Abstract
Pathogenic filamentous fungi pose a significant threat to global food security and human health. The limitations of available antifungal agents, including resistance and toxicity, highlight the need for developing innovative antifungal strategies. Antifungal proteins (AFPs) are a class of secreted small proteins that exhibit potent antifungal activity against filamentous fungi, yet the underlying mechanism remains partially understood. In this study, we investigate the molecular and cellular effects of two AFPs, PgAFP and AfAFP, on Aspergillus flavus, a representative filamentous fungus. These AFPs affect various fungal phenotypes and exert an intracellular effect by interacting with Ntp1, a fungi exclusive protein modulating diverse fungal traits. We find that Ntp1 amino acids 417–588 are critical for AFP binding and play a role in regulating growth, development, sporulation, sclerotia formation, toxin synthesis, and pathogenicity. Results generated from this study will help to control pathogenic fungi.
Similar content being viewed by others
Introduction
Plant pathogenic fungi are a significant source of biotic stress in plants, causing severe diseases in various crops worldwide, including rice blast, corn smut, and aflatoxin contamination1. These diseases cause significant reductions in crop yield, quality, and safety, and can cause notable economic and societal impacts. For example, the crop industry loses an estimated 10–23% of crops pre-harvest and 10–20% post-harvest due to pathogenic fungi2. Filamentous fungi are the most prevalent group of pathogenic fungi that produce conidia and mycelia, are widely distributed in field and post-harvest environments, and can parasitize plants, animals, and humans. Managing fungal infections remains challenging due to their inherent resistance to conventional antifungal agents, including fungicides and pesticides. The excessive and often improper use of these chemicals has led to the emergence of resistant fungal strains, which pose a serious threat to human and plant health1. This underscores the need for innovative antifungal strategies. Natural products have become an important source of bioactive compounds for preventing and controlling fungal diseases in animals and plants, and have sparked a surge in the discovery and development of antifungal substances.
Antifungal proteins (AFPs) are a group of secretory proteins predominantly synthesized by filamentous ascomycetes, particularly within the genera Aspergillus and Penicillium3. AFPs have also been identified in other fungi4. These proteins are typically categorized into several principal classes according to their species of origin5: Aspergillus giganteus antifungal protein (AFP)6, Penicillium chrysogenum antifungal protein (PAF)7, and Neosartorya fischeri anti-yeast protein (NFAP2)4. While most AFPs have been isolated and purified directly from fungal culture filtrate, their high-yield recombinant expression in prokaryotic systems is not well understood8. AFPs are characterized by several key features: (1) they have a low molecular mass (typically 50–100 amino acid residues); (2) they are highly basic (which aids in electrostatic interactions with negatively charged components of fungal cells), heat stable, and rich in cysteine; and (3) they exhibit potent, broad-spectrum antifungal activity against a variety of human and plant pathogenic fungi9,10,11. Additionally, they show minimal cytotoxicity towards mammalian cells in culture6,12. Because of these properties, AFPs are promising candidates for antifungal drug development across diverse fields, including biology, food production, and agriculture.
Although the potential of AFPs has been recognized, the precise mechanisms underlying the antifungal activity of AFPs remain incompletely understood. Previous studies revealed a high degree of sequence diversity among AFPs, yet they share a conserved three-dimensional structure, which is characterized by a cationic loop and a hydrophobic region, both of which are implicated in protein-protein interactions13,14. The γ-core motif, a conserved sequence at the N-terminus that is essential for the mature protein’s antifungal activity, interacts with negatively charged lipids on the fungal cell membrane14,15,16. AFPs exert their antifungal effects through diverse means, including (1) interactions with fungal cell wall and plasma membrane components, regarding which several studies reported that AFPs bind to fungal chitin and other cell wall components, inhibiting their synthesis and disrupting cell wall integrity17,18,19; (2) penetration and induction of regulated cell death, in which AFPs are rapidly and efficiently taken up by fungal cells at very low concentrations and then trigger a regulated cell death program in the target fungus20; (3) direct targeting of fungal organelles, where AFPs were shown to target host cells and nuclei of susceptible fungi, as demonstrated by co-localization and DNA-binding assays21; (4) modulation of fungal signaling pathways, where the cell wall integrity (CWI) pathway in A. niger17, the Pkc/mitogen-activated protein kinase (Mpk) cascade in Aspergillus nidulans22 and in Penicillium digitatum23, and the cAMP/protein kinase A (Pka) pathway in A. nidulans22 were shown to be moduled by AFPs; (5) factors affecting AFP activity, where high concentrations of cations, particularly sodium and potassium ions, can attenuate antifungal activity24. Furthermore, adding Ca2+ ions to culture medium was shown to counteract AFP toxicity, potentially by triggering the Ca2+ signaling pathway25,26.
While AFPs are known to be effective against fungal infections, their intracellular targets and precise mechanisms of action remain largely unexplored. Several pioneering studies revealed specific changes in transcriptomic profiles and protein expression patterns in fungi exposed to AFPs8,27,28,29, but more research is needed to elucidate the molecular mechanisms behind the growth-inhibiting effects of AFPs. This requires identifying and validating endogenous binding partners within their native cellular context, using techniques such as co-immunoprecipitation, mass spectrometry, and functional assays. Aspergillus flavus, a plant-pathogenic ascomycete that produces aflatoxin and causes aspergillosis, is an excellent model for studying AFP-fungi interactions.
In this study, we produce recombinant P. chrysogenum AFP (PgAFP) and Aspergillus fumigatus (AfAFP) and confirm their biotoxicity against A. flavus. Mass spectrometry assay identifies 38 potential cellular targets of PgAFP and AfAFP, which include Ntp1 which is only present in fungi. Ntp1 appears to be essential for regulating various vital processes of A. flavus, including growth, development, sporulation, sclerotia formation, toxin biosynthesis, and pathogenicity. We reveal that amino acid residues 417–588 of Ntp1 are critical for the interaction between Ntp1 and AFPs. These findings set a foundation for future studies aiming at deepening our understanding of the antifungal mechanisms of AFPs, identifying additional intracellular targets, and developing antifungal therapeutics and strategies to effectively counteract detrimental fungal infections.
Results
Structural and functional characterization of PgAFP and AfAFP and inhibition of A. flavus
As identified in the introduction, the nomenclature for AFPs has evolved with their continued discovery5,30. To deepen our understanding of the mechanisms by which AFPs inhibit fungal growth, we focused on two specific proteins from Penicillium and Aspergillus, designated PgAFP and AfAFP, respectively. PgAFP, secreted by Penicillium, effectively inhibits a variety of filamentous fungi under conventional conditions31. AfAFP was identified in A. flavus through sequence alignment methods (Fig. 1A). Both proteins were expressed in an E. coli recombinant system for two purposes: first, to produce a significant quantity of the AFPs for downstream experiments, such as inhibition assays, and second, to engineer and incorporate affinity tags into the AFPs for use in capturing interacting proteins.
A The alignment of PgAFP and AfAFP, only the mature forms of the two AFPs were expressed in E. coli. B SDS-PAGE analysis of Trx-6His-PgAFP purified using Ni-NTA column chromatography. Lane M: molecular weight marker; lane 1: precipitate post-IPTG induction; lane 2: supernatant post-IPTG induction; lanes 3–6: elution with 300 mM imidazole buffer; (C) SDS-PAGE analysis of tag-free PgAFP obtained through PreScission protease digestion of Trx-6His-PgAFP. Lane M: molecular weight marker; lane 1: Trx-6His-PgAFP prior to digestion (control); lane 2: tag-free PgAFP and Trx-6His tag post-digestion. D The cation-exchange chromatography profile of the purification of tag-free PgAFP. The sky-blue line indicates the protein 280 nm absorption values, while the gray line corresponds to the ionic strength in the elution buffer. E The gel filtration chromatography profile of the purified Tag-free PgAFP was performed. Here, the sky-blue line denotes the protein 280 nm absorption values. F SDS-PAGE analysis of PgAFP was performed to determine its molecular weight and purity. Lane M: protein marker; lane 1: control sample (Trx-6His-PgAFP); lane 2: purified PgAFP protein. G SDS-PAGE analysis of AfAFP was performed to determine its molecular weight and purity. H HPLC analysis was conducted to determine the hydrophobicity of PgAFP and AfAFP, with the black line and sky-blue line representing the protein 280 nm absorption values for PgAFP and AfAFP, and the red line indicating the percentage of phase B. I The inhibitory effects of PgAFP and AfAFP on WT strains of A. flavus was assessed by measuring the relative dry weight of the mycelia, defined as the ratio of mycelial mass in the protein-treated group to that of the WT control group. Data were presented as mean ± SEM. The experiments described above included three biological replicates. J The structural alignment of AfAFP and PgAFP revealed a β-barrel fold consisting of five β-sheets. K The structural analysis of recombinant PgAFP and AfAFP revealed the strategic placement of six cysteine residues, forming internal disulfide bonds. Source data are provided as a Source Data file.
By implementing a combined approach using codon optimization with a fusion partner (Supplementary Fig. 1), we successfully expressed both the soluble recombinant PgAFP and AfAFP in an E. coli recombinant system. The target protein was purified using Ni-NTA affinity chromatography (Fig. 1B). After the Trx-6His fusion tag was removed (Fig. 1C), the protein was isolated by binding to a RESOURCE S cation exchange chromatography column; the intrinsic basic protein properties at pH 7.4 in a low ionic strength Tris-HCl buffer prevented other protein contaminants from also binding (Fig. 1D). The homogeneity and purity of PgAFP were then confirmed via gel filtration chromatography (Fig. 1E), and the results demonstrated high protein purity (Fig. 1F). A similar purification methodology was applied to AfAFP, as delineated in Fig. 1G. The mass spectrometric data and peptide sequences, detailed in Supplementary Figs. 2 and 3 and Supplementary Data 1 and 2, validated the designed peptide sequence. Although the mass spectrometric data and peptide sequences indicated some potential residues were modified by acetylation or phosphorylation, these modifications were observed at low coverage percentages and were therefore considered negligible. We further applied liquid chromatography-tandem mass spectrometry (LC-MS/MS) under non-reducing conditions to analyze the formation of disulfide bridges in PgAFP and AfAFP (Supplementary Data 3). Additionally, a polarity analysis conducted using high-performance liquid chromatography (HPLC) revealed that the hydrophobicity of AfAFP was lower than that of PgAFP, as evidenced by its shorter retention time (Fig. 1H).
Antimicrobial assays conducted in vitro demonstrated that the discovered AfAFP effectively inhibited A. flavus at concentrations ranging from 2–4 μM, outperforming PgAFP, which required higher concentrations to achieve a similar level of inhibition (Fig. 1I). Although NMR and crystal structures of PgAFP from P. chrysogenum were previously determined12,32, we further investigated the differences between these two AFPs expressed in E. coli expression systems. We obtained the X-ray crystallography structure of PgAFP (PDB accession 9IID) (refer to Supplementary Data 4 for data collection and refinement statistics and Supplementary Fig. 4). The crystal structure of PgAFP was compared to the AlphaFold2 predicted model of AfAFP after multiple unsuccessful attempts to crystallize AfAFP (Fig. 1J). The structural alignment revealed a similarity between AfAFP and PgAFP, despite their relatively low sequence identity (Fig. 1A, J). In both protein structures, the six cysteine residues were demonstrated to have a crucial role in stabilizing the protein conformation (Fig. 1K). The LC-MS/MS data indicated that these cysteine residues formed multiple disulfide bond configurations (Supplementary Data 3). Two potential explanations for this phenomenon were proposed. First, during an LC-MS/MS analysis of disulfide bridges, the proteins were fragmented into various peptide segments, which were then reassembled based on the observed disulfide linkages. This reassembly process may have led to multiple disulfide bond arrangements. Alternatively, the PgAFP crystal structure and the predicted crystal structure of AfAFP suggested that the spatial arrangement of these cysteine residues may have facilitated the random formation of diverse disulfide networks. Under physiological conditions, the close proximity of these cysteine residues may result in the formation of various disulfide bridges.
