Abstract
Irreversible damage of the lung epithelium in idiopathic pulmonary fibrosis (IPF) patients causes high mortality worldwide, with no lung repair approaches available currently. Here we show that in murine and monkey models, the KRT5+ P63+ progenitor cells in airway basal layer can enter the alveolar area post fibrotic injury. Aided with an automated culture system, we clone and characterize airway basal progenitor cells from 44 donors with various lung conditions. Transplantation of human progenitor cells into the mouse lung efficiently re-epithelializes the injured alveolar area, forms new respiratory tract and saccule-like structures, which ameliorates fibrotic lesions and improves survival of mice. Mechanistically, the engrafted human progenitor cells do not function by differentiating into mature alveolar cells in mouse lung; instead, they differentiate into saccular cells expressing multiple tight junction proteins such as CLDN4, which help the lung to re-establish epithelial barriers. Furthermore, by cloning P63+ airway basal progenitors from larger mammals and birds, we construct multiple lung-chimerism animals and uncover the evolutionarily conserved roles of these progenitor cells in lung repair. Overall, our data highlight the fate of airway basal progenitor cells in fibrotic lung and provide a potential therapeutic strategy for pulmonary diseases that lack inherent recovery mechanisms.
Similar content being viewed by others
Introduction
Lung epithelium is exposed to the atmosphere and is critical for the orchestration of initial responses to infectious organisms, toxins, and allergic stimuli, and most importantly maintenance of normal gas exchange function. To support efficient gas exchange, the lung must maintain an epithelial barrier between the atmosphere and fluid-filled tissues to avoid infiltration of cells or tissues into the alveolar area. IPF is a severe respiratory disease characterized by a progressive decline in pulmonary gas exchange function, often leading to patient mortality within 3–5 years post-diagnosis1,2. The lungs of IPF patients typically exhibit a loss of alveolar epithelium accompanied by collapse of the alveolar structure. This loss of epithelial intactness in IPF results in the inability to effectively close the site of injury with newly differentiated epithelial cells, thus compromising the maintenance of an effective physical barrier. Instead, the architectures would be replaced by hyperproliferating fibroblasts and inflammatory cells, ultimately leading to hazardous consequences. Therefore, the impaired epithelium repair processes are currently recognized as central events in IPF, which, however, can hardly be rescued by conventional anti-fibrotic drugs targeting fibroblast cells (e.g., Pirfenidone or Nintedanib)3,4. This encouraged the development of new therapeutic strategies aiming to restore the gas exchange function of IPF lung5.
Stem/progenitor cells possess significant potential to reconstitute the lost lung epithelium and restore lung function. Following lung epithelium injury, various types of epithelial stem and progenitor cells in the airway and alveoli can be activated to initiate tissue repair6,7,8,9,10,11,12,13,14,15,16,17,18,19,20. Among them, a population of stem/progenitor cells, which line the basal layer of the airway epithelium and express the P63 and KRT5 genes, have been extensively studied. In bronchi, P63+ KRT5+ airway basal progenitor cells are responsible for the homeostasis maintenance and regeneration of airway epithelium. Interestingly, upon severe lung damage, these progenitor cells in the airway could be mobilized and migrate into the more distal alveolar area. Their exact behavior and function in the alveolar area are relatively controversial. Some studies believed that airway basal progenitor cells in the alveolar region have the potential to regenerate type II and type I alveolar epithelium to facilitate the lung repair process21,22,23,24; while in recent years others argued that these airway basal progenitor cells in the alveolar region tend to form dysplastic structures such as “basaloids”, bronchiolization, or honeycomb cyst structures within the space25,26,27,28,29,30, which would impair the lung repair process and even directly drive the fibrotic process. In addition, anatomic and functional differences may exist between airway basal progenitor cells among different species, such as mice versus humans. In the current study, we tried to systematically evaluate the function of human airway basal progenitor cells in the alveolar area and compare them with progenitor cells from other animal species, in order to explore the potential therapeutic approaches based on airway basal progenitor cell transplantation.
Results
Activation of airway basal progenitors in injured mouse and non-human primate lung
Firstly, we studied the P63+ KRT5+ airway basal progenitor cells in animal models. In the mouse lung, following injury induced by the pro-fibrosis drug bleomycin, we observed the persistent presence of Krt5+ cells in the alveolar area (Supplementary Fig. 1a), in consistency with previous reports16. To confirm whether some of the Krt5+ cells in the alveolar space were originated from resident basal cells, we utilized the Krt5 CreERT2-Gt (ROSA)26Sortm4(ACTB-tdTomato-EGFP) mouse line, which allowed efficient lineage labeling of airway basal cells before bleomycin-induced injury by tamoxifen administration. At 30 days post-injury, we observed GFP+ cells distributed in the airway epithelium. We also observed GFP+ cells with thin, flat morphology lining the alveolar area, which however do not express known alveolar cell markers (Supplementary Fig. 1b). Although the number of GFP+ cells in the alveolar space was relatively scarce, this data indicated that at least some of the pre-existing airway basal progenitor cells could be activated and enter the alveolar area after fibrotic lung injury. This finding was different from observations in an influenza model22 and consistent with previous reports showing that mouse Krt5+ cells could not efficiently regenerate mature alveolar epithelial cells. It also suggested that other Krt5− cell populations could give rise to Krt5+ cells following lung injury20,31,32,33.
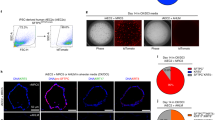
To investigate the relevance of this observation in mice to higher animals, here we used a cynomolgus macaque as a study model of pulmonary fibrosis. The lung architecture of non-human primates is almost identical to humans. Unlike mice, the airway basal progenitor cells in monkeys and humans are distributed throughout the major airways down to the distal respiratory bronchioles but absent from the alveolar area in healthy monkeys. By administering bleomycin directly into the monkey lung, we observed extensive injury and fibrosis in the monkey lung at 71 days post-bleomycin administration, as observed through CT imaging and confirmed by histological staining (Fig. 1a, b). Similar to that in mice, we observed the expansion of airway basal progenitor cells in the alveolar region of the injured monkey lung (Fig. 1c, d and Supplementary Fig. 1c). By utilizing the previously established progenitor cell cloning method22,34, we isolated P63+ KRT5+ progenitor clones from the injured monkey lung, and their number was approximately 6-fold more than that from control lung (Fig. 1e). Altogether, these data confirmed that in mice and primates, some endogenous airway basal progenitors located in the airway can be activated and mobilized into the alveolar area after lung damage.
a Representative images of consecutive CT scans showing the presence of local lesions (red arrow) before a 71 days post first bleomycin injured in the macaque. b Masson’s trichrome staining for collagen deposition (blue) on histological section of macaque monkey lung 71 days post bleomycin injury. Scale bar, 80 μm. c Immunofluorescence images of KRT5+ pods in monkey lung injured by bleomycin with anti-KRT5 (red) staining and DNA counterstain (DAPI, blue). Scale bar, 100 mm. d Quantification of macaque KRT5+ cells numbers in the alveolar area of control or injured macaque monkey lung (n = 6 technical replicates). Data are presented as mean ± SEM. Statistical analysis was performed using a two-tailed t test (***P < 0.001). e Colony forming numbers of macaque KRT5+ P63+ cells derived from control or injured macaque monkey lung by in vitro culture (n = 3 biological replicates). Data are presented as mean ± SEM. Statistical analysis was performed using a two-tailed t test (****P < 0.0001). f Representative lung CT images of sampling and damaged area of the IPF patients. Green circle: sampling area; red circle: damaged area. g Left, Brightfield image of human airway basal progenitor cells clones. Right, immunostaining of airway basal progenitor cell clones with anti-KRT5 (green), anti-P63 (red) and DNA counterstain (DAPI, blue). Scale bar, 100 μm. h The automated progenitor cell culture system. Left, an overview of the system; Right, a 6-axis robotic arm mimicking the human arm movements. i Representative images of human airway basal progenitor cells cultured by junior workers or by automated cell culture system. Scale bar,100 μm. j Unsupervised principal component analysis (PCA) of whole genome transcriptome of airway basal progenitor cells derived from healthy donors or patients. Each dot represented the cell clone derived from an individual donor. The dotted line indicated the clones which were transcriptomically similar to healthy donors. k Correlation analysis of gene expression level with pulmonary function of patients. The shaded area represents the 95% confidence interval around the regression line, reflecting the standard error of the fitted values.
Clone airway basal progenitors from relatively healthy zone of patient lungs
Next, to investigate airway basal progenitor cells in humans, we cloned progenitors from both healthy donors and patients with various lung diseases as previously reported22,34. For those patients, to obtain progenitor cells less affected by diseases, tissues were collected from 3-5th orders of the airway of the relatively healthy zone of the lung and grown for progenitor cell culture. To avoid sampling pathological tissues from IPF patients35, bronchoscopic brushing sampling was performed exclusively in the airway from the radiologically healthy upper lobes, as determined by CT imaging prior to sampling (Fig. 1f) and confirmed by direct observation under bronchoscope. Airway basal progenitor cells cloned from the bronchoscopic brushing samples form colonies with P63 and KRT5 expression (Fig. 1g). Similar sampling and cell clone strategy was applied for patients with chronic obstructive pulmonary disease (COPD) and non-cystic fibrosis bronchiectasis (Supplementary Fig. 1d). The cells cloned from various donors exhibited generally similar morphology (Supplementary Fig. 1e). To minimize the batch effect associated with manual cell culture and improve working efficiency, we developed an automated cell cloning system. This system utilized a 6-axis robotic arm to mimic the manual handling of cells (Fig. 1h and Supplementary Fig. 1f). The verification data demonstrated that the airway basal progenitors cultured using the robotic system exhibited equally good quality as junior workers (Fig. 1i).
