Abstract
Parainfluenza virus 3 (PIV3) infection poses a substantial risk to vulnerable groups including infants, the elderly, and immunocompromised individuals, and lacks effective treatments or vaccines. This study focuses on targeting the hemagglutinin-neuraminidase (HN) protein, a structural glycoprotein of PIV3 critical for viral infection and egress. With the objective of targeting these activities of HN, we identified eight neutralizing human monoclonal antibodies (mAbs) with potent effects on viral neutralization, cell-cell fusion inhibition, and complement deposition. Three epitopes on PIV3 HN were delineated and one epitope, Site 2, elicits a mAb with cross-neutralizing ability against PIV1 and PIV3. Cryo-EM revealed the cross-neutralizing mAb utilizes a long CDR3 loop to bind inside the pocket of the sialic acid binding site. Additionally, we resolved the structure of a non-protective mAb binding to Site 1 near the HN:F-interaction site. The potent Site 2-directed mAb demonstrated clinical efficacy in hamsters, reducing viral replication prophylactically and therapeutically. These findings advance our understanding of PIV3 immunity and underscore the significance of targeting HN for clinical therapeutic development against PIV3.
Similar content being viewed by others
Introduction
Human parainfluenza viruses (PIVs) are clinically prevalent viruses consisting of four types circulating globally each year1,2. Although viral isolates have been detected in people of all ages, PIVs primarily cause severe disease in infants and young children, the elderly, the immunocompromised, and those with preexisting comorbidities, such as lung transplants or asthma3,4,5. With clinical manifestations ranging from mild upper respiratory distress to bronchitis and pneumonia, PIV infection can lead to hospitalization and death in these high-risk populations6. PIV type 3 (PIV3) is the second leading cause of bronchitis and pneumonia in children after respiratory syncytial virus (RSV)7,8. Enhanced respiratory disease caused by PIV3 infection typically afflicts young children during their first 2 years of life. Nearly 60% of children show serological evidence for PIV3 infection before the age of 5, with reinfection occurring throughout life3,8,9,10. Prior to the COVID-19 pandemic, PIV3 accounted for approximately 60% of detected PIVs; however, post-pandemic PIV3 detection increased to approximately 99%, subsequently increasing its pervasiveness and the risk for advanced respiratory disease hospitalizations11,12. With no approved therapy or vaccine, PIV3 continues to spread and cause severe respiratory complications in susceptible populations.
PIV3 is a negative sense, single-stranded RNA virus in the family Paramyxoviridae. The genome encodes six viral proteins, with two surface glycoproteins initiating viral infection: the fusion (F) protein and the hemagglutinin-neuraminidase (HN) protein13,14. The HN protein attaches to the host cell by interacting with its cognate receptor, sialic acid, which in turn initiates F to refold, beginning the fusion process15,16,17,18,19,20. The PIV3 F protein has similar functions to other class I fusion proteins, in that it initiates fusion between the virion and the host cell membrane, subsequently allowing the virus to enter and infect the host cell15,16,17,18,19,20.
PIV3 HN is a type II membrane protein that exists as a covalently linked dimer composed of a stalk and head region16,21,22. In addition to activating the F protein for viral fusion, the head domain of PIV3 HN contains an active site necessary for both sialic acid binding and cleavage, making it necessary for viral entry as well as viral release, and therefore an excellent target for mAb development15,16,23,24. It is hypothesized that, following receptor binding to sialic acid, HN may activate F through a loop that extends into a hydrophobic pocket within the apex of F, lowering the activation barrier and triggering F to undergo conformational changes that fuse the viral and cell membranes together15,25. After viral replication is complete within the host cell cytoplasm and viral progeny is preparing to bud from the cell, HN cleaves sialic acid from carbohydrate chains, releasing viral progeny from the host cell26. These functions of HN create multiple targets for clinical intervention, with the potential to prevent receptor binding, viral fusion, or egress. While the F protein is essential for viral infection, the diverse activities of HN present opportunities for more robust treatments.
Isolation of PIV3 F-specific mAbs that neutralize PIV3 has identified several antigenic sites. For example, mAb PIA174 binds across all three protomers at the apex of PIV3 F, blocking HN interaction and subsequent fusion27,28. Recently, cross-binding and cross-neutralizing mAbs targeting PIV1 and PIV3 F proteins have been identified, namely mAb 3 × 1, which protects from PIV3 challenge in vivo29. PIV1 commonly causes croup in infants and young children, and serological evidence has shown it to be the second-most frequently circulating PIV2. PIV1 and PIV3 HN proteins share 49% sequence homology, which suggests the potential for cross-reactive mAb binding.
With PIV3 circulation persisting, the lack of treatments and vaccines against PIVs leaves the susceptible populations entirely vulnerable to infection and enhanced disease. Here, we have isolated and characterized human mAbs against the PIV3 HN protein. Using these mAbs, we have successfully mapped three distinct epitopes on the HN protein, assessed the functional characteristics of mAbs binding to each epitope including fusion inhibition and complement deposition, determined the structural features of two epitopes found on the PIV3 HN protein, and evaluated potent neutralizing mAbs as prophylactic and therapeutic treatments in vivo. One mAb decreased PIV3 infection in vivo and binds to the sialic acid active site on HN, presumably preventing binding and fusion of the virus. This mAb introduces promising insights into HN-targeted treatments and vaccines for the clinical realm.
Results
Eight neutralizing human mAbs were isolated against the head domain of the PIV3 HN protein
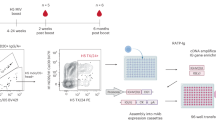
We isolated human peripheral blood mononuclear cells (PBMCs) from blood bank leukocyte reduction filters and sorted antigen-specific B cells using fluorescent streptavidin tetramers conjugated to the head region of the PIV3 HN protein (Supplementary Table S1). CD19+/IgA-/IgD-/IgM-/PIV3 HN+ B cells were single-cell sorted onto an irradiated NIH/3T3 cell feeder layer expressing human CD40L, a human B cell activating factor, and human interleukin-21 (hIL-21) with added CpG to stimulate B cell expansion and differentiation into antibody-secreting cells as previously described (Supplementary Figs. S1, S2)30,31. The antibody-containing supernatant was evaluated for positive binding to recombinant PIV3 HN protein by enzyme-linked immunosorbent assay (ELISA), and RNA from positive clones was extracted, sequenced, and cloned into an expression vector (Supplementary Table S2). Eight mAbs were recombinantly expressed and evaluated for binding to recombinant PIV3 HN head protein by ELISA (Fig. 1A and Supplementary Table S3). We also evaluated the neutralizing activity of each mAb against PIV3 using a plaque reduction neutralization assay (PRNT) (Fig. 1B). All eight mAbs displayed HN binding activity, with effective concentrations (EC50) less than 50 ng/mL (Fig. 1C). In addition, all mAbs were able to neutralize PIV3, with mAbs PIV3HN-03, PIV3HN-05, PIV3HN-09, PIV3HN-11, and PIV3HN-15 being the most potent, displaying inhibitory concentrations (IC50) lower than 100 ng/mL (Fig. 1C). Clinically available mAb therapies encompass mAbs with IC50 values close to 100 ng/mL, corroborating the clinical relevancy of the identified mAbs in this study32,33,34,35,36.
A ELISA binding curves data displays mAb binding with high affinity to the head domain of HN. Data points are the average of four technical replicates from one experiment and are representative of two biological replicates. Data are presented as mean values +/− SD. B Plaque reduction neutralization test curves for the eight mAbs. Data points are the average of three technical replicates from one experiment and are representative of two biological replicates. Data are presented as mean values +/− SD. C Summary of the half maximal effective concentration (EC50) values and half maximal inhibitory concentration (IC50) values calculated from (A) and (B). D PIV3-infected LLC-MK2 cells were stained with anti-PIV3 HN mAbs PIV3HN-05, and PIV3HN-09 conjugated to PE. Anti-F mAb PIA174 was used as a positive control, and a mAb against human metapneumovirus, MPV467, was used as a negative control. Both anti-HN mAbs and anti-F mAbs were detected in virally infected cells. Data represent the results from one technical replicate and are representative of two biological replicates. E Epitope binning was performed via biolayer interferometry (BLI) by loading His-tagged HN protein onto an anti-HIS sensor, and then associating the sensor with one mAb followed by a second mAb. Created in BioRender. Miller, R. (2024) https://BioRender.com/x68o050. F Three distinct epitopes on the PIV3 HN protein were discovered from the competitive binding to HN by the mAbs. Data indicate the binding of the second antibody to the sensor in the presence of the first antibody. Values in black boxes describe mAbs with high levels of competition (≤ 30), whereas values in white boxes describe mAbs that do not compete for binding (≥ 70).
