Abstract
The heterodimeric Rab3GAP complex is a guanine nucleotide exchange factor (GEF) for the Rab18 GTPase that regulates lipid droplet metabolism, ER-to-Golgi trafficking, secretion, and autophagy. Why both subunits of Rab3GAP are required for Rab18 GEF activity and the molecular basis of how Rab3GAP engages and activates its cognate substrate are unknown. Here we show that human Rab3GAP is conformationally flexible and potentially autoinhibited by the C-terminal domain of its Rab3GAP2 subunit. Our high-resolution structure of the catalytic core of Rab3GAP, determined by cryo-EM, shows that the Rab3GAP2 N-terminal domain binds Rab3GAP1 via an extensive interface. AlphaFold3 modelling analysis together with targeted mutagenesis and in vitro activity assay reveal that Rab3GAP likely engages its substrate Rab18 through an interface away from the switch and interswitch regions. Lastly, we find that three Warburg Micro Syndrome-associated missense mutations do not affect the overall architecture of Rab3GAP but instead likely interfere with substrate binding.
Similar content being viewed by others
Introduction
Different membrane trafficking pathways mediate the movement of proteins and other macromolecules between different subcellular compartments, which is critical to the maintenance of organelle identity and the execution of complex processes from protein secretion to protein glycosylation1,2. These pathways employ a common mechanism that involves selecting and packaging cargo into a membrane-bound transport vesicle at the donor organelle, moving the cargo-laden vesicle along the cytoskeletal filaments, and docking and fusion of the incoming transport vesicle with the membrane of the acceptor organelle where the content is delivered1. The different steps of membrane trafficking rely on the coordinated activity of specialized factors. These factors include adapter proteins that are involved in cargo selection, coat proteins that mediate vesicle budding, motor proteins that move transport vesicles, tethers that link the incoming vesicle to the acceptor membrane, and soluble N-ethylmaleimide-sensitive factor activating receptors (SNAREs) that bring two membranes close, reducing the energy barrier for fusion1,3,4. Fundamental to these membrane trafficking steps are members of the Ras superfamily of GTPases, including the Arf, Arl, and Rab subfamilies. The coordinated activation and inactivation of these GTPases control the targeted recruitment of their effectors which is critical in providing spatiotemporal control of membrane transport1.
Amongst the different families of small GTPases, the Rab family GTPases are considered master regulators of membrane trafficking5,6. With 11 members in yeast and over 60 members in human cells, different Rabs are uniquely distributed across distinct membrane compartments. Like other members of the Ras superfamily, Rabs switch between a GDP-bound inactive state and a GTP-bound active state, as well as between a cytoplasmic form and a membrane-bound form through their prenylated C-termini and the action of chaperone-like GDP dissociation inhibitors (GDI). Upon activation and localization to the proper membrane compartments, Rab GTPases coordinate different transport steps by recruiting diverse effectors5,6. The activation of Rab GTPases is controlled by guanine nucleotide exchange factors (GEF) that catalyze the exchange of bound GDP with GTP and GTPase activating proteins (GAP) that accelerate the intrinsically slow GTP hydrolysis rate of the Rab5,6. Some GEFs also play a role in assisting recruitment of their substrate Rabs to specific membrane compartments7. While almost all known Rab GAPs belong to the TBC (Tre-2/Bub/Cdc16) domain family, Rab GEFs are divergent in primary sequence and structure5,8. Interestingly, despite the diversity in composition and structure of Rab GEFs, all GEFs characterized structurally thus far were observed to utilize a similar strategy to promote nucleotide exchange of their substrate Rabs8. This involves binding the nucleotide-bound Rab at its switch I/switch II and interswitch regions and inducing conformational changes that open the nucleotide-binding pocket and hinder the binding of a magnesium ion, leading to the release of the bound nucleotide5. After this, the GEF forms a high-affinity complex with the nucleotide-free Rab. The higher cellular concentration of GTP compared to GDP results in the conversion of the Rab to the GTP-bound form and displacement of the GEF9.
Rab3GAP is a heterodimeric complex composed of the 130 kDa Rab3GAP1 and the 150 kDa Rab3GAP2 subunits. Originally isolated from rat brain tissues and identified to be the GAP for brain-specific Rab3, Rab3GAP was later shown to exhibit GEF activity on Rab18, a Rab that is ubiquitously expressed in all human tissues and is one of the six primordial Rabs found in the last eukaryotic common ancestor10,11,12,13. Subsequent studies revealed that mutations in genes encoding Rab3GAP1, Rab3GAP2, and Rab18 cause Warburg Micro Syndrome (WMS), an autosomal recessive genetic disorder characterized by postnatal growth retardation, microcephaly, congenital cataracts, optical atrophy, spastic paraplegia, hypogonadism, and delayed motor and intellectual development14,15,16,17,18,19,20. These observations suggest that activating Rab18 might be the primary function for Rab3GAP in most cell types. Rab3GAP facilitates the recruitment of Rab18 to lipid droplets (LDs), the endoplasmic reticulum (ER), and the Golgi apparatus12,21,22,23. Rab18 has been implicated in LD metabolism, establishing and/or maintaining ER morphology, ER-to-Golgi trafficking, autophagy, secretion, peroxisome regulation, and viral assembly12,24,25,26,27. Rab18 has also been shown to promote the formation of ER-LD contact sites by recruiting the NAG-RINT1-ZW10 (NRZ) tethering complex and ER-associated SNAREs, Syntaxin18, Use1, and BNIP1, to facilitate lipid transfer between these organelles25. Apart from the findings that both subunits are required for the Rab18 GEF activity of Rab3GAP and that three WMS-associated point mutations disrupt this activity, little is known about the biochemical and structural properties of Rab3GAP12. Furthermore, how this heterodimeric complex engages its cognate substrate and promotes GDP-to-GTP exchange and how WMS-associated point mutations affect this critical function remain obscure due to the lack of sequence and structural similarities to other Rab GEFs. Lastly, what allows Rab3GAP to perform two seemingly opposite functions in Rab3 inactivation and Rab18 activation is still a mystery.
In this work, we characterize the biochemical and structural properties of human Rab3GAP using a combination of negative stain electron microscopy (EM) and cryo-EM, in vitro GEF assays, hydrogen-deuterium exchange mass spectrometry (HDX-MS), and in silico structural modeling. We find that full-length Rab3GAP is conformationally flexible and adopts a range of open-to-closed conformations. We observe that Rab3GAP exhibits enhanced Rab18 GEF activity in the presence of a membrane. We also find that core Rab3GAP composed of Rab3GAP1 and RabGAP2 devoid of its C-terminal domain, shows increased Rab18 GEF activity, raising the possibility that the full-length complex is autoinhibited. We subsequently determine the high-resolution structure of core Rab3GAP by cryo-EM and validate the observed intersubunit interface by hydrogen-deuterium exchange mass spectrometry (HDX-MS). Furthermore, by combining AlphaFold3 modeling, targeted mutagenesis in conjunction with in vitro GEF assays, confocal microscopy, and WMS disease mutation mapping, we find that Rab3GAP likely binds its substrate Rab18 through a platform opposite of the critical switch and interswitch regions. Collectively, our work provides biochemical and structural insights on human Rab3GAP and how this complex may engage its substrate Rab18, and presents a molecular framework to predict how WMS disease mutants interfere with Rab18 binding and activation.
Results
Human Rab3GAP adopts a flexible bilobal overall architecture
To facilitate biochemical and structural analyses, we established a baculovirus-insect cell-based method to reconstitute the dimeric complex of human Rab3GAP1-Rab3GAP2. In brief, we constructed a plasmid encoding a Rab3GAP1-Rab3GAP2 two-gene cassette using the biGBac system and then generated baculoviruses for infecting and co-expressing the two Rab3GAP subunits in Sf9 cells28. Reconstituted human Rab3GAP was subsequently purified using anti-FLAG affinity chromatography and size exclusion chromatography. Rab3GAP elutes at a volume corresponding to the predicted mass for a ~300 kDa complex, which is consistent with the previously proposed 1:1 subunit stoichiometry for this complex (Fig. 1a)11,12. We next examined the ability of the reconstituted complex to promote nucleotide exchange by carrying out an in vitro GEF assay on recombinant human Rabs loaded with the fluorescent GDP analog 3-(N-methyl-anthraniloyl)-2-deoxy-GDP (Mant-GDP) (Fig. 1b). The fluorescent signal of Mant-GDP bound to a Rab is substantially stronger than free Mant-GDP, allowing nucleotide dissociation to be monitored as a decrease in fluorescence29. We found that recombinant Rab3GAP promotes GDP release of its cognate substrate Rab18 but not Rab11a (Supplementary Fig. 1). We also validated the previous finding that both subunits of Rab3GAP are required for this Rab18 GEF activity as Rab3GAP1 subunit alone cannot promote nucleotide exchange (Fig. 1c)12. By measuring Mant-GDP release at different concentrations of Rab3GAP, we determined that Rab3GAP has a catalytic efficiency of 2.5 × 103 M−1 s−1, a value that is comparable to those reported for other Rab GEFs (Fig. 1d)30,31,32,33,34.
