Abstract
Activation of endogenous full-length utrophin, a dystrophin homolog, presents an attractive therapeutic strategy for Duchenne muscular dystrophy (DMD), regardless of mutation types and loci. However, current dCas9-based activators are too large for efficient adeno-associated virus delivery, and the feasibility and durability of such treatments remain unclear. Here, we develop a muscle-targeted utrophin activation system using the compact dCasMINI-VPR system, termed MyoAAV-UA. Systemic administration of MyoAAV-UA in male mdx mice leads to substantial upregulation of utrophin at the sarcolemma, resulting in significant improvements in skeletal muscle function and a slowing of heart function deterioration. These benefits remain observable at six months post-treatment. In male nonhuman primates, systemic administration of MyoAAV-UA increases utrophin expression by twofold in skeletal muscle, with no significant side effects observed. Furthermore, MyoAAV-UA upregulates utrophin and utrophin-glycoprotein complexes in induced pluripotent stem cell-derived myotubes from DMD patients. In conclusion, these findings demonstrate the potential of MyoAAV-UA as a therapeutic approach for DMD.
Similar content being viewed by others
Introduction
Duchenne muscular dystrophy (DMD) is the most prevalent childhood-onset genetic neuromuscular disorder, affecting ~1 in every 3500–5000 male live-born infants1. Boys with DMD typically begin to experience difficulties climbing stairs and develop waddling gate symptoms around the ages of 2–42. Most become wheelchair-dependent ~12 years old and ultimately face fatal cardiorespiratory failure in their early twenties3. Unfortunately, there is currently no cure for DMD4.
DMD is caused by mutations in the dystrophin gene (DMD), with thousands of mutations identified5. Research on DMD treatment has primarily focused on restoring dystrophin functions. While several strategies, such as antisense oligonucleotides (ASOs), Premature Termination Codon, and micro/mini-dystrophin gene therapies, have advanced from the bench to the clinic, remaining issues include limited patient coverage, suboptimal therapeutic efficacy, high costs, and immunogenicity issues6,7.
Utrophin shares ~85% homology with dystrophin8. In muscles of DMD patients, utrophin is re-expressed on the sarcolemma, enhancing early muscle function8,9. Thus, theoretically, upregulation of utrophin expression and ensuring its membrane localization is considered a promising approach for all DMD patients. Currently, there are two main approaches to overexpressing utrophin: activating endogenous full-length utrophin and delivering exogenous synthetic micro-utrophin genes or proteins10. While micro-utrophin delivered through AVs or AAVs shows efficacy in DMD mice and dogs11,12,13, it is unclear whether the truncated micro-utrophin fully replicates the function of the full-length protein. In contrast, activation of endogenous full-length utrophin may offer functional completeness, applicability to all dystrophin mutation types and loci, and non-immunogenicity14.
Pharmacological activation of endogenous utrophin showed alleviated muscle-related symptoms in DMD mouse models15. The small molecule utrophin modulator Ezutromid was shown to be safe and well-tolerated in Phase 1 trials. In the Phase 2 study (NCT02858362), treatment for 24 weeks led to increased utrophin expression and a significant reduction in muscle damage in DMD patients. However, after 48 weeks of treatment, no significant clinical efficacy difference was observed between the Ezutromid treatment group and the placebo group, which may be due to metabolism15,16,17. CRISPR activation (CRISPRa) systems, which use nuclease-deactivated dCas proteins fused with transcriptional or epigenetic effectors, show precise upregulation of targeted genes and could be used to activate endogenous utrophin18. However, delivering second-generation dCas9 activators (e.g., dCas9-VPR/MPH/SAM/SPH/SunTag) in vivo is challenging due to the AAV payload limit (~4.7 kb)19. This limitation necessitates a two-component delivery approach20,21. Additionally, the feasibility, effectiveness, and long-term sustainability of systemic CRISPRa treatment remain underexplored. To address these, in this study, we developed a highly efficient muscle-directed CRISPR activator based on dCasMINI-VPR22 and achieved single AAV delivery, termed single MyoAAV-delivered utrophin activator (MyoAAV-UA), and then assessed its efficacy, safety, and long-term sustainability. Our results show that MyoAAV-UA effectively activates endogenous full-length utrophin across multiple species, including mice, cynomolgus monkeys, and humans. Upregulation of endogenous full-length utrophin ameliorates disease symptoms with long-term efficacy in Dmd mice, exhibits efficiency and safety in cynomolgus monkeys, and upregulates utrophin-glycoprotein complex (UGC) in induced pluripotent stem cell (iPSC)-derived myoblasts from DMD patients, indicating that MyoAAV-UA is a promising therapeutic strategy for all DMD patients.
Results
MyoAAV-UA effectively activates utrophin in different Dmd mouse models
To evaluate the efficacy of dCasMINI-VPR in activating utrophin (Fig. 1a), 18–20 sgRNAs located 2000 bp upstream of the transcription start site were designed to target the promoter region of the Utrn/UTRN gene in mice, cynomolgus monkeys, or humans, respectively (Supplementary Data 1). Mouse neuroblastoma cells (Neuro 2a), cynomolgus monkey skeletal muscle cells (MSkM), and human embryonic kidney 293 T cells (HEK293T) were transfected with dCasMINI-VPR containing different sgRNAs. RT-qPCR results revealed significant utrophin upregulation across all tested species (Supplementary Fig. 1).
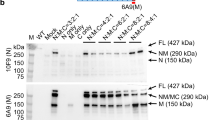
a Schematic diagram of the dCasMINI-VPR construct used for utrophin activation. b–d RT-qPCR analysis showing high efficiency of combined sgRNAs in activating utrophin mRNA in Neuro 2a cell line (b), cynomolgus monkey skeletal muscle cells (MSkM) (c), and HEK293T cell line (d). Data are normalized to control (mock-sgRNA). n = 3 technical replicates in each group. e Schematic of the MyoAAV-UA expression cassette and its application in Dmd mice, NHPs, and humans. f Representative immunoblots of dystrophin and utrophin in quadriceps muscles from WT group, mdx mice treated with saline, mdx mice treated with MyoAAV-UA, DmdΔEx50h mice treated with saline, and DmdΔEx50 mice treated with MyoAAV-UA. Vinculin was used as a loading control. g Representative immunofluorescence staining of dystrophin (green), utrophin (red), and DAPI (blue) in quadriceps muscles from different groups. Scale bar, 100 µm. h Quantification of utrophin protein levels in quadriceps from different groups were normalized to vinculin (n = 6 mice in each group). i Quantification of utrophin mRNA expression levels in quadriceps muscles from different groups, normalized to Gapdh (n = 6 mice in each group). f, g The experiment was repeated with six biologically independent replicates with similar results. h, i Data are normalized to WT (n = 6 biologically independent experiments in each group). b–d, h, i Data are presented as mean ± s.d. Statistical analysis was performed using one-way ANOVA followed by Dunnett’s post hoc test. Source data are provided as a Source Data file.
Studies have shown that using multiple sgRNAs to target both the promoter and enhancer regions of the same gene can significantly enhance target gene activation, improving specificity and efficiency19,23,24. The compact nature of the dCasMINI system provides sufficient space to accommodate multiple sgRNAs, enabling the simultaneous targeting of multiple sites to enhance the activation of utrophin. We tested combinations of the most effective sgRNAs and found that certain sgRNA pairs, especially M8 + M12 (mouse), Rh13 + Rh17 (monkey), and H1 + H10 (human), resulted in utrophin activation levels of 3.04-fold, 8.75-fold, and 3.92-fold, respectively, exhibiting superior activation efficiency compared to single sgRNAs (Fig. 1b–d). These results demonstrate the effectiveness of the dCasMINI-VPR system in achieving high utrophin activation in vitro.
To achieve specific and robust activation of endogenous utrophin in muscle tissues, we employed the SPc5-12 promoter, known for its cardiac and skeletal muscle specificity25,26, to drive the expression of the dCasMINI-VPR system and fused it with screened dual sgRNAs targeting the Utrn/UTRN gene promoter. Furthermore, to save space and efficiently express multiple sgRNAs targeting the UTRN gene promoter, we fused effective dual sgRNA combinations using tRNA under a single human U6 promoter27,28. This integrated compact vector (4.6 kb) was then delivered using a muscle-specific serotype, MyoAAV29, creating a potent muscle utrophin activation system referred to as MyoAAV-UA (Fig. 1e).
We then evaluated the effectiveness of MyoAAV-UA in activating endogenous utrophin in two well-established Dmd mouse models with different mutations: one with a nonsense mutation in exon 23 (mdx mice) and another with an exon 50 deletion (DmdΔEx50 mice)30,31. Both of these mouse models exhibit characteristic DMD muscle pathology and no dystrophin protein expressed (Fig. 1f, g). We administrated MyoAAV-UA, containing M8 and M12, via intramuscular injection into the quadriceps (QD) muscles of mdx and DmdΔEx50 mice at a dose of 1 × 1011 vg. Muscle samples were collected two weeks post-treatment. Both mdx mice (3.73-fold) and DmdΔEx50 mice (2.43-fold) showed a significant upregulation of utrophin protein expression compared to the WT group (Fig. 1f, h). The mRNA levels of utrophin were 5.29-fold higher in mdx muscles and 3.75-fold higher in DmdΔEx50 muscles compared to the WT group (Fig. 1i). Immunostaining showed that utrophin was upregulated along the sarcolemma in both mdx and DmdΔEx50 mice, closely resembling the distribution of dystrophin in the WT group (Fig. 1g).
