Abstract
Channelrhodopsins (ChRs) are light-gated ion channels and invaluable tools for optogenetic applications. Recent developments in multicolor optogenetics, in which different neurons are controlled by multiple colors of light simultaneously, have increased the demand for ChR mutants with more distant absorption wavelengths. Here we report the 2.7 Å-resolution cryo-electron microscopy structure of a ChR from Klebsormidium nitens (KnChR), which is one of the most blue-shifted ChRs. The structure elucidates the 6-s-cis configuration of the retinal chromophore, indicating its contribution to a distinctive blue shift in action spectra. The unique architecture of the C-terminal region reveals its role in the allosteric modulation of channel kinetics, enhancing our understanding of its functional dynamics. Employing a rational approach, we developed mutants with blue-shifted action spectra. Finally, we confirm that UV or deep-blue light can activate KnChR-transfected precultured neurons, expanding its utility in optogenetic applications. Our findings contribute valuable insights to advance optogenetic tools and enable refined capabilities in neuroscience experiments.
Similar content being viewed by others
Introduction
Channelrhodopsins (ChRs) are light-gated ion channels in the eyespot of green algae. Channelrhodopsin-1 and -2, derived from Chlamydomonas reinhardtii (CrChR1 and CrChR2), were the first ChRs to be discovered and characterized, and the latter has been extensively utilized for optogenetic applications1. These proteins conduct cations such as H+, Na+, K+, and Ca2+2,. Cation channelrhodopsins with sequence homology to ChR2 are denominated as chlorophyte CCRs3. X-ray structures of a ChR chimera of C1C2 and CrChR2 have unveiled intricate details regarding the molecular architecture4, shedding light on the photoactivation and ion conduction pathway. ChRs have been employed to elicit action potentials in light-insensitive cells and tissues with unparalleled spatiotemporal precision, thereby launching a nascent research discipline, optogenetics3. ChR variants have been iteratively engineered to improve their functionalities5. Furthermore, genome mining has expedited the discovery of novel ChRs and their applications for optogenetics.
The mining and engineering of ChRs has facilitated multicolor optogenetics6,7,8,9,10,11,12,13,14,15,16,17. This technology leverages red and blue opsins to concurrently regulate two distinct neurons. It is particularly useful in synaptic communication, as an invaluable asset for probing the intricacies of complex neural circuits. Among the red opsins, ChrimsonR6 is commonly employed, and the recently discovered ChRmine18 harbors the potential to surpass ChrimsonR. In the realm of blue opsins, popular mutant variants of CrChR2 have been adopted5 alongside recently identified ChRs such as sdChR and Chronos6. Blue-shifted ChRs are crucial for coupling with an optical readout using for example a genetically encoded calcium sensor. Optical stimulation and readout must be well separated in their wavelengths to avoid interference.
A rational approach to creating color variants has been applied, producing the blue-shifted color variant C1C2GA19. The variant is based on the changes in the energy gaps between the stable state and the activated one, mostly caused by the twisting of β-ionone ring20. Former studies suggest that the rotation can yield a 40-degree rotation was proved to change the wavelength from 430 to 480 nm, which is a 7 kcal/mol energy change21. Additionally, an 11 kcal/mol blue shift was reported in a rotation from 0 (fully conjugated) to 90 (fully deconjugated) degrees21, and a 6 kcal/mol excitation energy increase resulting from 6-s-trans to 6-s-cis conformation were reported22. Nevertheless, many challenges remain for altering the conformation of retinal in closed state ChRs to achieve blue opsins in multicolor optogenetics.
To minimize crosstalk with red opsins or optical readouts, the activation of blue opsins with blue light (405 nm) is imperative9,17, as red opsins are less sensitive to this wavelength. This limitation may generate inadequate excitation by low levels of blue opsin expression, consequently impeding the utility of multicolor optogenetics. In light of these considerations, a blue opsin with shorter wavelength excitation is keenly desired for use in multicolor optogenetics.
Among the most pronounced blue-shifted ChRs is a recently discovered ChR derived from the filamentous terrestrial alga Klebsormidium nitens, designated as KnChR23. KnChR comprises a 7-transmembrane rhodopsin domain that contains a channel pore, followed by a C-terminal moiety that encodes a peptidoglycan binding domain known as FimV. KnChR exhibits higher Ca2+ permeability and about 10-fold higher metal cation permeability relative to H+, as compared with CrChR2. KnChR exhibits maximal sensitivities at 430 and 460 nm, with the former making KnChR one of the most notable blue-shifted ChRs to date, with the potential to serve as a pioneering framework for exploring the molecular mechanisms governing ChR color tuning. Notably, the rate of channel closure is impacted by the C-terminal moiety, and the truncation of this moiety led to over 10-fold prolongation of the channel’s open lifetime. Two pivotal arginine residues, R287 and R291, play a crucial role in modulating the photocurrent kinetics. However, little is known about how the C-terminal residues modulate the channel activity and the mechanisms governing blue-shifted excitation, as well as their optogenetic utility. Here, we perform structural and functional analyses of knChR, illuminating how the blue-shifted excitation occurs and the influence of the C-terminal on channel kinetics.
