Abstract
Animal migrations are extensive, ubiquitous, and in decline. To effectively protect migratory species, it is often crucial to identify the interconnected sets of sites they rely upon. Gaps between primary ecological research and synthesised information that is useful to policymakers has limited effective conservation of long-distance migrants, particularly in the marine realm. By synthesising 1304 references to identify 1787 sites and develop model migratory networks for 109 species, we show the minimum extent of marine megafauna connectivity across the global oceans. Our analyses underscore the importance of transboundary cooperation for migratory species conservation at scales larger than current regional structures afford and provide a free online system that will enable policymakers to efficiently summarise how marine migrants use and connect their jurisdictions.
Similar content being viewed by others
Introduction
From rock art depicting ungulate herds on the move, to Aristophanes suggesting one ‘buy some light clothing’ when the swallows return, humans have long observed the seasonal comings and goings of animals. Modern telemetry devices have now recorded humpback whales (Megaptera novaeangliae) swimming from the coast of Brazil to Antarctica1, leatherback turtles (Dermochelys coriacea) travelling 20,000 km from Indonesia to Oregon2, and Arctic Terns (Sterna paradisaea) undertaking the longest known migration: up to 80,000 km each year from pole to pole3. While these animals demonstrate the phenomenon at its most extreme, perhaps an even more remarkable element of migration is its ubiquity. From Monarch butterflies to blue whales, animals of all sizes, life history strategies, and taxonomic groups take part in migrations4. Unfortunately, the routes and resources critical for numerous migrations are under threat, and many populations of migratory species are in decline4,5,6. Indeed, nearly half of all marine migratory species are listed as threatened, near threatened, or data deficient7.
Effective conservation of these migratory species requires us to understand not only the scale of their movements, but also their transboundary nature7,8,9,10. Migratory movements connect life stages that occur in disparate areas of the planet. As animals move across vast areas, they encounter a diverse suite of threats governed by disjointed sets of policy instruments that impede coordinated protection9,11. Failures of existing mechanisms to comprehensively protect migrants are evident both in management outcomes (migratory fish stocks are overfished at twice the rate of those that remain in a single jurisdiction12) and conservation shortfalls (91% of migratory birds are inadequately protected across their life cycle13). The unprecedented rate of declines in migratory populations5,7,14, and global attention on the goals of the Kunming-Montreal Global Biodiversity Framework (GBF), have resulted in urgent calls to understand the role of migratory species in ecological communities—before those communities are degraded beyond recognition. However, we can neither understand a species’ ecological roles nor assess any protections provided to a migratory population if we do not understand their migratory cycles.
Animal movement data repositories such as Movebank15, Birdlife International’s Seabird Tracking Database16, and regional or taxonomic data aggregation projects (e.g. the Retrospective Analysis of Antarctic Tracking Data (RAATD)17, Animal Telemetry Network18, ZoaTrack19, OBIS-SEAMAP20) have grown exponentially over the last two decades and are incredibly useful resources for researchers considering migratory connectivity21. While making tracking data available through such repositories is critical, the jump from raw data to usable knowledge still requires synthesis and communication12,22. There are also questions around the effectiveness of data archiving policies to truly ensure raw datasets are accessible21. To address these challenges, a new type of knowledge repository has been developed to fill the gap between raw data repositories and managers, planners, and policymakers. The development of Important Bird and Biodiversity Areas23, Important Marine Mammal Areas24 and Important Shark and Ray Areas25 all signal a shift to aggregating actionable knowledge that can more easily be directly consumed by management and policy processes. These knowledge repositories are complementary to, and frequently reliant upon, raw data repositories. While these taxonomic knowledge repositories have made steady headway in identifying critical areas for megafauna populations, the connectivity generated by movement between the locations has remained in the shadows.
In this work, we aim to summarise not only the known key use areas for a core set of marine migrants, but also the critical links between them. Here we present a platform hosting a freely accessible, cross-taxa, global dataset describing migratory species connectivity in the ocean (MiCO, accessible at: www.mico.eco), which contains networks for over 100 species, representing fishes, marine mammals, sea turtles and seabirds. We present a preliminary estimate of the previously unquantified (minimum) scale and extent of migratory links across the world’s oceans.
Results
This study builds upon a dataset of migration 'sites' and 'routes' identified in Kot et al.26. Defining ‘migration’ itself is an object of extensive discussion27. Here, we use it to mean movements of at least 100 km between key habitat areas, using the limit from the Global Registry of Marine Species28. This captures both seasonal migratory movements between breeding and non-breeding areas29, and within-season foraging movements for some central-place foragers.
