Abstract
Steroidal alkaloids play a crucial role in plant defense and exhibit distinct stereochemistry at C25, forming either the tomato-type (25S) or eggplant-type (25R) isomers. Here, we uncover the molecular mechanisms shaping this stereochemical diversity. Phylogenetic analysis of GLYCOALKALOID METABOLISM 8 (GAME8) cytochrome P450 hydroxylases across the Solanaceae family revealed two distinct clades producing either 25S or 25R isomers. Ancestral GAME8 likely favored 25R, with gene duplications giving rise to 25S-producing enzymes in more recent Solanum species. In S. nigrum and S. dulcamara, multiple GAME8 copies generate mixed isomeric profiles. Notably, in wild S. cheesmaniae from the Galápagos, mutations in GAME8 have driven a shift from 25S back to the ancestral 25R, suggesting reverse evolution. Our findings highlight how GAME8 evolution has shaped alkaloid diversity in the genus Solanum, demonstrating a complex interplay between enzyme function, genetic variation, and evolutionary adaptation.
Similar content being viewed by others
Introduction
The Solanaceae family, commonly known as the nightshade family, is a diverse and evolutionarily significant group of flowering plants that includes over 2700 species distributed across ~100 genera1,2. This family exhibits remarkable ecological and morphological diversity, encompassing major crop species such as tomato (Solanum lycopersicum), potato (Solanum tuberosum), eggplant (Solanum melongena), peppers (Capsicum spp.), and tobacco (Nicotiana spp.), as well as numerous wild relatives. Phylogenetic studies suggest that Solanaceae originated in the early Eocene, with subsequent rapid diversification driven by major geological and climatic shifts3,4. Within the family, the Solanum genus represents one of the most species-rich lineages, undergoing extensive adaptive radiation across different habitats, particularly in South America5. Evolutionary innovations in specialized metabolism, including SGAs, played a key role in plant defense mechanisms, contributing to the ecological success of Solanum species2. Understanding the evolutionary trajectories of metabolic pathways within Solanaceae provides valuable insights into how biosynthetic diversity and stereochemical specialization emerged within this family.
The significance of chirality in natural products cannot be overstated, as it plays a critical role in determining the plethora of biological activities and interactions of these molecules6,7,8,9. Chirality often controls the efficacy and safety of pharmaceuticals, as well as the functionality of agrochemicals. For instance, the two enantiomers of a drug might have vastly different effects in the human body, with one being therapeutic and the other potentially harmful4,10,11. The significance of stereochemical diversity extends to steroidal glycoalkaloids (SGAs); secondary metabolites found predominantly in the Solanum genus, including the crop plants tomato, potato, and eggplant12. SGAs exhibit a wide range of activities, including antifungal, antibacterial, and anti-nutritional effects13,14,15. They are composed of sugar moieties linked to a steroidal aglycone with multiple chiral centers. Only one of these stereocenters exists in either the 25S or 25R forms, with diverse Solanum species favoring different configurations (Supplementary Fig. 1). For example, tomato (Solanum lycopersicum) and potato (Solanum tuberosum) predominantly produce SGAs with 25S configuration, while eggplant (Solanum melongena) makes the 25R isomer15.
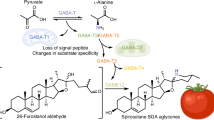
The biosynthesis of SGAs follows a highly structured metabolic pathway, beginning with the modification of cholesterol as the core scaffold for steroidal aglycones. This pathway involves multiple classes of enzymes, including Cellulose Synthase Like M, cytochromes P450, glycosyltransferases and dioxygenases12,16,17,18,19,20,21,22,23,24. The cytochrome P450 enzyme GAME8 (GLYCOALKALOID METABOLISM 8, CYP72A208) has been reported to hydroxylate the terminal methyl group in the side chain of cholesterol, leading to the formation of a chiral center at C2512. However, the enzymes responsible for the production of the two different enantiomers at C25 remained elusive. Additionally, the evolutionary mechanisms driving stereochemical diversity among Solanum species remain poorly understood, underscoring GAME8 as a key subject for further research. The glycosylation of SGAs scaffolds, catalyzed by UDP-glycosyltransferases such as GAME1, GAME17, GAME18, and GAME2, is a crucial downstream step in SGA biosynthesis, as it significantly impacts the compounds’ solubility, stability, and bioactivity12. These glycosylation reactions occur following the formation of the C25 chiral center, meaning that the stereochemistry of the scaffold (i.e., the aglycone) directly influences the structure and function of the final glycoalkaloid products. The presence of 25S or 25R stereochemistry in aglycones, therefore, could dictate the biological activity of SGAs, potentially shaping their ecological roles in plant defense
Here, we sought to elucidate the molecular mechanisms and evolutionary pathways responsible for the stereochemical diversity of SGAs within the Solanum genus. We investigated the C25 chirality in steroidal aglycones across various Solanum species and further explored the stereospecificity and potential evolutionary divergence of the GAME8 enzymes. Our findings highlight the crucial role of GAME8 enzymes in generating distinct SGAs stereochemistry. The evolution of chirality in SGAs biosynthesis has occurred through diverse mechanisms, including gene duplication, neofunctionalization, and reverse evolution, the latter being observed in a specific wild tomato species in the Galápagos Islands. This study reveals how enzymatic and evolutionary mechanisms drive the production of isomeric metabolite variants, tailored to function in particular ecological niches.
Results
Stereodiversity of steroidal alkaloid aglycones in the genus Solanum
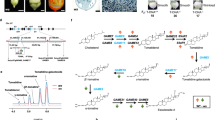
Previous studies showed that certain Solanum species produce steroidal alkaloid aglycones with 25S configuration (e.g., tomato and potato) while others produce 25R isomers (e.g., eggplant)15,25,26 (ig. 1a). We sought to explore this structural variation at the C25 position and conducted a comprehensive analysis of steroidal aglycone composition in diverse species from the genus Solanum covering the M (Morelloid and Dulcamaroid), Petota, Tomato, Old World and Spiny Solanum clades (Fig. 1a, c and Supplementary Figs. 2, 3). Thus, we extracted SGAs from these plants employed acid hydrolysis to remove the sugar moieties and isolate the aglycones. Subsequently, we used standards such as tomatidenol (25S), solasodine (25R), and their reduced counterparts tomatidine (25S) and soladulcidine (25R) to compare and identify the aglycones present in the samples (Fig. 1b, and Supplementary Fig. 4). Our findings revealed distinct patterns in the chirality of the aglycones among the different Solanum species. In tomato leaves, we predominantly observed the 25S isomer, with only a small proportion of the 25R isomer (up to 10%). In contrast, eggplant seed samples exclusively contained the 25 R isomer (Fig. 1c, d and Supplementary Fig. 2j). We also observed that spiny Solanum (including Old World species) produce only 25R isomers while species from Petota and the Tomato clades produce predominantly 25S compounds as shown previously in literature27. This variation in chirality at C25 suggests species-specific enzyme activities and potentially different biological activities or functions of these aglycones.
a Phylogenetic tree of genus Solanum with chirality of C25 stereocenter marked in purple, red, and purple-red stripes denoting species accumulating 25R, 25S, or both stereoisomers. b Chemical structures of aglycones most frequently found in the genus Solanum—configurations at C25 are marked either with purple (25R) or red (25S) dots. c Total Ion Chromatograms (TICs) of tomato leaf and eggplant seed extracts. Most abundant SGAs and saponins are annotated. d Extracted Ion Chromatograms (EICs) for α-tomatine (EIC 1034.5530), dehydrotomatine (EIC 1032.5537), solamargine (EIC 868.5053), and solasonine (EIC 884.5002). Metabolites with different chirality at C25 are appropriately annotated either 25R or 25S. Images of tomato, potato, and eggplant are from BioRender.
GAME8 enzymes dictate steroidal glycoalkaloid chirality of SGAs in Solanum species
According to the current knowledge of SGA biosynthesis, we hypothesized that the different configurations of C25 (R/S) observed in this metabolite class from various Solanum species are most likely due to the activity of GAME8, a cytochrome P450 responsible for hydroxylating the terminal methyl group(s) in the side chain of the cholesterol molecule (Fig. 2b). Although both methyl groups (C26 and C27) appear identical, this is not the case. Hydroxylation of either carbon creates a chiral center, and depending on which methyl group is modified, either the 25S or 25R isomer will form. These groups are annotated as pro-S or pro-R due to their potential to generate the respective chirality. To test this hypothesis, we cloned GAME8 genes from tomato (SlGAME8) and eggplant (SmGAME8) and co-expressed them with other SGAs pathway genes (namely, SlGAME15–Cellulose synthase-like M, SlGAME6–CYP72A188, SlGAME11–2-Oxoglutarate-dependent dioxygenase, SlGAME12–transaminase, and SlGAME4–CYP88B1) in Nicotiana benthamiana to generate the basic aglycones and confirm the chirality of the GAME8 products. Analysis of N. benthamiana extracts showed that SlGAME8 expression leads to production of almost only tomatidenol (25S), while SmGAME8 yields exclusively solasodine (25R) (Fig. 2c).
a Phylogenetic analysis of GAME8 coding sequences (CDSs) from multiple Solanaceae species. CYP72B1 (brassinosteroid C26 hydroxylase) from tomato was used as an outgroup, and GAME6 CDSs (cholesterol-GlcA C22 hydroxylase) were used to stabilize the tree. Two clades with distinct stereo specificities are marked with either red or purple braces. Red and purple disks designate C25 stereo specificity of in planta tested (i.e., in N. benthamiana) GAME8s; yellow triangles label duplicated proteins with divergent activities clustering to different clades, while blue and green triangles label proteins with opposite activities clustering in one clade. Pale purple disk designates GAME8 enzyme lacking C26/C27 hydroxylase activity in planta. b Proposed SGA aglycones biosynthetic pathway with C25 chirality marked either in red (25S) or purple (25R). c Extracted Ion Chromatograms depicting solasodine (25S) and tomatidenol (25R) production by selected Solanum GAME8 enzymes in N. benthamiana. d Simplified chemical structures of 22S,26(27)-dihydroxycholesterol-d7 glucuronides with C25 configuration assigned. e Extracted ion chromatograms depicting accumulation of isotopically labeled 22S,26(27)-dihydroxycholesterol-d7 glucuronides (25S or 25R) confirming GAME8 activity as C26 or C27 hydroxylase. Retention time difference between deuterium-labeled and non-labeled metabolites is due to the deuterium isotope effect.
