Abstract
Protein ubiquitylation is maintained by a dynamic balance of the conjugation and deconjugation of ubiquitin. It remains unclear how deubiquitylation-stabilized substrates are directed for degradation. Branched ubiquitin chains promote substrate degradation through the proteasome. TRIP12 and UBR5 are HECT-type E3 ubiquitin ligases, which are specific for lysine 29 (K29) and lysine 48 (K48) linkages, respectively. Here, we show that the deubiquitylase (DUB) OTUD5 is cooperatively modified by TRIP12 and UBR5, resulting in conjugation of K29/K48 branched ubiquitin chains and accelerated proteasomal degradation. TRIP12–OTUD5 antagonism regulates TNF-α–induced NF-κB signaling. Mechanistically, OTUD5 readily cleaves K48 linkages, but does not affect K29 linkages. Consequently, K29 linkages overcome OTUD5 DUB activity to facilitate UBR5-dependent K48-linked chain branching. This mechanism is applicable to other OTUD5-associated TRIP12 substrates. Thus, the combination of DUB-resistant and proteasome-targeting ubiquitin linkages promotes the degradation of deubiquitylation-protected substrates, underscoring the role of branched ubiquitin chains in shifting the ubiquitin conjugation/deconjugation equilibrium.
Similar content being viewed by others
Introduction
The ubiquitin–proteasome system regulates numerous cellular functions. Ubiquitin moieties are linked through the N-terminus, one of the seven lysine (K) residues, or certain serine/threonine residues, to form polyubiquitin chains. The functional diversity of different ubiquitin linkages is referred to as the ubiquitin code, which regulates protein degradation, inflammatory signaling, and endocytosis1.
Heterotypic ubiquitin chains, which consist of two or more ubiquitin linkage types, serve as important biological signals, adding complexity to the ubiquitin code2. Among them, branched ubiquitin chains, in which one ubiquitin moiety is modified with two or more ubiquitin molecules through different linkages, function as a priority signal for proteasome-mediated protein degradation3,4. K11/K48 branched ubiquitin chains are generated by anaphase-promoting complex/cyclosome (APC/C) or by UBR5/UBR4, and regulate cell cycle progression, gene transcription, and quality control of misfolded proteins3,5. K63-linked ubiquitin chains are further modified with either K48-linked or K48/K11-linked chains to assemble K48/K63-linked6,7 or K11/K48/K63-linked8 branched chains, respectively; these convert nonproteolytic K63-linked chains into proteasomal degradation signals. K29/K48 branched ubiquitin chains are assembled during ERAD pathways9, PROTAC-induced targeted degradation10, or ubiquitin-fusion degradation (UFD)11,12 to promote proteasomal degradation. Defects in enzymes involved in the regulation of branched ubiquitin chains cause diseases13, underscoring the biological importance of this emergent type of the ubiquitin code.
Several mechanisms have been proposed to explain how branched ubiquitin chains promote proteasomal degradation. The most prevalent view is that ubiquitin chain branching increases ubiquitin signal density irrespective of linkage type and enhances proteasome recruitment14. Indeed, substrates modified with either K11/K48 or K11/K48/K63 branched chains preferentially associate with the proteasome3,8. Additionally, branched ubiquitin chains containing K48 linkages are preferentially recognized by the proteasome-associating deubiquitylase UCH37, which debranches K48 linkages for proteasomal degradation15,16. Branched chains also promote association with the p97 segregase/unfoldase3,17. Another mechanism is that a previously formed ubiquitin chain serves as a foundation for K48-linked branching, contributing to efficient K48-linked chain assembly8,10.
These studies indicate that branched ubiquitin chains containing K48 linkages serve as superior signals for proteasomal degradation, irrespective of linkage types. However, the specific roles of each linkage type within a branched chain remain poorly characterized. For example, the K29 linkage is the fourth-most abundant linkage type in mammalian cells18,19 and, most K29 linkages lie within heterotypic chains containing K48 linkages20. Nonetheless, the specific function of K29 linkages within the branched chains remains unknown. Given that homotypic ubiquitin linkages have specific functions and dictate specific signaling pathways, it remains to be studied whether, and how, cognate linkage types within branched ubiquitin chains exert specific functions to promote protein degradation.
Ubiquitylation is reversibly regulated by deubiquitylases (DUBs), adding plasticity and robustness to the ubiquitin system21. DUBs fall into seven classes according to their catalytic domains: USP, OTU, MJD, UCH, JAMM, MINDY, and ZUFSP families22. Among them, OTU-family DUBs often exhibit ubiquitin linkage–type specificity23. DUBs associate with client proteins and prevent excessive ubiquitylation, protecting them from proteasomal degradation. Given their catalytic nature, it is unclear how deubiquitylation-protected proteins, e.g., DUBs and DUB-associated proteins, are destined for proteasomal degradation when cells need to decrease their protein levels.
In this work, we use proteomics screening to identify substrates ubiquitylated by TRIP12, the HECT-type E3 ubiquitin ligase that specifically assembles K29-linked ubiquitin chains and adds K29 branched linkages off the K48-linked chains10, and identify the OTU-family DUB, OTUD5. We find that OTUD5 is cooperatively modified by TRIP12 and UBR5 to assemble K29/K48 branched ubiquitin chains. TRIP12 and OTUD5 reciprocally regulate NF-κB signaling. We further explore the interplay between these three enzymes to understand how branched chains promote substrate degradation. Notably, OTUD5 readily cleaves K48 linkages but weakly cleaves K29 linkages. Consequently, OTUD5 DUB activity counteracts UBR5 but TRIP12 overcomes the deubiquitylation, facilitating branched chain formation and OTUD5 proteasomal degradation. Certain other TRIP12 substrates associated with OTUD5 are also targeted for degradation by TRIP12 and UBR5, similarly. Our results indicate that a combination of DUB-resistant and proteasome-destined ubiquitin codes efficiently target DUB-protected substrates for proteasomal degradation. These findings highlight an important role for two specific ubiquitin linkages that together, act as a robust degradation signal for DUB-protected proteasomal substrates.
Results
TRIP12 promotes OTUD5 ubiquitylation and degradation
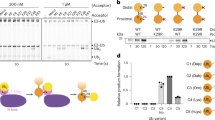
Although candidate approaches have identified substrates modified with branched ubiquitin chains4, there is no versatile methodology to systematically identify these proteins. In this study, we focused on TRIP12. Because K29 linkages lie mostly within K29/K48 heterotypic chains20, we hypothesized that TRIP12 substrates may be modified with K29/K48 branched chains. Moreover, these expected proteasomal substrates enable proteome-wide screening of candidate proteins using quantitative proteomics from cell lysates. Thus, we used tandem-mass-tag (TMT)-based quantitative proteomics to identify proteins accumulated after TRIP12 knockdown. We obtained 35 candidate proteins accumulated under TRIP12 knockdown in HT1080 cells (Fig. 1a and Supplementary Data 1). Gene ontology analysis revealed the enrichment of proteins related to chromatin regulation and RNAPII-mediated transcription (supplementary Fig. 1a), consistent with previously known roles of TRIP1224. Among the candidates, we focused on OTUD5 because endogenous OTUD5 was accumulated after TRIP12 knockdown using two independent siRNAs (Fig. 1b). Consistently, a downstream target of OTUD5, UBR5, was also slightly accumulated (Supplementary Data 1). We confirmed that OTUD5 protein accumulation upon TRIP12 knockdown was not attributable to their mRNA levels (Fig. 1c); although OTUD5 mRNA levels were marginally increased under TRIP12 knockdown (1.3-fold) (Fig. 1c), it was unlikely to cause OTUD5 protein accumulation (~5-fold) (Fig. 1b).
a TMT-based quantitative proteomics. HT1080 cells were transfected either with scrambled siRNA or siTRIP12 for 72 h. Total lysates were subjected to TMT analysis (two biological replicates). b HT1080 cells were transfected with either scrambled siRNA (Scr) or two independent siRNAs targeting TRIP12 for 72 h, and endogenous OTUD5 was analyzed by western blotting. The right panel shows quantification of band intensities (OTUD5/Actin). Two-sided ANOVA, three biological replicates. c mRNA expression levels were analyzed by quantitative RT-qPCR using HT1080 cells 72 h after the knockdown. Two-sided ANOVA, four biological replicates. d Cycloheximide (CHX) chase assay was performed using HEK293T cells transfected with the indicated siRNAs for 72 h and treated with CHX for 0, 7, or 14 h. The lower panel shows quantification of band intensities (OTUD5/Actin, two biological replicates). e HEK293T cells were transfected with the indicated expression vectors (2.5 μg FLAG-TRIP12 and/or 5 μg HA-OTUD5) for 48 h, and co-immunoprecipitation was performed using anti-FLAG antibody. f TRIP12 promotes OTUD5 ubiquitylation in cells. HEK293T cells were transfected with the indicated expression plasmids for 48 h, and treated with MG132 (20 μM, 2 h) before cell lysis. Ubiquitylated proteins were enriched using TUBE2-conjugated agarose from cell lysates. The ubiquitylated OTUD5 was stained using anti-OTUD5 antibody. g Ubiquitylation of endogenous OTUD5 is regulated by TRIP12. HT1080 cells were transfected with either scrambled or TRIP12 siRNAs for 72 h, and treated with 20 μM MG132 for 2 h before cell lysis. h TRIP12 ubiquitylates OTUD5 in vitro. OTUD5 (1 μg) was subjected to an in vitro ubiquitylation reaction with E1 (100 ng), E2 (100 ng UBCH5c or UBCH7), ubiquitin (10 μg), and TRIP12 (1 μg). OTUD5 was immunopurified after the reaction, and ubiquitylated OTUD5 was analyzed using anti-ubiquitin antibody. Source data are provided as a Source Data file.
OTUD5 belongs to the OTU family of deubiquitylases23. OTUD5 regulates several pathways, including NF-κB signaling pathways and transcriptional regulation through chromatin modification25. OTUD5 also regulates DNA damage response and immune signaling through association with UBR526,27. OTUD5 preferentially cleaves K48- and K63-linked chains28; however, its deubiquitylation specificity toward long or complex ubiquitin chains has not been well-characterized. The relationship between OTUD5 and TRIP12 is also unknown.
We subsequently analyzed the half-life of OTUD5 protein. TRIP12 knockdown massively slowed OTUD5 turnover (Fig. 1d). TRIP12 interacted with OTUD5 during co-immunoprecipitation (Fig. 1e). We subsequently investigated whether TRIP12 promoted ubiquitylation of OTUD5 in cell and in vitro. Using TUBE2 (tandem ubiquitin-binding entity 2), which serves as a pan-ubiquitin chain binder when four repeats of the UBA1 domains from HR23A are tandemly fused29, we found that OTUD5 pan-ubiquitylation was enhanced by the expression of wild-type, but not catalytically inactive (C1959A), TRIP12 (Fig. 1f). On the other hand, TRIP12 did not affect ubiquitylation of other tested OTU family deubiquitylases such as OTUD3 or OTUD4 (supplementary Fig. 1b). We also confirmed that TRIP12 knockdown decreased ubiquitylation of endogenous OTUD5 under proteasomal inhibition (Fig. 1g). In an in vitro ubiquitylation assay using purified OTUD5 and TRIP12 proteins (supplementary Fig. 1c), OTUD5 was ubiquitylated by TRIP12 (Fig. 1h). Collectively, these results indicate that TRIP12 interacts with OTUD5 and promotes its ubiquitylation and degradation.
