Abstract
Adenine base editing can convert sickle hemoglobin (HbS, βΕ6V) to G-Makassar hemoglobin (HbG, βE6A), a naturally occurring variant that is clinically asymptomatic. However, the quality and functionality of purified HbG and of mature HbGG and HbGS red blood cells (RBC) has not been assessed. Here, we develop a mouse model to characterize HbG. Purified HbG appears normal and does not polymerize under hypoxia. The topology of the hemoglobin fold with the βΕ6Α mutation is similar to HbA in the oxy and deoxy states. However, RBC containing HbGS are dehydrated, showing altered function and increased sickling under hypoxia. Blood counts and mitochondrial retention measures place HbGS RBCs as intermediate in severity between HbAS and HbSS, while organ function is comparable to HbAS. HbGG resembles HbAA for most metrics. Our results highlight the importance of functionally assessing the mature red cell environment when evaluating novel gene editing strategies for hematologic disorders.
Similar content being viewed by others
Introduction
Sickle cell disease (SCD) is a monogenic disorder caused by a single point mutation in the beta-globin gene (HBB), producing an abnormal hemoglobin (HbS) that polymerizes under hypoxia, resulting in rigid, poorly deformable red blood cells (RBCs)1,2. Clinically, this causes vaso-occlusion, severe pain crises, organ damage, and early mortality3,4,5. While pharmacologic therapies, including hydroxyurea and newer second line therapies, and chronic transfusion therapy may modify the symptoms of SCD, none are considered curative6,7,8. Allogenic hematopoietic cell transplantation may be curative for individuals with SCD; but matched sibling donors, which produce the best outcomes, are available to only ~10% of individuals with SCD9. Haploidentical transplants from a parent may be possible depending on the age of the patient. Although outcomes are improving, the risk of graft versus host disease remains10,11. Autologous transplant with gene modified cells eliminates the donor barrier, avoids graft versus host disease, and can directly address the underlying pathology of SCD by increasing the amount of functional hemoglobin relative to HbS12,13. In December of 2023, the FDA approved genetic medicines exa-cel and lovo-cel for individuals with SCD 12 years of age and over. Recently, preclinical data demonstrated that gene editing therapies utilizing adenine base editing can achieve potent, uniform, and durable fetal hemoglobin induction14,15 without requiring double strand breaks in the genome. In addition to inducing fetal hemoglobin, adenine base editing technology can be used to directly edit the sickle point mutation. While the sickle point mutation in the hemoglobin β subunit (βE6V) cannot be directly corrected to wild-type (βE6) by an adenine base editor (ABE), it can be converted to a naturally occurring hemoglobin variant, known as HbG-Makassar (HbG, βE6A). This approach presents a promising alternative base editing option to treat SCD, as it has the potential to convert the pathogenic HbS to a non-pathogenic form of hemoglobin.
The HbG variant was described in 1969; individuals heterozygous and homozygous for HbG were identified in Southeast Asia16. Clinical characterization of HbAG individuals across Southeast Asia did not reveal a hematological phenotype that correlated with HbG expression17,18. Recently, a larger study involving individuals with heterozygous HbG/beta thalassemia in Malaysia indicated that HbG behaves as a benign hemoglobin variant19. An HbGG individual was reported to have normal red cell indices and was also pregnant at the time of the report, although mildly increased red cell fragility was noted20. In a more recent study, the conversion of HbS to HbG through ex vivo base editing in a SCD mouse model showed the potential of this gene editing approach to correct the anemia and splenomegaly associated with SCD although red cell biophysical properties were not tested21. Other investigations into the biochemical and biophysical properties of recombinant HbG hemoglobin demonstrated the non-sickling, nonpathogenic properties of the HbG variant22,23,24,25.
However, no compound heterozygous individuals with HbGS have been identified, so HbG’s potential interaction with HbS is not known. This may be due to low population frequency of HbG and/or the fact that HbG and HbS cannot be distinguished from each other using gel electrophoresis or high-performance liquid chromatography26. The biophysical properties, including deformability and sickling, of mature RBCs containing HbGS and HbGG have not been described. Furthermore, the lack of an HbG protein crystal structure prevents assessing how the βE6A mutation affects the globin protein fold and its function.
While HbG appears to have normal function, compound heterozygotes of non-pathogenic Hb variants can result in a disease state when combined with HbS. For example, HbC (βΕ6Κ) is recognized as a non-sickling variant, and HbCC individuals are in general asymptomatic27,28. With HbSC RBCs containing 50% HbS and 50% HbC, it could be assumed that HbSC would be phenotypically similar to HbAS, which is 60% HbA, 40%HbS. However, HbSC is a disease state due to the impact of HbC on red cell hydration29,30. The effect of HbC on HbS can only be observed in the mature RBC; dehydration increases the mean corpuscular hemoglobin concentration (MCHC), increasing the risk of HbS polymerization and amplifying the effect of the 50% HbS contained in these cells. Thus, to further biologically de-risk a base editing strategy predicated on the direct conversion of HbS to HbG-Makassar, it is crucial to understand the impact of both HbGG and HbGS on red blood cell function in the mature RBC.
In this work, we generate transgenic human mouse models expressing human HbGG and HbGS from a well-established Townes mouse model and characterize the purified HbG protein as well as the mature RBC biophysical properties. Functional and structural characterization of HbG, coupled with RBC functional assays and organ characterization, reveal that purified HbG as well as homozygous HbGG RBCs have characteristics resembling that of functional hemoglobins. Interestingly, when HbG was present with HbS in an RBC environment, HbGS RBCs have a phenotype intermediate in severity between HbAS and HbSS as observed by various hematological indices. In addition to highlighting the importance of functionally assessing the mature RBC environment when evaluating novel gene editing strategies for hematological disorders, our work provides evidence that supports the direct bi-allelic correction of HbS to HbG using base editing as a viable and promising approach to treat individuals suffering with sickle cell disease.
Results
Generation of Townes HbG-Makassar animal model
To evaluate the function of HbG containing RBCs, we generated a knock-in mouse model utilizing the Townes mice that carry several human hemoglobin genes, replacing the endogenous mouse hemoglobin genes (Fig. 1)31. We targeted the Glu6 of HBB to install HbG-Makassar mutation as well as a silent mutation in the Pro5 amino acid residue, replicating our base editing strategy32. Townes mice expressing HbG hemoglobin were then crossed with HbAS Townes mice to generate HbGS animals.
Gene architecture of the chromosome 7 locus in the Townes mouse containing Human HBB genes63. Endogenous mouse genes are highlighted with teal bars with their respective gene names. Human HBG1 (5675 bp) and human HBB (4113 bp) tandem sequences, containing intact human genomic 5′ promoter and 3′ flanking regions highlighted in yellow are replacing murine Hbb-bmaj and Hbb-bmin. Intended knock in nucleotide substitutions to generate Makassar β globin (E6A, codon underlined) in exon 1 of HBB are highlighted in red. The gRNA spacer regions used to generate KI are shown in blue and green. Silent, synonymous mutations introduced with DNA donor oligo template to prevent re-cutting are indicated in bold black.
