Abstract
The accessory protein Vpx from the red-capped mangabey or mandrill SIV (SIVrcm/mnd-2) lineage has been reported to increase HIV-1 infection in resting CD4+ T cells without affecting SAMHD1, a known target of Vpx in HIV-1 infection. This indicates that Vpx, in addition to SAMHD1, circumvents other restriction factors for lentiviruses. To identify potential restriction factors, this study examined cellular proteins interacting with Vpxrcm and found that keratin-72 (KRT72), an intermediate filament (IF) protein expressed in resting CD4+ T cells, is a host antiviral factor targeted by Vpx. Vpxrcm/mnd-2 lineages could strongly promote KRT72 degradation, resulting in increased HIV-1 infection in resting CD4+ T cells. We discovered that KRT72 restricts HIV-1 replication by sequestering incoming HIV-1 capsids in cytoplasmic IFs. With KRT72, the capsid cores of HIV-1 become attached to IFs, and their trafficking toward the nucleus is inhibited. In contrast, without KRT72, HIV-1 capsids are transported to the nucleus, leading to high levels of integrated HIV-1 DNA. Thus, KRT72 is a Vpx-counteracted antiviral factor that binds the incoming capsids to cytoplasmic IFs, restricting HIV-1 infection in resting CD4+ T cells.
Similar content being viewed by others
Introduction
HIV-1 replication is restricted at different postentry steps in resting CD4+ T and myeloid cells1,2,3,4,5,6,7,8,9. A major barrier in postentry steps is the block that limits reverse transcription (RT) of the incoming viral RNA genome2,4,10,11. Supplying the accessory lentiviral protein X (Vpx) to HIV-1-infected myeloid or resting CD4+ T cells with increased viral RT allows the progression of the HIV-1 replication cycle in these primary target cells12,13,14. In this context, the deoxynucleotide triphosphate (dNTP) triphosphohydrolase SAM and HD domain-containing protein 1 (SAMHD1) has been identified as a target of Vpx, and SAMHD1 depletes the dNTP pool to inhibit HIV-1 RT15,16,17,18,19,20. However, recent findings indicate the existence of Vpx targets other than SAMHD1 in resting CD4+ T cells21,22,23,24,25. Vpx proteins from the second Vpx+ lentiviral lineage, represented by simian immunodeficiency viruses (SIV) SIVrcm and SIVmnd-2, promote HIV-1 infection in CD4+ T cells but have no effect on SAMHD1 and the dNTP substrate concentration, suggesting that Vpxrcm and Vpxmnd-2 overcome another restriction factor in CD4+ T cells21. In addition to SAMHD1, Vpx from the major SIV strain in rhesus macaque (mac) counteracts this unknown restriction factor21. Vpxmac boosts HIV-1 infection in resting CD4+ T cells from a patient with Aicardi–Goutières syndrome who did not express SAMHD121. These findings indicate that lentiviral Vpx proteins antagonize a second SAMHD1-independent host restriction factor that acts at the RT level or as a potent barrier to the nuclear import of viral capsids, decreasing late RT21. Unlike Vpxmac, the Vpxrcm and Vpxmnd-2 lineage cannot enhance HIV-1 infection in primary macrophages21, suggesting that this unidentified restriction factor targeted by Vpx is expressed in resting CD4+ T cells but not macrophages.
To investigate this potential restriction factor in resting CD4+ T cells, this study established a coimmunoprecipitation (CoIP) assay to pull down the overexpressed FLAG-tagged Vpxrcm protein in resting CD4+ T cells treated with virus-like particle (VLP)-Vpxrcm. Keratin-72 (KRT72), a type II keratin, is highly expressed in resting CD4+ T cells associated with Vpxrcm when the proteasome is inhibited. During virus entry, KRT72 inhibits HIV-1 infection by sequestering incoming HIV-1 capsid cores in the cytoplasmic intermediate filament (IF) network it forms, preventing their transport into the nucleus, resulting in late RT and nuclear 2-long-terminal repeat (LTR) circular DNA downregulation. Vpxrcm and Vpxmnd-2, which affect the SAMHD1 protein, promote KRT72 degradation and improve HIV-1 infection in resting CD4+ T cells. These findings indicate that KRT72 is a host restriction factor in resting CD4+ T cells and that lentiviral Vpx protein degrade KRT72 to counteract this restriction, suggesting a novel potential anti-HIV-1 therapeutic strategy.
Results
KRT72 is a partner of Vpxrcm that is specifically expressed in resting CD4+ T cells
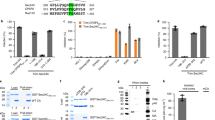
As Vpxrcm and Vpxmnd-2 overcome a SAMHD1-independent block to improve HIV-1 RT in early-phase HIV-1 replication in resting CD4+ T cells21, the objective was to explore whether using immunoprecipitation (IP) assays to pull down the Vpxrcm protein allowed the precipitation of an unknown restriction factor in resting CD4+ T cells when the proteasome is inhibited. This study adopted a similar strategy to improve the chances of isolating potential FLAG-tagged Vpx-interacting cellular proteins in resting CD4+ T cells, as used in a previous study to isolate FLAG-Gag-interacting cellular proteins in resting CD4+ T cells3. Supplementing a FLAG-tagged Vpx expression vector with VLP-Vpx may increase the possibility of isolated cellular proteins interacting with Vpx. In particular, virion-incorporated FLAG-tagged Vpx has the same effect as nontagged Vpx in promoting HIV-1 infection in resting CD4+ T cells11,21. A FLAG-tagged Vpxrcm expression vector was electroporated into resting CD4+ T cells treated with VLPs that carry FLAG-tagged Vpxrcm (VLP-Vpxrcm) with or without MG132, a proteasome inhibitor. Cells were lysed and subjected to IP assays with an anti-FLAG antibody to precipitate FLAG-tagged Vpxrcm from expression vectors and VLP (Fig. 1a). Liquid chromatography–mass spectrometry (LC–MS) was used to identify cellular proteins interacting with FLAG–Vpxrcm. Two independent experiments observed 61 cellular proteins in IPs with or without treatment with MG132 (Supplementary Table 1). Vpxrcm and Vpxmnd-2 do not promote HIV-1 infection in primary macrophages21; therefore, this potential restriction factor should not be expressed in macrophages. Using the above selection principles of the number of peptides >10 and fold change >2, KRT72 emerged as the only candidate among these candidate proteins. KTR1, KRT2, and KTR9 were observed in IPs; however, their levels did not show differences with or without MG132, and KRT1, KRT2, and KRT9 were not expressed in CD4+ T cells. Therefore, they were excluded from this selection. Compared to the entire gene expression profile of macrophages, KRT72 is a cellular protein found in resting CD4+ T cells but not macrophages (Supplementary Fig. 1a). Therefore, KRT72, a type II keratin that forms a complex filament system in the cytoplasm of eukaryotes26, is a likely candidate for a factor with which Vpxrcm interacts. The cytoskeleton of metazoan cells is mainly composed of three filamentous networks: microfilaments, microtubules, and IFs27. IFs generally form a cytoplasmic network that extends from the nucleus to the cell membrane, but their density is frequently highest in the subcortical cytoplasm near the nuclear envelope. IF proteins are expressed in a highly cell type-specific manner; keratins include acidic (type I) and basic (type II) keratins encoded by two gene families containing >50 genes in humans and mice, respectively28,29. Type I (28 members) and type II (26 members) keratins are proteins that form 10 nm filaments, which act as strict obligate heteropolymers. Keratins play an important role in cell–matrix interactions by stabilizing focal adhesion sites and contributing to generating traction force27.
a Schematic representation of KRT72 interaction with VLP-VPxrcm in resting CD4+ T cells. Resting CD4+ T cells were electroporated with a FLAG-tagged Vpx expression vector derived from SIVrcm. Six hours after electroporation, cells were treated with VLP-Vpxrcm for 8 h and treated with or without MG132 (0.75 µM) for 6 h. Cells were lysed for anti-FLAG IP to precipitate cellular Vpxrcm-interacting proteins. b, c KRT72 is expressed in resting CD4+ T cells. Total RNA was extracted from primary monocytes, MDMs, MDDCs, PMA-treated or untreated THP-1, and CD4+ T cells stimulated with or without CD3/CD28 and IL-2 for 72 h in established cell lines (293T, HeLa, and Jurkat cells). KRT72 transcript levels were measured using qPCR and normalized to GAPDH protein levels (b). Western blotting assessed KRT72 and GAPDH protein levels (c). ***P < 0.001 (two-tailed, unpaired Student’s t-test), and P-value is 0.0004. Data are the mean ± standard error of the mean (SEM) of three independent experiments. d–f KRT72 is degraded by Vpx in a proteasome-dependent manner. Resting CD4+ T cells were electroporated (d) or not with FLAG-KRT72 (e, f) and treated with VLP particles with or without Vpx from SIVmac239, SIVmnd-2, SIVrcm, or HIV-2Rod with or without MG132. Western blotting assessed exogenous FLAG-KRT72, endogenous KRT72, SAMHD1, and GAPDH protein levels using specific antibodies (d, e). Total RNA was extracted for qPCR to measure KRT72 transcript levels normalized to GAPDH protein levels (f). n.s. not significant (two-tailed, unpaired Student’s t-test) and P-values are 0.53, 0.84, 0.47, respectively. Data are the mean ± SEM of three independent experiments. Western blotting data are representative of three independent experiments. Source data are provided as a Source Data file.
Subsequently, we determined the expression of KRT72 in various host cells. KRT72 is highly expressed in resting CD4+ T cells (Fig. 1b, c), expressed to some extent in hair follicle stem cells (Supplementary Fig. 1b) but not stimulated CD4+ T lymphocytes, primary macrophages, dendritic cells, 293T cells, and Jurkat cells. KRT72 expression was also rapidly reduced when resting CD4+ T cells were stimulated (Supplementary Fig. 1c, d). KRT72 transcript and protein levels decreased rapidly upon stimulation. Interferons (IFN-α, -β, and -γ) did not induce KRT72 expression in primary CD4+ T cells (Supplementary Fig. 1e, f), suggesting that KRT72 is not a downstream molecule used by IFNs against HIV-1. Therefore, KRT72 is likely a protein interacting with Vpx that is highly expressed in resting CD4+ T cells.
Vpx proteins degrade KRT72
Next, we examined whether Vpx could target the KRT72 protein for degradation. First, VLP-encapsidated FLAG-tagged Vpx protein levels were determined (Supplementary Fig. 1g). Data demonstrated that these Vpx proteins derived from SIVmac239, SIVmnd-2, SIVrcm, and HIV-2Rod displayed similar levels inside VLPs (Supplementary Fig. 1h). To allow Vpx encapsulation in HIV-1 virions, this study generated an HIV-1 variant (HIV-1*NL4-3-Luc.R-E-) that can trans-package Vpx proteins via a Vpx-interaction motif in Gag p615. HIV-1* virions were purified, and the packaged different SIV-derived Vpx protein levels were examined. In Supplementary Fig. 1i, Vpx proteins of SIV were efficiently encapsidated into HIV-1* particles at similar levels.
Subsequently, we electroporated a FLAG-KRT72 expression vector in resting CD4+ T cells and then treated those cells with VLP-Vpx proteins from mac239, mnd-2, rcm, and HIV-2. The exogenous KRT72 level decreased with all Vpx proteins (Fig. 1d). Endogenous SAMHD1 was only degraded by Vpx from mac239 and HIV-2Rod; however, Vpxrcm and Vpxmnd-2 did not have such a degrading effect on SAMHD1. A decreased level of exogenous KRT72 protein was unlikely due to the reduced level of the transcript, as they came from this electroporated exogenous vector, which uses the cytomegalovirus (CMV) promoter to generate the exogenous FLAG-tagged KRT72 RNA. Using MG132 to inhibit proteasome activity could abrogate exogenous KRT72 or endogenous SAMHD1 protein degradation induced by these Vpx proteins. In addition, we directly examined endogenous KRT72 protein levels with these Vpx proteins. KRT72 was degraded by all Vpx proteins (Fig. 1e), whereas SAMHD1 was degraded only by Vpx from mac239 and HIV-2Rod but not Vpxrcm and Vpxmnd-221. MG132 could inhibit KRT72 or SAMHD1 degradation. These Vpx proteins did not affect KRT72 transcripts (Fig. 1f). Therefore, these findings suggested that KRT72 is a novel target for proteasomal degradation by Vpx in resting CD4+ T cells. Importantly, it is expected that mac239 and HIV-2 Vpx proteins degrade KRT72 and SAMHD1 proteins in a proteasome-dependent fashion, as mentioned in a previous study; that is, mac239 and HIV-2 Vpx proteins, in addition to SAMHD1, antagonize a second SAMHD-independent cellular restriction in resting CD4+ T cells21.
