Abstract
Integrin clusters facilitate mechanical force transmission (mechanotransduction) and regulate biochemical signaling during cell adhesion. However, most studies have focused on rigid substrates. On fluid substrates like supported lipid bilayers (SLBs), integrin ligands are mobile, and adhesive complexes are traditionally thought unable to anchor for cell spreading. Here, we demonstrate that cells spread on SLBs coated with Invasin, a high-affinity integrin ligand. Unlike SLBs functionalized with RGD peptides, integrin clusters on Invasin-SLBs grow in size and complexity comparable to those on glass. While actomyosin contraction dominates adhesion maturation on stiff substrates, we find that on fluid SLBs, integrin mechanotransduction and cell spreading rely on dynein pulling forces along microtubules perpendicular to the membranes and microtubules pushing on adhesive complexes, respectively. These forces, potentially present on non-deformable surfaces, are revealed in fluid substrate systems. Supported by a theoretical model, our findings demonstrate a mechanical role for microtubules in integrin clustering.
Similar content being viewed by others
Introduction
Integrin-mediated adhesion is critical to fundamental cellular processes such as cell migration1, differentiation2, and the development of tissues and organs3. Integrin clusters serve as communication hubs transmitting mechanical forces between cells and substrates. During integrin-mediated mechanotransduction, forces generated by cells and transmitted to a substrate regulate biochemical signaling as a function of substrate stiffness4.
In recent years, studies on mechanotransduction have generated a wealth of information and conceptual shifts in our understanding of cell adhesion5. A plethora of “adhesome proteins” connecting integrins with the cytoskeleton were identified, and their role in mechanotransduction and cell adhesion was established6. The majority of these studies, however, were performed on substrates with immobilized integrin ligands, such as glass or deformable 2D-gels7. The actin cytoskeleton plays a crucial mechanical role in adhesion reinforcement on stiff substrates8. Actin polymerization is required to form small nascent adhesions (NAs)9, and actomyosin contraction promotes their growth into large and dense focal adhesions (FAs) connected to stress fibers10. However, much less is known about adhesion on soft substrates like 3D matrices11 or the plasma membrane of other cells12, which are relevant to the interaction between immune and target cells13,14, where integrin ligands are not immobilized but embedded in fluid membranes.
Supported lipid bilayers (SLBs) are convenient model membranes for mimicking the fluid characteristic of plasma membranes to study cell-cell adhesion15,16,17,18. Previous studies of SLBs functionalized with canonical RGD peptides have shown that cells neither spread, nor develop large and dense integrin adhesions like FAs and stress fibers19,20. The current understanding is that mobile integrin-ligand complexes cannot serve as anchoring points on fluid membranes to promote cell spreading, as on stiff substrates10, and NAs cannot be reinforced through mechanotransduction to promote strong adhesion on fluid substrates21. However, previous studies have not addressed the effect of the integrin receptor-ligand affinity which may significantly differ from that of RGD peptides and could regulate cell adhesion22,23.
To test the role of integrin receptor-ligand affinity directly, we used SLBs functionalized with a high-affinity ligand, the Yersinia bacterial protein Invasin24, that binds to a subset of β1-integrins, including the fibronectin receptor α5β1 (Supplementary Methods). On these SLBs, we seeded mouse embryonic fibroblasts (MEF) expressing a recombinant integrin β1-subunit labeled with a Halotag at its ectodomain, instead of the endogenous β1-integrin25. This construct labeled with membrane impermeable Halotag-dyes enabled us to study β1-integrins primarily at the cell surface, excluding signals from intracellular integrins otherwise observed with genetic labeling. We used confocal microscopy to detect β1-integrin clustering during the time course of adhesion. We quantified cluster areas and integrin densities using fluorescent SLB calibration standards18,26 and compared them between Invasin-SLBs and RGD-SLBs. Thus, we found evidence of mechanotransduction occurring on Invasin-SLBs, leading to FA-like β1-integrin adhesions. We attributed this phenomenon to the vertical forces generated by microtubules and dyneins on integrin clusters, and we developed a theoretical model supporting this mode of mechanotransduction. Additionally, we found that cell spreading occurred on Invasin-SLBs but not on RGD-SLBs, in a receptor-ligand affinity dependent fashion. Finally, we observed that microtubules play a significant role in mechanotransduction on these SLBs.
Results
Cell “trembling” and spreading on SLBs depend on ligands
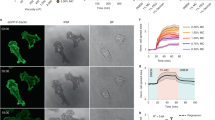
We functionalized SLBs with RGD peptides or Invasin and confirmed their fluidity by fluorescence recovery after photobleaching (FRAP) (Supplementary Fig. S1). We then added MEF cells on these bilayers and we observed, through brightfield microscopy, both “trembling” cells with fluctuating edges, and “adherent” cells with immobile edges (Fig. 1A; Supplementary movies 1-2). The fraction of the latter increased over time, reaching 80% on RGD- and Invasin-SLBs after 45 minutes (Fig. 1B). Adherence was more rapid on RGD- than on Invasin-SLBs (Fig. 1B), possibly due to the higher effective RGD densities on SLBs (20,000 RGD/µm2 vs. 600 Invasin/µm2) and/or the larger range of integrin types that RGD binds to, which could facilitate integrin activation27. This hypothesis was tested by enhancing cell adhesion with manganese (Mn2+), a known integrin activator28. Notably, while Mn2+ did not alter adhesion dynamics on RGD-SLBs, it significantly accelerated adhesion rates on Invasin-SLBs to levels comparable to those observed on RGD-SLBs (Fig. 1B). While similar fluctuations of the cell edge were previously reported on glass29, we found that the proportion of adherent cells on glass remained constant and similar for both ligands at comparable concentration (400 nM), as used for the SLBs, with and without Mn2+ (Supplementary Figs. S2A-C). These observations suggest that, under given conditions, ligand’s ability to activate integrins is a more critical factor for cell adhesion on SLBs than on glass.
A Top: schematic illustration of a cell adhering on an SLB coated with a ligand of β1-integrin (RGD, or Invasin) (Created in BioRender. Mikhajlov, O. (2025) https://BioRender.com/m84x622). Bottom: brightfield images of an adherent (left) and a trembling (right) cell on Invasin-SLB with corresponding kymographs describing cell edge movements over 2 minutes. Scale bars: 10 µm; 1 µm. B Time evolution of fractions of adherent cells on RGD- (blue) and Invasin-SLBs (red) in the presence (Mn + , full line) or absence (Mn-, dashed line) of Mn2+. Line scatter plot, mean, SEM. These results are based on: RGD-SLBs (Mn-): 0-15’: Ncells = 65; Nexp = 3. 15’-30’: Ncells = 72; Nexp = 4. 30’-45’: Ncells = 98; Nexp = 4. 45’-1h: Ncells = 59; Nexp = 3. Invasin-SLBs (Mn-): 0-15’: Ncells = 88; Nexp = 3. 15’-30’: Ncells = 94; Nexp = 3. 30’-45’: Ncells = 92; Nexp = 3. 45’-1h: Ncells = 93; Nexp = 4. RGD-SLBs (Mn + ): 0-15’: Ncells = 42; Nexp = 4. 15’-30’: Ncells = 71; Nexp = 5. 30’-45’: Ncells = 113; Nexp = 4. 45’-1h: Ncells = 44; Nexp = 4. Invasin-SLBs (Mn + ): 0-15’: Ncells = 74; Nexp = 3. 15’-30’: Ncells = 89; Nexp = 3. 30’-45’: Ncells = 85; Nexp = 3. 45’-1h: Ncells = 75; Nexp = 3. SLBs without ligands (Ctrl-): 45’-1h: Ncells = 103; Nexp = 3. Source data are provided as a Source Data file. C Brightfield images of cells of different morphological shapes adhered on an RGD-SLB (blue) and Invasin-SLBs (red). Projected areas and circularities are indicated as “Area” and “Circ.”. Scale bars: 10 µm. D, E Trembling (“Tr”, plots on grey background area) and adherent (“Ad”, 45 min – 1 h after seeding) cells in the presence (Mn + ) or absence (Mn-) of Mn2+ adhering on RGD- (blue) or Invasin-SLBs (red). Data from trembling cells on: RGD-SLBs: Ncells = 44; Nexp = 3. Invasin-SLBs: Ncells = 57; Nexp = 3. And from cells adhered on: RGD-SLBs (Mn-): Ncells = 48; Nexp = 3. RGD-SLBs (Mn + ): Ncells = 102; Nexp = 5. Invasin-SLBs (Mn-): Ncells = 75; Nexp = 3. Invasin-SLBs (Mn + ): Ncells = 78; Nexp = 5. Source data are provided as a Source Data file. D Cell circularity. Statistical significance (two-tailed Mann-Whitney tests): trembling vs. adhered on RGD-SLBs (Mn-): p-value = 0.0770; trembling vs. adhered on Invasin-SLBs (Mn-): p-value < 0.0001; adhered on RGD-SLBs Mn- vs. Mn + : p-value = 0.0598; adhered on Invasin-SLBs Mn- vs. Mn + : p-value = 0.0001; adhered on RGD- vs. Invasin-SLBs (Mn-): p-value < 0.0001; adhered on RGD- vs. Invasin-SLBs (Mn + ): p-value = 0.0001. Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. E Cell projected area in μm2. Statistical significance (two-tailed Mann-Whitney tests): trembling vs. adhered on RGD-SLBs (Mn-): p-value = 0.6103; trembling vs. adhered on Invasin-SLBs (Mn-): p-value < 0.0001; adhered on RGD-SLBs Mn- vs. Mn + : p-value = 0.2583; adhered on Invasin-SLBs Mn- vs. Mn + : p-value < 0.0001; adhered on RGD- vs. Invasin-SLBs (Mn-): p-value = 0.0032; adhered on RGD- vs. Invasin-SLBs (Mn + ): p-value < 0.0001. Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines.
We also examined cell spreading on SLBs to assess adhesion strength30. We measured the “projected areas” of the cells on the SLB surface and their “circularities”, which characterize cell shape irregularity (Fig. 1C). Consistent with an early adhesion stage, all “trembling” cells on both ligands displayed small, round areas, irrespective of Mn2+ treatment (Supplementary Fig. S3A, B). Moreover, in agreement with previous studies on fluid substrates19, we found that fibroblasts did not spread significantly on RGD-SLBs, maintaining small projected areas below 200 µm2 and a high degree of roundness (circularity around 1), regardless of Mn2+ treatment (Fig. 1D, E). In contrast, cells spread significantly more on Invasin-SLBs, with median projected areas 1.5- and 2-fold higher than on RGD-SLBs for Mn2+-untreated and treated cells, respectively (Fig. 1E). Moreover, 35% of Mn2+-untreated and 75% of Mn2+-treated cells on Invasin-SLBs developed multiple protrusions and irregular shapes (circularity <0.8), with projected areas twice and three times larger than those of trembling cells, respectively (Fig. 1C).
These findings suggest that although integrin activation by Mn2+ may not be critical on RGD-SLBs, it accelerates cell adhesion and significantly enhances spreading on Invasin-SLBs. The different role of manganese in the two systems can be explained by the much lower density of Invasin than of RGD on these SLBs, and by the fact that RGD binds to a much larger range of integrins than Invasin.
β1-integrin clusters are denser and larger on Invasin- than on RGD-SLBs
We used confocal microscopy to study the clustering of β1-integrins labeled with Alexa488-Halotag ligands at the cell-SLB interface during the first hour of adhesion. We observed isotropic β1-integrin clusters on both RGD- and Invasin-SLBs, which were morphologically distinct from FAs or actin-dependent podosome-like structures mainly composed of β3-integrins (Prof. Cheng-Han Yu, private communication)20. Through fluorescence calibration, we generated integrin density maps (Supplementary Fig. S4) and employed image segmentation algorithms to detect and quantify integrin clusters by their areas (σ) and densities of integrins (ρ) (Methods). Specifically, we segmented integrin density maps with a threshold of 300 integrins/µm2, corresponding to the minimal spacing of 58 nm between integrin-ligand pairs observed during mechanotransduction on glass31.
