Abstract
Fusarium crown rot (FCR) is a serious underlying disease to threaten wheat yield and quality recently. Here, we identify a catalase antioxidant enzyme (TaCAT2) through genome wide association study (GWAS) and whole-exome sequencing (WES) in two nested bi-parental populations. We verify the function of TaCAT2 regulating wheat FCR resistance by genetic transformation. Moreover, we screen a sucrose non-fermenting-1-related protein kinase alpha subunit (TaSnRK1α) interacting with TaCAT2, and subsequently find that TaSnRK1α phosphorylates TaCAT2. We next identify an FCR-resistance haplotype TaCAT2Ser214, and confirm that Ser214 of TaCAT2 is a key phosphorylation site for TaSnRK1α. We also find that TaSnRK1α results in higher protein accumulation in TaCAT2Ser214 than in TaCAT2Thr214, which possibly contribute to scavenging ROS (reactive oxygen species) in TaCAT2Ser214 wheat plants. Furthermore, the function of TaSnRK1α regulating FCR resistance is verified by genetic transformation. Taken together, we propose a TaSnRK1α-TaCAT2 model to mediate FCR resistance by scavenging the ROS in wheat plants.
Similar content being viewed by others
Introduction
Fusarium crown rot (FCR), mainly caused by the pathogen F. pseudograminearum (F.pg), is one of the most serious underlying diseases of wheat and barley1. The F.pg is distributed worldwide and can chronically survive in the soil or within stubble to cause disease, thereby resulting in serious production reduction2,3. More recently, how to effectively control wheat FCR has been listed as one of the top ten industrial issues at the 24th Annual Meeting of China Association for Science and Technology in 2022 (https://www.cast.org.cn). In addition, various toxins (deoxynivalenol, DON; nivalenol, NIV) and secondary metabolites accumulate in the plant stems or grains during the F.pg infection, which are seriously harmful to humans and livestock after consumption4,5. Screening resistant germplasm to prevent damage from FCR disease has been widely carried out, but no immune or highly resistant accession has been reported so far6,7,8. Most modern varieties widely used in wheat production are susceptible or highly susceptible to FCR. Therefore, the development of resistant wheat varieties to control the spread of FCR disease is necessary.
The application of resistance genes is an effective strategy for the development of resistant varieties by marker-assisted selection3,9. Several genetic loci have been reported to confer resistance to FCR. Multiple researchers reported a large-effect quantitative trait loci (QTL) for wheat FCR on chromosome 3B in different environments7,8,10. Two major QTLs on 5D and 2D for FCR were identified in the population Wylie/Sumai311. An important QTL on 4B also significantly affected FCR resistance8,11. Moreover, we previously reported a TaDIR-B1 gene to control FCR resistance, possibly through regulating the lignin content12. A Fusarium head blight resistance gene Fhb7 also conferred FCR resistance in wheat13. In general, the FCR-resistant loci are widely distributed on 21 pairs of chromosomes of wheat, but a restricted number of genes have been reported so far. Therefore, it is important to discover FCR resistance genes in wheat3,9,10.
Reactive oxygen species (ROS), including hydrogen peroxide (H2O2), superoxide (O2−), hydroxyl radical (HO−), and singlet oxygen (1O2), are essential signaling molecules by interaction with other signaling networks during pathogens invasion into plants14,15. Many reports suggested that an oxidative burst usually confers resistance in many plant-pathogen interactions during a hypersensitive response in the case of a biotrophic pathogen attack16,17,18. However, the death of plant cells induced by excess ROS provides an advantage in host colonization to necrotrophic pathogens, thereby leading to host susceptibility19,20. Several studies reported that the ROS burst possibly provided an advantage for necrotrophic pathogens to acquire nutrients from the dead cells of the host plant, consequently enhancing the pathogens’ pathogenicity21,22. To date, F.pg is considered either a hemi-biotroph pathogen or necrotrophic pathogen that is associated with FCR23,24,25. The fungal hyphae infect the seedling leaf sheaths often through natural openings like stomata and then occupy the leaf sheath tissue and cross to the neighbor cells through infection structures. During infection, toxins have also been produced and act as virulence factors to bring harm to hosts26. No matter the short biotrophy stage or primary necrotrophy stage, the cellular redox state during pathogen invasion affects the pathogen-host interaction. Catalase (CAT) is the critical enzyme that catalyzes the degradation of H2O2 into water and molecular oxygen to regulate the cellular redox state27. CAT is crucial in maintaining ROS homeostasis to modulate the necrotrophic fungal pathogen Rhizoctonia cerealis28. CAT could regulate ROS to mediate plant resistance, and the overexpression of CAT gene in plants resulted in increased resistance to B. cinerea strain B05.1029.
With the increased availability of chromosome or near chromosome-scale wheat genome sequence, GWAS (genome-wide association study) is becoming an effective technique for identifying essential genes controlling complex wheat traits. In this study, we identified a TaCAT2 gene controlling wheat FCR resistance by integrating GWAS, WES (whole-exome sequencing), and transcriptome analysis. We subsequently verified the function of TaCAT2 in eliminating excess ROS and contributing to enhanced FCR resistance. We then screened a TaSnRK1α (sucrose non-fermenting-1-related protein kinase alpha subunit) that interacted with TaCAT2. We further identified the key TaCAT2 phosphorylation site Ser214 by TaSnRK1α. We revealed the differential protein level between TaCAT2Ser214 and TaCAT2Thr214 that was possibly caused by the phosphorylation of TaSnRK1α. Therefore, our results identified a very important FCR resistance gene, TaCAT2, and provided new insight into further understanding the regulatory mechanism of wheat FCR resistance.
Results
Identification of the TaCAT2 gene potentially affecting wheat FCR resistance
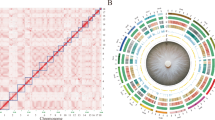
In this study, a genome-wide association study (GWAS) using the Wheat 660 K SNP array was performed for FCR disease index (DI) in 243 Chinese wheat accessions over two years. The distribution of significant SNPs indicated that an important genetic locus on chromosome 6A was clustered into a 14-Mb interval from 10 Mb to 24 Mb in the genome database of Chinese Spring (http://plants.ensembl.org/Triticum_aestivum/Tools/Blast?db=core) (Fig. 1a and Supplementary Data 1). Based on haplotype analysis on 6A (Supplementary Fig. 1), we crossed the FCR-resistant cultivar Jinmai1 with two FCR-susceptible cultivars, Yunong 805 and Zhengmai 082, to create two F4 nested recombinant inbred lines (RIL) populations Jinmai1/Yunong805 (RIL-JY) and Jinmai1/Zhengmai082 (RIL-JZ) (Supplementary Fig. 2). We scored the DI of FCR in the two RIL populations after inoculation by Fp-IV, and subsequently selected extreme phenotype offspring lines to develop FCR-resistant pools (FCR-RPo) and FCR-susceptible pools (FCR-SPo) (Supplementary Fig. 2). Then we genotyped the extreme phenotype pools and parents with three replicates for each population by whole-exome sequencing (WES). The differential SNPs were mainly clustered into an interval of 22–24 Mb on chromosome 6A, covering 53 annotated genes in the genome of Chinese Spring after excluding background interference of parents (Fig. 1b, c Supplementary Figs. 3, 4, Supplementary Data 2). Sequence analysis indicated that 15 of the 53 annotated genes showed polymorphic variants at the amino acid level or in the promotor region between the FCR-RPo vs FCR-SPo and Jinmai1 vs Yunong805/Zhengmai082 (Fig. 1d and Supplementary Data 3).
a Manhattan plot revealed that the most significant locus identified by GWAS analysis was distributed on chromosome 6A. b, c The WES assay identified the significant loci in the interval of 22–24 Mb on 6A for FCR resistance, containing 53 annotated genes. d Sequence analysis indicated that 15 of the 53 annotated genes showed polymorphism between the FCR-RPo vs FCR-SPo and Jinmai1 vs Yunong805/Zhengmai082. Red and blue boxes represent genes with and without polymorphism, respectively. e Analysis of differential expression genes (DEG) in the 22–24 Mb on 6A by BSR-Seq. iFCR-RPo, FCR resistance pool with inoculation; iFCR-SPo, FCR susceptible pool with inoculation; nFCR-RPo, FCR resistance pool without inoculation; nFCR-SPo, FCR susceptible pool without inoculation. f The TaCAT2 gene was overlapped by the integration of GWAS, WES, BSR-Seq, and VIGS. g The schematic comparison of gene structures between TaCAT2-R and TaCAT2-S. h Development of co-dominant marker to distinguish two TaCAT2 haplotypes. Lanes 1 to 10 indicated amplification with TaCAT2 marker in 10 lines from nested bi-parental populations. The experiment was independently repeated three times. i Comparison of FCR disease index between TaCAT2-R and TaCAT2-S in two nested bi-parental populations. j, k Haplotype analysis of TaCAT2 and comparison of FCR disease index among TaCAT2-R, TaCAT2-M, and TaCAT2-S in the test wheat accessions. TaCAT2-R, resistance haplotype; TaCAT2-M, medium-resistance haplotypes; TaCAT2-S, susceptible haplotype. The data are presented as the means ± SE from three biological replicates. *p < 0.05, **p < 0.01 (two-tailed Students’s t-test). Source data are provided as a Source Data file.
