Abstract
Autophagy, a conserved catabolic process implicated in a diverse array of human diseases, requires efficient fusion between autophagosomes and lysosomes to function effectively. Recently, SNAP47 has been identified as a key component of the dual-purpose SNARE complex mediating autophagosome-lysosome fusion in both bulk and selective autophagy. However, the spatiotemporal regulatory mechanisms of this SNARE complex remain unknown. In this study, we found that SNAP47 undergoes acetylation followed by deacetylation during bulk autophagy and mitophagy. The acetylation status of SNAP47 is regulated by the acetyltransferase CBP and the deacetylase HDAC2. Notably, the spatiotemporal regulatory dynamics of SNAP47 acetylation differ between bulk autophagy and mitophagy due to distinct regulation on the activity of acetyltransferase and deacetylase. Acetylated SNAP47 inhibits autophagosome-lysosome fusion by indirectly impeding SNARE complex assembly. Mechanistically, deacetylated SNAP47 recruits HOPS components to autophagic vacuoles independently of STX17 and STX17-SNAP47 interaction, while acetylated SNAP47 inhibits this recruitment, consequently leading to the failure of SNARE complex assembly. Taken together, our study uncovers a SNAP47 acetylation-dependent regulatory mechanism governing autophagosome-lysosome fusion by modulating the recruitment of HOPS to autophagic vacuoles without involving STX17, SNAP47-STX17 interaction and ternary SNARE complex formation.
Similar content being viewed by others
Introduction
Autophagy is a highly conserved lysosome-dependent degradation process, vital for quality control and cellular homeostasis by eliminating surplus or impaired organelles, protein aggregates, and intracellular pathogens1,2. Dysfunction in autophagy has been implicated in numerous human diseases, including cancer, neurodegenerative disorders, cardiovascular disorders, and metabolic syndromes3,4,5.
In response to diverse stimuli, autophagosome formation is initiated, which subsequently undergoes fusion with lysosomes for the degradation of substrates6,7,8. The STX17-SNAP29-VAMP7/VAMP8 SNARE complex mediates the fusion process between autophagosomes and lysosomes during bulk autophagy9,10. Incomplete fusion between autophagosomes and lysosomes in STX17 deficient cells leads to the discovery of a secondary SNARE complex YKT6-SNAP29-STX7 complex in mammalian cells11. YKT6 is also involved in autophagosome-lysosome fusion in Drosophila and Saccharomyces cerevisiae, but functions through different mechanisms12,13,14.
Recently, we identified the STX17-SNAP47-VAMP7/VAMP8 complex as a dual-role SNARE complex responsible for autophagosome-lysosome fusion in both selective autophagy and bulk autophagy15. SNAP47 is recruited to phagophores and autophagosomes through its Pleckstrin homology domain, independently of STX17, via coincident binding to ATG8s and PI(4,5)P215. Subsequently, it assembles with STX17 and VAMP7/VAMP8 to form a functional ternary SNARE complex mediating autophagosome-lysosome fusion15. Nevertheless, the precise spatiotemporal regulatory mechanisms governing this SNARE complex remain unknown.
In this study, we found that SNAP47 undergoes acetylation and subsequent deacetylation in response to the induction of bulk autophagy or mitophagy. The acetylation status of SNAP47 is regulated by the acetyltransferase CBP and the deacetyltransferase HDAC2. The spatiotemporal regulation of SNAP47 acetylation varies between bulk autophagy and mitophagy, primarily because of the differential control of acetyltransferase and deacetylase activities. Deacetylation of SNAP47 indirectly enhances SNARE complex assembly and promotes autophagosome-lysosome fusion by facilitating HOPS recruitment to autophagic vacuoles, independently of STX17 and the SNARE complex assembly. Collectively, our findings elucidate a SNAP47 acetylation-mediated spatiotemporal regulatory mechanism governing autophagosome-lysosome fusion without involving STX17, SNAP47-STX17 interaction and ternary SNARE complex formation.
Results
Identification of acetylation sites on SNAP47
To investigate the spatiotemporal regulatory mechanism on the function of SNAP47 in autophagosome-lysosome fusion, we found that both endogenous and exogenous SNAP47 was acetylated (Fig. 1a and Supplementary Fig. 1a) and the level of SNAP47 acetylation increased in the presence of the HDAC family inhibitor trichostatin A (TSA), in contrast to sirtuin family inhibitor nicotinamide (NAM) (Fig. 1b, c). In addition, the acetylation of SNAP47 did not show further increase in the presence of both TSA and NAM compared to TSA alone (Fig. 1b, c). The acetylation level of SNAP47 decreased in bulk autophagy induced by starvation with Earle’s Balanced Salt Solution (EBSS) or mitophagy induced by oligomycin and antimycin A (OA), respectively (Fig. 1d, e), suggesting SNAP47 is acetylated and undergoes deacetylation during bulk autophagy or mitophagy. Treatment with thapsigargin, puromycin, and hypoxia—inducers of ER-phagy, aggrephagy, and mitophagy, respectively—also resulted in a lesser reduction of SNAP47 deacetylation compared to OA and EBSS treatment (Supplementary Fig. 1b). A similar deacetylation pattern was observed in STX17(Supplementary Fig. 1c). Mass spectrometry analysis identified seven acetylation sites of SNAP47 at residues K58, K131, K156, K259, K286, K301, and K377, respectively (Fig. 1f, g and Supplementary Fig. 1d–j). These seven lysine residues were found to be highly conserved across species (Fig. 1h), indicating the possible conserved acetylation sites in different eukaryotes.
a Immunoblot analysis of Flag-SNAP47 acetylation in HEK293T cells (n = 3 independent experiments). b, c Immunoblot analysis of Flag-SNAP47 acetylation in HEK293T cells treated with or without TSA alone, NAM alone or a combination of TSA and NAM (b). The levels of Ace-lys were quantified by ImageJ software and normalized with Flag-SNAP47 (c). d, e Immunoblot analysis of Flag-SNAP47 acetylation in HEK293T cells stably expressing Parkin with or without EBSS or OA (oligomycin 2.5 µM and antimycin-A 250 nM) treatment (d). The levels of Ace-lys were quantified by ImageJ software and normalized with Flag-SNAP47 (e). f, g The schematic diagram of SNAP47 acetylation sites (f). Structure model of human SNAP47 (UniProt entry: 501A0A087X0B7) generated by AlphaFold 2. The acetylation sites in SNAP47 are highlighted in red, and the PH domain, SNARE motif, and second SNARE motif are colored blue, green, and laurel green, respectively (g). h Alignment of amino acid sequences of SNAP47 from various species, with amino acids numbered based on the human sequence. Yellow texts highlight the amino acids corresponding to acetylated lysine residues of SNAP47 identified through mass spectrometry. i–l Characterization of the critical acetylation sites of SNAP47 through acetylation analysis of Flag-SNAP47. HEK293T cells were transfected with WT or lysine substitution mutants of Flag-SNAP47 (i). Twenty-four hours after transfection, cells were treated with or without TSA for 16 h (k). The levels of Ace-lye were quantified by ImageJ software and normalized with Flag-SNAP47 (j, l). Data in (c–l) are presented as mean ± SD, n = 3 independent experiments, statistical significance was assessed by a one-way ANOVA (c–j) or two-way ANOVA (l). P values are indicated in the figure. Source data are provided as a Source Data file.
