34+ HSPCs (hHSPCs)28,30(Fig. 5A). A dim, but consistent expression of the fluorescent reporters was observed by flow cytometry in the conditions targeted with the promoter-less cassettes at the CX3CR1 locus, likely reflecting the activity of the CX3CR1 promoter in this heterogenous cell population when cultured in vitro (Fig. 5B, C and Supplementary Fig. 6B). Reporter gene expression in the AAVS1 condition was consistent with previous data28 (Fig. 5B, C). Analysis of targeting efficiency in the edited cells by ddPCR showed an efficient integration (up to 45% of targeted alleles) of all the tested cassettes (Fig. 5D), resulting in transcriptional and protein downregulation in HSPCs edited at the CX3CR1 locus (Fig. 5E, F). Importantly, in the CX3CR1 intron condition, targeted insertion allowed regulated transgene expression and CX3CR1 knocking-out only in HDR-edited cells, while NHEJ-indels did not significantly impact CX3CR1 expression, thus reproducing a haploinsufficiency condition.
A Experimental scheme. After thawing, hHSPCs were kept in prestimulation media enriched with a cytokine cocktail for 2 days. Then, cells were nucleofected with sgRNAs delivered as RNP complexes with CRISPR/Cas9 and transduced with the AAV6 donors. Cells were then kept in culture and analyzed for NHEJ, HDR, FACS, and gene expression to measure the efficiency of editing at the CX3CR1 locus. B Representative FACS plots of the hHSPCs nucleofected with CRISPR/Cas9 RNP with the chosen sgRNAs and transduced with the AAV6. GFP expression is indicative of the activity of the promoter (promoterless constructs) or efficiency of targeted integration (AAVS1 safe harbor control). C Percentage of GFP assessed by FACS in hHSPCs edited with CRISPR/Cas9 + AAV6 vectors in the tested and control conditions. D Percentage of targeted alleles assessed by ddPCR in the edited hHSPCs in the tested and control conditions. E CX3CR1 gene expression in edited hHSPCs in the tested conditions, shown as fold on mock. F CX3CR1 protein expression assessed by FACS in edited hHSPCs in the tested conditions shown as fold on mock. C–F Individual replicates and mean values ± SD are shown, 3 independent experiments. Source data are provided as a Source Data file.
Engraftment and differentiation of CX3CR1-edited hHSPCs in serial transplants
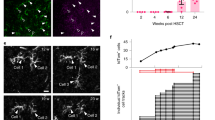
The functional features of the CX3CR1 edited hHSPCs were tested in vivo by transplantation into busulfan myeloablated nonobese diabetic (NOD)-severe combined immunodeficiency (SCID)-gamma (NSG) immunodeficient recipients. As the human cell engraftment in the CNS of NSG transplant recipients is generally extremely low due to the lack of adequate stimuli supporting the human cell engraftment and differentiation, edited cells were administered by combined IV and ICV delivery20. A pool of CD34+ hHSPCs was edited as described above in two independent experiments. Unedited (mock) and AAVS1-edited hHSPCs were used as controls (Supplementary Fig. 7A). The presence of human cells and their differentiation in the recipients were assessed by longitudinal blood cytofluorimetric and molecular analyses. At every time point of analysis, a lower engraftment of the edited hHSPCs as compared to the mock-treated cells was observed, as expected28 (Fig. 6A). Interestingly, the difference in engraftment between mice transplanted with mock-treated versus edited hHSPCs was less significant in the case of cells edited at the CX3CR1 locus as compared to AAVS1-edited hHSPCs (Fig. 6A). On average, 40% of the engrafted human cells in the AAVS1-edited group expressed the transgene, and this fraction remained stable over time in the peripheral blood, indicating efficient and successful editing of repopulating hHSPCs (Supplementary Fig. 7B). Notably, mice transplanted with the CX3CR1 edited cells showed overall low levels of fluorescent reporter expression in the human cells circulating in peripheral blood, with the highest expression at the earliest time point of observation, when the majority of the graft consists of myeloid cells31. This data may suggest that the integrated promoter-less cassette, also in vivo, could be expressed with a specific pattern that possibly reflects the hematopoietic reconstitution dynamics.
A Frequency of mock and edited (at CX3CR1 or at the AAVS1 safe harbor locus) hCD45+ cells detected by flow cytometry in the peripheral blood of NSG recipient mice at 4, 8, 10, and 12 weeks post-transplant. Mean values ± SD are shown. B Frequency of mock and edited hCD45+ cells in the bone marrow of primary and secondary recipients evaluated at 12 weeks post-transplant. Mean and individual values ± SEM are shown. C Percentage of engrafted hCD45+ cells expressing CX3CR1 in BM and brain of mice transplanted with mock or edited hHSPCs. Mean values ± SEM are shown. D GFP expression (left panel) and its relative mean fluorescent intensity (MFI) (right panel) in hCD45+ engrafted in the BM and brain of mice transplanted with CX3CR1 edited (CX exon or CX intron) or safe harbor edited (AAVS1) hHSPCs. Mean and individual values ± SEM are shown. E Percentage of targeted alleles (HDR) assessed by ddPCR in tissues retrieved from the mice transplanted with CX3CR1 edited (CX exon or CX intron) or safe harbor edited (AAVS1) hHSPCs, compared with targeting efficiency retrieved in the infused cell product (input). The human albumin gene was used for normalization. Individual replicates and mean values ± SEM are shown. F Progeny of CX3CR1 (top) or safe harbor (bottom) edited hHSPCs engrafted in the brain of transplant recipients and differentiated into MLCs. The green signal shows the expression of the GFP transgene in edited cells driven by the endogenous CX3CR1 locus (top) or the PGK promoter (bottom). Engrafted cells identified by hNuclei (red signal) express the Iba-1 marker (pink signal) as endogenous/recipient microglia cells. G Branching analysis performed on brain engrafted MLCs edited at the CX3CR1 or safe-harbor loci. The edited cells were identified through the expression of GFP. The cumulative length of branches, complexity index (CI), and covered environment area (CEA) are shown. Mean values ± SEM are reported. H Correlation between intersection radii and sum intersection parameters obtained from Sholl analysis performed on CX3CR1 or AAVS1-edited cells engrafted in the brain of transplanted mice. The vertical and horizontal lines divide the graphs into four quadrants, to describe the cells according to different grades of morphologic complexity, i.e., UR upper right quadrant, for very complex cells characterized by a high sum of intersections and a high number of intersecting radii; LL lower left quadrant, for cells with lower complexity; UL (upper left) and LR (lower right) quadrants for cells displaying intermediate complexity between the LL and the UR quadrants. In the dotted square, the histogram represents the percentage of cells retrieved in each of the four quadrants to quantify the data. In all panels, Mock and AAVS1, n = 6; CX exon, n = 9; CX intron, n = 8. Two independent experiments. Two-way ANOVA with multiple comparison by Tukey’s test in (A, C), with significant effects of treatment (****p < 0.0001) and time points (****p < 0.0001) in (A) and of treatment (****p < 0.0001) and tissue (*p < 0.05); the results of the multiple comparisons are indicated. One-way ANOVA with multiple comparison by Kruskal–Wallis test was performed in (B, D, E, G). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The absence of statistical test information means not significant (ns) except for (D, panel on the right, GFP MFI) were the following comparisons (AAVS1 BM vs CX exon Br, AAVS1 BM vs CX intron Br, CX exon BM vs CX exon Br, CX exon BM vs CX intron Br, CX intron BM vs CX exon Br, CX intro BM vs CX intron Br, AAVS1 Br vs CX exon Br, AAVS1 Br vs CX intron Br) had a ***p < 0.001. In (H), distribution of ratio analysis was performed: ****p < 0.0001 in the LL and UR quadrants. Images were acquired via Zeiss 980 Confocal acquisition, 20X and 40X, Z-stack. n > 80 cells, n = 3 mice/group. Source data are provided as a Source Data file.
