Abstract
Lysine methyltransferase KMT5C catalyzes deposition of trimethylation on histone H4 lysine 20 (H4K20me3), an epigenetic marker usually associated with gene repression and maintenance of heterochromatin. KMT5C is widely expressed in a variety of tissues, however, its functional role in liver has not been explored. Here, we show Kmt5c is a fasting- and glucagon-induced gene in liver which regulates hepatic gluconeogenesis. Loss of KMT5C in hepatocytes results in downregulated gluconeogenic gene expression and compromised glucose output during fasting. KMT5C fosters gluconeogenesis through decreasing ubiquitination-mediated PGC-1α degradation, which is unexpectedly independent of its methyltransferase activity. In fact, KMT5C impedes the E3 ligase RNF34 binding to the C-terminal of PGC-1α and subsequent ubiquitination-associated degradation. The diabetic mice models and patients show elevated KMT5C levels in the livers, and KMT5C knockdown beneficially reduces gluconeogenesis and fasting blood glucose levels. In conclusion, the present study identifies KMT5C as a hepatic gluconeogenesis regulator by affecting PGC-1α stability.
Similar content being viewed by others
Introduction
The liver is a central hub to maintain blood glucose levels during starvation. There are two major pathways of hepatic glucose production, namely glycogenolysis and gluconeogenesis. Hepatic glycogen stores are progressively exhausted within hours of fasting, then gluconeogenesis contributes more for glycaemia homeostasis with prolonged fasting1,2. Hyperglycaemia and insulin resistance are hallmarks of type 2 diabetes mellitus (T2DM). In this pathological setting, liver glycogen content and glycogenolysis are lower, but gluconeogenesis is abnormally enhanced to exaggerate both the fasting and postprandial hyperglycaemia3,4. Thus, understanding the regulation of gluconeogenesis and its role in disorders of diabetes is of critical importance.
Glucagon secreted by pancreatic α cells is one of the most important hormones to activate hepatic gluconeogenesis. Firstly, glucagon-dependent PKA signaling phosphorylates CREB promptly to increase its transcriptional activity and bind to the coactivators or histone-modifying enzymes, such as CREB-regulated transcription coactivator 2 (CRTC2), CREB binding protein (CREBBP) and lysine acetyltransferase 2B (KAT2B)5. This leads to a more accessible chromatin environment around gluconeogenic genes to other transcriptional factors which promotes transcription of gluconeogenic program. Then, the CREB cascade induces the expression of a secondary wave of transcriptional factors that mediate a delayed fasting response. Pgc-1α is one of the secondary transcriptional coactivators downstream of CREB-CRTC2 in fasting livers6. Induced PGC-1α activates hepatocyte nuclear factor-4α (HNF4α) and forkhead box O (FOXO) 1, and, therefore, boosts the gene transcription of key gluconeogenic enzymes during prolonged fasting7,8. In addition to the transcriptional regulation, acetylation and phosphorylation on the PGC-1α protein have also been extensively investigated in nutrient control of glucose homeostasis9,10,11,12. Notably, PGC-1α is rapidly turned over whose half-life time only last 0.3 h13. Thus, modulation of its protein stability represents another important mechanism to manipulate hepatic gluconeogenesis.
KMT5C is a histone H4 lysine 20 methyltransferase that participates in a plethora of biological processes14, including heterochromatin formation, genomic integrity, and gene expression regulation. All these functional roles are closely related to the repressive H4 lysine 20 trimethylation (H4K20me3) mark imprinted by KMT5C. However, there have been no any reports to clarify its duty in hepatic metabolism. Here, we highlight that KMT5C enhances hepatic gluconeogenesis in a methyltransferase-independent manner. KMT5C competes against an E3 ubiquitin ligase RNF34 to interact with PGC-1α and increases the PGC-1α level by retarding its proteasomal degradation. In light of aberrantly increased KMT5C levels in livers of prediabetic and diabetic mice and human patients, KMT5C may serve as a potential therapeutic target to decrease blood glucose levels in T2DM patients.
Results
Loss of KMT5C decreases gluconeogenic gene expression in hepatocytes
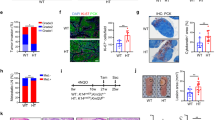
Our previous data showed KMT5C is ubiquitously expressed in various tissues15, however, the functional role of hepatic KMT5C has not been defined. The liver is of prime importance in the modulation of systemic carbohydrate and lipid fluxes. We wondered whether KMT5C participated in the energy metabolism process in livers. Interestingly, fasting for 16 h elevated the hepatic KMT5C level, and refeeding reversed the KMT5C level to the basal level (Fig. 1a). A significant increase in the level of KMT5C was observed in the livers of ob/ob, db/db and high-fat diet-fed mice as compared to lean mice (Fig. 1b–d). Concurrently, the levels of the H4K20me3 mark were correctly connected with the KMT5C level in these mice (Fig. 1a–d). Hepatocytes constitute about 60% of the total cell population in the liver tissue, with non-parenchymal cells (NPCs) accounting for the other 40%16. We isolated hepatocytes and NPCs fractions from mouse livers17 and verified that KMT5C indeed existed abundantly both in hepatocytes and NPCs (Fig. 1e).
a–d Western blot (top) and densitometry analyses (bottom) were conducted to determine KMT5C and H4K20me3 protein levels in liver tissues. a Eight-week-old male C57BL/6 J mice were sacrificed at fed, 16 h fasting, and 4 h refeeding states (n = 3 for KMT5C, n = 4 for H4K20me3). b Control and ob/ob mice at fed state (2-month-old male mice, n = 5 each group). c Control and db/db mice at fed state (2-month-old male mice, n = 3 for KMT5C, n = 4 for H4K20me3). d Male normal chow and HFD-induced obese mice at fed state. HFD has been fed for 12 weeks. (n = 4 for KMT5C, n = 6 for H4K20me3). e The KMT5C protein level in non-parenchymal cells (NPC) and hepatocytes isolated from male mice livers at the age of 2 months. (n = 2 for each group). f–i Western blot analysis of indicated proteins in primary hepatocytes (f, h) and protein quantifications (g, i). f Cre or control GFP adenovirus-infected primary hepatocytes isolated from 12-week-old male Kmt5cflox/flox mice (n = 4 per group). h primary hepatocytes were isolated from 12-week-old male WT and KO mice and were stimulated with or without 100 nM glucagon for 14 h (n = 3 per group). j, k Glucose production was measured in the culture medium of primary hepatocytes isolated from 12-week-old male WT and KO mice, using pyruvate and lactate (j) (n = 3 per group) or glycerol (k) (n = 4 per group) as substrates. a–d, g, i–k data present mean ± s.e.m. a–e n presents biologically independent animals. f–k, n represents biologically independent cell samples. b–d, g, j, k two-tailed unpaired Student’s t test; a, i one-way ANOVA. All experiments here were repeated independently three times with similar results. Source data are provided as a Source Data file.
Then, we knocked down Kmt5c by transducing the lentiviral small hairpin RNA (shRNA) against Kmt5c into HepG2 cells and analyzed the gene expression profile involved in hepatic glucose and lipid metabolism. The diminished H4K20me3 levels indicated efficient knockdown of Kmt5c (Supplementary Fig. 1a). Losing KMT5C expression dramatically reduced the mRNA levels of Pc (pyruvate carboxylase), Pck1 (phosphoenolpyruvate carboxykinase 1) and G6pc (glucose-6-phosphatase, catalytic) which were the key enzymes of gluconeogenesis (Supplementary Fig. 1b). As for genes responsible for glycogen metabolism, glycolysis, fatty acid β-oxidation and lipogenesis, there was no difference between the control and Kmt5c knockdown cells (Supplementary Fig. 1b). Similar gene expression pattern was also shown in the primary hepatocytes (Fig. 1f–i). Loss of Kmt5c in primary hepatocytes was manipulated by infecting Cre recombinase adenovirus to Kmt5cflox/flox hepatocytes (Fig. 1f, g) or by isolation of liver cells from the wild-type and liver-specific Kmt5c knockout mice (Fig. 1h, i). The glucose production assay in vitro also revealed the Kmt5c knockout liver cells bore a compromised ability to produce glucose with sodium lactate and sodium pyruvate or glycerol as substrates (Fig. 1j, k).