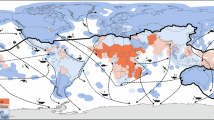
Kot et al.26 reviewed literature on ecological connectivity generated by the movements of 173 marine species across 4 taxonomic groups, and we expanded the dataset following that publication. Of the species reviewed, 109 had relevant geospatial information that described connectivity (i.e. at minimum, two identifiable locations/sites linked by the movement of at least one individual), including 40 seabirds (938 sites), 34 marine fish (1198 sites), 28 marine mammals (21 of these being cetaceans) (1430 sites) and 7 sea turtles (1910 sites). After extracting information from these papers on site locations, routes (i.e. connections between sites; n = 5498), and behaviours (e.g. breeding, wintering, foraging, etc.), we synthesised these into summary metasites (n = 1787) and created interactive connectivity networks for each species (e.g. Fig. 1) which were analysed to assess geographic and taxonomic trends in connectivity. Similar numbers of metasites were generated across taxonomic groups (fishes, 521; marine mammals, 401; sea turtles, 513; seabirds, 352 (Fig. 2)). Note that the 'routes', and therefore metaroutes, were recorded using start and end locations of migratory movements, and do not describe a specific path travelled. We found migratory links exist between almost every region in the world (Fig. 3).
The number of combined sites (#sites) is noted for each circular metasite. Colours indicate known behaviour associated with each metasite (e.g. breeding, feeding). Red arrows indicate known links between locations, with the number adjacent to the arrow indicating the minimum number of known individuals connecting the two metasites. The inset shows the information available when clicking on a particular metasite. The crimson shading indicates the IUCN range map for this species, highlighting range areas which lack connectivity information. View the dynamic versions of this and over 100 other network models at mico.eco/system (including an option to change viewing colours).
Marginal density plots show the latitudinal and longitudinal distribution of metasites across four taxonomic groups: fishes (blue), marine mammals (red), sea turtles (green) and seabirds (orange). Base map is from rnaturalearth69. Source data and code to reproduce this map can be found at 10.5281/zenodo.14873514.
Shades indicate the key regional groups: Africa (red), the Americas (green), Asia (orange), Europe (blue), Oceania/Antarctica (purple), and Areas Beyond National Jurisdiction (grey). Note that because start-end locations of movement were summarised rather than complete telemetry tracks, these diagrams do not include links to regions through which animals transited, and as such represent minimum linkages. Data and code to reproduce chord diagrams can be found at https://zenodo.org/records/14873514 (https://doi.org/10.5281/zenodo.14873514). Species silhouettes from PhyloPic (https://www.phylopic.org), individual image contributors are: Wandering albatross, Alexandre Vong (2023, CC0 1.0); Green turtle, Edwin Price (2023, CC0 1.0); Humpback whale, Guillame Dera (2023, CC0 1.0); Albacore tuna, Felix Vaux (2021, CC0 1.0).
On average, countries were connected to 27.9 ± 20.8 (mean ± sd, median = 22) other EEZs or to ABNJ by migratory marine species. The most highly connected countries were the United States and France, with routes linking them to 115 and 114 other national waters, respectively. This is likely due to their high number of overseas territories and large investments in sampling effort, rather than any ecological driver. Ectotherms (fishes, sea turtles) tended to connect tropical regions to each other, with numerous routes recorded, particularly in the equatorial Americas. On the other hand, endotherms (seabirds and marine mammals) had more routes moving from the tropics to the poles, or even pole to pole (Fig. 3). Across all groups, there were extensive inter- and intra-regional movements, emphasizing the importance of both regional and global governance structures in migratory species conservation and management.
The system includes migratory information collected using over 16,000 animal-borne tags (conservative estimates: 6658 deployed on fishes, 2468 on marine mammals, 3367 on sea turtles and 3521 on seabirds). Most animals measured are of unknown sex for all taxa except sea turtles, for which females are most commonly recorded (Fig. 4A). Sea turtles also, along with seabirds, have more data collected from breeding adults than any other life stage, while fish and marine mammals have mostly migratory (or observation) information from animals of unknown life stages (Fig. 4B).
A Individuals by sex. B Individuals by life stage. Counts include both the number of individuals carrying tags as well as some sites for which the local population size was counted or estimated. For a list of species included in this figure, see Supplementary Table 1. Data and code to reproduce this figure can be found at https://zenodo.org/records/14873514 (https://doi.org/10.5281/zenodo.14873514).
Discussion
The network models described here constitute a freely accessible, cross-taxa, global dataset describing migratory connectivity in the ocean for over 100 species, representing fishes, marine mammals, sea turtles and seabirds. They provide an initial baseline of information on the (minimum) extent of connectivity generated by megafauna across the world’s oceans. While the models are not comprehensive, even for the species reviewed, the dataset already reveals and confirms interesting gaps and biases in data collected on migratory species. In particular, we found latitudinal differences in the distribution of sites among taxonomic groups that reflect both ecological and anthropogenic influences. For example, the clumped distribution of sea turtle research near the equator is driven by the largely tropical distribution of this taxon30. On the other hand, the distribution of seabird sites identified was biased towards polar regions, under-representing the numerous tropical species within this group. This is reflective of seabird research broadly, which has a comparative lack of study sites in the tropics—underrepresenting some of the most species-rich and human-impacted areas of the globe31. Lower levels of breeding synchrony in tropical seabirds, and thus reduced efficiencies in tag deployment and collection compared to polar species, are likely at least partly responsible for the reduced tagging in the tropics32. Given likely declines in close to half of the seabird populations of the Great Barrier Reef 33, this underrepresentation of tropical seabirds may mean we are losing connections between regions before they are even identified.
