Abstract
The shrinkage of glaciers and the vanishing of glacier-fed streams (GFSs) are emblematic of climate change. However, forecasts of how GFS microbiome structure and function will change under projected climate change scenarios are lacking. Combining 2,333 prokaryotic metagenome-assembled genomes with climatic, glaciological, and environmental data collected by the Vanishing Glaciers project from 164 GFSs draining Earth’s major mountain ranges, we here predict the future of the GFS microbiome until the end of the century under various climate change scenarios. Our model framework is rooted in a space-for-time substitution design and leverages statistical learning approaches. We predict that declining environmental selection promotes primary production in GFSs, stimulating both bacterial biomass and biodiversity. Concomitantly, predictions suggest that the phylogenetic structure of the GFS microbiome will change and entire bacterial clades are at risk. Furthermore, genomic projections reveal that microbiome functions will shift, with intensified solar energy acquisition pathways, heterotrophy and algal-bacterial interactions. Altogether, we project a ‘greener’ future of the world’s GFSs accompanied by a loss of clades that have adapted to environmental harshness, with consequences for ecosystem functioning.
Similar content being viewed by others
Introduction
Predicting the impacts of climate change on biodiversity has become a mainstay in ecological research1,2. Despite the intricate relationships between microorganisms and climate, climate change microbiology is still in its infancy, which is particularly true for forecasting responses of entire microbiomes to climate change3. This is surprising, given the predictive power encoded in microbiomes, which integrate past and current environmental conditions and drive key ecosystem functions4. Despite this, relatively few studies have predicted climate impacts on microbiome structure and function—mostly in the ocean5,6 and soils7,8. Today, no such study exists for stream and river microbiomes, which contrasts their relevance for global biogeochemical cycling9 and ecosystem services10.
Glacier-fed streams (GFSs) initiate the flow of water for some of the world’s largest river systems and provide water resources to large human populations11, but are also most vulnerable to climate change. The shrinking of glaciers not only threatens water availability but also fundamentally alters the GFS environment, putting their ecological communities at risk. Recent studies on invertebrates suggest that species adapted to the harsh environmental conditions of GFSs become increasingly imperilled as glaciers disappear, and their distributions are reduced to cold-water refugia12,13. This biodiversity at higher trophic levels is to a large extent sustained by microbial biofilms that coat the GFS streambeds and regulate key ecosystem processes, including metabolism and nutrient cycling14. The genomic repertoire of biofilm-dwelling microorganisms allows them to cope with the highly selective GFS environment (e.g. near-freezing water temperatures, high UV radiation, and ultra-oligotrophy), and to seize opportunities when they become available at punctuated periods of the year15,16. Recent studies based on space-for-time substitution approaches suggest that climate change-induced glacier shrinkage may stimulate organic matter decomposition by microorganisms in GFSs17,18 and shift microbial energetics from chemolithoautotrophy to heterotrophy15,17. However, predictions of how the GFS microbiome structure and function will change under projected climate change scenarios are still lacking.
Here, we develop a hierarchical, machine-learning-based modelling framework to forecast how the structure and function of the global GFS microbiome will change in response to climate change under different greenhouse gas emissions scenarios (Shared Socioeconomic Pathways; SSP19) for the years 2070–2100. We leverage 2333 bacterial metagenome-assembled genomes (MAGs, strain-level resolution, 99% average nucleotide identity) and 6226 KEGG orthologs, alongside glaciological and environmental data, from GFSs sampled by the Vanishing Glaciers project across the Caucasus Mountains, the European Alps, the Andes, Himalayas, Pamir and Tien Shan, Rwenzori Mountains, Scandinavian Mountains, New Zealand Southern Alps, and Southwest Greenland (Fig. 1A). In each of the 164 GFSs, we captured the deglaciation history since the end of the Little Ice Age period, mirroring our forecasting horizon (Supplementary Fig. 1). Our forecasts show how the global GFS microbiome, including its biomass and biodiversity, as well as structure and function will shift under future climate scenarios.
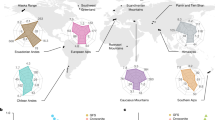
A Shown is a world map showing the mountain ranges and the number (in circles) of glacier-fed streams sampled per mountain range. Made with Natural Earth. Free vector and raster map data @ naturalearthdata.com. B Scatter plots of present-day (red) and future projections (blue) of algal biomass (chlorophyll a) and streamwater turbidity, C algal biomass and bacterial cell abundance, D algal biomass and the Shannon index of bacterial communities, and E algal biomass and the within community mean nearest taxon distance (α-MNTD) of the bacterial communities. The current and future states of each GFS are linked with grey lines. Two-sided tests for correlations between predicted changes were all significant (p < 0.001, Spearman rho = −0.96, 0.98, 0.69, and 0.97 for panels B–E, respectively). Source data are provided as a Source Data file.
Results
Climate-induced changes in the glacier-fed stream environment
The structure and function of stream biofilms are shaped by a suite of environmental factors (e.g. flow-induced hydraulics, temperature, resources), which are changing predictably as glaciers shrink14. In fact, glacier shrinkage alters flow and temperature regimes in GFSs, as well as sediment loads, with implications for turbidity and resources (e.g. phosphorus, nitrogen, organic carbon)17,20,21,22. Given the influence the environmental template has over the GFS microbiome23, we first modelled the future GFS environment (2070–2100), considering SSP climate change scenarios, of which we here report the results for SSP3 (results for SSP1 and SSP5 are provided in Supplementary Information). Our modelling framework is rooted in glaciological and climatological projections based on the Global Glacier Evolution Model (GloGEM)24 and Climatologies for Earth’s Land Surface Areas (CHELSA)25 at high spatial resolution, respectively. Combining these projections with measured data of the GFS environment along the chronosequence (i.e. leveraging differences between up- and downstream reaches for each GFS) (“Methods”, Supplementary Fig. 1), we first predict how the GFS environment will change in response to glacier retreat by the end of the century.
Our predictions suggest that median streamwater temperature and electrical conductivity (a proxy for ion concentration) will increase by 306.7% (IQR: 87.9–633.1%) and 88.2% (IQR: 51.6–130.7%), respectively, while median streamwater turbidity will decrease by 44.4% (IQR: 31.6–71.7%) across all study GFSs (Supplementary Fig. 2). Median concentrations of soluble reactive phosphorus and dissolved inorganic nitrogen will decrease by 14.1% (IQR: 9.6–27.3%) and 11.5% (IQR: 5.2–16.3%), respectively, and pH will be lowered by 2.8% (IQR: 1.6–4.2%) (Supplementary Fig. 2). Response curve analyses suggest that the magnitude of changes depend on glacier area (Supplementary Fig. 3). Consistent with previous studies and conceptual models20,26, these findings suggest reduced erosion and weathering capacity of shrinking glaciers, leading to decreases in streamwater fine suspended sediments and soluble reactive phosphorus concentrations. Changes in these selective constraints are likely to have significant impacts on the microbiome.
