Abstract
SARS-CoV-2 main protease, Mpro, is responsible for processing the viral polyproteins into individual proteins, including the protease itself. Mpro is a key target of anti-COVID-19 therapeutics such as nirmatrelvir (the active component of Paxlovid). Resistance mutants identified clinically and in viral passage assays contain a combination of active site mutations (e.g., E166V, E166A, L167F), which reduce inhibitor binding and enzymatic activity, and non-active site mutations (e.g., P252L, T21I, L50F), which restore the fitness of viral replication. To probe the role of the non-active site mutations in fitness rescue, here we use an Mpro triple mutant (L50F/E166A/L167F) that confers nirmatrelvir drug resistance with a viral fitness level similar to the wild-type. By comparing peptide and full-length Mpro protein as substrates, we demonstrate that the binding of Mpro substrate involves more than residues in the active site. Particularly, L50F and other non-active site mutations can enhance the Mpro dimer-dimer interactions and help place the nsp5-6 substrate at the enzyme catalytic center. The structural and enzymatic activity data of Mpro L50F, L50F/E166A/L167F, and others underscore the importance of considering the whole substrate protein in studying Mpro and substrate interactions, and offers important insights into Mpro function, resistance development, and inhibitor design.
Similar content being viewed by others
Introduction
Since its emergence in late 2019, the COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has had immense health, economic, and social impacts globally. With new infection rates remaining high1, SARS-CoV-2 continues to pose an urgent health threat. The main protease (Mpro) of SARS-CoV-2 is a homo-dimeric cysteine protease responsible for cleaving the viral polyproteins into its mature substituents, including Mpro itself, during viral replication2,3. Mpro is a validated drug target and is inhibited by nirmatrelvir (PF-07321332)4, which is the active agent in Pfizer’s oral COVID-19 drug, Paxlovid (nirmatrelvir/ritonavir combination)5,6. While Paxlovid is one of the most effective COVID-19 treatments to date7,8, Mpro has demonstrated that it is prone to mutations in and outside the active site, many of which have resulted in SARS-CoV-2 variants showing in vitro resistance to nirmatrelvir9,10,11,12, as well as several in immunocompromised patients13,14,15,16,17.
Recent studies have shown that the SARS-CoV-2 virus harboring a Mpro triple mutant, L50F/E166A/L167F, is highly resistant to nirmatrelvir while demonstrating similar fitness of replication as the wild-type (WT) virus in cell culture and animal models18,19,20. The triple mutant was originally identified during viral passaging with an experimental probe compound, ALG-09716, but was later found to harbor significant cross-resistance to nirmatrelvir18. The L50F mutation is located away from the Mpro active site and is not directly involved with binding to nirmatrelvir or the nsp4-5 peptide substrate commonly used in biochemical analysis10,21,22. Yet, L50F is able to rescue the reduced viral fitness caused by the active site substitutions (e.g., E166A, E166V)10,22, and has been observed in clinical mutants (e.g., E166V/L50F)13. Biochemical analysis has indicated that compensatory mutations outside the active site, such as L50F, have little impact on the hydrolysis of the viral nsp4-5 peptide substrate, and improve the enzymatic activity of the active site mutants, such as E166V, by only two-fold (from 3% of WT activity for E166V to 6% for L50F/E166V), yet they fully restore the viral fitness in cell-based studies23. The discrepancy between the biochemical data and viral replication assay remains a puzzle and impedes our ability to understand Mpro resistance mutations and develop new inhibitors.
The majority of the in vitro biochemical studies to date have only used small peptide substrates of SARS-CoV-2 corresponding to the nsp4-5 Mpro cleavage site on the viral polyproteins, which corresponds to the Mpro N-terminal self-cleavage site. Whereas the peptides can capture the enzyme-substrate (ES) interactions inside the active site, we hypothesize that there may be important protein-protein interactions outside the active site in the ES complex. In this study, we have constructed inactive C145A Mpro fusion proteins to mimic the natural protein substrate, as Mpro cleaves its own termini out of the viral polyproteins. The biochemical data and crystal structures demonstrate the ability of non-active site mutations, such as L50F, to promote the formation of the ES complex and fully restore the enzymatic activity of resistant mutants.
Results
To determine the effect of the different Mpro mutations on substrate binding and nirmatrelvir resistance, we characterized the L50F/E166A/L167F triple mutant through enzymatic assays and X-ray crystallography. Using the nsp5-6 cleavage sequence containing FRET peptide and the full-length Mpro as substrates, we determined the catalytic activity of the L50F/E166A/L167F triple mutant and compared the results with the WT (hCoV-19/Wuhan/WIV04/2019), as well as corresponding single and double mutants. Additionally, the crystal structures of Mpro L50F/E166A/L167F and Mpro L50F were determined to be 2.23 and 2.21 Å resolutions (Supplementary Table 1), respectively, to illustrate the underlying molecular interactions.