At steady-state hematopoiesis, 12 weeks post-transplant, mice were euthanized to collect hematopoietic organs and brain after extensive perfusion. CD34+ human progenitor cells were isolated from their BM to perform secondary transplantations (by IV cell delivery) into busulfan-conditioned NSG recipients (Supplementary Fig. 7A). At sacrifice of all the primary and secondary recipients 12 weeks after transplant, the progeny of the transplanted human cells was detected in hematopoietic organs (BM, spleen, thymus) and brain (Fig. 6B and Supplementary Fig. 7C, D). Overall, and consistent with peripheral blood data and the literature, also in tissues the edited hHSPCs showed a lower engraftment than the mock-treated cells. As in peripheral blood, also in the bone marrow the hHSPCs edited at the CX3CR1 locus showed a higher engraftment than the hHSPCs edited at the AAVS1 locus and, particularly for the hHSPCs edited in the intron, more similar to the mock-treated cells (Fig. 6B). A trend towards a better engraftment of the CX3CR1 edited cells was also observed in the brain, despite significance could not be reached likely as engraftment was overall very low in all study animals20 (Supplementary Fig. 7C). In most of the mice transplanted with CX3CR1 intron-edited hHSPCs the thymus was present and repopulated by the human cells, at odds with the other groups (Supplementary Fig. 7D). Engrafted human cells were also characterized for their immunophenotype, showing a multilineage differentiation in all the experimental groups (Supplementary Fig. 7E). Low percentages of CX3CR1+ human cells were retrieved in hematopoietic organs, while more than 50% of the brain-associated human cells expressed CX3CR1 (Fig. 6C), with fewer CX3CR1+ cells in the CX3CR1-edited hHSPC recipients as compared to the AAVS6 edited condition, possibly reflecting the modulation of protein expression consequent to the editing process. Mice transplanted with AAVS1-edited hHSPCs showed a homogeneous pattern of transgene expression (with approximately 40% positive cells) in all tested tissues (Fig. 6D). On the contrary, and consistent with the CX3CR1 expression, transgene expression in mice transplanted with hHSPCs edited at the CX3CR1 locus with the promoter-less cassette was relatively low (Fig. 6D), but not in the brain-associated human cells in which the highest transgene expression was observed (Fig. 6D). Interestingly, the mean fluorescence intensity (MFI) of the reporters in brain engrafted human cells was higher in mice transplanted with CX3CR1 edited hHSPCs as compared to AAVS1-edited cells, where the expression of the transgene is driven by the PGK promoter (Fig. 6D). This data indicates that the promoter-less cassette allows a higher than PGK and specific expression of a targeted integrated transgene under the control of the endogenous CX3CR1 promoter also in vivo, in tissues.
We then evaluated the targeting efficiency in vivo by performing a ddPCR designed at the vector-to-genome junction region to detect the integrated cassette in tissues collected from the transplanted mice. Targeting efficiency was good in all the groups in hematopoietic organs at sacrifice (Fig. 6E). We confirmed a multilineage editing in both myeloid (hCD13) and lymphoid (hCD19) sorted populations from the spleen in all the groups of transplanted mice (Supplementary Fig. 7F), as well as a good targeting efficiency, stable over time, in the peripheral blood of transplant recipients (Supplementary Fig. 7G). This was also proven in the BM of secondary recipients in all the groups, indicating a good efficiency of the editing procedure in the stem compartment (Supplementary Fig. 7H). Interestingly, despite the relatively low human engraftment observed in the brain due to the challenging mouse-into-human setting in the NSG model, a higher targeted integration was retrieved in the brain myeloid progeny of the hHSPCs edited at the CX3CR1 locus, especially with the construct targeting CX3CR1 intron, as compared to the other tissues (Fig. 6H). This data suggests an enrichment of the CX3CR1 edited (and haploinsufficient) cells in the brain as compared to hematopoietic organs.
To strengthen this observation, we examined the frequency of biallelic versus monoallelic editing in vivo by analyzing targeting in the cell progeny obtained by colony-forming cell (CFC) assay from the infused HSPCs and the BM retrieved from the transplanted mice at sacrifice. This assay allows analyzing the targeted integration of a transgene expression cassette at the clonal level by screening colonies by means of an ‘IN-OUT’ PCR approach30. In the CFC progeny of the infused HSPCs, we observed a similar frequency of mono and biallelic targeting, with 21% of not targeted Colony Forming Units (CFUs) in the CX intron approach versus 31% in the exon approach. Instead, in the CFU progeny of the BM retrieved from the transplanted mice, which reflects the frequency of edited progenitors that had engrafted in vivo, a trend towards a higher proportion of monoallelic versus biallelic targeting was observed compared to the infused product, especially with the CX intron approach, suggesting a possible advantage of the monoallelic targeting in vivo (Supplementary Fig. 7I, J).
Next, we performed an immunofluorescence analysis on brain tissue slices to characterize the brain progeny of the transplanted hHSPCs. Engrafted human cells showed a microglia-like morphology, with a good extent of ramifications, positivity for microglia markers, such as Iba-1, and expression of the transgene (Fig. 6F). To better characterize the morphology of the brain engrafted cells edited at CX3CR1 or safe-harbor locus, we performed branching and Sholl analyses, using the same approach employed in the mouse-into-mouse transplantation setting described above. Interestingly, CX3CR1 edited hHSPC progeny cells showed a greater extent of ramifications and, in general, a more complex morphology and a greater complexity of ramifications as compared to control AAVS1-edited cells (Fig. 6G). Similarly, the analysis showed that MLCs derived from CX3CR1 edited hHSPC covered a larger surface and displayed more arborizations as compared to control AAVS1-edited cell progeny (Fig. 6H; Supplementary Movie 1 - AAVS1 edited MLC and Supplementary Movie 2 - CX3CR1 edited MLC). Thus, in vivo, differentiation toward a microglia-like phenotype of hHSPCs is favored by CX3CR1 haploinsufficiency.
Discussion
In the context of experiments aimed at identifying the population mostly contributing to brain myeloid cell turnover after HSPC transplantation, we identified CX3CR1 as a unique locus that could be exploited to design HSC gene therapy approaches endowed with a unique therapeutic potential for the treatment of NMDs and NDs. Haploinsufficiency of the CX3CR1 locus of murine HSPCs resulted in an enhanced capability of the engineered cells and their progeny to engraft in the CNS and replace endogenous microglia with gene-modified MLCs upon transplantation. Editing and targeted addition at the CX3CR1 locus of human HSPCs instead favored their and their progeny myeloid/microglia differentiation in the CNS, and allowed obtaining a robust and regulated expression of the integrated cassette in transplant-derived cells, with a strong potential of exerting therapeutic effects in the CNS.