To evaluate whether MyoAAV-UA has tissue-specific delivery and dose-dependent effects, 2-week-old mdx mice were systemically injected with saline or varying doses of MyoAAV-UA (2 × 1012 vg/kg, 2 × 1013 vg/kg, and 5 × 1013 vg/kg). Muscle tissue and multiple organ samples were collected four weeks post-treatment for analysis. Viral genome copies are significantly higher in skeletal muscle and heart tissues compared to non-muscle organs (e.g. liver, spleen, kidney, brain, and lung), indicating the muscle specificity of MyoAAV-UA. A dose-dependent increase in viral genome copies was observed across all tissues (Supplementary Fig. S2a–e). Consistently, a dose-dependent increase in dCasMINI expression was detected (Supplementary Fig. S2f). Notably, increasing the injection dose from 2 × 1013 to 5 × 1013 vg/kg led to a significant increase in utrophin expression, whereas increase in utrophin expression between 2 × 1012 and 2 × 1013 vg/kg was less pronounced (Supplementary Fig. S3a–d and S4a). Histological staining of skeletal muscle and heart tissue sections were corroborated this dose-dependent effect, the 5 × 1013 vg/kg dose showing the most substantial improvement in myofiber morphology and fibrosis (Supplementary Fig. S3e, S4b, c, and S5). Based on the robust utrophin expression achieved at the highest dose (5 × 1013 vg/kg), this dose was selected for subsequent experiments.
Together, these results demonstrate the high muscle specificity of MyoAAV-UA and its efficacy in activating utrophin expression in various muscles of mdx mice.
The potential off-target effects of MyoAAV-UA were examined eight weeks post-injection. MyoAAV-UA carrying mock-sgRNA (MyoAAV-mock) or Utrn-targeted sgRNA (MyoAAV-UA) was systemically administered into mdx mice. RNA-seq analysis revealed no significant difference in the expression of Utrn or other endogenous genes between the mdx-MyoAAV-mock and mdx-saline groups (Supplementary Fig. S6a), suggesting no significant off-target effects of the system. The utrophin mRNA expression in the mdx-saline group and mdx-MyoAAV-mock group was higher than in the WT group (Supplementary Fig. S6b, c). Importantly, utrophin mRNA expression in the mdx-MyoAAV-UA group was significantly higher than in the mdx-MyoAAV-mock, mdx-saline, and WT groups (Supplementary Fig. S6d–f), confirming that MyoAAV-UA specifically and effectively targets utrophin.
Together, these results demonstrate the high efficacy of MyoAAV-UA in activation of endogenous utrophin in a dose-dependent manner, with high muscle specificity and no detectable off-target effects.
Activation of endogenous utrophin alleviates dystrophic phenotypes in mdx mice with a long-term effect
We next evaluated the effectiveness of the MyoAAV-UA in alleviating dystrophic phenotypes. Two-week-old mdx mice were systemically administrated with 5 × 1013 vg/kg MyoAAV-UA (Fig. 2a). At eight weeks post-MyoAAV-UA injection, utrophin protein levels increased by 3.04-fold in biceps brachii (BB) and 3.10-fold in diaphragms (DIA), while utrophin mRNA levels significantly increased by over 4.21-fold in BB and 2.56-fold in DIA (Fig. 2b–g). Six months post-injection, the MyoAAV-UA group continued to exhibit significantly higher utrophin expression in muscles compared to the mdx-saline group, with a more than 1.87-fold increase in BB and 1.91-fold increase in DIA (Fig. 2c–e). RT-qPCR showed a 1.58-fold increase in utrophin mRNA in BB and a 1.87-fold increase in the DIA (Fig. 2f, g). However, a significant decline in viral genome copies is observed at 6 months compared to 8 weeks in both QD and heart tissues, suggesting reduced viral presence over time. Similarly, dCasMINI expression levels also decrease at 6 months, consistent with the reduction in viral genome copies (Supplementary Fig. S7).
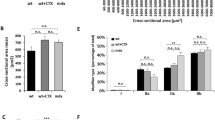
a Experimental timeline depicts MyoAAV-UA systemic injection and subsequent muscle biopsy time points in mdx mice. b, c Immunoblots of utrophin and dystrophin in biceps brachii (BB) and diaphragm (DIA) from different groups at 8 weeks (b) and 6 months (c) post-MyoAAV-UA injection. Vinculin was used as a loading control. d, e Quantification of utrophin protein levels in BB (d) and DIA (e) were normalized to vinculin (n = 6 mice in each group). f, g Quantification of utrophin mRNA expression level in BB (f) and DIA (g) at 8 weeks and 6 months post-MyoAAV-UA injection, normalized to Gapdh (n = 6 mice in each group). h, k Representative immunofluorescence staining of utrophin in BB, QD, and DIA muscles at 8 weeks and 6 months post-MyoAAV-UA injection. Scale bar, 100 µm. i, l Representative H&E staining of BB, QD, and DIA muscles at 8 weeks and 6 months post-MyoAAV-UA injection. Scale bar, 100 µm. j, m Representative Sirius Red staining of BB, QD, and DIA muscles at 8 weeks and 6 months post-MyoAAV-UA injection. Scale bar, 100 µm. n Statistical analysis of centrally nucleated myofibers in BB and QD (n = 6 mice in each group). o, p Statistical analysis of the fibrotic area percentage, indicated by red in Sirius Red-stained sections of BB and QD (o), and DIA (p) (n = 6 mice in each group). q Serum creatine kinase levels in different groups at 8 weeks and 6 months post-MyoAAV-UA injection (n = 6 mice in each group). b, c, h–m the experiment was repeated with six biologically independent replicates with similar results. d–g, n–q data are presented as mean ± s.d (n = 6 biologically independent experiments). Statistical analysis was performed using one-way ANOVA followed by Dunnett’s post hoc test. BB biceps brachii, QD quadriceps, DIA diaphragm. Source data are provided as a Source Data file.
Utrophin was prominently observed on the sarcolemma of BB, QD, and DIA muscles eight weeks after MyoAAV-UA administration (Fig. 2h and Supplementary Fig. S8a, c, e). These muscles exhibited reduced inflammatory cell infiltration, decreased centrally nucleated myofibers percentage, improved myofiber alignment and integrity, and decreased fibrosis (Fig. 2h–j, n–p, Supplementary Fig. S9 and S10a, b). The MyoAAV-UA-treated group showed a significant reduction in embryonic myosin heavy chain fibers (Supplementary Fig. S11a) and decreased levels of MuRF1 and Desmin (Supplementary Fig. S11b, c), indicating reduced muscle damage. Immunofluorescence staining confirmed the restoration of the utrophin-glycoprotein complex (UGC) in MyoAAV-UA-treated mdx mice, with α-sarcoglycan and syntrophin co-localized with utrophin at the sarcolemma, resembling the WT group (Supplementary Fig. S12a, b). However, neuronal nitric oxide synthase, a marker of the dystrophin-glycoprotein complex (DGC), was not localized to the sarcolemma in the MyoAAV-UA-treated group, confirming the UGC-specific composition (Supplementary Fig. S12c). Taken together, these findings demonstrate that MyoAAV-UA restores UGC components, such as α-sarcoglycan and syntrophin, while reducing muscle damage and inflammation, exerting a protective and stabilizing effect on muscle tissue.
Whole transcriptome analysis revealed significant differences between the mdx and WT groups, while the MyoAAV-UA treated group exhibited a gene expression profile closely resembling that of the WT group, indicating muscle function recovery post-treatment (Supplementary Fig. S13a). Gene Ontology (GO) analysis revealed a close association between upregulated utrophin expression in the MyoAAV-UA-treated group and the enrichment of transcripts related to muscle development, DGC construction, myotube fusion, and extracellular matrix composition (Supplementary Fig. S13b). Weighted gene co-expression network analysis (WGCNA) further identified clusters of genes with similar expression patterns, showing significant correlations between the MyoAAV-UA and WT groups, particularly within the cyan and red modules. In contrast, no correlative modules were found between the mdx-saline group and the other two groups (Supplementary Fig. S13c). Random Forest analysis revealed a significant increase in the ranking of the turquoise module in the MyoAAV-UA group, which is associated with muscle cell development and the negative regulation of protein degradation (Supplementary Fig. S13d-f).
