Abstract
Despite advances in treatments, multiple myeloma (MM) remains an incurable cancer where relapse is common. We developed a circulating tumor DNA (ctDNA) approach in order to characterize tumor genomics, monitor treatment response, and detect early relapse in MM. By sequencing 412 specimens from 64 patients with newly diagnosed or relapsed/refractory disease, we demonstrate the correlation between ctDNA and key clinical biomarkers, as well as patient outcomes. We further extend our approach to simultaneously track CAR-specific cell-free DNA (CAR-cfDNA) in patients undergoing anti-BCMA CAR T-cell (BCMA-CAR) therapy. We demonstrate that ctDNA levels following BCMA-CAR inversely correlate with relative time to progression (TTP), and that measurable residual disease (MRD) quantified by peripheral blood ctDNA (ctDNA-MRD) was concordant with clinical bone marrow MRD. Finally, we show that ctDNA-MRD can anticipate clinical relapse and identify the emergence of genomically-defined therapy-resistant clones. These findings suggest multiple clinical uses of ctDNA for MM in molecular characterization and disease surveillance.
Similar content being viewed by others
Introduction
Therapeutic advances including novel immunotherapies have improved outcomes of patients with multiple myeloma (MM)1,2,3,4. Despite this, most patients eventually experience disease relapse, and subsequent therapies become increasingly less effective. Disease biology also undergoes dramatic change through sequential gain of genetic mutations, copy number alterations, and epigenetic modifications. These changes lead to clinical manifestations of extramedullary disease and/or oligo/non-secretory disease, which pose challenges in clinical disease monitoring5,6,7,8. Chimeric antigen receptor (CAR) T-cell therapy demonstrates great promise in patients who otherwise have dismal outcomes from conventional therapy. However, virtually all patients relapse eventually, and there are few established biomarkers predictive of clinical outcomes9,10, and the mechanisms of resistance are largely unknown. Thus, there is an unmet need to identify a predictive biomarker, and to understand the mechanisms of resistance to this novel immunotherapy.
Currently, assessment of genomic aberrations including MRD is performed using bone marrow (BM) aspirate, thus limiting its frequency and utility. Furthermore, MM is a heterogeneous disease with the involvement of different clones in different anatomical sites11, which poses challenges for assessing genomic information of the whole tumor. Therefore, improved tools to overcome spatial heterogeneity and monitor disease are crucial for developing personalized therapeutic approaches.
Circulating tumor DNA (ctDNA) has emerged as a promising minimally invasive assay for diagnosing and monitoring various cancers, including several hematologic malignancies12,13,14,15. As ctDNA is shed into the blood from tumor cells throughout the body carrying tumor-related genomic information, it can be distinguished from non-tumor-derived cell-free DNA (cfDNA) and provides multifaceted genomic information from the whole tumor. Moreover, ctDNA has myriad potential uses, including molecular genotyping, treatment response assessment, and quantification of MRD using a blood draw. Various methodologies have been used to assess ctDNA including initial studies in MM16,17,18,19,20,21. However, these studies focus on either whole exome sequencing (WES) with relatively limited depth of sequencing, or targeted deep sequencing with a small number of genes, limiting the number of genetic alterations captured per case and the utility for applications in MRD22,23. Improved approaches are therefore greatly needed.
To address these unmet needs, we developed a robust and ultra-sensitive CAPP-Seq (cancer personalized profiling by deep sequencing)24,25 hybrid capture approach for ctDNA in MM. Our approach allows genomic characterization, disease burden quantification, treatment response monitoring, and assessment of mechanisms of therapeutic resistance (Fig. 1). We demonstrate the potential applications of ctDNA in patients treated with CAR T-cell therapies to enable MRD detection, outcome prediction, and understanding of genomic determinants of therapeutic resistance.
Schematic depicting the usage of cell-free DNA assay for disease characterization, treatment response monitoring, CAR T-cell tracking, and early relapse detection. By following sequential samples over the course of treatments, clonal dynamics, as well as treatment resistance mechanisms can be revealed.
Results
Developing CAPP-Seq for MM ctDNA detection
To profile genomic alterations in MM from ctDNA, we designed a targeted CAPP-Seq panel for MM utilizing publicly available WGS and WES data26,27,28,29, including well-characterized coding genes harboring frequent mutations in MM. Furthermore, as the sensitivity for detecting residual tumor-derived DNA depends on having multiple mutations available per case25,30, we included frequently observed hypermutated immunoglobulin loci and other non-coding genomic regions identified in previously reported WGS data29, which had higher rates of mutations compared to canonical drivers mutations in coding regions (Fig. 2A). These regions had a significant overlap with diffuse large B-cell lymphoma (DLBCL) cases (Fig. S1) where these mutations were demonstrated as targets of activation-induced cytidine deaminase (AID)31. As AID is also indicated in aberrant somatic hypermutation in MM32, the inclusion of these regions allows us to maximize the sensitivity for ctDNA-MRD detection from plasma25,30. This panel is described in Table S1.
A A bar plot showing the distribution of single nucleotide variants in multiple myeloma from WGS data29. The genome was divided into 1000-bp bins, and the fraction of patients with a variant in each bin was calculated. Both frequently mutated driver coding genes (navy) and most frequently mutated genomic regions (pink) are highlighted. B, C Number of SNVs identified from genotyping samples (tumor for esophageal and lung cancers; tumor or blood plasma for MM or DLBCL), as well as allele frequencies of ctDNA from pre-treatment samples in order to demonstrate ctDNA detection in MM. Data are presented as mean values ± 95% CI. The p-values were calculated using a two-tailed Wilcoxon rank sum test. p-values were B 8.2e−14 (esophageal vs MM), 2e-15 (lung vs MM), 0.0003 (DLBCL vs MM); C 1.4e-9 (esophageal vs MM), 1.8e-8 (lung vs MM), 0.28 (DLBCL vs MM). ****P < 0.0001, ***P < 0.001, and P > 0.0001. D Number of SNVs identified by our CAPP-Seq assay compared to clinical NGS assay for hematological malignancies for samples that had paired data in our cohort (n = 11). The p-value was calculated by a two-sided Wilcoxon rank-sum test. E The ctDNA detection limits are shown for coding genes only and coding + non-coding genomic regions based on the number of mutations detected in our cohort. Data are presented as mean values ± SD. The p-value was calculated using the Wilcoxon rank sum test, showing 6.9e-16.
