Abstract
Nuclear speckles are membraneless organelles that associate with active transcription sites and participate in post-transcriptional mRNA processing. During the cell cycle, nuclear speckles dissolve following phosphorylation of their protein components. Here, we identify the PP1 family as the phosphatases that counteract kinase-mediated dissolution. PP1 overexpression increases speckle cohesion and leads to retention of mRNA within speckles and the nucleus. Using APEX2 proximity labeling combined with RNA-sequencing, we characterize the recruitment of specific RNAs. We find that many transcripts are preferentially enriched within nuclear speckles compared to the nucleoplasm, particularly chromatin- and nucleus-associated transcripts. While total polyadenylated RNA retention increases with nuclear speckle cohesion, the ratios of most mRNA species to each other are constant, indicating non-selective retention. We further find that cellular responses to heat shock, oxidative stress, and hypoxia include changes to the phosphorylation and cohesion of nuclear speckles and to mRNA retention. Our results demonstrate that tuning the material properties of nuclear speckles provides a mechanism for the acute control of mRNA localization.
Similar content being viewed by others
Introduction
Specific proteins and long noncoding RNAs form biomolecular condensates called nuclear speckles1. In particular, the proteins SON and SRRM2 act as scaffolds for their formation, while the long non-coding RNA MALAT1 is highly enriched2,3,4. Splicing factors are proposed to cycle between speckles and active splicing sites in response to changes in their phosphorylation5,6. Messenger RNAs (mRNAs) also localize to speckles and have been shown to rapidly exchange between nuclear speckles and the nucleoplasm7. Recently, RNA sequencing after proximity-based labeling showed differential speckle association among transcripts8. Although nuclear speckles lack the well-defined layered structure of nuclear condensates like nucleoli, the two scaffold proteins SON and SRRM2 tend to be more enriched within the centre of speckles, while MALAT1, mRNAs, and splicing factors tend to be located towards the periphery9. Within the nucleus, nuclear speckles associate with sites of active transcription and boost transcription of proximal genes10,11,12.
So far, few studies have explored how the morphology and material properties of nuclear speckles relate to their proposed functions. Nuclei contain around 20-40 nuclear speckles, with estimated areas of around 2 µm2 13,14. Inhibition of transcription and splicing have both been reported to increase the size of nuclear speckles15,16,17,18. Transcriptional inhibition also increases their mobility within the nucleus and the exchange of splicing factors between speckles and the nucleoplasm15,18. Recently, expression of a synthetic arginine-rich mixed-charge domain in cells was reported to increase the cohesion of nuclear speckles, namely the extent to which speckle components interact with each other and unmix from the surrounding nucleoplasm, based on a decrease in the exchange of proteins between speckles and the nucleoplasm19. At the same time, the mixed charge domain increased the enrichment of polyA FISH signal in the nucleus. The material properties of nuclear speckles could therefore be important in the control of mRNA localization, but it is unclear whether this mechanism of regulation is actively controlled under physiological conditions.
Nuclear speckles undergo a cycle of dissolution during early mitosis through the activity of kinases CLK1 and DYRK3. They then re-form between late metaphase and telophase13,20,21. Most research has focused on the activity of kinases in the control of condensates, and we have little insight into the effects of dephosphorylation by phosphatases. In general, two families of serine-threonine phosphatases, PP1 and PP2A, control an estimated 90% of dephosphorylation events within mammalian cells22. The two families share some overlapping substrates; however, the catalytic phosphatases of each family also show sequence-based preferences23 and gain additional specificity through association with regulatory subunits24. The PP1 regulatory subunits PPP1R8 and PPP1R10 have been found within nuclear speckles25,26. In addition, both catalytic PP1 and PP2A are associated with functional splicing27,28, suggesting potential links to the regulation of nuclear speckles. In C. elegans, a PP2A phosphatase homolog was found to counteract the dissolution of P granules mediated by a DYRK-family kinase, MBK229. However, which phosphatases counteract kinase activity to regulate nuclear speckles in mammalian cells is unknown.
Various perturbations can modulate the phosphorylation of nuclear speckle-associated proteins. For example, hypoxia results in increased CLK1 expression and phosphorylation of splicing factors30. On the other hand, phosphorylation of splicing factors decreases in response to heat shock, leading to differential splicing, export, and increased translation of the CLK1 transcript, important for stress recovery31,32.
Here, we hypothesized that differential phosphorylation of nuclear speckle-associated proteins affects the material properties of the structures, not only in mitotic cells during dissolution, but also in interphase cells to affect mRNA localization. Indeed, we found that overexpression of PP1 phosphatases or drug inhibition of DYRK3 or CLK1 increases nuclear speckle cohesion and mRNA enrichment in speckles. Sequencing of speckle-enriched RNAs suggested global, non-selective retention of mRNAs following dephosphorylation of nuclear speckles, with few genes disproportionately retained. Both heat shock and oxidative stress increased nuclear speckle cohesion and nuclear mRNA retention, consistent with lower phosphorylation, while hypoxia produced the opposite effects. Our results point to a mechanism for rapid and dynamic control of mRNA localization through modulation of the material properties of an RNA-protein condensate.
Results
PP1 phosphatases counteract the effects of DYRK3/CLK1 and increase the cohesion of nuclear speckles
Nuclear speckles comprise protein and RNA components, including the scaffold proteins SON and SRRM2 and polyadenylated (polyA) RNA (Fig. 1a). We first investigated which phosphatases can maintain nuclear speckle structure during kinase overexpression. For this, we overexpressed the kinase DYRK3, previously shown to dissolve nuclear speckles at high concentration during interphase21, and co-overexpressed GFP-tagged phosphatases in HeLa cells. Among 72 phosphatases tested from a previously published library33, we found that upon overexpression of the PP1 catalytic subunit PPP1CC, a higher concentration of DYRK3 was necessary to induce speckle dissolution, an effect not seen with overexpression of a PP2A phosphatase (PPP2CA) (Fig. 1b, Supplementary Fig. 1a, Supplementary Data 1). Inhibiting PP1 and PP2A phosphatases with the small molecule calyculin A also led to nuclear speckle dissolution, consistent with a role for PPP1CC in maintaining nuclear speckles (Supplementary Fig. 1b). Based on these results, we hypothesized that PPP1CC and DYRK3 share similar substrates within nuclear speckles.
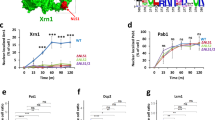
a PolyA FISH and immunofluorescence imaging shows colocalization of mRNA and nuclear speckle proteins, representative of 68 cells imaged. b Above: overexpression of mCh-DYRK3 dissolves nuclear speckles, an effect counteracted by co-overexpression of GFP-PPP1CC. Shaded regions in line plot correspond to standard deviations, while dots correspond to the mean per bin. Below: staining for SON in example cells at the same level of mCh-DYRK3 expression (indicated by dotted line, where p = 3E-5 with GFP-PPP1CC and p = 0.1 GFP-PPP2CA compared to controls, by two-sided Mann-Whitney U test). A total of n = 80 mCh-DYRK3, 129 mCh-DYKR3 + GFP-PPP1CC, and 133 mCh-DYRK3 + GFP-PPP2CA cells were analyzed. c Pulling down GFP-DYRK3 and GFP-PP1-NIPP1 enriches for overlapping nuclear speckle proteins. Highlighted points indicate proteins pulled down with L2FC > 2 and p < 0.05 (two-sided t-test) for both genes compared to GFP (see Supplementary Data 2 for exact values). d Overexpression of GFP-PP1-NIPP1 or YFP-PPP1CC reduces phospho-SR within the nucleus, while overexpression of GFP-DYRK3 or GFP-CLK1 increases phospho-SR. Left: example images. Right: quantification across nuclei, with kinases in yellow, active phosphatases in blue, and controls (GFP and mutant catalytic phosphatase) in grey. Log2FC was calculated relative to the mean pSR intensity for untransfected cells in the same well (n = number of transfected cells). P values were calculated by two-sided t-test with comparison to GFP, and Bonferroni-corrected for multiple testing. Box plots within violins show median values and interquartile ranges (IQR), with whiskers indicating 1.5*IQR. Green points indicate mean shifts in total SR expression for three replicates with overexpression of each of the same constructs. e Differential phosphorylation across scaffold protein SRRM2 with PP1 phosphatase overexpression. Shading corresponds to p value (likelihood ratio test, Supplementary Data 2). Below: arginine (R) and serine (S) enrichment. f Fluorescence recovery after photobleaching of SRRM2-mCh shows increased recovery with GFP-DYRK3 overexpression and decreased recovery (a higher immobile fraction) with YFP-PPP1CC overexpression compared to controls. Dots show the mean per time point and error bars show standard deviations. g Examples of FRAP recovery representative of the measurements included in f. Scale bars correspond to 10 µm.
Previous research established that the regulatory subunit NIPP1, or PPP1R8, retargets PP1 catalytic subunits from the nucleolus and elsewhere in the cell to nuclear speckles26 (Supplementary Fig. 1c). We therefore used a GFP-tagged construct of the catalytic subunit PPP1CC fused to NIPP1 (PP1-NIPP1) to enrich for protein targets of PP1 within nuclear speckles. Following immunoprecipitation-mass spectrometry (IP-MS), we compared proteins enriched with IP of GFP-PP1-NIPP1 to those enriched with GFP-DYRK3. Many serine/arginine-rich (SR) splicing factors, as well as the nuclear speckle scaffold proteins SON and SRRM2, interacted with both phosphatase and kinase (Fig. 1c, Supplementary Data 2). We used an antibody (1H4) that detects phospho-epitopes in serine/arginine-rich proteins to confirm that overexpression of either DYRK3 or CLK1 increases phosphorylation of SR regions (pSR) in splicing factors, while PP1-NIPP1 or PPP1CC overexpression decreases pSR in immunofluorescence imaging, beyond changes in SR protein expression (Fig. 1d, Supplementary Fig. 1d). A catalytically impaired mutant version of PP1 joined to NIPP1 (GFP-PP1m-NIPP134) showed no effect on pSR.
To confirm whether PP1-NIPP1 overexpression also increases dephosphorylation of the scaffold proteins of nuclear speckles, we performed phosphoproteomics after IP of GFP-PP1-NIPP1 or GFP-PP1m-NIPP1. Many sites (148 with log2 fold change ≥ 2, p < 0.05) within SRRM2 showed differential phosphorylation depending on whether the fully active or mutant catalytic phosphatase was overexpressed (Fig. 1e, Supplementary Data 1). We detected only five sites in the second scaffold protein of nuclear speckles, SON, that showed significant differences in phosphorylation depending on fully active or mutant phosphatase overexpression35 (Supplementary Fig. 1e). Fifty PP1-regulated sites across proteins were also differentially phosphorylated by DYRK3 compared to DYRK3 inhibited by small compound inhibitor GSK626616, with 12 of those sites within SRRM2 (Supplementary Fig. 1e, f). However, we note that the dissolution of nuclear speckles in response to overexpressed DYRK3 compared to increased enrichment of DYRK3 in nuclear speckles upon inhibition21 could affect which interactions we detect. Additional PP1 regulatory subunits may also be involved in recruiting PP1 to nuclear speckles and modulating the activity of the catalytic subunit (e.g. Saitoh et al.25) although we did not further investigate differences in holoenzyme activity.
To then study the effects of phosphorylation on the material properties of nuclear speckles, we used fluorescence recovery after photobleaching (FRAP), a method to estimate the exchange rate of components between the bleached area and the surrounding phase and thus the extent to which structures behave as “liquid-like” compartments. Overexpression of GFP-DYRK3 led to higher mean recovery (by 11% compared to GFP alone) from photobleaching of fluorescent signal in mCherry (mCh)-tagged SRRM2 condensates, suggesting increased mobility of SRRM2 and decreased cohesion. Meanwhile, overexpression of YFP-PPP1CC or its fellow PP1 catalytic subunits, GFP-PPP1CA and GFP-PPP1CB, increased the cohesion of SRRM2-mCh and decreased mean recovery from photobleaching (YFP-PPP1CC by 34% compared to GFP) (Fig. 1f, g, Supplementary Fig. 1g). A GFP-PP2A family phosphatase did not show similar effects. Despite enrichment of PP1 phosphatases within the nucleolus in addition to nuclear speckles, mCh-PP1 overexpression did not affect the cohesion of GFP-nucleolin, showing that PP1-mediated regulation is not universal across condensates (Supplementary Fig. 1h). Proteomics data, immunofluorescence staining, phosphoproteomics, and FRAP experiments thus establish that PPP1CC dephosphorylates nuclear speckle-associated proteins to counteract the effects of DYRK3 and increase the cohesion of the nuclear speckles.