The extensive migratory information revealed within the MiCO system only scratches the surface of the true connectivity of the global oceans. However, it is important to note that the system does not claim to show all oceanic migratory connectivity: rather, it shows us the minimum connectivity that has been measured. In this way, gaps within the system can be informative. Because the species included in MiCO are all known migrants12, absences of migratory links shown in the system do not indicate that no migration occurs, but rather may highlight opportunities to address data gaps. The system includes the functionality to visualise migratory links overlaid upon the best available range maps for each species, as a coarse indicator of these potential gaps (Fig. 1). As an example, there is little information available for the Spiny dogfish Squalus acanthias, which is known to migrate—the lack of data indicates further research, or better access to and aggregation of, existing data is critical.
The existence of biases in biological datasets is well-known26,34, and in our system, these are observed in both geography and taxonomy. Specifically, geographic biases mimicking the wealth and distribution of university researchers were evident (Fig. 2). Data on migratory connectivity have been collected from locations (mostly the USA, UK, and Europe) that broadly reflect the distribution of ecological research published worldwide35. Further, there are sex and age class biases within taxonomic groups themselves. This is almost entirely due to logistical constraints around capturing and tracking migratory marine species: for example, satellite tags are almost exclusively deployed on nesting female turtles (Fig. 4A), while breeding seabirds are often easy to access but difficult to visually determine sex in the field, leading to high numbers of adult breeders of unknown sex in our database (Fig. 4B). To capture complete migratory connectivity information within species, demographic gaps must be identified and accounted for (either via targeted data collection, or appropriate modelling of population-level uncertainty).
Much discussion has been had on the importance of collating and sharing animal tracking data, and possible mechanisms for doing so20,36,37,38,39. As evidence mounts that the impacts of deploying biologging tags can be non-trivial40,41, justifying the costs to animals is increasingly important. Indeed, national standards (e.g. the Australian code for the care and use of animals for scientific purposes42) explicitly call for balancing the potential effects to animal wellbeing against the benefits of intrusive data collection. Examples such as the North Atlantic Current and Evlanov Sea basin (NACES) MPA, in which 2188 seabird tracks contributed to Birdlife International’s Seabird Tracking Database were used to identify a critical foraging area in the Atlantic Ocean, show the potential policy outcomes that can be achieved for highly mobile species when researchers commit to sharing data43,44. However, the NACES MPA is the exception not the rule, and a lack of accessible information on marine migratory connectivity continues to hamper the development of environmental assessments and conservation planning that meaningfully address the needs of migrants.
The need for easily accessible geographic information on migrations has been made explicit in the mandate given to the Convention on Migratory Species (CMS) to develop a Global Atlas of Animal Migration. MiCO supports delivery of the Atlas and illuminates and summarises the fundamental problem that led to the creation of CMS: no country can manage its migratory species alone. Beyond informing CMS, the networks identified through this literature review provide diverse opportunities to integrate migratory connectivity into policy. The use of these models to inform environmental impact assessments may radically alter their geographic scope, as cumulative impacts across many species’ migratory cycles can now be feasibly included. Further, these models provide an ecological analogue to economics and trade models that underpin many current studies on telecoupled effects between human and natural systems, e.g. refs. 45,46. Additionally, the role of migratory species as vectors of disease transmission, including zoonotic diseases, has recently been highlighted47. However, our understanding of transmission pathways has been drastically limited by the lack of access to synthesised information on how migratory species connect the world. Delivery of these models of migratory connectivity through the MiCO system could enable epidemiological assessments of the role of migratory species contemporaneously with outbreaks of e.g. highly-pathogenic avian influenza, which is currently decimating seabird colonies and other wildlife48.
Finally, and perhaps most importantly, networks identified through this literature review can support ocean governance structures to better consider migratory connectivity. Currently, governance of fisheries (a primary source of mortality for turtles49, sharks50, seabirds51, and marine mammals52) in areas beyond national jurisdiction (ABNJ, i.e. >200 nautical miles from shore) occurs on a regional basis, with insufficient cooperation among regions to address, e.g. pole-to-pole migrations. Our networks can support interested parties to better understand with which Regional Fisheries Management Organisations (RFMOs) species from their jurisdiction are interacting, thus opening the door for engagement with these bodies from non-fishing nations. Similarly, Regional Seas Organizations (fora where countries cooperate to tackle shared environmental issues) are largely limited to national jurisdictions and operate on one side or another of an ocean basin. Information contained within our system can support these groups to identify target species and regions for collaborative management of shared biodiversity.
