Abstract
Lesional focal epilepsy (LFE) is a common and severe seizure disorder caused by epileptogenic lesions, including malformations of cortical development (MCD) and low-grade epilepsy-associated tumors (LEAT). Understanding the genetic etiology of these lesions can inform medical and surgical treatment. We conducted a somatic variant enrichment mega-analysis in brain tissue from 1386 individuals who underwent epilepsy surgery, including 599 previously unpublished individuals with ultra-deep ( > 1600x) targeted panel sequencing. Here we confirm four known associations (BRAF, SLC35A2, MTOR, PTPN11), support eight associations without prior statistical support (FGFR1, PIK3CA, AKT3, NF1, PTEN, RHEB, KRAS, NRAS), and identify novel associations for two genes, DYRK1A and EGFR. Both novel genes show specific histopathological phenotypes, interact with LFE genes and pathways, and may represent promising candidates as biomarkers and potentially druggable targets.
Similar content being viewed by others
Introduction
Lesional focal epilepsy (LFE) is a common disorder with an estimated prevalence of 2.70 per 1000 persons (95% CI: 1.12–3.81)1. Individuals with LFE suffer from uncontrolled seizures, low quality of life, and mortality twice that of the general population2. New diagnostic and therapeutic options are urgently needed. Around half of all cases with LFE requiring surgery are associated with malformations of cortical development (MCD), including focal-cortical dysplasia (FCD), or low-grade epilepsy-associated tumors (LEAT)3,4. Across all epileptogenic brain lesions, ~65% of cases lack detectable genetic abnormalities5,6. This diagnostic gap suggests that additional genes likely contribute to LFE.
Recent cohort studies (n ~ 100–500 tissue samples) have started to elucidate the genetic etiology of LFE, pointing to mainly somatic variants in 19 genes as the cause in 15-80% of individuals, depending on the type of lesion, sampling strategy, and sequencing technology7,8,9,10. Thus, some gene-disease associations have been very well characterized (e.g., MTOR and FCD type II) while other lesions have less clear genotype-phenotype correlations where candidate genes were reported in a limited number of cases. Establishing formal statistical support for their association with LFE, one of the strongest criteria for assessing gene-disease validity by the Clinical Genome Resource (ClinGen11), would aid future integration into clinical genetic tests and open avenues for targeted therapies, including repurposing FDA-approved drugs.
In this study, we present a mega-analysis pooling raw data of somatic variants in brain tissues from 1386 individuals who underwent epilepsy surgery, including 599 previously unpublished individuals. Individuals either received ultra-deep ( > 1600x) targeted panel sequencing (n = 599) or deep ( > 300x/>350x) whole-exome sequencing (n = 787). This represents the largest somatic variant detection study in epilepsy to date, enabling a comprehensive somatic variant enrichment analysis using dNdScv. Here, we confirm four previously established gene-disease associations (BRAF, SLC35A2, MTOR, PTPN11), provide statistical support for eight associations (FGFR1, PIK3CA, AKT3, NF1, PTEN, RHEB, KRAS, NRAS), and identify novel associations for two genes, DYRK1A and EGFR. Building upon the statistical results, we support the plausibility of these novel associations with histopathological reviews and comprehensive in silico analyses including structural modeling and interaction analysis. Our study offers large-scale statistical support to inform diagnostic panel design and identifies potential diagnostic biomarkers and druggable targets for experimental follow-up studies.
Results
Somatic variant enrichment analysis reveals disease associations for twelve known and two novel genes
This study includes data from three cohorts: (i) A new cohort using targeted panel sequencing of brain tissue from 599 individuals (MCD, n = 206; LEAT, n = 207; controls, n = 186; Supplementary Data 3). Panel sequencing was done to achieve ultra-deep (>1600x) coverage, and panel design is explained in the Methods; (ii) Our previously published study on deep (>350x) whole-exome sequencing (WES) of brain tissue from 474 individuals (MCD, n = 223; LEAT, n = 154; controls, n = 97)8; and (iii) Data from literature and collaborators on deep WES in 313 individuals (MCD, n = 311; LEAT, n = 1; controls, n = 1; Fig. 1, Supplementary Data 4)9. Diagnostic yield was 33.4–35.8% (Supplementary Fig. 1).
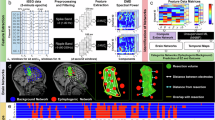
a Study overview and analysis workflow. b DYRK1A and EGFR are novel genes associated with MCD. c EGFR is a novel gene associated with LEAT. Results of the mega-analysis are shown as global dNdScv Q-values (see “Methods“) versus a gene-based collapsing test of relative enrichment (odds ratio, OR) of samples with deleterious variants in LFE versus control pathology samples (Fisher’s exact test; Supplementary Data 5a) and as distribution of observed unadjusted global P values (QQ plots). dNdScv Q-values have been adjusted for multiple testing using the Benjamini-Hochberg method. Excess driver variant ratios are shown for missense and nonsense variants for each gene, denoting if gene effects are specific to certain variant types. Category definitions for each group are given in Supplementary Data 5b.
Out of 1386 brain samples, 1006 samples had at least one somatic single-nucleotide variant after filtering (MCD, n = 614; LEAT, n = 251; controls, n = 141; Supplementary Data 4). We tested these samples for somatic variant enrichment relative to the neutral mutational rate with dNdScv12 and: (i) replicated associations8 for four LFE genes (global Q < 0.05; BRAF, SLC35A2, MTOR, PTPN11); (ii) validated eight established LFE genes without previous statistical support (any Q < 0.05; FGFR1, PIK3CA, AKT3, NF1, PTEN, RHEB, KRAS, NRAS); and (iii) identified two novel genes: DYRK1A and EGFR (global Q < 0.05; Supplementary Data 5a and b). Eleven EGFR or DYRK1A carriers had other variants in genes previously associated with their specific histopathology (Supplementary Data 6). DYRK1A was significantly enriched in MCD (incl. FCD type II; global Q = 0.02; missense dN/dS ratio 27.52, P = 1.60 × 10−5). EGFR was significantly enriched in MCD (incl. FCD type II; global Q = 0.007; missense dN/dS ratio 23.92, P = 5.02 × 10−6) and LEAT (global Q = 0.032; missense dN/dS ratio 47.37, P = 4.11 × 10−5) (Fig. 1).
