Abstract
Native cells possess membrane-bound subcompartments, organelles, such as mitochondria and lysosomes, that intercommunicate and regulate cellular functions. Extensive efforts are directed to develop synthetic cells, or protocells, that replicate these structures and functions. Among these approaches, phase-separated coacervate microdroplets composed of polymers, polysaccharides, proteins, or nucleic acids are gaining interest as cell-mimicking systems. Particularly, compartmentalization of the synthetic protocell assemblies and the integration of functional constituents in the containments allowing signaling, programmed transfer of chemical agents, and spatiotemporal controlled catalytic transformations across the protocell subdomains, are challenging goals in developing artificial cells. Here, we report the assembly of compartmentalized, phase-separated cyanuric acid/polyadenine coacervate microdroplets. Hierarchical, co-centric compartmentalization is achieved through the dynamic and competitive spatiotemporal occupation of pre-engineered barcode domains within the polyadenine microdroplet framework by invading DNA strands. By encoding structural and functional information within these DNA-invaded compartments, the light-triggered, switchable reconfiguration of compartments, switchable catalytic reconfiguration of containments, and reversible aggregation/deaggregation of the compartmentalized microdroplets are demonstrated.
Similar content being viewed by others
Introduction
Recent research efforts are directed towards the development of synthetic containments emulating the structure and functions of native cells, protocells1,2. Diverse microstructures, such as liposomes3,4, polymersomes5,6, dendrosomes7, microcapsules8, were suggested as model systems for native cells. Within these efforts, phase-separated coacervate microdroplets consisting of polymers9, polysaccharides10, proteins11 or nucleic acids12 attract growing interest as cell-mimicking containments. The encapsulation of payloads, such as drugs or enzymes in these reservoirs was applied for controlled drug delivery8,13 and biocatalysis14,15,16. The assembly of nucleic acid-based phase separated microdroplet frameworks is particularly promising due to the structural and functional information embedded in nucleic acid scaffolds17,18. This includes dictated base-paring controlled hybridization and strand displacement processes19,20, the triggered switchable reconfiguration of DNA strands by auxiliary stimuli, such as the formation and dissociation of G-quadruplexes21,22, metal-ion/ligand bridged stabilization and dissociation of mismatched strands23,24, the triggered formation and separation of aptamer-ligand complexes25 or the light-induced stabilization/destabilization of duplex nucleic acids by photoisomerizable intercalators26,27,28, providing means to control the structures of the phase-separated microdroplets. Furthermore, integration of catalytic nucleic acids, DNAzymes29,30, or enzymes, such as nickase, endonuclease or ligases31,32,33 within the nucleic acid microdroplets introduce means to control the sizes, shapes and stiffness of the containments. Evidently, phase-separated microdroplets could find broad applications as sensors34, and confined microenvironments for catalysis14. Indeed, different methods to assemble nucleic acid-based phase-separated microdroplets were reported, including the crosslinking of four-arm or three-arm nanostar subunits, modified at their ends with self-complementary single strand tethers35,36. The effects of the structure of the nanostar constituents on the sizes and shapes of the microdroplets and their subsequent phase transitions into dispersed states or hydrogels were explored37,38. In particular, the interaction of the microdroplets with auxiliary enzymes resulting in the temporal growth of vacuole droplets, revealing dynamic motility39, or temporal degradation was reported35. Moreover, the separation of crosslinked phase-separated microdroplets by auxiliary nucleic acid strands, e.g., miRNAs allowed the use of the microdroplets as functional sensing containments34. Also, the ligase-stimulated polymerization of two pre-engineered nucleic acid reaction modules, exhibiting complementary tethers, led, in the presence of ATP and an endonuclease enzyme, to the phase-separated assembly of microdroplets accompanied by the catalyzed dissipative separation40. An alternative method to assemble microcompartmentalized DNA condensates included the dynamic oligomerization of lipophilic cholesterol-modified barcode-functionalized cross-shaped DNA core units and the controlled invasion of the framework with patterning strands of variable lengths41. A dynamic reaction-diffusion mechanism was suggested to guide the patterned compartmentalization of the condensates. Beyond the phase-separation of DNA-based coacervates, the triggered assembly of crosslinked oligonucleotide frameworks, and particularly oligoadenine frameworks42, was addressed. These included the pH-stimulated aggregation of oligoadenine frameworks43 and the crosslinking of rosette-stabilized oligoadenine/cyanuric acid complexes into hydrogel matrices44,45. In addition, formation of oligoadenine/cyanuric acid fibril structures46, and their photo-triggered pH-induced, dynamic reconfiguration into dissipative microtubules was reported47.
Native cells include in their envelope membrane sub-membrane protected compartments or organelles, such as mitochondria or lysosomes in which fuel (ATP)-generating machineries or cell signaling apoptotic processes proceed48,49. Other membrane-less intracellular compartments emerging from macromolecular liquid-liquid phase separation were reported50. Examples include nucleoli which generate ribosomes in the nucleus51, centrosomes nucleating microtubules52, Cajal bodies generating spliceosomes53 or stress granules formed at different cellular stress conditions54. The cell functions are, then, driven by programmed spatiotemporal communication between the cell sub-constituents and compartments, involving signaling, programmed transfer of chemical agents, and progress of dictated catalytic processes. These complex cellular features are, as yet, unprecedented in artificial model systems and emulating such spatiotemporal processes in protocells is a challenging holy-grail.
In the present study, we introduce the assembly of oligoadenine/cyanuric acid (CA) supramolecular triplex-crosslinked soft condensed DNA microdroplets (CDMDs) and their use as functional protocells for the assembly and dynamic intercommunication of hierarchical sub-microcompartments. The dynamic, kinetically-controlled hierarchical hybridization of invading strands with barcode tethers associated with the polyadenine/CA condensates leads to the patterned compartmentalization of the microdroplets. A kinetic model assuming a competitive association/dissociation invasion of the strands, resulting in cooperative patterning of the compartmentalized microdroplets, is provided. By engineering of photoactive ortho-nitrobenyl phosphate protecting groups55 into the invading strands, light-triggered dynamic reconfiguration of microcompartments compositions through strand displacement processes are demonstrated, and photoinduced dynamic and switchable transitions between single microcompartmentalized CDMDs and bi-microcompartmentalized CDMDs are demonstrated. Moreover, by engineering of functional duplex invading strands coupled to light responsive fuel strands, cyclic switchable light-induced aggregation/deaggregation of bi-microcompartmentalized and single microcompartmentalized CDMDs are demonstrated. Also, by the light-triggered reconfiguration of the microdroplet compartments, catalytic DNAzymes are formed, leading to sequestered catalytically-dictated, hierarchically-guided, reconfigured compartmentalized microdroplet. Moreover, the assembly of DNAzyme-functionalized CDMDs is exploited to develop a cyclic switchable, DNAzyme-guided reconfiguration of the single/double microcmompartmentalized condensates.
