Abstract
Oxidative modifications can disrupt protein folds and functions, and are strongly associated with human aging and diseases. Conventional oxidation pathways typically involve the free diffusion of reactive oxygen species (ROS), which primarily attack the protein surface. Yet, it remains unclear whether and how internal protein folds capable of trapping oxygen (O2) contribute to oxidative damage. Here, we report a hidden pathway of protein damage, which we refer to as O2-confined photooxidation. In this process, O2 is captured in protein cavities and subsequently converted into multiple ROS, primarily mediated by tryptophan residues under blue light irradiation. The generated ROS then attack the protein interior through constrained diffusion, causing protein damage. The effects of this photooxidative reaction appear to be extensive, impacting a wide range of cellular proteins, as supported by whole-cell proteomic analysis. This photooxidative mechanism may represent a latent oxidation pathway in human tissues directly exposed to visible light, such as skin and eyes.
Similar content being viewed by others
Introduction
Protein integrity in living organisms is susceptible to nonenzymatic posttranslational modifications, such as oxidation, which occur under both physiological and pathological conditions1,2. Protein quality control systems, primarily involving the proteasomal and lysosomal pathways, facilitate the removal of modified proteins that are misfolded and dysfunctional3,4. Nevertheless, elevated oxidative stress resulting from redox imbalance can lead to the accumulation of defective proteins, causing cellular dysfunction and contributing to aging and diseases like neurodegenerative disorders and cancers4,5,6,7,8,9,10.
The main processes of protein oxidation begin with the conversion of dissolved O2 into reactive oxygen species (ROS), such as superoxide radical (O2•–), hydrogen peroxide (H2O2), hydroxyl radical (•OH), and singlet oxygen (1O2)6,10,11. These ROS then freely diffuse within cells or solutions, primarily leading to the oxidation of easily accessible protein surface residues12,13,14,15,16,17. Consequently, exposed surfaces of proteins are particularly prone to oxidative damage12,13,14,15,16,17. Endogenous ROS are typically produced as natural byproducts of cellular metabolism like aerobic respiration18,19,20. Exposure to light, especially in the ultraviolet-visible (UV-VIS) spectrum, can also generate ROS through conventional photochemical reactions, such as electron transfer (type I) or energy transfer (type II) mechanisms, facilitated by endogenous photosensitizers21,22.
It is worth noting that intricate protein structures can contain internal spaces capable of accommodating small gas molecules like oxygen (O2), leading to direct O2 binding without the need for transition metal ions23,24,25,26. Given that O2 is substantially dissolved in human tissues up to ∼13% (cf. ∼21% in the atmosphere)27, the O2-binding events may significantly impact protein integrity in cells. However, the specific effects of O2 binding on protein oxidation and potential damage remain elusive.
In this work, we describe a distinct pathway of protein damage that we refer to as O2-confined photooxidation, where O2 enters the internal spaces of proteins and ultimately oxidizes the buried residues through light-induced ROS generation. Our findings contrast with the prevailing view that stable protein folding protects inner residues from oxidative attacks12,13,14,15,16,17. Using a single-molecule tweezer approach under blue light irradiation, we find that contrary to expectations, oxidative protein damage is facilitated even in its folded state, which conceals a substantial portion of oxidizable atoms. The primary cause of this unusual protein damage appears to be the significant oxidation of protein core residues near the trapped O2 molecules within protein cavities, as supported by mass spectrometry analysis, molecular dynamics (MD) simulations, and protein cavity analysis. The oxidation of buried residues likely arises from the attack by powerful ROS, such as 1O2 and •OH, which are converted from the trapped O2, as supported by spectrophotometric assays, electron paramagnetic resonance (EPR) spectroscopy, density functional theory (DFT) calculations, and Monte Carlo analysis. Proteomic and structural analyses of oxidized proteins in HeLa cells suggest that this photooxidative reaction may affect a broad range of cellular protein classes, beyond the specific model proteins used here. By integrating experimental, computational, and bioinformatic approaches, this study unveils a hidden pathway of protein damage via O2-confined photooxidation, offering significant implications for redox proteostasis.
Results
Declining protein foldability during cyclic unfolding
This study was prompted by the unexpected observation of a rapid decline in protein foldability under blue light irradiation (λpeak = 447 nm; Supplementary Fig. 1 for the full spectrum), using a recently developed single-molecule tweezers28 (Fig. 1a and “Methods”). This tweezer approach demonstrated high stability, enabling the capture of ∼1000 reversible unfolding events in a single protein28. We employed this method to repetitively unfold and refold maltose-binding protein (MBP), a widely-used model protein in protein folding studies, over hundreds of cycles.
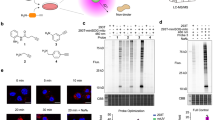
a Schematics of our single-molecule magnetic tweezers. All single-molecule experiments were performed under blue light (λpeak = 447 nm) except for the IR-light condition (λpeak = 860 nm) and the light-off condition in (e). b Representative force-extension curves at different numbers of unfolding cycles. Foldability trend over time under different pulling conditions (c), under different oxidative stress conditions (d), or under different light conditions (e) (n = 5–12 molecules). The symbols V and T represent the magnet speed and the waiting time between unfolding cycles (V = 0.3 mm/s and T = 2.5 min for the control condition). f Measurement of primary reactive oxygen species (ROS; H2O2, •OH, O2•–, and 1O2) in the foldability measurement condition (see “Methods”). The data are normalized to those in no-light condition and presented as mean ± SD (n = 3 independent samples for each condition). One-way ANOVA with post hoc Tukey HSD test (*** for p < 0.001, n.s. for p > 0.05). g Foldability trend over time under long incubation conditions in specific protein states. In the U-state condition, the U state is maintained at 35 pN for 90 min in every unfolding cycle (∼95% of total experimental time at the U state). In the N-state condition, the N state is maintained at 1 pN for the initial 6 h before the cyclic unfolding (n = 5–12 molecules). h Proportion of water-exposed atoms in the N and U states for three different atom groups (n = 30 data points from three MD simulation trajectories; mean ± SD). Relevant source data are provided as a Source Data file.
During each unfolding cycle, the force was applied in the range of 1 pN to 50 pN and then immediately relaxed back to 1 pN, followed by a 2.5-min waiting period to allow for complete refolding (Fig. 1b and Supplementary Figs. 2 and 3). In the initial several cycles, two distinct unfolding steps were clearly observed: a partial unfolding at lower forces followed by the complete unfolding of the remaining structure at higher forces (Fig. 1b and Supplementary Fig. 3a, b). This unfolding pattern is consistent with previous studies29,30, where the partial and complete unfolding steps correspond to the unfolding of the external α-helices at the C-terminus and the more stable core structure, respectively (Fig. 1a and Supplementary Figs. 2e and 3c).
As the number of unfolding cycles increased, however, the unfolding pattern became progressively inconsistent and difficult to define (Fig. 1b). This observation reflects a decrease in foldability to the native state. To quantitatively assess the declining foldability during the cyclic unfolding, we analyzed the proportion of normal unfolding patterns (i.e., the unfolding of the external α-helices followed by the unfolding of the core structure) at each number of unfolding cycles, based on data from multiple molecules (Fig. 1c and Supplementary Fig. 3d). The protein foldability was found to decline to zero during ∼200 cycles, corresponding to a decay with a time constant (τ) of 207 ± 7 min. Throughout all the cycles, refolding was driven by relaxing the stretched state of the identical fully unstructured polypeptide at 50 pN, which serves as the starting point for folding into the tertiary protein structure. Thus, our result suggests a gradual increase in defects at the level of amino acid residues.
Unusual photooxidative damage as main cause of protein foldability decay
Given the same initial state of the unstructured polypeptide for each refolding attempt, the question arises as to what factors contribute to the observed foldability decay. To elucidate the underlying causes, we first examined the potential influence of the repetitive pulling process itself (Supplementary Fig. 3a). We compared the trends in the foldability upon a large change in only one of the two pulling parameters: the magnet speed modulating the pulling speed and the waiting time between pulling cycles. For all conditions, we found that the decay rates of foldability were comparable with τ = 207–226 min (Fig. 1c). This result indicates that the declining foldability during the cyclic unfolding is irrelevant to the repetitive process of pulling and unfolding.
We hypothesized that the accumulation of oxidative modifications causes a gradual decay in protein foldability over time. To test this hypothesis, we assessed the effect of varying levels of oxidative stress on protein foldability (Fig. 1d and Supplementary Figs. 3a and 4). Indeed, a vacuum-degassed or N2 gas-bubbled buffer with reduced O2 concentration delayed the foldability decay with τ = ∼456 or 357 min, respectively. In contrast, when the protein was exposed to an O2 gas-bubbled buffer with an elevated O2 concentration, the foldability was dramatically reduced to ∼53% even at the first unfolding. 1% H2O2, a relatively mild ROS, also largely impaired the foldability to ∼26% at the first unfolding, with an accelerated decay rate of τ = ∼22 min. We also observed a delay in the foldability decay under no-light or infrared (IR)-light conditions (Fig. 1e). During the cyclic unfolding experiment, we turned the light off for the initial 6 h (no data collected during this period due to the absence of light for imaging) and then turned it back on to see how the foldability decay was affected by blue light. We found that compared to the light exposure condition, the foldability level was enhanced under the no-light condition, similar to what was observed with the vacuum-degassed buffer (Fig. 1e). Under IR light with λpeak = 860 nm, even at higher power, the foldability decay rate also slowed to a similar extent as in the vacuum-degassed buffer (Fig. 1e and “Methods”). These results collectively indicate that the foldability decay is primarily attributed to photooxidative damage under blue light irradiation.
In cellular and tissue environments, various biological oxidants, such as ROS, are generated during metabolic reactions like the electron transport chain reaction18,19,20. However, no such reactions occur in our in vitro experiments. Furthermore, there are no exogenous photosensitizers, transition metal ions like Fe2+ essential for Fenton’s reaction, or high-energy ionizing radiation. As expected, the background levels of primary ROS—such as H2O2, •OH, O2•–, and 1O2—were negligible under our photoirradiation condition, similar to those observed under no-light condition (Fig. 1f and “Methods”).
Under this condition of negligible ROS levels, we observed an unexpected finding—a rapid foldability decay even in the fully folded state (N state), which conceals numerous oxidizable atoms. Despite maintaining the N state at 1 pN for the initial 6 h (no data collected during this period due to the absence of cyclic unfolding), the foldability level still exhibited a significant decrease, similar to the control result with τ = ∼207 min (Fig. 1g). The foldability decrease in the N-state condition was even slightly faster than that in the U-state condition (τ = ∼293 min), where the fully unstructured polypeptide state (U state) was maintained at 35 pN for an extended period of 90 min during each unfolding cycle (∼95% of total experimental time at the U state) (Fig. 1g). This result contradicts the generally accepted notion that the compact, native state (N state), with a large portion of oxidizable atoms buried, is less vulnerable to oxidative damage compared to the unstructured polypeptide state (U state), where all oxidizable atoms are exposed to water12,13,14,15,16,17. Indeed, the proportion of water-exposed, oxidizable atoms in the N state was estimated as 2–3 times lower than in the U state, regardless of analyzed atom groups (Fig. 1h and Supplementary Fig. 5). This unusual photooxidative damage under the negligible ROS background appears puzzling, implying that there may be a hidden, unknown pathway of protein oxidation.
Substantial oxidation near trapped O2 molecules within protein cavities
We thus conceived an alternative oxidation pathway that could account for the observed results in the single-molecule tweezer experiments—dissolved O2 molecules become trapped within the native protein structure, promoting oxidative damage through close interactions within the diffusion-suppressing, protein core region. According to this oxidation pathway, amino acid residues inside the protein are expected to undergo more pronounced oxidation than those on the protein surface. To pinpoint the positions of oxidized residues, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis for our model protein, MBP, exposed to blue light (“Methods” and Supplementary Table 1). From a volcano plot, we identified 18 oxidized residues under blue light irradiation, which were statistically significant compared to no-light condition (Fig. 2a and Supplementary Fig. 6). Indeed, a large portion of the identified residues are located in the protein core region, accounting for 64% of their side chain atoms (Fig. 2b). This high proportion aligns with the prediction from the alternative oxidation scenario.
a Volcano plot obtained from LC-MS/MS analysis. The volcano plot shows oxidized residues of maltose-binding protein (MBP) under blue light irradiation, which are statistically significant compared to no-light condition (p value (p) < 0.05, fold change (FC) > 1.5). p values were calculated using Student’s one-tailed t test. The amino acid residues are denoted as 3-letter code. The residue numbers indicate those following the signal peptide sequence. b Structural distribution of the oxidized residues. c Barcode plot showing the binding events between MBP and O2 molecules from molecular dynamics (MD) simulations. The blue-colored regions indicate the bindings between the protein and O2, sustained within 3.5 Å for longer than 10 ns. d All distributed positions of trapped O2 molecules in simulation #1. Only the O2 molecules trapped for a specific time range (t) are represented. e All distributed center positions of protein cavities with Dcavity > Do in simulation #1. The Dcavity and Do represent the diameter of cavities and the kinetic diameter of O2 (3.46 Å), respectively.