The results above prompted us to comprehensively assay a large number of GAME8 enzymes, focusing on their sequence similarity and C25 stereospecificity. To accomplish this, we mined available genomes and transcriptomes of Solanum and Solanaceae species to find as many genes as possible encoding GAME8 enzymes (Supplementary Data 1). We collected sequences from tomato and potato clades, Old World spiny solanum, M clade, and other Solanaceae species such as Jaltomata, Iochroma, Petunia, Capsicum, Cestrum, Lycium, Anisodus, and Nicotiana. Phylogenetic analysis of GAME8 nucleotide and protein sequences revealed two distinct clades: one containing sequences similar to the tomato GAME8 and with a second clade represented by sequences that are similar to eggplant GAME8 (Fig. 2a and Supplementary Fig. 5). To determine if enzymes from different clades generate aglycones with opposite chirality at C25, we cloned or synthesized twenty of these enzymes and transiently expressed them in N. benthamiana (Fig. 2c). Analysis of the aglycones showed a perfect match with the phylogenetic clustering. All tested sequences from the “eggplant” clade produced aglycones with the 25R configuration, while almost all tested enzymes from the “tomato” clade produced the 25S product (Fig. 2a, c).
To confirm that GAME8 from tomato and eggplant specifically modifies one of the terminal methyl groups, we conducted an in planta (i.e., transient expression in N. benthamiana) experiment using deuterium-labeled cholesterol, with all hydrogens replaced at those positions. Analysis of GAME8 products, following modifications of cholesterol by GAME15 and GAME6, revealed that 22S-hydroxycholesterol glucuronide serves as the substrate for GAME8 enzymes. The reaction produces two distinct isomers, with SlGAME8 generating the 25S isomer and SmGAME8 producing the 25R isomer (Fig. 2d, e, and Supplementary Fig. 6).
Duplication and neofunctionalization shape SGA chirality in Solanum
In the same experiments portrayed above, we observed that in some species, there are two or even three different GAME8 genes located in the same chromosomal region, and their protein products cluster into two clades: one similar to eggplant and the other to the tomato clade. Among these species were Solanum nigrum and Solanum dulcamara (also termed bittersweet nightshade), belonging to the M clade in the genus Solanum (Fig. 1a). We checked if these species indeed produce molecules with different chirality at C25. Analysis of leaves, stems, and roots of S. nigrum revealed that these tissues accumulate exclusively one isomer of the steroidal saponin uttroside B (25R) (these tissues do not accumulate SGAs) (Fig. 3a). However, in S. nigrum root, we observed an additional peak that corresponds to 25-epi-uttroside B (25S) (Fig. 3a and Supplementary Fig. 7). We obtained similar results for S. dulcamara: its leaves accumulated SGAs composed exclusively of solasodine (25R) as an aglycone (Fig. 3b and Supplementary Fig. 4). In the stem, we detected predominantly solasodine-based SGAs but also minute amounts of tomatidenol-based alkaloids (Fig. 3b and Supplementary Fig. 4). Finally, S. dulcamara roots accumulated comparable levels of tomatidenol and solasodine-derived SGAs (Fig. 3b and Supplementary Fig. 4). To confirm that metabolite composition corresponds with GAME8 transcript levels, we performed expression analysis and observed that in S. nigrum, tomato-like GAME8 is expressed only in roots, and in S. dulcamara, mostly in the root, while the eggplant-like GAME8 was expressed in all tissues, explaining the observed chemical composition (Fig. 3c, d). Finally, we examined both GAME8 enzymes from S. dulcamara and S. nigrum and confirmed their activity and stereospecificity in N. benthamiana (Fig. 3e, f). This demonstrated that the phylogenetic analysis predicted correctly the functionality of GAME8 enzymes in Solanum species from the M clade.
a Extracted Ion Chromatograms (EICs) depicting accumulation of the steroidal saponins uttroside B (25R) and epi-uttroside B (25S) in leaf, stem, and root of Solanum nigrum. b Liquid chromatography–mass spectrometry (LC-MS) analysis of SGA aglycones accumulated in leaf, stem, and root of Solanum dulcamara. c, d Quantitative PCR (qPCR) analysis of relative expression levels of GAME8-1 and GAME8-2 in S. nigrum (c) and S. dulcamara tissues (d), mean values (center line) with SD (whiskers) are shown (n = 3 of independent replicates for each tissue). e, f EICs illustrating solasodine (25S) and tomatidenol (25R) production by GAME8-1 and GAME8-2 enzymes from S. nigrum (e) and S. dulcamara (f) in N. benthamiana. g Chromosomal regions of various Solanaceae species with GAME8 genes marked in purple (25R), red (25S) or yellow (inactive) depending on their enzymatic activity and stereospecificity.
Intrigued by the existence of multiple copies of GAME8 in a single genome and located in a similar chromosomal region, we examined whether other genomes contain GAME8 (multi)duplication or this is exclusive to S. nigrum and S. dulcamara. Screening the available Solanaceae genomes, we found species with only one, two identical (or almost identical), or two to three different copies of GAME8. Cestrum, Nicotiana, Petunia, Anisodus, and Lycium have only one copy of GAME8, and in all cases, they clustered with eggplant-type enzymes. The genome of Capsicum annuum contains two copies of GAME8; one producing the 25R isomer and one inactive with premature stop codon. The genome of eggplant contains two similar copies of GAME8, and both copies encode almost identical proteins (96.08% identical) with 25R stereospecificity (Figs. 2c, 3g). Potato and tomato genomes also contain two identical copies. In tomato, two GAME8 proteins are 100% identical, while in potato GAME8 proteins are 99.80% identical (differ in a single residue), but in this case, their products display 25S stereospecificity (Figs. 2c, 3g). This analysis suggested that during evolution, the first GAME8 enzyme was encoded by a single copy gene and catalyzed the formation of the 25 R product—as seen in Cestrum imbricatum. Later, this single copy underwent duplication (observed in the Old World and spiny Solanum species) and, in some cases, duplication was followed by neofunctionalization, as observed in the M clade (i.e., S. nigrum and S. dulcamara). Furthermore, we found that in the potato and tomato clades, only two copies of the GAME8 gene encoding almost identical proteins with 25S stereospecificity were retained (Fig. 3g).
Structure-guided mutagenesis shifts tomato GAME8 stereospecificity
To understand the structure-function relationship in GAME8 enzymes, we used Alphafold2 to model the 3D structures of SlGAME8 and SmGAME8. Using Chimera28, we superimposed both proteins and observed that the overall fold was almost identical (Fig. 4a), except for a twelve amino acid residues loop present in SmGAME8 (Fig. 4a). We also observed that this ~12 amino acids are present in all 25R producing GAME8 proteins but missing in enzymes with 25S activity (Supplementary Fig. 8). We removed these 12 amino acids from SmGAME8 and inserted them into SlGAME8 (Supplementary Fig. 8). However, these changes did not affect the stereospecificity of the GAME8 enzymes (Supplementary Fig. 9). We then used molecular docking to understand how the substrate of GAME8 proteins (22S-hydroxycholesterol glucuronide) interacts with amino acids in the active site of the enzyme. Combining docking data with multiple sequence alignment of several GAME8 proteins with either 25S or 25R stereospecificity revealed eight highly conserved amino acids around the active site that differ between tomato- and eggplant-like GAME8 proteins (Fig. 4b, c). We used LigPlot+ to predict molecular interactions between the protein and its substrate29. This analysis confirmed interactions for at least half of the selected residues. Others, though not detected, may play a steric role, influencing the positioning of key amino acids (Supplementary Fig. 10). To test if the conserved amino acids are responsible for enzymes stereospecificity, we decided to replace all eight amino acids in SlGAME8 with the corresponding ones from SmGAME8. This modification of SlGAME8 resulted in a complete change of stereospecificity, with the mutated version producing exclusively solasodine, the 25R isomer of tomatidenol (Fig. 4d).
a Superposition between SlGAME8 (orange) and SmGAME8 (blue) and molecular docking of 22S-hydroxycholesterol glucuronide in the enzymes active site. Amino acids within 10 Å from the substrate metabolite are visualized, as well as twelve amino acid loop present only in SmGAME8. b GAME8 active site with designated differential amino acid between SmGAME8 and SlGAME8. Red arrow indicates substrate attack from the side of heme molecule. c Multiple sequence alignment of active sites’ amino acid sequences from multiple GAME8 enzymes with 25S (red) or 25R (purple) stereospecificity (for more details see Supplementary Data 1). Amino acid substitutions are indicated with arrows. d Extracted ion chromatograms depicting production of tomatidenol by SlGAME8 WT and solasodine by SmGAME8 and SlGAME8-mut (8 amino acids replaced) in N. benthamiana.
