Abstract
Balanced self-renewal and differentiation of stem cells are crucial for maintaining tissue homeostasis, but the underlying mechanisms of this process remain poorly understood. Here, from an RNA interference (RNAi) screen in adult Drosophila intestinal stem cells (ISCs), we identify a factor, Pax, which is orthologous to mammalian PXN, coordinates the proliferation and differentiation of ISCs during both normal homeostasis and injury-induced midgut regeneration in Drosophila. Loss of Pax promotes ISC proliferation while suppressing its differentiation into absorptive enterocytes (ECs). Mechanistically, our findings demonstrate that Pax is a conserved target gene of the Hippo signaling pathway in both Drosophila and mammals. Subsequent investigations have revealed Pax interacts with Yki and enhances its cytoplasmic localization, thereby establishing a feedback regulatory mechanism that attenuates Yki activity and ultimately inhibits ISCs proliferation. Additionally, Pax induces the differentiation of ISCs into ECs by activating Notch expression, thus facilitating the differentiation process. Overall, our study highlights Pax as a pivotal component of the Hippo and Notch pathways in regulating midgut homeostasis, shedding light on this growth-related pathway in tissue maintenance and intestinal function.
Similar content being viewed by others
Introduction
Adult epithelial tissues experience constant damage from environmental stress, leading to cell dysfunction and death1,2. In the intestine, resident intestinal stem cells (ISCs) repair damage by proliferating and differentiating into specialized cells, maintaining tissue integrity3,4,5. In the Drosophila midgut, ISCs mainly undergo asymmetric division to give rise to a renewed ISC and a nondividing progenitor cell known as enteroblast (EB) or pre-enteroendocrine cell (pre-EE)5,6,7,8. Subsequently, these progenitor cells differentiate into either absorptive enterocytes (ECs) with large size as well as polyploid nuclei via endoreplication, or hormone-producing enteroendocrine cells (EEs) with small nuclei, respectively (Fig. 1a)9,10,11,12. Upon injury, such as dextran sodium sulfate (DSS) or bacterial infection, ISC proliferation is accelerated, with occasional symmetric division observed to timely replace damaged cells and maintain homeostasis13,14,15,16.
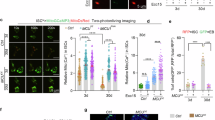
a Schematic diagram of the ISC lineage and intestinal epithelium in Drosophila adult midguts. ISC intestinal stem cell, EB enteroblast, EC absorptive enterocyte, Pre-EE Precursor enteroendocrine, EE enteroendocrine. Esg+ Dl+ cells are ISCs. Esg+ Su(H)+ cells are EBs. Esg+ Dl+ Pros+ cells are pre-EEs. Pdm1+ or Pros+ labels mature ECs or EEs, respectively. b–d’ Adult midguts from flies fed with either Glucose (b–b’) or DSS (c–c’) for 3 days, followed by 3 days of recovery (d–d’), were stained with Pax (red) and DAPI (nuclei, blue). Flies expressing EsgGal4; tubGal80ts (Esgts>) were used. 5% Glucose solution with or without 3% DSS was fed to the flies. ISCs and EBs/pre-EEs were marked by EsgGal4-driven GFP expression. e–f” Adult midguts of Esgts>Ctrl (e–e”) and Esgts>Pax RNAi (V107789) (f–f”) flies were immunostained with p-H3 (gray), Dl (red) and DAPI (nuclei, blue). Phospho-Histone 3 (p-H3) marks mitotic cells derived from ISCs. ISCs and EBs/pre-EEs were marked by EsgGal4-driven GFP expression. White arrows indicate proliferative ISCs marked by p-H3. g Quantification of p-H3+ cells of adult midguts of the indicated genotypes of (e–f”) (n = 12,12). The counting of p-H3+ cells was conducted across the entire midgut. h Quantification of Dl+ cells of adult midguts of the indicated genotypes of (e–f”) (n = 12,12). i–j” Adult midguts of Ctrl (i–i”) and homozygous allele of Pax20 (j–j”) were dissected and immunostained with Arm+Pros (green), p-H3 (red) and DAPI (nuclei, blue). Midguts were dissected 5 days after eclosion. k Quantification of p-H3+ cells of adult midguts of (i–j”) (n = 12,12). The counting of p-H3+ cells was conducted across the entire midgut. l–o’ Adult midguts of Esgts>Ctrl (l–l’, n–n’) and Esgts > HA-Pax (m–m’, o–o’) were treated with Glucose or DSS for 3 days before gut dissection. Midguts were immunostained with p-H3 (red) and DAPI (nuclei, blue). A 5% Glucose solution with or without 3% DSS was fed to the flies. ISCs and EBs/pre-EEs were marked by EsgGal4-driven GFP expression. White arrows indicate proliferative ISCs marked by p-H3. p Quantification of p-H3+ cells of adult midguts of the indicated genotypes of (l–o’), n = 10,15,10,10. The counting of p-H3+ cells was conducted across the entire midgut. q–r” Adult midguts of MyoIA-Gal4; tubGal80ts (MyoIAts)>Ctrl (q–q”) and MyoIAts >Pax RNAi (r–r”) were dissected and immunostained with Dl (red), p-H3 (gray) and DAPI (nuclei, blue). ECs are marked with MyoIA-GFP (green). White arrows indicate proliferative ISCs marked by p-H3. s Quantification of p-H3+ cells of adult midguts of the indicated genotypes of (q–r”) (n = 14, 14). The counting of p-H3+ was conducted across the entire midgut. t Quantification of the percentage of Dl+ cells in adult midguts of the indicated genotypes of (q–r”) (n = 11, 11). u–v” Adult midguts containing MyoIAts>Ctrl (u–u”) and MyoIAts >Pax RNAi (v–v”) were immunostained with Phalloidin (red) and DAPI (nuclei, blue). ECs are marked with MyoIA-GFP (green). Three independent experiments were performed, and the error bars are mean ± SEM. In each box plot, the center line indicates the median, the edges of the box represent the first and third quartiles, and the whiskers extend to the minimum and maximum values. P values of significance (indicated with asterisks, NS no significance P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****p < 0.0001) were calculated by two-tailed Student’s t-test (g, h, k, s, t) and one-way ANOVA with Tukey’s test (p). Scale bars: 30 µm. Confocal images were taken from the boundary region between R4c and R5a of the posterior midgut.
ISC divisions yield four daughter cell types: ISC-EB (asymmetric, 70%), ISC-pre-EE (asymmetric, 10%), ISC-ISC (symmetric, 10%), and EB-EB (symmetric, 10%)11. During mitosis, spindle orientation determines whether ISCs divide symmetrically or asymmetrically6,17. Symmetric division occurs when spindles align parallel to the basement membrane, whereas asymmetric division is characterized by spindles aligning perpendicular to it18,19. Apico-basal and planar cell polarities actively influence spindle orientation across various organisms20. The Par complex localization to the apical daughter cell is crucial for ISC asymmetric division18,19. Integrins facilitate asymmetric division by anchoring the basal daughter cell to the basement membrane, guiding Par complex localization to the apical pole. Reduction of integrins leads to spindle reorientation parallel to the basement membrane, thereby inducing ISC-ISC duplication18,20. During anaphase of both symmetric and asymmetric ISC division, daughter cell fate is dictated by Notch pathway activity, which is itself regulated by Dpp and BMP pathway11,16,21. Heightened Notch activation in ISC promotes its differentiation into EB or EB-EB duplication (asymmetric or symmetric division), while reduced activation favors ISC-ISC duplication or pre-EE differentiation11,18,19,22,23. The pre-EEs undergo further asymmetric division, forming one class I EE cell and one class II EE cell, the latter specified by Notch signaling7,8,19.
Numerous studies have shown that intestinal homeostasis is tightly regulated under physiological and stressed conditions through the cooperative activity of conserved growth control and cytokines signaling pathways, including Notch, BMP, Wingless, JAK-STAT, and EGFR pathways2,16,24,25,26,27,28. Particularly, the Hippo pathway has been shown to play a critical role in regulating midgut homeostasis and regeneration29,30,31,32. This evolutionarily highly conserved tumor suppressor pathway, originally discovered in Drosophila, is vital in controlling organ growth by modulating cell proliferation, differentiation, and survival33,34,35,36,37,38,39. Dysfunction of the Hippo pathway has been implicated in various homeostatic disorders such as human cancers40,41,42,43. The Hippo pathway comprises a core kinase cascade known as Hippo (Hpo)-Warts (Wts)44,45,46. The Hpo kinase phosphorylates and activates the Wts kinase with the assistance of scaffold proteins Salvador (Sav)44,45,46. Activated Wts subsequently interacts with Mob as a tumor suppressor (Mats) to provoke the phosphorylation of the transcriptional coactivator Yorkie (Yki). This phosphorylation event prevents Yki from translocating into the nucleus and forming a complex with the transcription factor Scalloped (Sd), thereby inhibiting downstream signaling transduction47,48,49. Yki regulates the expression of target genes involved in cell proliferation, cell cycle progression, and apoptosis, such as Drosophila inhibitor of apoptosis protein1 (Diap1), cell cycle regulator CyclinE (CycE), and microRNA Bantam50,51,52.
The inactivation of the Hippo signaling in either midgut precursor cells or differentiated EC cells has been found to trigger the proliferation of ISCs by provoking the activity of Yki. Loss of the Hippo signaling or overexpression of Yki in ISC/EB cells induces the expression of targets in the Hippo pathway as well as the JAK-STAT ligands Unpaireds (Upds), which leads to a cell-autonomous proliferation of ISCs29,31. However, the inactivation of the Hippo signaling in EC cells results in an increased expression of multiple EGFR and JAK-STAT pathway ligands that activate the EGFR and JAK-STAT pathways in ISCs to trigger their proliferation, indicating a unique non-cell-autonomous role of the Hippo pathway in blocking the proliferation of ISCs30,31,32,51,53,54. In addition, Yki is considered one of the important sensors for injury-induced midgut regeneration under stress conditions, such as bacterial infection and exposure to tissue-damaging reagents26,29,32. This perspective complements the role of JNK, which is systematically required, particularly in cases involving extensive damage31,32,55,56,57.
Despite the pivotal role of Yki in regulating intestinal homeostasis and regeneration, deletion of Yki in ISC under homeostatic condition does not result in a strong loss of ISC. This prompts us to consider how the activity of Yki is switched during the processes of intestinal homeostasis and injury repair in Drosophila.
Paxillin (Pax), a multidomain adapter protein, is the main component of focal adhesions (FAs), plays an important role in the transduction of extracellular signals into intracellular responses upon Integrin-ECM engagement58,59,60,61,62. Pax recruits diverse signal proteins which participate in intracellular signaling cascades. Activation of these pathways ultimately leads to actin cytoskeleton reorganization and assembly/disassembly of FAs, which are essential for cell adhesion, morphological change, migration, and signaling transduction63,64,65,66,67. Dysregulation and mutations in Pax have been linked to the occurrence, invasion, and metastasis of various tumors68,69,70,71. Pax consists of multiple conserved protein interaction domains found in both mammalian and Drosophila, including five leucine-rich motifs (LD1-5) located at the amino-terminal of Pax that are necessary for binding with focal adhesion kinase (Fak), integrin-linked kinase (ILK), and Vinculin65,72,73,74. Additionally, the carboxyl-terminal of Pax contains four zinc finger-like LIM (LIM1-4) domains that mediate interaction with tubulin65. Integrins play a critical role in regulating proliferation and self-renewal in the ISC lineage by maintaining the asymmetric localization of the Par complex, thus ensuring the asymmetric division of ISCs18. However, the role of Pax in maintaining intestinal homeostasis and midgut regeneration remains underexplored.
In this study, using a genetic screen, we identify Pax, as a target gene and component of the Hippo pathway, acts in downstream of Wts and plays a crucial role in constraining ISC proliferation while promoting their differentiation. Specifically, loss of Pax leads to hyperproliferation of ISC in a manner dependent on Yki/Sd complex. Our investigations further reveal that Pax, as a common target of mammalian and Drosophila the Hippo pathway, interacts with Yki and promotes the cytoplasmic localization of Yki, thus forming a feedback loop to inactivate Yki in ISCs/EBs under both homeostasis and regeneration conditions. In addition, Pax promotes the activity of the Notch signaling pathway by upregulating the expression of Notch, thereby promoting the differentiation of ISCs into ECs. In summary, our findings highlight Pax as a signaling effector that fine-tunes the balance between ISC self-renewal and differentiation through its positive regulation of the Hippo and Notch pathways, implying the central role of Pax in maintaining tissue homeostasis and facilitating regeneration.
