Abstract
The mycobacterial ABC transporter IrtAB features an ABC exporter fold, yet it imports iron-charged siderophores called mycobactins. Here, we present extensive cryo-EM analyses and DEER measurements, revealing that IrtAB alternates between an inward-facing and an outward-occluded conformation, but does not sample an outward-facing conformation. When IrtAB is locked in its outward-occluded conformation in nanodiscs, mycobactin is bound in the middle of the lipid bilayer at a membrane-facing crevice opening at the heterodimeric interface. Mutations introduced at the crevice abrogate mycobactin import and in corresponding structures, the crevice is collapsed. A conserved triple histidine motif coordinating a zinc ion is present below the mycobactin binding site. Substitution of these histidine residues with alanine results in a decoupled transporter, which hydrolyzes ATP, but lost its capacity to import mycobactins. Our data suggest that IrtAB imports mycobactin via a credit-card mechanism in a transport cycle that is coupled to the presence of zinc.
Similar content being viewed by others
Introduction
Iron is an essential micronutrient that is locked away by host organisms to suppress the growth of pathogenic invaders1. Microorganisms such as the causative agent of tuberculosis (TB), Mycobacterium tuberculosis, have therefore evolved highly specific iron scavenging systems relying on secreted small molecules called siderophores, which bind ferric iron (Fe3+) with extraordinarily high affinity2. In mycobacteria, the most important class of siderophores are called mycobactins, which are produced either in a secreted form called carboxymycobactin (cMBT) or a membrane-bound form called mycobactin (MBT)3. MBT and cMBT share the same iron-binding core and only differ in their modification with either a shorter dicarboxylate (cMBT) or a longer fatty acid (MBT). Biosynthesis of mycobactins occurs in the cytoplasm, and they are exported to the periplasm via the proton-driven transporters MmpL4 and MmpL5 in a process that also requires the respective accessory protein MmpS4 and MmpS54,5,6. Proteins facilitating mycobactin-transport across the outer mycobacterial membrane are yet to be identified. The final steps of the mycobactin cycle are the uptake of iron-loaded Fe-(c)MBT across the inner membrane followed by the reductive release of ferrous iron (Fe2+) in the cytoplasm, processes that are both accomplished by the unusual ABC transporter IrtAB7,8.
IrtAB belongs to the ubiquitous ATP-binding cassette (ABC) transporter family9. Typical ABC transporters consist of two transmembrane domains (TMDs), which are highly variable at the structural and sequence level, and conserved nucleotide-binding domains (NBDs). Substrate translocation across the lipid bilayer is mediated by the TMDs and is coupled to the movement of the NBDs, which dimerize and disengage as a result of ATP binding and hydrolysis, respectively. In the case of IrtAB, the entire transporter is composed of two polypeptide chains (IrtA and IrtB), which heterodimerize to build the fully assembled transporter composed of two TMDs and two NBDs8. As a specialty, the N-terminus of the IrtA chain encodes for a siderophore interaction domain (SID), which is located in the cytoplasm and mediates mycobactin reduction and iron release8,10.
IrtAB has been initially identified to play a key role in the mycobactin pathway based on the observation that its transcription is under the control of the iron regulator protein IdeR11, and its genetic deletion resulted in a pronounced virulence defect in a TB mouse model7. Further work revealed that IrtAB imports iron-bound mycobactin into the cytoplasm10. This function, however, appeared puzzling since IrtAB belongs to the subfamily of type I ABC exporters (also categorized as type IV ABC transporter), which are generally known to export substrates such as for example, drugs out of the cell12. Our lab solved a combined X-ray and cryo-EM structure of IrtAB of the thermophilic species Mycobacterium thermoresistibile, which confirmed the ABC exporter fold8. Functional elucidation with purified and membrane-reconstituted IrtAB showed that the transporter is capable of Fe-cMBT import in proteoliposomes in an ATP-dependent fashion8. The cryo-EM structure of inward-facing (IF) IrtAB revealed that the SID is ideally positioned to reduce MBT8. Recently, an outward-occluded (OFoccl) cryo-EM structure of IrtAB of M. tuberculosis (MTB) was described, which was obtained by introducing glutamate to glutamine substitutions at the Walker B motifs of IrtA and IrtB13.
Here, we thoroughly characterized the unusual ABC transporter IrtAB by determining a cryo-EM structure in a complex with Fe-MBT and by combining functional and biochemical assays with DEER measurements to formulate a novel mycobactin transport mechanism.
Results
Cryo-EM analysis of IrtAB in nanodiscs
IrtAB of M. thermoresistibile (MTHERM) exhibits robust ATPase activity in nanodiscs8. Full-length wild-type (WT)-IrtAB was therefore reconstituted in nanodiscs and analyzed by cryo-EM. In the presence of ATP-γ-S-Mg, we determined a cryo-EM structure of inward-facing IrtAB at a global resolution of 3.67 Å (dataset #1, Supplementary Table S1 and Supplementary Fig. 1) and observed the binding of two ATP-γ-S molecules at the respective nucleotide binding sites (Supplementary Fig. 2a). The inward-facing cryo-EM structure was found to be highly similar to the previously solved X-ray structure of MTHERM-IrtAB determined with the transporter purified in β-DDM8 and the cryo-EM structure of inward-facing MTB-IrtAB determined in digitonin13 (Supplementary Fig. 3). This can be explained by interactions of conserved hydrophobic C-terminal residues of the IrtB-NBD with the IrtA-NBD, which restrict the conformational freedom of the NBDs (Supplementary Fig. 4a).
Upon incubation with ATP-Mg and vanadate, cryo-EM analysis (dataset #2, Supplementary Table S1 and Supplementary Fig. 5) revealed the structure of an outward-occluded (OFoccl) conformation determined at a global resolution of 3.14 Å (Fig. 1). Particles corresponding to the IF conformation were found as well, but could not be resolved to high resolution (Supplementary Fig. 5). We observed density for two ADP-vanadate molecules at the fully closed NBD dimer interface (Supplementary Fig. 2b). The extracellular wings at the TMDs remained closed, as first described for the ABC transporter McjD14 and later for MsbA15,16, TmrAB17, AtAtm318 and PCAT119 (Supplementary Fig. 3). As a consequence, the TMDs surround a large internal cavity (Fig. 1c) that is sealed from both sides of the membrane, enclosing a volume of 5005 Å3, which is large enough to fit up to two iron bound mycobactins simultaneously (Supplementary Fig. 6). Of note, the recently determined OFoccl structures of MTB-IrtAB and yersiniabactin importer YbtPQ feature a similarly sized cavity13,20.
a Domain organization of IrtAB. The siderophore interaction domain (SID, blue) is N-terminally positioned to IrtA (teal). IrtB (pink) forms a separate open reading frame. b Cryo-EM maps of full-length IrtAB reconstituted in nanodisc or purified in LMNG detergent. Inward-facing (IF) and outward-occluded (OFoccl) maps were determined from distinct datasets. At the periplasmic side of OFoccl IrtAB, a non-proteinaceous density (contoured at 6 σ) is observed, which likely corresponds to a phospholipid (nanodisc sample) or an LMNG molecule (detergent sample). The SID (blue density) was generally less well resolved. c The IF cavity is accessible from the inner leaflet of the lipid bilayer. The OFoccl conformation features a large cavity that is sealed from both sides of the membrane. In the OFoccl conformation, a lateral membrane-facing crevice spanning across the whole lipid bilayer is formed between TMH2IrtA and TMH5IrtB.
Cryo-EM analysis of IrtAB in LMNG
Considering that nanodiscs might bias the conformational ensemble of ABC transporters16, the purification protocol was optimized using LMNG as a detergent. In contrast to IrtAB purified in β-DDM or β-DM8, LMNG-purified IrtAB exhibits ATPase activity, which was stimulated up to 24-fold by MBT and up to 5-fold by cMBT (Supplementary Fig. 2e). Full-length IrtAB in LMNG was analyzed by cryo-EM in the presence or absence of ATP-Mg vanadate. Again, we observed the IF conformation in the absence of nucleotides (global resolution of 3.42 Å) and the OFoccl conformation in the presence of ATP-Mg vanadate (global resolution of 3.23 Å) (Fig. 1b and Supplementary Fig. 7,8). However, no additional conformations were identified. Distance comparisons between conserved residues in the coupling helices of the TMDs and the NBDs showed that the structures determined in LMNG differed only marginally when compared to their respective counterparts determined in nanodiscs (Supplementary Fig. 3).
The SID is highly mobile in outward-occluded IrtAB
The four structures determined for full-length WT-IrtAB in nanodiscs and LMNG allowed us to analyze the flexibility of the SID in the context of different membrane mimics and alternate conformations. When IrtAB was purified in LMNG, the SID was generally better resolved than upon reconstitution in nanodiscs (Fig. 1b). Likewise, density for the SID was found to be less defined when IrtAB adopts its outward-occluded conformation. We performed a 3D variability analysis using the inward-facing structure determined in LMNG and observed a swinging movement of the SID along the membrane in front of the inward-facing opening of the IrtA chain (Supplementary Fig. 4b).
Mycobactin binds at a membrane-facing crevice at the heterodimer interface
The cryo-EM analyses of full-length WT-IrtAB were mostly carried out in the presence of Fe-MBT or Fe-cMBT at a concentration that results in maximal stimulation of ATPase activity (Supplementary Table S1). However, in these datasets, we were unable to identify non-proteinaceous cryo-EM density corresponding to mycobactin. Instead, we observed the binding of a lipid of unclear identity (in case of nanodisc-reconstituted IrtAB) or an LMNG molecule to a hydrophobic pocket located at the periplasmic end of a crevice spanning the almost the heterodimer interface between helices TMH2IrtA and TMH5IrtB of OFoccl IrtAB (Fig. 1b). The membrane-facing crevice and its lipid/detergent-filled hydrophobic pocket at the periplasmic end is a unique feature of OFoccl IrtAB; in inward-facing IrtAB, the pocket is closed and devoid of bound lipid (Figs. 1b, 2a).
a Cryo-EM structures of inward-facing IrtAB (left panel, Dataset #1, PDB 9FXC) and outward-occluded 2xEQ-IrtAB (right panel, Dataset #5, PDB 9FW3), both obtained in nanodiscs. Shown are the surface representations of the modeled structures. IrtA is colored teal, IrtB magenta, and bound Fe-MBT in orange sticks. The membrane boundaries are indicated in gray planes. b Cryo-EM map of outward-occluded 2xEQ-IrtAB (Dataset #5, PDB 9FW3) with non-proteinaceous densities contoured at 5 σ and colored in orange (accounted for Fe-MBT) or in gray (likely stemming from lipids and the nanodiscs belt protein). c Detailed depiction of Fe-MBT binding site at the membrane-facing crevice formed between TMH2IrtA and TMH5IrtB. Side chains interacting with Fe-MBT are highlighted as sticks.