We then turned to all-atom MD simulations to investigate the potential trapping of O2 molecules within the protein (“Methods”). Sustained and close interactions between MBP and O2 within 3.5 Å for over 10 ns (or 5 ns) were observed in three 1-μs MD trajectories (Fig. 2c), where O2 molecules were predominantly located inside the protein (Fig. 2d, Supplementary Figs. 7 and 8 and Supplementary Movie 1). The O2-trapping events were also found to be energetically feasible, based on free energy calculations (Supplementary Figs. 9–11 and “Methods”). Notably, the oxidized residues are in close proximity to the trapped O2 molecules (<3.5 Å and >10 ns), within a range of 2.3–4.2 Å in their minimum distances (Supplementary Fig. 7e). The O2-trapping areas also largely overlap with the protein cavity areas analyzed from the MD trajectories, mostly within 2 Å (Fig. 2e, Supplementary Figs. 7 and 8 and “Methods”). These cavities are small spaces within the protein but are large enough to capture O2 (Dcavity > Do = 3.46 Å, the kinetic diameter of O2)31,32. Thus, the results of LC-MS/MS analysis, MD simulations, and protein cavity analysis collectively support the alternative oxidation pathway, which involves the trapping of O2 within the protein cavities.
The primary O2-trapping areas (Supplementary Figs. 9 and 10 and Supplementary Tables 2 and 3) are deeply located from the protein surface (Supplementary Fig. 12a, b), and contain a large portion of hydrophobic residues even more than the protein core region (Supplementary Fig. 12c). A hydropathy analysis shows that the O2-trapping areas also exhibit larger hydrophobicity than nearly all successive sequence blocks with an arbitrary 15-residue window (Supplementary Fig. 12d–f). This analysis indicates that the distant residues in the sequence space would form the hydrophobic O2 pockets. Additionally, the O2-trapping times in MBP are comparable to or even longer than the binding times of its natural substrate, maltose (Supplementary Figs. 13–17 and Supplementary Movies 1 and 2). These results suggest that the capture of nonpolar O2 molecules is mainly mediated by hydrophobic interactions within the cavity-forming areas.
Trp-mediated ROS generation under blue light irradiation
O2 is recognized as kinetically stable33, and thus direct oxidation by O2 is unlikely to occur, even with the local confinement and close contacts. It is noteworthy that under ultraviolet (UV) light, aromatic amino acids such as Trp can function as endogenous photosensitizers capable of generating ROS21,22,34. However, the UV absorption spectrum of MBP at ∼280 nm, primarily mediated by Trp, is almost entirely separated from our blue light spectrum at ∼450 nm (Supplementary Fig. 1a). Intriguingly, a recent study has shown that when O2 is captured within a close distance of ∼3.5 Å in a photosensitizing polymer, it can be converted to 1O2 through a distinct photochemical mechanism involving spin-flip-based electron transfers35. Hence, using DFT calculations, we first examined whether the blue light irradiation in our experiments could induce the generation of 1O2 through this mechanism and identified which residue is involved.
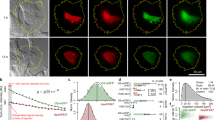
We conducted mixed-reference spin-flip time-dependent DFT calculations35,36 for each residue-O2 pair in four O2-trapping subareas exhibiting relatively tight O2 binding (Fig. 3a–d, Supplementary Fig. 10, Supplementary Table 3 and “Methods”). In subarea 1, only the Trp62-O2 pair showed noticeable absorption intensity in the blue light region at ∼445 nm (Fig. 3b), closely matching the peak wavelength of 447 nm used in our experiments. Likewise, the Trp129-O2 pair in subarea 3 also exhibits similar absorption behavior at ∼480 nm (Supplementary Fig. 18). The ground electronic state of the Trp62-O2 is the T0 state (1Trp62-3O2), with each π* orbitals of O2 singly occupied (Fig. 3d). The two lowest triplet excited states (TCT; Trp62•+-O2•–) are nearly degenerate, corresponding to the charge transfer (CT) excitation from Trp62 to the two respective π* orbitals of O2 (Fig. 3c, d). The energy level of ∼2.7 eV for the T0 → TCT transition suggests that the Trp62-O2 can be excited by blue light at ∼450 nm, which is equivalent to ∼2.8 eV in energy (Fig. 3c). Additionally, the singlet excited states (SCT) are also nearly degenerate, and their energy level of ∼2.8 eV is very close to that of the TCT states (Fig. 3c, d). This close match in the excited-state energy levels indicates that efficient intersystem crossing (ISC) from the TCT to SCT states can occur, involving an electron spin-flip (Fig. 3d). The SCT states can then undergo internal conversion to the S3 state (1Trp62-1O2(1Σg+)) followed by the S1/S2 states (1Trp62-1O2(1Δg)), resulting in the generation of 1O2 (Fig. 3d).
a O2-trapping subareas with relatively tight O2 binding (see “Methods”). In each subarea, the colored residues represent those whose atoms are located within 3.5 Å from O2 atoms during the binding periods. b Absorption spectra of residue-O2 pairs in O2-trapping subarea 1 from density functional theory (DFT) calculations. The residues are denoted as 3-letter code. The residue numbers indicate those following the signal peptide sequence. The inset shows the Trp62-O2 structure with the maximum oscillator strength. c Relative energy diagram for singlet and triplet states of Trp62-O2 pair. The molecular orbitals are shown for the transition from T0 to TCT state. d Molecular orbital diagrams and overall pathway for 1O2 generation. e ABDA assay for 1O2 detection (n = 3 independent samples for each condition; mean ± SD). The gray-shaded box indicates the region of absorbance for amino acids other than Trp. f Electron paramagnetic resonance (EPR) measurement using TEMP for 1O2 detection. The EPR spectra are averaged from triplicates for each condition and are expressed in arbitrary unit (a.u.). The inset shows the EPR intensity for each condition (n = 3 independent samples for each condition; mean ± SD). One-way ANOVA with post hoc Tukey HSD test (***p = 0.0004, **p = 0.007, and *p = 0.03). g DHR123 assay for the detection of type I reactive oxygen species (ROS), such as O2•–, H2O2, and •OH (n = 3 independent samples for each condition; mean ± SD). The gray-shaded box indicates the region of absorbance for amino acids other than Trp. h HPF assay which is more specific to •OH (n = 3 independent samples for each condition; mean ± SD). Relevant source data are provided as a Source Data file.
We confirmed the Trp-mediated 1O2 production using a spectrophotometric assay and EPR spectroscopy (“Methods”). For Trp dissolved in H2O containing ABDA as a 1O2 probe37,38, we observed a gradual decrease in absorbance of ABDA at 400 nm under blue light irradiation, indicating 1O2-induced ABDA oxidation (Fig. 3e and Supplementary Fig. 19). In contrast, other tested amino acids (aromatic: Phe, charged: Glu, hydrophobic: Ala) did not show noticeable changes in the ABDA absorbance (Fig. 3e and Supplementary Fig. 19). The same trend was also observed in D2O, which increases the lifetime of 1O2 (ref. 38), albeit with a slightly lower extent of change for Trp (Fig. 3e). This result may be attributed to a higher rate of oxidative degradation of Trp by 1O2 due to its increased lifetime22, resulting in a reduced amount of intact Trp available for 1O2 generation. In the EPR measurements using TEMP, commonly used for 1O2 detection37,39, we observed the greatest intensity of the characteristic trilinear signals of TEMPO (the oxidized product of TEMP) under the Trp+/light+ condition, compared to the control groups under Trp+/light− or Trp−/light+ conditions (Fig. 3f). The EPR measurement samples contained no other exogenous molecules that could react with TEMP, such as peroxymonosulfate and peracetic acid39. Thus, the possibility of false positives is unlikely under our measurement conditions, especially given the suitable controls. The two independent spectrophotometric and EPR experiments indicate that Trp can generate 1O2 under blue light, consistent with the DFT calculations.
The spin-flip-based electron transfer pathway occurs only during transient, short-distance interactions between Trp and O2, inherently limiting the detection of a relevant absorbance peak in the blue light region. On the other hand, the direct absorption of Trp without O2 interactions in the UV region shows a tail extending into the blue light region, despite the very low absorption level—only ∼1/2000 of the UV peak at 450 nm (Supplementary Fig. 1b). Thus, we cannot entirely rule out the involvement of the conventional electron transfer (type I) pathway through direct Trp excitation. However, the conventional energy transfer (type II) pathway would still remain highly unlikely due to the very low, if any, photoluminescence of Trp in the blue light region, as well as the large spectral separation in the energy transfer to O2, which extends into the IR light region (760–1300 nm)37,40,41,42.
In particular, the type I pathway generates multiple types of ROS, such as O2•–, H2O2, and •OH. We confirmed Trp-mediated production of type I ROS using two spectrophotometric assays: the less specific DHR123 assay, which provides broad detection coverage of type I ROS, including O2•–, H2O2, and •OH (refs. 43,44,45), and the more specific HPF assay for •OH, which is approximately 90–360 times more sensitive than for O2•– and H2O2 (ref. 46) (Fig. 3g, h and “Methods”). In the DHR123 assay, Trp showed a more prominent increase in the DHR123 fluorescence at 530 nm under blue light compared to other tested amino acids, suggesting that type I ROS can also be generated by Trp (Fig. 3g). The spin-flip-based electron transfer pathway also generates O2•– as an intermediate, which may have contributed to the result of the DHR123 assay. In the HPF assay, only Trp exhibited a dramatic increase in the HPF fluorescence at 515 nm under blue light, suggesting that type I ROS, more specifically •OH, can also be generated by Trp (Fig. 3h).
Although the HPF assay can also respond to peroxynitrite (ONOO–), it is six times more specific to •OH than to ONOO– (ref. 46). Additionally, ONOO– is primarily generated in more complex systems such as living cells47. In cellular environments, nitric oxide (NO) and O2•– are produced during arginine metabolism and cellular respiration18, respectively. These two ROS then react to form ONOO– (ref. 18). In contrast, our in vitro system is very simple, containing only deionized water and an amino acid, with no catalytic or enzymatic reactions. Given the higher specificity of the HPF assay for •OH and the simplicity of our detection system, it is highly likely that the HPF assay results are primarily due to •OH. All our experiments and calculations described in this section collectively support the possibility of blue light-induced generation of various ROS, including 1O2 and •OH, near Trp residues through multiple photochemical processes.
Positional correlation between Trp and oxidized residues
Since Trp residues seem to be primarily involved in ROS production under blue light, the oxidized residues of the model protein MBP detected in the LC-MS/MS analysis are likely in close proximity to them. Despite this apparent proximity manifested in the residue’s structural distribution (Fig. 4a), we also conducted Monte Carlo analysis on a 1-μs MD trajectory of MBP to quantitatively assess the degree of proximity (“Methods”). By randomly sampling residues from each amino acid type in MBP (n = 1–8, with n = 8 representing the benchmark number of Trp residues), we calculated the average distance between the sampled residues and the oxidized residues across all possible combinations. This process was iterated to construct probability density functions for the average distance (PDFavg; n = 103 for each sampling number of residues). Remarkably, Trp emerged as the closest residue to the oxidized residues among all amino acid types (Fig. 4b and Supplementary Fig. 20).
a Structural distribution of tryptophan (Trp) residues and oxidized residues in maltose-binding protein (MBP). The residue numbers indicate those following the signal peptide sequence. b–d Monte Carlo analysis for the proximity between Trp and oxidized residues (see “Methods”). The b, c show the probability density functions of average distance from oxidized residues to each residue type (PDFavg) (b) and to arbitrary residues sampled from the entire protein (c), respectively. The data for other residue types in b are distinguishably presented in Supplementary Fig. 20. In c, the occurrence probability of the proximity between Trp and oxidized residues by chance is indicated for each case of increasing sampling numbers of arbitrary residues, up to the total number of benchmark Trp residues in MBP (n = 8). The d shows the occurrence probabilities of the proximity by chance for all residue types, which have a larger number of residues than the total number of benchmark Trp residues in MBP. The proportion of each residue in the protein is shown in Supplementary Fig. 12c. The residues are denoted as 3-letter code. e Minimum distances from oxidized residues to Trp residues. f, g Maximum end-to-end distances (f) and total travel lengths (g) of trapped O2 molecules within the protein obtained from molecular dynamics (MD) simulations.