To further explore the candidate genes, we performed a BSR-Seq (bulked segregation RNA sequencing) assay for the extreme phenotype pools before and after inoculation by Fp-IV. The results indicated that most of the identified genes participated in defense to stress/pathogen attack or associated with photosynthesis process (Supplementary Fig. 5). The BSR-Seq results showed that 21 common expression genes (> 0.1 TPM) were identified on 6 A in the region of 22 ~ 24 Mb while 3 (TaCAT2, TraesCS6A02G041700; TaLBP, TraesCS6A02G042600; TaHMA, TraesCS6A02G043000) of them showed significantly differential expression between the FCR-RPo vs FCR-SPo (Fig. 1e and Supplementary Data 4). Furthermore, the virus-induced gene silencing (VIGS) experiments mediated by the barley stripe mosaic virus (BSMV) in the FCR-resistance cultivar Jinmai1 were performed for TaCAT2 and TaLBP, while not for TaHMA that could not be obviously detected at the transcription level in the stem of wheat plants. Results indicated that the FCR resistance of the silenced-TaCAT2 plants were obviously decreased, while the silenced-TaLBP plants showed no significant difference when compared to the control plants (Fig. 2a–c and Supplementary Fig. 6). Therefore, TaCAT2 (TraesCS6A02G041700) was regarded as a candidate gene for further analysis (Fig. 1f).
a–e TaCAT2-silenced plants showed significantly decreased FCR resistance by VIGS (virus-induced gene silencing). The FCR phenotype (a); The relative expression levels (b); The average disease index (c); The activity of CAT enzyme (d); The H2O2 content (e) of wild type (WT), BSMVγ and BSMVTaCAT2 plants. BSMVTaCAT2 represented the VIGS experiments conducted in the FCR-resistance cultivar Jinmai1. f–k Tetraploid Kronos EMS mutants (cat2) with premature stop codon of TaCAT2 showed decreased FCR resistance. Mutation site of TaCAT2 in red box (f); Comparison of phenotype and disease index for FCR between WT and cat2 in greenhouse and field (g, h, i); Detection of H2O2 content by a Hydrogen Peroxide Assay Kit (j) and DAB (3,3’-diamino benzidine hydrochloride) staining (k) in WT and cat2 plants, respectively. WT, wild Kronos; cat2, the tetraploid Kronos EMS mutants with premature stop codon of TaCAT2; Seedling, the FCR resistance investigated in the greenhouse; Adult, the FCR resistance investigated in the field. Red boxes included the white heads that were caused by FCR root infection but not by secondary floral infections. l–q TaCAT2-OE lines significantly enhanced wheat FCR resistance. Relative expression levels of TaCAT2 gene in wild-type KN199 and overexpression lines (O.E.#) (l); Comparison of phenotype and disease index for FCR between WT and overexpression lines in the greenhouse and field (m, n, o); Detection of H2O2 content by a Hydrogen Peroxide Assay Kit (p) and DAB staining (q) in WT and overexpression lines, respectively. The wheat leaf bases were sampled for staining DAB after infection by Fp-IV for four weeks in the greenhouse. Three biological replicates were used for each group, and each replicate contained five plants. Transcript levels were examined by qRT‒PCR with TaActin as an endogenous control. The data are presented as the means ± SE from three biological replicates. *p < 0.05, **p < 0.01 (two-tailed Students’s t-test). Source data are provided as a Source Data file.
Phylogenetic analysis using TaCAT and its homoeologous proteins in wheat and the proximal species (https://ensembl.gramene.org/index.html) showed that these CAT proteins were classified into three clades (Supplementary Figs. 7 and 8a). Analysis of exon-intron structure showed that the intron digit of CATs ranged from 1 to 7 (Supplementary Fig. 8b), indicating a diversification evolutionary in its gene family. Further analysis revealed a total of 10 conserved motifs in CATs, and most of them displayed relatively consistent motif compositions (Supplementary Fig. 8c). Analysis of the conserved function domains indicated that all the CAT proteins possessed a catalase core domain (PF00199) and most of them contained a catalase-related immune-responsive domain (PF06628) (Supplementary Fig. 8d), implying a relative conservation of CAT gene family. In addition, ten CAT genes in wheat were identified to classify into three clades (Supplementary Fig. 7, Supplementary Data 5). The measurement results of enzymatic activity indicated a relatively higher value in TraesCS6A02G041700 (TaCAT2) and TraesCS7A02G549800 (TaCAT3) than in TraesCS5A02G498000 (TaCAT1) (Supplementary Fig. 9).
Association of TaCAT2 variants with FCR resistance in Chinese wheat cultivars
To explore the polymorphism of TaCAT2 identified in this study (Supplementary Fig. 10), we cloned TaCAT2 from three parents (Jinmai1, Yunong 805, and Zhengmai 082), respectively. TaCAT2 possessed a 215-bp InDel (insertion/deletion) and 36 SNPs at the DNA level but only had two deduced amino acids differences (Ser214Thr and Lys418Arg) at the protein level between resistant and susceptible parents (Fig. 1g, j Supplementary Fig. 11). We then developed a co-dominant marker to distinguish TaCAT2 alleles (Fig. 1h), and wheat lines with TaCAT2-S (including the 215-bp insertion) showed significantly higher DI than those with TaCAT2-R (without the 215-bp insertion) in both RIL-JY and RIL-JZ populations (Fig. 1i). Furthermore, to illustrate the association of TaCAT2 variants with FCR resistance, we sequenced the TaCAT2 gene in 243 Chinese wheat cultivars. Results indicated that 3 variants of TaCAT2 formed 8 haplotypes in the test cultivars (Fig. 1j). Cultivars with TaCAT2-R exhibited a significantly more resistant FCR than those with TaCAT2-S (Fig. 1k), and the enzymatic activity of TaCAT2-R was significantly higher than TaCAT2-S (Supplementary Fig. 12). After adding TaCAT2 variants into the Wheat 660 K SNP assay, we re-ran GWAS and found that the TaCAT2 marker explained 12.2%–12.7% of the phenotype variance (Supplementary Fig. 13). These results suggested that TaCAT2 was closely associated with wheat FCR resistance.
Silencing of the TaCAT2 significantly decreased wheat FCR resistance
To verify the function of TaCAT2, we performed VIGS experiments in the FCR-resistance cultivar Jinmai1 (TaCAT2-R). In the silenced-TaCAT2 plants, the expression level of TaCAT2 was significantly decreased by qRT-PCR (Fig. 2b) and the TaCAT enzyme activity was remarkably reduced (Fig. 2d) 14 days after BSMV inoculation. The silenced-TaCAT2 plants possessed significantly higher DI than controls after inoculation by Fp-IV (Fig. 2a, c). Additionally, the silenced-TaCAT2 plants in FCR-susceptible cultivar Yunong268 (TaCAT2-S) also possessed relatively higher DI than controls after inoculation by Fp-IV (Supplementary Fig. 14). H2O2 content was significantly increased in the silenced-TaCAT2 plants (Fig. 2e). These results possibly implied that TaCAT2 positively regulated wheat FCR resistance.
To further illustrate the influence of silencing TaCAT2 on FCR resistance, we screened a mutant K3868 (a premature stop codon at the 133 AA position of TaCAT2; Fig. 2f) in the EMS-mutagenized tetraploid wheat Kronos libraries. To exclude other mutation sites, we backcrossed K3868 with wild type twice to obtain BC2 (cat2) mutant. After inoculation by Fp-IV, homozygous cat2 mutants had a significantly higher DI than the wild type in both greenhouse and field (Fig. 2g and Supplementary Fig. 15). At the grain-filling stage, white-heads caused by severe FCR infection were observed in the mutant plants, implying that cat2 mutant was more susceptible to FCR compared with the wild type (Fig. 2h). H2O2 content and fungal biomass were significantly increased in the cat2 mutant (Fig. 2j and Supplementary Fig. 16a). After treatment by DAB (3,3’-diamino benzidine hydrochloride), dark-brown spots caused by H2O2 accumulation were sharply increased in the cat2 mutant (Fig. 2k). These results indicated that silencing of TaCAT2 resulted in decreased FCR resistance possibly through mediating the accumulation of ROS in wheat plants.
Overexpression of TaCAT2 significantly enhanced wheat FCR resistance
To further confirm the function of TaCAT2 in FCR resistance, we constructed a wheat overexpression (OE) vector pUbi::LGY-OE3 containing the full-length cDNA of the TaCAT2-R from Jinmai1 and transferred it into an FCR-susceptible cultivar KN199 with TaCAT2-S. After positive detection, we quantified expression levels of TaCAT2 in positively transgenic lines by qRT-PCR, selected two highly expressed plants to self-cross into T2 generation, and further inoculated them by Fp-IV (Fig. 2l). Scoring results showed that TaCAT2-overexpression sharply enhanced FCR resistance in the greenhouse at the seedling stage (Fig. 2m, o). In field conditions, TaCAT2-OE lines boosted FCR resistance in the adult plants compared with the wild type (Fig. 2n, o). In addition, the plant height, grain size, and thousand-kernel weight of the TaCAT2-OE lines showed no significant difference from WT (Supplementary Fig. 17). H2O2 content and fungal biomass were significantly decreased in the overexpressed plants (Fig. 2p, q and Supplementary Fig. 16c). The above data suggested that TaCAT2 positively regulated wheat FCR, possibly through scavenging ROS without penalty of agronomic traits.