In total, we cloned airway basal progenitor cells from 44 donors, including 6 healthy donors and 12 patients with IPF, 13 with COPD, and 13 with bronchiectasis for transcriptomic analysis. The demographic information of patients was briefly listed in Supplementary Table 1. Unsupervised principal component analysis (PCA) of the whole transcriptome revealed that in general, there were no statistically significant differences between healthy donors and patients with COPD (P-value = 0.11), IPF (P-value = 0.16), or bronchiectasis (P-value = 0.33). However, great heterogeneity was observed among donors. Although most patient clones exhibited similar transcriptomic profiles to healthy controls, there were approximately 25% of patient clones demonstrated distinct differences, particularly in 4 IPF clones and 4 COPD clones (Fig. 1j and Supplementary Fig. 1g), consistent with previous reports about variant clones in patients26,28. Furthermore, when all patients were pooled together for analysis, we observed that the P63 expression level of cells showed a significant negative correlation with the Forced expiratory volume in 1 second (FEV1) of patients (Fig. 1k), suggesting that obstruction or inflammation of the human airways may upregulate the P63 gene expression in the airway basal progenitor cells, in line with previous observations in mice30. No significant correlation between the P63 level and the Diffusing capacity of the lung for carbon monoxide (DLCO) was observed, suggesting that the alveolar damage of the patient lung had less direct influence on the airway basal progenitor cells in the airway.
We also noticed that compared to healthy donors, the COPD progenitor cell clones exhibited high expression of genes associated with cortisone responses (Supplementary Fig. 1h). This observation may be attributed to the long-term inhaled cortisone therapy typically administered to COPD patients, which led to alterations in the gene expression profile of the progenitor cells. It is worth noting that the COPD clones did not exhibit high expression of known pro-inflammatory variant cell signature genes, such as the CXCL family chemokines, interleukins, and interferons26 (Supplementary Fig. 1i), consistent with their origination from relatively healthy zone of lung. In the IPF patients, the progenitor cell clones demonstrated high expression of genes involved in anti-microbial responses, which could be linked to the frequent upper airway infections observed in IPF patients36(Supplementary Fig. 1j). Importantly, these IPF cells cloned from healthy upper lobes did not exhibit high expression of previously reported pro-fibrotic variant cell signature genes, indicating that they were unlikely to be pathogenic and were likely to be relatively normal27,28,37(Supplementary Fig. 1k).
Engrafted cells generated saccule-like structures
To delineate the behavior of human airway basal progenitors in the alveolar area, we transplanted GFP-labeled human airway basal progenitors into the lungs of immune-deficient mice that had been subjected to a bleomycin-induced injury (Fig. 2a). Different from the previous intratracheal delivery method, we employed syringe ventilation on the mice to ensure effective distribution of the progenitor cells throughout the distal airway. The identity of the engrafted human cells in the murine alveolar area was subsequently confirmed through immunostaining using antibodies that specifically recognized human antigens (Fig. 2b and Supplementary Fig. 2a). The transplanted cells, labeled with GFP-expression lentivirus, were detectable through immunostaining on histological sections at both 8- and 35-days post-transplantation (dpt). Importantly, these engrafted cells were exclusively localized in the damaged or repairing lobes of the lung, with no presence observed in the healthy lobes (Fig. 2c and Supplementary Fig. 2b, c). We also confirmed that no engraftment would be detected if cells were transplanted into healthy mouse lungs (Supplementary Fig. 2d, e).
a Direct fluorescence image of mouse lung transplanted with human GFP-labeled human airway basal progenitor cells showing engraftment, captured under a stereomicroscope. Scale bar, 1 mm. b Immunofluorescence images of engrafted human cells in mouse lung with human-specific antibodies anti-HuLamin (red) immunostaining. Nuclei counterstain, DAPI (blue). Scale bar, 1 mm. c IHC staining of engrafted cells (brown) in mouse lung 8 (left) or 35 days (right) post-transplantation (dpt) with anti-GFP antibodies. Scale bar, 1 mm. Enlarged inset: high magnification views of the representative pod and saccular structures. Scale bar, 100 μm. d Mean linear intercept (MLI) of native human (hAlveoli) and murine alveoli (mAlveoli) as well as human saccules (Sac) (n = 13, 6, 23 technical replicates from 3 biological replicates). Data are presented as mean ± SEM. e Wall thickness of native human (hAlveoli) and murine alveoli (mAlveoli) as well as human saccules (Sac) (n = 50, 50, 275 technical replicates from 3 biological replicates). Data are presented as mean ± SEM. f 3D visualization showing the connection of human pods and saccules(sac) with mouse airway. 3D reconstruction was performed based on anti-GFP IHC stained serial sections of mouse lungs, which were transplanted with human airway basal progenitor cells and analyzed on 8 dpt (left) or 35 dpt (right), respectively. g Schematic diagram showing the process of lung respiratory tract growth and saccule formation from mouse airway. h H&E staining (left) and anti-GFP IHC staining (right) of engrafted human cells on a lung tissue section for spatial transcriptomic analysis. Green circle: saccule zone, blue circle: pod zone. Scale bar, 300 μm. i Spatial visualization of human cells in lung section which was noted using human reference genome (GRCh38) by spatial transcriptomic analysis. Scale bar, 1 mm. j Spatial visualization of various gene expression in lung sections by spatial transcriptomic analysis. Green circle: saccule zone, blue circle: pod zone. Scale bar, 300 μm.
On 8 dpt, the engrafted cells exhibited large-scale engraftment within the alveolar area. The majority of these cells displayed a cuboidal morphology and formed structures resembling “pods”. These pods-like arrangements closely resembled the endogenous Krt5+ pods observed in fibrotic mouse lungs, as well as the KRT5+ pods found in the lungs of patients with IPF38. On 35 dpt, the engrafted cells underwent a notable transformation, adopting a flattened morphology and organizing into acinar structures like “saccules” within the alveolar area (Fig. 2c). Notably, these saccular structures presented a single-layered morphology distinct from the previously described “bronchiolization” structures characterized by bi-layered cuboidal or columnar cells8,39. The mean linear intercept (MLI) of the saccules was 92.55 μm (IQR: 76.92, 108.1), which was a smaller size than human alveoli (mean: 193.40, IQR: 162.4, 232.5) but larger than mouse alveoli (mean: 59.14, IQR: 53.57, 64.56) (Fig. 2d). In addition, the median minimal thickness of the saccule walls was 10.83 μm (IQR: 8.91, 14.22). This measurement closely approximated the thickness observed in human alveoli (median 8.29, IQR: 7.28,12.47) and surpassed that of mouse alveoli (median: 5.16, IQR: 4.23, 6.25) (Fig. 2e).
In order to understand the spatial distribution of engrafted cells, we performed a 3-dimensional (3D) visualization of the mouse lungs. We found that the pods and saccules generated by human cells were predominantly connected with the mouse bronchi, forming new respiratory tracts originating from the mainstem bronchi (Fig. 2f). On 8 dpt, the majority of the pods were distributed within 500 μm surrounding the main bronchi. By 35 dpt, most of the structures within the 500 μm radius had transitioned into saccules, and the pods extended to more distal areas (500–2000 μm from the main bronchi) (Supplementary Fig. 2f). These findings supported a working model suggesting that the “pod-to-saccule” transition occurs in a directional manner from the proximal to distal airway, with undifferentiated airway basal progenitor cells persisting at the migrating tips (Fig. 2g). Intriguingly, this proximal-distal gradient of cell morphology change pattern closely might mirror the sacculation developmental process observed during fetal lung development8,40,41, suggesting that the re-epithelialization process recapitulates, to some extent, the lung developmental process.
Barrier function analysis of airway basal progenitor cell post transplantation
To further investigate the characteristics and potential function of the saccule structure, we performed a high-resolution spatial transcriptomic analysis on the 35 dpt lung section. Immunohistochemistry (IHC) staining confirmed the location of human pods and saccules formed by engrafted cells (Fig. 2h), which was corroborated by sequencing analysis of human reference genome number count (Fig. 2i). Spatial transcriptomic analysis revealed distinct gene expression patterns in the saccular zone compared to the pod zone. The pod zone exhibited much higher levels of P63 and KRT5 gene expression than the saccular zone. In contrast, the saccular zone exhibited a higher expression level of tight junction gene CLDN4. Previous studies showed that in the human lung, CLDN4 was highly expressed in both type I alveolar epithelial cells (AEC1) and type II alveolar epithelial cells (AEC2), whose overexpression could augment alveolar epithelial barrier function and contribute to clearance of alveolar fluid42. Meanwhile, the saccular zone also expressed high levels of other tight junction genes, including CLDN3, CLDN7, ZO-2, and JAM, which allow the assembly of tight junction complexes. The saccular zone also highly expressed water channel gene AQP5, which could regulate fluid homeostasis and therefore allow the formation of an intact barrier together with tight junction proteins in the saccular zone43. (Fig. 2j and Supplementary Fig. 2g, h).
