Abstract
Most bacteria lack membrane-enclosed organelles and rely on macromolecular scaffolds at different subcellular locations to recruit proteins for specific functions. Here, we demonstrate that the optogenetic CRY2-CIB1 system from Arabidopsis thaliana can be used to rapidly direct proteins to different subcellular locations with varying efficiencies in live Escherichia coli cells, including the nucleoid, the cell pole, the membrane, and the midcell division plane. Such light-induced re-localization can be used to rapidly inhibit cytokinesis in actively dividing E. coli cells. We further show that CRY2-CIBN binding kinetics can be modulated by green light, adding a new dimension of control to the system. Finally, we test this optogenetic system in three additional bacterial species, Bacillus subtilis, Caulobacter crescentus, and Streptococcus pneumoniae, providing important considerations for this system’s applicability in bacterial cell biology.
Similar content being viewed by others
Introduction
Compartmentalizing biological processes at specific subcellular locations for specialized functions is a general strategy employed by cells across all domains of life. Eukaryotic cells often achieve subcellular localization using membrane-enclosed organelles. Bacteria cells generally lack membranous organelles but can also achieve subcellular localization by directing molecules to different scaffolds made of protein, DNA, RNA, or the plasma membrane to form stable assemblies and/or phase-separated condensates1. These subcellular scaffolds underlie several fundamental cellular processes, including partitioning of cellular content, chromosome organization, segregation, cell elongation and division, and appendage growth and receptor clustering1,2. Controlling the recruitment of proteins to subcellular scaffolds and/or modulating scaffolds’ assembly states has proven to be a valuable tool for probing and manipulating their functions3.
Previously, chemically induced protein dimerization platforms such as rapamycin-based FKBP-FRB4, coumermycin-based GyrB5 and gibberellin-based6 systems have been employed to manipulate subcellular localizations of various proteins in both bacteria7 and eukaryotic8 cells. While relatively fast and robust, these methods lack precise spatial targeting ability and are often irreversible on fast timescales4. In contrast, optogenetic-based systems use light-modulated binding of a photoreceptor either with itself or with its associated protein-ligand and are reversible9,10; the application of light activation can be precisely defined in space to target specific cell subpopulations and even within a single cell’s volume11,12,13,14,15,16. Currently available light-induced protein-protein interaction systems have been extensively characterized in mammalian cell systems3 and have kinetic rates on timescales ranging from milliseconds to hours17. Despite the widespread use of optogenetic systems in eukaryotic cells, their use in bacteria is only beginning to emerge (reviewed in ref. 3). The major difficulty of their implementation resides in the bacteria cell’s 1000-fold smaller cellular volume compared to that of a mammalian cell18. Such a small cytoplasmic volume imposes strict requirements on the concentration and kinetic ranges of the optogenetic systems in order to achieve a high signal contrast and spatial resolution18,19.
One particular system, the CRY2/CIB1 system from Arabidopsis thaliana, can be induced by a low dose of blue light (peak activation wavelength at 450 nm) and exhibits rapid heterodimerization kinetics with tight and reversible binding19,20. These properties make it an attractive candidate for use in model bacterial systems. The system is composed of the N-terminal domain (amino acids 1-170) of AtCIB1 (from here on denoted as CIBN) and the photolyase homology region21 (amino acids 1-498) of AtCRY2 (from here on denoted as CRY2)19,22. Here we show that this optogenetic system can be used to rapidly target proteins to major subcellular compartments in living Escherichia coli cells (Fig. 1a, left). We further demonstrate that such an optogenetically controlled system can inhibit cytokinesis in living E. coli cells. We observed that the association and dissociation kinetics of the system can be modulated by green light, adding a new dimension of control to suit different synthetic biological systems. Lastly, we tested the system in three additional bacterial model organisms, B. subtilis, C. crescentus and S. pneumoniae, providing important considerations for the applicability of this system in bacterial cell biology.
a CRY2 is targeted to different subcellular localizations in live E. coli cells: the chromosomal DNA, cell pole, inner membrane, and the midcell division plane. At each subcellular location, CIBN is fused to a bait protein at the target either at the C or N terminus, and CRY2 is fused to the N terminus of mCherry as a universal prey reporter for CIBN’s light (488 nm)-dependent association with CIBN-Target (inset box). The association is reversible in dark. b CIBN and CRY2 fusion proteins can be expressed co-transcriptionally (left) or independently (right) to suit different experimental needs.
Results
General considerations for using the CRY2-CIBN system in Escherichia coli cells
Previous studies have shown that CIBN can be tagged at either its N- or C-terminus without impeding its binding to CRY2, whereas CRY2 functions best with its N-terminus free23. Therefore, in the following experiments, we fused CIBN at its N- or C-terminus to an array of proteins that localize to a subcellular compartment of interest to serve as the bait for the recruitment platform. We then fused a fluorescent reporter, mCherry, to the C-terminus of CRY2 to serve as a universal prey (Fig. 1a, right). Using the same CRY2-mCherry reporter to various subcellular recruitment platforms allows us to compare recruitment kinetics between different systems.
Next, to attain the desired control of CRY2/CIBN expression levels inside small bacterial cells in a variety of experimental scenarios, we exogenously expressed the CRY2-CIBN fusion proteins either co-transcriptionally from a single plasmid where the two fusions were coupled under the control of a lac-inducible promotor, or independently from a two-plasmid system where the CRY2 and CIBN fusions were under the control of arabinose- and lac-inducible promoters, respectively (Fig. 1b, Supplementary Data 1). Whether the one- or two-plasmid expression system was employed depended on specific considerations for each experiment outlined below. We found that the flexibility in modulating the expression levels of the CRY2 and CIBN fusion proteins is important for minimizing light-independent background interaction19 while maintaining a fast light-dependent recruitment speed (discussed below).
Rapid, reversible, and light-dependent recruitment of cytoplasmic protein to chromosomal DNA
In bacterial cells, the nucleoid is the major subcellular location for all DNA-related processes. The ability to optically control chromosomal DNA topology (e.g., the formation of DNA loops or topological domains) or the binding of transcription factors on specific DNA sequences has proven useful to study transcription dynamics and gene expression19,24,25 in mammalian and bacterial cells24,26,27,28,29. Therefore, we chose the chromosomal DNA as the first platform to develop a light-dependent recruitment assay of cytoplasmic proteins.
To visualize the light-dependent chromosomal recruitment process, we used an E. coli strain harboring a 240X tetO array sequence inserted near oriC in its chromosome30 (Fig. 2a). The presence of tandem tetO arrays enhances the mCherry signal strength and facilitates the characterization of the recruitment kinetics. We then fused CIBN to the C-terminus of TetR, the tetracycline repressor that binds tightly to its cognate tetO operator sequence31 and co-transcriptionally expressed the TetR-CIBN and CRY2-mCherry reporter fusions exogenously from a single plasmid (Fig. 1c, left). The use of the coupled expression system helps to maintain a low and near equimolar expression of the two fusions proteins for the 1:1 binding ratio of TetR-CIBN and CRY2-mCherry, effectively minimizing the background signal that would arise from excess CRY2-mCherry reporter. In the absence of the blue (488 nm) activation light, CRY2-mCherry remained homogenously distributed in the cytoplasm as expected for freely diffusing CRY2-mCherry in its unbound state (Fig. 2b, left). Upon exposure to 488 nm activation light pulses (30 ms pulses at 84.6 W/cm2 delivered every 5 seconds over 200 seconds), we observed the rapid formation of CRY2-mCherry foci at locations representative of individual oriC sites in almost all cells in the view field (96 ± 1.3 %, μ ± s.e.m., mean ± standard error of the mean, N = 3 independent experiments with 1359 cells in total, Fig. 2b, right, Fig. 2c, top, Supplementary Movie 1) across a large range of CRY2-mCherry expression levels in individual cells. Notably, the cytoplasmic distribution of CRY2-mCherry remained unchanged in control cells exposed to the 488 nm light, but that did not contain the 240X tetO array (Fig. 2c, middle panel, Supplementary Movie 2), or in cells that contained the tetO array but not exposed to the 488 nm activation light (Fig. 2c, bottom panel, Supplementary Movie 3). Quantification of the CRY2-mCherry signal (percentage of fluorescence increase compared to that before activation) at foci positions over time showed that 90% recruitment was reached within 85 seconds (τ0.9 = 85 ± 9 seconds, μ ± s.e.m., N = 3 independent experiments with 442 cells in total, τ0.9 was used to show the extent of reaction completion as the curve was not well described by an exponential function, Fig. 2d). These results demonstrate that the formation of CRY2-mCherry foci is rapid and both light- and target-dependent.
a Schematic depicting the relocalization of cytoplasmic CRY2-mCherry to TetR-CIBN bound tetO sites near OriC to from foci after activation with blue light. b CRY2-mCherry is uniformly distributed throughout the cell prior to blue light exposure (left panel). After blue light exposure CRY2-mCherry rapidly relocalizes to form foci (right panel). Images are representative of N = 3 independent experiments. Scale bar = 2 µm. c Single cell time course images demonstrating that the recruitment of CRY2-mCherry to DNA foci occurs only when blue light and the tetO array are both present (top row) but not in the absence of the tetO target (middle row) or blue light activation (bottom row). Images are representative of N = 3 independent experiments. Scale bar = 2 µm. d Averaged percent increase (black dots) of CRY2-mCherry signal at DNA foci (N = 3 independent experiments of 1359 cells in total) demonstrating that 90% recruitment is reached within 85 seconds. Shaded region indicated the s.e.m. e Example trace of the mean CRY2-mCherry signal at DNA foci (gray dots; N = 1 experiment of 89 cells) after activation with blue light (first blue section), relaxation in the dark (middle gray section), and after a second activation sequence (second blue region) demonstrating that CRY2/CIBN disassociation at DNA foci is reversible with a time constant of ~ 9 minutes. Error bars indicated s.e.m. f Schematic depicting the relocalization of cytoplasmic CRY2-mCherry to CIBN-GFP-PopZ foci at the cell pole after blue light activation. Scale bar = 2 µm. g CRY2-mCherry is uniformly distributed throughout the cell prior to blue light exposure (left panel). After blue light exposure, CRY2-mCherry rapidly relocalizes to the cell poles and form foci (right panel). Image is representative of N = 3 independent experiments. h Single cell time course images demonstrating that the recruitment of CRY2-mCherry to the cell pole occurs only when both blue light and the CIBN are present (top row) but not in the absence of the CIBN target (middle row) or blue light (bottom row). Images are representative of N = 3 independent experiments. Scale bar = 1 µm. i Averaged percent increase (black dots) of CRY2-mCherry signal at cell pole foci (N = 3 experiments with 315 cells in total) demonstrating that 90% recruitment is reached within ~8 seconds. Shaded region indicates the s.e.m.