KRT72 restricts HIV-1 infection in resting CD4+ T cells
To explore the potential anti-HIV activity of KRT72, RNA interference (RNAi) strategies were used to silence KRT72 expression in resting CD4+ T cells, allowing us to evaluate the ability of the protein to restrict HIV-1 infection. Primary CD4+ T cells were first activated to promote the transduction of lentiviral vectors carrying short hairpin RNA (shRNA) for KRT72 (Supplementary Fig. 2a). The interleukin (IL)-2 concentration was gradually reduced, and cells were analyzed to detect HIV-1 infection after they returned to a quiescent state by determining the CD69, CD25, HLA-DR, and CellTrace violet levels (Supplementary Fig. 2b, c). This experimental setting challenged equal numbers of postactivated resting CD4+ T cells generated from five independent healthy donors with VSV-G-pseudotyped HIV-1 for 48 h with or without efavirenz (EFV). KRT72 silencing (Supplementary Fig. 2h) significantly increased HIV-1 replication only without EFV (Supplementary Fig. 2d–g). Viral late RT (Supplementary Fig. 2f) was restored without KRT72, whereas viral early RT (Supplementary Fig. 2e) was unaffected with or without KRT72. Viral nuclear 2-LTR circular DNA also increased without KRT72 (Supplementary Fig. 2g), suggesting that KRT72 is an inhibitor of late-phase HIV-1 RT.
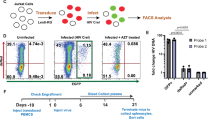
Furthermore, KRT72 small interfering RNAs (siRNAs) were directly electroporated in resting CD4+ T cells infected with VSV-G-pseudotyped HIV-1 with or without EFV (Fig. 2a–f). Using siRNAs to deplete KRT72 (Fig. 2f) increased HIV-1 infection in resting CD4+ T cells (Fig. 2a). Data revealed increased levels of HIV-1 late RT (Fig. 2c), integrated DNA (Fig. 2d), and 2-LTR circular DNA (Fig. 2e), whereas early RT (Fig. 2b) remained almost intact without KRT72. Importantly, when siRNA targeted to the 3′-untranslated region (UTR) of the KRT72 transcript was applied, inhibition of HIV-1 infection was rescued in siRNA-mediated KRT72-silenced primary resting CD4+ T cells electroporated with an exogenous KRT72 expression vector. Exogenous KRT72 expression was not affected by this siRNA because it did not affect KRT72 expression derived from its expression vector, into which the open reading frame (ORF) of KRT72 was cloned. KRT72 silencing did not result in the activation of resting CD4+ T cells by measuring CD69, CD25, HLA-DR, and CellTrace violet levels (Supplementary Fig. 3a, b), suggesting that enhanced HIV-1 replication is not due to cell stimulation.
a–f KRT72 knockdown restores HIV-1 infection in resting CD4+ T cells. Resting CD4+ T cells were electroporated with KRT72 or control siRNA with or without a nontagged KRT72 expression vector. Twenty-four hours after electroporation, cells were spinoculated with HIV-1NL4-3.Luc.R-E- (VSV-G) with or without EFV treatment (300 nM) and cultured with or without stimulation with CD3/CD28 plus IL-2. At 3 days postinfection (dpi), cells were lysed to measure luciferase activity (a), and genomic and circular viral DNA was extracted for qPCR to assess early RT (b), later RT (c), HIV-1 integration (d), and 2-LTR circular DNA (e). Data are the mean ± SEM of three independent donors. ***P < 0.001; **P < 0.01; *P < 0.05; n.s. (two-tailed, unpaired Student’s t-test). Aliquoted cells were also analyzed using flow cytometry to measure the surface levels of CD69, CD25, HLA-DR, and CellTrace violet (see Supplementary Fig. 3). Western blotting assessed KRT72, SAMHD1, and GAPDH protein levels using specific antibodies (f). Western blotting data are representative of three independent experiments. Source data are provided as a Source Data file.
To validate these results derived from VSV-G-pseudotyped HIV-1 reporter virus, we challenged resting CD4+ T cells with wild-type HIV-1NL4-3 in the presence and absence of KRT72 (Fig. 3a–e). KRT72 knockdown (Fig. 3c) increased the number of Gag+ cells (Fig. 3a, b). Importantly, the remaining levels of the KRT72 protein were inversely proportional to the HIV-1 Gag levels produced, suggesting that KRT72 is an inhibitor of WT HIV-1. This study also determined that KRT72 depletion increased late RT (Fig. 3d) and nuclear 2-LTR circular DNA (Fig. 3e) levels. KRT72 silencing increased late RT and 2-LTR circular DNA levels and viral late RT after infection. In general, KRT72 silencing promoted WT HIV-1 infection in resting CD4+ T cells. This inhibition of WT HIV-1 infection was rescued in endogenous KRT72-silenced resting CD4+ T cells (Fig. 3f–j) electroporated with siRNA targeting the 3′-UTR of the KRT72 transcript with an exogenous KRT72 expression vector. Exogenous KRT72 expression was not affected by this siRNA against the 3′-UTR of KRT72 (Fig. 3j). Reintroduction of exogenous KRT72 expression rescued this inhibition of WT HIV-1 Gag+ cells (Fig. 3f, g) and viral late RT (Fig. 3h) and 2-LTR circular DNA (Fig. 3i) in resting CD4+ T cells. These findings suggested that KRT72 inhibits HIV-1 infection in resting CD4+ T cells.
a–e KRT72 suppresses HIV-1 late RT and 2-LTR circular DNA levels in resting CD4+ T cells. Resting CD4+ T cells were electroporated with KRT72 or control siRNA. Twenty-four hours after electroporation, cells were spinoculated with HIV-1NL4-3 with or without EFV (300 nM). At 48 hpi, cells were analyzed by flow cytometry (a). FACS data are the mean ± SEM of three independent donors (b). Western blotting assessed KRT72, SAMHD1, and GAPDH protein levels using specific antibodies (c). At 24, 48, 72, and 96 hpi, genomic and circular viral DNA was extracted for qPCR to assess late RT (d) and 2-LTR circular DNA (e), respectively. Data are the mean ± standard deviation (SD) of three triplicates and are representative of three independent experiments. ***P < 0.001; **P < 0.01; *P < 0.05 (two-tailed, unpaired Student’s t-test), and P-values are 0.009, 0.003, 0.0044 respectively. f–j Resting CD4+ T cells were electroporated with siRNAs for KRT72 or control with or without an untagged KRT72 expression vector. Twenty-four hours after electroporation, cells were spinoculated with HIV-1NL4-3 with or without EFV (300 nM). At 2 dpi, cells were analyzed by flow cytometry (f). FACS data are the mean ± SEM of three independent donors (g). Genomic and circular viral DNA was extracted for qPCR to evaluate later RT (h) and 2-LTR circular DNA (i), respectively. Data are the mean ± SEM of three independent donors. ***P < 0.001; **P < 0.01; *P < 0.05; n.s. (two-tailed, unpaired Student’s t-test), and P-values are 0.0385, 0.0003, 0.0003, 0.0002, 0.0092, 0.0082, 0.0003, respectively. Western blotting assessed KRT72, SAMHD1, and GAPDH protein levels using specific antibodies (j). Western blotting data are representative of three independent experiments. Source data are provided as a Source Data file.
Vpx improves HIV-1 and HIV-2 infection by promoting KRT72 degradation
This study explored whether Vpxrcm and Vpxmnd-2 enhanced HIV-1 replication by reducing KRT72. Without VLP-Vpx treatment, KRT72 or SAMHD1 knockdown (Fig. 4a, lanes 1–3) increased HIV-1 replication in resting CD4+ T cells (Fig. 4b). With VLP-Vpxmac239 treatment, neither the siRNA targeting KRT72 nor SAMHD1 (Fig. 4a, lanes 4–6) resulted in upregulated HIV-1 replication. The Vpxmac239 protein severely degraded KRT72 and SAMHD1 proteins; therefore, treatment with their siRNAs was superfluous and did not affect their protein levels with Vpxmac239. Vpxrcm and Vpxmnd-2, unlike Vpxmac239, strongly promoted KRT72 degradation but not SAMHD1 (Fig. 4a, lanes 7–12). Consequently, using siRNA to deplete SAMHD1 enhanced HIV-1 replication, even with Vpxrcm and Vpxnmd2. In contrast, KRT72 siRNA treatment did not increase HIV-1 replication with Vpxrcm and Vpxmnd-2 because it already resulted in KRT72 protein degradation (Fig. 4a, lanes 7–12, and Fig. 4b). In general, because KRT72 is degraded by all Vpx proteins and KRT72 siRNA is redundant for KRT72 silencing with these Vpx proteins, it is not surprising that KRT72 siRNA treatment did not increase HIV-1 infection. As Vpxrcm or Vpxmnd-2 did not affect SAMHD1, SAMHD1 silencing using RNAi is expected to increase HIV-1 infection even with Vpxrcm or Vpxmnd-2 (Fig. 4a, lanes 7–12, and Fig. 4b). In contrast, because Vpxmac239 degraded KRT72 and SAMHD1, it is unsurprising that treatment with siRNAs to either of these genes did not enhance HIV-1 infection (Fig. 4a, lanes 4–6). To substantiate the results derived from VLP-Vpx, the HIV-1* variant (capable of trans-packaging Vpx proteins via a Vpx-interaction motif in Gag p6) carrying different Vpx proteins (Supplementary Fig. 1i) was also used to infect resting CD4+ T cells with or without KRT72 or SAMHD1. Vpx from SIVmac239 or SIVrcm could promote HIV-1 replication in resting CD4+ T cells (Fig. 4c), and both Vpx proteins promoted KRT72 degradation (Fig. 4d, lane 1 vs. 4 and 7). SAMHD1 decreased only with Vpxmac239 but not Vpxrcm (Fig. 4d, lane 1 vs. 4 and 7). With Vpxmac239 or Vpxrcm, KRT72 silencing did not promote HIV-1 replication (Fig. 4c), as both Vpx proteins reduced KRT72 protein levels, and KRT72 siRNA treatment would result in redundant reduction (Fig. 4d, lane 4 vs. 5 and 7 vs. 8). In contrast, SAMHD1 depletion could increase HIV-1 replication only with Vpxrcm because Vpxrcm did not affect SAMHD1 (Fig. 4d, lane 1 vs. 7). With Vpxmac239, SAMHD1 siRNA did not promote HIV-1 replication, as SAMHD1 siRNA could not reduce its protein with Vpxmac239 (Fig. 4d, lane 4 vs. 6). Therefore, these findings were consistent with those derived from HIV-1* virions. Thus, Vpxrcm and Vpxmnd-2 enhance HIV-1 infection via KRT72 degradation.
a, b Vpx promotes KRT72 degradation to restore HIV-1 infection in resting CD4+ T cells. Resting CD4+ T cells were pretreated with VLP with or without Vpx derived from SIVmac239, SIVrcm, and SIVmnd-2 isolates. Cells were electroporated with siRNAs for KRT72, SAMHD1, or control. Twenty-four hours after electroporation, cells were spinoculated with HIV-1NL4-3.Luc.R-E- (VSV-G) with or without EFV (300 nM). At 3 dpi, cells were lysed for Western blotting to assess KRT72, SAMHD1, and GAPDH protein levels using specific antibodies (a) or measure luciferase activity (b). c, d CD4+ T cells were electroporated with siRNAs targeting KRT72, SAMHD1, or control. Twenty-four hours after electroporation, cells were spinoculated with HIV-1*NL4-3-Luc.R-E- (VSV-G) carrying or not carrying Vpx proteins derived from SIVmac239 or SIVrcm with or without EFV (300 nM). At 3 dpi, cells were lysed to measure luciferase activity (c) or determine KRT72, SAMHD1, and GAPDH protein levels using their respective antibodies (d). ***P < 0.001, **P < 0.01, *P < 0.05, n.s. (two-tailed, unpaired Student’s t-test). Data are the mean ± SEM of three or five independent experiments. e, f Vpxmac293 mutants L25A, H39A, and W56A but not Q76A promote KRT72 degradation to enhance HIV-1 infection in resting CD4+ T cells. Resting CD4+ T cells were spinoculated with 100 ng HIV-1*NL4-3-Luc.R-E- (VSV-G) carrying or not carrying WT or mutant FLAG-tagged (L25A, H39A, W56A, or Q76A) Vpxmac293 with or without EFV treatment (300 nM). At 2 dpi, cells were lysed to measure luciferase activity (e) or for Western blotting to assess KRT72, SAMHD1, Vpx, and GAPDH protein levels using specific antibodies (f). **P < 0.01; n.s. (two-tailed, unpaired Student’s t-test). Data are the mean ± SEM of three independent experiments. Western blotting data are representative of three independent experiments. Source data are provided as a Source Data file.
Notably, Vpxmac239 mutants L25A, H39A, and W65A, which neither affect SAMHD1 nor elevate dNTP levels, enhanced HIV-1 infection in primary resting CD4+ T cells21. Whether these mutants could degrade KRT72 to promote HIV-1 infection was investigated. These mutants, including L25A, H39A, and W65A, except Q76A, enhanced HIV-1* infection (Fig. 4e, f). L25A, H39A, and W65A mutants degraded KRT72, but none affected SAMHD1. WT Vpxmac239 degraded SAMHD1 and KRT72; however, the Q69A mutant that could not interact with CRL4DCAF121 did not affect KRT72, suggesting that Vpx-mediated KRT72 degradation is dependent on the interaction with CRL4DCAF1. Therefore, Vpxrcm and Vpxmnd-2 enhance HIV-1 infection via KRT72 degradation.