To determine which are the major acetylation sites of acetyltransferase(s), we individually introduced non-synonymous mutations to convert the seven lysine sites identified by mass spectrometry into arginine residues to mimic the deacetylation status of SNAP47. Assessment of SNAP47 acetylation levels in cells harboring each of the mutated acetylation sites revealed a notable decrease in SNAP47 acetylation in the K58R, K259R and K377R mutants regardless of the presence of TSA (Fig. 1i-l). We then generated a SNAP47-3KR triple mutant (i.e., harboring K58R, K259R, and K377R) which exhibited almost complete abolition of SNAP47 acetylation (Fig. 1k, l). These findings suggest that the primary acetylation sites of SNAP47 are residues K58, K259, and K377.
CBP and HDAC2 have antagonistic effects on SNAP47 acetylation
To identify the acetyltransferase(s) responsible for the acetylation of SNAP47, the five most common acetyltransferases related to cell metabolism were overexpressed. We observed that only CBP (CREB binding protein) overexpression led to increased acetylation levels of SNAP47. Other acetyltransferases, including its paralogue p300, did not show significant effects (Fig. 2a, b). Knockdown of CBP correspondingly reduced SNAP47 acetylation, which was restored by complementation with wild-type (WT) CBP (Fig. 2c, d). Additionally, treatment with CTB, a p300/CBP activator, or C646, their inhibitor, respectively promoted and suppressed SNAP47 acetylation (Fig. 2e, f). These findings suggest that CBP is responsible for SNAP47 acetylation.
a, b Acetylation of Flag-SNAP47 in HEK293T cells overexpressing the indicated individual histone acetyltransferases (a). Quantification of Ace-lys by ImageJ software and normalized with Flag-SNAP47 (b). c, d Flag-SNAP47 Acetylation in HEK293T cells transfected with CBP-HA after incubation with CBP siRNA (c). Quantification of Ace-lys by ImageJ software and normalized with Flag-SNAP47 (d). e, f Flag-SNAP47 acetylation in HEK293T cells treated with CBP activator CTB or inhibitor C646 (e). Quantification of Ace-lys by ImageJ software and normalized with Flag-SNAP47 (f). g Co-IP analysis of interactions between Flag-SNAP47 and CBP-HA in HEK293T cells (three independent experiments). h, i In vitro acetylation assay using purified GST-SNAP47 and CBP-HA immunoprecipitated from HEK293T cells in the presence of C646 or CTB (h). Quantification of Ace-lys by ImageJ software and normalized with GST-SNAP47 (i). j, k Acetylation of Flag-SNAP47 WT or 3KR (K58R, K259R and K377R) in HEK293T cells with or without CBP-HA (j). LE: low exposure; HE: high exposure. Quantification of Ace-lys by ImageJ software and normalized with Flag-SNAP47 (k). l, m In vitro acetylation assays using purified GST-SNAP47 WT or 3KR and CBP-HA immunoprecipitated from HEK293T cells (l). Quantification of Ace-lys by ImageJ software and normalized with GST-SNAP47 (m). n–q Flag-SNAP47 acetylation in HEK293T cells transfected with or without CBP-HA treated with EBSS, with or without TSA (n), or with NAM alone or both NAM and TSA (p). Quantification of Ace-lys by ImageJ software and normalized with Flag-SNAP47 (o, q). r–u Flag-SNAP47 acetylation in HEK293T cells expressing Parkin with or without CBP-HA treated with OA, with or without TSA (r), or with NAM alone or both NAM and TSA (t). Quantification of Ace-lys by ImageJ software and normalized with Flag-SNAP47 (s, u). Data in (b–u) are presented as mean ± SD, n = 3 independent experiments, statistical significance was assessed by a one-way ANOVA (b–m) or two-way ANOVA (o–u). P values are indicated in the figure. Source data are provided as a Source Data file.
Based on the observed relationship between CBP and SNAP47, we investigated whether SNAP47 is a direct substrate of CBP. Co-immunoprecipitation (Co-IP) results showed an in vivo interaction between SNAP47 and CBP (Fig. 2g). In in vitro acetylation assays, where recombinant GST-SNAP47 was incubated with immunoprecipitated CBP-HA in the presence of acetyl-CoA, we observed the acetylation of SNAP47 by CBP (Fig. 2h, i). Furthermore, C646 inhibited SNAP47 acetylation, while CTB promoted it (Fig. 2h, i). Importantly, the acetylation of SNAP47 was significantly reduced in both in vivo and in vitro when using the SNAP47-3KR triple mutant (Fig. 2j–m). These results collectively confirm that CBP directly catalyzes the acetylation of SNAP47.
Furthermore, we observed a marked decrease in SNAP47 acetylation upon starvation, whereas its acetylation significantly increased in the presence of TSA with or without exogenous expression of CBP (Fig. 2n, o). However, NAM failed to reverse this reduction (Fig. 2p, q). Similarly, Histone 3 acetylation decreased during EBSS starvation, and this reduction was reversed by NAM, but not by TSA, regardless of exogenous CBP expression (Fig. 2n–q). This suggests that the acetylation activity of CBP towards SNAP47 increases during starvation induced by EBSS, yet the deacetylation of SNAP47 is more pronounced, resulting in reduced levels of SNAP47 acetylation. Similarly, the acetylation of both SNAP47 and Histone 3 was dramatically reduced during OA-induced mitophagy (Fig. 2r–u). Interestingly, the acetylation levels of SNAP47 and Histone 3 did not reverse in the presence of TSA or both TSA and NAM (Fig. 2r–u), indicating a decreased acetylation activity of CBP towards SNAP47 during OA-induced mitophagy.
TSA increased the acetylation of SNAP47, suggesting that HDACs are responsible for SNAP47 deacetylation (Figs. 1 and 2). To identify which HDAC deacetylates SNAP47, we co-expressed all HDACs with SNAP47 and found that HDACs interacted with SNAP47 to varying extents (Fig. 3a). However, only HDAC2 markedly reduced the acetylation of SNAP47 (Fig. 3b, c). Knockdown of HDAC2, but not HDAC1, significantly increased the acetylation of SNAP47, and this enhancement was reversed by re-expression of HDAC2 (Fig. 3d-g). However, HDAC2 knockdown failed to increase the acetylation of SNAP47-3KR, which mimicked the deacetylation status of SNAP47 (Fig. 3h, i). A similar increase and decrease in acetylation of both endogenous SNAP47 and STX17 were observed in CBP knockdown and HDAC2 knockdown cells, respectively (Supplementary Fig. 1a). Altogether, these results suggest that CBP and HDAC2 regulate the acetylation or deacetylation of SNAP47, respectively.
a Co-IP analysis of interactions between the indicated Flag-tagged individual histone deacetylases with HA-SNAP47 in HEK293T cells (n = 3 independent experiments). b, c Acetylation of Flag-SNAP47 in HEK293T cells expressing Flag-SNAP47 and the indicated HA-tagged individual histone deacetylases (b). The levels of Ace-lys were quantified by ImageJ software and normalized with Flag-SNAP47 (c). d, e Flag-SNAP47 Acetylation in HEK293T cells expressing Flag-SNAP47 after 48 h incubation with siRNAs against HDAC1 or HDAC2 (d). The levels of Ace-lys were quantified by ImageJ software and normalized with Flag-SNAP47(e). f, g Flag-SNAP47 Acetylation in HEK293T cells expressing Flag-SNAP47 transfected with HDAC2-HA after 48 h incubation with HDAC2 siRNA (f). The levels of Ace-lys were quantified by ImageJ software and normalized with Flag-SNAP47 (g). h, i Acetylation of Flag-SNAP47 WT or 3KR in HEK293T cells incubated with or without HDAC2 siRNA (h). The levels of Ace-lys were quantified by ImageJ software and normalized with Flag-SNAP47 (i). Data in (c– i) are presented as mean ± SD, n = 3 independent experiments, statistical significance was assessed by a one-way ANOVA (c–g) or two-way ANOVA (i). P values are indicated in the figure. Source data are provided as a Source Data file.