Our first serendipitous observation at the basis of our work was that murine HSPCs haplo-insufficient at the Cx3cr1 locus show a unique transplant-associated phenotype characterized by enhanced engraftment and differentiation capability towards MLCs as compared to WT cells in the CNS of standard and competitive-transplant recipients18. In the more challenging and translationally relevant setting of human HSPC transplantation into immunodeficient mice, characterized by conditions suboptimal for the generation of myeloid chimerism in the brain, editing of the CX3CR1 locus was associated with a more robust maturation of the transplanted hHSPCs towards bona fide MLCs, as compared to cells edited at other loci. Likely because of the limitations of the chimeric model, differences in CNS engraftment of the CX3CR1 edited versus control cells did not reach statistical significance. Thus, the data indicated that CX3CR1 haploinsufficiency confers a quantitative and qualitative advantage to HSPCs in reconstituting the brain myeloid compartment of transplant recipients that could be exploited for translational goals.
CX3CR1 is one of the main microglia signature genes, active in early microglial precursors, and expressed throughout adulthood32,33. Microglia precursors develop into CD45+ c-kitlow CX3CR1- immature cells that then mature into CD45+ c-kit− CX3CR1+ cells, making CX3CR1 one of the first genes turned on during microglia ontogenesis33,34. As we have previously shown that newly appearing MLCs in transplanted mice are generated through a process that closely resembles the physiologic postnatal microglia maturation18, we can hypothesize that the CX3CR1 locus could participate as well in the process of microglia repopulation following the HSPC transplant, also considering its role in the control of cell proliferation and migration16 and that perturbations of CX3CR1 expression levels could modulate the process as well. Notably, a similar effect was also observed in hematopoietic organs, thus suggesting a possibly wider role of this molecule in the hematopoietic reconstitution process.
Interestingly, transcriptomic analysis performed on MLCs sorted from competitively transplanted mice revealed significant expression of microglia genes, including ApoE, in clusters enriched in Cx3cr1−/+ cells, while expression of genes associated to an active process of early myeloid differentiation and proliferation characterized clusters enriched in Cx3cr1+/+ cells, thus suggesting an actively occurring process of maturation and differentiation in the latter and, likely, a more advanced stage of maturation in the former. Moreover, upregulation of specific pathways associated with microglia differentiation and migration, including Cdc42-associated genes, was observed in a cell cluster enriched in Cx3cr1−/+ cells (cluster 4), that was also phenotypically confirmed at protein level. Cx3cr1-induced signal transduction pathway can activate Cdc42 via Syk and PI3K, that are ultimately required for macrophage chemotaxis towards Cx3cl135. Cdc42 signal transduction pathway has been also associated with myelopoiesis and HSC engraftment, as dysregulation of Cdc42 results in disorganized actin structure in hematopoietic cells and defective engraftment in stem cell transplant protocols36,37. Of note, HSPCs from Cdc42−/− mice show defective migration and adhesion, which is associated with abnormal F-actin assembly, homing, and engraftment/retention in the BM38. Thus, Cx3cr1 haploinsufficiency could perturb Cdc42-associated pathways and ultimately result in cytoskeleton rearrangements, increased cell motility, and cell cycle progression during the microglia reconstitution process following HSPC transplant, a phenomenon that could explain our findings.
This serendipitous discovery opened for us an interesting scenario where this genomic locus could be exploited to enhance the ability of HSPCs to engraft and repopulate the hematopoietic system and, more importantly, the CNS myeloid compartment of transplant recipients. Moreover, since CX3CR1 is robustly expressed by microglia in neuropathological conditions and hence considered a possible relevant therapeutic target in neurodegenerative disorders39, we envisaged to design a CRISPR-based targeted gene addition strategy at the human HSPC CX3CR1 locus inserting a wild-type copy of a disease-causing gene that could also allow obtaining a regulated and robust therapeutic transcript expression by transplant-derived MLCs. To challenge the applicability of this strategy, we developed new tools for CRISPR/Cas9-based gene editing and AAV6 based gene addition at the human CX3CR1 locus allowing for the generation of a population of edited CX3CR1 haploinsufficient/knocked out HSPCs that could express a therapeutic transcript under the control of the CX3CR1 promoter. As control, we chose the AAVS1 locus, a well-validated safe harbor29.
Targeted insertion at the chosen intronic region of the CX3CR1-expressing cell line resulted in a regulated expression of the reporter gene and concomitant CX3CR1 knock-out only in HDR-edited cells, while NHEJ-INDELs did not impact CX3CR1 expression. Since only a minor fraction of engrafting cells will contain biallelic integrations, this strategy will mostly result in the generation of the desired haplo-insufficient cells. These results were confirmed on hHSPCs, where integration of the reporter cassette resulted in a dim, but consistent expression of the fluorescent reporters, reliably reflecting the activity of the CX3CR1 promoter in this heterogenous cell population when cultured in vitro. Molecular analysis confirmed a good efficiency of targeted integration, resulting in CX3CR1 transcriptional and protein downregulation, reproducing a haploinsufficiency condition. These findings were also maintained in secondary transplant recipients, indicating that CX3CR1 editing occurred in long-term repopulating HSCs and that its modulation could impact the functional properties of these cells and their progeny. Remarkably, the higher targeted integration retrieved in the brain myeloid progeny of the transplanted CX3CR1-edited hHSPCs, as compared to what was observed in the cells that repopulated the hematopoietic organs of the primary and secondary recipients, suggests an enrichment of the edited cells within the CNS-engrafted progeny of the transplanted HSPCs. This enrichment was not seen in the AAVS1 group, thus indicating in CX3CR1 downregulation the most likely mechanism leading to this enrichment. Of note, the branching study revealed that the CX3CR1-edited cells engrafted in the brain more robustly differentiated towards human MLCs, thus confirming that the reduction of CX3CR1 expression also in human cells could favor the maturation and differentiation of the transplanted HSPCs and their progeny in the CNS towards microglia.
Importantly, the targeted integration of a promoterless cassette at the CX3CR1 locus of HSPCs allowed specific, regulated, and robust transgene expression in their progeny engrafted (in hematopoietic organs and) in the CNS. Indeed, the transgene integrated at the CX3CR1 locus was expressed consistently with the activity of the endogenous promoter, with substantially higher expression in CNS-associated transplant progeny MLCs as compared to similarly edited cells engrafted in hematopoietic organs and to cells edited at the AAVS1 locus, where the integrated cassette contained a conventional PGK promoter to drive transgene expression. The latter finding is of particular relevance as the PGK promoter is currently used in HSPC gene therapy clinical applications for neurodegenerative/neurometabolic conditions1,2,4,5. Overall, the robust, lineage-specific and regulated therapeutic transcript expression coupled with the faster pace of transplant progeny cell engraftment in the CNS that can be achieved with our CX3CR1 editing and targeted gene addition approach could be particularly advantageous in neurodegenerative/neurometabolic conditions. As a matter of fact, despite HSPC gene therapy has shown unprecedented clinical benefits in neurometabolic patients treated at a very early disease stage or when pre-symptomatic, symptomatic patients cannot benefit from the treatment because of the progression of their disease before therapeutic cells could exert their effects in the CNS. Our approach, by shortening and fostering the microglia reconstitution process after engineered HSPC transplantation, could expand the temporal window for treatment and guarantee robust therapeutic gene expression.