Because Vpx is not physically present in HIV-1, this study also examined WT and Vpx-defective HIV-2 infection of resting CD4+ T cells. KRT72 depletion only increased Vpx-defective but not WT HIV-2 infection (Supplementary Fig. 4a, b). Late RT was also increased with WT HIV-2 compared to its Vpx-defective counterpart. This result was consistent with previous data (Fig. 1d–f) that VLP-Vpx carrying the Vpx protein derived from HIV-2 was used to treat resting CD4+ T cells. KRT72 siRNA treatment did not increase HIV-2 infection because KRT72 siRNA is redundant for KRT72 silencing with Vpx (Supplementary Fig. 4b). Without Vpx, neither SAMHD1 nor KRT72 was degraded. Therefore, KRT72 depletion using siRNA could partially rescue Vpx-defective HIV-1 infection as SAMHD1 was unaffected. Moreover, we were unable to detect the produced HIV-1 virions in the culture media, suggesting that the late stage of HIV-2 replication is blocked in resting CD4+ T cells and that KRT72 functions as an inhibitor during the early stage of HIV-2 replication. Because the HIV-2Rod Vpx mutant that only degrades KRT72 but not SAMHD1 proteins was unknown, this study took advantage of the above SIVmac239 mutant Vpx proteins that only degraded KRT72 but not SAMHD1. The vpx gene of HIV-2Rod was replaced with WT or SIVmac239 mutant Vpx genes, including L25A, H39A, W56A, and Q76A, to generate the HIV-2-Vpxmac293 virus. These HIV-2-Vpxmac293 virions (L25A, H39A, W56A, and Q76A) were used to infect resting CD4+ T cells. Mutant L25A, H39A, and W56A HIV-2-Vpxmac293 infection resulted in KRT72 but not SAMHD1 degradation, whereas WT resulted in SAMHD1 and KRT72 degradation (Supplementary Fig. 4c, d). In sharp contrast, the Q76A mutant HIV-2 induced neither SAMHD nor KRT72 degradation, suggesting that this degradation depends on CRL4DCAF1. These findings strengthened our proposal that Vpx from HIV-2 and SIVmac239 could degrade SAMHD1 and KRT72 to promote infection in resting CD4+ T cells. Therefore, Vpx could promote KRT72 degradation to restore HIV-1 and HIV-2 infection.
KRT72 restricts HIV-1 infection in a SAMHD1-independent manner
After confirming that KRT72 acted as a Vpx-targeted HIV-1 inhibitor in resting CD4+ T cells, we wanted to determine whether this inhibition was independent of SAMHD1, as suggested by previous findings21. To achieve this, HIV-1-infected resting CD4+ T cells were treated with dNTP precursors (dNs) to inhibit the antiviral activity of SAMHD1. Simultaneously, we utilized siRNA to deplete KRT72 in HIV-1-infected cells. In the absence of dNTP, silencing of SAMHD1 increased HIV-1 infection (Supplementary Fig. 5a–e). SAMHD1 siRNA treatment did not promote HIV-1 infection in the presence of dNTPs (Supplementary Fig. 5a), suggesting that dNTPs abolished SAMHD1-mediated inhibition. However, in the presence and absence of dNTPs, silencing KRT72 resulted in a similarly increased level of HIV-1 infection of 5.8- and 6.09-fold, respectively, suggesting that dNTP was not related to the anti-HIV-1 activity of KRT72. We also examined viral late RT and 2-LTR circular DNA levels and found that silencing KRT72 resulted in their upregulation in the absence or presence of dNTPs (Supplementary Fig. 5b, c). However, SAMHD1 silencing only increased late RT and 2-LTR circular DNA in the absence of dNTP treatment, indicating that dNTP treatment abrogated the anti-HIV activity of SAMHD1. Furthermore, we also analyzed the surface markers CD69, CD25, and HLA-DR in these cells. As shown in Supplementary Fig. 5d, resting CD4+ T cells were not activated by siRNA or dNTP treatment, suggesting that the increase in HIV-1 infection was not due to cell stimulation. Moreover, neither the level of SAMHD1 nor KRT72 protein was affected by dNTP treatment (Supplementary Fig. 5e). Therefore, collectively, these findings indicate that the anti-HIV-1 activity of KRT72 was not dependent on SAMHD1.
KRT72 is a postentry inhibitor of HIV-1 in target cells
Subsequently, we investigated the molecular mechanism by which KRT72 inhibits HIV-1 replication. First, KRT72 did not inhibit HIV-1 production in producer cells (Supplementary Fig. 6a), indicating that KRT72 can restrict HIV-1 in the early replication phase. This study also examined whether KRT72 inhibits virion entry using the BlaM-Vpr-based viral entry assay (Supplementary Fig. 6b–e). KRT72 did not affect HIV-1 entry but significantly inhibited late RT levels. Thus, this restrictive effect of KRT72 to HIV-1 should occur after viral entry. 293 T cells were infected with VSV-G-pseudotyped HIV-1, and KRT72 restricted HIV-1 replication in a dose-dependent manner (Fig. 5a). Viral late RT and nuclear 2-LTR circular DNA was significantly inhibited with KRT72. In contrast, early RT levels were not significantly affected by KRT72. Next, overexpressed MX2 and KRT72 proteins were compared to investigate the KRT72-mediated suppression of HIV-1, as SAMHD1 did not affect HIV-1 in dividing cells such as activated CD4+ T or 293T cells15,30. KRT72 and MX2 inhibited HIV-1 replication at different doses of virion particles used to infect target cells (Fig. 5b, c). Integrated nuclear viral and 2-LTR circular DNA decreased with KRT72 and MX2. However, late RT was reduced with KRT72 but not MX2 expression in target cells, indicating that KRT72, unlike MX2, adopts a different molecular mechanism to restrict HIV-1 replication. Afterward, we examined KRT72 with replication-competent HIV-1 in HeLa-P4 cells expressing the CD4 receptor and coreceptors CXCR4 and CCR5. KRT72 inhibited different HIV-1 isolates BaL, AD8, or 89.6, resulting in late viral RT downregulation, but early RT remained almost intact (Supplementary Fig. 6f–h). This study determined the inhibitory effect of KRT72 on HIV-1 infection in stimulated CD4+ T cells with or without KRT72. KRT72 inhibited HIV-1 replication by reducing late RT and 2-LTR DNA, whereas early RT was not significantly affected (Fig. 5d). To support this inhibitory effect, whether exogenous KRT72 could affect HIV-1 spread in stimulated CD4+ T lymphocytes was also examined. KRT72 restricted the spread of CXCR4-tropic HIV-1 in dividing CD4+ T cells (Supplementary Fig. 6i–k). KRT72 expression did not visibly affect primary CD4+ T lymphocyte proliferation (Supplementary Fig. 6i), indicating that this inhibitory effect was due to the presence of KRT72.
a 293T cells were transfected with FLAG-tagged KRT72 or mock expression constructs at the indicated doses. At 24 hpi, 50 ng HIV-1NL4-3.Luc.R-E- was used to infect cells (VSV-G). At 2 dpi, cells were lysed to measure the luciferase reporter activity, and total DNA was extracted for qPCR to measure early RT, late RT, and 2-LTR circular DNA. **P < 0.01; *P < 0.05; n.s. (two-tailed, unpaired Student’s t-test) and P-values are 0.0006, 0.65, 0.0008, and 0.0006, respectively. Data are the mean ± SEM of three independent experiments. b, c 293T cells were transfected with FLAG-tagged KRT72, MX2, or mock expression constructs. At 24 hpi, cells were infected with 1, 5, 10, 25, 50, or 100 ng HIV-1NL4-3.Luc.R-E- (VSV-G) with or without EFV (300 nM). At 2 dpi, cells were lysed and the activity of the luciferase reporter was measured. Genomic and circular viral DNA were extracted for qPCR to assess HIV-1 late RT, HIV-1 integration, and 2-LTR circular DNA, respectively, and the results obtained from 1 ng HIV-1 infection with mock were set at a relative level of 1. Data are the mean ± SD of three triplicates and are representative of three independent experiments (b). Western blotting was conducted to assess exogenous protein expression using specific antibodies (c). d KRT72 inhibits HIV-1 infection in primary CD4+ T cells. Stimulated CD4+ T cells were electroporated with FLAG-tagged KRT72 or mock expression vectors at the indicated doses. Twelve hours after electroporation, cells were infected with 10 ng HIV-1NL4-3.Luc.R-E- (VSV-G). At 2 dpi, cells were lysed to measure the luciferase reporter activity, and total DNA was extracted for qPCR to measure early RT, late RT, and 2-LTR circular DNA. **P < 0.01; *P < 0.05; n.s. (two-tailed, unpaired Student’s t-test), and P-values are 0.0002, 0,79, 0.0005, and 0.0019, respectively. Data are the mean ± SEM of three independent experiments. Western blotting data are representative of three independent experiments. Source data are provided as a Source Data file.
Similarly, KRT72 inhibited the spread of CCR5-tropic HIV-1 in nondividing primary macrophages (Supplementary Fig. 6l–n). This study measured the lactate dehydrogenase (LDH) release from these lentiviral-transduced or nontransduced macrophages as a measure of toxicity. KRT72 did not lead to any significant toxicity in macrophages (Supplementary Fig. 6n), suggesting that cell toxicity does not induce poor HIV-1 proliferation with KRT72. KRT72 inhibited viral late RT in primary macrophages using VSV-G-pseudotyped reporter HIV-1 for a single-cycle infection (Supplementary Fig. 6o, p), indicating that KRT72 employs a similar molecular mechanism to inhibit HIV-1 in CD4+ T cells and macrophages. Therefore, KRT72 inhibits HIV-1 in primary CD4+ T cells and macrophages.
KRT72 restricts HIV-1 spread in CD4+ T cells of HIV-1-infected individuals ex vivo
To substantiate the notion that KRT72 inhibits HIV-1 infection, total CD4+ T cells were first isolated from two untreated HIV-1-infected individuals (see their backgrounds in Supplementary Table 2), and CD4+ T cells were stimulated for lentiviral expression vector transduction (Supplementary Fig. 7a). After the produced virus was washed out, cells were divided into two groups for lentiviral expressing KRT72-green fluorescent protein (GFP) or GFP with puromycin. KRT72 inhibited HIV-1 spread in these CD4+ T cells ex vivo (Supplementary Fig. 7c, d). No obvious influence on CD4+ T-cell proliferation was observed among KRT72-expressing CD4+ T cells (Supplementary Fig. 7b), and decreased viral titers were likely due to exogenous KRT72 expression. This study also investigated whether KRT72 restricts HIV-1 spread in CD4+ T cells isolated from four antiretroviral therapy (ART)-treated HIV-1-infected individuals (people Living with HIV) whose viral loads were <50 copies/mL (see their backgrounds in Supplementary Table 2). After their CD4+ T cells were stimulated, lentiviral expression vectors of KRT72 were transduced into these cells. Exogenous KRT72 did not affect their CD4+ T-cell proliferation (Supplementary Fig. 7e), whereas KRT72 inhibited HIV-1 spread (Supplementary Fig. 7f, g). Therefore, KRT72 inhibits HIV infection in CD4+ T cells ex vivo.
Vpx counteracts KRT72 to promote HIV-1 and HIV-2 infection
The objective was to unravel whether Vpx could restore HIV-1 replication in KRT72-overexpressing cells. First, 293T cells were transfected with a KRT72-expressing vector to mimic similar levels in resting CD4+ T cells. Next, VLP-Vpxrcm was used to treat KRT72-expressing 293T cells, and VLP-Vpxrcm could promote KRT72 but not MX2 degradation (Supplementary Fig. 8a, b). SAMHD1 was not examined because it does not affect HIV in dividing cells such as 293T cells. Two types of HIV-1* virions (NL4-3-Luc.R-E- and NL4-3-ΔEnv-EGFP) carrying different Vpx proteins were used to infect KRT72-expressing 293T cells, and all Vpx proteins could increase HIV-1 replication with KRT72 (Supplementary Fig. 8c, f). In contrast, none of the Vpx proteins significantly affected HIV-1 replication without KRT72. All Vpx proteins degraded KRT72 (Supplementary Fig. 8d, g). The mCherry monitor protein was not affected by Vpx (Supplementary Fig. 8g), suggesting that Vpx targets KRT72 in 293T cells. Total RNA was extracted to determine KRT72 transcripts. No Vpx affected KRT72 transcripts derived from expression vectors (Supplementary Fig. 8e). Thus, Vpx could also rescue HIV-1 replication in KRT72-overexpressing cell lines when KRT72 expression is similar to those in resting CD4+ T cells. Next, when mutant SIVmac293 Vpx (L25A, H39A, W56A, and Q76A) with exogenous KRT72 in 293T cells was examined, we obtained similar results as in resting CD4+ T cells. Mutant and WT KRT72, except Q76A mutant, induced KRT72 degradation and promoted HIV-1 infection (Supplementary Fig. 8h, i), whereas the mCherry protein was unaffected. This result was reproduced when WT HIV-1* was used to infect KRT72-expressing 292T cells (Supplementary Fig. 8j, k), suggesting that the SIVmac239 Vpx mutant could degrade KRT72 in target cells.
Moreover, WT or Vpx-defective HIV-2 infection of KRT72-expressing 293T cells was examined. When a KRT72 expression vector at 0.5 or 1.0 μg was used, KRT72 could not inhibit WT but inhibited Vpx-defective HIV-2 infection (Supplementary Fig. 9a). KRT72 but not MX2 was degraded only with Vpx (Supplementary Fig. 9b). WT HIV-2 was inhibited by KRT72 when a KRT72 expression vector was transfected at 1.5 μg, indicating that Vpx could not counter such high exogenous KRT72 levels. KRT72 expression vectors at 1.0 μg were electroporated in stimulated CD4+ T cells, and these KRT72-expressing cells were infected with HIV-2 or HIV-2Vpx- virions. Similar results were obtained, as Vpx present in WT HIV-2 promoted KRT72 degradation to enhance infection (Supplementary Fig. 9c, d) and HIV-2-Vpxmac293 infected KRT72-expressing 293T cells as in resting CD4+ T cells. WT, L25A, H39A, and W56A, except Q76A mutant, SIVmac293 Vpx resulted in KRT72 degradation to promote HIV-2 infection (Supplementary Fig. 9e, f). Therefore, Vpx can counteract KRT72 to promote HIV-1 and HIV-2 infection.
