Abstract
Myelomatous bone disease is a complication characterized by lytic bone lesions, reduced bone formation, bone pain, and increased fracture risk. Understanding these underlying mechanisms is crucial for developing effective therapeutic approaches. Here we show the role of enhancer of zeste homolog 2 (EZH2) in bone lesions induced by myeloma cells. Our research reveals that cytokines produced by myeloma-associated adipocytes activate the expression of EZH2 in myeloma cells. Furthermore, we find that EZH2 forms a transcriptional repression complex with transcription factor AP2α. This complex promotes trimethylation at lysine 27 of histone H3 (H3K27me3) in the promoter region of the tumor suppressor gene EMP1, resulting in transcriptional silencing. EMP1 silencing leads to increased myeloma cell proliferation and the concomitant secretion of osteolytic cytokines that contribute to bone destruction. Importantly, EZH2 inhibitors effectively treat myeloma-induced osteolytic lesions. Thus, targeting EZH2 represents a potential therapeutic strategy for preventing and managing myeloma bone disease.
Similar content being viewed by others
Introduction
Multiple myeloma is a malignancy arising from malignant plasma cells in the bone marrow that remains an incurable disease. Osteolytic bone disease is a prominent feature of myeloma, affecting over 80% of patients and causing various complications such as pathological fractures, intense bone pain, spinal cord compression, and hypercalcemia. The detrimental consequences of bone destruction greatly impair the quality of life of affected individuals and contribute to poor survival outcomes1.
Bone is a dynamic tissue that continuously remodels itself through the coordinated actions of osteoclasts, which resorb bone, and osteoblasts, which form new bone. Osteoclasts originate from cells of hematopoietic monocytic precursors upon stimulation of two essential factors, the macrophage colony stimulating factor (M-CSF) and receptor activation of NF-κB ligand (RANKL)2. M-CSF interacts with its receptor, colony-stimulating factor 1 receptor (c-Fms), to activate specific signaling pathways that promote the proliferation and survival of osteoclast precursors3. RANKL binds to RANK on the surface of osteoclast precursor cells, triggering the activation of nuclear factor of activated T-cells, cytoplasmic 1 protein (NFATc1). This activation subsequently upregulates the expression of genes involved in osteoclast differentiation, such as tartrate-resistant acid phosphatase (TRAP), calcitonin receptor (CALCR), and cathepsin K (CTSK)2. The osteoblast, another crucial player in the bone remodeling cycle, originates from mesenchymal stem cells (MSCs). The differentiation of osteoblasts is regulated by the activation of core-binding factor α-1/runt-related transcription factor 2 (RUNX2) and osterix. This activation induces the expression of genes associated with osteoblast differentiation, including bone gamma-carboxyglutamic acid-containing protein (BGLAP), alkaline phosphatase (ALP), and collagen type I α1 (COL1A1)4. The equilibrium of bone remodeling processes is frequently disturbed in a variety of malignancies, including myeloma and solid tumors like breast and lung cancer1,5. Myeloma cells can exert direct effects on osteoblasts and osteoclasts via cell-to-cell contact6,7, and also impact them by releasing cytokines, or metabolic byproducts2,5,8,9. Moreover, myeloma cells can indirectly modulate the differentiation of osteoblasts and osteoclasts by affecting other cell types10,11,12.
EZH2 (Enhancer of zeste homolog 2), a core member of the PcG (Polycomb group) gene family, plays a crucial role in regulating the trimethylation of the lysine 27 residue (H3K27me3) on histone H3, thereby repressing gene transcription13. Under normal physiological conditions, EZH2 plays crucial regulatory roles in individual development, stem cell differentiation, self-renewal14, cell cycle maintenance15, cellular autophagy and apoptosis16, DNA damage repair and aging17, as well as cell fate determination18. Additionally, activation of EZH2 expression is observed in numerous tumors and plays an important regulatory role in tumorigenesis and tumor progression. For example, EZH2 promotes the proliferation and invasion of breast cancer cells19. In lung cancer, EZH2 inhibits E2F transcription factor 1 (E2F1)-regulated apoptosis, invasion, metastasis, and drug resistance20,21. In melanoma, EZH2 knockout mouse models exhibit reduced lymph node and lung metastasis22. In pancreatic cancer models, overexpression of EZH2 promotes epithelial-mesenchymal transition (EMT) and angiogenesis23,24. In ovarian cancer, EZH2 inhibits the expression of chemokine CXCL9, thereby regulating the infiltration of CD8+ T cells into the tumor25. In myeloma, EZH2 is closely associated with poor prognosis in patients26. Additionally, EZH2 promotes myeloma cell proliferation by regulating the IRF4-Myc signaling axis27. Our preliminary work also revealed that myeloma cells integrin receptor that binds to marrow adipocytes, promoting the expression of EZH2 in adipocytes through the PI3K (phosphoinositide 3-kinase)/Akt signaling and nuclear factor kappa B (NF-κB) signaling. These reprogrammed adipocytes acquire the ability to induce bone lesions11. However, it remains unclear whether EZH2 contributes to bone lesions formation.
Through a combination of in vitro, in vivo, and patient sample studies, we have reported that EZH2 has a unique role in myeloma-induced bone disease. We found that tumor necrosis factor α (TNFα) and Interleukin-6 (IL6) secreted by myeloma-associated adipocytes activate EZH2 in myeloma cells. The transcription factor AP2α interacts directly with EZH2 to form a transcriptional repression complex. This complex promotes the trimethylation of the lysine 27 residue on histone H3 at the promoter region of the tumor suppressor gene EMP1, leading to transcriptional silencing. Inhibition of EMP1 results in increased proliferation of myeloma cells and up-regulation of osteoclastogenic cytokines, ultimately promoting osteolytic bone lesions. Interestingly, in both in vitro and in vivo models, EZH2 inhibitors significantly improved myeloma-induced osteolytic bone lesions. Our findings not only provide insights into the mechanism of cancer-induced bone destruction but also suggest a potential therapeutic strategy for tumor patients with osteolytic bone lesions by targeting EZH2.
Results
EZH2 enhances bone lesions induced by myeloma cells
To understand whether EZH2 may affect myeloma cell-induced bone lesions, we compared its expression levels in myeloma cells of patients from a published dataset (GEO: GSE755). EZH2 expression was higher in the myeloma cells of patients with bone lesions than in those without (Fig. 1a). Furthermore, we found a strong positive correlation between EZH2 expression in myeloma cells and the number of bone lesions in 30 patients (Fig. 1b). Representative images of x-ray scanning and magnetic resonance imaging showed more lytic lesions in the skull (Fig. 1c) and spine (Fig. 1d) of patient with high bone lesion numbers (P1) than in patient with low lesion numbers (P2). Bone marrow aspirates immunohistochemistry results further demonstrated that EZH2 and CD138 (myeloma cells surface marker) expression was higher in P1 than in P2 (Fig. 1e, f). These results indicate an association between EZH2 and myeloma bone disease.
a In a published GEO dataset (GSE755), EZH2 mRNA levels were compared in primary myeloma cells from 37 patients without bone lesions (BL = 0) and 136 patients with bone lesions (BL ≥ 1). Data are means ± SD. p value was determined by unpaired two-tailed t test. b Primary myeloma cells were isolated from bone marrow aspirates of 30 newly diagnosed patients, some with and some without bone lesions. Shown are correlation coefficient of the mRNA levels of EZH2 and numbers of bone lesion in patients (n = 30). The correlation was evaluated using Pearson coefficient with two-tailed p value. r, correlation coefficient. Representative images of magnetic resonance imaging and x-ray scanning for lytic lesions in the skull (c) and spine (d) in patient 1 (P1: 25 bone lesions) and patient 2 (P2: 4 bone lesions). Yellow arrow, bone lesion. Scale bar, 1 cm. e, f Representative images of immunohistochemical staining and densitometry analysis show the expression of CD138 and EZH2 in the biopsy segment of two patients (P1 and P2) (n = 6 areas/patient). e is representative of three independent experiments. Scale bar, 30 µm. p values were determined by unpaired two-tailed t test. Source data are provided as a Source data file.