We applied CAPP-Seq to bone marrow and blood plasma samples from 64 patients with plasma cell disorders including newly diagnosed multiple myeloma (NDMM, n = 11), treatment-naïve monoclonal gammopathy of undetermined significance (MGUS, n = 3), treatment-naïve smoldering myeloma (SMM, n = 3), and relapsed/refractory MM (RRMM, n = 47) (Table 1). A median of 83 single-nucleotide variants (SNVs, range 0–246) at an average allele frequency (AF) of 2.7% (range 0–51%) were detected per case. Compared to solid (lung and esophageal) malignancies where CAPP-Seq has been previously applied and established as a useful tool for tracking ctDNA-MRD, we detected a greater number of SNVs for tumor tracking in MM33,34,35 (Fig. 2B, C), which directly implies an improved sensitivity compared to these diseases. Indeed, while previous studies in DLBCL identified a higher number of SNVs per case compared to MM, we did observe a similar level of ctDNA in pretreatment samples (Fig. 2C), indicating that ctDNA is a viable analyte in MM.
Importantly, our approach identifies and tracks more mutations than approaches focused on coding genes alone, thus increasing the sensitivity for ctDNA-MRD detection. To demonstrate this, we compared the performance of our panel with an in-house hematological mutational assay covering 164 genes (Stanford Tumor Actionable Mutation Panel for Hematopoietic and Lymphoid Neoplasms; Heme-STAMP) performed on the BM for cases where paired data was available (n = 11). We saw significantly higher numbers of detected SNVs by our panel compared to Heme-STAMP, suggesting that a current clinical next-generation sequencing-based assay focusing on coding mutations does not detect an adequate number of mutations (Fig. 2D). Correspondingly, the limit of detection (LOD) significantly improved with increasing number of SNVs detected by incorporating non-coding genomic regions, as opposed to considering coding SNVs alone (Fig. 2E). Importantly, we saw a high degree of concordance (92%, 11/12 mutations) in the overlapping regions of the genome between these two panels, indicating the robustness of our genotyping approach, as well as implying that the increased number of mutations seen is driven by selecting enriched regions of the genome (Table S2). We additionally evaluated the specificity of our assay for both mutational genotyping and monitoring ctDNA-MRD, thus establishing the performance of panel-wide genotyping and the 95% specificity of our monitoring assay (Fig. S2, Supplemental Materials).
Genomic characterization of MM via ctDNA
We genotyped the disease of each patient in our cohort via bone marrow aspirate or cfDNA derived from blood plasma (Fig. 3A). 63 cases (98%) of cases had detectable SNVs. Among detected SNVs, 73% were in immunoglobulin loci (IGH, IGK, and IGL), 20% were in noncoding genomic regions, and 7% were in non-Ig coding genes (Fig. 3B), consistent with our expected enrichment of AID-related mutations. Indeed, detected mutations in noncoding genomic regions include intronic areas of BCL6 (73% of patients), LPP/BCL6 super-enhancer region (48%), and BCL7A (36%), which is concordant with previous reports29,36,37. Mutations in immunoglobulin loci were detected in 90% (IGH), 84% (IGL), and 64% (IGK) of patients, respectively. Single base substitution mutational signatures in these immunoglobulin and noncoding regions demonstrated canonical signatures related to AID activity (SBS84 and SBS85), as well as non-canonical signatures previously associated with AID activity (SBS37, SBS39)31,38 (Fig. 3C). The prevalence of AID-related mutations demonstrates that our approach captures biologically relevant mutations in addition to conventionally reported driver genes, increasing the number of mutations available to track MRD and thereby increasing sensitivity for disease detection. By using conventionally reported driver genes, only 45/64 cases (70%) had detected mutations. The improved performance for disease monitoring considering both coding and non-coding alterations is highlighted in a case vignette (Fig. 3D). This case had developed oligo-secretory disease with high levels of ctDNA, with quantitative levels tracking with treatment response. Notably, after ctDNA levels declined due to disease response, even when coding genes became undetectable (red arrows in Fig. 3D) at a point of low disease burden, multiple mutations in non-coding and AID-related regions remained detectable. In addition to improvements in disease detection, tracking non-coding variants allows assessment of clonal heterogeneity between ctDNA and bone marrow, where detected clonal composition evolves over time. This effect is highlighted by tracking SNVs in non-coding regions in blood plasma (Fig. S3).
A A bar plot depicting the number of SNVs detected from genotyping samples (bone marrow aspirate, n = 20; or plasma, “cfDNA”, n = 44). The number of non-coding single-nucleotide variants (SNVs), inclusive of immunoglobulin (Ig) loci, are shown in aqua; the number of coding SNVs not including Ig loci are shown in red. B A pie-chart showing the breakdown of a total of 5145 SNVs detected in Fig. 3A. IGH immunoglobulin heavy chain, IGK immunoglobulin kappa light chain, IGL immunoglobulin lambda light chain. C A bar plot showing the mutational signatures of frequently detected intronic genes (BCL7A, BCL6, LPP/BCL6 super-enhancer region), immunoglobulin loci, and intergenic regions as compared to frequently detected coding mutations in driver genes (KRAS, NRAS, TP53). SBS37, 39, 84, and 85 are highlighted as being associated with AID activity. D An example of tracking coding and non-coding mutations for disease monitoring in a case. Red arrows point to timepoints when coding genes became undetectable but non-coding regions and immunoglobulin loci remained detectable. E An oncoprint showing the coding mutations detected in our cohort (n = 64). Only coding SNVs detected in at least two patients are shown. MGUS/SMM vs MM is identified by the color bar at the bottom. MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor-κB. Source data are provided as a Source Data file. F Venn diagram showing concordance of single nucleotide variants detected from bone marrow (BM) and plasma across 22 paired samples.
In addition to these immunoglobulin genes and non-coding mutations, we identified frequent SNVs in coding genes including KRAS (23% of patients), NRAS (14%), BRAF (13%), and TP53 (16%), which is concordant with prior reports27,28,39. Of note, the rate of TP53 mutation was significantly higher than in Walker et al.39. However, this potentially reflects the fact that Walker et al. only included NDMM, as opposed to our cohort, which also contains RRMM (Figs. 3E and S4 and S5).