Notably, six months post-treatment, the MyoAAV-UA-treated group still exhibited elevated utrophin expression on muscle cell membranes, reduced inflammatory cell infiltration, decreased fibrosis, and improved myofiber alignment and integrity (Fig. 2k–p, Supplementary Fig. S8b, d, f, S9, S10c, d). Serum creatine kinase (CK) levels, an important marker of muscle injury, were significantly decreased to WT levels at eight weeks post-injection and continued to show significant reductions at six months (Fig. 2q). Muscle strength and exercise tolerance, critical indicators of patient condition32 were assessed through behavioral evaluations at 4 weeks, 8 weeks, and 6 months post-MyoAAV-UA injection. No significant changes in body weight or mortality were observed within six months post-treatment (Supplementary Fig. S14a, b). The MyoAAV-UA-treated group showed significant improvements in grip strength and Rotor-Rod test performance at 8 weeks, with these improvements persisting at 6 months (Supplementary Fig. S14c, d). Improvements in locomotor activity were observed at 8 weeks post-treatment but not at 6 months (Supplementary Fig. S14e, f). Collectively, these findings demonstrate that MyoAAV-UA is a potent activator of muscular utrophin, effectively alleviating dystrophic muscle phenotypes while exhibiting long-term efficacy and safety.
Activation of endogenous utrophin slows down heart function deterioration in mdx mice
Heart failure remains one of the leading causes of mortality in DMD patients33. To evaluate the effects of utrophin activation on cardiac phenotypes, we analyzed hearts at eight weeks and 6 months post-MyoAAV-UA injection. At 8 weeks, the utrophin protein level increased by 2.17-fold, while at 6 months, it increased by 1.35-fold (Fig. 3a, b). Similarly, the utrophin mRNA level showed a significant increase of 4.54-fold at 8 weeks and 2.26-fold at 6 months (Fig. 3c). Immunofluorescent staining showed significantly increased utrophin expression on cardiomyocyte membrane at 8 weeks, with sustained expression at 6 months (Fig. 3d). Hematoxylin and Eosin (H&E), Sirius Red staining, and immunological staining revealed a notable reduction in inflammatory cell infiltration and dramatically decreased fibrosis at 6 months in the MyoAAV-UA-treated group, showing similar morphology to WT (Fig. 3e–g and Supplementary Fig. S15).
a Representative immunoblots of utrophin in the hearts from different groups at 8 weeks and 6 months post-MyoAAV-UA injection. Vinculin was used as a loading control. b Quantification of utrophin protein levels in the heart was normalized to vinculin (n = 6 mice in each group). c Quantification of utrophin mRNA expression level in the hearts from different groups, normalized to Gapdh (n = 6 mice in each group). d Representative immunofluorescence staining of utrophin (red) in the hearts of different groups. Scale bar, 100 µm. e Representative H&E staining of heart tissues at 8 weeks and 6 months post-MyoAAV injection. Scale bar, 100 µm. f Representative Sirius Red staining of heart tissues at 8 weeks and 6 months post-MyoAAV injection. Scale bar, 100 µm. g Statistical analysis of the fibrotic area percentage, indicated by red in Sirius Red-stained sections of the hearts (n = 6 mice in each group). h Transcriptomic analysis of relative utrophin mRNA expression in the hearts from different groups (n = 6 mice in each group). i Gene Ontology (GO) analysis shows pathways enriched with differential gene expression in different groups. Significant terms (Benjamini–Hochberg adjusted P < 0.05) were identified via a one-sided hypergeometric test (over-representation analysis) with FDR correction for multiple comparisons. j, k Gene Set Enrichment Analysis (GSEA) of gene expression related to cardiac muscle contraction (j), and cardiac conduction (k). a, d–f The experiment was repeated with six biologically independent replicates with similar results. b, c, g, h Data are presented as mean ± s.d. (n = 6 biologically independent samples). Statistical analysis was performed using one-way ANOVA followed by Dunnett’s post hoc test. Source data are provided as a Source Data file.
We further conducted a transcriptome analysis of the myocardium six months post-administration to further assess the myocardial protective effect associated with utrophin activation. In the myocardium of the mdx-MyoAAV-UA group, utrophin expression remained significantly elevated (Fig. 3h), consistent with the utrophin mRNA qPCR findings. GO differential gene enrichment analysis demonstrated that the differential genes between the mdx-MyoAAV-UA group and the WT group were mainly enriched in pathways related to virus receptor activation, virus response-related pathways, and positive regulation of cell-cell adhesion, promoting tighter connections between cells (Fig. 3i). In contrast, the myocardial transcriptome of the mdx-saline group showed significant differences in pathways related to heart processes, cardiac muscle contraction, and cardiac conduction, and extracellular matrix composition and fibrous connective tissue formation compared to the WT and MyoAAV-UA treatment groups (Fig. 3i). Gene Set Enrichment Analysis (GSEA) of key pathways, such as myocardial contraction and cardiac conduction, revealed significant differences were observed between the mdx-saline group and the WT group as well as the MyoAAV-UA groups. Remarkably, gene expression in MyoAAV-UA group was very similar to that in WT group (Fig. 3j, k). These results demonstrated that MyoAAV-UA-mediated utrophin upregulation significantly ameliorates both skeletal muscle and cardiac pathology and function.
Activation of endogenous utrophin in nonhuman primates (NHPs) by MyoAAV-UA demonstrates efficacy and safety
Although dosages of 1 × 1014 vg/kg AAV are commonly used in mouse studies, clinical trials have shown that this dosage can cause severe liver and kidney damage, as well as life-threatening complications34. Based on the demonstrated efficacy of MyoAAV-UA at 5 × 1013 vg/kg in mice, we evaluated its safety and efficacy in NHPs35. Four 1.5-year-old male cynomolgus monkeys were selected, with two receiving 5 × 1013 vg/kg MyoAAV-UA and the remaining two receiving saline (Fig. 4a). The sgRNA pair H1 and H10 was selected based on their high activation efficiency in HEK 293 T cell lines (Fig. 1d), and they are also identical between humans and NHPs, achieving a 3.32-fold activation efficiency in MSkM (Fig. 1c). Oral immunosuppressant prednisone was administered five days before injection, following the dosage regimen (Fig. 4a) adapted from the AVXS-101-CL-303 clinical trial (NCT03306277 on the ClinicalTrials.gov database), a gene therapy designed to treat spinal muscular atrophy (SMA)36. A significant increase in dCasMINI expression was observed in GM and BB muscles four weeks post-injection (Supplementary Fig. S16a). Neutralizing antibody (NAb) titers increased after MyoAAV-UA injection, peaking at Day 28 and stabilizing thereafter (Supplementary Fig. S16b). Utrophin protein expression in the GM and BB increased by 2.17- and 2.16-fold separately compared to pre-administration levels (Fig. 4b–d). Average utrophin mRNA expression in GM and BB exceeded a 2.96- and 2.97-fold separately (Fig. 4e). Immunofluorescence staining showed significantly utrophin localization along the sarcolemma rather than a few scattered distributions prior to treatment (Fig. 4f and Supplementary Fig. S16c). Histological staining indicated normal muscle morphology after MyoAAV-UA treatment, with no presence of CD3+ and CD8+ T cells detected (Fig. 4g and Supplementary Fig. S16d). These findings suggest that utrophin, as an endogenous protein, did not aberrantly activate the immune system, preventing inflammatory reactions in the muscles.
a Timeline of experimental procedures in NHPs. b, c Immunoblots of utrophin and dystrophin in various muscles before and 4 weeks post-MyoAAV-UA injection (n = 2 monkeys in each group). Vinculin was used as a loading control. d Quantification of utrophin protein levels was normalized to vinculin. e Quantification of utrophin mRNA expression levels before and 4 weeks post-MyoAAV-UA injection, normalized to GAPDH. f Representative immunofluorescence staining of dystrophin (green), utrophin (red), and DAPI (blue) before and 4 weeks post-MyoAAV-UA injection. Scale bar, 100 µm. g Representative H&E staining, Sirius Red staining, and DAB staining of CD3+ and CD8+ T cells before and 4 weeks post-MyoAAV-UA injection. Scale bar, 100 μm. h–k Measurement of interferon gamma (IFN-γ) (h), alanine aminotransferase (ALT) (i), aspartate aminotransferase (AST) (j), and platelets (PLT) (k) before and after MyoAAV-UA injection. n = 2 monkeys in each measurement group, with the mean value shown. BB biceps brachii, QD quadriceps, GM gastrocnemius. b, c, f, g The experiment was repeated with three technical replicates with similar results. d, e n = 2 monkeys in each group, with data points shown. The bar represents the mean value. Source data are provided as a Source Data file.
We next evaluated the safety of MyoAAV-UA administration (Supplementary Fig. S16e–o). Bone mineral density (BMD) and bone mineral content (BMC) exhibited an upward trend in all four monkeys, indicating a low risk of osteoporosis or fractures (Supplementary Fig. 16e, f). None of the monkeys showed significant weight loss within two months post-viral injection (Supplementary Fig. S16g). Their body temperatures remained within the normal range during the first week post-viral injection (Supplementary Fig. S16h). Interferon (IFN-γ) levels in monkey serum were measured to assess the inflammatory response. IFN-γ levels rose from day 7 post-viral injection, peaked at day 21, and returned to normal by day 42 (Fig. 4h). High doses of AAV typically lead to acute liver dysfunction in humans and NHPs. Serum biochemical assays indicated that liver function markers alanine aminotransferase (ALT) and aspartate aminotransferase (AST) transiently increased but remained below 500 U/L 3 days post-MyoAAV-UA injection, returning to normal within two weeks (Fig. 4i, j). Thrombocytopenia, a common adverse reaction post-AAV injection, showed a transient decrease in platelets post-MyoAAV-UA administration, returning to normal within one week (Fig. 4k). Other parameters such as blood creatinine, gamma-glutamyl transferase (GGT), cholesterol (CHO), and blood glucose showed no significant changes pre- and post-administration (Supplementary Fig. S16i–l). Ultrasound evaluations of vital organs before and four weeks post-MyoAAV-UA administration detected no abnormalities such as enlargement, nodules, cystic changes, or abnormal blood flow signals (Supplementary Fig. S16m). Brain magnetic resonance imaging (MRI) showed clear brain structures with no abnormal signals on T1-weighted images pre- and post-administration. T2-weighted images revealed no abnormal bleeding foci or brain tissue edema (Supplementary Fig. S16n). Chest X-rays showed symmetrical thoracic cages in all four monkeys, normal ribs and intercostal spaces, clear lung textures, and no substantial infiltration or abnormalities in the lung hilum, mediastinal shadow, heart size, or morphology. The diaphragm appeared smooth, with sharp bilateral costophrenic angles, and no pleural effusion was evident (Supplementary Fig. S16o).