Biodiversity shifts with the greening of glacier-fed streams
Benthic microbial biomass is key to stream ecosystem functioning, as it fuels the food web and regulates ecosystem energetics and nutrient cycling14. Our projections suggest significant increases in benthic chlorophyll a (339.7%; IQR: 183–852.2%), a proxy for algal biomass, and bacterial abundance (88.5%; IQR: 60.4–150.2%) until the end of the century (Fig. 1B, C). This projection corroborates recent evidence suggesting that GFSs become ‘greener’ because of the fading capacity of glaciers to generate fine sediments, thereby reducing light limitation in GFSs17. In fact, fine suspended sediments render GFSs turbid, which reduces light availability for photosynthesis and increases physical abrasion, a notion that is supported by a negative correlation between projected values of streamwater turbidity and benthic chlorophyll a (rho = −0.96, p < 0.001). Yet, the projected benthic biomass of GFSs is still low compared to high-alpine streams without glacial influence27, which points to the persistence of constraints in GFSs other than those related to flow and turbidity.
Despite the harsh GFS environment, resident biofilms host a distinct and diverse microbiome15,23,28. Our projections show that this diversity (expressed as Shannon H’) will increase by 6.2% (IQR: 4.7–8.9%) under SSP3 (Fig. 1D). Positive relationships of both, bacterial Shannon diversity (rho = 0.69, p < 0.01) and bacterial abundance (rho = 0.98, p < 0.001), with chlorophyll a supports the notion that increased primary production sustains higher microbial biodiversity. Underpinning mechanisms may include elevated environmental stability and energy availability, the latter being a well-known driver of biodiversity29. Hence, we predict that future increases in algal biomass, and thus resource availability, will impact GFS microbiome diversity.
Both deterministic and stochastic assembly processes imprint upon the phylogenetic structure of microbial communities30. Environmental filtering, for instance, can lead to phylogenetic clustering. In GFSs, deterministic environmental selection favours microdiverse clades, which greatly contribute to microbiome diversity31. We posit that weaker selective constraints under future climates will alter microbial community assembly processes. To explore this emerging property, we modelled the mean nearest taxon distance and mean phylogenetic distance, reflecting relatedness at shallow and deep phylogenetic branching, respectively. We find median values of mean nearest taxon distance to significantly (Generalized Additive Model, GAM p < 0.001) increase by 3.5% (IQR: 2.2–5.8%, Fig. 1E), suggesting terminal phylogenetic clustering will diminish in the future GFS microbiome. Correlation between predicted mean nearest taxon distance and chlorophyll a (rho = 0.97, p < 0.001) suggests increased resource availability may contribute to the reduction of selective constraints. While the ability to use different carbon substrates can promote divergence within microdiverse clades32, microdiversity in GFSs is concentrated in clades utilising chemolithotrophic energy pathways31. Reduced environmental selection may erode microdiversity in GFSs, with yet unknown consequences for the stability and resilience of the GFS microbiome33. Similarly, the mean phylogenetic distance is predicted to increase by 3.2% (IQR: 1.7–6.3%), pointing towards deeper-branching changes to the future GFS microbiome phylogeny. In line with the predicted increase in alpha diversity, we attribute such a deeper-branching expansion of the phylogeny to the establishment of novel taxa and lineages in future GFSs.
Climate change shifts strain distributions
Species distribution models are commonly used to forecast climate-change impacts on the structure and diversity of ecological communities34. To predict the impacts of climate change, and integrate glaciological and environmental controls on the GFS microbiome, we built individual models for each of the 2333 strain-level MAGs in a strain distribution model framework (“Methods”, Supplementary Fig. 1). For this, we used a combination of climatic, glaciological, and mineralogical data, as well as forecasts of streamwater physico-chemistry. Overall, these models predicted strain abundance with satisfactory accuracy (median cross-validation R2 = 0.25; IQR: 0.13–0.36; Supplementary Fig. 4).
We first assessed the importance of covariates selected as predictors to identify the main drivers of strain abundances (Supplementary Fig. 5). Across all strains, streamwater electrical conductivity, pH, and temperature were the most important predictors, along with latitude. Distance to the glacier, annual snow cover, and bioclimatic variables (reduced by Principal Component Analysis) were also identified as important predictors (Supplementary Fig. 5). Interestingly, chlorophyll a did not rank among the strongest predictors of strain abundances. Together, these predictors highlight the direct controls of glacial meltwaters and underlying bedrock geology on the abundance of microorganisms in GFSs. Moreover, phylogenetically closely related strains shared similar predictors compared to less related strains (Spearman correlation, rho = −0.15, p < 0.0001, Supplementary Fig. 6). This apparent niche conservatism supports our modelling approach.
Next, using forecasts of these predictors, we assessed future abundance distributions for each strain. The majority (i.e. 64.7%) of the 2333 strains are expected to increase in abundance, with only 5.3% remaining unchanged (Fig. 2A and Supplementary Table 3). This overall gain in abundance aligns with our independent forecast of increasing bacterial abundance. Overall, Gamma- and Alphaproteobacteria, which numerically dominate the present-day GFS microbiome15, are projected to experience the largest increase in abundance (Fig. 2C). This increase in abundance can, at least partially, be attributed to the large number of strains in these classes. However, strains that currently occur at low abundance (i.e. lower half of the abundance distribution) are projected to increase disproportionately more in abundance compared to strains that are currently abundant in GFS microbiomes (Fig. 2B). Among them, the class of Gemmatimonadetes, which is known to form close associations with freshwater algae35 and Paceibacteria, known for their parasitic or symbiotic lifestyles36, revealed the largest relative increase in abundance. We hypothesise that Paceibacteria may be promoted by a larger size of the host pool, putatively related to elevated microbial biomass in future GFSs.
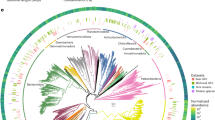
A Phylogenetic tree depicting the log2 fold-change between current and future projections of MAG (n = 2333) abundances. Symbol size represents current relative abundances. Taxonomic affiliation is provided as a coloured inner ring. Phylogenetic clades predicted to decrease in abundance are highlighted using red edges, the ones predicted to increase in abundance in blue (not significant in black). B Shown are the distributions of log2 fold-change and C total absolute change in abundance for the 11 most abundant classes (same as in A). Source data are provided as a Source Data file.