Liver-specific KMT5C knockout mice show impaired gluconeogenesis in vivo
Liver-specific Kmt5c deficient mice were generated by crossing Kmt5c floxed mice to the Albumin-Cre transgenic line (Supplementary Fig. 2a). Quantitative PCR reassured that Alb promoter-driven Cre recombinase specifically reduced hepatic Kmt5c expression and H4K20me3 level (Supplementary Fig. 2b–d). The mice deficient Kmt5c in the liver (KO; Kmt5cflox/flox with Alb-Cre+) had the same growth curve of body weight and equivalent food intake as the control wild type (WT; Kmt5cflox/flox) littermates (Supplementary Fig. 2e, f). The liver weight and the cellular and tissue structure, as revealed by hematoxylin and eosin (H&E) staining were comparable between the genotypes (Supplementary Fig. 2g, h).
Gluconeogenesis is a distinctive function of hepatocytes to supply the need for plasma glucose between meals by de novo synthesis of glucose from non-carbohydrate precursors. Therefore, we starved the mice for 24 h to induce gluconeogenesis in a physiological setting. The KO mice had lower fasting blood glucose levels but alike random blood glucose levels relative to the WT mice (Fig. 2a). Accordingly, the KMT5C ablation reduced the protein and mRNA levels of gluconeogenic genes (Fig. 2b and Supplementary Fig. 2i), without altering the expression of other glucose and lipid metabolism genes and triglyceride contents in the liver (Supplementary Fig. 2i, j). The circulating free fatty acids and glycerol were possibly contributing to the gluconeogenesis process. However, the WT and KO mice possessed comparable levels of serum non-esterified fatty acids (NEFA) and glycerol after overnight fasting (Supplementary Fig. 2k, l). As expected, the KO mice, no matter at the age of 2 months, 4 months, or 11 months, produced less glucose from pyruvate (pyruvate tolerance test, PTT), glycerol (glycerol tolerance test, GlTT) as well as upon stimulation by glucagon (glucagon tolerance test, GcTT) (Fig. 2c–g). It is well established that blood glucose homeostasis is tightly controlled by hormones. The observed lower fasting blood glucose level in the KO mice was not due to the aberrant hormone secretion since deficiency of KMT5C in livers did not influence the serum insulin, glucagon, or corticosterone levels (Fig. 2h–j). The scarcity of hepatic KMT5C did not affect the systemic insulin sensitivity either, as indicated by the comparable glucose and insulin tolerance between genotypes (Supplementary Fig. 2m–p). In summary, hepatic Kmt5c knockout appears to reduce gluconeogenesis by suppressing gluconeogenic gene expression.
a Blood glucose levels were measured from 2-month-old (left) (n = 7 each group, female) and 11-month-old (right) (n = 7 each group, male) WT and KO mice under feeding or 24-hour fasting condition. b Western blot analysis (left) and protein quantification (right) of indicated proteins in the livers of 12-week-old male WT and KO mice after 24 h fasting (n = 5 each group). c, d Pyruvate tolerance test was performed in 2-month-old (c) (n = 7 each group, female) and 11-month-old mice (d) (n = 7 each group, male). e, f Glucagon tolerance test was performed in mice of (c) and (d) (n = 7 each group, female for c, male for d). g Glycerol tolerance test was performed in 4-month-old WT and KO mice (n = 6 each group, male). h–j Serum insulin (h) (n = 7 each group), glucagon (i) (n = 7 each group), and corticosterone levels (j) (n = 5 each group) in 12-week-old male WT and KO mice under fed and 24 h fasting conditions. a–j data present mean ± s.e.m; n shown here presents biologically independent animals. a–j two-tailed unpaired Student’s t test. All experiments here were repeated independently three times with similar results. Source data are provided as a Source Data file.
KMT5C stimulates gluconeogenesis independent of its methyltransferase activity
Next, we queried whether an increased KMT5C level in hepatocytes, imitating the higher KMT5C level in obese mice (see Fig. 1b–d), could stimulate gluconeogenesis. Overexpressing Kmt5c in primary hepatocytes by lentivirus-mediated transduction elevated the protein levels of PCK1 and G6PC (Fig. 3a). KMT5C catalyzes the generation of trimethylation of histone H4 lysine 20 (H4K20me3) on gene locus, that mediates gene repression and maintenance of heterochromatin. The catalytic activity of KMT5C is usually required for its regulation of gene expression. Surprisingly, the KMT5C G140Y/W174A mutant (namely Mut) that lost methyltransferase activity15 was still able to stimulate gluconeogenic gene expression as well as glucose output in the primary hepatocytes, which was indistinguishable from the KMT5C (Fig. 3a, b). In addition, the KMT5C and KMT5C Mut recovered the PCK1 level in the primary KO hepatocytes to a similar degree (Fig. 3c). A-196 is a substrate-competitive inhibitor of KMT5B and KMT5C18. By using A-196 to treat primary hepatocytes, the H4K20me3 level was reduced in a dose-dependent manner, however, the PCK1, G6PC and PC levels were not affected (Fig. 3d).
a Representative western blot (left) (n = 2 per group) and densitometry analyses (right) (n = 4 per group) of indicated proteins in primary hepatocytes. Primary hepatocytes isolated from 12-week-old C57BL/6 mice were infected with lentiviruses encoding either Kmt5c or Kmt5c Mut for 48 h, followed by western blot analysis. b Glucose production using pyruvate and lactate as substrates was measured in the culture medium of primary hepatocytes treated as in (a) (n = 3 per group). c Western blot analysis (top) and protein quantifications (bottom) in primary hepatocytes (n = 3 per group). Primary hepatocytes isolated from 12-week-old female WT and KO mice were infected with Kmt5c or Kmt5c Mut lentiviruses for 48 h, followed by western blot analysis. d Western blot (left) and densitometry analyses (right) of indicated proteins in primary hepatocytes (n = 3 per group). Primary hepatocytes isolated from 12-week-old C57BL/6 male mice were treated with A-196 for 72 h. e Eight-week-old C57BL/6 male mice were injected with AAV8-TBG-Ctrl (n = 6), AAV8-TBG-KMT5C (n = 7), or AAV8-TBG-Mut KMT5C (n = 8) viruses. Blood glucose levels were measured 3 weeks after virus infusion underfeeding and 16 h fasting conditions. f, g PTT (f) (n = 6 each group, male) and GcTT (g) (AAV-Ctrl, n = 6; AAV-KMT5C, n = 7; AAV-Mut, n = 7, male) were performed in mice of (e). h, i Quantitative PCR (h) (AAV-Ctrl, n = 6; AAV-KMT5C, n = 7; AAV-Mut, n = 8, male) and western blot (i) (AAV-Ctrl, n = 5; AAV-KMT5C, n = 5; AAV-Mut, n = 4, male) detected gluconeogenic gene expression in mice of (e). The densitometry analysis was shown in (i, right). Mice were fasted for 16 h before the assay. j, k Representative KMT5C (j) (WT + IgG, n = 2; WT + αKMT5C, n = 5; KO + IgG, n = 2; KO + αKMT5C, n = 5, male) and H4K20me3 (k) (WT + IgG, n = 2; WT + αH4K20me3, n = 4; KO + IgG, n = 2; KO + αH4K20me3, n = 5, male) ChIP assays performed in the WT and KO mice livers after 24 h fasting. Zfp560 and Rdna loci were used as positive controls for H4K20me3 enrichment. a–k data present mean ± s.e.m. a–d n represents biologically independent cell samples; e–k n presents biologically independent animals. a–k statistical analysis was performed using one-way ANOVA. All experiments here were repeated independently three times with similar results. Source data are provided as a Source Data file.
Then, the overexpression of either Kmt5c or Kmt5c Mut in mouse livers was achieved by intravenous injection of adeno-associated virus 8 (AAV8) coding for the expression of KMT5C from the hepatocyte-specific thyroxine-binding globulin (TBG) promoter (Supplementary Fig. 3a, b). The transient overexpression of KMT5C for 3 weeks in adult mouse livers significantly increased the H4K20me3 level in the livers but did not cause any alteration in mice body weight, liver mass, liver triglyceride contents, and hepatic histology (Supplementary Fig. 3c–g). Although enhanced hepatic KMT5C expression did not alter the gluconeogenesis gene or other major metabolic gene expression in the liver at fed state (Supplementary Fig. 3h, i), it elevated fasting blood glucose levels, augmented the ability of gluconeogenesis in response to both starvation and glucagon by upregulating the expression of key gluconeogenic enzymes (Fig. 3e–i). All these effects did not connect with the KMT5C methyltransferase activity, considering that the KMT5C Mut was fully competent to stimulate gluconeogenesis and the related gene expression in vivo (Fig. 3e–i). Alike to the KMT5C KO mice, the serum insulin, glucagon or corticosterone levels, and glucose tolerance were similar among groups (Supplementary Fig. 3j–n).