Different strategies have been proposed to favor microglia replacement in murine models40,41,42, and recent studies allowed identifying key variables affecting the occurrence and efficiency of the process43. These works showed that emptying microglia niches and administering a myeloablative conditioning regime before transplant favor the engraftment of donor-derived cells. Moreover, as shown by Xu and colleagues40, also the cell source for transplant could modulate the cell replacement process. These variables also affect the transcriptional profile exhibited by MLCs originated by the hematopoietic cell transplant in different experimental conditions: the transplantation of peripheral blood or total bone marrow cells following administration of CSF1R inhibitors and irradiation results in cells showing macrophage-like transcriptional characteristics, whereas MLCs derived from HSPCs transplanted in recipients pre-treated with myeloablative busulfan show a transcriptional profile and morphological characteristics superimposable to microglia18,20. Importantly, these strategies may equally benefit from the use of CX3CR1-edited HSPCs. This would be of particular relevance for the development of more effective and safer protocols for microglia reconstitution by HSPC transplantation44 in NDs, for which this approach has already shown to be promising in murine models45,46,47.
Based on this, the development of a simple, efficient, and very specific treatment platform for NMDs based on an empowered HSPC-targeted gene addition could also reduce potential safety concerns associated with side effects related to vector integration8,9,10,11, as high-sensitivity safety workflows are being quickly developed and applied prior to clinical translation, therefore maximizing the chances for successful clinical implementation and long-term safety of site-specific genome editing therapies48,49.
In conclusion, based on indications strongly supporting a quantitative and qualitative advantage in the post-transplant maturation of CX3CR1 deficient HSPCs towards MLCs, we designed a promoterless, splice-trapping cassette allowing for efficient editing and targeted gene addition at the CX3CR1 locus, enhancing the ability of HSPCs to repopulate the CNS myeloid compartments of transplant recipients with mature microglia-like cells, obtaining specific, regulated and robust transgene expression in the hematopoietic system and in the CNS. We envision to apply this platform for the treatment of disorders characterized by a rapid progression of neurodegeneration where CX3CR1 edited and engineered cells could express at high levels a therapeutic transcript in the myeloid progeny of the transplanted cells.
Methods
Mouse studies
All experiments and procedures involving animals were performed with the approval of the Dana Farber Animal Facility Institutional Animal Care and Use Committee (IACUC 15-031). Mice have been housed in appropriate rooms with controlled temperature, humidity, and automated lighting, ensuring a standard light/dark circadian cycle. Wild-type C57BL/6J mice or C57BL6/Ly5.1 mice (hereafter called CD45.2 or CD45.1, respectively) were obtained from Jackson Lab or Charles River. NOD-SCID-IL2Rg−/− (NSG) female mice (Stock No: 005557) were obtained from The Jackson Laboratory.
Cx3cr1GFP/+ (referred to as Cx3cr1−/+) mice were generated by crossing Cx3cr1GFP/GFP (referred to as Cx3cr1−/−) obtained from The Jackson Laboratory (Stock. No 005582) with wild type CD45.2 mice (referred as Cx3cr1+/+). For all transplantation experiments, mice were randomly distributed to each experimental group. Female animals were used as recipients due to their better response to busulfan conditioning and homogeneity in the engraftment. Female and male mice were used as donors.
Isolation, transduction, and transplantation of murine hematopoietic cells
Seven/eight-week-old wild type, Cx3cr1−/+ or Cx3cr1−/− mice were euthanized with CO2, and the BM was harvested by crushing bones. After BM lysis, HSPCs were purified by Lin- selection using the Lineage Cell Depletion Kit (Miltenyi, #130-090-858) with the autoMACS™ magnetic separation, following the manufacturer’s instruction.
Sorting experiments
GFP+ and GFP- cells were sorted from the Lin- pool isolated from Cx3cr1−/+ mice using the BD FACSAria II high-speed cell sorter. Collected cells were freshly transplanted IV into busulfan (4 doses 25 mg/kg) conditioned CD45.1 recipients at a 1:1 donor/recipient ratio. Mice also received 1.0 × 106 CD45.1 BMNC IV 5 days post-transplant for hematopoietic rescue. Forty-five days post-transplant mice were sacrificed, and BM and brain were collected for cytofluorimetric analysis.
Standard transplantation experiments
Isolated Cx3cr1+/+ or Cx3cr1−/+ Lin- were transplanted IV into busulfan-conditioned CD45.1 recipients (1.0*106/mouse) after 12–16 h of culture in StemSpan medium supplemented with cytokines as previously described50. Mice were sacrificed at 45, 90, and 180 days post-transplant to collect hematopoietic organs and brain for flowcytometric analysis.
Competitive experiments
Isolated Cx3cr1+/+, Cx3cr1−/+, and Cx3cr1−/− Lin- were transduced with LVs, for 12–16 h at multiplicity of infection (MOI) 100 51. The following LVs were used:
pCCLsin.cPPT.humanPGK.BlueFluorescentProtein.Wpre (BFP-LV) for Cx3cr1−/+ or Cx3cr1−/−HSPCs and pCCLsin.cPPT.humanPGK.mCherryProtein.Wpre (mCherry-LV) for Cx3cr1+/+ HSPCs. A fraction of the transduced cells was cultured for 10 days in vitro50 to assess transgene expression by cytofluorimetric analysis. Transduced cells were injected via the tail vein or directly in the CNS by means of ICV injection into seven/eight-week-old conditioned CD45.1 female mice as previously described20.
For IV group a total of 1.0 × 106 cells/mouse (0.5 × 106 Cx3cr1−/+ or Cx3cr1−/− BFP+ HSPCs + 0.5 × 106 Cx3cr1+/+ mCherry+ HSPCs) was injected. For ICV injection, a total of 0.3 × 106cells/mouse (0.15 × 106 Cx3cr1−/+ BFP+ HSPCs + 0.15 × 106 Cx3cr1+/+ mCherry+ HSPCs) was injected. ICV transplanted mice received also 1.0 × 106 CD45.1 BMNC IV 5-day post-transplant for hematopoietic rescue.
Trans-well migration assay
A trans-well migration assay was performed using HSPCs isolated from naïve wild type and Cx3cr1−/+ mice. 2 × 105 purified HSPCs were seeded in the upper chamber of 5-μm-pore filters trans-well plate, while in the bottom chamber Fractalkine (FLK), Cx3cr1 ligand, was added at different concentrations (100 and 200 ng/mL). The percentage of migrating cells was analyzed after 24 h. As a control, in another chamber SDF-1 (50 nM) was added, as HSPCs are expected to chemotactically migrate in the presence of this chemokine25. Additionally, the same assay was performed with RAW 264.7, a murine macrophage cell line expressing high levels of Cx3cr1 and, therefore, expected to migrate in the presence of FLK.
Cell lines and primary cell culture
HEK293T cells used for AAV and LV productions were cultured in Dulbecco’s Modification of Eagle’s Medium (DMEM) 4.5 g/L glucose or Iscove’s modified Dulbecco’s medium (IMDM; Corning), respectively, supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2% glutamine. To choose and test the tools for gene editing at the CX3CR1 locus, we used RPMI 8226 (ATCC #CCL-155™), a suitable transfection host cell line expressing CX3CR1 gene, and K562 (ATCC #CCL-243™), not expressing CX3CR1, as negative control. RPMI 8226 were cultured in RPMI medium supplemented with 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2% glutamine. K562 were cultured in Iscove’s modified Dulbecco’s medium (IMDM; Corning) supplemented with 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2% glutamine. Both cell lines were cultured in a 5% CO2 humidified atmosphere at 37 °C.
