Abstract
The transcriptional repressor FOXN3 plays a key role in regulating pulmonary inflammatory responses, which are crucial in the development of pulmonary fibrosis. However, its specific regulatory function in lung fibrosis remains unclear. Here, we show that FOXN3 suppresses pulmonary fibrosis by inhibiting Smad transcriptional activity. FOXN3 targets a substantial number of Smad response gene promoters, facilitating Smad4 ubiquitination, which disrupts the association of the Smad2/3/4 complex with chromatin and abolishes its transcriptional response. In response to pro-fibrotic stimuli, NEK6 phosphorylates FOXN3 at S412 and S416, leading to its degradation. The loss of FOXN3 inhibits β-TrCP-mediated ubiquitination of Smad4, stabilizing the Smad complex’s association with its responsive elements and promoting transcriptional activation, thus contributing to the development of pulmonary fibrosis. Notably, we found a significant inverse expression pattern between FOXN3 and Smad4 in clinical pulmonary fibrosis cases, underscoring the importance of the NEK6-FOXN3-Smad axis in the pathological process of pulmonary fibrosis.
Similar content being viewed by others
Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive, and irreversible respiratory disease characterized by inflammation and extensive lung remodeling1,2,3,4. Its origin is unknown, and it has a median survival rate of 3–5 years. Currently, there are only two drugs, pirfenidone and nintedanib, which show moderate efficacy in slowing down disease progression. Apart from these medications, there are no other pharmacologic treatments available. The development of IPF is characterized by abnormal alveolar repair and the proliferation of activated fibroblasts known as myofibroblasts. This leads to the accumulation of abnormal extracellular matrix (ECM) and tissue remodeling2,5,6. Although the exact cause of IPF is not fully understood, there is evidence suggesting that excessive epithelial-mesenchymal transition (EMT) plays a critical role in its development7,8. It has been found that approximately one-third of fibroblasts in pulmonary fibrosis originate from epithelial cells. During the EMT process, the levels of E-cadherin decrease in epithelial cells, while N-cadherin and α-smooth muscle actin (α-SMA) increase in mesenchymal cells. Myofibroblasts, which are derived from epithelial cells through EMT, undergo rapid proliferation and contribute to the excessive production of extracellular matrix9,10,11,12.
The transforming growth factor-β (TGF-β) and its related polypeptides comprise the largest cytokine family, playing a crucial role in regulating various cellular processes including proliferation, differentiation, adhesion, and apoptosis13,14,15,16. These TGF-β-like factors exert their signaling effects through two types of transmembrane receptors: the TGF-β type I receptor and the type II receptor. In addition, the intracellular substrates known as Smad proteins are involved in mediating these signaling responses. Upon binding with its ligand, the receptors become activated and proceed to phosphorylate regulatory Smad (R-Smad) proteins, specifically Smad2 or Smad3. These phosphorylated R-Smads then form complexes with Smad4 and translocate to the nucleus, where they regulate the transcription of a diverse range of genes17,18,19,20,21. Thus, the Smad4 protein serves as a common signaling molecule shared by the entire TGF-β superfamily, and it plays a critical role in transcriptional activation. A growing body of evidence has shown that the TGF-β/Smad signaling pathway plays a crucial role in the development of pulmonary fibrosis. Abnormal activation of this pathway can result in the excessive secretion of pro-fibrotic factors, including collagen proteins, fibronectin (FN1), and α-SMA (also known as Acta2)22,23,24,25. In addition, activation of the TGF-β/Smad signaling pathway induces the production of mesenchymal marker proteins such as N-cadherin and Vimentin, thereby promoting the process of EMT26,27,28,29.
In this study, we present compelling evidence supporting the inhibitory role of FOXN3 in the progression of pulmonary fibrosis by establishing a lung-specific FOXN3 knockout mouse model and a FOXN3 transgenic mouse model for FOXN3 overexpression. Our results demonstrate that FOXN3 negatively regulates the transcriptional activity of Smad signaling by promoting the ubiquitination of Smad4, which leads to the dissociation of the Smad2/3/4 complex from chromatin in an unstimulated state. However, in response to pro-fibrotic stimuli, FOXN3 is directly phosphorylated by the kinase NEK6, leading to phosphorylation-dependent degradation. This degradation subsequently prevents the ubiquitination of Smad4, stabilizing the association of the Smad complex with its target gene elements for transcriptional activation and ultimately contributing to the development of pulmonary fibrosis. Thus, this study provides new insights into the molecular basis of the pathological development of pulmonary fibrosis and identifies potential therapeutic targets for its treatment.
Results
FOXN3 suppresses the development of pulmonary fibrosis
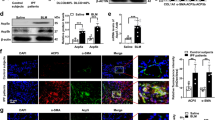
In our previous studies, we have shown that FOXN3, a transcriptional repressor, plays a crucial role in the pulmonary inflammatory response30, which is known to contribute to the development of pulmonary fibrosis. However, the specific involvement of FOXN3 in pulmonary fibrosis has not been fully understood. To investigate this further, we generated a mouse model with conditional knockout (KO) of FOXN3 (Foxn3fl/fl). This was accomplished by administering an adenovirus encoding Cre-recombinase through intratracheal delivery (Fig. 1a, b). Three days after the adenoviral infection, the mice were exposed to Bleomycin (BLM) treatment to induce pulmonary fibrosis. The analysis, conducted using trichrome staining, revealed a noteworthy acceleration of pulmonary fibrosis in the FOXN3 KO mice when compared to the mice that did not undergo Cre-recombinase treatment (Fig. 1c). This was accompanied by increased pulmonary inflammatory injury, as demonstrated by hematoxylin and eosin (H&E) histological staining (Fig. 1d). In addition, an enzyme-linked immunosorbent assay (ELISA) demonstrated a significant elevation in the levels of pro-inflammatory cytokines in the bronchoalveolar lavage (BAL) fluid obtained from the lungs of FOXN3-KO mice (Fig. 1e). To validate the inhibitory role of FOXN3 in lung fibrosis, we evaluated the levels of pro-fibrotic factors in the mouse lungs. Our results demonstrated that the disruption of FOXN3 led to a significant upregulation of these pro-fibrotic factors (Fig. 1f). Furthermore, the disruption of FOXN3 resulted in a marked increase in the total collagen content, as evidenced by hydroxyproline measurements (Fig. 1g). To further support our in vivo findings, we conducted siRNA-mediated knockdown of FOXN3 in A549 cells and assessed the RNA levels of pro-fibrotic regulators. As expected, FOXN3 knockdown resulted in a significant increase in the expression of these pro-fibrotic regulators (Fig. 1h, i). These collectively findings reveal that FOXN3 plays an inhibitory role in lung fibrosis.
a A quantitative PCR assay was conducted to validate the efficiency of FOXN3 KO in the lungs of Foxn3fl/fl mice following the administration of Cre-recombinase (n = 4). b Western blot analysis was conducted to validate the efficiency of FOXN3 KO in the lungs of Foxn3fl/fl mice following the administration of Cre-recombinase (n = 3). c Histological trichrome staining analysis comparing the fibrotic response in the lungs of Foxn3fl/fl mice with or without the administration of Cre-recombinase, in the presence or absence of BLM induction. Scale bars, 100 μm. d H&E staining analysis showing the inflammatory response in the lungs of Foxn3fl/fl mice with or without the administration of Cre-recombinase, in the presence or absence of BLM induction. Scale bars, 100 μm. e The Enzyme-linked immunosorbent assay (ELISA) was conducted to measure the levels of TNFα and IL-6 in the bronchoalveolar lavage (BAL) fluid obtained from the lungs of Foxn3fl/fl mice with or without the administration of Cre-recombinase following BLM treatment (n = 5). f Quantitative PCR analysis was conducted in the lungs of mice to evaluate the impact of lung-specific FOXN3 knockout on the expression levels of pro-fibrotic factors (n = 4). g A hydroxyproline assessment was performed to evaluate the collagen content in the lungs of Foxn3fl/fl mice with or without the administration of Cre-recombinase, in the presence or absence of BLM induction (n = 4). h Quantitative PCR analysis was conducted in A549 cells with or without siRNA-mediated FOXN3 knockdown to detect the changes in the RNA levels of pro-fibrotic factors. The cells were collected 48 hrs post-transfection. Representative data from three independent experiments (n = 3). i A western blot assay was conducted on the total cell lysate (TCL) of A549 cells to assess the efficiency of FOXN3 protein knockdown. The cells were collected 48 hrs post-transfection. Representative blot from three independent experiments (n = 3). The data (a) and (e) were assessed by two-tailed Student’s t test, and the data (f–h) were assessed by one-way ANOVA. All data are shown as the mean ± SD.
To validate the functional significance of FOXN3 in the progression of pulmonary fibrosis, we conducted additional investigations on a loxp-stop-loxp (LSL) knock-in (KI) mouse model (Foxn3LSL). This model allows for the conditional overexpression of FOXN3 exclusively in the lungs of mice through the induction of Cre-recombinase (Fig. 2a, b). The results from both single allele and double allele KI mice demonstrate that the conditional overexpression of FOXN3 in the lungs of mice effectively reduced the fibrotic response, as demonstrated by the significant decrease in fibrotic tissue observed in the trichrome staining assay (Fig. 2c). In addition to the reduced fibrotic response, our study also demonstrated significant suppression of the inflammatory response in the lungs of mice with conditional overexpression of FOXN3, as demonstrated by histological H&E staining and BAL fluid assays (Fig. 2d–f). To further investigate the impact of highly expressed FOXN3, we conducted quantitative PCR analysis to assess its influence on the levels of pro-fibrotic factors. Our findings consistently showed that FOXN3 overexpression critically suppressed the expression of these pro-fibrotic factors both in the lungs of mice and in primary pulmonary fibroblasts isolated from the KI mice, as depicted in Fig. 2g, h. These results align with the observed decrease in collagen content in the lungs of mice with FOXN3 overexpression, as shown in Fig. 2i. Taken together, these findings provide compelling evidence demonstrating that FOXN3 plays a negative regulatory role in pulmonary fibrosis.
a Quantitative PCR analysis showing the RNA expression levels of WT and Foxn3LSL KI mice following BLM induction (n = 4). b Western blot analysis showing FOXN3 protein levels in the lungs of WT and Foxn3LSL KI mice (n = 4). c Histological trichrome staining analysis comparing the fibrotic response in the lungs of WT and Foxn3LSL KI mice following BLM induction. Scale bars, 100 μm. d H&E staining analysis showing the inflammatory response in the lungs of WT and Foxn3LSL KI mice following BLM induction. Scale bars, 100 μm. e Enzyme-linked immunosorbent assays (ELISA) showing the levels of TNFα in the BAL fluid from the WT and Foxn3LSL KI mice upon BLM induction (n = 5). f ELISA analysis showing the levels of IL-6 in the BAL fluid from the WT and Foxn3LSL KI mice upon BLM induction (n = 5). g Quantitative PCR analysis showing the RNA levels of the pro-fibrotic factors in the lungs of WT and Foxn3LSL KI mice following BLM induction (n = 4). h Quantitative PCR analysis showing the RNA levels of the pro-fibrotic factors in primary pulmonary fibroblasts isolated from WT and Foxn3LSL KI mice following BLM stimulation (n = 3). i Hydroxyproline measurements comparing the collagen content in the lungs of WT and Foxn3LSL KI mice, with or without BLM stimulation (n = 4). The data (e and f) were assessed by two-tailed Student’s t test and the data (a) and (g–i) were assessed by one-way ANOVA. All data are shown as the mean ± SD.