We also found that 30% of the strains will decrease in abundance (Fig. 2B), further pointing to the reorganisation of the GFS microbiome. Unlike covariates associated with strains predicted to increase in abundance, annual snow cover duration, distance to the glacier, bioclimatic variables, and streamwater temperature were the best predictors for strains that will decrease in abundance (Supplementary Fig. 5B). In line with the predicted pruning of phylogenetically clustered clades, this suggests that bacteria particularly well adapted to the cryospheric influence in GFS are facing a reduction in abundance in the future. This is underlined by the fact that changes in abundance were more similar for phylogenetically closely related strains than for distantly related strains (i.e. phylogenetic signal in log2-fold abundance change: lambda = 0.88, p < 0.001, Fig. 2A and Supplementary Table 4). The finding that phylogenetically related strains decrease in abundance reflects their shared evolutionary history and suggests that fundamental ecological niches in GFSs will undergo major transformations as glaciers shrink. This raises concerns, as it may mean that climate change imperils entire clades in GFSs rather than individual strains. This concern is substantiated by the fact that 26.6% of the strains projected to decrease in abundance were members of monophyletic clades, of which all members are projected to decrease in abundance (Supplementary Table 5). Notably, some of the largest clades (e.g. Ferruginibacter, Lacisediminihabitans, Acetobacteraceae) are hallmarks of cryospheric ecosystems37,38,39. Taken together, these findings suggest that the microbiome of the world’s GFSs will experience a profound compositional and phylogenetic restructuring with climate change. Although overall diversity of the GFS microbiome may expand due to the availability of novel niches associated with increased algal biomass the loss of microdiversity in putatively well-adapted clades likely has important ramifications for community stability and resilience.
Genomic traits associated with decreased abundance
To gain more mechanistic insights into the projected trajectories of strains, we next linked genomic traits to predicted changes in abundance. We found that strains predicted to decrease in abundance have generally smaller genomes (GAM, fixed effect estimate = −0.2 mbp, p < 0.01) but encode more KEGG (Kyoto Encyclopaedia of Genes and Genomes) ortholog groups (KOs; GAM, mean difference = 4.02, p < 0.001), compared to strains that will increase or stay invariant in abundance (Fig. 3A). Elevated metabolic diversity (i.e. higher KO numbers), despite reduced genome size, points towards genome optimisation, which can be explained by reduced KO redundancy. Indeed, estimating KO redundancy as the ratio of unique KOs to all KOs on genomes, we found that strains predicted to decrease in abundance have reduced KO redundancy (GAM, mean difference = −0.0136, p < 0.001).
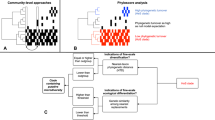
A Shown is the relationship between genome size and the total number of KOs for MAGs predicted to decrease in abundance (blue, n = 700) and for MAGs predicted to increase or remain unchanged in abundance (red, n = 1633). The lines reflect GAM fits with a linear effect of ln(genome size) on the total number of KOs for both groups of MAGs, accounting for completeness, contamination and N50 of the genomes. The slopes of these relationships differ significantly (two-way ANOVA, p < 0.001, slopeDecrease = 127.3 ± 1.1, slopeOthers = 118.6 ± 0.7) suggesting that MAGs predicted to decrease in abundance have optimized genomes (i.e. more KOs despite smaller genomes). B Displayed are the proportion of KEGG orthologous groups (KOs) associated with carbohydrate metabolism and energy metabolism in the genome of the MAGs that are predicted to decrease in abundance (blue, n = 700) and all other MAGs (red, n = 1633). The statistically significant difference was assessed in a generalised additive model, taking the completeness, N50 and contamination of the MAGs into account as tensor products smoothed cubic splines with interactions (GAM, for both p < 0.001). On average, MAGs projected to decrease in abundance had an additional 4.02 KOs categorized as “Carbohydrate metabolism” (deviance explained = 10.1%, F = 5755, meanothers = 43.5 ± 0.5) and an additional 3.44 KOs categorized as “Energy metabolism” (deviance explained = 13.9%, F = 3857, meanothers = 31.2 ± 0.4). The boxplots display the interquartile range as a box, and the median as a horizontal line. The whiskers extend to 1.5x the interquartile range after the box, and outliers are represented as dots and defined as data points above and below this threshold. Source data are provided as a Source Data file.
Together, this suggests that despite a broad metabolic repertoire, smaller genomes are favoured under current climatic and environmental conditions in GFSs, putatively reflecting the adaptation to ultra-oligotrophy and the unstable GFS environment. This notion is supported by the observation of a large degree of mixotrophy among GFS bacteria, supposedly enabling them to exploit varying energy sources (e.g. solar radiation and organic carbon15). However, these traits may no longer be favoured in GFSs under a future climate.
Given the predicted decline in abundance of entire clades, we next focused on KOs associated with strains that are projected to decrease in abundance. To this end, we constructed random forest classifiers and used a leave-one-out approach accounting for phylogenetic structure (“Methods”), identifying 408 KOs that characterise these strains (p < 0.05, feature importance quantile > 0.95). KOs associated with biofilm formation and cold adaptation were particularly prevalent (Supplementary Table 6). These include, for instance, the sec-independent protein translocase (TatB) which has previously been linked to cold-shock in Shewanella oneidensis40, and DNA topoisomerase I (topA), whose mutants become cold sensitive in Escherichia coli 41.
Using enrichment analysis, we found KEGG categories related to carbohydrate metabolism (Fisher test, OR = 2.20, p < 0.001) and energy metabolism (Fisher test, OR = 1.80, p < 0.001) to be significantly enriched among strains predicted to decrease in abundance (Fig. 3B and Supplementary Table 7). The prevalence of these KEGG categories may be linked to changing glacial and terrestrial carbon sources within catchments42,43. In fact, as GFSs become ‘greener’, carbon sources may shift towards more predictable algal-derived organic carbon17. Consequently, MAGs encoding chemolithoautotrophic metabolisms may become less prevalent whereas metabolisms such as aerobic chemoorganoheterotrophy may become more prevalent15. These results thus provide evidence for an alleviation of the multi-faceted selective constraints on future GFS microbiomes, ultimately altering their function.
Alteration of the functional potential
The predicted compositional reorganisation of the GFS microbiome is expected to affect its metabolic repertoire. Besides resolving genomic traits and functions associated with strains that will decrease or increase in abundance in the future, we modelled shifts in microbiome functional potential by building 6226 individual models of KO abundances. Compared to the strain-resolved models, these models yielded generally lower predictability (median cross validation R2 = 0.01, IQR: 0.0009–0.053), which is intuitive because of the invariance of core functions. However, 45.7% of KOs are predicted to increase, whereas 38.4% were predicted to decrease in abundance (Fig. 4). Pathways with the largest proportions of KOs forecasted to increase in abundance included photosynthesis (88% of KOs in this pathway are predicted to increase) and photosynthesis antennae (88%). We posit that this putatively reflects the predicted increase in light availability because of reduced turbidity in future GFSs. Several more specific metabolic pathways are also expected to increase in abundance, including selenocompounds metabolism (61.5%), alanine, aspartate and glutamate metabolism (62.7%), taurine and hypotaurine metabolism (60%), and nitrogen metabolism (58.7%) (Fig. 4). Pathways enriched in KOs predicted to decrease in abundance, and thus expected to play a reduced role in the future GFS microbiome, were related to protein processing (62.1%), sphingolipid metabolism (61.9%), replication and repair (60.4%), thiamine metabolism (59.1%), peptidoglycan biosynthesis (56.7%), fatty acid degradation (54.5%), and cell growth (51.7%) (Fig. 4). Taken together, the eminence of microbiome functional turnover suggests a substantial functional reorganisation of the future GFS microbiome with potential impacts on ecosystem functioning (e.g. decomposition and nutrient cycling).
