Abstract
Quality improvement, clinical research, and patient care can be supported by medical predictive analytics. Predictive models can be improved by integrating more patient records from different healthcare centers (horizontal) or integrating parts of information of a patient from different centers (vertical). We introduce Distributed Cross-Learning for Equitable Federated models (D-CLEF), which incorporates horizontally- or vertically-partitioned data without disseminating patient-level records, to protect patients’ privacy. We compared D-CLEF with centralized/siloed/federated learning in horizontal or vertical scenarios. Using data of more than 15,000 patients with COVID-19 from five University of California (UC) Health medical centers, surgical data from UC San Diego, and heart disease data from Edinburgh, UK, D-CLEF performed close to the centralized solution, outperforming the siloed ones, and equivalent to the federated learning counterparts, but with increased synchronization time. Here, we show that D-CLEF presents a promising accelerator for healthcare systems to collaborate without submitting their patient data outside their own systems.
Similar content being viewed by others
Introduction
Medical predictive analytics can support quality improvement, clinical research, and eventually improve patient health status1. For example, machine learning was leveraged to better understand the Coronavirus Disease 2019 (COVID-19) pandemic and discover actionable factors2. To improve the performance of modeling approaches and to identify medication-outcome associations for diseases, these approaches need to use a large number of patient records. This can be accomplished by integrating data “horizontally”, e.g., when multiple patients have the same type of data from different institutions, or by expanding the collection of clinical information by integrating the data “vertically”, e.g., a patient has clinical data in one hospital and vaccination data elsewhere, from multiple healthcare systems. Many institutions collect COVID-19 data; however, a higher number of records is needed to increase statistical power, especially when the data are imbalanced. On the other hand, many of these patients may have primary care physicians (PCPs) as well as a large portion of their Electronic Health Records (EHRs) in another hospital. Therefore, the capability to use data in higher volume (i.e., horizontally-partitioned) or with a wider/deeper range of variables (i.e., vertically-partitioned) across institutions is also critical for collaborative model improvement that responds rapidly to pandemics. Existing solutions such as centralized, federated3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18, or decentralized privacy-preserving19,20,21,22,23,24,25,26,27,28 methods possess potential privacy risks29, security issues30,31,32, and typically focus only on horizontally-partitioned data19,20,21,22,23,24,25,26,27,28 (SUPPLEMENTARY NOTES Section 1).
We hypothesized that a completely decentralized cross-institutional learning method would be capable of incorporating horizontally- and vertically-partitioned data while still protecting patients’ privacy and conforming with policies/regulations. A solution should ideally include the following features: (1) Data. Ideally, the method should be able to increase the number of data records in the horizontal scenario; it should also be capable of extending the collection of clinical variables in the vertical scenario; and, more importantly, patients’ privacy should be preserved by not disseminating their sensitive data outside each institution. (2) Sites. Additionally, the method should allow the institutions of the research network consortium to collaboratively improve the predictive model; it should also adopt autonomous peer-to-peer topology to avoid single-point-of-control; and the compute loads for each participating site should be fair. (3) Processes. The core federated learning algorithms should exhibit performance equivalent to their centralized learning counterparts; the underlying distributed systems should engage the community to allow long-term sustainability; and the whole learning process should be recorded immutably for future auditing and dispute resolution. (4) Models. Finally, the learned models should be source-verifiable by each site to ensure provenance/trustworthiness; they should also be shared transparently to enable identification of potentially tampered models; and model storage should be scalable. Here, we introduce D-CLEF, which incorporates both horizontally- and vertically-partitioned data without patient-level record dissemination, supports a fully-distributed and computationally-fair model building across institutions, adopts centralization-equivalent algorithms and community-backed distributed platforms with an immutable audit trail, and constructs trustworthy, transparent and scalable predictive models.
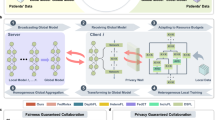
Conceptually, two major cross-institutional data partitioning scenarios are horizontal (Fig. 1a) and vertical (Fig. 1b). If each hospital database contains enough patients and covariates, it would be feasible to locally train a predictive model for each respective institution (Fig. 1c). Improving the predictive model requires collecting more data from multiple institutions, either horizontally or vertically. Once the data are collocated in a central database, a “global” model can be trained (Fig. 1d); however, this also brings privacy risks such as re-identification29. Federated learning methods3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18 build a global model by only disseminating the “local” models instead of horizontally- or vertically-partitioned data, thus protecting patients privacy (Fig. 1e). That said, the central learning server still presents a single-point-of-failure/control 30,31,32.
a Horizontally-partitioned structural clinical data from multiple healthcare sites. In this scenario, each site contains data from different patients, while the covariates are standardized across all sites. b Vertically-partitioned data from sites. In this scenario, each site contains different parts of the covariates for the same set of patients. c Siloed learning. For both horizontal and vertical scenarios, each site may train their own “local” models. However, such models could suffer from smaller sample size, as well as incomplete covariate information. d Centralized learning. Intuitively, the data can be disseminated to a cloud or on premise central repository, to train a “global” model with a larger sample size and a complete covariate set. Nevertheless, transferring sensitive data could present a privacy risk of patients’ data being re-identified. e Concept of federated learning. To build a model with more patients and covariates, federated learning algorithms allow sites to exchange only partially-trained machine learning models without disseminating patients’ data. However, a central server is still required to moderate the learning process, and such a server is a vulnerable single-point-of-control. f Principle of D-CLEF. By decentralizing the modeling process while keeping data locally, D-CLEF protects patients’ privacy without a single-point-of-control. More importantly, D-CLEF handles both horizontal and vertical data partitioning scenarios, thus can support wider biomedical applications where data is distributed in either way. g Conceptual design of D-CLEF, which contains two permissioned networks: the modeling and the storage ones. The former is a blockchain-based network for model training purposes, and the latter is a distributed file system to share the models. Each D-CLEF site contains three nodes: local computing, modeling, and storage. The data are only used within the local computing node and never disseminated to either of the networks.
In this work, to mitigate these shortcomings, we developed D-CLEF, which adopts blockchain30,31,32 to support decentralized training on local data in horizontal and vertical scenarios (Fig. 1f). Each D-CLEF site contains a local computing node to access data, a modeling node connecting to a permissioned (i.e., not publicly-available, to further protect patients’ privacy) blockchain for model training processes, and a storage node connecting to a permissioned distributed file system33 for model storage (Fig. 1g). The advantages of the design of D-CLEF are summarized in terms of data, sites, processes, and models (Supplementary Fig. 1a). The modeling network stores training details, while the actual model contents are stored in the storage network (Supplementary Fig. 1b). D-CLEF uses smart contracts30, which are programs stored and run on a blockchain network, to execute and record the model construction process (Supplementary Fig. 1c). The use of blockchain and smart contracts ensures the provenance/transparency of the models and the immutable learning process audit trail, while the integration with a distributed file system improves the scalability in terms of the model size. D-CLEF is also sustainable by adopting a blockchain and a distributed file system developed and maintained by the community. By keeping the data locally within each institution and learning the global model in a fully-distributed way, D-CLEF not only protects patients’ privacy, but also avoids the risks of having a central server (Supplementary Fig. 2a). Meanwhile, each D-CLEF site serves as a “virtual” server in each model learning iteration in a round-robin way to ensure computational fairness across institutions (Supplementary Fig. 2b). Furthermore, the adoption of the algorithms that were mathematically-proven to be equivalent to the centralized learning methods further guarantees the performance of D-CLEF (Supplementary Fig. 3a), for horizontal (Supplementary Fig. 3b) and vertical (Supplementary Fig. 3c) scenarios. We select three clinical datasets for evaluation: COVID-19 pandemic, total hip arthroplasty surgery, and myocardial infarction disease, to demonstrate the wide potential adoption of D-CLEF in various use cases, then compared D-CLEF models with siloed, centralized, and existing federated learning models for horizontal and vertical partitions. Compared to existing healthcare analytics technologies that implement federated learning, blockchain, or distributed file systems, D-CLEF is innovative because it not only combines all the above-mentioned technologies, but also supports horizontal data partitioning, vertical data partitioning, and fully distributed learning (Supplementary Data 1). These technical novelties can advance the field of cross-institutional privacy-preserving model learning.