Results and discussion
Assembly of phase-separated microdroplets
Figure 1A depicts schematically the self-assembly of phase separated triplex-stabilized cyanuric acid (CA)/polyadenine (pA) crosslinked condensed DNA microdroplets (CDMDs). Annealing of polyadenine strands tethered to barcode nucleic strands (pA-Bs) leads, in the presence of a delicate concentration balance of Mg2+ ions, CA and an optimized adenine base concentration in the pA strand, Fig. 1A, Panel I, to the formation of pA-Bs DNA condensates in a continuous aqueous environment. (For conserving the integrity of the strands upon formation of the CDMDs, see Figure S1, and accompanying discussion.) The preparation process of the condensed DNA microdroplets is schematically depicted in Panel II and involves the primary formation of rosette CA-stabilized pA-triplexes45,46, followed by their aggregation into fibers and their crosslinked coagulation into CDMDs (The three assembly steps are further detailed in Figure S2 and accompanied discussions.). For circular dichroism (CD) and scanning electron microscope (SEM) imaging following the triplex CA/pA-Bs and their fibril/coagulation, see Figure S3A and Figure S3B and accompanying discussion. Panel III depicts the phase diagram showing the threshold concentrations and concentration ranges of the constituents at which the soft DNA condensate microdroplets, CDMDs, are formed. The confocal microscopy image of the resulting fibers generated by the coagulated pA-Bs strand (pA-22 bases) is displayed in Fig. 1A, Panel IV and the confocal microscopy image of the CDMDs is shown in Fig. 1A, Panel V. The yield of the CDMDs corresponded to ca. 90% and the average size of the CDMDs corresponded to ca. 12 µm. (For the method estimating the yield of CDMDs and the histogram of size distribution of the CDMDs, see Figure S3C). For further discussion on the feature of the CDMDs, see Figure S4 and accompanying discussion.
A Panel I and II-Schematic phase-separation of pA-Bs-tethered strand and CA into CDMDs through the assembly of pA-Bs/CA triplex structures, their oligomerization into fibril intermediates that aggregate into the Bs-tethered pA/CA CDMDs. Panel III-Phase diagram corresponding to the concentrations of the constituent generating the CDMDs. Panel IV-Confocal brightfield image of the intermediate pA-Bs/CA fibrils. Panel V-Confocal bright-field image of the phase-separated CDMDs. (Indistinguishable images were obtained in n = 3 independent experiments of Panels IV and V) B Panel I-Schematic dynamic invasion of the Bs1-tethered CDMDs with red fluorophore-labeled S5. Panel II-Schematic dynamic invasion of the Bs1-functionalized CDMDs with blue fluorophore-labeled S22. Panel III-Competitive dynamic invasion of the Bs1 CDMDs with S5 and S22. C Temporal dynamic invasion of the Bs1-tethered CDMDs with: Panel I-red fluorophore-labeled S5 strand; Panel II-blue fluorophore-labeled S22 strand; Panel III-competitive invasion with red fluorophore-labeled S5 and blue fluorophore-labeled S22 (Indistinguishable images were obtained in n = 3 independent experiments). Parts of Fig. 1A, Panel I were generated using Servier Medical Art, provided by Servier, licensed under CC BY 4.0.
Temporal compartmentalization of microdroplet by invading strands
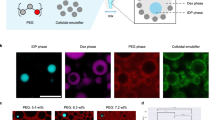
The resulting pA-Bs coagulated CDMDs are functionalized with the barcode Bs strand, Bs1 composed of a pre-engineered sequence that can hybridize with auxiliary invading strands composed of variable lengths of complementary base sequences. The dynamic interaction of the barcode-functionalized CDMDs with different fluorophore-modified invading strands, and the kinetically-controlled spatiotemporal formation of confined domains in the CDMDs is exemplified in Fig. 1B. Subjecting the CDMDs to the red-fluorescent Cy5-labled invading strand S5 (5-complementary bases to the barcode Bs1) results in rapid occupation of the entire CDMDs volume through hybridization to the barcode tether, Panel I. Subjecting the CDMDs to the blue-fluorescent Cy3-labled invading strand S22 (composed of 22-complementary bases to the barcode tethers) leads, however, to a substantially slower spatiotemporal occupation of the CDMDs, Fig. 1B, Panel II. After a short time scale, only a narrow rim confined compartment is formed. The compartment is progressively extended, and the CDMDs are fully invaded by the S22 strand only after 72 h. Subjecting the CDMDs to the mixture of invading strands S5 and S22 results in the kinetically controlled competitive hybridization of the invading strands with the barcode tethers, leading to the spatiotemporal compartmentalization of the CDMDs consisting of the rapidly occupied S5-functionalized core CDMDs surrounded by the slower S22 stabilized domain (blue fluorescence), Fig. 1B, Panel III. Figure 1C depicts the temporal occupation of the CDMDs by the short hybridization (S5) invading strands, Panel I, long hybridization (S22) invading strands, Panel II, and the mixture of S5 and S22, Panel III. While the short hybridization S5 saturates the CDMDs within 30 min, the long hybridization S22 invading strand saturates the CDMDs within a timescale of 72 h. That is, the equilibration of the barcode domain is strongly affected by the hybridization lengths of the invading strands with the barcodes. Subjecting the CDMDs to the mixture of the two strands results in, however, the rapid exchange of the fast generated S5 compartment with the long hybridization strand (S22), and the CDMDs are fully saturated by S22 within a time-interval of 104 min, (as compared to the 72 h shown in Panel II). That is, the mixture of the short/long hybridization invading strands results in the short strand enhanced occupation of the CDMDs by the S22. (For the quantitatively temporal invading rates of the long hybridization sequence S22 and the short hybridization sequence S5 and the competitive invading rates of the strands S22 and S5, resulting in the enhanced invading of the CDMDs by S22 and the displacement of S5, see Figure S5.) This enhancement is attributed to the unique toehold-mediated strand displacement phenomena19,20,56, as schematically depicted in Figure S6. While the equilibration of the short strand S5 with the barcode domains is fast, allowing the rapid occupation of the CDMDs, the equilibration of the long hybridization strand with the complementary barcode domains is slow (no toehold-mediated equilibration). Subjecting the CDMDs to the mixture of the two invading strands allows the rapid toehold-mediated and propagated exchange of S5 with the S22, as schematically presented in Figure S6. The S5 and S22 strands exhibit a common base-sequence complementary to the barcode sequence. As a result, the short hybridization strand S5 rapidly occupies the barcode region, allowing the subsequent interaction with the long hybridization strand S22 while facilitating the toehold-mediated propagation process, leading to the propagated invasion by the S5 and exchange of S5 by S22, forming an energetically stabilized duplex, within a relatively short time-interval. For further experiments and discussion supporting the suggested mechanism, see Figure S7, Figure S8, Figure S9 and accompanying discussions. For a detailed kinetic model evaluating quantitatively the temporal pattern formation by the competitive invasion of the CDMDs, see Figure S10 ~ Figure S22, Table S6 ~ Table S10 and accompanying discussion. It should be noted, however, that upon removal of the invading strands S5 and S22 from the surrounding solution of the CDMDs, the spatiotemporal, kinetically-dictated, compartmentalized CDMDs retain metastable two compartment configurations for a time-interval of two days, after which slow dynamic exchange of invading strands S5 and S22 proceeds, leading after 3 days to scrambled mixture of the invading strands within the droplets. (For further experiments probing the dynamic exchange of the strands within the S5/S22 strand compartment, see Figure S23 and accompanying discussion. Also, for enzyme-promoted strand invasion of the CDMDs, see Figure S24 and accompanying discussion, supporting information).