To rule out the possibility of coincidence in this statistical analysis, we compared the average distances calculated for Trp with those for arbitrary residues sampled from the entire protein, regardless of amino acid type (Fig. 4c; n = 106 for each sampling number of residues). As the number of sampled residues increases, the average distance for Trp approaches the top 0.2% in the PDFavg constructed from the arbitrary residue sampling (Fig. 4c). This indicates only a 0.2% chance that the close proximity of oxidized residues to Trp is due to random chance. In general, hydrophobic residues tend to show relatively close proximity due to their prevalence in the protein core region, where the oxidized residues are largely located (Supplementary Fig. 20). However, the probabilities of these proximities occurring by chance range from 7% to 48%, with Trp being a notable exception (Fig. 4d). This Monte Carlo analysis indicates that the close proximity between Trp and oxidized residues is unlikely to occur accidentally. Instead, it suggests a probable correlation in their positions, reconciling Trp-mediated ROS generation with the subsequent oxidation of nearby residues. In addition, the individual oxidized residues are located within ∼2 nm of the nearest Trp residues (Fig. 4e). This distance range falls within the maximum end-to-end distances and total travel lengths of trapped O2, reaching up to ∼3 nm and tens of nm, respectively (Fig. 4f, g and Supplementary Movie 1). These estimated diffusion distances for constrained O2 are roughly considered to be the maximum diffusion ranges of constrained ROS if no reaction occurs en route.
The positional correlation between Trp and oxidized residues was also examined using an additional suitable protein. We selected human carbonic anhydrase II (CA II) because it has cavity structures that can accommodate O2 molecules, albeit to a lesser extent than MBP, as well as several Trp residues in its interior (Fig. 5a and “Methods”). Indeed, the positions of trapped O2 molecules, which closely interact with CA II within 3.5 Å over 10 ns (or 5 ns), largely overlap with the protein cavity areas, both analyzed from a 1-μs MD trajectory (Fig. 5a, b and “Methods”). Additionally, all three oxidized residues under blue light, identified by LC-MS/MS analysis, are predominantly located in the protein core region, with 91% of their side chain atoms residing in this area (Fig. 5a–c, Supplementary Fig. 21a, Supplementary Table 1 and “Methods”). The oxidized residues are in close proximity to both the trapped O2 molecules (<3.5 Å and >10 ns) and Trp residues, with minimum distances of 0.25–0.35 nm and 0.5–1.4 nm, respectively (Fig. 5d). Moreover, the range of proximity to Trp falls within the maximum end-to-end distances of trapped O2 (and roughly ROS as well; see above), reaching up to ∼2.3 nm (Fig. 5e).
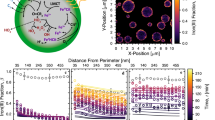
a Structural distribution of oxidized residues, tryptophan (Trp) residues, protein cavities, and trapped O2 molecules in human carbonic anhydrase II (CA II). Only the O2 molecules trapped for a specific time range (t) are represented. The Dcavity and Do represent the diameter of cavities and the kinetic diameter of O2 (3.46 Å), respectively. b Close-up views of the regions surrounding the oxidized residues. c Proportion of oxidized residues’ side-chain atoms located in the protein core and surface regions. d Minimum distances from oxidized residues to trapped O2 molecules or Trp residues. e Maximum end-to-end distances of trapped O2 molecules within the protein obtained from molecular dynamics (MD) simulations. f, g Monte Carlo analysis for the proximity between Trp and oxidized residues (see “Methods”). The f shows the probability density functions of average distance from oxidized residues to each residue type (PDFavg). The data for other residue types in f are distinguishably presented in Supplementary Fig. 21b. Trp* represents a subset of Trp residues (Trp97, Trp192, Trp209, and Trp245), which are responsible for the leftmost peak in the PDFavg for Trp (see the main text for more details). The PDFavg shown in blue is constructed from the sampling of Trp*. The g shows the occurrence probabilities of the proximity by chance for all residue types, which have a larger number of residues than the number of benchmark Trp* residues.
Similar to our analysis of MBP, we performed Monte Carlo analysis on a 1-μs MD trajectory of CA II to evaluate how unlikely the proximity of oxidized residues to Trp is by random chance (“Methods”). The identification of only three oxidized residues may indicate that only a small number of Trp residues are involved in residue oxidation in CA II. This conjecture is reasonable because unlike in MBP, certain Trp residues (Trp5, Trp16, and Trp123) in CA II are relatively distant from the trapped O2 molecules and the cavity areas (Fig. 5a). Indeed, when sampling from all Trp residues (n = 7), the overall proximity between Trp and oxidized residues is not particularly prominent, comparable to other types of hydrophobic residues (Fig. 5f and Supplementary Fig. 21b). However, there is an additional, close peak in the PDFavg for Trp, which exactly matches the peak of the PDFavg for Trp*, the other group of Trp residues (Trp97, Trp192, Trp209, and Trp245) (Fig. 5f). These Trp residues are the closest to the oxidized residues and are also close to the trapped O2 molecules and the cavity areas (Fig. 5a, b). Notably, the proximity of oxidized residues to Trp* has an exceptionally low probability of occurring by chance (0.2%), consistent with observations in MBP (Fig. 5f, g, Supplementary Fig. 21c and “Methods”). These results with CA II further support the role of Trp as a key residue in ROS generation and nearby residue oxidation.
Potential universality of O2-confined photooxidation
Synthesizing all our results up to this point, we propose an O2-confinement pathway for oxidative protein damage (O2-confined photooxidation), consisting of three main steps (Fig. 6): (1) the confinement of O2 molecules within protein cavities, (2) Trp-mediated ROS production under blue light irradiation, and (3) the oxidation of nearby residues by ROS through constrained diffusion within the protein. As we elucidate this photooxidation pathway based on experimental and computational results from two specific model proteins (MBP and CA II), an additional important question arises—how widespread could this oxidative reaction be in proteins? To address this, we performed whole-cell proteomic analysis of oxidized proteins in HeLa cells, as well as structural analysis of the human and E. coli proteomes.
Dissolved oxygen (O2) is trapped within protein cavities containing tryptophan (Trp) as an endogenous photosensitizer. Upon exposure to blue light (400–500 nm), the trapped O2 is converted to reactive oxygen species (ROS), such as singlet oxygen (1O2) and hydroxyl radical (•OH), through interactions with Trp. The generated ROS oxidizes nearby residues through constrained diffusion within the protein. The protein structure represents maltose-binding protein (MBP), one of the model proteins studied in this work.
According to the oxidation mechanism, the O2-confined photooxidation is expected to occur in proteins with Trp-containing cavities larger than the size of O2, facilitating both the capture of O2 and Trp-mediated ROS generation. Using the AlphaFold Protein Structure Database48, we found that the majority of whole proteomes in human and E. coli cells contain cavity structures (n \(\ge\) 1 for the number of cavities), accounting for ∼92% of distinct proteins analyzed (Fig. 7a and “Methods”). While the proportions decrease for proteins with Trp-containing cavities larger than the kinetic diameter of O2 (Do = 3.46 Å)32, they still remain significant, comprising ∼31% of the proteins (Fig. 7a). This trend also holds for a subset of proteomes with experimentally determined structures deposited in the Protein Data Bank (∼95% for those with cavities and ∼42% for those with large Trp-containing cavities) (Fig. 7a). This structural feature of protein cavities appears to be closely linked to the conformational flexibility required for protein function. For example, analyses using the PANTHER classification system show that specific protein classes, such as those with catalytic or transport activities, tend to have relatively large cavities (Dcavity > Do) in high proportions, regardless of the presence of Trp residues (Supplementary Figs. 22 and 23 and “Methods”). These structural analyses of cellular proteomes suggest that the O2-confined photooxidation may have a widespread impact on protein integrity in human and bacterial cells, potentially affecting approximately one in three proteins.
a Proportion of distinct proteins with cavity structures (n \(\ge\) 1 for the number of cavities) in protein structure databases of AlphaFold DB and PDB. The Dcavity, Do, and Trp indicate the diameter of cavities, the kinetic diameter of O2 (3.46 Å), and tryptophan, respectively. b Volcano plot obtained from whole-cell proteomic analysis using LC-MS/MS. The volcano plot shows oxidized proteins in HeLa cells under blue light, which are statistically significant compared to no-light condition (p value (p) < 0.05–0.01 with decrement of 0.01, fold change (FC) > 1.5–3.5 with increment of 0.5). p values were calculated using Student’s one-tailed t test. The areas with varying criteria of p and FC are highlighted with different colors. c Change in proportion of proteins with cavity structures against oxidation level and statistical significance. d Change in proportion of proteins with Trp-containing cavities (Dcavity > Do) against oxidation level and statistical significance across specific protein categories (shown for n \(\ge\) 3). The data for oxidized proteins in c and d correspond to those selected for p < 0.05–0.01 with decrement of 0.01 and FC > 1.5–3.5 with increment of 0.5 in the volcano plot. The data in d are obtained based on the PANTHER classification system.
We further investigated whether O2-confined photooxidation could contribute to the overall oxidation levels of human proteins in a cellular context. To this end, we conducted whole-cell proteomic analysis using LC-MS/MS on HeLa cells (“Methods” and Supplementary Table 1). We first identified oxidized proteins in HeLa cells under blue light irradiation, which were statistically significant compared to no-light condition (Fig. 7b). Intriguingly, as the extent of oxidation increased, the proportion of proteins with large Trp-containing cavities (Rtcc) increased significantly, reaching up to 22% (Fig. 7b, c and Supplementary Fig. 24). However, this trend was not evident in the other protein subsets, such as those with cavities or large cavities (Fig. 7c). These results suggest that proteins with large Trp-containing cavities (Dcavity > Do), which can capture O2 and facilitate Trp-mediated ROS generation, are more susceptible to photooxidative damage. This structure-oxidation correlation implies that the O2-confined photooxidation could be a primary oxidation pathway in human cells under blue light. Utilizing the PANTHER classification system49, we further classified the oxidized proteins based on molecular function, protein class, biological process, cellular anatomical entity, and primary subcellular structure (Fig. 7d and “Methods”; Supplementary Table 4 for classification terminology definitions). Except for a few protein categories, the majority exhibited the same structure-oxidation trend, with ΔRtcc values ranging from approximately +10% to +65% (Fig. 7d).
Despite the overall structure-oxidation trend, the detailed oxidation pattern of cellular proteins appears to be more complex. Deviations from this trend likely reflect the influence of cellular microenvironments characterized by differences in dissolved O2 levels, molecular composition, subcellular organization, and metabolic processes. For instance, a few protein categories in HeLa cells show the opposite trend, with negative ΔRtcc values—including metabolite interconversion enzymes, protein-binding activity modulators, proteins at the cell periphery, and proteins involved in developmental processes, multicellular organismal processes, or cell projections (Fig. 7d and Supplementary Table 4 for classification terminology definitions). For these protein categories, the O2-confined photooxidation may be considered a minor reaction, possibly due to the effects of their specific microenvironments.
Discussion
We present a unique pathway of oxidative protein damage, which we refer to as O2-confined photooxidation, integrating findings from a wide array of experimental and computational approaches. The initial step, O2 capture within protein cavities, is supported by MD simulations, cavity analysis, and LC-MS/MS analysis. The subsequent step, Trp-mediated ROS generation under blue light, is supported by spectrophotometric assays, EPR spectroscopy, DFT calculations, and Monte Carlo analysis. The final step, residue oxidation in the protein interior, is supported by LC-MS/MS analysis and single-molecule tweezer experiments. This oxidation process stands in sharp contrast to conventional oxidation pathways, which involve the free diffusion of ROS in cell or solutions, primarily leading to the oxidation of easily accessible surface residues of proteins12,13,14,15,16,17.
The O2-trapping areas of the model proteins MBP and CA II were identified based on MD simulation trajectories, reflecting the protein’s flexibility and motional dynamics. The locations of these O2-trapping areas closely match the protein cavities large enough to capture O2. While the positional match supports the validity of the simulations and relevant analyses, a well-designed spectroscopic approach could experimentally locate prominent O2-trapping sites. Since O2 molecules are paramagnetic, their presence in the protein core region can be detected via paramagnetic enhancement of the protein’s site-specific spin relaxation rate23,24,25,26.
The oxidized residues of MBP identified in the LC-MS/MS analysis encompass a broad range of amino acid types, including those with aromatic rings and alkyl side chains. This result is likely attributed to various ROS, such as 1O2 and •OH, possibly generated through multiple photochemical pathways (spin-flip and type I), though it remains unclear which pathway is more dominant. The oxidation sensitivity may also be influenced by the distinct environment within the protein, where the diffusion of ROS is restricted. Specifically, the rugged structural shapes of local protein regions may constrain the diffusion paths and modulate the apparent reaction rates for individual residues15,50.
Given the proposed oxidation mechanism, Trp-containing cavity structures are likely to promote oxidative protein damage through O2-confined photooxidation under blue light. This insight allows us to predict inherent vulnerability to blue light-induced damage based on structural analysis. Due to thermal fluctuations in protein conformation, proteins may experience more frequent and broadly distributed O2 trapping, facilitating the photooxidation process. Conversely, this work also provides key considerations for designing more robust proteins with greater resistance to oxidative damage. Reducing the size and number of protein cavities and/or replacing Trp with light-insensitive residues can prevent the O2-confined photooxidation, thereby enhancing overall oxidation resistance.