Results
Structure determination
To examine the process of the blue-shifted excitation and channel modulation, we performed a cryo-electron microscopy (cryo-EM) single-particle analysis of KnChR. We used KnChR-697 (Supplementary Fig. 1a), which consists of 697 amino acids, including the long C-terminal region with the modulation site of the channel activity and the FimV domain. First, we expressed KnChR-697 in insect cells and purified it in detergent micelles (Supplementary Fig. 1b). However, during the cryo-EM analysis, only the top view was visible in the 2D class averages (Supplementary Fig. 1c). This limitation was attributed to either the orientation bias or the deleterious effects of the micelles. Consequently, we reconstituted purified KnChR into MSP2N2 nanodiscs (Supplementary Fig. 2a) and performed the cryo-EM structural analysis. The nanodisc reconstitution eliminated the orientation bias, and the side view became visible (Supplementary Fig. 2b). Finally, we determined the structure of KnChR in the ground state at a nominal resolution of 2.7 Å (Fig. 1a, Supplementary Table 1, Supplementary Fig. 2b).
a Overall structure of the nanodisc-reconstituted KnChR dimer. The cryo-EM density maps are shown with the two protomers in different colors. b Ribbon representation of KnChR. Each KnChR protomer is depicted in a cartoon model, with the N-terminal region in blue and the C-terminal region in red. The intermolecular disulfide bridge, formed by C28, is shown as a yellow stick. c Superimposition of KnChR and CrChR2 (PDB ID: 6EID). d Characteristic interactions in the N-terminal region.
Like other chlorophyte CCRs, KnChR forms a dimer and comprises transmembrane helices24 (TMs) 1–7, as well as N- and C- terminal regions (Fig. 1b). We successfully modeled residues 27 to 291. Notably, the C-terminal FimV domain was disordered (Supplementary Figs. 1a, 2b, 3a–d), indicating its inherent structural flexibility. This observation aligns with the previous report23 that the FimV truncation does not alter the channel activity. The retinal chromophore is covalently bound to K254, forming the retinal Schiff base (RSB)2. The local resolution surrounding the central retinal is ~2.6 Å, while the N- and C-terminal regions exhibit a comparatively lower resolution of 3.8 Å (Supplementary Fig. 3a, b). The rhodopsin domain of KnChR has 34.5% identity and 69.3% homology with CrChR2 and superimposed well on CrChR24 (PDB ID: 6EID), with a root mean square deviation of 1.15 Å4 (Fig. 1c). The N-terminal region extends to the center of the dimer interface, and C28 of each protomer forms a disulfide bond with its counterpart. These structural features represent the typical architecture of chlorophyte CCRs2 (Supplementary Figs. 3b, 4a–e)4,19,25,26.
Remarkably, in contrast to other ChRs25,26, the N-terminal region adopts an elongated conformation and does not contain any α-helices (Supplementary Fig. 4a–e), and is stabilized by extensive electrostatic interactions between histidine residues (H37, H38, H41, and H99) and two glutamate residues (E39, E42, and E240) (Fig. 1d). Moreover, aromatic residue clusters constitute the dimer interface under the C28-C28 disulfide bond (Supplementary Fig. 3b). These structural features of the N-terminal region are unique to KnChR.
Process of blue-shifted excitation
We first attempted to model all-trans retinal (ATR) into the density (Fig. 2a) but encountered difficulty in confidently fitting the β-ionone ring. Alternatively, 6-s-cis retinal was clearly fitted into the density (Fig. 2b, c, Supplementary Fig. 5a, b). In ATR, the β-ionone ring lies in the same plane as the polyene chain, due to the fixed planar conformation of the C6–C7 bond (Fig. 2b, d). Conversely, in 6-s-cis retinal, the β-ionone ring undergoes a rotation of ~140°, enforced by torsion around the C6–C7 bond, thus deviating from planarity2,19,27 (Fig. 2a, c).
Chemical structure of the chromophore, RSB, and the atom numbering of ATR (a) and 6-s-cis retinal (b). c–f, Close-up views of the chromophore binding pocket around the β-ionone ring. Surrounding residues are shown as sticks with transparent cpk models. Distances between retinal and I158 in KnChR (g), I160 in CrChR2 (PDB ID: 6EID) (h), and I199 in C1C2GA (PDB ID: 4YZI) (i). Double-ended arrows indicate the distances. j Superimposition of KnChR, CrChR2 (PDB ID: 6EID), and C1C2GA (PDB ID:4YZI), focused on the interaction between retinal and I158 in KnChR.
Among ChRs, the C6–C7 bond is known to be easily twisted. In CrChR2, the C17 methyl group plunges into the cavity between T159 and G163 (Fig. 2d). Instead of the G163 in CrChR2, the G202A mutation in the engineered mutant C1C2GA (PDB ID:4YZI)19 and A140 in HcKCR2 (PDB ID: 8H87)26 force the retinal to twist (6-s-cis retinal configuration) (Fig. 2e, f), to avoid the conflict with the C17 methyl group in ATR, thus referred to as “Non-G rule25”. KnChR also follows this rule, and A161 can enforce the torsion around the C6–C7 bond (Fig. 2c), shrinking the π-conjugation, and accordingly leading to a blue shift of the absorption spectra of KnChR.