a Analytical gel filtration elution profile of Rab3GAP on an ENrich™ SEC 650 column and representative SDS-PAGE gel of the reconstituted complex stained with Coomassie Blue. Analytical gel filtration and SDS-PAGE gels were performed in biological triplicate (n = 3). b Schematic of in vitro GEF assays to assess nucleotide exchange activity towards Rabs using the fluorescent GDP analog, Mant-GDP. c GEF assays on Rab18 with Rab3GAP1 and Rab3GAP1/2. Nucleotide exchange was detected by measuring fluorescent decrease in reactions containing 0 nM GEF (Mock) or 300 nM GEF with 4 µM Mant-GDP loaded Rab18 and 100 µM GTPγS. Data are presented as mean with error bars showing SEM for assays performed in technical triplicate (n = 3). d Determination of catalytic efficiency (kcat/Km) using reactions with Rab3GAP1 + 2 (0–300 nM), 4 µM Mant-GDP loaded Rab18 and 100 µM GTPγS. Reactions were conducted in technical triplicate (n = 3). Data are presented as mean with error bars showing SEM and kcat/Km was calculated as described previously34. e 2D class averages showing the general architecture of Rab3GAP and schematic representation of core Rab3GAP (pink) and the flexible arm (orange) adopting closed, V-shaped, and extended conformations. Source data are provided as a Source Data file.
Having successfully reconstituted Rab3GAP, we next examined the overall architecture of this complex by negative stain single-particle EM. Class averages generated from our two-dimensional (2D) analysis revealed that Rab3GAP is composed of an extended but rigid arm region attached to the head of a tadpole-shaped body (Fig. 1e). The gallery of class averages revealed that the arm can rotate around the junction point at the head of up to 180° with respect to the body and this results in a range of architectures from fully clasped V-shaped to fully opened I-shaped.
The C-terminal domain of Rab3GAP2 confers flexibility to Rab3GAP
The conformational heterogeneity of Rab3GAP posed technical challenges to high-resolution cryo-EM analysis. To mitigate this problem, we investigated the basis of the conformational flexibility of the arm region by first examining the AlphaFold predicted structural models of human Rab3GAP1 and Rab3GAP2. Rab3GAP2 is predicted to be composed of two globular domains that show low predicted alignment error (PAE) joined by an unstructured linker (Fig. 2a and Supplementary Fig. 2a, b). Based on the predicted Rab3GAP2 structural model, we designed two expression constructs encoding either Rab3GAP2N (N-terminal domain: 1–544) or Rab3GAP2C (C-terminal domain: 545–1393) and then prepared two biGBac vectors encoding Rab3GAP1 and one of the Rab3GAP2 domains. We found that Rab3GAP1 is only capable of binding the N-terminal domain of Rab3GAP2, and we were able to purify the reconstituted Rab3GAP1-Rab3GAP2N complex (Supplementary Fig. 2c). This truncated complex eluted as a monodisperse peak with 1:1 subunit stoichiometry in size exclusion chromatography (Fig. 2b). We next subjected the Rab3GAP1-Rab3GAP2N complex to an in vitro GEF assay and found that this complex retains the ability to promote Rab18 nucleotide exchange, revealing that the C-terminal domain of Rab3GAP2 is not required for GEF activity (Fig. 2c). In light of this finding, we hereafter refer the Rab3GAP1-Rab3GAP2N complex as “core Rab3GAP” and Rab3GAP1-Rab3GAP2 as “full-length Rab3GAP”. We next analyzed core Rab3GAP by negative stain EM. Our 2D analysis showed that this truncated complex is structurally homogeneous and retains only the tadpole-shaped body without the flexible arm (Fig. 2d). In summary, we were able to identify that the Rab3GAP2 C-terminal domain confers conformational flexibility to full-length Rab3GAP, and this domain is not required for Rab18 GEF activity.
a Schematic of Rab3GAP1 with GAP domain annotated, and Rab3GAP2 domain 1 and Rab3GAP2 domain 2 connected by a flexible linker. b Analytical gel filtration elution profile of core Rab3GAP on an ENrich™ SEC 650 column and representative SDS-PAGE gel of the reconstituted complex stained with Coomassie Blue. Analytical gel filtration and SDS-PAGE gels were performed in biological triplicate (n = 3). c GEF assays on core Rab3GAP. Nucleotide exchange was detected by measuring fluorescent decrease in reactions containing 0 nM GEF (Mock) or 300 nM GEF with 4 µM Mant-GDP loaded Rab18 and 100 µM GTPγS. Data are presented as mean with error bars showing SEM for assays performed in technical triplicate (n = 3). d Representative 2D class averages comparing the general architecture of core Rab3GAP to full-length Rab3GAP. Schematic showing only a single conformation for core Rab3GAP compared to the open and closed conformation of full-length Rab3GAP, with Rab3GAP1 in blue and Rab3GAP2 in pink. Source data are provided as a Source Data file.
Rab3GAP GEF activity is enhanced by membrane-anchored Rab18
Having confirmed that core Rab3GAP represents the minimal machinery necessary to catalyze Rab18 nucleotide exchange, we next compared the GEF activity of core Rab3GAP to full-length Rab3GAP. Intriguingly, we found that the core complex exhibits enhanced Rab18 GEF activity, with its catalytic efficiency (5.5 × 103 M−1 s−1) more than twice as high as the full-length complex (Fig. 3a, b). This raises the possibility that the activity of full-length Rab3GAP is autoinhibited by the C-terminal domain of Rab3GAP2. Previous studies showed that many Rab GEFs show enhanced rates when their substrates are presented on a membrane34,35. To determine if Rab3GAP activity is affected by a membrane, we incubated Mant-GDP-loaded, C-terminally His-tagged Rab18 with 100 nm-diameter lipid vesicles containing Ni-NTA conjugated lipids and then carried out an in vitro GEF assay using the resulting membrane-anchored Rab18 as a substrate (Fig. 3c). We found that the GEF activity of both full-length and core Rab3GAP increased more than ten-fold when Rab18 was presented on a membrane, with a catalytic efficiency of 6.6 × 104 M−1 s−1 for FL Rab3GAP and 4.4 × 104 M−1 s−1 for core Rab3GAP (Fig. 3d). Finally, we carried out systematic GEF assays using Rab18 anchored to vesicles with different lipid compositions mimicking different cellular organelles (Fig. 3e). Ordinary one-way ANOVA showed that varying the lipid composition of the vesicles does not result in significant differences in nucleotide exchange rates (kobs), suggesting that the enhancement in GEF activity is not induced by a specific type of lipid (Fig. 3f).
a In vitro GEF assays with full-length Rab3GAP or core Rab3GAP to detect activity towards Rab18. Nucleotide exchange was detected by measuring fluorescent decrease in reactions containing 0 nM GEF (Mock) or 300 nM GEF with 4 µM Mant-GDP loaded Rab18 and 100 µM GTPγS. Data are presented as mean with error bars showing SEM for assays performed in technical triplicate (n = 3). Data from Fig. 2c was used to generate this plot. b Determination of catalytic efficiency (kcat/Km) using reactions with full-length Rab3GAP or core Rab3GAP (0–300 nM), 4 µM Mant-GDP loaded Rab18 and 100 µM GTPγS. Reactions were conducted in technical triplicate (n = 3). Data are presented as mean with error bars showing SEM and kcat/Km was calculated as described previously34. Data from Fig. 1d was used to generate this plot. c Schematic of Mant-GDP based GEF activation assay with C-terminally His-tagged Rab18 anchored to NiNTA-containing lipid vesicles. d Evaluation of membrane regulation for full-length or core Rab3GAP activity towards Rab18. Catalytic efficiency (kcat/Km) was calculated using reactions with full-length Rab3GAP or core Rab3GAP (0–15 nM), 4 µM Mant-GDP loaded Rab18, 0.2 mg mL−1 liposomes (75% PC, 20% PE, 5% 18:1 DGS-NTA(Ni), 100 nm) and 100 µM GTPγS. Reactions were conducted in technical triplicate (n = 3). Data are presented as mean with error bars showing SEM and kcat/Km was calculated as described previously34. e Comparison between various membrane compositions with respect to regulation of Rab3GAP activity. Nucleotide exchange was detected by measuring fluorescent decrease in reactions containing 300 nM GEF, 4 µM Mant-GDP loaded Rab18, 100 µM GTPγS and 100 nm extruded liposomes with various lipid composition (Supplementary Table 2) at 0.2 mg mL−1. Data are presented as mean with error bars showing SEM for assays performed in technical triplicate (n = 3). f Data from e was fit to a non-linear one-phase exponential decay model to determine the rate of nucleotide exchange (kobs). Data are presented as mean with error bars showing SEM. p values were generated using an ordinary one-way Anova (ns indicates p > 0.05). Source data are provided as a Source Data file.