We suspected that the flexible SID limits cryo-EM map resolution, and made a IrtAB construct lacking the SID. ATPase activity measurements with full-length IrtAB and IrtABΔSID purified in LMNG revealed that the deletion of the SID causes only minor changes in the maximally stimulated ATPase activity (Supplementary Fig. 2g), which differs within the typical error range of protein concentration determination underlying such assays. In order to trap IrtABΔSID more efficiently in its NBD-closed conformation, we mutated the Walker B glutamate of both NBDs into glutamine (E815QIrtA-E493QIrtB, henceforth denoted the 2xEQ mutation), which has been shown previously to render IrtAB inactive8. Purified 2xEQ-IrtABΔSID was then reconstituted into nanodiscs and ATP-Mg as well as Fe-MBT were added before grid preparation (dataset #5, Supplementary Table 1 and Supplementary Fig. 9). The same preparation without added Fe-MBT served as negative control (dataset #6, Supplementary Table 1 and Supplementary Fig. 10). In these samples, we only found 2D and 3D classes corresponding to the outward-occluded conformation, which could be further split into two similarly sized subclasses (Supplementary Fig. 9). In the samples with mycobactin present, one of this subclasses featured a strong non-proteinaceous density close to a membrane-facing crevice present at the heterodimer interface between helices TMH2IrtA and TMH5IrtB of OFoccl IrtAB (Fig. 2b). Fe-MBT could be placed unambiguously into the density. Importantly, the density was not present in the control samples prepared in parallel devoid of mycobactin (Supplementary Fig. 11). Relative to the lipid bilayer, the mycobactin binding site is located at the level of the hydrophobic core (Fig. 2). Hence, during sample preparation, Fe-MBT most likely entered the crevice at the periplasmic entrance and slit down to its binding site with its bulky head group partially exposed to the lipid bilayer and partially shielded by the crevice formed between TMH2IrtA and TMH5IrtB. Curiously, an additional density featuring an α-helix not stemming from IrtAB was found to interact with Fe-MBT, which most likely corresponds to a part of the MSP protein stabilizing the lipid patch of the nanodiscs (Fig. 2b). The same density feature was also found the control dataset devoid of Fe-MBT (Supplementary Fig. 11). It is possible that this α-helix blocks Fe-MBT during its passage and was necessary to trap the substrate at this position.
With a buried surface area of 486 Å2 (determined by PDBePISA server), IrtAB interacts extensively with Fe-MBT mostly via van der Waals contacts (Fig. 2c). A notable residue is Q249IrtB, whose side chain is also engaged in a hydrogen bond with Fe-MBT. While the density for the iron-chelating moiety of Fe-MBT is strong, the tail of Fe-MBT is much less resolved, and thus assumes an ensemble of different conformations.
Mutational analysis of the mycobactin binding site
To functionally analyze the mycobactin binding site, we mutated Q249IrtB to alanine, leucine, phenylalanine, or arginine. All four variants could be purified in LMNG with yields and size exclusion chromatography profiles similar to WT-IrtAB. We measured ATPase activities of the purified variants in the presence and absence of 5 µM Fe-MBT, a mycobactin concentration at which WT-IrtAB is strongly stimulated (Fig. 3 and Supplementary Fig. 2g, h). Strikingly, the basal ATPase activities of variants Q249AIrtB, Q249LIrtB, and Q249FIrtB were highly elevated as compared to WT-IrtAB, and could not be further stimulated by Fe-MBT (Fig. 3b). Of further note, stimulated ATPase activities of Q249LIrtB and Q249FIrtB were almost around twice as high as those of WT-IrtAB. Our data suggest that Q249IrtB, as found in WT-IrtAB, triggers ATPase activity only if it interacts with mycobactin, and accordingly suppresses ATPase activity when the natural substrate is absent. This “break” is evidently released when Q249IrtB is substituted by alanine, leucine, or phenylalanine. By stark contrast, the Q249RIrtB variant exhibited ATPase activities very similar to WT-IrtAB.
a The membrane-facing crevice opens up between TMH2IrtA and TMH5IrtB in the OFoccl conformation. IrtA is colored teal, and IrtB is magenta. In the middle part of the crevice, Fe-MBT is bound (Dataset #5, PDB 9FW3, OFoccl structure). Residue Q249rtB interacts with Fe-MBT, and was functionally characterized. b ATPase activity of purified Q249AIrtB, Q249LIrtB, Q249FIrtB, and Q249RIrtB variants with WT-IrtAB as control in the presence or absence of 5 µM Fe-MBT. Data points correspond to technical triplicates. c In vivo uptake of 55Fe-cMBT in M. smegmatis cells unable to produce their own siderophores and lacking the genomic copy of irtAB. Cells were complemented with WT-IrtAB (positive control), ATPase-deficient 2xEQ-IrtAB (negative control), and the four mycobactin binding site variants. Note that M. smegmatis cells exhibit IrtAB-independent Fe-cMBT uptake. Data points correspond to technical triplicates. a, b For both functional assays, representative data of at least two biological replicates are shown. Source data are provided as a Source Data file. d Structural comparison of Fe-MBT bound 2xEQ-IrtAB (Dataset #5, PDB 9FW3) and the 2xEQ-IrtAB-Q249RIrtB variant (Dataset #7, PDB 9G2S) unable to bind Fe-MBT owing to crevice collapse.
Next, the variants were used to complement a previously described M. smegmatis triple knockout (TKO) strain, which lacks two genes for siderophore biosynthesis and its endogenous irtAB gene8. As controls, complementation was carried out with WT-IrtAB (homolog from M. thermoresistibile throughout) and ATPase-deficient 2xEQ-IrtAB. Cells were grown under low iron conditions and radioactively labeled 55Fe3+-cMBT was added before incubating the cells for 30 min at 37 °C to facilitate IrtAB-mediated cMBT uptake (Fig. 3c and Supplementary Fig. 12, where further details are provided). In this assay, a clear difference in terms of cMBT uptake between cells complemented with WT-IrtAB and 2xEQ-IrtAB is consistently observed. However, cells complemented with 2xEQ-IrtAB exhibit basal uptake levels, which we previously showed to occur independently from IrtAB via an alternative uptake pathway8 (Supplementary Fig. 12). Cells complemented with the variant Q249AIrtB exhibited near WT-IrtAB transport activity, while variants Q249LIrtB and Q249FIrtB showed a partial loss of cMBT uptake (Fig. 3c). Hence, the polar interactions between Q249IrtB and Fe-MBT do not appear to be critically important for transport.
Intriguingly, the variant Q249RIrtB completely lost its capacity to import Fe-cMBT and only exhibited the IrtAB-independent uptake seen also for ATPase deficient 2xEQ-IrtAB (Fig. 3c). This finding was puzzling, because in terms of basal and stimulated ATPase activities, Q249RIrtB is very similar to WT-IrtAB. Hence, the single Q249RIrtB substitution resulted in a decoupled transporter, which hydrolyzes ATP, but lost its capacity to import mycobactins. To gain structural insights, the Q249RIrtB substitution was introduced into the 2xEQ-IrtAB background for cryo-EM analyses. In nanodiscs, cryo-EM analyses of this variant were unfortunately not successful. Therefore, we determined its structure in LMNG in the presence of ATP-Mg and Fe-MBT (dataset #7, Supplementary Table 1 and Supplementary Fig. 13). The best-resolved map revealed local conformational rearrangements resulting in a collapse of the entire membrane-facing crevice (Fig. 3d). Importantly, this striking difference cannot be explained by differences between IrtAB prepared in nanodiscs or LMNG, because the OFoccl conformation of other cryo-EM structures determined in LMNG feature an open membrane-facing crevice (Fig. 1b).
Mutational analysis of the membrane-facing crevice
Fe-MBT likely slides along the membrane-facing crevice formed at the IrtA/IrtB interface to reach its binding site as observed in the mycobactin-bound structure of OFoccl IrtAB (Fig. 2). At its periplasmic entry site, the crevice is filled with a lipid, bound to a hydrophobic cavity (Fig. 1b). To block Fe-MBT passage at the lower end of the hydrophobic cavity, we mutated A256IrtB into bulky or charged residues, namely leucine, phenylalanine or arginine (Fig. 4a). When purified in LMNG, the three variants exhibited elevated basal ATPase activity, with the A256RIrtB showing more than ten-fold higher ATP hydrolysis than WT-IrtAB (Fig. 4b). The basal activities of the three A256IrtB variants were found to be further stimulated by Fe-MBT. Subsequently, we performed the radioactive 55Fe3+-cMBT uptake assay to assess the transport capacity of the variants. Variants A256LIrtB and A256FIrtB were fully transport-competent (Fig. 4c). In contrast, the variant A256RIrtB completely lost its capacity to import cMBT, hence exhibiting a decoupled transport phenotype akin to the above-described Q249RIrtB variant.
a The membrane-facing crevice opens up between TMH2IrtA and TMH5IrtB in the OFoccl conformation. IrtA is colored teal, and IrtB is magenta. In the upper part of the crevice, a hydrophobic pocket is formed, which is occupied with a lipidic moiety (Dataset #5, PDB 9FW3, OFoccl structure). Residue A256IrtB, located below the lipid-binding pocket, is shown as a green stick and was functionally characterized. b ATPase activity of purified A256LIrtB, A256FIrtB, and A256RIrtB variants with WT-IrtAB as control in the presence or absence of 5 µM MBT. Data points correspond to technical triplicates. c In vivo uptake of 55Fe-cMBT in M. smegmatis cells unable to produce their own siderophores and lacking the genomic copy of irtAB. Cells were complemented with WT-IrtAB (positive control), ATPase-deficient 2xEQ-IrtAB (negative control), and the three lateral crevice variants. Note that M. smegmatis cells exhibit IrtAB-independent Fe-cMBT uptake. Data points correspond to technical triplicates. b, c For both functional assays, representative data of at least two biological replicates are shown. Source data are provided as a Source Data file. d Structural comparison of 2xEQ-IrtAB with bound Fe-MBT (Dataset #5, PDB 9FW3) and 2xEQ-IrtAB_ A256RIrtB (Dataset #8, PDB 9G2T, Arg256IrtB pointing to protein inside) unable to bind Fe-MBT and exhibiting a collapsed membrane-facing crevice.