Next, our goal was to address whether KRT72 exerts broad antiviral activity by challenging KRT72-overexpressing cells with VSV-G-pseudotyped SIV, equine infectious anemia virus (EIAV), and murine leukemia virus (MLV) reporter virus (Supplementary Fig. 9g–i). KRT72 inhibited the replication of all these viruses in target cells. Although KRT72 strongly inhibited EIAV (Supplementary Fig. 9h) and MLV (Supplementary Fig. 9i) replication in target cells, KRT72 had a relatively mild inhibitory effect on SIV infection (Supplementary Fig. 9g). Most likely, SIV Vpx proteins can compromise overexpressed KRT72-mediated inhibition of transfected KRT72 vectors in low dosages, which require future investigation. In contrast, EIAV and MLV did not have Vpx proteins to diminish the antiviral activity of exogenous KRT72 in target cells. Therefore, KRT72 may have broad antiviral activity against retroviruses.
Subsequently, we cloned all type II keratins (Supplementary Fig. 10a) and examined their potential restriction of HIV-1 activity. Only KRT72 in type II keratins had a clear inhibitory effect on HIV-1 infection compared to the positive control MX231,32, a genuine host restriction factor inhibiting HIV-1 capsid translocation into the nucleus (Supplementary Fig. 10b, c). Interestingly, KRT74 also had a mild HIV-1 inhibitory effect; however, its expression was significantly lower than that of KRT72 when we compared the expression of types I and II keratins in stimulated and resting CD4+ T cells (Supplementary Fig. 10d). Only KRT72 and KRT73 in the keratins were similarly highly expressed in the resting (but not stimulated) CD4+ T cells. However, KRT73 had no effect on HIV-1 infection (Supplementary Fig. 10b). These data support that KRT72 functions as an inhibitor of proteins within the KRT family during the postentry stage of HIV-1 in target cells.
KRT72 sequesters incoming HIV-1 capsid cores in the cytoplasm
After confirming that mCherry-tagged KRT72 exhibits comparable antiviral activity to FLAG-tagged or nontagged KRT72 (Supplementary Fig. 11a), the subcellular localization of mCherry-tagged KRT72 and MX2 was examined to determine the mechanism by which KRT72 inhibits HIV-1. mCherry-tagged or nontagged KRT72 typically forms a cytoplasmic network that stretches from the nucleus to the cell membrane, and MX2 is primarily localized to the nuclear membrane (Supplementary Fig. 11b, c). Next, cytoplasmic and nuclear fractions were separated, and KRT72 was observed to be mainly located in the cytoplasm. In contrast, MX2 was located in the cytoplasm and nucleus (Supplementary Fig. 11d). Of course, this study did not exclude the possibility that KRT72 could still be connected with the nucleus to some extent, but the main KRT72 should be in the cytoplasm. Therefore, we proposed that KRT72 is mainly located in the cytoplasm, suggesting that KRT72 may inhibit HIV-1 trafficking toward the nucleus.
In particular, the correct choreography uncoating of HIV-1 capsid cores is essential for RT and nuclear imports33,34, and postentry trafficking of the core across the cytoplasm and into the nucleus is also critical for HIV-1 uncoating and RT34,35,36,37. Thus, it was hypothesized that KRT72 may form a filamentous network in the cytoplasm, sequestering HIV-1 cores to inhibit their trafficking to the nucleus and reducing viral RT and nuclear import. To verify these findings, whether KRT72 colocalizes with incoming HIV-1 cores was further investigated. Ectopically expressed mCherry-KRT72 proteins colocalized preferentially with INmNG-labeled HIV-1 cores in the cytoplasm at 6 and 9 h postinfection (hpi) (Supplementary Fig. 12a, b; Fig. 6a, b). The entry of INmNG-labeled cores into the nucleus was efficiently observed at 6 and 9 hpi without KRT72. In contrast, viral cores were sequestered in the KRT72 filament network in the cytoplasm and were nearly incapable of entering the nucleus when KRT72 was present. Without VSV-G, no obvious signals were detected from labeled HIV-1 cores, indicating that HIV-1 particle cell phagocytosis did not occur in this system. The number of viral cores in the cytoplasm and nucleus were determined with or without KRT72 (Fig. 6c). KRT72 significantly reduced the number of nuclear viral cores and increased the number of cytoplasmic viral cores (Fig. 6c), whereas the total number of viral cores was similar with and without KRT72. In Supplementary Fig. 12c, when the cell membrane was drawn by a white dot line, KRT72 was mainly localized in the cytoplasm colocalized with viral cores. To substantiate these results, WT HIV-1 was used to infect target cells, and an anti-p24 antibody was used to directly label incoming WT HIV-1 cores at 12 hpi with raltegravir to prevent the possibility that the anti-p24 antibody can label the Gag produced during HIV-1 infection. In Supplementary Fig. 12d, e, KRT72 was colocalized with incoming HIV-1 cores in the cytoplasm, and KRT72 inhibited the trafficking of viral cores to move to the nucleus. KRT72 similarly increased the number of cytoplasmic HIV-1 cores while decreasing the number of nuclear cores (Supplementary Fig. 12f). KRT72 inhibited viral cores from entering the nucleus. Therefore, KRT72 sequesters HIV-1 cores in the cytoplasm during the early stages of infection.
a–c KRT72 restricts HIV-1 cores trafficking toward the nucleus. 293T cells were transfected with mCherry-tagged KRT72 or mock expression constructs. At 24 hpi, cells were infected with HIV-1 labeled with INmNG with or without VSV-G. At 6 hpi (see Supplementary Fig. 12a, b) or 9 hpi, cells were analyzed for capsid colocalization with KRT72 and nuclei were stained with DAPI. Bar, 10 μm. Images (a) and quantification (b) of KRT72-mCherry signals associated with INmNG-labeled from 70 cells. Error bars, mean ± SEM. Micrograph data are representative of three independent experiments. Quantitative results of INmNG-labeled puncta (c) in the cytoplasm or nucleus per cell based on the analysis of 30 cells with or without KRT72. Data are the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01; ****P < 0.0001; n.s. (two-tailed, unpaired Student’s t-test), and P-values in (c) are 0.96, 0.0023, and 0.0107, respectively. d, e KRT72 binds to HIV-1 cores during HIV-1 infection. 293T cells were transfected with a FLAG-tagged KRT72 expression construct. At 24 hpi, cells were infected with HIV-1NL4-3.Luc.R-E- (VSV-G); at 3 hpi, cells were washed thrice with PBS, lysed, and treated with or without benzonase for IP assays with an anti-FLAG antibody to precipitate KRT72 (d) or an anti-p24 antibody to precipitate incoming capsids (e). Western blotting was performed to detect FLAG-KRT72 and p24CA using specific antibodies. f, g KRT72 binds to HIV-1 cores in HIV-1-infected resting CD4+ T cells. Resting CD4+ T cells were spinoculated with HIV-1NL4-3 with raltegravir at 300 nM; at 12 h after spinoculation, cells were washed thrice with PBS, lysed, and treated with or without benzonase for IP assays with anti-KRT72 (f), anti-p24 (g), or IgG. Western blotting was performed to detect KRT72 and p24CA using specific antibodies. Western blotting data are representative of three independent experiments. Source data are provided as a Source Data file.
KRT72 interacts with HIV-1 incoming cores
To understand whether the KRT72 protein binds to HIV-1 capsid cores, KRT72 ectopically expressing 293T cells were infected with HIV-1 for 3 h, and CoIP assays were used to precipitate KRT72 (Fig. 6d) or viral cores (Fig. 6e). KRT72 and incoming HIV-1 cores precipitated one another in a reciprocal manner. This interaction was independent of cellular nucleic acids, as it could occur with benzonase. In contrast, neither KRT71 nor KRT73 from the KRT family that lacked anti-HIV-1 activity could bind to viral cores (Supplementary Fig. 13a, b), indicating that KRT72 is only one member binding to viral cores. Importantly, this study sought to determine whether this interaction also occurred in HIV-1-infected resting CD4+ T cells. Through CoIP assays, KRT72 interacted with HIV-1 incoming cores in a nucleic acid-independent manner (Fig. 6f, g), suggesting that endogenous KRT72 associates with HIV-1 cores in resting CD4+ T cells. In particular, in these experiments, raltegravir was used to inhibit probably productive infection to produce Gag progeny; therefore, the detected capsid proteins (CA) in pulldown precipitates should come from capsid cores incoming from HIV-1. Therefore, KRT72 can bind to incoming cores during the early stages of infection. Next, this study determined whether this interaction between KRT72 and viral cores was important for the anti-HIV activity of KRT72. This binding of KRT72 to HIV-1 cores is critical for its antiviral activity (Supplementary Fig. 13c–i). The KRT72 N-terminal mutant (Δ1–124) (Supplementary Fig. 13c), which did not interact with HIV-1 cores (Supplementary Fig. 13f, g), could not inhibit HIV-1 infection in target cells (Supplementary Fig. 13d). In contrast, similar to WT KRT72, the KRT72 mutant (Δ438–511) (Supplementary Fig. 13c), which interacted with viral cores (Supplementary Fig. 13h, i), also caused HIV-1 inhibition (Supplementary Fig. 13d), suggesting that KRT72 binding to HIV-1 cores is essential for its antiviral activity. Unlike WT KRT72, HIV-1 cores could efficiently traffic into the nucleus with the KRT72 N-terminal mutant (Δ1–124) (Supplementary Fig. 13j, k), confirming that this mutant does not have a binding activity to viral cores.
Furthermore, fate-of-capsid assays were also used to examine the interaction between WT KRT72 and its mutant (Δ1–124) and HIV-1 cores in viral-infected 293T cells parallel to MX2. WT KRT72 and MX2 interacted with HIV-1 cores in target cells (Supplementary Fig. 14a). In contrast, the KRT72 N-terminal mutant (Δ1–124) did not interact with HIV-1 cores. Without HIV-1, KRT72 was not found in pellets. To determine whether KRT72 could bind to isolated HIV-1 cores in cell lysates, WT HIV-1 particles were purified, and their envelopes were removed before exposing viral cores to KRT72, its mutant (Δ1–124), and MX2. In Supplementary Fig. 14b, ectopically expressed KRT72 and MX2 proteins in cell lysates bound to isolated HIV-1 cores, whereas the KRT72 mutant (Δ1–124) did not exhibit this interaction, indicating that KRT72, like MX2, can bind to HIV-1 capsids in target cells, a bona fide HIV-1 capsid-interacting host restriction factor31,32. As KRT72 inhibits MLV (Supplementary Fig. 9i), this study also examined the interaction between KRT72 and incoming MLV cores during viral infection. Like MX2, KRT72 showed binding activity to MLV cores (Supplementary Fig. 14c). Therefore, KRT72 may recognize and bind to HIV-1 capsids to exert its inhibitory effect on HIV-1.
To substantiate these results, CA mutants (T54A, Q63A/Q67A, G89V, N57A, P90A, and A92E) were generated in HIV-1 according to previous studies38,39,40,41,42,43 to determine a CA mutant HIV-1 insensitive to KRT72 inhibition for subsequent experiments. WT or these CA mutant HIV-1 variants were used to infect 293T cells with KRT72. All CA mutants (T54A, Q63A/Q67A, G89V, or P90A) interacted with KRT72, except N57A and A92E (Supplementary Fig. 15a). As fate-of-capsid assay was performed, confirming that the CA A92E mutant cannot associate with KRT72 in infected cells (Supplementary Fig. 15b). This study examined HIV-1 replication in these CA mutants with or without KRT72 and discovered that KRT72 inhibited T54A, Q63A/Q67A, G89V, and P90A CA mutant HIV-1 variants as well as WT HIV-1, whereas KRT72 did not show any inhibitory effect against A92E and N57A CA mutant HIV-1 replication (Supplementary Fig. 15c). The N75A CA mutant HIV-1 exhibited almost no replication ability, as reported previously, as the cores were not transported to the nucleus41. In contrast, A92E mutant but not WT cores could efficiently enter the nucleus even with KRT72 (Supplementary Fig. 15d, e). The A92E mutant did show a colocalization with KRT72. When nuclear and cytoplasmic fractions were separated, A92E mutant cores could transition into the nucleus, whereas WT viral cores were sequestered in the cytoplasm with KRT72 (Supplementary Fig. 15f). However, N57A mutant cores could only be found in the cytoplasm regardless of whether or not KRT72 is present. Therefore, A92E is a CA-insensitive mutant to KRT72 inhibition. Subsequently, the A92E CA mutant HIV-1 was used to infect resting CD4+ T cells to explore endogenous KRT72 antiviral activity. Vpxmnd-2 promoted WT HIV-1 replication (Supplementary Fig. 15g) by promoting KRT72 degradation but not SAMHD1 (Supplementary Fig. 15h). However, Vpxmnd-2 lost its ability to increase A92E CA mutant HIV-1 infection, but it still promoted KRT72 degradation. The A92E CA mutant HIV-1 also showed a higher infection rate than WT HIV-1 with KRT72. This A92E mutant exhibited similar infection dynamics in resting CD4+ T cells with or without KRT72 (Supplementary Fig. 15g), suggesting that the A92E CA mutant HIV-1 could overcome the KRT72 barrier in resting CD4+ T cells. The N57A CA mutant HIV-1 replication was inefficient in resting CD4+ T and 293T cells. To verify this result, cytoplasmic and nuclear fractions were separated from HIV-1-infected resting CD4+ T cells, and Vpxmnd-2 increased the nuclear accumulation of the WT HIV-1 core but not A92E CA mutant cores (Supplementary Fig. 15i). In contrast, cores of the A92E mutant HIV-1 accumulated in the nucleus at similar levels regardless of treatment with or without VLP-Vpxmnd-2. Again, N57A mutant HIV-1 cores were unable to enter the nucleus. Therefore, KRT72 may interact with and sequester HIV-1 cores in the cytoplasm in resting CD4+ T cells.