At 45 minutes after cell seeding, we observed that the clusters on Invasin-SLBs were larger and denser compared to those on RGD-SLBs, independent of Mn2+ (Fig. 2B-D). We did not observe differences on Invasin-SLBs in the presence or absence of Mn2+ (Fig. 2B). Median densities of integrins per cell (ρ) were approximately 100 integrins/µm2 for trembling cells for both ligands, independent of Mn2+ (Fig. 2B; Supplementary Fig. S5). For the adherent cells, however, the median densities were significantly higher, reaching up to 160 integrins/µm2 and 450 integrins/µm2 on RGD- and Invasin-SLBs, respectively. Importantly, ρ was significantly higher on Invasin-SLBs, attaining levels comparable to those observed on glass32, which were never reached on RGD-SLBs (Fig. 2B). We also evaluated the surface area of dense integrin clusters (σ300) corresponding to ρ > 300 integrins/µm2 (Fig. 2C) and found that clusters on Invasin-SLBs were significantly larger than on RGD-SLBs, regardless of Mn2+ treatment. Moreover, individual integrin clusters were over twice as dense on Invasin-SLBs compared to RGD-SLBs with median values equal to 257 integrins/µm2 and 117 integrins/µm2, respectively (Fig. 2D). Independent of Mn2+ treatment, σ300 per cell was significantly higher on Invasin-SLBs, with 23% of clusters larger than the theoretical diffraction limit (180 nm), versus only 9% on RGD-SLBs (Fig. 2E).
A Main panels: β1-integrin density maps, using the “physics” LUT of ImageJ below (Methods), showing β1-integrin cluster organization in cells adhering on SLBs coated with RGD (upper left) and Invasin (upper right). Images are taken at the focal plane of the SLBs. Lower left corners: corresponding brightfield images. Zoomed panels (white squares on the main panels): regions with β1-integrin clusters (black arrows). Bottom: β1-integrin density profiles along the white rectangles in the zoomed panels. Scale bars: 5 µm (main panels); 1 µm (zoomed panels). B-E) RGD-SLB (blue), Invasin-SLB (red). Data from trembling cells on: RGD-SLBs: Ncells = 44; Nexp = 3. Invasin-SLBs: Ncells = 57; Nexp = 3. And from cells adhered on: RGD-SLBs (Mn-): Ncells = 48; Nexp = 3. RGD-SLBs (Mn + ): Ncells = 102; Nexp = 5. Invasin-SLBs (Mn-): Ncells = 75; Nexp = 3. Invasin-SLBs (Mn + ): Ncells = 78; Nexp = 5. Source data are provided as a Source Data file. B Top: schematic illustration of integrin clustering at the cell-SLB interface (Created in BioRender. Mikhajlov, O. (2025) https://BioRender.com/y70w219). Bottom: Mean β1-integrin density per cell in trembling (“Tr”) and adherent (“Ad”) cells in the presence (Mn + ) or absence (Mn-) of Mn2+ adhering on RGD- (blue) or Invasin- SLBs (red). Statistical significance (two-tailed Mann-Whitney tests): trembling cells (Tr, Mn-) on RGD vs. Invasin-SLBs: p-value = 0.2174; cells adhering on RGD- vs. Invasin-SLBs in the absence of Mn2+ (Ad, Mn-): p-value < 0.0001; cells adhering on RGD- vs. Invasin-SLBs in the presence of Mn2+ (Ad, Mn + ): p-value < 0.0001; trembling vs. adhering cells on RGD-SLBs: p-value < 0.0001; trembling vs. adhering cells on Invasin-SLBs: p-value < 0.0001; cells adhering on RGD-SLBs in the presence vs. absence of Mn2+: p-value = 0.0332; cells adhering on Invasin-SLBs in the presence vs. absence of Mn2+: p-value = 0.5838. Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. C Total area of β1-integrin clusters of density higher than 300 β1-integrins/µm2 per cell (σ300) in the presence (Mn + ) or absence (Mn-) of Mn2+ in cells adhering on RGD- or Invasin-SLBs. Statistical significance (two-tailed Mann-Whitney tests): cells adhering on RGD- vs. Invasin-SLBs in the absence of Mn2+ (Mn-): p-value < 0.0001; cells adhering on RGD vs. Invasin-SLBs in the presence of Mn2+ (Mn + ): p-value < 0.0001. Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. D Histograms displaying the distribution of β1-integrin densities in individual clusters of Mn2+ treated cells adhering on RGD- (blue) or Invasin-SLBs (red). The x-axis represents the β1-integrin density, while the y-axis indicates the frequency of clusters with corresponding densities. Statistical significance (two-tailed Mann-Whitney tests): p-value < 0.0001. Data from 10,813 clusters on Invasin and 16,115 clusters on RGD. E Areas of individual β1-integrin clusters with densities exceeding 300 β1-integrins/µm2 in Mn2+-treated cells adhering on RGD (blue)- or Invasin (red)-coated SLBs. The data highlights differences in the sizes of high-density β1-integrin clusters between the two adhesion substrates. Statistical significance (two-tailed Mann-Whitney tests): p-value < 0.0001. 75-percentile of σ300 per cell distribution: 8-9 µm2 for Invasin-SLBs and 0.1-0.15 µm2 for RGD-SLBs. DL – theoretical diffraction limit. Data from 347 clusters for cells adhering on Invasin and 9203 clusters for cells adhering on RGD. Box-and-whisker plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines.
To investigate the potentially inhibitory effect of RGD crowding on integrin clustering, we reduced the surface fraction of RGD to 0.1% (comparable to Invasin concentration on SLBs). This led to a noticeable decrease in β1-integrin cluster density and in the total area of clusters, compared to SLBs with 2% surface fraction of RGD (Supplementary Fig. S6A, B). Additionally, cell spreading maintained the same low level (Supplementary Fig. S6C, D).
Since the affinity of Invasin to β1-integrins is two-orders of magnitude larger than that of RGD33,34, we hypothesized that the difference in cluster density could be due to the difference in affinity. Therefore, we modulated the affinity of Invasin for β1-integrins by treating cells with antibodies that block or activate β1-integrins specifically, in contrast to Mn2+, which targets all integrins (Methods). We observed a significant increase in β1-integrin density in cells treated with activating antibodies, as compared to control conditions without the antibody (Supplementary Fig. S7A). Conversely, upon cell treatment with blocking antibodies, the density dropped down to the baseline level corresponding to non-adhering cells in the presence of EDTA, a chelating agent for divalent ions. Altogether, these results confirm that ligand–β1-integrin affinity is an important parameter for β1-integrin clustering.
Focal adhesion proteins are recruited at β1-integrin clusters
Integrin clusters grow and mature during cell adhesion on rigid substrates, recruiting focal adhesion (FA) proteins in response to mechanotransduction driven by actomyosin contractility6,35. To determine whether β1-integrin adhesive clusters could also mature on SLBs, we quantified the recruitment of FA proteins (Fig. 3A, B). “Early adhesion proteins” like talin, kindlin-2, paxillin, vinculin and “late adhesion proteins” like VASP and zyxin were recruited to β1-integrin clusters both on Invasin- and RGD-SLBs (Supplementary Fig. S8A-E). Except for talin, all FA proteins showed a higher relative enrichment on Invasin-SLBs compared to RGD-SLBs (relative to their mean levels in the cell). This suggests that higher mechanical forces are at play on Invasin-bound cells. Particularly, the strong recruitment of kindlin-2 on Invasin-SLBs may indicate an enhanced stabilization of the β1-integrin-talin bond, capable of sustaining greater forces36.
All cells are Mn2+-treated and imaged 45 min – 1 h after seeding in the chamber. A Main panels: fluorescence multi-channel images showing β1-integrin (cyan) and paxillin (magenta) cluster organization in cells adhering on SLBs coated with RGD (left) and Invasin (right). Images are taken as max intensity z projections (in Fiji) of the volume with 1 µm height (z coordinate) centered at the focal plane of the SLB. Lower left corners: corresponding brightfield images. Medium panels: zooms corresponding to the white squares on the main panels, showing regions with β1-integrin and paxillin clusters. Bottom: β1-integrin and paxillin intensity profiles along the lines (white rectangles in the zoomed panels). Scale bars: 5 µm (main panels); 1 µm (zoomed panels). B Relative enrichment of focal adhesion (FA) proteins in β1-integrin clusters in cells on RGD- (blue) and Invasin-SLBs (red). It is calculated as an increase in fluorescence of a FA protein in β1-integrin clusters compared to the mean cell fluorescence normalized by the mean cell fluorescence. These measurements were conducted on fixed cells to examine FA proteins, with β1-integrin clusters identified using a threshold of 100 arbitrary units (‘arb. units’) (Methods). Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. Data from: Talin: Ncells = 87, Nexp = 4 (RGD-SLBs); Ncells = 65, Nexp = 4 (Invasin-SLBs). Kindlin-2: Ncells = 42, Nexp = 3 (RGD-SLBs); Ncells = 87, Nexp = 4 (Invasin-SLBs). Paxillin: Ncells = 53, Nexp = 3 (RGD-SLBs); Ncells = 128, Nexp = 3 (Invasin-SLBs). Vinculin: Ncells = 59, Nexp = 4 (RGD-SLBs); Ncells = 56, Nexp = 3 (Invasin-SLBs). VASP: Ncells = 42, Nexp = 4 (RGD-SLBs); Ncells = 66, Nexp = 4 (Invasin-SLBs). Zyxin: Ncells = 61, Nexp = 4 (RGD-SLBs); Ncells = 76, Nexp = 4 (Invasin-SLBs). Statistical significance for cells on RGD- vs. Invasin-SLBs (two-tailed Mann-Whitney tests): Talin: p-value = 0.1030. Kindlin-2: p-value < 0.0001. Paxillin: p-value = 0.0002. Vinculin: p-value = 0.0131. VASP: p-value = 0.0147. Zyxin: p-value < 0.0001. Source data are provided as a Source Data file. C, D Experiments focused on F-actin and microtubule recruitment to β1-integrin clusters performed on live cells in which the clusters were quantitatively defined using a threshold of 100 integrins/μm2 (Methods). D Relative enrichment of the actin in β1-integrin clusters in cells on RGD- (blue) and Invasin-SLBs (red). Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. Data from Ncells = 58, Nexp = 3 for RGD and Ncells = 75, Nexp = 3 for Invasin. Statistical significance (two-tailed Mann-Whitney tests): p-value < 0.0001. Source data are provided as a Source Data file. E Relative enrichment of the microtubules in β1-integrin clusters in cells on RGD- (blue) and Invasin-SLBs (red). Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. Data from Ncells = 53, Nexp = 3 for RGD and Ncells = 54, Nexp = 3 for Invasin. Statistical significance (two-tailed Mann-Whitney tests): p-value < 0.0001. Source data are provided as a Source Data file. E Fluorescence multi-channel images showing β1-integrin (yellow), actin (magenta) and microtubules (cyan) in cells adhering on SLBs coated with RGD (blue frame, top) and Invasin (red frame, bottom). Images are taken in the focal plane of the SLB. β1-integrin density, actin and microtubules intensity profiles along the lines (black rectangles in the zoomed panels) are plotted for both cells. Scale bars: 5 µm, 1 µm.