Functional characterization of purified Hb G-Makassar
To determine the impact of βE6A mutation in the function of HbG, we performed comparative oxygen binding studies with purified HbA, HbS, and HbG. To eliminate confounding results due to the source of isolation we first compared functional parameters between HbA and HbS isolated from human blood and from Townes mice. Purified HbA and HbS have similar p50 values and hill coefficients irrespective of the purification source utilized (Supplementary Fig. 1). Oxygen equilibrium curves show HbG has similar functional parameters as those observed for HbA and HbS, indicating that the βE6A mutation has a negligible impact on the ability of HbG to bind and release oxygen cooperatively (Fig. 2a). To assess how the βE6A mutation affects hemoglobin polymerization33, we performed comparative polymerization assays with purified HbS, HbG, and HbA. As previously reported using recombinant material isolated from bacteria, HbG did not polymerize within the designated assay time frame, (Fig. 2b)23. No polymerization was observed even when the HbG concentration was increased to a level that induced a 40 s delay time in polymerization for HbS (compare Fig. 2b and Supplementary Fig. 2). Following the characterization of the polymerization behavior of pure fractions of HbG, the polymerization potential of hemoglobin mixtures was assessed to model the possible monoallelic editing outcome resulting in HbGS. Notably, the polymerization delay times associated with HbG:HbS protein mixtures were similar to the ones observed for HbA:HbS mixtures (compare Fig. 2c, d). Plotting the logarithm of the inverse delay time as a function of the logarithm of HbS concentration used in the assays models how the polymerization rate of HbS is affected by the presence of other hemoglobin variants (Fig. 2e). Interestingly, the lines for each mixture composition are not significantly different (p = 0.51) suggesting that when HbG is present in an in vitro mixture with HbS, similarly to HbA, it does not exacerbate the polymerization behavior of HbS.
a Oxygen binding affinity comparison between HbA, HbS, and HbG. Traces shown represent a single curve of data collected in quadruplicates. Numbers shown represent the average and standard deviations of Hill coefficients and p50 values calculated from each individual trace. Name in parenthesis indicates source utilized for isolation (Townes-mice blood, h blood-human blood). In vitro polymerization kinetics of purified (b) HbG at various concentrations, (c) HbA:HbS and (d) HbG:HbS mixtures at various HbS percentages at a total hemoglobin concentration of 2 mg/mL. e The relationship between the log of reciprocal delay time and HbS concentration of binary mixtures of either HbA (blue circles) or HbG (red squares) with HbS is shown. Extra sum-of-squares F test was performed to compared both data sets. HbS percentages used for each mixture are shown above their respective data points. Source data are provided as a Source Data file.
To assess the impact of βE6A on the hemoglobin structure, we determined the 1.94-Å and 2.24-Å resolution X-ray crystal structures of HbG in the R- or liganded state and T- or unliganded state (Supplementary Table 1). The R-state structure contained one copy of the functional heterotetramer (α1β1–α2β2) in the asymmetric unit (Fig. 3a, b). A heme cofactor is seen in all four subunits and its Fe center is coordinated with a water molecule as an axial ligand (Fig. 3a). This structure shows a high degree of similarity to the R2-state structure of HbA (PDB ID 1BBB)34 as indicated by the RMSD of 0.390-Å and 0.218-Å for all Cα atoms in the α and β subunits, respectively (Fig. 3b). The T-state structure was solved with two copies of the functional heterotetramer in the asymmetric unit. Analysis of the electron density map revealed there are no non-proteinogenic axial ligands bound to the heme iron in both α- and β-subunits, indicating we captured deoxygenated or unliganded state (Fig. 3c). The T-state hemoglobin is characterized by the formation of a large cavity in the center of the heterotetramer, which is formed due to the rearrangement of two αβ dimers when the hemoglobin is unliganded or deoxygenated (compare Fig. 3b, d). The T-state structures of HbG and HbA (PDB ID 2DN2)34 as well as HbS (PDB ID 2HBS)35 also revealed high conformational similarity with an RMSD of 0.352-Å and 0.421-Å for all of Cα atoms, respectively (Fig. 3d and Supplementary Fig. 3). Taken together, our results show the βE6A mutation does not alter the topology of the hemoglobin structure nor the ability of HbG to bind and release oxygen as observed in the characterization presented above.
a, c Simulated annealing composite omit maps (2Fo-Fc) are contoured to 1.0 σ for the heme cofactor in the HbG-Makassar tetramer. Heme cofactor is shown as red sticks and the composite omit map is shown as blue mesh. The presence (a) and absence (c) of the electron density for the water molecule coordinated to the heme iron suggest the protein is in its relaxed and tense states, respectively. b, d Ribbon representation of superimposed structures of HbG-Makassar and HbA in its R2 (b; HbA PDB id: 1BBB) and T state (d; HbA PDB id:2DN2). The α and β protomers are shown in teal and purple for HbG-Makassar and gray for HbA, respectively. Hemes are shown as red sticks and the βE6A substitution found in HbG-Makassar is shown as light purple spheres.
Characterization of red blood cells expressing HbG
Following the biochemical and structural characterization of purified HbG, we next wanted to assess the functional parameters of HbG in the mature red cell, and the RBCs as a component of whole blood. Conventional hematology lab values showed that the blood indices of HbGG mice resembled those of HbAA mice, while HbGS indices more closely resembled the indices of HbSS than HbAS mice. Mice with HbGG had median hemoglobin levels of 11.3 g dL-1 (6.3 to 12.5 g dL-1), compared to HbAA mice with median hemoglobin levels of 12.2 g dL-1 (5.5 to 12.6 g dL-1) and HbAS with median hemoglobin levels of 11.7 g dL-1 (10.9 to 15.7 g dL-1). The Hb values of mice with HbGS (median 8.5 g dL-1) were not significantly different (p = 0.70) from mice with HbSS (median 6.9 g dL-1). HbAS mice had higher Hb levels than HbGS mice (median Hb 11.7 and 8.5 g dL-1, respectively; p = 0.0003, Fig. 4a). The white blood cell counts (WBC) of HbAA and HbGG mice were not significantly different (median 5.1 × 103 µL-1 and 7.4 × 103 µL-1, respectively; p > 0.999). WBC of HbGS mice were comparable to the WBC of HbSS mice (median 25.4 × 103 µL-1 and 26.9 × 103 µL-1, respectively; p > 0.999), suggesting similar amounts of inflammation. HbGS mice had a significantly higher WBC count than HbAS mice (median 25.47 × 103 µL-1 and 8.1 × 103 µL-1, respectively; p = 0.001, Fig. 4b).
Complete blood count was performed on peripheral blood across all genotypes. The box plots show median (middle line), 25th percentile (upper line), 75th percentile (lower line), and whiskers to include all observations. a Hemoglobin levels (***p = 0.0003), (b) white blood cell (WBC) counts (**p = 0.001), (c) absolute reticulocyte counts (*p = 0.03), and (d) mean corpuscular values (MCV) (*p = 0.02, ****p < 0.0001). Data were analyzed using the two-sided Kruskal-Wallis test with p values adjusted for multiple comparisons. Blue-HbAA, red-HbGG, green-HbAS, purple-HbGS, and orange-HbSS. For sample size information please see Supplementary Table 5. Source data are provided as a Source Data file.
HbAA and HbGG mice had similar absolute reticulocyte counts (ARC), suggesting that the RBC of HbGG mice had similar lifespans and rates of hemolysis as that of HbAA mice (median 829.9 × 103 cells µL-1 and 1099 × 103 cells µL-1, respectively; p = 0.45). The ARC from HbGS mice was significantly higher when compared to HbAS (median 2632 × 103 cells µL-1 and 1015 × 103 cells µL-1; p = 0.03) mice but not to HbSS. (median 2632 ×103 cells µL-1 and 2391 × 103 cells µL-1; p > 0.99) (Fig. 4c). Mean corpuscular volume (MCV) was significantly lower in RBC from HbGG mice compared to the RBC of HbAA mice (median 29.7 and 34.5 fL, respectively; p = 0.02). The MCV of HbGS RBC was significantly lower than that of HbSS mice (median 31.1 and 41.5 fL, respectively; p < 0.0001) (Fig. 4d), indicating that HbG contributes to red cell dehydration. Of note, RBC lifespan as suggested by reticulocyte count are similar between HbAA and HbGG at steady state but may be different in a physiologically stressed or dehydrated state. Additionally, coinheritance of a hemoglobinopathy (HbGG or HbSS converted to HbGG by gene therapy) and an otherwise benign RBC condition that impacts RBC hydration (i.e., PIEZO1 mutations) could worsen HbGG lifespan, leading to a more severe clinical presentation, including ineffective erythropoiesis.
