Abstract
Proteases are defined by their nucleophile but require additional residues to regulate their active sites, most often arranged as catalytic triads that control the generation and resolution of acyl-enzyme intermediates. Threonine N-terminal nucleophiles represent a diverse family of proteases and transferases that possess two active site nucleophiles, the side chain hydroxyl and the free amino-terminus, and require autocatalytic cleavage of their N-terminal propeptides. Here we provide evidence that the proteasome, which mediates intracellular protein degradation and contains three different threonine protease subunits, utilizes a unique catalytic pentad mechanism. In addition to the previously defined lysine/aspartate pair which regulates threonine’s side chain, a second serine/aspartate pair appears to regulate threonine’s amino-terminus. The pentad is required for substrate proteolysis and assembly-coupled autocatalytic cleavage, the latter triggered by alignment of the full pentad upon fusion of two half-proteasome precursors. A similar pentad mechanism was required by the ornithine acetyltransferase Arg7, suggesting that this may be a general property of threonine N-terminal nucleophiles. Finally, we show that two patient-derived proteasome mutations compromise function of the serine/aspartate unit in yeast, suggesting that defective pentad function may underlie some human proteasomopathies.
Similar content being viewed by others
Introduction
The major families of proteases are defined by their active site nucleophile and include serine proteases, cysteine proteases, aspartyl proteases, and metalloproteases. Threonine proteases are a relatively recent addition, having been recognized in the 1990s1,2,3, and are part of the larger family of so-called N-terminal nucleophiles that includes both proteases and various transferase enzymes. As the name indicates, N-terminal nucleophiles require the catalytic threonine to be exposed at the protein’s N-terminus and, accordingly, such enzymes are synthesized as inactive precursors with N-terminal propeptides. Mature enzymes are generated by propeptide cleavage, which is typically autocatalytic.
The best characterized threonine protease is the proteasome, the multisubunit complex responsible for most non-lysosomal protein degradation. Protein degradation occurs within its central core particle (CP)4, a barrel-shaped complex distinguished from conventional proteases by having three different proteolytic subunits that cleave after hydrophobic (β5), basic (β2), and acidic (β1) residues, respectively, thus accounting for the proteasome’s exceptionally broad substrate repertoire.
In addition to the active site nucleophile, proteases typically require additional residues for proteolysis. In most serine and cysteine proteases, the critical residues are arranged in spatial proximity as a catalytic triad with the two additional residues, often histidine and aspartate, serving as an acid-base system to control the protonation state of the nucleophile5,6. In a serine/histidine/aspartate protease, for example, histidine serves as a proton acceptor, polarizing serine’s side chain hydroxyl to facilitate nucleophilic attack on the substrate, resulting in a tetrahedral acyl-enzyme intermediate. Collapse of this intermediate releases the N-terminal substrate fragment, resetting the protonation status of histidine which can serve again as a proton acceptor to polarize a water molecule for nucleophilic attack on the remaining acyl-enzyme intermediate. Resolution of this intermediate releases the cleaved substrate protein and resets the protease active site.
N-terminal nucleophiles possess two chemically reactive groups: the side chain hydroxyl and the free amino group. Given this arrangement, there was initial speculation that they might not even require a catalytic triad, since it was thought that threonine’s N-terminus might directly regulate the protonation state of the side chain hydroxyl7,8,9. However, it was eventually recognized that such enzymes do utilize a catalytic triad which, in the case of the proteasome, consists of threonine, lysine, and aspartate2,3,10,11. This triad is thought to operate analogously to that of most serine and cysteine proteases, as described above, with lysine being the proton acceptor/donor for threonine’s side chain hydroxyl and aspartate stabilizing the lysine.
Autocatalytic activation of the proteasome is coupled to CP assembly12,13, an elegant arrangement that protects cells from the potentially harmful consequences of premature or promiscuous protein degradation. The CP consists of four stacked heteroheptameric rings with an α7β7β7α7 arrangement, and it is the fusion of two αβ half-CP precursors that triggers activation. In recent work we showed that, prior to half-CP fusion, the catalytic triad residues are not aligned due to flexibility in multiple proteins segments that eventually form the β-β-ring interface at the CP midline14. Mutual stabilization of these complementary loops upon CP fusion rigidifies these regions, bringing the triad residues into alignment which facilitates autocatalytic activation.
The flexibility of the protein loop controlling the triad residues prior to CP fusion was the initial clue to the proteasome’s mechanism of activation14. But the active site subunits harbor a second unstructured loop with a complementary unstructured region in their respective midline β-subunit partners (Fig. 1). Here we provide evidence that this second unstructured loop controls a second catalytic triad including a conserved serine and aspartate that apparently regulate the threonine active site’s free amino group, rather than the side chain hydroxyl, a model initially proposed by Groll and colleagues15. We refer to this constellation of residues as a catalytic pentad and demonstrate a similar arrangement in the ornithine acetyltransferase Arg7, suggesting that this may be a general feature of threonine N-terminal nucleophile enzymes. Finally, we show that two different patient-derived proteasome mutations compromise function of the Ser/Asp pair in yeast, suggesting that defects in this regulatory mechanism may explain some human proteasomopathies.
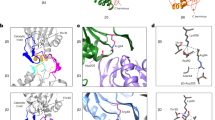
Shown are the atomic models of wild-type CP (PDB: 5CZ4) with corresponding unresolved regions from the pre-15S assembly intermediate (PDB: 7LS6) shown in various colors. Panel (a) shows the β2/β6 interface. One unresolved loop in β2 appears to regulate the canonical threonine/lysine/aspartate catalytic triad, suggesting that stabilization of this loop may be a key feature of autocatalytic activation14. However, β2 contains a separate unresolved loop whose function was unknown, and which contains a highly conserved aspartate residue with a nearby conserved serine residue, both in close proximity to the active site threonine. A similar arrangement was seen for the β5/β3 pair (b).