KRT72 binds directly to HIV-1 cores
Furthermore, we also explored whether the recombinant KRT72 protein (Supplementary Fig. 16a) directly bound to the capsid cores of HIV-1 or CA (Supplementary Fig. 16c) in vitro. To this end, biochemical experiments were conducted to investigate the effects of recombinant His-KRT72 protein binding on incoming HIV-1 cores in cell lysate and preassembled CA tubes in vitro. The WT recombinant KRT72 protein, but not the recombinant KRT72 N-terminal mutant (Δ1–124) and HIV-1 integrase cofactor LEDGF/p75, bound directly to incoming viral cores (Supplementary Fig. 16b), indicating that KRT72 could recognize the incoming HIV-1 cores. In particular, this recombinant LEDGF/p75 protein can enhance HIV-1 integrase strand transfer activity in vitro in a previous study44; however, it could not bind to incoming viral cores.
To understand KRT72 interaction with HIV-1 cores in vitro, His-tagged recombinant CA was purified (Supplementary Fig. 16c) and allowed to form CA tubes in vitro (Supplementary Fig. 16d). The GST-KRT72 recombinant protein but not its N-terminal mutant (Δ1–124) bound to CA tubes only with the assembly buffer (Supplementary Fig. 16e). Without this buffer, any CA binding to KRT72 cannot be detected, indicating that KRT72 preferentially recognizes and binds to HIV-1 capsid cores but not CA. As KRT72 did not show an inhibitory effect on the A92E mutant CA HIV-1 infection, whether KRT72 binds to CA tubes of the A92E mutant in vitro was examined. Using electron microscopy, this A92E mutant CA can form CA tubes in vitro as WT CA did (Supplementary Fig. 16d). Through biochemical experiments, T54A, N57A, Q63A/Q67A, G89V, P90, A92E mutant CA, and WT CA can form CA tubes in vitro (Supplementary Fig. 16f), as reported previously43. Only WT CA tubes precipitated by ultracentrifugation were detected with the assembly buffer. Without the assembly buffer, CA was not detected in the pellets, indicating that WT CA can form tubes only with the assembly buffer. Mutant CA can form tubes such as WT CA in vitro because ultracentrifugation can successfully precipitate these CA tubes. After confirming that mutant CA can form tubes, CA tubes interacting with recombinant KRT72 protein were examined in vitro. KRT72 could not bind to A92E and N57A mutant CA tubes but bound to WT CA tubes (Supplementary Fig. 16g), and KRT72 bound to T54A, Q63A/Q67A, G89V, and P90A CA tubes. This was expected as KRT72 interacted with these mutant HIV-1 cores in cells during infection (Supplementary Fig. 15a). Therefore, KRT72 recognizes and directly binds to HIV-1 cores.
KRT72 captures incoming HIV-1 capsids from microtubules
In particular, the cytoplasmic movement of HIV-1 capsid cores toward the nucleus depends on actin microfilaments and microtubules together with host factors involved in motor proteins associated with the cytoskeleton and microtubules, such as dynein and kinesin33,45. The anti-HIV-1 activity of KRT72 was examined using chemical inhibitors of microfilaments and microtubules. With inhibitors of microtubules (nocodazole) or microfilament actin (cytochalasin D) at the concentrations used, the inhibitory effect of KRT72 against HIV-1 was not significantly affected (Supplementary Fig. 17a, b), indicating that KRT72 antiviral activity may not be closely related to cell microfilaments and microtubules. Then, whether KRT72 captures HIV-1 cores from microtubules in the cytoplasm of HIV-1-infected cells was examined. Without KRT72, bicaudal D homology 2 (BICD2), a core-associated dynein adapter used by HIV-1 to transport to the nucleus33,46,47,48, bound to HIV-1 cores in 293T cells (Supplementary Fig. 17c, d). With KRT72, the interaction between BICD2 and HIV-1 cores was abolished, as predicted. BICD2 did not bind to HIV-1 cores when KRT72 was bound to them, suggesting that KRT72 can capture HIV-1 cores from microtubules when HIV-1 cores use microtubules for trafficking toward the nucleus. Importantly, whether KRT72 also prevented the interaction between HIV-1 cores and microtubules in HIV-1-infected resting CD4+ T cells was also determined. As expected, using VLP-Vpxrcm to degrade the KRT72 protein could promote the association between HIV-1 cores and BICD2 (Supplementary Fig. 17e, f), suggesting that HIV-1 cores may hijack microtubules for trafficking without KRT72. Therefore, KRT72 may capture HIV-1 incoming cores from microtubules and sequester them in the cytoplasm, inhibiting their trafficking toward the nucleus.
Discussion
Consistent with previous reports that HIV-1 cannot efficiently enter the nucleus in resting CD4+ T cells1,2,4,15,16,49,50 and indicates a barrier to nuclear import in resting CD4+ T cells, the functional analysis of KRT72 antiviral activity provides insights into cell type specificity and mode of action of postentry restriction factors for HIV-1 core trafficking toward to the nucleus. In particular, MX2, an HIV-1 nuclear import inhibitor, cannot function in resting CD4+ T cells51, confirming the existence of an unidentified restriction factor that inhibits HIV-1 nuclear import. Recent findings suggested that in addition to SAMHD1, Vpx from the main SIV rhesus macaque could counteract a second host restriction factor, likely a nuclear import inhibitor, to improve HIV-1 infection21. More importantly, Vpx proteins from a second SIV lineage (SIVrcm and SIVmnd-2), which did not affect SAMHD1 and dNTP pools, could also strongly enhance HIV-1 infection in resting CD4+ T cells. In contrast, this improvement in HIV-1 infection did not occur in primary macrophages with Vpxrcm and Vpxmnd-2, indicating that they can overcome a previously uncharacterized restriction factor for lentiviruses and that this factor is specific to resting CD4+ T cells but not macrophages. KRT72 is not expressed in macrophages; therefore, it is expected that Vpx proteins from SIVrcm and SIVmnd-2 cannot enhance HIV-1 replication in macrophages.
In this study, IP assays in conjunction with proteomic analysis were used to identify KRT72, a type II keratin highly expressed in resting CD4+ T cells, as a restriction factor to limit HIV-1 infection targeted by Vpx. Vpxrcm and Vpxmnd-2 proteins could largely degrade KRT72 but not SAMHD1, which promoted HIV-1 infection in resting CD4+ T cells. In contrast, Vpx from SIV rhesus macaque and HIV-2 could simultaneously degrade KTR72 and SAMHD1 to improve HIV-1 infection. Probably, these Vpx variants discriminating to target KRT72 and SAMHD1 are based on their different sequences. For example, these Vpx variants may have a consensus sequence that allows them to target KRT72, resulting in degradation. As Vpx is not physiologically related to HIV-1, this study also explored HIV-2 infection in resting CD4+ T cells with or without Vpx and found that KRT72 depletion can only restore Vpx-defective but not WT HIV-2 infection. Like HIV-1, HIV-2 late RT is also inhibited by KRT72.
Further evidence demonstrated that KRT72 restricts HIV-1 infection in resting CD4+ T cells in a SAMHD1-independent manner, inhibiting viral late RT and nuclear 2-LTR circular DNA levels but with early RT remaining intact. Unlike the finding of Baldauf et al.21 that early and late RT were upregulated with Vpxmnd-2 or Vpxrcm, early RT was not affected by KRT72, which requires further investigation to elucidate whether Vpxmnd-2 or Vpxrcm can overcome postentry barriers other than KRT72 in resting CD4+ T cells. This study investigated the molecular mechanism by which KRT72 inhibits HIV-1 and found that KRT72 sequesters incoming HIV-1 cores in the IF network in the cytoplasm, reducing late RT, integrated DNA, and 2-LTR circular DNA. This was expected as interference in the trafficking of HIV-1 cores could inhibit viral RT levels and nuclear 2-LTR circular DNA34,35,36,37. Several studies supported that uncoating occurs gradually as HIV-1 undergoes RT and is trafficked toward the nucleus52,53,54. Accumulated evidence showed that capsid cores remain intact or almost intact long after entering the cells and even once in the nucleus; subsequently, uncoating and RT occurred in the nucleus55,56,57. Probably, KRT72-mediated inhibition of core trafficking could influence viral uncoating, resulting in a reduction in viral RT. The intact early RT with KRT72 may reflect that RT initiation can be accomplished in the cytoplasm, which does not require capsid uncoating. Therefore, whether KRT72 sequestration affects HIV-1 uncoating is an interesting question, which warrants further investigation. This study did not investigate how KRT72 and SAMHD1 interact and elegantly choreograph their individual effects to implement their anti-HIV-1 activities in resting CD4+ T cells, as the antiviral activity of KRT72 is independent of SAMHD1. If uncoating and RT occur in the nucleus in resting CD4+ T cells, the restriction of KRT72 may occur earlier than SAMHD1, which is a necessary study to understand HIV-1 infection in resting CD4+ T cells. Thus, the KRT72 filamentous network is expected to sequester incoming HIV-1 cores, inhibiting their trafficking to the nucleus and causing the reduction of RT and 2-LTR circular DNA.
Importantly, only KRT72 had an obvious inhibitory effect on HIV-1 infection in target cells in type II keratins. KRT74 also had an inhibiting effect on HIV-1, albeit one that was significantly weaker than that of KRT72. KRT74 is not highly expressed in resting or stimulated CD4+ T cells, so it is unlikely to contribute to the HIV-1 resistance of resting CD4+ T cells. This study examined whether KRT72 binds to incoming cores in vivo and in vitro, and biochemical experiments revealed that KRT72 binds directly to HIV-1 capsid cores in vitro but not CA and that the antiviral activity of KRT72 is based on its association with HIV-1 cores. In sharp contrast, other keratins (KRT71 and KRT73) could not bind to HIV-1 cores. This study also investigated relevant CA mutations in infected cells and in vitro. The mutant HIV-1 virus-containing A92E substitution was fully resistant to KRT72. In contrast, HIV-1 mutants with substitutions T54A, Q63A/Q67A, G89V, and P90A were sensitive to KRT72 restriction. In accordance with these cellular assays, purified KRT72 did not interact with A92E or N57A CA tubes but with T54A, Q63A/Q67A, G89V, and P90A CA tubes. These findings explained the specificity of the interaction between KRT72 and HIV-1 capsids, indicating that KRT72 recognizes and binds to HIV-1 cores to exert its antiviral activity.
HIV-1 infection depends on efficient intracytoplasmic transport of incoming viral cores to the target nucleus. Evidence suggested that the cytoplasmic transport of HIV-1 cores is dependent on the microtubule network, and this movement is facilitated by the microtubule motor dynein33,45. In particular, using chemical inhibitors of microfilaments and microtubules, KRT72 may capture viral cores from cytoplasmic microtubule-associated protein to prevent HIV-1 from taking advantage of microtubules for trafficking to the nucleus. KRT72 depletion promotes the binding of HIV-1 cores to microtubules in resting CD4+ T cells, indicating that KRT72 partially explains why the HIV-1 genome can almost not enter the nucleus in resting CD4+ T cells. Of course, this requires future investigations using RNAi to deplete the critical components of microfilaments or microtubules to examine the anti-HIV activity of KRT72. Overall, this study indicated that KRT72 is a restriction factor in the early phase of HIV-1 replication targeted by Vpx in resting CD4+ T cells. Importantly, because KRT72 could restrict HIV-1 spread in CD4+ T cells of ART-treated or untreated HIV-1-infected individuals ex vivo, KRT72 might be used as an effective agent combating HIV-1. These findings provided new insights into the defenses of resting CD4+ T cells against HIV-1 infection and suggested a study direction for novel anti-HIV therapeutics.
Methods
Ethics statement
This study was approved by the Research and Ethics Committee of The First Hospital of China Medical University (approval no. 2021-328-2 on June 24 2021). Blood samples were obtained from healthy donors following the National Health and Medical Research Council guidelines. Written informed consent was obtained from each healthy donor before the study. Peripheral blood mononuclear cells (PBMCs) were isolated from healthy volunteers (n = 32; age range: 22–45 years). Exclusion criteria included recent infection (within the past 4 weeks), autoimmune disorders, chronic inflammatory diseases, or the use of immunosuppressive medications. Blood samples were collected in EDTA tubes and processed within 4 h of collection.
