Abstract
Lung cancer remains one of the most common malignancies, and its brain metastases significantly worsen the prognosis for patients. Current treatments for lung cancer face many challenges, including poor drug accumulation and the inability to simultaneously control primary and metastatic tumors. Here, we show that the mRNA-binding protein insulin-like growth factor 3 is crucial for non-small cell lung cancer progression and metastasis. We construct an inhalable nanoliposome system to co-deliver osimertinib and DNA plasmid for gene knockdown. Upon inhalation, these nanoparticles efficiently penetrate pulmonary barriers and accumulate in lungs by mimicking natural lung surfactants. Within tumor cells, released osimertinib inhibits tumor growth, while the DNA triggers the production of engineered exosomes that can travel to the brain to suppress tumors. This strategy effectively inhibits both primary and metastatic tumors while enhancing antitumor immune responses. This work suggests that this inhalable nanomedicine offers a safe and versatile strategy for cancer therapy.
Similar content being viewed by others
Introduction
Non-small cell lung cancer (NSCLC), accounting for 80-85% of lung cancer cases, is a leading cause of human death globally1,2. The majority of deaths occur in patients with metastatic disease, with 30%–50% of patients eventually developing brain metastases1,2. These patients have a poor prognosis, and conventional treatments including surgery, radiotherapy, and chemotherapy, haven’t shown effective in improving survival outcomes. While targeted therapies, such as epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs), have demonstrated initial promise, the emergence of drug resistance after repeated administration inevitably limits their effectiveness3,4,5. Therefore, there is an urgent need to develop therapeutic strategies to treat brain metastases in NSCLC patients to overcome this critical clinical challenge6.
N6-methyladenosine (m6A) is the most prevalent internal modification of messenger and non-coding RNA in eukaryotes7,8. It plays a critical role in tumor progression by regulating RNA transcription, splicing, processing, translation, and degradation through writer, eraser, and reader proteins9. Previous studies have shown that insulin-like growth factor II mRNA-binding protein 3 (IGF2BP3) is overexpressed in multiple cancer types, including breast, renal, and lung cancers, and is relevant to tumor cell proliferation, invasion, and resistance to chemotherapy10,11,12. Bioinformatics analyses have revealed a positive correlation between high expression levels of IGF2BP3 and poor survival in NSCLC patients, particularly those with metastases. Zhang et al. have identified IGF2BP3 as a key driver of acquired resistance to EGFR inhibitors in NSCLC, underscoring its potential as a therapeutic target13. However, no small molecule inhibitors of IGF2BP3 are currently available, making gene interference the most promising approach for modulating this m6A reader.
Extracellular vesicles, particularly exosomes, are increasingly utilized as delivery vehicles for gene therapies, particularly natural RNA, owing to their excellent biocompatibility, low immunogenicity, natural protection of internal bioactive molecules, and ability to penetrate biological barriers14. Additionally, their biological origin imparts specific properties, such as enhanced tissue homing and tumor targeting, selective uptake by recipient cells, and modulation of the tumor microenvironment15. Currently, exosomes for disease treatment are primarily produced in vitro. However, their large-scale manufacturing and clinical translation still face significant challenges, such as heterogeneity, the lack of standardized isolation and purification protocols, limited production scalability, and high costs16. Recent studies have shown that endogenous cells can specifically package siRNAs and miRNAs into exosomes. This inspired a promising strategy to instruct endogenous cells to produce exosomes loaded with RNAs directly in vivo17,18.
Another major challenge in NSCLC treatment is the unsatisfied delivery efficiency of drugs to the lungs and metastatic sites through conventional administration routes, such as oral or intravenous delivery, which could influence the therapeutic efficacy and increase the risk of systemic adverse effects19. Non-invasive pulmonary inhalation therapy offers a direct and localized route to the lungs, increasing drug concentrations in the target tissue20. However, this approach still faces critical challenges including the stability of particles during nebulization and the deep penetration through both cellular and extracellular barriers in lung tissue12,21. One of the main reasons why many inhaled particles have failed to deliver drugs into target cells is their interaction with lung surfactants22. Recent studies have explored leveraging lung surfactants as drug carriers due to their biocompatibility and safety. These surfactants can spread along the gas-liquid interface and transport cargo by reprocessing and redistribution, which enable sustained and efficient drug delivery to deep lung tissues23,24.
In this work, we develop an inhalable nanoparticle with a biomimetic surface. It utilizes an in vivo engineering exosome-based siRNA delivery strategy for the treatment of NSCLC patients with brain metastases. The nanoparticles, referred to as MPDOLs, are constructed as follows. First, the cationic poly (β-amino ester) (PβAE) is synthesized to compress the DNA plasmid and form a stable core25. Next, liposomes loaded with osimertinib (OST) are prepared using the film dispersion method, and the PβAE/DNA (PD) is then loaded into the aqueous lumen of the liposomes26. Finally, a layer of stem cell membrane with high expression of pulmonary surfactant protein B (SP-B) is fused onto the surface of the liposomes. Upon inhalation, the MPDOLs penetrate the lung tissues with the help of SP-B and are endocytosed by the tumor cells. The OST and DNA plasmids in MPDOLs are then released from the endo-lysosome into the cytoplasm, where OST directly binds to the intracellular kinase domains of EGFR family proteins, inhibiting their phosphorylation and downstream signaling pathways, thereby suppressing NSCLC cell progression and proliferation. Meanwhile, the DNA plasmid depletes IGF2BP3 expression in lung cancer cells and stimulates the production of exosomes loaded with IGF2BP3 siRNA and expressing rabies virus glycoprotein (RVG) on their surface to guide the brain targeting27. This multifunctional nanotherapeutic platform effectively suppresses both in situ and brain metastatic tumor progression in NSCLC mice models, which also demonstrates enhanced anti-tumor immune responses (Fig. 1). Leveraging a highly efficient and low-toxic inhalation-based delivery strategy, it offers a promising approach for pulmonary drug delivery and in vivo generation of exosomes for epigenetic modulation in anti-tumor therapy.
After nebulization, MPDOLs deposited and penetrated the tumor tissues with the help of SP-B. Upon cellular internalization, MPDOLs released OST and DNA plasmids to the cytoplasm where the OST inhibited the EGFR signaling pathway, while DNA plasmid suppressed IGF2BP3 expression and triggered the self-assembly and secretion of RVG-EXOs(si) for brain metastasis tumor treatment.
Results
IGF2BP3 was widely expressed in NSCLC and increased after OST treatment
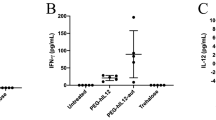
We first analyzed the expression of m6A modulators in NSCLC tumor tissues using The Cancer Genome Atlas (TCGA) dataset (Fig. 2a). Among these modulators, IGF2BP3 emerged as one of the most significantly upregulated m6A-related genes in metastatic tumors compared to primary tumors (Fig. 2b). Further analysis was done using the Gene Expression Profiling Interactive Analysis (GEPIA) to verify the elevated expression of IGF2BP3 in two kind of lung tumors and its association with poor survival (Fig. 2c, d). Consistent with these bioinformatics findings, the protein level of IGF2BP3 in lung tumor tissue was much higher than the adjacent tissue in NSCLC patients (Fig. 2e, f). Additionally, immunohistochemical evidence and quantitative real-time PCR (qRT-PCR) assay revealed elevated IGF2BP3 expression in the brain metastasis regions in NSCLC mouse models (Fig. 2g, h). Importantly, while long-term OST treatment (5 mg kg−1) partially reduced tumor burden, it also slightly upregulated IGF2BP3 expression, which aligned with the recent studies reporting that IGF2BP3 mediated acquired resistance to NSCLC TKIs13. In the established Lewis lung cancer (LLC) cell line with acquired resistance to OST(LLC-OR), the half-maximum inhibitory concentration (IC50) values increased from 2.03 µM to 29.30 µM, with a higher mRNA expression of IGF2BP3 (Supplementary Fig. 1). In normal LLC cells, IGF2BP3 mRNA expression increased after prolonged stimulation with OST (Fig. 2i). These findings suggest that targeting IGF2BP3 may enhance the sensitivity of tumor cells to OST and inhibit brain metastasis in NSCLC treatment. Since no small molecule inhibitors of IGF2BP3 are available, we designed and screened the siRNA sequence (S: GCCGUCUCAUUGGUAAAGATT, AS: UCUUUACCAAUGAGACGGCTT) to achieve effective interference of IGF2BP3 (Fig. 2j, k).
a Expression distribution of m6A modulator genes in normal and NSCLC tissues based on TCGA database analysis (n = 1017 NSCLC tumor tissues and n = 686 normal tissues). b Molecular characterization of m6A regulators in NSCLC with and without metastasis using TCGA data (n = 520 without metastasis and n = 485 with metastasis). In box plots, the center line represents the median, the bounds of the box indicate the interquartile range from the 25th to 75th percentile, and whiskers extend to the 1.5× interquartile range. Outliers beyond this range are plotted individually. c The IGF2BP3 gene expression profiles in human NSCLC within the GEPIA database (n = 483 lung adenocarcinoma tumor tissues and n = 59 normal tissues, n = 486 lung squamous cell carcinoma tumor tissues and n = 50 normal tissues). d The correlation between IGF2BP3 expression and survival rate of NSCLC patients within the GEPIA database (n = 481 patients). e Immunohistochemical staining of IGF2BP3 in human NSCLC tissues and adjacent tissues and f quantitative analysis of relative IGF2BP3-positive expression (n = 4 patients), scale bar, 50 μm. g Immunohistochemical staining of IGF2BP3 in NSCLC tissues from mice, scale bars, 20 μm. h qRT-PCR analysis of IGF2BP3 mRNA levels in NSCLC and normal tissues from mice (n = 3 mice). i qRT-PCR analysis of IGF2BP3 mRNA levels and j, k western blot analysis of IGF2BP3 protein levels in LLC cells treated with OST and IGF2BP3 siRNA (n = 4 independent experiments). Data are shown as mean ± SD. a and b were analyzed by two-sided Wilcox test. d was assessed using the log-rank (Mantel-Cox) test. h, i and k were calculated using the one-way ANOVA with a Tukey post hoc test. c and f were analyzed by a two-sided t-test. Source data are provided as a Source Data file.