Validation of ctDNA single nucleotide variants and copy number alterations
To assess the performance of blood plasma as a surrogate for tumor mutational genotyping, we compared mutations observed in the blood plasma with paired BM samples in 22 patients where BM samples were available and evaluable. The correlation between the allele fraction of each SNV detected in the BM and cfDNA is shown in Fig. S6, where 17/22 (77%) of cases demonstrated a significant positive correlation between tumor and plasma. For these samples, among SNVs detected in the BM, 60% (1360/2266) were also detectable in the plasma (Fig. 3F). As expected, we also observed that the sensitivity for detecting BM-derived SNVs improved as the burden of tumor DNA in the plasma increased (Fig. S7A). Overall, in samples where the mean ctDNA fraction was ≥0.3%, the sensitivity of detecting BM-derived SNVs was 82%. The frequency of overlapping mutations suggests that in most cases of MM, where the ctDNA amount is above this level, BM-derived SNVs are reliably detected from the plasma. In addition to BM-derived SNVs, we detected an additional 483 SNVs in the plasma across these 22 patients that were not present in the BM, including KRAS, NRAS, TP53, and BRAF mutations, suggesting ctDNA may capture spatial tumor heterogeneity not present in BM biopsies. While our assay does include deep sequencing of matched normal to exclude germline alleles and clonal hematopoiesis, it is possible these alleles could arise from another malignancy. However, none of these patients had second malignancies documented at the time of assessment, making this an unlikely source of these mutations. When considering mutations that were shared between BM and plasma vs those private to plasma, mutations in driver genes such as NRAS, KRAS, and TP53 were more likely to be shared compared with other mutations, suggesting these mutations may occur in early pathogenesis and are more clonal (Fig. S7B).
Given the importance of structural variants in MM disease biology, we also assessed the performance of our assay for identifying copy number alterations (CNAs) directly from the blood plasma. Detection of CNAs from plasma has been historically challenging, with prior methods focused on low-pass WGS reporting successful genotyping of arm-level alterations only at tumor fractions greater than 3%40. Here, we employed our previously described method, CANARy (Copy number ANomaly Assessment and RecoverY), a computational framework to use on- and off-target sequencing reads from targeted sequencing data to identify genome-wide copy number alterations (Fig. S8A)34,41. We compared the performance of CANARy for identifying CNAs with historical clinical fluorescence in situ hybridization (FISH) from BM for key CNAs (del(17p), gain(1q) and del(13q)). As with SNV genotyping, we observed that the sensitivity of detection improved as plasma AF increased. For samples with an AF ≥ 2.5%, CANARy demonstrated sensitivity of 80%, 80%, and 100% for detecting del(17p), gain(1q), and del(13q), respectively (Fig. S8B, C). This performance is comparable to dedicated LP-WGS methods from cfDNA42, suggesting that CNAs detected using CANARy are concordant with BM FISH tests and that sensitivity improves with higher plasma tumor burden. Additionally, unlike FISH assay, which requires specific probes for chromosome regions of interest, CANARy assesses cfDNA CNAs across the whole genome, leading to the discovery of additional alterations. This analysis can be potentially useful in the research and discovery settings; however, we note that this assay should not replace the current gold standard bone marrow-based FISH test for clinical assessment.
ctDNA levels are associated with adverse risks and disease burden of myeloma
We next quantified ctDNA levels (Supplemental Methods) in patients with plasma cell neoplasms and correlated with clinical disease features. Among 64 patients, there were 70 samples that were collected at diagnosis or at relapse. Among them, ctDNA was detected in 96% (67/70) with a median of 2.0 log10 haploid genome equivalents per mL of plasma (hGE/mL). Levels of ctDNA were higher in MM compared to MM-precursors such as MGUS or SMM (2.10 vs 1.09 log hGE/mL, p = 0.028; Fig. 4A), as well as RRMM compared to NDMM (2.17 vs 1.15 log he/mL, p = 0.043; Fig. 4B) and patients with a history of extramedullary disease compared to those without the extramedullary disease (2.24 vs 1.41 log he/mL, p = 0.026; Fig. 4C). There was no significant difference if a patient had R-ISS I vs II/III (p = 0.050), oligo/non-secretory disease (p = 0.96), or high-risk cytogenetics (p = 0.058) in our cohort (Fig. S9A–C). We next assessed ctDNA levels correlating with clinical disease burden measurements. We observed a significant correlation between ctDNA and M-spike (rho = 0.55, p = 1.4 × 10−6; Fig. 4D), as well as free light chain difference (dFLC) (rho = 0.69, p = 2.2 × 10−16; Fig. 4E). Notably, in patients with oligo/non-secretory disease, where the blood-based assessment of disease burden is challenging clinically, we detected comparable ctDNA levels to patients with the secretory disease (1.3 vs 1.6 logs hGE/mL, p = 0.18 Figs. 4D and S9B).
A–C Levels of ctDNA grouped by disease type. MGUS monoclonal gammopathy of undetermined significance, SMM smoldering myeloma, MM multiple myeloma, ND newly diagnosed, RR relapsed/refractory, EMD extramedullary disease, hGE/mL human genome equivalent per mL of plasma, ND in x-axes, non-detected. Data are presented as mean ± SD. p-values were calculated by a two-sided Wilcoxon rank-sum test. D, E Correlation of ctDNA levels and clinical biomarkers (M-spike and difference between involved and uninvolved free light chains) by linear regression based on secretory disease status. Correlation coefficients and p-values were calculated using a two-tailed Spearman’s rank-order test. F Waterfall plot of quantified molecular response (log10-transformed fold-change in ctDNA from day 0 to day 28 post-treatment). Cases are categorized and color-coded according to the best response as assessed by IMWG criteria. The p-value was calculated by a two-sided Wilcoxon rank-sum test. VGPR very good partial response, PR partial response, SD stable disease, MR minimal response, PD progressive disease.