The deformability and point of sickling (PoS) of RBCs from animals of each genotype was assessed using oxygen gradient ektacytometry. The elongation index minimum (EImin) measures deformability under hypoxia; RBCs from HbAA and HbAS mice had better deformability under low oxygen tension than HbGG (p < 0.0001) and HbGS (p < 0.0001) mice, respectively. HbGS RBCs had better deformability than that of HbSS (p < 0.0001, Fig. 5a). The median EImin values of HbAA, HbGG, HbAS, HbGS, HbSS were 0.56, 0.47, 0.53, 0.40, and 0.15, respectively. The elongation index maximum, (EImax), which measures the RBC deformability under normoxia, demonstrates that HbAA and HbAS RBCs have better deformability when oxygenated than HbGG (p = 0.0001) and HbGS (p < 0.0001), respectively (Fig. 5b). Interestingly, HbGS RBCs had a slightly worse deformability when compared to HbSS (p < 0.04). The median EImax values of HbAA, HbGG, HbAS, HbGS, HbSS were 0.57, 0.49, 0.54, 0.44, and 0.49, respectively. HbAA, HbAS, and HbGG do not sickle and therefore do not have PoS values. HbGS mice had lower point of sickling (p < 0.0001) compared to HbSS (median 19.3 and 24.8 mmHg, respectively), typically below a physiologic threshold of pO2 > 15% in the bone marrow reference (Fig. 5c). Visualized sickling under chemical hypoxia produced results similar to the PoS obtained using oxygen gradient ektacytometry, with no sickling in HbGG RBCs. HbGS shows a trend toward decreased sickling compared to HbSS (Fig. 5d). Whole blood from animals with HbGS, HbAS and HbSS show comparable hematocrit-viscosity ratio (HVR), indicating similar oxygen carrying capacity of RBCs with these genotypes. HbAA had a significantly higher HVR (viscosity measured at 225 s-1) compared to HbGG (p = 0.001, median 14.4 and 6.8, respectively, Fig. 5e).
The box plots show median (middle line), 25th percentile (upper line), 75th percentile (lower line), and whiskers to include all observations. HbG related RBC dehydration impacts deformability. a, b Oxygen gradient ektacytometry (Oxygenscan) data collected using the LORRCA device showing (a) elongation minimum (EImin) (****p < 0.0001), and (b) elongation maximum (EImax) (*p = 0.04, ***p = 0.0001, ****p < 0.0001) (c) PoS for all genotypes that showed a sickling phenotype (****p < 0.0001). d Quantification of sickled cells via imaging following chemical hypoxia. e Hematocrit viscosity ratio (HVR) at shear rate of 225 (1/s) (**p = 0.0012). f Dense red blood cell percentages (**p = 0.001,***p = 0.00021). Data were analyzed using two-sided nonparametric tests (Median test for panel a, Mann-Whitney test for panel (c), and Kruskal-Wallis test for the rest. The p-values for the panels a, b, and d–f were adjusted for multiple comparisons). Blue-HbAA, red-HbGG, green-HbAS, purple-HbGS, and orange-HbSS. For sample size information please see Supplementary Table 5. Source data are provided as a Source Data file.
Given the low MCV and concern for RBC dehydration of RBCs containing HbG, we assessed dense red blood cell percentages using an ADVIA hematology analyzer36. High red blood cell density can contribute to HbS polymerization, subsequent sickling37, and clinical complications such as vaso-occlusive crises and avascular necrosis in humans with SCD38,39,40,41. However, due to their short lifespan, low hemoglobin levels and high numbers of reticulocytes, HbSS mouse RBCs are typically not dense; nor are HbAS or HbAA42. However, HbG-containing RBCs, whether HbGS or HbGG, exhibited 5% to 14.8% dense red blood cells (Fig. 5f).
RBCs from mice with the HbGG genotype had significantly more mitochondrial retention compared to the RBCs of HbAA mice (median 15.7% and 6.3%, respectively; p = 0.002, Fig. 6a). Retention of mitochondria is seen in immature erythroid precursors and ineffective erythropoiesis; to assess if the differences in mitochondrial retention on the Makassar mice were due to ineffective erythropoiesis, we characterized erythroid maturation in the mouse bone marrow (Fig. 6b and Supplementary Fig. 4). Here, we saw the same distribution of erythroid maturation in the bone marrow of mice with HbAA and HbGG genotypes (Fig. 6c), suggesting that HbGG mice do not have ineffective erythropoiesis, as they do not demonstrate an increase in erythroid progenitors in the bone marrow compared to HbAA. HbGS erythroid maturation was delayed at the basophilic stage compared to HbAS (median 1.29 vs 0.34%, p = 0.014, Fig. 6d) while erythroid maturation was comparable between HbGS and HbSS, suggesting that HbGS mice have ineffective erythropoiesis (Fig. 6e). In SCD, ineffective erythropoiesis is driven by apoptosis of erythroid precursors in the BM; this occurs under normoxia, so HbS polymerization is not required. It is likely that this occurs in HbGS mice as well, based on spleen and erythroid maturation findings. Clinical implications of ineffective erythropoiesis include exacerbation of anemia and iron overload.
The box plots (panels a, c–e) show median (middle line), 25th percentile (upper line), 75th percentile (lower line), and whiskers to include all observations. a Mitochondrial retention (mean fluorescence intensity, MFI) measured using MitoTracker Deep Red+ flow panel (**p = 0.002). The data was analyzed with the two-sided median test and p-values were adjusted for multiple comparisons. b MitoTracker and TER119+ representative flow histogram. c–e Erythroid cell maturation in the bone marrow was measured as % TER119+ cells in each population (Pro – RBC, I -VI) divided by comparisons of interest (c) HbAA vs HbGG, (d) HbAS vs HbGS (*p = 0.014, **p = 0.004), (e) HbGS vs HbSS. Each maturation stage of the comparison group was compared using the two-sided Mann-Whitney test and p values adjusted for multiple comparisons. For sample size information please see Supplementary Table 5. Source data are provided as a Source Data file.
Organ pathology
We next assessed organ pathology in mice of all genotypes, at 24–28 weeks of age. Liver weight per total mass was comparable across genotypes HbAA, HbGG, HbAS, and HbGS, and in all cases was less than HbSS mice (median values: HbAA 5.78%, HbGG 5.83%, HbAS 5.68%, HbGS 6.44%, HbSS 8.75%) (Fig. 7a). Spleen weight per total mass was comparable between HbAA and HbGG mice (median 0.51% and 1.07%, respectively, p = 0.24). The mass ratio of HbGS spleens was significantly larger than HbAS (median 1.57% and 0.69%, respectively, p = 0.012), but smaller when compared to HbSS (median 1.57% and 4.78%, respectively, p = 0.012, Fig. 7b). There was no significant difference in glomerular sclerosis between HbGG and HbAA mice (p > 0.99). The glomerular scores of HbGS were intermediate in severity between HbAS and HbSS; however, this difference was not statistically significant (median values HbAS 17, HbGS 43, and HbSS 59.5, Fig. 7c, d).
The box plots (panel a–c) show median (middle line), 25th percentile (upper line), 75th percentile (lower line), and whiskers to include all observations. a Liver size to body mass ratio (**p = 0.002). b Spleen size to body mass ratio (*p = 0.012). c Total sclerotic score across genotypes using glomerular sclerosis index. d Representative images of glomerular sclerosis for each genotype. Data for panels a-c were analyzed using the two-sided median test and p values were adjusted for multiple comparisons. The yellow bar in the bottom right corner of images is a 50 μm scale bar. Blue-HbAA, red-HbGG, green-HbAS, purple-HbGS, and orange-HbSS. For sample size information please see Supplementary Table 5. Source data are provided as a Source Data file.