SNAP47 acetylation inhibits autophagosome-lysosome fusion in bulk autophagy and mitophagy
To explore the involvement of SNAP47 acetylation in mitophagy, we investigated mitochondrial protein degradation in cells expressing either the SNAP47-3KQ or SNAP47-3KR mutant, which respectively mimic the acetylated or deacetylated state of SNAP47. In cells expressing the SNAP47-3KQ acetylation mimic, we observed a significant inhibition of mitochondrial protein degradation, as indicated by the levels of mitochondrial proteins PHB2, COX2, and TOM20, while cells expressing SNAP47-3KR showed a slight increase in mitochondrial protein degradation (Fig. 4a–d). Additionally, we utilized the mito-Keima assay, which detects changes in emission wavelength upon pH alteration, to monitor mitophagic flux to lysosomes16. We found that the mito-Keima shift indicative of mitophagy was markedly inhibited in cells expressing the SNAP47-3KQ acetylation mimic compared to those expressing SNAP47-WT and SNAP47-3KR (Fig. 4e, f). These findings suggest that deacetylation of SNAP47 is essential for autophagosome-lysosome fusion in OA-induced mitophagy.
a–d WT, SNAP47 KO, and Flag-SNAP47 (WT, 3KR or 3KQ) complemented SNAP47 KO HeLa/Parkin cells treated with or without OA for 18 h and immunoblotting was then performed with the indicated antibodies (a). Quantification of normalized TOM20, PHB2 and COX2 by ImageJ software (b–d). e, f WT, SNAP47 KO, and Flag-SNAP47 (WT, 3KR or 3KQ) complemented SNAP47 KO HeLa/Parkin stably expressing mito-Keima cells were treated with or without OA for 18 h. Living cells were then imaged for mito-keima 448 nm and 552 nm laser excitation by confocal microscope (e). Scale bar, 10 μm. Quantification of the relative ratio of fluorescence intensity (552 nm: 448 nm) by ImageJ software (f). n = 3, 50 cells from three independent experiments were quantified. g–i WT, SNAP47 KO, and Flag-SNAP47 (WT, 3KR or 3KQ) complemented SNAP47 KO HeLa cells were treated with or without EBSS for 2 h and immunoblotting was then performed with the indicated antibodies (g). The intensity of LC3-II and p62 bands was normalized to actin (h, i). j, k WT, SNAP47 KO, and Flag-SNAP47 (WT, 3KR or 3KQ) complemented SNAP47 KO HeLa cells stably expressing RFP-GFP-LC3 were cultured in EBSS for 2 h (j). Scale bar, 5 μm. Quantification of the percentage of RFP+GFP- LC3 puncta among total LC3 puncta (k). n = 3, 100 cells from three independent experiments. Data in (b–k) are presented as mean ± SD, n = 3 independent experiments, statistical significance was assessed by a two-way ANOVA (b–i) or one-way ANOVA (k). P values are indicated in the figure. Source data are provided as a Source Data file.
Similarly, in line with the role of SNAP47 in OA-induced mitophagy, the SNAP47-3KQ acetylation mimic significantly impeded the degradation of LC3 and p62, along with RFP-GFP-LC3 acidification, as indicated by GFP quenching due to the low lysosomal pH and stable RFP, compared to SNAP47-WT and SNAP47-3KR in starvation-induced autophagy (Fig. 4g–k). These results suggest that deacetylation of SNAP47 is necessary for autophagosome-lysosome fusion in starvation-induced autophagy.
SNAP47 acetylation inhibits the SNARE complex assembly indirectly
To investigate how SNAP47 acetylation affects autophagosome-lysosome fusion, we first examined whether SNAP47 acetylation affects SNARE complex assembly. The results from Co-IP experiments revealed that the SNAP47-3KR mutant displayed increased interactions with STX17 and VAMP7/VAMP8, whereas the SNAP47-3KQ acetylation mimic exhibited significantly reduced interactions in SNAP47 KO cells (Fig. 5a, b), indicating that SNAP47 acetylation inhibits the formation of the STX17-SNAP47-VAMP7/VAMP8 complex in vivo. In addition, the interactions of SNAP47-WT or SNAP47-3KR with STX17 and VAMP7/VAMP8, but not SNAP47 3KQ, increase during OA-induced mitophagy (Supplementary Fig. 2a).
a, b Co-IP analysis of the endogenous HOPS subunits, STX17, VAMP7 and VAMP8 with HA-SNAP47 (WT, 3KR or 3KQ) in SNAP47 KO HeLa/Parkin cells (a). This result is part of Supplementary Fig S2a. The proteins levels were quantified by ImageJ software and normalized with HA-SNAP47 (b). c, d HeLa cells stably expressing GFP-SNAP47 (WT, 3KR or 3KQ) were cultured in EBSS for 2 h, then stained with antibody against LC3 (c). Scale bar, 5 μm. The percentage of GFP-SNAP47-positive LC3 puncta in total LC3 puncta is shown (d). n = 3, 100 cells from three independent experiments. e, f HeLa/Parkin cells stably expressing mito-BFP and GFP-SNAP47 (WT, 3KR or 3KQ) were treated with OA for 4 h, then stained with with antibody against LC3 (e). Scale bar, 5 μm. The percentage of GFP-SNAP47+ LC3+ mito-BFP+ puncta in total LC3+ mito-BFP+ puncta is shown (f). n = 3, 100 cells from three independent experiments. g Schematic for the experimental procedures used in the reconstituted fusion reactions. h, i The effect of SNAP47 acetylation on fusion activity between proteoliposomes reconstituted with STX17 and VAMP8 (h). neg.ctrl. denotes no addition of SNAP29 and SNAP47. Statistics of the liposome fusion in (h) at 2400 s was shown (i). j, k The in vitro interaction of purified GST-SNAP47 (WT or mutants) with His-STX17 and VAMP8 (j) or His-STX17 and His-VAMP7 (k). l Schematic for the experimental procedures used in the SNARE complex assembly assay. m-o Assembly of the SNARE complex containing STX17, SNAP47 (WT, 3KR or 3KQ), and VAMP7/VAMP8 analyzed by FRET assays (m, n). Statistics of EPR (relative proximity ratio of FRET) at 2,500 s were shown (o). Data in (b–o) are presented as mean ± SD, n = 3 independent experiments, statistical significance was assessed by a two-way ANOVA (b, o) or one-way ANOVA (d, f). P values are indicated in the figure. Source data are provided as a Source Data file.