ctDNA dynamics to track treatment response in myeloma
Having demonstrated the prognostic performance of baseline ctDNA levels, we next explored the utility of ctDNA to predict therapeutic responses. We considered 31 patients with a subsequent sample 28 days after initiation of therapy where clinical evaluation according to International Myeloma Working Group (IMWG) criteria was also available (CAR T, n = 21; non-cellular therapy, n = 10). We evaluated whether the change in ctDNA level on day 28 (D28) from pre-treatment correlated with the best response (Fig. 4F). The treatments for patients in this cohort are summarized in Table S3. The log-fold change (LFC) in ctDNA at D28 was significantly greater in patients who achieved a partial response (PR) or better, compared to patients with less than a PR (median LFC −2.1 vs 0.03, p = 0.002). Interestingly, one significant outlier to this trend was observed (CAR-06), achieving a very good partial response (VGPR) by clinical criteria on D28, while observing a more than 100-fold increase in ctDNA. This patient relapsed on D82 after CAR with the new extramedullary disease, demonstrating that in some cases, ctDNA may serve as a more accurate assessment of treatment response compared to standard criteria. M-spike and difference in free light chains (dFLC) were also correlated with disease response in this cohort, although numerous cases were unevaluable by one or both of these methods (Fig. S10).
ctDNA levels are prognostic for time to progression after CAR T-cell therapy in myeloma
We then applied CAPP-Seq to serial samples from 36 patients with RRMM undergoing anti-BCMA CAR T-cell (BCMA-CAR) therapy. Patients were treated as part of the standard of care and received either ide-cel (n = 30) or cilia-cel (n = 6) (Table S4). Genotyping of each patient’s myeloma was performed using BM or blood plasma. We then tracked ctDNA-MRD levels and mutational profiles in serial samples for each patient throughout the course of therapy (Supplemental Methods). 31 of 36 (86%) patients had sufficient SNVs for evaluation and ctDNA monitoring. Notably, among the five patients with insufficient mutations for evaluation, four were assessed for clonal immunoglobulin sequences from BM by next-generation sequencing (NGS; ClonoSeq, Supplemental Methods). Three out of four patients failed to identify any dominant clone, demonstrating low disease burden. This highlights the importance of identifying mutations from cfDNA or BM at diagnosis or relapse when there is measurable disease for further disease monitoring and therapeutic response.
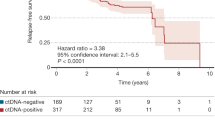
The median time to progression (TTP) and follow-up of the evaluable study cohort (n = 31) were 6.6 and 22.2 months, respectively; 23 patients experienced disease progression during follow-up. We categorized patients into those with progression within 90 days of CAR infusion (early progressors) and those without progression in that timeframe (non-early progressors). 12/18 (67%) of non-early progressors cleared ctDNA by a median of 21 days post-CAR, whereas only 1/8 (13%) early progressors had undetectable ctDNA by D28 (Fig. 5A). On D28, non-early progressors had significantly lower ctDNA levels compared to early progressors (−1 vs 1.97 median log hGE/mL; p = 5 × 10−4, Fig. 5B). Similar results were seen between D60 and 90. Moreover, ctDNA levels at any time points after D28 were significantly associated with relative TTP from that particular time point (Fig. 5C), and better correlated compared to M-spike or dFLC (Fig. S11A–B), suggesting a prognostic value of ctDNA monitoring after BCMA-CAR.
A A Swimmer plot of the individual patients receiving BCMA-CAR therapy and their follow-up, overlaid by ctDNA levels. Early progressors; patients who progressed within 3 months following BCMA-CAR. Non-early progressors; patients who did not progress within 3 months. B Dynamics of ctDNA after CAR T-cell infusion. Lines indicate the median and interquartile range. p-values on day 28 and day 60–90 were calculated by a two-sided Wilcoxon rank-sum test. C A scatter plot showing a correlation between ctDNA levels on ≥28 days post-CAR and relative TTP from individual time points. Data are presented as linear regression with 95% CI. The p-value was calculated using Spearman’s rank-order test. D Correlation of clinical bone marrow MRD and ctDNA at matched time points. Clinical bone marrow MRD was performed using NGS (ClonoSeq, n = 12) or NGF (n = 4). p-Value was calculated using a two-tailed Sperman’s rank-order test on samples that had the ClonoSeq test (n = 12). The line indicates y = x. E Kaplan–Meier analysis of progression-free survival in patients stratified by ctDNA-MRD positivity on day 90. The p-value was calculated using the log-rank test. F Correlation of CAR-cfDNA with an absolute count of CAR T-cells by flow cytometry at matched time points. Data are presented as linear regression with 95% CI. The p-value was calculated by two-tailed Spearman’s rank-order test.
Clinical MRD assessment correlates with ctDNA levels
We assessed whether ctDNA levels correlate with clinical BM MRD measurements. Among our CAR cohort, 28 patients had clinical NGS performed to identify dominant clonal immunoglobulin sequences (ClonoSeq) prior to CAR-T therapy, including seven patients who already had identified dominant clones during prior lines of therapy. Thirteen out of 28 (46%) failed clonal identification on initial assessment; 4 of these patients had clonal sequences identified after assessment of additional archival samples. Overall, the 19/28 patients (68%) with dominant sequences identified by ClonoSeq had a median bone marrow plasma cells (BMPCs) of 52.5% (5–95%) whereas those who failed identification (9/28) had a median BMPCs of 0% (0-5%). This highlights a challenge in identifying dominant sequences in relapsed/refractory patients from BM, as these patients have lower BMPCs (median BMPC 21% in NDMM vs 5% in RRMM in the study cohort). In the remaining patients, MRD assessment was done by next-generation flow cytometry when feasible (NGF, n = 4, Supplemental Methods). In contrast, as discussed above, our cfDNA-based assay identified trackable variants in 31/36 (86%) cases to allow disease monitoring.
Overall, 16 patients had paired ctDNA-MRD and BM-MRD (NGS, n = 12, NGF, n = 4) to allow direct comparison between the two anatomical compartments. NGS-based BM-MRD levels significantly correlated with ctDNA-MRD (rho = 0.92, p = 0.00003; Fig. 5D). ctDNA-MRD negativity on D90 was significantly associated with TTP by Kaplan–Meier analysis (p = 0.0008, Fig. 5E). Kaplan–Meier analysis of clinical BM MRD vs TTP was not statistically significant, although this analysis is limited by cohort size (Fig. S11C). When patients were categorized by BM MRD and ctDNA, TTP was significantly different among ClonoSeq + /ctDNA + vs ClonoSeq − /ctDNA− vs NGF− /ctDNA+ groups (Fig. S11D).