In silico and histopathological assessment support novel gene-disease associations for DYRK1A and EGFR with LFE
Next, we investigated gene-disease plausibility with four different approaches. First, we conducted sequence- and structure-based in silico analysis to assess variant deleteriousness. Somatic variants in DYRK1A and EGFR identified in our study were more likely to be located in missense-intolerant regions and had higher pathogenicity scores compared to variants from public databases (Fig. 2a, b; “Methods” and Supplementary Fig. 2). On structural analysis, somatic variants in DYRK1A were located in functionally essential protein regions (Fig. 2c), and one recurrent variant (p.R316C) was found to disrupt an autophosphorylation site critical for kinase function (Fig. 2d)13. Putative mechanisms for additional variants are shown in Supplementary Fig. 3.
a Variants in DYRK1A from our mega-analysis (n = 10) compared to public databases, including (likely) pathogenic (n = 24) and (likely) benign variants (n = 52) from ClinVar, and gnomAD (n = 287), across different predictors of deleteriousness (CADD_PHRED, p = 0.227, p < 0.002, p = 0.009, respectively; EVE, p = 0.103, p = 0.0011, p = 0.24, respectively; REVEL, p = 0.16, p < 0.0001, p < 0.0001, respectively) and by their distribution in missense-intolerant regions (MTR, p = 0.076, p < 0.0001, p = 0.00016, respectively). Paired one-sided Wilcoxon test: ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, ns: p > 0.05. b Variants in EGFR from our mega-analysis (n = 17) compared to public databases including (likely) pathogenic (n = 29) and (likely) benign variants (n = 10) from ClinVar, and gnomAD (n = 604), across different predictors of deleteriousness (CADD_PHRED, p = 0.003, p < 0.0001, p < 0.0001, respectively; EVE, p = 0.838, p = 0.00021, p < 0.0001, respectively; REVEL, p = 0.406, p = 0.00015, p < 0.0001, respectively) and by their distribution in missense-intolerant regions (MTR, p = 0.03, p = 003, p < 0.0001, respectively). Paired one-sided Wilcoxon test: ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, ns: p > 0.05. Data are presented as box plots that indicate median (center), and the interquartile range (IQR; bounds of box) up to 1.5 IQR (whiskers). Adjustments for multiple testing were done with the Holm-Bonferroni method. c Variants in DYRK1A mapped on the Dyrk1A structure (PDB-ID: 7FHS – chain A). Essential-3D sites are shown in pink. d Variants in EGFR mapped on protein structure fragments of the epidermal growth factor receptor (EGFR). *Variants observed in multiple samples across our cohorts. †Variants present in COSMIC at significance tiers 1–3.
Second, we conducted a histopathological review for each DYRK1A and EGFR carrier as the knowledge of an underlying genetic etiology can improve histopathological classification4,14. The initial diagnosis was confirmed in each case. Notably, every DYRK1A carrier was positive for pS6 (Ser240/444), demonstrating Akt/mTOR pathway activation specifically in dysplastic neurons and balloon cells of FCD type IIB but also in dysplastic neurons of ganglioglioma cases (GG; Supplementary Fig. 4, 5). Interestingly, EGFR-associated LEAT showed atypical nodular growth and spread into subarachnoid spaces, and one GG had markedly pronounced proliferative growth (Supplementary Fig. 6, Supplementary Data 7). Thus, both DYRK1A and EGFR carriers have specific histopathological phenotypes.
Third, interactions with established pathways may explain the role of DYRK1A and EGFR. We noted structural and functional interactions with LFE genes on gene-gene network analysis (Supplementary Fig. 7). Next, we analyzed functional readouts across 15,847 genes in 423 cell lines from DepMap15. Both DYRK1A and EGFR were functionally co-dependent with established LFE genes (Supplementary Fig. 8). These cell-line effects were selective and correlated across RNAi and CRISPR systems (Supplementary Fig. 9). Clusters of functionally similar genes were enriched for the Ras/Raf/MAPK, ErbB, and PI3K/Akt pathways (Supplementary Fig. 10). These interactions align with the previous literature: Signaling between EGFR and mTOR is well-established16,17, and recent evidence suggests a similar interaction between DYRK1A and mTOR18.
Fourth, we classified each variant in line with the standards for the classification of pathogenicity of somatic variants in cancer19. The criteria used include population frequency, functional and in silico data, and somatic frequency. We acknowledge that these criteria are not fully applicable to non-cancer phenotypes, but they may represent a semi-quantitative and approximate measure of pathogenicity, complementary to the indirect experimental evidence from histopathology. The majority of carriers with variants in either candidate gene had (likely) oncogenic variants (EGFR: 13/22, DYRK1A: 12/18), with only three carriers of likely benign variants, the rest (EGFR: 8/22, DYRK1A: 4/18) carrying variants of uncertain significance (Supplementary Data 8). A single variant (EGFR p.T354M with mild malformation of cortical development with oligodendroglial hyperplasia, MOGHE; sample 179863) was known by OncoKB to be likely neutral. Multiple variants in EGFR were recurrent, and 12 samples carried variants previously assigned tier 1–3 significance in the cancer mutation census (COSMIC)20. Therefore, we conclude that it is likely that variants in DYRK1A or EGFR are contributory in the majority of cases.