In conclusion, we provide global-scale projections of the GFS microbiome for climate change scenarios. Environmental constraints that vary little across GFSs worldwide, but impose strong selective forces on the microbiome, allowed us to forecast changes in the environment and linked to this, of microbiome structure and function. Overall, we found that climate-induced glacier shrinkage relieves GFS microbiomes from environmental constraints, putting at risk entire clades of microorganisms that have adapted to the extreme environment. Our projections also reveal that microbiome functions will shift, particularly towards light acquisition pathways and translation, which is in line with the ‘greening’ of GFSs. We acknowledge potential caveats inherently related to species distribution models, including potentially missing variables and overfitting. We argue that a priori knowledge of the GFS environment and its microbiome, as well as cross-validation and ensemble modelling collectively make our projection architecture robust. Ultimately, future expeditions and long-term observations will allow a better understanding of climate change impacts on the microbiome of a rapidly vanishing ecosystem.
Methods
Study sites and sample collection
GFSs were sampled by the Vanishing Glaciers project between January 2019 and July 2022 (Supplementary Fig. 1 and Supplementary Table 1), and the resulting stream parameters’ dataset is also described in ref. 15 was used for this study (physicochemical parameters, stream water characteristics, benthic chlorophyll-a and bacterial abundance). Our global sampling included the European Alps, Scandinavian Mountains, Himalayas, Pamirs and Tian Shan, Ecuadorian and Chilean Andes, Southwest Greenland, Alaska Range, Caucasus, Rwenzori Mountains, and the New Zealand Southern Alps. For the sake of comparability, GFSs were predominantly sampled in spring or autumn during ‘windows of opportunities’16, which are characterised as periods either directly preceding or following peak glacier melt. For each GFS, two reaches were sampled: one as close as possible to the glacier terminus (median distance to glacier snout = 76 m, IQR = 29–301), and one located further downstream (median distance = 706 m, IQR = 336–1280). This approach allowed us to capture changes in glacier influence over the two stream reaches, creating a space-for-time substitution design. Space-for-time substitutions are well suited for communities shaped by environmental selection, but require communities to be part of the same ecological trajectory. This constrained the location of downstream reaches to be situated close to the terminal moraine, in many mountain regions hallmarks of the last glacial maximum (Little Ice Age). Because of the poorly developed soils and lack of vegetation in catchments above the terminal moraine, the influence of glacial meltwater (e.g. low temperature, turbidity, inorganic nutrient and organic carbon biogeochemistry) determines the environmental template in these reaches. At both, up- and downstream reach, we sampled three separate sediment patches (<10 m distance) with flame-sterilised devices. At each patch, we collected sandy (250 μm–3.15 mm; Retsch) sediments from the upper 5 cm of the benthic zone. Sediment samples were transferred to sterile cryovials, immediately flash-frozen in liquid nitrogen, and subsequently stored at −80 °C prior to and following shipping to Switzerland for DNA extraction and biomass analyses.
Streamwater and sediment physicochemical parameters
We measured streamwater temperature, pH, specific conductivity, and turbidity using a portable MultiLine Multi 3630 IDS (WTW) metre. Streamwater samples for nutrient analyses were filtered through pre-combusted GF/F filters (Whatman) into acid-washed Nalgene HDPE bottles and frozen within 48 h. Samples for the determination of DOC concentration were filtered identically, but into pre-combusted glass vials and kept at 4 °C. A LaChat QuikChem 8500 flow injection analyser was used to measure concentrations of ammonium (N-NH4+; QuikChem method 10-107-06-3-D), nitrate (N-NO3−, QuikChem method 10-107-05-1-C), and soluble reactive phosphorus (P-SRP; Method 10-115-01-1-M). Inorganic forms of nitrogen were combined to quantify dissolved inorganic nitrogen (DIN). A Sievers M9 TOC Analyser (GE) was used to measure DOC concentrations. Major cation and anion concentrations were measured from sterile-filtered (0.2 μm, Millipore) streamwater samples (stored in the dark at 4 °C) using a Metrohm 930 Compact IC flex system. Benthic sediment mineralogy was determined using an X-TRA ThermoARL Diffractometer (errors varied between 5% and 10% for the phyllosilicates and 5% for grain minerals). Relative abundances of the main mineral groups (clays, quartz, feldspar, and calcite) were computed from counts of mica, chlorite, amphibole, feldspars, calcite, and quartz.
Benthic chlorophyll a and bacterial abundance
We measured benthic chlorophyll a concentrations as a proxy for benthic algal biomass. For this, we added 90% ethanol to sediment samples, and incubated the samples in a hot water bath (78 °C) for 10 min, followed by incubation in the dark at 4 °C for 24 h. Then, samples were vortexed, and centrifuged and the supernatant was read in a high-sensitivity plate reader (BioTek Synergy H1) at 436/680 ex/em wavelengths. Chlorophyll a concentrations were calculated from standard curves obtained using a spinach standard (Sigma Aldrich) and normalised to a dry mass of sediment. Bacterial abundance was determined using flow cytometry as described previously31. Briefly, 2.5–3 g of sediment were fixed using a paraformaldehyde/glutaraldehyde mixture and kept frozen. In the laboratory, sodium pyrophosphate (final concentration of 0.025 mM) was amended and microbial cells were detached from sediments using vigorous shaking and sonication (Sonifier 450, Branson, 1 min, 60% duty cycle, output 5). Extracts were briefly spun, the supernatant 10-fold diluted and stained using SYBR Green and analysed on a NovoCyte flow cytometer (ACEA Biosciences) equipped with a 488 nm laser. Results were corrected for dilutions and normalised to sediment dry mass.