To confirm our enrichment strategy of binding to soluble HN domains selected for antibodies capable of binding to full-length HN protein, we assessed the binding of two mAbs, PIV3HN-05 and PIV3HN-09, to PIV3-infected cells via flow cytometry. Following a 48 hr infection of LLC-MK2 cells with PIV3, cells were stained with phycoerythrin (PE)-conjugated mAbs, which were tested alongside the anti-PIV3 F positive control mAb PIA17428 and the anti-human metapneumovirus F negative control mAb MPV46730. mAbs PIV3HN-05, PIV3HN-09, and PIA174 stained the infected cells while mAb MPV467 did not (Fig. 1D). This supported our hypothesis that mAbs targeting HN head domains retain the ability to bind to full-length HN and can identify PIV3-infected cells similarly to mAbs targeting F.
Three distinct binding epitopes were discovered on the PIV3 HN protein
We next determined the binding epitope groups of the mAbs on the head domain of the PIV3 HN protein using competitive biolayer interferometry (BLI) (Fig. 1E). Hexahistidine-tagged PIV3 HN was loaded onto an anti-HIS sensor prior to associating with one mAb. Following an increase in binding, the sensor was associated with a second mAb and competition for the same epitope was identified from the lack of binding after the second mAb association. Three distinct epitopes on the PIV3 HN protein were mapped using this method, termed Sites 1–3 (Fig. 1F). Site 2 and Site 3 contained two mAbs that cross-competed, PIV3HN-03 and PIV3HN-04, suggesting these two epitopes are in proximity on the PIV3 HN protein. Site 1 contained the two least potent neutralizing mAbs, PIV3HN-12 and PIV3HN-13, while Site 2 contained the three most potent neutralizing mAbs, PIV3HN-03, PIV3HN-05, and PIV3HN-11, suggesting that Site 2 plays an important role in viral infection and replication. No correlations were found between epitope and V(D)J gene usage of mAbs (Supplementary Tables S4, S5).
PIV3 HN mAbs targeting all three epitopes can inhibit fusion
The PIV3 HN protein has several proposed functions, one of which is activating the F protein to initiate a conformational change from the pre-fusion to the post-fusion conformation after binding to sialic acid15,37,38. To determine the fusion-inhibiting activity of each mAb with respect to the three epitopes on the PIV3 HN protein, independent from receptor binding, Vero-SLAM cells were transfected with plasmids encoding the PIV3 F and PIV3 HN proteins along with green fluorescent protein (GFP). After treating with individual mAbs at decreasing concentrations, syncytia were visualized under a fluorescent cell imaging microscope. mAb PIA174 was used as a positive control since this mAb targets the PIV3 F protein, binds at the apex of the protein, and has been shown to inhibit fusion28. Half of the mAbs, including PIV3HN-04, PIV3HN-09, PIV3HN-11, and PIV3HN-13, successfully prevented syncytia formation at the highest concentration of 100 µg/mL, with several inhibiting fusion at a lower concentration of 10 µg/mL (Fig. 2). Fusion inhibition was not site-specific, but rather mAbs in each epitope group were able to inhibit fusion, while others in the same groups had no effect on syncytia formation. In addition, the fusion inhibition activity was not correlated to the neutralization potency of the mAbs (Fig. 1B, C).
Fusion inhibition of mAbs from each site was assessed by transfecting Vero-SLAM cells with either HN or F and GFP 3 h prior to adding mAb. Syncytia formation was visualized 24 h following transfection and fusion inhibition was evaluated by comparison to the PBS-treated control. Previously identified anti-F mAb PIA174 was used as a positive control. PBS-treated cells were used as a negative control to show standard levels of syncytia. The results shown are images from one technical replicate from one experiment and are representative of 3 biological replicates. The scale bar represents 100 µm.
mAbs binding to Site 2 on the PIV3 HN protein induce complement deposition
Although mAbs can prevent viruses from functioning by binding to a functional epitope via the fragment antigen binding (Fab) region, they can also recruit complement proteins in circulation to bind to the Fc region via the classical pathway39,40. Complement deposition can lead to the formation of the membrane attack complex or can recruit innate cells to kill the infected cell through phagocytosis or cytotoxicity (Fig. 3A)41,42. To elucidate the efficacy of mAbs to each site in deposing complement, we stained PIV3 HN-conjugated fluorospheres with individual mAbs complexed with the guinea pig complement C3b protein and detected C3b signal using flow cytometry as previously reported43. Beads that had high levels of fluorescence indicated high levels of complement deposition when contrasted to the complement-only control. mAbs binding to Site 2 were highly effective in complement deposition compared to mAbs binding to Sites 1 and 3 (Fig. 3B). To corroborate these experiments, we sought to determine the neutralizing activity of each mAb in the presence of complement, as the previous neutralization assays were conducted without added complement. As expected, we detected decreases in IC50 values for mAbs binding to Site 2 and Site 3, but not those binding to Site 1 (Fig. 3C).
A Complement deposition was assessed due to its importance in aiding mAbs in preventing viral replication. Complement has several functions, including formation of the membrane attack complex (MAC), and initiation of cell phagocytosis or cellular cytotoxicity. Created in BioRender. Miller, R. (2024) https://BioRender.com/l30r012. B HN-coated beads were stained with individual mAbs before introducing the C3 complement protein. Deposition of the C3b protein to HN-coated beads stained with mAbs was measured by comparison to C3b deposition alone. Data points are the results of three technical replicates from one experiment, and the data are representative of two biological replicates. Data are presented as mean values +/− SD. Statistical significance was calculated using a one-sided ANOVA for multiple comparisons (**P < 0.0086). C PRNT was performed with added guinea pig complement at a 1:1000 dilution. All except two mAbs had increased neutralization potency with added complement, indicated by a decreased IC50 value. Each data point is the IC50 value from one neutralization experiment with or without complement, and the data are representative of two biological replicates. D One mAb from each epitope was evaluated for hemagglutination inhibition (HI) using guinea pig red blood cells. Data are the results from one experiment representative of two biological replicates. E One mAb, PIV3HN-05, cross-neutralized parainfluenza virus type 1 (PIV1) in vitro with an IC50 value of 180 ng/mL. The PIV1/3 cross-neutralizing mAb, 3 × 1, was used as a positive control. Data points are the average of three technical replicates from one experiment and are representative of two biological replicates. Data are presented as mean values +/− SD.
mAbs from all sites inhibit hemagglutination of guinea pig red blood cells
To determine if the mAbs can inhibit functional activity of the PIV3 HN protein, a hemagglutination inhibition (HI) assay was performed using guinea pig red blood cells (RBCs) by co-incubating serial dilutions of mAbs targeting each epitope (PIV3HN-05, PIV3HN-09, and PIV3HN-13) with 4 HA units of PIV3 before adding to guinea pig RBCs. After a 1, 2 h incubation, the minimal inhibitory concentration (MIC) of each mAb to inhibit hemagglutination activity was measured by the last dilution of mAb that displayed HI activity. All three mAbs displayed the ability to inhibit the HA activity of the PIV3 HN protein, suggesting that mAbs targeting multiple epitopes of the PIV3 HN protein are able to prevent virus attachment (Fig. 3D and Supplementary Fig. S3).
The Site 2-specific mAb PIVHN-05 cross-neutralizes PIV1
Parainfluenza virus type 1 (PIV1) commonly causes croup in young children and is the second most prevalent circulating parainfluenza virus after PIV33. The two viral seasons occur separately and encompass the calendar year, necessitating a need for a viral therapy that can target both viruses44. The HN protein from PIV3 and PIV1 share 49% sequence homology. One PIV1/3 cross-neutralizing mAb, 3 × 1, has been identified and targets the F protein29; however, given the similarity between the two PIV HN proteins, we sought to identify anti-HN mAbs that can cross-neutralize PIV1 and PIV3 viruses. We assessed the neutralizing activity of all eight mAbs against PIV1, and only mAb PIVHN-05 successfully neutralized PIV1 with an IC50 value of 180 ng/mL (Fig. 3D).