DYRK1A and EGFR are potential biomarkers and therapeutic targets in LFE
Although the precise mechanism by which DYRK1A and EGFR are involved in the etiology of LFE remains unresolved, both genes act on potentially druggable pathways and thus represent established direct and indirect therapeutic targets. For DYRK1A, novel inhibitors are available21. For EGFR, the majority of individuals in our cohort had known oncogenic variants with experimental evidence of gain-of-function, which can be targeted by FDA-approved inhibitors (Supplementary Data 8)22. Overall, at least one interventional trial is currently ongoing for 13/14 genes for which we have shown statistical support, and 10/14 genes have known target-drug associations (Supplementary Fig. 11).
Discussion
This work introduced the largest study on somatic variant detection in epilepsy. We demonstrated statistical support for 14 genes associated with LFE, which provides strong evidence of gene-disease validity and will guide future integration into clinical genetic tests. We further identified two novel gene-disease associations with LFE: DYRK1A and EGFR. These genes accounted for 9/364 (2.5%) and 11/364 (3.0%) of cases, respectively.
DYRK1A, which in our study was enriched in MCD, is critical for early mammalian development in general23, and oligodendrocyte progenitor development in particular24. Germline variants in DYRK1A are known to cause epilepsy and other neurodevelopmental disorders25 through dysregulation of ERK/MAPK and mTOR signaling26. We found histopathological evidence of mTOR pathway activation in every DYRK1A carrier in line with prior evidence of direct interaction between DYRK1A and mTOR signaling18,26. The same mTOR pathway activation was also seen in dysplastic neurons of both DYRK1A-associated GG cases (Supplementary Fig. 4, 5). While the exact mechanism by which DYRK1A may cause LFE remains to be resolved, DYRK1A alteration and the resulting mTOR pathway activation may serve as both a potential diagnostic biomarker and potential treatment target pending further experimental validation.
EGFR has previously been identified in other cancers, including lung cancer27 and glioblastoma multiforme28. Given this, we suspected that EGFR may also be mutated in lower-grade gliomas such as LEAT. Interestingly, previously published associations between EGFR and CNS tumors were primarily driven by gene amplifications (Supplementary Fig. 12), while we found missense variation in LEAT. This association was specific to GG and was not found for DNET (Supplementary Data 3). This may suggest a distinct and specific disease mechanism. The exact mechanism remains to be resolved and may range from monogenic driver mutations towards a second-hit or oligogenic model as in PTPN11-altered CNS tumors29 or cerebral cavernous malformations30. Further, indirect evidence from transcriptome studies has previously implicated EGFR-mediated signaling in a subgroup of LEAT with adverse clinical outcomes, including earlier recurrence31. We have demonstrated that our EGFR carriers had more malignant growth patterns and proliferative activity. These features closely resemble the hallmarks of GG associated with adverse clinical outcomes, which we have previously linked to alterations in PTPN11 and other RAS-/MAP-kinase pathway genes29. Together, this suggests that EGFR alteration may be another potential prognostic biomarker in LEAT.
The enrichment of somatic EGFR variants in both MCD (FCD type II, MOGHE) and LEAT (GG) was intriguing (Supplementary Data 3). Prior gene-disease associations in LFE were considered relatively specific for single histopathological groups. However, mounting evidence suggests that MCD and LEAT have shared developmental characteristics16. This phenotypic spectrum is best seen in PIK3CA-associated lesions, which appear markedly different based on cell type, organ, and variant allelic fraction (VAF)32; and in MTOR-associated lesions, which range from balloon cells in FCD type II to hemimegalencephaly based on developmental stage33. Indeed, EGFR has been identified in DNA methylation and RNA sequencing studies of FCD type II34,35. Perhaps most interestingly, EGFR was found to be expressed in MCD organoids and treatment with the EGFR inhibitor afatinib decreased lesion burden22. Thus, our observation of EGFR-mutated MCD is in line with prior evidence and further supports the potential phenotypic overlap between MCD and LEAT.
Variants in eleven EGFR or DYRK1A carriers co-occurred with variants in genes previously associated with their specific histopathology. The co-occurrence of multiple somatic variants in cancers36, vascular malformations30, and non-cancer epilepsy lesions37 has been previously established. In each of these examples, multiple co-occurring variants were shown to have an impact on phenotype. Thus, we do not believe that the presence of other (possible) driver variants in the same samples as EGFR or DYRK1A necessarily implies a lower likelihood of pathogenicity by itself.
Taken together, the gene-disease associations of DYRK1A and EGFR with LFE each are consistent with previous evidence. We have demonstrated how EGFR and DYRK1A variants identified in this study overlap with known pathogenic germline variants in ClinVar (Supplementary Fig. 2) and are recurrent among somatic variants in COSMIC (Supplementary Data 8). We have shown that additional evidence from structural modeling, histopathology, and network and pathway interactions each independently support these novel gene-disease associations. Thus, DYRK1A and EGFR may represent promising candidate biomarkers and therapeutic targets pending further validation. Of note, more experimental work is required to elucidate whether the mechanisms in LFE are the same as for other diseases already associated with these genes. Only then can these findings safely and effectively be translated to the clinical care of individuals with LFE. Overall, our findings expand the genetic spectrum of LFE and highlight unique treatment opportunities for future clinical trials.