Results
D-CLEF for prediction of COVID-19 mortality
Our first use case is to predict mortality among 15,297 patients diagnosed with COVID-19 (Fig. 2a) from five University of California (UC) Health medical centers: UC San Diego (UCSD), UC Irvine (UCI), UC Los Angeles (UCLA), UC Davis (UCD), and UC San Francisco (UCSF), to look for beneficial or harmful impact in eight datasets (X for overall, X1–X2 for horizontal, and X3–X7 for vertical scenarios). Across the entire dataset, 1072 (7.0%) patients died. The categories of covariates included in this data were “patient and COVID-19 information” (e.g., “patient age group”, “vaccine manufacturer”, etc.) and “drug prescribed” (e.g., “ibuprofen”, “albuterol”, etc.) (Supplementary Data 2). The data were derived from the UC Health COVID-19 Research Data Set (CORDS) limited data set34 (METHOD Section 1.1). Relevant biological variables for human subjects (age group, sex, race, and ethnicity) were included and addressed as covariates for the predictive models (Supplementary Figs. 4a–d, Supplementary Data 3–6).
a The UC CORDS data (dataset X) with n = 15,279 patients and m = 100 covariates, including patient and COVID-19 information (dataset X1) and drugs prescribed (dataset X2). The patients were from five UC Health medical centers (datasets X3–X7). b Overview of the setup on dataset X to estimate the performance of D-CLEF. c Main results of the horizontal scenario on dataset X, comparing D-CLEF with siloed and centralized LR models. d Main results of D-CLEF in the vertical scenario on dataset X, compared with siloed/centralized LR models. e Main overall comparison on dataset X, using two-sided, two-sample Wilcoxon signed rank-test. All box plots are derived from 30 trials, with median as center line, upper and lower quartiles as box limits, 1.5x interquartile range as whiskers, and outliers as points.
We compared D-CLEF’s performance versus siloed and centralized models on dataset X (Fig. 2b) using the full Area Under the receiver operating characteristic Curve (AUC)35 as our major performance metric (SUPPLEMENTARY NOTES Section 2). We compared AUCs between methods using the two-sided, two-sample Wilcoxon signed rank-test36 (“METHOD” Section 5) in our evaluation process (SUPPLEMENTARY NOTES Section 4). For the horizontal scenario, the results demonstrated that D-CLEF performed similarly to centralized Logistic Regression (LR) learning, and that it outperformed all siloed LR models (Fig. 2c) and provided prediction performance with a smaller interquartile range. Although our algorithms were mathematically proven to be equivalent to the centralized counterparts, the different performances happen because of the regularization setting (SUPPLEMENTARY NOTES Section 2). For the vertical scenario, D-CLEF provided the same-level results as the centralized LR model, with a higher AUC score than the siloed LR models (Fig. 2d). Although vertical siloed learning results were tested using only partial clinical covariates, we still compared their results with D-CLEF and centralized learning, to demonstrate the benefits of being able to use a wider coverage of covariates. Lastly, we compared the horizontal and vertical D-CLEF models with the centralized and ensemble ones (“METHOD” Section 2.7). In general, the vertical D-CLEF and the centralized models performed better than the horizontal D-CLEF model statistically; however, the actual differences in AUC scores were relatively small (Figs. 2e, Supplementary Data 7–8). The ensemble models in general performed at a similar level when compared to D-CLEF without ensemble. Next, we tested the horizontal D-CLEF method on datasets X1–X2 (Fig. 3), and the results (Supplementary Data 9) showed that D-CLEF performed similarly to centralized LR learning, which was in general close to or better than the siloed LR models (SUPPLEMENTARY NOTES Section 5). Vertical D-CLEF was also evaluated on five datasets X3–X7 (Fig. 4), and the results (Supplementary Data 10) showed that D-CLEF generally performed better when compared to the centralized and siloed LR models (SUPPLEMENTARY NOTES Section 6).
We also implemented and compared D-CLEF with its federated learning counterpart (“METHOD” Section 2.7). Since the predictive performances are the same, we focused on evaluating the runtime difference of the two implementations. For both horizontal and vertical learning, the results (Supplementary Data 11–12) showed that in general D-CLEF required about 10% more runtimes per iteration than their federated implementations (SUPPLEMENTARY NOTES Section 7). In essence, we demonstrated that for both horizontal and vertical scenarios, D-CLEF is applicable for pandemic patient outcome prediction at a comparable level with the centralized learning while outperforming siloed learning, with an additional cost of synchronization time per learning iteration.
D-CLEF to identify prolonged hospitalization after major surgery
We constructed a second use case to predict prolonged hospital length of stay following total hip arthroplasty (THA) for 960 patients at UCSD (Fig. 5a), defined as greater than or equal to the average 3-day hospitalization20,23,24 (“METHOD” Section 1.2). Across the entire dataset, 267 (27.8%) patients had a prolonged hospital stay. The categories of covariates included in this data were “demographic, lab test results, and preoperative information”, “osteoarthritis and surgery Information”, as well as “comorbidities” (Supplementary Data 13). This use case included six datasets (Y for overall, Y1–Y3 for horizontal, and Y4–Y5 for vertical scenarios) and considered demographic variables of male, geriatric age, and English speaker (Supplementary Figs. 4e, Supplementary Data 14). We then used dataset Y (Fig. 5b) to evaluate D-CLEF (SUPPLEMENTARY NOTES Section 8). In the horizontal scenario, D-CLEF’s predictive capability was at a similar level as that of centralized LR learning, while being better than both siloed LR models (Fig. 5c). The vertical scenario results demonstrated that D-CLEF performed slightly better than the centralized LR model, which outperformed the siloed LR ones (Fig. 5d). Overall, vertical D-CLEF statistically outperformed centralized and horizontal D-CLEF, with comparable AUC scores (Figs. 5e, Supplementary Data 15–16). The D-CLEF ensemble models provided a similar level of performance. We also used datasets Y1–Y3 (Fig. 6) to evaluate D-CLEF in the horizontal scenario, and the results demonstrated that D-CLEF in general provided similar predictive capability to the centralized LR model and was better/steadier than that of siloed LR models (Supplementary Data 17). For the vertical scenario, D-CLEF was tested on datasets Y4–Y5 (Fig. 7) and again provided a higher predictive capability when compared to centralized and siloed LR learning (Supplementary Data 18). We also evaluated D-CLEF’s runtimes in this use case and compared it to federated learning. For both horizontal/vertical scenarios, the runtimes per iteration results (Supplementary Data 19–20) demonstrated that D-CLEF in general required about 10% more than its federated counterpart (SUPPLEMENTARY NOTES Section 9). In short, at a cost of increased synchronization time, D-CLEF could perform predictions in the surgical data space at a comparable level with the centralized method and outperformed in both horizontal/vertical scenarios.
a The UCSD Total Hip Arthroplasty (THA) surgery data (n = 960 and m = 34, dataset Y). The covariates include demographics, labs, and preoperative information (dataset Y1), osteoarthritis and surgery information (dataset Y2), as well as comorbidities (dataset Y3). It was split horizontally into two patient sets to simulate two UCSD hospitals in the San Diego area (datasets Y4 and Y5). Demo: Demographics, Preop: Preoperative. b Overview of the setup on dataset Y to evaluate D-CLEF. c Main results of the horizontal scenario on dataset Y. d Main results of the vertical scenario on dataset Y. e Main overall comparison of the centralized, D-CLEF, and ensemble models on dataset Y, using two-sided, two-sample Wilcoxon signed rank-test. All box plots are derived from 30 trials, with median as center line, upper and lower quartiles as box limits, 1.5x interquartile range as whiskers, and outliers as points.
a Evaluation setup on dataset Y1, Y2, and Y3. The dataset Y1 training data contains covariates C1 only and was partitioned equally and horizontally into two patient sets. These horizontally partitioned data sets were used to build two siloed models and a D-CLEF horizontal one. A centralized model was also trained for comparison. Then, all models were evaluated using the testing data. The same process was conducted on dataset Y2 (for covariates C2) and Y3 (for covariates C3). b–d Main results for horizontal modeling on dataset Y1, Y2 and Y3. All box plots are derived from 30 trials, with median as center line, upper and lower quartiles as box limits, 1.5x interquartile range as whiskers, and outliers as points.
a Evaluation setup on dataset Y4 and Y5. The dataset Y4 training data was partitioned vertically into three covariate sets. These vertically partitioned data sets were used to construct three siloed models as well as a D-CLEF vertical one. Then, a centralized model was also built for comparison. All models were evaluated on the testing data. The same process was conducted on dataset Y5. b–c Main results for vertical modeling on dataset Y4 and Y5. All box plots are derived from 30 trials, with median as center line, upper and lower quartiles as box limits, 1.5x interquartile range as whiskers, and outliers as points.