In addition to its rapid association kinetics, the CRY2-CIBN system is also reversible in the absence of blue light19. When we allowed the CRY2-CIBN system to relax in the absence of blue light following the complete localization of CRY2-mCherry to oriC foci (Fig. 2e, first blue-shaded region), we observed an almost complete reversion of CRY2-mCherry foci back to a uniform cytoplasmic distribution after ~40 minutes (Figs. 2e, Supplementary Fig. 1). The decay curve was well described by an exponential function with a relaxation time constant of ~10 minutes (τrev = 10 ± 2 min, μ ± s.e.m., N = 3 independent experiments with 142 cells in total, Fig. 2e, shaded gray region). This relaxation time scale is consistent with the reversion times measured previously in mammalian cells17,19. Additionally, relaxed cells could be activated again17,19 to re-form the same oriC foci with nearly identical recruitment rates and final plateau (Fig. 2e, second blue-shaded region and Supplementary Movie 4). In summary, these results demonstrate that the CRY2-CIBN system can be used to induce reversible recruitment of a cytoplasmic protein to chromosomal DNAs rapidly and efficiently in live E. coli cells.
Rapid, light-dependent recruitment of cytoplasmic protein to the cell pole
In bacteria, asymmetric cell division is often achieved by partitioning proteins to each incipient daughter cell via cell pole localized protein scaffolds32. In symmetrically dividing bacteria such as E. coli, the two poles are also differentiated in age, and proteins can be targeted specifically to the old cell pole to produce a dimorphic cell population33,34. Furthermore, the cell pole can be engineered into an optically controlled apical protein sink to present an inert space away from the cytoplasmic environment35.
To develop a light-triggered cell pole recruitment platform, we took advantage of a protein called PopZ from Caulobacter crescentus. PopZ forms a stable, liquid droplet-like matrix enriched at the old cell pole when heterologously expressed in E. coli cells36,37,38 (Fig. 2f). To visualize the cell pole localization of PopZ, we fused CIBN-GFP to the N terminus of PopZ and placed the fusion gene under a lac-inducible promoter on its own plasmid (Fig. 1b, right). After optimizing the expression of the PopZ fusion, we observed large, stable CIBN-GFP-PopZ foci at the cell poles of nearly every cell (Supplementary Fig. 2). We then expressed the CRY2-mCherry fusion under the control of an arabinose-inducible promoter from a separate vector (Fig. 1b, right). Here, we used the two-plasmid system for independent expression control of the two fusion proteins, because the liquid droplet nature of the PopZ foci can accommodate a very high expression level of CIBN-GFP-PopZ, which ensures a stable cell pole localization and provide a sufficient number of binding sites for CRY2-mCherry.
As shown in Fig. 2g and Supplementary Movie 5, following a single 100 ms blue light activation pulse (P488 = 8.5 W/cm2), we observed the rapid formation of CRY2-mCherry foci at the cell poles. Importantly, in cells exposed to blue light only expressing GFP-PopZ, but not CIBN-GFP-PopZ (Fig. 2h, middle panel and Supplementary Movie 6), or in cells without blue light activation (Fig. 2h, bottom panel and Supplementary Movie 7), we observed unchanged, homogenous distribution of CRY2-mCherry signal in the cytoplasm throughout the time course of the experiment. Quantification of the percent increase in CRY2-mCherry signal at the cell poles revealed that 90% recruitment was achieved within ~8 seconds (τ0.9 = 8.3 ± 0.9 seconds, μ ± s.e.m., N = 3 independent experiments with 315 cells in total, Fig. 2i). Note that in this system, the recruitment of cytoplasmic CRY2-mCherry was sped up to ~10-fold compared to that in the DNA recruitment assay. The faster kinetics are most likely driven by the higher expression levels of both CIBN-GFP-PopZ and CRY2-mCherry in the system.
Formation of inner membrane-associated hetero-clusters of CRY2-CIBN upon light activation
In many cell types, the inner membrane (IM) is the site of diverse signaling pathways. In bacteria, the IM also serves as the assembly site for large macromolecular structures such as the divisome39, the flagellum40, or chemotaxis receptor clusters41. In mammalian cells, synthetic activation of signaling cascades can be achieved using cytoplasmic CRY2-tagged proteins that are recruited to membrane-anchored CIBN16,19,23,42,43. To develop a light-dependent membrane recruitment assay in bacteria, we fused CIBN-GFP to an amphipathic helix from Bacillus subtilis (BsMTS)44 and expressed it from a lac-inducible promotor independently from the arabinose-inducible CRY2-mCherry on a second plasmid. We used an exogenous membrane anchor to avoid potential interference with endogenous E. coli membrane proteins. Notably, the BsMTS-GFP-CIBN fusion showed uniform membrane localization as expected (Supplementary Fig. 3a, left). Upon blue light activation (50 ms pulses at 84.6 W/cm2 delivered every 5 s), we observed the formation of dynamic CRY2-mCherry puncta that co-localized with CIBN-GFP-BsMTS puncta along the IM (Supplementary Fig. 3b, & Supplementary Movie 8). The distributions of these puncta were distinct from the near uniform distribution of CIBN-GFP-BsMTS along the IM surface before activation with blue light (Supplementary Fig. 3a, left), or that of directly membrane-targeted CRY2-GFP-BsMTS (Supplementary Fig. 3a, right). We reason that the puncta formation along the IM of CIBN-GFP-BsMTS and CRY2-mCherry upon light activation is likely indicative of the formation of individual membrane-tethered CIBN-CRY2 heteroclusters as previously predicted12,23,45. Despite the many strategies we employed, we could not achieve uniform co-localization of CRY2-mCherry and CIBN on the IM of E. coli cells (Supplementary Table 1, Reference 46). This ability is in contrast to what was observed previously in mammalian cells12,19. Further experiments are required to investigate this phenomenon.
Rapid, light-dependent recruitment of cytoplasmic proteins to the midcell
In almost all bacteria studied to date, the essential process of cytokinesis is mediated by the formation of a large macromolecular complex, termed the divisome, at the future division plane of the cell47. In E. coli, the divisome comprises more than 30 proteins, recruited to the midcell by the essential tubulin homolog, FtsZ39,48. FtsZ polymerizes at the midcell to assemble into a ring-like structure, and new data suggests that the dynamics of the Z-ring are essential to coordinate the action of the divisome for cell wall constriction and septum morphogenesis during cell division49,50,51. Achieving a means to rapidly and reversibly deliver proteins to the divisome and/or modulate the assembly of the divisome at different cell division stages would provide a new way to probe the dependence of varying divisome proteins and their functions on the assembly state of the divisome specifically at the division site52.
To achieve optically controlled delivery of proteins to the cell division plane, we first tested the ability to recruit CRY2-mCherry to the Z-ring by creating a C-terminal fusion of CIBN to ZapA (Fig. 3a). ZapA is a conserved Z-ring-associated protein, which has served as a faithful Z-ring marker in previous studies53,54,55,56. Similar to the pole-recruitment platform, we expressed the ZapA-CIBN fusion independently from the CRY2-mCherry reporter using the two-plasmid expression system to optimize their expression levels separately. Upon exposing cells to a single 100 ms pulse of 488 nm activation light (P488 = 8.5 W/cm2), we observed rapid midcell localization of CRY2-mCherry fluorescence (τ0.9 = 9.3 ± 0.15 seconds, μ ± s.e.m., N = 3 independent experiments with 443 cells in total, Fig. 3b-d, Supplementary Movie 9). Similar to what was observed in the chromosomal DNA and cell pole recruitment assays, the midcell localization of CRY2-mCherry was dependent on the blue activation light (Fig. 3c). These results demonstrate that cytoplasmic proteins can be recruited to the Z-ring at the midcell rapidly and specifically using the CRY2-CIBN system.