Site 1 mAb PIV3HN-13 binds near the dimer interface of the PIV3 HN protein
Based on the studies above, we determined that Site 2-targeting mAb PIV3HN-05 is highly neutralizing (IC50 < 50 ng/mL), induces complement deposition, and cross-neutralizes PIV1. In contrast, mAbs binding Site 1 (PIV3HN-12 and PIV3HN-13) are weakly neutralizing (IC50 > 300 ng/mL), inhibit fusion, and lack complement deposition activity. While these two epitopes are discrete on the PIV3 HN protein (Fig. 1F), we sought to further elucidate these epitopes. For this purpose, we determined structures of one mAb from two epitopes, PIV3HN-13 targeting Site 1 and PIV3HN-05 targeting Site 2. We determined an X-ray crystal structure of the Site 1-binding mAb PIV3HN-13 to 2.4 Å by complexing the PIV3 HN protein and PIV3HN-13 Fab together and confirming successful complexation through size exclusion chromatography (SEC) and negative stain electron microscopy (nsEM) (Supplementary Fig. S4). We identified that mAb PIV3HN-13 binds at the PIV3 HN dimer interface (Fig. 4A, Supplementary Fig. S5, and Supplementary Table S6). There appear to be no steric interactions between the PIV3HN-13 Fab and the PIV3 HN sialic acid active site, found at amino acids 275 to 280 on the HN head domain (Fig. 4A)15. When fit into a map of the previously proposed PIV3 F-HN interaction (EMD-27550)15, which occurs shortly after HN activation at the cell membrane, mAb PIV3HN-13 appears to bind distal to the interaction site (Fig. 4B). Interestingly, mAb PIV3HN-13 still inhibited cell-cell fusion, which may be attributed to IgG flexibility or other mAb recruitment functions. We assessed the specific interactions found between the PIV3HN-13 Fab and PIV3 HN and found numerous hydrogen bonds and a hydrophobic pocket between the Fab and antigen (Fig. 4C). Within the heavy chain complementarity determining region 3 (CDRH3), a hydrophobic pocket is formed with three beta hairpins on PIV3 HN (Fig. 4C, top). Surrounded by hydrogen bonds within the CDRH2 and CDRH3, this interface likely creates a hydrophobic attraction to the Site 1 epitope. Along with CDRH2 hydrogen bonds, which are detailed below, one hydrogen bond can be found in the CDRH3 between PIV3HN-13 Fab residue F113 and PIV3 HN residue Y428 (3.0 Å). This hydrophobic pocket possibly strengthens the antibody-antigen interaction and stabilizes the HN protein, which may inhibit further processes such as PIV3 F protein activation. CDRH2 and heavy chain framework region 3 (FRH3) contain the majority of the hydrogen bonds between PIV3HN-13 and PIV3 HN (Fig. 4C, middle). Several hydrogen bonds are present between the CDRH2 and the beta hairpin on PIV3 HN, including Fab to HN residues G58 to S122 (3.1 Å), and S64 to L124 (2.7 Å), respectively. In addition, S64 on the CDRH2 forms two additional hydrogen bonds with a second beta-sheet on HN at residues K430 and Y432 (3.1 Å and 3.1 Å, respectively). Another residue on the CDRH2, S59, forms separate hydrogen bonds with residue N123 on HN, bonding with both the amine and the hydroxyl atoms (2.8 Å and 3.3 Å, respectively). CDRH2 residue T65 forms a hydrogen bond with HN residue K430, the second bond to form with this HN residue (2.9 Å). Of note, one hydrogen bond is formed between the CDRH2 region of mAb PIVHN-13 and the second HN monomer, at residues H63 and N139, respectively (3.2 Å). In addition, FRH3 residue Y66 forms two hydrogen bonds with HN residues H427 and Y432 (2.7 Å and 2.8 Å, respectively). A final FRH3 residue Y67 forms a bond with N429 on HN (2.9 Å). Although few, the light chain of PIV13 also contains several hydrogen bonds with the HN protein, including those in the light chain CDR3 (CDRL3) and the FRL1 (Fig. 4C, bottom). These bonds include Fab residue D1 binding to both amine groups on HN residue R47 (3.4 Å and 3.1 Å, respectively), along with Fab residue A107 binding to HN residue Y428 (2.8 Å). In total, we observed 15 hydrogen bonds between the PIV3HN-13 Fab and the PIV3 HN protein, along with the hydrophobic pocket within the CDRH3 and several beta hairpins in PIV3 HN.
A X-ray crystal structure displaying PIVHN-13 Fab binding to the dimer interface of the head domain of the PIV3 HN protein was determined to be 2.4 Å. PIV3 HN monomers are shown in steel blue and sea green. The PIVHN-13 heavy chain is shown in brick red, while the light chain is shown in salmon. The previously proposed interaction site between the PIV3 F and PIV3 HN proteins is highlighted in gold and the sialic acid active site is highlighted in magenta. B Structures of the PIVHN-13 Fab, the PIV3 HN dimer, and the pre-fusion PIV3 F (PDB: 6MJZ) protein were fit into the PIV3 F-HN interaction map (EMB: 27550). Site 1 on PIV3 HN contains no interactions with the PIV3 F-PIV3 HN interaction site. C Interactions between the heavy and light chain of PIVHN-13 Fab and both PIV3 HN monomers were identified. Top: CRDH3 forms a hydrophobic pocket surrounded by hydrogen bonds between CRD2 residues Gly58 and Ser64 with PIV3 HN residues Ser122 and Leu124, respectively. Hydrophobic regions are highlighted with yellow translucent surfaces, with decreasing hydrophobicity depicted in teal. Middle: CDRH2 and FRH3 in the Fab heavy chain form various hydrogen bonds with the PIV3 HN monomer. All hydrogen bonds are formed with one monomer except for one between His63 on the heavy chain and Asn139 on the second HN monomer. Bottom: Hydrogen bonds between the light chain of the Fab and one HN monomer are found in the CDRL3 and FRL1. Hydrogen bonds are shown in black dotted lines. Oxygen atoms are colored red, and nitrogen atoms are colored blue.
The Site 2 mAb PIV3HN-05 binds toward the apex of PIV3 HN, distal to the F-interaction sites
To determine the binding site of mAb PIV3HN-05, we utilized cryo-electron microscopy (cryo-EM) (Supplementary Fig. S5). To enable larger particle formation, we leveraged the discrete binding epitopes and the X-ray structure determined above and generated a complex containing both Fabs PIV3HN-05 and PIV3HN-13 in complex with the PIV3 HN protein (Supplementary Fig. S4). After particle picking and heterogeneous refinement, we obtained an electron density map containing a PIV3 HN dimer with four Fabs bound that was further processed to a global resolution of 3.57 Å (Supplementary Fig. S6 and Table S7). The density in the map was clear for the PIV3HN-13 Fab and the PIV3 HN protein, however, the density for the PIV3HN-05 Fab contained some disordered regions in portions of the variable region. The PIV3HN-05 Fab binds at the apex of the HN head domain directly within the sialic acid binding site and distal to the binding site of PIV3HN-13, with one Fab binding to one PIV3 HN monomer (Fig. 5A). The CDRH3 of mAb PIV3HN-05 fits deep into the binding pocket of the HN active site and distal to the PIV3 F-HN interaction site, indicating the mechanism of protection by mAb PIV3-05 likely occurs through blocking receptor binding rather than blocking the HN-F protein interaction (Fig. 5B). Importantly, the electron density for the CDRH3 was clearly visible in the cryoEM map (Supplementary Fig. S7) allowing us to focus solely on the high-resolution interactions between PIV3HN-05 Fab and PIV3 HN. No other interactions besides the CDRH3 were observed with the experimentally built structure. However, since the variable region was missing portions in the electron density, we leveraged Alphafold3 to predict a structure of the PIV3HN-PIV3HN-05 interaction. The Alphafold3 model of PIV3HN-05 Fab with PIV3 HN aligns well with the solved structure of the CDRH3 of the PIV3HN-05 Fab, and also suggested minimal interactions between the PIV3HN-05 Fab and the PIV3 HN protein outside of the CDRH3 (Fig. 5C). Based on our experimentally determined structure, there are two hydrogen bonds between the PIV3HN-05 CDRH3 and the HN active site, respectively: S111D with C97 and Y112C with T358 (both 2.8 Å). In addition, hydrophobic interactions are seen within the binding site made by residues on HN previously identified to form the HN active site, seemingly locking the CDRH3 within the active site binding pocket (Fig. 5D)21. When fit to a previously solved structure of PIV3 HN complexed with a sialic acid analog, difluorosialic acid (DFSA), the CDRH3 occupies the same space of DFSA, corroborating the hypothesis of PIV3HN-05 blocking sialic acid binding45. Like other viral proteins containing hemagglutinin (HA) and neuraminidase (NA) activity, such as influenza virus HA and NA and Newcastle disease virus HN, residues containing sialic acid binding and catalytic activity on PIV3 HN lies at the apex of the head domain21. We elucidated that Site 2 is within the sialic acid site of the HN protein, leading us to hypothesize that PIV3HN-05 and other mAbs binding to Site 2, may block receptor binding or prevent viral release by inhibiting HN from interacting with sialic acid.