Preparation and characterization of MPDOLs for co-delivery of DNA and OST
We developed a biomimetic nanomedicine named MPDOLs for the co-delivery of DNA plasmids encoding IGF2BP3 siRNA and a brain-targeting tag, along with OST. The MPDOLs were composed of PβAE-DNA complexes, lipid shells, and mesenchymal stem cell (MSC) membranes with highly expressed SP-B protein. The sequences of oligonucleotides are shown in Supplementary Table 1 and the preparation process of MPDOLs is illustrated in Fig. 3a. First, a DNA plasmid structure was constructed using the classical design strategy and was engineered to express miRNA precursors based on a cytomegalovirus promoter, which was also replaced with the sequence encoding IGF2BP3 siRNA to facilitate siRNA production. A sequence encoding the RVG was added downstream of the cytomegalovirus promoter and linked to the N-terminal end of the exosome membrane protein CD6328. Successful plasmid construction was confirmed by gene sequencing (Supplementary Fig. 2).
a Schematic illustration of the MPDOL fabrication process. b Agarose gel electrophoresis showing plasmid DNA complexation by PβAE at different weight ratios. c Size distribution and morphological images of PDs, OLs, PDOLs, and MPDOLs, scale bar, 200 nm. d Fluorescence microscopy image of MSCs transfected by LV-Sftpb with GFP for 24 h, scale bar, 100 μm. e Western blot analysis confirming high SP-B expression in SP-B+ MSC cells. f The emission spectrum of the FRET pair generated by Dil-labeled membranes and DiO-labeled liposomes in MPDOLs. g SDS-PAGE analysis of membrane proteins in MPDOLs. h Release kinetics of OST from MPDOLs in two different pH environments (n = 3 independent samples). i Representative in vivo imaging of LLC tumor-bearing mice after inhalation of DiR, DiR-labeled PDOLs, and DiR-labeled MPDOLs, and j quantitative analysis of fluorescence radiant efficiency in major organs (n = 3 mice). k Immunofluorescence images of lung sections from mice administered with DiR, DiR-labeled PDOLs, and DiR-labeled MPDOLs, scale bar, 500 μm. l Flow cytometry analysis showing cell percentage that internalizes MPDOLs in CD45− cells, lymphocytes, and macrophages in total cells of the lung tissues (n = 3 mice). Data are shown as mean ± SD. j was calculated using the one-way ANOVA with a Tukey post hoc test in each kind of organ. Source data are provided as a Source Data file.
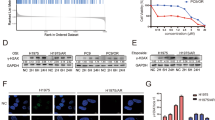
Next, PβAE was synthesized to complex with plasmid DNA and form an inner core (PD). The structure of PβAE was confirmed by 1H nuclear magnetic resonance (NMR) spectroscopy (Supplementary Figs. 3 and 4). PD nanoparticles were constructed at various weight ratios (1:1, 2.5:1, 5:1, 10:1, 20:1, and 40:1) to evaluate the complexing ability of PβAE with plasmid DNA. Agarose gel electrophoresis results indicated that plasmid DNA could be fully complexed when the weight ratio was at least 10:1 (Fig. 3b). To minimize cell cytotoxicity, the 10:1 weight ratio was chosen for the subsequent NSCLC cell transfection experiments. The LLC cells were treated with PD nanoparticles composed of the DNA plasmids carrying the green fluorescent protein (GFP) tag. Flow cytometry showed that the GFP positive cell percentage was over 80%, which was comparable to the performance of commercial transfection agent jetPRIME (Supplementary Fig. 5). At this weight ratio, PD nanoparticles had a mean size of 50.47 ± 5.36 nm, a positive zeta potential of 26.97 ± 3.85 mV, and a spherical morphology (Fig. 3c). Meanwhile, the liposome shell was prepared using a thin film dispersion method to encapsulate OST. Obtained OST liposomes (OLs) had a mean size of 141.47 ± 3.07 nm and a zeta potential of 6.65 ± 0.37 mV. Subsequently, the PD particles were added to the equal volume of OLs to form PDOLs, with a mean size of 182.63 ± 6.04 nm (Fig. 3c). The similar zeta potentials of PDOLs (6.74 ± 0.73 mV) and OLs confirmed the successful encapsulation of PD nanoparticles into PDOLs.
In addition, lentiviral transfection was employed to establish an MSC line stably expressing SP-B protein and GFP fluorescence was visibly detected 48 h post-transfection14 (Fig. 3d). Stable clones were selected following puromycin treatment for 7 days and western blot experiments confirmed the high expression of SP-B (Fig. 3e). After coating with the cell membrane, the size of the resulting MPDOLs increased slightly to 195.20 ± 4.56 nm, and the zeta potential shifted from 6.74 mV to −4.28 mV. The fusion of MSC membranes with liposomes was further validated by fluorescence resonance energy transfer (FRET) experiments21. When the MSC membrane and liposome were near to allow fusion, the absorption wavelength of DiO in the liposomes excited the fluorescent group of Dil in the membrane through energy-efficient resonance. This resulted in fluorescence characteristic spectral changes, including weakened DiO emission fluorescence and appeared Dil emission fluorescence (Fig. 3f). The proteomic analysis further validated the retention of membrane proteins in MPDOLs (Fig. 3g).
The encapsulation efficiency of the OST and DNA in MPDOLs was analyzed by the high-performance liquid chromatography (HPLC) method and a commercial DNA quantification kit. The encapsulation efficiency of OST reached 81.37 ± 2.9%, corresponding to a drug loading capacity of 6.72 ± 0.3%. The mass ratio of DNA and OST encapsulated in MPDOLs was determined to be around 1:1. We further evaluated the release profile of OST under two different pH to mimic the normal physiological conditions and the acidic environment in tumor cells. Free OST showed rapid release, with over 40% released within the first hour of incubation. In contrast, the release of OST encapsulated in liposomes exhibited a slower release profile at pH = 7.4, extending over several days. At pH = 5.0, 79.85% of the OST in MPDOLs was released within 12 h, indicating that MPDOLs could rapidly release drugs in endo-lysosomes of tumor cells (Fig. 3h). Additionally, the stability of DNA encapsulated in MPDOLs was assessed by incubation with fetal bovine serum (FBS) for 48 h. While free DNA was completely degraded under these conditions, DNA within MPDOLs remained visible, demonstrating the ability of MPDOLs to maintain structural integrity and protect DNA cargo in biological environments (Supplementary Fig. 6).
After atomization with a commercially available portable nebulizer, the droplets containing MPDOLs were approximately 3 µm in diameter, making them suitable for efficient deposition. Importantly, the nebulization process had no significant influence on the physicochemical characteristics of MPDOLs, including size, zeta potential, morphology, and encapsulation efficiencies (Supplementary Fig. 7). The tolerance of MPDOLs to severe shear during nebulization made them ideal candidates for inhalation therapy. The above results supported that MPDOLs have the potential to be stable and effective nanocarriers delivering OST and DNA plasmids.
MPDOLs exhibited superior distribution and retention capabilities in lung tissues
Inhaled medicine offers higher local drug concentrations and direct access to target cells in pulmonary lesions compared to systemic administration routes, which often suffer from first-pass metabolism and systemic side effects19. In this study, we used the commercially available vibrating mesh nebulizer for administration29. To investigate the in vivo distribution of MPDOLs, mice were separately treated with free DiR, DiR-labeled PDOLs, or DiR-labeled MPDOLs, and their fluorescence biodistribution was monitored over 24 h using the in vivo imaging system. The fluorescence intensity in the lungs of the MPDOL-treated group was approximately twice that of the free DiR group (Fig. 3i, j). The superior retention of MPDOLs in lung tissues can be attributed to the function of the MSC membrane enriched with SP-B protein. As a critical component of pulmonary surfactant, SP-B could reduce alveolar surface tension, and facilitate drug transport by interfacial interactions. In the MPDOL-treated group, the fluorescence intensity in the lungs peaked at 12 h post-inhalation and gradually declined over time (Fig. 3i). Fluorescence microscopy of lung tissue sections further confirmed the enhanced penetration and retention of MPDOLs, with higher accumulation in bronchioles and alveoli compared to PDOLs (Fig. 3k). Given the complexity of the lung tumor microenvironment, consisting of a variety of cells, extracellular matrix, and secretions, we next investigated the main cell populations that internalize MPDOLs via flow cytometry. The results showed that MPDOLs predominantly localized to CD45⁻ malignant cells, accounting for approximately 70% of the total cell population, followed by 20% of macrophages (Fig. 3l). These findings suggested that MPDOLs effectively penetrated the lung tissues and were mainly endocytosed by malignant cells, demonstrating their potential for enhanced pulmonary drug delivery.
MPDOLs demonstrated enhanced cellular uptake and endosomal escape capability in vitro
We then evaluated the intracellular fate of MPDOLs (Fig. 4a). The uptake of MPDOLs in LLC cells was investigated by labeling the lipid layer with coumarin 6 (C6). Both flow cytometry analysis and confocal microscopy results revealed a time-dependent fluorescence signal in the MPDOL-treated group. Higher mean fluorescence intensity (MFI) of FITC compared to the C6 group indicated enhanced cellular uptake (Fig. 4b, c, and Supplementary Fig. 8). Nanoparticles are known to enter cells via multiple pathways, including clathrin-mediated endocytosis, caveolae-mediated endocytosis, lipid raft-mediated endocytosis, and macropinocytosis30. Different endocytosis inhibitors were then used to determine the MPDOL internalization pathway in LLC cells. The results indicated that chlorpromazine (CPZ, a clathrin-mediated endocytosis inhibitor) and genistein (GEN, a caveolae-mediated uptake inhibitor) inhibited intracellular MFI, whereas methyl-β-cyclodextrin (m-β-CD, a lipid raft inhibitor) and amiloride hydrochloride (AMI, a macropinocytosis inhibitor) showed no effect, which indicated MPDOLs were internalized primarily via clathrin- and caveolae-mediated endocytosis (Fig. 4d).
a Diagram illustrating the intracellular fate of MPDOLs and their roles in exosome production in LLC cells. b Flow cytometry analysis and c quantitative analysis of the MFI of C6-labeled MPDOLs internalized by LLC cells (n = 3 independent experiments). d Effect of different endocytosis inhibitors on MPDOL internalization in LLC cells (n = 3 independent experiments). e Confocal microscopy showing lysosomal escape of MPDOLs in LLC cells after 4, 8, and 12 h of incubation, scale bar, 10 μm. f Acridine orange staining of lysosomal damage induced by MPDOLs, scale bar, 40 μm. g qRT-PCR analysis of IGF2BP3 mRNA levels (n = 3 independent experiments) and h western blot analysis of IGF2BP3 protein expression in LLC cells treated with different formulations. i Cell viability of LLC cells treated with free OST, PDs, and MPDOLs at various concentrations for 48 h (n = 4 independent experiments). j The total apoptosis rate of LLC cells after incubation with different preparations for 48 h measured by flow cytometry (n = 3 independent experiments). Data are presented as mean ± SD. Violin plot of d depicting data distribution with the minimum and maximum values, median (center), interquartile range (box bounds), and whiskers extending to 1.5× IQR. g and j were calculated using the one-way ANOVA with a Tukey post hoc test. Source data are provided as a Source Data file.