To delve deeper into how SNAP47 acetylation hinders the formation of the STX17-SNAP47-VAMP7/VAMP8 complex, we investigated its impact on SNAP47 localization to autophagosomes. Compared to SNAP47-WT both SNAP47-3KR and SNAP47-3KQ mutants displayed no discernible differences in colocalization with LC3 or with both LC3 and mito-BFP (BFP tagged mitochondrial targeting sequence of COX4) during starvation-induced autophagy or OA-induced mitophagy, respectively (Fig. 5c–f). This suggests that SNAP47 acetylation does not regulate complex formation via its localization to autophagosomes. Additionally, in vitro liposome fusion assays indicated that neither SNAP47-3KR nor SNAP47-3KQ obviously affected fusogenic activity in STX17-SNAP47-VAMP8 complex formation (Fig. 5g–i). These results indicate that SNAP47 acetylation likely indirectly regulates influencing SNARE complex formation. In line with this inference, the results from both in vitro pull-down assays and in vitro SNARE assembly assays demonstrated that neither SNAP47-3KR nor SNAP47-3KQ markedly affected SNARE assembly (Fig. 5j–o). Thus, SNAP47 acetylation indirectly inhibits SNARE complex assembly.
SNAP47 recruits the HOPS complex to autophagic vacuoles
Next, we sought to investigate how SNAP47 acetylation affects SNARE assembly in vivo. Given the critical role of the HOPS complex in tethering autophagosomes to endolysosomes, thereby facilitating subsequent SNARE complex assembly17,18,19,20,21, we aimed to ascertain whether SNAP47 is involved in recruiting the HOPS complex to autophagosomes. Initially, we observed co-localization between GFP-SNAP47 and mCherry-tagged components of the HOPS complex (VPS11, VPS16, VPS18, VPS33A, VPS39, and VPS41), as well as LC3 and a few of these puncta also exhibited limited co-localization with LAMP2 (Supplementary Fig. 3a). These findings suggest that SNAP47 primarily co-localizes with components of the HOPS complex on autophagosomes, with only a minor fraction associating with autolysosomes. Co-IP results showed that SNAP47 interacted not only with all exogenous HOPS complex components, but also with endogenous components (VPS11, VPS16, VPS18, VPS33A, and VPS41), whereas SNAP29 did not exhibit such interactions. (Fig. 6a, b). In vitro pull-down assays revealed a significant and direct interaction between SNAP47 and VPS39 (Fig. 6c). Notably, SNAP47 still interacts with endogenous HOPS components (VPS11, VPS16, VPS18, VPS33A, and VPS41) in SNAP47/STX17 double knockout cells (DKO) (Supplementary Fig. 2b). Further, the SNAP47-2QR (Q143R/Q390R) mutant, which disrupts the STX17-SNAP47-VAMP7/VAMP8 interaction due to alterations in critical amino acids for the 0-layer of the SNARE complex, did not significantly affect the interactions between SNAP47 and HOPS components in SNAP47 KO cells (Fig. 6d-f and Supplementary Fig. 2c). These results suggest that SNAP47 interacts with HOPS independently of STX17 and the SNARE complex formation. Additionally, the interactions of SNAP47-WT or SNAP47-2QR with endogenous HOPS components (VPS11, VPS16, VPS18, VPS33A, and VPS41) in SNAP47 KO and SNAP47/STX17 DKO cells increased in starvation-induced autophagy and OA-induced mitophagy, respectively (Supplementary Fig. 2b, c).
a Co-IP analysis of interactions between the Flag-tagged HOPS subunits with HA-SNAP47 or HA-SNAP29 in HEK293T cells. PI line is for indicating the position of HA-SNAP47 and HA-SNAP29. n = 3 independent experiments. b Co-IP analysis of interactions between the endogenous HOPS subunits with Flag-SNAP47 or Flag-SNAP29 in HEK293T cells. n = 3 independent experiments. c In vitro interactions of purified GST-SNAP47 with His-tagged HOPS subunits. n = 3 independent experiments. d The central ionic layer of SNAP47 was identified through sequence alignment, participating residues glutamine-143 and glutamine-390 in SNAP47 are indicated by red color. The PH domain, SNARE motif, and second SNARE motif are colored by blue, green, and laurel-green, respectively. e, f Co-IP analysis of the endogenous HOPS subunits, STX17, VAMP7 and VAMP8 with HA-SNAP47 (WT or 2QR-Q143R/Q390R) in SNAP47 KO HeLa/Parkin cells (e). This result is part of Supplementary Fig S2c. The proteins levels were quantified by ImageJ software and normalized with HA-SNAP47 (f). g–j WT and SNAP47 KO HeLa/Parkin cells were treated with OA for 4 h. Following separation via OptiPrep membrane flotation, autophagosome-enriched fractions (light-density fractions 3–5) were analyzed by immunoblotting using the indicated antibodies (g). The proteins levels were quantified by ImageJ software and normalized with LC3 (h–j). k–n WT, SNAP47 KO, and SNAP47 WT or Flag-SNAP47 LIR mutant complemented SNAP47 KO HeLa/Parkin cells were treated with OA for 4 h. Following separation via the OptiPrep membrane flotation, autophagosome-enriched fractions (light-density fractions 3–5) were analyzed by immunoblotting using the indicated antibodies (k). The proteins levels were quantified by ImageJ software and normalized with LC3 (l–n). Data in (f–n) are presented as mean ± SD, n = 3 independent experiments, statistical significance was assessed by a two-way ANOVA. P values are indicated in the figure. Source data are provided as a Source Data file.
As the localization of HOPS components is obscured by the strong signal in the cytoplasm, we employed gradient fractionation assays to investigate the impact of SNAP47 on the localization of HOPS components to autophagosomes11,19. The results revealed a decrease in the amount of HOPS components in the autophagosome-enriched fractions (i.e., fractions 3–5) of SNAP47 knockout (KO) cells compared to WT cells (Fig. 6g–j). Similarly, the SNAP47 LIR mutant, which exhibits a dramatic decrease in localization to autophagosomes15, also led to a decreased level of HOPS components in the autophagosome-enriched fractions (Fig. 6k-n). Further, both SNAP47 and STX17 are capable of recruiting HOPS subunits in SNAP47/STX17 DKO cells, respectively (Supplementary Fig. 2d), suggesting SNAP47 can recruit HOPS components independently of STX17, and vice versa. In SNAP47/SNAP29 DKO cells, both SNAP47-WT and SNAP47-2QR mutants recruit HOPS components to similar levels during starvation-induced autophagy and OA-induced mitophagy (Supplementary Fig. 2e, f), suggesting SNAP47 can recruit HOPS components independently of the SNAP47-STX17 interaction and SNARE complex formation. Collectively, these findings suggest that SNAP47 contributes to the recruitment of HOPS components to autophagosomes independently of STX17, SNAP47-STX17 interaction, and ternary SNARE complex formation.