Interfering with the phosphorylation and cohesion of nuclear speckles correlates with mRNA retention
To assess how changes in the cohesion of nuclear speckles mediated through phosphorylation could affect mRNA regulation, we stained for bulk mRNA using polyA FISH. Overexpression of mCh-PPP1CC increased the polyA signal within nuclear speckles and the ratio of nuclear-to-cytoplasmic polyA RNA, while a PP2A-family phosphatase did not (Fig. 2a, b, Supplementary Fig. 2a, b). By contrast, the dissolution of nuclear speckles through DYRK3 overexpression correlated with a decrease in nuclear polyA RNA (Fig. 2c). Similar correlations were observed by dissolving speckles through overexpression of GFP-CLK1 (as in Sacco-Bubulya and Spector36) or the cargo-binding domain of TNPO3, which interacts with SR-rich regions37 (Supplementary Fig. 2c). Altogether, this shows that increasing the cohesion of nuclear speckles correlates with higher mRNA retention within the nucleus.
a Example images showing differential polyA FISH intensity with mCh-PPP1CC and mCh-DYRK3 overexpression. mCherry images were scaled separately for mCh-PPP1CC and mCh-DYRK3, with outlines shown for transfected cells. SON and polyA FISH intensities were normalized based on untransfected cells in the same well. b Quantification for the ratio of nuclear to cytoplasmic polyA intensities for data in a and Supplementary Fig. 2 A. The top 5% of cells based on mCherry intensity were considered for each condition. P values were calculated by two-sided t-tests, with Bonferroni correction for multiple testing. c Nuclear polyA intensity negatively correlates with the dissolution of nuclear speckles through GFP-DYRK3 overexpression (Pearson correlation coefficient = −0.25). The x-axis indicates a measure of increasing dissolution (1 minus the proportion of nuclear area occupied by nuclear speckles). d Example images and quantification showing polyA intensity in nuclear speckles in hiPSCs under control conditions (DMSO), inhibition of DYRK3 (GSK626616), inhibition of transcription (DRB), and inhibition of splicing (Pladienolide-B). e Recovery from photobleaching in endogenously tagged 2xGFP-SON hiPSCs increased with transcriptional or splicing inhibition but decreased with DYRK3 inhibition. Dots show the mean per time point and error bars show standard deviations. f Overexpression of a positively charged arginine-rich mixed charge domain (GFP-R-MCD + 02) leads to the accumulation of polyadenylated RNA in nuclear speckles. Solid and dashed lines represent mean values, shaded area shows 95% confidence interval. 34 - 2697 cells were used per bin (n total inducible GFP = 5115, n total GFP-R-MCD + 02 = 2920). g Nuclear PolyA FISH intensity and 5-EU nuclear intensity after 30 min of incubation with 5-EU and 2 h of chase are elevated depending on the expression level of a positively charged arginine-rich mixed charge domain (GFP-R-MCD + 0.2). Solid lines represent mean values, shaded area shows 95% confidence interval. 121 - 12664 single cells were used per bin (n total GFP = 17,888, n total GFP-R-MCD + 02 = 14,464). Scale bars correspond to 10 µm. Box plots within the violin plots shown in (b) and (d) indicate median value and IQR, with whiskers indicating 1.5*IQR.
We further measured an increase in polyA RNA signal within nuclear speckles by inhibiting DYRK3 using the small molecule inhibitor GSK626616 in human induced pluripotent stem cells (hiPSCs) expressing endogenously tagged GFP-SON. Under DYRK3 inhibition, speckles became larger and rounder (Fig. 2d). Previous reports described similar effects on the morphology of speckles under transcriptional or splicing inhibition15,16,17,18. We therefore compared DYRK3 inhibition to the inhibition of transcription (through CDK9 binding) using DRB and the inhibition of splicing (through SF3B1 binding) using Pladienolide-B to determine whether all exerted similar effects on nuclear speckle cohesion and RNA regulation. PolyA RNA retention in nuclear speckles increased with DYRK3 inhibition and splicing inhibition, but decreased under transcriptional inhibition, likely due to ongoing mRNA export. The nucleus as a whole similarly showed an increase in polyA retention upon DYRK3 or splicing inhibition, but we measured no change within the nucleoplasm (Supplementary Fig. 2d). Meanwhile, the cohesion of nuclear speckles increased under DYRK3 inhibition, as measured using FRAP, but decreased in response to transcriptional or splicing inhibition (Fig. 2e). Although GSK626616 and Pladienolide-B share effects on the morphology of nuclear speckles as well as on polyA RNA retention, the material properties of the speckles differ under the two inhibitors. Importantly, these differences suggest that increased nuclear mRNA retention (as with splicing inhibition) does not increase nuclear speckle cohesion.
We next generated an inducible hiPSC line for orthogonal modulation of nuclear speckle cohesion (Supplementary Fig. 2e). Expressing a positively charged arginine-rich mixed charge domain (R-MCD + 0.2) in a polyclonal population of hiPSCs increased nuclear and nuclear speckle mRNA intensity, as previously reported19 (Fig. 2f, g, Supplementary Fig. 2f). We also observed an increase of polyA FISH signal in the nucleoplasm, but no change in the cytoplasm (Supplementary Fig. 2f). To further verify that the observed increase in nuclear intensity represents transcript retention, we then used 5-EU to label nascent transcripts. A 30-minute pulse of 5-EU incorporation, followed by a 2-hour chase showed that newly generated mRNA remains within the nucleus for an extended time when cohesion increases through R-MCD + 0.2 expression (Fig. 2g). Notably, R-MCD + 0.2 overexpression led to a decrease in 5-EU incorporation (Supplementary Fig. 2g), as previously reported, consistent with feedback between nuclear retention of mRNA and transcription rate38. Thus, increasing the cohesion of nuclear speckles either through dephosphorylation or expression of a mixed charge domain correlates with higher bulk mRNA retention in the nucleus.
Characterization of the nuclear speckle transcriptome by APEX2-seq
To explore if bulk enrichment of polyA RNA upon increased cohesion reflected changes in nuclear speckle RNA composition, we characterized the nuclear speckle transcriptome. Until recently, the localization of specific mRNAs to nuclear speckles had been characterized for only a few genes39,40,41. Proximity-based labelling followed by RNA sequencing has now enabled transcriptomic profiling of distinct subcellular compartments42,43, including nuclear speckles8.
We used a proximity-based labelling approach to characterize the nuclear speckle transcriptome of hiPSCs. To avoid the effects of plasmid overexpression, we used CRISPR/Cas9 to tag the endogenous nuclear speckle scaffold protein SON with the engineered ascorbate peroxidase APEX2. As a control, a second cell line was generated by inserting APEX2 fused to a nuclear localization signal (NLS) into the genome at a safe-harbour locus to assess the overall nuclear transcriptome (Fig. 3a). After inducing the APEX2 enzymatic activity by addition of biotin-phenol and hydrogen peroxide, we detected biotinylated RNA using a dot-blot assay (Supplementary Fig. 3a). Staining for biotin using fluorescent streptavidin showed distinct localization of the streptavidin signal to nuclear speckles in the APEX2-SON cell line, and to the nucleus in the APEX2-NLS cell line (Fig. 3b). We note that we observe an enrichment of APEX2-NLS in the nucleolus compared to the nucleoplasm. The small size of APEX2-NLS with GFP ( ~ 59 kDa) might allow entry into the nucleolus, as has been shown for dextran particles of sizes smaller than 70 kDa44. Partitioning into the nucleolus through binding to RNA has also been shown for the same NLS (SV40) joined to GFP45. However, we do not observe a strong accumulation of biotinylated molecules in the region of the nucleolus after inducing enzymatic activity, suggesting that the controls do represent the general nuclear fraction of RNA (Fig. 3b).
a illustration of APEX2-tagged hiPSCs and biotinylation reaction. b Addition of hydrogen peroxide leads to biotinylation of biomolecules near nuclear speckles (APEX2-SON hiPSCs, left) or in the nucleoplasm (APEX2-NLS hiPSCs, right). Two technical replicates were collected. c APEX2-seq experimental outline. APEX2 hiPSCs were treated with drugs prior to RNA biotinylation. 5 biological replicates were collected per condition. d Pseudocolor images (see methods) of smFISH signal for two gene targets found to differentially localize to nuclear speckles by APEX2 sequencing in the control condition. P-values were calculated using a one-sided Mann–Whitney U test. Box plots show median values and IQR, with whiskers indicating 1.5*IQR. APEX2-seq and smFISH show overall positive correlation (Pearson correlation coefficient = 0.71, p value = 4.98e-06, two sided). Shaded area indicates 95% confidence interval. e Genes enriched in nuclear speckles display a larger proportion of lncRNA and snoRNA/scaRNA/snRNA than nuclear speckle depleted and neither genes. f Network representation of GSEA. Terms associated with ribosome, mitochondria and electron transport chain and the spliceosome are enriched in nuclear speckles. g Overview of feature categories used to build UMAP and train MLR model. Number of features in each category are indicated in brackets. h UMAP of transcript feature space. Nuclear speckle depleted/enriched transcripts occupy distinct regions. Gene sets found to be nuclear speckle enriched by GSEA show similar distributions. i Features from (g) were used to train a MLR model to predict nuclear speckle enrichment of protein-coding genes. Scatterplot shows random samples of 600 transcripts on test sets aggregated over 10-fold CV. j Combinatorial training of MLR model. The mean R2 score of ten models is shown (error bars indicate standard deviation, points indicate the score of individual models). k Importance analysis of features used to predict nuclear speckle enrichment. Color indicates Pearson correlation with nuclear speckle enrichment. l TPM and nuclear speckle enrichment of protein-coding transcripts show a negative correlation (Pearson correlation coefficient = -0.38, p value < 2.225e-308, two-sided). m Comparison of APEX2-seq nuclear speckle enrichment and enrichment in different subcellular compartments based on fractionation sequencing. Scale bar corresponds to 10 µm.
We collected RNA from both cell lines following 2-hour treatment with DYRK3 inhibitor (GSK626616) or DMSO (control) to evaluate changes in the nuclear speckle transcriptome in response to increased nuclear speckle cohesion. We also included treatments with DRB and Pladienolide-B to compare the effect of increased nuclear speckle cohesion to transcriptional inhibition and splicing inhibition (Fig. 3c).
We started by analysing baseline transcript enrichment in nuclear speckles under control conditions. Differential gene expression analysis revealed 2080 genes that showed significant transcript enrichment and 1349 genes that showed transcript depletion from nuclear speckles ( | log2 fold change | > 0.5 and padj <0.05) (Supplementary Fig. 3b, Supplementary Data 3). We then designed single-molecule FISH (smFISH) probes against a subset of hits for validation (Supplementary Fig. 3c). Enrichment measured by smFISH (quantified as the ratio of mean FISH signal in nuclear speckles over the remaining nucleoplasm) correlated well with enrichment detected by APEX2 sequencing (Pearson correlation coefficient = 0.71, p value = 4.98e-06) (Fig. 3d, Supplementary Fig. 3e). Based on sequencing, transcripts enriched in nuclear speckles included a higher proportion of long non-coding RNAs and a category of RNAs including small nuclear RNAs (snRNAs), small Cajal body-specific RNAs (scaRNAs) and small nucleolar RNAs (snoRNAs) (Fig. 3e). Association of snRNAs with nuclear speckles is well established46. Specific snRNAs are processed by scaRNAs in Cajal bodies47 or snoRNAs in the nucleolus48,49,50. There is increasing evidence that the nucleolus and Cajal bodies, besides their physical association, overlap in functions51 and that scaRNAs and snoRNAs function beyond their respective compartments52. Consistent with past reports suggesting that some stem cells lack Cajal bodies53, we found that over half of cells in a population of hiPSCs did not show coilin-enriched foci (Supplementary Fig. 3f). Therefore, the RNA species normally associated with Cajal bodies in other cell types may show alternative localization to nuclear speckles in hiPSCs. We further hypothesize that snoRNAs and scaRNAs may be transiently recruited to nuclear speckles through ongoing interactions with snRNAs. We also found enrichment of NEAT1, a long-noncoding RNA known to reside in paraspeckles, in the nuclear speckle fraction. Paraspeckles and nuclear speckles are closely associated nuclear bodies54, which could explain the co-enrichment of some paraspeckle components, although the functional overlap between NEAT1 and MALAT1 has also been proposed55. Our FISH results confirm close association and even overlap between NEAT1 lncRNA and the nuclear speckle marker SC35 (Supplementary Fig. 3g).
Gene set enrichment analysis showed that genes with transcripts enriched in nuclear speckles were associated with ribosomal proteins and oxidative phosphorylation, whereas genes with transcripts depleted from nuclear speckles were associated with a diverse set of terms, among them cytoskeletal and cytoplasmic localization terms (Fig. 3f).
We next compared our APEX2-seq dataset to the previously published proximity labeling dataset from Barutcu et al. 8. In this study, nuclear speckle-enriched genes were identified by expressing APEX2 fused to three splicing factors in HEK293T cells. We found overlaps of 354, 436, and 268 for genes enriched with SRSF1, SRSF7, and RNSP1, respectively (Supplementary Fig. 3d). Our pairwise comparisons of gene sets enriched or depleted in the SRSF1, SRSF7 and RNSP1 datasets showed an average Jaccard index of 0.226 between the different markers, whereas we found an average Jaccard index of 0.077 for the three markers compared to our dataset. This indicates that the overlap between RNAs in proximity with different markers for nuclear speckles is relatively low, and that there might be substantial differences in nuclear speckle RNA composition between different cell types.