To determine the functional role of myeloma expressing EZH2 in lytic bone lesions in vivo, we knocked down its expression in RPMI8226 myeloma cells using small hairpin RNAs (shRNAs) against human EZH2 (shEZH2) and over expressed EZH2 complementary DNA (EZH2) in MM.1S myeloma cells (Supplementary Fig. 1). Next, we injected shEZH2 RPMI8226 cells into mouse femurs and observed fewer lytic lesions and slower tumor growth than control RPMI8226 cells (shCtrl). Conversely, EZH2 MM.1S cells showed more femoral lesions and faster tumor growth than control MM.1S (Vec) cells did (Fig. 2a–c). To assess the impact of myeloma-expressed EZH2 on osteoclast-mediated bone resorption in vivo, we quantified the serum levels of procollagen type I N-terminal propeptide (PINP), which serves as a marker of bone formation, and C-telopeptide of type I collagen (CTX-1), which indicates bone resorption. Our findings revealed that the shEZH2 and Vec groups exhibited elevated PINP levels and reduced CTX-1 levels in comparison to the shCtrl and EZH2 groups (Fig. 2d, e). We also stained myeloma-bearing mouse femurs for TRAP and Toluidine blue. Percentages of bone volume/total volume (BV/TV) were assessed by microcomputed tomography, percentages of bone surface eroded by osteoclasts (ES/BS) and percentages of bone surface covered with osteoclasts (Oc. S/BS) were assessed by TRAP staining (Supplementary Fig. 2), percentage of osteoid surface (OS/BS) and bone surface lined with osteoblasts (Ob. S/BS) were assessed by Toluidine blue staining. Bone formation rate was measured by calcein staining. Bone histomorphometric analysis demonstrated lower percentages of BV/TV (Fig. 2f), higher percentages of ES/BS (Fig. 2g), percentages of Oc. S/BS (Fig. 2h) and a lower percentage of OS/BS (Fig. 2i), Ob. S/BS (Fig. 2j) and bone formation rate (Fig. 2k, l) in mice injected with myeloma cells expressing high levels of EZH2 (shCtrl or EZH2) than in those injected with low EZH2 myeloma cells (shEZH2 or Vec). To provide additional evidence for our findings, we employed a humanized mouse model of intrahepatic injection of patient-derived CD138+ primary myeloma cells (Fig. 2m). The primary myeloma cells used here are consistent with P1 and P2 shown in Fig. 1. We extracted serum from mice and performed ELISA analysis to determine the concentrations of human λ chains, an indicator of myeloma burden. The results revealed a higher level of human λ chains in the mouse serum injected with high EZH2 expression primary myeloma cells (P1) than low EZH2 expression primary myeloma cells (P2) (Fig. 2n). After the mice were euthanized, myeloma cells were isolated from the bone marrow of PDX mice using anti-CD138 antibody-coated magnetic beads. The expression of EZH2 in myeloma cells of group P1 remains higher than that in group P2 (Fig. 2o). In addition, bone histomorphometry analysis showed that P1 cells caused more femoral lesions (Fig. 2p, Supplementary Fig. 3), lower bone BV/TV (Fig. 2q), higher ES/BS% (Fig. 2r), and lower OS/BS% (Fig. 2s) than low EZH2 expression (P2). The immunohistochemical analysis of multiple tissues in PDX mice also pointed out that myeloma cells were found in the bone marrow and spleen of the mice, but less in the liver (Supplementary Fig. 3). Collectively, these findings unveil a pivotal role of EZH2 expression in myeloma cells, promoting the development of lytic bone lesions and tumor progression in both patients and murine models of myeloma.
6–7 weeks old female NSG mice were intrafemorally injected with myeloma cell lines RPMI8226 [nontargeted shRNA (shCtrl), and EZH2 shRNA (shEZH2)] or MM.1S [control vector (Vec), and EZH2 cDNA (EZH2)]. The mice not receiving myeloma cells (No MM) served as controls. After six weeks of cells injection, shown are the representative x-ray images of lytic lesions (a) (n = 5 mice/group). Yellow arrow, bone lesion. Scale bar, 1 mm. Another batch of NSG mice were intrafemorally injected with the same combination of myeloma cell lines. After four weeks of injections, shown are the representative images of bioluminescent signals (b) and microcomputed tomography images (c), concentrations of PINP (d) or CTX-1 (e) in mouse sera, percentages of BV/TV (f), ES/BS (g), Oc.S/BS (h), OS/BS (i), and Ob.S/BS (j). Scale bar, 1 mm. k, l Bone formation rate (BFR/BS) was measured by calcein injection, and the bone sections were imaged and analyzed. Shown are representative images and summarized data of bone formation in mouse femurs (n = 5 mice/group). Scale bar: 20 μm. m Schematic diagram showing primary patient-derived mouse model generation and analyzes performed. n ELISA analysis shown the concentrations of human λ chains in mouse serum. o After the mice were euthanized, myeloma cells were isolated from the bone marrow of PDX mice using anti-CD138 antibody-coated magnetic beads. EZH2 mRNA levels were compared between P1 and P2 groups (n = 5 mice/group). p Shown are the representative microcomputed tomography images in PDX mouse model. Scale bar, 0.5 mm. Shown are the percentages of BV/TV (q), ES/BS (r), and OS/BS (s) in PDX mouse model. The P1 and P2 cells used here are from the same source as those in Fig. 1b. d–k, q–s: p values were determined using one-way ANOVA with Tukey’s test; (n, o): p values were determined by unpaired two-tailed t test. Data are means ± SD. Source data are provided as a Source data file.
There are two additional questions we are particularly interested in: first, whether the expression of EZH2 is also activated in other stromal cells of the bone marrow microenvironment apart from myeloma cells; second, whether the up-regulation of EZH2 expression in myeloma cells is associated with bone marrow stromal cells. Based on data analysis from the GEO database, we examined EZH2 expression in several types of stromal cells isolated from the normal bone marrow microenvironment or myeloma microenvironment. The results indicated that there was no significant difference in EZH2 expression among stromal cells of the myeloma microenvironment, including fibroblasts (Supplementary Fig. 4a), macrophages (Supplementary Fig. 4b), and mesenchymal stem cells (Supplementary Fig. 4c). However, EZH2 expression was activated in adipocytes (Supplementary Fig. 4d). Our previous studies indicated that myeloma cells activate EZH2 in bone marrow adipocytes, leading to alterations in the secretion of adipokines by adipocytes, ultimately resulting in bone damage, and the expression of TNFα and IL6 is activated in this process11. Myeloma cells were treated with conditioned medium from myeloma-associated adipocytes or normal adipocytes. The results demonstrated that, in comparison to normal adipocytes, myeloma-associated adipocytes exhibit a significantly enhanced activation of the NF-κB pathway and up-regulation of EZH2 expression (Supplementary Fig. 5a, b). Furthermore, inhibition of NF-κB can suppress the up-regulation of EZH2 (Supplementary Fig. 5c). Further evidence demonstrated that TNFα and IL6 can activate the expression of EZH2 (Supplementary Fig. 5d). Additionally, neutralizing antibodies against TNFα or IL6 can also partially reverse the activation of the NF-κB pathway and the up-regulation of EZH2 (Supplementary Fig. 5e, f). Similarly, the NF-κB pathway inhibitor can also block NF-κB pathway activation and reduce the expression of EZH2 (Supplementary Fig. 5g,h). Our findings indicate that there is a reciprocal interaction between myeloma cells and adipocytes, leading to the activation of EZH2 gene expression in both cell types.
EZH2 enhances RANKL-induced osteoclastogenesis and inhibits osteoblastogenesis through osteolytic cytokines in myeloma cells
Maintenance of bone remodeling is dependent on a delicate equilibrium between the resorption mediated by osteoclasts and the formation facilitated by osteoblasts2. In order to investigate the potential regulatory role of EZH2 in maintaining this balance, we initially evaluated its effect on osteoclast differentiation. In the context of RANKL stimulation, the interaction between osteoclast precursors (preOCs) and myeloma cells with high EZH2 expression (shCtrl or EZH2) significantly amplified the generation of multinucleated tartrate-resistant acid phosphatase-positive (TRAP+) cells (Fig. 3a, b), TRAP 5b secretion (Fig. 3c), and the expression of osteoclast differentiation markers (Fig. 3d, e) relative to cocultures with myeloma cells expressing low EZH2 levels (shEZH2 or Vec). Collectively, these observations highlight the pivotal role of EZH2 in promoting osteoclastogenesis in myeloma cells. To further explore the impact of EZH2 on osteoblastogenesis, we conducted coculture experiments using osteoblast progenitors (MSCs) in osteoblast differentiation medium alongside myeloma cells. MSCs cultured independently in the same medium were used as positive controls. Notably, cocultures with myeloma cell lines expressing low EZH2 (Vec or shEZH2) exhibited enhanced osteoblast maturation (Fig. 3f, g), elevated alkaline phosphatase (ALP) activity (Fig. 3h), and upregulated expression of osteoblast differentiation-associated genes (Fig. 3i, j) compared to those with high EZH2 expression (EZH2 or shCtrl). These findings suggest that EZH2 in myeloma cells functions as a repressor of osteoblastogenesis.
Precursors of osteoclasts were co-cultured with myeloma cell lines RPMI8226 (shCtrl, shEZH2) or MM.1S (Vec, EZH2) in osteoclast medium. Shown are the numbers (a, b) of multinuclear (≥3) TRAP+ cells, TRAP 5b levels (c), and relative expression of the TRAP, CALCR, and CTSK genes in precursors of osteoclasts (d, e) (n = 3 biological replicates). Scale bars, 100 μm. MSCs were cocultured with myeloma cell lines RPMI8226 (shCtrl, shEZH2) or MM.1S (Vec, EZH2) in osteoblast medium. Shown are representative images (f) and summarized data of Alizarin Red S staining (g), ALP activity (h), and the relative expression of BGLAP, ALP, and COL1A1 genes in MSCs (i, j) (n = 3 biological replicates). Scale bar: 0.4 cm. Data are means ± SD. All p values were determined using one-way ANOVA with Tukey’s test. Source data are provided as a Source data file.