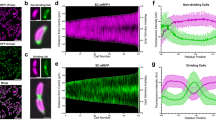
a Schematic depicting the relocalization of cytoplasmic CRY2-mCherry to the ZapA-CIBN ring present at midcell after induction of CRY2/CIBN binding with blue light. b An example image of cells showing CRY2-mCherry’s diffusive cytoplasmic localization before blue light activation (left) and midcell localization after blue light activation (right). Image is representative of N = 3 independent experiments. Scale bar = 2 µm. c Single cell time course images demonstrating that CRY2-mCherry recruitment to midcell occurs only after activation with blue light. Scale bar = 1 µm. d Averaged percent increase (black dots) of CRY2-mCherry signal at midcell demonstrating that 90% recruitment is reached within 9 seconds (N = 3 experiments with 443 cells). Shaded region indicates the s.e.m. e Schematic depicting the LInC assay on both short and long time scales. Recruitment of CRY2-MinC to Z-rings via ZapA-CIBN by blue light activation results in instant destabilization of the Z-ring at a short time scale and cell division inhibition at a long time scale. In the absence of blue light cells grow and divide like WT cells. f Short timescale LInC Assay. Cells harboring the LInC system exhibited diffusive Z-rings (arrow heads) after blue light activation delivered every 10 s for 5 min. Scale bar = 2 µm. The projected fluorescence intensity of ZapA-mCherry (Z-ring) along the cell long-axis before (gray) and after a 5-minute blue light induction (blue) for the cell labeled with a white arrow in (f) is plotted in (g) to demonstrate the significant decondensation of the Z-ring (p = 0.032, determined by a two-sample KS-test). h Short timescale Z-ring destabilization by the LInC system is dependent on cell length. The percent reduction of ZapA-mCherry intensity (μ ± s.e.m, Supplementary Table 2) at midcell in cells expressing the LInC system before (N = 3 experiments with 1465 cells in total) and after (N = 3 experiments with 1658 cells in total) a 5-minute period of blue light activation (blue datapoints) or darkness (gray datapoints) plotted as a function of cell length. i Long timescale LInC assay. Time-lapse imaging shows that cells ectopically expressing CRY2-MinC only (Control, top row) grew and divided normally while cells expressing both CRY2-MinC and ZapA-CIBN (LInC, bottom row) filamented when both were exposed to 100 ms pulses of blue and green light every 5 minutes for a 12-h period. Cells harboring only CRY2-MinC (right panel, top) or both CRY2-MinC and ZapA-CIBN (right panel, bottom row) but were not exposed to blue light divided normally during the same period (right most panel). White arrow heads indicate the position of cells that are enlarged in the insets. Images are representative of N = 2 independent experiments. Scale bar = 2 µm. j Smoothed and averaged long axis projections of the Z-ring intensity measured by ZapA-mCherry at the 12-h timepoint after blue light activation demonstrated significant (p = 6.36 x 10−6, determined by a two-sample KS-test) widening of Z-ring in cells harboring the LInC system (FWHM = 221 nm, N = 2 experiments with 16 cells in total) compared with those in the Control cells (FWHM = 102 nm, N = 2 experiments with 33 cells in total).
Light-dependent destabilization of the Z-ring and inhibition of cell division
We next used the midcell recruitment platform to develop a Light-induced Inhibition of Cytokinesis (LInC) assay in live E. coli cells (Fig. 3e). In the LInC assay, we kept the same ZapA-CIBN fusion as the bait, but fused a Z-ring antagonist, MinC, to CRY2 to generate a CRY2-MinC fusion. MinC inhibits FtsZ polymerization in vitro. It prevents Z-ring assembly at the cell poles by forming a high-to-low concentration gradient from the cell pole to the midcell with its membrane binding partner ATPase MinD57. When highly overexpressed by itself, MinC antagonizes Z-ring assembly at the midcell as well57,58,59. We verified that the CRY2-MinC fusion is fully functional as it complemented a minC deletion background (Supplementary Fig. 4). Therefore, by recruiting CRY2-MinC to FtsZ-associated ZapA-CIBN, we expect to increase the local concentration of MinC at the Z-ring, which may result in the destabilization of the Z-ring and subsequent inhibition of cell division (Fig. 3e). To visualize the progress of Z-ring destabilization in the LInC assay, we tagged the endogenous chromosomal copy of ZapA with mCherry as the Z-ring marker53,54.
We first established growth and expression conditions under which cells ectopically expressing the LInC system (ZapA-CIBN and CRY2-MinC) exhibited a near-WT midcell Z-ring morphology and underwent successful cell division cycles in the absence of blue light (Supplementary Note 1). Notably, these cells were moderately longer than uninduced cells due to the presence of higher than WT CRY2-MinC concentrations (L = 3.4 ± 0.4 μm, μ ± s.d., N = 3 independent experiments with 1465 cells in total compared to L = 2.3 ± 0.06 μm, μ ± s.d., N = 2 independent experiments with 379 cells in total for uninduced cells). We then imaged the Z-ring morphology of these cells before and after exposing them to 100 ms blue light pulses (P488 = 84.6 W/cm2) every 10 seconds for 5 minutes (Fig. 3f). Before the activation light exposure, we quantified that on average ~46% of all the cells harbored clear ZapA-mCherry localization at midcell, consistent with previous reports that the Z-ring only assembles after a significant portion of the chromosome is segregated60. About 53% of these cells (53.3 ± 3.97%, μ ± s.e.m., N = 3 independent experiments with 1658 cells in total) exhibited considerably broader and more diffuse ZapA-mCherry midcell fluorescence upon exposure to blue light compared to that before light exposure (Fig. 3f & g), indicating the destabilization of the Z-ring. In uninduced cells or in cells without blue light activation, we observed significantly lower, ~40% (40.3 ± 15.0%, μ ± s.e.m., N = 2 independent experiments with 379 cells in total) or ~44%, (44.1 ± 1.68%, μ ± s.e.m., N = 3 independent experiments with 1433 cells in total), cells exhibiting diminished ZapA-mCherry midcell fluorescence after a 5 minutes, indicative of the natural disassembly process of the Z-ring during cell division. Interestingly, the extent of the Z-ring destabilization appeared to be dependent on the cell division stage (Figs. 3h, Supplementary Table 2). We observed that after Z-ring assembly, cells of relatively short lengths (4-8 µm in length, indicative of earlier cell division stages) showed significant destabilization of the Z-ring (Fig. 3h, blue) compared to cells of longer lengths, or cells of similar lengths but without the activation light (Fig. 3h, gray). Previous studies have shown that when the divisome matures and cell wall constriction initiates, the Z-ring becomes denser and denser until it finally and rapidly disassembles toward the end of the cell cycle61,62. Therefore, the LInC assay may destabilize the Z-ring effectively only before the Z-ring becomes too dense to be effectively depolymerized by MinC.
While we achieved efficient and rapid Z-ring destabilization in cells judged by their widened Z-ring morphology and the reduction in the midcell localization percentage of FtsZ, it remained unclear whether these destabilized Z-rings were indeed defective in cell division. To address this question, we monitored cell division for an extended period in cells harboring the LInC system after induction with blue light. As shown in Fig. 3i, when we subjected LInC cells to 100 ms pulses of 488-nm activation light (P488 = 8.5 W/cm2) and 561-nm imaging light (P561 = 8.7 W/cm2) every 5 minutes for a twelve-hour period, we observed that the majority of these cells ceased division and grew into long filaments instead. On average, ~76% (75.7 ± 12.3, μ ± s.e.m., N = 3 experiments with 33 cells) were unable to divide during the 12 h period. Additionally, the average division time (τLInC = 355 ± 84.6 minutes, μ ± s.e.m., N = 3 experiments of 33 cells in total) of the remaining ~24% (24.3 ± 12.3, μ ± s.e.m., N = 3 experiments of 31 cells in total) cells that were able to divide eventually was nearly 1.5 times longer than the mean division time of the control group where the ZapA-CIBN bait fusion was absent (τcontrol = 232 ± 31.2 minutes, μ ± s.e.m., N = 3 experiments of 31 cells in total). These observations indicated a considerable deficiency of LInC cells in division. The inhibited cell division also correlated with significantly destabilized Z-rings. In all long, filamentous cells, the midcell fluorescence of ZapA-mCherry exhibited a wider spread, in contrast to the sharp midcell localization prior to light exposure (Fig. 3j). Note that the inhibition of cell division in these cells was not caused by photodamage due to the continuous illumination of the 488 nm activation light and the 561 nm imaging light, because cells under the same induction and illumination condition but ectopically expressing only CRY2-MinC grew and divided as wild type cells not exposed to light (Fig. 3i, top row). Furthermore, the inhibition was 488 nm light-dependent because cells expressing both CRY2-MinC and ZapA-CIBN without blue light grew normally as WT cells (Fig. 3i, bottom row rightmost panel). Taken together, these results show that the CRY2-CIBN system can be used to destabilize the Z-ring rapidly and inhibit cell division in living E. coli cells in a light-dependent manner.
CRY2-CIBN complex formation is sensitive to green light
Previous studies have shown that the light sensitivity of CRY2 is imparted by its flavin adenine dinucleotide (FAD) cofactor through a photocycle52,53 (Fig. 4a). In this cycle, the redox state of CRY2’s FAD is thought to mediate a conformational change in CRY2 to form a signaling-active state that is competent to bind CIB63. Briefly, the fully oxidated FADox cofactor is found in the resting inactive state of CRY2. Following photoexcitation by blue light, FADox accepts an electron from a nearby tryptophan followed by protonation from an aspartic acid to become the semi-reduced, stable semiquinone radical (FADHo). This redox state is thought to trigger a conformational change in CRY2 (denoted as CRY2*), allowing it to bind to CIB64,65. To complete the photocycle, semi-reduced FADHo is reduced slowly to become FADH-, reversing the conformation change, after which it is oxidated rapidly back to its fully oxidized resting state (FADox). Previous studies have indicated that the semiquinone form FADHo is the only green-absorbing species in FAD’s photocycle, and that green light can deplete FADHo in vitro65. In short-day entrained plants, green light delays flowering, likely due to the increased reduction of FADHo to FADH- under green light exposure54,55. However, previous characterization of CRY2-CIBN complex formation in vivo in heterologous expression systems showed no sensitivity to green light19.