Commercial cord blood CD34+ human HSPCs were purchased frozen from AllCells and were thawed using thymocyte thawing media (TTM) medium, modified from ref. 51. TMM medium was prepared with RMPI medium supplemented with 30% FBS, 1%Pen/Strep, 10 ug/ml DNase I (Sigma), and 20 U/ml heparin. Cord blood CD34+ cells were gently thawed in pre-warmed TMM medium and left in the water bath at 37 °C for 1 h. Cells were then spun down (300×g 5 min) and seeded at the concentration of 5 × 105 cells/ml in a cytokine-enriched medium for prestimulation. Serum-free StemSpan SFEM II medium (StemCell Technologies, #09605) was supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin, 100 ng/ml hSCF (PeproTech), 100 ng/ml hFlt3-L (PeproTech), hTPO 100 ng/mL (PeproTech), hIL-6 (PeproTech) and SR1 (StemRegenin1; 0.75 μM, CellagenTech). Cells were left in prestimulation for 2 days before the editing procedure. Cord blood CD34+ cells were cultured in a 5% CO2 humidified atmosphere at 37 °C in low oxygen conditions.
LV and AAV6 production
Third-generation SIN Lentiviral vectors were produced and titered as previously described in ref. 50. For in vitro experiments, AAV6 vectors were produced using the AAVpro® Purification Kit (Takara Bio #6666) according to manufacturer instructions and titered as previously described in ref. 30. For in vivo experiments, AAV6 vectors were produced using iodixanol gradients and ultracentrifugation by the Boston Children’s viral vector core.
CRISPR/Cas9 Gene editing in cell lines and in human hematopoietic stem cells
Sequences of the gRNAs were designed using an online tool52 and selected for predicted specificity score and on-target activity. AAV6 donor templates were generated from a construct containing AAV2 inverted terminal repeats (ITRs) as previously reported29. Vector maps were designed with SnapGene software v5.0.7 (from GSL Biotech; available at snapgene.com).
For experiments in cell lines, RNP complexes were assembled by incubating at 1:1.5 molar ratio Streptococcus pyogenes (Sp)Cas9 protein (integrated DNA technologies-IDT) with pre-annealed synthetic Alt-R® crRNA:tracrRNA (IDT) for 15’ at room temperature together with 0.1 nmol of Alt-R® Cas9 Electroporation Enhancer (IDT), added prior to electroporation according to manufacturer’s instructions. Both RPMI 8226 and K562 were nucleofected (FF120 program, Lonza 4D-Nucleofector) with the SF Cell 4 d Nucleo Kit (Lonza #V4XC-2032) using 50 pM of the different sgRNAs combined with CRISPR/Cas9 in the form of RNP complexes. AAV6 transduction (20,000MOI) was performed right after electroporation, maintaining the cells at a density of 0.25 × 106 cells/ml. After 5 h, cells were diluted in the proper medium and moved to a bigger well to maintain a cell density of 0.5–0.7 × 106 cells/ml overnight. The following day (~12–16 h after), cells were washed with PBS and seeded for expansion for molecular characterization.
For experiments in primary hematopoietic stem cells (CD34), RNP complexes were assembled by incubating at RT for 5′ (Sp)Cas9 protein (IDT) with synthetic single guide RNAs (sgRNA) chemically modified (with 2′-O-methyl at 3 first and last bases, 3′ phosphorothioate bonds between first 3 and last 2 bases and addition of 80-mer SpCas9 scaffold to create a single guide RNA) obtained from Synthego. Per 300,000–500,000 cells in in vitro experiments, nucleofection strips from LONZA Kit S (P3 Primary Cell 4D-Nucleofector™ X #V4XP-3032) were used, mixing 6 μg of Cas9 protein with 3.2 μg of sgRNA at room temperature for 5’. For in vivo experiments, the Lonza 4D nucleocuvette cuvettes from LONZA Kit L (P3 Primary Cell 4D-Nucleofector™ X #V4XP-3012) were used, scaling up the amounts of cells and reagents of five folds.
After thawing, cells were put in prestimulation for 2 days in the cytokine-enriched medium. Cells were then washed with PBS, counted, and nucleofected with the assembled RNP complexes (DZ100 program, Lonza 4D-Nucleofector). AAV6 transduction was performed right after electroporation at 20 K MOI, maintaining the cells at a density of 0.25 × 106 cells/ml. After 5 h, cells were diluted with the cytokine-enriched medium and moved to a bigger well to maintain a cell density of 0.5–0.7 × 106 cells/ml overnight. The following day (~12–16 h after), cells were washed with PBS and seeded for expansion for flowcytometric analysis and molecular characterization or were prepared for transplantation.
Molecular characterization of edited cells
TIDE analysis
Four to five days after electroporation, a fraction of cells (~105) was collected. DNA was extracted using DNA Extraction QE Buffer (Biosearch Technologies #QE09050) and amplified using different primer assays according to the target genomic region. A list of primers and thermal protocols is available in supplementary material. After amplification, the PCR product was run on 1.5% Agarose gel (Tte Laboratories Inc., #BMAG01), and DNA was extracted from the gel with GeneJET Gel Extraction Kit (Life Technologies, # K0692). Purified DNA was then sequenced (SANGER seq) to evaluate the percentage of Insertions and Deletions (INDELs) with the TIDE (Tracking of Indels by Indels by DEcomposition) software53 (Brinkman et al., 2014).
Targeted integration (HDR)
To evaluate the efficiency of integration by homology direct repair (HDR), DNA was extracted with the QIAamp DNA Micro Kit (QIAGEN) from cells expanded in vitro for 10–14 days. About 20–50 ng of genomic DNA were analyzed using the QX200 Droplet Digital PCR System (Bio-Rad) according to the manufacturer’s instructions. Different assays of primers and probes were designed on the junction between the vector sequence and the targeted locus. Human albumin was used for normalization. A list of primers and thermal protocols is available in supplementary material.
Gene expression
To evaluate the expression of the CX3CR1 gene, RNA was extracted with RNeasy Plus Micro Kit (Qiagen) from cells expanded in vitro for 7–10 days. cDNA was synthetized with Maxima Reverse Transcriptase (Thermo Fisher # EP0742) and CX3CR1 expression was quantified using TaqMan® Assays Hs01922583_s1 and human GAPDH Hs02786624_g1 as housekeeping reference. About 1 ng RNA equivalent was analyzed using the QX200 Droplet Digital PCR System (Bio-Rad). Poisson statistical analysis of the numbers of positive and negative droplets yields absolute quantification of the target sequence.
Flow cytometry
CX3CR1 Monoclonal Antibody (2A9-1) eBioscience™ was used to assess protein expression by flow cytometry in cells expanded in vitro for 7 days. Cells were collected and resuspended in 100 ul of MACS buffer with 2:100 FcR Blocking Reagent (Miltenyi, #130-059-901) and incubated 10’ at 4 °C to avoid aspecific binding of the antibodies. Cells were then incubated with the antibody for 20’ (labeling procedure) at 4 °C. After washing, cells from different tissues were resuspended in MACS buffer (300–400 μl). Vital dye (7AAD) was added, and samples were analysed at BD LSR Fortessa. Results were analysed by FlowJo 10.8.0 software.
Transplantation of edited human hematopoietic stem cells in immunodeficient recipients
Edited human HSPCs were transplanted into 7–8 weeks old NSG females conditioned with busulfan (16.25 mg/kg per 4 days). Cells were administered IV (0.5 × 106/mouse) and ICV (0.3 × 106/mouse) as previously described20. Mice were then provided with syngeneic BMNC for hematopoietic rescue 4 days post-transplant.