In planta evidence identifies GAME8 as key for C25 stereochemistry
To further confirm that GAME8 is responsible for stereochemistry at C25, we used CRISPR-Cas9 to generate SlGAME8 knockout lines (SlGAME8-ko) of tomato hairy roots. Analysis of SGAs and their aglycones confirmed that SlGAME8 is indispensable for the production of α-tomatine in tomato hairy roots (Fig. 5a, b). We next generated tomato hairy root lines overexpressing the eggplant GAME8 in the SlGAME8-ko background (Fig. 5a, b, and Supplementary Fig. 11). Analysis of the SlGAME8-ko simultaneously overexpressing SmGAME8 hairy roots showed the production of the 25R isomer of α-tomatine with soladulcidine as an aglycone (Fig. 5a, b, e). We also analyzed these lines for the accumulation of SGA pathway intermediates and detected high accumulation of cholesterol glucuronide and 22S-hydroxycholesterol glucuronide in the SlGAME8-ko line itself, while in the combined SlGAME8-ko and SmGAME8-overexpressing line, levels of intermediates were similar to the control sample, i.e., hairy roots expressing GFP alone (Fig. 5c, d). This suggested that SmGAME8 utilized these intermediates for SGA aglycone formation. Additionally, we generated transgenic lines of Solanum nigrum expressing SlGAME8, which accumulated up to 30% of the steroidal saponin 25-epi-utroside B (25S isomer), compared to the 3% produced in wild-type plants (Fig. 5g, h). Altogether, these results from modified plants demonstrated that GAME8 is the key enzyme conferring stereochemistry at the C25 position of SGAs and steroidal saponins.
a LC-MS analysis of SGA (α-tomatine) in either tomato hairy roots with knocked out SlGAME8 (SlGAME8-ko), or expressing the eggplant SmGAME8 in the tomato SlGAME8-ko background. Hairy roots expressing GFP only were used as a control. b LC-MS analysis of aglycones (tomatidine and soladulcidine) in tomato hairy roots with knocked out SlGAME8 (SlGAME8-ko), or expressing SmGAME8 in SlGAME8-ko background. c, d LC-MS analysis of intermediates cholesterol-GlcA (c) and 22S-hydroxycholesterol-GlcA (d) in the same hairy root lines. Identity of metabolites was assigned based on accurate mass and our previous studies24. e Percentage of 25 R α-tomatine in control and SmGAME8 expressing tomato hairy roots (n = 14 of independent replicates for each genotype, P = 2.32756E-45). f Extracted ion chromatograms (EICs) showing accumulation of the steroidal saponin uttroside B and 25-epi-uttroside B in WT and SlGAME8 expressing Solanum nigrum leaves. g Percentage of 25-epi-uttroside B in WT and SlGAME8 expressing S. nigrum (n = 5 of independent replicates for each genotype, P = 1.11608E-07). Asterisks indicate statistically significant differences as compared to control or WT determined by two-sided Student’s t-test. Box plots show all data points, center line represents median value, bounds correspond to upper and lower quartile, and whiskers represent min and max values.
Stereochemical diversity of alkaloids across wild tomato populations
We questioned whether wild tomato accessions as cultivated ones, produce the 25S isomers or if their SGAs also exist in the 25R configuration. To answer this, we profiled SGAs from eleven wild tomato species and two accessions of the domesticated Solanum lycopersicum (Fig. 6a, g). Nine wild species and both domesticated accessions produced almost exclusively α-tomatine with 25S chirality (Fig. 6a). Nonetheless, S. pennellii and S. cheesmaniae accumulated considerable quantities of the 25R isomer, in fact, S. pennellii produced predominantly “eggplant” type α-tomatine (Fig. 6a). We confirmed that these are indeed 25R isomers by analyzing the aglycones of those SGAs (Fig. 6b). The presence of the 25R isomer in S. pennellii was unexpected, as both of its GAME8 enzymes cluster with other tomato-like enzymes on the phylogenetic tree (Fig. 2a). We cloned both copies of SpGAME8 and ScheGAME8 and tested their stereospecificity. We observed that SpGAME8-1 produces exclusively the 25R isomer while SpGAME8-2 produces almost only the 25S counterpart (Fig. 6c). On the other hand, S. cheesmaniae GAME8 was able to produce both isomers (Fig. 6c). Comparison of amino acid sequences revealed that S. pennellii GAME8 enzymes differ by only six amino acids, with four of them located in the active site of the enzyme (Fig. 6d and Supplementary Fig. 12). These four amino acids were among the previously described eight highly conserved amino acids in the active site that differ between tomato- and eggplant-like GAME8 proteins. Additionally, we discovered that S. cheesmaniae GAME8 has 3 amino acids mutated in the active site (counterparts of those in S. pennellii), potentially explaining its capability to produce both 25S and 25R products (Fig. 6d). Analysis of SpGAME8-1 and SpGAME8-2 transcript levels in S. pennellii leaves showed that the expression of SpGAME8-2 (25S) was only 30% of that recorded for SpGAME8-1 (25R) and this correlated with the levels of different α-tomatine isomers in the same tissue (Fig. 6e, f). Finally, we analyzed a set of S. pennellii Introgression Lines (IL 6-1, IL 6-2, IL 6-2-2, IL 6-3 and IL 6-4) generated in the M82 cultivated tomato background30 and observed that lines with S. pennellii introgression in chromosome 6 (IL 6-2 and IL 6-2-2) in which GAME8 genes are located, produced predominantly 25R α-tomatine. The results demonstrated the role of GAME8 in determining stereodiversity among wild tomato species (Fig. 6h).
a Extracted ion chromatograms (EICs) showing accumulation of α-tomatine C25 isomers in domesticated and wild tomato species. Arrows indicate 25S and 25R isomer. b LC-MS analysis of SGA aglycones (tomatidine and soladulcidine) in S. lycopersicum, S. pennellii, and S. cheesmaniae leaves. c Production of SGA aglycone isomers (tomatidenol and solasodine) by S. pennellii GAME8-1 or GAME8-2, or S. cheesmaniae GAME8 in N. benthamiana leaves analyzed by LC-MS. As controls we used GAME8 genes from tomato and eggplant. d Multiple sequence alignment of active sites’ amino acid sequences from GAME8 enzymes with 25S (red) or 25R (purple) or mixed stereospecificity. SgalGAME8 corresponds to GAME8 enzyme from Solanum galapagense (see Fig. 7 for more details). Differential amino acids are highlighted in yellow and pale blue. e qPCR expression analysis of SpGAME8 genes in S. pennellii leaf (n = 8 of independent measurements for each gene, P = 4.70744E-15). f Analysis of relative concentration of 25S and 25R α-tomatine isomers in S. pennellii leaf (n = 7, independent measurement, P = 2.02513E-08). Comparing (e, f), we can see that levels of gene expression correlate with metabolite content in the same tissue. g Map of northwestern part of South America showing origins of wilds tomato accessions used in this study (Fig. 6a); the blue star on the map represents Lima, the capital of Peru. h EICs showing accumulation of α-tomatine C25 isomers in selected S. pennellii introgression lines (ILs) and in parental species. Arrows indicate 25S and 25R isomer. Asterisks indicate statistically significant differences determined by two-sided Student’s t-test (*** −P < 0.001). Box plots show all data points, center line represents median value, bounds correspond to upper and lower quartile, and whiskers represent min and max values.