D-CLEF for prediction of myocardial infarction
The third use case involved prediction of myocardial infarction (Fig. 8a) with sample size = 1253. We used the derived data20,21,37 originally collected at Edinburgh, UK3,4,27,28,38,39 (“METHOD” Section 1.3), with covariates of “symptoms” and “electrocardiogram (ECG) Information” (Supplementary Fig. 4f, Supplementary Data 21–22). Across the entire dataset, 274 (21.9%) patients suffered a myocardial infarction. There were five datasets (Z for overall, Z1–Z2 for horizontal, and Z3–Z4 for vertical scenarios) in this use case. We leveraged dataset Z (Fig. 8b) to evaluate D-CLEF (SUPPLEMENTARY NOTES Section 10). Horizontally, D-CLEF’s performance was at a similar level as that of centralized LR one, while being slightly better than siloed LR models (Fig. 8c). Vertically, D-CLEF again provided similar performance to the centralized LR one and outperformed both siloed LR models (Fig. 8d). In general, horizontal and vertical D-CLEF could predict at a similar level of the centralized method statistically, with relatively small AUC score differences (Figs. 8e, Supplementary Data 23–24). The ensemble of D-CLEF also provided a similar level of results. Next, datasets Z1–Z2 were used to evaluate further the horizontal scenario (Fig. 9), showing that D-CLEF in general provided similar/better predictive capability when compared to the centralized and siloed LR models (Supplementary Data 25). Also, D-CLEF was tested on datasets Z3–Z4 (Fig. 10) for the vertical scenario, and again performed similar to or better than centralized/siloed LR models (Supplementary Data 26). For runtime evaluation, the results (Supplementary Data 27–28) showed that the runtimes per iteration D-CLEF in general needed 10% more time than the federated model (SUPPLEMENTARY NOTES Section 11). In effect, D-CLEF could predict outcomes related to internal medicine at a similar level with the centralized method (and outperformed siloed ones) in both horizontal/vertical scenarios at a cost of additional runtime.
a The Edin data (n = 1,253 and m = 9, dataset Z) include symptoms (dataset Z1) and electrocardiogram (ECG) information (dataset Z2) as the covariates. It was also split horizontally into two patient sets for simulation (datasets Z3 and Z4). ECG: Electrocardiogram. b Overview of the setup on dataset Z to test D-CLEF. c Main results of the horizontal scenario on dataset Z. d Main results of the vertical scenario, comparing the two vertically siloed, the centralized, and the D-CLEF vertical model, on dataset Z. e Main overall comparison of the centralized, D-CLEF, and ensemble models on dataset Z, using two-sided, two-sample Wilcoxon signed rank-test. All box plots are derived from 30 trials, with median as center line, upper and lower quartiles as box limits, 1.5x interquartile range as whiskers, and outliers as points.
a Evaluation setup on dataset Z1 and Z2. The dataset Z1 training data (for covariates C1) was partitioned equally and horizontally into two patient sets, which were used to build two siloed models and a D-CLEF horizontal one. We also built a centralized model for comparison, and all models were evaluated using the testing data. The same process was conducted on dataset Z2 (for covariates C2). b–c Main results for horizontal modeling on dataset Z1 and Z2. All box plots are derived from 30 trials, with median as center line, upper and lower quartiles as box limits, 1.5x interquartile range as whiskers, and outliers as points.
a Evaluation setup on dataset Z3 and Z4. The dataset Z3 training data was partitioned vertically into two covariate sets, which were used to construct two siloed models as well as a D-CLEF vertical one. Then, a centralized model was also built for comparison, and all models were evaluated on the testing data. The same process was conducted on dataset Z4. b–c Main results for vertical modeling on dataset Z3 and Z4. All box plots are derived from 30 trials, with median as center line, upper and lower quartiles as box limits, 1.5x interquartile range as whiskers, and outliers as points.
Discussion
Preserving privacy and security of personal information has become a crucial challenge in modern society, especially for studies involving health data. Re-identification risks and data breaches require policies and regulations for data sharing across healthcare and research institutions. While policies/regulations may not solve the problem in the era of machine learning, advanced technologies that work together with policies are important to address privacy and security concerns. In the case of collaborative predictive modeling across institutions, D-CLEF can protect privacy/security when the patients’ healthcare records are distributed horizontally or vertically across multiple medical institutions. By combining fair-computational federated learning, decentralized blockchain, and distributed file system technologies, D-CLEF can provide model trustworthiness, transparency, and scalability, as well as system sustainability/auditability. In general, the predictive performance results for horizontal and vertical D-CLEF were similar to the centralized solution, outperformed the siloed ones, could incorporate different machine learning algorithms to potentially improve prediction capability, and were identical to the federated learning counterparts. However, D-CLEF required a modest increase in synchronization time.
Platforms that allow privacy-protecting horizontal and vertical integration of data across multiple institutions are particularly well suited to the study of rare clinical entities (including orphan diseases) and/or infrequent events that cannot be studied within one or even a handful of institutions. Meanwhile, both organizations and patients see loss of control of data as the most significant barrier to performing multisite research40. When asked about their perspectives on research, patients are more willing to share data with their home compared to other non-profit and for-profit institutions41. For organizations, sharing data across national boundaries is especially complicated due to differing standards and protections42. Our study demonstrates that D-CLEF may provide a solution across a wide range of healthcare disciplines, including pandemic-related research, surgical outcomes, and internal medicine.
Utilization of technology, such as D-CLEF, that helps leverage data from diverse institutions can potentially address several ethical issues associated with risk prediction models. In addition to patient data privacy and security, additional ethical concerns include fairness/bias and reproducibility/generalizability43. Fairness/bias focuses on whether predictive models have algorithmic bias towards specific social groups, such as race/ethnicity or socioeconomic status44. One approach to reduce such bias is utilizing diverse training data for models, which would necessitate incorporating data from various geographical regions, healthcare settings, and demographic backgrounds. Because of privacy and security issues within healthcare institutions, fostering diverse data via the sharing between multiple sites is challenging. Furthermore, the ethical principles of reproducibility and generalizability are also dependent on diverse training datasets45. D-CLEF offers one technologic solution to address building fair and unbiased models that may be generalizable.
The limitations of D-CLEF include the following. First, moving towards real-world deployment of D-CLEF in healthcare institutions may require privacy and security hardening in terms of algorithms (e.g., by using differential privacy46 to further protect patients from being identified) and infrastructure (e.g., by incorporating trusted execution environments based confidential computing47), and therefore warrants further investigation. Second, extending D-CLEF to incorporate different data modalities (e.g., patients’ genomic information, medical/radiological images, or personal health data, etc.) and outcomes (e.g., multi-class for mutually-exclusive, or multi-label for non-mutually-exclusive, prediction of multiple possible outcomes) will require further exploration. Third, refactoring the underlying D-CLEF algorithms would be required to incorporate complex global models (e.g., Multi-Layer Perceptron (MLP)48, eXtreme Gradient Boosting (XGB)49, Convolutional Neural Network (CNN)50, Long Short-Term Memory (LSTM)51, or Transformers52) for horizontal and vertical decentralized learning. After which, a thorough scalability evaluation (e.g., in terms of the number of sites, the data size on each site in the horizontal scenario, or the number of covariates on each site in the vertical scenario) and improvement (e.g., using mapping53 or multi-contract54 architectures), as well as comprehensive hyper-parameter optimization, will require further study. Fourthly, using the principals of D-CLEF to decentralize the training of medical-based language models for leveraging unstructured clinical notes across multiple healthcare institutions is a future direction that could potentially improve the use of these diverse and under-utilized notes. Finally, as with any multifactorial analysis seeking to identify predictive algorithms, associations identified through D-CLEF will require hypothesis-based evaluations to evaluate causality. This challenge could ultimately become one of D-CLEF’s strengths: the multidisciplinary richness of the data and the potential to study much larger populations provides unique opportunities to conduct secondary analyses that rigorously test causal relationships.