Cryo-EM structure of human core Rab3GAP
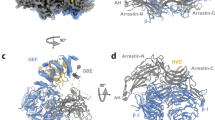
Knowing that core Rab3GAP retains Rab18 GEF activity and is structurally homogeneous, we next pursued high-resolution structural analysis of this complex using cryo-EM. By performing PEGylation on core Rab3GAP, we were able to minimize aggregation at high protein concentrations which we observed in our initial vitrification trials36. We subsequently acquired a cryo-EM dataset of 19,822 movies. The class averages obtained from the 2D analysis showed that the tadpole-shaped core Rab3GAP contains a knob-shaped region for the head connected to a central body density that carries an extended curved tail (Supplementary Fig. 3). The curved tail is blurry, suggesting that core Rab3GAP contains another conformationally flexible region. Local refinement to this flexible tail region however did not improve the resolution of the tail region. Following multiple rounds of 3D classification and local refinement to improve the rigid region of the core, we obtained a final density map with an average resolution of 3.37 Å from a final stack of 170,636 particles (Fig. 4a and Supplementary Fig. 3). We could visualize side chain densities in the central core but the map region proximal to the tail is smeared and has a lower local resolution (Fig. 4b, c). We next iteratively fit the AlphaFold2-predicted models of the Rab3GAP1 and Rab3GAP2 subunits into the density map and then further optimized the fit and refined the structural model (Fig. 4d). The flexible tail, which was excluded in local refinement, comprised residues 332 to 781 of Rab3GAP1 and could not be accurately modeled. Our final structural model consists of residues 19 to 331 and 782 to 981 for Rab3GAP1 and residues 2 to 544 for Rab3GAP2. We were unable to model the first N-terminal 18 residues, residues 242 to 263, 902 to 929 of Rab3GAP1, and the N-terminal residue, residues 27 to 60, 328 to 363, and 437 to 456 of Rab3GAP2 due to the lack of or poorly defined densities (Fig. 4b, d and Supplementary Fig. 3).
a Side views of the locally refined cryo-EM density map of the core Rab3GAP solved to 3.39 Å resolution from 170,636 particles. Map was processed using Phenix. b Local resolution of the cryo-EM map estimated in cryoSPARC. c Cryo-EM density and model for selected regions near the interface of the obtained map. d Atomic model and structural architecture of the core Rab3GAP complex. Rab3GAP1 and Rab3GAP2(1–544) are displayed in blue and pink, respectively.
Our refined structure revealed the Rab3GAP1 consists of two structural domains (Fig. 4d). Domain 1, which formed the central body density seen in the 2D class averages, comprises an αβ domain. Interestingly, Domain 1 forms a curved β-sheet from both the Rab3GAP1 N-terminal (58–316) and C-terminal region (880–981). This curved β-sheet is composed of β1 to β6, followed by the last 3 β-strands of the C-terminal region and then β7 to β9. Aside from the β3-β4 pair, the extended β-strands of Domain 1 are antiparallel. Domain 2 contains a group of five mapped α-helices that form the extended stalk that leads to the unmapped tail density. Foldseek analysis of our model revealed that Rab3GAP1 shows structural similarities to Zwilch, a subunit of the ROD–Zwilch–ZW10 (RZZ) complex that is a component of the outer kinetochore and mediates recruitment of the dynein and Mad1-Mad237,38,39. On the other hand, Rab3GAP2 N-terminal domain represents the head of the tadpole-shaped complex and forms a canonical seven-bladed β-propeller with each blade having four antiparallel β-strands. This subunit also contains three α-helices (residues 15 to 21, 74 to 78 and 221 to 241 of Rab3GAP2) that are located distal to the interfacial region of the complex. Relative to Rab3GAP1, the β-propeller is perched onto the curved β-sheet of Rab3GAP1 along its edge. Sandwiched between the β-propeller and the curved β-sheet in the subunit interaction region are α-helices from each subunit (residues 142 to 159 and 22 to 37 of Rab3GAP1 and 277 to 287 of Rab3GAP2).
The Rab3GAP1-Rab3GAP2N subunit interaction interface
The binding of Rab3GAP1 to Rab3GAP2N results in the burial of 2063 Å2 of surface area. The interaction between Rab3GAP1 and Rab3GAP2N is held together by hydrophobic packing, hydrogen bonds from 8 residue pairs (T19 to K423, E24 to K423, H170 to N286, E191 to K282, V193 to S276, K215 to D279, P950 & G457, and Y951 to R197) (Supplementary Fig. 4a) and electrostatic interactions (Supplementary Fig. 4b). The negative electrostatic potential of Rab3GAP1’s interface is composed of E24, E142, E180, D189 and the C-term has complementary charges to the positive electrostatic potential of Rab3GAP2’s R197, R377, R415, and K423 (Supplementary Fig. 4b). To determine if the observed interface in our cryo-EM-derived structure of core Rab3GAP is consistent with that of full-length Rab3GAP, we conducted HDX-MS analysis. HDX-MS measures the exchange of protein backbone amide hydrogens with deuterated solvent, with amide exchange rates acting as a useful surrogate for the stability of protein secondary structure. HDX-MS therefore is useful in mapping both direct protein-protein interactions, and allosteric changes in protein conformation40,41.
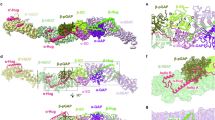
As we could only purify the Rab3GAP1 subunit but not the Rab3GAP2 subunit, we designed HDX-MS experiments to measure conformational differences between Rab3GAP1 alone and Rab3GAP1 in the presence of Rab3GAP2. In brief, we incubated Rab3GAP1 and full-length Rab3GAP in deuterated buffer and measured deuterium incorporation over 5 time points (3, 30, 300, 3000 s at 18 °C and 3 s at 0 °C, which is referred to as 0.3 s). For Rab3GAP1 the HDX-MS coverage spanned 83.6% of the primary sequence, with the full raw deuterium incorporation data for all generated peptides provided in the source data. The HDX-MS experiment comparing Rab3GAP1 alone and Rab3GAP1 in the fully assembled Rab3GAP complex had multiple significant decreases in H/D exchange (defined as having >5%, >0.4 Da and p < 0.01 in an unpaired two-tail t test at any time point), with the largest changes occurring at the N-terminus of Rab3GAP1 (residues 25–32, 37–53, 129–166, 169–191, 193–208, and 211–229) and the C-terminus of Rab3GAP1 (948–966, 973–977, and 978–987) (Fig. 5a–c). Although some of these regions were not modeled in our cryo-EM-derived structural model, this finding agrees with the observed Rab3GAP2N interaction interface, which is made up of both terminal regions of Rab3GAP1 according to our structure. We also observed small significant decreases in deuterium incorporation in regions distal to the subunit interaction interface (Fig. 5b, Supplementary Fig. 5 and Source Data). These changes could potentially result from conformational changes that Rab3GAP1 undergoes after binding Rab3GAP2 or from obstruction from the C-terminal domain of Rab3GAP2.
a Rab3GAP1 regions showing significant differences in deuterium exchange (defined as >5%, >0.4 Da, and p < 0.01 in an unpaired two-tail t-test at any time point) upon complex formation with Rab3GAP2 are highlighted on the cryo-EM structure. The differences in deuterium exchange are indicated by the legend in (a). b Sum of the number of deuteron difference of Rab3GAP1 upon complex formation with Rab3GAP2, analyzed over the entire deuterium exchange time course for Rab3GAP1. Each point is representative of the center residue of an individual peptide. Peptides that met the significance criteria described in (a) are colored red. Experiments were performed in technical triplicate (n = 3). Each point represents a single peptide, and data are presented as the sum of the mean number of deuteron difference across all 5 time points (n = 3) and error bars represent the sum of standard deviations across all 5 time points (n = 3 for each time point). Domain schematic above depicts Rab3GAP1 architecture, with the thin box representing unmodelled residues in (a). c Selected deuterium exchange time courses of Rab3GAP1 peptides that showed significant decreases and increases in exchange. Data are presented as mean values with error bars representing SD from experiments performed in technical triplicate (n = 3). A full list of all peptides and their deuterium incorporation are provided as a Source Data file.