FOXN3 impedes the process of epithelial-mesenchymal transition
Given the well-established understanding of the crucial role of epithelial-mesenchymal transition (EMT) in pulmonary fibrosis7,8,31,32, it was reasonable to hypothesize that FOXN3 might play a role in the regulation of EMT. In order to test our hypothesis, we conducted invasion, migration, and wound healing assays simultaneously. Intriguingly, the lentivirus-mediated overexpression of FOXN3 significantly inhibited the invasive and migratory capabilities of A549 cells (Supplementary Fig. 1a–c). In line with these observations, quantitative PCR analysis demonstrated significant reductions in the RNA expression levels of mesenchymal marker genes such as Vimentin and N-Cadherin in A549 cells overexpressing FOXN3, while the levels of the epithelial marker E-Cadherin were elevated. (Supplementary Fig. 1d). To support our findings, we conducted an analysis on primary pulmonary fibroblasts obtained from Foxn3LSL KI mice and compared them with fibroblasts from WT littermates. Interestingly, the primary pulmonary fibroblasts from the Foxn3LSL KI mice exhibited a significant decrease in both RNA and protein levels of the mesenchymal markers compared to those from the WT littermates (Supplementary Fig. 1e, f). To validate the inhibitory effect of FOXN3 on EMT, we performed siRNA-mediated FOXN3 knockdown in A549 cells. Subsequently, invasion, migration, and wound healing assays were conducted. As anticipated, the FOXN3-depleted A549 cells displayed enhanced invasive and migrational abilities compared to the mock control-treated A549 cells (Supplementary Fig. 2a–c). This was accompanied by a notable upregulation of mesenchymal markers and a downregulation of the epithelial marker gene, E-Cadherin (Supplementary Fig. 2d, e). These findings offer significant evidence supporting the inhibitory capacity of FOXN3 in the regulation of EMT during pulmonary fibrosis development.
FOXN3 suppresses the transcriptional activity of Smad signaling
To investigate the underlying molecular mechanisms by which FOXN3 suppresses lung fibrosis, we performed transcriptome sequencing on lung tissues with or without conditional overexpression of FOXN3. Our analysis identified 1460 downregulated genes and 2201 upregulated genes (Fig. 3a). Gene ontology analysis revealed a strong association of FOXN3 with immune and inflammatory responses, as well as collagen-containing matrix organization, which are major factors modulating pulmonary fibrosis (Fig. 3b). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed significant enrichment of the TGF-β signaling pathway (Fig. 3c). Further investigation showed that the lung-specific overexpression of FOXN3 led to a significant reduction in the expression of TGF-β/Smad signaling downstream-regulated genes (Fig. 3c). This suggests that FOXN3 may be involved in regulating Smad signaling, which is a critical signaling cascade in the pathogenesis of pulmonary fibrosis23,33. To explore the potential regulatory mechanism, we initially assessed the effect of FOXN3 on the activation of Smad signaling induced by TGF-β, a well-known activator of Smad signaling. Notably, TGF-β stimulation significantly downregulated FOXN3 while upregulating Smad4 (Fig. 3d), suggesting a potential role for FOXN3 in modulating Smad signaling activation. Furthermore, siRNA-mediated knockdown of FOXN3 led to an additional increase in Smad4 expression levels (Fig. 3d), while the phosphorylation levels of Smad2/3 showed no significant changes upon FOXN3 depletion (Fig. 3d). To validate these observations in vivo, we assessed the levels of these proteins in the lungs of WT and Foxn3LSL/LSL mice. We observed a significant decrease in Smad4 protein expression in the lungs with conditional overexpression of FOXN3 (Fig. 3e), whereas the phosphorylation levels of Smad2/3 showed no significant changes (Fig. 3e). To confirm the involvement of FOXN3 in controlling the stability of the Smad4 protein, we conducted additional experiments using cycloheximide (CHX) treatment to determine the half-life of Smad4 following FOXN3 depletion. Consistent with our previous findings, we observed that the deficiency of FOXN3 resulted in a prolonged half-life of the Smad4 protein (Supplementary Fig. 3a, b), underscoring the role of FOXN3 in modulating Smad4 stability. We also measured the RNA levels of Smad4 and found no significant changes in mouse lungs with FOXN3 overexpression or in A549 cells with FOXN3 knockdown (Supplementary Fig. 3c, d).
a A volcano plot illustrating the differential expression of genes in the lungs of mice following FOXN3 overexpression. b Gene ontology analysis was performed on the RNA sequencing data obtained from the lungs of both WT and Fxon3LSL KI mice. GO enrichment analysis of the differentially expressed genes was conducted using the Wallenius non-central hypergeometric distribution, which accounts for gene length bias. A p-value threshold of ≤ 0.05 was established for enrichment significance. c KEGG and heatmap analyses of the RNA sequencing data were conducted to illustrate the expression changes of Smad downstream-regulated genes. d Immunoblot analysis was conducted to assess the effect of siRNA-mediated FOXN3 knockdown on the levels of Smad proteins in both A549 and primary ATII cells upon TGF-β treatment (10 ng/ml, 16 hr). The cells were treated with 0.05% fetal bovine serum (FBS) for 8 h prior to TGF-β treatment and collected 48 hrs post-siRNA transfection. TCL: total cell lysate. e Immunoblot analysis was conducted to assess the protein expression levels of Smads and FOXN3 in the lungs of both WT and Fxon3LSL KI mice (n = 4). (f–h) ChIP assays were conducted in A549 cells with or without TGF-β treatment (10 ng/ml, 16 hr) to investigate the effect of siRNA-mediated FOXN3 knockdown on the transcriptional recruitment of Smad proteins to the promoters of their target genes. The cells were collected 48 hrs post-transfection. Representative data from three independent experiments (n = 3). i ChIP assays were conducted in A549 cells with or without TGF-β treatment (10 ng/ml, 16 hr) to examine the distribution of FOXN3 on the promoters of Smad target genes. Representative data from three independent experiments (n = 3). The data (f–h) were assessed by one-way ANOVA, and the data (i) was assessed by a two-tailed Student’s t test. All data was shown as the mean ± SD. The blotting data (d) are representative of two independent experiments and were quantified using ImageJ software.
Subsequently, we investigated whether the disruption of FOXN3 affects the transcriptional activity mediated by Smad proteins through chromatin immunoprecipitation (ChIP) assays. Our results demonstrate that the stimulation of recombinant TGF-β significantly enhances the recruitment of Smad proteins to its responsive elements (Fig. 3f–h), while concomitantly reducing the binding of FOXN3 to these Smad-targeted promoters (Fig. 3i). Importantly, we observed that the absence of FOXN3 substantially enhanced the binding affinity of Smad4, along with phosphorylated Smad2 and Smad3, to Smad response elements (Fig. 3f–h), suggesting that FOXN3 disruption notably influences the transcriptional response facilitated by Smad proteins. These findings collectively indicate that FOXN3 restricts the transcriptional response driven by TGF-β/Smad signaling, possibly through the regulation of Smad4 abundance.
FOXN3 facilitates the β-TrCP-mediated Smad4 ubiquitination and subsequent degradation
In our previous study, we discovered that β-TrCP, a direct E3 ligase of Smad4, interacts with FOXN3/hnRNPU complex to regulate a non-canonical NF-κB signaling cascade30. Based on this finding, we hypothesize that FOXN3 may also be involved in the β-TrCP-mediated ubiquitination and subsequent proteasomal degradation of Smad4. To test this hypothesis, we initially performed a co-immunoprecipitation (Co-IP) assay to investigate the association between Smad4 and the FOXN3/hnRNPU complex. As illustrated in Fig. 4a, b, our findings reveal a distinct interaction between exogenous or endogenous FOXN3 and Smad4, as well as with β-TrCP and hnRNPU. Subsequently, we treated A549 cells with recombinant TGF-β to activate Smad signaling and then examined the association of FOXN3 with Smad4. Our findings unveiled that following TGF-β treatment, FOXN3 was dissociated from the hnRNPU/Smad4 complex (Fig. 4c), thus confirming the potential role of FOXN3 as a repressor in the activation of TGF-β/Smad signaling. Therefore, we investigated whether FOXN3 is required for β-TrCP-mediated Smad4 ubiquitination and degradation. Our ubiquitination assay indicated that enforced FOXN3 overexpression enhanced Smad4 ubiquitination and degradation in the presence of β-TrCP, while loss of β-TrCP abolished ubiquitin-mediated Smad4 degradation induced by FOXN3 overexpression (Fig. 4d–f). Consistently, FOXN3 depletion strongly inhibited endogenous Smad4 ubiquitination (Supplementary Fig. 4a).