Systematic conservation planning and environmental assessments are common and critical tools used in conservation but have rarely been designed to address migratory connectivity (c.f 53.), instead more commonly focusing on terrestrial or larval connectivity54,55. Impressive global movement syntheses have been published for multiple key taxa (e.g. Pacific predators9, albatrosses56, fishes57, and sea turtles58) that demonstrate the complex and extensive connectivity of marine migrants, however, these analyses do not provide easily accessible, actionable knowledge for policymakers engaged at the national or species level. A lack of accessible information has led conservation planners to use broad habitat models for migratory species that cover most of the globe, with no record of the relative importance of different areas. Protection of a proportion (e.g. refs. 59,60) of a species' range is unlikely to be effective for migrants, as this is unlikely to capture key habitats across a species’ migratory cycle and life history stages, nor ensure persistence of safe routes between these habitats to maintain population connectivity. The freely-accessible, cross-taxa connectivity models presented in this study provide a critical resource that can enable inclusion of the functional connectivity generated by marine migrations in conservation planning, and thus the delivery of a system of area-based management tools that are well-connected with respect to migratory species.
We are at a critical moment to scale up efforts to conserve biodiversity, as the world comes together to meet the new goals and targets set through the GBF. Recognition of the importance of connectivity to the conservation of biodiversity has resulted in the inclusion of connectivity directly (or indirectly) in a quarter of the goals and more than a fifth of the targets under this framework61. Despite this significant focus on connectivity, the connectivity generated by marine megafauna remains unassessed and unaddressed under the GBF, as there are no suggested indicators for either marine connectivity or migratory connectivity62. The GBF is likely to drive a huge expansion of area-based management as the world aims for 30% of the globe to be 'effectively conserved and managed through ecologically representative, well-connected and equitably governed systems of protected areas and other effective area-based conservation measures [OECMs].' Unless we can generate and incorporate information on migratory connectivity into the developing network of MPAs and OECMs, it is entirely feasible that we will protect 30% of the planet (with concomitant costs to industry) without providing effective protection to the species that society values (and legislates to protect) the most. If society seeks to preserve the incredible ebb and flow of animal migrations for future generations, then we must underpin national and international governance structures with an understanding of how migratory populations connect us and the ecosystems upon which we rely.
Methods
Data
The MiCO system (https://mico.eco/system) is an online tool that collects, displays, and summarises information about marine migrations to support policymakers. It was developed to describe regional to global-scale connectivity generated by migratory marine megavertebrates, with an initial focus on species utilising areas beyond national jurisdiction. Tutorials on the capabilities of the online system are available at mico.eco/system-help.
The list of species to be evaluated in this study was selected from the initial high-seas species considered by MiCO12 and focused on those listed in CMS appendices or managed by an RFMO, excluding sturgeon, sea ducks and taxonomic families with limited representation. While our approach maximises the utility of the system for specific policy arenas, we acknowledge that species listed under CMS or managed by RFMOs represent a fraction of all migratory marine species. As such, and despite the breadth of work undertaken here, connectivity for more than two-thirds of migratory marine megafauna remains unassessed.
For this study, a total of 5476 sites were identified from the literature across all taxonomic groups: 1910 sea turtle sites, 1430 marine mammal sites, 1198 fish sites, and 938 bird sites. Sites were identified based on the authors’ description of either processed data (26.7% of studies) or tracks (73.3% of studies), and were usually associated with a specific behaviour. If no behaviour was noted in the study, the site was listed as an 'observation'. Reviewers collecting data from the literature identified the centroid of the activity described. While identification of the exact centroid for a given activity was an inherently subjective process, training of reviewers to ensure standardisation, as well as quality control of datasheets by the review manager, increased reproducibility and confidence in the results. A buffer was applied to each centroid to describe the area used. To focus the system on provision of regional to global scale information on connectivity, we used 1° as the smallest buffer for sites, and applied a 5° or 10° buffer to larger sites to allow for differentiation among the size of areas identified in different studies and to increase the efficiency of reviewers assessing the literature. Using such a coarse resolution for sites also decreases the sensitivity of the sites to potential errors in deciding on the exact location of the centroid. These sites made up the base dataset used in this study. The data collected for those sites were subjected to quality control processes to ensure all identified literature was included, reviewer responses were standardised, and georeferencing of sites conformed with site names and species distributions (Supplementary Fig. 1).
