Abstract
Riboswitches are ligand-responsive gene-regulatory RNA elements that perform key roles in maintaining cellular homeostasis. Understanding how riboswitch sensitivity to ligand (EC50) is controlled is critical to explain how highly conserved aptamer domains are deployed in a variety of contexts with different sensitivity demands. Here we uncover roles by which RNA folding dynamics control riboswitch sensitivity in cells. By investigating the Clostridium beijerinckii pfl ZTP riboswitch, we identify multiple mechanistic routes of altering expression platform sequence and structure to slow RNA folding, all of which enhance riboswitch sensitivity. Applying these methods to riboswitches with diverse aptamer architectures and regulatory mechanisms demonstrates the generality of our findings, indicating that any riboswitch that operates in a kinetic regime can be sensitized by slowing expression platform folding. Our results add to the growing suite of knowledge and approaches that can be used to rationally program cotranscriptional RNA folding for biotechnology applications, and suggest general RNA folding principles for understanding dynamic RNA systems in other areas of biology.
Similar content being viewed by others
Introduction
Riboswitches are RNA sensors that regulate gene expression based on the cellular concentration of their cognate ligand1. Riboswitches comprise two functional domains, a ligand-binding aptamer domain (AD) and a gene-regulatory expression platform (EP), which interface to enable ligand-dependent cis-regulation of downstream genes2. Across bacteria, riboswitches are integrated into diverse gene-regulatory circuits that maintain cellular homeostasis by sensing a wide variety of metabolites, ions, enzyme cofactors, amino acids, uncharged tRNAs, and other signaling molecules3,4. Because they control central housekeeping functions within pathogenic organisms, there is growing exploration of riboswitches as antibiotic targets5,6,7,8. Riboswitches have also served as excellent model systems to study RNA structure/function relationships9,10,11,12,13,14,15, and increased understanding of riboswitch mechanisms has enabled them to be engineered in biotechnology applications16,17,18, most notably as biosensors for environmental19,20 and human health biomarkers21,22.
A central goal of riboswitch biology is to understand the molecular mechanisms used by these ubiquitous RNA sensors to make gene-regulatory decisions. While much attention has been focused on understanding how properties of the AD, such as the KD of ligand binding, govern riboswitch function23, recent studies have found that ligand-binding interactions alone are poor predictors of riboswitch regulatory potential24,25,26,27. These findings reveal shortcomings in our current understanding of the biochemical principles underlying riboswitch functional properties, namely sensitivity, quantified as the concentration of ligand required for 50% gene expression induction or EC50, and dynamic range, quantified as the fold-change of gene expression between ON and OFF states.
Broadly, it has been well known that transcriptional dynamics, including transcription speed and RNAP pausing, can affect riboswitch function by altering the RNA cotranscriptional folding landscape28,29,30,31,32. However, recent work is showing that the RNA folding kinetics of the EP itself can influence function as well. For example, we recently showed that introducing kinetic barriers to EP folding via internal strand displacement resulted in quantitative increases in the dynamic range of the Clostridium beijerinckii pfl (Cbe pfl) ZTP riboswitch33. However, it is an open question whether EP folding can control riboswitch sensitivity, or if sensitivity is solely governed by the ligand-binding AD.
Here, we investigate this question through the detailed study of several classes of transcriptional riboswitches. Transcriptional riboswitches regulate transcription termination34, which requires these riboswitches to fold into a ligand-binding competent AD structure, sample the environment for ligand, and undergo ligand-dependent global structural rearrangements to produce a genetic decision, all during active transcription35. Transcriptional riboswitches operate in a kinetic regime, meaning that they feature a ligand-binding time window that kinetically constrains the ligand:AD binding interaction before the gene-regulatory decision is made23,29,30,36. This means that all transcriptional riboswitches must execute their genetic decision within the ms-s timeframe before RNAP escapes the termination site37, which generally results in these riboswitches having EC50 values much higher than would be expected based on AD:ligand equilibrium binding alone (KD)30. Given this feature, we hypothesize that increasing the ligand-binding time window of transcriptional riboswitches by altering EP folding kinetics could serve to enhance the sensitivity (lower the EC50) of these kinetically operated riboswitches to more closely approach KD.
In this work, we first investigate this hypothesis through an in vivo functional mutagenesis approach applied to the Cbe pfl ZTP riboswitch. Cbe pfl is an ideal model system, as its switching mechanism features a well-characterized 7–10 nt ligand-binding time window extending from the initial formation of the ligand binding-competent AD to the nucleation of the EP via the closing of an RNA hairpin loop that ultimately results in the formation of an intrinsic terminator hairpin via strand displacement (Fig. S1a)11,38,39. Strand displacement is a process of nucleic acid structural rearrangement characterized by the simultaneous disruption of an existing helix and the formation of a new, mutually exclusive helix40. In strand displacement nomenclature, the existing helix is composed of a substrate strand bound by an incumbent strand. The strand displacement reaction then happens when an invader strand associates with the substrate strand by evicting the incumbent strand via successive base pair exchanges in a ‘break-one-form-one’ manner41. Strand displacement is made more efficient when the incumbent can initiate this process by pairing with a free ‘toehold’ region that seeds the displacement reaction. In the case of Cbe pfl, the base of the P3 hairpin acts as the initial substrate-incumbent duplex, and the 3’ portion of the EP terminator acts as the invader; when the invader strand disrupts the P3 stem of the apo-AD, the terminator hairpin can fold and attenuate transcription (Fig. S1)39. We found that altering the length and sequence of the Cbe pfl EP terminator loop finely controls riboswitch sensitivity. Extending this single-stranded region decreases EC50, as does the presence of A-A stacking interactions near the 3’ end of this domain.
Unexpectedly, we discover a functional tradeoff between sensitivity and dynamic range for this riboswitch, which we show by systematic mutational analysis to be mediated by A-A stacking within the EP loop region. Functional characterization of 3,289 different EP loop sequences using a high-throughput FACS-seq approach showed that this tradeoff is inherent to the Cbe pfl EP architecture. Applying a synthetic approach, we design a remote toehold EP architecture that escapes this functional tradeoff, enabling simultaneous enhancement of sensitivity and dynamic range. Bioinformatic investigation of natural ZTP riboswitch EPs further reveals that a diversity of structural approaches are used in nature to tune ZTP riboswitch sensitivity, all involving changes to the EP that are predicted to alter EP folding pathway kinetics.
We next seek to generalize our findings by investigating other riboswitches, beginning with a ZTP riboswitch from Pectobacterium carotovorum that regulates translation. The introduction of kinetic barriers to EP nucleation in this riboswitch enhances sensitivity, indicating that translational ZTP riboswitches can also operate in a kinetic, cotranscriptional regime. Synthetic transcriptional variants of this riboswitch are also sensitized by similar EP changes, approaching the same limit of EC50 independent of regulatory mechanism.
Finally, we apply principles learned from ZTP riboswitches to rationally design EP kinetic barrier mutations for a range of transcriptional riboswitches from other classes, including the pbuE and yxjA purine riboswitches and the crcB fluoride riboswitch. These mutations all result in sensitization, with trends matching those found in the ZTP system. Surprisingly, we find that there is a limit to the EC50 that can be achieved with kinetically driven switches, suggesting that in vivo cotranscriptionally folded ADs access structural states that have weaker ligand affinities than those characterized with refolded RNA in vitro.
Taken together, these results suggest a general principle by which EP folding kinetics can control riboswitch sensitivity for diverse kinetically-operated riboswitches. This finding may help explain the diversity of natural riboswitch EPs, give insights into riboswitches as antibiotic targets, and provide routes to rationally engineering riboswitches for a range of applications. Given that diverse non-coding RNAs have also been shown to utilize kinetically driven folding pathways similar to riboswitches42, these results add to our generalized understanding of how dynamic RNA folding can impact RNA function43.
Results
Cbe pfl terminator loop length and sequence tune riboswitch sensitivity
The Cbe pfl ZTP riboswitch AD ligand binding pocket is formed by a pseudoknot between the P3 stemloop and an upstream helix-junction-helix motif (Fig. 1a)44. This ligand-binding competent structure is mutually exclusive with the formation of the EP, which consists of a long intrinsic terminator stem involving all of the nucleotides from P339 (Figs. S1b, c). The cotranscriptional folding pathways of three ZTP riboswitches have been biophysically characterized11,38,39. Together, these studies indicate that the ligand-binding window opens shortly after the 3’ end of the P3 stemloop emerges from the RNAP exit channel, remains open during transcription of the downstream ~7–10 nt (including the EP loop sequence), and closes when the first 2–3 nts of the EP invader domain nucleate the strand displacement of P3, after which the AD loses the ability to bind ligand38 (Fig. S1a). Therefore, the time constraint posed by this short ligand-binding window should be determined by both the amount of time required to transcribe the EP loop sequence between P3 and the invader domain, and the rate of the RNA folding step that nucleates EP strand displacement. We hypothesized that both of these factors could be affected by EP loop length, since longer loops will take longer to transcribe and face an increased entropic cost to close45.