Next, we investigated whether engrafted human cells could contribute to injured mouse lung function. First, we evaluated the epithelium intactness of the mouse lung. Analysis of the epithelial cell marker CDH1 (E-Cadherin) and CLDN4 indicated that the fibrotic honeycomb zone was devoid of epithelium, especially the epithelium with tight junction gene expression. In contrast, the transplantation of human airway basal progenitor cells efficiently re-epithelialized the lung in the saccular zone with CLDN4+ epithelial cells (Fig. 3a). In addition, the human saccular zone exhibited very low expression levels of mouse fibroblast markers (Col1a1 and Fn1) or immune cell markers (Cd45 and Cd11b) (Fig. 3b, c and Supplementary Fig. 3a). In contrast to the saccular zone, some of the human pod zones still expressed high levels of mouse fibroblast genes similar to the damaged area (Fig. 3c). These observations were further confirmed by histological analysis (Supplementary Fig. 3b). Interestingly, in comparison to the honeycomb area, significantly higher expression levels of the mouse type II alveolar cell marker SPC were detected in the human saccular zone (Fig. 3c). Abundant expression of the mouse alveolar capillary marker gene Car444 was also observed in the human saccular zone (Fig. 3c), which was consistent with the immunostaining data for the blood vessel markers CD31 and CD34 (Fig. 3d). These data indicated that the engrafted human airway basal progenitor cells could efficiently re-epithelialize the murine alveolar area.
a Spatial visualization of the mouse (red) and human (green) epithelial cell marker CDH1 (E-cadherin) and CLDN4 gene expression in different lung zones. Yellow circle: damaged honeycombing zone; red circle: normal alveolar tissue zone; green circle: saccular zone; blue circle: pod zone. Scale bar, 1 mm. b Spatial visualization of mouse fibrotic marker Collagen1 (Col1a1) gene expression in different lung zones. High magnification indicated the Collagen1 signal in pod and saccule zones. Scale bar, 1 mm. c Expression level of multiple mouse marker genes in different lung zones. Normal, normal alveolar area; Damage, damaged honeycombing zone; Sac, saccular zone (n = 5 technical replicates). d Immunofluorescence images of mouse capillary vessels in the saccular zone with anti-GFP (green), anti-CD31 (red), and anti-CD34 (red) antibodies co-staining and DNA counterstain (DAPI, blue). Scale bar, 50 μm. e Barrier function assays of airway basal progenitor cells in vitro. Left, schematic diagram of the barrier function transwell assays; middle, TEER assay of monolayers formed by airway basal progenitor cells or A549 cells before and after Triton X-100 treatment (n = 6 biological replicates); right, assay of fibroblast migration through monolayers formed by airway basal progenitor cells or A549 cells (n = 6 biological replicates). f Quantification of the inflammatory area of mouse lung with or without airway basal progenitor cell transplantation (n = 8, 12, 15 technical replicates from 3 biological replicates). g Ashcroft score of lung fibrosis and quantification of the relative fibrotic area by Masson’s trichrome staining of mouse lungs with or without airway basal progenitor cell transplantation (n = 8, 12, 15 technical replicates from 3 biological replicates). h O2 partial pressure to CO2 partial pressure ratio of mouse arterial blood with or without human airway basal progenitor cell transplantation (n = 9 biological replicates). i Survival curve of mice challenged by bleomycin with or without airway basal progenitor cell treatment (n = 3, 4, 5 biological replicates). Statistical analysis was performed using the Log-rank (Mantel-Cox) test. c, e–h All data are presented as mean ± SEM. Statistical analysis was performed using one-way ANOVA with Tukey’s post hoc test and two-way ANOVA with Sidak’s multiple comparisons test (for the middle panel of E) (**P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, no significant difference).
We also used transwell assays to study the barrier function of human airway basal progenitor cells in vitro. The progenitor cells were grown as monolayers on the semi-permeable membranes of the well until reaching 100% confluency (Fig. 3e). The transepithelial electrical resistance (TEER) technique was then applied to measure the barrier integrity. The data showed that the TEER value of human airway basal progenitors was approximately 360 Ω/cm2, which was 5-fold higher than the human alveolar epithelial cancer cell line A549. In another transwell assay, we applied human fibroblasts MRC5 on the epithelial cell monolayer in the upper chamber and counted the number of fibroblast cells migrated to the lower chamber 24 hours afterward. The results showed that in contrast to the A549 monolayer cells, the progenitor cell monolayer almost completely blocked the fibroblast invasion into the lower chamber (Fig. 3e). Altogether, the above findings suggested that the epithelial barriers established by airway basal progenitor cells have potent function to block the progression of fibrosis.
To determine whether re-epithelialization by airway basal progenitor cell transplantation could benefit the mouse lung, we analyzed the fibrotic status of the mouse lung 3 weeks post-transplantation. The data showed that cell transplantation significantly decreased the inflammatory area of lung (Fig. 3f) and the level of lung fibrosis (Fig. 3g and Supplementary Fig. 3c). Although pulmonary function analysis in mice indicated no significant improvement in mouse lung vital capacity, inspiratory capacity or forced expiratory volume, the mouse arterial blood analysis showed statistically significant increase in the O2 partial pressure/CO2 partial pressure ratio compared to the placebo control (Fig. 3h and Supplementary Fig. 3d). Survival curve analysis showed that cell transplantation effectively avoided the death of mice with lethal dose of bleomycin injury (Fig. 3i). Altogether these findings suggested that the re-epithelialization of denuded or fibrotic lung tissue by airway basal progenitors could serve as a “band-aid” solution to facilitate the lung recovery process.
Fate mapping of engrafted human airway basal progenitors by single-cell transcriptomic analysis
To gain molecular insights into the fate specification process of human airway basal progenitor cells in the lung, we sorted the engrafted GFP+ cells at different time points using flow cytometry (Supplementary Fig. 4a), followed by 10x Genomics single-cell RNA sequencing analysis. Unsupervised clustering of the cells resulted in the identification of 7 distinct populations (Fig. 4a–c and Supplementary Fig. 4b). At 8 dpt, the major human cell populations in the lung included quiescent KRT5+ P63+ airway basal progenitors, proliferating KRT5+ P63+ airway basal progenitors and KRT5+ P63− cells. By 35 dpt, the proportion of quiescent KRT5+ P63+ airway basal progenitors remained stable, but the proportion of proliferating KRT5+ P63+ airway basal progenitors dramatically decreased to only 1.77% of total cells. There was a slight increase in the number of SCGB1A1+ human secretory cells at 35 dpt, and their presence in murine bronchioles was confirmed by immunostaining (Fig. 4d). In addition, a population of human ciliated cells was detected in sequencing data, and their distribution in murine bronchioles was confirmed by immunostaining as well (Fig. 4d). The ratio of differentiated secretory cells to ciliated cells was unbiased when compared to the human lung single-cell atlas45,46, which is a characteristic of normal airway basal progenitor cells25.
a Single-cell RNA-sequencing cellular cluster map of GFP+ human cell from 8 (left) and 35 (right) dpt mouse lungs. The pie plots showed the percentages of distinct cell clusters. b Feature plots showing the expression of multiple marker genes in distinct single-cell clusters of engrafted human cells. c Feature plots showing the expression of genes signatures in distinct single-cell clusters of engrafted human cells. d Immunofluorescence images of engrafted human cells with anti-GFP (green), ciliated cell marker anti-acetylated-tubulin (red), Club cell marker anti-CC10 (red) or tight junction marker anti-CLDN4 (red) antibodies staining. Scale bar, 50 μm. e Gene Ontology enrichment analysis of the differentially expressed genes identified in CLDN4+ CLDN3+ saccular cells. f Pseudo-time trajectory projected onto a UMAP of transplanted human cells using the quiescent KRT5+ P63+ airway basal progenitors as a starting point. Pseudo-time values are color-coded. g Density of each human cell population ordered by pseudo-time as computed by reversed graph embedding approach of Monocle3. h Schematic diagram showing the putative differentiation paths starting from KRT5+ P63+ airway basal progenitor cells predicted by the pseudo-time analysis. i Schematic diagram showing the putative re-epithelialization process in the fibrotic lung after human airway basal progenitor transplantation.
Consistent with the spatial transcriptomic data mentioned above, there were a large cluster of cells expressing tight junction gene CLDN4, which remained the most abundant human cell population throughout the period from 8 dpt to 35 dpt. Immunostaining confirmed the expression of CLDN4 protein in engrafted human cells forming saccular structure (Fig. 4d). This population of CLDN4+ cells also highly expressed other tight junction genes such as JAM and CLDN7 but did not express CLDN3 (Fig. 4b and Supplementary Fig. 4b). Interestingly, this population of CLDN4+ CLDN3− saccular cells also exhibited high expression of VEGFA, suggesting their potential contribution to the capillary vascularization (Supplementary Fig. 4b)47.
A small population of further differentiated CLDN4+ saccular cells started to express CLDN3 and ZO-2. These CLDN4+ CLDN3+ saccular cells also expressed sodium channel gene CLIC5/CLIC3 and water channel gene AQP5, which was consistent with the spatial sequencing data above. The proportion of CLDN4+ CLDN3+ saccular cell population increased threefold from 8 dpt to 35 dpt (Fig. 4a, b). Gene ontology analysis demonstrated that the CLDN4+ CLDN3+ saccular cells population highly expressed genes associated with tight junction, endothelial development, and the establishment of the endothelial barrier, suggesting their potential role in the reconstitution of the blood-air interface (Fig. 4e). Pseudo-time analysis showed that the CLDN4+ CLDN3+ saccular cells population was derived from the CLDN4+ CLDN3− cells. Secretory cells and ciliated cells were also found to be derived from CLDN4+ CLDN3− cells as well (Fig. 4f, g), which was consistent with previous reports48,49. The cell differentiation model was summarized in Fig. 4h. Of note, no RAGE+ or SPC+ cells were detected among the populations analyzed, suggesting that the engrafted human progenitors cannot fully differentiate into mature alveolar cells in mouse lungs (Supplementary Fig. 4b, c), in contrast to the previous claim in mouse-to-mouse transplantation model22.