To further support the 6-s-cis retinal configuration in KnChR, we made the double mutant A157T/A161G, replacing these residues with their homologs in CrChR2. In our patch-clamp recordings, this double mutant exhibited a 20-nm red shift in its action spectrum (λmax = 470 nm) compared with the wild type (λmax = 450 nm) (Supplementary Fig. 6), indicating that the A157T/A161G mutation reverts the 6-s-cis conformation toward 6-s-trans. These data suggests that both A157 and A161 play crucial roles in stabilizing the 6-s-cis conformation and thereby imparting the pronounced blue-shift in KnChR.
In the rotated conformation, the C18 methyl group plunges into the cavity between P224 and F227 (Fig. 2c), instead of the C16 methyl group in CrChR2 (Fig. 2d). The rotation also moves the C17 methyl group to the cavity of the C18 methyl group in ATR. The rotation of the C17 methyl group thus avoids steric conflict between the C17 methyl group and A161. On the other hand, the side chain of A157 in KnChR fills less cavity than T159 in CrChR2 and thus avoid conflict with the C16 methyl group (Fig. 2c), as T198G in C1C2GA (Fig. 2e) and A136 in HcKCR2 (Fig. 2f).
Upon comparing the other residues that constitute the retinal binding site (Supplementary Fig. 7a, b), I158 in KnChR exhibited a unique structural feature (Fig. 2g), although it is highly conserved among the chlorophyte CCRs. In CrChR2, the distance between the homologous I160 and the retinal is 5.5 Å (Fig. 2h)1, suggesting the absence of a direct interaction. This holds true for C1C2GA with the 6-s-cis retinal configuration (Fig. 2i)19. However, in KnChR, the I158 side chain diverges more toward the 6-s-cis retinal (Fig. 2g), resulting in a distance of 3.8 Å to C9 of the retinal, indicative of a direct interaction (Fig. 2j). While the underlying cause of this feature remains elusive, it may be attributed to structural disparities specific to KnChR. The additional van der Waals interaction with the retinal is hypothesized to result from an electronic effect between an occupied orbital in the CH3 group of the isoleucine and an empty π-orbital with a large lobe on C9 (Fig. 2g). This interaction is attributed to steric hindrance, which typically influences both ground and excited states equally. Consequently, it is postulated that this interaction elevates the energy level of the excited state, leading to a larger energy gap between the ground and excited states and thereby inducing the observed blue shift.
Ion pathway
In chlorophyte CCRs, the ion pathways are formed by TM2, 3, 6, and 7 and are constrained by three constriction sites that limit ion permeation: the intracellular (ICS), central part of the membrane (CCS), and extracellular (ECS) constriction sites. The ion pathway in KnChR adheres to this pattern (Fig. 3a), except for the ECS. The extracellular volume (EV) in KnChR is not divided by the ECS, in contrast to CrChR2. In CrChR24, Q117, R120, and H249 primarily constitute the ECS, dividing the extracellular volume (EV) into EV1 and EV2 (Fig. 3b). In KnChR, Q117 is replaced with P115, resulting in a unified EV (Fig. 3a). The single EV is similarly observed in C1C2, indicating the ion pathway for the extracellular space. One of the prominent differences between the structures of KnChR and CrChR2 is the interaction around E94 (E97 in CrChR2) (Fig. 3b). Both glutamate residues form electrostatic interactions with lysine residues K90 and K93 in KnChR and CrChR2, respectively, and form water-mediated hydrogen bonds with arginine residues R118 and R120 in KnChR and CrChR2, respectively. Furthermore, in KnChR, E94 forms a direct hydrogen bond with Y239, forming an extensive hydrogen bonding network with surrounding water molecules (Fig. 3b, Supplementary Fig. 3d). Taken together, E94 in KnChR has more intramolecular interactions than E97 in CrChR2. The KnChR E94 mutation retarded the on- and off-kinetics in the patch clamp measurement (Fig. 3c), whereas minimal kinetic effects were reported for the ChR2 E97 mutation28. These results highlight the distinct role of KnChR E94 in the channel gating.
a Water accessible cavities are illustrated in the KnChR structure, showing the putative ion pathway. The three constriction sites for the inner, central, and extracellular gates are enclosed within squares. Water molecules are indicated by red spheres. b Comparison of the constriction sites of KnChR (left panels) and CrChR2 (PDB ID: 6EID) (right panels), for the inner (upper panels), central (middle panels), and outer (lower panels) gates. The constituent residues are shown as sticks. Water molecules are indicated by red spheres. Black dashed lines indicate hydrogen bonds. c Representative photocurrent traces of the C-terminal truncated variant of KnChR (KnChR-272) and mutants. We used KnChR-272 as a template for the mutant, because KnChR-272 exhibits higher channel activity than KnChR-697 owing to the absence of the C-terminal region. The cells were illuminated with light (λ = 470 nm) during the time indicated by blue bars. The membrane voltage was clamped from −60 to +40 mV for every 20-mV step. d Comparison of photocurrent amplitudes of KnChR-272 and mutants at −60 mV. In the bar graph, gray bars indicate the amplitude from the peak photocurrent (Ip), and open bars indicate the amplitude from the steady-state photocurrent (Iss). n = 6 cells. Source data are provided as a Source Data file. e The channel-closing kinetics of KnChR-272 and mutants after illumination cessation (τ-off) at −60 mV. n = 6 cells. All data in (d, e) are expressed as mean ± SEM and were evaluated with the Mann-Whitney U test for statistical significance. It was judged as statistically insignificant when p > 0.05. *p < 0.05, **p < 0.01 *** p < 0.005. Source data are provided as a Source Data file.