We have presented a well-powered study based on rigorous statistical evidence supported by expert histopathological review, in silico modeling, and expression patterns in non-lesional cell lines. However, this study does not provide strong nor direct experimental confirmation of pathogenicity and cannot definitively elucidate the underlying disease mechanisms. Of note, we prioritized specificity over sensitivity of the somatic variant enrichment analysis by including only variants with a VAF > 0.02 (2%) in the somatic variant enrichment analysis. The choice of threshold was based on previous credible intervals8. We used sequencing technology aimed at reducing the impact of sequencing artifacts (UMI-based calling38) and acknowledge that many variants of interest are likely below this threshold39 but cannot confidently include ultra-low VAF variants in the absence of paired samples. Thus, our analysis may have missed genes with later-stage brain somatic variation and genes not included in the ultra-deep targeted sequencing panel. Exploring the ultra-low VAF genetic spectrum of LFE remains for future work with paired samples or single-cell approaches40. Our enrichment analysis focused on somatic variants and thus was not designed for gene-disease associations with a germline or two-hit mechanism (e.g., DEPDC5, NPRL2, NPRL3, TSC1, TSC2). Again, investigating these specific associations will require further studies with paired samples. Other known or expected gene-disease associations may be absent due to insufficient sample size, and further studies on even larger cohorts may identify less prevalent causal genes.
Methods
The study protocol was approved by the institutional review boards of the Cleveland Clinic Epilepsy Center (IRB approval ID 20-151) and the University of Erlangen, Germany (IRB approval ID 193_18B). All participants provided written informed consent for study participation. Study participants did not receive compensation. In this somatic variant enrichment mega-analyses, no individual-level clinical or demographic data (including sex and/or gender) were considered in study design or analysis.
Study cohorts
In this international multi-center study, we recruited a whole-exome sequencing (WES) cohort of 474 individuals and a panel sequencing cohort of 599 individuals. Each of these individuals underwent resective epilepsy surgery for drug-resistant focal epilepsy. All individuals had previously received comprehensive presurgical epilepsy evaluation followed by a multidisciplinary patient management conference where the surgical strategy was approved. Formalin-fixed paraffin-embedded (FFPE) surgical brain tissue samples were obtained from each individual.
Cases were defined as having a histopathological diagnosis of long-term epilepsy-associated tumor (LEAT) or malformation of cortical development (MCD) including any focal-cortical dysplasia (FCD type I-III). For our analysis, FCD type IIA and type IIB were pooled. Control brain tissues were derived from individuals with focal epilepsy who either had histopathologically confirmed non-lesional epilepsy or epilepsy-associated lesions with a low monogenic etiology probability, thus likely not carrying overgrowth disorder or cancer driver variants that are predominantly involved in MCD or LEAT. Such lesional epilepsy types included environmental or acquired causes (i.e., ischemic or hemorrhagic stroke, acute or chronic trauma), immune-related causes (i.e., infectious or autoimmune encephalitis), or hippocampal sclerosis (HS). Somatic variants have been implicated in HS – however, there is no evidence for statistical enrichment of somatic variants in HS8,41. Further information on control phenotypes and the rationale behind their inclusion is provided in Supplementary Data 1. Histopathological reviews of all samples were performed by an experienced neuropathologist (I.B.) using the International League Against Epilepsy (ILAE) consensus classification of focal cortical dysplasia4 and the 2016 World Health Organization Classification of Tumors of the Central Nervous System42.
DNA extraction
Genomic DNA was extracted from FFPE brain tissue for all individuals. The DNeasy Blood and Tissue Kit (Qiagen) was used according to the manufacturer’s protocol.
Sequencing cohorts
The panel sequencing cohort consisted of 599 individuals with lesional focal epilepsy or control pathologies who received targeted ultra-deep ( > 1600x) panel sequencing. We chose panel sequencing since we aimed to achieve high coverage capable of detecting variants with low VAF while maintaining costs that would allow for the sequencing of a large study cohort. This was further supported by the low number of previous genes (see below), previous evidence for lower gene complexity with a small tail distribution of expected gene associations, and the observation that LFE genes were limited to a few established pathways40.
Panel design was based on: (i) 19 established LFE genes (defined as MTOR, SLC35A2, AKT3, PIK3CA, RHEB, TSC1, TSC2, NPRL2, NPRL3, DEPDC5, PTEN, BRAF, FGFR1, MYB, MYBL1, PTPN11, NRAS, KRAS, and NF1); (ii) genes with dNdScv p < 0.005 in our previous study8; (iii) genes with >1 somatic variant or dNdScv p < 0.05 in our previous study8 and among: (a) published candidate brain tumor genes (n = 100 from PubMed search, keywords: glioma, angiocentric glioma, dysembryoplastic neuroepithelial, ganglioglioma, multinodular and vacuolating neuronal, papillary glioneuronal, polymorphous low-grade neuroepithelial); (b) developmental disorders genes (n = 285, from previous gene discovery43 and an additional PubMed search, keywords: developmental and epileptic encephalopathy, neurodevelopmental disorder); (c) COSMIC Cancer Gene Census Tier 1 cancer-driving genes (n = 570); (d) Genes enriched with somatic mutations in tumor or CNS tumor samples from cBioPortal44; (e) genes with cancer driver mutations with OncodriveFML p < 0.00545; (f) established epilepsy genes (n = 206)46,47; (g) evolutionarily constrained and brain-expressed genes (n = 1146; in-house database).
The WES cohort consisted of 474 individuals with lesional focal epilepsy (LFE) or control pathologies who underwent bulk-tissue deep ( > 350x) whole-exome sequencing for somatic variant detection. This cohort was previously published8.
To calculate diagnostic yield, we used the set of 19 established LFE genes outlined above and our two novel gene-disease associations (DYRK1A, EGFR).
Sequencing and variant calling
Library preparation was conducted using Agilent SureSelect Custom Enrichment Kit, and libraries underwent paired-end sequencing on Illumina HiSeq 4000 Sequencing Systems according to the manufacturer’s protocol. Data processing followed GATK (Genome Analysis Toolkit) Best Practices48. Paired-end FASTQ files were aligned to the GRCh37/hg19 human reference genome using the Burrows-Wheeler Aligner (BWA-MEM, version 0.7.17) and sorted by read group using samtools (version 1.16.1). The merged BAM files were marked for duplicate reads using Picard (version 2.8.14). We performed indel realignment and base quality score recalibration with GATK (version 4.1.9.0).