Ex vivo HbG-Makassar gene editing in CD34+ cells
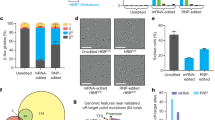
To characterize the ex vivo base editing outcomes that install the Makassar hemoglobin variant in HbSS hCD34+ cells, we used an engineered NGC-PAM inlaid adenine base editor (IBE), IBE 12.232 to convert HbS to HbG (Fig. 8a). In both mobilized and non-mobilized homozygous HbSS CD34+ cells we achieved >58% Makassar editing at 48 hours post-EP with mRNA encoding our base editor and guide RNA targeting HbS. Makassar editing continued to increase to >80% as cells differentiated (Fig. 8b). Ultra-high-performance liquid chromatography (UHPLC) was used to quantify HbG relative to HbS in erythroid cells. HbG protein levels correlated closely with those of DNA editing rates; HbS was reduced to <16% of total beta globins (Fig. 8c). To understand the distribution of editing outcomes that gave >80% bulk HbG-Makassar editing, we single cell cloned CD34+ cells 2 days post-EP to obtain allelic editing on a clonal level. Around 75% ± 7% of cells were bi-allelically Makassar edited, 14.2% ± 6% were mono-allelically edited, 6.7% ± 1.7% were Makassar bi-allelically edited with one allele harboring Makassar and rare bystander edits32, and 2.5% ± 0.98% were completely unedited (Fig. 8d). Similar allelic editing frequencies were also observed from single BFU-E colonies obtained through a colony forming unit assay that were sequenced (Supplementary fig. 5). This distribution of allelic editing is similar to those obtained using a different NRCH-PAM ABE and guide RNA pair to install the Hb-G Makassar variant21.
a Protospacer and PAM (blue) sequence used for base editing approach to target the HbS allele to convert sickle valine to alanine with target adenines highlighted. b NGS of non-mobilized (n = 2 donors) and mobilized peripheral blood (n = 1 donor) HbSS CD34+ cells 48 h post-electroporation and d14 in vitro erythroid differentiated (IVED) cells indicating highly efficient Makassar editing and low percentage non-synonymous bystander editing. c Globin quantification of relative abundance of Makassar and HbS globins by UHPLC of bulk IVED cultures from (b). Two independent IVED cultures were generated for the mobilized donor and quantified for globin abundance. d Allelic breakdown of editing outcomes from single IVED clones derived non-mobilized (n = 2 donor) and mobilized (n = 1 donor) shown in (a). For non-mobilized donor (n = 172 and 128 single cell colonies) and for mobilized HbSS donor (n = 83 single cell colonies), single colonies were analyzed respectively. e Globin abundance of bulk (n = 1 donor) and single IVED colonies (n = 18 single cell colonies) of a HbAS donor and mono-allelically edited Makassar HbSS IVED colonies (n = 14 colonies). ND not detected, SC single colonies, bulk d18 IVED editing measured at day 18 following IVED. Source data are provided as a Source Data file. Data is presented as mean ± standard deviation.
To further characterize the impact of the HbGS heterozygous state derived from primary hCD34+ cells, we isolated single erythroid colonies derived in vitro from HbSS CD34+ cells that were base edited and assessed colonies that were confirmed by NGS at the genomic level to be heterozygous HbGS for expression of beta globins. The percent globin protein expression detected by UHPLC in these HbGS cells displayed a similar 60:40 ratio of HbG-Makassar globin to HbS globin seen in HbGS Townes mice and to individuals with sickle trait (HbAS) (compare Supplementary Fig. 6 and Fig. 8e). Altogether, our ex vivo editing data suggests that installation of the HbG-Makassar variant can be achieved in CD34+ cells with high efficiency and can lead to >90% of red blood cells that no longer express HbS protein or achieve therapeutically relevant reduced levels of HbS.
Discussion
There has been significant progress in gene-edited cell therapy options for individuals with SCD, making “one-and-done” therapies a possibility. HbF induction through disruption of the BCL11A erythroid enhancer element is an FDA approved gene editing strategy, providing clear benefit for patients receiving this therapy. Induction of HbF through various alterations of the gamma globin promoter are also in clinical trials. In addition, prime editing approaches, with the potential to correct the sickle mutation to wild-type, are being evaluated preclinically. These strategies create known, non-mutant hemoglobins, either HbF or HbA; taking advantage of the vast literature available on the effect of these hemoglobins in a mature RBC when present with HbS.
Converting HbS to HbG-Makassar through adenine base editing is a promising alternative therapeutic approach12,15,21,32 This approach has the additional benefit of eliminating both sickle allele and the associated pathogenic protein, leading to minimal residual HbS of <15%. We and others have shown exceptionally high editing efficiency, particularly in the bi-allelic conversion of HbS to HbG-Makassar using ABEs21,32. However more in-depth characterization of the functionality of isolated HbG protein and within its native RBC environment is needed to truly assess the therapeutic potential of converting HbS to HbG-Makassar. Whereas the hematological indices of an HbGG individual have been described in the literature20; the current available data is limited, and does not include tests of red cell function. Among the values reported in humans with HbGG, it is noteworthy to observe that the normal Hb and increased reticulocyte count (reported as percentage, not ARC), is consistent with our HbGG mouse model results. Interestingly, the young HbGG individual described had 6.9% HbF; our adult HbGG mouse model produced no detectable HbF (Supplementary Fig. 6).
In this study, we demonstrated that human HbG purified from the Townes mice has functional (p50 and Hill coefficients) and structural properties similar to human HbA. Purified HbG is a non-sickling variant that does not polymerize like HbS, even under hypoxia conditions, and results are in alignment with previous studies23,24,25. HbG does not polymerize because the β6Α substitution in HbG is believed to increase the critical concentration needed for polymerization33. Considering the functional similarities between HbG and HbA, it was anticipated that HbG:HbS mixtures would polymerize to the same extent as HbA:HbS in vitro. Indeed, our polymerization assays revealed HbG did not exacerbate the polymerization of HbS when present in mixtures. Other studies looking at polymerization of mixtures of HbS and other non-sickling variants like HbC have shown similar results43. This observation suggests that the β6A mutation in HbG is not prone to make additional contacts in polymers being formed by hybrid HbGS mixtures. Our studies with purified HbG suggests that the benefits of having the βE6A substitution in the Makassar variant in mitigating hemoglobin polymerization does not come at the expense of affecting its functional parameters when it comes to oxygen binding, thus providing a direct benefit to SCD patients.
Interestingly, we noted abnormal deformability in HbGG even though sickling was absent in HbGG RBCs. Poor deformability in the absence of sickling has been observed previously in non-pathogenic hemoglobin variants in the homozygous state, such as HbCC43,44. In the case of HbCC, poor deformability is attributed to HbC activation of the Gardos channel, causing RBC dehydration30. Individuals with HbC have high levels of dense red blood cells, similar to the mice with HbGG RBCs. In the absence of HbS, increased red cell density from RBC dehydration found when HbC is present may cause a mild hemolysis, but is otherwise not clinically significant. However, HbSC becomes a disease state due to RBC dehydration potentiating sickling30. HbSS individuals also have increased %DRBC, but this may be due to a different mechanism, of gradual dehydration due to cycles of sickling and unsickling, and loss of RBC membrane. In HbSS, older RBCs are denser; this is why in the mouse model, HbSS RBC are not dense; RBC lifespan is shorter than in the human. The rheological properties of HbGG RBCs resembled that of HbAA RBCs, except for metrics affected by dehydration, such as %DRBC.
HbGG erythroid maturation was not significantly different from HbAA; further evidence for normal erythropoiesis in HbGG mice is supported by similar numbers of erythroid precursors in the bone marrow and similar spleen size, both of which was not significantly different from HbAA mice. In cases of ineffective erythropoiesis, the spleen becomes a site of red cell production, resulting in enlargement45. Our assessment of organ function continued with liver and kidney; again, HbSS mice exhibited typical pathology of liver enlargement; there was no significant difference between the liver sizes of HbAA, HbAS, HbGS and HbGG mice (Fig. 7a). Although the glomerular sclerosis score demonstrated a rising trend across genotypes with the highest score in HbSS, differences did not achieve statistical significance, likely due to the small sample size (N = 3).
HbSS RBCs are known to be rigid and poorly deformable under normoxia due to membrane abnormalities. Deformability worsens with hypoxia-induced polymerization of HbS46,47. Dehydration of the RBC increases the likelihood of polymerization. HbGS hematologic indices and measures of red cell function were often intermediate between that of HbAS and HbSS. Given that, like HbC, HbG has lower solubility than HbA25, does not sickle, and appears to cause red cell dehydration, we hypothesize that the intermediate red cell function (between HbAS and HbSS) that we observe in HbGS is due to dehydration of the RBC, possibly through the Gardos channel. A direct comparison between a HbGS and HbSC mouse RBC was not possible, as a HbSC mouse is not available. However, based on our experience with human HbSC rheology, it appears that HbGS RBC are better quality than that of HbSC; possibly due to less dehydration, or the fact that HbS: HbC is 50:50, whereas the ratio of HbG to HbS is 60:40. In the setting of ex vivo base editing, the HbGS state represents a less frequent (<15%) editing outcome, with bi-allelic editing by far predominating (>70%). A minor population of RBCs are likely to have a 60:40 ratio of HbG:HbS, and even these RBCs will exhibit significant functional improvement over the original HbSS RBCs. Overall, we expect to significantly improve the sickle cell phenotype in >90% of cells with a base editing approach.