To investigate this finding at the structural level, the A256RIrtB mutation was combined with the ATPase-deficient 2xEQ mutant. While determining a structure in nanodiscs was not successful for this mutant, we achieved to obtaine highly resolved cryo-EM structures in LMNG in the presence of ATP-Mg (dataset #8, Supplementary Table 1 and Supplementary Fig. 14). Data analysis revealed two main classes. The first class closely resembles the OFoccl structure of 2xEQ-IrtAB and shows density for the newly introduced R256IrtB residue pointing outward and thereby interrupting the crevice. Density for LMNG was found to be weaker than in other OFoccl structures (Supplementary Fig. 15). The second class features the R256IrtB residue pointing towards the occluded cavity (Supplementary Fig. 14). As a consequence, the entire lateral crevice is collapsed (Fig. 4d) and no detergent molecule is bound at the location where it is present in OFoccl structures showing the open crevice (Supplementary Fig. 15).
In conclusion, our structural analyses clearly show that the A256RIrtB mutation blocks the pathway from the periplasm to the mycobactin binding site by either directly interrupting the lateral crevice or indirectly causing large conformational rearrangements that result in a complete crevice collapse.
A conserved triple-histidine (HHH)-motif binds zinc
At the bottom of the lateral crevice, below the mycobactin binding site, we observed a strong non-proteinaceous density in both conformations (Fig. 5a, Supplementary Fig. 16). The density is coordinated by three histidine residues (HHH-motif) that are conserved among mycobacterial homologs of IrtAB (Fig. 5b). An analogous motif is also present in the yersiniabactin importer YbtPQ21, with the third histidine being an aspartate (HHD-motif) (Fig. 5b and Supplementary Fig. 16). Based on the chemical environment and the geometrical arrangement, we assumed it is a transition metal ion with a preference for soft Lewis base ligands in a tetrahedral arrangement, i.e., Zn2+, Cd2+, Co2+ or Ni2+ 22. To identify the coordinated metal ion, we performed Inductively Coupled Plasma Mass Spectrometry (ICP-MS) analysis with metals extracted from WT-IrtAB or 3xHtoA-IrtAB, in which the three histidines were substituted by alanines to prevent metal coordination. In order to avoid metal cross-contamination, the transporters were purified using a nanobody-functionalized resin capturing IrtAB via the C-terminal GFP-tag, instead of Ni2+-NTA affinity chromatography. In the first ICP-MS experiment, we measured the signals of 75 elements extracted from purified WT-IrtAB and compared them to the buffer control. Thereby, we discovered that purified WT-IrtAB contains elevated levels of Zn2+- and Ni2+- ions. Next, purified WT-IrtAB or 3xHtoA-IrtAB were incubated with either 200 µM ZnSO4, 200 µM NiSO4, or buffer, and unbound metal was removed using a desalting column. The concentration of the eluting transporter was determined by absorption measurements, and the metal content was quantified by ICP-MS. This experiment confirmed our initial finding that WT-IrtAB carries along Zn2+- and Ni2+- ions during purification and further revealed that metal co-purification does not occur in the 3xHtoA-IrtAB control (Fig. 5c). Upon pre-incubation with ZnSO4 or NiSO4, WT-IrtAB was fully saturated with Zn2+-ions (one Zn2+-ion per IrtAB heterodimer), whereas it could not be completely saturated with Ni2+-ions. We observed background binding of Zn2+ and Ni2+-ions to the 3xHtoA-IrtAB control upon pre-incubation, but bound metal concentrations reached only around 10 % of the concentrations bound to WT-IrtAB, clearly demonstrating that the HHH-motif is chiefly responsible for metal binding. In addition, in the case of WT-IrtAB, around 10 % Zn2+ remained bound in the Ni2+- soaking experiment, whereas Zn2+- soaking fully replaced Ni2+ (Fig. 5c), showing that the HHH-motif prefers to bind Zn2+ over Ni2+.
a Three histidine residues are localized in the cavity at the bottom of the lateral crevice and coordinate a metal atom. b The HHH motif is highly conserved among mycobacterial homologs of IrtAB, and it is also present as a slightly deviant HHD motif in YbtPQ, a yersiniabactin importer from uropathogenic E. coli. c ICP-MS analysis of WT-IrtAB and 3xHtoA-IrtAB in the absence of added metals or pre-soaked with 200 µM Zn2+- or Ni2+-ions. d Zn2+-titration curves using the zinc-sensitive FluoZin-3 (FZ-3) fluorescent probe. FZ-3 concentration was 200 nM for all measurements. Titrations were performed in buffer control or in the presence of equimolar WT-IrtAB (200 nM), 3xHtoA-IrtAB (200 nM), or the strong zinc-chelator EGTA (200 nM). EC50 values correspond to the Zn2+ concentration at which half-maximal florescence of ZN-3 is reached. Source data are provided as a Source Data file.
To obtain semi-quantitative insights into the binding affinity for Zn2+, WT-IrtAB, 3xHtoA-IrtAB, and the strong Zn2+-chelator EGTA (reported KD for Zn2+ of 4.5 × 10−10 M23) were titrated with increasing concentrations of Zn2+ in the presence of FluoZin-3, which becomes fluorescent upon Zn2+ chelation and has a reported KD for Zn2+ of 15 nM24. First, the Zn2+ concentration needed to reach half-maximal fluorescence of 200 nM FluoZin-3 was determined to be 44.2 nM (EC50 value) (Fig. 5d). Then, Zn2+-titration curves were recorded wherein 200 nM WT-IrtAB, 3xHtoA-IrtAB or EGTA were present at an equimolar ratio to FluoZin-3. The corresponding EC50 of FluoZin-3 for Zn2+ shifted to higher values for WT-IrtAB (EC50 = 86.4 nM) and EGTA (EC50 = 109.5 nM) but was basically unaltered when 3xHtoA-IrtAB was present (EC50 = 48.6 nM). Since the EC50 increases by a factor of two in the equimolar presence of WT-IrtAB as compared to the buffer control, the affinity of the HHH-motif for Zn2+ is in the same range as the one of FluoZin-3 (i.e., double-digit nanomolar) (Fig. 5d).
The HHH-motif is essential for energy coupling
In the next step, we characterized the functional role of the HHH-motif. We found that the MBT-stimulated ATPase activity of the LMNG-purified IrtAB variants was not affected by mutations introduced at the HHH-motif (Fig. 6a). This also demonstrates that mutations in this conserved motif did not affect protein folding. Curiously, the H439AIrtA variant and even more so the 3xHtoA variant showed slightly elevated basal ATPase activities (Fig. 6a). We then performed an in vivo uptake assay with radioactively labeled 55Fe-cMBT in M. smegmatis (Fig. 6b). The H444AIrtA variant was fully capable of mycobactin import, which can be explained by the conservation pattern of the HHH-motif because the third histidine is the least conserved and appears as an aspartate in YbtPQ21. In contrast, the single variant H393AIrtA exhibited a partial loss of transport activity, while the single variant H439AIrtA, as well as the 3xHtoA variant, lost their ability to import 55Fe3+-cMBT. Hence, disruption of the zinc coordination motif decouples ATP hydrolysis from mycobactin import.
a ATPase activity of purified HHH-motif variants with WT-IrtAB as a control in the presence or absence of 5 µM MBT. Data points correspond to technical quadruplicates. b In vivo uptake of 55Fe-cMBT in M. smegmatis cells unable to produce their own siderophores and lacking the genomic copy of irtAB. Cells were complemented with WT-IrtAB (positive control), ATPase-deficient 2xEQ IrtAB (negative control), and the individual and combined HHH-motif variants. Note that M. smegmatis cells exhibit IrtAB-independent Fe-cMBT uptake. Data points correspond to technical triplicates. b, c For both functional assays, representative data of at least two biological replicates are shown. Source data are provided as a Source Data file. c Structures of WT-IrtAB under ATP turnover conditions (dataset #9) with Fe-MBT (orange sticks) placed where it was found in nanodiscs (dataset #5) for reference. The HHH motif (green) is highlighted in sticks and the membrane boundaries are indicated in gray planes. d Superimposition of inward-facing 3xHtoA-IrtAB (gray, dataset #11) with inward-facing WT-IrtAB (colored, dataset #9).
Conformational landscape under ATP turnover conditions
To potentially detect additional conformations, we performed cryo-EM analysis of IrtAB under ATP turnover conditions. To this end, we pre-incubated LMNG-purified WT-IrtABΔSID (20 µM) with ATP-Mg (2 mM) in the presence or absence of Fe-MBT (20 µM) at 25 °C for 30 s before grid freezing and recorded two large cryo-EM datasets (datasets #9 and #10, Supplementary Table 1 and Supplementary Figs. 17, 18). Regardless of whether Fe-MTB was present or not, the cryo-EM analysis only revealed the IF and the OFoccl conformations to be present. The corresponding density maps reached global resolutions of 3.21-3.76 Å and 2.78-3.00 Å, respectively. When examining the densities of the bound nucleotides of the OFoccl structure in more detail, the γ-phosphate of ATP was clearly found to be present, suggesting that it represents a pre-hydrolysis state (Supplementary Fig. 2d). In both datasets, the number of particles corresponding to the OFoccl conformation was higher than the number of particles corresponding to the IF conformation, clearly showing that the OFoccl conformation is populated under hydrolysis conditions (Supplementary Fig. 19). The high number of particles corresponding to the OFoccl conformation, i.e., 402’468 and 283’513 for the Fe-MBT and the mycobactin-free dataset, respectively (Supplementary Table 1), would have made it highly feasible to detect the structurally related OFopen conformation, but we did not observe it.
Cryo-EM structure of 3xHtoA-IrtAB under turnover conditions
To obtain molecular insights into the uncoupled transport phenotype of 3xHtoA-IrtAB, we performed a cryo-EM analysis using the LMNG-purified, SID-less construct in the presence of 20 µM MBT under turnover condition (dataset #11, Supplementary Table 1 and Supplementary Fig. 20), resulting in maps at global resolutions of 3.2 Å and 3.0 Å for IF and OFoccl conformations, respectively (Fig. 6c). Intriguingly, the IF/OFoccl equilibrium of 3xHtoA-IrtAB is prominently shifted towards the IF conformation as compared to WT-IrtAB (Supplementary Fig. 19). Compared to the WT-IrtAB sample analyzed under the same experimental conditions (dataset #10), the OFoccl models are basically identical (RMSD: 0.453 Å across 1126 residues), apart from the mutated residues. By contrast, we noted a fine difference between the nearly identical IF structures, namely a rearrangement of the unwound stretch of TMH5IrtB and concomitant detachment from the 3xHtoA motif (Fig. 6d). Of note, TMH5IrtB is part of the domain swap helices which are prominently broken in the IF structure of IrtAB, but straighten during the transition to the OFoccl conformation, thereby representing one of the most dynamic regions of IrtAB (Supplementary Fig. 21). Further, TMH5IrtB together with TMH2IrtA delineates the lateral crevice that opens up during the IF-OFoccl transition (Fig. 2). Our structural analysis of the 3xHtoA variant therefore suggests an allosteric coupling of the HHH-motif with dynamic events occurring at the lateral crevice that are essential for mycobactin import.