Somatic single-nucleotide variants (SNVs) and indels were called with MuTect2 (GATK v4.1.9.0). We created a Panel of Normals (PoN) by merging public resources from the GATK resource bundle with our own whole-exome data from an additional 124 resected brain tissue samples. MuTect2 was used with this PoN at standard parameters, and results were filtered for a minimum unique read count ≥3, minimum alt reads required on both forward and reverse strands ≥1, and a minimum median distance of variants from the end of reads ≥5. We also applied UMI-VarCal2 (version 2.6.0), a novel calling algorithm designed for Illumina-targeted sequencing data that uses unique molecular identifiers (UMI) to increase sensitivity for low-frequency variants while reliably rejecting artefactual variants38.
Candidate somatic variant calls were further filtered by the following criteria: (i) Consensus call by both MuTect2 and UMI-VarCal2; (ii) the variant passed caller-specific quality control confidence filters (MuTect2 PASS, UMI-VarCal2 CERTAIN or STRONG); (iii) the variant was supported by >3 alternate reads at a total read depth of >100; (iv) the variant was either absent or present at an allele frequency of less than 3.26×10−5 in eleven large population databases: gnomAD, UKBB, TOPMed, DiscovEHR, HRC, Kaviar, 2KJPN, Wellderly, GoNL, ABraOM, GME, and cg6949,50,51,52,53,54,55,56,57,58,59,60. This maximum credible population allele frequency was calculated based on an estimated prevalence of 6.52±1.89 in 100,00061, allelic heterogeneity = 0.1, genetic heterogeneity = 1, and penetrance = 0.162; (v) the variant was present at a variant allelic fraction (VAF) of <0.30 to reduce the likelihood of a germline variant call; (vi) the variant was present at a VAF of >0.005 (for candidate disease-causing variants) or >0.02 (for the somatic variant enrichment mega-analysis; to prevent a batch effect bias from the different sequencing methodologies of the pooled cohorts), where the minimum thresholds were based on previously published credible intervals63; (vii) The variant was present in less than 10% of batch samples to reduce sequencing artifacts, as highly recurrent somatic variants would not be expected, except for BRAF V600E, which was not filtered. This filtering procedure resulted in a final set of 5046 calls from the WES cohort and 544 calls from the panel sequencing cohort that were included in the mega-analysis (section “Mega-analysis”). Likely deleterious somatic variants were identified using the following criteria: (i) exonic non-synonymous SNVs or protein-truncating variants; and (ii) REVEL score >0.75 for missense variants only64.
After germline and somatic variant calling, we conducted an additional post hoc quality control step in order to reduce the likelihood of sequencing variants in the final set of calls. Quality control metrics were gathered with CollectHsMetrics and CollectVariantCallingMetrics (GATK v4.1.9.0), again following GATK Best Practices. Samples were removed if they had an excess of somatic variants two standard deviations over the cohort mean (‘hypermutators’, n = 27) or if they had fewer bases with >100x coverage two standard deviations below the cohort mean (‘low coverage’, n = 20), or both (n = 2).
Mega-analysis
Somatic variant calls from the WES cohort (5046 calls) and panel cohort (544 calls) were pooled with data from collaborators (PI: Stéphanie Baulac; 1607 calls) and previously published cohort studies (Chung et al. 557 calls)9. For the data from Chung et al., only calls from WES samples were used to avoid resequencing bias. All variant calls were filtered for a common minimum VAF threshold of 0.02 (2%) to reduce systematic bias and the likelihood of sequencing artifacts. We detected genes under positive selection in somatic evolution with dNdScv, a set of maximum-likelihood methods to estimate the excess or deficit of driver variant types with respect to the background variation12,65. All analysis was done with the dNdScv R package v.0.1.0 at default parameters. The significance threshold was set at α = 0.05 with post hoc correction for multiple testing of 122 genes with the Benjamini-Hochberg method.
Somatic variant enrichment analysis with dNdScv was done separately for subsets based on histopathology (sub-)group: For example, enrichment in MCD was tested by using variant calls from all samples that had MCD (incl. FCD type I, FCD type II, MOGHE, and others), while enrichment in FCD type II was tested by using only variant calls from samples that had FCD type II. Enrichment analysis is, therefore, a control-free approach that tests across different levels of specificity based on the subset definition: (i) All lesional focal epilepsy samples; (ii) major categories (i.e., MCD or LEAT); and (iii) subcategories (e.g., FCD type II or GG).
Case-control testing was done for confirmatory purposes and to estimate the Odds Ratio. For this gene-based collapsing test, we carried out Fisher’s exact test for relative enrichment (odds ratio) of the number of carriers of deleterious somatic variants in LFE samples versus healthy brain controls. Only carriers and controls in the panel cohort subsample were used, as the numbers of non-carriers were not available for the other cohorts.
All variants in novel genes were visually inspected using the Integrative Genomics Viewer (IGV) to assess strand bias, read quality, and local alignment quality66.
Sequence- and structure-based in silico analysis
We assessed pathogenicity by variant annotation with pathogenicity scores (REVEL, CADD_PHRED, EVE)64,67,68, regional missense constraint (MTR)69, local protein disorder (IUPRED3)70, and functional domains (UniProt)71. Variants from this mega-analysis were compared to variants from ClinVar, HGMD, and gnomAD49,72,73. Variant scores were tested for significant differences by paired one-sided Wilcoxon test adjusted for multiple testing with the Holm-Bonferroni method.