Results
Structural aspects of proteasome autocatalytic activation
We previously obtained cryo-EM structures of a pre-fusion assembly intermediate known as pre-15S which contains five β-subunits (β2-6)16. This intermediate showed a small number of unresolved regions in complementary segments that later form the β-β-midline interface. Upon half CP fusion, these interrelated segments undergo a disordered-to-ordered transition14. In the active site subunit β2, the two unresolved loops precisely matched the three disordered segments in β2’s future midline partner, β6 (Fig. 1a). One of these unresolved β2 loops contained the triad residue Asp46 and was immediately adjacent to the triad Lys62. Furthermore, Lys62 was rotated ~180° away from the active site threonine14. These findings suggested that the catalytic triad residues are misaligned prior to CP fusion, and the precise alignment of the β2/β6 midline interface upon half-CP fusion drives autocatalytic activation through stabilization of these catalytic residues. A similar arrangement was seen for the active site subunit β5 and its midline partner β3 (Fig. 1b).
However, the second unresolved loop in β2 and β5 could not be readily explained by this mechanism. This is because these loops were present on the opposite side of the threonine active sites, and therefore not positioned to regulate the canonical catalytic triads (Fig. 1a, b). Each of these unresolved loops harbored a highly conserved Asp residue with a conserved Ser residue nearby (β2-Ser158/Asp195 or β5-Ser206/Asp243), and each Ser/Asp pair was present very close to the active site threonine (Fig. 1a, b; Supplementary Fig. 1).
The Ser/Asp residues are required for autocatalytic activation and proteolysis by β2
The conventional triad residues in β2, Asp46 and Lys62, were hydrogen-bonded to each other and to the active site threonine’s side chain hydroxyl group (Fig. 2a; Supplementary Table 1). An analogous arrangement was present for the second set of residues, Ser158 and Asp195 (Fig. 2a), although hydrogen bonding was via threonine’s free amino group rather than its side chain.
a Structural arrangement of the proposed pentad residues as seen in mature wild-type CP (PDB: 5CZ4). Lys62, Asp46, and Thr30 represent the canonical catalytic triad, while Ser158 and Asp195 represent additional residues proposed to complete the catalytic pentad. Dashed lines, hydrogen bonds. b Phenotypic analysis of the indicated mutants, expressed from low-copy centromeric plasmids harboring the endogenous promoter/terminator elements, in a strain where endogenous β2 is under the control of the pGAL1 promoter. In glucose-containing media, expression of the endogenous β2 locus is repressed and plasmid-derived β2 is the sole source of this protein. Plates were cultured at 30 °C for the indicated number of days. c Purified CP from wild-type and mutant strains, as analyzed using SDS-PAGE followed by Coomassie staining. Asterisk, a break-down product of α7. Note that Pba1, Pba2, and immature β5 co-migrate as a single band. d Analysis of wild-type and mutant CP by native gel electrophoresis followed by immunoblotting with antibodies against α5, Pba1/2, and immature (propeptide-bearing) β5. e Proteasome activity assays using active site-specific fluorescent substrate probes. Background fluorescence has been subtracted and relative activity has been normalized to untreated controls. Individual points represent biologic duplicates. f Levels of transcription factor Rpn4, which senses cellular proteasome capacity, in whole cell extracts from wild-type and mutant strains, as determined by SDS-PAGE and immunoblotting. GAPDH, loading control. Similar results were obtained in 5 (b), 4 (c, d) and 2 (f) independent experiments, respectively.
To determine the physiologic significance of this arrangement, we mutated Ser158 and Asp195. The S158A mutant showed impaired growth, even in the absence of stress, consistent with a significant defect in proteasome function (Fig. 2b, compare left and middle panels). In contrast, the D195A mutant was altogether inviable (Fig. 2b). This was surprising because even mutation of β2’s threonine active site is not lethal15, and therefore implied a major defect in proteasome assembly. We generated the more conservative D195N mutation, which retains hydrogen bonding capacity. This mutant was viable but showed a severe growth defect (Fig. 2b).
A defect in regulating the active site threonine should compromise not only substrate proteolysis but also autocatalytic activation. A defect at this stage would be expected to stabilize the preholoproteasome which has undergone half-CP fusion but remains catalytically inactive and therefore retains the β-subunit propeptides as well as assembly chaperones Ump1, Pba1, and Pba217,18,19. We affinity purified CP from these mutants using a genomically integrated C-terminal TEV-Protein A tag on β4. In all cases, the dominant species was mature 20S CP (Fig. 2c, d) which was expected since the proteasome is essential for viability. However, the S158A and D195N mutants showed accumulation of preholoproteasome, which is larger than 20S and whose identity was confirmed by immunoblot analysis showing the presence of Pba1/2 and immature β5 (Fig. 2d), the latter detected using an antibody that specifically recognizes β5’s propeptide. The D195N mutant also showed accumulation of sub-20S intermediates (Fig. 2d), consistent with an additional pre-fusion assembly defect that may contribute to its stronger phenotype.
To determine whether these findings were specific to the S158 and D195 residues, we mutated a neighboring residue, Ser198, which is also in close proximity to the active site threonine (Supplementary Fig. 2A), located just three residues away from D195, and highly conserved (Fig. S1). We purified CP from the S198A mutant and found no evidence of preholoproteasome accumulation (Supplementary Fig. 2B). Further examination of the β2 structure did not reveal any other candidate polar residues in close proximity (<5 Å) to the threonine active site.