Taken together, the above data indicated that human airway basal progenitor cells might not be able to differentiate into mature alveolar cells. In the alveolar area, most of the progenitors will gradually lose their P63 and KRT5 gene expression, give rise to CLDN4+ cells with tight junction assembled, and a small proportion of them can further differentiate into AQP5+ /CLIC5+ saccular cells expressing even more tight junction markers such as CLDN3. The working model of human airway basal progenitor fate commitment and epithelial barrier function establishment were summarized in Fig. 4i.
Clone airway basal progenitor cells from other mammals and birds
In order to understand the evolutionary conservation of lung re-epithelialization mechanism, we cloned the P63+ airway basal progenitor cells from the airways of adult mouse, goat, canine, pig, and monkey. Progenitor cells from all species could be long-term passaged in our culture system except the one from monkey for unknown reason (Supplementary Fig. 5a). The progenitor cells from the other species were labeled with GFP using lentivirus and then transplanted into bleomycin-injured mouse lungs (Fig. 5a). The data showed that the goat clones exhibited the highest engraftment in the mouse lungs, reaching a chimeric ratio of up to 40% (Fig. 5b), in comparison to less than 10% for human clones. Immunostaining with GFP antibody revealed that almost the entire damaged mouse lobe was replaced by goat tissue (Fig. 5c). At 35 dpt, the engrafted goat cells formed saccule structures that three-dimensionally connected to the mouse airways (Fig. 5d). Morphologically, the regenerated goat saccular wall had 2-3 layers of cells, which was thicker than the wall of mouse alveoli but similar to native goat alveoli (Fig. 5e, f). The size of the goat saccules was also much larger than mouse alveoli and similar to goat alveoli. The lung-injured mice transplanted with goat clones exhibited prominent improvement in pulmonary inflammation and fibrosis and were close to complete recovery of their pulmonary gas exchange function (Fig. 5g–j and Supplementary Fig. 5b). Comparatively, the porcine and canine clones had lower chimeric ratios in mouse lungs. The canine saccules exhibited morphology similar to human alveoli, while the porcine saccules showed expression of the classical type 1 alveolar cell marker RAGE (Supplementary Fig. 5c–e).
a Brightfield and direct fluorescence images of mouse lungs post-transplantation with GFP-labeled mouse, goat, porcine, and canine airway basal progenitor cells. Scale bar, 1 mm. b Cell engraftment efficiency of goat, porcine, and canine clones in mouse lung 8 days and 35 days post-transplantation (n = 3). Data are presented as mean ± SEM. Statistical analysis was performed using two-tailed t tests to compare the 8dpt and 35dpt groups within each species. (*P < 0.05, **P < 0.01, ***P < 0.001). c Engrafted goat cells in mouse lungs by anti-GFP (green) immunostaining with DNA counterstain (DAPI, blue). Upper, without cell transplantation; lower, with goat airway basal progenitor cell transplantation. Scale bar, 1 mm. d 3D reconstruction of anti-GFP IHC stained serial sections of mouse lung, which were transplanted with goat airway basal progenitor cells (red) and analyzed on 35 dpt. The blue color indicated mouse bronchi, red color indicated goat cells. e Immunofluorescence images of engrafted goat cells in mouse lung by anti-GFP (green), anti-AQP5 (red) immunostaining, and DNA counterstain (DAPI, blue). Scale bar, 100 μm. f H&E staining of the native goat alveoli section. Scale bar, 100 μm. g Quantification of the inflammatory area of mouse lung with or without goat airway basal progenitor cell transplantation (n = 4, 6, 6 technical replicates from 3 biological replicates). h–j Arterial blood gas analysis of murine arterial blood with or without goat airway basal progenitor cell transplantation. Each dot indicated an individual mouse (n = 3, 4, 4 biological replicates). g–j All data are presented as mean ± SEM. Statistical analysis was performed using one-way ANOVA with Tukey’s post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, no significant difference). k H&E staining of the native pigeon alveoli section. Left, scale bar, 40 μm. Right, scale bar, 80 μm. l Immunofluorescence images of native pigeon lung with anti-α-SMA (green), and air capillary marker anti-CK7 (red) immunostaining and DNA counterstain (DAPI, blue). Scale bar, 50 μm. m Immunofluorescence images of native pigeon airway with anti-KRT8 (green), anti-P63 (red) staining, and DNA counterstain (DAPI, blue). Scale bar, 50 μm. n Immunofluorescence images of engrafted pigeon cells in mouse lungs with bird-specific antibody anti-chicken α-Actin (green) and air capillary marker anti-CK7 (red) co-staining. Scale bar, 20 μm.
Unlike mammalian lungs, avian lungs do not have a typical alveolar structure. Instead, birds utilize tiny parabronchi and CK7+ air capillaries for efficient air exchange (Fig. 5k, l), supporting their high oxygen needs during fast flying. Similar to most mammals, the major airways of birds also have airway basal progenitor cells (Fig. 5m). With a modified culture medium supplemented with chicken serum and egg white, we successfully cloned P63+ airway progenitor cells from the secondary bronchi of adult pigeons (Supplementary Fig. 5a). After intrapulmonary transplantation, the pigeon progenitor cells exhibited some engraftment in mouse lung, as shown by staining of the lung tissue by a validated bird-specific α-Actin antibody. The engrafted pigeon cells gave rise to some air capillary-like respiratory unit structures and expressed the bird air capillary marker CK7 (Fig. 5n and Supplementary Fig. 5f). Of note, the bird-to-mouse engraftment ratio was quite low, probably due to the evolutionary gap between the two animal classes. All the data presented above demonstrated that the P63+ progenitor-based re-epithelialization process may be an evolutionarily conserved mechanism among tetrapods during the water-to-land transition50.
Safety evaluation of human airway basal progenitor cells by intrapulmonary transplantation
As human airway basal progenitor cells showed potential to facilitate the lung repair process, we investigated the pre-clinical safety of patient-derived airway basal progenitor cell in animals by performing long-term toxicity, tumorigenicity, and allergenicity evaluations. In previous studies, the safety and efficacy of intrapulmonary cell transplantation were evaluated in short-term experiments in rodents and monkeys51,52. Here, in the long-term toxicity experiment, the data showed that all mouse organs remain normal, and most tested blood/urine parameters remained stable within 57 days post-cell transplantation. A few exceptions showed significant alterations, including increased monocyte counts, decreased serum urea, increased serum glucose, and increased serum calcium levels (Supplementary Figs. 6a, b, and 7a). These alterations might be related to changes in immune and metabolic status following xeno-transplantation. Tumorigenicity assays showed obvious tumor formation in immune-deficient nude mice within 6 months after HeLa cancer cell line xenograft subcutaneously, but none after progenitor cell xenograft (Supplementary Fig. 7b, c). The allergenicity assays showed that the positive control drug (mercaptobenzothiazole) led to an allergenic response in guinea pigs, while progenitor cell treatment did not (Supplementary Fig. 8a). These data altogether indicated that transplantation of patient-derived airway basal progenitor cells in mice would not lead to obvious deleterious change of lung or other organs.
Discussion
Re-epithelialization refers to covering of wound with new epithelium, which is a common critical process for injury recovery of various organs53,54. Impaired re-epithelialization is associated with non-healing wounds and is linked to chronic progressive pulmonary diseases such as emphysema and IPF8,55,56.In the current study, we observed that transplantation of airway basal progenitor cells was effective in re-establishing the epithelial surface of injured lungs. Furthermore, we found that re-epithelialization could control the progression of inflammation and fibrosis, leading to improved gas exchange in murine models of lung fibrosis. Importantly, although the re-epithelialization process could not provide permanent benefits as true regeneration, the beneficial effects of progenitor transplantation in mice appeared to be persistent.
The current study highlights the function of P63+ airway basal progenitor cells in the alveolar area, which has been a controversial issue for decades. Now, it is getting clear that the potential role of airway basal progenitor cells in either lung long-term regeneration, temporary repair, or even tissue remodeling may involve multiple mechanisms57,58. Interestingly, a recent study identified a population of bleomycin injury-induced P63+ progenitor cells in mice which originate from airway secretory cells and could further differentiate into mature alveolar epithelial cells59. As we know, the intrapulmonary airways in mice rarely have P63+ basal progenitor cells, therefore, it could be necessary for mice to mobilize and convert the airway secretory cells into P63+ progenitors for the lung repair process. In contrast, the intrapulmonary airways in humans, however, harbor a highly abundant population of P63+ airway basal progenitors, which could directly contribute to the lung repair process according to our findings. Of note, our current single-cell transcriptomic data and high-resolution spatial transcriptomic data showed that the human airway basal progenitor cells could not give rise to mature RAGE+ type I alveolar cells or SPC+ type II alveolar cells. The possible reasons could be: (1) Unlike the progenitor cells in fetal or infant lung, the adult P63+ progenitor cells might have lost their potential to fully differentiate into mature alveolar epithelial cells. (2) The fibrotic context in mouse lungs (e.g., the TGF-β enriched microenvironment) might not support the full maturation of progenitor cells. Future studies aiming to rejuvenate the adult progenitor to its infancy status or engineering the lung microenvironment could have a positive effect on the maturation of transplanted progenitor cells.
Our data also confirmed that the previously known KRT5+ P63+ “pods” were likely to be non-functional dysplastic structures in the lung, but their progeny could form functional saccular structures. This finding helps to explain previous studies showing that P63+ cells had little beneficial effect on lung recovery within 18 days of their appearance in the alveolar space27,30, as the maturation from pods to saccular structures generally takes 3 weeks according to our observations. Furthermore, our study indicated that the saccules, instead of pods, exhibited a high expression level of multiple tight junction genes, including CLDN4, CLDN3, CLDN7, and ZO-1/2 demonstrated barrier function to resist inflammatory cell and fibroblast invasion. Moreover, the barrier function of P63+ cells has been reported in previous studies12,60,61, and here we further investigated the underlying mechanisms and their differentiation lineages at single-cell resolution. These results were consistent with a previous study demonstrating rat airway basal progenitor cells could efficiently re-epithelialize the decellularized lung scaffold62.