The spatiotemporal kinetic control over hierarchical confined compartments in the CDMDs guided by the hybridization lengths of the invading strands and the possibility to retain the compartmentalized structures by washing-off of the solution solubilized invading strands, suggested, that these functions can be employed to enhance the functional complexity of the microcompartments. Specifically, the kinetic control over the structure and composition of the invading strands and the time-intervals allowing the dynamic exchange of microcompartments, can be utilized to tailor emerging functionalities, Fig. 2. This is exemplified in Fig. 2A with temporally, dynamically controlled hierarchical assembly of different compartmentalized CDMDs. Subjecting the Bs1-functionalized phase-separated CDMDs to four strands, S5 (modified with the red fluorescence-Cy5), S10 (modified with the green fluorescence-FAM), S15 (invading strand lacking any fluorophore) and S22 (modified with the blue fluorescence-Cy3), leads to competitive hierarchical dynamic compartmentalization of the CDMDs. After a time-interval of 20 min, the kinetically, hybridization-guided hierarchical four compartment CDMDs were formed. After 40 min, the green invading strands displace the red fluorescent core strand due to enhanced hybridization efficiency, leading to green/black/blue compartmentalized CDMDs. After 50 min, the non-fluorescent invading strands S15 displace the green-fluorescent strand to yield the S15/S22 two-compartment CDMDs, and after a time-interval of 65 min, the S22 invading strands displace the less stable S15/barcode hybrid to yield the most stable S22/barcode hybrid CDMDs, for the zoom out images and quantitative temporal formation of the respective compartmentalized CDMDs, see Figure S25. Upon removal of the invading strands from the surrounding solution of the CDMDs, the spatiotemporal, kinetically-dictated, quadra-confined domains of the CDMDs retain a metastable four-compartment configuration for a time-interval of 2 days, after which slow dynamic exchange of invading strands S5 and S10 proceeds, leading after 3 days to tri-confined domains within the droplets, see Figure S26. As stated, each of these metastable containment structured CDMDs configuration can be isolated for a time interval of ca. 2 days by exclusion of the invading auxiliary strands from the bulk surrounding solution. The complexity of compartmentalized CDMDs is even further enhanced by extending the length of the barcode tethers associated with the coagulated CDMDs, Bs2, and applying a mixture of seven invading strands exhibiting controlled hybridization efficacies for spatiotemporal guided hierarchical, formation and dynamic formation of the compartmentalized domains, Fig. 2B. The spatiotemporal formation of a seven compartment nanostructured CDMD after challenging the CDMDs with the seven invading strands in 20 min is observed, Panel II, and the 3D reconstructed confocal microscopy image clearly shows the seven co-centered structure within CDMDs, Figure S27, followed by the dynamic reconfiguration to the six-compartment CDMDs consisting of the S15 core surrounded by co-centered S22 (blue), S27 (red), S35 (green), S42 (black) and S50 (blue) compartments, after 30 min, the five hierarchical structured CDMDs composed of the blue core of S22 surrounded by the green S35, black S42 and blue S50 compartments, generated after a time-interval of 45 min, the four co-centered structure composed of the red S27 core, surrounded by the green S35, black S42 and blue S50 compartments, after a time-interval of 60 min, and finally the blue S50 stabilized CDMDs formed after 100 min. (For the spatiotemporally and quantitatively dynamic reconfiguration of the hepta-confined compartments, see Supplementary Movie 1, Figure S28, Figure S29 and accompanying discussion.) It should be noted that the microcompartmentalized patterned CDMDs structures, and all other patterned CDMDs described in the study, were reproduced in three (or more) independent replicated experiments with an accuracy of <5%.
A Panel I-Schematic competitive assembly and reconfiguration of compartmentalized CDMDs using four different invading strands.Panel II-Temporal fluorescence confocal microscopy images of compartmentalized CDMDs generated by the dynamically-modulated competitive interaction of four invading strands with the pA-Bs1 barcoded phase-separated droplets (Indistinguishable images were obtained in n = 3 independent experiments). B Panel I-Schematic competitive assembly and reconfiguration of compartmentalized CDMDs using seven different invading strands. Panel II-Temporal fluorescence confocal microscopy images of compartmentalized CDMDs generated by the dynamically-modulated competitive interaction of seven invading strands with the pA-Bs2 barcoded phase-separated droplets (Indistinguishable images were obtained in n = 3 independent experiments).
The advances of the present study as compared to the previous art describing invasive patterning of microdroplet condenstaes41 should be noted. Beyond introducing an alternative compartmentalization mechanism involving a competitive association/dissociation hopping invasion process, dynamic functionalities associated with the microcompartmentalized condensates are addressed. These include light-triggered dynamic exchange of constituents between the compartments, and emerging switchable catalytic DNAzyme functions within the microcompartments, processes emulating native cell properties in the protocells.
Light-triggered switchable reconfiguration of CDMDs
The dynamic reconfiguration of multi-compartment CDMDs, guided by the duplex stabilities of the invading strands with the barcodes, suggested that encoding triggering elements into the strands could enhance the spatiotemporal programmability of the compartments. This is exemplified in Fig. 3 with the light-triggered dynamic reconfiguration of metastable multi-structured containments CDMDs. In this system, auxiliary light signals trigger the compartmentalized CDMDs to perform a strand-guided dynamic reconfiguration into a new metastable structure. The phase-separated pA-Bs1 CDMDs were challenged with three invading strands that included each an o-nitrobenzyl phosphate ester bridging photocleavable functionality37, where one invading strand, P10, is labeled with a blue fluorophore and a 10-base complementary domain towards the Bs1 strand, the second invading strand consists of a photocaged hairpin P15 functionalized with a red fluorescence label and a tether composed of 15-complementary bases to the barcode tethers Bs1, and the third invading strand P22 consists of a photocaged hairpin modified with the green fluorescent probe and a tether composed of 22-base complementary to the barcode Bs1. Subjecting the CDMDs to the three invading constituents results in the competitive hierarchical formation of the hybridization-guided three-compartment CDMDs composed of the blue fluorescence modified core of P10 surrounded by the red P15-modified containment and the exterior green fluorescent P22-labeled containment, state K. The confocal fluorescence microscopy images of the hierarchical three containments CDMDs using the respective fluorescence channels and the merge image are displayed in Fig. 3B, confirming the formation of the parent three-compartment structure Bs1-P10/P15/P22, state K. Irradiation of the CDMDs in state K with UV light, λ = 365 nm, unlocks all photoprotective o-nitrobenzyl phosphate ester units yielding the Bs1-P10 fragment consisting of the free blue fluorescence labeled strand P10’, the fragmented Bs1-toehold modified strand P15’ hybridized with the red fluorescent strand R15, and the fragmented Bs1 toehold-modified strand P22’ hybridized with the green fluorescent-labeled strand R22. The released blue fluorescent strand P10’ is, however, pre-engineered to displace R15 associated with the parent red fluorescent compartment to yield the blue fluorescent structure P10’-P15’-Bs1 as the second reconfigured compartment. The R15 displaced strand is engineered, however, to displace the toehold-modified exterior compartment R22-P22’-Bs1 to yield the exterior compartment composed of the red fluorescent R15-P22’-Bs1 structure, where the released green fluorescent label acts as background fluorescent constituent. That is, the UV irradiation of the photocaged blue-red-green fluorescent compartmentalized structure, state K, undergoes a dynamic reconfiguration into a three compartments structure consisting of a non-fluorescent core (black) surrounded by blue fluorescent compartment (of P10’-P15’-Bs1) that is further surrounded by the green fluorescent compartment (R15-P22’-Bs1), state L. The light induced dynamic transitions of the three compartments MD structure in state K to state L were imaged by confocal fluorescence microscopy, Fig. 3B. Imaging the CDMDs by the respective channels reveals that upon irradiation of CDMDs in state K the core MD compartment turned from blue to non-fluorescent, the surrounding green fluorescent confinement turned to a red fluorescent compartment (comprised of the P10’-P15’-Bs1 structure) and a green exterior compartment, (exchanging the red fluorescent compartment with the R15-P22’-Bs1 structure).