Evolution of Solanum cheesmaniae GAME8 drives SGA diversity on the Galápagos Islands
To explore the chemical and genetic diversity of SGAs in S. pennellii and S. cheesmaniae more deeply, we analyzed the composition and stereochemistry of multiple accessions of each species (40 and 35, respectively). Accessions of S. pennellii, though originating from various environments, produced similar mixtures of C25 isomers of SGAs (Supplementary Fig. 13). In contrast, different accessions of S. cheesmaniae produced varying levels of 25S and 25R α-tomatine isomers (Fig. 7a). We found that all accessions of S. cheesmaniae can be grouped into three distinct populations based on their SGA stereochemistry: the first with almost exclusively 25S isomer (93%; termed “C25R-low”), the second with 80% 25S isomer and 20% 25 R (“C25R-medium”), and the third with almost 50% 25R α-tomatine (“C25R-high”; Fig. 7a, b). All analyzed accessions of S. cheesmaniae are native to the Galápagos Islands but were collected from different locations spread over multiple islands (Fig. 7c). We found that the “C25R-low” population, producing mostly 25S α-tomatine, consisted of accessions collected from eastern and the central islands (i.e., San Cristóbal, Santa Fé, and Santa Cruz), while the “C25R-medium” and “C25R-high” populations, possessing higher levels of 25R α-tomatine, contained accessions exclusively from the western islands (i.e., Fernandina and Isabela; Fig. 7b). We also observed a correlation between the age of an island and the amount of the 25R isomer. Older (1 million years and more) eastern islands are home to S. cheesmaniae accessions producing primarily the 25S isomer, while the younger western islands (less than 0.5 My) host accessions with higher levels of 25R SGAs31 (Fig. 7b). This suggests that originally S. cheesmaniae produced the 25S isomer, and during evolution GAME8 lost its stereospecificity, and began producing the second isomer. To confirm this hypothesis, we sequenced GAME8 genes from twenty-five accessions of S. cheesmaniae to find causal mutations and tested their stereospecificity through transient expression in N. benthamiana plants (Fig. 7d, e). Sequence analysis confirmed that the “C25R-medium” and “C25R-high” populations from the western islands had accumulated mutations resulting in amino acid substitutions within the active site, leading to altered stereospecificity (Fig. 7d, e and Supplementary Figs. 14, 15). C25R-medium accessions carried mutation resulting in one amino acid substitution [GAME8(Ile401Thr)], while C25R-high accessions had three amino acids substitutions [GAME8(Phe398Cys, Ile401Thr, Tyr402Cys)] (Fig. 7d, and Supplementary Figs. 14, 15). Solanum cheesmaniae is one of two tomato species native to the Galápagos Islands; the second, Solanum galapagense, is also dispersed over multiple islands, mostly central and western ones (Fig. 7b). We analyzed 21 accessions of S. galapagense, but all of them displayed identical SGA composition and produced almost exclusively 25S α-tomatine (Fig. 7a). Notably, these findings are consistent with previous studies on the genetic diversity of wild tomato species from the Galápagos Islands32.
a Extracted ion chromatograms (EICs) showing accumulation of α-tomatine C25 isomers in selected accessions (LA0746, LA1402, and LA1412) of S. cheesmanie originating from different islands. Red, yellow, and purple annotations correspond do accessions accumulating 7%, 20%, or 40% of the 25R isomer, respectively. b Relative amount of 25 R α-tomatine isomer (center lines and whiskers correspond to mean values and SD, respectively) in 35 accessions of S. cheesmaniae and 21 accessions of S. galapagense. Red, yellow, and purple triangles and circles correspond do accessions with different amounts of the 25 R isomer. c Map of Galápagos archipelago. Red triangles mark locations of S. galapagense accessions. Red, yellow, and purple disks indicate locations of S. cheesmaniae accessions accumulating 7%, 20%, and 45% of 25R α-tomatine, respectively. Different hues of blue represent age of the islands. Increasing color depth indicates older age of the island (My – million years). Adapted from Štefka31. d Phylogenetic analysis and multiple sequence alignment of active sites’ amino acid sequences of selected GAME8 enzymes from multiple accessions of S. cheesmaniae. Red, yellow, and purple indicate percentage of 25R α-tomatine accumulated and the origin of analyzed accessions. An arrow indicates the evolutionary trajectory of S. cheesmaniae GAME8. e EICs illustrating in planta production (i.e., by transient expression in N. benthamiana) of solasodine (25R) and tomatidenol (25S) by GAME8 enzymes from selected accessions of S. cheesmaniae. Scatter plots represent mean value with SD, all data points are shown.
25R stereospecificity as the Ancestral State of GAME8 Enzymes in Solanaceae
To further investigate the possibility of reverse evolution in species such as Solanum cheesmaniae, we first sought to determine the ancestral state of GAME8 within the Solanaceae family. Understanding the evolutionary trajectory of this enzyme is essential for interpreting shifts in stereospecificity among Solanum species. To achieve this, we compiled a diverse collection of GAME8 coding sequences from various Solanaceae species. This dataset included representatives from early-diverging lineages that split from the family long before the emergence of the Solanum genus, such as Cestrum, Petunia, Nicotiana, Lycium, and Anisodus. Additionally, we compiled sequences from multiple Solanum species, encompassing both 25R- and 25S-producing lineages. Using multiple sequence alignment and a Maximum Likelihood phylogenetic approach, we constructed a phylogenetic tree that allowed us to infer ancestral GAME8 sequences at key divergence points. To improve the accuracy of our evolutionary inferences, we calibrated the tree using publicly available divergence times from TimeTree533 (Fig. 8a). Based on this phylogenetic reconstruction, we identified six critical nodes corresponding to major clade divergences within Solanaceae and selected these for further functional analysis (marked in Fig. 8a).
a Maximum Likelihood (ML) phylogenetic tree of GAME8 genes from diverse Solanaceae species. The tree was calibrated using divergence times from TimeTree5 – calibration points indicated by green stars. Six key divergence nodes (126, 127, 128, 129, 130, and 113) were selected for ancestral sequence reconstruction. Purple and red circles indicate 25 R or 25S stereospecificity at each node, respectively. Genes in bold were tested for their activity, and results are shown in (b). b Functional analysis of ancestral GAME8 sequences and GAME8 genes from early-diverging species (i.e., Cestrum imbricatum, Anisodus acutangulus, and Lycium ruthenicum) expressed in Nicotiana benthamiana via Agrobacterium-mediated infiltration. LC-MS analysis (Extracted Ion Chromatograms for m/z 414.3374) showing production of 25R (solasodine) or the 25S (tomatidenol) isomer. Solasodine and tomatidenol were used as standards, SlGAME8 and SmGAME8 served as positive cotrols, while WT, infiltration media, or GAME15, 6, 11, 4, and 12 infiltrated plants served as negative controls. c ML phylogenetic analysis of full-length GAME8 genes, including introns, showing that 25R-producing genes form a distinct ancestral clade, with 25S-specific GAME8 genes diverging later in certain Solanaceae lineages. Bootstrap support values are indicated at branch nodes.
To empirically test the stereospecificity of these ancestral GAME8 sequences, we synthesized and expressed their predicted protein-coding genes in Nicotiana benthamiana using Agrobacterium-mediated transient expression (Fig. 8b). Additionally, we cloned and tested the activity of GAME8 genes from Cestrum imbricatum, Anisodus acutangulus, and Lycium ruthenicum, which represent early-diverging Solanaceae species (Fig. 8). LC-MS analysis of N. benthamiana leaf extracts revealed that GAME8 proteins from these three species predominantly produce the 25R isomer in planta. Furthermore, analysis of N. benthamiana leaves infiltrated with sequences reconstructed through ancestral state inference showed that all predicted ancestral GAME8 proteins produced the 25R metabolite except for one, node 113, which yielded nearly equimolar amounts of both the 25R and 25S isomers.
To further investigate the relationship between gene structure and function, we performed multiple sequence alignment (MSA) of full-length GAME8 genes, including introns, to identify structural differences that may have contributed to functional divergence. Maximum Likelihood phylogenetic analysis of these sequences revealed that genes encoding 25R-producing enzymes cluster together, supporting the hypothesis that 25R stereospecificity is the ancestral state. Additionally, we observed that GAME8 genes encoding 25S-specific enzymes diverged from the 25R-producing clade, suggesting that the shift to 25S stereochemistry arose later in Solanaceae evolution (Fig. 8c). Together, these findings provide strong molecular and evolutionary evidence that the ancestral GAME8 enzyme in Solanaceae was stereospecific for the 25R isomer. The transition to 25S stereochemistry in certain Solanum lineages likely occurred through gene duplication, followed by functional divergence, ultimately shaping the metabolic diversity observed in modern species. Another possible explanation, which we cannot definitively exclude, is that GAME8 underwent multiple duplication events, with some copies being lost over time.
Discussion
Chirality, or the presence of enantiomers, is a fundamental aspect of chemical diversity in nature, profoundly influencing biological activities and ecological interactions34. In plants, chiral compounds play crucial roles in defense mechanisms, pollinator attraction, interactions with microorganisms, and medicinal properties. For instance, enantiomers of pyrrolizidine alkaloids exhibit varying toxicity levels to herbivores, impacting plant defense strategies35,36. Similarly, the chiral properties of floral scent compounds like linalool affect pollinator behavior, influencing pollination success37. Plants produce a wide range of compounds with potential medicinal properties, where chirality can affect the pharmacokinetics and pharmacodynamics of these compounds in human and animal systems. For example, ephedrine, a compound used to manage and treat hypotension, exists as four different chiral isomers, each of them displays different biological activity and stability38. Additionally, chirality influences the environmental fate of plant metabolites, affecting their persistence in soils and water bodies, with the phytotoxicity of allelopathic compounds being enantioselective, influencing plant competition and community dynamics38. SGAs’ chirality could significantly impact their biological functions, especially in plant defense against herbivores and pathogens. The stereochemistry at the C25 position determined by GAME8 enzymes could possibly dictate the compounds’ toxicity and efficacy. Our findings reveal how evolutionary changes in GAME8 enzymes contribute to the stereochemical diversity of SGAs, potentially enhancing the adaptive capabilities of Solanum species.
Although it is difficult to prove, some evidence suggests that SGAs with different C25 chirality could potentially influence plant-pest interactions to varying degrees. For instance, a study reported that an accession of Solanum dulcamara predominantly accumulating C25R SGAs was more susceptible to slug herbivory compared to an accession with a higher percentage of C25S isomers39. However, there has not been enough research conducted to fully support the hypothesis that C25 chirality significantly affects plant resistance to pests.