TRIP12 and UBR5 cooperatively assemble K29/K48 branched ubiquitin chains
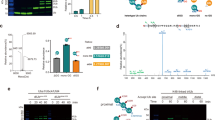
We explored whether OTUD5 was modified by TRIP12 with K29-linked chains. OTUD5 was immunopurified after an in vitro ubiquitylation reaction with TRIP12, then subjected to mass spectrometric quantification using parallel reaction monitoring (Ub-AQUA/PRM)30. We found that OTUD5 was specifically modified with K29-linked chains by TRIP12 (Fig. 2a). K29 specificity was confirmed using single-lysine ubiquitin mutants in the same in vitro ubiquitylation assay (Fig. 2b). To analyze modification with K29-linked chains in cells, we used GST-fused TRABID–NZF1 as a K29 binder. TRABID–NZF1 is specific for K29 and K33 linkages20,31, but OTUD5 is not modified with K33-linked chains; we, therefore, used it as a K29 binder here. As shown in Fig. 2c, OTUD5 modification with K29-linked chains was massively increased by wild-type TRIP12 but not by catalytically inactive TRIP12 (Fig. 2c).
a TRIP12 assembles K29-linked ubiquitin chains onto OTUD5 in vitro. OTUD5 ubiquitylated in vitro using TRIP12 and UBCH7 (as in Fig. 1h) was subjected to PRM-based absolute quantification. Two biological replicates were used. b An in vitro ubiquitylation assay was performed as in Fig. 1h using single-lysine mutant ubiquitin and UBCH7. c TRIP12 promotes K29-linked ubiquitylation of OTUD5 in cells. HEK293T cells were transfected as indicated, for 48 h, and treated with MG132 (20 μM, 2 h) before cell lysis. Proteins modified with K29-linked chains were enriched using K29-binder. d Absolute quantification of ubiquitin linkages conjugated on OTUD5. HEK293T cells were transfected with FLAG–HA–OTUD5 and HA–TRIP12, and OTUD5 was immunopurified. e TMT-based quantitative proteomics from control or UBR5 knockdown HT1080 cells. Two biological replicates were used. f HT1080 cells were transfected with the indicated siRNAs for 72 h, and CHX chase assay was performed. g–j OTUD5 was cooperatively modified with K29- and K48-linked ubiquitin chains. HEK293T cells were transfected with the indicated siRNAs for 72 h and expression plasmids for 48 h, respectively, and treated with MG132 (20 μM, 2 h) before cell lysis. Cell lysates were subjected to ubiquitin pulldown using either TUBE2, TRABID-NZF1, or K48TUBE as indicated. k Endogenous OTUD5 was immunoprecipitated from HT1080 cells and subjected to UbiCRest analysis. The ubiquitin chains were analyzed using Ub-AQUA/PRM. High-molecular-weight (HMW, >150 kDa) and low-molecular-weight (LMW, 75–150 kDa) polyubiquitylated OTUD5 were separated by SDS-PAGE. Two biological replicates. l Both TRIP12 and UBR5 are required for the degradation of endogenous OTUD5. HT1080 cells were transfected with the indicated siRNAs for 72 h. Cell lysates were subjected to western blotting. The right panel shows quantification of band intensities (OTUD5/Actin, three biological replicates). m Schematic model. TRIP12 and UBR5 cooperatively assemble K29/K48 branched ubiquitin chains onto OTUD5. The K29/K48 branched chains are indispensable for efficient degradation of OTUD5. Source data are provided as a Source Data file.
We subsequently quantified ubiquitin chain linkages conjugated to OTUD5, immunopurified from cells. OTUD5 was modified with 69% of K48 linkages and 19% of K29 linkages, both of which increased in the presence of TRIP12 (Fig. 2d). Because TRIP12 specifically assembles K29-linked chains10, the results raised the possibility that another E3(s) assembled K48-linked chains. Previous studies report that OTUD5 interacts with UBR5, a HECT-type K48-linkage specific E3, in the regulation of DNA damage response and NF-κB signaling27,32. Therefore, we conducted a TMT analysis to identify UBR5-regulated substrates. Surprisingly, OTUD5 was identified as a protein accumulated after UBR5 knockdown (Fig. 2e and Supplementary Data 2). Endogenous OTUD5 turnover was consistently delayed by UBR5 knockdown (Fig. 2f), suggesting that OTUD5 degradation was regulated by both TRIP12 and UBR5.
These results implied that TRIP12 and UBR5 cooperatively assembled K29/K48 branched ubiquitin chains. UBR5 has been shown to participate in the formation of K48/K637 or K11/K483 branched ubiquitin chains by specifically assembling K48 linkages. Therefore, we verified cooperative actions of TRIP12 and UBR5 in OTUD5 pan-ubiquitylation using TUBE2. UBR5-dependent ubiquitylation of OTUD5 was almost completely diminished with TRIP12 knockdown (Fig. 2g, lanes 2 and 4), whereas TRIP12-dependent ubiquitylation was partially decreased by UBR5 knockdown (Fig. 2h, lanes 2 and 4, and supplementary Fig. 1d). These results showed that UBR5 action is dependent on TRIP12.
We analyzed OTUD5 modifications with K29- or K48-linked chains using K29 binder (TRABID–NZF1) or K48–TUBE, respectively33. UBR5 expression promoted both K29- and K48-linked ubiquitylation of OTUD5, which was almost completely diminished by TRIP12 knockdown (Fig. 2i, lanes 2, 4, 6, and 8). However, the TRIP12-dependent K29-linked ubiquitylation of OTUD5 was partially decreased by UBR5 knockdown, whereas TRIP12-dependent increase of K48-linked chains was massively diminished by UBR5 knockdown (Fig. 2j, lanes 2, 4, 6, and 8).
To investigate whether endogenous OTUD5 was modified with K29/K48 branched ubiquitin chains, we immunopurified endogenous OTUD5 using anti-OTUD5 antibody; the conjugated ubiquitin chains were then cleaved by the engineered K48-specific DUB OTUB1*34. The reciprocal experiment to specifically cleave K29 linkages was technically challenging at this stage because K29/K33-linkage-specific TRABID can also cleave other linkages such as K63 linkage in vitro34. We found that, after the immunopurification and OTUB1* treatment, pan-ubiquitylation was partially retained (Fig. 2k). Mass spectrometric quantification confirmed that OTUB1* treatment reduced K48 linkage abundance (Fig. 2k). After OTUB1* treatment, the abundance of high-molecular-weight K29 linkages was decreased, whereas that of low-molecular-weight K29 linkages was almost retained (Fig. 2k) This suggests a model in which most of the K29 linkages lie in the proximal side of the chains from which K48 linkages branch off, whereas a portion of the K48 linkages is further modified by K29 linkages. Collectively, these results suggest that OTUD5 is modified with K29 linkages by TRIP12 and then with K48 linkages by UBR5, resulting in the conjugation of K29/K48 branched ubiquitin chains in which the majority of K48-linkages are added to the distal sides of K29-linked chains.
We investigated whether TRIP12 and UBR5 cooperatively regulated the degradation of endogenous OTUD5. Knockdown of either TRIP12 or UBR5 accumulated OTUD5 (Fig. 2l and supplementary Fig. 1e). Importantly, simultaneous knockdown of TRIP12 and UBR5 did not show further OTUD5 accumulation (Fig. 2l, lane 5). This indicates that both TRIP12 and UBR5 are required for OTUD5 degradation; that is, TRIP12 or UBR5 alone are insufficient for degradation. This supports the cooperative ubiquitylation model (Fig. 2m).
TRIP12–OTUD5 antagonism regulates NF-κB signaling
We investigated whether TRIP12 antagonizes OTUD5 cellular function. RNA sequencing was performed to identify pathways altered by TRIP12 or OTUD5 or double knockdown (Fig. 3a and Supplementary Data 3). We hypothesized that changes in gene expression caused by OTUD5 accumulation under TRIP12 knockdown, may be canceled by simultaneous knockdown of OTUD5. Therefore, we isolated genes whose expression levels were 1) downregulated <0.67-fold by TRIP12 knockdown compared to scramble and 2) recovered to >1.5-fold by TRIP12/OTUD5 double knockdown compared with TRIP12 single knockdown (Fig. 3b). Gene ontology analysis revealed that the GO term “inflammatory response” was accumulated in such a gene set (Fig. 3c). However, GO terms related to PI3-kinase and ERK/MAPK cascade were accumulated in a gene set changed by TRIP12 single knockdown (Fig. 3c). These results suggested that TRIP12–OTUD5 antagonism may regulate the NF-κB inflammatory signaling pathway.
a RNA-sequencing analysis. HT1080 cells were transfected with the indicated siRNAs for 72 h and total RNAs were isolated. Three biological replicates were used. b Protein-coding genes whose expression levels are down-regulated (<0.67-fold) by TRIP12 knockdown (siTRIP12 vs. scramble) and restored (>1.5-fold) by OTUD5 double knockdown (siTRIP12+siOTUD5 vs. siTRIP12). c Gene ontology (GO) analysis of genes isolated in (b). The P value stands for the adjusted P-value, which is corrected by the Benjamini-Hochberg method. For gene ontology analysis using DAVID, the raw P values, which were one-sided and not adjusted, were used. d NF-κB signal transduction is down-regulated in TRIP12 knockdown cells. HT1080 cells transfected with the indicated siRNAs for 72 h were treated with TNF-α for the indicated duration, and total lysates were subjected to western blotting. e NF-κB signaling is antagonistically regulated by TRIP12 and OTUD5. Cells were treated as in (d). In the lowest panel, total cell lysates (20 μg) were loaded to detect OTUD5 and OTUD5 band intensities were quantified. f Expression of NF-κB target genes is antagonistically regulated by TRIP12 and OTUD5. HT1080 cells were treated with 0.1 ng/mL TNF-α for 3 h, and RT-qPCR was performed. Four biological replicates were used. g HEK293T cells were transfected with FLAG-HA-OTUD5 plasmids for 48 h, and immunoprecipitation was performed using an anti-FLAG antibody. The association of endogenous TRAF2 with FLAG-HA-OTUD5 was detected. h K63 ubiquitylation of TRAF2 in response to TNF-α stimulation is antagonistically regulated by TRIP12 and OTUD5. HT1080 cells were treated with TNF-α as indicated, and cell lysates were subjected to K63TUBE pulldown to enrich proteins modified with K63-linked ubiquitin chains. i Schematic model. TRIP12 and UBR5 down-regulate the steady-state protein level of OTUD5. Accumulation of OTUD5 under TRIP12/UBR5 deficiency repressed TNF-α-induced NF-κB signaling partly, through deubiquitylation of TRAF2. Source data are provided as a Source Data file.
We also conducted RNA-sequencing of TRIP12, UBR5, or HOIP (a control, as HOIP and HOIL-1L generate M1- and Ser/Thr-linked heterotypic ubiquitin chains35) knockdown cells (supplementary Fig. 2a–c and Supplementary Data 3). Genes up/down-regulated in these knockdown cells were only partially overlapped, consistent with the notion that they regulate several biological pathways (e.g., TRIP12 regulates cell cycle progression and chromatin remodeling24, while UBR5 regulates DNA replication and protein quality control36,37).