At the CCS part, ChR2 forms a characteristic hydrogen bonding network with E90, K93, E123, and D253, whereas only Y91 interacts with D250 in KnChR4. In the central part of KnChR, two carboxylates E121 and D250 serve as counterions, thereby stabilizing the positive charge of the RSB (Fig. 3b), as in other chlorophyte CCRs. In CrChR2, E90, one of the key determinants of H+ selectivity, forms water-mediated hydrogen bond interactions with E123 and D253, constituting the CCS4. In KnChR, the side chain of the homologous residue E87 diverges from the counterions and engages in direct and water-mediated hydrogen bonds with N255 (Fig. 3b, Supplementary Fig. 3d). The KnChR E87Q mutant exhibits slower channel-closure kinetics in the patch-clamp measurement (Fig. 3c), suggesting that E87 is responsible for the reprotonation of the RSB.
On the intracellular side of KnChR, E79, H261, and R265 form the ICS (Fig. 3b), dividing the intracellular volume (IV) into two discrete parts (IV1 and IV2) as in CrChR24. IV1 is further divided into IV1 and IV1” by E79 and V80. The E79Q mutation slightly retarded the off-kinetics in the patch-clamp measurement (Fig. 3c), indicating that this residue affects the channel gating. IV2 is smaller in KnChR compared to CrChR24, because of the electrostatic interaction above IV2, featuring E139, E269, and K272. Thus, the differences in ICS, CCS, and ECS between KnChR and CrChR2 (Fig. 3a, b) are likely to contribute to the distinct ion permeabilities of these channels.
Insight into the C-terminal region
We illuminated the density corresponding to the C-terminal region modulates the channel activity (Supplementary Fig. 3a–c) and modeled the residues up to R291 (Fig. 4a, b). The C-terminal region extends clockwise from TM7 to TM5, forming extensive interactions with TM5 (Fig. 4c, d), a unique structural feature in ChRs (Supplementary Fig. 4f–j)4,19,25,26. Among the C-terminal residues, R287 exhibits the most significant impact on channel modulation, since the R287A mutation prolongs the lifetime of the channel-opening and increases the current amplitude23. According to the density map, there are two possible positions of R287, forming electrostatic interactions with E196 and E285 (Fig. 4c). Thus, we measured the photocurrents of the R287E and E196R mutants, in which the electrical charge is inverted (Fig. 4e). The photocurrent amplitude of the R287E mutant was elevated by ~220%, and the kinetics of channel-closing (τoff) slowed down from 25 ms to 160 ms, confirming the importance of the residue for channel gating (Fig. 4f, g). By contrast, only a small effect on τoff was observed in the E196R mutant, although the current amplitude was increased (Fig. 4f–h). These results indicate that not only the electrostatic interaction but also a polar interaction could be responsible for the channel modulation.
a Water accessible cavities, viewed from the intracellular side. The C-terminal region is highlighted. b The residues in the C-terminal region are shown as sticks with transparent CPK models. c Close-up view of the c-terminal region. Based on density, R287 was modeled in two ways, including an alternative conformation. Black two-way arrows indicate distances between two atoms. d Close-up view of R291 and POPC. Black dashed lines indicate hydrogen bonds. e Representative photocurrent traces of KnChR-697 and mutants. The cells were illuminated (λ = 470 nm) during the time indicated by blue bars. The membrane voltage was clamped from −60 to +40 mV for every 20-mV step. f Comparison of photocurrent amplitudes of KnChR-697 and mutants at −60 mV. In the bar graph, gray bars indicate the amplitude from the peak photocurrent (Ip), and open bars indicate the amplitude from the steady-state photocurrent (Iss). n = 5 to 6 cells. Source data are provided as a Source Data file. g The channel-closing kinetics of KnChR-697 and mutants after illumination cessation (τ-off) at −60 mV. n = 5 to 6 cells. All data in (f, g) are expressed as mean ± SEM and were evaluated with the Mann-Whitney U test for statistical significance. It was judged as statistically insignificant when p > 0.05. It was judged as statistically insignificant when p < 0.05. *p < 0.05, **p < 0.01 *** p < 0.005. Source data are provided as a Source Data file.
Compared to R287, R291 is exposed to the membrane environment and poorly interacts with the other part of KnChR (Fig. 4c), while the R291A mutation also prolongs the lifetime of the channel-opening and increased the current amplitude. However, R291 interacts with both the head group and ester region of 1-Palmitoyl-2-oleoylphosphatidylcholine (POPC), which is located outside of TM5 and TM6 (Fig. 4d, Supplementary Fig. 3c). The interaction of R291 with the lipid may anchor the C-terminal region, which is essential for the channel modulation.
Moreover, Y282 forms van der Waals interaction with R287 and the main chain of F199 (Fig. 4c). The Y282A mutant exhibited a relatively small but a significant change in τoff. These interactions between the C-terminus and TM5 would be involved in channel modulation.