As suggested by previous literature8,56,57, intron retention was higher in nuclear speckle-associated mRNA compared to the nucleus, with at least twice as many retained introns detected in speckles compared to the nucleus depending on the splicing analysis tool used (Supplementary Fig. 3h). Consistent with the results of Barutcu et al. 8, introns retained in speckles also showed higher GC content (mean ~49% vs. ~45% for randomly selected introns). However, differences in intron length between introns retained in speckles and the nucleus were inconsistent across splicing analysis tools (Supplementary Fig. 3i).
Next, we aimed to predict nuclear speckle enrichment of single transcripts. We performed differential transcript enrichment analysis and bioinformatically integrated transcript features from various sources. These features included general sequence features (e.g., GC content, number of exons), transcript expression level (transcripts per million, TPM), putative RNA binding protein (RBP) binding motifs, kinetic rates, and promoter motifs (Fig. 3g, Supplementary Data 4). Among these features, we find exon length positively correlated with nuclear speckle enrichment, while transcript expression level (TPM) anticorrelated with nuclear speckle enrichment (Supplementary Fig. 3j). Embedding these features into a uniform manifold approximation and projection (UMAP)58,59 reveals that transcripts enriched in or depleted from speckles occupy distinct regions within this feature space. Genes belonging to the enriched GO categories associated with the ribosome and mitochondria display similar distribution patterns, suggesting that transcript similarities may influence their association with nuclear speckles (Fig. 3h).
To assess the predictive power of these features, we trained a multilinear regression model to predict nuclear speckle enrichment on a single transcript level. Using this approach, we achieve an average coefficient of determination (R2) of 0.41 ( ± 0.008, 95% CI) (Fig. 3I, j). We trained the model on all possible combinations of feature categories. A model trained on minimal sets of general sequence features, transcript expression level and either RBP motif (R2 = 0.38 ± 0.0123) or rate features (R2 = 0.39 ± 0.0092) performed similarly to a model incorporating all the feature sets, suggesting that these are the most determining factors in speckle localization (Fig. 3j). A model based on sequence features alone (general, RBP, and promoter motifs) reached an average R2 of 0.26 ( ± 0.013), while a model based solely on transcript expression level reached an average R2 of 0.23 ( ± 0.0066).
We then measured the importance of individual features by training multiple iterations of the model and calculating the frequency with which a feature appeared in the top loadings of the top principal components. By a large margin, transcript expression level (TPM) appears as the most important feature in this analysis (Fig. 3k, Supplementary Fig. 3j). Interestingly, although expression level showed an inverse correlation with nuclear speckle enrichment (Fig. 3k, l), extending this analysis to all subtypes of RNA revealed a subset of highly expressed nuclear speckle-enriched transcripts composed mainly of snoRNAs, scaRNAs, and snRNAs (Supplementary Fig. 3k).
To assess whether the enrichment of transcripts in nuclear speckles relative to the nucleus correlates with the enrichment of transcripts in the nucleus relative to the cytoplasm, we biochemically fractionated hiPSCs into chromatin-associated nucleus, nucleus, and cytoplasm fractions and performed RNA sequencing. We found that nuclear speckle-associated transcripts tend to be enriched in the chromatin fraction compared to the nucleus and cytoplasm fractions, and to a lesser extent, in the nucleus compared to the cytoplasm (Pearson correlation coefficient of 0.41, 0.44 and 0.24, respectively) (Fig. 3m). We likewise found a positive correlation between nuclear speckle enrichment and nuclear enrichment (quantified as ratio of nuclear over cytoplasmic FISH signal) in our smFISH dataset (Pearson correlation coefficient = 0.65, p value = 2.08-05, Supplementary Fig. 3l).
Collectively, these results offer a comprehensive overview of transcript localization relative to nuclear speckles in hiPSCs. Statistical modeling showed that transcript expression level is a strong determinant of nuclear speckle enrichment. Contrary to sequence features, the transcript expression level is a variant feature, suggesting that nuclear speckle enrichment of transcripts could differ between cell types and tissues. The general enrichment of speckle-associated transcripts within the nuclear fraction is consistent with a role for speckles in storing nuclear-detained mRNAs. Interestingly, this may be more important for transcripts that are less highly expressed, suggesting tighter buffering of their translational availability.
An increase in nuclear speckle cohesion correlates with proportional enrichment of mRNA
We next investigated whether the increased recruitment of bulk polyadenylated RNA we observed with increased speckle cohesion (see Fig. 2a–d) reflects the differential retention of specific transcripts. We therefore compared APEX2 sequencing results of DYRK3-inhibited cells to our DMSO-treated controls. Surprisingly, few genes (n = 24) significantly changed in speckle enrichment under DYRK3 inhibition relative to other genes, whereas 759 and 1031 genes changed under transcriptional and splicing inhibition, respectively (log2 fold change | > 0.5, padj <0.1) (Fig. 4a). To reconcile this finding with the bulk enrichment of polyadenylated RNA, we hypothesized that enrichment of RNA is nonspecific, and that most genes are retained proportionally in nuclear speckles upon increased cohesion (Fig. 4b). Here, we did not include spike-in controls, as the biotinylation reaction and enrichment of biotinylated RNA could introduce technical variability between experiments prior to spike-in addition. We were therefore unable to quantify absolute changes in transcript abundances between samples from our APEX2 sequencing data. We note that generation and addition of a biotinylated spike-in before enrichment could help quantification in future experiments, although would still not control for variability from an initial biotinylation reaction.
a Volcano plots showing differentially expressed genes in nuclear speckles vs. nucleoplasm in different drug treatments. Annotations highlight genes for which smFISH probes were designed. P-values (Benjamini-Hochberg-corrected for multiple testing) were calculated using a two-sided Wald test. b Schematic illustration of proportional enrichment of RNA in nuclear speckles upon increase in speckle cohesion. c Pseudocolor images and quantification of FISH signal for two gene targets found to be nuclear speckle depleted (DUSP22 [p-value = 4.9e-18], MALAT1 [p value < 1e-300], EXOSC1 [p value = 1.2e-90]) or enriched (YJU2 [p value = 3.8e-175], AMOTL2 [p value < 1e-300], TRERF1 [p value = 4.5e-14]) in the indicated drug condition. Density plots show single cell log2FoldChange of nuclear speckle vs. nucleoplasm FISH signal. d Correlation of nuclear speckle enrichment in response to drug treatment in APEX2-seq vs. smFISH. Red dots highlight genes found to significantly change ( | log2FoldChange | > 0.5, padj <0.1) in APEX2-seq. Pearson correlation coefficient (calculated over all data points, two-sided) is displayed. Shaded area indicates 95% confidence interval. e Distributions of FISH drug vs. DMSO nuclear speckle log2FoldChange. Colors indicate genes identified to be nuclear speckle enriched, depleted or neither upon drug treatment based on APEX2-seq. P-values were calculated with a two-sided Welch’s t-test. Genes belonging to the “neither” class display a bias towards being more enriched in nuclear speckles upon GSK626616 treatment in smFISH. f Pseudocolor images and quantification of FISH signal of pooled probe sets representing gene targets found to be nuclear speckle depleted (p value = 1.7e-04), enriched (p value = 5.2e-47) or neither (p value = 1.6e-14) upon GSK626616 treatment. Density plots show single cell mean nuclear speckle FISH signal. g Example images and single-cell quantification showing changes in translational rates in response to two inhibitors of CLK1 compared to DMSO, based on incorporation of O-Propargyl Puromycin (OPP) into nascent peptides. Box plots in shown in c, e, f and g show median values and IQR, with whiskers indicating 1.5*IQR. P values in (c) and (F) were calculated using a one-sided Mann–Whitney U test. Scale bars correspond to 10 µm.
To test proportional enrichment, we used smFISH to quantify absolute RNA levels across compartments. As for baseline speckle enrichment, we found a positive correlation (Pearson correlation of 0.49-0.76) between APEX2 sequencing and smFISH (Fig. 4c, d, Supplementary Fig. 4a–c). Under DYRK3 inhibition, we observed a median log2FoldChange in nuclear speckle enrichment of 0.184 for gene targets that did not show disproportionate changes based on APEX2 sequencing. Inhibition of transcription or splicing did not result in the enrichment of these gene targets in nuclear speckles (Fig. 4e). This indicates a small general shift into nuclear speckles upon increased cohesion but not splicing inhibition. To further explore this, we designed three smFISH pools, each targeting around 50 random genes identified as enriched, depleted or left unchanged (neither) upon DYRK3 inhibition based on APEX2 sequencing results. Because few genes changed significantly when using more conservative thresholds, we defined a relaxed threshold of significance for enriched and depleted pools ( | log2 fold change | > 0.3, padj <0.6) and a very stringent threshold for the neither pool ( | log2 fold change | <0.05, padj > 0.99) (Supplementary Fig. 4d). As anticipated, the enriched and depleted smFISH pools showed an increase (log2 fold change=0.5) and decrease (log2 fold change = −0.1), respectively, in localization of signal to nuclear speckles upon DYRK3 inhibition. However, the smFISH pool representing gene targets that showed no disproportionate changes in enrichment in APEX2 sequencing showed increased smFISH signal in nuclear speckles (log2 fold change=0.27) (Fig. 4f). Notably, treatment with CLK-IN-T3, a CLK inhibitor, led to a similar increase in SON cohesion and enrichment of polyadenylated RNA in nuclear speckles as treatment with GSK626616, and we also observed non-specific enrichment when performing FISH with mixed smFISH pools (Supplementary Fig. 4e-g). This further supports the notion of a general shift of the nuclear RNA population into nuclear speckles upon increasing speckle cohesion. While accumulation of FISH signal in polyA FISH could also be attributed to changes in polyA tail length or polyA tail masking, the pooled FISH results confirm an increase of RNA abundance in nuclear speckles upon inhibition of DYRK3. We note that while GSK626616 was found to primarily affect DYRK family kinases, increasing effects on CLK and HIPK family kinases occur at higher concentrations60.
Previous work has also described an association between higher retention of transcripts within speckles and intron retention within transcripts61,62. Changes in the association between specific transcripts and nuclear speckles could therefore result from decreased splicing efficiency under drug perturbations. Based on splicing analysis of the RNA-seq data, we found that a substantial subset (175/711) of genes more enriched in speckles in response to splicing inhibition also showed measurable increases in intron retention (Supplementary Fig. 4h). With GSK626616, however, an order of magnitude more genes (425) showed changes in intron retention than disproportionate changes in nuclear speckle association (24, |log2 fold change | > 0.5, p < 0.1). The relatively few and bi-directional splicing changes that we find in response to DYRK3 inhibition are thus a poor explanation for the broad retention of transcripts in speckles. Thus, while our results under baseline conditions and splicing inhibition are consistent with a role for intron retention in the recruitment of specific transcripts to nuclear speckles, splicing analysis suggests the global shift in transcript retention under DYRK3 inhibition is not driven by perturbed splicing.
Increased nuclear retention of transcripts implies decreased translational availability. Single-molecule FISH data shows that under DYRK3 kinase inhibition, the cytoplasmic concentration of most transcripts decreases and the nuclear-to-cytoplasmic ratio increases (Supplementary Fig. 4i). We further verified the effects of nuclear phosphorylation perturbations on translation. Inhibition of the nuclear-localized kinase, CLK1, led to a decrease in overall translation rates compared to controls (Fig. 4g). Inhibition of DYRK3 did not decrease overall translation in this assay, although we note that cytoplasmic DYRK3 is known to regulate the mTOR pathway60 (Supplementary Fig. 4j).
Taken together, our data supports a model in which RNA localizes to nuclear speckles relative to nuclear speckle cohesion. Our APEX2 sequencing results indicate that this relationship is rarely gene-specific but instead affects a large fraction of the nuclear RNA population equally, leading to proportional changes in the enrichment of different RNAs as speckles become more gel-like. Measuring translation rates suggests that increased retention of mRNA in the nucleus correlates with decreased translation.