DEER analyses of IrtAB
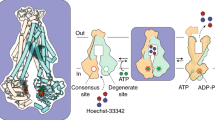
To study the conformational space of IrtAB by an orthogonal method, we performed DEER measurements on selected spin-labeled variants. We created three pairs of cysteine mutants to be labeled with the paramagnetic nitroxide spin label MTSL using the program MMM to simulate inter-label distances25 on the IF and OFoccl structures of IrtAB as well as on an IrtAB homology model based on OFopen TM287/288 (PDB: 6QUZ)26. Two spin-labeling pairs were placed at the periplasmic side on TMH1IrtA and TMH3IrtA or TMH1IrtB and TMH6IrtB (Fig. 7). The inter-spin distances of these pairs are expected to remain largely constant during the transition from the IF to the OFoccl conformation, but would become around 20 Å longer in the OFopen conformation if the extracellular wings open up. As a control, an intracellular spin-labeling pair was placed on TMH5IrtB and TMH6IrtB. This pair is expected to become around 20 Å shorter when IrtAB transits from the IF to the OFoccl conformation but is invariant regardless of whether IrtAB assumes an OFoccl or OFopen conformation (Fig. 7).
a Spin-labeling positions are shown between intracellular pair N305CIrtB-S221CIrtB (green) and extracellular pairs S51CIrtB-S153CIrtB (blue) or A362CIrtA-T579CIrtA (orange) for cryo-EM structures (IF and OFoccl conformation) or a homology model based on the OFopen TM287/288 structure. IrtA is colored teal, and IrtB is magenta, and cysteines are shown as spheres. Dotted lines connect the thiol-groups. b Distances between nitroxide spin-labels (MTSSL) were simulated based on cryo-EM structures (for IF and OFoccl conformation, shaded gray and red areas, respectively) or the homology model (shaded blue) shown in (a). Measured distance distributions are shown as solid lines. The distance distributions and their corresponding uncertainties (shown as gray-shaded areas around the curves corresponding to a 95 % confidence bound) were determined using a neural network analysis (ref. 53).
Spin-labeled IrtAB purified in LMNG was measured under three conditions: (i) WT-IrtAB, apo; (ii) WT-IrtAB in the presence of ATP-Mg and MBT and (iii) 2xEQ-IrtAB in the presence of ATP-Mg. For the intracellular pair, we observed distance shortening of the 2xEQ-IrtAB/ATP-Mg sample with respect to the WT-IrtAB/apo sample (Fig. 7), which indicates complete NBD dimerization and intracellular gate closure. In contrast, in the presence of ATP-Mg and Fe-MBT (i.e., under turnover conditions), we observed only minor changes with respect to the apo-state peak, indicating a prevalent inward-facing conformation with separated NBDs. Curiously, our cryo-EM analyses suggested a strong population of the OFoccl conformation under basically identical turnover conditions, showing that cryo-EM and DEER analyses can lead to different conclusions. Potential reasons for such discrepancies are different freezing conditions and the introduction of spin labels in the case of DEER experiments, which may have both influenced the conformational space of IrtAB.
For the two extracellular pairs, we did not observe any sign of distance change that would indicate periplasmic gate opening (Fig. 7). Hence, the DEER measurements support a transport cycle of IrtAB that alternates between IF and OFoccl conformations but does not sample the OFopen conformation.
IrtAB is highly asymmetric
When comparing the IF and OFoccl structures of IrtAB by dissecting them according to transmembrane helix (TMH) bundles moving as rigid bodies in type IV ABC transporters (Supplementary Fig. 21), we observed that the most prominent structural rearrangements occur in the domain swap helices TMH4-5 of IrtA and IrtB, which differ by RMSD values of 2.7 Å and 4.6 Å, respectively. These RMSD values go way beyond what is observed for analogous rigid body movements of any other type IV ABC transporter of known structure undergoing the IF to OFoccl transition (Supplementary Fig. 22). Overall, the conformational transition of IrtAB is highly asymmetric (Supplementary Fig. 21) and is associated with unusually strong rotation of the NBDs (Supplementary Fig. 23), which underpin the inability of IrtAB to assume an outward-facing conformation. Of note, previous type IV ABC transporter structures have revealed functionally relevant α-helical breaks in the TMH4-5 region, namely for C. elegans ABCB127 and Cyanidioschyzon merolae ABCB128. Another notable example is a break of TMH4 of E. coli MsbA caused by the inhibitor G90729. Finally, inward-facing YbtPQ features a coil-to-helix transition in TMH4 of YbtP upon yersiniabactin binding21.
Size reduction of the occluded cavity does not impact mycobactin import
To assess whether the outward-occluded cavity of IrtAB is directly involved in mycobactin import, we decreased its volume by introducing four bulky tryptophane residues into the cavity wall (4xW-IrtAB). Intriguingly, 4xW-IrtAB retained its capacity to import mycobactin in the radioactive cMBT uptake assay (Fig. 8), providing indirect evidence that mycobactin does not pass through the central cavity of IrtAB.
a Model of OFoccl IrtAB wherein four small cavity residues were mutated to tryptophane (4xW-IrtAB). b ATPase activity of purified 4xW-IrtAB with WT-IrtAB as the control in the presence or absence of 5 µM MBT. Data points correspond to technical quadruplicates. c In vivo uptake of 55Fe-cMBT in M. smegmatis cells unable to produce their own siderophores and lacking the genomic copy of irtAB. Cells were complemented with WT-IrtAB (positive control), ATPase-deficient 2xEQ-IrtAB (negative control), and 4xW-IrtAB. Note that M. smegmatis cells exhibit IrtAB-independent Fe-cMBT uptake. Data points correspond to technical triplicates. b, c For both functional assays, representative data of at least two biological replicates are shown. Source data are provided as a Source Data file. d Side-by-side comparison of the occluded cavity of WT-IrtAB (experimental structure) and 4xW-IrtAB (model).
Discussion
Our data suggest that IrtAB most likely catalyzes the import of Fe-MBT via a credit-card mechanism (Fig. 9), wherein the transporter provides a hydrophilic groove for the polar substrate head group and shields it during translocation, and the lipophilic part remains embedded in the lipid bilayer30,31. We provide here direct experimental evidence that Fe-MBT binds to the outside of the TMDs of IrtAB (state 2 in Fig. 9). Since the structure was determined in nanodiscs, Fe-MBT must have penetrated the lipid bilayer to reach the observed mycobactin binding site. The prominent membrane-facing crevice at the IrtA/IrtB interface constitutes the most plausible pathway for Fe-MBT translocation. In strong support of this notion, we identified two mutations placed at the crevice or directly at the mycobactin binding site, which both prevent mycobactin import without affecting protein folding. Structural analyses of these variants revealed that they result in the collapse of the membrane-facing crevice.
How Fe-MBT continues its journey from the middle of the membrane to the cytoplasm leaves room for alternative interpretations. One possibility is a trap-and-flip mechanism, wherein mycobactin would first enter the large internal cavity of outward-occluded IrtAB. Transition to the inward-facing conformation would then release mycobactin to the inner leaflet of the cytoplasmic membrane15,32,33, where it can be reduced by the SID to release Fe2+8 (transition via states 3a and 3b in Fig. 9). This translocation route would require further opening of the membrane-facing crevice to provide access of mycobactin to the large cavity, which is, considering the size of the mycobactin head group, not plausible. Further, the 4xW-IrtAB variant retains transport activity (Fig. 8) even with its four bulky tryptophane residues pointing into the cavity, which speaks against a trap-and-flip mechanism.
In a more likely scenario, the path of mycobactin continues at the outside of IrtAB’s TMDs (transition via state 3 in Fig. 9). The crevice collapses when the transporter transits to its inward-facing conformation (Fig. 2), thereby squeezing the bound mycobactin towards the cell interior. Concomitantly, TMH5IrtB breaks and thereby provides space for the mycobactin head group to reach the cytosol. In the structure of 3xHtoA-IrtAB, the conformation of the loop connecting the broken helices of TMH5IrtB is altered (Fig. 6). It is plausible to assume that alterations in this region prevent Fe-MBT release when IrtAB transits to its inward-facing conformation, thereby explaining why 3xHtoA-IrtAB lacks transport activity. One may also speculate that this region only permits Fe-MBT passage when the HHH-motif is complexed with a zinc ion, though we do not provide structural evidence for this notion in this work.
Our suggested route of transport shares analogies to credit-card mechanisms described for scramblases34,35, with the notable difference that the credit-card swipe is split into two parts and that conformational changes invoked by ATP binding and hydrolysis are required to push the head group of the substrate towards the cytosol. In support of this notion, ATPase deficient 2xEQ-IrtAB is heavily impaired in mycobactin import. Among ABC transporters, IrtAB shares commonalities with PglK, a type IV ABC transporter responsible for the flipping of lipid-linked oligosaccharides in bacteria36. PglK was suggested to flip its amphiphilic substrate by catalyzing the translocation of the polar head group while the lipidic moiety remains embedded in the lipid bilayer without trapping the entire substrate in the central TMD cavity. Other aspects of the transport mechanism, however, differ between IrtAB and PglK, namely the direction of transport (import versus export) and the conformations required to drive transport (i.e., alternating between OFoccl and IF conformations for IrtAB and between OFoccl and OFopen conformations for PglK).
A remarkable biochemical observation of our study is the identification of three uncoupled IrtAB variants (Q249RIrtB, A256RIrtB, and 3xHtoA). In purified form, these variants exhibit basal ATPase activities, which can be further stimulated by Fe-MBT, suggesting that they are still capable of sensing Fe-MBT. At the same time, these variants completely lack transport activity, owing to structural changes in the membrane-facing crevice as discovered in three corresponding cryo-EM structures. This raises the question about the molecular mechanism by which Fe-MBT triggers ATP hydrolysis. Since we have not found a variant whose basal activity is similarly low as WT-IrtAB, but which cannot be longer stimulated by Fe-MBT, there must be an unidentified binding site responsible for Fe-MBT sensing. Another recurrent pattern was the elevation of the basal ATPase activity for many studied variants. Especially Q249AIrtB, Q249LIrtB, and Q249FIrtB cannot be further stimulated by Fe-MBT. Curiously, these fully stimulated variants still import Fe-MBT, suggesting that they wastefully hydrolyze ATP in the absence of available Fe-MBT, but are not strongly affected when it comes to Fe-MTB transport itself. In other words, the residues lining the membrane-facing crevice of WT-IrtAB, and in particular Q249IrtB, strongly suppress the basal ATPase activity, and thereby allow for the capacity to strongly stimulate ATPase activity when Fe-MBT is present.