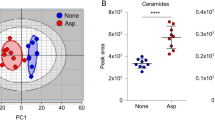
Sphingolipids within the fungal plasma membrane are pivotal regulators of pathogenicity33, and phosphatidylinositol constitutes a small membrane-associated molecule unique to filamentous fungi34. AFPs feature a positively charged motif within the γ-core region and exhibit positively charged residues on their surface, both of which are pivotal to their antifungal toxicity35. This toxicity manifests either by disrupting plasma membrane integrity or through interactions with target molecules35. AfAFP has five positively charged residues within its γ-core region, which are hypothesized to serve as potential sites for membrane interaction. In contrast, PgAFP contains only three such residues (Fig. 1J). Furthermore, the surface of AfAFP presents a greater number of solvent-exposed positively charged residues compared to PgAFP (Fig. 2A). This disparity in charge distribution may underlie the lower antifungal efficacy of PgAFP relative to AfAFP. Additionally, we assessed the ability of the heat-treated recombinant AFPs to inhibit A. flavus conidia production in both corn and peanut seeds (Fig. 2B–E). SDS-PAGE analysis confirmed the remarkable stability of these protein samples, which exhibited no degradation after exposure to heat treatment (72 °C for 72 h) or treatment with hydrolase enzymes (Supplementary Fig. 5A–D).
A Illustration of the electrostatic surface potentials of PgAFP and AfAFP, respectively, with their orientations aligned for direct comparisons. Negatively charged regions are depicted in red, positively charged regions in blue, and neutral regions in white. The graphical representations were visualized using the PyMol APBS electrostatics plugin. Both proteins displayed predominantly positive charges on their front surfaces. However, on the back surface, PgAFP exhibited multiple negatively charged regions involving the N-terminal residues Gly8, Glu9, Cys10, Thr16, and Thr18, and the C-terminal residues Asp41, His44, Glu46, Thr58, Pro59, and Val60. In contrast, AfAFP only featured a negatively charged region known as the “oxygen hole,” comprising residues Ala35, Asn36, Arg38, Thr40, Cys57, and Lys58. The molecular weights of PgAFP and AfAFP were determined using mass spectrometry, and their isoelectric points were calculated using SnapGene software (version 6.0). The electrostatic surfaces were computed using Adaptive Poisson–Boltzmann Solver (APBS) software (version 3.4.1)72. B, C Inhibitory effect of heat-treated rAFPs on conidia production by A. flavus in corn and peanut seeds. (D, E) Statistical analysis of conidia production by A. flavus in (B, C). Data were presented as mean ± SEM (n = 3). Statistical significance was determined by one-way ANOVA and indicated by ** for P < 0.01. All experiments were performed in triplicate. Source data are provided as a Source Data file.
AFPs reduce chitin content, accumulate in the cytoplasm, and interact with fungal protein
In filamentous fungi, the cell wall is composed of chitin, chitosan, glucans, and glycoproteins, with chitin playing a pivotal role in maintaining structural integrity and facilitating morphological transitions36. Our observations indicate that treatment with AFPs leads to a reduction in the chitin content of wild-type strains of A. flavus, suggesting that these proteins have an inhibitory effect on chitin biosynthesis (Supplementary Fig. 6A). This inference is further substantiated by the results of staining AFP-treated cells with the chitin-binding dye calcofluor white (CFW) (Supplementary Fig. 6B). In the absence of AFPs, fluorescence intensity was markedly diminished following treatment with either AfAFP or PgAFP. However, the addition of calcium ions restored CFW fluorescence intensity in A. flavus treated with PgAFP or AfAFP to levels comparable to those of the untreated wild-type control (Supplementary Fig. 6C). Moreover, deletion of genes such as crz1, which encodes a transcription factor implicated in calcium homeostasis, or knockout of chitin synthase genes (Δchs3, Δchs5, or the double knockout Δchs3Δchs5) in A. flavus, resulted in decreased CFW fluorescence intensity (Supplementary Fig. 6D, E). These gene deletion mutants also exhibited reduced chitin content (Supplementary Fig. 6F), which was further diminished upon AFP treatment, reaching levels lower than those observed in the control strain (i.e., deletion mutants without AFP treatment) (Supplementary Fig. 6G–I). These findings indicated that AFPs disrupted the cell wall integrity by reducing chitin levels, and they had more pronounced effects on cells with impaired cell walls.
Building upon our previous experiments demonstrating that AFPs reduced the chitin content in fungal cell walls, thereby disrupting cell wall integrity, we extended our investigation to elucidate the intracellular pathways activated by AFPs and potential molecular targets. Both AFPs were conjugated to the green fluorescent dye fluorescein isothiocyanate (FITC) to track their intracellular localization within Aspergillus flavus cells using confocal microscopy (Supplementary Fig. 5E and 5F). Importantly, FITC labeling did not compromise the antifungal efficacy of the AFPs, which continued to effectively inhibit fungal growth (Supplementary Fig. 5G). Within 2 h of incubation, both proteins successfully traversed the cellular membrane barrier and localized to the cytosol. At 2 and 8 h post-uptake, the AFPs predominantly accumulated within the cytoplasm (Supplementary Fig. 5E, F). Viability assays with propidium iodide (PI) staining revealed a progressive increase in cell death, which was particularly pronounced 8 h post-treatment (Supplementary Fig. 5E, F). Collectively, these findings suggested that AFPs reduced the cell wall chitin content, were internalized by fungal cells, and accumulated in the cytoplasm where they ultimately induced cell death.
To identify the intracellular targets of AFPs in A. flavus, we used tandem affinity purification and mass spectrometry (TAP-MS). We engineered recombinant PgAFP and AfAFP to include Flag and Strep-tag II epitopes (Fig. 3A and Supplementary Fig. 7A, B). We obtained purified PgAFP-Flag-Strep protein (PgAFP-F-S) (Fig. 3B–F). However, AfAFP-Flag-Strep protein (AfAFP-F-S), tagged with thioredoxin, predominantly formed inclusion bodies. To enhance the solubility of AfAFP-F-S, we introduced an additional solubilizing glutathione S-transferase (GST) tag (Supplementary Fig. 7B). After cleaving the GST and Trx tags with 3C protease, we found that separating the AfAFP-F-S protein from the protein mixture using a RESOURCE S cation exchange chromatography column was challenging (Fig. 3C). Therefore, we instead successfully isolated the fraction containing the AfAFP-F-S protein and GST tag using the following method. A sample containing the AfAFP-F-S protein and GST tag was applied to a Superdex 75 column. We collected fractions based on UV absorption peaks, concentrated them, and then performed a second purification using the same column (Fig. 3E). The PgAFP-F-S and AfAFP-F-S proteins exhibited a clean background on SDS-PAGE (Fig. 3F) and were verified by mass spectrometry (Supplementary Data 5 and 6). The secondary mass spectra data of the proteins confirmed the presence of the Flag and Strep II motifs at their C-termini, which was different from AFPs lacking these tags (Supplementary Figs. 2 and 3). To ensure that the peptide epitopes did not induce potential steric hindrance, we used AlphaFold2 to predict the AFP conformations with these two tags (Fig. 4A–C). Our predictions indicated that the tags did not affect the protein structure, given the flexibility of the C-terminal regions. Furthermore, antifungal experiments confirmed that the AFPs with Flag and Strep tags maintained their antifungal activity (Fig. 3H).
A The alignment of PgAFP and AfAFP, both tagged with Flag and Strep, was meticulously delineated. B Purification and tag removal of recombinant proteins Trx-6His-PgAFP-F-S and Trx-6His-AfAFP-F-S-GST were performed using Ni-NTA affinity chromatography, followed by PreScission protease-mediated cleavage to excise the Trx-6His and GST tags. Lane M: molecular weight marker; lane 1: Trx-6His-PgAFP-F-S pre-cleavage; lane 2: Trx-6His-PgAFP-F-S post-cleavage, yielding Trx-6His tag and PgAFP-F-S; lane 3: Trx-6His-AfAFP-F-S-GST pre-cleavage; lane 4: Trx-6His-AfAFP-F-S-GST post-cleavage, yielding Trx-6His tag, GST tag, and AfAFP-F-S. C SDS-PAGE analysis of PgAFP-F-S and AfAFP-F-S following purification by cation-exchange chromatography. Lane M: molecular weight marker; lane 1: mixture of PgAFP-F-S protein and Trx-6His pre-purification on a cation exchange column; lane 2: cation exchange column flow-through fraction, containing PgAFP-F-S protein; lane 3: PgAFP-F-S protein eluted from a cation exchange column; lane 4: mixture of PgAFP-F-S protein, GST, and Trx-6His pre-loading on a cation exchange column; lane 5: cation exchange column flow-through fraction, containing AfAFP-F-S protein; lane 6: AfAFP-F-S protein eluted from a cation exchange column. The elution was further refined by dual gel filtration to achieve highly purified AfAFP-F-S protein. D, E Top: Cation-exchange chromatography profiles of PgAFP-F-S and AfAFP-F-S, along with their associated tags. Bottom: Gel filtration chromatography profiles of the purified PgAFP-F-S and AfAFP-F-S proteins. The sky-blue line indicates the protein 280 nm absorption values, while the gray line corresponds to the ionic strength in the elution buffer. F SDS-PAGE analysis of the PgAFP-F-S and AfAFP-F-S proteins purified by gel filtration chromatography is shown. Lane M: molecular weight marker; lane 1: purified PgAFP-F-S protein; lane 2: purified AfAFP-F-S protein. G HPLC analysis was conducted to determine the hydrophobicity of PgAFP-F-S and AfAFP-F-S. The black and sky-blue lines represent the 280 nm absorbance values of PgAFP-F-S and AfAFP-F-S, respectively, while the red line denotes the percentage of phase B. H Inhibitory effects of PgAFP-F-S and AfAFP-F-S on wild-type (WT) strains of A. flavus were assessed. Data are presented as mean ± SEM (n = 3). All experiments were conducted in biological triplicates. Source data are provided as a Source Data file.
A The overall crystal structure of PgAFP, obtained through recombinant expression methods, is presented as a gray-colored cartoon representation. The predicted three-dimensional structure of AfAFP is shown. B, C The three-dimensional structures of PgAFP-Flag-Strep and AfAFP-Flag-Strep fusion proteins are illustrated, showing the molecular architecture. D Tandem affinity purification coupled with mass spectrometry (TAP-MS) was used with PgAFP-Flag-Strep and AfAFP-Flag-Strep as bait protein to isolate potential interacting proteins of AFPs in A. flavus. A total of 76 and 58 candidate protein targets were identified, respectively. A subsequent comparative analysis of the proteomes identified 38 proteins potentially influenced by the AFPs. E The 38 candidate interacting proteins of AFPs in A. flavus were systematically categorized. F, G Analysis of the binding interactions between Ntp1546–588. E The 38 candidate interacting proteins of AFPs in A. flavus were systematically categorized. F, G Analysis of the binding interactions between Ntp1546–588 (colored with gray) and the Flag-Strep tagged variants of PgAFP (yellow) and AfAFP (yellow) suggested that the Flag-Strep tag did not contribute to the recognition process of Ntp1 by the AFPs. Furthermore, the predicted protein-protein interaction structures are categorized by four levels of confidence intervals. High confidence: regions with pLDDT > 90, indicating high modeling accuracy. Moderate confidence: regions with pLDDT between 70 and 90, generally reflect accurate backbone predictions. Low confidence: regions with pLDDT between 50 and 70, which should be interpreted cautiously. Structural disorder: regions with pLDDT < 50, suggesting potential structural disorder.
