Abstract
N-terminal acetyltransferases including NAA10 catalyze N-terminal acetylation, an evolutionarily conserved co- and post-translational modification. However, little is known about the role of N-terminal acetylation in cardiac homeostasis. To gain insight into cardiac-dependent NAA10 function, we studied a previously unidentified NAA10 variant p.(Arg4Ser) segregating with QT-prolongation, cardiomyopathy, and developmental delay in a large kindred. Here, we show that the NAA10R4S variant reduced enzymatic activity, decreased NAA10-NAA15 complex formation, and destabilized the enzymatic complex N-terminal acetyltransferase A. In NAA10R4S/Y-induced pluripotent stem-cell-derived cardiomyocytes (iPSC-CMs), dysregulation of the late sodium and slow delayed rectifier potassium currents caused severe repolarization abnormalities, consistent with clinical QT prolongation. Engineered heart tissues generated from NAA10R4S/Y-iPSC-CMs had significantly decreased contractile force and sarcomeric disorganization, consistent with the pedigree’s cardiomyopathic phenotype. Proteomic studies revealed dysregulation of metabolic pathways and cardiac structural proteins. We identified small molecule and genetic therapies that normalized the phenotype of NAA10R4S/Y-iPSC-CMs. Our study defines the roles of N-terminal acetylation in cardiac regulation and delineates mechanisms underlying QT prolongation, arrhythmia, and cardiomyopathy caused by NAA10 dysfunction.
Similar content being viewed by others
Introduction
N-terminal acetyltransferases (NATs) catalyze protein N-terminal acetylation (Nt-acetylation), an evolutionarily conserved co-translational and post-translational modification that regulates protein degradation, protein-protein interactions, membrane targeting, and protein folding1,2. In up to 80% of mammalian proteins, following the initiation of translation, NATs irreversibly transfer an acetyl group from acetyl-CoA directly to the initiating methionine (iMet) of the nascent polypeptide or to the exposed second amino acid after iMet removal by methionine aminopeptidases. N-terminal acetyltransferase A (NatA), one of seven mammalian NATs (NatA, NatB, NatC, NatD, NatE, NatF, and NatH) with distinct substrate specificities2, preferentially acetylates G, S, A, T, V, or C residues exposed at protein N-termini after iMet removal3,4. By sequence analysis, NatA can modify up to 40% of expressed mammalian proteins, although there are few functionally validated targets. Despite the diversity of potentially affected signaling pathways, little is known about the effects of Nt-acetylation in the heart.
Originally discovered in yeast5,6, and conserved in humans7, NatA contains a catalytic subunit, NAA10, and a regulatory subunit, NAA15. Pathogenic variants in NAA10 cause NAA10-related syndrome, a rare multi-system disorder characterized by developmental delay, hypotonia, QT prolongation, arrhythmias, and increased mortality often before 1 year of age8. In addition to QT prolongation and sudden death, NAA10 variants are associated with hypertrophic cardiomyopathy and congenital heart defects, including atrial septal defects, ventricular septal defects, and tetralogy of Fallot8,9,10,11,12. Located on the X-chromosome, pathogenic NAA10 variants are inherited in an X-linked pattern, with severe clinical phenotypes manifesting predominantly in males13,14. Female carriers present with more variable manifestations that range from mild to severe developmental delay with or without cardiac involvement11,13,15. Patients with cardiac involvement more commonly have NAA10 variants within the N-terminally located NAA15 interaction or catalytic domains, whereas those with neurodevelopmental delays generally have variants widely distributed on NAA1016,17, suggesting the importance of NatA complex formation for cardiac homeostasis.
Recent large-scale exome sequencing projects of patients with congenital heart disease identified several inherited and de novo NAA15 variants18. Cardiomyocytes derived from human induced pluripotent stem cells (iPSC-CMs) with a single missense mutation (NAA15R276W/WT) or NAA15 haploinsufficiency (NAA15WT/-) demonstrated minor contractile defects under loaded conditions, whereas iPSCs lacking NAA15 had poor viability and did not readily differentiate into iPSC-CMs18. These data support the importance of NatA function in a broad range of cardiovascular diseases, but the underlying mechanisms and cardiac targets remain undefined.
Despite NAA10-related syndrome pointing to a key role of NAA10 and Nt-acetylation in human cardiac homeostasis, little is known about the underlying disease mechanisms. Here, we addressed this question by using iPSC disease modeling, bioengineering, cellular electrophysiology, optogenetics, and quantitative proteomics. We identify the cardiac-specific targets dysregulated by NAA10 dysfunction and, together with the fact that NAA15 variants cause cardiac phenotypes18, this study provides mechanistic insights into the roles of Nt-acetylation in cardiac homeostasis and disease.
Results
Identification of previously unidentified NAA10 variant
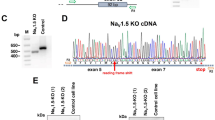
We evaluated a four-generation kindred with multiple members who had QT prolongation and sudden cardiac death (SCD) at a young age (Fig. 1a). Clinical genetic testing of the individuals with phenotypic QT prolongation for variants in congenital long QT syndrome (cLQTS)-related genes revealed a variant of unknown significance in KCNH2 p.R164H (ClinVar, VCV000067508.8) that did not segregate with the disease phenotype (Fig. 1a, Table 1). Expanded genetic testing for cardiomyopathies and skeletal myopathies in patients III:1 and III:8 revealed no known variants. However, in addition to QT prolongation and arrhythmia, male patients had significant neurodevelopmental delay and mild peripheral myopathy. These extra-cardiac phenotypes suggested a multi-system disorder. Extended genetic testing for variants associated with developmental delay identified a previously unidentified variant, NAA10 c.10 C > A, p.R4S, that co-segregated with clinical manifestations in an X-linked pattern in family members available for genetic testing (Fig. 1a, b). Male family members with the NAA10R4S/Y variant had significant QT prolongation and T-wave abnormalities (Patients III:1, III:8, and IV:4, Fig. 1c, Supplementary Fig. 1). The NAA10R4S/Y variant lies within the N-terminal NAA15 interaction domain necessary for the formation of the NatA complex19 (Fig. 1d). Consistent with severe QT prolongation, patient III:8 developed frequent episodes of ventricular tachycardia despite high-dose beta-blocker therapy (nadolol 2.8 mg/kg/day), necessitating insertion of an implantable cardiac defibrillator (ICD) (Fig. 1e). All his episodes of ventricular arrhythmia occurred during rest or with only minimal physical activity. Additionally, this patient developed progressive heart failure with an ejection fraction <25%, LV dilation, and symptoms consistent with class IV NYHA heart failure20(Fig. 1f). Despite aggressive medical and supportive therapies, he died from complications of heart failure at 17 years of age. Histopathology of the post-mortem LV samples demonstrated prominent interstitial fibrosis, and hypertrophic cardiomyocytes with bizarre hyperchromatic nuclei and highly irregular nuclear contours as in hypertrophic cardiomyocytes secondary to increased DNA ploidy21,22 (Fig. 1g). These clinical data support the identification of a previously unidentified NAA10 variant associated with severe QT prolongation, ventricular arrhythmias, and dilated cardiomyopathy.
a A four-generation family pedigree identified predominantly male patients with QT prolongation, developmental delay, and early mortality in a family referred for “gene-negative long QT syndrome”. Clinical sequencing revealed that the NAA10 p.R4S variant (red) to segregate with the clinical phenotype, whereas the variant of unknown significance in KCNH2 (p.R164H, shown in green) did not segregate with QT prolongation. In generation III, both male subjects (III:9, III:11) died as infants. A female (III:6) died at the age of 26 years from cardiac arrest prior to the availability of testing for NAA10. b Sanger sequencing of the proband (III:8). c ECG for patient III:8 with QT prolongation (QTc = 545 msec) and inverted T-waves. d A schematic of the exon structure of the NAA10 gene overlaid with the p.R4S variant and its relationship to the domain structure of the NAA10 protein. e Tracing from an implantable cardiac defibrillator showing short-long-short coupling prior to initiation of Torsades de Pointes (TdP). f M-mode echocardiography of patient III:8 showing a calculated ejection fraction (EF) of 22.3%. g Cardiac micrographs from patient III:8 post-mortem. The upper panel shows Masson’s Trichrome staining (scale bar: 100 µm), and the lower panel shows hematoxylin and eosin staining (scale bar: 50 µm) of the left ventricle. Arrowheads indicate abnormal appearing nuclei. Abbreviations for panel A: CM Cardiomyopathy, d. age of death, Myo Peripheral skeletal myopathy, NDD Neurodevelopmental delay, ↑QT QTc prolongation, SCD Sudden cardiac death, and VT Recurrent ventricular tachycardia.
The R4S mutation impairs NatA catalytic activity
The arginine at position 4 (R4) is highly conserved, suggesting a key role in NAA10 function (Fig. 2a). In silico analysis of existing high-resolution structures of the NAA10-NAA15 complex23,24 predicted that the R4S variant would impair local hydrogen bonding and ionic interactions destabilizing NAA10 (Fig. 2b). To measure the effect of NAA10-R4S on NAA10-NAA15 interaction, we performed co-immunoprecipitation assays in transfected HEK293 cells. NAA10-R4S co-immunoprecipitated 50% less NAA15 than wild-type NAA10 (Fig. 2c). Furthermore, in the HEK293 cell models, we detected that the protein level of NAA10-R4S expressed was lower than that of the wild-type protein (Supplementary Fig. 2a), leading us to suspect increased protein degradation. To assess protein half-life, we inhibited protein synthesis with cycloheximide and measured protein levels of expressed WT and mutant NAA10 proteins. NAA10-R4S displayed more rapid protein degradation than NAA10-WT when normalized to vinculin (Fig. 2d, e).
a NAA10 protein sequence alignment across multiple species shows an evolutionarily conserved arginine (arrowhead). b In silico prediction (DynaMut2) suggests that the R4S mutation affects NAA10 stability and potential interactions with NAA15. c HEK293 cells co-transfected with NAA10-FLAG and NAA15 were immunoprecipitated with anti-FLAG antibody. Binding capacity was quantified. (WT = 1.0 ± 0.0 vs R4S = 0.5 ± 0.1; independent transfection experiments, n = 4 (right panel); p = 0.0267; two-tailed paired t-test). d HEK293T cells expressing NAA10WT-FLAG and NAA10R4S-FLAG were treated with cycloheximide for 0, 2, 4, and 6 hours, and lysates were analyzed by western blotting with fluorescent secondary antibodies for vinculin and FLAG. MW: molecular weight. e Quantification of d, with normalization of NAA10/vinculin ratio to initial NAA10 protein levels at multiple time points: 2 h (WT: 0.98 ± 0.08 vs R4S: 0.43 ± 0.02, p < 0.0001), 4 h (WT: 0.65 ± 0.10 vs R4S: 0.17 ± 0.02, p < 0.0001), and 6 h (WT: 0.40 ± 0.03 vs R4S: 0.13 ± 0.01, p = 0.0204); independent transfection experiments, n = 3; p = 0.0001; two-way ANOVA). f Schematic of ThioGlo4 assay to quantify the enzymatic activity of NAA10. Created in BioRender. Yoshinaga, D. (2025) https://BioRender.com/z86v428. g Representative traces of ThioGlo4 reaction over time indicating NAA10 activity. NAA10-WT or NAA10-R4S enzyme concentrations were 100 nM. No enzyme was added to the control sample. h Initial reaction velocity was measured by ThioGlo4 assay at varying NAA10 concentrations, with significant differences at 100 nM (p = 0.0205) and 200 nM (p = 0.0020) (n = 6, duplicates from 3 independent experiments; p < 0.0001; mixed-effects analysis). i The total amount of CoA released 30 minutes after initiation of the enzymatic reaction was significantly reduced at 100 nM (p = 0.0205) and 200 nM (p = 0.0020) (n = 6, duplicates from 3 independent experiments; p < 0.0001; mixed-effects analysis). Data are presented as mean ± SEM. ****p < 0.0001, **p < 0.01, *p < 0.05. Source data are provided as a Source Data file.
As the catalytic subunit of NatA, NAA10 catalyzes the transfer of acetyl-CoA to protein N-termini, releasing free CoA1. Addition of the ThioGlo4 reagent reacts with the free CoA, forming a fluorescent adduct25 (Fig. 2f, Supplementary Fig. 2b). We exploited this reaction to measure the effects of the NAA10-R4S variant on NAA10 function by incubating purified NAA10-WT and NAA10-R4S with a synthetic peptide substrate containing the N-terminal sequence of EEEI, which specifically measures NAA10 function26,27. Monitoring ThioGlo4-CoA fluorescence over time showed that purified NAA10-R4S protein had significantly lower catalytic activity than NAA10-WT protein (Fig. 2g, h). Increasing protein concentration failed to overcome the enzymatic defect of NAA10-R4S (Fig. 2i). Collectively, these data show that the mutation NAA10-R4S destabilizes the NatA complex, increases NAA10 protein degradation, and directly impairs NAA10 enzymatic activity.