TaCAT2 interacted with TaSnRK1α in vivo and in vitro
To explore the mechanism of TaCAT2 regulating FCR resistance, we used the TaCAT2-R protein as a bait to screen the cDNA library that was constructed with infected wheat plants by Fp-IV using yeast two-hybrid (Y2H) (Supplementary Data 6). A candidate protein TraesCS1A02G350500 annotated as TaSnRK1α (sucrose non-fermenting-1-related protein kinase alpha subunit) from the Y2H screening results was selected since TaSnRK1α as a protein kinase was previously reported to positively modulate Fusarium head blight resistance in wheat30. The interaction of TaCAT2 and TaSnRK1α was confirmed in the yeast cell by Y2H (Fig. 3a) and in the tobacco leaves by the split luciferase complementation experiments (Fig. 3b). Furthermore, an in vitro pull-down assay revealed the interaction between TaCAT2 and TaSnRK1α using the purified TaCAT2-GST and TaSnRK1α-His recombinant proteins from E. coli (Fig. 3c). These results indicated that TaCAT2 interacted with TaSnRK1α in vivo and in vitro. In addition, fragment interaction indicated that TaSnRK1α interacted with the catalase domain (containing the Ser214Thr site) but not the immune-responsive domain (containing the Lys418Arg site) in TaCAT2 (Fig. 3d).
a TaCAT2 interacted with TaSnRK1α in the yeast cell by Y2H. Different concentrations of the yeast transformants were grown on medium lacking SD/Trp-Leu and SD/Trp-Leu-His-Ade plates, respectively; AD-T + BD-53 and AD-T + BD-lam were used as positive control and negative control, respectively. b TaCAT2 interacted with TaSnRK1α in Nicotiana. benthamiana leaves by firefly luciferase complementation assay. SnRK1α-cLUC/PAP6-nLUC was positive control, and empty vectors were negative controls. c TaCAT2 interacted with TaSnRK1α by in vitro GST Pull-down. Marked protein mixtures (Input) or proteins co-purified with TaCAT2-His from the mixtures (pull-down assay) were detected with anti-His and anti-GST antibodies by western blot, respectively. The experiment was independently repeated three times. d The TaSnRK1α interacted with the catalase domain at the N-terminal covering the Ser214Thr site but did not interact with the immune-responsive domain at the C-terminal by Y2H. Different concentrations of the yeast transformants were grown on a medium lacking SD/ Trp-Leu and SD/Trp-Leu-His-Ade plates; AD-T + BD-53 was the positive control, and AD-T + BD-lam was the negative control. e, f TaCAT2Ser214 was phosphorylated by TaSnRK1α in vitro and in vivo, respectively. Immunoblots detected that the phosphorylation level of TaCAT2Ser214 with the addition of both TaSnRK1α and GRIK1 showed a stronger signal than that with the individual addition of TaSnRK1α or GRIK1 in vitro (e). Immunoblots detected that the phosphorylation level of TaCAT2Ser214 was increased after adding TaSnRK1α using an anti-Flag antibody and Pan Phospho-S/T antibody in vivo (f). g, h Differential phosphorylation levels were detected among TaCAT2Ser214, TaCAT2Thr214 and TaCAT2Ala214 after phosphorylation by TaSnRK1α in vitro and in vivo, respectively. Immunoblots detected higher phosphorylation levels of TaCAT2Ser214 than TaCAT2Thr214 and TaCAT2Ala214 by TaSnRK1α and GRIK1 with Pan Phospho-S/T antibody in vitro; Different variants of TaCAT2 were detected with anti-His antibody; TaSnRK1α and GRIK1 were detected with anti-GST antibody (g). Immunoblots detected higher phosphorylation level of TaCAT2Ser214 than TaCAT2Thr214 and TaCAT2Ala214 by TaSnRK1α with Pan Phospho-S/T antibody in vivo; Different variants of TaCAT2 were detected with anti-Flag antibody; TaSnRK1α was detected with anti-GFP antibody (h). All experiments were independently repeated three times.
To determine the localization place of TaCAT2 and TaSnRK1α, subcellular localization in tobacco leaves was performed by transiently transforming with Ubi::GFP-TaCAT2 or Ubi::GFP-TaSnRK1α, respectively. Results showed that TaCAT2 was mainly localized in the cytoplasm, membrane, and peroxisome, while TaSnRK1α was mainly localized in the cytoplasm and membrane (Supplementary Fig. 18). Furthermore, a bimolecular fluorescent complementary (BiFC) in wheat protoplast was performed, and results showed that TaCAT2 interacted with TaSnRK1α in the cytoplasm (Supplementary Fig. 19).
TaSnRK1α phosphorylated TaCAT2 in vitro and in vivo
As a typical protein kinase, TaSnRK1α usually performs the function through the phosphorylation of Ser/Thr residues of substrate proteins31. To explore whether TaSnRK1α could directly phosphorylate TaCAT2, we performed in vitro and in vivo phosphorylation experiments. Previous studies have reported that total extraction from SnRK1α in vitro could not directly phosphorylate substrate, but GRIK1 (geminivirus Rep interacting kinases) was able to activate SnRK1α to phosphorylate substrates31,32. Therefore, we added GRIK1 into TaSnRK1α to detect the phosphorylation level of TaCAT2 in vitro. Immunoprecipitation results indicated that TaCAT2 with individual addition of GRIK1 or TaSnRK1α showed very faint signals, but TaCAT2 with the addition of both TaSnRK1α and GRIK1 showed a strong signal (Fig. 3e). In addition, in vivo assay also showed a stronger band in TaCAT2 + TaSnRK1α than in individual TaCAT2 (Fig. 3f). These results indicated that TaSnRK1α could phosphorylate TaCAT2 in vitro and in vivo.
As the potential phosphorylation site of 214Ser/Thr was identified in the above-mentioned TaCAT2-R (TaCAT2Ser214) and TaCAT2-S (TaCAT2Thr214) proteins, respectively, we mutated serine (Ser) into alanine (Ala) at this site in the TaCAT2-R protein to generate phosphorylation-dead mutant. We further compared the phosphorylation level among three variants (TaCAT2Ser214, TaCAT2Thr214, and TaCAT2Ala214). Results indicated that TaCAT2Ser214 displayed relatively more intense bands than TaCAT2Thr214 and mutant TaCAT2Ala214 both in vitro and in vivo after phosphorylation by TaSnRK1α (Fig. 3g, h). These results indicated that the 214Ser/Thr was a key phosphorylation site in TaCAT2 by TaSnRK1α, and possibly resulted in the difference of FCR resistance in wheat plants.
TaSnRK1α increased the protein level of TaCAT2
To illustrate whether TaSnRK1α affects the protein level of TaCAT2, we purified TaCAT2-Flag and TaSnRK1α-His proteins in tobacco leaf cells, respectively. After the addition of TaSnRK1α-His, the protein level of TaCAT2 was gradually enhanced with the increase of TaSnRK1α-His (Fig. 4a), indicating that TaSnRK1α could prevent the degradation of TaCAT2.
a Immunoblots detected that TaSnRK1α increased the protein level of TaCAT2Ser214 in tobacco leaf cells; TaCAT2Ser214 was detected with anti-Flag antibodies, while TaSnRK1α was detected with anti-His antibody. b Immunoblots detected that TaSnRK1α increased more the protein level of TaCAT2Ser214 than those of TaCAT2Thr214 and TaCAT2Ala214 in tobacco leaf cells; TaCAT2Ser214, TaCAT2Thr214 and TaCAT2Ala214 were detected with anti-Flag antibody, and TaSnRK1α was detected with anti-GFP antibody. c Immunoblots detected higher protein level of TaCAT2Ser214 than TaCAT2Thr214 and TaCAT2Ala214 in wheat protoplasts of two-week-old leaves from Fielder and TaSnRK1α-OE plants; TaCAT2Ser214, TaCAT2Thr214 and TaCAT2Ala214 were detected with anti-GFP antibody. d TaSnRK1α enhanced the function of TaCAT2 scavenging ROS by DAB staining in tobacco leaves. TaSnRK1α enhanced the ability of TaCAT2 scavenging ROS but showed more efficiency on TaCAT2Ser214 than on TaCAT2Thr214. Different treatments (I-VI) represent MOCK + 35S::GFP, P.s.t. DC3000 + 35S::GFP, P.s.t.DC3000 + 35S::TaCAT2Ser214, P.s.t.DC3000 + 35S::TaCAT2Ser214 + 35S::TaSnRK1α, P.s.t.DC3000 + 35S::TaCAT2Thr214 and P.s.t.DC3000 + 35S::TaCAT2Thr214 + 35S::TaSnRK1α, respectively. DC3000, as a control, was used to induce ROS. e The relative DAB staining intensity of each treated leaf. Different letters indicate significant differences between each treated group, which were determined by one-way ANOVA at p < 0.01. All experiments were independently repeated three times.