a Secondary structure schematic of the Cbe pfl riboswitch immediately preceding EP nucleation. The polyA terminator loop (purple) closes to allow the invader domain (gold) to initiate strand displacement of the P3 stemloop (red). Ligand binding kinetically competes with this process to determine the riboswitch genetic decision. b EC50 values for riboswitch variants with homopolymeric loops of varying lengths. c Normalized dose response curves of riboswitch variants with varying polyA loop lengths. d Vfold3D models of P3 with un-nucleated 7nt polyA or polyU terminator loops. PolyA (purple, left) exhibits regular stacking that separates the invader nucleotides (gold) from the substrate nucleotides (red) in the P3 stem, whereas polyU (teal) exhibits no regular order. e EC50 values for riboswitch variants with length 7 polyA loops in which one residue has been mutated to either C (red) or G (blue). Nucleotide coordinates are relative to the 3’ end of the loop. Solid horizontal line indicates EC50 value for WT polyA, with dashed horizontal lines indicating the standard error. Cartoon insets illustrate how A→C mutations (top) will disrupt local stacking, while A→G mutations allow local stacking interactions (bottom). f Fold change of the mutants in (e). Solid horizontal line indicates fold change of WT polyA, with dashed horizontal lines indicating the standard error. Data in panel (c) indicates normalized dose response curves over n = 9 replicates, with points indicating mean and error bars indicating standard deviation. Dose response curves were normalized by dividing all data points for each mutant by (Fmax – Fmin) of the fitted curve, with standard deviation calculated from the 9 normalized replicate values for each concentration. Data in panels (b), (e), and (f) are determined as described in Methods, from dose response curves including n = 9 biological replicates at each of 8 concentrations. Error bars in (b), (e), and (f) indicate standard error of the fit parameter. Source data are provided as a Source Data file.
To test this hypothesis, we varied the 7 nt polyA loop of Cbe pfl (Fig. 1a) from 3 nt to 16 nt, and altered the sequence from polyA to polyU and polyC. We measured dose-response curves in vivo, and fit them to the Hill equation (see “Methods”), allowing us to characterize sensitivity (EC50), and fold change (ratio of the maximum gene expression activation to the minimum) for each variant. We observed that extending homotypic EP loops enhances riboswitch sensitivity (Fig. 1b–c), with polyA loops resulting in lower EC50 relative to polyU/C loops of the same length (Fig. 1b). Unsurprisingly, loop extension also resulted in increases in uninduced expression (leak, Fmin) (Figs. S4a–c), since longer loops decrease hairpin stability46, thus affecting termination efficiency47 (Fig. S4d).
While any loop sequence should slow initial EP nucleation as a function of length by increasing the entropic cost of loop closure, we reasoned that polyA stacking interactions within the loop may slow the rate of loop closure further by introducing an additional energetic barrier (Fig. 1d)48,49,50. To test this interpretation, we performed a mutational scan of the 7 nt polyA loop in which each residue was mutated to C, which should destabilize neighboring A-A stacking interactions51,52. We observed that A→C mutations near the 3’ end of the loop region resulted in desensitization (Fig. 1e). Performing the same mutational scan with A→G mutations, which should maintain neighboring stacking interactions, showed that these mutations did not affect sensitivity (Fig. 1e), supporting our hypothesis that stacking interactions within the EP loop enhance riboswitch sensitivity by slowing loop closure kinetics.
We further tested these trends by chimerically replacing the 7A loop of Cbe pfl with sequence variants of the 12 nt A-rich domain from the B. subtilis queC preQ1 riboswitch (Bsu queC) found by NMR to have A-A stacking interactions that rigidify the domain51, and observed similar trends (Fig. S5). These results support the conclusion that polyA stacking within the loop region, especially near the 3’ end, extends the ligand-binding window by slowing strand displacement nucleation.
Surprisingly, the A→C disruptions to stacking in the 7A polyA loop that resulted in desensitization also resulted in marked increases in dynamic range, while A→G mutations had neither effect (Fig. 1f). The same relationship was observed in the Bsu queC mutants (Figs. S5b, c), suggesting that the effects of disruptions to A-A stacking on sensitivity and fold-change are somehow coupled in an apparent tradeoff relationship.
Taken together, these results support our hypothesis that EP loop length and sequence composition controls riboswitch function, likely through posing kinetic barriers (entropic or energetic) to loop closure that extend the ligand-binding window. However, disrupting EP loop stacking in Cbe pfl has the apparently coupled effect of decreasing sensitivity and increasing dynamic range, which we sought to investigate further.
PolyA loop stacking mediates tradeoff between sensitivity and dynamic range
To interrogate the mechanistic basis by which disruptions to A-A stacking interactions within the Cbe pfl EP terminator loop result in apparently coupled increases in EC50 and fold change, we performed A→C mutational scans on a variety of polyA loop lengths, and observed similar phenotypic patterns regardless of overall loop length (Fig. 2a–c). Consistently, A→C mutations near the 3’ end of the loop had the greatest effects in simultaneously decreasing sensitivity and increasing fold change (Fig. 2b, c). Enhancements in fold change were caused by dramatic position-dependent increases in ON-state gene expression for these mutants (Figs. S6a–f). Previous work in the ZTP system showed that disfavoring the EP strand displacement process by disrupting the C100:G108 nucleating base pair results in similar dramatic increases in ON-state gene expression33, leading us to hypothesize that disrupting A-A stacking interactions within the terminator loop may also disfavor downstream strand displacement.
a Secondary structure schematic of A→C mutational scans in which riboswitch variants with different length polyA loops were mutated at each position within the loop from A to C to disrupt local A-A stacking. Positional coordinates refer to the distance from the 3’ end of the loop. b EC50 measurements for mutational scans of different length terminator loops into which single A→C mutations have been made at every position. c Fold change of the mutational scans in (b). d EC50 measurements of riboswitch variants with polyA loops of varying lengths into which either no mutations have been made (polyA), the third to last residue relative to the 3’ end of the loop has been mutated to C (−3C), the initial invading residue G108 has been mutated to C (G108C), or both mutations have been made (−3C G108C). e Fold change of the mutational scans in (d). f Vfold3D models of the nucleated Cbe pfl terminator loop featuring either a length 7 or length 3 polyA loop. The longer loop allows continuous stacking of A residues within the 3’ end of the loop and the G108 invader residue, while the shorter loop does not allow enough slack for stacking to occur in this conformation. Additional loop length models in Fig. S7. Data in (b), (c), (d) and (e) are determined as described in Methods, from dose response curves including n = 9 biological replicates at each of 8 concentrations. Error bars in (b), (c), (d) and (e) indicate standard error of the fit parameter. Source data are provided as a Source Data file.
To test this hypothesis, we extended loop length in the presence of the G108C mismatch mutation (G108C), an A→C mutation at the -3 position relative to the 3’ end (-3C), or both -3C and G108C (-3C G108C). While all of the loop extensions resulted in similar desensitization compared to perfect polyA across loop lengths (Fig. 2d), we observed key differences in the fold change phenotypes between these mutational series (Figs. 2e, S6g–i). While G108C enhances fold change regardless of loop length as expected, -3C mutations do not enhance fold change for loops shorter than 6 nt (Fig. 2e). At the same time, perfect polyA loops shorter than 6 nt show enhanced fold change relative to longer loops, caused by large increases in ON-state gene expression (Fig. 2e, S4a). Taken together, these findings suggest that a minimum loop length of around 6 nts is required for 3’ A-A stacking to suppress fold change.
To try to understand why a minimum loop length is required, we performed 3D visualizations of the nucleated terminator hairpin with polyA loops of varying lengths using Vfold3D. While this modeling approach is limited in its ability to assess the molecular dynamics of single stranded regions, the visualizations it produced suggest a putative stacking platform of A residues within the 3’ side of the terminator loop, which was not present in loops shorter than 6 nt (Figs. 2f, S7). If present, stacked residues within this 3’ region could align with G108 to provide an energetic backstop to reverse branch migration, analogous to the ‘cooperative toehold’ approach in DNA nanotechnology53, increasing the overall forward rate of strand displacement and decreasing ON-state gene expression. On the other hand, if loops are too short, the visualizations suggest that there is insufficient slack to align multiple parallel purine rings below G108 (Figs. 2f, S7), potentially explaining the experimental observation that the fold change of short polyA loop variants is not affected by -3C mutations (Fig. 2e). Importantly, we observe that -3C in the G108C background results in no additional increase in fold change regardless of loop length (Fig. 2e). This apparent epistasis of G108C over -3C is consistent with the hypothesis that a 3’ A-A stacking platform in the terminator loop stabilizes G108 to enhance strand displacement efficiency.
Previous studies have found that toeholds, base-paired regions preceding strand displacement initiation sites, are potent enhancers of strand displacement efficiency41. To test if a toehold would suppress riboswitch fold change in the same manner as 3’ A-A stacking, we performed a mutational series in which a length 13 polyA loop was progressively shortened with A→U mutations, converting loop sequence into toehold base pairs, either in the A:U or U:A orientation (Fig. S8a). Both 5’ and 3’ mutational series resulted in progressive desensitization, as initial loop nucleation should be made much easier by the presence of a toehold region (Fig. S8b). However, we observed that while 5’ A→U mutations, which preserve 3’ A-A stacking, decreased fold change (Figs. S8c, e), 3’ A→U mutations that break 3’ A-A stacking greatly increased fold change (Figs. S8c, d). These results suggest that 3’ A-A stacking preceding G108 also enhances strand displacement in the context of toehold-mediated strand displacement, extending our model of stacking-mediated branch migration enhancement. In a complementary experiment, we added toehold base pairs to a length 7 polyA loop in either the A:U or U:A orientation, which revealed similar trends (Fig. S8f–l). Together, these data indicate that even in the context of toehold-mediated strand displacement, stacking interactions immediately preceding the invader domain can enhance branch migration and suppress dynamic range.