SNAP47 deacetylation promotes its interactions with the HOPS complex components
Given that SNAP47 plays a role in recruiting HOPS components to autophagosomes (Fig. 6), we investigated how acetylation of SNAP47 affects this recruitment process. Co-IP experiments revealed that the acetylation variants of SNAP47, namely SNAP47-3KR and SNAP47-3KQ, exhibited increased and decreased interactions, respectively, with both endogenous and HA-tagged HOPS components (Figs. 7a–g and 5a). Additionally, we observed a decrease in the levels of HOPS subunits in autophagosome-enriched fractions (i.e., fractions 3–5) in SNAP47-3KQ cells compared to SNAP47-WT, whereas SNAP47-3KR notably increased the abundance of HOPS components in fraction 5 (Fig. 7h–k). Collectively, these findings suggest that deacetylation of SNAP47 promotes the recruitment of HOPS components to autophagosomes.
a–g Co-IP analysis of interactions between Flag-SNAP47 (WT, 3KR or 3KQ) with HA-tagged HOPS subunits in HEK293T cells (a–f). The levels of HA-tagged HOPS subunits were quantified by ImageJ software and normalized with Flag-SNAP47 (g). h–k SNAP47 KO, and SNAP47 WT, SNAP47 3KR or SNAP47 3KQ mutant complemented SNAP47 KO HeLa/Parkin cells were treated with OA for 4 h. Following separation via the OptiPrep membrane flotation, autophagosome-enriched fractions (light-density fractions 3–5) were analyzed by immunoblotting using the indicated antibodies (h). The proteins levels were quantified by ImageJ software and normalized with LC3 (i–k). Data in (g–k) are presented as mean ± SD, n = 3 independent experiments, statistical significance was assessed by a two-way ANOVA. P values are indicated in the figure. Source data are provided as a Source Data file.
Discussion
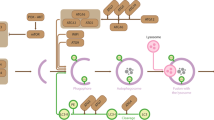
STX17-SNAP47-VAMP7/VAMP8 complex is a recently identified dual-role SNARE complex responsible for autophagosome-lysosome fusion in both selective autophagy and bulk autophagy. However, the spatiotemporal regulation mechanism on this SNARE is unknown. In this study, we found SNAP47 is acetylated and undergoes deacetylation during autophagy. The acetylation status of SNAP47 is regulated by acetyltransferase CBP and deacetylase HDAC2. The spatiotemporal regulatory dynamics of SNAP47 acetylation differ between bulk autophagy and mitophagy. This disparity arises from distinct regulation of acetyltransferase and deacetylase activity. SNAP47 acetylation negatively regulates autophagosome-lysosome fusion in bulk autophagy and mitophagy by indirectly inhibiting SNARE complex assembly via inhibiting HOPS components recruitment to autophagic vacuoles. Our study uncovers SNAP47 acetylation-based spatiotemporal regulation mechanism on autophagosome-lysosome fusion (Fig. S4).
The HOPS complex was initially characterized for its role in facilitating homotypic fusion among lysosomes or heterotypic fusion between late endosomes and lysosomes in yeast22,23,24,25,26,27. Subsequently, it was discovered to tether autophagosomes to lysosomes, thereby facilitating autophagosome-lysosome fusion17,18,20,28,29,30. It has been proposed that the HOPS complex promotes autophagosome-lysosome fusion through interaction with STX1717,18. In this study, we observed interactions between all HOPS components and SNAP47, and further demonstrated that SNAP47 contributes to the localization of HOPS components on autophagosomes independently of STX17 and the SNARE complex assembly. This suggests that both STX17 and SNAP47 functions in recruiting HOPS components to autophagosomes. Additionally, STX17 may have a minor role in SNAP47’s recruitment of HOPS components, as the interaction between SNAP47 and HOPS components appears slightly reduced in the absence of STX17. Recent studies have proposed that mammalian HOPS components can be categorized into two sub-complexes: HOPS-2 (VPS39/VPS11) and HOPS-4 (VPS16/VPS18/VPS33A/VPS41). These sub-complexes cooperate with specific Rab GTPase pairs on opposing vesicles to facilitate membrane tethering31. However, we observed all HOPS components on autophagosomes, suggesting that two different assembly forms of the HOPS complex (HOPS-2/HOPS-4 and HOPS-6) coexist in vivo.
Autophagosome-lysosome fusion is a critical step for fulfilling autophagic function. Our previous study identified a default SNARE complex (STX17-SNAP47-VAMP7/VAMP8) for both bulk autophagy and selective autophagy. In this study, we revealed another layer of spatiotemporal regulation based on post-translational modification. Since autophagy has been implicated in many human diseases, further study is warranted to investigate the relationship between autophagosome-lysosome fusion mediated by this SNARE complex and various human diseases. It could provide a mechanistic basis for developing targeted therapeutic interventions for diseases related to dysfunction of autophagy.
Methods
Antibodies
Antibodies used in this study were as follows: Mouse monoclonal anti-SNAP47 (Santa Cruz Biotechnology, sc-514428, Lot#C1615, western blot (WB) 1:1,000), Rabbit polyclonal anti-Acetylated-lysine (Cell Signaling Technology, 9441s, Lot#14, RRID:AB_331896, WB1:1,000), Rabbit polyclonal anti-CBP (Cell Signaling Technology, 7389s, Lot#5, RRID:AB_11015158, WB 1: 2000), Mouse monoclonal anti-VPS11 (Santa Cruz Biotechnology, sc-515094, Lot#H2517, RRID:AB_2687986, WB 1:1,000), Mouse monoclonal anti-VPS41 (Santa Cruz Biotechnology, sc-377118, Lot#A0719, RRID:AB_2687987, WB 1:1,000), Rabbit polyclonal anti-VPS16 (Proteintech, 17776-1-AP, Lot#00096872, RRID:AB_2217085, WB 1:2,000), Rabbit polyclonal anti-VPS18 (Proteintech, 10901-1-AP, Lot#00084835, RRID:AB_2273089, WB 1:1,000), Rabbit polyclonal anti-VPS33A (Proteintech, 16896-1-AP, Lot#00045470, RRID:AB_2214916, WB 1:3,000), Rabbit polyclonal anti-COX2 (ABclonal, A11522, Lot#1150910201, RRID:AB_2758591, WB 1:2,000), Rabbit polyclonal anti-HDAC1 (ABclonal, A19571, Lot#4000000050, RRID:AB_28626751, WB 1:4,000), Rabbit polyclonal anti-HDAC2 (ABclonal, A2084, Lot#1000100101, RRID:AB_764104, WB 1:4,000), Rabbit polyclonal anti-PHB2 (Proteintech, 12295-1-AP, Lot#00049480, RRID:AB_2164779, WB 1:3,000), Rabbit polyclonal anti-VAMP7 (Cell Signaling Technology, 14811, Lot#1, RRID:AB_2798625, WB1:1,000), Rabbit monoclonal anti-VAMP8 (Abcam, ab76021, Lot#GR3190214-2, RRID:AB_1310798, WB 1:1,000), Rabbit polyclonal anti-STX17 (Sigma, HPA001204, Lot#G107062, RRID:AB_1080118, WB 1:1,000), Rabbit polyclonal anti-Flag (Sangon Biotech, D110005, Lot#G928AA0012-0200, WB 1:10,000), Rabbit polyclonal anti-HA (Sigma-Aldrich, H6908, Source#0000086963, RRID:AB_260070, WB 1:10,000), Mouse monoclonal anti-LAMP2 (Santa Cruz Biotechnology, sc-18822, Lot#L3216, RRID:AB_626858, IF 1:500), Rabbit polyclonal anti-LC3 (Sigma, L7543, Lot#084M4798V, RRID:AB_796155, WB 1:10,000, IF 1:500), Rabbit polyclonal anti-Actin (Service Bio, GB11001, Lot#Ls192617, RRID: AB_2801259. WB 1:5,000), Rabbit polyclonal anti-6×His (Sangon Biotech, D110002, Lot#F515AA0012, WB 1:1,000), Mouse monoclonal anti-SQSTM1/p62 (Cell Signaling Technology, 88588, Lot#1, RRID: AB_2800125, WB 1:5,000), fluorescein (FITC) AffiniPure goat anti-mouse IgG (H + L) (Jackson, 115-095-003, Lot#136596, RRID: AB_2338589, IF 1:500), fluorescein (FITC) AffiniPure goat anti-rabbit IgG (H + L) (Jackson, 111-095-003, Lot#133027, RRID: AB_2337972, IF 1:500), Cy3 AffiniPure goat anti-mouse IgG (H + L) (Jackson, 115-165-003, Lot#117093, RRID: AB_2338680, IF 1:500), Cy3 AffiniPure goat anti-rabbit IgG (H + L) (Jackson, 111-165-003, Lot#128284, RRID: AB_2338000, IF 1:500), Alexa FluroTM647 goat anti-mouse IgG (H + L)(Invitrogen, A21235, Lot#2482945, RRID: AB_2535804, IF 1:500), Alexa FluroTM 647 goat anti-Rabbit IgG (H + L) (Invitrogen, A21244, Lot#2433883, RRID: AB_2535812, IF1:500), goat anti-mouse IgG(H + L)-HRP (Southern Biotech, 1036-05, Lot#D1912-SL7113, RRID: AB_2794348, WB 1:500), goat anti-rabbit IgG(H + L)-HRP (Southern Biotech, 403005, Lot#A2718-TA21, RRID: AB_2687483, WB 1:500).