Prolonged repolarization in NAA10 R4S/Y-iPSC-CMs
To model the effects of NAA10-R4S and gain mechanistic insight into NAA10 function in cardiomyocytes, we created induced pluripotent stem cell lines (iPSCs) from patients III:1 and III:8 through somatic-cell reprogramming28. Isolated iPSC clones were positive for the NAA10R4S variant and had normal karyotypes and markers of pluripotency (Supplementary Fig. 3a-c). A single patient-derived clone from patient III:8 was selected for further studies and was designated as pNAA10R4S/Y. To control for genetic background, we also introduced the NAA10R4S variant into a male WT iPSC line by genome editing. This genome-edited isogenic line was designated as eNAA10R4S/Y (Supplementary Fig. 3d). We also sequenced predicted off-target genome editing sites and did not detect any additional mutations (Supplementary Fig. 3e). We differentiated NAA10 mutant and WT-iPSC-lines into iPSC-cardiomyocytes (CMs) using established small molecule differentiation protocols29. We confirmed robust cardiomyocyte differentiation of all three iPSC lines by cardiac troponin T (cTnT) immunostaining followed by flow cytometry (Supplementary Fig. 3f) and no significant difference in the percentage of atrial or ventricular subtypes (Supplementary Fig. 4). Western blotting of both pNAA10R4S/Y- and eNAA10R4S/Y-iPSC-CMs showed that the mutant lines expressed significantly less NAA10 protein compared to the control line (Fig. 3a, b), consistent with decreased stability of NAA10-R4S.
a Western blot analysis of whole-cell lysates from differentiated WT-, patient-derived (pNAA10R4S/Y)-, or genome-edited (eNAA10R4S/Y)-iPSC-CMs using NAA10, vinculin or cardiac troponin T antibodies. MW: molecular weight. b NAA10 protein levels, normalized to vinculin, were quantified: WT = 1.00 ± 0.04, pNAA10R4S/Y = 0.60 ± 0.06 (p < 0.0001), and eNAA10R4S/Y = 0.38 ± 0.06 (p < 0.0001); duplicates in 8 independent differentiation experiments; p < 0.0001; one-way ANOVA. c Differentiated iPSC-CMs were plated on multi-electrode arrays (MEAs, upper panel) and transduced with adenovirus for the channelrhodopsin ChR2-GFP (lower panel) for optical pacing. Scale bar: 750 μm d Representative MEA recordings of WT- (black), pNAA10R4S/Y- (red), and eNAA10R4S/Y- (blue) iPSC-CMs optically paced at 1 Hz with 488 nm light (blue triangles). Correspondingly colored dashed lines indicate field potential duration (FPD). e Quantified FPD paced at 1 Hz: WT = 156.9 ± 6.8 ms, pNAA10R4S/Y = 351.9 ± 8.1 ms (p < 0.0001), and eNAA10R4S/Y = 289.3 ± 4.6 ms (p < 0.0001); n = 22, 26, and 42, independent replicates; n = 6, 3, and 5, independent differentiation experiments; p < 0.0001; Kruskal-Wallis test. f Representative whole-cell recordings of single iPSC-CMs under current clamp conditions at 1 Hz pacing. g Action potential duration (APD) at 90% repolarization: WT = 290.1 ± 52.2 ms, pNAA10R4S/Y = 500.8 ± 35.9 ms (p = 0.0092), eNAA10R4S/Y = 464.8 ± 49.6 ms (p = 0.0282); n = 8, 11, and 12, independent experiments; n = 7, 6, and 7, independent differentiation experiments; p = 0.0128; one-way ANOVA. h The NAA10R4S variant did not affect peak action potential amplitude (APA) or mean diastolic potential (MDP): APA = 97.7 ± 3.9, 103.7 ± 3.8, 108.0 ± 2.9 mV; MDP = −67.0 ± 1.5, −64.0 ± 3.2, −62.7 ± 2.0 mV in WT-, pNAA10R4S/Y-, eNAA10R4S/Y-iPSC-CMs, respectively (p = 0.1538, 0.5086; one-way ANOVA). Sample sizes are annotated in each graph (same for panels g and h). Data are presented as mean ± SEM. ns = p > 0.1, *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001. Source data are provided as a Source Data file.
Prolonged repolarization is a hallmark of NAA10-related syndrome and was prominent in the affected males in the NAA10-R4S pedigree (Table 1). To determine if NAA10R4S/Y-iPSC-CMs recapitulate this phenotype, we used multi-electrode arrays (MEAs) to measure the field potential duration (FPD) of iPSC-CM monolayers, which correlates with the ECG-based QT interval30. Since FPD depends on iPSC-CM beat rates31, we optically paced iPSC-CMs at a fixed rate by transducing them with adenovirus expressing the channelrhodopsin ChR2 fused to GFP (Ad-ChR2-GFP, Fig. 3c) and stimulating with 488 nm light at 1 Hz (Fig. 3d, blue triangles). Under these conditions, pNAA10R4S/Y- and eNAA10R4S/Y-iPSC-CMs had dramatically prolonged FPDs compared to WT-iPSC-CMs (Fig. 3d, e).
To evaluate the effect of NAA10-R4S on cardiomyocyte repolarization more directly, we performed single-cell electrophysiology experiments on iPSC-CMs. After establishing a whole-cell current clamp configuration, we injected depolarizing currents at 1 Hz to elicit membrane action potentials (APs). iPSC-CM evoked APs had a characteristic ventricular-like morphology (Fig. 3f). The action potential duration (APD) measured at 90% and 50% of peak repolarization (APD90 and APD50) was significantly longer in NAA10-mutant lines compared to WT-iPSC-CMs, while the APD20 was not consistently prolonged in the mutant lines (Fig. 3f, g, and Supplementary Fig. 5a, b). There were no differences in action potential amplitude (APA) or maximum diastolic potential (MDP) between WT- and NAA10R4S/Y-iPSC-CMs (Fig. 3h), suggesting the observed APD prolongation did not arise from differences in differentiation or maturation32. In addition, NAA10R4S/Y-iPSC-CMs demonstrated increased upstroke velocity (dV/dt max) but without differences in input resistance or membrane capacitance (Supplementary Fig. 5c-e). To gain additional insight into the risk of arrhythmia imposed by NAA10 dysfunction, we electrically paced iPSC-CMs loaded with the membrane voltage dye FluoVolt at progressively shorter coupling intervals to determine possible changes in APD restitution (Supplementary Fig. 6a)33. Under these conditions, NAA10R4S/Y-iPSC-CMs demonstrated a steeper APD restitution curve and a longer pacing capture rate (Supplementary Fig. 6b, c) consistent with increased arrhythmogenic risk34,35. Finally, to confirm that loss of NAA10 function directly impacts cardiomyocyte repolarization, we transfected WT-iPSC-CMs with NAA10-specific siRNA and measured the FPD by MEA. Consistent with the NAA10R4S/Y-iPSC-CMs, NAA10 knock-down significantly prolonged the FPD in WT-iPSC-CMs (Supplementary Fig. 7). Collectively, these data demonstrate that the NAA10R4S variant causes severe repolarization abnormalities and increased markers of arrhythmia in iPSC-CMs.
NAA10 dysfunction dysregulates both sodium and potassium currents
To identify potential target proteins that might be affected by NAA10 dysregulation and cause repolarization abnormalities, we exploited the discovery that N-terminal protein sequence predicts the likelihood of Nt-acetylation. Therefore, we analyzed the N-terminal sequences of all proteins associated with cLQTS and their respective probabilities of modification by NatA (Table 2). This analysis suggested that multiple cardiac ion channels and related proteins known to influence cardiac repolarization could be NatA targets and, therefore, affected by NAA10 dysfunction. Among these candidates were genes that contribute to the voltage-gated sodium current (INa; SCN5A), the slow delayed rectifier potassium current (IKs; KCNQ1), the rapid delayed rectifier potassium current (IKr; KCNH2), and the inward calcium current (ICa-L; CACNA1C)36. We systematically evaluated the effect of NAA10R4S on these currents.
SCN5A gain-of-function variants cause LQT type III (LQT3)37 through at least two mechanisms: (1) alterations in voltage-dependent activation/inactivation and (2) increased late sodium current. To investigate the first mechanism, in the presence of the calcium channel blocker nifedipine (10 μM), depolarizing steps from a hyperpolarized holding potential of -100 mV in single iPSC-CMs38 evoked stereotypical rapidly activating and inactivating currents consistent with INa (Fig. 4a). Peak inward sodium currents were significantly increased in pNAA10R4S/Y- and eNAA10R4S/Y-iPSC-CMs by as much as 2.5-fold compared to WT-iPSC-CMs (Fig. 4a, b upper panel). While increased peak INa current density is associated with heart failure and other forms of NaV1.5-mediated cardiovascular disorders, it is not a recognized mechanism for cLQTS39. Instead, differential alterations in the voltage-dependent activation and inactivation of INa that increase the “window current” are associated with LQT3. While there was a small shift to more hyperpolarizing potentials in both the activation and inactivation curves in NAA10-mutant iPSC-CMs, there was no increase in the net activation probability (Fig. 4b lower panel, Supplementary Patch-clamp Data Table).
a Sodium current (INa) traces from iPSC-CMs. b Increased peak INa in NAA10-mutant iPSC-CMs (upper). Voltage-dependent activation and inactivation curves (lower). (n = 14, 8, and 10; independent differentiation experiments, n = 4, 3, 6 in WT-, pNAA10R4S/Y-, and eNAA10R4S/Y-iPSC-CMs; p = 0.0265; two-way repeated-measures ANOVA). c Late INa (INaL) traces elicited by tetrodotoxin (TTX) blockade (gray). d INaL normalized to peak INa: WT = 0.03 ± 0.005 %, vs pNAA10R4S/Y = 0.06 ± 0.01 % (p = 0.0268), and eNAA10R4S/Y = 0.07 ± 0.02 % (p = 0.0105); n = 13, 8, and 6; n = 7, 5, 3, independent differentiation experiments; one-way ANOVA. e Slow delayed rectifier potassium current (IKs) traces. f IKs current-voltage relationship (n = 5, 8, and 10; n = 5, 7, and 4, independent differentiation experiments in WT-, pNAA10R4S/Y-, and eNAA10R4S/Y-iPSC-CMs; p = 0.0009; two-way repeated-measures ANOVA). g Rapid delayed rectifier potassium current (IKr) traces. h IKr-tail current demonstrated no differences (n = 6, 7, and 7; n = 3, 3, and 3, independent differentiation experiments in WT-, pNAA10R4S/Y-, and eNAA10R4S/Y-iPSC-CMs; p = 0.4440; two-way repeated-measures ANOVA). Membrane capacitance: WT = 49.8 ± 6.8 pF, pNAA10R4S/Y = 46.8 ± 12.7 pF, eNAA10R4S/Y = 43.7 ± 6.6 pF. i. Immunoblot for membrane NaV1.5, confirming separation with Transferrin receptor (TFRC)-positive and β-actin-negative blot. j Membrane NaV1.5 normalized to TFRC: WT = 1.0 ± 0.0, pNAA10R4S/Y = 3.2 ± 1.1 (p = 0.0414), eNAA10R4S/Y = 2.3 ± 0.3 (p = 0.0164); n = 6; Kruskal-Wallis test. k Schematic of KCNQ1 with cMyc epitope (extracellular) and EGFP (C-terminus). Created in BioRender. Yoshinaga, D. (2025) https://BioRender.com/b59q327. l Flow cytometry analysis of EGFP and cMyc-Alexa 647 intensity in KCNQ1-transfected cells. m cMyc intensity normalized to EGFP: siNC = 1.0 ± 0.04 vs siNAA10 = 0.6 ± 0.01; biological replicates, n = 3; p = 0.0075; two-tailed unpaired t-test. Data are presented as mean ± SEM. ns = p > 0.1, *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001. Source data are provided as a Source Data file.
SCN5A variants that increase the late sodium current (INaL) also cause LQT3 and have also been associated with heart failure40. To investigate the effects of the NAA10R4S variant and associated NAA10 dysfunction on INaL, we stimulated iPSC-CMs with long-depolarizing steps (800 ms) at baseline and in the presence of 30 µM tetrodotoxin (TTX) to normalize for background membrane leak41. Consistent with a LQT3-type phenotype there was a > 2-fold increase in INaL in pNAA10R4S/Y- and eNAA10R4S/Y-iPSC-CMs as compared to WT-iPSC-CMs (Fig. 4c, d) suggesting a direct biophysical effect of NAA10 on NaV1.5 channel activity.
KCNQ1 variants that cause LQT type I (LQT1) reduce IKs, while KCNH2 variants that cause LQT type II (LQT2) reduce IKr. We isolated IKs by subtracting the current traces before and after administering the specific IKs blocker, HMR1556 (Fig. 4e). Activating voltage steps demonstrated a significant reduction in IKs current density in pNAA10R4S/Y- and eNAA10R4S/Y-iPSC-CMs within the physiologic depolarization range (Fig. 4e, f). Next, we elicited IKr by subtracting the current traces before and after administering specific IKr blocker, E4031. This maneuver did not reveal significant differences in IKr in NAA10R4S/Y-iPSC-CMs compared to WT-iPSC-CMs (Fig. 4g, h, and Supplementary Fig. 5f). These data suggest that NAA10R4S may affect KCNQ1 and reduce IKs repolarizing current.
To probe the underlying mechanism of how NAA dysfunction alters ion channel function, we first determined if the NAA10R4S variant affected target ion channel expression. Quantitative reverse transcription PCR (qPCR) of total RNA isolated from WT and NAA10-mutant iPSC-CMs did not show significant alterations in the levels of SCN5A or other ion channel transcripts (Supplementary Fig. 8). Changes in ion channel surface expression from alterations in membrane trafficking or channel retention at the cell surface are common regulatory mechanisms. Therefore, we performed surface biotinylation of WT and mutant iPSC-CMs followed by streptavidin-mediated isolation of membrane-localized proteins. Western blot analysis revealed significantly greater surface expression of NaV1.5 in pNAA10R4S/Y- and eNAA10R4S/Y-iPSC-CMs compared to WT cells (Fig. 4i, j). The increase in NaV1.5 surface localization at the plasmalemma was comparable in magnitude and direction to the changes in INa density in NAA10-mutant lines determined by INa whole-cell recordings (Fig. 4a,b).