In clathrin- and caveolae-mediated endocytosis, lysosomal escape is a critical step that affects the subcellular bioavailability of nanomedicines31. To examine the intracellular fate of MPDOLs in LLC cells, LysoTracker was used to label endo-lysosomes, and GFP-labeled MPDOLs were visualized via confocal microscopy32. After incubation for 4–8 h, the GFP signal was mostly trapped in lysosomes. Over time, the colocalization decreased, with the GFP signal gradually diffusing into the cytoplasm. The Pearson correlation coefficient decreased from 0.897 to 0.528 at 12 h, indicating successful lysosomal escape (Fig. 4e). This phenomenon can be explained from several perspectives. Release experiments demonstrated that the acidic environment of endo-lysosomes triggered the rupture of MPDOLs, and facilitated the release of OST and PDs. The degradable PβAE can be protonated under acidic conditions, leading to an influx of chloride ions to maintain ionic balance and causing osmotic swelling of lysosomes, thus resulting in the physical rupture of membranes33. To further confirm this, LLC cells were stained with acridine orange and cells incubated with MPDOLs showed a marked decrease in red fluorescence as compared to the controls, indicating increased lysosomal membrane permeability (Fig. 4f).
MPDOLs downregulated IGF2BP3 expression and induced cytotoxicity in vitro
Encouraged by the in vitro uptake results of MPDOLs, the therapeutic effect of MPDOLs was further investigated in LLC cells. To assess the impact of MPDOLs on IGF2BP3 expression, qPCR and western blot analysis revealed that both MPDOLs and PDs strongly downregulated their expression (Fig. 4g, h, Supplementary Fig. 9). The cytotoxicity assay demonstrated a dose-dependent response, with IC50 values for OST, PDs, and MPDOLs being 2.03, 2.36, and 1.23 μM, respectively (Fig. 4i). Subsequently, flow cytometry with Annexin V-FITC and propidium iodide (PI) staining was performed to identify cell apoptosis, and the MPDOL-treated group exhibited the highest apoptosis rate at 48 h, accounting for 17.6% of the total population (Fig. 4j and Supplementary Fig. 10). These results supported that MPDOLs can effectively deliver OST and DNA into the tumor cytoplasm, leading to the downregulation of IGF2BP3 and subsequently inhibiting LLC cell growth.
Additionally, we examined the expression of EGFR and phosphorylated-EGFR(p-EGFR) following different treatments in LLC cells (Supplementary Fig. 11). PDs and MPDOLs reduced total EGFR expression, while OST and MPDOLs led to a slight decrease in p-EGFR levels. EGFR inhibition was not significantly observed in the OST-only group, consistent with previous reports that OST suppresses tumor cell proliferation by inhibiting EGFR phosphorylation34. The mechanism by which IGF2BP3 inhibition affects total EGFR levels remains unclear and warrants further investigation.
MPDOLs induced the production of RVG-EXOs(si), enabling brain-targeted systemic delivery
Next, we investigated whether plasmid transfection via MPDOLs could direct siRNA loading into exosomes. Following MPDOL transfection in LLC cells, the cell culture supernatant was collected to extract exosomes, which were named by RVG-EXOs(si). Nanoparticle tracking analysis (NTA) revealed that exosomes had a mean diameter of 132.5 ± 4.6 nm, with morphological characterization showing a flat disc-like structure and a characteristic lipid bilayer (Fig. 5a, b). Heat shock protein 70 (HSP70) and ALIX, the specific exosome markers, were determined by western blotting (Fig. 5c)35,36. A Flag epitope tag was used to label the RVG-CD63 because of the lack of an anti-RVG antibody. After transfection of LLC cells with MPDOLs composed of the Flag tag plasmid, the enrichment of Flag in the exosomes extracted from cell culture medium supernatant was observed, which confirmed the expression of the guidance protein. Furthermore, the similar density of ALIX bands across samples confirmed consistent exosome production levels (Fig. 5d). PCR analysis demonstrated the increase in IGF2BP3 siRNA content in exosomes generated from cells transfected with MPDOLs (Fig. 5e). When LLC cells were incubated with isolated RVG-EXOs(si) (5 μg mL−1 of total protein), both mRNA and protein levels of IGF2BP3 were knocked down (Fig. 5f, Supplementary Fig. 12). The in vitro therapeutic effects of RVG-EXOs(si) were further investigated. RVG-EXOs(si) exhibited dose-dependent cytotoxicity, whereas control exosomes from normal LLC cell supertenant (EXOs) showed negligible effects on cancer cell growth, indicating that RVG-EXOs(si) effectively loaded IGF2BP3 siRNA and retained its biological function(Fig. 5g). To assess the ability of RVG-EXOs(si) to cross the blood-brain barrier (BBB), the in vitro BBB model was constructed using bEnd.3 cells to form tight junctions37. Dil-labeled RVG-EXOs(si) were added to the upper chamber of the trans-well system (Fig. 5h)38. Fluorescence intensity and the microscope images of LLC cells in the lower chamber confirmed that RVG enhanced the ability of exosomes to penetrate the BBB (Fig. 5i, j).
a Size distribution and b morphological characterization of RVG-EXOs(si), scale bar, 150 nm. c Western blot analysis of exosome markers ALIX and HSP70 in RVG-EXOs(si), and d Flag expression in exosomes transfected with MPDOLs containing RVG-CD63-Flag fusion plasmids. e Relative IGF2BP3 siRNA content in RVG-EXOs(si) compared with regular EXOs (n = 3 independent experiments). f qRT-PCR analysis of IGF2BP3 mRNA level in LLC cells treated with RVG-EXOs(si) and regular EXOs (n = 3 independent experiments). g Cell viability of LLC cells treated with RVG-EXOs(si) or EXOs at various total protein concentrations for 48 h (n = 4 independent experiments). h Schematic illustration of the trans-well system used to evaluate BBB penetration abilities of Dil-labeled RVG-EXOs(si). i Representative fluorescence images and j quantitative fluorescence intensity analysis in the lower chambers after incubation with RVG-EXOs(si) (n = 4 independent biological samples), scale bar, 200 μm. k Ex vivo imaging of major organs and k quantitative fluorescence intensity of DiD in main organs (brains, hearts, livers, spleens, lungs, kidneys) 12 h after intravenous administration of free DiD, DiD-labeled EXOs, and DiD-labeled RVG-EXOs(si) isolated from cell culture medium (n = 3 mice). Data are presented as mean ± SD. e and j were analyzed by a two-sided t-test. f and data from the same tissue of l were calculated using the one-way ANOVA with a Tukey post hoc test. Source data are provided as a Source Data file.
To investigate the in vivo production and function of RVG-EXOs(si), C57BL/6 mice bearing LLC tumors in the lungs and brains were inhaled with MPDOLs. After 72 h, plasma was obtained to isolate the exosomes. Extracted exosomes had a disc-like morphology and a particle size of 135.0 ± 8.0 nm (Supplementary Fig. 13a). Marker proteins including ALIX and HSP70, were detected via western blotting (Supplementary Fig. 13b). The plasmid-encoded RVG was fused to CD63, and thus the apparent elevation in the CD63 content of the exosomes produced by the MPDOL transfection implied a successful high expression of the RVG protein (Supplementary Fig. 13c). qRT-PCR analysis showed higher IGF2BP3 siRNA levels in RVG-EXOs(si) compared with regular EXOs isolated from mice plasma, and absolute quantitative PCR declared that 100 μL of exosomes from plasma contained 68.3 ± 2.08 pg of IGF2BP3 siRNA (Supplementary Fig. 13d). Previous studies also reported siRNA concentrations of approximately 1500 fM in plasma exosomes following intravenous plasmid DNA injection, which is similar to our reported concentration18.
Subsequently, we also investigated the tissue targeting ability of RVG-EXOs(si) produced in vivo or ex vivo. Tumor-bearing mice received intravenous injections of DiD, DiD-labeled EXOs, and DiD-labeled RVG-EXO(si). Organ fluorescence was imaged 12 h post-administration. RVG-EXOs(si) produced by genetically engineered tumor cells exhibited significantly higher fluorescence signals in brain tissue compared to regular EXOs, with lower accumulation in the liver (Fig. 5k, l). For exosomes isolated from plasma, the fluorescence distribution in other major organs was similar in both EXO and RVG-EXO(si) groups, while free DiD accumulated in the liver and spleen (Supplementary Fig. 14). In each group, the other four mice were sacrificed to isolate the brain tissues for the immunofluorescence staining, and five regions were randomly selected from brain tissue sections for the quantification of the DiD integrated density for each mouse. Brain sections of the RVG-EXO(si) group also revealed increased DiD integrated density (Supplementary Fig. 15), confirming the brain-targeting ability of RVG.
