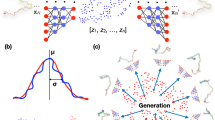
Abstract
Identifying transitional states is crucial for understanding protein conformational changes that underlie numerous biological processes. Markov state models (MSMs), built from Molecular Dynamics (MD) simulations, capture these dynamics through transitions among metastable conformational states, and have demonstrated success in studying protein conformational changes. However, MSMs face challenges in identifying transition states, as they partition MD conformations into discrete metastable states (or free energy minima), lacking description of transition states located at the free energy barriers. Here, we introduce Transition State identification via Dispersion and vAriational principle Regularized neural networks (TS-DAR), a deep learning framework inspired by out-of-distribution (OOD) detection in trustworthy artificial intelligence (AI). TS-DAR offers an end-to-end pipeline that can simultaneously detect all transition states between multiple free minima from MD simulations using the regularized hyperspherical embeddings in latent space. The key insight of TS-DAR lies in treating transition state structures as OOD data, recognizing that they are sparsely populated and exhibit a distributional shift from metastable states. We demonstrate the power of TS-DAR by applying it to a 2D potential, alanine dipeptide, and the translocation of a DNA motor protein on DNA, where it outperforms previous methods in identifying transition states.
Similar content being viewed by others
Introduction
Understanding the transition states of protein conformational changes, which typically encompass a collection of conformations located at the saddle points of free energy barriers that separate different free energy basins, is crucial for gaining insights into various biological processes, including protein folding, misfolding, gene expression, etc. This understanding also facilitates drug design and enzyme engineering. For example, a recent study1 suggests that a single point mutation can rescue RNA polymerase II from pausing at an epigenetically modified DNA base by destabilizing the transition state during the conformational change of translocation. This insight into transition states deepens our understanding of how epigenetic modifications interfere with gene expression. Moreover, understanding transition states could aid in designing drug compounds that bind to these states, thereby effectively inhibiting the functions of targeted proteins. However, due to the low populations and transient features of these transition state structures, it remains challenging to directly investigate them at atomic resolutions using experimental techniques.
Molecular dynamics (MD) simulations can serve as a powerful approach to complement experimental methods in studying protein conformational changes, as they enable the elucidation of conformational dynamics in a high spatial and time resolution. However, all-atom MD simulations typically operate at femtosecond time-step, posing a challenge in capturing protein conformational changes that usually occur in milliseconds or longer. Markov state models (MSMs)2,3,4,5,6,7,8,9,10,11,12,13,14 have effectively addressed this challenge by integrating multiple short MD trajectories to predict long-time scale dynamics. Despite the significant success of MSM in studying conformational changes over the past decade15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35, identifying transition state structures remains challenging. In MSMs, dynamic processes are modeled as a series of Markovian transitions among metastable conformational states (or free energy minima) at discrete time intervals (called lag times). Each MD conformation, including those in the transition state region, is therefore assigned to a specific metastable state, complicating the unraveling of transition states.
For an MSM containing a large number of small states (or microstates), one approach to identifying transition state structures within the MSM pipeline is to compute the committor probabilities of these small microstates. Microstates with an equal probability (committor equals 0.5) of reaching the initial and final metastable states can be identified as transition states between the two. However, the efficacy of this method relies significantly on the quality and accuracy of constructing the corresponding microstate MSMs. Additionally, it can only identify transition states between pairs of metastable states one at a time. Recently, a deep learning-based approach, MaxEnt-VAMPNets36, has been developed to identify transition states structures to facilitate the adaptive sampling. This approach utilizes the state assignment probabilities output from VAMPnets37 to calculate the Shannon entropy for each MD conformation. It assumes that the conformations with higher Shannon entropy values are more likely to be located at low-probability regions (i.e., the summit of free energy barriers). However, the state assignment probabilities from VAMPnets37 represent the basis functions that can best linearly reconstruct the system’s slowest dynamic modes38. It is not guaranteed that they can precisely reflect the probabilities of MD conformations transitioning in or out of metastable states.
In the past decades, a number of other MD simulation-based methods have been developed for identifying the transition states of conformational dynamics. For instance, transition path sampling (TPS)39,40,41 directly uncovers transition states using the committor function derived from the transition path ensemble generated through Monte Carlo sampling. Additionally, a deep reinforcement learning approach, integrating an efficient path sampling method called enhanced sampling of reactive trajectories (ESoRT)42,43, has been employed to identify transition states by framing the problem as a shooting game44. Rather than directly capturing transition states, more approaches have been developed to extract the optimal reaction pathways45,46,47,48,49,50,51,52 or calculate committor functions53,54,55,56, both of which greatly assist in transition states identification. However, despite the robustness and promise of these methods, they may pose challenges in terms of their high computational cost, or the requirement for prior knowledge and accurate characterization of the initial and final states.
Recent advancements from the out-of-distribution (OOD) detection within trustworthy artificial intelligence (AI) offer potential solutions to the challenges of simultaneously identifying all transition states between multiple free-energy minima from MD simulations. OOD detection57, an important task for trustworthy AI, has emerged and attracted increasing attention in recent years. The major issue targeted by OOD detection is that the model trained on a specific closed-world dataset, i.e., the in-distribution (ID) data, may make overconfident and wrong predictions on unknown examples, i.e., the OOD data, from the open-world. Therefore, a reliable deep learning model should be able to perform a binary ID v.s. OOD detection classification task, and reject OOD data points. This is especially important when applying deep learning models to safety-critical applications58,59,60,61,62,63 such as self-driving cars and rare disease detection tasks. OOD detection, which has not been previously employed in the study of biomolecular conformational changes, bears significant potential in identifying transition states. This is because the conformations at the transition states, located at free energy barriers, are sparsely-populated and exhibit out-of-distribution characteristics in contrast to conformations within the densely populated free energy basins (the ID data). Consequently, these transition states can be considered as OOD data.
Recently, Ming et al. developed a Compactness and DispErsion Regularized learning framework (CIDER)64 for detecting OOD images. This was achieved by harnessing latent hyperspherical embeddings that are effectively regularized through the joint optimization of compactness loss and dispersion loss. Specifically, the hyperspherical embeddings (\({{\rm{z}}}\in {{\mathbb{R}}}^{d}\)) refers to a set of points in \(d\)-dimensional Euclidean space that are located at a constant distance from the center, collectively defining a (\(d-1\))-dimensional hypersphere, as illustrated in Fig. 1c. Following this, the compactness loss encourages the tightening of image samples in each class on the hypersphere, while the dispersion loss promotes large angular distances between class prototypes. Consequently, OOD samples are expected to lie between class prototypes and can be detected based on cosine similarity-based measures. The concept introduced by CIDER serves as inspiration for our development of a deep learning approach aimed at detecting transition state structures in protein conformational changes. In the context of biomolecular dynamics, the metastable free energy basins, akin to class prototypes, should be separated from one another, while the transition state structures, representing OOD samples, are expected to reside in between them. However, a major challenge in directly applying CIDER to protein dynamics is that CIDER was developed under supervised learning settings, requiring labeled data for computing class prototypes. In contrast, protein conformations in MD trajectories typically lack these ground-truth labels. VAMPnets37 offers a potential solution by bridging the gap between unsupervised learning and the need for labeled data. Furthermore, the VAMP-2 loss function, which is designed to capture the slowest dynamic modes of the system based on the separation of timescales, can facilitate the compression of metastable conformations within each energy basin, playing a similar role to the compactness loss in CIDER.
a Overview of the TS-DAR framework. Similar to VAMPnets37, TS-DAR takes transition pairs \({{{\rm{x}}}}_{t}\), \({{{\rm{x}}}}_{t+\tau }\) from simulation trajectories as input, generates the Softmax outputs, and estimates the VAMP-2 loss. Meanwhile, TS-DAR introduces an L2-norm/scale layer to create the hyperspherical embeddings at the penultimate layer. These embeddings, combined with the pseudo state assignments obtained from the Softmax outputs, are used to estimate the dispersion loss. The framework then optimizes the neural networks using a combined loss function of the VAMP-2 loss and the dispersion loss, weighted by a constant \(\beta\). b Utilization of the L2-norm/scale layer to confine original feature embeddings \(\widetilde{{{\rm{z}}}}\) at the penultimate layer within a hypersphere of radius \(\gamma\), producing the hyperspherical embeddings \({{\rm{z}}}\). c Identification of the transition states in the hyperspherical latent space. The VAMP-2 loss enhances data compactness within each metastable state, while the dispersion loss encourages the centers of different metastable states to be far apart across the hypersphere.