Site aggregation
To address duplication of sites identified in different studies of the same species, we developed a site aggregation process (Supplementary Fig. 2). Aggregation of sites into 'metasites' had three objectives: 1) aggregate information across multiple sites that identify a species’ activities in the same location; 2) maintain information on large-scale (>500 km) connections generated by animal movements; 3) maintain the resolution of information on critical reproductive life history stages; 4) provide a minimum level of known animals sampled within a site and moving between sites. To address these objectives, we developed a five-step process for site aggregation that was run on the sites and routes for each species in the MiCO System.
Data prep
The site aggregation process iteratively selected a site and identified sites that should be aggregated with it based on a series of rules. Because the process iteratively selected sites to examine, the groupings of sites that were aggregated together were dependent on the order of the sites in the dataset. We use this inherent prioritisation to maintain information at a high resolution for reproductive sites. All reproductive sites were examined first and were ordered from smallest (1°) to largest (10°). All remaining sites were evaluated after the reproductive sites based on size, with the largest sites (10°) considered first. This approach for non-reproductive sites was chosen to limit overlap between non-reproductive sites and generate the most parsimonious network.
Site section
We iteratively selected the next site (the 'Lookup Site') within the sorted set of sites to evaluate which sites should be aggregated with it. All sites in the sorted site dataset were evaluated to determine if their centroid fell within the radius (buffer) of the lookup site. Those sites that met this criterion ('Nearby Sites') were grouped for further consideration (Supplementary Fig. 3).
The sites identified in Kot et al.26 have very coarse resolutions (1–10° radii). When aggregating sites together with 10° sites, it is possible that sites that were >1000 km away could be selected for inclusion. To ensure that large-scale connectivity between such sites was not lost in the aggregation process, routes and connections were projected to a metre-level Azimuthal Equidistant projection (centred at 0° N, 0° E) and any routes or connections >500 km long were not included in the aggregation process for a given Lookup Site. That is to say, any Nearby Site that included a route or connection to a Lookup Site >500 km away from it was removed from consideration for grouping ('Removed Site') with the Lookup Site. To account for the removal of the site, any sites that were: a) greater than 1° from the Lookup Site centroid, and b) within the buffer of the Removed Site, and c) closer to the Removed Site centroid than the Lookup Site centroid were also removed so that they could later be aggregated with the Removed Site (see Supplementary Figs. 2 and 3 for the decision flowchart).
Metasite centroid definition
To develop a metasite from the Lookup Site and any Nearby Sites that were not removed during site selection, an iterative subprocess was implemented. In this subprocess, the new metasite centroid was calculated as the average latitude and longitude from those remaining sites, and the buffer from the Lookup Site was used as the metasite buffer. Any sites with centroids that did not intersect the metasite buffer were removed, and a new metasite centroid was calculated repeatedly until all sites intersected the metasite buffer. Any sites included in the new metasite were removed from consideration for aggregation into subsequent metasites.
Data duplication check
Information on the sampling technology, temporal scope of site use, and species behaviour within the site was collated from individual sites included within the new metasite. Information on number of individuals, sex, and life history stage was summed. Given that many tracking studies either reuse or build on existing datasets, we implemented a three-step process to identify potentially duplicate data and ensure that our sample size estimates were a minimum estimate of known connectivity in a site, or travelling between sites (for routes and connections).
The first step was to identify duplicate sites. All references used in the Kot et al.26 literature review included a unique reference ID (ZoteroID). If sites had the same ZoteroID and identical latitudes and longitudes, they were considered duplicate sites. In this situation, only the site with the greatest number of individuals was included when summing any count of individuals.
Sites that had a route between them and were aggregated together in a metasite were considered the same site, so as not to double count animals moving within a metasite. Again, only the site with the greatest number of individuals was included when summing any count of individuals.
Finally, if sites included in the same metasite met both of the following conditions, they were assumed to have used the same data and only one site was included:
-
a.
The sites came from references with at least one author with the same surname, and
-
b.
The temporal scope (i.e. months and years) identified for one site was a subset of the other site
As above, if these conditions were met, only the site with the greatest number of individuals was included when summing any count of individuals.
Metasite aggregation
Following the data duplication check, the new metasite was added to a database of metasites for the species under consideration. Site identification codes (IDs) used to define connections were then replaced with the corresponding metasite IDs, and all connections that joined the same metasites were aggregated into metaconnections. If metaconnections subsequently connected a metasite to itself, they were removed. If site information was not included when summing numbers of individuals in a metasite (due to data duplication), the individuals on associated routes/connections were not included when calculating the same statistics. Final model outputs (points and polyline feature layers within file geodatabases) were then generated from the metasite information and made available via the MiCO System. All products in the MiCO System are freely distributed under the CC-BY copyright.
Network models
Network models were generated in R using the igraph63 package version 2.1.1, using metasites as nodes and the metaroutes as edges. The radius of a given metasite was used as the radius for the node, and the weight of the edges was based on the number of individuals associated with the metaroute. Nodes were coloured based on the activities occurring in the metasite.