To detect whether TaSnRK1α could differentially affect the accumulation of TaCAT2Ser214, TaCAT2Thr214, and mutant TaCAT2Ala214 proteins, we transiently co-overexpressed vector 35S::TaSnRK1α-GFP with 35S::TaCAT2Ser214-Flag, 35S::TaCAT2Thr214-Flag and 35S::TaCAT2Ala214-Flag in tobacco leaf cells, respectively. After the addition of TaSnRK1α, protein abundances of three TaCAT2 variants were obviously increased, and the protein abundance of TaCAT2Ser214 was relatively higher than those of TaCAT2Thr214 and TaCAT2Ala214 (Fig. 4b). Subsequently, we transiently overexpressed vectors 163::TaCAT2Ser214-GFP, 163::TaCAT2Thr214-GFP and 163::TaCAT2Ala214-GFP in protoplasts from WT and TaSnRK1α-OE wheat plants, respectively. Results showed that TaSnRK1α promotes protein accumulations of three TaCAT2 variants, and TaCAT2Ser214 displayed relatively higher protein abundance than TaCAT2Thr214 and TaCAT2Ala214 in wheat protoplast (Fig. 4c). These results implied that TaSnRK1α had a more effective influence on TaCAT2Ser214 than TaCAT2Thr214.
TaSnRK1α enhanced the function of TaCAT2 in ROS scavenging
To illustrate whether TaSnRK1α enhances the function of TaCAT2 scavenging ROS, we detected H2O2 accumulation in tobacco leaves using different combinations containing 35S::GFP, 35S::TaCAT2Ser214, 35S::TaCAT2Thr214 and 35S::TaSnRK1α, respectively. The results of DAB staining showed that ROS accumulation was significantly decreased with the addition of TaCAT2Ser214 and TaCAT2Thr214, indicating the ability of TaCAT2 to scavenge ROS (Fig. 4d, e). After adding TaSnRK1α, ROS accumulation level was significantly lower in TaCAT2Ser214 than in TaCAT2Thr214, indicating a more effective influence on TaCAT2Ser214 than TaCAT2Thr214 by TaSnRK1α (Fig. 4d, e). These results suggested that the TaSnRK1α enhanced the function of TaCAT2 scavenging ROS, thereby resulting in an improved FCR resistance in wheat plants.
TaSnRK1α positively modulated wheat FCR resistance
To verify the phenotypic effect of TaSnRK1α on FCR resistance, we screened an EMS mutant K331 with premature stop codon of TaSnRK1α from the EMS-mutagenized tetraploid wheat Kronos library (Fig. 5a). To exclude other mutation sites, we backcrossed K331 with wild type twice to create a BC2 line (snrk1α). After inoculation by Fp-IV, the homozygous snrk1α showed a significantly decreased FCR resistance in both seedling and adult stages (Fig. 5b–d), and possessed significantly increased accumulation of H2O2 (Fig. 5e, f) and fungal biomass (Supplementary Fig. 16b) compared with wild type, suggesting that the mutation of TaSnRK1α reduced FCR resistance in the tetraploid wheat.
a–f Tetraploid Kronos EMS mutants (snrk1α) with premature stop codon of TaSnRK1α showed decreased FCR resistance. Mutation site of SnRK1α gene in red box (a); Comparison of phenotype and disease index for FCR between WT and snrk1α in greenhouse and field (b, c, d); Detection of H2O2 content by a Hydrogen Peroxide Assay Kit (e) and DAB (3,3’-diamino benzidine hydrochloride) staining (f) in WT and snrk1α plants, respectively. WT, wild type; snrk1α, the EMS mutants; Seedling, the FCR resistance investigated in the greenhouse; Adult, the FCR resistance investigated in the field. Red boxes included the white heads that were caused by FCR root infection but not by secondary floral infections. g–i TaSnRK1α-OE lines significantly enhanced wheat FCR resistance. Relative expression levels of TaSnRK1α gene in wild-type Fielder and overexpression lines (O.E.#) (g); Comparison of phenotype and disease index for FCR between WT and overexpression lines in greenhouse and field (h, i, j); Detection of H2O2 content by a Hydrogen Peroxide Assay Kit (k) and DAB staining (l) in WT and overexpression lines, respectively. The wheat leaf bases were sampled for staining DAB after infection by Fp-IV for four weeks in the greenhouse. Three biological replicates were used for each group, and each replicate contained five plants. Transcript levels were examined by qRT-PCR with TaActin as an endogenous control. The data are presented as the means ± SE from three biological replicates. **p < 0.01 (two-tailed Students’s t-test). Source data are provided as a Source Data file.
To further confirm the function of TaSnRK1α, we transferred the vector pUbi::LGY-OE3 containing TaSnRK1α into Fielder (TaCAT2-R) for overexpression (Fig. 5g). Two positive lines with high expression by qRT-PCR were self-crossed into T2 generation. After infection by Fp-IV, TaSnRK1α-OE lines displayed significantly enhanced FCR resistance compared with the WT in both the greenhouse and field (Fig. 5h–j). In addition, the expression level of TaCAT2 was significantly increased (Supplementary Fig. 20), and the accumulation of H2O2 and fungal biomass was significantly decreased in the TaSnRK1α-OE lines in the greenhouse (Fig. 5k, l and Supplementary Fig. 16d). These results indicated that TaSnRK1α positively modulated wheat FCR resistance possibly through scavenging H2O2.
In summary, we hypothesized that the FCR pathogen infected wheat plants accompanied by the pathogenic factors biosynthesis and ROS production, which accelerates plant cell death and F. pg reproduction. However, in the resistance cultivar harboring TaCAT2-R, TaSnRK1α contributed to the accumulation of TaCAT2-R protein through phosphorylation, and thereby enhanced TaCAT2 scavenging ROS, finally improving wheat FCR resistance (Fig. 6).
In wheat varieties harboring TaCAT2-R, TaSnRK1α phosphorylated TaCAT2-R protein and thereby enhanced the protein level of TaCAT2, resulting in increasingly scavenging ROS to enhance FCR resistance after infection of Fp-IV into wheat plants. However, in wheat varieties harboring TaCAT2-S, the phosphorylation level and protein level of TaCAT2-S were relatively weak, resulting in an excessive ROS to decrease FCR resistance.
Discussion
As one of the most severe soil-borne diseases, FCR not only causes enormous yield and economic losses but also causes trichothecene mycotoxins (e.g. deoxynivalenol) contamination that constitute significant health problems for humans and animals if consumed in respective food or feed products33,34,35. More recently, the FCR incidence area has rapidly increased and reached 6.6 million hectares only in Huanghuai valley of China during the past two years. However, no immune or highly resistant wheat germplasm has been identified so far, and most modern varieties are highly susceptible to FCR3,36. Moreover, few resistant genes have been discovered, though many QTL were mapped on different chromosomes9,36,37. A receptor-like kinase gene TaRLK-6A was found to enhance FCR resistance by regulating the expression of defense-related genes in wheat38. A cell wall invertase gene (TaCWI) was reported to modulate resistance to FCR and sharp eyespot in common wheat through mediating cell wall thickness and components39. Another receptor-like kinase TaCRK-7A was identified to mediate wheat FCR resistance by inhibiting F. pg growth40. Nevertheless, these genes showed limited roles to prevent damage from FCR in wheat resistance breeding. In this study, we identified a novel FCR resistance gene TaCAT2 through multiple approaches. We also discovered TaCAT2 showing interaction with TaSnRK1α, and further verified the functions of both TaCAT2 and TaSnRK1α positively modulating FCR resistance by the genetic transformation in wheat. Therefore, this study provided valuable genes for the improvement of wheat FCR resistance by pyramiding breeding.
Catalases (CATs) have been identified in a large number of living plants and were involved in organism growth, development, and response to the regulation of environment stimuli41,42. CATs are a gene family found across a diversity of plants, including Arabidopsis thaliana, Oryza sativa, Zea mays, Hordeum vulgare, Secale cereale, Sorghum bicolor, Triticum aestivum, etc., and these CATs are classified into three clades based on phylogenetic relationship (Supplementary Figs. 7 and 8)43,44. In Arabidopsis, AtCATs play important roles in catalyzing the decomposition of H2O2 to control ROS homeostasis45. The expression of AtCAT1 was mediated by MAPK and was induced by ABA (abscisic acid), but did not respond to circadian rhythms. AtCAT2, induced by light and cold, was able to regulate the accumulation of H2O2. AtCAT3 was highly expressed during the whole development stage in plants and was regulated by CPK8 to participate in response to drought stress46. In rice, OsCATA and OsCATC as stress-responsive members could be activated and phosphorylated by STRK1 to improve salt and oxidative tolerance47,48. In wheat, ten CAT genes were identified to classify into three clades49, and analysis of gene structure and domains revealed relative conservation of TaCAT2 gene family. In this study, a key phosphorylation site Ser214 of TaCAT2 was associated with FCR resistance, generating a relatively superior allele TaCAT2-HapA. However, as FCR resistance was regulated by multiple genes, three haplotypes with Ser214 (HapB, HapC, HapD) exhibited a middle resistance to FCR, possibly due to their relatively limited number (Supplementary Fig. 21). The measurement results of the enzymatic activity indicated a relatively higher value in TaCAT2-R than in TaCAT2-S and TaCAT1 (Supplementary Figs. 9 and 12).