Overall, the C-terminal region faces another spacious IV, rather than IV2 (Figs. 3a, 4a). This voluminous IV is also present in CrChR2, but is not involved in the ion pathway (Fig. 3b)4. This observation implies that it exerts allosteric modulation, potentially curtailing conformational shifts and facilitating rapid inactivation in response to light. This interaction of the C-terminal region with TM5 distinguishes KnChR from other ChRs, and thus represents a unique structural feature (Supplementary Fig. 4f–j)4,19,25,26.
Blueshift color-tuning and optogenetic characterization
ChRs with shorter wavelength excitation are optimal for multicolor optogenetics with dual light applications. Consequently, we explored blue-shifted mutants of KnChR (Fig. 5a, Supplementary Fig. 8a–c). Initially, we focused on A157/A161, which are responsible for the 6-s-cis configuration (Fig. 5b). We surmised that the A157G mutant, which mimics the T198G/G202A mutation in C1C2GA, would reduce the steric interference with the C16 methyl group and further favor the 6-s-cis conformation. Consistently, the A157G mutation (KnChortRG) showed a 10-nm blue-shifted action spectrum (λmax= 440 nm) in the patch-clamp recording, as compared with that of the wild-type (λmax= 450 nm) (Fig. 5a, c).
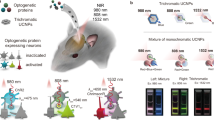
a The λmax values of KnChR-272 and KnChR mutants. We used KnChR-272 as a template for the mutant, because KnChR-272 exhibits higher channel activity than KnChR-697 owing to the absence of the C-terminal region, which is suitable for the optogenetic application. b Close-up view of I129 and surrounding residues, shown as sticks. c Action spectra of I129Q and A157G. The wavelength dependency of the photocurrent from KnChR-272 (black) and mutants (blue) is depicted. Membrane voltage was clamped at −60 mV. n = 5 to 6 cells. d Representative responses of a KnChR-272 expressing neuron. The action potentials evoked by 385, 423, 469, and 511 nm wavelengths of light. The 50 ms light pulse was performed as indicated by the colored time points. Light intensities were 1.4 mW/mm2. e KnChR-272 expressing neurons were excited by four wavelengths of light at the same light intensities. To compare the success rate of optical stimulation, spike probabilities (total numbers of evoked action potentials / total numbers of light stimulation) are compared. n = 5 cells. Data in (e) are expressed as mean ± SEM. Source data are provided as a Source Data file.
Subsequently, we focused on the L/Q switch, which is known for its role in color-tuning29. For the proteorhodopsin group, the presence of leucine and glutamine at position 105, referred to as the L/Q switch, regulates the spectra between the green- and blue-absorbing types, respectively. The homologous residue I129 in KnChR is surrounded by bulky hydrophobic residues and is positioned proximal to the Schiff base, at a distance of 4.9 Å (Fig. 5b). The I129Q mutation (KnChortRQ) showed a 30 nm blue-shifted action spectrum (λmax = 420 nm) (Fig. 5a, c). I129Q could form a polar interaction with the Schiff base and thereby lower the energy at the ground state, which would form the basis of this blue-shift.
We finally tested the optogenetics applicability of KnChR, particularly with short-wavelength light. Precultured cortical neurons were transfected with KnChR, and optical stimulations were performed. As shown in Fig. 5d, 50 ms light-pulse successfully triggered action potentials. As expected from the action spectrum of KnChR, 423 and 469 nm light are most effective, exhibiting ~90–100% of spike probability (Fig. 5e). But less probability was observed with 511 nm light (50%), which is consistent with action spectrum exhibiting less activity at 510 nm (Fig. 5c). Notably, 385 nm light was able to excite action potentials with relatively good efficiency (70%). These results indicate that KnChR could be used as an optogenetics tool, particularly with UV or deep-blue light. The action spectra of KnChR shows essentially no activity at longer wavelength light than 550 nm (Fig. 5c). These demonstrations above indicate that coupling KnChR with an optical readout such as R-GECO or RCaMP (excitation at ~575 nm) would be feasible30.
Discussion
In this study, we performed structural and functional analyses of the blue-shifted channelrhodopsin KnChR. Compared to typical channels and GPCR-G-protein complexes31, the cryo-EM structural analysis of KnChR was challenging, because KnChR has fewer features such as large domains in the outer membrane region. We succeeded in the structural determination by using nanodiscs32. The distinctive C-terminal region covers the intracellular loops of the protein, implying the potential allosteric modulation of conformational changes upon light irradiation. Furthermore, our findings revealed that the 6-s-cis retinal configuration serves as a key determinant of blue-shifted excitation. Employing a rational design approach, we created the KnChR mutants with the most pronounced, blue-shifted excitation (KnChortRG and KnChortRQ), thus harboring the potential for efficacious dual-light utilization in optogenetics. This engineering strategy holds promise for applications to other ChRs, thereby extending the horizons of optogenetic utility.