While integrin-mediated mechanotransduction on glass typically involves the actin cytoskeleton, microtubules also play a crucial role in stabilizing and regulating protein turnover within FAs37,38. We first assessed the association of F-actin to β1-integrin clusters in MEF cells using lifeact-mScarlet (Fig. 3C), by measuring its relative enrichment in these clusters for both ligands; however, the actin stress fibers typically associated with mechanotransduction were absent (Fig. 3C, E). Interestingly, we found that F-actin was more enriched at the β1-integrin adhesion clusters on RGD- than Invasin-SLBs (Fig. 3C). This finding contrasts with the difference in the recruitment of FA proteins at these structures and suggests that actin does not drive FA maturation on Invasin-SLBs. We then investigated the role of the microtubule cytoskeleton using EMTB-iRFP. We found that microtubules were organized in similar networks at the adhesion interface on SLBs and glass (Supplementary Fig. S9A, B). However, the relative increase in the microtubule signal at the β1-integrin clusters was nearly twice as high on Invasin- compared to RGD-SLBs (Fig. 3D), suggesting their involvement in maturation. Moreover, we observed that adaptor proteins linking talin to microtubules, such as KANK139 and ELKS40, were also recruited to these clusters (Supplementary Fig. S9D, E).
Dynein pulling along vertical microtubules leads to growth of integrin clusters
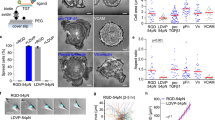
During mechanotransduction, integrin clusters are exposed to cellular mechanical forces that are significantly higher in magnitude if applied against rigid substrates10. On fluid SLBs, the lateral components of the forces are negligible compared to those on glass and are balanced only by the resistive forces due to membrane viscosity41. In comparison, SLBs can sustain larger forces normal to the surface, potentially leading to the maturation of integrin clusters. These normal forces might cause joint local deformations in both the SLBs and cell plasma membranes. We observed such deformations while imaging β1-integrin clusters above the SLB focal plane (Fig. 4A; zoom 1; 0 <z < 1.5 µm). Similar to the clusters observed at SLB surfaces (Fig. 4A; zoom 2; z = 0), clusters within the cell volume had a larger total area in cells on Invasin-SLBs compared to those on RGD-SLBs (Supplementary Fig. S10A). Some clusters were associated with membrane tubes pulled out of the SLBs and with the locally vertical actin and microtubules linked to those tubes (Fig. 4A; zoom 2). The proportion of cells with tubes and the number of tubes per cell were significantly higher on Invasin than on RGD (Fig. 4B). These results are consistent with the notion that integrin clusters on Invasin-SLBs are exposed to greater mechanical forces than those on RGD-SLBs, leading to their growth in size and density, the higher recruitment of FA proteins, and the formation of more tubes.
All cells are Mn2+-treated and imaged 45 min – 1 h after seeding in the chamber on RGD- or Invasin- SLBs. A Top panel: fluorescence multi-channel image of a MEF cell showing the SLB labeled with a TR-DHPE lipid, β1-integrin with Halotag-Alexa488, actin with lifeact-mScarlet and microtubules (MT) with EMTB-iRFP. A brightfield image of the cell is in the upper left corner of the merged image. Middle panels: schematic illustrations of a cell with integrin clusters associated with a membrane tube pulled from the SLB (left) and resided on the surface of the SLB (right) (Created in BioRender. Mikhajlov, O. (2025) https://BioRender.com/q92n558). Bottom panels (zoom): regions corresponding to white squares 1 and 2 on the main panels. Top: x-y. Bottom: x-z section corresponding the lines in x-y. Scale bars: 10 µm (main panel); 2 µm (zoomed panel xy); 0.5 µm (zoomed panel xz). B Histograms of the number of detected tubes per cell in MEF cells adhering on RGD- (blue) and Invasin-coated (red) SLBs. Data from: Ncells = 102, Nexp = 5 (RGD-SLBs); Ncells = 78, Nexp = 5 (Invasin-SLBs). Statistical significance (two-tailed Mann-Whitney tests): p-value = 0.0006. Source data are provided as a Source Data file. C, D Data from cells adhering on Invasin-coated SLBs in the presence of drugs: DMSO: Ncells = 78, Nexp = 5. Y-27632: Ncells = 72, Nexp = 3. CK666: Ncells = 102, Nexp = 4. SMIFH2: Ncells = 81, Nexp = 3. Nocodazole (NZ): Ncells = 86, Nexp = 3. Ciliobrevin D (CBD): Ncells = 69, Nexp = 4. Source data are provided as a Source Data file. C Fraction of MEF cells with detected membrane tubes. Bar plots: mean, SEM. Statistical significance (Kruskal-Wallis test followed by Dunn’s multiple comparisons tests): Y-27632 vs. DMSO: p-value = 0.0009. CK666 vs. DMSO: p-value = 0.5692. SMIFH2 vs. DMSO: p-value > 0.9999. Nocodazole (NZ) vs. DMSO: p-value < 0.0001. Ciliobrevin D (CBD) vs. DMSO: p-value < 0.0001. D Total area of β1-integrin clusters of density higher than 300 β1-integrins/µm2 per cell (σ300). Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. Statistical significance (Kruskal-Wallis test followed by Dunn’s multiple comparisons tests): Y-27632 vs. DMSO: p-value < 0.0001. CK666 vs. DMSO: p-value = 0.0782. SMIFH2 vs. DMSO: p-value = 0.0166. Nocodazole (NZ) vs. DMSO: p-value < 0.0001. Ciliobrevin D (CBD) vs. DMSO: p-value < 0.0001. E Schematic illustration of theoretical model. Free receptors and ligands form bound complexes (bonds) at a rate \({k}_{{on}}\), and bonds dissociate into ligand-receptor pairs at a rate \({k}_{{off}}\). The concentration of bonds is \({\phi }_{b}\), and the space left vacant by the bonds is partially occupied by free receptors and ligands with concentrations \({\bar{\phi }}_{r}(1-{\phi }_{b})\) and \({\bar{\phi }}_{l}(1-{\phi }_{b})\), respectively. Microtubules apply a constant vertical force, \(f\), to each bond, which can stretch the adaptor proteins and generate attractive inter-bond interactions above an activation force scale, \(f > {f}_{{activ}}\). The interaction strength, \(\chi (f)\), describes the effective attractive interaction between bonds for a given force. F Phase diagram of the system. The parameter space of bond chemical potential, \({\mu }_{b}\), and microtubule vertical force, \(f\), is subdivided into regions where the theoretical model predicts a dense/dilute phase co-existence (clustering), or a single homogeneous phase (no clusters). The clustering thresholds are indicated as dashed-black and solid-pink lines for two values of the adhesome-mediated attraction, \({\chi }_{0}=0\) and \({\chi }_{0}=15\), respectively. Under each condition, clustering is predicted above the corresponding threshold curve, with clustering being driven mostly by membrane interactions in the grey region, and by adhesome interactions in the pink region. The dotted horizontal lines indicate our \({\mu }_{b}\) estimates for Invasin and RGD (at two concentrations). The parameter values \({f}_{{activ}}=2{pN}\), \({f}_{{elast}}=20{pN}\), \({f}_{{break}}=50{pN}\), \(\frac{{E}_{{activ}}}{{k}_{B}T}={\chi }_{0}=15\), \(\frac{{E}_{{elast}}}{{k}_{B}T}=200\), and \(r=25\) were used in the derivation of these results.
To elucidate the origin of the vertical forces pulling on integrin clusters, we tested how inhibitors of cytoskeleton polymerization and associated motors affect the proportion of cells with tubes and the number of tubes per cell in the case of Invasin-SLBs (Fig. 4C; Supplementary Fig. S10B). We found that the inhibition of formins and the Arp2/3 complex, which nucleate actin polymerization using SMIFH2 and CK666, respectively, had no effect on the frequency of tube formation and little to no effect on integrin clustering (Fig. 4C, D; Supplementary Fig. S10B, C). Preventing actomyosin contractility by Rho kinase inhibition (Y-27632) decreased the percentage of cells with tubes by half and the number of tubes per cell by 60%, and led to a significant decrease in β1-integrin cluster size and density (Fig. 4D; Supplementary Fig. S10C). Altogether, these results suggest that the forces associated with actin polymerization mediated by formins or Arp2/3 are not critical for integrin clustering through the vertical deformations of the plasma membrane. However, the effects of Y-27632 suggest that actomyosin contractility might play a role in integrin clustering on SLBs, possibly through another mechanism of mechanotransduction related to the described local “pinching” of nascent adhesion clusters19,42.
Next, we investigated whether microtubules and associated molecular motors were involved in the tubular deformations of SLBs, as they were reported to play a role in FA mechanosensing on rigid substrates43,44. We observed dense and large integrin clusters at the SLB surface and on tubes associated with microtubules (Figs. 3D; 4A; Supplementary Fig. S9B). When microtubules were depolymerized with nocodazole (NZ) or when dynein activity was blocked with ciliobrevin D (CBD), the fraction of cells in which tubes were detected and the number of tubes per cell decreased significantly (Fig. 4C; Supplementary Fig. S10B). We observed similar effects in HeLa cells on Invasin-SLBs after dynein inhibition with CBD or silencing of the dynactin subunit p150glued (Supplementary Fig. S11B, D). These findings suggest that the vertical forces applied to β1-integrin clusters depend on microtubules and are driven by dynein activity. Finally, NZ and CBD treatment significantly decreased the total area of β1-integrin clusters and their density (Fig. 4D and Supplementary Fig. S10C). Notably, the effect of NZ on cell adhesion to fluid substrates is opposite to that observed on rigid substrates, where it leads to an increase in the size and density of FAs due to enhanced actomyosin contractility45,46.
To further characterize the role of integrin-ligand affinity on integrin clustering in the case of fluid substrates, we propose a mean-field theoretical model that focuses on the interplay of binding affinity, substrate fluidity, and vertical forces from microtubules. The fluidity of the SLB enables the lateral mobility of ligands, which distinguishes our theory from previous models of ligand-receptor clustering on rigid substrates.
In our model, the integrin receptors and associated ligands are distributed in two parallel lipid bilayers representing the cell membrane and the SLB, with an average distance \(h\) between them (Fig. 4E, side view). They form bound receptor-ligand complexes, termed ‘bonds’, by stretching vertically to bridge the distance between the two membranes. This stochastic process is characterized by binding and unbinding rates, \({k}_{{{\rm{on}}}}\) and \({k}_{{{\rm{off}}}}\), which depend crucially on the binding affinity of the receptor-ligand pair, as well as other factors detailed in the Supplementary Information. The bonds occupy an area fraction \({\phi }_{b}\) of the cell-SLB interface, while free receptors and ligands partially fill the space unoccupied by bonds through lateral diffusion in the membranes, resulting in a net area fraction \({\phi }_{l}={\bar{\phi }}_{l}\left(1-{\phi }_{b}\right)\) of ligands, and \({\phi }_{r}={\bar{\phi }}_{r}\left(1-{\phi }_{b}\right)\) of receptors (Supplementary Fig. S12).
Physically, we interpret the process of integrin clustering as the phase separation of dense regions of bonds (high \({\phi }_{b}\)) from a dilute background (low \({\phi }_{b}\)). Thus, the model can be summarized by the condition of equal chemical potential between the dense and dilute phases, given by
where the effective, dimensionless chemical potential, \({\mu }_{b}=\frac{\epsilon }{{k}_{B}T}+{{\mathrm{ln}}}\left({\bar{\phi }}_{l}{\bar{\phi }}_{r}\right)\), is a control parameter fixed by the integrin-ligand binding energy, \(\epsilon\), and the availability of free ligands and receptors, \({\bar{\phi }}_{l}{\bar{\phi }}_{r}\). The second control parameter is the average vertical force, \(f\), exerted by microtubules on the bonds, while the bond density \({\phi }_{b}\) is a variable obtained by solving Eq. (1). This equation provides two stable solutions for the bond density (i.e., dense and dilute phases) when the left-hand side of the equation is non-monotonic in \({\phi }_{b}\), and the effective chemical potential, \({\mu }_{b}\), is sufficiently large (Supplementary Fig. S13). We thus generate a phase diagram in the \(\left(f,{\mu }_{b}\right)\) space which indicates the parameter regimes compatible with integrin clustering (shaded regions in Fig. 4F).