These results suggest that there are two separable mechanistic bases by which disrupting stacking within the 3’ end of the loop simultaneously alters sensitivity and fold change: i) disrupting stacking increases the rate of loop closure, shortening the ligand-binding window time and thus decreasing sensitivity; and ii) disrupting stacking decreases EP strand displacement efficiency, thus increasing fold change by increasing ON-state gene expression.
Remote toehold EP architecture uncouples tradeoff between sensitivity and dynamic range
To evaluate whether the apparent tradeoff between sensitivity and fold change is an inherent relationship for the Cbe pfl EP architecture, or mainly a result of the polyA EP loop, we randomized the loop region and performed FACS-seq54,55,56 to construct dose response curves for 3289 length 7 nt loop variants (Fig. S9). Characterizing variants at this scale allowed us to construct a functional landscape, mapping riboswitches onto EC50 and fold change axes (Fig. S10a). This landscape revealed a tradeoff between sensitivity and dynamic range (Fig. S10a), extending our model to suggest that this tradeoff between EC50 and fold change is inherent to the Cbe pfl EP loop-only architecture.
Based on this observation, we next sought to redesign the EP architecture to separate these two phenotypes by including a remote toehold. Remote toehold systems use a design concept from DNA nanotechnology consisting of a toehold connected to an invader by an extendible tether that slows down strand displacement by imposing an entropic cost on the invader:substrate duplex, allowing fine-tuning of strand displacement kinetics57. We applied the remote toehold architecture in the context of the ZTP riboswitch by modifying the EP to consist of a synthetic toehold hairpin separated from the AD and invader by variable-length polyA tethers on both sides (Fig. 3a, b). Extending either one or both of these tethers should simultaneously impose two penalties: a kinetic penalty on the initial nucleation of P3 strand displacement, thus enhancing sensitivity, and an entropic penalty on the invader:substrate duplex, enhancing dynamic range53,57.
a Schematic of the remote toehold EP design, in which the polyA loop of Cbe pfl is replaced by a synthetic hairpin connected to the AD and invader by single stranded extendable polyA tethers. b Vfold3D model of a nucleated terminator featuring a remote toehold extension with a 5A 5’ tether and a 1A 3’ tether (5:1). c Dose responses of select remote toehold variants with varying length 5’ tethers, including the 5:1 design shown in (b). Vertical lines indicate EC50 values of the curve with the corresponding color. d EC50 measurements for remote toehold variants with varying length 5’ and 3’ tethers. n:1 indicates the length of the 5’ tether has been extended, with the length of the 3’ tether fixed at 1A. 1:m indicates the length of the 3’ tether has been extended, with the length of the 5’ tether fixed at 1A. n:m indicates the length of both tethers have been simultaneously extended. e Fold change measurements for variants in (d). f EC50 and fold change data from (d) and (e) plotted against each other. Increasing opacity represents longer overall tether length. Data in panel (c) are dose response curves over n = 9 biological replicates, with points indicating mean and error bars indicating standard deviation. Data in panels (d), (e), and (f) are determined as described in Methods, from dose response curves including n = 9 biological replicates at each of 8 concentrations. Error bars in (d), (e), and (f) indicate standard error of the fit parameter. Source data are provided as a Source Data file.
Extending these tethers on either or both sides results in dual enhancement of sensitivity and dynamic range (Fig. 3c–e). Increasing tether length enhances sensitivity as predicted (Fig. 3d), and fold change is enhanced by increases in ON-state gene expression, especially for 5’ tether extensions, up to a point where increasing tether length causes baseline leak expression to increase (Fig. S11). This results in the remote toehold architecture variants showing a Pareto front-type of tradeoff with better sensitivity and fold-change than the loop variants characterized by FACS-seq (Figs. 3f, S10b).
These findings led us to conclude that the apparent tradeoff between sensitivity and dynamic range in Cbe pfl is not inherent to the ZTP riboswitch itself, but rather arises from the WT Cbe pfl EP loop-only architecture that involves the same loop nucleotides in both the initial EP nucleation attempts and the downstream branch migration process needed to form a complete terminator. Redesigning the EP architecture allowed us to achieve dual improvement in sensitivity and dynamic range that was precluded in the loop-only architecture.
Natural ZTP riboswitch EPs use multiple approaches to enhance sensitivity
The amenability of Cbe pfl to drastic EP changes motivated us to investigate whether natural ZTP riboswitch EPs feature architectures beyond the loop-only EP common to the model ZTP switches previously characterized11,38,39,58. To do so, we bioinformatically identified all transcriptional ZTP riboswitches annotated in the Rfam entry RF01750 with a similar invader:P3 strand displacement fold as Cbe pfl (287 sequences; see “Methods”). Using secondary structure prediction of the spacer regions between P3 and the invader domain, we classified EPs into three architectural subtypes: 1) unstructured loop-only like Cbe pfl (158 sequences), 2) remote toehold EPs separated from the AD or invader by mismatches or internal loops (86 sequences), or 3) direct toehold EPs including 2 or more base pairs preceding the invader:P3 junction that could serve as toeholds of EP strand displacement (43 sequences) (Fig. 4a, b).
a The EPs of 1755 putative ZTP riboswitches were analyzed for the presence of a downstream polyU tract (730/1755), the presence of a consensus P3 stem (695/730), and the presence of an invader domain that pairs with P3 and competes with the pseudoknotted region for base pairs (287/695). These 287 EP candidates were manually classified into 3 categories based on the spacer region between P3 and the invader as indicated in panel (b): loop only, remote toehold, and direct toehold. A complete table of the 287 EP candidates and their classifications is provided in the Source Data file. (c) Secondary structure schematic of a panel of loop only, direct toehold, and remote toehold EPs (color coding as in panels (a) and (b) selected for further functional characterization. Illustrated regions were chimerically inserted in place of the Cbe pfl polyA terminator loop. The Rsp spacer region is a remote toehold because the invader includes a G108∆ deletion, so G108 was deleted from the invader region in the Rsp chimera. A table of the full sequences of these riboswitches and their genomic loci can be found in Table S1. d In vivo fluorescence data for chimeric riboswitches ordered according to spacer length, with Cbe indicating the WT Cbe pfl sequence. (e) EC50 measurements of chimeric ZTP riboswitches shown in (d). f Fold change of chimeric ZTP riboswitches shown in (e). Bars in panel (d) indicate average Fluorescence/OD600 values over n = 9 biological replicates. Data in (e) and (f) are determined as described in Methods, from dose response curves including n = 9 biological replicates at each of 8 concentrations. Error bars in (e) and (f) indicate standard error of the fit parameter. Source data are provided as a Source Data file.
Loop-only EPs were found to be similar in length to the Cbe pfl loop on average (Fig. S12a), and biased toward being enriched for purines, with many loops possessing 100% purine content (Fig. S12b), suggesting that other ZTP riboswitches may also employ stacking-mediated sensitivity enhancement like Cbe pfl. Direct toehold EPs are shorter in overall length and significantly enriched for AU content relative to remote toehold EPs, and rarely exceed 5 bp in length (Figs. S12c–e), suggesting that strong direct toeholds are disfavored relative to weaker toeholds. Additionally, we observed a strong bias in the orientation of direct toehold base pairs, with the three nearest pairs to the invasion site being significantly enriched for R-Y orientation relative to Y-R (Figs. S12f, g). Based on our observation that U-A toeholds result in very poor dynamic range relative to A-U toeholds (Figs. S8c, h, j), we reasoned that these ZTP EPs are under purifying selection to avoid suppressing ON-state gene expression.
To test how these natural EP configurations contribute to riboswitch function, we constructed chimeric riboswitches by grafting the spacer regions from various natural sequences in place of the 7 nt polyA loop in Cbe pfl. We selected 4 loops longer than 7 nt, a relatively long direct toehold, and 3 remote toeholds of varying lengths (Fig. 4c). We observed that all of the spacer regions supported efficient switching by the chimeric riboswitches (Fig. 4d), and that the length of the spacer region was correlated with riboswitch sensitivity (Fig. 4e). Mechanistically, these extremely long spacer regions may delay EP nucleation by simply consuming transcription time. Average transcription speeds of RNAP have been measured between 50–100 nt/s59, meaning that the longest natural ZTP spacer regions may extend the ligand-binding window by an entire second before the invader domain emerges from RNAP. As in the case of the synthetic remote toehold (Fig. 3), sensitivity enhancement by natural remote toeholds did not result in suppression of fold change (Fig. 4f), supporting our earlier conclusion that the remote toehold architecture uncouples the sensitivity-dynamic range tradeoff observed in the loop-only architecture. In fact, the Dba chimera has better sensitivity and fold change than any of the synthetic remote toehold variants.
Together, these analyzes reveal that the principles uncovered by functional mutagenesis of the Cbe pfl ZTP riboswitch can help explain the sequence diversity of transcriptional ZTP riboswitch EPs, and that these natural riboswitches have evolved to exploit multiple EP architectural solutions to tune riboswitch sensitivity.