The Rab18 binding site is distal to the switch and interswitch regions
We next investigated how Rab3GAP interacts with its cognate substrate Rab18. Despite numerous efforts, we were unable to reconstitute a core Rab3GAP-Rab18 complex. Unlike other previously characterized Rab-GEF pairs, Rab3GAP does not appear to form a high-affinity complex with nucleotide-free Rab18. We thus turned the computational approach of AlphaFold3 to predict the structure of core Rab3GAP in a complex with Rab18 (Fig. 6a and Supplementary Fig. 6a, b). All five models generated show Rab18 positioned at the edge of the subunit interaction interface of Rab3GAP. Each model has an ipTM score of 0.8, indicating confident high-quality predictions42. Critically, the PAE of Rab18 relative to the core Rab3GAP interface was 5 Å or less, further highlighting the confidence of these models (Supplementary Fig. 6a). Surprisingly, these models show that Rab3GAP does not appear to directly engage the switch I, switch II, and interswitch regions of Rab18 as had been observed for other Rab GEFs5,8. Instead, the unstructured N-terminus and α1 of Rab3GAP1 and the loops connecting adjacent β-sheets from the β-propeller of Rab3GAP2 contact the α5, α6 and β6 located on the opposite side of the switch regions of Rab18 (Fig. 6a, b). A cluster of nonpolar residues on Rab18 (L137, L146, F147 and I148) and along the N-terminal region of Rab3GAP1 (F12, I14 and F17) are conserved and may be involved in mediating hydrophobic interaction between Rab3GAP1 and its substrate (Supplementary Fig. 7a–c).
a AlphaFold3 prediction of the interaction between Rab18 and core Rab3GAP. Rab3GAP1 in light blue, Rab3GAP2(1–544) in pink, Rab18 in cyan with switch I, switch II and p-loop colored in red, orange, and yellow, respectively. b Zoomed in view of AlphaFold3 model showing predicted electrostatic interactions at the Rab18-Rab3GAP binding interface. Residues predicted to form salt bridges (Rab3GAP2 R426 with Rab18 E164, Rab3GAP1 E13 and E31 with Rab18 R133 and R141 respectively) are shown as sticks and labeled on the structure. c In vitro GEF assays with full-length Rab3GAP to detect activity against Rab18 with mutations suspected of disrupting the Rab3GAP binding interface. Nucleotide exchange was detected by measuring fluorescent decrease in reactions containing 0 nM GEF (Mock) or 400 nM GEF with 4 µM Mant-GDP loaded Rab18 (WT, R133A, R141A or E164A) and 100 µM GTPγS. Data are presented as mean with error bars showing SEM for assays performed in technical triplicate (n = 3). d Data from Fig. 4c was fit to a non-linear one-phase exponential decay model to determine the rate of nucleotide exchange (kobs). Data are presented as mean with error bars showing SEM. p values were generated using a two-tailed Student’s t tests (***p < 0.01). e Nucleotide exchange was detected by measuring fluorescent decrease in reactions containing 0 nM GEF (Mock) or 300 nM GEF with 4 µM Mant-GDP loaded Rab18 and 100 µM GTPγS. Data are presented as mean with error bars showing SEM for assays performed in technical triplicate (n = 3). f GTPase activity was measured using the Promega GTPase-Glo assay with reactions containing 0 nM GAP (Mock) or 500 nM GAP, 5 µM GTP and 8 µM GST-Rab3a. Data are presented as mean with error bars showing SEM for assays performed in technical triplicate (n = 3), with p values generated using a two-tailed Student’s t tests (*p < 0.05). Source data are provided as a Source Data file.
According to the predicted models, residues R133, R141, E164 of Rab18, which are distal to the switch and interswitch regions, form salt bridges with Rab3GAP1 E13 and E31 and Rab3GAP2 R426 (Fig. 6b). To validate the predicted interaction interface, we generated site-specific mutants targeting three Rab18 residues (R133A, R141A, E164A) that are predicted to mediate interaction with Rab3GAP. We loaded these three mutant Rab18 with Mant-GDP and then used them for in vitro GEF assays with full-length Rab3GAP. Consistent with the AlphaFold3 generated structural models of Rab3GAP-Rab18, we found that the Rab18 R133A and R141A mutations significantly impair the ability of Rab3GAP to promote nucleotide exchange, while the Rab18 E164A mutation almost completely abolished nucleotide exchange (Fig. 6c, d). Previous studies showed that Rab3GAP regulates Rab18 localization12. To further examine the effects of the E164A mutation on Rab18’s localization, we transfected HeLa cells with construct encoding wild-type or E164A mutant GFP-Rab18 and then monitored their localization by fluorescent microscopy. The Rab18 E164A mutant showed a more diffuse and disorganized localization pattern compared to WT Rab18, supporting the hypothesis that Rab3GAP binds Rab18 through an interface opposite of the nucleotide-binding domain (Supplementary Fig. 8). Unlike the highly conserved switch and interswitch regions, α5, α6 and β6 are substantially less conserved between Rabs. Furthermore, multiple sequence alignment showed that the residues critical to Rab18 activation by Rab3GAP (R133, R141 and E164) are not conserved among Rab family members (Supplementary Fig. 9), explaining how Rab3GAP might selectively regulate Rab18 despite significant structural homology within this family of small GTPases.
As GEFs typically remodel the Rab switch regions to induce nucleotide exchange, we explored the possibility that another region of Rab3GAP may be responsible for engaging the Rab18 switches. Rab3GAP1 contains a putative GAP domain that is located in a conformationally flexible tail region and was previously shown to bind the switch regions of Rab310,43. To determine if this domain participates in Rab18 nucleotide exchange, we generated a core Rab3GAP construct that encodes Rab3GAP1 devoid of the GAP domain (residues 618–748) (Supplementary Fig. 10a) and purified this truncated complex, which we will refer to as Rab3GAPΔGAP. In vitro GEF assays demonstrated that Rab3GAPΔGAP still retains Rab18 GEF activity (Fig. 6e). By contrast, in vitro GAP assays showed that this Rab3GAPΔGAP is unable to catalyze GTP hydrolysis of Rab3a above basal levels (Fig. 6f). These results suggest that the Rab3GAP1 GAP domain is not involved in Rab18 nucleotide exchange and that the GAP and GEF activities of Rab3GAP are mapped to distinct locations of the complex.
Warburg micro syndrome mutations likely affect substrate binding
Previous studies showed that three WMS-associated missense mutations (Rab3GAP1 T18P, Rab3GAP1 E24V, Rab3GAP2 R426C) disrupt Rab3GAP’s Rab18 GEF and membrane-targeting activities12. We generated expression constructs encoding these mutations and purified the three full-length Rab3GAP complexes carrying these mutations. We conducted negative stain EM analysis on these Rab3GAP mutants and our 2D analysis showed that the three WMS mutations did not alter the overall architecture of Rab3GAP (Fig. 7a). Using in vitro GEF assays, we validated that three mutant Rab3GAP complexes show dramatically decreased rates of Rab18 nucleotide exchange compared to WT (Fig. 7b).
a SDS-PAGE gel of purified Rab3GAP complex containing Rab3GAP1 T18P, Rab3GAP1 E24V or Rab3GAP2 R426C mutations stained with Coomassie Blue. 2D class averages comparing the general architecture of WT Rab3GAP to complexes containing T18P, E24V or R426C point mutations. SDS-PAGE gel were performed in biological triplicate (n = 3). b In vitro GEF assays with clinical Rab3GAP WMS mutants to detect activity towards Rab18. Nucleotide exchange was detected by measuring fluorescent decrease in reactions containing 0 nM GEF (Mock) or 300 nM GEF with 4 µM Mant-GDP loaded Rab18 and 100 µM GTPγS. Data are presented as mean with error bars showing SEM for assays performed in technical triplicate (n = 3). c Zoomed in view showing Rab3GAP WMS mutants mapped onto the cryo-EM structure of core Rab3GAP. Residues mutated in WMS are shown as sticks and the predicted position of the T18P mutation is circled in black. d Zoomed in view of the AlphaFold3 generated model showing the WMS mutants mapped to the predicted Rab18 binding interface. Residues mutated in WMS and residues predicted to interact electrostatically with WMS residues are shown as sticks and labeled. Rab3GAP1 in light blue, Rab3GAP2(1–544) in pink and Rab18 in cyan. Source data are provided as a Source Data file.
Using our cryo-EM-derived structure, we mapped the locations of these mutation sites. Although Rab3GAP1 T18 was not modeled in our structure due to lack of density, the other two mutation sites (Rab3GAP1 E24 and Rab3GAP2 R426) are located near the edge of the intersubunit interaction interface (Fig. 7c). Our structure shows that RabGAP1 E24 engages in electrostatic interaction with R407 of Rab3GAP2 to stabilize the Rab3GAP1 α1 helix. On the other hand, Rab3GAP2 R426 is located in an accessible region proximal to the intersubunit interaction interface. When we examined the AlphaFold3-predicted model of the Rab3GAP-Rab18 complex, we observed that the three WMS mutation sites are located within the Rab18 binding pocket and that these missense mutations could potentially disrupt interactions with Rab18 (Fig. 7d). In particular, T18 is part of the Rab3GAP1 unstructured N-terminal region located at the center of the substrate binding interface. Substitution with a proline residue at this site would induce a kink in this region and may disrupt the flexibility needed to accommodate substrate binding. On the other hand, the Rab3GAP1 E24 is located in the α1 helix of this subunit that forms part of the Rab18 binding site. As this residue is predicted to engage in an ionic interaction with R407 of Rab3GAP2 to stabilize the α1 helix, substitution with a valine residue may disrupt this local structure critical to substrate binding. Lastly, Rab3GAP2 R426 is predicted to engage in an electrostatic interaction with E164 of Rab18 and replacement with a cysteine interferes with substrate binding. Indeed, as shown earlier, an E164A mutation to Rab18 prevented nucleotide exchange by Rab3GAP.