Secondary transplantation was performed upon injection of 2 × 106 beads-purified human CD34+ cells (CD34 MicroBead Kit, Miltenyi #130-046-702) harvested from the BM of primary engrafted NSG mice euthanized at 12 weeks post-transplant. Mice were then provided with syngeneic BMNC for hematopoietic rescue 5 days post-transplant. Transplanted mice were monitored by bleeding once/month and euthanized after 12 weeks as previously described.
Evaluation of peripheral engraftment in the blood
Human CD45+ cell engraftment and the presence of edited cells were evaluated by bleeding mice periodically from the tail vein. 0.1 ml of blood was obtained per sample and collected into tubes containing 10 μl EDTA solution (45 ng/ml) to avoid coagulation of the blood during the blood draw. Cells of donor origin were detected by FACS analysis after lysis of erythrocytes with ACK (10 min at room temperature) and specific staining as specified below. The remaining cells were used to extract DNA with QIAamp DNA Micro Kit (QIAGEN) to perform molecular analysis as specified below. A list of flow cytometry antibodies is available in supplementary material.
Mouse tissue collection and processing for flow cytometry and histology
According to the time and the experimental settings, mice were euthanized under deep anesthesia (Ketamine/Xylazine mix) by extensive intracardiac perfusion with cold PBS for 15 min after clumping the femur. Hematopoietic organs and the brain were then collected and differentially processed.
BM cells were harvested by flushing the tibias and femurs with PBS 2%FBS. Spleen and thymus were mechanically disaggregated on a cell strainer (40 μm) in PBS 2%FBS (15 ml for spleen, 5 ml for thymus). About 500 μl of homogenate tissues were centrifuged at 900 × g for 5′ and then resuspended in 100 μl of blocking solution (MACS buffer with 2:100 FcR Blocking Reagent Miltenyi #130-059-901 and 1:100 CD16/CD32 Blocking Assay, BD Biosciences #553142) and incubated 10 min at 4 °C to avoid aspecific binding of antibodies. Cells were then incubated with specific antibodies for 20 min (labeling procedure) at 4 °C. After washing, cells from different tissues were resuspended in MACS buffer (300–400 μl). Vital dye (DAPI or 7AAD) was added, and samples were analysed at BD LSR Fortessa. Results were analysed by FlowJo 10.8.0 software. A list of flow cytometry antibodies is available in supplementary material.
The brain was removed, and the two hemispheres were differently processed. For immunofluorescence analysis, one hemisphere was fixed for 24 h in 4% PFA, embedded in OCT compound, and stored at −80 °C, after equilibration in sucrose gradients (from 10 to 30%) supplemented with 0,02% of sodium azide to avoid contamination. For flow cytometry analysis, cells were obtained by mechanic disaggregation of one brain hemisphere in 1.5 ml in EBSS medium and processed with a papain-based digestion procedure according to the Neural Tissue Dissociation Kit (Miltenyi, #130-092-628). After washing with EBSS medium, the digested suspension was enriched in myeloid cells with Percoll gradient (700×g for 15 min, no break). The cell suspension was then washed with PBS 2%FBS, put in a blocking solution, and stained with specific antibodies as described above for flowcytometric analysis. For the ICV experiment, the brain hemisphere containing the injection site was analysed by FACS analysis, while the contralateral part was used for immunofluorescence analysis.
From all the tissues, a fraction of the cell suspension was stored at −80 °C. DNA was then extracted with the QIAamp DNA Micro Kit (QIAGEN) to perform molecular characterization and evaluate editing efficiency in vivo as described above. A list of primers and thermal protocols is available in supplementary material.
IN-OUT PCR
The frequency of biallelic vs monoallelic editing was assessed by analyzing the in vitro CFU progeny of the infused CD34+ HSPCs and the in vitro CFU progeny of the BM retrieved from the transplanted mice at sacrifice. This assay enables the analysis of clonal genotypes by PCR screening of colonies for targeted integration of a transgene expression cassette30. 800 CD34+ HSPCs and 0.15 × 106 total bone marrow nucleated cells retrieved from transplanted mice at sacrifice were seeded in an enriched methylcellulose-based medium (StemCell Technologies, Cat#H4435) and single-picked after 14 days of in vitro culture. DNA was extracted with DNA Extraction QE Buffer (Biosearch Technologies, Cat# QE09050).
To identify biallelic vs monoallelic editing, we used an “IN-OUT” PCR approach, in which one primer was designed in the targeted genomic locus outside the region of the homology arm (Outside LHA), and the other primer was located inside the transgene cassette (on YFP). A third primer, located in the genomic region on the opposite side of the sgRNA target site from the primer outside the homology arm (on RHA), was also included for identifying alleles without integration. After amplification, DNA was loaded on a 1% Agarose gel, and bands were visualized at ChemiDoc 1708279 (Bio-Rad). A list of primers and thermal protocols is available in supplementary material and methods.
Immunofluorescence analysis
Brains embedded in OCT were serially cut in the sagittal planes on a cryostat in 18-μm sections. Brain sections were obtained from the contralateral side of cell injection for intracerebroventricularly transplanted mice.
Tissue slides were washed twice with PBS, air dried, and blocked with 0.3% Triton and 10% FBS for 1 h at room temperature. Then sections were incubated overnight at 4 °C with primary antibodies diluted in PBS, 0.3% Triton, 10% FBS as follows: rabbit anti-Iba-1 (Wako) 1:250; chicken anti-GFP (Abcam) 1:100; rabbit anti-cherry (Abcam) 1:100; anti-human nuclei (Sigma Aldrich) 1:100. The secondary antibodies goat IgG anti-Chicken Alexa Fluor 488, goat IgG anti-Rabbit Alexa Fluor 488, 546, or 633, goat IgG anti-Rat Alexa Fluor 546 or 633, goat IgG anti-Mouse Alexa Fluor 546 (Molecular Probes, Invitrogen) were diluted 1:500 in PBS, 1% FBS and incubated with sections for 90 min at room temperature. Nuclei were stained with DAPI (Roche) at 1:30 in PBS. Slices were washed in PBS, air dried, and mounted with Fluorsafe Reagent (Calbiochem). Not transplanted mice were used as negative controls for the reporter transgene staining. Incubation with a secondary antibody alone was performed to exclude the background signal. Samples were analyzed with a confocal microscope (Zeiss and Leica TCS SP2; Leica Microsystems Radiance 2100; Bio-Rad; Leica SPE confocal) (λ excitation = 488, 586, 660).
Fluorescent signal was processed by Lasersharp 2000 software. Images were imported into ImageJ software and processed by using automated level correction. For the reconstruction of brain sections, we used a fluorescence microscope Delta Vision Olympus Ix70 for the acquisition of the images, which were then processed by the Soft Work 3.5.0 software. Images were then imported into the Adobe Photoshop CS 8.0 software and reconstructed.
For branching analysis, a Macro was developed in the lab with ImageJ software to perform a standardized analysis on multiple individual cells (>50 per group) from different transplanted mice (n = 3/group). Branching results obtained on fluorescent markers were confirmed on Iba-1stained cells, independently from the fluorescent signal reporter.
Automated Sholl analysis22 was performed by applying the same strategy, to study the radial distribution of microglia branches around the cell body. The sum intersections and a number of intersection radii were selected as parameters to characterize donor-derived cells, in terms of complexity and spatial extension of the cell arborizations, respectively.
