Abstract
Fetal hematopoiesis takes place in the liver before colonizing the bone marrow where it will persist for life. This colonization is thought to be mediated by specification of a microenvironment that selectively recruits hematopoietic cells to the nascent bone marrow. The identity and mechanisms regulating the specification of this colonization niche are unclear. Here we identify a VCAM1+ sinusoidal colonization niche in the diaphysis that regulates neutrophil and hematopoietic stem cell colonization of the bone marrow. Using confocal microscopy, we find that colonizing hematopoietic stem and progenitor cells (HSPC) and myeloid cells selectively localize to a subset of VCAM1+ sinusoids in the center of the diaphysis. Vcam1 deletion in endothelial cells impairs hematopoietic colonization while depletion of yolk-sac-derived osteoclasts disrupts VCAM1+ expression, and impairs neutrophil and HSPC colonization to the bone marrow. Depletion of yolk-sac-derived myeloid cells increases fetal liver hematopoietic stem cell numbers, function and erythropoiesis independent of osteoclast activity. Thus, the yolk sac produces myeloid cells that have opposite roles in fetal hematopoiesis: while yolk-sac derived myeloid cells in the bone marrow promote hematopoietic colonization by specifying a VCAM1+ colonization niche, a different subset of yolk-sac-derived myeloid cells inhibits HSC in the fetal liver.
Similar content being viewed by others
Introduction
In mammals, the process of blood cell production, also known as hematopoiesis, takes place in different organs through embryonic and fetal development. In mice, the first hematopoietic cells are produced in the yolk sac at around embryonic day 7.0 (E7.0)1,2. Erythro-myeloid progenitors from the yolk sac subsequently appear at around E8.5, and generate different subsets of tissue-resident myeloid cells3 that can persist postnatally (including Kupffer cells4,5 in the liver, osteoclasts6 in the bone marrow) and into adulthood (microglia7 in the brain). The first definitive hematopoietic stem and progenitor cells (HSPC) are specified in the dorsal aorta at E10.258,9. These HSPCs will seed the fetal liver to establish definitive hematopoiesis in the fetus8. At E16.5 hematopoiesis will then progressively migrate to the developing bone marrow where it will reside for life10. How the nascent bone marrow microenvironment recruits definitive hematopoietic cells from the liver and how this microenvironment is specified during fetal bone marrow development are not well established. A study from the Hirschi group showed that colony-forming activity (a surrogate assay for HSPC function) was specifically enriched in the diaphysis but not the metaphysis of the developing bones10. The Adams lab also showed that the trochanter artery localizes to the center of the fetal diaphysis and produces Wnt ligands that support HSC expansion11. Together these studies supported the concept that the fetal bone marrow contains spatially restricted microenvironments that selectively recruit hematopoietic cells from the liver to the bone marrow. In this work, we use imaging analyses combined with mouse genetic fate mapping and loss of function experiments to investigate the identity of these niches and how they are specified. We show that yolk-sac-derived osteoclasts control the specification of a VCAM1+ vascular niche in the diaphysis to selectively recruit definitive hematopoietic cells to the fetal bone marrow.
Results
The fetal bone marrow does not support terminal hematopoiesis and grows by recruitment of mature myeloid cells
Recent studies in adult mice have shown that bone marrow hematopoiesis has a unique spatial anatomy. This is characterized by an even distribution of multipotent stem and progenitor cells and enriched near megakaryocytes. In contrast, lineage-specific progenitors and immature cells cluster together in discrete production sites for each major blood lineage that associate with specific blood vessels12,13. To investigate how this anatomical organization arises during development, we imaged the fetal bone marrow at E18.5. This is the time point where large numbers of hematopoietic cells are evident in the bone marrow10. In contrast with the adult femur, the E18.5 femur had much lower frequencies of Lin-CD117+ progenitors. These progenitors mapped almost exclusively to the center of the diaphysis and did not localize near megakaryocytes, as megakaryocytes were absent (Fig. 1a and Supplementary Fig. 1a). Additionally, Ter119+ erythroid cells were detected primarily inside the blood vessels and not in the bone marrow proper (Fig. 1a and Supplementary Fig. 1b). This indicated a lack of terminal megakaryopoiesis and erythropoiesis in the fetal bone marrow. We then examined all other major steps of hematopoiesis by flow cytometry. Despite the high frequencies of myeloid cells—particularly mature neutrophils—we found an almost complete absence of monocytic, dendritic, and lymphoid progenitors (< 3 cells per femur), as well as immature cell subsets within each lineage, indicating that the fetal bone marrow does not support terminal differentiation (Fig. 1b-d, Supplementary Fig. 1c, and Supplementary Fig. 2). These findings agree with those from a study by the McKinney-Freeman lab which showed that the fetal bone marrow does not support stem and multipotent progenitors until birth, indicating that further maturation of the microenvironment is required14. FACS analyses also showed that virtually all cells in the E18.5 bone marrow were myeloid cells (Fig. 1d). The low progenitor numbers suggested that the fetal bone marrow expands by recruiting hematopoietic cells from the periphery rather than local production from progenitors. To test this hypothesis, we compared E17.5 and E18.5 bones. In this 24 h period, the number of hematopoietic cells increased by ~60,000 cells (Fig. 1e). However, we only detected 0-1 multipotent Lin-Sca1+CD117+ and ~20 Lin-Sca1-CD117+ oligopotent progenitors at E17.5 (Fig. 1e). These progenitors would have to divide 9 times over 24 hours (faster than yeast) without exhausting themselves to account for this expansion. Together, these experiments indicate that the fetal bone marrow grows by the recruitment of definitive hematopoietic cells.
a Representative images of twelve-week-old (adult, positive control) or E18.5 femur stained with the indicated markers. The right image shows a zoomed-in region showing extravasated Ter119+ erythroid cells and the location of Lin-CD117+ hematopoietic progenitors (blue spots for easier visualization). b Representative flow cytometry plots and quantifications showing a lack of CD11b+CD117+Ly6G+ immature neutrophils (IN) and increased CD11b+CD117-Ly6G+ mature neutrophils (MN) in the E18.5 bone marrow. Each dot shows the frequencies detected by pooling long bones (tibia, femur, humerus) from one litter. c Quantifications of granulocyte progenitors (GP), monocyte dendritic cell progenitors (MDP), monocyte progenitors (MOP), and common lymphoid progenitors (CLP) in the average E18.5 femur from one litter. d Cellular composition of the E18.5 femur; note the absence of erythroid cells. Each dot corresponds to one fetus. e Number of total hematopoietic cells and Lin-CD117+Sca1- (LK) and Lin-Sca1+CD117+ (LSK) HSPC in the femurs from E17.5 (n = 13) and E18.5 (n = 15) fetuses. Each dot corresponds to one femur from one fetus in 2 independent litters. f The left image shows a whole-mounted E18.5 femur from a Cx3cr1-gfp fetus stained with the indicated markers. The right image is a map of a representative 5 µm optical section from the image on the left where each indicated cell type has been replaced with a dot of the same diameter as the cell of interest. Osteoclasts (CD11bdim CD61+ Ly6C- F4/80- CX3CR1dim CD31-), macrophages (CD11bdim CD61- Ly6C- F4/80+ CX3CR1+ CD31-), Ly6C- monocytes (CD11b+ CD61- Ly6c- F4/80+/- CX3CR1+ CD31-), Ly6C+ monocytes (CD11b+ CD61- Ly6c+ F4/80+/- CX3CR1+ CD31-) and neutrophils (CD11b+ CD61- Ly6c+ F4/80- CX3CR1- CD31-). g The dot plot shows the Y position (along the anteroposterior axis of the bone, relative to the geometric center of each section) of all myeloid cell types found in three representative 5 µm thick optical sections of 4 femurs from 4 different E18.5 Cx3cr1-gfp fetuses as shown in (f). For HSPC, the data shows the position of all HSPCs found in 3 whole-mounted femurs from 3 different E18.5 wild-type fetuses as shown in (a). Based on the differential localization of myeloid cells, we defined the metaphysis as the region of the bone that is >600 µm from the center of the bone. The interdiaphysis is the region that is more than 400 µm -but less than 600 µm—from the bone center. The rest of the bone is considered the central diaphysis. Each dot corresponds to one cell. Total cell numbers of each myeloid cell population in 4 femur sections from 4 E18.5 Cx3cr1-gfp fetuses: osteoclasts n = 122, 109, 145, 170; macrophages n = 96, 33, 108, 84; Ly6C+ monocytes n = 324, 468, 427, 510; Ly6C- monocytes n = 158, 169, 192, 194; neutrophils (Neu) n = 1386, 1142, 1726, 1401; and HSPC (Lin-CD117+) n = 56, 75, 75 from 3 femur sections from 3 E18.5 wild-type fetuses. h The plots show the number of each indicated cell type per 100 µm in each bone marrow region (M=metaphysis). Each dot corresponds to one section from one E18.5 fetus. Statistical differences were calculated using one-way ANOVA. For panels b to e each dot corresponds to one femur and statistical differences were calculated using two-way Student’s t-test if the distributions were normal and Mann-Whittney if not normal. Source data are provided as a Source Data file.