Taken together, the systemic delivery of MyoAAV-UA containing homologous sgRNAs for both humans and NHPs effectively activates utrophin in NHP skeletal muscles with no significant side effects observed.
Activation of utrophin upregulations UGC in iPSC-derived myoblasts from DMD patients
To evaluate the potential application of MyoAAV-UA in DMD patients, we isolated peripheral blood mononuclear cells (PBMCs) from two DMD patients with DMD mutation: NM_004006; c. 790 G > T p. G264* and △Exon 49–50 (Fig. 5a) and established induced pluripotent stem cells (iPSCs) (Fig. 5b and Supplementary Fig. S17a). H9 cells were used as a healthy control. These DMD-iPSCs and H9 cells were then differentiated into skeletal muscle cells (Fig. 5c, d and Supplementary Fig. S17b, c). We have identified the combination of H1 and H10 as the most effective sgRNA pair within the human UTRN promoter region (Fig. 1d). MyoAAV-UA, including sgRNAs H1 and H10, was added to DMD patients’ iPSC-derived myoblasts. Five days post-infection, both the G264* and △Exon 49-50 iPSC-derived myotubes demonstrated a significant upregulation of utrophin protein expression, with a 3.45-fold increase for the G264* mutation and a 3.79-fold increase for the exon 49–50 deletion mutation (Fig. 5e–g), and mRNA level was increased to 3.63-fold (G264*) and 4.34-fold (△Exon 49-50), respectively (Fig. 5h). Immunofluorescent staining showed upregulation of α-sarcoglycan and γ-sarcoglycan, suggesting restoration of UGC in DMD patient’s iPSC-derived myoblasts (Fig. 5i, j).
a Sanger sequencing of the iPSC genome from DMD patients. b Phase-contrast images showing iPSC generated from DMD patient’s PBMC. Scale bar, 100 µm. c Schematic diagram illustrating the timeline of myotube differentiation in vitro. d Representative immunofluorescence staining of myosin heavy chain (MHC) (green) and dystrophin (red) in myotubes differentiated from H9 or DMD patient iPSC. Scale bar, 200 µm. e, f Representative immunoblots showing utrophin and dystrophin protein levels in myotubes derived from H9, DMD, and DMD treated with MyoAAV-UA. Vinculin was used as a loading control. g Quantification of utrophin protein levels in myotubes from H9, DMD, and DMD treated with MyoAAV-UA, normalized to vinculin. h Quantification of utrophin mRNA expression levels in myotubes from H9, DMD, and DMD treated with MyoAAV-UA, normalized to GAPDH. i, j Representative immunofluorescence staining of α-sarcoglycan (red), γ-sarcoglycan (red), MHC (green), and DAPI (blue) in myotubes differentiated from the respective groups. Scale bar, 100 µm. d–f, i, j The experiment was repeated with three technical replicates with similar results. g, h n = 3 technical replicates, with data points shown. The bar represents the mean value. Source data are provided as a Source Data file.
Collectively, these findings demonstrated the effectiveness of the MyoAAV-UA system in upregulating the utrophin expression and restoring UGC in iPSC-derived myoblasts from DMD patients, indicating the potential clinical applications.
Discussion
Previous strategies for activating gene expression, including ZFN37, pharmacological modulators, or other activation methods, have encountered various challenges, such as low efficiency, complexity in design, or poor target specificity38,39. The secondary dCas9 activation approach presents an ideal solution, yet its large size limits its packaging, and splitting it significantly reduces efficacy.
To address this issue, we developed MyoAAV-UA, which employed dCasMINI-VPR, a miniaturized version of the CRISPRa activation tool, to achieve the activation and dual sgRNAs to be efficiently packaged within a single AAV vector for delivery. We identified highly efficient sgRNA combinations in vitro that significantly enhance activation efficiency compared to single sgRNAs20. By utilizing a muscle-specific promoter (SPc5-12) and the MyoAAV serotype, we achieved muscle-targeted activation within skeletal and cardiac muscles. Compared to previous activation systems13,20,40,41, MyoAAV-UA achieves effective systemic muscle delivery and utrophin activation with a dosage requirement of only 5 × 1013 vg/kg, thereby reducing AAV-associated toxicity. This integrated system also enhances efficiency compared to dual-AAV reconstructed system20, highlighting its significant value for future clinical translation.
In Dmd mouse models, MyoAAV-UA successfully activated utrophin in both mdx and Dmd△Ex50 mice, achieving utrophin upregulation in muscles with different mutation types and proper membrane localization. Following systemic administration, this upregulation alleviated dystrophic symptoms, including muscle degeneration and fibrosis. The improvements can be observed at six months. Transcriptomic analyses revealed a strong correlation between utrophin upregulation and the restoration of muscle structure, particularly in cardiac tissue. These findings align with recent studies that highlight the upregulation of utrophin in improving the physiological function of DMD hiPSC-derived cardiomyocytes in vitro through dCas9-VP6442. Notably, high utrophin expression and significant pathological improvements were observed in the myocardium at six months, contrasting with severe fibrosis in untreated subjects. Given that cardiomyocytes are post-mitotic, the upregulated utrophin and recovery of cardiomyocyte pathology are likely to be maintained for long-term. Additionally, transcriptome analysis indicates that utrophin upregulation is not only positively correlated with UGC reconstruction in skeletal muscle and extracellular matrix adhesion function, but also highly associated with extracellular matrix organization and cell-cell adhesion in cardiac muscle. Many DMD patients develop heart failure in later stages, suggesting that our treatment might delay heart failure onset and reduce mortality43,44.
We observed varying efficiency of utrophin activation between mdx and Dmd△Ex50 mice. This variation may be due to the different DMD gene mutations present in these models. Similarly, DMD patients with different mutation types also exhibit significant differences in utrophin expression45. The underlying mechanisms of these differences remain unclear. Previous research suggests that large deletions spanning exons 10–60 of the DMD gene can impede utrophin localization to the muscle cell membrane, leading to severe muscle wasting45. Further investigation is required to fully understand these mechanisms. Previous clinical trials with AAV have indicated severe adverse effects under high-dose conditions. Systemic administration doses exceeding 2 × 1014 vg/kg lead to liver function failure and fatalities, raising safety concerns46. NHPs are essential in translational medicine research due to their genetic, physiological, and behavioral similarities to humans, which make them valuable models for studying human diseases and testing potential therapies before clinical trials47. In this study, MyoAAV-UA containing homologous sgRNAs between humans and NHPs, was administered systemically at a dose of 5 × 1013 vg/kg in NHPs. Results demonstrated that MyoAAV-UA achieved more than a twofold activation of utrophin in cynomolgus monkey muscles without severe toxic side effects, highlighting its efficacy and favorable safety profile in large animals. The distribution of utrophin in the healthy cynomolgus monkeys appears scattered along the sarcolemma, which contrasts with the homogeneous sarcolemmal expression seen in dystrophic mice. The scattered utrophin expression observed in our study is due to the absence of dystrophic pathology in the monkeys. This may due to the sarcolemma is already occupied by dystrophin, so the upregulated utrophin will only appear in certain regions of the sarcolemma, resulting in a scattered staining pattern. Furthermore, we evaluated MyoAAV-UA efficiency in iPSC-derived myotubes from DMD patients, demonstrating a ~3.5-fold upregulation of utrophin. Consistent with findings of Tejvir S. Khurana and colleagues48, our study also demonstrated that utrophin overexpression led to increased sarcolemmal α-sarcoglycan and γ-sarcoglycan staining, aligning with improved restoration of the UGC. These results show the clinical potential of MyoAAV-UA for DMD treatment.
While the mdx mouse model is widely used to study DMD, it does not fully recapitulate the progressive muscle fibrosis observed in human patients30,31. In particular, the cardiac manifestations in mdx mice are less severe compared to those in human DMD patients49,50. Although utrophin upregulation led to improvements in both skeletal and cardiac muscle in our models, future studies using more suitable models, such as NHP models and rat models, would provide better insights into cardiac outcomes. We observed a reduction in therapeutic effects over time, based on our findings, we expect that the protective benefits would also diminish over time. Given that 6 months represents a relatively long period in the lifespan of a mouse, this timeframe might be considered as a substantial therapeutic window in humans. In future studies, it will be crucial to develop strategies to enhance the durability of utrophin expression.