For cryo-EM studies, nanodiscs are frequently used for the structural determination of ion channels and transporters. In general, nanodisc reconstitution diminishes the unwanted densities of lipids and detergents, while concurrently improving the protein stability. This enhancement facilitates particle alignment during the cryo-EM data processing. This methodology has also been effectively utilized for determining the structures of relatively small transporters. For microbial rhodopsins, nanodisc reconstitution has been used in the cryo-EM structural analyses of the ChRmine trimer33 and the Kin4B8 pentamer34. We adopted this approach for KnChR, and ultimately determined the cryo-EM structure of the nanodisc-reconstituted KnChR. It is noteworthy that the molecular weight of the KnChR dimer, as extrapolated from the structural model, is 60.8 kDa, underscoring its status as one of the smallest membrane proteins for which cryo-EM structures have been determined. We initially evaluated the nanodisc reconstitution efficiency on a small scale with various membrane scaffold proteins (MSPs) (Supplementary Fig. 2a). The results showed that MSP2N2 is the best for the reconstruction, as it creates nanodiscs with a larger diameter than those of MSP1E3, MSP1E3D1, and MSP1D135. Since MSP1E3D1 was previously employed in the structural analysis of the ChRmine trimer, it is somewhat remarkable that MSP2N2, boasting a larger diameter, successfully facilitated the structure determination of the KnChR dimer. Accordingly, in the structural analyses of moderately-sized membrane proteins, the selection of the appropriate type of MSP warrants careful consideration.
X-ray crystallography has been the pioneering method for the structural elucidations of microbial rhodopsins since the late 1990s36. Subsequently, the architectures of GPR37, ChRmine33, Kin4B834, and GtCCRs38 have been determined by cryo-EM, a technique that obviates the necessity for crystallization. Notably, the success of this structural analysis of KnChR indicates that the structural examination of all microbial rhodopsins has become fundamentally feasible, given their propensity to form dimers or additional multimeric configurations. Specifically, several X-ray crystal structures of dimeric ChRs used in optogenetics have reported24,39, yet a plethora of structures remain unsolved, encompassing optogenetically useful variants, disparate subtypes within a family, and ChRs engineered by machine learning. The acquisition of these structural data would substantively aid in deciphering and engineering the multifaceted diversity of ChRs.
Methods
Ethical approval
This study was carried out in accordance with regulations by the institutional animal care and use committee of the Nagoya Institute of Technology (approval number: 2021009). All methods are reported in accordance with the ARRIVE guidelines.
Expression of KnChR for structure analysis
The KnChR gene was subcloned into a pFastBac vector, modified to include a C-terminal EGFP-His8 tag, followed by a tobacco etch virus (TEV) protease recognition site. The construct was expressed in sf9 cells using the pFastBac baculovirus system. The baculovirus was used to infect the sf9 cells when the cells were grown in suspension to a density of 3.5 × 106 cells mL−1, and the cells were then cultured at 27 °C for 42 h. All-trans-retinal was supplemented to a final concentration of 10 µM in the cell medium, 24 h after the infection.
Purification of KnChR with GDN
The cells were harvested by centrifugation at 5,000 × g for 12 min. The pellets were first disrupted in solubilization buffer, containing 150 mM NaCl, 20 mM Tris-HCl, pH 8.0, 10% glycerol, 5.2 μg/ml aprotinin, 2.0 μg/ml leupeptin, 1.4 μg/ml pepstatin A, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1% n-Dodecyl-β-D-maltoside (DDM) (Calbiochem), and 0.2% cholesteryl hemisuccinate (CHS), using a glass dounce homogenizer. Both the supernatant and insoluble fractions were then collected and solubilized in the same solubilization buffer for 30 min at 4 °C. The supernatant was separated from insoluble material by ultracentrifugation at 180,000 × g for 20 min, and incubated with TALON resin (Clontech) for 30 min. The resin was washed with ten column volumes of buffer, containing 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.05% glyco-diosgenin (GDN) (Anatrace), and 15 mM imidazole. The receptor was eluted in buffer, containing 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.05% GDN, and 200 mM imidazole. TEV protease was added to the eluate, and the mixture was dialysed against buffer (20 mM Tris–HCl, pH 8.0, 300 mM NaCl, and 0.01%GDN). The cleaved GFP and the protease were removed with Ni-NTA superflow resin (QIAGEN). The flow-through fraction was collected and concentrated using an Amicon Ultra 100 kDa molecular weight cutoff centrifugal filter unit (Merck Millipore). The concentrated sample was then subjected to size-exclusion chromatography on a Superose™ 6 Increase 10/300 GL column (GE Healthcare Life Sciences), in buffer containing 150 mM NaCl, 20 mM Tris-HCl, pH 8.0, and 0.01% GDN. Peak fractions were pooled and frozen in liquid nitrogen (Supplementary Fig. 1b).
