Abstract
While seasoned organic chemists can often predict suitable catalysts for new reactions based on their past experiences in other catalytic reactions, developing this ability is costly, laborious and time-consuming. Therefore, replicating this remarkable expertize of human researchers through machine learning (ML) is compelling, albeit that it remains highly challenging. Herein, we apply a domain-adaptation-based transfer-learning (TL) approach to photocatalysis. Despite being different reaction types, the knowledge of the catalytic behavior of organic photosensitizers (OPSs) from photocatalytic cross-coupling reactions is successfully transferred to ML for a [2+2] cycloaddition reaction, improving the prediction of the photocatalytic activity compared with conventional ML approaches. Furthermore, a satisfactory predictive performance is achieved by using only ten training data points. This experimentally readily accessible small dataset can also be used to identify effective OPSs for alkene photoisomerization, thereby showcasing the potential benefits of TL in catalyst exploration.
Similar content being viewed by others
Introduction
Machine learning (ML) is gaining increasing attention in catalysis research as an efficient tool for catalyst exploration and design1,2,3,4,5,6,7,8. Although seasoned experts in organic synthesis can often predict effective catalysts, even for new reactions, by drawing on the knowledge gained from previous experiences with other reactions, imitating this ability of human researchers through ML would significantly save experimental efforts and time. Transfer learning (TL) is an ML technique where knowledge gained from one task or domain is applied to the improvement of the predictive performance of ML on a different but related task or domain9,10,11. While related concepts have been elegantly applied in catalysis research12,13,14,15,16,17, further development of TL methods in this field is required. It would be ideal if the acquired knowledge could be shared across different types of organic reactions, but the feasibility of this potentially promising protocol has yet to be sufficiently demonstrated. In particular, the effectiveness of TL in predicting the catalytic activity of photosensitizers as an attractive tool for organic chemists18,19,20 remains unclear.
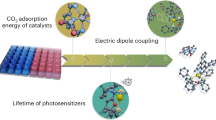
Herein, we demonstrate the pivotal role of domain adaptation (DA)9,21,22,23, a TL technique that has been underestimated in catalysis research, in improving the predictive performance of ML for the estimation of the catalytic behavior of organic photosensitizers (OPSs) and in identifying promising OPSs for a specific reaction using relatively small datasets of seemingly distinct photoreactions (Fig. 1a). ML techniques have already been used for the prediction of photocatalytic behavior, including redox catalysis and energy transfer (EnT) in reactions such as hydrogen evolution24,25, singlet-oxygen generation26, and the nickel/photocatalytic synthesis of phenols27. However, the construction of regression models for an accurate prediction of the catalytic behavior of various π-conjugated organic molecules potentially serving as OPSs without using extensive training data remains challenging25,27. In contrast to these conventional ML approaches in photocatalysis, we hypothesized that sharing the knowledge of the catalytic behavior of OPSs, even in photoreactions seemingly distinct from a target photoreaction, could enhance the prediction accuracy if OPSs play a similar role in each reaction. Thus, we collected data on the catalytic behavior of OPSs for nickel/photocatalytic C–O, C–S, and C–N bond-forming reactions as well as a [2+2] cycloaddition reaction and assessed the effectiveness of sharing the as-acquired knowledge using TrAdaBoostR2, an instance-based DA method28. Transferring knowledge from the domain of photocatalytic cross-coupling reactions led to a more accurate prediction of the photocatalytic behavior in the target domain, i.e., the [2+2] cycloaddition. Moreover, our concept proved to be valid even when using only ten training data points, and this experimentally accessible small dataset of ten OPSs could be employed to propose promising OPSs in an alkene photoisomerization reaction through DA. The present study thus demonstrates that combining the existing database of catalysts with DA provides a promising tool for sharing knowledge across different types of reactions, such as the cross-coupling, cycloaddition, and alkene isomerization reactions, to discover effective catalysts, highlighting the potential utility of DA in assisting organic synthesis research.
Results
Conventional ML in [2+2] cycloaddition
First, we examined the catalytic behavior of 100 OPSs in the photocatalytic [2+2] cycloaddition (henceforth referred to as CA) of 4-vinylbiphenyl (1), which has been reported as an EnT reaction29,30. Our OPS dataset consists of the 60 OPSs previously used in our study27, together with 40 newly prepared OPSs. In contrast to the previous set of 60 neutral OPSs containing electron-donor and -acceptor groups (D–A-type OPSs), the updated dataset includes not only D–A-type OPSs (e.g., OPS1), but also π–π*-type OPSs (e.g., OPS75), n–π*-type OPSs (e.g., OPS86), and cationic OPSs (e.g., OPS99). The OPSs used in this study are shown in Figs. S1 and S2. The investigation of the photocatalytic behavior of OPSs revealed that several D–A-type OPSs (OPS1, OPS7, OPS10, OPS11, OPS12, OPS13, OPS44, and OPS67) are effective, affording the desired product (2) in > 70% yield after 3 h in this photoreaction. In contrast, π–π*-type, n–π*-type, and cationic OPSs exhibited very low catalytic activity (Fig. 2).
Descriptors for the 100 OPSs were generated using density functional theory (DFT) calculations and Python toolkits. The DFT-derived descriptor set, which is denoted as DFT descriptors or simply DFT in the context of ML, contains the HOMO (EHOMO) and LUMO (ELUMO) energy levels based on the optimized ground-state geometry calculated at the B3LYP-D3/6-31G(d) level. Additionally, the vertical excitation (absorption) energies of the lowest singlet (E(S1)) and triplet (E(T1)) excited states, the corresponding vertical singlet–triplet splitting (ΔEST = E(S1) – E(T1)), and the oscillator strengths of the lowest singlet excitation (f(S1)) were obtained from single-point quantum chemical calculations using time-dependent DFT (TD-DFT) with the Tamm–Dancoff approximation (TDA). The molecular geometries were prepared through the ground-state geometry optimizations at the B3LYP-D3/6-31G(d) level (for details, see the Computational details section in the Supplementary Information). The TD-DFT/TDA calculations for the S1 and T1 states were performed at the M06-2X/6-31+G(d) level with the PCM model for toluene solutions27,31. Furthermore, to clearly differentiate the change in properties between ground and excited states for various compounds including D–A-type and π–π*-type OPSs, the difference in dipole moments between these two states of OPSs (ΔDM) was calculated as a descriptor. ΔDM was determined by conducting single-point calculations for the ground and excited states at the PCM(toluene)-M06-2X/6-31+G(d) level. Furthermore, we employed four descriptor sets generated from SMILES of OPSs, including the RDKit descriptor (RDKit), the MACCSKeys (MK), the Mordred, and the Morgan fingerprint (MF). Since some of these descriptor sets consisted of over several hundred features, we also prepared descriptors reduced in dimension using principal component analysis (PCA), and the resulting descriptor sets contained 12 (RDKit_pca), 12 (MK_pca), nine (Mordred_pca), and 29 (MF_pca) features, respectively.