Heat shock, oxidative stress, and hypoxia modulate nuclear speckle phosphorylation and mRNA retention
To explore a physiological role for the material properties of nuclear speckles, we asked whether environmental perturbations could affect mRNA retention in speckles through changes in phosphorylation. Previous research established that CLK1 expression and SR phosphorylation change in response to heat shock and hypoxia30,32,63. We first exposed hiPSCs to these conditions to confirm measurable differences in SR phosphorylation in immunofluorescence. Heat shock (43 °C for 1 h) decreased SR phosphorylation, while hypoxia (0.2% oxygen for 24 h) had the opposite effect. We further considered oxidative stress (0.5 mM sodium arsenite for 1 h), which decreased pSR, similar to heat shock (Fig. 5a, Supplementary Fig. 5a). Using FRAP, we found that conditions in which SR phosphorylation decreased also showed higher cohesion of nuclear speckles, while hypoxia led to lower cohesion (Fig. 5b, Supplementary Fig. 5b).
a Phosphorylation of SR sites decreases in the nucleus under heat shock (43°C, 1 h) or arsenite treatment (0.5 mM, 1 h) and increases with hypoxia (0.2% O2, 24 h) compared to control. b Fluorescence recovery of 2xGFP-SON-tagged nuclear speckles decreases under heat shock and arsenite treatment and increases under hypoxia compared to control. Dots indicate the mean per time point and error bars show standard deviations. c PolyA RNA retention in nuclear speckles (segmented based on immunofluorescence staining for SON) increases with heat shock and arsenite treatment but decreases with hypoxia compared to control. d Example images of polyA FISH data quantified in c. Stress granules were segmented out using a pixel classifier and are here shown in black. e Example images of 5-EU staining for the three experiments in f showing transcription levels are not significantly altered by heat shock or hypoxia and decrease with arsenite stress. f Quantification of median nuclear 5-EU signal for one to two replicate wells in three experiments under different perturbations (p values calculated by two-sided Mann–Whitney U test, with Bonferroni correction). g Dissolution of nuclear speckles by GFP-CLK1 overexpression is reversed by heat shock. Dots indicate the mean per bin and shaded regions correspond to standard deviations. h Mean nuclear pSR and speckle polyA FISH intensities across cells in each imaged well reveal an inverse correlation (by two-sided Pearson correlation coefficient) between phosphorylation (modulated by DYRK3 and CLK1 inhibitors) and speckle polyA RNA under hypoxia and control conditions. arb. units = arbitrary units. i Example images of PP1, GFP-SON, and NPM1 for data in (j) and Supplementary Fig. 5M. j PP1 and GFP-SON pixel correlations across nuclei under different conditions. k Schematic representation of the relationship between nuclear speckle phosphorylation, cohesion, and mRNA retention. Scale bars correspond to 10 µm. Box plots within violins in a and c show median value and IQR, with whiskers indicating 1.5*IQR.
These changes in cohesion extended to the retention of polyadenylated RNA. Hypoxia decreased polyA FISH signal in both nuclear speckles and the cell as a whole. Heat shock and oxidative stress increased the polyA FISH signal within nuclear speckles, and to a lesser extent, the nucleoplasm. We also observed mRNA accumulation within stress granules in the cytoplasm under heat shock and oxidative stress (Fig. 5c, d, Supplementary Fig. 5c–f). Notably, changes in nuclear speckle retention did not reflect the effects of changed transcription rates based on 5-EU staining (Fig. 5e, f, Supplementary Fig. 5g). Oxidative stress inhibited RNA production and heat shock did not significantly affect transcription in iPSCs, as previously reported in HeLa cells (Zhang and Kleiner, 2019). By contrast, hypoxia increased transcription. Taken together, our data suggests that differential retention of mRNA under these conditions depends on nuclear speckle properties.
We then investigated whether we could rescue the observed changes in mRNA retention by modulating nuclear speckle cohesion. Similar to the prevention of DYRK3-mediated speckle dissolution by PPP1CC (see Fig. 1b), 1 hour of heat shock reversed the dissolution of speckles normally observed with high levels of GFP-CLK1 overexpression (Fig. 5g). However, in cells where dissolution did occur, heat shock failed to increase the nuclear retention of polyA RNA (Supplementary Fig. 5h). Treating hiPSCs kept under hypoxic conditions for 24 h with either CLK1 or DYRK3 inhibitors showed recovery of nuclear speckle polyA RNA levels after 2 h of treatment (Supplementary Fig. 5i-j). Mean nuclear pSR and mean speckle polyA RNA showed a strong negative correlation (R = -0.88, p = 1.5×10-4) across drug treatments under hypoxia and 20% O2 (Fig. 5h). Similar to kinase inhibition, R-MCD + 0.2 overexpression also restored nuclear speckle polyA RNA levels during hypoxia, showing that increasing cohesion is sufficient for the rescue of RNA retention (Supplementary Fig. 5k).
Previous reports find changes in CLK1 expression under environmental perturbations30,32,63, leaving unexamined a role for phosphatase availability. Although the involvement of regulatory subunits remains to be further elucidated, we observed enrichment of PP1 in nuclear speckles relative to the rest of the nucleus during oxidative stress and heat shock, but higher correlation with a nucleolar marker under hypoxia (Fig. 5I, j, Supplementary Fig. 5l). Overall, our results suggest that differences in mRNA localization in response to environmental perturbations can be regulated through changes in the cohesion of nuclear speckles, modulated through differential kinase and phosphatase activity at nuclear speckles.
Two of the conditions we considered (oxidative stress and heat shock) also induced stress granules. Stress granules are membraneless organelles, like nuclear speckles, and show accumulation of mRNAs during stress64. To determine whether similar transcripts are recruited to stress granules as retained in nuclear speckles, we next compared the nuclear speckle transcriptome to previously published data on the stress granule transcriptome65. We found that these two gene sets were largely mutually exclusive: 120 genes were depleted from nuclear speckles but enriched in stress granules, and 74 genes were enriched in nuclear speckles but depleted from stress granules (Supplementary Fig. 5m). By contrast, only three genes were enriched in both compartments and only 12 depleted from both. This mutual exclusivity could be related to the nuclear or cytoplasmic localization of transcripts and suggests a model wherein more nuclear transcripts tend to be retained within nuclear speckles, while more cytoplasmic transcripts are retained within stress granules in response to stress.
Discussion
Here, we examine a possible role for nuclear speckles in the dynamic control of mRNA localization. We characterize the continuous regulation of nuclear speckles through a phosphorylation-dephosphorylation cycle. As previously demonstrated, nuclear speckles have multiple subcompartments66 and the dynamics of nuclear speckle components differ18,37. Not all speckle proteins show changed dynamics in response to altered phosphorylation (Supplementary Fig. 6). Shifting the balance towards dephosphorylation increases the cohesion of the speckle scaffold proteins SRRM2 and SON in particular, as well as mRNA retention. Although dephosphorylation of SRSF proteins has been implicated in mRNA export67, our data shows lower nuclear polyA RNA levels when CLK1 or DYRK3 dissolve nuclear speckles. This suggests that export continues for many mRNAs under hyperphosphorylation. Earlier models speculated that kinases localize to condensates, while phosphatase activity occurs in the surrounding cytoplasm or nucleoplasm68. However, our results indicate that PP1 phosphatases localize to nuclear speckles and show enrichment under conditions associated with increased condensate cohesion. This suggests that both phosphorylation and dephosphorylation occur inside or in the immediate vicinity of condensates.
mRNA enrichment within speckles varies by transcript. Using an established proximity labeling and sequencing approach for RNA, we characterized the nuclear speckle transcriptome in hiPSCs. Overall, we found that transcripts enriched in nuclear speckles tend to be more retained within the nucleus. We observed effects linking increased cohesion to nuclear mRNA retention in both HeLa cells and hiPSCs. However, differences between the baseline hiPSC speckle transcriptome and the Hek293T cell speckle transcriptomes published by Barutcu et al. 8 suggest that enrichment of individual mRNAs may differ among cell types. As we describe the nuclear speckle transcriptome in pluripotent stem cells, it will be interesting in the future to compare over the course of cellular differentiation into various lineages. Transcript expression levels, which vary between cell types and tissues, were anti-correlated with nuclear speckle enrichment. A predictive model excluding expression levels as a feature and based only on invariant sequence features showed limited prediction strength. Transcripts also exhibit differential localization within nuclear speckles69, which could influence results of proximity-based labelling approaches depending on which nuclear speckle marker is used. This may further differentiate the nuclear speckle transcriptome we describe from previously reported ones.
The retention of mRNA within the nucleus under stress conditions has previously been described in yeast, which lack nuclear speckles. There, nuclear export acts as the dominant factor in nuclear retention70,71. In mammalian cells, our results suggest that the material properties of nuclear speckles can be tuned to retain mRNA in the nucleus. Similar to a role proposed for stress granules, these condensates therefore sequester mRNA away from the translation machinery during stress64,72. While stress granules may not exclude all translational machinery73, though, we show decreased translation under conditions with higher mRNA retention. In the acute heat stress and oxidative stress conditions we examined, we found small shifts in enrichment for overall polyA RNA, but the changes we observe under dephosphorylating conditions suggest widespread effects across genes. Heat shock has also previously been reported to affect splicing, with one paper reporting both increased and decreased intron retention and another reporting splicing inhibition74,75. Thus, intron retention could contribute to increased nuclear speckle retention for specific subsets of genes, although this remains to be confirmed.
Environmental perturbations have also been shown to affect the phosphorylation of splicing factors through changes in the expression of CLK1, as well as decreased activity of CLK1 in vitro in response to heat shock30,31,32. In the case of hypoxia, another study reported the dissolution of nuclear speckles through a decrease in SRSF6 expression76. Here, we find that speckles remain intact under hypoxia in hiPSCs, although the material properties of the structures shift towards a more liquid-like state in tandem with increased phosphorylation of SR-rich proteins. Our study addresses only short perturbations that result in reversible changes in phosphorylation. Further investigation is necessary to determine whether longer-term changes related to the accumulation of neurodegenerative disease-related transcripts and proteins within nuclear speckles also promote mRNA retention across the transcriptome and decreased translation77,78,79. Based on our results, showing a link between phosphorylation, nuclear speckle scaffold protein cohesion, and mRNA nuclear retention, we propose a means through which transient changes in the nuclear retention of transcripts occur in response to stress and other environmental perturbations in mammalian cells.
Methods
Cell culture
HeLa-FlpIn-Trex cells were a kind gift from Ivan Dikic (Goethe University, Frankfurt) and HEK293T cells were from ATCC (CRL-3216, Molsheim Cedex). HeLa and HEK cells were maintained in DMEM supplemented with 10% FBS and 5% L-glutamine.
hiPSCs were obtained from the Allen Cell Collection of the Coriell Institute for Medical Research (GM25256, AICS‑0094‑024). hiPSCs were maintained according to the standard operating procedures from the Allen Cell Institute (SOP: WTC culture v1.7) with minor adaptations. In short, hiPSCs were maintained in mTeSR Plus (STEMCELL Technologies, 100-0276). For passaging, hiPSCs were washed with DPBS and treated with Accutase (Thermo Fisher, A1110501) until they detached. Accutase treatment was quenched by dilution in DPBS. Cells were seeded on Geltrex (Thermo Fisher, A1413301) coated dishes and the growth media was supplemented with 10 µM Y-27632 (Hello Bio, HB2297) for 24 h after replating.
Cells were grown in a humidified incubator at 37 °C under 5% CO2 unless otherwise noted.
Immunofluorescence
Cells were fixed with 4% PFA for 15-30 min and permeabilized with 0.5% Triton-X for 30 min at room temperature. Cells were then blocked for 1 h at room temperature and stained with primary antibody for 1 h (HeLa cells) or 2 h (hiPSCs), washed in PBS, and stained with secondary antibody for 1 h (HeLa cells) or 2 h (hiPSCs).
Antibodies
1H4/pSR (Merck MABE50, clone 1H4, 1:100 or 1:150), B23/NPM1 (Sigma B0556, clone FC82291, 1:600), G3BP (abcam ab56574, clone 2F3, 1:400), HIF-1α (abcam ab179483, clone EPR16897, 1:500), PPP1CA (abcam ab137512, polyclonal, 1:250), SC35 (Sigma S4045, clone SC35, monoclonal, 1:1000), SON (abcam ab121759, polyclonal, 1:500), 16H3/SR (Merck MABE126, clone 16H3, 1:100).
Plasmids
GFP-PP1-NIPP1 and GFP-PP1m-NIPP1 plasmids were a kind gift from the Bollen Lab22. The cargo-binding domain of TNPO3 (CBDT) plasmid was a kind gift from the Shav-Tal Lab37. GFP-phosphatase plasmids were a kind gift from the Bodenmiller Lab33. pCMV-hyPBase was a kind gift from the Wellcome Trust Sanger Institute80. pECFP(C1)-NIPP1 (Addgene plasmid # 44226; http://n2t.net/addgene:44226; RRID:Addgene_44226), pEGFP(C1)-PP1alpha (Addgene plasmid # 44224; http://n2t.net/addgene:44224; RRID:Addgene_44224), pEGFP(N3)-PP1beta (Addgene plasmid # 44223; http://n2t.net/addgene:44223; RRID:Addgene_44223), and pEYFP(C1)-PP1gamma (Addgene plasmid # 44230; http://n2t.net/addgene:44230; RRID:Addgene_44230) were gifts from Angus Lamond & Laura Trinkle-Mulcahy. AICSDP-82:SON-mEGFP was a gift from Allen Institute for Cell Science (Addgene plasmid # 133964; http://n2t.net/addgene:133964; RRID:Addgene_133964). AICSDP-42:AAVS1-mTagRFPT-CAAX was a gift from Allen Institute for Cell Science (Addgene plasmid # 107580; http://n2t.net/addgene:107580; RRID:Addgene_107580). APEX2-NLS was a gift from Alice Ting (Addgene plasmid # 124617; http://n2t.net/addgene:124617; RRID:Addgene_124617)81. XLone-GFP was a gift from Xiaojun Lian (Addgene plasmid # 96930; http://n2t.net/addgene:96930; RRID:Addgene_96930)82.