In this work, we present an end-to-end approach called Transition State identification via Dispersion and vAriational principle Regularized neural networks (TS-DAR) for detecting transition state structures of protein conformational changes from MD simulations. This approach utilizes the hyperspherical latent representations of the biomolecular conformations. The loss function comprises two terms: VAMP-2 loss and dispersion loss. By minimizing the VAMP-2 loss, the MD conformations within the same metastable state (or free energy basin) are compacted in the hyperspherical latent space. The dispersion loss further ensures the metastable state centers are uniformly distributed across the hypersphere. Consequently, all transition state conformations, located between free energy basins, could be simultaneously and automatically identified in the latent space. In TS-DAR, the key advantage of introducing hyperspherical latent space is that its geometry effectively prevents the dispersion term from pushing data infinitely apart, thereby ensuring a uniform distribution of free energy basins across the hypersphere. We have demonstrated the efficacy of our method by applying it to three systems: the 2D Müller potential, alanine dipeptide and the translocation of a DNA motor protein along double-strand DNA (dsDNA).
Results and discussion
Hyperspherical latent representations at the penultimate layer in TS-DAR
The schematic representation of TS-DAR’s model architecture is shown in Fig. 1a. Different from VAMPnets37, which directly employs two parallel encoders to utilize transition pairs of MD conformations to produce the SoftMax probabilities of state assignments, TS-DAR introduces a enhancement in its model architecture. Specifically, it incorporates an additional L2-norm/scale layer at the penultimate layer to extract the hyperspherical latent representations of MD conformations (Fig. 1a). These hyperspherical latent representations can be effectively regularized by the joint optimization of VAMP-2 loss and dispersion loss, enabling the robust transition states identification on the latent hypersphere (Fig. 1c). In particular, the L2-norm/scale layer consists of two parts. The feature vectors at the penultimate layer are first divided by their L2-norms, and then rescaled by a scaling factor \(\gamma\) (Fig. 1b). As a result, the feature embeddings at the penultimate layer are successfully confined on a hypersphere of radius \(\gamma\), referring to as the hyperspherical latent representations. To illustrate the hyperspherical representations of MD trajectories, we utilize a 2D Müller potential65 with three minima as an example (Fig. 2a). In the latent space of the trained TS-DAR model for the Müller potential (Fig. 2b), all MD conformations perfectly lie on a hypersphere. Three distinct free energy basins can be clearly identified in this hypersphere, with dashed lines denoting the mean vectors of each basin in Fig. 2b. Furthermore, these three free energy basins in the hypersphere (Fig. 2d) correspond clearly to the three energy basins of the Müller potential (see basin 1–3 in Fig. 2a). Pseudo-code for the TS-DAR algorithm is provided in SI Fig. S1, and additional details on training the TS-DAR model can be found in the Methods section.
a The 2D-Müller potential. b Projections of molecular dynamics (MD) conformations onto the latent hyperspherical space, and the free energy (\(-{\mbox{k}}T{\mathrm{ln}}P(\theta )\)) is displayed, where \({\mbox{k}}T\) is the Boltzmann constant and temperature, \(P\left(\theta \right)\) corresponds to the probabilities of MD conformations at the polar angle \(\theta\) on the hypersphere. The dashed lines indicate the mean vectors of each of the three metastable states. c Visualization of the output state assignments (state 1 to 3) of the TS-DAR model overlaid on the Müller potential. d The same as (b) except that the state assignments rather than potential of mean force in the latent space are displayed. e and f are the same as (c) and (d), respectively, except that the results of a control experiment that only includes the VAMP-2 loss37 when training the TS-DAR model are shown. Source data are provided as a Source Data file.
Utilizing VAMP-2 loss to capture the slowest dynamics within the hypersphere
Like in VAMPnets37, the minimization of the VAMP-2 loss (\({{{\mathcal{L}}}}_{{\mbox{vamp}}}\) in Eq. 1) ensures our TS-DAR model can capture the slowest dynamic modes underlying the conformational changes of interest.
where \({\bar{{{\rm{C}}}}}_{01}\) represents the time-lagged correlation matrix, \({\bar{{{\rm{C}}}}}_{00}\) and \({\bar{{{\rm{C}}}}}_{11}\) denote the time-instantaneous covariance matrices at time \(t\) and \(t+\tau\) (\(\tau\) is the lag time). The details for computing these correlation matrices are presented in the Methods section. According to this property, the SoftMax outputs from TS-DAR can provide optimal state assignments that are aligned with the free energy basins, allowing the on-the-fly labeling of MD conformations during the training. In addition, with the help of the VAMP-2 loss function term, the hyperspherical latent representations in the trained TS-DAR model can retain all relevant kinetic geometries and compact the conformations on the hypersphere in terms of their kinetic metastability. This is demonstrated by the latent space representation of the Müller potential (Fig. 2d), where the three identified free energy minima and their order of connection correspond precisely with the arrangements of the three energy basins of the Müller potential (Fig. 2c). Additionally, we observe that the hyperspherical latent space effectively condenses MD conformations into three distinct clusters, each corresponding to a specific metastable free energy basin. For instance, the largest free energy basin (basin 1 in Fig. 2a) has been more significantly compressed compared to the other two basins due to its large metastability, resulting in a small but deep free energy minimum in the latent space (Fig. 2b). We attribute this observation to the presence of a sole fully connected layer extending from the latent bottleneck to the outputs, compelling the latent representations to optimally capture the slowest dynamics through the VAMP-2 loss. This design of the penultimate layer for representation learning has already exhibited considerable promise in diverse deep learning fields ranging from computer vision to natural language processing66,67. Finally, to elucidate the distinct roles of the VAMP-2 and dispersion terms in the loss function, we conducted a control experiment by omitting the dispersion loss. The aforementioned observations remain the same despite the absence of the dispersion loss (Fig. 2e, f). However, it is noteworthy that the three free energy basins exhibit uneven distribution in the latent space when the dispersion loss is excluded. Further discussion on this point will be presented in the next section.
Implementing dispersion loss for uniform latent distribution of metastable state centers and defining an OOD score for transition state identification
The dispersion loss was initially introduced by us and has successfully been employed in OOD detection for image classification tasks64. In TS-DAR, we introduce the dispersion loss aiming to encourage the state centers (i.e., free energy minima) to be uniformly distributed across the hypersphere by maximizing the angular distances between these centers. For example, in the presence of the dispersion loss (Fig. 2d), the centers of 3 free energy basins of the Müller potential (labeled as state 1–3) are well separated and uniformly distributed in the latent space. In sharp contrast, without the dispersion loss (Fig. 2f), the three state centers exhibit an uneven distribution in the latent space. The dispersion loss is defined as follows:
where \(C\) corresponds to the number of states, \({{{\mathbf{\mu }}}}_{c}\) is a unit vector, representing the mean direction of all conformations (state center) in state \(c\), and \(\sigma\) is a scaling hyperparameter and specifically defined as 0.1. To compute the dispersion loss, it is necessary to first estimate the state center vectors \({\left\{{{{\mathbf{\mu }}}}_{c}\right\}}_{c=1}^{C}\). For the robustness and the efficiency of training, we employ an exponential-moving-average (EMA)68 method to estimate \({\left\{{{{\mathbf{\mu }}}}_{c}\right\}}_{c=1}^{C}\) on-the-fly and update them frequently during the training (see the Methods section and SI Fig. S1 for details).