Kinetics of EP nucleation also control translational ZTP riboswitch sensitivity
Natural ZTP riboswitch remote toehold architectures reminded us of the layout of Pectobacterium carotovorum rhtB (Pca rhtB), a translational ZTP riboswitch previously characterized by Breaker and colleagues, which controls translation initiation by using the start codon of the rhtB gene as part of its EP invader domain (Fig. 5a)58. Translational riboswitches are generally thought to operate as thermodynamic switches29,36, although growing evidence is suggesting that some riboswitches may control translation initiation in a kinetic regime60,61. If true, such translational riboswitches should feature ligand-binding time windows, and therefore should be able to be sensitized by slowing EP nucleation. We therefore sought to test this prediction through functional characterization of Pca rhtB EP variants using the principles learned about tuning sensitivity from studying transcriptional ZTP riboswitches.
a Schematic of the Pca rhtB ZTP riboswitch, which regulates translation initiation. In this mechanistic model based on findings of Breaker and colleagues58, a ribosome binding site (RBS) is located within the distal portion of the EP, while the strand displacement switching mechanism is initiated by the G of the start codon as the invading nucleotide. Arrows indicate the positions where optional polyA bulges were inserted: proximal (purple, 77-An-78) or distal (teal, 80-An-81). b Single-round in vitro transcription reactions of the Cbe pfl and Pca rhtB riboswitches in the presence or absence of ZMP (1 mM). Filled triangles indicate the terminated length, and open triangles indicate full-length product based on the length of the linear template DNA. Source gel images of 2 replicates can be found in Source Data. c EC50 measurements of proximal and distal bulge insertion mutants. d Schematic of the synthetic EP designed to convert the Pca rhtB riboswitch to transcriptional regulation (Pca TX). The native RBS was scrambled, and the first 6 codons of the rhtB gene were replaced by a synthetic invader domain coupled to a polyU tract designed to form a functional intrinsic terminator. Arrow indicates the position where optional polyA bulges were inserted. e EC50 measurements of Pca rhtB (Pca TL) and Pca TX bulge variants. f EC50 measurements of chimeric riboswitches Cbe-Pca TL and Cbe-Pca TX which consisted of the EPs of Pca TL and Pca TX grafted onto the AD of Cbe pfl. Data in panels (c), (e), and (f) are determined as described in Methods, from dose response curves including n = 9 biological replicates at each of 8 concentrations. Error bars in (c), (e), and (f) indicate standard error of the fit parameter. Source data are provided as a Source Data file.
We first confirmed that Pca rhtB does not regulate transcription elongation in a ligand-dependent fashion, like Cbe pfl or other transcriptional ZTP riboswitches, by showing that it does not function in an in vitro transcription reaction (Fig. 5b). We next tested the predicted switching mechanism of the Pca rhtB EP by mutating key regions, confirming their importance for switching function (Fig. S13a, S14).
To investigate whether the kinetics of EP nucleation affect riboswitch sensitivity, we introduced polyA bulges near the invader:P3 junction to slow the formation of the predicted start codon-sequestering structure. These polyA bulges were either placed proximal to the strand displacement initiation site, to convert the EP to a remote toehold architecture, or 1 nt distal, to extend the A80:C103 mismatch into an internal loop (Fig. 5a). We observed that proximal polyA bulge extension enhanced sensitivity in a manner similar to remote toehold leash extension, while distal bulge extension also enhanced sensitivity though to a lesser extent (Fig. 5c). As expected, these bulge extension series, which disrupt the region responsible for mediating switching function (Fig. S13a), resulted in length-dependent decreases in fold change caused by increases in riboswitch leak (Figs. S14a–c).
We next sought to confirm that these same polyA bulges would enhance sensitivity in a transcriptional version of this riboswitch that should operate in a kinetic regime. To do so, we converted the Pca rhtB EP to a transcriptional regulatory mechanism (Pca TX) by scrambling the native RBS, extending the native invader domain to fully complement P3, and replacing the 9 codons of the rhtB gene with a polyU tract followed by a synthetic RBS (Fig. 4d). We confirmed by single round IVT that this modified riboswitch functions by regulating transcription termination (Figs. S13b, S14). Interestingly, we found that the Pca TX variant was more sensitive than the wildtype translational version, and importantly was also further sensitized by the insertion of a polyA bulge on the 5’ side of the toehold (Fig. 4e).
To ensure that no unknown features of the Pca rhtB AD were confounding our mutagenesis approach, we constructed chimeric versions of these riboswitches with the Pca rhtB AD replaced by the distantly-related Cbe pfl AD to create translational (Cbe-Pca TL) and transcriptional (Cbe-Pca TX) versions, and observed very similar sensitization patterns (Fig. 4f, S13B). Interestingly, for both Pca rhtB and Cbe pfl ADs, their respective transcriptional and translational versions approach the same EC50 value as the polyA bulge is extended, suggesting that as EP nucleation is slowed, EC50 values approach a common minimum value that does not rely on EP mechanism. All four of these mutational series resulted in expected length-dependent increases in riboswitch leak leading to decreases in fold change (Figs. S14d–i).
Together, these results demonstrate that the sensitivity of translational ZTP riboswitches can be enhanced through EP architecture changes that slow EP nucleation kinetics, generalizing our understanding of kinetically driven riboswitches to include examples of translational regulation.
Delaying EP nucleation sensitizes diverse riboswitches
Finally, we sought to extend our understanding of how EP nucleation kinetics tune riboswitch sensitivity to other riboswitch classes that feature diverse aptamer structures and direct toehold EP architectures, including riboswitches for purines and fluoride (Fig. 6a–c).
a Schematic of the Bsu pbuE* leader truncation variant of the pbuE transcriptional ON riboswitch62. A weak direct toehold facilitates nucleation of the EP, in which an invader (gold) strand displaces P1 (black and red) to form the terminator hairpin. Arrow indicates the position of polyA bulge extension (d) EC50 measurements of pbuE polyA bulge variants. b Schematic of the Bce crcB fluoride transcriptional ON riboswitch. A direct toehold facilitates nucleation of the EP, in which an invader (gold) strand displaces the pseudoknot (black and red) to form the terminator hairpin. Boxes show terminator extension inserts appended to the end of the terminator hairpin. c Schematic of the Bsu yxjA transcriptional OFF riboswitch. A weak toehold comprising P4 and an internal loop facilitates nucleation of the EP, in which an invader (gold) strand displaces P1 (black and red) to form the central helix. The terminator loop (teal) can close to allow a second invader (blue) to displace the central helix (red and gold) and form the terminator hairpin. The second strand displacement process gives this switch OFF logic. Boxes show terminator loop extension variants, and two types of P4 spacer mutations: P4 truncations with different lengths added back, and P4 extensions. e EC50 measurements of crcB terminator extension variants. f EC50 values of yxjA terminator loop extension mutants. g EC50 measurements of yxjA P4 spacer mutations. Data in panels (d), (e), (f), and (g) are determined as described in Methods, from dose response curves including n = 9 biological replicates at each of 8 concentrations. Error bars in (d), (e), (f), and (g) indicate standard error of the fit parameter. Source data are provided as a Source Data file.
The model B. subtilis pbuE riboswitch senses adenine through an AD consisting of a three-way junction, and features a weak toehold connecting the AD and invader (Fig. 6a). We created a remote toehold architecture by inserting a polyA bulge at the strand displacement initiation site (Fig. 6a), and observed that polyA bulge length correlated with enhanced riboswitch sensitivity and fold change in a manner reminiscent of the remote toehold extensions in ZTP (Figs. 6d, Fig. S15a, b). Importantly, these polyA insertions disrupt a polyU tract previously reported to be an important regulator of sensitivity and dynamic range via putative RNAP pausing62,63, although we observe improved sensitivity and dynamic range for all polyA bulge variants relative to WT pbuE (Figs. 6d, S15a, b).
The Bacillus cereus crcB fluoride riboswitch uses an AD consisting of an H-type pseudoknot, and features a 7 bp direct toehold spacer region (Fig. 6b). We extended this direct toehold by chimerically grafting on the long ZTP spacer regions characterized in Fig. 4c, as well as a long (63 nt) spacer region from the Enterococcus pallens eno (Epa) fluoride riboswitch identified using similar bioinformatic analysis as described above (Fig. 6b, Methods). These spacer region extensions substantially sensitized the crcB fluoride riboswitch in a length-dependent manner (Fig. 6e), resulting in commensurate length-dependent decreases in fold change corresponding to increasing riboswitch leak (Figs. S15c, d).
We next sought to apply our findings to transcriptional riboswitches that feature an OFF regulatory logic. In previous work, we developed a synthetic OFF variant of the Cbe pfl ZTP riboswitch by inserting an 8 nt ‘flipping domain’ that inverts the EP regulatory logic (Fig. S16a)33. To test if this OFF-switch could also be sensitized by delaying EP nucleation, we extended the polyA loop and observed length-dependent sensitization similar to what was observed for the WT ON-switch (Figs. S16b, c).