Fluorescence recovery after photobleaching (FRAP) experiments
All FRAP experiments were performed on a Leica SP8 Falcon microscope using a 63 × 1.3 NA, glycerol, Plan-Apochromat objective. SRRM2-mCh was transiently overexpressed in HeLa TREx cells in combination with various GFP- or YFP-tagged plasmids. For FRAP during drug treatments and environmental perturbations, hiPSCs were endogenously tagged with 2xGFP at the SON locus as described below. Photobleaching was performed during 50–70 min of DRB treatment, 90–110 min of GSK626616 treatment, 70–85 min of Pladienolide-B treatment, or after 40–85 min of arsenite (500 µM) treatment or 50–105 min of heat shock (43 °C).
Proteomics and phosphoproteomics
Hek293T cells were seeded in 10-cm dishes to reach 70% confluency at the time of transfection. Triplicate technical replicates were gathered per condition. Cells were transfected with 5 µg DNA (EGFP-C1, EGFP-DYRK3, GFP-PP1-NIPP1, or GFP-PP1m-NIPP1) using GeneJuice. 24 h post-transfection, cells were washed twice in PBS and lysed in 450 µl lysis buffer (25 mM Tris HCl pH 7.4, 125 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, 1% Triton-X, 2x protease inhibitor cocktail, and 2x phosphatase inhibitor cocktail in milliQ-H2O) for 30 min on ice, harvested by scraping, then centrifuged at 17,000 g for 10 min at 4 °C. 25 µl anti-GFP magnetic agarose beads (Chromotek) per sample were equilibrated by washing 3x with 500 µl lysis buffer. Supernatants from the cellular lysates were then added to the beads and rotated at 4 °C for 1 h. Beads were washed 2x with 500 µl lysis buffer, and once with 125 µl wash buffer (1 mM Tris HCl, 125 mM NaCl, 1 mM MgCl2 in milliQ H2O).
For each sample, the anti-GFP beads with 100 µl of 10 mM Tris/2 mM CaCl2, pH 8.2 and re-suspended in 45 µl digestion buffer (triethylammonium bicarbonate (TEAB), pH 8.2), reduced with 5 mM TCEP (tris(2-carboxyethyl) phosphine) and alkylated with 15 mM chloroacetamide. Proteins were on-bead digested using 500 ng Sequencing Grade Trypsin (Promega). The digestion was carried out at 37 °C overnight. The supernatants were transferred to new tubes and the beads were washed with 150 µl trifluoroacetic acid (TFA) buffer (0.1% TFA, 50% acetonitrile) and combined with the first supernatant. For controls without phosphoenrichment, 10% of the samples were dried to completeness and re-solubilized in 20 µl of MS sample buffer (3% acetonitrile, 0.1% formic acid).
For the enrichment of phosphopeptides, the residual 90% of the samples were dried almost to completeness ( ~ 5 µl). The phosphopeptide enrichment was performed using a KingFisher Flex System (Thermo Fisher Scientific) and Ti-IMAC MagBeads (ReSyn Biosciences). Beads were conditioned following the manufacturer’s instructions, consisting of 3 washes with 200 µl of binding buffer (80% acetonitrile, 0.1 M glycolic acid, 5% TFA). Each sample was dissolved in 200 µl binding buffer. The beads, wash solutions and samples were loaded into 96 well deep well plates and transferred to the KingFisher. Phosphopeptide enrichment was carried out using the following steps: washing of the magnetic beads in binding buffer (5 min), binding of the phosphopeptides to the beads (30 min), washing the beads in wash 1-3 (binding buffer, wash buffer 1 and 2, 3 min each) and eluting peptides from the beads (50 µl 1% NH4OH, 10 min). The phosphopeptides were dried to the completeness and re-solubilized with 10 µl of 3% acetonitrile, 0.1% formic acid for MS analysis.
LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) equipped with a Digital PicoView source (New Objective) and coupled to a nanoAcquity UPLC (Waters Inc.). Solvent composition at the two channels was 0.1% formic acid for channel A and 0.1% formic acid, 99.9% acetonitrile for channel B. Column temperature was 50 °C. For each sample, 4 μl of peptides were loaded on a commercial Symmetry C18 trap column (5 µm, 180 µm x 20 mm, Waters Inc.) connected to a BEH300 C18 column (1.7 µm, 75 µm x 150 m, Waters Inc.). The peptides were eluted at a flow rate of 300 nl/min with a gradient from 5 to 35% B in 60 min, 35 to 60% B in 5 min and the column was washed at 80% B for 10 min before equilibrating back to 5% B.
The mass spectrometer was operated in data-dependent mode (DDA) using Xcalibur, with spray voltage set to 2.5 kV and heated capillary temperature at 275 °C. Full-scan MS spectra (350 − 1500 m/z) were acquired at a resolution of 70’000 at 200 m/z after accumulation to a target value of 3’000’000 and a maximum injection time of 100 ms, followed by HCD (higher-energy collision dissociation) fragmentation on the ten most intense signals per cycle. Ions were isolated with a 1.2 m/z isolation window and fragmented by higher-energy collisional dissociation (HCD) using a normalized collision energy of 25 %. HCD spectra were acquired at a resolution of 35’000 or 70’000 and a maximum injection time of 125 or 250 ms for phospho-enriched and non-enriched samples, respectively. The automatic gain control (AGC) was set to 3000 ions. Charge state screening was enabled and singly and unassigned charge states were rejected. Only precursors with intensity above 25’000 or 12’000 for phospho-enriched and non-enriched samples, respectively, were selected for MS/MS. Precursor masses previously selected for MS/MS measurement were excluded from further selection for 40 s, and the exclusion window tolerance was set at 10 ppm. The samples were acquired using internal lock mass calibration on m/z 371.1010 and 445.1200.
The mass spectrometry proteomics data were handled using the local laboratory information management system (LIMS)83.
Proteomics analysis
The acquired raw MS data were processed by MaxQuant (version 1.6.2.3), followed by protein identification using the integrated Andromeda search engine84. Spectra were searched against the Uniprot Homo sapiens reference proteome (taxonomy 9606, canonical version from 2019-07-09), concatenated to its reversed decoyed fasta database and common protein contaminants. Carbamidomethylation of cysteine was set as fixed modification, while methionine oxidation, phosphor (STY) and N-terminal protein acetylation were set as variable. Enzyme specificity was set to trypsin/P allowing a minimal peptide length of 7 amino acids and a maximum of two missed cleavages. MaxQuant Orbitrap default search settings were used. The maximum false discovery rate (FDR) was set to 0.01 for peptides and 0.05 for proteins. Label-free quantification was enabled and a 2-minute window for match between runs was applied. In the MaxQuant experimental design template, each file is kept separate in the experimental design to obtain individual quantitative values.
Data was then processed using R (v3.6.3). Results were first filtered to exclude reverse database hits, potential contaminants, and proteins identified only by site. Protein groups were then filtered for entries for ≥ 2 replicates in any condition under comparison. Missing LFQ intensities were imputed with random noise simulating the detection limit of the mass spectrometer (a log-normal distribution with 0.25x the standard deviation of the measured, logarithmized values, down-shifted by 1.8 standard deviations). Sample differences were then tested with the t.test function in R.
For analysis of differential phosphorylation from phosphoproteomics data, significance was calculated for single sites (phosphopeptides and matched unphosphorylated peptides from input samples) by comparing generalized linear models with and without an interaction term for phosphorylation status and condition using a likelihood ratio test.
5-EU and pulse-chase
Induction of R-MCD ( + 0.2)-GFP and inducible GFP hiPSCs was started 24 h prior to the start of the experiment by supplementing the growth medium with 2 µg/ml doxycycline. Then, the cells were pulsed with fresh growth medium containing 20 µM CX5461 (MCE, HY-13323) and 1 mM 5-EU for 30 min. After the pulse, the cells were washed twice with a growth medium and then kept in a growth medium containing 1 mM Uridine for up to four hours. A new batch of cells was pulsed every hour and all cells were fixed at the same timepoint.
For 5-EU measurements under environmental perturbations, cells were similarly treated with 20 µM CX5461 and 1 mM 5-EU for 30 min, then fixed.
CLICK reactions to detect 5-EU were performed after fixation and permeabilization by washing cells twice in TBS, then adding a solution of 2 mM CuSO4, 10 µM AlexaFluor 647 Azide Triethylammonium Salt (ThermoFisher A10277), and freshly added 100 mM sodium ascorbate in TBS, and incubating in the dark at room temperature for 30 min before washing in PBS and proceeding with additional stainings.
PolyA FISH
Cells were fixed and permeabilized as for immunofluorescence imaging. Cells were then washed twice with FISH wash buffer (10% formamide, 2x saline-sodium citrate (SSC)), then incubated 1 h at 37 °C in pre-hybridization buffer (100 mg/ml dextran sulfate, 7.5% formamide, 1.5x SSC). Half the volume was then aspirated and 2x pre-warmed hybridization buffer (100 mg/ml dextran sulfate, 10% formamide, 2x SSC, 800 nM dT-30-Atto488, dT-30-Cy3, or dT-30-Cy5 oligomer) added in equal volume for overnight incubation at 37 °C. The following day, cells were washed twice for 30 min at 37 °C with pre-warmed FISH wash buffer, then washed with 2x SSC and finally PBS before proceeding with immunofluorescence staining and imaging.
Design of smFISH probes
Target selection for genes under baseline condition (DMSO) was performed using raw DESeq2 results without input normaliztion. Probes for smFISH were designed as branched DNA probes for signal amplification. Twelve primary probes were designed per target gene using PaintSHOP85, using the hg38 newBalance (isoform flattened) probe set with default settings or OligoMiner86,87. When more than twelve probes were found for a target, the probes with the highest on_target value (PaintSHOP) or lowest melting temperatures (OligoMiner) were chosen. For three gene targets, fewer than 12 probes were available (EXOSC1: 8, H2BC11: 8, POU5F1: 6). For pooled probes, around 47 or 48 gene targets were selected per pool (enriched pool: 48, depleted pool: 47, neither pool: 48) based on relaxed thresholds of significance (enriched and depleted pools: |log2 fold change | > 0.3, padj <0.6, neither pool: |log2 fold change | <0.05, padj > 0.99). The primary probes were then extended by color-specific barcodes to which four secondary probes could bind. Primer binding sequences for probe purification were also included. The signal was amplified in this manner up to quaternary probes. Finally, fluorophore labelled probes (label probes) were bound to quaternary probes. Color-specific barcodes were derived from orthogonal 25mer barcode sequences designed previously88. The labelled probes were based on sequences used in the amplification method for smFISH signals described previously89. Raw probe sequences as generated by PaintSHOP and OligoMiner, as well as sequences extended with barcodes, are listed in Supplementary Data 5. Probe sequences for amplification probes (secondary, tertiary, quaternary and label probes) are listed in Supplementary Data 6. Primary probes were ordered from Twist Bioscience or Integrated DNA Technologies (IDT) as oligo pools. Amplification probes were ordered from Microsynth.
Purification of oligo pools
FISH probes were purified from oligo pools in three steps. First, oligo pools were amplified by PCR using primers to introduce T7 RNA polymerase promoter sequences and the amplicon was cleaned up using Zymo DNA Clean & Concentrator-25 (Zymo Research, D4033). The resulting amplicon was used as starting material for in vitro transcription using HiScribe T7 High Yield RNA Synthesis Kit (NEB, E2040S). Finally, the RNA product was used for reverse transcription using Maxima H Minus Reverse Transcriptase (Thermo Fisher, #EP0752) and RNA was removed by alkaline lysis.