Taken together our results indicate that the HbG variant contributes to RBC dehydration, impacting its deformability. However, in the absence of HbS, this dehydration does not create a disease state in homozygous HbG. However, HbGS RBCs may not be functionally optimal given the observed hematological indices and red cell function characteristics. Our findings demonstrate the importance of functionally assessing the mature RBCs when considering novel editing strategies for hematological conditions, as defects in HbG in combination with HbS were not detectable by in vitro polymerization assays of purified HbG proteins or RBC sickling assays performed on the erythroid precursors from cultures of Makassar edited HSCs14,21. Altogether, the Makassar direct replacement strategy of pathogenic HbS, through adenine base editing, offers another promising therapeutic option for patients with sickle cell disease.
Methods
The research presented in this manuscript complies with all relevant ethical regulations. All mice were maintained and studied according to the National Institute of Health Guide for the Care and Use of Laboratory Animals following an approved protocol 202000101 by Emory University Institutional Care and Use of Animals Committees. Mobilized peripheral blood stem cell (PBSC) from Sickle Cell Disease (SCD) patients were collected under BCH protocol IRB-P00034825. Written informed consent was obtained for research purposes only. Non-mobilized HbSS CD34+ cells were obtained under an Emory approved IRB protocol IRB00034535 for research use in the present project and written informed consent was provided by the donors.
Experimental Design
All experimental mice used in this study were bred in-house at Emory and genotypes (HbAA, HbAS, HbSS, HbGS, or HbGG) were verified using Sanger sequencing (Genewiz). Supplemental Table 5 contains the numbers of mice and statistical analyses used for each figure. Each mouse was weighed at the time of sacrifice and euthanized via cervical dislocation using isoflurane anesthesia. Peripheral blood and organs (kidneys, spleen, and liver) were collected to characterize hematology and rheology, RBC sickling, mitochondrial retention, erythroid maturation, and organ pathology. Mice could not be randomized to experimental groups, but experimenters were blinded to genotype during the experimentation and analysis.
Generation of animal models
A humanized beta globin locus mouse (JAX stock 013071 B6;129-Hbbtm2(HBG1,HBB*)Tow/Hbbtm3(HBG1,HBB)TowHbatm1(Hba)Tow/J referred to as the Townes mouse model) with the Makassar allele was created at the Jackson Laboratory by knocking-in the Makassar point mutation using CRISPR/Cas9 and donor oligos (Supplementary Table 2) to generate WT/Makassar heterozygotes (HbAG) which were backcrossed to generate HbGG homozygotes. HbGS genotypes were subsequently generated by crossing HbAG to HbSS and HbAS Townes mice. Genotypes were determined by Sanger sequencing. We analyzed the red cell function of HbAA, HbAS, HbSS, HbGG, and HbGS mice. The mice were group-housed in individually ventilated cages on corncob in a barrier facility, given ad libitum reverse-osmosis–purified chlorinated water and irradiated rodent diet (LabDiet: 5053 - PicoLab® Rodent Diet 20), and maintained on a 12:12-h light: dark cycle.
Purification and characterization of hemoglobin variants
Whole blood carrying the desired hemoglobin variant was collected from appropriate sources (Supplementary Table 3) and resuspended in 100 mL of IEX binding buffer (10 mM sodium phosphate dibasic pH 6.5). Cells were homogenized (5600 psi, 1 passage) and lysate clarified via centrifugation (4 °C, 36000 x g, 45 min). Proteins were purified using a method described previously to separate hemoglobin variants using a MonoS HR16/10 column48. Recombinant hemoglobin HbG for structural studies was purified as described previously49,50. Purified proteins were stored at –80 °C. Success in isolating individual hemoglobin variants was assessed via mass spectrometry (Supplementary Fig. 7).
Intact mass measurement of hemoglobin variants by LC-MS
The intact mass and sequence identification of purified hemoglobin proteins were determined by Liquid Chromatography-Mass Spectrometry to verify its molecular identity. Purified proteins (n = 1) were diluted to 0.5 mg mL-1 and analyzed on a UHPLC system with a binary pump equipped with a 2.1 × 150 mm C18 column. Mobile phase A was 0.1% v trifluoroacetic acid (TFA) in water, and mobile phase B was 0.08% v TFA in acetonitrile, with a flow rate of 0.25 mL min-1. Mass detection was performed with an in-line Q Exactive HF mass spectrometer (Thermo Fisher Scientific) acquiring intact mass data in the range from 500 to 2000 Da. Spectrum was analyzed using the BioPharma FinderTM software (Thermo Fisher Scientific, version 4.1).
O2 binding measurements of purified hemoglobin variants
Oxygen equilibrium curves were measured for each purified hemoglobin at 30 μM (heme) concentration using a HEMOX analyzer (TCS Scientific Corp.) per manufacturer’s recommendations at 37 °C. Each deoxy/oxy cycle was repeated three times. To reduce methemoglobin formation during data acquisition, samples were supplemented with the Hayashi enzymatic reduction system51. Oxygen equilibration curves were analyzed using the HEMOX analyzer data software yielding p50 values (partial pressure of oxygen at which hemoglobin is half-saturated) and n50 (Hill coefficient). Values reported for the p50 and Hill coefficients consist of the average and standard deviation of 4 individual measurements.
Polymerization kinetics
Polymerization assays were performed as previously described33. Briefly, 100 μL of sample at various concentrations (2, 1.5, 1.0, 0.5 mg mL-1) in polymerization buffer (1.8 M potassium phosphate buffer, 5 mg mL-1 sodium dithionite) was dispensed on a pre-chilled 96 well plate. The plate was then loaded onto an Omega Spectrostar plate reader coupled with an atmospheric-controlled unit where hypoxic conditions (<1% oxygen) were kept by flushing the instrument with nitrogen gas. Polymerization delay times were measured by monitoring the optical density of the samples at 37 °C and at a 700 nm wavelength as a function of time. Data was acquired for 1000 seconds. Polymerization of hemoglobin mixtures was performed as described above with slight modifications. HbAS and HbGS mixtures of hemoglobins were made at a constant 2 mg mL-1 total hemoglobin with HbS ratios of 0%, 25%, 50%, and 75%. Mixtures were made in polymerization buffer and polymerization delay times were measured as described above.
Structural characterization of HbG
All crystallization conditions were prepared and optimized using a Mosquito robot (SPT Labtech) at 20 °C. For the HbG R2-state structure, drops were prepared by mixing 0.5 μL of HbG isolated from Townes mice (13 mg mL-1 in 20 mM Tris-HCl pH 7.5, 150 mM NaCl, and 1 mM TCEP) and 0.5 μL of reservoir solution (0.1 M TRIS pH 8.0; 26-36% (v/v) PEG 6,000) and equilibrated against 70 μL of reservoir solution. The crystals were transferred to a cryoprotectant solution (0.1 M Tris pH 8, 36% PEG 6,000, 18% glycerol) and flash-cooled in liquid nitrogen. For HbG in its T-state, the crystallization condition was identified and optimized in an MBraun anaerobic glovebox. Drops were prepared by mixing 0.5 μL of recombinant HbG isolated from E. coli (25 mg mL-1 in 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM TCEP, and 20 mM sodium dithionite) and 0.5 μL of reservoir solution (0.055 M Citric acid, 0.045 M Bis-Tris propane, pH 4.5, 22% PEG 3,350), and equilibrated against 70 μL of reservoir solution. The crystals were transferred to a cryoprotectant solution (0.055 M Citric acid, 0.045 M Bis-Tris propane, pH 4.5, 24% PEG 3,350, 20% glycerol), and flash-cooled in liquid nitrogen.