Moreover, we also showed that the P63+ airway basal progenitors cloned from other mammals and even birds could also regenerate air sac-like structures in mice, which more or less recapitulated their native morphology in corresponding species. These data suggest that the airway basal cell-based lung re-epithelialization process may be a conserved mechanism throughout the evolution of tetrapods. Considering that xeno-transplantation of specific pathogen-free animal cells or organs is becoming a realistic therapeutic strategy63, our discovery of goat airway basal progenitor cells with extraordinary lung re-epithelialization function, proposed the possibility of xeno-transplantation therapy in the future, if the inter-species immune barrier could be properly overcome. However, it must be admitted that, currently, the more realistic strategy to help patients with irreversible lung damage might be autologous airway basal progenitor cell transplantation64. Our data here also confirmed the 3-month long-term safety of progenitor cell intrapulmonary transplantation, which warrants their test in clinical trials. Future co-administration using other transplantable pluripotent stem cell progenies, mesenchymal stem cells, or organoids65,66,67,68,69,70,71,72 together with airway basal progenitor cells might further improve the potential therapeutic effect of airway basal progenitor cells, offering hope for achieving full regeneration of injured human lung. Moreover, one limitation of the current work is the lack of detailed medical history and drug use information of the patients (e.g., corticosteroid use in COPD patients and immunosuppressive medications in IPF patients), which although might not impact the major conclusion of the current study, should be addressed in further studies with larger sample size and more comprehensive patient clinical information.
Methods
Human tissue
The study was approved by the institutional review board of Tongji University (TJAA06221101) and Shanghai East Hospital Human Cell Clinical Research Ethics Committee approval (EC. D(BG).016.01.1), in accordance with the Declaration of Helsinki. Written informed consent from each participant was obtained before the tissue sampling procedure. The participants recruited for cell isolation included 6 healthy volunteers with no history of pulmonary diseases and 13 patients with bronchiectasis, 13 patients with COPD, and 12 patients with IPF, which were diagnosed following the ATS/ERS guidelines. The group comprised 32 males and 12 females. The reported races were 43 Chinese and 1 Spanish. The collection of 3-5th order bronchi tissue was performed using bronchoscopic brushing.
Mouse
6 ~ 10-week-old male NOD-SCID mice were obtained from Shanghai SLAC Laboratory Animal Co. Ltd. and were housed under specific pathogen-free conditions within the animal care facility at the Center of Laboratory Animal, Tongji University in Shanghai, China (TJAA06221101). The animals were maintained in a well-ventilated environment with controlled conditions, including a room temperature of 20–26 °C, relative humidity of 40–70%, and a 12 h light/dark cycle. All animal experiments followed guidelines set by the NIH, and the Institutional Animal Care and Use Committee of Tongji University provided oversight for the animal experiments, ensuring compliance. Pulmonary fibrosis was induced in NOD-SCID mice by intratracheally administering 3 U/kg body weight of bleomycin (Selleckchem, USA) in a sterile phosphate-buffered saline (PBS) volume of 40 μL. Similarly, C57BL/6 J mice were subjected to 6 U/kg body weight of bleomycin (Selleckchem, USA) in a sterile PBS volume of 40 μL. After 7 days of bleomycin treatment, airway basal progenitor cells suspended in 40 μL saline were intratracheally transplanted into the mouse lungs, with each mouse receiving 1.5 million cells. Afterward, positive syringe ventilation was applied on the mouse trachea using a 20 mL syringe. Approximately 150 mL air was given to the mouse lung to facilitate cell distribution into the distal lung. The control group received an equivalent volume of sterile PBS. At specific time points, mice were sacrificed, and their lung tissues were extracted for analysis.
For the pre-clinical safety study, male and female C57BL/6 J mice (6–8 weeks old) purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (China), were maintained in SPF animal facilities of JOINN Laboratories infrastructure facilities (Suzhou) in compliance with Animal Welfare Act and Regulations (Public Law 99-198) promulgated by USDA. The protocols were approved by the institutional ethical committee under statement number ACU18-667. A total of 80 male and female mice were equally divided into two groups: a cell treatment group receiving P63+ cells from 3 donors or a control group that received normal saline. Mice in the cell treatment group received two deliveries of P63+ cells at a dose level of 6 × 107 cells/kg/delivery on day 0 and day 28. Changes in fur, skin, limbs, mouth, nose, and eyes; abnormal behavior in physical, physiological, or neurological activities; and changes in reactivity to handling or sensory stimuli, if any, were recorded daily. Bodyweight and food/water intake were recorded weekly. Twenty mice from each group were sacrificed and subjected to multiple examinations by the end of the administration period (day 30), and the other mice were sacrificed and subjected to examination by the end of the observation period (day 57). A whole set of the examination includes hematologic profiling, lymphocyte subset counting, blood coagulation test, serum biochemistry analysis, and quantification of serum immunoglobulin, complement, and inflammatory cytokines. The gross necropsy and histopathological examination of organs, which included the brain, heart, lungs, trachea, kidneys, liver, spleen, testis, and bone marrow, were carried out.
Monkey
A 60-month-old adult Cynomolgus macaque (M. fascicularis) sourced from Guangdong Qianyan Biological Science and Technology Co. Ltd. was utilized for this study. The macaque was housed at JOINN Laboratories infrastructure facilities in Suzhou, in accordance with the Animal Welfare Act and Regulations (Public Law 99-198) enforced by the United States Department of Agriculture (USDA). The animal was housed in a standard animal facility, with room temperature controlled between 18 °C and 26 °C, relative humidity maintained at 40–70%, and a 12 h light/dark cycle. The study protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Joinn Laboratories (Suzhou) (ACU18-1164).
To generate a monkey model of lung fibrosis, a monkey lung was injected twice with bleomycin (14.4 mg in total) into a specific point of the bronchial segment of the left lung using a flexible fiber-optic bronchoscope for infant use. The procedure was carried out by board-certified respiratory physicians. To assess lung damage, the macaque lung was monitored using small animal micro-CT before and after the injury. Afterward, the left lung of the monkey was collected for histopathological analysis. The right lung of this monkey was collected as well for control.
Guinea pigs
Four to Six-week-old male and female Hartley guinea pigs, weighing from 252 to 304 g, were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. and were kept in specific pathogen-free conditions within the animal care facility at JOINN Laboratories infrastructure facilities in Suzhou, in accordance with the Animal Welfare Act and Regulations (Public Law 99-198) enforced by USDA. The animals were maintained in a well-ventilated environment with controlled conditions, including a room temperature of 20–26 °C, relative humidity of 40–70%, and a 12 h light/dark cycle. The protocols were approved by the institutional ethical committee under statement number ACU18-844. For the experiment, 40 Hartley guinea pigs were randomly divided into three groups, including negative control (10 guinea pigs), positive control (10 guinea pigs), and P63+ cell group (20 guinea pigs). Animal skin was challenged with saline (0.2 ml per guinea pig), mercaptobenzothiazole (0.2 g/ml, 0.2 ml per guinea pig), and cell suspension (3 × 107 cell/mL, 0.2 ml per guinea pig). 14- and 21-days post initiation of sensitization, guinea pigs received the provocation challenge, with the dosage of 0.1 g/mL mercaptobenzothiazole or 1 × 107 cell/mL cell suspension, respectively. At 1-, 2- and 3-days post provocation, skin erythema, edema, and other abnormal reactions were observed and scored.
Goat, canine, pig and pigeon
All sampling procedures of this study strictly followed the “Guidelines on the Ethical Treatment of Experimental Animals” established by the Ministry of Science and Technology, China, and were approved by the IACUC of Regend Therapeutics (Shanghai) (S2021-010-10). We declare that this study complied with the Convention on Biological Diversity. Tissue samples were obtained from 4 species of adult animals, including: canine (Canis lupus familiaris), died from a traffic accident; goat (Capra aegagrus hircus), farmed and slaughtered for nutritional purposes; pig (Sus scrofa domestica), farmed and slaughtered for nutritional purpose; pigeon (Columba livia domestica), farmed and slaughtered for nutritional purpose. The aforementioned animals were raised in normal daily conditions in Shanghai.
Clone P63+ airway basal progenitor cells
To isolate human airway basal progenitor cells, the brushing samples were collected from the 3-5th level bronchi tissue of donors. After removing the sputum, the tiny tissues were directly digested in a dissociation buffer consisting of DMEM/F12 (Gibco, USA), 1 mg/mL protease (Sigma, USA), 0.005% trypsin (Gibco, USA), and 10 ng/mL DNase I (Sigma, USA). The specimens were incubated at 37 °C for one hour with gentle rocking. Alternatively, airway tissues collected from adult animals were dissected from lung tissue, cut into small pieces, and digested in the same dissociation buffer overnight at room temperature. The dissociated cells were passed through a 70 μm Nylon mesh (Falcon, USA) to remove aggregates and then washed twice with a cold F12 medium. Cell viability was assessed by trypan blue dye exclusion. The cell suspension was seed into the flask with lethally irradiated 3T3 (CRL-1658, ATCC) fibroblast feeder cells and cultured in a pharmaceutical-grade medium, including DMEM/F12, 10% FBS (HyClone), Pen/Strep, amphotericin, and growth factor cocktail as previously described22,73. For pigeon cell, the cultivation temperature was adjusted to 39 °C, supplemented with an additional 5% chicken serum (Sbjbio) and 5% sterile egg white. Other mammalian cells were cultured following the human cell culture protocol. Airway basal progenitor cell clones emerged within 3–10 days post-plating and were passaged every 5 to 7 days and passaged at a 1:7 ratio by trypsin or TrypLE. After 3–4 passages, airway basal progenitor cell clones were subjected to immunofluorescence staining for KRT5 and P63. GFP+ cells were labeled by pHIV-EGFP lentivirus infection and sorted by FACS.