A Cryo-EM map of PIV3HN-05 Fab complexed with HN, with PIV3HN-05 Fab binding to the apical surface of HN. B When fit into a map of F (PDB: 6MJZ) and HN interaction (EMB: 27550), the PIV3HN-05 CDRH3 does not spatially interact with the active site of HN but fits directly within the HN active site. C Overlay of the Alphafold3 predicted structure (gray) and experimentally determined structure (HC: indigo, LC: mauve) of PIV3HN-05 Fab complexed to HN. HN monomers are shown in steel blue and sea green. D Interactions of the PIV3HN-05 CDRH3 with the PIV3 HN protein in the sialic acid binding pocket. Two hydrogen bonds are indicated with black dotted lines. Hydrophobic regions are highlighted with yellow translucent surfaces, with decreasing hydrophobicity depicted in teal. HN residues associated with sialic acid receptor interactions are labeled. Difluorosialic acid atoms fit into the HN model (PDB: 4WEF) and are depicted in black.
mAb targeting HN the active site can prevent viral replication in Syrian hamster lungs prophylactically and therapeutically
The targeted binding of mAb PIV3HN-05 to the sialic acid binding site on HN, along with its potent neutralization, complement deposition, and PIV1 cross-neutralization, led us to assess PIV3HN-05 as a prophylactic and therapeutic treatment in vivo. In addition to PIV3HN-05, we tested the efficacy of mAb PIV3HN-12, which binds to structurally solved Site 1 and inhibits fusion, as well as mAb PIV3HN-09, which binds to Site 3 and is another potent neutralizer of PIV3. For this purpose, we leveraged the Syrian golden hamster, which supports viral replication of PIV3 in the lungs and nasopharynx46,47. In Study 1, hamsters (n = 10) were treated intraperitoneally with 10 mg/kg of one mAb at each site 24 h before PIV3 viral challenge (Fig. 6A). We also included an isotype control mAb treated group using mAb PhtD7, which targets the pneumococcal histidine triad protein D48, a PBS treated group, and a group of hamsters that were administered PBS as a treatment and as a mock PIV3 infection. Four days after the intranasal PIV3 challenge, lung samples were collected, and viral load was quantified via plaque assay. Compared to the PBS-treated and PIV3 infected group (1 × 105 PFU/mL/g), mAbs PIV3HN-05 (1 × 103 PFU/mL/g) and PIV3HN-09 (5 × 103 PFU/mL/g), binding to Sites 2 and 3, respectively, effectively decreased PIV3 replication in the lungs, while the Site 1-specific mAb, PIV3HN-12 (7 × 104 PFU/mL/g), did not (Fig. 6B). The success of the prophylactic mAb treatment of PIV3HN-05 and PIV3HN-09 led us to evaluate both mAbs as a more clinically relevant therapeutic treatment delivered 24 hrs after PIV3 challenge (Fig. 6C). mAb PIVHN-05 was able to decrease viral load in the lungs (1 × 103 PFU/mL/g) compared to the PBS-treated group (2 × 105 PFU/mL/g) (Fig. 6D). mAb PIV3HN-09 was also able to reduce lung viral titers (3 × 103 PFU/mL/g) compared to the PBS-treated group to a lesser, but still significant, degree (Fig. 6D).
A Syrian hamsters (n = 10) were treated with 10 mg/kg of one mAb binding to each identified site on HN 24 h prior to the intranasal PIV3 challenge. Lungs were collected four days following the viral challenge. Created in BioRender. Miller, R. (2024) https://BioRender.com/w46b621. B Plaque assays revealed decreased viral load in the lungs with prophylactically PIV05- and PIV09-treated hamsters. Data are presented as mean values. Statistical significance was calculated using a one-sided ANOVA for multiple comparisons (***P = 0.0002, *P = 0.02). C Syrian hamsters (n = 5) were treated 24 hours after the intranasal PIV3 challenge. Lungs were collected four days following the viral challenge. Created in BioRender. Miller, R. (2024) https://BioRender.com/y64f007. D Viral load in the lungs of PIV05 was significantly lower than in the mock-treated group. Data are presented as mean values. Statistical significance was calculated using a one-sided ANOVA for multiple comparisons (**P = 0.007, *P = 0.04).
In summary, this paper showcases a specific application of mAb therapy in combating PIV3 infection, specifically with the success of mAb PIV3HN-05 in reducing viral replication in the lungs likely by blocking HN receptor binding. Beyond its immediate impact on PIV3 treatment, the introduction of mAb therapy targeting different activities of HN holds broader implications for the development of treatments against Paramyxoviruses and other respiratory viruses through the assessment of structural targets and mAb functional versatility.
Discussion
Due to the potential for severe PIV3 disease in infants, young children, the elderly, and the immunocompromised, there exists an unfulfilled need for effective vaccines and therapeutics against PIV3. With the recent advancements of other pediatric virus treatments and vaccines, such as for RSV, this paper not only identifies potential mAb treatments to be further assessed for their clinical effectiveness but also introduces new insights into the PIV3 HN protein and its potential as a vaccine target.
The current understanding of B cell immunity to PIV3 infection is lacking, and these studies help fill gaps in the current knowledge as we have identified human mAbs with high levels of potency in both binding affinity and neutralization. Our assessment of mAbs to three discrete epitopes as treatments led us to understand the potential of mAbs to Site 2, particularly mAb PIV3HN-05, which was shown to reduce viral replication in the lungs of hamsters. Along with this, we have defined the structural interactions between antibody and protein for two mAbs targeting discrete epitopes.
Of the three identified epitopes, Sites 2 and 3 of PIV3 HN contain the most potent neutralizing mAbs, and mAbs binding to Site 2 are highly effective in complement deposition, followed by those binding to Site 3. Indeed, the addition of complement increased the neutralizing potency of mAbs binding to Site 2 and Site 3, which may involve Fc-effector functions such as cellular phagocytosis or cellular cytotoxicity. In addition, while not specific to any binding epitope or correlated to neutralization potency, a subset of mAbs was able to inhibit fusion even at low concentrations, although the mechanism for this inhibition remains to be determined. Of all the mAbs, only the Site 2-specific mAb PIV3HN-05 demonstrated cross-neutralizing activity against PIV1, the second most prevalent of the PIVs. Thus, now mAbs to both PIV3 HN and to PIV3 F28,29 have been identified, and there likely remain additional epitopes capable of eliciting cross-reactive mAbs. However, the mechanism by which these mAbs cross-react between the two viruses remains to be determined.
In addition to the efficacy studies, we determined structures of mAbs binding Sites 1 and 2 on the PIV3 HN protein. The Site 1-binding mAb PIV3HN-13 binds at the PIV3 HN dimer interface, with a hydrophobic pocket within the CDRH3 creating an energetically attractive interface for PIV3 HN binding and numerous hydrogen bonds within the CDRH2 likely strengthening the bond between the Fab and antigen. The cryo-EM map and solved CDRH3 structure of the protective mAb PIV3HN-05 identified that Site 2 lies at the apex of PIV3 HN directly inside the sialic acid binding site and distal from both Site 1 and the F interaction site toward the base of the HN head domain. With the promising functional activities of this mAb and the clear resolution of the CDRH3 secured inside the active site of HN, we pursued mAb PIV3HN-05 as a treatment in vivo.
We assessed the prophylactic efficacy of mAbs targeting each of the three discovered epitopes in a whole-body system using a model of PIV3 infection in Syrian golden hamsters. The Site 1 mAb PIV3HN-12 did not influence lung viral titers, while the Site 3 mAb PIV3HN-09 was somewhat effective in Study 1 but not in Study 2. However, mAb PIV3HN-05 successfully decreased the lung viral load in hamsters and was effective whether it was administered prophylactically or therapeutically. Based on these data, and the fact that mAb PIV3HN-05 binds to the sialic acid binding site, is potently neutralizing, effective at complement deposition, and cross-neutralizes PIV1, PIV3HN-05 is marked with high potential as a clinical treatment for PIV3 infection and possibly for PIV1 infection. However, the activity of PIV3HN-05 against PIV1 infection in vivo still needs to be assessed in future studies.
Overall, these studies provide new immunological insights into the areas of focus for HN-directed therapies, through way of potential receptor binding inhibition, antibody-mediated immunity, in vivo efficacy, and structural interactions. Additional in vivo experiments can be conducted to study the effectiveness of a mAb cocktail treatment by combining multiple neutralizing mAbs binding to different epitopes and inhibiting various HN functions, testing modifications of Fc domains to increase complement deposition, and determining the efficacy of mAb PIV3HN-05 against PIV1. Structural insights regarding mAbs binding to Site 2 will also need to be further assessed, due to the disordered density of the cryo-EM map generated for the PIV3HN-05 Fab/PIV3 HN structure. Site 3 will need to be further studied to complete the structural assessment of the three epitopes defined against HN, which may lead to future investigations on neutralizing mechanisms of mAbs and their effectiveness in vivo. It is also highly likely additional epitopes on the PIV3 HN protein will be identified as additional human mAbs are isolated. In summary, these data and future data may lead to the development of effective vaccines and therapeutics against PIV3 infection and other PIVs, and prompt additional research surrounding the functional aspects of the PIV3 HN protein as well as HN proteins from other PIVs.
Methods
Study design
The overall objective of this study was to identify novel mAbs against the HN protein on PIV3 and assess their neutralizing capabilities, protection in vivo, and structural characteristics. These goals were completed using experiments surrounding binding analysis, virus neutralization, in vivo treatment, and structural analysis, all of which are detailed below in the Methods. mAbs were labeled as potent binders and neutralizers by EC50 values and IC50 values below 50 ng/mL and 200 ng/mL, respectively. All procedures involving hamsters were performed in accordance with guidelines of the Institutional Animal Care and Use Committee of the University of Georgia under the Animal Use Protocol code A2022 08-003-Y2-A3.