Finally, we investigated the B. subtilis yxjA purine riboswitch, which uses an AD similar to pbuE and which previous work demonstrated to possess two competing strand exchange processes (Fig. 6c)64. We wondered if, like pbuE, the nucleation of the first strand displacement process (central helix) determines the ligand-binding time window, or if rather the nucleation of the terminator hairpin is the ligand-binding window time-determining step. To interrogate this question, we first varied the length of the terminator hairpin loop from 4A to 22A, which had a strong negative effect on dynamic range (Figs. S15e, f), but had little effect on sensitivity (Fig. 6f). Next, we manipulated the long remote toehold that nucleates the central helix. We either shortened this region by deleting the 5’ tether and the P4 stem, or we extended this region by grafting the spacers from ZTP and fluoride riboswitch EPs onto the end of P4. We observed that the shortened P4∆ mutants became less sensitive than WT, and the extended variants all became more sensitive, with EC50 correlating with the overall length of the spacer region (Fig. 6g, S15g, h). These results confirm that the initial strand displacement step that forms the central helix determines the ligand-binding time window, as in pbuE, and that the second strand displacement step, which allows the terminator to form, occurs after the riboswitch has committed to produce a given regulatory outcome.
Taken together, these results demonstrate that the principle of tuning riboswitch sensitivity through EP sequence/structural changes that control EP nucleation kinetics are generalizable, and can be used to sensitize any riboswitch that operates in a kinetic regime.
Minimum achievable EC50 from in vivo kinetic riboswitches differs from in vitro aptamer:ligand KD values
Early discoveries in the riboswitch field demonstrated a large discrepancy between in vitro AD:ligand KD measurements and transcriptional riboswitch EC50 measurements, leading to the general acceptance of the model that EC50 is elevated in these riboswitches because the kinetic constraint of transcription prevents the AD from reaching equilibrium with its ligand29,30. Having discovered and developed a set of approaches to extend the AD:ligand interaction time and sensitize various riboswitch classes, we asked whether these enhancements in sensitivity allow EC50 to approach AD:ligand KD as interaction time increases. Interestingly, for the various methods of sensitization we employed, EC50 seems to asymptotically approach some minimum value for each riboswitch class as the kinetic barrier to EP nucleation increases (Figs. 1b, 3d, 4e, 5e, f, 6b, 6d, 6g, S16c). However, comparing the minimum achievable EC50 values with published KD values reveals a remaining 2-200x discrepancy (Table 1). To ensure that this discrepancy was not caused by membrane effects limiting intracellular ligand concentration in our in vivo assay, we compared our in vivo EC50 measurements to literature values for in vitro EC50, which showed that WT Cbe pfl and Bce crcB replicate in vitro EC50 values almost exactly (Table 1). However, the observed in vivo EC50 for pbuE exceeds the in vitro EC50 by ~10x (Table 1), indicating that membrane permeability may confound our in vivo EC50 measurements for 2AP-sensing riboswitches.
Focusing our attention on Cbe pfl and Bce crcB, we hypothesized that minimum EC50 may be elevated relative to in vitro KD because the cotranscriptional ensembles of these riboswitches feature abundant kinetically trapped states only capable of forming partial ligand contacts within the timeframe of transcription. An immediate upshot of this model is that aptamers with more extensive ligand interactions will have more possible partially bound states of lower affinity than the refolded structure, resulting in a larger discrepancy between cotranscriptional sensitivity and KD. This model is consistent with the observed trend that for Bce crcB, which features relatively simple ligand-binding interactions65, only a ~ 2x minimum EC50:KD ratio is observed (Table 1), while for ZTP riboswitches, which feature more numerous and elaborate ligand-binding interactions66, a ~ 50x minimum EC50:KD ratio is observed (Table 1). These findings emphasize the importance of other factors in limiting transcriptional riboswitch sensitivity besides the kinetic constraint of ligand-binding windows.
Discussion
Riboswitches are highly conserved gene regulatory elements controlling housekeeping and emergency-response functions in bacteria1. Because of their key homeostatic roles, riboswitches must be finely tuned to allow the commensurate amount of gene expression in response to the intracellular concentrations of their cognate ligands. The demanding sensitivity and specificity requirements of ADs seem to be why they are among the most highly conserved RNA domains in all of biology67. This high conservation of ADs could explain the observed natural diversity of EPs. As the more evolutionarily malleable riboswitch domain, EPs enable nearly invariant ADs to regulate entirely different steps of gene expression, and allow ADs with nM affinity to operate in the μM or mM sensitivity ranges68,69. However, the mechanisms by which EP sequence changes tune riboswitch function remain relatively unexplored.
We previously observed that the EP of the Cbe pfl ZTP riboswitch features an invader domain that can quantitatively reprogram riboswitch dynamic range by altering the kinetics of internal strand displacement33. Other work has shown that the length of the pbuE EP toehold can tune dynamic range62, and that nuanced EP sequence features can tune the sensitivity and dynamic range of the Pseudomonas aeruginosa guanidine-II translational riboswitch70. In this work, we identify EP nucleation as a key determinant of riboswitch sensitivity in Cbe pfl, and describe three approaches used by natural ZTP riboswitches to delay EP nucleation and tune sensitivity: purine-rich loops, remote toeholds, and long spacer regions. Together, these findings are forming a complete sequence-structure-function understanding of each component of the ZTP riboswitch EP (Fig. S17), which reveals the outsized role of EP folding in determining all aspects of riboswitch function69.
Importantly, we have shown that principles for tuning riboswitch sensitivity and fold change through manipulating EP folding pathways are general across diverse riboswitches that control different aspects of gene expression. Specifically, we characterized a translational ZTP riboswitch, Pca rhtB, and showed that it is sensitized by the introduction of kinetic barriers to EP nucleation, indicating that this riboswitch operates in a kinetic regime. This finding supports other work showing that some translational riboswitches are kinetically controlled, and are not all thermodynamic switches60. While it is possible that transcription-translation coupling may contribute to the observed kinetic constraint for ligand binding for Pca rhtB71, we did observe similar sensitization patterns when the EP was converted into a transcriptional mechanism. In principle, any translational switch could be forced to operate in a kinetic regime if the EP-nucleated state includes a kinetic trap that blocks re-formation of the ligand-binding competent AD structure, as is true in the case of ZTP riboswitches38. For thermodynamically-operated translational riboswitches, ready interchange between ON and OFF states will be possible even in the presence of the EP, as is true in the case of adenine translational riboswitches29,72.
We extended our findings from the ZTP system to a panel of diverse riboswitches with different ADs and different regulatory logic (ON vs OFF), and show that kinetic barriers that delay EP nucleation result in riboswitch sensitization in all of these cases, demonstrating the generality of our findings. A deeper comparison of observed sensitivity improvements across these diverse riboswitches raises an intriguing question – is there a fundamental limit to the sensitivity of a kinetically driven riboswitch? While we were able to achieve 2.5-12x enhancements in sensitivity for various riboswitches, for most classes the EC50 values of the best performing variants asymptotically approach levels that remain 1–2 orders of magnitude greater than published AD:ligand KD values (Table 1). Moreover, very similar minimum EC50 values are obtained for kinetically driven transcriptional and translation riboswitches that use the same AD (Fig. 5e, f), suggesting that minimum EC50 is a property set by the binding interaction between the ligand and the AD, but is also fundamentally limited by the constraints of a kinetically driven process. This could be due to two reasons: (1) There is a limit to the ability to extend the ligand-binding window time, perhaps due to the fundamental instability of kinetic, transient, out-of-equilibrium RNA folds, and thus there is no chance to ever approach the equilibrium AD:ligand interactions observed in KD measurements of refolded RNAs; or (2) The AD ligand-binding interactions that occur during the out-of-equilibrium transient cotranscriptional RNA folding processes of kinetically driven switches are fundamentally different, with lower affinity than those characterized in vitro with refolded RNA. If the latter model, originally proposed by Crothers and colleagues30, is true, it could give insights into what is needed to develop drugs that target transient cellular RNA structures73.
Conceptually, kinetically-operated riboswitches function more like dimmer switches or fuses than digital switches30,74. This study highlights the key role of RNA folding kinetics, especially those of EPs, in governing the functional properties of riboswitches in terms of sensitivity (EC50) and dynamic range (fold change). Taken together with many discoveries characterizing the nuanced ways in which transcriptional dynamics can reciprocally control RNA folding28,30,31,75,76,77,78, these results emphasize how natural systems exploit the biophysical properties of cotranscriptional RNA folding to generate riboswitch variants that meet the needs of diverse genetic contexts43. This work also begins to generalize our understanding of riboswitch tuning strategies across diverse classes and across different modes of regulation. We anticipate these studies to guide further development of riboswitch-based biotechnologies18,19,20,21,22, RNA-targeting drugs including antibiotics5,6,7,8, and add to our general understanding of how out-of-equilibrium RNA folding dynamics impacts RNA function.
Methods
Cloning and plasmid construction
Riboswitch reporter plasmids were prepared as described previously33. Briefly, plasmids were constructed on a p15A plasmid backbone with a reporter cassette comprising a J23119 σ70 consensus promoter, riboswitch variant, ribosome binding site, and superfolder green fluorescent protein (sfGFP) coding sequence. Riboswitch mutants were generated by inverse polymerase chain reaction (iPCR) followed by blunt end ligation. Sequences were confirmed by Sanger sequencing (Quintara Biosciences), and plasmid stocks were screened for oligomerization by agarose gel electrophoresis. Approximately 20% of clones generated by the iPCR method were visibly oligomerized and discarded. Correct constructs were re-transformed into 10-beta chemically competent cells (NEB), from which clones were used to prepare glycerol stocks (25% glycerol, stored −70°C) and minipreps for data collection. Sequences of all reporter plasmids can be found in the Source Data file along with Addgene accession numbers for select constructs.