In summary, as a completely decentralized cross-institutional learning method, D-CLEF can support collaborative privacy-preserving modeling across multiple healthcare institutions with horizontally- or vertically-partitioned data. Meanwhile, D-CLEF can also keep patients’ data protected to conform with policies and regulations. D-CLEF allows healthcare systems to collaborate with other systems with similar (horizontal) or complementary (vertical) healthcare records while addressing modeling concerns related to privacy and equal distribution of resources. Furthermore, D-CLEF has the potential to be adapted for other healthcare predictive modeling tasks (i.e., predicting different types of patient outcomes), or even beyond the medical field (e.g., distributed learning of overall customer preferences without sharing of proprietary behavior data).
Methods
Data preprocessing
This retrospective study utilized the University of California COVID Research Data Set (UC CORDS). UC CORDS is an Institutional Review Board (IRB)-approved database containing de-identified clinical data for patients from the University of California Health System who received COVID testing34. Our research complies with all relevant ethical regulations, and the use of all datasets was exempt from University of California San Diego (UCSD) Human Research Protections Program (HRPP) IRB requirements under category 45 CFR 46.104(d) (4) Secondary research (# 804237), on May 16, 2022. The informed consent was not required because the study presented no more than minimal risk to the individuals whose information was accessed, the project did not affect individual’s clinical care, adequate subject/data confidentiality protection measures were used, and there was no information to contact those under study. Also, the datasets used in this study were de-identified.
University of California (UC) health COVID-19 data (dataset X)
We derived a de-identified data set from the limited UC Health COVID-19 Research Data Set (CORDS), collected as of September 13, 202134. To predict the mortality of patients diagnosed with COVID-19, our inclusion criteria include: (i1) known medical center from one of the five UC Health medical centers: UCSD, UC Irvine (UCI), UC Los Angeles (UCLA), UC Davis (UCD), and UC San Francisco (UCSF); (i2) 0 years ≤ patient’s age ≤ 89 years; and (i3) used at least one of top-100 frequent medications (to increase generalizability). On the other hand, the exclusion criteria include: (e1) missing age, drug, breakthrough, or death information; (e2) unspecified, mixed, or other types of vaccine manufacturer; and (e3) unclear vaccination status. We selected a set of variables selected by our expert: patient information (age group, gender, race, and ethnicity), COVID-19 related information (vaccine manufacturer, vaccination status at infection, and breakthrough case), as well as drugs prescribed (top-100 most frequently used ones out of 5066 in our data). Vaccine manufacturer is “N/A” if the patient was not vaccinated. For the vaccine manufacturer, we focused on three Centers for Disease Control (CDC)-approved vaccines (i.e., Pfizer, Moderna, and Janssen) at the time we retrieved the data. We then split the patients into five sets based on their medical centers (UCSD, UCI, UCLA, UCD, or UCSF) for evaluating horizontal methods (Supplementary Data 2). Next, we extracted 118 variables from the patient demographics (12 variables using dummy coding for gender, race, and ethnicity), COVID-related information (6 variables also using dummy coding), medications administered (top 100 most frequent ones administered after positive COVID-19 diagnosis, converted to binary variables using multi-label encoding). Then, per medical center, we averaged the ages in each age group to obtain the estimated age, removed variables with the same value across all patients, and combined duplicated variables. Additionally, we excluded the male gender, because of its high collinearity with female gender due to very small percentage ( ~ 0.025%, as shown in the Supplementary Fig. 4b) of unknown gender (which was also removed). After data preprocessing the number of patients was n = 15,279, and the number of covariates was m = 100 (10 patient information, 6 COVID-19 information, and 84 medication ones). There were about 22,000 patients excluded because they did not meet our inclusion criteria. The excluded patients in general were demographically similar to the included ones (Supplementary Figs. 4a–d, Supplementary Data 3–6), with fewer elderly patients (i.e., age groups of 61 + ), relatively fewer patients in all races except white and unknown, and relatively fewer Hispanic or Latina ethnicity patients. Also, we grouped the covariates into two categories: patient and COVID-19 information (16 covariates) and drug prescribed (84 covariates), to simulate the vertically split data (Supplementary Data 2).
UCSD total hip arthroplasty (THA) surgery data (dataset Y)
We used the UCSD THA surgery data derived in previous publications20,23,24, which contains n = 960 patients and m = 34 covariates. This dataset is Health Insurance Portability and Accountability Act (HIPAA)-deidentified. To predict the longer hospital length of stay (i.e., > expected 3 days) for the unilateral primary THA surgery, the covariates include 3 demographics (male sex, patient’s age ≥ 65 years, and non-English speaker), 1 lab (obesity body mass index > 30 kg/m2), 2 preoperative (metabolic equivalents < 4 and opioid use), 4 operative-side osteoarthritis grades (mild, moderate, severe, and avascular necrosis), 6 contralateral hip description (no osteoarthritis, mild osteoarthritis, moderate osteoarthritis, severe osteoarthritis, previous surgery, and avascular necrosis), 1 anesthesia (general—versus neuraxial), 3 surgical approach (posterior, anterolateral, and anterior), and 14 comorbidities (chronic kidney disease, chronic obstructive pulmonary disease, congestive heart failure, coronary artery disease, hypertension, diabetes mellitus, obstructive sleep apnea, dialysis, psychiatric history— depression / anxiety / bipolar disease, active smoker, asthma, thrombocytopenia—platelets < 150000/uL, anemia, and dementia). All covariates were dummy-coded as binary ones, and we checked to ensure there are no covariates with the same value across all patients nor duplicated covariates. We split the patients randomly into two sets in a stratified way (i.e., keeping the positive/negative ratio) to simulate two UCSD hospitals in the San Diego area (La Jolla and Hillcrest) for horizontal methods’ evaluation (Supplementary Data 13). Then, we grouped the covariates into three categories: demographics, labs, and preoperative information (6 covariates), osteoarthritis and surgery-related information (14 covariates, including operative-side osteoarthritis grades, contralateral hip description, anesthesia, and surgical approach covariates), and comorbidities (14 covariates), to evaluate vertical methods (Supplementary Data 13).
Edinburgh myocardial infarction data (dataset Z)
We adopted the Edinburgh myocardial infarction data3,4,27,28,38,39 derived in existing literatures20,21,37. The data are publicly available (DATA AVAILABILITY) and contain n = 1,253 patients and m = 9 covariates. To predict the presence of disease, the covariates include symptoms (5 variables, including hypoperfusion, nausea, sweating, pain in left arm, and pain in right arm) and ECG information (4 variables, including new Q waves, T wave inversion, ST elevation, and ST depression). All covariates were binary without covariates with the same value across all patients and without duplicated covariates. We split the patients randomly into two sets in a stratified way to evaluate horizontal methods (Supplementary Data 21), and then grouped the covariates into two categories (symptoms and ECG information) to simulate vertical data splitting (Supplementary Data 21).
Distributed cross-learning for equitable federated models (D-CLEF) framework and computational algorithms
D-CLEF incorporated four technologies: horizontal federated learning, vertical federated learning, blockchain distributed ledger, and distributed file sharing. D-CLEF, based on mathematically-proven federated learning algorithms, aims at enabling collaborative predictive modeling across multiple healthcare institutions, whether the data were split horizontally (i.e., different patients from each institution with the same set of covariates across all institutions) or vertically (i.e., same set of patients across all institutions with different part of covariates in each institution). D-CLEF’s learning process does not disseminate observation-level health data and therefore protects patients’ privacy. Compared to existing horizontal/vertical federated learning methods, D-CLEF does not require a centralized moderating server, thus increasing autonomy of each site while reducing single-point-of-control. Additionally, by utilizing blockchain and distributed file system technologies, D-CLEF ensures the models’ provenance, transparency, and scalability, as well as the immutability of the learning process audit trail. A complete list of the desirable technical features of D-CLEF is illustrated in Supplementary Fig. 1a.