To further explore the relationship between the KMT5C catalytic activity and gluconeogenic gene expression, a chromatin immunoprecipitation (ChIP) assay was carried out with the KMT5C or H4K20me3 antibody. Endogenous KMT5C, AAV-expressed HA-KMT5C, or its catalytic dead mutant associated with Pck1 and G6pc promoters in fasted C57BL/6 mouse livers (Fig. 3j, o). As reported19,20, KMT5C imprinted H4K20me3 on the promoter regions of Zfp560, and the genomic DNA loci coding ribosomal RNAs in the WT mice livers, and deleting Kmt5c as shown in the KO mice dramatically reduced the abundance of H4K20me3 modification on their promoters (Fig. 3k). However, there was few H4K20me3 mark enriched on Pck1 and G6pc promoters in the both WT and KO mice (Fig. 3k). Thereby, KMT5C apparently regulated hepatic gluconeogenesis in a methyltransferase-independent manner.
KMT5C extends the half-life time of PGC-1α by reducing its ubiquitination
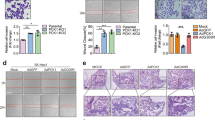
Transcriptional regulation plays an important part in gluconeogenesis modulation. Transcriptional factors (TFs) respond to hormone or nutrient hints to turn on or off gluconeogenic gene expression. KMT5C affected gluconeogenic gene expression by adjusting their mRNA levels, so we assumed transcriptional (co)factors might be involved in the KMT5C regulation pathway. We detected the protein levels of transcriptional (co)factors, including PGC-1α, CRTC2, FOXO1, CREB1, and HNF4α, which were well-established regulators of gluconeogenesis, in the KMT5C knockout and overexpression livers or hepatocytes. PGC-1α was moderately but significantly downregulated by KMT5C deficiency (Fig. 4a and Supplementary Fig. 4a). In contrast, overexpressing KMT5C raised the level of PGC-1α (Fig. 4b and Supplementary Fig. 4b). When we infected lentivirus carrying Pgc-1α to the cultured knockout hepatocytes, PGC-1α perfectly restored the expression of PCK1 and G6PC (Supplementary Fig. 4c). Moreover, AAV-mediated PGC-1α expression in the KO mice liver rehabilitated PCK1 and G6PC levels, and repaired the defect in gluconeogenesis shown in the KO mice (Fig. 4c–e), while the glucose and insulin tolerance did not alter by the AAV-PGC-1α expression (Supplementary Fig. 4d, e). In addition, KMT5C and PGC-1α brought about the synergistic effect on PCK1 and G6PC expression in primary hepatocytes (Fig. 4f). When knocking down Pgc-1α, however, KMT5C failed to upregulate the expression of PCK1 and G6PC (Fig. 4g), implying that the existence of PGC-1α was essential for the role of KMT5C to foster gluconeogenesis.
a Western blot analysis (top) (n = 5 each group) and protein quantification (bottom) of indicated proteins in the livers from 12-week-old female WT and KO mice after 24 h fasting. b Western blot (top) and densitometry analyses (bottom) were conducted in mouse livers expressing AAV-KMT5C or AAV-Mut (AAV-Ctrl, n = 5; AAV-KMT5C, n = 5; AAV-Mut, n = 4, male). Mice were fasted for 16 h before the assay. c AAV8-TBG-Ctrl or AAV8-TBG-Pgc-1α viruses were infused into the WT and KO mice. Four weeks later, the mice were starved for 24 h followed by western blot (top) analysis. The quantification of proteins was shown at bottom (WT + AAV-Ctrl, n = 5; KO + AAV-Ctrl, n = 4; KO + AAV-Pgc-1α, n = 5, 12-week-old male mice). d, e PTT (d) (n = 7 each group, male) and GcTT (e) (n = 7 each group, male) were performed in mice of (c) in the 3rd weeks after AAV infusion. f Representative western blot (left) and densitometry analysis (right) in primary hepatocytes expressing lentiviral Pgc-1α, Kmt5c, or Kmt5c Mut (n = 3 per group). Western blot analysis was performed 48 hours after lentivirus-mediated gene expression. g Lentiviruses carrying the shRNA targeting Pgc-1α infected Kmt5c-overexpressing primary hepatocytes for 48 h. Indicated proteins were detected by western blot analysis (n = 2 per group). h Co-immunoprecipitation analysis was conducted to assess the interaction between PGC-1α and KMT5C in liver tissues. Mice were starved for 16 h following the immunoprecipitation with PGC-1α antibody in the hepatic nuclear extracts. i The binding affinity between PGC-1α and KMT5C was determined by microscale thermophoresis (MST) assay (n = 4 per group). Inset: thermophoretic movement of fluorescently labeled proteins. Kd represents the dissociation constant. j Immunoblot analysis was conducted to assess the ubiquitination level of PGC-1α in Kmt5c-overexpressed primary hepatocytes. Primary hepatocytes were infected with lentivirus expressing vector, KMT5C, or Mut for 48 h, followed by treatment with 10 µM MG132 for 6 h. Then the ubiquitination of PGC-1α was analyzed. k Immunoblot to detect the ubiquitination level of PGC-1α in primary hepatocytes. Primary hepatocytes isolated from WT and KO mice were treated with 10 µM MG132 for 6 hours, followed by immunoprecipitation and western blot analysis. a–f, i data present mean ± s.e.m. a–e, n presents biologically independent animals; f, g, i, n represents biologically independent cell samples. a two-tailed unpaired Student’s t test; b–f one-way ANOVA. All experiments here were repeated independently three times with similar results. Source data are provided as a Source Data file.
PGC-1α is a critical transcriptional coactivator in gluconeogenesis. Next, we examined how KMT5C affected the PGC-1α level. Given the fact of the invariable Pgc-1α mRNA levels in the KMT5C knockout or overexpressing hepatocytes (Supplementary Fig. 4f–h), we speculated that KMT5C modifies the protein level of PGC-1α post-transcriptionally and may interact with each other. In fact, both KMT5C and KMT5C Mut were detected to attach to PGC-1α when they were co-expressed by plasmid transfection in HEK293T cells (Supplementary Fig. 4i). Deficiency of the N-terminal, internal or C-terminal part of KMT5C did not impair its association with PGC-1α (Supplementary Fig. 4j), indicating more than one region in KMT5C mediated interaction with PGC-1α. More importantly, the endogenous KMT5C also existed in the PGC-1α immunoprecipitations from mouse liver tissues, indicating hepatic KMT5C and PGC-1α were in the same protein complex (Fig. 4h). A microscale thermophoresis (MST) assay was performed to quantify the dissociation constant (Kd) of 1.733 μΜ for KMT5C binding PGC-1α (Fig. 4i). The proteasome inhibitor MG132 could increase the PGC-1α level in the KMT5C KO hepatocytes (Supplementary Fig. 4k), which suggested the ubiquitin-proteasome system may be involved in tuning down the PGC-1α level in KO cells. The presence of both KMT5C and KMT5C Mut markedly reduced the ubiquitination of PGC-1α, on the contrary, the absence of KMT5C increased the ubiquitination of PGC-1α in primary hepatocytes (Supplementary Fig. 4j, k). Therefore, it was not surprised to see that the half-life time of PGC-1α was shortened in the KMT5C KO liver cells as compared to in the WT hepatocytes (Supplementary Fig. 4l).