Results
Pax restrains ISC proliferation in both homeostatic and stress conditions
We performed an RNA interference (RNAi) screen in Drosophila aiming to identify regulators of Yki whose disruption could suppress hyperproliferative phenotypes caused by ectopically expressed Yki during wing and eye development75. There are studies have demonstrated that Yki/YAP (the homolog of Yki in mammals) induces the expression of its negative regulators, including LATS1/2 (the homolog of Wts in mammals), NF2 (the homolog of Merlin in mammals), ex, and Kibra, to establish a feedback mechanism during developmental growth control76,77,78,79. Therefore, we conducted Sd ChIP-seq80 and TEAD4 ChIP-seq (ChIP Atlas) to identify genes regulated by both factors. We then compared these genes with published Yki/YAP/TEADs (the homolog of Sd in mammals) ChIP and RNA-seq data81,82,83,84,85,86,87,88. Subsequently, we identified 25 candidates as potential targets of the Hippo pathway (Supplementary Table 1). Next, we collected RNAi lines for these candidates from our RNAi library and examined their phenotypes in adult wings and eyes using MS1096, GMR, and GMR-Yki drivers. Among the candidates, we identified RNAi lines (V107789; NIG31794r-c-1) targeting Pax, which showed a significantly enhanced overgrowth phenotype, while Pax overexpression reduced it (Supplementary Fig. 1a–d and Supplementary Table 1). Importantly, co-expression of Yki and Pax RNAi resulted in further upregulation of the Hippo signaling targets diap1 and ex compared to Yki overexpression alone (Supplementary Fig. 1e). Loss of Pax in conjunction with Yki overexpression induced wing shrinkage (Supplementary Fig. 1c, black arrows), further confirming the excessive proliferation. We generated a polyclonal antibody against Pax (targeting amino acids 1–320) and confirmed RNAi efficiency through immunostaining with this antibody, complemented by real-time PCR and western blot analyses (Supplementary Fig. 1f–n). These findings suggest that Pax might function as a regulator of Yki.
Considering the essential role of Yki in maintaining homeostasis and promoting regeneration in the adult Drosophila midgut, we sought to investigate the potential involvement of Pax in regulating midgut homeostasis. Firstly, we examined the expression pattern of endogenous Pax in both larval and adult midgut. During larval development, adult midgut precursors (AMPs) undergo multiple divisions, culminating in the formation of imaginal midgut islets resembling imaginal disks89,90, as identified by Armadillo (Arm) immunostaining. In the adult midgut, we employed the EsgGal4, UAS-GFP; tubGal80ts (Esgts) driver to label ISCs and EBs/pre-EEs with GFP. Additionally, we distinguished EEs through Pros staining and ECs via polyploid nuclei91,92,93. Our observations reveal that Pax expression is enriched in ISCs/EBs/pre-EEs (GFP+ cells) and EEs of the adult midgut, as well as in AMPs of the larval midgut, but minimal expression in ECs (Supplementary Fig. 2a–d”).
Interestingly, the expression of Pax was upregulated in precursor cells after a continuous DSS treatment for 3 days (Fig. 1b–c’ and Supplementary Fig. 2e). However, after 3 days of recovery time, Pax expression returned to baseline (Fig. 1d–d’ and Supplementary Fig. 2e). To further explore the important role of Pax in midgut regeneration, we used whole midgut cells and isolated Esg-GFP+ cells using flow cytometry (FACS) to assess Pax expression with or without DSS treatment. As shown in Supplementary Fig. 2f, g, DSS treatment significantly increased Pax expression, which reverted to baseline after the recovery period. In summary, these findings indicate that Pax is enriched in ISCs/EBs/pre-EEs/EEs and imply its potential role in regulating ISCs proliferation and differentiation under homeostatic and stress conditions.
To investigate whether Pax functions as a regulator in tissue homeostasis, we expressed Pax transgenic RNAi lines (V107789 and NIG31794r-c-1) under the control of Esgts. Notably, the knockdown of Pax in precursor cells resulted in significant increases in both the number of Esg-GFP+ and Dl+ cells compared to the control (Fig. 1e–f”, h and Supplementary Fig. 2h–i”). Concurrently, the number of phospho-histone positive (p-H3+) cells, a specific marker for mitotic cells, also increased (Fig. 1g), suggesting that Pax reduction promotes ISCs proliferation. However, there were no discernible differences in the number of p-H3+ cells between the wild-type and Pax-overexpressing groups in aged fruit flies (Supplementary Fig. 2j–p). These findings suggest the regulation of proliferation in ISCs differs between aged and young fruit flies.
We further generated a Pax null allele mutation strain Pax20 by using the CRISPR-Cas9 system (Supplementary Fig. 3a and Supplementary Table 5). Immunostaining and western blot confirmed the absence of any Pax signal in the Pax20 homozygous midgut (Supplementary Fig. 3b, o–p”), while the p-H3+ cell numbers were apparently increased in homozygous Pax20 mutant midguts (Fig. 1i–j”, k). To further elucidate Pax’s role in the adult midgut, we generated GFP-marked homozygous stem cell clones for the Pax20 using the MARCM system. As expected, the clone size of Pax mutant was bigger than wild-type clones under both normal and DSS treatment conditions, suggesting that Pax is crucial for maintaining ISC homeostasis (Supplementary Fig. 3c–j). Additionally, the deletion of Pax significantly facilitated more Dl+ cell compared to wild-type clones (Supplementary Fig. 3k–n”), indicating that Pax deletion enhances ISCs proliferation. However, no obvious undergrowth phenotype was observed when UAS-Pax transgene was expressed in precursor cells (Fig. 1l–m’, p). In contrast, overexpression of Pax in precursors remarkably decreased both Esg-GFP and p-H3+ cell number after continuous 3 days DSS treatment (Fig. 1n–o’, p), suggesting that the overexpression of Pax in precursor cells inhibits ISC proliferation under injury condition. We also examined the effect of Pax on the development of Notch mutant ISC-like tumors, which are induced by inhibition of ISC differentiation93,94. Consistently, Pax overexpression markedly suppressed the formation of intestinal tumors induced by Notch depletion (Supplementary Fig. 3q–r”), suggesting that Pax restricts ISC-like tumor formation. These observations demonstrate that Pax plays a crucial role in maintaining ISC proliferation under both normal and stress conditions.
In addition to the cell-autonomous regulation of ISC proliferation by Pax, we also assessed the potential role of Pax in regulating non-cell-autonomous proliferation of ISCs. Firstly, we employed FACS to isolate Esg-GFP+ or MyoIA-GFP+ cells from whole midgut cells induced by Esgts or MyoIAts driver, which specifically drives expression in terminally differentiated ECs10. As shown in Supplementary Fig. 3s–u, w–x”, droplet digital PCR (ddPCR), immunostaining, and western blot assays revealed that Pax was expressed in ECs, albeit at lower levels compared with Esg-GFP+ cells, consistent with our staining results (Supplementary Fig. 2a–d”). Moreover, the expression of Pax in single-cell sequencing data from previously published studies corroborates these observations (Supplementary Fig. 3v)95. Subsequently, we used the MyoIAts driver to specifically express Pax RNAi in EC cells, resulting in significant promotion of ISC proliferation, as indicated by the increased number of Dl+ and p-H3+ cells (Fig. 1q–t). Moreover, reducing Pax expression in ECs resulted in gut hypertrophy characterized by multi-layered intestinal epithelium (Fig. 1u–v”), possibly a consequence of hyperproliferation or abnormal differentiation of precursors. Given that ECs interact with ISCs and modulate their proliferation96, this suggests a non-cell-autonomous role of Pax in regulating ISC proliferation during midgut homeostasis. In the mammalian gastrointestinal (GI) tract, anoikis—a form of cell death induced by loss of basement membrane (BM) interaction—contributes to epithelial renewal maintenance. Focal adhesion component Integrin engagement is critical for inhibiting this programmed cell death97,98. We investigated whether Pax loss in ECs induces cell death, but found that knockdown of Pax did not trigger EC cell death (Supplementary Fig. 3y–z’), suggesting that changes in adhesion or mechanical properties induced by Pax loss in ECs do not stimulate such effects in Drosophila midgut. Taken together, these results indicate that Pax is essential in controlling both cell-autonomous and non-cell-autonomous proliferation of ISCs.
Pax plays a critical role in EB cell differentiation
To investigate the impact of Pax on the terminal differentiation of ISCs, we conducted lineage tracing experiments using EsgGal4 tubGal80ts UAS-GFP; UAS-flp Act > CD2>Gal4 (Esgts F/O). This system enables labeling and tracing precursors and their terminally differentiated cells via expressing GFP55. As shown in Fig. 2a–c”, a 3-day induction of Pax loss resulted in a suppression of EC-like GFP+ cells compared with the control group. Conversely, overexpression of Pax increased the population of renewed ECs. The number of Pros+ EEs and Pros+ Esg-GFP+ pre-EEs remained unaffected by either Pax knockdown or overexpression (Fig. 2g and Supplementary Fig. 4a–c”, g). Following a 5-day induction, we observed that the renewal of ECs (GFP+ and Pdm1+) in wild-type midguts (Fig. 2d–d”, h) was significantly lower compared to the overexpression of Pax (Fig. 2f–f”, h), with the latter showing midguts filled with renewed ECs. In contrast, Pax RNAi significantly decreased the number of renewed ECs (Fig. 2e–e”, h). In addition, using the Esgts system to overexpress Pax in precursors also resulted in an increase in ECs population, while knockdown of Pax decreased them (Supplementary Fig. 4d–f”, h), suggesting that Pax stimulates the differentiation of ECs.
a–f” Adult midguts of EsgGal4 tubGal80ts UAS-GFP; UAS-flp Act > CD2>Gal4 (EsgtsF/O)>Ctrl (a–a”, d–d”), EsgtsF/O>Pax RNAi (b–b”, e–e”) and EsgtsF/O > HA-Pax (c–c”, f–f”) were induced for 3 days (a–c”) or 5 days (d–f”). Midguts were immunostained with Arm + Pros (red) (a–f”) and DAPI (nuclei, blue). ECs are marked by Pdm1 (red) (d–f”). ISCs/EBs and EEs/ECs are marked by GFP. White arrows at (a–c”) mark Pros+ pre-EE/EE cells. White arrows at (d–f”) mark Pdm1+ and GFP+EC cells. g Quantification of Pros+ pre-EE/EE cells in the whole midgut from the indicated genotypes in (a–c”) (n = 11, 12, 11). The counting of Pros+ EE cells was conducted across the entire midgut. h Quantification of GFP+ and Pdm1+ EC cells in the same region of midgut from the indicated genotypes in (d–f”) (n = 11, 11, 11). i–k” Adult midguts of Esgts>Ctrl (i–i”), Esgts>Pax RNAi (j–j”) and Esgts > HA-Pax (k–k”) were immunostained with NICD (red) and DAPI (nuclei, blue). ISCs and EBs/pre-EEs were marked by EsgGal4-driven GFP expression. l–n’ Adult midguts of NRE-lacZ;Esgts>Ctrl (l–l’), NRE-lacZ; Esgts>Pax RNAi (m–m’) and NRE-lacZ;Esgts > HA-Pax (n–n’) were immunostained with β-gal (red), HA (blue) and DAPI (nuclei, gray). EsgGal4-driven GFP marks ISCs and EBs/pre-EEs. o Quantification of lacZ+ EB cells in the same region of midgut from the indicated genotypes in (l–n’) (n = 10,10,10). p–q”’ Adult midguts of Esgts>Flag-Pax (p–p”’) and Esgts>Flag-Pax; Notch RNAi (q–q”’) were immunostained with Pdm1 (red), Flag (blue), and DAPI (nuclei, gray). EsgGal4-driven GFP marks ISCs and EBs/pre-EEs. r Quantification of the percentage of Pdm1+ EC cells in the same region of midgut from the indicated genotypes in (p–q”’) and (Supplementary Fig. 6a–b”) (n = 15,12,11,11). s Total RNA for real-time PCR was collected from whole midguts of the indicated genotypes: Esgts>Ctrl and Esgts > HA-Pax. Midguts were analyzed 5 days post-induction (n = 3,3). The relative mRNA level was normalized with that of Esg-GFP. t Total RNAs for real-time PCR were collected from whole midguts of the indicated genotypes: Esgts>Ctrl and Esgts > HA-Pax. Midguts were analyzed 5 days post-induction (n = 3,3). The relative mRNA level was normalized with that of Esg-GFP. u Total RNA for real-time PCR was collected from whole midguts of the indicated genotypes: Esgts>Ctrl and Esgts>Pax RNAi. Midguts were analyzed 5 days post-induction (n = 3,3). The relative mRNA level was normalized with that of Esg-GFP. Three independent experiments were performed, and the error bars are mean ± SEM. In each box plot, the center line indicates the median, the edges of the box represent the first and third quartiles, and the whiskers extend to the minimum and maximum values. P values of significance (indicated with asterisks, NS no significance P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****p < 0.0001) were calculated by two-tailed Student’s t-test (s–u) and one-way ANOVA with Tukey’s test (g, h, o, r). Scale bars: 30 µm. Confocal images were taken from the basal layer of the posterior midgut.