The fetal bone marrow is spatially organized with myeloid cells selectively enriched in different regions
The flow cytometry analyses in Fig. 1d,e indicated that the E18.5 bone marrow is comprised of neutrophils, monocytes, macrophages, and very rare Lin-CD117+ HSPC. The bone marrow also contains osteoclasts that are not detected by FACS due to their large size and fragility. To quantify osteoclasts, we performed confocal microscopy. A recent study indicated that CD61 specifically stains osteoclasts in developing bones15. We confirmed this and validated that CD61 is a marker for osteoclasts (Supplementary Fig. 3) which, when combined with F4/80, CD11b, Ly6C and Ly6G stains in Cx3cr1-gfp reporter mice16, can detect all myeloid cell subsets in the fetal bone marrow (Supplementary Fig. 4). Imaging analyses demonstrated that osteoclasts are distributed throughout the E18.5 bone marrow cavity. In contrast, Ly6C- monocytes and macrophages are selectively enriched in a 200 µm region immediately adjacent to the metaphysis that we named interdiaphysis. Ly6C+ monocytes, neutrophils, and Lin-CD117+ HSPC selectively localize to the center of the diaphysis (Fig. 1f–h). Thus, each myeloid cell type shows preferential localization within the fetal bone marrow.
Kinetics of myeloid cell colonization
The differential enrichment of myeloid cell populations in the central diaphysis is surprising (Fig. 1d, with all cells tallied in Fig. 2a), as all regions of the bone marrow contain large numbers of vessels that are similarly perfused (Figs. 1a and 2a). Possible explanations for the zonation of myeloid cells in the E18.5 bone marrow are differences in the time point at which each cell colonizes the bone marrow and/or the existence of specific regions that retain each cell type. Previous studies have shown that a significant fraction of osteoclasts are derived from erythro-myeloid progenitors that are specified in the yolk sac around E8.56,17. We fate-mapped yolk-sac-derived cells by mating Csf1r-MeriCreMer mice18 with Ai14Tdtomato mice19 to generate Csf1r-MeriCreMer:iTdTomato fetuses. In this model, a tamoxifen-inducible Cre is under the control of the Csf1r promoter. Administration of tamoxifen at E8.5 results in recombination and TdTomato expression in yolk-sac-derived myeloid cells (ref. 4,20,21 and Fig. 2b) but not HSC-derived cells, as HSC are not specified until E10.58,22,23,24,25,26,27,28. In agreement with other studies6,17 we found that over 60% of osteoclasts at E18.5 were yolk-sac-derived, whereas less than 1% of all other myeloid cells originated in the yolk sac (Fig. 2b,c, Supplementary Fig. 5). We then systematically mapped myeloid cell localization from E16.5 to E18.5. At E16.5 the osteoclasts were evenly distributed throughout the marrow, with all other cell types localizing to the center of the diaphysis (Fig. 2d and Supplementary Fig. 6a). This result indicates that all myeloid cell subsets colonize the bone marrow as soon as it is specified at E16.5 and that the observed myeloid cell zonation (Fig. 1f–h) is not due to differences in the time of colonization. At E17.5 the number of hematopoietic cells in the bone marrow dramatically increased due to large increases in recruitment of neutrophils and Ly6C+ monocytes, which colonize the bone marrow at much faster rates than other populations (Fig. 2d,e and Supplementary Fig. 6b). This expansion is spatially organized at E17.5 and takes place in the center of the diaphysis with recruitment also evident in the diaphyseal vessels that project toward the interdiaphysis. Importantly, at E17.5, most macrophages and Ly6C- monocytes localized to the interdiaphysis. This spatial distribution was maintained as the bone grows at E18.5 (Fig. 2d and Supplementary Fig. 6b). Collectively, these results show that the myeloid patterning is not due to differences in time of colonization. Instead, all myeloid cells -except osteoclasts- are recruited first to the center of the diaphysis and progressively separate as the bone grows.
a the graphs show the number of hematopoietic cells detected in Fig. 1g (left) and percentage of perfused vessels (right) defined by the presence of Ter119+ red blood cells in the vessel lumen of the E18.5 bone marrow. Each dot corresponds to one femur section from one fetus. b Experimental scheme for fate mapping yolk-sac-derived cells using Csf1r-MeriCreMer and Ai14tdTomatoTom/Tom mice. The right panel shows the percentage of the indicated cells that are yolk-sac-derived (TdTomato+) by flow cytometry. c Representative images and quantifications of TdTomato+ yolk-sac-derived osteoclasts (CD61+). Each dot corresponds to one fetus in one experiment. d The left panels show whole-mount images and representative maps from 5 µm optical sections of E16.5, E17.5 and E18.5 femurs showing progressive colonization by the indicated myeloid cells. The cell diameter and cell type are represented by the diameter and color of the spot. The right panels are representative images showing progressive upregulation of VCAM1 in sinusoidal and perisinusoidal cells in the central diaphysis as the bone marrow matures. e number of the indicated cells in the E16.5, E17.5 and E18.5 femur sections shown in d. Each dot corresponds to the average of 1 femur from 3 fetuses. Data are presented as mean values +/- SD. f Quantifications of the VCAM1+ area (black line) and VCAM1 intensity (red line) in the samples shown in c. n = 3 femur sections from 3 fetuses per time point. Data are presented as mean values +/- SD. g Representative zoomed-in optical section and map of an E18.5 femur in d showing that myeloid cells selectively map to VCAM1+ sinusoids in the central diaphysis and not to VCAM1- sinusoids. h Graph showing the distance of a respective myeloid cell population (one dot = one cell) in 3 E16.5 sections from each fetus to the closest VCAM1+ sinusoid. Statistical differences were calculated using the Kruskal-Wallis multiple comparisons test. i microscopy images showing expression of VCAM1 in CD31+ vascular endothelial and PDGFRβ stromal cells. j experimental scheme for conditional Vcam1 deletion in endothelial cells using Cdh5-CreERT:Vcam1flox/flox mice pulsed with tamoxifen at E15.5 (prior to colonization of definitive hematopoiesis). k, l quantifications (k) and raw images (l) showing colonization by Ly6C + CD11b+ cells (monocytes and neutrophils) and VCAM1 expression in CD31+ cells in femur sections from Cdh5-CreERT:Vcam1flox/flox mice (KO) or littermate controls (LC). n = 5 for both conditions. Each dot corresponds to one section from one fetus. For all panels, P values were calculated using two-tailed Mann-Whittney analyses. Source data are provided as a Source Data file.