After we obtained the regularized hyperspherical latent representations through the joint optimization of VAMP-2 and dispersion loss, the metastable states are compacted and uniformly separated. As a result, transition states in between these metastable states will share equal angular distances to their nearest state centers (Fig. 2d). This prompts us to define an OOD score based on the cosine similarity to quantify the angular distances in the hyperspherical latent space (Eq. (3)), and thus help identify all the transition states automatically and simultaneously.
where \({{\rm{z}}}\) represents the hyperspherical latent embedding of the conformation, \({{\mathcal{U}}}\) denotes as \(\left[{{{\mathbf{\mu }}}}_{1},{{{\mathbf{\mu }}}}_{2},\ldots,{{{\mathbf{\mu }}}}_{C}\right]\). In light of the provided definition, the OOD score ranges from a minimum of \(0\), with higher values indicating increased out-of-distribution characteristics, thereby highlighting the structures in transition states. As shown in Fig. 3b, MD conformations situated at two different transition state regions (between basins 1 & 2, and between basins 2 & 3 of the Müller potential in Fig. 2a) are identified simultaneously, as they display equal and the largest OOD scores in the latent space. We then selected MD conformations with OOD scores larger than a threshold as transition state structures. As shown in Fig. 3c, the predicted conformations at various OOD threshold choices all exhibit committor probabilities with an average of 0.5 for both TS12 and TS23 (see the Methods section for the analytical solution of committor for the Müller potential). Since transition state structures theoretically should have committor probabilities of 0.5 (i.e., equally likely to reach the product or return to the reactant), these findings suggest that TS-DAR can robustly predict the transition state structures. In addition, with the increase in the OOD threshold from 0.225 to 0.425, the number of predicted transition state structures decreases from ∼1,000 to ∼300 for TS12, and from ∼3,500 to ∼300 for TS23 (Fig. 3c). This provides a way to tune the OOD threshold to obtain a desired number of transition state structures.
a Hyperspherical latent representations with out-of-distribution (OOD) scores obtained from a 3-state TS-DAR model are shown. Dashed lines point to the centers of metastable states. b Molecular dynamics (MD) conformations with their OOD scores obtained from TS-DAR are overlaid with the Müller potential. c The average committor probabilities (error bar denotes the standard deviation) of transition state structures for TS12 and TS23 selected by TS-DAR with different threshold of the OOD score (top). The sample sizes (n) corresponding to each data point reflect the number of selected MD conformations for TS12 and TS23 at different OOD score thresholds, as shown in the bottom panel. These sample sizes are: [1154, 1011, 890, 776, 657, 560, 474, 384, 304] for TS12 and [3668, 3181, 2678, 2228, 1817, 1435, 1068, 693, 337] for TS23. d, e MD conformations with committor probabilities obtained from the 1000-state Markov state model (MSM) overlaid with the Müller potential. The white regions represent the source and sink states, which are chosen as the highest populated states in basin 1 and 2 (d) and the highest populated states in basin 2 and 3 (e), respectively. For further details, please refer to the SI Sec. 4. f MD conformations with Shannon entropy obtained from 3-states MaxEnt-VAMPNets36 are overlaid with the Müller potential. The black dashed lines in (b, d–f) correspond to iso-committor lines with the committor probability of 0.5 obtained from the analytical solutions. For further details, please refer to the Methods section. Source data are provided as a Source Data file.
Previous studies57 in the trustworthy AI field have utilized other similarity-based metrics in the latent representations for OOD detection. However, these methods may not be ideally suited for direct application in identifying transition states for protein conformational changes. For example, one can directly detect OOD samples at the state boundaries on the hypersphere and treat them as transition states structures. However, this method may not be well-suited for identifying transition states in our TS-DAR model. This is because the VAMP-2 loss in TS-DAR is designed to optimize the slowest dynamic modes, rendering state boundaries less sensitive and thereby limiting its ability to accurately pinpoint transition state conformations. As shown in SI Fig. S3b, MD conformations located at the state boundaries in the Müller potential do not accurately correspond to true transition states. Recent approaches64,69 including CIDER aim to detect low-density regions in the latent space as OOD samples, which may serve as potential transition state structures in our context. However, these methods may overlook the transition states separated by relatively low free energy barriers or incorrectly classify the low-density regions as transition states. For example, choosing an appropriate density threshold to concurrently identify two transition states of the Müller potential is challenging. With a density threshold set at \(\exp (-7.2{\mbox{k}}T)\), only the transition state between states 1 and 2 can be identified (SI Fig. S4c). Reducing the threshold to \(\exp (-6{\mbox{k}}T)\) enables the identification of the transition state between states 2 and 3. However, in this case, the transition state between states 1 and 2 becomes poorly defined (SI Fig. S4d).
TS-DAR outperforms MSM’s committor probabilities and MaxEnt-VAMPNets in identifying transition states for the Müller potential
The Müller potential exhibits three potential energy basins, with two transition states, positioned between them (Fig. 2a). We trained our TS-DAR model by specifying 3 metastable states. The validation curves are depicted in Fig. S2a, and more training details are in the Methods section. With the trained model, we first plotted the OOD scores of all MD conformations in the latent hyperspherical space (Fig. 3a). As shown in Fig. 3b, the MD conformations positioned between the state centers consistently align with those located at the summit of the energy barriers. This facilitates the straightforward identification of transition state structures. In the three-state model, both transition states situated between the three energy basins are simultaneously captured (Fig. 3b). If one’s interest lies solely in the transition state associated with the highest energy barrier (TS12), we show that a two-state TS-DAR model can also accurately identify this transition state (SI Fig. S5).
We next demonstrate that our TS-DAR outperforms two previously developed methods, MaxEnt-VAMPNets36 and MSM’s committor probabilities (MSM-committor)15, in identifying the transition states for the Müller potential. For MaxEnt-VAMPNets, the VAMPnets model was trained with three states (see SI Sec. 3 for details). Regarding committor probabilities, the Müller potential was first discretized into 1,000 microstates using k-centers clustering. Committor probabilities were then computed based on the 1,000-state MSM by specifying the source and sink states (see SI Section 4 for details). As shown in Fig. 3f, MaxEnt-VAMPNets mistakenly identified the energy basin 1 instead of the energy barrier as the transition state region, characterized by high Shannon entropy. We anticipate that this misidentification occurs because the Shannon entropy computed from VAMPnets’ outputs lacks direct physical connections with the true uncertainty measures of MD data, either in terms of transitioning in or out of energy basins. Regarding MSM-committor, its performance is heavily dependent on the selection of the source and sink states (SI Fig. S6a, c). When the source and sink states are small (each containing only one microstate), MSM-committor can identify the correct transition state region for TS12, despite assigning relatively low committer probabilities of ∼0.3 for true transition state structures obtained from the analytical solution (Fig. 3d). Strikingly, for TS23, MSM-committor erroneously predicts a broad transition state region including even the entire energy basin 1 (Fig. 3e). In this scenario, microstates in the energy basin 1 that are far away from the sink and source states are wrongly categorized as transition states. These remote states might have weak connections with sink and source states, and therefore share an equal probability of transitioning to either, leading to the misidentification as shown in Fig. 3e. When the source and sink states are relatively large (e.g., containing 100 and 10 most populated microstates for TS12 and TS23, respectively), MSM-committor can successfully uncover the correct regions for both transition states (SI Fig. S6b, d).
Transition states for alanine dipeptide
Alanine dipeptide consists of 10 heavy atoms, with its conformational changes often visualized through two backbone torsion angles, \(\phi\) and \(\psi\) (Fig. 4a, b). To identify its transition states, we trained 3 TS-DAR models for alanine dipeptide using the Cartesian coordinates of its 10 heavy atoms as input features, and the number of metastable states was set to be 2, 3, and 4, respectively (see Fig. 4c and refer to the Methods section for additional details). In the 2-state TS-DAR model, we successfully identified the transition state (TS13) located on the highest free energy barrier (Fig. 4d). This free energy barrier corresponds to the slowest dynamic transition between \(\beta\) (free energy basin 1 in Fig. 4b) and \({\alpha }_{{\mbox{L}}}\) (free energy basin 3 in Fig. 4b) metastable states of alanine dipeptide. In the 3-state model, we identified additional transition state regions situated between free energy basins 1 and 2 (TS12). It is noteworthy that since the torsion angles exhibit periodicity every 2π, there are two transition state regions that separate basins 1 and 2 \(({\alpha }_{{\mbox{R}}})\) (Fig. 4e). In the 4-state model, all the transition states were simultaneously identified, with additional transition states (TS34) detected corresponding to the third slowest dynamic mode, separating the two free energy basins (3 and 4) located on the right-hand side of Fig. 4f. Among the three transition states, TS13 (located at the highest free energy barrier) is the least sampled by our MD simulations, with only 6 conformations found in this region (SI Fig. S7a). In contrast, TS12 is the most sampled, and TS-DAR can provide a varying number of MD conformations in this region by changing the OOD threshold: the number of candidate transition state structures ranges from ∼30 to ∼7,000 with the OOD threshold decreases from 0.4 to 0.2 (SI Fig. S7b). To further assess the robustness of TS-DAR and validate its computational efficiency with respect to MD sampling, we also trained our TS-DAR models on a series of down-sampled datasets, with the total MD sampling of 3, 30, 90, 300, and 750 ns (the full dataset). As shown in SI Fig. S8a–d, our 3-state TS-DAR models successfully identified TS12 across all down-sampled datasets, yielding consistent results with that obtained from the full dataset (Fig. 4e). Strikingly, as illustrated in SI Fig. S8a, TS-DAR was able to identify TS12 with just 3 ns of total sampling. For TS13, it requires TS-DAR 90 ns sampling to properly identify this transition state, located at the highest free energy barrier. In a previous study70, TPS was applied to investigate the transition state TS12, which required a total of ∼77 ns of MD simulation sampling.