Purification of KnChR with LMNG
We performed detergent screening to enhance the purification efficiency, and found that Lauryl Maltose Neopentyl Glycol (LMNG) (Anatrace) is best for KnChR solubilization. After the cells were harvested, the pellets were disrupted in solubilization buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 5.2 μg/ml aprotinin, 2.0 μg/ml leupeptin, 1.4 μg/ml pepstatin A, 1 mM PMSF, 1% LMNG, and 0.2% CHS, using a glass dounce homogenizer. Both the supernatant and insoluble fractions were then collected and solubilized in the same solubilization buffer for 2 h at 4 °C. The supernatant was separated from insoluble material by ultracentrifugation at 180,000 × g for 20 min, and incubated with GFP nanobody-coupled resin at 4 °C for 2 h. To cleave the tags, TEV protease was added, and the sample was rotated for 12 h at 4 °C. The collected eluate was reloaded on the Ni-NTA superflow resin, with 20 mM imidazole, to trap the cleaved EGFP-His tags and the overloaded TEV protease. The flow-through fraction was collected, concentrated, and then loaded onto a Superose™ 6 Increase 10/300 GL column, in buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 0.01% LMNG. Protein-containing fractions were pooled and frozen in liquid nitrogen.
Nanodisc reconstitution
To determine whether KnChR can be reconstituted in nanodiscs, the protein in LMNG micelles was mixed with a film of lipid SoyPC (initially dissolved in chloroform, L-a-Phosphatidylcholine (40% Lecithin) Soy Extract (40%) Lot#: SPC40-10 Avanti Order #: 179171) resuspended with 1% cholic acid and several MSPs, at a molar ratio of 1:200:4, respectively, and incubated at 4 °C for 1 h. The LMNG detergent was removed by adding Bio-Beads SM2 (Bio-Rad) to 40 mg mL−1, followed by gentle agitation. The Bio-Beads were replaced with fresh ones after 2 h, and this second batch (equal amount) was incubated overnight at 4 °C. The Bio-Beads were then removed, and the sample was purified by size-exclusion chromatography on a Superose™ 6 Increase 10/300 GL column, equilibrated with buffer containing 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl (Supplementary Fig. 2a).
For the cryo-EM grid preparation of the KnChR, the protein was reconstituted in nanodiscs composed of SoyPC and 2N2, as described above. After the Bio-Beads were removed, the solution was ultracentrifuged and then purified by size-exclusion chromatography. The peak fractions of the protein were collected and concentrated to 4 mg mL−1, using an Amicon Ultra 100 kDa molecular weight cutoff centrifugal filter unit.
Sample vitrification
A 3 µl portion of the sample was loaded onto glow-discharged holey carbon grids (Quantifoil Cu/Rh 300 mesh R1.2/1.3), which were then plunge-frozen in liquid ethane, using a Vitrobot Mark IV (Thermo Fischer Scientific).
For KnChR purified in GDN, data collections were performed on a 200 kV Talos Arctica G2 microscope (Thermo Fisher Scientific) equipped with a BioQuantum K2 imaging filter and a Falcon 3EC direct electron detector (Gatan), using the EPU software (Thermo Fisher’s single-particle data collection software). Images were obtained at a dose rate of about 71.939 e Å−2, with a defocus range from −0.8 to −1.8 μm. The total exposure time was 4.25 s, with 50 frames recorded per micrograph. A total of 5022 videos were collected. All acquired movies in super-resolution mode were binned by 2, dose-fractionated, and subjected to beam-induced motion correction implemented in cryoSPARC v3.340.
For KnChR reconstituted in nanodiscs, data collections were performed on a 300 kV Titan Krios microscope (Thermo Fisher Scientific) equipped with a BioQuantum imaging filter and a K3 direct electron detector (Gatan), using the EPU software (Thermo Fisher’s single-particle data collection software). Images were obtained at a dose rate of about 49.26 e Å−2, with a defocus range from −0.6 to −1.6 μm. The total exposure time was 2.11 s, with 48 frames recorded per micrograph. A total of 8554 videos were collected. All acquired movies in super-resolution mode were binned by 2, dose-fractionated, and subjected to beam-induced motion correction implemented in cryoSPARC v3.340.
Cryo-EM single particle analysis
The contrast transfer function (CTF) parameters were estimated by using patch CTF estimation in cryoSPARC v3.340. Particles were initially picked from a small fraction with Gaussian blob picking and subjected to 2D classification.
For KnChR purified in GDN, class averages showing reasonable features of the KnChR dimer in various orientations were selected as templates, and particles were then picked using the templates. For each full dataset, extracted particles were down sampled to 3.32 Å, followed by several rounds of 2D classification to remove ‘junk’ particles. Particles from these class averages generated an Ab-initio model in cryoSPARC. Non-uniform refinement with the ab initio model as a reference was performed using C2 symmetry. Finally, the 87,443 particles in the best class were reconstructed using non-uniform refinement, resulting in a 3.45 Å resolution reconstruction, with the gold-standard Fourier shell correlation (FSC = 0.143) criteria. The processing strategy is described in Supplementary Fig. 1c.
For KnChR reconstituted in nanodiscs, class averages showing reasonable features of the KnChR dimer in various orientations, especially those with side views, were selected as templates for the training of the Topaz particle picking program41 and particles were then picked accordingly. Particles from these class averages were used to generate an Ab-initio model in cryoSPARC. For each full dataset, extracted particles were down-sampled, followed by several rounds of heterogeneous refinement to remove ‘junk’ particles. Multiple rounds of Local CTF refinement and Non-uniform refinement were performed using cryoSPARC. Finally, the 164,260 particles in the best class were reconstructed using Non-uniform refinement and local refinement, resulting in a 2.69 Å resolution reconstruction, with the gold-standard Fourier shell correlation (FSC = 0.143) criteria. The processing strategy is described in Supplementary Fig. 2b.