In addition to changes in NaV1.5 membrane trafficking, we hypothesized that decreased surface expression of KCNQ1 was the mechanism for impaired IKs density in pNAA10R4S/Y- and eNAA10R4S/Y-iPSC-CMs (Fig. 4e, f). Indeed, disruption of ion channel trafficking of KCNQ1 is a well-recognized mechanism for LQT142. Therefore, to determine the impact of NAA10 dysfunction on KCNQ1 trafficking, we inserted a cMyc epitope within the first extracellular linker of the channel43 and fused EGFP at the C-terminus (Fig. 4k). We expressed the cMyc-KCNQ1-EGFP construct into HEK cells previously transfected with NAA10-targeted siRNA to knockdown NAA10 protein levels (Supplementary Fig. 9a, b). Live-cell staining with anti-cMyc antibody and subsequent flow-cytometry43,44 demonstrated a significant reduction in the cMyc/EGFP ratio, indicating diminished KCNQ1 surface expression (Fig. 4l, m, and Supplementary Fig. 9c, d). While limited by low expression levels of KCNQ1 in iPSC-CMs requiring overexpression in HEK cells, these data support the hypothesis that impaired trafficking of KCNQ1 caused by NAA10 is the mechanism for reduced IKs in NAA10 mutant iPSC-CMs.
Gain-of-function variants in CACNA1C are associated with LQTS type VIII (LQT8) by increasing the inward calcium (Ca2+) current ICa-L45. To determine if changes in ICa-L were underlying APD prolongation in NAA10 mutant iPSC-CMs, we recorded ICa-L by inhibiting NaV1.5 channels with an elevating holding potential (-40 mV) in the recording bath. With perforated patch recordings to prevent current rundown, we did not observe significant differences in total current density or L-type Ca2+ channel properties in NAA10R4S/Y-iPSC-CMs (Supplementary Fig. 5g, h, Supplementary Patch-clamp Data Table).
Taken together, these data indicate that QT prolongation and risk of arrhythmia in NAA10-related syndrome are caused by a combination of increased INaL and reduced IKs.
NAA10R4S/Y-iPSC-CMs have structural and contractile abnormalities
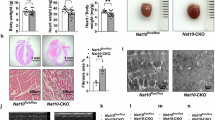
Patients with NAA10-related syndrome have variable heart disease phenotypes ranging from hypertrophic cardiomyopathy to congenital heart disease46. Our patient III:8 died from complications related to severe dilated cardiomyopathy (DCM), which has not been previously reported as associated with NAA10-related syndrome. To investigate possible cardiomyopathic effects of the NAA10 p.R4S variant and related NAA10 dysfunction, we analyzed the effect of NAA10 p.R4S on iPSC-CM structural assembly. We used micro-contact printing to deposit rectangular fibronectin islands with the 7:1 aspect ratio characteristic of adult human ventricular cardiomyocytes (Fig. 5a)47,48,49,50,51. Plating iPSC-CMs on these patterned substrates induces sarcomere alignment, structural integrity, and features of cellular maturity when compared to unpatterned cells (Fig. 5b). High-resolution confocal imaging of iPSC-CMs stained for sarcomeric alpha-actinin (SAA) and subjected to objective, computational image analysis50 demonstrated decreased sarcomeric packing density (SPD) and increased sarcomere length in NAA10R4S/Y-iPSC-CMs (Fig. 5c, d), consistent with a DCM-like phenotype51.
a Depiction of micro-contact printing with polydimethylsiloxane (PDMS) stamps of fibronectin for spatially restricted patterning of iPSC-CMs. Rectangle patterns have a length-to-width ratio of 7:1 (scale bar: 100 μm). b Confocal micrographs of WT-, pNAA10R4S/Y-, or eNAA10R4S/Y-iPSC-CMs, stained for sarcomeric alpha-actinin (SAA) as either unpatterned (left panels, scale bar: 20 μm) or patterned on micro-contact printed substrates (right panels, scale bar: 10 μm). c A customized Matlab program calculated sarcomeric packing density (SPD) from single micro-patterned iPSC-CMs. SPD reflects sarcomere expression and alignment in a certain area. Therefore, lower SPD values correspond to a more disordered sarcomeric structure. SPD was quantified as 0.44 ± 0.01 for WT, 0.37 ± 0.01 for pNAA10R4S/Y (p < 0.0001), and 0.30 ± 0.01 for eNAA10R4S/Y (p < 0.0001) (n = 153, 66, and 43, independent experiments; n = 3, 3, and 3, independent differentiation experiments; p = <0.0001; Kruskal-Wallis test). d Sarcomere length was measured in single micro-patterned iPSC-CMs and quantified as 1.65 ± 0.01 μm for WT, 1.83 ± 0.03 μm for pNAA10R4S/Y (p < 0.0001), and 1.83 ± 0.03 μm for eNAA10R4S/Y (p < 0.0001) (n = 152, 56, and 36, independent experiments; n = 3, 3, and 3, independent differentiation experiments; p < 0.0001; Kruskal-Wallis test). e Brightfield images of engineered heart tissues (EHTs) created from isogenic control (WT) and eNAA10R4S/Y-iPSC-CMs on PDMS “pillars” after transduction with ChR2-GFP adenovirus for optical pacing (scale bar: 1 mm). f Representative traces of developed contractile force of EHTs calculated from video data of PDMS pillar displacement in response to optical pacing with 488 nm light at 1 Hz. g Quantification of contractile force calculated over 10 beats (WT = 0.46 ± 0.04 mN vs eNAA10R4S/Y = 0.019 ± 0.03 mN; n = 17, and 15, independent experiments; n = 3, and 3, independent differentiation experiments; p < 0.0001; two-tailed unpaired t-test). Data are presented as mean ± SEM. The number of cells or tissues (n) is annotated on each graph. ****p < 0.0001. Source data are provided as a Source Data file.
To further investigate the effects of NAA10 dysfunction on the contractile force, we generated 3D-engineered heart tissues (EHTs) from WT and eNAA10R4S/Y-iPSC-CMs52. Cells embedded in extracellular matrix were molded around two silicone pillars and transduced with Ad-ChR2-GFP to facilitate optical pacing (Fig. 5e). After tissue formation and maturation, EHTs were optically paced at 1 Hz. The contractile force was measured based on pillar displacement (Fig. 5f). Despite long-term culture for up to 28 days, eNAA10R4S/Y-EHTs generated minimal force, which was significantly lower than WT-EHTs (Fig. 5f, g). These data collectively identify structural defects and a severe contractile defect that was not observed in a previously reported model of NatA dysfunction caused by an NAA15 mutant18.
Abnormal Ca2+ handling in NAA10R4S/Y-iPSC-CMs
Homeostasis of intracellular Ca2+ levels is critical for normal cardiomyocyte function. Indeed, excess diastolic Ca2+ is associated with impaired cardiomyocyte contraction and relaxation53. To determine if Ca2+ handling defects underlie impaired contractility in NAA10R4S/Y-iPSC-CMs, we used the ratiometric calcium indicator Fura-2 to record Ca2+ transients in patterned iPSC-CMs. With pacing at a physiological rate of 1 Hz (60 beats per minute), there was a significant increase in diastolic Ca2+ levels while there was no difference at slower rates (Fig. 6a, b). Further, while the Ca2+ transient magnitude did not differ among the cell lines, the Ca2+ transient relaxation coefficient, a measure of the rate of cytosolic Ca2+ clearance, was significantly elevated in the NAA10R4S/Y-iPSC-CMs compared to controls (Fig. 6c, d). While other measures of impaired Ca2+ clearance were present in NAA10R4S/Y-iPSC-CMs, including prolongation of peak to baseline time, parameters associated with Ca2+ influx were unchanged (Supplementary Fig. 10a-f). Collectively, these data suggested that intracellular Ca2+ clearance was significantly disrupted in NAA10R4S/Y-iPSC-CMs.
a Micro-patterned iPSC-CMs loaded with Fura-2 were electrically paced at 1 Hz. Diastolic Ca2+ levels are indicated by arrows between the baseline and dashed lines. b End-diastolic Ca2+ levels were measured at 0.5 Hz and 1 Hz pacing (0.5 Hz: 0.044 ± 0.022, 0.085 ± 0.014, 0.054 ± 0.009; 1 Hz: 0.038 ± 0.007, 0.229 ± 0.029, 0.176 ± 0.017 in WT-, pNAA10R4S/Y-, and eNAA10R4S/Y-iPSC-CMs). Significant differences were observed at 1 Hz for WT vs. pNAA10R4S/Y (p < 0.0001) and WT vs. eNAA10R4S/Y (p < 0.0001) (n = 3, 3, and 3, independent differentiation experiments; p < 0.0001; mixed-effects analysis). c The baseline to peak height of Ca2+ transients paced at 1 Hz (WT = 23.6 ± 2.3 % vs pNAA10R4S/Y = 29.0 ± 2.8 %, and eNAA10R4S/Y = 32.4 ± 3.1 %; n = 15, 18, and 26, independent experiments; n = 3, 3, and 3, independent differentiation experiments; p = 0.2913; Kruskal-Wallis test). d Ca2+ transient relaxation coefficient τ from the same samples in c: WT = 0.38 ± 0.05, pNAA10R4S/Y = 0.62 ± 0.03 (p < 0.0001), and eNAA10R4S/Y = 0.55 ± 0.02 (p = 0.0008) (p < 0.0001; one-way ANOVA). e Immunoblot of RyR2, SERCA2a (SERCA), phospholamban (PLN), NCX1, and vinculin. f Quantification of RyR2, SERCA, PLN, NCX1, and the SERCA/PLN ratio: RyR2 (1.0 ± 0.1, 6.3 ± 0.6, and 3.8 ± 0.9), SERCA (1.0 ± 0.1, 1.6 ± 0.2, and 1.4 ± 0.1), PLN (1.0 ± 0.1, 2.5 ± 0.2, and 2.0 ± 0.2), NCX1 (1.0 ± 0.1, 0.8 ± 0.2, and 0.7 ± 0.3), and SERCA/PLN (1.0 ± 0.1, 0.8 ± 0.1, and 0.7 ± 0.1) in WT-, pNAA10R4S/Y-, and eNAA10R4S/Y-iPSC-CMs; n = 3, 8, 8, 4 independent differentiation experiments for RyR2, SERCA, PLN, and NCX1; p = 0.0017, 0.0053, <0.0001, 0.0045, and 0.0029; Kruskal-Wallis test) Values were log2-transformed and visualized in a heatmap. Data are presented as mean ± SEM. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, ns = p > 0.05. Source data are provided as a Source Data file.
During each Ca2+ transient, Ca2+ is cleared from the cytosol by reuptake into the sarcoplasmic reticulum (SR) via the SR Ca-ATPase 2a (SERCA) and by Ca2+ efflux across the cell membrane via the sodium/calcium exchanger (NCX1)53. SERCA is inhibited by phospholamban (PLN) binding. To determine if SERCA or PLN levels were affected by NAA10 dysfunction, we performed western blot analysis of whole-cell lysates from WT-, pNAA10R4S/Y-, and eNAA10R4S/Y-iPSC-CMs. We observed a significant decrease in the SERCA/PLN ratio and NCX1 level (Fig. 6e, f).
Taken together, these data support the hypothesis that NAA10 dysfunction alters cardiomyocyte Ca2+ signaling by impairing Ca2+ reuptake and increasing diastolic Ca2+.
Rescue of NAA10 disease phenotypes
In our NAA10-related syndrome iPSC models, decreased NAA10 protein level and catalytic activity correlated with cardiomyocyte dysfunction, suggesting that restoration of NAA10 activity may reverse the disease phenotypes. To test this hypothesis, we generated adenovirus that expresses WT NAA10 along with the self-labeling protein HaloTag (Ad-NAA10, Fig. 7a). We transduced cultures of NAA10R4S/Y- iPSC-CMs with either Ad-NAA10 or control adenovirus expressing LacZ (Ad-LacZ) (Fig. 7b). Western blot analysis of iPSC-CM lysates 48 hours after transduction demonstrated robust expression over baseline levels (Fig. 7c). The endogenous protein migrated at a different size from exogenously expressed NAA10 (Fig. 7c, gray arrow).
a iPSC-CMs were transduced with adenoviruses encoding LacZ or NAA10-P2A-HaloTag. Created in BioRender. Yoshinaga, D. (2025) https://BioRender.com/p38t891. b Incubation with HaloTag dye indicates the expression of NAA10 (left lower) compared to LacZ (left upper). Scale bars: 100 μm. Halo-positive cells showed 84.3 ± 0.7 % of cells were transduced (n = 3) (right). c Western blot of whole-cell lysates from pNAA10R4S/Y- or eNAA10R4S/Y-transduced with Ad-LacZ or Ad-NAA10 and probed with anti-NAA10. The faint lower band represents native NAA10 expression (gray triangle). The intensity of NAA10 was quantified and normalized to vinculin (n = 1). MW: molecular weight. d Representative MEA recordings of iPSC-CM monolayers co-transduced with Ad-ChR2-GFP and either Ad-LacZ or Ad-NAA10, optically paced at 1 Hz (blue arrowheads). e Field potential duration (FPD) 48 hours post-transduction. NAA10 replacement significantly shortened FPD in pNAA10R4S/Y (299.8 ± 13.9 ms vs 255.4 ± 13.0 ms, p = 0.0015) and eNAA10R4S/Y (247.6 ± 6.4 vs 195.2 ± 5.7 ms, p = 0.0003), while it did not in WT (176.8 ± 13.0 ms vs 149.4 ± 10.3 ms, p = 0.0978). (three independent differentiation experiments; mixed-effects analysis). f MEA monolayers were measured at baseline and treated with the specific INaL blocker GS967. GS967 significantly shortened FPD in WT (167.7 ± 4.6 ms vs 154.1 ± 2.4 ms, p = 0.0379), pNAA10R4S/Y (352.2 ± 7.1 ms vs 272.5 ± 4.5 ms, p < 0.0001), and eNAA10R4S/Y (300.1 ± 6.6 ms vs 251.5 ± 5.6 ms, p < 0.0001). (three independent differentiation experiments; p = 0.0001; mixed-effects analysis). g Micro-patterned iPSC-CMs transduced with Ad-NAA10 were stained for sarcomeric alpha-actinin (SAA) to quantify changes in sarcomeric organization (scale bar: 10 μm). Automated analysis of single iPSC-CMs on micro-patterned substrates for h. sarcomeric packing density (Ad-NAA10(-) = 0.30 ± 0.01 vs Ad-NAA10(+) = 0.35 ± 0.02; three independent differentiation; p = 0.0734; two-tailed unpaired t-test) and i. Sarcomere length (Ad-NAA10(-) = 1.83 ± 0.03 vs Ad-NAA10(+) = 1.67 ± 0.04; three independent differentiation; p = 0.0063; two-tailed unpaired t-test). Data are presented as mean ± SEM. ****p < 0.0001, ***p < 0.001, **p < 0.01, ns = p > 0.1. Source data are provided as a Source Data file.