The purified double-tagged AFPs were incubated with A. flavus. This process is fully detailed in the Methods Section. For the purposes of this experiment, the Flag-Strep tag was chemically synthesized to function as the control. Three experimental groups were established: AfAFP-F-S, PgAFP-F-S, and F-S, to facilitate comparative analysis. Proteomic analysis was performed using LC-MS/MS, which identified 38 putative protein interactors (as detailed in Fig. 4D,E and Supplementary Data 7). Subsequent efforts focused on the generation of A. flavus deletion mutants corresponding to the genes encoding the 38 proteins. Despite attempts to disrupt all 38 AFP-interacting protein (AFPIP) genes, we were unable to obtain viable mutant transformants for 19 of the genes, suggesting that these genes were essential for A. flavus viability. Consequently, our comprehensive analysis centered on elucidating the functions of the remaining 19 genes that were successfully deleted in A. flavus. This included evaluating colony growth, sporulation, sclerotial development, aflatoxin synthesis, pathogenicity, and stress response mechanisms, as detailed in Supplementary Data 8.
Ntp1 proteins are fungal-specific regulators with diverse functions
Using tandem affinity purification coupled with mass spectrometry, Ntp1 was identified as one of 38 AFPIPs. Interestingly, although the ntp1 gene and its corresponding protein sequence were available in NCBI databases (Fig. 5A, and Supplementary Figs. 8 and 9), Ntp1 is largely unannotated in current bioinformatics repositories, suggesting a unique and potentially unprecedented role in A. flavus. Disrupting the ntp1 gene markedly enhanced the organism’s resistance to both recombinant PgAFP and AfAFP (Fig. 5B,C). To further substantiate the interaction between Ntp1 and AFPs, fluorescence co-localization studies were conducted (Fig. 5D, E). Red fluorescent dye-labeled AFPs co-localized with Ntp1-GFP, producing orange signals in the merged images (Fig. 5D, E). This confirms their intracellular co-localization, providing robust evidence of their interaction. Red fluorescent dye-labeled AFPs were also able to inhibit fungal growth, as shown in Fig. 5F. This was further validated through co-immunoprecipitation assays using anti-HA antibodies, which reinforced the significance of Ntp1 as an intracellular target of AFPs within A. flavus (Fig. 5G, H). Collectively, these observations suggested that the ntp1 gene encoded a pivotal intracellular effector protein integral to the function of AFPs.
A Sequence analysis of the unannotated antifungal interaction protein (Ntp1) was performed using the Pfam database, which provided insights into the protein families, domains, and other characteristic features. Using this, the design of Ntp1 truncated mutants was meticulously detailed, and identified the amino acid ranges of each segment: part 1 (1–284), part 2 (285–416), part 3 (417–588), and part 4 (589–708). This figure was created using Microsoft PowerPoint 365. B, C Inhibitory effects exerted by PgAFP and AfAFP on Ntp1 mutant strains of A. flavus were determined by measuring the mycelial dry weight. Data are presented as mean ± SEM (n = 3). All the experiments described above were conducted in triplicate. D, E Confocal laser microscopy was used to visualize the interactions between fluorescently labeled AFPs and Ntp1-GFP in A. flavus mycelium. Specifically, tetramethylrhodamine isothiocyanate-labeled PgAFP (PgAFP–TRITC) and AfAFP (AfAFP–TRITC) were used. Fungal spores germinated into mycelia over 18 h and were subsequently treated with 3 mg/mL of the aforementioned proteins for various intervals, ranging from 0 to 8 h. Representative images from confocal fluorescence microscopy of fungal germlings, including untreated controls and those treated with the two AFPs, were captured. Individual fluorescence images were recorded for the AFPs and Ntp1-GFP at their respective excitation and emission wavelengths. To enhance visualization, merged images are presented in both darkfield and brightfield modes. All micrographs include a scale bar corresponding to 10 μm. F Inhibitory effects of PgAFP–TRITC and AfAFP–TRITC on the A. flavus WT strain. Data are presented as mean ± SEM (n = 3). All the experiments described above were conducted in triplicate. G, H Co-immunoprecipitation of HA-tagged Ntp1 with AfAFP and PgAFP. Top: Western blot with anti-HA antibodies using whole-cell lysates of A. flavus expressing HA-Npt1. Middle: Western blots of immunoprecipitates (with anti-HA antibody) from the corresponding lysates. Bottom: Strep-Tactin beads loaded with purified AfAFP-F-S, PgAFP-F-S, and F-S proteins, respectively. The co-immunoprecipitation experiments were performed three times, each producing similar results. Source data are provided as a Source Data file.
By examining multiple protein databases, we found that Ntp proteins are exclusive to fungi and absent in mammalian systems. Along with their potential involvement in the AFP-mediated mechanism, they have emerged as promising targets for antifungal therapies. However, the precise functions of Ntp proteins remain largely unclear. A Pfam analysis of Ntp1 delineated five distinct structural domains, including two putative metallo-dependent phosphatase-like domains (residues 48–99 and 129–297) located in the N-terminal region. Another notable domain with an unknown function, DUF2433 (residues 285–414), can serve as a fungal-specific marker as it is absent in bacteria and animals (Fig. 5A and Supplementary Fig. 9). Additionally, two subsequent domains (residues 417–548 and 589–708) lacked homologous sequences in the Pfam database (Supplementary Fig. 9), suggesting unique structural features. This structural analysis underscored the significance of generating Ntp1 truncated mutants to elucidate the functional role of the protein. By segmenting the amino acid sequence into defined regions—part 1 (residues 1–284), part 2 (285–416), part 3 (417–588), and part 4 (589–708)—we engineered three Ntp1 deletion mutants (Δntp1285–708, Δntp1417–708, and Δntp1589–708), each lacking specific regions of the protein (Fig. 5A and Supplementary Fig. 9). These mutants provided critical insights into the essential functional regions of Ntp1 and allowed us to assess whether the absence of particular domains impaired recognition and interaction with the AFPs.
To pinpoint the precise Ntp1 domain responsible for interacting with the AFPs, we performed co-immunoprecipitation assays using PgAFP-F-S and AfAFP-F-S as investigative bait. The truncated HA-Δntp1285–708 protein failed to precipitate either AFP, while another variant, HA-Δntp1417–708, showed minimal interaction with the AFPs (Fig. 6A, B). This suggested that the deletion of residues 285–708 or 417–708 significantly impaired the binding ability of Ntp1. Conversely, the HA-Δntp1589–708 truncated version of Ntp1 exhibited enhanced binding relative to the other mutants. In our molecular docking studies, we performed predictive modeling on the full-length structures of Ntp1 with the two AFPs, and on the 417–588 region of Ntp1 with both AFPs. Although initial models posited the 417–588 sequence of Ntp1 as a potential binding site for AFPs, the structural reliability of the models was called into question by less-than-ideal pLDDT, pTM, and ipTM scores obtained from AlphaFold2 and AlphaFold3 beta. Therefore, we refined our focus on docking AFPs to the 546–588 section of the 417–588 region of Ntp1. This specific docking demonstrated relatively satisfactory results when analyzed using AlphaFold2 multimer. The docking structures of the Ntp1546–588-PgAFP and Ntp1546–588-AfAFP complexes revealed that the β3 sheet of PgAFP and AfAFP acted as the primary interface for Ntp1, with binding characterized by hydrophobic interactions and hydrogen bonds (Fig. 6C). In contrast, the β2 and β1 sheets, despite containing the γ-core motif known for antifungal properties, were not imperative for the interaction between AFP and Ntp1 (Fig. 6C). Additionally, the presence of the Flag and Strep-tag II epitopes at the C-terminus did not impede the protein-protein recognition process (Fig. 4F, G). To further elucidate how Ntp1 modulated the antifungal activity of the AFPs, we assessed the susceptibility of each deletion mutant to PgAFP and AfAFP. As expected, the Δntp1285–708, Δntp1417–708, and Δntp1 mutants exhibited increased resistance to both AFPs compared to the WT and the Δntp1589–708 mutant strains (Fig. 5B, C). Thus, these observations and molecular simulations compellingly suggested that the segment spanning amino acids 417–588 was the principal AFP-binding domain of Ntp1 and strongly modulated resistance to AFPs.
A, B Analysis of the interaction between AfAFP and PgAFP with various Ntp1 mutant proteins. Top: Western blot detection with anti-HA antibodies in whole-cell lysates of A. flavus expressing HA-tagged Ntp1 mutant proteins. Middle: Western blots of immunoprecipitates with anti-HA antibody from the corresponding lysates. Bottom: Strep-Tactin beads loaded with purified AfAFP-F-S, PgAFP-F-S, and F-S proteins. C In silico docking simulations using AlphaFold2 multimer to analyze interactions between Ntp1546–588 and PgAFP with AfAFP. The simulations revealed that for Ntp1546–588 and PgAFP, there were three hydrogen bonds (two between Ntp1V579 and PgAFPV25, and one between Ntp1F581 and PgAFPN23) and one salt bridge (between Ntp1K588 and PgAFPE7). For Ntp1546–588 and AfAFP, there were four hydrogen bonds (two between Ntp1V579 and AfAFPF26, one between Ntp1F581 and AfAFPD24, and another between Ntp1N582 and AfAFPK23). The visual representations of PgAFP and AfAFP are depicted as cartoons in yellowish brown and light red, respectively, while the structure of Ntp1546–588 is shown in light blue. The predicted protein-protein interaction structures are also categorized by four levels of confidence intervals. D Growth phenotypes of WT, Δntp1285–708, Δntp1417–708, and Δntp1589–708 strains on YGT medium. E Colony diameter measurements of the strains. F Microscopic view of conidiophore development in the WT, Δntp1285–708, Δntp1417–708, and Δntp1589–708 strains. G Spore counts of the strains. H Sclerotia formation phenotypes of WT, Δntp1285–708, Δntp1417–708, and Δntp1589–708 strains on CM medium. I The amount of sclerotia generated by these strains. J Aflatoxin production in WT, Δntp1285–708, Δntp1417–708, and Δntp1589–708 mutant strains, as detected by thin-layer chromatography (TLC). K Optical density-based semi-quantification of aflatoxin levels corresponding to (J). Data in (E, G, I, K) are presented as mean ± SEM (n ≥ 3). Statistical significance was determined by one-way ANOVA and indicated by *** for P < 0.001. All experiments were performed in triplicate. Source data are provided as a Source Data file.
