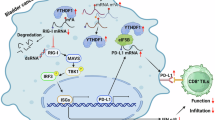
Abstract
The N6-methyladenosine binding protein YTHDF1, often upregulated in cancer, promotes tumor growth and hinders immune checkpoint blockade treatment. A comprehensive understanding of the molecular mechanisms governing YTHDF1 protein stability is pivotal for enhancing clinical response rates and the effectiveness of immune checkpoint blockade in cancer patients. Here, we report that USP5 interacts with YTHDF1, stabilizing it by removing K11-linked polyubiquitination. Insulin activates mTORC1, phosphorylating USP5 and promoting its dimerization, which binds to and protects YTHDF1 from degradation. Conversely, the CUL7-FBXW8 E3 ligase promotes YTHDF1 degradation. Deficiency in YTHDF1 or USP5 increases PD-L1 expression and suppresses immune-related gene expression, facilitating immune evasion. Combining USP5 inhibition with anti-PD-L1 therapy enhances anti-tumor immunity, suggesting USP5 as a potential biomarker for patient stratification. This study reveals a ubiquitination-dependent regulation of YTHDF1, proposing USP5 inhibition alongside PD-(L)1 blockade as a promising cancer treatment strategy.
Similar content being viewed by others
Introduction
Cancer arises from numerous genetic and epigenetic alterations, which can be either inherited or acquired somatically. The treatment landscape for various cancers has undergone a revolution with the development of immunotherapies, now considered standard clinical tools1. Currently, immunotherapeutic targets encompass cytotoxic T lymphocyte-associated protein 4, programmed cell death protein 1 (PD-1), and programmed cell death ligand 1 (PD-L1)2, among others. Unfortunately, not all cancer patients respond to immune checkpoint blockade (ICB) agents3,4, necessitating the identification of combinational agents targeting immune pathways5. This study aims to improve both the efficacy and response rate of ICB therapies.
N6-methyladenosine (m6A) is the most prevalent internal modification in mammalian messenger RNA (mRNA), which controls transcript fate through its reader proteins6,7,8. One such reader, YTH N6-methyladenosine RNA binding protein 1 (YTHDF1), enhances mRNA translational efficiency9,10. The expression of YTHDF1 is meticulously regulated at both transcriptional and post-translational levels. Notably, aberrant YTHDF1 expression is observed in various human cancers, including hepatocellular carcinoma (HCC)11,12,13,14. Studies involving Ythdf1-deficient (Ythdf1-/-) mice reveal elevated antigen-specific CD8+ T cell anti-tumor responses15. Additionally, genetic depletion of YTHDF1 has been shown to enhance anti-tumor immunity and improve responses to immune checkpoint inhibitors (ICIs)16,17,18. Consequently, there is a growing interest in the potential pharmacological inhibition of YTHDF1 to improve the efficacy of ICB. However, the identification of specific chemical inhibitors or drugs that modulate YTHDF1 function remains an ongoing challenge.
The ubiquitin proteasome system plays critical roles in diverse cellular processes, encompassing immune response, metabolism, and cell cycle progression19. Ubiquitination and deubiquitination is a reversible process that is tightly controlled by ubiquitin E3 ligases and deubiquitinating proteases (DUBs)20,21. While ubiquitin E3 ligases attach ubiquitin chains to their target proteins, DUBs can cleave and remove ubiquitin chains from substrate proteins. Dysregulation of DUBs is implicated in numerous human diseases, including cancer, emphasizing the potential of DUBs as targets for cancer therapy22,23. Recent evidence underscores the crucial impact of DUBs on anti-tumor immunity, primarily through the stabilization of pivotal checkpoints or key regulators of T-cell functions24. Although ubiquitination of YTHDF1 has been suggested25, the regulatory mechanisms and functional effects of this ubiquitination remain largely unknown.
In this report, we present evidence demonstrating that ubiquitin specific peptidase 5 (USP5) removes K11-linked polyubiquitination of YTHDF1 on multiple residues within the YTH domain. Conversely, the Cullin 7-F-Box and WD repeat domain containing 8 (CUL7-FBXW8) promotes ubiquitination of YTHDF1. mTORC1 activation triggers USP5 dimerization and inhibits FBXW8 from binding to YTHDF1, establishing the USP5-YTHDF1 axis as a regulator of the expression of multiple immune response-related genes that modulate the anti-tumor immune response. Genetic ablation or pharmacological inhibition of USP5, remarkably, reprograms the tumor microenvironment. This reprogramming leads to increased antigen presentation, priming of anti-tumor T cell immunity, and heightened efficacy of immunotherapy. Our findings unveil a ubiquitination-dependent regulation of YTHDF1 function and propose a strategy for enhancing the efficacy of PD-1/PD-L1 blockade.
Results
USP5 deubiquitinates and stabilizes the YTHDF1 onco-protein
As resistance to targeted therapies is often linked to the accumulation of the therapeutic target26,27,28, it becomes crucial to unravel the regulatory mechanisms governing YTHDF1 protein stability and to ascertain whether disruptions in YTHDF1 abundance contribute to cellular resistance to therapy. Remarkably, we observed a gradual decrease in the abundance of the YTHDF1 protein under cycloheximide (CHX) culturing conditions in multiple cell lines (Supplementary Fig. 1a–c). Notably, when cells were treated with proteasome inhibitor MG132, but not lysosome inhibitor chloroquine, significant accumulation of endogenous YTHDF1 (Supplementary Fig. 1a, d), predominantly by extending its protein half-life (Supplementary Fig. 1b, c). The covalent conjugation of ubiquitin is a pivotal step in the proteasome-mediated degradation of target proteins. Supporting this, we observed polyubiquitination of YTHDF1 in both in vitro and in vivo ubiquitination assays (Fig. 1a and Supplementary Fig. 1e), indicating that YTHDF1 protein stability is regulated through the ubiquitin-proteasome pathway.
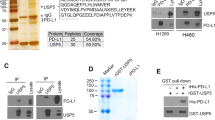
a Immunoblot (IB) analysis of whole-cell lysates (WCL) and anti-Flag immunoprecipitates (IP) from HEK293T cells transfected with GFP-Ub and Flag-YTHDF1. b YTHDF1 polyubiquitination could largely be detected in cells transfected with indicated constructs. c IB analysis of WCL and His immunoprecipitate from HEK293T cells transfected with indicated constructs. d PLC/PRF/5 cells were immunoprecipitated with either anti-USP5 or anti-YTHDF1 antibody and then analyzed by IB. e IB and IP products analysis of USP5-YTHDF1 interaction in HEK293T cells expressing HA-USP5 WT or the indicated truncated YTHDF1 mutants. f, g IB analysis of YTHDF1 levels in HEK293T cells expressing HA-USP5 (DNA content of 250 ng or 500 ng) or indicated plasmids. h IB and QRT-PCR analysis of YTHDF1 from Hepa1-6 cells with Usp5 knockout. n = 3. i, j IB analysis of WCL from Hepa1-6 cells with the depletion of Usp5 or HEK293T cells transfected with indicated constructs for 36 h. Cells were treated with 100 μg/ml CHX at indicated time points. The YTHDF1 levels was quantified by the ImageJ software. k IB analysis of WCL and IP products from HEK293T cells transfected with indicated constructs. Cells were treated with 20 μM MG132 for 8 h. l Effects of Usp5 knockout in Hepa1-6 cells on Ythdf1 K11-linked polyubiquitination were evaluated by IB. m Effects of WP1130 on USP5-mediated YTHDF1 K11-linked polyubiquitination. Cells expressing indicated plasmids were treated with different doses of WP1130. n IB analysis of proteins labeled with puromycin using anti-puromycin antibody upon Usp5 depletion with or without expressing Ythdf1. o, p Assessment of subcutaneous tumor formation from PLC/PRF/5 cells after depletion of USP5, or YTHDF1, or stably expressing YTHDF1 with endogenous USP5 knockdown. Tumor weight was measured at the endpoint of the study. Tumor growth was measured at the indicated time points. n = 5. *p < 0.05, t-test. q Kaplan-Meier analysis revealed a relationship between YTHDF1 and USP5 expression and overall survival in HCC patients. All data are presented as mean ± SEM. All IB data are representative of three independent experiments. Source data are provided as a Source Data file.
Ubiquitination plays a pivotal role in regulating protein function, stability, trafficking, and protein-protein interactions through eight possible linkages of polyubiquitin chains19,29. To explore the impact of individual lysine residues on ubiquitin in YTHDF1 polyubiquitination, we introduced mutations in each lysine residue. Notably, only the K11R (Lys11 to Arg) ubiquitin mutation resulted in the inhibition of YTHDF1 polyubiquitination (Fig. 1b and Supplementary Fig. 1f). To confirm YTHDF1 K11-linked polyubiquitination, we employed a K11-only ubiquitin, where only the K11 lysine residue was retained (with the other six lysine residues mutated to arginine). Although K6, K27, K29, K33, K48, and K63 ubiquitin exhibited some minor impact, K11-ubiquitin promoted the strongest chain formation on YTHDF1, similar to the level observed with wild-type (WT) ubiquitin (Supplementary Fig. 1g). In vivo ubiquitination assays further confirmed K11-linked polyubiquitination of YTHDF1 (Supplementary Fig. 1h). These data strongly suggest the occurrence of K11-linked polyubiquitination on YTHDF1.
The human genome contains ~90 deubiquitinating enzymes (DUBs)30. To identify the potential DUBs for YTHDF1, we overexpressed individual DUBs in a screening effort to discover regulators of YTHDF1 degradation. Among the tested DUBs, USP5 emerged as the sole interacting partner with YTHDF1 (Fig. 1c and Supplementary Fig. 2a, b). Moreover, we performed immunoprecipitation (IP) assay using anti-Flag antibody using cell lysates from HEK293T cells transfected with Flag-YTHDF1 or empty vector (EV) as a negative control. The IP samples were analyzed by mass spectrometry (MS) to identify potential YTHDF1-interacting proteins (Supplementary Fig. 2c and Supplementary data 1). Of note, USP5 was detected in the IP sample from YTHDF1 expressing cells, but not the control cells (Supplementary Fig. 2c). This interaction was further validated in the cytoplasm through reciprocal co-immunoprecipitation (Co-IP) assays with endogenous USP5 and YTHDF1, as well as through immunofluorescence (IF) staining assays (Fig. 1d and Supplementary Fig. 2d, e). In addition, USP5 directly interacts with YTHDF1 in vitro as shown in the GST pull-down assay (Supplementary Fig. 2f). To delineate the regions mediating USP5-YTHDF1 interaction, we mapped the interaction to the C-terminal fragment of USP5 and a C-terminal fragment (aa 191-559) of YTHDF1 (Fig. 1e and Supplementary Fig. 2g). Docking simulation of the protein-protein interacting interface between USP5 and YTHDF1 was gain further structural insight (Supplementary Fig. 2h).
USP5 is known to cleave various polyubiquitin linkages, including K6, K11, K29, K48, and K6331,32. To evaluate the functional significance of the physical interaction between USP5 and YTHDF1 in inhibiting YTHDF1 ubiquitination, we overexpressed WT USP5 in multiple cell lines, resulting in a dose-dependent increase in YTHDF1 protein abundance. In contrast, expression of the catalytically inactive mutant USP5 [C335A (Cys335 to Ala)] abolished this effect (Fig. 1f, g and Supplementary Fig. 2i). Conversely, depletion of USP5 using short hairpin RNAs (shRNAs) or CRISPR/Cas9 led to a significant decrease in YTHDF1 protein levels, and this process could be efficiently blocked by MG132 (Fig. 1h and Supplementary Fig. 2j). Importantly, YTHDF1 mRNA levels, total m6A RNA methylation levels and other YT521-B homology (YTH) domain protein levels, including YTHDF2, and 3 and YTHDC1 and YTHDC2, remained unchanged (Fig. 1h and Supplementary Fig. 2j–l). The half-life of YTHDF1 was markedly shortened or extended in USP5-depleted or overexpressed cells (Fig. 1i, j and Supplementary Fig. 2m, n), this is accompanied by an increase or decrease in YTHDF1 ubiquitination levels (Fig. 1k, l and Supplementary Fig. 2o, p). Interestingly, compared with WT USP5, the half-life of YTHDF1 was shortened in cells overexpressing the truncated forms of USP5 (Supplementary Fig. 2q). Moreover, in vitro deubiquitination assays demonstrated that YTHDF1 ubiquitylation decreased after the overexpression of USP5 WT, but not the USP5 C335A mutant (Supplementary Fig. 2r). Consistently, treatment with the USP5 inhibitor WP1130 induced YTHDF1 ubiquitination and degradation (Fig. 1m and Supplementary Fig. 2s–u). Strikingly, the surface sensing of translation (SUnSET) assay revealed a significant decrease in protein synthesis in USP5- or YTHDF1-deficient cells (Fig. 1n and Supplementary Fig. 2v). However, USP5-deficient cells re-expressing YTHDF1 restored protein synthesis to WT level (Fig. 1n). Notably, ectopic expression or depletion of YTHDF1 had no effect on USP5 expression (Supplementary Fig. 2w). Thus, these findings validated USP5 as a bona fide deubiquitinating enzyme that removes K11-linked polyubiquitination from YTHDF1.