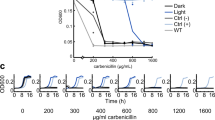
a Schematic of CRY2 photoactivation pathway highlighting the redox states of its flavin (FAD) cofactor along the path. Breifly, the fully oxidized FADox is photoexcited by a blue light photon and accepts an electron from the nearby tryptophan (Trp397) to form a stable, semi-reduced neutral FADHo. This formation induces a conformational change in CRY2, which allows it to bind to CIBN. In the absence of green light, the semi-reduced FADHo slowly decays to a fully reduced state (FADH-). As FADHo is the only species in the cycle that can absorb green light, it is likely that green light speeds up the further reduction of FADHo to FADH-, which prevents CRY2 to associate or remain associated with CIBN. b The percent enrichment of cell pole-recruited CRY2-Halo shows a stepwise dependence on the intensity of the 100 ms blue (488 nm) light pulse used to trigger CRY2/CIBN complex formation. (N = 2 experiments with > 85 individual cells for each condition, error bars = s.d.) c The maximal fold enrichment of cell pole-recruited CRY2-Halo under varying green light intensities relative to that in the absence of green light showed a green light dependent reduction (left, N = 2 experiments with > 100 individual cells in total for each green light condition respectively, error bars = s.e.m. via bootstrapping), while the 90% recruitment time (τ0.9) under varying green light intensities relative to that in the absence of green light was not significantly altered (right, N = 2 experiments, error bars = s.e.m. via bootstrapping) indicating that green light can modulate the levels of activated CRY2 available for complex formation with CIBN (p-values determined by a two-tailed t-test). d The change in the final fold reduction of cell pole-recruited CRY2-Halo dissociation under green light exposure relative to that in the absence of green light showed no change (left, N = 4 experiments with 91 cells in total, error bars = s.d.), while the reversion time (τrev) under green light exposure relative to that in the absence of green light showed a considerable but statistically insignificant reduction (right, N = 4 experiments with 62 cells in total, error bars = s.d., p = n.s., determined by a two-tailed t-test).
We reason that if the association/dissociation kinetics of the CRY2-CIBN system are indeed sensitive to green light, we could use green light (561 nm) in combination with blue activation light to fine-tune the kinetic behavior of the CRY2-CIBN system. To investigate this possibility, we swapped the mCherry reporter with Halo66, which can be covalently labeled with a bright, photostable far-red organic dye67 (JF646), allowing us to avoid using green light to image the reporter. We verified that the extent of CRY2-CIBN association indicated by the percentage increase in cells expressing the Halo-labeled system exhibited a blue-light activation dose dependence (Fig. 4b) as expected19, and that the final association plateau could be further increased using subsequent multiple pulses of activation to recruit additional CRY2-Halo to the pole (Supplementary Fig. 5). These results indicate that CRY2-Halo can be activated similarly as CRY2-mCherry by a 100 ms blue pulse and that the final pole-enrichment of CRY2 is dependent on the available level of activated CRY2, which can be modulated by the intensity of the activation pulse.
Next, to investigate the influence of green light on CRY2-CIBN association, we monitored CRY2-CIBN complex formation at the cell poles using a single 100 ms activation pulse of 8.5 W/cm2 blue light followed by continuous illumination of green light of different intensities (561 nm, from 8.71 to 970 W/cm2) accompanying the imaging 647 nm light (streamed acquisition using a 50 ms exposure time for a total of 15 seconds). We found that the amount of CRY2 recruited to the cell pole decreased significantly with increasing green light intensity, accompanied by slight changes in the 90% recruitment value (Fig. 4c, left and right panels, respectively, Supplementary Fig. 6, Supplementary Table 3). At the highest green light intensity (970 W/cm2), the pole-recruited CRY2 fraction was reduced to ~50% compared to that in the absence of the green light. The decreased level of recruitment but largely unaffected time constant is most consistent with decreased levels of activated CRY2* under the green light illumination condition; the green light potentially drives activated CRY2* back to the inactivated state even before it forms a complex with CIBN. Hence, the green light could modulate the level of activated CRY2* and subsequently the amount of CRY2*-CIBN complex formation in live E. coli cells.
To investigate whether green light might also modulate CRY2-CIBN’s dissociation kinetics, we light-induced CRY2-CIBN complex formation using blue light and then monitored the dissociation kinetics of the complex in the presence or absence of green light (561 nm, 473 W/cm2, 100 ms pulses every 3 min, Fig. 4d). We found that while the fold of reduction of CRY2-CIBN complex formation indicated by the final dissociation plateau (Supplementary Fig. 7) did not appear to change dramatically, we observed a considerable but statistically insignificant decrease in the time constant of the dissociation reaction from 18.3 ± 8.2 minutes (N = 4 independent experiments with 91 cells in total) in the absence of green light to 12.7 ± 5.7 minutes (μ ± s.e.m., N = 4 independent experiments with 62 cells in total) in the presence of the green light, (Figs. 4d, left and right panel respectively, Supplementary Fig. 7, Supplementary Table 3). This process is possibly through the photoreduction of FADHo within the CRY2*-CIBN complex. These observations suggest that while the green light might also act upon activated CRY2* in complex with CIBN to drive it back to the inactivated form, the speeding up the dissociation of the CRY2*-CIBN complex is not significant. Further experiments are needed to verify the effect.
Adapting CRY2-CIBN to other bacterial species
The rod-shaped monoderm bacteria B. subtilis, the ovoid-shaped monoderm pathogen S. pneumoniae, and the crescent-shaped diderm bacteria C. crescentus are three widely used model organisms for bacterial cell biology and genetic studies. Testing whether the CRY2-CIBN system could also be adapted to these organisms will provide first-of-its-kind information on the feasibility of developing new optogenetic tools in species other than E. coli. As we describe below, for each species, we tested a variety of expression, labeling and imaging conditions (Supplementary Notes 2–4). Among these variables, we discovered that species-specific codon optimization and expression levels were the most critical. For all the studies presented below, we used species-codon-optimized fusion proteins. We obtained varied degrees of adaptation in these species, none as robust as we observed in E. coli. Nevertheless, our observations are important for further explorations of the CRY2-CIBN system in different bacterial species.
For B. subtills, we employed the Z-ring recruitment platform (Fig. 5a) as the Z-ring assembly pathway in B. subtilis is well established. We fused a B. subtilis codon-optimized CRY2 to the C-terminus of tdTomato and placed the fusion gene under the control of an IPTG inducible promoter at the chromosomal thrC locus (thrC::Phyperspank-CRY2Bs-tdTomato) in strain SC757 (see Supplementary Note 2 for full genotype and strain construction). We then expressed a zapA-CIBN fusion gene under the control of the native zapA promoter (PzapA-zapA-CIBN) at the amyE locus of the chromosome. Similar to E. coli, ZapABs localize to the midcell concurrently with FtsZ68. When we grew B. subtilis cells expressing both the CRY2-tdTomato and ZapA-CIBN fusions in M9+ medium and exposed them to sequential pulses of blue and green light (50 ms pulses of 488 nm light at 1.28 W/cm2 and 500 ms pulses of 561 nm light at 1.63 W/cm2, delivered every 3 seconds), we observed blue light-dependent septal localization of CRY2-mCherry over the course of 8 minutes in 16 ± 6% cells (μ ± s.e.m.; N = 4 independent experiments with 810 cells in total) (Fig. 5b, c). The recruitment of CRY2-tdTomato to midcell localized ZapA-CIBN was slower than that in E. coli, with an initial lag period of ~150 sec followed by a largely linear increase phase to ~15% over 8 min (Fig. 5d). Further optimization of the expression and imaging conditions of the system using cells grown in casein hydrolysate (CH) media, a rich media used for conventional B. subtilis imaging experiments, produced variable degrees of midcell localization of CRY2-mCherry with similar kinetics (Supplementary Fig. 8a–c).
a Schematic depicting the re-localization of cytoplasmic CRY2-tdTomato to ZapA-CIBN at midcell after activation with blue light. b Z-projections of cells undergoing blue light pulsing for the indicated number of frames. Consecutive 50 ms blue and 500 ms green light pulses were delivered every 3 seconds for a period of 8 minutes. The yellow triangles indicate midcell recruitment observed in a z-projection of the last 20 frames. Images are representative of N = 3 independent experiments. c Example showing blue light dependent localizing CRY2-tdTomato to midcell for a single cell over a 360 second time period. d Averaged percent increase (black dots) with s.e.m. (red) of CRY2-tdTomato signal at midcell demonstrating that light-dependent recruitment to midcell (N = 71 cells from three independent biological replicates). Scale bar = 1.6 µm.
For C. crescentus, we used the PopZ localization platform, as the cell pole in C. crescentus is an important location for asymmetric cell division and stalk growth69,70,71. We expressed C. crescentus codon optimized mCherry-CRY2 fusion under the control of a xylose-inducible promoter (PXyl-mCherry-CRY2Cc) and a CIBN-GFP-PopZ fusion under the control of an orthogonal vanillate-inducible (Pvan-CIBN-GFP-popZ) promoter (see Supplementary Data 2). CIBN-GFP-PopZ localized to the cell pole in greater than 90% of cells even in the absence of vanillate induction due to the leaky expression of the vector (Supplementary Fig. 9a). However, despite our best effort in optimizing expression levels, growth conditions and imaging conditions, (Supplementary Note 3), we only observed that maximally in ~30% of cells, diffusive, cytoplasmic mCherry-CRY2 relocated to cell poles. The pole localization depends on CIBN-GFP-PopZ expression but is independent of vanillate induction or blue light activation (Supplementary Figs. 9a, b). The relatively low efficiency and light independence in CRY2-mCherry’s pole localization are likely due to the less optimal folding of CRY2 and the small cell volume of C. crescentus cells respectively, as we observed heterogenous expression of CRY2-mCherry across cell populations (Supplementary Fig. 9a). This example highlights the challenge in optimizing the expression level of the CRY2-CIBN system in smaller bacterial cells such as C. crescentus.