VCAM1 defines a “colonization niche” for HSC-derived cells that expands from the central diaphysis as the bone marrow matures
The fact that macrophages, monocytes, neutrophils and HSPC (which are produced by HSC in the fetal liver vs. osteoclasts that are also yolk-sac-derived; Fig. 2b,c) first colonize the vessels in the central diaphysis (Fig. 2d) but not the metaphysis—despite vessels in both regions being equally perfused (Fig. 2a)—suggested the existence of a unique microenvironment that mediates recruitment of HSPC and myeloid cells to this region. Numerous studies have demonstrated that neutrophil and HSPC homing and retention in the adult bone marrow after myeloablation and transplantation are mediated by leukocyte adhesion molecules, including VCAM1, E and P selectins, and other adhesion molecules and cytokines29,30,31. scRNAseq analyses of human fetal bone marrow revealed high levels of Vcam132. Based on this we profiled VCAM1 through fetal bone marrow development. We found that VCAM1 was expressed -exclusively- in the center of the diaphysis at E16.5. At E17.5 (when HSPC, monocytes, and neutrophils colonize the bone marrow in earnest) VCAM1 expression in sinusoids and perisinusoidal regions dramatically increased (Fig. 2d, f). VCAM1 expression upregulated further at E18.5 in nearly all sinusoids in the diaphysis but was largely excluded from the metaphyseal vessels (Fig. 2d). Importantly, all myeloid cell types -except osteoclasts- were selectively recruited to VCAM1+ sinusoids (Fig. 2d, g, h). The endothelial cells in bone marrow sinusoids frequently associate with PDGFRβ+ stromal cells, known to play a role in establishing fetal hematopoiesis33. FACS analyses showed that VCAM1 was expressed both in PDGFRβ+ stromal cells and CD31+ endothelial cells but that the increase in VCAM1 expression from E16.5 to E18.5 was mediated mainly by increases in VCAM1+ endothelial cell numbers (Supplementary Fig. 6c, d). To examine the role of VCAM1 in mediating recruitment,t we first deleted Vcam1 in the non-endothelial stromal cells by crossing Prx1-Cre mice34 with Vcam1flox mice31 to generate Prx1-Cre:Vcam1flox/flox mice. This resulted in an almost complete loss of VCAM1 in the stroma but not in vascular endothelial cells, and yet there were no changes in hematopoietic cell recruitment to the bone marrow (Supplementary Fig. 6e-g). To test the role of VCAM1 in endothelial cells, we crossed Cdh5-CreERT mice35 with Vcam1flox mice to generate Cdh5-CreERT:Vcam1flox/flox fetuses, and pulsed pregnant Vcam1flox/flox dams with tamoxifen at E15.5, before definitive hematopoietic cells colonize the bone marrow. Even though this treatment only reduced endothelial VCAM1 by ~20%, the Cdh5-CreERT:Vcam1flox/flox fetuses showed reductions of neutrophil colonization by 50%, with clear loss of hematopoietic recruitment in diaphyseal regions where vessels lost VCAM1 expression (Fig. 2j-l). These results formally demonstrate that VCAM1+ endothelial cells in the diaphyseal sinusoids provide a hematopoietic recruitment niche in the bone marrow.
Ablation of yolk-sac-derived myeloid cells disrupts vessel growth and the VCAM1+ recruitment niche
Since osteoclasts are known to promote angiogenesis36, we hypothesized that yolk-sac-derived osteoclasts regulate the establishment of the VCAM1+ sinusoids. To selectively ablate yolk-sac-derived myeloid cells we injected C57BL/6 J pregnant dams at E6.5 with anti-CD115 depleting antibody (Fig. 3a). This is an established protocol that ablates myeloid cells derived from the yolk sac while sparing hematopoietic cells produced by HSC, as HSC are specified later4,21,37,38,39. We then compared anti-CD115 (depleted, αCD115) and anti-IgG (control) litters at E18.5. anti-CD115 treatment led to loss of osteoclasts (Fig. 3a-c) and overall reduction in osteoclast size (Supplementary Fig. 7a). This size change is likely due to the replacement of the large yolk-sac-derived osteoclasts by smaller osteoclasts (which will be derived from HSC) throughout the bone marrow. In the anti-CD115 group, there was a ~ 2-fold reduction in the number of neutrophils but no other myeloid cells (Fig. 3b, d). FACS analyses demonstrated an almost complete loss of VCAM1 expression in the bone marrow endothelium and overall loss of endothelial cells and PDGFRβ+ stromal cells (Fig. 3e). Imaging analyses showed selective loss of VCAM1 and perturbed vessel growth in the diaphysis (Supplementary Fig. 7b-d). Unexpectedly, depletion of yolk-sac-derived myeloid cells led to de novo expression of VCAM1 in the metaphysis (Fig. 3b, f) and this correlated with de novo recruitment of neutrophils to this region (Fig. 3g). These results indicate that yolk-sac-derived myeloid cells promote vessel growth and VCAM1 expression in the diaphysis and suppress VCAM1 expression in the metaphysis, and that VCAM1 expression is sufficient to direct neutrophil colonization to specific regions of the bone marrow.
a Experimental design depleting yolk-sac-derived myeloid cells as previously reported4,21,37,38,39. b Representative images of whole-mounted E18.5 femurs and maps of the indicated regions. c Number of CD61+ osteoclasts after depletion of yolk-sac-derived myeloid cells. Each dot corresponds to one femur section from one E18.5 fetus. d Number of indicated cells per section; n = 5 sections from 5 E18.5 fetuses. e Cell numbers and percent VCAM1+ cells of CD45-Ter119-CD31+CD105+ vascular endothelial cells (VEC) or CD45-Ter119-CD31-PDGFRβ+ stromal cells (PDGFRβ+). Each dot corresponds to one E18.5 fetus from two independent litters. f, g Zoomed-in image from b and quantifications showing de novo misexpression of VCAM1 and subsequent recruitment of neutrophils (Ly6C+) in the metaphysis of anti-CD115-treated fetuses. For VCAM1 quantifications; IgG n = 5 and αCD115 n = 4 sections from femurs of different E18.5 fetuses in each group. For metaphyseal neutrophil quantifications; n = 5 femur sections from five E18.5 fetuses in each group. h Number of total hematopoietic cells along with Lin-Sca1+CD117+ multipotent progenitor numbers and frequency in the femur of anti-CD115- or IgG-treated E18.5 fetuses quantified by FACS. Each dot corresponds to one mouse. i Percentage of donor (CD45.2+) cells in the peripheral blood of CD45.1+ recipients at the indicated time points after transplantation of all the hematopoietic cells found in one femur of anti-CD115- or IgG-treated E18.5 fetuses along with 1 ×105 CD45.1+ competitor cells purified from adult mice. Each dot corresponds to one mouse, n = 7 IgG and 8 anti-CD115 recipients in two experiments. Statistics shown are for total engraftment at 16 weeks. j Representative pentachrome stain and quantification of bone marrow area (not occupied by cartilage or bone) in E18.5 femur from IgG or αCD115 groups. Each dot corresponds to one femur from one fetus. For all panels, p values are shown, and statistical differences were calculated using a two-tailed Student’s t-test if the distributions were normal or Mann-Whitney if not normal. Source data are provided as a Source Data file.
Due to very low stem and progenitor numbers at E18.5 it was not feasible to quantify recruitment of these cells using imaging. We thus examined HSPC recruitment using FACS analyses by pooling long bones from multiple fetuses. We confirmed the loss of multipotent Lin-CD117+Sca1+ (LSK) cells after the ablation of yolk-sac-derived cells (Fig. 3h). To functionally confirm the depletion of HSC we performed competitive transplants. Long-term reconstitution potential was not detectable in mice transplanted with all the hematopoietic cells found in a femur from the anti-CD115-treated mice, whereas 3 out of 7 recipients showed multilineage donor cell engraftment when transplanted with the IgG control bone marrow (Fig. 3i and Supplementary Fig. 8a, b) indicating that ablation of yolk-sac-derived myeloid cells results in impaired HSPC colonization of the fetal bone marrow.
Because osteoclasts are responsible for bone resorption and maintain the size of the bone marrow cavity40,41, an alternative explanation for reduced bone marrow colonization after loss of yolk-sac-derived myeloid cells would be a reduced bone marrow cavity in the anti-CD115-treated mice. However, we found that anti-CD115 treatment did not lead to reductions in bone length or the size of the bone marrow cavity, indicating that reduced colonization is not mediated by loss of available bone marrow space (Fig. 3j and Supplementary Fig. 8c). Since complete loss of osteoclasts in CMV-Cre:Rankfl/fl mice (Supplementary Fig. 3k, henceforth RankΔ/Δ) results in a reduced bone marrow cavity at E18.5 (Supplementary Fig. 3h, i) these results also indicate that the osteoclasts that persist after anti-CD115-treatment (Fig. 3c), which are likely derived from HSC and not the yolk sac (Fig. 2c), are sufficient to generate a bone marrow cavity after anti-CD115 treatment.