Next, we aimed to unravel the biological role of USP5 in regulating YTHDF1 stability. YTHDF1 has previously been identified as a key player in cell proliferation and tumorigenesis11,12,13,14. In line with these findings, our observations revealed that, compared to the control, the depletion of either YTHDF1 or USP5 significantly hindered colony formation and tumorigenesis (Fig. 1o, p and Supplementary Fig. 3a–c). Importantly, the stable expression of YTHDF1 in USP5-depleted cells restored the colony formation and tumorigenesis (Fig. 1o, p and Supplementary Fig. 3a–c). Consequently, we noted that, in comparison to the control, the depletion of YTHDF1 counteracted the inhibitory effects of the USP5 inhibitor WP1130 on colony formation (Supplementary Fig. 3d). Consistent with these findings, the levels of YTHDF1 and USP5 were significantly higher in tumors from HCC patients, exhibiting a positive correlation in 23 tumor tissues (Supplementary Fig. 3e–g). Immunohistochemistry (IHC) analyses further revealed that higher levels of USP5 and YTHDF1 correlate with poorer overall survival (Fig. 1q and Supplementary Fig. 3h, i). Taken together, these data strongly suggest that USP5 promotes cancer progression primarily by inhibiting YTHDF1 K11-linked polyubiquitination and degradation in the context of HCC (Supplementary Fig. 3j).
USP5 deubiquitinates YTHDF1 on multiple lysine residues to confer oncogenicity
To further elucidate the molecular mechanisms underlying USP5-mediated deubiquitination of YTHDF1, we found that the fragment (aa 389-559) of YTHDF1, particularly the YTH domain, was both necessary and sufficient for YTHDF1 ubiquitination in cells (Fig. 2a and Supplementary Fig. 4a, b). Consistently, the removal of K11-linked polyubiquitin chains on YTHDF1 was essential for its interaction with USP5, primarily through the C-terminal domain on USP5, leading to YTHDF1 deubiquitylation and stability (Fig. 2b, c). Previous mass spectrometry analyses identified 12 lysine residues as potential ubiquitination sites on YTHDF1 (Supplementary Fig. 4c; www.phosphosite.org). K11-linked polyubiquitination and degradation of YTHDF1 were impeded when all 12 lysine residues or four lysine residues (K469, K473, K500, and K527) in the 389-559 aa fragment were simultaneously mutated to arginine (R) (Fig. 2d), which substantially prolonged the half-life of YTHDF1 (Fig. 2e). Notably, we found that USP5-mediated removal of polyubiquitination and degradation of YTHDF1 were blocked if all four lysine residues (K469, K473, K500, and K527) were mutated to arginine (R) (Fig. 2f). In addition, compared to PLC/PRF/5-sgUSP5 cells expressing WT YTHDF1, cells expressing the ubiquitination-deficient YTHDF1 mutants (4KR or 12KR) failed to promote YTHDF1 polyubiquitination (Fig. 2g). Moreover, these four lysine residues are conserved across species (Supplementary Fig. 4c). Notably, none of the single mutations blocked YTHDF1 ubiquitination (Supplementary Fig. 4d). Furthermore, the single or multiple mutations in YTHDF1 affect its binding affinity to USP5 to various degrees (Supplementary Fig. 4e). These results indicate that the removal of K11-linked polyubiquitination on multiple sites by USP5 may be evolutionarily conserved and crucial for the regulation of YTHDF1 function.
a IB analysis of WCL and anti-Flag IPs from HEK293T cells transfected with indicated constructs and treated with the 20 μM MG132 for 8 h. b In vivo ubiquitination assay of YTHDF1 in HEK293T cells expressing Flag-YTHDF1 WT and indicated truncated YTHDF1 mutants in the presence or absence of ectopic USP5 expression. Cells were treated with 20 μM MG132 for 8 h. c IB analysis of the protein levels of the indicated truncated YTHDF1 mutants in HEK293T cells expressing increasing amounts of HA-USP5. d IB analysis of WCL and IP derived from HEK293T cells transfected with indicated constructs. Cells were treated with 20 μM MG132 for 8 h. e IB analysis of WCL derived from HEK293T cells transfected with indicated constructs. 36 h post-transfection, cells were treated with 100 μg/ml CHX at indicated time points. The YTHDF1 protein abundance was quantified by the ImageJ software. f In vivo ubiquitination assays of WCL and anti-Flag IPs derived from HEK293T cells transfected with plasmids expressing the indicated proteins. Cells were treated with 20 μM MG132 for 8 h. g In vivo ubiquitination assay of USP5 knockout in PLC/PRF/5 cells transfected with WT Flag-YTHDF1 or indicated mutant constructs. YTHDF1 polyubiquitination was evaluated by IB analysis. Cells were treated with 20 μM MG132 for 8 h. h Colony formation assays of PLC/PRF/5 cells stably expressing YTHDF1-WT or -4KR mutant with endogenous YTHDF1 knockout. n = 3 per group. **p < 0.01. t-test. i, j Assessment of subcutaneous tumor formation from PLC/PRF/5 cells stably expressing YTHDF1-WT or -4KR mutant with endogenous YTHDF1 knockout. Tumor weight was measured at the endpoint of the study. In vivo tumor growth was measured at the indicated time points and tumors were dissected at the endpoint. n = 6 per group. **p < 0.01, t-test. All data are presented as mean ± SEM. All IB data are representative of two independent experiments. Source data are provided as a Source Data file.
In an effort of assessing the impact of YTHDF1 K11-linked polyubiquitination on cell proliferation and tumorigenic potential, we found that the cells with the 4KR mutations exhibited elevated colony formation and tumorigenic potential as compared to their WT counterparts (Fig. 2h–j). These findings collectively indicate that USP5-mediated K11-linked YTHDF1 deubiquitylation on multiple lysine residues enhances the oncogenic function of YTHDF1 in vitro and in vivo.
Insulin/mTORC1 regulates USP5 function and inhibits YTHDF1 degradation
Protein homeostasis, comprised of protein synthesis, modification, localization, and degradation processes, is intricately influenced by stimuli from microenvironment such as signaling molecules, metabolites, and pathogens33. We next asked what the feeding signal is for the stabilization of YTHDF1 by USP5. In response to insulin, but not other growth factors that we examined, or hepatitis B virus X (HBx), YTHDF1 protein levels were elevated in a dose-dependent manner, while USP5 levels remained constant (Fig. 3a and Supplementary Fig. 5a, b). Notably, under serum-starved conditions, ubiquitinated YTHDF1 was primarily K11-linked, and the levels of this linkage were significantly reduced upon insulin stimulation (Fig. 3b and Supplementary Fig. 5c). Furthermore, YTHDF1 K11-linked polyubiquitination was significantly reduced by insulin stimulation between 5 and 45 min, but slightly increased at 60 and 90 min (Supplementary Fig. 5d). Strikingly, depletion of USP5 by shRNA or CRISPR/Cas9-mediated knockout completely blocked the insulin-mediated elevation of YTHDF1 protein abundance (Fig. 3c and Supplementary Fig. 5e, f).
a IB analysis using the indicated antibodies in PLC/PRF/5 and Hepa1-6 cells treated with increasing concentrations of insulin for 30 min. b Reduced YTHDF1 polyubiquitination upon growth factor stimulation. HEK293T cells transfected with ubiquitin constructs were lysed for anti-YTHDF1 IP and IB. c The WT and Usp5-KO cells were treated with insulin (100 ng/ml) or Torin1 (1.0 μM). After 30 min or 24 h, cells were collected and analyzed by IB. d IB analysis of YTHDF1 protein levels in PLC/PRF/5 cells expressing the indicated constructs. e Knockdown of USP5 inhibits mTOR or RPTOR-induced accumulation of YTHDF1. The WT and USP5-KD PLC/PRF/5 cells were transfected with Flag-mTOR or HA-RPTOR constructs for 36 h. f IB analysis of YTHDF1 immunoprecipitate or WCL from PLC/PRF/5 cells transfected with Flag-YTHDF1. Cells were then serum starved for 16 h and stimulated with insulin (100 ng/ml) for 30 min. g IB analysis of WCL and IP derived from HEK293T cells transfected with various HA-USP5 constructs as well as Flag-USP5. Cells were pretreated with 1.0 μM Torin1 for 24 h. h IB analysis of WCL and IP derived from HEK293T cells transfected with Flag-mTOR together with the HA-USP5 constructs for 36 h. i In vivo ubiquitination analysis of YTHDF1 in PLC/PRF/5 cells expressing the indicated HA-USP5 constructs. Cells were treated with 20 μM MG132 for 8 h. j In vivo phosphorylation assay of HA-USP5 WT and S149A mutant in HEK293T cells with or without ectopic Flag-mTOR expression. k The S149 mutant disrupted USP5 dimerization process in cells. IB analysis of WCL and IP derived from HEK293T cells transfected with constructs indicated. l Co-IP analysis of USP5/YTHDF1 interaction in HEK293T cells with expressing the indicated constructs. m A simplified model depicting the regulatory mechanism of USP5 by insulin signaling pathways. The active mTORC1 phosphorylates USP5 at S149, which promotes its dimerization and stabilizes YTHDF1, and then feedback positively regulates mTORC1 activity by increasing RPTOR mRNA translation. All data are representative of two independent experiments. Source data are provided as a Source data file.
The insulin signaling cascade involves the PI3K, AKT, and mTOR signaling pathways34. Under serum-starved or normal culture conditions, Wortmannin (a PI3K inhibitor), AKTi-1/2 (an AKT inhibitor), Torin1 (an mTOR inhibitor), and rapamycin (an mTORC1 inhibitor) all abrogated insulin- or USP5-mediated accumulation of YTHDF1 (Fig. 3c and Supplementary Fig. 5e–h). Consistently, Wortmannin, AKTi-1/2, or Torin1 were not able to reduce YTHDF1 protein abundance in USP5-deficient cells, indicating that mTOR activity, as determined by downstream events, remained unaffected (Fig. 3c and Supplementary Fig. 5e, f, i, j). Moreover, Rapamycin and Torin1 treatment decreased YTHDF1 protein abundance in a time- or dose-dependent manner (Supplementary Fig. 5k–m). Importantly, Torin1 could not gradually decrease YTHDF1 abundance in USP5-deficient cells (Supplementary Fig. 5n–q). These results suggest that insulin acts through mTOR activation to elevate YTHDF1 abundance, and this function is dependent on USP5. As a result, USP5 deletion largely abolished the decrease of YTHDF1 ubiquitination in cells induced by growth factor stimulation (Supplementary Fig. 5r). The enhanced interaction of YTHDF1 with mTOR components needs further investigation (Supplementary Fig. 5r, s). Thus, USP5 likely responds to insulin stimulation downstream of mTOR pathway.
mTOR nucleates two functionally distinct complexes, mTOR complex 1 (mTORC1) and mTORC2. The conserved components of mTORC1 include mTOR, mLST8, and RPTOR, whereas mTORC2 consists of mTOR, mLST8, SIN1, and RICTOR35,36. In alignment with the idea that YTHDF1 abundance decreases upon mTOR inhibitor treatment, depletion of mTOR or RPTOR by shRNA led to a marked decrease in the protein level of YTHDF1 (Supplementary Fig. 6a). In contrast, the ectopic expression of mTORC1 and its downstream substrate S6K2, but not mTORC2, S6K1, or S6K kinase inactive mutations, resulted in a significant elevation of YTHDF1 protein abundance (Fig. 3d and Supplementary Fig. 6b, c). The elevation of YTHDF1 protein abundance occurred largely through the extension of its protein half-life (Supplementary Fig. 6d, e) and was accompanied by a decrease in YTHDF1 ubiquitination (Supplementary Fig. 6f). Importantly, mTORC1-mediated increase of YTHDF1 abundance could be efficiently blocked by USP5 silencing (Fig. 3e). Additionally, tuberous sclerosis complex 1 (TSC1) and tuberous sclerosis complex 2 (TSC2) are tumor suppressors and specifically suppress mTORC1 activity37. Ectopic expression or knockdown of TSC2, but not TSC1, decreased or increased the protein abundance of YTHDF1, respectively, without affecting USP5 levels (Supplementary Fig. 6g–j).