Next, we investigated whether TRIP12 regulated TNF-α–induced NF-κB signaling. TNF-α treatment promoted phosphorylated I-κB, IKK, and p65 with a concomitant decrease in the total I-κB level (Fig. 3d, lanes 1–6). With TRIP12 knockdown, these TNF-α responses were down-regulated (Fig. 3d, lanes 7–12). The results indicate that TRIP12 is an enhancer of TNF-α–induced NF-κB signaling.
We next verified TRIP12–OTUD5 antagonism in TNF-α–induced NF-κB signaling. TNF-α–induced phospho-I-κB elevation was decreased or increased by TRIP12 or OTUD5 knockdowns, respectively (Fig. 3e, lanes 4–9). Such elevation of phospho-I-κB under OTUD5 knockdown was partially canceled by the simultaneous knockdown of TRIP12 and OTUD5 (Fig. 3e, lanes 10–12), with concomitant accumulation of OTUD5 (compare lanes 7–9 and 10–12 in anti-OTUD5 loading (++)). Consistently, the expression of NF-κB target genes (IL6, IL11, ADAM8, and IRAK2 (supplementary Fig. 2d)) in presence of TNF-α was reciprocally regulated by TRIP12 and OTUD5 (Fig. 3f).
To identify OTUD5 target(s) during NF-κB activation, we analyzed FLAG–OTUD5 associated protein identified by LC-MS. Among the putative interactants related to ‘signaling’ in KEGG pathways (Supplementary Data 4), we focused on TRAF2 because 1) TRAF2 modification with K63-linked chains serves as a hub for downstream NF-κB signaling pathways38 and 2) OTUD5 deubiquitylates the K63-ubiquitylated TRAF3, another TRAF-family protein32. Indeed, we found that TRAF2 associated with OTUD5 in cells (Fig. 3g). We explored whether OTUD5 regulated deubiquitylation of TRAF2. We found that K63-ubiquitylation of TRAF2 was increased by OTUD5 knockdown and decreased by TRIP12 knockdown (Fig. 3h). It cannot be ruled out that OTUD5 may also cleave K48 linkages modified on TRAF2 or on other substrates. Collectively, these results suggest a model in which TRIP12 regulates the steady-state level of OTUD5 protein, whose accumulation under TRIP12 depletion represses TNF-α–induced NF-κB signaling partly through TRAF2-deubiquitylation (Fig. 3i).
OTUD5 efficiently cleaves K48 linkages but not K29 linkages
The data presented thus far indicate that OTUD5 is cooperatively modified by K29-specific TRIP12 and K48-specific UBR5, resulting in the conjugation of K29/K48-linked branched ubiquitin chains, and the biological importance of TRIP12–OTUD5 antagonism is observed in TNF-α–induced NF-κB signaling. Nonetheless, how branched ubiquitin chains consisting of K29 and K48 linkages promote substrate degradation remains unclear. Therefore, we further explored the interplay of TRIP12, UBR5, and OTUD5 in the regulation of branched ubiquitin chain assembly.
We hypothesized that OTUD5 DUB activity may counteract TRIP12 and/or UBR5. To test this, we first analyzed the ubiquitin chain linkage preference of OTUD5 DUB activity. Diubiquitins of eight linkage types were subjected to an in vitro deubiquitylation assay with OTUD5. OTUD5 readily cleaved K11-, K48-, and K63-linked chains, whereas K27, K29, and K33 linkages were resistant to OTUD5 (Fig. 4a). OTUD5 showed modest activity toward K6-linked chains and did not cleave M1-linked chains.
a OTUD5 efficiently cleaves K11-, K48-, and K63-linked diubiquitin. Diubiquitins (1.0 μg) of eight linkage types were incubated with OTUD5 (0.5 μg) for the indicated duration in an in vitro deubiquitylation assay. The right panel shows quantification of band intensities. b In vitro deubiquitylation assay as in (a) using the indicated tetraubiquitin as substrates. c (i) A schematic workflow for the preparation of K29-Ub4. Enzymatically synthesized Ub4 includes cyclic K29-Ub4 and K29/K48 branched/mixed Ub4 as well as K29-Ub4. Enzymes were used to identify the linkage type of Ub4, and fractions with a high purity of K29-Ub4 were collected. See also online methods. (ii) A schematic workflow for the preparation of K29/K48-Ub8. K29/K48-Ub8 was synthesized from K29-Ub4 and Ub (K48R). The byproduct K29/K48-Ub15~, synthesized by the reaction between K29-Ub4, was removed using cation-exchange chromatography. See also online methods. d In vitro deubiquitylation assay. (i, ii, iii) Either K29-linked Ub4, K48-linked Ub4, or K29/K48-linked Ub8 (1.0 μg) was incubated with OTUD5 (0.5 μg) for the indicated duration. In (iv), the sample of K29/K48-linked Ub8 incubated with OTUD5 for 4 h was stained with anti–K48-linkage antibody. (v), possible architectures of Ub5 in (iv). e In vitro deubiquitylation assay as in (d) with the indicated architectures of Ub2 or Ub3. The graph (iii) shows quantification of four independent experiments (Fig. 4e (i), (ii), and supplementary Fig. 4c). Two-sided ANOVA was used. Source data are provided as a Source Data file.
Because OTUD5 prefers longer ubiquitin chains as substrates28, we characterized the linkage specificity of OTUD5 using K11-, K29-, K48-, or K63-linked tetra-ubiquitin. Again, OTUD5 readily cleaved K48- or K63-linked tetraubiquitin and, to a lesser extent, K11-linked tetraubiquitin but exhibited minimal activity toward K29-linked tetraubiquitin (Fig. 4b, d (i)).
We subsequently analyzed whether OTUD5 deubiquitylates K29/K48-linked branched ubiquitin chains. To imitate the architecture of branched ubiquitin chains conjugated to OTUD5, i.e., distal K48 linkages branching off the proximal K29 linkages, we inserted monoubiquitins into the four K48 residues within a K29-linked tetraubiquitin, resulting in a K29/K48-linked branched octamer (Fig. 4c and supplementary Fig. 3a–f). When cleaved by OTUB1*, octa-ubiquitin separated into Ub4 and Ub1, confirming the architecture with K48 linkages inserted into K29-Ub4 (supplementary Fig. 4a). Using these ubiquitin chains, we conducted a timecourse deubiquitylation assay with OTUD5. Using the K29/K48-linked branched octamer as a substrate, we found that Ub5 and Ub4 were accumulated after the cleavage (Fig. 4d (iii)). Staining with a K48-linkage-specific antibody revealed that the Ub5, but not Ub4 and shorter chains, contained K48 linkages (Fig. 4d (iv)). This suggests that K29-linked Ub4 with a single K48 branched/mixed linkage is partially resistant to OTUD5 DUB activity (Fig. 4d (v)). We also conducted a similar DUB assay with the titration of different amounts of OTUD5 (supplementary Fig. 4b), confirming the results shown in Fig. 4d.
To gain insight into architecture that may repress OTUD5 DUB activity, we investigated whether the cleavage of a K48 linkage is repressed when it is either branched with the K29-linkage or tandemly-mixed at the distal or proximal side of the K29 linkages (Fig. 4e and supplementary Figs. 4c–e and 5a–c). We observed a modest, but reproducible repression of OTUD5 DUB activity towards K48 linkages in all three types of K29 mixed linkages, i.e., K29/K48 branched Ub3, K29/K48 tandemly-mixed Ub3, and K48/K29 tandemly-mixed Ub3 (Fig. 4e and supplementary Fig. 4c–d). We subjected equal amounts of (Fig. 4e) or equimolar (supplementary Fig. 4e) OTUD5 to the DUB assays to obtain consistent results. Although a precise understanding of the underlying mechanism depends on detailed structural analyses, our results suggest that the K29 linkage is not only resistant by itself to OTUD5, but also modestly stabilizes the mixed/branched K48 linkage.
Collectively, these results indicate that OTUD5 readily cleaves K48 linkages but only weakly cleaves K29 linkages, supporting the proposed model where OTUD5 counteracts the assembly of K48-linked ubiquitin chains conjugated by UBR5.
OTUD5 counteracts UBR5-mediated K48 ubiquitylation, overcome by TRIP12
To investigate whether OTUD5 counteracted ubiquitin chain assembly, we conducted an in vitro ubiquitylation/deubiquitylation assay using wild-type or catalytically inactive (C224R) OTUD5 (OTUD5–C224R). We purified FLAG–HA–OTUD5 protein from cells treated with the E1 inhibitor TAK-243, preventing contamination of ubiquitylated forms of OTUD5. We introduced a Cys-to-Arg point mutation (C224R) based on a previous report that Cys-to-Ser mutation, but not Cys-to-Arg, increases interaction with ubiquitin39. We analyzed the elongation of free ubiquitin chains assembled either by UBR5 or TRIP12, to assess the activity of these E3s, irrespective of substrate properties. We found that TRIP12-dependent polyubiquitin chain formation was unaltered irrespective of co-incubation with wild-type or C224R OTUD5 (Fig. 5a, right panel). However, UBR5-dependent polyubiquitylation was less efficient when co-incubated with wild-type OTUD5 than with C224R OTUD5 (Fig. 5a, left panel), leaving oligo-ubiquitylated species (~15 to 37 kDa). Notably, most of the mono-ubiquitin remained non-reacted in our assay. The data indicate that UBR5-dependent ubiquitin chain assembly is counteracted by OTUD5.
a OTUD5 DUB activity counteracts UBR5- (not TRIP12-) ubiquitin chain assembly. In vitro ubiquitylation/deubiquitylation was performed with 0.25 μg OTUD5 (wild-type or C224R) and UBR5 or TRIP12 (3 h). b OTUD5 ubiquitylation by UBR5 is counteracted by OTUD5 DUB activity. OTUD5 (wild-type or C224R) (0.25 μg) was incubated (3 h) with (i) UBR5 or (ii) TRIP12 for in vitro ubiquitylation/deubiquitylation, as in (a). OTUD5 was then immunopurified using anti-HA antibody, and ubiquitylated OTUD5 visualized using anti-ubiquitin antibody. c Ubiquitin chain linkages in (b) (0.25 μg OTUD5 and 50 ng UBR5/TRIP12) (i and ii) were quantified using PRM. Signature peptide abundance (fmol) for each linkage type was obtained from two independent experiments. d (i) UBR5_UBA interaction with ubiquitin chains. Diubiquitin chains (eight linkage types) were incubated with recombinant Halo-UBR5_UBA fused protein; associated chains were analyzed by western blotting. (ii) Band intensities in (i) were quantified; UBR5_UBA pulldown to input ratio is shown. (iii) Similar pulldown analysis using K29-linked Ub2, Ub3, and Ub4. e Purified OTUD5–SUE1 (SUE1 sequence fused at N-terminus) was incubated with E1, UBE2E, and K29-Ub2 for in vitro ubiquitylation. OTUD5–SUE1 modification was visualized using western blotting or CBB. Asterisk indicates proteins unrelated to OTUD5. f Unmodified OTUD5 or OTUD5–SUE1 modified with K29-Ub2 (e) was immobilized on anti-HA affinity resin and subjected to in vitro ubiquitylation with E1, UbcH7, Ub, or UBR5. After washing, resin-bound OTUD5 was denatured and subjected to western blotting using anti-UbK48 (Apu.2) antibody. g Turnover of catalytically inactive OTUD5 bypasses TRIP12 requirement. HEK293T cells were transfected with siRNAs (72 h) and then with either WT or C224R OTUD5 (48 h). Cells were treated with cycloheximide (50 μg/mL, 14 h); total cell lysates were subjected to western blotting. The lower panel shows quantification of band intensities (anti-HA/Actin, two biological replicates). h Schematic model. OTUD5 deubiquitylates K48-linked ubiquitin chains conjugated by UBR5. K29-linked ubiquitin chains conjugated by TRIP12 are resistant to OTUD5 DUB activity. UBR5 assembles branched K48-linked chains off previously formed K29-linked chains (more efficient than de novo assembly of K48-linked chains). Source data are provided as a Source Data file.
