Abstract
Thousands of nucleus-encoded chloroplast proteins are synthesized as precursors on cytosolic ribosomes and posttranslationally imported into chloroplasts. Cytosolic accumulation of unfolded chloroplast precursor proteins (e.g., under stress conditions) is hazardous to the cell. The global cellular responses and regulatory pathways involved in triggering appropriate responses are largely unknown. Here, by inducible and constitutive overexpression of ClpD-GFP to result in precursor protein overaccumulation, we present a comprehensive picture of multilevel reprogramming of gene expression in response to chloroplast precursor overaccumulation stress (cPOS), reveal a critical role of translational activation in the expression of cytosolic chaperones (heat-shock proteins, HSPs), and demonstrate that chloroplast-derived reactive oxygen species act as retrograde signal for the transcriptional activation of small HSPs. Furthermore, we reveal an important role of the chaperone ClpB1/HOT1 in maintaining cellular proteostasis upon cPOS. Together, our observations uncover a cytosolic heat shock-like response to cPOS and provide insights into the underlying molecular mechanisms.
Similar content being viewed by others
Introduction
Chloroplasts are endosymbiotic organelles that provide oxygen, which is required by most organisms on Earth, and energy for plant growth as well as for other living organisms. The chloroplast genome of higher plants typically contains 80–100 protein-coding genes, whereas the bulk of genes from the ancestral cyanobacterial genome have been transferred to the nucleus1,2. The nucleus-encoded proteins are synthesized on cytosolic ribosomes and targeted to chloroplasts to exert their function. Consequently, both the biogenesis and operation of chloroplasts rely heavily on the import of proteins (precursor proteins) from the cytosol3. The majority of the nucleus-encoded chloroplast proteins possess an N-terminal chloroplast transit peptide (cTP) which is recognized by the receptor subunits of the translocon at the outer envelope membrane of chloroplasts (TOC) and cleaved off upon successful import4,5.
Owing to their posttranslational targeting mode, nascent chloroplast precursor proteins are transiently exposed to the cytosol before being imported into chloroplasts. Under environmental stresses or genetic perturbations that reduce import capacity, precursor proteins tend to accumulate in the cytosol6,7. The unfolded precursors are proteotoxic to the cell and are likely eliminated by the ubiquitin–proteasome system (UPS)6,8. For example, in the gun1 (a key regulator of plastid retrograde signaling and protein import7,9) clpc1 (the chaperone subunit of the Clp protease, which also affects import10) double mutant, and in the gun1 single mutant upon treatment with lincomycin (an inhibitor of chloroplast translation) or norflurazon (an inhibitor of carotenoid biosynthesis), the chloroplast precursor proteins overaccumulate, leading to the upregulation of the UPS and cytosolic chaperones7. These findings suggest that sophisticated mechanisms exist to cope with the aberrant accumulation of chloroplast precursor proteins.
By using protein import-related mutants, the overexpression of individual mitochondrial-targeted proteins, or the inducible expression of “clogging” proteins, multiple pathways involved in the mitochondrial precursor protein response in yeast and mammals have been identified11,12,13. Under nonstress conditions, the ribosome-associated quality control pathway for mitochondrial polypeptides (mitoRQC) governs the cytosolic synthesis of mitochondrial proteins14, and the mitochondrial protein translocation-associated degradation (mitoTAD) pathway and the Pth2-DSK2 pathway function as monitoring systems that continuously clear arrested precursor proteins from the import channel15,16. The overaccumulation of mitochondrial precursors causes cytosolic proteostasis stress (mPOS)17 and induces a systematic response to rebalance cellular proteostasis (known as the unfolded protein response activated by mistargeting of proteins, UPRam)18,19. These responses include upregulation of the UPS and chaperones, and repression of cytosolic ribosome biogenesis and translation, which collectively promote cell survival. Upon precursor accumulation, a surveillance mechanism, namely the mitochondrial compromised protein import response (mitoCPR), is activated, which induces Cis1 expression and Msp1-dependent precursor clearance from the import channel by the proteasome20, thus releasing the import capacity and protecting mitochondrial function. Unimported mitochondrial proteins can also be translocated to the nucleus21 or engage an endoplasmic reticulum (ER) membrane-associated protein, Ema19, for degradation22. In contrast to the responses to mitochondrial precursor protein accumulation in yeast and mammals, the cellular responses to aberrant chloroplast precursor accumulation in plants and the underlying regulatory mechanisms are largely unknown.
Here, by constitutive or induced overexpression of ClpD/ClpD-GFP, a chaperone subunit of the chloroplast Clp protease that we previously identified to be prone to precursor accumulation7, we studied the cellular events that set in upon chloroplast precursor overaccumulation (cPOS). Our data unveil distinct mechanisms regulating the expression of different classes of genes and emphasize the important role of translational regulation on the expression of cytosolic chaperones (heat-shock proteins) in response to cPOS. We revealed the retrograde signaling role of chloroplast-derived reactive oxygen species (cpROS) and identified ClpB1/HOT1 as an important factor in maintaining cellular proteostasis upon cPOS.
Results
Precursor granules form in close proximity to chloroplasts
Mitochondrial import stress can be triggered by overloading the import machinery through the overexpression of individual (nucleus-encoded) mitochondrial proteins such as Cox5a, TIM50, Psd1, or Ccp120. In the case of chloroplast nuclear-encoded proteins, the precursor of ClpD (preClpD) can readily be detected (a larger molecular mass band above the mature form of ClpD on immunoblots with total cellular proteins but not with proteins from isolated chloroplasts)7, and is a potential candidate for triggering chloroplast import stress. To investigate the effects of chloroplast precursor accumulation in relation to the dynamics and subcellular localization of the overaccumulated precursors, we constructed inducible ClpD and ClpD-GFP overexpression lines (Supplementary Table 1). Interestingly, preClpD accumulated highly at 12 h of induction but then quickly declined to levels that were hardly detectable at 36 h (Supplementary Fig. 1a). In contrast, although starting to significantly accumulate only after 1 day of induction, the ClpD-GFP fusion protein was much more stable. The preClpD-GFP continued to accumulate until the end of the induction experiments (4 days; Fig. 1a) and declined slowly even after the removal of the induction reagent (Supplementary Fig. 1b). The ClpD-GFP fluorescence signals show a diffuse distribution within chloroplasts that corresponds well with chlorophyll autofluorescence after 1 day of induction (Fig. 1b–d). However, after 2 days, preClpD-GFP significantly accumulated and formed punctate granules in close proximity to chloroplasts (Fig. 1b, c), which was further confirmed by visualizing the boundaries of the chloroplasts with differential interference contrast imaging (DIC; Supplementary Fig. 1g). This is very similar to the mitochondrial precursor granules observed in yeast (named MitoStore23) and human cells24. This phenomenon was observed not only in the toc75III background (t-i-ClpD-GFP; Fig. 1b–d and Supplementary Fig. 1g), which is the major isoform of the channel-forming subunit of the TOC apparatus25,26,27, but also in the wild type (WT; C-i-ClpD-GFP) and gun1 (g-i-ClpD-GFP) backgrounds (Supplementary Fig. 1c–f, h, i). Some GFP signals were overlapping with chlorophyll autofluorescence, indicating that the import was only partially repressed. The accumulation of precursor granules correlates with the chloroplast import capacity in that more granules accumulated in t-i-ClpD-GFP plants than in C-i-ClpD-GFP and g-i-ClpD-GFP plants (Fig. 1e). Importantly, under abiotic stress conditions such as heat and high light stresses, chloroplast precursors also accumulate in untransformed plants, especially in import-related mutants (Fig. 1f and Supplementary Fig. 2a). Additionally, when cpGFP (chloroplast-targeted GFP by transit peptide from the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, RuBisCo)28 was introduced into the tic40-4 mutant background (cpGFP tic40-4), pre-cpGFP granules also accumulated weakly under heat stress (Supplementary Fig. 2b). These results indicate that the accumulation of precursors is physiologically relevant.
a Immunoblots showing preClpD-GFP accumulation in the t-i-ClpD-GFP line detected by anti-GFP antibodies. The numbers below the immunoblot represent the accumulation of preClpD-GFP relative to mature ClpD-GFP (in %). Seven-d-old seedlings were induced for the days indicated and whole seedlings were harvested for analyses. Actin served as loading control. pre: preClpD-GFP, mat: mature ClpD-GFP. b Representative images showing that preClpD-GFP granules closely associated with chloroplasts (see also Supplementary Fig. 1g). Seven-d-old t-i-ClpD-GFP seedlings were induced for 1 or 2 days, the newly emerged true leaves were analyzed. The boxed areas in the middle panel are magnified and shown below. Scale bars, 20 μm for the upper panel, and 5 μm for the middle and bottom panels. c Fluorescence intensity plots along the magenta lines in (b) measured by using Fiji software. d Colocalization of GFP and chlorophyll fluorescence of the boxed area in (b) analyzed by Coloc 2 in Fiji software. PCC, Pearson’s colocalization coefficient; MCC, Manders’ colocalization coefficient (tM1 and tM2 are the proportion of colocalized signal to GFP and chlorophyll fluorescence, respectively). e Proportion of cells containing preClpD-GFP granules in different transgenic lines at 2 days of induction. Data are presented as means ± s.d. (n = 10 images from 2 independent experiments). Two-tailed Student’s t-tests were performed to determine the significance of differences. f PreClpD accumulation after 2 days of heat stress at 37 °C. Newly emerged true leaves were harvested for analyses. The numbers below the immunoblot represent the accumulation of preClpD relative to the mature ClpD (in %). Pre, preClpD; mat, mature ClpD; asterisks, nonspecific cross-reactions. Actin served as loading control. g The maximum photochemical efficiency of PS II (Fv/Fm) of t-i-ClpD-GFP and untransformed toc75III seedlings after 4 days of induction. Scale bars, 1 cm. h Light-response curves of chlorophyll a fluorescence parameters after 2 days of induction. Y(II), quantum yield of PSII; ETR (II), electron transfer rate of PSII. The data are means ± s.d. (n = 30 biological replicates). In (f–h), 6-d-old seedlings were used for induction. Source data are provided as a Source Data file.
The maximum quantum yield of photosystem II (PSII; Fv/Fm) of the t-i-ClpD-GFP line was 71.2% relative to that of the untransformed toc75III mutant and 69.3% to that of the WT after induction (Fig. 1g and Supplementary Fig. 2c–e). The light‒response curves of chlorophyll a fluorescence revealed a pronounced decrease in PSII yield and electron transfer rate with increasing light intensity (Fig. 1h and Supplementary Fig. 2f). These results suggest that accumulation of chloroplast precursor proteins negatively affects photosynthetic activity.
Accumulation of chloroplast precursor proteins impair plant growth
To investigate the effects of precursor accumulation on plant development, we constitutively overexpressed ClpD in the WT, gun1, and toc75III backgrounds (Supplementary Table 1). PreClpD can be readily detected in the transgenic lines in the gun1 (g-ClpD) and toc75III (t-ClpD) backgrounds, but not in the WT (C-ClpD) background (Supplementary Fig. 3a). Consequently, the growth of g-ClpD and t-ClpD seedlings was significantly retarded (Supplementary Fig. 3b, c). Additionally, as previously reported, g-ClpD seedlings exhibited an albino cotyledon phenotype7 (Supplementary Fig. 3b). Importantly, the phenotype is not caused by the accumulation of mature ClpD inside chloroplasts, because the C-ClpD lines, although overaccumulating mature ClpD, do not exhibit this phenotype (Supplementary Fig. 3a, b).