Furthermore, YTHDF1-deficient cells exhibited a dampened lysosomal translocation of mTORC1 (Supplementary Fig. 6k–m) and ectopic expression of YTHDF1 enhances mTOR and RPTOR interacting activity (Supplementary Fig. 6n). However, the GST pull-down assay suggested that YTHDF1 indirectly interacts with mTOR in vitro (Supplementary Fig. 6o). Moreover, YTHDF1-deficient cells exhibited a decrease in RPTOR protein levels but did not change other mTOR signaling events that we examined (Supplementary Fig. 6p, q). In contrast, ectopic expression of YTHDF1 led to an elevation of mTORC1 activity and increased RPTOR abundance (Supplementary Fig. 6r–t). RPTOR is responsible for targeting mTORC1 to the lysosome and is a prerequisite for mTORC1 activation38,39. Strikingly, RPTOR mRNA was modified by m6A and potentially targeted by YTHDF19. The expression of a YTHDF1 mutant, with two point-mutations (K395A and Y397A) in the YTH domain abolishing its m6A-binding activity40, did not enhance RPTOR protein levels (Supplementary Fig. 6u). This suggests that YTHDF1-regulated RPTOR translation is m6A modification-dependent. Moreover, YTHDF1 is a major substrate of USP5, and it regulates mTORC1 activity through a positive feedback loop. The re-expression of YTHDF1 completely abolished the inhibitory effect on mTORC1 in USP5-deficient cells (Supplementary Fig. 6v).
mTORC1 phosphorylates USP5 to promote its dimerization and function
To understand how insulin/mTORC1 signaling triggers USP5-mediated YTHDF1 deubiquitylation, we initially observed that treatment of cells with insulin enhanced the interaction between USP5 and YTHDF1 (Fig. 3f and Supplementary Fig. 7a). Moreover, overexpression of mTOR or TSC2 resulted in increased or decreased interaction between USP5 and YTHDF1, respectively, (Supplementary Fig. 7b, c).
Previous studies showed that USP5 localizes in the nucleus, where USP5 is involved in the regulation of DNA-repair or heat-induced stress granules41,42. However, recent studies suggested that USP5 largely localizes in the cytoplasm, where USP5 interacts with NLRP3 to govern inflammasome activation41,43. To further investigate the mechanism by which insulin/mTORC1 signaling promotes USP5 binding to YTHDF1, we examined the cellular localization of USP5 after insulin treatment. We found that insulin did not impact the nuclear-cytoplasmic shuttling of USP5 (Supplementary Fig. 7d, e). Consistently, Torin1 treatment or mTOR overexpression also didn’t alter USP5 cellular localization (Supplementary Fig. 7f–i). Interestingly, overexpression of mTOR led to an increase in USP5 dimerization (Supplementary Fig. 7j). In contrast, Torin1 treatment inhibited USP5 dimerization (Fig. 3g and Supplementary Fig. 7k). mTOR is a highly conserved serine/threonine protein kinase. Indeed, mTOR interacted with USP5 and effectively phosphorylated the N-terminal fragment of USP5 (Fig. 3h and Supplementary Fig. 7l). Torin1 blocked this phosphorylation (Supplementary Fig. 7m). Moreover, ectopic expression of S6K2, but not S6K1 or S6K kinase inactive mutations, specifically promoted USP5 protein phosphorylation in cells (Supplementary Fig. 7n). Notably, S6K2 and S6K2 K99R, but not S6K1 and its mutants, interacted with YTHDF1 or USP5 (Supplementary Fig. 7o).
Differential phosphorylation of several peptides in USP5 has been identified using mass spectrometry44 (Supplementary Fig. 8a). Mutation of the phosphorylation site at Ser149 (Ser149 to Ala) with a well-characterized mTOR substrate motif, abolished the ability of USP5 to elevate YTHDF1 protein abundance, significantly shortened the half-life of YTHDF1 (Supplementary Fig. 8b, c), and promoted YTHDF1 ubiquitination in cells (Fig. 3i and Supplementary Fig. 8d). Importantly, the phosphorylation-null USP5 (S149A) was not efficiently phosphorylated by mTORC1 (Fig. 3j), and its phosphorylation was efficiently blocked by the ectopic expression of TSC2 (Supplementary Fig. 8e). Like the enzymatic-dead C335A mutant, both the phosphorylation-null USP5 (S149A) and the phosphomimic USP5 (Ser149 to Asp, S149D) failed to promote YTHDF1 stability, protein synthesis, and colony formation capacity (Supplementary Fig. 8f–i). Compared with USP5-WT, the S149A or S149D mutations abolished its dimerization (Supplementary Fig. 8j). Consistent with these findings, the ectopic expression of mTORC1 failed to enhance USP5 dimerization and reduce binding to YTHDF1 in the two USP5 mutants (Fig. 3k, l and Supplementary Fig. 8k–m). These results support a model in which mTORC1 phosphorylates USP5 at S149, thereby increasing dimerization, recruiting it to YTHDF1, and antagonizing its degradation in an enzyme activity-dependent manner (Fig. 3m). The molecular mechanism behind the enhanced binding of dimerized USP5 to YTHDF1 warrants further investigation.
CUL7-FBXW8 ubiquitinates YTHDF1 to promote its degradation
To identify the E3 ubiquitin ligase responsible for YTHDF1 ubiquitination and degradation, we employed MLN4924, an inhibitor of cullin-based ubiquitin E3 ligases. The treatment resulted in elevated endogenous YTHDF1 protein levels, suggesting the involvement of a cullin-based ligase in its degradation (Fig. 4a). Subsequent investigation revealed an interaction between YTHDF1 and a member of the cullin family, Cullin 7 (CUL7), which also promoted YTHDF1 degradation (Fig. 4b and Supplementary Fig. 9a, b). Further experiments demonstrated the dose-dependent effect of CUL7 on YTHDF1 protein degradation, and this process could be efficiently blocked by MG132 (Supplementary Fig. 9c, d). Ectopic expression of CUL7 led to YTHDF1 ubiquitination and reduced protein half-life (Supplementary Fig. 9e–g), while endogenous CUL7 depletion resulted in a significant increase in YTHDF1 protein levels and abolished YTHDF1 ubiquitination (Supplementary Fig. 9h, i). This was accompanied by an extension of its half-life (Supplementary Fig. 9j).
a IB analysis of YTHDF1 protein levels in Huh7 and PLC/PRF/5 cells after treatment of 20 μM MG132, 10 μM Bortezomib, or 5 μM MLN4924 for 8 h. b IB analysis of YTHDF1 protein levels in PLC/PRF/5 cells transfected with an empty vector (EV) or vectors encoding various Myc-tagged Cullin proteins. c IB analysis of YTHDF1 protein levels in PLC/PRF/5 cells expressing increasing amount of T7-FBXW8. Cells were treated with 20 μM MG132 for 8 h. d FBXW8 and USP5 control K11-linked ubiquitination of YTHDF1. IB analysis of IP and WCL derived from HEK293T cells transfected with indicated constructs. e IB and IP products analysis of FBXW8-YTHDF1 interaction in HEK293T cells expressing T7-FBXW8 WT or the indicated truncated YTHDF1 mutants. Cells were treated with 20 μM MG132 for 8 h before harvesting. f IB and IP products analysis of FBXW8-YTHDF1 interaction in HEK293T cells expressing Flag-YTHDF1 or the indicated truncated FBXW8 mutants. Cells were treated with 20 μM MG132 for 8 h before harvesting. g In vivo ubiquitination assay of YTHDF1 in HEK293T cells expressing Flag-YTHDF1 or the indicated truncated YTHDF1 mutants in the presence or absence of ectopic FBXW8 expression. Cells were treated with 20 μM MG132 for 8 h before harvesting. h In vivo ubiquitination assay of YTHDF1 in HEK293T cells expressing Flag-YTHDF1 in the presence ectopic FBXW8 expression or the indicated truncated FBXW8 mutants. Cells were treated with 20 μM MG132 for 8 h before harvesting. i IB analysis of YTHDF1 protein levels in PLC/PRF/5 cells expressing the indicated mTOR components. j FBXW8 antagonism USP5-YTHDF1 interaction. IB and IP products analysis of USP5/YTHDF1 or FBXW8/YTHDF1 interaction in HEK293T cells expressing Flag-YTHDF1 or HA-USP5, plus with or without expressing increasing amounts of T7-FBXW8. k IB and IP products analysis of USP5/YTHDF1 or FBXW8/YTHDF1 interaction in HEK293T cells with or without ectopic mTOR expression. l A model illustrating the roles of mTORC1 pathway in governing USP5/FBXW8-controlled YTHDF1 K11-linked poly-ubiquitination. All data are representative of two independent experiments. Source data are provided as a Source data file.
The WD40 repeat-containing F-box protein FBXW8 serves as the sole substrate-targeting subunit of the CUL7 E3 complex, with its C-terminal region responsible for substrate recognition and interaction45. Upon ectopic expression of FBXW8, YTHDF1 protein degradation occurred in a dose-dependent manner (Fig. 4c and Supplementary Fig. 10a). The FBXW8-mediated degradation of YTHDF1 was efficiently blocked by MG132 (Fig. 4c and Supplementary Fig. 10a), indicating a role of the ubiquitin-proteasome pathway in this process. FBXW8 demonstrated interaction with YTHDF1, promoting K11-linked polyubiquitination and shortening the half-life of YTHDF1 protein in cells (Fig. 4d and Supplementary Fig. 10b–e). Conversely, depletion of FBXW8 resulted in a significant increase in YTHDF1 protein levels and abolished YTHDF1 ubiquitination, with a significant extension of its protein half-life (Supplementary Fig. 10f–h). To unravel the molecular mechanism governing FBXW8-mediated ubiquitination of YTHDF1, we mapped the region(s) responsible for FBXW8-YTHDF1 interaction. The C-terminal fragment of FBXW8 and the YTH domain of YTHDF1 were identified as critical for their binding (Fig. 4e, f and Supplementary Fig. 10i–k). FBXW8 promoted degradation and elevated K11-linked polyubiquitination of YTHDF1 in a manner dependent on the FBXW8 recognition motif (Fig. 4e, g and Supplementary Fig. 10l). Further investigation revealed that the C-terminal region of FBXW8 was both necessary and sufficient for YTHDF1 ubiquitination in cells (Fig. 4f, h). Deletion of the F-box or WD motif did not affect FBXW8’s interaction with YTHDF1 but enhanced YTHDF1 protein stability to different levels as compared to WT-FBXW8 (Supplementary Fig. 10j–n). This indicates that both motifs are necessary for FBXW8’s activity. In line with the importance of YTHDF1 ubiquitination sites in controlling YTHDF1 homeostasis, the deficiency in YTHDF1 ubiquitination abolished FBXW8-mediated protein ubiquitination and degradation (Supplementary Fig. 10o, p).
Upon insulin stimulation, mTORC2 phosphorylates FBXW8 at S86 to regulate its protein stability and cytosolic localization46. Overexpression of mTORC2 components led to an elevation in FBXW8 abundance, while ectopic expression of mTORC1 components resulted in a significant decrease in FBXW8 protein levels (Fig. 4i and Supplementary Fig. 11a). Consistently, depletion of endogenous mTOR or RPTOR led to a marked increase in FBXW8 protein abundance (Supplementary Fig. 11b, c). Similar results were observed upon mTOR inhibitor treatment (Supplementary Fig. 11d). Given that USP5 removes the K11-linked ubiquitin chain on YTHDF1, we examined whether USP5 can also remove FBXW8-mediated K11-linked ubiquitination on YTHDF1. Ectopic expression of USP5 dramatically reduced FBXW8-promoted K11-linked ubiquitination of YTHDF1 in cells (Fig. 4d). This reduction in YTHDF1 ubiquitination was attributed to the induced binding of USP5 to YTHDF1, antagonizing FBXW8 binding (Fig. 4j). Interestingly, the antagonism between FBXW8 and USP5 interaction with YTHDF1 was affected by mTORC1 signaling (Fig. 4k and Supplementary Fig. 11e–h). These findings suggest that USP5-mediated deubiquitination, which counters FBXW8-induced ubiquitination, is physiologically regulated by growth factor signaling to control YTHDF1 ubiquitination (Fig. 4l).