With the dataset for 100 OPSs in hand, we evaluated the predictive performance for an estimation of the catalytic activity of OPSs using random forest (RF) as an ML algorithm (Table 1). To mitigate the influence of variations in the partition pattern between training and test datasets on the predictive performance, we examined 100 different partition patterns (50 compounds (50%) for the training data, and 50 compounds (50%) for the test data) and compared the average (Avg R2), maximum (Max R2), and standard deviation (Std R2) of the R2 scores on the test set. Consequently, RF models employing descriptors obtained from SMILES yielded poor predictive performance, with Avg R2 values dropping below 0.25 (Table 1, entries 1–8). The DFT descriptors showed a marginal improvement in R2 scores, but an Avg R2 was only 0.27 with a Max R2 of 0.55 (Table 1, entry 9). Concatenating the DFT descriptors with RDKit_pca did not result in a good predictive performance (Table 1, entry 10; Avg R2 = 0.23), which stands in sharp contrast to our previous findings (for the investigation of other combinations, see Table S8)27. Moreover, alternative ML algorithms such as Lasso, support vector machine (SVM), and XGBoost (XGB) could not improve the R2 scores (Table 1, entries 11–13).
DA between [2+2] cycloaddition and cross-coupling reactions
Although using large, well-qualified training datasets, e.g., high-throughput experiment datasets, provides excellent predictive performance1,32,33, collecting a vast amount of high-quality experimental data for target reactions may not always be practical for organic chemists. In contrast, the diversity of organic reactions, including photoreactions, could be advantageous in ML for organic synthesis because extracting relatively similar reactions from a previously constructed database containing various reactions and subsequently sharing the acquired knowledge can offer an alternative approach, i.e., TL, to enhance the predictive performance.
We then attempted to improve the performance of ML in predicting the catalytic behavior of OPSs in CA using data on other photocatalytic reactions. Previously, we developed an ipso-substitution of aryl halides with water for the synthesis of phenol derivatives catalyzed by an OPS and an inorganic nickel salt (OPS/Ni)27. Many cross-coupling reactions facilitated by photosensitizers and Ni catalysts, including our case, have been suggested to involve an EnT process to some extent20,34,35,36,37. The cross-coupling reaction is an apparently different type of organic transformation from CA. However, considering the potentially similar role of OPSs as EnT catalysts in these reaction systems, we hypothesized that knowledge transfer from cross-coupling reactions could effectively enhance the prediction of photocatalytic activity in CA. Therefore, we collected experimental data on the catalytic behavior of OPSs in the OPS/Ni-catalyzed synthesis of phenols using 4-bromobenzonitrile (3a; reaction time = 1.5 h: CO_a, 7.5 h: CO_b), 4-bromobiphenyl (3b, CO_c), methyl 4-bromo-3-methylbenzoate (3c, CO_d), and 4-chlorobenzonitrile (3 d, CO_e) (Fig. 3a). In addition, we diversified our dataset by collecting data on the catalytic behavior of OPSs in C–S bond and C–N bond-forming reactions with 3a (CS and CN, respectively)37,38. We also performed a Pearson correlation analysis to determine trends in the catalytic activity, i.e., the yield of each product (2, 4a–4c, 5, and 6) (Fig. 3b). Although the correlation coefficients between CA and CO_a, CO_b, CO_c, CO_d, CO_e, and CS, respectively, were moderate (0.52–0.64), CA and CN showed a relatively strong correlation (0.76).
First, we investigated the impact of simply increasing the data volume on prediction accuracy. To this end, we utilized the entire data from cross-coupling experiments comprising a total of 700 data points, as the training dataset (Fig. 6a, Method B). In subsequent investigations, we used one-hot encoding (OHE) to distinguish the types of reactions. The predictive performance of the Lasso, SVM, RF, and XGB models was evaluated using all the nine descriptor sets previously tested (Table 1). Although the prediction accuracy improved, the highest Avg R2 achieved was only 0.41 (XGB/MF_pca model). Next, we incorporated a dataset originally used for training in CA into the cross-coupling dataset, resulting in a combined training dataset of 750 data points (Fig. 6a, Method C). This approach was expected to serve as a simple TL model, facilitating the sharing of information among CA and others. Consequently, predictive performance improved further, with the best Avg R2 reaching 0.52 (XGB/DFT model). The detailed results of these ML investigations are provided in the Supplementary Information (Tables S11 and S12).
Meanwhile, we envisioned that using TL methods that more clearly distinguish between the source and target domains could further improve predictive performance. To achieve this, supervised DA, such as TrAdaBoostR2 (TrAB), was applied, using data on cross-coupling experiments as the source domain. In TrAB, the source and target domains are combined similarly to the aforementioned attempt (Fig. 6a, Method C), but this method decreases the weights of instances with large prediction errors in the source domain at each step of the boosting process, while those of instances with large errors in the target domain are increased (Fig. 1b). This approach enables more efficient knowledge transfer compared to simply combining the datasets, potentially leading to enhanced predictive performance for the target domain. An additional potential advantage of TrAB is its effectiveness even when using a smaller dataset as the source of knowledge than that used for deep-learning-based TL methods such as fine-tuning.