Climatic data
Climatology data at high spatial resolution was obtained from the CHELSA database (version 2.1)25. CHELSA provides both climatic and bioclimatic data typically used in species distribution modelling approaches at high spatial resolution (~1 km) based on a global downscaling approach. The database is based on an ERA-Interim climatic reanalysis and contains future projections of climatology for Shared Socioeconomic Pathways (SSP)19. The data was obtained using Python and the database’s API, and processed with the rasterio (v1.3.8) and gdal (v3.7.0) Python libraries44,45. GPS coordinates of all sampling locations were used to identify the corresponding grid cells of the (bio-)climatic dataset. The specific sampling months were used for monthly parameters. For future projections, five different institutional models were downloaded and combined by averaging (gfdl-esm4, ukesm1-0-ll, mpi-esm1-2-hr, ipsl-cm6a-lr, and mri-esm2-0). Because values in the database were only available up to 2010, linear extrapolation based on the 1981–2010 period (per month) was performed to obtain data matching the 2019–2022 sampling period (Supplementary Fig. 7). In addition, climatic data for the 2070–2100 time period for three SSPs scenarios corresponding to different greenhouse gas emission trajectories were obtained. These scenarios included SSP1, SSP3, and SSP5, encompassing a range of potential future climate outcomes. We report median change and interquartile ranges for the site-specific changes for all parameters. To test for significant shifts, we conducted Wilcoxon tests of the future projections minus the present conditions. All statistical analyses were performed using R (v4.3.1)46 and the tidyverse R package suite (v2.0.0)47. Figures were prepared using the ggplot2 (v3.4.2)48 and ggridges R packages (v0.5.4)49.
Glaciological data
The future evolution of all mountain glaciers is available through the Global Glacier Evolution Model (GloGEM)24. The model is initialised with present-day glacier extents and computes changes in snow accumulation and melt, as well as changes in glacier length based on an ensemble of Global Circulation Models using different greenhouse-gas emission scenarios. The model has been calibrated to match observed mass changes at the scale of every individual glacier globally50. For this study, we extracted the distance of sampling locations to the glacier terminus, as well as the area of the glacierized surface for the time period 2070–2100. We report median change and interquartile ranges for the site-specific changes for all parameters. To test for a significant shift, we conducted Wilcoxon tests of the future projections minus present-day observations.
Metagenomics
Metagenomes were sequenced for 155 sediment samples covering all visited mountain ranges except the Alaska Range (these samples were collected last by the Vanishing Glacier Project and were not included in metagenomic sequencing runs). DNA extraction, purification, library preparation, sequencing and metagenome assembly steps were performed as described in ref. 51. Briefly, 5 g of GFS sediments were treated using an optimised phenol:chloroform-based extraction method subsequently followed by an ethanol precipitation step. This protocol yielded on average 50 ng of DNA per sample which was used for library preparation using the NEBNext Ultra II FS library kit, which included 6 PCR cycles. Sequencing library quality was screened using Qubit (Invitrogen) and Bioanalyzer (Agilent). Subsequently, libraries of app. Sixty samples were pooled and sequenced at the Functional Genomics Centre Zurich on a NovaSeq (Illumina, 150 bp) using S4 flowcells.
The metagenomic sequence data generated for ref. 15. was used, and was processed using the Integrated Meta-omic Pipeline workflow (version 3.0; commit# 9672c874)52. Briefly, adaptor trimming from reads is followed by an iterative assembly using MEGAHIT 53 and metaFlye 54. Reads from each individual sample were assembled into contigs and subjected to multi-coverage binning to obtain MAGs. For each individual assembly, we mapped the reads of the 5 geographically closest samples (Euclidean distances of gps coordinates) using BWA-mem (v0.7.17)55. To reduce computation time, we removed sequences in the assembly of <1.5 kbp. Subsequently, 10% of the pre-processed reads were randomly selected before mapping with seqtk (v1.3)56. We then used MetaBAT2 (v2.15)57, CONCOCT (v1.1.0)58 and MetaBinner (v1.4.3)59 using default parameters to obtain bins. DAS Tool (v1.1.4)60 was then employed to generate a non-redundant set of bins using a score threshold of 0.3. The quality of bins was assessed using CheckM2 (v1.0.1)61. Bins from all samples (including the ones generated by IMP3) with completeness of more than 50% (n = 12,599 bins) were selected for further analyses. We used MDMCleaner (v0.8.3)62 to reduce contamination. Finally, after rerunning CheckM2 to get final estimates of completeness and contamination, we used dRep (v3.2.2)63 to dereplicate bins using minimum completeness of 70 % and maximum contamination of 10% and an ANI of 99%. This led to 2868 MAGs (strain-level dereplication). Functional annotation of the MAGs was performed with eggNOG-mapper (v2.1.9)64 against eggNOG v5.0 after obtaining coding regions with prodigal (v2.6.3)65. The coverage of the MAGs was estimated by mapping the reads of the samples to the genomic contigs using CoverM (v0.6.1, available at https://github.com/wwood/CoverM) using the trimmed_mean parameter. We normalized the coverage by similarly mapping the reads on the recA gene (K03553). After removing low-abundance strains (prevalence lower than 20% at a 10× recA coverage threshold), 2333 were selected for the strain distribution modelling. GTDB-Tk (v 2.1)66 was used to determine the taxonomy of the MAGs. We used the concatenated alignment of 120 ubiquitous single-copy proteins created by GTDB-Tk to de novo generate a phylogenetic tree using FastTree2 under the WAG model of protein evolution with gamma-distributed rate heterogeneity.
Modelling environmental parameters and microbial biomass
Models of streamwater temperature, turbidity, electrical conductivity, pH, soluble reactive phosphorus (SRP), and dissolved inorganic nitrogen (DIN) were built using climatic, bioclimatic, glaciological and geological parameters as covariates, chosen by automated feature selection (Supplementary Fig. 3). The same modelling approach was used to model biomass parameters (chlorophyll-α, bacterial abundance) and diversity metrics (Shannon H’, mean nearest taxon distance, mean phylogenetic distance). Response variables and covariates were log-transformed where necessary to improve the normality of residuals, adding a constant equal to half of the smallest non-zero value. Before model fitting, all variables were z-transformed (i.e. scaled to mean and units standard deviation). All generalised additive models (GAM) models were created using the Gaussian family function and the bam function of the mgcv package (v1.9_0)67. GAMs have been successfully used to model and predict future changes in environmental sciences and ecology, especially for spatially structured data7,68,69, and have also been applied to stream ecosystems70,71. Here, GAMs were used to model stream parameters and biomass in order to account for spatial autocorrelation at the regional scale using a spatial spline (formula: s (latitude, longitude, bs = ‘sos’, m = 1, k = −1)). Feature selection was performed by building individual GAM models with a spline for each covariate (k = 3, bs = ‘ts’) along with the spatial spline and were then ranked by −log(p-value) of the covariate spline. The top three variables were retained for a final model. This procedure was repeated on each of the nine models for each cross-validation fold. A final model was then built with the spatial spline and a spline for the three top variables with the following parameters: k = 3, bs = ‘ts’. These parameters allowed for non-linear relationships while the small number of knots and the penalisation on the spline were added to control for smoothness and to avoid overfitting.