Study participants
People living with HIV (PLWH) (n = 4, under the standard of care antiretroviral treatment) with viral loads of <50 copies/mL and untreated HIV-1+ participants (n = 2) (Supplementary Table 2) were enrolled in this study. PBMCs from participants were prepared using Ficoll-Hypaque density gradient centrifugation. CD4+ T cells were isolated from PBMC by negative selection with a human CD4+ T-cell enrichment cocktail (Stem Cell Technologies). Ethical approval was obtained from the Research and Ethics Committee of The First Hospital of China Medical University (approval no. 2021-328-2 on June 24 2021), and written informed consent for participation was obtained from all participants.
Cells and culture reagents
HeLa, 293T, TZM-bl, and Jurkat cells were grown and maintained in Dulbecco’s modified Eagle’s medium (Gibco) or RPMI-1640 medium (Gibco). Both media were supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/mL penicillin, and 100 mg/mL streptomycin. Plasmids were transfected into 293T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. PBMCs obtained from healthy blood donors were purified by Ficoll-Hypaque gradient centrifugation. CD4+ T cells or monocytes were isolated from PBMC by negative selection with human CD4+ T cells or a CD14+ enrichment cocktail (Stem Cell Technologies). To stimulate CD4+ T cells, the CD3/CD28 magnetic beads activator (Invitrogen) was added to the culture medium for 2 days with IL-2 (50 U/mL; Biomol) according to the manufacturer’s instructions. Isolation and culture of monocytes, monocyte-derived macrophages (MDMs), and monocyte-derived dendritic cells (MDDCs) were carried out as described previously58,59. Monocytes were stimulated with 10 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF; R&D) and 50 ng/mL recombinant human macrophage CSF (R&D) for 7 days to generate MDMs. MDDCs were generated by incubating CD14-purified monocytes in Iscove’s modified Dulbecco’s medium (Gibco) supplemented with 10% FBS, 2 mM l-glutamine, 100 IU/mL penicillin, 100 mg/mL streptomycin, 10 mM HEPES, 1% nonessential amino acids, 1 mM sodium pyruvate, 10 ng/mL GM-CSF, and 50 ng/mL IL-4 (Miltenyi Biotec). On day 4, two-thirds of the culture medium was replaced with fresh medium containing GM-CSF and IL-4. Immature MDDCs were harvested and used for experiments on day 6.
Chemical reagents
Nocodazole, cytochalasin D, and MG132 were purchased from Sigma-Aldrich.
Plasmids
KRT72, Vpx derived from SIVmac239, SIVrcm, and SIVmnd-2, and HIV-2Rod expression vectors were purchased from OriGene, and their ORFs were de novo cloned in the pCMV-3Tag-2A vector (Addgene). The luciferase-expressing Env-defective NL4-3.Luc.R-E-, EGFP-expressing Env-defective NL4-3-ΔEnv-EGFP, SIVmac239-luc, HIV-2Rod-luc, and MLV-luc were provided by Dr. Guangxia Gao60. HIV-1 proviral NL4-3, BaL, and 89.6 vectors were obtained through the National Institutes of Health AIDS Reagent Program. Dr. Eric Freed gifted the HIV-1 proviral vectors of pNL-AD8. Vectors to make Vpx(−) VLP were gifted by Dr. Nathaniel Landau11. To allow Vpx to enter HIV-1 virions, HIV-1* variants (NL4-3-Luc.R-E-, NL4-3-ΔEnv-EGFP, and NL4-3) were generated by introducing the amino acid motif of SIVmac239 p6 that mediates the Vpx packaging. The SIV p6 fragments were DPAVDLLKNYMQSFRFGEETTTPSQKQEPIDKELYPLASLRSLFGSDPSSQ and inserted into NL4-3-Luc.R-E-, NL4-3-ΔEnv-EGFP, and NL4-3 by SpeI and Sbfl digestion.
To construct the CA point mutation of the HIV-1 virus plasmid, the CA mutated amino acid motif was generated, CA point mutations mediated by the motif were T54A/N57A/Q63A/G89V/P90A/A92E, and NL4-3-Luc.R-E-* was inserted by enzymatic digestion of BssHII and SpeI. To construct Vpx-defective HIV-2Rod viral plasmids (HIV-2Vpx-), HIV-2-luc enzymatically digested BsmBI and AarI to remove a 914 bp fragment containing tat and vpx genes and inserted a 575 bp fragment without vpx gene, which was amplified from HIV-2-luc containing a tat gene by BsmBI and AarI digestion. To construct the HIV-2 viral plasmid with Vpxmac239 point mutations, a 914 bp fragment containing tat and vpx genes removed from HIV-2-luc by BsmBI and AarI digestion. The point mutated SIVmac239 Vpx sequences (WT/L25A/H39A/W56A/Q76A) were introduced by PCR respectively, and these PCR fragments containing tat and vpx (WT/L25A/H39A/W56A/Q76A) genes were inserted into the above HIV-2-luc by BsmBI and AarI digestion to construct HIV-2-luc-Vpxmac239. The shRNA-mediated silencing of KRT72 (RHS4430-200280629 or RHS4430-200282567 targeting ORF; RHS4430-200211840 targeting 3′-UTR) or control (Catalog #RHS4346) was achieved by introducing a microRNA-adapted shRNA lentivirus into postactivated resting CD4+ T cells.
RNAi in resting CD4+ T cells
Resting CD4+ T cells were individually isolated using a resting CD4+ T-cell isolation kit (Stem Cell Technologies) according to the manufacturer’s instructions. Resting CD4+ T cells (4 × 106/mL) were stimulated with the CD3/CD28 magnetic beads activator (Invitrogen) and IL-2 (50 U/mL), as described previously44. Activated cells were transduced on day 3 by spinoculation with VSV-G-pseudotyped lentiviral vectors carrying pLKO.1-puro-control-shRNA or pLKO.1-puro-shKRT72 in a medium containing 40 U/mL IL-2. Transduced cells were pooled on day 5 and cultured at 2 × 107/mL with 30 U/mL IL-2 and 0.5 µg/mL puromycin. The IL-2 concentration was reduced to 15 U/mL on day 7. Viable puromycin-resistant cells were purified on day 8 by Ficoll gradient centrifugation and resuspended in 5 mL fresh medium containing 15 U/mL IL-2. Fifty percent of the culture medium was replaced with fresh medium on day 10 to reduce the final IL-2 concentration to 7.5 U/mL. The culture medium was replaced on day 12 with fresh medium to reduce the final IL-2 concentration to 3.75 U/mL. The IL-2 concentration was adjusted to 1.25 U/mL on day 13, and the total number of cells and living cells were counted and equalized before HIV-1 infection. Cells aliquoted were stained to examine the surface markers CD25, CD69, and HLA-DR and analyzed by flow cytometry. Stealth-grade siRNA (5′-UGUAGCUAUAACUGCUUGAGGCGCC or 5′-ACUGCAGCUGCAGACGUCAAGACCA targeting ORF and 5′-GUUGUCUUUGCAUUUCAGGTT targeting 3′-UTR) against KRT72 and controls (catalog no. RHS4346) were purchased from Invitrogen. Differentiated THP-1 cells and MDDCs were directly transfected with siRNA using Lipofectamine 3000 (Thermo Fisher Scientific, Bremen, Germany) to achieve siRNA-mediated silencing. CD4+ T cells were stimulated and electroporated with siRNA using an Amaxa system with human T-cell nucleofector solution and Program U-14 (Amaxa).
Identification of KRT72 interaction with Vpxrcm in resting CD4+ T cells
Resting CD4+ T cells isolated from three healthy donors were electroporated with a FLAG-tagged Vpx expression vector derived from SIVrcm. Twenty-four hours after electroporation, cells were treated with VLP-Vpxrcm with or without 1.5 μM MG132 treatment for 8 h. Cells were lysed with IP lysis buffer [50 mM Tris-HCl (pH 7.2), 50 mM NaCl, 1% NP-40, 1 mM EDTA, 2% glycerol, 1× protease inhibitor cocktail, complete]. Cell lysates from the three donors were pooled. Cell lysates were incubated on ice for 30 min before centrifugation at 14,000 × g at 4 °C for 10 min. The supernatants were transferred to new tubes, and the pellets were combined with cold IP lysis buffer before sonication and a second round of centrifugation at 14,000 × g for 10 min at 4 °C. The supernatants obtained from the two extraction steps were pooled and incubated with a mixture of protein A/G Dynabeads (Thermo Fisher Scientific) pretreated overnight with an anti-FLAG antibody at 4 °C. The IP products were washed with cold IP buffer and phosphate-buffered saline (PBS) with Tween 20 (PBST) 5–10 times (500 μL/wash). The IP products were reduced in 20 mM dithiothreitol (DTT; Sigma-Aldrich) at 95 °C for 5 min before alkylation in 50 mM iodoacetamide (Sigma-Aldrich) in the dark for 30 min. After alkylation, the samples were transferred to a 10 kDa centrifugal spin filter (Millipore) and sequentially washed thrice with 200 μL of 8 M urea and twice with 200 μL of 50 mM ammonium bicarbonate via centrifugation at 14,000 × g. Enzyme digestion was performed by adding trypsin (Promega) at a 1:50 ratio (enzyme/substrate, m/m) in 200 μL of 50 mM ammonium bicarbonate at 37 °C for 16 h. The peptides were recovered by transferring the filter to a new collection tube and centrifugation at 14,000 × g. The filter was washed twice with 100 μL of 50 mM NaHCO3 to increase peptide yield. The peptides were desalted using a StageTip. MS experiments were performed on a nanoscale ultra-high-performance LC (HPLC) system (EASY-nLC1000; Proxeon Biosystems, Odense, Denmark) connected to an Orbitrap Q-Exactive instrument equipped with a nanoelectrospray source (Thermo Fisher Scientific). The peptides were dissolved in 0.1% FA with 5% CH3CN and separated on an analytical column reverse-phase HPLC (75 μm × 15 cm) packed with 2-m C18 beads (Thermo Fisher Scientific) and eluted using a gradient of 5% to 40% acetonitrile in 0.5% formic acid for 2 h at a flow rate of 250 nL/min. The spray voltage was 2.5 kV, and the ion transfer capillary temperature was 275 °C. A full tandem MS (MS/MS) cycle consisted of one full MS scan [resolution, 70,000; automatic gain control (AGC) value, 1 × 106; maximum injection time, 50 ms] in profile mode over a mass range of m/z 300 and 1800 and fragmentation of the 10 most intense ions via high-energy collisional dissociation with normalized collision energy at 28% in centroid mode (resolution, 17,500; AGC value, 1 × 105; maximum injection time, 100 ms). The dynamic exclusion window was also set at 40 s, and a microscan was acquired for each MS and MS/MS scan. Unassigned ions and those with charges of 1+ and >7+ were rejected for MS/MS, and lock mass correction utilizing a background ion (m/z 445.12003) was applied61. Raw data were processed using Proteome Discoverer (PD) version 2.1, and the reviewed Swiss-Prot human proteome database was searched using MS/MS spectra. All searches were carried out with a precursor mass tolerance of 7 ppm and a fragment mass tolerance of 20 millimass units, with oxidation (Met) (+15.9949 Da) and acetylation (protein N-terminus) (+42.0106 Da) as variable modifications and carbamidomethylation (+57.0215 Da) as fixed modification, and two trypsin-missed degradations were allowed. Only peptides of a minimum of six amino acids in length were considered. PD filtered the identification of peptides and proteins to control the false discovery rate at <1%. At least one unique peptide was required for protein identification.
HIV-1 spinoculation
Resting CD4+ T cells (0.5 million) were incubated with 100 or 500 ng p24 virus and centrifuged at 1200 × g for 2 h at 25 °C in a 96-well U-bottomed plate.
Enzyme-linked immunosorbent assay (ELISA)
ELISA measured the p24 protein in the culture supernatants (ABL Corporation) according to the manufacturer’s instructions. Test Samples are mixed with Disruption Buffer to inactivate the virus and to release HIV-1 p24 into solution to enable detection. The microtiter wells of a 96-well plate are coated with two murine monoclonal antibodies that react with unique epitopes on HIV-1 p24. When HIV-1 p24 Standard solutions or tissue culture Test Samples are added to the wells, an immune complex forms with the plate-bound antibodies and the p24 in solution. Unbound materials are then thoroughly washed away. Conjugate Solution, containing peroxidase-conjugated human anti-p24 polyclonal antibodies, is then added. The conjugated antibodies complex with the captured HIV-1 p24. After washing the wells to remove the unbound conjugated antibodies, Peroxidase Substrate is added to the wells. The enzyme-substrate reaction results in a blue color change. Upon adding Stop Solution, the blue color changes to yellow, and the absorbance is measured at 450 nm. There is a linear relationship between the absorbance at 450 nm and the amount of HIV-1 p24 bound to the well. The concentration of HIV-1 p24 in Test Samples can be determined from linear regression analysis of the standard curve.
Luciferase detection assay
Luciferase activity was quantified as relative luminescence units in cell lysates (Promega) according to the manufacturer’s instructions. The TZM-bl indicator cell line was used for the quantitative analysis of HIV-1 infectivity using luciferase as a reporter, and the luminescence units of the background of TZM-bl cells were subtracted from each data point.