Learning on horizontally partitioned data
For horizontal learning scenario, we adopted the Grid binary LOgistic Regression (GLORE) algorithm, which was based on Newton-Raphson method and was mathematically proven to be equivalent as centralized LR3. In the horizontal scenario, each site contains a different set of patients with the same m covariates. In GLORE, all global model coefficients with size = (m + 1), including an intercept term, are first initialized as zeroes. In each iteration, each site uses the global model to compute its local gradient vector with size = (m + 1) and local variance-covariance matrix with size = (m + 1) x (m + 1), and then sends this local partial model (including both the vector and the matrix) to the central server; then, the server combines the partial models from all sites to update the global model and sends the updated global model back to each site. The iteration continues until convergence of the global model. The original GLORE code was implemented in Java using the National Institute of Standards and Technology (NIST) JAMA library v1.0.3, thus we integrated the code directly into D-CLEF, which is also primarily developed in Java.
Learning on vertically partitioned data
We incorporated the VERTIcal Grid lOgistic regression (VERTIGO) algorithm8 for the vertical learning scenario. VERTIGO was based on Newton’s method and was also proven to be equivalent as centralized LR mathematically. For vertically partitioned data, each site contains a different set of covariates with the same n patients, who are already “linked” by unique identifiers. These identifiers are pseudo-generated instead of real ones such as Medical Record Number (MRN). In this case, techniques for horizontal modeling (e.g., GLORE) to decompose the global model learning would not be feasible, because each site possesses the information for different covariates (thus each site cannot update the coefficients for all covariates locally). Therefore, VERTIGO converts the primal problem (i.e., finding a global model containing coefficients for all covariates) to a dual one (i.e., finding a global dual-form model containing coefficients for all patients). Since each site possesses the information for the same patients, each site can update the coefficients for all patients locally. Also, a covariate of 1 s is added to one of the sites for computing the intercept term. In VERTIGO, the first step is to obtain a global gram matrix as the kernel trick to solve the dual problem. To do this, each site first computes a local gram matrix (with size = n × n) by using dot products of its local data and sends the matrix to the server. Then, the server combines the local gram matrices using element-wise addition to a global gram matrix. After the dot product computation, it is very difficult to reverse engineer and obtain the patient-level data. Next, all global dual-form model coefficients with size = n are initialized as zeroes. In each iteration, each site uses the global dual-form model to compute its local dual-form model with size = n and sends this partial model to the server; then, the server combines the partial dual-form models from all sites to update the global dual-form model and sends the updated global dual-form model back to each site. The iteration continues until convergence of the global dual-form model. Then, each site uses the global dual-form model to compute a part of global primal-form model coefficients corresponding to the covariates (and the intercept term, if added by the site) available in the local data and sends that part of coefficients again to the server. Finally, the server simply concatenates all parts of coefficients to form a complete global primal-form model, and then sends the global model to each site. The original VERTIGO8 was implemented in MATLAB, while an updated version VERTIGO-CI14 was implemented in Python. To integrate VERTIGO into D-CLEF, we re-implemented it using Java and the JAMA library, referring to both VERTIGO and VERTIGO-CI algorithms. One of the most time-consuming steps of VERTIGO is to invert a large n x n matrix at the central server side with the time complexity of O(n3)8, and therefore in our implementation we computed matrix inversion via the jBLAS library55 v1.2.6 snapshot with native system package to expedite the computational speed.
D-CLEF modeling network using blockchain distributed ledger
For both horizontal and vertical learning scenarios, we eliminate the need for a “physical” centralized server (as required for federated learning) by using the fair compute load approach23,24,27,28, which asks each site to serve as the “virtual” server in each learning iteration. This design not only preserves the prediction capability when compared to federated learning, but also ensures computing fairness for each participating site because they take turns to contribute computation cost as a virtual server (in addition to the computational cost as a client). We also adopted the maximum iteration design20,21,23 by adding a “time-to-leave” counter to cap the number of iterations (and thus the cost of model transferring, which is critical for peer-to-peer communications). Based on the above-mentioned fair compute load and maximum iteration, we adopted blockchain distributed ledger31 as the D-CLEF modeling network. Blockchain is distributed (i.e., no single intermediary server), immutable (i.e., very hard to be changed), transparent (i.e., everyone can see everything on the chain), highly-available (i.e., no single-point-of-failure because of full-redundancy), and source-verifiable (i.e., clear provenance because there is no “root” user)30,31,32. Several major blockchain platforms also support smart contract, a computer program stored and executed on blockchain56,57, to provide the above-mentioned benefits to computer codes in addition to data. Compared to other decentralized systems and strategies, blockchain brings the benefits and addresses many current challenges in distributed networks. Specifically, we adopted Ethereum56, which is an open-source and community-based blockchain platform that can be configured as both public/permissionless (thus have a huge momentum to improve because of its large financial user base) and private/permissioned (thus suitable for cross-institution beyond-financial applications like our study) blockchain networks30. We used the Go-Ethereum (Geth) v1.9.1, and configured it as a permissioned blockchain network. We used Proof-of-Authority (PoA) consensus protocol that is round-robin-based58. This is because in a permissioned network, a non-compute-intensive PoA protocol could improve efficiency and reduce energy cost59 when compared to computing/energy-intensive protocols (e.g., Proof-of-Work, or PoW, used in Bitcoin60) required for permissionless blockchain networks. Therefore, we adopted the Clique61 PoA protocol with period = 0 and epoch = 30000. On the Ethereum permissioned blockchain network, we further adopted smart contracts to store detailed training information (related to process, sites, data, and models) of horizontal or vertical learning. Meanwhile, the horizontal/vertical model learning as well as the access to the data only happens within the local computing node, and therefore no patients’ data will be disseminated on blockchain through the smart contracts. The list of the training details and its isolation from the local computing node are shown in Supplementary Fig. 1b. Specifically, we adopted Solidity62, one of the most popular smart contract languages for Ethereum, v0.8.19. We developed two Solidity smart contracts: the Model Contract (one for each site, to store the training details) and the Catalog Contract (only one for all sites, to manage site names and Model Contract deployment addresses on blockchain). Finally, we used the Web3j CLI v1.4.1 and its dependency/preliminary libraries to manage the blockchain and deploy/run smart contracts on the blockchain from the D-CLEF local computing. The Model Learner and the two D-CLEF smart contracts are outlined in Supplementary Fig. 1c. Our design of smart contracts are unified for both horizontal GLORE and vertical VERTIGO learning methods and are extensible to incorporate other learning algorithms (in the Model Contract) or even to exchange other training-related information (as smart contracts to be indexed by the Catalog Contract).
D-CLEF sharing network using distributed file system
Although most of the training details can be recorded directly on blockchain via smart contracts, the local or global models themselves (i.e., model contents) are in general much larger than the rest of the information. Moreover, the size of the model contents scales with the data size, while the rest of the information only requires a fixed size of on-chain storage space. For horizontal learning, the largest on-chain storage requirement is for the local variance-covariance matrix with size = (m + 1) x (m + 1), where m is the number of covariates in the data. On the other hand, for vertical learning the largest storage space needed on blockchain is for the gram matrix with size = n x n, where n is the number of patients in the data. For a typical predictive task, n is usually at least a few times larger than m, to avoid model overfitting and to be more focused on few actionable covariates. This is also true for all our evaluation datasets as shown in Figs. 2a, 5a, and 8a. Although existing studies enabled the storage of large amount of data directly on blockchain by using a splitting-and-merging method63,64,65, the prolonged time required for disseminating the large matrices to every site (because each site will have a copy of the whole blockchain data31) could present an efficiency issue for data transferring. Therefore, we adopted distributed file system33 as the D-CLEF sharing network, to store and retrieve the model contents efficiently without the need of a centralized intermediating repository. Specifically, we adopted InterPlanetary File System (IPFS), which is a popular protocol to serve as the “disk” of blockchain in various application fields66. We used the Kubo IPFS v0.20.0, which is one of the most widely adopted implementations of IPFS, and configured it as a permissioned distributed file system. To store the horizontal and vertical local/global models, we first serialized and concatenated the 1-dimensional (1D) vector (e.g., gradient vector) and 2-dimensional (2D) matrix data (e.g., variance-covariance matrix) as a single byte array. Then, we compressed the byte array using the DEFLATE lossless data compression algorithm67 to further reduce the amount of data to be transferred. Next, we share the compressed byte array via IPFS and obtain a Content Identifier (Content ID) which is a hash value to uniquely identify the stored byte array across the IPFS network. The Content ID is then stored on the blockchain together with other training details, as shown in Supplementary Fig. 1b. We developed two D-CLEF storage network components: the Model Sharing Peer (one for each site, to store model contents) and the Model sharing connector (only one for all sites, to connect the distributed file sharing peers using a pre-shared key; only peers with the key can participate in the network). The two D-CLEF sharing network components are shown in Supplementary Fig. 1c. Our design of model storage not only supports both horizontal GLORE and vertical VERTIGO methods, but also is extensible for other horizontal/vertical learning methods because their arbitrary data structures can be serialized, concatenated, compressed, and stored on IPFS to enable distributed model sharing. Specifically, although the space complexities of the horizontal and vertical D-CLEF are O(m2) and O(n2) respectively (which is already larger than the O(m) of centralized LR), significantly larger and more complex models may contain even more parameters68 and thus require larger storage space as well as data transferring bandwidth. Therefore, the use of a distributed file system is essential to the scalability of D-CLEF in terms of model size.