Cells and virus
LLC-MK2 cells were obtained from ATCC (CCL-7) and grown in Opti-MEM (Gibco, Cat #31985070) supplemented with 2% FBS (Gibco, Cat #26140079) at 37 °C, 5% CO2. To split, cells were washed twice with PBS (Corning, Cat #21040CV) and trypsinized with 0.25% trypsin-EDTA (Corning, Cat #25053CI) and neutralized with Opti-MEM, 2% FBS media. PIV3 strain C243 was obtained from IRR (VR-93). PIV1 strain C35 was obtained from IRR (VR-94). The virus was diluted into virus medium (Opti-MEM, 5 µg/mL trypsin-EDTA, and 1% antibiotic-antimycotic) and added to LLC-MK2 cells grown at an MOI of 0.01. Virus was propagated at 37 °C, 5% CO2 for 3–5 days until CPE was observed. The virus was subsequently harvested using a sucrose freeze/thaw method as previously described49. The virus-containing sucrose solution was aliquoted, flash-frozen in liquid nitrogen, and stored at − 80 °C until further use.
PIV3 HN head protein construct design, expression, and purification
A plasmid construct for the head domain of PIV3 HN was designed based on the wild-type PIV3 strain C243 HN sequence (GenBank JN089924.1) with the N-terminal cytoplasmic tail and transmembrane domains removed and a Hexahistidine tag added. Plasmid containing the HN head domain insert was transformed into DH5a E. coli competent cells prior to plasmid isolation and transfection. The recombinant protein was expressed in Expi293F cells (ThermoFisher, Cat #A14527) as previously described30. Culture supernatant was filtered through a 0.45 µm filter before purifying through a HisTrap Excel pre-packed column (Cytiva, Cat #17371206) per the manufacturer’s instructions. Purified protein was buffer exchanged into PBS and stored at − 80 °C until use.
Isolation of peripheral blood mononuclear cells from healthy human donors
Peripheral blood mononuclear cells (PBMCs) from Subject 1 were obtained through study recruitment to the University of Georgia Clinical and Translational Research Unit and written informed consent was obtained. This study was approved by the University of Georgia Institutional Review Board as PROJECT00002304. PBMCs from Subjects 2-3 were obtained from leukocyte reduction filters supplied by Shepheard Community Blood Center in Augusta, Georgia as discarded samples. Each filter was gently washed with PBS and the flow-through, which contains the leukocytes, was collected in a 50 mL conical tube. In a separate tube, 13 mL of warmed Ficoll-Paque (Cytiva, Cat #17144002) was added to a separate 50 mL conical tube. The cells were gently added to the top of the Ficoll layer and then centrifuged at 300 × g for 30 min at 4 °C with a slow acceleration and deceleration rate. The PBMC layer below the PBS and above the Ficoll was gently aspirated from the tube and added to a separate tube containing Dulbecco’s Modified Eagle’s Medium (DMEM) (Corning, Cat #10014CV) and centrifuged at 300 × g for 5 min. The supernatant was discarded, and the cell pellet was resuspended in 10 mL RBC lysis buffer (155 mM NH4Cl, 12 mM NaCO3, 0.1 mM EDTA). The cells were centrifuged at 300 × g for 5 min, the supernatant was discarded, and the PBMCs were washed with DMEM. Finally, the cells were resuspended in ClonaCell-HY Medium A (StemCell Cat #03801) supplemented with 10% DMSO, transitioned to − 80 °C, and stored in LN2 until further use.
Antigen-specific flow sorting with memory B cells against the PIV3 HN protein
One day before B cell sorting, gamma-irradiated NIH/3T3 cells (ATCC, CRL-1658) modified to express human CD40L, human interleukin-21 (IL-21), and human B-cell activating factor (BAFF) (a gift from Deepta Bhattacharya), were diluted in StemCell Medium A (0.5 million cells/mL) and supplemented with the TLR agonist CpG (phosphorothioate-modified oligodeoxynucleotide ZOEZOEZZZZZOEEZOEZZZT; Invitrogen) (6 µg/mL) as previously described31. The diluted cells were added to the inner wells of tissue culture-grade 384 well plates (50 µL/well), excluding the outer two rows on each side, which contained sterile diH2O (100 µL/well) to prevent evaporation of media containing the sorted B cells. Prior to antigen-specific sorting, PIV3 HN was biotinylated as per the manufacturer’s instructions (ThermoFisher, Cat #21455). Streptavidin-conjugated PE (Invitrogen, Cat #S866) and streptavidin-conjugated BV605 (BioLegend, Cat #405229) were slowly added to biotinylated HN at a fluorophore to protein molar ratio of 4:1 and stored on ice away from light. PBMCs (106 cells) were suspended in FACS buffer (PBS, 2% FBS, 2% goat serum, 0.5 mM EDTA) (1 mL) and Fc-blocked with Human TruStain FcX (BioLegend, Cat #422301) (5 µL/million cells) on ice for 30 min. A staining mixture containing anti-human CD19-APC (BioLegend, Cat #302212), IgM-FITC (Southern Biotech, Cat #2020-02), IgD-FITC (Southern Biotech, Cat #2030-02), GhostDye Red 780 (Tonbo Biosciences, Cat #130865T100) or DAPI (ThermoFisher, Cat #62248) (live/dead stains), BT-SA conjugated PIV3 HN-PE and BT-SA conjugated PIV3 HN-BV605 was prepared and kept on ice away from light. Cells were centrifuged at 400 × g for 5 min to pellet the cells and washed with FACS buffer once before resuspending in 30 µL FACS buffer. Single-stain controls were prepared with the cells and the remainder of the cells were added to the staining mixture and incubated on ice for 30 min away from light. Cells were washed once with FACS buffer and resuspended in FACS buffer. Antigen-specific live B cells (CD19 +, IgM-, IgD-, PE +, BV605 +) were single-cell sorted into each well containing the 3T3 cell monolayer and incubated for 13 days at 37 °C, 5% CO2. Wells containing B cell clones were fed with Medium A (50 µL/well) 7 days after flow sorting.
Enzyme-linked immunosorbent assay against the PIV3 HN protein
For both PBMC screening following B cell sorting and binding curves for transfected mAbs, 384-well high binding plates (Greiner, Cat #781061) were coated with PIV3 HN, and the assay was performed as previously described30. The OD405 values were measured on a Biotek plate reader using Gen5 3.11, and EC50 values were calculated using GraphPad Prism9.
RNA extraction and DNA amplification of RNA from antigen-positive B cells
RNA extraction was performed with the Qiagen RNeasy Micro Kit (Qiagen, Cat #74004) according to the manufacturer’s instructions. In brief, B cells from positive wells were isolated by gently scraping and pipetting the base of the well and removing the culture prior to lysis and extraction. RNA samples were immediately translated into DNA or stored at − 80 °C until use. RNA was translated and amplified into separate heavy chains and light chains using the OneStep RT-PCR kit according to the manufacturer's instructions (Qiagen, Cat # 210212). Briefly, RNA was thawed on ice and combined with 5X buffer, dNTP mix, enzyme mix, nuclease-free water, and a primer mix for heavy chains, kappa chains, or lambda chains, as previously described50. The reaction was performed on a Thermal Cycler (50 °C – 30 mins, 95 °C – 15 mins, 40 cycles of (94 °C – 40 s, 55 °C – 60 s, 72 °C – 60 s), 72 °C – 10 min). The reaction product was amplified in a second PCR. DNA product from the previous PCR was combined with 2X DreamTaq Master Mix (ThermoFisher, Cat #K1081), nuclease-free water, and the heavy chain, kappa chain, or lambda chain primer mix. The reaction was performed on a Thermal Cycler (94 °C – 5 min, 40 cycles of (94 °C – 30 s, 57 °C – 30 s, 72 °C – 60 s), 72 °C – 10 min). The presence of heavy chain and light chain DNA was confirmed on a 1% agarose gel. PCR products were purified using the E.Z.N.A Cycle Pure Kit (Omega Bio-Tek, Cat #D6492-01) according to the manufacturer’s instructions before Sanger sequencing. Finally, after the sequences of each heavy chain and light chain were determined, samples were cloned using the appropriate primer based on the sample’s V and J genes. DNA was combined with 2X DreamTaq Master Mix, primer mix, and nuclease-free water as previously described50. The reaction was performed using a Thermal Cycler (94 °C – 4 min, 40 cycles of (94 °C – 30 s, 58 °C – 30 s, 72 °C – 45 s), 72 °C – 8 min). DNA product was confirmed on a 1% agarose gel and purified using the E.Z.N.A Cycle Pure Kit prior to ligating into an expression vector.