MPDOLs effectively and safely inhibit tumor growth
To evaluate the in vivo anti-tumor efficacy of MPDOLs, orthotopic and brain metastatic lung cancer tumor models were established in C57BL/6J mice using luciferase-transfected LLC cells (LLC-Luc). MPDOLs, PDOLs, PDs, OLs, and phosphate buffer solution (PBS) were inhaled by mice every four days for a total of five times, while OST was given by intragastric administration (Fig. 6a). Body weights were recorded every three days, and tumor progression was monitored weekly using bioluminescence imaging (Fig. 6b). Bioluminescence signals increased rapidly in the PBS group, accompanied by significant weight loss after day 21, indicating tumor progression (Fig. 6b, c). Free OST showed minimal therapeutic efficacy, while OLs and PDs produced limited tumor inhibition. In contrast, MPDOL inhalation significantly suppressed tumor growth and prolonged survival. Mice treated with MPDOLs demonstrated the longest survival, with 80% remaining alive at 70 days, underscoring its superior therapeutic effect (Fig. 6d). On day 28, the diffuse distribution of tumor shadows was observed via micro-computed tomography (micro-CT), and eight mice were euthanized in each group for further analysis. Their blood samples, lung tissue, brain tissue, and other important organs were taken for flow cytometry, cytokine assay, and histological evaluation (HE). Micro-CT revealed reduced lung tumor burden in the MPDOL group, with lower lung density and wet weights compared to PBS and OST groups (Fig. 6e, f and Supplementary Fig. 16). Ex vivo luminescence imaging further showed reduced tumor burden in both the lungs and brains in MPDOL-treated group. While OLs primarily reduced lung tumor burden, PDs exhibited greater efficacy against brain metastases (Fig. 6g, h, and Supplementary Fig. 17). This suggested the potential of producing exosomes mediated by DNA plasmid in vivo to treat distant tumors. HE analyses revealed that MPDOL treatment reversed lung tumor pathology and reduced tumor cell proliferation was indicated by decreased Ki67 staining in lung and brain tissues (Supplementary Figs. 18–20). In the brain metastasis model, PBS-treated mice exhibited large tumor burdens, while MPDOL treatment suppressed brain metastases to nearly undetectable levels (Fig. 6i and Supplementary Fig. 17). The MPDOL, and PDOL treatment significantly reduced Ki67-positive areas in brain tissue sections, which offered a promising therapeutic approach for combating both primary lung tumors and metastatic brain lesions (Supplementary Fig. 20).
a Experimental timeline for inhalation treatment. Orthotopic lung cancer was established by intrathoracic injection of 5 × 105 LLC-Luc cells on day −7. 4 × 104 LLC-Luc cells were injected intracranially into the mice on day 0. From day 8, mice received different treatments, including the inhalation of MPDOLs, PDOLs, PDs, OLs, and PBS every 4 days for a total of 5 cycles. The mice given OST by oral gavage were set as the positive control group. Eight mice in each group were randomly euthanized on day 28 for organ collection, and the remaining ten mice were monitored until day 70. b In vivo bioluminescence imaging of lung tumors and brain tumors in mice. c Mice weight changes (n = 8 mice) and d survival curves for each treatment group (n = 10 mice). e Mean lung density analyzed by micro-CT imaging and f isolated lung weights on day 28 after treatments (n = 5 mice). g Ex vivo bioluminescence of tumor cells in lungs and h brains on day 28 post-treatment (n = 3 mice). i Representative HE staining images of brain sections from different treated mice, scale bar, 1 mm. The data are expressed as mean ± SD. Violin plot of f depicting data distribution with the minimum and maximum values, median (center), interquartile range (box bounds), and whiskers extending to 1.5× IQR. Mice weights in c were analyzed using the two-way ANOVA and survival data in d were analyzed by log-rank (Mantel-Cox) test with the PBS group. Colored lines represent different groups, with statistical significance indicated by corresponding colors for p values. e–h were calculated using the one-way ANOVA with a Tukey post hoc test. Source data are provided as a Source Data file.
Quantification of the IGF2BP3 protein expression was then performed blindly. The distribution range and IGF2BP3-positive cell ratio in the lung tumors in the MPDOL group were reduced (Supplementary Fig. 19). Furthermore, the observed reduction in IGF2BP3 levels in brain metastases indicated successful delivery of in vivo-produced exosomes to brain tissue, enabling IGF2BP3 siRNA to exert its biological function. We also analyzed the expression of p-EGFR and total EGFR in the brain tissues. The results showed that there was no significant difference in the expression of total EGFR among various treatment groups, while the expression of p-EGFR was significantly decreased in MPDOL, PDOL, PD, and OL groups (Supplementary Figs. 21 and 22).
Systemic toxicity remains a critical consideration for drug delivery systems39. The toxicity of MPDOLs in vivo was assessed following repeated inhalation. No acute injuries resulting in immediate death were observed in any of the groups. HE images revealed no damage or necrosis in major organs, including the heart, liver, spleen, or kidneys, in the MPDOL-treated group. In contrast, the liver in the OST group showed inflammatory cell infiltration, hemorrhage, and edema (Supplementary Fig. 23). Furthermore, the serum biochemical analysis supported the safety of MPDOLs, as key liver and kidney function indicators—blood urea nitrogen (BUN), alanine aminotransferase (ALT), and aspartate aminotransferase (AST)—remained within normal ranges (Supplementary Table 2). Collectively, these findings underscored the favorable biocompatibility of MPDOLs and suggested that this gene therapy-based nanomedicine platform is a safe and effective strategy for lung cancer treatment.
The treatment of MPDOLs reprogrammed the anti-tumor immune response
The immunomodulatory effects of MPDOLs on the orthotopic lung tumor microenvironment were systematically evaluated40. Treatment with MPDOLs resulted in significant increases in the proportion of CD8+ T cells (Fig. 7a). Elevated interferon-gamma (IFN-γ) levels were associated with CD8⁺ T cell-mediated tumor cell killing, while regulatory T cells (Tregs) are known to play an anti-tumor immunosuppressive role41. Notably, MPDOL treatment reduced the percentage of Tregs by threefold and increased the proportion of IFN-γ⁺CD8⁺ T cells from 2.16% to 9.16% compared to the PBS group (Fig. 7b, c). The percentage of natural killer (NK) cells was increased in the MPDOL group, further contributing to anti-tumor immune response (Fig. 7d). The tumor immune microenvironment was also characterized by a reduced proportion of M2-like macrophages, and an increased proportion of pro-inflammatory M1-like macrophages, which may be attributed to the knockout of IGF2BP3, and was consistent with previous findings (Fig. 7e, f)42,43. Cytokine profiling via enzyme-linked immunosorbent assay (ELISA) revealed that serum levels of IFN-γ and tumor necrosis factor-alpha (TNF-α) were significantly elevated, while levels of interleukin-6 (IL-6), transforming growth factor-beta 1 (TGF-β1), and interleukin-10 (IL-10) were decreased in the MPDOL-treated group compared to the PBS group44 (Fig. 7g). The above results indicated that MPDOL treatment elicited anti-tumor immune responses and ultimately promoted cytotoxic immune effects against tumor cells45,46.
a Flow cytometry and quantification of CD8+ T cells (CD45+CD3+CD8+), b cytotoxic T cell (CD45+CD3+CD8+IFN-γ+), c Treg cells (CD45+CD4+CD25+Foxp3+) and d NK cells (CD45+CD3−NK1.1+) in the lung tumor microenvironment following different treatments (n = 4 mice). e Quantification of M2-like macrophages (CD45+CD11b+F4/80+CD206+) and f M1-like macrophages (CD45+CD11b+F4/80+iNOS+) in the lung tumor microenvironment (n = 4 mice). g Relative cytokine levels in the serum including TNF-α, IFN-γ, IL-6, IL-10, and TGF-β1 (n = 3 mice). Data are presented as mean ± SD. Heatmap of (g) showing the average cytokine expression levels and the color scale ranges from low (white) to high (blue) expression. a–f were calculated using the one-way ANOVA with a Tukey post hoc test. Source data are provided as a Source Data file.
MPDOLs downregulated Myc and influenced the Warburg effect
Transcriptomic analysis was performed to elucidate the therapeutic mechanism of MPDOLs on LLC cells. RNA sequencing identified 1203 differentially expressed genes (DEGs) (|log2(Fold Change)| > 1, p < 0.05) after MPDOLs treatment, with 597 downregulated and 606 upregulated (Fig. 8a). Gene Ontology (GO) enrichment analysis revealed that the downregulated genes were associated with biological processes and pathways linked to cell proliferation, cell invasion, DNA damage repair, and cancer-related biological processes and pathways. Subsequently, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis highlighted pathways implicated in the observed biological effects. Cytokine-cytokine receptor interaction (30 DEGs) and the mitogen-activated protein kinase (MAPK) signaling pathway (38 DEGs) were significantly enriched in the MPDOL-treated group compared to the control group (Fig. 8b). To investigate the potential interaction and central hub genes, protein-protein interaction (PPI) network data were imported into Cytoscape software to intersect the top ten genes from five cytoHubba algorithms (MNC, EPC, Degree, Closeness, and BottleNeck). As shown in Fig. 8c, Vegfa, Jund, Kdr, Ptgs2, Fos, and Myc were identified as hub genes, which were downregulated in MPDOL compared to the controls. Of these, Myc, Fos, and Ptgs2 showed |log₂(Fold Change)| values exceeding 2, while Vegfa, Jund, and Kdr exhibited |log₂(Fold Change)| values greater than 1 (Fig. 8a). Myc had the highest absolute abundance among these genes. Meanwhile, it has been reported that the enrichment of Myc target genes observed after knocking down IGF2BP3 and Myc is critical for tumor progression and drug resistance in NSCLC47,48,49. Given its central role, Myc was selected as a target of interest to further explore its involvement in the therapeutic response to MPDOLs.
a Volcano plot illustrating differentially expressed genes of LLC cells in the MPDOL treatment group compared to the control group. Genes are categorized as unaltered (blue, log₂(Fold Change) < 1 or p > 0.05), upregulated (red, log₂(Fold Change) > 1 and p < 0.05), and downregulated (green, log₂(Fold Change) < −1 and p < 0.05). b TOP 20 significantly regulated pathways in the MPDOL group via KEGG functional enrichment analysis. c Top 10 hub genes identified through PPI analysis using 5 algorithms. d Western blot analysis of Myc protein expression in LLC cells with different treatments (n = 4 independent experiments). e The glucose consumption, f lactic acid concentration, and g NAD+ levels of LLC cells after different treatments (n = 4 independent experiments). h Immunofluorescence staining of HK2 and i GLUT1 in LLC cells after different treatments, scale bar, 15 μm. j Schematic illustration of proposed synergistic mechanisms of MPDOL treatment. Data are presented as mean ± SD. DEseq2 was used to obtain the P-value, and statistical significance was determined using the Wald test (two-sided) in a. Multiple testing correction was applied using the Benjamini-Hochberg method to control the false discovery rate. KEGG pathway enrichment analysis was performed using clusterProfiler, and statistical significance was assessed using Fisher’s exact test (two-sided). Multiple testing correction was performed using the Benjamini-Hochberg method in b. d–g were calculated using the one-way ANOVA with a Tukey post hoc test. Source data are provided as a Source Data file.