Protein structures were gathered from the Protein Data Bank (PDB)74 for a comprehensive structural analysis. For the Epidermal Growth Factor Receptor (EGFR) protein, the following structural fragments were used: a dimeric extracellular module bound to EGF (PDB-ID: 7YSE), a transmembrane helix in the N-terminal dimer conformation (PDB-ID: 5LV6), a transmembrane helix in the C-terminal dimer conformation (PDB-ID: 2M0B), and an asymmetric dimer of the kinase domain (PDB-ID: 6DUK). Additionally, the structure of the dual specificity YAK1-related kinase protein, DYRK1A (PDB-ID: 7FHS), was collected. Precise mapping of the identified variants onto the respective protein structures and the generation of protein structure figures were performed with the PYMOL molecular visualization system75.
Immunohistochemistry
All tumors with available tissue were confirmed as GG using a routine immunohistochemical protocol: Panel with Cluster of Differentiation 34 (CD34, Mouse Monoclonal, Clone QBEnd-10, Dako, California, USA); Protein 16 (p16/CDKN2A protein, Mouse Monoclonal, Clone G175-405, BD Bioscience, California, USA); Isocytrate Dehydrogenase 1 (IDH1, Monoclonal Mouse, Clone H09, Dianova, Hamburg, Germany); ATP-dependent helicase ATRX (ATRX, Mouse Monoclonal, Clone BSB-108, Bio SB, California, USA); Microtubule Associated Protein 2 (MAP2, Mouse Monoclonal, Clone C, Riederer Lab, Lusanne, Switzerland); Glial Fibrillary Acidic Protein (GFAP, Polyclonal Rabbit, Z0334, Dako, California, USA); Ki67 Protein (Ki67 Rabbit Monoclonal, Clone SP6); Protein 53 (p53, Mouse Monoclonal, Clone DO-7, Dako, California, USA).
The rationale for staining tumor samples follows the current WHO classification for Central Nervous System Tumors (5th Edition) and the diagnostic requirements for gliomas42. Low-grade glioneuronal tumors are a heterogeneous cohort of lesions. One of these lesions is the GG with frequent BRAF V600E mutations, which are positive for CD3476,77, whereas a homozygous deletion of CDKN2a (p16 or FISH analysis) is a characteristic marker for pleomorphic xanthoastrocytoma. Stainings against MAP2 are used to demonstrate the neuronal differentiated subpopulation in the group of LEATs and are, therefore, a crucial diagnostic marker.
Additional information on the amount, dilution, and validation of all antibodies is available in the Reporting Summary.
Functional analysis
We annotated all variants included in the mega-analysis using ANNOVAR, COSMIC, and OncoKB20,78,79. To examine cancer cell line dependencies of established and novel genes, we used data from 15,847 genes in 423 cell lines available from DepMap, data release 23Q215. Gene characteristics including selectivity, i.e., the difference in gene essentiality across cell lines, and efficacy, i.e., gene essentiality in sensitive cell lines, and the clustering algorithm ECHODOTS were previously described80. We analyzed gene-gene interaction by network analysis in STRING v.12.0 at default parameters81. Pathway enrichment on GO Molecular Function, GO Biological Process, and KEGG was calculated with EnrichR82,83,84.
Statistics and reproducibility
Statistical analyses and data visualization were performed in R version 4.3.1 (2023-06-16).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study, including somatic variant calls, are available within the paper and its Supplementary Information. Raw sequencing data generated from previously unpublished individuals (panel cohort) are not openly available due to informed consent limitations, which did not include specific language permitting public raw data sharing (IRB 12-1000; IRB 20-151). These data are securely stored in controlled access data storage at the Cleveland Clinic Epilepsy Biorepository and Data Registry, Cleveland Clinic, OH, USA. To access the raw sequencing data, requests should be directed to the corresponding author, Dennis Lal, PhD (Address: JJL 445-5, 1133 John Freeman Blvd, Department of Neurology, McGovern Medical School, UTHealth, Houston, TX 77030, US; Email: dennis.lal@uth.tmc.edu). Requests will be reviewed promptly, with a response provided within one month of receipt. Data sharing will be contingent upon an institutional data use agreement, which will not impose prior restrictions on the use of the data. Our previously published WES cohort is subject to the same data availability restrictions. The referenced external dataset by Chung et al. is available on the NIMH Data Archive under study number 1484 “Comprehensive multi-omic profiling of somatic mutations in malformations of cortical development”. This study used cancer cell line dependency data from Dependency Map (DepMap), data release 23Q2, which is available for community use on FigShare (https://figshare.com/articles/dataset/DepMap_23Q2_Public/22765112).
Code availability
The code used in this study is available on GitHub (https://github.com/christianbosselmann/LFE) and Zenodo (https://doi.org/10.5281/zenodo.13983287).
References
Fiest, K. M. et al. Prevalence and incidence of epilepsy: A systematic review and meta-analysis of international studies. Neurology 88, 296–303 (2017).
Ben-Menachem, E., Schmitz, B., Kälviäinen, R., Rhys, T. & Klein, P. The burden of chronic drug-refractory focal onset epilepsy: Can it be prevented? Epilepsy Behav. 148, 109435 (2023).
Blumcke, I. et al. Histopathological findings in brain tissue obtained during epilepsy surgery. N. Engl. J. Med. 377, 1648–1656 (2017).
Najm, I. et al. The ILAE consensus classification of focal cortical dysplasia: An update proposed by an ad hoc task force of the ILAE diagnostic methods commission. Epilepsia 63, 1899–1919 (2022).
Oegema, R. et al. International consensus recommendations on the diagnostic work-up for malformations of cortical development. Nat. Rev. Neurol. 16, 618–635 (2020).
Straka, B. et al. Genetic testing for malformations of cortical development: A clinical diagnostic study. Neurol. Genet. 8, e200032 (2022).
Lai, D. et al. Somatic variants in diverse genes leads to a spectrum of focal cortical malformations. Brain 145, 2704–2720 (2022).
López-Rivera, J. A. et al. The genomic landscape across 474 surgically accessible epileptogenic human brain lesions. Brain 146, 1342–1356 (2023).