Our model suggests two mechanisms for clustering: attraction between adaptor proteins at the adhesion site47,48,49 and co-operative ligand-receptor binding that minimizes bond deformation between the membranes50,51,52. To elucidate these mechanisms, we explain the physical meaning of the terms in Eq. (1), which are derived and interpreted in greater detail in the Supplementary Information (SI). The first term on the left-hand side of Eq. (1) represents the entropy of mixing bonds at the two-dimensional cell-SLB interface. The second term takes into account the increase of the unbinding rate with the value of the vertical force, with a typical force scale \({f}_{break}\) required for bond rupture. Phase separation is driven by the third and fourth terms, which describe adhesome-driven and deformation-driven attraction, respectively. The “adhesome” term \(\chi \left(f\right){\phi }_{b}\) always favours clustering and its magnitude depends on the microtubule force. Beyond a typical “activation” force, \({f}_{{activ}}\), adaptor proteins undergo conformational changes and increase the effective attraction between bonds (e.g., via the exposure of vinculin binding sites on talin53,54) as illustrated in Fig. 4E. The onset and extent of adhesome-dominated clustering (pink-shaded region, Fig. 4F) are determined by \({f}_{{activ}}\) and \({\chi }_{0}=\chi \left(f\gg {f}_{{activ}}\right)\). The “elastic” term \({\mu }_{{el}}\left({\phi }_{b},f\right)\) represents the elastic energy cost of bond deformation across the inter-membrane distance, \(h=h\left({\phi }_{b}\right)\). For small forces that do not break the bonds (\(f < {f}_{{break}}\)), as is relevant to the experiments (see estimates in SI), binding cooperativity favours clustering in the following way. An increase in the local concentration of bonds brings the membranes closer together, which lowers the elastic energy cost of forming new bonds. Below the activation threshold of the adaptor proteins (\(f < {f}_{{activ}}\)), clustering is enabled by this membrane-mediated binding cooperativity (grey-shaded region, Fig. 4F).
The predictions of our theoretical model are consistent with our experimental observations of cell adhesion on SLBs, since ligand-receptor pairs with higher binding affinity form denser clusters (Supplementary Fig. S14) and can sustain larger vertical forces (i.e., the shaded region in Fig. 4F extends to larger \(f\) when \({\mu }_{b}\) is higher). Our estimate of the effective chemical potential for the Invasin-SLBs experiment (\({\mu }_{b}\approx 5\)) is well within the clustering region, suggesting dense clusters even in the absence of microtubule forces, whereas the estimate for RGD-SLBs with 2% RGD area fraction (\({\mu }_{b}\approx 1\)) lies very close to the clustering threshold, indicating more dilute clusters, as observed in the experiments. Finally, our estimate for 0.1% RGD area fraction (\({\mu }_{b}\approx -2\)) lies within the no-clustering phase for weak forces, and it supports clusters only for forces greater than approximately 2 pN. As such strong forces are expected to be relatively rare, we predict sparse clusters, consistent with the experiments (Supplementary Fig. S14).
Altogether, these findings suggest that mechanotransduction on SLBs results from the pulling activity of the dynein motors on microtubules normal to the cell-SLB interface that can resist mechanical forces (Supplementary Fig. S9C), leading to the growth and maturation of integrin clusters. Higher microtubule enrichment and higher frequency of tube formation on Invasin- than on RGD-SLBs suggest that mechanotransduction at integrin clusters on SLBs depends on integrin-ligand affinity, in contrast with glass. This is consistent with the notion that a longer lifetime of the integrin-ligand bonds supports more efficient mechanotransduction.
Integrin clusters are pushed to the cell periphery in a dynein- and microtubules-dependent manner
Cell spreading correlates with integrin clustering due to mechanotransduction, in the case of glass and high viscosity RGD-SLBs41. Unexpectedly, we observed that cells formed protrusions on low viscosity Invasin-SLBs, leading to their symmetrical spreading without polarization (Fig. 5A). We found that high integrin densities within clusters correlated with high spreading area and low cell circularity in Mn2+-treated cells on Invasin-SLBs (Fig. 5B and Supplementary Fig. S15). Moreover, the presence of blocking antibodies significantly reduced cell spreading and triggered a strong reduction in integrin density, whereas activating antibodies enhanced integrin density within clusters but did not increase cell spreading, suggesting a saturation (Supplementary Fig. S7B, C).
All cells are Mn2+-treated MEF cells and imaged during the first hour after seeding to the chamber with Invasin-coated SLBs. A Composite (brightfield and fluorescence channels) images of spread cells on Invasin-SLBs showing β1-integrin clusters (green). Scale bars: 10 µm. B Cell projected area in μm2 (top left) and circularity (bottom left) of cells with β1-integrins clusters densities: ρ > 500 (magenta); 300 <ρ < 500 (green); and ρ < 300 integrins/µm2 (black). Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. (Right): Representative composite images (brightfield and fluorescence channels) of spread cells on each density class, showing β1-integrin clusters (green). Scale bars: 10 µm. Data from: ρ > 500: Ncells = 157, Nexp = 15. 300 <ρ < 500: Ncells = 63, Nexp = 15. ρ < 300: Ncells = 85, Nexp = 16. Statistical significance (Kruskal-Wallis test followed by Dunn’s multiple comparisons tests): ρ > 500 vs. 300 <ρ < 500: p-value = 0.006 (projected area), p-value = 0.1192 (circularity). ρ > 500 vs. ρ < 300: p-value < 0.0001 (projected area), p-value < 0.0001 (circularity). 300 <ρ < 500 vs. ρ < 300: p-value = 0.0003 (projected area), p-value = 0.0022 (circularity). Source data are provided as a Source Data file. C A schematic representation of the central and peripheral zones in a spreading cell (Created in BioRender. Mikhajlov, O. (2025) https://BioRender.com/b18d794). Dark green represents the cell periphery, which is the band parallel to the cell border. Light green represents the cell center, which is the part of the cell excluding the periphery. The two zones are chosen to have approximately the same area, depending on the cell’s circularity. For perfectly round cells, the zones have exactly equal areas. For cells with circularity <0.8, the width of the periphery zone set to 20% of the square root of the projected cell area (Methods). D Ratios of area fractions of β1-integrin (green) and paxillin (red) clusters in the periphery zone (Pperiphery) to the center zone (Pcenter) for spread (projected area > 450 µm2) and non-spread (projected area <450 µm2) fixed cells. Clusters were detected automatically using a Renyi’s entopy thresholding algorithm (Methods). Box-and-whisker plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. Data from: β1-integrin: Ncells = 115, Nexp = 3. Paxillin: Ncells = 119, Nexp = 3. Statistical significance (two-tailed Mann-Whitney tests): Spread vs. non spread: p-value = 0.0015 (β1-integrin), p-value = 0.0001 (paxillin). Source data are provided as a Source Data file. E Average distance between integrin clusters and the cell border normalized by average cluster-to-border distance for uniform integrin distribution for cells with high (ρ > 500 integrins/µm2) and low (ρ < 300 integrins/µm2) integrin density (Methods). A value above (or below) 1 describes an integrin cluster distribution skewed towards the center (or periphery) of the cell, relative to the uniform distribution. Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. Data from: High integrin density: Ncells = 24, Nexp = 3. Low integrin density: Ncells = 23, Nexp = 3. Statistical significance (two-tailed Mann-Whitney tests): p-value = 0.0035. Source data are provided as a Source Data file. F Projected area of cells with and without detected tubes (in μm2). Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. Data from Ncells = 78, Nexp = 5. Statistical significance (two-tailed Mann-Whitney tests): p-value = 0.0006. Source data are provided as a Source Data file. G Comparative illustration of integrin cluster distribution for cells treated with various drugs (CK666, SMIFH2, Y27632, NZ, CBD) and in non-treated cells (DMSO control). β1-integrin density maps are shown using the “physics” LUT of Fiji (calibrated bar on the side). Images are taken at the SLB plane. Corresponding brightfield images are shown in the upper right corners. Scale bars: 10 µm. (H–J) Data from cells adhering on Invasin-coated SLBs in the presence of drugs: DMSO: Ncells = 78, Nexp = 5. Y−27632: Ncells = 72, Nexp = 3. CK666: Ncells = 102, Nexp = 4. SMIFH2: Ncells = 81, Nexp = 3. Nocodazole (NZ): Ncells = 86, Nexp = 3. Ciliobrevin D (CBD): Ncells = 69, Nexp = 4. Source data are provided as a Source Data file. H Area fractions of β1-integrin clusters in the periphery zone of cells (φperiphery). Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. Statistical significance (Kruskal-Wallis test followed by Dunn’s multiple comparisons tests): Y-27632 vs. DMSO: p-value < 0.0001. CK666 vs. DMSO: p-value > 0.9999. SMIFH2 vs. DMSO: p-value > 0.9999. Nocodazole (NZ) vs. DMSO: p-value = 0.002. Ciliobrevin D (CBD) vs. DMSO: p-value < 0.0001. I Cell projected area in μm2. Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. Statistical significance (Kruskal-Wallis test followed by Dunn’s multiple comparisons tests):Y-27632 vs. DMSO: p-value > 0.9999. CK666 vs. DMSO: p-value = 0.0132. SMIFH2 vs. DMSO: p-value < 0.0001. Nocodazole (NZ) vs. DMSO: p-value < 0.0001. Ciliobrevin D (CBD) vs. DMSO: p-value < 0.0001. J Cell circularity. Violin plots with the median represented by a central line and the interquartile range (25th-75th percentiles) indicated by the upper and lower lines. Statistical significance (Kruskal-Wallis test followed by Dunn’s multiple comparisons tests): Y-27632 vs. DMSO: p-value = 0.0044. CK666 vs. DMSO: p-value = 0.0167. SMIFH2 vs. DMSO: p-value < 0.0001. Nocodazole (NZ) vs. DMSO: p-value < 0.0001. Ciliobrevin D (CBD) vs. DMSO: p-value < 0.0001.
Next, we measured and compared cluster areas at the periphery and center of the cell (regions of approximately the same area (Fig. 5C; Methods)) for two classes of cell morphology termed “spread” (projected area > 450 μm2) and “non-spread” (projected area <450 μm2). We found that integrin and paxillin clusters in spread cells were located preferentially at the cell periphery compared to the cell center, in contrast with the clusters in non-spread cells (Fig. 5D). We found similar results when comparing “non-round” (circularity <0.8) to “round” cells (circularity > 0.8) (Supplementary Fig. S16A). Furthermore, integrin clusters were located closer to the cell border in cells exhibiting “dense” integrin clusters (ρ > 500 integrins/µm2) compared to cells in which the clusters were “not dense” (ρ < 300 integrins/µm2) (Fig. 5E). These results suggest a relation between the growth of adhesion clusters, their movement to the cell periphery and cell spreading.