Discussion
The biochemical and structural data on human Rab3GAP reported here contribute to building the knowledge base of the divergent and enigmatic family of Rab GEF regulators. Our cryo-EM structure of the core human Rab3GAP confirmed that this Rab18 GEF shares no structural homology with other Rab GEFs with known structures8. Instead, we found that the Rab3GAP1 subunit of this heterodimeric complex shares structural similarity to Zwilch, a component of the mitotic RZZ complex that is involved in spindle assembly. Similar to Rab3GAP1, which binds the N-terminal β-propeller of Rab3GAP2 via its β-sheet structure, the recent cryo-EM structure of the RZZ complex showed that Zwilch similarly interacts with Rod within this complex44. Notably, the β-propeller from Rod associates with the curved β-sheet structure of Zwilch. Zwilch and Rod have been proposed to be Rab18 effectors but the molecular basis of how Zwilch and Rod binds Rab18 requires further investigation45.
Although human Rab3GAP is structurally unique compared to other Rab GEFs, this heterodimeric GEF appears to share two features found in several Rab GEFs. The first feature is the membrane-dependent enhancement of GEF activity. For example, it has been shown that the GEF activity of SH3BP5 is dependent on its substrate Rab11 being anchored to a membrane34. In the same vein, we demonstrated that membrane presentation of Rab18 resulted in a greater than 10-fold increase in GEF activity for both full-length and core Rab3GAP. The second feature is fine-tuning GEF activity by autoinhibition. The catalytic efficiency of core Rab3GAP is more than twice that of full-length Rab3GAP, leading us to speculate that the C-terminus of Rab3GAP2 is an autoinhibitory domain. In comparison, the catalytic activity of the Rab5 GEF Rabex-5, which is autoinhibited and has low inherent GEF activity over its substrate, is increased 2- to 3-fold through binding the Rab5 effector Rabaptin-531 or through truncations31,46. Based on our structural data showing the conformational flexibility of full-length Rab3GAP, we speculate that the Rab3GAP2 C-terminal domain suppresses Rab3GAP’s GEF activity by obstructing the substrate binding site. Future studies should focus on determining the basis of the enhanced GEF activity in the presence of membrane-anchored Rab18 and to delineate the potential autoinhibitory mechanism of Rab3GAP.
A major question concerning the function of Rab3GAP is why both subunits are required for its GEF activity. Our cryo-EM structure revealed that the two subunits form a stable complex by engaging in an extensive interaction interface. Our subsequent AlphaFold3 modeling predicted that the edge of this intersubunit interaction interface forms the substrate site where Rab18 binds through a platform composed of its α5 and α6 helices and β6 strand. This predicted model was validated by targeted mutagenesis in conjunction with in vitro GEF assays and localization studies. Consistent with this in silico-generated model, our multiple sequence alignment (MSA) analysis showed that the Rab3GAP and Rab18 residues predicted to interact with one another are found to be conserved (Supplementary Fig. 7a–c). However, as illustrated in a structural overlay of the AlphaFold3-predicted model of Rab3GAP-Rab18 with other representative solved structures of Rab GEFs (PDB code: 3TW8, 2OT3 and 6DJL) bound to their respective Rab substrate, this mode of GEF-Rab interaction has not been previously observed (Supplementary Fig. 11). Other Rab GEFs engage their substrates at the switch I, II and interswitch regions to induce structural rearrangement, reducing the affinity for the bound nucleotide5,8,32,34,47. Without directly engaging the switch regions, how does Rab3GAP promote Rab18 nucleotide exchange? We initially speculated that Rab3GAP1’s flexible tail region, which contains the GAP domain, may assist in promoting nucleotide exchange by engaging the switch regions of Rab18. However, this idea was refuted by our finding that a GAP domain deficient Rab3GAP retains GEF activity towards Rab18 (Fig. 6e). A systematic mutagenesis study on the small GTPase Gsp1/Ran uncovered the presence of allosteric sites away from the switch region and found that disturbances at these sites induced a preference for GTP binding over GDP binding48. Interestingly, the structural alignment of Rab18 with Gsp1/Ran shows that some of the corresponding allosteric sites map to regions where Rab18 binds Rab3GAP, with multiple sites located in α6 of the small GTPase. These findings raise the possibility that Rab3GAP could promote nucleotide release by modulating one or more of these putative allosteric sites in Rab18. Nevertheless, resolving the mystery surrounding Rab3GAP’s GEF mechanism would necessitate a high-resolution structural analysis of Rab3GAP in complex with Rab18.
Rab3GAP is the only known dual-function Rab regulator in eukaryotic cells. What allows Rab3GAP to serve both as a GAP for Rab3a and a GEF for Rab18 remains unclear. Previous studies demonstrated that Rab3GAP1 alone exhibits Rab3a GAP activity and identified a putative GAP domain located in the C-terminal region of this Rab3GAP subunit43. Our cryo-EM structure of core Rab3GAP revealed that the GAP domain is located in the flexible tail of Rab3GAP1 and is not involved in the interaction with Rab3GAP2. We found that removing the GAP domain in core Rab3GAP abolished its GAP activity toward Rab3a but had no effect on its GEF activity towards Rab18 (Fig. 6e, f). These findings suggest that the two catalytic activities are present in different locations of this complex. AlphaFold3 modeling of the Rab3GAP1-Rab3a complex showed that the GAP domain of Rab3GAP1 binds to the switch and interswitch region of its substrate (Supplementary Fig. 10a). In agreement with mutational analysis of previous studies, AlphaFold predictions are consistent with a mechanism similar to that of Ras and Rho GAPs with R728 of Rab3GAP1 and Q81 of Rab3a providing critical arginine and glutamine fingers to stabilize the transition state during GTP hydrolysis (Supplementary Fig. 10b)43. It is not known if Rab3GAP has preferential substrate specificity in neuronal cells where both Rab3 and Rab18 are expressed. Future studies could focus on mechanistic investigation of the GAP activity of Rab3GAP and further delineate the functional relationships between these two distinct catalytic activities.
Mutations to Rab3GAP1 and Rab3GAP2 cause the multisystem disorder WMS12,14,17,20. Data from our biochemical and structural studies show that three WMS-associated missense mutations do not affect the assembly or the overall architecture of Rab3GAP. Instead, these mutations disrupt Rab3GAP function by altering the Rab18 binding site. Our cryo-EM structure of core Rab3GAP, the Rab3GAP-Rab18 structural model predicted by AlphaFold3, and the biochemical platform to reconstitute human Rab3GAP offers experimental tools to predict and validate the potential impact of numerous variants of unknown significance of Rab3GAP1 and Rab3GAP2 subunits on the biochemical and structural properties of Rab3GAP.
Methods
Plasmids
Full-length human Rab3GAP1 (HsCD00860097) and Rab3GAP2 (HsCD00867180) genes were purchased from DNASU. Rab3GAP1 was subcloned into a pLIB vector containing an N-terminal TSP tag and Rab3GAP2 was subcloned into a pLIB vector containing a C-terminal 3x-FLAG tag. Site-directed mutagenesis was carried out on pLIB-TSP-Rab3GAP1 to generate T18P and E24V mutations and on pLIB-Rab3GAP2-3x-FLAG to generate R426C and 1–544 mutations. The biGBac protocol was used to assemble various combinations of both Rab3GAP1 and Rab3GAP2 into pBIG1A28. Rab3a, Rab11a and Rab18 were subcloned into pOPTGcH vectors containing a TEV-cleavable GST tag at the N-terminus and a 6x-His tag at the C-terminus.
Protein expression
Baculovirus containing Rab3GAP was produced from the appropriate vector (Supplementary Table 1) using the Baculovirus Expression Vector System. Optimized amounts of baculovirus were used to infect Sf9 cells at a density between 1.5 and 4 × 106 cells mL−1, and cells were harvested 72 to 96 h post infection. Cell pellets were flash frozen in liquid nitrogen and stored at −70 °C until use. Rabs were expressed in BL21 C41(DE3) E. coli induced with 0.5 mM isopropyl B-d-1-thiogalacpyranoside [IPTG] (GoldBio) for 4 h at 37 °C. Cells were then harvested, flash frozen in liquid nitrogen and stored at −70 °C.