Next, we investigated the mechanism by which EZH2 modify bone cell differentiation. One of the major mechanisms by which myeloma cells induce bone damage is through the secretion of osteolytic cytokines, which enhances osteoclast differentiation and inhibits osteoblast differentiation28. A published RNA sequencing dataset (GSE103567) was used to examine the cytokine expression profiles in myeloma cells treated with or without the EZH2 inhibitor GSK343. Using the criteria of a fold change greater than 2 between the two groups, we identified four down-regulated genes (CCL5, IL16, CSF1, and DKK1) (Fig. 4a). The RNA sequencing results shows knocking down the expression of EZH2 in myeloma cell lines also found similar results (Supplementary Fig. 6). Quantitative PCR analysis confirmed the downregulation of CCL5 (C-C Motif Chemokine Ligand 5), IL16 (Interleukin 16), CSF1 (Colony Stimulating Factor 1), and (Dickkopf-related protein 1) DKK1 in EZH2 knockdown cells (shEZH2) compared with control cells (shCtrl) (Fig. 4b). Additionally, compared to control cells (Vec), the expression of these four cytokines was significantly elevated in EZH2 overexpression cells (EZH2) (Fig. 4c). It is worth mentioning that the RNA sequencing results demonstrated that knocking down EZH2 expression in RPMI8226 cells suppressed the expression of CCL5, DKK1, and CSF1, but the change in IL16 expression was not significant. We believe that this discrepancy may primarily be attributed to the low signal value of IL16 or potential errors introduced by the limited sample size. ELISA results also indicated that EZH2 enhanced the expression of these four cytokines (Fig. 4d–g). Moreover, in Fig. 1b, primary myeloma cells were isolated from bone marrow aspirates of 30 newly diagnosed patients. Correlation analysis revealed a strong positive association between EZH2 expression and the expression of these four cytokines in these patients (Fig. 4h–k). Previous studies have demonstrated that CCL529, IL1630, and CSF12 promote osteoclast differentiation, while DKK1 inhibits osteoblast differentiation2. Addition of antibodies against CCL5, IL16 or CSF1 to preOCs co-incubated with EZH2 overexpressed myeloma cells (EZH2) reduced the numbers of TRAP+ cell (Fig. 4l). Likewise, the utilization of antibodies against CCL5 or DKK1 partially restored the suppression of osteoblast differentiation induced by EZH2 overexpressed myeloma cells (EZH2) (Fig. 4m). Similarly, addition of CCL5, IL16 or CSF1 recombinant protein to preOCs co-incubated with EZH2 knockdown myeloma cells (shEZH2) increased the numbers of TRAP+ cell (Fig. 4n), and CCL5 or DKK1 recombinant protein enhanced the suppression of osteoblast differentiation induced by EZH2 knockdown myeloma cells (shEZH2) (Fig. 4o). These experiments demonstrate that these cytokines have biological effects on osteoblast and osteoclast function. Therefore, EZH2 in myeloma cells regulates the differentiation of osteoclasts and osteoblasts by modulating the expression of these cytokines.
a A published GEO dataset (GSE103567) analysis shows the expression of cytokines in RPMI8226 cell treated with or without GSK343 (4 μM). b, c The expression of CCL5, IL16, CSF1 and DKK1 in RPMI8226 (shCtrl, shEZH2) or MM.1S (Vec, EZH2) cells (n = 3 biological replicates). d–g ELISA analysis shows the secretion of CCL5, IL16, CSF1 and DKK1 in RPMI8226 (shCtrl, shEZH2) or MM.1S (Vec, EZH2) (n = 3 biological replicates). Correlation coefficient of the levels of CCL5 (h), IL16 (i), CSF1 (j) or DKK1 (k) and levels of EZH2 (n = 30). The cells used here are from the same source as those in Fig. 1b. The correlation was evaluated using Pearson coefficient with two-tailed p value. r, correlation coefficient. l The numbers of multinuclear TRAP+ cells among precursors of osteoclasts cultured with MM.1S cells (Vec or EZH2) in the presence of blocking antibodies against CCL5, IL16, CSF1 or DKK1 (1 μg/ml) (n = 3 biological replicates). m The Alizarin red S staining of MSCs cultured in osteoblast medium with MM.1S cells (Vec or EZH2) cells in the presence of antibodies against CCL5, IL16, CSF1 or DKK1 (1 μg/ml) (n = 3 biological replicates). n The numbers of multinuclear TRAP+ cells among precursors of osteoclasts cultured with RPMI8226 cells (shCtrl or shEZH2) in the presence of CCL5 (20 ng/ml), IL16 (2 ng/ml), CSF1 (10 ng/ml) or DKK1 (20 ng/ml) (n = 3 biological replicates). o The Alizarin red S staining of MSCs cultured in osteoblast medium with RPMI8226 cells (shCtrl or shEZH2) cells in the presence of CCL5 (20 ng/ml), IL16 (2 ng/ml), CSF1 (10 ng/ml) or DKK1 (20 ng/ml) (n = 3 biological replicates). Data are means ± SD. b–g: p values were determined by unpaired two-tailed t test; l–o: p values were determined using one-way ANOVA with Tukey’s test. Source data are provided as a Source data file.
EZH2 restricts EMP1 expression through up-regulation of promoter histone methylation levels in myeloma cells
Our next objective was to elucidate the mechanisms through which EZH2 promotes the proliferation of myeloma cells and the expression of these cytokines. There are reports indicated that EZH2 can inhibit the transcription of numerous tumor suppressor genes31. To identify the specific tumor suppressor genes regulated by EZH2 in myeloma cells, we compared the expression levels of several classic tumor suppressor in myeloma cells treated with or without EZH2 inhibitors from published datasets (GEO: GSE109673 and GSE155135). The results revealed that EMP1 (Epithelial membrane protein 1) gene in myeloma cells was significantly activated following the inhibition of EZH2 in these cells (Fig. 5a, b and Supplementary Fig. 7a, b). EMP1 belongs to the peripheral myelin protein 22 kDa (PMP22) gene family. EMP1 has been widely described as a tumor suppressor in various cancers, where it inhibits cell growth and metastasis, induces apoptosis, and preventing angiogenesis32. However, their role and regulatory mechanisms in myeloma remain unclear. Similarly, through analysis of published GEO datasets (GEO: GSE2658 and GSE5900), we found that EMP1 expression was much lower in myeloma cells compared with normal plasma cells (Fig. 5c). Furthermore, we found a strong negative correlation between EZH2 and EMP1 expression in myeloma cells (Fig. 5d). Western blot analysis revealed that compared to normal plasma cells, EZH2 expression was upregulated in primary myeloma cells, while the expression of EMP1 was suppressed (Fig. 5e). In myeloma cell lines, knockdown of EZH2 activates the expression of EMP1, while conversely, overexpression of EZH2 inhibits the expression of EMP1 (Fig. 5f). EZH2 and SUZ12 are the core subunits of polycomb repressive complex 2 (PRC2), a transcriptional repressor11. The only known histone methyltransferase activity of PRC2 targets the regulation of histone H3 lysine 27 trimethylation11. To demonstrate the specificity of this methylation, we treated myeloma cells with 3-deazaneplanocin A (DZNep), a PRC2 inhibitor, and found inhibition of EZH2 expression and reduced levels of H3K27me3, resulting in the up-regulation of EMP1 expression (Fig. 5g). Immunofluorescence analysis of bone marrow samples from myeloma patients indicated that high expression of EZH2 in myeloma cells is often accompanied by low levels of EMP1 (Fig. 5h and Supplementary Fig. 8a–d). We queried H3K27me3 and EZH2 enrichment with a published genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) result33. We found that EZH2 inhibitors reduced the enrichment of H3K27me3 and EZH2 in the EMP1 promoter of myeloma cells (Fig. 5i). Additionally, ChIP PCR analysis showed that EZH2 inhibition reduced the enrichment of EZH2 and H3K27me3 in the EMP1 promoter (Fig. 5j–l). Furthermore, we found a negative correlation between EMP1 expression and the number of bone lesions in 30 patients (Fig. 5m), and EMP1 expression was lower in the myeloma cells of patients with bone lesions than in those without (Fig. 5n). These results indicate that EZH2 upregulates histone methylation of EMP1 at H3K27me3 and thereby downregulates EMP1 expression.