a An Anti-Flag Co-IP assay was conducted in HEK293T cells transfected with Flag-tagged Smad4 to investigate the association of Smad4 with endogenous β-TrCP, hnRNPU, and FOXN3. The cells were collected 48 h post-transfection. TCL: total cell lysate. b An anti-Smad4 Co-IP assay was performed in A549 cells to detect the Smad4 association with endogenous β-TrCP, hnRNPU, and FOXN3. c An anti-Flag Co-IP assay was conducted to detect the interaction between Flag-tagged FOXN3 and both Smad4 and hnRNPU in A549 cells following TGF-β treatment (10 ng/ml, 16 hr) at the indicated time points. The cells infected with lentivirus expressing Flag-tagged FOXN3 were treated with 0.05% FBS for 8 h prior to TGF-β treatment and then collected 48 h post-infection. d The effect of FOXN3 overexpression on Flag-tagged Smad4 ubiquitination in HEK293T cells was detected by a ubiquitination assay. The cells transfected with indicated combinations of HA-tagged FOXN3, Flag-tagged Smad4, Myc-tagged ubiquitin, and shRNA targeting β-TrCP were treated with MG-132 (20 μM, 4 h) and collected 48 h post-transfection. e Western blot analysis was carried out in A549 cells to examine the impact of FOXN3 overexpression on the stability of Smad4 in the presence or absence of β-TrCP. Lentiviral infection was used to transduce HA-tagged FOXN3 and β-TrCP shRNA into the cells, which were collected 48 h post-infection. f Western blot analysis was carried out in primary pulmonary fibroblasts to examine the impact of FOXN3 overexpression on the stability of Smad4 in the presence or absence of β-TrCP. Lentiviral infection was utilized to transduce HA-tagged FOXN3 and β-TrCP shRNA into the cells, which were collected 48 hrs post-infection. g A Co-IP assay was conducted to assess the impact of lentivirus-mediated FOXN3 overexpression on the association of endogenous Smad4 with hnRNPU and β-TrCP in A549 cells. Before collection, the cells infected with lentivirus expressing Flag-tagged FOXN3 were treated with MG-132 (20 μM, 4 h) and collected 48 hrs post-infection. h An anti-Flag Co-IP assay was performed to detect the effect of FOXN3 forkhead domain deficiency on the interaction between Flag-tagged Smad4 and both hnRNPU and β-TrCP in HEK293T cells. Prior to collection, the cells transfected with indicated combinations of HA-tagged WT FOXN3 or its forkhead-deficient mutant, Flag-tagged Smad4, and Myc-tagged β-TrCP (ΔF) were treated with MG-132 at a concentration of 20 μM for 2 h and collected 48 hrs post-transfection. i Quantitative PCR analysis showing the RNA levels of pro-fibrotic factors in A549 cells overexpressing FOXN3 WT or its forkhead domain-deficient mutant through lentiviral infection. The cells were collected 48 hrs post-infection. The data (i) was assessed by one-way ANOVA and was shown as the mean ± SD. The blotting data (a–h) are representative of two independent experiments and were quantified using ImageJ software.
To gain further insight into the biochemical function of FOXN3 in β-TrCP-mediated Smad4 ubiquitination, we conducted experiments to assess the impact of FOXN3 on the interaction between β-TrCP and Smad4. Our findings demonstrated that the enforced overexpression of FOXN3 significantly enhanced the association of Smad4 with both the endogenous β-TrCP and the exogenously introduced F-Box domain-deficient β-TrCP variant [β-TrCP (ΔF)] (Fig. 4g and Supplementary Fig. 4b), which is recognized for its improved substrate binding capability34. Conversely, disruption of FOXN3 led to resistance in Smad4’s interaction with the β-TrCP/hnRNPU complex (Supplementary Fig. 4c). To further confirm the role of FOXN3 in Smad4 ubiquitination, we investigated the effect of the FOXN3 forkhead domain on Smad4 ubiquitination. This domain is known to be essential for FOXN3’s transcriptional repressive ability30. Our co-immunoprecipitation (Co-IP) assay revealed that the deficiency of the FOXN3 forkhead domain resulted in minimal association with Smad4 (Supplementary Fig. 4d). Significantly, the forkhead domain-deficient FOXN3 mutant exhibited a weaker interaction between Smad4 and β-TrCP or hnRNPU compared to WT FOXN3 (Fig. 4h). This resulted in a critical impediment of Smad4 ubiquitination and degradation facilitated by β-TrCP (Supplementary Fig. 4d, e) and the subsequent activation of Smad protein-mediated transcriptional response (Fig. 4i and Supplementary Fig. 4f). Collectively, these observations offer compelling evidence for the necessity of FOXN3 in facilitating the interaction between Smad4 and β-TrCP, leading to the ubiquitin-mediated degradation of Smad4 and the consequent inactivation of the Smad transcriptional response.
FOXN3-mediated Smad4 ubiquitination disrupts the Smad complex’s association with its response elements
As documented in our prior report, FOXN3 is exclusively associated with chromatin, with chromatin-free FOXN3 being undetectable30. To confirm this characteristic in A549 cells, primary alveolar type II (ATII) cells, and pulmonary fibroblasts, we conducted a step-wise fractionation assay to isolate both the chromatin-free (low-salt extracted fraction, LSF) and chromatin-bound (low-salt extracted nuclei, LSEN) fractions (Fig. 5a), as previously described30. Western blot analysis revealed that FOXN3 is almost fully associated with chromatin in these cell types (Fig. 5b). In contrast, the small nuclear ribonucleoprotein (snRNP) complex component HEXIM1, a well-established chromatin-free nuclear protein, was entirely localized in the LSF (Fig. 5b). This distribution indicates that the chromatin-free and chromatin-bound components were efficiently separated. Based on these findings, we hypothesized that the ubiquitination of Smad4 mediated by FOXN3 occurs in a chromatin-associated manner. To investigate this hypothesis, we performed cleavage under target and tagmentation (CUT&Tag) analysis to assess the genomic distribution of both FOXN3 and Smad4. Our experiments identified 51,490 peaks associated with the anti-FOXN3 antibody and 37,198 peaks associated with the anti-Smad4 antibody, with 19,868 overlapping peaks characterized (Fig. 5c). Surprisingly, further analysis revealed that the promoters of 12,880 genes were co-targeted by both FOXN3 and Smad4 (Fig. 5d), demonstrating overlapping genomic landscapes and peak distributions (Fig. 5e, f). KEGG analysis consistently indicated that many signaling pathways, including the TGF-β signaling pathway, were co-enriched (Fig. 5g), suggesting that FOXN3 and Smad4 co-regulate numerous biochemical processes. Notably, FOXN3 and Smad4 exhibited remarkably similar peak locations on the promoters of representative Smad response genes (Fig. 5h), providing further evidence of their physical interaction and functional connection.
a Flowchart depicting the experimental procedure of step-wise fractionation. The flowchart was created in BioRender. LIAN, J. (2024) https://BioRender.com/r01u561. b Step-wise analysis was performed to assess the subcellular distribution of FOXN3 and Smad proteins. c Venn diagrams illustrating the overlapping peaks of FOXN3 and Smad4 in A549 cells, as determined via CUT&Tag data analysis. d Venn diagrams illustrating the overlapping genes co-targeted by FOXN3 and Smad4 at the promoters in A549 cells, as determined via CUT&Tag data analysis. e Density distributions (read counts per million mapped reads) of FOXN3 and Smad4 peaks at the transcription start site (TSS) are shown. f Genomic distribution of the transcriptional targets of FOXN3 and Smad4 in A549 cells, as determined via CUT&Tag data analysis. g KEGG analysis of the CUT&Tag data for anti-FOXN3 and anti-Smad4. h The binding profiles of FOXN3 and Smad4 to representative transcriptional targets of Smads are displayed. i Step-wise fractionation analysis was performed in A549 cells to detect the effect of FOXN3 overexpression on the enhanced association of the Smad complex with chromatin triggered by TGF-β treatment (10 ng/ml, 16 hr). The cells infected with lentivirus expressing HA-tagged FOXN3 or a mock control were treated with 0.05% FBS for 8 h prior to TGF-β treatment and collected 48 h post-infection. j A step-wise fractionation assay was conducted in primary ATII cells to assess the impact of FOXN3 knockdown on the association of the Smad complex with chromatin. The cells were collected 48 h post-siRNA transfection. k A stepwise fractionation assay was conducted in HEK293T cells to evaluate the impact of the Smad4 R100T mutant on the association of the Smad complex with chromatin. Cells transfected with Flag-tagged WT Smad4 or its R100T mutant were treated with TGF-β (10 ng/ml for 16 h). Prior to TGF-β treatment, the cells were exposed to 0.05% FBS for 8 h. Subsequently, a ubiquitination assay was performed to confirm the ubiquitination levels of Flag-tagged Smad4 and its mutant (lower panel). The cells were treated with MG132 (20 μM, 2 hr) before collection 48 h post-transfection. The blotting data (b, i–k) are representative of two independent experiments and were quantified using ImageJ software.
The genomic colocalization of FOXN3 and Smad4 prompted us to investigate whether FOXN3-mediated ubiquitination of Smad4 disrupts the association of the Smad2/3/4 complex from chromatin, thereby inhibiting Smad transcriptional activity. To explore this hypothesis, we first conducted a fractionation assay to assess the subcellular distribution of the Smad complex with or without FOXN3 overexpression. Our results indicated that forced overexpression of FOXN3 significantly disrupted the enhanced association of Smad2/3 with chromatin induced by TGF-β stimulation, along with the accelerated degradation of Smad4 (Fig. 5i). Conversely, the knockdown of FOXN3 resulted in a stronger association of the Smad2/3/4 complex with chromatin (Fig. 5j), which aligns well with our previous ChIP results indicating an enhanced association of the Smad complex with its target gene elements upon FOXN3 depletion (Fig. 3f–h). To further support the idea that Smad4 ubiquitination influences the association of the Smad complex with its response elements, we generated a Smad4 mutant, R100T (where arginine at position 100 is replaced by threonine), which has been demonstrated to significantly enhance Smad4 ubiquitination35. We then investigated the effect of this highly ubiquitinated Smad4 on the stability of the Smad complex in relation to chromatin. As expected, the R100T mutant resulted in a much weaker association of the Smad complex with chromatin compared to the WT form (Fig. 5k). These findings collectively suggest that FOXN3-mediated ubiquitination of Smad4 contributes to the dissociation of the Smad2/3/4 complex from chromatin, ultimately leading to the disruption of the Smad transcriptional response and the degradation of Smad4.
HnRNPU is required for FOXN3-mediated inhibition of Smad transcriptional response
According to Fig. 4g, the overexpression of FOXN3 not only increased the association of Smad4 with β-TrCP but also with hnRNPU, which is known for its scaffold-like protein function. We hypothesized that hnRNPU might be necessary for the interaction between Smad4 and β-TrCP for ubiquitination. To test this hypothesis, we first examined the effect of hnRNPU on Smad4 stability. Western blot analysis revealed that the absence of hnRNPU prevented the degradation of Smad4 induced by FOXN3 overexpression (Supplementary Fig. 5a). In addition, silencing hnRNPU abolished the enhanced interaction triggered by FOXN3 overexpression (Supplementary Fig. 5b). Consistently, the half-life experiment of Smad4 showed that knockdown of hnRNPU significantly prolonged its stability (Supplementary Fig. 5c). To confirm the essential role of hnRNPU in this process, we used a mutant form of hnRNPU with a deletion of amino acids 238–550 (Δ238–550), which does not bind to FOXN330. We assessed the impact of this variant on β-TrCP-mediated ubiquitination and degradation of Smad4. As expected, FOXN3 failed to enhance ubiquitin-mediated Smad4 degradation in the absence of this specific amino acid region (Supplementary Fig. 5d, e). Consistent with this observation, the Δ238–550 hnRNPU mutant impeded the heightened interaction between Smad4 and β-TrCP triggered by FOXN3 overexpression (Supplementary Fig. 5f). Notably, step-wise fractionation analysis revealed that the Δ238–550 mutant stabilized the association of the Smad2/3/4 complex with chromatin (Supplementary Fig. 5g, h), leading to enhanced Smad transcriptional activity (Supplementary Fig. 5i, j). In conclusion, these findings underscore the critical role of hnRNPU in FOXN3-mediated Smad4 ubiquitination and the subsequent inactivation of the Smad transcriptional response through the dissociation of the Smad complex.