Reagents and treatment
The chemicals were used as follows unless indicated otherwise: oligomycin (Santa Cruz Biotechnology, sc-203342; 2.5 μM) and antimycin-A (Santa Cruz Biotechnology, sc-202467A; 250 nM) was added to the medium for 2 h, 4 h or 18 h. Trichostatin A (TSA, MCE, 58880-19-6; 400 nM) was added to the culture medium for 16 h. Nicotinamide (NAM, MCE, 98-92-0; 5 mM) was added to the medium for 8 h. CTB (Sigma-Aldrich, C6499; 50 μM) was added to the medium for 6 h. C646 (MCE, 328968-36-1; 10 μM) were added to the medium for 4 h. For starvation treatment, cells were washed three times with PBS (Hyclone) and then incubated with EBSS (Solarbio, H2020) for the indicated time.
Cell culture and transfection
HEK293T and HeLa cells were generous gifts from Dr. Qing Zhong (Shanghai Jiao Tong University, Shanghai, China). HEK293T and HeLa cells were cultured in DMEM (Hyclone) supplemented with 10% FBS (Gibco) and 1% Penicillin-Streptomycin Solution (Beyotime) at 37 °C with 5% CO2. HeLa cells were transiently transfected using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. Transient transfection of plasmid in HEK293T cells was performed using PEI according to the manufaturer’s protocol. The stealth RNAi oligonucleotides were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. 2 d after transfection, cells were harvested for analysis.
The following siRNAs were used: CBP,5′-AAUCAACUCCUGUGUCGUCUUUU-3′, 5′-AAUCCACAGUACCGAGAAAUGUU-3′; HDAC1,5′-CGUUCUUAACUUUGAACCAUA-3′; HDAC2,5′-CAGUCUCACCAAUUUCAGAAA-3′. CRISPR guide RNA (gRNA) sequences targeting the human SNAP47 and STX17 genes were cloned into the lentiCRISPR v2-Blast vector (Addgene #83480). The target sequences for human STX17 were 5′-TCTGGATAGCTGGTTCAAGA-3′, and for human SNAP47, 5′-TCCATCACCATCCTGGAGAA-3′. Expression plasmids for SNAP47, SNAP29, PCAF, GCN5, and TIP60 were constructed by cloning the corresponding genes from human cDNA (prepared from 293 T cells) into the pMRXIP vector, which includes enhanced GFP, 3 × Flag, or 3 × HA tags for eukaryotic expression. The cDNAs of SNAP47, VAMP7, and VAMP8 were cloned into pGEX-4T-1 or pET-28a vectors for prokaryotic expression. The CBP-HA, p300-HA, VPS11-HA, VPS16-HA, VPS18-HA, VPS33A-HA, VPS39-HA, and VPS41-HA constructs were kindly provided by Dr. Qiming Sun from Zhejiang University. The HDAC family of deacetylases was generously gifted by Dr. Wei Liu, also from Zhejiang University.
Immunostaining assays
Cells grown on coverslips were washed with PBS and fixed in 4% paraformaldehyde in PBS for 15 min at room temperature. After washing three times with PBS, Cells were permeabilized with 0.1% saponin in PBS for 10 min, blocked with 10% FBS in PBS for 1 h. Then, the cells were incubated with primary antibodies for 1 h or overnight at 4 °C. After washing three times with PBS, cells were incubated with appropriate secondary antibodies at room temperature for 1 h, and washed three times with PBS. The slides were mounted and images were acquired under a laser scanning confocal microscope (FV3000, Olympus).
Mito-Keima mitophagy assay
HeLa SNAP47 KO cells, which stably expressed Parkin, were infected with a lentivirus carrying the mito-Keima vector. Following this, the cells were treated with or without OA in fresh growth medium for 18 h, and were then analyzed using confocal microscopy. Live cells were cultured in glass-bottom dishes. After OA treatment, the cells were scanned, and images were acquired using a laser scanning confocal microscope (FV3000, Olympus) with argon lasers (at 448 nm for mito-Keima at neutral pH and 552 nm for mito-Keima at acidic pH). Ratiometric analysis (552 nm:448 nm) was performed using ImageJ software.
Immunoprecipitation assays
HEK293T cells were washed in ice-cold PBS and lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 ug/ml Aprotinin, 1 ug/ml Pepstatin A, 1 ug/ml Leupeptin, 200 mM NaF, 200 mM Na3VO3) for 30 min at 4 °C. After centrifugation at 12,000 g for 10 min at 4 °C, the supernatants were subjected to immunoprecipitation using anti-Flag M2 (A2220; Sigma-Aldrich) or anti-HA Affinity Gel (A2095; Sigma-Aldrich) for 12 h at 4 °C. Precipitated immunocomplexes were washed three times in lysis buffer and boiled in 2× sample buffer. Then samples were subjected to SDS-PAGE and analyzed by Western Blotting.
Western Blotting
Cells were harvested and lysed with sodium dodecyl sulfate (SDS) buffer. Samples extracted from cells were subjected to SDS-PAGE electrophoresis and immobilized on a polyvinylidene fluoride (PVDF) membrane (BIO-RAD, 162-0177). After blocking with 5% nonfat milk in PBST, membrane was incubated with the primary antibodies, followed incubated with HRP-conjugated secondary antibody for 1 h at room temperature.
Recombinant protein purification and in vitro acetylation assay
GST-SNAP47 was expressed in Escherichia coli BL21. Bacteria were treated with 0.4 mM IPTG at 16 °C to induce protein expression and were harvested and resuspended in PBS containing 0.5% Triton X-100, 2 mM EDTA, and 1 mM PMSF, followed by ultrasonication. The recombinant SNAP47 proteins were purified using by glutathione-Sepharose 4B beads (GE healthcare, 17-0756-04), eluted with glutathione at 4 °C for 4 h to release the proteins.