Next, we characterized the effect of Ad-NAA10 on NAA10-R4S mutant and wild-type iPSC-CMs. We transduced iPSC-CM monolayers plated on MEAs with either Ad-LacZ or Ad-NAA10. Cells were also transduced with Ad-ChR2-GFP. Under optical pacing at 1 Hz, Ad-NAA10 did not significantly affect wild-type iPSC-CMs, suggesting that NAA10 overexpression is well tolerated (Fig. 7d, e). Meanwhile, Ad-NAA10 significantly shortened FPD in NAA10-R4S mutant iPSC-CMs (Fig. 7d, e). To assess the contribution of increased INaL to the repolarization abnormalities in pNAA10R4S/Y- or eNAA10R4S/Y-iPSC-CMs, we treated MEA-plated monolayers with the selective late current sodium channel blockers GS967 and ranolazine41. GS967 and ranolazine significantly shortened FPDs of NAA10R4S/Y-iPSC-CMs but did not completely normalize them to WT values (Fig. 7f; Supplementary Fig. 11). In contrast, mexiletine, a class IB antiarrhythmic which also has effects on potassium channels54, did not significantly change the FPD (Supplementary Fig. 11).
Furthermore, to determine if NAA10 overexpression reverses the observed structural defects in NAA10R4S/Y-iPSC-CMs, we transduced iPSC-CMs with Ad-NAA10 and plated them on microcontact-printed substrates (Fig. 7g). Following immunostaining and confocal imaging, unbiased computational image analysis demonstrated significant shortening of the mean sarcomere length, although SPD was not fully restored (Fig. 7h, i), suggesting a partial rescue of structural phenotypes by NAA10 supplementation. Collectively, these data demonstrate that gene replacement may be a viable therapeutic strategy for NAA10-related syndrome.
Global Nt-acetylation was disrupted in NAA10R4S/Y-iPSC-CMs
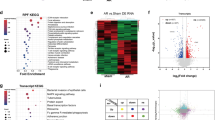
Although a substantial majority of proteins are predicted to be Nt-acetylated by NatA, there are no validated targets of Nt-acetylation in cardiomyocytes. To identify Nt-acetylated proteins in iPSC-CMs, we adapted two complementary proteomic techniques: Hydrophobic Tagging-Assisted N-Termini Enrichment (HYTANE) and LysN amino terminal enrichment (LATE) to identify N-terminal acetylated proteins with smaller sample inputs by mass spectrometry55. These complementary methods enabled us to identify a total of 985 Nt-acetylated proteins by enhancing the enrichment of Nt-peptides through the depletion of internally acetylated peptides (Supplementary Fig. 12 and Supplementary Data 1 and Supplementary Data 2). Consistent with predictions and previous reports56, the majority of Nt-acetylated proteins were substrates for NatA (Fig. 8a, b). The remaining subset of identified proteins were predicted to be substrates for NatB or NatC/E/F based on the alpha-peptide sequence (Fig. 8c and Supplementary Data 1 and Supplementary Data 2).
a The first amino acids enriched by Nt-peptide enrichment. b Sequence logos of the first 5 amino acids of NatA-subject peptides. c Sequence logos of the first 5 amino acids of peptides which were not subject to the cleavage of methionine aminopeptidase. d Representative mass spectra of Nt-acetylated MYH7. e Gene Ontology of Biologic Processes (GOBP) terms for enriched Nt-acetylated proteins. f Scatter plot of the Nt-acetylated proteins with the relative ratio of Nt-acetylated peptides normalized by total protein. Each protein was ranked according to the order of the relative ratio. Proteins known as causative to heart diseases are highlighted in red. g The normalized relative ratio of Nt-acetylated peptides from f., showing a significant reduction in Nt-acetylation in eNAA10R4S/Y-iPSC-CMs (−0.25 ± 0.08; mean ± SEM; p = <0.0001; one sample Wilcoxon test). h Volcano plot of total proteins. The cut-off value was set to 0.5 or −0.5 for log2(fold change), and 1.0 for log10(FDR). Upregulated proteins with statistical significance were highlighted in red, and downregulated proteins were highlighted in blue. i Representative gene set enrichment analysis (GSEA) plot of the running enrichment score for an upregulated GOBP term (GO:0033275, actin_myosin_filament_sliding). j Representative GSEA plot of the running enrichment score for a downregulated GOBP term (GO:0006096, glycolytic_process). k The results of GSEA for GOBP terms. GSEA was performed by a one-sided permutation test. The Benjamini-Hochberg method was applied to correct multiple comparisons. GOBP terms with an adjusted P-value (Padj) less than 0.05 are listed. Gene ranks depict the ranked list metric, and NES represents normalized enrichment score. ****p < 0.0001. Source data are provided as a Source Data file.
By mass-spectrometry, we identified multiple cardiac-associated proteins that were Nt-acetylated by NatA, including MYH7, MYH6, TNNI3, and DES (Fig. 8d and Supplementary Data 1 and Supplementary Data 2). In addition to cardiac-disease-associated proteins, Gene Ontology of Biological Processes (GOBP) analysis demonstrated enrichment of Nt-acetylated proteins associated with metabolic processes and RNA regulation and processing (Fig. 8e)
To determine if the NAA10-R4S variant impacts NatA enzymatic activity in cardiomyocytes, we normalized Nt-acetylated peptides by mass spectrometry to internal peptides for each identified protein and ranked them by the degree of Nt-acetylation (Fig. 8f and Supplementary Data 3). While not all originally identified Nt-acetylated proteins could be normalized, we demonstrated that 69% of NatA targets showed a reduction in Nt-acetylation (Fig. 8g and Supplementary Data 3). Interestingly, parallel coordinate plots for Nt-acetylated peptides and internal peptides of the same proteins demonstrated that 67% of the internal peptides were upregulated overall while Nt-peptides were downregulated (Supplementary Fig. 13). Proteins associated with nuclear envelope structure or transport, including RAN and MATR3, and structural proteins, including FHL1 and CFL2, demonstrated decreased Nt-acetylation in NAA10R4S/Y-iPSC-CMs (Fig. 8f). These data support the hypothesis that the NAA10-R4S variant significantly impairs the function of NatA and reduces global Nt-acetylation in affected iPSC-derived CMs.
In addition to identifying differentially Nt-acetylated proteins in our NAA10R4S/Y-iPSC-CMs, we sought to investigate the impact of NatA dysfunction on total protein expression. Quantitative mass-spectrometry of total proteins isolated from WT and eNAA10R4S/Y-iPSC-CMs demonstrated differential expression of 84 peptides (56 proteins) (Fig. 8h and Supplementary Data 4 and Supplementary Data 5). Proteins associated with the sarcomere, including MYBPC3, MYL4, MYH7, and MYL2, were upregulated while those involved with metabolic pathways (PKM, PGK1, and ALDOC) were down-regulated (Fig. 8h). Indeed, gene set enrichment analysis (GSEA) revealed that the biological process related to cardiac muscle was upregulated while metabolic processes and developmental cell growth was downregulated in NAA10R4S/Y-iPSC-CMs (Fig. 8i, j, k). Collectively, proteomic analysis reveals that the NAA10-R4S variant impairs NatA function, reduces protein Nt-acetylation, and induces dysregulation of pathways related to cardiomyocyte structure and glycolytic processes.
Discussion
In this study, we gained previously unrecognized mechanistic insights into the function of NAA10 and Nt-acetylation in cardiac homeostasis. Capitalizing on the identification of a previously unidentified NAA10 variant in an extended pedigree, we created and characterized an iPSC-CM model of NAA10 dysfunction to show that Nt-acetylation by NAA10 is required for normal regulation of INaL and IKs, Ca2+ clearance, as well as for normal sarcomere assembly and contraction.
From our clinical cohort, we identified a NAA10 variant that segregated with the disease phenotype supporting its pathogenicity. A KCNH2 variant was also present in some family members, but it did not co-segregate with the cardiac phenotypes, and there is little evidence supporting its pathogenicity. Consistent with other reports of NAA10-related syndrome, we observed variable clinical manifestations in gene-positive patients, with a predominance of SCD in male patients. However, at least one terminal cardiac event occurred in a carrier female (III:6). While initially described in males, an increasing number of NAA10-related syndrome cases have been described in heterozygous females13,15. The reported phenotypes are highly variable and range from asymptomatic to severe, often manifesting with a predominantly neurological or cardiac phenotype. It is presumed that skewed X-chromosome inactivation with tissue-specific expressivity contributes to the variable phenotypes seen in female patients9,57. In addition to the previously described impact of NAA10 dysfunction on the cardiovascular system, one of our patients (III:8) also developed severe dilated cardiomyopathy. It is possible that his presentation represents a natural progression of the disease given his relatively older age (17 years old), although confirmation requires study of additional affected patients. Combined with our experimental data, our clinical cohort broadens the potential cardiac phenotypes associated with NAA10-related syndrome. Like other NAA10 syndrome patients with cardiac involvement, our identified variant is near the extreme N-terminus of NAA10 and within the NAA15 interaction domain. Our biochemical data demonstrate that the p.R4S mutation not only induces NAA10 protein degradation and directly inhibits its Nt-acetylation activity but also destabilizes NAA10’s interaction with NAA15. Our results parallel an early report of NAA10-related syndrome, where several other pathogenic variants induced NAA10 protein degradation and impaired catalytic function11,26,27,57. However, the authors did not directly investigate the effects of the p.Y43S variant on the interaction between NAA10 and NAA15, nor did they study the impact of NAA10 dysfunction on cellular physiology. Our data show that the NAA10 p.R4S is a pathogenic variant; we detail the molecular mechanisms that lead to NAA10 dysfunction and delineate the consequences of NAA10 dysfunction on cardiomyocyte physiology. For future investigation, this system can also be applicable to investigate NAA10 dynamics in heterozygous females.
To investigate the effects of NAA10 dysfunction, we developed iPSC models by reprogramming somatic cells from affected patients and created an isogenic iPSC line by introducing the same variant into a control line via genome editing. Our iPSC lines are accurate models of NAA10 dysfunction based on several lines of evidence. First, NAA10 protein levels in mutant iPSC-CMs are reduced by more than 50%, consistent with reduced protein stability. Second, NAA10R4S/Y-iPSC-CMs have repolarization abnormalities as both monolayers and single cells, consistent with QT prolongation. NAA10R4S/Y-iPSC-CMs also demonstrate steeper APD restitution as well as longer pacing capture rates with larger AP dispersion, which can lead to unidirectional conduction block and reentry58. Third, single iPSC-CMs and EHTs derived from NAA10R4S/Y lines had profound defects in sarcomere formation and contractility. These data strongly support the conclusion that our NAA10-mutant iPSC models accurately recapitulate the clinical phenotype of severe arrhythmogenesis and cardiomyopathy as seen in our clinical cohort.
Our clinical and experimental data indicate that NAA10 is required for normal cardiac repolarization. To define the physiological mechanisms, we systematically examined each major ionic current underlying the cardiac action potential and found that NAA10 dysfunction alters both INa and IKs. This unique combination induces significant repolarization abnormalities consistent with a high-risk clinical phenotype (QT > 500 msec, recurrent TdP), similar to patients with complex heterozygosity and missense variants in both KCNQ1 and SCN5A59. The male NAA10 p.R4S patients had ventricular arrhythmias predominantly during periods of rest and despite high-dose beta-blocker therapy. These are typical clinical features of pathogenic SCN5A variants and LQT3 and support an essential role for increased INaL in abnormal repolarization in this disorder. This is further supported by the shortening of the FPD in response to GS967 and ranolazine, both selective INaL blockers41,60. In contrast, mexiletine, a class 1B sodium channel blocker, failed to shorten the FPD in MEA recordings of eNAA10R4S/Y-iPSC-CMs. In addition to inhibiting INaL, mexiletine also inhibits IKr and can prolong the APD in iPSC-CMs61. Since NAA10 dysfunction impairs the remaining repolarizing potassium current, IKs, mexiletine’s effect on IKr likely accounts for its ineffectiveness in normalizing the repolarization of NAA10R4S/Y-iPSC-CMs. Removal of mexiletine’s off-target effects against IKr has been the subject of recent research to improve mexiletine as a selective therapy for LQT3 patients61, and our data suggest that this selectivity will also benefit NAA10-syndrome patients. In our study, we did not observe significant changes in ICaL, although a recent report describes an increase in ICaL in patient-derived NAA10 mutants62. In that report, no other cardiac currents were examined, so it is difficult to determine the relative contribution of ICaL versus the defects in INaL and IKS as observed in our models. Finally, our phenotypic and mechanistic data in the setting of NAA10 knockdown support the conclusion that a more generalized disruption of NAA10 function favors the mechanisms delineated in our NAA10 p.R4S model.