DA methods are broadly categorized into feature-based and instance-based approaches. While instance-based DA methods, including TrAB, aim to address differences in data distribution between the source and target domains by weighting or selecting samples, feature-based DA methods focus on aligning the feature distributions between source and target domains by transforming or mapping them into a shared feature space39. Therefore, in addition to TrAB, we tested the feature augmentation (FA) and correlation alignment (CORAL) as feature-based methods, as well as balanced weighting (BW) as an alternative option of instance-based DA. For the implementation of these DA models, we used the ADAPT library, which is an open-source Python toolkit40. After a brief investigation of the estimators of DA models and descriptors, we found that the combined use of light gradient boosting machine (LGBM) and the DFT descriptors outperformed other options (Tables S13 and S14).
When we employed data from all cross-coupling reactions as the source domain, the instance-based DA methods resulted in the substantial improvement compared with the others tested (Table 2, entries 1 and 4; TrAB: Avg R2 = 0.65, BW: Avg R2 = 0.63). In contrast, feature-based DA methods were less effective in enhancing prediction accuracy (Table 2, entries 2 and 3; CORAL: Avg R2 = 0.08, FA: Avg R2 = 0.55). For both source and target domains, the descriptors included DFT-based properties of OPSs and OHE-based reaction recognition, with the prediction target being the reaction yield, meaning that the input and output structures are very similar. Thus, although cross-coupling and cycloaddition reactions are different reaction types, our protocol is likely based on a homogeneous domain shift from the viewpoint of ML. In such cases, instance-based DA, which places greater emphasis on individual data points, could be more effective than feature-based DA. The difference in Avg R2 values between BW and TrAB was not substantial. However, TrAB iteratively and dynamically adjusts the weights of instances, enabling more efficient adaptation to the target characteristics. This likely contributes to the slightly improved predictive performance of TrAB.
Next, we examined the influence of the source domains on the predictive performance. We constructed new source domains based on the Pearson correlation coefficients of the entire dataset. The correlation coefficients of CO_a, CO_b, CO_c, CO_d, CO_e, CS, and CN with respect to CA are 0.59, 0.61, 0.61, 0.52, 0.63, 0.64, and 0.76, respectively. Accordingly, we designed S1 (CO_e, CS, CN), S2 (CN), and S3 (CS, CN) to include photoreaction datasets with high correlation coefficients, while S4 (CO_a, CO_d) and S5 (CO_a, CO_b, CO_c, CO_d) consisted of those with low correlation coefficients. The source domain, which consists of reactions with similar trends in catalytic behavior (S1), provided favorable results in improving the predictive performance (Table 2, entry 5; Avg R2 = 0.68). However, the predictive performance was not improved when using source domains that included a smaller number of data such as S2 and S3 (Table 2, entries 6 and 7; S2: Avg R2 = 0.50, S3: Avg R2 = 0.64). In addition, datasets with poor similarity in catalytic behavior were less effective as source domains (Table 2, entries 8 and 9; S4: Avg R2 = 0.49, S5: Avg R2 = 0.53). Therefore, both a high similarity of the tendency in the photocatalytic activity and data diversity are important features for providing effective source domains. The difference in predictive performance associated with the correlation coefficient was particularly pronounced when the data size of the source domain was small, as observed in the comparison between S3 and S4 (S3: Avg R2 = 0.64, S4: Avg R2 = 0.49). Meanwhile, despite the contamination of less useful datasets such as those from CO_a and CO_d, the decline in predictive performance was mitigated when the source domain contained all cross-coupling data (Avg R2 = 0.65). As mentioned earlier, TrAB decreases the weights of less useful instances in the source domain. Given this mechanism, it is reasonable that a source domain comprising diverse data would be more tolerant of the inclusion of ineffective data, aligning well with our findings discussed above.
To further enhance the prediction accuracy, we revised the descriptors using S1 as the source domain. First, we eliminated ELUMO and f(S1), which were found to deteriorate the predictive performance. Next, we used Featuretools, an open-source Python toolkit for feature engineering, to design a more effective descriptor set41. We converted DFT-derived descriptors into percentiles, i.e., P(EHOMO), P(E(S1)), P(E(T1)), P(ΔEST), and P(ΔDM). Subsequently, we performed multiplication, division, addition, and subtraction operations on pairs of these percentile descriptors, thereby generating 50 additional descriptors. After assessing the effectiveness of each descriptor, we identified P(E(S1)) * P(E(T1)), P(ΔDM) * P(E(S1)), P(ΔDM) * P(EHOMO), P(E(S1)) + P(EHOMO), P(ΔEST) − P(E(S1)), and P(ΔEST) / P(EHOMO) as effective descriptors. The new descriptor set, which is denoted as DFT_FE, led to an Avg R2 of 0.74 and a Max R2 of 0.88 (Table 2, entry 10). Fig. 4a, b show violin plots illustrating the distribution of R2 scores and an example of 2D scattering plots depicting the relationship between experimental and predicted yields, respectively, which clearly demonstrate the improvement in the predictive performance when employing the DA method. It is worth noting that compared with conventional RF models with the DFT descriptors (Table 1, entry 9), the predictive performance of the constructed TrAB models was improved in all 100 runs (Table S19) and the influence of training data, i.e., Std R2, became small (TrAB/DFT_FE: 0.09, RF/DFT: 0.15). We have previously clarified that unlike the cycloaddition reactions, OPS/Ni-catalyzed cross-coupling reactions may involve not only an EnT process but also oxidative- and reductive-quenching processes27, which underscores that the roles of OPSs in these reactions are not identical. Nevertheless, a knowledge transfer from substitution-type organic transformation (source domain) to addition reaction (target domain) was successfully achieved by combining various types of cross-coupling reactions as the source domain.