Phosphorylation of FOXN3 at S412 and S416 facilitates its ubiquitin-mediated degradation by β-TrCP
Through sequence analysis, we identified a conserved motif [aa 411–416 (DSGXXS)] in the FOXN3 C-terminal region (Fig. 6a), which is well-known as a degron crucial for β-TrCP substrate recognition and subsequent degradation36,37. Immunoprecipitation assays demonstrated that both a Δ411–416 FOXN3 deletion mutant and a S412,416 A double conversion mutant (where S412 and S416 are both substituted with alanine) exhibited significantly disrupted interactions with β-TrCP (Fig. 6b, c). This disruption aligns with phosphorylation-dependent substrate binding by β-TrCP. To ascertain if FOXN3 directly serves as a substrate for β-TrCP, we conducted ubiquitination assays. These experiments revealed that β-TrCP effectively induced the ubiquitination of WT FOXN3, while the S412,416 A mutant showed marked resistance to ubiquitination (Fig. 6d). In addition, knockdown of β-TrCP markedly reduced FOXN3 ubiquitination (Fig. 6e), further confirming FOXN3 as a direct substrate for β-TrCP.
a Sequence alignment across various species reveals a conserved DSGYAS motif located within the C-terminal domain of FOXN3. b A Co-IP assay was conducted in HEK293T cells to assess the interaction between Myc-tagged β-TrCP and Flag-tagged WT FOXN3 or FOXN3 lacking the DSGYAS motif (Δ411 − 416). The cells transfected with the indicated combinations were collected 48 hrs post-transfection. c Immunoblot analysis of Myc-tagged β-TrCP immunoprecipitated by Flag-tagged WT FOXN3 or its S412,416 A mutant form in HEK293T cells. The cells transfected with the indicated combinations were collected 48 hrs post-transfection. d A ubiquitination assay was conducted in HEK293T cells to examine the effect of β-TrCP overexpression on the ubiquitination of WT FOXN3 or its S412,416 A mutant. The cells transfected with Flag-tagged FOXN3, Myc-tagged β-TrCP, and HA-tagged ubiquitin were collected 48 h post-transfection. e A ubiquitination assay was conducted in HEK293T cells to examine the effect of β-TrCP knockdown on the ubiquitination of FOXN3. The cells transfected with Flag-tagged FOXN3, HA-tagged ubiquitin, and shRNA targeting β-TrCP were collected 48 hrs post-transfection. f A FOXN3 deletion-mapping assay was carried out in HEK293T cells to identify the specific region accountable for its ubiquitination. The cells transfected with the indicated various mutants of Flag-tagged FOXN3, Myc-tagged β-TrCP, and HA-tagged ubiquitin were collected 48 h post-transfection. g Ubiquitination analysis of WT FOXN3 and its different mutant forms in HEK293T cells to pinpoint the specific site responsible for FOXN3 ubiquitin conjugation. The cells transfected with the indicated various mutants of Flag-tagged FOXN3, Myc-tagged β-TrCP, and HA-tagged ubiquitin were collected 48 h post-transfection. h Immunoblot analysis of the linkage of FOXN3 ubiquitination in HEK293T cells expressing the indicated combinations of Myc-tagged β-TrCP and HA-tagged ubiquitin mutants (K48O or K63O). The cells were collected 48 h post-transfection. i Immunoblot analysis of the linkage of FOXN3 ubiquitination in HEK293T cells expressing the indicated combinations of Myc-tagged β-TrCP, HA-tagged ubiquitin mutant (K63O or K63R) and WT FOXN3 or its K114R mutant. The cells were collected 48 h post-transfection. j Immunoblot analysis was performed in primary pulmonary fibroblasts to assess the impact of WT β-TrCP or its inactive ΔF mutant on the expression of FOXN3. The cells infected with lentivirus expressing WT β-TrCP or its inactive ΔF mutant were collected 48 h post-infection. k Immunoblot analysis was performed in BEAS-2B cells to assess the impact of WT β-TrCP or its inactive ΔF mutant on the expression of WT FOXN3 or its K114R mutant. The indicated combinations were overexpressed in BEAS-2B cells using lentivirus infection, which were collected 48 hrs post-infection. The blotting data (b–k) are representative of two independent experiments and were quantified using ImageJ software.
We then proceeded to identify the sites of ubiquitin conjugation on FOXN3. Deletion mapping assays revealed that the removal of the forkhead domain (aa 114–199) almost completely halted β-TrCP-mediated ubiquitination of FOXN3 (Fig. 6f), suggesting that ubiquitin primarily attached to the forkhead domain. To pinpoint the specific sites responsible for ubiquitin conjugation, we systematically mutated all lysine residues within the FOXN3 forkhead domain and conducted ubiquitination assays. The outcomes demonstrated that the K114R mutant (in which the Lys 114 residue was changed to Arg) significantly impaired FOXN3 ubiquitination by β-TrCP (Fig. 6g), highlighting K114 as the primary ubiquitination site on FOXN3. Furthermore, we investigated the chain linkage of FOXN3 ubiquitination by utilizing ubiquitin mutants K48O (where all Lys residues except Lys48 were converted to Arg) and K63O (where all Lys residues except Lys63 were converted to Arg). Immunoblot analysis unveiled that β-TrCP primarily facilitated K63-linked polyubiquitination of FOXN3 (Fig. 6h). Notably, a ubiquitin mutant bearing solely the K63R modification exhibited resistance to β-TrCP-mediated FOXN3 ubiquitination (Fig. 6i). To assess whether FOXN3 ubiquitination by β-TrCP led to its degradation, we overexpressed WT β-TrCP and its inactive mutant β-TrCP (ΔF) in primary pulmonary fibroblasts and BEAS-2B cells to evaluate their impact on FOXN3 stability. Our observations indicated that WT β-TrCP, but not β-TrCP (ΔF), promoted the degradation of WT FOXN3. Conversely, the K114R mutant displayed substantial resistance to FOXN3 degradation by β-TrCP (Fig. 6j, k). These collective findings suggest that phosphorylation of S412 and S416 on FOXN3 promotes its K63-linked ubiquitination and subsequent degradation by β-TrCP.
Phosphorylation-dependent FOXN3 degradation exacerbates pulmonary fibrosis by activating Smad signaling
To examine the influence of S412 and S416 phosphorylation-dependent FOXN3 degradation on Smad signaling activation and its involvement in regulating pulmonary fibrosis, we generated a FOXN3 knock-in (KI) mouse model (Foxn3KI/KI). In this model, FOXN3 was engineered to carry non-synonymous serine to alanine mutations at both the S379 and S383 sites. It is noted that the homologous sites in humans are S412 and S416. Post BLM administration, we isolated the lungs of mice and assessed the phosphorylation levels of FOXN3 S379 and S383. Immunoblot assay indicated a crucial blockage of S379,383 phosphorylation in the KI mice compared to WT mice (Fig. 7a). The attenuation of S379,383 phosphorylation resulted in increased total FOXN3 protein levels, which caused a marked decrease of Smad4 protein expression (Fig. 7a). To analyze the ablation of S379,383 phosphorylation on pulmonary fibrotic phenotype, we performed trichrome staining analysis and observed that the Foxn3KI/KI mice displayed an improved fibrosis phenotype relative to their WT littermates (Fig. 7b). This finding is consistent with the reduced inflammatory injury observed when the S379,383 phosphorylation of FOXN3 was blocked (Fig. 7c). Furthermore, the analysis of BAL fluid also revealed a reduced secretion of pro-inflammatory cytokines in the lungs of the Foxn3KI/KI mice (Fig. 7d, e). To further support the inhibitory role of FOXN3 phosphorylation on fibrotic response, we performed quantitative PCR analysis and discovered that the expression levels of pro-fibrotic factors were lower in Foxn3KI/KI mice compared to WT mice (Fig. 7f). In addition, we measured the pulmonary collagen content using a hydroxyproline assessment assay and observed a decreased collagen content in Foxn3KI/KI mice relative to WT mice (Fig. 7g). To further validate these observations, we isolated primary pulmonary fibroblasts from both KI and WT mice and assessed the levels of Smad4 protein along with its downstream pro-fibrotic regulators. Consistently, fibroblasts derived from KI mice showed significantly reduced levels of Smad4 protein compared to those from WT mice (Supplementary Fig. 6a), resulting in a crucial suppression of downstream pro-fibrotic factors regulated by Smads (Fig. 7h). Immunostaining assays further confirmed the inhibitory impact of disrupting FOXN3 S379,383 phosphorylation on Smad4 protein expression (Supplementary Fig. 6b, c). These findings lead us to conclude that phosphorylation of FOXN3 at S379 and S383 in mice (equivalent to S412 and S416 in humans) results in the loss of its inhibitory effect on pulmonary fibrosis by activating the Smad signaling pathway.
a A Western blot assay was performed to assess the alterations in FOXN3 and Smad4 expression levels in the lungs of WT or Foxn3KI/KI mice harboring double mutations at S379 and S383 (n = 3). b Histological trichrome staining analysis comparing the fibrotic response in lungs of WT and Foxn3KI/KI mice, with or without BLM stimulation. Scale bars, 100 μm. c H&E staining analysis showing the inflammatory response in lungs of WT and Foxn3KI/KI mice, with or without BLM stimulation. Scale bars, 100 μm. d, e ELISA analysis was performed to detect the levels of TNFα (d) and IL-6 (e) in the BAL fluid obtained from the lungs of BLM-induced WT or Foxn3KI/KI mice (n = 4). f A quantitative PCR assay showing the RNA levels of pro-fibrotic factors in lungs of WT and Foxn3KI/KI mice, with or without BLM stimulation (n = 4). g A hydroxyproline assessment was conducted to evaluate the collagen content in the lungs of WT and Foxn3KI/KI mice, with or without BLM stimulation (n = 4). h Quantitative PCR analysis was performed to examine the impact of FOXN3 phosphorylation on the expression levels of pro-fibrotic factors in primary pulmonary fibroblasts isolated from WT and Foxn3KI/KI mice following BLM treatment (n = 3). The data (d, e, and h were assessed by two-tailed Student’s t test, and the data (f and g) were assessed by one-way ANOVA. All data are shown as the mean ± SD.