To further explore whether Pax impacts the renewal of ECs and EEs, we employed EsgGal4 tubGal80ts UAS-GFP; UAS-H2B::RFP (EsgREDDMts) lineage tracing method, which enables the dynamics of precursor cells at single-cell to tissue levels with spatial and temporal resolution, distinguishing Esg+ mother cells (GFP+RFP+) from progeny (RFP+)99. As shown in Supplementary Fig. 4i–n, o, overexpression of Pax accelerated the replenishment of ECs (new ECs, RFP+ EC cells) following a temperature shift of 1–2 weeks, while reducing Pax delayed the turnover of EC. However, neither Pax overexpression nor knockdown affected EE differentiation (new EEs, RFP+ EEs) (Supplementary Fig. 4i–n, p), indicating that Pax facilitates ECs differentiation, not EEs.
Next, we used the Su(H)GBEGal4 tubGal80ts; UAS-GFP (NREts) system and DlGal4 tubGal80ts; UAS-GFP (Dlts) system to assess Pax’s influence on ISC differentiation. These systems allow for specific Gal4 expression in EBs or ISCs/pre-EEs, respectively100. We observed that overexpression of Pax in EBs promoted differentiation into ECs (Supplementary Fig. 5a, b, g). Conversely, reducing Pax in EBs inhibited this process (Supplementary Fig. 5c, g). Furthermore, Pax overexpression in EBs increased the proportion of EB cells expressing the EC marker Pdm1 (GFP+ Pdm1+), suggesting enhanced premature differentiation into ECs (35%) compared to normal conditions (15%) (Supplementary Fig. 5a, b, g). Consistently, using Dlts driver to modulate Pax levels in ISC/pre-EEs cells caused a consistent change in the differentiation of ECs (Supplementary Fig. 5i–k, o). However, regardless of whether Pax was overexpressed or reduced in precursors, EE populations remained unaffected (Supplementary Fig. 5d–f, h, l–n, p), indicating that Pax favors EC differentiation by both the process of accelerating the differentiation of EBs into ECs and by promoting ISC differentiation into EBs. Importantly, we performed immunostaining for Pdm1 and quantified Pdm1+ ECs, medium-size nucleus of ISCs/EBs, and mini-size nucleus of pre-EEs/EEs in Pax20 MARCM clones. As shown in Supplementary Fig. 5w-ab, Pax mutants exhibited larger clone sizes compared to wild-type clones, along with an increase in the number of ISCs/EBs, a reduction in ECs, and no change in pre-EEs/EEs compared with wildtype, consistent with our aforementioned findings. In summary, our data demonstrate that Pax suppresses ISC proliferation while promoting its differentiation into ECs.
Next, we investigated the potential mechanisms through which Pax promotes ISC differentiation into EBs. On the one hand, some studies emphasize the critical role of the Par complex and Integrins in the asymmetric division of ISCs18,19,20. In this context, we investigated whether Pax knockdown or overexpression affects the distribution of the Par complex component Protein kinase C (Pkc) in dividing ISCs labeled by p-H3. However, rabbit-derived antibodies p-H3 and Pkc did not yield satisfactory results. Notably, Pax overexpression reduced Dl+ ISCs while maintaining Esg-GFP+ cell numbers (Supplementary Fig. 5q–t). Additionally, Pax overexpression in ISCs/EBs increased the EB cell population (Supplementary Figs. 4, 5a–p), suggesting a shift towards symmetric EB-EB duplication. Immunofluorescence analysis revealed predominant co-localization of Pax with Integrin at the basement membrane in ISCs and EBs (Supplementary Fig. 5u–v”), suggesting Pax’s critical role in anchoring the basal daughter cell and guiding ISC asymmetric division.
On the other hand, the symmetric divisions of ISC give rise to two distinct daughter cell types: ISC-ISC and EB-EB. During the symmetric division, low Notch signaling leads to ISC-ISC production. Conversely, a high Notch leads to EB-EB production11. Therefore, we investigated the relationship between Pax and Notch signaling. As shown in Fig. 2i–k”, knockdown of Pax in precursors caused decreased NICD+ cells, indicative of decreased Notch intracellular domain (NICD) activity, whereas overexpression of Pax increased their abundance. Next, we co-expressed Pax or Pax RNAi with Notch signaling reporter Gbe-Su(H)m8-lacZ (NRE-lacZ), which integrates enhancer sequences from E(spl)m8 containing Grainyhead (Grh) and Su(H) binding sites101,102. Expectedly, overexpression of Pax significantly upregulated NRE-lacZ levels (Fig. 2l–n’, o), implying that Pax activates Notch signaling activity in precursors of adult midgut. Importantly, the increase in Pdm1+ ECs due to Pax overexpression was dramatically abolished upon Notch pathway inactivation through Notch knockdown (V100002) (Fig. 2p–q”’, r and Supplementary Fig. 6a–b”), indicating a genetic epistasis of Pax over the Notch pathway. However, overexpression of Pax in wild-type or Notch-deleted ISCs-like clones did not influence the proportion of pre-EEs (Esg+/Pros+) (Supplementary Fig. 6c–g). These results demonstrate that overexpression of Pax triggers symmetric division of ISCs into EB-EB through facilitating Notch pathway activity.
To gain further insights into the relationship between Pax expression and Notch signaling activity in the midgut, we performed Notch signaling components transcriptional comparisons between wild-type midguts and those overexpressing Pax or Pax RNAi. Of note, we observed elevated transcription of Notch after overexpression of Pax (Fig. 2s), whereas Pax RNAi conversely decreases it (Fig. 2u). Given that the NICD binds with the transcription factor suppressor of hairless (Su(H)) to activate target genes that direct ISC differentiation in the midgut103, we next investigated whether Pax regulates the known Notch target genes, including Hey, vestigial (vg), and various E(spl)-C genes in precursors102. As expected, overexpression of Pax in precursors significantly increased the transcription of these target genes (Fig. 2t). Conversely, Pax knockdown decreased their expression (Fig. 2u). Based on these results, we conclude that Pax stimulates the expression of Notch to activate the Notch pathway, thereby promoting symmetric division of ISC into EB-EB and triggering its premature differentiation into ECs.
Pax is regulated by the Hippo signaling pathway
Through combining publicly available RNA-seq and ChIP-seq datasets for Yki and Sd81,82,83,84,85,86,87,88, we have identified the potential of the Hippo signaling pathway target genes, among which Pax was included. RT-qPCR experiments further validated that Yki regulated the transcription of Pax in the wing disc (Supplementary Fig. 7a). To further confirm this observation, we employed four approaches to modulate the Hippo pathway in precursors and ECs: (1) overexpression of Yki, (2) introduction of constitutively active Sd (Sd-GA), an activated form of Scalloped in which the Gal4 activation domain was fused to Sd C-terminus49, (3) knockdown of hpo using RNAi line (BS33614), and (4) knockdown of Yki using RNAi line. As shown in Fig. 3a–f’, the inactivation of the Hippo signaling in precursor cells and ECs effectively promoted Pax expression, while the knockdown of Yki decreased it (Supplementary Fig. 7d–e”).
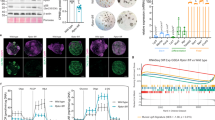
a–f’ Adult midguts of Esgts>Ctrl (a–a’), Esgts>Yki(b–b’), Esgts > HA-Sd-GA(c–c’), Esgts >Hpo RNAi (d–d’), MyoIAts>Ctrl (e–e’), and MyoIAts>Hpo RNAi (f–f’) were immunostained with Pax (red), Arm + Pros (blue) and DAPI (nuclei, gray). GFP marks ISCs/EBs/pre-EEs or ECs. g–i’ Adult midguts of Pax-GFP;Esgts>Ctrl (g–g’), Pax-GFP;Esgts>Yki (h–h’), Pax-GFP; Esgts > HA-Sd-GA (i–i’) were immunostained with Flag/HA (red) and DAPI (nuclei, blue). GFP marks Pax+ cells. j, k Total RNAs for real-time PCR were collected from whole midguts of the indicated genotypes: Esgts>Ctrl, Esgts>Yki and Esgts>Hpo RNAi, MyoIAts>Ctrl, MyoIAts>Yki and MyoIAts>Hpo RNAi. Midguts were analyzed 5 days post-induction (n = 3 for each group). The relative mRNA level was normalized with that of Esg-GFP in (j). l Plot of ChIP peaks of Pax regulated by Yki. Transcription units of Pax are in purple. Regions called as Yki-Sd peaks are identified by black boxes. m Relative enrichment of Pax on binding regions compared to IgG in control (blue bars) and overexpression of HA-Sd group (red bars) were analyzed by ChIP and real-time PCR. n Relative activity of the luciferase reporter plasmids carrying Sd-binding-region on Pax (B1, B2, B3) were transfected into S2 cells with the indicated constructs. Three independent experiments were performed, and the error bars are mean ± SEM. P values of significance (indicated with asterisks, NS no significance P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****p < 0.0001) were calculated by two-tailed Student’s t-test (j, k, m, n). Scale bars: 30 µm. Confocal images were taken from the boundary region between R4c and R5a of the posterior midgut.
Subsequent experiments using FACS to sort Esg-GFP+ cells in the Yki or Sd overexpressed midgut were employed to further verify the regulation of Pax by the Hippo signaling pathway. As shown in Supplementary Fig. 7b, overexpression of Yki or Sd increased the proportion of Esg-GFP+ cells (12.7 and 9.75%, respectively) compared with wild-type (5.49%), followed by elevated Pax transcription (Supplementary Fig. 7c), indicating that the Hippo pathway modulates Pax expression in the adult midgut.
To demonstrate the universality of this regulation, we additionally inactivated the Hippo pathway in wing discs or eye discs using the AG4 flip-out clone system to induce overexpression of Yki, Sd-GA, or wts RNAi. As shown in Supplementary Fig. 8a–c”’, g–j”, inactivation of the Hippo signaling in wing or eye discs consistently elevated Pax expression in GFP+ clones with larger clone sizes. Additionally, we employed Sd or Yki RNAi transgenes driven by hedgehog-Gal4 (hhGal4) to specifically activate UAS transgenes in the posterior (P) compartment. Consistently, the knockdown of Yki or Sd resulted in reduced Pax expression in P-compartment cells and a decrease in compartment size (Supplementary Fig. 8d–f”). These findings strongly support that the expression of Pax is controlled by the Hippo pathway.
Next, in order to visualize the transcriptional activity of Pax in vivo, we employed a gene trap technique using BS43941 (referred to as Pax-GFP), which exhibited a progenitor-specific expression pattern, mirroring the endogenous Pax expression in the adult midgut (Fig. 3g–g’). We observed that Pax-GFP responded to the Hippo signaling activity: inactivation of the Hippo pathway by overexpressing Yki and Sd-GA promoted Pax-GFP expression (Fig. 3h–i’), suggesting that Yki and Sd regulate the transcription of Pax in vivo. The RT-qPCR analysis consistently demonstrated upregulation of Pax mRNA levels upon Yki overexpression or Hpo RNAi in precursors and ECs (Fig. 3j, k), whereas reducing Yki decreased Pax expression (Supplementary Fig. 8k), confirming Pax as a downstream target gene of Yki. A more detailed analysis of the binding regions of Yki on the Pax gene showed four predicted regions (B1, B2, B3, B4) within the 5’ UTR and 3’UTR of Pax83 (Fig. 3l). Three (B1, B2, B3) contained the potential Hippo response elements (CATTCC, HREs) sites. Chromatin immunoprecipitation qPCR (ChIP-qPCR) assays showed dramatic enrichment of Sd in B1, B2, and B3 regions of Pax, indicating direct interaction between Sd and Pax in these regions (Fig. 3m). To further confirm that Yki/Sd promotes the transcription of Pax, Yki/Sd-binding regions (B1, B2 and B3) of Pax were cloned into upstream of the luciferase gene. As shown in Fig. 3n, the activities of these reporters were markedly increased with Yki/Sd complex overexpression. However, deletion of potential Hippo HRE sites (ΔB1, ΔB2, ΔB3) resulted in no change in luciferase activity upon Yki and Sd overexpression (Supplementary Fig. 8l). These findings collectively demonstrate that Yki/Sd complex directly regulates Pax expression via binding to the enhancer regions B1, B2, and B3.