CAR-cfDNA levels correlate with flow cytometry enumeration
In addition to tracking tumor DNA, we designed our assay to detect and track CAR-derived cfDNA molecules, similar to prior work35 (Supplemental Methods). We compared CAR-cfDNA levels with flow cytometry (FC) enumeration at matched time points. FC and cfDNA quantification of CAR T-cell levels significantly correlated (rho = 0.73, p = 2.2e-16; Fig. 5F). Interestingly, levels of CAR detected in the BM cells and plasma cfDNA were also significantly correlated (Fig. S12A).
The median time to reach a peak level of CAR-cfDNA was 14 days for both ide-cel and cilia-cel, with a median peak concentration of 2.57 (log hGE/mL) for ide-cel, and 2.18 (log hGE/mL) for cilia-cel (Fig. S12B, C). CAR-cfDNA levels and ctDNA burden were inversely correlated (rho = −0.26, p = 0.0024 Fig. S12D), demonstrating the relationship between CAR expansion and response.
Clonal selection detected by ctDNA upon relapse after CAR T-cell therapy
In addition to providing MRD detection from ctDNA, our approach allows assessment of clonal selection and emergence of genetic alterations. At the time of relapse after BCMA-CAR, we observed the emergence of new alterations in ctDNA, including both SNVs and CNAs. However, the emergence of new CNAs was more recurrent. In particular, we did not discover any emergent SNVs or frameshift mutations in TNFRSF17/BCMA (i.e., the target antigen of the CAR T-cell) associated with therapeutic resistance. In contrast, 4/23 patients (17%) had an emergent copy number loss of TNFRSF17 (Fig. S13A). One of the cases (CAR-10) is depicted in Fig. 6A–C, which had an initial response to ide-cel with a concurrent decrease in ctDNA level and a clinical VGPR. This patient then experienced disease progression on D120 (Fig. 6A). Notably, ctDNA remained detectable and started to rise on D90, which was 30 days prior to clinical relapse, with concurrent copy number loss of TNFRSF17 (Fig. 6A, B). Moreover, a second focal deletion of TNFRSF17 was observed in the targeted sequencing data (Fig. 6B insert). This biallelic loss of TNFRSF17 was confirmed in WGS and CAPP-Seq of the pre-treatment and relapsed BM aspirate, respectively (Fig. S13B, C). Furthermore, the loss of BCMA expression by tumor cells was confirmed by immunohistochemistry (IHC) of the relapsed BM specimen (Fig. 6C). There was another case (CAR-13), who also developed focal loss of TNFRSF17 (Fig. S14). This suggests that ctDNA may be usable in detecting clonal selection that leads to relapse earlier than current clinical biomarkers, as well as detecting focal genomic loss of the target antigen.
A A case of early detection of relapse by ctDNA, showing the dynamics of ctDNA and CAR-cfDNA (upper panel), as well as the dynamics of TNFSRSF17 (BCMA) z-score (lower panel). Open points reflect non-significant q-score indicating the BCMA deletion was not detectable. B Genome-wide copy number profile inferred from cfDNA WGS of the patient depicted in A). Emergent chromosome 16 deletion is magnified to show further focal deletion in TNFRSF17 (BCMA), which was detected by CAPP-Seq. Segmentation was performed using the DNAcopy R package. CNR, copy number ratio. C Immunohistochemistry of the bone marrow core of the patient depicted in A prior to BCMA-CAR (left) and upon relapse (right) demonstrating loss of BCMA. D The volcano plot shows a clonal selection of somatic copy number alterations at individual gene levels in patients who relapsed (n = 23). Positive z-score reflects clonal selection in patients at relapse. p-values were calculated using a two-sided Wilcoxon rank-sum test. Copy number alterations that were significantly clonally selected (p < 0.05) are highlighted in green for amplifications, and red for deletions. E Kaplan–Meier analysis of progression-free survival in patients with and without underlying copy number loss of BCMA prior to treatment. F Forest plot of variable effects on time to progression (n = 36). The plot depicts hazard ratios ± SD calculated by a univariate Cox proportional hazards regressions model, with significant values shown in red (p < 0.05). Error bars reflect a 95% confidence interval.
Across all patients with pretreatment and relapse samples available, we observed recurrent clonal selection of multiple genes upon relapse, including deletion of TNFRSF17 (n = 4), amplification of MYC (n = 4), deletion of EGR1 (n = 4), deletion of AVL9 (n = 3), and deletion of KRAS (n = 5) (Fig. 6D).
In addition to assessing the emergence and clonal selection of alterations, we assessed the prognostic significance of baseline CNAs and SNVs for outcomes after BCMA-CAR. Five of 36 patients (14%) had underlying copy number loss of TNFRSF17 identified prior to CAR infusion; 4 of these cases had a monoallelic loss, while only one had a biallelic loss. Notably, there was a discordance between ctDNA assessment and bone marrow assessment, likely representing spatial heterogeneity (Table S5). Interestingly, these patients had significantly shorter TTP after treatment with BCMA-CAR T-cells compared to those who had normal copy numbers prior to BCMA-CAR therapy (p = 0.0073 Fig. 6E).
Levels of ctDNA and molecular features are prognostic for outcomes after BCMA-directed CAR-T therapy
Having applied this multi-faceted approach to cfDNA after BCMA-CAR, we then applied univariate Cox proportional hazards regression models to assess the impact of key patient and disease features on TTP. As expected, higher ctDNA levels on D28 and D90 were prognostic for inferior TTP (D28, HR = 1.9, p = 2.5 × 10−4; D90, HR = 2.7, p = 7.5 x 10−4) (Fig. 6F).
Finally, we assessed the prognostic significance of specific genetic alterations genome-wide prior to treatment on outcomes following BCMA-CAR. Interestingly, there were no SNVs that predicted TTP (Fig. S15A). In contrast, CNAs in multiple genes were associated with adverse outcomes to BCMA-CAR. These included amplification of BRAF and MYC, as well as deletion of FAM46C and CDKN2C, which are well-described oncogenes and tumor suppressors, respectively, that are implicated in MM pathology (Fig. S15B, C). Indeed, both this pre-treatment CNA analysis, as well as clonal selection analysis implicate MYC as a key determinant of resistance to MM immunotherapy. These findings suggest that our tool can assess ctDNA quantity and CNAs simultaneously, and has utility for predicting clinical outcomes after BCMA-CAR therapy.