Furthermore, we examined the robustness of the models regarding the number of data points in the training sets. When the size of the training data in the target domain was reduced to 40, 30, or 20, the decline in R2 scores and the variability in predictive performance based on the used training data decreased considerably in the TrAB model (Fig. 4c), thus demonstrating higher robustness than conventional approaches. Considering the robustness of the models constructed through TrAB, we conducted DA using ten OPSs as training data in the target domain (Fig. 4d). In this survey, histogram-based gradient boosting (HGB) was found to be the most effective estimator. Ten OPSs for training data were selected on the basis of the predictive performance from 100 partition patterns, identifying OPS5, OPS9, OPS20, OPS23, OPS27, OPS31, OPS34, OPS44, OPS59, and OPS83 as the most effective. The use of this training set achieved an R2 score on the test set of 0.73 when combined with DFT_FE, and the detailed results of this preliminary investigation are described in the Supplementary Information (Tables S23 and S24). Following additional feature engineering, a new descriptor set, referred to as DFT_FE2, was developed, resulting in an improved R2 score of 0.83. DFT_FE2 consists of EHOMO, E(S1), E(T1), f(S1), ΔDM, P(ΔDM) * P(E(S1)), P(ΔDM) * P(ΔEST), P(ΔEST) * P(f(S1)), P(ΔDM) + P(ΔEST), P(ΔEST) + P(E(S1)), P(ΔDM) – P(f(S1)), and P(f(S1)) – P(EHOMO). The yields of 2 obtained using five OPSs out of the seven OPSs providing more than 70% yield of product 2 were accurately predicted. In contrast, OPS13 and OPS67, which exhibited relatively poor photocatalytic activity in other reactions but showed high activity in CA, resulted in inaccurate predictions.
Although the use of the above-mentioned ten OPSs resulted in satisfactory predictive performance, the learning curve obtained from the evaluation of 100 different training-test splits highlighted the difficulty of ensuring generalization ability when using only ten OPSs as a training set. In the TrAB/DFT_FE2 model trained on ten data points, the Max R2 value was not significantly different from cases using a larger training dataset (50–20 data points: 0.87–0.82, ten data points: 0.83), but the Avg R2 value declined substantially (50–20 data points: 0.71–0.62, ten data points: 0.54) (Fig. S5 and Table S22). These findings indicate that, even in a TL-based approach, careful selection of appropriate training data is essential for achieving good predictive performance with such a limited training dataset. While this approach might not be ideal from a data science perspective, identifying an experimentally accessible small training dataset that is useful for ML-based screening can be a practical strategy in organic chemistry.
DA in alkene photoisomerization
Given the limitations of TL using extremely small training datasets mentioned above, we further examined whether this TL strategy could still be effective by applying it to the prediction of potent OPSs in another photoreaction involving an EnT pathway. We tested the catalytic activity of the aforementioned ten OPSs (OPS5, OPS9, OPS20, OPS23, OPS27, OPS31, OPS34, OPS44, OPS59, and OPS83) in the photocatalytic (E)- to (Z)-isomerization of trans-stilbene (7), which involves an EnT pathway42, and attempted to predict the top five OPSs through DA (Fig. 5). A correlation analysis revealed that the catalytic activity tendencies among the ten OPSs experimentally tested in the alkene photoisomerization were similar (Pearson correlation coefficient ≥ 0.5) to those in CO_e, CS, CN, and CA. Consequently, two source domains were prepared: one consisting of the combined data of only cross-coupling reactions (CO_e, CS, and CN; denoted as S1) and the other of the combined data of cross-coupling and cycloaddition reactions (CS, CN, and CA; denoted as S6). The photocatalytic behavior of the remaining 90 OPSs was then predicted using these two source domains and DFT_FE2 as the descriptor set. In both cases, OPS1, OPS7, OPS11, and OPS12 were selected, and they exhibited remarkably high catalytic activity (91%–96%). As the last remaining top performers, OPS10 and OPS67 were selected when using the source domains S1 and S6, respectively, affording cis-stilbene (7’) in 95% and 84% yields. Overall, the selected OPSs have similar structures, i.e., they are all cyanoarene-based compounds that bear carbazolyl groups or diarylamino groups. Although the ML model constructed with S1 showed larger errors between experimental and predicted yields than that constructed with S6 (S1: MAE = 8.4, S6: MAE = 3.8), all the proposed OPSs afforded 7’ in yields of > 90%. It is noteworthy that these OPSs (OPS1, OPS7, OPS10, OPS11, OPS12, and OPS67) were still identified as top performers even when using a training dataset of eight OPSs excluding the highly active OPS9 and OPS44, while the predictions were less accurate (Figs. S14 and S15; S1: MAE = 14.8, S6: MAE = 9.6).
Meanwhile, the TrAB/DFT_FE model proposed OPS24, OPS25, OPS32, and OPS36 as top performers, which were not ranked among the top five OPSs with DFT_FE2. However, these OPSs gave unsatisfactory experimental yields of 7’ (OPS24:16%, OPS25: 31%, OPS32: 71%, OPS36: 77%). Moreover, when using DFT_FE, the errors between experimental and predicted yields were much larger (S1: MAE = 30.1, S6: MAE = 10.0) than those obtained with DFT_FE2. While DFT_FE2 underperformed DFT_FE in terms of generalization ability (Fig. S5 and Table S22), it proved useful in selecting superior OPSs for the specific task (CA): DFT_FE2 delivered better results in the more practical catalyst exploration, utilizing the experimentally accessible number of OPSs.
When the RF model with the DFT descriptors was used to predict the top five OPSs using the same ten OPSs as a training set, OPS41, OPS43, OPS63, OPS64, and OPS65 were selected. Unfortunately, these OPSs were ineffective (1%–21%), and the errors between experimental and predicted yields were large (MAE = 66.6). In addition, although this alkene photoisomerization is considered to involve an EnT process from the photoexcited OPS to alkene 7, the RF model selected wrong answers, OPSs with strong reducing properties38,43, as was confirmed by feature-importance and SHAP analyzes (for the SHAP analysis, see Fig. S19b). It is worth noting that such a critical misunderstanding in the selection of OPSs using the simple RF model could be prevented by sharing the knowledge, namely by using a DA-based TL strategy, even among seemingly different photoreactions, allowing the successful identification of OPSs with very high catalytic activity.