Together, our results indicate that yolk-sac-derived myeloid cells control definitive HSPC and myeloid cell colonization of the developing bone marrow through the specification of a VCAM1+ vascular recruitment niche.
Yolk-sac-derived myeloid cells are negative regulators of fetal liver hematopoiesis
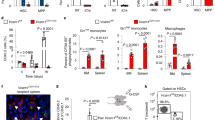
The liver is the main organ of hematopoiesis during fetal development42,43,44,45,46,47. The identity of the niches that support and regulate fetal liver hematopoiesis is unclear. Different studies have proposed that portal vessels, hepatocytes, stellate cells, endothelial cells, and macrophages all function as HSPC niches43,44,45,46,47,48,49,50. Since the fetal liver also contains yolk-sac-derived myeloid cells (including fetal liver macrophages, or Kupffer cells4,5 - Fig. 2b), and depletion of macrophages perturbs fetal liver hematopoiesis48,49,50, we examined the effect of anti-CD115 treatment in the E18.5 fetal liver.
Unexpectedly, depletion of yolk-sac-derived myeloid cells -using the anti-CD115 protocol- dramatically increased fetal liver cellularity (30 million additional cells when compared to IgG controls, Fig. 4a, b). We used flow cytometry to systematically quantify stepwise differentiation for all major blood lineages (Supplementary Fig. 9). These revealed that the expansion in cellularity was due -almost exclusively- to increased erythropoiesis, especially of CD71brightTer119bright late erythroblasts (Fig. 4c). Importantly, we found no reductions in neutrophils or HSC (Fig. 4d, e), indicating that reduced neutrophil and HSC recruitment to the fetal bone marrow after anti-CD115 treatment (Fig. 3d, h) was not due to impaired production of these cells in the liver.
a Experimental design for depletion of yolk-sac-derived myeloid cells and examination of HSC function in primary and secondary competitive transplantation experiments in the E18.5 fetal liver. b–e Number of the indicated cells in the fetal liver of mice treated as in a Each dot corresponds to one fetus in two different litters. CFU-E Colony forming unit erythrocyte progenitors, Ery Erythroblasts, Retics Reticulocytes, RBC erythrocytes, Neutros Mature Neutrophils LT-HSC Lin-Sca1+CD117+Flt3-CD48-CD150+ Long term hematopoietic stem cells, LSK Lin-Sca1+CD117+. f Frequency of donor CD45.2+ cells in the blood of B6-SJL (CD45.1+) recipients transplanted with 50 Lin-Sca1+CD117+Flt3-CD48-CD150+ CD45.2+ LT-HSC from fetal livers and 2×105 CD45.1+ competitor cells as shown in a. Note that the Lineage panel does not contain CD11b, as this molecule is expressed in fetal HSC45 (in contrast to adult HSC). The dot shows the mean, and the error bars the SEM. n = 6 and 7 recipients in 2 independent experiments. P values: All cells 4 weeks = 0.0298, B cells 4 weeks = 0.0087, B cells 8 weeks = 0.0291, T cells 8 weeks = 0.0041. g The table shows the fraction of CD45.1+ secondary recipients that showed peripheral blood engraftment (defined as more than 0.1% CD45.2+ donor cells in peripheral blood); the right graph shows the frequency of donor cells for the same mice. Statistical differences were calculated using a one-tailed Chi-square test. Unless otherwise indicated for all other panels: p values are shown, each dot corresponds to one mouse, and statistical differences were calculated using a two-tailed Student’s t-test if the distributions were normal or Mann-Whitney if not normal. Source data are provided as a Source Data file.
These analyses also demonstrated that the number of Lin-Sca1+CD117+Flt3-CD48+CD150+ LT-HSC and Lin-Sca1+CD117+ LSK doubled after yolk-sac-derived cell depletion (Fig. 4e). To rigorously assess HSC function, we performed competitive transplantation experiments. We FACS-purified 50 Lin-Sca1+ CD117+Flt3-CD41-CD48+CD150+ HSC from the liver of anti-CD115 or IgG-treated fetuses. We transplanted these CD45.2+ HSC along with 200,000 CD45.1+ competitor cells into lethally irradiated CD45.1+ recipients. These experiments revealed 2-fold increases in short-term T cell and B cell engraftment after transplantation (Fig. 4f). To further examine the ability of HSC to self-renew functionally, we performed secondary competitive transplantation experiments. We transplanted 100 45.2+ HSC from the primary recipients along with 2x105 competitor cells into irradiated secondary recipients (Fig. 4a). Twelve out of fourteen (85.7%) of secondary recipients transplanted with the HSC first purified from the anti-CD115 depleted fetuses displayed multilineage engraftment (≥0.1% donor cells in peripheral blood) at 16 weeks after transplantation. In contrast, only three out of eight (37.5%) of the secondary recipients transplanted with HSC from the IgG controls displayed engraftment (Fig. 4g). This experiment demonstrates increased HSC function and self-renewal in the fetal livers from the anti-CD115-depleted fetuses. Together our results demonstrate that yolk-sac-derived myeloid cells are negative regulators of HSC and erythropoiesis in the fetal liver. Since the depletion of liver macrophage populations has different effects on fetal liver hematopoiesis48,49,50, our results also suggest that multiple myeloid cell subsets function in the liver to regulate blood production in the fetus.
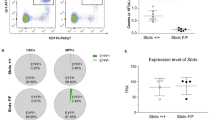
Mice lacking osteoclasts do not display changes in fetal liver hematopoiesis
Flow cytometric analyses found no differences in fetal liver HSC and mature erythroid cell numbers between the anti-CD115 and IgG control groups at E14.5, indicating that these cells expand in the liver during the E14.5 to E18.5 period (Fig. 5a). Since BM colonization is impaired at E18.5 in the anti-CD115 group due to the loss of the VCAM1 niche (Fig. 3), these results raised the possibility that the increased HSC and erythroid cells in the E18.5 liver after anti-CD115 treatment were due to lack of trafficking to the bone marrow. To test for this possibility, we crossed CMV-Cre mice51 with Rankflox/flox mice52 to generate RankΔ/Δ fetuses, which lack osteoclasts (Supplementary Fig. 3h-k), as RANK-RANK ligand signaling is necessary for osteoclast formation40,41. RankΔ/Δ fetuses recapitulated previously published data on Rank-/- mice41, showing reduced bone marrow cavity and reduced colonization by hematopoietic cells (Fig. 5c). These mice also showed complete loss of VCAM1 expression, strongly suggesting that the VCAM1+ colonization niche is specified by osteoclasts (Fig. 5d, e). If the accumulation of HSC and erythroid cells in the anti-CD115-treated E18.5 liver (Fig. 4c, e) was due to a lack of trafficking to the bone marrow, then we predicted that HSC numbers, function, and erythroid cell numbers should also be increased in the RankΔ/Δ fetal liver. In contrast, we found no differences in HSC function or numbers of any cell population assessed in the RankΔ/Δ fetal liver when compared to littermate controls (Fig. 5f-h). These results demonstrate that HSC and erythroid cell accumulation in the anti-CD115-treated liver are not due to an inability to colonize the bone marrow. These results also indicate that yolk-sac-derived myeloid cells in the bone marrow are positive regulators of fetal bone marrow hematopoiesis by controlling the specification of the colonization niche, whereas a different yolk-sac-derived cell negatively regulates fetal liver hematopoiesis.
a Numbers of the indicated cells in the E14.5 fetal liver from fetuses from dams treated with IgG or anti-CD115 as in Fig. 4a. LT-HSC = Lin-Sca1+CD117+Flt3-CD48-CD150+ Long-term hematopoietic stem cells, CFU-E Colony forming unit erythrocyte progenitors, Ery Erythroblasts, Retics Reticulocytes, RBC erythrocytes. b Breeding strategy to generate fetuses lacking osteoclasts (RankΔ/Δ). c Representative pentachrome-stained E18.5 femur sections and quantifications from RankΔ/Δ mice and littermate controls showing reduced bone marrow cavity (control n = 10, RankΔ/Δ= 5) and hematopoietic cells per femur (control n = 11, RankΔ/Δ= 4) in RankΔ/Δ mice. d, e Representative images and quantification showing lack of VCAM1 expression and loss of Ly6C+ monocyte and neutrophil recruitment to the E18.5 RankΔ/Δ marrow. f, g Number of indicated cells in the E18.5 fetal liver from RankΔ/Δ or littermate control fetuses. h Percentage of CD45.2+ cell engraftment after transplantation of 50 Lin-Sca1+CD117+Flt3-CD48-CD150+ CD45.2+ LT-HSC purified from RankΔ/Δ or littermate controls. The dots show the mean, and the error bars the SEM. n = 6 ctrl and 4 RankΔ/Δ recipients in 2 independent experiments. For all other panels and unless otherwise indicated: p values are shown, each dot corresponds to one fetus, and statistical differences were calculated using a two-tailed Student’s t-test if the distributions were normal or Mann-Whitney if not normal. Source data are provided as a Source Data file.