GEO dataset (GSE109673) shows gene expression in RPMI8226 (a) or MM.1S (b) cells treated with or without EZH2 inhibitor OR-S1 (1 μM). c EZH2 mRNA levels in patient-derived myeloma cells (Pt MM) (n = 559) compared to normal plasma cells (n = 22) (GSE2658 and GSE5900). d Correlation of mRNA levels of EZH2 and EMP1 in 30 patients. e Western blotting shows EZH2 and EMP1 expression in normal plasma cells (n = 4) and Pt MM (n = 4). f Expression of EMP1 in RPMI8226 (shCtrl, shEZH2) or MM.1S (Vec, EZH2) cells. g Western blotting shows the expression of EZH2, EMP1 and H3K27me3 in myeloma cells treated with or without DZNep (1 μM). The samples derive from the same experiment but different gels for each protein and processed in parallel (e, g). h Immunofluorescent staining of patient samples with DAPI and antibodies against EZH2, CD138 and EMP1. Scale bar, 10 μm. i ChIP-seq results (GSE214669) for EZH2 and H3K27me3 enrichment in EMP1 gene promoter of MM.1S cells treated with or without EZH2 inhibitors C27 or MS177 (2.5 μM). j–l ChIP assay showing EZH2 and H3K27me3 enrichment in EMP1 gene promoter of Pt MM cells (n = 5) (j), RPMI8226 (k) or MM.1S (l) cells treated with or without DZNep (1 μM). m Correlation of levels of EMP1 and numbers of bone lesion in 30 patients. n EMP1 levels were compared in malignant plasma cells without bone lesions (BL = 0, n = 5) and with bone lesions (BL ≥ 1, n = 25). The cells used in h, m, and n are from the same source as those in Fig. 1b. e–h, k, l are representative of three independent experiments. Data are means ± SD. The correlation was evaluated using Pearson coefficient. r, correlation coefficient. c, f, n: p values were determined by unpaired two-tailed t test; j–l: p values were determined using one-way ANOVA with Tukey’s test. Source data are provided as a Source data file.
Another issue that needs to be proven is whether the up-regulation of EMP1 expression in myeloma cells can reverse the proliferation of myeloma cells and bone damage caused by EZH2. We first co-overexpressed EZH2 and EMP1 in myeloma cells (Supplementary Fig. 9a). The results indicated that overexpression of EMP1 can reverse the accelerated proliferation of myeloma cells (Supplementary Fig. 9b), enhanced osteoclast differentiation (Supplementary Fig. 9c), and inhibited osteoblast differentiation caused by the overexpression of EZH2 (Supplementary Fig. 9d). Additionally, overexpression of EMP1 can also significantly reverse the up-regulation of osteolytic cytokines caused by the overexpression of EZH2 (Supplementary Fig. 10a, b). Correlation analysis showed negative association between EMP1 expression and the expression of these four cytokines in these patients (Supplementary Fig. 10c–f). On the other hand, overexpressing EMP1 solely in myeloma cells can also significantly inhibit their proliferation (Supplementary Fig. 11a, b). We examined the profile of EMP1-mediated signaling pathways and observed down-regulation of phosphorylated IκBα, but not extracellular signal-regulated kinase 1/2 (ERK1/2), p38 mitogen-activated protein kinase (MAPK), and Akt (Supplementary Fig. 11c). These results suggest that EMP1 regulates the growth of myeloma cells and the expression of osteolytic cytokines by inhibiting the NF-κB pathway in myeloma cells, and further studies are needed to elucidate the specific regulatory mechanisms.
AP2α bridges the interaction between the EZH2 and EMP1 promoter
In mammalian cells, PRC2 proteins require a linker to facilitate their association with specific DNA regions for the regulation of DNA methylation, as they are unable to bind to DNA directly11. To identify this linker, immunoprecipitates of HA tagged EZH2 protein in RPMI8226 cells transfected with an HA-EZH2 plasmid using Coomassie blue staining and mass spectrometry. The analysis identified the transcription factor AP2α as a specific binding partner of EZH2 (Fig. 6a and Supplementary Data 1). To investigate the interaction between AP2α and PRC2, we performed co-immunoprecipitation assays. HEK293T cells were co-transfected with plasmids expressing AP2α and EZH2 or SUZ12. Immunoprecipitation was performed, and the immunoprecipitates were analyzed for the presence of EZH2, SUZ12, or AP2α proteins. The results demonstrated that AP2α specifically interacted with EZH2, whereas no interaction was observed with SUZ12 (Fig. 6b). Subsequently, we performed co-immunoprecipitation experiments by combining lysates from HEK293T cells expressing HA-tagged EZH2 (HA-EZH2) or HA-SUZ12 with lysates of AP2α-expressing HEK293T cells. The immunocomplexes were pulled down using an anti-HA antibody. Notably, our findings demonstrated a direct binding interaction between AP2α and EZH2 (Fig. 6c). To investigate the presence of endogenous AP2α/PRC2 complex in myeloma cells, we performed immunoprecipitation assays using an anti-EZH2 antibody on myeloma cell lysates. Finally, we examined the lysates for the presence of SUZ12 or AP2α (Fig. 6d). Our results confirmed the presence of SUZ12 and AP2α proteins in the immunoprecipitates, indicating the presence of the AP2α/PRC2 complex in myeloma cells. Additionally, by immunoprecipitation using an anti-AP2α antibody, we detected EZH2 proteins in the immunocomplexes (Fig. 6d), further supporting the interaction between AP2α and the PRC2 complex (Fig. 6d). Moreover, immunofluorescence staining provided further evidence of the interaction between EZH2 and AP2α in myeloma cells (Fig. 6e). According to previous studies, the primary sequence of the AP2α protein contains three relatively conserved domains responsible for protein-protein interactions (Fig. 6f), referred to as the activation domain (Act), basic domain (Bas) and dimerization domain (Dim)34. To determine which AP2α domain interacts with EZH2, we selectively knocked out these three domains respectively (ΔAct, ΔBas or ΔDim) (Fig. 6g). Pulldown assay showed that AP2α interacted with EZH2 through Dimerization domain (Fig. 6h). The immunofluorescence results from the bone marrow sections also corroborated the interaction between EZH2 and AP2α in primary myeloma cells (Fig. 6i).
a Coomassie blue staining for HA pulldown of EZH2 complex in RPMI8226 cells transfected with HA-EZH2 plasmid. The proteins identified are indicated on the right. b Co-immunoprecipitation of EZH2 or SUZ12 with AP2α in HEK293T cells. c Pull-down of HA-EZH2 or HA-SUZ12 with AP2α in HEK293T cells. d Co-immunoprecipitation of EZH2 or AP2α in myeloma cells. e Immunofluorescent staining of myeloma cells with DAPI and antibodies against EZH2 and AP2α. Scale bar, 2 μm. f Schematic of the truncations including ΔAct, ΔBas, and ΔDim fragments. g Western blotting shows truncations of AP2α in HEK293T cells. h Pull-down of Myc-EZH2 with truncations of HA-AP2α in HEK293T cells. i Immunofluorescent staining of patient samples with DAPI and antibodies against CD138, EZH2, or AP2α (n = 5). Scale bar, 10 μm. j Schematic of the EMP1 promoter luciferase reporter. Solid boxes: promoter region; red crosses: mutations site. k The luciferase assay of EMP1 promoter. ns, not significant. l ChIP assay shows AP2α enrichment in EMP1 promoter in patient cells (left panel) (n = 5) or cell lines (right panel) (n = 5 biological replicates). m Western blotting shows the expression of AP2α in cells transfected with AP2α shRNAs and non-target control (shCtrl). The samples derive from the same experiment but different gels for each protein and processed in parallel (b–d, g, h, m). n, o ChIP assay shows enrichment of SUZ12, EZH2, and H3K27me3 in EMP1 gene promoter in shCtrl or shAP2α transfected Pt MM (n = 5), RPMI8226 or MM.1S cells. p Formation of TRAP+ cells and Alizarin red S staining for osteoblast differentiation, cultured with shCtrl or shAP2α myeloma cells. q Schematic of the recruitment of PRC2 complex by AP2α to the EMP1 promoter. a–e, g, h, k, m, o, p are representative of three independent experiments. Data are means ± SD. k, l: p values were determined by unpaired two-tailed t test; n–p: p values were determined using one-way ANOVA with Tukey’s test. Source data are provided as a Source data file.
To determine whether AP2α binds to EMP1 promoter, we examined a 1.3-kilobase region around the EMP1 transcriptional start site. According to the promoter sequence, we predicted one AP2α binding site (−528 base pairs [bp]) in the EMP1 promoter (Fig. 6j). We generated two truncated forms of the EMP1 promoter. Deletion mapping identified an AP2α binding site between −558 and −458 bp in the EMP1 promoter (Fig. 6k). Mutating a putative motif at −528 bp in the EMP1 promoter from GCCN4GG to GATN4TA (Fig. 6j) confirmed these loci as AP2α binding sites (Fig. 6k). A ChIP assay using an anti-AP2α antibody confirmed the enrichment of AP2α in the promoter region of EMP1 around −528 bp in myeloma cells (Fig. 6l). To further determine whether EZH2 protein binds to the EMP1 promoters via AP2α, we knocked down AP2α expression in myeloma cells using shAP2α (Fig. 6m) and observed decreased enrichment of EZH2, SUZ12 or H3K27me3 in the EMP1 promoter (Fig. 6n, o) and up-regulation of EMP1 expression (Supplementary Fig. 12a). Additionally, knockdown of AP2α expression can inhibit the proliferation of myeloma cells (Supplementary Fig. 12b) and the expression of osteolytic cytokines (Supplementary Fig. 12c). Moreover, knockdown of AP2α expression can weaken the promotion of osteoclast differentiation and the inhibition of osteoblast differentiation induced by myeloma cells (Fig. 6p). To determine the functional role of AP2α in lytic bone lesions in vivo, we injected AP2α knockdown myeloma cells into mouse femurs and observed higher percentages of BV/TV (Supplementary Fig. 12d), lower percentages of Oc. S/BS (Supplementary Fig. 12e), and higher percentages of Ob. S/BS (Supplementary Fig. 12f) in mice injected with AP2α knockdown myeloma cells than in those injected with control myeloma cells. These results demonstrate that AP2α plays a crucial role in recruiting the PRC2 complex to the EMP1 promoter, thereby promoting myeloma proliferation and bone damage (Fig. 6q).