Deleting Ntp1 significantly impacted A. flavus growth and development. All three mutants displayed reduced colony sizes and conidiation compared to the WT strain. The colony diameters of the Δntp1285–708 and Δntp1417–708 strains were comparable to that of the full-length deleted strain (Δntp1 strain), but smaller than that of the WT strain. However, the Δntp1589–708 strain exhibited a partially restored colony phenotype relative to the Δntp1 strain (Fig. 6D, E). Microscopic observations showed that the deletion strains had smaller and sparser conidial heads and conidiophores than the WT strain, particularly Δntp1285–708 and Δntp1417–708 (Fig. 6F). The spore numbers of the Δntp1, Δntp1285–708, and Δntp1417–708 strains were noticeably lower than those of the WT strain (Fig. 6G). In contrast, the sporulation of the Δntp1589–708 strain was similar to that of the WT strain, as shown in Fig. 6H, I. Aflatoxin production, a major concern due to its toxicity, showed an unexpected pattern. Thin-layer chromatography results indicated that the aflatoxin production of the Δntp1285–708, Δntp1417–708, and Δntp1 strains were notably higher than that of the WT strain, while the aflatoxin production of the Δntp1589–708 strain was similar to that of the WT strain (Fig. 6J, K), implying a regulatory role for the 417–588 region in aflatoxin biosynthesis. The altered virulence of the various mutant strains was reflected in the spore numbers of infected peanut and maize seeds. The spore numbers of the Δntp1285–708 and Δntp1417–708 truncated mutant strains were similar to those of the Δntp1 deletion mutant strain, but considerably lower than those of the WT strain (Fig. 7A–D). Moreover, the Δntp1285–708 and Δntp1417–708 truncated mutant strains synthesized aflatoxins at a rate comparable to that of the Δntp1 deletion mutant strain during peanut and maize infestation, which was markedly higher than that of the WT strain (Fig. 7E–H). Based on the results presented above, we hypothesized that part 3 (417–588) could be the main regulatory functional region of Ntp1.
A, B Phenotypic observations of WT, Δntp1285–708, Δntp1417–708, and Δntp1589–708 mutant strains colonizing on peanut and corn seeds. (C, D) Comparison of the spore yields by the strains on infected peanut and corn seeds. E, F Toxin biosynthesis by mutant strains on infected peanut and corn kernels was analyzed by TLC. G, H The optical density of toxin accumulated in infected peanuts in (E) and corn kernels in (F). Data in (C, D, G, H) are presented as mean ± SEM (n = 3). Statistical significance was evaluated using one-way ANOVA, with *** indicating P < 0.001. All experiments were performed in triplicate. Source data are provided as a Source Data file.
Discussion
This study delved into the intricate interplay between AFPs and the filamentous fungus A. flavus. We demonstrated that AFPs significantly reduced the chitin content in the fungal cell wall and were subsequently internalized into the fungal cell. Furthermore, we identified an effector protein, Ntp1, which was shown to bind to AFPs and modulate various fungal characteristics. Our findings shed light on the multifaceted mode of action of AFPs against A. flavus, suggesting potential antifungal drug development strategies. The mode of action is illustrated in Fig. 8.
The cell wall and the cell membrane (the two outer layers) are completely different cellular structures. Upon disrupting the cell wall barrier, AFPs significantly reduce the chitin content within the fungal cell wall. The Ch3 and Ch5 genes are responsible for chitin synthesis, which is critical for maintaining cell wall integrity. However, the calcium/Crz pathway confers resistance to AFPs. Following the reduction of chitin, AFPs are internalized into the fungal cell across the cell membrane phospholipid bilayer. The mechanism by which AFPs traverse the membrane requires further exploration. Ntp1 was identified as a novel intracellular effector protein for AFPs but is not present in human or bacterial genomes. The region spanning amino acids 417–588 of Ntp1 is essential for binding AFPs and mediating their antifungal effects on A. flavus. The next step of our research is to discover and identify more unknown AFPIPs. This figure was created using Microsoft PowerPoint 365, with design enhancements by Copilot and Figdraw.
Cysteine-stabilized antifungal proteins, particularly abundant in Eurotiales fungi30,37, have promising potential due to their antifungal properties. However, the endogenous synthesis of these proteins is frequently constrained by low expression rates and a reliance on specific environmental triggers. For example, AnAFP from A. niger was detectable solely after extended periods of carbon deprivation, and its transcriptional surge did not coincide with the peak of protein expression37. Similarly, AfpB from Penicillium digitatum did not exhibit a parallel increase in protein levels despite a hundredfold increase in mRNA when expressed constitutively3. The transcription of the afp gene was shown to be influenced by various environmental stimuli, including pH, heat treatment, and chemical molecules38,39, which presents substantial obstacles for achieving consistent protein expression, purification, and functionality.
The heterologous expression of cysteine-rich antifungal proteins from filamentous ascomycetes in E. coli systems presents a multifaceted challenge. Recently, it was reported that an initial attempt to express eight different AFPs in E. coli resulted in the successful expression of only a single protein8. In our preliminary experiments, the recombinant AFPs in E. coli systems predominantly formed inclusion bodies, markedly hindering their expression8. We postulated that two principal impediments were encountered: first, a codon usage disparity between eukaryotic genes and prokaryotic systems; second, the intrinsic properties of AFPs, which are small and cysteine-rich, potentially causing suboptimal expression and improper protein folding in prokaryotic hosts. To surmount these challenges, codon optimization was performed to modify synonymous codons to better reflect the codon usage bias of the host organism. Additionally, a Trx tag was added to promote correct protein folding.
Using optimized bacterial recombinant systems, we successfully purified the PgAFP and AfAFP proteins from E. coli expression systems and achieved substantial yields of soluble and stable proteins that exhibited potent antimicrobial activity against A. flavus. The E. coli prokaryotic platform provided several advantages for AFP production: (1) efficiency and cost-effectiveness, i.e., compared to native fungal hosts, bacterial systems have faster growth rates and lower production costs, rendering them exceptionally conducive to large-scale AFP production; (2) genetic manipulation, i.e., recombinant systems support convenient AFP gene manipulation through techniques such as site-directed mutagenesis and fusion tags. These modifications also further improved the solubility and stability of the recombinant proteins, enhancing their suitability for downstream applications.
Most AFPs have been observed to disrupt the cell wall barrier of filamentous fungi. Previous studies hinted at an interaction between AFPs and chitin17, a major component of fungal cell walls. Despite our study did not detect direct interactions between AFPs (AfAFP and PgAFP) and chitin, both proteins exhibited significant inhibitory effects on chitin synthesis. Our findings aligned with previous report on PAF and A. nidulans22, which proposed that AFPs may preferentially target cell wall remodeling enzymes, as opposed to exerting a direct effect on chitin itself. In contrast, in Fusarium acnes and Aspergillus oryzae, knocking out the class III and V chitin synthase genes (chs3, chs5) increased resistance to AFPs17. Similarly, in Penicillium digitatum, mutants deficient in chs5 exhibited a slight increase in chitin levels. However, this change did not correspond to an increased susceptibility to AFPs compared to the original strains40. This highlights the intricate and multifaceted nature of fungal defense mechanisms against AFPs.
Calcium ions (Ca2+) have emerged as pivotal modulators of the interactions between AFPs and fungal cells. Our results demonstrated that calcium supplementation mitigated the antifungal effects of AfAFP and PgAFP, and effectively reversed the chitin accumulation induced by these proteins. This observation was consistent with the established role of calcium in CWI pathway activation, which enhanced chitin synthesis and fortified fungal cell walls against antifungal agents, such as caspofungin41,42 and other antifungal protein43,44. Furthermore, our study indicated that the outcome of deleting the crz1 gene corroborated with the findings reported in the existing literature and emphasized the critical function of crz1 in conferring resistance to AFPs in yeast cells25. Intriguingly, our data also indicated that AFPs penetrated the cell wall barrier and induced intracellular cell death, as evidenced by PI staining. This implied that the cell wall is not the exclusive target of AFPs17,18,19,20,22,23, and that AFPs may have multiple modes of action that compromise fungal viability. The influence of Ca2+ ions on the antifungal activity of AFPs can also be explained by Ca2+ ions competing with cationic AFPs to bind negatively charged sites on the fungal cell membrane or to components within the lipid bilayer, thereby interfering with the interaction between AFPs and the fungal cell.
Quantitative proteomic analysis has become an indispensable technique for unraveling the molecular intricacies of antimicrobial peptides. By identifying differentially expressed proteins, researchers can infer potential signaling cascades affected by these agents (e.g., amphotericin B) within host cells27,45,46. However, traditional mass spectrometry-based proteomics, despite providing comprehensive insights into cellular dynamics, are limited in their ability to resolve mechanisms involving direct communication between exogenous AFPs and intracellular proteins in fungi. This gap in knowledge significantly hinders our understanding of non-cell-membrane or wall-targeting mechanisms of AFPs, particularly their entry modes and intracellular killing mechanisms. While AFPs can disrupt fungal cell walls, cross the cell membrane phospholipid bilayer, and are easily taken up by fungal cells20, the finer details of their intracellular actions remain elusive.
To address this critical gap in our understanding, our pioneering study introduced an innovative methodology using an exogenous tandem affinity tag technique to capture potential endogenous AFP-interacting proteins in filamentous fungi. Using this technique, we isolated putative AFP-binding proteins within the model organism A. flavus. Through a comparative analysis of protein profiles in response to specific AFPs (PgAFP and AfAFP), we gained crucial insights into the intricate interplay between these proteins and fungal cellular components. This innovative approach revealed a number of potential AFP-binding proteins, suggesting the presence of a network of interactions more intricate than previously understood. During the MS identification phase, we used a Flag-Strep tag protein as a control to minimize nonspecific adherence to potential AFP-binding proteins. We refined our analysis by focusing on the intersection of proteins consistently identified across multiple comparative assessments of binding protein data for both PgAFP-F-S and AfAFP-F-S proteins. In the next phase, we will elucidate the precise functions of the 38 potential AFP-binding proteins in A. flavus, including their direct interactions with AFPs and their potential regulation of various phenotypes of this filamentous fungus. Our research provides a gateway to a deeper understanding of the multifaceted antifungal mechanisms orchestrated by AFPs.
While antimicrobial peptide research has thoroughly investigated bacterial antagonism using modalities such as membrane disruption, nucleic acid binding, and protein synthesis inhibition47, the exploration of AMPs targeting intracellular enzymes and proteins in fungi is notably limited. The discovery of a proline-rich peptide, spanning 18–20 amino acids, that impedes DnaK ATPase activity in E. coli48,49, and the plant-derived osmotin, ~240 residues in length, that triggers apoptosis in yeast via a G-protein coupled receptor-like mechanism50, are exceptions rather than the norm. However, the sequences of these peptides are markedly distinct from the AFPs used in our investigation, and the methods used to detect them, including immobilization of the peptides on Sepharose-4B beads to capture intracellular constituents and membrane fractions in lysate, contrasted with our tandem affinity purification strategy. Our method involved incubating fungi with AFPs followed by target protein identification. This distinction highlights the inherent challenges in pinpointing AFP effector proteins under physiological conditions, which is vital for elucidating their complex antifungal actions and developing efficacious antifungal agents.