Vector digestion and ligation
Expression vectors were digested prior to DNA ligation as previously described50. Heavy chain, kappa chain, and light chain vectors were used (AbVec-hIgG1 (FJ475055), AbVec-hIgKappa (FJ475056), and PBR322-based Ig-lambda expression vector (FJ517647), respectfully). Digested vectors were separated on a 1.5% agarose gel (100 V – 5 min, 75 V – 75 min) and bands at 6000 bp were cut out and purified using the E.Z.N.A. Gel Extraction Kit (Omega Bio-Tek, Cat #D2500-01) according to the manufacturer’s instructions. Heavy chain and light chain DNA inserts were combined with the linearized vector, NEBuilder HiFi DNA Assembly Master Mix (New England BioLabs, Cat #E2621S). Assembled vectors were stored at − 20 °C until transformations were performed.
Plasmid transformation into E. coli competent cells
Following vector ligation, assembled vectors were transformed into E. Coli DH5a competent cells as previously described50. Colony PCR was performed as described above to confirm successful transformation. Individual colonies were selected for expansion into LB cultures with ampicillin (5 mL) and shaken for 8–12 h at 37 °C. DNA plasmids were extracted from cultures as described previously. Plasmids were sequenced to confirm mAb transformation, and plasmids were re-transformed into DH5a cells (1 µL) as described above. The bacteria were cultured to a final volume of 250 mL and DNA plasmids were extracted using the E.Z.N.A Plasmid Maxi Kit (Omega Bio-Tek, Cat # D6922-04) according to the manufacturer’s instructions. Plasmids were sterile-filtered in preparation for mammalian cell transfection.
Plasmid transfection into HEK-293F and ExpiCHO cells
mAbs were transfected in either Freestyle293 or ExpiCHO cultures. For HEK293 transfections, the volume of mAb plasmids, including both heavy chains and light chains, were calculated based on the final culture volume (1 µg/mL), combined with Opti-MEM media and PEI MAX transfection reagent, and added to Expi293F cells as mentioned above. ExpiCHO transfections were conducted using ExpiCHO-S cells (ThermoFisher, Cat #A29127) and the ExpiFectamine CHO transfection kit (ThermoFisher, Cat #A29129) under the Max Titer protocol as described by the manufacturer.
mAb purification
Six days after plasmid transfection in HEK293 cells, or 14 days after plasmid transfection in ExpiCHO cells, the culture was centrifuged at 6000 × g for 10 min. The supernatant was filtered through a 0.45 µm filter. PBS was run through a HiTrap Protein G prepacked column (Cytiva, Cat #17040503) according to the manufacturer’s instructions. Eluted mAbs were buffer exchanged with PBS. Concentrations of each mAb were taken on a Denovix system using the IgG setting. An SDS-PAGE was performed to confirm successful transfection of mAbs under non-reducing and reducing conditions and stained with Coomassie Blue.
PIV3 mAb plaque reduction neutralization assay (PRNT)
PIV3 mAbs were serially diluted in Opti-MEM and incubated with PIV3 in a 1:1 ratio for 1 hr at room temperature. The mAb/virus mixture was added to LLC-MK2 (Opti-MEM, 2% FBS) monolayer in a 24-well tissue culture plate (50 µL/well) and rocked for 1 hr at 37 °C, 5% CO2. Cells were overlaid with warm 0.75% methylcellulose in Opti-MEM media supplemented with 5 µg/mL trypsin-EDTA and 1% antibiotic-antimycotic and incubated for 4 days at 37 °C, 5% CO2. Following incubation, cells were fixed with 10% neutral buffered formalin for 30 min at room temperature, washed 3 times with water, and blocked with blocking solution (2% milk blocking solution) supplemented with 2% goat serum in 0.05% PBS-Tween-20 for 1 hr at room temperature. Plates were washed 3 times with water, and the primary antibody (PIA17428 for PIV3, 3 × 1 for PIV129) was added (5 µg/mL in blocking buffer, 200 µL/well) for 1 hr at room temperature. Plates were washed 3 times with water, and HRP-conjugated secondary antibody (goat anti-human IgG Fc HRP, Southern Biotech Cat #2048-05) was added to the wells at a 1:2000 dilution (200 µL/well) for 1 hr at room temperature. Plates were washed 5 times with water and developed with TrueBlue substrate (SeraCare, Cat #5510-0030) (120 µL/well) and rocked for 10 min at room temperature before washing once with water and allowed to dry. Immunostained plaques were manually counted with a stereomicroscope. IC50 values were calculated using GraphPad Prism9.
Flow cytometry with PIV3-infected cells
Fluorophore-conjugated antibody was prepared by incubating biotinylated mAb with streptavidin-conjugated PE (Invitrogen, Cat #S866) for 1 h on ice 24 hours prior to cell staining. LLC-MK2 cells were grown in a T75 flask (Opti-MEM, 2% FBS) at 37 °C, 5% CO2 to 80% confluency prior to infection. Cells were washed twice with PBS before infecting with PIV3 (MOI = 0.01). Cells were incubated with the virus for 1 h at 37 °C, 5% CO2, rocking every 10 mins. Virus culture media (Opti-MEM, 5 µg/mL trypsin-EDTA, 1% antibiotic-antimycotic) was added to the flask after 1 h, and cells were infected for 48 hours. After the infection was complete, cells were digested with Versene (Gibco, Cat # 15040-066) at 37 °C, 5% CO2 for 30–45 mins until cells were no longer attached to the flask. Cells were washed twice with FACS and incubated in FACS buffer on ice for 30 mins. Cell aliquots were individually stained with PE-conjugated mAb (PE-PIV05, PE-PIV09, PE-PIA174, PE-MPV467) on ice away from light for 30 mins. Cells were washed twice with FACS buffer and resuspended in FBS. Cells positive for PE were analyzed on the NovoCyte Quanteon Flow Cytometer and analyzed in FlowJo.
Biolayer interferometry competition assay
PIV3 HN protein monomer and human mAbs were diluted at 100 µg/mL in Octet buffer (PBS, 0.02% Tween-20, 0.1% BSA). After obtaining a baseline in the Octet buffer for 60 s, PIV3 HN was loaded onto HIS1K Biosensors (Sartorius, Cat #18-5120) for 120 s. mAbs were primarily associated with HN for 300 s, then the biosensors were placed directly into wells with mAbs for 300 seconds for comparative competition. Biosensors were regenerated with glycine (0.01 M, pH 2.7) 5 times for 6 s before reusing. The degree of competition was calculated by dividing the total binding (nm) of the second mAb by the total binding (nm) of the first mAb * 100. Higher values indicate lower competition, whereas lower values indicate high levels of competition.
Fusion inhibition assay
Vero-SLAM cells51 were transfected with plasmids encoding PIV-HN, PIV-F, and GFP at a 3:3:1 ratio with JetOptimus transfection reagent (PolyPlus, Cat #101000006) as per manufacturer’s instructions. Three hours following transfection, the media was replaced, and antibody or PBS was added to the cells at the indicated concentration. Syncytia was visualized 24 h following transfection and images were taken using a Zoe microscope (Bio-Rad) (20 × magnification).
Hemagglutination inhibition assay
For the HAI experiment, 25 µL of 4 HA units of HPIV3 were incubated with 25 µL two-fold serially diluted antibodies in PBS (starting with 20 µg/mL of each antibody) for 1 h at room temperature. Then, 50 µL of 0.5% guinea pig RBCS (Innovative Research Inc.) was added to the virus-antibody mixture and allowed to settle for 1-2 h at room temperature. The minimal inhibitory concentration of each antibody was detected as the lowest concentration that inhibits the HA activity of 4 HA units of PIV3.
Complement deposition flow cytometric assay
Antibody-dependent complement deposition was performed as previously described43. In brief, biotinylated HN antigen was coupled with FluoSpheres NeutrAvidin beads (Invitrogen, Cat #F8776) at a 1:1 ratio of antigen (µg): beads (µL). Beads are then washed twice in 5% PBS-BSA and resuspended at 1:100 of starting bead volume in 0.1% BSA. MAbs were then incubated at 1–10 µg/mL with 10 µL of antigen-specific beads for 2 h at 37 °C. Bead-mAb complexes are then washed twice in 0.1% BSA. Guinea pig complement (MP Biomedicals, Cat #8642836) was then diluted in R-10 buffer (RPMI-1640 + 10%FBS) at a 1:50 dilution. The complement and the bead-mAb complex were incubated for 15 min and washed twice in PBS. A 1:100 dilution in PBS of secondary anti-C3 antibody (ICL, Cat #GC3-90P-Z) (fluorescein-conjugated goat anti-guinea pig complement C3) was then incubated at RT for 15 min. Complexes were then washed twice in PBS and resuspended in a final volume of 150 µL PBS. Samples were read on the Novocyte Quanteon.
Complement PRNT
A PRNT was performed as described above with the addition of a guinea pig complement (MP Biomedicals, Cat #8642836). After serially diluting mAb into Opti-MEM, guinea pig complement was added at a 1:1000 volume ratio. PIV3 virus was added to the complement + mAb mixture and the assay was performed as stated above.