A Schematic competitive assemble of hierarchical compartmentalized CDMDs composed of three photo-protected invading strands and the photo-triggered dynamically-modulated reconfiguration of the compartmentalized CDMDs. B Confocal fluorescence microscopy images of the CDMDs generated upon subjecting the pA-Bs1 CDMDs to the three fluorophore-labeled photo-protected invading strands for a time interval of 30 min, in the absence of external irradiation. Images correspond to the fluorescence channels of the fluorophore labels associated with the invading strands and the respective merge image. Fluorescence microscopy images of the UV (λ = 365 nm) irradiated deprotected CDMDs (3 min of irradiation) allowing to dynamically reconfigure for a time-interval of 10 min. Images correspond to the fluorescence channels of the fluorophore labels associated with the compartment constituents and the respective merge image (Indistinguishable images were obtained in n = 4 independent experiments).
The photo-triggered reconfiguration of the microcompartments associated with the CDMDs was then applied to develop light-stimulated switchable compartmentalization of the microdroplets as outlined in Fig. 4. The barcode-functionalized CDMDs were subjected to the duplex S1/L1 (20 bp), where strand S1 is modified with the blue fluorescent fluorophore (Cy3) and L1 is functionalized with the red fluorescent probe (Cy5). The toehold sequence x associated with S1 acts as an invading strand recognizing the barcode Bs1 and the L1-strand associated with the duplex domain includes an overhang that upon uncaging yields a 10 nt invading strand domain. Invasion of the CDMD with S1/L1 yields a microcompartment exhibiting violet fluorescence at the boundary of the CDMD (overlay of blue + red fluorescence), Fig. 4A. Interaction of S1/L1-Bs1 compartmentalized CDMDs with the bis-o-nitrobenzyl phosphate photoresponsive fuel strand F1 that is fully complementary to S1, being a part of the S1/L1-Bs1 duplex associated with the CDMD boundary, results in the toehold-mediated displacement of L1, leading to the blue fluorescent boundary of F1/S1-Bs1. The displaced strand L1 includes, however, a short invader strand complementarity to the barcode resulting in a bi-compartment CDMD revealing outer blue fluorescence and inner red fluorescence. Photodeprotection of the o-nitrobenzyl phosphate ester caging units results in the formation of fragmented F “waste” strands, being displaced and stabilized by the retro-hybridization of L1 with S1-Bs1, recovering the violet fluorescent microcompartment consisting of the S1/L1-Bs1 outer functionalized boundary. (Note that the fuel displacement of S1-L1-Bs1 yields the outer compartment comprising blue-fluorescent F1-S1-Bs1 unit and the released red fluorescent fragment L1 acting as short 10 nt invading strand forming the inner red fluorescent compartment. The photo-deprotection of the outer compartment F1-S1-Bs1 fragment constituent F1 into “waste” product and generates a hybridization vacancy allow the formation of a 20 base pair stabilized blue/red fluorescent constituent S1-L1-Bs1 comprising the outer compartment. That is, the blue/red bi-compartmentalized intermediate CDMDs dynamically recover the parent single compartment, violet fluorescence, due to enhanced hybridization of L1 to the outer compartment.) By the cyclic treatment of the S1/L1-Bs1 microcompartmentalized CDMDs with the fuel strand F1 followed by photodeprotection of F1, a switchable dynamic reconfigured CDMDs composed of violet single compartmentalized CDMDs and blue/red bi-compartmentalized CDMDs are formed, Fig. 4A. Figure 4B depicts the fluorescence confocal microscopy images of the S1/L1-Bs1 functionalized CDMDs subjected to cyclic interactions with the fuel strand F1/light deprotection signal followed by treatment with F1. The dynamically switchable red fluorescence intensities of the inner L1-modified microcompartment of the CDMDs are displayed in Fig. 4C.
A Switchable F1-strand triggered reconfiguration of single microcompartmentalized CDMDs invaded domains consisting of the S1-L1 (blue-red) duplex invading strands, state M, into the red and blue bi-compartmentalized patterned CDMDs composed of blue F1-S1 and Red-L1, state N, and the light-stimulated recovery of state N to state M by photocleavage of the light-responsive F1 strand in state N. B Confocal fluorescence microscopy images of the microcompartmentalized CDMDs upon cyclic treatment of the S1-L1 modified condensates with F1 and UV light (λ = 365 nm) (Indistinguishable images were obtained in n = 4 independent experiments). C Switchable red-fluorescence intensities of the inner patterned domain upon reversible treatment with F1 or light triggers. Data are presented as mean values ± SD, error bars are derived from n = 4 experiments.
Light switchable aggregation/deaggregation of CDMDs
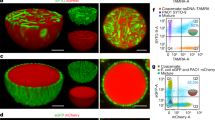
Furthermore, the concept of switchable light-induced reconfiguration of the single compartmentalization condensates into bi-compartmentalized particles and back, was extended to stimulate a light-induced switchable aggregation/deaggregation of the DNA condensates, accompanied by dynamically reconfigured single microcompartments in the separated particle and bi-microcompartmentalized domains in the aggregated condensates. This is exemplified Fig. 5A. The barcode (Bs1) of polyA/CA condensates are interacted with the blue fluorophore/red fluorophore labeled duplex invading strand L2/S2 to yield single microcompartmentalized condensates, state P. The photoresponsive fuel strand F2 leads to displacement of the S2/L2-Bs1 supramolecular structure yielding the F2-S2-Bs1 duplex strand, labeled with the blue fluorophore, and to the separated F2-L2 duplex acting as an inactive strand that occupies inner barcode domains of the condensate. As a result, a bi-microcompartmentalized particle is formed composed of an exterior domain of S2-Bs1 (blue) and an inner domain of the shorter invading duplex F2-L2-Bs1 (red) domain (condensates in state Q). The S2 strand associated with the exterior invading strand S2-Bs1 is, however, pre-engineered to include a self-complementary sequence, leading to the aggregation of the condensates in state Q. Photochemical deprotection of the caging units associated with F2 leads to fragmented waste strand while separating the strand L2. The released strand L2 rehybridizes with the invading strand S2 and concomitantly separates the self-complementary S2-Bs1 bridging duplex, leading to the parent single microcompartmentalized condensates, state P. Thus, by treatment of condensates in state P with the fuel strand F2 and subsequent UV irradiation, cyclic transitions between the aggregated bi-compartmentalized condensates and the separated single-compartmentalized particles proceeds. Figure 5B depicts the confocal fluorescence images of the reversibly switched microcompartmentalized violet condensate and the blue/red bi-compartmentalized aggregated CDMDs. Figure 5C depicts the switchable average sizes of the condensate undergoing aggregation of the condensates and back (i), and the concomitant red fluorescence intensities upon switchable transition between the separated condensates in state P and the aggregated condensates in state Q, (ii). Beyond demonstrating the light-induced switchable dynamic reconfiguration of microcompartmentalized condensate protocells, the system introduces accompanying dynamic switchable aggregation/deaggregation functions of the protocells.
A Formation of separated single microcompartmentalized CDMDs by invasion of the barcode modified CDMDs with blue/red duplexes S2-L2, state P. Subjecting CDMDs in state P to photoresponsive fuel strand F2 results in the displacement of S1-L2-Bs1 units leading to the aggregation of CDMDs through self-complementary domain of S2 accompanied by the reconfigured red and blue patterned bi-compartmentalized in the aggregated CDMDs, state Q. UV light-degradation of F2 in the aggregated CDMDs lead to the dynamic separation of the CDMDs into revealing single patterned microcompartments. B Confocal fluorescence microscopy images corresponding to the formation of separated CDMDs, state P, by invading the CDMDs with strand S2-L2, and the switchable patterned bi-compartmentalized blue/red aggregated CDMDs, state Q, separated into the single patterned microcompartmentalized CDMDs, state P (Indistinguishable images were obtained in n = 5 independent experiments). C Switchable size-changes (i) and red fluorescence changes (ii) of inner microcompartment domain upon reversible F2 and light-induced aggregation/deaggregation of the CDMDs. Data are presented as mean values ± SD, error bars are derived from n = 5 experiments.