USP5 deficiency triggers tumor cell-intrinsic immune responses to remodel anti-tumor immunity
In a recent study, the deficiency of Ythdf1 has been linked to enhanced antigen-specific CD8+ T cell anti-tumor responses17,18. Building upon the aforementioned findings, we aimed to investigate the mechanism connecting the inhibition of USP5 to the augmentation of anti-tumor T cell immunity. Our investigation focused on the impact of tumor cell-intrinsic USP5 or YTHDF1 on the infiltration of T cells in the tumor microenvironment. IHC staining results from 90 HCC specimens revealed higher levels of CD4+ and CD8+ tumor-infiltrating lymphocytes (TILs) associated with better survival in cases with USP5 or YTHDF1 low expression, but not in cases with high expression (Fig. 5a and Supplementary Fig. 12a, b). Using the Tumor Immune Dysfunction and Exclusion (TIDE) tool data sets47, we observed that a higher level of cytotoxic T lymphocytes (CTLs) correlates with improved survival in multiple cancer types with low expression of USP5, but not in cases with USP5 high expression (Supplementary Fig. 12c). This correlation was also evident in cohorts with breast cancer, lung cancer, and colorectal cancer (Supplementary Fig. 12d). These data suggest that tumor-intrinsic Usp5 promotes tumor growth likely through inducing tumor immune evasion. To determine which cell type contributes to the tumor immunosurveillance, we depleted NK cells, CD4+ T cells, or CD8+ T cells using neutralizing antibodies in Hepa1-6 tumor models. The results showed that the depletion of CD8+ T cells, but not CD4+ T cells or NK cells, abolished the differences in tumor growth between WT and Usp5-KO Hepa1-6 tumors (Fig. 5b). These findings suggest that USP5 regulates tumor evasion from CD8+ T cell-mediated immune surveillance.
a Kaplan-Meier analysis of USP5 and CD4 or CD8 expression and overall survival of 90 HCC patients. b A total of 5.0 × 106 WT or Usp5-KO Hepa1-6 cells were subcutaneously implanted in C57BL/6 mice pretreated with anti-IgG, anti-NK1.1, anti-CD4, and anti-CD8 antibodies (n = 6). Tumor growth was monitored at the indicated time points. ***p < 0.001 by two-way ANOVA. c IB analysis of lysates derived from dissected xenografts formed by Usp5-depleted H22 or Hepa1-6 cells. d, e IB analysis of Pd-l1 in the WT and Usp5-KO Hepa1-6 cells transfected with Flag-Ythdf1 constructs for 36 h or treated with 2.5 μM WP1130 for 24 h. f Activated T cells and tumor cells were co-cultured in 24-well plates for 4 days and surviving tumor cells were visualized by crystal violet staining. Relative fold ratios of surviving cell intensities are shown. n = 3. **p < 0.01. t-test. g Correlation between the fold changes of differentially expressed genes in Ythdf1 and Usp5 knockout Hepa1-6 cells. h The top 5 terms in GO analysis of the immune-related genes suppressed by both Usp5 and Ythdf1 knockout Hepa1-6 cells. n = 3. i GSEA and heatmap showing the differential expression of genes in Fig. 5g. n = 3. j mRNA levels of indicated genes from sgUsp5 or sgCtrl Hepa1-6 cells were analyzed using QRT-PCR. n = 3. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. t-test. k The mRNA and protein levels in Usp5 and Ythdf1 knockout Hepa1-6 cells are measured by RNA-Seq and proteomics. l, m Usp5 or Ythdf1 depletion affects tumor growth in mouse xenograft in NOD/SCID and C57BL/6 mice (n = 8). Tumor growth was monitored at the indicated time points, and tumor volume were measured at the endpoint (l). Tumor image and tumor weight are presented (m). ***p < 0.001 by two-way ANOVA. All data are presented as mean ± SEM. All IB data are representative of two independent experiments. Source data are provided as a Source Data file.
The induction of PD-L1 has been linked to the suppression of the functional activity of CD8+ TILs48. Bioinformatic analysis (http://cis.hku.hk/TISIDB49) revealed that high mRNA levels of YTHDF1 or USP5 in HCC were inversely correlated with several immunosuppressive genes, including PD-L1 (Supplementary Fig. 12e). In line with this observation, tumor samples with USP5 depletion exhibited higher levels of PD-L1 protein (Fig. 5c). Notably, USP5 was found to negatively regulate PD-L1 expression in a YTHDF1-dependent manner (Fig. 5d and Supplementary Fig. 12f–h). Treatment with the USP5 inhibitor WP1130 led to an increase in endogenous PD-L1 expression in a time- and high dose-dependent manner (Fig. 5e and Supplementary Fig. 12i, j). Intriguingly, USP5 deficiency induces nuclear and membrane enrichment of PD-L1, but not the cytosolic level of PD-L1 (Supplementary Fig. 12k–m). To further validate the role of the USP5/YTHDF1 axis in PD-L1-mediated immunosuppression, we investigated T cell-mediated cancer cell killing in Hepa1-6 cells with depleted Usp5 or Ythdf1. The results indicated that the suppression of USP5 or YTHDF1 in cancer cells significantly improved T cell activity (Fig. 5f and Supplementary Fig. 13a, b), and this effect was further enhanced by PD-1 mAb treatment (Fig. 5f). This result is consistent with the results obtained from USP5 inhibitor WP1130 treatment experiment (Supplementary Fig. 13c). WP1130 treatment in Ythdf1 deficient Hepa1-6 cells improved T cell responsiveness (Supplementary Fig. 13d, e). These findings suggest that the inhibition of USP5 or YTHDF1 in cancer cells may lead to elevated PD-L1 expression levels, contributing to the suppression of T cell activity.
We then conducted a comprehensive transcriptomic analysis to gain insights into the signaling pathways regulated by USP5 or YTHDF1 in cancer cells. The results demonstrated a significant impact of Usp5 or Ythdf1 inhibition on gene expression in Hepa1-6 cells (Fig. 5g and Supplementary Fig. 13f). Intriguingly, the deficiency of USP5 or YTHDF1 in cancer cells led to the downregulation of a set of genes associated with immune responses, particularly those involved in interferon-gamma (IFNγ)-mediated signaling, inflammation-mediated signaling, and innate immune signaling pathways (Fig. 5h, i and Supplementary Fig. 13g). This observation was further validated by qRT-PCR, which confirmed the reduction in the expression of IFNγ or inflammation signaling-related genes upon USP5 depletion (Fig. 5j). Notably, several IFN-inducible T-cell chemo-attractants (Ccl2, Ccl9, Ccl20) exhibited significant downregulation in Hepa1-6 cells treated with sgUsp5 (Fig. 5j and Supplementary Fig. 13h). To search for the differential YTHDF1-binding mRNAs in Usp5 knockout Hepa1-6 cells, we performed RIP-seq to map RNA transcripts bound by YTHDF1 in both the sgCtrl and sgUsp5 cells. Through this analysis, we found 528 candidate YTHDF1-binding mRNAs that are affected by USP5 knockout (Supplementary Data 2). We performed Gene Ontology (GO) analysis on the 528 YTHDF1-binding mRNAs and found that transcripts involved in immune responses were highly enriched (Supplementary Fig. 13i, j). Furthermore, the proteomic profiles also supported the heightened engagement of the innate immune signaling pathway in Usp5- or Ythdf1-null tumor cells, establishing consistency between our proteomic and transcriptomic datasets (Fig. 5k and Supplementary Data 3). Further analysis of the RNA-seq data set showed that a panel of genes involved in the MHC-I-mediated antigen processing and presenting were enriched in the Usp5-deficient Hepa1-6 cells (Supplementary Fig. 13k). Moreover, bioinformatic analysis demonstrated that most of the genes in the MHC-I pathways are significantly upregulated in liver adenocarcinoma patients with low USP5 expression, as compared with high USP5 expression (Supplementary Fig. 13l). We further confirmed that Usp5 depletion significantly elevated cell surface H2Kd/H2Dd levels, but expression of Ythdf1 WT or 4KR mutations abolished this phenotype in Hepa1-6 cells (Supplementary Fig. 13m, n). Furthermore, this phenotype was also observed by USP5 inhibitor WP1130 treatment (Supplementary Fig. 13m, o). These findings propose a model wherein the lower expression of USP5 or YTHDF1 in cancer cells may elevate a panel of immune response genes and T-cell chemo-attractants, in part, increases the antigen processing and presentation, which might support cytotoxic T cells to eliminate cancer cells.
Based on our in vitro findings that YTHDF1 and USP5 regulate cellular processes potentially related to tumor immune responses, we investigated whether the inhibition of USP5 or YTHDF1 could induce anti-tumor immunity in vivo. To explore this possibility, we utilized mouse syngeneic tumor models by subcutaneously inoculating Hepa1-6 cells into immunodeficient and immunocompetent mice. Upon inoculation into immunocompetent mice, the growth of implanted tumors was noticeably delayed after the depletion of Ythdf1 or Usp5 compared to immunodeficient hosts in vivo (Fig. 5l, m and Supplementary Fig. 13p, q). This result suggests that inhibiting USP5 or YTHDF1 in tumor cells elicits potent anti-tumor T cell immunity in vivo, rather than affecting tumor cell autonomous growth to restrain tumor burden.
Targeting USP5 potentiates anti-PD-L1 immunotherapy in murine HCC models
Recent studies have revealed a correlation between the response to PD-1/PD-L1 blockade and PD-L1 expression levels in tumor cells50,51. The USP5-YTHDF1 axis negatively regulates tumor cell PD-L1 accumulation (Fig. 5c). Based on our mechanism studies, we hypothesized that inhibition of USP5 might synergize with anti-PD-1/PD-L1 therapy to elicit an enhanced therapeutic effect. Notably, genetic depletion of Usp5 sensitized Hepa1-6 tumors to anti-PD-L1 immunotherapy in immunocompetent C57BL/6 mice (Fig. 6a, b). Pharmacologic inhibition of USP5 might synergize with PD-1/PD-L1 blockade for cancer treatment; therefore, we examined this hypothesis using the syngeneic mouse HCC tumor model treated with PD-L1 antibody and/or USP5 inhibitor WP1130 (Fig. 6c). Compared to the anti-PD-L1 or WP1130 treatment group, the combination of the USP5 inhibitor and anti-PD-L1 markedly retarded tumor growth and dramatically improved overall survival rates (Fig. 6d, e). Analysis of infiltrated immune cells demonstrated that the USP5 inhibitor combined with anti-PD-L1 treatment significantly increased the percentage of CD8+ T cells (Fig. 6f, g and Supplementary Fig. 14a). To further address whether the USP5 inhibitor affects the activation of tumor-infiltrating CD8+ T cells and the profile of exhausted T cells, we detected the T-cell activation marker, Granzyme B (GzmB), and the exhausted T-cell marker, Tim-3, on infiltrated CD8+ T cells in the syngeneic Hepa1-6 and HCC mouse tumor models. Our results showed that the USP5 inhibitor combined with anti-PD-L1 treatment significantly elevated the expression of GzmB and reduced the expression of Tim-3 on infiltrated CD8+ T cells (Fig. 6f, g and Supplementary Fig. 14a). In addition, significant upregulation of Pd-l1 and downregulation of Ythdf1 were also observed in tumor tissues treated with the USP5 inhibitor or combined treatment compared with control treatment (Fig. 6f). Our study suggests that the combination of a USP5 inhibitor and anti-PD-1/PD-L1 treatment is an effective strategy for human cancers.
a Tumor growth of sgCtrl and sgUsp5 Hepa1-6 cells in C57BL/6 mice with anti-IgG mAb or anti-PD-L1 mAb treatments. n = 10. b Kaplan-Meier survival curves for four treatment groups demonstrate the improved efficacy of PD-L1 mAb after knockout Usp5. **P < 0.01. c A schematic model illustrating the treatment plan for mice bearing subcutaneous Hepa1-6 or H22 tumors. Male C57BL/6 mice were implanted with Hepa1-6 or H22 cells subcutaneously and treated with four arms: control antibody (anti-IgG mAb) treatment, anti-PD-L1 mAb treatment, USP5 inhibitor WP1130 treatment, and anti-PD-L1 mAb plus USP5 inhibitor combination treatment. d Hepa1-6 and H22 implanted tumor-bearing mice were enrolled in different treatment groups as indicated. Tumor volumes of mice treated with control antibody, anti-PD-L1 mAb, the USP5 inhibitor WP1130, or combined therapy were measured every 3 days and plotted individually. n = 10 mice per group. e Kaplan-Meier survival curves for each treatment group demonstrate the improved efficacy of combining PD-L1 mAb with the USP5 inhibitor WP1130. *** P < 0.001. f Immunohistochemical (IHC) analysis of Cd8, Granzyme B (GzmB), Tim-3, Ythdf1 and Pd-l1 expression in Hepa1-6 tumors after indicated treatments. g Representative multiplex immunohistochemistry (mIHC) images of Cd8 (Gray), GzmB (Green), Tim-3 (Red), and DAPI nuclear staining (blue) in Hepa1-6 tumors after indicated treatments. All data are presented as mean ± SEM. Source data are provided as a Source Data file.