Our data also point to an important role for NAA10 in cardiomyocyte sarcomere assembly and contractile function. In general, monogenic causes of cLQTS are not associated with decreased contractility or heart failure63. A potential exception to this convention is that selected SCN5A variants are associated with DCM and QT prolongation in some patients64. We likewise observed increased INaL in our iPSC models of NAA10 dysfunction. The resulting increased sodium flux could contribute to NAA10-mediated contractile dysfunction by elevating diastolic Ca2+ levels through the acceleration of the reverse mode of the sodium-calcium exchanger, NCX1. In addition, the deceleration of the diastolic Ca2+ decay observed in NAA10 mutant cells may also contribute to the reduction in contractile force as observed in heart failure and hypertrophic cardiomyopathy65,66. Additionally, we investigated changes in Ca2+ handling proteins that might underlie or exacerbate dysregulation of intracellular Ca2+ signaling. Proteins responsible for Ca2+ reuptake were downregulated, including NCX1 and SERCA/PLN, further supporting impairment in Ca2+ handling caused by NAA10 dysfunction. Indeed, a recent report mirrors this mechanistic relationship with significant contractile dysfunction in the setting of impaired Ca2+ reuptake in isolated human myocardium65. In addition to contractile dysfunction, impaired Ca2+ signaling is also a mechanism for arrhythmogenesis in many cardiovascular disorders, often associated with the dysregulation of RyR2 channel function. When RyR2 dysfunction is the underlying etiology, there is an increase in delayed afterdepolarizations (DADs) as a cellular marker for increased arrhythmogenesis, such as in catecholaminergic polymorphic ventricular tachycardia (CPVT). While most CPVT patients do not have QT prolongation or other ECG abnormalities at baseline37, there are case reports of patients with RyR2 variants presenting with QT prolongation, suggesting a potential mechanistic crossover between RyR2 dysfunction and increased APD67. However, despite extensive single-cell and multicellular recordings, we never observed any DADs, suggesting that RyR2 dysfunction is not a primary mechanism for increased arrhythmogenic risk in our models.
We also observed abnormal sarcomere structure in NAA10 mutant iPSC-CMs, indicating that contractile dysfunction in this disorder is likely multifactorial. Supporting these functional analyses, our mass-spectrometry experiments revealed significantly reduced global Nt-acetylation levels in NatA-target proteins. In a fraction of isolated proteins from NAA10-mutant iPSC-CMs, reduced Nt-acetylation levels were associated with total protein downregulation, likely through N-degron pathway-mediated protein degradation68. However, in the remaining fraction of proteins with decreased Nt-acetylation, corresponding internal peptides were increased, suggesting a translationally mediated feedback system to maintain overall protein levels69. Affected proteins were enriched in pathways related to glycogen metabolism and regulation of cardiac muscle hypertrophy. Further supporting the effects of NatA disruption on cardiac dysfunction and sarcomere disarray, analysis of differentially expressed proteins revealed enrichment of hypertrophy-related proteins, including MYBPC3 and MYH7, and downregulation of critical proteins in the glycolysis pathway. While fatty acid oxidation is the primary metabolic pathway in the adult heart, the glycolytic pathway is upregulated early in cardiac hypertrophy, likely as an adaptive response. In the setting of NAA10 disruption, downregulation of key glycolytic proteins, including pyruvate kinase M1/2 (PKM) and phosphoglycerate kinase 1 (PGK1), may be maladaptive, further contributing to worse cardiomyocyte dysfunction. Therefore, the impairment of NAA10 and Nt-acetylation negatively impacts cardiomyocyte structure and function through the dysregulation of multiple integrated pathways contributing to severe cardiac dysfunction as seen in our clinical cohort.
Our iPSC lines represent previously uncharacterized models of NAA10 dysfunction, demonstrating both repolarization and severe contractile abnormalities. A previous report by Ward et al. modeled NAA15 dysfunction in iPSC-CMs but only detected minor contractile impairment18. This could represent incomplete inhibition of the NatA complex, or differential targets caused by individual subunit dysregulation. There was no electrophysiological assessment of NAA15-mutant iPSC-CMs, precluding a comparison of the effects of NAA15 versus NAA10 dysfunction on the cardiomyocyte action potential. However, given that NAA10-R4S affects both NAA10 function and its association with NAA15, the severity of effects may correlate with a hierarchy of NatA complex formation > NAA10 function > NAA15 function, suggesting that cardiac phenotypes are correlated with the degree of Nt-acetylation for specific target proteins18,70. In our study, the NAA10-R4S mutation reduced NAA10 protein stability, its binding to NAA15, and NatA activity. Our proteomic data demonstrated a significant reduction in Nt-acetylation, supporting the hypothesis that disruption of Nt-acetylation of NatA-targeted proteins is the mechanism for the observed phenotypes in our iPSC models. However, while NAA10 predominantly functions as an N-acetyltransferase, there are reports of NAA10 functioning independently of NatA as a lysine acetyltransferase (KAT) and non-catalytic regulator2. Although controversy remains regarding the non-NAT functions of NAA1071, we cannot completely exclude the possibility that these alternative mechanisms may underlie the cardiac phenotypes observed with NAA10 dysfunction. Further investigation is necessary to understand the effect of distinct NAA10 or NAA15 variants on cardiomyocyte function.
iPSC disease modeling has dramatically shortened the arc of therapeutic translation. The repurposing of already approved drugs72 and development of targeted gene therapies are just two therapeutic development pathways that have benefited from iPSC modeling of cardiac disorders53. To determine if NAA10-related syndrome would be amenable to targeted gene therapy, we developed an adenovirus vector to overexpress WT NAA10 in the NAA10-mutant iPSC-CMs. This therapy partially rescued key pathogenic features of NAA10 dysfunction. Incomplete rescue may be due to transduction efficiency, NAA10 expression levels, and dominant negative effects of the mutant endogenous allele15,57. The short duration of our experiment compared to the turnover rate of key Nt-acetylation targets could also have limited the therapeutic effect, given that Nt-acetylation is an irreversible PTM. Further optimization and testing in animal models of NAA10 dysfunction will be critical for effective translation.
Our detailed physiological characterization of iPSC models of NAA10 dysfunction defines the essential roles of protein Nt-acetylation in cardiomyocyte function and delineates the pathogenic mechanisms by which NAA10 dysfunction leads to QT prolongation, risk of sudden death, and contractile dysfunction. These data underscore the importance of Nt-acetylation as a critical biological process in the heart and other organs.
Methods
Ethics Declaration
Studies involving patient-derived materials were conducted in accordance with protocols approved by the Boston Children’s Hospital Institutional Review Board (IRB). Informed written consent was obtained from all participating patients. Specific permissions were also provided in the consent form for the publication of relevant clinical information from participating patients including family history, age, sex, and clinical status as part of a retrospective review study in an open access manner.
Patient Data
The proband was identified as part of a large family originally diagnosed with “gene-negative” long QT syndrome. Retrospective clinical data were collected after enrollment into an IRB-approved protocol at Boston Children’s Hospital. All patient-related information was contained in an encrypted password-protected database (REDCap) with only de-identified samples available to researchers. A certified genetic counselor conducted a four-generation family history assessment. Initial genetic testing for Long QT syndrome included CACNA1C, CALM1, CALM2, CALM3, KCNE1, KCNH2, KCNJ2, KCNQ1, SCN5A, TRDN, AKAP9, ANK2, CAV3, KCNE2, KCNJ5, SCN4B, and SNTA1 (Invitae, test code 02211). Testing for cardiomyopathies included up to 121 genes (Invitae, test code 02251) with the NAA10-R4S variant identified on the neurodevelopmental disorders panel (Invitae, test code 728434). Echocardiography, electrocardiography, and remote device monitoring were performed as standard of care at Boston Children’s Hospital and Dartmouth Hitchcock Medical Center. Postmortem examination with cardiac histopathological evaluation was performed by the Cardiac Registry service at Boston Children’s Hospital. For the purpose of this study, females are defined as persons with two X chromosomes, and males are defined as persons with one X chromosome and one Y chromosome, as NAA10 is an X-linked gene.
Generation of iPSC lines
Patients who consented to participate in this study supplied peripheral blood mononuclear cells (PBMCs) for somatic cell reprogramming into iPSCs. This protocol was IRB-approved at Boston Children’s Hospital, and participation was offered to patients with NAA10 variants as part of a larger study on inherited arrhythmia syndromes. Patients were recruited through electrophysiology clinics at both Boston Children’s Hospital and Dartmouth Hitchcock Medical Center. Consent was obtained by the study coordinator. Variant sequencing, pluripotency marker staining, and karyotype analysis performed at regular intervals ensured iPSC model integrity.
Gene editing of iPSCs
For genome editing, variant-specific sgRNAs were designed with CRISPOR73, and synthesized using EnGen sgRNA Synthesis Kit (E3322; New England Biolabs, Ipswich, MA, USA). The HDR donor template (Integrated DNA Technologies, Coralville, IA, USA) was designed to include synonymous mutations to avoid further digestion by Cas9 (Materials and Methods Table 1).
A doxycycline-inducible WT-iPSC line (WTC-Cas9)74, which was previously generated from a wild-type human male iPSC line (Coriell Institute: # GM25256), was used for gene-editing75. Briefly, 16 hours before nucleofection, doxycycline was administered at a final concentration of 2 µg/mL. 5 µg of sgRNA and 5 µg of HDR template were transfected into 1 × 106 doxycycline-treated WTC-Cas9 iPSCs using Human Stem Cell Nucleofector Kit (VPH-5012; Lonza Bioscience, Basel, Switzerland) and 4D-Nucleofector (Lonza Bioscience). The next day, the medium was replenished with fresh mTeSR1 without doxycycline, and the single cells were seeded sparsely to a 10 cm dish a few days later. Thereafter, colonies were selected for sequencing. Off-target sites were predicted by CRISPOR73, and the sequence was analyzed by Sanger sequencing (Materials and Methods Table 1).
Differentiation and purification of iPSC-CMs
iPSCs were differentiated into CMs using ‘GiWi’ method29. Briefly, 2 days after iPSCs were seeded onto a 12-well plate, the medium was replaced with RPMI / B27 minus insulin (Thermo Fisher Scientific, Waltham, MA, USA) containing 6 µM CHIR99021 (StemCell Technologies, Vancouver, Canada) (Day 0). On Day 2, the medium was replaced with RPMI/B27 minus insulin containing 5 µM IWP2 (Tocris Bioscience, Bristol, UK). On Day 4, the medium was replaced with RPMI/B27 minus insulin. On Day 6 and every 3 to 4 days thereafter, RPMI/B27 medium was replenished. CMs were purified in glucose-depleted lactate-supplemented medium76. Differentiated CMs on Days 50 through 90 were used for experiments.
For large-scale differentiation, iPSCs were differentiated in the DASGIP® Parallel Bioreactor Systems (Eppendorf)77. In short, iPSCs were cultured in T80 cell culture flasks (Life Technologies, # 178905) pretreated with 1:100 (v/v) diluted Geltrex (Life Technologies, # A1413302) at an initial seeding density of 15,000 cells/cm2. Cells were maintained in E8 medium (Life Technologies, # A1517001) with daily medium change until 90% cell confluency was reached. For dissociation, T80 flasks were washed once with phosphate-buffered saline (PBS), incubated with 5 mL Versene (Life Technologies, # 1540066) for 15 to 20 min at 37 °C and dissociation was stopped by adding 5 mL E8 medium. 50 million single iPSCs were resuspended in 50 mL E8 medium supplemented with 10 μM of ROCK inhibitor. The bioreactor vessel was removed from the bioreactor system and placed under the laminar flow. Then, 50 mL E8 ( + 10 μM Y-27632) and 50 mL of the iPSC solution were added, resulting in a final volume of 100 mL per vessel. Cells were agitated at a speed of 60 rpm, passed with 21% O2 and 5% CO2 by 10 sL/h overlay gassing, and maintained at 37 °C. The next day, the diameter of spontaneously formed embryoid bodies (EBs) was measured to estimate the time of differentiation start. If critical diameter (100-300 µm) was reached, cardiac differentiation was induced by a complete medium change to RPMI 1640 with B27 supplemented with 7 μM CHIR99021 (day 0). After 24 h (day 1), the complete medium was changed to basic medium, and cells were incubated for an additional 24 h. On day 2, the complete medium was changed again to basic medium containing 5 µM IWR-1 for 48 h. On day 4, the complete medium was changed to basic medium, and cells were incubated for an additional 72 h. From day 7, cells were cultured in a basic medium supplemented with 1:1000 (v/v) insulin (Sigma-Aldrich, # I9278), followed by 50% medium refreshments on days 9, 11, and 13. Finally, cells were dissociated on day 15 for 3-4 h, depending on EB size and density, using Collagenase II (Worthington, # LS004176)52, and frozen using a controlled rate freezer (Grant, CRF-1).
Electrophysiology
Whole-cell patch clamp recordings
Cultured iPSC-CMs were dissociated with Accutase and plated sparsely onto Geltrex-coated 11 mm coverslips. Single iPSC-CMs were analyzed 3 to 6 days after dissociation. Single iPSC-CMs were recorded under different conditions to acquire each parameter (Materials and Methods Table 2)78,79. Perforated patch recordings were performed for action potential (AP) analysis and the L-type Ca2+ current (ICaL). APs were elicited at a pacing rate of 1 Hz by 5-ms depolarizing current injections of 400–600 pA. Perforated patch was applied to prevent rundown in ICaL recording80. The ruptured patch technique was used for INa, IKs, and IKr recordings. Series resistance and cell capacitance were compensated to ~60 % for all the voltage clamp experiments. To measure INa, starting from a holding potential of -100 mV, 40 ms of depolarizing pulses from -100 mV to 90 mV were applied in 10-mV increments. For the INa steady-state inactivation, following 400 ms of prepulses with 10-mV increments from -110 mV to -20 mV, 40 ms of 0 mV pulse was applied. For IKs, test pulses were applied for 5 s with 20 mV increments from -20 mV to 40 mV from a holding potential of -40 mV. For IKr, test pulses were applied for 4 s with 5 mV increments from -35 mV to 10 mV from a holding potential of -40 mV. To measure ICaL, starting from a holding potential of -80 mV, 3-s long -50 mV prepulse was applied and then 300 ms of depolarizing pulses from -50 mV to 50 mV were applied in 10-mV increments. For the ICaL steady-state inactivation, from -40mV holding potential, test pulses were applied for 2 s with 10 mV increments ranging from -80 mV to 10 mV followed by 10-ms-long -40 mV pulse, and then 0 mV pulse was applied for 250 ms. For AP analysis, iPSC-CMs exhibiting an APD90 / APD50 ratio of less than 1.4 were defined as ventricular type81. INaL, IKs, and IKr were defined as currents specifically sensitive to 30 µM TTX, 1 µM HMR1556, and 1 µM E4031 respectively. The current traces were subtracted before and after the drug administration to elicit those specific currents. Pipettes were pulled from thick-walled borosilicate glass capillaries (1B150F-4; World Precision Instruments, FL, USA) for AP and ICaL, and from thin-walled capillaries (TW150-4; World Precision Instruments) for INa, IKs, and IKr. The resistance of the pipettes for INa recording was 1–2 MΩ. For the other recordings, the pipettes with 3–5 MΩ were used. β-escin (25 μM) was applied in the recording solution to create the perforated patch configuration82. Access resistance was 10–25 MΩ for perforated patch recording and <5 MΩ for ruptured patch recording. dPatch®, and SutterPatch® (Sutter Instrument, CA, USA) were used for data acquisition. All the data were acquired from at least three independent experiments using different biological replicates.