Additional methods
Two summary exports were made from the file geodatabases: one, a subset of all metaroutes which had source and destination metasites in different EEZs; and two, a list of all metasites and their associated metadata. All subsequent analyses were undertaken in R version 4.3.064, using the packages tidyverse65 version 2.0.0, circlize66 version 0.4.16, jsonlite67 version 1.8.8, and ggExtra68 version 0.10.1. Metasites were allocated to a region of the UN geoscheme based on the latitude and longitude of the metasite centroid. These were geographic allocations rather than political, for example, the British Virgin Islands was allocated to the Caribbean rather than to Northern Europe. Twenty-three individual breeding stages (specific to species groups) were summarised into four key life stage categories: ‘Breeding Adult’, ‘Non-breeding Adult’, ‘Juvenile’, and ‘Unknown’. Adjacency matrices were constructed from the data on metaroutes that crossed EEZs, and were used to generate chord diagrams.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available as follows. Network models generated in this study are available on the open-source system, http://MiCO.eco, and file geodatabases for all underlying species data can be downloaded from this site under a CC-BY Open License. Raw literature review datasheets are available at UQ eSpace data repository: https://doi.org/10.48610/aa77f3c. Summary datasets, including all data and code required to reproduce figures, are available at github.com/lkbentley/mico, https://doi.org/10.5281/zenodo.14873514.
Code availability
Code to reproduce analyses and figures is publicly available at github.com/lkbentley/mico, https://doi.org/10.5281/zenodo.14873514. Code utilised in the back end of the mico.eco system is available to replicate analyses at https://github.com/danielcdunn/mico/tree/main/OutsideMiCOAggregation, https://doi.org/10.48610/aa77f3c.
References
Zerbini, A. et al. Satellite-monitored movements of humpback whales Megaptera novaeangliae in the Southwest Atlantic Ocean. Mar. Ecol. Prog. Ser. 313, 295–304 (2006).
Benson, S. R. et al. Post-nesting migrations of Leatherback Turtles (Dermochelys coriacea) from Jamursba-Medi, Bird’s Head Peninsula, Indonesia. Chelonian Conserv. Biol. 6, 150–154 (2007).
Egevang, C. et al. Tracking of Arctic terns Sterna paradisaea reveals longest animal migration. Proc. Natl Acad. Sci. USA 107, 2078–2081 (2010).
Bowlin, M. S. et al. Grand challenges in migration biology. Integr. Comp. Biol. 50, 261–279 (2010).
Wilcove, D. S. & Wikelski, M. Going, going, gone: is animal migration disappearing. PLOS Biol. 6, e188 (2008).
Hazen, E. L. et al. Predicted habitat shifts of Pacific top predators in a changing climate. Nat. Clim. Change 3, 234–238 (2013).
Lascelles, B. et al. Migratory marine species: their status, threats and conservation management needs. Aquat. Conserv. Mar. Freshw. Ecosyst. 24, 111–127 (2014).
Marra, P. P., Hunter, D. & Perrault, A. M. Migratory connectivity and the conservation of migratory animals. Environ. Learn. 41, 317 (2011).
Harrison, A.-L. et al. The political biogeography of migratory marine predators. Nat. Ecol. Evol. 2, 1571–1578 (2018).
Davis, K. L. et al. Breeding season management is unlikely to improve population viability of a data-deficient migratory species in decline. Biol. Conserv. 283, 110104 (2023).
Ban, N. C. et al. Systematic conservation planning: a better recipe for managing the high seas for biodiversity conservation and sustainable use. Conserv. Lett. 7, 41–54 (2014).
Dunn, D. C. et al. The importance of migratory connectivity for global ocean policy. Proc. R. Soc. B. 286, 20191472 (2019).
Runge, C. A. et al. Protected areas and global conservation of migratory birds. Science 350, 1255–1258 (2015).
Both, C., Bouwhuis, S., Lessells, C. M. & Visser, M. E. Climate change and population declines in a long-distance migratory bird. Nature 441, 81–83 (2006).
Kays, R. et al. The Movebank system for studying global animal movement and demography. Methods Ecol. Evolut. 13, 419–431 (2022).
Carneiro, A. P. B. et al. The BirdLife Seabird Tracking Database: 20 years of collaboration for marine conservation. Biol. Conserv. 299, 110813 (2024).
Ropert-Coudert, Y. et al. The retrospective analysis of Antarctic tracking data project. Sci. Data 7, 94 (2020).
Block, B. A. et al. Toward a national animal telemetry network for aquatic observations in the United States. Anim. Biotelemetry 4, 6 (2016).
Dwyer, R. G. et al. An open Web-based system for the analysis and sharing of animal tracking data. Anim. Biotelemetry 3, 1 (2015).
Halpin, P. N. et al. OBIS-SEAMAP: The World Data Center for marine mammal, sea bird, and sea turtle distributions. Oceanography 22, 104–115 (2009).