Next we assayed the proteasome’s proteolytic activity using fluorescent substrates that specifically report on each of the proteasome’s three active sites. Both S158A and D195A mutants showed a near total loss of β2 activity (Fig. 2e). In the case of S158A, this defect was highly specific with no detectable loss of β1 or β5 activity. The D195N mutant showed partial loss of β1 and β5 activities as well; however, this is likely secondary to the accumulation of sub-20S intermediates (Fig. 2d), which are catalytically inactive.
Finally, we looked at the cellular response to these mutations. The transcription factor Rpn4 senses cellular proteasome capacity and controls proteasome abundance in response to stress20,21. Cellular Rpn4 levels were strongly elevated in both mutants, consistent with a major defect in proteasome function (Fig. 2f). Altogether, these results establish a role for Ser158 and Asp195 in both autocatalytic activation and substrate proteolysis.
A similar pentad arrangement in β5 and β1
The analogous residues in β5 are arranged in a nearly identical manner, with Ser206 and Asp243 being hydrogen bonded to each other and the active site threonine (Fig. 3a; Supplementary Table 1). The D243A mutant showed a growth defect, even in the absence of exogenous stress, and this phenotype was exacerbated by concurrent S206A mutation (Fig. 3b). Purification of CP from the double mutant showed preholoproteasome accumulation (Fig. 3c), consistent with a defect in autocatalytic activation, and near-total loss of β5’s proteolytic activity, which was specific as β2 and β1 both showed nearly wild-type levels of function (Fig. 3d).
a Structural arrangement of the proposed pentad residues as seen in mature wild-type CP (PDB: 5CZ4). Lys108, Asp92, and Thr76 represent the canonical catalytic triad, while Ser206 and Asp243 represent additional residues proposed to complete the catalytic pentad. Dashed lines, hydrogen bonds. b Phenotypic analysis of the indicated mutants, expressed from low-copy centromeric plasmids harboring the endogenous promoter/terminator elements in a strain where endogenous β5 is under the control of the pGAL1 promoter. In glucose-containing media, expression of the endogenous β5 locus is repressed and plasmid-derived β5 is the sole source of this protein. Plates were cultured at 30 °C for 3 days. c Analysis of wild-type and mutant CP by native gel electrophoresis followed by immunoblotting with antibodies against α5, immature (propeptide-bearing) β5, and Pba1/2. d Proteasome activity assays using active site-specific fluorescent substrate probes. Background fluorescence has been subtracted and relative activity has been normalized to untreated controls. Individual points represent biologic duplicates. e Turnover of immature β5, as determined by cycloheximide chase analysis of whole cell extracts. Analysis was by SDS-PAGE followed by immunoblotting with an antibody that specifically recognizes immature β5. GAPDH, loading control. Asterisk, non-specific band. Similar results were obtained in 4 (b) and 2 independent experiments (c and e), respectively.
Using our β5-propeptide-specific antibody16,17, we directly monitored maturation of β5 by cycloheximide chase assay. In wild-type cells, immature β5 rapidly disappeared, consistent with efficient CP biogenesis (Fig. 3e). In contrast, the D243A and S206A/D243A mutants showed much slower turnover (Fig. 3e), again consistent with a defect in CP biogenesis. Indeed, the defect was strong enough to increase the steady-state (time = 0) levels of immature β5.
Βeta1 showed the same arrangement of pentad residues (Fig. 4a; Supplementary Table 1). In contrast to β2 and β5, phenotypic defects could not be identified under non-stress conditions (Fig. 4b). This likely reflects the known physiologic importance of the active sites: β5 > β2 > β122. Nevertheless, phenotypes were readily apparent for the D185A mutant upon canavanine treatment, which causes widespread protein misfolding, and this phenotype was further exacerbated by the S148A mutation (Fig. 4b). The S148A/D185A mutant showed a modest but reproducible increase in preholoproteasome abundance (Fig. 4c), and a near-total loss of β1’s proteolytic activity, which was highly specific for that subunit (Fig. 4d).
a Structural arrangement of the proposed pentad residues as seen in mature wild-type CP (PDB: 5CZ4). Lys52, Asp36, and Thr20 represent the canonical catalytic triad, while Ser148 and Asp185 represent additional residues proposed to complete the catalytic pentad. Dashed lines, hydrogen bonds. b Phenotypic analysis of the indicated mutants, expressed from low-copy centromeric plasmids harboring the endogenous promoter/terminator elements in a strain where endogenous β1 is under the control of the pGAL1 promoter. In glucose-containing media, expression of the endogenous β1 locus is repressed and plasmid-derived β1 is the sole source of this protein. Plates were cultured at 30 °C for 2 (no drug) and 7 days (canavanine, 1.5 μg/mL). c Analysis of wild-type and mutant CP by native gel electrophoresis followed by immunoblotting with antibodies against α5, immature (propeptide-bearing) β5, and Pba1/2. d Proteasome activity assays using active site-specific fluorescent substrate probes. Background fluorescence has been subtracted and relative activity has been normalized to untreated controls. Individual points represent biologic duplicates. e Structures of the helix-loop-helix motif responsible for the spatial proximity of the Ser/Asp pair within the catalytic pentad (PDB: 5CZ4). Similar results were obtained in 2 independent experiments (b, c).