Next, we investigated whether OTUD5 DUB activity counteracted OTUD5 ubiquitylation by TRIP12 or UBR5. After in vitro ubiquitylation/deubiquitylation of wild-type or C224R OTUD5 with TRIP12/UBR5, OTUD5 was purified using anti-HA immunoprecipitation. Notably, UBR5 ubiquitylation of wild-type OTUD5 was less effective than that of C224R OTUD5 (Fig. 5b (i)). However, wild-type and C224R OTUD5 were ubiquitylated by TRIP12 at similar levels (Fig. 5b (ii)). To validate ubiquitin linkage specificity, we subjected this reaction to mass spectrometric quantification. As shown in Fig. 5c (i), assembly of K48 linkages onto OTUD5 by UBR5 was approximately 6-fold more abundant on the C224R mutant than on the wild-type OTUD5 (Fig. 5c (i)). However, efficiency of K29-linkage formation by TRIP12 was similar in both wild-type and C224R OTUD5 (Fig. 5c (ii)). These results reveal that OTUD5 DUB activity counteracts UBR5-dependent K48-linked chain assembly conjugated to OTUD5, whereas K29-linked chain formation by TRIP12 is resistant to OTUD5.
Based on results thus far, we postulated the following model: while K48-linked ubiquitylation by UBR5 is restricted by OTUD5, TRIP12 assembles K29-linked chains, which are resistant to OTUD5 DUB activity. UBR5 then adds K48 branched linkages off the K29-linked chains, more efficiently than de novo chain assembly.
UBR5 has a UBA domain, which may mediate K48 branched ubiquitylation off the K11-linked3 or K63-linked chains7. Therefore, we investigated whether the UBR5 UBA associated with K29-linked diubiquitin. Although UBR5 UBA did not exhibit a clear linkage-type specificity, it associated with K29-linked diubiquitin with the highest affinity among other linkage types except for the K27 linkage (extremely low abundant19, may not contribute to OTUD5 proteasomal degradation) (Fig. 5d (i) and (ii)). UBR5 UBA associated with longer K29-linked chains more strongly (Fig. 5d (iii)).
We next investigated whether UBR5 assembled branched K48-linked chains off the K29-linked chains in vitro. For this, OTUD5 was modified with K29-linked diubiquitin using a SUE1 strategy40. We first confirmed successful OTUD5 modification with monoubiquitin at its N-terminus, using the SUE1 sequence (supplementary Fig. 6a), although the SUE1 sequence insertion within native ubiquitylation sites did not work in our experiments. We then modified the OTUD5 N-terminus with K29-linked Ub2 (Fig. 5e). Wild-type OTUD5 or K29Ub2-modified OTUD5 were subjected to UBR5-mediated K48-linked ubiquitylation in vitro. We found that K29Ub2-modified OTUD5 was further conjugated with K48-linked ubiquitin chains by UBR5 more efficiently than wild-type OTUD5 (Fig. 5f). This supports our model where modification of K29-linked chains changes the equilibrium between ubiquitylation by UBR5 and deubiquitylation by OTUD5.
Finally, we tested the ubiquitylation/deubiquitylation competition in living cells. We hypothesized that, if K29-linked ubiquitylation is required to counteract OTUD5 DUB activity, then degradation of catalytically inactive OTUD5 should bypass TRIP12 requirement. To test this, wild-type or OTUD5-C224R were expressed in either control or TRIP12-knockdown cells, and cycloheximide chase assay was performed. As shown in Fig. 5g, the turnover of wild-type OTUD5 was delayed by TRIP12 knockdown, as expected (Fig. 5g, lanes 1–2 and 5–6). By contrast, OTUD5-C224R was degraded even in the TRIP12-knockdown cells (Fig. 5g, lanes 3–4 and 7–8). Thus, degradation of catalytically inactive OTUD5 does not require TRIP12, suggesting that K29-linked ubiquitylation is required to overcome OTUD5 DUB activity. Supporting our model, the turnover of OTUD5-C224R is dependent on UBR5 (supplementary Fig. 6b). Consistently, K48-ubiquitylation of OTUD5-C224R in the presence of UBR5 did not require TRIP12 (supplementary Fig. 6c).
Collectively, these results indicate that TRIP12 conjugates K29-linked ubiquitin chains onto OTUD5, which is resistant to OTUD5 DUB activity; then UBR5 associates with the K29-linked chains and assembles branched K48-linked chains off the K29 linkages (Fig. 5h).
Proteomics analysis reveals that certain TRIP12/UBR5–co-regulated substrates are protected by OTUD5
We hypothesized that certain other substrates of TRIP12/UBR5 may also be regulated by similar mechanisms. Therefore, we attempted to identify substrates regulated by both TRIP12 and UBR5 and analyze their protein turnover rates. Re-analyzing the TMT-based quantitative proteomics data to identify substrates of either TRIP12 (Fig. 1) or UBR5 (Fig. 2), we found that certain proteins were enriched in both TRIP12- and UBR5-knockdown cells (Fig. 6a (i) and (ii)). We analyzed endogenous protein levels of selected substrates identified in Fig. 6a and found that their abundances were indeed co-regulated by TRIP12 and UBR5 (Fig. 6b). Gene ontology analysis revealed enrichment of proteins related to snRNA regulation, transcription, and chromatin regulation in the siTRIP12/siUBR5 overlapped proteins (supplementary Fig. 7a–e and Supplementary Data 5).
a TMT-based quantitative proteomics using HT1080 cells. (i) siTRIP12 vs. scrambled siRNA, (ii) siUBR5 vs. scrambled siRNA, (iii) cycloheximide treatment for 6 h vs. vehicle control. Ratio of protein expression (Log2) and P-values (−Log10) from two biological replicates are shown. b Validation of proteins co-regulated by TRIP12 and UBR5. HT1080 cells were transfected with siRNAs for 72 h, and cell lysates were subjected to western blotting. c ARMC7 and ZGPAT, substrates co-regulated by TRIP12/UBR5, associate with OTUD5. HEK293T cells were transfected with the indicated expression vectors, and co-immunoprecipitated. d ARMC7 or ZGPAT complexes exhibit DUB activity in vitro. HEK293T cells were transfected with either FLAG–ARMC7 or FLAG–ZGPAT, together with HA–OTUD5, and the anti-FLAG immunocomplexes were incubated for 6 h with either K48- or K29-linked diubiquitin in an in vitro deubiquitylation assay. The complexes preferentially cleave K48-linked chains over K29-linked chains. OTUD5 inclusion in the ARMC7 or ZGPAT immunocomplex is shown in Fig. S3b. e ARMC7 is modified with K29- and K48-linked ubiquitin chains in a manner dependent on wild-type TRIP12. HEK293T cells were transfected with the indicated plasmids (1 μg of HA-ARMC7 and/or FLAG-TRIP12) for 48 h, and treated with MG132 (20 μM, 2 h) before cell lysis. Cell lysates were subjected to TUBE pulldown as indicated. f ARMC7 is cooperatively modified with TRIP12-dependent K29-linked chains and UBR5-dependent K48-linked chains. HEK293T cells were treated as in (e). g TRIP12 is dispensable for the turnover of endogenous ARMC7 when OTUD5 is knocked down. HEK293T cells were transfected with the indicated siRNAs for 72 h, and cycloheximide chase assay was performed for 0, 7, or 14 h. h Schematic model. Not only OTUD5 itself, the degradation of OTUD5-associated proteins are also regulated by the interplay of E3s TRIP12/UBR5 and DUB OTUD5. Source data are provided as a Source Data file.
To characterize TRIP12/UBR5 substrates, we analyzed protein turnover rates by applying TMT proteomics to HT1080 cells treated with either vehicle or cycloheximide for 6 h (Fig. 6a (iii)). While typical proteasomal substrates such as CDKN1A (p21), HIF-1α, and MCL1 were turned over at short timecourses, the substrates we found as TRIP12/UBR5-co-regulated proteins, including OTUD5, were relatively long-lived proteins (Fig. 6a (iii) and b).
We hypothesized that these long-lived substrates co-regulated by TRIP12/UBR5 may be protected by OTUD5 through deubiquitylation. Indeed, we found that ARMC7 and ZGPAT, whose protein levels are co-regulated by TRIP12 and UBR5 (Fig. 6b), stably associated with OTUD5 in cells (Fig. 6c). Moreover, FLAG–ARMC7 or FLAG–ZGPAT immunocomplex, which contained OTUD5 (supplementary Fig. 6d), exhibited DUB activity toward K48-linked diubiquitin (Fig. 6d). However, their DUB activity toward K29-linked diubiquitin was much weaker (Fig. 6d), suggesting that K29-linked ubiquitin chains were relatively resistant to DUB activity of ARMC7–OTUD5 or ZGPAT–OTUD5 complexes.
We further characterized ARMC7 ubiquitylation and degradation. K29- and K48-linked ubiquitylation of ARMC7 was induced by the expression of wild-type, but not that of the catalytically inactive, TRIP12 (Fig. 6e). Moreover, TRIP12-dependent K48-linked ubiquitylation of ARMC7 was decreased by UBR5 knockdown (Fig. 6f). In contrast, K29-linked ubiquitylation was retained after the knockdown but the size of ubiquitylated ARMC7 decreased, presumably due to the absence of K48 branched linkages (Fig. 6f). These results suggest that ARMC7 is ubiquitylated and degraded by a mechanism similar to that for OTUD5. Importantly, while the turnover of ARMC7 was delayed by TRIP12 knockdown (Fig. 6g, lanes 1–6), turnover of ARMC7 in the OTUD5 knockdown cells did not require TRIP12 (Fig. 6g, lanes 7–12). Therefore, TRIP12-mediated K29-ubiquitylation is required to counteract OTUD5 DUB activity, similar to the OTUD5 degradation mechanism.
Collectively, these results demonstrate the generality of the OTUD5–TRIP12/UBR5 antagonistic model in which the degradation of both OTUD5 and OTUD5-associated proteins requires TRIP12-mediated K29/K48 branched ubiquitin chains. With this mechanism, deubiquitylation of K48 linkages is overcome by altering the ubiquitylation/deubiquitylation equilibrium (Fig. 6h).