Data collections were performed at the Highly Automated Macromolecular Crystallography (AMX) beamline of the National Synchrotron Light Source II. Diffraction data were processed using XDS52 and scaled using AIMLESS53. The crystal structures were determined by molecular replacement techniques implemented in Phaser54 using coordinates of the human hemoglobin structure (PDB ID 2DN2 or PDB ID 2DN1)55. Following molecular replacement, simulated annealing was performed to remove model bias using PHENIX.refine56. All models were refined by iterative rounds of model building and the addition of water molecules was performed using Coot57. Non-crystallographic symmetry restraints, TLS (translation, libration, and screw), and positional and B-factor refinement were used on all structures. The data collection and refinement statistics are summarized in Supplementary Table 1. The residues visualized in the structures from 141 and 146 residues for the α and β subunit respectively, are listed in Supplementary Table 4.
Hematology measurements
Whole blood, collected in EDTA, was used for all rheological devices and measurements. Complete blood counts were obtained using hematology analyzers Element HT5 (HESKA, Loveland, CO, USA) and ADVIA 2120i (Siemens, Malvern, PA, USA).
Sickling assay
RBCs were stained with Hoechst 33342 and subjected to 2% sodium metabisulfite (MBS) by volume as previously described58. Images were collected by light microscopy at baseline and every minute for thirty minutes post-MBS addition. Number of sickled RBCs, reported as a percentage of the total were quantified by two individuals blinded to the RBC genotype.
Rheology measurements
Red cell deformability (elongation index, EI) was measured using oxygen gradient ektacytometry using the laser-assisted optical rotational red cell analyzer (LORRCA, RR Mechatronics, Zwagg, Netherlands) with the oxygenscan test under normoxic (EImax) and hypoxic (EImin) conditions as previously described59.
Viscosity was measured using a Beckman cone and plate viscometer. An aliquot of 500 μL of whole blood was run through a multipoint viscosity test starting at 6 rpm with a shear rate of 45 (1 s-1) then raised to 30 rpm at a shear rate of 225 (1 s-1). The two average values of viscosity across those conditions were then recorded, and the hematocrit to viscosity ratio (HVR) calculated. As viscosity rises past a transition point, typically an Hb of 15 in a healthy individual, and an Hb of 10 in an individual with SCD, oxygen carrying capacity begins to be impaired due to decreased blood flow. In SCD, viscosity rises more precipitously with increase in Hb than in a healthy individual; however, Hb is lower; therefore, the HVR is more informative than whole blood viscosity; higher HVR is favorable, indicating less viscosity rise with increase in Hb/hematocrit.
Assessment of mitochondrial retention
RBC mitochondrial retention was measured by flow cytometry using MitoTracker Deep Red (Invitrogen, cat# M46753) on washed RBCs as previously described60. Briefly, an aliquot of 5 μL of EDTA anticoagulated whole blood was washed once with staining buffer (Dulbecco’s Phosphate-Buffered Saline without Ca2+ and Mg2 + , 2% fetal bovine serum, 2 mM EDTA). Blood was centrifuged at 500 x g for 5 minutes at room temperature and supernatant was discarded. Red blood cells were stained in 100 μL of 300 nM MitoTracker Deep Red (Invitrogen, cat# M46753)) and 0.4 μg of TER119-BUV395 (BD Biosciences Cat# 563827) in staining buffer, prewarmed to 37 °C. Cells were incubated in the dark for 45 min, then washed three times with PBS. Cells were then fixed in 0.4% PFA on ice for 10 minutes before washing three times. The flow panels were run on a FACSymphony A5 and A3 flow cytometers. Analysis was then performed through FlowJo v10 software.
Measure of erythroid maturation
In a sterile environment, femurs, tibias, and humeri were extracted from mice of each genotype. The compact bone was cut at the caps to expose the bone marrow and spun into a solution of bone marrow harvest media (RPMI, 10% heat inactivated FBS, 20 U mL-1 DNase, 4 U mL-1 heparin) at 10,000 rpm for 30 s. Supernatant was then removed, and the pellets resuspended in 1 mL of DPBS. Samples were then counted via hemocytometer at a 200x dilution. The erythroid maturation panel was made up of Ter119 (BD Biosciences cat# 563827), CD44+ (Biolegend cat# 103012), Annexin V (Biotium cat# 29004), and LIVE/DEAD fixable viability dye (Life Technologies, cat# L34992). Ter119 and CD44+ were used to characterize the erythroid population with Annexin V measured apoptosis, as previously described61. Briefly, 1 × 10^6 cells were washed in 1× Annexin Binding Buffer (Biotium Cat# 99902). Dead cells were identified using the Live/Dead Fixable Near IR (780) Viability dye at a 1:1000 dilution. Samples were incubated at room temperature for 15 min, in the dark. Without washing, cells were stained with TER119-BUV395 (0.4 μg/106 cells) and CD44-APC (0.35 μg/106 cells) were added and incubated for an additional 20 min in the dark. Cells were washed twice with 1× Annexin Binding Buffer, Annexin V was then added, and cells were incubated in 100 μL for 15 min in the dark. Samples were analyzed on the cytometer within one hour of staining for Annexin. The flow panels were run on a FACSymphony A5 and A3 flow cytometers. Analysis was performed using FlowJo v10 software.
Pathology
Mice were weighed at the time of sacrifice. Kidneys, spleens, and livers were harvested from each of the five genotypes and washed in PBS prior to being weighed. Kidneys were preserved in formalin before paraffin embedding. The paraffin blocks were sliced using a microtome to obtain 5 μm thick sections. Sections were stained with hemolysin and eosin and imaged under a Keyence microscope. Images were scored for glomerular sclerosis by two physicians blinded to genotype as previously described62.
Statistical analysis and Reproducibility
Comparisons across genotypes (HbAA vs HbGG, HbAS vs HbGS, and HbGS vs HbSS) were performed using a two-sided nonparametric test (Kruskal-Wallis or Median test) with statistically significant values being selected at adjusted p < 0.05; all performed using StataNow 18.5 (College Station) or GraphPad Prism 10 (Boston). The Median test was used when the distribution of the groups was found to be unequal as per the Kolmogorov-Smirnov test. Mann-Whitney test was used instead of the Kruskal-Wallis test for comparison of two groups. Statistical analysis for data obtained on FlowJo was completed on Prism. Non-linear regression curves for the polymerization of protein mixtures were compared using the extra sum-of-squares F test using Prism 10. The sample size as well as the statistical tests used for comparisons are shown in Supplementary Table 5. Sample size was derived using data from a pilot sample of three mice. No data was excluded from the analyses, the experiments were not randomized, and the investigators were not blinded.
mRNA production for ABE editors used in CD34+ cells
All adenine base editor mRNA was generated as previously described. Inlaid Base editor 12.2 (IBE12.2) was cloned into a plasmid encoding a dT7 promoter50. PCR amplification of the mRNA template was used in subsequent in vitro transcription. The NEB HiScribe High-Yield Kit was used as per the instruction manual but with full substitution of N1-methyl-pseudouridine for uridine and co-transcriptional capping with CleanCap AG (Trilink). Reaction cleanup was performed by lithium chloride precipitation.
CD34+ cell culture and electroporation
Mobilized peripheral blood from HbAS patients was obtained and enriched for Human CD34+ HSPCs and frozen in single-use aliquots (HemaCare, M001F-GCSF/MOZ-2). The CD34+ cells were cultured in X-VIVO 10 (Lonza) containing 1% v Glutamax (Gibco), 100 ng mL-1 of TPO (Peprotech), SCF (Peprotech) and Flt-3 (Peprotech) and cultured for 48 h prior to electroporation. Electroporation of hCD34+ cells was conducted with MaxCyte Atx with OC25x3 cassettes and HSC-3 program, as previously described32. Electroporations were conducted with 50 nM of in vitro transcribed mRNA encoding base editor IBE12.2 and 5 μM of sgRNA targeting the HbS allele in 25 µL final volume reactions.