The preClpD protein was not detected in the C-ClpD lines (Supplementary Fig. 3a). This indicates that the import capacity cannot easily be saturated by the overexpression of a single protein in the WT background. Compared with preClpD, preClpD-GFP accumulates constantly at high levels upon induction (Fig. 1a and Supplementary Fig. 1a). To determine the impact of precursor accumulation on plant growth and investigate the corresponding cellular responses while excluding the effects of the gun1 (in g-ClpD) and toc75 (in t-ClpD) mutations, we constructed ClpD-GFP overexpression lines in the WT background and screened the transgenic lines for plants that accumulate the precursor protein (Supplementary Table 1). PreClpD-GFP accumulated in 3-d-old seedlings of ClpD-GFP overexpressing lines #5 (to 9% of the mature ClpD) and #6 (to 8% of the mature ClpD). However, on day 6, preClpD-GFP was nearly undetectable in the weaker overexpressor (line #6), whereas it remained highly abundant in the stronger overexpressor (line #5; Fig. 2a). In line with this observation, the line #5 seedlings presented a pale-green and growth-retardation phenotype (Fig. 2b), and the chlorophyll content and fresh weight were significantly lower than those of the WT (Supplementary Fig. 4a, b). Similar to the inducible lines (Fig. 1b, c and Supplementary Fig. 1c–i), preClpD-GFP granules formed around the chloroplasts, and these chloroplasts were smaller than neighboring chloroplasts, which are less affected and are not associated with precursor granules (Supplementary Fig. 4c, d).
a PreClpD-GFP accumulation in 3-d-old and 6-d-old constitutive ClpD-GFP overexpressing lines (lines #6 and #5). The numbers below the ClpD-GFP immunoblot represent the accumulation of preClpD-GFP relative to mature ClpD-GFP (in %). Actin served as loading control. pre, preClpD-GFP; mat, mature ClpD-GFP. b Representative images of 3-d-old and 6-d-old WT, ClpD-GFP line #6, and line #5 seedlings. Scale bar, 1 cm. c Immunoblots showing reduced accumulation of photosynthesis-related proteins in ClpD-GFP line #5 compared with the WT and line #6. LHC, light-harvesting complex; b6f, cytochrome b6f complex; ATP, ATP synthase; Chl, enzymes involved in chlorophyll biosynthesis pathway. Asterisks indicate nonspecific bands (cross-reactions). Actin served as loading control. d Quantification of the immunoblots for the selected proteins shown in (c). Data are means ± s.e. (n = 3 biological replicates). Two-tailed Student’s t-tests were performed to determine the significance of differences between the WT and the ClpD-GFP lines #5 or #6. e BN‒PAGE analysis showing the reduction of photosynthetic complexes in ClpD-GFP line #5 on day 6. Equal amounts of thylakoid proteins were loaded. In (a, c–e), whole seedlings of 3 d or 6 d were harvested for analyses. f TEM images showing the disturbed chloroplast ultrastructure in 6-d-old seedlings of ClpD-GFP line #5. The boxed areas are magnified and shown in the right panels of both timepoints. Scale bars, 1 μm for the left images and 0.5 μm for the right magnified images. Source data are provided as a Source Data file.
As additional controls, we overexpressed the ClpD transit peptide alone-fused with GFP7 (ClpDTP-GFP; Supplementary Fig. 5 and Supplementary Table 1) and an N-terminally truncated ClpD-GFP that lacked the transit peptide (ΔTP-ClpD-GFP; Supplementary Fig. 6 and Supplementary Table 1). The ClpDTP-GFP fusion protein was efficiently imported into chloroplasts without the accumulation of detectable precursors (Supplementary Fig. 5a, b) and the plants were indistinguishable from the WT plants (Supplementary Fig. 5c, d). The ΔTP-ClpD-GFP protein was diffusely distributed in the cytosol and did not form chloroplast-associated granules (Supplementary Fig. 6a, b). Although the fresh weight of the ΔTP-ClpD-GFP transgenic seedlings was mildly reduced (78% of that of the WT, compared with 59% of line #5 relative to the WT), they did not exhibit a pale-green phenotype (Supplementary Fig. 6c–e). These results indicate that the formation of preClpD-GFP granules and their negative effects rely on the sorting of the proteins in a cTP-dependent manner and their routing toward the chloroplast import pathway.
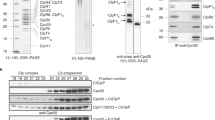
Under mitochondrial import stress in yeast and human cells, the precursors stall in the import machinery20,29. To examine whether preClpD-GFP stalls in the TOC complex, we performed coimmunoprecipitation (co-IP) with seedlings from the ΔTP-ClpD-GFP, ClpD-GFP (line #5), and t-i-ClpD-GFP lines (Supplementary Fig. 7a). Although weakly detected in the ΔTP-ClpD-GFP immunoprecipitates (possibly because of nonspecific binding), Toc75 substantially coprecipitated with (pre)ClpD-GFP from t-i-ClpD-GFP plants, in which a significant amount of preClpD-GFP accumulated (Supplementary Fig. 7a). In the ClpD-GFP line #5, the amount of copurified Toc75 was only slightly greater than that in the ΔTP-ClpD-GFP line, possibly because the accumulation of ClpD-GFP protein was much lower (Supplementary Fig. 7a). These results suggest that a fraction of accumulated preClpD-GFP stalls in the import machinery.
The abundance of all analyzed subunits of the photosynthesis-related protein complexes was strongly reduced in the ClpD-GFP line #5 plants, including chloroplast-encoded core subunits (PsbA, PsbC, PsbD for PSII; PsaA for PSI; PetA for the cytochrome b6f complex; and AtpA, AtpB for ATP synthase) and the nucleus-encoded subunits of the light-harvesting complex (LhcB1 and LhcA4; Fig. 2c, d). As expected, this resulted in a significant reduction in the corresponding photosynthetic complexes and supercomplexes (Fig. 2e). In contrast, although decreased on day 3, the accumulation of the subunits from the Clp protease complex (ClpC, ClpP4, and ClpR3) in ClpD-GFP line #5 was comparable to that in line #6 and the WT on day 6 (Supplementary Fig. 7b, c), suggesting that the pale-green phenotype of line #5 on day 6 was not caused by a reduction of Clp subunits. Interestingly, GUN5 (the H subunit of Mg-chelatase which also acts as a regulator of tetrapyrrole biosynthesis (TPB)-mediated plastid retrograde signaling30) and Tic40 were increased in line #5, but not in line #6. The structure of the thylakoid membrane network was noticeably altered in the line #5 chloroplasts on day 3, and more extensive aberrations were observed on day 6, with large empty areas being observable (Fig. 2f). Consistent with this ultrastructural evidence of photodamage, increased amounts of reactive oxygen species (ROS) accumulated in the line #5 seedlings (Supplementary Fig. 4e).
Surprisingly, homozygous ClpD-GFP line #5 plants showed phenotypic segregation in the homozygous T4 progeny (Supplementary Fig. 4f). The appearance of the pale-green phenotype in the offspring correlated with preClpD-GFP accumulation (Supplementary Fig. 4f, g). The steady-state levels of ClpD-GFP mRNA in the green segregants were substantially lower than those in the pale-green plants (Supplementary Fig. 4h), indicating that the transgene was partially silenced in these green plants. Taken together, these findings further support the conclusion that the phenotype correlates with the accumulation of preClpD-GFP protein.
Overall, the phenomena observed here are similar to those observed upon mitochondrial precursor overaccumulation stress (mPOS) in yeast17. We therefore refer to them as chloroplast precursor overaccumulation stress (cPOS).
Upregulation of cytosolic translation and PQC in response to cPOS
To reveal the response of plant cells to cPOS, we conducted proteomic analyses on 3-d-old and 6-d-old ClpD-GFP line #5 seedlings, with the WT and the ΔTP-ClpD-GFP lines used as controls. Compared with those in the WT seedlings, 69 increase and 17 decrease accumulated proteins were identified in the ΔTP-ClpD-GFP seedlings on day 3, and 6 increase and 36 decrease accumulated proteins were identified on day 6 (Supplementary Fig. 6f and Supplementary Data 1). Gene Ontology (GO) enrichment analysis indicated that the term “oxoacid metabolic process” was enriched for the proteins whose abundance increased on day 3. No other GO terms were enriched in any of the other sets of differentially accumulated proteins. Clearly distinct from the ΔTP-ClpD-GFP line, 166 increase and 105 decrease accumulated proteins were identified on day 3, and 217 increase and 231 decrease accumulated proteins were identified on day 6 in ClpD-GFP line #5 (Fig. 3a and Supplementary Data 1). The peptides derived from the cTP region (P-TPs), indicating the cytosolic accumulation of chloroplast precursors (given that cTP is cleaved off and degraded upon successful import), accumulated to a higher level on day 6 in ClpD-GFP line #5 than in the WT (Fig. 3b), which was proven by immunoblotting analyses of ClpD and glutamine-tRNA reductase (GluTR; Fig. 3c). These proteins include many photosynthesis-related proteins and metabolic enzymes (Supplementary Data 2), indicating that the import of essential chloroplast proteins was affected because the import capacity was reduced by ClpD-GFP overexpression.
a Volcano plot illustrating the differentially expressed proteins between the WT and ClpD-GFP line #5 on day 3 (left) or day 6 (right) of seedling development. Proteins with a Student’s t-test (two-tailed) p < 0.01 and fold change >1.5 between genotypes were identified as differentially accumulated proteins. PS photosynthesis, Ribo ribosome, 26S 26S proteasome. b Histogram showing intensity-based fractions of peptides mapped to transit peptide regions (P-TPs, indicating precursor accumulation in the cytosol) relative to the total peptide intensities of the proteins from which the P-TP was derived. Data are means ± s.e. (n = 3 biologically independent samples). c Immunoblot analyses showing the preClpD-GFP and preGluTR accumulated in 6-d-old ClpD-GFP #5 seedlings. The whole seedlings were harvested for analyses. The numbers below the immunoblots represent the accumulation of precursors relative to mature proteins (in %). d Interaction network of increase accumulated proteins in ClpD-GFP line #5 on day 6 showing proteins related to the proteasome, ribosome, translation regulation, and chaperone system. The interaction network was generated by STRING31 (including physical and functional interactions) and visualized by Cytoscape87. e Immunoblot analyses showing the increased abundance of 26S proteasomal subunits (RPT2a and RPT5a), the cytosolic chaperones HSP70 (cy70) and HSP90.1 (cy90.1), a 40S ribosomal subunit (US3Z), and polyubiquitinated proteins in 6-d-old ClpD-GFP line #5 seedlings. cp70, chloroplast HSC70. Actin served as loading control. f Quantification of selected proteins from the immunoblots shown in (e). Data are means ± s.e. (n = 3 biological replicates). In (b, f), two-tailed Student’s t-tests were performed to determine the significance of differences between the WT and ClpD-GFP lines #5 or #6. g Polysome profiles of 6-d-old WT and ClpD-GFP line #5 seedlings. h Genome-wide aggregated normalized P-site counts in the WT and ClpD-GFP line #5 from Ribo-seq data. The regions 9 nt upstream and 18 nt downstream of the start codon, and 9 nt downstream and 18 nt upstream of the stop codon are shown. Source data are provided as a Source Data file.