Discussion
While BM-based MRD is well established in MM, minimally invasive approaches to disease detection and characterization remain an unmet need in MM. Prior studies have demonstrated the feasibility and utility of liquid biopsy in MM via ctDNA detection. However, these early approaches have had challenges in the detection of low-level MRD due to limited sensitivity15,16,18,19,43,44,45.
In this study, we designed a comprehensive targeted gene panel to characterize and track ctDNA via peripheral blood in patients with MM. By performing CAPP-Seq, we show that an abundance of tumor markers (i.e., a median of over 80 SNVs per case), are detected using this panel for disease tracking, and ctDNA level was associated with disease burden and adverse risk factors in MM. Notably, ctDNA levels were comparable in secretory vs oligo/non-secretory patients, making ctDNA a potential blood biomarker in patients who have lost their circulating protein-based biomarker. In addition, we demonstrate that changes in ctDNA levels after 1 month of therapy corresponded with the best clinical response following both cellular and non-cellular therapies.
Recently, novel immunotherapies including anti-BCMA CAR T-cell therapy have emerged as promising entities in treating RRMM. However, there are no established predictive biomarkers for treatment response or duration of response after CAR-T. Our study demonstrates that ctDNA tracks with disease burden and detects MRD at a comparable sensitivity to conventional BM-based MRD, which requires an invasive procedure. Additionally, ctDNA levels at any given time after 28 days following BCMA-CAR are associated with TTP from that time point. By incorporating oligonucleotides targeted to CAR vectors, we demonstrate simultaneous detection of CAR-cfDNA and ctDNA, enabling tracking of the expansion and persistence of CAR T-cells along with therapeutic response. We show that peak CAR-cfDNA levels are associated with TTP in our ide-cel cohort.
Another application of our assay is to explore the genomic mechanisms of resistance to therapy. Resistance to CAR T-cell therapy is thought to be multifactorial, including tumor intrinsic factors, CAR T-cell fitness, and tumor microenvironment factors46. Emergent BCMA loss with or without BCMA mutation is known to be one of these mechanisms of relapse after anti-BCMA therapy9,47. It is also reported in 6–7% of ND or RRMM patients, who never had an exposure to BCMA-targeted therapy48,49. Our assay detected clonal selection after BCMA-CAR, where a clone harboring loss of BCMA was selected and became dominant, with a concurrent rise in ctDNA. This ctDNA rise and clonal selection were observed prior to biochemical relapse as assessed by clinical myeloma laboratory markers, suggesting that ctDNA may be useful for early disease relapse detection, as well as revealing resistance mechanisms. In our cohort, we observed that 5/36 patients (14%) had underlying BCMA loss prior to BCMA-CAR, 1 of whom later had a clonal evolution with a focal BCMA loss resulting in loss of both alleles. Additionally, three patients acquired emergent BCMA loss upon relapse. Those who had underlying BCMA loss had shorter TTP compared to those who did not. We note that these patients were also enriched for underlying amplification of MYC, which is implicated in conferring poor prognosis50, and reported as an emergent CNV upon relapse after CAR-T47. In our cohort, we also noted the selection of clones harboring amplification of MYC, or deletion of EGR1, AVL9, or KRAS upon relapse after BCMA-CAR. Future studies will be needed to determine if these alterations in genes outside of TNFRSF17 (BCMA) represent specific mechanisms of resistance, or are simply markers of more aggressive disease biology.
While there are multiple uses of ctDNA-based assessment in MM as outlined above, it is important to recognize the limitations of this assay. While we have demonstrated the ability to detect both SNVs and CNAs using this approach, the sensitivity for CNA detection is lower, as described in previous ctDNA approaches40. Furthermore, we did not explore the detection of translocations using our approach, which is a key recurrent type of alteration in MM. Indeed, we envision ctDNA assays will supplement—rather than replace—current bone marrow assays, which will remain important to establish a pathological diagnosis and to assess structural alterations such as CNAs and translocations using gold-standard FISH. Indeed, approaches such as this are likely to find initial use in research and discovery settings—such as the exploration of mechanisms of response and resistance to therapy—prior to gaining use in the clinic. The strengths of our approach are to non-invasively characterize MM, to monitor residual disease, and to assess resistance mechanisms to therapy as patients undergo treatment. Additional limitations of our study include a small cohort size from a single center and a short follow-up. Further investigation in a larger cohort will allow further analysis such as rigorous subgroup analysis of patients undergoing ide-cel vs cilta-cel. A larger cohort may also provide deeper insights via longitudinal therapeutic monitoring and may reveal additional genomic mechanisms of resistance. Future studies validating pre-treatment BCMA loss and association with clinical outcomes, which has significant clinical implications, will be needed.
Taken together, we demonstrate that a non-invasive ctDNA assay can inform multifaceted genomic information that is useful for genotyping, disease monitoring, early detection of disease relapse, and detecting mechanisms of treatment resistance. Overall, this study indicates a strong potential to improve the clinical management of MM using personalized ctDNA analysis.
Methods
Patients
Patients with MM or other plasma cell disorders who were treated under standard-of-care at Stanford Cancer Center were enrolled in this study between February 2017 to July 2024. A total of 64 patients had samples collected and were included in this study. Seventeen patients had newly diagnosed and/or treatment-naïve disease, and 47 patients had relapsed/refractory disease. Thirty-six patients subsequently underwent CAR T-cell therapy under the standard of care (idecabtagene vicleucel, ide-cel; n = 30; ciltacabtagene autoleucel, cilta-cel; n = 6). The study was approved by the Stanford Institutional Review Board (protocol #43375) in accordance with the Declaration of Helsinki, and all patients provided written informed consent. There was no compensation to the participants. Additional information can be found in the Supplemental Methods.