USP5 expression defines the clinical outcome in anti-PD-1/PD-L1 immunotherapy as a biomarker
To determine the translational significance of these findings, we investigated the relationship between USP5 expression and immunotherapy response in cancer patients. Bioinformatic analysis showed that USP5 mRNA was elevated in various tumors compared with normal tissues (Supplementary Fig. 14b). Higher USP5 levels correlate with poorer overall survival rates and potentially weaker response for PD-1 antibody therapy (Supplementary Fig. 14c, d). To directly evaluate the relationship between USP5 expression and immunotherapy responses in cancer patients, we collected data from a cohort of HCC (n = 14) and lung cancer patients (n = 15) enrolled in a basket trial of anti-PD-1 treatment (Fig. 7a, b). We separated cancer patients into two groups based on their tumoral USP5 levels. We found markedly lower levels of tumoral USP5 in cancer patients with partial response (PR) or stable disease (SD) compared to that in cancer patients with progressive disease (PD) (Fig. 7c, d). The patients with low levels of USP5 had a higher overall response rate (ORR) compared to patients with high USP5 levels (ORR: 66.67% vs 14.29% with HCC or 83.33% vs 33.33% with LUAD) (Fig. 7e, f). These data from immunotherapy-treated cancer patients support the clinical relevance of our experimental findings from mouse tumor models with USP5 deficiency (Fig. 6). Of note, we employed HCC patient-derived xenograft (PDX) models to investigate whether the ablation of USP5 enhances tumor sensitivity to anti-PD-1 inhibitors (Fig. 7g). Consistent with our earlier observations, PDX tumors originated from HCC191017 exhibited markedly higher sensitivity to anti-PD-1 treatment compared to those from HCC180328 (unpublished data). Additionally, HCC191017 tumors treated with PD-1 antibodies showed a decrease in PD-1 expression and an increase in USP5 and YTHDF1 expression (Fig. 7h). In contrast, there was no significant difference observed in the HCC180328 group (Fig. 7h). Taken together, these findings underscore the potential of targeting USP5 in tumor cells as a valuable strategy for anti-PD-(L)1 immunotherapy.
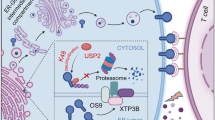
Representative USP5 protein staining of tumor sections (top: 100×, bottom: 400×) (left) and CT scans (right) from liver cancer patients (a) or lung cancer patients (b) undergoing anti-PD-1 treatment. CT scans of patient tumors are shown. c, d Waterfall plot depicting the responses to anti-PD-1 treatment based on the best change in the sum of target lesions compared to baseline in cancer patients with low USP5 or high USP5 expression. Each bar represents one patient, and the colors correspond to the response to anti-PD-1 treatment (PR: partial response, SD: stable disease, PD: progressive disease). Dotted black lines indicate the response as described by RECIST1.1. e, f Pie charts illustrating the response fractions for each group of patients with USP5-low and USP5-high expression in tumor cells. g IB analysis of indicated proteins derived from two fresh HCC samples (HCC180328 and HCC191017) before the establishment of the humanized mouse model. h IB analysis of indicated proteins in a subcutaneous humanized mouse model after 21 days of treatment with anti-IgG or anti-PD-1. i A working model for targeting USP5 to sensitize tumors to PD-1/PD-L1 blockade. USP5 downregulates PD-L1 and enhances the immunosuppressive response, leading to resistance to PD-1/PD-L1 blockade (Left Panel). Inhibition of USP5 by WP1130 upregulates PD-L1 and reduces the immunosuppressive response to sensitize the tumor to anti-PD-1/PD-L1 immunotherapy (Right Panel). All IB data are representative of two independent experiments. Source data are provided as a Source Data file.
Discussion
Ubiquitination/de-ubiquitination is a reversible process that is controlled by DUBs and ubiquitin E3 ligases. Our study illuminates the dynamic regulatory roles of K11-linked polyubiquitination on multiple lysine residues of YTHDF1, orchestrated by USP5 and CUL7-FBXW8. Additionally, we unveil an unexpected regulatory axis from mTORC1 to YTHDF1 through USP5 phosphorylation and dimerization. The USP5-YTHDF1 axis serves as a tumor-intrinsic checkpoint, limiting anti-tumor immunity and positioning USP5 as a promising target for immunotherapy. Notably, the remarkable ability of USP5 inhibition to transform a PD-(L)1 blockade-resistant tumor into a PD-(L)1 blockade-responsive tumor suggests its potential to enhance the efficacy of anti-PD-(L)1 cancer therapy, thus converting ‘cold’ tumors into ‘hot’ ones (Fig. 7i).
YTHDF1, a crucial m6A reader protein governing translation, is implicated in tumor progression and predicts poor prognosis in cancer patients11,13,17,18. Our findings highlight the post-translational regulation of YTHDF1 abundance. Specifically, USP5 interacts with YTHDF1, and enhances YTHDF1 stability through deubiquitylation. This is independent of YTHDF1 gene transcription. Within the YTHDF1 protein complex, CUL7-FBXW8 emerges as another YTHDF1-associated protein responsible for polyubiquitylating YTHDF1, leading to its proteasomal degradation. USP5 binds to the same region of YTHDF1 as FBXW8, inhibiting the YTHDF1-FBXW8 interaction and preventing FBXW8-mediated YTHDF1 polyubiquitylation and degradation. Meanwhile, we do not exclude the possibility that USP5-FBXW8-YTHDF1 form a transient ternary complex or other mechanisms that affect their interactions. The post-translational modification including ubiquitination on the YTHDF1 or FBXW8 might alter their conformation to affect their interactions. Additionally, insulin stimulates mTORC1 to phosphorylate USP5 at S149, decreasing FBXW8 expression. Consequently, USP5 is recruited to the YTHDF1 complex, counteracting its degradation.
While abnormal USP5 expression was observed in various carcinomas, its role in anti-tumor immunity and immunotherapy remained unclear. Our study revealed that YTHDF1 or USP5 deficiency triggers potent anti-tumor immunity, marked by increased T cell infiltration and immunogenicity. Furthermore, targeting USP5 or YTHDF1 in tumor cells induces cellular responses, regulating the expression of pro-inflammatory and immune response-related genes. This promotes anti-tumor T cell immunity and fosters immune inflammation in the local tumor microenvironment, rendering the tumor more sensitive to ICB therapy in an HCC mouse model. Despite the elevated levels of PD-L1, a key inhibitor of T cell responses, in YTHDF1 or USP5 KO tumors, the combination of USP5 abrogation and PD-L1 blockade demonstrates a striking effect on controlling tumor growth. Our findings suggest that USP5 inhibition overcomes hepatoma resistance to PD-(L)1 blockade by increasing tumor immunogenicity and T cell infiltration47,52,53,54. Consistent with mouse models, the analysis of cancer patient data reveals an inverse correlation between USP5 expression and CD8+ T cell infiltration and prognosis, highlighting the therapeutic potential of targeting USP5 in combination with PD-(L)1 blockade for cancer treatment. The association of high USP5 expression with poor prognosis across various cancers suggests that USP5 expression may also serve as an informative biomarker.
While our study is valuable in shedding light on the role of YTHDF1 in enhancing the efficacy of ICB through the targeting of tumoral USP5 and promoting tumor immunogenicity, we acknowledge certain limitations. The modest number of cancer patients enrolled in our study restricts the validation of USP5’s predictive role in immunotherapy efficacy. Despite these limitations, our research significantly contributes to the understanding of how YTHDF1 amplifies ICB efficacy by targeting tumor-associated USP5, thereby enhancing tumor immunogenicity. Moreover, we uncover that USP5-YTHDF1 axis governs PD-1 homeostasis in T cells, modulating tumor immunotherapy. This proof-of-concept study not only offers molecular insights into the mechanisms by which the USP5-YTHDF1 axis triggers anti-tumor immunity but also proposes the targeting of tumor cell-intrinsic USP5 for more effective immunotherapy. However, detailed mechanisms behind the cell-intrinsic effects of the USP5-YTHDF1 axis in cancer patients remain to be defined.
In conclusion, our study lays the foundation of developing a combinatorial approach involving USP5 inhibitors and anti-PD-(L)1 for cancer therapy. Considering the overarching role of the USP5/YTHDF1 axis in regulating various immune responses and PD-L1 expression, the targeting of USP5 in conjunction with anti-PD-(L)1 holds promise as a broadly applicable and innovative strategy in cancer immunotherapy.
Methods
Cell culture, transfection, lentiviral and drug treatments
HEK293T, PLC/PRF/5, Hep3B, H22, Hepa1-6, HeLa, and NIH/3T3 were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM); H1299 cells were cultured in Roswell Park Memorial Institute (RPMI-1640) medium (Gibco); A549 cells were cultured in F-12 medium (Gibco) containing 10% fetal bovine serum (FBS), 100 units of penicillin and 100 mg ml−1 streptomycin in a sterile 37 °C incubator with a humidified 5% CO2 atmosphere. Cells were authenticated by short tandem repeat profiling. Immediately upon receipt, cells were expanded and then frozen to be revived every 3–4 months. All cell lines were validated to be free of mycoplasma contamination. Transfection was performed using Neofect DNA transfection reagent (Neofect) or effectene transfection reagent (QIAGEN) according to the manufacturer’s protocol. For lentiviral infection, lentiviruses were produced by cotransfection of HEK293T cells with recombinant lentivirus vectors and lentivirus packaging plasmids pMD2.G, pRSV-Rev, pMDLGpRRE using a calcium phosphate precipitation-based method, then the virus particles were collected for infection. Cells were selected with Puromycin (1 μg ml−1).
For serum starvation, 32 h post transfection, cells were washed with PBS twice and cultured in FBS-free DMEM for 14–16 h. To initiate growth factor signaling, the medium was added with EGF (Proteintech AF-100-15), FGF (Peprotech 100-18B), IL-6 (Cell Signaling Technology 8904), HGF (Peprotech 100-39) or insulin (Invitrogen 41400-045) for indicated period of times.
For drug treatments, MG132 (S2619), WP1130 (S2243), AKTi-1/2 (S7776), Rapamycin (S1039), Wortmannin (S2758), and Torin1 (S2827) were purchased from Selleck Chemicals. CHX (N11534) and chloroquine (C6628) were purchased from Sigma-Aldrich. Medium containing a final concentration were then used to replace the original medium and cells were cultured in the presence of drugs for the indicated times were described in detail in the text.
Generation of YTHDF1 and USP5 knock-out cell lines using CRISPR/Cas9 system
sgYTHDF1-human (Santa Cruz Biotechnology sc-412379) and sgYthdf1-mouse (Santa Cruz Biotechnology sc-432834) cell lines (PLC/PRF/5 and Hepa1-6) were generated using a YTHDF1-specific CRISPR/Cas9/EGFP plasmids. sgUsp5-mouse Hepa1-6 and H22 cell lines were generated using a pLentiCRISPR-Usp5-sgRNA purchased from GeneChem (Shanghai, China). The selection of single cell clones was performed by serial dilution in 96-well plates, followed by immunoblot analysis of YTHDF1 or USP5 to confirm knockout efficiency of multiple selected clones.