More than 82% (87 out of 105 on day 3, 195 out of 231 on day 6) of the decreased proteins were chloroplast localized and enriched in various processes of photosynthesis (Supplementary Figs. 8a–c and 9a). Compared with that on day 3, the number of downregulated photosynthesis-related proteins was largely increased on day 6 (from 37 to 101; Fig. 3a and Supplementary Data 1). This is consistent with the pale-green phenotype and the reduced abundance of photosynthetic complexes (Fig. 2b–e). Adding 1% or 2% sucrose to the growth medium partially rescued the retarded growth phenotype of the line #5 plants, thus further confirming that they suffer from reduced photosynthetic capacity (Supplementary Fig. 9b, c).
The proteins whose abundance increased were enriched in protein quality control (PQC)-related processes such as “protein folding”, “ribosome biogenesis”, “translation”, and “proteasomal protein catabolic process” (Supplementary Fig. 8a–c). Protein interaction network analysis (including physical and functional interactions predicted by STRING database31) revealed pronounced overrepresentation in functional categories related to cytosolic ribosomes and translation regulation, chaperones, and the proteasome (Fig. 3d). This finding was further verified by immunoblot analyses of representative proteins or subunits associated with these cellular processes (Fig. 3e, f), in contrast to no visible changes of these proteins in ClpDTP-GFP plants (Supplementary Fig. 5a).
The numbers of differentially accumulated ribosomal proteins at days 3 and 6 were comparable (31 vs. 32; Fig. 3a and Supplementary Data 1). However, on day 3 most of the affected ribosomal proteins were decreased in abundance (26 out of 31; Supplementary Fig. 8d, e), among which 21 were chloroplast 70S ribosomal subunits (Supplementary Fig. 8f), suggesting that chloroplast translation decreased in ClpD-GFP line #5 plants on day 3. In contrast, the abundance of differentially accumulated ribosomal proteins on day 6 were mainly increased (20 out of 32; Supplementary Fig. 8d, e), and all of these increased ribosomal proteins were cytosolic 80S ribosomal proteins, with 13 belonging to the 60S subunit and 7 to the 40S subunit (Fig. 3d–f and Supplementary Data 1). Moreover, 8 translation regulators, including eukaryotic translation initiation factor 2α (eIF2α), elongation factor 1α (EF-1α), and factors involved in ribosome assembly and aminoacyl-tRNA synthetases, were also increased (Fig. 3d). This is consistent with the enrichment of the GO terms “ribosome biogenesis” and “translation” on day 6 for the increase accumulated proteins (Supplementary Fig. 8c), reflecting an increased translation capacity in the cytosol upon cPOS. This conclusion was further substantiated by polysome profile analyses (Fig. 3g and Supplementary Fig. 10a) and the global increase in P-site counts (representing the increase in total cellular translation32,33) in ClpD-GFP line #5, as revealed by ribosome profiling analysis (see below; Fig. 3h and Supplementary Fig. 10b). The increase of cytosolic ribosomal subunits and translation capacity in response to cPOS is in stark contrast to the repressed translation upon mitochondrial import stress in yeast17,18,19.
Transcriptional and translational remodeling upon cPOS
To characterize the molecular outcome of enhanced cytosolic translation and its role in the cPOS response, we carried out unbiased ribosome profiling analyses (Ribo-seq), combined with RNA-seq, on 6-d-old ClpD-GFP line #5 and WT seedlings. More than 96.4% and 85.8% of the reads from RNA-seq and Ribo-seq, respectively, were uniquely mapped, with a total mapping ratio >98.2% in both datasets (Supplementary Table 2). The high quality of the Ribo-seq data32,34 is further evidenced by the high coding sequence (CDS) mapping ratio, the 28–36 nt fragment length distribution, the abrupt appearance of a footprint signal 3 nt upstream of the start codon, a rapid decline in the signal at the stop codon, and most importantly, a strong 3-nt periodicity (Supplementary Fig. 11), which represents the codon-long stepwise movement of the ribosomes along the mRNA. None of these features were observed in the RNA-seq data (Supplementary Fig. 11). We thus obtained datasets for the transcriptome and translatome of two highly reproducible biological replicates (Supplementary Fig. 12a, b).
The global transcript abundance and translation output (represented by ribosome footprints) within the same samples were largely correlated, with a Pearson’s correlation coefficient (r) > 0.88 in both the WT and ClpD-GFP line #5 (Supplementary Fig. 12c). However, in agreement with previous comparisons35,36,37, the correlation between the changes in transcript accumulation and the changes in translation output was relatively poor (r = 0.195; Supplementary Fig. 12d), suggesting the existence of a layer of regulation at the translational level. Indeed, we identified 630 upregulated and 294 downregulated genes at the transcript level in ClpD-GFP line #5 in comparison with the WT. In contrast, 978 upregulated and 699 downregulated genes at the translation output level (excluding the organelle genome-encoded genes) were identified (Fig. 4a, c and Supplementary Data 3 and 4). A direct comparison between translation output and transcript abundance, referred to as translation efficiency (TE), identified 405 upregulated and 298 downregulated genes (Fig. 4e and Supplementary Data 5). These results provide further support for strong translational regulation in response to cPOS. Interestingly, among the transcriptionally downregulated genes, 59.5% (175 out of 294) were followed by corresponding alterations at the level of translation output, but only 31.3% (197 out of 630) of the transcriptionally upregulated genes were also translationally upregulated (Supplementary Fig. 12e).
a, c, e Volcano plots illustrating the differentially expressed genes at the transcript level (a), translation output level (c, revealed by ribosome footprints), or with significantly changed TE (e) between the WT and ClpD-GFP line #5. Organelle-encoded genes were not included. b, d, f GO term enrichment analyses of the differentially expressed genes at the transcript level (b), translation output level (d), or with significantly changed TE (f) between the WT and ClpD-GFP line #5. Significantly enriched GO terms were identified by two-tailed Fisher’s exact test with FDR (<0.05) for multiple testing corrections and an enrichment fold >3.
Consistent with the increased protein accumulation of cytosolic ribosomal subunits and translation regulators (Fig. 3d and Supplementary Data 6) and the increased translation capacity (Fig. 3g, h), GO terms related to ribosome biogenesis and rRNA processing and/or maturation were enriched for the transcriptionally upregulated genes (Fig. 4b). This transcriptional upregulation was undermined by translational repression, which led to a mild increase in translation output (Supplementary Fig. 13a, b and Supplementary Data 6), indicating a tight control to the expression of these genes. A similar phenomenon was also observed for the cytosolic ribosomal subunits, except for the 40S subunit Rps5 (Supplementary Fig. 13c, d), which was upregulated through translational activation. However, the relatively minor increase in translation output resulted in a significant increase in protein abundance (Supplementary Fig. 13e and Supplementary Data 6), possibly indicating a slow protein turnover rate of ribosomal proteins, which is consistent with a previous determination of the protein degradation rate by 15N progressive labeling of developing Arabidopsis leaves38 (Supplementary Fig. 13f and Supplementary Data 7). The proportion of slow-turnover chloroplast ribosomal proteins was also much higher (57.6%; 19 out of 33) than that of the other protein groups. For example, 11.5% (14 out of 122) of photosynthesis-related proteins had a slow degradation rate, and 13.1% (161 out of 1228) of all proteins the degradation rate of which has been determined38 (Supplementary Fig. 13f and Supplementary Data 7). This likely explains our observation that, although 21 chloroplast ribosomal proteins were decreased at the protein level on day 3 in ClpD-GFP line #5, 18 of them accumulated to comparable levels to those in the WT on day 6 (Supplementary Fig. 8d, f).
A subset of genes related to organellar RNA metabolism were upregulated at the transcriptional level upon cPOS. The GO terms “plastid transcription”, “cytidine to uridine editing”, and “RNA modification” were enriched (Fig. 4b). We identified 101 transcriptionally upregulated pentatricopeptide repeat (PPR) genes, accounting for ~22.5% of all PPR genes in Arabidopsis39. Many of these genes have been well studied for their function in plastid and/or mitochondrial RNA editing40 (Supplementary Fig. 14a, b and Supplementary Data 8). We then examined the editing efficiency of 6 known plastid RNA editing sites, including clpP-559, rpl23-89, ndhD-2, ndhG-50, rpoA-200, and rpoC1-497. The results revealed significantly increased editing efficiencies for clpP-559 and rpoC1-497 and decreased editing efficiencies for the other sites (Supplementary Fig. 14c). For mitochondria, we analyzed 9 editing sites from the cox2 and orfX genes, which were increased for cox2-138 and decreased for all orfX sites (Supplementary Fig. 15). These results suggest that the RNA editing was affected in both plastids and mitochondria upon cPOS.
Next, we investigated the regulation of photosynthesis-related genes. Proteomic analysis identified 101 decrease accumulated photosynthesis-related proteins in 6-d-old ClpD-GFP line #5 seedlings, of which 82 were nucleus-encoded (Fig. 5a and Supplementary Data 9). The correlation between the changes in transcript accumulation and the changes in translation output (as determined by ribosome profiling) of these 82 genes was relatively high (r = 0.67) between ClpD-GFP line #5 and the WT (Fig. 5b, c). A similar trend was observed for other photosynthesis-related genes, including the small subunit of RuBisCo (RbcS2B), LhcB2.1, GOLDEN2-LIKE1 (GLK1) and GLK2, which encode key transcriptional activators of many photosynthesis-related genes41 (Fig. 5d). In contrast, the correlation between the changes in translation output and the changes in protein abundance was rather poor (r = 0.03), with a large decrease at the protein levels (Fig. 5b, c). By calculating the superfold of the change in protein abundance over the change in translation output, 69 of the 82 downregulated photosynthesis-related proteins (84.1%) exhibited a superfold change of more than a 1.5-fold decrease (Fig. 5a and Supplementary Data 9), suggesting a strong downregulation by posttranslational mechanisms. These proteins could be degraded either by chloroplast-localized proteases (upon or after import) or by cytosolic proteolytic systems, especially the upregulated UPS (Fig. 3d, e), when their precursors accumulate in the cytosol. Indeed, from 12 of these proteins, P-TPs were detected in our mass spectrometric analyses (Fig. 5a, marked with asterisks, and Supplementary Data 2), including several LhcB proteins (LhcB1.2, LhcB1.5, LhcB5, and LhcB6), PsbP-1, RbcS3B, the B subunit of ATP synthase (PDE334), and enzymes from the Calvin‒Benson cycle.
a Hierarchical clustering heatmap showing the log2 FC (line #5/WT) of the transcripts, footprints, and TEs of the 82 significantly decreased nucleus-encoded photosynthesis-related proteins identified by comparative proteomic analyses. Genes with a protein FC (line #5/WT) over translation output (represented by footprints) FC (line #5/WT) of less than 1.5-fold are marked in green. Proteins whose P-TPs were detected (cf. Supplementary Data 2) are marked with asterisks. b, c Scatter plot showing the correlation between the FC of transcripts and the FC of footprints (b), and between the FC of footprints and the FC of proteins (c) for these 82 genes (cf. Supplementary Data 9). Pearson’s correlation coefficients (r) are indicated. d Normalized distribution of RNA-seq and Ribo-seq reads over representative photosynthesis-related genes in the WT and ClpD-GFP line #5 plants. The fold change and the associated FDR at the transcript, footprint, and TE levels are shown.