Coupled light/DNAzyme-stimulated dynamic compartmentalization of microdroplets
Moreover, beyond the light-induced guided reconfiguration of the composition of CDMDs through dynamic toehold-mediated stand displacement transitions, catalytically guided reconfiguration of the compartments was realized. This is exemplified in Fig. 6 with the Mg2+-ion-dependent DNAzyme-stimulated dynamic reconfiguration of three microcompartmentalized patterned CDMDs. The Bs1 barcoded CDMDs are subjected to three invading strands consisting of a blue fluorophore labeled strand D1 conjugated to a short invading tether I1 through a photocaged unit, a hairpin D2 modified at one stem end with a red fluorophore and conjugated at the other stem end with a medium length invading strand I2, where the loop domain of the hairpin is caged by the o-nitrobenzyl phosphate-photocaging unit, and a third functional invading strand SD composed of the longest hybridization domain I3, complementary to the Bs1 barcode, tethered to a quencher (Q)-modified tether. The sequence I3 tethered to the quencher-modified strand is modified with the green fluorophore, yet the spatial proximity between the green fluorophore and the quencher leads to intra strand quenching of the green fluorophore. The fluorophore-quencher strand is further modified with a ribonucleobase rG. The hybridization lengths of the invading strands associated with the constituent D1, D2, SD leads, upon treatment of the CDMDs with the mixture of invading strands, to a three microcompartmentalized patterned CDMD assembly shown in Fig. 6A, state R. The deprotection of D1 yields D1’ lacking affinity to the core barcode compartment, yet it displaces strand Dz from the fragmented Dz-D2’-Bs1 compartment, where the displaced Dz reveals binding affinity towards the SD-Bs1 stabilized microcompartment. That is, immediately after exposure of the CDMDs in state R to UV light and deprotection of D1 and D2, state S is formed, where the core is non-fluorescent and the compartment D2’-Bs1 is displaced by the blue fluorescent strand surrounded by the red strand Dz, state S. The binding of Dz to the Dz-SD-Bs1 microcompartment assembles, however, the Mg2+-ion-dependent DNAzyme, resulting in the cleavage of the barcode-linked quenched constituent generating the Dz (red fluorescence)-SD’ (green fluorescence)-Bs1 microcompartment (merged yellow fluorescence), state T. These dynamic light-induced/DNAzyme-driven temporal reconfiguration of state R to state S and state T are imaged by confocal fluorescence imaging at time intervals of the transitions, Fig. 6B, Panel Y and Panels Z1 and Z2. Immediately, after photochemical deprotection of state R, the CDMDs in state S are formed, Fig. 6B, Panel Y. Subsequently, the temporal DNAzyme driven transition of the CDMDs in state S to state T is observed, Fig. 6B, Panels Z1 and Z2. (After 10 min of deprotection, full red and partial green fluorescence in the exterior microcompartment is observed, and after 60 min a full yellow fluorescence of the merged red/green fluorescence of the constituents is detected.)
A Competitive compartmentalization of CDMDs with two photocaged fluorophore-labeled strands D1, D2 and a fluorophore-quencher-modified strand SD, yielding CDMDs in state R. The light-induced deprotection of state R dynamically reconfigure CDMDs in state R to the compartmentalized CDMDs in state S that undergoes reconfiguration into Mg2+-ion-dependent DNAzyme-modified compartmentalized CDMDs undergoing catalyzed dynamic reconfiguration into CDMDs of state T. B Temporal fluorescence confocal microscopy images following the dynamic reconfiguration of the compartmentalized CDMDs upon the light/DNAzyme guided transitions across state R (Panel X)→state S(Panel Y)→state T(Panels Z1 and Z2) (Indistinguishable images were obtained in n = 4 independent experiments).
Switchable DNAzyme-catalyzed reconfiguration of CDMDs
The DNAzyme-driven dynamic reconfiguration of the CDMDs microcompartments, was further applied to develop CDMDs revealing cyclic, switchable, DNAzyme-mediated reconfiguration of the patterned microcompartments, Fig. 7A. The barcode Bs1-modified CDMDs, were subjected to the duplex D3/E, where D3 is functionalized with the blue fluorescent dye (Cy3) and strand E is modified with the red fluorescent label (Cy5). Strand D3 hybridized with E includes a loop domain and a single-strand toehold tether t, as invading domain (22 bases). Interaction of the CDMDs with the duplex D3/E results in the t-invading strand-guided formation of an outer microcompartment and an inner vacant non-fluorescent core. The outer microcompartmentalized CDMDs composed of the D3/E blue/red fluorophore leads to an overlayed violet fluorescent microcompartment, state U. Treatment of the CDMDs in state U with the fuel strand F3 results in the displacement of strand E from the duplex D3/E-Bs1, generating free E and D3/F3-Bs1 as outer microcompartment. The released strand E labeled with the red fluorophore was pre-engineered to include a short invading sequence domain complementary to the barcode Bs1. As a result, the invading strand E migrates into an inner core domain forming bi-microcompartmentalized CDMDs, state V. The fuel strand F3 displacing strand E was, however, pre-designed to include a ribonucleobase, rA, that upon hybridization into the supramolecular loop duplex D3/F3-Bs1 structure yields the Pb2+-ion-dependent DNAzyme oligonucleotide complex. Subjecting CDMDs in state V to Pb2+-ion activates the Pb2+-ion-dependent DNAzyme cleavage of F3, leading to “waste” fragmented strands separated from constituent D3-Bs1. The free strand D3 induces then the separation of E from the inner compartment and the guided recovery of the energetically-stable single microcompartmentalized CDMDs, in state U. That is, by the cyclic treatment of CDMDs in state U with F3, dynamic reconfiguration of CDMDs in state U to state V proceeds and the subsequent treatment of CDMDs in state V with Pb2+-ion recovers state U. Figure 7B depicts the fluorescence confocal microscopy images of the CDMDs upon the cyclic F3/Pb2+-ion guided reconfiguration of CDMDs in state U (imaged by a single violet microcompartmentalized domain) to the bi-compartmentalized CDMDs, in state V, (imaged by distinct blue/red fluorescent compartments), and back. Figure 7C shows the cyclic fluorescence intensities of the inner microcompartment upon the F3/Pb2+-ion guided reconfiguration of CDMDs, state U, to CDMDs, state V and back.
A Switchable dynamic DNAzyme-mediated patterning of micro-compartmentalized CDMDs by invading the CDMDs with a blue/red fluorophore functionalized duplex D3/E. Subjecting the single patterned microcompartmentalized CDMDs to fuel strand F3 results in the displacement of D3/E and to the dynamic reconfiguration of the CDMDs, state U, into red and blue patterned bi-compartmentalized CDMDs, state V, where the constituent of the blue compartment is composed of a Pb2+-ion-dependent inactive DNAzyme loop constituent D3F3 and the red compartment includes the red strand E. Subjecting CDMDs in state V to Pb2+-ion activates the DNAzyme towards cleavage of F3 resulting the dynamic recovery of the single compartmentalized CDMDs in state U, through the rehybridization of E with strand D3. B Confocal fluorescence microscopy images corresponding to the formation of the single patterned microcompartment CDMDs, state U, through the invasion of the barcode modified condensates with D3/E. Subjecting the CDMDs in state U to F3 results in the patterned bicomaprtmentalized CDMDs in state V. Activation of the DNAzyme units in state V with Pb2+-ion results in the dynamic reconfiguration of state V to the single patterned microcompartmentalized CDMDs in state U (Indistinguishable images were obtained in n = 4 independent experiments). C Switchable red fluorescence intensities of the inner red patterned domain upon reversible treatment of the CDMDs with F3 and the subsequent Pb2+-ion-activated DNAzyme units transforming CDMDs in state V to state U. Data are presented as mean values ± SD, error bars are derived from n = 4 experiments.