KMT5C competes against RNF34 to retard ubiquitination-mediated PGC-1α degradation
To investigate the molecular mechanism by which KMT5C decreased PGC-1α ubiquitination, we focused on the two reported E3 ubiquitin ligase for PGC-1α, RNF34 (ring finger protein 34) and FBXW7 (F-box and WD repeat domain containing 7)21,22. We first verified that expressing either RNF34 or FBXW7 decreased the PGC-1α level in HepG2 cells (Supplementary Fig. 5a). However, KMT5C could combat RNF34, but not FBXW7, to reverse the reduction of PGC-1α (Supplementary Fig. 5a). Both KMT5C and its mutant could recover the reduction in Gal4-Pgc1α activity caused by appearance of RNF34 in the reporter assay (Supplementary Fig. 5b). Moreover, liver tissue and hepatocytes expressed a much higher level of endogenous Rnf34 than Fbxw7 (Supplementary Fig. 5c). Thus, we explored whether RNF34-mediated PGC-1α ubiquitination got involved in the PGC-1α reduction in the KMT5C KO cells. Immunoprecipitation assay revealed that both KMT5C and RNF34 interacted with the C-terminal (amino acid 565 to 797 fragment) of PGC-1α (Fig. 5b, c), implying that they might compete with each other to bind to PGC-1α. Increased amount of KMT5C really dampened RNF34 association with PGC-1α (Fig. 5d). As expected, KMT5C greatly extended the half-life time of PGC-1α in the presence of RNF34 in HEK293T cells (Fig. 5e). Knocking down RNF34 expression fully restored the declined PGC-1α level and reconstituted the depressed glucose production in the primary KO liver cells (Fig. 5f, g). Even when we knocked down hepatic RNF34 level in the KO mice by infusion of AAV8-TBG-shRNF34 virus (Supplementary Fig. 5d, e), the suppressed capacity of gluconeogenesis was revived, as demonstrated by the increased fasting blood glucose level, restored pyruvate and glucagon tolerance, and recovered PCK1, PC expression with no alternation in serum insulin levels (Fig. 5h–l). However, KMT5C overexpression could not further upregulate PGC-1α and PCK1 levels in the RNF34 knockdown cells, suggesting KMT5C and loss of RNF34 do not exert a synergistic effect on PGC-1α and its target gene expression (Supplementary Fig. 5f, g).
a HEK293T cells were co-transfected with plasmids as indicated for 48 h, followed by treatment with 10 µM MG132 for 6 h. Immunoprecipitation was then performed. b, c The truncated HA-Pgc-1α plasmids and Flag-Kmt5c (b) or Flag-Rnf34 (c) were transfected into HEK293T cells. Immunoprecipitation was conducted by HA antibody after transfection for 36 h. d The indicated plasmids were transfected into HEK293T cells. Immunoprecipitation was conducted to show the interaction among PGC-1α, RNF34, and KMT5C (upper). The level of PGC-1α associated with RNF34 was quantified (lower panel, n = 3). e The indicated plasmids were transfected into HEK293T cells. At 36 h after transfection, cells were treated with CHX (50 μM) for the indicated time, followed by western blot analysis. f Western blot (left) and densitometry analyses (right) of indicated proteins in primary hepatocytes (n = 3 per group). Primary hepatocytes isolated from 2-month-old WT and KO mice were infected with shRnf34 or control lentivirus for 48 h following the western blot analysis. g Glucose production using pyruvate and lactate as substrates was measured in the primary hepatocytes. The cells were treated as in (f) (n = 3 per group). h TBG promoter-driven AAV-shRnf34 and AAV-Scr were injected into 2-month-old WT and KO mice. After three weeks, blood glucose levels were measured under both fed (n = 7 each group, male) and 16-hour fasting conditions (n = 7 each group, male). i, j PTT (i) (n = 6 each group, male) and GcTT (j) (n = 6 each group, male) assay in the mice of (h). k Representative western blot analysis (top) and densitometry analyses (bottom) in the livers of the indicated mice after 16 h fasting (WT + Scr, n = 3; KO + Scr, n = 3; WT + shRnf34, n = 4; KO + shRnf34, n = 4, male). Mice were treated as in (h). l Serum insulin levels in mice of (h) underfeeding and 16-hour fasting condition (WT + Scr, n = 7; KO + Scr, n = 7; WT + shRnf34, n = 6; KO + shRnf34, n = 7, male). m Immunoprecipitation assay to detect interaction among PGC-1α, RNF34, and KMT5C in the mouse liver. Liver nuclear extracts were prepared from fed and 24h-fasting mice. The PGC-1α protein complex was immunoprecipitated using PGC-1α antibody, followed by western blot analysis with a similar amount of PGC-1α in input and IP samples. n Immunoprecipitation assay to detect interaction among PGC-1α, RNF34, and KMT5C proteins in primary hepatocytes. Primary hepatocytes isolated from C57BL/6 mice were treated with 100 nM glucagon for 16 h, followed by immunoprecipitation using PGC-1α antibody. KMT5C and RNF34 levels in the precipitates were analyzed by western blot. d, f–l, data present mean ± s.e.m. d, f, g n represents biologically independent cell samples; (h–l) n presents biologically independent animals. Significance analysis was performed using one-way ANOVA. All experiments here were repeated independently three times with similar results. Source data are provided as a Source Data file.
Fasting induced hepatic KMT5C (see Fig. 1a) and PGC-1α expression, but had no impact on the RNF34 level (Supplementary Fig. 5h). The increased KMT5C in the fasted mouse livers predominated over RNF34 to attach to PGC-1α (Fig. 5m), which could partly contribute to the increased PGC-1α protein level during starvation. Similarly, glucagon treatment increased the KMT5C level in primary hepatocytes (Supplementary Fig. 5i), and more endogenous KMT5C interacting with PGC-1α was observed in primary hepatocytes after glucagon stimulus (Fig. 5n). As a transcriptional coactivator, PGC-1α collaborates with distinct transcription factors to regulate gluconeogenesis or fatty acid oxidation in hepatocytes. By immunoprecipitation assay, we found KMT5C was confined into the HNF4α transcriptional machinery in hepatocytes, but not in the PPARα or FOXO1 complex (Supplementary Fig. 5j), which may explain only gluconeogenesis but not fatty acid β-oxidation was downstream of KMT5C in hepatocytes. In addition, RNF34 inhibited PGC-1α-mediated activation of Pck1 and G6pc promoters when assayed in luciferase reporters. Whereas, KMT5C or the mutant KMT5C could combat RNF34 to restore the activities of Pck1 and G6pc promoters (Supplementary Fig. 5k, l). These combined results displayed that KMT5C modulated the PGC-1α level by competing with RNF34 to limit PGC-1α ubiquitination and degradation.
Targeting KMT5C retrieves disordered gluconeogenesis in diabetic mice
Type 2 diabetes is characterized by hepatic insulin resistance and increased hepatic glucose production due to glucagon-induced gluconeogenesis and lack of suppression of glycogenolysis23,24. Genetically diabetic db/db mice showed elevated levels of gluconeogenic enzymes25 and KMT5C in livers (see Fig. 1c). Knocking down the KMT5C in the livers of db/db mice by adenovirus injection decreased fasting glucose levels, suppressed gluconeogenesis and downregulated hepatic gluconeogenic gene and PGC-1α levels (Fig. 6a–d). Furthermore, decreasing KMT5C level tended to improve glucose intolerance without interference in insulin tolerance in db/db mice (Fig. 6e–g). db/db mice with decreased hepatic KMT5C expression had equal amounts of food intake, similar body weight, liver mass, liver triglyceride levels, and histology as compared to the mice administrated with the control virus (Supplementary Fig. 6a–e).
a Five-week-old db/db mice were injected with adenoviruses encoding murine Kmt5c shRNA via the tail vein. Two weeks later, blood glucose levels were measured underfed (n = 6 in each group, male) and 16 h fasting conditions (n = 6 each group, male). b, c PTT (b) (n = 6 per group, male) and GcTT (c) (n = 6 each group, male) were performed in the mice treated as in (a). d Representative western blot analysis (left) and protein quantifications (right) in the indicated mice after 16 h fasting (n = 6 for each group, male mice). e–g GTT (e) (db/db+Scr, n = 6; db/db+shKmt5c, n = 7, male) and ITT (g) (n = 6 each group, male) were performed in the mice described in (a). The quantification of GTT was shown in (f). h Five-week-old WT and KO male mice were fed a high-fat diet for 12 weeks. Blood glucose levels were then measured under fed conditions (n = 7 in each group, male mice) and after 24-hour fasting (n = 7 in each group, male mice). i–l PTT (i), GcTT (j), GTT (k), and ITT (l) assay were performed in the mice of (h) (n = 7 each group, male mice). m Pearson correlation analysis for normalized Kmt5c mRNA levels versus fasting plasma glucose levels in human subjects (n = 19). n, o Relative Kmt5c mRNA level (n) (Non-T2DM, n = 11; T2DM, n = 8) and representative KMT5C protein level (o) (Non-T2DM, n = 6; T2DM, n = 7) in the human liver specimens. a–o data present mean ± s.e.m; n represent biologically independent animal/human samples. a–l, n, o two-tailed unpaired Student’s t test. m Pearson correlation analysis. Experiments in (a–l) were repeated independently three times with similar results. Source data are provided as a Source Data file.