In vivo bulk fluorescence reporter assay
Riboswitch functional assays were performed as described previously33. Briefly, E. coli BW25113 (Keio parent) was transformed with reporter plasmids, and colonies were used to inoculate 300 μL overnight cultures in LB media. 4 μL of overnight cultures were then used to inoculate 200 μL subcultures in M9 enriched media (1x M9 salts, 1 mM thiamine hydrochloride, 0.4% glycerol, 0.2% casamino acids, 2 mM MgSO4, 0.1 mM CaCl2, 34 μg/ml chloramphenicol) containing the indicated amount of ligand. For ZTP riboswitch assays, 5-aminoimidazole-4-carboxyamide-ribonucleoside (Z) (Millipore Sigma) in DMSO was used; for purine riboswitch assays 2-aminopurine (2AP) (Millipore Sigma) in DMSO was used. For fluoride riboswitch assays, reporter plasmids were transformed into E. coli JW0619 (∆crcB Keio), and subcultures were treated with NaF (Millipore Sigma) in water. Subcultures were shaken at 1000 rpm and 37°C in a benchtop incubator for 6 hr, after which sfGFP fluorescence and OD600 were measured on a Biotek Synergy H1 (Agilent) microplate reader operated with BioTek Gen5 (Agilent) software. All in vivo reporter assays were performed with three experimental replicates, each performed in triplicate with three separate colonies (biological replicates) for a total of nine data points (n = 9) per reporter construct. To obtain sensitivity and dynamic range information, each construct was treated with a range of eight ligand concentrations. Therefore, each reported EC50 and fold change value is the result of 72 independent measurements comprising three experimental replicates, triplicate biological replicates, and eight different ligand conditions.
Fluorescence data was processed by subtracting average media blank measurements from both OD600 and fluorescence measurements, and dividing fluorescence values by OD600 values for each sample containing well. Dose response data from the 72 independent measurements was analyzed by performing least squares fitting (code available at79) to the collection of data points using the Hill equation:
where [L] is the concentration of ligand supplemented to the media, the fit parameters Fmin and Fmax account for baseline expression (leak) and the theoretical maximum gene expression under saturating ligand concentrations (ON-state), and the fit parameter EC50 is the concentration of supplemented ligand required to induce half-maximum fluorescence. The Hill coefficient n was heuristically set based on the riboswitch class: n = 1.33 for ZTP riboswitches, n = 1.13 for pbuE riboswitches, n = −1 for OFF ZTP riboswitches, n = 2.22 for fluoride riboswitches, and n = −1.41 for yxjA riboswitches. We performed this procedure to ensure all of the error of the fits was contained in the EC50 and fold change values. See Figs. S2, 3 for more detail about the fitting procedure. EC50 values were extracted from fitting to the 72 data points. Fold change was calculated by dividing the fitted Fmax by Fmin, with standard error propagation:
All fluorescence data and the resulting fits are available in the Source Data file. Where reported, EC50, Fmin, Fmax, and fold change are from fits to these 72 independent measurements with errors reported as standard error of the fits or propagated errors as described. All plots were generated using DataGraph 5.3.
Vfold3D structure modeling
Un-nucleated and nucleated Cbe pfl EP subdomains were modeled by submitting subsequences of the riboswitch and expected secondary structure to the Vfold3D webserver80. Resulting models were analyzed using ChimeraX 1.7 (UCSF)81.
Bioinformatic analysis of ZTP riboswitch EPs
1755 putative ZTP AD sequences from Rfam entry RF01750 were used to query the relevant genomes in GenBank for the downstream 200 bp. These putative riboswitch-containing loci were tested for the presence of a polyU tract (6 Ts in an 8 nt sliding window) and an Rfam consensus P3 stem. Secondary structure prediction (ViennaRNA 2.6.3) was performed on the subdomains extending from P3 to the polyU tract to ensure the formation of a single terminator stem-loop structure. The EP was examined for the presence of a region complementary to the 3’ side of the Rfam consensus P3 stem, which was marked as a putative invader. Terminator-containing EPs were manually classified using NUPACK secondary structure prediction to validate invader predictions and annotate EP architecture (loop-only, direct toehold, remote toehold). All bioinformatic code was written using ChatGPT 3.5 (OpenAI), and is available at https://github.com/LucksLab/Bushhouse_Riboswitch_Sensitivity_2024.
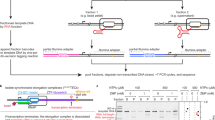
Single round in vitro transcription functional assay
Transcription termination assays were performed as described previously33. Briefly, 30 s single round IVT reactions were performed with 400 μM NTPs in the presence or absence of the indicated ligand concentration. Reaction products were isolated by phenol-chloroform extraction and DNase digestion, run on denaturing polyacrylamide gels, stained with 1x SYBR Gold Nucleic Acid Gel Stain (Invitrogen), and imaged for quantification. Imaging was performed on a Bio-Rad ChemiDoc Touch running Image Lab Touch 3.0. Quantification analysis was performed in Bio-Rad Image Lab 6.1. IVT assays were performed in duplicate.
FACS-seq high throughput assay
A FACS-seq high throughput assay was developed based on existing methods54,55,56. A library of Cbe pfl reporter plasmids with random terminator loop sequences was generated by performing iPCR cloning with a hand-mixed random primer (IDT). Ligated PCR products were electroporated into E. coli BW25113 and plated on several LB agar plates overnight at 37°C. The following morning, plates were transferred to the benchtop for approximately 8 hr, after which colonies were combined by repeatedly pipetting 10 mL of LB across the surface of each plate and resuspending colonies with sterile spreader arms. The combined resuspended library was vortexed briefly, passed through a 100 μM filter (CELLTREAT) to remove cell aggregates, and used to inoculate a 5 mL LB overnight culture shaken at 37 C. The following morning, 100 μL of overnight culture was used to inoculate 5 mL subcultures in M9 enriched media containing the appropriate amount of Z. Subcultures were shaken at 37 C for 6 hr, after which they were passed through a 100 μM filter (CELLTREAT), diluted 50x in 1x PBS, and placed on ice for 2 hr. Library subcultures were preliminarily characterized using a BD Accuri™ C6 Plus flow cytometer to examine library fluorescence as a function of supplied Z (Fig. S9b). Flow cytometry data was analyzed using FlowCal 1.2.
A sample of each subculture was sorted into 4 bins based on FITC fluorescence intensity (Fig. S9b). Sorting was performed using a BD FACSMelody™ Cell Sorter running BD VACSChorus 3.0 at the Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Core Facility at Northwestern University. Cell sort counts for each bin of each subculture were recorded and used in the data analysis procedure below (Source Data). After sorting, cells were placed on ice for 2 hr, after which the entire volume of sorted cells for each bin, as well as 1 mL of unsorted diluted subculture for each ligand condition, were used to inoculate separate 5 mL LB cultures, which were shaken overnight at 37°C and miniprepped.
Sequencing libraries for each sorted bin were generated by two-step PCR using a Phusion® DNA Polymerase (NEB) kit. In the first round, for each sample, a 100 μL PCR was assembled using 1 μg of input plasmid template and 400 nM of both Round 1 primers (DZB.C08, DZB.C09) which were designed to install 5 nt of random sequence on both sides of the amplified region to increase sequence diversity during initial sequencing cycles. After 3 rounds of PCR, reactions were placed briefly on ice, and 0.5 μL of ExoI (NEB) was mixed into each reaction. Reactions were incubated at 37°C for 30 min to digest Round 1 primers, and 80°C for 20 min to denature ExoI. Following ExoI deactivation, Round 2 primers (Illumina TruSeq Adapters) were added to each reaction to 400 nM, and PCR was resumed for 10 additional cycles. A unique i7 index was used for each sorted bin (Source Data). PCR products were purified from template plasmid and primers by two-step bead purification using SPRI beads (Cytiva) and quantified using Qubit 3.0 High Sensitivity dsDNA kit (Invitrogen). Indexed libraries across ligand and bin conditions were pooled in equimolar ratios and submitted to the NUSeq Core Facility for sequencing on a full flowcell of a NextSeq 500 (Illumina) instrument for paired-end 75 × 75 cycles, with an expected 58 bp of overlap between read 1 and read 2. The entire FACS-seq experiment from library construction to sequencing was performed in duplicate. Sequences of oligos used for sequencing library preparation can be found in the Source Data file.
For each ligand condition, the FACS-seq protocol generated 4 pairs of paired-end sequencing files, each containing the reads from one of the four sorted bins. Sequencing reads were analyzed to generate mean fluorescence estimates for each loop sequence in each ligand condition54. Dose response curves were then fit across ligand conditions to determine functional properties (EC50, fold-change) for each loop sequence (Fig. S9). Data was processed as follows:
Paired-end sequence files for each index were first filtered to remove reads with Qscore < 30 and reads in which the 58 bp of overlapping sequence in the forward and reverse reads did not match. Reads that failed to match the riboswitch template sequence or contained ambiguous base calls (‘N’) were also discarded.