EZH2 inhibitors reduce bone lesions and osteoclastogenesis and enhances osteoblastogenesis in vitro and in vivo
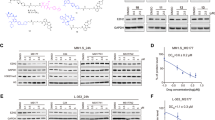
In terms of clinical application, extensive research has been conducted on various EZH2 inhibitors as potential therapeutic agents for cancer treatment. These inhibitors are being tested in clinical trials and have shown promising prospects for use in cancer therapy13,35. To develop a therapeutic approach, we asked whether inhibiting EZH2 can prevent myeloma-induced osteolytic bone lesions. In in vitro experiments, we pre-treated RPMI8226 cells with or without EZH2 inhibitor (DZNep or GSK343). Fresh culture medium was replaced after 24 h, and an additional 48 h later, conditioned medium was collected for co-culture experiments. The data showed that EZH2 inhibitors suppressed the enhanced osteoclast differentiation of myeloma cells, while promoting the inhibited osteoblast differentiation of myeloma cells (Fig. 7a, b). In in vivo experiments, RPMI8226 cells were intravenously injected into mice, followed by intraperitoneal administration of EZH2 inhibitors three times per week for a duration of three weeks, beginning one week after cell injection. We observed that EZH2 inhibitors significantly reduced the tumorigenic potential of RPMI8226 cells in xenograft models (Fig. 7c). Microcomputed tomography (Fig. 7d) and bone histomorphometric analysis (Fig. 7e) showed higher percentages of BV/TV (Fig. 7f), lower percentages of ES/BS and Oc.s/BS (Fig. 7g, h), higher percentages of OS/BS and Ob.s/BS (Fig. 7i, j), and lower human λ chain levels (Fig. 7k) in mice injected with EZH2 inhibitors. Additionally, EZH2 inhibition can significantly reduce the concentration of osteolytic cytokines in the serum of myeloma-bearing mice (Fig. 7l) and upregulate the expression of EMP1 in myeloma cells (Fig. 7m).
a, b We pre-treated myeloma cells (RPMI8226 or MM.1S) with DZNep (1 μM) or GSK343 (4 μM) and collected conditional medium. Shown are the formation of TRAP+ cells from preOCs and Alizarin red-S staining for osteoblast differentiation from MSCs, cultured with different myeloma cells conditional medium (30% volume ratio). Addition of PBS or no addition of conditional medium served as a control (n = 4 biological replicates). 6–7 weeks old female NSG mice were intravenously injected with luciferase-labeled RPMI8226 cells, and intraperitoneal administration of DZNep or GSK343 (100 μg/kg body weight) three times per week for a duration of two weeks, beginning two weeks after cell injection. c Representative images and summarized data of bioluminescent signals in NSG mice (n = 5 mice/group). d Shown are the representative microcomputed tomography images. NSG mice left femurs were extracted, fixed, TRAP- or toluidine blue-stained, and analyzed (n = 5 mice/group). Scale bar: 0.5 mm. e Representative images of TRAP- or toluidine blue-stained femurs of mice (n = 5 mice/group). Scale bar: 50 μm. Black arrow, osteoblast. Shown are the percentages of BV/TV (f), ES/BS (g), Oc.S/BS (h), OS/BS (i), Ob.S/BS (j), and human λ chain levels (k) (n = 5 mice/group). l ELISA analysis shown the concentrations of human CCL5, IL16, CSF1 and DKK1 in mouse serum (n = 5 mice/group). m After treatment, myeloma cells were also isolated from the right femurs of NSG mice using anti-CD138 antibody-coated magnetic beads. EMP1 mRNA levels were compared in isolated myeloma cells treated with or without EZH2 inhibitors (n = 5 mice/group). n Depiction of EZH2-mediated signaling pathways in myeloma cells and adipocytes. MM, multiple myeloma. Data are means ± SD. All p values were determined using one-way ANOVA with Tukey’s test. Source data are provided as a Source data file.
In order to further expand the clinical relevance of our findings, we also investigated whether our conclusions are applicable to other solid tumors that also induce osteolytic bone lesions. By analyzing the TCGA database, we observed increased EZH2 expression in several malignancies, such as breast, colon, kidney, and liver cancer (Supplementary Fig. 13a–d). To further confirm our findings, we knocked down or overexpressed EZH2 in breast cancer cell line MCF7 (Supplementary Fig. 14a), we observed decreased growth in EZH2 knockdown group compared to control group (Supplementary Fig. 14b), and increased growth in EZH2 overexpression group compared to control group (Supplementary Fig. 14b). Similar to myeloma cells, MCF7 cells expressing high levels of EZH2 (shCtrl or EZH2) induced more TRAP+ cells formation (Supplementary Fig. 14c) than cocultured with low EZH2 MCF7 cells (shEZH2 or Vec). Cocultured with low EZH2-expressing MCF7 cells (Vec or shEZH2) had comparatively more mature osteoblasts (Supplementary Fig. 14d) than those with high levels of EZH2 (EZH2 or shCtrl). These findings may have broader implications for understanding the mechanisms underlying the development of bone lesions caused by these and other types of tumors.
Discussion
Osteolytic bone lesions are a prominent characteristic of the majority of patients with myeloma. By disturbing the intricate equilibrium between bone formation and resorption, myeloma cells contribute to the development of debilitating osteolytic bone lesions. Although several studies have focused on how EZH2 promotes myeloma cell proliferation, its role in the development of myeloma-induced osteolytic bone lesions remains unclear. Our research revealed that cytokines derived from myeloma-associated adipocytes activate EZH2 in myeloma cells. The transcription factor AP2α facilitates binding of the EZH2 complex to the promoter region of the tumor suppressor gene EMP1. This binding induces the trimethylation of lysine 27 residues on histone H3, leading to the suppression of EMP1 expression. The inhibition of EMP1 consequently triggers myeloma cell proliferation and the secretion of osteolytic cytokines (Fig. 7n). We also conducted clinical studies to investigate the potential correlation between the expression level of EZH2 in myeloma cells and extent of osteolytic bone lesions in newly diagnosed patients. These studies provided additional evidence supporting our conclusion. Thus, this study revealed a mechanism that explains how EZH2 induces tumor proliferation and bone destruction. Our research indicated that EZH2 can serve as a dual therapeutic target for both myeloma and osteolytic bone lesions.
In contrast to previous investigations, our study delved into the role of EZH2 in the context of the bone marrow microenvironment, specifically in relation to the progression of myeloma and bone destruction. Our analysis revealed that both myeloma cells and bone marrow adipocytes exhibit heightened activation of EZH2. Moreover, we observed that myeloma cells were capable of reprogramming adipocytes, resulting in the secretion of increased levels of TNFα and IL6. These inflammatory cytokines not only contribute to bone damage, but also upregulate the expression of EZH2 in myeloma cells. More importantly, the EZH2 inhibitory complex in myeloma cells suppressed the expression of the tumor suppressor gene EMP1 through trimethylation of the lysine 27 residue on histone H3. Consequently, the progression of myeloma and development of osteolytic bone damage were facilitated. The interplay between myeloma cells and bone marrow adipocytes, with EZH2 serving as a central regulatory factor, forms a malignant regulatory loop that perpetuates the progression of myeloma and the occurrence of bone damage. Targeting EZH2 as a therapeutic strategy provides the potential for a dual impact, addressing both myeloma cells and associated bone destruction. By disrupting this regulatory loop, intervention aimed at EZH2 offers a promising approach to mitigate the myeloma progression and alleviate the burden of osteolytic bone disease.
The mechanism by which the PRC2 complex binds to DNA in mammalian cells is still not fully understood as it is not capable of directly binding to chromatin. Additional mediators such as transcription factors are necessary to facilitate this interaction. One of these, AP2α binds to the consensus sequence 5′-GCCN4GG-3′ in the promoter region36. This study demonstrated the function of AP2α as a bridge between the PRC2 complex and EMP1 gene promoter. On one hand, AP2α can bind to the PRC2 complex, and on the other hand, it can directly bind to specific sites in the promoter of the EMP1 gene. AP2α is expressed in various mammalian cells. The interaction between PRC2 and AP2α, as well as the recruitment of the PRC2/ AP2α complex to AP2α-targeted genes, may represent a widespread mechanism of gene silencing in eukaryotic systems.
Despite the significant insights gained from this study, there are noteworthy limitations that should be acknowledged. First, while our findings demonstrate the promising efficacy of EZH2 inhibitors in animal models, it is important to exercise caution regarding their translation to human diseases. These animal models operate within the murine bone marrow microenvironment, which may differ in critical aspects of the human marrow environment. Hence, the effectiveness of EZH2 inhibitors in the context of the human marrow remains uncertain and warrants further investigation. Second, there is a need to elucidate the molecular mechanisms underlying the inhibitory effect of EMP1 on myeloma cell proliferation and its regulation of the expression of key osteolytic cytokines. While our current study provides evidence for the role of EZH2 in EMP1 repression and the subsequent promotion of myeloma progression, extensive research is required to comprehensively understand the intricacies of this regulatory pathway. Efforts to address these limitations will contribute to a more thorough understanding of potential therapeutic strategies targeting EZH2 in myeloma-induced bone disease. Such investigations will aid in the development of effective treatments that can be translated into the clinical settings for the benefit of patients.