FCR is caused by multiple Fusarium species and shows disease symptoms on roots, coleoptiles, leaf sheaths, and stem bases with light brown discoloration initially that turns dark brown or black later and causes whitehead in mature wheat26. During infection, the toxins biosynthesis, secondary metabolites biosynthesis (SMB), and ROS production were significantly associated with pathogenesis50. ROS serves as a crucial factor in reinforcing the plant cell wall and stimulating phytoalexin production during the plant defense response. However, an excess of ROS can damage essential cellular components in plant19,28. Reduced ROS accumulation significantly enhances resistance against S. sclerotiorum by reducing the host tissue necrosis that was required for necrotrophy colonization51. ROS was reported to act as a virulence factor in the infection of a necrotrophic pathogen B. cinerea, and a high level of ROS was present in the infected tissue, which accelerated the growth of the necrotrophic pathogen21. In Arabidopsis, the CAT2 was reported to detoxify ROS in the photosynthetic tissues52. In this study, the TaCAT we identified was a member of the CAT2 family (Supplementary Data 5). We detected significant changes in fungal biomass and H2O2 in the TaCAT2-mutant and TaCAT2-OE plants (Supplementary Fig. 16). This indicates that the overexpression of TaCAT2 enhances wheat FCR resistance, possibly through scavenging ROS to prevent the reproduction of pathogen Fp-IV in wheat plants. It should be noted that the expression of TaCAT2 was increased in the early 2 days after inoculation, and then decreased from 2 days to 7 days and increased from 7 days to later (Supplementary Fig. 22). It may indicate that when the plants suffered from fungal infection, a highly ROS level in the short early stage (2 day to 7 day) can restrict the fungal growth and a low ROS level in the late stage (7 day) can restrict the nutrient supply for the fungal. Additionally, we also investigated the sharp eyespot in TaCAT2-OE plants, and found that the TaCAT2-OE plants showed relatively stronger resistance than the controls. Meanwhile, TaCAT2-OE did not exhibit an obviously negative effect on important agronomic traits in wheat (Supplementary Fig. 17). These results indicate that TaCAT2 possesses a great potential to improve soil-borne disease resistance in wheat breeding programs.
As a kind of protein kinase, sucrose non-fermenting-1-related protein kinase (SnRK) has been reported to play a key role in multiple metabolic regulations and stress responses31,53. SnRK can be divided into three subfamilies, SnRK1, SnRK2, and SnRK3, and SnRK1 could further form a heterotrimeric complex containing the catalytic subunit SnRK1α and regulatory subunits SnRK1β, and SnRK1γ54,55. In rice, overexpression of SnRK1α sharply improved broad-spectrum disease resistance by enhancing the defense response mediated by jasmonate56. In wheat, TaSnRK1α could enhance resistance to deoxynivalenol (DON) produced by fusarium fungi57; TaSnRK1α interacted with TaFROG to avoid the degradation from an orphan protein Osp24 in F. graminearum and thus resulted in increased resistance to Fusarium head blight30. In this study, TaSnRK1α was identified to interact with TaCAT2 and phosphorylate TaCAT2 in vivo and in vitro with the help of GRIK1. The expression pattern of TaSnRK1α was the same as TaCAT2, and the fungal biomass and ROS accumulation were higher in the mutant plants and lower in the overexpression lines compared to wild-type plants, respectively (Supplementary Figs. 16 and 22). These results indicated that the overexpression of TaSnRK1α improved FCR resistance, possibly through phosphorylation enhancing the TaCAT2 protein stability that decreased the accumulation of ROS in wheat plants. However, we only verified phosphorylation experiments in E. coli and tobacco leaves. In addition, TaSnRK1α interacting with TaCAT2-R, TaCAT2-S and TaCATs paralogous (TaCAT1 and TaCAT3) implies a general interaction of TaCAT family with TaSnRK1α (Supplementary Fig. 23). Therefore, TaSnRK1α effectively assists TaCAT2 to scavenge ROS and could be considered as an important gene to improve wheat FCR resistance in pyramiding breeding.
In conclusion, we have identified an important wheat FCR resistance gene TaCAT2, and brought forth a TaSnRK1α-TaCAT2 model mediating FCR resistance by preventing ROS burst during infection of F.pg into wheat plants. TaSnRK1α, as a protein kinase, could phosphorylate and increase the protein level of the resistant haplotype TaCAT2-R, thereby resulting in a low ROS level to enhance wheat FCR resistance. However, the susceptible haplotype TaCAT2-S could not be phosphorylated adequately by TaSnRK1α, and excessive ROS burst finally contributed to wheat FCR susceptibility. This study provides a new insight into the regulation of FCR resistance by TaCAT2 and paves the way for understanding the molecular mechanism of FCR resistance genes in wheat.
Methods
Plant materials and FCR inoculation
A total of 243 wheat accessions were selected as an association panel for FCR resistance investigation and GWAS analysis. Seeds were harvested in Yuanyang Scientific Research and Education Center of Henan Agricultural University (N35.05°, E113.96°) during the cropping seasons 2017-2018 and 2018-2019.
Based on haplotype analysis on 6A from GWAS results in our study, an FCR-resistant cultivar Jinmai1 (DI = 30.95) possessed resistant haplotype (Hap_1A+Hap_2B) on 6A and two FCR-susceptible varieties Yunong 805 (DI = 74.71) and Zhengmai 082 (DI = 80.16) possessed susceptible haplotype (Hap_1C+Hap_2E) on 6A. Then we crossed Jinmai1 with Yunong 805 and Zhengmai 082, respectively, and developed two F4 nested segregation populations by the single seed descent method from 2017 to 2020 at Yuanyang. The Jinmai1/Yunong805 (RIL-JY) and Jinmai1/Zhengmai082 (RIL-JZ) populations were consisted of 267 and 143 lines, respectively.
A highly aggressive Chinese F. pseudograminearum isolate Fp-IV was routinely cultured on potato dextrose agar (PDA) at 25 °C. Millets grains were used as the pathogen medium to inoculate wheat seedlings. In greenhouse, materials were planted with three replicates, as each one contained 12 seeds. Seeds were sowed in 7 × 7 × 7 cm plastic containers with sterilized soil and inoculated with a 0.5 % colonized millet grains to soil ratio after seedlings grew to 3 cm long3. All seedlings were grown under 16 h/8 h day/night conditions at 25/20 °C day/night temperatures with 60% ~ 80% relative humidity and watered every two days. The severity of FCR was evaluated 28 days after inoculation by Fp-IV. The FCR resistance of each accession was scored using a disease index (DI) of 0–9 levels according to the methods of Yang3. In the field, all test accessions, along with boiled millets infected by Fp-IV were planted in a 0.2 × 1.5 m row with an inoculation ratio of 1:1 for scoring FCR resistance. Each accession was planted with two replicates. The FCR severity was investigated at the wheat grain-filling stage using the DI of 0–4 levels according to the method of Zhou35.
After scoring, we selected extreme phenotype offspring lines to develop FCR-resistant pools (FCR-RPo) and FCR-susceptible pools (FCR-SPo). All selected lines were sampled in greenhouse before and after inoculation by Fp-IV at 5 days for further analysis. The FCR-RPo (DI < 35) and FCR-SPo (DI > 65) were composed of 30 lines, respectively, with three replicates.
Genome-wide association study
All 243 accessions were genotyped by the wheat 660 K SNP array in Beijing CapitalBio Technology Company (http://cn.capitalbio.com/). The quality control of genotyping data was carried out with the PLINK software with threshold of -maf 0.02 and -geno 0.1 (http://zzz.bwh.harvard.edu/plink/tutorial.shtml)58. GWAS analysis was proceeded with R software by using the mixed linear model (PCA + K) in GAPIT packages59. The variance-covariance kinship matrix was calculated using the VanRaden method. A modified Bonferroni correction was used for calculating the significant level, and the threshold for P value was set at 10-3 60.
Whole-exome sequencing (WES)
The FCR-RPo, FCR-SPo and parents were used to conduct the WES program with three replicates by Tcuni Technologies (Chengdu, China). The gDNA library was prepared by a KAPA Hyper Pre Kit and was amplificated using KAPA HiFi HotStart ReadMix. The target regions were captured by hybridizing the gDNA sample library with the SeqCap EZ probo pool and recovered by the SeqCap Hybridization and Wash Kit. After the captured DNA was amplificated, these samples were sequenced by the Illumina HiSeq Nova platform to generate 150-bp paired-end reads61.
A total of 30 Gb data were obtained for each replicate. Raw sequencing reads were processed by ‘Fastap’ (version0.12.4) to filter low-quality reads and adapters. BWA (version0.7.16) mem was used to align the high-quality reads in IWGSC RefSeq V1.1 genome with default parameters. Samtools (version 1.9) was used to order and remove reads of PCR duplications. After that, the raw cohort vcf was worked out with GATK (version 4.0.10.1). The minimum mapping quality parameter was set as 30 for only high-quality alignment reads that were used to call variants. The bcftools (version 1.9) were performed as variants quality filtering with QUAL > 30 and DP > 5. The variants were annotated by customized database, including the IWGSC V1.1 HC/LC gene and its own annotated gene62.