It is worth mentioning that GAME8 not only plays a crucial role in the biosynthesis of steroidal glycoalkaloids but is also indispensable for the production of multiple steroidal saponins in Solanaceae plants. Previous studies have shown that plants like Datura stramonium, Capsicum annuum, Atropa belladonna, Petunia hybrida, Nicotiana rustica, N. tabacum, and Solanum melongena produce steroidal saponins with aglycones displaying C25R chirality40. In contrast, tomato and potato plants accumulate in certain tissues steroidal saponins with C25S chirality, which aligns with our phylogenetic studies and functional analysis of GAME8 activity in some of these species40. While our study provides an explanation for the stereodiversity at the C25 position of SGAs and steroidal saponins in Solanaceae plants, this knowledge cannot be easily extrapolated to the hundreds of monocot species (such as Veratrum and Paris polyphylla) that produce steroidal saponins or alkaloids with aglycones displaying either C25S or C25R chirality. Although aglycones with the C25R configuration (such as diosgenin and its derivatives) appear to be more prevalent, we cannot overlook the saponins that utilize yamogenin (the C25S isomer) and its derivatives. It is also intriguing that certain monocot plants produce saponins with both C25R and C25S chirality. Cytochrome P450 CYP94D108 form Paris polyphylla was shown to catalyze C26 hydroxylation in diosgenin (25R) biosynthesis41, however, it shares only 19.6% and 17.8% identity with GAME8 enzymes from tomato and eggplant, respectively. Moreover, there is no known cytochrome from monocot plants that is involved in production of 25S isomer yamogenin. Since no protein in monocots shows more than 50% similarity to the GAME8 protein, we cannot assume that GAME8-like enzyme is involved in their biosynthesis. Additional research is required to determine whether C25 stereodiversity in monocot plants arises from the activity of a single protein that has diverged similarly to GAME8 or from the activity of two unrelated enzymes.
Reverse evolution, or the reversion to ancestral traits, challenges the conventional view of evolution as a strictly forward process. It occurs through mechanisms such as genetic and phenotypic reversions due to selective pressures. This concept highlights the flexibility of evolutionary processes and the potential for species to re-adapt to “past” environmental conditions42. Solanum cheesmaniae, a wild tomato species native to the Galápagos Islands, exemplifies how reverse evolution could shape diversity. Our research indicates that populations from older islands predominantly produce the 25S isomer of α-tomatine, while those from younger islands display increased levels of the “ancestral” 25R isomer. This pattern suggests that ongoing evolutionary shifts in the GAME8 enzyme function are influenced by differing ecological pressures across the islands.
The geographical isolation of Galápagos Islands leads to a limited gene flow between populations on different islands, resulting in divergent evolution and the emergence of unique traits within populations32,43,44,45. Environmental variations, including climate, soil type, altitude, and ecological niches, drive the adaptation of S. cheesmaniae populations to these differing conditions, leading to morphological and, as we reported here, chemical diversity46,47. Selective pressures such as herbivory, pathogen attack, and competition with other plants could drive the evolution of defensive compounds like SGAs, with variation in composition and stereochemistry as a response to these pressures. Random mutations and genetic drift also might contribute to genetic diversity, leading to genetic differentiation between populations48. Furthermore, hybridization and introgression could shape genetic variation in Galápagos tomato plants49. Solanum cheesmaniae exhibits considerable genetic diversity, evident in its morphological and physiological traits, resulting from plant adaptation to various microenvironments across the Galápagos archipelago46. Island biogeography, with varying environmental conditions and unique microclimates, further drives the adaptation and diversification of this species. If environmental conditions on the Galápagos Islands resemble those of the past, selective pressures could favor ancestral traits, potentially altering the enzymatic pathways involved in SGA biosynthesis. The proposed occurrence of reverse evolution and emergence of individuals producing 25S and 25R isomers could significantly affect the biodegradation of SGAs in S. cheesmaniae, influencing their ecological impact in terms of food web dynamics and habitat formation. Microbial degradation by soil bacteria and fungi, facilitated by microbial enzymes such as glycosidases and oxidases, plays a significant role in the SGA breakdown process50,51,52. Soil composition and temperature can also affect the rate and extent of SGA degradation.
Building on the concept of reverse evolution, our study explores mutations in the GAME8 enzyme across different species. Based on phylogenetic analysis and reconstitution of ancestral state, we consider GAME8 with 25R stereospecificity as an ancestral protein (e.g., from Cestrum imbricatum, Petunia inflata, Lycium ruthenicum, Anisodus acytangulus, Iochroma cyaneum, or Capsicum annuum). In addition, we can observe a recurring pattern of up to eight mutations that alter the enzyme’s stereospecificity. In the spiny and Old World Solanum species, GAME8 has retained all eight ancestral amino acids and maintains the same stereospecificity. In the M clade, we find that one of the GAME8 copies retained the ancestral structure and activity, while the other copies have all eight amino acids mutated to produce the 25S isomer. In contrast, both copies of GAME8 in tomato species are identical, each having all eight amino acids mutated from the ancestral enzyme, resulting in the production of the 25S isomer. Interestingly, in S. pennellii, one of the GAME8 proteins has partially reverted to its ancestral form, with three out of four mutated amino acids returning to those found in the ancestral GAME8. Similarly, in S. cheesmaniae, we observe a gradual restoration of ancestral-type amino acids in the enzyme’s active site (Supplementary Fig. 16). Populations that accumulate moderate amounts of the 25R isomer of tomatine show a single amino acid substitution that matches the eggplant form. In contrast, populations with high levels of the 25R isomer exhibit three mutated amino acids, two of which have reverted to the ancestral configuration (Supplementary Fig. 16). These observations suggest that there is strong selective pressure driving specific changes in the enzyme’s stereospecificity, further illustrating how reverse evolution can influence the diversity of plant metabolites.
The existence of two nearly identical GAME8 enzymes in Solanum pennellii that produce different isomers is challenging to explain. However, this duplication and subsequent neofunctionalization may be a response to the significant environmental pressures that this species faces in its arid habitats in Peru. These habitats include rocky and sandy slopes, dry riverbeds, and mountainous areas, often at elevations ranging from 500 to 3000 m above sea level. The emergence of two GAME8 enzymes and their persistence in all analyzed accessions suggest that one of the copies evolved shortly after S. pennellii and S. habrochaites diverged47. The presence of both SGA isomers may have provided S. pennellii with an advantage, enabling it to thrive in the dry regions of coastal Peru, while other tomato species migrated north or east along river valleys to more humid environments47. Additionally, the neofunctionalization of GAME8 is not the only example of how this species has evolved to enhance its chemical diversity. Mutations accumulated in a ripening-related alcohol acyltransferase SpAAT1 have resulted in a more versatile and efficient enzyme compared to the tomato counterpart, underscoring its role in the variations in volatile profiles between the species53.
Our study exposes the intricate relationship between GAME8 enzyme evolution, genetic diversity, and the stereochemical composition of SGAs in Solanum species. Understanding these dynamics enhances our comprehension of plant secondary metabolite biosynthesis and adaptation, highlighting the pivotal role of particular chemical reactions, such as the one performed by the GAME8 enzyme in shaping chemical diversity. Moreover, the concept of reverse evolution offers valuable insights into how historical environmental conditions and genetic reversion can influence current and future biodiversity, particularly in unique ecosystems like the Galápagos Islands.
Methods
Chemicals
Unless otherwise specified, chemicals used in this study were purchased from Sigma-Aldrich. Standard of tomatidine/tomatidenol was purchased from Sigma-Aldrich (Cat# T2909), solasodine was from ChemFaces (Cat# CFN90200), and soladulcidine from Toronto Research Chemicals (Cat# S676530).
Plant material
Tomato (Solanum lycopersicum cultivar Micro-Tom and M82), potato (Solanum tuberosum), pepper (Capsicum annuum), Solanum pennellii, Solanum habrochaites, Solanum cheesmaniae, Solanum nigrum, Solanum americanum, Solanum dulcamara, Solanum villosum, Solanum melongena, Solanum quitoense, Solanum torvum, Solanum pyracanthos plants were grown in a greenhouse. Nicotiana benthamiana was grown in a growth room with 24 °C: 18 °C, day: night temperatures and a 16/8 h, light: dark photoperiod cycle.
LC-MS analysis of steroidal glycoalkaloids
Briefly, 100 mg of frozen powdered plant tissue was extracted with 300 µl of 80% methanol +0.1% formic acid, briefly vortexed, and then sonicated for 20 min at room temperature. Extracts were centrifuged for 10 min at 14,000 × g and filtered through 0.22 µm filters. At least three biological replicates (n = 3) from sample were used for metabolic analysis. A SYNAPT-G2 qTOF mass spectrometer (Waters) coupled to ACQUITY UPLC system (Waters) was used for liquid chromatography-high-resolution mass spectrometry (LC-HRMS) analysis. MassLynx software v.4.1 (Waters) was used to control the instrument and calculate accurate masses and elemental compositions. Separation of metabolites was performed on a 100 mm × 2.1 mm i.d. (internal diameter), 1.7 µm UPLC BEH C18 column (Waters Acquity). The mobile phase consisted of 0.1% formic acid in acetonitrile:water (5:95, v/v; phase A) and 0.1% formic acid in acetonitrile (phase B). The flow rate was 0.3 ml min−1, and the column temperature was kept at 35 °C. The following linear gradient was used for analysis of SGAs: from 100 to 72% phase A over 22 min, from 72% to 0% A over 14 min, then held at 100% phase B for 2 min; and then returned to the initial conditions (100% phase A) within 0.5 min and conditioning at 100% phase A for 1.5 min. Total run time was 40 min. In some instances, high-resolution mass measurements were performed on Orbitrap Exploris240 operated in full-scan mode (m/z range: 90–1350) with the following parameters: capillary—3.5 kV, vaporizer temperature—300°, ion transfer tube—325 °, sheath gas—45, Aux gas—10, Sweep gas—1. Scan parameters: Resolution 120k, scan time 100 ms, normalized AGC target 100%, RF lens—70%. Data was processed using FreeStyle 1.8 SP2 (ThermoScientific). In this experiment, we used two types of negative controls—blank (80% methanol +0.1% formic acid) and extracted blank (80% methanol +0.1% formic acid without plant material but following the extraction protocol). We also used a-tomatine standard to confirm identity of molecules. Identity of other SGAs was determined based on MS2 spectra and literature data.