Currently, few targeted therapies are available for patients with myeloma bone disease. The most commonly used treatment for tumor-induced bone damage in clinical practice is bisphosphonate drugs. While they can inhibit osteoclast activity, they do not restore osteoblast activity2. Moreover, bisphosphonate drugs only show partial efficacy and are associated with side effects such as gastrointestinal discomfort, kidney damage, and osteonecrosis of the jaw2. Additionally, the anti-bone resorption drug denosumab (RANKL monoclonal antibody) has been approved for market use, but its phase III clinical trial results only demonstrated moderate anti-bone resorption activity and it does not aid in restoring osteoblast activity37. The antibody-based drug BHQ880 (DKK1 monoclonal antibody), which targets the osteolytic cytokine DKK1 derived from myeloma cells, did not show the ability to promote bone formation in phase I/II clinical trial results38. We believe that targeting EZH2 is a potential strategy for the treatment of myeloma-induced bone disease. Moreover, it is worth noting that other types of malignancies, such as breast and lung cancers, frequently metastasize to the bone, resulting in the formation of osteolytic lesions. Therefore, our findings may have significant implications beyond myeloma and contribute to a better understanding of the mechanisms underlying the development of osteolytic lesions in these cancers as well.
Methods
Ethic approvals
This study was approved by the Ethics Committee of Xiamen University and all protocols conformed to the Ethical Guidelines of the World Medical Association Declaration of Helsinki. Signed informed consent was obtained from all participating individuals prior to participation in the study. Mouse studies were approved by the Institutional Animal Care and Use Committee of Xiamen University. All protocols conformed to the Guidelines for Ethical Conduct in the Care and Use of Nonhuman Animals in Research.
Cell lines and primary myeloma cells
The myeloma cell lines ARP-1 and CAG were supplied by Dr. Zhiqiang Liu’s laboratory at Shandong First Medical University and Shandong Academy of Medical Sciences. The MCF7 (#HTB-22), HEK293T (#ACS-4500), RPMI8226 (CCL-155), and MM.1S (CRL-2974) cell lines were obtained from the American Type Culture Collection (ATCC). Primary myeloma cells were extracted from the bone marrow aspirates of myeloma patients using magnetic beads coated with anti-CD138 antibodies (Miltenyi Biotec). All patient samples were collected from the First Affiliated Hospital of Xiamen University. In our study, a total of 30 myeloma patient bone marrow samples were collected, including 17 male patients (56.7%) and 13 female patients (43.3%), with an age range of 49 to 77 years. It is compliant with the Guidance of the Ministry of Science and Technology (MOST) for the Review and Approval of Human Genetic Resources. Myeloma cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS), while MCF7 and HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS.
Plasmid construction, antibodies, and reagents
Human HA-tagged EZH2, MYC-tagged EZH2, HA-tagged SUZ12, HA-tagged AP2α, EMP1 and AP2α plasmids were subcloned into a plvx-AcGFP1-N1 vector (NovoPro, #V012707). The primers used for plasmid construction and amino acid deletion fragment of AP2α are listed in Supplementary Table 1. pRRL lentiviral luciferase vectors were purchased from Addgene (#120798). Except where specified, all chemicals were purchased from Sigma-Aldrich, and all antibodies for western blot analysis were purchased from Cell Signaling Technology. shRNAs against EZH2, AP2α and non-target controls were purchased from Sigma-Aldrich.
Stable cell line construction
Transient transfections of HEK 293T cells were conducted using polyethyleneimine (PEI) (Polysciences, Warrington) in OPTI-MEM medium (Life Technologies) at a 1:4 DNA-to-PEI ratio. Viral particles were then produced in HEK 293T cells co-transfected with 4 μg pMD2.G (#12259) and 6 μg psPAX2 (#12260) packaging plasmids (Addgene), and together with 8 μg lentiviral expressing vectors encoding target genes, including human EZH2, EMP1, pLKO.1 vector encoding shRNAs targeting EZH2 or AP2α. The viral supernatant was collected 48 h post-transfection and concentrated 100-fold using Polyethylene glycol 8000 (Sigma-Aldrich). Myeloma cells were infected by spinfection with the concentrated viral particles in the presence of polybrene at 800 × g for 30 min at 37 °C. The medium was replaced 12 h post-infection, and the cells were cultured for an additional 48 h before further processing. The stable cell lines were selected by adding puromycin (2 μg/mL) (#540222, Sigma-Aldrich) into the culture medium. MCF-7 cells were infected with lentivirus for 12 h at 37 °C with 5 mg/mL polybrene. Cells were selected in DMEM with 10% FBS and 2 μg/mL puromycin for two weeks. To establish myeloma cell lines RPMI8226 (Vec, EZH2) or MM.1S (Vec, EZH2) with human EMP1 forced expression, cells were infected twice with lentiviruses expressing human EMP1. To track cancer cells in mice, myeloma cell lines RPMI8226 (shCtrl, shEZH2) or MM.1S (Vec, EZH2) were infected twice with lentiviruses with forced luciferase expression.
Collection of adipocyte-conditioned medium in vitro
Bone marrow mesenchymal stem cells were seeded on the back of the Transwell insert, induced them to differentiate into mature adipocytes11, and then seeded myeloma cells in the insert. The pore size of the insert membrane at 0.4 µm was chosen, which allows physical contact but at the same time the cells can’t penetrate through. After 3 days of co-culture, the myeloma cells were removed by collagenase digestion, allowing the treated adipocytes to fall to the bottom of the culture well, and then the adipocytes were replated. After 3 days, the supernatant was removed and replaced with fresh medium for an additional 3-day culture period. The supernatant obtained is the required adipocyte-conditioned medium.
In vitro osteoblast and osteoclast formation and function assays
MSCs were extracted from bone marrow samples. Subsequently, these MSCs were differentiated into mature osteoblasts utilizing a specialized osteoblast differentiation medium, following the protocol outlined in our prior study2. Alizarin Red S staining (Sigma-Aldrich) was employed to evaluate the osteogenic capacity of the resultant osteoblasts. In parallel, human monocytes were carefully isolated from peripheral blood mononuclear cells and subjected to culture conditions conducive to the development of osteoclast precursors. These precursors were then exposed to a culture medium supplemented with M-CSF (25 ng/ml) and a low dose of RANKL (10 ng/ml). Additionally, co-culture experiments were conducted, wherein the precursors were either cultured alone or in the presence of myeloma cells for a duration of 7 days. The successful formation of mature osteoclasts was subsequently confirmed through TRAP staining, utilizing a leukocyte acid phosphatase kit (Sigma-Aldrich). For the purpose of investigating the gene expression profiles associated with the differentiation processes of both osteoclasts and osteoblasts, a co-culture period of 2 days was established as the standard timeframe.
Western blot analysis
After collection, cells were disrupted using a 1× lysis buffer (#9803, Cell Signaling Technology). The resulting cell lysates were then processed through SDS-PAGE, followed by transfer onto a nitrocellulose membrane. Subsequently, the membrane was probed with antibodies against GAPDH (#5174, 1:2000), EZH2 (#5246, 1:1000), SUZ12 (#3737, 1:1000), AP2α (#3215, 1:1000), phospho-IκBα (#2859, 1:1000), IκBα (#4812, 1:1000), H3K27me3 (#9733, 1:1000), phospho-ERK1/2 (#4370, 1:1000), ERK1/2 (#4695, 1:1000), phospho-p38 (#4511, 1:1000), p38 (#8690, 1:1000), phospho-Akt (#4060, 1:1000), Akt (#9272, 1:1000), Myc tag (2276, 1:2000) and HA tag (#3724, 1:2000) (Cell Signaling Technology). EMP1 antibody was purchased from Abcam (#ab230445, 1:1000).
Real-time quantitative PCR
Total RNA extraction was conducted utilizing the RNeasy kit (QIAGEN). A quantity of 1 µg of the total RNA was subsequently utilized for reverse transcription (RT) employing the SuperScript II RT-PCR kit (Invitrogen). Quantitative PCR analysis was carried out with the SYBR Green Master Mix (Life Technologies), using the QuantStudio 3 Real-Time PCR System (Life Technologies). The PCR protocol consisted of an initial denaturation at 95 °C for 10 min, succeeded by 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 60 s. GAPDH was employed as the endogenous control for normalization. The specific primers utilized in the study are detailed in Supplementary Table 2.
Cell viability and ELISA analysis
For viability assays, cells were plated at 1 × 104 cells/well in triplicate. Assays were performed using Cell Counting Kit-8 (CCK-8) (#CK04, Dojindo). ELISA kits were purchased from R&D system and Immunodiagnostic Systems.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections of bone marrow biopsy samples obtained from myeloma patients were deparaffinized and stained. Slides were stained with anti-CD138 (LS-B9360, LifeSpan BioSciences, 1:100) and EZH2 (#5246, Cell Signaling Technology, 1:50) antibodies using an EnVision System (#K5361, DAKO) following the manufacturer’s instructions and counterstained with hematoxylin.