Bulk segregant RNA sequencing (BSR-Seq)
The FCR-RPo, FCR-SPo before and after inoculation by Fp-IV at 5 days were used for the BSR-Seq program, respectively, with three replicates. RNA-Seq was conducted by OE Biotech. Co., Ltd. (Shanghai, China) using an Illumina HiSeq TM 2500 platform. Quality control was assessed by raw RNA-Seq reads using the software Trimmomatic v0.36 to obtain clean reads63. Approximately 11 Gb clean reads were generated for each sample. Bowtie2 v2.2.3 was used to map clean reads in the Chinese Spring genome (IWGS Ref Seqv1.1) to obtain gene locations and the specific sequence characteristics. The reads were re-assembled using StringTie 1.3.3b. FPKM value of each gene was calculated by Cufflinks 2.2.164,65, and read counts were obtained by Htseq-count 0.9.166. The differentially expressed genes (DEGs) were detected using the DESeq1.18.0 R package functions SizeFactors and nbinomTest. The threshold for significant DEG was set as p < 0.05 and fold change >2. The software snpEff v4-1g was used to annotate and predict the effects of variants on genes.
Barley stripe mosaic virus (BSMV)-mediated gene silencing
To silence the TraesCS6A02G041700 (TaCAT2) gene and TraesCS6A02G042600 (TaLBP) gene, 153-bp and 168-bp fragments of cDNA sequence designed by Primer 3 were amplified from Jinmai1 to construct into the recombinant vector pSL038-1 for the virus-induced gene silencing (VIGS) experiments, respectively. Each vector (α, β, γ, γ-PDS, and recombinant γ-gene) was linearized and transcribed to RNA in vitro and mixed with a ratio of 1:1:167 for inoculating Jinmai1 that was highly sensitive to BSMV. The BSMV:PDS (phytoene desaturase) was used as a positive control. Ten days after infection with Fp-IV, the mixture RNA virus was added to the FES buffer to inoculate onto the second leaves of seedlings. After staying in darkness for 24 h, all plants subsequently were grown at 23–25 °C with 60% ~ 80% relative humidity. After 2 weeks, FCR resistance was investigated, and the corresponding tissues were collected to measure the physiological indicators. RNA was extracted from leaves for qRT-PCR to assess silencing efficiency. The primers used in this section were listed in Supplementary Data 7.
Ethyl methanesulfonate mutants of TaCAT2 gene and TaSnRK1α gene
Seeds of the tetraploid wheat Kronos were mutagenized by the team of Jorge Dubcovsky through chemical mutagen ethyl methanesulfonate (EMS)68. The mutant lines K3868 (G to A: stop gained effect at 133 AA of TaCAT2) and K331 (G to A: stop gained effect at 386 AA of TaSnRK1α) were screened. The confirmed mutants by sequencing were backcrossed with wild type two times to obtain BC2 mutants (cat2, snrk1α) to eliminate background interference.
Overexpression of the TaCAT2 and TaSnRK1α genes
The CDS sequences of TaCAT2 and TaSnRK1α from Jinmai1 were inserted into the LGY-OE3 vector containing Ubi-promoter, and then were introduced to the Agrobacterium tumefaciens EHA105 strain. Fusion vectors were subsequently transferred into immature embryos of KN199 and Fielder, respectively. Positive transgenic lines were detected using gene-specific primers by sequencing, and the qRT-PCR assay was conducted to quantify expression levels of TaCAT2 and TaSnRK1α gene in positively transgenic lines. More than 10 independent transgenic lines were obtained for each of TaCAT2 and TaSnRK1α, and two independent lines with over 15-fold increased expression for each gene were self-crossed into T2 generation for further analysis.
Fungal biomass analysis by real-time PCR
To measure the relative fungal biomass, the total DNA of wheat stems from EMS mutants, overexpressed lines, and respective wild-type plants that were inoculated with Fp-IV for 0 day, 7 days, 14 days, and 21 days were extracted by the CTAB method, respectively. 100 ng DNA was used in the quantitative real-time PCR in a real-time PCR instrument using 2XHieff® Robust PCR Master Mix (Yeasen) with F.pg-Tubulin specific primers (Supplementary Data 7). The wheat tubulin gene was used as the endogenous control to normalize the samples. The 2−ΔΔCt analysis method was used to calculate the relative gene expression. Each sample was conducted with three replicates.
Quantification of ROS accumulation
To detect ROS content, we used 3,3’-diamino benzidine hydrochloride (DAB) to measure the peroxide in wheat plants and tobacco leaves. Wheat seedlings for DAB staining and the H2O2 content were sampled from the leaf base after Fp-IV infection for four weeks. Tobacco for DAB staining was sampled from the four-week-old leaves 2 days after infiltration. Samples were treated in vacuum infiltration for 3 min and were placed in 1 mg/mL DAB buffer (Sangon Biotech, Shanghai, China) for 8 h at 22 °C with light. Then samples were de-stained at 80 ~ 90 °C until the chlorophyll was removed in 70% ethanol. After that, the samples were observed under a bright-field type microscope to evaluate the ROS content69. In addition, we also used a commercial Hydrogen Peroxide Assay Kit (S0038; Beyotime) to measure H2O2 concentration in samples according to the instruction of manufacturer70.
Yeast two-hybrid assays (Y2H)
The different tissues of Jiyanmai 7 after infection by Fp-IV were sampled for cDNA library construction by OE Biotech (Shanghai, China). Different tissues (root, stem, leaf sheath, leaf, young ear, and ear at wheat filling stage) were sampled from five independent wheat plants for RNA extraction, respectively. The CDS of TaCAT2 recombined into pGBKT7 vector was used as a bait to screen interaction proteins in the wheat cDNA library. Yeast colonies with β-galactosidase activity on SD/Trp-Leu-His-Ade were selected as candidate clients. The client genes were further sequenced. The full-length cDNA of TaSnRK1α was amplified and cloned into the pGADT7 vector. Four types of TaCAT (TaCAT2-S, TaCAT3, TaCAT1 and TaCAT2-R) and TaCAT2-R containing the N-terminal with the Ser214Thr site and the C-terminal with the Lys418Arg site were amplified and cloned into the pGBKT7 vector, respectively. Bait and prey vectors were co-transformed into yeast strain AH109 and were screened on SD/Leu-Trp and SD/Trp-Leu-His-Ade to verify their interactions.
Firefly luciferase complementation assay
To further confirm the interaction, TaCAT2 was inserted into pCAMBIA1300-cLUC vector, and TaSnRK1α was inserted into pCAMBIA1300-nLUC vector, respectively. The recombinant vectors were transformed into Agrobacterium strain GV3101. Different vector combinations were co-transfected into the lower epidermis of the four-week-old Nicotiana benthamiana leaves. The SnRK1α-cLUC/PAP6-nLUC was positive control71, and the TaCAT2-cLUC/nLUC and TaSnRK1α-nLUC/cLUC were negative controls. After incubation in 24 h/24 h dark/light, 1 mM/L luciferin (E1065, Promega) was sprayed onto the lower leaf epidermis. After dark-adaption for 5 min, LUC activities of co-infiltrated leaves were observed72.
Pull-down assays in vitro
The CDS of TaCAT2 and TaSnRK1α were cloned into pGEX-6p-1 and pET-28a-His vectors, respectively, and then expressed in E. coli BL21 strain. Mixture recombinant vectors with TaSnRK1α were induced by 0.05 mM isopropyl β-D-1-thiogalactopyranoside for expression at 16 °C, respectively. To detect interactions of TaCAT2 with TaSnRK1α, equal amounts of TaCAT2-GST with TaSnRK1α-His and empty GST with TaSnRK1α-His fusion proteins were incubated at 4 °C for 2 h and then were mixed with glutathione for affinity purification, respectively, and were subsequently detected by western blot. Presences of TaCAT2-GST and GST empty vector were detected by anti-GST (1:5000, M20007, Mouse, Abmart) antibody, and TaSnRK1α-His with anti-His (1:5000, M20001, Mouse, Abmart) antibody. An anti-mouse IgG HPR (AU100302, Abmart) was used as the secondary antibody.
Subcellular localization and bimolecular fluorescence complementation assay
CDS sequences without stop codons of TaCAT2 and TaSnRK1α were cloned into the 35S::GFP vector to generate 35S::GFP-TaCAT2 and 35S::GFP-TaSnRK1α, respectively. The fusion constructs with peroxisome, plasma and membrane markers were mixed with 1:1 ratio, respectively, and then were introduced into four-week-old tobacco leaf cells. After incubation for 48 h post agroinfiltration, the mCherry and GFP fluorescence signals were examined using confocal microscopy (Carl Zeiss LSM170).
For bimolecular fluorescence complementation (BiFC) assay, the CDS of TaCAT2, TaGFP and TaSnRK1α were cloned into pSAT1-cEYFP and pSAT1-nEYFP vectors to generate cEYFP-TaCAT2, cEYFP-TaGFP, nEYFP-TaGFP and nEYFP-TaSnRK1α, respectively. The fusion constructs and empty vector were introduced into two-week-old wheat leaf protoplasts. cEYFP-TaCAT2+nEYFP-GFP and nEYFP-TaSnRK1α + cEYFP-GFP were used as negative controls. After incubation for 20 h at 25 °C in the darkness, the GFP and YFP fluorescence signals were examined using confocal microscopy (Carl Zeiss LSM170).