For S. pneumoniae, we also employed the Z-ring recruitment platform. We constructed various bait (ZapA-CIBN or FtsZ-CIBN) and prey fusions (CRY2-mCherry, CRY2-Halo, or CRY2-tdTomato) and tested a variety of codon optimization strategies, genetic backgrounds, and growth and imaging conditions. However, despite our best efforts, we were unable to observe any midcell recruitment of CRY2 fusions (see Supplementary Note 4 & Supplementary Table 5). The underlying reason is unclear, but it may involve poor folding of expressed CRY2-CIBN domains in S. pneumoniae, blockage of CRY2-CIBN by interactions with pneumococcal proteins, absorbance of blue light by proteins in this bacterium, and/or limited availability of FAD cofactor for CRY2 binding.
Discussion
The landscape of optogenetic systems to induce protein-protein interactions inside living cells is constantly growing3,72. Optogenetic systems are especially useful for live-cell imaging studies due to their rapid induction, reversible binding and spatial precision73,74. Here we demonstrate that the Arabidopsis thaliana CRY2-CIBN system (Fig. 1a), used extensively in mammalian cell systems, can be adapted to target a protein cargo to different subcellular structures including the chromosomal DNA, cell pole, inner membrane and the division plane at the midcell in living E. coli cells (Fig. 1b). During the optimization process we found that the expression levels of CRY2-CIBN system in bacteria cells can be varied to achieve recruitment halftimes ranging from the seconds to minutes timescales similar to those observed in mammalian cells19,23. We then demonstrate that the CRY2-CIBN system can also be implemented to rapidly destabilize the cytokinetic ring and inhibit cell division in E. coli (Fig. 3), providing a rapid and spatially specific tool to manipulate bacterial cell division.
During our characterization of the CRY2-CIBN system in E. coli, we found that both the association and dissociation of CRY2 with CIBN can be modulated by green light (Fig. 4c). This effect not only enables another layer of system tuning akin to the blue light dose-dependent activation behavior (Fig. 4b) that we and others19,25 have observed, but also provides direct evidence that green light inactivates CRY2 likely by acting on the semi-reduced FADHo cofactor directly to reverse the active conformation of CRY2, hence lowering the amount of CRY2* available for complex formation and also speeding up the dissociation kinetics of previously formed CRY2*-CIBN complex. These results are consistent with previous experiments where green light exposure rapidly converts CRY2’s cofactor from FADHo to FADox in vitro and inhibits CRY2/CIB1 activity in vivo65.
In adapting the CRY2-CIBN system to other bacterial species, however, we observed that it was most successful in B. subtilis, less efficient in C. crescentus, and did not work in S. pneumoniae (Fig. 5, Supplementary Fig. 9, Supplementary Notes 3 and 4). These observations highlight the feasibility and also challenges in implementing the optogenetic system in bacterial species other than E. coli, as specific codon optimization, cytoplasmic folding environment, and cell physiology need to be carefully considered on a species-by-species and need-specific basis.
In mammalian cells the CRY2-CIBN system has already been employed to optogenetically control cellular pathways including Cre recombination71, Raf/M.E.K./Erk23, TGF-β72, NF-\({\kappa }\)B15, Wnt/β-catenin11, and RhoA11. Most of these systems involve the binding of two proteins to initiate a signaling cascade. In bacteria, we envision that the CRY2-CIBN system can be similarly applied to a large number of processes to enable optogenetic control. For example, most of the two-component signaling (T.C.S.) systems73 in bacteria employ a histidine kinase that, upon stimulation, will recruit and phosphorylate its cognate response regulator activating downstream signaling73. These protein pairs could be tagged and controlled similarly to eukaryotes to allow for pathway regulation in real time. Furthermore, a number of pre-existing chemically inducible systems could be transformed into optogenetic systems by modifying their dimerization platforms. Potential candidate systems could include split ClpXP adapters for optical control of protein degradation7, split T7 RNAP for optical control of gene expression24 and split Cas9 for optical control of gene editing75.
Methods
E. coli bacterial strains, plasmids, and growth conditions
All E. coli strains used for the cell pole recruitment, Z-ring recruitment, and cytokinesis inhibition experiments were derivatives of BW25113 (CGSC #7636) strain and were grown in M9+ Glucose minimal media (recipe is provided in Supplementary Table 4). The DNA recruitment background strain was previously constructed by the Sherratt Lab30 (CGSC #12294) and was grown in EZ Rich Defined Media (EZRDM, Teknova Bio.) as described in Supplementary Table 4. Antibiotic requirements are specified in Supplementary Table 4 and the corresponding concentrations for each were chloramphenicol (150 μg/mL), streptomycin (50 μg/mL) and gentamycin (5 μg/mL). Inducer concentrations for respective strains and plasmid combinations are outlined in Supplementary Table 4 and described in more detail below. All bacterial cultures were grown in tubes covered in foil to minimize contact with ambient light. Plasmids expressing exogenous CRY2-CIBN fusion proteins in E. coli with their associated sequences are described in Supplementary Data 1.
Recruitment of CRY2-mCherry to the DNA via TetR-CIBN in E. coli
Cultures were started from a single colony of the strain RM187 and grown in 6 mL EZ Rich Defined Medium (EZRDM, Teknova Bio.) containing chloramphenicol and gentamycin at 25 °C with shaking until the cells entered early log-phase (OD600 between 0.1 and 0.2, ~19 h). When the culture entered early log-phase the cells were induced with 60 μM IPTG and supplemented with 240 nM anhydrous-tetracycline (ATC) to avoid a replication block induced by binding of TetR-CIBN similarly as previously described. The cells were induced for 2 h at 25 °C with shaking. At the end of the induction period the cells were centrifuged for 10 minutes at 1200 × g and the pellet was resuspended in 6 mL of EZRDM containing chloramphenicol and 240 nM ATC and grown for an additional 30 minutes at 25 °C while shaking after which it was, again, centrifuged for 10 minutes at 1200 × g and resuspended in 6 mL EZRDM containing chloramphenicol and gentamycin and allowed to grow an additional 2.5 h at 25 °C while shaking to allow the mCherry fluorophore to mature. Following the outgrowth period, the cells were prepared for imaging by centrifuging 1 mL of the culture at 11,000 × g for 1 minute and then resuspending the pellet in 50 µL of fresh EZRDM. Following this, 0.5 µL of the resuspension was then placed on a gel pad and the pad and prepared for imaging as specified in the “Live cell imaging” section below.
Recruitment of CRY2-mCherry to the cell pole via CIBN-GFP-PopZ and the Z-ring via ZapA-CIBN in E. coli
Cultures were started from a single colony of the strain of interest and grown in M9+ Glucose minimal medium containing chloramphenicol and streptomycin at 37 °C overnight until the cells reached stationary phase. The overnight culture was then diluted 1:100 in fresh of M9+ Glucose containing chloramphenicol and streptomycin and was allowed to grow at 37 °C until it reached log-phase (OD600 between 0.1 and 0.2, ~3 h) at which point the culture was induced with 0.4% arabinose and either 40 μM IPTG (ZapA fusion) or 100 μM IPTG (PopZ fusion). Following a 1-h induction at 37 °C the cells were prepared for imaging by centrifuging 1 mL of the culture at 11,000 × g for 1 minute and then resuspending the pellet in 70 µL of fresh M9+ Glucose. Following this, 0.5 µL of the resuspension was then placed on a gel pad and the pad and prepared for imaging as specified in the “Live cell imaging” section below. The cells on the gel pad were then assembled into the imaging chamber and allowed to equilibrate on the microscope in the dark for 2 h at ambient room temperature (RT).
Recruitment of CRY2-HaloTag to the cell pole via CIBN-GFP-PopZ using variable green (561 nm) light in E. coli
Cultures were started from a single colony of the strain of interest and grown in M9+ Glucose minimal medium containing chloramphenicol and streptomycin at 37 °C overnight until the cells reached stationary phase. The overnight culture was then diluted 1:100 in fresh of M9+ Glucose containing chloramphenicol and streptomycin and was allowed to grow at 37 °C until it reached log-phase (OD600 between 0.1 and 0.2, ~3 h) at which point the culture was induced. Following a 1-hour induction at 37 °C the cells were prepared for imaging by centrifuging 1 mL of the culture at 11,000 × g for 1 minute and then resuspending the pellet in 100 µL of fresh M9+ Glucose with 1 µM Janelia Fluor® 646 HaloTag® ligand (Promega Corp.) and allowed to incubate at RT for 2 h covered in foil. Following this, 0.5 µL of the resuspension was then placed on a gel pad and the pad and prepared for imaging as specified in the “Live cell imaging” section below.
Rapid Z-ring decondensation experiments in E. coli
Cultures of strain RM077 and RM078 were started from a single colony and grown in LB at 37 °C overnight until the cells reached stationary phase. The overnight LB culture was then diluted 1:200 in 3 mL of M9+ Glucose and grown overnight at room temperature (RT, 25 °C) until they reached log-phase (OD600 between 0.1 and 0.2) at which point the culture was induced with 10 µM IPTG and 0.2% arabinose. Following a 2-h induction at RT. Cells were prepared for imaging by centrifuging 1 mL of the culture at 11,000 × g for 1 minute and then resuspending the pellet in 100 µL of fresh M9+ Glucose. Following resuspension, 0.5 µL of the resuspension was transferred to a 3% M9+ Glucose agarose gel pad containing 10 µM IPTG and 0.2% arabinose for imaging.