Overall, MyoAAV-UA efficiently activates full-length utrophin in mice, NHPs, and humans. Utrophin upregulation significantly alleviated dystrophic symptoms in mdx mice, and no significant side effects were observed in NHPs, highlighting MyoAAV-UA as a promising approach for DMD treatment and providing support for its clinical translation.
Methods
Ethics statements
The experimental procedures involving mice were conducted in strict compliance with the regulations governing animal welfare ethics established by the Experimental Animal Center of Kunming University of Science and Technology. These procedures were thoroughly reviewed and approved by the Animal Ethics Committee (Approved No. PZWH (Yunnan) K2022-0019). All cell experiments involving DMD patients were approved by the Medical Ethics Committee of the Third Medical Center of the Chinese PLA General Hospital (Approved No. KY2121-001). Informed consent forms, approved by the Ethics Committee, were signed by the parents or guardians of underage participants. All procedures involving NHPs were conducted in strict compliance with the ethical requirements for experimental animals outlined by the Laboratory Animal Ethics Committee of Kunming University of Science and Technology (Approval Number: KUST202301010).
Mice
mdx (C57BL/10ScSn-Dmdmdx/J) mice were generously provided by Professor Yafeng Song’s laboratory at the Institute of Sport and Health Science, Beijing Sport University. DmdΔEx50 (C57BL/10-Dmd<Del50 > ) mouse model lacking exon 50 of the Dmd gene was constructed using the CRISPR/Cas9 system31. Wildtype (C57BL/10 J) mice and immunodeficient NOD-SCID IL2Rγnull mice used in this study were purchased from the Yunnan University Experimental Animal Center. All mice were housed in a specific pathogen-free facility under controlled conditions. The temperature of the room was maintained at 22 ± 1 °C, and the humidity was controlled at 50 ± 10%. Light and dark were alternated for 12 hours and the light period was from 7:00 am to 7:00 pm. Mice were provided with sufficient food and water ad libitum, and their bedding was changed regularly to maintain a clean and comfortable environment. Body weight was monitored weekly. DMD is an X-linked recessive genetic disorder, therefore, only male mdx/Dmd and WT mice were used for the experiments. Therapeutic agents were administered to all selected mice at 2 weeks of age. CK levels, in vivo functional assays, western blot analysis, and immunohistochemistry were performed at various time points as outlined in the manuscript. The subcutaneous tumor formation experiment, using immunodeficient mice for teratoma studies, began when the mice were 4 weeks old and analyzed at 8 weeks old. For both the wildtype, treatment, and control groups, six mice were used per group as biological replicates for statistical analysis.
NHPs
DMD is an X-linked recessive genetic disorder that primarily affects males. In this study, four healthy 1.5-year-old male cynomolgus monkeys were selected as the research subjects. These animals showed no signs of infection or other adverse conditions. The titers of NAbs against MyoAAV in their serum were all below 1:5. The monkeys were randomly assigned to two groups: the MyoAAV injection group and the saline control group. All monkeys used in the experiments were housed in the animal facility of the Institute of Primate Translational Medicine at Kunming University of Science and Technology. The room temperature was maintained at 22 ± 1 °C with approximately 50% humidity. The indoor lighting schedule was fixed from 8:00 a.m. to 8:00 p.m. The monkeys had ad libitum access to water and were provided three meals daily, consisting of fruits and a balanced compound feed.
Cell lines, cell culture, and transfection
The HEK293T (SCSP-502) and Neuro2a cell lines (SCSP-5035) were obtained from the National Collection of Authenticated Cell Cultures and cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS) at 37 °C in a humidified incubator with 5% CO2. Plasmid transfection was performed using Lipofectamine 3000 (Thermo Fisher, # L3000015), and cells were collected for RNA extraction 48 hours post-transfection. Skeletal muscle cells from cynomolgus monkeys were isolated and cultured following the protocol described in Methods in Molecular Biology51.
PBMCs from two male DMD patients were isolated from the peripheral blood using 15 ml SepMate tubes (STEMCELL, #15450) and Lymphoprep density gradient medium (STEMCELL, #07801). All cell culture-related experiments involving DMD patient’s PBMCs were conducted in compliance with the Chinese PLA General Hospital Ethics Committee guidelines (Approval number KY2121-001). iPSCs were generated from PBMCs using the Invitrogen Sendai virus kit (Invitrogen, #A16518). The healthy control group cell line H9 (SCSP-307) were obtained from the National Collection of Authenticated Cell Cultures in China. Differentiation of iPSCs into skeletal muscle was performed following the flowchart outlined in Fig. 5c. The medium used at different culture stages are listed in Supplementary Table S1. For myotube formation, myoblasts were grown in 12-well plates to 90% confluence before switching to myotube differentiation medium. Two days post-differentiation, MyoAAV-UA (2 × 1011 viral particles) was added to each well. The medium was replaced 24 hours after transduction, and cells were harvested five days post-infection for cellular immunostaining, RNA and protein extraction, and subsequent analysis.
Teratoma assay
iPSC clones were detached by digesting with 50 μl of 20 mg/ml type IV collagenase in 1 ml of medium at 37 °C for 10 minutes. The culture dish was gently tapped to release the clones, and the supernatant was transferred to a 15 ml tube. After settling for 20 seconds, the supernatant was discarded, and the cells were washed twice with DPBS. The clones were further digested with 1 ml of Accutase (STEMCELL, #07920) at 37 °C for 10 minutes, neutralized with 1 ml of medium, centrifuged at 300 × g for 5 minutes, and resuspended. After counting, 1 × 106 cells were mixed with 100 μl of Matrigel (Corning, #354277) in a 1.5 ml tube. The cell suspension was injected subcutaneously into NOD-SCID IL2Rγnull mice, ensuring no blood in the syringe. Tumor formation was monitored, and when the teratoma size reached <1 cm3, the tumor was excised, fixed, and stained to evaluate the pluripotency of the iPSCs.
Plasmid construction
The sgRNAs were designed based on the PAM site sequence TTTR using the CRISPR RGEN Tools (www.rgenome.net), as detailed in Supplementary Data 1. To construct the dCasMINI-VPR-U6-crRNA scaffold vector, the dCasMINI-VPR sequence was obtained from the plasmid pHR-PGK-SV40_NLS-dCasMINI-V4-VPR-c-Myc_NLS-mCherry-WPRE (Addgene: pSLQ9926), and the DNA fragment was subcloned into an AAV vector. The dCasMINI and VPR elements were under the control of a CMV promoter, and the human U6 promoter was connected to the activation element to control the sgRNA and its crRNA. ClonExpress MultiS One Step Cloning Kit (Vazyme, #c113-01) was used for ligation. For the construction of the vector containing a muscle-specific promoter and dual sgRNAs, pX601-AAV-SPc5-12-dCasMINI-VPR-Cmyc-U6-M8-M12 and pX601-AAV-SPc5-12-dCasMINI-VPR-Cmyc-U6-H1-H10 were designed as integrated vectors for MyoAAV packaging. These plasmids included a muscle-specific promoter (SPc5-12) and dual sgRNAs targeting the utrophin gene.
AAV packaging and injection in Dmd mice
MyoAAV was packaged by PackGene Biotech (Guangzhou, China). Two-week-old male mdx mice and DMDΔEx50 mice were administered the virus either through local injection or tail vein intravenous injection, respectively. Control mice (2-week-old male C57BL/6 mice) were injected with an equivalent volume of saline. A minimum of six mice were used for each experimental condition.
Immunosuppressant administration and intravenous MyoAAV-UA injection in NHPs
The oral administration of immunosuppressant regimen was adapted from the AVXS-101-CL-303 clinical trial (ClinicalTrials.gov identifier: NCT03306277), a Phase 3 study evaluating the efficacy and safety of Onasemnogene abeparvovec (AVXS-101) in patients with Type 1 SMA. In the present study, prednisolone was administered orally at 1 mg/kg/day starting five days before the injection and continued for 30 days. The dose was then tapered to 0.5 mg/kg/day for two weeks, followed by 0.25 mg/kg/day for an additional two weeks. Two healthy 1.5-year-old male monkeys received a systemic dose of MyoAAV-UA at 5 × 1013 vg/kg, packaged by PackGene Biotech (Guangzhou, China). The injection was administered through the saphenous vein in the right lower limb, with the virus diluted in 30 ml of saline and infused over 30 minutes, followed by a 10 ml saline flush. The control group consisted of two healthy age-matched male monkeys, which received the same volume of saline infused at the same rate as the MyoAAV-UA group. Post-injection, all monkeys were monitored for at least 30 minutes, with no abnormal signs observed during this period.