Conceptually, it would be rather simple to convert an ABC exporter to an ABC importer. Namely, the OFopen conformation would need to encompass a high-affinity binding pocket for the substrate, which would collapse when the transporter transits to the IF conformation, thereby resulting in substrate release inside the cell. Indeed, inversion of transport directionality has been observed for several single mutants of EfrCD to import Hoechst instead of exporting it ref. 37, as well as for ABCB1 carrying 14 alanine substitutions in the TMDs exhibiting drug import activity38. For IrtAB, however, we were unable to detect the OFopen conformation in our cryo-EM and DEER measurements. Instead, we provide here evidence for a transport mechanism that functions by alternating between the IF and OFoccl conformation (Fig. 9). As shown by dissecting IrtAB into structural elements typically moving as rigid bodies in type IV ABC transporters (Supplementary Fig. 21), the most dramatic conformational distortions occur at the domain swap helices TMH4IrtB and TMH5IrtB, which are broken in the IF conformation and straighten during the IF to OFoccl transition. These are exactly the key conformational changes that finally establish the lateral crevice at the IrtA/IrtB interface, thereby forming an alternative periplasmic gate for mycobactins that strikingly differs from the canonical gate opening as first observed for Sav1816 and being the hallmark of the OFopen conformation17,26,39. Of note, even in the closely related OFoccl structure yersiniabactin importer YbtPQ20, the crevice is not apparent (Supplementary Fig. 16), suggesting that IrtAB’s import route might be unique among siderophore importers of the same fold.
We discovered that IrtAB harbors a highly conserved Zn2+-binding site that is required to couple ATP hydrolysis to active import. Our data suggest that zinc coordination fulfills a regulatory role, likely at the release step when IrtAB transits to its inward-facing conformation (transition via step 3 in Fig. 9). The binding affinity of the HHH-motif for Zn2+ was found to be in the double-digit nanomolar range, thus allowing for exchange with free zinc present within the cell. In a physiological context, the HHH-motif might ensure that IrtAB-mediated Fe-(c)MBT uptake only takes place when there is enough zinc present in the cell. Transcriptome and proteome analyses of M. tuberculosis have revealed several layers of cross-signaling between zinc and iron uptake systems, with the ESX-3 secretion machinery playing a key role for both micronutrients40,41,42. Mediated by transcriptional regulators, zinc uptake in Aspergillus fumigatus was found to be dependent on iron availability under zinc-replete conditions, thereby preventing a toxic imbalance of iron and zinc43. Iron and zinc are both essential constituents for co-factors of enzymes and biomolecules playing a role in electron transport chains, but they become toxic when taken up in excess, in particular in the case of free intracellular Fe2+ catalyzing the Fenton reaction44. During the infection cycle of mycobacteria, the invading pathogen is confronted with highly divergent Zn2+/Fe3+ ratios. While iron availability is limited at all times, the zinc concentration varies from low in the blood and in the mycobacteria-specific case, where it is scavenged by calprotectin, to very high in the macrophage phagosome, where bursts of zinc are released to intoxicate mycobacteria45. To counteract these bursts, mycobacteria upregulate detoxifying Zn2+ exporters belonging to the family of P-type ATPases, that organize into metal efflux platforms46. These mechanisms coordinating the uptake of iron and zinc operate at the transcriptional level and are, therefore, comparatively slow. By contrast, the zinc-binding site we discovered in IrtAB might provide a much faster reaction to respond to sudden changes in Zn2+/Fe3+ ratios. Why and in which physiological contexts this rapid regulation mechanism is crucial will be subject to future studies.
Methods
Strains, media, constructs, and cloning
E.coli MC1061 was used for cloning and expression of proteins. Cloning and expression precultures were performed in Luria Broth (LB) liquid medium or LB agar plates. For protein expression in E. coli, terrific broth (TB) was used. For mycobactin and carboxymycobactin production, M. smegmatis mc2155 wild-type and M. smegmatis ∆fxBC strains were used, respectively. For radioactive uptake experiments, M. smegmatis ∆fxbA∆mbtD∆irtAB (TKO) complemented with the respective IrtAB variant was used. For mycobacterial cultures in rich medium, 7H9 with OADC supplement was used for liquid cultures and 7H10 with OADC supplement for agar plates. Low-iron minimal medium for the production of mycobactins was prepared exactly as described previously8. Antibiotics used for protein expression and cloning in E.coli were ampicillin 100 µg/ml, chloramphenicol 25 µg/ml, and apramycin 50 µg/ml. When working with M. smegmatis, apramycin was used at a concentration of 25 µg/ml.
M. thermoresistible irtAB previously cloned into the pINIT vector8 was used for the generation of point mutants (Q249LIrtB, Q249FIrtB, Q249RIrtB, Q249AIrtB, A256LIrtB, A256FIrtB, A256RIrtB) with primer pairs listed in Supplementary Table S2. Cystein-free IrtAB was created by mutating the native C199AIrtA by using primer pair #16. Afterwards, DEER labeling pairs N305CIrtB-S221CIrtB, S51CIrtB-S153CIrtB, A362CIrtA-T579CIrtA were introduced into cysteine-free IrtAB, by using primer pairs #17-#18, #19-#20, #21-#22, respectively, in two separate quickchange cloning reactions. The Walker B mutations E815QIrtA and E493QIrtB to obtain a 2xEQ-IrtAB background were introduced on the three existing cysteine pair mutants using primer pairs #23 and #24 in two separate cloning steps. For the functional analysis of the triple histidine motif, single mutations H393AIrtA, H439AIrtA, and H444AIrtA were introduced with primer pairs #8, #9, and #10, respectively. The 3xHtoA mutant was created on IrtAB-H393AIrtA with primer pair #11 introducing both H439AIrtA and H444AIrtA mutations. For the structural analysis of the 3xHtoA mutant, the truncated 3xHtoA devoid of the SID was cloned from the pINIT vector by using primers #25 and #26-rev. 4xW-IrtAB mutant was created with subsequent quickchange reactions using primers #12-#15 in Table S2.
For protein expression, pBXC3GH (Addgene #47070) plasmid was used to express GFP-tagged protein for ICP-MS measurements, fluorescent titrations, and nanodisc reconstitution to generate datasets #1, #2, #5, and #6. For ATPase assays and structure determination in LMNG (datasets #3-4 and #9-11), the pBXC3H (Addgene #47068) plasmid was used, which adds a His-tag at the C-terminus of IrtB. For radioactive 55Fe3+-cMBT uptake assays in M. smegmatis ∆fxbA∆mbtD∆irtAB, point mutants were subcloned from pINIT vectors into the mycobacterial complementation vector pFLAG (Addgene #110095)47.
Protein expression and purification
Proteins were produced in and purified from E. coli MC1061 using a TB medium containing 100 μg/ml ampicillin. Cells were inoculated 1:50 from an overnight LB culture and grown for 2 h at 37 °C while shaking in baffled flasks. After temperature adjustment to 25 °C over 1 h, expression was induced with 0.02 % (w/v) L-arabinose for overnight expression. Cells were collected by centrifugation at 9000 × g for 15 min at 4 °C. The pellet was resuspended in TBS (20 mM Tris-HCl pH 7.5, 150 mM NaCl) containing DNase I (Thermo Scientific). Resuspended cells were lysed with a Microfluidizer M-110P (Microfluidics) with three passes at 25 kPa. The lysate was first centrifuged for 30 min at 8000 × g at 4 °C to remove cell debris, and the supernatant was further centrifuged at 170’000 g to pellet membrane fraction (Beckman-Colter). The pelleted membrane vesicles were resuspended in TBS buffer, flash-frozen, and stored at − 80 °C until further use.
IrtAB-specific sybody SybNL5 was previously selected as crystallization chaperone8,48. SybNL5 was expressed in the pBXNPHM3 vector overnight. Cells were collected and lysed with a microfluidizer, then the remaining cell debris was removed by low-spin centrifugation. Lysate was purified over a Ni-NTA column and eluted protein-MBP complex was cleaved with 3 C enzyme during dialysis. His-tagged MBP and 3 C enzyme were removed by reapplying the sample on a Ni-NTA column. The flow-through was concentrated and further purified by size exclusion chromatography (SEC) on a Superdex 200 Increase 10/300 GL (GE Healthcare) column. As SybNL5 did not hinder the ATPase activity of IrtAB (Supplementary Fig. 2h), it was used as fiducial marker datasets #1 and #9-11.
Depending on the experimental use, IrtAB was either purified via C-terminal His-tag with Ni2+-affinity chromatography or via C-terminal GFP-tag with in-house prepared GFP-nanobody (PDB 3K1K) resin. For structure determination in detergent-solubilized samples and ATPase assays, membrane vesicles were thawed and solubilized in 1.3 % β-DDM for 2 h during head-over rotation. Solubilized membrane proteins were separated by 30 min ultracentrifugation at 170’000 g. The supernatant was supplemented with 20 mM imidazole and loaded onto Ni-NTA resin (Qiagen) for batch-binding at 4 °C for at least an hour with gentle stirring. Subsequently, the resin was washed on a column with 20 column volumes (CV, typically 100 ml) wash buffer (50 mM imidazole pH 7.5, 200 mM NaCl, 10% glycerol, and 0.01 % LMNG). IrtAB was eluted with 2 CV elution buffers (200 mM imidazole, pH 7.5, 200 mM NaCl, 10 % glycerol, and 0.01% LMNG). The eluted protein was concentrated to 1–3 mg/ml and 560 µl aliquots were frozen for subsequent separation by SEC in TBS buffer supplemented with 0.01 % (w/v) LMNG for ATPase assays, datasets #3-4 or 0.005 % (w/v) LMNG for datasets #7-11.
For cryo-EM analyses in nanodisc, IrtAB was purified as described above, but only β-DDM (0.03 %, w/v) was used during Ni-affinity purification and the first round of SEC (0.03 %, w/v). Then, IrtAB was reconstituted into nanodiscs as previously described8 with minor modifications as follows. For samples of datasets #1-2, IrtAB:MSP1D1E3:lipids ratio of 1:8:240 was used, whereas for datasets #5 and #6, a very tight nanodisc with 1:3:90 ratio was assembled. After removal of Bio-Beads (SM-2, Bio-Rad), reconstituted IrtAB was cleaved by 3 C enzyme and subsequently concentrated using a 100 kDa cut-off filter (Amicon). This way, most of the cleaved GFP, 3 C enzyme, and empty nanodiscs were removed from the sample prior to size-exclusion chromatography. The nanodiscs-reconstituted protein was finally separated on a Superose 6 Increase 10/300 GL column (GE Healthcare) in TBS buffer and peak fractions were pooled and concentrated for subsequent cryo-EM sample preparation in nanodisc.