D-CLEF site architecture
Each D-CLEF site contains three nodes (local computing, modeling, and storage). The local computing node takes data as input for the Model Learner (either horizontal or vertical), and then uses the pre-shared catalog address to setup Ethereum blockchain within the modeling node and then connect to the Catalog Contract, which in turn provides the names and the Model Contract addresses of all participating sites. On the other hand, the local computing node also uses the pre-shared key to set up IPFS within the storage node and then connect to the Model Sharing Connector, which in turn sets up the Model Sharing Peer. During the modeling process, the local computing node stores the model contents to IPFS to obtain a Content ID, and then records the training details (including the Content ID) to the Model Contracts of the local site. Meanwhile, the local computing nodes also query the Model Contracts of the other sites to obtain the training details as well as the Content IDs from other sites, and then retrieve other sites’ model contents via the Content IDs. The D-CLEF system architecture is depicted in Supplementary Fig. 2a.
Workflow and algorithms
The overall workflow for D-CLEF is shown in Supplementary Fig. 2b, and the relationship with the local computing, the modeling network, and the storage network majorly involved in each step is also specified. The workflow starts from the main D-CLEF algorithm (D-CLEF, algorithm A1 in Supplementary Fig. 3a) that executes either horizontally (D-CLEF-H, algorithm A2 in Supplementary Fig. 3b) or vertically (D-CLEF-V, algorithm A3 in Supplementary Fig. 3c) learning algorithm. We standardize these steps to make D-CLEF a unified framework for both horizontal and vertical learning scenarios. The global/local models can be either primal-form or dual-form. For horizontal learning, the initialization step is trivial (by setting all global model coefficients to zeroes) and the finalization step simply returns the learned global model. In contrast, for vertical learning the initialization step also involves computing/transferring/combining the local gram matrices, and the finalization step converts the global dual-form model to its primal-form. We also adopted utility libraries such as the Apache Commons CSV v1.9.0, the Apache Commons Math v1.2, as well as the Java Utilities v1.0.0, in our implementation.
Centralized, ensemble, and federated implementations for comparison
To evaluate the prediction performance of D-CLEF, we adopted LR with Ridge estimator for L2 regularization on integrated data (i.e., combined from horizontally- or vertically-partitioned data) to serve as the centralized comparing counterpart. Specifically, we used the implementation from the Weka Dev v3.9.1 library. We also compared D-CLEF with MLP48, XGB49, CNN50 and LSTM51 using Weka DeepLearning4J v1.7.2 and Weka XGBoost v0.2.0 libraries. Since CNN and LSTM were originally designed for spatially-related or sequential data, we transformed our three datasets into either two-dimensional (2D) or one-dimensional (1D) formats (SUPPLEMENTARY NOTES Section 2). Specifically, for CNN we tested on both 2D and 1D data (CNN-2D and CNN-1D models, respectively), and for LSTM we used 1D data. This would allow a site, should they desire, to combine the benefits of a global federated model with a more complex local model. For the ensembled prediction score, we averaged the predicted scores from D-CLEF with the scores from either siloed MLP, XGB, CNN-2D, CNN-1D, or LSTM model (to still preserve patients’ privacy).
On the other hand, to gauge the runtime performance of D-CLEF, we also implemented a federated version of our method using Apache ActiveMQ, an open-source message broker. We set up the ActiveMQ Java Message Service (JMS) queue server on one of the sites, to simulate the real-world situation that a central server usually resides within one of the collaborating sites. This configuration also provides a fair comparison with the distributed version of D-CLEF, by using the same number of computational resources (i.e., both versions use the exact same number of computers for each horizontal/vertical evaluation scenario). For this message-based federated version, each site submit both training details and model contents together as a single message to the server and retrieve other sites’ information also from the server. The training details and model contents are serialized as a byte array with message compression, which is similar to the distributed version of D-CLEF to be compared fairly. Since the core algorithms for both federated and distributed versions are the same, the prediction performance and the learning iteration were also the same, and therefore we focused on the runtime comparison only. We adopted Apache ActiveMQ v5.15.15 and its dependency/preliminary libraries, with a configuration of frame size = 1 GB with non-transacted auto-acknowledging sessions.
Simulation settings
In our experiments, each site was set up on a separate instance (instead of simulating multiple sites on one instance), to represent a real-world usage scenario as well as to obtain accurate time measurements. We ran multiple experiments in parallel using these 10 instances. We built a total of 366 models (162 for the UC Health COVID-19, 114 for the UCSD THA surgery, and 90 for the Edinburgh myocardial infarction datasets). With 30 trials for each model, our evaluation included 10,980 trials in total. This corresponds to 1,395 h of computation (968 for UC Health COVID-19, 258 for UCSD THA surgery, and 169 for Edinburgh myocardial infarction).
Inclusion & ethics
The research included local researchers (R.A.G., J.K., and R.T.S) throughout the research process. The research locally relevant, which has been determined in collaboration with local partners. The roles and responsibilities were agreed amongst collaborators ahead of the research and a capacity-building plan for local researchers was discussed. This research would not have been severely restricted or prohibited in the setting of the researchers, and the study has been approved by a local ethics review committee (i.e., UCSD HRPP IRB Exemption Category 4, # 804237, approved on May 16, 2022). The animal welfare regulations, environmental protection and biorisk-related regulations in the local research setting were not applicable to this study. The research does not result in stigmatization, incrimination, discrimination or otherwise personal risk to participants, and does not involve health, safety, security or other risk to researchers. The benefit sharing measures in case biological materials, cultural artefacts or associated traditional knowledge has been transferred out of the country are not applicable to this research. We have taken relevant local and regional research into account in citations.
Statistics & reproducibility
No statistical method was used to predetermine the sample size of each dataset. The sample sizes were chosen based on the available samples in each dataset (15,279 for dataset X, 960 for dataset Y, and 1253 for dataset Z), which are sufficient for predictive modeling purposes. We repeated our experiments for n = 30 independent permuted trials, and for each trial we reset the blockchain and the distributed file system networks, to verify the reproducibility of our experimental findings. We used a fixed set of random seeds (different per trial) to increase the reproducibility of our experiments. The exclusion of data in dataset X and the reasons are described in “METHOD” Section 1.1. We further evaluated the AUC values with two-sided, two-sample Wilcoxon signed-rank tests36 to assess statistical significance between the two D-CLEF methods (i.e., horizontal and vertical), compare D-CLEF versus the centralized LR approach, and also compare D-CLEF ensembled with other machine learning algorithms. We employed n = 30 trials with a p-value < 0.05 indicating a statistically significant difference. The investigators who conducted this study were not blinded to allocation during evaluation experiments and outcome/result assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The University of California Health COVID-19 Research Data Set (UC CORDS, dataset X) is not publicly available due to institutional privacy restrictions. The data is incorporated into the UC Data Discovery Platform and is available to UC Health researchers via an internal process developed for granting access to this data, and there is currently no limitation of the data access time period. Access can be obtained by UC Health affiliated researchers with permission from the Center for Data-Driven Insights and Innovation (CDI2, https://www.ucop.edu/uc-health/departments/center-for-data-driven-insights-and-innovations-cdi2.html). The timeframe of response to requests and availability of data will be determined by CDI2. The links to the request forms are only available to UC Health researchers. The researcher needs to be affiliated with UC Health to access the data. The data usage via Data Use Agreement (DUA) is restricted to UC Health researchers. The UC San Diego (UCSD) Total Hip Arthroplasty (THA) surgery data (dataset Y) are available to researchers under restricted access governed by UCSD policy and US/California law. Access to the data requires approval by the UCSD Institutional Review Board (IRB), approval by the UCSD Health Data Oversight Committee (HDOC), and the successful execution of a DUA between UCSD and the researcher receiving the data. Researchers seeking access should contact Dr. Rodney A. Gabriel (ragabriel@health.ucsd.edu) to undertake this process. The timeframe of response to requests and availability of data will be determined by UCSD. The time period and allowed use of the data by the recipient researcher will be determined by UCSD and outlined in the data use agreement. The Edinburgh myocardial infarction data (dataset Z) used in this study are available in the Zenodo database under accession code: 10.5281/zenodo.1492820 (https://doi.org/10.5281/zenodo.1492820)37. Source data are provided with this paper.