Approximately 30–40% of proteins in human and E. coli cells possess Trp-containing cavities large enough to capture O2 molecules. This prevalent structural feature hints at the universality of O2-confined photooxidation, further supported by the whole-cell proteomic and structural analyses of HeLa cells. Although the results and interpretations are limited to relatively highly expressed proteins (Supplementary Fig. 24a), the number of proteins detected and analyzed (2915 proteins) is sufficient to assess an overall structure-oxidation trend—the higher the oxidation extent, the greater the proportion of proteins with Trp-containing cavities large enough to capture O2. What drives this trend, which is not intuitively obvious? The O2-confined photooxidation described here offers a plausible explanation based on its oxidation mechanism. Hence, the findings—the prevalence of cavity structures in proteins and the observed structure-oxidation correlation—suggest that O2-confined photooxidation could have a widespread impact on cellular proteins.
It has been reported that blue light irradiation can lead to tissue aging and disorders, particularly in visible light-penetrating tissues such as the skin and eyes51,52,53,54,55. These photodamage effects are thought to result from elevated oxidative stress caused by blue light-induced ROS generation through endogenous photosensitizers51,56,57,58,59,60. The O2-confined photooxidation we propose here introduces an additional layer of oxidation. This pathway involves the initial capture of O2, thereby bypassing antioxidant defense systems that target only freely diffusing ROS, not O2 itself. This photooxidation process may thus represent a hidden pathway within oxidative damage networks, contributing to overall protein damage in tissues directly exposed to visible light. The O2-confinement pathway should be considered particularly significant, given the high levels of blue light emitted by LED lighting and electronic displays55.
Methods
Protein expression and purification
Maltose-binding protein (MBP) without the signal peptide sequence was expressed and purified as in previous single-molecule tweezer studies29,30 (Supplementary Fig. 2a for its amino acid sequence). The SpyTag, a short peptide tag, was introduced at the N- and C-terminal ends for the DNA handle attachment. The corresponding gene block was cloned into the pET24a vector. The plasmid vectors were inserted into E. coli BL21(DE3) cells (Thermo Fisher, EC0114) using heat shock transformation (40 s, 42 °C). The cells were cultured in 1 L of LB medium with kanamycin (25 µg/ml) at 37 °C. Around OD600 = 1.0, the protein was overexpressed by 0.8 mM IPTG for 4 h at 30 °C. The cells were pelleted by centrifugation (5993 × g, 10 min, 4 °C), resuspended in 50 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM TCEP, 10% glycerol, 1 mM PMSF, and then lysed using Emulsiflex C3 at ∼17,000 psi. After another round of centrifugation (34,811 × g, 30 min, 4 °C), 1 ml Ni-IDA resin (His60 Ni-Superflow Resin, Clontech) was added to the supernatant and incubated for 1 h. The resin was washed with 50 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM TCEP, 10% glycerol, 20 mM imidazole. The protein was eluted from the resin with 50 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM TCEP, 10% glycerol, 300 mM imidazole (Supplementary Fig. 2b). The eluted protein was stored at −80 °C in aliquots.
Human carbonic anhydrase II (CA II) was expressed and purified following a procedure adapted from previous studies61,62 (Supplementary Fig. 2a, d). The gene block for CA II was cloned into the pET24a vector, which was then introduced into E. coli BL21(DE3) cells using the heat shock transformation. The cells were cultured in 1 L of LB medium with kanamycin (25 µg/ml) at 37 °C, followed by induction with 0.4 mM IPTG at OD600 = 0.6 and the addition of 1 mM zinc sulfate. The cells were further cultured for 16 h at 16 °C and then harvested by centrifugation (5993 × g, 10 min, 4 °C). The cell pellets were re-suspended in 100 mM Tris-HCl, pH 7.6, 200 mM sodium sulfate, 1 mM TCEP, 1 mM PMSF, and then lysed using EmulsiFlex C3 at 15,000–17,000 psi. After another round of centrifugation (34,811 × g, 30 min, 4 °C), 1 ml Ni-IDA resin was added to the supernatant and incubated for 1 h. The resin was washed with 100 mM Tris-HCl, pH 7.6, 200 mM sodium sulfate, 20 mM imidazole. The protein was eluted from the resin with 100 mM Tris-HCl, pH 7.6, 200 mM sodium sulfate, 400 mM imidazole. The eluted protein was further purified using HiLoad 16/600 Superdex 75 column (Cytiva) equilibrated in 50 mM Tris, pH 7.8, 150 mM NaCl. The zinc ion was removed from CA II to prevent potential metal-induced complexity in this work. To this end, 0.8 mg/ml of purified CA II was incubated in a chelation buffer (25 mM MOPS, pH 7.0, 100 mM pyridine-2,6-dicarboxylic acid) for 8 h at 20 °C (refs. 61,62,63). The protein sample was desalted using PD-10 desalting column (Cytiva) with 50 mM Tris-HCl, pH 7.8.
DNA handle conjugation to protein
Two types of 1022-bp DNA handles were attached to MBP for the single-molecule tweezer experiments28,64. The DNA constructs, modified at one end with amine group and at the other end with either azide or 2×biotin, were generated by PCR using λ DNA template (NEB, N3011S)28,64. ∼8 ml PCR product (azide-DNA:2xbiotin-DNA = 1:1) was purified using HiSpeed Plasmid Maxi kit (Qiagen) in 1 ml NaHCO3 (pH 8.3). The amine group of the purified DNA constructs was further modified to maleimide by incubating the sample with 1 mM SM(PEG)2 (Thermo Scientific Pierce) for 20 min at ∼23 °C. The sample was subsequently purified using Econo-Pac 10DG Desalting Column (Bio-Rad) in 1.5 ml of 0.1 M sodium phosphate (pH 7.3) with 150 mM NaCl. ∼0.5 µM of the DNA sample were incubated with ∼20 µM of a SpyCatcher construct for 2 h at ∼23 °C for the molecular conjugation through the functional groups of maleimide and cysteine thiol28,64. Unconjugated proteins were removed using anion exchange chromatography (HiTrap Q HP column, Cytiva) with a gradient mode of 0–1 M NaCl in 20 mM Tris-HCl (pH 7.5). The DNA peak fractions, ∼30% of which corresponds to SpyCatcher-conjugated DNA, was concentrated to ∼100 nM for the conjugated construct. One µl of ∼20 µM MBP was incubated with 10 µl of the DNA sample for 2 h at ∼23 °C, for the DNA handle attachment to MBP at the N- and C-terminal ends through the SpyTag-SpyCather binding (Supplementary Fig. 2c). A covalent, isopeptide bond is spontaneously formed in the bound complex and its unfolding is not observed in our force range up to 50 pN (ref. 28) (Supplementary Fig. 3e). The final sample was diluted to make the hybrid molecular construct with azide and 2xbiotin at both termini to be ∼200 pM and stored at −80 °C in aliquots.
Preparation of single-molecule sample chambers
The surfaces of coverslips (VWR, No. 1.5, 24 × 50 and 24 × 40 mm) were cleaned using KOH and Piranha solution28,64. The bottom coverslip (24 × 50 mm) was passivated with polyethylene glycol (PEG) polymers of methyl-PEG and biotin-PEG at a molar ratio of 100:1 (refs. 28,64). These two surface-treated coverslips were combined to construct a single-molecule sample chamber with a channel volume (CV) of ∼10 µl (\(\equiv\) 1 CV)28,64. One µl of polystyrene beads coated with streptavidin was washed with 0.1 M sodium phosphate, pH 7.4, 150 mM NaCl, 0.1% Tween 20 (refs. 28,64). One CV of the bead sample was injected into the sample chamber and incubated for 2–5 min at ∼22 °C for the surface binding. The polystyrene beads bound to the surface were used to correct the thermal drift of the microscope stage. A total of 100 mg/ml of bovine serum albumin (BSA) was injected into the chamber and incubated for 5 min at ∼22 °C for additional surface passivation. The chamber was washed with 50 mM Tris-HCl, pH 7.4, 150 mM NaCl (Buffer A). Ten µl of ∼200 pM MBP-DNA construct sample was mixed with 1 µl of 0.2 µM traptavidin (TTV) for 15 min at ∼22 °C. One CV of the MBP-DNA-TTV construct sample (100–200 pM) was injected into the chamber and incubated for 5 min at ∼22 °C for the surface binding. To block unoccupied biotin-binding sites of TTV, 1 CV of a biotin-labeled oligonucleotide (30 nucleotides; 10 µM in Buffer A) was injected into the chamber and incubated for 5 min at ∼22 °C. The chamber was washed again with Buffer A. One µl of DBCO-coated magnetic beads was washed and resuspended with Buffer A28,64. One CV of the bead sample was injected into the chamber and incubated for 1 h at ∼25 °C. The beads are covalently attached to the surface-tethered molecular constructs through dibenzocyclooctyne (DBCO)-azide conjugation. The sample chamber was finally washed with 50 mM HEPES, pH 7.6, 100 mM KCl, 5 mM MgCl2 (Buffer B), which was used in previous single-molecule forced unfolding studies on MBP29,30.
Single-molecule tweezer experiments
The single-molecule tweezer experiments were conducted at ∼22 °C using a custom-built magnetic tweezer apparatus28,65,66. The assembled sample chamber was placed on an inverted microscope (Olympus, IX73) equipped with a motorized sample stage (ASI, MS-2000 XY Automated Stage). The imaging spots were illuminated using a blue light-emitting diode (LED; Thorlabs, M455L4, λpeak = 447 nm; 9.2 mW/cm2; Supplementary Fig. 1 for the full spectrum) or an infrared LED (Thorlabs, M850LP1, λpeak = 860 nm; 26.3 mW/cm2). The force applied to a tethered magnetic bead was pre-calibrated using an inverted pendulum model of the bead-molecular construct28. The extension (end-to-end distance) of the protein-DNA construct was determined by tracking changes in bead diffraction patterns captured by a charge-coupled device (CCD) camera (JAI, CM-040 GE)28. For the control experiments, the force-scanning range for each unfolding cycle was 1–50 pN with waiting times between the cycles as 2.5 min at 1 pN and 1 s at 50 pN. Especially for the experiments involving a prolonged incubation in the U state, the maximum force was set to 35 pN for 90 min in every unfolding cycle. This force level is sufficiently high to induce the unfolding and is relatively more resistant to bead detachment during the waiting time. Ninety-three % were fully unfolded during the stretching phase before reaching 35 pN, whereas 7% were fully unfolded at 35 pN within 0.66 ± 0.16 s (mean ± SD). The total incubation time for the U state was estimated to be ∼95% of the total experiment time. In the N-state condition, the N state was maintained at 1 pN for the initial 6 h before the cyclic unfolding. The foldability curves in Fig. 1 were smoothed using a moving-average window of three data points to reduce fluctuation noise and obtain more accurate decay time constants. The curves for the control condition and other conditions of the same large number of data points per unit time were binned into groups of five data points for clearer visualization and comparison.
Analysis of force-extension curve data
The force-extension curves were median-filtered for the extension (half-window size = 5) and smoothed for the force (half-window size = 10). The unfolding forces and step sizes extracted from the force-extension curves were analyzed using the worm-like chain (WLC) model67, FLp/kBT = \(l\)/L + (1–\(l\)/L)–2/4 – 1/4, where F is the applied force, kB is the Boltzmann constant, T is the absolute temperature, \(l\) is the protein extension, Lp is the persistence length, and L is the contour length. The contour length was estimated as the number of amino acid residues multiplied by the average residue-residue distance of 0.38 nm (ref. 68), followed by correction with protein structure factors (Supplementary Fig. 2e). The persistence length was estimated as 0.29 ± 0.01 (SE) nm for the external α-helices at the C-terminus and 0.51 ± 0.01 (SE) nm for the fully unstructured polypeptide from the WLC model, which aligns with a range of protein persistence lengths69,70. The slight variance in persistence length for each case is likely due to the difference in residue composition within the unstructured portions69,71. The most probable unfolding forces for the external α-helices and the core structure were measured as 13.9 ± 0.2 pN and 25.3 ± 0.7 pN (peak value ± SE), respectively. The foldability at each unfolding cycle was estimated by the proportion of normal unfolding with the distinctive pattern, i.e., the unfolding of the external α-helices followed by the core structure, obtained from multiple molecules.