Effects of distal mutations on Mpro activity using protein substrates
Mpro cleaves the viral polyproteins at 11 sites, including those at its own two termini. We posit that the rate-limiting step in the processing of the viral polyproteins by Mpro is the self-cleavage of the protease from the viral polyprotein, namely the Mpro N-terminal nsp4-5 sequence, and the C-terminal nsp5-6 sequence (Fig. 1a). A recent study has shown that Mpro self-cleavage is intra-molecular at its N-terminus (nsp4-5), and inter-molecular at its C-terminus (nsp5-6), involving a Mpro dimer functioning as enzyme acting on another Mpro dimer serving as substrate (Fig. 1b)24. The mature N-terminus is located near the active site and at the dimer interface, crucial to the stability of the dimer and the active site25,26. The C-terminal cleavage liberates the protease from the membrane-bound nsp6 protein, enabling it to freely access the other cleavage sites on the polyproteins. We hypothesize that the C-terminal cleavage would be slower than the N-terminal cleavage because it is inter-molecular rather than intra-molecular, thus limited by diffusion rates and protein concentrations. This may also make the cleavage at its C-terminus (nsp5-6) more susceptible to inhibitors such as nirmatrelvir. Hence, the most effective resistance mutations may enhance the enzymatic activity at this site. It is consistent with the observation that a common resistance mutation, T304I, occurs at the Mpro C-terminus cleavage site. However, the mechanism of T304I is not entirely clear. Moreover, the enzymatic activity of Mpro in digesting the nsp5-6 sequence has not been characterized for any of the resistance mutants.
a Overview of the Mpro dimer (cyan/orange) showing nirmatrelvir (yellow) bound in the active site and the locations of mutations of interest in the active site (red spheres, e.g., E166/L167) and distal to the active site (blue spheres, e.g., L50) (PDB 8DCZ). Mpro is also named nsp5 on viral polyproteins. b Mpro self-cleavage at the C-terminus. E: Mpro as enzyme, S: Mpro as substrate. The polyprotein chain after Mpro is represented by the curved line. c Schematic of how the fluorescence gel-based assay functions. A protein substrate is constructed by using the catalytically inactive C145A Mpro conjugated with a bacterial protein PBP3, serving as a reporter for quantification purposes after it reacts with the fluorescent inhibitor Bocillin. A schematic image for the assay was created in BioRender. Kohaal, N. (2025) https://BioRender.com/n42a748. d Rate of activity for the C145A Mpro-PBP3 and the C145A/L50F Mpro-PBP3 substrates, in comparison to the peptide substrates. The rate of activity for the C145A/T21I Mpro-PBP3 is shown in Supplementary Fig. 1a. e Cleavage reaction of C145A-PBP3 by multiple Mpro mutants. The cleavage reaction of C145A/L50F Mpro-PBP3 by the same Mpro mutants is shown in Supplementary Fig. 1b. Source data for Fig. 1d and e are provided as a Source Data file. Rates are the average of two replicates.
To determine the effect that the nirmatrelvir-resistant triple mutant L50F/E166A/L167F had on Mpro activity, we employed a fluorescence electrophoresis-based assay using a Mpro protein substrate in which the catalytically inactive Mpro C145A protein was linked to Clostridium difficile Penicillin Binding Protein 3 (PBP3) by the first six residues of the nsp6 N-terminus (Fig. 1c), which, together with the Mpro C-terminal residues, corresponds to the nsp5-6 cleavage sequence. The majority of the nsp6 protein is membrane-embedded and thus not included in the fusion construct. A fluorescently labeled penicillin, Bocillin, was used to covalently react with the catalytic serine of PBP3 and monitor the ability of the Mpro enzyme to cleave at the nsp5-6 junction of the fusion protein. By monitoring the fluorescence intensity of the band representing the cut nsp6-linked PBP3 on a gel, we were able to determine the activity of the Mpro mutants (Fig. 1d, eand Supplementary Figs. 1–3). The concentrations for both the Mpro enzyme and substrate Mpro fusion protein were fixed in our experiments, and the initial velocity of the reaction was determined and compared. At high substrate concentrations, interactions among the fusion protein substrate molecules appeared to sequester the cleavage site and result in a unique substrate inhibition. Therefore, we were unable to vary the substrate concentrations and obtain kcat/Km values of the reaction. In parallel to the protein substrate assay, we also characterized the enzymatic activity and nirmatrelvir inhibition of the Mpro L50F/E166A/L167F triple mutant using the conventional FRET substrates containing the 12-residue nsp5-6 or nsp4-5 cleavage sequence.