To further explore the role of Myc, we analyzed its protein levels in LLC cells. The results demonstrated that MPDOLs downregulated the protein expression of Myc (Fig. 8d). It has been reported that most tumor cells rely on glycolysis, commonly referred to as the Warburg effect. As a key regulator of glycolysis, Myc influences the metabolic process by driving glucose oxidation into pyruvate, yielding two molecules of adenosine triphosphate (ATP) and promoting the oxidized form of nicotinamide adenine dinucleotide (NAD⁺) to reduced nicotinamide adenine dinucleotide (NADH)50,51. The pyruvate is predominantly converted to lactate. We quantified several glycolysis-related metabolites in LLC cells following MPDOL treatment. Both glucose uptake and lactate production, the latter being the terminal product of glycolysis, were significantly reduced (Fig. 8e, f). Additionally, an increase in NAD⁺ levels was observed in the MPDOL group (Fig. 8g). Mechanistically, we identified that the expression of two rate-limiting enzymes in glycolysis, glucose transporter 1 (GLUT1) and hexokinase 2 (HK2), was downregulated, which may be the reason why the Warburg effect was affected (Fig. 8h, i). Besides, the content of lactic acid in the tumor microenvironment usually affects the anti-tumor immune regulation, which was also complementary to the results of anti-tumor immune response in vivo (Fig. 8j)46.
Discussion
In this study, we developed an inhalable multifunctional nanoparticle for the treatment of primary and metastatic NSCLC. With the help of SP-B, inhaled MPDOLs demonstrated enhanced retention in lung tissues. Upon delivery to tumor cells, OST inhibited EGFR N-linked glycosylation, while the DNA depleted IGF2BP3 protein and induced exosome generation, facilitating efficient delivery of IGF2BP3 siRNA to brain metastases. The synergistic approach achieved effective gene silencing and improved the therapeutic efficacy for NSCLC with brain metastases.
Our findings demonstrated that repeated OST administration in NSCLC models hasn’t downregulated IGF2BP3. Bioinformatic analyses further confirmed increased IGF2BP3 expression in metastases. Therefore, interfering with IGF2BP3 via DNA plasmids or siRNA in conjunction with OST might be a therapeutic strategy for the management of NSCLC. Our hypothesis has driven research to develop drug delivery systems for this combination therapy.
Efficient delivery of therapeutic agents to target tissues is critical for treating lung diseases. Non-invasive inhalation helps to achieve direct drug accumulation to the lung, which is better than other delivery routes such as intravenous administration. Meanwhile, inspired by the efficacy of surfactant replacement therapy in the treatment of lung diseases, we engineered a liposome with MSC-derived cell membranes highly expressing SP-B, which facilitated the adsorption at the lung interface and enhanced intrapulmonary retention. Tumor bioluminescence imaging and immunofluorescence images confirmed the effective delivery of agents to the lungs via MPDOLs. This surfactant-inspired nanomedicine offered a versatile platform that could be extended to other pulmonary diseases, such as idiopathic pulmonary fibrosis and acute lung injury.
To date, exosome-mediated delivery systems have gained attention for small nucleic acid drugs. However, current clinical applications of the exosomes still face some challenges, including the disruption of the RNA biological activity and high costs for exosome purification. Here, we aim to use gene editing to produce and self-assemble siRNAs in host cells, and then transport them into endogenously produced exosomes. By regulating the expression of exosome surface proteins (e.g., RVGs) via gene editing, exosomes achieved brain targeting to minimize the off-target side effects, which highlights the clinical potential of this safe and biocompatible strategy. In addition, the flexibility of the liposome composition allows this strategy to expand to other combination therapies. The modular design of DNA plasmids accommodates various genet sequences, enabling the integration of switchable siRNAs or additional target proteins. This adaptability supports the potential of MPDOLs as a broad platform for personalized medicine and improving patient outcomes.
It’s worth pointing out that despite the excellent therapeutic efficacy of MPDOLs, the clinical translation remains challenging. Comprehensive toxicology, safety, and efficacy studies in large animal models are needed, alongside investigations into the pharmacokinetics of siRNA production and metabolism. In addition, scalability is another crucial step to take into account for clinical translation. The large-scale production cost of such complex biomimetic liposomes may be high, and the nanostructures are difficult to maintain during the actual production process or applications. Therefore, further investigation of the proposed MPDOLs is essential for clinical translation.
In conclusion, as a non-invasive inhalation therapy and autologous exosome production platform, MPDOLs stand out in terms of safety, biocompatibility, compliance, and efficiency for NSCLC therapy. While further optimization and research are required, it could open up an exciting field of precision medicine.
Methods
Ethical statement
Our research complies with all relevant ethical regulations of Shandong University. All animal experiments are approved and conducted following the Ethics Committee of Scientific Research of Cheeloo College of Medicine, Shandong University, China, which agreed to 20% weight loss as a humane endpointity (Approval No. 230041). We confirmed that the maximal tumor burden of mice was not exceeded. All the clinical specimens were approved by the Ethics Committee of the Second Hospital of Shandong University (Approval No. KYLL-2020(KJ)P-0099), and informed consent was obtained from all subjects (2 of male and 2 of female) in accordance with the Helsinki Declaration.
Materials
The IGF2BP3 siRNA (Sense: 5′- GCCGUCUCAUUGGUAAAGATT -3′, antisense:5′- UCUUUACCAAUGAGACGGCTT -3′) was synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The lentivirus expressing surfactant protein B (LV-Sftpb) was prepared by Shanghai Genechem Biotechnology Co., Ltd (Shanghai, China). OST, dodecylamine, and bisphenol A glycerol diacrylate, 4- (2-aminomethyl) morpholine were bought from Sigma-Aldrich (St Louis, USA). Lecithins was purchased from AVT Pharmaceutical Tech Co. Ltd. (Shanghai, China). Protease Inhibitor Cocktail was from Sangon Biotech (Shanghai, China). Luria-Bertani medium broth medium was from Hopebiol (Qingdao, China). jetPEI was purchased from Polyplus-transfection (Illkirch, France). The RNA-Quick Purification Kit was from YiShan Biological Technology Co., Ltd (Shanghai, China). cDNA Synthesis Kit and miRNA 1st strand cDNA synthesis kit were from Accurate Biotechnology (Hunan) Co., Ltd (Changsha, China). SYBR Green PCR Master Mix was from Yeasen Biotechnology (Shanghai, China). The BCA assay kit was from Absin (Shanghai, China). Specific primers used for RT-PCR assays were from Generabio and GenePharma Co, Ltd. (Shanghai, China). Alexa Fluor 488-labeled Goat Anti-Rabbit IgG (H+L), Immunostaining Fix Solution, Immunostaining Blocking Buffer, and Enhanced Immunostaining Permeabilization Buffer were from Beyotime Biotechnology (Shanghai, China). FBS was from VivaCell (Shanghai, China) and Inner Mongolia Opcel Biotechnology Co., Ltd, (Beijing, China). Penicillin-streptomycin (PS) solution, trypsin-EDTA solution, red blood cell lysis solution, DAPI, DNase, PicoGreen dsDNA quantitative detection kit, and PBS were obtained from Solarbio Science & Technology Co., Ltd. (Beijing, China). The Cell Counting Kit-8 (CCK-8) was purchased from APExBIO technology (Houston, USA). The Annexin V-FITC/PI apoptosis detection kit was purchased from Vazyme Biotech Co., Ltd. and Jiangsu KeyGEN BioTECH (Nanjing, China). Luciferin potassium salt, and D-PBS, were purchased from Dalian Meilun Biotechnology Co., Ltd. (Dalian, China). Anti-GLUT1 (ab115730), anti-HK2 (ab209847), anti-c-Myc (ab32072), anti CD63 (ab217345), anti-ALIX (ab275377), anti-HSP70 (ab181606), anti-phosphorylated EGFR (ab40815) antibodies were purchased from Abcam (Toronto, Canada). Anti-IGF2BP3(sc-390639), and anti-SP-B(sc-133143) antibodies were from Santa Cruz (California, US). Anti-Flag(F1804) was bought from Sigma-Aldrich (St Louis, USA) and the anti-EGFR (18986-1-AP) antibody was from Proteintech (Wuhan, China). Lung dissociation kits were obtained from Miltenyi Biotec. (Bergisch Gladbach, Germany). Percoll was purchased from GE Health (Uppsala, Sweden). Intracellular Fixation & Permeabilization Buffer Set and Foxp3/Transcription Factor Staining Buffer Set were from eBioscience (San Diego, USA). Fluorochrome-coupled antibodies used for flow analysis were from BioLegend, Inc. (San Diego, USA), eBioscience (San Diego, USA), and BD (Franklin Lakes, USA). ELISA kits for IFN-γ, TGF-β, IL-10, and IL-6 were purchased from BOSTER Biological Technology Co., ltd (Wuhan, China), and ELISA kits for TNF-α were purchased from Lianke Bio (Hangzhou, China). Other reagents and solvents were purchased from Sigma-Aldrich (St Louis, USA) or Merk (Darmstadt, Germany).
Mice
C57BL/6J mice (20 ± 2 g, 5–6 weeks, male) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Animals were housed at 20–22 °C with 12 h:12 h light: dark cycles at 50–60% humidity. For the orthotopic NSCLC model, LLC-Luc cells were digested, centrifuged, resuspended in cooled PBS, and mixed with Matrigel. A small incision was made in the epidermis and muscle on the side of the thorax. A total of 5 × 10 5 LLC-Luc cells were intrathoracic injected into the lung using a syringe at the 8th and 9th intercostal spaces22. The incision was sealed using veterinary skin adhesive and all operations were performed in a sterile environment52,53.
To establish the brain metastasis model of lung cancer, 4 × 10⁴ LLC cells were implanted into the striatum (coordinates: X = −2, Y = −1, Z = −3) at a rate of 1 μL min−1 using a stereotaxic apparatus. Mice were used for further studies from 7 days post-implantation.
For in vivo nebulized delivery in the anti-tumor efficacy experiments, five mice per group were placed in one chamber for whole-body exposure, and the dose was set at 5 mg kg−1. The nebulization system was connected to the nebulizer, with an oxygen flow rate of 15 standard cubic feet per hour directing aerosol into the chamber. Nebulization continued until no aerosol was observed in the chamber for 10 min. For the biodistribution assay of MPDOLs, mice were placed in the 3D-printing nose-only exposure restraints. The nebulization rate did not exceed 100 μL min−1, with each mouse exposed for 9–11 min20.
Cells
Murine LLC cells were obtained from Procell Life Science and Technology Co., Ltd. (CL-0140, Wuhan, China). The LLC-Luc cell line was purchased from Tongpai Biotechnology Co., Ltd. (FS-LLC-luc, Shanghai, China). MSCs were sourced from Immocell Biotechnology Co., Ltd. (Xiamen, China). The mouse brain microvascular endothelial cell line (bEnd.3) was a gift from the Department of Pharmacology, School of Pharmaceutical Sciences, Shandong University. LLC, LLC-Luc and bEnd.3 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and 1% PS, while the MSC cell was maintained in DMEM-F12 supplemented with 10% FBS and 1% PS. The cell lines were certified by the manufacturers and all cell lines used in this study were confirmed mycoplasma-negative by routinely examining. All cells were cultivated in an environment with a 5% CO2 atmosphere at 37 °C. Mycoplasma contamination testing for all cell lines was negative.