These results indicate that the Ser/Asp pairs in β2, β5, and β1 are arranged in close proximity to the active site threonines, required for autocatalytic activation, and required for site-specific substrate proteolysis. The proximity of the Ser/Asp residues appears to be a consequence of a helix-loop-helix arrangement common to all β-subunits. In the active site subunits, Ser is present at the beginning of the first helix and Asp is present near the end of the second helix (Fig. 4e). Furthermore, the Ser and Asp residues are spaced exactly 37 residues apart in all active site subunits (Supplementary Fig. 1), suggesting that precise arrangement of this motif is essential for proteasome function.
The ornithine acetyltransferase Arg7 utilizes a similar catalytic pentad
We sought to determine whether this catalytic pentad arrangement was specific to the proteasome or might be a general feature of threonine N-terminal nucleophiles. We focused on Arg7, an ornithine acetyltransferase that functions in arginine biosynthesis23. Such enzymes are conserved from bacteria to humans and, despite their biosynthetic role, utilize a catalytic mechanism involving acyl-enzyme intermediates, similar to proteases. In addition, Arg7 requires autocatalytic cleavage to expose its active site Thr21524,25.
Structural data for the yeast ornithine acetyltransferase Arg7 is lacking, but there is a crystal structure of the closely related bacterial ortholog, ArgJ, from Streptomyces clavuligerus26. In this structure, ArgJ has been cleaved to expose its N-terminal threonine but the large N-terminal fragment remains non-covalently bound to the rest of the enzyme. We identified a group of five residues that were arranged in a strikingly similar manner to those seen in the proteasome, with a Lys/Asp pair on one side of the threonine nucleophile, and a Ser/Asp pair on the other side (Fig. 5a). Both pairs were hydrogen bonded to the active site, as in the proteasome, and all 5 residues showed strong evolutionary conservation (Fig. 5b). Remarkably, the Lys/Asp pair derives from Arg7’s N-terminal propeptide, a very different arrangement from the proteasome where the propeptides are thought to be degraded or released.
a Structural arrangement of the proposed pentad residues as seen for Streptomyces clavuligerus ornithine acetyltransferase (PDB: 2VZK). For clarity, residue numbering is after yeast Arg7. Dashed lines represent hydrogen bonds. b Evolutionary conservation of the proposed catalytic pentad residues. c Phenotypic analysis of the indicated mutants, expressed from low-copy centromeric plasmids harboring the endogenous promoter/terminator elements in the arg7Δ mutant, which is strongly (although not fully) auxotrophic for arginine. Plates were cultured at 30 °C for 2 days. d Maturation of wild-type and mutant Arg7-3xHA proteins, as determined by analysis of whole cell extracts using SDS-PAGE followed by immunoblotting with anti-HA antibody. GAPDH, loading control. e Expression of recombinant wild-type and mutant Arg7 in bacteria, as analyzed by SDS-PAGE followed by Coomassie staining. Gene expression is induced by IPTG. The dimer seen for mature Arg7 reflects autocatalytic cleavage into two comparably sized polypeptides. Similar results were obtained in 2 (b) and 3 independent experiments (d, e), respectively.
Arg7 null mutants grow poorly in media lacking arginine24. We therefore mutated each of the pentad residues to alanine and assayed their ability to restore growth to arg7Δ (Fig. 5c). The catalytically dead T215A mutant phenocopied the null mutant, as expected. Interestingly, the other four mutants were as deficient as the T215A mutant, indicating that all five residues are required for Arg7 function (Fig. 5c).
Next, we sought to determine whether these same residues were also required for autocatalytic activation. To monitor Arg7 protein levels by immunoblot, we inserted a C-terminal 3xHA tag at the endogenous locus, while maintaining upstream promoter sequences. The tagged strain grew comparably to wild-type on media lacking arginine, indicating that the tag did not compromise protein function (Supplementary Fig. 3). In the wild-type strain, both the mature C-terminal fragment and the uncleaved immature Arg7 protein were detected, with most of the protein having matured (Fig. 5d). The T215A mutant showed exclusively the immature form, as expected. The remaining four mutants showed a striking defect in maturation with all or nearly all Arg7 present as the immature form (Fig. 5d). Mutation of an unrelated but nearby serine residue, S247A, did not impair Arg7 maturation (Supplementary Fig. 2 C), again indicating specificity for the pentad residues identified.
We repeated the Arg7 maturation assay by cloning and expressing recombinant Arg7 in bacteria. Upon induction of protein expression, essentially all wild-type Arg7 was present as a cleaved dimer (Fig. 5e). In contrast, all five mutants again showed a major defect in autocatalytic activation (Fig. 5e). Thus, the distinctive pentad arrangement is required for the function of two unrelated threonine N-terminal nucleophiles and may be a general feature of the enzyme family.
Defects in second triad function may explain some human proteasomopathies
An emerging family of human diseases is caused by inherited mutations in the CP13,27. These proteasomopathies are characterized by immune dysregulation, dysmorphic features, and cognitive impairment. More than 40 mutations have been identified, but their specific effects on proteasome function and their broader effects on cellular physiology are largely unknown.
In addition to the constitutive proteasome which is conserved from yeast to humans, higher organisms also possess an immunoproteasome that incorporates distinct active site subunits (β5i, β2i, β1i) which are adapted for generation of antigenic peptides for presentation by major histocompatibility complex-I molecules28. Given the specialized role and limited tissue distribution of immunoproteasomes, mutations in their genes are likely to be better tolerated than corresponding mutations in the constitutive proteasome.