In our experiments, we focused on the L50F/E166A/L167F triple mutant and the corresponding single and double mutants (e.g., E166A/L167F), as this triple mutant is one of the first nirmatrelvir resistant mutants identified with a similar fitness of replication as the WT. T21I was also included in our analysis due to its prevalence in resistant mutants10,15. Compared with the experiments using the peptide substrates (nsp4-5 and nsp5-6 FRET peptides), L50F exhibited significantly larger effects on the Mpro enzymatic activity when protein substrates were used (Mpro C145A and C145A/L50F fusion proteins) (Fig. 1d). In our experiments, Mpro L50F was found to be 3.1 times more active than the WT in cleaving the C145A Mpro protein substrate, while it was 1.9- and 1.7-fold more active than the WT in cleaving the nsp5-6 and nsp4-5 FRET substrates, respectively. For comparison, the L50A mutant was also constructed to investigate the contribution of the original leucine side chain to the enzyme activity. The L50A mutant was slightly less active in cleaving the C145A and C145A/L50F Mpro protein substrates (0.\(7\)-fold), while showing slightly more activity than the WT (1.4-fold) in the peptide substrate assays. The E166A and L167F single mutants and the E166A/L167F double mutant, all had reduced enzymatic activity in cleaving the nsp5-6 peptide and protein substrates (0.2 to 0.7-fold of WT). In comparison, a more profound reduction of enzymatic activity was observed in cleaving the nsp4-5 FRET peptide substrate (0.03 to 0.1-fold of WT), suggesting the nsp5-6 cleavage sequence is more relevant in explaining the restoration of the fitness of replication. This difference between the peptide substrates appears to have largely originated from the L167F mutation, which showed 0.3- and 0.05-fold activity of the WT for the nsp5-6 and nsp4-5 peptides, respectively. The most striking observation from our studies was the ability of L50F to rescue the reduced enzymatic activity of the E166A/L167F double mutant (0.2-fold WT) with the L50F/E166A/L167F triple mutant (1.6-fold WT) exhibiting slightly better activity than WT in cleaving the C145A Mpro protein substrate. In comparison, L50F was less effective in rescuing the reduced enzymatic activity of the E166A/L167F double mutant in cleaving the nsp5-6 (0.2-fold to 0.4-fold WT) and nsp4-5 (0.03-fold to 0.03-fold) FRET peptide substrates. Since the L50F mutation may affect protein-protein interactions when present on either the enzyme or protein substrate, we also investigated the reactions using the C145A/L50F mutant in the fusion substrate protein. Overall, the results were similar to that of the C145A substrate, suggesting that L50F exerts its influence on the reaction mainly through the enzyme rather than the substrate (Fig. 1b). Collectively, our results showed that the L50F mutant can rescue the reduced enzymatic activity of the E166A/L167F mutant to a similar level as the WT protein when the Mpro protein with the nsp5-6 cleavage sequence was used as the substrate. Compared to the low activity of the triple mutant in hydrolyzing the nsp4-5 peptide substrate assay (3% of WT), our results using the Mpro protein substrates highlight the differences between the two assays and the importance of using protein substrates in studying Mpro resistance mutations.
Compared with L50F, T21I did not seem to affect the enzyme activity significantly in the peptide assays, consistent with previous studies23. In addition, its activity was similar to the WT in our protein substrate assays mimicking the C-terminal cleavage, using either the C145A or C145A/L50F substrates (Fig. 1d). Compared with the C145A and C145A/L50F substrates, the C145A/T21I substrate also exhibited similar cleavage rates for the WT and various mutants (Supplementary Fig. 1). These results suggest that T21I likely affects Mpro activity through a different mechanism.
L50F/E166A/L167F triple mutant crystal structure
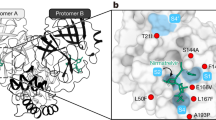
The Mpro triple mutant crystallized in the P21 space group, and the crystal diffracted to 2.23 Å resolution, with four copies of the protein in the asymmetric unit, and each biological dimer forming a dimer of dimers together with another symmetry-related Mpro dimer from adjacent asymmetric unit (Fig. 2a). Interestingly, the side chain of the mutated F50 residue of one Mpro protomer resides at this dimer-dimer interface and nestles into a hydrophobic pocket that is formed by V212, L220, T257, and I259 residing on helices of an adjacent Mpro protomer from a different dimer. Moreover, the C-terminus of this Mpro protomer from the adjacent dimer enters the active site inside the Mpro protein harboring the aforementioned F50 from the bottom of the catalytic pocket, similar to what is observed in the complex structure of Mpro with peptide substrates27. These observations suggest that the triple mutant crystal structure has captured the product complex of Mpro self-cleavage and that the L50F mutation may promote protein-protein interactions facilitating the positioning of the C-terminal substrate peptide in the active site for cleavage. It is also possible that the aromatic fluorophore of the FRET peptide substrate may mimic some of these inter-molecular interactions with F50, leading to enhanced substrate binding and increased reaction rate.
a Mpro triple mutant dimer of a dimer (dark green/green, magenta/salmon), showing P1–P6 residues (magenta stick) of the C-terminus from the substrate dimer (post cleavage, a.k.a, product) bound in the active site of the enzyme dimer (green). Zoomed-in view shows the interactions of the enzyme F50 (green) within the substrate hydrophobic pocket (magenta). Substrate residues are noted in red text. b Close-up view of the binding pose of P1–P6 bound in the triple mutant active site (substrate shown in magenta and enzyme shown in green). The N-terminus from an adjacent protomer is noted in orange. Substrate residues are noted in red text. c Movement of the 166–168 backbone in the triple mutant with P1–P6 bound in the active site (magenta/green) vs. Mpro C145A with nsp5/6 substrate bound (cyan/light purple) (PDB 7MB5). Substrate residues are noted in red text, and the N-terminus from an adjacent monomer is noted in orange. d Binding pose of nirmatrelvir (white, PBD 8DCZ) superimposed into the triple mutant binding pocket. The N-terminus from an adjacent protomer is noted in orange.