Acid hydrolysis of SGA and extraction of aglycones
In 2 ml Eppendorf tube 250 µl of methanolic leaf extract was mixed with 250 µl of 2 M HCl and incubated at 75 ° for 2 h. After cooling down, 150 µl of 4 M NaOH was added yielding a bit basic solution that was subsequently extracted 4 times with 1 ml of hexane. Pooled hexane fractions were evaporated at room temperature under stream of nitrogen and dissolved in 200 µl of EtOH containing 1% formic acid. Samples were centrifuged for 10 min at 17,000 × g and placed in LC-MS vials and directly analyzed on LC-MS.
LC-MS analysis of aglycones of steroidal glycoalkaloids
Extraction of tomatidine and dihydroxycholesterols: four leaf disks (diameter = 8 mm) were collected from two different tobacco leaves, placed in a 2 ml tube together with 2 metal balls and snap-frozen in liquid nitrogen. Tissues were powdered using bead beater for 2 min at 22 Hz (RETSCH MM 400 Mixer Mill). Ground tissue was mixed with 0.5 ml of 20% KOH in 50% EtOH, briefly vortexed, and incubated at 65 ° for 1 h. Each sample was extracted trice with 0.6 ml of hexane. Pooled hexane fractions were evaporated, and residue was resuspended in 0.1 ml of EtOH. Samples were analyzed on the LC-MS on the same day. LC-MS was performed on a Xevo TQS quadrupole mass spectrometer (Waters) coupled to an ACQUITY UPLC system (Waters). MassLynx software v.4.1 (Waters) was used to control the instrument. Some runs were performed on Orbitrap Exploris240 (ThermoScientific). Separation of metabolites (SGA aglycones) was performed on a 100 mm × 2.1 mm i.d. (internal diameter), 1.7 µm UPLC BEH C18 column (Waters Acquity). The mobile phase consisted of 0.1% formic acid in acetonitrile:water (5:95, v/v; phase A) and 0.1% formic acid in acetonitrile (phase B). The flow rate was 0.3 ml min−1, and the column temperature was kept at 35 °C. The following linear gradient was used for analysis of tomatidenol, tomatidine, solasodine and soladulcidine in N. benthamiana agroinfiltrated leaves: from 85 to 55% phase A over 13 min, from 55% to 0% A over 0.5 min, then held at 100% phase B for 3.5 min; and then returned to the initial conditions (85% phase A) within 0.5 min and conditioning at 85% phase A for 2.5 min. MS settings: For LC-MS analysis, Xevo TQS quadrupole mass spectrometer (Waters) was configured to perform multiple reaction monitoring scans with the following parameters: capillary −3.0 kV, source temperature −150°, sampling cone −20 V, desolvation temperature −350°, cone gas flow C, –150 (L/hr), desolvation gas flow −650.0 L/hr, nebulizer gas flow 7.0 bar, Tomatidenol/solasodine: MRM1: 414.30 > 253.25, collision energy 25.0 MRM2: 414.30 > 271.20, collision energy 25.0 Tomatidine/soladulcidine: MRM1: 416.40 > 255.25, collision energy 25.0 MRM2: 416.40 > 273.30, collision energy 25.0. Metabolites were identified by comparing the retention times and mass fragments of standard compounds—tomatidine, tomatidenol, solasodine, and soladulcidine. Relative quantification of metabolites was carried out using the TargetLynx (Waters) program.
Purification of 25S and 25R α-tomatine isomers from tomato
Isolation of pure stereoisomers of α-tomatine was performed as described previously with some modifications54. The system consisted of an Agilent 1290 Infinity II UPLC system equipped with a quaternary pump, autosampler, diode array detector, a Bruker/Spark Prospekt II LC–SPE system (Spark), and Impact HD UHR-QqTOF MS (Bruker) connected via a Bruker NMR MS Interface (BNMI-HP). MS spectra, in positive mode, were acquired between m/z 150 and 1300. The calibration was done with a 10 mM sodium trifluoroacetate (Sigma-Aldrich) automatically, which is introduced at the beginning and the end of each chromatographic run. Separation was done on a XBridge LC column (BEH C18, 5 μm particle size and 250 × 4.6 mm; Waters). The chromatographic conditions were as following; a flow rate of 0.6 ml min−1 starting with a solvent composition of 80% A (5% ACN + 0.1% FA) and 20% B (100% ACN + 0.1% FA) with a linear gradient to 74% A at 22 min, followed by another linear gradient to 100% B at 23 min 100% B hold for 3.5 min followed linear gradient to 80% A at 27 and hold for 1 min for equilibration. SGAs were collected on solid phase extraction (SPE) cartridges in preset time windows. For this trapping process, a makeup flow of 1.5 ml min−1 water was added to the eluent before it passed through the SPE cartridges to increase the retention of analytes on the cartridges. For the trapping 10 × 2 mm SPE cartridges filled with GP resin were used. Each cartridge was loaded five times with the same compounds, ten cartridges were used for trapping one isomer.
Analysis of GAME8 sequences from Solanaceae species
To identify GAME8 sequences across different species, genomic and transcriptomic data were systematically mined. We utilized BLAST to search for GAME8 homologs in all Solanaceae genomes and transcriptomes available in NCBI SRA database. Initial searches identified candidate sequences or genomic regions with GAME8 genes, which were further validated through manual short read assembly or gene annotation in particular chromosomal region using Softberry FGENESH software55. The resulting sequences were aligned and analyzed to ensure high-confidence identification of GAME8 orthologs across species.
Multiple sequence alignment and phylogenetic analysis
Protein sequences were aligned using the Muscle algorithm, and phylogenetic tree was inferred with RAxML rapid bootstrapping and subsequent maximum likelihood search56. The likelihood of final tree was evaluated and optimized under the gamma model of rate heterogeneity across sites. Gamma model parameters were estimated up to an accuracy of 0.1000000000 log likelihood units. One thousand bootstrap replicates using the fast bootstrap option of RAxML were performed. The substitution matrix used was DAYHOFF. The phylogenetic tree was visualized with iTOL57.
Cloning of GAME8 from Solanaceae species
Total RNA was isolated from plants tissues using TRIZol method—tomato (Solanum lycopersicum, leaf), potato (Solanum tuberosum, leaf), pepper (Capsicum annuum, leaf), Solanum pennellii (leaf, stem, root), Solanum habrochaites (leaf), Solanum cheesmaniae (leaf), Solanum nigrum (leaf, stem, root), Solanum americanum (leaf, stem, root), Solanum dulcamara (leaf, stem, root), Solanum villosum (leaf), Solanum melongena (leaf), Solanum quitoense (leaf), Solanum torvum (leaf). cDNA synthesis was performed according to the manufacturer’s instructions (High-Capacity cDNA Reverse Transcription Kit, Applied Biosystems™). Phusion High-Fidelity DNA Polymerase (New England Biolabs) was used for all PCR amplification steps according to the manufacturer’s instructions. Genes were directly cloned into pDBG3α1 vector using the One Step Cloning Kit (C112-02, Vazyme) according to manufactures instructions. Oligonucleotide primers were purchased from Sigma-Aldrich. DNA excised from agarose gels was purified using the Gel/PCR extraction kit (Hy-Labs). E. coli TOP10 cells (Invitrogen) were used for plasmid isolation before transformation into other heterologous hosts. Plasmid DNA was isolated from E. coli cultures using the Plasmid Mini Extraction Kit (Hy-Labs). For a list of primers used for cloning, see Supplementary Data 2.
Sequences of GAME8 genes from Jaltomata sinuosa, Iochroma cyaneum, Petunia inflata, Cestrum imbraticum, Lycium ruthenicum, and Anisodus acutangulus were synthesized by Twist Bioscience.
Mutagenesis of GAME8 genes
Sequences of SlGAME8 gene encoding protein with additional twelve amino acids from eggplant, SmGAME8 gene lacking nucleotides encoding twelve twelve-amino-acid acid loop, codon-optimized SmGAME8 used for overexpression in tomato hairy roots with knocked out SlGAME8 were ordered and synthesized by Twist Bioscience. For nucleotide and amino acid sequences, see Supplementary Data 1.