Interestingly, when the changes in protein accumulation were compared with the changes in translation output for the chloroplast-encoded genes (Supplementary Fig. 16), the translation output change was more pronounced than the protein abundance change (44 out of 56 proteins that we quantified via mass spectrometry). Among these proteins, 36 (64.3%) exhibited a superfold change of more than 1.5-fold, and for 30 of the 36 proteins, the decrease in protein abundance was smaller than the decrease in translation output. This was especially the case for ribosomal protein-coding genes (Supplementary Fig. 16), which is consistent with the slow turnover rate of ribosomal proteins (Supplementary Fig. 13f).
Precursors are usually unfolded proteins that are proteotoxic to the cell and induce an upregulation of cytosolic chaperones in the yeast mPOS17,19. The GO terms related to “chaperone-mediated protein complex assembly”, “protein folding/refolding”, and “response to heat” were enriched at the footprint and TE levels but were less pronounced at the transcript level (Fig. 4b, d, f), indicating a strong control by translational activation. The genes in these GO terms were mostly molecular chaperones, including the genes encoding HEAT SHOCK PROTEIN 70 (HSP70), HSP90, the HSP100/ClpB family of chaperones, and many small HSPs (sHSPs; Supplementary Data 10 and Supplementary Fig. 17). These results suggest that, compared with relatively mild upregulation at the transcript level (cf. Figs. 6a and 7a)19, translational regulation plays a major role in chaperone induction in response to cPOS. Importantly, this translational activation of HSPs was not observed in the high-light treated virilizer1 (vir1) mutant (encoding a N6-methyladenosine writer of mRNA), which exhibited similar reductions in chlorophyll content and photosynthetic complex abundance35 (Supplementary Data 11), indicating that it represents a specific cellular response to overaccumulated chloroplast precursors.
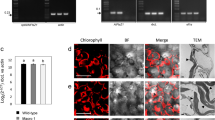
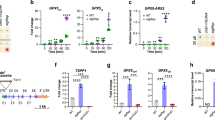
a Heatmap illustrating the differentially expressed sHSP genes at the transcriptional or translational level that belong to the GO terms “response to oxidative stress” or “response to hydrogen peroxide”. The ROS-scavenging enzymes in different subcellular compartments are also shown. b RT‒qPCR analyses demonstrating that the transcriptional upregulation of sHSP genes (17.4, 17.6A, 17.6C, 21, 23.5, 23.6) in ClpD-GFP line #5 is suppressed by the ROS scavenging reagent GSH. HSP18.5 and the transcripts encoding two large HSPs (HSP90C and mtHSC70-1) were used as negative controls. c Representative images showing ROS burst in the chloroplasts of the inducible ClpD overexpressing line C-i-ClpD-10 at 12 h of induction. Newly emerged true leaves from 6-d-old seedlings were stained with the H2DCFDA fluorescent dye for ROS visualization. Scale bar, 20 μm. d RT‒qPCR analyses showing the transcriptional upregulation of HSP17.4, HSP17.6A, and HSP17.6C from the samples in (c) (C-i-ClpD-10) and an additional line (C-i-ClpD−2). In (b, d), data are mean ± s.d. (n = 3 biological replicates). Two-tailed Student’s t-tests were performed to determine the significance of differences. Source data are provided as a Source Data file.
a Heatmap showing the translational upregulation of cytosolic large HSPs in response to precursor accumulation. b–d ClpD-GFP line #5 is sensitive to the HSP90 inhibitor GDA and the HSP70 inhibitor VER. Representative images (b), chlorophyll contents (c), and fresh weights (d) of 6-d-old seedlings are shown. Data are mean ± s.d. (n = 3 and n = 17 biological replicates in (c, d), respectively). For fresh weight quantification, each replicate was an average of 10 seedlings. In (c, d), the percentage of line #5 relative to that of the WT is indicated. Scale bar in (b), 1 cm. e Candidate proteins associated with preClpD-GFP identified by co-IP-MS using cytosolic fractions from ClpD-GFP line #5 or ΔTP-ClpD-GFP seedlings. f Colocalization assay of HOT1-mCherry with preClpD-GFP. The preClpD-GFP and HOT1-mCherry colocalized granules were indicated with arrows. The purple arrow-marked granules were enlarged on the right. Five-d-old #5/HOT1-mCherry seedlings were used for analyses. Scale bar, 5 μm, and 1 μm for the bottom-right enlarged panel. Chl, chlorophyll. g Representative images of 6-d-old WT, hot1-3, ClpD-GFP line #5, and hot1−3 #5 (hot1 #5) seedlings. Scale bar, 0.5 cm. h, i Chlorophyll (Chl) contents (h) and chlorophyll a/b ratios (i) of the seedlings shown in (g). Data are means ± s.d. (n = 3 biological replicates). Two-tailed Student’s t-tests were performed to determine the significance of difference between line #5 and hot1 #5. j Polyubiquitinated proteins accumulate to higher levels in hot1 #5 than in line #5. Whole seedlings of 6-d-old were harvested for analyses. The numbers below the ClpD-GFP immunoblot represent the accumulation of preClpD-GFP relative to mature ClpD-GFP (in %). Actin served as loading control. Source data are provided as a Source Data file.
Chloroplast ROS are required for the transcriptional activation of sHSPs
The reduced import of essential nucleus-encoded subunits inhibited photosystem biogenesis in ClpD-GFP line #5 seedlings (Fig. 2c–e) and caused hydrogen peroxide (H2O2) accumulation (Supplementary Fig. 4e). The GO term “response to hydrogen peroxide” was highly enriched at the footprint and TE levels (Fig. 4d, f), including many sHSP genes, the H2O2 scavenging enzymes catalase 1 (CAT1) and CAT3, and HEAT SHOCK FACTOR A2 (HSFA2, a key transcription activator of HSPs upon heat stress42,43; Fig. 6a and Supplementary Data 10). While high levels of ROS are detrimental, ROS also act as important signaling molecules for chloroplasts to communicate with the nucleus and mitochondria44,45. In addition to the strong translational activation of sHSPs (Fig. 6a and Supplementary Fig. 17), many sHSPs, including cytosolic HSP17.4, HSP17.6A, HSP17.6C, plastid HSP21, and mitochondrial HSP23.5 and HSP23.6, were also transcriptionally upregulated (Fig. 6a). The transcriptional induction of these sHSPs was significantly inhibited by treatment with the antioxidant reduced glutathione (GSH), indicating that ROS are necessary for their induction (Fig. 6b). In contrast, the upregulation of plastid HSP90C and mitochondrial HSC70-1 (mtHSC70-1) was not affected by GSH treatment (Fig. 6b), suggesting a specific role of ROS in the transcriptional induction of these sHSPs.
Importantly, these ROS are chloroplast-derived (cpROS) and can be readily detected in the chloroplasts of inducible ClpD-expressing lines after 12 h of induction (Fig. 6c and Supplementary Fig. 18), the time point at which preClpD accumulates at high levels (Supplementary Fig. 1a). Consistent with the observations made in the constitutive ClpD-GFP line #5, sHSP genes were highly upregulated in the inducible lines at 12 h of induction (Fig. 6d). Together, our findings suggest that cpROS serves as a retrograde signal for the transcriptional induction of sHSPs upon cPOS.
ClpB1/HOT1 plays a crucial role in maintaining cellular proteostasis in cPOS
In addition to sHSPs, large HSP family proteins, including HSP70s, HSP90s, and HSP100s/ClpBs, are constitutively expressed and play essential roles during development and under stress46,47,48. Cytosolic HSP70-5, HSP70-8, HSP90.1, HSP70/90 cochaperones (including HSP40/DnaJ proteins), and HSP101/ClpB1 (also known as HOT1)49 were largely upregulated at the translation output and TE levels, suggesting that they are activated mainly by translational upregulation (Fig. 7a and Supplementary Fig. 17). In contrast, the levels of HSP70/90/100 from other cellular compartments are either unchanged or decreased (plastid, ER), or only mildly increased (mitochondria). In particular, the ClpD-GFP line #5 seedlings are hypersensitive to the cytosolic HSP70 inhibitor VER155008 (VER) and the HSP90 inhibitor geldanamycin (GDA; Fig. 7b–d), suggesting a potential role for HSP70/90 in the cPOS response. To verify the physiological relevance of the phenomena observed in ClpD-GFP overexpressing lines, we analyzed several protein import-related mutants, including ppi1, toc75III, and tic40-4 (Supplementary Fig. 19). Precursors of ClpD and GluTR were detected in these mutants, and increased accumulation of 26S proteasome subunits (RPT2a and RPT5a) and cytosolic HSP70 and HSP90 was also observed (Supplementary Fig. 19).
Having shown that precursor-containing granules accumulate in the cytosol (Fig. 1b, Supplementary Figs. 1c, d and 4c), we next wanted to dissect the composition and cellular regulation of the granules. To this end, we conducted co-IP analyses with cytosolic fractions from the ClpD-GFP #5 and ΔTP-ClpD-GFP lines (Supplementary Fig. 20a). In addition to highly enriched ClpD-GFP precursors (represented by the identification of P-TPs), the precursors of several other chloroplast proteins were also identified (Supplementary Fig. 20b). Interestingly, many translation-related proteins have been identified, including eIF4G1, eIF2α, several 80S ribosomal subunits, RACK1C, a highly conserved WD40 domain protein found in all eukaryotes that is associated with the head of the 40S ribosome which controls translation initiation in an mRNA specific manner50,51,52,53, and LOS4, an RNA helicase that is essential for mRNA export from the nucleus under both normal temperature and heat stress54 (Fig. 7e). Νοtably, eIF2α and RACK1C were also increased at the protein level in ClpD-GFP line #5 (Fig. 3d), raising the possibility that they play a role in gene-specific localized translation at the precursor granules. Importantly, the identification of translation-related components does not come from the co-IP of these proteins with nascent ClpD polypeptide chains that stalled on the ribosomes because of ClpD overexpression. This is evidenced by no ribosome stalls being observed on ClpD transcripts (Supplementary Fig. 21a) and by control experiments with the ΔTP-ClpD-GFP line, which accumulates comparable levels of ClpD proteins to those of the ClpD-GFP line #5 plants (Supplementary Figs. 6a and 21b).
Notably, the HSP100 family chaperone ClpB1/HOT1 (hereafter HOT1) was identified to associate with preClpD-GFP (Fig. 7e). HOT1 is a key molecular chaperone involved in plant thermotolerance and participates in plant development under nonstress conditions55,56,57. Expression of HOT1-mCherry in ClpD-GFP line #5 (#5/HOT1-mCherry; Supplementary Table 1) indicated that HOT1 partially colocalized with preClpD-GFP in preClpD-GFP granule-containing cells (Fig. 7f and Supplementary Fig. 22a). Introduction of the hot1-3 allele58 into the ClpD-GFP line #5 significantly aggravated the phenotype of the line #5 plants (Fig. 7g–i and Supplementary Fig. 22b, c). The preClpD-GFP levels did not show a visible difference between lines #5 and hot1 #5, but polyubiquitinated proteins accumulated to higher levels in hot1 #5 (Fig. 7j). These results are consistent with the function of HOT1 disaggregase, which does not participate in preClpD-GFP degradation but is important for maintaining cytosolic proteostasis upon cPOS.