PCR primers for oligo pool amplification:
Pair 1:
Forward: GTTGGTCGGCACTTGGGTGC
Reverse: CCACCGGATGAACCGGCTTT
Pair 2:
Forward: CGATGCGCCAATTCCGGTTC
Reverse: CAACCCGCGAGCGATGATCA
smFISH
hiPSCs were grown on 96-well plates (Greiner, 655090) and fixed with 4% PFA (EMS, 15710) in PBS for 15 min at room temperature. When pooled probe sets were used, to ensure even entry of probes, cells were dissociated and seeded as single cells before the experiment. The sample was then washed with PBS and permeabilized with 0.5% Triton X-100 (Sigma Aldrich, X100) in PBS for 30 min at room temperature. After another PBS wash, if pooled probe sets were used, the sample was treated with Protease QS (Thermo Fisher, QVP0011) diluted 1:2000 in PBS for 10 min at room temperature while shaking. Protease treatment was stopped by washing the sample with protease stop buffer (Thermo Fisher, QVP0011) twice and once with PBS. The sample was then incubated with 40% wash buffer (40% Formamide, 2x SSC, 0.001% tween20 in RNase-free H2O) for one hour at 65 °C. The sample was then incubated with primary probes diluted to a final concentration of 2 nM per probe in primary probe hybridization solution (10% Dextran, 40% Formamide, 2x SSC, 0.01% yeast tRNA, murine RNase inhibitor, 0.001% tween20 in RNase free H2O) for 16 h at 37 °C. The sample was washed three times with 40% wash buffer and once with 30% wash buffer (30% Formamide, 2x SSC, 0.001% tween20 in RNase-free H2O) for 6 min at 37 °C. The sample was then incubated with secondary probes diluted to 5 nM in probe solution (10% Dextran, 40% Formamide, 2x SSC, 0.001% tween20 in RNase-free H2O) for 30 min at 37 °C. After incubation with secondary probes, the sample was washed three times with 30% wash buffer for 6 min at 37 °C. Probe addition and washing were repeated in the same manner for tertiary and quaternary branching probes. The sample was then washed with PBS and incubated with label probes diluted to 0.5 µM in PBS for 1 h at 37 °C protected from light.
Nucleofection of hiPSCs
Y-27632 was added to hiPSC growth media 1–6 h prior to nucleofection at a final concentration of 10 µM. Cells were then passaged as previously described and 8 × 105 cells were resuspended in 100 µl of nucleofector solution (Lonza, VPH-5012). Depending on the experiment, plasmids, sgRNA and Cas9 were added to the resuspended cells. The cell suspension was then transferred to a nucleofection cuvette and nucleofection was performed using Lonza Nucleofector 2b using program A-023. After nucleofection, 500 µl of warm growth medium was added to the nucleofection cuvette and cells were transferred to a Geltrex-coated well of a 6-well plate containing pre-equilibrated (37 °C, 5% CO2) mTeSR plus supplemented with 10 µM Y-27632. Y-27632 concentration in the growth medium was halved to 5 µM after 24 h, and it was removed completely after 48 h of plating.
Design of sgRNA and Donor Plasmids for APEX2-SON and APEX2-NLS hiPSCs
We used crRNA sequences specified by the Allen Cell Institute to guide Cas9 to the SON locus and the AAVS1 locus. We ordered sgRNAs incorporating the crRNA and tracrRNA sequences from Sigma Aldrich. We used plasmid AICSDP-82:SON-mEGFP as template for the APEX2-SON donor plasmid and AICSDP-42:AAVS1-mTagRFPT-CAAX as a template for the APEX2-NLS donor plasmid. These plasmids already contained the homology arms needed for downstream CRISPR/Cas9 genome editing. AICSDP-42:AAVS1-mTagRFPT-CAAX additionally contained the CAG promoter to allow stable and consistent transgene expression in hiPSCs90,91. The APEX2 enzyme sequence and NLS motif were PCR amplified from plasmid APEX2-NLS. The APEX2 sequence was inserted in front of mEGFP in AICSDP-82:SON-mEGFP to create the APEX2-mEGFP-SON donor plasmid. The APEX2-NLS sequence was inserted along with the mEGFP sequence from AICSDP-82:SON-mEGFP into AICSDP-42:AAVS1-mTagRFPT-CAAX, replacing mTagRFPT-CAAX, to create the mEGFP-APEX2-NLS donor plasmid. All donor plasmids were assembled by Gibson assembly using NEB Gibson Assembly Master Mix (NEB, E2611L).
crRNA sequences
SON: CTGCTCGATGTTGGTCGCCA
AAVS1: GGGGCCACTAGGGACAGGAT
Cell line generation
The protocols for CRISPR/Cas9 genome engineering in hiPSCs were adapted from published protocols92,93. To generate the 2xGFP-SON, APEX2-NLS, and APEX-SON cell lines, hiPSCs were nucleofected as described above with 2 µg of donor plasmid and 1.5 µl each of 10 µM sgRNA and 10 µM Cas9 (Sigma Aldrich, CAS9PROT-50UG). Once cells reached confluency, they were passaged, resuspended in phenol red free mTeSR1 (STEMCELL, 05876) and sorted by FACS for GFP positive cells.
To generate doxycycline inducible R-MCD ( + 0.2)-GFP and GFP cell lines, hiPSCs were nucleofected as described above with 1 µg of XLone-R-MCD ( + 0.2)-GFP or XLone-GFP plasmid along with 1 µg of PiggyBack transposase plasmid (pCMV-hyPBase). XLone-R-MCD ( + 0.2)-GFP plasmid was generated by PCR amplification of R-MCD ( + 0.2) domain from pmEGFP-N1-R-MCD( + 0.2) plasmid (kind gift from Dr. Gregory Jedd) and cloned into XLone-GFP plasmid using Gibson assembly kit (NEB, E2611S). Two days after nucleofection, selection was started by adding blasticidin (Santa Cruz, sc-495389) at a concentration of 10 μg/ml. Blasticidin selection was carried out for multiple passages before cells were frozen in liquid nitrogen for long-term storage.
Drug treatment
Drug treatment with GSK626616 (5 µM), DRB (75 µM) and Pladienolide-B (2 µM) was carried out in DMEM/F-12 medium without serum for two hours at 37 °C and 5% CO2. For DYRK3 and CLK1 inhibition in hypoxia, GSK626616 (5 µM), TG003 (100 µM), or DMSO were likewise diluted in serum-free medium (pre-equilibrated overnight under hypoxic conditions) and added to cells for two hours after 22 h in hypoxia (0.2% O2, 5% CO2, 37 °C). Inhibition with CLK-IN-T3 (1 µM) was carried out in complete medium for 8 h (OPP experiment) or 12 (FISH experiments) hours.
OPP
Translation was quantified based on the incorporation of O-propargyl puromycin (OPP) in nascent peptides94. Cells were incubated in media containing 30 µM OPP (with drugs) for 45 min before fixation and detection of OPP through a CLICK reaction, as described above for 5-EU.
Stresses and environmental perturbations
Heat shock was performed at 43 °C for 1 h in a cell culture incubator maintained at 5% CO2. Oxidative stress was induced for 1 h using 500 µM sodium meta-arsenite dissolved in media. To induce hypoxia, cell media was exchanged for media pre-equilibrated under hypoxic conditions (0.2% O2), and cells were maintained at 0.2% O2/5% CO2 in a humidified atmosphere at 37 °C in a hypoxia workstation (Baker-Ruskinn).
APEX2 enzymatic reaction
The protocol for the APEX2 enzymatic reaction was adapted from Fazal et al. 42,43 and Padron et al. 43. APEX2-tagged hiPSCs were washed with DPBS (Thermo Fisher, 14190136) and then incubated in DPBS containing 0.5 mM biotin-phenol (Iris Biotech, LS-3500.1000) and 0.005% digitonin (Sigma Aldrich, D141) for 3 min at room temperature. To trigger the enzymatic reaction, hydrogen peroxide (Sigma Aldrich, 1.07209) was added to a final concentration of 0.5 mM and the dish was tilted for 1 min at room temperature. To stop the reaction, cells were washed once with a quenching solution (5 mM Trolox, 10 mM sodium ascorbate, 10 mM sodium azide in DPBS) and three times with wash solution (5 mM Trolox, 10 mM sodium ascorbate in DPBS).
RNA extraction and enrichment of biotinylated RNA
The protocol for extraction and enrichment of biotinylated RNA was adapted from Fazal et al. 42,43 and Padron et al. 43 hiPSCs were lysed by adding RNA lysis buffer (Zymo Research, R1060-1-50) directly to the cell culture dish. The cells were scraped into a solution and total RNA was purified using a Zymo Quick-RNA Miniprep kit (Zymo Research, R1054). Of the isolated total RNA, 5 µl was set aside as an input sample. To purify biotinylated RNA, we used Pierce Streptavidin Magnetic Beads (Thermo Fisher, 88816). First, 30 µl of beads per sample were resuspended in Binding and Washing (B&W) buffer (5 mM Tris-HCL (pH 7.5), 1 mM EDTA, 2 M NaCl) by vortexing, and then washed three times with B&W buffer. The beads were then resuspended in Solution A (0.1 M NaOH, 0.05 M NaCl) and incubated for 2 min. Then the beads were washed twice with Solution B (0.1 M NaCl) and resuspended in 100 µl Solution B. An equal volume of total RNA sample was added, and the samples were incubated for 2 h at 4 °C on a rotator to allow the biotinylated RNA to bind to the beads. The beads were then washed three times with B&W buffer and resuspended in 54 µl of RNase-free H2O. A 3X proteinase buffer was prepared (For 1 ml: 300 µl PBS, 300 µl 20% N-Lauryl sarcosine sodium solution (Sigma Aldrich, L7414), 60 µl 0.5 M EDTA, 15 µl 1 M DTT, 225 µl RNase free H2O), and 33 µl of this buffer was added to the beads together with 10 µl Recombinant Proteinase K Solution (20 mg/ml, Thermo Fisher, AM2548) and 3 µl RiboLock RNase inhibitor (Thermo Fisher, EO0381). The beads were then incubated for 1 h at 42 °C and then for 1 h at 55 °C on a shaker. Finally, biotinylated RNA was purified using RNA Clean & Concentrator-5 kit (Zymo Research, R1013).
For biotinylated RNA, five biological replicates were collected per cell line and drug condition. For input RNA, three biological replicates were collected per cell line and drug condition.
Subcellular fractionation of hiPSCs
The protocol used for subcellular fractionation of hiPSCs was adapted from Mayer & Churchman, 2017. Throughout the protocol, samples were kept at 4 °C and handled under RNase-free conditions. Briefly, hiPSCs were grown to confluency on 10 cm dishes and were first washed with and then scraped into DPBS. Cells were then pelleted by centrifugation for 3 min at 211 g and resuspended in lysis buffer (0.15% NP-40, 150 mM NaCl, 25 µM α-amanitin, 10 U SUPERase.IN (Thermo Fisher, AM2696), 1x cOmplete protease inhibitor mix, EDTA free (Sigma Aldrich, 11873580001)). The lysate was layered onto a sucrose buffer (10 mM Tris-HCl (pH 7.0), 5 M NaCl, 25% (w/v) sucrose, 25 µM α-amanitin, 10 U SUPERase.IN, 1x cOmplete protease inhibitor mix, EDTA free) and centrifugated for 10 min at 16,000 g. The supernatant representing the cytoplasmic fraction was collected, and the remaining pellet was washed twice with nuclei wash buffer (1 mM EDTA, 25 µM α-amanitin, 40 U SUPERase.IN, 1x cOmplete protease inhibitor mix, EDTA free prepared in PBS). The nuclear pellet was then resuspended in glycerol buffer (20 mM Tris-HCl (pH 8.0), 75 mM NaCl, 0.5 mM EDTA, 50% (v/v) glycerol, 0.85 mM DTT, 25 µM α-amanitin, 10 U SUPERase.IN, 1x cOmplete protease inhibitor mix, EDTA free) to which nuclei lysis buffer (1% NP-40, 20 mM HEPES (pH 7.5), 300 mM NaCl, 1 M urea, 0.2 mM EDTA, 1 mM DTT, 25 µM α-amanitin, 10 U SUPERase.IN, 1x cOmplete protease inhibitor mix, EDTA free) was added. After 5 min of incubation, the suspension was centrifugated at 18500 g for 2 min. The supernatant representing the nuclear fraction was collected and the remaining pellet was washed with PBS and centrifugated for 1 min at 1150 g. The supernatant was discarded, and the pellet representing the chromatin fraction was resuspended in 50 µl of PBS. The chromatin fraction was then incubated with TRIzol (Life Technologies, 15596) and chloroform for 5 min at room temperature. The sample was centrifuged, and the upper aqueous phase was collected.
To isolate RNA, 3.5 sample volumes of RLT buffer (Qiagen, 79216) were added to chromatin, nuclei and cytoplasm fractions. After mixing, 2.5 volumes of ice-cold 75% ethanol was added. RNA was then cleaned up using the RNeasy Mini kit (Qiagen, 74104). A total of 5 biological replicates were collected.
Library preparation and RNA sequencing
For APEX2-sequencing samples, libraries were prepared using SMARTer Stranded Total RNA-Seq Kit v2 - Pico Input Mammalian (Takara, 634411). Single-end sequencing was performed on the Illumina NovaSeq platform with a sequencing depth of 80 million reads and a read length of 100 bp.
For subcellular fraction sequencing samples, libraries were prepared using the Truseq Stranded mRNA kit (Illumina, 20020594). Single-end sequencing was performed on the Illumina NovaSeq platform with a sequencing depth of 200 million reads and a read length of 100 bp.
Library preparation and RNA sequencing was performed by the Functional Genomics Center Zurich (FGCZ).
Processing of RNAseq data
Trimmomatic v0.3995 was used to trim adapter sequences from raw reads with the following settings: ILLUMINACLIP:TruSeq3-SE:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36. Quality checks were carried out before and after trimming with FastQC v0.11.9. Trimmed reads were mapped to the human reference genome (hg38, GRCh38.p14 primary genome assembly) using GENCODE v40 gene annotations with STAR v2.7.3a96. Count tables were generated using the featureCounts97 function of the R package Rsubread v2.10.598.