Prophylactic/therapeutic treatment and viral challenge of Syrian hamsters
Four- to six-week-old male and female Syrian hamsters (Charles River Laboratories #049) were single-housed in the animal biosafety level 2 (ABSL-2) facility at the University of Georgia College of Veterinary Medicine. Hamsters were randomly grouped and given their respective mAb prophylactic treatment (10 mg/kg) through an intraperitoneal (IP) injection one day before the viral challenge. 24 h after mAb prophylaxis, hamsters were anesthetized with 4% isoflurane for 2 min before infecting with PIV3 strain C243 intranasally (IN) (106 PFU/mL, 50 µL). Hamsters were monitored before placing back in the cage. Four days after infection, hamsters were humanely euthanized by asphyxiation under CO2. Lungs were collected and gently washed with sterile PBS, then weighed, added to cold Medium A (3 mL), and homogenized for viral titration. Unless immediately used, lung samples were stored in 1% SPG buffer at − 80 °C following flash freezing in LN2.
PIV3 viral titration from Syrian hamster lungs
Immediately following sample collection, lung homogenates were serially diluted in cold Opti-MEM media and added to LLC-MK2 (Opti-MEM, 2% FBS) monolayer in a 24-well tissue culture plate (50 µL/well) and rocked for 1 hour at 37 °C, 5% CO2. The remainder of the plaque assay was performed as described above. Viral titers were calculated as log10(PFU/mL/g) and analyzed in GraphPad Prism9.
Assembly of Fab-antigen complex
Fab was digested using the Pierce Fab Preparation Kit (ThermoFisher, Cat #44985) prior to complexing with HN. Pure Fab and antigen were isolated through size exclusion chromatography (SEC) as described below. Fab and antigen were combined in a 2:1 molar ratio in a low salt buffer (120 mM NaCl, 20 mM Tris, pH 7.5) and incubated overnight at 4 °C. The Fab-antigen complex was isolated through SEC prior to use.
Size exclusion chromatography
Individual HN protein, Fab, and Fab-antigen complexes were isolated through SEC on a Superdex S200 10/300 (Cytiva, Cat #28990944) in column buffer (120 mM NaCl, 20 mM Tris, pH 7.5) based on their molecular weight elution profiles. The desired elute was concentrated prior to use.
PIV3HN-13 Fab:HN crystal formation
Complexes were formed by combining a 1:1.2 molar ratio of PIV3HN to Fab. This solution was incubated at 4 °C overnight and was then run over a Superdex 200 Increase 10/300 GL column. The peak coordinating with a complex was isolated and concentrated to 9 mg/ml and was then screened using a sitting drop method in which a 0.5 µL protein solution to 0.5 µL crystal condition was combined. Crystals formed between 1–4 weeks. PIV3HN-13 Fab:HN crystallized in a solution of 0.1 M Ammonium Sulfate, 0.1 M MES pH 6.5, and 15% (w/v) PEG4000 and was flash frozen with no cryo-protectant. PIV3HN-13 Fab crystallized from the Molecular Dimensions 1 + 2 kit in 0.2 M Ammonium Sulfate, 0.1 M MES pH 6.5, and 30% w/v PEG5000 MME.
X-ray Crystallography data collection, processing, and refinement
Data was collected at the Advanced Photon Source (APS) beamlines 22-ID. Data Processing was performed using HKL-2000 (v 719.2) and CCP4 suite (v 8.0.005). Molecular Replacement was performed Phaser-MR (full-featured) out of the Phenix suite of programs. To phase the complexes, homology models were generated using the SWISS-MODEL PIV3 HN model based on the previously solved structure (PDB: 1V3B). After phasing structures went through multiple rounds of refinement in Coot (v 0.9.8.3) and Phenix (v1.20.1).
Negative-stain electron microscopy
The size-excluded Fab-antigen complex was diluted into HyPure water (Cytiva, Cat #SH30538.02) to a final concentration of 15 µg/mL. Glow discharged, carbon-coated copper grids were stained with the diluted complex (5 µL) for 3 min, then washed twice with HyPure water. The grid was washed once with 1% uranyl formate solution, then stained in 1% uranyl formate solution for 45 s. The excess stain was removed with a Kim wipe and the grid air dried. Negative-stain electron micrographs were imaged on the JEOL JEM1011 transmission electron microscope equipped with a high-contrast 2K-by-2K AMT mid-mount digital camera to obtain 2D images of the Fab-antigen complex before assessing for structural interactions through cryo-electron microscopy.
Cryo-electron microscopy
Fabs 05 + 13:HN complex, at a concentration of 1.15 mg/mL, respectively, were applied to Quantifoil 2/1 (300 mesh) grids previously glow-discharged for 15 s at 15 mAmp currents on both sides. Grids were blotted for 3 s with 100% humidity and plunge-frozen in liquid ethane using a FEI Vitrobot Mark IV instrument. Cryo-EM data were collected on a Krios G2 (Thermo Fisher) equipped with a K2 summit DED camera (Gatan, Pleasanton, CA.). Cryo-EM movies were acquired using a nominal magnification of 48,544 x with a pixel size of 1.03 Å. Movies were recorded as 40-frame videos in counting mode, with a defocus range from − 0.8 to − 2.6 µm. Data were processed in cryoSPARC according to Supplementary Fig. S552. The X-ray structure of the PIV3HN-13-PIV3 HN complex was used as a template to fit into the cryoEM density, the CDRH3 of the PIV3HN-05 Fab was manually built into the cryo-EM map, and Alphafold3 was used to generate an initial model for the remainder of the PIV3HN-05 Fab, which was fit into the cryo-EM map in ChimeraX53,54. The model was then manually refined in COOT followed by refinement in Phenix55,56.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The X-ray crystallography structure of PIV3HN-13 generated by this study has been deposited to the Protein Data Bank with accession code 9B2W. The cryo-EM structure of PIV3HN-05 generated by this study has been deposited to the Protein Data Bank with accession code 9DZQ. The cryo-EM map of PIV3HN-05 generated by this study has been deposited to the Electron Microscopy Data Base with accession code EMB-47333 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-47333). The previously published structures were accessed through the Protein Data Bank with accession codes 6MJZ (pre-fusion PIV3 F), 4WEF (PIV3 HN), and 1V3B (PIV3 HN). The previously published map of PIV3 F-HN interaction was accessed through the Electron Microscopy Data Base with accession code EMB-27550. Additional raw data are available at https://doi.org/10.6084/m9.figshare.27904791. Source data are provided in this paper.
Code availability
No unique code was generated.
References
DeGroote, N. P. et al. Human parainfluenza virus circulation, United States, 2011–2019. J. Clin. Virol. 124, 104261 (2020).
Human Parainfluenza National Trends - NREVSS | CDC [Internet]. https://www.cdc.gov/surveillance/nrevss/human-paraflu/natl-trend.html (2023).
Branche, A. R. & Falsey, A. R. Respiratory viral infections: Parainfluenza virus infection. Semin Respir. Crit. Care Med. 37, 538 (2016).
Cortez, K. J. et al. Outbreak of human parainfluenza virus 3 infections in a hematopoietic stem cell transplant population. J. Infect. Dis. 184, 1093–1097 (2001).
Matsuse, H. et al. Naturally occurring Parainfluenza virus 3 infection in adults induces mild exacerbation of asthma associated with increased sputum concentrations of cysteinyl Leukotrienes. Int. Arch. Allergy Immunol. 138, 267–272 (2005).
Pecchini, R. et al. Parainfluenza virus as a cause of acute respiratory infection in hospitalized children. Braz. J. Infect. Dis. 19, 358 (2015).
Schmidt, A. C. Progress in respiratory virus vaccine development. Semin Respir. Crit. Care Med. 32, 527 (2011).
Counihan, M. E., Shay, D. K., Holman, R. C., Lowther, S. A. & Anderson, L. J. Human parainfluenza virus-associated hospitalizations among children less than five years of age in the United States. Pediatr. Infect. Dis. J. 20, 646–653 (2001).
Wang, L. et al. Epidemiology and clinical severity of the serotypes of human parainfluenza virus in children with acute respiratory infection. Virol. J. 20, 1–7 (2023).
Weinberg, G. A. & Edwards, K. M. Goldman’s Cecil Medicine. (2012).
Kim, H. M. et al. Recent increase in the detection of human parainfluenza virus during the coronavirus disease-2019 pandemic in the Republic of Korea. Virol. J. 19, 1–12 (2022).
Feng, Z. et al. A multicentre study on the incidence of respiratory viruses in children with community-acquired pneumonia requiring hospitalization in the setting of the zero-COVID policy in China. Arch. Virol. 168, 1–8 (2023).
Khayyata, S. H. & Farver, C. Parainfluenza Virus. Viruses and the Lung: Infections and Non-Infectious Viral-Linked Lung Disorders. https://www.ncbi.nlm.nih.gov/books/NBK560719/ (2022).