Taken together, our results reported here suggest that cPOS induces a heat shock-like response that strongly upregulates the translation of HSPs to maintain cellular proteostasis.
Discussion
Owing to the high proteotoxicity of unfolded proteins, the precursors of both mitochondrial and chloroplast proteins are restricted to very low levels. The resulting poor detectability has hampered the dissection of cellular quality control and clearance pathways. Low precursor accumulation is achieved by transcriptional repression via retrograde signaling, as observed in the import-defective ppi2 mutant59, and through efficient degradation of precursors by the UPS, as observed for both chloroplasts and mitochondria6,15,18,20. Multiple surveillance and response pathways have been identified for the mitochondrial precursor accumulation response in yeast14,15,16,18,20. In this work, by designing different ClpD-GFP/ClpD overexpression systems, we elucidated the cellular responses to chloroplast precursor accumulation (Fig. 8). Overaccumulation of preClpD-GFP/preClpD resulted in a pale-green and growth-retarding phenotype (Fig. 2b and Supplementary Fig. 3b, c) and decreased in size and disrupted ultrastructure of the chloroplasts (Supplementary Fig. 4c and Fig. 2f). These phenotypes are likely caused by impaired import of essential proteins for chloroplast biogenesis (Figs. 2c–e, 3b, c and Supplementary Fig. 9a). Possibly the ClpD-GFP is a slow-translocating protein, similar to the proteins used in yeast mitochondrial import stress studies19,20. In yeast and human cells, the precursors were shown to stall in the import machinery and clog the import channel20,29. The preClpD-GFP possibly also stalls in the TOC complex, as revealed by the strong coprecipitation of Toc75 with (pre)ClpD-GFP in the t-i-ClpD-GFP line in which a large amount of preClpD-GFP accumulated after induction (Supplementary Fig. 7a). The overloading of relative import machinery by ClpD-GFP overexpression, consequently, represses the translocation of other chloroplast proteins (Fig. 3b, c and Supplementary Data 2).
Limiting the chloroplast import capacity by ClpD-GFP overexpression reduces the translocation of essential chloroplast proteins, thus inhibiting chloroplast development (e.g., repressing photosystem and ribosome biogenesis, altering RNA editing, and inducing cpROS production). Chloroplast-derived ROS act as retrograde signal to transcriptionally activate sHSP expression. Aberrantly accumulated precursor proteins disequilibrate cytosolic proteostasis. In response, the cell (i) transcriptionally upregulates genes related to ribosome biogenesis, thus increasing the abundance of cytosolic ribosomes and translation capacity; (ii) activates sHSPs and large HSPs mainly by translational upregulation; (iii) HOT1 and sHSPs bind to precursors and form precursor granules close to chloroplasts, and also facilitating the refolding of other misfolded cytosolic proteins; and (iv) upregulates the UPS to degrade faulty proteins. cPOS likely also affects the homeostasis of mitochondria, as evidenced by mitochondrial HSPs becoming significantly upregulated and RNA editing being altered.
The accumulation of chloroplast ribosomal proteins decreased (Fig. 3a and Supplementary Fig. 8d, f), which in turn caused a reduced translation output in the chloroplasts of ClpD-GFP line #5 (Supplementary Fig. 16), a phenomenon that was also observed upon mitochondrial import defects in mammals60. While the P-TPs accumulated to comparable levels in 3-d-old WT and ClpD-GFP line #5 seedlings, they significantly decreased in the WT but not in the line #5 on day 6 (Fig. 3b). Chloroplast biogenesis is actively ongoing from day 3 onward, and the highly abundant proteins of the photosynthetic apparatus start to accumulate28. Consequently, the import load on day 3 of seedling development is also high in the WT. While the limiting import capacity can be resolved as seedling development proceeds in the WT, as evidenced by decreased P-TP accumulation on day 6, the import capacity remains limited in line #5 plants. Precursor accumulation is indeed correlated with import capacity. PreClpD can be readily detected in the transgenic lines in the gun1 (g-ClpD) and toc75III (t-ClpD) backgrounds but not in the WT (C-ClpD) background (Supplementary Fig. 3a). In addition, the precursors of ClpD and GluTR accumulated in the untransformed import-related mutants but not in the WT under both unstressed and abiotic stress conditions (Supplementary Figs. 2a and 19).
Similar to mPOS19, the UPS components and cytosolic chaperones were significantly upregulated in ClpD-GFP line #5 seedlings (Fig. 3d–f and Supplementary Data 1). Increased accumulation of cytosolic HSP70/90 and proteasomal subunits was also observed in the gun1 clpc1 double mutant7 and other import-related mutants in which chloroplast precursor proteins overaccumulated (Supplementary Fig. 19), indicating that this phenomenon occurred rather than under ClpD-GFP overexpression but represents a common cellular response to chloroplast precursor accumulation stress. Our present study revealed a critical role for the HSP100/ClpB family chaperone HOT1, a key molecular chaperone involves in plant thermotolerance and participates in plant development under nonstress conditions55,56,57, in the cPOS response. Interestingly, the HSP100 family chaperones HSP104 in yeast23 and HSPH1 (HSP105) in human cells24 also participate in the formation of precursor granules of mitochondria. Disruption of HOT1 significantly aggravated the phenotype of ClpD-GFP line #5 (Fig. 7g and Supplementary Fig. 22b, c) and caused increased accumulation of polyubiquitinated proteins (Fig. 7j). However, the accumulation of preClpD-GFP did not differ between lines #5 and hot1 #5, indicating that, as a disaggregase, HOT1 does not participate in the turnover of preClpD-GFP, but rather functions in the maintenance of cellular proteostasis by affecting the homeostasis of other proteins.
In contrast to the observations of the yeast mPOS response18,19, the protein abundance of a number of cytosolic ribosomal proteins and translation regulators significantly increased in cPOS (Fig. 3d and Supplementary Data 1). This phenomenon was accompanied by transcriptional upregulation of genes related to ribosome biogenesis (Fig. 4b), increased polysome profiles (Fig. 3f), and enhanced global translation (as revealed by global increases in P-site counts; Fig. 3h and Supplementary Fig. 10b) upon cPOS. Our data revealed that the upregulated expression of HSPs occurred mainly through translational activation, suggesting a mechanism that is distinct from that operating in mPOS in yeast. The enriched GO terms for upregulated genes at the TE level were related mainly to the cellular response to various stresses (Fig. 4f), suggesting that increased translation capacity promotes protein synthesis with some selectivity. This conclusion is in line with the increased abundance of eIF2α and RACK1 (Fig. 3d), which participate in mRNA-specific translation under mitochondrial stress and various other environmental stress conditions51,53,61,62. Notably, eIF2α also participates in the integrated stress response (ISR) triggered by mitochondrial dysfunction in mammalian cells62.
MitoStores provide deposits for mitochondrial precursors and suppress the toxic potential of aberrantly accumulated precursors, especially when the upregulation of the UPS is interrupted23. These deposited precursors can be released and imported into the mitochondria after the import clogger is removed. HSP104 and HSP42 were shown to be essential for MitoStore formation. In addition, HSP26 and HSP110 (Sse1) have been shown to associate with HSP104 under import stress23. The precursors (represented by P-TP identification) of other chloroplast proteins were identified in the ClpD-GFP precursor granules (Supplementary Fig. 20b). Notably, HOT1 (and BOB1/HSP20) were copurified with preClpD-GFP (Fig. 7e), suggesting a possibly conserved role to yeast HSP104 in the formation of precursor granules. The HSP100 and HSP20 family chaperones HSPH1/HSP105 and HSPB1/HSP27 have also been shown to associate with mitochondrial precursor granules in human cells24. Considering the similarities to MitoStore, we named the chloroplast precursor granules as ChloroStore. Importantly, the chloroplast precursor granules were observed not only in ClpD-GFP line #5 but also in cpGFP tic40-4 (Supplementary Fig. 2b), cpGFP gun1-101, and ClpDTP-GFP gun1-101 lines7 under different abiotic stress conditions, such as under lincomycin treatment which represses chloroplast biogenesis by inhibiting chloroplast translation. In addition to HOT1, many translation-related proteins were copurified with preClpD-GFP (Fig. 7e), including eIF2α, RACK1, and LOS4, and several 80S ribosomal subunits, suggesting that localized translation may occur near the precursor granules. Whether this local translation mainly involves in the synthesis of HSPs needs to be elucidated. Notably, these translation-related proteins were not detected in MitoStores23, indicating that different regulatory mechanisms in chloroplast precursor homeostasis.
sHSPs function independently of ATP and provide immediate protection by selectively binding to unfolded proteins, thus preventing aggregation and facilitating subsequent refolding by other (ATP-dependent) chaperones63,64. In addition to translational induction, the activation of sHSPs is also dependent on transcriptional upregulation, given their very low transcript levels under nonstress conditions (Supplementary Data 3). Similar to the UPRmt (ref. 65), this transcriptional upregulation of sHSPs is dependent on cpROS retrograde signaling from chloroplasts. cpROS were readily detected in chloroplasts after 12 h induction of ClpD overexpression (Fig. 6c and Supplementary Fig. 18b), the time point at which preClpD highly accumulated (Supplementary Fig. 1a). Our results suggest a crucial function of cpROS-mediated retrograde signaling in cPOS. However, compared with transcriptional upregulation, translational activation is much stronger not only for sHSPs but also for large HSPs and their cochaperones (including DnaJ proteins; Figs. 6a and 7a). A previous study showed that cpROS repress cytosolic translation via eIF2α phosphorylation by the GCN2 kinase upon a dark-to-light shift66. However, heat stress, which also triggers plastid ROS production67, represses eIF2α phosphorylation66, indicating that the cpROS generated under different stress conditions may function differently. Whether the cpROS generated in cPOS (which, as shown in this work, triggers a heat shock-like response) directly participate in translational activation remains to be investigated.
The mitoTAD pathway monitors the translocase of the outer membrane of the mitochondria (TOM) complex under nonstress conditions15. The AAA+ ATPase CDC48 associates with the TOM complex through Tom70 via the Ubx2 interaction and mediates the extraction of precursors from the TOM complex for proteasomal degradation15. The CDC48 complex works together with SP2 to extract TOC subunits from the outer envelope membrane for SP1-mediated proteasomal degradation in the chloroplast-associated protein degradation (CHLORAD) pathway68,69. However, compared with the increased accumulation of TOC components, the accumulation of LhcB4.1 does not increase in the sp1 ppi2 double mutant in comparison to the ppi2 single mutant70. Therefore, more detailed analyses are needed to determine whether the CHLORAD pathway is involved in the degradation of precursors clogging in the TOC complex.
In summary, our findings reported here elucidate the systematic cellular responses to chloroplast precursor protein accumulation. We have shown that precursor accumulation triggers a heat shock-like response that helps to rebalance cytosolic proteostasis. Our results also uncover a crucial role of translational regulation in cPOS (Fig. 8). Recent reports have shown that a mild increase in chloroplast precursor proteins caused by the introduction of an RPN8a (encoding a 19S proteasomal subunit) mutation into the ppi2 mutant increases the accumulation of functional photosynthetic complexes70. Moreover, the efficient degradation of the precursors of the harmful chloroplast protein TT3.2 confers heat stress tolerance in rice71. These findings, together with our data reported here, suggest potential strategies for crop improvement by manipulating the homeostasis of chloroplast precursor proteins.