Next, the WT and Kmt5c KO mice were fed a high-fat diet (HFD) for 12 weeks to induce obesity. The deficiency of KMT5C in the liver did not interfere with the body weight gain. The liver weight, the hepatic triglyceride contents, and histological morphology were comparable between the WT and KO mice after the HFD feeding (Supplementary Fig. 6f–i). The HFD-induced obese KO mice still kept lower hepatic glucose output and even showed improved glucose and insulin tolerance (Fig. 6h–l). Consistently, the reduced G6PC and PGC-1α levels were observed in the HFD-fed KO mice (Supplementary Fig. 6j). Moreover, the hepatic Kmt5c mRNA level in humans positively correlated with the fasting blood glucose level (Fig. 6m). Both the mRNA and protein levels of KMT5C were significantly greater in livers from type 2 diabetic patients than from the non-diabetes controls (Fig. 6n, o). Thus, these results indicate that decreasing hepatic KMT5C expression could improve glucose homeostasis in prediabetic and diabetic conditions.
CREB1 transcriptionally regulates Kmt5c expression in hepatocytes
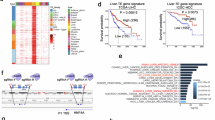
Hepatic KMT5C itself was upregulated during fasting or in diabetic mouse models and human subjects. We queried the signaling cascade responsible for KMT5C induction in these physiological and pathological situations. The induction of hepatic Kmt5c was about a 2-fold increase in its mRNA level after 18 to 24 h starvation (Fig. 7a), indicating transcriptional regulation played an important role in Kmt5c expression during fasting. Much of the gluconeogenesis during fasting and diabetes is mediated by glucagon-dependent gene transcription5. The protein level of KMT5C was increased by glucagon stimulus in primary hepatocytes (see Supplementary Fig. 5i). We wondered how glucagon cascade activates KMT5C expression. CREB1 is a major glucagon-activated transcriptional factor. It binds to cAMP response elements (CREs, TGACGTCA) or to a half palindrome (CGTCA or TGACG)5 to stimulate target gene expression. There are 2 CRE half-sites existing in the Kmt5c promoter. ChIP assay showed that CREB1 was enriched on the proximal CRE site (Fig. 7b). Overexpressing or knocking down CREB1 in primary hepatocytes increased or reduced Kmt5c expression on the mRNA and protein levels (Supplementary Fig. 7a–d). Among the CREB1-associated transcriptional cofactors, only interference of Crtc2 led to dramatic KMT5C downregulation in cultured hepatocytes (Supplementary Fig. 7e–g). The transcriptional activation effects on Kmt5c expression by CREB1 and CRTC2 have also been confirmed in vivo. Transient expression shRNAs targeting Creb1 or Crtc2 through hydrodynamics-based plasmid transfection to mouse liver suppressed Kmt5c induction upon fasting signal (Fig. 7c–f). By contrast, overexpressing Creb1 in mouse liver dramatically elevated the Kmt5c level at the fed state (Fig. 7g–i). In summary, we identified KMT5C as a regulator of hepatic gluconeogenesis. The glucagon-CREB-CRTC axis triggers both Kmt5c and Pgc-1α transcription, and KMT5C consecutively facilitates PGC-1α protein stabilization by decreasing its ubiquitination and degradation. Loss of methyltransferase activity does not impair KMT5C’s capability to inspire gluconeogenesis, since this functional role of KMT5C is accomplished through competitive advantage over RNF34 to bind to PGC-1α (Fig. 7j).
a Kmt5c mRNA levels in the fasted mice livers. Eight-week-old mice were fasting for 6, 12, 18, and 24 h, followed by quantitative PCR analysis (n = 4 for each group, male mice). b Primary hepatocytes were infected with HA-Creb1 lentivirus for 48 h, followed by chromatin immunoprecipitation (ChIP) using an HA antibody. The enrichment of the Kmt5c promoter was quantified by quantitative PCR analysis (n = 3 per group). c–f Quantitative PCR analysis (c, d) (n = 5 per group, male mice) and representative western blot analysis (f) (n = 4 per group, male mice) in the livers of mice. Ten-week-old mice were infused with shRNA plasmids against Creb1, Crtc2 through tail vein injection. Then the mice were fasted for 16 h following the gene expression analysis. The quantification of (f) was shown in (e). g–i Quantitative PCR (g) (n = 6 per group, male mice) and western blot analysis (i) (n = 6 per group, male mice) in the livers of mice. Ten-week-old mice were infused with pCMV vector or pCMV-Creb1 plasmids via tail vein injection. The mice were euthanized 16 h after injection at a fed state. The quantification of (i) was shown in (h). j A model for KMT5C regulating gluconeogenesis. CRTC2 cooperates with CREB1 to activate Kmt5c transcription in response to glucagon. KMT5C retards RNF34-mediated PGC-1α ubiquitination and subsequent degradation to promote gluconeogenesis. a–e, g, h data present mean ± s.e.m. a, c–i n represent biologically independent animal; (b) n represents biologically independent cell samples. a one-way ANOVA; (b–e, g, h) two-tailed unpaired Student’s t test. All experiments were repeated independently three times with similar results. Source data are provided as a Source Data file.
Discussion
Glucose is the principal metabolic substrate for most tissues to produce energy. Normal blood glucose levels are safeguarded within a narrow range by a series of hormones. Among them, glucagon is defined as a counter-regulatory hormone for insulin to raise blood glucose levels. Recently, FOXO1-S273 phosphorylation by p38α MAPK in answer to glucagon stimuli has been reported to increase FOXO1 protein stability, promoting glucose production in both health and disease26. The glucagon-CREB-KMT5C axis that reduces PGC-1α ubiquitination could be another case of protein stability regulation on transcriptional cofactors downstream of glucagon.
PGC-1α is a short-lived protein whose level is determined by its ubiquitination and subsequent proteasome-mediated degradation. RNF34 and FBXW7 are the two identified ubiquitin E3 ligases that mediate PGC-1α degradation either in brown adipocytes or in neurons21,22. Our data showed that the expression level of RNF34 extremely predominated over FBXW7 in livers. Knocking down hepatic RNF34 dramatically increased fasting blood glucose level, enhanced pyruvate tolerance as well as upregulated PGC-1α protein level (Fig. 5), indicating RNF34 is an essential PGC-1α partner in charge of its turnover in livers. However, glucagon or fasting-induced KMT5C broke the harmony of PGC-1α turnover during fed by occupying the C-terminus (amino acid 565-797) of PGC-1α to expel RNF34, therefore retarded its degradation. Previous studies showed that truncated PGC-1α 1-565 fragment was stable, and MG132 treatment could not further increase its protein level27, indicating the C-terminal was important for PGC-1α degradation. This is in agreement with RNF34 binding to the C-terminal of PGC-1α. As a CREB1-regulated gene, the increased Kmt5c mRNA and protein levels were observed in fasting mouse liver. Considering Pgc-1α transcription is also in the control of CREB28, the upregulated transcription and increased protein stability after prolonged starvation maximizes the PGC-1α induction for gluconeogenesis to accommodate the systemic glucose scarcity. Intriguingly, it has been demonstrated that KMT5C increases the transcription and translation of Pgc-1α in a methyltransferase activity-dependent manner in brown adipocytes15,29. Together with the results here, KMT5C may affect PGC-1α expression in diverse manners that are cell-type and context-dependent.
PGC-1α serves as a molecular switch of cell metabolism, including gluconeogenesis, fatty acid β-oxidation, and ketogenesis, by coordinating versatile transcriptional factors. In this study, PGC-1α reduction elicited by loss of KMT5C did not affect the gene expression involved in fatty acid β-oxidation, which is usually downstream of the PPARα-PGC-1α transcriptional complex. The observation that KMT5C did not interact with PPARα but specifically associated with HNF4α, indicates the KMT5C-PGC-1α complex may only exist on gluconeogenic gene promoters due to their association with HNF4α.