Protein purification
Sf9 cell pellets expressing Rab3GAP were resuspended in buffer A (50 mM Tris pH 7.4, 150 mM NaCl, 5% glycerol, 0.1% Tween-20, 2 mM phenylmethylsulfonyl fluoride [PMSF] and Complete ethylenediaminetetraacetic acid (EDTA) free protease inhibitor). Cells were lysed with four cycles of sonication for 30 s using the Branson Sonicator 450 set to duty cycle 40% and output control 4. The lysate was centrifuged at 125,000 × g for 40 min at 4 °C. Supernatant containing Rab3GAP1 was incubated with Strep-Tactin XT 4Flow resin (IBA) and supernatant containing all other Rab3GAP constructs was incubated with Anti-FLAG M2 affinity gel (Sigma-Aldrich) equilibrated with buffer B (50 mM Tris pH 7.4, 150 mM NaCl and 5% glycerol). The resin was washed with buffer B and eluted with 50 mM biotin in buffer B for Strep-Tactin purifications and 250 µg mL−1 3x FLAG peptide in buffer B for Anti-FLAG M2 purifications. The eluate was concentrated using an Amicon 100 kDa concentrator (Millipore) and applied to an ENrich SEC650 10 × 300 column (Bio-Rad) equilibrated with buffer C (20 mM HEPES pH 7.5, 150 mM NaCl and 1 mM TCEP). Fractions containing desired proteins were pooled, concentrated, flash frozen and stored at −70 °C.
E. coli C41(DE3) cell pellets expressing Rabs were resuspended in buffer D (20 mM Tris pH 8, 100 mM NaCl, 2 mM B-mercaptoethanol (BME) and 2 mM PMSF). Cells were lysed with four cycles of sonication for 60 s at duty cycle 50% and output control 5. The lysate was centrifuged at 20,000 × g for 40 min at 4 °C. The supernatant was incubated with glutathione resin (GenScript) equilibrated with buffer E (20 mM Tris pH 8, 100 mM NaCl and 2 mM BME). The resin was washed with buffer E and eluted with 30 mM glutathione in buffer E. TEV protease was added to the eluate and dialyzed in buffer F (20 mM Tris pH 8, 100 mM NaCl, 10 mM BME and 5 mM EDTA) overnight at 4 °C. The protein was then applied to a HiPrepQ FF 16/10 column (GE Healthcare) equilibrated with buffer E and gradient elution with buffer G (20 mM Tris pH 8, 1 M NaCl and 2 mM BME) was performed to isolate target proteins. Glutathione resin in buffer E was used to remove GST, and cleaved Rabs were collected in the flowthrough. The flowthrough was concentrated, flash frozen in liquid nitrogen and stored at −70 °C.
Lipid vesicle preparation
Nickelated lipid vesicles were prepared by combining chloroform stocks of phosphatidylcholine (egg yolk PC, Avanti), phosphatidylethanolamine (egg yolk PE, Sigma), phosphatidylserine (porcine brain PS, Avanti), L-α-phosphatidylinositol (soy PI, Avanti) and 18:1 DGS NTA(Ni) (Avanti) according to Supplementary Table 2. Argon gas was used to evaporate residual chloroform followed by overnight desiccation under vacuum. Liposome buffer (20 mM HEPES pH 7.5, 150 mM NaCl) was used to resuspend the lipid film at a final concentration of 1.5 mg mL−1. The mixture was vortexed vigorously for 20 min then sonicated for 15 min with a Branson 1510 sonicator. Vesicles were flash frozen in liquid nitrogen then warmed in a water bath for a total of 10 freeze-thaw cycles. Vesicles were extruded 21 times through a 100 nm polycarbonate membrane (Avanti) and stored at −70 °C.
In vitro GEF assay
His-tagged Rabs were purified as described above in protein purification. EDTA was added at a final concentration of 5 mM and incubated at 25 °C for 30 min. Mant-GDP (Thermo Scientific) was added at a fivefold molar excess and incubated at 25 °C for 1 h. MgCl2 was added at a final concentration of 10 mM and incubated at 25 °C for 1 h. Rabs were loaded onto a Superdex 75 Increase column (GE Healthcare) equilibrated with buffer H (20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, and 1 mM MgCl2). Fractions containing Mant-GDP loaded Rabs were pooled, concentrated, flash frozen in liquid nitrogen at stored at −70 °C. Ten µL reactions were prepared with a final concentration of 4 µM Mant-GDP loaded Rab, 0–300 nM Rab3GAP complex and 100 µM GTPγS in buffer H. Rab and membrane (0–0.2 mg mL−1) were aliquoted into a Corning 384-well low volume polystyrene microplate (4514). Rab3GAP complex and GTPγS were added to the plate and a BioTek Synergy HTX Multimode Reader was used to measure the fluorescent signal for 1 h at 25 °C (excitation h = 366 nm; emission h = 443 nm). GraphPad Prism 7 Software was used to analyze data and kcat/Km analysis was performed as described by Delprato et al. Curves were fit to a non-linear one-phase exponential decay model according to the following equation, I(t) = (Io − If)*exp(−kobs) + If (GraphPad Software). I(t) is the emission intensity at time t, Io is the initial intensity, If is the intensity at t = inf. Catalytic efficiency, kcat/Km, was obtained by the slope of a linear least squares fit to kobs = kcat/Km*[GEF] + ki, with ki being kobs in the absence of GEF.
In vitro GAP assays
GAP activity was measured using the Promega GTPase-Glo Assay with GST-tagged Rab3a and Rab3GAP1, core Rab3GAP or Rab3GAPΔGAP. Twenty µL reactions were prepared in a white CELLSTAR 96-well plate with a final concentration of 5 µM GTP, 0.5 mM DTT, 8 µM GST-Rab3a and 0 nM (Mock) or 500 nM GAP in Promega GTPase/GAP buffer and incubated at 25 °C for 1 h. Twenty µL of 1x GTPase-Glo reagent with 10 mM ADP in GTPase-Glo buffer was added to each reaction and incubated at 25 °C for 30 min, followed by 40 µL detection reagent and incubated at 25 °C for 10 min. Luminescence was measured using a BioTek Synergy HTX Multimode Reader. Reactions were performed in triplicate.
HDX-MS sample preparation
HDX reactions comparing Rab3GAP1 apo to Rab3GAP1 + Rab3GAP2 were carried out in 50 µL reaction volumes containing 8 pmol of Rab3GAP1 or Rab3GAP1 + Rab3GAP2. Prior to the exchange reactions, 1.5 mM EDTA was added to each sample and samples were incubated on ice for 1 h. Exchange reactions were initiated by the addition of 46 µL of D2O buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 94.34% D2O (V/V)) to 4 µL of protein mixture (final D2O concentration of 86.79%). The reactions proceeded for 0.3 s (3 s on ice), 3, 30, 300, or 3000 s at room temperature, before being quenched with ice cold acidic quench buffer resulting in a final concentration of 0.6 M guanidine-HCl and 0.9% formic acid post quench. All conditions and timepoints were created and run in independent triplicate. Samples were flash frozen immediately after quenching and stored at −80 °C.
Protein digestion and MS/MS data collection
Protein samples were rapidly thawed and injected onto an integrated fluidics system containing a HDx-3 PAL liquid handling robot and climate-controlled (2 °C) chromatography system (LEAP Technologies), a Dionex Ultimate 3000 UHPLC system, as well as an Impact HD QTOF Mass spectrometer (Bruker). The full details of the automated LC system are described previously49. The Rab3GAP1 ± Rab3GAP2 samples were run over one immobilized pepsin column (Trajan; ProDx protease column, 2.1 mm × 30 mm PDX.PP01-F32) at 200 µL min−1 for 3 min at 8 °C. The resulting peptides were collected and desalted on a C18 trap column (ACQUITY UPLC BEH C18 1.7 µm column, 2.1 mm × 5 mm; Waters 186004629). The trap was subsequently eluted in line with an ACQUITY 1.7 μm particle, 100 mm × 1 mm C18 UPLC column (Waters), using a gradient of 3–35% B (Buffer A 0.1% formic acid; Buffer B 100% acetonitrile) over 11 min immediately followed by a gradient of 35–80% over 5 min. Mass spectrometry experiments acquired over a mass range from 150 to 2200 m/z using an electrospray ionization source operated at a temperature of 200 °C and a spray voltage of 4.5 kV.
Peptide identification
For the Rab3GAP1 apo and Rab3Gap1 + Rab3GAP2 experiment, peptides were identified from the non-deuterated samples of Rab3GAP1 using data-dependent acquisition following tandem MS/MS experiments (0.5 s precursor scan from 150–2000 m/z; twelve 0.25 s fragment scans from 150–2000 m/z). MS/MS datasets were analyzed using PEAKS7 (PEAKS), and peptide identification was carried out by using a false discovery-based approach, with a threshold set to 0.1% using a database of purified proteins and known contaminants. The search parameters were set with a precursor tolerance of 20 ppm, fragment mass error 0.02 Da, charge states from 1–8, leading to a selection criterion of peptides that had a −10logP score of 26.6.
Mass analysis and measurement of deuterium incorporation
HD-Examiner Software (Sierra Analytics) was used to automatically calculate the level of deuterium incorporation into each peptide. All peptides were manually inspected for correct charge state, correct retention time, appropriate selection of isotopic distribution, etc. Deuteration levels were calculated using the centroid of the experimental isotope clusters. Results are presented as relative levels of deuterium incorporation and the only control for back exchange was the level of deuterium present in the buffer (86.79%). Differences in exchange in a peptide were considered significant if they met all three of the following criteria: >5% change in exchange, >0.4 Da difference in exchange, and a p value < 0.01 using a two-tailed Student’s t test. Samples were only compared within a single experiment and were never compared to experiments completed at a different time with a different final D2O level. The data analysis statistics for all HDX-MS experiments are in the source data according to the guidelines set out previously50. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository51 with the dataset identifier PXD033072.