Automated cell culture system
XLotus-I, an automated cell culture system prototype, was co-design and manufactured by Hair Biomedical and Regend Therapeutics. The machine was composed by an operation unit (including a 6-axis robotic arm, liquid filling table, and bottle opener), a culture unit (including CO2 incubator, phase-contrast microscope), a control unit (including programmable logic controller, and electrical module), and a storage unit (including storage rack, incubation table, injection pump, centrifuge). The operation protocol for cell culture, passage, and harvest were executed by a robotic arm using the pre-installed software, operated by trained staff through the touch screen of the control unit. All procedures of progenitor cell cloning, from tissue digestion to eventually cell harvest could be completely performed by the system according to the order input from the touch screen.
In vitro transwell assay
Airway basal progenitors and A549 (CCL-185, ATCC) were prepared for transwell assays by culturing up to 90–100% confluence in 8 μm transwell (Falcon, 353097) with an inoculation quantity of 6 × 104/per well. Next, the cells were cultured in a serum-free medium for 24 h with FGF10 (50 ng/mL, PeproTech), transferrin (5 μg/mL, PeproTech), hepatocyte growth factor (20 ng/mL, PeproTech), and 5% bovine serum albumin. TEER was measured using the Millicell ERS-2 (MERCK, MERS00002). For measurements, the electrode was overlaid by 400 µL DMEM medium. For the fibroblasts migration assay, 3 × 104 red fluorescent protein labeled MRC5 (CCL-171, ATCC) cells was added to the upper chamber, and 10 ng/mL TGF-β was added into the lower chamber to promote chemotaxis. The number of MRC5 cells migrated to the lower chamber was counted 24 hours afterward by direct fluorescence observation. Red fluorescent protein-positive MRC5 cells were labeled by pHIV-tdTomato lentivirus infection.
Lineage tracing experiments
Krt5 CreERT2-Gt (ROSA)26Sortm4(ACTB-tdTomato-EGFP) mice (The Jackson Laboratory, USA) were used to perform lineage tracing. For induction of Cre-ERT2 protein, 6 ~ 10-week-old mice were administrated with tamoxifen (3 mg, four times) dissolved in corn oil via an intra-peritoneal route every 2 days. Seven days after the last injection of tamoxifen, mice underwent lung injured by bleomycin. After 30 days, the mice were sacrificed, and their lung tissues were extracted for analysis.
Arterial blood gas measurements
For blood gas analysis, mice were anesthetized, and blood samples were collected from the carotid aorta using polypropylene syringes. The syringes contained 60 IU of dry, electrolyte-balanced heparin (PICO70; Radiometer Medical, Copenhagen, Denmark) to prevent coagulation. The collected blood samples were then analyzed for partial oxygen pressure (Po2), partial carbon dioxide pressure (Pco2), and oxygen saturation (So2) using an ABL90 Flex Blood Gas Analyzer (Radiometer Medical). For the experimental group, lung engraftment of GFP+ cells were assessed under a stereo fluorescence microscope. Only animals with successful GFP+ cell engraftment were included in the blood gas analysis.
Measurement of mouse lung function and hydroxyproline
To measure the mouse lung function, the mice were weighed and anesthetized with pentobarbital injection. When achieving the required level of anesthesia, surgical scissors were used to expose the neck skin, and blunt dissection with surgical forceps separated the connective tissue around the trachea, exposing the trachea. A small incision of about 1 mm was made in the exposed tracheal area with surgical scissors, and the tracheal cannula was inserted into the incision, secured with a cotton thread. The animal was then connected to the small animal ventilator (flexiVent, Scirecq) via an endotracheal cannula and mechanically ventilated at a respiratory rate of 150 breaths/min, a tidal volume of 10 mL/kg and a PEEP set at 3 cm H2O. In order to measure the inspiratory capacity (IC), vital capacity (VC), and forced expiratory volume of 50 ms (FEV50) in mice, we employed a rigorous method using the automated algorithm, which was carefully integrated into a mouse-specific mechanical scanning script. This script can be automatically repeated to ensure analysis accuracy and operational consistency, thereby promoting accurate assessment of lung function.
To evaluate the extent of tissue fibrosis, the total collagen content was measured using a hydroxyproline assay kit (Nanjing Jiancheng Bioengineering Institute, China).
Histopathology
For histopathological analysis, the lung tissue samples were fixed overnight at 4 °C using 4% paraformaldehyde and subsequently embedded in either paraffin or OCT based on specific requirements. The blocks were then sliced into 5 ~ 7 μm thick sections using a microtome (Leica Microsystems, Germany) at different planes. These sections were placed on poly-lysine-coated glass slides and stored at room temperature or 4 °C until further use. Hematoxylin and eosin (H&E), as well as Masson’s trichrome (Solarbio, Beijing) staining, was performed following standard protocols. The area of interest in each section was outlined and quantified using a light microscope (Leica Microsystems, Germany) equipped with an image-analysis system (Version 4.12.0, Leica Microsystems, Germany). The area of fibrosis and inflammation in Masson’s trichrome-stained sections and H&E-stained sections was quantified using ImageJ version 1.54 d (National Institutes of Health, USA). Standard Ashcroft score74 was also graded to assess the fibrosis of the lungs.
Immunostaining
For immunostaining, section slides underwent antigen retrieval in citrate buffer (Ph 6, Sigma-Aldrich, USA) heated in a microwave oven for 20 min. The following antibodies were utilized for immunostaining: GFP (1:500, ab6673, Abcam), STEM121 (1:200, Y40410, TAKARA), HuLamin (1:500, ab108595, Abcam), KRT5 (1:500, MA5-14473, Thermo Fisher), P63 (1:200, ab735, Abcam), SCGB1A1 (1:300, sc-365992, Santa Cruz), SFTPC (1:200, 10774-1-AP, Proteintech), AQP5 (1:300, ab92320, Abcam),CLDN4 (1:300, ab53156, Abcam), FOXJ1 (1:200, 14-9965-80, Thermo Fisher), RAGE (1:500, ab216329, Abcam), Ki67 (1:200, 550609, BD), CD31 (1:100, sc-1506, Santa Cruz), CD34 (1:1000, ab81289, Abcam), acetylated-α-Tubulin (1:1000, ab24610, Abcam), Alpha-SMA (1:200, 14395-1-AP, Proteintech), Cytokeratin 7 (1:200, sc-53263, Santa Cruz) and Anti-chicken Alpha-actin (1:200, A2543, Sigma-Aldrich).
For immunofluorescence staining, Alexa-conjugated Donkey 488/594 secondary antibodies (1:200, Life Technologies, USA) along with DAPI (Roche, USA) were used. The tissue slides underwent auto-fluorescence removal and were mounted using mounting media (Vectashield, Vector Labs, USA). Slides were observed under a fluorescent microscope (Olympus) or confocal microscope (Leica STELLARIS 5). On the other hand, immunohistochemistry staining involved the use of HRP-conjugated secondary antibodies (1:500, Abcam) and a DAB substrate kit (8059, CST) for detection. A nuclear counterstain was performed using hematoxylin.
3D visualization
Mouse lung 3D reconstruction was performed using the 3D Slicer version 4.11 (National Institutes of Health, USA) based on IHC staining images. After harvesting, the tissues were pre-fixed in 4% paraformaldehyde and subsequently processed for paraffin embedding. Serial sections with a thickness of 5 ~ 7 μm were prepared from the paraffin-embedded lung tissue, followed by IHC-DAB staining targeting the GFP antigen. Serial section image files in JPEG format were used as input for the reconstruction process.
Bulk RNA-sequencing and bioinformatics
Total RNA was extracted from P63+ cells using TRIzol Reagent (Invitrogen, Life Technologies, USA) following the manufacturer instructions. Subsequently, the extracted RNA was treated with DNase I (Invitrogen, Life Technologies, USA) to remove any contaminating DNA. The cDNA library construction and sequencing, the BGI-NSG platform was utilized. The sequencing data obtained was then subjected to filtering using SOAP nuke75. The filtering process involved the following steps: (1) Removing reads containing sequencing adapters, (2) Removing reads with a low-quality base ratio (base quality less than or equal to 15) higher than 20%, and (3) Removing reads with an unknown base (‘N’ base) ratio higher than 5%. Following the filtering steps, clean reads were obtained and stored in FASTQ format for further analysis. The clean data were mapped to the reference genome (Hgrc38) by HISAT76 (v2.1.0). The expression level of genes was calculated by RSEM (v1.2.8)77 and FPKM (Fragments Per Kilobase per Million) of each gene was calculated based on the length of the gene and read counts mapped to this gene.
PCA and differential expression analysis was performed using the DESeq2 R package (1.38.3), followed by clustering analysis using k-means. Significance analysis of PCA is performed using the permutational multivariate analysis of variance (PERMANOVA) method by the pairwiseAdonis R package. Adjusted P-value of 0.05 and an absolute fold change of 2 were set as the threshold for significant differential expression. Z-scores were calculated for FPKM values of each gene in bulk RNA sequencing data, representing the related expression of the genes. Pearson’s correlation scores were calculated to assess the relationship between the expression levels of the genes and the lung function index. Gene Ontology (GO) enrichment analysis of differentially expressed genes were implemented by the ClusterProfiler R package78. GO terms with a P-value < 0.05 were considered significantly enriched by differentially expressed genes and the results were visualized by the enrichplot R package using dot plots.