Code availability
The D-CLEF software is available in the Zenodo database under accession code: 10.5281/zenodo.14052533 (https://doi.org/10.5281/zenodo.14052533)69. Also, the data preprocessor to preprocess/split the raw data for horizontal and vertical D-CLEF is available in the Zenodo database under accession code: 10.5281/zenodo.14052494 (https://doi.org/10.5281/zenodo.14052494) 70.
References
Deist, T. M. et al. Infrastructure and distributed learning methodology for privacy-preserving multi-centric rapid learning health care: euroCAT. Clin. Transl. Radiat. Oncol. 4, 24–31 (2017).
Leung N. H., et al. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 26, 676–680 (2020).
Wu, Y., Jiang, X., Kim, J. & Ohno-Machado, L. Grid Binary LOgistic REgression (GLORE): building shared models without sharing data. J. Am. Med. Inform. Assoc. 19, 758–764 (2012).
Wang, S. et al. EXpectation Propagation LOgistic REgRession (EXPLORER): distributed privacy-preserving online model learning. J. Biomed. Inform. (JBI) 46, 480–496 (2013).
Yan, F., Sundaram, S., Vishwanathan, S. & Qi, Y. Distributed autonomous online learning: Regrets and intrinsic privacy-preserving properties. IEEE Trans. Knowl. Data Eng. 25, 2483–2493 (2013).
El Emam, K. et al. A secure distributed logistic regression protocol for the detection of rare adverse drug events. J. Am. Med. Inform. Assoc. 20, 453–461 (2013).
Fienberg S. E., Fulp W. J., Slavkovic A. B., Wrobel T. A. “Secure” Log-linear And Logistic Regression Analysis Of Distributed Databases. in International Conference on Privacy in Statistical Databases. 277–290 (Springer, 2006).
Li, Y., Jiang, X., Wang, S., Xiong, H. & Ohno-Machado, L. VERTIcal Grid lOgistic regression (VERTIGO). J. Am. Med. Inform. Assoc. 23, 570–579 (2015).
Gascón, A. et al. Secure tasets. IACR Cryptol. ePrint Arch. 2016, 892 (2016).
Mohassel P., Zhang Y. SecureML: A System For Scalable Privacy-preserving Machine Learning. in IEEE Symposium on Security and Privacy (SP). 19–38 (IEEE) (2017).
Slavkovic A. B., Nardi Y., Tibbits M. M. Secure Logistic Regression of Horizontally and Vertically Partitioned Distributed Databases. in Seventh IEEE International Conference on Data Mining Workshops (ICDMW). 723–728 (IEEE) (2007).
Nardi Y., Fienberg S. E., Hall R. J. Achieving both valid and secure logistic regression analysis on aggregated data from different private sources. J. Privacy and Confidentiality 4, (2012).
Aono, Y., Hayashi, T., Phong, L. T. & Wang, L. Privacy-preserving logistic regression with distributed data sources via homomorphic encryption. IEICE TRANSACTIONS Inf. Syst. 99, 2079–2089 (2016).
Kim J., Li W., Bath T., Jiang X., Ohno-Machado L. VERTIcal Grid lOgistic regression with Confidence Intervals (VERTIGO-CI). in AMIA Annual Symposium Proceedings. 355 (American Medical Informatics Association, 2021).
Heinze‐Deml, C., McWilliams, B. & Meinshausen, N. Preserving privacy between features in distributed estimation. Stat 7, e189 (2018).
Shen M., Zhang J., Zhu L., Xu K. & Tang X. Secure SVM training over vertically-partitioned datasets using consortium blockchain for vehicular social networks. IEEE Trans. Veh. Technol. 69, 5773–5783 (2020).
Gao D., et al. Privacy-preserving heterogeneous federated transfer learning. in IEEE International Conference on Big Data (Big Data). 2552–2559 (IEEE) (2019).
Her, Q. L. et al. A query workflow design to perform automatable distributed regression analysis in large distributed data networks. eGEMs 6, 11 (2018).
Kuo T.-T., Hsu C.-N., Ohno-Machado L. ModelChain: Decentralized Privacy-Preserving Healthcare Predictive Modeling Framework on Private Blockchain Networks. In: ONC/NIST Use of Blockchain for Healthcare and Research Workshop) (2016).
Kuo, T.-T., Gabriel, R. A., Cidambi, K. R. & Ohno-Machado, L. EXpectation Propagation LOgistic REgRession on permissioned blockCHAIN (ExplorerChain): decentralized online healthcare/genomics predictive model learning. J. Am. Med. Inform. Assoc. (JAMIA) 27, 747–756 (2020).
Kuo, T.-T. The anatomy of a distributed predictive modeling framework: online learning, blockchain network, and consensus algorithm. J. Am. Med. Inform. Assoc. Open (JAMIA Open) 3, 201–208 (2020).
Chen X., Ji J., Luo C., Liao W., Li P. When Machine Learning Meets Blockchain: A Decentralized, Privacy-preserving and Secure Design. in IEEE International Conference on Big Data (Big Data). 1178–1187 (IEEE) (2018).
Kuo, T.-T., Gabriel, R. A. & Ohno-Machado, L. Fair compute loads enabled by blockchain: sharing models by alternating client and server roles. J. Am. Med. Inform. Assoc. (JAMIA) 26, 392–403 (2019).
Kuo, T.-T., Kim, J. & Gabriel, R. A. Privacy-Preserving Model Learning on Blockchain Network-of-networks. J. Am. Med. Inform. Assoc. (JAMIA) 27, 343–354 (2020).
Kim, H., Kim, S.-H., Hwang, J. Y. & Seo, C. Efficient privacy-preserving machine learning for blockchain network. IEEE Access 7, 136481–136495 (2019).
Warnat-Herresthal, S. et al. Swarm Learning for decentralized and confidential clinical machine learning. Nature 594, 265–270 (2021).
Kuo, T.-T. & Pham, A. Quorum-based model learning on a blockchain hierarchical clinical research network using smart contracts. Int. J. Med. Inform. (IJMI) 169, 104924 (2022).
Shao X., Pham A., Kuo T.-T. WebQuorumChain: a web framework for quorum-based health care model learning. Informatics in Medicine Unlocked, 2024, 101590 (2024).
Rocher, L., Hendrickx, J. M. & de Montjoye, Y.-A. Estimating the success of re-identifications in incomplete datasets using generative models. Nat. Commun. 10, 3069 (2019).
Kuo, T.-T., Zavaleta Rojas, H. & Ohno-Machado, L. Comparison of blockchain platforms: a systematic review and healthcare examples. J. Am. Med. Inform. Assoc. (JAMIA) 26, 462–478 (2019).
Kuo, T.-T., Kim, H.-E. & Ohno-Machado, L. Blockchain distributed ledger technologies for biomedical and health care applications. J. Am. Med. Inform. Assoc. (JAMIA) 24, 1211–1220 (2017).
Lacson, R., Yu, Y., Kuo, T.-T. & Ohno-Machado, L. Biomedical blockchain with practical implementations and quantitative evaluations: a systematic review. J. Am. Med. Inform. Assoc. 31, 1423–1435 (2024).
Huang, H., Lin, J., Zheng, B., Zheng, Z. & Bian, J. When blockchain meets distributed file systems: an overview, challenges, and open issues. IEEE Access 8, 50574–50586 (2020).
UCBRAID. COVID-19 Clinical Data Sets for Research. Preprint at https://www.ucbraid.org/cords.