CD34+ isolation from HbSS donors
Mobilized peripheral blood CD34+ cells from individuals with SCD were generously provided by the Manis Lab (BCH). Non-mobilized HbSS CD34+ cells were obtained from red cell exchange bags collected under an Emory approved IRB protocol for research use in the present project and written informed consent was provided by the donors. Peripheral blood mononuclear cells (PBMCs) were isolated using density centrifugation by Ficoll-Paque (GE healthcare) per manufacturer’s protocols of apheresis product. RBCs were removed using GlyA (StemCell Technologies RBC depletion kit); CD34+ cells were isolated by magnetic separation with UltraPure human CD34+ positive enrichment kit with LS columns (Miltenyi Biotech).
Erythrocyte differentiation post ABE electroporation
CD34+ cells underwent three phase in vitro erythroid differentiation (IVED) 48 h post electroporation as previously described15. Briefly, 48 h post-electroporation, hCD34+ cells were pelleted and resuspended in Iscove’s Modified Dulbecco’s Medium (IMDM; ATCC) containing 5% human serum, 330 μg mL-1 transferrin (Sigma), 10 μg mL-1 human insulin (Sigma), 2 U mL-1 heparin sodium (Sigma), 3 U mL-1 EPO (PeproTech), 100 ng mL-1 SCF (PeproTech), 5 μg mL-1 IL3 and 50 μM hydrocortisone (Sigma) at 2 × 104 cells per mL. On day 7, cells were pelleted and resuspended in the same IMDM media without IL3 and hydrocortisone at a concentration of 2 × 105 cells per mL. On day 11, cells were pelleted and resuspended in IMDM medium containing without SCF at 1 × 106 cells per mL. On day 14, the cells were pelleted and resuspended in the same IMDM medium at 5 × 106 cells per mL. Differentiated red blood cells were collected on day 18 for UHPLC processing. Single cell IVED clones were obtained by limiting dilution of CD34+ cells 48 h post-electroporation into 96 well U-bottom plates that were confirmed by NGS to be the genotypes of interest.
Ultra-high-performance liquid chromatography (UHPLC) Analysis
UHPLC analysis was performed as previously described15. The separation conditions were further optimized for the separation of HbG from HbS. A reverse-phase column at a temperature of 75 °C was used. Mobile phases were 0.1% v trifluoroacetic acid (TFA) in water (A) and 0.08% v TFA in acetonitrile (B) with a flow rate of 0.25 mL min-1. A gradient of 38–48%B 0–23 min; 48–99%B 10–23–26 min; and 99–38%B to 26–28 min was applied. Sample injection volume was 10 μL. UV spectra at a wavelength of 220 nm with a data rate of 5 Hz were collected throughout the analysis.
Genomic DNA extraction and NGS
Genomic DNA from cells was isolated using the Quick Extract (Lucigen) per manufacturer’s recommended protocol. Genomic DNA samples were amplified and prepared for high throughput sequencing as previously reported32.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The X-ray crystal structures described in this study have been deposited in the Worldwide Protein Data Bank under accession codes 9AV9 and 9AYZ. Published X-ray crystal structures used for structural comparison, 1BBB and 2DN2 are available in the Worldwide Protein Data Bank. NGS raw data described in this study has been deposited in NCBI under accession number PRJNA1198049. Source data are provided with this manuscript. Materials are available upon request. Source data are provided with this paper.
References
Noguchi, C. T. Polymerization in erythrocytes containing S and non-S hemoglobins. Biophys. J. 45, 1153–1158 (1984).
Brittenham, G. M., Schechter, A. N. & Noguchi, C. T. Hemoglobin S polymerization: primary determinant of the hemolytic and clinical severity of the sickling syndromes. Blood 65, 183–189 (1985).
Platt, O. S. et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N. Engl. J. Med. 330, 1639–1644 (1994).
Elmariah, H. et al. Factors associated with survival in a contemporary adult sickle cell disease cohort. Am. J. Hematol. 89, 530–535 (2014).
Jiao, B. et al. Long-term survival with sickle cell disease: a nationwide cohort study of Medicare and Medicaid beneficiaries. Blood Adv. 7, 3276–3283 (2023).
Locatelli, F., Corbacioglu, S., Hobbs, W., Frangoul, H. & Walters, M. C. Defining curative endpoints for sickle cell disease in the era of gene therapy and gene editing. Am. J. Hematol. 99, 430–438 (2024).
Ali, M. A. et al. Efficacy and safety of recently approved drugs for sickle cell disease: a review of clinical trials. Exp. Hematol. 92, 11–18.e11 (2020).
Chou, S. T., Hendrickson, J. E. & Fasano, R. M. Transfusion therapy for sickle cell disease: what’s new? Blood Adv. 7, 2551–2553 (2023).
Justus, D., Perez-Albuerne, E., Dioguardi, J., Jacobsohn, D. & Abraham, A. Allogeneic donor availability for hematopoietic stem cell transplantation in children with sickle cell disease. Pediatr. Blood Cancer 62, 1285–1287 (2015).
Pawlowska, A. B. et al. HLA haploidentical stem cell transplant with pretransplant immunosuppression for patients with sickle cell disease. Biol Blood Marrow Transplant 24, 185–189 (2018).
Saraf, S. L. et al. Haploidentical peripheral blood stem cell transplantation demonstrates stable engraftment in adults with sickle cell disease. Biol. Blood Marrow Transplant 24, 1759–1765 (2018).
Abraham, A. A. & Tisdale, J. F. Gene therapy for sickle cell disease: moving from the bench to the bedside. Blood 138, 932–941 (2021).
Leonard, A., Tisdale, J. & Abraham, A. Curative options for sickle cell disease: haploidentical stem cell transplantation or gene therapy? Br. J. Haematol. 189, 408–423 (2020).
Mayuranathan, T. et al. Potent and uniform fetal hemoglobin induction via base editing. Nat. Genet. 55, 1210–1220 (2023).
Gaudelli, N. M. et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900 (2020).
Blackwell, R. Q., Oemijati, S., Pribadi, W., Weng, M. I. & Liu, C. S. Hemoglobin G Makassar: beta-6 Glu leads to Ala. Biochim Biophys Acta 214, 396–401 (1970).
Saechan, V., Nopparatana, C., Nopparatana, C. & Fucharoen, S. Molecular basis and hematological features of hemoglobin variants in Southern Thailand. Int. J. Hematol. 92, 445–450 (2010).
Panyasai, S., Fucharoen, G. & Fucharoen, S. Hemoglobin variants in northern thailand: prevalence, heterogeneity and molecular characteristics. Genet Test Mol. Biomarkers 20, 37–43 (2016).
Esa, E. et al. Clinical and haematological characteristics of 38 individuals with Hb G-Makassar in Malaysia. EJHaem 4, 940–948 (2023).
Sangkitporn, S., Rerkamnuaychoke, B., Sangkitporn, S., Mitrakul, C. & Sutivigit, Y. Hb G Makassar (beta 6:Glu-Ala) in a Thai family. J. Med. Assoc. Thai 85, 577–582 (2002).
Newby, G. A. et al. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature 595, 295–302 (2021).
Baudin-Chich, V. et al. Enhanced polymerization of recombinant human deoxyhemoglobin beta 6 Glu–Ile. Proc. Natl. Acad. Sci. USA 87, 1845–1849 (1990).
Pagnier, J., Bihoreau, M. T., Baudin, V., Edelstein, S. J. & Poyart, C. Polymerization and solubility of recombinant hemoglobins alpha 2 beta 2 6 Glu->Ala (Hb Makassar) and alpha 2 beta 2 6 Glu->Ala, 23 Val->Ile]. C R Acad. Sci. III 316, 431–436 (1993).
Bihoreau, M. T. et al. Steric and hydrophobic determinants of the solubilities of recombinant sickle cell hemoglobins. Protein Sci. 1, 145–150 (1992).
Adachi, K. et al. Effects of beta 6 amino acid hydrophobicity on stability and solubility of hemoglobin tetramers. FEBS Lett. 315, 47–50 (1993).