Investigations into applicability and limitations
To evaluate the applicability and limitations of the DA strategy (Fig. 6), we assessed the predictive performance for each cross-coupling reaction (CO_a, CO_b, CO_c, CO_d, CO_e, CS, and CN). First, we compared the performance of TrAB with that of RF without the source-domain dataset (Fig. 6a, Method A), both using the DFT descriptors. In TrAB, the source domain consisted of either all data from photoreactions except for the target reaction or data from three photoreactions with high correlation coefficients to the target. We observed that the DA competence of these source domains was consistently superior to that of the source domain consisting of three photoreactions with trends in catalytic activity dissimilar to each target (Table S28). In all cases, Method A significantly underperformed TrAB in the prediction accuracy. While the TrAB/DFT model consistently delivered moderate to high prediction accuracy in CO_a, CO_b, CO_c, CO_d, CO_e, and CN (TrAB: Avg R2 = 0.64–0.85, Method A: Avg R2 = 0.26–0.49), its predictive performance was poor for CS (Avg R2 = 0.43). Method A also showed the extremely poor predictive performance for CS (Avg R2 = 0.07), suggesting that DA may not always provide sufficient improvements, particularly for tasks with inherently elusive characteristics, such as CS.
a Comparison of predictive performance among TrAB and other methods when datasets of cross-coupling reactions were used as target domains. The RF/DFT model was applied in Method A, while the XGB/DFT model was used in Methods B and C. b Comparison of prediction accuracy between data included in the source domain and those excluded from the source domain (target: CA, source domain: S1, descriptor set: DFT_FE).
Next, we tested XGB models trained on 700 data points from the photoreactions excluding the target (Fig. 6a, Method B), as well as on a dataset of 750 data points, in which the aforementioned 700 data points were combined with the training dataset of the target reaction (Fig. 6a, Method C). These XGB models were also combined with the DFT descriptors. Compared to the predictive performance of TrAB, that of Method B was inferior in all cases (Method B: Avg R2 = 0.16–0.67), while it was comparable when CS, for which TrAB also demonstrated the insufficient performance, was the target (TrAB: Avg R2 = 0.44, Method B: Avg R2 = 0.42). The primary difference between Method C and TrAB lies in the ability of TrAB to apply sample weighting (Fig. 1b), which more effectively differentiates the source and target domains. In the case of C–O bond-forming reactions where the constructed database contains a sufficient number of reactions with tendencies in photocatalytic activity comparable to the target, TrAB consistently delivered favorable results (CO_a: Avg R2 = 0.78, CO_b: Avg R2 = 0.76, CO_c: Avg R2 = 0.85, CO_d: Avg R2 = 0.72, CO_e: Avg R2 = 0.80). In contrast, although Method C performed well for CO_a, CO_b, and CO_c (CO_a: Avg R2 = 0.74, CO_b: Avg R2 = 0.72, CO_c: Avg R2 = 0.77), its predictive performance fell significantly short of TrAB for CO_d and CO_e (CO_d: Avg R2 = 0.42, CO_e: Avg R2 = 0.65). Additionally, Method C showed limited effectiveness for CS and CN, with its performance being comparable to, or even worse than, Method B, which did not utilize training data from the target (Method B/CS: Avg R2 = 0.42, Method B/CN: Avg R2 = 0.45, Method C/CS: Avg R2 = 0.31, Method C/CN: Avg R2 = 0.48). Overall, while Method C demonstrated good predictive performance in some instances, it never outperformed TrAB in any scenario. Moreover, TrAB consistently exhibited more stable and higher performance than the others in all cases, effectively addressing the instability observed in approaches that relied simply on the increased data volume.
Subsequently, we evaluated the predictive performance for OPSs that were not included in the source domain (Fig. 6b). Extrapolative predictions have been a persistent challenge in ML applications for catalytic reactions. For instance, in ML research on C–N bond-forming reactions conducted by Dreher and Doyle1, successful extrapolative predictions were achieved for isoxazole additives, whereas those for aryl halides proved to be highly challenging. Similarly, Zhang and Hong developed a graph neural network-based approach, demonstrating improved performance for the same task44. Nevertheless, extrapolative predictions for aryl halides and bases remained challenging even in this approach. Following this context, we randomly excluded 30 OPSs from the source-domain datasets while retaining them in the target-domain dataset, to assess the performance of our proposed strategy under similar extrapolation conditions. In this investigation, CA was selected as the target, with DFT_FE and S1 employed as the descriptor set and the source domain, respectively. Consequently, the predictive performance for OPSs included in the source domain was satisfactory (Avg R2 = 0.76), whereas it was significantly lower for those excluded from the source domain (Avg R2 = 0.28). Similar trends were observed even when patterns of OPSs excluded from the source domain were different (Tables S29–S48). These results indicate that the success of this strategy relies heavily on the coverage of OPSs provided by the experimental database used for the source domain. Therefore, continuous efforts by organic chemists to expand and refine the database are essential for further strengthening the proposed DA-based TL strategy.
Discussion
Insights on the selection of source-domain datasets
The improvements in prediction accuracy observed in this study were likely due to the successful construction of source-domain datasets with trends in photocatalytic activity similar to the target reaction. The photocatalytic cross-coupling, cycloaddition, and alkene isomerization reactions utilized in our study are considered to involve EnT processes, resulting in this similarity in catalytic activity trends. Thus, first and foremost, constructing an appropriate database that accounts for reaction mechanisms is essential for establishing an effective source domain.
As an alternative approach for selecting source-domain datasets, we found that a simple metric, i.e., the Pearson correlation coefficient, could be used to differentiate between effective and ineffective datasets to some extent. This is derived from the observation that effective source-domain datasets generally exhibited higher correlation coefficients to the target reaction than ineffective datasets (Table 2 and S28). Notably, this approach is applicable even with limited knowledge of the reactions. For clarity, we primarily used the source domain constructed based on the correlation coefficients of the entire dataset, but this metric-based approach also proved effective when the source domain was constructed using only the correlation coefficients among OPSs in the training data (Table S21 and Figs. S8, S9, and S16), which aligns more closely with practical scenarios. However, the difference in correlation coefficients was not always sufficient to fully distinguish between effective and ineffective datasets. Moreover, in several cases, the DA competence of a source domain comprising all photoreactions other than the target was comparable to, or even outperformed, that of a source domain consisting solely of selected photoreactions biased toward trends in photocatalytic activity similar to the target (Table 2 and S28).