Cell-free assay
To detect whether TaSnRK1α could affect the protein level of TaCAT2, the CDS of TaCAT2Ser214 was ligated into pCambia2306 vector to construct TaCAT2Ser214-Flag vector, and TaSnRK1α was cloned into pET-28a to construct TaSnRK1α-His vector, respectively. The constructed vector containing TaCAT2Ser214 gene was transferred into Agrobacterium tumefaciens and infiltrated into the lower epidermis of four-week-old Nicotiana benthamiana leaves by infiltration, and then were incubated in darkness for 12 h for total proteins extraction. The purified protein was divided into four equal parts, and each reaction system contained 500 μg total protein in 2 mM ATP with different concentrations of TaSnRK1α (0 μg, 50 μg, 100 μg, 150 μg) for incubation at 25 °C for 1.5 h. Anti-actin antibodies (Abbkine, Lot number ABM40122) and anti-His (Abmart, lot number M20001) antibodies were used as loading controls before protein abundance assays. The reaction mixtures were detected by western blot for the remaining TaCAT2 Ser214-Flag proteins with an anti-Flag antibody (Abmart, Lot number M20008) after the reaction.
Transient expression of proteins
As Ser/Thr214 was a key differential site of TaCAT2, we performed site-directed mutagenesis and generated a mutant TaCAT2Ala214 (Ser214Ala, TCC/GCC) using the Fast Site-Directed Mutagenesis Kit (Tiangen, China, Catalog No. KM101). Positive mutants were verified by sequencing. In the Nicotiana benthamiana transient expression experiments, the CDS of TaCAT2Ser214, TaCAT2Ala214, TaCAT2Thr214 and TaSnRK1α were ligated into the pCambia2306 and 35S::GFP vectors to construct TaCAT2Ser214-Flag, TaCAT2Ala214-Flag, TaCAT2Thr214-Flag and TaSnRK1α-GFP vectors, respectively. These constructed vectors were co-infiltrated into the lower epidermis of four-week-old Nicotiana benthamiana leaves through A. tumefaciens-mediated transient transformation by infiltration, and then were incubated in darkness for 48 h for total protein extraction in tobacco leaves. Proteins were separated by SDS-PAGE and transferred to a PVD membrane (IPVH00010, Millipore), and further were detected by anti-GFP and anti-Flag antibodies (Abmart, Lot number M20008). Anti-actin antibodies (Abbkine, Lot number ABM40122) were as controls for protein loading.
In the wheat protoplasts transient expression experiments, the CDS of TaCAT2Ser214, TaCAT2Ala214, and TaCAT2Thr214 were ligated into the 163::TaCAT2-GFP vector and further transiently transfected into the protoplasts of two-week-old leaves in wild-type Fielder and TaSnRK1α-OE plants, respectively. Proteins of transfected protoplast were immunoprecipitated and analyzed by immunoblots with an anti-GFP antibody (Abcam, lot number 502) and anti-actin antibody (Abbkine, Lot number ABM40122).
Protein phosphorylation
In the in vitro phosphorylation experiments, the full-length cDNA of TaCAT2 (TaCAT2Ser214, TaCAT2Ala214, and TaCAT2Thr214), TaSnRK1α and GRIK1 were inserted into the pET-28a-His, pGEX-6p-1-GST, and pGEX-6p-1-GST vectors, respectively. Each recombination vectors were transformed into a BL21 strain, and proteins were induced by 0.05 mM isopropyl β-D-1-thiogalactopyranoside for expression at 16 °C. Then the released proteins were removed by glutathione agarose resin (P2262, Beyotime) and the His-tag protein purification kit (P2226, Beyotime) to purify. TaCAT2 was phosphorylated by TaSnRK1α after adding GRIK1. The reaction system was a total volume of 100 μL (2 μL 1 M Tris-HCL, 1 μL 0.6 M MgCL2, 0.1 μL 1 M CaCl2, 0.5 μL 10 mM ATP, 0.2 μL 1 M DTT, 20 μg TaCAT2, and corresponding calculated water, 5 μg TaSnRK1α and 5 μg GRIK1). After the reaction, the products were separated by SDS-PAGE, and the phosphorylation levels were detected by Pan Phospho-Serine/Threonine antibody (T91067, Abmart).
In the in vivo differential phosphorylation experiments, the cDNA of TaCAT2Ser214, TaCAT2Ala214, and TaCAT2Thr214 with Flag tag were inserted into pCambia2306 vectors, respectively, and TaSnRK1α was inserted into the 35S::GFP vectors, and GV3101 carrying the above constructs were co-infiltrated into four-week-old Nicotiana benthamiana leaves. The infiltrated leaves were harvested after 48 h darkness and total proteins were extracted. The TaCAT2 proteins extracted from the above extracts were immunoprecipitated with an anti-Flag antibody (Abcom, Lot number AF0036) and Protein A/G PLUS-Agarose (Santa Cruz, Lot number sc-2003). After washing three times, eluted proteins were detected by immunoblotting with anti-Flag (Abmart, Lot number M20008). Then the anti-phosphorylation antibodies (T91067, Abmart) were used to detect the phosphorylation degree of TaCAT2 in vivo and the differential phosphorylation level of TaSnRK1α on TaCAT2Ser214, TaCAT2Ala214, and TaCAT2Thr214.
Phylogenetic tree of CAT genes
The nucleotide and protein sequences of the CAT gene family in wheat and relative species were retrieved from the Ensemble plant database (https://ensembl.gramene.org/index.html). The phylogenetic tree was constructed using MEGA-X software by the neighbor-joining (NJ) method with the bootstrap of 1000 replications. The exon-intron structures of these CAT genes were retrieved according to the Gene Structure Display Server (GSDS 2.0) (http://gao-lab.org/). The conserved protein motifs were analyzed by the MEME software (http://meme-suite.org/meme/). The conserved domains of CAT proteins were identified using the Hidden Markov Model (HMMER) software (https://www.ebi.ac.uk/Tools/hmmer/). The molecular weight (MW) and theoretically isoelectric point (pI) were identified by the ExPASY Compute pI/Mw tool (https://web.expasy.org/compute_pi/). The subcellular localization of TaCAT2 proteins was predicted using the PSORT.
Statistical analysis
The significant differences among different treatments were statistically determined by ANOVA comparison and followed by a Student’s t-test if ANOVA analysis was significant at p < 0.05 or p < 0.01.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data supporting the findings of this work are available within the paper and its supplementary information files. Detailed sequence data of WES and BSR-Seq are deposited in NCBI database under accession numbers PRJNA1228985 and PRJNA1209998, respectively. The plant materials and datasets generated and analyzed during the present study are available from the corresponding author upon request. Source data are provided in this paper.
References
Zhang, X. et al. Survey of Fusarium spp. causing wheat crown rot in major winter wheat growing regions of China. Plant Dis. 99, 1610–1615 (2015).
Duan, S. et al. Integrated transcriptome and metabolite profiling high lights the role of benzoxazinoids in wheat resistance against Fusarium rown rot. Crop J. 10, 407–417 (2022).
Yang, X. et al. Investigation and genome-wide association study for Fusarium crown rot resistance in Chinese common wheat. BMC Plant Biol. 19, 153 (2019).
Beccari, G. et al. Development of three fusarium crown rot causal agents and systemic translocation of deoxynivalenol following stem base infection of soft wheat. Plant Pathol. 67, 1055–1065 (2018).
Obanor, F. & Chakraborty, S. Aetiology and toxigenicity of Fusarium graminearum and F. pseudograminearum causing crown rot and head blight in Australia under natural and artificial infection. Plant Pathol. 63, 1218–1229 (2014).
Bovill, W. D. et al. Pyramiding QTL increases seedling resistance to crown rot (Fusarium pseudograminearum) of wheat (Triticum aestivum). Theor. Appl. Genet. 121, 127–136 (2010).
Martin, A. et al. Markers for seedling and adult plant crown rot resistance in four partially resistant bread wheat sources. Theor. Appl. Genet. 128, 377–385 (2015).
Poole, G. J. et al. Identification of quantitative trait loci (QTL) for resistance to Fusarium crown rot (Fusarium pseudograminearum) in multiple assay environments in the Pacific Northwestern US. Theor. Appl. Genet. 125, 91–107 (2012).
Liu, C. & Ogbonnaya, F. C. Resistance to Fusarium crown rot in wheat and barley: a review. Plant Breed. 134, 365–372 (2015).
Zheng, Z. et al. Fine mapping of a large-effect QTL conferring Fusarium crown rot resistance on the long arm of chromosome 3B in hexaploidy wheat. BMC Genomics 16, 850 (2015).
Zheng, Z., Kilian, A., Yan, G. & Liu, C. QTL conferring Fusarium crown rot resistance in the elite bread wheat variety EGA Wylie. PLoS ONE 9, e96011 (2014).
Yang, X. et al. A loss-of-function of the dirigent gene TaDIR-B1 improves resistance to Fusarium crown rot in wheat. Plant Biotechnol. J. 19, 866–868 (2021).
Wang, H. et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368, eaba5435 (2020).
Naveed, Z. A., Wei, X., Chen, J., Mubeen, H. & Ali, G. S. The PTI to ETI continuum in phytophthora-plant interactions. Front. Plant Sci. 11, 593905 (2020).
Yun, B. et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268 (2011).
Bellegarde, F. et al. The chromatin factor HNI9 and elongated hypocotyl5 maintain ROS homeostasis under high nitrogen provision. Plant Physiol. 180, 582–592 (2019).