Discussion
Different ubiquitin linkages exert specific roles in regulating biological pathways. Specific functions of ubiquitin linkages within heterotypic chains are less understood compared to those in homotypic chains4. In this study, we demonstrated a mechanism in which a combination of DUB-resistant and proteasome-targeting ubiquitin linkages acts as a ubiquitin code for substrate degradation. In this model, K29 linkages play a critical role in proteasomal degradation as a “DUB-resistant ubiquitin chain”, facilitating K48-linkage assembly. Supporting our observation, an in vitro reconstitution analysis indicated that K29/K48 branched chains are a less effective proteasomal signal than the homotypic K48-linked chains of the same length40, implying that K29/K48 branched chains may be involved in proteasomal degradation under a cellular environment. We presume that K29/K48 branched chains serve as a superior degradation signal under ubiquitylation/deubiquitylation equilibrium. DUB degradation is the most exaggerated case as DUBs are likely to cleave ubiquitin chains conjugated onto themselves. However, we speculate that such equilibrium is common within cells. We have, therefore found a critical role for the branched ubiquitin chains in shifting the equilibrium between ubiquitin conjugation and deconjugation. Considering reported functions of branched ubiquitin chains in recruitment of the proteasome or p973,17 and efficient chain assembly8, identification of other linkage-specific roles with regard to branched ubiquitin chains should be studied further.
The present study revealed a sophisticated interplay between TRIP12, UBR5, and OTUD5. TRIP12 exclusively assembles K29-linked ubiquitin chains and adds K29-linked branches onto K48-linked chains10. UBR5 is a K48-specific E3 and adds K48 branches to assemble K11/K48- or K48/K63-linked branched chains3,7,41. On the other hand, OTUD5 preferentially cleaves K11-, K48-, and K63-linked chains but only scarcely acts toward K29 linkages. These properties of the three enzymes constitute a complex “rock–paper–scissors”-like interplay.
OTUD5 protects UBR5 from self-ubiquitylation26,27, suggesting that OTUD5 antagonizes UBR5-mediated K48 ubiquitylation. Thus, OTUD5 may regulate its own stability by antagonizing UBR5-mediated ubiquitylation. This equilibrium favors ubiquitin chain assembly and subsequent OTUD5 degradation when TRIP12 interacts with OTUD5; UBR5 assembles K48-linked ubiquitin chains onto previously formed K29-linked chains more efficiently than de novo ubiquitylation (Fig. 5), presumably through ubiquitin interaction of the UBA domain (Fig. 5d and ref. 7). Moreover, we found that mixing of K29-linkages (both branched or tandemly-mixed) modestly represses the cleavage of K48-linkages by OTUD5 (Fig. 4). Thus, TRIP12-generated K29-linkage besides protecting itself from OTUD5-mediated deubiquitylation, also 1) facilitates UBR5-dependent assembly of K48-linked chains and 2) modestly protects K48-linkages from disassembly as forms of mixed/branched linkages. Collectively, we show how an interplay of three enzymes with different ubiquitin linkage specificities fine-tunes substrate degradation through editing of complex ubiquitin architectures.
Despite the important role demonstrated for K29/K48 branched ubiquitin chains in substrate degradation, several questions remain regarding the assembly and architecture of the branched chains. First, whether TRIP12 constitutively associates with OTUD5 is unknown. Because UBR5 appears to stably associate with OTUD526,27, and because TRIP12 possesses phosphorylation sites (https://www.phosphosite.org), we presume that TRIP12 may associate with OTUD5 under certain cellular conditions, triggering rapid OTUD5 degradation to fulfill cellular demand.
Second, our model suggests complex ubiquitin architectures consisting of K29 and K48 linkages. The data on a catalytically-inactive OTUD5 (Fig. 5) suggest that the ubiquitylation of OTUD5 by TRIP12 and UBR5 appears as if they act sequentially because OTUD5 DUB activity negates UBR5 activity, which is different from typical chain initiation (priming)-elongation model42,43. Since UBR5 can stably associate with OTUD526, UBR5 may transiently ubiquitylate OTUD5 with K48-linked chains. However, they may be vulnerable because of deubiquitylation by OTUD5. Consequently, ubiquitin architectures with K29 linkages as foundation and K48 linkages as branches may survive in cells due to E3s/DUBs competition and serve as a productive signal for proteasomal degradation. Thus, we presume that the architectures of branched ubiquitin chains can differ depending on the substrates and contexts.
We note that our UbiCRest analysis could not technically distinguish TRIP12 addition of branched K29-linkages onto the most proximal ubiquitin moiety of the previously assembled K48-linked chains. Given that TRIP1210 (and UFD412) efficiently adds branched K29-linkages off K48-linkages, TRIP12 may occasionally use remaining short K48-linked chains for seeding K29-linkage assembly. Nonetheless, our data collectively indicate that the majority of UBR5-mediated K48-ubiquitylation depends on TRIP12 (Figs. 5h and 6h).
Given their catalytic nature, it is enigmatic how proteins protected by deubiquitylation (e.g., DUB-associating proteins and DUBs themselves) are destined to proteasomal degradation. Conceivably, DUBs can protect themselves from ubiquitylation. However, DUBs also need to be turned over to fulfill cellular proteostatic maintenance. In addition, cells may need to decrease certain DUB protein levels in response to cellular changes. Thus, inducing DUB degradation through ubiquitylation can be viewed as a paradox. In this study, we identified an elegant mechanism to counteract deubiquitylation that allows a DUB, OTUD5, to be efficiently targeted for degradation. In previous studies, certain DUBs such as BAP1 and USP4 were found to counteract their self-ubiquitylation via their DUB activities; however, whether/how the equilibrium favors DUB degradation remains unclear44,45. Collectively, our results indicate that balancing mechanisms to ensure proper DUB turnover may be more widespread than previously recognized. Additional studies are required to reveal whether other DUBs are regulated by similar mechanisms.
The biological importance of the TRIP12–OTUD5 interplay was demonstrated in the regulation of NF-κB signaling. In response to TNF-α, certain substrates including RIP1, TRAF2, and cIAP1 are modified with K63-linked chains by enzymatic activity of cIAP1, which recruits LUBAC and induces a phosphorylation cascade that eventually activates NF-κB–mediated gene induction46. We showed that TRIP12 positively regulates TNF-α–induced NF-κB signaling. Because the simultaneous knockdown of OTUD5 partially restores NF-κB signaling under TRIP12 knockdown, the TRIP12–OTUD5 antagonism may account for the regulatory function of TRIP12 in NF-κB signaling. We identified TRAF2 as a deubiquitylation substrate of OTUD5. However, we could not rule out that other, unknown substrate(s) are also regulated by OTUD5 in response to TNF-α. In any event, TRIP12 regulates NF-κB activity by limiting the steady-state OTUD5 protein level before TNF-α stimulation, unlike other E3s regulating NF-κB signaling such as LUBAC, TRAFs, and cIAP1, which are transiently activated to directly ubiquitylate the NF-κB cascade regulators through K63- or M1-linked chains46. Whether the TRIP12–OTUD5 axis also regulates other pathways should be studied further.
We have shown the generality of the combinatorial ubiquitin code with other TRIP12 substrates, suggesting that certain OTUD5-protected substrates are also regulated by DUB-resistant K29-linked ubiquitin chains. The TRIP12 substrates we analyzed, ARMC7 and ZGPAT, are not DUBs but associate with OTUD5 and exhibit DUB activity toward K48-linked chains, as multiprotein complexes. Notably, while OTUD5 cleaves several linkage types, OTUD5 mutations found in human patients with developmental disorders affect DUB activity toward K48 linkage28. This finding implies the physiological importance of OTUD5 in regulating the stability of associating proteins. We also presume that TRIP12 may cooperate with E3s such as UBR5 or CUL2/CUL410 to regulate yet unknown substrates.
More generally, we presume that such a mechanism is potentially applicable to other DUBs. In a previous analysis of linkage specificity of ~90 DUBs in vitro47, a majority of DUBs were found to prefer K48 linkages over K29 linkages (64 out of 73 DUBs prefer K48 over K29; 2 prefer K29 over K48; and 7 have similar preferences47). Although the K48 and K63 residues lie in the flexible loop regions of ubiquitin, the K29 residue lies within the α-helix structure. Therefore, the K29 linkage is intrinsically more difficult to be accessed by the DUB catalytic center, rendering it a DUB-resistant linkage. Considering these observations, we propose a role of the branched ubiquitin chains in promoting the degradation of DUB-protected substrates under ubiquitylation/deubiquitylation equilibrium.
Methods
Plasmids, siRNAs, and chemicals
Mammalian expression vectors for human TRIP12 and UBR5, and Halo-UBR5(UBA), have been described previously7,10. ARMC7 and ZGPAT cDNA clones were obtained from Promega (Flexi ORF clones). Escherichia coli expression vector for OTUB1* (an engineered OTUB1) (#65441) and mammalian expression vector for OTUD5 (#22610) were obtained from Addgene34,48. Site-directed mutagenesis was performed using PrimeStar Max (Takara) to generate FLAG–HA–OTUD5_C224R, FLAG–TRIP12_C1959A, FLAG–ARMC7, HA–ARMC7, and FLAG–ZGPAT.
Ambion Silencer Select siRNAs for TRIP12 (#1: s17810, #2: s17809), OTUD5 (s226854), UBR5 (#1: s28026, #2: s60863), ARMC7 (s35975), HOIP (RNF31; s30108), and a scrambled siRNA (Silencer Select Negative Control #1: 4390843) were purchased from Thermo Fisher Scientific.
PR619 (LifeSensors), MG132 (Santa Cruz Biotechnology), iodoacetamide (Sigma-Aldrich), cycloheximide (Sigma-Aldrich), and protease inhibitor cocktails (Nacalai Tesque, 03969-34) were purchased from the indicated manufacturers.
Antibodies
The following antibodies were used: anti-TRIP12 (Proteintech, 25303, 1:1,000), anti–p-I-kB (Cell Signaling Technology, 9246 s, 1:1,000), anti–p-IKK (Cell Signaling Technology, 2078 s, 1:1,000), anti-I-κB (Cell Signaling Technology, 4812 s, 1:1,000), anti–p-p65 (Cell Signaling Technology, 3033, 1:1,000), anti-HA (Cell Signaling Technology, 3724 s, 1:1,000), anti-FLAG (SIGMA, 7425, 1:1,000, or Cell Signaling Technology, 14793 s, 1:1,000), anti-TRAF2 (Abcam, ab-126758, 1:1,000), anti-OTUD5 (Cell Signaling Technology, 20087, 1:1,000), anti-ARMC7 (NOVUS, NBP1-82129, 1:1,000), anti-ZGPAT (Proteintech, 23203, 1:1,000), anti-GTF2B (Proteintech, 16467, 1:1,000), anti-MEAF6 (Themo Fisher Scientific, PA5-40704, 1:1,000), anti-ZBED1 (Proteintech, 26140, 1:1,000), anti-PRDM2 (Proteintech, 27718, 1:1,000), anti-NR2F2 (COUP-TFII) (Cell Signaling Technology, 6434 s, 1:1,000), anti-DHX32 (Proteintech, 68306-1-lg, 1:1,000), anti-Rad18 (Cell Signaling Technology, 9040 s, 1:1,000), anti–β-Actin (Santa Cruz Biotechnology, sc-47778, 1:1,000), anti-ubiquitin (P4D1, Santa Cruz Biotechnology, sc-8017, 1:1,000), anti-OTUD3 (Proteintech, 29622, 1:1000), anti-OTUD4 (Proteintech, 25070, 1:1000), anti-UBR5 (Santa Cruz Biotechnology, sc-515494, 1:1000), and anti-K48ubiquitin (Abcam, ab-140601, 1:1,000). For immunoprecipitation, anti-FLAG (Sigma-Aldrich, M2, A2220), anti-HA (Sigma-Aldrich, A2095), and anti-OTUD5 (Cell Signaling Technology, 20087, 1:1,000) antibodies were used.