CBP-HA protein was purified from HEK293T cells 48 h after transfection by immunoprecipitation with anti-HA affinity beads (A2095; Sigma-Aldrich). For in vitro acetylation assay, GST-SNAP47 protein (10 μg)was incubated with CBP-HA immunoprecipitated from cell lysate in the presence of acetyl-coenzyme A (4 μg; Sigma-Aldrich,10101893001) and 10 μl 5 × HAT assay buffer(250 mM Tris–HCl, pH 8.0, 50% glycerol, 0.5 mM EDTA, 5 mM dithiothreitol) in a total volume of 50 μl. The contents were mixed gently and placed in a 30 °C shaking incubator for 4 h. Then 2 × sodium dodecyl sulfate (SDS) buffer (50 μl) was added to the reaction and the mixture was boiled for 5 min. The reaction products were separated by SDS-PAGE and immunoblotted with anti-acetyl-lysine.
In vitro binding assay
Genes were cloned into pGEX-4T-1 or pET-28a vector for expression in E. coli BL21 (DE3). The recombinant proteins were purified by glutathione-sepharose resin or Ni-affinity resin. In GST pull-down assays, GST and GST tagged proteins were applied to GST resin, then were incubated with 1 μg His-tagged proteins in binding buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.5% NP40) supplemented with protease inhibitor cocktail (Roche) for 2 h at 4 °C. After three washes, proteins were eluted and dissolved in sample buffer for SDS–PAGE and immunoblotting.
Liposome Fusion
Lipids were mixed at the proper ratio as indicated below to a final concentration of 1 mM. Donor liposome (reconstituted with full-length VAMP7 or VAMP8) contains 75.5 % POPC, 20 % POPE, 3 % PI-3-P, and 1.5 % DiI (molar ratio). Acceptor liposome (reconstituted with full-length Syx17) contains 75.5 % POPC, 20 % POPE, 3 % PI-3-P, and 1.5 % DiD (molar ratio). Lipid mixtures were dried under nitrogen flow and further incubating in vacuum for 1 h at room temperature in the dark. Lipid films were resuspended in TBS150 supplied with 0.2 mM TCEP and 1% 3-[(3-Cholamidopropyl) dimethylammonio]-1-propane sulfate (w/v, CHAPS) (VWR, #0465). Purified proteins were added to resuspended lipids with a protein-to-lipid ratio of 1:500. After incubation on the ice for 20 min, lipid-protein mixtures were desalted using PD-10 desalting column. Prepared proteoliposomes were stored at 4 °C in the dark before using. Liposome fusion assays were carried out using FluoDia T70 fluorescence plate reader (PTI) equipped with 530/10 excitation filter, 580/10 and 667/10 emission filters at 37 °C. Donor and acceptor liposomes were mixed at a concentration of 100 μM (total lipids) with addition of 2 μM recombinant SNAP-29 or SNAP-47. Donor (DiI) and acceptor (DiD) fluorescence were monitored every 20 s. Liposome fusion signals were interpreted as the FRET efficiency between the donor (DiI) and acceptor (DiD): Equation (1)
Where the \({I}_{{DiD}}\) and \({I}_{{DiI}}\) are the fluorescence intensities of DiD and DiI under the 530/10 excitation filter, respectively. All the experiments were independently repeated for three times.
FRET assay for SNARE complex assembly
Purified STX17 SNARE domain (residues 161−231, E198C), VAMP7 SNARE domain (residues 123−187, E152C), and VAMP8 SNARE domain (10−74, E39C) were conjugated with 3-fold molar ratio of ATTO647 maleimide (ATTO-TEC GmbH, #AD 550-45) and ATTO550 maleimide (ATTO-TEC GmbH, #AD 647N-45), respectively, according to the manufacturer’s instruction. The conjugated mixtures were desalted by using PD-10 desalting column (Cytiva, #17085101) with TBS300 buffer. The concentrations of fluorescent conjugated proteins were analyzed by using UV-visible spectrophotometer (Shimazu UV-2450) according to the manufacturer’s instruction.
SNARE complex assembly assays were carried out using FluoDia T70 fluorescence plate reader (PTI) equipped with 530/10 excitation filter, 580/10 and 667/10 emission filters at 37 °C. ATTO647N-STX17, ATTO550-VAMP7, and ATTO550-VAMP8 were added to 0.5 μM; SNAP47(WT or mutant) was added to 2 μM. The raw FRET efficiency, proximity ratio (EPR), which indicates SNARE complex assembly, was calculated according to Equation (2):
Where IATTO647N and IATTO550 are the raw fluorescent intensities of acceptor (ATTO647N) and donor (ATTO550). Each experiment was repeated 3 times independently.
In-Gel Digestion
Bands of interest were excised, cut into small pieces (1 mm × 1 mm), rinsed with water and 2 × 30 min washed in 50 mM ammonium bicarbonate (AmBic) at 37 °C. The gel pieces were then shrunk in 100 % acetonitrile (ACN) and shaken for 5 min. Solvent was removed, and gel pieces rehydrated in 10 mM DTT in 50 mM AmBic, followed by 40 min incubation at 60 °C. The gel pieces were again shrunk in 100 % ACN and shaken for 30 min. Solvent was removed, and gel pieces rehydrated in 55 mM iodoacetamide in 50 mM AmBic, followed by 40 min incubation at room temperature. The gel pieces were again shrunk in 100 % ACN and shaken for 15 min. The solvent was removed, gel pieces briefly rinsed with 50 mM AmBic and rehydrated in a small volume (10 μL) of 50 mM AmBic supplemented with Trypsin at 37 °C for 18 hrs. Peptides were stage-tip purified, dried, and reconstituted in 10 μL of 0.1 % formic acid prior to analysis.
Protein acetylation identification by LC-MS/MS
The peptides were first separated with a Vanquish™ Neo UHPLC LC system using trap-elute mode and then emitted into a Thermo Scientific Orbitrap Ascend Tribrid mass spectrometer (Thermo Fisher, San Jose) for identification. C18 column (Thermo Scientific™ Acclaim™ PepMap™ 100 C18, 75 μm * 25 cm, 2 μm, 100 Å) was used for the LC separation, and the column temperature was maintained at 55 °C.
The mobile phases A and B were water and 80% acetonitrile containing 0.1% formic acid, respectively. The elution gradient was set as follows: 0–3 min = 4% B to 4.5% B with a flow rate of 0.7 uLmin-1, 3–71 min = 4.5% B to 20% B with a flow rate of 0.7 to 0.3 uLmin-1, 71–111 min = 20% B to 35% B with a flow rate of 0.3 uLmin-1, 111–112 min = 35% B – 55% B with a flow rate of 0.3 uLmin-1, 112–113 min = 55% B to 99% B with a flow rate from 0.3–0.7 uLmin-1 and 113–120 min = 99% B with a flow rate of 0.7 uLmin-1. For single-shot proteomics with data-dependent acquisition (DDA), the MS1 scan resolution was set to 240, 000 and maximum injection time was set to 507 ms. For the ITMS2 scans the ion trap scan rate was set to Turbo, the isolation window was set to 1.6, the AGC target was set to custom, the normalized ACG target was set to 300%, and maximum injection time was set to auto.