Mohamad, A. S., Hamzah, R., Selvaratnam, V., Yegapan, S. & Sathar, J. Human hemoglobin G-Makassar variant masquerading as sickle cell anemia. Hematol. Rep. 10, 7210 (2018).
Itano, H. A. & Neel, J. V. A new inherited abnormality of human hemoglobin. Proc. Natl. Acad. Sci. USA 36, 613–617 (1950).
Kaplan, E., Zuelzer, W. W. & Neel, J. V. Further studies on hemoglobin C. II. The hematologic effects of hemoglobin C alone and in combination with sickle cell hemoglobin. Blood 8, 735–746 (1953).
Morgan, J. L., Bowles, R. M. & Harris, J. S. Hemoglobin C: report of the homozygous condition and of combinations with normal and sickle cell hemoglobin. Pediatrics 15, 185–190 (1955).
Nagel, R. L., Fabry, M. E. & Steinberg, M. H. The paradox of hemoglobin SC disease. Blood Rev. 17, 167–178 (2003).
Ryan, T. M. et al. Human sickle hemoglobin in transgenic mice. Science 247, 566–568 (1990).
Chu, S. H. et al. Rationally designed base editors for precise editing of the sickle cell disease mutation. CRISPR J. 4, 169–177 (2021).
Adachi, K. & Asakura, T. Nucleation-controlled aggregation of deoxyhemoglobin S. Possible difference in the size of nuclei in different phosphate concentrations. J. Biol. Chem. 254, 7765–7771 (1979).
Silva, M. M., Rogers, P. H. & Arnone, A. A third quaternary structure of human hemoglobin A at 1.7-A resolution. J. Biol. Chem. 267, 17248–17256 (1992).
Harrington, D. J., Adachi, K. & Royer, W. E. Jr. The high resolution crystal structure of deoxyhemoglobin S. J. Mol. Biol. 272, 398–407 (1997).
Mohandas, N. et al. Automated quantitation of cell density distribution and hyperdense cell fraction in RBC disorders. Blood 74, 442–447 (1989).
Du, E., Diez-Silva, M., Kato, G. J., Dao, M. & Suresh, S. Kinetics of sickle cell biorheology and implications for painful vasoocclusive crisis. Proc. Natl. Acad. Sci. USA 112, 1422–1427 (2015).
Bartolucci, P. et al. Erythrocyte density in sickle cell syndromes is associated with specific clinical manifestations and hemolysis. Blood 120, 3136–3141 (2012).
Rakotoson, M. G. et al. Biological parameters predictive of percent dense red blood cell decrease under hydroxyurea. Orphanet J. Rare Dis. 10, 57 (2015).
Vekilov, P. G. Sickle-cell haemoglobin polymerization: is it the primary pathogenic event of sickle-cell anaemia? Br. J. Haematol. 139, 173–184 (2007).
Warth, J. A. & Rucknagel, D. L. Density ultracentrifugation of sickle cells during and after pain crisis: increased dense echinocytes in crisis. Blood 64, 507–515 (1984).
Alvarez-Argote, J. et al. Pathophysiological characterization of the Townes mouse model for sickle cell disease. Transl. Res. 254, 77–91 (2023).
Murphy, J. R. Hemoglobin CC disease: rheological properties or erythrocytes and abnormalities in cell water. J. Clin. Invest. 47, 1483–1495 (1968).
Berkowitz, L. R. & Orringer, E. P. Cell volume regulation in hemoglobin CC and AA erythrocytes. Am. J. Physiol. 252, C300–C306 (1987).
Kean, L. S. et al. Comparison of mechanisms of anemia in mice with sickle cell disease and beta-thalassemia: peripheral destruction, ineffective erythropoiesis, and phospholipid scramblase-mediated phosphatidylserine exposure. Exp. Hematol. 30, 394–402 (2002).
Lu, M., Rab, M. A., Shevkoplyas, S. S. & Sheehan, V. A. Blood rheology biomarkers in sickle cell disease. Exp. Biol. Med. (Maywood) 245, 155–165 (2020).
Ballas, S. K. & Connes, P. Rheological properties of sickle erythrocytes in patients with sickle-cell anemia: The effect of hydroxyurea, fetal hemoglobin, and alpha-thalassemia. Eur. J. Haematol. 101, 798–803 (2018).
Moo-Penn, W. F. & Jue, D. L. Separation of hemoglobin variants by ion-exchange chromatography on Monobead resins. J. Chromatogr. 318, 325–332 (1985).
Shen, T. J. et al. Production of unmodified human adult hemoglobin in Escherichia coli. Proc. Natl. Acad. Sci. USA 90, 8108–8112 (1993).
Elmer, J., Harris, D. & Palmer, A. F. Purification of hemoglobin from red blood cells using tangential flow filtration and immobilized metal ion affinity chromatography. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 879, 131–138 (2011).
Hayashi, A., Suzuki, T. & Shin, M. An enzymic reduction system for metmyoglobin and methemoglobin, and its application to functional studies of oxygen carriers. Biochim. Biophys. Acta 310, 309–316 (1973).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Park, S. Y., Yokoyama, T., Shibayama, N., Shiro, Y. & Tame, J. R. 1.25 A resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms. J. Mol. Biol. 360, 690–701 (2006).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Wu, Y. et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med. 25, 776–783 (2019).
Rab, M. A. E. et al. Oxygen gradient ektacytometry-derived biomarkers are associated with vaso-occlusive crises and correlate with treatment response in sickle cell disease. Am. J. Hematol. 96, E29–E32 (2021).
Moriconi, C. et al. Retention of functional mitochondria in mature red blood cells from patients with sickle cell disease. Br. J. Haematol. 198, 574–586 (2022).
An, X. & Chen, L. Flow Cytometry (FCM) Analysis and Fluorescence-Activated Cell Sorting (FACS) of Erythroid Cells. Methods Mol. Biol. 1698, 153–174 (2018).
Kasztan, M. et al. Hyperfiltration predicts long-term renal outcomes in humanized sickle cell mice. Blood Adv. 3, 1460–1475 (2019).
Ryan, T. M., Ciavatta, D. J. & Townes, T. M. Knockout-transgenic mouse model of sickle cell disease. Science 278, 873–876 (1997).
Acknowledgements
We would like to thank Dr. Nico Tjandra from the National Heart, Lung, and Blood Institute at the NIH for providing the expression plasmids for recombinant hemoglobin and Damien Fenske-Corbiere from Beam Therapeutics for NGS support. The authors thank the Drennan Lab at MIT for providing access to equipment needed to perform anaerobic crystallographic studies. This research used resources of the National Synchrotron Light Source II; a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Center for BioMolecular Structure (CBMS) is primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant (P30GM133893), and by the DOE Office of Biological and Environmental Research (KP1605010).
Author information
Authors and Affiliations
Contributions
M.A.O. designed, executed, analyzed, and supervised all biochemical studies. P.F. designed, executed, and analyzed all structural experiments. C.X. performed biochemical and structural studies. S.H.C., E.B., D.L. M.Z., J.J., B.C. performed ex vivo primary human CD34 isolation, editing and downstream in vitro experiments. V.W. and R.J. performed all LC-MS and UHPLC globin analytic assays. A.V. performed animal blood collections. S.J.L. supervised biochemical and structural work. A.J.H. and G.C. supervised the project and reviewed manuscript. Z.K. oversaw mouse breeding, molecular biology with B.H., M.A.P., and J.Z., pathology with Y.Z., B.H. and M.A.P., and rheology assays with J.D., E.N.E., J.Z., C.K.K., and B.H. assisting. A.P., B.H., and K.G. performed statistical analysis. V.S. designed the red cell function and mouse pathology experiments. J.M. provided mobilized HbSS material. M.A.O, Z.K., S.J.L., S.H.C., A.P., K.G., and V.S. wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
All authors from Beam Therapeutics disclose a conflict of interest and are shareholders of Beam Therapeutics. The remaining authors declare no competing interest.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kostamo, Z., Ortega, M.A., Xu, C. et al. Base editing HbS to HbG-Makassar improves hemoglobin function supporting its use in sickle cell disease. Nat Commun 16, 1441 (2025). https://doi.org/10.1038/s41467-025-56578-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56578-3