The models were validated using 10-fold cross-validation, meaning that several times the models were trained on 90% of the glacier-fed streams, and tested on the remaining data. The performance of the models (measured as the cross-validation R2) was computed by pooling the results of all 10-folds of the cross-validation72. The choice of parameters and ensemble modelling were chosen to control for potential overfitting. Moreover, we selected only three covariates for each strain distribution model to further avoid overfitting. The shape of the smoothed splines was then inspected and all had reasonable smoothing (Supplementary Fig. 3). To further improve the robustness of the approach, we performed ensemble modelling by averaging the predictions of GAMs built on each cross-validation fold using an elastic net linear regressor. Different alpha values of 0, 0.5, and 1 were tested and the best one was retained based on the error reported by the cv.glmnet function of the glmnet R package (v4.1_7)73. Using 5-fold cross-validation, the function automatically computes the best lambda value. We avoided autocorrelation at the GFS level (as two reaches were sampled from each GFS) by randomly sampling one of the two reaches before creating each of the sub-models. By fitting nine separate models to predict each cross-validation fold, we allowed a fraction to represent local variations in the data, and stacking multiple models allowed for all samples to be included in the final model (since only one sample per GFS is included in each model). The final predictions were the mean of the predictions made by all ten ensemble models trained on 9-folds.
Response curves were obtained using predictions for all sampling points. The consistency of the stream parameter models created for each SSP scenario was assessed by comparing the selected features and their corresponding response curves (Supplementary Fig. 3). We measured model performance as cross-validation R2 comparing predicted and observed data for the held-out folds (Supplementary Table 2). Statistics comparing present and future stream parameters were obtained in the same way as statistics for the climatic dataset. Correlation between predicted stream parameters was assessed using Spearman correlations across all sites.
Models of strain and KO abundances
Models of strain abundance (normalised using the recA gene coverage) and KO abundance were built using climatic, glaciological, and mineralogical data, as well as stream parameter forecasts as covariates, chosen by feature selection (one set of features for each strain). To reduce overfitting and collinearity, we reduced the dimensionality of bioclimatic variables using principal component analysis (PCA) implemented in the prcomp R function, and retained the first six dimensions, as they represented more than 95% of the variation (Supplementary Fig. 8). Strain and KO abundances, as well as covariates were log-transformed and z-scaled as described above. Models were created with the same approach as for the stream parameters models, using GAMs stacked with an elastic net, the only difference being that spatial splines were not used; instead latitude, absolute latitude, and longitude, along with elevation and the slope of the stream, were added as potential covariates in the feature selection process. For each model, four variables were selected with the same feature selection procedure as for the stream parameters models. Model performance (predictive power) was assessed using the R2prediction pooled over all ten of the cross-validation folds34.
Statistical analyses
Phylogenetic signal in log2 fold-change in future abundance and in R2prediction was measured by using the “lambda” method of the phytools R package (v1.5_1)74. We report median change and interquartile ranges for the site-specific changes for all parameters. To test for a significant shift, we conducted Wilcoxon tests of the future projections minus the present observations. Evenness was assessed by comparing present and future median predictions of abundance for all strains, and by showing that the relationship had a slope less than one (Supplementary Fig. 9). The largest monophyletic sub-trees composed only of representatives predicted to decrease under the SSP3 scenario were identified using R packages phytools (v.1.5_1)74 and ape (v5.7_1)75. The phylogenetic tree was plotted using R packages ggtree (v3.8.0)76 and ggtreeExtra (v1.10.0)77.
We used Spearman correlations between the number of shared covariates and phylogenetic distance of pairs of MAGs to assess phylogenetic signal in covariates (i.e. whether phylogenetically similar MAGs share more covariates than phylogenetically distant MAGs). Similarly, to assess taxonomic similarities, we split MAGs based on taxonomy and used the Kruskal–Wallis test to evaluate whether covariates have similar importance in all classes. This was computed using the kruskal.test R function, and plots were created to show the distribution of median relative ranks across taxa with the ggridges R package (v0.5.4)49.
Random forest classifiers were used to identify KOs associated with strains that decrease in abundance. To this end, we accounted for phylogenetic structure by splitting the data into ten phylogenetic clusters, and training models on nine of them while assessing the importance on the one left out. Random forest classifiers were created using the ranger R package (v0.15.1)78, using random grid search (n = 50) to find hyperparameter settings (Supplementary Table 8). Feature importance (i.e. the importance of KOs) was estimated using the method developed by ref. 79. We considered KOs with a p-value < 0.05 and in the upper 95th percentile of the importance values in at least one of the phylogenetic clusters as significant. We considered “top” KOs that were significant in at least eight out of the ten phylogenetic clusters (n = 21), and for these, descriptions were gathered on the KEGG website (https://www.genome.jp/kegg/) (Supplementary Table 6).
Enrichment analysis was carried out at the level of KEGG categories to identify categories overrepresented in the set of significant KOs. This was done with the fisher.test in R and p-values were corrected using the Bonferroni method. We only considered positively significant categories with the thresholds: p-value < 0.05 and odds ratio (OR) > 1. To compare the number of KOs among genomes of strains predicted to decrease in abundance with “others”, we used GAMs considering the completeness, contamination, and N50 of these genomes and their interactions with a tensor (k = 3, bs = ’cs’). GAMs (gaussian family model) were created for the counts of KOs in the MAGs. Weights were added by multiplying the inverse of the number of MAGs for each phylogenetic cluster (such that all phylogenetic clusters obtained equal weights), the completeness of the genomes (to give less weight to KO absences owing to incomplete genomes), and the mean relative abundance. These models were fit using the bam function in the mgcv R package (v1.9_0)67. Using the same approach, we tested for differences between the two groups of bacterial genomes (i.e. “decreasing in abundance” vs “others”) in the total number of KOs, genome length and KO redundancy (unique KOs/total KOs). We further regressed the number of KOs on the genomes against genome length (accounting for completeness, contamination and N50 with a tensor; function ‘te’ with parameters k = 3 and bs = ’cs’). For these models, we tested the difference between the strains predicted to decrease in abundance and “others” by fitting fixed effects (and an interaction for the KO number ~ genome length model) with ANOVAs as implemented in the stats R package included in the base R package (v4.3.1)46 and reported the estimated means and standard errors.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The climatic and glaciological data generated in this study has been deposited on Zenodo alongside the metagenomic-derived and environmental datasets under the (https://doi.org/10.5281/zenodo.10409762). All sequencing raw data and MAGs are deposited in NCBI under bioproject PRJNA781406. Source data are provided with this paper.
Code availability
The code is available on this GitHub repository: https://github.com/Mass23/CrystalBall and was published on Zenodo under the (https://doi.org/10.5281/zenodo.14497117). To fully reproduce the study, the conda environment included in the envs/needs to be installed and the data copied from Zenodo in the directory. Then the scripts 1_Create_data.py and 2_Analyse.R need to be run sequentially. All data tables, stat tables and figures will be generated automatically.
References
Bellard, C., Bertelsmeier, C., Leadley, P., Thuiller, W. & Courchamp, F. Impacts of climate change on the future of biodiversity. Ecol. Lett. 15, 365 (2012).