Light induced inhibition of cytokinesis experiments in E. coli
Cultures were started from a single colony of the strain of interest (see Supplementary Table 4) and grown in LB containing chloramphenicol and/or streptomycin (depending on the strain background, see Supplementary Table 4) at 37 °C overnight until the cells reached stationary phase. The overnight culture was then diluted 1:100 in fresh of M9+ Glucose containing chloramphenicol and streptomycin and was grown overnight at 25 °C until it reached log-phase (OD600 between 0.1 and 0.2) at which point the culture was induced. Following a 2-h induction at 25 °C the cells were washed into fresh M9+ Glucose media lacking inducer and allowed to outgrow at 24 °C for 2 h. After the outgrowth, the cells were prepared for imaging by centrifuging 1 mL of the culture at 11,000 × g for 1 minute and then resuspending the pellet in 40 µL of fresh M9+ Glucose. Following this, 0.5 µL of the resuspension was then placed on a gel pad and the pad and prepared for imaging as specified in the “Live cell imaging” section below. Control cells were placed in a chamber, wrapped in foil, left at RT in next to microscope for the duration of the experiment and imaged after the experiment ended.
B. subtilis culturing, sample preparation, and imaging
Cultures were started from a single colony of the strain SC757 grown on LB plates at 30 °C for 16–20 h. We note all the following culturing, sample preparation, and induction steps occurred in the dark to avoid pre-activation caused by ambient light.
For imaging in M9+ media (glucose minimal media with vitamins, see Supplementary Table 4), single colonies from fresh plates were then inoculated into M9+ with 100 µM IPTG to induce CRY2-tdTomato for 6–7 h at 30 °C shaking. When the culture entered early log phase between OD600 0.1 and 0.3, 500 μL of the cultures were harvested by centrifuging for 5 minutes at 1652 × g and the pellet resuspended in 50 μL of M9+ by vortexing. 1 µL of the resuspended culture was spotted on 3% (w/v) M9+ agarose pad supplemented with 100 µM IPTG. The sample was allowed to equilibrate at the imaging temperature of 25 °C for 1 h prior to imaging.
For imaging in B. subtilis casein hydrolysate (CH) media76 (B. subtilis rich media commonly used for imaging77), single colonies from fresh plates were inoculated into CH media with 50 µM IPTG to induce CRY2-tdTomato for 16–20 h at 25 °C shaking in 10-fold dilution series. 500 µL of the cultures in early log phase between OD600 0.1 and 0.3, were harvested by centrifuging for 5 minutes at 1652 × g and the pellet resuspended in 50 µL of CH media by vortexing. 1 µL of the resuspended culture was spotted on 3% (w/v) CH agarose pad supplemented with 50 µM IPTG. The sample was allowed to equilibrate at the imaging temperature of 25 °C for 1 h prior to imaging.
To image the cells, cells were placed into focus using the 561 nm imaging laser that was also used for image acquisition at 1.63 W/cm2 measured 1 cm from the objective. The 488 nm laser power used to activate CRY2-CIBN system was 1.28 W/cm2 measured 1 cm from the objective in an area 200 × 200 pixels (160 nm/pixel). Every 3 seconds, cells were pulsed with blue light for 500 ms to activate the system followed by 50 ms green light to acquire image for a total of 160 frames (8 min).
C. crescentus culturing, sample preparation, and imaging
Cultures were started from single colony of EG3979 and EG3988 grown on PYE supplemented with both 5 µg/mL gentamycin and 25 µg/mL kanamycin or only kanamycin, respectively, at 30 °C for 48–72 h. Colonies from fresh plates were then inoculated into PYE supplemented with appropriate antibiotics (1 µg/mL gentamycin and/or 5 µg/mL kanamycin) and grown overnight to an OD600 of ~0.3–0.6. Cultures were diluted to an OD600 of ~0.08 in 2 mL of appropriate media supplemented with 0.3% xylose and 0.5 mM vanillate for EG3979 or just xylose for EG3988 and induced for 2 h. Control samples were left growing in appropriate media lacking inducers. All culture tubes were protected from ambient light.
500 µL of cells were concentrated 10X by spinning for 5 minutes at 1652 × g, removing 450 µL of media, and resuspending in left over 50 µL, 1 µL was spotted on PYE agarose pad containing gentamycin and kanamycin for EG3979 or just kanamycin for EG3988. Samples were allowed to equilibrate at the imaging temperature of 25 °C for 30 minutes prior to image acquisition at 25 °C.
Live cell imaging
Live cell imaging was performed on a custom-built optical setup routed to an Olympus IX71 inverted microscope with a 100 × 1.49 NA oil-immersion objective (Olympus Inc.). The light was focused onto the chip of an EMCCD camera (iXon Ultra 897, Andor Technology) with a final pixel size of 160 × 160 nm. The imaging focal plane was controlled by a piezo-driven stage (ASI, Eugene, OR). The EMCCD camera, lasers and shutters were controlled by MetamorphTM software (Molecular Devices).
Excitation light was provided by solid state 488 nm (Coherent OBIS), 561 nm (Toptica Photonics) or 647 nm (Coherent OBIS) lasers. The fluorescence emission signal was collected using either a ZET 488/561 nm (Chroma Technology) or ZET 488/561/647 nm (Chroma Technology) dual-band dichroic depending on the imaging conditions used. For two-color experiments, the emission light from the RFP and GFP channels were split using a 525/50 (Chroma Technology) and 650/50 (Chroma Technology) filter set mounted inside of an Optosplit II beam-splitter system (Cairn Research) prior to focusing on the EMCCD camera chip. The 488 nm laser power used to activate the CRY2-CIBN system was the same as what was used to image GFP fusions at 8.08 W/cm2 measured 1 cm from the objective for PopZ and ZapA experiments and 80.8 W/cm2 measured 1 cm from the objective for the TetR experiments measured using a Thor Labs Inc. Power Meter). The 561 nm laser power used to image the mCherry fusions was 8.71 W/cm2 measured 1 cm from the objective for the PopZ and ZapA experiments and 93.3 W/cm2 measured 1 cm from the objective for the TetR experiments unless otherwise noted in the text.
For two-color experiments using the 647 nm laser, the emission light from the RFP and GFP channels were split using a 556 nm long-pass filter (T556lpxr, Chroma Technology) in the Optosplitter followed by 700/75 (Chroma Technology) filter for the RFP emission light prior to focusing on the EMCCD camera chip.
To avoid activation of the CRY2-CIBN system by the LED bright-field lamp (LDB100F System, Prior Scientific Inc.), we placed a 715 nm long-pass filter (RG715, Thor Labs Inc.) or a 665 nm long-pass filter (ET665lp, Chroma Technology) filter after the condenser and the BF lamp intensity was adjusted such that the measured 488 nm light after the filter was measured to be ~200 nW at 1 cm from the sample plane.
Gel pads used to support the living bacteria cells were prepared by melting 3% w/v solution of agarose powder (SeaPlaqueTM GTGTM, Lonza Scientific) and either EZRDM or M9+ Glucose for 0.5–1 h at 70 °C. The melted gel was then transferred to a gasketed, cover-glass (FCS2, Bioptechs Inc.) and allowed to polymerize while sandwiched between the cover-glass and a cleaned coverslip (40 CIR-1, VWR Inc.) for 4 h at 25 °C (room temperature). 0.5 μL of the cells to be imaged were placed on the gel pad and allowed to dry ( ~ 3 minutes) before a fresh coverslip was placed on top and the imaging chamber (FCS2, Bioptechs) was assembled.
Generally, the blue light intensities and pulse sequences used to activate CRY2-CIBN in various imaging configurations were chosen to minimize phototoxicity of cells to blue light while maximizing the amount of activated CRY2. Whenever possible, we tried to minimize both the intensity and the number of blue pulses we delivered over the course of an acquisition. However, if the observed level of complex formation was not sufficient under a particular blue light induction scheme, we increased the number of pulses and or the intensity of blue light used to activate the system accordingly.
For the DNA recruitment experiments, the cells were irradiated with 30 ms pulses of 488 nm and 561 nm light delivered consecutively every 5 seconds using a 30 ms exposure time for each.
For the cell pole and Z-ring recruitment experiments the cells were first imaged for 2.5 seconds with 561 nm light after which they were irradiated with 488 nm light for 100 ms and then imaged with 56 nm light by streaming for 15 seconds using a 50 ms exposure time.
For the cell pole recruitment experiments where the green (561 nm) light was varied: the cells were first imaged for 2.5 seconds with both 647 nm and 561 nm light after which they were irradiated with 488 nm light for 100 ms and then imaged with 647 nm and 561 nm light by streaming for 15 seconds using a 50 ms exposure time.
For the fast Z-ring decondensation experiments, cells were initially imaged with 561 nm light after which they were irradiated with 50 ms pulses of 488 nm light delivered consecutively every 10 seconds for a 5-minute period. The cells were then imaged again with 561 nm light at the end of the 5-minute period.
For the Light-induced Inhibition of Cytokinesis (LInC) experiments, cells were irradiated with alternating 100 ms pulses of 488 nm and 561 nm every five minutes for a period of 12 h.
Quantifying the percentage increase in the DNA recruitment experiments in E. coli
To quantitatively characterize the accumulation of CRY2-mCherry at chromosomal sites, we calculated the increase in the Weber contrast78 of single cells. This calculation was used because chromosomal DNA spots were diffusive in both the lateral and azimuthal directions (relative to the z-axis of our objective) over the course of the experiments. This type of movement made it difficult to track individual spots and accurately estimate their intensities using the method we applied to our cell pole and midcell platforms (see above). Since the accumulation of CRY2-mCherry from a uniform distribution to a single concentrated location inside the cell body is equivalent to an increase in the Weber Contrast of a single cell image, we estimated the accumulation using the following method. First, we segmented the cells with observable spots in the last frame of the time lapse movie. There are normally 1 or 2 spots in a segmented cell region. Then we subtracted the background intensity of each image using ImageJ79. Finally, the normalized contrast for a single cell, j, in frame i is defined as:
Where \({C}_{{norm}}(i,j)\) reflects the extended accumulation of CRY2-mCherry in the DNA spot and \({I}_{\max i,j}\) and \({I}_{\min i,j}\) are the intensity of the brightest and darkest pixels in the cells, respectively. The contrast is normalized by the mean intensity, 〈Ii,j〉, of all the pixels in this region considering cell-to-cell variations in expression levels. Figures 2d and e were calculated using this method with the y-axis in Fig. 2e converted to percent increase \((\%{Inc})\) by:
for ease of comparison with other experiments.