Gas sparging and vacuum degassing
A vial containing 5 ml Buffer B was sealed with a rubber septum and parafilm to block air flow. For the gas sparging, a gasbag filled with either nitrogen or oxygen gas was connected to a 2 ml syringe with a needle. The needle linked to the gasbag was inserted into the buffer solution through the septum/parafilm. Another needle was inserted into the air above the buffer through the septum/parafilm, allowing the gas from the gasbag to replace the dissolved gas by the pressure difference. The gas sparging process was maintained at ∼22 °C for 30 min. For the vacuum degassing, a vial containing 5 ml Buffer B was connected to a vacuum pump for 1 h. To facilitate the degassing, the vial was rapidly frozen using liquid nitrogen and then thawed during the vacuum degassing. The freezing and thawing was repeated three times. After the gas sparging or vacuum degassing, the concentration of dissolved molecular oxygen (O2) and buffer pH were immediately measured (Supplementary Fig. 4).
ROS measurement for single-molecule experiment samples
The levels of four primary reactive oxygen species (ROS; hydrogen peroxide (H2O2), hydroxyl radical (•OH), superoxide (O2•–), singlet oxygen (1O2)) were measured after 10 min incubation at ∼22 °C in three conditions: the foldability measurement condition (Buffer B with 0.12 mg/ml magnetic beads under blue light), negative control (no-light condition), and positive control (ROS-generating condition). Absorbance or fluorescence was obtained as a signal from 200 µl samples using a microplate reader. The measurement for each condition was conducted in triplicate. The values for the three conditions were normalized to that of the negative control. Peroxidase (POD)-catalyzed oxidation of N,N-diethyl-p-phenylenediamine (DPD) by H2O2 was measured for the assessment of H2O2 level (DPD assay)72. One mM H2O2 was used for the positive control. After 10 min incubation, each 200 μl sample was mixed with 80 μl of 0.1 M sodium phosphate (pH 6.0), 224 μl deionized (DI) water, 10 μl of 1% w/v DPD (Merck, 07672), and 10 μl of 0.1% w/v POD (Thermo Fisher, 31490). The H2O2-induced generation of the radical cation of DPD (DPD•+) was manifested by an increase at OD510 (refs. 72,73). Oxidation of hydroxyphenyl fluorescein (HPF) by •OH was measured for the assessment of •OH level (HPF assay)46. Five μM HPF (Cayman Chemical, 10159) was dissolved in each buffer solution. A Fenton’s reagent (100 μM FeSO4, 1 mM EDTA, 0.3% H2O2) was used for the positive control. The fluorescence from oxidized HPF by •OH was measured with λex = 485 nm and λem = 525 nm (refs. 35,74). Oxidation of dihydrorhodamine 123 (DHR123) by O2•– was measured for the assessment of O2•– level (DHR123 assay)35,42,43. Five μM DHR123 (Thermo Fisher, D23806) was dissolved in each buffer solution. A total of 0.5 μM of an iridium-complex photosensitizer (TIr3)42 was used for the positive control. TIr3 was reported to generate O2•– under blue light irradiation42. The fluorescence from the oxidized DHR123 (rhodamine 123) by O2•– was measured with λex = 485 nm and λem = 525 nm (ref. 43). Oxidation of 9,10-anthracenediylbis(methylene)dimalonic acid (ABDA) by 1O2 was measured for the assessment of 1O2 level (ABDA assay)38. A total of 100 μM ABDA (Merck, 75068) was dissolved in each buffer solution. A total of 0.5 μM TIr3 was used for the positive control as TIr3 was also reported to generate 1O2 under blue light irradiation42. 1O2-induced ABDA oxidation was manifested by a decease at OD400 (refs. 38,42). The absorbance for the positive control was corrected by that without ABDA since TIr3 itself has absorbance around 400 nm (ref. 42).
ROS measurement for free amino acids
Spectrophotometric assays were used to detect ROS in solutions of free amino acids under blue light: 1O2 using the ABDA assay38, type I ROS such as O2•–, H2O2, and •OH using the DHR123 assay43,44,45, and •OH using the HPF assay46. 25 mM of each amino acid in H2O (DI water; or D2O) and 1–10 mM ROS-detection probe in DMSO (10 mM ABDA, 1 mM DHR123, or 1 mM HPF) were mixed at a ratio of 99:1 (v/v). The mixtures were then exposed to blue light (λpeak = 446 nm; Supplementary Fig. 1 for the full spectrum) for 6 min. The absorbance or fluorescence was measured using a microplate reader: absorbance at 400 nm for the ABDA assay, fluorescence with λex = 485 nm and λem = 515 nm for the DHR123 assay, and fluorescence with λex = 485 nm and λem = 530 nm for the HPF assay. The values from each assay were normalized to those of the no-light condition (t = 0 min). Each measurement was conducted in triplicate.
Sample preparation for LC-MS/MS
The procedure of sample preparation for liquid chromatography-tandem mass spectrometry (LC-MS/MS) was modified from a previous study75. Ten µg MBP was added to either the same volume of DI water upon O2- or Ar-bubbling. For control groups not exposed to light, the protein samples in the O2- or Ar-bubbled condition was kept in a dark chamber for 30 min. Each group corresponds to a high or low concentration of dissolved O2 (Supplementary Fig. 4) and show little difference in oxidation based on a volcano plot (Supplementary Fig. 6a). For the experimental group subjected to light exposure, the protein sample in the O2-bubbled condition was irradiated with a blue LED (λpeak = 446 nm; 16 mW/cm2; Supplementary Fig. 1 for the full spectrum) for 30 min. Every sample was further purified using SDS-free polyacrylamide gel electrophoresis (native PAGE) for LC-MS/MS, which does not induce protein denaturation. To prevent undesirable protein oxidation caused by ammonium persulfate during gel running, 0.01% (v/v) thioglycolic acid, an antioxidant, was added to sample loading buffer76,77. N2-bubbled DI water was used in making a 7.5% SDS-free bis-acrylamide gel and gel-running buffer. The electrophoresis was carried out for 1.5 h at 160 V, maintaining a temperature of 4 °C. Following washing with LC-grade water, the gel was stained using ImperialTM Protein Stain (Thermo Scientific, 24617). After destaining with LC-grade water, the gel section containing the major band was cut into ∼1 mm3 cubes using a razor blade and transferred to a non-treated 96-well plate. The gel cubes were further destained with MeOH, 50 mM ammonium bicarbonate (ABC), and 40% acetonitrile (ACN) (aq), and then dehydrated with ACN. Extra washing and dehydration steps were carried out with LC-grade water, 40% ACN (aq), and ACN. One µg/µl trypsin in MS-grade acetic acid was diluted to 0.01 µg/µl using a 9:1 (v/v) mixture of 50 mM ABC and ACN (Buffer X). Twenty-five µl of the trypsin solution and 75 µl of Buffer X were added to the dehydrated gel cubes. After a 5-min wait, the 96-well plate was sealed with parafilm and incubated for ∼18 h at 37 °C. The resulting tryptic peptides of MBP were eluted from the gel cubes by 0.1% formic acid/50% ACN (aq), and then transferred to Eppendorf Protein LoBind tubes. Additional elution was performed using 0.1% formic acid/99.9% ACN. The eluted samples were dried using SpeedVac at 60 °C, resuspended in 0.1% trifluoroacetic acid (TFA) (aq), and subjected to clean-up using PierceTM C18 Tips (Thermo Scientific, 87782). The tips were wet and then equilibrated by aspirating and discarding 50% ACN (aq) followed by 0.1% TFA (aq). The peptide suspension was slowly and repetitively aspirated and dispensed (\(\times\)10). After rinsing the tip with 0.1% TFA/5% ACN (aq), the peptides were eluted by slowly aspirating and dispensing 0.1% formic acid/80% ACN (aq). The eluted peptides were transferred to fresh LoBind tubes, dried using SpeedVac at 60 °C, and resuspended in 0.1% formic acid (aq) for LC-MS/MS analysis. All experiments were conducted in triplicate. The other studied protein, CA II, underwent the same procedure of sample preparation for LC-MS/MS.
LC-MS/MS analysis
After the in-gel tryptic digestion for MBP, the resulting tryptic peptides were analyzed by LC-MS/MS. The tryptic digest was separated through online reversed-phase chromatography using a peptide trap column Acclaim PepMap 100 C18 (Thermo Fisher Scientific; 3 μm particle size, 75 μm diameter, 2 cm length) and an analytical column Acclaim PepMap RSLC C18 (Thermo Fisher Scientific; 3 μm particle size, 75 μm diameter, 15 cm length). The procedure was followed by electrospray ionization at a flow rate of 300 nl/min. The samples were eluted using a gradient of 3–50% solution B (80% ACN/0.1% formic acid (aq)) for 60 min and 50–80% solution B for 10 min, and then the columns were washed with 100% solution B for 10 min. The chromatography system was coupled in-line with an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was operated in the data-dependent mode with a 120,000-resolution MS1 scan (375–1500 m/z), an AGC target of 5e5, and a maximum injection time of 50 ms. Peptides above a threshold of 5e3 and with a charge of 2–7 were selected for fragmentation with dynamic exclusion. The dynamic exclusion duration was set to be 15 s with 10-ppm mass tolerance.
For CA II, LC‒MS/MS analysis was performed using UltiMate 3000 RSLCnano system (Thermo Fisher Scientific Inc.) coupled to an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific Inc.). Mobile phase A and B were 100% DI water containing 0.1% formic acid and 100% ACN containing 0.1% formic acid, respectively. Peptide separation was achieved using a 90-min LC gradient at a flow rate of 400 nl/min. The Orbitrap Eclipse Tribrid mass spectrometer operated in data-dependent mode during separation. MS data were acquired with the following parameters: Full scan MS1 spectra (375–1500 m/z) were obtained in the Orbitrap, with a maximum ion injection time of 50 ms, a resolution of 120,000, and a standard mode automatic gain control (AGC) target. MS2 spectra were acquired in the Orbitrap mass analyzer at a resolution of 30,000, applying high-energy collision dissociation (HCD) with 30% normalized collision energy and an AGC target value of 5.0e4, with a maximum ion injection time of 54 ms. The previously fragmented ions were excluded for 60 s.
LC-MS/MS data analysis
The raw MS data were converted to mzML files using MSConvert (v3.0.20279), and MS/MS spectra were searched with MSFragger (v3.8)78 through FragPipe (v20.0). Oxidations listed in Supplementary Table 1 were specified as variable modifications, allowing up to three variable modifications per peptide. Carbamidomethylation of cysteine was set as a fixed modification. For MBP, precursor and fragment ion mass tolerances were set to 10 ppm and 0.4 Da, respectively, and isotope error was set to 0/1. For CA II, precursor and fragment ion mass tolerances were set to 10 ppm and 20 ppm, respectively, with isotope error set to 0/1, and MSFragger’s built-in mass calibration option was used. The protein database consisted of MBP (or CA II) and 118 contaminant sequences, appended with decoy sequences. Protein digestion parameters were set to fully enzymatic cleavage using the “stricttrypsin” enzyme setting (allowing up to two missed cleavages), peptide lengths of 7–50 amino acids, and enabled N-terminal methionine clipping. PSM validation was performed using Philosopher (v5.0), with FDR calculations based on a target-decoy approach. PSM probabilities were modeled in PeptideProphet using the default closed-search settings. The oxidation level for each amino acid residue was quantified as the oxidative modification count for the residue divided by the total detected peptide count containing the residue. All the LC-MS/MS experiments and analysis were performed in triplicate with biologically independent samples in each control and experimental group. The volcano plots were generated to compare the oxidation level between two groups. In the volcano plots, the fold change values were obtained based on the mean oxidation level for each residue from the triplicate data. Residues of no detection or zero oxidation in all triplicate data were omitted in the volcano plots because the fold change cannot be defined. Significantly oxidized residues compared to the control group were selected as those with p value < 0.05 and fold change > 1.5 (ref. 79). All the details—identified peptides, sequence coverage, oxidized sites, modification site occupancy, replicate data, and more—are provided in Source Data.