Sample collection and processing
Peripheral blood samples were collected in Cell-Free DNA Collection Tubes (Roche, Pleasanton, CA) or Cell-Free DNA Blood Collection Tube (BCT) (Streck, La Vista, NE) and processed within 7 days by centrifugation at 1800 g for 10 min. Plasma and plasma-depleted whole blood were subsequently isolated and cryopreserved at −70 °C as previously described34. Additional information can be found in the Supplemental Methods.
Designing of a targeted gene panel for multiple myeloma
In order to detect genomic alterations in MM, a custom 570 kb oligonucleotide panel (IDT, Coralville, IA) was designed using publicly available whole genome sequencing (WGS) and WES data26,27,28,29. This panel includes genomic regions known to be recurrently mutated or of known biological significance in MM. In addition to the MM panel, supplemental custom oligonucleotide panels were designed to target ide-cel and cilia-cel transgenes, T-cell receptor genes, and genes known to be implicated in immune evasion. Coding genes captured in this panel are shown in Table S1. We define coding mutations as non-synonymous gene mutations, whereas non-coding mutations are used to refer to both synonymous gene mutations and mutations in non-coding or intergenic regions.
Statistical analysis
Statistical analyses were performed using R (v.4.0.3), MATLAB (R2020b), and GraphPadPrism (v.9.0.0). Analyses for significant differences between the two groups were assessed using the Wilcoxon rank-sum test. Survival probabilities were estimated using the Kaplan–Meier method, and the survival of the two groups was compared using a log-rank test. We considered time to progression (TTP) wherein an event was defined as disease progression or relapse as the survival endpoint. ctDNA detection was determined with our previously described Monte-Carlo-based ctDNA detection index25. The statistical significance of Pearson or Spearman correlation was determined using t statistics. Regression analysis of covariates was conducted by Cox proportional hazards modeling, with p-values assessed using the log-likelihood test. For Cox regression analyses, continuous variables were used as input and standardized using Fisher’s z-score transformation. Units were defined as follows: ctDNA, log10(haploid genome equivalents per milliliter; hGE/mL), CAR-cfDNA, log10(genomes/mL).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The genotyping data generated in this study have been deposited in Figshare (https://doi.org/10.6084/m9.figshare.24803172). Due to patient confidentiality and restrictions from patient consent forms, the raw genomic data are protected and are not available per the institutional review board. The processed genomic data generated in this study are provided in the Supplementary Information/Source Data file. For additional inquiries, please contact the corresponding authors at dkurtz@stanford.edu. Source data are provided with this paper.
References
Kumar, S. K. et al. Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia 28, 1122–1128 (2014).
Braunlin, M., Belani, R., Buchanan, J., Wheeling, T. & Kim, C. Trends in the multiple myeloma treatment landscape and survival: a U.S. analysis using 2011–2019 oncology clinic electronic health record data. Leuk. Lymphoma 62, 377–386 (2021).
Binder, M. et al. Mortality trends in multiple myeloma after the introduction of novel therapies in the United States. Leukemia 36, 801–808 (2022).
Corre, J. et al. Improved survival in multiple myeloma during the 2005–2009 and 2010–2014 periods. Leukemia 35, 3600–3603 (2021).
Munshi, N. C. & Avet-Loiseau, H. Genomics in multiple myeloma. Clin. Cancer Res. 17, 1234–1242 (2011).
Manier, S. et al. Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 14, 100–113 (2017).
Patel, U. H., Drabick, J. J., Malysz, J. & Talamo, G. Nonsecretory and light chain escape in patients with multiple myeloma. Clin. Lymphoma Myeloma Leuk. 18, e515–e519 (2018).
Pawlyn, C. & Morgan, G. J. Evolutionary biology of high-risk multiple myeloma. Nat. Rev. Cancer 17, 543–556 (2017).
Munshi, N. C. et al. Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N. Engl. J. Med. 384, 705–716 (2021).
Desai, K. et al. Baseline and early post-infusion biomarkers associated with optimal response to idecabtagene vicleucel (ide-cel) in the KarMMa-3 study of triple-class–exposed (TCE) relapsed and refractory multiple myeloma (RRMM). J. Clin. Oncol. 41, 8031 (2023).
Rasche, L. et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat. Commun. 8, 268 (2017).
Kurtz, D. M. et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J. Clin. Oncol. 36, 2845–2853 (2018).
Fernández-Miranda, I. et al. Monitoring of circulating tumor DNA predicts response to treatment and early progression in follicular lymphoma: results of a prospective pilot study. Clin. Cancer Res. 29, 209–220 (2023).
Mutter, J. A. et al. Circulating tumor DNA profiling for detection, risk stratification, and classification of brain lymphomas. J. Clin. Oncol. 41, 1684–1694 (2023).
Dhakal, B. et al. Assessment of molecular residual disease using circulating tumor DNA to identify multiple myeloma patients at high risk of relapse. Front Oncol. 12, 786451 (2022).
Mithraprabhu, S. et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia 31, 1695–1705 (2017).
Guo, G. et al. Genomic discovery and clonal tracking in multiple myeloma by cell-free DNA sequencing. Leukemia 32, 1838–1841 (2018).
Manier, S. et al. Whole-exome sequencing of cell-free DNA and circulating tumor cells in multiple myeloma. Nat. Commun. 9, 1691 (2018).
Gerber, B. et al. Circulating tumor DNA as a liquid biopsy in plasma cell dyscrasias. Haematologica 103, e245–e248 (2018).
Kis, O. et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat. Commun. 8, 15086 (2017).
Vij, R. et al. Deep sequencing reveals myeloma cells in peripheral blood in majority of multiple myeloma patients. Clin. Lymphoma Myeloma Leuk. 14, 131–139.e131 (2014).
Mazzotti, C. et al. Myeloma MRD by deep sequencing from circulating tumor DNA does not correlate with results obtained in the bone marrow. Blood Adv. 2, 2811–2813 (2018).
Pugh, T. J. Circulating tumour DNA for detecting minimal residual disease in multiple myeloma. Semin Hematol. 55, 38–40 (2018).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med 20, 548–554 (2014).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Walker, B. A. et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 132, 587–597 (2018).
Bolli, N. et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 5, 2997 (2014).
Lohr, J. G. et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 25, 91–101 (2014).
Chapman, M. A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011).
Scherer, F. et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci. Transl. Med. 8, 364ra155 (2016).