AAV genome copy numbers and vector genome quantification in tissue samples
Genomic DNA was extracted from tissues using the Wizard® Genomic DNA Purification Kit. DNA concentration and purity were measured using a Nanodrop 2000 spectrophotometer, and samples were diluted to 100 ng/µl. AAV viral copy number in tissues were quantified using qPCR with SYBR Green Technology and QX200™ AutoDG™ Droplet Digital™ PCR. A 22 µL reaction mixture was prepared with QX200 ddPCR EvaGreen Supermix and 100 nM primers. DNA samples were diluted to 50 ng/µl and pre-treated with DNaseI. The 22 µL reaction mixture was transferred to a 96-well plate, sealed, and subjected to PCR. Results were automatically analyzed using Quanta software. The Number of vector genomes (dCasMINI) per diploid genome (Gapdh) was determined using qPCR (Supplementary Table S2). Absolute quantification of dCasMINI and GAPDH/Gapdh in each sample was achieved using standard curves generated by amplifying plasmids containing dCasMINI or GAPDH/Gapdh sequences. These standard curves were used to calculate the ratio of vector genomes to diploid genomes.
Blood collection and serum separation
The forearm or hind leg skin of the monkey was shaved and disinfected with iodine. Using a vacuum blood collection tube (BD, #367983), 3 ml of blood was collected from either the brachial vein or small saphenous vein. The blood collection tube was left undisturbed for 30 minutes to allow clot formation. Afterward, the tube was centrifuged at 3000 × g for 10 minutes. The pale-yellow serum layer was carefully aspirated using a sterile, enzyme-free pipette and stored at −80 °C for long-term use.
ELISA determination of mouse serum CK
Serum CK in mice was measured using the CK Activity Assay Kit (Colorimetric) (Abcam, #ab155901), following the manufacturer’s instructions for accurate quantification.
MyoAAV NAb assay
The MyoAAV4E-CMV-β-galactosidase virus (1 × 1013 vg/ml) was packaged by Guangzhou PackGene Biotech. For each well in a 96-well plate, a total of 109 virus genome copies were calculated. Mix 10 µl of the virus stock with 5500 µl of diluent (2% FBS + 97% DMEM + 1% PS). Perform gradient dilution (e.g., 1:2, 1:4, 1:8) and mix 55 µl of each dilution with 55 µl of virus diluent. Incubate the mixtures at 37 °C for 1 hour. Add the mixture to a 96-well plate seeded with HEK293T cells at 60% confluence. Each sample is tested in duplicate. Include three positive control wells containing the virus diluent only and three negative control wells containing basal medium only. Incubate the plate at 37 °C with 5% CO2 for 72 hours. After incubation, add 100 µl of X-Gal cell lysis solution to each well, let it sit for 10 minutes, and pipette gently to disperse the cells. Transfer 50 µl of the lysate to a 96-well plate, add 50 µl of β-galactosidase assay reagent (Beyotime, # C0602), and incubate at 37 °C for 1 hour. Add 150 µl of reaction termination solution to each well and measure the absorbance at 420 nm using a plate reader. The NAby titer for each sample is determined by identifying the dilution at which the OD value is half the average of the positive control wells. This absorbance corresponds to the sample’s NAb activity.
Blood biochemical index testing
After sampling, serum samples are stored in a 4 °C refrigerator and delivered to the biochemical testing department of Kunming Biotech International Co., Ltd. on the same day. Blood biochemical index (ALT, AST, Creatinine, GGT, CHO, Glucose) testing is conducted using the Roche C511 automated biochemical analyzer.
Whole blood cell analysis
Anticoagulated whole blood is collected using EDTA-K2 vacuum blood collection tubes (BD, #367863). 1 ml of blood collected from the experimental monkeys was transferred to the biochemical testing department of Kunming Biotech International Co., Ltd. Within 2 hours, red blood cells, white blood cells, platelets, etc., were analyzed using the Sysmex XN-1000 automated hematology analyzer from Sysmex Corporation.
Mice tissue procurement and storage
Under regulations governing animal welfare, euthanasia was performed on the mice to minimize pain and distress. Subsequently, the heart, gastrocnemius (GS) muscle, QDs, diaphragm, liver, kidneys, lungs, brain, and spleen were collected and further processed. The muscles were wrapped with yellow beeswax and flash-frozen in liquid nitrogen-precooled isopentane for 30 seconds to maintain muscle cell morphology. They were then immediately transferred to dry ice and transported to a −80 °C freezer for storage. The remaining tissues were embedded in OCT and placed on dry ice for pre-cooling embedding. All samples were thawed for at least 1 hour in a −20 °C freezer before sectioning to preserve tissue morphology. The sections were promptly stored in a light-protected −20 °C freezer. Samples for Western blot analysis and RNA analysis were collected in 1.5 ml RNase-free EP tubes, sealed tightly, and flash-frozen in liquid nitrogen for preservation. They were then transferred to a −80 °C freezer for long-term storage.
RNA analysis and Sanger sequencing
The total RNA of cells and tissues was extracted using TRIzol (Invitrogen, # 15596026), and 1 μg of RNA was reverse-transcribed to cDNA with the PrimeScript™ (Takara, # RR047) RT kit. The TransStart® Green qPCR SuperMix Kit (Transgen, # AQ101-01) is used to quantify the relative mRNA expression of genes. Sanger sequencing was performed by Tsingke Biotechnology Co., Ltd. The primers are shown in Supplementary Table S2.
Transcriptome analysis
The mouse GS muscle was quickly frozen with liquid nitrogen, and the tissue block was ground in liquid nitrogen to prevent protein and RNA from being degraded. The total tissue RNA was extracted with TRIzol and the mRNA was enriched by oligonucleotide (dT) magnetic beads to construct a cDNA library. Nanodrop 2000 was used to determine the concentration of nucleic acid. Illumina sequencing platform was used for high-throughput sequencing, and the sequencing read length was PE150. Custom R scripts were used to perform further TPM (transcripts per million mapped reads) normalization and quality control. Downstream plots used the ggplot252. Sequencing data were matched to Mus musculus genome data (GRCm39) using hisAT2 (Version 2.2.1). Transcriptome assembly using a hybrid of long and short reads with StringTie53. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 (biomedcentral.com). GO pathway enrichment analysis elucidates the relationship between utrophin gene expression and the regulation of associated pathways54. The WGCNA package provides R functions for weighted correlation network analysis to be used for finding clusters (modules) of highly correlated genes55. The Random Forest package provides a Rinter-face to the Fortran programs by Breiman and Cutler (available at http://www.stat.berkeley.edu). To perform the Gene Set Variation Analysis, the GSEABase package (version 1.44.0) was used to load the gene set file, which was downloaded and processed from KEGG database (https://www.kegg.jp/). To assign pathway activity estimates to individual cells, we applied GSEA using standard settings, as implemented in the GSEA package (version 1.30.0). The differences in pathway activities scored per cell were calculated with the LIMMA package (version 3.38.3)56.
Western blotting
Myocytes were digested with 0.05% trypsin and collected by centrifugation at 300 × g. Mouse or NHP muscle tissue was frozen in liquid nitrogen and ground into powder. RIPA lysis buffer with PMSF was added to extract protein from the cell pellet or tissue. The mixture was lysed on ice for 40 minutes, vertexing every five minutes. Protein supernatant was obtained by centrifuging at 10,000 × g for 10 minutes at 4 °C. Proteins were separated by SDS-PAGE and transferred to Immobilon-P® membranes. Membranes were blocked with 5% skim milk, incubated with antibodies overnight, washed, and detected using ECL substrate. Band intensity was analyzed with ImageJ. The primary antibodies are shown in the reporting summary.
Histological analysis
All muscles embedded in yellow beeswax were cryo-sectioned at 8 µm. Hematoxylin-eosin staining (Solarbio #G1120) and Sirius red staining (Solarbio, #G1472) were conducted according to the manufacturer’s instructions. DAB (3,3’-Diaminobenzidine) staining for muscle sections was conducted following the protocol outlined in the Beyotime Peroxidase (BSP) Immunohistochemistry Kit (Beyotime #P0202).
Immunofluorescence staining
Cultured cells, skeletal muscles (QDs, brachialis, and diaphragm), and hearts were cryo-sectioned at 8 µm and fixed with 4% paraformaldehyde for 10 min. Sections were blocked and permeabilized with 3% BSA and 10% FBS containing 0.2% TritonX-100 for 2 hours at room temperature. Sections and dishes were then incubated with primary antibody overnight at 4 °C, washed with 1× PBST three times, five minutes each, followed by donkey anti-rabbit/mouse IgG H&L secondary antibody for 2 hours at room temperature. Use TureBlack (Biotium #23007) to reduce autofluorescence in tissue sections according to the post-treatment with TrueBlack following immunostaining. Slides were mounted with an antifade mounting medium containing DAPI (Abcam, #ab104139). Immunofluorescence images were taken with Nikon AX and Leica SP8 confocal microscope. The primary antibodies are shown in the reporting summary.
Morphometric analysis
Three groups of mice, C57BL/6 (wildtype) and mdx randomized to either injection with saline or MyoAAV-UA, were studied by investigators blinded to specimen identification (n = 6 mice for each group). Three groups of mice brachialis, and QDs were stained with H&E screened with a light microscope. six muscle sections from different mice per group were evaluated for the centrally nucleated fibers. Areas at the myotendinous junctions were excluded from the measurements as they are rich in both mdx and controls. In total, 7439 fibers were evaluated for centrally nucleated fibers percentage. And the percentage of fibrotic area in brachialis, QDs, diaphragm, and heart sections staining with Sirius Red Staining. six muscle sections from different mice per group were evaluated for fibrotic area percentage in ImageJ software, each muscle section was measured in 3 mm2 per muscle. The area of interest (AOI) tool is employed to manually outline all complete muscle fibers. In the HSI mode, using the H/S/I curve tool, the muscle fibers are labeled as red, while the interstitium is left unlabeled. Subsequently, the total area of muscle fibers within the AOI is measured. Six sections of the brachialis and QDs muscles are randomly selected for statistical analysis in each group of mice.