Studies of GAME8 activity in Nicotiana benthamiana
Tomato GAME4, GAME12, GAME11 genes were cloned into pDBG3α1 or pDBG3α2 vectors with SlUbi10 promoter and terminator using GoldenBraid cloning system24. GAME6, GAME15, and all GAME8 genes were directly cloned into pDBG3α1 (with SlUbi10 promoter and terminator) vector using the One Step Cloning Kit (C112-02, Vazyme) according to manufactures instructions. Constructs were transformed into Agrobacterium tumafaciens (GV3101) electrocompetent cells. Transformants were grown on LB plates containing 50 μg ml−1 kanamycin and 50 μg ml−1 gentamicin at 28 °C. Then, 10 ml of LB medium supplemented with antibiotics was inoculated with a single colony and grew O/N at 28 °C. Cells were centrifuged at 3000 × g for 10 min, and supernatant was removed. Pellet was resuspended in 5 ml of infiltration medium (100 mM MES buffer, 2 mM Na3PO4•12H2O, 100 μM acetosyringone) and centrifuged again. Pellet was resuspended again in 10 ml of infiltration medium and incubated at room temperature for 2 h. Agrobacterium suspensions consisting of one strain or multiple strains (OD600 = 0.3 for each strain) were infiltrated into the underside of N. benthamiana leaves with a needleless 1 ml syringe. Plants were grown for 4–5 weeks under a 16 h light cycle before infiltration. Leaves were harvested 4 days postinfiltration, frozen in liquid nitrogen, and stored at −80 °C for later processing. Biological replicates consisted of several leaves all from different tobacco plants. As negative controls, we performed two additional experiments, i.e., one in which we infiltrated only media and another in which we expressed all GAME genes except GAME8. In both cases, no SGA aglycones were detected in the control plants, confirming that GAME8 is essential for production and establishing the stereochemistry of these steroidal compounds (Fig. 8).
Isotopic labeling of 22S,26(27)-dihydroxycholesterol glucoronide
To confirm that GAME8 is hydroxylating cholesterol either in position C26 or C27 we used labeled cholesterol-d7 (all hydrogens at terminal methyl groups C26, C27, and in position C25 replaced with deuterium-2H). Nicotiana benthamiana plants were infiltrated with mix of A. tumefaciens cells transformed with mix of GAME genes (SlGAME15, SlGAME6, and SlGAME8 or SmGAME8). After 2 days post-infiltration, the same leaves were additionally infiltrated with an infiltration solution containing cholesterol-d7 at C = 100 μg/ml. After 3 days, tissue was collected and stored in −80° for LC-MS analysis. Conversion of cholesterol-d7 by GAME15 and GAME6 to 22S-hydroxycholesterol-d7 glucuronide (m/z = 584.4186) would allow GAME8 to further convert it to either 22S,26-dihydroxycholesterol-d7 glucuronide (SlGAME8, m/z = 599.4072) or 22S,27-dihydroxycholesterol-d7 glucuronide (SmGAME8, m/z = 599.4072). During this reaction on of the deuterium atoms would be replaced with a hydroxyl group, resulting in mass 1 Da smaller compared to hydroxylation product in any other position of 22S-hydroxycholesterol-d7 glucuronide (m/z = 600.4134).
Gene expression analysis: quantitative PCR of SnGAME8s and SdGAME8s
Expression levels of SnGAME8 and SdGAME8 were analyzed as described before24. Briefly, gene expression analysis was performed with three or more biological replicates for each plant tissue. RNA isolation was performed by the TRIZol method (Sigma-Aldrich). DNase I (Sigma-Aldrich)-treated RNA was reverse transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Gene-specific oligonucleotides were designed with Primer BLAST software (NCBI). The TRANSLATION ELONGATION FACTOR ALPHA 1 (EF1-α) gene was used as an endogenous control. Oligonucleotides used are listed in Supplementary Data 2.
Protein structure prediction and molecular docking
Protein 3D structure modeling was done with Alphafold258. Molecular docking of 22S-hydroxycholesterol glucuronide to enzymes active site was performed with Autodock Vina59,60. Structures of SmGAME8 and SlGAME8 were superimposed and visualized with Chimera28. Molecular interactions were predicted with LigPlot+29.
Generation of transgenic tomato hairy roots
Transgenic hairy roots were generated as described previously61. More details regarding plasmids used can be found in Supplementary Fig. 11. For identifying SmGAME8-overexpressing hairy roots we used GFP as a marker (Supplementary Fig. 11). To achieve efficient editing of SlGAME8 while still being able to overexpress the very similar eggplant gene, we used a codon-optimized SmGAME8 ORF that did not contain target sequences for gRNAs targeting SlGAME8.
Analysis of SGAs in wild tomato species
LC-MS analysis of α-tomatine isomers in wild tomato accessions was conducted as described before. For extraction and mass spec analysis, we used leaf tissue of following species grown in a greenhouse Solanum lycopersicum MT, Solanum lycopersicum M82, Solanum corneliomulleri, Solanum chmielewskii, Solanum pimpinellifolium, Solanum neoricki, Solanum arcanum, Solanum huaylasense, Solanum chilense, Solanum habrochaites, Solanum peruvianum, Solanum pennellii, Solanum cheesmaniae.
Accessions of Solanum cheesmaniae, Solanum galapagense, and Solanum pennellii were grown in a greenhouse. For full list of analyzed accessions, see Supplementary Data 3.
Analysis of SGAs in introgression lines (ILs) of tomato
Solanum pennellii introgression lines were grown in a growth room as described before. LC-MS analysis of α-tomatine isomers was performed as described in LC-MS analysis paragraph.
Analysis of S. cheesmaniae GAME8 sequences and their functional characterization
Gene cloning, sequencing, and functional characterization in N. benthamiana were performed as described before. Twenty-five accessions were selected for GAME8 amplification and sequencing (Supplementary Fig. 14). Selected GAME8s with different mutations were used for functional characterization and LC-MS analysis of SGA aglycone stereochemistry.
Analysis of the Ancestral State of GAME8
To investigate the evolutionary history of GAME8, we performed a comprehensive ancestral state reconstruction using multiple nucleotide sequences from diverse Solanaceae species. We compiled a dataset of GAME8 nucleotide sequences from various Solanaceae species and conducted multiple sequence alignments to identify conserved and variable regions. Alignments were performed using MUSCLE. Maximum Likelihood phylogenetic analysis was then conducted using MEGA11 with the best-fitting evolutionary model, allowing us to infer relationships between GAME8 homologs and identify key divergence events in the gene’s evolutionary trajectory. Ancestral sequence reconstruction was carried out in MEGA11, using ML-based inference to predict the most likely ancestral sequences at different evolutionary nodes. We focused on six major divergence points, selecting predicted ancestral GAME8 sequences from these nodes for further analysis. These reconstructed sequences were synthesized (Twist Bioscience) then tested in planta for their potential enzymatic activity to determine how stereospecificity evolved over time. To further validate the ancestral state of GAME8, we identified, synthesized, and cloned GAME8 sequences from early-diverging Solanaceae species, including Cestrum imbricatum, Lycium ruthenicum, Petunia inflata, and Anisodus acutangulus. These species represent lineages that split before the diversification of the Solanum genus. By expressing these genes and analyzing their enzymatic products, we aimed to determine whether they preferentially produce the 25S or 25R isomer.
Statistics and reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyzes and the experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Source data are provided with this paper.
References
Olmstead, R. G. & Bohs, L. A summary of molecular systematic research in Solanaceae: 1982-2006. Acta Hortic. 745, 255–268 (2007).
Barboza, G. E. et al. Solanaceae: Solanaceae Juss., Gen. pl.: 124 (1789), nom. cons. in Flowering Plants. Eudicots 295–357 (Springer International Publishing, Cham, 2016).
Wilf, P., Carvalho, M. R., Gandolfo, M. A. & Cúneo, N. R. Eocene lantern fruits from Gondwanan Patagonia and the early origins of Solanaceae. Science 355, 71–75 (2017).
Yang, S. et al. Distinction of chiral penicillamine using metal-ion coupled cyclodextrin complex as chiral selector by trapped ion mobility-mass spectrometry and a structure investigation of the complexes. Anal. Chim. Acta 1184, 339017 (2021).
Huang, J. et al. Nuclear phylogeny and insights into whole-genome duplications and reproductive development of Solanaceae plants. Plant Commun. 4, 100595 (2023).
Gal, J. The discovery of stereoselectivity at biological receptors: Arnaldo Piutti and the taste of the asparagine enantiomers–history and analysis on the 125th anniversary: Piutti, the asparagine enantiomers, sweet taste, and receptors. Chirality 24, 959–976 (2012).
Fouad, H A., de Souza Tavares, W. & Zanuncio, J C. Toxicity and repellent activity of monoterpene enantiomers to rice weevils (Sitophilus oryzae). Pest Manag. Sci. 77, 3500–3507 (2021).
Pinto, C., Cidade, H., Pinto, M. & Tiritan, M. E. Chiral flavonoids as antitumor agents. Pharmacology 14, 1267 (2021).
Oliveira, R. V., Simionato, A. V. C. & Cass, Q. B. Enantioselectivity effects in clinical metabolomics and lipidomics. Molecules 26, 5231 (2021).
Smith, R. L. & Mitchell, S. C. Thalidomide-type teratogenicity: structure-activity relationships for congeners. Toxicol. Res. 7, 1036–1047 (2018).
Melamud, A., Kosmorsky, G. S. & Lee, M. S. Ocular ethambutol toxicity. Mayo Clin. Proc. 78, 1409–1411 (2003).