Our investigation characterized an intracellular effector, Ntp1, that interacts with AFPs. Ntp1 comprises a putative serine/threonine phosphatase domain and a domain of unknown function (DUF2433), the latter of which is predominantly observed in fungal species. Both domains are homologous with the fungal glutaminase GtaA family. Additionally, Ntp1 contains two intrinsically disordered regions that merit further structural and functional analysis (Fig. 5A). Confocal microscopy-based co-localization analysis revealed that the red fluorescence signal from AFPs-TRITC and the green fluorescence signal from Ntp1-GFP have a few overlapping spots (Fig. 5D, E). Several factors may account for this observation, including (1) Multiplicity of AFP interactions: LC-MS analysis identified 38 potential AFP-interacting proteins, suggesting that AFPs could engage with multiple intracellular targets beyond Ntp1, leading to reduced co-localization signals; (2) Potential labeling interference: Although TRITC dyes had a minimal impact on AFP activity (Fig. 5F and Supplementary Fig. 5G), chemical labeling could partially obscure critical interaction sites between AFP and Ntp1; (3) Steric hindrance from fusion tags: Size of GFP tag fused to Ntp1 is substantially larger than AFP, potentially hindering or altering native interaction dynamics between AFP and Ntp1. To substantiate the direct interaction between AFP and Ntp1, we conducted inhibition assays using truncated Ntp1 variants and Co-IP experiments. The inhibition assays revealed that the Ntp1 region encompassing amino acids 285–588 was responsive to AFPs. Complementary Co-IP analyzes further refined this interaction, pinpointing the 417–588 amino acid segment of Ntp1 as the specific binding locus for AFPs—a finding corroborated by protein-protein docking simulations. Functional assays and phenotypic studies further demonstrated that the 417–588 region was critical for A. flavus growth, development, and secondary metabolism, whereas the 1–416 sequence appeared non-essential. Interestingly, increased cell death, as evidenced by PI staining, was observed starting at 4 h, with co-localization of AFP and Ntp1 signals emerging between 2 and 4 h post-treatment. Attempts to crystallize the Ntp1-AFP complexes for structural elucidation were impeded by precipitation issues during purification. We thus propose that Ntp1 functions as an intracellular transporter for AFPs and that their binding may inactivate or destabilize Ntp1, thereby contributing to fungal cell death12. Determining whether Ntp1 primarily acts as an initial AFP-binding molecule or as a secondary interaction partner within fungal cells requires further investigation. These investigations may include super-resolution video imaging for dynamic real-time fluorescence tracing, along with additional experiments focusing on earlier AFP interaction events, ultimately clarifying Ntp1’s specific functional role. Moreover, elucidating the precise mechanisms and interactions between Ntp1 and AFPs is crucial for verifying its role as a versatile intracellular target that potentially governs multiple pathways in A. flavus.
The uniqueness of Ntp1, which is exclusive to fungi and is absent in humans, likely contributes to the selective antifungal efficacy of AFPs. This specificity may explain why most AFPs are capable of inhibiting mold proliferation without affecting human cells12. In contrast, the receptor for the plant defense protein osmotin is reported to be the plasma membrane protein PHO36, which has a mammalian counterpart that functions as a receptor for the hormone adiponectin50. Despite the lack of sequence homology between osmotin and adiponectin, these two proteins have demonstrated analogous biological activities across species50. Additionally, certain antimicrobial peptides are known to protect humans from infections by exerting antimicrobial actions and eliciting immune responses51. Consequently, it is essential to conduct further research to determine whether these two AFPs trigger an immune response in humans, thereby aiding in the defense against foreign pathogens.
Methods
Strains and culture conditions
Plasmid DNA preparation and recombinant protein expression were performed using E. coli strains DH5α and BL21(DE3), respectively. In this study, the wild-type A. flavus (WT) strain (Δku70) was maintained in our laboratory52. The CA14PTs strain (Δku70; ΔpyrG) used for constructing other mutant strains was kindly provided by Dr. Perng-Kuang Chang53. The growth of the A. flavus strains was evaluated on YGT media (comprising 6 g/L yeast extract, 20 g/L glucose, 1 mL/L trace elements, and 1.5% agar) at 37 or 28 °C. Sclerotia production of the strains was assessed on CM media (complete medium containing 6 g/L yeast extract, 6 g/L peptone, 10 g/L sucrose, and 1.5% agar), and aflatoxin biosynthesis was examined in a YES liquid culture (yeast extract/sucrose medium; 2% yeast extract, 150 g/L sucrose, and 1 g/L MgSO4 7H2O) at 28 °C. The trace element solution (100 mL) consisted of 2.2 g of ZnSO4•7H2O, 1.1 g of H3BO3, 0.5 g of MnCl2•4H2O, 0.5 of g FeSO4•7H2O, 0.17 g of CoCl2•5H2O, 0.16 g of CuSO4•5H2O, 0.005 g of (NH4)6Mo7O24•5H2O, and 4.45 g of Na2EDTA54.
Mutant strain construction
A homologous recombination strategy was used to generate a deficient strain, as described in Supplementary Fig. 10A55. In brief, the upstream and downstream flanking regions of the target gene and the A. fumigatus pyrG nutritional marker were amplified using specific primers, as listed in Supplementary Data 9. The fragments were then purified using a gel extraction kit and subsequently fused using overlap extension PCR to construct a homologous recombination cassette. The final fusion PCR product was further purified and transformed into CA14PTs strain protoplasts. The protoplasts were prepared by digesting the cell wall of freshly collected mycelium using an enzymatic solution containing 20 mM NaH2PO4 (pH 5.8), 20 mM CaCl2, 1.2 M NaCl, Snailase (7.5 g/mL), lysozyme (7.5 g/mL), lywallzyme (7.5 g/mL), and 3 mg/mL driselase. The mixture was incubated at 29 °C with shaking at 180 rpm for 1–1.5 h. The efficiency of the enzymatic lysis of the mycelium’s cell wall was assessed under a microscope after 1 h. The truncated mutation strains of Npt1, HA-Npt1, and GFP-Npt1 were generated in a similar manner. Putative positive transformants were validated using diagnostic PCR. The procedure included amplifying the target gene and the upstream and downstream regions of the fusion fragments, using the mutant strain’s genomic DNA as the PCR template56. The truncated mutation strains of Npt1 were confirmed through Southern blot analysis (Supplementary Fig. 10B). The genomic DNA of each strain was extracted using a Solarbio fungi genomic DNA extraction kit. The extracted DNA was then digested with the restriction enzymes BamHI and XbaI, as shown in Supplementary Fig. 10C. The digested genomic products were separated on agarose gels and subsequently transferred to a nylon membrane for Southern hybridization. The DNA probe for hybridization was amplified using primers detailed in Supplementary Data 9. The probe was then purified using an agarose gel DNA extraction kit and labeled with a Roche DIG-High Prime DNA labeling and detection starter kit II. Following prehybridization and hybridization, the DIG-labeled single-stranded DNA probe was hybridized to target DNA within a mixture of various DNA sequences, forming a stable double-stranded DNA structure. After washing the membrane to remove any non-hybridized probes, immunological detection was performed using anti-DIG-AP. Finally, the membrane was exposed to a Lumi-Film imager using a G:BOX Chemi XT4 imaging system (Syngene).
Protein expression, purification, and mass spectrometry analysis
Full-length PgAFP includes the signal peptide, propeptide, and mature peptide regions. However, an E. coli prokaryotic intracellular expression system lacks the ability of the fungal extracellular secretion system to cleave the signal and propeptide regions. Consequently, we chose to express solely the mature form of PgAFP in the E. coli system. Using SignalP 6.0 and BLAST analysis, we identified the sequence of the mature AfAFP for subsequent recombinant expression. The genes encoding AfAFP and PgAFP were produced by using full gene synthesis with codon optimization and subsequently cloned into a pET-32a expression vector. The resulting constructs included a 6His tag and a Trx tag at the N-terminus, followed by a PreScission protease recognition site before the target protein. To produce the PgAFP-Flag-Strep or AfAFP-Flag-Strep-GST proteins, a pET-32a vector was also used. These constructs were engineered using two strategically placed PreScission protease sites for efficient tag removal during protein purification to yield the proteins PgAFP-F-S or AfAFP-F-S. In the case of PgAFP-Flag-Strep, the PreScission protease sites were positioned between the Trx tag and the Flag tag, and between PgAFP and the 6His tag. Similarly, for AfAFP-Flag-Strep-GST, the PreScission protease sites were located between the Trx tag and the Flag tag, and between AfAFP and the GST tag. These arrangements ensured the precise removal of tags during purification and enhanced the purity of the final protein product.
The plasmids harboring the coding sequences of PgAFP and AfAFP were subjected to sequence validation. Upon verification, the plasmids were individually transformed into E. coli BL21(DE3). The transformed cells were propagated in 10 mL of LB medium supplemented with 100 µg/mL ampicillin, and incubated at 37 °C with 180 rpm agitation for 12 h. Subsequently, cultures expressing either PgAFP or AfAFP were scaled up to 4 L of fresh LB medium with 100 µg/mL ampicillin and maintained under identical conditions until the optical density at 600 nm reached 0.5. Cells were then collected, resuspended in pre-cooled lysis buffer (comprising 50 mM Tris-HCl, 500 mM NaCl, and 20 mM imidazole, adjusted to pH 7.4), and disrupted by sonication on ice. The lysates were clarified by centrifugation at 12,000 × g for 20 min at 4 °C, which was repeated three times. The supernatant, devoid of cellular debris, was applied to a Ni–NTA affinity column. A stepwise elution was performed using buffers with increasing concentrations of imidazole (from 20 to 300 mM) to selectively bind and subsequently elute the His-tagged fusion protein57. The fusion tag was cleaved by incubating the protein with PreScission protease 3C at 4 °C for 16 h, followed by post-dialysis of the eluted fractions against a buffer composed of 50 mM Tris-HCl and 10 mM NaCl. The proteolytic reaction mixture, comprising the fusion protein, cleaved tag, and native protein, was further subjected to ion-exchange chromatography using a RESOURCE S cation exchange chromatography column. A linear gradient of NaCl was used for elution in an ӒKTA pure™ chromatography system, which yielded highly purified PgAFP and AfAFP. Size-exclusion chromatography was performed for the final purification using a Superdex 75 10/300 GL column, which was equilibrated and eluted with a buffer of 50 mM Tris-HCl and 150 mM NaCl at pH 7.458. The chromatographic profiles indicated that both proteins existed in monomeric states. The recombinant proteins PgAFP-Flag-Strep and AfAFP-Flag-Strep were purified following a similar protocol, followed by the removal of the affinity tags. The recombinant proteins PgAFP, AfAFP, PgAFP-Flag-Strep, and AfAFP-Flag-Strep were resolved by electrophoresis and analyzed by mass spectrometry to confirm their identities.
Protein crystallization, data collection, and structure analysis
PgAFP crystal growth conditions were screened by sitting drop vapor diffusion in 96-well plates using a Phoenix crystallization robot with purified PgAFP protein (28 mg/mL) at 16 °C. The optimal condition for protein crystal growth was determined to be a 0.1 M Tris buffer with a pH of 8.0 and 3.2 M ammonium sulfate using a 1:1 ratio of protein and crystallization buffer. The crystals were cryoprotected by adding 10–25% glycerol to the corresponding crystallization solution. The diffraction data were collected at the Shanghai Synchrotron Radiation Facility and processed with HKL2000 software59. The phase solution of the crystallographic structure was determined via the molecular replacement technique, using the Protein Data Bank entry 7BAE as the search template32. This meticulous process required multiple rounds of adjustment to ensure the fidelity of the model to the observed electron density. Further refinement and validation of the crystallographic data were performed using Collaborative Computational Project Number 4 and the Phenix software package (Phenix1.21)60,61. The final protein structure (PDB accession 9IID) and the structural comparisons were generated using the PyMOL molecular graphics system (PyMOL2.5)62.
Protein model prediction
Structures of PgAFP and AfAFP, the engineered variants PgAFP-Flag-Strep and AfAFP-Flag-Strep, and the AFP-Ntp1 complex were computed using two structure prediction tools. The first tool, ColabFold v1.5.5, used the AlphaFold2 algorithm available through Google Colaboratory63. The second tool was AlphaFold3, an online prediction website recently launched by DeepMind64. For each AFP and AFP-Ntp1 complex, a series of five homology models were produced and refined. The most accurate model was selected based on the per-residue confidence score (pLDDT), which assessed the predicted structure’s reliability at the amino acid level. This selection was further guided by the predicted template modeling (pTM) score and the interface predicted template modeling (ipTM) score65,66. The finalized structures were rendered using the PyMOL molecular graphics system, which displayed the pLDDT values as color-coded indicators on the structural representations.