Quantifying the RFP and/or GFP signal in cell pole and Z-ring recruitment experiments
A variation on a previously published custom MATLAB script80 used to quantify FRAP data was created to measure the GFP and RFP signals in the live-cell recruitment experiments. Briefly, cells were manually selected and cropped using a maximum-intensity projection image of the entire recruitment stream (created in ImageJ) to avoid any selection bias. The max intensity image was also used to manually crop both the total cell area as well as the recruitment site area (either Z-ring or cell pole). For the Z-ring experiments, the selected area was cropped manually using a rectangular selection. For the cell pole experiments, the PopZ region was cropped using a fixed circle with a 2-pixel radius. To calculate the percent increase at target loci, we first calculated the fraction \({Frac}\left(t\right)\) of the fluorescence intensity \({I}_{{target}}\left(t\right)\) at the target site at a given time t versus that of the whole cell \({I}_{{cell}}\left(t\right)\):
We then converted this fraction to percent increase \((\%{Inc})\) by:
The percentage increase curves of all cells were then averaged to obtain the mean and the associated standard error.
We noticed that our experimental setup introduced a slight variation in the time between activation with blue light and acquisition with either green or far-red light. This variation was because in some experiments (Figs. 2i and 3d), the CRY2-CIBN association kinetics were too fast that the microscope hardware speed of switching blue-light activation to green-light was not fast enough to capture the true time 0 after switching, and hence the first datapoint did not start at true time 0. To account for this delay, we created a custom MATLAB script to determine the true time delay between activation and acquisition frames using the imaging metadata obtained by Metamorph. We then used the actual time of switching when aggregating data and/or making measurements. Therefore, some of the time traces did not start at precisely time 0.
Quantifying the Halo signal in cell pole dissociation experiments in E. coli
Individual well-isolated cells were cropped from whole field of view time-course image stacks starting 5 minutes after CRY2-CIBN association was activated using blue light. The individual cell image stacks were then registered in x-y space using the ImageJ StackReg81 plug-in to minimize the shifting of the cells over long timescales due to growth. The depletion of cell pole foci was then measured using the Weber contrast-based methodology employed for the TetR recruitment platform described above. The resulting traces for individual cells were averaged together and the exponential decay curve was fit to
to extract the dissociation time constant (τoff).
Quantification of Z-ring decondensation in E. coli
Cells were segmented using the MiSiC deep learning-based cell segmentation algorithm82. After segmentation, the resulting cell masks were fed into MicrobeJ83 to quantify the mean ZapA-mCherry intensity at across the long axis of the cell and cell lengths. All cells were visually inspected to remove any cells that lacked a Z-ring at the start of the experiment. The midcell localization fraction of the Z-ring was measured by dividing the integrated ZapA-mCherry fluorescence intensity within a 3-pixel window (480 nm width) about the midcell maximum (approximate Z-ring position) by the total integrated ZapA-mCherry fluorescence intensity of the whole cell both before and after a 5-minute blue light induction. The change in the midcell localization fraction before and after blue light induction was calculated, normalized by the midcell fraction before induction and multiplied by a factor of 100 to obtain the percent change in Z-ring intensity at midcell. The percentage of cells that both harbored a Z-ring prior to induction with blue light and showed a reduction in the midcell fraction of ZapA-mCherry intensity at midcell greater than 4% were deemed as “decondensed”. This decondensation threshold was applied to every cell in each experimental population to determine the fraction of cell that underwent significant decondensation in each experimental condition. Bootstrapping was employed to estimate the standard error of the mean. Briefly, a subpopulation of cells (500 for the induced cells and 300 for the uninduced cells) were randomly sampled and used to calculate the percentage of cells that underwent decondensation. This calculation was repeated 100 times to obtain the standard error of the mean.
Quantification of inhibition of cytokinesis in E. coli
Cells were manually tracked throughout the duration of the experiment to determine if and when a successful division occurred. Cells were cropped and the average midcell intensity across the long-axis of the cells (long-axis projection) was calculated using a custom MATLAB script.
Quantification of maximal enrichment and reduction of CRY2-mCherry and CRY2-HaloTag
In order to reduce the effects of noise on the maximal fold enrichment and maximal fold reduction of CRY2-HaloTag signal at the cell poles (i.e., estimation of the plateau value), the final plateau value was calculated by taking the mean of the last ten datapoints of individual cell trajectories.
Statistics and reproducibility
All experimental biological replicates (N) were noted in the main text along with the number of data points used in the analysis where applicable. All error bars and significance values were calculated using MATLAB by a statistical test specified in the figure legend. p-values < 0.05 were accepted as statistically significant. The different significance levels indicated as stars in figures correspond to *p value < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 or n.s. where no significant difference was observed.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets generated during and/or analyzed during the current study are provided as a Source Data file. Source data are provided with this paper.
References
Surovtsev, I. V. & Jacobs-Wagner, C. Subcellular organization: a critical feature of bacterial cell replication. Cell 172, 1271–1293 (2018).
Thanbichler, M. & Shapiro, L. Getting organized — how bacterial cells move proteins and DNA. Nat. Rev. Microbiol 6, 28–40 (2008).
Kolar, K., Knobloch, C., Stork, H., Žnidarič, M. & Weber, W. OptoBase: a web platform for molecular optogenetics. Acs Synth. Biol. 7, 1825–1828 (2018).
Inobe, T. & Nukina, N. Rapamycin-induced oligomer formation system of FRB–FKBP fusion proteins. J. Biosci. Bioeng. 122, 40–46 (2016).
Farrar, M. A., Olson, S. H. & Perlmutter, R. M. [31] Coumermycin-induced dimerization of GyrB-containing fusion proteins. Methods Enzymol. 327, 421–IN5 (2000).
Miyamoto, T. et al. Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nat. Chem. Biol. 8, 465–470 (2012).
Davis, J. H., Baker, T. A. & Sauer, R. T. Small-molecule control of protein degradation using split adaptors. Acs Chem. Biol. 6, 1205–1213 (2011).
Voß, S., Klewer, L. & Wu, Y.-W. Chemically induced dimerization: reversible and spatiotemporal control of protein function in cells. Curr. Opin. Chem. Biol. 28, 194–201 (2015).
Losi, A., Gardner, K. H. & Möglich, A. Blue-light receptors for optogenetics. Chem. Rev. 118, 10659–10709 (2018).
Goglia, A. G. & Toettcher, J. E. A bright future: optogenetics to dissect the spatiotemporal control of cell behavior. Curr. Opin. Chem. Biol. 48, 106–113 (2018).
Bugaj, L. J., Choksi, A. T., Mesuda, C. K., Kane, R. S. & Schaffer, D. V. Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252 (2013).
Taslimi, A. et al. An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5, 4925 (2014).
Kawano, F., Suzuki, H., Furuya, A. & Sato, M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun. 6, 6256 (2015).
Guntas, G. et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl Acad. Sci. 112, 112–117 (2015).
DeFelice, M. M. et al. NF-κB signaling dynamics is controlled by a dose-sensing autoregulatory loop. Sci. Signal 12, eaau3568 (2019).
Benedetti, L. et al. Light-activated protein interaction with high spatial subcellular confinement. Proc. Natl Acad. Sci. 115, 201713845 (2018).
Hallett, R. A., Zimmerman, S. P., Yumerefendi, H., Bear, J. E. & Kuhlman, B. Correlating in vitro and in vivo activities of light-inducible dimers: a cellular optogenetics guide. Acs Synth. Biol. 5, 53–64 (2016).
Xiao, J. & Dufrêne, Y. F. Optical and force nanoscopy in microbiology. Nat. Microbiol 1, 16186 (2016).
Kennedy, M. J. et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 (2010).
Shao, K. et al. The oligomeric structures of plant cryptochromes. Nat. Struct. Mol. Biol. 27, 480–488 (2020).
Sancar, A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103, 2203–2238 (2003).
Liu, H. et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in arabidopsis. Science 322, 1535–1539 (2008).
Duan, L. et al. Understanding CRY2 interactions for optical control of intracellular signaling. Nat. Commun. 8, 547 (2017).
Pu, J., Zinkus-Boltz, J. & Dickinson, B. C. Evolution of a split RNA polymerase as a versatile biosensor platform. Nat. Chem. Biol. 13, 432–438 (2017).
Yamada, M., Suzuki, Y., Nagasaki, S. C., Okuno, H. & Imayoshi, I. Light control of the tet gene expression system in mammalian cells. Cell Rep. 25, 487–500.e6 (2018).
Castillo-Hair, S. M., Baerman, E. A., Fujita, M., Igoshin, O. A. & Tabor, J. J. Optogenetic control of Bacillus subtilis gene expression. Nat. Commun. 10, 3099 (2019).
Jayaraman, P. et al. Blue light-mediated transcriptional activation and repression of gene expression in bacteria. Nucleic Acids Res 44, 6994–7005 (2016).
Olson, E. J., Hartsough, L. A., Landry, B. P., Shroff, R. & Tabor, J. J. Characterizing bacterial gene circuit dynamics with optically programmed gene expression signals. Nat. Methods 11, 449–455 (2014).