Cell culture, treatment, and transfection
Human HEK293T and HT1080 cells were obtained from the American Type Culture Collection (ATCC) and maintained (37 °C, 5% CO2) in DMEM high-glucose medium (Sigma-Aldrich), supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich), penicillin–streptomycin (100 U/mL, Gibco, 15140148), sodium pyruvate (1 mM, Gibco, 11360070), and MEM nonessential amino acids (1×, Gibco, 11140050).
Transfection was performed using Lipofectamine 3000 reagent (Thermo Fisher Scientific) for plasmids and Lipofectamine RNAiMax (Thermo Fisher Scientific) for siRNAs. Transfected cells were used for experiments, 36–48 h (plasmids) or 72 h (siRNA) after transfection. For cycloheximide chase analysis, cells were treated with 50 μg/mL of cycloheximide for 0, 7, or 14 h before cell lysis. For ubiquitylation analysis, cells were treated with 20 μM of MG132 for 2 h, before cell lysis. For TNF-α signaling analysis, cells were treated with 5, 20, or 40 ng/mL of TNF-α (R&D SYSTEMS, 210-TA) for 5, 10, or 20 min before cell lysis.
Cell lysis, immunoprecipitation, and enrichment of ubiquitin chains
K29-, K48-, or K63-linked ubiquitin chains and their conjugated proteins were enriched using GST-TRABID-NZF1 (UBPBio, #J4470), FLAG K48TUBE HF (LifeSensors, UM607), or FLAG K63TUBE (LifeSensors, UM604), respectively. Cells were lysed in 1 mL of lysis buffer (10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 1% NP40, 0.5% Triton X-100, and 10% glycerol) supplemented with 20 μM PR619, 10 mM iodoacetamide, and a protease inhibitor cocktail and were sonicated using Handy Sonic (TOMY Seiko). GST-tagged TRABID NZF (40 μg), 1 μg of FLAG-tagged K48TUBE (LifeSensors, UM607), or 1 μg of FLAG-tagged K63TUBE (LifeSensors, UM604) were added to cell lysates to capture K29-, K48-, or K63-linked chains, respectively, and were precipitated using Glutathione Sepharose (GE Healthcare, 17-0756-01) or anti-FLAG affinity resin.
For co-immunoprecipitation, cells were lysed in a lysis buffer [10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 1% NP-40, and 10% glycerol] supplemented with a protease inhibitor cocktail. Soluble cell lysates were incubated with the indicated antibodies (4 °C, 4 h). For immunoprecipitation under denaturing conditions, cells were lysed in a urea-containing lysis buffer [10 mM Tris-HCl (pH, 7.5), 150 mM NaCl, 0.5 mM EDTA, 1% NP-40, 10% glycerol, and 6 M urea] supplemented with 20 μM MG132, 10 μM PR619, 10 mM iodoacetamide, and a protease inhibitor cocktail, and then extensively sonicated using Handy Sonic. Soluble lysates were diluted 10-fold in lysis buffer before immunoprecipitation.
Mass-spectrometric analyses
For absolute quantification of ubiquitin linkages (Ub-AQUA/PRM), analytes were separated using short-run SDS-PAGE and the gel pieces were washed in 50 mM AMBC/30% acetonitrile (ACN) for 2 h, in 50 mM AMBC/50% ACN for 1 h, and dehydrated in 100% ACN for 15 min. Proteins were digested (37 °C, 16 h) with 20 ng/mL sequence-grade trypsin (Promega) in 50 mM AMBC/5% ACN, pH 8.0. After trypsin digestion, a mixture of AQUA peptides (25 fmol/injection) previously reported by Ohtake et al.6 was added to the extracted peptides; the concentrated peptides were diluted with 20 μL of 0.1% trifluoroacetic acid (TFA) containing 0.05% H2O2. For liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis, an Easy-nLC 1200 (Thermo Fisher Scientific) was connected inline to an Orbitrap Fusion LUMOS (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Thermo Fisher Scientific). Peptides were separated on a C18 analytical column (IonOpticks, Aurora Series Emitter Column, AUR2-25075C18A 25 cm × 75 mm × 1.6 mm, FSC C18 with a nanoZero fitting) using a 55 min gradient (solvent A, 0.1% FA; and solvent B, 80% ACN/0.1% FA). Targeted acquisition of MS/MS spectra was performed in the parallel reaction monitoring mode, using the Xcalibur software 2.2. Peptides were fragmented using higher-energy collisional dissociation (HCD) with a normalized collision energy of 28. Data were processed using the PinPoint software (Thermo Fisher Scientific). The MS2 fragment ions used for quantification have been described elsewhere6.
For tandem mass tag (TMT) proteomics, peptides (25 µg at 0.5 µg/µL) from each sample were labeled with 0.25 mg of TMTpro mass tag (TMT) labeling reagent (Thermo Fisher Scientific) for 60 min. Combined samples were then fractionated into eight fractions using the High pH Reversed-Phase Peptide Fractionation Kit (Thermo Fisher Scientific). An EASY-nLC 1200 was connected inline to an Orbitrap Fusion Lumos equipped with a FAIMS-Pro ion mobility interface (Thermo Fisher Scientific). Peptides were separated on an analytical column (IonOpticks, same specifications as described earlier) using a 4 h gradient (0–28% acetonitrile over 240 min). Peptide ionization was performed using a Nanospray Flex Ion Source (Thermo Fisher Scientific), and FAIMS-Pro was set to three phases (−40, −60, and −80 CV). Raw files were analyzed using Sequest HT in Proteome Discoverer 2.4 (Thermo Fisher Scientific). TMT-based protein quantification was performed using the Reporter Ion Quantifier node in Proteome Discoverer 2.4.
RNA isolation, quantitative PCR, and RNA-sequencing
Total RNA was extracted using an RNeasy Mini Kit (Qiagen, 74104); reverse transcription was performed using total RNA (500 ng) and PrimeScript RT Master Mix (Takara, RR036A). Quantitative RT-PCR assays were performed using the QuantStudio 3 Real-Time PCR system (Applied Biosystems) with the TB Green Fast qPCR Mix (Takara, RR430) or the PowerUp SYBR Green Master Mix for qPCR (Thermo Fisher Scientific). Sequences of gene-specific primers were as follows:
Actin: [forward] 5′-ATTGGCAATGAGCGGTTC-3′, [reverse] 5′-CGTGGATGCCACAGGACT-3′, TRIP12: [forward] 5′-TTCAGATTGGTGGACCTTCC-3′, [reverse] 5′-GGCTACAACTGGGTCGATGT-3′, OTUD5: [forward] 5′-GAGCTGCATGCTGAATTGGG-3′, [reverse] 5′-AGTCATTCAGACTGGCGCAC-3′, IL-6: [forward] 5′-TACCCCCAAGGAGAAGATTCC-3′ [reverse] 5′-TTTTCTGCCAGTGCCTCTTT-3′, ADAM8: [forward] 5′-GAGGGTGAGCTACGTCCTTG-3′, [reverse] 5′-CAGCCGTATAGGTCTCTGTGT-3′, IRAK2: [forward] 5′-GAAATCAGGTGTCCCATTCCAG-3′, [reverse] 5′-TGGGGAGGTCGCTTCTCAA-3′, IL-11: [forward] 5′-CTGGGCTAGGGCATGAACTG-3′, [reverse] 5′-CTGGGACTCCAAGTGCAAGA-3′, TBP: [forward] 5′-GAGCCAAGAGTGAAGAACAGTC-3′, [reverse] 5′-GCTCCCCACCATATTCTGAATCT-3′.
RNA sequencing was performed by Macrogen using a Nova-Seq 6000 (Illumina). A library was prepared using the TruSeq stranded mRNA LT Sample Prep Kit (Illumina). Gene ontology analyses were performed using the DAVID functional annotation tool (https://davidbioinformatics.nih.gov/home.jsp).
In vitro ubiquitylation assay
Full-length FLAG-HA-OTUD5 was purified from 293 T cells treated with 10 μM MLN7243 for 2 h before cell lysis, using anti-FLAG antibody (Sigma-Aldrich, M2) and was eluted using 200 ng/μL of FLAG peptide (Sigma-Aldrich). FLAG-TRIP12 and FLAG-UBR5 were purified from 293 T cells using anti-FLAG antibody (Sigma-Aldrich, M2) and eluted using 200 ng/μL of FLAG peptide. Recombinant E1 (Sigma-Aldrich), UbcH5c (UBE2D3) (Boston Biochem), UBCH7 (Boston Biochem), and wild-type ubiquitin (LifeSensors) were obtained from indicated suppliers. In vitro ubiquitylation assays and in vitro ubiquitylation/deubiquitylation competition assays were performed (37 °C, 4 h or the indicated number of hours) with 5 μg ubiquitin, 50 ng E1, and 50 ng UBCH7 or UbcH5c (UBE2D3) as indicated, along with the indicated amount of TRIP12, UBR5, or OTUD5, in a 20 μL reaction [reaction buffer: 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM ATP, and 0.5 mM DTT].
For OTUD5 modification with K29-linked ubiquitin using site-directed ubiquitylation40, full-length SUE1-FLAG-HA-OTUD5 (SUE1 sequence inserted into the N-terminus) was purified from 293 T cells using anti-FLAG antibody and eluted with 200 ng/mL FLAG peptide. In vitro ubiquitylation was performed (37 °C, 0–72 h) with 400 ng E1, 300 ng UBE2E1 (K136R), 50 ng OTUD5, 480 ng K29-linked diubiquitin, or 240 ng or 120 ng ubiquitin in 7 μL reaction buffer [50 mM Tris-HCl (pH 8.0), 15 mM MgCl2, 0.6 mM DTT, 5 mM ATP]. Ubiquitin modification was detected by western blotting using anti-OTUD5 antibody or CBB stain (BIO CRAFT).
DUB (deubiquitylation) assay and UbiCRest
For the Ubi-CRest assay, OTUD5 was immunoprecipitated from HT1080 cells using an anti-OTUD5 antibody under nondenaturing conditions [10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 0.5% Triton X-100, 1% NP-40, 10% glycerol, 10 mM iodoacetamide, and 20 μM PR-619) to preserve ubiquitin chain conformation. Beads were washed thrice with lysis buffer, twice with buffer A [50 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 10% glycerol], and once with 50 mM Tris-HCl (pH 7.5). Purified OTUD5 was incubated with 1 μg of OTUB1* [50 mM Tris-HCl (pH 7.5) and 10 mM DTT] or buffer (2 h, 37 °C) in a 10 μL reaction volume. After the reaction, proteins were denatured with LDS loading buffer (Thermo Fisher Scientific) and heated (70 °C, 10 min).