Determination of disulfide bond linkages and protein sequences of AFPs
Samples of PgAFP and AfAFP were submitted to BioteckPack Company (Beijing, China) for disulfide bridge identification. Each sample (50 μL) was mixed with 50 μL of 50 mM ammonium bicarbonate (NH4HCO3). To this mixture, 1 μL of 100 mM iodoacetamide was added to a final concentration of 1 mM. The reaction was incubated in the dark at room temperature for 40 min to ensure effective alkylation. Following this, 5 μL of a trypsin enzyme solution (0.25 μg/μL) was added, and the digestion proceeded at 37 °C for 16 h. Peptide desalting was performed using a C18 desalting column, after which the samples were vacuum-dried at 45 °C and reconstituted for analysis. High-resolution LC-MS/MS data were acquired using a U3000/Q Exactive system. Data analysis was conducted with BioPharma Finder 5.1 software, which identified multiple disulfide-linked peptides. Data processing parameters: Protease (Trypsin); Variable Modifications (Deamidation, Oxidation, Carbamidomethylation, Disulfide bond); Mass Accuracy (10 ppm).
To determine the protein sequences, samples of PgAFP and AfAFP—including their tagged variants—were submitted to Applied Protein Technology in Shanghai for comprehensive analysis. The proteins were first digested with trypsin, then analyzed by LC-MS/MS using an EASY-nLC 1000 ultra-high-performance liquid chromatography system coupled with a high-resolution Q Exactive mass spectrometer (Thermo Fisher). The mass spectrometric data were processed with Mascot 2.2.2 to qualitatively identify the target peptides. Raw mass spectrometry files were searched against the corresponding databases, with parameters set for uniprotkb_Penicillium_chrysogenum and uniprotkb_Aspergillus_flavus, ultimately yielding the identified protein sequences. The search parameters were as follows:Peptide Mass Tolerance: 20 ppm; Fragment Mass Tolerance: 0.1 Da; Filter: Score ≥ 20. These samples were prepared with dithiothreitol to reduce disulfide bonds, which differed from the non-reducing conditions used for disulfide bridge identification.
Analysis of antifungal activity of AFPs through the dry weight measurement of fungal biomass
Sterilized and dried 1.5 mL Eppendorf tubes were weighed prior to use. Each tube was filled with 1 mL of YGT liquid medium. Gradient concentrations of PgAFP and AfAFP proteins, including fluorescence-labeled and affinity-tagged variants, were added to the medium. To achieve a final spore concentration of 1 × 104 spores/mL, 1 μL of a spore suspension containing 1 × 107 spores/mL was introduced into each tube. The fungal strains included both Ntp1 mutants and the wild-type strain. Cultures were incubated at 37 °C with continuous shaking at 180 rpm for 48 h. Following incubation, the cultures were centrifuged at 6200 × g for 5 min to harvest the mycelia, which were then washed thoroughly with double-distilled water to remove residual medium. The tubes were sealed with film, punctured with small holes to allow moisture to evaporate, pre-cooled at −80 °C for 30 min, and subsequently freeze-dried for 24 h. The dry weight of the mycelia was then accurately measured.
Protein stability analysis
The stability and susceptibility to proteolytic degradation of PgAFP and AfAFP were assessed by incubating the proteins under diverse thermal conditions and time periods followed by gel electrophoresis. The experimental parameters included incubations at 29 °C for 24, 48, and 72 h, which is the optimal growth temperature of A. flavus; at 37 °C for equivalent durations to reflect hot outdoor conditions; at 42 °C, which is the maximum growth threshold of A. flavus; and at 50 °C to simulate high soil temperatures. Each temperature was evaluated using the three time periods. In vitro proteolysis assays were also conducted, in which trypsin was introduced to the AFP samples at different weight-to-weight ratios (1:400, 1:300, 1:200, 1:100, 1:80, 1:60, 1:40, and 1:20). The reaction proceeded for 12 h at a constant temperature of 37 °C.
The PgAFP and AfAFP samples were incubated for 10 h at 29 °C, 37 °C, and 42 °C, and then added to a 107 spores/mL spore solution (final concentration of AFPs was 30 μM). A volume of 20 μL of the mixture was applied to the surface of sterilized peanuts and corn, both cultivated in China. The peanuts and corn were placed on filter paper and incubated at 29 °C (optimal growth temperature for A. flavus) in the dark for 5 days. Images of the experimental results were captured. The cultivated peanuts and corn were rinsed with water. The spore solution was collected and shaken well. The number of spores in the spore solution was counted using a hemocytometer.
Reverse-phase HPLC for assessing the hydrophobicity of AFPs
To evaluate the hydrophobic properties of AFPs, a reverse-phase high-performance liquid chromatography method was used. AFPs were dissolved in phosphate-buffered saline (PBS) at pH 7.8 to achieve a final concentration of 1 mg/mL. The prepared protein solution was injected into a C18 reverse-phase chromatography column (InertSustain®C18, 5 μm particle size, 4.6 × 150 mm dimensions). The mobile phase consisted of two components: phase A, double-distilled water (ddH2O) containing 0.1% trifluoroacetic acid (TFA), and phase B, acetonitrile with 0.1% TFA. The elution was performed using a gradient program, in which the proportion of phase B increased from 10 to 50–60% over 20 min. The chromatographic run was initiated, and proteins were monitored using a UV detector set to 280 nm. Chromatograms were recorded, documenting both retention times and peak areas for each protein sample. The retention time on the C18 column served as an indicator of each protein’s hydrophobicity, with longer retention times corresponding to greater hydrophobic interactions.
Determination of the chitin content using a colorimetric assay
This protocol was performed in accordance with the method for determining fungal biomass through dry weight measurements, as outlined in the preceding paragraph. Cell walls were disrupted using a lysis buffer comprising 50 mM Tris-HCl, 1 mM EDTA, 300 mM β-mercaptoethanol, and 2% sodium dodecyl sulfate (SDS) at pH 8.0 added to the desiccated mycelium. The mixture was then heated at 100 °C for 45 min in a water bath.
To quantify the chitin content, the desiccated samples were reconstituted in ddH2O to 25 mg/mL. Equal volumes of concentrated hydrochloric acid were added to each sample, which was then incubated in a dry block heater at 100 °C for 17 h to hydrolyze the chitin into glucosamine.
The subsequent colorimetric assay entailed filtering the hydrolyzed mixtures to obtain the supernatant. The pH of the supernatant was then adjusted to 9.5 using NaOH. A 200-μL aliquot of the supernatant was combined with 400 μL of acetylacetone buffer (1.5 mL acetylacetone in 50 mL of 1.25 mol/L sodium carbonate, freshly prepared) and incubated at 90 °C for 1 h. The solution was cooled to ambient temperature and 4 mL of dimethylaminobenzaldehyde buffer (1.33 g of dimethylaminobenzaldehyde in a solution of 25 mL absolute ethanol and concentrated hydrochloric acid) was added. The mixture was incubated for an additional 1 h at room temperature. The chitin content was determined by measuring the absorbance at 530 nm using a spectrophotometer. The results were meticulously recorded and analyzed against a glucosamine standard curve.
To generate the glucosamine standard curve, standard solutions of glucosamine at concentrations of 2, 4, 6, 8, and 10 mg/mL were prepared. A 200 μL sample from each standard solution was transferred into a 1.5 mL centrifuge tube, to which 400 μL of acetylacetone buffer was added. Following incubation at 90 °C for 1 h and subsequent cooling to room temperature, 4 mL of anhydrous ethanol and 400 μL of dimethylaminobenzaldehyde buffer were added. The solutions were then incubated at room temperature for 1 h. The absorbance at 530 nm was measured with a spectrophotometer, and the data were recorded to construct the standard curve.
Chitin content analysis of the cell wall by CFW staining
To visualize the chitin content of the A. flavus cell wall, fluorescent brightener 28 (CFW) dye was used22,67. The fungus was co-cultured with the antifungal protein in YGT medium at 37 °C for 12 h. Then, CFW was added to the medium at a final concentration of 10 mg/mL and incubated for 5 min. The mycelium was washed with PBS at least three times and then examined by confocal microscopy for CFW staining, which fluoresced under a 355 nm excitation wavelength.
Subcellular localization of FITC-labeled AfAFP and PgAFP in A. flavus
AfAFP and PgAFP were labeled with FITC (Anaspec, Fremont, CA, USA) by incubating 4 mL of protein with 100 μL of 20 mM FITC at room temperature in the dark for 8 h27. The FITC-labeled protein was dialyzed against a buffer containing 20 mM Tris and 10 mM NaCl, pH 7.4. After germinating fungal spores into mycelia over an 18-h period, the mycelia were treated with FITC-labeled PgAFP and AfAFP at a concentration of 3 mg/mL. The mycelia were washed with PBS three times and stained with PI at a final concentration of 5 μg/mL for 5 min in the dark to detect cell death. The uptake of the FITC-labeled proteins by A. flavus cells was examined at 30 min, 1, 2, 4, and 8 h by using laser scanning confocal microscopy. FITC was excited at 482 nm with emission at 500 nm. PI was excited at 500 nm with emission at 625 nm.
Subcellular localization of TRITC-labeled AfAFP and PgAFP in A. flavus
AfAFP and PgAFP were dialyzed against a sodium carbonate solution (pH 9.0) and concentrated at 1 mg/mL. Each protein was labeled with tetramethylrhodamine isothiocyanate (TRITC; Merck, Darmstadt, Germany) dissolved in dimethyl sulfoxide at 1 mg/mL using a dye-to-protein mass ratio of 3:100. The labeling reaction proceeded at room temperature for ~8 h. After reacting, the TRITC-labeled proteins were dialyzed against 20 mM Tris-HCl buffer containing 10 mM NaCl (pH 7.4) to remove unreacted dye and then concentrated to the desired concentration.
A. flavus Ntp1-GFP was pre-cultured for 18 h in YGT medium. Fungal hyphae were then incubated in fresh YGT medium supplemented with either AfAFP-TRITC or PgAFP-TRITC at a final concentration of 3 mg/mL in the dark for 30 min, 1, 2, 4, and 8 h. Following incubation, the hyphae were washed three times with sterile PBS to remove unbound proteins.
The subcellular localization of AfAFP and PgAFP within the hyphae and GFP expression were examined using laser scanning confocal microscopy to monitor the fluorescence distribution. For imaging TRITC-labeled proteins, excitation was at 550 nm with emission at 570 nm. For Ntp1-GFP visualization, excitation was at 488 nm with emission at 507 nm.