Multi-electrode array with optogenetics
Single iPSC-CMs were isolated by incubating collagenase-B (Roche, Roswell, GA, USA, 1 mg/mL) for 15 minutes and then 0.25% Trypsin or Accutmax (Innova Cell Technologies, San Diego, CA, USA) for 5 minutes for dissociation81. Cell suspensions (3 × 104 cells in 5 µL) were placed onto fibronectin-coated multi-electrode array (MEA) plates (CytoView MEA; Axion BioSystems, Atlanta, GA, USA). After 3 days, the CMs were infected with the crude adenovirus expressing Channelrhodopsin-2 fused to green fluorescent protein (GFP) (Ad-ChR2-GFP) to enable optical stimulation83. The blue light was delivered with Lumos 24 (Axion BioSystems). The center wavelength is 475 nm with the range of wavelength between 440 nm and 530 nm. The light intensity was set to ~0.98 mW/mm2. Four or more days after the infection, field potentials (FP) were recorded using Maestro Edge (Axion BioSystems). FP signals were digitally sampled at 12.5 kHz and the system bandwidth was 0.01 Hz–5 kHz. iPSC-CMs were stimulated by using a multi-well light stimulation system (Lumos 24; Axion BioSystems)83. Specifically, CMs were stimulated at the rate of interest (1 Hz, 2 Hz, or 3 Hz) for 40 beats, and the final 30 beats were averaged. FP duration (FPD) was defined as the interval between a spike and a subsequent positive deviation. This parameter was automatically measured with the Cardiac Software Module on the system. All the data were acquired from at least three independent biological replicates.
Optical recording of action potentials for APD restitution
Cells were dissociated using collagenase-B and Accutmax and seeded onto Geltrex-coated coverslips. After three days, cells were stained with 1:1000 FluoVolt membrane voltage-sensitive dye (Invitrogen, F10488) in Tyrode’s solution for 30 min at 37 °C. Thereafter, cells were washed twice with Tyrode’s solution and imaged using IonOptix system (IonOptix). The cells were stimulated with platinum field stimulation electrodes (IonOptix) and the stimulation was triggered by a custom-made TTL controller to run the standard dynamic method33. Specifically, cells were paced with a cycle length (CL) of 1000 ms for 30 beats until steady-state APD was reached. Thereafter, the CL was sequentially set as 660 ms, 500 ms, 450 ms, 400 ms, and 350 ms, after which the CL was progressively decreased by 20 to 10 ms until it reached to 150 ms. The CL was reduced after 12 beats. The signal was digitally sampled at 1 kHz and analyzed using Clampfit (Axon Instruments).
Immunofluorescence
Samples were washed with cold Ca2+-free PBS for 5 minutes before being fixed with 4% paraformaldehyde for 10 minutes at 4 °C and permeabilized with 0.1% Triton X-100 in PBS for 10 minutes at room temperature. Blocking was performed with 3% bovine serum albumin in PBS. Primary antibodies and secondary antibodies were sequentially incubated for 2 hours at room temperature, followed by serial washes with PBS and DAPI in the mounting media. Details of the antibodies are listed in Materials and Methods Table 3. The coverslips with stained samples were mounted with ProLong Diamond Antifade Mount (Invitrogen, 2273639), and after 24 hours of incubation in darkness at room temperature, imaging was performed. Confocal microscopy (Olympus FV3000R) with a 60x oil immersion objective was used.
Micro-contact patterning and sarcomere analysis
Using Computer-Aided Design (CAD), the single-cell pattern was designed as a series of rectangles with a 7:1 aspect ratio (105 μm by 15 μm) surrounded by a 220 μm thick boundary. The design was then converted to photolithography masks (CAD/Art Services Inc.). At the Center for Nanoscale Systems (Harvard University), silicon wafers (Wafer World) with a diameter of 3 inches were cleaned with a nitrogen gun and then spin-coated with photoresist SU8-3005 (MicroChem Corp.), followed by baking cycles of 1 minute at 65 °C and 2 minutes at 95 °C. Polymerization via UV-light exposure through the photomasks with the desired single-cell patterns was then executed for 20 seconds. The baking cycles were repeated, and the post-UV wafers were developed in propylene glycol methyl ether acetate (PGMEA, Sigma) for no more than 1 minute under vigorous agitation. After development, the wafer was desiccated overnight with a small amount of silane (United Chemical) to prevent Polydimethylsiloxane (PDMS) from binding to it permanently. Once the wafers were prepared, Polydimethylsiloxane (PDMS, Sylgard 184; Dow Corning) prepared at a 10:1 ratio (base: curing agent) was placed on the wafer, covering its entire surface, and baked for 24 hours at 65 °C. The next day, stamps were cut without damaging the wafer and sonicated in 70% ethanol for use in patterning.
Glass coverslips (12 mm, VWR, 48366-252) were spin-coated at custom recipes with a 1:1 ratio of Polydimethylsiloxane (PDMS) elastomer (Sylgard 184, The Dow Chemical Company, Midland, MI, USA) and dielectric gel (Sylgard 527, The Dow Chemical Company). PDMS 527 was prepared by combining Part A and Part B of the kit in a 1:1 ratio. Coverslips coated and incubated at 65 °C for 48 hours. Thereafter, stamps containing rectangular-shaped islands with a 7:1 aspect ratio47 were first coated for 1 hour with Fibronectin (Sigma Aldrich, F0895) (50 µg/ml) diluted in Geltrex (1:200, Life Technologies, A1413302). Fibronectin aliquots of 1 mg/ml were prepared in PBS and stored at -20 °C. In the meantime, PDMS-coated coverslips were exposed to UV Ozone (Jelight) for 8 minutes, and the patterning process was performed by placing the dried stamps onto the coverslips. The coverslips were immersed in 1% Pluronic F-127 (Sigma-Aldrich, P2443) for less than 10 minutes to block the portions of the coverslips not coated with fibronectin, followed by washing three times with room-temperature PBS.
iPSC-CMs were seeded onto the micropatterned coverslips 3 days before immunofluorescence imaging. Cells were stained with FITC-conjugated α-sarcomeric actinin, and Alexa-647 conjugated Phalloidin, followed by staining with Hoechst 33342.
To assess sarcomere alignment in micropatterned iPSC-CMs, we used an unbiased algorithm developed by the Disease Biophysics Group incorporating the ImageJ Plugin (Orientation J) and a custom-made MATLAB (Mathworks) script for structural analysis of single cells49. Briefly, a Sarcomere Packing Density (SPD) reflects the degree of spatial organization of the sarcomeres quantifying the immunosignal localized in a regular lattice and the periodicity of the positive structures, respectively, of their orientations. This means that the poorly formed sarcomeres that are not periodically spaced demonstrate a reduced SPD value, ranging from 1 to 0.
EHT generation
Engineered heart tissues (EHTs) were generated as described previously52 with some minor modifications. Briefly, 3D-differentiated 0.8 x 106 iPSC-CMs were used to generate each EHT. Cells were transduced with Ad-ChR2-GFP two to three days after casting. We modified the standard EHT culture medium (EHT-medium in the referenced literature52) by replacing DMEM with RPMI 1640 plus B27 minus insulin, removing 10% heat-inactivated horse serum, and reducing aprotinin concentration to 5 µg/mL. EHT contraction was recorded as described below starting from day 7, and functional analysis was performed from day 27 to day 33.
Functional assessment of EHTs
EHTs in a 24-well plate were placed in a stage-top incubator and maintained at 37 °C, 5% CO2. EHTs were optically paced with a custom-built illumination system positioned above the plate and recorded from below at 30 frames per second through a 561 nm long-pass filter (Semrock BLP02-561R-32) using an 8 mm f/1.4 lens (ThorLabs MVL8M1) mounted on a Basler acA1920 camera. The light intensity was 0.1 mW/mm2 (measured at the point of the EHT, with Thorlabs light meter #PW160). The center frequency was 460–465 nm. EHT post movement was tracked post hoc using the multi-template matching FIJI plugin and subsequently analyzed using a custom Python script84. Twitch force measurements were subsequently measured by applying post deflection to the beam bending theory for a known Young’s modulus of the posts, as described in detail elsewhere85. The custom code is available on GitHub with appropriate version control, https://github.com/AI-pECG/EHT.
Quantitative PCR
Cells were washed once with ice-cold PBS and lysed in TRIzol. Total RNA was extracted by centrifugation and RNA samples were isolated. Complementary DNA (cDNA) was made using a reverse transcriptase kit (Superscript III, Invitrogen). We quantified the total cDNA for each sample and normalized the concentration. Quantitative PCR was performed on a 96-well thermocycler (BioRad) at an annealing temperature of 55 °C with validated gene-specific primers. Ct values were compared to a housekeeping gene (GAPDH), and the fold-change was calculated and compared to control samples for each gene transcript.
Western blot
Cells were lysed with mTOR lysis buffer (120 mM NaCl, 40 mM HEPES, 40 mM NaF, 1 mM EDTA, 10 mM β-Glycerophosphate disodium, 0.3% CHAPS, pH 7.5 with NaOH) containing 1% Triton X-100 and Halt protease and phosphatase inhibitor (Life Technologies 78442). The concentration of the protein was measured with a BCA Protein Assay kit (Thermo scientific 23225), and 3 µg of protein in each lane was analyzed by SDS-PAGE and immunoblotting. Blots were incubated with primary antibodies and secondary antibodies sequentially for 2 hours at room temperature or overnight at 4 °C. Protein signals were detected using an enhanced chemiluminescent substrate (Bio-Rad), and images were captured using Azure 300 (Azure Biosystems, Dublin, CA, USA) and analyzed using ImageJ. The antibodies are listed in Supplemental Table 3.
Knockdown of NAA10
NAA10 was knocked down with siRNA. NAA10 siRNA (Thermo Fisher, s15744) and negative-control siRNA (Thermo Fisher, 4390843) were transfected with Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, 13778075), following the protocol provided by the manufacturer.
Expression plasmids
NAA10WT-3×FLAG cloned into pUC-GW-Kan vector was synthesized by a manufacturer (GENEWIZ, South Plainfield, NJ, USA). Then, NAA10 WT-3xFLAG was cloned into pcDNA3.1(+) vector, and NAA10R4S-3xFLAG/pcDNA3.1(+) was generated by site-directed mutagenesis with In-Fusion cloning (Takara Bio, Kusatsu, Japan). NAA15 expression plasmid (HG19640-UT, Sino Biological) was also cloned into pcDNA3.1(+) vector, and Myc tag was added using In-Fusion cloning. For c-Myc-tagged KCNQ1, we purchased Kv7.1/pcDNA3.1 (Addgene, #111452) and introduced c-Myc epitope (EQKLISEEDL) into the extracellular S1-S2 linker in-frame after nucleotide position 438 between E146 and Q14740.
Cycloheximide chase experiment
2.5 µg of plasmids were transfected into HEK293T cells on a 6-well plate with Lipofectamine 3000. 48 hours after the transfection, the medium of each well was replaced with 2 mL culture medium containing 50 µg/mL cycloheximide (Sigma, 01810). Cells were harvested at 0, 2, 4, and 6 hours after the cycloheximide administration. The cells harvested at 0 hour were not treated with cycloheximide. Harvested cells were centrifuged at 4 °C for 15 seconds with 14,000× g, and the cell pellets were washed with ice-cold PBS, centrifuged again, and stored at -80 °C after the supernatant was discarded. After all the samples were harvested, western blot was performed, and the membrane was stained with anti-DDDDK tag antibody (Abcam, ab1257) and anti-Vinculin antibody (Santa Cruz). For secondary antibodies, IRDye680 RD donkey anti-goat (LI-COR Biosciences, Lincoln, NE, USA) and IRDye800 CW donkey anti-mouse (LI-COR Biosciences) were used. Imaging was performed with LI-COR Odyssey Infrared Imaging System (LI-COR Biosciences).
NAA10-NAA15 binding assay
The NAA10- and NAA15-expression plasmid was transfected into HEK293 cells with Lipofectamine 3000. 48 hours later, the cells were harvested and lysed with mTOR lysis buffer. The lysate was incubated with anti-FLAG magnetic beads overnight at 4 °C. The protein was eluted with FLAG peptide and analyzed with SDS-PAGE. Immunoblotting was performed as described above. The protein was quantified using ImageJ.
Protein synthesis
50 µg of the NAA10 expression plasmid was transfected into HEK293 cells on a 15 cm dish using polyethylenimine (PEI) (Sigma 408727). Four dishes were prepared for each plasmid, and after lysing the cells the cell lysate was filtered with a 0.22 µm filter, and pre-cleared with mouse IgG agarose. Thereafter, FLAG-tagged NAA10 was pulled down with FLAG tag using anti-FLAG magnetic beads (Sigma, M8823). The protein was eluted with FLAG peptide and concentrated using Amicon Ultra-4 Centrifugal Filter Unit (Millipore, UFC8010).