Loss of Pax activates EGFR and JAK-STAT signaling in Drosophila midguts
Given that the EGFR and JAK-STAT pathways are the crucial mitotic pathways that drive ISC proliferation in the midgut26,104,105, we performed epistatic assays to determine whether Pax mediates these pathways to regulate ISC proliferation. To monitor the EGFR pathway activity, we examined the protein levels of dpErk, the diphosphorylated active form of MAPK106. In control guts, dpErk signals were slightly detected in precursor cells, whereas loss of Pax in ECs or precursor cells resulted in significantly increased dpErK signals in precursor cells (Fig. 4a–d’). In addition, the knockdown of Pax in ECs effectively upregulated the mRNA levels of EGFR ligands vein (vn), Keren (Krn), and spitz (spi)107 (Fig. 4e), further confirming EGFR activation.
a–d’ Adult midguts of Esgts>Ctrl (a–a’), Esgts>Pax RNAi (b–b’), MyoIAts>Ctrl (c–c’), and MyoIAts>Pax RNAi (d–d’) were immunostained with dpErk (red) and DAPI (nuclei, blue). GFP marks ISCs/EBs/pre-EEs (a–b’) or ECs (c–d’). e Relative mRNA levels of EGFR ligands spi, Krn, and vn were analyzed by real-time PCR. Total RNA was collected from whole midguts of the indicated genotypes: MyoIAts>Ctrl, and MyoIAts>Pax RNAi (n = 3,3). f–m Adult midguts of Upd-lacZ;Esgts>Ctrl (f), Upd-lacZ;Esgts>Pax RNAi (g), Upd-lacZ;MyoIAts>Ctrl (h), Upd-lacZ;MyoIAts>Pax RNAi (i), 10xStat-GFP;Esgts>Ctrl (j), 10xStat-GFP;Esgts>Pax RNAi (k), Stat-lacZ; MyoIAts>Ctrl (l), Stat-lacZ;MyoIAts>Pax RNAi (m) were immunostained with β-gal (red), GFP (green) and DAPI (nuclei, blue). Note that Upd and Stat are specifically expressed in precursors. n Relative mRNA levels of JAK-STAT ligands, including Upd, Upd2, Upd3, and target Scos36E were analyzed by real-time PCR. Total RNAs were collected from whole midguts of the indicated genotypes: MyoIAts>Ctrl, and MyoIAts>Pax RNAi (n = 3,3). Three independent experiments were performed, and the error bars are mean ± SEM. P values of significance (indicated with asterisks, NS no significance P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****p < 0.0001) were calculated by two-tailed Student’s t-test (e, n). Scale bars: 30 µm. Confocal images were taken from the boundary region between R4c and R5a of the posterior midgut.
Moreover, knockdown of Pax in either precursors cells or ECs activated JAK-STAT signaling, as indicated by the elevated expressions of JAK-STAT ligand Upd (Upd-lacZ), Stat-lacZ, and 10XStat-GFP, which is a multimerized Stat92E reporter driving GFP expression108 (Fig. 4f–m). Consistently, mRNA levels of Unpaired (Upd), Upd2, Upd3, and the JAK-STAT target Socs36E were remarkably elevated upon reducing Pax (Fig. 4n). However, knockdown of Pax in precursors only slightly increased the expression of Upds and EGFR ligands (Supplementary Fig. 9a, b), which aligns with previous research findings30. To further analyze the genetic interaction between Pax and JAK-STAT or EGFR pathway in the adult midgut, we co-expressed Pax RNAi with Stat RNAi or Vn RNAi to inactivate JAK-STAT or EGFR pathway in precursors. The results revealed that reducing Stat or Vn effectively suppressed the hyperproliferation induced by the loss of Pax, indicating that Pax acts upstream of the JAK/STAT and EGFR pathways (Supplementary Fig. 9c–g). Taken together, these findings underscore Pax’s role in regulating ISC proliferation by exerting control over EGFR and JAK-STAT signaling pathways.
Pax operates downstream of Wts to establish a feedback regulation of the Hippo pathway
Several signaling pathways, including Hh, Wnt, BMP, JNK, and the Hippo pathways regulate ISC proliferation by modulating the production of EGFR and JAK-STAT pathway ligands during midgut homeostasis and in response to injury. These pathways exhibit fine-tuned activity through various feedback loops16,21,29,30,32,109,110,111,112. We investigated whether Pax acts as both a target gene and regulator of Yki to control ISC proliferation via feedback mechanisms. As expected, the knockdown of yki in ECs dramatically suppressed the elevated expression of JAK-STAT and EGFR pathway ligands following Pax knockdown (Fig. 5a), indicating that Pax-mediated regulation of EGFR and JAK-STAT signaling relies on the Hippo pathway.
a Total RNA for real-time PCR was collected from whole midguts of the indicated genotypes: MyoIAts>Ctrl, MyoIAts>Pax RNAi, MyoIAts>Yki RNAi and MyoIAts>Pax RNAi;Yki RNAi (n = 3 for each group). b–e’ Adult midguts containing GFP-positive MARCM clones of control (b–b’), wtsx1 (c–c’), HA-Pax (d–d’) and wtsx1 in the presence of Pax (e–e’). Midguts were dissected 6 days after clone induction and immunostained with DAPI (nuclei, blue). Clones are marked by GFP. f Quantification of the cell number per clone in adult midguts from the indicated genotypes in (b–e’) at 2, 4, and 6 days after clone induction (n = 41, 72, 63, 67, 48, 92, 42, 80, 45, 74, 65, 29). g–n’ Adult midguts of bantam-lacZ;Esgts>Ctrl (g–g’), bantam-lacZ;Esgts>Pax RNAi (h–h’), Diap1-lacZ;Esgts>Ctrl (i–i’), Diap1-lacZ;Esgts>Pax RNAi (j–j’), bantam-lacZ;MyoIAts>Ctrl (k–k’), bantam-lacZ;MyoIAts>Pax RNAi (l–l’), Diap1-lacZ;MyoIAts>Ctrl (m–m’) and Diap1-lacZ;MyoIAts>Pax RNAi (n–n’) were immunostained with β-gal (red) and DAPI (nuclei, blue). Note, Bantam is specifically expressed in precursor cells and EEs, and Diap1 is upregulated in both precursors (GFP- lacZ+, white arrows) and ECs (GFP+ lacZ+, orange arrows) when Pax is knocked down in ECs. o–p Relative mRNA levels of Hippo signaling targets Diap1 and ex were analyzed by real-time PCR. Total RNA was collected from whole midguts of the indicated genotypes: Esgts>Ctrl, Esgts>Pax RNAi, MyoIAts>Ctrl, and MyoIAts>Pax RNAi (n = 3 for each group). The relative mRNA level was normalized with that of Esg-GFP in (o). Three independent experiments were performed, and the error bars are mean ± SEM. P values of significance (indicated with asterisks, NS no significance ≥0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****p < 0.0001) were calculated by two-tailed Student’s t-test (o, p) and one-way ANOVA with Tukey’s test (a, f). Scale bars: 30 µm. Confocal images were taken from the boundary region between R4c and R5a of the posterior midgut.
To further elucidate the genetic interaction between Pax and the Hippo pathway, GFP+ clones of wts mutant allele wtsx1 were generated by using the MARCM system. Clones were induced in parallel in adult flies raised at 25 °C for 2, 4, or 6 days after induction. We observed that overexpression of Pax in wtsx1 clones that exhibited the overgrowth phenotype dramatically reduced the clone size (Fig. 5b–e’, f), Conversely, knockdown of Pax in wtsx1 clones exhibiting overgrowth substantially increased the clone size (Supplementary Fig. 10a–e), indicating functional downstream effects of Pax. Moreover, we examined the expression of the Hippo pathway activity reporters, Diap1-lacZ and bantam-lacZ49,51. Diap1 has been reported to play a vital role in regulating EBs’ apoptosis113, while the microRNA bantam has been shown to stimulate ISCs proliferation75. Consistently, reducing Pax either in precursors or ECs inhibited the Hippo signaling as indicated by elevated expression of Diap1-lacZ and bantam-lacZ (Fig. 5g–n’). Consistently, loss of Pax in precursors or ECs caused a significant increase in the mRNA levels of Diap1 and ex, whereas yki expression remained unaffected (Fig. 5o, p and Supplementary Fig. 10f, g), indicating that loss of Pax enhances the signaling output of the Hippo pathway. In addition, the GFP+ clone size analysis of Pax20 null allele mutant, generated by the MARCM system, showed larger sizes compared with wild-type clones, accompanied by significantly increased expression of Diap1 (Supplementary Fig. 10h–i”), indicating that the loss of Pax in the precursor cells inhibits the activity of the Hippo pathway. In summary, these findings highlight a feedback loop between Pax and the Hippo pathway is crucial for maintaining intestinal homeostasis in Drosophila.
Pax regulates midgut homeostasis dependently on Yki-Sd
To probe the relationship between the Hippo pathway and Pax in the regulation of ISC proliferation, we ectopically activated the Hippo signaling by knocking down yki (V40497, 301 + 302) or sd (254 + 255) in ISCs/EBs, concurrently with Pax knockdown using the Esgts system. Genetic interaction analysis indicated that co-expression of Pax RNAi with either yki or sd RNAi in precursors effectively suppressed the increased p-H3+ cell numbers induced by Pax RNAi alone (Fig. 6a–e and Supplementary Fig. 11a–f’), suggesting that loss of sd or yki suppresses the Pax RNAi-induced ISC proliferation.
a–d’ Adult midguts of Esgts>Ctrl (a–a’), Esgts>Pax RNAi-1 (b–b’), Esgts>Pax RNAi−1;Yki RNAi-1 (c–c’) and Esgts>Pax RNAi-1;Sd RNAi (d–d’) were immunostained with p-H3+(red) and DAPI (nuclei, blue). ISCs and EBs/pre-EEs are marked with Esg-GFP (green). White arrows indicate the p-H3 signal. e Quantification of p-H3+ cells of adult midguts of the indicated genotypes of (a–d’) and (Supplementary Fig. 11a–f’) (n = 10,13,15,10,11,10,10,10,10). The counting of p-H3+ cells was conducted across the entire midgut. f–i’ Adult midguts containing GFP-positive MARCM clones of Ctrl (f–f’), sdΔB1 (g–g’), Pax RNAi (h–h’), sdΔB1 in the presence of Pax RNAi (i–i’) were dissected 3 days after clone induction and immunostained with Arm+Pros (red) and DAPI (nuclei, blue). Dotted lines mark the edges of the clone region. j Quantification of the cell number per clone in adult midguts from the indicated genotypes in (f–i’) at 3 days after clone induction (n = 30, 70, 33, 35). k–n’ Adult midguts containing GFP-positive MARCM clones of Ctrl (k–k’), ykiB5 (l–l’), Pax RNAi (m–m’), ykiB5 in the presence of Pax RNAi (n–n’) were dissected 3 days after clone induction and immunostained with Arm + Pros (red) and DAPI (nuclei, blue). Dotted lines mark the edges of the clone region. o Quantification of the cell numbers per clone in adult midguts from the indicated genotypes in (k–n’) at 3 days after clone induction (n = 35, 40, 46, 80). p–s’ Relative wing discs containing GFP-positive Flip-out clones of Ctrl (p–p’) and Pax RNAi (q–q’), Yki RNAi (r–r’), and Pax RNAi;Yki RNAi (s–s’) were dissected 3 days after clone induction and immunostained with DAPI (nuclei, blue). t Quantification of cell number of clones in panels (p–s’) (n = 14 for each genotype). Three independent experiments were performed, and the error bars are mean ± SEM. In each box plot, the center line indicates the median, the edges of the box represent the first and third quartiles, and the whiskers extend to the minimum and maximum values. P values of significance (indicated with asterisks, NS no significance P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****p < 0.0001) were calculated by one-way ANOVA with Tukey’s test (e, j, o, t). Scale bars: 30 µm (a–d’, f–i’, k–n’), 60 µm (p–s’). Confocal images were taken from the boundary region between R4c and R5a of the posterior midgut.
To further support our findings, a separate MARCM clone approach independent of yki or sd RNAi was employed by using yki or sd mutant allele ykib5 or sdΔb1. Clone sizes were compared by measuring the number of GFP+ cells per clone. As expected, Pax RNAi clones were bigger than the controls, whereas the sdΔB1 mutant clones with Pax RNAi dramatically inhibited overgrowth phenotype in clone size compared with Pax deletion alone (Fig. 6f–j). Similarly, the deletion of yki in MARCM mutant clones also decreased the larger clone size induced by Pax RNAi (Fig. 6k–o). Consistently, Pax knockdown leads to excessive proliferation of clones in wing discs, which is suppressed upon further Yki knockdown (Fig. 6p–t), suggesting that Pax acts upstream of Yki/Sd. In addition, co-expressing Pax RNAi and Yki RNAi in ECs suppressed the excessive ISC proliferation induced by Pax loss (Supplementary Fig. 11g–k), underscoring the reliance of Pax-regulated non-autonomous ISC proliferation on Yki.