In addition to the intermolecular interactions, intramolecular interactions between the C-terminus and the Q256-containing helix of the same protomer are also observed and help the placement of the substrate peptide. Specifically, these intramolecular interactions create a new well-defined S3 site to accommodate T304, which forms contacts with the helix backbone surrounding Q256 (Fig. 2b–d). These interactions were not present in previous complex structures with various peptide substrates, and they help explain the resistance mutation of T304I, as an isoleucine side chain can enhance the non-polar contacts between residue 304 and the Q256 side chain and backbone atoms. Interestingly, Q256L was also observed in the viral passage assays10 and can enhance the intramolecular interactions with the peptide backbone groups surrounding G302. Both the T304I and Q256L mutations may thus stabilize the conformation of the Mpro C-terminus that is required for its proper placement in the active site of another Mpro molecule for cleavage.
These interactions are also responsible for some active site conformational differences between the triple mutant and the WT. When comparing the triple mutant to a previously published Mpro C145A complex structure where the peptide substrate is also observed in the active site27, a distinct widening of the active site pocket is observed (Fig. 2c). It appears that the Q256-containing helix nudges the C-terminal peptide towards the backbone of residues 166–168 in the active site of the adjacent protomer (which the P3–P5 substrate residues normally stack against) and that they consequently also flex outwards, with the most pronounced shifts occurring at F167 and P168. This outward shift of the backbone atoms of residues 166–168 affords extra room in the active site and allows the substrate binding orientation to shift slightly from the previous complex structure at the P2–P6 positions. The shift of the backbone also appears to place residue A166 close to the S1 residue of the other Mpro protomer in the same dimer, which would likely cause steric clashes between the WT E166 side chain and the S1 residue. This suggests that the E166A mutation can facilitate the conformational change observed in the triple mutant, as the smaller alanine side chain prevents such potential steric clashing. It may also explain the synergy observed between the L50F and the E166A/L167F mutations. Specifically, L50F improved the activity of the E166A/L167F mutant by 6-fold in the L50F/E166A/L167F triple mutant, in comparison to the 3-fold activity difference between the L50F single mutant and WT28.
Aside from L50F, the triple mutant structure further illustrates the contribution of L167F to the binding of the nsp5-6 substrate. In this structure, V303 is sandwiched between F305 from the same Mpro molecule and F167 from the active site of the neighboring protein (Fig. 2b). It would form significantly more interactions than those involving the equivalent T303 in the nsp4-5 substrate which also has L305 in the placement of F305. These interactions may be the reason for the difference in the L167F activity change vs. WT when using the two peptide substrates (Fig. 1d). In addition, as shown by a recently published structure of the E166A/L167F mutant29, when the C-terminus is not bound in the active site, the 166–171 loop is positioned overall similarly to that in the unbound WT structure, with F167 placed in the area that V303 occupies in our triple mutant structure. The interactions of V303 with the bulkier F167, in comparison to L167, would also cause a larger shift in the 168–171 position than in the WT.
L50F single mutant crystal structure
In the 2.21 Å resolution crystal structure of the Mpro L50F single mutant, the F50 side chain is also involved in extensive intermolecular hydrophobic interactions that help place the C-terminus of the adjacent protomer in the active site. However, some interesting differences exist compared with the triple mutant structure. First, when the two interacting dimers are constructed using crystallographic symmetry, there is a pseudo two-fold symmetry with each dimer placing the C-terminus of one protomer in the active site of a protomer from the other dimer (Fig. 3a and Supplementary Fig. 4). In both dimer-dimer interfaces of the L50F single mutant structure, F50 is involved in similar hydrophobic interactions with P252, F294, and V297, although additional contacts are made with P293 and I249 at the chain B dimer interface (Fig. 3a). These hydrophobic interactions are different from those in the triple mutant structure involving V212, L220, T257, and I259 (Fig. 2a). Second, the Mpro substrate C-terminus adopts a non-canonical conformation in the active site. Typically, the substrate enters from the bottom of the catalytic pocket with the substrate P3–P5 positions placed along the enzyme backbone from E166-P168 and the P4 side chain in the S4 pocket. However, in the L50F structure, the substrate enters from the side of the active site, with the substrate P3–P5 side chains horizontal to the enzyme active site (Fig. 3b). In this new conformation, the P2 side chain (F305) is placed in the canonical S2 pocket. While Q306 resides near the S1 pocket, its position and conformation are slightly different from those in the triple mutant structure (Fig. 3c), with the terminal carboxylate group placed outside the oxyanion hole. Thus, this conformation does not represent the product conformation immediately after peptide bond cleavage. Nevertheless, the P2 F305 and P1 Q306 adopt the superimposable configurations in both structures suggesting that P1-P2 substituents are more important than P3–P5 substituents in forming the ES complex. It is unclear whether substrate peptides entering the active site through this alternative conformation can be properly cleaved by the enzyme, although it is possible that C145 may not be able to access the scissile peptide bond if the alternative peptide conformation is maintained. We hypothesize that while the protein-protein interactions observed in the L50F single mutant structure can help place the C-terminal cleavage site in the enzyme catalytic center, this state does not represent the productive configuration required for the peptide bond cleavage, and additional conformational changes may be required for the reaction to proceed. In fact, in a previously published L50F structure (PDB 8DKZ)21 where a similar dimer of dimers configuration was observed and the substrate C-terminal peptide adopts the typical conformation with Q306 properly nestled in the S1 pocket, F50 only forms limited interactions with V212 of the adjacent substrate Mpro. There are significantly fewer contacts than observed in our L50F single mutant or L50F/E166A/L167F triple mutant structures. It is possible that our L50F single mutant structure represents the initial encounter between the enzyme and substrate protein as facilitated by F50, and the previously published L50F structure (8DKZ) resembles the dimer of dimer configuration in the productive ES complex following some thermal motions after the initial encounter. It is consistent with the synergy between the L50F and E166A mutation and suggests that the optimal interactions involving the C-terminal peptide (as observed in the canonical binding mode) and F50 (as observed in our L50F and L50F/E166A/L167F structures) may not co-exist in the presence of the E166 side chain. It is also worth mentioning that similar dimer of dimers have also been observed in Mpro WT or C145A mutant crystal structures (PDB 7E5X, 7KHP)30,31 in addition to the aforementioned previously published L50F structure (8DKZ), where the C-terminus of one dimer is placed in the active site of the other dimer. The average surface area buried at the dimer interface is 1811 Å2 and 1375 Å2 for our L50F and L50F/E166A/L167F mutant structures respectively, in comparison to 902 Å2 (7E5X), 1090 Å2 (7KHP), and 866 Å2 (8DKZ) in these previously published structures, again highlighting the enhanced protein-protein interactions promoted by the L50F mutation revealed in our structures (Supplementary Fig. 5).