Methods
Plant material and growth conditions
All Arabidopsis (Arabidopsis thaliana) lines used in this study were of the Columbia-0 (Col-0) background. The gun1-101, toc75III-3, hot1-3, tic40-4, ppi1, ClpDTP-GFP, C-ClpD, and g-ClpD lines have been previously described7,58,72,73. The cpGFP line28 was crossed into the tic40-4 mutant to generate the cpGFP tic40-4 line. For plant growth under aseptic conditions, the seeds were surface-sterilized with 1.2% NaClO for 10 min, washed five times with sterile water, sown on half-strength Murashige & Skoog (MS) medium74 containing 1% (w/v) of sucrose, and stratified at 4 °C for 48 h in the dark prior to germination. For plant growth conditions, unless specified otherwise, the seeds were germinated and grown under long-day conditions of 16 h light (120 µmol photos m−2 s−1)/8 h dark at 22 °C. For the heat stress treatment, 6-d-old seedlings were incubated at 37 °C for the time indicated. For the induction of ClpD or ClpD-GFP in the inducible lines (i-ClpD or i-ClpD-GFP), a beaker containing ethanol (absolute) was put into the growth chamber next to the plant materials for evaporated ethanol induction for the time indicated. For GSH treatment, 4-d-old WT and constitutive ClpD-GFP expression line #5 seedlings were transferred to half-strength MS medium supplemented with DMSO or 500 µM GSH (MedChemExpress, HY-D0187) and cultivated for 2 days. For the cytosolic HSP90 inhibitor geldanamycin (GDA) and HSP70 inhibitor VER155008 (VER) treatments, the WT and ClpD-GFP line #5 were grown at half-strength MS medium supplemented with 40 µM GDA (MedChemExpress, HY-15230), 40 µM VER (MedChemExpress, HY-10941), or corresponding amount of DMSO for 6 days.
Construction of transgenic vectors and generation of transgenic plants
To produce inducible ClpD or ClpD-GFP transgenic lines in the WT (Col-0), gun1, and toc75III backgrounds, the complete CDS of ClpD was amplified from the pEZR(H)-LN-ClpD-GFP vector7 either unfused (with stop codon) or fused (without stop codon) with GFP. The obtained PCR products were subsequently cloned into the intermediate vector pUC-del-alcAN by using NovoRec® plus One step PCR Cloning Kit (Novoprotein, NR005). Then the expression cassette containing the alcA promoter and ClpD or ClpD-GFP CDS was cleaved using the AscI restriction enzyme and closed into the binary pBin-ΔalcR vector for the ethanol-induced expression of ClpD or ClpD-GFP. To produce constitutive (CaMV 35S promoter) ClpD-GFP expression lines in the WT background, the pEZR(H)-LN-ClpD-GFP vector7 was used for the transformation of Col-0 plants. To constitutively express (CaMV 35S promoter) HOT1-mCherry in ClpD-GFP line #5, the complete CDS (without stop codon) of HOT1 was amplified from the cDNA of Col-0 seedlings and cloned into pC231-N1-35S binery vector fused with mCherry by using NovoRec® plus One step PCR Cloning Kit (Novoprotein, NR005). The generated pC231-N1-35S-HOT1-mCherry construct was used for the transformation of ClpD-GFP line #5 plants. The constructs were transformed into Agrobacterium tumefaciens strain GV3101 and introduced into Arabidopsis by using the floral dip method75. The constitutive ClpD-GFP expression line #5 was crossed with the hot1-3 mutant to produce the ClpD-GFP expression line under the hot1-3 background (hot1 #5). All primers used for cloning are listed in Supplementary Table 3.
RNA extraction and RT‒qPCR analyses
Total RNA was extracted by using an RNAprep Pure Plant Kit (Tiangen, DP432) and genomic DNA contamination was eliminated by DNase treatment. First-strand cDNA was synthesized by using the Fastking RT kit including a gDNase treatment for a second round of genomic DNA elimination (Tiangen, KR116). Quantitative PCR (qPCR) was performed with a CFX384 Touch Real-Time PCR Detection System (Bio-Rad) using SYBR Green detection (MIKX, MKG800-10). The actin gene was used as an internal control and relative expression values were calculated for each target gene including the calculation of the amplification efficiencies of the different primers76. All primers used for the qPCR analyses are listed in Supplementary Table 3.
Protein extraction and western blot analyses
Total cellular proteins were extracted according to a published procedure77. In brief, 100 mg of plant materials was ground in liquid nitrogen and 500 μL of extraction buffer (0.7 M sucrose, 0.5 M Tris-HCl (pH 9.4), 50 mM EDTA, 100 mM KCl, 2% (v/v) β-mercaptoethanol, and 2× protease inhibitor) was added. After vortexing, an equal amount of phenol (Tris-saturated) was added, and the mixture was vortexed and centrifugated. The total cellular proteins were precipitated from the supernatant with 2.5× volumes of 0.1 M ammonium acetate in methanol overnight at −20 °C. After centrifugation, the protein pellet was washed with 0.1 M ammonium acetate in methanol, air-dried, dissolved in 1% SDS, and quantified by using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). Protein samples were separated by SDS‒PAGE and blotted onto PVDF membranes (Amersham Hybond P 0.45 µm, 10600023) using a standard method. Immunoblotting was performed with specific antibodies using the enhanced chemiluminescence system (SparkJade, ED0015). The signals were detected with a ChemiDoc Imaging system (Bio-Rad). Antibodies against ClpD and Toc75 were kindly provided by Dr. Adrian Clarke (Gothenburg University, Gothenburg, Sweden) and Dr. Qihua Ling (CAS Center for Excellence in Molecular Plant Science, Shanghai, China), respectively. All other antibodies were obtained from commercial suppliers (GFP from Clonetech; actin and ubiquitin from Sigma; PsbA, PsbD, AtpA, AtpB, LhcB1, LhcA4, PetA, GluTR, Tic40, cytosolic HSP70 and HSP90.1, and chloroplast HSP70 and Rps1 from Agrisera; PsaA, RPT2a, RPT5a, VAR1, VAR2, PsbC, PORB, GUN5, and CHLM from Orizymes Biotechnology; 40S ribosome subunit US3Z from Cell Signaling Technology). The band intensities from the immunoblots were quantified by using Image Lab software (Bio-Rad) when necessary.
Chlorophyll contents and chlorophyll fluorescence measurements
The chlorophyll contents and chlorophyll a/b ratios were determined using the N, N-dimethylformamide (DMF) method78. In brief, 0.1 g of plant material was collected and directly immersed into 10 mL of DMF solution in the dark at 4 °C for 24 h followed by spectrophotometric measurements at OD664, OD647, and OD625, after which the chlorophyll concentrations and a/b ratios were calculated. Chlorophyll a fluorescence was recorded by using the MAXI version of the IMAGING-PAM (Heinz Walz GmbH). The plants were dark-adapted for 0.5 h before the maximum quantum yield of photosystem II (Fv/Fm) was determined. Subsequently, the light-response curves were measured.
Confocal laser-scanning microscopy
GFP and RFP fluorescence, and the H2DCFDA fluorescence for ROS detection were observed by confocal laser-scanning microscopy (TCS SP8, Leica; or Fluoview FV3000, Olympus) using an argon laser for excitation. The excitation wavelength for GFP and H2DCFDA was 488 nm, and that for mCherry was 561 nm. Emission was detected with a 495–530 nm filter for GFP and H2DCFDA, and a 570–622 nm filter for mCherry. For the detection of chlorophyll fluorescence, a 650–702 nm filter was used. Line-scan analysis of relative fluorescence intensity was performed by measuring pixel intensity across an indicated line by using Fiji software79.
Transmission electron microscopy
For TEM analyses, cotyledons from 3-d and 6-d WT and line #5 seedlings were fixed with 2.5% glutaraldehyde in 50 mM sodium cacodylate (pH 7.4) containing 5 mM CaCl2 under vacuum until the leaf pieces were immersed into the solution. Afterward, the samples were incubated at 4 °C overnight. Postfixation with 1% OsO4 and 0.8% K3Fe(CN)6 in 50 mM cacodylate buffer (pH 7.4) was carried out for 2 h at 4 °C, followed by dehydration in an ethanol series, infiltration with a graded series of epoxy resins in epoxy propane, and embedding in Epon 812 resin. Ultrathin sections (50–70 nm) were cut with diamond knives. For TEM observation, ultrathin sections were stained with 2% uranyl acetate and lead citrate80. Images were acquired with a Talos L120C G2 TEM (Thermo Fisher Scientific).
Blue-native polyacrylamide gel electrophoresis (BN–PAGE)
BN-PAGE was performed as previously described81 with essential modifications. A native 5–13.5% (3% C) gradient polyacrylamide gel was used. The same volume of 5% gel containing 6.2% (w/v) glycerol and 13.5% gel containing 25% (w/v) of glycerol were mixed gradually by using a gradient gel maker. The samples were loaded on the basis of the total thylakoid proteins (50 μg). Thylakoids were pelleted at 7500 × g for 5 min at 4 °C, washed with 100 μL of 50BTH40G buffer (50 mM Bis-Tris, pH 7.5, 40% (w/v) glycerol) and resuspended in 10 μL of BN-PAGE solution buffer (25 mM Bis-Tris, pH 7.5, 20% (w/v) glycerol, and 1% n-dodecyl-β-D-maltoside). The resuspension was incubated on ice in the dark for 20 min and centrifugated at 12,000 × g for 10 min at 4 °C. The supernatants were resuspended in 1 μL of BN-PAGE sample buffer (0.5 M 6-amino-caproic acid, 0.1 M Bis-Tris, pH 7.0, 37.5% glycerol, 5% Coomassie brilliant blue G250) and run on a 5–13.5% native gel. The BN–PAGE was run at a constant current of 6 mA.
Polysome profile analysis
Polysome profile analyses were performed according to Mustroph et al.82. In brief, 4 g of 6-d-old WT and ClpD-GFP line #5 seedlings were collected, snap-frozen, and ground with liquid nitrogen. The samples were dissolved in 3 mL of precooled polysome extraction buffer (0.2 M Tris-HCl, pH 9.0; 0.2 M KCl; 25 mM EGTA; 35 mM MgCl2; 1% Brij-35; 1% Triton X-100; 1% NP40; 1% Tween 20; 1% sodium deoxycholate; 1% polyoxyethylene (10) tridecyl ether; 5 mM DTT; 1 mM PMSF; 50 μg/mL cycloheximide; 50 μg/mL chloramphenicol; 400 U/ml RNase inhibitor), and centrifugated at 14,000 × g for 15 min at 4 °C. The supernatant was filtered through two layers of Miracloth (Millipore, 475855), incubated on ice for 10 min, and centrifugated at 14,000 × g for 15 min at 4 °C. Half of the supernatant of each sample was transferred to a new tube and puromycin treatment was performed as a control (puromycin release ribosomes from the mRNA). The samples were loaded onto a sucrose gradient (10–60% sucrose in 40 mM Tris-HC1, pH 8.4; 20 mM KCl; 10 mM MgCl2) and centrifugated at 150,000 × g for 3.5 h at 4 °C (Beckman Optima XPN-100). The polysome absorbance profiles were recorded at 260 nm and the fractions were collected by a position gradient fractionator (Biocomp 152).