Quantification of HIV-1 and HIV-2 early and late RT products
HIV-1 or HIV-2 producer 293T cells were washed twice with PBS to remove the transfected HIV-1 or HIV-2 plasmid before infection. The viral stocks obtained were treated for 1 h at 37 °C with DNase I (Takara)18. DNA was extracted with a DNeasy Blood & Tissue Kit (Qiagen). The samples were quantified for total HIV cDNA against a serially diluted standard template amplified in the equivalent background amount of the negative control (uninfected) cell genomic DNA. Quantitative polymerase chain reaction (qPCR) was performed to quantify early or late viral RT of HIV-1 or HIV-2 with the following primers:
early HIV-1 RT forward 5′-GCCTCAATAAAGCTTGCCTTGA-3′ and early RT reverse 5′-TGACTAAAAGGGTCTGAGGGATCT-3′
or late HIV-1 RT forward 5′-AGCAGGAACTACTAGTACCC-3′ and late RT reverse 5′-TTGTCTTATGTCCAGAATGC-3′, as described previously44; and late HIV-2 RT forward 5′-CAGGATTTCAGGCACTCTCAGA -3′ and late RT reverse 5′-TGCTTGATGGTCGCCCACA-3′, as described previously62.
Alu and 2-LTR PCR
Alu-PCR was performed as described previously to quantify viral integrants44. Initial amplification used dilutions of the integration standard and samples from infected cells.
The integration standard was prepared using 2 × 107 CEM-SS cells infected with a high-titer, randomly integrated, replication-incompetent VSV-G-pseudotyped virions (HIV-1NL4-3/ΔEnv/GFP; 1 µg p24 Gag) by spinoculation, allowing a single round of infection. Acutely infected cells contained unintegrated HIV-1 DNA and integrated proviral DNA. The amount of unintegrated DNA per cell decreased with each cell division, whereas proviral DNA replicated alongside cellular genomic DNA. As reported previously, unintegrated DNA levels became undetectable after 30 days in culture44. GFP+-transduced cells were selected by fluorescence-activated cell sorting (FACS), and Southern blotting was used to verify the elimination of unintegrated HIV-1 DNA. Given that all DNA was integrated, there was ~1 integration event per cell present in this integration standard cell line, and 106 integration standard cells/mL were lysed in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.2 mM CaCl2, 0.001% sodium dodecyl sulfate (SDS), 0.001% Triton X-100, and 1 mg/mL proteinase K (Sigma-Aldrich). Cell lysates were digested overnight at 58 °C, and protease was heat-inactivated for 15 min at 95 °C. Aliquots of standard integration DNA were stored at −80 °C. Serial dilutions of standard integration DNA were made in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.001% SDS, and 0.001% Triton X-100 supplemented with 1 µg poly(rA)/mL to reduce nonspecific adsorption of DNA on the walls of the reaction vessel. DNA from the integration standard cells was diluted with uninfected PBMC DNA at 2 µg/mL to keep the number of Alu sites per reaction constant. Conditions of low infection frequency could be mimicked by diluting the integration standard in uninfected PBMC DNA, allowing the accurate measurement of integration to 0.5 provirus copies in 10,000 cellular genomes. Therefore, the method can measure the relatively lower number of integrated provirus copies in resting CD4+ T cells. The initial nonkinetic preamplification used dilutions of standard integration DNA and dilutions of unknown samples. The following primers were used for genomic DNA preamplification: Alu forward 5′-GCCTCCCAAAGTGCTGGGATTACAG-3′ and HIV-1 Gag reverse 5′-GCTCTCGCACCCATCTCTCTCC-3′, with 100 nM Alu forward primer, 600 nM Gag reverse primer, and 1 μL gDNA for every 20 μL PCR solution. The thermocycler (Eppendorf Mastercycler Pro Cycler) was programmed with a 2 min hot start at 94 °C, followed by 30 rounds of denaturation at 93 °C for 30 s, annealing at 50 °C for 1 min, and extension at 70 °C for 1 min 40 s. Using 2 μL of the product from the preamplification step, a second round of RT-qPCR was performed to quantify HIV-1 integration. The primer sequences were as follows: LTR forward 5′-GCCTCAATAAAGCTTGCCTTGA-3′ and LTR reverse 5′-TCCACACTGACTAAAAGGGTCTGA-3′. Small nongenomic DNA was isolated from cells using a Qiagen Miniprep kit to quantify HIV-1 2-LTR circular DNA. Primers MH535 (5′-AACTAGGGAACCCACTGCTTAAG-3′) and MH536 (5′-TCCACAGATCAAGGATATCTTGTC-3′) were used for amplification. The Applied Biosystems QuantStudio 7 RT-PCR system was utilized for all reactions. The thermal program was 2 min at 50 °C, followed by a 10 min hot start at 95 °C and 40 cycles of 95 °C for 15 s and 60 °C for 60 s. GAPDH was used as an internal control in which the total amount of DNA was normalized.
Reverse Transcriptase Assay
HIV-2 virions were purified and the level of virion-associated reverse transcriptase was measured according to the manufacturer’s instructions (Reverse Transcriptase Assay, colorimetric, Roche).
Measurement of viral infectivity
The p24 concentrations in culture supernatants were determined using an ELISA (ABL Corporation). The infectiousness of HIV-1 produced in culture supernatants was determined by infecting TZM-bl reporter cells. Virus infectivity was quantified using luciferase as a reporter. The luciferase activity in the cell lysate was quantified in relative luminescence units (Promega), and the background luminescence of the control cells (uninfected) TZM-bl was subtracted from each data point.
CoIP
293T or HeLa cells (5.0 × 106) were lysed with IP lysis buffer [50 mM Tris-HCl (pH 7.2), 50 mM NaCl, 1% NP-40, 1 mM EDTA, 2% glycerol, and 1× protease inhibitor cocktail, complete]. Cell lysates were incubated on ice for 30 min and centrifuged at 12,000 × g for 10 min at 4 °C. The supernatants were transferred to fresh tubes, and the pellets were mixed with cold IP lysis buffer before sonication, followed by a second round of centrifugation at 12,000 × g for 10 min at 4 °C. The supernatants obtained from the two extraction steps were pooled and incubated with mixed protein A/G Dynabeads (Invitrogen) pretreated overnight with antibodies at 4 °C. IP products were washed with cold IP and PBST buffers 5–10 times (500 µL/wash).
Western blotting and antibodies
Western blotting was performed using a standard method for detecting cellular proteins. The following antibodies were used in this study: polyclonal rabbit anti-KRT72 (Abcam; ab127030), polyclonal rabbit anti-BICD2 antibody (Abcam; ab237616), rabbit anti-GAPDH (Thermo Fisher Scientific, Cat.PA1-987), mouse monoclonal anti-FLAG (Sigma-Aldrich), rabbit polyclonal anti-p24 (Abcam, Cat.ab63913), mouse immunoglobulin-horseradish peroxidase (Abcam, Cat.6789), cross-adsorbed Alexa Fluor 488-labeled goat anti-rabbit secondary antibody (Thermo Fisher Scientific, Cat.A32731), cross-adsorbed Alexa Fluor 488- or 555-labeled goat anti-mouse secondary antibody (Thermo Fisher Scientific, Cat.A32723, Cat.A32727), rabbit and mouse immunoglobulin G (IgG) Trueblot (eBioscience), and rabbit or mouse IgG isotype control (Abcam, Cat. ab171870, Cat.ab18413).
Expression and purification of recombinant proteins
All recombinant proteins were expressed in BL21(DE3) cells at 37 °C and induced for 3.5 h with 1 mM isopropyl β-D-1-thiogalactopyranoside, as described previously44. Cell pellets were lysed by ultrasonication and clarified by centrifugation at 12,000 × g for 30 min. His-IN and His-p24CA were purified on a HisTrap HP column using a 250 mM imidazole gradient. On an SP-Sepharose 4B column, GST-KRT72 was purified with 25 mM reduced glutathione (Sigma-Aldrich). All recombinant proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and confirmed by Western blotting with specific antibodies [mouse monoclonal anti-His (Cell Signaling) and anti-GST (Abcam)].
Fate-of-capsid assay for HIV
293T cells were transfected with Flag-tagged KRT72, MX2, or mock (GFP-3FLAG). At 24 hpi, cells were infected with 1 µg HIV-1NL4-3.Luc.R-E- (VSV-G) with polybrene (5 μg/mL). With raltegravir, cells were incubated on ice for 30 min and infected with HIV-1. Cells were harvested by adding 1 mL trypsin-EDTA, incubated on ice for 30 s, and inactivated with culture medium. Cells were washed thrice with ice-cold PBS, treated with 2.5 mL hypotonic lysis buffer [10 mM Tris-HCl (pH 8.0), 10 mM KCl, 1 mM EDTA, and protease inhibitors], incubated on ice for 15 min, and homogenized with a Dounce homogenizer. Cell lysates were centrifuged at 12,000 × g for 10 min at 4 °C to separate the cytosolic fraction from the nuclear pellet (input). The supernatant was spun down in 50% sucrose in STE buffer [10 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 1 mM EDTA] for 2 h at 100,000 × g at 4 °C in an SW55Ti Beckman Coulter rotor. After centrifugation, 1 mL solution from the centrifuge tube was collected as the supernatant. Sucrose was removed, and the pellet was resuspended in 10× whole-cell lysate buffer (pellet solution).
Fate-of-capsid assay for MLV
293T cells were transfected with FLAG-tagged MX2, KRT72, or mock (GFP-3FLAG). At 24 hpi, cells were treated with the VSV-G-pseudotyped MLV with polybrene (5 μg/mL) on ice for 30 min, washed by PBS to remove input MLV virions, and returned to 37 °C. The infection was allowed to proceed for 6 h, and cells were washed thrice with ice-cold PBS, treated with hypotonic lysis buffer [10 mM Tris-HCl (pH 8.0), 10 mM KCl, 1 mM EDTA, and protease inhibitors], incubated on ice for 15 min, and homogenized with a Dounce homogenizer. Cell lysates were centrifuged at 12,000 × g for 4 min at 4 °C to separate the cytosolic fraction from the nuclear pellet (input). The supernatant was spun down in 50% sucrose in STE buffer [10 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 1 mM EDTA] for 2 h at 100,000 × g at 4 °C in an SW55Ti Beckman Coulter rotor. After centrifugation, 1 mL solution from the centrifuge tube was collected as the supernatant. The sucrose was removed, and the pellet was resuspended in 10× whole-cell lysate buffer (pellet solution). Soluble capsids in the supernatant and particulate capsids in the pellet were analyzed by Western blotting using specific antibodies.
HIV-1 core binding assay
The supernatant containing the HIV-1NL4-3.Luc.R-E- (with NL4-3-Env) virus was collected at 48 hpi, filtered, concentrated by ultracentrifugation, and spun through a solution of 20% (w/v) sucrose in PBS using an SW32Ti Beckman Coulter rotor for 3 h at 32,000 × g and 4 °C. The pasted viral particles were resuspended in STE buffer [10 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 1 mM EDTA] and passed through a layer of 1% Triton X-100 in a linear sucrose gradient (30%–70%) in STE buffer using an SW70Ti Beckman Coulter rotor for 2 h at 52,000 × g and 4 °C. Fractions of 1 mL were collected from the top of the gradient, divided into 0.2 mL aliquots, used for binding assays or frozen in flash, and stored at −80 °C. Then, 100 μL of each fraction were collected and lysed in 10× whole-cell lysate buffer for Western blotting analysis. HeLa cells expressing FLAG-MX2, KRT72, SAMHD1, or mock were lysed in hypotonic buffer [10 mM Tris-HCl (pH 7.4), 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM DTT in PBS]. The clarified lysates (input) were incubated with the fractions containing the core diluted in STE buffer with bovine serum albumin (10 μg/mL) for 2 h on ice. The mixture was spun on 30% sucrose in STE buffer for 1 h at 100,000 × g at 4 °C in an SW55Ti Beckman Coulter rotor. The supernatant was resuspended in 10× whole-cell lysate buffer (“supernatant”), and the pellet was resuspended in 10× whole-cell lysate buffer (“pellet”) for Western blotting analysis.
HIV-1 core in vitro binding assays
WT or CA mutant tubes were assembled by incubating 42.5 μM His-CA recombinant protein in a buffer containing 25 mM Tris-HCl (pH 8.0) and 2 M NaCl overnight on ice, as described previously42. The indicated quantities of the tested recombinant proteins were added to the preformed CA tubes, which were incubated for 30 min at 4 °C. The mixtures were centrifuged at 21,000 × g for 15 min at 4 °C. The supernatant was discarded, and the pellets were washed thrice and analyzed by SDS-PAGE and Western blotting to determine the presence of the protein.
Microscopy
Cells were placed on poly-l-lysine-coated glass slides, and the images were collected using a Zeiss LSM 980 confocal microscope with Airyscan 2. HeLa cells, MDMs, or MDDCs were fixed and stained primarily with anti-p24 that was probed with an Alexa Fluor 488 conjugated goat anti-rabbit or anti-mouse secondary antibody (Imag-iT Fixation/Permeabilization Kit, Invitrogen) according to the manufacturer’s instructions.
Colocalization analysis
The colocalization coefficient of mCherry-KRT72 or control signals with HIV-1 core signals from INmNG (VSV-G-pseudotyped HIV-1) or anti-p24 antibody (WT HIV-1) staining was calculated by the colocalization module of Carl Zeiss Micro-imaging Zen software Blue version 3.3. Infected cells were analyzed for capsid colocalization with mCherry-KRT72, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Quantification of KRT72-mCherry signals was associated with INmNG-labeled or anti-p24 antibody staining puncta from 70 cells.
qPCR
Total RNA was extracted from cells using TRIzol (Invitrogen) according to the manufacturer’s instructions. The obtained RNA was dissolved in 100 µL DPEC-H2O, and 1 µg purified RNA was treated with DNase I (amplification grade; Invitrogen) for 10–15 min at room temperature according to the manufacturer’s instructions. RNA was immediately primed with oligo(dT) and reverse transcribed using SuperScript III RT (Invitrogen). Real-time PCR analysis was performed using the ΔΔCT method. Results were normalized to the amplification results for GAPDH (internal control). The primers used in this study are shown in Supplementary Table 2.