Roche, D. G., Kruuk, L. E. B., Lanfear, R. & Binning, S. A. Public data archiving in ecology and evolution: How well are we doing? PLOS Biol. 13, e1002295 (2015).
Holmes, J. & Clark, R. Enhancing the use of science in environmental policy-making and regulation. Environ. Sci. Policy 11, 702–711 (2008).
Donald, P. F. et al. Important Bird and Biodiversity Areas (IBAs): the development and characteristics of a global inventory of key sites for biodiversity. Bird. Conserv. Int. 29, 177–198 (2019).
di Sciara, G. N. et al. Place-based approaches to marine mammal conservation. Aquat. Conserv.: Mar. Freshw. Ecosyst. 26, 85–100 (2016).
Hyde, C. A. et al. Putting sharks on the map: a global standard for improving shark area-based conservation. Front. Mar. Sci. 9 (2022).
Kot, C. Y. et al. Synthesizing connectivity information from migratory marine species for area-based management. Biol. Conserv. 283, 110142 (2023).
Dingle, H. & Drake, V. A. What is migration? BioScience 57, 113–121 (2007).
Riede, K. Conservation and modern information technologies: the global register of migratory species (GROMS). J. Int. Wildl. Law Policy 3, 152–165 (2000).
Webster, M. S., Marra, P. P., Haig, S. M., Bensch, S. & Holmes, R. T. Links between worlds: unraveling migratory connectivity. Trends Ecol. Evolut. 17, 76–83 (2002).
Davenport, J. Temperature and the life-history strategies of sea turtles. J. Therm. Biol. 22, 479–488 (1997).
Mott, R. & Clarke, R. H. Systematic review of geographic biases in the collection of at-sea distribution data for seabirds. Emu Austral Ornithol. 118, 235–246 (2018).
Bernard, A., Rodrigues, A. S. L., Cazalis, V. & Grémillet, D. Toward a global strategy for seabird tracking. Conserv. Lett. 14, e12804 (2021).
Woodworth, B. K. et al. Trends in seabird breeding populations across the Great Barrier Reef. Conserv. Biol. 35, 846–858 (2021).
Di Marco, M. et al. Changing trends and persisting biases in three decades of conservation science. Glob. Ecol. Conserv. 10, 32–42 (2017).
Martin, L. J., Blossey, B. & Ellis, E. Mapping where ecologists work: biases in the global distribution of terrestrial ecological observations. Front. Ecol. Environ. 10, 195–201 (2012).
Kranstauber, B. et al. The Movebank data model for animal tracking. Environ. Model. Softw. 26, 834–835 (2011).
Hays, G. C. et al. Translating marine animal tracking data into conservation policy and management. Trends Ecol. Evolut. 34, 459–473 (2019).
Newman, P., Dwyer, R. G., Belbin, L. & Campbell, H. A. ZoaTrack—an online tool to analyse and share animal location data: user engagement and future perspectives. Aust. Zool. 41, 12–18 (2020).
Rutz, C. Register animal-tracking tags to boost conservation. Nature 609, 221–221 (2022).
Wilson, R. P. & McMahon, C. R. Measuring devices on wild animals: what constitutes acceptable practice? Front. Ecol. Environ. 4, 147–154 (2006).
Bodey, T. W. et al. A phylogenetically controlled meta-analysis of biologging device effects on birds: Deleterious effects and a call for more standardized reporting of study data. Methods Ecol. Evol. 9, 946–955 (2018).
NHMRC. Australian Code for the Care and Use of Animals for Scientific Purposes, 8th Edition. (2013).
Davies, T. E. et al. Multispecies tracking reveals a major seabird hotspot in the North Atlantic. Conserv. Lett. 14, e12824 (2021).
Davies, T. E. et al. Tracking data and the conservation of the high seas: opportunities and challenges. J. Appl. Ecol. 58, 2703–2710 (2021).
Johnson, J. A. et al. Investing in nature can improve equity and economic returns. Proc. Natl Acad. Sci. USA 120, e2220401120 (2023).
Hulina, J. et al. Telecoupling framework for research on migratory species in the Anthropocene. Elementa: Sci. Anthropocene 5, 5 (2017).
Altizer, S., Bartel, R. & Han, B. A. Animal migration and infectious disease risk. Science 331, 296–302 (2011).
Rijks, J. M. et al. Mass mortality caused by highly pathogenic influenza A(H5N1) virus in sandwich terns, the Netherlands, 2022. Emerg. Infect. Dis. 28, 2538–2542 (2022).
Putman, N. F., Hawkins, J. & Gallaway, B. J. Managing fisheries in a world with more sea turtles. Proc. R. Soc. B 287, 20200220 (2020).
Dulvy, N. K. et al. You can swim but you can’t hide: the global status and conservation of oceanic pelagic sharks and rays. Aquat. Conserv.: Mar. Freshw. Ecosyst. 18, 459–482 (2008).