Chung, C. et al. Comprehensive multi-omic profiling of somatic mutations in malformations of cortical development. Nat. Genet 55, 209–220 (2023).
Baldassari, S. et al. Dissecting the genetic basis of focal cortical dysplasia: a large cohort study. Acta Neuropathol. 138, 885–900 (2019).
Rehm, H. L. et al. ClinGen — the clinical genome resource. N. Engl. J. Med. 372, 2235–2242 (2015).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21 (2017).
Becker, W. & Sippl, W. Activation, regulation, and inhibition of DYRK1A. FEBS J. 278, 246–256 (2011).
Blümcke, I. et al. Toward a better definition of focal cortical dysplasia: An iterative histopathological and genetic agreement trial. Epilepsia 62, 1416–1428 (2021).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576.e16 (2017).
Blumcke, I. et al. Neocortical development and epilepsy: Insights from focal cortical dysplasia and brain tumours. Lancet Neurol. 20, 943–955 (2021).
Fan, Q.-W. et al. EGFR signals to mTOR through PKC and independently of Akt in glioma. Sci. Signal. 2, ra4–ra4 (2009).
Wang, P. et al. DYRK1A interacts with the tuberous sclerosis complex and promotes mTORC1 activity. eLife 12, RP88318 (2024).
Horak, P. et al. Standards for the classification of pathogenicity of somatic variants in cancer (oncogenicity): Joint recommendations of Clinical Genome Resource (ClinGen), Cancer Genomics Consortium (CGC), and Variant Interpretation for Cancer Consortium (VICC). Genet Med. 24, 986–998 (2022).
Tate, J. G. et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019).
Kaltheuner, I. H. et al. Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation. Nat. Commun. 12, 6607 (2021).
Eichmüller, O. L. et al. Amplification of human interneuron progenitors promotes brain tumors and neurological defects. Science 375, eabf5546 (2022).
Bellmaine, S. F. et al. Inhibition of DYRK1A disrupts neural lineage specification in human pluripotent stem cells. eLife https://elifesciences.org/articles/24502 (2017).
Pijuan, I. et al. Impaired macroglial development and axonal conductivity contributes to the neuropathology of DYRK1A-related intellectual disability syndrome. Sci. Rep. 12, 19912 (2022).
van Bon, B. W. et al. (University of Washington, Seattle, Seattle (WA), 1993).
Levy, J. A., LaFlamme, C. W., Tsaprailis, G., Crynen, G. & Page, D. T. Dyrk1a mutations cause undergrowth of cortical pyramidal neurons via dysregulated growth factor signaling. Biol. Psychiatry 90, 295–306 (2021).
Sharma, S. V., Bell, D. W., Settleman, J. & Haber, D. A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 7, 169–181 (2007).
Hu, C. et al. Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias. Nature 602, 518–522 (2022).
Hoffmann, L. et al. Ganglioglioma with adverse clinical outcome and atypical histopathological features were defined by alterations in PTPN11/KRAS/NF1 and other RAS-/MAP-Kinase pathway genes. Acta Neuropathol. 145, 815–827 (2023).
Ren, A. A. et al. PIK3CA and CCM mutations fuel cavernomas through a cancer-like mechanism. Nature 594, 271–276 (2021).
Delev, D. et al. Long-term epilepsy-associated tumors: Transcriptional signatures reflect clinical course. Sci. Rep. 10, 96 (2020).
Canaud, G., Hammill, A. M., Adams, D., Vikkula, M. & Keppler-Noreuil, K. M. A review of mechanisms of disease across PIK3CA-related disorders with vascular manifestations. Orphanet J. Rare Dis. 16, 306 (2021).
D’Gama, A. M. et al. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann. Neurol. 77, 720–725 (2015).
Dixit, A. B. et al. Genome-wide DNA methylation and RNAseq analyses identify aberrant signalling pathways in focal cortical dysplasia (FCD) type II. Sci. Rep. 8, 17976 (2018).
Luo, Y. et al. Identification of epidermal growth factor receptor as an immune-related biomarker in epilepsy using multi-transcriptome data. Transl. Pediatr. 12, 681–694 (2023).
Skoulidis, F. & Heymach, J. V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 19, 495–509 (2019).
Pelorosso, C. et al. Somatic double-hit in MTOR and RPS6 in hemimegalencephaly with intractable epilepsy. Hum. Mol. Genet 28, 3755–3765 (2019).
Sater, V. et al. UMI-VarCal: A new UMI-based variant caller that efficiently improves low-frequency variant detection in paired-end sequencing NGS libraries. Bioinformatics 36, 2718–2724 (2020).
Sim, N. S. et al. Precise detection of low-level somatic mutation in resected epilepsy brain tissue. Acta Neuropathol. 138, 901–912 (2019).
Boßelmann, C. M., Leu, C. & Lal, D. Technological and computational approaches to detect somatic mosaicism in epilepsy. Neurobiol. Dis. 184, 106208 (2023).
Khoshkhoo, S. et al. Contribution of Somatic Ras/Raf/Mitogen-activated protein kinase variants in the hippocampus in drug-resistant mesial temporal lobe epilepsy. JAMA Neurol. 80, 578–587 (2023).
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 131, 803–820 (2016).
Kaplanis, J. et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature 586, 757–762 (2020).
de Bruijn, I. et al. Analysis and visualization of longitudinal genomic and clinical data from the AACR Project GENIE biopharma collaborative in cBioPortal. Cancer Res 83, 3861–3867 (2023).
Mularoni, L., Sabarinathan, R., Deu-Pons, J., Gonzalez-Perez, A. & López-Bigas, N. OncodriveFML: A general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 17, 128 (2016).
Lindy, A. S. et al. Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia 59, 1062–1071 (2018).
Heyne, H. O. et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet 50, 1048–1053 (2018).
Auwera, G. A. V. de & O’Connor, B. D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. (O’Reilly, Beijing Boston Farnham Sebastopol Tokyo, 2020).