Itkin, M. et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341, 175–179 (2013).
Wang, L.-H. et al. Review on toxicology and activity of tomato glycoalkaloids in immature tomatoes. Food Chem. 447, 138937 (2024).
Faria-Silva, C., de Sousa, M., Carvalheiro, M. C., Simões, P. & Simões, S. Alpha-tomatine and the two sides of the same coin: An anti-nutritional glycoalkaloid with potential in human health. Food Chem. 391, 133261 (2022).
Zhao, D.-K., Zhao, Y., Chen, S.-Y. & Kennelly, E. J. Solanum steroidal glycoalkaloids: structural diversity, biological activities, and biosynthesis. Nat. Prod. Rep. 38, 1423–1444 (2021).
Itkin, M. et al. GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 23, 4507–4525 (2011).
Umemoto, N. et al. Two cytochrome P450 monooxygenases catalyze early hydroxylation steps in the potato steroid glycoalkaloid biosynthetic pathway. Plant Physiol. 171, 2458–2467 (2016).
Nakayasu, M. et al. A dioxygenase catalyzes steroid 16α-hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiol. 175, 120–133 (2017).
Moehs, C. P., Allen, P. V., Friedman, M. & Belknap, W. R. Cloning and expression of solanidine UDP-glucose glucosyltransferase from potato. Plant J. 11, 227–236 (1997).
McCue, K. F. et al. Metabolic compensation of steroidal glycoalkaloid biosynthesis in transgenic potato tubers: using reverse genetics to confirm the in vivo enzyme function of a steroidal alkaloid galactosyltransferase. Plant Sci. 168, 267–273 (2005).
McCue, K. F. et al. The primary in vivo steroidal alkaloid glucosyltransferase from potato. Phytochemistry 67, 1590–1597 (2006).
McCue, K. F. et al. Potato glycosterol rhamnosyltransferase, the terminal step in triose side-chain biosynthesis. Phytochemistry 68, 327–334 (2007).
Akiyama, R. et al. The biosynthetic pathway of potato solanidanes diverged from that of spirosolanes due to evolution of a dioxygenase. Nat. Commun. 12, 1–10 (2021).
Jozwiak, A. et al. A cellulose synthase–like protein governs the biosynthesis of Solanum alkaloids. Science 386, eadq5721 (2024).
Särkinen, T., Bohs, L., Olmstead, R. G. & Knapp, S. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol. Biol. 13, 214 (2013).
Iijima, Y. et al. Steroidal glycoalkaloid profiling and structures of glycoalkaloids in wild tomato fruit. Phytochemistry 95, 145–157 (2013).
Patel, P. et al. Updates on steroidal alkaloids and glycoalkaloids in Solanum spp.: Biosynthesis, in vitro production and pharmacological values. in Bioactive Natural Products vol. 69 99–127 (Elsevier, 2021).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Laskowski, R. A. & Swindells, M. B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011).
Eshed, Y. & Zamir, D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162 (1995).
Štefka, J., Hoeck, P. E. A., Keller, L. F. & Smith, V. S. A hitchhikers guide to the Galápagos: co-phylogeography of Galápagos mockingbirds and their parasites. BMC Evol. Biol. 11, 284 (2011).
Pailles, Y. et al. Genetic diversity and population structure of two tomato species from the Galapagos Islands. Front. Plant Sci. 8, 138 (2017).
Kumar, S. et al. TimeTree 5: An expanded resource for species divergence times. Mol. Biol. Evol. 39, msac174 (2022).
Mori, K. Chirality in the natural world: chemical communications. Chirality Nat. Appl. Sci. 241, 260 (2002).
Lin, G., Cui, Y.-Y., Liu, X.-Q. & Wang, Z.-T. Species differences in the in vitro metabolic activation of the hepatotoxic pyrrolizidine alkaloid clivorine. Chem. Res. Toxicol. 15, 1421–1428 (2002).
Schramm, S., Köhler, N. & Rozhon, W. Pyrrolizidine alkaloids: biosynthesis, biological activities and occurrence in crop plants. Molecules 24, 498 (2019).
Reisenman, C. E., Christensen, T. A., Francke, W. & Hildebrand, J. G. Enantioselectivity of projection neurons innervating identified olfactory glomeruli. J. Neurosci. 24, 2602–2611 (2004).
Rice, J., Proctor, K., Lopardo, L., Evans, S. & Kasprzyk-Hordern, B. Stereochemistry of ephedrine and its environmental significance: Exposure and effects directed approach. J. Hazard. Mater. 348, 39–46 (2018).
Calf, O. W. et al. Gastropods and insects prefer different Solanum dulcamara chemotypes. J. Chem. Ecol. 45, 146–161 (2019).
Kintia, P. K. Chemistry and biological activity of steroid saponins from Moldovian plants. Adv. Exp. Med. Biol. 404, 309–334 (1996).
Christ, B. et al. Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat. Commun. 10, 1–11 (2019).
Porter, M. L. & Crandall, K. A. Lost along the way: the significance of evolution in reverse. Trends Ecol. Evol. 18, 541–547 (2003).
Barnett, J. R. et al. Evidence of fruit syndromes in the recently diverged wild tomato clade opens new possibilities for the study of fleshy fruit evolution. Plants People Planet 5, 948–962 (2023).
Fenstemaker, S., Sim, L., Cooperstone, J. & Francis, D. Solanum galapagense-derived purple tomato fruit color is conferred by novel alleles of the anthocyanin fruit and atroviolacium loci. Plant Direct 6, e394 (2022).
Li, N. et al. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nat. Genet. 55, 852–860 (2023).
Darwin, S. C., Knapp, S. & Peralta, I. E. Taxonomy of tomatoes in the Galápagos Islands: native and introduced species of Solanum section Lycopersicon (Solanaceae). Syst. Biodivers. 1, 29–53 (2003).
Pease, J. B., Haak, D. C., Hahn, M. W. & Moyle, L. C. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14, e1002379 (2016).
Schiffman, J. S. & Ralph, P. L. System drift and speciation. Evolution 76, 236–251 (2022).
Gibson, M. J., Torres, M. de L., Brandvain, Y. & Moyle, L. C. Introgression shapes fruit color convergence in invasive Galápagos tomato. Elife 10, e64165 (2021).
Jensen, P. H., Jacobsen, O. S., Henriksen, T., Strobel, B. W. & Hansen, H. C. B. Degradation of the potato glycoalkaloids–alpha-solanine and alpha-chaconine in groundwater. Bull. Environ. Contam. Toxicol. 82, 668–672 (2009).
Hennessy, R. C. et al. Discovery of a bacterial gene cluster for deglycosylation of toxic potato steroidal glycoalkaloids α-chaconine and α-solanine. J. Agric. Food Chem. 68, 1390–1396 (2020).
Song, F. et al. A novel endophytic bacterial strain improves potato storage characteristics by degrading glycoalkaloids and regulating microbiota. Postharvest Biol. Technol. 196, 112176 (2023).
Goulet, C. et al. Divergence in the enzymatic activities of a tomato and Solanum pennellii alcohol acyltransferase impacts fruit volatile ester composition. Mol. Plant 8, 153–162 (2015).
Jozwiak, A. et al. Plant terpenoid metabolism co-opts a component of the cell wall biosynthesis machinery. Nat. Chem. Biol. 16, 740–748 (2020).
Solovyev, V., Kosarev, P., Seledsov, I. & Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7, 1–12 (2006).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Eberhardt, J., Santos-Martins, D., Tillack, A. F. & Forli, S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python bindings. J. Chem. Inf. Model. 61, 3891–3898 (2021).
Ron, M. et al. Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 166, 455–469 (2014).
Acknowledgements
Molecular graphics and analyzes performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311. The seeds of Solanum cheesmaniae, S. galapagense and S. pennellii were developed by and obtained from the UC Davis/C.M. Rick Tomato Genetics Resource Center and maintained by the Department of Plant Sciences, University of California, Davis, CA 95616. We would like to thank Dr. Kate Ostevik for her valuable discussions on gene evolution and ancestral state reconstruction. A.A. is the incumbent of the Peter J. Cohn Professorial Chair. Adelis Foundation (AA). Leona M. and Harry B. Helmsley Charitable Trust (AA). Jeanne and Joseph Nissim Foundation for Life Sciences (AA). Tom and Sondra Rykoff Family Foundation (AA). Raymond Burton Plant Genome Research Fund (AA). National Center for Genome Editing in Agriculture grant 20-01-0209 (AA).
Author information
Authors and Affiliations
Contributions
A.J. and A.A. conceived of the project. M.A. perform work on Solanum pennellii, Solanum cheesmaniae, and Solanum galapagense. A.J., M.A., J.C., and S.P. were responsible for cloning. R.V. generated transgenic tomato hairy root cultures. M.P. generated transgenic Solanum nigrum plants. H.P. assisted with cultivation and analysis of wild tomato accessions. A.J., S.M., and I.R. were responsible for LC-MS analysis and metabolite identification. A.J. conducted all the remaining experiment and analyzed the data. A.J. and A.A. wrote the paper with the assistance and input of other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Zhenhua Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jozwiak, A., Almaria, M., Cai, J. et al. Enzymatic twists evolved stereo-divergent alkaloids in the Solanaceae family. Nat Commun 16, 5341 (2025). https://doi.org/10.1038/s41467-025-59290-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59290-4