a L50F single mutant dimer of a dimer (yellow/wheat, blue/light blue), showing P1–P6 (light blue) of the C-terminus bound in the chain B (wheat) active site. The zoomed-in view shows the interactions of the enzyme F50 with the substrate hydrophobic pocket. A view of the chain A interactions can be found in Supplementary Fig. 4. Mutation is noted in red text. b Binding of P1–P6 in the L50F chain B active site. The substrate protomer is shown in light blue, and the enzyme protomer is shown in wheat. The mutation is noted in red text. c Comparison of the binding modes of P1–P6 in the L50F single mutant (light blue/wheat) vs. the L50F/E166A/L167F triple mutant (magenta/green). Mutations are noted in red text.
In addition to L50F, the T21I and P252L mutations are two of the most frequently observed pathways leading to nirmatrelvir resistance in combination with active site mutations. The observation of P252 in the dimer-dimer interface of the L50F single mutant structure indicates that this residue may also affect protein-protein interactions in the ES complex. In the triple mutant structure, T21 also finds itself positioned on the edge of a hydrophobic pocket at the dimer-dimer interface, involving L67 from the same protomer and L232 and M235 from the other adjacent protomer (Supplementary Fig. 6). The effect of the T21I mutation on the protein-protein interactions at this interface may be difficult to predict and possibly not significant due to its peripheral location. Our analysis also indicates that T21I may not affect the C-terminal cleavage when present either on the enzyme or the substrate. However, we hypothesize that T21I may affect protein-protein interactions in other ES complexes, including possibly involving nsp4 residues at the N-terminus. In addition, other resistance mutations from viral passage assays are also found at the dimer-dimer interface of the triple mutant structure, such as A191V, A193P, Q256L, T304I, and P252L (Supplementary Fig. 6)10. Together with the protein-protein interactions enhanced by L50F, these observations lend support to the likely biological relevance of this dimer-dimer interface and suggest the compatibility of these mutations with one another in conferring resistance.
As SARS-CoV-2 continues to evolve and mutate, it will be important to understand the mechanisms behind Mpro resistance to our current best treatments and identify the mutations that are critical to this resistance, as any variants that emerge with such mutations will be of particular concern to the global health community and necessitate careful monitoring. Our results demonstrate that distal protein-protein interactions, away from the active site, contribute significantly to Mpro substrate binding, and mutations at these locations can dramatically alter Mpro activity and the fitness of viral replication. This study provides a structural explanation for the role of L50F in promoting Mpro protein-protein interactions and restoring the reduced enzymatic activity caused by active site drug-resistant mutants such as E166A/L167F. Aside from Mpro self-cleavage, it is possible that mutations like L50F can also enhance the interactions between Mpro and other nsp proteins on the viral polyproteins. Our results, including those comparing nsp5-6 and nsp4-5 peptide substrates, highlight the need to investigate the effects of resistance mutations on specific enzyme-substrate interactions inside and outside the active site for different cleavage sites, rather than using one generic peptide substrate. Similar to our studies of the C-terminal self-cleavage by Mpro, it will be important to analyze the resistant mutations’ impact on the N-terminal self-cleavage, as well as the other cleavage sites on the viral polyproteins, as previously examined for the WT enzyme32,33.