ThioGlo4 assay
ThioGlo4 assay was modified from a previous protocol25. The custom peptide as a substrate for NAA10 (EEEIA24: EEEIAALRWGRPVGRRRRPVRVYP) was synthesized by Biomatik (Kitchener, Ontario, Canada). Briefly, the mixture of 15 µM ThioGlo4 (MilliporeSigma, 59550410MG), 150 mM NaCl, 25 mM HEPES (pH 7.5), and 0.001% Triton X-100 was reacted with 0.5, 1, 2, 5, and 10 µM of CoA (Sigma, C4780), respectively, for standard curve. Additionally, 15 µM ThioGlo4, 50 µM acetyl CoA (Sigma, A2181), 50 µM substrate (EEEEIA24), 150 mM NaCl, 25 mM HEPES (pH 7.5), and 0.001% Triton X-100 were reacted with 25, 50, 100, 200 nM of purified NAA10, respectively. All experiments were performed at 25 °C in duplicate, and the fluorescence intensity was measured with an excitation wavelength of 400 nm and an emission wavelength of 465 nm using a FlexStation®3 multi-mode microplate reader. The fluorescence was continuously recorded for 30 minutes with an interval of 30 seconds. The baseline was subtracted, and initial velocity was calculated using SoftMax Pro Software.
Adenovirus generation
NAA10-P2A-HaloTag was cloned based on pH6HTC His6HaloTag® T7 Vector (Promega, Madison, WI, USA). The NAA10-P2A-HaloTag sequence was inserted into pENTR/D-TOPO vector (Invitrogen) and thereafter into pAd/CMV/V5/DEST vector (Invitrogen) following the manufacturer’s protocol. The vector was digested with PacI, and the PacI-digested vector was transfected into HEK293A cells with Lipofectamine 3000 on a 6-well plate. Thereafter, the virus was amplified by infecting HEK293A cells on a 10 cm dish. The crude stock was used in experiments and was designated as Ad-NAA10.
Ca2+ imaging
Single iPSC-CMs were seeded on PDMS-coated micro-patterned coverslips. After 3 days, the coverslips were incubated with 5 µM Fura-2 (ThermoFisher, F14185) at 37 °C with 5% CO2 for 20 minutes. We washed cells for 10–15 minutes in an extracellular buffer and placed them in a custom chamber with platinum field stimulation electrodes (IonOptix). Bath temperature was maintained between 36–37 °C by an in-line heater circulator and a closed-loop temperature controller. The extracellular buffer contains (in mM) NaCl 140, KCl 5.4, MgCl2 1.2, CaCl2 1.8, HEPES 10, Glucose 10, and sodium pyruvate 2, with pH 7.4. We imaged samples using the IonOptix Calcium imaging system installed on a modified Olympus IX71. During imaging, cells were stimulated with bipolar electrical pulses at a duration of 8 ms and a voltage of 20–30 V. We paced iPSC-CMs at both 0.5 Hz and 1 Hz to determine the relative change in diastolic Ca2+ levels. The end-diastolic level averaged for five transients during each pacing frequency was compared to the baseline before pacing. The data were analyzed using IonWizard software (IonOptix).
Cell-surface Biotinylation
After washing with PBS (+) (PBS containing 0.2 mM CaCl2 and 1.5 mM MgCl2), cells were incubated with 0.5 mg/mL Sulfo-NHS-SS-Biotin (Thermo) in PBS (+) for 1 hour on ice with occasional shaking. Thereafter, the biotinylating reaction was quenched by washing 3 times with PBS (+) containing 100 mM glycine. Cells were lysed with mTOR buffer, and part of the lysate was saved as total protein. The rest of the lysate was incubated with Pierce Streptavidin Magnetic Beads (Thermo) at 4 °C overnight on a rotator. The magnetic beads were collected on a magnetic stand, and the protein sample was eluted by incubating with SDS-PAGE, reducing sample buffer at 96–100 °C for 5 minutes.
Flow cytometry
HEK cells overexpressing c-Myc-tagged KCNQ1 plasmid were dissociated into singlets with Accumax, and stained with 1:100 Alexa Fluor® 647 anti-c-Myc antibody (BioLegend, 62680) or 1:100 Alexa Fluor® 647 conjugated isotype mouse IgG1 antibody (BioLegend, 40013) diluted in FACS buffer (PBS(-) containing 0.5% BSA and 0.1% NaN3) at room temperature for one hour in a dark room. Then, the cells were fixed with 200 µL of 4% PFA and incubated at 4 °C for 20 minutes. No permeabilization procedure was conducted. The cells were washed and resuspended in FACS buffer. The data were acquired on a benchtop SH800S cell sorter (Sony, Tokyo, Japan) and analyzed using FlowJo (BD Biosciences).
Nt-peptides enrichment
Nt-peptide enrichment was performed following the procedures described below56. Briefly, iPSC-CMs were scraped from plates, and after being washed three times with PBS, they were resuspended in 150 mM NaCl with 20 mM HEPES (pH 7.5). The cells were lysed by three cycles of freezing and thawing. 100 µg of protein from WT-iPSC-CMs and eNAA10R4S/Y-iPSC-CMs, respectively, was used for the downstream analysis.
Hydrophobic Tagging-Assisted N-termini Enrichment (HYTANE)
After being resuspended, the protein samples were reduced with 5 mM DTT and alkylated with 12.5 mM CAA. Thereafter, the leftover CAA was quenched with another dose of DTT. The samples from WT were labeled with 40 mM C12H2 formaldehyde (light) and 20 mM sodium cyanoborohydride, and those from eNAA10R4S/Y were labeled with 40 mM C13D2 formaldehyde (heavy) and 20 mM sodium cyanoborohydride by incubating at 37 °C overnight. The reaction was quenched with 100 mM glycine for 1 h at 37 °C. Then, the light- and heavy-labeled samples were mixed and diluted four times using 20 mM HEPES pH 8.0. The mixed samples were digested with trypsin (Cell Signaling Technology) overnight at room temperature. Next, the tryptic digestion was quenched by adding formic acid and then desalted with an OASIS-HLP cartridge (Waters). Part of the sample was kept for total protein analysis, and the remaining sample underwent downstream undecanal-based enrichment. The desalted samples were lyophilized and stored at -80 °C until they were used for the downstream analysis.
LysN Amino Terminal Enrichment (LATE)
After being resuspended, the protein samples were reduced with 5 mM DTT and alkylated with 12.5 mM CAA. Thereafter, the leftover CAA was quenched with another dose of DTT. The samples were digested with LysN at 1:100 w/w and incubated at 37 °C for 12 h. The digestion was quenched with 6% glacial acetic acid. The digested peptides were labeled with 40 mM C13D2 formaldehyde (heavy) and 20 mM sodium cyanoborohydride by incubating at 37 °C for 10 min. The labeling reaction was terminated by incubating with 100 mM glycine in 2% acetic acid. Next, light- and heavy-labeled samples were mixed and desalted using an OASIS-HLP cartridge. A portion of the sample was set aside for total protein analysis, and the remaining sample was used for the downstream undecanal-based enrichment.
Undecanal-based enrichment
The lyophilized samples were resuspended in 100 mM HEPES (pH 7.0). The undecanal labeling was done by incubating with 1 mg undecanal and 20 mM sodium cyanoborohydride at 50 °C for two hours while adding another dose of sodium cyanoborohydride after the first hour of incubation. The undecanal-labeled samples were centrifuged, and the supernatant was dried with SpeedVac. The dried samples were resuspended in 2% acetonitrile and 0.1% formic acid and desalted with an OASIS-HLP cartridge. Finally, the Nt-peptides were eluted with 60% acetonitrile, and lyophilized for downstream LC/MS analysis52. LC/MS was performed at Taplin Mass Spectrometry Facility at Harvard Medical School.
Analysis of Mass Spectrometry Data
The acquired data was analyzed using Trans Proteomic Pipeline (v. 6.3.3), where Comet (release 2022.01 rev.2) was used for sequence database search86. The detailed parameter settings are described in Materials and Methods Table 4. The database was downloaded from UniProt. The results were validated by PeptideProphet at a false discovery rate of peptide identification less than 1%. Relative peptide quantification was conducted using XPRESS. Searches were performed against the Homo sapiens proteome (UniProt ID: 9606, downloaded in November 2023). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE87 partner repository with the dataset identifier PXD053715. Further bioinformatic analysis of the Nt-peptides was conducted using Peptide-matcher (https://github.com/OKLAB2016/peptide-matcher) and Parse-and-unite (https://github.com/OKLAB2016/parse-and-unite), both available from the GitHub repository created by Oded Kleifeld Lab. The detailed parameter settings for data analysis are listed in Materials and Methods Table 4. Logos were created using WebLogo88. For total protein analysis, output data was further processed with the Perseus software (version 2.0.9.0). Over-representation analysis was performed with DAVID (v2024q1)89,90. Gene Set Enrichment Analysis was conducted using the R-package fgsea (https://github.com/ctlab/fgsea). The R programming environment (R v4.3.1) was used for data processing and visualization.
Bioinformatics
A multiple sequence alignment of the NAA10 sequence was performed with Clustal Omega, and the conservation annotation was conducted with Jalview91,92. DynaMut2 was used to predict the impact of the mutation on protein dynamics and stability93. Information on protein structure was obtained from Protein Data Bank (PDB), and the human NatA amino-terminal acetyltransferase complex (PDB code: 6C9M)24, was used for the analysis.
Statistics
Prism 9 (GraphPad, San Diego, CA, USA) was used for statistical analysis. Data were presented as mean ± SEM. Normality was tested with the Shapiro-Wilk test. Student’s t-test was used to compare two groups for normally distributed samples. For more than three groups, one-way analysis of variance (ANOVA), followed by Dunnett’s comparison test, was used. Two-way ANOVA followed by Tukey’s multiple comparisons test was used for samples with two independent variables. For repetitive measurements, a two-way repeated measures ANOVA followed by Dunnett’s comparison test, or mixed-effects analysis followed by Šídák’s multiple comparisons test was performed. For samples without normal distribution, Mann-Whitney test or Kruskal-Wallis test, followed by Dunn’s multiple comparisons test, was conducted for two- or multi-group comparisons. Statistics applied to each data set were denoted in the corresponding figure legend. Results were considered statistically significant at p < 0.05.
Image generation
Illustrations in Figs. 2f, 4k, 7a, and Supplementary Fig. 9a were created with BioRender under the appropriate license. Figure 5a was created by Michael Rosnach using in-house software.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Electrophysiology and other physiology data are available in the Dryad database with the following link: https://doi.org/10.5061/dryad.280gb5mvz.The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD053715. Additional clinical and sequencing data is available upon request with the appropriate approvals as per NIH data-sharing guidelines. Inquires for additional data should be made to Vassilios Bezzerides by email: vassilios.bezzerides@cardio.chboston.org. The expected timeframe for a response to reasonable requests is one week. Source data are provided with this paper.
Code availability
Custom code was developed for the analysis of EHT contractility. This code is available on Github at the following link: https://github.com/AI-pECG/EHT. Analysis algorithms for sarcomere alignment50 are available on the following website: http://diseasebiophysics.seas.harvard.edu/.
References
Ree, R., Varland, S. & Arnesen, T. Spotlight on protein N-terminal acetylation. Exp. Mol. Med. 50, 1–13 (2018).
Aksnes, H., Ree, R. & Arnesen, T. Co-translational, Post-translational, and Non-catalytic Roles of N-Terminal Acetyltransferases. Mol. Cell 73, 1097–1114 (2019).
Heathcote, K. C. et al. N-terminal cysteine acetylation and oxidation patterns may define protein stability. Nat. Commun. 15, 5360 (2024).
Arnesen, T. et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA. 106, 8157–8162 (2009).
Mullen, J. R. et al. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J 8, 2067–2075 (1989).
Park, E. C. & Szostak, J. W. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J 11, 2087–2093 (1992).
Arnesen, T. et al. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem. J 386, 433–443 (2005).
Rope, A. F. et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Hum. Genet. 89, 28–43 (2011).
Bader, I. et al. Severe syndromic ID and skewed X-inactivation in a girl with NAA10 dysfunction and a novel heterozygous de novo NAA10 p.(His16Pro) variant - a case report. BMC Med. Genet. 21, 153 (2020).
Cheng, H. et al. Phenotypic and biochemical analysis of an international cohort of individuals with variants in NAA10 and NAA15. Hum. Mol. Genet. https://doi.org/10.1093/hmg/ddz111 (2019).
Casey, J. P. et al. NAA10 mutation causing a novel intellectual disability syndrome with Long QT due to N-terminal acetyltransferase impairment. Sci. Rep. 5, 16022 (2015).
Støve, S. I. et al. A novel NAA10 variant with impaired acetyltransferase activity causes developmental delay, intellectual disability, and hypertrophic cardiomyopathy. Eur. J. Hum. Genet 26, 1294–1305 (2018).
Saunier, C. et al. Expanding the Phenotype Associated with NAA10-Related N-Terminal Acetylation Deficiency. Hum. Mutat. 37, 755–764 (2016).
Shishido, A. et al. A Japanese boy with NAA10-related syndrome and hypertrophic cardiomyopathy. Hum Genome Var 7, 23 (2020).
Popp, B. et al. De novo missense mutations in the NAA10 gene cause severe non-syndromic developmental delay in males and females. Eur. J. Hum. Genet. 23, 602–609 (2015).
Esmailpour, T. et al. A splice donor mutation in NAA10 results in the dysregulation of the retinoic acid signalling pathway and causes Lenz microphthalmia syndrome. J. Med. Genet. 51, 185–196 (2014).
Lyon, G. J. et al. Expanding the phenotypic spectrum of NAA10-related neurodevelopmental syndrome and NAA15-related neurodevelopmental syndrome. Eur. J. Hum. Genet. 31, 824–833 (2023).