Bioinformatics analysis
The resources of m6A-related genes were from the study by Xu et al. on the molecular characterization of m6A regulators across 33 cancer types. STAR-counts data and corresponding clinical information for NSCLC tumors were obtained from the TCGA database, comparing the expression levels between healthy individuals and NSCLC patients. We then extracted data in TPM format and performed normalization using the log2(TPM+1) transformation. Statistical analysis was conducted using R software. Survival analysis for NSCLC patients with high or low IGF2BP3 expression was conducted using the Genotype-Tissue Expression dataset, as well as the GEPIA database.
Analysis of IGF2PB3 expression in mice and patients
Four pairs of NSCLC tissues and adjacent tissues were collected from patients at the Second Hospital of Shandong University for IGF2BP3 immunohistochemical staining.
C57BL/6J mice bearing orthotopic and brain metastasis lung cancer were treated with OST (5 mg kg−1) or PBS via intragastric administration every two days for 15 times. Lungs, brains, and tumors were collected after treatment and subjected to IGF2BP3 evaluation.
To evaluate the effects of OST on IGF2BP3 mRNA and protein expression, LLC cells were treated with OST (1 μM) or IGF2BP3 siRNA(60 nM) for 72 h. mRNA expression and protein levels were subsequently analyzed using qRT-PCR and western blot.
Construction of DNA plasmid
The DNA plasmid was designed based on the GV658 vector of Shanghai Genechem. It was constructed by integrating a guiding RVG tag with the IGF2BP3 siRNA sequence and inserting this sequence into a 166-bp pre-miR-155 backbone at the extra-exosomal N-terminus of CD6318. The plasmid composition is detailed in Supplementary Table 1. The plasmids were transformed into E. coli and cultured in Luria-Bertani medium. After shaking for 14 h, E. coli was harvested for purifying plasmid DNA following the protocol of the Omega Plasmid DNA Mini or Maxi Kit. The purified plasmids were sequenced to ensure the gene sequence was correct. DNA concentration and quality were assessed using a Nanodrop 2000c Spectrophotometer (Thermo Fisher Scientific, USA).
Construction of genetically engineered MSCs
Recombinant lentiviruses carrying mouse SP-B plasmids and a puromycin resistance gene were engineered by Genechem. The sequence of SP-B Plasmids is shown in Supplementary Table 2. MSCs were transduced with the purified lentivirus at a multiplicity of infection of 20 and selected using 5 μg/mL puromycin to establish stable cell lines expressing SP-B. Successful transfection was confirmed by immunofluorescence microscopy (GFP-tag signal) and western blot analysis (SP-B protein expression). The SP-B-expressing MSCs were cultured and harvested to isolate cell membranes using hypotonic lysis and centrifugation (700 × g, 10 min; 14,000 × g, 30 min)32.
Preparation of MPDOLs
PβAE was synthesized via the Michael addition reaction of diethyl acetylene dicarboxylate and an amine25. The chemical structure of PβAE was confirmed on a 500-MHz NMR spectrometer (Bruker, USA). Plasmid DNA was diluted to a final concentration of 50 μg mL−1 in acetate buffer and mixed with an equal volume of PβAE solution by vortexing for 15 min to form PD nanoparticles. To optimize the formulation, PD nanoparticles were prepared at varying PβAE-to-DNA weight ratios, and the optimal ratio was determined by agarose gel electrophoresis to assess DNA condensation(110 V, 30 min). The resulting mixture was then dialyzed using Pur-A-Lyzer Mini Dialysis tubes or dialysis cassette for 2 h at 4 °C. The PD nanoparticles were subsequently used to transfect LLC cells and GFP fluorescence was assessed via flow cytometry.
The liposome shell was prepared by the thin film dispersion method to encapsulate the OST. Lecithins, cholesterol, and OST were dissolved in chloroform at a mass ratio of 8:1:1. The solvent was evaporated to form a lipid film at 25 °C, which was hydrated with PBS to obtain OLs.
To assemble MPDOLs, PD nanoparticles were added to an equal volume of the OL solution and incubated at 40 °C for 30 min. Subsequently, MPDOLs (5 mg mL−1) were mixed with SP-B+ MSC membrane solution (1 mg mL−1) under ultrasonication for 1 min at 25 °C and extruded through 200-nm filters 12 times to ensure uniformity. The mixture was then dialyzed at 4 °C for purification and all the preparation processes were done in laminar flow. The nanomedicine was concentrated through the Ultra Centrifugal Filter (50 kDa MWCO).
Characterization of MPDOLs
The particle size and zeta potential of different nanoparticles were characterized via the Zetasizer Nano ZS system (Malvern, U.K.), and the morphology was visualized by transmission electron microscopy (TEM, JEM 1200EX, JEOL, Japan). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed to evaluate protein composition, while FRET experiments were conducted to investigate membrane fusion between PDOLs and MSC membranes. In the FRET experiment, cell membranes were labeled with Dil, and PDOLs were prepared with DiO in the dark. Emission spectra were recorded using a multimode microplate reader at an excitation wavelength of 440 nm21,30.
The drug loading of OST was determined by HPLC (SHIMADZU, Japan) on a reversed-phase C18 column. The mobile phase consisted of a 65% aqueous solution (5 mmol L−1 sodium dihydrogen phosphate, 0.3% triethylamine, pH = 2.8) and 35% acetonitrile. The encapsulation efficiency of plasmid DNA was analyzed by a PicoGreen dsDNA kit. Serum stability was evaluated by incubating MPDOLs with 10% FBS and DNase I at 37 °C, followed by electrophoresis on a 1% agarose gel at 110 V for 30 min with YeaRed staining.
To analyze the release behavior of OST, MPDOLs were placed in dialysis bags and put in 10 mL of PBS containing 0.5% of Tween 80 in pH 5.0 or 7.4, with continuous shaking at 100 rpm. As a control, OST was also loaded into dialysis bags. In addition, particle size, zeta potential, encapsulation efficiency, and morphology of MPDOLs were recharacterized after atomization with a commercially available portable nebulizer to evaluate the impact of shear force on liposome structure.
Cellular uptake and internalization
For the cellular uptake assay, LLC cells were seeded at a density of 1 × 105 cells per well in a 12-well plate. After overnight incubation, C6-loaded MPDOLs were added and incubated for 4, 8, and 12 h. The cells were washed with PBS, fixed, and stained with DAPI for confocal microscopy (Dragonfly 200, Andor Technology, US). The cells were also trypsinized, washed, and analyzed by flow cytometry (Accuri C6 Plus, BD Biosciences, USA).
To investigate the endocytosis pathway, LLC cells were pre-incubated with different inhibitors (CPZ, GEN, AMI, and M-βCD) for 30 min. The medium was then replaced with DMEM containing C6-loaded MPDOLs, and cells were collected after 1 h for flow cytometry analysis. LLC cells seeded in glass bottom dishes were treated by MPDOLs with DNA plasmids carrying GFP-tag for 4, 8, and 12 h. After washing with PBS, the cells were sequentially stained with Lysotracker and DAPI to visualize lysosomal escape. Additionally, LLC cells treated with MPDOLs for 12 h were stained with acridine orange hydrochloride to assess lysosomal damage.
Cell cytotoxicity, proliferation, and apoptosis assay
LLC cells were subjected to a gradually increasing concentration of OST (from 0.1 to 20 µM) to allow for the development of acquired resistance. Cells with acquired resistance OST(LLC-OR) were developed after 11 weeks in drug media.
Cell cytotoxicity was evaluated using the CCK-8 assay. LLC cells were seeded at a density of 8 × 103 cells per well in a 96-well plate and incubated overnight. The cells were then treated with various formulations (MPDOLs, PDs, OST), all containing equivalent amounts of OST (0.1–10 μM) or DNA plasmids (0.05–5 μg mL−1), for 48 h. Following the addition of 10 µL of CCK-8 reagent to each well, the cells were incubated for 2 h, and the optical density was measured using a microplate reader (Infinite F50, Tecan, Sweden) at 450 nm.
For apoptosis analysis, LLC cells were seeded and treated with different formulations for 48 h. The apoptosis rate was assessed using the Annexin V-FITC Apoptosis Detection Kit, according to the manufacturer’s instructions.
Biodistribution of MPDOLs
Free DiR, DiR-labeled PDOLs, and DiR-labeled MPDOLs (10 μg of DiR per mouse) were administrated to C57BL/6J mice bearing orthotopic and brain metastasis lung cancer via the 3D-printing nose-only exposure restraints connected to the nebulizer. Fluorescence imaging was performed at 1, 4, 8, 12, and 24 h using the IVIS Spectrum small animal imaging system (PerkinElmer, USA). After 24 h, mice were sacrificed, and major organs were collected to quantify signals. Lungs were frozen, sectioned, and stained with DAPI to visualize the liposome localization in the whole tissues. To investigate the main cell populations that internalize MPDOLs, flow cytometry was used to verify MPDOL uptake in vivo12.
Exosome isolation and characterization
Exosomes were isolated from cell culture medium and mouse plasma. After being transfected with MPDOLs for 24 h, LLC cells were cultured in a serum-free medium, and the culture medium was then collected. After inhalation of different nanomedicines for 3 days, mice were sacrificed to collect plasma samples. The collected samples were sequentially centrifuged (10 min at 400 × g, 10 min at 2000 × g, 60 min at 10,000 × g) and filtered through 220 nm filter membranes. Exosomes were then isolated twice by ultracentrifugation (1,10,000 × g, 70 min) using the Beckman Optimal XPN-100 (USA) or EXODUS system (Huixinbio, China). For plasma exosome isolation, 1 mL of plasma was diluted with 20–30 mL PBS, ultracentrifuged, and resuspended in 100 μL PBS to obtain exosomes. For cell culture supernatant, 30 mL of cell culture medium was ultracentrifuged and resuspended in 30 μL of PBS. Meanwhile, we used the Flag epitope tag to label the CD63-RVG during the construction of the plasmid to realize the visualization of the RVG tag. Flag-EXOs(si) were produced via a similar method.