Methods
Fluorescence gel assay fusion protein construct and purification
The Mpro C145A (C145A/L50F) and C. difficile PBP3 42-554 fusion protein was inserted into the pETGSTSumo vector. The cleavage site, SAVKRT, was inserted between Mpro and PBP3 42-554. The expression constructs were transformed into Rosetta (DE3) pLysS cells. A single colony was picked and grew in LB (Luria-Bertani) media supplemented with 50 µg/mL kanamycin and 35 µg/mL chloramphenicol at 37 °C overnight. The overnight culture was then added into 1 L media at 1:100 and incubated at 37 °C until the OD600 reached 0.4. Protein expression was induced using 0.5 mM IPTG at 20 °C overnight. Cells were harvested by centrifugation at 5000 × g for 10 min. The cell pellet was resuspended and disrupted by sonication in buffer A (20 mM Tris-HCl pH 8.0, 300 mM NaCl, 20 mM imidazole, 10 % glycerol), followed by ultracentrifugation at 45,000 × g for one hour. The supernatant was then loaded onto a HisTrap HP affinity column and eluted by linear gradient into buffer B (20 mM Tris-HCl pH 8.0, 300 mM NaCl, 500 mM imidazole, 10 % glycerol). The His tagged protein was pooled, and buffer was exchanged into cleavage buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, and 10 % glycerol). The Sumo tag was removed by ULP1 incubation at 4 °C overnight. The sample was then loaded onto a HisTrap HP column for cleanup. The flowthrough was further purified using a HiPrep 16/60 Sepharcyl S-300 HR size exclusion column in storage buffer (20 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM DTT).
Fluorescence gel assay
Purified untagged Mpro C145A-PBP3 42-554 was diluted in assay buffer (20 mM Tris-HCl pH 8.0, 200 mM NaCl) and labeled with 1.5-fold Bocillin at room temperature for 30 min. 10 µM labeled Mpro C145A-PBP3 42-554 was incubated with 10 µM of different Mpro mutants for the indicated times. The reaction was stopped by using 2x SDS-PAGE loading buffer without dye. The samples were then loaded onto Tris-Glycine 7.5% SDS-PAGE gels. The intensity of the protein bands was analyzed using a ChemiDoc XRS + and the ImageJ software. The initial rate was analyzed by SigmaPlot and reported as 10−7 M/min. Rates are given as mean±s.d. for two biologically independent replicates.
Mpro FRET enzymatic assay
The Mpro nsp4-5 and nsp5-6 FRET substrates were synthesized using the Fmoc solid peptide synthesis34. The sequences are: nsp4-5/Dabcyl-K-TSAVLQ ↓ SGFRKM-E(Edans)-CONH2; nsp5-6/Dabcyl-K-SGVTFQ ↓ SAVKRT-E(Edans)-CONH2.
Km and Vmax of SARS-CoV-2 Mpro mutants were performed using the optimized concentration (the concentration that allows the initial velocity to saturate in the testing substrate concentration range). The final concentrations of substrate range from 0.78–200 µM. The reaction was carried out in a total volume of 100 µL reaction buffer containing 20 mM HEPES pH 6.5, 120 mM NaCl, 0.4 mM EDTA, 4 mM DTT, and 20 % glycerol. The signal was detected using a BioTek Cytation 5 imaging reader (Agilent) with the excitation at 360/40 nm and emission at 460/40 nm. The reaction was monitored every 70 s. The initial velocity of proteolytic activity was determined by linear regression of the first 600–1000 s of the kinetic progress curves. The Km and Vmax were calculated by plotting the initial velocity against FRET substrate concentrations using the classic Michaelis-Menten equation (Y = Vmax*X/(Km + X), X = substrate concentration; Y = enzyme velocity) in the Prism 8 software.
For Ki determination, the optimized mutant SARS-CoV-2 protein concentration (the concentration that gives at least 1 h linear initial velocity) was mixed with 20 µM FRET substrate and various concentrations of nirmatrelvir in 100 µL volume of reaction buffer. The reactions were monitored every 70 s for 3 h. The initial velocity was determined for the first hour by linear regression. The Ki was determined by plotting the initial velocity against inhibitor concentrations using the Morrison equation for tight binding (Y = V0*(1 − ((((Et + X + (Ki*(1 + (S/Km)))) − (((Et + X +(Ki*(1 + (S/Km))))2̂) − 4*Et*X)0.5̂))/(2*Et))), X = inhibitor concentration; Y = enzyme velocity; Et = enzyme concentration; V0 = enzyme velocity in the absence of inhibitor). The reported values were the average of two replicates ± standard error with a 95% confidence interval calculated as SE = (upper limit − lower limit)/3.92.
Mpro mutagenesis, protein expression, and purification
Mpro mutants were generated using the QuikChange® II Site-Directed Mutagenesis Kit from Agilent (Catalog #200524), using pET-SUMO-Mpro (from strain BetaCoV/Wuhan/WIV04/2019) plasmid as the template.
Mpro mutant proteins were expressed and purified as previously described, with minor modifications. Plasmids were transformed into RosettaPlyss (DE3) competent cells, and bacterial cultures overexpressing the target proteins were grown in LB media containing 50 µg/mL of kanamycin and 35 µg/mL chloramphenicol at 37 °C. Expression of the target protein was induced at an OD600 of 0.6–0.8 by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. The cell culture was incubated at 20 °C for 12–16 h. Bacterial cultures were harvested by centrifugation (5000 × g, 10 min, 4 °C) and resuspended in His buffer A (20 mM Tris pH 8.0, 300 mM NaCl, 40 mM imidazole, 10 % glycerol). Bacterial cells were lysed by alternating sonication (10% amplitude, 10 s on/15 s off). The lysed cell suspension was clarified by centrifugation (45,000 × g, 60 min, 4 °C), and the supernatant was loaded onto a HisTrap HP column. The column was thoroughly washed with 60 mM imidazole in lysis buffer. The protein was eluted by linear gradient, 1–100 %, using His buffer B (20 mM Tris pH 8.0, 300 mM NaCl, 500 mM imidazole, 10 % glycerol). The eluted protein was pooled and buffer exchanged into cleavage buffer (20 mM Tris pH 8.0, 100 mM NaCl, 10 % glycerol), and then incubated with ULP1 protease at 4 °C overnight. The sample was then loaded onto a HisTrap HP column, and the flowthrough was collected and concentrated. The flowthrough was further purified using a HiPrep 16/60 Sepharcyl S-300 HR size exclusion column in storage buffer (20 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM DTT).