We also found a positive correlation between cell spreading and the presence of membrane tubes, consistent with mechanotransduction at integrin clusters on SLBs (Fig. 5F). Moreover, spread cells exhibited more tubes in the center than at the periphery of the cell (Supplementary Fig. S16B), indicating that integrin clusters in the center were more likely to be exposed to vertical pulling forces. This observation agrees with the recent findings of Brockman et al. on RGD-coated SLBs that integrin clusters primarily experience vertical forces in the center of the cell and tangential forces at the cell periphery55. Since we found that microtubules and dyneins were crucial for both the growth and densification of integrin clusters, as well as the formation of membrane tubes on SLBs (Fig. 4C, D and Supplementary Fig. S10C), we hypothesized that microtubules oriented parallel to the substrate also play a role in the localization of clusters at the cell periphery, in a similar manner to cells spreading on glass56,57. In line with this, we consistently observed the accumulation of microtubules at peripheral integrin clusters in spread cells on Invasin-SLBs (Fig. 3E (Invasin); Fig. 4A (zoom 2); Supplementary Fig. S9B). Cell treatment with NZ or CBD decreased integrin clustering (Fig. 4D and Supplementary Fig. S10C) and completely suppressed the localization of integrin clusters at the cell periphery (Fig. 5G, H). Notably, cells treated with NZ or CBD did not spread (Fig. 5I, J). We attribute this result to a combination of the inhibition of microtubules and dyneins pushing on adhesion clusters and an increase in actomyosin contractility, which may be connected to microtubule depolymerization46. Consistent with an antagonistic role of the actomyosin contractility in cell spreading, the inhibition of Rho kinase with Y27632 resulted in a higher proportion of spread cells58 (Fig. 5G, I, J). Similarly to cells on glass59 and RGD-SLBs19, the inhibition of formins by SMIFH2 impaired cell spreading on Invasin-SLBs without significant effects on cluster growth (Figs. 4C, D; 5G; I-J). These findings suggest that dynein motor activity along microtubules pushes large and mature β1-integrin clusters towards the cell periphery, which contributes to cell spreading on SLBs through the actin-rich protrusions emanating from these adhesive clusters.
Discussion
Our understanding of FAs is primarily derived from studies on rigid substrates such as glass, where mechanotransduction is mainly driven by actomyosin-dependent forces transmitted through actin fibers tangential to the basal membrane of the cell60. Living tissues, however, exhibit a wide range of stiffness5. Instead of being immobilized, integrin ligands can diffuse laterally with little frictional resistance in fluid cell membranes61,62 and may not exert the range of counter-forces required for “canonical” mechanotransduction. Indeed, when cells adhere on SLBs, the absence of actin stress fibers usually associated with FAs indicates that planar forces on SLBs are negligible compared to glass63,64.
Using Yersinia Invasin as a high-affinity β1-integrin ligand, we show that cells adhere, spread, and develop large and dense β1-integrin clusters on fluid substrates consisting of SLBs, in a similar way to FAs on glass (Fig. 2B)32. On SLBs, our experiments showed a direct correlation between the β1-integrin–ligand affinity and the density of clusters, in agreement with our theoretical model. Figure 6 summarizes our observations of cell adhesion on fluid substrates. Contrary to glass, actin-related forces tangential to the plasma membrane do not play a dominant role in integrin adhesion on SLBs due to the fluidity of the substrate. Furthermore, instead of enhancing adhesion and spreading through the stimulation of actomyosin activity, as observed on glass45, microtubule depolymerization strongly inhibits cell adhesion on SLBs. This is reminiscent of cells migrating on soft 3D extracellular matrix56,65. Microtubules were also shown to be essential for cell spreading on relaxed collagen networks and for the formation of dendritic extensions66. Here, we show that adhesion maturation on bilayers relies on microtubule-dependent forces perpendicular to the bilayer. We provide evidence of these forces by observing local SLB deformations and tubes connecting the bilayer and integrin clusters in the plasma membrane (Fig. 4A). The existence of perpendicular force components at integrin clusters was also reported on RGD-SLBs using DNA-FRET force probes55, but their origin was not identified. Notably, we observe a stronger recruitment of kindlin-2 to the integrin clusters on Invasin-SLBs compared to RGD-SLB, consistent with the reported role of kindlin-2 in the reinforcement of the integrin-ligand adhesion bonds36 and, hence, with a higher load transmitted to the integrin clusters.
Created in BioRender. Mikhajlov, O. (2025) https://BioRender.com/f98k213.
On glass, microtubules align vertically67 and associate with mature FAs at the cell periphery via several adaptor proteins40,43. Moreover, dyneins that stabilize the plus-ends of microtubules at the plasma membrane68 interact with integrin adhesion clusters through paxillin and other FA proteins69,70. Here, we present several pieces of evidence that support the role of microtubules in exerting forces on β1-integrin clusters which are normal to the SLB surface: (i) microtubules are enriched at integrin clusters on SLBs (Fig. 3D); (ii) microtubule-talin adaptor proteins KANK1 and ELKS are recruited at these clusters (Supplementary Fig. S9D, E); and (iii) some microtubules adopt a vertical orientation when associated with integrin adhesions (Fig. 4A).
Molecular motors can collectively pull tubes from membranes by distributing the load on a dynamically formed motor assembly, as shown in vitro for kinesins71. Forces on the order of 60 pN or less are necessary to pull tubes from SLBs on glass72. Dynein dimers exert forces on the order of 7 pN73 and can pull on integrin adhesion complexes. When sufficiently many motors accumulate at the extremity of a microtubule, they may produce the force required to generate tubes from the plasma membrane and the SLB attached to it through integrin-ligand links. Forces within this range induce conformational changes in adaptor proteins leading to integrin clustering which, according to our theoretical model, is more probable for high-affinity Invasin than for RGD.
The observed tubular deformations of the membranes locally connected to integrin clusters are mostly microtubule- and dynein-dependent (Fig. 4D) and are not only involved in the maturation of clusters, but could also be related to β1-integrin endocytosis74. Internalization of active β1-integrins proceeds via the clathrin-independent CLIC-GEEC (CG) pathway74. The early stage of adhesion actually involves nanoclustering of GPI-anchored proteins (GPI-APs) as well as vinculin activation for further maturation75. GPI-APs are also endocytosed though the same CG pathway76. The micrometer-long tubes observed in our study could thus be non-cleaved endocytic structures. Contrary to glass, a double membrane tube is formed during endocytosis on SLBs, with the SLB membrane in the inner layer surrounded by the cell plasma membrane as an outer layer (Figs. 4A; 6). Such double-layer tubes could be more resistant to scission (dynamin-independent in the case of active β1-integrin74), leading to the formation of stable tubes.
On rigid substrates, cells form protrusions to spread. In this process, mature FAs act as anchoring points against which growing actin-rich protrusions push. The ability of cells to spread on glass depends on the density of ligands on the surface, the size of integrin adhesions, and their connection to the cytoskeleton77. On SLBs, on the other hand, integrin adhesions can move laterally in the adhesion plane upon mechanical stimuli. On Invasin-SLBs, integrin clusters are dragged in the SLB plane towards the cell periphery in a microtubule and dynein-dependent manner (Fig. 3E). Since dyneins are involved in the maturation of the adhesions, which is essential for spreading, we cannot distinguish whether some dyneins also push microtubules linked to adhesions along the direction parallel to the surface towards the cell edge78, or whether microtubule dynamics are responsible for this. This movement is likely antagonized by an inward force due to the actomyosin contraction of the cell cortex79,80 (Figs. 3E; 5H; 6), leading to stalling of integrin clusters at the cell edge where they serve as anchoring centers from which actin-rich protrusions emanate66,81. The antagonistic effect of actomyosin contractility and microtubules may also be important in the densification of integrin clusters at the cell periphery, as we have found for well-spread cells (Fig. 5B-E; Supplementary Fig. S16A). Importantly, such dense clusters are not found on Y-27632-treated cells (Figs. 4D; 5G-H; Supplementary Fig. S10C). Cell protrusions on SLBs might result from a combination of pushing forces from actin and microtubule polymerization, and from microtubule pushing through dynein activity. The pushing forces are balanced against the centrosome to which microtubules are connected (Supplementary Fig. S9C). In contrast with cells adhering on glass where microtubules are essential for cell polarization82,83, cells do not polarize on SLBs and conserve their central symmetry despite the key role of microtubules, through a mechanism that remains to be characterized.
To summarize, both microtubules and actin play a role in the maturation of integrin clusters on SLBs. Microtubules and dynein motors exert vertical forces on integrin clusters, which result in the formation of tubes and are partially responsible for integrin clustering. At the same time, microtubules and dyneins exert lateral forces that push integrin clusters to the cell periphery. Cell spreading is determined by the interplay between the microtubule-driven pushing of integrin clusters and the actomyosin contractility at the cell edge. This interplay may also be responsible for the densification of integrin clusters at the cell periphery, as we found for well-spread cells.
In HeLa cells on SLBs, we also observed dynein/microtubule-dependent formation of tubes (Supplementary Fig. S11), suggesting that mechanotransduction associated with forces normal to the adhesion plane occurs in various cell types. Functionalized SLBs allow to study the early stages of cell adhesion on a fluid interface, which is relevant for cell-cell interactions (i.e., brain cells, immune cells, etc.)4, but not accessible on substrates like glass or gels. Moreover, functionalized SLBs reveal forces probably at play on stiff substrates that cannot be observed with immobilized ligands. Using this platform, we found a role for dynein motors and microtubules in integrin adhesion cluster growth and maturation on SLBs. In addition, our experiments and physical modeling show that cell adhesion on a fluid interface can be strongly modulated by the receptor-ligand affinity, in contrast with solid surfaces. These regulation levels might be used during selective adhesion of T- or B-cells, in which microtubules and dynein motors are known to be involved in clustering TCR and BCR receptors, respectively84,85.
Methods
Buffers and reagents
Cell Buffer was composed of 25 mM HEPES pH 7.3, 120 mM NaCl, 7 mM KCl, 1.8 mM CaCl2, 0.8 mM MgCl2, and 5 mM glucose. Small Unilamellar Vesicles (SUV) buffer was composed of 10 mM Tris pH 7.3; 120 mM NaCl. Lipids used included DOPC, DGS-NTA(Ni), and DOPE-RGD from Avanti Polar Lipids (Alabaster, AL, USA), and Marina Blue DHPE from Invitrogen. Microtubules were labeled with SiR-tubulin (Cytoskeleton #CY-SC002) according to the manufacturer’s dilution guidelines. MnCl2 (Sigma Aldrich) was added at 0.5 mM at the time of cell seeding into the imaging chamber. β1-integrin specific antibodies, including activating (9EG7, BD Pharmingen #553715) and blocking (MB1.2, Merck #MAB1997) types, were used at 5 µg/ml during adhesion experiments. Various inhibitors (Arp2/3 complex inhibitor CK-666 at 50 µM (Sigma Aldrich #182515), formin inhibitor SMIFH2 at 10 µM (Sigma Aldrich #S4826), ROCK inhibitor Y-27632 at 50 µM (Sigma Aldrich #SCM075), nocodazole at 10 µM (Sigma Aldrich #M1404), cytoplasmic dynein inhibitor Ciliobrevin D at 50 µM (Sigma Aldrich #250401)) were applied at the time of cell seeding into the imaging chamber. Control samples were treated with DMSO, ensuring that its concentration did not exceed 0.001% v/v.
Cell culture and cell lines
Mouse Embryonic fibroblast (MEF) cells, including the β1KO β1-Halotag and MEF β1KO β1-Halotag paxillin-mCherry lines, were gifts from David Calderwood’s lab at Yale University25. The MEF β1KO β1-Halotag LifeAct-mScarlet line was developed by lentiviral transduction of the β1KO β1-Halotag MEF line with a LifeAct-mScarlet construct on the pLVX expression vector (Clontech #632164). The MEF β1KO β1-Halotag EMTB-iRFP, created by lentiviral transduction with an EMTB-iRFP construct on the pLVX expression vector, was provided by Simon De Beco at Paris Diderot University. The HeLa β1-Halotag cell line was developed by lentiviral transduction of a wild-type HeLa line with an ecto-tag construct from David Calderwood’s lab25.