For samples analyzed by ICP-MS and preparations for zinc affinity measurements, WT-IrtAB and 3xHtoA-IrtAB were purified via the C-terminal GFP-tag to avoid potential metal contamination. GFP-affinity purification was performed by in-house prepared beads coated with NHS immobilized GFP-nanobody (PDB 3K1K). DDM-solubilized membrane vesicles were incubated with GFP-resin overnight at 4 °C with gentle agitation to allow batch-binding. Then, the resin was transferred into an empty column and washed with 20 CV TBS buffer containing 10 % glycerol and 0.03 % β-DDM (w/v). For elution, the resin was loaded onto a 5 mL tube and incubated for 2 h with gentle agitation at 4 °C in the presence of 3 C enzyme (400 µg). Subsequently, the resin was loaded onto an empty column, and cleaved protein was eluted with 2 CV TBS containing 10 % glycerol and 0.03 % β-DDM (w/v). The flow-through was flash-frozen and stored at – 80 °C until further use.
Cysteine-labeled IrtAB samples for DEER measurements were expressed and purified as the cryo-EM samples described above, with the exception, that all purification buffers starting with membrane solubilization contained 2 mM DTT. Before MTSL labeling, DTT was removed by desalting on a PD10 column (GE Healthcare), and subsequently, a 5-fold molar excess of MTSL ((1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl)methanethiosulfonate, Toronto Research) per cysteine was added for overnight labeling at 4 °C with gentle agitation. Excess of spin label was removed by SEC on a Superose 6 Increase 10/300 GL (GE Healthcare) column in TBS containing 0.01 % LMNG (w/v). Protein fractions were concentrated and used immediately for sample preparation.
ATPase assay
ATPase activity assay was performed with LMNG solubilized IrtAB as described in our previous work8 with modifications. In brief, frozen aliquots of Ni-affinity purified MTHERM-IrtAB mutants were thawed and further purified on a Superose 6 Increase 10/300 GL (GE Healthcare) column in TBS containing 0.01 % LMNG. Main peak fractions were pooled, and concentration was adjusted to a concentration of 3–60 nM, depending on the basal or stimulated activity of each mutant, in order to reach phosphate levels detected in the linear range of the assay. Mutants were incubated at 37 °C for 15 min with 2.5 mM ATP or 1.25 mM ATP and 10 mM MgSO4 in TBS supplemented with 0.01 % LMNG, in the presence or absence of 5 µM MBT. ATPase stimulation of LMNG solubilized IrtAB with MBT and cMBT was performed with 15 nM protein and was incubated for 60 min at 37 °C. For read-out, malachite green-molybdate solution was added as described previously to detect the release of inorganic phosphate by measuring the absorbance at 650 nm49,50. Buffer control was used for background subtraction. The assays were performed with technical triplicates or quadruplicates and in biological duplicates and representative data is shown.
Radioactive uptake assay
55Fe-cMBT uptake was measured in vivo as described previously8, with minor modifications. Strains were grown to stationary phase in iron-rich 7H9 medium, then inoculated 1:50 in iron-depleted minimal medium and grown to OD600 2.5-5. Before the experiment, OD600 was adjusted to the lowest OD600 of all strains in the experiment. For variants Q249LIrtB, Q249FIrtB, Q249RIrtB, Q249AIrtB, A256LIrtB, A256FIrtB, A256RIrtB, and triple histidine motif variants H393AIrtA, H439AIrtA, H444AIrtA and 3xHtoA, three aliquots of 0.5 ml of each culture (technical triplicates) were put on ice. To each aliquot, radioactively labeled 55Fe-cMBT, dissolved in a minimal medium, was added to reach a final concentration of around 10 μM. While still on ice, for each variant, 3 x 200 μl culture was filtered and washed on a 96-well vacuum manifold using a glass fiber filtermat, thus measuring background binding values as technical triplicates. Then, the bacterial cultures were transferred to a 37 °C heating block and incubated while shaking at 1000 rpm. 30-minute end-point measurements were performed the same way as the background time points. Kinetic measurement for 3xHtoA mutant was performed from 1 ml bacterial culture by taking 200 µl samples at each time point. Accumulated radioactive 55Fe-cMBT was quantified with MicroBeta2 scintillation counter (Perkin Elmer), by using a MeltiLex-B solid scintillator sheet melted onto the glass fiber filter sheet.
Siderophore production, desferration and radioactive labeling
Mycobactins used for structure determination and functional assays, were produced, extracted, and radioactively labeled exactly as described before8.
Metal analysis with ICP-MS sample preparation and analysis
For samples analyzed by ICP-MS, frozen aliquots of GFP-tag purified WT-IrtAB and 3xHtoA-IrtAB were further purified by size exclusion chromatography (SEC) on a Superose 6 Increase 10/300 (GE Healthcare) column in TBS supplemented with 0.03 % β-DDM. Protein fractions corresponding to IrtAB were pooled and concentrated to 4 µM. For initial metal content analysis, untreated WT-IrtAB as well as SEC buffer were extracted by adding 1 ml HNO3 (60 %, Merck 1.01518.1000, for trace analysis) and 1 ml H2O2 (30 %, Merck, for trace analysis) in a sonication bath at 40 °C for 1 h. The volume was adjusted to 4 ml with 2 % HNO3 solution prior to ICP-MS analysis.
ICP-MS measurements were performed with an Agilent QQQ 8800 Triple quad ICP-MS spectrometer, equipped with a standard x-lens setting, nickel cones, and a “micro-mist” quartz nebulizer. The feed was 0.1 ml/min, and the RF power was 1550 W. Tune settings were based on the Agilent General Purpose method and only slightly modified by an autotune procedure using an Agilent 1 ppb tuning solution containing Li, Y, Ce, Tl, and Co (Agilent). Values are reported as the average of 30 sweeps x 3 replicates. Elements were measured without gas as well as with He and H2 as collision gases. All solutions were prepared from 60 % HNO3 (Merck 1.01518.1000 ultrapure) and 18.2 MΩ Millipore water.
For the metal saturation experiment, concentrated protein samples were divided into three aliquots. Each aliquot was incubated with either 200 µM ZnSO4, 200 µM NiSO4, or SEC buffer for control. After 30 min of incubation on ice, the samples were split into two technical replicates and desalted on PD midiTrap G-25 (Cytiva) columns. The protein concentration of the flow-through was measured on a NanoDrop 2000c (Thermo Scientific), and the samples were sent for quantitative ICP-MS metal analysis to determine the amount of bound metals. Extraction and ICP-MS analyses were carried out as above with the additional step that external calibration curves were recorded using a serial dilution made from ICP multi-element standard solution IV (Merck 1.11355.0100) in 2 % HNO3. Zinc and nickel quantification was performed based on single-charged ions and was limited to the following isotopes: 58Ni, 60Ni, 66Zn, and 68Zn. The metal saturation experiment was performed in biological duplicates starting from protein purification, and representative technical duplicates of a biological replicate are shown.
Zinc affinity measurement
For zinc affinity measurements, frozen aliquots of GFP-tag purified WT-IrtAB or 3xHtoA-IrtAB were thawed and gel filtrated on a Superose 6 Increase 10/300 (GE Healthcare) column in TBS supplemented with 0.03 % β-DDM. Peak fractions were pooled and used directly for competition assays. WT-IrtAB, 3xHtoA-IrtAB, or EGTA were added to the mixture with 200 nM FluoZin-3, at a final concentration of 200 nM. The mixture was titrated with Zn(NO3)2 standard solution (1000 mg/l, Supelco) in 5–500 nM steps up to 10 uM concentration while stirring at 900 rpm. Temperature was set to 25 °C and the interval between each titration step was 50 s. Zinc saturation on FluoZin-3 was monitored by the change in fluorescence by LS-55 florescence spectrometer (Perkin Elmer). The excitation wavelength was set to 494 nm, and emission was detected at 516 nm, with 10 nm slit-width for both.
Fluorescence data points for WT- and 3xHtoA-IrtAB were derived from the titration curve, by averaging the fluorescence value of final 30 s of each titration step and plotting it against the Zn2+ concentration. For EGTA competition, data points are the average of three single florescence readouts after each titration step.
For plotting, the data points (Fi) were normalized against the minimum (Fmin) and maximum (Fmax) fluorescence (Fnorm = (Fi – Fmin)/(Fmax−Fmin)). Fnorm points were plotted against Zn2+ concentration and fitted by least square fit using the Hill equation (Fnorm = Bmax[Zn2+]^h/(EC50^h + [Zn2+]^h)) in GraphPad Prism, where h is the Hill slope and Bmax is the maximum specific binding and EC50 corresponds to the Zn2+ concentration at which Fnorm = 0.5*Bmax. Fluorescence titrations were performed in a single biological replicate.
DEER sample preparation and analysis
Inter-spin distance distribution predictions were performed with a rotamer-based approach in MMM25,51 and used to choose suitable positions for the labeling. DEER samples were prepared using detergent-purified, MTSL-labeled IrtAB at a final concentration of 20 µM in TBS buffer supplemented with 0.01% LMNG. 10% glycerol-d8 (Sigma Aldrich) was added as a cryoprotectant to each sample and a final sample volume of 40 µl was frozen in liquid nitrogen. For the WT-IrtAB sample under turnover conditions, additionally 2.5 mM ATP, 2.5 mM MgCl2, and 20 µM MBT were added, whereas the spin-labeled samples prepared on the 2xEQ-IrtAB background were supplemented with 2.5 mM ATP and 2.5 mM MgCl2. The ATPase activity of purified, spin-labeled IrtAB was highly similar to WT-IrtAB, both in the presence and absence of Fe-MBT.
DEER measurements were performed at 50 K on a Bruker ELEXSYS E580Q-AWG (arbitrary waveform generator) dedicated pulse Q-band spectrometer equipped with a 150 W TWT amplifier and a homemade resonator52. A 4-pulse DEER sequence with Gaussian, non-selective observer and pump pulses of 32 ns length (corresponding to 13.6 ns FWHM) with 100 MHz frequency separation was used. The pump position was always chosen as the maximum of the nitroxide signal. DEER experiments were performed using the dead-time free 4-pulse DEER sequence (π/2)obs-(d1)-(π)obs-(d1+T)-(π)pump-(d2-T)-(π)obs-(d2)-(echo) with a 16-step phase cycling. The initial value of the interpulse delay t1 was set to 400 ns. Deuterium ESEEM was suppressed by incrementing t1 eight times by 16 ns and summing the individual traces (nuclear modulation averaging). The shot repetition time (SRT) was set to 3 ms. Data were analyzed using the latest version of DEERNet53 available for Spinach54. Raw DEER data are shown (Supplementary Fig. 24).