Behavioral analysis
Mouse Gripper (Shanghai Mei Rui Sai Scientific Instrument Co., Ltd. #MC-RMG01) was used to measure the mice forelimb grasping force. The tester gently pulls the mouse’s tail with his hand to make the mouse’s hind limbs hang in the air while the forelimbs grasp the barbed wire to test its strength. The body of the mouse is parallel to the barbed wire. Each measurement was performed 10 times, and the average value was used for statistical analysis. The Rotarod-Lantency to fall experiment is used to evaluate the exercise endurance of mice (Shanghai Mei Rui Sai Scientific Instrument Co., Ltd. #MC-RMM02). Referring to the experimental design scheme of the Rotor-Rod of the Behavioral and Functional Neuroscience Laboratory of Stanford University, the mice were subjected to a one-week training, to ensure that all subjects learned the task to the same degree. Mice in each group were tested at the same speed as the running wheel, and the time for the mice to fall from the wheel was recorded, each mouse ran three rounds for statistical analysis. Each mouse was placed in the Open Field apparatus (40 × 40 cm) and recorded for 30 minutes at night. All mice will be pre-exposed to the open field environment for 10 minutes. The movement and distance of mice were analyzed by Smart v3.0 software.
IFN-γ assay
Following the instructions outlined in the Thermo Fisher Rhesus Macaque IFN-gamma ELISA Kit (Thermo, # EP8RB) protocol, we conducted the detection of IFN-γ levels in serum samples. After equilibrating the serum to room temperature, we diluted it twofold with 1× Assay Diluent before proceeding with the detection. The IFN-γ values in the serum were then calculated using the standard curve.
Ultrasound examination
Animals were administered 0.1 mg/kg of atropine and 0.05 ml/kg of Zoletil intramuscularly for anesthesia. The procedures were swiftly carried out after inducing anesthesia. A coupling agent was applied to the examination site to eliminate air obstruction between the probe and the skin, reducing friction between them. Suitable probes of the LOGIQ Fortis Pro ultrasound machine were selected for examining the carotid arteries, thyroid gland, liver, kidneys, heart, and other areas. Following the examination, animals were administered local muscle injection of atipamezole 0.05 ml/kg for reversal of anesthesia. Animal conditions were observed for at least 30 minutes, feeding and watering were resumed four hours after awakening from anesthesia.
Brain MRI
Animals were administered 0.1 mg/kg of atropine and 0.05 ml/kg of Zoletil intramuscularly for anesthesia. They were positioned in the right lateral decubitus position for the procedure. Using a SIEMENS Prisma 3.0 T MRI scanner, the head coil was selected for scanning. Imaging data included T1-weighted images and T2-weighted images. The basic parameters for data acquisition were as follows: 3D T1 sequence: TR/TE = 1400 ms/3.55 ms, inversion time = 900 ms, flip angle = 8 degrees, field of view = 135 × 135 mm2, matrix size = 224 × 224, slice thickness = 0.6 mm, voxel size = 0.6 × 0.6 × 0.6 mm3, sections = 160; acceleration factor = 2; NEX = 3. 3D T2 sequence: TR/TE = 2500 ms/520 ms, field of view = 115 × 115 mm2, matrix size = 192 × 192, slice thickness = 0.6 mm, voxel size = 0.6 × 0.6 × 0.6 mm3, sections = 128; acceleration factor = 2; NEX = 4.
Muscle sampling and analysis of NHPs
In a sterile operating room, cynomolgus monkeys were anesthetized with intramuscular injections of 0.1 mg/kg atropine and 0.05 ml/kg Zoletil. Injections were administered away from the surgical site. The skin of the legs or arms was shaved and disinfected with iodine. A sterile drape exposed the surgical area. Using sterilized instruments, the skin and fascia were incised, avoiding major blood vessels. Hemostatic forceps separated the muscle fascia, and the muscle was cut to obtain tissue for analysis. Hemostasis was achieved with electrocautery or ligature, and the layers were sutured. The area was disinfected and dressed. Postoperative awakening medication, atipamezole (0.05 ml/kg) was administered intramuscularly. Prior to AAV treatment, biopsies were collected from the right QDs of four cynomolgus monkeys. Four weeks post-treatment, biopsies were collected from both the left GS and left BB muscles of the MyoAAV-UA treated and saline control monkeys. The monkey’s recovery was monitored for at least 30 minutes, and feeding and watering were resumed four hours after awakening from anesthesia. Ceftriaxone (50 mg/kg/day) was administered for 3 days to prevent infection.
Bone density assessment (iDXA) and chest X-ray scan analysis
Following the anesthesia depicted before, animals were placed in a supine position for X-ray examination of the chest using the X-ray HPS-HF-4.0 system (SEDECAL, SPAIN) to analyze lung texture distribution and cardiac size. Whole-body scans were performed using the Lunar iDXA system (GE Healthcare) to measure total body fat, lean muscle mass, and fat distribution ratio. The analysis was conducted using the following software versions: DPXL 4.7e, Prodigy 8.80, and iDXA 10.40. Specific anatomical landmarks described previously were used to partition regions, including the arms, legs, and trunk. For quality control of soft tissue mass related to densitometry, scans were performed monthly using 8-liter bottles filled with methanol and water to simulate fat and non-fat soft tissue, respectively. iDXA and X-ray assessments were conducted in Kunming Biotech International Co., Ltd. MRIcroN version 4.0 software is used to analyze brain MRI images.
Temperature assay
Using the infrared electronic thermometer (CEM, #DT-8806H), align it with the chest skin of the cynomolgus monkey, ensuring that the distance between the detector and the skin does not exceed 2 centimeters. Take three consecutive measurements, record the values, and calculate the average.
Statistical analysis
For the mouse experiments, results were derived from n = 6 biological replicates. For the NHP experiments, n = 2 biological replicates were used. Myoblasts were differentiated from two independent iPSCs biological samples with different DMD gene mutation types and the H9 cell line. Statistical analysis was performed for data with a biological sample size greater than 3. One-way ANOVA followed by the Tukey post hoc test was used for data with three groups. Statistical graphs were plotted by GraphPad Prism (version 8.0.2) software. Sample sizes and p values are provided in the Figure legends and Source Data file. P values smaller than 0.05 were considered statistically significant. Western blotting quantification, centrally nucleated myofibers, and Sirius red-stained muscle fibrosis were analyzed using ImageJ software (version 1.53k). Sanger sequencing results were aligned using SnapGene software (version 4.2.4). Immunofluorescence staining images were analyzed using NIS element viewer (version 5.21) and Leica Application suite X (version 3.7.4). Histopathological staining images were analyzed with Olyvia software (version v3.2). Droplets digital PCR (ddPCR) events were analyzed using Quanta software (version 1.7.4). Absorbance measurements were performed using SpectraMax software (version 7.1). X-ray examination of the chest was conducted using the X-ray HPS-HF-4.0 system (SEDECAL, SPAIN). Bone density assessment were performed using DPXL (version 4.7e), Prodigy (version 8.80), and iDXA (version 10.40). Ultrasound was performed suing The LOGIQ Fortis Pro Ultrasound System.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that all relevant data supporting the findings of this study are included in this published article and its Supplementary Information files. Source data are provided in this paper. RNA-seq data have been deposited at GSA-Human accession: PRJNA917214. Source data are provided with this paper.
References
Duan, D., Goemans, N., Takeda, S., Mercuri, E. & Aartsma-Rus, A. J. N. R. D. P. Duchenne muscular dystrophy. Nat. Rev. Dis. Prim. 7, 13 (2021).
Bushby, K. et al. Diagnosis and management of Duchenne muscular dystrophy, part 2: implementation of multidisciplinary care. Lancet Neurol. 9, 177–189 (2010).
Babbs, A. et al. From diagnosis to therapy in Duchenne muscular dystrophy. Biochem. Soc. Trans. 48, 813–821 (2020).
Roberts, T. C., Wood, M. J. A. & Davies, K. E. Therapeutic approaches for Duchenne muscular dystrophy. Nat. Rev. Drug Discov. 22, 917–934 (2023).
Bladen, C. et al. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat. 36, 395–402 (2015).
Alfano, L. et al. Long-term treatment with eteplirsen in nonambulatory patients with Duchenne muscular dystrophy. Medicine 98, e15858 (2019).
Frank, D. E. et al. Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy. Neurology 94, e2270–e2282 (2020).
Clerk, A., Morris, G. E., Dubowitz, V., Davies, K. E. & Sewry, C. A. Dystrophin-related protein, utrophin, in normal and dystrophic human fetal skeletal muscle. J. Histochem. Cytochem. 25, 554–561 (1993).
Schofield, J., Houzelstein, D., Davies, K., Buckingham, M. & Edwards, Y. H. Expression of the dystrophin-related protein (utrophin) gene during mouse embryogenesis. Dev. Dyn. 198, 254–264 (1993).