All cell lines were cultured in Dulbecco’s Modified Eagles Medium (DMEM) with high glucose and GlutaMAX (ThermoFischer), supplemented with 10% fetal bovine serum (FBS, EuroBio) and 1% penicillin-streptomycin (ThermoFischer). Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2. Routine mycoplasma contamination checks were conducted using a PCR-based method86, confirming all cell lines to be free of contamination.
DNA plasmids for transfection and transduction
Plasmids encoding various mCherry-tagged proteins were used for transient transfections, including talin-mCherry (addgene #55137), vinculin-mCherry87, VASP-mCherry (addgene #55151), kindlin-2-mCherry (from Christof Hauck, Konstanz University), and zyxin-mCherry (from Danijela Vignjevic, Institut Curie).
For stable genetic modifications via lentiviral transductions, the following plasmids were used: β1-Halotag (from David Calderwood, Yale University)25, pLVX EMTB-iRFP (from Simon De Beco, Paris Diderot University), and pLVX LifeAct-mScarlet. The pLVX LifeAct-mScarlet plasmid was prepared by first conducting PCR on the LifeAct-GFPtag2 plasmid (Ibidi USA 60101) using primers 5’-TCTAGAGCTACTAACTTCAGCCTGCTG-3’ and 5’-CGGTGGATCCCCTTCTTCC-3’. The PCR product was then cloned into the pLVX vector (Clontech #632164), which had been digested with NotI and BamHI restriction enzymes (New England Biolabs), using the In-Fusion HD enzyme kit (Takara #102518). A second PCR was performed for the mScarlet insert using primers 5’-GAAGGGGATCCACCGATGGTGAGCAAGGGCGAGG-3’ and 5’-TTAGTAGCTCTAGACTTGTACAGCTCGTCCATGCC-3’. This PCR product was then cloned using the In-Fusion HD into the pLVX vector already containing LifeAct, which had been cut with BamHI and XbaI enzymes.
Transfection
Transient transfections were performed following a previously described protocol88: cells were resuspended at 2.5 × 107 cells/ml in 15 mM Hepes, pH 7.4, buffered medium. 200 μl of cell suspension was mixed with 50 μl of a solution containing 210 mM NaCl, 5 μg of plasmid DNA, and 30 μg of salmon sperm DNA solution (ThermoFisher #15632011). Cells were electroporated at 240 V and 950 μF in a 4 mm wide cuvette in a BioRad Gene Pulser. Cells were then quickly transferred in the same medium and incubated for 48 hours to allow for expression of the transfected plasmid before analysis.
Transduction
Lentiviral particles (LVs) for cell transduction were produced in HEK 293 T cells, maintained in DMEM (ThermoFisher) supplemented with 10% FBS (EuroBio), 1% penicillin/streptomycin, Sodium Pyruvate and Non-Essential Amino Acids solution (Gibco) at 37 °C in 5% CO2 humidified incubators. Plasmids for lentiviral components psPAX2 (Gag-Pol-Hiv1), pMD2-G (VSV-G) (addgene #12259), and the plasmid of interest were co-transfected using PEI MAX 40k transfection reagent (Tebu-Bio #24765-1) in a ratio of 4:1:4 (µg), following the manufacturer’s instructions. 48 hours post-transfection, LVs were collected, concentrated using a 100k Amicon column (Merck Millipore) and resuspended in up to 400 μL of PBS. Target cells were seeded at approximately 200,000 cells per well of a 6-well plate, transduced with 100 μL of the concentrated LVs and incubated for 72 hours. After transduction, cells expressing the target proteins were selected with 2 μg/mL puromycin and sorted using a SH800 FACS Cell Sorter (Sony).
Immunofluorescence
MEF cells were fixed in 100% ice-cold methanol at −20 °C for 5 minutes. Subsequently, cells were washed three times in PBS and thoroughly saturated in PBS containing 1 mg ml−1 BSA (blocking buffer (BB)). They were then incubated with primary antibodies: rabbit-anti-ELKS (1:200, from Stephanie Miserey-Lenkei, Institut Curie) or rabbit-anti-KANK1 (1:200, Atlas antibodies #HPA005539) for 45 minutes at room temperature. After three washing steps in BB, cells were incubated with the AlexaFluor555-conjugated goat-anti-rabbit secondary antibody (1:1000, Invitrogen #A21428) for 30 minutes at room temperature. After a series of washes: twice in BB, once in PBS, and once in deionized water, cells were mounted on coverslips with Abberior mounting medium and were examined under a fluorescence microscope to visualize the labeling of ELKS and KANK1.
siRNA silencing of Dynactin
p150Glued silencing in HeLa cells was achieved by the following siRNA sourced from Eurogentec: 5’-GGUAUCUGACACGCUCCU-3’ and 5’-UAGGAGCGUGUCAGAUAC-3’. A non-targeting siRNA ON-TARGETplus (Dharmacon, #D-001810-10-05), referred to as “siRNA scramble” was used as a control. siRNA transfections were performed using Lipofectamine RNAiMAX transfection reagent (ThermoFisher, #13778075) at a final concentration of 40 nM. The efficiency of the p150Glued silencing was verified by Western Blot 48 hours post-transfection.
Western Blot analysis of Dynactin silencing
Western blotting was conducted on protein extracts from siRNA-silenced HeLa cells to evaluate the knockdown efficiency of Dynactin (p150glued), as shown in Supplementary Fig. S11D. Cells silenced for Dynactin (p150glued) were lysed using RIPA buffer (ThermoFisher #8990) enriched with cOmplete protease inhibitor cocktail (Roche #11836170001) on ice. Western blot analysis was then conducted with mouse antibodies against p150glued (1:1000, BD Biosciences #610474) and β-actin (1:1000, GeneTex #GTX629630) as a loading control. Secondary antibodies used were goat-anti-mouse HRP (1:10,000, Sigma Aldrich #AP308P). The relative changes in the detected p150glued expression were quantified using the Fiji “Measure” plugin, applying the same region of interest (ROI) across the bands in one blot, normalized by the intensity of the corresponding β-actin band to ensure accurate protein load assessment. The raw images of the blots are provided within the Source Data.
Invasin preparation
The plasmid for Invasin expression, named pOM3474, was created by synthesizing and subcloning a DNA sequence coding for the terminal 474 amino acids of Invasin (inv474) with a 6x-Histidine tag and TEV cleavage site into the pMal p5x expression vector (addgene #150814). This construct enabled Invasin to be expressed as a fusion with periplasmic maltose-binding protein (MBP) in E. coli under an IPTG-inducible Ptac promoter.
For expression, E. coli lacking DegP protease was transformed with pOM3474 and cultured in 2YT medium with ampicillin, kanamycin, and 0.2% glucose. The culture was incubated at 30 °C until OD600 reached 0.6, and protein expression was induced with 0.5 mM IPTG for 4 hours at 30 °C.
For purification, amylose resin was used to affinity-purify MBP-Invasin, which was then cleaved from MBP using TEV protease. The purified Invasin could be optionally labeled with a reactive dye. Final purification was achieved by size exclusion chromatography using a Superdex 200 column, ensuring high purity of the Invasin protein. Additional details on Invasin preparation are available in Supplementary Methods.
Microscopy
A spinning disk confocal microscope was used to image fluorescent lipid bilayers and cell adhesions. The setup included a CSU-X1 Yokogawa spinning disk unit mounted on a Ti-E Nikon microscope with a motorized stage (MadCity Lab), a 100x NA1.45 objective, and a Photometrics 95B-sCMOS camera. Image acquisition was controlled by Metamorph software (Molecular Devices). The microscope was equipped with 4 Cobolt lasers from Hübner Photonics (405 nm at 100 mW, 488 nm–100 mW, 561 nm–50 mW and 633 nm–100 mW) for multi-channel fluorescence imaging (405, GFP, Cy3, and Cy5 channels). Additionally, the setup included wide-field imaging and a FRAP module. Consistent imaging conditions were maintained: 405 (15% power, 100 ms exposure), GFP (30%, 300 ms), Cy3 (30%, 300 ms), and Cy5 (30%, 300 ms). Z-stack images with a 0.3 µm step size centered on the SLB plane were captured for detailed analysis of cell-SLB interactions.
Supported Lipid Bilayers preparation
Supported Lipid Bilayers (SLBs) coated with Invasin were prepared in four steps following Galush et al.18: chamber preparation, Small Unilamellar Vesicles (SUVs) preparation, SLB formation, and SLB functionalization18. First, an imaging chamber was constructed using a cleaned and assembled 26 mm×76 mm coverglass and a #1.5 coverslip, joined by parafilm strips. The coverslip was thoroughly cleaned by sequential sonication in distilled water, 2% Hellmanex III, 1 M KOH, and distilled water, then dried under nitrogen. The coverglass was rinsed with ethanol and water and also dried with nitrogen. For the SUVs preparation, lipids were mixed in specific compositions (DOPC/DGS-NTA(Ni)/DHPE-MarinaBlue (94/2/4 mol/mol) and DOPC/DOPE-RGD/DHPE-MarinaBlue (94/2/4 or 95.9/0.1/4 mol/mol), dried, rehydrated, sonicated for 30 minutes, centrifuged at 20,000 RCF for 30 minutes, and filtered through a 0.22 μm filter (Millipore) to form SUVs at 0.5 mg/ml in the SUV Fusion Buffer (10 mM Tris pH 7.3, 120 mM NaCl). Then, the SUVs were incubated in the imaging chamber for 30 minutes to allow SUV fusion onto the coverslip. Unfused vesicles were washed away with SUV Fusion Buffer, and the SLB was further washed with Cell Buffer. The SLB was then passivated with a blocking solution (Cell Buffer with 0.1 mg/ml β-casein) for 15 minutes, followed by a final wash with Cell Buffer. For functionalization, the Ni-lipid-containing SLBs were incubated with 400 nM 6xHis-tagged Invasin in Cell Buffer for 1 hour at room temperature. Unbound Invasin was removed by two washes with Cell Buffer, with a 30-minute incubation between washes.
Fluidity control of SLBs by FRAP
The fluidity of SLBs was quantitatively assessed using Fluorescence Recovery After Photobleaching (FRAP). In this method, a defined region of the SLB was photobleached, and the recovery of fluorescence within that region was monitored over time. FRAP curves from multiple experiments were recorded and analyzed to determine the diffusion coefficients of the fluorescent lipids or proteins. Additional details on this procedure are available in Supplementary Methods.
Fluorescent calibration of SLBs
The procedure to accurately quantify the density of fluorescent species within the cell adhesion plane of SLBs was based on methods developed by Jay Groves’ laboratory18,26. Since the fluorescence signal measured by confocal microscopy is proportional to the number of fluorescent molecules in the confocal volume, the fluorescence intensity (measured in arbitrary units) can be converted into a density of bound molecules per unit area. This calibration was performed using SLBs with fluorescent lipids at known densities, which were compared with the fluorophore of interest, with both being imaged under identical conditions. Additional details on the fluorescence calibration are available in Supplementary Methods.
Cell adhesion experiments
Cells were serum-starved for 24 hours, detached with Versene solution (Sigma Aldrich) for 30 minutes at 37 °C, and labeled with 400 nM Alexa Fluor488 Halotag ligand (Promega) for 10–15 minutes at room temperature to target β1-Halotag. After labeling, cells were centrifuged at 1000 rpm, resuspended in Cell Buffer to remove Versene and excess dye, and filtered through a 40 µm Cell Strainer (Corning) to eliminate cell clumps. The cells were then introduced into the imaging chamber with SLB (time point zero) and sealed with mineral oil (Sigma Aldrich #M8410). Cells adhered to the SLB at 37 °C until the designated imaging time point (0, 15, 30 or 45 minutes). The chamber containing the cells was then mounted on a microscopy stage equipped with a Tokai Hit incubator set to 30 °C. The temperature was set to 30 °C due to technical limitations of the system, as maintaining a stable imaging environment at 37 °C led to thermal drifts that affected image quality. Cells were imaged for 15 minutes to observe their interactions with the bilayers. Fixed cell imaging was performed at room temperature using the same microscopy setup.