For in vitro deubiquitylation assays with OTUD5, FLAG-HA-OTUD5 expressed in 293 T cells were purified using an anti-FLAG antibody. Either Ub2, Ub4, or Ub8 (1.0 μg) was incubated with OTUD5 (0.5 μg or the indicated amount) (37 °C, 4 h or for the indicated duration) in the reaction buffer [100 mM Hepes (pH 7.5), 5 mM EGTA, 0.5% NP40, 10 mM EDTA, and 50 mM DTT].
Preparation of ubiquitin monomers and ubiquitin-related enzymes
In this study, the pET26b, pET28a, and pCold-I expression vectors modified to produce N-terminal His6-SUMO fusion proteins are referred to as pET26b-SUMO, pET28a-SUMO, and pCold-SUMO expression vectors, respectively. Ub-encoding gene was previously cloned into the pET26b expression vector49, and Ub gene mutations were generated by PCR. The gene encoding C-terminal His8-tagged E1 (residues 1-1058) was previously cloned into the pGEX-6P-1 expression vector50. The gene encoding AMFR-E2G2 (fusion of the RING domain of human AMFR (residues 227-298) and human E2G2 (residues 1-165)) was previously cloned into the pET28a-SUMO expression vector51. The gene encoding human OTUB1 (residues 2-271) was previously cloned into the pET26b-SUMO expression vector52. The gene encoding N-terminal His6-tagged GST fusion AMSH-LP (residues 264-436) was previously cloned into the pCold I expression vector49. The genes encoding Cezanne (residues 53-446, E287K E288K), UbcH5c (residues 1-147), and E3C (residues 693-1083) were previously cloned into the pCold-SUMO expression vector53. The genes encoding E2E1, Ubc9, and Sae2 were PCR amplified from a human cDNA library (Clontech). The amplified E2E1 and Ubc9 genes were cloned into the pET28a-SUMO expression vector, and the K136R and K14R mutations were introduced into the E2E1 and Ubc9 genes, respectively, by PCR. For the generation of the chimeric E1 (ChE1) gene, the gene encoding E1 (41-957) was fused to the 5’-terminus of the gene encoding Sae2 (450-549), and the A680T, N728Y, F729I, A788S, D853E, and T898S mutations were generated by PCR54. The ChE1 gene was cloned into the pGEX-6P-1 expression vector and the His10-tag was fused to the 3’-terminus of the ChE1 gene by PCR. The codon-optimized gene encoding LotA (residues 1-276) was synthesized (Integrated DNA Technologies) to improve its expression in E. coli, and cloned into the pET28a-SUMO expression vector. Ubs and Ub-related enzymes were overproduced in E. coli strain Rosetta 2 (DE3) cells (Novagen) at 15 °C. Cells overproducing Ubs were disrupted by sonication and incubated (15 min, 70 °C). The supernatant was passed over a Resource Q anion exchange column (Cytiva). Flow-through fractions were further purified by HiLoad 16/600 Superdex 75 size exclusion column (Cytiva). Cells overproducing Ub-related enzymes were disrupted by sonication and purified by nickel-nitrilotriacetic acid (Qiagen) and Resource Q. Purified samples were concentrated with Amicon Ultra-15 (Millipore), and stored at −80 °C until use.
Preparation of K29-Ub4 and K29/K48 branched Ub8
For synthesis of noncyclic K29-linked Ub tetramer (K29-Ub4), E1 (1 μM), UbcH5c (50 μM), E3C (18 μM), OTUB1 (5 μM), Cezanne (E287K, E288K; 2.8 μM), AMSH-LP (5 μM), and Ub (2.4 mM) were mixed in elongation buffer [50 mM Tris-HCl (pH 9.0) containing ATP (10 mM), MgCl2 (10 mM), and DTT (0.6 mM)], and incubated (30 °C) (Fig. 4c and Supplementary Fig. 3a). After 18 h, OTUB1 (30 μM), Cezanne (E287K, E288K; 2.8 μM), and AMSH-LP (5 μM) were added, followed by incubation (2 h, 37 °C). The reaction solution contained cyclic K29-linked Ub chains and a small amount of K48 linkage, in addition to noncyclic K29-linked Ub chains. To purify noncyclic K29-Ub4, the reaction solution was mixed with four volumes of 50 mM ammonium acetate buffer (pH 4.5) and applied to a Resource S cation-exchange column (Cytiva) pre-equilibrated with the same buffer. The synthesized Ub chains were eluted using a linear gradient of 100–400 mM NaCl in 50 mM ammonium acetate buffer (pH 4.5) (Supplementary Fig. 3b). Fractions corresponding to the front, middle, and back parts of the Ub4 peak were collected separately. The middle and back part- fractions contained only Ub4, whereas the front part fraction contained a small amount of Ub3 (Supplementary Fig. 3b). The presence of K48 linkage was confirmed by reaction with OTUB1 (5 μM), a K48-linkage–specific DUB. This assay showed that the front-part fraction was slightly cleaved by OTUB1, whereas the middle- and back-part fractions were barely cleaved (Supplementary Fig. 3b). Thus, the front-part fraction contained K48 linkages, whereas the middle- and back-part fractions contained less K48 linkage.
We subsequently tested whether K29-Ub4 was cyclic or noncyclic using ChE1 and Ubc954. ChE1 and Ubc9 specifically ubiquitinate the second Lys residue of the I–K–Q–E tetrapeptide sequence. Thus, noncyclic K29-Ub4 modifies EGFP with an additional IKQE sequence (EGFP-IKQE), whereas cyclic K29-Ub4 does not (Supplementary Fig. 3c). ChE1 (1 μM), Ubc9 (K14R; 15 μM), EGFP-IKQE (25 μM), and K29-Ub4 (20 μM) were mixed in the elongation buffer and incubated (16 h, 37 °C). The reaction solution was analyzed by SDS-PAGE (Supplementary Fig. 3d). The front and middle part fractions of the Ub4 peak modified the GFP-IKQE, whereas the back part fraction did not. Therefore, the front- and middle-part fractions mainly contained noncyclic K29-Ub4, whereas the back-part fraction contained cyclic K29-Ub4. The middle part fraction of the Ub4 peak (noncyclic K29-Ub4) was concentrated to ~150 μM and stored at −80 °C until use.
For K29/K48 branched Ub octamer (K29/K48-Ub8) synthesis, noncyclic K29-Ub4 (25 μM), E1 (1 μM), AMFR-E2G2 (10 μM), and Ub (K48R; 150 μM) were mixed in the elongation buffer and incubated (20 h, 37 °C) (Fig. 4c (ii) and Supplementary Fig. 3e). Because the elongation enzymes prefer the monomer Ub to the Ub chain, Ub (K48R) modifies four K48 residues of K29-Ub4 in the major reaction to synthesize K29/K48-Ub8. However, linking of K29-Ub4 to a K48 residue of K29-Ub4 also occurs in the minor reaction. In addition, Ub (K48R) and K29-Ub4 further modify other K48 residues of K29-Ub4, forming a K29/K48 branched Ub chain with more than 15-mer Ub (K29/K48-Ub15 ~ ). To purify K29/K48-Ub8, we mixed the reaction solution with four volumes of 50 mM ammonium acetate buffer (pH 4.5) and loaded them onto a Resource S cation exchange column pre-equilibrated with the same buffer. The synthesized K29/K48 branched chains were eluted with a linear gradient of 100–500 mM NaCl in 50 mM ammonium acetate buffer (pH 4.5) (Supplementary Fig. 3f). The fraction corresponding to the K29/K48-Ub8 peak was collected, concentrated to ~50 μM, and stored at −80 °C until use.
Preparation of mixed and branched Ub3
To prepare the K29/K48 branched Ub trimer (K29/K48-Ub3 branch), K29-Ub2 (Distal K29R K48R) was synthesized in the first reaction step (supplementary Fig. 5a (i)): E1 (1.5 μM), UbcH5c (75 μM), E3C (30 μM), OTUB1 (10 μM), Cezanne (E287K E288K; 1.2 μM), AMSH-LP (2 μM), Ub (D77; 400 μM), and Ub (K29R K48R; 500 μM) were mixed in elongation buffer and incubated (30 °C). The K29R and D77 mutations were introduced to prevent undesirable Ub chain elongation, and the K48R mutation was introduced to restrict the K48 linkage position in the second reaction step. After 18 h of incubation, OTUB1 (10 μM), Cezanne (E287K, E288K; 1.2 μM), and AMSH-LP (2 μM) were added and further incubated (2 h, 30 °C). The synthesized K29-Ub2 (Distal K29R K48R) was purified by Resource S cation exchange column. For synthesis of K29/K48-Ub3 branch, E1 (3 μM), AMFR-E2G2 (15 μM), K29-Ub2 (Distal K29R K48R; 50 μM), and Ub (K48R; 150 μM) were mixed in the elongation buffer and incubated (18 h, 37 °C) in the second reaction step (supplementary Fig. 5a (ii) and (iii)). The Ub monomer K48R mutation was introduced to prevent undesirable Ub chain elongation. The synthesized K29/K48-Ub3 was purified by Resource S cation exchange column. The K29/K48 tandemly-mixed Ub3 (K29/K48-Ub3 mix) was prepared in the same way as the K29/K48-Ub3 branch, except that Ub (K48R D77) and Ub (K29R) were used instead of the Ub (D77) and Ub (K29R K48R) in the first reaction step (supplementary Fig. 5b).
To prepare the K48/K29 tandemly-mixed Ub3 (K48/K29-Ub3 mix), K48-Ub2 (Proximal K11R K29R, Distal K11R K48R) was synthesized in the first reaction step (supplementary Fig. 5c (i)): E1 (1 μM), AMFR-E2G2 (15 μM), Ub (K11R K48R; 1200 μM), and Ub (K11R K29R D77; 1000 μM) were mixed in the elongation buffer and incubated (18 h, 37 °C). K48R and D77 mutations were introduced to prevent undesirable Ub chain elongation, and the K29R mutation was introduced to restrict the K29 linkage position in the second reaction step (Supplementary Fig. 5c (ii) and (iii)). The K29 chain elongation enzyme E3C, used in the second reaction step, also elongates K6, K11, K48, and K63 chains31. Since LotA and AMSH-LP are highly specific DUBs for K6 and K63 chains, respectively, they can remove undesirable linkages synthesized by E3C. However, K11-linkage-specific DUB Cezanne cannot be used to cleave the K11 linkage in the second reaction step, because it exhibits weak activity towards K48 chains20. Therefore, the K11R mutation was required to prevent the synthesis of the K11 linkage (supplementary Fig. 5c (ii)). The synthesized K29-Ub2 (Proximal K11R K29R, Distal K11R K48R) was purified by Resource S cation exchange column. For synthesis of K48/K29-Ub3 mix, E1 (2 μM), UbcH5c (50 μM), E3C (20 μM), AMSH-LP (4 μM), LotA (4 μM), K48-Ub2 (Proximal K11R K29R, Distal K11R K48R; 200 μM), and Ub (K11R K29R K48R; 500 μM) were mixed in the elongation buffer and incubated (18 h, 30 °C) (Supplementary Fig. 5c (ii) and (iii)). The Ub monomer K29R mutation was introduced to prevent undesirable Ub chain elongation. K11R and K48R mutations of the Ub monomer were introduced to prevent elongation of the K11 and K48 linkages because OTUB1 and Cezanne cannot be used in the second step. The synthesized K48/K29-Ub3 mix was purified by Resource S cation exchange column.