Label-free quantification by mass spectrometric analyses
For quantitative proteomic analyses, 3-d-old and 6-d-old WT, ClpD-GFP line #5, and ΔTP-ClpD-GFP seedlings with biological replicates of each were collected and samples of 100 mg of plant material from each sample were ground in liquid nitrogen and extracted with 200 μL of protein extraction buffer (0.7 M sucrose; 0.5 M Tris-HCl, pH 9.4; 50 mM EDTA; 0.1 M KCl; 2% β-mercaptoethanol (v/v) and 2× protease inhibitor, Roche) according to a published protocol77. After protein quantification, 40 μg of total cellular protein was digested with trypsin overnight at 37 °C at a 1:80 ratio (0.5 μg of trypsin) to the protein. The peptides were desalted with MonoSpin C18 columns (Shimadzu, 5010-21701).
For MS analysis, peptides were separated via a nanoflow HPLC (EASY-nLC 1200, Thermo Fisher Scientific) by using a homemade C18 analytical column that was 35 cm in length and 75 µm in inner diameter. A binary buffer system of buffer A (0.1% formic acid) and buffer B (80% acetonitrile plus 0.1% formic acid) was used with a linear gradient running from 8% to 98% buffer B over 135 min. The nLC was coupled with a high-resolution Orbitrap hybrid mass spectrometer (Orbitrap Exploris 480; Thermo Fisher Scientific). Proteins were identified based on the data-dependent acquisition of fragmentation spectra of multiple charged peptides. The scan was carried out with a 2 s cycle time between MS and MS/MS in the top-speed method. The orbitrap resolution (full-width half-maximum) of the full scan and MS/MS scan was set at 60,000 and 15,000, respectively.
The MaxQuant software (version 1.6.7.0) was used for protein identification and quantification83,84, and the sequences were searched against the UniProt Arabidopsis FASTA database UP000006548 (39,280 entries) using the Andromeda search engine85. The following settings were applied, fixed modification: carbamidomethyl (C); variable modifications: oxidized methionine (M) and protein N-terminal acetylation; the digesting protease was specified to trypsin with up to two missed cleavages allowed; minium peptide length was set to 7 as default; multiplicity was set to 1 for label-free quantitation; a time window of 0.7 min was set for “match between runs”; common contaminants (trypsin, keratin, etc.) were included during the database search, and unique and razor peptides were used for protein quantification. All other parameters were set to their defaults. The Perseus software was used for postdata and statistical analyses86. Proteins with a Student’s t-test p < 0.01 and a fold change >1.5 between genotypes were identified as differentially expressed proteins. The subcellular location was determined according to that of SUBA5 (SUBAcon; http://Suba.live). The interaction relationship networks were generated using STRING31 and visualized by Cytoscape87. The P-TPs were identified according to the previous publication7. Peptides that were completely localized within a transit peptide region and peptides whose N-terminus has ≥3 amino acids overlapping with the C-terminus of the transit peptide were defined as P-TPs.
For co-IP samples, immunoprecipitates were separated by SDS–PAGE and the gels were stained with colloidal Coomassie blue solution. Each lane was cut into several gel slices followed by in-gel tryptic digestion7. For MS analysis, a 60-min linear gradient was used. To identify the putative preClpD-GFP associated proteins, the following criteria were used: (i) were detected by MS in at least 2 biological replicates of the ClpD-GFP line #5 co-IP samples, and at least 2 unique peptides were identified in each sample; (ii) the LFQ protein intensity of the ClpD-GFP line #5/ΔTP-ClpD-GFP > 2; and (iii) the subcellular localization of the candidate was in the cytosol according to the SUBAcon in SUBA5.
RNA-seq
6-d-old WT and ClpD-GFP line #5 seedlings were collected and total RNA was extracted using a TRIzol reagent (Invitrogen, 15596018CN) according to the manufacturer’s instructions. RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies). The mRNA was enriched by using oligo (dT) beads, fragmented, and cDNA was synthesized by using the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, 7530). The purified double-strand cDNA fragments were end-repaired, and an A base was added and ligated to Illumina sequencing adapters. The ligation products were purified with the AMPure XP beads (1.0X), subjected to size selection by agarose gel electrophoresis, and amplified by PCR. Sequencing was performed on an Illumina HiSeq 2500 platform by Gene Denovo Biotechnology Co. (Guangzhou, China).
Low-quality reads and adapter sequences were filtered by fastp software (version 0.18.0)88. The reads mapped to ribosomal RNA (rRNA) were identified by the short-read alignment tool Bowtie2 (version 2.2.8)89 and removed. The clean reads were mapped to the TAIR10 Arabidopsis reference genome using HISAT2.2.490 and assembled by using StringTie v1.3.191,92 for each sample in a reference-based approach. For each transcription region, the fragment per kilobase of transcript per million mapped reads (FPKM) value was calculated by using RSEM software93. Differential gene expression analysis was performed using DESeq2 software94. Genes fulfilling the criteria of an FDR < 0.05 and a fold change > 2 were considered to be differentially expressed and GO enrichment analyses were performed (https://www.geneontology.org/).
Ribosome profiling (Ribo-seq)
6-d-old WT and ClpD-GFP line #5 seedlings were snap-frozen, ground with liquid nitrogen, and then dissolved in 400 µL of lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 100 µg/mL cycloheximide, and 1% (v/v) Triton X-100). The samples were incubated on ice for 10 min, and centrifugated at 20,000 × g for 10 min at 4 °C. For footprint preparation, 10 µL of RNase I (NEB) and 6 µL of DNase I (NEB) were added to 400 µL of lysate and incubated for 45 min at room temperature with gentle mixing. Afterward, 10 µL of SUPERase-In RNase inhibitor (Ambion, AM2696) was added to stop nuclease digestion. The digested samples were loaded onto preequilibrated size exclusion columns (Illustra MicroSpin S-400 HR Columns; GE Healthcare) and centrifugated at 600 × g for 2 min. Footprints were isolated by using an RNA Clean and Concentrator-25 kit (Zymo Research, R1018). To eliminate rRNAs, short (50–80 nt) antisense DNA probes complementary to rRNA sequences were added, followed by RNase H (NEB) and DNase I (NEB) treatment to remove rRNA and residual DNA probes95. Footprints were further purified using VAHTS RNA Clean magnetic beads (Vazyme, N412-02). Ribo-seq libraries were constructed using the NEBNext® Multiple Small RNA Library Prep Set for Illumina® (NEB, E7330L) and sequencing was performed on an Illumina HiSeq 2500 platform by Gene Denovo Biotechnology Co. (Guangzhou, China).
Raw reads containing more than 50% low-quality bases or 10% N bases were removed and adapter sequences were trimmed by fastp software. Reads with lengths between 20 and 40 nt were subjected to further analysis. The reads mapped to rRNAs, transfer RNAs (tRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and miRNAs were removed by using the short read alignment tool Bowtie289. The remaining reads were mapped to the TAIR10 Arabidopsis reference genome by using STAR software96. Three-nucleotide periodicity was plotted using the riboWaltz R package97. The read counts in the open reading frames of the coding genes were calculated by RSEM software93, and the FPKM values were calculated. Differentially translated genes were identified by using DESeq2 software. Genes fulfilling the criteria of an FDR < 0.05 and a fold change >2 were considered to be differentially translated and were subjected to GO enrichment analysis (https://www.geneontology.org/).
The translation efficiency (TE) was calculated as the ratio of the translating mRNAs (FPKM from Ribo-seq) over the total mRNAs (FPKM from RNA-seq) of a gene32. RiboDiff98 was used to identify the genes with TE significantly changed across sample groups. Genes fulfilling the criteria of an FDR < 0.05 and a fold change >2 were considered to be differential TE genes and were subjected to GO analysis (https://www.geneontology.org/). The aggregated normalized P-site counts were analyzed according to a published method99.
ROS staining
For 3,3′-diaminobenzidine (DAB; Sigma-Aldrich, D5637) staining of H2O2 (ref. 100), 6-d-old WT and ClpD-GFP line #5 seedlings were immersed in freshly prepared 1 mg/mL DAB staining solution (pH 3.8) and infiltrated under vacuum. The samples were subsequently incubated at 25 °C in the dark for 6 h, washed with buffer (acetic acid: glycerol: ethanol = 1:1:3, v/v/v) for at least 8 times, and stored in 95% (v/v) ethanol until imaging. To determine the subcellular localization of ROS production, 6-d-old WT, gun1, toc75, C-i-ClpD, g-i-ClpD, and t-i-ClpD lines were induced for 12 h. The newly emerged true leaves were infiltrated with 10 μM of 2′,7′-dichlorofluorescein diacetate (H2DCFDA; MedChemExpress, HY-D0940) solution and the fluorescence signals were analyzed 10 min after infiltration by confocal laser-scanning microscopy101.
Coimmunoprecipitation (co-IP)
For co-IP to examine whether preClpD-GFP stalls in the import machinery, whole seedlings from 5-d-old ΔTP-ClpD-GFP and ClpD-GFP #5 lines, and the true leaves from 6-d-old t-i-ClpD-GFP plants induced for another 4 days were harvested, ground in liquid nitrogen, and solubilized in co-IP buffer (0.5 M sucrose, 50 mM HEPES-KOH, pH 7.5 2 mM EDTA, 3 mM MgCl2, 1.5% n-dodecyl β-D-maltoside (w/v), and protease inhibitor cocktail) for 20 min on ice and centrifugated for 2 × 20 min at 20,000 × g at 4 °C. Subsequently, 25 µL of anti-GFP magnetic beads (Alpalife, KTSM1334) were added to the supernatant and incubated for 4 h at 4 °C with agitation to facilitate binding. The beads were then washed and the bound proteins were released by incubation in SDS‒PAGE sample buffer at 95 °C. The immunoprecipitates were analyzed by western blotting with specific antibodies.
For co-IP to isolate preClpD-GFP-associated cytosolic proteins, the crude cytosolic fractions from the ClpD-GFP line #5 and ΔTP- ClpD-GFP seedlings were used. Two grams of 6-d-old seedlings for each replicate were briefly homogenized with a blender in isolation buffer (30 mM HEPES-KOH of pH 7.5, 2 mM EDTA, 300 mM sorbitol, and 1% (w/v) of PVP-40), filled through 2 layers of Miracloth (Millipore, 475855), and centrifugated at 2500 × g for 5 min at 4 °C. The supernatant was further centrifugated at 20,000 × g for 20 min at 4 °C. For co-IP, 25 μL of anti-GFP magnetic beads (Alpalife, KTSM1334) were added to the cytosolic fractions. Immunoprecipitation and MS identification were performed according to a published method7.
Statistical analysis
Student’s t‐tests were performed using GraphPad Prism 6.01 software to identify the significance of the difference between two experimental groups. For proteomic analysis, Perseus software was used for postdata, and Student’s t‐tests were used to identify differentially accumulated proteins with a fold change > 1.5 and p < 0.01. For the RNA-seq and Ribo-seq experiments, DESeq2 software was used to identify differentially expressed genes with a fold change >2 and and FDR < 0.05. RiboDiff software was used to identify differential TE genes with a fold change >2 and an FDR < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry-based proteomic data generated in this study have been deposited in the PRIDE partner repository of the ProteomeXchange Consortium via the iProX partner repository under the dataset identifiers PXD051682 [https://www.iprox.cn/page/PSV023.html;?url=17140556959271g0j] and PXD051764 [https://www.iprox.cn/page/SSV024.html;url=17141394135138lLy]. The RNA-seq and Ribo-seq raw sequencing data generated in this study were deposited in the Gene Expression Omnibus (GEO) under the data identifier GSE265858 and GSE265859, respectively. All other data are available in the main text or the supplementary materials. Source data are provided with this paper.