Image processing
Cells were classified as “trembling” if their edges moved within five seconds while their centers of mass remained static in bright field images. Cell contours were manually traced in bright filed images using ImageJ’s ‘Polygonal Selection’ tool to calculate the projected area (A) and circularity index (Circularity), with \({Circularity}=\frac{4\pi A}{{P}^{2}}\), where P being the perimeter.
Integrin clusters were detected by first correcting for uneven illumination. This was done by normalizing the images against an ‘illumination map,’ which was created by capturing a fluorescent image of the SLB and normalizing it by its maximum intensity. This correction was performed by dividing the pixel intensities of the original image by those of the corresponding pixels in the ‘illumination map’, where values ranged from (least illuminated) to 1 (most illuminated). The corrected images were then transformed into integrin concentration maps using fluorescence calibration data. These maps were segmented to distinguish clusters from the background by setting an integrin density threshold, either manually or automatically using ImageJ’s ‘Renyi’s Entropy thresholding’ method89. When the density threshold was set manually to a value \({\rho }_{{set}}\), the total area of detected integrin clusters in the cell was computed and referred to as \({\sigma }_{{\rho }_{{set}}}\). When the automatic thresholding was applied, the integrin density identified by the algorithm was defined and referred to as \(\rho\).
Membrane tubes associated with β1-integrin clusters were identified using ImageJ’s “Reslice” function on the SLB channel. This function created cross-sectional images perpendicular to the SLB at the locations of integrin clusters. These cross-sections were then segmented using Renyi’s entropy thresholding to identify membrane tethers, with a tube being defined as any structure that exceeded 1.5 µm in the z-dimension.
To localize integrin clusters and membrane tubes relative to the cell’s adhesion plane, the cell was divided into ‘center’ and ‘periphery’ zones based on a scaled transformation of the cell contour. The periphery zone was defined as a band parallel to the cell border, while the center zone encompassed the remaining internal area. These zones were designed to have approximately equal areas, depending on the cell’s circularity. For perfectly round cells (circularity > 0.8), the center and periphery zones had exactly equal areas. For cells with circularity <0.8, the width of the periphery zone was set to 20% of the square root of the projected cell area.
The distances from integrin clusters to the cell border were calculated by measuring the shortest path from the edge of each cluster to the cell border. For each cell, these distances were then averaged using a weighted mean, where the weight for each cluster was based on the number of integrins it contained. This weighted mean distance was normalized against a reference value, which represents the average distance expected if clusters were uniformly distributed throughout the cell. To calculate this reference distance, the cell area was divided into a fine rectangular mesh. This mesh was used to simulate a uniform distribution of integrin clusters, and the average distance from these simulated clusters to the cell border was computed. The mesh was iteratively refined until the error in the calculated distances was reduced to below 1% in a self-convergence test.
The relative enrichment of focal adhesion (FA) proteins, actin, and microtubules in β1-integrin clusters was assessed by measuring the fluorescence signal intensity within the area of integrin clusters and comparing it to the average fluorescence signal across the entire cell. Relative enrichment was calculated as \(\frac{{I}_{{clusters}}-{I}_{{cell}}}{{I}_{{cell}}}\), where \({I}_{{clusters}}\) and \({I}_{{cell}}\) represent the mean fluorescence signals at β1-integrin clusters and across the entire cell, respectively. These measurements were conducted on fixed cells to examine FA proteins, with β1-integrin clusters identified using a threshold of 100 arbitrary units (‘arb. units’). For experiments focused on F-actin and microtubule recruitment to β1-integrin clusters, which were performed on live cells the clusters were quantitatively defined using a threshold of 100 integrins/μm2.
The programs used for image analysis described above are available through a public online repository90.
Statistics and reproducibility
Statistical analysis was performed using GraphPad Prism software, with a p-value of less than 0.05 considered statistically significant. Each statistical test was based on \({N}_{\exp }\) independent experiments, with \({N}_{\exp }\ge 3\). Each measurement was taken from distinct samples. Before hypothesis testing, data sets were assessed for normality using the D’Agostino and Pearson tests. For data sets that followed a normal distribution, two-tailed Student’s t-tests were used for comparisons between two conditions, while one-way ANOVA followed by Tukey’s tests was applied for comparisons involving more than two conditions. For non-normally distributed data, the Kruskal–Wallis test followed by Dunn’s multiple comparisons tests was used for more than two conditions, and the two-tailed Mann-Whitney test was applied for two-condition comparisons.
In this study, no statistical method was used to predetermine sample size. No data were excluded, except in cases where the SLB was perturbed during live-cell adhesion experiments. All presented micrographs (Figs. 1A, C, 2A, 3A, E, 4A, 5A, B, G, and Supplementary Figs. S1A, S8A-E, S9A-E, S11A) are representative of findings from at least three independent experiments and were consistently reproducible. This standard of reproducibility applies to all other data included in this publication.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw microscopy images supporting the findings of this manuscript are available on the Biostudies repository with accession S-BIAD1420. Source data are provided with this paper.
Code availability
All code for conducting image analysis and simulations related to the theoretical model in this submission are openly available on GitHub: https://github.com/mikhajlov-oleg/cell_adhesion-image-analysis (https://doi.org/10.5281/zenodo.14567205) and https://github.com/MariaTatuleaCodrean/Cell-adhesion-theory (https://doi.org/10.5281/zenodo.14591503).
References
Huttenlocher, A. & Horwitz, A. R. Integrins in Cell Migration. Cold Spring Harb. Perspect. Biol. 3, a005074 (2011).
Vogel, V. & Sheetz, M. P. Cell fate regulation by coupling mechanical cycles to biochemical signaling pathways. Curr. Opin. Cell Biol. 21, 38–46 (2009).
Bökel, C. & Brown, N. H. Integrins in Development: Moving on, Responding to, and Sticking to the Extracellular Matrix. Developmental Cell 3, 311–321 (2002).
Discher, D. E., Janmey, P. & Wang, Y.-L. Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 (2005).
Iskratsch, T., Wolfenson, H. & Sheetz, M. P. Appreciating force and shape — the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 15, 825–833 (2014).
Winograd-Katz, S. E., Fässler, R., Geiger, B. & Legate, K. R. The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 15, 273–288 (2014).
Sun, Q., Hou, Y., Chu, Z. & Wei, Q. Soft overcomes the hard: Flexible materials adapt to cell adhesion to promote cell mechanotransduction. Bioact. Mater. 10, 397–404 (2022).
Sun, Z., Guo, S. S. & Fässler, R. Integrin-mediated mechanotransduction. J. Cell Biol. 215, 445–456 (2016).
Choi, C. K. et al. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039–1050 (2008).
Bershadsky, A. D. et al. Assembly and mechanosensory function of focal adhesions: experiments and models. Eur. J. Cell Biol. 85, 165–173 (2006).
Yamada, K. M., Doyle, A. D. & Lu, J. Cell–3D matrix interactions: recent advances and opportunities. Trends Cell Biol. https://doi.org/10.1016/j.tcb.2022.03.002 (2022).
Huveneers, S. & de Rooij, J. Mechanosensitive systems at the cadherin-F-actin interface. J. Cell Sci. 126, 403–413 (2013).
Harjunpää, H., Llort Asens, M., Guenther, C. & Fagerholm, S. C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 10, (2019).
Rose, D. M., Alon, R. & Ginsberg, M. H. Integrin modulation and signaling in leukocyte adhesion and migration. Immunological Rev. 218, 126–134 (2007).
Grakoui, A. et al. The Immunological Synapse: A Molecular Machine Controlling T Cell Activation. Science 285, 221–227 (1999).
Dustin, M. L. et al. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell 94, 667–677 (1998).
Smith, A.-S., Sengupta, K., Goennenwein, S., Seifert, U. & Sackmann, E. Force-induced growth of adhesion domains is controlled by receptor mobility. Proc. Natl Acad. Sci. 105, 6906–6911 (2008).
Galush, W. J., Nye, J. A. & Groves, J. T. Quantitative fluorescence microscopy using supported lipid bilayer standards. Biophys. J. 95, 2512–2519 (2008).
Yu, C., Law, J. B. K., Suryana, M., Low, H. Y. & Sheetz, M. P. Early integrin binding to Arg-Gly-Asp peptide activates actin polymerization and contractile movement that stimulates outward translocation. Proc. Natl Acad. Sci. USA 108, 20585–20590 (2011).
Yu, C. et al. Integrin-matrix clusters form podosome-like adhesions in the absence of traction forces. Cell Rep. 5, 1456–1468 (2013).
Glazier, R. & Salaita, K. Supported lipid bilayer platforms to probe cell mechanobiology. Biochimica et. Biophysica Acta (BBA) - Biomembranes 1859, 1465–1482 (2017).
Springer, T. A. Adhesion receptors of the immune system. Nature 346, 425–434 (1990).
Langereis, J. D. Neutrophil integrin affinity regulation in adhesion, migration, and bacterial clearance. Cell Adh Migr. 7, 476–481 (2013).
Isberg, R. R. & Leong, J. M. Multiple β1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 60, 861–871 (1990).
Huet-Calderwood, C. et al. Novel ecto-tagged integrins reveal their trafficking in live cells. Nat. Commun. 8, 570 (2017).
Nye, J. A. & Groves, J. T. Kinetic Control of Histidine-Tagged Protein Surface Density on Supported Lipid Bilayers. Langmuir 24, 4145–4149 (2008).
Shattil, S. J., Kim, C. & Ginsberg, M. H. The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 11, 288–300 (2010).
Ni, H., Li, A., Simonsen, N. & Wilkins, J. A. Integrin activation by dithiothreitol or Mn2+ induces a ligand-occupied conformation and exposure of a novel NH2-terminal regulatory site on the beta1 integrin chain. J. Biol. Chem. 273, 7981–7987 (1998).
Galbraith, C. G., Yamada, K. M. & Galbraith, J. A. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science 315, 992–995 (2007).
Price, L. S., Leng, J., Schwartz, M. A. & Bokoch, G. M. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol. Biol. Cell 9, 1863–1871 (1998).
Cavalcanti-Adam, E. A. et al. Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly. Eur. J. Cell Biol. 85, 219–224 (2006).
Wiseman, P. W. et al. Spatial mapping of integrin interactions and dynamics during cell migration by image correlation microscopy. J. Cell Sci. 117, 5521–5534 (2004).
Van Nhieu, G. T. & Isberg, R. R. The Yersinia pseudotuberculosis invasin protein and human fibronectin bind to mutually exclusive sites on the alpha 5 beta 1 integrin receptor. J. Biol. Chem. 266, 24367–24375 (1991).
Akiyama, S. K. & Yamada, K. M. The interaction of plasma fibronectin with fibroblastic cells in suspension. J. Biol. Chem. 260, 4492–4500 (1985).
Kanchanawong, P. et al. Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 (2010).
Bodescu, M. A., Aretz, J., Grison, M., Rief, M. & Fässler, R. Kindlin stabilizes the talin·integrin bond under mechanical load by generating an ideal bond. Proc. Natl Acad. Sci. 120, e2218116120 (2023).
Stehbens, S. & Wittmann, T. Targeting and transport: How microtubules control focal adhesion dynamics. J. Cell Biol. 198, 481–489 (2012).
LaFlamme, S. E., Mathew-Steiner, S., Singh, N., Colello-Borges, D. & Nieves, B. Integrin and microtubule crosstalk in the regulation of cellular processes. Cell. Mol. Life Sci. 75, 4177–4185 (2018).
Bouchet, B. P. et al. Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. eLife 5, e18124 (2016).
Lansbergen, G. et al. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev. Cell 11, 21–32 (2006).