Davis J., Goadrich M. The Relationship Between Precision-Recall and ROC Curves. Proceedings of the 23rd International Conference on Machine Learning (ICML), 233–240 (2006).
McDonald J. Handbook of Biological Statistics. 3rd edn.(Sparky House Publishing: Baltimore, MD.). (2014).
Kuo T.-T., Gabriel R. A., Cidambi K. R., Ohno-Machado L. tsungtingkuo/explorerchain v1.0.0 (Version v1.0.0). Preprint at https://doi.org/10.5281/zenodo.1492820 (2018).
Kennedy, R., Fraser, H., McStay, L. & Harrison, R. Early diagnosis of acute myocardial infarction using clinical and electrocardiographic data at presentation: derivation and evaluation of logistic regression models. Eur. heart J. 17, 1181–1191 (1996).
Kuo, T.-T. & Pham, A. Detecting model misconducts in decentralized healthcare federated learning. Int. J. Med. Inform. (IJMI) 158, 104658 (2022).
Mazor, K. M. et al. Stakeholders’ views on data sharing in multicenter studies. J. Comp. effectiveness Res. 6, 537–547 (2017).
Kim, J. et al. Patient perspectives about decisions to share medical data and biospecimens for research. JAMA Netw. open 2, e199550–e199550 (2019).
Scheibner, J. et al. Data protection and ethics requirements for multisite research with health data: a comparative examination of legislative governance frameworks and the role of data protection technologies. J. Law Biosci. 7, lsaa010 (2020).
Weidener, L. & Fischer, M. Role of ethics in developing AI-based applications in medicine: insights from expert interviews and discussion of implications. Jmir ai 3, e51204 (2024).
Panch, T., Mattie, H. & Atun, R. Artificial intelligence and algorithmic bias: implications for health systems. J. Glob. Health 9, 010318 (2019).
Semmelrock H., Kopeinik S., Theiler D., Ross-Hellauer T., Kowald D. Reproducibility in machine learning-driven research. arXiv preprint https://doi.org/10.48550/arXiv.2307.10320 (2023).
Wu, X. et al. An adaptive federated learning scheme with differential privacy preserving. Future Gener. Computer Syst. 127, 362–372 (2022).
Mo, F. et al. Ppfl: Enhancing privacy in federated learning with confidential computing. GetMobile: Mob. Comput. Commun. 25, 35–38 (2022).
Dreiseitl, S. & Ohno-Machado, L. Logistic regression and artificial neural network classification models: a methodology review. J. Biomed. Inform. 35, 352–359 (2002).
Chen T., Guestrin C. XGBoost: a scalable tree boosting system. In proceedings of the 22nd ACM sigkdd international conference on knowledge discovery and data mining. 785–794 (Association for Computing Machinery).
LeCun Y., et al. Handwritten digit recognition with a back-propagation network. Advances in neural information processing systems 2, 396–404 (1989).
Hochreiter S. Long Short-term Memory. Neural Computation MIT-Press, (1997).
Vaswani A. et al. Attention is all you need. Advances in Neural Information Processing Systems, (2017).
Pham, A., Edelson, M., Nouri, A. & Kuo, T.-T. Distributed management of patient data-sharing informed consents for clinical research. Computers Biol. Med. 180, 108956 (2024).
Yu Y., et al. Distributed, immutable, and transparent biomedical limited data set request management on multi-capacity network. Journal of the American Medical Informatics Association, (2024).
Braun M. jBLAS Project. Preprint at https://github.com/jblas-project.
Buterin, V. A next-generation smart contract and decentralized application platform. white Pap. 3, 2–1 (2014).
Yu H., Sun H., Wu D., Kuo T.-T. Comparison of Smart Contract Blockchains for Healthcare Applications. in AMIA Annual Symposium. (American Medical Informatics Association, Bethesda, MD).
De Angelis S., et al. Pbft vs proof-of-authority: applying the cap theorem to permissioned blockchain. CEUR workshop proceedings 2058, (2018).
Singh P. K., Singh R., Nandi S. K., Nandi S. Managing Smart Home Appliances With Proof Of Authority And Blockchain. in Innovations for Community Services: 19th International Conference, I4CS, Wolfsburg, Germany Proceedings. 19, 221–232 (Springer, 2019).
Nakamoto S. Bitcoin: A peer-to-peer electronic cash system. Decentralized Bus Rev., 21260 (2008).
Szilágyi P. Ethereum Improvement Proposals (EIP) 225: Clique proof-of-authority consensus protocol. Preprint at http://eips.ethereum.org/EIPS/eip-225 (2017).
Dannen C. et al. Introducing Ethereum and solidity. Springer (2017).
Li, M. M. & Kuo, T.-T. Previewable contract-based on-chain x-ray image sharing framework for clinical research. Int. J. Med. Inform. (IJMI) 156, 104599 (2021).
Kuo, T.-T. et al. Blockchain-enabled immutable, distributed, and highly available clinical research activity logging system for federated COVID-19 data analysis from multiple institutions. J. Am. Med. Inform. Assoc. (JAMIA) 30, 1167–1178 (2023).
Tellew, J. & Kuo, T.-T. CertificateChain: decentralized healthcare training certificate management system using blockchain and smart contracts. J. Am. Med. Inform. Assoc. Open (JAMIA Open) 5, 1–9 (2022).
Kumar, S., Bharti, A. K. & Amin, R. Decentralized secure storage of medical records using blockchain and IPFS: a comparative analysis with future directions. Security Priv. 4, e162 (2021).
Deutsch P. RFC1951: DEFLATE compressed data format specification version 1.3. Preprint at https://www.rfc-editor.org/rfc/rfc1951.html (1996).
Zhang S., et al. Architectural complexity measures of recurrent neural networks. Advances in neural information processing systems 29, (2016).
Kuo T.-T., Gabriel R. A., Koola J., Schooley R. T., Ohno-Machado L. Distributed cross-learning for equitable federated models (D-CLEF). Preprint at https://doi.org/10.5281/zenodo.14052533 (2024).
Kuo T.-T., Gabriel R. A., Koola J., Schooley R. T., Ohno-Machado L. D-CLEF data preprocessor. Preprint at https://doi.org/10.5281/zenodo.14052494 (2024).
Acknowledgements
The authors were funded by the U.S. National Institutes of Health (NIH) (R01EB031030: T.-T.K., R.A.G., J.K., R.T.S.; RM1HG011558: T.-T.K., L.O.-M.; R01HL136835: T.-T.K., L.O.-M.; R01LM013712: L.O.-M.; R01HG011066: T.-T.K., L.O.-M.; U54HG012510: T.-T.K., L.O.-M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors thank the Center for Data-driven Insights and Innovation at UC Health (CDI2; https://www.ucop.edu/uc-health/departments/center-for-data-driven-insights-and-innovations-cdi2.html), for its analytical and technical support related to use of the UC Health Data Warehouse and related data assets, including the CORDS dataset. The authors would also like to thank Heidi Sofia, Xiaoqian Jiang, Jihoon Kim, Tyler Bath, and Wentao Li for helpful discussions; Mike Hogarth, Cora Han, Pagan Morris, Paresh Desai, Nguyen Trieu, and Mark Mooney for providing support for the UC CORDS and/or the UCSD THA dataset; Amy Sitapati, Nancy Herbst, Matteo D’Antonio, and Kai Post for technical and administrative support; and Andrew Greaves, Jit Bhattacharya, Justean Giger, Monica Falk, Alen Moxley, Christopher Lebron, and Reza Beykzadeh for providing support for the UCSD Health Secure Cloud iDASH 2.0.
Author information
Authors and Affiliations
Contributions
T.-T.K. contributed conceptualization, methodology, software, validation, formal analysis, investigation, resources, data curation, visualization, supervision, project administration, funding acquisition, and writing (original draft). R.A.G., J.K., and R.T.S. contributed validation, investigation, data curation, and writing (review & editing). L.O.-M. contributed conceptualization, validation, formal analysis, investigation, resources, visualization, supervision, project administration, funding acquisition, and writing (review & editing).
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kuo, TT., Gabriel, R.A., Koola, J. et al. Distributed cross-learning for equitable federated models - privacy-preserving prediction on data from five California hospitals. Nat Commun 16, 1371 (2025). https://doi.org/10.1038/s41467-025-56510-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56510-9