Garcia, R. A., Cabeza, M., Rahbek, C. & Araújo, M. B. Multiple dimensions of climate change and their implications for biodiversity. Science 344, 1247579 (2014).
Cavicchioli, R. et al. Scientists’ warning to humanity: microorganisms and climate change. Nat. Rev. Microbiol. 17, 569–586 (2019).
Correa-Garcia, S., Constant, P. & Yergeau, E. The forecasting power of the microbiome. Trends Microbiol. 31, 444–452 (2023).
Frémont, P. et al. Restructuring of plankton genomic biogeography in the surface ocean under climate change. Nat. Clim. Chang. 12, 393–401 (2022).
Zhang, Z. et al. Global biogeography of microbes driving ocean ecological status under climate change. Nat. Commun. 15, 4657 (2024).
Mod, H. K. et al. Predicting spatial patterns of soil bacteria under current and future environmental conditions. ISME J. 15, 2547–2560 (2021).
Verdon, V. et al. Can we accurately predict the distribution of soil microorganism presence and relative abundance? Ecography n/a, e07086.
Battin, T. J. et al. River ecosystem metabolism and carbon biogeochemistry in a changing world. Nature 613, 449–459 (2023).
Albert, J. S. et al. Scientists’ warning to humanity on the freshwater biodiversity crisis. Ambio 50, 85–94 (2021).
Immerzeel, W. W. et al. Importance and vulnerability of the world’s water towers. Nature 577, 364–369 (2020).
Brown, L. E. et al. Functional diversity and community assembly of river invertebrates show globally consistent responses to decreasing glacier cover. Nat. Ecol. Evol. 2, 325–333 (2018).
Wilkes, M. A. et al. Glacier retreat reorganizes river habitats leaving refugia for Alpine invertebrate biodiversity poorly protected. Nat. Ecol. Evol. 7, 841–851 (2023).
Battin, T. J., Besemer, K., Bengtsson, M. M., Romani, A. M. & Packmann, A. I. The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol 14, 251–263 (2016).
Michoud, G. et al. Mapping the metagenomic diversity of the multi-kingdom glacier-fed stream microbiome. Nat. Microbiol. 10, 217–230 (2025).
Busi, S. B. et al. Genomic and metabolic adaptations of biofilms to ecological windows of opportunity in glacier-fed streams. Nat. Commun. 13, 2168 (2022).
Kohler, T. J. et al. Global emergent responses of stream microbial metabolism to glacier shrinkage. Nat. Geosci. 17, 309–315 (2024).
Fell, S. C. et al. Fungal decomposition of river organic matter accelerated by decreasing glacier cover. Nat. Clim. Chang. 11, 349–353 (2021).
O’Neill, B. C. et al. A new scenario framework for climate change research: the concept of shared socioeconomic pathways. Clim.Change 122, 387–400 (2014).
Milner, A. M. et al. Glacier shrinkage driving global changes in downstream systems. PNAS 114, 9770–9778 (2017).
Zhang, T. et al. Warming-driven erosion and sediment transport in cold regions. Nat. Rev. Earth Environ. 3, 832–851 (2022).
Slemmons, K. E. H., Saros, J. E. & Simon, K. The influence of glacial meltwater on alpine aquatic ecosystems: a review. Environ. Sci. Process. Impacts 15, 1794–1806 (2013).
Ezzat, L. et al. Benthic biofilms in glacier-fed streams from scandinavia to the himalayas host distinct bacterial communities compared with the streamwater. Appl. Environ. Microbiol. 88, e00421-22 (2022).
Huss, M. & Hock, R. A new model for global glacier change and sea-level rise. Front. Earth Sci. https://doi.org/10.3389/feart.2015.00054 (2015).
Karger, D. N. et al. Climatologies at high resolution for the earth’s land surface areas. Sci. Data 4, 170122 (2017).
Ren, Z., Martyniuk, N., Oleksy, I. A., Swain, A. & Hotaling, S. Ecological stoichiometry of the mountain cryosphere. Front. Ecol. Evol. https://doi.org/10.3389/fevo.2019.00360 (2019).
Brandani, J. et al. Spatial patterns of benthic biofilm diversity among streams draining proglacial floodplains. Front. Microbiol. https://doi.org/10.3389/fmicb.2022.948165 (2022).
Wilhelm, L., Singer, G. A., Fasching, C., Battin, T. J. & Besemer, K. Microbial biodiversity in glacier-fed streams. ISME J. 7, 1651–1660 (2013).
Gaston, K. J. Global patterns in biodiversity. Nature 405, 220–227 (2000).
Stegen, J. C. et al. Quantifying community assembly processes and identifying features that impose them. ISME J. 7, 2069–2079 (2013).
Fodelianakis, S. et al. Microdiversity characterizes prevalent phylogenetic clades in the glacier-fed stream microbiome. ISME J. 16, 666–675 (2022).
Larkin, A. A. & Martiny, A. C. Microdiversity shapes the traits, niche space, and biogeography of microbial taxa. Environ. Microbiol. Rep. 9, 55–70 (2017).
García-García, N., Tamames, J., Linz, A. M., Pedrós-Alió, C. & Puente-Sánchez, F. Microdiversity ensures the maintenance of functional microbial communities under changing environmental conditions. ISME J. 13, 2969–2983 (2019).
Araújo, M. B. et al. Standards for distribution models in biodiversity assessments. Sci. Adv. 5, eaat4858 (2019).
Mujakić, I. et al. Common presence of phototrophic gemmatimonadota in temperate freshwater lakes. mSystems. https://doi.org/10.1128/msystems.01241-20 (2021).
Nelson, W. & Stegen, J. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. https://doi.org/10.3389/fmicb.2015.00713 (2015).
Carey, C. J. et al. Microbial community structure of subalpine snow in the Sierra Nevada, California. Arct., Antarct. Alp. Res. 48, 685–701 (2016).
Bourquin, M. et al. The microbiome of cryospheric ecosystems. Nat. Commun. 13, 3087 (2022).
Jaarsma, A. H. et al. Exploring microbial diversity in Greenland Ice Sheet supraglacial habitats through culturing-dependent and -independent approaches. FEMS Microbiol. Ecol. 99, fiad119 (2023).
Gao, H., Yang, Z. K., Wu, L., Thompson, D. K. & Zhou, J. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 188, 4560–4569 (2006).
Stupina, V. A. & Wang, J. C. Viability of Escherichia coli topA mutants lacking DNA topoisomerase I. J. Biol. Chem. 280, 355–360 (2005).
Hood, E., Fellman, J. B. & Spencer, R. G. M. Glacier loss impacts riverine organic carbon transport to the ocean. Geophys. Res. Lett. 47, e2020GL089804 (2020).
Robison, A. L., Deluigi, N., Rolland, C., Manetti, N. & Battin, T. Glacier loss and vegetation expansion alter organic and inorganic carbon dynamics in high-mountain streams. Biogeosciences 20, 2301–2316 (2023).