a A representative conformation of alanine dipeptide. b Projection of the free energy landscape for alanine dipeptide onto two backbone torsion angles, \(\phi\) and \(\psi\). c Hyperspherical representations in a 3D latent space (2D hypersphere) for the 4-state TS-DAR model, with dashed lines indicating the centers of metastable states. d–f TS-DAR models with 2, 3, and 4 states successfully identified transition states located at different free energy barriers. Notably, for visualization, the molecular dynamics (MD) conformations with large out-of-distribution (OOD) scores (shown in yellow) are highlighted. g–i MD conformations with committor probabilities obtained from the 1,000-state Markov state model (MSM) overlaid with the free energy landscape mapped onto two backbone torsion angles. Notably, for visualization, the conformations with the committor probabilities between 0.4 and 0.6 are highlighted. The white regions represent the source and sink states, utilized as input into the transition path theory for computing committor probabilities. For further details, please refer to the SI Sec. 4. j–l Shannon entropy values of MD conformations obtained from 2, 3 and 4-state MaxEnt-VAMPNets36 are shown. Source data are provided as a Source Data file.
We then compare the performance of TS-DAR with MSM-committor and MaxEnt-VAMPNets. As shown in Fig. 4g–i, three independent analyses using MSM-committor with different pairs of source and sink states successfully identify three transition state regions for alanine dipeptide, consistent with the findings of a single 4-state TS-DAR (Fig. 4e). However, the committor probabilities obtained from MSM-committor analysis still depend on the selection of the source and sink states (SI Fig. S9). In contrast, in MaxEnt-VAMPNets models, while MD conformations in the transition state regions TS12 and TS23 are assigned high Shannon entropy values, those in the free energy basins 1 and 2 also receive high Shannon entropy values. Consequently, this complicates the clear identification of TS12 and TS23 (Fig. 4j–l). For TS34, the 4-state MaxEnt-VAMPNets model correctly identifies this transition state (see Fig. 4l). In the 4-state TS-DAR model, we chose a three-dimensional latent space (2D hypersphere), and the centers of the 4 metastable states (indicated by the dashed lines, see Fig. 4c) form a tetrahedral geometry with the inter-state angular distance approximately equals to 109 degrees. In the implementation of the TS-DAR method, we recommend choosing a three-dimensional latent space (2D hypersphere) for models containing 4 or more states.
Transition states for the translocation of a motor protein on DNA
Bacillus cereus alkylpurine glycosylase D (AlkD) is a DNA motor protein that can translocate along the dsDNA, and repairs DNA damage to maintain genome integrity71. Understanding the translocation of AlkD on dsDNA is crucial for comprehending how it diffuses along the genomic DNA to locate sparse lesion sites efficiently and accurately within the entire genome. We employed TS-DAR to investigate the transition states of the diffusion dynamics of AlkD along a double-stranded DNA for the distance of one base pair. We have followed our previous study72 to select 684 pairwise distances as the input features (SI Fig. S10) and 4 states to train TS-DAR. In addition, we have also trained TS-DAR models on different input feature sets (see SI Sec. 2 for details) and shown that TS-DAR has exhibited robust convergence in terms of incorporating more features (SI Fig. S11). Figure 5a displays the hyperspherical latent representations of AlkD-DNA complexes obtained from the TS-DAR model, where four successively connected free energy basins (pointed by dashed lines) are uncovered and uniformly separated across the hypersphere. These four free energy basins are consistent with those revealed by MSMs in a previous study72 (SI Fig. S12). Specifically, free energy basin 1, 2, 3 correspond to the pre-translocation, an intermediate state exhibiting a rotation of AlkD on dsDNA, and post-translocation state, respectively (Fig. 5b). Notably, we didn’t show basin 4 as it represents a hyper-translocation state, where the AlkD has translocated along the dsDNA beyond one base pair. We identify three transition states with high OOD scores (>\(0.21\), see Fig. 5a and the Methods section for additional details) that separate adjacent free energy basins in the hyperspherical latent space.
a Hyperspherical representations in a 3D latent space of AlkD-DNA complex system from the TS-DAR model, with dashed lines indicating the center vectors of metastable states and the rectangle box outlining the selected transition states structures. b Representative conformations of the three basins and the transition state are shown from two different point of views. AlkD and dsDNA are shown as white and gray, respectively. The \({{\rm{\alpha }}}\)-helices that are in contact with the dsDNA are shown in individual colors and the phosphor atom of the A7 phosphate group is shown as a blue sphere. The average number of hydrogen bonds between a residue of AlkD and two adjacent nucleotides of the dsDNA were calculated for each basin and transition state. c The average number of hydrogen bonds between AlkD-dsDNA that are broken (black bars) and new hydrogen bonds that are formed (red bars), during the transition from basin 1 to TS12 (left) and from TS12 to basin 2 (right). d The same as (c) except that the average number of hydrogen bonds for the transition from basin 2 to TS23 and from TS23 to basin 3 is shown. e Interaction energies between AlkD and dsDNA for basin 1, TS12, and basin 2, are shown. The interaction energies are computed by considering AlkD residues and DNA ribonucleotides that are within 3 Å of each other in at least one molecular dynamics (MD) conformation. f The same as except that AlkD-dsDNA interaction energies for basin 2, TS23, and basin 3 are shown. The average and error bars in (c)–(f) were calculated via bootstrapping of the MD simulation trajectories 100 times. For each bootstrap sample, the mean of the variable (either number of hydrogen bonds or protein-DNA interactions) was calculated for each basin and transition state. The plotted average and error bar is the average and standard deviation of all the mean values from 100 bootstrap samples, respectively. Source data are provided as a Source Data file.
Previous MSM72 revealed a two-step translocation pathway of AlkD on dsDNA involving an intermediate state (Fig. 5b). However, it remains elusive why transitions from pre-translocation (basin 1) to the intermediate state (basin 2) constitute the rate-limiting step, which is an order of magnitude slower than the other step from basin 2 to basin 3 (∼17.8 μs v.s. ∼1.3 μs72). This issue arises because MSMs can only provide information on the free energy basins, while kinetic rates are determined by both free energy basins and the transition states. The transition states revealed by our TS-DAR successfully address this issue and elucidate molecular mechanisms responsible for the rate-limiting step of the AlkD translocation. Specifically, the transition state (TS12) separating basin 1 and basin 2 exhibits a partial rotation of AlkD on dsDNA (see the top view of Fig. 5b), resulting in an unfavorable interaction energy between the protein and dsDNA compared to both basin 1 and basin 2 (Fig. 5d). Further analysis reveals that this unfavorable interaction energy is associated with significant reconfigurations of hydrogen bonds between AlkD and dsDNA. During the transition from basin 1 to TS12, approximately 10 existing AlkD-dsDNA hydrogen bonds must be broken, while only about 5 new hydrogen bonds are formed (Fig. 5c). Consequently, there exists a net loss of ∼5 hydrogen bonds within TS12. For example, the rotational movement of AlkD disrupts the hydrogen bond initially formed between residue R43 and base 23T. Subsequently, a new hydrogen bond forms with an adjacent base, 22 G (SI Fig. S13a). Thus, TS12 presents a significant free energy barrier that AlkD must overcome. In contrast, in the transition state TS23, AlkD forms a similar number of hydrogen bonds with the dsDNA compared to basins 2 and 3 (Fig. 5e), along with a comparable AlkD-dsDNA interaction energy (Fig. 5f). This results in a relatively fast transition from the intermediate state to the post-translocation state (basin 3), involving the translation of AlkD on dsDNA (see the side view of Fig. 5b). Transition states identified through our TS-DAR offer insights into comprehending the rate-limiting step in AlkD’s translocation on dsDNA and underscore the important role of hydrogen bonds in governing the dynamics of this conformational change.