NEK6-mediated FOXN3 degradation activates the Smad transcriptional response
Given the pivotal role of FOXN3 S412 and S416 phosphorylation in the regulation of pulmonary fibrosis, we utilized the “Scansite” predictor to pinpoint the upstream kinase responsible for this phosphorylation mechanism. This analysis unveiled NIMA-related kinase 6 (NEK6) as a potential candidate kinase for the phosphorylation of S412 and S416. In immunoprecipitation experiments conducted in HEK293T cells, it was observed that NEK6 significantly enhanced the interaction between β-TrCP and WT FOXN3, but not the S412,416 A mutant (Fig. 8a). Notably, NEK6 remarkably elicited the phosphorylation of WT FOXN3, as evidenced by an antibody recognizing either phosphorylated S412 or S416, while the S412,416 A mutant remained unphosphorylated (Supplementary Fig. 7a). To ascertain if FOXN3 directly functions as a substrate for NEK6, we individually exposed bacterially expressed WT FOXN3 and the S412,416 A mutant to in vitro kinase reactions with NEK6. Intriguingly, the WT FOXN3 was readily phosphorylated by recombinant NEK6, whereas the S412,416 A mutant exhibited complete resistance to NEK6-mediated phosphorylation (Fig. 8b). To further refine the identification of the specific phosphorylation sites on FOXN3 targeted by NEK6, we created individual S412A and S416A mutations in FOXN3 and performed additional in vitro kinase assays. Immunoblot assays demonstrated that NEK6 had minimal phosphorylation activity towards the S412A mutant of FOXN3, while the S416A mutant displayed partial resistance to phosphorylation (Supplementary Fig. 7b), suggesting that S412 is the primary site targeted by NEK6 for FOXN3 phosphorylation. Consistent with these findings, the disruption of FOXN3 S412 phosphorylation almost entirely abolished its interaction with β-TrCP, while the S416A mutant exhibited only partial impairment in its association with β-TrCP (Supplementary Fig. 7c). These results collectively indicate that S412 is the predominant site responsible for NEK6-mediated FOXN3 phosphorylation and subsequent interaction with β-TrCP.
a A Co-IP assay was conducted in HEK293T cells to detect the impact of NEK6 overexpression on the interaction between Flag-tagged WT FOXN3 or its S412,416 A mutant and Myc-tagged β-TrCP. The cells transfected with the indicated combinations were collected 48 hrs post-transfection. b Bacterially expressed NEK6 and either WT FOXN3 or its S412,416 A mutant were subjected to an in vitro kinase assay, followed by immunoblot analysis of FOXN3 S412 and S416 phosphorylation with a phosphospecific antibody. c A ubiquitination assay was performed in HEK293T cells to detect the impact of NEK6 overexpression on the ubiquitination of WT FOXN3 or its S412,416 A mutant. The cells transfected with the indicated combinations of Flag-tagged FOXN3, HA-tagged NEK6, Myc-tagged β-TrCP, and HA-tagged ubiqutin were treated with MG-132 (20 μM, 4 h) prior to collection 48 hrs post-transfection. d A ubiquitination assay was performed in HEK293T cells to detect the impact of lentivirus-mediated NEK6 overexpression on the ubiquitination of endogenous Smad4. The cells transfected with Flag-tagged WT FOXN3 or its S412,416 A mutant and HA-tagged NEK6 were treated with MG-132 (20 μM, 4 hr) prior to collection 48 hrs post-transfection. e Immunoblot analysis in HEK293T cells showing the effect of NEK6 overexpression on the levels of FOXN3 and Smad4. The cells transfected with the indicated combinations of Flag-tagged Smad4, HA-tagged FOXN3, HA-tagged NEK6, and Myc-tagged β-TrCP were collected 48 hrs post-transfection. f A Co-IP assay was conducted in HEK293T cells to detect the effect of NEK6 overexpression on the interaction between Flag-tagged Smad4 and Myc-tagged β-TrCP in the presence of WT FOXN3 or its S412,416 A mutant. The cells were treated with MG-132 (20 μM, 2 hr) prior to collection 48 hrs post-transfection. g Immunoblot analysis was performed to assess the impact of NEK6 knockdown on the levels of indicated proteins in primary ATII cells in response to TGF-β treatment (10 ng/ml, 16 hr). The cells were treated with 0.05% FBS for 8 h prior to TGF-β treatment and then collected 48 h post-siRNA transfection. h Immunoblot analysis was conducted in primary pulmonary fibroblasts isolated from WT or Foxn3 KI mice to demonstrate the effects of NEK6 knockdown on the levels of FOXN3, its phosphorylation, and Smad4. All cells were treated with 0.05% FBS for 8 h, followed by TGF-β treatment (10 ng/ml, 16 hr), and then collected 48 h post-siRNA transfection. i, j Step-wise fractionation analysis was performed in both A549 (i) and primary ATII (j) cells to assess the effect of NEK6 knockdown on the association of Smad complex with chromatin. All cells were treated with 0.05% FBS for 8 h, followed by TGF-β treatment (10 ng/ml, 16 hr), and then collected 48 h post-siRNA transfection. k, l Quantitative PCR analysis was conducted in both A549 (k) and primary ATII (l) cells to assess the effect of NEK6 knockdown on Smad transcriptional activity. All cells were treated with 0.05% FBS for 8 h, followed by TGF-β treatment (10 ng/ml, 16 hr), and then collected 48 h post-siRNA transfection. m A proposed working model elucidates the transcriptional activation of Smad signaling through NEK6-mediated FOXN3 degradation. The schematic representation was created in BioRender. LIAN, J. (2024) https://BioRender.com/w42l861. The data (k and l) were assessed by a two-tailed Student’s t test. All data are shown as the mean ± SD. The blotting data (a–j) are representative of two independent experiments and were quantified using ImageJ software.
To confirm that the ubiquitination of FOXN3 by β-TrCP depends on NEK6-mediated phosphorylation, ubiquitination assays were conducted. The results demonstrated that WT FOXN3, as opposed to the phosphorylation-resistant mutant S412,416 A, underwent β-TrCP-mediated ubiquitination and subsequent degradation in the presence of NEK6 overexpression (Fig. 8c, e). This ubiquitin-mediated degradation of FOXN3 significantly impeded the ubiquitination of Smad4 (Fig. 8d), leading to the transcriptional activation of Smad signaling and an increase in Smad4 protein levels (Supplementary Fig. 7d and Fig. 8e), possibly due to the enhanced association of the Smad complex with its target gene elements, as previously demonstrated in Fig. 5. This reduced ubiquitination level of Smad4 was attributed to its disrupted interaction with β-TrCP (Fig. 8f). Consistent with these findings, silencing NEK6 using siRNA significantly impeded the increased phosphorylation levels of S412 and S416 induced by TGF-β stimulation. This resulted in upregulated levels of FOXN3 and downregulated levels of Smad4 in both primary ATII cells and WT primary fibroblasts (Fig. 8g, h). However, this effect was not observed in primary fibroblasts carrying the S412,416 A mutation (Fig. 8h). To further validate the hypothesis that NEK6-mediated phosphorylation of FOXN3 at S412 and S416 promotes its degradation, thereby stabilizing the association of Smad2/3/4 complex with chromatin and activating the Smad transcriptional response, we employed a stem-wise fractionation assay to evaluate the effect of NEK6 depletion on the levels of the chromatin-bound Smad complex. Our results indicated that the disruption of NEK6 led to decreased association of the Smad2/3/4 complex with chromatin (Fig. 8i, j), ultimately suppressing Smad transcriptional activity (Fig. 8k, l). These findings illustrate that, in response to pro-fibrotic stimuli such as TGF-β, FOXN3 is phosphorylated by the kinase NEK6, leading to its degradation. The loss of FOXN3 blocks β-TrCP-mediated ubiquitination of Smad4, thereby enhancing the association of the Smad complex with chromatin and promoting Smad transcriptional activation (Fig. 8m).
FOXN3 expression exhibits a significant negative correlation with clinical pulmonary fibrosis
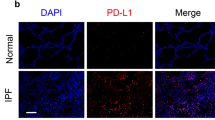
To investigate the relationship between FOXN3 and Smad4 in pulmonary fibrosis, we conducted experiments on mice exposed to the pro-fibrotic agent BLM. In our investigation, although the RNA levels of FOXN3 and Smad4 remained relatively stable (Supplementary Fig. 8a), We observed a significant reduction in total FOXN3 protein expression in the lungs of mice with BLM-induced fibrosis (Supplementary Fig. 8b–d). This finding is consistent with in vitro data indicating that FOXN3 levels decrease in response to TGF-β stimulation (Fig. 3d). Furthermore, there was a discernible rise in Smad4 protein expression levels and the phosphorylation status of FOXN3 at S379 and S383 (Supplementary Fig. 8b–d), as evidenced by immunoblot and immunostaining assays. These results suggest a significant relationship between the phosphorylation-dependent degradation mechanism of FOXN3 and the elevated levels of Smad4 protein, which reflect the activation of the Smad transcriptional response in the context of pulmonary fibrosis.
To impart clinical significance to our findings, we examined 20 clinical samples from patients diagnosed with pulmonary fibrosis. Trichrome staining was conducted to validate the presence of fibrotic lesions in the samples (Fig. 9a). In accordance with the BLM-induced pulmonary fibrosis model, the protein levels of Smad4 and FOXN3 exhibited contrasting patterns, with significantly decreased FOXN3 levels and increased Smad4 levels (Fig. 9b–d). These findings underscore the pivotal role of the FOXN3-Smad axis in the progression of lung fibrosis.
a–c Histological immunohistochemical staining analysis was performed to assess the expression levels of FOXN3 (b) and Smad4 (c) in fibrotic lesion areas and corresponding adjacent normal areas of one patient diagnosed with lung fibrosis, with scale bars set at 100 μm. Trichrome staining was carried out simultaneously to confirm the presence of the fibrotic lesions (a), with scale bars set at 100 μm. A representative image (a) was taken from at least three different areas for each sample. d Quantitative analysis of b and c was shown by immunoreactive score (IRS) (n = 20). e A proposed working model clarifies the activation of Smad signaling in response to pro-fibrotic stimuli. The schematic representation was created in BioRender. LIAN, J. (2024) https://BioRender.com/d99u033. The data (d) was assessed by a two-tailed Student’s t test and is shown as the mean ± SD.