Interestingly, the simultaneous expression of Yki and Pax suppressed the hyperproliferation of ISCs induced by the overexpression of Yki, resulting in downregulation of Yki targets Diap1 and ex (Supplementary Fig. 12a–f). This suggests that Pax inhibits Yki activity in adult midgut. To further uncover the relationship between Pax and Yki, we co-expressed Pax with Sd-GA or co-expressed Pax RNAi with Yki separately. Overexpressing Pax has no effect on the increased ISC proliferation induced by Sd-GA overexpression, since Sd-GA constitutively activates Sd independently of Yki (Supplementary Fig. 12g–j’, n). Conversely, knocking down Pax further enhanced ISC proliferation induced by Yki overexpression (Supplementary Fig. 12k–n). Altogether, these findings underscore that Pax mediates ISC proliferation by restricting Yki activity.
Pax forms a complex with Yki and promotes its cytoplasm localization
Given that the nuclear translocation of Yki is a crucial step in the Hippo signaling47,49,114,115,116, we detected whether Pax modulates the cytoplasmic-nuclear shuttling of Yki in S2 cells. The overexpressed wild-type Yki were partially localized in the cytoplasm, while co-expression with Pax induced more cytoplasmic translocation of Yki (Fig. 7a–b’, g). Surprisingly, a truncation of Yki (Yki-N) that lacks C-terminal 195–418 amino acids (Yki-C) exhibited a dispersive distribution in S2 cells, whereas co-expression with Pax further promoted its cytoplasmic localization (Fig. 7c–d’, g). However, Yki-C is mainly distributed in the cytoplasm, and co-expression with Pax did not alter its localization significantly (Fig. 7e–f’, g). These results suggest that Pax may promote cytoplasmic localization of Yki through binding to its N-terminal domain, thereby inactivating it. Additionally, we assessed the activity of the 3xSd luciferase reporter, which reflects Sd-Yki transcriptional activity49. Co-expression of full-length Yki and Sd synergistically boosted the luciferase activity. However, Pax inhibited this enhancement, indicating that Pax suppresses Sd-Yki transcriptional activity (Fig. 7h). Consistent with this, remarkably cytoplasmic localization of endogenous Yki was also detected upon expression of Pax in precursors (Fig. 7i–j” and Supplementary Fig. 13c) or in flip-out clones expressing Pax using the AG4 driver (Supplementary Fig. 13a–c). Conversely, the knockdown of Pax in precursor cells resulted in increased Yki translocation into the nuclei (Fig. 7k–k” and Supplementary Fig. 13d–f). In addition, exogenous Yki is predominantly distributed in the nuclei, whereas co-expression with Pax significantly resulted in the cytoplasmic localization of Yki (Fig. 7l–m” and Supplementary Fig. 13c). Taken together, these results indicate that Pax restricts ISC proliferation by promoting cytoplasmic localization of Yki.
a–f’ S2 cell expressing Myc-Yki with or without HA-Pax (a–b’), Myc-Yki-N with or without HA-Pax (c–d’) and Myc-Yki-C with or without HA-Pax (e–f’) were immunostained with Myc (red), HA (green) and DAPI (Nuclei, blue). g Quantification of the percentage of nucleocytoplasmic distributions in (a–f’) (n = 3 for each group, counting 100, 125, 112 S2 cells). h Analysis 3xSd luciferase activity of S2 cells after overexpression of Pax. 3xSd luciferase is a dual reporter reflecting Sd-Yki transcriptional activity with 3xSd-binding sites. The indicated constructs were transfected into S2 cells. i–k” Adult midguts of Esgts >Ctrl (i–i”), Esgts > HA-Pax (j–j”), Esgts>Pax RNAi (k–k”) were immunostained with Yki (red), Arm + Pros (blue) and DAPI (nuclei, gray). ISCs and EBs/pre-EEs are marked with GFP. White arrows indicate the Yki signal. l–m” Adult midguts containing GFP-positive flip-out clones of act > CD2>Gal4; UAS-Dicer2, UAS-GFP (AG4; Dcr2GFP)>Flag-Yki (l–l”) and AG4;Dcr2GFP > HA-Pax; Flag-Yki (m–m”) were dissected 2 days after clone induction and immunostained with Flag (red), Arm + Pros (blue) and DAPI (nuclei, gray). Note that exogenous Yki is localized in the nucleus of precursors/EEs/ECs. White arrows indicate the Flag-Yki signal. n S2 cells were transfected with plasmids expressing Myc-Yki with or without Flag-Pax, Flag-Pax-N, and Flag-Pax-C. o S2 cells were transfected with plasmids expressing Flag-Pax with or without Myc-Yki, Myc-Yki-N, and Myc-Yki-C. p Co-IP analysis confirms that endogenous Pax interacts with endogenous Yki using Pax antibody. q S2 cells were transfected with plasmids expressing Flag-Pax or Flag-Pax-dLDs with Myc-Yki. Three independent experiments were performed, and the error bars are mean ± SEM. P values of significance (indicated with asterisks, NS no significance P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****p < 0.0001) were calculated by one-way ANOVA with Tukey’s test (g, h). Scale bars: 5 µm in (a–f’), and 20 µm in (i–m”). Confocal images were taken from the boundary region between R4c and R5a of the posterior midgut in (i–m”).
Next, we investigated whether exist the interaction between Yki and Pax. Co-immunoprecipitation (Co-IP) assays were performed in S2 cells expressing Myc-Yki and Flag-Pax, we observed that Yki formed a complex with Pax (Fig. 7n, o). Furthermore, using Pax antibody to pull down endogenous Yki further supported their interaction (Fig. 7p). To map the binding regions of Yki and Pax, we constructed the truncated variants of both proteins (Supplementary Fig. 13g). Co-IP results indicated that Flag-Pax-N, but not Flag-Pax-C, interacted with Myc-Yki (Fig. 7n). Similarly, Myc-Yki-N, but not Myc-Yki-C, interacted with Flag-Pax (Fig. 7o), suggesting that the binding of Yki and Pax is mediated by their N-termini. However, the removal of any LD motif of Pax disrupted the interaction between Yki and Pax (Fig. 7q), and decreased Yki-Sd transcriptional activity (Supplementary Fig. 13h). Furthermore, the overexpression of PaxdLD1 did not change Yki localization (Supplementary Fig. 13i–l), highlighting the critical role of all five LD motifs of Pax in interacting with and modulating Yki activity.
According to Goulas and colleagues' research, they found that cell-matrix adhesion receptors Integrin are localized basolaterally within ECs and cortically around the ISCs/EBs with an enrichment basally contacting the basement membrane, regulating ISC proliferation and self-renewal18. We investigated whether Pax induced more Yki co-localization with Integrin. As shown in Supplementary Fig. 14a–d”, overexpression of Pax notably facilitated Yki’s co-localization with Integrin, followed by more cytoplasm localization of Yki, suggesting that Pax promotes the cytoplasm localization of Yki via sequestering Yki at membrane localization of cell-matrix adhesion. Altogether, these findings indicate that Pax forms a feedback loop with Yki, facilitating cytoplasmic localization of Yki and thereby constraining its activity.
The conservation of Pax’s role as a regulator of Yki activity was investigated in mammalian cells. Previous studies have demonstrated that high cell density in mammalian cells activates LATS1/2, resulting in the phosphorylation and inactivation of YAP, while low cell density promotes the nuclear localization of YAP114. We either overexpressed or reduced the expression of PXN (the homolog of Pax in mammals) in Hela cells, and subsequently examined the subcellular localization of endogenous YAP under both high and low density conditions. As shown in Supplementary Fig. 14e–l”, YAP localization remained unchanged regardless of PXN overexpression or knockdown. Moreover, biochemical assays revealed no physical interaction between PXN and YAP (Supplementary Fig. 14m). However, overexpression of PXN significantly increased p-YAP (S127) levels and decreased the expression of the Hippo pathway target gene CYR61 (Supplementary Fig. 14n). In addition, deletion of LATS1/2 observably upregulated the expression of PXN (Supplementary Fig. 14o). These findings suggest that while PXN/Pax suppresses YAP/Yki activity in both mammals and Drosophila, the underlying mechanisms are not conserved.
Pax restricts Yki activity by promoting its cytoplasmic localization to regulate ISCs proliferation and regeneration
We finally examined whether the cytoplasmic localization of Yki, stimulated by Pax, is capable of modulating ISCs proliferation and regeneration. To test this, we induced the expression of Pax in GFP+ clones of wts mutant allele wtsx1 using the MARCM system to monitor whether the attenuated ISC proliferation induced by Pax is caused by cytoplasmic localization of Yki. As shown in Fig. 8a–e, wtsx1 clones exhibited the overgrowth phenotype and nuclear localization of Yki. In contrast, overexpression of Pax in GFP+ wtsx1 mutant clones dramatically reduced the clone size and induced cytoplasmic localization of Yki, indicating that the inhibitory effect of Pax on midgut hyperproliferation is achieved through elevating the cytoplasmic localization of Yki. To further validate our observations, we examined whether the suppressed midgut regeneration through overexpressed Pax is triggered by cytoplasmic localization of Yki. As shown in Fig. 8f–j, overexpression of Pax significantly promoted the retention of Yki in the cytoplasm of precursors compared with the control group under both normal and DSS-induced injury conditions. Altogether, these data demonstrate that Pax inhibits ISC proliferation and injury-induced regeneration through guiding cytoplasmic localization of Yki, thereby ensuring normal adult midgut homeostasis and damage repair.
a–d’ Adult midguts containing GFP-positive MARCM clones of FRT82 (Ctrl) (a–a’), wtsx1 clones (b–b’), overexpressing Pax clones (c–c’) and Pax overexpression plus wtsx1 clones (d–d’) were immunostained with Yki (red), HA (blue), and DAPI (nuclei, gray). Midguts were dissected 4 days after clone induction at 25 ˚C. Dotted lines mark the edges of the clone region. Yellow arrows mark the GFP+ clone region, and white arrows mark the GFP- region. e Quantification of the percentage of nucleocytoplasmic distributions in panels (a–d’) (n = 3 for each group, counting 123, 125, 112 Yki+ cells). f–i’ Adult midguts of Esgts>Ctrl (f–f’, h–h’) and Esgts > HA-Pax (g–g’, i–i’) flies treated with Glucose (Glu) or DSS for 3 days before dissection. Midguts were immunostained with Yki (red), DAPI (nuclei, gray), and HA (blue). ISC and EB/pre-EE were marked by EsgGal4-driven GFP expression. Arrows indicate HA+ and Yki+ cells. 5% Glucose solution with or without 3% DSS was fed to the flies. j Quantification of the percentage of nucleocytoplasmic distributions in panels (f–i’) (n = 3 for each group, counting 100, 125, 112 Yki+ cells). k Proposed model illustrating how Pax regulates Hippo and Notch signaling to suppress ISC proliferation and promote its differentiation in adult Drosophila midgut. Three independent experiments were performed, and the error bars are mean ± SEM. P values of significance (indicated with asterisks, NS no significance P ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****p < 0.0001) was calculated by one-way ANOVA with Tukey’s test (e, j). Scale bars: 20 µm. Confocal images were taken from the boundary region between R4c and R5a of the posterior midgut.
Discussion
The regulation of adult stem cell proliferation and differentiation in the Drosophila midgut is essential for maintaining tissue homeostasis, facilitating injury-induced regeneration, and preventing tumorigenesis. The Hippo signaling pathway has been extensively studied for its critical role in regulating midgut homeostasis and regeneration. However, the precise mechanisms governing the regulation of key signaling components, such as Yki, and their coordination of ISC proliferation and differentiation remain unknown. In this study, we focused on investigating the role of Pax function as a potential regulator of Yki and as a target of the Hippo pathway in a feedback loop to regulate homeostasis and regeneration of midgut. Through genetic approaches, we have identified Pax as a target of the Hippo pathway in the adult Drosophila midgut. Notably, loss of Pax promotes ISC proliferation through two mechanisms: autonomously inhibiting the Hippo pathway in ISCs (where Pax forms a complex with Yki to promote its cytoplasmic localization), and suppressing the Hippo signaling in EC cells, leading to increased expression of multiple ligands of the EGFR and JAK-STAT pathways. These ligands activate EGFR and JAK-STAT signaling in ISCs, thereby initiating their proliferation. Additionally, Pax overexpression in precursors facilitates Notch expression to activate the Notch pathway, thereby promoting the symmetric division of ISCs into EB-EB and enhancing its premature differentiation into ECs. Consequently, the resultant increase in ISCs and ECs ultimately induces dysplasia, a precursor to carcinoma characterized by abnormal cell proliferation, differentiation, as well as disorganized architecture4,117 (Fig. 8k). Our findings emphasize the Pax as a key mediator in ISC proliferation and differentiation under homeostatic and stress conditions. Overall, our study enriches the understanding of the complex regulatory network involving the Notch/Hippo pathway and Pax, offering potential therapeutic targets to enhance tissue regeneration and suppress cancers.