In this study, we found the hepatic KMT5C expression was induced by glucagon stimulus. Hyperglucagonemia is observed in prediabetic and T2DM patients30. Glucagon-stimulated hepatic glucose production has been linked to hyperglycemia during T2DM. In humans and mice with diabetes, the KMT5C level in livers was aberrantly higher than in the controls, which could be owing to hyperglucagonemia. Knockdown or deleting Kmt5c lowered the blood glucose levels by reducing PGC-1α-mediated gluconeogenic gene expression, without exacerbating liver steatosis. In view of the fundamental role of KMT5C and H4K20me3 modification in genomic integrity, targeting KMT5C-PGC-1α interaction by inhibitor may not affect the level of H4K20me3, but potentially increases ubiquitination-mediated PGC-1α degradation, which offers an alternative avenue to improve glucose homeostasis in diabetes.
Another interesting finding is the independence from KMT5C methyltransferase activity to promote hepatic gluconeogenesis, representing a novel working model for KMT5C. Actually, many histone-modifying enzymes have been reported to possess non-catalytic functions. The KMT5C paralog KMT5B was revealed catalytically independent roles that cause a detachment of nucleosomal DNA from the histone octamer and promote chromatin condensation31. EZH2, the canonical H3K27me3 methyltransferase in Polycomb Repressive Complex 2 (PRC2), can also recruit non-PRC2 factors and exert methyltransferase-independent transcriptional activation32. Histone methyltransferase G9a showed translation-regulatory function by promoting METTL3 expression at translational level33. A deep comprehension of the uncoupling of catalytic and non-catalytic roles of histone-modifying enzymes would shed light on new therapeutic strategies to target their dysregulation in diseases.
Methods
Animal studies
Animal experiments were accomplished according to procedures approved by the Fudan University Shanghai Medical College Animal Care and Use Committee. Kmt5c floxed mice were created by Beijing Biocytogen Co, Ltd. Albumin-Cre mice, db/db, and ob/ob mice were purchased from the Shanghai Model Organisms Center, Inc. (Shanghai, China). All mice were generated in the C57BL/6 background and were housed with 12 h light and 12 h dark cycle and with a chow diet (Shanghai SLAC Animal Inc, #P1101F; 12% energy from fat, 63% energy from carbohydrate) feeding ad libitum. For diet-induced obesity, 6-week mice were fed a high-fat diet (HFD) containing 60% energy from fat (Research Diet, #D12492,) for 12 weeks. Generally, experiments were conducted in 2-3-months mice, and mice were fasted 16 h to 24 h for gluconeogenic gene expression analysis unless specified. The mice were euthanized through carbon dioxide inhalation. For the TG content measurement in the liver, 50 mg of liver tissues were analyzed by a kit following the manufacture protocol (Applygen, E1025). Serum insulin (Mercodia, 10-1113-01), corticosterone (Nanjing Jiancheng, H205-1-1), and glucagon levels (Sangon biotech, HY-P0082) were measured by ELISA. Serum free fatty acid (Nanjing Jiancheng, A042-2-1), glycerol (Nanjing Jiancheng, F005-1-1) were measured by commercial kits. For histone extraction from liver tissues, 20 mg of liver tissue was homogenized according to the manufacturer’s protocol (EpiQuik, OP-0006-100). For hydrodynamics-based plasmid infusion to mouse liver, plasmids were purified using the endotoxin removal kit (Vazyme, DC202) and stored in saline. Mice were injected with 10 µg of the specified plasmid DNA in 1.5 ml of saline via the tail vein within 5 s. Subsequently, the mice were either fasted or fed for 16 h followed by the gene expression analysis.
Human studies
The human subjects (aged 28–76) were recruited from the Hangzhou First People’s Hospital. Liver samples were collected from surgical patients and divided into non-diabetic (n = 11) and type 2 diabetes groups (n = 8) according to clinic diagnosis. The detailed patient information is shown in Supplementary Table 1. Subjects who had type 1 diabetes, chronic viral hepatitis, hypothyroidism, excessive use of alcohol, use of drugs that may affect glucose metabolism, abnormal renal function, or infectious diseases were excluded. The human study was approved by the human research ethics committee of Hangzhou First People’s Hospital and performed following the declaration of Helsinki. Written informed consent was obtained from each subject.
GTT, PTT, GlTT, GcTT, and ITT
Mice were fasted for 16 h (GTT, PTT), 4 h (GITT), or 6 h (GcTT and ITT). Then glucose, sodium pyruvate, glycerol, glucagon, or insulin were administered through intraperitoneal injection for GTT, PTT, GlTT, GcTT, and ITT. Doses were 2 g/kg glucose, 2 g/kg sodium pyruvate, 2 g/kg glycerol, 10 μg/kg glucagon, and 0.5 μ/kg insulin for studies in non-diabetic models; 2 g/kg glucose, 2 g/kg sodium pyruvate, 10 μg/kg glucagon, and 0.75 μ/kg insulin for studies in HFD-fed mice; 0.5 g/kg glucose, 0.5 g/kg sodium pyruvate, 5 μg/kg glucagon, and 1 μ/kg insulin for studies in the db/db mice. Blood glucose levels were measured from tail bleeds at the indicated time after injection.
Histological analysis
Fresh mouse livers were dissected, fixed in formalin, and subsequently embedded in paraffin. Tissue sections were then cut and subjected to hematoxylin and eosin staining to observe cellular morphology (Shanghai Runner Bio Technology Co., Ltd).
Mouse primary hepatocyte isolation and cell culture
Primary hepatocyte isolation was performed using a previously described method17. After the mice had been anesthetized, the inferior vena cava was cannulated. The liver was perfused with Krebs-Ringer buffer (120 mM NaCl, 24 mM NaHCO3, 5 mM HEPES, 5.6 mM Glucose, 4.8 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, pH 7.4) with EGTA (1 mM) for 3 min, followed by continuous perfusion with the same buffer containing CaCl2 (5 mM) and 0.05% collagenase (Sigma, C5138) for 5 min. After digestion, hepatocytes were filtrated through a 100 μm cell strainer, purified by percoll (GE) purification solution, and collected after centrifugation (50 × g for 3–5 min at 4 °C). Hepatocytes and debris will be pelleted, while most NPCs will remain suspended. The supernatant containing the hepatocyte-depleted NPCs was transferred into a new tube. NPCs were pelleted by centrifugation at 500 × g for 5–7 min. Hepatocyte and NPCs pellets were lysed in lysis buffer followed by western blot or RNA analysis. Or hepatocytes were suspended in a culture medium and seeded (2 × 105 per well in a 12-well plate) into a culture plate or into a 6 cm dish coated with mouse tail collagen (Corning, C5138). HepG2 (ATCC, HB-8065) and HEK293T (ATCC, CRL-11268) cells were cultured in DMEM (Thermo) containing 10% fetal bovine serum (Hyclone, #30396.03) and 1% penicillin/streptomycin (Hyclone, #SV30010). The cell lines were maintained at 37 °C in a humidified environment with 5% CO2 and were assessed for mycoplasma contamination periodically to ensure the negative results.
Immunoprecipitation
The indicated plasmid or lentivirus constructs were expressed in HEK293T, HepG2, or primary hepatocytes with or without 100 nM glucagon treatment for 16 h. Total cell lysates were collected and incubated with PGC-1α antibody (Novus, NBP1-04676), Flag beads, or HA-beads (Smart-Lifesciences, SA068001, SA042001) for 3 h. Then, beads were washed for 3 times with wash buffer [200 mM NaCl, 0.5% Triton-X-100, 5% glycerol, 50 mM Tris-HCl (pH 7.5), 1 mM PMSF]. The immunoprecipitates were separated by SDS-PAGE for western blot.
For immunoprecipitation assay in the liver tissues, nuclear fractions were extracted, and PGC-1α protein levels were quantified by Western blot. The immunoprecipitates containing the same amount of PGC-1α protein were used to detect RNF34 and KMT5C.
For ubiquitination assay, HEK293T cells or primary hepatocytes with indicated plasmid or lentivirus transduction were treated with MG132 (5 μM) for overnight and then lysed in lysis buffer [100 mM NaCl, 0.5% Triton-X-100, 5% glycerol, 50 mM Tris-HCl (pH 7.5), 1 mM PMSF and protease inhibitor mixture cocktail, 5 μM MG132]. Immunoprecipitation assay was performed as described.