Tandem affinity purification and mass spectrometry analyzes
The wild-type A. flavus strain was grown in YGT medium at 37 °C for 24 h. The mycelia were then harvested and incubated with the purified PgAFP-F-S protein, AfAFP-F-S protein, and F-S (Flag-Strep tag) peptide synthesized by Sangon Biotech (Sangon, Shanghai, China) for another 24-h in YGT medium68. For mass spectrometry analyzes, Each treatment with PgAFP-F-S protein, AfAFP-F-S protein, and F-S peptide was conducted separately, with three independent replicates per condition (n = 3). Following incubation, the mycelia were washed with PBS buffer three times. Total protein was extracted from the A. flavus mycelial cells by grinding them in liquid nitrogen using a total protein extraction kit (inhibitor-free). The main components of the protein extraction kit were 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 1% Triton X-100, without protease and phosphatase inhibitors, which allowed the original protein-protein interactions to be maintained. The cell lysate was centrifuged at 4700 × g for 20 min at 4 °C and the supernatant was incubated with Strep-Tactin magrose(Cat.20495ES08, Yeasen Inc.) overnight. The bound proteins were eluted with desthiobiotin and further purified using anti-Flag affinity beads. Proteins isolated on the anti-Flag antibody-coated MagBeads (Cat. # 20565ES76, Yeasen Inc.) underwent SDS-PAGE, and the distinct bands were excised for analysis. The comprehensive LC-MS/MS process was performed by the Shanghai Biowing biotechnology Co.LTD (Shanghai, China). In brief, the gel pieces were washed twice with ultrapure water, followed by incubation with decolorizing solution (50% acetonitrile, 25 mM ammonium bicarbonate) at 37 °C for 10 min. Subsequently, the gel pieces were dehydrated using anhydrous acetonitrile. Reduction was achieved by treating the gel pieces with 20 mM dithiothreitol (DTT) at 37 °C for 1 h. Upon cooling to room temperature, an equal volume of 60 mM iodoacetamide (IAA) was added, and the alkylation reaction was conducted at room temperature in the dark for 30 min. The gel pieces were then dehydrated, dried, and subjected to overnight digestion with trypsin at 37 °C. Each sample fraction was subjected to nanoLC-MS/MS analysis as following: Peptide samples were prepared by dissolving them in 10 μL of solution A (water with 0.1% formic acid). For the first setup, the solution was loaded onto a C18-reversed phase analytical column in the EASY-nano-LC system at 2 μL/min. Chromatographic separation was then performed at 600nL/min using a 60-min linear gradient that ramped solution B (acetonitrile with 0.1% formic acid) from 3% to 32%. The eluent was directly transferred to an Orbitrap Fusion Lumos mass spectrometer, with the ion source maintained at a spray voltage of 2.3 kV. The raw MS/MS data were processed using Proteome Discoverer for database searching against the UniProt database. Search parameters included fixed modifications—Carbamidomethylation of cysteine—and variable modifications such as oxidation of methionine and N-terminal acetylation. The analysis allowed for up to two missed cleavage sites, with a peptide mass tolerance of 10 ppm and a fragment mass tolerance of 0.05 Da.
Peptide identification was confirmed through statistical methods, and proteins with significant changes were identified. The proteomics data from the mass spectrometry analysis were submitted to the ProteomeXchange Consortium via the iProX partner repository under the dataset identifier PXD049263 (http://proteomecentral.proteomexchange.org)69,70.
Immunoprecipitation analysis of target protein
The peptides AfAFP-F-S and PgAFP-F-S, purified in our laboratory, along with commercially sourced F-S peptides, were subjected to overnight incubation at 4 °C with Strep-Tactin beads. Prior to incubation, the beads were repeatedly washed with PBS to eliminate non-specifically bound peptides. The mycelia from our genetically engineered strains, which expressed HA-Ntp1 and the truncated variants HA-Δntp1285–708, HA-Δntp1417–708, and HA-Δntp1589–708, were frozen and disrupted using RIPA buffer to extract total protein. The lysates containing the HA-tagged proteins from the four mutant strains were then incubated with the peptide-loaded Strep-Tactin beads for 2 h at 4 °C. Following incubation, the beads underwent a triple wash with PBS to remove unbound proteins. The affinity-bound proteins were eluted using desthiobiotin and the eluates were resolved by SDS-PAGE. The resolved proteins were then transferred onto PVDF membranes and probed with an anti-HA antibody (HA-Tag (C29F4) Rabbit mAb, Cat. 3724, Cell Signaling Technology, Inc.). The HA antibody is used at a 1:1000 dilution. Specific signals were detected and quantified using a G:BOX Chemi XT4 imaging system (Syngene, UK)71.
Analysis of vegetative growth and formation of conidia and sclerotia
The colony morphology and growth rate were studied using YGT medium57. The colony diameter was evaluated57 using YG solid media plates inoculated with a spore suspension at a concentration of 107 spores/mL. The plates were then incubated at 37 °C for 4 days in the dark. Post-incubation, photographs were obtained to document the colony morphology, and the colony diameters were measured. Conidia production was compared57 using YGT solid media plates similarly inoculated with 107 spores/mL and incubated at 37 °C for 4 days in the dark. Following incubation, the plates were washed with sterile water to collect the spores. The conidial count was determined using a hemocytometer. Sclerotia production56 was analyzed using fresh spore suspensions of both mutant and WT strains at 107 spores/mL, inoculated onto the center of a 10 mL CM solid medium plate, and incubated in the dark at 37 °C for 7 days. The aerial mycelium and conidia were removed by washing with 75% ethanol. The plates were then photographed. The sclerotia morphology was captured using a stereomicroscope, and the sclerotia were quantified from the magnified images based on a quarter section of the colony.
Thin-layer chromatography analysis
To extract aflatoxin from the cultured liquid medium, fresh spore suspensions of various A. flavus strains, including WT and gene-edited strains, at a concentration of 107 spores/mL were inoculated into 10 mL of YES liquid medium. The cultures were incubated in the dark at 29 °C for 5 days. Subsequently, 7 mL of the post-cultivation medium was collected, and an equivalent volume of dichloromethane was added to each sample. After thorough mixing, the resulting mixture was shaken at 180 rpm for 30 min and then immediately centrifuged. A 5 mL aliquot of the dichloromethane layer (the lower layer) was extracted for subsequent analysis.
Thin-layer chromatography (TLC) analysis was performed using dried samples re-dissolved in 1 mL of dichloromethane. For the analysis, 10 μL of each dissolved sample was applied to silica gel plates. A developing solvent with a dichloromethane to acetone ratio of 9:1 was used57. Toxin bands were observed under a UV light imaging system.
Pathogenicity analysis
A pathogenicity analysis of deletion and truncated strains on peanut and maize seeds, which were treated with AFP, was conducted following method57. To prevent germination, embryos were removed from both types of seeds. Uniformly sized seeds were selected and sterilized by washing with 0.5‰ sodium hypochlorite for 3 min, followed by rinsing with sterile water to remove any sodium hypochlorite residue. Subsequently, the seeds were soaked in 75% ethanol for 15 s, washed several times with sterile water, and soaked in sterile water for 5 min to completely remove the ethanol. The sterilized seeds were then placed on sterilized filter paper to remove excess water. The pathogenicity testing of the WT strain used PgAFP and AfAFP subjected to heat treatment at 29, 37, and 42 °C for 10 h. The heat-processed proteins were then added to a fresh spore suspension containing 106 spores/mL, with a final AFP concentration of 30 µM. A 20 µL aliquot of the spore suspension was applied to the surface of the sterilized peanut and maize seeds. The seeds were placed on filter paper in Petri dishes and incubated at 29 °C in the dark for 5 days. For testing the pathogenicity of the Ntp1 deletion and truncated strains, sterilized peanut or maize seeds were immersed in a spore suspension with 103 spores/mL for 30 min. Filter paper was placed on Petri dishes, 1 mL of sterile water was added, and the seeds were placed on the filter paper. The seeds were incubated at 29 °C in the dark for 7 days57.
For the spore count analysis, 20 mL of sterile water was added to the cultured seeds, shaken in a tube, and the spore suspension was diluted before counting using a hemocytometer. For metabolite extraction following co-cultivation, dichloromethane was added to the spore suspension (prepared with sterile water and cultured infected seeds, as described above) at a 1:1 ratio. The resulting mixture was incubated at 37 °C with continuous shaking at 180 rpm for 10 min. A TLC analysis of aflatoxins was conducted following the method described above.
Statistical analysis and reproducibility
The samples were chosen through a random selection process to ensure an unbiased and representative dataset. Random sampling minimizes selection bias and increases the generalizability of the findings. All representative experiments, such as micrographs and co-immunoprecipitations, were conducted with a minimum of three independent replicates to ensure the reliability and reproducibility of the findings. Data analysis was conducted using GraphPad Prism 10.0 and OriginPro 2023, with statistical significance assessed using one-way ANOVA, and statistical significance was assessed by means of one-way ANOVA. All analyzes were performed on a minimum of three biological replicates unless otherwise specified.
Reporting summary
Further information on research design is available in Nature Portfolio Reporting Summary linked to this article.
Data availability
Crystallographic structure of PgAFP has been deposited in the RCSB Protein Data Bank (PDB) under accession 9IID. Mass spectrometry-based proteomics data related to AFP-interacting proteins have been submitted to ProteomeXchange Consortium via the iProX partner repository under the accession PXD049263 or iProX project ID IPX000809900069,70. Sequence and annotation of PgAFP [https://www.ncbi.nlm.nih.gov/protein/D0EXD3.1?report=genbank&log$=prottop&blast_rank=1&RID=X5YSNKWN013] and AfAFP [https://www.ncbi.nlm.nih.gov/protein/KOC12588.1?report=genbank&log$=prottop&blast_rank=1&RID=X5YZRW1B013] are available at NCBI Protein Database. Ntp1 homologs across different fungal species are also available at NCBI Protein Database: Aspergillus flavus [https://www.ncbi.nlm.nih.gov/protein/KAJ1717178.1], Aspergillus fumigatus [https://www.ncbi.nlm.nih.gov/protein/KAL3411886.1], Fusarium falciforme [https://www.ncbi.nlm.nih.gov/protein/XP_053004632.1], Penicillium citrinum [https://www.ncbi.nlm.nih.gov/protein/XP_056506444.1], Trichoderma harzianum [https://www.ncbi.nlm.nih.gov/protein/PNP50238.1]. Source data are provided with this paper, which is also available at Figshare [https://figshare.com/articles/dataset/Source_date_NC_xlsx/28600874?file=53009507]. Source data are provided with this paper.
References
Stukenbrock, E. & Gurr, S. Address the growing urgency of fungal disease in crops. Nature 617, 31–34 (2023).
Steinberg, G. & Gurr, S. J. Fungi, fungicide discovery and global food security. Fungal Genet. Biol. 144, 103476 (2020).
Garrigues, S., Gandia, M. & Marcos, J. F. Occurrence and function of fungal antifungal proteins: a case study of the citrus postharvest pathogen Penicillium digitatum. Appl. Microbiol. Biotechnol. 100, 2243–2256 (2016).
Toth, L. et al. NFAP2, a novel cysteine-rich anti-yeast protein from Neosartorya fischeri NRRL 181: isolation and characterization. AMB Express 6, 75 (2016).
Huber, A. et al. Two small, cysteine-rich and cationic antifungal proteins from Penicillium chrysogenum: a comparative study of PAF and PAFB. Biochim. Biophys. Acta Biomembr. 1862, 183246 (2020).
Nakaya, K. et al. Amino acid sequence and disulfide bridges of an antifungal protein isolated from Aspergillus giganteus. Eur. J. Biochem. 193, 31–38 (1990).
Marx, F. et al. Cloning, structural organization and regulation of expression of the Penicillium chrysogenum paf gene encoding an abundantly secreted protein with antifungal activity. Gene 167, 167–171 (1995).
Chen, Y. P., Li, Y., Chen, F., Wu, H. & Zhang, S. Characterization and expression of fungal defensin in Escherichia coli and its antifungal mechanism by RNA-seq analysis. Front. Microbiol. 14, 1172257 (2023).
Szappanos, H. et al. The Penicillium chrysogenum-derived antifungal peptide shows no toxic effects on mammalian cells in the intended therapeutic concentration. Naunyn Schmiedebergs Arch. Pharmacol. 371, 122–132 (2005).
Palicz, Z. et al. In vivo application of a small molecular weight antifungal protein of Penicillium chrysogenum (PAF). Toxicol. Appl. Pharmacol. 269, 8–16 (2013).