Immunoprecipitation and pull-down assays
Cells were lysed and incubated on ice for 15 min. The total protein lysate was immunoprecipitated with an agarose-immobilized antibody at 4 °C overnight. After washing 6 times, the beads were resuspended in 30 μl of 1× SDS buffer. After boiling for 5 min, pull-down samples were run on SDS-PAGE gel along with a 5% input sample and transferred to a PVDF membrane for immunoblotting. IgG was used as a control and total cell lysates were used as input controls. For the pull-down assay, HEK293T cells were transfected with either HA or Myc-tag protein plasmid. Lysates of the cells were pulled down with anti-HA agarose (#26181, Thermo Fisher Scientific) or anti-c-Myc agarose (#20168, Thermo Fisher Scientific) and further incubated with cell lysates transfected with full length or truncated AP2α. The immunoprecipitates and whole cell lysates (WCL) were immunoblotted. Cells that were not transfected with HA-EZH2, HA-SUZ12 or Myc-EZH2 plasmids served as controls.
ChIP assay
Cells were fixed in 4% formaldehyde and sonicated to prepare chromatin fragments. Chromatin samples were immunoprecipitated with antibodies against SUZ12 (#3737, Cell Signaling Technology, 1:100), EZH2 (#5246, Cell Signaling Technology, 1:100), H3k27me3 (#9733, Cell Signaling Technology, 1:50), AP2α (#3215, Cell Signaling Technology, 1:100), and control immunoglobulin G (IgG) at 4 °C for 3 h. Immunoprecipitates and total chromatin inputs were reverse cross-linked, DNA was isolated and analyzed using PCR with primers targeting the promoter regions of EMP1. The primer sequences used are listed in Supplementary Table 3.
Immunofluorescent staining
Myeloma cells were fixed with 4% formaldehyde and permeabilized with 0.3% Triton X-100, then cells were blocked with 2% goat serum. After stained with anti-EZH2 (#5246, Cell Signaling Technology, 1:200) and AP2α (#sc-12726, Santa cruz, 1:25) antibodies overnight at 4 °C, cells were incubated with FITC-conjugated Goat anti-Mouse IgG (H + L) (#AS001, Abclonal, 1:200) and Cy3-conjugated Goat anti-Rabbit IgG (H + L) (#AS007, Abclonal, 1:200) for 60 min at room temperature and nucleus counterstaining with DAPI. Formalin-fixed, paraffin-embedded sections of bone marrow biopsy samples obtained from myeloma patients were deparaffinized and stained. Slides were stained with anti-CD138 (#AF2780, R&D system, 1:100), EZH2 (#MA5-18108, Thermo Fisher Scientific, 1:25), AP2α (#3215, Cell Signaling Technology, 1:100) or EMP1 (#ab230445, Abcam, 1:50) overnight at 4 °C. After incubation, samples were stained with FITC-conjugated Goat anti-Mouse IgG (H + L) (#AS001, Abclonal, 1:200), Cy3-conjugated Goat anti-Rabbit IgG (H + L) (#AS007, Abclonal, 1:200) and Cy5-conjugated Mouse Anti-Goat IgG (#K0038M-Cy5, Solarbio, 1:200) for 60 min at room temperature and nucleus counterstaining with DAPI. Immunofluorescence images were acquired with an IX71 confocal microscope system (Olympus). Mean fluorescence intensity (MFI) was quantified using ImageJ software (v 2.1.0/1.53c). The mean fluorescence intensity of each region per individual was obtained by initially computing the ratio of the total fluorescence intensity of all visual fields to the total area of the corresponding fields in the indicated region. The mean fluorescence intensity for each respective signal was determined per slide in at least six areas per tumour.
Whole-transcriptome sequencing
RNAs were isolated from RPMI8226 (shCtrl, shEZH2) and then purified with the VAHTSTM DNA Clean Beads (Vazyme). RNA-seq libraries were synthesized with VAHTS Universal V6 RNA-seq Library Prep Kit (Vazyme). Pooled libraries were quantified using Qubit TM dsDNA HS Assay Kit (ThermoFisher), examined for size distribution using the Fragment Analyzer (Advanced Analytical), and sequenced in MGI Tech DNBSEQ-T7 Sequencer.
Mass spectrometry
Lysates of the HA-EZH2 transfected RPMI8226 cells were pulled down with anti-HA agarose and subjected to SDS-PAGE gel. Gel bands stained with Coomassie blue were excised separately, alkylated, and digested with trypsin. The tryptic peptides were then analyzed using a nano-LC/MS/MS (Thermo Fisher Scientific) coupled with an 1100 HPLC system (Agilent Technologies). The MS/MS spectra were searched using the SEQUEST software program with the BioWorks Browser (version 3.3.1; Thermo Fisher Scientific) against the NCBI database. RPMI8226 cells transfected with an empty vector were used as controls.
Luciferase assay in vitro
The construct covers the full-length (Luc-EMP1: −1.258 kb to 62 bp), two truncated forms (Luc-EMP1-Δ1: −558 bp to 62 bp; Luc-EMP1-Δ2: −458 bp to 62 bp), and a mutated form (Luc-EMP1-mut: GCCN4GG to GATN4TA) made around −528 bp upstream of the starting codon in the promoter of the EMP1 gene were subcloned into the pGL2 vector, and their transcriptional activities in HEK293T cells were examined using a Dual-Luciferase Reporter Assay System (#E1910, Promega) according to the manufacturer’s instructions. The luciferase activity of Luc-EMP1 constructs was set to 1. The primers used in the subcloning are listed in Supplementary Table 4.
In vivo mouse experiments, measurement of tumor burden, radiography and bone histomorphometry
Mouse studies were approved by the Institutional Animal Care and Use Committee of Xiamen University. 6–7 weeks old female NSG mice purchased from the Shanghai Model Organisms Center, Shanghai, China, were maintained in Xiamen University Animal Care-accredited facilities. All mice were housed in the specific pathogen-free (SPF) room under controlled temperature (20–26 °C) and humidity (40–70%) conditions with 12 h light/dark cycle. NSG mice were assigned randomly to groups and intravenously or intrafemorally injected with luciferase-expressing myeloma cells (RPMI8226 or MM.1S) (5 × 105 cells/mouse). For the primary patient-derived xenograft mouse model, newborn (1–3 days) NSG mice were assigned randomly to groups and intrahepatically injected with primary myeloma cells (1 × 106 cells/mouse) isolated from myeloma patients. Tumor progression was monitored by levels of mouse serum human λ chains. Human λ chains ELISA Kits were purchased from Bethyl Laboratories (#E88-116). The tumor burden of all tumor-bearing mice involved in this study were controlled within 20 μg/ml, which is in accordance with the permission from the ethics committee of Xiamen University. The maximal tumor burden was not exceeded. For the bioluminescent signal assay, mice were imaged using an IVIS Spectrum in vivo imaging system from PerkinElmer, following the manufacturer’s recommendations. The end of the experiments was decided based on our and other previous studies2,10,39,40. In brief, x-ray analysis was performed 6 weeks after injection of tumor cells to facilitate the observation of cortical bone surface damage. Additionally, the human λ chain levels in mice serum were required to be below 20 μg/ml. For the other myeloma cell line mouse model, micro-CT analysis, bioluminescent analysis, and bone histomorphometry analysis were conducted 4 weeks after myeloma cell injection, with serum human λ chain levels maintained below 10 μg/ml. At this time point, the damage to the trabecular bone in mice can be conveniently observed. For the primary patient-derived xenograft mouse model, due to the slower growth rate of primary patient myeloma cells, micro-CT analysis and bone histomorphometry analysis were performed 18 weeks after myeloma cell injection, with serum human λ chain levels below 10 μg/ml. To examine lytic bone lesions, radiographs were scanned with a Bruker In-Vivo Xtreme imaging system. μ-CT was conducted with the Scanco μCT-40 system (55-peak kilovoltage and 145-μA x-ray source). Mouse femurs were scanned at 16-μm resolution, and further analyzed with Microview (Parallax Innovations) software. To assess dynamic histomorphometric indices, mice were administered two injections of 20 mg/kg calcein (Sigma Aldrich) at 6 and 3 days before dissection. Bone tissues were fixed in 10% neutral-buffered formalin and decalcified, and sections of them were stained with toluidine blue or TRAP following standard protocols. Both analyzes were performed using the BIOQUANT OSTEO (v18.2.6) software program (BIOQUANT Image Analysis Corporation).