For AlkD, the transition state regions revealed by our TS-DAR (SI Fig. S14c) align with those identified through two separate MSM-committor analyses (TS12 in SI Fig. S15c and TS23 in SI Fig. S15d). The committor probabilities obtained from the MSM-committor analysis vary slightly depending on the selection of the source and sink states (SI Fig. S15c–h). The 4-state MaxEnt-VAMPNets model assigned high Shannon entropy values to the free energy basins 1 and 2, erroneously identifying them as transition state regions (SI Fig. S16). In TS-DAR, as the OOD threshold increases from 0.2 to 0.4, the number of transition state structures decreases from approximately 150 to about 20 for TS12, and from roughly 850 to around 100 for TS23 (SI Fig. S14a, b). This enables TS-DAR to provide the desired number of candidate structures for a given transition state by adjusting the OOD threshold.
In this study, we show that the hyperspherical latent representations from TS-DAR model’s penultimate layer serve as a good reduced kinetic space for understanding the slow dynamics of protein conformational changes. Previous methods, such as tICA73,74, and SRVs75, which are rooted in the variational approach, aim to identify decorrelated orthogonal collective variables (CVs) for dimensionality reduction. However, these methods may hinder the comprehensive understanding of intricate collaborative dynamic motions. In contrast, alternative approaches like RC flow76 are specifically designed to reveal latent kinetic manifolds that preserve full-state kinetic information. Nevertheless, these methods pose greater challenges in terms of training and may struggle to discover clearer state boundaries. Here, we highlight the robustness of our hyperspherical latent representations for capturing the relevant reduced kinetics and underscores that these representations benefit from two perspectives. Firstly, the utilization of penultimate layer of a deep neural network for representation learning is simple and robust. This design choice facilitates the learned latent representations to be directly regularized through the optimization of the loss function defined on the output layer, such as VAMP-2 score in our framework. Secondly, and of more significance, representation learning on a unit sphere helps the model better capture and describe kinetics. Previous works in the field of deep learning77,78,79,80,81,82 have demonstrated that the hyperspherical latent space performs better than traditional Euclidean space in applications ranging from variational autoencoders to convolutional neural networks for image classification tasks. In our specific context of biomolecular dynamics, the hyperspherical latent space is particularly important, offering greater capacity for describing more complex kinetic geometries including specific kinetic symmetries and periodicity of the kinetic data. In the future, we anticipate that the hyperspherical latent representations in TS-DAR will have broad applications in the study of biomolecular dynamics. For example, it can provide a good platform for analyzing the parallel transition pathways of complex dynamic systems on the hypersphere. Furthermore, recent developments76,83 have utilized autoencoder or normalizing flow architectures to learn a reduced kinetic model in the latent space, such as Brownian dynamics, by introducing specific dynamical assumptions or constraints in the loss function. We expect that the penultimate hyperspherical latent space in TS-DAR can be deployed for learning a continuous dynamic model by integration of more physical-driven designs of loss functions.
Our TS-DAR framework can also be applied in various fields beyond its original scope. For example, MaxEnt-VAMPNets can accelerate phase space exploration by iteratively launching parallel, short, unbiased MD trajectories from high-uncertainty regions, identified through the Shannon entropy from VAMPnets outputs. Similarly, TS-DAR holds significant promise for adaptive sampling of biomolecular dynamics. The OOD scores generated by TS-DAR can serve as a robust metric to select the initial seeds (i.e., the conformations with the highest OOD scores) for adaptive sampling. This anticipated robustness of OOD scores from TS-DAR for adaptive sampling can be attributed to two key reasons. First, as demonstrated in this work, OOD scores enable the automatic and simultaneous identification of TS conformations of biomolecular dynamics. TS, positioned at the free energy barriers, serve as ideal candidates for initiating trajectories that can effectively explore the configuration space. Second, OOD scores measures the degree of the distributional shift away from the free energy minima. Under-sampled regions that exhibit distributional shifts away from well-explored metastable states can thus be easily detected by OOD scores. As such, OOD score can serve as an effective metric to detect the high-uncertainty regions for adaptive sampling in future studies.
Despite the robustness of TS-DAR, we summarize the potential limitations when applying our method. First, TS-DAR is trained on MD simulation datasets, so it is essential that the original MD trajectories capture the transitions between free energy basins and sample the transition state regions. Second, the input number of states (N) for TS-DAR cannot be too large. Similar to VAMPnets, a too large N will result in empty assignments of states and unstable training. Additionally, too many states can overcrowd the hypersphere latent space, making it difficult to distinguish between conformations in the transition state and those in the free energy basins (metastable states) using angular distance measures.
In this work, we introduce TS-DAR, a deep learning approach designed for detecting transition states from MD simulations by utilizing hyperspherical embeddings in the latent space. Inspired by recent advancements in trustworthy AI for identifying OOD data, TS-DAR treats transition state structures as OOD data. This approach allows TS-DAR to discern transition state conformations that separate multiple metastable states, facilitated by the introduction of a dispersion loss function term. Moreover, the hyperspherical embeddings of MD conformations in TS-DAR retain all relevant kinetic geometries and are compacted in terms of their kinetic metastability through the incorporation of a VAMP-2 loss function term. Collectively, our TS-DAR method establishes an end-to-end pipeline capable of simultaneously and automatically identifying all transition states across multiple free energy barriers underlying protein conformational changes. To demonstrate the efficacy of TS-DAR, we apply it to the 2D Müller potential, alanine dipeptide, and the translocation of a DNA motor protein on dsDNA. Across all three systems, we show that our TS-DAR outperforms MSM-committor and MaxEnt-VAMPNets in terms of efficiency and accuracy in simultaneously identifying all transition states for protein conformational changes. We anticipate that TS-DAR can find widespread application in identifying transition states for protein conformational changes.
Methods
Remove-mean time-instantaneous and time-lagged correlation matrices
Given a set of basis functions \({{\mathcal{{X}}}}={\left[{{{\mathcal{X}}}}_{1},{{{\mathcal{X}}}}_{2},\ldots,{{{\mathcal{X}}}}_{m}\right]}^{{{\rm{T}}}}\), and a MD trajectory of length \(T\) (\(\left\{{{{\rm{x}}}}_{1},\ldots,{{{\rm{x}}}}_{T}\right\}\)), the remove-mean time-instantaneous and time-lagged correlation matrices are defined as follows:
where \({{\rm{X}}}\) and \({{\rm{Y}}}\) are two \(T-\tau\) by \(m\) matrices, defined as \({\left[{{\mathcal{X}}}\left({{{\rm{x}}}}_{1}\right),\ldots,{{\mathcal{X}}}\left({{{\rm{x}}}}_{T-\tau }\right)\right]}^{{{\rm{T}}}}\) and \({\left[{{\mathcal{X}}}\left({{{\rm{x}}}}_{\tau+1}\right),\ldots,{{\mathcal{X}}}\left({{{\rm{x}}}}_{T}\right)\right]}^{{{\rm{T}}}}\) (\(\tau\) is the lag time). \({{{\rm{\pi }}}}_{0}\) and \({{{\rm{\pi }}}}_{1}\) are mean vectors of \({{\rm{X}}}\) and \({{\rm{Y}}}\), which equal to \(\frac{1}{T-\tau }{{{\rm{X}}}}^{{{\rm{T}}}}1\) and \(\frac{1}{T-\tau }{{{\rm{Y}}}}^{{{\rm{T}}}}1\), respectively. In our method, we parameterized the basis functions, \({{\mathcal{X}}}\), using the SoftMax outputs of two parallel networks with shared parameters from TS-DAR.
Exponential-moving-average (EMA) to estimate the metastable state centers
We denote the hyperspherical embeddings of conformations as \({\left\{{{{\rm{z}}}}_{i}\right\}}_{i=1}^{N}\), where \({{{\rm{z}}}}_{i}\in {{\mathbb{R}}}^{d}\), and \(\left\{{\mathrm{1,2}},\ldots,C\right\}\) as the metastable state indices, the metastable state centers correspond to vectors \({\left\{{{{\mathbf{\mu }}}}_{c}\right\}}_{c=1}^{C}\) that can be computed via a EMA manner:
where \({\widetilde{y}}_{i}\in \left\{{\mathrm{1,2}},\ldots,C\right\}\) represents the state index of the conformation \(i\), refers to as the pseudo-labels obtained from the Softmax outputs of TS-DAR neural network. \(\theta\) is the state center update factor and specifically selected as 0.5.