Palicz, Z. et al. Application of a low molecular weight antifungal protein from Penicillium chrysogenum (PAF) to treat pulmonary aspergillosis in mice. Emerg. Microbes Infect. 5, e114 (2016).
Huber, A. et al. New antimicrobial potential and structural properties of PAFB: a cationic, cysteine-rich protein from Penicillium chrysogenum Q176. Sci. Rep. 8, 1751 (2018).
Campos-Olivas, R. et al. NMR solution structure of the antifungal protein from Aspergillus giganteus: evidence for cysteine pairing isomerism. Biochemistry 34, 3009–3021 (1995).
Utesch, T. et al. A computational modeling approach predicts interaction of the antifungal protein AFP from aspergillus giganteus with fungal membranes via its gamma-core motif. mSphere 3, e00377–18 (2018).
Yount, N. Y. & Yeaman, M. R. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA 101, 7363–7368 (2004).
Lacerda, A. F., Vasconcelos, E. A., Pelegrini, P. B. & Grossi de Sa, M. F. Antifungal defensins and their role in plant defense. Front. Microbiol. 5, 116 (2014).
Hagen, S., Marx, F., Ram, A. F. & Meyer, V. The antifungal protein AFP from Aspergillus giganteus inhibits chitin synthesis in sensitive fungi. Appl. Environ. Microbiol. 73, 2128–2134 (2007).
Theis, T., Marx, F., Salvenmoser, W., Stahl, U. & Meyer, V. New insights into the target site and mode of action of the antifungal protein of Aspergillus giganteus. Res. Microbiol. 156, 47–56 (2005).
Munoz, A., Marcos, J. F. & Read, N. D. Concentration-dependent mechanisms of cell penetration and killing by the de novo designed antifungal hexapeptide PAF26. Mol. Microbiol. 85, 89–106 (2012).
Bugeda, A. et al. The antifungal protein AfpB induces regulated cell death in its parental fungus Penicillium digitatum. mSphere 5, e00595–20 (2020).
Moreno, A. B., Martínez Del Pozo, A. & San Segundo, B. Biotechnologically relevant enzymes and proteins. Antifungal mechanism of the Aspergillus giganteus AFP against the rice blast fungus Magnaporthe grisea. Appl. Microbiol. Biotechnol. 72, 883–895 (2006).
Binder, U., Oberparleiter, C., Meyer, V. & Marx, F. The antifungal protein PAF interferes with PKC/MPK and cAMP/PKA signalling of Aspergillus nidulans. Mol. Microbiol. 75, 294–307 (2010).
Gandia, M., Garrigues, S., Hernanz-Koers, M., Manzanares, P. & Marcos, J. F. Differential roles, crosstalk and response to the Antifungal Protein AfpB in the three Mitogen-Activated Protein Kinases (MAPK) pathways of the citrus postharvest pathogen Penicillium digitatum. Fungal Genet Biol. 124, 17–28 (2019).
Theis, T., Wedde, M., Meyer, V. & Stahl, U. The antifungal protein from Aspergillus giganteus causes membrane permeabilization. Antimicrob. Agents Chemother. 47, 588–593 (2003).
Ouedraogo, J. P., Hagen, S., Spielvogel, A., Engelhardt, S. & Meyer, V. Survival strategies of yeast and filamentous fungi against the antifungal protein AFP. J. Biol. Chem. 286, 13859–13868 (2011).
Binder, U., Bencina, M., Eigentler, A., Meyer, V. & Marx, F. The Aspergillus giganteus antifungal protein AFPNN5353 activates the cell wall integrity pathway and perturbs calcium homeostasis. BMC Microbiol. 11, 209 (2011).
Delgado, J., Owens, R. A., Doyle, S., Asensio, M. A. & Nunez, F. Impact of the antifungal protein PgAFP from Penicillium chrysogenum on the protein profile in Aspergillus flavus. Appl. Microbiol. Biotechnol. 99, 8701–8715 (2015).
Giner-Llorca, M., Locascio, A., Del Real, J. A., Marcos, J. F. & Manzanares, P. Novel findings about the mode of action of the antifungal protein PeAfpA against Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 107, 6811–6829 (2023).
Ropero-Pérez, C. et al. Transcriptomic profile of Penicillium digitatum reveals novel aspects of the mode of action of the antifungal protein AfpB. Microbiol. Spectr. 11, e0484622 (2023).
Sonderegger, C. et al. The evolutionary conserved γ-Core motif influences the anti-candida activity of the Penicillium chrysogenum Antifungal Protein PAF. Front. Microbiol. 9, 1655 (2018).
Rodriguez-Martin, A. et al. Characterization of the novel antifungal protein PgAFP and the encoding gene of Penicillium chrysogenum. Peptides 31, 541–547 (2010).
Guagnini, F., Huber, A., Alex, J. M., Marx, F. & Crowley, P. B. Porous assembly of an antifungal protein mediated by zinc and sulfonato-calix[8]arene. J. Struct. Biol. 213, 107711 (2021).
Zhu, X. M. et al. The biological functions of sphingolipids in plant pathogenic fungi. PLoS Pathog. 19, e1011733 (2023).
Bennion, B. et al. Glycosphingolipids of the model fungus Aspergillus nidulans: characterization of GIPCs with oligo-alpha-mannose-type glycans. J. Lipid Res. 44, 2073–2088 (2003).
Batta, G. et al. Functional aspects of the solution structure and dynamics of PAF-a highly-stable antifungal protein from Penicillium chrysogenum. FEBS J. 276, 2875–2890 (2009).
Garcia-Rubio, R., de Oliveira, H. C., Rivera, J. & Trevijano-Contador, N. The fungal cell wall: candida, cryptococcus, and Aspergillus species. Front. Microbiol. 10, 2993 (2019).
Meyer, V. & Jung, S. Antifungal peptides of the AFP family revisited: are these cannibal toxins? Microorganisms 6, 50 (2018).
Meyer, V. & Stahl, U. The influence of co-cultivation on expression of the antifungal protein in Aspergillus giganteus. J. Basic Microbiol. 43, 68–74 (2003).
Hegedus, N., Sigl, C., Zadra, I., Pocsi, I. & Marx, F. The paf gene product modulates asexual development in Penicillium chrysogenum. J. Basic Microbiol. 51, 253–262 (2011).
Gandia, M., Garrigues, S., Bolos, B., Manzanares, P. & Marcos, J. F. The myosin motor domain-containing chitin synthases are involved in cell wall integrity and sensitivity to antifungal proteins in Penicillium digitatum. Front. Microbiol. 10, 2400 (2019).
Munro, C. A. et al. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 63, 1399–1413 (2007).
Walker, L. A. et al. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4, e1000040 (2008).
Binder, U. et al. Protein kinase A signaling and calcium ions are major players in PAF mediated toxicity against Aspergillus niger. FEBS Lett. 589, 1266–1271 (2015).
Binder, U., Chu, M., Read, N. D. & Marx, F. The antifungal activity of the Penicillium chrysogenum protein PAF disrupts calcium homeostasis in Neurospora crassa. Eukaryot. Cell 9, 1374–1382 (2010).
Miao, J. et al. iTRAQ-based quantitative proteomic analysis of the antimicrobial mechanism of peptide F1 against escherichia coli. J. Agric Food Chem. 63, 7190–7197 (2015).
Truong, T. et al. Comparative ploidy proteomics of candida albicans biofilms unraveled the role of the AHP1 gene in the biofilm persistence against amphotericin B. Mol. Cell Proteom. 15, 3488–3500 (2016).
Zhang, Q. Y. et al. Antimicrobial peptides: mechanism of action, activity and clinical potential. Mil. Med. Res. 8, 48 (2021).
Otvos, L. Jr. et al. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 39, 14150–14159 (2000).
Kragol, G. et al. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 40, 3016–3026 (2001).
Narasimhan, M. L. et al. Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol. Cell 17, 171–180 (2005).
Hancock, R. E., Haney, E. F. & Gill, E. E. The immunology of host defence peptides: beyond antimicrobial activity. Nat. Rev. Immunol. 16, 321–334 (2016).
Yang, K. et al. The high-affinity phosphodiesterase PdeH regulates development and aflatoxin biosynthesis in Aspergillus flavus. Fungal Genet. Biol. 101, 7–19 (2017).
Chang, P. K., Scharfenstein, L. L., Wei, Q. & Bhatnagar, D. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods 81, 240–246 (2010).
Wang, Y. et al. N-Myristoyltransferase, a potential antifungal candidate drug-target for Aspergillus flavus. Microbiol. Spectr. 11, e0421222 (2023).
Yang, K. et al. Cyclase-associated protein cap with multiple domains contributes to mycotoxin biosynthesis and fungal virulence in Aspergillus flavus. J. Agric. Food Chem. 67, 4200–4213 (2019).
Zhang, F. et al. The MAP kinase AflSlt2 modulates aflatoxin biosynthesis and peanut infection in the fungus Aspergillus flavus. Int. J. Food Microbiol. 322, 108576 (2020).
Wang, Y. et al. Molecular and structural basis of nucleoside diphosphate kinase–mediated regulation of spore and sclerotia development in the fungus Aspergillus flavus. J. Biol. Chem. 294, 12415–12431 (2019).
Wang, S. et al. Expression, purification, crystallization and preliminary crystallographic analysis of nucleoside Diphosphate kinase (NDK) from Aspergillus flavus. Chin. J. Struct. Chem. 35, 1708–1713 (2016).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Winter, G. et al. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr. D Struct. Biol. 74, 85–97 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D 75, 861–877 (2019).
Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.8 (2015).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 636, E4 (2024).
Zhang, Y. & Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins 57, 702–710 (2004).
Xu, J. & Zhang, Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26, 889–895 (2010).
Delgado, J., Owens, R. A., Doyle, S., Asensio, M. A. & Nunez, F. Increased chitin biosynthesis contributes to the resistance of Penicillium polonicum against the antifungal protein PgAFP. Appl. Microbiol. Biotechnol. 100, 371–383 (2016).
Kaneko, A. et al. Tandem affinity purification of the Candida albicans septin protein complex. Yeast 21, 1025–1033 (2004).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217 (2019).
Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 50, D1522–D1527 (2022).
Wang, S. et al. The regulatory role of the Aspergillus flavus core retromer complex in aflatoxin metabolism. J. Biol. Chem. 298, 102120 (2022).
Jurrus, E. et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27, 112–128 (2018).
Acknowledgements
This work was generously supported by the National Natural Science Foundation of China (Grant No. 32370205 awarded to S.W.), the Natural Science Foundation of Fujian Province (Grant No. 2022J01141 awarded to Y.W.), and the Fund for Outstanding Doctoral Dissertation from Fujian Agriculture and Forestry University (awarded to Sen Wang). X-ray data were collected at the Shanghai Synchrotron Radiation Facility. We thank Zunyan Li for providing insightful suggestions during the manuscript revision process.
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.W., Sen Wang, and H.X.; Investigation: Sen Wang, Y.W., Y.C., H.X., Y.L., R.L., W.Z., and X.N.; Writing—Original Draft: Y.W.; Writing—Review & Editing: Y.W. and S.W.; Validation: Y.C., C.X., and X.C.; Formal Analysis: Y.C., C.X., and R.L.; Supervision: S.W. Funding Acquisition: S.W., Y.W., and Sen Wang.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jose Marcos and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Y., Wang, S., Chen, Y. et al. The role of Npt1 in regulating antifungal protein activity in filamentous fungi. Nat Commun 16, 2850 (2025). https://doi.org/10.1038/s41467-025-58230-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58230-6