Description of %D and #D graphs
The raw HDX data are shown in two different formats. To allow for visualization of differences across all peptides, we utilized number of deuteron difference (#D) plots (Fig. 5b). The raw peptide deuterium incorporation graphs for a selection of peptides with significant differences are shown in Fig. 5c and Supplementary Fig. 5, with the raw data for all analyzed peptides in the source data. These plots show the total difference in deuterium incorporation over the entire H/D exchange time course, with each point indicating a single peptide.
Negative stain electron microscopy and image processing
Negative-stained specimens were prepared as previously described52. Briefly, FL Rab3GAP and core Rab3GAP from their respective peak fraction of size exclusion chromatography were adsorbed onto a carbon-coated copper grid for 0 s, washed with ddH2O and stained with 0.75% uranyl formate (Electron Microscopy Sciences) for 30 s. Using the Talos L120C Transmission Electron Microscope (Thermo Fisher Scientific) equipped with a Ceta camera, micrographs were collected with an accelerating voltage of 120 kV, nominal magnification of ×49,000 and a defocus of 2 µm. The micrographs were imported and processed using CryoSPARC v4.0.353. A small subset of particles was manually picked and extracted with a box size of 100 pixels. 2D class averages were created from these particles to be used to generate templates for particle picking. FL Rab3GAP was subjected to multiple rounds of 2D classification to curate particle quality. A 2D classification was used to determine the overall architecture of core Rab3GAP.
Cryo-EM sample preparation and data collection
C-Flat 2/1 grids were subjected to a 25 s glow discharge at 15 mA using a Pelco easiGlow glow-discharger. Following this, 3 µL of purified truncated Rab3GAP1/2, at a concentration of 0.3 mg mL−1, was adsorbed onto the grids and quickly plunge frozen into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific), with a blot force of 5 and 2 s blot time at 100% humidity and 4 °C. To assess the quality of the ice and particle quality, grids were screened using the 200 kV Glacios transmission electron microscope, equipped with a Falcon 3EC direct electron detector (DED). A total of 19,822 movies were collected using the Titan Krios transmission electron microscope (Thermo Fisher Scientific) operating at 300 kV, equipped with a Falcon IV DED and Selectris energy filter. The movies were obtained at a nominal magnification of ×215,000, corresponding to a pixel size of 0.59 Å, and with a total dose of 50 e- Å−2 over 59 frames.
Cryo-EM data processing and model building
The cryo-EM data in this study was processed using cryoSPARC v.4.0.3. The movies were subjected to patch motion correction with a Fourier cropping by a factor of 2 and the CTF parameters of the micrographs were generated using patch CTF estimation with default settings. Initially, a subset of movies was selected for template-free 2D class averaging to generate templates for particle picking. These templates were employed to pick 15,244,538 particles from the whole dataset. Particles were extracted with a box size of 640 pixels, Fourier cropped to a box size of 160 pixels, and were subjected to multiple rounds of 2D classification to discard poorly defined particles, resulting in the selection of high-quality particles for further processing. Particles from the best classes were re-extracted with a 640-pixel box size to be utilized for multiclass ab initio reconstruction and heterogeneous refinement using two classes. These particles were used to carry out per-particle local-motion correction with a 1280-pixel box size, downsized to 640 pixels. Particles were then subjected to additional rounds of 2D classification, ab initio reconstruction, homogeneous refinement, and non-uniform-refinement, which generated a global reconstruction with an overall resolution of 3.39 Å, as assessed by the Fourier shell correlation 0.143 criterion. Local refinements of the rigid region of the complex using masks were performed to further improve the map quality to be resolved at 3.37 Å. Local resolutions were estimated using the local resolution estimation tool in cryoSPARC and auto-sharpening was performed in PHENIX54. Structural models of Rab3GAP1 and Rab3GAP2 from the AlphaFold Protein Structure Database were next fit into the cryo-EM density map using Chimera55. This was followed by iterative rounds of refinement in Phenix.real_space_refine and manual model building in COOT56. Structural and refinement statistics is summarized in Supplementary Table 3.
Bioinformatics
Protein structure and binding interfaces were predicted for core Rab3GAP with and without Rab18 using AlphaFold3 with default settings except for minimization of the top-ranked structure57,58. Prediction confidence metrics were reported as pTMscore, iPTMscore, PAE and pLDDT. The cryo-EM structure of core Rab3GAP was used as input for Foldseek to identify structurally related proteins37. Clustal Omega Multiple Sequence Alignment59 was used to align the sequences of human Rabs, followed by ESPript 3.0 analysis to identify conserved regions60.
Cell culture
HeLa cells were cultured in DMEM high glucose with L-glu Na-pyruvate (Sigma-Aldrich, D6429) supplemented with 10% (v/v) fetal bovine serum (FBS) (Sigma-Aldrich, F1051) at 37 °C and 5% CO2. Cells are routinely checked for mycoplasma contamination.
Immunofluorescence
HeLa cells were seeded at 2.5 × 105 cells per well onto #1.5 coverslips 24 h prior to co-transfection with 0.25 μg of pcDNA3.1-GFP-Rab18 (Genscript) using Lipofectamine 3000 (Invitrogen, L3000008) according to the manufacturer’s protocol. Transfection reagent was removed after 5 h and replaced with fresh media and allowed to incubate for another 19 h. Cells were rinsed 2x with PBS and then fixed in 3% PFA + 0.2% Glutaraldehyde for 15 min at room temperature. Coverslips were washed 3x with PBS before permeabilization with 0.2% Triton X-100 for 5 min at room temperature. Coverslips were washed 3x with PBS before incubating with 1 mg mL−1 sodium borohydride for 10 min at room temperature. Coverslips were washed 3x with PBS and then blocked using 2.5% Bovine serum albumin (Sigma-Aldrich) for 1 h at room temperature. Nuclei were stained with 0.5 mg mL−1 DAPI (Invitrogen, D1306) for 5 min. Cells were washed 3x with PBS and then coverslips were mounted using ProLong Glass Antifade (Invitrogen, P36980). Fixed samples were imaged on a 63x/1.40 NA objective (oil immersion, HC PL APO Leica Microsystems) on a Leica STELLARIS 5 LIAchroic inverted scanning confocal microscope. GFP signal was acquired using a 488 nM laser line and the power HyD S detector (Leica Microsystems). Brightness was adjusted in ImageJ/Fiji.
Statistical analysis
For all GAP and GEF assays, experiments were performed in technical triplicate with mean ± SEM shown in figures. Statistical analysis between conditions was performed using a two-tailed Student’s t test or ordinary one-way ANOVA (GraphPad Prism). For HDX-MS assays, experiments were performed in technical triplicated with mean ± SD shown in figures. Statistical analysis between conditions was performed using a two-tailed Student’s t test. The following legend is used for statistical significance: *p < 0.05 and ns p > 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The 3D reconstruction of core Rab3GAP is available at the Electron Microscopy Data Bank under accession code EMD-43655. The atomic coordinates for core Rab3GAP are available at the Protein Data Bank under accession code 8VYB. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository51 with the dataset identifier PXD033072. All other data are available from the corresponding author upon request. Source data are provided with this paper.
References
Derby, M. C. & Gleeson, P. A. New insights into membrane trafficking and protein sorting. Int. Rev. Cytol. 261, 47–116 (2007).
Linders, P. T. A., Peters, E., Ter Beest, M., Lefeber, D. J. & van den Bogaart, G. Sugary logistics gone wrong: membrane trafficking and congenital disorders of glycosylation. Int. J. Mol. Sci. 21, 1–28 (2020).
Yarwood, R., Hellicar, J., Woodman, P. G. & Lowe, M. Membrane trafficking in health and disease. Dis. Model Mech. 13, dmm043448 (2020).
Li, G. Rab GTPases, membrane trafficking and diseases. Curr. Drug Targets 12, 1188 (2011).
Müller, M. P. & Goody, R. S. Molecular control of Rab activity by GEFs, GAPs and GDI. Small GTPases 9, 5–21 (2018).
Hutagalung, A. H. & Novick, P. J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119 (2011).
Cabrera, M. & Ungermann, C. Guanine nucleotide exchange factors (GEFs) have a critical but not exclusive role in organelle localization of Rab GTPases. J. Biol. Chem. 288, 28704 (2013).
Toma-Fukai, S. & Shimizu, T. Structural insights into the regulation mechanism of small GTPases by GEFs. Molecules 24, 3308 (2019).
Traut, T. W. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 140, 1–22 (1994).
Fukui, K. et al. Isolation and characterization of a GTPase activating protein specific for the Rab3 subfamily of small G proteins. J. Biol. Chem. 272, 4655–4658 (1997).