Database searching
The generated MS/MS spectra were searched against the human SNAP47 protein with an uniport number of Q5SQN1 using the SEQUEST searching engine in Proteome Discoverer 2.5 software. The search criteria were as follows: full tryptic specificity was required, two missed cleavage was allowed, carbamidomethyl on cysteine was set as the fixed modifications, oxidation on methionine was set as the variable modification, acetylation on lysine was set as the variable modification. Precursor ion mass tolerances were set at 10 ppm, and the fragment ion mass tolerance was set to 0.6 Da for all MS2 spectra acquired. Peptide spectral matches (PSM) were validated using the Percolator provided by Proteome Discoverer software based on q-values at a 1% false discovery rate (FDR). The MS/MS spectra were manually checked to confirm the exact acetylation sites according to the b- or y-characteristic fragments.
Statistical analysis and reproducibility
Statistical analysis was carried out on the data from at least three independent experiments using Prism 8 (GraphPad software). Data were presented as mean ± SD, and statistical significance was determined by one-way ANOVA or two-way ANOVA. n values and P values are indicated in the figure. Purposeful statistical subjects varied across experiments, and each experiment are independently repeated three times. No data were excluded from the analysis. Source data are provided as a Source Data file.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are available in the main text or the supplementary materials. The proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD052239. Source data are provided with this paper.
References
Lamb, C. A., Yoshimori, T. & Tooze, S. A. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774 (2013).
Morishita, H. & Mizushima, N. Diverse cellular roles of autophagy. Annu. Rev. Cell Dev. Biol. 35, 453–475 (2019).
Mizushima, N., Levine, B., Cuervo, A. M. & Klionsky, D. J. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008).
Klionsky D. J. et al. Autophagy in major human diseases. EMBO J. 40, e108863 (2021).
Moehlman, A. T. & Youle, R. J. Mitochondrial quality control and restraining innate immunity. Annu. Rev. Cell Dev. Biol. 36, 265–289 (2020).
Nakamura, S. & Yoshimori, T. New insights into autophagosome-lysosome fusion. J. Cell Sci. 130, 1209–1216 (2017).
Yu, L., Chen, Y. & Tooze, S. A. Autophagy pathway: cellular and molecular mechanisms. Autophagy 14, 207–215 (2018).
Zhao, Y. G., Codogno, P. & Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 22, 733–750 (2021).
Itakura, E., Kishi-Itakura, C. & Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 (2012).
Takáts, S. et al. Autophagosomal syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201, 531–539 (2013).
Matsui, T. et al. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol. 217, 2633–2645 (2018).
Takáts, S. et al. Non-canonical role of the SNARE protein Ykt6 in autophagosome-lysosome fusion. PLoS Genet 14, e1007359 (2018).
Bas, L. et al. Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J. Cell Biol. 217, 3656–3669 (2018).
Gao, J., Reggiori, F. & Ungermann, C. A novel in vitro assay reveals SNARE topology and the role of Ykt6 in autophagosome fusion with vacuoles. J. Cell Biol. 217, 3670–3682 (2018).
Jian, F. et al. The STX17-SNAP47-VAMP7/VAMP8 complex is the default SNARE complex mediating autophagosome-lysosome fusion. Cell Res. 34, 151–168 (2024).
Katayama, H., Kogure, T., Mizushima, N., Yoshimori, T. & Miyawaki, A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 18, 1042–1052 (2011).
Takats, S. et al. Interaction of the HOPS complex with syntaxin 17 mediates autophagosome clearance in Drosophila. Mol. Biol. Cell 25, 1338–1354 (2014).
Jiang, P. et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell 25, 1327–1337 (2014).
Uematsu, M., Nishimura, T., Sakamaki, Y., Yamamoto, H. & Mizushima, N. Accumulation of undegraded autophagosomes by expression of dominant-negative STX17 (syntaxin 17) mutants. Autophagy 13, 1452–1464 (2017).
Jia, R., Guardia, C. M., Pu, J., Chen, Y. & Bonifacino, J. S. BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy 13, 1648–1663 (2017).
Shen, Q. et al. Acetylation of STX17 (syntaxin 17) controls autophagosome maturation. Autophagy 17, 1157–1169 (2021).
Wada, Y., Ohsumi, Y. & Anraku, Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J. Biol. Chem. 267, 18665–18670 (1992).
Rieder, S. E. & Emr, S. D. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell 8, 2307–2327 (1997).
Hickey, C. M. & Wickner, W. HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol. Biol. Cell 21, 2297–2305 (2010).
Baker, R. W. et al. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114 (2015).
Sato, T. K., Rehling, P., Peterson, M. R. & Emr, S. D. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell 6, 661–671 (2000).
Seals, D. F., Eitzen, G., Margolis, N., Wickner, W. T. & Price, A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl Acad. Sci. USA 97, 9402–9407 (2000).
Cheng, X. et al. Pacer is a mediator of mTORC1 and GSK3-TIP60 signaling in regulation of autophagosome maturation and lipid metabolism. Mol. Cell 73, 788–802 e787 (2019).
Marwaha, R. et al. The Rab7 effector PLEKHM1 binds Arl8b to promote cargo traffic to lysosomes. J Cell Biol 216, 1051–1070 (2017).
Zhen, Y. & Li, W. Impairment of autophagosome-lysosome fusion in the buff mutant mice with the VPS33A(D251E) mutation. Autophagy 11, 1608–1622 (2015).
Zhang, S. et al. C9orf72-catalyzed GTP loading of Rab39A enables HOPS-mediated membrane tethering and fusion in mammalian autophagy. Nat. Commun. 14, 6360 (2023).
Acknowledgements
We thank Dr. Wei Liu (Zhejiang University) for sharing the HDAC family of deacetylases. We thank Dr. Qiming Sun (Zhejiang University) for sharing CBP-HA, p300-HA, VPS11-HA, VPS16-HA, VPS18-HA, VPS33A-HA, VPS39-HA, and VPS41-HA constructs. We thank the Center for Precision Medicine Multi-Omics Research, Institute of Advanced Clinical Medicine, Peking University for the quantitative mass spectrum analysis. The work was supported by grants from NSFC (92254302, 91854116, 32170685 and 31771529 to Y.R.). The work was partially supported by the Fundamental Research Funds for the Central Universities (5003510089 and 2023BR028 to Y.R.).
Author information
Authors and Affiliations
Contributions
F.J. and Y.R. conceived and designed the experiments. F.J. carried out the biological and biochemical experiments. S.W., C.M. and Y.R. designed and S.W. carried out the in vitro liposome fusion experiments and SNARE assembly assay. Y.C. and W.T. performed the mass spectrometry analysis. F.J., S.W., C.M., SX.W., Y.L., and Y.R. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Geert van den Bogaart, Thanh Nguyen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jian, F., Wang, S., Tian, W. et al. Deacetylated SNAP47 recruits HOPS to facilitate autophagosome-lysosome fusion independent of STX17. Nat Commun 16, 543 (2025). https://doi.org/10.1038/s41467-025-55906-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-55906-x
This article is cited by
-
Autophagy in adult stem cell homeostasis, aging, and disease therapy
Cell Regeneration (2025)