Whole-cell proteomic analysis by LC-MS/MS
The whole-cell proteomic data for HeLa cells (ATCC, CCL-2) by LC-MS/MS were utilized as control groups in our previous report80 and were re-analyzed here for our purpose. The cell line was authenticated by the commercial vendor ATCC. Example morphology, STP profiling, Karyotype, or other information about the cell line are provided in a website (https://www.atcc.org/products/ccl-2). The mass spectrometry data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository under the accession code PXD038746. HeLa cells were cultured on 100 mm cell culture dishes with DMEM supplemented with FBS and antibiotics at 37 °C in a humidified chamber containing 5% CO2. The cells in the experimental group were irradiated with a blue LED (λpeak = 446 nm; 16 mW/cm2; Supplementary Fig. 1 for the full spectrum) for 10 min, while those in the control group were kept in the dark chamber. The cells were collected and lysed by RIPA buffer with 1% protease cocktail inhibitor added. Cell debris were removed by centrifugation of 16,000 × g for 10 min at 4 °C. Tryptic peptides were obtained by following the protocol of on-filter digestion using S-trapTM (PROTIFI), as follows. In brief, protein suspension of ∼100 μg was diluted with 10% SDS solubilization buffer containing 100 mM triethylammonium bicarbonate (pH 7–8, adjusted by phosphoric acid). The solution was centrifuged at 13,000 × g for 10 min and the supernatant was collected. To prevent self-crosslinking of proteins, the reduction and alkylation of cysteines were conducted by adding 20 mM dithiothreitol for 10 min at 95 °C followed by adding 40 mM iodoacetamide for 30 min at room temperature in a dark chamber. The solution was centrifuged at 13,000 × g for 10 min and the supernatant was collected. Twelve % phosphoric acid and S-trap binding buffer (90% aqueous methanol containing a final concentration of 100 mM triethylammonium bicarbonate, pH 7.1) was added to the supernatant. The solution was transferred to an S-trap mini spin column for tryptic digestion and elution. The column was washed with S-trap binding buffer, the proteins on column were digested by trypsin for overnight at 37 °C, and the tryptic peptides were then sequentially eluted with 50 mM Tris, 0.2% formic acid, and 50% ACN/0.2% formic acid solution. The samples were fractionated using Pierce High pH Reversed-Phase Peptide Fractionation Kit (Thermo Scientific, #84868) according to the provided protocol. A total of eight fractionated samples were dried using a speed-vac to obtain the peptide powder. Dry tryptic peptides were analyzed using a Q Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source. A C18 reverse-phase HPLC column (500 mm × 75 μm ID) was used to separate pure analyte from the crude peptide suspension. An ACN/0.1% formic acid gradient of 2.4–24% was used as eluent at a flow rate of 300 nl/min. For MS/MS analysis, precursor ion scan MS spectra (m/z 400–2000) were acquired with an internal lock mass. The 20 most intense ions were isolated via high-energy collision-induced dissociation.
The LC-MS/MS spectra data were analyzed using Sequest Sorcerer platform (Sagen-N Research) and a modification search tool, MODplus (v1.02)81. In brief, the oxidized proteome search was conducted in two stages. In the first stage, only oxidation on Met was considered. In the second stage, unidentified spectra from the first stage were re-analyzed for various types of oxidations, including mono-, di-, and trioxidation on various residues (Supplementary Table 1). The oxidation level for each protein was obtained by summing the intensities for the oxidative modifications of its residues. All the LC-MS/MS experiments and analysis were performed in triplicate with biologically independent samples in each control and experimental group. The volcano plots were generated to compare the oxidation level between two groups. In the volcano plots, the fold change values were obtained based on the mean oxidation level for each protein from the triplicate data. Proteins of no detection or zero oxidation in all triplicate data were omitted in the volcano plots because the fold change cannot be defined. Significantly oxidized proteins compared to the control group were selected as those with criteria of increasing stringency, ranging from p value < 0.05 and fold change > 1.5 to p value < 0.01 and fold change > 3.5. Refer to the last subsection of “Methods” for bioinformatic and structural analyses for oxidized proteins.
All-atom molecular dynamics (MD) simulation
All MD trajectories were generated using CHARMM36m force field (charmm36-mar2019)82, which were implemented in the GROMACS molecular dynamics software package (version 2021.4). The input files of simulation systems were generated using CHARMM-GUI83. The initial coordinates of MBP and CA II were obtained from crystal structures (PDB ID 1ANF and 3D92). The systems were solvated with TIP3P water molecules84. K+ and Cl– ions were added to neutralize the systems and maintained at ∼150 mM. The force field and structure files of O2 molecule were generated using CHARMM-GUI Ligand Reader & Modeler85. The force field and structure files of maltose was generated using Open Babel86 and SwissParam87. Using a Gromacs module of gmx insert-molecules, twenty O2 or one maltose molecule were inserted into the simulation systems. The van der Waals (vdW) interactions were calculated with a cutoff distance of 12 Å and smoothly switched off at 10–12 Å by a force-switch function88. The long-range electrostatic interactions were calculated using the particle-mesh Ewald method with a cutoff distance of 12 Å and a mesh size of 1.2 Å (ref. 89). During the equilibration run, the NVT ensemble was applied with 1-fs time step for 125 ps. The temperature was maintained at 303.15 K using the Nosé–Hoover temperature coupling method with τt = 1 ps (ref. 90). The production run was performed with 2-fs time step in the NPT ensemble. The pressure was maintained at 1 bar using the isotropic Parrinello-Rahman method with τp = 5 ps and a compressibility of 4.5 × 10–5 bar–1 (ref. 91). The production times were 1 μs for the protein/O2 and MBP-only systems and 15 μs for the MBP/maltose system. The MD simulation trajectory data were analyzed using Gromacs modules and custom-built Python codes. After the centering of MBP (or CA II) in each MD simulation system, the translational and rotational movements of the protein were eliminated in the MD trajectories using “-fit rot+trans” option of gmx trjconv module. Visual Molecular Dynamics (VMD) was used for the graphical visualization and movie generation.
Estimation of protein surface and core atoms
The solvent-accessible surface (SAS) of MBP and CA II was analyzed using Gromacs solvent-accessible surface area (SASA) module. The SAS was generated with a probe radius (rprobe) of 1.4 Å in every 100 ns of a MD simulation. 1.4 Å is a typically used value for rprobe, which approximates the radius of a water molecule. The distances between the atom centers and SAS (datom-SAS) were calculated for all protein atoms. By subtracting the vdW radius (rvdW)92 and the probe radius from datom-SAS, the distances between the vdW surface of atoms and the solvent-excluded surface (SES) (dvdW-SES) were calculated for all protein atoms. The relative frequencies of dvdW-SES were calculated for all protein atoms, all side-chain atoms, and highly oxidizable side-chain atoms93. For each case, the protein surface or core atoms were classified based on dvdW-SES values smaller or larger than 0.1 Å, respectively (Supplementary Fig. 5).
Position and distance analyses of trapped O2 molecules and protein cavities
The minimum distance between atoms of MBP (or CA II) and O2 molecules was extracted from the MD trajectories in every 1 ns. We collected all time points, during which the distance remains less than a cutoff distance for longer than a cutoff time span. 3.5 Å was used as the cutoff distance to include only the close binding. The cutoff distance value of 3.5 Å has been used to determine the atoms involved in protein-ligand interactions94,95. Ten ns or 5 ns was used as the cutoff time span to exclude a large number of transient interactions and only consider more sustained binding events. In this analysis, the first and the last time frames for each duration were excluded to analyze the time regions of complete O2 trapping within the proteins. The positions of O2 molecules as a center coordinate of O2 atoms were extracted from all the collected time points and superimposed onto the protein structures. The maximum end-to-end distances and total travel lengths of individual trapped O2 molecules were also calculated for all O2-trapping time regions. For the protein cavity analysis, the atom coordinates of the proteins were extracted as PDB files from the MD trajectories in every 1 ns. For every PDB file, the center positions and diameters of protein cavities were obtained using Voronoia with a grid distance of 0.2 Å (ref. 31). We only considered the cavities with a diameter larger than the kinetic diameter of O2 (3.46 Å)32. The center positions of the protein cavities were superimposed onto the protein structures.
Determination of predominant O2-trapping (sub)areas
To determine the predominant O2-trapping areas (Supplementary Fig. 9 and Supplementary Table 2), the minimum distance between atoms of MBP and O2 molecules was first extracted in every 1 ns from a MD simulation. We collected all time points, during which the distance remains less than a cutoff distance for longer than a cutoff time span. 3.5 Å was used as the cutoff distance to include only the close binding. The cutoff distance value of 3.5 Å has been used to determine the atoms involved in protein-ligand interactions94,95. Ten ns was used as the cutoff time span to exclude a large number of transient interactions and only consider more sustained binding events. All protein residues of which the atoms are less than the cutoff distance from O2 atoms were extracted from the binding time points in every 1 ns. From a histogram of total counted times for the extracted residues, the top 32.9 ± 1.7% (mean ± SD; n = 3 simulations) of the residues were selected for the predominant O2-binding residues, covering 90% of the total counted times of all extracted residues. The selected residues can be considered as the residues that bind to O2 as predominantly as 90% of the total binding times. The O2-binding residues comprise the O2-trapping areas in the N- and C-terminal lobes of MBP. We conducted this analysis for three MD simulations and determined the common O2-trapping areas. To determine the O2-trapping subareas that more tightly bind to O2 (Supplementary Fig. 10 and Supplementary Table 3), only the binding time regions longer than 100 ns were analyzed (7.6% of all binding time regions). All protein residues of which the atoms are less than the cutoff distance from O2 atoms were extracted from the binding time points in every 1 ns. The residues tightly binding to O2 were selected from the highest counted residue by the cutoff number of residues, from a histogram of total counted time for each extracted residue. The cutoff number was defined as the maximum number of residues binding to O2 at a certain time point. From three MD simulations, total 15 cases with >100-ns binding times were analyzed and classified into four distinct O2-trapping subareas based on residue overlap.
Free energy calculation for O2-trapping events
We conducted the free energy calculations using the alchemical transformation method applied to the generated MD trajectories. The initial structures were sampled from the MD trajectories, where O2 is trapped within the O2-trapping subareas of MBP for the longest dwell times (Supplementary Table 3). For the alchemical transformation, a dummy O2 molecule was introduced into the surrounding solution. The simulations employed 20 windows with different λ values. Over the range between λ = 0 and λ = 1, the interactions of the trapped O2 molecule within the complex were decreased, while those of the dummy O2 were recovered. Within each window, the systems were equilibrated in the following sequence: (1) energy minimization, utilizing the steepest descent algorithm for a maximum of 5000 steps; (2) 100-ps NVT equilibration, using Langevin dynamics96 with a reference temperature of 300 K; (3) 100-ps NPT equilibration, employing the Berendsen algorithm97. Throughout the two equilibration steps, position restraints were applied to the trapped O2 molecule. A production run of 10 ns was conducted using the Parrinello-Rahman pressure coupling scheme91. The multiple Bennett acceptance ratio (MBAR) method98 was employed to estimate the free energies accompanied with the alchemical transformations (Supplementary Fig. 11).
Depth and hydrophobicity analyses for predominant O2-trapping (sub)areas
The depths of the O2-trapping (sub)areas of MBP from the protein surface (solvent-excluded surface) were calculated from three MD simulations in every 100 ns (Supplementary Fig. 12). The distances between the protein surface and atom centers (atom depths) were calculated using Michel Sanner’s Molecular Surface (MSMS) for each time point99. The atom depths were averaged for all amino acid residues comprising the O2-trapping (sub)areas (area depths). The area depths were further averaged over the analyzed time points. The Moon-Fleming hydrophobicity scale for amino acid residues100 was utilized in the hydrophobicity analyses for the predominant O2-trapping (sub)areas (Supplementary Fig. 12). The hydrophobicity scale is expressed as the free energy difference in stability (ΔΔG) between the wild-type transmembrane (TM) protein OmpLA and each amino acid variant at the position A210 (e.g., for cysteine, ΔΔG = ΔGWT–ΔGA210C). In this scale, more hydrophobic amino acid has more negative ΔΔG. The hydrophobicity for each area was estimated as the average ΔΔG of comprising amino acid residues from three MD simulations. The hydropathy plots for MBP as well as bovine rhodopsin (as an example protein with clearly segmented TM and water-soluble regions) were also analyzed using the hydrophobicity scale. The hydrophobic regions of the rhodopsin in the hydropathy plot match those of the protein’s TM domains provided in OPM database (PDB ID 1U19).
Analysis for O2-trapping times and maltose-binding times
Representative MD trajectories and barcode plots for the O2-trapping (sub)areas of MBP were obtained through the same procedure used to determine the predominant O2-trapping areas, but only with residues of each structural region considered (Supplementary Figs. 14–16). The O2-trapping times for each structural region were extracted from the barcode plots from three 1-μs MD simulations (Supplementary Fig. 13). The maltose-binding times were obtained only for the specific binding to the maltose-binding site (Supplementary Figs. 13 and 17). The comprising residues of the maltose-binding site (D14, K15, W62, D65, R66, E111, E153, Y155, W230, W340) were selected from previous structural studies70,101. The structure of maltose (45 atoms) is relatively large and complex compared to O2, so the binding criteria based on the minimum distance between atoms can be misleading in determining the specific/fit binding to the binding site. Instead, the distance between the center of mass of the selected residues and maltose was utilized for the binding criteria102 and calculated at every 1 ns from a 15-μs MD simulation. The corresponding barcode plot was generated for the time regions during which the distance remains less than 8 Å for longer than 10 ns. The 8-Å cutoff distance can distinguish the specific and nonspecific bindings (Supplementary Fig. 17c). Transient interactions shorter than 10 ns were excluded, same as in the analysis of O2-trapping times. Representative O2-trapping events and maltose-binding events are visualized in Supplementary Movies 1 and 2.