Cryo-EM sample preparation
For the sample recorded as dataset #1, 5 mM ATP-γ-S, 5 mM MgCl2, and 1.5 molar excess of SybNL5 were added to nanodisc reconstituted full-length WT-IrtAB (15 µM) prior to blotting. The sample was blotted on a glow-discharged QuantiFoil R1.2/1.3 Au 200 grid for 2.5–3.5 s at 10 °C in 90% relative humidity, then frozen in liquid ethane in a Leica GP2 plunge-freezer.
Orthovanadate-trapped nanodisc sample yielding dataset #2 was reconstituted as described above, but the lipid mixture was spiked with 50 µM Fe-MBT before incubation with Bio-Beads. Prior to blotting, 2.5 mM ATP, 2.5 mM MgCl2, and 5 mM orthovanadate were added to nanodiscs-reconstituted full-length WT-IrtAB (15 µM). Orthovanadate solution was preheated at 98 °C for 10 min before addition to the sample. Then, the sample was blotted on a glow-discharged QuantiFoil R1.2/1.3 Cu 200 grid for 2.5–3.5 s and plunged into liquid ethane on a Leica GP2 plunge-freezer.
LMNG solubilized full-length WT-IrtAB (20 µM) giving rise to dataset #3 was frozen without additives. The sample was blotted on a glow-discharged QuantiFoil R 1.2/1.3 Au 200 mesh for 3-4 s and plunged in liquid ethane on a Leica GP2 plunge-freezer at 11 °C and 70% relative humidity.
Sample of dataset #4 contained LMNG solubilized full-length WT-IrtAB (20 µM), which was frozen upon adding 100 µM Fe-MBT, 2.5 mM ATP, 2.5 mM MgCl2 and 5 mM orthovanadate. The sample was frozen on glow discharged QuantiFoil R 1.2/1.3 Au 200 mesh grids and blotted for 3-4 s and plunged in liquid ethane on a Leica GP2 plunge-freezer operated at 10 °C and 95% relative humidity.
ATP-trapped, nanodiscs-reconstituted 2xEQ-IrtABΔSID with or without Fe-MBT (dataset #5 and #6, respectively) were plunged on QuantiFoil R1.2/1.3 Au 300 grids, blotted for 5.5-6 s at 4 °C in 100% relative humidity and frozen in liquid ethane-propane mixture using a Vitrobot IV Mark plunge-freezer (Thermo Fisher Scientific). Grids of dataset #5-11 were glow-discharged under a 39 Pa air atmosphere, at 15 mA current for 60 s with a Pelco easiGlow glow-discharger (Ted Pella). Mycobactin containing sample (dataset #5) was pre-incubated with 20 µM Fe-ATP for 5 min on ice, then prior to plunging, 5 mM ATP-Mg was added, and the sample and incubated at 25 °C for 30 s. A control sample was handled the same way, except for the addition of Fe-MBT (dataset #6).
LMNG-purified 2xEQ-IrtABΔSID containing additionally the Q249RIrtB or A256RIrtB substitutions (datasets #7 and #8, respectively), were plunged on glow-discharged QuantiFoil R1.2/1.3 Au 300 grids, blotted for 5.5-6 s at 4 °C in 100% relative humidity using a Vitrobot IV Mark plunge-freezer (Thermo Fisher Scientific). Before plunging, 20 µM mycobactin and 5 mM ATP-Mg were added to the samples and preincubated at 25 °C for 30 s.
For the turnover samples, WT-IrtABΔSID (datasets #9 and #10) or 3xHtoA- IrtABΔSID (dataset #11) were purified in TBS supplemented with 0.005% LMNG, and 1.5 molar excess of SybNL5 was added to form a complex before the final size-exclusion step. The peak fractions corresponding to the IrtAB-SybNL5 complex were pooled and concentrated to 20 µM. Prior to blotting, each sample was preincubated for 30 s at 25 °C in a thermocycler in the presence of 2 mM ATP, 2 mM MgCl2, and either 20 µM mycobactin (datasets #9 and #11) or no substrate as control (dataset #10). Turnover samples were dispensed on glow-discharged QuantiFoil R1.2/1.3 Au 300 grids and blotted for 5-6 s (blotting force 20) at 4 °C and 100% relative humidity, then freeze in liquid ethane-propane mixture, using a Vitrobot Mark IV (Thermo Fisher Scientific) plunge-freezer.
Cryo-EM data collection and processing
Datasets #1-6 and #9-11 were collected on a Titan Krios G3i (Thermo Fisher Scientific) operated at 300 kV. Micrographs were recorded with a 100 μm objective aperture and a post-column BioQuantum energy filter with a 20 eV slit and a K3 direct electron detector (Gatan) in super-resolution mode at a nominal magnification of 130,000 ×. Movies in datasets #1-4 were saved without binning with a physical pixel size of 0.65 Å (0.325 Å in super-resolution mode) and datasets #5-6 and #9-11 were collected with 2-times binning within EPU, allowing for faster data collection, with a pixel size of 0.65 Å in super-resolution mode in LZW-compressed file format. Datasets #7-8 were collected on Glacios 2 equipped with Falcon 4i detector (Thermo Fisher Scientific) operated at 200 kV. Micrographs were recorded with 100 μm objective aperture without an energy filter at a nominal magnification of 190’000x. Movies of these two datasets were saved with a physical pixel size of 0.72 Å in LZW-compressed TIFF file format. Data acquisition was automatized with EPU v2.9-v3.6 (Thermo Fisher). Data acquisition parameters (total dose and defocus range) are listed for each dataset in Supplementary Table 1.
Movies of datasets #1-4 were corrected for beam-induced motion in MotionCor2 and simultaneously binned to 0.65 Å pixel size. Motion-corrected micrographs were imported into CryoSPARC v3.0, v3.1, or v3.2 (dataset #1-2, dataset #4 and dataset #3, respectively) for processing. CTF values were estimated with the Patch CTF method and micrographs were rejected based on relative ice thickness, full frame motion, and estimated CTF resolution. The initial volume for template picking was generated on a smaller subset of particles by blob picking. Template-picked particles were extracted with binning and purified in silico through several rounds of 2D classification. Clean particles were subjected to ab initio reconstruction and reconstructions with promising features were subjected to heterogeneous refinement to further classify particles in 3D. Finally, maps showing defined, high-resolution features were refined using a non-uniform refinement algorithm with global CTF refinements correcting for beam-tilt55.
Turnover datasets #5-11 were fully processed in CryoSPARC v3.2-v4.5. Pre-processing of movies was performed on the fly win CryoSPARC Live, and generously template-picked particles were extracted. Large amounts of particles were purified in silico with 2D classification, then bona fide particles were subjected to ab initio reconstruction, several rounds of heterogeneous refinements, and finally non-uniform refinement with global CTF refinements correcting for data collection with beam tilt. In the case of datasets #6-8, conformational heterogeneity was further resolved with 3D classification, and final classes were refined with non-uniform algorithm.
3D Variability Analysis (3DVA) of WT-IrtAB in LMNG (dataset #3) was performed in cryoSparc v3.3.156. The final set of particles from the last non-uniform refinement was used to solve 5 modes filtered at 6 Å resolution. The mode resolving the SID swinging movement was displayed in linear mode with 20 frames.
Model building and refinement
The IF model was built based on the crystallographic model of IrtAB (PDB: 6TEJ) and was fitted into the cryo-EM maps of dataset #1 with Dock in Map tool in Phenix. After visual inspections in Coot the model was polished in ISOLDE57. This model was then used for model building of the other IF structures. The OFoccl models were initially built manually in Coot by using the inward-facing IrtAB crystal structure as a template and finally polished in ISOLDE. Mg2+, Zn2+, waters, and other ligands such as ATP-γ-S, ADP, and ATP were added from the CCP4 monomer library in Coot and fine-tuned in ISOLDE. Geometry restraints for ADP-orthovanadate ligands were generated in the eLBOW tool of Phenix based on isomeric SMILES from the AOV monomer listed in the Ligand Expo ligand library58. The initial 3D structure of Fe-MBT was built using the Molefacture module in VMD59. The initial structure was energy-minimized for 2000 steepest descent steps. Then, Fe-MBT was placed in a POPE membrane and was equilibrated for 100 ns using Gromacs60. A few snapshots from the end of equilibration were taken and converted into the CIF format. One of the snapshots, fitting the EM density the best, was used for the final modeling by generating geometry restraints on the Grade2 v1.6.0 webserver (Global Phasing). For ADP-orthovanadate and Fe-MBT, the polishing step in ISOLDE was skipped since vanadate and iron were not supported by the force field. Before deposition, each model was refined in real space in Phenix against the final maps of the respective datasets.
Structure analysis
RMSD and distance calculations were performed in Chimera v1.15 and ChimeraX v1.5. Figures were prepared in ChimeraX v1.5-861,62,63. Cavity volumes were calculated by a locally implemented 3 V Voss Volume Voxelator, using a 9 Å outer probe radius and an inner probe radius of 1.87 Å, which corresponds to the size of a single methyl group to visualize lipid accessible space64,65. SID movements were visualized and calculated from the 3DVA. The angle was calculated by fitting the crystallographic model of the SID (PDB ID: 6TEK) in the first and last frames and measuring rotation between the fitted models in ChimeraX. Conformational state distribution analysis was performed by ab initio reconstruction of bona fide particles with 5 classes because the differences between IF and OFoccl particles can be discerned at this low resolution.
Docking of mycobactin molecules in the cavity was performed with the Autodock Vina tool within Chimera v1.15 using default parameters66. Iron(III)-bound M. smegmatis mycobactin was drawn in MarvinSketch (Chemaxon https://www.chemaxon.com)) from the isomeric SMILES on PubChem (CID: 135499491). The model of IrtAB and mycobactin was prepared for docking with the Dock Prep tool with default settings, and with the exception of mycobactin, the charge assignment was performed with Gasteiger parameters.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Structural models have been deposited to the Protein Data Bank (PDB) with the accession codes 9FXC, 9G2K, 9G2L, 9G2M, 9FW3, 9G2P, 9G2S, 9G2T, 9GL3, 9G2V, 9G2X, 9G2Y, 9G2Z, 9G36 and 9G37. The cryo-EM data has been deposited in the Electron Microscopy Data Bank (EMDB) with the accession codes EMDB-50848, EMDB-50977, EMDB-50978, EMDB-50979, EMDB-50820, EMDB-50980, EMDB-50982, EMDB-50983, EMDB-51435, EMDB-50985, EMDB-50987, EMDB-50988, EMDB-50989, EMDB-50992 and EMDB-50993. Plasmids and other data that support the findings of this study are available from the corresponding authors upon request. Source data are provided in this paper.