Tissue homeostasis should be strictly and precisely regulated, and the following facts demonstrate the critical role of the Pax-Yki loop and Pax-Notch regulation pattern mediated ISCs proliferation and differentiation in maintaining intestinal homeostasis. First, knocking down Pax expression in both ISCs and ECs leads to a remarkable increase in ISC proliferation (Fig. 1e–f”, q–r”), whereas overexpression of Pax significantly inhibits ISC proliferation in MARCM clone of wtsx1 or DSS-induced midgut regeneration (Figs. 1n–o’, 5e–e’). Second, our investigation revealed a crucial feedback regulation between the Hippo signaling pathway and Pax in both ISC and EC. We have demonstrated that Pax, as a target gene of the Hippo signaling pathway (Fig. 3a–f’, m), interacts with Yki to sequester it in the cytoplasm (Fig. 7i–m”, o), thereby exerting control over the proliferation of ISCs during homeostasis and injury-induced regeneration through suppressing Yki activity (Fig. 8a–j). Third, Pax promotes the differentiation of ISC toward the EC lineage through regulating Notch expression and activation, thereby triggering symmetric division of ISC into EB-EB (Fig. 2 and Supplementary Fig. 5). Last, the overgrowth phenotype induced by overexpression of Yki is dramatically reduced when Pax is overexpressed in adult Drosophila wings or eyes (Supplementary Fig. 1a–d), indicating that Pax exerts a universal regulation on Yki activity in Drosophila. These findings pinpoint that Pax as a component of the Hippo signaling pathway, contributes to the maintenance of intestinal homeostasis by precisely balancing the proliferation and differentiation of intestinal stem cells.
The Hippo pathway is extracellularly regulated by mechanical stimuli and diffusible chemical substances. These signals are largely sensed by receptors, such as G-protein coupled receptors (GPCRs) and adherence complexes embedded in the plasma membrane118,119,120. Focal adhesions (FAs) serve as crucial mechanosensory elements in cells, regulating Yes-associated protein homolog (YAP) (orthologous of Drosophila Yki) activity in response to mechanical signals121,122,123. FAs are dynamic multiprotein complexes comprised of Integrins, Vinculin, Paxillin, Zyxin, focal adhesion kinase (Fak), and tyrosine kinase Src121. Numerous studies have highlighted the role of integrins in activating YAP through the SRC and FAK123,124, but the function of Paxillin in this process remains unclear. Additionally, several studies have demonstrated that FAs exert feedback on the Hippo pathway125. Notably, YAP overexpression results in increased transcription of genes encoding FA components such as Vinculin and Zyxin, which reciprocally promote YAP activity through the elevated interaction between FAs and cytoskeleton126. Similarly, our findings uncover that Yki regulates the expression of FAs component Pax (Fig. 3m), which in turn suppresses Yki activity through a negative feedback regulatory mechanism in adult Drosophila midguts (Fig. 7h). Integrins have been demonstrated to play a crucial role in maintaining the asymmetric division of ISCs18,19. Consistently, we observed that Pax predominantly co-localizes with Integrin at the basement membrane in ISCs/EBs (Supplementary Fig. 5u–v”), and overexpression of Pax facilitates the symmetric division of ISCs into EB-EB (Supplementary Fig. 5a–t), suggesting that Pax is also essential for the assignment of asymmetric cell fate during ISC mitosis.
Our results, obtained from both in vivo and in vitro analyses, have elucidated that Pax promotes the differentiation of ISC toward the EC lineage through regulating Notch expression and activation (Fig. 2). Further investigation of the regulatory role of Pax on Notch transcription would offer more comprehensive insights into its mechanisms. Previous investigations have emphasized the dynamic shuttling ability of Pax in the cytoplasm and nucleus, enabling it to function as a transcription factor that regulates the expression of imprinted gene Histocompatibility 9 (H9)127,128,129,130. We indeed observed that Pax could also be stained within the nuclei of ISCs/EBs (Supplementary Fig. 2a–b”’), which may directly interact with the promoter region of Notch, thereby modulating its transcription.
Paxillin is highly conserved between species (57% identity between Drosophila and humans)131. Based on our findings (Supplementary Fig. 14n, o), which demonstrate that PXN is a Hippo pathway target and PXN considerably suppresses YAP activity in mammals, we propose that the modulation of Pax-regulated midgut homeostasis could represent a shared function for maintaining tissue homeostasis and regeneration in mammals. However, the potential mechanism may unique between Drosophila and mammals, as PXN does not interact with YAP and does not affect YAP localization (Supplementary Fig. 14e–l”). The dysfunction of Pax in Drosophila midgut would lead to intestinal Dysplasia (Fig. 1f–f”, Fig. 2c–c”), a precursor to carcinoma in the mammalian and Drosophila gastrointestinal tract. This phenotype resembles the tumorigenic effects caused by abnormal activity of Pax in mammals70,71. Additionally, it is worth noting that the activity of YAP is closely associated with the occurrence and development of colorectal cancer132. However, currently, there are no clinically approved YAP inhibitors that are both effective and specific132,133. This underscores the need for further research and development to identify and design new compounds that could selectively target YAP activity while maintaining therapeutic efficacy. Given the crucial role of the Hippo pathway and Pax in Dysplasia and cancer development134, along with the precise regulation facilitated by the Pax-Yki negative feedback loop, Paxillin has the potential to be a safer therapeutic target for restricting YAP activity. Our findings also provide clues to understanding the role of Pax in other types of cancers.
Methods
Plasmids, transgenic flies, Pax 20 mutant
The full-length pax DNA fragment (pax-RA: 1746 bp, PXN-iso1: 1776 bp) was amplified from Drosophila or Human cDNA by PCR. The DNA fragments mentioned above were cloned in the pUAST-Flag, pUAST-HA, and pcDNA3.1 vectors according to the standard protocols. Pax N and C are truncated pax variants with 1–341 bp and 342–581 bp, respectively. PaxdLD1, dLD2, dLD3, dLD4, and dLD5 are truncated Pax deleting 25-38aa/127-140aa/165-178aa/251-264aa/321-334aa, respectively. For overexpressed pax transgenic fly generation, a pUAST vector with attB sequence inserted upstream of the UAS-binding sites was used to construct pUAST-attB-Flag-Pax or pUAST-attB-HA-Pax plasmids. The shRNA of PXN was cloned in the pLKO.1-shPXN-EGFP vector. All plasmids were verified by DNA sequencing. Transgenic flies carrying these constructs were generated.
Pax20 mutant flies were generated using the CRISPR-Cas9 system described previously135. We conducted a sequence alignment of Pax and used the second and third exons as templates to design three sgRNAs (see supplementary information). Following microinjection and sequencing-based screening, Pax20 was identified as a genetic null allele of Pax, resulting from a 344-base deletion causing a frameshift mutation. The homozygous mutant flies are viable and fertile.
Drosophila stocks and genetics
The fly stocks used in this study are listed in Supplementary Table 6.
MARCM clone analysis
GFP-positive mutant clones were generated using the MARCM system136. Flies were crossed and raised at 25 °C. F1 adult flies with related genotypes undergoing heat shock at 37 °C for 1 h to induce clones at 2-day-old flies. Then, flies were raised at 25 °C for 2, 4, or 6 days before dissection. Clones of more than 10 midguts were used for analyze in each group.
Temperature-controlled expression
The flies using Esgts, MyoIAts, EsgREDDMts, NREts, and Dlts were crossed and cultured at 21 °C to suppress Gal4 activity. After one day, F1 adult flies with appropriate genotypes were then shifted to 29 °C for a 5, 7, or 14-day incubation to induce inactivation of Gal80ts and expression of the UAS transgenes or RNAi. 10 female adult flies at least were dissected for each genotype, and used for immunostaining, microscopy, and statistical analysis. For the intestinal stem cell lineage tracing experiment, we used the inducible lineage tracing Esgts F/O system. Flies were crossed and cultured at 21 °C. F1 adult flies with correct genotypes were subjected to heat shock at 37 °C for 40 min to induce clones at the age of 2 days. Then, flies were shifted to 29 °C for 3 or 5 days to induce the expression of transgenes.
DSS feeding experiments
Female adult flies, which has cultured at 29 °C for 2 days, were used to perform DSS-treated feeding experiments. Flies were cultured in an empty vial containing a piece of 9 cm2 chromatography paper wet with or without 3% dextran sulfate sodium in 5% glucose solution for 3 days at 29 °C.
Immunostaining
Female flies were used for gut immunostaining in all experiments. The entire intestine or wing discs were dissected and fixed in 4% formaldehyde for 1 h (disc: 30 min). Samples were washed three times in PBS plus 0.1% Triton X-100 (PBS-T). S2 cells were fixed in 1% formaldehyde and washed three times in PBS plus 0.1% Triton X-100 (PBS-T). The guts and S2 cells were incubated in the primary and secondary antibodies diluted in a solution containing PBS-T. Fluorescently labeled samples were counterstained with DAPI for visualization of DNA. Images were captured with a Leica TCS SP8 confocal microscope and assembled in Adobe Illustrator.
Cell culture, transfection, immunoprecipitation, and lentiviral infection
S2 Drosophila cells from American Type Culture Collection (ATCC) Ref: CRL-1963 were used for related experiments. S2 cells were cultured in Drosophila Schneider’s Medium with 10% fetal bovine serum and 100 U/ml of penicillin and Streptomycin. Plasmid transfection was carried out using Calcium Phosphate Transfection (2 M CaCl2, 2xHBS (HBS consisted of 50 mM Hepes and 1.5 mM Na2HPO4, 280 mM NaCl and 20 mM KCl, adjusted to pH 7.05), ddH2O) and X-tremeGENE HP DNA Transfection Reagent according to manufacturer’s instructions. A ubiquitin-Gal4 plasmid was co-transfected with pUAST expression vectors for all transfection experiments. Immunoprecipitation and Western blot analyses were performed according to standard protocols as previously described49.
Hela and HEK-293T cells were obtained from ATCC. Huh-7 cells were obtained from the Japanese Collection of Research Bioresources (JCRB). These cells were maintained in Dulbecco modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin and streptomycin antibiotics at 37 °C in 5% CO2 (v/v). All cell lines have been tested for mycoplasma contamination. Lentiviral infection was done as follows: HEK-293T cells were co-transfected with pLKO.1-shPXN-EGFP vector and packaging plasmids. The progeny viruses released from HEK-293T cells were collected and used to infect HeLa cells. The pLKO.1 and pcDNA3.1 plasmids were transfected with the Lipofectamine 3000 reagent.
Real-time PCR
Total RNAs were extracted from ten midguts of 5-day-old female flies or 50 wing discs from third instar larvae with indicated genotypes using RNA isolator Total RNA Extraction Reagent, and the cDNAs were synthesized using HiScript III RT SuperMix for qPCR kit. Real-time PCR was performed using the AceQ Universal SYBR qPCR Master Mix reagent with the Lightcycler R System. Results were repeated for three independent biological replicates. Unless otherwise, RpL32 was used as a normalized control. Relative quantification of mRNA levels was calculated using the 2−ΔΔCT method.
Droplet digital PCR
Total RNA was isolated from MyoIA-GFP+ cells using RNA isolator Total RNA Extraction Reagent, and the cDNAs were synthesized using HiScript III RT SuperMix for qPCR kit according to the manufacturer’s instructions. The assay was performed using the Bio-Rad QX200 Droplet Digital System (Bio-Rad Laboratories). About 1 μL cDNA samples, 1 μL primer mixture, 10 μL 2× ddPCR EvaGreen supermix, and 8 μL RNase-free water were mixed with 70 μL of QX200 Droplet Generation Oil for EvaGreen into the oil wells. Droplets were then generated by a QX200 droplet generator device (Bio-Rad). The cycling conditions were as follows: 10 min at 95 °C, 40 cycles of denaturation at 95 °C for 30 s, annealing and extension at 60 °C for 1 min, and final step at 90 °C for 5 min. Droplets were read in the droplet reader and analyzed using QuantaSoft (Bio-Rad Laboratories).