Therefore, drawing definitive conclusions about the optimal method for selecting source-domain datasets based solely on our observations remains challenging, while our findings offer helpful insights into the criteria for selecting the source-domain datasets. A viable approach to addressing this issue is to construct a source domain comprising more diverse photoreactions with various substrate–product examples, taking into account the ability of TraAdaBoostR2 to mitigate the influence of uninformative data in the source domain. This approach is particularly effective when the source-domain datasets are appropriately selected based on organic-chemistry insights or validated using metrics such as the correlation coefficient. Nonetheless, future studies need to investigate whether such a sample-weighting mechanism would remain effective when the source domain includes reactions with trends in catalytic activity significantly different from those of the target EnT reaction, such as photoredox reactions.
Summary of the work
Through a combination of experimental and computational investigations, we have demonstrated how DA-based TL can be effectively employed for the prediction of the catalytic behavior of OPSs. Compared with conventional approaches, e.g., the RF model with the DFT-derived descriptor set, the present TrAdaBoostR2-based DA strategy enhances the predictive performance. Specifically, it improved the average R2 score from 0.27 to 0.74, the maximum R2 score from 0.55 to 0.88, and the standard deviation of R2 scores from 0.15 to 0.09 when predicting the photocatalytic activity of OPSs in the [2+2] cycloaddition using data obtained from OPS/Ni-catalyzed cross-coupling reactions, which are organic transformations that are apparently different from the target reaction. Furthermore, our study showcased that the use of only ten OPSs as training data in the target domain resulted in satisfactory predictive performance (R2 = 0.83), and this experimentally readily accessible small dataset proved instrumental in proposing effective OPSs in the alkene photoisomerization. Meanwhile, although our approach also demonstrated stable performance even when targeting photocatalytic cross-coupling reactions, it does not appear to be well-suited for predicting OPSs that are not included in the source domain. Our results not only illustrate that constructing appropriate databases informed by organic-chemistry knowledge can strengthen ML-driven catalyst exploration even for newly tested reactions, but also set a precedent for more effectively applying ML in other areas where data scarcity is a barrier. Further investigations to assess the applicability of the DA-based TL method to a broader range of photoreactions, including photoredox reactions, are currently in progress in our laboratory.
Methods
Procedure for collecting data on CA
4-Vinylbiphenyl 1 (180.3 mg, 1.00 mmol), a photosensitizer (2.5 μmol, 0.25 mol%), and CH2Cl2 (1 mL) were added to a test tube under air at room temperature. After the mixture was degassed by three freeze-pump-thaw cycles, the test tube was put in PhotoRedOx Box and the reaction was carried out under visible light irradiation (λ = 450 nm) for 3 h at room temperature. The solvent was evaporated and the yield was determined by 1H NMR spectroscopy with 1,3,5-trimethoxybenzene as an internal standard.
Procedures for collecting data on CO_a, CO_b, CO_c, CO_d, and CO_e
A substrate (3a–3d, 0.500 mmol), DABCO (84.1 mg, 0.750 mmol, 1.5 equiv.), a photosensitizer (2.5 μmol, 0.5 mol%), H2O (72.1 μL, 4.00 mmol, 8 equiv.), and NiBr2•DME in NMP or DMI (5 mM, 2 mL, 2 mol%) were added to a test tube under air at room temperature. After the mixture was degassed by three freeze-pump-thaw cycles, the test tube was put in PhotoRedOx Box and the reaction was carried out under visible light irradiation (λ = 450 nm) for 1.5 h–24 h at room temperature. Water (15 mL) was added and the mixture was extracted with Et2O (15 mL × 3). The organic phase was dried over Na2SO4, filtered, and evaporated. The yield was determined by 1H NMR spectroscopy with 1,3,5-trimethoxybenzene as an internal standard.
Procedure for collecting data on CS
4-Bromobenzonitrile 3a (91.0 mg, 0.500 mmol), a photosensitizer (2.5 μmol, 0.5 mol%), 1-octanethiol (174 μL, 0.84 g/mL, 0.999 mmol, 2 equiv.), pyridine (80.7 μL, 0.98 g/mL, 1.00 mmol, 2 equiv.), and NiBr2•DME in DMA (5 mM, 2 mL, 2 mol%) were added to a test tube under air at room temperature. After the mixture was degassed by three freeze-pump-thaw cycles, the test tube was put in PhotoRedOx Box and the reaction was carried out under visible light irradiation (λ = 450 nm) for 0.5 h at room temperature. Water (15 mL) was added and the mixture was extracted with Et2O (15 mL x 3). The organic phase was dried over Na2SO4, filtered, and evaporated. The yield was determined by 1H NMR spectroscopy with 1,3,5-trimethoxybenzene as an internal standard.
Procedure for collecting data on CN
4-Bromobenzonitrile 3a (91.0 mg, 0.500 mmol), a photosensitizer (2.5 μmol, 0.5 mol%), pyrrolidine (145 μL, 0.86 g/mL, 1.75 mmol, 3.5 equiv.), and NiBr2•DME in NMP (5 mM, 2 mL, 2 mol%) were added to a test tube under air at room temperature. After the mixture was degassed by three freeze-pump-thaw cycles, the test tube was put in PhotoRedOx Box and the reaction was carried out under visible light irradiation (λ = 450 nm) for 3 min at room temperature. Water (15 mL) was added and the mixture was extracted with Et2O (15 mL x 3). The organic phase was dried over Na2SO4, filtered, and evaporated. The yield was determined by 1H NMR spectroscopy with 1,3,5-trimethoxybenzene as an internal standard.
Protocol for comparing model performance
In the performance comparison, 10–50% of the entire dataset was used for training, while the remaining data was used for testing. To construct domain-adaptation-based prediction models, we tested feature augmentation, correlation alignment, balanced weighting, and TraAdaBoostR2. These models were implemented using the ADAPT library40. Additionally, we evaluated prediction models based on Lasso, support vector machine, random forest, and XGBoost. To mitigate the impact of variations in the splitting pattern between the training and test datasets on predictive performance, we performed 100 different training-test splits and compared the average, maximum, and standard deviation of the R2 scores.