Antibodies
The following antibodies were used in immunoblotting. Anti-Vinculin (1:2000 dilution, Cat# V9131, RRID:AB_477629), anti-Flag-Tag (1:2000 dilution, Cat# F1804, RRID:AB_262044) were purchased from Sigma-Aldrich. Anti-HA-Tag (1:1000 dilution, Cat# 3724, RRID:AB_1549585), anti-GFP-Tag (1:1000 dilution, Cat# 2956, RRID:AB_1196615), anti-T7-Tag (1:1000 dilution, Cat# 13246, RRID:AB_2798161), anti-Myc-Tag (1:1000 dilution, Cat# 2278, RRID:AB_490778), anti-His-Tag (1:1000 dilution, Cat# 12698, RRID:AB_2744546), anti-ubiquitin (1:1000 dilution, Cat# 3936, RRID:AB_331292), anti-PD-L1 (1:1000 dilution, Cat# 13684, RRID:AB_2687655), anti-YTHDF1 (1:1000 dilution, Cat# 86463, RRID:AB_2923055), anti-YTHDF2 (1:1000 dilution, Cat# 80014 RRID:AB_2923056), anti-mTOR (1:1000 dilution, Cat# 2983, RRID:AB_2105622), anti-Phospho-mTOR (Ser2448) (1:1000 dilution, Cat# 5536, RRID:AB_10691552), anti-4E-BP1 (1:1000 dilution, Cat# 9644, RRID:AB_2097841), anti-Phospho-4E-BP1 (Thr37/46) (1:1000 dilution, Cat# 2855, RRID:AB_560835), anti-p70 S6 (1:1000 dilution, Cat# 9202, RRID:AB_331676), anti-Phospho-p70 S6 (Thr389) (1:1000 dilution, Cat# 97596, RRID:AB_2800283), anti-Acetyl-CoA Carboxylase 1 (1:1000 dilution, Cat# 4190, RRID:AB_10547752), anti-Phospho-Acetyl-CoA Carboxylase (Ser79) (1:1000 dilution, Cat# 11818, RRID:AB_2687505), anti-AKT (1:1000 dilution, Cat# 4691, RRID:AB_915783), anti-Phospho-Akt (Ser473) (1:1000 dilution, Cat# 4060, RRID:AB_2315049), anti-SIN (1:1000 dilution, Cat# 12860, RRID:AB_2798048), anti-GβL (1:1000 dilution, Cat# 3274, RRID:AB_823685), anti-Raptor (1:1000 dilution, Cat# 2280, RRID:AB_561245), anti-RICTOR (1:1000 dilution, Cat# 2114, RRID:AB_2179963), anti-DEPTOR (1:1000 dilution, Cat# 11816, RRID:AB_2750575), anti-DYKDDDDK (1:1000 dilution, Cat# 14793, RRID:AB_2572291), anti-USP1 (1:1000 dilution, Cat# 8033, RRID:AB_10858879), anti-USP15 (1:1000 dilution, Cat# 66310, RRID:AB_2799706), Rabbit (DA1E) mAb IgG XP® Isotype Control (Cat# 3900, RRID:AB_1550038), Mouse (G3A1) mAb IgG1 Isotype Control (Cat# 5415, RRID:AB_10829607), anti-Lamin B1 (1:1000 dilution, Cat# 13435, RRID:AB_2737428), anti-GAPDH (1:1000 dilution, Cat# 2118, RRID:AB_561053) were purchased from Cell Signaling Technology. Anti-Ubiquitin K11-linkage (1:1000 dilution, Cat# MABS107-1, RRID:AB_120851823), anti-puromycin (1:1000 dilution, Cat# MABE343, RRID:AB_2566826) were purchased from Millipore. Anti-α-tubulin (1:1000 dilution, Cat# sc-23948, RRID:AB_628410), anti-USP5 (1:1000 dilution, Cat# sc-390943, RRID:AB_2934285), anti-LAMP2 (1:1000 dilution, Cat# sc-18822, RRID:AB_626858), anti-ubiquitin (1:1000 dilution, Cat# sc-8017, RRID:AB_628423) were purchased from Santa Cruz Biotechnology. Anti-FBXW8 (1:1000 dilution, Cat# PA5-116803, RRID:AB_2901434), anti-Cullin 7 (1:1000 dilution, Cat# PA5-22313, RRID:AB_11154447) were purchased from Invitrogen. Anti-USP3 (1:1000 dilution, Cat# ab229348), anti-USP5 (1:1000 dilution, Cat# ab244290) were purchased from Abcam. Anti-Phosphoserine/threonine (1:1000 dilution, Cat# 612548, RRID:AB_399843) and anti-YTHDF1 (1:1000 dilution, Cat# NBP2-7-6361) were purchased from BD Biosciences and NOVUS. Anti-USP28 (1:1000 dilution, Cat# 4217, RRID:AB_1581796), anti-YTHDF3 (1:1000 dilution, Cat# 25537-1-AP, RRID:AB_2847817), anti-YTHDC1 (1:1000 dilution, Cat# 14392-1-AP, RRID:AB_2878052), and anti-YTHDC2 (1:1000 dilution, Cat# 27779-1-AP, RRID:AB_AB_2880970) were purchased from Proteintech. Anti-Pd-l1 (1:2000 dilution, Cat# HA722184) and anti-PD-L1 (1:2000 dilution, Cat# HA721176) were purchased from HUABIO. Below-described secondary antibodies were diluted at 1:5,000 in 5% non-fat milk for immunoblotting analysis. Secondary antibodies HRP Goat anti-Mouse IgG (H + L) Secondary Antibody (1:5000 dilution, Cat# 31430, RRID:AB_228307) and HRP Goat anti-rabbit IgG (H + L) Secondary Antibody (1:5000 dilution, Cat# 32460, RRID:AB_1185567) were purchased from Thermo Fisher Scientific.
Plasmids and shRNAs
Expression vectors Myc-Cullin constructs (Cullin 1, 2, 3, 4A, 4B, 5, and 7), GFP-Ubiquitin (Ub), HA-Ub (WT), HA-Ub (K6R), HA-Ub (K11R), HA-Ub (K27R), HA-Ub (K29R), HA-Ub (K33R), HA-Ub (K48R), HA-Ub (K63R), 6×His-Ub (WT), 6×His-Ub (K6) only, 6×His-Ub (K11) only, 6×His-Ub (K27) only, 6×His-Ub (K33) only, 6×His-Ub (K48) only, and 6×His-Ub (K63) only constructs were described previously55. Flag-HA-tagged USP family members of DUBs, Flag-USP5, Flag-TSC1, Flag-TSC2, Flag-mTOR, HA-RPTOR, HA-RICTOR, HA-SIN, HA-GβL, HA-S6K2, HA-S6K2 K99R, HA-Ub K11R and HA-Ub K11 only were purchased from Addgene. Flag-YTHDF1, 6×His-YTHDF1, Flag-Ythdf1, HA-USP5, HA-Usp5, HA-S6K1, 3×HA-TMEM192, and T7-FBXW8 cDNA were amplified and cloned into the pcDNA3.1 vector. Flag-YTHDF1 (aa 1-190), Flag-YTHDF1 (aa 191-388), Flag-YTHDF1 (aa 389-529), 6×His-YTHDF1 (aa 1-388), 6×His-YTHDF1 (aa 389-529), 6×His-YTHDF1 (Δ aa 359-523), HA-USP5 C-terminal domain, HA-USP5 N-terminal domain, HA-USP5 Δzf-UBP, HA-USP5 ΔUBA, HA-USP5 Δzf-UBP + ΔUBA, T7-FBXW8 C-terminal domain, T7-FBXW8 N-terminal domain, T7-FBXW8 ΔF-box, T7-FBXW8 ΔWD1, T7-FBXW8 ΔWD2, T7-FBXW8 ΔWD3, T7-FBXW8 ΔWD4, T7-FBXW8 ΔWD5, and Flag-PD-L1 were generated by sub-cloning the corresponding cDNAs into the pcDNA3.1 or pCMV vector. Flag-YTHDF1 constructs (K12R, K191R, K240R, K266R, K370R, K372R, K380R, K387R, K469R, K473R, K500R, K527R, K469/473R, K469/500R, K469/527R, K473/500R, K473/527R, K500/527R, K469/473/500R, K469/473/527R, K473/500/527R, 4KR, 12KR), Flag-Ythdf1 4KR, HA-USP5 constructs (Y44A, S149A, S149D, T250A, S564A, Y568A, T577A, S783A), T7-FBXW8 S86A, HA-S6K1 S39A, and HA-S6K1 T421E/S434E/S441E/T444E/S447E mutants were generated using the Quick Change Q5 Site-Directed Mutagenesis Kit (NEBaseChanger) according to the manufacturer’s instruction. The pLKO.1-puro lentiviral MISSION shRNA constructs targeting endogenous gene expression. pLKO-shUSP5 (TRCN0000004069, TRCN0000004068), pLKO-shUsp5 (TRCN0000030738), pLKO-shYTHDF1 (TRCN0000062771), pLKO-shYthdf1 (TRCN0000348503), pLKO-shmTOR (TRCN0000039785), pLKO-shRPTOR (TRCN0000039772), pLKO-shTSC1 (TRCN0000010453), pLKO-shTSC2 (TRCN0000013290), pLKO-shFBXW8 (TRCN0000004323), pLKO-shCUL7 (TRCNTRCN0000006480, TRCN0000006483), and a non-targeting (NT) control shRNA (TRC1/1.5) were from Sigma-Aldrich. pLenti-HA-YTHDF1 WT, pLenti-HA-YTHDF1 4KR, pLenti-HA-Ythdf1, pLenti-HA-USP5 and pLenti-HA-Usp5 were generated in this study. All the constructs were confirmed by DNA sequencing. Details of plasmid construction are available upon request.
Immunoblotting (IB) and immunoprecipitation (IP) assays
Cells were lysed in IP lysis buffer (Thermo Fisher Scientific 87788) or RIPA lysis buffer (Thermo Fisher Scientific 89901) supplemented with protease inhibitors (Complete Mini, Roche A32965) and phosphatase inhibitors (Roche A32957). Protein concentrations were measured by the Bio-Rad protein assay reagent. Equal amounts of protein were resolved by SDS-PAGE and immunoblotted with indicated antibodies. For IP assay, total cell lysates were incubated with primary antibodies (2–3 μg) overnight at 4 °C. Protein A/G Sepharose beads (Thermo Fisher Scientific 78610) were then added and the incubation was continued for 3–4 h before 4 × wash with IP lysis buffer before being resolved by SDS-PAGE and immunoblotted with indicated antibodies. Immunoreactive bands were visualized by enhanced chemiluminescence (Thermo Fisher Scientific 34096).
GST pull-down assay
Human YTHDF1 was subcloned into the pGEX-4T-1 construct (Addgene 70087). Recombinant glutathione S-transferase (GST) tagged YTHDF1 was purified from Escherichia coli BL21. GST pull-down assays were performed by incubating 3 μg GST-YTHDF1 immobilized on the glutathione-Sepharose resin (Thermo Fisher Scientific 21516) with HA-USP5 or Flag-mTOR at 4 °C for 3 h. GST pull-down products were washed five times with wash buffer and eluted by 2 × SDS-PAGE loading buffer. Protein samples were resolved by SDS-PAGE and subjected to IB analysis.
Protein half-life assays
To measure the half-life of YTHDF1 protein in cells, a CHX based assay was performed following our previously described experimental procedure55,56. Cells were transfected or treated under indicated conditions. For half-life studies, CHX (100 μg ml−1) was added to the medium. At indicated time points thereafter, cells were harvested and protein abundances were determined by IB analysis.
Ubiquitination assays in cells
Cellular ubiquitination assays were performed as described previously55. In brief, cells were transfected with plasmids expressing indicated proteins then cultured for 36-48 h. Cells were further treated with 20 μM MG132 for 6 or 8 h before lysed with IP lysis buffer. IP was performed using antibodies against the tagged proteins. Immunoprecipitants were washed five times with IP lysis buffer before being resolved by SDS-PAGE and immunoblotted with antibodies against the tags on ubiquitin.
In vitro deubiquitination assay
HEK293T cells were transfected together with the 6×His-YTHDF1 and GFP-Ub expression plasmids for 48 h. The cells were treated with MG132 for 8 h and lysed. 6×His-YTHDF1 protein was purified by nickel-nitrilotriacetic acid (Ni-NTA) matrices (QIAGEN 31314). HEK293T cells were transfected with HA-USP5 WT or C335A plasmids, and HA-USP5 WT or C335A proteins were purified by HA-tag Protein IP Assay Kit with Magnetic Beads (Beyotime P2185M). Recombinant USP5 proteins were subsequently incubated with Ub-YTHDF1 in deubiquitination reaction buffer (50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol) at 37 °C for 30 min. The ubiquitination status of YTHDF1 was evaluated by IB analysis.
Protein extraction and digestion
For protein extraction, cell samples (sgCtrl, sgUsp5, sgYthdf1; n = 1) were sonicated three times on ice using a high intensity ultrasonic processor (Scientz) in lysis buffer (8 M urea, 1% protease inhibitor, 1% phosphatase inhibitor). The insoluble debris was removed by centrifugation at 12,000 × g at 4 °C for 10 min and the supernatant was retained for proteomic experiment. The protein concentration was determined with BCA kit according to the manufacturer’s instructions.
For protein digestion, the protein solution was reduced with 5 mM dithiothreitol for 30 min at 56 °C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in the darkness. The protein sample was then diluted by adding 100 mM triethyl ammonium bicarbonate (TEAB) to urea concentration less than 2 M. All centrifugations were performed at 12,000 × g at 25 °C. Proteins were digested with trypsin for 18 h at a radio of 1:50 (w/w), followed by another 4 h with digestion at a radio of 1:10 (w/w) in 50 mM TEAB. After digestion, peptides were desalted and subjected to further analysis.
LC-MS/MS proteomic analysis
For proteomic analysis, the fractionated peptides were resolved using buffer A (0.1% formic acid and 2% acetonitrile in water) and a quarter of each fraction was separated using a home-made reversed-phase analytical column (75 μM × 250 mm). The gradient was set as follows: 6–24% in buffer B (0.1% formic acid in acetonitrile) over 70 min; 24–35% buffer B in 14 min; 80% buffer B in 3 min with twice. All at a constant flow rate of 450 nL/min on a nanoElute UHPLC system (Bruker Daltonics). Proteomic analyses were performed by the timsTOF Pro mass spectrometer (Bruker Daltonics). The electrospray voltage applied was 1.60 kV. Precursors and fragments were analyzed at the TOF detector, with a MS/MS scan range from 100 to 1700 m/z. The timsTOF Pro was operated in parallel accumulation serial fragmentation (PASEF) mode. Precursors with charge states 0 to 5 were selected for fragmentation, and 10 PASEF-MS/MS scans were acquired per cycle. The dynamic exclusion was set to 30 s. The resulting MS/MS data were processed using Proteome Discoverer (v2.4.1.15) and Maxquant (v1.6.15.0) search engine. Tandem mass spectra were searched against the SwissProt database (musculus_10090_SP_20210721.fasta 17089 entries) concatenated with reverse decoy database. Trypsin/P was specified as cleavage enzyme allowing up to 4 missing cleavages. FDR was adjusted to <1%. The relative quantitative values of peptides in different samples are obtained by centralizing the signal intensity values in different samples.