The size and concentration of exosomes resuspended in PBS were characterized using the nanoparticle tracking analyzer (ZetaView, Particle Metrix, Germany), and the morphology was visualized after negative staining with 1% uranyl acetate solution. For protein analysis, exosomes resuspended in RIPA lysis buffer were subjected to western blotting to check the presence of the exosome markers CD63, HSP70, ALIX, or the Flag expression. The exosomes resuspended in TRIzol reagent were used to isolate total RNA and analyze the amounts of siRNA using the stem-loop RT-PCR method.
The biological effect of RVG-EXOs(si)
CCK-8 was used to evaluate the cytotoxicity of the isolated RVG-EXOs(si)54. To evaluate the capacity of exosomes to penetrate the BBB, 1.5 × 105 bEnd.3 cells were seeded into the upper chambers of 24-well trans-well inserts (0.4 μm pore size) and cultured for 7 days until the trans-endothelial electrical resistance (TEER) exceeded 200 Ω·cm². Subsequently, LLC cells were seeded into the lower chamber, and a new medium containing Dil-labeled exosomes was added to the upper chamber37. After 12 h, the inverted microscope (Ti2-U, Nikon, Japan) and microplate reader (Infinite F50, Tecan, Sweden) were used to assess the fluorescence intensity in LLC cells to evaluate exosome penetration across the TEER.
The brain-targeting ability of RVG-EXOs(si) in vivo was assessed using an IVIS imaging system. Isolated RVG-EXOs(si) were sterilized, quantified, labeled by DiD, and diluted in PBS before tail vein injection. After 12 h, the mice were euthanized, and major organs were harvested for imaging. Brain tissues were then preserved in a sucrose solution, sectioned, and stained to evaluate exosome localization.
Anti-tumor efficacy in NSCLC orthotopic tumor model and brain metastasis model
Tumor-bearing mice were randomly divided into six groups. Five groups received inhalation treatments with MPDOLs, PDOLs, OLs, PDs, or PBS (the dose of OST is 5 mg/kg) every four days for a total of five administrations, as outlined in Fig. 6a. The sixth group received 5 mg kg−1 OST via intragastric administration on the same schedule. The weights and survival time of mice were recorded every three days, while tumor progression was monitored weekly using bioluminescence imaging. Micro-CT scanning (GX2, PerkinElmer Quantum, Japan) was performed to evaluate mean lung density and tumor development. Following imaging, eight mice from each group were euthanized to collect the important tissues and blood samples. Ex vivo luciferase imaging of lungs and brains, as well as measurements of lung wet weights, were conducted for quantitative assessment. Serum biochemical indicators were analyzed using commercial ALT, AST, and BUN kits to evaluate the liver and kidney functions. Fixed samples were for HE and immunohistochemical staining, and the other lung tumors were collected for flow cytometry analysis.
Flow cytometry
Isolated lungs were dissociated into single cells using the Lung dissociation kit. The cell suspensions were passed through a 70 μm filter, purified with Percoll, and lysed using red blood cell lysis buffer. Samples were blocked with anti-mouse CD16/32 for 30 min before staining. For flow cytometric analysis, two staining panels were prepared. One panel included Fixable Viability Stain 700 (FVS-700), PerCP-Cy5.5 anti-CD45, BV421 anti-CD3, FITC anti-CD4, APC anti-CD8a, PE-CF594 anti-CD25, and APC-CY7 anti-NK1.1. Cells were permeabilized by the Foxp3/Transcription Factor Staining Buffer Set before staining for anti-Foxp3 and anti-IFN-γ. The second panel included live/dead staining, anti-CD45-Percp-Cy5.5, anti-CD11b-FITC, and anti-F4/80-PE, followed by fixation, permeabilization, and intracellular staining for BV421 anti-CD206 and APC anti-iNOS. After staining, the cells were washed and analyzed via a flow cytometer (FACS Celesta, BD Biosciences). Single-stained controls, unlabeled cells, and fluorescence-minus-one controls were employed for gating and compensation. Gating strategies are shown in Supplementary Figs. 24 and 25. Data analysis was performed using FlowJo software.
Histopathology and immunohistochemistry
Excised lungs and brains from mice administrated with dye-labeled preparations or exosomes were frozen, sectioned, and stained under dark conditions. Other organs were fixed in 4% paraformaldehyde, embedded in paraffin, sectioned using a microtome, and stained for HE pathological analysis.
Excised lung tumors and brain tumors were analyzed for IGF2BP3 expression via immunohistochemistry. Tumor cell proliferation was evaluated by immunohistochemical staining for Ki67 on 5 μm paraffin-embedded sections. Additionally, IGF2BP3 expression was analyzed in tumor tissues, lungs, and brains. Quantification of histological and immunohistochemical results was performed by the experienced pathologist using Image J.
Cytokine quantification
Blood samples were collected to isolate the serum and mice were euthanized to harvest important tissues. The collected blood was placed in sterile Eppendorf tubes at 4 °C for at least 1 h, followed by centrifugation at 1000 × g for 15 min. The upper yellow serum layer was carefully transferred to a new tube and centrifuged again to ensure purity. ELISA kits were used to determine the level of IFN-γ, TNF-α, TGF-β, IL-6, and IL-10 according to the manufacturer’s instructions.
SDS-PAGE and western blotting
Cells treated with nanoparticles or exosomes were lysed in a cooled RIPA buffer. After centrifugation, the supernatants were mixed with protein loading buffer and heated to 95–100 °C for 5–10 min. 10 or 20 μg of protein sample was loaded into acrylamide gels for electrophoretic separation and transferred onto polyvinylidene fluoride membranes. The membrane was dyed with Coomassie blue G-250 and then decolorated overnight for visualization of total protein. For specific proteins, the membranes were then blocked with 5% (w/v) skim milk at room temperature for 2 h, followed by overnight incubation at 4 °C with primary antibodies. Membranes were then incubated with secondary antibodies at room temperature for 1 h. The chemiluminescent substrate was finally applied, and protein bands were visualized using a digital gel image analysis system (4800, Tanon, China). Uncropped and unprocessed scans of the important blots are shown in the Source Data file.
RNA isolation and qRT-PCR
Total RNA was isolated from cultured cells or mouse tissues using the RNA-Quick Purification Kit or TRIzol reagent. For IGF2BP3 mRNA expression analysis, total RNAs were reverse transcribed to first-strand complementary DNA (cDNA) using a cDNA synthesis kit. IGF2BP3 siRNA and miRNAs (reference gene miR-16) were quantified by miRNA 1st strand cDNA synthesis kit and a customized stem-loop RT primer. Following the manufacturer’s instructions, the SYBR RT-PCR kit was performed for the mean CT analysis on a LightCycler480 II instrument (Roche, Germany).
RNA sequencing
Total RNA was extracted from LLC cells treated with MPDOLs or PBS using a TRIzol reagent. The quality of the RNA was assessed using an Agilent 2100 Bioanalyzer. RNA libraries were prepared using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA). The index-coded samples were clustered on a cBot Cluster Generation System with the TruSeq PE Cluster Kit v3-cBot-HS (Illumina, USA). After cluster generation, libraries were sequenced using Illumina Novaseq technology to obtain 150 bp paired-end reads. Reads were aligned using HISAT2, and featureCounts v1.5.0-p3 was employed to count the number of reads mapped to each gene. Differential expression analysis was conducted using the DESeq2 R package (version 1.16.1). GO and KEGG pathway enrichment analyses were performed on differentially expressed gene sets using the clusterProfiler package. PPI network analysis was performed using the STRING database, and the interaction data were imported into Cytoscape software to identify central hub genes by intersecting the top 10 genes from five cytoHubba algorithms. Library preparation and sequencing were performed by Genechem.
Molecular mechanism study of the treatment in vitro
LLC cells were seeded on cover slides, and treated with PBS, OST, PDs, or MPDOLs for 48 h (corresponding concentration of 2 μM OST). Then, they were fixed, permeabilized, and stained with specific antibodies. The antibodies for immunofluorescence included anti-GLUT1, anti-HK2, and Alexa Fluor 488 anti-rabbit IgG. Nuclei were stained with DAPI. Images were acquired using a confocal microscope (LSM-980, Zeiss, Germany). Glucose uptake and lactate production are key indicators of the Warburg effect. They were quantified using a glucose uptake assay kit and a lactate assay kit, respectively, following the manufacturer’s instructions.
Figures and artwork
Some graphic elements (including lung, MPDOL, red cell, tumor cell, subcellular structures, exosomes, and brain in Fig. 1; Fig. 3a; Fig. 4a; exosomes and tumor cells in Fig. 5h; lung and brain in Fig. 6a) were created using Cinema 4D. And other graphical elements were created using PowerPoint.
Statistical and Reproducibility
Biological replicates were employed in all studies unless otherwise specified. Representative images in Figs. 2g, 3k, and 6i were selected from three mice. Representative images in Figs. 3c, g, 4e, f, and 8h, i were obtained from three independently prepared nanoparticle samples or agents per group, which were analyzed simultaneously. Figures 3b, d, e, 5a–d were selected from three independent experiments.
The statistical analyses were carried out using GraphPad Prism 9.0 software. Data are presented as mean ± SD. The two groups were compared using the student’s t-test (two-tailed unpaired). For multiple group comparisons, one-way analysis of variance (ANOVA) followed by Tukey’s or Dunnett’s post hoc tests were used. Mice weights were analyzed using the two-way ANOVA and survival data were compared by log-rank (Mantel-Cox) test with the PBS group. A p-value of <0.05 was considered statistically significant. No statistical method was used to predetermine the sample size. No data were excluded from the analyses. All experiments were conducted in a randomized manner, and investigators were blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The 1H NMR spectroscopy data of PβAE are provided in Supplementary Fig. 4 and the raw data is included in the source data file. The RNA sequencing data generated in this study have been deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE289979) under accession number GSE289979. All other data supporting the findings of this study are available within the article, supplementary information, or source data files. Source data are provided with this paper.
References
Rahman, P. et al. Clinical and genomic predictors of brain metastases (BM) in non-small cell lung cancer (NSCLC): an AACR Project GENIE analysis. J. Clin. Oncol. 39, 2032 (2021).
Peters, S., Paz-Ares, L., Herbst, R. S. & Reck, M. Addressing CPI resistance in NSCLC: targeting TAM receptors to modulate the tumor microenvironment and future prospects. J. Immunother. Cancer 10, e004863 (2022).
Huang, J. et al. Targeted drug/gene/photodynamic therapy via a stimuli-responsive dendritic-polymer-based nanococktail for treatment of EGFR-TKI-resistant non-small-cell lung cancer. Adv. Mater. 34, e2201516 (2022).