References
Bock, R. Witnessing genome evolution: experimental reconstruction of endosymbiotic and horizontal gene transfer. Annu. Rev. Genet. 51, 1–22 (2017).
Stegemann, S., Hartmann, S., Ruf, S. & Bock, R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl. Acad. Sci. USA 100, 8828–8833 (2003).
Jarvis, P. & López-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802 (2013).
Chu, C.-C., Swamy, K. & Li, H.-M. Tissue-specific regulation of plastid protein import via transit-peptide motifs. Plant Cell 32, 1204–1217 (2020).
Sun, Y. & Jarvis, R. P. Chloroplast proteostasis: import, sorting, ubiquitination, and proteolysis. Annu. Rev. Plant Biol. 74, 259–283 (2023).
Lee, S. et al. Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21, 3984–4001 (2009).
Wu, G. Z. et al. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants 5, 525–538 (2019).
Lee, D. W. et al. Arabidopsis BAG1 functions as a cofactor in Hsc70-mediated proteasomal degradation of unimported plastid proteins. Mol. Plant 9, 1428–1431 (2016).
Koussevitzky, S. et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719 (2007).
Flores-Pérez, U. et al. Functional analysis of the Hsp93/ClpC chaperone at the chloroplast envelope. Plant Physiol. 170, 147–162 (2016).
Gao, L. L., Hong, Z. H., Wang, Y. & Wu, G. Z. Chloroplast proteostasis: a story of birth, life, and death. Plant Commun. 4, 100424 (2023).
Song, J., Herrmann, J. M. & Becker, T. Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell Biol. 22, 54–70 (2021).
Maruszczak, K. K., Ayyamperumal, S. & Chacinska, A. Defective mitochondrial import as a challenge for cellular protein homeostasis. FEBS Lett. 597, 1555–1568 (2023).
Izawa, T., Park, S. H., Zhao, L., Hartl, F. U. & Neupert, W. Cytosolic protein Vms1 links ribosome quality control to mitochondrial and cellular homeostasis. Cell 171, 890–903 e818 (2017).
Martensson, C. U. et al. Mitochondrial protein translocation-associated degradation. Nature 569, 679–683 (2019).
Schulte, U. et al. Mitochondrial complexome reveals quality-control pathways of protein import. Nature 614, 153–159 (2023).
Wang, X. & Chen, X. J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 524, 481–484 (2015).
Wrobel, L. et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524, 485–488 (2015).
Boos, F. et al. Mitochondrial protein-induced stress triggers a global adaptive transcriptional programme. Nat. Cell Biol. 21, 442–451 (2019).
Weidberg, H. & Amon, A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science 360, eaan4146 (2018).
Shakya, V. P. et al. A nuclear-based quality control pathway for non-imported mitochondrial proteins. Elife 10, e61230 (2021).
Laborenz, J. et al. The ER protein Ema19 facilitates the degradation of nonimported mitochondrial precursor proteins. Mol. Biol. Cell 32, 664–674 (2021).
Kramer, L. et al. MitoStores: chaperone-controlled protein granules store mitochondrial precursors in the cytosol. EMBO J 42, e112309 (2023).
Kim, M. et al. Immunoproteasome-specific subunit PSMB9 induction is required to regulate cellular proteostasis upon mitochondrial dysfunction. Nat. Commun. 14, 4092 (2023).
Baldwin, A. et al. A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol. 138, 715–733 (2005).
Jin, Z. et al. Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes. Cell 185, 4788–4800 (2022).
Liu, H., Li, A., Rochaix, J. D. & Liu, Z. Architecture of chloroplast TOC-TIC translocon supercomplex. Nature 615, 349–357 (2023).
Wu, G. Z. et al. Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol. 176, 2472–2495 (2018).
Kim, J. et al. ATAD1 prevents clogging of TOM and damage caused by un-imported mitochondrial proteins. Cell Rep. 43, 114473 (2024).
Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A. & Chory, J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98, 2053–2058 (2001).
Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Calviello, L. et al. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods 13, 165–170 (2016).
Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M. & Weissman, J. S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012).
Zhang, M. et al. N6-methyladenosine RNA modification regulates photosynthesis during photodamage in plants. Nat. Commun. 13, 7441 (2022).
Merchante, C. et al. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163, 684–697 (2015).
Juntawong, P., Girke, T., Bazin, J. & Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 111, E203–E212 (2014).
Li, L. et al. Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell 29, 207–228 (2017).
Lurin, C. et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103 (2004).
Li, M. et al. Plant editosome database: a curated database of RNA editosome in plants. Nucleic Acids Res. 47, D170–D174 (2019).
Waters, M. T. et al. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21, 1109–1128 (2009).
Charng, Y. Y. et al. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262 (2007).
Friedrich, T. et al. Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat. Commun. 12, 3426 (2021).
Chan, K. X., Phua, S. Y., Crisp, P., McQuinn, R. & Pogson, B. J. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53 (2016).
Li, M. & Kim, C. Chloroplast ROS and stress signaling. Plant Commun. 3, 100264 (2022).
Han, S. H., Park, Y. J. & Park, C. M. HOS1 activates DNA repair systems to enhance plant thermotolerance. Nat. Plants 6, 1439–1446 (2020).
Queitsch, C., Sangster, T. A. & Lindquist, S. Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624 (2002).
Morán Luengo, T., Mayer, M. P. & Rüdiger, S. G. D. The Hsp70-Hsp90 chaperone cascade in protein folding. Trends Cell Biol. 29, 164–177 (2019).
Hong, S. W. & Vierling, E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc. Natl. Acad. Sci. USA 97, 4392–4397 (2000).
Chang, I. F., Szick-Miranda, K., Pan, S. & Bailey-Serres, J. Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol. 137, 848–862 (2005).
Shor, B., Calaycay, J., Rushbrook, J. & McLeod, M. Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J. Biol. Chem. 278, 49119–49128 (2003).
Sengupta, J. et al. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol. 11, 957–962 (2004).
Majzoub, K. et al. RACK1 controls IRES-mediated translation of viruses. Cell 159, 1086–1095 (2014).
Gong, Z. et al. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17, 256–267 (2005).
Queitsch, C., Hong, S. W., Vierling, E. & Lindquist, S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12, 479–492 (2000).
Qin, F., Yu, B. & Li, W. Heat shock protein 101 (HSP101) promotes flowering under nonstress conditions. Plant Physiol. 186, 407–419 (2021).
Zhang, S. S. et al. Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in Arabidopsis. Plant Cell 29, 1007–1023 (2017).
Hong, S. W. & Vierling, E. Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J. 27, 25–35 (2001).
Kakizaki, T. et al. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 151, 1339–1353 (2009).
Quiros, P. M. et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017).
Harding, H. P. et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 (2003).
Guo, X. et al. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 579, 427–432 (2020).
Haslbeck, M., Franzmann, T., Weinfurtner, D. & Buchner, J. Some like it hot: the structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 12, 842–846 (2005).
Adriaenssens, E. et al. Small heat shock proteins operate as molecular chaperones in the mitochondrial intermembrane space. Nat. Cell Biol. 25, 467–480 (2023).
Sutandy, F. X. R., Gossner, I., Tascher, G. & Munch, C. A cytosolic surveillance mechanism activates the mitochondrial UPR. Nature 618, 849–854 (2023).
Lokdarshi, A. et al. Light activates the translational regulatory kinase GCN2 via reactive oxygen species emanating from the chloroplast. Plant Cell 32, 1161–1178 (2020).
Czarnocka, W. & Karpinski, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 122, 4–20 (2018).
Ling, Q. et al. Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363, eaav4467 (2019).
Ling, Q., Huang, W., Baldwin, A. & Jarvis, P. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338, 655–659 (2012).
Grimmer, J. et al. Mild proteasomal stress improves photosynthetic performance in Arabidopsis chloroplasts. Nat. Commun. 11, 1662 (2020).
Zhang, H. et al. A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science 376, 1293–1300 (2022).
Huang, W., Ling, Q., Bédard, J., Lilley, K. & Jarvis, P. In vivo analyses of the roles of essential Omp85-related proteins in the chloroplast outer envelope membrane. Plant Physiol. 157, 147–159 (2011).
Jarvis, P. et al. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103 (1998).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497 (1962).
Clough, S. J. & Bent, A. F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Cahoon, E. B., Shanklin, J. & Ohlrogge, J. B. Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc. Natl. Acad. Sci. USA 89, 11184–11188 (1992).
Moran, R. Formulae for determination of chlorophyllous pigments extracted with n,n-dimethylformamide. Plant Physiol. 69, 1376–1381 (1982).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Maclean, A. E. et al. Absence of complex I is associated with diminished respiratory chain function in European mistletoe. Curr. Biol. 28, 1614–1619 e1613 (2018).
Schagger, H., Cramer, W. A. & von Jagow, G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220–230 (1994).
Mustroph, A., Juntawong, P. & Bailey-Serres, J. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol. Biol. 553, 109–126 (2009).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 13, 2513–2526 (2014).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Morlan, J. D., Qu, K. & Sinicropi, D. V. Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PLoS ONE 7, e42882 (2012).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Lauria, F. et al. riboWaltz: Optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput. Biol. 14, e1006169 (2018).
Zhong, Y. et al. RiboDiff: detecting changes of mRNA translation efficiency from ribosome footprints. Bioinformatics 33, 139–141 (2017).
Hang, R. et al. Arabidopsis HOT3/eIF5B1 constrains rRNA RNAi by facilitating 18S rRNA maturation. Proc. Natl. Acad. Sci. USA 120, e2301081120 (2023).
Wu, J. et al. Deficient plastidic fatty acid synthesis triggers cell death by modulating mitochondrial reactive oxygen species. Cell Res. 25, 621–633 (2015).
Liu, X. et al. EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. Plant Cell 29, 3051–3067 (2017).
Acknowledgements
We are grateful to Dr. Xiao-Fan Zhang (Shanghai Jiao Tong University) for the help with the bioinformatic analysis to identify the P-TPs; Dr. Jian-Xiang Liu (Zhejiang University, China) for providing the hot1-3 seeds. This research was supported by grants from the National Natural Science Foundation of China (NSFC; 32070299, 32270285, and 32470290), the 111 project from the Ministry of Education of China (B23033), the Shanghai Collaborative Innovation Center of Agri-Seeds (ZXWH2150201/022), the Shanghai Pujiang Program (20PJ1405600), and the Partner Group Program of Max Planck Society to G.-Z.W.
Author information
Authors and Affiliations
Contributions
G.-Z.W. conceived and designed this research. Z.-H.H. performed most of the experiments. L.Z. screened and identified some of the transgenic lines. L.-L.G. and T.S. constructed the vectors for transformation. Z.Z., L.K., and X.-N.W. performed the mass spectrometry analyses. G.-Z.W., Z.-H.H., and R.B. wrote the manuscript, with input from the coauthors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hong, ZH., Zhu, L., Gao, LL. et al. Chloroplast precursor protein preClpD overaccumulation triggers multilevel reprogramming of gene expression and a heat shock-like response. Nat Commun 16, 3777 (2025). https://doi.org/10.1038/s41467-025-59043-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59043-3