Mpro crystallization and structure determination
The Mpro mutants were crystallized as previously described9. Briefly, Mpro mutants were diluted to 5 mg/mL in storage buffer. Crystals were grown by hanging drop in 25 % PEG 3350, 0.1 M Na/K tartrate, 0.005 M MgCl2, by mixing 1.5 µL of the protein solution with 1.5 µL of the crystallization condition. Crystals grew after 1–3 days of incubation at 20 °C. Crystals were transferred into a cryoprotectant solution (crystallization condition supplemented with 20% glycerol) and flash-frozen in liquid nitrogen.
X-ray diffraction data were collected at cryogenic temperature (100 K) at the Structural Biology Center (SBC) 19-ID beamline at the Advanced Photon Source (APS) in Argonne, IL, using a Pilatus3 6 M detector and a wavelength of 0.97918 Å. Data were processed with HKL-3000 and CCP4, and PHASER was used for molecular replacement using a previously solved SARS-COV-2 Mpro structure (PDB 6WTT) as the reference model. Model building and structure refinement was completed using the CCP4 suite35, Coot36, and the PDB REDO server (pdb-redo.edu)37. All images were generated using the PyMOL Molecular Graphics System (Schrödinger, LLC).
Full crystallographic statistics are provided in Supplementary Table 1, and images of representative electron density for each structure are provided in Supplementary Figs. 7 and 8. The Ramachandran statistics are 98.8% in the favored region, 0.9% in the allowed region, and 0.2% in the outlier region for PDB 8U4Y (Mpro L50F); 98.9% in the favored region, 0.8% in the allowed region, and 0.4% in the outlier region for PDB 8U25 (Mpro L50F/E166A/L167F).
Buried surface area calculation
The total solvent accessible surface area (SASA) was computed for isolated dimers, and the specific SASA was restricted against the adjacent dimer. The dimer-dimer buried surface area (ddBSA) was then calculated for each dimer as the difference between these two values and then averaged. The VMD program’s implementation of the maximal speed molecular surfaces algorithm (measure SASA command, restrict option)38,39 with a probe radius of 1.4 Å was used for this analysis. All non-protein moieties were stripped from the coordinate files. Dimers of dimer complexes were generated using symmetry operations in PyMOL if not part of the asymmetric unit.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The X-ray crystal structures generated in this study have been deposited in the Protein Data Bank (PDB) under accession codes 8U4Y (Mpro L50F, https://doi.org/10.2210/pdb8U4Y/pdb) and 8U25 (Mpro L50F/E166A/L167F, https://doi.org/10.2210/pdb8U25/pdb). Source data are provided in this paper.
Materials availability
Materials used in this study will be made available under an appropriate materials transfer agreement.
Code availability
No code was used for this study.
References
Rubin, R. As COVID-19 Cases Surge, Here’s What to Know About JN.1, the Latest SARS-CoV-2 “Variant of Interest”. Jama 331, 382–383 (2024).
Hu, Q. et al. The SARS-CoV-2 main protease (M(pro)): Structure, function, and emerging therapies for COVID-19. MedComm 3, e151 (2022).
Jin, Z. et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293 (2020).
Owen, D. R. et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 374, 1586–1593 (2021).
Joyce, R. P., Hu, V. W. & Wang, J. The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): an orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations. Med. Chem. Res. 31, 1637–1646 (2022).
Tanne, J. H. Covid-19: FDA authorises pharmacists to prescribe Paxlovid. BMJ 378, o1695 (2022).
Najjar-Debbiny, R. et al. Effectiveness of Paxlovid in reducing severe coronavirus disease 2019 and mortality in high-risk patients. Clin. Infect. Dis. 76, e342–e349 (2023).
Khunte, M., Kumar, S., Salomon, J. A. & Bilinski, A. Projected COVID-19 mortality reduction from Paxlovid rollout. JAMA Health Forum 4, e230046–e230046 (2023).
Hu, Y. et al. Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir. ACS Cent. Sci. 9, 1658–1669 (2023).
Iketani, S. et al. Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. Nature 613, 558–564 (2023).
Zhu, Y. et al. In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance. Sci. Adv. 10, eadl4013 (2024).
Sasi, V. M. et al. Predicting antiviral resistance mutations in SARS-CoV-2 main protease with computational and experimental screening. Biochemistry 61, 2495–2505 (2022).
Zuckerman, N. S., Bucris, E., Keidar-Friedman, D., Amsalem, M. & Brosh-Nissimov, T. Nirmatrelvir resistance—de novo E166V/L50V mutations in an immunocompromised patient treated with prolonged nirmatrelvir/ritonavir monotherapy leading to clinical and virological treatment failure—a case report. Clin. Infect. Dis.78, 352–355 (2023).