Dias, M. P. et al. Threats to seabirds: a global assessment. Biol. Conserv. 237, 525–537 (2019).
Read, A. J., Drinker, P. & Northridge, S. Bycatch of marine mammals in US and global fisheries. Conserv. Biol. 20, 163–169 (2006).
Reynolds, M. D. et al. Dynamic conservation for migratory species. Sci. Adv. 3, e1700707 (2017).
Correa Ayram, C. A., Mendoza, M. E., Etter, A. & Salicrup, D. R. P. Habitat connectivity in biodiversity conservation: a review of recent studies and applications. Prog. Phys. Geogr.: Earth Environ. 40, 7–37 (2016).
Fontoura, L. et al. Protecting connectivity promotes successful biodiversity and fisheries conservation. Science 375, 336–340 (2022).
Beal, M. et al. Global political responsibility for the conservation of albatrosses and large petrels. Sci. Adv. 7, eabd7225 (2021).
Palacios-Abrantes, J., Reygondeau, G., Wabnitz, C. C. C. & Cheung, W. W. L. The transboundary nature of the world’s exploited marine species. Sci. Rep. 10, 17668 (2020).
Kot, C. Y. et al. Network analysis of sea turtle movements and connectivity: a tool for conservation prioritization. Divers. Distrib. 28, 810–829 (2022).
Jones, K. R. et al. Area requirements to safeguard Earth’s marine species. One Earth 2, 188–196 (2020).
Brito-Morales, I. et al. Towards climate-smart, three-dimensional protected areas for biodiversity conservation in the high seas. Nat. Clim. Chang. 12, 402–407 (2022).
Xu, H. et al. Ensuring effective implementation of the post-2020 global biodiversity targets. Nat. Ecol. Evol. 5, 411–418 (2021).
UNEP-WCMC. Indicators for the Kunming—Montreal Global Biodiversity Framework. https://www.post-2020indicators.org/.
Csárdi, G. et al. Igraph: network analysis and visualization in R.(https://doi.org/10.5281/zenodo.7682609). See https://CRAN.R-project.org/package=igraph (2024).
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing (2020).
Wickham, H. et al. Welcome to the tidyverse. J. Open Source Softw. 4, 1686 (2019).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. Circlize implements and enhances circular visualization in R. Bioinforma. 30, 2811–2812 (2014).
Ooms, J. The jsonlite Package: A Practical and Consistent Mapping Between JSON Data and R Objects. arXiv:1403.2805 [stat.CO], (2014).
Attali, D. & Baker, C. GgExtra: Add Marginal Histograms to ‘Ggplot2’, and More ‘ggplot2’ Enhancements. (2023).
South, A., Michael, S. & Massicotte, P. rnaturalearthdata: World Vector Map Data from Natural Earth Used in ‘rnaturalearth’. (2025).
Acknowledgements
The authors thank the Migratory Connectivity in the Ocean (MiCO) project partners, collaborators, and advisory board members for sharing their expertise and support. We would also like to thank Amalia Alberini, Hannah Blondin, Andre Boustany, Maggie Chory, Jesse Cleary, Claire Atkins-Davis, Alejandro Herrero Palacio, Elizabeth Mason, Ella Watkins, Meredith Whitten, and Colyer Woolston for their valuable contributions. Primary support was from the Global Ocean Biodiversity Initiative (GOBI) grant from the International Climate Initiative (IKI) (16_IV_049_Global_A_Global Ocean Biodiversity Initiative GOBI: DCD & PNH). The German Federal Ministry for the Environment, Nature Conservation, Nuclear Safety and Consumer Protection (BMUV) supports this initiative on the basis of a decision adopted by the German Bundestag. Additional funding provided under an Australian Research Council Discovery Project (DP240101026: DCD, PNH & ALH).
Author information
Authors and Affiliations
Contributions
Conceptualization: D.C.D., C.C., A.L.H., P.N.H., C.Y.K. Methodology: D.C.D., C.C., C.Y.K., A.L.H., S.P., S.E.D., G.O.C. Software: E.F., B.D. Investigation: D.C.D., S.E.D., E.I.H., C.Y.K., D.N., G.O.C., S.P., L.K.B. Visualization: L.K.B. Funding acquisition: D.C.D., P.N.H. Project administration: C.C., C.Y.K., D.C.D. Supervision: D.C.D., P.N.H. Writing—original draft: L.K.B., D.C.D. Writing—review & editing: L.K.B., D.N., E.F., C.C., S.E.D., B.D., E.I.H., A.L.H., C.Y.K., S.P., P.N.H., D.C.D.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bentley, L.K., Nisthar, D., Fujioka, E. et al. Marine megavertebrate migrations connect the global ocean. Nat Commun 16, 4089 (2025). https://doi.org/10.1038/s41467-025-59271-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59271-7