Zhang, S., Cheng, Q., Luo, C., Qin, Y. & Chen, M. Human Parainfluenza virus type 3 matrix protein reduces viral RNA synthesis of HPIV3 by regulating inclusion body formation. Viruses 10, 125 (2018).
Marcink, T. C. et al. Subnanometer structure of an enveloped virus fusion complex on viral surface reveals new entry mechanisms. Sci. Adv. 9, https://doi.org/10.1126/sciadv.ade2727 (2023).
Marcink, T. C. et al. Human parainfluenza virus fusion complex glycoproteins imaged in action on authentic viral surfaces. PLoS Pathog. 16, e1008883 (2020).
Porotto, M., Murrell, M., Greengard, O. & Moscona, A. Triggering of human Parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J. Virol. 77, 3647–3654 (2003).
Chang, A. & Dutch, R. E. & Paramyxovirus Fusion and Entry. Multiple paths to a common end. Viruses 4, 613–636 (2012).
Xu, R. et al. Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo. mBio 4, https://doi.org/10.1128/mbio.00803-13 (2013).
Porotto, M. et al. Spring-loaded model revisited: Paramyxovirus fusion requires engagement of a receptor binding protein beyond initial triggering of the fusion protein. J. Virol. 85, 12867–12880 (2011).
Lawrence, M. C. et al. Structure of the haemagglutinin-neuraminidase from human Parainfluenza virus type III. J. Mol. Biol. 335, 1343–1357 (2004).
Moscona, A. Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease. J. Clin. Invest. 115, 1688–1698 (2005).
Zhang, L. et al. Infection of ciliated cells by human Parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 79, 1113–1124 (2005).
Tappert, M. M., Porterfield, J. Z., Mehta-D’Souza, P., Gulati, S. & Air, G. M. Quantitative comparison of human Parainfluenza virus hemagglutinin-neuraminidase receptor binding and receptor cleavage. J. Virol. 87, 8962–8970 (2013).
Palermo, L. M. et al. Human Parainfluenza virus infection of the airway epithelium: Viral hemagglutinin-neuraminidase regulates fusion protein activation and modulates infectivity. J. Virol. 83, 6900–6908 (2009).
Porotto, M., Greengard, O., Poltoratskaia, N., Horga, M. A. & Moscona, A. Human Parainfluenza virus type 3 HN-receptor interaction: Effect of 4-Guanidino-Neu5Ac2en on a neuraminidase-deficient variant. J. Virol. 75, 7481–7488 (2001).
Boonyaratanakornkit, J. et al. Protective antibodies against human parainfluenza virus type 3 infection. MAbs. 13, https://doi.org/10.1080/19420862.2021.1912884 (2021).
Stewart-Jones, G. B. E. et al. Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1–4. Proc. Natl. Acad. Sci. USA 115, 12265–12270 (2018).
Cabán, M. et al. Cross-protective antibodies against common endemic respiratory viruses. Nat. Commun. 14, 1–15, (2023).
Banerjee, A. et al. Structural basis for ultrapotent antibody-mediated neutralization of human metapneumovirus. Proc. Natl. Acad. Sci. USA 119, e2203326119 (2022).
Bar-Peled, Y. et al. A potent neutralizing site III-specific human antibody neutralizes human metapneumovirus in vivo. J. Virol. 93, 342–361 (2019).
Pace, C. S. et al. Anti-CD4 monoclonal antibody ibalizumab exhibits breadth and potency against HIV-1, with natural resistance mediated by the loss of a V5 glycan in envelope.J. Acquir. Immune. Defic. Syndr. 62, 1–9 (2024).
Ejemel, M. et al. A cocktail of human monoclonal antibodies broadly neutralizes North American rabies virus variants as a promising candidate for rabies post-exposure prophylaxis. Sci. Rep. 12, 1–11 (2022).
Wu, H. et al. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J. Mol. Biol. 368, 652–665 (2007).
Cagigi, A. et al. Vaccine generation of protective ebola antibodies and identification of conserved B-cell signatures. J. Infect. Dis. 218, S528–S536 (2018).
Ahani, B. et al. Molecular and phenotypic characteristics of RSV infections in infants during two nirsevimab randomized clinical trials. Nat. Commun. 14, 1–10 (2023).
Porotto, M., Fornabaio, M., Kellogg, G. E. & Moscona, A. A second receptor binding site on human Parainfluenza virus type 3 hemagglutinin-neuraminidase contributes to activation of the fusion mechanism. J. Virol. 81, 3216–3228 (2007).
Takimoto, T., Taylor, G. L., Connaris, H. C., Crennell, S. J. & Portner, A. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J. Virol. 76, 13028–13033 (2002).
Cooper, N. R. & Nemerow, G. R. The role of antibody and complement in the control of viral onfections. J. Invest. Dermatol. 83, S121–S127 (1984).
Dunkelberger, J. R. & Song, W. C. Complement and its role in innate and adaptive immune responses.Cell Res. 20, 34–50 (2010).
Xie, C. B., Jane-Wit, D. & Pober, J. S. Complement membrane attack complex: New roles, mechanisms of action, and therapeutic targets. Am. J. Pathol. 190, 1138–1150 (2020).
Goldberg, B. S. & Ackerman, M. E. Antibody-mediated complement activation in pathology and protection. Immunol. Cell Biol. 98, 305–317 (2020).
Fischinger, S. et al. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods 473, 112630 (2019).
Human Parainfluenza Viruses (HPIV) Seasons | CDC. https://www.cdc.gov/parainfluenza/seasons.html (2023).
Streltsov, V. A., Pilling, P., Barrett, S. & McKimm-Breschkin, J. L. Catalytic mechanism and novel receptor binding sites of human parainfluenza virus type 3 hemagglutinin-neuraminidase (hPIV3 HN). Antivir. Res 123, 216–223 (2015).
Garg, R. et al. Vaccination with a human parainfluenza virus type 3 chimeric FHN glycoprotein formulated with a combination adjuvant induces protective immunity. Vaccine 35, 7139–7146 (2017).
Greer, C. E. et al. Long-term protection in hamsters against human Parainfluenza virus type 3 following mucosal or combinations of mucosal and systemic immunizations with chimeric alphavirus-based replicon particles. Scand. J. Immunol. 66, 645–653 (2007).
Huang, J. et al. Broadly reactive human monoclonal antibodies targeting the pneumococcal histidine triad protein protect against fatal pneumococcal infection. Infect. Immun. 89, https://doi.org/10.1128/iai.00747-20 (2021).
Caidi, H., Harcourt, J. L. & Haynes, L. M. RSV growth and quantification by microtitration and qRT- PCR assays. Methods Mol. Biol. 1442, 13–32, (2016).
Guthmiller, J. J., Dugan, H. L., Neu, K. E., Lan, L. Y. L. & Wilson, P. C. An efficient method to generate monoclonal antibodies from human B cells. Methods Mol. Biol. 1904, 109–145, (2019).
Ono, N. et al. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75, 4399–4401 (2001).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Meng, E. C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Casañal, A., Lohkamp, B. & Emsley, P. Current developments in Coot for macromolecular model building of electron cryo‐microscopy and crystallographic data. Protein Sci. 29, 1069 (2020).
Acknowledgements
We thank A. McCormick for the assistance with mAb development, J. Huang for assisting with project approach, A. Pena for the assistance with protein and mAb expression, K. Nagashima for assisting with structural assessment, C. Page for the assistance with manuscript organization, D. Williams and Arizona State University Erying Materials Center for acquiring cryo-EM data, and the staff of the University of Georgia College of Veterinary Medicine Animal Resources for assistance with animal studies. This work was funded by the National Institutes of Health under contracts R01AI143865 and R56AI181850.
Author information
Authors and Affiliations
Contributions
R.J.M. contributed to project investigation, including B cell sorting and mAb development. I.A.D., N.A., and J.J.M. contributed to structural analysis. A.G.B. and R.G.D. contributed to protein and antibody expression and neutralization assays. M.A.B. contributed to fusion inhibition assays. R.J.M., A.G.B., and R.G.D. performed ELISAs and animal study plaque assays. M.A.E. contributed to the HAI assays. I.A.D. contributed to X-ray crystallography data acquisition and analysis. R.J.M. contributed to negative stain EM imaging and analysis. N.A. and J.J.M. contributed to cryo-EM data analysis. R.J.M. and A.D.G. conducted animal studies. R.J.M. and J.J.M. wrote the original manuscript draft. All authors reviewed and edited the manuscript. J.J.M. supervised the project and reviewed all data.
Corresponding author
Ethics declarations
Competing interests
J.J.M. is an inventor on a patent application related to anti-parainfluenza virus monoclonal antibodies.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Miller, R.J., Durie, I.A., Gingerich, A.D. et al. The structural basis of protective and nonprotective human monoclonal antibodies targeting the parainfluenza virus type 3 hemagglutinin-neuraminidase. Nat Commun 15, 10825 (2024). https://doi.org/10.1038/s41467-024-55101-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55101-4