Chromatin immunoprecipitation-PCR (ChIP-PCR)
ChIP assays were performed using S2 cells. Cells were crosslinked with 1% formaldehyde diluted in PBS buffer for 15 min at 37 °C and stopped by adding Glycine to a final concentration of 0.125 M. Next, fixed cells were washed for 10 min by using ice-cold PBS three times. Cells were sheared to 200–600 bp fragments in 1 ml sonication buffer (50 mM Hepes-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8.0, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and proteinase cocktail) with a Bioruptor sonicator. After centrifugation, lysates were incubated with 4 mg antibody of HA or normal mouse IgG (sc-2025, Santa Cruz) overnight; then protein A/G PLUS agarose beads were then added and incubated in antibody/antigen complexes for 2 h on a rotator at 4 °C. Beads were washed with the ChIP wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, pH 8.0, 150 mM NaCl, and 20 mM Tris-Cl, pH 8.0), and then washed with ChIP final wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, pH 8.0, 500 mM NaCl, and 20 mM Tris-Cl, pH 8.0). Next, genomic DNA was eluted with elution buffer (1% SDS and 100 mM NaHCO3) at 65 °C for 30 min. The Eluted NDA fragments were incubated with 200 mM NaCl at 65 °C for 4 h. Then, 0.5 M EDTA and 0.25 mg/ml proteinase K were added to incubation with eluted DNA. The final mixture was incubated at 55 °C for 2 h to digest the protein. Genomic DNA was purified with a QIAQIUCK PCR Purification Kit. Purified DNA was performed with ChIP-PCR.
Luciferase reporter assay
Luciferase assays were performed in S2 cells with the pGL3-3 x sd promoter luciferase reporter and pGL3-Pax promoter luciferase reporter. To generate the Pax-luc-B1, Pax-luc-B2, or Pax-luc-B3 reporter genes, 3 DNA segments covering Sd/Yki-binding region of Pax were subcloned between BglII and KpnI sites of the pGL3-Promoter vector (Pax-luc-B1: −7103 to −6392, Pax-luc-B2: −3415 to −2729, Pax-luc-B3: +11512 to +12101), respectively. S2 cells were transfected with indicated reporters (1 ug) and copia-renilla luciferase reporter (0.005 ug) to normalize for transfection efficiency. For luciferase assays in overexpression plasmid-transfected cells, cells were transfected with the indicated plasmids and reporter plasmid together for 48 hr. The reporter assay was performed using the Dual-Luciferase reporter assay system. Dual-Luciferase measurements were performed in triplicate using a GloMax-Multi JR Single-Tube Multimode Reader.
Single-cell suspension preparation and FACS
FACS purification of specific cell types from the Drosophila gut were performed according to the method described previously137. The lines of Esgts > GFP or MyolAts > GFP were used to harvest Esg-GFP or MyolA-GFP cells in this study. Briefly, about 150 midguts of female flies aged 5–7 days old with foregut and hindgut portion removed were dissected in cold PBS within 1 h and digested in 1 mg/ml Elastase (Sigma, #E0258) solution at 25 °C for 1 h. Dissociated cells were pelleted at 400× g for 25 min, re-suspended in PBS with 0.2% BSA, filtered with 70 mm filters (BD Falcon), and sorted using a FACS Aria III sorter (BD Biosciences). GFP was used to sort the Esg-GFP and MyolA-GFP cell population, while cells from w1118 midgut was used to set the negative fluorescence gate of the GFP panel. GFP+ cells were sorted into PBS solution with 0.2% BSA, and after pelleting (400× g for 25 min) to collect total RNA using RNA isolator total RNA extraction reagent.
Quantification and statistical analysis
Unless otherwise indicated, the images shown in this study and the respective quantifications focused on the posterior midgut boundary between R4c and R5a, where the stem cells have been previously shown to be more active than those in other regions138,139,140. Midgut images were taken at ×40 magnification within the boundary between the R4c and R5a region with the same microscope settings. A region of interest (ROI) of about 20,000 μm² were applied in which all cells were DAPI labeled. Images were analyzed by using ImageJ to measure the fluorescence positive cells of interest. Statistical analyses were carried out with Prism 7 (GraphPad), and data from at least three independent experiments were combined. The unpaired two-tailed Student’s t-test was used for the comparison of two independent samples, and one-way ANOVA with Tukey’s test was used to determine statistical significance with multiple comparisons between three or more independent groups. p Value is indicated by asterisks in the Figures: ns p ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data were statistically present as average with the standard error of the mean (mean ± SEM).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper.
References
Morrison, S. J. & Spradling, A. C. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611 (2008).
Guo, Z., Lucchetta, E., Rafel, N. & Ohlstein, B. Maintenance of the adult Drosophila intestine: all roads lead to homeostasis. Curr. Opin. Genet Dev. 40, 81–86 (2016).
Antonello, Z. A., Reiff, T. & Dominguez, M. Mesenchymal to epithelial transition during tissue homeostasis and regeneration: patching up the Drosophila midgut epithelium. Fly 9, 132–137 (2015).
Li, H. & Jasper, H. Gastrointestinal stem cells in health and disease: from flies to humans. Dis. Model Mech. 9, 487–499 (2016).
Perochon, J., Carroll, L. R. & Cordero, J. B. Wnt signalling in intestinal stem cells: lessons from mice and flies. Genes https://doi.org/10.3390/genes9030138 (2018).
Morrison, S. J. & Kimble, J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068–1074 (2006).
Beehler-Evans, R. & Micchelli, C. A. Generation of enteroendocrine cell diversity in midgut stem cell lineages. Development 142, 654–664 (2015).
Chen, J. et al. Transient Scute activation via a self-stimulatory loop directs enteroendocrine cell pair specification from self-renewing intestinal stem cells. Nat. Cell Biol. 20, 152–161 (2018).
Pasco, M. Y., Loudhaief, R. & Gallet, A. The cellular homeostasis of the gut: what the Drosophila model points out. Histol. Histopathol. 30, 277–292 (2015).
Jin, Z. et al. The Drosophila ortholog of mammalian transcription factor Sox9 regulates intestinal homeostasis and regeneration at an appropriate level. Cell Rep. 31, 107683 (2020).
Joly, A. & Rousset, R. Tissue adaptation to environmental cues by symmetric and asymmetric division modes of intestinal stem cells. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21176362 (2020).
Guo, X., Lv, J. & Xi, R. The specification and function of enteroendocrine cells in Drosophila and mammals: a comparative review. FEBS J. 289, 4773–4796 (2022).
Clevers, H. A gutsy approach to stem cells and signalling: an interview with Hans Clevers. Dis. Model Mech. 6, 1053–1056 (2013).
Jiang, H. & Edgar, B. A. Intestinal stem cell function in Drosophila and mice. Curr. Opin. Genet Dev. 22, 354–360 (2012).
Zhai, Z., Boquete, J. P. & Lemaitre, B. A genetic framework controlling the differentiation of intestinal stem cells during regeneration in Drosophila. PLoS Genet. 13, e1006854 (2017).
Tian, A. & Jiang, J. Intestinal epithelium-derived BMP controls stem cell self-renewal in Drosophila adult midgut. Elife 3, e01857 (2014).
Venkei, Z. G. & Yamashita, Y. M. Emerging mechanisms of asymmetric stem cell division. J. Cell Biol. 217, 3785–3795 (2018).
Goulas, S., Conder, R. & Knoblich, J. A. The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell 11, 529–540 (2012).
Guo, Z. & Ohlstein, B. Stem cell regulation. Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency. Science https://doi.org/10.1126/science.aab0988 (2015).
Lu, M. S. & Johnston, C. A. Molecular pathways regulating mitotic spindle orientation in animal cells. Development 140, 1843–1856 (2013).
Guo, Z., Driver, I. & Ohlstein, B. Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J. Cell Biol. 201, 945–961 (2013).
Jiang, H., Grenley, M. O., Bravo, M. J., Blumhagen, R. Z. & Edgar, B. A. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8, 84–95 (2011).
Al Hayek, S. et al. Steroid-dependent switch of OvoL/Shavenbaby controls self-renewal versus differentiation of intestinal stem cells. EMBO J. 40, e104347 (2021).
Lin, G., Xu, N. & Xi, R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature 455, 1119–1123 (2008).
Beebe, K., Lee, W. C. & Micchelli, C. A. JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev. Biol. 338, 28–37 (2010).
Jiang, H., Tian, A. & Jiang, J. Intestinal stem cell response to injury: lessons from Drosophila. Cell Mol. Life Sci. 73, 3337–3349 (2016).
Kux, K. & Pitsouli, C. Tissue communication in regenerative inflammatory signaling: lessons from the fly gut. Front. Cell Infect. Microbiol. 4, 49 (2014).
Tian, A. et al. Damage-induced regeneration of the intestinal stem cell pool through enteroblast mitosis in the Drosophila midgut. EMBO J. 41, e110834 (2022).
Karpowicz, P., Perez, J. & Perrimon, N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development 137, 4135–4145 (2010).
Ren, F. et al. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc. Natl Acad. Sci. USA 107, 21064–21069 (2010).
Shaw, R. L. et al. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137, 4147–4158 (2010).
Staley, B. K. & Irvine, K. D. Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr. Biol. 20, 1580–1587 (2010).
Pan, D. The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 (2010).
Zhao, B., Li, L., Lei, Q. & Guan, K. L. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 (2010).
Halder, G. & Johnson, R. L. Hippo signaling: growth control and beyond. Development 138, 9–22 (2011).
Yin, M. & Zhang, L. Hippo signaling: a hub of growth control, tumor suppression and pluripotency maintenance. J. Genet Genom. 38, 471–481 (2011).
Irvine, K. D. Integration of intercellular signaling through the Hippo pathway. Semin. Cell Dev. Biol. 23, 812–817 (2012).
Calses, P. C., Crawford, J. J., Lill, J. R. & Dey, A. Hippo pathway in cancer: aberrant regulation and therapeutic opportunities. Trends Cancer 5, 297–307 (2019).
Dey, A., Varelas, X. & Guan, K. L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 19, 480–494 (2020).
Bergmann, C. et al. Polycystic kidney disease. Nat. Rev. Dis. Prim. 4, 50 (2018).
Austin, K. M. et al. Molecular mechanisms of arrhythmogenic cardiomyopathy. Nat. Rev. Cardiol. 16, 519–537 (2019).
Sun, M. et al. New insights into the Hippo/YAP pathway in idiopathic pulmonary fibrosis. Pharm. Res. 169, 105635 (2021).
Russell, J. O. & Camargo, F. D. Hippo signalling in the liver: role in development, regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 19, 297–312 (2022).
Tapon, N. et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478 (2002).
Harvey, K. F., Pfleger, C. M. & Hariharan, I. K. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 (2003).
Wu, S., Huang, J., Dong, J. & Pan, D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 (2003).
Goulev, Y. et al. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18, 435–441 (2008).
Wu, S., Liu, Y., Zheng, Y., Dong, J. & Pan, D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388–398 (2008).
Zhang, L. et al. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377–387 (2008).
Huang, J., Wu, S., Barrera, J., Matthews, K. & Pan, D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 (2005).
Nolo, R., Morrison, C. M., Tao, C., Zhang, X. & Halder, G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16, 1895–1904 (2006).
Dong, J. et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007).
Thompson, B. J. & Cohen, S. M. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126, 767–774 (2006).
Houtz, P. et al. Hippo, TGF-beta, and Src-MAPK pathways regulate transcription of the upd3 cytokine in Drosophila enterocytes upon bacterial infection. PLoS Genet. 13, e1007091 (2017).
Jiang, H. et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009).
Biteau, B., Hochmuth, C. E. & Jasper, H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell 9, 402–411 (2011).
Loudhaief, R. et al. Apoptosis restores cellular density by eliminating a physiologically or genetically induced excess of enterocytes in the Drosophila midgut. Development 144, 808–819 (2017).
Chen, G. C. et al. Regulation of Rho and Rac signaling to the actin cytoskeleton by paxillin during Drosophila development. Mol. Cell Biol. 25, 979–987 (2005).
Conway, W. C. et al. Paxillin modulates squamous cancer cell adhesion and is important in pressure-augmented adhesion. J. Cell Biochem. 98, 1507–1516 (2006).
Lopez-Colome, A. M., Lee-Rivera, I., Benavides-Hidalgo, R. & Lopez, E. Paxillin: a crossroad in pathological cell migration. J. Hematol. Oncol. 10, 50 (2017).
Ibata, N. & Terentjev, E. M. Development of nascent focal adhesions in spreading cells. Biophys. J. 119, 2063–2073 (2020).
Legerstee, K. & Houtsmuller, A. B. A layered view on focal adhesions. Biology https://doi.org/10.3390/biology10111189 (2021).
Turner, C. E., Glenney, J. R. Jr. & Burridge, K. Paxillin: a new vinculin-binding protein present in focal adhesions. J. Cell Biol. 111, 1059–1068 (1990).
Liu, S. & Ginsberg, M. H. Paxillin binding to a conserved sequence motif in the alpha 4 integrin cytoplasmic domain. J. Biol. Chem. 275, 22736–22742 (2000).
Brown, M. C. & Turner, C. E. Paxillin: adapting to change. Physiol. Rev. 84, 1315–1339 (2004).
Deakin, N. O. & Turner, C. E. Paxillin comes of age. J. Cell Sci. 121, 2435–2444 (2008).
Deakin, N. O., Pignatelli, J. & Turner, C. E. Diverse roles for the paxillin family of proteins in cancer. Genes Cancer 3, 362–370 (2012).