A patient-derived mutation, G201V, has been previously identified in β5i29. This residue is conserved in the constitutive human proteasome and yeast, where it corresponds to Gly205 (Fig. 6A; Supplementary Fig. 1). This residue is the immediate neighbor of the pentad residue Ser206, suggesting that the mutation might impair function of the Ser/Asp pair of the proposed catalytic pentad. We therefore sought to determine the consequences of this mutation in our yeast system. The corresponding G205V mutation was lethal (Fig. 6B). A more conservative G205A mutation was viable, showing a mild phenotype even in the absence of stress, and a strong phenotype when exposed to proteotoxic stress (Fig. 6B). We purified CP from this mutant which showed strong accumulation of preholoproteasome, consistent with a defect in autocatalytic activation (Fig. 6C). Interestingly, we observed two distinct preholoproteasome species, one containing both immature β5 and Pba1/2, and the other containing only immature β5 (Fig. 6C; Supplementary Fig. 4); the latter species may represent a later assembly intermediate on the path to mature 20S. The G205A mutant was strongly deficient in β5 catalytic activity, but without defects in the activities of β2 or β1 (Fig. 6D). Finally, we measured the rate of β5 maturation in cells by cycloheximide chase assay. The G205A mutant showed a strong defect in β5 maturation (Fig. 6E). These results suggest that G205 is critical for the function of the Ser/Asp pair of the catalytic pentad. In all species examined to date, that Ser residue is flanked by Gly on either side (Supplementary Fig. 1), an arrangement that may endow the Ser residue with local flexibility, which may be critical for its function. A similar Gly-Ser-Gly arrangement is also seen for β2 and β1 (Supplementary Fig. 1).
A Sequence alignment of human immunoproteasome β5i and yeast β5. The pentad serine residue is in cyan. B Phenotypic analysis of the indicated mutants, expressed from low-copy centromeric plasmids harboring the endogenous promoter/terminator elements in a strain where endogenous β5 is under the control of the pGAL1 promoter. In glucose-containing media, expression of the endogenous β5 locus is repressed and plasmid-derived β5 is the sole source of this protein. Plates were cultured at 30 °C for 2 (galactose), 1 (glucose), and 5 (canavanine 1.5 μg/mL) days. C Analysis of wild-type and mutant CP by native gel electrophoresis followed by immunoblotting with antibodies against α5, immature (propeptide-bearing) β5, and Pba1/2. D Proteasome activity assays using active site-specific fluorescent substrate probes. Background fluorescence has been subtracted and relative activity has been normalized to untreated controls. Individual points represent biologic duplicates. E Turnover of immature β5, as determined by cycloheximide chase analysis of whole extracts. Analysis was by SDS-PAGE followed by immunoblotting with the antibody that specifically recognizes immature β5. GAPDH, loading control. Asterisk, non-specific band. Similar results were obtained in 3 (B) and 2 independent experiments (C and E), respectively.
A separate patient-derived mutation, G201R, has been recently identified in β2i30. Again, this residue was conserved in both the constitutive human proteasome and in yeast (Gly191) (Fig. 7A). It was located near the end of the second helix in the helix-loop-helix motif responsible for positioning of the Ser/Asp pair and, indeed, a substitution at that position would be predicted to disrupt the Ser/Asp pair (Fig. 7B). We introduced the corresponding mutation, G191R, in yeast and it was lethal, as was the more conservative G191V substitution (Fig. 7C). A G191A mutation was viable, showed no growth defect under basal conditions, but did show a growth defect upon exposure to proteotoxic stress (Fig. 7C). Upon native gel analysis, the purified CP appeared structurally unremarkable without evidence of accumulation of preholoproteasome or of sub-20S species (Fig. 7D). Nevertheless, there was a significant and specific defect in β2’s proteolytic activity (Fig. 7E), although to a lesser extent than seen with β2-S158A and D195N mutants (Fig. 2e). Thus, Gly191 appears important for activity of the Ser/Asp pair.
A Sequence alignment of human immunoproteasome β2i and yeast β2. The pentad aspartate residue is in cyan. B Position of the corresponding G191 residue in yeast CP. An amino acid side chain at this position would be predicted to disrupt the Ser/Asp pair of the catalytic pentad. C Phenotypic analysis of the indicated mutants, expressed from low-copy centromeric plasmids harboring the endogenous promoter/terminator elements in a strain where endogenous β2 is under the control of the pGAL1 promoter. In glucose-containing media, expression of the endogenous β2 locus is repressed and plasmid-derived β2 is the sole source of this protein. Plates were cultured at 30 °C for 2 (galactose and glucose) and 7 (canavanine, 2 μg/mL) days. D Analysis of wild-type and mutant CP by native gel electrophoresis followed by immunoblotting with α5 antibody. E Proteasome activity assays using active site-specific fluorescent substrate probes. Background fluorescence has been subtracted and relative activity has been normalized to untreated controls. Individual points represent biologic duplicates. Similar results were obtained in 3 (C) and 2 independent experiments (D), respectively.
Discussion
Evidence supporting a catalytic pentad model for autocatalytic activation and proteolysis by the proteasome
The concept of a catalytic pentad mechanism for the proteasome was first proposed by Groll and colleagues15 (Supplementary Fig. 5-6), but experimental testing of this model has been limited to date. The model makes two main predictions: mutation of the Ser/Asp residues should compromise both autocatalytic activation and substrate proteolysis. Our data strongly support the model as we show that the Ser/Asp pair is required by all three active site subunits for both autocatalytic activation and substrate proteolysis.