Immunoprecipitation coupled with mass spectrometry (MS) analysis to identify YTHDF1 interacting protein
HEK293T cells transfected with pCMV3-3 × Flag-YTHDF1 or pCMV3-empty vector (EV) were lysed with IP lysis buffer supplemented with protease inhibitors (Thermo Fisher Scientific A32965) and phosphatase inhibitors (Roche A32957). The supernatants of cell lysates were incubated with anti-Flag-beads (Sigma-Aldrich M8823) in a rotating incubator for 6 h at 4 °C. The beads were washed five times with IP lysis buffer. The samples (n = 1) prepared from IP were sent to the PTM Biolabs Inc (Hangzhou, China) for the mass spectrometry analysis and data processing.
Real-time RT-PCR analysis
Total RNA was extracted using Trizol reagent (TakaRa 9109). cDNA was synthesized using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas K1622), and qRT-PCR was performed with SYBR Select Master Mix (TakaRa RR420A). Averages and standard deviations calculated from at least 3 independent experiments were shown in the figures. The housekeeping gene GAPDH was used as the reference gene in all RT-PCR analyses. The primer information is shown: hYTHDF1 Forward: 5′-ACCTGTCCAGCTATTACCCG-3′, Reverse: 5′-TGGTGAGGTATGGAATCGGAG-3′, hGAPDH Forward: 5′-TGGTATCGTGGAAGGACTC-3′, Reverse: 5′-AGTAGAGGCAGGGATGATG-3′. mGAPDH Forward: 5′-AATGGATTTGGACGCATTGGT-3′, Reverse: 5′-TTTGCACTGGTACGTGTTGAT-3′, mCCL2 Forward: 5′-TAAAAACCTGGATCGGAACCAAA-3′, Reverse: 5′-GCATTAGCTTCAGATTTACGGGT-3′, mCCL9 Forward: 5′-CCCTCTCCTTCCTCATTCTTACA-3′,Reverse: 5′-AGTCTTGAAAGCCCATGTGAAA-3′
mCCL20 Forward: 5′-ACTGTTGCCTCTCGTACATACA-3′, Reverse: 5′-GAGGAGGTTCACAGCCCTTTT-3′, mGBP4 Forward: 5′-AGACTTTCCGTGAGGAAGGAT-3′, Reverse: 5′-TTCCACAAGGGAATCACCATTTT-3′, mGBP8 Forward: 5′-CACACCCCACTAAACCAGAGC-3′, Reverse: 5′-TTCCATCAGGATTTGGTGAAGAC-3′, mEND1 Forward: 5′-ATGGAATCCCGAGGAAAGTCA-3′, Reverse: GCCTGAGGCACCTTGGTATC-3′, mGBP3 Forward: 5′-CTGACAGTAAATCTGGAAGCCAT-3′, Reverse:5′-CCGTCCTGCAAGACGATTCA-3′, mTGTP2 Forward: 5′-TGGGACCACTAACTTCACACC-3′, Reverse: 5′-GGCCAGTTGTGCATCATTTTC-3′, mTLR2 Forward: 5′-CTCTTCAGCAAACGCTGTTCT-3′, Reverse: 5′-GGCGTCTCCCTCTATTGTATTG-3′, mCXCL16 Forward; 5′-ACCCTTGTCTCTTGCGTTCTT-3′, Reverse: 5′-CAAAGTACCCTGCGGTATCTG-3′, mRAB20 Forward: 5′-AAGCCGGATGGGAAGATCGTA-3′,Reverse: 5′-GGAAGCGACGCTCCATGTA-3′, mSTX11 Forward: 5′-TGTCCAGGAGCTATGACCAG-3′, Reverse: 5′-TCGGTCTCGAACACTATGTCC-3′ mSPN Forward: 5′-ACGCCCCATCAGGGTAACA-3′,Reverse: 5′-CTTCTCCTCCGAGGCATAGC-3′, mCXCL5 Forward: 5′-GTTCCATCTCGCCATTCATGC-3′, Reverse: 5′-GCGGCTATGACTGAGGAAGG-3′, mTRAF3 Forward: 5′-CAGCCTAACCCACCCCTAAAG-3′,Reverse: 5′-CGGCACTTCTCGCACTTGT-3′, mRAET1D Forward: 5′-CTGAGCTATGGATACACCAACG-3′, Reverse: 5′-AGCACTTCACTTCATCTGCTG-3′, mGPRC5B Forward: 5′-TTGTGATGGCGCTCATCTACG-3′, Reverse: 5′-ACCTTCCACCGTTTGAACTTG-3′.
RNA extraction and sequencing analysis
Hepa1-6 cells infected with sgUsp5, sgYthdf1, or sgControl (sgCtrl) were collected for RNA extraction using the Total RNA Extractor (Trizol) kit (B511311, Sangon Biotech, China). To eliminate genomic DNA contamination, the total RNA samples underwent treatment with RNase-free DNase I. High-quality RNA samples were then sent to Sangon Biotech (Shanghai) Co., Ltd. for library preparation and sequencing. For RNA-seq library preparation, 1 μg of RNA was used as input material, and VAHTSTM mRNA-seq V2 Library Prep Kit for Illumina was employed. Subsequently, paired-end sequencing was conducted on the NovaSeq sequencers (Illumina, San Diego, CA). The sequencing reads were aligned to the mouse genome (gencode vM26; GRCm39) using STAR (v2.7.10a)57. Gene read counts were extracted using salmon (v1.9.0)58, and differential gene expression analysis was performed using DESeq2 (v1.32.0)59. Further analysis included gene set enrichment tests and the construction of plots using Webgestalt 201960 based on significantly differentially expressed genes (Padjusted <0.05).
RNA immunoprecipitation assay
Hepa1-6 cells infected with sgUsp5 or sgCtrl were seeded in a 10 cm dish at 70–80% confluence. A total of 5 μg of YTHDF1 (Proteintech 17479-1-AP) antibody and a corresponding control rabbit IgG were conjugated to protein G magnetic beads (InvitrogenTM 10004D) by incubation for 4 h at 4 °C, followed by washing three times and incubation with lysate of Hepa1-6 cells in RNA immunoprecipitation (RIP) buffer (Epibiotek R18019) at 4 °C overnight. After washing with RIP buffer three times, beads were resuspended in 80 μL PBS, followed by DNA digestion at 37 °C for 15 min and incubation with 50 μg of proteinase K at 37 °C for 15 min. RIP RNA samples were collected and subjected to strand-specific library preparation. The libraries were then sequenced using the Illumina high-throughput sequencing platform (NovaSeq 6000) at PE150 read length. The sequencing data was processed using Bioptic Qsep100. DEGseq was used to analyze the differences.
In vivo RNA methylation assays
For quantification of m6A levels, 200 ng total RNA extracted from Hepa1-6 cells with Ythdf1 or Usp5 knockout were used to measure the cellular m6A levels using the EpiQuik™ m6A RNA methylation quantification kit (colorimetric) (P-9005, Epigentek, Farmingdale, NY, USA) according to the manufacturer’s instructions.
Surface sensing of translation (SUnSET) assay
SUnSET assay was conducted as previously described61. In brief, cells were incubated with puromycin (10 μg ml-1) for 30 min and then washed with ice-cold PBS and lysed using RIPA lysis buffer. Equal amount of cell lysates was submitted to immunoblot using anti-puromycin antibody to detect protein synthesis.
Flow cytometry
Cells were washed once in PBS and stained using the PE conjugated Pd-l1 or APC conjugated H2Kd/H2Dd for 30 min at 4 °C. After staining, samples were fixed for 30 min at 4 °C using the eBioscience™ Fixation/Permeabilization kit. After washing once in PBS, cells were analyzed and data were acquired on Beckman CYTOFLEX and Beckman CytExpert Software 2.3. Results were analyzed by the software FlowJo. All gates were set based on isotype-matched control antibody. Gating strategies used for flow cytometry staining was provided in Supplementary Fig. 15.
Subcellular fractionation assays
Nuclei and cytosol proteins were isolated using the nuclear and cytoplasmic extraction kit (Thermo Fisher Scientific 78833) according to the manufacturer’s instructions. Nuclear proteins (60 μg) and cytosolic proteins (14 μg) were used in immunoblotting analyses.
Immunofluorescence (IF) analysis
Immunofluorescence analyses were performed as described previously55. Briefly, cells were fixed and incubated with primary antibodies, Alexa Fluor dye-conjugated secondary antibodies, and 4’, 6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, D9542) according to standard protocols. Images were captured using Leica confocal microscope.
Colony formation assays
The indicated tumor cells (PLC/PRF/5 and Hepa1-6) were plated in 24-well plates (500 or 1000 cells per well) and maintained for up to 10–16 days. Visible colonies were washed with PBS and fixed with 10% acetic acid/10% methanol for 30 min, and then stained with 0.4% crystal violet/20% ethanol. After washing with distilled water and air-dried, the colonies were quantified and analyzed.
Ex vitro T cell coculture assay
CD8+ T cells were isolated from splenic single-cell suspension using the Mouse CD8 (Ly2) MicroBeads isolation kit (130-117-044 Miltenyi Biotec). Cells were then activated with mouse anti-CD3/CD28 antibodies (11453D Gbico) for 2 days supplemented with human IL-2 (50 U/ml) (CK24 Novoprotein). Activated T cells were plated at 2 × 106 cells per well in a 24-well plate or 2 × 104 cells per well in a 96-well plate. Tumor cells were then added at a ratio of 10:1. Activated T cells and tumor cells were co-cultured with or without anti-PD-1 antibody (or WP1130) in 24-well plates for 2 or 4 days and then surviving tumor cells were visualized by crystal violet staining. Activated T cells and tumor cells were co-cultured in 96-well plates for 24 h and tumor death rate was determined by the Live-Death Cell Imaging Kit (R37601 Invitrogen™), following the manufacturer’s instructions. Activated T cells and tumor cells were co-cultured in 96-well plates for 24 h and Caspase3/7 activation was determined using the Caspase-Glo3/7 Assay (G8092 Promega), following the manufacturer’s instructions.
Mouse xenograft assays
The study is compliant with all relevant ethical regulations regarding animal research. All procedures performed in studies involving mouse experiments were in accordance with the ethical standards of the Second Affiliated Hospital of Chongqing Medical University. Ethics Committee (Chongqing, China) and with the 1964 Helsinki declaration (Approval IACUC-SAHCQMU-2024-00010). The animals were randomly allocated to different experimental groups. Hepa1-6 (5 × 106), H22 (2.5 × 106) or PLC/PRF/5 (1 × 106) cells were suspended in 100 μl of DMEM medium and mixed with Matrigel (Corning, 354234, 1:1) and injected subcutaneously into both flanks of 6–8 week old male C57BL/6 mice or 4–6 week old male NOD/SCID mice as described previously62. Tumor size was measured every 3 days after implantation with a caliper, and the tumor volume was determined with the formula: L × W2 × (π/6), where L is the longest diameter and W is the shortest diameter. At the experimental endpoints, mice were sacrificed until the tumor burden less than 20 mm and in vivo solid tumors were dissected and tumor weights were measured and recorded.
In vivo experimental therapy in mouse model
Subcutaneous xenografts were established by injecting with the H22 and Hepa1-6 mouse HCC cell lines into 6–8 week-old male C57BL/6 mice. For experiments with CD8+ T cell, CD4+ T cell, or NK cell depletion, mice were intraperitoneally injected with 200 μg of anti-CD8 (clone 2.43, Bio X Cell), anti-CD4 (clone GK1.5, Bio X Cell), or anti-NK1.1 (clone PK136, Bio X Cell), respectively, on days 0 and 7 after tumor inoculation. For the H22 and Hepa1-6 C57BL/6 mouse models, the mice were randomly allocated into four groups: control antibody (RTK4530, Biolegend), PD-L1 mAb (clone 10 F.9G2, Bio X Cell), USP5 inhibitor WP1130, and WP1130 plus PD-L1 mAb. For Usp5 knockout Hepa1-6 C57BL/6 mouse model, mice were randomly allocated two groups (control antibody and PD-L1 mAb). The control and PD-L1 mAb treatments were conducted by intraperitoneal injection (200 μg/mouse in 88 μl HBSS saline buffer) and WP1130 treatment was given by intraperitoneal injection with a dosage of 15 mg/kg (in a 100 μl suspension of DMSO/PEG300 (1:1)) every 3 days for a total of 7 injections. For survival studies, animals were monitored for tumor volumes for 80–90 days, until tumor volume exceeded 1000 mm3, or until tumor became ulcerated with ulcer diameter reaching 1.0 cm. Statistical analysis was conducted using the GraphPad Prism software (GraphPad Software). Kaplan-Meier curves and corresponding Gehan-Breslow-Wilcoxon tests were used to evaluate statistical differences between groups in survival studies.