The loop sequences from all remaining reads were extracted and counted based on the index, bin and ligand conditions, resulting in a table of read counts for each loop sequence, \(i\), in each bin \(j\), in each ligand condition,\(k\), \({read\; coun}{t}_{i,j,k}\). The FACS sorting process results in different numbers of sorted cells in each bin and ligand condition. We therefore normalized read counts according to:
where index_totalj.k and cellsj.k are the number of reads and the number of cells sorted for a given bin j and ligand condition k, respectively, and index_totalmax and cellsmax are the maximum number of total reads and the number of cells sorted across bins and ligand conditions, respectively. The factor \(\left(\frac{{cell}{s}_{\max }\,}{{cell}{s}_{j,k}}\right)\) reduces the signal from bins that have a low number of sorted cells which would normally be over-represented, while the factor \(\left(\frac{{inde}{x}_{-}{tota}{l}_{j,k}}{{inde}{x}_{-}{tota}{l}_{\max }}\right)\) boosts the signal from bins that had lower numbers of reads sampled during sequencing which would normally be under-represented.
For each loop sequence, mean fluorescence at each ligand condition (\({F}_{i,k}\)) was estimated by calculating the weighted average of the normalized read counts across the four sorted bins:
where Bj denotes the geometric mean fluorescence value of the bin j, calculated using the upper, \({U}_{j}\), and lower, \({L}_{j}\), bounds of the gate applied during cell sorting (Source Data):
Mean fluorescence values were calculated for each loop sequence in each ligand condition, converted out of log space, and values of both experimental replicates were pooled before fitting dose-response curves using Eq. (1) as described above. Data quality criteria were applied to remove poor fits and loop sequences with sparse data. Loop sequences with reads in fewer than 3 bins for any ligand concentration in both replicates were excluded, as were loop sequences where the ratio of the standard error of the fit parameters to the parameters themselves (\(\frac{{{SE}}_{{EC}50}}{{{EC}}_{50}}\) or \(\frac{{{SE}}_{{fold\; change}}}{{fold\; change}}\)) exceeded 1, or if the extracted EC50 value exceeded 500 μM. Of the 4912 loop sequences with reads in ≥ 3 bins at each ligand concentration in at least one replicate, 3289 passed the data quality criteria and were used for subsequent analysis. Density plots were generated by performing a gaussian kernel density estimation on the fold change and EC50 measurements of the 3289 characterized loops. All FACS-seq data analysis code was written using ChatGPT 3.5 (OpenAI) and is available79.
Statistics & reproducibility
For each riboswitch variant, in vivo fluorescence data was the result of three independent experiments, with three biological replicates in each experiment (n = 9 biological replicates). In vitro transcription assays were performed in duplicate. No statistical method was used to predetermine sample size. No data were excluded from the analyzes. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. FACS-seq raw sequencing data generated in this study have been deposited in the Sequence Read Archive database under accession code PRJNA1087340. The flow cytometry data generated in this study have been deposited in the FlowRepository database under accession code FR-FCM-Z7HM.
Code availability
All code has been released on Zenodo79.
References
Mccown, P. J., Corbino, K. A., Stav, S., Sherlock, M. E. & Breaker, R. R. Riboswitch diversity and distribution. RNA 23, 995–1011 (2017).
Winkler, W., Nahvi, A. & Breaker, R. R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002).
Breaker, R. R. The biochemical landscape of riboswitch ligands. Biochemistry 61, 137–149 (2022).
Scull, C. E., Dandpat, S. S., Romero, R. A. & Walter, N. G. Transcriptional riboswitches integrate timescales for bacterial gene expression control. Front. Mol. Biosci. 7, 607158 (2021).
Blount, K. F. & Breaker, R. R. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 24, 1558–1564 (2006).
Ellinger, E. et al. Riboswitches as therapeutic targets: promise of a new era of antibiotics. Expert Opin. Therapeutic Targets 27, 433–445 (2023).
Howe, J. A. et al. Selective small-molecule inhibition of an RNA structural element. Nature 526, 672–677 (2015).
Traykovska, M., Otcheva, L. A. & Penchovsky, R. Targeting TPP Riboswitches Using Chimeric Antisense Oligonucleotide Technology for Antibacterial Drug Development. ACS Appl. Bio Mater. 5, 4896–4902 (2022).
Freida, K. L. & Block, S. M. Direct observation of cotranscriptional folding in an adenine riboswitch. Science 338, 397–400 (2012).
Helmling, C. et al. Rapid NMR screening of RNA secondary structure and binding. J. Biomolecular NMR 63, 67–76 (2015).
Hua, B. et al. Real-time monitoring of single ZTP riboswitches reveals a complex and kinetically controlled decision landscape. Nat. Commun. 11, 4531 (2020).
Suddala, K. C. et al. Direct observation of tRNA-chaperoned folding of a dynamic mRNA ensemble. Nat. Commun. 14, 5438 (2023).
Watters, K. E., Strobel, E. J., Yu, A. M., Lis, J. T. & Lucks, J. B. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat. Struct. Mol. Biol. 23, 1124–1131 (2016).
Zhang, K. et al. Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 Å resolution. Nat. Commun. 10, 5511 (2019).
Porter, E. B., Marcano-Velázquez, J. G. & Batey, R. T. The purine riboswitch as a model system for exploring RNA biology and chemistry. Biochimica et. Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1839, 919–930 (2014).
Braselmann, E. et al. A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells. Nat. Chem. Biol. 14, 964–971 (2018).
Weigand, J. E. & Suess, B. Aptamers and riboswitches: perspectives in biotechnology. Appl. Microbiol. Biotechnol. 85, 229–236 (2009).
Hallberg, Z. F., Su, Y., Kitto, R. Z. & Hammond, M. C. Engineering and in vivo applications of riboswitches. Annu. Rev. Biochem. 86, 515–539 (2017).
Espah Borujeni, A., Mishler, D. M., Wang, J., Huso, W. & Salis, H. M. Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Res. 44, 1–13 (2016).
Thavarajah, W. et al. Point-of-use detection of environmental fluoride via a cell-free riboswitch-based biosensor. ACS Synth. Biol. 9, 10–18 (2019).
Harbaugh, S. V. et al. Engineering a synthetic dopamine-responsive riboswitch for in vitro biosensing. ACS Synth. Biol. 11, 2275–2283 (2022).
Vezeau, G. E., Gadila, L. R. & Salis, H. M. Automated design of protein-binding riboswitches for sensing human biomarkers in a cell-free expression system. Nat. Commun. 14, 2416 (2023).
Serganov, A. & Patel, D. J. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu. Rev. Biophysics 41, 343–370 (2012).
Tran, B. et al. Parallel discovery strategies provide a basis for riboswitch ligand design. Cell Chem. Biol. 27, 1241–1249.e4 (2020).
Kaiser, C., Schneider, J., Groher, F., Suess, B. & Wachtveitl, J. What defines a synthetic riboswitch? – Conformational dynamics of ciprofloxacin aptamers with similar binding affinities but varying regulatory potentials. Nucleic Acids Res. 49, 3661–3671 (2021).
Parmar, S. et al. Mechanistic analysis of Riboswitch ligand interactions provides insights into pharmacological control over gene expression. Nat. Commun. 15, 8173 (2024).
Fullenkamp, C. R. et al. Machine learning-augmented molecular dynamics simulations (MD) reveal insights into the disconnect between affinity and activation of ZTP riboswitch ligands. Preprint at bioRxiv https://doi.org/10.1101/2024.09.13.612887 (2024).
Widom, J. R. et al. Ligand modulates cross-coupling between riboswitch folding and transcriptional pausing. Mol. Cell 72, 541–552.e6 (2018).
Lemay, J.-F. et al. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 7, e1001278 (2011).
Wickiser, J. K., Winkler, W. C., Breaker, R. R. & Crothers, D. M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 18, 49–60 (2005).
Perdrizet, G. A., Artsimovitch, I., Furman, R., Sosnick, T. R. & Pan, T. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc. Natl Acad. Sci. 109, 3323–3328 (2012).
Yadav, R., Widom, J. R., Chauvier, A. & Walter, N. G. An anionic ligand snap-locks a long-range interaction in a magnesium-folded riboswitch. Nat. Commun. 13, 207 (2022).
Bushhouse, D. Z. & Lucks, J. B. Tuning strand displacement kinetics enables programmable ZTP riboswitch dynamic range in vivo. Nucleic Acids Res. 51, 2891–2903 (2023).
Roberts, J. W. Mechanisms of bacterial transcription termination. J. Mol. Biol. 431, 4030–4039 (2019).
Mironov, A. S. et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756 (2002).
Coppins, R. L., Hall, K. B. & Groisman, E. A. The intricate world of riboswitches. Curr. Opin. Microbiol. 10, 176–181 (2007).
Aboul-ela, F., Huang, W., Abd Elrahman, M., Boyapati, V. & Li, P. Linking aptamer-ligand binding and expression platform folding in riboswitches: prospects for mechanistic modeling and design. WIREs RNA 6, 631–650 (2015).
Binas, O., Schamber, T. & Schwalbe, H. The conformational landscape of transcription intermediates involved in the regulation of the ZMP-sensing riboswitch from Thermosinus Carboxydivorans. Nucleic Acids Res. 48, 6970–6979 (2020).