Protocol for identifying effective photosensitizers in alkene photoisomerization
First, the catalytic activity of ten organic photosensitizers (OPS5, OPS9, OPS20, OPS23, OPS27, OPS31, OPS34, OPS44, OPS59, and OPS83) was investigated in alkene isomerization. Two source domains, consisting of photoreactions that exhibit catalytic activity trends similar to this reaction, were constructed (S1: CO_e, CS, and CN, S6: CS, CN, and CA). Using the above-mentioned ten photosensitizers as the training dataset and each of the prepared source domains, we constructed TraAdaBoostR2-based models to predict the catalytic activity of the remaining 90 photosensitizers. The catalytic activity of the predicted top five performers was then experimentally tested. The experimental procedures for alkene photoisomerization are described in the following section.
Experimental procedure for alkene photoisomerization
trans-Stilbene 7 (90.1 mg, 0.500 mmol), a photosensitizer (2.5 μmol, 0.5 mol%), and CH2Cl2 (2 mL) were added to a test tube under air at room temperature. After the mixture was degassed by three freeze-pump-thaw cycles, the test tube was put in PhotoRedOx Box and the reaction was carried out under visible light irradiation (λ = 450 nm) for 0.5 h at room temperature. The solvent was evaporated and the yield was determined by 1H NMR spectroscopy with 1,3,5-trimethoxybenzene as an internal standard.
Data availability
The data supporting the findings of this study are available within the Supplementary Information, and a file summarizing SMILES, DFT-based properties and yields of OPSs are available at the GitHub repository (https://github.com/Naoki-Noto/P2-20231212/tree/main/data) and Zenodo45. All data are available from the corresponding author upon request. Source data are provided with this paper.
Code availability
All codes necessary for the research are available at the GitHub repository (https://github.com/Naoki-Noto/P2-20231212) and Zenodo45.
References
Ahneman, D. T., Estrada, J. G., Lin, S., Dreher, S. D. & Doyle, A. G. Predicting reaction performance in C–N cross-coupling using machine learning. Science 360, 186–190 (2018).
Zahrt, A. F. et al. Prediction of higher-selectivity catalysts by computer-driven workflow and machine learning. Science 363, eaau5631 (2019).
Toyao, T. et al. Machine learning for catalysis informatics: recent applications and prospects. ACS Catal. 10, 2260–2297 (2020).
Shields, B. J. et al. Bayesian reaction optimization as a tool for chemical synthesis. Nature 590, 89–96 (2021).
Yang, L.-C., Li, X., Zhang, S.-Q. & Hong, X. Machine learning prediction of hydrogen atom transfer reactivity in photoredox-mediated C–H functionalization. Org. Chem. Front. 8, 6187–6195 (2021).
Xu, S. et al. Self-improving photosensitizer discovery system via Bayesian search with first-principle simulations. J. Am. Chem. Soc. 143, 19769–19777 (2021).
Samha, M. H. et al. Predicting success in Cu-catalyzed C–N coupling reactions using data science. Sci. Adv. 10, eadn3478 (2024).
Dai, L. et al. Harnessing electro-descriptors for mechanistic and machine learning analysis of photocatalytic organic reactions. J. Am. Chem. Soc. 146, 19019–19029 (2024).
Pan, S. J. & Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2009).
Weiss, K., Khoshgoftaar, T. M. & Wang, D. A survey of transfer learning. J. Big Data 3, 9 (2016).
Tan, C. et al. A survey on deep transfer learning. In Artificial Neural Networks and Machine Learning – ICANN 2018 270–279 (ICANN, 2018).
Reid, J. P. & Sigman, M. S. Holistic prediction of enantioselectivity in asymmetric catalysis. Nature 571, 343–348 (2019).
Shim, E. et al. Predicting reaction conditions from limited data through active transfer learning. Chem. Sci. 13, 6655–6668 (2022).
Singh, S. & Sunoj, R. B. A transfer learning protocol for chemical catalysis using a recurrent neural network adapted from natural language processing. Digital Discov. 1, 303–312 (2022).
Zhang, Z.-J. et al. Data-driven design of new chiral carboxylic acid for construction of indoles with C-central and C–N axial chirality via cobalt catalysis. Nat. Commun. 14, 3149 (2023).
Wang, S. et al. Transfer learning aided high-throughput computational design of oxygen evolution reaction catalysts in acid conditions. J. Energy Chem. 80, 744–757 (2023).
Schlosser, L., Rana, D., Pflüger, P., Katzenburg, F. & Glorius, F. EnTdecker – a machine learning-based platform for guiding substrate discovery in energy transfer catalysis. J. Am. Chem. Soc. 146, 13266–13275 (2024).
Romero, N. A. & Nicewicz, D. A. Organic photoredox catalysis. Chem. Rev. 116, 10075–10166 (2016).
Strieth-Kalthoff, F., James, M. J., Teders, M., Pitzer, L. & Glorius, F. Energy transfer catalysis mediated by visible light: principles, applications, directions. Chem. Soc. Rev. 47, 7190–7202 (2018).
Chan, A. Y. et al. Metallaphotoredox: The merger of photoredox and transition metal catalysis. Chem. Rev. 122, 1485–1542 (2022).
Ben-David, S., Blitzer, J., Crammer, K. & Pereira, F. Analysis of representations for domain adaptation. In Advances in Neural Information Processing Systems 19 137–144 (MIT Press, 2007).
Dai, W., Yang, Q., Xue, G.-R. & Yu, Y. Boosting for transfer learning. In Proc. 24th International Conference on Machine Learning 193–200 (ICML, 2007).
Daumé III, H. Frustratingly easy domain adaptation. In Proc. 45th Annual Meeting of the Association of Computational Linguistics 256–263 (ACL, 2007).
Bai, Y. et al. Accelerated discovery of organic polymer photocatalysts for hydrogen evolution from water through the integration of experiment and theory. J. Am. Chem. Soc. 141, 9063–9071 (2019).
Li, X. et al. Combining machine learning and high-throughput experimentation to discover photocatalytically active organic molecules. Chem. Sci. 12, 10742–10754 (2021).