A key question concerns the specificity of the Ser/Asp residues in controlling enzymatic activity, rather than a less specific structural role. Several lines of evidence support the notion of a specific function in catalytic regulation. First is the striking similarity between the Ser/Asp pair and the canonical Lys/Asp pair: both pairs are arranged in almost mirror image configuration around the active site threonine, are hydrogen bonded to the active site, are chemically capable of regulating the active site’s protonation state, and are strictly conserved across all three proteolytic subunits and across evolutionary time. Second, both the Lys/Asp and Ser/Asp pairs are regulated similarly during CP assembly and undergo an unstructured-to-structured transition at the moment of half-CP fusion (Fig. 1). This analogous regulation of the second unstructured loop could not be explained by our previous model in which we proposed that autocatalytic activation occurs through the alignment of the Lys/Asp/Thr triad14. But this dynamic structural configuration would seem perfectly arranged to control autocatalytic activation via simultaneous alignment of all 5 residues upon half-CP fusion. Third, mutation of an unrelated but equally well-positioned Ser residue in β2 did not impair autocatalytic activation (Supplementary Fig. 2), indicating specific functions of the Ser/Asp residues. An early study in Thermoplasma acidophilum, which utilizes a single β-subunit for its proteasomes, supports these findings: a D166A mutation (corresponding to the Asp residue studied here) showed essentially complete loss of β-subunit processing and proteolytic activity and this was also specific as an unrelated D105A mutation showed no such defects3.
The value of the catalytic pentad concept is that it helps explain key features of the proteasome and related threonine N-terminal nucleophile enzymes, and why they are so different from traditional proteases. First, it helps explain why these enzymes have their threonine active site exposed at the N-terminus of the protein. That is because unlike all other protease types, the active site utilizes two nucleophiles: threonine’s side chain hydroxyl and threonine’s free amino group. The importance of the free amino group is especially apparent from early observations that N-terminal acetylation, which would occur on threonine’s free amino group, inactivates the proteasome22,31. If there are two nucleophiles, then it stands to reason that both nucleophiles should be regulated in this cyclic fashion to ensure their continual activity, as described here. Second, it helps explain how the propeptides can be autocatalytically cleaved, which is essential to expose the N-terminal threonine in the first place. According to the model originally proposed by Groll and colleagues, the canonical Lys/Asp/Thr triad would be capable of initiating autocatalysis but would not be able to complete that process since initial attack by Lys on the propeptide results in the Lys residue becoming covalently bound to the propetide (Supplementary Fig. 5). So another nucleophile must be used to resolve that intermediate. This is the proposed role of threonine’s free N-terminus which polarizes a water molecule for this purpose, and which in turn requires tight regulation of its own protonation state by the Ser/Asp pair.
Use of a catalytic pentad in other N-terminal nucleophiles
In addition to the proteasome, the family of threonine N-terminal nucleophiles is broad and includes asparaginases, gamma-glutamyltransferases, ornithine acetyltransferases, and others9, raising important questions about the generality of the mechanism described here. We showed that a strikingly similar arrangement of residues was required for both autocatalytic activation and catalysis by the ornithine acetyltransferase Arg7. Indeed, the very nature of an N-terminal nucleophile may necessitate this type of arrangement, suggesting that catalytic pentads may be a defining feature of this class of enzymes. However, in addition to threonine N-terminal nucleophiles, there are also serine and cysteine N-terminal nucleophiles9, and so an important question for future work is whether these enzymes also require the pentad mechanism.
This work also raises interesting questions regarding Arg7’s mechanism of autocatalytic activation. The ability of Arg7 to self-activate upon expression in bacteria makes it unlikely that higher order assembly with other proteins triggers activation, as for the proteasome. It may be that translation-coupled protein folding triggers activation through alignment of the catalytic pentad. Alternately, activation may be triggered post-translationally through still to be discovered regulatory pathways. A major challenge is the lack of structural information on the pre-activation conformation of Arg7, which represents an important goal for future work.
Mechanistic basis of human proteasomopathies
In recent years, mutations in the proteasome have been recognized as the cause of an emerging family of human diseases30,32,33,34. There are >40 mutations in 7 different CP subunits or assembly chaperones, raising questions of how individual mutations cause proteasome dysfunction. Our work here has two limitations, namely that we modeled these patient-derived immunoproteasome mutations in yeast and that in both cases we were forced to study more conservative substitutions since the original mutations proved lethal in yeast. Nevertheless, our results suggest that defects in the function of the pentad’s Ser/Asp pair might be relevant to some proteasomopathies, likely through dual compromise of autocatalytic activation and substrate proteolysis as shown here. A human mutation has also been identified in the aspartate of the canonical catalytic triad (β2i-D56H)30, suggesting that both the Lys/Asp and Ser/Asp pairs may be dysregulated in human disease. In turn, the human mutations greatly informed our fundamental mechanistic studies as they suggested unexpected contributors to regulation of the pentad mechanism. A detailed understanding of the molecular pathogenesis of these diseases may lead the way to therapeutic approaches, not just for patients with inherited proteasomopathies, but perhaps for other diseases characterized by insufficient proteasome biogenesis or activity.
Methods
Strains, plasmids, and antibodies
Yeast strains and plasmids are listed in Supplementary Tables 2 and 3, respectively. Yeast strains were generated by standard homologous recombination. Plasmids were generated by standard restriction cloning in low-copy centromeric vectors utilizing the genes’ endogenous promoter and terminator sequences. Plasmids were verified by sequencing and, for wild-type plasmids, by complementation of the corresponding null (or expression-repressed) strains. Point mutations were introduced by site-directed mutagenesis and verified by sequencing.
To analyze β5, β2, and β1, respectively, the endogenous promoters were replaced by the pGAL1 promoter, which confers expression in galactose-containing media but is repressed by glucose. Wild-type or mutant plasmids were then expressed in these strains; upon promoter shut-off in glucose-containing media, the plasmid provides the sole source of expression for that gene. For CP purification, a C-terminal TEV-Protein A tag was inserted into the endogenous β4 locus.