High-resolution spatial transcriptomic analysis
Mouse lung tissue was perfused with 2% paraformaldehyde and fixed overnight at 4 °C. The fixed tissue samples were then embedded in Tissue-Teck OCT. Using a Leica CM1950 cryostat, tissue sections with a thickness of 10 μm were obtained and attached to Stereo-seq chips immediately. The spatial transcriptomics data was obtained using the STOmics technology79. Briefly, the tissue sections were adhered to the Stereo-seq chip and heated for 3 min at 37 °C using a slide warmer. The chip was then incubated in − 20 °C methanol for 30 min and stained with a nucleic acid dye (Thermo Fisher, Q10212). The stained chip was imaged using a Ti-7 Nikon Eclipse microscope. The cDNA products were released and purified from the Stereo-seq chips after permeabilization for 26 min and reverse transcription. Indexed cDNA libraries were constructed following the manufacturer’s protocol. Finally, the cDNA libraries were sequenced on an MGI DNBSEQ-Tx sequencer, with 50 bp for read 1 and 100 bp for read 2. After sequencing, the raw data of in-situ spatial transcriptomics were processed, and unsupervised clustering was performed as described previously79. Briefly, the expression profile matrices were generated using SAW, a pipeline available on GitHub (https://github.com/BGIResearch/SAW). The sequences were filtered to remove invalid CID sequences with a 1-base mismatch tolerance and low-quality UMI sequences with a quality score lower than 10. The remaining sequences were aligned to the reference genome (GRCh38 and GRCm38) using STAR75 to generate expression matrices. Finally, the raw bin 1 matrix was aggregated into the final bin 50 matrix and assigned coordinates for each bin. Normalization of the total UMI counts of the genes was performed based on the region area to enable comparison of gene expression across different regions. Spatial gene expression was visualized using the BGI stereomap system (binsize = 100). For better visualization, ST dots were superimposed manually on high-resolution ssDNA fluorescence images.
Single-cell sorting, sequencing and bioinformatic analysis
The lungs with engraftment of GFP+ cells were minced and then digested in an enzymatic mix on a shaker at 37 °C for 1 h. The dissociated cells were filtered through a 100 μm cell strainer, and the erythrocytes were removed using Red Blood Cell Lysis Buffer. Engrafted cells were sorted from the cell suspension based on GFP fluorescence using a BD FACS Arial flow cytometry. These sorted cells were then barcoded using the 10 × Chromium Controller (10 × Genomics) and scRNA-seq libraries were generated using the Chromium Single-Cell 3’v3 Reagent Kit (10 × Genomics). The resulting sequencing libraries were loaded onto an Illumina NovaSeq with 2 × 150 paired-end kits at Novogene China. The raw sequencing reads were processed using the Cell Ranger v.3.1.0 pipeline from 10 × Genomics. This involved demultiplexing the reads, aligning them to the human GRCh38 genome, and quantifying UMI counts per gene per cell to generate a gene-barcode matrix. The data from different samples were aggregated and normalized to the same sequencing depth, resulting in a combined gene-barcode matrix.
Post-processing steps were performed using Seurat (4.0.2)80. The following criteria were then applied: gene number between 200 and 6500 and mitochondrial gene percentage < 0.25. Genes were filtered out if only detected in less than three cells. Finally, a filtered gene-barcode matrix of 8 dpt and 35 dpt samples was integrated with Seurat to remove batch effects across the different samples. In parameter settings, the first 30 dimensions of canonical correlation analysis (CCA) and PCA were used. The filtered gene-barcode matrix was then normalized using a global-scaling normalization method called ‘LogNormalize’ with a scale factor of 10,000. A subset of highly variable genes was identified for downstream analysis, and a linear transformation (ScaleData) was applied as a pre-processing step. PCA was performed to reduce the dimensionality of the data using the highly variable genes as input in the Seurat function RunPCA. The top 30 significant principal components (PCs) were selected for two-dimensional uniform manifold approximation and projection (UMAP). Function FindCluster in Seurat was used to identify cell clusters. Contaminated mouse cell clusters were removed according to the number of Count_RNA, and the other cells were re-clustered using the same parameter mentioned above in the clustering step. The function Featureplot in Seurat was utilized to generate gene expression visualization and the R package pheatmap was utilized to generate all heatmap graphs in this analysis. The top 50 differentially expressed genes (DEGs) of epithelium cluster in the previously published human single-cell landscape were extracted as distinct signatures. The AddModuleScore function was performed to assigned scores to cells in our dataset based on their similarity to the signatures. To infer the cluster and lineage relationships between the cell types identified, Monocle381 was applied to our data sets. UMAP embeddings and cell clusters generated from Seurat were used as input, and trajectory graph learning and pseudotime measurement through reversed graph embedding were performed with Monocle3. Finally, the cells were ordered along the trajectory.
Statistics and reproducibility
Statistical analyses were conducted using GraphPad Prism 9 software. Two-sample comparisons were performed using an unpaired two-tailed Student’s t test or a Mann–Whitney U test, depending on the results of a normality test, while one-way or two-way Analysis of Variance (ANOVA) was performed to larger datasets utilizing multiple comparisons in GraphPad Prism. A p-value of less than 0.05 (P < 0.05) was considered statistically significant. The level of statistical significance is indicated by asterisks as follows: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. The n numbers for each experiment are provided in the text or figures. All experiments and micrographs described were replicated at least three times independently with biological replicates showing similar results. Except one biological sample was used for single-cell RNA-seq, spatial transcriptomics, and non-human primate study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings from this study are available within the manuscript and its Supplementary Information. RNA-seq of P63+ progenitor cells from different donors generated during this study have been deposited at GEO under accession code GSE260777. Single-cell mRNA sequencing and spatial transcriptomics data of transplanted P63+ progenitor cells generated during this study have been deposited at GEO under accession codes GSE260778 and GSE260779 separately. Source data are provided in this paper.
Code availability
The majority of the analysis was carried out using published and freely available software and pre-existing packages mentioned in the methods. No custom code was generated. R scripts used to analyze data and generate figures are available upon request to W.Z.
References
Raghu, G. et al. Diagnosis of idiopathic pulmonary fibrosis. An official ATS/ERS/JRS/ALAT clinical practice guideline. Am. J. Respir. Crit. Care Med. 198, e44–e68 (2018).
Raghu, G. et al. Idiopathic pulmonary fibrosis (an Update) and progressive pulmonary fibrosis in adults: An official ATS/ERS/JRS/ALAT clinical practice guideline. Am. J. Respir. Crit. Care Med. 205, e18–e47 (2022).
Weiss, D. J., Kolls, J. K., Ortiz, L. A., Panoskaltsis-Mortari, A. & Prockop, D. J. Stem cells and cell therapies in lung biology and lung diseases. Proc. Am. Thorac. Soc. 5, 637–667 (2008).
Stolz, D. et al. Towards the elimination of chronic obstructive pulmonary disease: a Lancet Commission. Lancet 400, 921–972 (2022).
Camelo, A., Dunmore, R., Sleeman, M. A. & Clarke, D. L. The epithelium in idiopathic pulmonary fibrosis: breaking the barrier. Front. Pharmacol. 4, 173 (2014).
Rawlins, E. L. et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534 (2009).
Lee, J. H. et al. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 156, 440–455 (2014).
Basil, M. C. et al. Human distal airways contain a multipotent secretory cell that can regenerate alveoli. Nature 604, 120–126 (2022).
Liu, Q. et al. Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat. Genet. 51, 728–738 (2019).
Whitsett, J. A., Kalin, T. V., Xu, Y. & Kalinichenko, V. V. Building and regenerating the lung cell by cell. Physiol. Rev. 99, 513–554 (2019).
Alysandratos, K. D., Herriges, M. J. & Kotton, D. N. Epithelial stem and progenitor cells in lung repair and regeneration. Annu. Rev. Physiol. 83, 529–550 (2021).
Kathiriya, J. J., Brumwell, A. N., Jackson, J. R., Tang, X. & Chapman, H. A. Distinct airway epithelial stem cells hide among club cells but mobilize to promote alveolar regeneration. Cell Stem Cell 26, 346–358 (2020).
Konkimalla, A., Tata, A. & Tata, P. R. Lung regeneration: Cells, models, and mechanisms. Cold Spring Harb. Perspect. Biol. 14, https://doi.org/10.1101/cshperspect.a040873 (2022).
Basil, M. C. et al. The cellular and physiological Basis for lung repair and regeneration: Past, present, and future. Cell Stem Cell 26, 482–502 (2020).
Hogan, B. L. et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 (2014).
Jain, R. et al. Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat. Commun. 6, 6727 (2015).
Kadur Lakshminarasimha Murthy, P. et al. Human distal lung maps and lineage hierarchies reveal a bipotent progenitor. Nature 604, 111–119 (2022).
Kim, C. F. et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835 (2005).
Reyfman, P. A. et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 199, 1517–1536 (2019).
Zacharias, W. J. et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255 (2018).
Kumar, P. A. et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 147, 525–538 (2011).
Zuo, W. et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature 517, 616–620 (2015).
Vaughan, A. E. et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625 (2015).
Xi, Y. et al. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat. Cell Biol. 19, 904–914 (2017).
Carraro, G. et al. Single-cell reconstruction of human basal cell diversity in normal and idiopathic pulmonary fibrosis lungs. Am. J. Respir. Crit. Care Med. 202, 1540–1550 (2020).