MD simulation datasets
The analytical form of the Müller potential65 (Fig. 2a) is as follows:
where \(\left({A}_{1},\ldots,{A}_{4}\right)=\left(-10,-5,-8.5,0.75\right)\), \(\left({a}_{1},\ldots,{a}_{4}\right)=\left(-1,-1,-6.5,0.7\right)\), \(\left({b}_{1},\ldots,{b}_{4}\right)=\left(0,0,11,0.6\right)\), \(\left({c}_{1},\ldots,{c}_{4}\right)=\left(-10,-10,-6.5,0.7\right)\), \(\left({\bar{x}}_{1},\ldots,{\bar{x}}_{4}\right)=\left(1,0,-0.5,-1\right)\), \(\left({\bar{y}}_{1},\ldots,{\bar{y}}_{4}\right)=\left(0,0.5,1.5,1\right)\). We performed a Brownian dynamics simulation (time step equals \(2\times {10}^{-4}\), damping factor equals 1) to sample this Müller potential at the temperature of \(0.9\). A reflective boundary condition is adopted: \({x}_{1}\in \left[-1.5,1.2\right]\), \({x}_{2}\in \left[-0.2,2\right]\). The simulation trajectory contains \(3\times {10}^{5}\) frames with the saving interval of 0.01. For alanine dipeptide, we obtained the MD simulation dataset from a previous study84. It contains three 250-ns MD trajectories, with a saving interval of 1 ps. As a result, the entire dataset contains 750,000 MD conformations. All conformations were aligned to the first frame according to the minimal root mean square deviation. The input features consist of the \(x\), \(y\) and \(z\) coordinates of the 10 heavy atoms (totally 30 input features). For the translocation of a DNA motor protein (AlkD) on DNA system, we obtained the MD simulation dataset from a previous study72. It contains 200 50-ns and 100 45-ns MD trajectories, with a saving interval of 20 ps. As a result, the entire dataset contains 725,300 MD conformations. The input features consist of the pairwise distances of 38 atoms, which are constituted by phosphate atoms of five base pairs in the center region of dsDNA and heavy atoms of five protein helixes within 12 Å of nucleotides. See SI Fig. S10 for illustration.
Analytical solution of committor probabilities for the Müller potential
For this 2D-potential, the forward committor function (\(q\left({{\rm{x}}}\right)\)) can be mathematically obtained by solving the backward Kolmogorov equation with Dirichlet boundary conditions:
where \(V\) is the potential energy function, \(\beta=\frac{1}{{\mbox{k}}T}\) is the inverse temperature, \(\nabla=\left(\frac{\partial }{\partial {x}_{1}},\ldots,\frac{\partial }{\partial {x}_{n}}\right)\) is the gradient operator and \(\triangle={\sum }_{i=1}^{n}\frac{{\partial }^{2}}{\partial {x}_{i}^{2}}\) is the Laplace operator. \(A\) and \(B\) are the source and sink states, \(\partial A\) and \(\partial B\) represent the boundary of the two states, respectively. We used finite element method (FEM) to solve the Eq. (7) for the 2D Müller potential by the solver FreeFem + +85. Specifically, we selected the regions with potential energy below 5.5 \({\mbox{k}}T\) in basin 1 and 2 as the source and sink states to compute the committor function for TS12, and the regions with potential energy below 3.9 \({\mbox{k}}T\) in basin 2 and 3 as the source and sink states to compute the committor function for TS23.
Training details of the TS-DAR models
We specified the scaling factor (\(\gamma\)) of the hyperspherical embeddings as 1, the scaling hyperparameter in dispersion loss as 0.1, the weight (\(\beta\)) of dispersion loss as 0.01 for Müller potential and alanine dipeptide datasets, 0.05 for AlkD-DNA dataset, the state center update factor (\(\theta\)) as 0.5, and the lag time (\(\tau\)) as 1 time step for Müller potential, 1 ps for alanine dipeptide, 8 ns for AlkD-DNA dataset. In addition, we established a criterion for determining the dimensionality (denote as \(d\)) of the latent hyperspherical embeddings (i.e., (\(d-1\))-sphere): \(d=3\), if number of states is more than 3, otherwise, \(d=2\). For the specific training hyperparameters of TS-DAR on Müller potential, alanine dipeptide and AlkD-DNA datasets, please refer to SI Sec. 1 for details.
The validation of \(\beta\)
The selection of \(\beta\) is trivial in TS-DAR model. However, choosing a proper magnitude of \(\beta\) is important for the fully optimization of both VAMP-2 and dispersion losses. In this work, we performed the ablation tests on \(\beta\) within Müller potential, alanine dipeptide and AlkD-DNA systems by training the TS-DAR model with different magnitudes of \(\beta\). The determination of the magnitude of \(\beta\) is guided by the following two criteria: (1) The dispersion loss can converge to the minimum boundary. (2) There is no significant deviation in VAMP-2 loss before and after integrating the dispersion loss optimization. Please see SI Fig. S17 for details.
The selection of transition states structures based on the OOD scores
With obtaining OOD scores from TS-DAR model that effectively distinguish between transition and metastable states, there are multiple ways for selecting transition state structures based on these scores for practical applications. We recommend that users define the OOD threshold based on the desired number of transition state structures they want to obtain. This strategy is particularly useful for adaptive sampling. Alternatively, one can define a threshold as the following:
where \({\theta }^{\star }\) represents the angle between two nearest neighbor state center vectors on the hypersphere after the optimization of TS-DAR model. In this work, we successfully selected the transition states structures of AlkD translocation along a DNA system (Fig. 5) by applying the threshold of ∼0.21 defined by Eq. (8).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All the MD simulation datasets and the training models of TS-DAR in this study are available in the Zenodo repository at https://zenodo.org/records/1383558086. Source data are provided as a Source Data file. Source data are provided with this paper.
Code availability
MSM related study were conducted with MSMBuilder87,88 (version msmbuilder2022-3.8.5), freely available at https://github.com/msmbuilder/msmbuilder2022. Biological analysis for AlkD dataset was performed using GROMACS89 version 2019.6 package. TS-DAR is built based on PyTorch90. The source code of TS-DAR and associated documentation are available at https://github.com/xuhuihuang/ts-dar91.
References
Konovalov, K. A. et al. A comprehensive mechanism for 5-carboxylcytosine-induced transcriptional pausing revealed by Markov state models. J. Biol. Chem. 296, 100735 (2021).
Prinz, J.-H. et al. Markov models of molecular kinetics: generation and validation. J. Chem. Phys. 134, 174105 (2011).
Chodera, J. D., Singhal, N., Pande, V. S., Dill, K. A. & Swope, W. C. Automatic discovery of metastable states for the construction of Markov models of macromolecular conformational dynamics. J. Chem. Phys. 126, 155101 (2007).
Husic, B. E. & Pande, V. S. Markov state models: from an art to a science. J. Am. Chem. Soc. 140, 2386–2396 (2018).
Bowman, G. R., Pande, V. S. & Noé, F. in An Introduction to Markov State Models and Their Application to Long Timescale Molecular Simulation (eds. Bowman, G. R., Pande, V. S. & Noé, F.) 1–6 (Springer Netherlands, 2014).
Pan, A. C. & Roux, B. Building Markov state models along pathways to determine free energies and rates of transitions. J. Chem. Phys. 129, 064107 (2008).
Buchete, N.-V. & Hummer, G. Coarse master equations for peptide folding dynamics. J. Phys. Chem. B 112, 6057–6069 (2008).
Wang, W., Cao, S., Zhu, L. & Huang, X. Constructing Markov State Models to elucidate the functional conformational changes of complex biomolecules. WIREs Comput. Mol. Sci. 8, e1343 (2018).
Huang, X., Bowman, G. R., Bacallado, S. & Pande, V. S. Rapid equilibrium sampling initiated from nonequilibrium data. Proc. Natl Acad. Sci. 106, 19765–19769 (2009).
Malmstrom, R. D., Lee, C. T., Van Wart, A. T. & Amaro, R. E. Application of molecular-dynamics based Markov state models to functional proteins. J. Chem. Theory Comput. 10, 2648–2657 (2014).
Morcos, F. et al. Modeling conformational ensembles of slow functional motions in Pin1-WW. PLOS Comput. Biol. 6, e1001015 (2010).