Flow cytometry
CD4+ T cells were cultured and treated with CellTrace (CellTrace Cell Proliferation Kit; Invitrogen) according to the manufacturer’s instructions and stained in FACS buffer with αCD69-PE (BD Pharmingen), αCD25-BB515 (BD Horizon), or HLA-DR-APC (BioLegend). For HIV-1 p24 intracellular detection, cells were fixed in 1% paraformaldehyde and stained in FACS buffer with the anti-p24-FITC (Beckman Coulter) HIV-1 core antigen and KC57 clone (BD Cytofix) according to the manufacturer’s instructions. Data were collected on a SONY ID7000 and analyzed using FlowJo.
Cytotoxicity assays
The culture supernatant of lentiviral-transduced or untransduced MDMs was collected to analyze LDH release. Cytotoxicity was quantified colorimetrically with the Pierce LDH cytotoxicity kit (Thermo Fisher Scientific), where LDH activity was measured by subtracting A490 nm from A680 nm. Lysis control wells were treated with 10× lysis buffer for 1 h. The cytotoxicity percentage was calculated with the formula as described previously63:
where LDHlenti-MDM or LDHMDM is the amount of LDH activity in the supernatant of lentiviral-treated or untreated cells, and LDHMaximum is the amount in the supernatant of the lysis control. LDH activity in the cell-free culture medium was subtracted from each value before normalization.
BlaM-Vpr-based viral entry assay
HIV-1 particles incorporating a fusion protein between Vpr and BlaM reporter protein were produced by NL4-3-Luc-R+-E- cotransfection with an expression vector encoding BlaM-Vpr. The produced viruses were quantified by p24 ELISA, and target cells were incubated with 100 ng p24 viruses at 37 °C for 4 h to allow viral entry. After washing thrice with Hank’s balanced salt solution (HBSS; Thermo Fisher Scientific), cells were resuspended and loaded with 1 μM CCF2-AM dye (Thermo Fisher Scientific), a fluorescent substrate for BlaM, in HBSS containing 1 mg/mL Pluronic F-127 surfactant (Thermo Fisher Scientific) and 0.001% acetic acid for 1 h at RT and washed twice with HBSS. The BlaM reaction corresponding to the cleavage of intracellular CCF2 dye by BlaM-Vpr was developed for 14 h at RT in HBSS supplemented with 10% FBS. Cells were washed thrice with PBS and fixed in a 1.2% paraformaldehyde solution. Fluorescence was monitored at 520 and 447 nm by flow cytometry using SONY ID7000.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 6.0. Unless otherwise noted, the unpaired two-tailed Student’s t-test was used for statistical comparison between groups.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
A summary of the report for this article is available as a Supplementary Information file. RNA sequencing (RNA-seq) data used in this study (Supplementary Fig. 1a) were retrieved from The Sequence Read Archive (SRA) using accession codes PRJNA533941 and PRJNA522052. Proteomics dataset for the identification of KRT72 that interacts with Vpxrcm has been deposited in the ProteomeXchange Consortium under accession code PXD059264. Data supporting the conclusions of this study are available within the article and its Supplementary Information files upon reasonable request from the corresponding authors. Source data are provided in this paper. Source data are provided with this paper.
References
Stevenson, M., Stanwick, T. L., Dempsey, M. P. & Lamonica, C. A. HIV-1 replication is controlled at the level of T cell activation and proviral integration. Embo J. 9, 1551–1560 (1990).
Zack, J. A. et al. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61, 213–222 (1990).
Liang, G. et al. Membrane metalloprotease TRABD2A restricts HIV-1 progeny production in resting CD4(+) T cells by degrading viral Gag polyprotein. Nat. Immunol. 20, 711–723 (2019).
Pan, X., Baldauf, H. M., Keppler, O. T. & Fackler, O. T. Restrictions to HIV-1 replication in resting CD4+ T lymphocytes. Cell Res. 23, 876–885 (2013).
Bergamaschi, A. et al. The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A-DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection. J. Virol. 83, 4854–4860 (2009).
Bakri, Y. et al. The maturation of dendritic cells results in postintegration inhibition of HIV-1 replication. J. Immunol. 166, 3780–3788 (2001).
Goujon, C. et al. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J. Virol. 82, 12335–12345 (2008).
Goujon, C. et al. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4, 2 (2007).
Kaushik, R., Zhu, X., Stranska, R., Wu, Y. & Stevenson, M. A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe 6, 68–80 (2009).
Fujita, M. et al. Vpx is critical for reverse transcription of the human immunodeficiency virus type 2 genome in macrophages. J. Virol. 82, 7752–7756 (2008).
Sunseri, N., O’Brien, M., Bhardwaj, N. & Landau, N. R. Human immunodeficiency virus type 1 modified to package Simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J. Virol. 85, 6263–6274 (2011).
Pertel, T., Reinhard, C. & Luban, J. Vpx rescues HIV-1 transduction of dendritic cells from the antiviral state established by type 1 interferon. Retrovirology 8, 49 (2011).
Srivastava, S. et al. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLOS Pathog. 4, e1000059 (2008).
Yu, X. F., Yu, Q. C., Essex, M. & Lee, T. H. The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophage. J. Virol. 65, 5088–5091 (1991).
Baldauf, H. M. et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18, 1682–1687 (2012).
Descours, B. et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9, 87 (2012).
Hrecka, K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011).
Laguette, N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011).
Lahouassa, H. et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 (2012).
Lim, E. S. et al. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204 (2012).
Baldauf, H. M. et al. Vpx overcomes a SAMHD1-independent block to HIV reverse transcription that is specific to resting CD4 T cells. Proc. Natl. Acad. Sci. USA 114, 2729–2734 (2017).
Singh, S. P., Raja, S. & Mahalingam, S. Viral protein X unlocks the nuclear pore complex through a human Nup153-dependent pathway to promote nuclear translocation of the lentiviral genome. Mol. Biol. Cell 31, 304–317 (2020).
Chougui, G. et al. HIV-2/SIV viral protein X counteracts HUSH repressor complex. Nat. Microbiol. 3, 891–897 (2018).
Yurkovetskiy, L. et al. Primate immunodeficiency virus proteins Vpx and Vpr counteract transcriptional repression of proviruses by the HUSH complex. Nat. Microbiol. 3, 1354–1361 (2018).
Fink, D. L. et al. HIV-2/SIV Vpx antagonises NF-kappaB activation by targeting p65. Retrovirology 19, 2 (2022).
Langbein, L., Rogers, M. A., Praetzel, S., Winter, H. & Schweizer, J. K6irs1, K6irs2, K6irs3, and K6irs4 represent the inner-root-sheath-specific type II epithelial keratins of the human hair follicle. J. Investig. Dermatol. 120, 512–522 (2003).
Etienne-Manneville, S. Cytoplasmic intermediate filaments in cell biology. Annu. Rev. Cell Dev. Biol. 34, 1–28 (2018).
Schweizer, J. et al. New consensus nomenclature for mammalian keratins. J. Cell Biol. 174, 169–174 (2006).
Magin, T. M., Vijayaraj, P. & Leube, R. E. Structural and regulatory functions of keratins. Exp. Cell Res. 313, 2021–2032 (2007).
Herrmann, A. et al. The SAMHD1-mediated block of LINE-1 retroelements is regulated by phosphorylation. Mob. DNA 9, 11 (2018).
Goujon, C. et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 24, 559–562 (2013).
Kane, M. et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502, 563–566 (2013).
Campbell, E. M. & Hope, T. J. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 13, 471–483 (2015).
Yamashita, M. & Engelman, A. N. Capsid-dependent host factors in HIV-1 infection. Trends Microbiol. 25, 741–755 (2017).
Lukic, Z., Dharan, A., Fricke, T., Diaz-Griffero, F. & Campbell, E. M. HIV-1 uncoating is facilitated by dynein and kinesin 1. J. Virol. 88, 13613–13625 (2014).
Pawlica, P. & Berthoux, L. Cytoplasmic dynein promotes HIV-1 uncoating. Viruses 6, 4195–4211 (2014).
Jayappa, K. D. et al. Human immunodeficiency virus type 1 employs the cellular dynein light chain 1 protein for reverse transcription through interaction with its integrase protein. J. Virol. 89, 3497–3511 (2015).
Selyutina, A. et al. Cyclophilin A prevents HIV-1 restriction in lymphocytes by blocking human TRIM5alpha binding to the viral core. Cell Rep. 30, 3766–3777.e6 (2020).
Lee, K. et al. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7, 221–233 (2010).
Yamashita, M., Perez, O., Hope, T. J. & Emerman, M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLOS Pathog. 3, 1502–1510 (2007).
Fischer, D. K. et al. CA mutation N57A has distinct strain-specific HIV-1 capsid uncoating and infectivity phenotypes. J. Virol. 93, e00214–e00219 (2019).
Rebensburg, S. V. et al. Sec24C is an HIV-1 host dependency factor crucial for virus replication. Nat. Microbiol. 6, 435–444 (2021).
Lu, M. et al. Dynamic allostery governs cyclophilin A-HIV capsid interplay. Proc. Natl. Acad. Sci. USA 112, 14617–14622 (2015).
Liang, G. et al. CTNNBL1 restricts HIV-1 replication by suppressing viral DNA integration into the cell genome. Cell Rep. 38, 110533 (2022).
Gaudin, R., de Alencar, B. C., Arhel, N. & Benaroch, P. HIV trafficking in host cells: motors wanted! Trends Cell Biol. 23, 652–662 (2013).
Carnes, S. K., Zhou, J. & Aiken, C. HIV-1 engages a dynein-dynactin-BICD2 complex for infection and transport to the nucleus. J. Virol. 92, 10–1128 (2018).
Naghavi, M. H. HIV-1 capsid exploitation of the host microtubule cytoskeleton during early infection. Retrovirology 18, 19 (2021).
Hoogenraad, C. C. & Akhmanova, A. Bicaudal D family of motor adaptors: linking dynein motility to cargo binding. Trends Cell Biol. 26, 327–340 (2016).
Pierson, T. C. et al. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J. Virol. 76, 4138–4144 (2002).
Pierson, T. C. et al. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76, 8518–8531 (2002).
Albanese, M. et al. Rapid, efficient and activation-neutral gene editing of polyclonal primary human resting CD4(+) T cells allows complex functional analyses. Nat. Methods 19, 81–89 (2022).
Hulme, A. E., Perez, O. & Hope, T. J. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. USA 108, 9975–9980 (2011).
Perez-Caballero, D., Hatziioannou, T., Zhang, F., Cowan, S. & Bieniasz, P. D. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J. Virol. 79, 15567–15572 (2005).
McDonald, D. et al. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159, 441–452 (2002).
Burdick, R. C. et al. HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. USA 117, 5486–5493 (2020).
Dharan, A., Bachmann, N., Talley, S., Zwikelmaier, V. & Campbell, E. M. Nuclear pore blockade reveals that HIV-1 completes reverse transcription and uncoating in the nucleus. Nat. Microbiol. 5, 1088–1095 (2020).
Zila, V. et al. Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 184, 1032–1046.e1018 (2021).
Wang, S. et al. Neuropilin-1, a myeloid cell-specific protein, is an inhibitor of HIV-1 infectivity. Proc. Natl. Acad. Sci. USA 119, e2114884119 (2022).
Zhao, L. et al. Vpr counteracts the restriction of LAPTM5 to promote HIV-1 infection in macrophages. Nat. Commun. 12, 3691 (2021).
Wang, Q., Zhang, X., Han, Y., Wang, X. & Gao, G. M2BP inhibits HIV-1 virion production in a vimentin filaments-dependent manner. Sci. Rep. 8, 32736 (2016).
Wisniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Meissner, M. E. et al. Differential activity of APOBEC3F, APOBEC3G, and APOBEC3H in the restriction of HIV-2. J. Mol. Biol. 434, 167355 (2022).
Ellis, M. J. et al. A macrophage-based screen identifies antibacterial compounds selective for intracellular Salmonella Typhimurium. Nat. Commun. 10, 197 (2019).
Acknowledgements
We thank the laboratory members for contributing to this study and Jingjing Chen for technical support. This study was supported by the National Science Fund for Distinguished Young Scholars (grant no. 82325034), the National Key R&D Program (grant no. 2021YFC2301900), the Youth Program of the National Natural Science Foundation of China (no. 82202506), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2023-PT320-01), and the Liaoning Revitalization Talents Program.
Author information
Authors and Affiliations
Contributions
G.L. directed and conceived the research. Y.H., M.X., J.O., L.Z., T.M., X.Z., and R.W. performed the experiments and analyzed the data. H.S. provided intellectual advice on experimental design. G.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, Y., Xu, M., Ouyang, J. et al. Keratin-72 restricts HIV-1 infection in resting CD4+ T cells by sequestering capsids in intermediate filaments. Nat Commun 16, 2998 (2025). https://doi.org/10.1038/s41467-025-58218-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58218-2