Discussion
In this groundbreaking study, we have successfully identified FOXN3, a recognized inflammatory regulator30, as a key player in pulmonary fibrosis. This significant finding was achieved through the utilization of a conditional lung-specific FOXN3 knockout model and a transgenic mouse model with specific overexpression of FOXN3. Our in vivo investigations have provided compelling evidence supporting the role of FOXN3 as a critical fibrotic regulator. Specifically, our observations indicate that tissue-specific FOXN3 knockout increased the development of pulmonary fibrosis, whereas the transgenic mouse model with lung-specific FOXN3 overexpression robustly inhibited the fibrotic response induced by BLM, a widely recognized inducer of pulmonary fibrosis. These discoveries illuminate the vital role of FOXN3 in repressing fibrosis and offer valuable insights into potential therapeutic strategies for the management of pulmonary fibrosis.
Extensive research has unequivocally confirmed the pivotal role of the inflammatory response in the progression of lung fibrosis38,39,40. In our prior study, we illustrated that FOXN3 acts as a significant transcriptional repressor, effectively impeding the activation of a non-canonical NF-κB signaling pathway through its interaction with hnRNPU30. This repression subsequently hampers the onset of pulmonary inflammatory responses. The findings from our ongoing study offer compelling evidence that FOXN3 also exerts a suppressive influence on the inflammatory response triggered by BLM. Therefore, it would be highly intriguing to investigate whether the FOXN3-mediated non-canonical inflammatory cascade is implicated in the evolution of this fibrotic process and contributes to the development of pulmonary fibrosis.
Our research has expanded upon the known biochemical function of FOXN3 to unveil a novel regulatory role in suppressing the Smad transcriptional response by facilitating the dissociation of the Smad complex from chromatin. Our observations from CUT&Tag analysis indicate that, in an unstimulated state, FOXN3 targets the promoters of numerous Smad response genes and inhibits the Smad transcriptional response by modulating the ubiquitination of the Smad4 protein. Ubiquitinated Smad4 facilitates the dissociation of the Smad2/3/4 complex from its response elements and promotes the degradation of Smad4, effectively abolishing the Smad transcriptional response and ultimately suppressing pulmonary fibrosis (Fig. 9e). However, in response to pro-fibrotic stimuli, FOXN3 is phosphorylated by the kinase NEK6 and undergoes phosphorylation-dependent degradation. This process prevents the ubiquitination of Smad4 and stabilizes the association of Smad complexes with its responsive elements, consequently enhancing Smad transcriptional activity and contributing to the pathogenesis of pulmonary fibrosis (Fig. 9e). However, it remains to be determined whether Smad4 ubiquitination alone is sufficient to induce the dissociation of the Smad complex from chromatin and abrogate Smad transcriptional response. Future studies will be necessary to investigate whether other biochemical events coordinate with Smad4 ubiquitination in this process.
Interestingly, the FOXN3-mediated enhancement of the interaction between β-TrCP and Smad4 is dependent on hnRNPU, a recognized scaffold protein. Disruption of hnRNPU hampers the recruitment of Smad4 to β-TrCP, significantly diminishing β-TrCP-mediated Smad4 ubiquitination. Moreover, our findings indicate that the amino acid region 238–550 of hnRNPU, crucial for its interaction with FOXN3, is essential for β-TrCP-mediated Smad4 ubiquitination and the suppression of Smad transcriptional activity. These results underscore the indispensable role of FOXN3 in this intricate biochemical process. Based on the aforementioned observations, we propose that hnRNPU functions as a scaffold protein, providing an anchoring site for FOXN3, which is essential for recruiting Smad4 to β-TrCP for ubiquitination. This process contributes to the inactivation of the Smad transcriptional response by disrupting the association of the Smad complex with chromatin. This research adds to the growing body of evidence that underscores the diverse roles of hnRNPU as a scaffold protein in various biochemical processes. Furthermore, in alignment with our earlier finding that the forkhead domain of FOXN3 is essential for its capacity to suppress NF-κB activity, the absence of this domain confers significant resistance to the ubiquitination of Smad4, thereby inducing the Smad transcriptional activation. This highlights that FOXN3’s inhibitory role in Smad signaling activation functions via a chromatin-dependent mechanism.
Through sequence analysis of the FOXN3 protein, we have identified two previously unreported potential phosphorylation sites within the well-known motif (DSGYAS), widely recognized as a degron crucial for β-TrCP substrate recognition and subsequent degradation36,37. In an in vitro kinase assay, we demonstrated that the direct kinase NEK6 phosphorylates these two serine residues. This phosphorylation event is essential for the degradation of FOXN3 by β-TrCP, which is necessary for stabilizing the association of the Smad complex with its target gene elements, thereby influencing Smad transcriptional activity. A knock-in (KI) mouse model with double serine residue mutations established through genetic substitution provides robust evidence for the critical role of S412 and S416 phosphorylation in the development of pulmonary fibrosis. While both FOXN3 and Smad4 can be direct substrates of β-TrCP, they do not compete in recognizing β-TrCP. Instead, FOXN3 assists in recruiting Smad4 to the β-TrCP/hnRNPU complex for ubiquitination. Upon exposure to pro-fibrotic stimuli such as BLM or TGF-β, NEK6 triggers the phosphorylation of FOXN3 at S412 and S416, leading to its degradation. This, in turn, suppresses Smad4 ubiquitination and stabilizes the association of the Smad complex with its target gene elements, as well as enhances Smad4 stability, thereby activating its transcriptional response and contributing to pulmonary fibrosis (Fig. 9e). Analysis of clinical specimens from fibrotic regions in patients diagnosed with pulmonary fibrosis demonstrated a substantial decrease in FOXN3 protein levels but an increase in Smad4 expression. This emphasizes the importance of the NEK6-FOXN3-Smad axis in the pathogenesis of pulmonary fibrosis. Therefore, targeting this axis could represent a promising strategy for the clinical treatment of pulmonary fibrosis.
Methods
Cell culture and isolation
HEK293T cells BEAS-2B cells and mouse pulmonary fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM). Human primary ATII cells were cultured in Dulbecco’s modified Eagle’s medium/F12 (DME/F12). A549 cells were cultured in an F-12K medium. Both culture media were supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Millipore). Cultures were maintained at 37 °C in a humidified 5% CO2 incubator. The HEK293T, BEAS-2B, and A549 cell lines were obtained from the American Type Culture Collection (ATCC). The human primary ATII cells were obtained from iCell Bioscience (Shanghai, China). For the isolation of pulmonary fibroblasts, we followed the detailed procedure previously reported41,42.
Antibodies and chemical reagents
The primary antibodies were used: For WB: anti-hnRNPU (1:1000, 14599-1-AP, Proteintech), anti-HA (1:2000, 51064-2-AP, Proteintech), anti-Vimentin (1:1000, 10366-1-AP, Proteintech), anti-N-Cadherin (1:1000, 22018-1-AP, Proteintech), anti-E-Cadherin (1:1000, 20874-1-AP, Proteintech), anti-Smad4 (1:1000, 10231-1-AP, Proteintech), anti-FOXN3 (1:1000, 25399-1-AP, Proteintech), anti-Myc (1:1000, 16286-1-AP, Proteintech), anti-Flag (1:1000, 20543-1-AP, Proteintech), anti-GAPDH (1:2000, 2118, Cell Signaling Technology), anti-β-TrCP (1:1000, 4394, Cell Signaling Technology), anti-Smad4 (1:1000, 46535, Cell Signaling Technology), anti-FOXN3 (1:1000, ab129453, Abcam), anti-phospho-Smad2/3 (1:500, 8828, Cell Signaling Technology), anti-Smad2/3 (1:1000, 8685, Cell Signaling Technology), anti-NEK6 (1:1000, sc-374491, Santa Cruz), anti-Lamin B1 (1:1000, 12987-1-AP, Proteintech) and anti-phospho-FOXN3 S412/416 (1:1000). The rabbit polyclonal phospho-specific antibody targeting S412/S416 was generated using the double-phosphorylated peptide (LGDSpGYASpQHK) as the immunogen; For IHC: anti-FOXN3 (1:200, ab129453, Abcam), anti-Smad4 (1:200, AF300076, AiFang biological) and anti-phospho-FOXN3 S412/416 (1:200); For IP: anti-Smad4 (10231-1-AP, Proteintech), anti-FOXN3 (ab129453, Abcam) and anti-rabbit IgG (30000-0-AP, Proteintech). 3–5 μg of the primary antibody was used for each sample; For CUT&Tag: anti-Smad4 (10231-1-AP, Proteintech) and anti-FOXN3 (ab129453, Abcam). 2 μg of the primary antibody was used for each sample; For ChIP-qPCR: anti-Smad2 (12570-1-AP, Proteintech), anti-Smad3 (66516-1-Ig, Proteintech), anti-Smad4 (10231-1-AP, Proteintech) and anti-FOXN3 (ab129453, Abcam). 3–5 μg of primary antibody was used for each sample. The chemical reagents were used, anti-DYKDDDDK magnetic beads (A36798) and Anti-HA magnetic beads (88836) were purchased from Thermo Fisher Scientific. The following chemical reagents were used: Cycloheximide (HY-12320, MedChemExpress), MG132 (HY-13259, MedChemExpress), and TGF-β (HY-P70543, MedChemExpress).
Plasmid constructions
Flag- or HA-tagged FOXN3, NEK6, and hnRNPU were constructed by cloning human complementary DNAs into a pLV-EF1α lentiviral vector. Flag- or Myc-tagged Smad4 was constructed by cloning human complementary DNAs into a pCMV6 vector. HA- or Myc-tagged ubiquitin was cloned into a pCMV6 vector for transient transfection. The K48R and K63R ubiquitin mutants were generated using site-directed mutagenesis. The K48O and K63O ubiquitin mutants were obtained by cloning the synthesized oligonucleotides into a pCMV6 vector. Flag-tagged FOXN3 S412,416 A mutant was generated by site-directed mutagenesis. In addition, Flag- or HA-tagged truncation peptides hnRNPU (Δ550–818) and FOXN3 (ΔForkhead) were generated using the COP-QuickChange (COP-QC) protocol, as previously described43. All constructs generated in this study were verified by DNA sequencing.