Cell sorting from hematopoietic organs of transplanted NSG mice
To evaluate the gene editing efficiency of the hematopoietic compartment in vivo, engrafted cells were sorted from the spleen of transplanted NSG mice as previously described54. Briefly, spleens were crushed, and the cell suspension was filtered with a 40-μm cell strainer with cold MACS buffer. The homogenate was then lysed with ACK lysis buffer. After washing with MACS buffer, cells were stained with the dedicated anti-human antibody cocktail for cell lineage sorting. hCD45, hCD3, hCD19, and hCD13 antibodies were used. Dead cells were marked with 7AAD staining. Lymphoid and myeloid populations were sorted with the BD FACSAria II high-speed cell sorter. After sorting, cells were pelleted and stored at −80 °C to proceed with DNA extraction and ddPCR analysis for HDR quantification as described above.
Single-cell data
Single-cell dataset generation
Single cell RNA-Seq was provided by the Single Cell Core at Harvard Medical School, Boston, MA using the 10X Genomics technology. Briefly, MLC sorted from competitively transplanted mice were isolated, and single-cell suspensions were prepared for each sample. Cells were then encapsulated in droplets containing unique barcodes and reverse transcription reagents, followed by library preparation and sequencing.
We generated two libraries for microglia-like cells – MLCs: one for the Cx3cr1+/+ cells and one for the Cx3cr1−/+MLCs. Four animals were competitively transplanted with Cx3cr1+/+ mCherry+ HSPCs plus Cx3cr1−/+ BFP+ HSPCs. At sacrifice, 15 days post-transplant, brains were processed to obtain a single cell suspension, and all the mice were pooled to obtain one sample to be stained and sorted as 7AAD−/CD11b+/mCherry+ and 7AAD-/CD11b+/BFP+. A total of 2.435 mCherry+ cells and a total of 2.634 BFP+ cells were sorted and processed for single-cell barcoding in the 10X Genomics platform.
Data processing and quality control
The raw sequencing data was processed using the Cell Ranger software (version 4.0.0)55 to obtain gene expression matrices for each sample. The resulting matrices were then imported into the Seurat package (version 4.0.4)56 for quality control and downstream analysis. Cells with a low number of detected genes (<350) and high mitochondrial gene content (>15%) were filtered out. Cells with a total number of reads less than 3500 and more than 35,000 were also removed from the dataset. The filtering thresholds have been set based on the inspection of the data distributions and literature.
We employed the ScType algorithm57 for automated cluster annotation across the entire single-cell dataset, setting “Brain” as the reference tissue. ScType effectively assigned putative cell types to each cluster. All the smaller clusters displaying association with various other brain cell types, suggesting potential sample contamination, were excluded from downstream analyses to ensure the integrity of the dataset.
Normalization and scaling
The expression data were normalized and scaled using the SCTransform58 function in Seurat. This method applies a regularized negative binomial regression to model the count data and correct for technical noise and batch effects. In the normalization process we included as covariates quantitative indexes reflecting cell cycle status (S.Score, G2M.Score), and percentage of cumulative mitochondrial gene expression. Gene Cx3cr1 was removed from the set of genes to avoid biases related to haploinsufficiency. The resulting scaled data was used for all downstream analyses.
Principal component analysis
Principal component analysis (PCA) was performed on the scaled data using the RunPCA function in Seurat. The top 12 principal components were selected based on their contribution to the total variance in the data. The resulting principal components were used as input for UMAP dimensionality reduction.
Uniform manifold approximation and projection (UMAP)
UMAP (McInnes et al. preprint59), was performed on the PCA-reduced data using the RunUMAP function in Seurat. This method projects high-dimensional data into a low-dimensional space while preserving the global structure of the data. The resulting UMAP plot was used for visualization and cell type identification.
Clusters identification
Based on the distance matrix calculated using PCA scores, the k-nearest neighbors graph (k = 20, Seurat default) has been calculated. Clusters were identified using the Louvain algorithm. We tested different resolution values (0.6; 1.2) and finally set it at 0.8 based on the robustness of the clustering generated.
Cell type identification
We identified cell cluster marker genes with the FindAllMarkers function in Seurat, which performs differential expression analysis between each cell cluster and the remaining cells in the dataset. We tested SCT normalized data using the t-test option and setting the minimum fraction (min.pct) to 0.1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw and processed scRNAseq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) repository under accession code GSE279132. Source data are provided with this paper.
References
Biffi, A. et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341, 1233158 (2013).
Sessa, M. et al. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: an ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet 388, 476–487 (2016).
Eichler, F. et al. Hematopoietic stem-cell gene therapy for cerebral adrenoleukodystrophy. N. Engl. J. Med. 377, 1630–1638 (2017).
Gentner, B. et al. Hematopoietic stem- and progenitor-cell gene therapy for Hurler syndrome. N. Engl. J. Med. 385, 1929–1940 (2021).
Fumagalli, F. et al. Lentiviral haematopoietic stem-cell gene therapy for early-onset metachromatic leukodystrophy: long-term results from a non-randomised, open-label, phase 1/2 trial and expanded access. Lancet 399, 372–383 (2022).
Cartier, N. et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 326, 818–823 (2009).
Cartier, N. et al. Lentiviral hematopoietic cell gene therapy for X-linked adrenoleukodystrophy. Methods Enzymol. 507, 187–198 (2012).
Chetty, K. & Booth, C. Gene therapy for primary immunodeficiencies: up-to-date. Expert Opin. Biol. Ther. 21, 529–538 (2021).
Goyal, S. et al. Acute myeloid leukemia case after gene therapy for sickle cell disease. N. Engl. J. Med. 386, 138–147 (2022).
Hsieh, M. M. et al. Myelodysplastic syndrome unrelated to lentiviral vector in a patient treated with gene therapy for sickle cell disease. Blood Adv. 4, 2058–2063 (2020).
Tucci, F., Galimberti, S., Naldini, L., Valsecchi, M. G. & Aiuti, A. A systematic review and meta-analysis of gene therapy with hematopoietic stem and progenitor cells for monogenic disorders. Nat. Commun. 13, 1315 (2022).
Gentner, B. et al. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci. Transl. Med. 2, 58ra84 (2010).
Bosch, M. E. & Kielian, T. Neuroinflammatory paradigms in lysosomal storage diseases. Front. Neurosci. 9, 1–11 (2015).
Subhramanyam, C. S., Wang, C., Hu, Q. & Dheen, S. T. Microglia-mediated neuroinflammation in neurodegenerative diseases. Semin Cell Dev. Biol. 94, 112–120 (2019).
Pawelec, P., Ziemka-nalecz, M., Sypecka, J. & Zalewska, T. The impact of the CX3CL1/CX3CR1 axis in neurological disorders. Cells 9, 2277 (2020).
Wolf, Y., Yona, S., Kim, K. W. & Jung, S. Microglia, seen from the CX3CR1 angle. Front. Cell Neurosci. 7, 26 (2013).
Jung, S. et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol. 20, 4106–4114 (2000).
Capotondo, A. et al. Brain conditioning is instrumental for successful microglia reconstitution following hematopoietic stem cell transplantation. Proc. Natl Acad. Sci. USA 109, 15018–15023 (2012).
Peviani, M. et al. An innovative hematopoietic stem cell gene therapy approach benefits CLN1 disease in the mouse model. EMBO Mol. Med. 15, e15968 (2023).