Wu, R., Song, Y., Wu, S. & Chen, Y. Promising therapeutic approaches of utrophin replacing dystrophin in the treatment of Duchenne muscular dystrophy. Fundam. Res. 2, 885–893 (2022).
Gilbert, R. et al. Adenovirus-mediated utrophin gene transfer mitigates the dystrophic phenotype of mdx mouse muscles. Hum. Gene Ther. 10, 1299–1310 (1999).
Odom, G., Gregorevic, P., Allen, J., Finn, E. & Chamberlain Microutrophin delivery through rAAV6 increases lifespan and improves muscle function in dystrophic dystrophin/utrophin-deficient mice. Mol. Ther. 16, 1539–1545 (2008).
Song, Y. et al. Non-immunogenic utrophin gene therapy for the treatment of muscular dystrophy animal models. Nat. Med. 25, 1505–1511 (2019).
Tinsley, J. et al. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat. Med. 4, 1441–1444 (1998).
Soblechero-Martin, P., Lopez-Martinez, A., de la Puente-Ovejero, L., Vallejo-Illarramendi, A. & Arechavala-Gomeza, V. Utrophin modulator drugs as potential therapies for Duchenne and Becker muscular dystrophies. Neuropathol. Appl. Neurobiol. 47, 711–723 (2021).
Guiraud, S., Roblin, D. & Kay, D. E. The potential of utrophin modulators for the treatment of Duchenne muscular dystrophy. Expert Opin. Orphan Drugs 6, 179–192 (2018).
Wilkinson, I. V. L. et al. Characterisation of utrophin modulator SMT C1100 as a non-competitive inhibitor of firefly luciferase. Bioorg. Chem. 94, 103395 (2020).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Chavez, A. et al. Comparison of Cas9 activators in multiple species. Nat. Methods 13, 563–567 (2016).
Liao, H. K. et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell 171, 1495–1507.e1415 (2017).
Böhm, S. et al. A gene therapy for inherited blindness using dCas9-VPR-mediated transcriptional activation. Sci. Adv. 6, eaba5614 (2020).
Xu, X. et al. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol. Cell 81, 4333–4345.e4334 (2021).
Clark, T. et al. CRISPR activation screens: navigating technologies and applications. Trends Biotechnol. 42, 1017–1034 (2024).
Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10, 973–976 (2013).
Li, X., Eastman, E. M., Schwartz, R. J. & Draghia-Akli, R. Synthetic muscle promoters: activities exceeding naturally occurring regulatory sequences. Nat. Biotechnol. 17, 241–245 (1999).
Malerba, A. et al. Dose-dependent microdystrophin expression enhancement in cardiac muscle by a cardiac-specific regulatory element. Hum. Gene Ther. 32, 1138–1146 (2021).
Shiraki, T. & Kawakami, K. A tRNA-based multiplex sgRNA expression system in zebrafish and its application to generation of transgenic albino fish. Sci. Rep. 8, 13366 (2018).
Yuan, Q. & Gao, X. Multiplex base- and prime-editing with drive-and-process CRISPR arrays. Nat. Commun. 13, 2771 (2022).
Tabebordbar, M. et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell 184, 4919–4938.e4922 (2021).
McGreevy, J. W., Hakim, C. H., McIntosh, M. A. & Duan, D. Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Dis. Model. Mech. 8, 195–213 (2015).
Amoasii, L. et al. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl. Med. 9, https://doi.org/10.1126/scitranslmed.aan8081 (2017).
Salmaninejad, A. et al. Common therapeutic advances for Duchenne muscular dystrophy (DMD). Int. J. Neurosci. 131, 370–389 (2021).
Fayssoil, A., Abasse, S. & Silverston, K. Cardiac involvement classification and therapeutic management in patients with duchenne muscular dystrophy. J. Neuromuscul. Dis. 4, 17–23 (2017).
Duan, D. Lethal immunotoxicity in high-dose systemic AAV therapy. Mol. Ther. 31, 3123–3126 (2023).
Huang, M., Yang, J., Li, P. & Chen, Y. Embryo-engineered nonhuman primate models: progress and gap to translational medicine. Research 2021, 9898769 (2021).
Ogbonmide, T. et al. Gene therapy for spinal muscular atrophy (SMA): a review of current challenges and safety considerations for onasemnogene abeparvovec (Zolgensma). Cureus 15, e36197 (2023).
Sander, J. D. et al. In silico abstraction of zinc finger nuclease cleavage profiles reveals an expanded landscape of off-target sites. Nucleic Acids Res. 41, e181 (2013).
Altucci, L. & Rots, M. G. Epigenetic drugs: from chemistry via biology to medicine and back. Clin. Epigenetics 8, 56 (2016).
Petersen, B. Basics of genome editing technology and its application in livestock species. Reprod. Domest. Anim. 52, 4–13 (2017).
Strimpakos, G. et al. Novel adeno-associated viral vector delivering the utrophin gene regulator Jazz counteracts dystrophic pathology in mdx mice. J. Cell Physiol. 229, 1283–1291 (2014).
Pisani, C. et al. Utrophin up-regulation by artificial transcription factors induces muscle rescue and impacts the neuromuscular junction in mdx mice. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 1172–1182 (2018).
Andrysiak, K. et al. Upregulation of utrophin improves the phenotype of Duchenne muscular dystrophy hiPSC-derived CMs. Mol. Ther. Nucleic Acids 35, 102247 (2024).
Kamdar, F. & Garry, D. J. Dystrophin-deficient cardiomyopathy. J. Am. Coll. Cardiol. 67, 2533–2546 (2016).
Meyers, T. A. & Townsend, D. Cardiac pathophysiology and the future of cardiac therapies in Duchenne muscular dystrophy. Int. J Mol. Sci. 20, https://doi.org/10.3390/ijms20174098 (2019).
Guiraud, S. & Davies, K. Utrophin correlates with disease severity in Duchenne muscular dystrophy. Med 4, 220–222 (2023).
Agarwal, S. High-dose AAV gene therapy deaths. Nat. Biotechnol. 38, 910 (2020).
Ren, S. et al. Profound cellular defects attribute to muscular pathogenesis in the rhesus monkey model of Duchenne muscular dystrophy. Cell 187, 6669–6686.e6616 (2024).
Sengupta, K. et al. Genome editing-mediated utrophin upregulation in Duchenne muscular dystrophy stem cells. Mol. Ther.-Nucleic Acids 22, 500–509 (2020).
Duan, D. Challenges and opportunities in dystrophin-deficient cardiomyopathy gene therapy. Hum. Mol. Genet. 15 Spec No 2, R253–261 (2006).
Schultz, T. I., Raucci, F. J. Jr. & Salloum, F. N. Cardiovascular disease in Duchenne muscular dystrophy: overview and insight into novel therapeutic targets. JACC Basic Transl. Sci. 7, 608–625 (2022).
Quattrocelli, M. et al. Mouse and human mesoangioblasts: isolation and characterization from adult skeletal muscles. Methods Mol. Biol. 798, 65–76 (2012).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Shumate, A., Wong, B., Pertea, G. & Pertea, M. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS Comp. Biol. 18, e1009730 (2022).
Hu, J. et al. Siglec15 shapes a non-inflamed tumor microenvironment and predicts the molecular subtype in bladder cancer. Theranostics 11, 3089–3108 (2021).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
We thank Dr. Yafeng Song from the Institute of Sport and Health Science, Beijing Sport University, for kindly providing us with mdx mice, and Dr. Shuaiwei Ren for his advice on cell culture and plasmid construction. We appreciate the support and assistance provided by the Advanced Imaging Platform and Laboratory Animal Center of the Institute of Primate Translational Medicine, Kunming University of Science and Technology. This work was supported by the National Natural Science Foundation of China (81930121 and 82125008 to Y. Chen), the National Science and Technology Innovation 2030 Major Program (2021ZD0200900 to Y. Chen), the National Key Research and Development Program of China (2023YFA1801800 and 2018YFA0801403 to Y. Chen), the Natural Science Foundation of Yunnan Province (FWCY-ZD2024006 to Y.L.; 202001BC070001 and 202102AA100053 to W.J.).
Author information
Authors and Affiliations
Contributions
Y. Chen, W.J., S.W., and R.W. conceived and designed the research. R.W., P.L., Y. Chang, X.A., L.C., and Y.Z. performed molecular and behavioral experiments. R.W., P.L., and W.S. conducted all the cynomolgus’s experiments. X.W. analyzed RNA-seq data. R.W., J.L., P.X., and Y.L. performed histochemical staining. S.Z. is responsible for collecting and isolating PBMCs from DMD patients in the hospital. R.W., S.Z., and W.G. completed stem cell identification, culture, and differentiation experiments. W.J., S.L., Z.W., and J.W. provided guidance on the experimental design and performed data analysis. R.W., P.L., Y. Chen, S.W., and W.G. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Massimiliano Runfola, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, R., Li, P., Xiao, P. et al. Activation of endogenous full-length utrophin by MyoAAV-UA as a therapeutic approach for Duchenne muscular dystrophy. Nat Commun 16, 2398 (2025). https://doi.org/10.1038/s41467-025-57831-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57831-5