Differential gene expression analysis
Genes with less than 10 counts in any of the samples were removed prior to the analysis. Differential Gene Expression (DGE) analysis was performed with DESeq2 v1.36.099 using the default two-sided Wald test and Benjamini-Hochberg correction for multiple testing. To quantify enrichment in nuclear speckles vs. nucleoplasm in DMSO treatment, we tested for the influence of the cell line factor. Thresholds of |log2FoldChange | > 0.5 and padj <0.05 were used. To quantify enrichment in nuclear speckles vs. Nucleoplasm in drug-treated vs. DMSO-treated cells we tested for the influence of the interaction term treatment:cell line. Thresholds of |log2FoldChange | > 0.5 and padj <0.1 were used. Results of DESeq2 analyses are listed in Supplementary Data 7 and Supplementary Data 3.
Differential transcript expression analysis
Transcript quantification was performed using Salmon v0.12.0100 with the --numGibbsSamples option set to 30 to generate Gibbs samples. Differential transcript expression analysis was performed using Fishpond v2.2.0101.
Normalization with input samples
Unless otherwise indicated, DESeq2 results with input normalization were used for all analysis. Log2FoldChanges for input sample nuclear speckle enrichment was quantified with DESeq2 as described above. DGE results for nuclear speckle enrichment were normalized by fitting a linear regression model with log2FoldChanges of input samples as the independent variable and log2FoldChanges of biotinylated samples as the dependent variable. The residuals of the linear model were used as corrected log2FoldChange.
Gene set enrichment analysis
Gene Set Enrichment analysis was performed with the GSEA software v4.2.3102,103. GSEA was run in pre-ranked mode with default settings and using KEGG, Reactome and GO:CC, GO:BP and GO:MF gene sets. As ranking metric, we used input normalized log2FoldChange in case of nuclear speckle enrichment in DMSO condition and -log10(padj) * sign(log2FoldChange) in case of nuclear speckle enrichment in drug vs. DMSO conditions.
Splicing analysis
Differences in splicing between the transcripts enriched in nuclear speckles or the nucleoplasm were assessed in DMSO control samples using three tools: iREAD104, VAST v2.5.1105,106, and MAJIQ v 2.4.dev3 + g85d0781.d20220721107.
For VAST, analysis was run starting with untrimmed reads, as recommended, with thresholds set for detection in ≥1 samples with ≥ 10 reads, with a minimum probability ≥ 0.95. Residual differences in retention were calculated using input samples from each cell line and a residual difference of >0.15 used for plotting. Control introns (n = 10,000) were selected at random from all detected introns for feature comparisons.
For MAJIQ, detection thresholds were set for observation of alternative splicing in ≥ 1 sequencing replicate with ≥ 5 reads per junction (prior-minreads) and ≥ 2 reads per experiment (minreads), along with the default probability threshold for local splice variation of 0.2. Control introns (n = 3633) were introns with absolute differential percent spliced in index <0.1 and probability <0.2. For comparison between drug-treated cells and controls (Supplementary Fig. 5G), splicing differences were calculated using MAJIQ for SON-enriched samples under drug treatments compared to DMSO.
Network visualization of GSEA results
Network visualizations of GSEA results were made using Cytoscape108 v3.9.1 and EnrichmentMap109. Only Nodes with gene set sizes between 29 and 496 and NES smaller than -2 or greater than 1.8 are displayed. Annotations were generated using the AutoAnnotate plugin110 and manually adapted. Annotated groups were positioned manually.
Prediction of transcript-level nuclear speckle enrichment
Feature categories used for the linear model were collected from the following sources. General sequence features: custom Python script using GENCODE v43 primary assembly gff3 annotations. As part of the general sequence features, we also included the GGACU m6A motif density and the AGCCC nuclear localization motif111 density. RNA binding protein features: oRNAment database112. Kinetic rates:113. Promoter motifs: The Eukaryotic Promoter Database EPD114. TPM: Salmon quantification (Supplementary Data 4).
Features for training were filtered to only include protein-coding transcripts. Features representing motif counts were transformed into density. Features that contained 0 values for more than 75% of the data were removed. All features were log transformed, missing values were imputed by the mean of the feature and features were z-scored. Principal Component Analysis (PCA)115 was performed on the resulting feature set and the number of principal components explaining 99% of the variance were retained (n = 414). All transcripts annotated to the same gene were only allowed to be in either the test or the train set. The reported coefficients of determination were calculated by averaging results of 10-fold cross-validation. Preprocessing and model training were done in Python (v3.9.7) using scikit-learn (v1.3.0).
Feature importance scoring
The linear regression model was trained for one hundred iterations with different test (15% of the data) and train (85% of the data) subsets. The top ten loadings of the ten principal components with the highest absolute coefficients were extracted for each iteration. The frequency at which a feature occurs in this subset of features was taken as a measure of feature importance. For example, a feature occurring in the top 10 loadings of every single top 10 PC will have a frequency of occurrence of 1.
UMAP representation
Transcript features were preprocessed as described above and were used as the input for the UMAP algorithm as implemented in Python58,59.
Immunofluorescence and FISH microscopy
Microscopy images were acquired on a CellVoyager 7000 microscope (Yokogawa) equipped with an enhanced CSU-W1 spinning disk (Microlens-enhanced dual Nipkow disk confocal scanner, wide view type) and Andor cSMOS cameras or on a CellVoyager 8000 microscope (Yokogawa) equipped with CSU-W1 spinning disk and ORCA-Flash4.0 V3 cameras. Images were acquired using a 60x Nikon water immersion objective (NA = 1.2) or 40x Nikon air objective (NA = 0.95) on the CellVoyager 7000, and using a UPLSAP60xW customized Yokogawa objective (NA = 1.2) on the CellVoyager 8000. Z-stacks with a 1 μm spacing were acquired per site, spanning the whole height of the sample (12–20 μm). For subsequent processing, maximum intensity projections (MIPs) were performed for each site, except where noted.
Image processing
Nuclear speckles were segmented based on SC35 or SON intensity images using the pixel classifier functionality of Ilastik v1.4.0116,117. Nuclei were segmented based on DAPI intensity images using Cellpose118. Image processing was performed on a computing cluster (ScienceCloud) (https://www.zi.uzh.ch/en/teaching-and-research/science-it/computing/sciencecloud.html) provided by the Service and Support for Science IT (S3IT) facility of the University of Zurich (UZH) using TissueMAPS (https://github.com/pelkmanslab/TissueMAPS), Fractal (https://fractal-analytics-platform.github.io/) and custom Python scripts. Pearson correlation coefficients were calculated in python using scipy.stats.
Pseudocoloring of smFISH images
Intensity values of smFISH images were mapped to different color gradients depending on the subcellular localization of the signal. Signal overlapping with nuclear speckle segmentation (FISH ∩ nuclear speckles, based on SON-mEGFP or SC35 antibody staining) was mapped to an orange gradient, while signal not overlapping with speckles (FISH \ nuclear speckles) was mapped to a blue gradient. Intensities for the same smFISH target were always scaled the same way across both gradients. Outlines of nuclei (segmented based on DAPI) and nuclear speckles (segmented based on SC35) are indicated by white overlays. Color gradients were applied in Python using the microfilm package v0.2.1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. Raw sequencing data has been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE240892. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE119 partner repository under accession code PXD055775. Given the number of imaging experiments performed and the volume of data, we will arrange to upload or share this data on specific request.
Code availability
Analysis was performed using publicly available software noted in the methods. Custom Python scripts used to extract transcript sequence features, perform multilinear regression analysis and perform proteomics analyses were deposited on Github [https://github.com/adrtsc/nuclear_speckles_pelkmans_code] and Zenodo [[https://zenodo.org/records/14082902]120. Python scripts developed to create branched DNA probe sequences from oligominer and paintshop outputs were deposited on Github [https://github.com/adrtsc/fools]. Additional custom scripts for analysis and plotting are available on request.
References
Spector, D. L. & Lamond, A. I. Nuclear Speckles. Cold Spring Harb. Perspect. Biol. 3, a000646 (2011).
Hutchinson, J. N. et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 8, 1–16 (2007).
Ilık, İ. A. et al. Son and srrm2 are essential for nuclear speckle formation. Elife 9, 1–48 (2020).
Miyagawa, R. et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 18, 738 (2012).
Cao, W., Jamison, S. F. & Garcia-Blanco, M. A. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA 3, 1456–1467 (1997).
Mermoud, J. E., Cohen, P. T. W. & Lamond, A. I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 13, 5679–5688 (1994).
Politz, J. C. R. et al. Rapid, diffusional shuttling of poly(A) RNA between nuclear speckles and the nucleoplasm. Mol. Biol. Cell 17, 1239–1249 (2006).
Barutcu, A. R. et al. Systematic mapping of nuclear domain-associated transcripts reveals speckles and lamina as hubs of functionally distinct retained introns. Mol. Cell 82, 1035–1052.e9 (2022).
Fei, J. et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J. Cell Sci. 130, 4180–4192 (2017).
Hu, Y., Plutz, M. & Belmont, A. S. Hsp70 gene association with nuclear speckles is Hsp70 promoter specific. J. Cell Biol. 191, 711–719 (2010).
Kim, J., Venkata, N. C., Hernandez Gonzalez, G. A., Khanna, N. & Belmont, A. S. Gene expression amplification by nuclear speckle association. J. Cell Biol. 219, e201904046 (2020).
Zhang, L. et al. TSA-seq reveals a largely conserved genome organization relative to nuclear speckles with small position changes tightly correlated with gene expression changes. Genome Res 31, 251–264 (2021).
Galganski, L., Urbanek, M. O. & Krzyzosiak, W. J. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res 45, 10350–10368 (2017).
Zhang, Q. et al. Coordinated Dynamics of RNA Splicing Speckles in the Nucleus. J. Cell Physiol. 231, 1269–1275 (2016).
Kim, J., Han, K. Y., Khanna, N., Ha, T. & Belmont, A. S. Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J. Cell Sci. 132, jcs226563 (2019).
Misteli, T. & Spector, D. L. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors 7, 1559–1572. https://doi.org/10.1091/mbc.7.10.1559 (1996).
O’Keefe, R. T., Mayeda, A., Sadowski, C. L., Krainer, A. R. & Spector, D. L. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124, 249–260 (1994).
Rino, J. et al. A Stochastic View of Spliceosome Assembly and Recycling in the Nucleus. PLoS Comput Biol. 3, e201 (2007).
Greig, J. A. et al. Arginine-Enriched Mixed-Charge Domains Provide Cohesion for Nuclear Speckle Condensation. Mol. Cell 77, 1237–1250.e4 (2020).
Colwill, K. et al. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15, 265 (1996).
Rai, A. K., Chen, J. X., Selbach, M. & Pelkmans, L. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216 (2018).
Bollen, M., Peti, W., Ragusa, M. J. & Beullens, M. The extended PP1 toolkit: Designed to create specificity. Trends Biochem Sci. 35, 450–458 (2010).
Hoermann, B. et al. Dissecting the sequence determinants for dephosphorylation by the catalytic subunits of phosphatases PP1 and PP2A. Nat. Commun. 11, 1–20 (2020).
Bertolotti, A. The split protein phosphatase system. Biochem. J. 475, 3707–3723 (2018).
Saitoh, N. et al. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 15, 3876–3890 (2004).
Trinkle-Mulcahy, L., Sleeman, J. E. & Lamond, A. I. Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J. Cell Sci. 114, 4219–4228 (2001).
Shi, Y., Reddy, B. & Manley, J. L. PP1/PP2A Phosphatases Are Required for the Second Step of Pre-mRNA Splicing and Target Specific snRNP Proteins. Mol. Cell 23, 819–829 (2006).
Trinkle-Mulcahy, L. et al. Nuclear organisation of NIPP1, a regulatory subunit of protein phosphatase 1 that associates with pre-mRNA splicing factors. J. Cell Sci. 112, 157–168 (1999).
Wang, J. T. et al. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife 3, e04591 (2014).
Jakubauskiene, E., Vilys, L., Makino, Y., Poellinger, L. & Kanopka, A. Increased serine-arginine (SR) protein phosphorylation changes pre-mRNA splicing in hypoxia. J. Biol. Chem. 290, 18079–18089 (2015).
Haltenhof, T. et al. A Conserved Kinase-Based Body-Temperature Sensor Globally Controls Alternative Splicing and Gene Expression. Mol. Cell 78, 57–69.e4 (2020).
Ninomiya, K., Kataoka, N. & Hagiwara, M. Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. J. Cell Biol. 195, 27–40 (2011).
Lun, X. K. et al. Analysis of the Human Kinome and Phosphatome by Mass Cytometry Reveals Overexpression-Induced Effects on Cancer-Related Signaling. Mol. Cell 74, 1086–1102.e5 (2019).