Nagano, F. et al. Molecular cloning and characterization of the noncatalytic subunit of the Rab3 subfamily-specific GTPase-activating protein. J. Biol. Chem. 273, 24781–24785 (1998).
Gerondopoulos, A. et al. Rab18 and a Rab18 GEF complex are required for normal ER structure. J. Cell Biol. 205, 707 (2014).
Klöpper, T. H., Kienle, N., Fasshauer, D. & Munro, S. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol. 10, 71 (2012).
Handley, M. T. et al. Mutation spectrum in RAB3GAP1, RAB3GAP2, and RAB18 and genotype–phenotype correlations in Warburg micro syndrome and Martsolf syndrome. Hum. Mutat. 34, 686–696 (2013).
Khalesi, R. et al. Novel manifestations of Warburg micro syndrome type 1 caused by a new splicing variant of RAB3GAP1: a case report. BMC Neurol. 21, 180 (2021).
Alavi, O. et al. Two novel Warburg micro syndrome 1 cases caused by pathogenic variants in RAB3GAP1. Hum. Genome Var. 8, 39 (2021).
Morris-Rosendahl, D. J. et al. New RAB3GAP1 mutations in patients with Warburg micro syndrome from different ethnic backgrounds and a possible founder effect in the Danish. Eur. J. Hum. Genet. 18, 1100 (2010).
Bem, D. et al. Loss-of-function mutations in RAB18 cause Warburg micro syndrome. Am. J. Hum. Genet. 88, 499 (2011).
Handley, M. T. et al. Warburg micro syndrome is caused by RAB18 deficiency or dysregulation. Open Biol. 5, 150047 (2015).
Aligianis, I. A. et al. Mutation in Rab3 GTPase-activating protein (RAB3GAP) noncatalytic subunit in a kindred with Martsolf syndrome. Am. J. Hum. Genet. 78, 702–707 (2006).
Ozeki, S. et al. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118, 2601–2611 (2005).
Martin, S., Driessen, K., Nixon, S. J., Zerial, M. & Parton, R. G. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280, 42325–42335 (2005).
Dejgaard, S. Y. et al. Rab18 and Rab43 have key roles in ER-Golgi trafficking. J. Cell Sci. 121, 2768–2781 (2008).
Dejgaard, S. Y. & Presley, J. F. Rab18: new insights into the function of an essential protein. Cell Mol. Life Sci. 76, 1935–1945 (2019).
Xu, D. et al. Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 217, 975–995 (2018).
Takáts, S. et al. The Warburg micro syndrome-associated Rab3GAP-Rab18 module promotes autolysosome maturation through the Vps34 Complex I. FEBS J. 288, 190–211 (2021).
Hantan, D., Yamamoto, Y. & Sakisaka, T. VAP-B binds to Rab3GAP1 at the ER: its implication in nuclear envelope formation through the ER-Golgi intermediate compartment. Kobe J. Med. Sci. 60, 48–56 (2014).
Weissmann, F. et al. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc. Natl Acad. Sci. USA 113, E2564–E2569 (2016).
Kanie, T. & Jackson, P. K. Guanine nucleotide exchange assay using fluorescent MANT-GDP. Bio Protoc. 8, e2795 (2018).
Harris, N. J. et al. Biochemical insight into novel Rab-GEF activity of the mammalian TRAPPIII complex. J. Mol. Biol. 433, 167145 (2021).
Lauer, J. et al. Auto-regulation of Rab5 GEF activity in Rabex5 by allosteric structural changes, catalytic core dynamics and ubiquitin binding. Elife 8, e46302 (2019).
Wu, X. et al. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc. Natl Acad. Sci. USA 108, 18672–18677 (2011).
Esters, H. et al. Vps9, Rabex-5 and DSS4: proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J. Mol. Biol. 310, 141–156 (2001).
Jenkins, M. L. et al. Structural determinants of Rab11 activation by the guanine nucleotide exchange factor SH3BP5. Nat. Commun. 9, 1–13 (2018).
Cherfils, J. & Zeghouf, M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309 (2013).
Zhang, Z., Shigematsu, H., Shimizu, T. & Ohto, U. Improving particle quality in cryo-EM analysis using a PEGylation method. Structure 29, 1192–1199.e4 (2021).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 1–4 https://doi.org/10.1038/s41587-023-01773-0 (2023).
Mosalaganti, S. et al. Structure of the RZZ complex and molecular basis of its interaction with Spindly. J. Cell Biol. 216, 961 (2017).
Pereira, C. et al. Self-assembly of the RZZ complex into filaments drives kinetochore expansion in the absence of microtubule attachment. Curr. Biol. 28, 3408–3421.e8 (2018).
James, E. I., Murphree, T. A., Vorauer, C., Engen, J. R. & Guttman, M. Advances in hydrogen/deuterium exchange mass spectrometry and the pursuit of challenging biological systems. Chem. Rev. 122, 7562 (2022).
Masson, G. R., Jenkins, M. L. & Burke, J. E. An overview of hydrogen deuterium exchange mass spectrometry (HDX-MS) in drug discovery. Expert Opin. Drug Discov. 12, 981–994 (2017).
Yin, R., Feng, B. Y., Varshney, A. & Pierce, B. G. Benchmarking AlphaFold for protein complex modeling reveals accuracy determinants. Protein Sci. 31, e4379 (2022).
Clabecq, A., Henry, J. P. & Darchen, F. Biochemical characterization of Rab3-GTPase-activating protein reveals a mechanism similar to that of Ras-GAP. J. Biol. Chem. 275, 31786–31791 (2000).
Raisch, T. et al. Structure of the RZZ complex and molecular basis of Spindly‐driven corona assembly at human kinetochores. EMBO J. 41, e110411 (2022).
Gillingham, A. K., Sinka, R., Torres, I. L., Lilley, K. S. & Munro, S. Toward a comprehensive map of the effectors of Rab GTPases. Dev. Cell 31, 358 (2014).
Zhang, Z. et al. Molecular mechanism for Rabex-5 GEF activation by Rabaptin-5. Elife 2014, 2687 (2014).
Delprato, A. & Lambright, D. G. Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat. Struct. Mol. Biol. 14, 406–412 (2007).
Mathy, C. J. P. et al. A complete allosteric map of a GTPase switch in its native cellular network. Cell Syst. 14, 237–246.e7 (2023).
Stariha, J. T. B., Hoffmann, R. M., Hamelin, D. J. & Burke, J. E. Probing protein–membrane interactions and dynamics using hydrogen–deuterium exchange mass spectrometry (HDX-MS). Methods Mol. Biol. 2263, 465–485 (2021).
Masson, G. R. et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 16, 595 (2019).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543 (2022).
Ohi, M., Li, Y., Cheng, Y. & Walz, T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 (2004).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679 (2022).
Madeira, F. et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50, W276–W279 (2022).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Acknowledgements
This work was supported by a Project Grant from the Canadian Institutes of Health Research to C.K.Y. (PJT-168907) and a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada to C.K.Y. (AWD-007855). Grids were prepared and data collected at the High Resolution Macromolecular Electron Microscopy (HRMEM) facility at the University of British Columbia (https://cryoem.med.ubc.ca). We thank Claire Atkinson, Joeseph Felt, Liam Worrall and Natalie Strynadka. HRMEM is funded by the Canadian Foundation for Innovation and the British Columbia Knowledge Development Fund. Fluorescent images were collected and/or image analysis for this work was performed in the University of British Columbia Life Sciences Institute Imaging Core Facility, RRID: SCR_023783. We thank Guang Guo for support with image analysis. Molecular graphics and analyses performed with UCSF Chimera (developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311), ChimeraX (developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases), and PyMOL Molecular Graphics System (Schrödinger, LLC).
Author information
Authors and Affiliations
Contributions
G.M.J.F., K.M.N., H.R.S., S.N., A.L.S., J.E.B., and C.K.Y. designed research; G.M.J.F., K.M.N., H.R.S., S.N., A.L.S., M.A.H.P., X.W., M.L.J., and M.G. performed research; G.M.J.F., K.M.N., H.R.S., S.N., A.L.S., M.A.H.P., J.E.B., and C.K.Y. analyzed data; and G.M.J.F., K.M.N., and C.K.Y. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
J.E.B. reports personal fees from Scorpion Therapeutics and Reactive Therapeutics and research contracts from Novartis and Calico Life Sciences. All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Daniel Deredge who co-reviewed with Juliet Obi; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fairlie, G.M.J., Nguyen, K.M., Nam, SE. et al. Biochemical and structural characterization of Rab3GAP reveals insights into Rab18 nucleotide exchange activity. Nat Commun 16, 479 (2025). https://doi.org/10.1038/s41467-025-55828-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-55828-8
This article is cited by
-
Biallelic null RAB3GAP1 variants impair cortical development and autophagy in Warburg Micro syndrome: evidence from fetal brain tissue and patient fibroblasts
Acta Neuropathologica Communications (2025)