Zhang, B. W. et al. Simulating replica exchange: markov state models, proposal schemes, and the infinite swapping limit. J. Phys. Chem. B 120, 8289–8301 (2016).
Konovalov, K. A., Unarta, I. C., Cao, S., Goonetilleke, E. C. & Huang, X. Markov state models to study the functional dynamics of proteins in the wake of machine learning. JACS Au 1, 1330–1341 (2021).
Liu, B., Qiu, Y., Goonetilleke, E. C. & Huang, X. Kinetic network models to study molecular self-assembly in the wake of machine learning. MRS Bull. 47, 958–966 (2022).
Noé, F., Schütte, C., Vanden-Eijnden, E., Reich, L. & Weikl, T. R. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc. Natl Acad. Sci. 106, 19011–19016 (2009).
Bowman, G. R., Voelz, V. A. & Pande, V. S. Taming the complexity of protein folding. Curr. Opin. Struct. Biol. 21, 4–11 (2011).
Da, L.-T. et al. A jump-from-cavity pyrophosphate ion release assisted by a key lysine residue in T7 RNA polymerase transcription elongation. PLOS Comput. Biol. 11, e1004624 (2015).
Da, L.-T., Wang, D. & Huang, X. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J. Am. Chem. Soc. 134, 2399–2406 (2012).
Da, L.-T. et al. Bridge helix bending promotes RNA polymerase II backtracking through a critical and conserved threonine residue. Nat. Commun. 7, 11244 (2016).
Silva, D.-A. et al. Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc. Natl Acad. Sci. 111, 7665–7670 (2014).
Malmstrom, R. D., Kornev, A. P., Taylor, S. S. & Amaro, R. E. Allostery through the computational microscope: cAMP activation of a canonical signalling domain. Nat. Commun. 6, 7588 (2015).
Kohlhoff, K. J. et al. Cloud-based simulations on Google Exacycle reveal ligand modulation of GPCR activation pathways. Nat. Chem. 6, 15–21 (2014).
Deng, N.-J., Dai, W. & Levy, R. M. How kinetics within the unfolded state affects protein folding: an analysis based on markov state models and an ultra-long MD trajectory. J. Phys. Chem. B 117, 12787–12799 (2013).
Wan, H., Ge, Y., Razavi, A. & Voelz, V. A. Reconciling simulated ensembles of apomyoglobin with experimental hydrogen/deuterium exchange data using bayesian inference and multiensemble markov state models. J. Chem. Theory Comput. 16, 1333–1348 (2020).
Buch, I., Giorgino, T. & De Fabritiis, G. Complete reconstruction of an enzyme-inhibitor binding process by molecular dynamics simulations. Proc. Natl Acad. Sci. 108, 10184–10189 (2011).
Lawrenz, M., Shukla, D. & Pande, V. S. Cloud computing approaches for prediction of ligand binding poses and pathways. Sci. Rep. 5, 7918 (2015).
Silva, D.-A., Bowman, G. R., Sosa-Peinado, A. & Huang, X. A Role for Both Conformational Selection and Induced Fit in Ligand Binding by the LAO Protein. PLOS Comput. Biol. 7, e1002054 (2011).
Plattner, N. & Noé, F. Protein conformational plasticity and complex ligand-binding kinetics explored by atomistic simulations and Markov models. Nat. Commun. 6, 7653 (2015).
Klem, H., Hocky, G. M. & McCullagh, M. Size-and-shape space gaussian mixture models for structural clustering of molecular dynamics trajectories. J. Chem. Theory Comput. 18, 3218–3230 (2022).
Wang, B., Sexton, R. E. & Feig, M. Kinetics of nucleotide entry into RNA polymerase active site provides mechanism for efficiency and fidelity. Biochim. Biophys. Acta (BBA)—Gene Regulatory Mech. 1860, 482–490 (2017).
Khaled, M., Gorfe, A. & Sayyed-Ahmad, A. Conformational and dynamical effects of Tyr32 phosphorylation in K-ras: molecular dynamics simulation and Markov state models analysis. J. Phys. Chem. B 123, 7667–7675 (2019).
Barros, E. P., Demir, Ö., Soto, J., Cocco, M. J. & Amaro, R. E. Markov state models and NMR uncover an overlooked allosteric loop in p53. Chem. Sci. 12, 1891–1900 (2021).
Feng, J., Selvam, B. & Shukla, D. How do antiporters exchange substrates across the cell membrane? An atomic-level description of the complete exchange cycle in NarK. Structure 29, 922–933.e3 (2021).
Son, C. Y., Yethiraj, A. & Cui, Q. Cavity hydration dynamics in cytochrome c oxidase and functional implications. Proc. Natl Acad. Sci. 114, E8830–E8836 (2017).
Jiang, H. et al. Markov state models reveal a two-step mechanism of miRNA loading into the human argonaute protein: selective binding followed by structural re-arrangement. PLOS Comput. Biol. 11, e1004404 (2015).
Kleiman, D. E. & Shukla, D. Active learning of the conformational ensemble of proteins using maximum entropy VAMPNets. J. Chem. Theory Comput. 19, 4377–4388 (2023).
Mardt, A., Pasquali, L., Wu, H. & Noé, F. VAMPnets for deep learning of molecular kinetics. Nat. Commun. 9, 5 (2018).
Wu, H. & Noé, F. Variational approach for learning Markov processes from time series data. J. Nonlinear Sci. 30, 23–66 (2020).
Bolhuis, P. G., Chandler, D., Dellago, C. & Geissler, P. L. Transition path sampling: throwing ropes over rough mountain passes, in the dark. Annu. Rev. Phys. Chem. 53, 291–318 (2002).
Dellago, C., Bolhuis, P. G. & Geissler, P. L. in Advances in Chemical Physics (eds Prigogine, I. & Rice, S. A.) 1–78 (Wiley, 2002).
Jung, H., Okazaki, K.-I. & Hummer, G. Transition path sampling of rare events by shooting from the top. J. Chem. Phys. 147, 152716 (2017).
Zhang, J., Yang, Y. I., Yang, L. & Gao, Y. Q. Dynamics and kinetics study of “in-water” chemical reactions by enhanced sampling of reactive trajectories. J. Phys. Chem. B 119, 14505–14514 (2015).
Zhang, J. et al. Rich dynamics underlying solution reactions revealed by sampling and data mining of reactive trajectories. ACS Cent. Sci. 3, 407–414 (2017).
Zhang, J. et al. Deep reinforcement learning of transition states. Phys. Chem. Chem. Phys. 23, 6888–6895 (2021).
Jónsson, H., Mills, G. & Jacobsen, K. W. in Classical and Quantum Dynamics in Condensed Phase Simulations 385–404 (World Scientific, 1998).
E, W., Ren, W. & Vanden-Eijnden, E. String method for the study of rare events. Phys. Rev. B 66, 052301 (2002).
E, W., Ren, W. & Vanden-Eijnden, E. Finite temperature string method for the study of rare events. J. Phys. Chem. B 109, 6688–6693 (2005).
E, W. & Vanden-Eijnden, E. Towards a theory of transition paths. J. Stat. Phys. 123, 503–523 (2006).
Maragliano, L., Fischer, A., Vanden-Eijnden, E. & Ciccotti, G. String method in collective variables: minimum free energy paths and isocommittor surfaces. J. Chem. Phys. 125, 024106 (2006).
Pan, A. C., Sezer, D. & Roux, B. Finding transition pathways using the string method with swarms of trajectories. J. Phys. Chem. B 112, 3432–3440 (2008).
Roux, B. String method with swarms-of-trajectories, mean drifts, lag time, and committor. J. Phys. Chem. A 125, 7558–7571 (2021).
He, Z., Chipot, C. & Roux, B. Committor-consistent variational string method. J. Phys. Chem. Lett. 13, 9263–9271 (2022).
Lai, R. & Lu, J. Point cloud discretization of fokker-planck operators for committor functions. Multiscale Modeling Simul. 16, 710–726 (2018).
Khoo, Y., Lu, J. & Ying, L. Solving for high-dimensional committor functions using artificial neural networks. Res. Math. Sci. 6, 1 (2018).
Li, Q., Lin, B. & Ren, W. Computing committor functions for the study of rare events using deep learning. J. Chem. Phys. 151, 054112 (2019).