Buglak, A. A. et al. Quantitative structure-property relationship modeling for the prediction of singlet oxygen generation by heavy-atom-free BODIPY photosensitizers. Chem. Eur. J. 27, 9934–9947 (2021).
Noto, N., Yada, A., Yanai, T. & Saito, S. Machine-learning classification for the prediction of catalytic activity of organic photosensitizers in the nickel(II)-salt-induced synthesis of phenols. Angew. Chem. Int. Ed. 62, e202219107 (2023).
Pardoe, D. & Stone, P. Boosting for regression transfer. In Proc. 27th International Conference on Machine Learning 863–870 (ICML, 2010).
Liu, Z. et al. Aggregation-enabled intermolecular photo[2+2]cycloaddition of aryl terminal olefins by visible-light catalysis. CCS Chem. 1, 582–588 (2019).
Golfmann, M., Glasgow, L., Giakoumidakis, A., Golz, C. & Walker, J. C. L. Organophotocatalytic [2+2] cycloaddition of electron-deficient styrenes. Chem. Eur. J. 29, e202202373 (2023).
Sun, H., Zhong, C. & Brédas, J. L. Reliable prediction with tuned range-separated functionals of the singlet–triplet gap in organic emitters for thermally activated delayed fluorescence. J. Chem. Theory Comput. 11, 3851–3858 (2015).
Schwaller, P., Vaucher, A. C., Laino, T. & Reymond, J. L. Prediction of chemical reaction yields using deep learning. Mach. Learn.: Sci. Technol. 2, 015016 (2021).
Liu, Z., Moroz, Y. S. & Isayev, O. The challenge of balancing model sensitivity and robustness in predicting yields: a benchmarking study of amide coupling reactions. Chem. Sci. 14, 10835–10846 (2023).
Welin, E. R., Le, C., Arias-Rotondo, D. M., McCusker, J. K. & MacMillan, D. W. C. Photosensitized, energy transfer-mediated organometallic catalysis through electronically excited nickel(II). Science 355, 380–385 (2017).
Kim, T., McCarver, S. J., Lee, C. & MacMillan, D. W. C. Sulfonamidation of aryl and heteroaryl halides through photosensitized nickel catalysis. Angew. Chem. Int. Ed. 57, 3488–3492 (2018).
Lu, J. et al. Donor–acceptor fluorophores for energy-transfer-mediated photocatalysis. J. Am. Chem. Soc. 140, 13719–13725 (2018).
Kudisch, M., Lim, C.-H., Thordarson, P. & Miyake, G. M. Energy transfer to Ni-amine complexes in dual catalytic, light-driven C–N cross-coupling reactions. J. Am. Chem. Soc. 141, 19479–19486 (2019).
Du, Y. et al. Strongly reducing, visible-light organic photoredox catalysts as sustainable alternatives to precious metals. Chem. Eur. J. 23, 10962–10968 (2017).
Farahani, A., Voghoei, S., Rasheed, K. & Arabnia, H. R. A brief review of domain adaptation. In Advances in Data Science and Information Engineering: Proceedings from ICDATA 2020 and IKE 2020 877–894 (Springer, 2021).
de Mathelin, A. et al. ADAPT: Awesome domain adaptation python toolbox. Preprint at http://arxiv.org/abs/2107.03049 (2021).
Kanter, J. M. & Veeramachaneni, K. Deep feature synthesis: Towards automating data science endeavors. In 2015 IEEE Int. Conf. on Data Sci. Adv. Analytics (DSAA) 1–10 (IEEE, 2015).
Fabry, D. C., Ronge, M. A. & Rueping, M. Immobilization and continuous recycling of photoredox catalysts in ionic liquids for applications in batch reactions and flow systems: catalytic alkene isomerization by using visible light. Chem. Eur. J. 21, 5350–5354 (2015).
Pan, X. et al. Mechanism of photoinduced metal-free atom transfer radical polymerization: experimental and computational studies. J. Am. Chem. Soc. 138, 2411–2425 (2016).
Li, S.-W., Xu, L.-C., Zhang, C., Zhang, S.-Q. & Hong, X. Reaction performance prediction with an extrapolative and interpretable graph model based on chemical knowledge. Nat. Commun. 14, 3569 (2023).
Noto, N. et al. Transfer learning across different photocatalytic organic reactions, P2-20231212. https://zenodo.org/records/15058999 (2025).
Acknowledgements
This research was supported by the JSPS/MEXT Grants-in-aid for Transformative Research Areas (A) Digi-TOS (22H05356 [N.N.], 21H05221 [R.Kojima]), the JSPS/MEXT Grants-in-aid for Early-Career Scientists (23K13744 [N.N.]), the JSPS/MEXT Grant-in-Aid for Scientific Research(A) (24H00449 [T.Y.]), the JSPS/MEXT Grants-in-aid for: Transformative Research Areas (A) Green Catalysis Science, International Leading Research, and Specially Promoted Research, KAKENHI (23H04904, 22K21346, and 23H05404 [S.S.]), the JST CREST (JPMJCR22L2 [S.S.]), and the Deutsche Forschungsgemeinschaft (DFG) within the IRTG 2678 (GRK 2678 − 437785492 [T.R.]).
Author information
Authors and Affiliations
Contributions
N.N. primarily performed computational and experimental studies, receiving advice from M.H., R.Kojima, O.G.M., T.Y, and S.S. on aspects of machine learning, quantum chemical calculations, and experimental design. R.Kunisada and T.R. provided experimental support. The work was directed by N.N. and supervised by S.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Xin Hong, Chuanqi Tan and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
.Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Noto, N., Kunisada, R., Rohlfs, T. et al. Transfer learning across different photocatalytic organic reactions. Nat Commun 16, 3388 (2025). https://doi.org/10.1038/s41467-025-58687-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58687-5
This article is cited by
-
Roadmap for transforming heterogeneous catalysis with artificial intelligence
Nature Catalysis (2026)
-
Transfer learning from custom-tailored virtual molecular databases to real-world organic photosensitizers for catalytic activity prediction
Communications Chemistry (2025)