Hensel, Z., Weng, X., Lagda, A. C. & Xiao, J. Transcription-factor-mediated dna looping probed by high-resolution, single-molecule imaging in live E. coli cells. Plos Biol. 11, e1001591 (2013).
Lau, I. F. et al. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol 49, 731–743 (2003).
Ramos, J. L. et al. The TetR family of transcriptional repressors. Microbiol Mol. Biol. R. 69, 326–356 (2005).
Lasker, K. et al. Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nat Microbiol 1–12. https://doi.org/10.1038/s41564-019-0647-7 (2020).
Molinari, S. et al. A synthetic system for asymmetric cell division in Escherichia coli. Nat. Chem. Biol. 15, 917–924 (2019).
Mushnikov, N. V., Fomicheva, A., Gomelsky, M. & Bowman, G. R. Inducible asymmetric cell division and cell differentiation in a bacterium. Nat. Chem. Biol. 15, 925–931 (2019).
Lim, H. C. & Bernhardt, T. G. A PopZ‐linked apical recruitment assay for studying protein–protein interactions in the bacterial cell envelope. Mol. Microbiol 112, 1757–1768 (2019).
Laloux, G. & Jacobs-Wagner, C. Spatiotemporal control of PopZ localization through cell cycle–coupled multimerization. J. Cell Biol. 201, 827–841 (2013).
Bowman, G. R. et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134, 945–955 (2008).
Ebersbach, G., Briegel, A., Jensen, G. J. & Jacobs-Wagner, C. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134, 956–968 (2008).
McQuillen, R. & Xiao, J. Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu Rev. Biophys. 49, 1–33 (2020).
Macnab, R. M. How bacteria assemble flagella. Annu Rev. Microbiol 57, 77–100 (2003).
Sourjik, V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol 12, 569–576 (2004).
Katsura, Y. et al. An optogenetic system for interrogating the temporal dynamics of Akt. Sci. Rep. -uk 5, 14589 (2015).
Mühlhäuser, W. W. D., Hörner, M., Weber, W. & Radziwill, G. Light-regulated protein kinases based on the CRY2-CIB1 System. Methods Mol. Biol. Clifton N. J. 1596, 257–270 (2017).
Szeto, T. H., Rowland, S. L., Habrukowich, C. L. & King, G. F. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J. Biol. Chem. 278, 40050–40056 (2003).
Che, D. L., Duan, L., Zhang, K. & Cui, B. The dual characteristics of light-induced cryptochrome 2, homo-oligomerization and heterodimerization, for optogenetic manipulation in mammalian cells. Acs Synth. Biol. 4, 1124–1135 (2015).
McQuillen, R. J. et al. Optogenetic Control of Protein Organization in Living Escherichia coli Cells. (Johns Hopkins University, 2022).
Goehring, N. W. & Beckwith, J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15, R514–R526 (2005).
Lutkenhaus, J. & Du, S. E. coli Cell Cycle Machinery. in Prokaryotic Cytoskeletons. 84, 27–65 (Springer, Cham, 2013).
Yang, X. et al. A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat. Microbiol. https://doi.org/10.1038/s41564-020-00853-0 (2021).
McCausland, J. W. et al. Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism. Nat. Comms. https://doi.org/10.1038/s41467-020-20873-y (2021).
McQuillen, R. & Xiao, J. Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-Ring. Annu Rev. Biophys. 49, 309–341 (2020).
Goehring, N. W., Gueiros-Filho, F. & Beckwith, J. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Gene Dev. 19, 127–137 (2005).
Galli, E. & Gerdes, K. FtsZ-ZapA-ZapB interactome of Escherichia coli. J. Bacteriol. 194, 292–302 (2012).
Buss, J. et al. In vivo organization of the FtsZ‐ring by ZapA and ZapB revealed by quantitative super‐resolution microscopy. Mol. Microbiol 89, 1099–1120 (2013).
Fu, G. et al. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). Plos One 5, e12680 (2010).
Buss, J. et al. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. Plos Genet 11, e1005128 (2015).
Rowlett, V. W. & Margolin, W. The bacterial min system. Curr. Biol. 23, R553–R556 (2013).
Hu, Z. & Lutkenhaus, J. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 182, 3965–3971 (2000).
Justice, S. S., García‐Lara, J. & Rothfield, L. I. Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol. Microbiol 37, 410–423 (2000).
Romberg, L. & Levin, P. A. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu Rev. Microbiol 57, 125–154 (2003).
Coltharp, C., Buss, J., Plumer, T. M. & Xiao, J. Defining the rate-limiting processes of bacterial cytokinesis. Proc. Natl Acad. Sci. 113, E1044–E1053 (2016).
Lan, G., Daniels, B. R., Dobrowsky, T. M., Wirtz, D. & Sun, S. X. Condensation of FtsZ filaments can drive bacterial cell division. Proc. Natl Acad. Sci. 106, 121–126 (2009).
Liu, B., Liu, H., Zhong, D. & Lin, C. Searching for a photocycle of the cryptochrome photoreceptors. Curr. Opin. Plant Biol. 13, 578–586 (2010).
Bouly, J.-P. et al. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J. Biol. Chem. 282, 9383–9391 (2007).
Banerjee, R. et al. The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J. Biol. Chem. 282, 14916–14922 (2007).
Los, G. V. et al. Halotag: a novel protein labeling technology for cell imaging and protein analysis. Acs Chem. Biol. 3, 373–382 (2008).
Grimm, J. B., Brown, T. A., English, B. P., Lionnet, T. & Lavis, L. D. Synthesis of Janelia fluor halotag and SNAP-tag ligands and their use in cellular imaging experiments. 1663, 179–188 (2017).
Gueiros-Filho, F. J. & Losick, R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Gene Dev. 16, 2544–2556 (2002).
Skerker, J. M. & Laub, M. T. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2, 325–337 (2004).
Tsokos, C. G. & Laub, M. T. Polarity and cell fate asymmetry in caulobacter crescentus. Curr. Opin. Microbiol. 15, 744–750 (2012).
Barrows, J. M. & Goley, E. D. Synchronized swarmers and sticky stalks: caulobacter crescentus as a model for bacterial cell biology. J. Bacteriol. 205, e00384–22 (2023).
Lindner, F. & Diepold, A. Optogenetics in bacteria – applications and opportunities. Fems Microbiol Rev. 46, fuab055 (2021).
Repina, N. A., Rosenbloom, A., Mukherjee, A., Schaffer, D. V. & Kane, R. S. At light speed: advances in optogenetic systems for regulating cell signaling and behavior. Annu Rev. Chem. Biomol. 8, 13–39 (2017).
Tischer, D. & Weiner, O. D. Illuminating cell signalling with optogenetic tools. Nat. Rev. Mol. Cell Bio 15, 551–558 (2014).
Boekel, J. et al. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33, 139–142 (2015).
Sterlini, J. M. & Mandelstam, J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113, 29–37 (1969).
Kim, E. Y., Tyndall, E. R., Huang, K. C., Tian, F. & Ramamurthi, K. S. Dash-and-recruit mechanism drives membrane curvature recognition by the small bacterial protein SpoVM. Cell Syst. 5, 518–526.e3 (2017).
Pelli, D. G. & Bex, P. Measuring contrast sensitivity. Vis. Res. 90, 10–14 (2013).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Yang, X. et al. GTPase activity–coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 (2017).
Thevenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. Ieee T Image Process 7, 27–41 (2021).
Panigrahi, S. et al. MiSiC, a general deep learning-based method for the high-throughput cell segmentation of complex bacterial communities. eLife. https://doi.org/10.7554/eLife.65151 (2021).
Ducret, A., Quardokus, E. M. & Brun, Y. V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol 1, 16077 (2016).
Acknowledgements
We would like to thank T. Bernhardt and J. Buss for the BW25113 (zapA-mCherry) strain. Additionally, we would like to thank N. Yehya for helpful insight and discussions. This work was supported by NIH R01 GM086447, NIH R35 GM136436 (to J.X.), NIH T32 GM008403 (to R.M.), NIH F32 GM150262 (to A.J.P.), NIH R35 GM131767 to (M.E.W.), NIH R35 GM136221 (to E.D.G.), NIH T32 GM144272 (to E.L.S.). K.S.R. was funded by the Intramural Research Program of the NIH, the National Cancer Institute, the Center for Cancer Research.
Author information
Authors and Affiliations
Contributions
R.M. constructed and characterized all the E. coli strains and fusions and proposed and carried out all E. coli experiments as well as quantified and interpreted all E. coli data unless otherwise noted. C.H.B. assisted in developing methodologies for fold enrichment calculations with R.M. and J.X and assisted in analysis of CRY2-CIBN green light effects. X.Y. developed the Normalized Contrast method used to analyze the DNA localization data and performed the data analysis using this method. J.W.M., together with R.M., designed strains for the light induced inhibition of cytokinesis assay. K.E.B. and A.J.P. performed experiments in S. pneumoniae while H.C.T.T. and Y.M.H. constructed strains with oversight by M.E.W. on experimental design. E.L.S. constructed strains in C. crescentus while E.L.S. and A.J.P. performed experiments with oversight by E.D.G. on experimental design. S.C. constructed B. subtilis strains while A.J.P. performed experiments with oversight by K.S.R. on experimental design. A.J.P. and R.M. performed data analysis on data acquired from B. subtilis or C. crescentus experiments. J.X. planned and directed the project and, together with R.M. and A.J.P., designed experiments, interpreted the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial or non-financial interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
McQuillen, R., Perez, A.J., Yang, X. et al. Light-dependent modulation of protein localization and function in living bacteria cells. Nat Commun 15, 10746 (2024). https://doi.org/10.1038/s41467-024-54974-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54974-9