Rao, W. et al. Regenerative metaplastic clones in COPD lung drive inflammation and fibrosis. Cell 181, 848–864 (2020).
Jaeger, B. et al. Airway basal cells show a dedifferentiated KRT17(high)Phenotype and promote fibrosis in idiopathic pulmonary fibrosis. Nat. Commun. 13, 5637 (2022).
Wang, S. et al. Cloning a profibrotic stem cell variant in idiopathic pulmonary fibrosis. Sci. Transl. Med. 15, eabp9528 (2023).
Xie, T. et al. Abnormal respiratory progenitors in fibrotic lung injury. Stem Cell Res. Ther. 13, 64 (2022).
Weiner, A. I. et al. ΔNp63 drives dysplastic alveolar remodeling and restricts epithelial plasticity upon severe lung injury. Cell Rep. 41, 111805 (2022).
Ray, S. et al. Rare SOX2(+) airway progenitor cells generate KRT5(+) cells that repopulate damaged alveolar oarenchyma following influenza virus infection. Stem Cell Rep. 7, 817–825 (2016).
Kathiriya, J. J. et al. Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5(+) basal cells. Nat. Cell Biol. 24, 10–23 (2022).
Yang, Y. et al. Spatial-temporal lineage restrictions of embryonic p63(+) progenitors establish distinct stem cell pools in adult airways. Dev. Cell 44, 752–761 (2018).
Ma, Q. et al. Regeneration of functional alveoli by adult human SOX9(+) airway basal cell transplantation. Protein Cell 9, 267–282 (2018).
McDonough, J. E. Ready and waiting: Where early-stage idiopathic pulmonary fibrosis fibroblasts are primed to be activated. Am. J. Respir. Cell Mol. Biol. 66, 1–2 (2022).
Molyneaux, P. L. & Maher, T. M. The role of infection in the pathogenesis of idiopathic pulmonary fibrosis. Eur. Respir. Rev. 22, 376–381 (2013).
Habermann, A. C. et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, eaba1972 (2020).
Smirnova, N. F. et al. Detection and quantification of epithelial progenitor cell populations in human healthy and IPF lungs. Respir. Res. 17, 83 (2016).
Taylor, M. S. et al. A conserved distal lung regenerative pathway in acute lung injury. Am. J Pathol. 188, 1149–1160 (2018).
Treutlein, B. et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014).
Hurley, K. et al. Reconstructed single-cell fate trajectories define lineage plasticity windows during differentiation of human PSC-derived distal lung progenitors. Cell Stem Cell 26, 593–608 (2020).
Wray, C. et al. Claudin-4 augments alveolar epithelial barrier function and is induced in acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 297, L219–227 (2009).
Kasper, M. & Barth, K. Potential contribution of alveolar epithelial type I cells to pulmonary fibrosis. Biosci. Rep. 37, https://doi.org/10.1042/bsr20171301 (2017).
Gillich, A. et al. Capillary cell-type specialization in the alveolus. Nature 586, 785–789 (2020).
Sikkema, L. et al. An integrated cell atlas of the lung in health and disease. Nat. Med. 29, 1563–1577 (2023).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Vila Ellis, L. et al. Epithelial vegfa specifies a distinct endothelial population in the mouse lung. Dev. Cell 52, 617–630 (2020).
Rock, J. R. et al. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 8, 639–648 (2011).
Mori, M. et al. Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development 142, 258–267 (2015).
Cupello, C. et al. Lung evolution in vertebrates and the water-to-land transition. Elife 11, https://doi.org/10.7554/eLife.77156 (2022).
Zhou, Y. et al. Stable long-term culture of human distal airway stem cells for transplantation. Stem Cells Int. 2021, 9974635 (2021).
Shi, Y. et al. Distal airway stem cells ameliorate bleomycin-induced pulmonary fibrosis in mice. Stem Cell Res. Ther. 10, 161 (2019).
Chu, G. Y. et al. Stem cell therapy on skin: Mechanisms, recent advances and drug reviewing issues. J. Food Drug Anal. 26, 14–20 (2018).
Huang, S. et al. Lgr6 marks epidermal stem cells with a nerve-dependent role in wound re-epithelialization. Cell Stem Cell 28, 1582–1596 (2021).
White, E. S., Lazar, M. H. & Thannickal, V. J. Pathogenetic mechanisms in usual interstitial pneumonia/idiopathic pulmonary fibrosis. J. Pathol. 201, 343–354 (2003).
Kanai, N., Yamato, M. & Okano, T. Cell sheets engineering for esophageal regenerative medicine. Ann. Transl. Med. 2, 28 (2014).
Fernanda de Mello Costa, M., Weiner, A. I. & Vaughan, A. E. Basal-like progenitor cells: A review of dysplastic alveolar regeneration and remodeling in lung repair. Stem Cell Rep. 15, 1015–1025 (2020).
Planer, J. D. & Morrisey, E. E. After the storm: Regeneration, repair, and reestablishment of homeostasis between the alveolar epithelium and innate immune system following viral lung injury. Annu. Rev. Pathol. 18, 337–359 (2023).
Lv, Z. et al. Alveolar regeneration by airway secretory-cell-derived p63(+) progenitors. Cell Stem Cell 31, 1685–1700 (2024).
Erjefält, J. S., Sundler, F. & Persson, C. G. Epithelial barrier formation by airway basal cells. Thorax 52, 213–217 (1997).
Basil, M. C., Alysandratos, K. D., Kotton, D. N. & Morrisey, E. E. Lung repair and regeneration: Advanced models and insights into human disease. Cell Stem Cell 31, 439–454 (2024).
Greaney, A. M. et al. Platform effects on regeneration bypulmonary basal cells as evaluated by single-Cell RNA sequencing. Cell Rep. 30, 4250–4265 (2020).
Aschheim, K. & DeFrancesco, L. Xenotransplantation: how close are we? Nat. Biotechnol. 41, 452–460 (2023).
Wang, Y. et al. Autologous transplantation of P63(+) lung progenitor cells for chronic obstructive pulmonary disease therapy. Sci. Transl. Med. 16, eadi3360 (2024).
Louie, S. M. et al. Progenitor potential of lung epithelial organoid cells in a transplantation model. Cell Rep. 39, 110662 (2022).
Toonkel, R. L., Hare, J. M., Matthay, M. A. & Glassberg, M. K. Mesenchymal stem cells and idiopathic pulmonary fibrosis. Potential for clinical testing. Am. J. Respir. Crit. Care Med. 188, 133–140 (2013).
Wang, D., Morales, J. E., Calame, D. G., Alcorn, J. L. & Wetsel, R. A. Transplantation of human embryonic stem cell-derived alveolar epithelial type II cells abrogates acute lung injury in mice. Mol. Ther. 18, 625–634 (2010).
Dye, B. R. et al. Human lung organoids develop into adult airway-like structures directed by physico-chemical biomaterial properties. Biomaterials 234, 119757 (2020).
Huang, S. X. et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 32, 84–91 (2014).
Tan, Q., Choi, K. M., Sicard, D. & Tschumperlin, D. J. Human airway organoid engineering as a step toward lung regeneration and disease modeling. Biomaterials 113, 118–132 (2017).
Serrano-Mollar, A. et al. Safety and tolerability of alveolar type II cell transplantation in idiopathic pulmonary fibrosis. Chest 150, 533–543 (2016).
Weiner, A. I. et al. Mesenchyme-free expansion and transplantation of adult alveolar progenitor cells: steps toward cell-based regenerative therapies. NPJ Regen. Med. 4, 17 (2019).
Wang, X. et al. Cloning and variation of ground state intestinal stem cells. Nature 522, 173–178 (2015).
Ashcroft, T., Simpson, J. M. & Timbrell, V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J. Clin. Pathol. 41, 467–470 (1988).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287 (2012).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 (2022).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Acknowledgements
We thank Prof. Tao Ren for his aid with the monkey experiments. We thank Profs. Jieming Qu, Shiyue Li, Min Zhou, Zuojun Xu, Xiaotian Dai, Zili Meng, and Qiang Li for helping with experimental materials. We thank Dr. Yuhua Sun, Yi Wang, and engineers from Haier Biomedical, China for help with the automated system. We thank Profs. Ying Xi and Pengfei Sui for helpful discussion of the manuscript. We also thank Haier Biomedical, Inc. for the co-development of an automated cell cloning system. This work was funded by the National Key Research and Development Plan (2024YFA1108900 [to T.Z.] and 2024YFA1108500 [to W.Z.]); National Science Fund for Excellent Young Scholars (82122038 [to W.Z.]); Shanghai Science and Technology Talents Program (19QB1403100 [to W.Z.]); Jiangsu Province Science and Technology Special Project (Key Research and Development Plan for Social Development) (BE2023727 [to W.Z.]); National Biopharmaceutical Technology Research Project Funding (NCTIB2023XB01011 [to W.Z.]).
Author information
Authors and Affiliations
Contributions
W.Z. and T.Z. designed the study. Y.Z., Y.Q.Z., W.P.Z., M.Z.L., X.P.Z., J.D., Q.W.M., and Y.J.W. performed the experiments and collected the data. Z.L.G. collected the clinical samples and patient information. W.Z., Y.Z., Y.Q.Z., W.P.Z., and Z.L.G. analyzed and interpreted the data. T.Z. and Y.Z.Z participated discussion of the data. W.Z., Y.Z., Y.Q.Z., and W.P.Z. drafted the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Chen Ting, Argyrios Tzouvelekis, and the other anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, Y., Zhou, Y., Zhang, W. et al. Cloned airway basal progenitor cells to repair fibrotic lung through re-epithelialization. Nat Commun 16, 1303 (2025). https://doi.org/10.1038/s41467-025-56501-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56501-w