Hirotsu, Y. et al. Multidrug-resistant mutations to antiviral and antibody therapy in an immunocompromised patient infected with SARS-CoV-2. Med 4, 813–824 (2023).
Yamamoto, C. et al. Nirmatrelvir resistance in an immunocompromised patient with persistent coronavirus disease 2019. Viruses 16, 718 (2024).
Nooruzzaman, M. et al. Emergence of transmissible SARS-CoV-2 variants with decreased sensitivity to antivirals in immunocompromised patients with persistent infections. Nat. Commun. 15, 7999 (2024).
Tamura, T. J. et al. Emerging SARS-CoV-2 resistance after antiviral treatment. JAMA Netw. Open 7, e2435431–e2435431 (2024).
Jochmans, D. et al. The substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro are selected by a protease inhibitor in vitro and confer resistance to nirmatrelvir. mBio 14, e0281522 (2023).
Abdelnabi, R. et al. Nirmatrelvir-resistant SARS-CoV-2 is efficiently transmitted in female Syrian hamsters and retains partial susceptibility to treatment. Nat. Commun. 14, 2124 (2023).
Kiso, M. et al. In vitro and in vivo characterization of SARS-CoV-2 strains resistant to nirmatrelvir. Nat. Commun. 14, 3952 (2023).
Chen, S. A. et al. SARS-CoV-2 M(pro) protease variants of concern display altered viral substrate and cell host target galectin-8 processing but retain sensitivity toward antivirals. ACS Cent. Sci. 9, 696–708 (2023).
Zhou, Y. et al. Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system. Sci. Adv. 8, eadd7197 (2022).
Duan, Y. et al. Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir. Nature 622, 376–382 (2023).
Aniana, A. et al. Insights into the mechanism of SARS-CoV-2 main protease autocatalytic maturation from model precursors. Commun. Biol. 6, 1159 (2023).
Nashed, N. T. et al. Autoprocessing and oxyanion loop reorganization upon GC373 and nirmatrelvir binding of monomeric SARS-CoV-2 main protease catalytic domain. Commun. Biol. 5, 976 (2022).
Kovalevsky, A. et al. Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors. J. Biol. Chem. 299, 104886 (2023).
Shaqra, A. M. et al. Defining the substrate envelope of SARS-CoV-2 main protease to predict and avoid drug resistance. Nat. Commun. 13, 3556 (2022).
Hattori, S.-i. et al. Structural and virologic mechanism of the emergence of resistance to Mpro inhibitors in SARS-CoV-2. Proc. Natl. Acad. Sci. USA 121, e2404175121 (2024).
Kovalevsky, A. et al. Effects of SARS-CoV-2 main protease mutations at positions L50, E166, and L167 rendering resistance to covalent and noncovalentinhibitors. J. Med. Chem. 67, 18478–18490 (2024).
Zhao, Y. et al. Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2. Proc. Natl. Acad. Sci. USA 119, e2117142119 (2022).
Lee, J. et al. Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site. Nat. Commun. 11, 5877 (2020).
Yadav, R. et al. Biochemical and structural insights into SARS-CoV-2 polyprotein processing by Mpro. Sci. Adv. 8, eadd2191 (2022).
Narwal, M., Armache, J. P., Edwards, T. J. & Murakami, K. S. SARS-CoV-2 polyprotein substrate regulates the stepwise M(pro) cleavage reaction. J. Biol. Chem. 299, 104697 (2023).
Cady, S. D., Wang, J., Wu, Y., DeGrado, W. F. & Hong, M. Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy. J. Am. Chem. Soc. 133, 4274–4284 (2011).
Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Joosten, R. P., Long, F., Murshudov, G. N. & Perrakis, A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220 (2014).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–38 (1996).
Sanner, M. F., Olson, A. J. & Spehner, J. C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38, 305–320 (1996).
Acknowledgements
This research was supported by the National Institutes of Health (NIH) grant AI158775 to J.W. and Y.C. Results shown in this report are derived from work performed at the Structural Biology Center funded by the U.S. Department of Energy (DOE), Office of Biological and Environmental Research, and operated for the DOE Office of Science at the Advanced Photon Source by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We thank the scientists and staff at SBC for their assistance with X-ray diffraction data collection. The schematic image for the assay in Fig. 1c was created using BioRender.com (https://BioRender.com/n42a748).
Author information
Authors and Affiliations
Contributions
E.M.L., J.W., and Y.C. wrote the manuscript; X.Z., A.F., and L.M.C.J. carried out the gel assay; H.T. performed the FRET enzymatic assay; E.M.L. and A.J.B. crystallized proteins; E.M.L. and N.K. solved and deposited protein structures; J.J.M. carried out computational analysis of the crystal structures.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lewandowski, E.M., Zhang, X., Tan, H. et al. Distal protein-protein interactions contribute to nirmatrelvir resistance. Nat Commun 16, 1266 (2025). https://doi.org/10.1038/s41467-025-56651-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-56651-x
This article is cited by
-
A novel, covalent broad-spectrum inhibitor targeting human coronavirus Mpro
Nature Communications (2025)