In conclusion, the study introduced barcode-functionalized cyanuric acid/polyadenine triplex stabilized phase-separated microdroplets acting as reservoirs for the kinetically-controlled organization of fluorescent nucleic acid compartments, and eventually catalytically active microcompartments. The spatiotemporal compartmentalization of the microdroplets was controlled by the hybridization domain lengths associated with the nucleic acid sequences invading the droplets and the temporal dynamic toehold-mediated displacement and exchange of the nucleic acids comprising the compartments. Moreover, by applying o-nitrobenzyl phosphate-caged invading nucleic acid strands, the light-triggered and subsequent kinetically-controlled dynamic reconfiguration of the nucleic acid compartments was demonstrated. Furthermore, by engineering functional duplex invading strands and photoresponsive o-nitrobenzyl phosphate protected fuel strands, light-induced switchable microcompartmentalization of CDMDs was accomplished, and cyclic switchable, dynamic aggregation/deaggregation of single/double compartmentalized condensates was demonstrated. Particularly, the spatiotemporal compartmentalization of the microdroplets led to the assembly of catalytic DNAzymes in the compartments that stimulated sequential dynamic reconfiguration of the compartmentalized microdroplets. By appropriate engineering of the DNAzyme-modified CDMDs, the cyclic and switchable dynamic reconfiguration of microcompartmentalized condensates was achieved. The different dynamically modulated compartmentalized microdroplets emulate intracellular signaling pathways across submembrane organelles, such as mitochondria, lysosomes or liquid-liquid phase-separated entities. The study advances the development of protocell assemblies by providing means to tether enzymes, transcription machineries or receptor units to the compartments, thus enabling spatiotemporal control over biocatalytic cascades, gene expression or miRNA/siRNA agent formation in cell-like containment. At present, the study included single-type DNA condensates. By mixing different functional condensates, the intercommunication of CDMDs can be envisaged as models for intercellular signaling and communication.
Methods
Materials
Cyanuric acid (CA) was purchased from Sa’en Chemical Technology Co., Ltd. Sodium chloride (NaCl), Lead acetate (Pb(Ac)2) and magnesium chloride (MgCl2) were obtained from Sinopharm Chemical Reagents Co. Ltd, China., and Ribonuclease H (RNase H) were purchased from New England Biolabs (NEB). All chemical reagents utilized in the experiment were of analytical grade and did not undergo further purification. All nucleic acid sequences were obtained from Hippo Biotechnology Co., Ltd. (Beijing, China) and purified using high-performance liquid chromatography (HPLC). Detailed nucleic acid sequences are shown in Table S1-S5, supporting information. Quantification of all the nucleic acid sequences was performed using UV absorbance spectroscopy at a wavelength of 260 nm.
Buffer conditions
Unless otherwise specified, the solution for CDMDs preparation consisted of a TE buffer comprising 10 mM Tris(hydroxymethyl)aminomethane (pH=8.0) and 1 mM EDTA. In the washing steps and invading processes of CDMDs, a saline buffer containing 1 × TE, 100 mM NaCl, and 10 mM CA was employed, and the same buffer is used for switchable light-induced reconfiguration of CDMDs and DNAzyme-driven dynamic reconfiguration of the CDMDs. It is important to note that in magnesium or lead ion-dependent DNA enzyme experiments, the corresponding metal ions must be added to activate the DNA enzyme.
Synthesis of phase-separated CDMDs preparation
A standard protocol to prepare the phase-separated CDMDs was formulated. A mixture consisting of 10 mM CA, 30 mM MgCl2, and either 10 µM pA-Bs1 or 6.7 µM pA-Bs2 was prepared in a 200 µL Eppendorf tube, with a total volume of 30 µL. The samples were subjected to a temperature of 95 °C using a dry bath (Labnet International D1100 Accublock Digital) and incubated for 75 minutes until complete evaporation of the solution occurred. Subsequently, the samples were cooled from 95 °C to room temperature over a period of 2 hours. In order to isolate the phase-separated CDMDs that were formed, the solid residue obtained after evaporation was dissolved in ultrapure water (total 30 µL), resulting in the acquisition of a substantial quantity of uniformly dispersed CDMDs.
Purification of phase-separated CDMDs
Samples were subjected to a washing procedure wherein 150 µL of saline buffer was added to the Eppendorf tube housing the phase-separated CDMDs. The sample was allowed to stand for a duration exceeding 15 minutes, ensuring the settling of the phase-separated CDMDs at the bottom of the tube, following which 150 µL of the supernatant was removed. This washing process was performed three times for each sample. The purposes of washing the CDMDs were to eliminate any surplus components arising from uncertainties in concentration measurements and pipetting, and to eliminate excessive components employed during invading steps. This aspect holds significant importance in the context of switchable light-induced reconfiguration of CDMDs and DNAzyme-driven dynamic reconfiguration of the CDMDs, as an excessive amount of template can result in substrate cleavage occurring in solution, rather than being confined solely within the CDMDs.
Hierarchical confined compartmentalization of CDMDs
In the hierarchical four-compartment CDMDs experiment, phase-separated CDMDs were prepared using pA-Bs1 oligonucleotides according to the standard protocol and subsequently washed. These phase-separated CDMDs were utilized for the invading with four different DNA strands, S5 (modified with red fluorescence-Cy5), S10 (modified with green fluorescence-FAM), S15 (no modification), and S22 (modified with blue fluorescence-Cy3), which contain 5, 10, 15, and 22 complementary bases with barcode Bs1, separately. All strands were dissolved in 1 × TE buffer containing 10 mM MgCl2 and 10 mM CA. The final concentration of each single strand of DNA was 2 µM, except for the S5, which had a concentration of 4 µM. For the preparation of metastable four-compartment CDMDs, the excess invading strands were removed by washing the CDMDs three times to stop the temporally dynamic invasion.
In the hierarchical seven-compartment CDMDs experiment, phase-separated CDMDs were prepared using pA-Bs2 oligonucleotides in accordance with the established protocol, followed by thorough washing. Subsequently, these phase-separated CDMDs were employed for the purpose of invading with seven distinct DNA strands, S10, S15, S22, S27, S35, S42, and S50, containing 10, 15, 22, 27, 35, 42, and 50 complementary bases with barcode Bs2, separately. In details, S10 and S35 were subjected to modification using green fluorescence-FAM, S22 and S50 were modified with blue fluorescence-Cy3, and S27 was modified with red fluorescence-Cy5. While S15 and S42 were intentionally left unmodified (no fluorophore modification) to enable differentiation of neighboring compartments, as they exhibit similar fluorescence intensity to the background in confocal fluorescence microscopy images. These single strands were dissolved in 1 × TE buffer containing 10 mM MgCl2 and 10 mM CA, with a final concentration of 2 µM for each individual DNA strand. For the preparation of metastable seven-compartment CDMDs, the excess invading strands were removed by washing the CDMDs three times to stop the temporally dynamic invasion.