Prinz, J.-H., Held, M., Smith, J. C. & Noé, F. Efficient computation, sensitivity, and error analysis of committor probabilities for complex dynamical processes. Multiscale Modeling Simul. 9, 545–567 (2011).
Yang, J., Zhou, K., Li, Y. & Liu, Z. Generalized out-of-distribution detection: a survey. Int. J. Comput. Vis. 132, 5635–5662 (2024).
Amodei, D. et al. Concrete problems in AI safety. Preprint at https://arxiv.org/abs/1606.06565 (2016).
Dietterich, T. G. Steps Toward Robust Artificial Intelligence. AI Mag. 38, 3–24 (2017).
Leike, J. et al. AI safety gridworlds. Preprint at https://arxiv.org/abs/1711.09883 (2017).
Smuha, N. A. The EU approach to ethics guidelines for trustworthy. Artif. Intell. Comput. Law Rev. Int. 20, 97–106 (2019).
Shneiderman, B. Bridging the gap between ethics and practice: guidelines for reliable, safe, and trustworthy human-centered AI systems. ACM Trans. Interact. Intell. Syst. 10, Article 26 (2020).
Mohseni, S. et al. Taxonomy of machine learning safety: a survey and primer. ACM Comput. Surv. 55, 157 (2022).
Ming, Y., Sun, Y., Dia, O. & Li, Y. How to exploit hyperspherical embeddings for out-of-distribution detection? In The Eleventh International Conference on Learning Representations (ICLR, 2023).
Müller, K. & Brown, L. D. Location of saddle points and minimum energy paths by a constrained simplex optimization procedure. Theoretica Chim. Acta 53, 75–93 (1979).
Chen, T., Kornblith, S., Norouzi, M. & Hinton, G. A simple framework for contrastive learning of visual representations. In: Proc. 37th International Conference on Machine Learning Vol. 119 (eds Hal, D., I. I. I. & Aarti, S.) 1597–1607 (PMLR, Proceedings of Machine Learning Research, 2020).
Wang, T. & Isola, P. Understanding Contrastive Representation Learning through Alignment and Uniformity on the Hypersphere. In: Proc. 37th International Conference on Machine Learning Vol. 119 (eds Hal, D., I. I. I. & Aarti, S.) 9929–9939 (PMLR, Proceedings of Machine Learning Research, 2020).
Li, J., Xiong, C. & Hoi, S. MoPro: webly supervised learning with momentum prototypes. In International Conference on Learning Representations (ICLR, 2021).
Sun, Y., Ming, Y., Zhu, L. & Li, Y. Out-of-distribution detection with deep nearest neighbors. In Proc. 39th International Conference on Machine Learning 20827–20840 (PMLR, 2022).
Bolhuis, P. G., Dellago, C. & Chandler, D. Reaction coordinates of biomolecular isomerization. Proc. Natl Acad. Sci. 97, 5877–5882 (2000).
Jones, L. E. Jr. et al. Differential effects of reactive nitrogen species on DNA base excision repair initiated by the alkyladenine DNA glycosylase. Carcinogenesis 30, 2123–2129 (2009).
Peng, S. et al. Target search and recognition mechanisms of glycosylase AlkD revealed by scanning FRET-FCS and Markov state models. Proc. Natl Acad. Sci. 117, 21889–21895 (2020).
Schwantes, C. R. & Pande, V. S. Improvements in Markov state model construction reveal many non-native interactions in the folding of NTL9. J. Chem. Theory Comput. 9, 2000–2009 (2013).
Pérez-Hernández, G., Paul, F., Giorgino, T., De Fabritiis, G. & Noé, F. Identification of slow molecular order parameters for Markov model construction. J. Chem. Phys. 139, 015102 (2013).
Chen, W., Sidky, H. & Ferguson, A. L. Nonlinear discovery of slow molecular modes using state-free reversible VAMPnets. J. Chem. Phys. 150, 214114 (2019).
Wu, H. & Noé, F. Reaction coordinate flows for model reduction of molecular kinetics. J. Chem. Phys. 160, 044109 (2024).
Xu, J. & Durrett, G. Spherical Latent Spaces for Stable Variational Autoencoders 4503–4513 (Association for Computational Linguistics, Brussels, Belgium, 2018).
Davidson, T. R., Falorsi, L., De Cao, N., Kipf, T. & Tomczak, J. M. Hyperspherical variational auto-encoders. In: 34th Conference on Uncertainty in Artificial Intelligence 2018, UAI 2018 (eds Silva, R. & Globerson, A.) 856–865 (Association For Uncertainty in Artificial Intelligence (AUAI), 2018).
Bojanowski, P. & Joulin, A. Unsupervised learning by predicting noise. In: Proc. 34th International Conference on Machine Learning Vol. 70 (eds Doina, P. & Yee Whye, T.) 517–526 (PMLR, Proceedings of Machine Learning Research, 2017).
Mettes, P., Van der Pol, E. & Snoek, C. Hyperspherical prototype networks. Adv. Neural Inform. Process. Syst. 32 (2019).
Liu, W. et al. SphereFace: deep hypersphere embedding for face recognition. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 6738–6746 (IEEE, 2017).
Wang, F., Xiang, X., Cheng, J. & Yuille, A. L. NormFace: L2 hypersphere embedding for face verification. In Proc. 25th ACM international conference on Multimedia 1041–1049 (Association for Computing Machinery, 2017).
Wang, D., Wang, Y., Evans, L. & Tiwary, P. From latent dynamics to meaningful representations. J. Chem. Theory Comput. 20, 3503–3513 (2024).
Nüske, F. et al. Markov state models from short non-equilibrium simulations—analysis and correction of estimation bias. J. Chem. Phys. 146, 094104 (2017).
Hecht, F. New development in freefem. J. Numer. Math. 20, 251–266 (2012).
Liu, B. et al. Data for: exploring transition states of protein conformational changes via out-of-distribution detection in the hyperspherical latent space. Zenodo https://doi.org/10.5281/zenodo.13835580 (2024).
Beauchamp, K. A. et al. MSMBuilder2: modeling conformational dynamics on the picosecond to millisecond scale. J. Chem. Theory Comput. 7, 3412–3419 (2011).
Harrigan, M. P. et al. MSMBuilder: statistical models for biomolecular dynamics. Biophysical J. 112, 10–15 (2017).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015).
Paszke, A. et al. Pytorch: an imperative style, high-performance deep learning library. Adv. Neural Inform. Process. Syst. 32 (2019).
Liu, B. xuhuihuang/ts-dar: exploring transition states of protein conformational changes via out-of-distribution detection in the hyperspherical latent space. Zenodo https://doi.org/10.5281/zenodo.14037314 (2024).
Acknowledgements
X.H. acknowledges the support from the NIH/NIGMS under award number 1 R01GM147652 and the support from the Hirschfelder Professorship Fund from University of Wisconsin-Madison. Y.L. acknowledges the support by the AFOSR Young Investigator Program under award number FA9550-23-1-0184, National Science Foundation (NSF) Award No. IIS-2237037 & IIS-2331669, Office of Naval Research under grant number N00014-23-1-2643, and faculty research awards/gifts from Google and Meta. X.H. and Y.L. are also supported by the Research Forward Fund from the University of Wisconsin-Madison Office of the Vice Chancellor for Research with funding from the Wisconsin Alumni Research Foundation. X.D. is supported by the Jane Street Graduate Research Fellowship. We also acknowledge helpful discussions with Siqin Cao and Michael Kalin.
Author information
Authors and Affiliations
Contributions
B.L. developed the methodology, established the software, and produced the majority of the results. J.B. contributed to conceptual discussions and validated the results. I.U. conducted the biological analysis on AlkD-dsDNA system. X.D. also contributed to conceptual discussions. Y.L. contributed conceptually to the methodology development. X.H. initiated this study, contributed conceptually to the methodology development and provided supervision throughout the project. All authors contributed to writing the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Matteo Degiacomi, Johan Ulander and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, B., Boysen, J.G., Unarta, I.C. et al. Exploring transition states of protein conformational changes via out-of-distribution detection in the hyperspherical latent space. Nat Commun 16, 349 (2025). https://doi.org/10.1038/s41467-024-55228-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55228-4
This article is cited by
-
Memory kernel minimization-based neural networks for discovering slow collective variables of biomolecular dynamics
Nature Computational Science (2025)
-
Calculation of minimum energy pathways in transport proteins
Communications Chemistry (2025)