Bennett, M. et al. Molecular clutch drives cell response to surface viscosity. Proc. Natl Acad. Sci. 115, 1192–1197 (2018).
Ghassemi, S. et al. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc. Natl Acad. Sci. 109, 5328–5333 (2012).
Rafiq, N. B. M. et al. A mechano-signalling network linking microtubules, myosin IIA filaments and integrin-based adhesions. Nat. Mater. 18, 638–649 (2019).
Seetharaman, S. et al. Microtubules tune mechanosensitive cell responses. Nat. Mater. 21, 366–377 (2022).
Bershadsky, A., Chausovsky, A., Becker, E., Lyubimova, A. & Geiger, B. Involvement of microtubules in the control of adhesion-dependent signal transduction. Curr. Biol. 6, 1279–1289 (1996).
Chang, Y.-C., Nalbant, P., Birkenfeld, J., Chang, Z.-F. & Bokoch, G. M. GEF-H1 Couples Nocodazole-induced Microtubule Disassembly to Cell Contractility via RhoA. MBoC 19, 2147–2153 (2008).
Geiger, B. & Bershadsky, A. Exploring the neighborhood: adhesion-coupled cell mechanosensors. Cell 110, 139–142 (2002).
Schwarz, U. S. & Safran, S. A. Physics of adherent cells. Rev. Mod. Phys. 85, 1327–1381 (2013).
Braeutigam, A., Simsek, A. N., Gompper, G. & Sabass, B. Generic self-stabilization mechanism for biomolecular adhesions under load. Nat. Commun. 13, 2197 (2022).
Komura, S. & Andelman, D. Adhesion-induced lateral phase separation in membranes. Eur. Phys. J. E 3, 259–271 (2000).
Bihr, T., Seifert, U. & Smith, A.-S. Nucleation of Ligand-Receptor Domains in Membrane Adhesion. Phys. Rev. Lett. 109, 258101 (2012).
Fenz, S. F. et al. Membrane fluctuations mediate lateral interaction between cadherin bonds. Nat. Phys. 13, 906–913 (2017).
del Rio, A. et al. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009).
Yao, M. et al. The mechanical response of talin. Nat. Commun. 7, 11966 (2016).
Brockman, J. M. et al. Mapping the 3D orientation of piconewton integrin traction forces. Nat. Methods 15, 115–118 (2018).
Shakiba, D. et al. The Balance between Actomyosin Contractility and Microtubule Polymerization Regulates Hierarchical Protrusions That Govern Efficient Fibroblast-Collagen Interactions. ACS Nano 14, 7868–7879 (2020).
Dehmelt, L., Nalbant, P., Steffen, W. & Halpain, S. A microtubule-based, dynein-dependent force induces local cell protrusions: Implications for neurite initiation. Brain Cell Bio 35, 39–56 (2006).
Chen, B.-H., Tzen, J. T. C., Bresnick, A. R. & Chen, H.-C. Roles of Rho-associated Kinase and Myosin Light Chain Kinase in Morphological and Migratory Defects of Focal Adhesion Kinase-null Cells. J. Biol. Chem. 277, 33857–33863 (2002).
Iskratsch, T. et al. FHOD1 is needed for directed Forces and Adhesion Maturation during Cell Spreading and Migration. Dev. Cell 27, 545–559 (2013).
Burridge, K. & Wittchen, E. S. The tension mounts: Stress fibers as force-generating mechanotransducers. J. Cell Biol. 200, 9–19 (2013).
Marlin, S. D. & Springer, T. A. Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1). Cell 51, 813–819 (1987).
Elices, M. J. et al. VCAM-1 on activated endothelium interacts with the leukocyte integrin VLA-4 at a site distinct from the VLA-4/fibronectin binding site. Cell 60, 577–584 (1990).
Livne, A. & Geiger, B. The inner workings of stress fibers − from contractile machinery to focal adhesions and back. J. Cell Sci. 129, 1293–1304 (2016).
Tojkander, S., Gateva, G. & Lappalainen, P. Actin stress fibers – assembly, dynamics and biological roles. J. Cell Sci. 125, 1855–1864 (2012).
Bouchet, B. P. & Akhmanova, A. Microtubules in 3D cell motility. J. Cell Sci. 130, 39–50 (2017).
Rhee, S., Jiang, H., Ho, C.-H. & Grinnell, F. Microtubule function in fibroblast spreading is modulated according to the tension state of cell–matrix interactions. Proc. Natl Acad. Sci. USA 104, 5425–5430 (2007).
Stehbens, S. J. et al. CLASPs link focal adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16, 561–573 (2014).
Hendricks, A. G. et al. Dynein Tethers and Stabilizes Dynamic Microtubule Plus-Ends. Curr. Biol. 22, 632–637 (2012).
Rosse, C. et al. Binding of dynein intermediate chain 2 to paxillin controls focal adhesion dynamics and migration. J. Cell Sci. 125, 3733–3738 (2012).
Redwine, W. B. et al. The human cytoplasmic dynein interactome reveals novel activators of motility. eLife 6, e28257 (2017).
Leduc, C., Campàs, O., Joanny, J.-F., Prost, J. & Bassereau, P. Mechanism of membrane nanotube formation by molecular motors. Biochimica et. Biophysica Acta (BBA) - Biomembranes 1798, 1418–1426 (2010).
Gumí-Audenis, B. et al. Pulling lipid tubes from supported bilayers unveils the underlying substrate contribution to the membrane mechanics. Nanoscale 10, 14763–14770 (2018).
Gennerich, A., Carter, A. P., Reck-Peterson, S. L. & Vale, R. D. Force-induced bidirectional stepping of cytoplasmic dynein. Cell 131, 952–965 (2007).
Moreno-Layseca, P. et al. Cargo-specific recruitment in clathrin- and dynamin-independent endocytosis. Nat. Cell Biol. 23, 1073–1084 (2021).
Kalappurakkal, J. M. et al. Integrin Mechano-chemical Signaling Generates Plasma Membrane Nanodomains that Promote Cell Spreading. Cell 177, 1738–1756.e23 (2019).
Mayor, S., Parton, R. G. & Donaldson, J. G. Clathrin-Independent Pathways of Endocytosis. Cold Spring Harb. Perspect. Biol. 6, a016758 (2014).
Cavalcanti-Adam, E. A. et al. Cell Spreading and Focal Adhesion Dynamics Are Regulated by Spacing of Integrin Ligands. Biophysical J. 92, 2964–2974 (2007).
Mazel, T. et al. Direct observation of microtubule pushing by cortical dynein in living cells. Mol. Biol. Cell 25, 95–106 (2014).
Murrell, M., Oakes, P. W., Lenz, M. & Gardel, M. L. Forcing cells into shape: the mechanics of actomyosin contractility. Nat. Rev. Mol. Cell Biol. 16, 486–498 (2015).
Rossier, O. M. et al. Force generated by actomyosin contraction builds bridges between adhesive contacts. EMBO J. 29, (2010).
Manneville, J.-B., Jehanno, M. & Etienne-Manneville, S. Dlg1 binds GKAP to control dynein association with microtubules, centrosome positioning, and cell polarity. J. Cell Biol. 191, 585–598 (2010).
Etienne-Manneville, S. Microtubules in cell migration. Annu Rev. Cell Dev. Biol. 29, 471–499 (2013).
Etienne-Manneville, S. & Hall, A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell 106, 489–498 (2001).
Hashimoto-Tane, A. et al. Dynein-driven transport of T cell receptor microclusters regulates immune synapse formation and T cell activation. Immunity 34, 919–931 (2011).
Schnyder, T. et al. B Cell Receptor-Mediated Antigen Gathering Requires Ubiquitin Ligase Cbl and Adaptors Grb2 and Dok-3 to Recruit Dynein to the Signaling Microcluster. Immunity 34, 905–918 (2011).
Young, L., Sung, J., Stacey, G. & Masters, J. R. Detection of Mycoplasma in cell cultures. Nat. Protoc. 5, 929–934 (2010).
Valencia-Gallardo, C. et al. Shigella IpaA Binding to Talin Stimulates Filopodial Capture and Cell Adhesion. Cell Rep. 26, 921–932.e6 (2019).
Gautreau, A., Louvard, D. & Arpin, M. Morphogenic Effects of Ezrin Require a Phosphorylation-Induced Transition from Oligomers to Monomers at the Plasma Membrane. J. Cell Biol. 150, 193–204 (2000).
Kapur, J. N., Sahoo, P. K. & Wong, A. K. C. A new method for gray-level picture thresholding using the entropy of the histogram. Computer Vis., Graph., Image Process. 29, 273–285 (1985).
Mikhajlov, O. et al. Cell adhesion and spreading on fluid membranes through microtubules-dependent mechanotransduction. GitHub v1.0, (2025).
Acknowledgements
We thank all laboratory members and collaborators, particularly Jay Groves (UC Berkeley) and Julien Pernier (Institute for Integrative Biology of the Cell) for their help in production of bilayers, Luke Lavis (Janelia Farm) for providing fluorophores, David Calderwood (Yale University) for providing MEF cell lines, Christof Hauck (University of Konstanz), Simon De Beco (Paris Diderot University) and Danijela Vignjevic (Institut Curie) for providing genetic constructs used in the study, Stephanie Miserey-Lenkei (Institut Curie) for the help in immunofluorescence experiments, Rémi Fert and Eric Nicolau for the help at the mechanical workshop, Jost Enninga (Institut Pasteur), Anna Akhmanova, Ana-Suncana Smith (Friedrich-Alexander-Universität), Jean-Baptiste Manneville (Laboratoire Matière et Systèmes Complexes, Paris-Cité University), Kristine Schauer (Institut Gustave Roussy), Pierre Sens, Gaelle Boncompain, Alexandre Baffet, Ryszard Wimmer (all Institut Curie) for insightful discussions. We also acknowledge the experimental support of the Molecular Biology and Cells platform at Institut Curie. This work was supported by the Institut Curie, the Collège de France, the Institut National de la Santé et de la Recherche Médicale (Inserm) and the Centre National de la Recherche Scientifique (CNRS) and Grants ANR-20-CE15-0001-01 and ANR-21-CE35-0007-03. We further acknowledge the Nikon Imaging Centre at Institut Curie-CNRS, member of the French National Research Infrastructure France-BioImaging (ANR10-INSB-04). O.M. was funded by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 666003, the ARC foundation, the Labex CelTisPhyBio (Grant ANR-11-LABX-0038, ANR-10-IDEX-0001-02). R.M.A. acknowledges funding from Fondation pour la Recherche Médicale (FRM Postdoctoral Fellowship). P.B. and J.F.J. are members of the CNRS consortium Approches Quantitatives du Vivant, the Labex CelTisPhyBio (ANR-11-LABX0038) and Paris Sciences et Lettres (ANR-10-IDEX-0001-02).
Author information
Authors and Affiliations
Contributions
O.M., P.B. and G.T.V.N. conceived the study. P.B. and G.T.V.N. acquired funding and equally contributed to this work. O.M. designed and performed experiments, analyzed data, and wrote the original manuscript. A.S.M. and O.M. developed image analysis programs for integrin cluster. F.D.F, J.M. and O.M. performed cloning, expression, purification and labeling of Invasin. A.B. and F.T generated lentiviral constructs and cell lines used in the study. J.F.J. supervised the theoretical work performed by R.M.A. and M.T.C. who equally contributed to this work. O.M., G.T.V.N. and P.B. reviewed and edited the final manuscript. All authors approved the final manuscript prior to submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mikhajlov, O., Adar, R.M., Tătulea-Codrean, M. et al. Cell adhesion and spreading on fluid membranes through microtubules-dependent mechanotransduction. Nat Commun 16, 1201 (2025). https://doi.org/10.1038/s41467-025-56343-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56343-6