Purification of PGC-1α and KMT5C proteins
Full-length mouse Pgc-1α and Kmt5c were separately cloned into the PET-28a vector and transformed into Escherichia coli BL21(DE3) via induction with isopropyl β-D-thiogalactopyranoside. The recombinant his-PGC-1α and his-KMT5C proteins were purified using Ni-NTA beads (Smart Life Sciences, SA004005) and eluted with imidazole. The elution buffer was then passed through a centrifugal filter with a molecular cutoff of 10 kDa (Merk, UFC9010) at 500 × g for 30 min. Phosphate-buffered saline (PBS) was added for three consecutive washes. After protein quantification of his-KMT5C, thrombin was added to cleave his tag at 4 °C for 16 h, followed by filtration through a centrifugal filter with a molecular cutoff of 10 kDa. The purity and integrity of the purified protein were confirmed on an SDS-PAGE gel using Coomassie blue staining.
Microscale thermophoresis assay
Purified His-PGC-1α protein was labeled with a red-tris-NTA 2nd generation His-tag labeling kit (NanoTemper, #MO-L018, Germany). Then 50 nM labeled His-PGC-1α protein was mixed with twofold serial dilutions of purified KMT5C protein (the star concentration was 20 μM) in PBST buffer (PBS with 0.05% Tween 20). The samples were loaded in the capillaries (NanoTemper, #MO-K022, Germany) and measured by the Monolith X instrument (NanoTemper, Germany). Fnorm = F1/F0 (Fnorm: normalized fluorescence; F1: fluorescence after thermodiffusion; F0: initial fluorescence or fluorescence after T-jump). Kd was determined by the NanoTemper analysis software.
Dual-luciferase reporter assay
The 1 kb mouse Pck1 or G6pc promoter was cloned into the PGL3 vector and validated by DNA sequencing. HEK293T cells in 48-well plates were transfected with luciferase reporters, CMV-β-galactosidase plasmids, and the indicated expression constructs using PEI reagents (Life-iLab, AC04L091). The amounts of plasmid transfected are provided in the supplementary data. A luciferase assay was performed using the dual-luciferase reporter assay system after 48 h (Beyotime, RG029).
Plasmids
Ep300, Prmt5, Crtc2, Crebbp, Creb1, RNF34 short hairpin RNA (shRNA) lentiviral constructs were generated using psp108 vector (Addgene). Ep300 shRNA targeting sequence is: 5’-caggtacaagcaaagaatcaa-3’. Prmt5 shRNA targeting sequence is: 5’-GCACAGTTTGAGATGCCTTAT-3’. Crtc2 shRNA targeting sequence is: 5’-GAGGACTCATTCCGTAGTGAT-3’. Crebbp shRNA targeting sequence is: 5’-ATGCATCAGATTTGTGTTCTA-3’. Creb1 shRNA-1 targeting sequence is: 5’-CAGCAGCTCATGCAACATCAT-3’, shRNA-2 is: 5’-GCCTGAAAGCAACTACAGAAT-3’. Pgc-1α shRNA targeting sequence is: 5’-GGTGGATTGAAGTGGTGTAGC-3’. Rnf34 shRNA targeting sequence is: 5’-GGCACAGGTACAAAGTGAAAT-3’ (This sequence was used for AAV-shRnf34 construction). Kmt5c shRNA targeting sequence is: 5’- GAGAAGAATGAGCACTGTGAA-3’ (This sequence was used for shKmt5c adenoviruses construction). Full-length mouse Kmt5c cDNA and Kmt5c mutant G140Y/W174A were constructed as described previously15. Full-length mouse Rnf34, Pgc-1α, Creb1, the Kmt5c deletion mutants, and the truncated Pgc-1α cDNAs were amplified by PCR and cloned into the pcDNA3.3 vector and lentiviral entry vector pENTR1A (Addgene). The HA-Ub vector was a gift provided by Dr. Qi-Qun Tang, Fudan University.
Hepatic glucose production assay
Primary hepatocytes were isolated and cultured overnight in DMEM with 10% FBS. Cells were washed three times with PBS and incubated for 4 h in glucose production buffer (glucose-free DMEM containing 20 mM sodium lactate and 2 mM sodium pyruvate, without phenol red). For the glucose production assay using glycerol, cells were incubated in a glucose production buffer (glucose-free DMEM containing 5 mM glycerol, without phenol red) for 8 h. The medium was collected for glucose measurement (Applygen, E1010).
ChIP assay
CHIP assays were performed with antibodies against H4K20me3 (Abcam, ab9053), KMT5C (Abcam, ab91224), HA (Santa Cruz, sc-805X), or normal rabbit IgG (Sigma) as described previously15. Briefly, primary hepatocytes overexpressing HA-Creb1, HA-Kmt5c, or HA-Kmt5c mutant were fixed in 1% formaldehyde for 5 min at room temperature. 100 mg liver tissues from Kmt5c WT or KO mice were fixed and homogenized into a single-cell suspension using a glass homogenizer. Cells pellets were centrifuged at 1000 ×g for 5 min, suspended in low salt buffer [10 mM HEPES (pH 7.9), 10 mM KCl, 0.1 mM EDTA, and proteinase inhibitor], swelled on ice for 15 min, and added NP-40 to a final concentration of 0.6%. After centrifugation, the pellet was considered as the cellular nuclear fraction. The nuclear pellets were suspended in high salt buffer [20 mM HEPES (pH 7.9), 0.4 M NaCl, 10% glycerol, 1 mM EDTA, and proteinase inhibitor], incubated for 30 min at 4 °C, and collected after centrifuge.
The primary hepatocytes or nuclear fraction of livers were suspended in SDS lysis buffer and sonicated to shear the DNA into 100–1000 bp by ultrasonic processor (Cole Parmer). One percent of the chromatin sample was saved as input control. The sheared chromatin was immunoprecipitated with antibodies overnight. The immunoprecipitants were washed for 5 times and reversed crosslinked by heating at 65 °C overnight. The DNA samples were purified with QIAquick PCR Purification PCR kits and applied to quantification by QPCR.
Adeno-associated virus, adenoviruses and lentiviruses
Adeno-associated virus 8 constructs were generated and packaged by OBIO Tech (Shanghai). All the constructs were under the thyroxine binding globin (TBG) promoter to ensure their specific expression in the liver. AAV (2 × 1011 genome copies per mouse) was delivered into the mice via tail vein injection.
Adenoviruses were purified with cesium chloride ultracentrifugation, and virus tilters were assessed in HEK293T cells by calculating green fluorescent protein-positive cells. Cre or GFP adenovirus-infected primary hepatocytes isolated from Kmt5cflox/flox mice to generate Kmt5c knockout cells. Equivalent particles of adenoviruses (2 × 109 PFU per mouse) were injected into 8-week-old male db/db mice via the tail vein at 3-day intervals. GTT, ITT, PTT and GcTT were performed after injection twice. After the third injection, the mice were sacrificed for analysis.
For lentivirus production, lentiviral vectors and the packaging plasmids were co-transfected to HEK293T cells. The virus-containing medium was collected two days after transfection, passed through a 0.22 μM filter (millipore), overlaid on a 5 X PEG buffer at a 4:1 V/V ratio, and centrifuged at 4000 × g for 40 min at 4 °C. The viruses were resuspended with PBS and stored at − 80 °C.
Antibodies
The following primary antibodies were used: anti-HSP90 (13171-AP), anti-PGC-1α (66369-1-Ig for western blot), anti-PC (16588-1-AP), anti-Flag (66008-2-Ig), anti-α-Tubulin (66031-1-Ig), anti-Histone H4 (16047-1-AP), anti-Ubiquitin (10201-2-AP), from Proteintech; anti-KMT5C (WH140428), anti-FOXO1 (A25985), anti-CREB1 (A11064), anti-PCK1 (A2036), anti-G6PC (A16234), anti-ACLY (A22273) from Abclonal; Anti-PGC-1α (NBP1-04676 for IP), anti-RNF34 (NBP2-56413) from Novus bio; anti-HA (sc-805X), anti-HNF-4α (sc-374229), anti-CRTC2 (sc-271912) from Santa Cruz; anti-PPARα (ab314112) from abcam; anti-H4K20me3 (39672) from active motif. Secondary antibodies, anti-rabbit IgG (Vazyme, RA1008) and anti-mouse IgG (Vazyme, RA1009), were used for western blot following immunoprecipitation.