Light-activated dynamic reconfiguration of the three-compartment CDMDs
Oligonucleotide pA-Bs1 was employed in the preparation of phase-separated CDMDs following the standard procedure. Following the washing process, three distinct categories of invading strands were dissolved in 1 × TE buffer, supplemented with 10 mM MgCl2 and 10 mM CA, to facilitate the dynamic reconfiguration of multi-compartment stabilized metastable structure CDMDs. P10 single-stranded DNA was subsequently diluted to a concentration of 20 µM in 1 × TE buffer containing 10 mM CA and stored at a temperature of 4 °C for future utilization. The P15 and P22 hairpin DNA samples were diluted to a concentration of 5 µM in 1 × TE buffer and subjected to a rapid annealing program. This program involved heating the samples at 95 °C for 10 minutes, followed by immediate transfer to an ice bath for 15 minutes, and subsequent storage at 4 °C. The P10, P15, and P22 samples were then added to the phase-separated CDMDs, resulting in final concentrations of 4 µM, 2 µM, and 2 µM, respectively. Following competitive hierarchical formation of the hybridization-guided co-centered three hierarchical compartment CDMDs, the supernatant was carefully removed, and the phase-separated CDMDs were washed three times using 1 × TE buffer containing 10 mM CA, following the standard washing procedure for phase-separated CDMDs. Then, the samples were exposed to UV light (λ = 365 nm) irradiated for 3 minutes and subjected to laser scanning confocal microscopy images.
Switchable light-induced reconfiguration of the CDMDs
Oligonucleotide pA-Bs1 was used to prepare phase-separated CDMDs using a standardized procedure. After thorough washing, S1 and L1 single-stranded DNA were diluted 1:1 to a concentration of 4 µM in 1 × TE buffer containing 10 mM CA, and heated at 95 °C for 15 minutes before slowly lowering to room temperature for 2 h. The S1-L1 duplex was formed after annealing, and the annealed sample was stored at 4 °C. Similarly, the same procedure was performed on S2 and L2 to form the S2-L2 duplex. In washed, phase-separated CDMDs, annealing was added to form S1-L duplexes to a final concentration of 2 µM. After overnight equilibration, the supernatant was carefully removed and the phase-separated CDMDs was subjected to three wash steps using 1 × TE buffer supplemented with 10 mM MgCl2 and 10 mM CA, following the regular wash protocol. Subsequently, fuel DNA strand F1 or F2 was added, and after equilibration for 2 h, the supernatant was carefully removed and the phase-separated CDMDs was subjected to three washing steps using 1 × TE buffer supplemented with 10 mM MgCl2 and 10 mM CA, following for a conventional washing protocol, samples are exposed to UV light at a wavelength of 365 nm for 3 minutes and then collected at various time intervals for confocal microscopy imaging.
Dynamic reconfiguration of three-compartment CDMDs triggered by light and DNAzyme
Oligonucleotide pA-Bs1 was employed in the preparation of phase-separated CDMDs using a standardized procedure. Following a thorough washing, D1 and substrate SD single-stranded DNA were diluted to a concentration of 20 µM in 1 × TE buffer containing 10 mM CA and stored at 4 °C for subsequent utilization. Similarly, D2 hairpin DNA was diluted to a concentration of 5 µM in 1 × TE buffer and stored at 4 °C after undergoing a rapid annealing program. Within the phase-separated CDMDs, D1, D2, and SD were introduced to achieve final concentrations of 4 µM, 2 µM, and 2 µM, respectively. Following the establishment of a competitive assembly of three containment CDMDs, the supernatant was meticulously eliminated, and the phase-separated CDMDs were subjected to three washing steps using 1 × TE buffer supplemented with 10 mM MgCl2 and 10 mM CA, adhering to the conventional washing protocol. Subsequently, the samples were subjected to UV light irradiation at a wavelength of 365 nm for a duration of 3 minutes, and then incubated at a temperature of 37 °C to initiate DNAzyme cleavage and activate the temporal reconfiguration. Samples were collected at various time intervals for confocal microscopy imaging.
Similarly, D3 and E single-stranded DNA were diluted 1:1 to a concentration of 4 µM in 1 × TE buffer containing 10 mM CA and heated at 95 °C for 15 min and then slowly lowered to room temperature over 2 h. Annealing treatment, after annealing, store it at 4 °C for subsequent use. In the phase-separated CDMDs, introduce D3-E to reach 2 µM. After overnight equilibration, carefully remove the supernatant and perform three experiments on the phase-separated CDMDs. For washing steps, follow the regular washing protocol. Subsequently, fuel chain F3 at a final concentration of 2 µM was added, and samples were collected at different time intervals for confocal microscopy imaging. After D3-E fluorescence separation, Pb2+-ions at a final concentration of 500 nM were added to initiate Pb2+-ion dependent DNAzyme activity, and then Incubated at 37 °C, samples were collected at different time intervals for confocal microscopy imaging.
Note: Each of the experiments described in “Method” section was independently replicated for at least three times, and temporal fluorescence changes of the invading strands were recorded and analyzed. For each of these replicates, examples are provided in the supporting material, relevant experiment/all experiments were reproduced with an accuracy of 5% to the results presented in the main text.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data generated in this study are provided in the main text and Supplementary information, and from corresponding author(s) upon request. Source data are provided with this paper.
References
Buddingh’ BC, van Hest JCM. Artificial cells: synthetic compartments with life-like functionality and adaptivity. Acc. Chem. Res. 50, 769–777 (2017).
Jiang, W. et al. Artificial cells: past, present and future. ACS Nano 16, 15705–15733 (2022).
Li, Z. et al. Dynamic fusion of nucleic acid functionalized nano-/micro-cell-like containments: from basic concepts to applications. ACS Nano 17, 15308–15327 (2023).
Schoonen, L. & van Hest, J. C. M. Compartmentalization approaches in soft matter science: from nanoreactor development to organelle mimics. Adv. Mater. 28, 1109–1128 (2016).
Chandrawati, R. & Caruso, F. Biomimetic liposome- and polymersome-based multicompartmentalized assemblies. Langmuir 28, 13798–13807 (2012).
Discher, D. E. & Eisenberg, A. Polymer vesicles. Science 297, 967–973 (2002).
Nazemi, A. & Gillies, E. R. Dendrimersomes with photodegradable membranes for triggered release of hydrophilic and hydrophobic cargo. Chem. Commun. 50, 11122–11125 (2014).
Liao, W.-C. & Willner, I. Synthesis and applications of stimuli-responsive DNA-based nano- and micro-sized capsules. Adv. Funct. Mater. 27, 1702732 (2017).
Wang, F. et al. Progress report on phase separation in polymer solutions. Adv. Mater. 31, 1806733 (2019).
Tolstoguzov V. et al. Compositions and Phase Diagrams for Aqueous Systems Based on Proteins and Polysaccharides. In: International Review of Cytology (eds Walter H., Brooks D. E., Srere PA). Academic Press (1999).
André, A. A. M. & Spruijt, E. Liquid–liquid phase separation in crowded environments. Int. J. Mol. Sci. 21, 5908 (2020).
Udono, H., Gong, J., Sato, Y. & Takinoue, M. DNA droplets: intelligent, dynamic fluid. Adv. Biol. 7, 2200180 (2023).
Liao, W.-C. et al. The application of stimuli-responsive VEGF- and ATP-aptamer-based microcapsules for the controlled release of an anticancer drug, and the selective targeted cytotoxicity toward cancer cells. Adv. Funct. Mater. 26, 4262–4273 (2016).
Elani, Y., Law, R. V. & Ces, O. Vesicle-based artificial cells as chemical microreactors with spatially segregated reaction pathways. Nat. Commun. 5, 5305 (2014).
Gräfe, D., Gaitzsch, J., Appelhans, D. & Voit, B. Cross-linked polymersomes as nanoreactors for controlled and stabilized single and cascade enzymatic reactions. Nanoscale 6, 10752–10761 (2014).
Tan, H. et al. Heterogeneous multi-compartmental hydrogel particles as synthetic cells for incompatible tandem reactions. Nat. Commun. 8, 663 (2017).