Density functional theory (DFT) calculation
We performed the mixed-reference spin-flip time-dependent DFT (MR-SF-TDDFT) at the BHHLYP/6-31G* level of theory with a conductor-like polarizable continuum model (C-PCM), as implemented in the GAMESS program package103. Since a system with O2 contains with nearly degenerate π* orbitals which produce doubly degenerate states, the electron correlation effects are crucial for electronic structure calculations, which can be handled by the MR-SF-TDDFT calculation. The excitation energies for singlet and triplet states of amino acid residue-O2 pair geometries in each O2-trapping subarea of MBP were calculated to obtain the absorption spectra. Coordinates for the calculations were extracted from the MD trajectories in every 10 ns of the longest dwell-time regions for each area. The smooth absorption spectrum, I(λ), was obtained from I(λ) = (πΓ/2)Σi εosci Li(λ), where εosci is the oscillator strength of the ith configuration and Li(λ) is the normalized Lorentzian function centered at λi,abs with the broadening Γ, i.e., Li(λ) = 2Γ/(π(4(λ–λi,abs)2 + Γ2)). Forty nm was chosen for the broadening parameter.
Electron paramagnetic resonance (EPR) spectroscopy
We performed the EPR measurements using 2,2,6,6-tetramethylpiperidine (TEMP) (Thermo Scientific, 151951000), which is commonly used for 1O2 detection39. The oxidation of TEMP by 1O2 produces 2,2,6,6-tetramethylpiperidine-1-oxyl (TEMPO), which exhibits the characteristic trilinear EPR signal. 0.5 mM TEMP and 0.5 mM tryptophan (Trp) were dissolved in the mixed solvent of O2-bubbled DI water and O2-bubbled acetonitrile at a 1:1 ratio (v/v). For the experimental groups subjected to light exposure, the mixtures were incubated for 10 min, with exposure to blue light (λpeak = 446 nm, Supplementary Fig. 1 for the full spectrum), and then transferred to EPR capillary tubes. The EPR signals were acquired using EMXplus-9.5/12 spectrometer (Bruker). Ten scans were conducted for each sample with the following instrument parameters: sweep width, 70 G; power, 1.00 mW; modulation amplitude, 1.00 G; time constant, 81.92 ms; conversion time, 30 ms; sweep time, 30.72 s.
Monte Carlo analysis
The atom coordinates of MBP or CA II were extracted from a MD trajectory in every 10 ns and then averaged for each atom. The time-averaged coordinates of all side-chain atoms for each residue were further averaged to assign the residue position. The residues from each type were randomly sampled and the distances between the oxidized residues and the sampled residues were averaged over all possible combinations in each sampling. The numbers of sampled residues were 1–8 for MBP and 1–7 for CA II as the total number of benchmark Trp residues is eight for MBP and seven for CA II. The sampling number was 103 for each number of sampled residues. All possible cases were analyzed in case that its total number was less than the sampling number. The calculation was iterated to construct a probability density function for each residue type (PDFavg). This Monte Carlo analysis was also conducted for establishing the probability density distributions for arbitrary residues sampled from the entire protein (n = 8, the benchmark number of Trp residues for MBP; n = 4, the benchmark number of Trp* residues for CA II). The sampling number was 106 for each number of sampled residues. Likewise, all possible cases were analyzed in case that its total number is less than the sampling number. The mean value of the average distances analyzed for each residue type was used to calculate the occurrence probabilities for proximity by chance.
Bioinformatic and structural analyses for cellular proteomes
Protein structures for distinct proteins of the species H. sapiens and E. coli were obtained from PDB (updated by Feb. 2024) and AlphaFold DB (https://alphafold.ebi.ac.uk, AlphaFold DB v4, updated on Nov. 2022)48. The PDB provides experimentally determined protein structures for H. sapiens (total 8164 proteins collected) and E. coli (total 1674 proteins collected), while the AlphaFold DB provides computationally predicted protein structures for H. sapiens (total 20,504 proteins collected) and E. coli (total 4363 proteins collected). The HeLa cell proteome was determined based on protein-coding genes with expressed transcripts per million (TPM) larger than 0, which are available in the Gene Expression Omnibus (GEO) database (accession number GSE240542)104. The proteins detected in the whole-cell LC-MS/MS analysis represent relatively highly expressed genes in HeLa cells, as manifested in the TPM distribution (Supplementary Fig. 24a). All the detected proteins were obtained from AlphaFold DB, which provides their full protein structures (total 2915 proteins collected). Gene Ontology (GO) enrichment analysis was conducted for the H. sapiens proteins collected from AlphaFold DB and the HeLa cell proteins detected in the whole-cell LC-MS/MS analysis. Using the PANTHER classification system (http://www.pantherdb.org, PANTHER 18.0)49, each set of proteins was classified into subgroups regarding molecular function, protein class, biological process, and cellular anatomical entity. Although not all proteins were assigned to each subgroup due to limited databases of GO and PANTHER, the GO enrichment analysis was performed for large sets of proteins, ranging from 54% to 76%, specifically, 56–64% for molecular function, 68–76% for protein class, 60–71% for biological process, and 57–74% for cellular anatomical entity. The protein cavities in each protein structure were analyzed using Voronoia with a grid distance of 0.2 Å to obtain the cavity diameter and their constituting atoms31. The identified atoms were converted to amino acid residues to determine whether Trp residues are involved in forming the protein cavities. Although not all proteins were analyzed due to unknown errors in Voronoia, the cavity analysis was performed for the majority of protein structures, covering over 94% of them, specifically, 97.6% and 94.2% for the H. sapiens proteins collected from AlphaFold DB and PDB, 99.7% and 98.7% for the E. coli proteins collected from AlphaFold DB and PDB, and 97.1% for the HeLa cell proteins detected in the whole-cell LC-MS/MS analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data that support the findings of this study are available in the main text, Supplementary Information, and Source Data, as well as from corresponding author(s) upon request. The mass spectrometry data for single proteins have been deposited to the ProteomeXchange consortium via the jPOST repository under the accession codes PXD056895 for ProteomeXchange and JPST003425 for jPOST. The mass spectrometry data for whole-cell proteomics were deposited to the ProteomeXchange consortium via the PRIDE partner repository under the accession code PXD038746. Other raw data such as the MD simulation trajectory files and bioinformatic/structural analyses for cellular proteomes have been deposited to figshare, which are available in https://doi.org/10.6084/m9.figshare.24751755. Source data are provided with this paper.
Code availability
Analysis codes are available in Code Ocean [https://doi.org/10.24433/CO.6042424.v4].
References
Lee, J. M., Hammaren, H. M., Savitski, M. M. & Baek, S. H. Control of protein stability by post-translational modifications. Nat. Commun. 14, 201 (2023).
Harmel, R. & Fiedler, D. Features and regulation of non-enzymatic post-translational modifications. Nat. Chem. Biol. 14, 244–252 (2018).
Zhao, L., Zhao, J., Zhong, K., Tong, A. & Jia, D. Targeted protein degradation: mechanisms, strategies and application. Signal Transduct. Target Ther. 7, 113 (2022).
Reeg, S. & Grune, T. Protein oxidation in aging: does it play a role in aging progression? Antioxid. Redox Signal. 23, 239–255 (2015).
Kehm, R., Baldensperger, T., Raupbach, J. & Hohn, A. Protein oxidation-formation mechanisms, detection and relevance as biomarkers in human diseases. Redox Biol. 42, 101901 (2021).
Hawkins, C. L. & Davies, M. J. Detection, identification, and quantification of oxidative protein modifications. J. Biol. Chem. 294, 19683–19708 (2019).
Krisko, A. & Radman, M. Protein damage, ageing and age-related diseases. Open Biol. 9, 180249 (2019).
Liguori, I. et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 13, 757–772 (2018).
Liu, H. et al. Redox imbalance in the development of colorectal cancer. J. Cancer 8, 1586–1597 (2017).
Davies, M. J. Protein oxidation and peroxidation. Biochem. J. 473, 805–825 (2016).
Murphy, M. P. et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 4, 651–662 (2022).
Walker, E. J., Bettinger, J. Q., Welle, K. A., Hryhorenko, J. R. & Ghaemmaghami, S. Global analysis of methionine oxidation provides a census of folding stabilities for the human proteome. Proc. Natl Acad. Sci. USA 116, 6081–6090 (2019).
Walker, E. J. et al. Protein folding stabilities are a major determinant of oxidation rates for buried methionine residues. J. Biol. Chem. 298, 101872 (2022).
Valle-Orero, J. et al. Mechanical deformation accelerates protein ageing. Angew. Chem. Int. Ed. 56, 9741–9746 (2017).
Jensen, R. L., Arnbjerg, J., Birkedal, H. & Ogilby, P. R. Singlet oxygen’s response to protein dynamics. J. Am. Chem. Soc. 133, 7166–7173 (2011).
Sharp, J. S., Becker, J. M. & Hettich, R. L. Analysis of protein solvent accessible surfaces by photochemical oxidation and mass spectrometry. Anal. Chem. 76, 672–683 (2004).
Sharp, J. S., Becker, J. M. & Hettich, R. L. Protein surface mapping by chemical oxidation: structural analysis by mass spectrometry. Anal. Biochem. 313, 216–225 (2003).
Di Meo, S., Reed, T. T., Venditti, P. & Victor, V. M. Role of ROS and RNS sources in physiological and pathological conditions. Oxid. Med. Cell Longev. 2016, 1245049 (2016).
Schieber, M. & Chandel, N. S. ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462 (2014).
Turrens, J. F. Mitochondrial formation of reactive oxygen species. J. Physiol. 552, 335–344 (2003).
Pattison, D. I., Rahmanto, A. S. & Davies, M. J. Photo-oxidation of proteins. Photochem. Photobiol. Sci. 11, 38–53 (2012).
Davies, M. J. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 305, 761–770 (2003).
Kitahara, R. et al. Nuclear magnetic resonance-based determination of dioxygen binding sites in protein cavities. Protein Sci. 27, 769–779 (2018).
Kawamura, T. et al. Analysis of O(2)-binding sites in proteins using gas-pressure NMR spectroscopy: outer surface protein A. Biophys. J. 112, 1820–1828 (2017).
Kitahara, R., Yoshimura, Y., Xue, M., Kameda, T. & Mulder, F. A. Detecting O2 binding sites in protein cavities. Sci. Rep. 6, 20534 (2016).
Teng, C. L. & Bryant, R. G. Experimental measurement of nonuniform dioxygen accessibility to ribonuclease a surface and interior. J. Am. Chem. Soc. 122, 2667–2668 (2000).
Carreau, A., El Hafny-Rahbi, B., Matejuk, A., Grillon, C. & Kieda, C. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell. Mol. Med. 15, 1239–1253 (2011).
Kim, S., Lee, D., Wijesinghe, W. C. B. & Min, D. Robust membrane protein tweezers reveal the folding speed limit of helical membrane proteins. Elife 12, e85882 (2023).
Naqvi, M. M. et al. Protein chain collapse modulation and folding stimulation by GroEL-ES. Sci. Adv. 8, eabl6293 (2022).
Mashaghi, A. et al. Alternative modes of client binding enable functional plasticity of Hsp70. Nature 539, 448–451 (2016).
Staritzbichler, R., Ristic, N., Goede, A., Preissner, R. & Hildebrand, P. W. Voronoia 4-ever. Nucleic Acids Res. 49, W685–W690 (2021).
Mehio, N., Dai, S. & Jiang, D. E. Quantum mechanical basis for kinetic diameters of small gaseous molecules. J. Phys. Chem. A 118, 1150–1154 (2014).
Borden, W. T., Hoffmann, R., Stuyver, T. & Chen, B. Dioxygen: what makes this triplet diradical kinetically persistent? J. Am. Chem. Soc. 139, 9010–9018 (2017).
Chin, K. K. et al. Quantitative determination of singlet oxygen generated by excited state aromatic amino acids, proteins, and immunoglobulins. J. Am. Chem. Soc. 130, 6912–691 (2008).
Nam, J. S. et al. Singlet oxygen generation from polyaminoglycerol by spin-flip-based electron transfer. JACS Au 2, 933–942 (2022).
Lee, S., Filatov, M., Lee, S. & Choi, C. H. Eliminating spin-contamination of spin-flip time dependent density functional theory within linear response formalism by the use of zeroth-order mixed- reference (MR) reduced density matrix. J. Chem. Phys. 149, 104101 (2018).
Bregnhoj, M., Thorning, F. & Ogilby, P. R. Singlet oxygen photophysics: from liquid solvents to mammalian cells. Chem. Rev. 124, 9949–10051 (2024).
Entradas, T., Waldron, S. & Volk, M. The detection sensitivity of commonly used singlet oxygen probes in aqueous environments. J. Photochem. Photobiol. B Biol. 204, 111787 (2020).