To investigate the identity of these fetal liver cells, we performed flow cytometric analyses of E18.5 livers of Csf1r-MeriCreMer:iTdtomato fetuses to fate map TdTomato+ yolk-sac derived cells (Supplementary Fig. 10a). We found TdTomato+ yolk-sac-derived cells in the monocyte, neutrophil, erythroid and macrophage cell compartments, with phenotypically defined Kupffer cells being the most abundant TdTomato+ cells in the E18.5 fetal liver Supplementary Fig. 10b). Recent manuscripts have depleted macrophages (of which Kupffer cells are a subset) in the fetal liver using different methods and discovered hematopoiesis was perturbed in these depletion models, indicating that macrophages regulate fetal liver hematopoiesis48,49,50. To formally test the role of Kupffer cells in fetal hematopoiesis, we bred Clec4f-Cre-tdTomato mice53 with ROSA-iDTA54 mice (Supplementary Fig. 10c) to deplete Kupffer cells in E18.5 Clec4f-Cre:iDTA fetuses, as Clec4f-Cre-tdTomato is selectively expressed in Kupffer cells53. Despite ~3-fold fewer Kupffer cells (Supplementary Fig. 10d) we found no changes in HSC numbers or erythroid cells in the Clec4f-Cre:iDTA fetal livers (Supplementary Fig. 10e). These experiments indicate that Kupffer cells do not regulate E18.5 fetal liver hematopoiesis and that other rare subsets of yolk-sac-derived cells are responsible for negatively regulating HSC and erythroid cells at this stage of development.
Discussion
In this study we sought to identify the mechanisms regulating colonization of the fetal bone marrow by definitive (HSC-derived) hematopoietic cells. Using imaging approaches, we demonstrate that the fetal bone marrow is spatially organized by differential localization of myeloid cell subsets that selectively localize to VCAM1+ sinusoids in the diaphysis. Our results indicate that this VCAM1+ niche is both necessary and sufficient to promote recruitment, as loss of VCAM1 significantly reduced colonization, and misexpression of VCAM1 in the metaphysis resulted in de novo recruitment of neutrophils cells to that region. This VCAM1+ colonization niche is specified by yolk-sac-derived myeloid cells, as depletion of these cells abrogated the VCAM1+ niche, resulting in reduced leukocyte and HSPC recruitment to the bone marrow. We propose that the yolk-sac-derived myeloid cell that specifies the VCAM1 niche is an osteoclast, as these are the most abundant yolk-sac-derived myeloid cell in the bone marrow and genetic depletion of osteoclasts abolishes the VCAM1+ niche. The observation that depletion of yolk-sac derived osteoclasts -using anti-CD115- does not impact bone size but depletion of yolk-sac- and HSC-derived of osteoclasts -in Rank knockouts– raises the possibility that different subsets of osteoclasts might have unique functions in bone and bone marrow development. This would agree with a recent study that showed that vessel-associated osteoclasts control vascular development and growth plate homeostasis15. We also found that a different subset of yolk-sac-derived cells regulates erythropoiesis and HSC function in the fetal liver in an osteoclast and Kupffer cell-independent manner. Thus, different subsets of yolk-sac-derived myeloid cells differentially regulate hematopoiesis in two fetal organs.
Even though Kupffer cells are the most abundant yolk-sac-derived myeloid cell in the liver, genetic depletion of Kupffer cells did not impact fetal liver hematopoiesis. This was unexpected because recent studies have shown that depletion of fetal liver macrophages via pharmacological inhibition of CSF1R from E11.5-14.5 or Rank-Cre-mediated deletion of Pu.1 induced a myeloid bias in HSC, increased neutrophil production, and perturbed erythropoiesis49,50. Our results thus suggest that these phenotypes are mediated by macrophage populations distinct from Clec4f-Cre-targeted Kupffer cells.
An accompanying manuscript, using a different mouse model for ablating yolk-sac-derived myeloid cells, also shows a role for these cells in establishing the neonatal bone marrow microenvironment (see accompanying Percin et al manuscript55). This study shows that yolk-sac-derived myeloid cells regulate the expansion of CXCL12-producing cells to promote hematopoietic recruitment to the neonatal bone marrow. The findings from both independent studies thus demonstrate that yolk-sac-derived myeloid cells act on different subsets of non-hematopoietic cells to specify “colonization niches” in the bone marrow.
Our study also has some limitations: while our data demonstrates that yolk-sac-derived myeloid cells control the maturation of the microenvironment to promote neutrophil and HSPC recruitment to the bone marrow- our results also indicate that additional signals regulate myeloid cell colonization, as depletion of yolk-sac-derived osteoclasts did not impair colonization by monocytes or macrophages. Our fate-mapping experiments demonstrate that most yolk-sac-derived cells in the bone marrow are osteoclasts. However, tamoxifen-induced cre recombination is inherently variable, and we cannot exclude the possibility that rarer subsets of yolk-sac-derived cells are also present in the bone marrow at these fetal stages. We also did not identify the yolk-sac-derived cell(s) that regulates fetal liver HSC.
This study highlights the power of in situ imaging approaches to reveal the underlying biology of hematopoiesis within the developing fetus. They reveal a complex serial recruitment and spatially organized crosstalk between hematopoietic cells with different developmental origins and non-hematopoietic cells in different organs to regulate fetal hematopoiesis.
Methods
Mice
All mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Cincinnati Children’s Hospital Medical Center. All the mice (Mus musculus) used in the study were bred and housed in the Cincinnati Children’s Hospital Medical Center’s animal care facility in compliance with institutional and National Institutes of Health guidelines. Mice were maintained at the vivarium at Cincinnati Children’s Hospital Medical Center under a 14-hours light:10-hours darkness schedule, 30–70% humidity, 22.2 ± 1.1 °C, and specific-pathogen-free conditions. The following mouse strains were used: C57BL/6J-Ptprcb (CD45.2), B6.SJL-PtprcaPepcb/BoyJ (CD45.1), B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (Ai14tdTomato), FVB-Tg(Csf1r-cre/Esr1*)1Jwp/J (Csf1r-MeriCreMer), B6.C-Tg(CMV-Cre)1Cgn/J, B6.Cg-Tnfrsf11atm1.1Irw/J (Rankflox/flox), B6.Cg-Tg(Prrx1-cre)1Cjt/J, B6.129(C3)-Vcam1tm2Flv/J, C57BL/6J-Clec4f em1(cre)Glass/J, B6.129P2-Gt(ROSA)26Sor tm1(DTA)Lky/J and C57BL/6-Tg(Cdh5-cre/ERT2)1Rha.
Collection of fetal bone marrow and liver
Dam and sire mating pairs were examined every morning before 10:00 local time. Dams with a visible mating plug were considered pregnant at E0.5. At the indicated time points, the dams were euthanized by isoflurane overdose followed by cervical dislocation, and the fetuses harvested and euthanized by decapitation following IACUC-approved protocols. Fetal long bones (tibia, humeri, femur) were crushed and filtered through a 70 µm strainer before analysis. Fetal livers were pushed through a 100 µm strainer and again through a 70 µm strainer before analysis. Cell counts were obtained with a hemocytometer.