Lentivirus and adenovirus
Lentivirus infection was carried out using a pLV-EF1α-based core vector expressing the desired protein or shRNA, along with the psPAX2 packaging and pMD2.G envelope plasmids. These plasmids were co-transfected into HEK293T cells to produce lentiviral particles. The lentiviral infection procedure was conducted following previously established protocols44. The adenovirus encoding Cre-recombinase was purchased from the OBIO company (Shanghai, China).
RNAi knockdown
For shRNA knockdown experiments, the target sequences of shRNAs were inserted into a pHBLV-U6 lentiviral vector. The specific sequences of the shRNAs used are as follows: shFOXN3, 5’-GCCTGACATCCGATTAGAA-3’; shhnRNPU, 5’-AGATCATGGCCGTGGATATTT-3’; shβ-TrCP: 5’-TTACCAACATGGGCACATAAA-3’ (human); 5’-GAATTTGTAGAACACCT TATA-3’ and 5’-CGTCAGAGTGTGGGATGTAAA-3’ (mouse). The siRNA sequences utilized in this study are as follows: siFOXN3 #1, 5’-AGACGATGACCTCGACTTT-3’; siFOXN3 #2, 5’-GCCTGACATCCGATTAGAA-3’; siFOXN3 #3, 5’-CGACGACCACTATGAGTTT-3’. The siRNA targeting NEK6 were purchase from RIBOBIO company (siNEK6 #1, siG000010783A-1-5; siNEK6 #2, siG000010783B-1-5; siNEK6 #3, siB161216031201-1-5; Guangzhou, China).
Immunoprecipitation and ubiquitination assays
For immunoprecipitation, cells were lysed in a lysis buffer containing 50 mM Tris-Cl (pH 7.4), 250 mM NaCl, 1% Triton X-100, 1 mM DTT, protease and phosphatase inhibitor cocktail. The cell lysates were incubated with indicated antibodies for immunoprecipitation, followed by immunoblot analysis. For ubiquitination assay, cells were lysed in a 1% SDS buffer containing Tris-HCl (pH 7.5), 0.5 mM EDTA, 1 mM DTT. After boiling for 10 min, the cell lysates were diluted 10-fold with Tris-HCl buffer. After centrifuge, the supernatant was subjected to immunoprecipitation, followed by immunoblot analysis.
Hydroxyproline assay
The lung hydroxyproline was analyzed using the hydroxyproline colorimetric assay kit from Sigma, following the manufacturer’s instructions. Briefly, the lungs from both the control and experimental mice were dried until a constant weight was achieved. They were then hydrolyzed in 12 M HCl for 3 hours at 120 °C. The resulting digestions were reacted with the Chloramine T reagent and visualized using the DMAB reagent. The absorbance was measured at 560 nm using a microplate reader.
Bronchial alveolar lavage (BAL) fluid acquisition and analysis
The lungs of mice were lavaged three times with 0.8 ml of ice-cold PBS to collect the BAL fluid. The BAL fluid was then centrifuged at 4 °C to separate the cell-free supernatant. This supernatant was used directly to determine the levels of pro-inflammatory cytokines. The pro-inflammatory factors TNFα and IL-6 were detected using two ELISA assay kits from Invitrogen.
BLM-induced fibrosis model and adenovirus delivery
A dosage of 1.5 mg/kg of bleomycin (BLM) was dissolved in 50 μl of saline and intratracheally instilled into the lungs of mice. Three days after BLM induction, an administration of 5 × 108 PFU of adenovirus encoding Cre-recombinase (per mouse) was also performed through intratracheal delivery. The mice were sacrificed three weeks later, following adenoviral infection, in order to isolate their lungs for further studies.
Animal studies
This study involved male C57BL/6 mice aged 8–10 weeks. The Foxn3LSL KI mice, which carry a floxed stop cassette at the Hipp11 locus, were generously provided by Dr. Yinming Liang at Xinxiang Medical University. The FOXN3 KI mice with S379 and S383 mutations, as well as the Foxn3fl/fl mice, were created using the CRISPR/Cas9 gene-editing system, as previously detailed30. The Foxn3fl/fl and Foxn3LSL KI mice were validated through Southern blot analysis (Supplementary Fig. 9a). Furthermore, the FOXN3 KI mice carrying the S379 and S383 mutations were confirmed using Sanger sequencing (Supplementary Fig. 9b). The genotyping was verified by PCR analysis. All mice were housed under specific pathogen-free (SPF) conditions with a 12 h light/12-hour dark cycle at 21–24 °C and 40–60% humidity and were fed a standard chow diet. At the conclusion of the animal studies, all mice were euthanized using carbon dioxide inhalation. All animal studies were approved by the Institutional Review Board of Bengbu Medical University, and the manuscript complies with the ARRIVE guidelines for the reporting of animal experiments. The specific gRNA sequences for the Foxn3fl/fl mice are: gRNA-A1: GGTGTGCTGACTTGATTAGG – TGG; gRNA-A2: CCAA GGGCTGTCGTGGCCGA – GGG; gRNA-B1: TGCGGTGTGCTGACTTGATT – AGG; gRNA-B2: CCCAAGGGCTGTCGTGGC CG – AGG. For the S379 and S383 mutation KI mice, the gRNA sequences are: gRNA-A1: AGTGGCTTTCTCCACCGTGC – TGG; gRNA-A2: TGGA GGAAGTACGCCATCAA – GGG; gRNA-B1: GTGGCTTTCTCCACCGTGCT – GGG; gRNA-B2: GAAGTACGCCATCAAGG GGT – TGG.
Histological assays
For the histological staining assay, the lungs of mice were fixed in 4% paraformaldehyde and then embedded in paraffin. The embedded tissue was sectioned at a thickness of 4 μm. These sections were subsequently subjected to hematoxylin and eosin (H&E) staining or Masson trichrome staining analysis. The detailed procedures for staining were carried out as previously described30.
High-throughput mRNA sequencing
The mRNA sequencing experiment was conducted by GENEWIZ (Suzhou, China). In this experiment, total RNAs were isolated using Trizol reagent, and then library construction was performed following the standard Illumina protocol. The libraries were sequenced using the Illumina NovaSeq 6000 platform with the paired-end RNA-seq approach. For the analysis of the resulting data, a detailed method was employed, which was previously reported45. The raw data has been deposited in the Sequence Read Archive (SRA) database with accession numbers SRR31107669, SRR31107670, SRR31107671, SRR31107672, SRR31107673, and SRR31107674.
Quantitative real-time PCR analysis
Total RNAs were extracted from cells using Trizol reagent, following the instructions provided by the manufacturer. For reverse transcription, 1 μg of RNA was used, and the reverse transcription reaction was performed according to the instructions provided with a 5 × All-In-One RT Kit (G486, Applied Biological Materials). Quantitative real-time PCR was conducted using an EvaGreen qPCR Master Mix (G891, Applied Biological Materials). The relative gene expression levels were determined using the 2-ΔΔCT method. The primer sequences used for the gene amplification in this study can be found in the Supplementary Table 1 and 2.
Luciferase assay
A Firefly luciferase reporter plasmid containing a promoter with a Smad response element was combined with a Renilla luciferase reporter plasmid and the specified plasmids. These plasmids were then co-transfected into cells for further analysis. Luminescence was measured, and the data analysis for Smad transcriptional activity was performed following previously reported methods45.
Chromatin immunoprecipitation (ChIP) assay
Briefly, 1 × 106 A549 cells were cross-linked with 1% formaldehyde for 10 min at room temperature, then quenched using 125 mM glycine. The cells were lysed using a lysis buffer containing 50 mM Tris-HCl (pH7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5 mM PMSF, and the genomic DNAs were subjected to sonication to acquire fragments of broken DNA (100 to 500 bp). After centrifuge, the cleared cell lysates were subjected to immunoprecipitation with indicated antibodies, followed by quantitative PCR analysis. The detailed procedures were conducted as previously reported46. The primer sequences used in quantitative PCR analysis were shown in Supplementary Table 3.
CUT&Tag assay
CUT&Tag was conducted using the NovoNGS® CUT&Tag High-Sensitivity Kit. A549 cells were incubated at 37 °C or 4 °C, harvested, and centrifuged at 500 × g for 4 min. After gentle lysis and a 10 min incubation on ice, intact nuclei were collected by centrifugation. The nuclei were resuspended in PBS, cross-linked with formaldehyde for 2 min, and the reaction was quenched with glycine. Concanavalin A-coated magnetic beads were added and incubated for 15 min. The unbound supernatant was removed, and the beads were resuspended in a 1:50 dilution of the indicated primary antibody incubated overnight at 4 °C. Following a 1 h incubation with a secondary antibody, the nuclei were washed and incubated with the pA-Tn5 adapter complex for 1 h. Nuclei were resuspended in tagmentation buffer with Mg2+ and incubated at 37 °C for 1 h, then treated with SDS overnight at 37 °C. DNA was extracted using Tagment DNA Extract Beads, followed by PCR amplification and SPRI magnetic bead cleanup. A negative control omitted the primary antibody. The purified DNA was used for library preparation, quality-assessed on the Agilent Bioanalyzer 2100, and sequenced on an Illumina platform. The raw data have been deposited in the Sequence Read Archive (SRA) database with accession numbers SRR31070175 and SRR31070176.
Step-wise fractionation assay
In brief, 1 × 107 cells were swollen in buffer A (10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCl; 1 mM DTT; 0.5 mM PMSF). The swollen cells were extracted using buffer A containing 1% NP-40, followed by two additional rounds of extraction with a low-salt buffer A containing 0.5% NP-40 and 75 mM NaCl. The supernatants from all three rounds of extraction were combined and designated as the low-salt fraction (LSF). The low-salt extracted nuclei (LSEN) were lysed in an SDS-loading buffer for immunoblot analysis or subjected to the ubiquitination assay as described above.
Patient samples
Lung specimens from patients with pulmonary fibrosis were obtained in accordance with research ethics board approval from Bengbu Medical University and the Affiliate Hospitals. Informed written consent was obtained from all participants in the study. The samples collected included both lesion areas and corresponding adjacent normal areas. These samples were then subjected to Masson trichrome or immunohistochemical staining analysis, following previously described protocols30. The detailed clinical information of patients was shown in Supplementary Table 4.
Statistical analysis
All statistical analyses were conducted using GraphPad Prism software. The data are represented as mean ± SD calculated using GraphPad. A two-tailed Student’s t test was utilized to compare two groups, while a one-way ANOVA followed by Tukey’s post hoc test was employed for comparisons involving three or more groups. A p-value of less than 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the article and its Supplementary Information files. The raw RNA sequencing data have been deposited in the Sequence Read Archive (SRA) under the accession numbers SRR31107669, SRR31107670, SRR31107671, SRR31107672, SRR31107673, and SRR31107674. (https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=1177358). The raw data for CUT&Tag have been deposited in the SRA with the accession numbers SRR31070175 and SRR31070176 (https://www.ncbi.nlm.nih.gov/biosample?LinkName=bioproject_biosample_all&from_uid=1174104). Source data are provided in this paper.