Chen, S. et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 625, 92–100 (2023).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12, e1001779 (2015).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021).
Dewey, F. E. et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 354, aaf6814 (2016).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet 48, 1279–1283 (2016).
Glusman, G., Caballero, J., Mauldin, D. E., Hood, L. & Roach, J. C. Kaviar: An accessible system for testing SNV novelty. Bioinformatics 27, 3216–3217 (2011).
Yamaguchi-Kabata, Y. et al. Evaluation of reported pathogenic variants and their frequencies in a Japanese population based on a whole-genome reference panel of 2049 individuals. J. Hum. Genet 63, 213–230 (2018).
Erikson, G. A. et al. Whole-genome sequencing of a healthy aging cohort. Cell 165, 1002–1011 (2016).
Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet 46, 818–825 (2014).
Naslavsky, M. S. et al. Exomic variants of an elderly cohort of Brazilians in the ABraOM database. Hum. Mutat. 38, 751–763 (2017).
Scott, E. M. et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet 48, 1071–1076 (2016).
Carnevali, P. et al. Computational techniques for human genome resequencing using mated gapped reads. J. Comput. Biol. 19, 279–292 (2012).
López-Rivera, J. A. et al. Incidence and prevalence of major epilepsy-associated brain lesions. Epilepsy Behav. Rep. 18, 100527 (2022).
Whiffin, N. et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med 19, 1151–1158 (2017).
Yang, X. et al. Genomic mosaicism in paternal sperm and multiple parental tissues in a Dravet syndrome cohort. Sci. Rep. 7, 15677 (2017).
Ioannidis, N. M. et al. REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet 99, 877–885 (2016).
Greenman, C., Wooster, R., Futreal, P. A., Stratton, M. R. & Easton, D. F. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics 173, 2187–2198 (2006).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47, D886–D894 (2019).
Frazer, J. et al. Disease variant prediction with deep generative models of evolutionary data. Nature 599, 91–95 (2021).
Traynelis, J. et al. Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res 27, 1715–1729 (2017).
Erdős, G., Pajkos, M. & Dosztányi, Z. IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 49, W297–W303 (2021).
UniProt Consortium. UniProt: The universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2023).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018).
Stenson, P. D. et al. The human gene mutation database (HGMD®): optimizing its use in a clinical diagnostic or research setting. Hum. Genet 139, 1197–1207 (2020).
Berman, H. M. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000).
Schrödinger, L. L. C. The PyMOL molecular graphics system, version 1.8 (2015).
Koh, H. Y. et al. BRAF somatic mutation contributes to intrinsic epileptogenicity in pediatric brain tumors. Nat. Med. 24, 1662–1668 (2018).
Cases-Cunillera, S. et al. Heterogeneity and excitability of BRAFV600E-induced tumors is determined by Akt/mTOR-signaling state and Trp53-loss. Neuro Oncol. 24, 741–754 (2022).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Chakravarty, D. et al. OncoKB: A precision oncology knowledge base. JCO Precision Oncol. 1–16 https://doi.org/10.1200/PO.17.00011 (2017).
Shimada, K., Bachman, J. A., Muhlich, J. L. & Mitchison, T. J. shinyDepMap, a tool to identify targetable cancer genes and their functional connections from Cancer Dependency Map data. eLife 10, e57116 (2021).
Szklarczyk, D. et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 14, 128 (2013).
Gene Ontology Consortium et al. The gene ontology knowledgebase in 2023. Genetics 224, iyad031 (2023).
Ashburner, M. et al. Gene ontology: Tool for the unification of biology. Nat. Genet 25, 25–29 (2000).
Acknowledgements
This study was supported by the National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke (NINDS) under grant R01 NS117544 (Principal Investigator: D.L.). I.B., L.H. and K.K. received research funding from the German Research Foundation (DFG), project number 460333672 – CRC1540 Exploring Brain Mechanics (subprojects A02, C03), and Bl421/4-1. K.K. is supported by the Else Kröner-Fresenius-Stiftung (EKFS, project number 2021 EKEA.3.3). P.N. received research funding from the German Research Foundation (DFG), grant agreement number NU 50/13-1. St.B. received research funding from Program d’Investissements d’Avenir (ANR-18-RHUS-005) and Fondation pour la Recherche médicale (S.1800.FRM22). Sequencing was facilitated by the DFG-funded West German Genome Center (WGGC).
Author information
Authors and Affiliations
Contributions
C.M.B., C.L. and D.L. conceived the study and designed the experiments. C.M.B. and C.L. conducted all statistical analyses. T.B. contributed structural modeling. L.H. and I.B. conducted the histopathological review. Sa.B., M.C., R.C., K.K., H.H., D.D., K.R., C.G.B., T.K., T.P., T.H., K.B., L.F., R.M.B., St.B., P.N., and I.N. contributed samples and/or genetic sequencing results used in the mega-analysis. C.M.B. prepared the first draft of the manuscript, and all authors contributed to manuscript review and editing. C.L., D.L., and I.B. supervised the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Boßelmann, C.M., Leu, C., Brünger, T. et al. Analysis of 1386 epileptogenic brain lesions reveals association with DYRK1A and EGFR. Nat Commun 15, 10429 (2024). https://doi.org/10.1038/s41467-024-54911-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54911-w
This article is cited by
-
State of the art: glioma-associated epilepsy—bridging tumor biology and epileptogenesis
Neurological Research and Practice (2025)
-
Epilepsy, seizures and hyperexcitability—a challenge in neurology
Neurological Research and Practice (2025)
-
Hotspots of human mutation point to clonal expansions in spermatogonia
Nature (2025)
-
Human brain tissue with MOGHE carrying somatic SLC35A2 variants reveal aberrant protein expression and protein loss in the white matter
Acta Neuropathologica (2025)