Wang, F., Lu, C.-H. & Willner, I. From cascaded catalytic nucleic acids to enzyme–DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures. Chem. Rev. 114, 2881–2941 (2014).
Tan, L. H., Xing, H. & Lu, Y. DNA as a powerful tool for morphology control, spatial positioning, and dynamic assembly of nanoparticles. Acc. Chem. Res. 47, 1881–1890 (2014).
Simmel, F. C., Yurke, B. & Singh, H. R. Principles and applications of nucleic acid strand displacement reactions. Chem. Rev. 119, 6326–6369 (2019).
Zhang, D. Y. & Seelig, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 3, 103–113 (2011).
Dong, J., O’Hagan, M. P. & Willner, I. Switchable and dynamic G-quadruplexes and their applications. Chem. Soc. Rev. 51, 7631–7661 (2022).
Mergny, J.-L. & Sen, D. DNA quadruple helices in nanotechnology. Chem. Rev. 119, 6290–6325 (2019).
Miyake, Y. et al. MercuryII-mediated formation of thymine−HgII−thymine base pairs in DNA duplexes. J. Am. Chem. Soc. 128, 2172–2173 (2006).
Ono A., et al. Specific interactions between silver(i) ions and cytosine–cytosine pairs in DNA duplexes. Chem. Commun., 39, 4825–4827 (2008).
Biniuri, Y., Luo, G.-F., Fadeev, M., Wulf, V. & Willner, I. Redox-switchable binding properties of the ATP–aptamer. J. Am. Chem. Soc. 141, 15567–15576 (2019).
Kamiya, Y. & Asanuma, H. Light-driven DNA nanomachine with a photoresponsive molecular engine. Acc. Chem. Res. 47, 1663–1672 (2014).
Lubbe, A. S., Szymanski, W. & Feringa, B. L. Recent developments in reversible photoregulation of oligonucleotide structure and function. Chem. Soc. Rev. 46, 1052–1079 (2017).
Wang, C. et al. Photoresponsive DNA materials and their applications. Chem. Soc. Rev. 51, 720–760 (2022).
Breaker, R. R. & Joyce, G. F. A DNA enzyme that cleaves RNA. Chem. Biol. 1, 223–229 (1994).
McConnell, E. M. et al. Biosensing with DNAzymes. Chem. Soc. Rev. 50, 8954–8994 (2021).
Johnson, A. & O’Donnell, M. DNA ligase: getting a grip to seal the deal. Curr. Biol. 15, R90–R92 (2005).
Gupta, R., Capalash, N. & Sharma, P. Restriction endonucleases: natural and directed evolution. Appl. Microbiol. Biotechnol. 94, 583–599 (2012).
Bath, J., Green, S. J. & Turberfield, A. J. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed. 44, 4358–4361 (2005).
Gong, J., Tsumura, N., Sato, Y. & Takinoue, M. Computational DNA droplets recognizing miRNA sequence inputs based on liquid–liquid phase separation. Adv. Funct. Mater. 32, 2202322 (2022).
Saleh, O. A., Jeon, B.-j & Liedl, T. Enzymatic degradation of liquid droplets of DNA is modulated near the phase boundary. Proc. Natl Acad. Sci. 117, 16160–16166 (2020).
Do, S., Lee, C., Lee, T., Kim, D.-N. & Shin, Y. Engineering DNA-based synthetic condensates with programmable material properties, compositions, and functionalities. Sci. Adv. 8, eabj1771 (2022).
Agarwal, S., Osmanovic, D., Klocke, M. A. & Franco, E. The growth rate of DNA condensate droplets increases with the size of participating subunits. ACS Nano 16, 11842–11851 (2022).
Jeon, B.-j, Nguyen, D. T. & Saleh, O. A. Sequence-controlled adhesion and microemulsification in a two-phase system of DNA liquid droplets. J. Phys. Chem. B 124, 8888–8895 (2020).
Saleh, O. A., Wilken, S., Squires, T. M. & Liedl, T. Vacuole dynamics and popping-based motility in liquid droplets of DNA. Nat. Commun. 14, 3574 (2023).
Deng, J. & Walther, A. Programmable ATP-Fueled DNA coacervates by transient liquid-liquid phase separation. Chem 6, 3329–3343 (2020).
Leathers, A. et al. Reaction–diffusion patterning of DNA-based artificial cells. J. Am. Chem. Soc. 144, 17468–17476 (2022).
Hu, Y. & Willner, I. Oligo-adenine derived secondary nucleic acid frameworks: from structural characteristics to applications. Angew. Chem. Int. Ed. 63, e202412106 (2024).
Hu, Y., Ke, Y., Willne,r I. A pH-Cascaded DNA hydrogel mediated by reconfigurable A-motif Duplex, i-Motif Quadruplex, and T·A-T triplex structures. Adv. Funct. Mater., 33, 2304966 (2023).
He, M., Nandu, N., Uyar, T. B., Royzen, M. & Yigit, M. V. Small molecule-induced DNA hydrogel with encapsulation and release properties. Chem. Commun. 56, 7313–7316 (2020).
Lachance-Brais, C. et al. Small molecule-templated DNA hydrogel with record stiffness integrates and releases DNA nanostructures and gene silencing nucleic acids. Adv. Sci. 10, 2205713 (2023).
Avakyan, N. et al. Reprogramming the assembly of unmodified DNA with a small molecule. Nat. Chem. 8, 368–376 (2016).
Rizzuto, F. J. et al. A dissipative pathway for the structural evolution of DNA fibres. Nat. Chem. 13, 843–849 (2021).
Friedman, J. R. & Nunnari, J. Mitochondrial form and function. Nature 505, 335–343 (2014).
Luzio, J. P., Pryor, P. R. & Bright, N. A. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8, 622–632 (2007).
Hyman, A. A., Weber, C. A. & Jülicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014).
Boisvert, F.-M., van Koningsbruggen, S., Navascués, J. & Lamond, A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 (2007).
Mahen, R. & Venkitaraman, A. R. Pattern formation in centrosome assembly. Curr. Opin. Cell Biol. 24, 14–23 (2012).
Gall, J. G. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 4, 975–980 (2003).
Buchan, J. R. & Parker, R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 (2009).
O’Hagan, M. P. et al. Photocleavable ortho-nitrobenzyl-protected DNA architectures and their applications. Chem. Rev. 123, 6839–6887 (2023).
Zhang, D. Y., Turberfield, A. J., Yurke, B. & Winfree, E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318, 1121–1125 (2007).
Acknowledgements
This work is supported by the National Natural Science Foundation of China (Grant No. 22377112 to F. H., Grant No. 22090050 to F. X.), the National Key R&D Program of China (Grant No. 2021YFA1200400 to F. X.), the Joint NSFC-ISF Research Grant Program (Grant No. 22161142020 to F. X. and I. W.).
Author information
Authors and Affiliations
Contributions
F.X., I.W., and F.H. conceived the research. S.X. performed most of the experiments. Y.O. and Y.Q. participated in the systems design and data analysis. All authors participated in discussing the experimental results. D. H. commented on the theoretical model. D.S. and Y. Q. formulated the theoretical model and its adaptation to the experimental results. I. W., D.S., Y.Q., and Y.O. wrote the paper. All authors edited reviewed and commented on the manuscript. F. X., I. W., and F. H. supervised, mentored and participated in funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, S., Ouyang, Y., Qin, Y. et al. Spatiotemporal dynamic and catalytically mediated reconfiguration of compartmentalized cyanuric acid/polyadenine DNA microdroplet condensates. Nat Commun 16, 3352 (2025). https://doi.org/10.1038/s41467-025-58650-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-58650-4