References
Lederer, D. J. & Martinez, F. J. Idiopathic pulmonary fibrosis.N. Engl. J. Med. 378, 1811–1823 (2018).
Moss, B. J., Ryter, S. W. & Rosas, I. O. Pathogenic mechanisms underlying idiopathic pulmonary fibrosis. Annu. Rev. Pathol. 17, 515–546 (2022).
Richeldi, L., Collard, H. R. & Jones, M. G. Idiopathic pulmonary fibrosis. Lancet 389, 1941–1952 (2017).
Shenderov, K., Collins, S. L., Powell, J. D. & Horton, M. R. Immune dysregulation as a driver of idiopathic pulmonary fibrosis. J. Clin. Investig. 131, https://doi.org/10.1172/jci143226 (2021).
Lynch, D. A. et al. Diagnostic criteria for idiopathic pulmonary fibrosis: a Fleischner Society White Paper. Lancet. Respir. Med.6, 138–153 (2018).
Martinez, F. J. et al. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Primers 3, 17074 (2017).
Chapman, H. A. Epithelial-mesenchymal interactions in pulmonary fibrosis. Annu. Rev. Physiol. 73, 413–435 (2011).
Namba, T. et al. Induction of EMT-like phenotypes by an active metabolite of leflunomide and its contribution to pulmonary fibrosis. Cell Death Differ. 17, 1882–1895 (2010).
Fang, X. Z. et al. Mechanosensitive ion channel Piezo1 mediates mechanical ventilation-exacerbated ARDS-associated pulmonary fibrosis. J. Adv. Res 53, 175–186 (2022).
Lee, H. W., Jose, C. C. & Cuddapah, S. Epithelial-mesenchymal transition: Insights into nickel-induced lung diseases. Semin. Cancer Biol. 76, 99–109 (2021).
Xie, L. & Zeng, Y. Therapeutic potential of exosomes in Pulmonary Fibrosis. Front. Pharmacol. 11, 590972 (2020).
Ling, H., Xiao, H., Luo, T., Lin, H. & Deng, J. Role of ferroptosis in regulating the epithelial-mesenchymal transition in pulmonary fibrosis. Biomedicines 11, 163 (2023).
Crowl, J. T. et al. Tissue-resident memory CD8(+) T cells possess unique transcriptional, epigenetic and functional adaptations to different tissue environments. Nat. Immunol. 23, 1121–1131 (2022).
Lan, Y. et al. Colocalized targeting of TGF-beta and PD-L1 by bintrafusp alfa elicits distinct antitumor responses. J. Immunother. Cancer 10, e004122 (2022).
Yu, X. et al. The cytokine TGF-beta promotes the development and homeostasis of alveolar macrophages. Immunity 47, 903–912 (2017).
Zhang, Y., Alexander, P. B. & Wang, X. F. TGF-beta family signaling in the control of cell proliferation and survival. Cold Spring Harb. Perspect. Biol. 9, a022145 (2017).
Clarke, D. C. & Liu, X. Decoding the quantitative nature of TGF-beta/Smad signaling. Trends Cell Biol. 18, 430–442 (2008).
Lee, J. H. & Massague, J. TGF-beta in developmental and fibrogenic EMTs. Semin. Cancer Biol. 86, 136–145 (2022).
Liu, J., Jin, J., Liang, T. & Feng, X. H. To Ub or not to Ub: a regulatory question in TGF-beta signaling. Trends Biochem. Sci. 47, 1059–1072 (2022).
ten Dijke, P. & Hill, C. S. New insights into TGF-beta-Smad signalling. Trends Biochem. Sci. 29, 265–273 (2004).
Zhang, J. et al. The regulation of TGF-beta/SMAD signaling by protein deubiquitination. Protein Cell 5, 503–517 (2014).
Borok, Z. et al. Grp78 Loss in epithelial progenitors reveals an age-linked role for endoplasmic reticulum stress in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 201, 198–211 (2020).
Boutanquoi, P. M. et al. TRIM33 prevents pulmonary fibrosis by impairing TGF-beta1 signalling. Eur. Respir. J. 55, 1901346 (2020).
Louzada, R. A. et al. NADPH oxidase DUOX1 sustains TGF-beta1 signalling and promotes lung fibrosis. Eur. Respir. J. 7, 1901949 (2021).
Wang, J. et al. Nestin promotes pulmonary fibrosis via facilitating recycling of TGF-beta receptor I. Eur. Respir. J. 59, 2003721 (2022).
Astanina, E. et al. The TFEB-TGIF1 axis regulates EMT in mouse epicardial cells. Nat. Commun. 13, 5191 (2022).
Mishra, V. K. et al. Kruppel-like transcription factor KLF10 suppresses TGFbeta-induced epithelial-to-mesenchymal transition via a negative feedback mechanism. Cancer Res. 77, 2387–2400 (2017).
Sakai, S. et al. Long noncoding RNA ELIT-1 acts as a Smad3 cofactor to facilitate TGFbeta/Smad signaling and promote epithelial-mesenchymal transition. Cancer Res. 79, 2821–2838 (2019).
Song, C. & Zhou, C. HOXA10 mediates epithelial-mesenchymal transition to promote gastric cancer metastasis partly via modulation of TGFB2/Smad/METTL3 signaling axis. J. Exp. Clin. Cancer Res. 40, 62 (2021).
Zhu, X. et al. p38-mediated FOXN3 phosphorylation modulates lung inflammation and injury through the NF-kappaB signaling pathway. Nucleic Acids Res. 51, 2195–2214 (2023).
Andugulapati, S. B., Gourishetti, K., Tirunavalli, S. K., Shaikh, T. B. & Sistla, R. Biochanin-A ameliorates pulmonary fibrosis by suppressing the TGF-beta mediated EMT, myofibroblasts differentiation and collagen deposition in in vitro and in vivo systems. Phytomedicine 78, 153298 (2020).
Liu, W., Han, X., Li, Q., Sun, L. & Wang, J. Iguratimod ameliorates bleomycin-induced pulmonary fibrosis by inhibiting the EMT process and NLRP3 inflammasome activation. Biomed. Pharmacother. 153, 113460 (2022).
Su, J. et al. TGF-beta orchestrates fibrogenic and developmental EMTs via the RAS effector RREB1. Nature 577, 566–571 (2020).
Tsuchiya, Y. et al. Nuclear IKKbeta is an adaptor protein for IkappaBalpha ubiquitination and degradation in UV-induced NF-kappaB activation. Mol. Cell 39, 570–582 (2010).
Wan, M. et al. SCF(beta-TrCP1) controls Smad4 protein stability in pancreatic cancer cells. Am. J. Pathol. 166, 1379–1392 (2005).
Ci, Y. et al. SCF(beta-TRCP) E3 ubiquitin ligase targets the tumor suppressor ZNRF3 for ubiquitination and degradation. Protein Cell 9, 879–889 (2018).
Frescas, D. & Pagano, M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 (2008).
Reynolds, H. Y. Lung inflammation and fibrosis: an alveolar macrophage-centered perspective from the 1970s to 1980s. Am. J. Respir. Crit. Care Med. 171, 98–102 (2005).
Thannickal, V. J., Toews, G. B., White, E. S., Lynch, J. P. 3rd & Martinez, F. J. Mechanisms of pulmonary fibrosis. Annu. Rev. Med. 55, 395–417 (2004).
Ward, P. A. & Hunninghake, G. W. Lung inflammation and fibrosis. Am. J. Respir. Crit. Care Med. 157, S123–S129 (1998).
Bonniaud, P. et al. Connective tissue growth factor is crucial to inducing a profibrotic environment in “fibrosis-resistant” BALB/c mouse lungs. Am. J. Respir. Cell Mol. Biol. 31, 510–516 (2004).
Janssen, L. J., Farkas, L., Rahman, T. & Kolb, M. R. ATP stimulates Ca(2+)-waves and gene expression in cultured human pulmonary fibroblasts. Int. J. Biochem. Cell Biol. 41, 2477–2484 (2009).
Wang, H. et al. An efficient approach for site-directed mutagenesis using central overlapping primers. Anal. Biochem. 418, 304–306 (2011).
Zhu, X. et al. LncRNA NKILA regulates endothelium inflammation by controlling a NF-kappaB/KLF4 positive feedback loop. J. Mol. Cell. Cardiol. 126, 60–69 (2018).
Lian, J. et al. Extracellular vesicle-transmitted miR-671-5p alleviates lung inflammation and injury by regulating the AAK1/NF-kappaB axis. Mol. Ther. 31, 1365–1382 (2023).
Zhu, X. et al. LncRNA HOXA-AS2 represses endothelium inflammation by regulating the activity of NF-kappaB signaling. Atherosclerosis 281, 38–46 (2019).
Acknowledgements
This work received support from grants provided by the National Natural Science Foundation of China (81900392 to X.Z. and 82402634 to J.Y.) and the Translational Project in Clinical Medical Research (202304295107020074 to Y.Z.). The generation of the Foxn3LSL KI mice used in this study was supported by the 111 Program (D20036). The schematic diagrams for this study were produced under an academic license from BioRender.
Author information
Authors and Affiliations
Contributions
X.Z., W.L., Juntang. Lin, J.G., and E.J.M. designed and conceived the project. J.Y., Yingke Li, Yiming Li, X.L., and Q.H. performed most of the experiments. N.W. and Yiming Li. collected clinical samples and performed data analysis. T.Z., Yong Zhang, J.Z., and H.Y.L. assisted in designing the experiments. Yangxia Zhang and Jie Lian contributed to data analysis and the creation of the schematic representation. X.Z. wrote the manuscript, and all authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Corinne Dupuy, Amnon Schlegel, and the other anonymous9 reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yu, J., Li, Y., Li, Y. et al. Phosphorylation of FOXN3 by NEK6 promotes pulmonary fibrosis through Smad signaling. Nat Commun 16, 1865 (2025). https://doi.org/10.1038/s41467-025-56922-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56922-7
This article is cited by
-
Chitosan-functionalized mesoporous silica nanoparticles co-loaded with chrysin and quercetin: a potent strategy against lung cancer cells
Scientific Reports (2026)
-
Extracellular vesicles derived from menstrual blood-derived mesenchymal stem cells suppress inflammatory atherosclerosis by inhibiting NF-κB signaling
BMC Medicine (2025)