Western blot analysis
Tissue or cultured cells were lysed by cell lysis buffer (100 mM NaCl, 0.5% Triton-X-100, 5% glycerol, 50 mM Tris-HCl (pH 7.5), 1 mM PMSF, and protease inhibitor mixture cocktail). Supernatants were collected by centrifugation at 16000 × g for 10 min, followed by protein quantification using a BCA protein assay (Beyotime, P0011). Equal amounts of protein samples were separated by SDS-PAGE and then transferred to a PVDF membrane. The membrane was incubated with primary antibodies at 4 °C overnight, followed by incubation with secondary antibodies (YEASEN) for 1 h, and developed using a chemiluminescence (ECL) detection system.
RNA extraction and quantitative RT-PCR
Primary hepatocytes were treated with A-196 (MCE, HY-100201) for 72 h or glucagon for 12 h. Total RNA, including that from tissues and cultured cells, was isolated according to the standard method using a Trizol reagent (Vazyme, #R401-01). RNA was then reverse transcribed using HiScript II Q RT SuperMix (Vazyme, R222-01), followed by quantitative PCR (QPCR) using SYBR Green SuperMix (Vazyme, #Q711). The relative levels of gene expression were determined using the 2−ΔΔCt method after normalization to the housekeeping gene Rplp0. The primers used for qPCR are provided in Supplementary Table 2.
Statistics
Data are presented as mean ± sem. Statistical analysis was performed using GraphPad Prism v.8.0 (GraphPad software). Statistical significance was determined by using unpaired two-tailed Student’s t test for two-group comparisons and one-way ANOVA for multiple comparisons. At least three independent cell cultures were used for all in vitro analysis. No technical replicates were used.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All the data can be found in either the main text or the supplementary materials. Source data are provided in this paper.
References
Nordlie, R. C., Foster, J. D. & Lange, A. J. Regulation of glucose production by the liver. Annu. Rev. Nutr. 19, 379–406 (1999).
Pilkis, S. J. & Granner, D. Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu. Rev. Physiol. 54, 885–909 (1992).
Krssak, M. et al. Alterations in postprandial hepatic glycogen metabolism in type 2 diabetes. Diabetes 53, 3048–3056 (2004).
Magnusson, I., Rothman, D., Katz, L., Shulman, R. & Shulman, G. Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J Clin. Invest. 90, 1323–1327 (1992).
Goldstein, I. & Hager, G. L. The three Ds of transcription activation by glucagon: direct, delayed, and dynamic. Endocrinology 159, 206–216 (2018).
Koo, S. H. et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437, 1109–1111 (2005).
Rhee, J. et al. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc. Natl. Acad. Sci. USA 100, 4012–4017 (2003).
Puigserver, P. et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423, 550–555 (2003).
Song, T. et al. Prognosis in patients with serous and mucinous stage I borderline ovarian tumors. Int. J. Gynecol. Cancer 22, 770–777 (2012).
Lerin, C. et al. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 3, 429–438 (2006).
Li, X., Monks, B., Ge, Q. & Birnbaum, M. J. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature 447, 1012–1016 (2007).
Rodgers, J. T., Haas, W., Gygi, S. P. & Puigserver, P. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab. 11, 23–34 (2010).
Trausch-Azar, J., Leone, T. C., Kelly, D. P. & Schwartz, A. L. Ubiquitin proteasome-dependent degradation of the transcriptional coactivator PGC-1{alpha} via the N-terminal pathway. J. Biol. Chem. 285, 40192–40200 (2010).
Gabellini, D. & Pedrotti, S. The SUV4-20H histone methyltransferases in health and disease. Int. J. Mol. Sci. 23, https://doi.org/10.3390/ijms23094736 (2022).
Zhao, Q. et al. KMT5c modulates adipocyte thermogenesis by regulating Trp53 expression. Proc. Natl. Acad. Sci. USA 117, 22413–22422 (2020).
Su, Q. et al. Single-cell RNA transcriptome landscape of hepatocytes and non-parenchymal cells in healthy and NAFLD mouse liver. iScience 24, 103233 (2021).
Mohar, I., Brempelis, K. J., Murray, S. A., Ebrahimkhani, M. R. & Crispe, I. N. Isolation of Non-parenchymal Cells from the Mouse Liver. Methods Mol. Biol 1325, 3–17 (2015).
Bromberg, K. D. et al. The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity. Nat. Chem. Biol. 13, 317–324 (2017).
Magklara, A. et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell 145, 555–570 (2011).
Schmitz, K.-M., Mayer, C., Postepska, A. & Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269 (2010).
Wei, P., Pan, D., Mao, C. & Wang, Y. X. RNF34 is a cold-regulated E3 ubiquitin ligase for PGC-1α and modulates brown fat cell metabolism. Mol. Cell Biol. 32, 266–275 (2012).
Olson, B. L. et al. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 22, 252–264 (2008).
Basu, R., Schwenk, W. F. & Rizza, R. A. Both fasting glucose production and disappearance are abnormal in people with “mild” and “severe” type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 287, E55–E62 (2004).
Basu, R. et al. Pathogenesis of prediabetes: role of the liver in isolated fasting hyperglycemia and combined fasting and postprandial hyperglycemia. J. Clin. Endocrinol. Metab. 98, E409–E417 (2013).
Chan, T. M., Young, K. M., Hutson, N. J., Brumley, F. T. & Exton, J. H. Hepatic metabolism of genetically diabetic (db/db) mice. I. Carbohydrate metabolism. Am. J. Physiol. 229, 1702–1712 (1975).
Yang, W. et al. Hepatic p38α MAPK controls gluconeogenesis via FOXO1 phosphorylation at S273 during glucagon signalling in mice. Diabetologia 66, 1322–1339 (2023).
Sano, M. et al. Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor gamma coactivator 1alpha. J. Biol. Chem. 282, 25970–25980 (2007).
Herzig, S. et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413, 179–183 (2001).
Cui, X. et al. The histone methyltransferase SUV420H2 regulates brown and beige adipocyte thermogenesis. JCI Insight 9, https://doi.org/10.1172/jci.insight.164771 (2024).
Ichikawa, R. et al. Basal glucagon hypersecretion and response to oral glucose load in prediabetes and mild type 2 diabetes. Endocr. J. 66, 663–675 (2019).
Abini-Agbomson, S. et al. Catalytic and non-catalytic mechanisms of histone H4 lysine 20 methyltransferase SUV420H1. Mol .Cell 83, 2872–2883. (2023).
Wang, J. et al. EZH2 noncanonically binds cMyc and p300 through a cryptic transactivation domain to mediate gene activation and promote oncogenesis. Nat. Cell Biol. 24, 384–399 (2022).
Muneer, A. et al. Non-canonical function of histone methyltransferase G9a in the translational regulation of chronic inflammation. Cell Chem. Biol. 30, 1525–1541 (2023).
Acknowledgements
This work was supported by grants from National Key R&D Program of China (2018YFA0800401 to Q.T.), the National Natural Science Foundation of China (32171140 to D.P., 82400940 to Q.Zhao.), the Innovation Fund of Ministry of Education of China (2023GY006 to M.Z.), Program of Shanghai Academic Research Leader (22XD1400500 to D.P.), Zhejiang Provincial Natural Science Foundation of China (LQ23C070004 to Q.Zhao.), and the Special Funds of Taishan Scholars Project of Shandong Province (TSQN202312384 to Y.X.).
Author information
Authors and Affiliations
Contributions
Q.Zhao. performed most of the experiments and acquired data. X.C., Q.Zhu, and T.S. helped in animal experiments. F.L. isolated primary hepatocytes. H.L. and W.L. packaged adenovirus for this study. R.B., Y.X., Y.G., and M.Z. provided insightful and important discussion on the clinical issues. M.Z. shared important reagents and materials. D.P. designed the project, supervised the study and drafted the manuscript. Q.T. amended the manuscript. All the authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, Q., Cui, X., Zhu, Q. et al. Non-catalytic mechanisms of KMT5C regulating hepatic gluconeogenesis. Nat Commun 16, 1483 (2025). https://doi.org/10.1038/s41467-025-56696-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56696-y