Collection of adult bone marrow cells
Twelve-week-old C57BL/6 J male mice were anesthetized with isoflurane followed by cervical dislocation. For long bones, bone marrow cells were flushed out of the femurs with a 21-gauge needle in 1 ml of ice-cold PEB buffer (2 mM EDTA, 0.5% bovine serum albumin, 0.005% sodium azide in PBS) and filtered through a 70 μm strainer.
Tamoxifen treatment
Pregnant dams at E8.5 or E15.5 received an intraperitoneal injection containing 4-hydroxytamoxifen (75 μg/g of body weight Sigma-Aldrich # T5648-5G) and progesterone (37.5 μg/g body weight; Sigma-Aldrich # P0130-256) in corn oil. Progesterone was used to prevent fetal abortions due to the potential estrogenic effects of tamoxifen.
Depletion of yolk-sac-derived cells
Pregnant C57BL/6 J females were treated at E6.5 with a single intraperitoneal injection of 3 mg of anti-CSF1R mAb (Bio X Cell, clone AFS98 #BP0213) or rat IgG2a isotype control (Bio X Cell, clone 2A3 #BP0089) as previously described4,21,37,38,39.
Stromal Digestion
Protocol was modified from previous publications56,57. Briefly, the digestion buffer was made with 2 mg/ml collagenase type IV (Gibco, 17104-019) and 3 mg/ml dispase (Gibco, 17105-041) dissolved in room-temperature PBS. We harvested the adult bone marrow by flushing each tibia with 1 ml of digestion buffer into a 5 ml polypropylene snap-cap tube containing another 1 ml of digestion buffer. For fetal bones, long bones (femur, tibia, humerus) were dissected, washed with PBS 1x, and then crushed in a digestion buffer. We mixed the tubes vigorously by hand and incubated them at 37 °C for 5–7 min. After the first incubation, the tubes were mixed vigorously by hand and then incubated at 37 °C for another 5–7 min. After this second incubation, we collected the supernatant, taking care to leave any macroscopic clumps in the tube. We transferred the digested cells to a tube containing 5 ml of ice-cold PBS 2 mm EDTA 0.1% Bovine Serum Albumine 0.005% Sodium Azide (PEB). Then we added 1 ml of digestion buffer to the snap-cap tubes, and the process above was repeated until all macroscopic pieces of bone marrow had been digested. This was repeated no more than 5 times. For adult bone marrow, red blood cells were lysed once with RBC lysis buffer. All samples were filtered first through a 100-μm filter and then immediately spun down in a centrifuge. The cells were resuspended in 1 ml of ice-cold PEB, filtered in a 70μm filter, immediately spun down, and then used for subsequent analyses.
Flow cytometry
For FACS analyses, cells were stained with a cocktail with the indicated antibodies (Supplementary Data 1) used in each stain for 30 minutes, washed thrice with ice-cold PBS containing 2 mM EDTA and 0.5% Bovine Serum Albumin, and analyzed in a BD LSRFortessa™ Flow Cytometer (BD Bioscience) or purified using SH800S Cell Sorter (Sony Biotechnology). Dead cells and doublets were excluded based on FSC and SSC distribution and DAPI exclusion (Sigma-Aldrich, catalogue number D9542). The very low hematopoietic stem cell and progenitor numbers found in the E18.5 femur made rigorous detection of these cells in individual femurs challenging. Therefore, for each analysis, we stained and FACS-analyzed all cells found in the long bones from each litter and then plotted the values normalized to the cellularity of one individual femur. Supplementary Fig. 1 provides examples of gating strategies for the cell populations interrogated using flow cytometry.
Hematopoietic stem cell transplantation
B6.SJL-PtprcaPepcb/BoyJ (CD45.1+) recipient mice were myelo-ablated by two doses of 700 and 475 rads, three hours apart. For fetal bone marrow transplants the recipients were transplanted with all the hematopoietic cells in an E18.5 CD45.2+ femur and 1x105 total hematopoietic competitor cells from a CD45.1+ adult mouse. For fetal liver primary HSC transplants the recipients were transplanted with 50 Lin-Sca1+CD117+Flt3-CD48-CD150+CD45.2+ HSC purified from the liver and 2x105 total hematopoietic competitor cells from a CD45.1+ mouse. For secondary transplants we FACS-purified 100 CD45.2+ LT-HSC from the primary recipients and transplanted along with 2x105 total hematopoietic competitor cells from a CD45.1+ mouse. All recipients were myelo-ablated CD45.1+adult male mice. Multilineage reconstitution in the peripheral blood of the recipients was quantified by flow cytometry every four weeks with positive engraftment considered at >0.1% CD45.2+ cells.
Bone marrow and liver sectioning for immunostaining
For the detection of hematopoietic cells in the bone marrow (fetal and adult) and fetal liver, we modified a previously established protocol58. Tissues were fixed in 4% ice-cold PFA for six hours and washed in PBS. At this stage, femurs were decalcified overnight by rotating in the immersion of a solution of 10% w/v EDTA in PBS. Then, samples were cryoprotected by immersion in 20% w/v sucrose in dH2O and frozen in OCT (TissueTek). Samples were kept in -80 until sectioning.
For sectioning, tissues were removed from molds and placed in a frozen microtome. For bone marrow samples, the cavity was exposed by careful shaving of the bone surface until the central bone marrow is fully exposed. Similarly, a lobe of the fetal liver was shaved to expose a region of tissue within the liver for immunostaining. The sample is then removed from OCT, thawed in ice-cold PBS, blocked for 2 hours in PBS with 20% Goat Serum stained overnight with primary antibodies (Supplementary Data 1), washed thrice, and stained with secondary antibodies (Supplementary Data 1) for 4 hours.
For quantification of changes in the vasculature we modified a different protocol59. Fetal femurs were fixed overnight in 4% ice-cold PFA, washed in PBS, and decalcified overnight by immersion in a solution of 10% w/v EDTA in PBS. Bones were then mounted in agarose and sectioned with a vibratome (60 µm thick). For this protocol - blocking, staining and imaging was conducted using high-quality polymer chamber wells (Ibidi, 80426). The same reagents for staining were used as above.
Confocal Imaging acquisition
Confocal imaging was performed on whole-mounted tissue samples or vibratome sections in a Nikon AXR Inverted Confocal Microscope with the following specifications: high power 405 nm, 445 nm, 488 nm, 514 nm, 561 nm, 594 nm, 640 nm, and 730 nm solid-state diode lasers. All microscopes were equipped with a fully encoded scanning XY motorized stage, Piezo-Z nosepiece for high-speed Z-stack acquisition, resonant and galvanometric scanners, one high-quantum efficiency, low-noise Hamamatsu photomultiplier tube, and three high-quantum efficiency gallium arsenide phosphide photomultiplier tubes (GaAsP-PMTs) for overall 400-820 nm detection. For image acquisition, we used a LWD Lambda S 40XC water-immersion objective (Nikon, MRD77410) with a resonant scanner and 4X line averaging, 1024 x 1024 pixels resolution, and 0.3 μm Z-step.