Capotondo, A. et al. Intracerebroventricular delivery of hematopoietic progenitors results in rapid and robust engraftment of microglia-like cells. Sci. Adv. 3, e1701211 (2017).
Liang, K. et al. Regulation of dynamic behavior of retinal microglia by CX3CR1 signaling. Invest. Ophthalmol. Vis. Sci. 50, 4444–4451 (2009).
Verdonk, F. et al. Phenotypic clustering: a novel method for microglial morphology analysis. J. Neuroinflammation 13, 153 (2016).
Butovsky, O. & Weiner, H. L. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 19, 622–635 (2018).
Uyar, O. et al. Single-cell transcriptomics of the ventral posterolateral nucleus-enriched thalamic regions from HSV-1-infected mice reveal a novel microglia/microglia-like transcriptional response. J. Neuroinflammation 19, 81 (2022).
Vagima, Y. et al. Early sensing of Yersinia pestis airway infection by bone marrow cells. Front. Cell Infect. Microbiol. 2, 143 (2012).
Garin, A. et al. Two novel fully functional isoforms of CX3CR1 are potent HIV coreceptors. J. Immunol. 171, 5305–5312 (2003).
Garin, A., Pellet, P., Deterre, P., Debre, P. & Re, C. C. Cloning and functional characterization of the human fractalkine receptor promoter regions. Biochem. J. 368, 753–760 (2002).
Schiroli, G. et al. Precise gene editing preserves hematopoietic stem cell function following transient p53-mediated DNA damage response. Cell Stem Cell 24, 551–565.e8 (2019).
Lombardo, A. et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods 8, 861–869 (2011).
Bak, R. O., Dever, D. P. & Porteus, M. H. CRISPR/Cas9 genome editing in human hematopoietic stem cells. Nat. Protoc. 13, 358–376 (2018).
Konturek-Ciesla, A. & Bryder, D. Stem cells, hematopoiesis and lineage tracing: transplantation-centric views and beyond. Front. Cell Dev. Biol. https://doi.org/10.3389/fcell.2022.903528 (2022).
Butovsky, O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014).
Mizutani, M. et al. The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J. Immunol. 188, 29–36 (2012).
Kierdorf, K. et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273–280 (2013).
Park, H. & Cox, D. Syk regulates multiple signaling pathways leading to CX3CL1 chemotaxis in macrophages. J. Biol. Chem. 286, 14762–14769 (2011).
Wang, L., Yang, L., Filippi, M., Williams, D. A. & Zheng, Y. Genetic deletion of Cdc42GAP reveals a role of Cdc42 in erythropoiesis and hematopoietic stem / progenitor cell survival, adhesion, and engraftment. Blood 107, 98–105 (2006).
Williams, D. A., Zheng, Y. & Cancelas, J. A. Rho GTPases and regulation of hematopoietic stem cell localization. Methods Enzymol. 439, 365–393 (2008).
Yang, L. et al. Cdc42 critically regulates the balance between myelopoiesis and erythropoiesis. Blood 110, 3853–3861 (2007).
Subbarayan, M. S., Joly-Amado, A., Bickford, P. C. & Nash, K. R. CX3CL1/CX3CR1 signaling targets for the treatment of neurodegenerative diseases. Pharm. Ther. 231, 107989 (2022).
Xu, Z. et al. Efficient strategies for microglia replacement in the central nervous system. Cell Rep. 32, 108041 (2020).
Xu, Z., Zhou, X., Peng, B. & Rao, Y. Microglia replacement by bone marrow transplantation (Mr BMT) in the central nervous system of adult mice. STAR Protoc. 2, 100666 (2021).
Chadarevian, J. P. et al. Engineering an inhibitor-resistant human CSF1R variant for microglia replacement. J. Exp. Med. 220, e20220857 (2023).
Loeb, A., Pattwell, S., Meshinchi, S., Bedalov, A. & Loeb, K. R. Donor bone marrow derived macrophage engraftment into the central nervous system of allogeneic transplant patients. Blood Adv. 7, 5851–5859 (2023).
Sailor, K. A. et al. Hematopoietic stem cell transplantation chemotherapy causes microglia senescence and peripheral macrophage engraftment in the brain. Nat. Med. 28, 517–527 (2022).
Yoo, Y., Neumayer, G., Shibuya, Y., Marc-Daniel Mader, M. & Wernig, M. A cell therapy approach to restore microglial Trem2 function in a mouse model of Alzheimer’s disease. Cell Stem Cell 30, 1043–1053.e6 (2023).
Mishra, P. et al. Rescue of Alzheimer’s disease phenotype in a mouse model by transplantation of wild-type hematopoietic stem and progenitor cells. Cell Rep. 42, 112956 (2023).
Shibuya, Y. et al. Treatment of a genetic brain disease by CNS-wide microglia replacement. Sci. Transl. Med. 14, eabl9945 (2022).
Cromer, M. K. et al. Ultra-deep sequencing validates safety of CRISPR/Cas9 genome editing in human hematopoietic stem and progenitor cells. Nat. Commun. https://doi.org/10.1038/s41467-022-32233-z (2022).
Cromer, M. K. et al. Comparative analysis of CRISPR off-target discovery tools following ex vivo editing of CD34+ hematopoietic stem and progenitor cells. Mol. Ther. 31, 1074–1087 (2023).
Visigalli, I. et al. The galactocerebrosidase enzyme contributes to the maintenance of a functional hematopoietic stem cell niche. Blood 116, 1857–1866 (2010).
Denning, S. M., Tuck, D. T., Singer, K. H. & Haynes, B. F. Human thymic epithelial cells function as accessory cells for autologous mature thymocyte activation. J. Immunol. 138, 680–686 (1987).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Brinkman, E. K., Chen, T., Amendola, M. & Van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Ferrari, S. et al. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nat. Biotechnol. 38, 1298–1308 (2020).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Ianevski, A., Giri, A. K. & Aittokallio, T. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat. Commun. 13, 1246 (2022).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–47 (2019).
Acknowledgements
We are grateful to Drs Matt Porteus and Natalia Gomez-Ospina for their help in the initial editing strategy design. We acknowledge the DFCI flow cytometry core and DFCI confocal light microscopy core for technical support. We thank the Single Cell Core at Harvard Medical School, Boston, MA, for performing the single-cell RNA-Seq sample preparation. This work was supported by start-up funds, sponsored research, and grant # 101055262 (ERC-2021-ADG) to A.B.
Author information
Authors and Affiliations
Contributions
A.M. contributed to the experimental design, performed experimental setup and experiments, data analysis, and data interpretation, and drafted the manuscript and the figures; D.M. managed the mouse colony and performed experiments; D.P. performed single-cell RNA analysis and interpretation; M.P. and P.G. contributed to the experimental design, data interpretation, and revision of the manuscript; A.B. conceived the study, analysed and interpreted the data, retrieved the funding, and finalized the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.B., A.M., and P.G. are co-authors in the provisional patent application PCT/US23/22029. A.B. and A.M. are co-authors in the patent PCT/US20/45106. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Montepeloso, A., Mattioli, D., Pellin, D. et al. Haploinsufficiency at the CX3CR1 locus of hematopoietic stem cells favors the appearance of microglia-like cells in the central nervous system of transplant recipients. Nat Commun 15, 10192 (2024). https://doi.org/10.1038/s41467-024-54515-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54515-4