Wu, D. et al. A substrate-trapping strategy for protein phosphatase PP1 holoenzymes using hypoactive subunit fusions. J. Biol. Chem. 293, 15152–15162 (2018).
Hornbeck, P. V. et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43, D512–D520 (2015).
Sacco-Bubulya, P. & Spector, D. L. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156, 425 (2002).
Hochberg-Laufer, H. et al. Availability of splicing factors in the nucleoplasm can regulate the release of mRNA from the gene after transcription. PLoS Genet 15, e1008459 (2019).
Berry, S., Müller, M., Rai, A. & Pelkmans, L. Feedback from nuclear RNA on transcription promotes robust RNA concentration homeostasis in human cells. Cell Syst. 13, 454–470.e15 (2022).
Bahar Halpern, K. et al. Nuclear Retention of mRNA in Mammalian Tissues. Cell Rep. 13, 2653–2662 (2015).
Smith, K. P., Moen, P. T., Wydner, K. L., Coleman, J. R. & Lawrence, J. B. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 144, 617–629 (1999).
Wang, K. et al. Intronless mRNAs transit through nuclear speckles to gain export competence. J. Cell Biol. 217, 3912–3929 (2018).
Fazal, F. M. et al. Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell 178, 473–490.e26 (2019).
Padrón, A., Iwasaki, S. & Ingolia, N. T. Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. Mol. Cell 75, 875–887.e5 (2019).
Wei, M. T. et al. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 2017 9:11 9, 1118–1125 (2017).
Kitamura, A., Nakayama, Y. & Kinjo, M. Efficient and dynamic nuclear localization of green fluorescent protein via RNA binding. Biochem. Biophys. Res Commun. 463, 401–406 (2015).
Huang, S. & Spector, D. L. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc. Natl Acad. Sci. USA 89, 305 (1992).
Meier, U. T. RNA modification in Cajal bodies. RNA Biol. 14, 693–700 (2017).
Ganot, P., Jády, B. E., Bortolin, M.-L., Darzacq, X. & Kiss, T. Nucleolar Factors Direct the 2′-O-Ribose Methylation and Pseudouridylation of U6 Spliceosomal RNA. Mol. Cell Biol. 19, 6906–6917 (1999).
Lange, T. S. & Gerbi, S. A. Transient nucleolar localization of U6 small nuclear RNA in Xenopus laevis oocytes. Mol. Biol. Cell 11, 2419–2428 (2000).
Tycowski, K. T., You, Z. H., Graham, P. J. & Steitz, J. A. Modification of U6 Spliceosomal RNA Is Guided by Other Small RNAs. Mol. Cell 2, 629–638 (1998).
Trinkle-Mulcahy, L. & Sleeman, J. E. The Cajal body and the nucleolus: “In a relationship” or “It’s complicated”? RNA Biol. 14, 739–751 (2017).
Deryusheva, S. & Gall, J. G. scaRNAs and snoRNAs: Are they limited to specific classes of substrate RNAs? RNA 25, 17–22 (2019).
Liu, J. L. et al. Coilin Is Essential for Cajal Body Organization in Drosophila melanogaster. Mol. Biol. Cell 20, 1661 (2009).
Fox, A. H. et al. Paraspeckles: A Novel Nuclear Domain. Curr. Biol. 12, 13–25 (2002).
Zhang, B. et al. The lncRNA Malat1 Is Dispensable for Mouse Development but Its Transcription Plays a cis-Regulatory Role in the Adult. Cell Rep. 2, 111–123 (2012).
Dias, A. P., Dufu, K., Lei, H. & Reed, R. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 1, 1–10 (2010).
Wegener, M. & Müller-McNicoll, M. Nuclear retention of mRNAs – quality control, gene regulation and human disease. Semin Cell Dev. Biol. 79, 131–142 (2018).
McInnes, L., Healy, J. & Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction (2018).
McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 3, 861 (2018).
Wippich, F. et al. Dual Specificity Kinase DYRK3 Couples Stress Granule Condensation/Dissolution to mTORC1 Signaling. Cell 152, 791–805 (2013).
Martins, S. B. et al. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat. Struct. Mol. Biol. 18, 1115–1123 (2011).
Girard, C. et al. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 3, 1–12 (2012).
Shin, C., Feng, Y. & Manley, J. L. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427, 553–558 (2004).
Kedersha, N. & Anderson, P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc. Trans. 30, 963–969 (2002).
Khong, A. et al. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell 68, 808–820.e5 (2017).
Zhang, M. et al. SRRM2 phase separation drives assembly of nuclear speckle subcompartments. Cell Rep. 43, 113827 (2024).
Twyffels, L., Gueydan, C. & Kruys, V. Shuttling SR proteins: more than splicing factors. FEBS J. 278, 3246–3255 (2011).
Söding, J., Zwicker, D., Sohrabi-Jahromi, S., Boehning, M. & Kirschbaum, J. Mechanisms for Active Regulation of Biomolecular Condensates. Trends Cell Biol. 30, 4–14 (2020).
Paul, S. et al. Nuclear speckle-localized RNAs exhibit preferential positioning and orientation. bioRxiv 2022.10.17.512423 https://doi.org/10.1101/2022.10.17.512423 (2022).
Saavedra, C., Tuug, K. S., Amberg, D. C., Hopper, A. K. & Cole, C. N. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 10, 1608–1620 (1996).
Zander, G. et al. mRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature. 540, 593–596 (2016).
Moon, S. L. et al. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol. 21, 162–168 (2019).
Mateju, D. & Chao, J. A. Stress granules: regulators or by-products? FEBS J. 289, 363–373 (2022).
Boutz, P. L., Bhutkar, A. & Sharp, P. A. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 29, 63–80 (2015).
Shalgi, R., Hurt, J. A., Lindquist, S. & Burge, C. B. Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep. 7, 1362–1370 (2014).
de Oliveira Freitas Machado, C. et al. Poison cassette exon splicing of SRSF6 regulates nuclear speckle dispersal and the response to hypoxia. Nucleic Acids Res 51, 870–890 (2023).
Hung, C. L. K. et al. A patient-derived cellular model for Huntington’s disease reveals phenotypes at clinically relevant CAG lengths. Mol. Biol. Cell 29, 2809–2820 (2018).
Jain, A. & Vale, R. D. RNA phase transitions in repeat expansion disorders. Nature. 546, 243–247 (2017).
Lester, E. et al. Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron 109, 1675–1691.e9 (2021).
Yusa, K., Zhou, L., Li, M. A., Bradley, A. & Craig, N. L. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl Acad. Sci. USA 108, 1531–1536 (2011).
Kaewsapsak, P., Shechner, D. M., Mallard, W., Rinn, J. L. & Ting, A. Y. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. Elife 6, e29224 (2017).
Randolph, L. N., Bao, X., Zhou, C. & Lian, X. An all-in-one, Tet-On 3G inducible PiggyBac system for human pluripotent stem cells and derivatives. Sci. Rep. 7, 1–8 (2017).
Türker, C. et al. B-fabric: The Swiss army knife for life sciences. Advances in Database Technology - EDBT 2010 - 13th International Conference on Extending Database Technology, Proceedings 717–720 https://doi.org/10.1145/1739041.1739135 (2010).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Hershberg, E. A. et al. PaintSHOP enables the interactive design of transcriptome- and genome-scale oligonucleotide FISH experiments. Nat. Methods. 18, 937–944 (2021).
Beliveau, B. J. et al. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl Acad. Sci. USA 115, E2183–E2192 (2018).
Passaro, M. et al. OligoMinerApp: a web-server application for the design of genome-scale oligonucleotide in situ hybridization probes through the flexible OligoMiner environment. Nucleic Acids Res 48, W332–W339 (2020).
Xu, Q., Schlabach, M. R., Hannon, G. J. & Elledge, S. J. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl Acad. Sci. USA 106, 2289–2294 (2009).
Kishi, J. Y. et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Methods. 16, 533–544 (2019).
Luo, Y. et al. Stable enhanced green fluorescent protein expression after differentiation and transplantation of reporter human induced pluripotent stem cells generated by AAVS1 transcription activator-like effector nucleases. Stem Cells Transl. Med 3, 821–835 (2014).
Oceguera-Yanez, F. et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods 101, 43–55 (2016).
Haupt, A. et al. Endogenous protein tagging in human induced pluripotent stem cells using CRISPR/Cas9. J. Visual. Exp. 2018, e58130 (2018).
Roberts, B. et al. Systematic gene tagging using CRISPR/Cas9 in human stem cells to illuminate cell organization. Mol. Biol. Cell 28, 2854–2874 (2017).
Liu, J., Xu, Y., Stoleru, D. & Salic, A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl Acad. Sci. USA 109, 413–418 (2012).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Liao, Y., Smyth, G. K. & Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47, e47–e47 (2019).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14, 417–419 (2017).
Zhu, A., Srivastava, A., Ibrahim, J. G., Patro, R. & Love, M. I. Nonparametric expression analysis using inferential replicate counts. Nucleic Acids Res 47, e105–e105 (2019).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Li, H. D., Funk, C. C. & Price, N. D. IREAD: A tool for intron retention detection from RNA-seq data. BMC Genom. 21, 1–11 (2020).
Han, H. et al. Multilayered Control of Alternative Splicing Regulatory Networks by Transcription Factors. Mol. Cell 65, 539–553.e7 (2017).
Tapial, J. et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res 27, 1759–1768 (2017).
Vaquero-Garcia, J. et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife 5, e11752 (2016).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G. D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984 (2010).
Kucera, M., Isserlin, R., Arkhangorodsky, A. & Bader, G. D. AutoAnnotate: A Cytoscape app for summarizing networks with semantic annotations. F1000Research. 5, 1717 (2016).
Zhang, B. et al. A Novel RNA Motif Mediates the Strict Nuclear Localization of a Long Noncoding RNA. Mol. Cell Biol. 34, 2318–2329 (2014).
Benoit Bouvrette, L. P., Bovaird, S., Blanchette, M. & Lécuyer, E. oRNAment: a database of putative RNA binding protein target sites in the transcriptomes of model species. Nucleic Acids Res 48, D166–D173 (2020).
Ietswaart, R. et al. Genome-wide quantification of RNA flow across subcellular compartments reveals determinants of the mammalian transcript life cycle. Mol. Cell. 84, 2765–2784.e16 (2024).
Périer, R. C., Junier, T. & Bucher, P. The Eukaryotic Promoter Database EPD. Nucleic Acids Res 26, 353–357 (1998).
Maćkiewicz, A. & Ratajczak, W. Principal components analysis (PCA). Comput Geosci. 19, 303–342 (1993).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 16, 1226–1232 (2019).
Sommer, C., Straehle, C., Kothe, U. & Hamprecht, F. A. Ilastik: Interactive learning and segmentation toolkit. Proceedings - International Symposium on Biomedical Imaging 230–233 https://doi.org/10.1109/ISBI.2011.5872394 (2011).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods. 18, 100–106 (2020).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50, D543–D552 (2022).
Tschan, A. B. & McIntyre, A. B. R. Accompanying Code for the Publication ‘Phosphorylation of a nuclear condensate regulates cohesion and mRNA retention’. Zenodo resource at https://doi.org/10.5281/ZENODO.14082902 (2024).
Acknowledgements
We thank all members of the Pelkmans Lab for discussions and input, particularly Cheng-Han Yang and Quentin Szabo for their contributions to the development of the smFISH protocol, and Raffaella Gallo and Yixuan Fu for DYRK3 plasmids. We also thank Roland Wenger, Patrick Spielmann, and Agnieszka Jucht for assistance with hypoxia experiments. We thank the Functional Genomics Center Zurich for support in performing proteomics and sequencing experiments (in particular Catherine Aquino, Antje Dittmann, Tobias Kockmann, Laura Kunz, and Timothy Sykes), and the Center for Microscopy and Image Analysis for training for FRAP experiments and for FACS sorting. A.M. was supported by EMBO ALTF 455-2020, MSCA individual fellowship 101029269, and the University of Zurich (FK-20-119, 2024_Q1_CG_010). K.M was supported by EMBO ALTF 978-2019 and is supported by HFSP long term fellowship LT000086/2020-L. This work was supported by the Swiss National Science Foundation (grant 310030_192622, L.P.) and the University of Zurich.
Author information
Authors and Affiliations
Contributions
Conceptualization, A.M., L.P., A.T.; Methodology, K.F., A.M., L.P., A.T.; Investigation, A.M., K.M., A.R., A.T., S.W.; Writing, A.M., L.P., A.T.; Visualization, A.M., A.T.; Funding Acquisition, L.P.
Corresponding authors
Ethics declarations
Competing interests
L.P. consults for Dewpoint Therapeutics and has ownership interest in Sagimet Biosciences and Apricot Therapeutics. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
McIntyre, A.B.R., Tschan, A.B., Meyer, K. et al. Phosphorylation of a nuclear condensate regulates cohesion and mRNA retention. Nat Commun 16, 390 (2025). https://doi.org/10.1038/s41467-024-55469-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-55469-3