Mittler, R. ROS are good. Trends Plant Sci. 22, 11–19 (2017).
Wang, N. et al. Transcriptional repression of TaNOX10 by TaWRKY19 compromises ROS generation and enhances wheat susceptibility to stripe rust. Plant Cell 34, 1784–1803 (2022).
Lo Presti, L. et al. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545 (2015).
Unger, C., Kleta, S., Jandl, G. & Tiedemann, A. V. Suppression of the defence-related oxidative burst in bean leaf tissue and bean suspension cells by the necrotrophic pathogen Botrytis cinerea. J. Phytopathol. 153, 15–26 (2005).
Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227 (2005).
Heller, J. & Tudzynski, P. Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49, 369–390 (2011).
Duba, A., Goriewa-Duba, K. & Wachowska, U. A review of the interactions between wheat and wheat pathogens: Zymoseptoria tritici, Fusarium spp. and Parastagonospora nodorum. Int. J. Mol. Sci. 19, 1138 (2018).
Josefsen, L. et al. Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium graminearum. Autophagy 8, 326–337 (2012).
Sabburg, R., Obanor, F., Aitken, E. & Chakraborty, S. Changing fitness of a necrotrophic plant pathogen under increasing temperature. Glob. Change Biol. 21, 3126–3137 (2015).
Kang, R. et al. Expression of Fusarium pseudograminearum FpNPS9 in wheat plant and its function in pathogenicity. Curr. Genet. 66, 229–243 (2020).
Giri, M. K. et al. GBF1 differentially regulates CAT2 and PAD4 transcription to promote pathogen defense in Arabidopsis thaliana. Plant J. 91, 802–815 (2017).
Zhu, X. et al. The wheat NB-LRR gene TaRCR1 is required for host defence response to the necrotrophic fungal pathogen Rhizoctonia cerealis. Plant Biotechnol. J. 15, 674–687 (2017).
Yuan, H., Liu, W. & Lu, Y. Catalase2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe 21, 143–155 (2017).
Jiang, C. et al. An orphan protein of Fusarium graminearum modulates host immunity by mediating proteasomal degradation of TaSnRK1α. Nat. Commun. 11, 4382 (2020).
Jin, H. et al. Barley GRIK1-SnRK1 kinases subvert a viral virulence protein to upregulate antiviral RNAi and inhibit infection. EMBO J. 41, e110521 (2022).
Shen, W., Reyes, M. I. & Hanley-Bowdoin, L. Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol. 150, 996–1005 (2009).
Powell, J. J. et al. The Fusarium crown rot pathogen Fusarium pseudograminearum triggers a suite of transcriptional and metabolic changes in bread wheat (Triricum aestivum L.). Ann. Bot. 119, mcw207 (2016).
Kazan, K., Gardiner, D. M. & Manners, J. M. On the trail of a cereal killer: recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 13, 399–413 (2012).
Zhou, H. et al. Diversity of the Fusarium pathogens associated with crown rot in the Huanghuai wheat-growing region of China. Environ. Microbiol. 21, 2740–2754 (2019).
Jin, J. et al. Identification of a novel genomic region associated with resistance to Fusarium crown rot in wheat. Theor. Appl. Genet. 133, 2063–2073 (2020).
Pariyar, S. R. et al. Dissecting the genetic complexity of fusarium crown rot resistance in wheat. Sci. Rep. 10, 3200 (2020).
Qi, H. et al. TaRLK‐6A promotes Fusarium crown rot resistance in wheat. J. Integr. Plant Biol. 66, 12–16 (2024).
Lv, G., Zhang, Y., Ma, L. & Yan, X. A cell wall invertase modulates resistance to fusarium crown rot and sharp eyespot in common wheat. J. Integr. Plant Biol. 65, 1814–1825 (2023).
Wu, T. et al. The receptor-like kinase TaCRK-7A inhibits Fusarium pseudograminearum growth and mediates resistance to fusarium crown rot in wheat. Biology 10, 1122 (2021).
Mhamdi, A. et al. Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61, 4197–4220 (2010).
Su, T. et al. The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J. Integr. Plant Biol. 60, 591–607 (2018).
Hu, L., Yang, Y., Jiang, L. & Liu, S. The catalase gene family in cucumber: genome-wide identification and organization. Genet. Mol. Biol. 39, 408–415 (2016).
Raza, A. et al. Catalase (CAT) gene family in rapeseed (Brassica napus L.): genome-wide analysis, identification, and expression pattern in response to multiple hormones and abiotic stress conditions. Int. J. Mol. Sci. 22, 4281 (2021).
Xing, Y., Jia, W. & Zhang, J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54, 440–451 (2008).
Zou, J. et al. Arabidopsis CALCIUM-dependent protein KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27, 1445–1460 (2015).
Schmidt, R. et al. Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25, 2115–2131 (2013).
Zhou, Y. et al. The receptor-Like cytoplasmic kinase STRK1 phosphorylates and activates CATC, Thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 30, 1100–1118 (2018).
Zhang, Y. et al. Catalase (CAT) gene family in wheat (Triticum aestivum L.): evolution, expression pattern and function analysis. Int. J. Mol. Sci. 23, 542 (2022).
Jia, L. et al. A linear nonribosomal octapeptide from fusarium graminearum facilitates cell-to-cell invasion of wheat. Nat. Commun. 10, 922 (2019).
Ding, Y. et al. Sclerotinia sclerotiorum utilizes host-derived copper for ROS detoxification and infection. PLoS Pathog. 16, e1008919 (2020).
Kaurilind, E., Xu, E. & Brosché, M. A genetic framework for H2O2 induced cell death in Arabidopsis thaliana. BMC Genomics 16, 837 (2015).
Han, X. et al. SnRK1 phosphorylates and destabilizes WRKY3 to enhance barley immunity to powdery mildew. Plant Commun. 1, 100083 (2020).
Cho, Y., Hong, J., Kim, E. & Yoo, S. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 158, 1955–1964 (2012).
Margalha, L., Confraria, A. & Baena-González, E. SnRK1 and TOR: modulating growth-defense trade-offs in plant stress responses. J. Exp. Bot. 70, 2261–2274 (2019).
Seo, Y. et al. Towards establishment of a rice stress response interactome. PLoS Genet. 7, e1002020 (2011).
Perochon, A. et al. The wheat SnRK1α family and its contribution to Fusarium toxin tolerance. Plant Sci. 288, 110217 (2019).
Purcell, S. et al. Plink: a tool set for whole genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Lipka, A. E. et al. GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397–2399 (2012).
Li, H. et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 45, 43–50 (2013).
Zhang, L. et al. WheatGmap: a comprehensive platform for wheat gene mapping and genomic studies. Mol. Plant. 14, 187–190 (2021).
Dong, C. et al. Combining a new exome capture panel with an effective varBScore algorithm accelerates BSA-based gene cloning in wheat. Front. Plant Sci. 11, 1249 (2020).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Roberts, A., Trapnell, C., Donaghey, J., Rinn, J. L. & Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12, R22 (2011).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Anders, S., Pyl, P. T. & Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Zhang, N. et al. iTRAQ and virus-induced gene silencing revealed three proteins involved in cold response in bread wheat. Sci. Rep. 7, 7524 (2017).
Henry, I. M. et al. Efficient genome-wide detection and cataloging of ems-induced mutations using exome capture and next-generation sequencing. Plant Cell 26, 1382–1397 (2014).
Xu, Q. et al. An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 10, 5571 (2019).
Wang, J. et al. Arabidopsis BRCA1 represses RRTF1-mediated ROS production and ROS-responsive gene expression under dehydration stress. N. Phytol. 228, 1591–1610 (2020).
Zhang, L. et al. A TaSnRK1α modulates TaPAP6L-mediated wheat cold tolerance through regulating endogenous jasmonic acid. Adv. Sci. 10, 2303478 (2023).
Feng, Z. et al. Genetic analysis implicates a molecular chaperone complex in regulating epigenetic silencing of methylated genomic regions. J. Integr. Plant Biol. 63, 1451–1461 (2021).
Acknowledgements
This project was funded by the National Natural Science Foundation (U23A20191), National Key Research and Development Program (2023YFD1200403), National Natural Science Foundation of China Youth Science Fund Project (32401800), and Postdoctoral Science Foundation (2022M711066) of China.
Author information
Authors and Affiliations
Contributions
F.C. conceived and designed this study. X.Y., L.L.Z., J.J.W., X.N.Y., and M.J.Y. cloned the gene. X.Y., L.L.Z., L.R.Z., L.X.L., D.L., and N.Z. verified the gene function. X.Y., L.L.Z., and Y.R. scored FCR resistance. X.Y. and F.C. prepared the manuscript. All authors reviewed the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Donald Gardiner, Guixia Hao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, X., Zhang, L., Wei, J. et al. A TaSnRK1α-TaCAT2 model mediates resistance to Fusarium crown rot by scavenging ROS in common wheat. Nat Commun 16, 2549 (2025). https://doi.org/10.1038/s41467-025-57936-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57936-x
This article is cited by
-
Crown rot in wheat: pathogen biology, host responses, and management strategies
Stress Biology (2025)
-
Identification of a stable genetic locus and candidate genes for Fusarium head blight resistance on wheat chromosome 3BL
Theoretical and Applied Genetics (2025)