Immunohistochemistry (IHC) of HCC tissue microarray
Standard immunohistochemical (IHC) assays were performed as described previously55,56. Formalin-fixed and paraffin-embedded tissue microarrays of human HCC tissues were purchased from Shanghai OUTDO Biotech CO., LTD (HLivH180Su16; Shanghai, Chian). For IHC analysis, IHC staining was performed using a DAKO Autostainer (DAKO, Carpinteria, CA) with DAKO LSAB+ and diaminobenzadine as the chromogen. The primary antibodies used in this assay are: YTHDF1 (NBP2-94472, dilution 1:120), USP5 (ab244290, dilution 1:100), CD8 (85336, dilution 1:200), TIM-3 (83882, dilution 1:100). Images were taken by an Olympus camera and matched software. The overall score for each section was given by the multiplication of the intensity and the positive rate scores of stained cells as previously described63. Briefly, the extent of the staining, defined as the percentage of positive staining areas of tumor cells in relation to the whole tumor area, was scored on a scale of 0–4: 0; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. Staining intensity was scored on a scale of 0–3: 0, negative; 1, weak; 2, moderate; and 3, strong. The overall protein expression score (range 0–12) was calculated by multiplying the positive and intensity scores.
Multiplex immunohistochemical (mIHC) staining
mIHC staining using the Opal 4-Color Manufacturer’s IHC fluorescence Kit (AiFang biological Co., Ltd) was performed according to the manufacturer’s instructions. The tumor sections were blocked in Opal Antibody Diluent/Block for 12 min at room temperature. Primary antibodies were incubated for 1 h at 37 °C or overnight at 4 °C. Following TBST washing, the sections were incubated with secondary Abs Opal Polymer HRP Ms+Rb for 10 min at 37 °C. Sections were then washed in TBST and stained for 10 min with fluorescence staining diluted 1:100 in 1 × Plus Amplification Diluent. All the slides were stained with DAPI for 5 min at room temperature and imaged by Akoya Vectra3.
Patient samples
HCC cancer patient samples were from Department of Pathology, Xinqiao Hospital, Army Medical University, China. Fresh HCC samples were stored at −80 °C for future use. A total of 14 human HCC tissue samples from patients were collected pre-anti-PD-1 treatment at the Xinqiao Hospital, Army Medical University, Chongqing, China, and the total 15 human lung cancer tissue samples from patients were collected pre-anti-PD-1 treatment at the Second Affiliated Hospital of Zhejiang University, Zhejiang, China. Disease assessments were performed with the use of computed tomography (CT) or magnetic resonance imaging at baseline, until disease progression or discontinuation of treatment. The clinical objective response was determined as the investigator-assessed best response based on immune-related response evaluation criteria in solid tumors (irRECIST)64 using unidimensional measurements (CR: complete response, PR: partial response, SD: stable disease, PD: progressive disease). This study was conducted in accordance with the ethics principles of the Declaration of Helsinki and approved by the research and Ethics Committee of Xinqiao Hospital of Army Medical University (approval number: 2021-081-01) and Second Affiliated Hospital of Zhejiang University (approval number: IRB-2023-0507). All patients provided written informed consent.
Statistics and reproducibility
Unless explicitly mentioned, sample sizes were not determined using statistical methods, experiments lacked randomization, and investigators were not blinded during experiments and outcome assessments. Most experiments were conducted a minimum of three times to gather data for subsequent statistical analyses. Group variation was not estimated before experiments. Western blotting data included representative images from 2-3 biological replicate experiments. For quantitative analysis, original western blot images were quantified using ImageJ software to measure the band intensity for statistical analysis. The number of mice per group is specified in the corresponding figure legends, and no animals were excluded from the experiment. Quantitative data are presented as mean ± SEM. Statistical analyses were performed using GraphPad Prism 7 (GraphPad Software). Results were analyzed using a two-tailed unpaired or paired Student’s t-test, or two-way ANOVA, as appropriate. A P value below 0.05 was considered statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). Survival analysis utilized Kaplan-Meier survival curves, and comparisons were made using the log-rank test.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All reagents used in this study are either commercially available or can be made available from the corresponding authors upon reasonable request. RNA-seq and RIP-seq data supporting the findings of this study have been deposited in the Gene Expression Omnibus under accession codes GSE (GSE255922 and GSE278264). The proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) with the dataset identifier PXD05278 and PXD057917. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
Chen, D. S. & Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017).
Ribas, A. & Wolchok, J. D. Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018).
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017).
Restifo, N. P., Smyth, M. J. & Snyder, A. Acquired resistance to immunotherapy and future challenges. Nat. Rev. Cancer 16, 121–126 (2016).
Palmer, A. C. & Sorger, P. K. Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell 171, 1678–1691 (2017).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Frye, M., Harada, B. T., Behm, M. & He, C. RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018).
Shi, H. L., Wei, J. B. & He, C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019).
Wang, X. et al. N-6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Shi, H. et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Li, Q. et al. HIF-1 alpha-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct. Target. Ther. 6, 76 (2021).
Chen, Y. et al. Highly efficient enrichment of O-GlcNAc glycopeptides based on chemical oxidation and reversible hydrazide chemistry. Anal. Chem. 93, 16618–16627 (2021).
Chen, X. Y., Zhang, J. & Zhu, J. S. The role of m(6)A RNA methylation in human cancer. Mol. Cancer 18, 103 (2019).
Su, T. H. et al. Insufficient radiofrequency ablation promotes hepatocellular carcinoma metastasis through N6-methyladenosine mRNA methylation-dependent mechanism. Hepatology 74, 1339–1356 (2021).
Han, D. L. et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 566, 270–274 (2019).
Li, T. et al. Methionine deficiency facilitates antitumour immunity by altering m(6)A methylation of immune checkpoint transcripts. Gut 72, 501–511 (2023).
Bao, Y. et al. Targeting m(6)A reader YTHDF1 augments antitumour immunity and boosts anti-PD-1 efficacy in colorectal cancer. Gut 72, 1497–1509 (2023).
Lin, W. et al. Tumor-intrinsic YTHDF1 drives immune evasion and resistance to immune checkpoint inhibitors via promoting MHC-I degradation. Nat. Commun. 14, 265 (2023).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Harrigan, J. A., Jacq, X., Martin, N. M. & Jackson, S. P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78 (2018).
Clague, M. J., Urbe, S. & Komander, D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 20, 338–352 (2019).
Bonacci, T. & Emanuele, M. J. Dissenting degradation: deubiquitinases in cell cycle and cancer. Semin. Cancer Biol. 67, 145–158 (2020).
Lange, S. M., Armstrong, L. A. & Kulathu, Y. Deubiquitinases: from mechanisms to their inhibition by small molecules. Mol. Cell 82, 15–29 (2022).
Huang, X. et al. Deubiquitinating enzyme: a potential secondary checkpoint of cancer immunity. Front. Oncol. 10, 1289 (2020).
Akimov, V. et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 25, 631–640 (2018).
Gottesman, M. M. Mechanisms of cancer drug resistance. Annu Rev. Med. 53, 615–627 (2002).
Holohan, C., Van Schaeybroeck, S., Longley, D. B. & Johnston, P. G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13, 714–726 (2013).
Housman, G. et al. Drug resistance in cancer: an overview. Cancers 6, 1769–1792 (2014).
Ikeda, F., Crosetto, N. & Dikic, I. What determines the specificity and outcomes of ubiquitin signaling? Cell 143, 677–681 (2010).
Nijman, S. M. et al. A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 (2005).
Avvakumov, G. V. et al. Two ZnF-UBP domains in isopeptidase T (USP5). Biochemistry 51, 1188–1198 (2012).
Raasi, S., Varadan, R., Fushman, D. & Pickart, C. M. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 12, 708–714 (2005).
Urra, H. et al. Endoplasmic reticulum stress and the hallmarks of cancer. Trends Cancer 2, 252–262 (2016).
Li, S. J., Brown, M. S. & Goldstein, J. L. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl. Acad. Sci. USA 107, 3441–3446 (2010).
Zoncu, R., Efeyan, A. & Sabatini, D. M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 (2011).
Aylett, C. H. et al. Architecture of human mTOR complex 1. Science 351, 48–52 (2016).
Dibble, C. C. & Manning, B. D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 15, 555–564 (2013).
Sancak, Y. et al. Ragulator-rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010).
Chao, L. H. & Avruch, J. Cryo-EM insight into the structure of MTOR complex 1 and its interactions with Rheb and substrates. F1000Res 8, 14 (2019).
Xu, C. et al. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 290, 24902–24913 (2015).
Nakajima, S. et al. Ubiquitin-specific protease 5 is required for the efficient repair of DNA double-strand breaks. PLoS ONE 9, e84899 (2014).
Xie, X. et al. Deubiquitylases USP5 and USP13 are recruited to and regulate heat-induced stress granules through their deubiquitylating activities. J. Cell Sci. 131, jcs210856 (2018).
Cai, B. et al. USP5 attenuates NLRP3 inflammasome activation by promoting autophagic degradation of NLRP3. Autophagy 18, 990–1004 (2022).
Pan, C., Olsen, J. V., Daub, H. & Mann, M. Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol. Cell Proteom. 8, 2796–2808 (2009).
Xu, X. et al. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol. Cell 30, 403–414 (2008).
Kim, S. J. et al. mTOR complex 2 regulates proper turnover of insulin receptor substrate-1 via the ubiquitin ligase subunit Fbw8. Mol. Cell 48, 875–887 (2012).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018).
Juneja, V. R. et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J. Exp. Med 214, 895–904 (2017).
Ru, B. et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 35, 4200–4202 (2019).
Benci, J. L. et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 167, 1540–1554. e12 (2016).
Benci, J. L. et al. Opposing functions of interferon coordinate adaptive and innate immune responses to cancer immune checkpoint blockade. Cell 178, 933–948. e14 (2019).
Iwai, Y. et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 99, 12293–12297 (2002).
Hu, B. et al. IFNalpha potentiates anti-PD-1 efficacy by remodeling glucose metabolism in the hepatocellular carcinoma microenvironment. Cancer Discov. 12, 1718–1741 (2022).
Xiao, X. L. et al. ERK and USP5 govern PD-1 homeostasis via deubiquitination to modulate tumor immunotherapy. Nat. Commun. 14, 2859 (2023).
Shao, N. et al. KEAP1 mutations drive tumorigenesis by suppressing SOX9 ubiquitination and degradation. Adv. Sci. 7, 2001018 (2020).
Shao, N. et al. GASC1 promotes hepatocellular carcinoma progression by inhibiting the degradation of ROCK2. Cell Death Dis. 12, 253 (2021).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z. & Zhang, B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, 199–205 (2019).
Schmidt, E. K., Clavarino, G., Ceppi, M. & Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009).
Liu, C. et al. LSD1 stimulates cancer-associated fibroblasts to drive Notch3-dependent self-renewal of liver cancer stem-like cells. Cancer Res. 78, 938–949 (2018).
Liu, C. et al. Sox9 regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma. Hepatology 64, 117–129 (2016).
Nishino, M. et al. Developing a common language for tumor response to immunotherapy: immune-related response criteria using unidimensional measurements. Clin. Cancer Res. 19, 3936–3943 (2013).
Acknowledgements
The authors would like to express their gratitude to the laboratory of Prof. Qingfeng Chen at the Institute of Molecular and Cell Biology, Agency for Science, Technology, and Research (A*STAR), for generously sharing resources and tumor tissues of HCC PDX. Financial support for this work was provided by the National Natural Science Foundation of China (Grants No. 82372748 and No. 81972297 to C.L., and No. 32101024 to L.X). This work was also supported by the Chongqing Talent Project (No. cstc2022ycjh-bgzxm0042), and the Chongqing Young and Middle-Aged Medical Excellence Innovation Team Project (No. yxgdrctd20210101) to C.L. This work was also supported by the intramural funding from the Agency for Science, Technology and Research (A*STAR) of Singapore to F.X.
Author information
Authors and Affiliations
Contributions
C.L. directed the project, designed and wrote the manuscript; N.S., L.X., Y.L., M.I., L.Z., J.X., Belinda X.O., Q.Z., X.P., and X.Y., helped perform the experiments; Y.L.; Y.C. and X.X. collected human liver and lung cancer tissues, and statistics IHC results; Y.L. and Y.C. performed IHC staining of human and mouse tumor samples; N.S. and L.X. performed LC-MS/MS proteomic analysis and bulk RNA sequencing; N.S, L.X. and C.L. conducted data analysis; F.X. read and wrote the manuscript. All contributed to the editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Heng-Jia Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shao, N., Xi, L., Lv, Y. et al. USP5 stabilizes YTHDF1 to control cancer immune surveillance through mTORC1-mediated phosphorylation. Nat Commun 16, 1313 (2025). https://doi.org/10.1038/s41467-025-56564-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56564-9
This article is cited by
-
Ubiquitin-centered post-translational modification crosstalk orchestrates tumor immunity and immunotherapy response
Experimental Hematology & Oncology (2026)
-
Solasonine inhibits NSCLC malignant progression by regulating SLC7A11 ubiquitination degradation and inducing ferroptosis
Respiratory Research (2026)
-
Protein arginine methyltransferase 7-mediated arginine mono-methylation stabilizes SRY-box transcription factor 9 to promote non-small cell lung cancer progression
Molecular Biomedicine (2025)