Image analyses
Original images (.ND2 format file) were denoised by a built-in artificial intelligence algorithm (Denoise.AI) and stitched together using the NIS-Elements software (Nikon, version 5.20.02 and 5.30.03). The denoised and stitched .ND2 files were converted to Imaris (.IMS) files using Imaris software (Bitplane, version 9.5 to 9.9). Cells of interest were labeled with dots of the same diameter as the cell with the Imaris Spots function in manual mode, and then X, Y, and Z coordinates from the dots were automatically computed. Sinusoids (VCAM1+ or VCAM1-), were segmented based on the signal from the VCAM1 channel using the Imaris Surface function. For cells with irregular shapes (osteoclasts and macrophages) we measured the diameter of the minimum circle that contained the cell. Distances between cells and vessels were obtained with the Imaris surface-to-surface function. The distance from each cell to the center of the diaphysis was a point extrapolated from the Y coordinates as indicated above. For segmenting the metaphysis, intermetaphysis, and diaphysis within our bone marrow images, we first identified where the anterior and posterior ends of the marrow were, based on the CD31 signal. We define the center of the bone as the geometric point between these two ends. Then, we defined the metaphysis as the region of the bone that is >600 µm of the center of the bone. The interdiaphysis is the region that is more than 400 µm -but less than 600 µm - from the bone center. The rest of the bone is considered the central diaphysis. The y-coordinates of the manually annotated spots determined what region the cell was identified in. Vessel length and lumen measurements were extracted using point-to-point distance analysis on Imaris software. Vessel length was measured from branchpoint to branchpoint. Vessel lumen was measured based on the largest diameter within a vessel (as defined in vessel length). In Vcam1flox/flox mice, Ly6C + CD11b+ cells were calculated by generating a masked Ly6C channel for CD11b in Imaris, creating a surface based on a positive signal within the masked channel, and extracting the volume from that surface. This volume was then divided by the average volume of a neutrophil (as shown in Supplementary Fig. 3e) to obtain the total number of Ly6C + CD11b+ cells per image. This was then divided by the bone marrow volume, which was extracted from the manually annotated surface generated based on the CD31 signal that outlined the bone marrow tissue. In Cdh5-CreERT:Vcam1flox/flox mice, VCAM1 colocalization with CD31 was calculated using the color threshold function in ImageJ. In Prx1-Cre:VCAM1flox/flox mice, VCAM1 voxels were extrapolated from a surface based on a positive VCAM1 signal within the bone marrow in Imaris. Images shown are from a mask that removed VCAM1 signal within a surface based on the CD31 channel. Maximum pixel intensity values in Supplementary Fig. 3 were extracted from manually annotated spots in Imaris. Percentages were then calculated based on the following: 1) at or below the minimum intensity threshold was assigned the value of 0 (not real signal or background signal) while intensities at or above the maximum intensity threshold were considered saturated or maximum signal; and 2) Values that fell within this range were calculated by taking that value, dividing it by the maximum intensity signal, and multiplying by 100% to give a percent of maximum intensity. This was then applied to 10 different cells within each of the populations outlined in Supplementary Fig. 3. All software were installed in HP Z4 windows 10 x 64 workstations equipped with Dual Intel Xeon Processor W-2145, 192GB ECC-RAM, and an Nvidia Quadro RTX 5000 16GB GDDR6 Graphics Card.
Quantifications of vessel number, length, diameter, and perfusion
Bone marrow vessels were detected based on expression of CD31. Each vessel was defined as a continuous segment between two branching points. Vessel length was defined as the contiguous line from one branch point to another. Vessel lumen diameter was defined as the largest diameter within one vessel. Vessel perfusion was determined based on the presence or absence of Ter119+ erythroid cells within the lumen of an E18.5 fetal bone marrow sample.
Pentachrome stain and quantification of bone length and area size
Fetal femurs were fixed in 10% formalin overnight, washed thrice in PBS, and decalcified in 20% w/v in PBS overnight. Samples were dehydrated in 70% ethanol, embedded in paraffin, and sectioned (10 µm) in a microtome. Alternate consecutive sections were stained with hematoxylin/eosin or pentachrome solutions, and those determined to represent the central bone marrow used to quantify the size of the bone marrow cavity and bone length. Images of pentachrome-stained sections were acquired on a 3007 A Nikon 90i upright microscope. Image analysis was done using NIS-Elements GA3 processing to exclude the mucin and collagen signals within the marrow, with the resulting bone marrow area quantified. This process was inverted when quantifying the composition cartilage and bone of our RankΔ/Δ pentachrome images. For bone marrow length, we used NIS-Elements to measure the distance between center points of each end of marrow in our pentachrome sections.
Statistics
All statistical analyses were performed using Prism 9 (GraphPad Software). For graphs quantifying cells in different mice, we indicate the mean, and each dot corresponds to one mouse. For graphs showing distributions of cells, we indicate the mean, and each dot corresponds to one cell. Statistical analyses between two samples were performed by using the Student’s t-test if the data was normal and the Mann-Whitney test if the data was not normal. For statistical analysis between multiple samples, analyses were performed using two-way ANOVA followed by Sidak’s multiple comparisons test if the data was normal or the Kruskal-Wallis test if not normal.
Data reporting
No statistical methods were used to predetermine the sample size. Image analyses of the bone marrow are very time-consuming, and it is not possible to examine large numbers of samples. We have previously shown that 3 bones per condition allow the identification of a sufficient number of cells to detect changes in location and distribution in the bone marrow1. Based on this, we have striven to analyze 3 bones per condition and included additional bones when possible. All mice were included in the analyses. Mice were randomly allocated to different groups based on genotype and litter size. For all experiments, we aimed to have the same number of mice in the control and experimental groups. Investigators were not blinded to allocation during experiments and outcome assessment. This is because it was not possible to blind the investigator to the age ofthe fetal bone examined, as they are readily identified by size and cell distribution. Similarly, the depletion of yolk-sac-derived cells led to evident changes in cellular content and organization.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data is publicly available in the BioImage Archive60 under the BioImages accession number S-BIAD1761, including FACS data files (FCS and experiments), ND2 and IMS imaging files. Source data files are provided with this paper in.xls format and indicate the file locations within the data repository. Of note – FACS data files for Fig. 1 can be found in the Supplementary Fig. 2 folder of the repository, data in Supplementary Fig. 8 can be found in the Fig. 3 folder, and data in Supplementary Fig. 9 can be found in the Fig. 4 folder. All other data are available in the article and its Supplementary files or from the corresponding author upon request. Source data are provided with this paper.
Change history
04 June 2025
In this article ref. 55 was incorrect and should have been ‘Perçin, G., et al. Embryonic macrophages orchestrate niche cell homeostasis for the establishment of the definitive hematopoietic stem cell pool. Nat. Commun. 16, 4428 (2025)’.
References
Maximow, A. Untersuchungen über Blut und Bindegewebe: I. Die frühesten Entwicklungsstadien der Blut- und Bindegewebszellen beim Säugetierembryo, bis zum Anfang der Blutbildung in der Leber. Arch. F.ür. Mikrosk. Anat. 73, 444–561 (1908).
Palis, J. & Yoder, M. C. Yolk-sac hematopoiesis. Exp. Hematol. 29, 927–936 (2001).
Palis, J., Robertson, S., Kennedy, M., Wall, C. & Keller, G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126, 5073–5084 (1999).
Hoeffel, G. et al. C-Myb+ erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015).
Gomez Perdiguero, E. et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015).
Yahara, Y. et al. Erythromyeloid progenitors give rise to a population of osteoclasts that contribute to bone homeostasis and repair. Nat. Cell Biol. 22, 49–59 (2020).
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Müller, A. M., Medvinsky, A., Strouboulis, J., Grosveld, F. & Dzierzakt, E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity 1, 291–301 (1994).
Patel, S. H. et al. Lifelong multilineage contribution by embryonic-born blood progenitors. Nature 606, 747–753 (2022).
Coşkun, S. et al. Development of the fetal bone marrow niche and regulation of HSC quiescence and homing ability by emerging osteolineage cells. Cell Rep. 9, 581–590 (2014).
Liu, Y. et al. A specialized bone marrow microenvironment for fetal haematopoiesis. Nat. Commun. 13, 1327 (2022).
Zhang, J. et al. In situ mapping identifies distinct vascular niches for myelopoiesis. Nature 590, 457–462 (2021).
Wu, Q. et al. Resilient anatomy and local plasticity of naive and stress haematopoiesis. Nature 627, 839–846 (2024).
Hall, T. D. et al. Murine fetal bone marrow does not support functional hematopoietic stem and progenitor cells until birth. Nat. Commun. 13, 5403 (2022).
Romeo, S. G. et al. Endothelial proteolytic activity and interaction with non-resorbing osteoclasts mediate bone elongation. Nat. Cell Biol. 21, 430–441 (2019).
Jung, S. et al. Analysis of Fractalkine Receptor CX3 CR1 Function by Targeted Deletion and Green Fluorescent Protein Reporter Gene Insertion. Mol. Cell. Biol. 20, 4106–4114 (2000).
Jacome-Galarza, C. E. et al. Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature 568, 541–545 (2019).
Qian, B.-Z. et al. CCL2 recruits inflammatory monocytes to facilitate breast tumor metastasis. Nature 475, 222–225 (2011).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Gomez Perdiguero, E. et al. Tissue-resident macrophages originate from yolk sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015).