To generate the Arg7-3xHA plasmid (pJH538), a C-terminal 3xHA tag was first inserted into the endogenous ARG7 locus; this strain showed wild-type growth on media lacking arginine. The entire locus, including promoter and terminator sequences, was then cloned into the low-copy centromeric vector ycPlac33.
The Arg7 bacterial expression plasmid was created by cloning the open reading frame for Arg7 into pET-Duet-1 (EMD Millipore), which provides for an N-terminal 6x-histidine tag. Point mutations were introduced by site-directed mutagenesis and verified by sequencing.
Yeast were cultured in YPD (1% yeast extract, 2% Bacto peptone, and 2% dextrose), YPGal (1% yeast extract, 2% Bacto peptone, and 2% galactose), or in selective synthetic media containing either glucose or galactose as appropriate. Bacterial BL21 (DE3) cells were cultured in LB medium supplemented with the appropriate antibiotic for plasmid selection.
The following antibodies were used: Alpha516 (1:2500), Immature β516 (1:2500), Pba1/235 (1:2500), Rpn421 (1:2000), GAPDH (Sigma #G9545; 1:5000), and HA (Sigma #12013819001; 1:2000).
CP purification and analysis
CP was affinity purified using IgG resin (MP Biomedicals; ICN55961) with high-salt (500 mM NaCl) washes as previously described16. Purified CP was analyzed by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) followed by Coomassie staining, or by native gel analysis using 3-8% Tris-acetate gels (Invitrogen; EA0375BOX) followed by immunoblotting.
Proteasome activity assays were performed with site-specific fluorescent probes as previously described36,37: for β5, suc-LLVY-AMC (Bachem #4011369); for β2, boc-LRR-AMC (Bachem #4017087); and for β1, z-LLE-AMC (Bachem #4037381).
Phenotypic analyses
Overnight cultures of yeast were normalized by optical density to an OD600 of 0.2 and spotted in three-fold serial dilutions onto the indicated plates. Cells were cultured at 30 °C for the indicated number of days. Canavanine (Sigma #C9758) was used to induce proteotoxic stress. For analysis of Arg7, synthetic media lacking arginine was used.
Analysis of whole cell extracts
Whole cell extracts were prepared from exponentially growing cells using a lithium acetate/sodium hydroxide method as described38,39. For cycloheximide chase analyses, cycloheximide (Sigma #C7698) was added to a final concentration of 100 μg/mL at time 0. Extracts were analyzed by SDS-PAGE followed by immunoblotting.
Bacterial Arg7 expression assays
Wild-type and mutant Arg7 plasmids were transformed into E. coli BL21 (DE3) cells and cultured at 30 °C. Protein expression was induced in exponentially growing cells by addition of IPTG to a final concentration of 1 mM, followed by additional culture for 3 h at 30 °C. Cells were normalized by optical density (OD600) and extracts prepared by suspension in 1X Laemmli loading buffer followed by boiling for 5 min. Analysis was by SDS-PAGE followed by Coomassie staining.
Structural and sequence analyses
Structures were analyzed and illustrated using UCSF Chimera X40. These include wild-type yeast CP (PDB: 5CZ4), the pre-15S CP assembly intermediate (PDB: 7LS6), and Streptomyces clavuligerus ArgJ (PDB: 2VZK), which is the ortholog of yeast Arg7. Intersubunit distances were determined using Coot41. Sequence analyses were performed using Clustal Omega and then manually annotated.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Additional data generated in this study are provided in the Supplementary Information and Source Data files. Structures used in this study are available in the PDB database under accession codes 5CZ4, 7LS6, and 2VZK. Source data are provided with this paper.
References
Brannigan, J. A. et al. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378, 416–419 (1995).
Seemuller, E. et al. Proteasome from thermoplasma acidophilum: a threonine protease. Science 268, 579–582 (1995).
Seemuller, E., Lupas, A. & Baumeister, W. Autocatalytic processing of the 20S proteasome. Nature 382, 468–471 (1996).
Groll, M. et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386, 463–471 (1997).
Buller, A. R. & Townsend, C. A. Intrinsic evolutionary constraints on protease structure, enzyme acylation, and the identity of the catalytic triad. Proc. Natl Acad. Sci. USA 110, 653–661 (2013).
Kraut, J. Serine proteases: structure and mechanism of catalysis. Ann. Rev. Biochem 46, 331–358 (1977).
Duggleby, H. J. et al. Penicillin acylase has a single-amino-acid catalytic centre. Nature 373, 264–268 (1995).
Zhiryakova, D. et al. Do N-terminal nucleophile hydrolases indeed have a single amino acid catalytic center? FEBS J. 276, 2589–2598 (2009).
Linhorst, A. & Lübke, T. The human ntn-hydrolase superfamily: structure, functions and perspectives. Cells 11, 1592 (2022).
Schmidtke, G. et al. Analysis of mammalian 20S proteasome biogenesis: the maturation of beta-subunits is an ordered two-step mechanism involving autocatalysis. EMBO J. 15, 6887–6898 (1996).
Heinemeyer, W., Fischer, M., Krimmer, T., Stachon, U. & Wolf, D. H. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 272, 25200–25209 (1997).
Chen, P. & Hochstrasser, M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 86, 961–972 (1996).
Schnell, H. M., Walsh, R. M., Rawson, S. & Hanna, J. Chaperone-mediated assembly of the proteasome core particle - recent developments and structural insights. J. Cell Sci. 135, jcs259622 (2022).