Kurtz, D. M. et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-00981-w (2021).
Walker, B. A. et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat. Commun. 6, 6997 (2015).
Azad, T. D. et al. Circulating tumor DNA analysis for detection of minimal residual disease after chemoradiotherapy for localized esophageal cancer. Gastroenterology 158, 494–505.e496 (2020).
Chabon, J. J. et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 580, 245–251 (2020).
Sworder, B. J. et al. Determinants of resistance to engineered T cell therapies targeting CD19 in large B cell lymphomas. Cancer Cell 41, 210–225.e215 (2023).
Chakraborty, C. et al. Recurrent non-coding mutations in BCL7A may have significant functional consequence in multiple myeloma. Blood 134, 857 (2019).
Samur, M. K. et al. Landscape of recurrent mutations in non-coding genome with functional implications in newly-diagnosed multiple myeloma. Blood 132, 190 (2018).
Shen, J. C. et al. A high-resolution landscape of mutations in the. Proc. Natl Acad. Sci. USA 116, 24779–24785 (2019).
Walker, B. A. et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J. Clin. Oncol. 33, 3911–3920 (2015).
Adalsteinsson, V. A. et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017).
Palomino Mosquera, A. et al. Accurate detection of clinically actionable copy number variants in diverse hematological neoplasms by routine targeted sequencing: a comparative performance study. Blood 140, 10712–10713 (2022).
Wang, X. et al. PEcnv: accurate and efficient detection of copy number variations of various lengths. Brief Bioinform. https://doi.org/10.1093/bib/bbac375 (2022).
Auclair, D. et al. A next generation liquid biopsy approach for multiple myeloma. Blood 136, 33 (2020).
Long, X. et al. The utility of non-invasive liquid biopsy for mutational analysis and minimal residual disease assessment in extramedullary multiple myeloma. Br. J. Haematol. 189, e45–e48 (2020).
Deshpande, S. et al. Monitoring treatment response and disease progression in myeloma with circulating cell-free DNA. Eur. J. Haematol. 106, 230–240 (2021).
van de Donk, N. W. C. J., Themeli, M. & Usmani, S. Z. Determinants of response and mechanisms of resistance of CAR T-cell therapy in multiple myeloma. Blood Cancer Discov. 2, 302–318 (2021).
Lee, H. et al. Mechanisms of antigen escape from BCMA- or GPRC5D-targeted immunotherapies in multiple myeloma. Nat. Med. https://doi.org/10.1038/s41591-023-02491-5 (2023).
Samur, M. K. et al. Biallelic loss of BCMA as a resistance mechanism to CAR T cell therapy in a patient with multiple myeloma. Nat. Commun. 12, 868 (2021).
Da Via, M. C. et al. Homozygous BCMA gene deletion in response to anti-BCMA CAR T cells in a patient with multiple myeloma. Nat. Med. 27, 616–619 (2021).
Sekiguchi, N. et al. Impact of C-Myc gene-related aberrations in newly diagnosed myeloma with bortezomib/dexamethasone therapy. Int J. Hematol. 99, 288–295 (2014).
Acknowledgements
This study is supported by the National Cancer Institute (K08CA241076 to D.M.K.), the American Cancer Society-Stanford Cancer Institute IRG Pilot Grant (D.M.K.), the Gabrielle’s Angel Cancer Research Foundation Medical Research Award (D.M.K), the Leukemia Research Foundation New Investigator Research Grant (D.M.K.), and the Shanahan Family Foundations (D.M.K.), and CKD Family Fund (M.L.). H.H. is supported by the SITC-Merck Cancer Immunotherapy Clinical Fellowship. S.S. was supported by Stanford Clinical and Translational Science KL2 Career Development Award program (KL2 TR003143), Stanford Cancer Institute/American Cancer Society Pilot Grant 2022, and Doris Duke Charitable Foundation.
Author information
Authors and Affiliations
Contributions
H.H. and D.M.K. designed the study; H.H., D.I., and S.S. enrolled patients; K.T., M.C., and H.H. processed samples; H.H. and M.C. performed experiments; H.H., V.H., and S.S. collected clinical data; H.H., M.C., and A.M.M., and D.M.K. analyzed the data; B.Sa. designed CAR T-cell flow cytometry and S.Sy. conducted flow cytometry; O.S. performed IHC; H.S. provided Heme-STAMP sequencing data; H.H., M.C., and D.M.K. wrote the manuscript. B.S., G.D., T.Z., M.L., S.A., D.I., D.M., and M.S.K. critically reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
B.J.S.: consultancy: Foresight Diagnostics. M.L.: advisory board/consulting for Janssen, BMS, and Kite. D.M.: patent: Pharmacyclics supporting ibrutinib for chronic graft-vs-host disease. Research funding and consultancy: Pharmacyclics, Kite Pharma, Adaptive Biotechnologies, Novartis, Juno Therapeutics, Celgene, Janssen Pharmaceuticals, Roche, Genentech, Precision Bioscience, Allogene and Miltenyi Biotec. M.S.K: research funding: Nutcracker Therapeutics, CRISPR Therapeutics; Consultancy: Daiichi Sankyo, Myeloid Therapeutics. S.S.: research funding: Magenta Therapeutics, BMS, Allogene, Janssen; Consultancy: Magenta Therapeutics, BMS, Janssen, Sanofi, Oncopeptides, Takeda. D.M.K.: patents: D.M.K. reports issued patents pertaining to ctDNA-MRD detection assigned to Stanford University and licensed to Foresight Diagnostics. Consultancy: Foresight Diagnostics, Roche, Genentech, and Adaptive Biotechnologies. Stock and Equity Ownership: D.M.K. reports equity ownership in Foresight Diagnostics. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Sham Mailankody, Ariosto Siqueira Silva, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hosoya, H., Carleton, M., Tanaka, K. et al. Deciphering response dynamics and treatment resistance from circulating tumor DNA after CAR T-cells in multiple myeloma. Nat Commun 16, 1824 (2025). https://doi.org/10.1038/s41467-025-56486-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56486-6
This article is cited by
-
Zalcitabine induces ferroptosis in multiple myeloma through the TFAM–cGAS–STING–SLC7A11 axis
Journal of Translational Medicine (2026)