Ward, T. et al. Mechanisms of congenital heart disease caused by NAA15 haploinsufficiency. Circ. Res. 128, 1156–1169 (2021).
Liszczak, G. et al. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat. Struct. Mol. Biol. 20, 1098–1105 (2013).
Caraballo, C. et al. Clinical Implications of the New York Heart Association Classification. J. Am. Heart Assoc. 8, e014240 (2019).
Basso, C. et al. Cardiac hypertrophy at autopsy. Virchows Arch 479, 79–94 (2021).
Cunningham, K. S., Spears, D. A. & Care, M. Evaluation of cardiac hypertrophy in the setting of sudden cardiac death. Forensic Sci. Res. 4, 223–240 (2019).
Knorr, A. G. et al. Ribosome–NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation. Nat. Struct. Mol. Biol. 26, 35–39 (2018).
Gottlieb, L. & Marmorstein, R. Structure of Human NatA and Its Regulation by the Huntingtin Interacting Protein HYPK. Structure 26, 925–935.e8 (2018).
Ho, Y.-H., Chen, L. & Huang, R. Development of A Continuous Fluorescence-Based Assay for N-Terminal Acetyltransferase D. Int. J. Mol. Sci. 22, 594 (2021).
McTiernan, N. et al. NAA10 dysfunction with normal NatA-complex activity in a girl with non-syndromic ID and a de novo NAA10 p.(V111G) variant - a case report. BMC Med. Genet. 19, 47 (2018).
Ree, R. et al. A novel NAA10 p.(R83H) variant with impaired acetyltransferase activity identified in two boys with ID and microcephaly. BMC Med. Genet. 20, 101 (2019).
Churko, J. M., Burridge, P. W. & Wu, J. C. Generation of human iPSCs from human peripheral blood mononuclear cells using non-integrative Sendai virus in chemically defined conditions. Methods Mol. Biol. 1036, 81–88 (2013).
Lian, X. et al. Chemically defined, albumin-free human cardiomyocyte generation. Nat. Methods 12, 595–596 (2015).
Blinova, K. et al. International Multisite Study of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Drug Proarrhythmic Potential Assessment. Cell Rep 24, 3582–3592 (2018).
Izumi-Nakaseko, H. et al. Development of correction formula for field potential duration of human induced pluripotent stem cell-derived cardiomyocytes sheets. J. Pharmacol. Sci 135, 44–50 (2017).
Feyen, D. A. M. et al. Metabolic Maturation Media Improve Physiological Function of Human iPSC-Derived Cardiomyocytes. Cell Rep 32, 107925 (2020).
Goldhaber, J. I. et al. Action potential duration restitution and alternans in rabbit ventricular myocytes: the key role of intracellular calcium cycling: The key role of intracellular calcium cycling. Circ. Res. 96, 459–466 (2005).
Hothi, S. S. et al. Arrhythmogenic substrate and its modification by nicorandil in a murine model of long QT type 3 syndrome. Prog. Biophys. Mol. Biol. 98, 267–280 (2008).
Antoniou, C.-K. et al. QT prolongation and malignant arrhythmia: How serious a problem? Eur. Cardiol. 12, 112–120 (2017).
Grant, A. O. Cardiac ion channels. Circ. Arrhythm. Electrophysiol. 2, 185–194 (2009).
Schwartz, P. J. et al. Inherited cardiac arrhythmias. Nat Rev Dis Primers 6, 58 (2020).
Fatima, A. et al. The disease-specific phenotype in cardiomyocytes derived from induced pluripotent stem cells of two long QT syndrome type 3 patients. PLoS One 8, e83005 (2013).
Chadda, K. R., Jeevaratnam, K., Lei, M. & Huang, C. L.-H. Sodium channel biophysics, late sodium current and genetic arrhythmic syndromes. Pflugers Arch 469, 629–641 (2017).
Bennett, P. B., Yazawa, K., Makita, N. & George, A. L. Jr Molecular mechanism for an inherited cardiac arrhythmia. Nature 376, 683–685 (1995).
Potet, F., Vanoye, C. G. & George, A. L. Jr Use-Dependent Block of Human Cardiac Sodium Channels by GS967. Mol. Pharmacol. 90, 52–60 (2016).
Huang, H. et al. Mechanisms of KCNQ1 channel dysfunction in long QT syndrome involving voltage sensor domain mutations. Sci Adv 4, eaar2631 (2018).
Kanki, H., Kupershmidt, S., Yang, T., Wells, S. & Roden, D. M. A structural requirement for processing the cardiac K+ channel KCNQ1. J. Biol. Chem. 279, 33976–33983 (2004).
Bourdin, B., Segura, E., Tétreault, M.-P., Lesage, S. & Parent, L. Determination of the relative cell surface and total expression of recombinant ion channels using flow cytometry. J. Vis. Exp. https://doi.org/10.3791/54732 (2016).
Yazawa, M. et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 471, 230–234 (2011).
Valdivia, C. R. et al. Increased late sodium current in myocytes from a canine heart failure model and from failing human heart. J. Mol. Cell. Cardiol. 38, 475–483 (2005).
Bray, M.-A., Sheehy, S. P. & Parker, K. K. Sarcomere alignment is regulated by myocyte shape. Cell Motil. Cytoskeleton 65, 641–651 (2008).
McCain, M. L., Yuan, H., Pasqualini, F. S., Campbell, P. H. & Parker, K. K. Matrix elasticity regulates the optimal cardiac myocyte shape for contractility. Am. J. Physiol. Heart Circ. Physiol. 306, H1525–H1539 (2014).
Kuo, P.-L. et al. Myocyte Shape Regulates Lateral Registry of Sarcomeres and Contractility. Am. J. Pathol. 181, 2030–2037 (2012).
Pasqualini, F. S., Sheehy, S. P., Agarwal, A., Aratyn-Schaus, Y. & Parker, K. K. Structural phenotyping of stem cell-derived cardiomyocytes. Stem Cell Reports 4, 340–347 (2015).
Sun, N. et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 4, 130ra47 (2012).
Breckwoldt, K. et al. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 12, 1177–1197 (2017).
Eisner, D. A., Caldwell, J. L., Trafford, A. W. & Hutchings, D. C. The Control of Diastolic Calcium in the Heart. Circ. Res. 126, 395–412 (2020).
Gualdani, R. et al. Inhibition of hERG potassium channel by the antiarrhythmic agent mexiletine and its metabolite m-hydroxymexiletine. Pharmacol Res. Perspect 3, e00160 (2015).
Chen, L. et al. Hydrophobic tagging-assisted N-termini enrichment for in-depth N-terminome analysis. Anal. Chem. 88, 8390–8395 (2016).
Aksnes, H., Drazic, A., Marie, M. & Arnesen, T. First Things First: Vital Protein Marks by N-Terminal Acetyltransferases. Trends Biochem. Sci. 41, 746–760 (2016).
Wu, Y. & Lyon, G. J. NAA10-related syndrome. Exp. Mol. Med. 50, 85 (2018).
Tse, G., Chan, Y. W. F., Keung, W. & Yan, B. P. Electrophysiological mechanisms of long and short QT syndromes. Int. J. Cardiol. Heart Vasc. 14, 8–13 (2017).
Agudelo, W. A. et al. Structural Modelling of KCNQ1 and KCNH2 Double Mutant Proteins, Identified in Two Severe Long QT Syndrome Cases, Reveals New Insights into Cardiac Channelopathies. Int. J. Mol. Sci. 22, 12861 (2021).
Chorin, E. et al. Ranolazine for Congenital Long-QT Syndrome Type III. Circ. Arrhythm. Electrophysiol. 9, e004370 (2016).
McKeithan, W. L. et al. Reengineering an Antiarrhythmic Drug Using Patient hiPSC Cardiomyocytes to Improve Therapeutic Potential and Reduce Toxicity. Cell Stem Cell 27, 813–821.e6 (2020).
Belbachir, N. et al. Studying Long QT Syndrome Caused by NAA10 Genetic Variants Using Patient-Derived Induced Pluripotent Stem Cells. Circulation 148, 1598–1601 (2023).
Schwartz, P. J., Ackerman, M. J., George, A. L. Jr & Wilde, A. A. M. Impact of genetics on the clinical management of channelopathies. J. Am. Coll. Cardiol. 62, 169–180 (2013).
Shi, R. et al. The cardiac sodium channel mutation delQKP 1507-1509 is associated with the expanding phenotypic spectrum of LQT3, conduction disorder, dilated cardiomyopathy, and high incidence of youth sudden death. Europace 10, 1329–1335 (2008).
Sun, Z. et al. Synchronous force and Ca2+ measurements for repeated characterization of excitation-contraction coupling in human myocardium. Commun Biol 7, 220 (2024).
Helms, A. S. et al. Genotype-Dependent and -Independent Calcium Signaling Dysregulation in Human Hypertrophic Cardiomyopathy. Circulation 134, 1738–1748 (2016).
Tanaka, Y., Kawabata, M., Scheinman, M. M. & Hirao, K. Catecholaminergic polymorphic ventricular tachycardia with QT prolongation: Cpvt with qt prolongation. Pacing Clin. Electrophysiol. 38, 1499–1502 (2015).
Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA. 116, 358–366 (2019).
Linster, E. et al. Cotranslational N-degron masking by acetylation promotes proteome stability in plants. Nat. Commun. 13, 810 (2022).
Myklebust, L. M. et al. Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Hum. Mol. Genet. 24, 1956–1976 (2015).
Magin, R. S., March, Z. M. & Marmorstein, R. The N-terminal Acetyltransferase Naa10/ARD1 Does Not Acetylate Lysine Residues *. J. Biol. Chem. 291, 5270–5277 (2016).
Gaido, Reyes et al. An improved reporter identifies ruxolitinib as a potent and cardioprotective CaMKII inhibitor. Sci. Transl. Med. 15, eabq7839 (2023).
Concordet, J.-P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
Wang, G. et al. Efficient, footprint-free human iPSC genome editing by consolidation of Cas9/CRISPR and piggyBac technologies. Nat. Protoc. 12, 88–103 (2017).
Liu, X. et al. Increased reactive oxygen species-mediated Ca2+/calmodulin-dependent protein kinase II activation contributes to calcium handling abnormalities and impaired contraction in Barth syndrome. Circulation 143, 1894–1911 (2021).
Tohyama, S. et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 12, 127–137 (2013).
Prondzynski, M. et al. Efficient and reproducible generation of human iPSC-derived cardiomyocytes and cardiac organoids in stirred suspension systems. Nat. Commun. 15, 5929 (2024).
Ma, J. et al. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am. J. Physiol. Heart Circ. Physiol. 301, H2006–H2017 (2011).
Wu, J. et al. L-Type Calcium Channel Inhibition Contributes to the Proarrhythmic Effects of Aconitine in Human Cardiomyocytes. PLoS One 12, e0168435 (2017).
Verkerk, A. O. et al. Patch-Clamp Recording from Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes: Improving Action Potential Characteristics through Dynamic Clamp. Int. J. Mol. Sci. 18, 1873 (2017).
Yoshinaga, D. et al. Phenotype-Based High-Throughput Classification of Long QT Syndrome Subtypes Using Human Induced Pluripotent Stem Cells. Stem Cell Reports 13, 394–404 (2019).
Sarantopoulos, C., McCallum, J. B., Kwok, W.-M. & Hogan, Q. Beta-escin diminishes voltage-gated calcium current rundown in perforated patch-clamp recordings from rat primary afferent neurons. J. Neurosci. Methods 139, 61–68 (2004).
Clements, I. P. et al. Optogenetic stimulation of multiwell MEA plates for neural and cardiac applications. in Clinical and Translational Neurophotonics; Neural Imaging and Sensing; and Optogenetics and Optical Manipulation (eds. Madsen, S. J. et al) 9690 96902C (SPIE, 2016).
Thomas, L. S. V. & Gehrig, J. Multi-template matching: a versatile tool for object-localization in microscopy images. BMC Bioinformatics 21, 44 (2020).
Vandenburgh, H. et al. Drug-screening platform based on the contractility of tissue-engineered muscle. Muscle Nerve 37, 438–447 (2008).
Hanna, R. et al. In-depth characterization of apoptosis N-terminome reveals a link between caspase-3 cleavage and posttranslational N-terminal acetylation. Mol. Cell. Proteomics 22, 100584 (2023).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Rodrigues, C. H., Pires, D. E. & Ascher, D. B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 46, W350–W355 (2018).
Acknowledgements
We would like to thank Fujian Lu for supplying the adenovirus vector, Ping-Zhu Zhou for providing a mammalian expression vector, Joao Paulo from Harvard Medical School for technical advice on LC/MS, and Ross Tomaino from Taplin Mass Spectrometry Facility at Harvard Medical School for LC/MS. We would also like to thank Michael Rosnach from the Parker Lab for assistance with figures and schematics. V.J.B. was supported by funding from the National Institutes of Health K08HL140197 and 5R01HL164710. D.Y. was supported by funding from JSPS Overseas Research Fellowships, the Uehara Memorial Foundation, and the Mochida Memorial Foundation for Medical and Pharmaceutical Research.
Author information
Authors and Affiliations
Contributions
Conceptualization, D.Y. and V.J.B.; Methodology, D.Y. and V.J.B.; Investigation, D.Y., I.C., R.F., M.P., K.S., Y.T., J.M., J.M., D.W., R.B., C.K.C., B.B., S.U., and V.J.B.; Writing—Original Draft, D.Y. and V.J.B.; Writing—Review & Editing, M.P., K.S., K.K.P., W.T.P., and V.J.B.; Analysis, D.Y., J.M., and V.J.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yoshinaga, D., Craven, I., Feng, R. et al. Dysregulation of N-terminal acetylation causes cardiac arrhythmia and cardiomyopathy. Nat Commun 16, 3604 (2025). https://doi.org/10.1038/s41467-025-58539-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-58539-2