Wu, J. H., Chen, F., Yang, T. H. & Yu, H. Q. Unveiling singlet oxygen spin trapping in catalytic oxidation processes using in situ kinetic EPR analysis. Proc. Natl Acad. Sci. USA 120, e2305706120 (2023).
Strieth-Kalthoff, F. & Glorius, F. Triplet energy transfer photocatalysis: unlocking the next level. Chem 6, 1888–1903 (2020).
Strieth-Kalthoff, F., James, M. J., Teders, M., Pitzer, L. & Glorius, F. Energy transfer catalysis mediated by visible light: principles, applications, directions. Chem. Soc. Rev. 47, 7190–7202 (2018).
Nam, J. S. et al. Endoplasmic reticulum-localized iridium(III) complexes as efficient photodynamic therapy agents via protein modifications. J. Am. Chem. Soc. 138, 10968–10977 (2016).
Yu, L. et al. Photocatalytic superoxide radical generator that induces pyroptosis in cancer cells. J. Am. Chem. Soc. 144, 11326–11337 (2022).
Wrona, M., Patel, K. & Wardman, P. Reactivity of 2′,7′-dichlorodihydrofluorescein and dihydrorhodamine 123 and their oxidized forms toward carbonated nitrogen dioxide, and hydroxyl radicals. Free Radic. Biol. Med. 38, 262–270 (2005).
Henderson, L. M. & Chappell, J. B. Dihydrorhodamine 123—a fluorescent-probe for superoxide generation. Eur. J. Biochem. 217, 973–980 (1993).
Setsukinai, K., Urano, Y., Kakinuma, K., Majima, H. J. & Nagano, T. Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J. Biol. Chem. 278, 3170–3175 (2003).
Sugimoto, W., Miyoshi, D. & Kawauchi, K. Detection of intracellular reactive oxidative species using the fluorescent probe hydroxyphenyl fluorescein. Methods Mol. Biol. 2274, 207–215 (2021).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Mi, H., Muruganujan, A. & Thomas, P. D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386 (2013).
Jensen, R. L., Arnbjerg, J. & Ogilby, P. R. Reaction of singlet oxygen with tryptophan in proteins: a pronounced effect of the local environment on the reaction rate. J. Am. Chem. Soc. 134, 9820–9826 (2012).
Suitthimeathegorn, O., Yang, C., Ma, Y. & Liu, W. Direct and indirect effects of blue light exposure on skin: a review of published literature. Ski. Pharm. Physiol. 35, 305–318 (2022).
Yoo, J. A. et al. Blue light irradiation induces human keratinocyte cell damage via transient receptor potential vanilloid 1 (TRPV1) regulation. Oxid. Med. Cell Longev. 2020, 8871745 (2020).
Alaimo, A. et al. Toxicity of blue led light and A2E is associated to mitochondrial dynamics impairment in ARPE-19 cells: implications for age-related macular degeneration. Arch. Toxicol. 93, 1401–1415 (2019).
Arnault, E. et al. Phototoxic action spectrum on a retinal pigment epithelium model of age-related macular degeneration exposed to sunlight normalized conditions. PLoS ONE 8, e71398 (2013).
Nash, T. R. et al. Daily blue-light exposure shortens lifespan and causes brain neurodegeneration in Drosophila. NPJ Aging Mech. Dis. 5, 8 (2019).
Toh, K. et al. Chemoproteomic identification of blue-light-damaged proteins. J. Am. Chem. Soc. 144, 20171–20176 (2022).
Nakashima, Y., Ohta, S. & Wolf, A. M. Blue light-induced oxidative stress in live skin. Free Radic. Biol. Med. 108, 300–310 (2017).
Arthaut, L. D. et al. Blue-light induced accumulation of reactive oxygen species is a consequence of the Drosophila cryptochrome photocycle. PLoS ONE 12, e0171836 (2017).
Yoshida, A. et al. Blue light irradiation-induced oxidative stress in vivo via ROS generation in rat gingival tissue. J. Photochem. Photobiol. B 151, 48–53 (2015).
Godley, B. F. et al. Blue light induces mitochondrial DNA damage and free radical production in epithelial cells. J. Biol. Chem. 280, 21061–21066 (2005).
Kim, J. K. et al. Elucidating the role of metal ions in carbonic anhydrase catalysis. Nat. Commun. 11, 4557 (2020).
Kim, C. U. et al. Tracking solvent and protein movement during CO2 release in carbonic anhydrase II crystals. Proc. Natl Acad. Sci. USA 113, 5257–5262 (2016).
Avvaru, B. S. et al. Apo-human carbonic anhydrase II revisited: implications of the loss of a metal in protein structure, stability, and solvent network. Biochemistry 48, 7365–7372 (2009).
Lee, D. & Min, D. Single-molecule tethering methods for membrane proteins. Methods Enzymol. 694, 263–284 (2024).
Kim, S. & Min, D. Robust magnetic tweezers for membrane protein folding studies. Methods Enzymol. 694, 285–301 (2024).
Min, D. Folding speeds of helical membrane proteins. Biochem. Soc. Trans. 52, 491–501 (2024).
Wijesinghe, W. C. B. & Min, D. Single-molecule force spectroscopy of membrane protein folding. J. Mol. Biol. 435, 167975 (2023).
Min, D. et al. Unfolding of a ClC chloride transporter retains memory of its evolutionary history. Nat. Chem. Biol. 14, 489–496 (2018).
Stirnemann, G., Giganti, D., Fernandez, J. M. & Berne, B. J. Elasticity, structure, and relaxation of extended proteins under force. Proc. Natl Acad. Sci. USA 110, 3847–3852 (2013).
Bertz, M. & Rief, M. Ligand binding mechanics of maltose binding protein. J. Mol. Biol. 393, 1097–1105 (2009).
van Rosmalen, M., Krom, M. & Merkx, M. Tuning the flexibility of glycine-serine linkers to allow rational design of multidomain proteins. Biochemistry 56, 6565–6574 (2017).
Bader, H., Sturzenegger, V. & Hoigne, J. Photometric-method for the determination of low concentrations of hydrogen-peroxide by the peroxidase catalyzed oxidation of N,N-Diethyl-P-Phenylenediamine (Dpd). Water Res. 22, 1109–1115 (1988).
Zou, J. et al. Spectrophotometric determination of trace hydrogen peroxide via the oxidative coloration of DPD using a Fenton system. Chemosphere 224, 646–652 (2019).
Price, M., Reiners, J. J., Santiago, A. M. & Kessel, D. Monitoring singlet oxygen and hydroxyl radical formation with fluorescent probes during photodynamic therapy. Photochem. Photobiol. 85, 1177–1181 (2009).
Thoudam, T. et al. Enhanced Ca(2+)-channeling complex formation at the ER-mitochondria interface underlies the pathogenesis of alcohol-associated liver disease. Nat. Commun. 14, 1703 (2023).
Achilli, C., Ciana, A. & Minetti, G. Oxidation of cysteine-rich proteins during gel electrophoresis. J. Biol. Methods 5, e104 (2018).
Brewer, J. M. Artifact produced in disc electrophoresis by ammonium persulfate. Science 156, 256–257 (1967).
Kong, A. T., Leprevost, F. V., Avtonomov, D. M., Mellacheruvu, D. & Nesvizhskii, A. I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520 (2017).
Messner, C. B. et al. The proteomic landscape of genome-wide genetic perturbations. Cell 186, 2018–2034 e21 (2023).
Lee, C. et al. Oxidative photocatalysis on membranes triggers non-canonical pyroptosis. Nat. Commun. 15, 4025 (2024).
Na, S., Kim, J. & Paek, E. MODplus: robust and unrestrictive identification of post-translational modifications using mass spectrometry. Anal. Chem. 91, 11324–11333 (2019).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Kim, S. et al. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 38, 1879–1886 (2017).
O’Boyle, N. M. et al. Open Babel: an open chemical toolbox. J. Cheminform. 3, 33 (2011).
Zoete, V., Cuendet, M. A., Grosdidier, A. & Michielin, O. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 32, 2359–2368 (2011).
Steinbach, P. J. B. & Brooks, B. R. New spherical-cutoff methods for long-range forces in macromolecular simulation. J. Comput. Chem. 15, 667–683 (1994).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Hoover, W. G. Canonical dynamics—equilibrium phase-space distributions. Phys. Rev. A 31, 1695–1697 (1985).
Parrinello, M. R. A. polymorphic transitions in single-crystals—a new molecular-dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Bondi, A. Van Der Waals volumes + radii. J. Phys. Chem. 68, 441–44 (1964).
Xu, N., Chen, G. & Liu, H. Antioxidative categorization of twenty amino acids based on experimental evaluation. Molecules 22, 2066 (2017).
Khazanov, N. A. & Carlson, H. A. Exploring the composition of protein-ligand binding sites on a large scale. PLoS Comput. Biol. 9, e1003321 (2013).
Sylvetsky, N. Toward simple, predictive understanding of protein-ligand interactions: electronic structure calculations on Torpedo californica acetylcholinesterase join forces with the chemist’s intuition. Sci. Rep. 10, 9218 (2020).
Goga, N., Rzepiela, A. J., de Vries, A. H., Marrink, S. J. & Berendsen, H. J. Efficient algorithms for Langevin and DPD dynamics. J. Chem. Theory Comput. 8, 3637–3649 (2012).
Berendsen, H. J. C., Postma, J. P. M., Vangunsteren, W. F., Dinola, A. & Haak, J. R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Shirts, M. R. & Chodera, J. D. Statistically optimal analysis of samples from multiple equilibrium states. J. Chem. Phys. 129, 124105 (2008).
Sanner, M. F., Olson, A. J. & Spehner, J. C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38, 305–320 (1996).
Moon, C. P. & Fleming, K. G. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc. Natl Acad. Sci. USA 108, 10174–10177 (2011).
Spurlino, J. C., Lu, G. Y. & Quiocho, F. A. The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J. Biol. Chem. 266, 5202–5219 (1991).
Gu, S., Silva, D. A., Meng, L., Yue, A. & Huang, X. Quantitatively characterizing the ligand binding mechanisms of choline binding protein using Markov state model analysis. PLoS Comput. Biol. 10, e1003767 (2014).
Schmidt, M. W. et al. General atomic and molecular electronic-structure system. J. Comput. Chem. 14, 1347–1363 (1993).
Jin, H. et al. Systematic transcriptional analysis of human cell lines for gene expression landscape and tumor representation. Nat. Commun. 14, 5417 (2023).
Acknowledgements
This work was supported by the National Research Foundation of Korea (2020R1C1C1003937 to D.M.; 2021R1A2C2009504 to T.H.K.; RS-2024-00455131 to S.K.M.; 2022R1A2C2091815 to C.U.K.; 2021R1C1C1010943 and 2022R1A4A1033471 to J.M.C.), the National Cancer Center (HA22C010100 to T.H.K.), the Korea Basic Science Institute (C470000 to S.N.), the Institute for Basic Science (IBS-R022-D1 to K.M.), and POSCO TJ Park Foundation (POSCO Science Fellowship to J.M.C.).
Author information
Authors and Affiliations
Contributions
D.M. conceived and supervised the project. S.K. and D.M. designed the single-molecule tweezer experiments. S.K., V.W.S., and W.C.B.W. performed the single-molecule tweezer experiments. S.K. and D.M. analyzed the single-molecule experiment data. S.K., H.J., and C.U.K. purified the proteins. S.K., M.P., E.H., and C.L. performed the ROS measurements. E.K. and D.M. performed the MD simulations, Monte Carlo analysis, protein cavity analysis, and bioinformatic analysis. J.M.C. reviewed the MD simulation results. Y.G.E. and J.M.C. performed the free energy calculations. S.H.K. and S.K.M. performed the DFT calculations. S.K., G.Y., M.P., and T.H.K. performed the EPR measurements. M.P., B.G.K., and T.H.K. prepared the samples for the LC-MS/MS analysis. B.G.K., S.N., and K.M. performed the LC-MS/MS. S.K., E.K., M.P., B.G.K., S.N., T.H.K., and D.M. analyzed the LC-MS/MS data. S.K., E.K., M.P., S.K.M., T.H.K., and D.M. prepared the manuscript, with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yongli Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, S., Kim, E., Park, M. et al. Hidden route of protein damage through oxygen-confined photooxidation. Nat Commun 15, 10873 (2024). https://doi.org/10.1038/s41467-024-55168-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55168-z
This article is cited by
-
Photooxidative molecular damage under blue light
Experimental & Molecular Medicine (2026)
-
Molecular dynamics insights into the redox effects on PD-1/PD-L1 and PD-1/PD-L2 interactions
Scientific Reports (2025)
-
Cryo-EM structure of amyloid fibrils formed by full-length human αA-crystallin with pathogenic mutation R116C
Communications Chemistry (2025)
-
Single-molecule tweezers decode hidden dimerization patterns of membrane proteins within lipid bilayers
Nature Communications (2025)