Statistics & Reproducibility
Statistical significance was analyzed using the Graphpad (Version 9.0) program with two tailed unpaired Student’s t-tests for comparison of two groups, and one-way ANOVA with Tukey’s multiple comparisons test for comparison of more than two groups. p values less than 0.05 were considered statistically significant. All results were reproduced in at least three independent experiments. The determination of sample size, replication structure, and the chosen intermediate endpoint was informed by prior experience2,11. Initially, we calculated the sample size through a power analysis leveraging our existing insights into bone histomorphometry. We decided to include five mice in each group to maintain an 80% power for detecting variations in bone measurements across the groups, with the two-tailed type I error rate set at 0.05. Bone lesions in the study participants were characterized by radiologists who were blinded to our study. The investigators who performed the bench studies were blinded to the bone lesion status of the patients.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data associated with this study can be found in the paper or the supporting documents. Source data are provided with this paper. Uncropped scans of immunoblots and gels are provided in the Source data. The RNA-seq data generated in this study is available at NCBI Sequence Read Archive under accession code PRJNA1165696. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD056039. The processed mass spectrometry data are available at Supplementary Data 1. The raw and processed RNA-seq and ChIP-seq data accessed from other studies have been deposited in the NCBI GEO database (https://www.ncbi.nlm.nih.gov/gds/) with accession numbers GSE755, GSE24990, GSE143025, GSE176463, GSE196297, GSE113736, GSE113295, GSE155135, GSE103567, GSE109673, GSE2658, GSE5900, and GSE214669, respectively. Source data are provided with this paper.
References
Palumbo, A. & Anderson, K. Multiple myeloma. N. Engl. J. Med. 364, 1046–1060 (2011).
Liu, H. et al. Thymidine phosphorylase exerts complex effects on bone resorption and formation in myeloma. Sci. Transl. Med. 8, 353ra113 (2016).
Takegahara, N., Kim, H. & Choi, Y. Unraveling the intricacies of osteoclast differentiation and maturation: insight into novel therapeutic strategies for bone-destructive diseases. Exp. Mol. Med. 56, 264–272 (2024).
Giuliani, N., Rizzoli, V. & Roodman, G. D. Multiple myeloma bone disease: pathophysiology of osteoblast inhibition. Blood 108, 3992–3996 (2006).
Roodman, G. D. Biology of osteoclast activation in cancer. J. Clin. Oncol. 19, 3562–3571 (2001).
Liu, H. et al. Promotion of bone lesions through the myeloma integrin alpha6-mediated osteolytic signaling. Front. Oncol. 11, 692190 (2021).
Liu, Z. et al. Myeloma cells shift osteoblastogenesis to adipogenesis by inhibiting the ubiquitin ligase MURF1 in mesenchymal stem cells. Sci. Signal. 13, eaay8203 (2020).
He, J. et al. p38 MAPK in myeloma cells regulates osteoclast and osteoblast activity and induces bone destruction. Cancer Res. 72, 6393–6402 (2012).
Tian, E. et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N. Engl. J. Med. 349, 2483–2494 (2003).
Liu, H. et al. Osteocyte CIITA aggravates osteolytic bone lesions in myeloma. Nat. Commun. 13, 3684 (2022).
Liu, H. et al. Reprogrammed marrow adipocytes contribute to myeloma-induced bone disease. Sci. Transl. Med. 11, eaau9087 (2019).
Yang, J. et al. C-reactive protein promotes bone destruction in human myeloma through the CD32-p38 MAPK-Twist axis. Sci. Signal 10, eaan6282 (2017).
Kim, K. H. & Roberts, C. W. Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016).
Yin, X., Yang, S., Zhang, M. & Yue, Y. The role and prospect of JMJD3 in stem cells and cancer. Biomed. Pharmacother. 118, 109384 (2019).
Nutt, S. L., Keenan, C., Chopin, M. & Allan, R. S. EZH2 function in immune cell development. Biol. Chem. 401, 933–943 (2020).
Yao, Y. et al. Downregulation of enhancer of zeste homolog 2 (EZH2) is essential for the induction of autophagy and apoptosis in colorectal cancer cells. Genes 7, 83 (2016).
Ito, T., Teo, Y. V., Evans, S. A., Neretti, N. & Sedivy, J. M. Regulation of cellular senescence by polycomb chromatin modifiers through distinct DNA damage- and histone methylation-dependent pathways. Cell Rep. 22, 3480–3492 (2018).
Batool, A., Jin, C. & Liu, Y. X. Role of EZH2 in cell lineage determination and relative signaling pathways. Front. Biosci. 24, 947–960 (2019).
Kleer, C. G. et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl Acad. Sci. USA 100, 11606–11611 (2003).
Wu, Z. L. et al. Polycomb protein EZH2 regulates E2F1-dependent apoptosis through epigenetically modulating Bim expression. Cell Death Differ. 17, 801–810 (2010).
Liu, X. et al. LINC00665 induces acquired resistance to gefitinib through recruiting EZH2 and activating PI3K/AKT pathway in NSCLC. Mol. Ther. Nucleic Acids 16, 155–161 (2019).
Zingg, D. et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun. 6, 6051 (2015).
Ma, J., Zhang, J., Weng, Y. C. & Wang, J. C. EZH2-mediated microRNA-139-5p regulates epithelial-mesenchymal transition and lymph node metastasis of pancreatic cancer. Mol. Cells 41, 868–880 (2018).
Lu, C. et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell 18, 185–197 (2010).
Peng, D. et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527, 249–253 (2015).
Pawlyn, C. et al. Overexpression of EZH2 in multiple myeloma is associated with poor prognosis and dysregulation of cell cycle control. Blood Cancer J. 7, e549 (2017).
Ishiguro, K. et al. Dual EZH2 and G9a inhibition suppresses multiple myeloma cell proliferation by regulating the interferon signal and IRF4-MYC axis. Cell Death Discov. 7, 7 (2021).
Terpos, E., Ntanasis-Stathopoulos, I., Gavriatopoulou, M. & Dimopoulos, M. A. Pathogenesis of bone disease in multiple myeloma: from bench to bedside. Blood Cancer J. 8, 7 (2018).
Lee, J. W. et al. The HIV co-receptor CCR5 regulates osteoclast function. Nat. Commun. 8, 2226 (2017).
Chang, Y. et al. Synovial fluid interleukin-16 contributes to osteoclast activation and bone loss through the JNK/NFATc1 signaling cascade in patients with periprosthetic joint infection. Int J. Mol. Sci. 21, 2904 (2020).
Hernando, H. et al. EZH2 inhibition blocks multiple myeloma cell growth through upregulation of epithelial tumor suppressor genes. Mol. Cancer Ther. 15, 287–298 (2016).
Wang, Y., Zhang, L., Yao, C., Ma, Y. & Liu, Y. Epithelial membrane protein 1 promotes sensitivity to RSL3-induced ferroptosis and intensifies gefitinib resistance in head and neck cancer. Oxid. Med Cell Longev. 2022, 4750671 (2022).
Yu, X. et al. Dissecting and targeting noncanonical functions of EZH2 in multiple myeloma via an EZH2 degrader. Oncogene 42, 994–1009 (2023).
Eckert, D., Buhl, S., Weber, S., Jager, R. & Schorle, H. The AP-2 family of transcription factors. Genome Biol. 6, 246 (2005).
Duan, R., Du, W. & Guo, W. EZH2: a novel target for cancer treatment. J. Hematol. Oncol. 13, 104 (2020).
McPherson, L. A. & Weigel, R. J. AP2alpha and AP2gamma: a comparison of binding site specificity and trans-activation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 27, 4040–4049 (1999).
Terpos, E., Confavreux, C. B. & Clezardin, P. Bone antiresorptive agents in the treatment of bone metastases associated with solid tumours or multiple myeloma. Bonekey Rep. 4, 744 (2015).
Fulciniti, M. et al. Anti-DKK1 mAb (BHQ880) as a potential therapeutic agent for multiple myeloma. Blood 114, 371–379 (2009).
Lazzari, E. et al. Alu-dependent RNA editing of GLI1 promotes malignant regeneration in multiple myeloma. Nat. Commun. 8, 1922 (2017).
Li, Z. et al. Acetyl-CoA synthetase 2: a critical linkage in obesity-induced tumorigenesis in myeloma. Cell Metab. 33, 78–93e77 (2021).
Acknowledgements
We thank the Research Histology and Bone Histomorphometry Core Labs and the Preclinical Imaging Core at the Xiamen University. This research was supported by the National Natural Science Foundation of China (82170197 to H.L.), the Fujian Science Fund for Distinguished Young Scholars (2022D015 to H.L.), Shenzhen Science and Innovation Committee (JCYJ20220530143403007 to H.L.), Basic Scientific Research Projects of Public Welfare Research Institutes Affiliated to Fujian Province (2024R1036 to H.L.), and Guangdong Basic and Applied Basic Research Foundation (2024A1515010076 to H.L.).
Author information
Authors and Affiliations
Contributions
H.L. and Z.Q.L. designed all experiments and wrote the manuscript; R.L., Z.W.L., R.C., and Z.H.F. performed the experiments and statistical analysis; Z.H.F. provided patient samples. All authors have reviewed the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, R., Li, Z., Chen, R. et al. EZH2 serves as a viable therapeutic target for myeloma-induced osteolytic bone destruction. Nat Commun 16, 1206 (2025). https://doi.org/10.1038/s41467-025-56506-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56506-5
This article is cited by
-
Structural characteristics and SARs of EZH2 inhibitors
Molecular Diversity (2025)