Velez, B. et al. Mechanism of autocatalytic activation during proteasome assembly. Nat. Struct. Mol. Biol. 31, 1167–1175 (2024).
Huber, E. M. et al. A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome. Nat Commun 7, 10900 (2016).
Schnell, H. M. et al. Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis. Nat. Struct. Mol. Biol. 28, 418–425 (2021).
Walsh, R. M. et al. Structure of the preholoproteasome reveals late steps in proteasome core particle biogenesis. Nat. Struct. Mol. Biol. 30, 1516–1524 (2023).
Adolf, F. et al. Visualizing chaperone-mediated multistep assembly of the human 20S proteasome. Nat. Struct. Mol. Biol. 28, 2024.01.27.577538 (2024).
Kock, M. et al. Proteasome assembly from 15S precursors involves major conformational changes and recycling of the Pba1-Pba2 chaperone. Nat. Commun. 22, 6123 (2015).
Xie, Y. & Varshavsky, A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl Acad. Sci. USA 98, 3056–3061 (2001).
Guerra-Moreno, A. & Hanna, J. Induction of proteotoxic stress by the mycotoxin patulin. Toxicol. Lett. 276, 85–91 (2017).
Jäger, S., Groll, M., Huber, R., Wolf, D. H. & Heinemeyer, W. Proteasome beta-type subunits: unequal roles of propeptides in core particle maturation and a hierarchy of active site function. J. Mol. Bio 291, 997–1013 (1999).
Crabeel, M., Abadjieva, A., Hilven, P., Desimpelaere, J. & Soetens, O. Characterization of the Saccharomyces cerevisiae ARG7 gene encoding ornithine acetyltransferase, an enzyme also endowed with acetylglutamate synthase activity. Eur. J. Biochem 250, 232–241 (1997).
Abadjieva, A., Hilven, P., Pauwels, K. & Crabeel, M. The yeast ARG7 gene product is autoproteolyzed to two subunit peptides, yielding active ornithine acetyltransferase. J. Biol. Chem. 275, 11361–11367 (2000).
Marc, F., Weigel, P., Legrain, C., Glansdorff, N. & Sakanyan, V. An invariant threonine is involved in self-catalyzed cleavage of the precursor protein for ornithine acetyltransferase. J. Biol. Chem. 276, 25404–25410 (2001).
Elkins, J. M., Kershaw, N. J. & Schofield, C. J. X-ray crystal structure of ornithine acetyltransferase from the clavulanic acid biosynthesis gene cluster. Biochem J. 385, 565–573 (2005).
Poli, M. C. Proteasome disorders and inborn errors of immunity. Immunol. Rev. 322, 283–299 (2024).
Watanabe, A., Yashiroda, H., Ishihara, S., Lo, M. & Murata, S. The molecular mechanisms governing the assembly of the immuno- and thymoproteasomes in the presence of constitutive proteasomes. Cells 11, 1580 (2022).
Arima, K. et al. Proteasome assembly defect due to a proteasome subunit beta type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proc. Natl Acad. Sci. USA 108, 14914–14919 (2011).
van der Made, C. I. et al. Expanding the PRAAS spectrum: de novo mutations of immunoproteasome subunit β-type 10 in six infants with SCID-omenn syndrome. Am. J. Hum. Genet 111, 791–804 (2024).
Arendt, C. S. & Hochstrasser, M. Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. EMBO 18, 3575–3585 (1999).
Brehm, A. et al. Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J. Clin. Invest 125, 4196–4211 (2015).
Ansar, M. et al. Biallelic variants in PSMB1 encoding the proteasome subunit β6 cause impairment of proteasome function, microcephaly, intellectual disability, developmental delay and short stature. Hum. Mol. Genet 29, 1132–1143 (2020).
Kanazawa, N. et al. Heterozygous missense variant of the proteasome subunit β-type 9 causes neonatal-onset autoinflammation and immunodeficiency. Nat. Commun. 12, 6819 (2021).
Wani, P. S., Rowland, M. A., Ondracek, A., Deeds, E. J. & Roelofs, J. Maturation of the proteasome core particle induces an affinity switch that controls regulatory particle association. Nat. Commun. 6, 6384 (2015).
Velez, B. et al. Rational design of proteasome inhibitors based on the structure of the endogenous inhibitor PI31/Fub1. Proc. Natl Acad. Sci. USA 120, e2308417120 (2023).
Rawson, S. et al. Yeast PI31 inhibits the proteasome by a direct multisite mechanism. Nat. Struct. Mol. Biol. 29, 791–800 (2022).
Weisshaar, N., Welsch, H., Guerra-Moreno, A. & Hanna, J. Phospholipase Lpl1 links lipid droplet function with quality control protein degradation. Mol. Biol. Chem. 28, 716–725 (2017).
Jochem, M. et al. Targeted degradation of glucose transporters protects against arsenic toxicity. Mol. Biol. Chem. 39, e00559–18 (2019).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Acknowledgements
The authors thanks J. Roelofs (U. of Kansas Medical Center) for the Pba1/2 antibody. This work was supported by NIH grant R01-GM144367 (to J.H.).
Author information
Authors and Affiliations
Contributions
D.F., J.H., M.P., B.V., and L.A. performed the biochemical aspects of the work. A.R., S.R., and R.W. contributed to the structural analysis. D.F., A.R., E.F.P., and J.H. prepared the figures. J.H. and D.F. wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fung, D., Razi, A., Pandos, M. et al. Evidence supporting a catalytic pentad mechanism for the proteasome and other N-terminal nucleophile enzymes. Nat Commun 16, 2949 (2025). https://doi.org/10.1038/s41467-025-58077-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58077-x