Strobel, E. J., Cheng, L., Berman, K. E., Carlson, P. D. & Lucks, J. B. A ligand-gated strand displacement mechanism for ZTP riboswitch transcription control. Nat. Chem. Biol. 15, 1067–1076 (2019).
Hong, F. & Šulc, P. An emergent understanding of strand displacement in RNA biology. J. Struct. Biol. 207, 241–249 (2019).
Machinek, R. R. F., Ouldridge, T. E., Haley, N. E. C., Bath, J. & Turberfield, A. J. Programmable energy landscapes for kinetic control of DNA strand displacement. Nat. Commun. 5, 5324 (2014).
Wong, T. N., Sosnick, T. R. & Pan, T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc. Natl Acad. Sci. 104, 17995–18000 (2007).
Bushhouse, D. Z., Choi, E. K., Hertz, L. M. & Lucks, J. B. How does RNA fold dynamically? J. Mol. Biol. 434, 167665 (2022).
Jones, C. P. & Ferré-D’Amaré, A. R. Recognition of the bacterial alarmone ZMP through long-distance association of two RNA subdomains. Nat. Struct. Mol. Biol. 22, 679–685 (2015).
Flory, P. Principles of Polymer Chemistry. (Cornell University Press, 1953).
Groebe, D. R. & Uhlenbeck, O. C. Characterization of RNA hairpin loop stability. Nucleic Acids Res. 16, 11725–11735 (1988).
Gusarov, I. & Nudler, E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell 107, 437–449 (2001).
Brahms, J., Michelson, A. M. & Van Holde, K. E. Adenylate oligomers in single- and double-strand conformation. J. Mol. Biol. 15, 467–488 (1966).
Leng, M. & Felsenfeld, G. A study of polyadenylic acid at neutral pH. J. Mol. Biol. 15, 455–466 (1966).
Sutton, J. L. & Pollack, L. Tuning RNA flexibility with helix length and junction sequence. Biophysical J. 109, 2644–2653 (2015).
Eichhorn, C. D. et al. Unraveling the structural complexity in a single-stranded RNA tail: implications for efficient ligand binding in the prequeuosine riboswitch. Nucleic Acids Res. 40, 1345–1355 (2012).
Tang, T. T. L. & Passmore, L. A. Recognition of poly(A) RNA through its intrinsic helical structure. Cold Spring Harb. Symposia Quant. Biol. 84, 21–30 (2019).
Hu, P. et al. Cooperative toehold: a mechanism to activate DNA strand displacement and construct biosensors. Anal. Chem. 90, 9751–9760 (2018).
Peterman, N. & Levine, E. Sort-seq under the hood: implications of design choices on large-scale characterization of sequence-function relations. BMC Genomics 17, 206 (2016).
Kosuri, S. et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl Acad. Sci. 110, 14024–14029 (2013).
Townshend, B., Kennedy, A. B., Xiang, J. S. & Smolke, C. D. High-throughput cellular RNA device engineering. Nat. Methods 12, 989–994 (2015).
Genot, A. J., Zhang, D. Y., Bath, J. & Turberfield, A. J. Remote toehold: a mechanism for flexible control of DNA Hybridization Kinetics. J. Am. Chem. Soc. 133, 2177–2182 (2011).
Kim, P. B., Nelson, J. W. & Breaker, R. R. An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol. Cell 57, 317–328 (2015).
Roberts, J. W., Shankar, S. & Filter, J. J. RNA polymerase elongation factors. Annu. Rev. Microbiol. 62, 211 (2008).
Chauvier, A. et al. Monitoring RNA dynamics in native transcriptional complexes. Proc. Natl Acad. Sci. 118, e2106564118 (2021).
Eschbach, S. H., Hien, E. D., Ghosh, T., Lamontagne, A.-M. & Lafontaine, D. A. The Escherichia coli ribB riboswitch senses flavin mononucleotide within a defined transcriptional window. RNA 30, 1660–1673 (2024).
Drogalis, L. K. & Batey, R. T. Requirements for efficient ligand-gated co-transcriptional switching in designed variants of the B. subtilis pbuE adenine-responsive riboswitch in E. coli. PLOS ONE 15, e0243155 (2020).
Gong, S., Wang, Y. & Zhang, W. Kinetic regulation mechanism of pbuE riboswitch. J. Chem. Phys. 142, 015103 (2015).
Cheng, L. et al. Cotranscriptional RNA strand exchange underlies the gene regulation mechanism in a purine-sensing transcriptional riboswitch. Nucleic Acids Res. 50, 12001–12018 (2022).
Ren, A., Rajashankar, K. R. & Patel, D. J. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature 486, 85–89 (2012).
Ren, A., Rajashankar, K. R. & Patel, D. J. Global RNA fold and molecular recognition for a pfl riboswitch bound to ZMP, a Master Regulator of One-Carbon Metabolism. Structure 23, 1375–1381 (2015).
Breaker, R. R. Prospects for riboswitch discovery and analysis. Mol. Cell 43, 867–879 (2011).
Breaker, R. R. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 4, 1–15 (2012).
Olenginski, L. T., Spradlin, S. F. & Batey, R. T. Flipping the script: understanding riboswitches from an alternative perspective. J. Biol. Chem. 300, 105730 (2024).
Focht, C. M., Hiller, D. A., Grunseich, S. G. & Strobel, S. A. Translation regulation by a guanidine-II riboswitch is highly tunable in sensitivity, dynamic range, and apparent cooperativity. RNA 29, 1126–1139 (2023).
Chatterjee, S., Chauvier, A., Dandpat, S. S., Artsimovitch, I. & Walter, N. G. A translational riboswitch coordinates nascent transcription–translation coupling. Proc. Natl Acad. Sci. 118, e2023426118 (2021).
Blechar, J., de Jesus, V., Fürtig, B., Hengesbach, M. & Schwalbe, H. Shine–dalgarno accessibility governs ribosome binding to the adenine riboswitch. ACS Chem. Biol. 19, 607–618 (2024).
Childs-Disney, J. L. et al. Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 21, 736–762 (2022).
Baird, N. J., Kulshina, N. & Ferré D’Amaré, A. R. Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol. 7, 328–332 (2010).
Chauvier, A., Ajmera, P., Yadav, R. & Walter, N. G. Dynamic competition between a ligand and transcription factor NusA governs riboswitch-mediated transcription regulation. Proc. Natl Acad. Sci. 118, e2109026118 (2021).
Pan, T., Artsimovitch, I., Fang, X. W., Landick, R. & Sosnick, T. R. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl Acad. Sci. 96, 9545–9550 (1999).
Heilman-Miller, S. L. & Woodson, S. A. Effect of transcription on folding of the Tetrahymena ribozyme. RNA 9, 722–733 (2003).
Zhang, J. & Landick, R. A two-way street: regulatory interplay between RNA polymerase and nascent RNA structure. Trends Biochemical Sci. 41, 293–310 (2016).
Bushhouse, D., Fu, T. & Lucks, J. LucksLab/Bushhouse_Riboswitch_Sensitivity_2024: Publication Release. Zenodo https://doi.org/10.5281/ZENODO.14262103 (2024).
Xu, X., Zhao, P. & Chen, S.-J. Vfold: a web server for RNA structure and folding thermodynamics prediction. PLOS ONE 9, e107504 (2014).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Ceres, P., Trausch, J. J. & Batey, R. T. Engineering modular ‘ON’ RNA switches using biological components. Nucleic Acids Res. 41, 10449–10461 (2013).
Lemay, J. F. & LaFontaine, D. A. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA 13, 339–350 (2007).
Ceres, P., Garst, A. D., Marcano-Velázquez, J. G. & Batey, R. T. Modularity of select riboswitch expression platforms enables facile engineering of novel genetic regulatory devices. ACS Synth. Biol. 2, 463–472 (2013).
Baker, J. L. et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335, 233–235 (2012).
Acknowledgements
We thank John Marko and Petr Šulc for helpful discussions about polymer theory and RNA folding dynamics. We thank the Robert H. Lurie Comprehensive Cancer Center of Northwestern University in Chicago, IL, for the use of the Flow Cytometry Core Facility, which provided use of the BD Melody FACS sorter. Support for this work was provided by the National Institutes of Health grant R01GM130901 to J.B.L., the National Institutes of Health Training Grant T32GM008382 through the Northwestern University Molecular Biophysics Training Program to D.Z.B., the National Science Foundation Grant 2310382 to J.B.L., and the Northwestern University Rappaport Award for Research Excellence to D.Z.B. J.B.L. was also supported by a John Simon Guggenheim Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or NSF.
Author information
Authors and Affiliations
Contributions
Conceptualization, D.Z.B. and J.B.L.; Methodology, D.Z.B. and J.B.L.; Formal analysis, D.Z.B. and J.F., Investigation, D.Z.B. and J.F.; Writing–Original Draft, D.Z.B. and J.B.L.; Writing–Review & Editing, D.Z.B., J.F., and J.B.L.; Visualization, D.Z.B.; Software, D.Z.B. and J.F.; Supervision, J.B.L.; Funding Acquisition, J.B.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bushhouse, D.Z., Fu, J. & Lucks, J.B. RNA folding kinetics control riboswitch sensitivity in vivo. Nat Commun 16, 953 (2025). https://doi.org/10.1038/s41467-024-55601-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55601-3
This article is cited by
-
Beyond the niche - unlocking the full potential of synthetic riboswitches
Nature Communications (2025)