References
Kurthkoti, K. et al. The capacity of mycobacterium tuberculosis to survive iron starvation might enable it to persist in iron-deprived microenvironments of human granulomas. Mbio 8, e01092–17 (2017).
Ratledge, C. Iron, mycobacteria and tuberculosis. Tuberculosis 84, 110–130 (2004).
Ratledge, C. & Ewing, M. The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiology 142, 2207–2212 (1996).
Wells, R. M. et al. Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. Plos Pathog. 9, e1003120 (2013).
Zhang, L. et al. Comprehensive analysis of iron utilization by Mycobacterium tuberculosis. Plos Pathog. 16, e1008337 (2020).
Jones, C. M. et al. Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling. Proc. Natl. Acad. Sci. USA 111, 1945–1950 (2014).
Rodriguez, G. M. & Smith, I. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J. Bacteriol. 188, 424–430 (2006).
Arnold, F. M. et al. The ABC exporter IrtAB imports and reduces mycobacterial siderophores. Nature 580, 413–417 (2020).
Lewinson, O., Orelle, C. & Seeger, M. A. Structures of ABC transporters: handle with care. FEBS Lett. 594, 3799–3814 (2020).
Ryndak, M. B., Wang, S. S., Smith, I. & Rodriguez, G. M. The Mycobacterium tuberculosis high-affinity iron importer, IrtA, contains an FAD-binding domain. J. Bacteriol. 192, 861–869 (2010).
Rodriguez, G. M., Voskuil, M. I., Gold, B., Schoolnik, G. K. & Smith, I. ideR, an essential gene in Mycobacterium tuberculosis: Role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70, 3371–3381 (2002).
Thomas, C. et al. Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 594, 3767–3775 (2020).
Sun, S. et al. Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB. Protein Cell 2023 14, 448–458 (2023).
Choudhury, H. G. et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl. Acad. Sci. USA 111, 9145–9150 (2014).
Mi, W. et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233 (2017).
Galazzo, L. et al. The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells. Sci. Adv. 8, eabn6845 (2022).
Hofmann, S. et al. Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571, 580–583 (2019).
Fan, C. & Rees, D. C. Glutathione binding to the plant AtAtm3 transporter and implications for the conformational coupling of ABC transporters. Elife 11, e76140 (2022).
Lin, D. Y., Huang, S. & Chen, J. Crystal structures of a polypeptide processing and secretion transporter. Nature 523, 425–430 (2015).
Hu, W., Parkinson, C. & Zheng, H. Mechanistic insights revealed by YbtPQ in the occluded state. Biomolecules 14, 322 (2024).
Wang, Z. M., Hu, W. X. & Zheng, H. J. Pathogenic siderophore ABC importer YbtPQ adopts a surprising fold of exporter. Sci. Adv. 6, eaay7997 (2020).
Rulı́šek, L. R. & Vondrášek, J. Coordination geometries of selected transition metal ions (Co2+ , Ni2+ , Cu2+ , Zn2+ , Cd2+ , and Hg2+) in metalloproteins. J. Inorg. Biochem. 71, 115–127 (1998).
Lee, B. C., Chu, T. K., Dill, K. A. & Zuckermann, R. N. Biomimetic nanostructures: creating a high-affinity zinc-binding site in a folded nonbiological polymer. J. Am. Chem. Soc. 130, 8847–8855 (2008).
Gee, K. R., Zhou, Z. L., Qian, W. J. & Kennedy, R. Detection and imaging of zinc secretion from pancreatic beta-cells using a new fluorescent zinc indicator. J. Am. Chem. Soc. 124, 776–778 (2002).
Polyhach, Y., Bordignon, E. & Jeschke, G. Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 13, 2356–2366 (2011).
Hutter, C. A. J. et al. The extracellular gate shapes the energy profile of an ABC exporter. Nat. Commun. 10, 2260 (2019).
Jin, M. S., Oldham, M. L., Zhang, Q. & Chen, J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490, 566–569 (2012).
Kodan, A. et al. Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog. Proc. Natl. Acad. Sci. USA 111, 4049–4054 (2014).
Ho, H. et al. Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 557, 196–201 (2018).
Arndt, M. et al. Structural basis for the activation of the lipid scramblase TMEM16F. Nat. Commun. 13, 6692 (2022).
Roland, B. P. & Graham, T. R. Decoding P4-ATPase substrate interactions. Crit. Rev. Biochem. Mol. Biol. 51, 513–527 (2016).
Lambert, E., Mehdipour, A. R., Schmidt, A., Hummer, G. & Perez, C. Evidence for a trap-and-flip mechanism in a proton-dependent lipid transporter. Nat. Commun. 13, 1022 (2022).
Martinez-Molledo, M., Nji, E. & Reyes, N. Structural insights into the lysophospholipid brain uptake mechanism and its inhibition by syncytin-2. Nat. Struct. Mol. Biol. 29, 604 (2022).
Falzone, M. E. et al. Nimigean, and A. Accardi, Structural basis of Ca-dependent activation and lipid transport by a TMEM16 scramblase. Elife 8, e43229 (2019).
Paulino, C., Kalienkova, V., Lam, A. K. M., Neldner, Y. & Dutzler, R. Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature 552, 421–425 (2017).
Perez, C. et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433 (2015).
Meier, G. et al. Deep mutational scan of a drug efflux pump reveals its structure-function landscape. Nat. Chem. Biol. 19, 440–450 (2023).
Sajid, A. et al. Reversing the direction of drug transport mediated by the human multidrug transporter P-glycoprotein. Proc. Natl. Acad. Sci. USA 117, 29609–29617 (2020).
Dawson, R. J. & Locher, K. P. Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006).
Serafini, A., Pisu, D., Palu, G., Rodriguez, G. M. & Manganelli, R. The ESX-3 secretion system is necessary for iron and zinc homeostasis in Mycobacterium tuberculosis. PLoS ONE 8, e78351 (2013).
Siegrist, M. S. et al. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. USA 106, 18792–18797 (2009).
Dow, A. et al. Zinc limitation triggers anticipatory adaptations in Mycobacterium tuberculosis. PLoS Pathog. 17, e1009570 (2021).
Vicentefranqueira, R., Leal, F., Marin, L., Sanchez, C. I. & Calera, J. A. The interplay between zinc and iron homeostasis in Aspergillus fumigatus under zinc-replete conditions relies on the iron-mediated regulation of alternative transcription units of zafA and the basal amount of the ZafA zinc-responsiveness transcription factor. Environ. Microbiol. 21, 2787–2808 (2019).
Winterbourn, C. C. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol. Lett. 82-83, 969–974 (1995).
Botella, H. et al. Mycobacterial P-1-Type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10, 248–259 (2011).
Boudehen, Y. M. et al. Mycobacterial resistance to zinc poisoning requires assembly of P-ATPase-containing membrane metal efflux platforms. Nat. Commun. 13, 4731 (2022).
Arnold, F. M. et al. A uniform cloning platform for mycobacterial genetics and protein production. Sci. Rep. 8, 9539 (2018).
Zimmermann, I. et al. Synthetic single domain antibodies for the conformational trapping of membrane proteins. Elife 7, e34317 (2018).
Timachi, M. H. et al. Exploring conformational equilibria of a heterodimeric ABC transporter. ELife 6, e20236 (2017).
Hohl, M. et al. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl. Acad. Sci. USA 111, 11025–11030 (2014).
Jeschke, G. MMM: A toolbox for integrative structure modeling. Protein Sci. 27, 76–85 (2018).
Tschaggelar, R. et al. Cryogenic 35GHz pulse ENDOR probehead accommodating large sample sizes: Performance and applications. J. Magn. Reson. 200, 81–87 (2009).
Worswick, S. G., Spencer, J. A., Jeschke, G. & Kuprov, I. Deep neural network processing of DEER data. Sci. Adv. 4, eaat5218 (2018).
Hogben, H. J., Krzystyniak, M., Charnock, G. T. P., Hore, P. J. & Kuprov, I. Spinach – A software library for simulation of spin dynamics in large spin systems. J. Magn. Reson. 208, 179–194 (2011).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Punjani, A. & Fleet, D. J. 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. D Struct. Biol. 74, 519–530 (2018).
Moriarty, N. W., Grosse-Kunstleve, R. W. & Adams, P. D. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. Sect. D Biol. Crystallogr. 65, 1074–1080 (2009).
Humphrey, W., Dalke, A. & Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. Model. 14, 33–38 (1996).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015).
Pettersen, E. F. et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Voss, N. R. & Gerstein, M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 38, W555–W562 (2010).
Kawabata, T. & Go, N. Detection of pockets on protein surfaces using small and large probe spheres to find putative ligand binding sites. Proteins Struct. Funct. Bioinf. 68, 516–529 (2007).
Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Acknowledgements
We acknowledge the support of Piotr Szwedziak and Jannic Debus at the Center for Microscopy and Image Analysis, University of Zurich. We also acknowledge Zorana Krnetic and Sam Therampilly at the Department of Chemistry, University of Zürich, for the performance and technical description of ICP-MS measurements. We thank Cedric Hutter, Jennifer Earp, Alisa Garaeva, Sille Remm, Fabian Ackle, and all members of the Seeger lab for fruitful discussions. Work in the lab of MAS was supported by the European Research Council (ERC) (consolidator grant n° 772190), an SNSF Professorship of the Swiss National Science Foundation (PP00P3_144823), an SNSF Project grant (310030_188817) and a grant of the Novartis Foundation for Medical-Biological Research. Work in the lab of EB was supported by an SNSF project grant (310030E_210454 / 1). I.G. was supported by a Candoc fellowship at the University of Zurich (FK-19-028).
Author information
Authors and Affiliations
Contributions
I.G., F.M.A., and M.A.S. conceived the project. I.G. generated IrtAB variants, prepared all IrtAB samples, collected datasets #5-11, and performed data processing and model building for all datasets. A.R.M. generated the model for Fe-MBT. I.G. and N.P.L. performed ATPase activity and radioactive uptake assays. I.G. established and performed the ICP-MS measurements for metals and the fluorescent zinc binding assay. S.S. collected datasets #1-4 and provided cryo-EM training for I.G. L.G. performed DEER measurements with samples prepared by IG. All authors discussed the results. I.G. and M.A.S. wrote the paper. L.G., N.P.L., and E.B. edited the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Damian Ekiert, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gonda, I., Sorrentino, S., Galazzo, L. et al. The mycobacterial ABC transporter IrtAB employs a membrane-facing crevice for siderophore-mediated iron uptake. Nat Commun 16, 1133 (2025). https://doi.org/10.1038/s41467-024-55136-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55136-7