Gu, X., Heinrich, A., Li, S.-Y. & DeFalco, T. Testicular macrophages are recruited during a narrow fetal time window and promote organ-specific developmental functions. Nat. Commun. 14, 1439 (2023).
Medvinsky, A. & Dzierzak, E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86, 897–906 (1996).
Kumaravelu, P. et al. Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Dev. Camb. Engl. 129, 4891–4899 (2002).
North, T. et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Dev. Camb. Engl. 126, 2563–2575 (1999).
Yokomizo, T. et al. Requirement of Runx1/AML1/PEBP2alphaB for the generation of haematopoietic cells from endothelial cells. Genes Cells Devot. Mol. Cell. Mech. 6, 13–23 (2001).
Ganuza, M. et al. Murine foetal liver supports limited detectable expansion of life-long haematopoietic progenitors. Nat. Cell Biol. 24, 1475–1486 (2022).
Ema, H. & Nakauchi, H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 95, 2284–2288 (2000).
Mikkola, H. K. A. & Orkin, S. H. The journey of developing hematopoietic stem cells. Dev. Camb. Engl. 133, 3733–3744 (2006).
Papayannopoulou, T., Craddock, C., Nakamoto, B., Priestley, G. V. & Wolf, N. S. The VLA4/VCAM-1 adhesion pathway defines contrasting mechanisms of lodgement of transplanted murine hemopoietic progenitors between bone marrow and spleen. Proc. Natl Acad. Sci. 92, 9647–9651 (1995).
Frenette, P. S., Subbarao, S., Mazo, I. B., von Andrian, U. H. & Wagner, D. D. Endothelial selectins and vascular cell adhesion molecule-1 promote hematopoietic progenitor homing to bone marrow. Proc. Natl Acad. Sci. Usa. 95, 14423–14428 (1998).
Koni, P. A. et al. Conditional Vascular Cell Adhesion Molecule 1 Deletion in Mice. J. Exp. Med. 193, 741–754 (2001).
Jardine, L. et al. Blood and immune development in human fetal bone marrow and Down syndrome. Nature 598, 327–331 (2021).
Omatsu, Y., Seike, M., Sugiyama, T., Kume, T. & Nagasawa, T. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature 508, 536–540 (2014).
Logan, M. et al. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77–80 (2002).
Sörensen, I., Adams, R. H. & Gossler, A. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood 113, 5680–5688 (2009).
Xie, H. et al. PDGF-BB secreted by preosteoclasts induces angiogenesis during coupling with osteogenesis. Nat. Med. 20, 1270–1278 (2014).
Squarzoni, P. et al. Microglia Modulate Wiring of the Embryonic Forebrain. Cell Rep. 8, 1271–1279 (2014).
Jäppinen, N. et al. Fetal-derived macrophages dominate in adult mammary glands. Nat. Commun. 10, 281 (2019).
Lokka, E. et al. Generation, localization and functions of macrophages during the development of testis. Nat. Commun. 11, 4375 (2020).
Kong, Y.-Y. et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 397, 315–323 (1999).
Li, J. et al. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc. Natl Acad. Sci. 97, 1566–1571 (2000).
Christensen, J. L., Wright, D. E., Wagers, A. J. & Weissman, I. L. Circulation and Chemotaxis of Fetal Hematopoietic Stem Cells. PLoS Biol. 2, e75 (2004).
Khan, J. A. et al. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science 351, 176–180 (2016).
Lu, Y. et al. Spatial transcriptome profiling by MERFISH reveals fetal liver hematopoietic stem cell niche architecture. Cell Discov. 7, 1–17 (2021).
Lee, Y., Leslie, J., Yang, Y. & Ding, L. Hepatic stellate and endothelial cells maintain hematopoietic stem cells in the developing liver. J. Exp. Med. 218, e20200882 (2021).
Lee, Y., DiMaulo-Milk, E., Leslie, J. & Ding, L. Hematopoietic stem cells temporally transition to thrombopoietin dependence in the fetal liver. Sci. Adv. 8, eabm7688 (2022).
Helbling, P. M. et al. Tissue-scale dynamic mapping of Hematopoietic Stem Cells and supportive niche cells in the fetal liver. bioRxiv 2023.09.12.554625 https://doi.org/10.1101/2023.09.12.554625 (2023).
Gao, S. et al. Identification of HSC/MPP expansion units in fetal liver by single-cell spatiotemporal transcriptomics. Cell Res 32, 38–53 (2022).
Kayvanjoo, A. H. et al. Fetal liver macrophages contribute to the hematopoietic stem cell niche by controlling granulopoiesis. eLife 13, e86493 (2024).
Monticelli, S. et al. Early-wave macrophages control late hematopoiesis. Dev. Cell 59, 1284–1301.e8 (2024).
Schwenk, F., Baron, U. & Rajewsky, K. A. cre -transgenic mouse strain for the ubiquitous deletion of loxP -flanked gene segments including deletion in germ cells. Nucleic Acids Res 23, 5080–5081 (1995).
Rios, D. et al. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol. 9, 907–916 (2016).
Sakai, M. et al. Liver-Derived Signals Sequentially Reprogram Myeloid Enhancers to Initiate and Maintain Kupffer Cell Identity. Immunity 51, 655–670.e8 (2019).
Voehringer, D., Liang, H.-E. & Locksley, R. M. Homeostasis and Effector Function of Lymphopenia-Induced “Memory-Like” T Cells in Constitutively T Cell-Depleted Mice. J. Immunol. 180, 4742–4753 (2008).
Perçin, G. et al. Embryonic macrophages orchestrate niche cell homeostasis for the establishment of the definitive hematopoietic stem cell pool. Nat. Commun. 16, 4428 (2025).
Suire, C., Brouard, N., Hirschi, K. & Simmons, P. J. Isolation of the stromal-vascular fraction of mouse bone marrow markedly enhances the yield of clonogenic stromal progenitors. Blood 119, e86–e95 (2012).
Bowers, E. et al. Granulocyte-derived TNFα promotes vascular and hematopoietic regeneration in the bone marrow. Nat. Med. 24, 95–102 (2018).
Kusumbe, A. P., Ramasamy, S. K., Starsichova, A. & Adams, R. H. Sample preparation for high-resolution 3D confocal imaging of mouse skeletal tissue. Nat. Protoc. 10, 1904–1914 (2015).
Coutu, D. L., Kokkaliaris, K. D., Kunz, L. & Schroeder, T. Multicolor quantitative confocal imaging cytometry. Nat. Methods 15, 39–46 (2018).
Sarkans, U. et al. The BioStudies database-one stop shop for all data supporting a life sciences study. Nucleic Acids Res 46, D1266–D1270 (2018).
Acknowledgements
We thank the Bioanalysis Imaging Facility, the Research Flow Cytometry Facility in the Division of Rheumatology, Veterinary Services, and the Integrated Pathology Research Facility at Cincinnati Children’s Medical Center for experimental and technical assistance. Daniel Lucas is supported by R01 HL136529, R01 HL153229, R01 HL158616, R01HL160614 and U54 DK126108 and is a scholar of the Leukemia and Lymphoma Society. Jizhou Zhang was supported by The Edward P. Evans Foundation. Ty D. Troutman was supported in part by NIH P30 DK078392 of the Digestive Diseases Research Core Center in Cincinnati, and by Grant #PNC23-216751 from the American Association for the Study of Liver Diseases Foundation. Tony DeFalco is funded by R35GM119458 and R01HD094698. Data generated using an MA900 cell sorter was funded by NIH grant S10OD023410.
Author information
Authors and Affiliations
Contributions
D.L. conceptualized and managed the study. D.L., B.W., T.D.T., and T.D. designed experiments. B.W performed most experiments and analyses. S.H., M.K., X.G, S.K., A.T., J.Z., Q.W., A.S., J.M.K., assisted with sample and data collection and imaging analyses. D.L. and B.W. assembled the figures and wrote the manuscript, with editorial input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Weinhaus, B., Homan, S., Kincaid, M. et al. Differential regulation of fetal bone marrow and liver hematopoiesis by yolk-sac-derived myeloid cells. Nat Commun 16, 4427 (2025). https://doi.org/10.1038/s41467-025-59058-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59058-w