Tumbarello, D. A., Brown, M. C. & Turner, C. E. The paxillin LD motifs. FEBS Lett. 513, 114–118 (2002).
Deakin, N. O. & Turner, C. E. Distinct roles for paxillin and Hic-5 in regulating breast cancer cell morphology, invasion, and metastasis. Mol. Biol. Cell 22, 327–341 (2011).
Mackinnon, A. C. et al. Paxillin expression and amplification in early lung lesions of high-risk patients, lung adenocarcinoma and metastatic disease. J. Clin. Pathol. 64, 16–24 (2011).
Chen, D. L. et al. Abnormal expression of paxillin correlates with tumor progression and poor survival in patients with gastric cancer. J. Transl. Med. 11, 277 (2013).
Turner, C. E. & Miller, J. T. Primary sequence of paxillin contains putative SH2 and SH3 domain binding motifs and multiple LIM domains: identification of a vinculin and pp125Fak-binding region. J. Cell Sci. 107, 1583–1591 (1994).
Nikolopoulos, S. N. & Turner, C. E. Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions. J. Biol. Chem. 276, 23499–23505 (2001).
Schaller, M. D. Paxillin: a focal adhesion-associated adaptor protein. Oncogene 20, 6459–6472 (2001).
Huang, H. et al. Bantam is essential for Drosophila intestinal stem cell proliferation in response to Hippo signaling. Dev. Biol. 385, 211–219 (2014).
Cho, E. et al. Delineation of a Fat tumor suppressor pathway. Nat. Genet. 38, 1142–1150 (2006).
Hamaratoglu, F. et al. The tumour-suppressor genes NF2/Merlin and expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27–36 (2006).
Genevet, A., Wehr, M. C., Brain, R., Thompson, B. J. & Tapon, N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 18, 300–308 (2010).
Moroishi, T. et al. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 29, 1271–1284 (2015).
Ikmi, A. et al. Molecular evolution of the Yap/Yorkie proto-oncogene and elucidation of its core transcriptional program. Mol. Biol. Evol. 31, 1375–1390 (2014).
Nagaraj, R. et al. Control of mitochondrial structure and function by the Yorkie/YAP oncogenic pathway. Genes Dev. 26, 2027–2037 (2012).
Beyer, T. A. et al. Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 5, 1611–1624 (2013).
Oh, H. et al. Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep. 3, 309–318 (2013).
Oh, H. et al. Yorkie promotes transcription by recruiting a histone methyltransferase complex. Cell Rep. 8, 449–459 (2014).
Tremblay, A. M. et al. The Hippo transducer YAP1 transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation. Cancer Cell 26, 273–287 (2014).
Obier, N. et al. Cooperative binding of AP-1 and TEAD4 modulates the balance between vascular smooth muscle and hemogenic cell fate. Development 143, 4324–4340 (2016).
Rajbhandari, P. et al. Cross-cohort analysis identifies a TEAD4-MYCN positive feedback loop as the core regulatory element of high-risk neuroblastoma. Cancer Discov. 8, 582–599 (2018).
Wang, L. H. & Baker, N. E. Salvador-Warts-Hippo pathway regulates sensory organ development via caspase-dependent nonapoptotic signaling. Cell Death Dis. 10, 669 (2019).
Houtz, P., Bonfini, A., Bing, X. & Buchon, N. Recruitment of adult precursor cells underlies limited repair of the infected larval midgut in Drosophila. Cell Host Microbe 26, 412–425.e415 (2019).
Nawrot-Esposito, M. P. et al. Bacillus thuringiensis bioinsecticides induce developmental defects in non-target Drosophila melanogaster larvae. Insects https://doi.org/10.3390/insects11100697 (2020).
Brand, A. H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
McGuire, S. E., Le, P. T., Osborn, A. J., Matsumoto, K. & Davis, R. L. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765–1768 (2003).
Micchelli, C. A. & Perrimon, N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479 (2006).
Ohlstein, B. & Spradling, A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 (2006).
Nagai, H. et al. Nutrient-driven dedifferentiation of enteroendocrine cells promotes adaptive intestinal growth in Drosophila. Dev. Cell 58, 1764–1781.e1710 (2023).
Gervais, L. & Bardin, A. J. Tissue homeostasis and aging: new insight from the fly intestine. Curr. Opin. Cell Biol. 48, 97–105 (2017).
Edelblum, K. L., Yan, F., Yamaoka, T. & Polk, D. B. Regulation of apoptosis during homeostasis and disease in the intestinal epithelium. Inflamm. Bowel Dis. 12, 413–424 (2006).
Delgado, M. E., Grabinger, T. & Brunner, T. Cell death at the intestinal epithelial front line. FEBS J. 283, 2701–2719 (2016).
Antonello, Z. A., Reiff, T., Ballesta-Illan, E. & Dominguez, M. Robust intestinal homeostasis relies on cellular plasticity in enteroblasts mediated by miR-8-Escargot switch. EMBO J. 34, 2025–2041 (2015).
Zeng, X., Chauhan, C. & Hou, S. X. Characterization of midgut stem cell- and enteroblast-specific Gal4 lines in drosophila. Genesis 48, 607–611 (2010).
Furriols, M. & Bray, S. A model Notch response element detects suppressor of hairless-dependent molecular switch. Curr. Biol. 11, 60–64 (2001).
Guo, X., Huang, H., Yang, Z., Cai, T. & Xi, R. Division of labor: roles of Groucho and CtBP in Notch-mediated lateral inhibition that controls intestinal stem cell differentiation in Drosophila. Stem Cell Rep. 12, 1007–1023 (2019).
Bardin, A. J., Perdigoto, C. N., Southall, T. D., Brand, A. H. & Schweisguth, F. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development 137, 705–714 (2010).
Buchon, N., Broderick, N. A., Poidevin, M., Pradervand, S. & Lemaitre, B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200–211 (2009).
Jiang, L., Pearson, J. C. & Crews, S. T. Diverse modes of Drosophila tracheal fusion cell transcriptional regulation. Mech. Dev. 127, 265–280 (2010).
Gabay, L., Seger, R. & Shilo, B. Z. In situ activation pattern of Drosophila EGF receptor pathway during development. Science 277, 1103–1106 (1997).
Shilo, B. Z. Regulating the dynamics of EGF receptor signaling in space and time. Development 132, 4017–4027 (2005).
Bach, E. A. et al. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr. Patterns 7, 323–331 (2007).
Biteau, B., Hochmuth, C. E. & Jasper, H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell 3, 442–455 (2008).
Cordero, J. B., Stefanatos, R. K., Scopelliti, A., Vidal, M. & Sansom, O. J. Inducible progenitor-derived Wingless regulates adult midgut regeneration in Drosophila. EMBO J. 31, 3901–3917 (2012).
Li, Z., Zhang, Y., Han, L., Shi, L. & Lin, X. Trachea-derived dpp controls adult midgut homeostasis in Drosophila. Dev. Cell 24, 133–143 (2013).
Tian, A. et al. Injury-stimulated Hedgehog signaling promotes regenerative proliferation of Drosophila intestinal stem cells. J. Cell Biol. 208, 807–819 (2015).
Reiff, T. et al. Notch and EGFR regulate apoptosis in progenitor cells to ensure gut homeostasis in Drosophila. EMBO J. 38, e101346 (2019).
Zhao, B. et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007).
Hao, Y., Chun, A., Cheung, K., Rashidi, B. & Yang, X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509 (2008).
Wang, S. et al. Importin alpha1 mediates Yorkie nuclear import via an N-terminal non-canonical nuclear localization signal. J. Biol. Chem. 291, 7926–7937 (2016).
Pulusu, S. S. R. & Lawrance, I. C. Dysplasia and colorectal cancer surveillance in inflammatory bowel disease. Expert Rev. Gastroenterol. Hepatol. 11, 711–722 (2017).
Hansen, C. G., Ng, Y. L., Lam, W. L., Plouffe, S. W. & Guan, K. L. The Hippo pathway effectors YAP and TAZ promote cell growth by modulating amino acid signaling to mTORC1. Cell Res. 25, 1299–1313 (2015).
Panciera, T., Azzolin, L., Cordenonsi, M. & Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 18, 758–770 (2017).
Karaman, R. & Halder, G. Cell junctions in Hippo signaling. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a028753 (2018).
Kim, N. G. & Gumbiner, B. M. Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J. Cell Biol. 210, 503–515 (2015).
Elbediwy, A., Vincent-Mistiaen, Z. I. & Thompson, B. J. YAP and TAZ in epithelial stem cells: a sensor for cell polarity, mechanical forces and tissue damage. Bioessays 38, 644–653 (2016).
Mason, D. E. et al. YAP and TAZ limit cytoskeletal and focal adhesion maturation to enable persistent cell motility. J. Cell Biol. 218, 1369–1389 (2019).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Rausch, V. & Hansen, C. G. The Hippo pathway, YAP/TAZ, and the plasma membrane. Trends Cell Biol. 30, 32–48 (2020).
Nardone, G. et al. YAP regulates cell mechanics by controlling focal adhesion assembly. Nat. Commun. 8, 15321 (2017).
Woods, A. J., Kantidakis, T., Sabe, H., Critchley, D. R. & Norman, J. C. Interaction of paxillin with poly(A)-binding protein 1 and its role in focal adhesion turnover and cell migration. Mol. Cell Biol. 25, 3763–3773 (2005).
Gabory, A., Ripoche, M. A., Yoshimizu, T. & Dandolo, L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet. Genome Res. 113, 188–193 (2006).
Noh, K. et al. The hidden role of paxillin: localization to nucleus promotes tumor angiogenesis. Oncogene 40, 384–395 (2021).
Dong, J. M., Lau, L. S., Ng, Y. W., Lim, L. & Manser, E. Paxillin nuclear-cytoplasmic localization is regulated by phosphorylation of the LD4 motif: evidence that nuclear paxillin promotes cell proliferation. Biochem. J. 418, 173–184 (2009).
Turner, C. E. Paxillin and focal adhesion signalling. Nat. Cell Biol. 2, E231–E236 (2000).
Moroishi, T., Hansen, C. G. & Guan, K. L. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73–79 (2015).
Zeng, R. & Dong, J. The Hippo signaling pathway in drug resistance in cancer. Cancers https://doi.org/10.3390/cancers13020318 (2021).
Liu, W., Huang, X., Luo, W., Liu, X. & Chen, W. The role of paxillin aberrant expression in cancer and its potential as a target for cancer therapy. Int. J. Mol. Sci. https://doi.org/10.3390/ijms24098245 (2023).
Kondo, S. & Ueda, R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721 (2013).
Lee, T. & Luo, L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251–254 (2001).
Dutta, D. et al. Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep. 12, 346–358 (2015).
Buchon, N. et al. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3, 1725–1738 (2013).
Marianes, A. & Spradling, A. C. Physiological and stem cell compartmentalization within the Drosophila midgut. Elife 2, e00886 (2013).
O’Brien, L. E. Regional specificity in the Drosophila midgut: setting boundaries with stem cells. Cell Stem Cell 13, 375–376 (2013).
Acknowledgements
We would like to thank Dr. Jin Jiang, Dr. Erjun Ling, Dr. Rongwen Xi, Dr. Xiaohang Yang, Dr. Bruce A Hay, Dr. Haixia Jiang, Dr. Cui Tai, Vienna Stock Center, Bloomington Stock Center, NIG, Developmental Studies Hybridoma Bank for providing fly lines, antibodies and assay supports. This work is supported by grants from the National Key Research and Development Program of China (2019YFA0802001 to L.Z.), National Natural Science Foundation of China (No. 32030025 and 32293233, 32221002 to L.Z., 32370751 to Y.J) and Shanghai Municipal Science and Technology Major Project; Shanghai Leading Talents Program to L.Z.; National Key R&D Program of China (2021YFA0805805 to W.W).
Author information
Authors and Affiliations
Contributions
D.J.: Conceptualization; Data curation; Formal analysis; Validation; Investigation; Project administration; Methodology; Writing—original draft; Writing—review and editing. P.L.: Methodology; Validation; Resources. Y.L.: Methodology; Resources; Writing—review and editing. J.T.: Resources; Investigation. X.H.: Formal analysis; Resources. X.W.: Formal analysis; Methodology. W.W. and J.X.: Resources; Methodology. H.Z., X.L., and Y.C.: Resources. Y.J.: Supervision; Writing—review and editing; Funding acquisition. L.Z.: Conceptualization; Supervision; Writing—review and editing; Funding acquisition. All authors have read and approved the final submitted manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Armel Gallet and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, D., Li, P., Lu, Y. et al. A feedback loop between Paxillin and Yorkie sustains Drosophila intestinal homeostasis and regeneration. Nat Commun 16, 570 (2025). https://doi.org/10.1038/s41467-024-55255-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55255-1