Statistics and reproducibility
Statistical analysis was performed using GraphPad Prism 7. Two-tailed, one-way ANOVA test was used for biological replicates; data represent means from biological replicates. Sample size (n) for each experiment is indicated in the respective legends. For TMT-based proteomics screening, the P-value is calculated using two-sided, one-way ANOVA without adjustment (two biological replicates). For RNA-sequencing analysis, the raw P values, which were two-sided and not adjusted, were used (three biological replicates). For gene ontology analysis using DAVID, the raw P values, which were one-sided and not adjusted, were used. Gel blot images of Figs. 1e–h; 2b, f–j; 3d, e, h; 4a, b, d; 5a, b, d–f, 6b–g are representative of at least two independent experiments with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw datasets for RNA-sequencing analyses have been deposited to the GEO with the accession numbers GSE273273 and GSE286567. The proteomics datasets have been deposited to PRIDE with the accession code PXD054556. Source data are provided with this paper.
References
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem 81, 203–229 (2012).
Haakonsen, D. L. & Rape, M. Branching out: improved signaling by heterotypic ubiquitin chains. Trends Cell Biol. 29, 704–716 (2019).
Yau, R. G. et al. Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell 171, 918–933 e920 (2017).
Kolla, S., Ye, M., Mark, K. G. & Rape, M. Assembly and function of branched ubiquitin chains. Trends Biochem. Sci. 47, 759–771 (2022).
Oh, E. et al. Gene expression and cell identity controlled by anaphase-promoting complex. Nature 579, 136–140 (2020).
Ohtake, F., Saeki, Y., Ishido, S., Kanno, J. & Tanaka, K. The K48-K63 branched ubiquitin chain regulates NF-kB signaling. Mol. Cell 64, 251–266 (2016).
Ohtake, F., Tsuchiya, H., Saeki, Y. & Tanaka, K. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl Acad. Sci. USA 115, E1401–E1408 (2018).
Akizuki, Y. et al. cIAP1-based degraders induce degradation via branched ubiquitin architectures. Nat. Chem. Biol. 19, 311–322 (2023).
Leto, D. E. et al. Genome-wide CRISPR analysis identifies substrate-specific conjugation modules in ER-associated degradation. Mol. Cell 73, 377–389 e311 (2019).
Kaiho-Soma, A. et al. TRIP12 promotes small-molecule-induced degradation through K29/K48-branched ubiquitin chains. Mol. Cell 81, 1411–1424 e1417 (2021).
Liu, C., Liu, W., Ye, Y. & Li, W. Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nat. Commun. 8, 14274 (2017).
Mao, J. et al. Structural Visualization of HECT-E3 Ufd4 accepting and transferring Ubiquitin to Form K29/K48-branched Polyubiquitination on N-degron. bioRxiv, 2023.2005.2023.542033. https://doi.org/10.1101/2023.05.23.542033 (2023).
Haakonsen, D. L. et al. Stress response silencing by an E3 ligase mutated in neurodegeneration. Nature 626, 874–880 (2024).
Meyer, H. J. & Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921 (2014).
Deol, K. K. et al. Proteasome-Bound UCH37/UCHL5 Debranches Ubiquitin Chains to Promote Degradation. Mol. Cell 80, 796–809 e799 (2020).
Du, J. et al. A cryptic K48 ubiquitin chain binding site on UCH37 is required for its role in proteasomal degradation. Elife 11. https://doi.org/10.7554/eLife.76100 (2022).
Lange, S. M. et al. VCP/p97-associated proteins are binders and debranching enzymes of K48-K63-branched ubiquitin chains. Nat. Struct. Mol. Biol. https://doi.org/10.1038/s41594-024-01354-y (2024).
Ordureau, A., Munch, C. & Harper, J. W. Quantifying ubiquitin signaling. Mol. Cell 58, 660–676 (2015).
Ohtake, F. et al. Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO Rep. 16, 192–201 (2015).
Kristariyanto, Y. A. et al. K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin. Mol. Cell 58, 83–94 (2015).
Dewson, G., Eichhorn, P. J. A. & Komander, D. Deubiquitinases in cancer. Nat. Rev. Cancer 23, 842–862 (2023).
Lange, S. M., Armstrong, L. A. & Kulathu, Y. Deubiquitinases: From mechanisms to their inhibition by small molecules. Mol. Cell 82, 15–29 (2022).
Mevissen, T. E. et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184 (2013).
Brunet, M., Vargas, C., Larrieu, D., Torrisani, J. & Dufresne, M. E3 Ubiquitin Ligase TRIP12: Regulation, Structure, and Physiopathological Functions. Int. J. Mol. Sci. 21. https://doi.org/10.3390/ijms21228515 (2020).
Fu, L., Lu, K., Jiao, Q., Chen, X. & Jia, F. The Regulation and Double-Edged Roles of the Deubiquitinase OTUD5. Cells 12. https://doi.org/10.3390/cells12081161 (2023).
de Vivo, A. et al. The OTUD5-UBR5 complex regulates FACT-mediated transcription at damaged chromatin. Nucleic Acids Res. 47, 729–746 (2019).
Rutz, S. et al. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature 518, 417–421 (2015).
Beck, D. B. et al. Linkage-specific deubiquitylation by OTUD5 defines an embryonic pathway intolerant to genomic variation. Sci. Adv. 7. https://doi.org/10.1126/sciadv.abe2116 (2021).
Hjerpe, R. et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 (2009).
Ohtake, F., Tsuchiya, H., Tanaka, K. & Saeki, Y. Methods to measure ubiquitin chain length and linkage. Methods Enzymol. 618, 105–133 (2019).
Michel, M. A. et al. Assembly and specific recognition of k29- and k33-linked polyubiquitin. Mol. Cell 58, 95–109 (2015).
Kayagaki, N. et al. DUBA: a deubiquitinase that regulates type I interferon production. Science 318, 1628–1632 (2007).
Mori, Y. et al. Intrinsic signaling pathways modulate targeted protein degradation. Nat. Commun. 15, 5379 (2024).
Hospenthal, M. K., Mevissen, T. E. & Komander, D. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nat. Protoc. 10, 349–361 (2015).
Rodriguez Carvajal, A. et al. The linear ubiquitin chain assembly complex (LUBAC) generates heterotypic ubiquitin chains. Elife 10. https://doi.org/10.7554/eLife.60660 (2021).
Shearer, R. F., Iconomou, M., Watts, C. K. & Saunders, D. N. Functional roles of the E3 Ubiquitin Ligase UBR5 in cancer. Mol. Cancer Res 13, 1523–1532 (2015).
Mark, K. G. et al. Orphan quality control shapes network dynamics and gene expression. Cell 186, 3460–3475 e3423 (2023).
Chiu, Y. H., Zhao, M. & Chen, Z. J. Ubiquitin in NF-kappaB signaling. Chem. Rev. 109, 1549–1560 (2009).
Morrow, M. E. et al. Active site alanine mutations convert deubiquitinases into high-affinity ubiquitin-binding proteins. EMBO Rep. 19. https://doi.org/10.15252/embr.201745680 (2018).
Wu, X. et al. Structure-guided engineering enables E3 ligase-free and versatile protein ubiquitination via UBE2E1. Nat. Commun. 15, 1266 (2024).
Hehl, L. A. et al. Structural snapshots along K48-linked ubiquitin chain formation by the HECT E3 UBR5. Nat. Chem. Biol. 20, 190–200 (2024).
Scott, D. C. et al. Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell 166, 1198–1214 e1124 (2016).
Brown, N. G. et al. Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell 165, 1440–1453 (2016).
Mashtalir, N. et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol. Cell 54, 392–406 (2014).
Wijnhoven, P. et al. USP4 auto-Deubiquitylation promotes homologous recombination. Mol. Cell 60, 362–373 (2015).
Iwai, K., Fujita, H. & Sasaki, Y. Linear ubiquitin chains: NF-kappaB signalling, cell death and beyond. Nat. Rev. Mol. Cell Biol. 15, 503–508 (2014).
Takahashi, H. et al. A Human DUB protein array for clarification of linkage specificity of polyubiquitin chain and application to evaluation of its inhibitors. Biomedicines 8. https://doi.org/10.3390/biomedicines8060152 (2020).
Sowa, M. E., Bennett, E. J., Gygi, S. P. & Harper, J. W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009).
Sato, Y. et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362 (2008).
Sato, Y. et al. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat. Struct. Mol. Biol. 22, 222–229 (2015).
Men, Y. et al. ESCRT-I and PTPN23 mediate microautophagy of ubiquitylated tau aggregates. J Cell Biol, accepted (2025).
Sato, Y. et al. Molecular basis of Lys-63-linked polyubiquitination inhibition by the interaction between human deubiquitinating enzyme OTUB1 and ubiquitin-conjugating enzyme UBC13. J. Biol. Chem. 287, 25860–25868 (2012).
Sato, Y. et al. Structural basis for specific cleavage of Lys6-linked polyubiquitin chains by USP30. Nat. Struct. Mol. Biol. 24, 911–919 (2017).
Akimoto, G., Fernandes, A. P. & Bode, J. W. Site-specific protein ubiquitylation using an engineered, chimeric E1 activating enzyme and E2 SUMO conjugating enzyme Ubc9. ACS Cent. Sci. 8, 275–281 (2022).
Acknowledgements
We thank Keiji Tanaka for the discussions. This work was supported in part by JSPS KAKENHI (grant numbers JP21H02433 and JP23H04922 to F.O.; JP21H02418 and JP24H01899 to Yu.S.; and JP23H04921 to Ya.S), AMED-CREST (grant number 24gm1410007h0004 to F.O.), the Takeda Science Foundation (to F.O.), the Naito Foundation (to F.O.), AMED-PRIME (grant number JP21gm6410012 to A.E.), the MEXT Leading Initiative for Excellent Young Researchers (to Yu.S.), Tottori University Research Support Project for the Next Generation (to Yu.S.), and the Grant for International Joint Research Project of the Institute of Medical Science, the University of Tokyo (to Ya.S. and F.O.).
Author information
Authors and Affiliations
Contributions
F.O. designed the project and analyzed the data. M.M. performed most of the cell-based and in vitro DUB experiments. M.T. performed in vitro ubiquitylation assays. H.T. and Yu. S. synthesized and purified ubiquitin chains. R.C., S.T., and Y.A. assisted with biochemical experiments. F.O., Y.A., and A.E. performed mass spectrometric analyses. Ya.S. and T.T. provided reagents and advice. F.O. wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Martin Steger and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Morita, M., Takao, M., Tokuhisa, H. et al. Combinatorial ubiquitin code degrades deubiquitylation-protected substrates. Nat Commun 16, 2496 (2025). https://doi.org/10.1038/s41467-025-57873-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57873-9