Gillies, S. Rasterio: geospatial raster I/O for Python programmers (Rasterio, 2013).
GDAL Development Team. GDAL—Geospatial Data Abstraction Library, Version 3.7.0. (Open Source Geospatial Foundation).
R Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, 2023).
Wickham, H. et al. Welcome to the tidyverse. J. Open Source Softw. 4, 1686 (2019).
Wickham, H. Ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, New York, 2016).
Wilke, C. O. ggridges: Ridgeline Plots in ‘ggplot2’. 0.5.6 https://doi.org/10.32614/CRAN.package.ggridges (2017).
Hugonnet, R. et al. Accelerated global glacier mass loss in the early twenty-first century. Nature 592, 726–731 (2021).
Busi, S. B. et al. Optimised biomolecular extraction for metagenomic analysis of microbial biofilms from high-mountain streams. PeerJ 8, e9973 (2020).
Narayanasamy, S. et al. IMP: a pipeline for reproducible reference-independent integrated metagenomic and metatranscriptomic analyses. Genome Biol. 17, 260 (2016).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Kolmogorov, M. et al. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods 17, 1103–1110 (2020).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://doi.org/10.48550/arXiv.1303.3997 (2013).
Li, H. Toolkit for processing sequences in FASTA/Q formats (2024 r132). GitHub repository, https://github.com/lh3/seqtk.
Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019).
Alneberg, J. et al. Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146 (2014).
Wang, Z., Huang, P., You, R., Sun, F. & Zhu, S. MetaBinner: a high-performance and stand-alone ensemble binning method to recover individual genomes from complex microbial communities. Genome Biol. 24, 1 (2023).
Sieber, C. M. K. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018).
Chklovski, A., Parks, D. H., Woodcroft, B. J. & Tyson, G. W. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat. Methods 20, 1203–1212 (2023).
Vollmers, J., Wiegand, S., Lenk, F. & Kaster, A.-K. How clear is our current view on microbial dark matter? (Re-)assessing public MAG & SAG datasets with MDMcleaner. Nucleic Acids Res. 50, e76 (2022).
Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017).
Huerta-Cepas, J. et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol. Biol. Evol. 34, 2115–2122 (2017).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 119 (2010).
Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927 (2020).
Wood, S. mgcv: Mixed GAM Computation Vehicle with Automatic Smoothness Estimation. R Package Version 1.9-0. https://CRAN.R-project.org/package=mgcv (2023).
Colón-González, F. J., Fezzi, C., Lake, I. R. & Hunter, P. R. The effects of weather and climate change on dengue. PLOS Negl. Trop. Dis. 7, e2503 (2013).
Ravindra, K., Rattan, P., Mor, S. & Aggarwal, A. N. Generalized additive models: Building evidence of air pollution, climate change and human health. Environ. Int. 132, 104987 (2019).
Jowett, I. G., Parkyn, S. M. & Richardson, J. Habitat characteristics of crayfish (Paranephrops planifrons) in New Zealand streams using generalised additive models (GAMs). Hydrobiologia 596, 353–365 (2008).
Coleman, D., Bevitt, R. & Reinfelds, I. Predicting the thermal regime change of a regulated snowmelt river using a generalised additive model and analogue reference streams. Environ. Process. 8, 511–531 (2021).
Collart, F. & Guisan, A. Small to train, small to test: dealing with low sample size in model evaluation. Ecol. Inform. 75, 102106 (2023).
Tay, J. K., Narasimhan, B. & Hastie, T. Elastic net regularization paths for all generalized linear models. J. Stat. Softw. 106, 1–31 (2023).
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012).
Paradis, E. & Schliep, K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019).
Xu, S. et al. Ggtree: a serialized data object for visualization of a phylogenetic tree and annotation data. iMeta 1, e56 (2022).
Xu, S. et al. ggtreeExtra: compact visualization of richly annotated phylogenetic data. Mol. Biol. Evolution 38, 4039–4042 (2021).
Wright, M. N. & Ziegler, A. ranger: a fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 77, 1–17 (2017).
Altmann, A., Toloşi, L., Sander, O. & Lengauer, T. Permutation importance: a corrected feature importance measure. Bioinformatics 26, 1340–1347 (2010).
Acknowledgements
The Vanishing Glaciers project is supported by The NOMIS Foundation to T.J.B. We are most grateful to A. McIntosh and L. Morris in New Zealand, J. Abermann and T. Juul-Pedersen in Greenland, O. Solomina and T. Kuderina Maratovna in Russia, V. Crespo-Pérez and P. Andino Guarderas in Ecuador, J. Yde and S. Leth Jørgensen in Norway, S. Sharma and P. Joshi in Nepal, N. Shaidyldaeva- Myktybekovna and R. Kenzhebaev in Kyrgyzstan, J. Nattabi Kigongo, R. Nalwanga and C. Masembe in Uganda, M. Gonzlaléz and J. Luis Rodriguez in Chile, and C. Kuhle and P. Tomco in Alaska for logistical support. We particularly acknowledge the help from the porters and guides in Nepal, Uganda and Kyrgyzstan. T.J.K. was also supported by the Charles University project PRIMUS/22/SCI/001. S.B.B. was supported by the Swiss National Science Foundation grant CRSII5_180241 to T.J.B. The metagenomic preprocessing and assemblies used in this research were carried out at the HPC facilities of the University of Luxembourg (https://hpc.uni.lu). We acknowledge support from N. Deluigi, P. Pramateftaki and E. Oppliger for help in the laboratory, and A. Adde for providing advice on the modelling design.
Author information
Authors and Affiliations
Consortia
Contributions
M.B.: conceptualisation, methodology, data curation, formal analysis, visualisation, and writing—original draft preparation. H.P.: conceptualisation, investigation, visualisation, and writing—original draft preparation. S.B.B.: methodology, data curation, investigation, and formal analysis. G.M.: methodology, data curation, investigation, and formal analysis. T.J.K.: conceptualisation and writing—original draft preparation. A.L.R.: conceptualisation and writing—original draft preparation. M.S.: methodology. L.E.: conceptualisation and investigation. A.G.: conceptualisation and investigation. M.H.: methodology and formal analysis. S.F.: conceptualisation and investigation. T.J.B.: conceptualisation, methodology, investigation, and writing—original draft preparation, supervision, project administration, and funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Brian Lanoil, Birgit Sattler and Gilda Varliero for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bourquin, M., Peter, H., Michoud, G. et al. Predicting climate-change impacts on the global glacier-fed stream microbiome. Nat Commun 16, 1264 (2025). https://doi.org/10.1038/s41467-025-56426-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56426-4
This article is cited by
-
Genome-centric metagenomes unveiling microbial functional potential in a glacier river in the Mount everest
World Journal of Microbiology and Biotechnology (2026)
-
Post-glacial microbial succession and carbon sequestration processes: insights from recent research
Environmental Sciences Europe (2025)