Passaro, A., Jänne, P. A., Mok, T. & Peters, S. Overcoming therapy resistance in EGFR-mutant lung cancer. Nat. Cancer 2, 377–391 (2021).
Robichaux, J. P. et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature 2, 377–391 (2021).
Al Bakir, M. et al. The evolution of non-small cell lung cancer metastases in TRACERx. Nature 616, 534–542 (2023).
Uddin, M. B., Wang, Z. & Yang, C. The m6A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol. Cancer 20, 61 (2021).
Zhao, W. et al. Epigenetic regulation of m6A modifications in human cancer. Mol. Ther. Nucleic Acids 19, 405–412 (2020).
Huang, H. et al. Recognition of RNA N 6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 22, 1288 (2020).
Wan, W. et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N6-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol. Cancer 21, 60 (2022).
Yang, X. et al. m6A-dependent modulation via IGF2BP3/MCM5/Notch axis promotes partial EMT and LUAD metastasis. Adv. Sci. 10, e2206744 (2023).
Jiang, A. Y. et al. Combinatorial development of nebulized mRNA delivery formulations for the lungs. Nat. Nanotechnol. 19, 364–375 (2023).
Lin, Z. et al. Metabolic reprogramming driven by IGF2BP3 promotes acquired resistance to EGFR inhibitors in non–small cell lung cancer. Cancer Res 83, 2187–2207 (2023).
Salazar-Puerta, A. I. et al. Engineered extracellular vesicles derived from dermal fibroblasts attenuate inflammation in a murine model of acute lung injury. Adv. Mater. 35, e2210579 (2023).
You, Q. et al. m6A reader YTHDF1-targeting engineered small extracellular vesicles for gastric cancer therapy via epigenetic and immune regulation. Adv. Mater. 35, e2204910 (2023).
Liu, M., Hu, S., Yan, N., Popowski, K. D. & Cheng, K. Inhalable extracellular vesicle delivery of IL-12 mRNA to treat lung cancer and promote systemic immunity. Nat. Nanotechnol. 19, 565–575 (2024).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020).
Fu, Z. et al. In vivo self-assembled small RNAs as a new generation of RNAi therapeutics. Cell Res. 31, 631–648 (2021).
Meng, Q. F. et al. Inhalation delivery of dexamethasone with iSEND nanoparticles attenuates the COVID-19 cytokine storm in mice and nonhuman primates. Sci. Adv. 9, eadg3277 (2023).
Rotolo, L. et al. Species-agnostic polymeric formulations for inhalable messenger RNA delivery to the lung. Nat. Mater. 22, 369–379 (2023).
Chi, S. et al. Biomimetic nanocomposites camouflaged with hybrid cell membranes for accurate therapy of early-stage glioma. Angew. Chem. Int. Ed. 62, e202304419 (2023).
Wang, W. et al. Inhalable biomimetic protein corona-mediated nanoreactor for self-amplified lung adenocarcinoma ferroptosis therapy. ACS Nano 16, 8370–8387 (2022).
Herman, L., De Smedt, S. C. & Raemdonck, K. Pulmonary surfactant as a versatile biomaterial to fight COVID-19. J. Control. Release 342, 170–188 (2022).
Guagliardo, R., Pérez-Gil, J., De Smedt, S. & Raemdonck, K. Pulmonary surfactant and drug delivery: focusing on the role of surfactant proteins. J. Control. Release 291, 116–126 (2018).
Yan, X., Pan, Q., Xin, H., Chen, Y. & Ping, Y. Genome-editing prodrug: targeted delivery and conditional stabilization of CRISPR-Cas9 for precision therapy of inflammatory disease. Sci. Adv. 7, eabj0624 (2021).
Yuan, S. et al. Transforming cancer-associated fibroblast barrier into drug depots to boost chemo-immunotherapy in “shooting fish in a barrel” pattern. ACS Nano 17, 13611–13626 (2023).
Balyasnikova, I. V. et al. Indocarbocyanine nanoparticles extravasate and distribute better than liposomes in brain tumors. J. Control. Release 349, 413–424 (2022).
Garcia-Martin, R. et al. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 601, 446–451 (2022).
Lokugamage, M. P. et al. Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat. Biomed. Eng. 5, 1059–1068 (2021).
Nie, D. et al. Cancer-cell-membrane-coated nanoparticles with a yolk-shell structure augment cancer chemotherapy. Nano Lett. 20, 936–946 (2020).
Fu, X. et al. Precise design strategies of nanomedicine for improving cancer therapeutic efficacy using subcellular targeting. Signal Transduct. Target. Ther. 5, 262 (2020).
Wu, H. et al. Multifunctional biomimetic nanoplatform based on photodynamic therapy and DNA repair intervention for the synergistic treatment of breast cancer. Acta Biomater. 157, 551–565 (2023).
Smith, T. T. et al. In situ programming of leukaemia-specific t cells using synthetic DNA nanocarriers. Nat. Nanotechnol. 12, 813–820 (2017).
Pan, Z. et al. Cholesterol promotes EGFR-TKIs resistance in NSCLC by inducing EGFR/Src/Erk/SP1 signaling-mediated ERRα re-expression. Mol. Cancer 21, 77 (2022).
Gupta, D. et al. Amelioration of systemic inflammation via the display of two different decoy protein receptors on extracellular vesicles. Nat. Biomed. Eng. 5, 1084–1098 (2021).
Zhang, J. et al. Engineered neutrophil-derived exosome-like vesicles for targeted cancer therapy. Sci. Adv. 8, eabj8207 (2022).
Li, W. et al. BBB pathophysiology–independent delivery of siRNA in traumatic brain injury. Sci. Adv. 7, eabd6889 (2021).
Wang, R. et al. Bio-fabricated nanodrugs with chemo-immunotherapy to inhibit glioma proliferation and recurrence. J. Control. Release 354, 572–587 (2023).
Chenthamara, D. et al. Therapeutic efficacy of nanoparticles and routes of administration. Biomater. Res. 23, 20 (2019).
Elcheva, I. A. et al. IGF2BP family of RNA-binding proteins regulate innate and adaptive immune responses in cancer cells and tumor microenvironment. Front Immunol. 14, 1224516 (2023).
Chen, Y. et al. Targeting Xkr8 via nanoparticle-mediated in situ co-delivery of siRNA and chemotherapy drugs for cancer immunochemotherapy. Nat. Nanotechnol. 18, 193–204 (2023).
Jin, Y. et al. Targeting polarized phenotype of microglia via IL6/JAK2/STAT3 signaling to reduce NSCLC brain metastasis. Signal Transduct. Target Ther. 7, 52 (2022).
Pan, Z. et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer 21, 16 (2022).
Fu, X. et al. Combination of oxaliplatin and POM−1 by nanoliposomes to reprogram the tumor immune microenvironment. J. Control. Release 347, 1–13 (2022).
Propper, D. J. & Balkwill, F. R. Harnessing cytokines and chemokines for cancer therapy. Nat. Rev. Clin. Oncol. 19, 237–253 (2022).
Certo, M., Tsai, C. H., Pucino, V., Ho, P. C. & Mauro, C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 21, 151–161 (2021).
Zhang, Q. et al. EZH2/G9a interact to mediate drug resistance in non-small-cell lung cancer by regulating the SMAD4/ERK/c-Myc signaling axis. Cell Rep. 43, 113714 (2024).
Topper, M. J. et al. Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 171, 1284–1300 (2017).
Yue, S. W. et al. m6A-regulated tumor glycolysis: new advances in epigenetics and metabolism. Mol. Cancer 2, 137 (2023).
Sun, L. et al. Gold nanoparticles inhibit tumor growth via targeting the Warburg effect in a c-Myc-dependent way. Acta Biomater. 158, 583–598 (2023).
Rho, H., Terry, A. R., Chronis, C. & Hay, N. Hexokinase 2-mediated gene expression via histone lactylation is required for hepatic stellate cell activation and liver fibrosis. Cell Metab. 35, 1406–1423 (2023).
Li, H. Y. et al. The tumor microenvironment regulates sensitivity of murine lung tumors to PD-1/PD-L1 antibody blockade. Cancer Immunol. Res5, 767–777 (2017).
Fu, X. et al. Immunotherapeutic hydrogel for co-delivery of STAT3 siRNA liposomes and lidocaine hydrochloride for postoperative comprehensive management of NSCLC in a single application. Asian J. Pharm. Sci. 19, 100925 (2024).
Li, B., Hao, G., Sun, B., Gu, Z. & Xu, Z. P. Engineering a therapy-induced “immunogenic cancer cell death” amplifier to boost systemic tumor elimination. Adv. Funct. Mater. 30, 1909745 (2020).
Acknowledgements
The authors acknowledge the Pharmaceutical Biology Sharing Platform and Translational Medicine Core Facility of Shandong University. We also thank the Research Center for Basic Medical Science of Qilu Hospital affiliated with Shandong University for consultation and instrument availability that supported this work. This work was supported by the National Natural Science Foundation of China (grant number 22373059, G.L.) and the Natural Science Foundation of Shandong Province (grant number ZR2023MB082, G.L.).
Author information
Authors and Affiliations
Contributions
G.L. and X.F. conceived the study and designed the experiments. Y.S., X.Z., Z.T., and L.L. provided suggestions on methods and technical support for the project. X.F., H.W., Y.Z., X.C., and Z.T. performed the experiments. X.F., Y.L., X.W., and J.Z. analyzed and visualized the data. X.F. wrote the original draft. G.L., H.W., and S.Q. revised the manuscript. G.L. and H.Z. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks William Weiss, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fu, X., Shi, Y., Wu, H. et al. Inhalable liposomal delivery of osimertinib and DNA for treating primary and metastasis lung cancer. Nat Commun 16, 3336 (2025). https://doi.org/10.1038/s41467-025-58312-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58312-5
This article is cited by
-
Recent advances in nanomaterial-based precision medicine for orthotopic tumor therapy
Journal of Nanobiotechnology (2026)
-
IGF2BP3 in Multi-System Diseases: Molecular Mechanisms, Pathological Roles and Translational Insights
Cell Biochemistry and Biophysics (2026)
-
Inhalable Herbal Nano-Liposomes Co-Delivering Astragaloside IV and Polyphyllin VII Reprogram Immunosuppressive Microenvironment To Overcome Anti-PD-1 Resistance in NSCLC
Biological Procedures Online (2025)
-
Inhalable nanoparticle-based drug delivery system for non-small cell lung cancer therapy: promises and challenges
Saudi Pharmaceutical Journal (2025)