Model building and refinement
The quality of the map with the nanodisc-reconstituted protein was sufficient to build a model manually in Coot. The model building was facilitated by the AlphaFold-predicted42,43,44 KnChR model. We manually readjusted the model into the density map using Coot and refined it using phenix.real_space_refine (v.1.19)45,46. The final model of KnChR contained residues 27–291, retinal, 20 water molecules (Supplementary Table 1, Supplementary Fig. 3c), and one POPC (atoms C1–C22). Detailed parameters are listed in Supplementary Table 1. The 3D map was sharpened using Servalcat47 and the final model statistics were validated using comprehensive validation (cryo-EM) in PHENIX and provided in Supplementary Table 1. Structural analysis and figure preparations were performed with the USCF ChimeraX (V1.6.1)48 and CueMol (Ver2.2.3.443) software.
Expression plasmid for electrophysiology
The pKnChR-eYFP plasmid for expression in mammalian cells was described previously23. Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Agilent, CA, USA) or KOD -Plus- Mutagenesis Kit (Toyobo, Osaka, Japan). All constructs were verified by DNA sequencing (Fasmac Co., Ltd., Kanagawa, Japan).
Virus preparation
The AAV7m8 KnChR-Venus vector was produced by VectorBuilder (Yokohama, Japan).
Cell culture
ND7/23 cells, a hybrid cell line derived from neonatal rat dorsal root ganglia neurons fused with mouse neuroblastoma cells, were grown on collagen-coated coverslips in Dulbecco’s modified Eagle’s medium (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan), supplemented with 2.5 μM all-trans retinal and 5% fetal bovine serum, under a 5% CO2 atmosphere at 37 °C. The expression plasmids were transiently transfected by using FuGENE HD (Promega, Madison, WI, USA), according to the manufacturer’s instructions. Electrophysiological recordings were then conducted 16–36 h after the transfection. Successfully transfected cells were microscopically identified by eYFP fluorescence prior to the measurements.
Cortical neurons were isolated from embryonic day 16 Wistar rats (Japan SLC, Inc., Shizuoka, Japan) using Nerve-Cells Dispersion Solutions (Fujifilm Wako Pure Chemical Corporation) according to the manufacturer’s instructions, and grown in the neuron culture medium (FUJIFILM Wako Pure Chemical Corporation) under a 5% CO2 atmosphere at 37 °C. Cultured cortical neurons were infected at day in vitro (DIV) 7, using an adeno-associated virus (1 × 1010 GC/mL). Viral dilutions (1 μL) were added to cultured cortical neurons seeded on coverslips in 24-well plates, which were incubated at 37 °C. At DIV21, electrophysiological recordings were conducted using neurons identified by fluorescence under a conventional epifluorescence system.
Electrophysiology
All experiments were performed at room temperature (23 ± 2 °C). For whole-cell voltage clamp recording, photocurrents were recorded by using an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA, USA). Data were filtered at 5 kHz, sampled at 10 kHz, digitized by Digidata1550 (Molecular Devices, Sunnyvale, CA, USA), and stored in a computer. The pipette resistance was between 3–6 MΩ. The internal pipette solution for whole-cell voltage-clamp recording contained (in mM) 120 NaCl, 1 KCl, 2 CaCl2, 2 MgCl2, 5 EGTA, and 25 HEPES, adjusted to pH 7.2. The extracellular solution contained (in mM) 140 NaCl, 1 KCl, 2 CaCl2, 2 MgCl2, and 10 HEPES, adjusted to pH 7.2. For recording action spectra, the pipette solution contained (in mM) 126 Na aspartate, 0.5 CaCl2, 2 MgCl2, 5 EGTA, 25 HEPES, and 12.2 N-methyl D-glucamine, adjusted to pH 7.4 with citric acid. The standard extracellular solution contained (in mM) 150 NaCl, 1.8 CaCl2, 1 MgCl2, 10 HEPES, 10 N-methyl D-glucamine, and 5 glucose, adjusted to pH 7.4 with N-methyl D-glucamine. Irradiation at 470 nm was performed with a collimated LED (LCS-0470-03-22, Mightex, Toronto, Canada) controlled by computer software (pCLAMP10.7, Molecular Devices, Sunnyvale, CA, USA). Light power was measured directly through the objective lens of a microscope by a power meter (LP1, Sanwa Electric Instruments Co., Ltd., Tokyo, Japan). All action spectra were measured at the same light intensity in the range of 390 nm to 650 nm by an OSG xenon light source (Hamamatsu Photonics, Hamamatsu, Japan).
For whole-cell current clamp recording, action potentials were recorded by using an amplifier IPA (Sutter Instrument, Novato, CA, USA) under a whole-cell patch clamp configuration. Data were filtered at 5 kHz, sampled at 10 kHz, and stored in a computer (SutterPatch, Sutter Instrument, Novato, CA, USA). The pipette resistance was between 5–10 MΩ. The internal pipette solution contained (in mM) 125 K-gluconate, 10 NaCl, 0.2 EGTA, 10 HEPES, 1 MgCl2, 3 MgATP, 0.3 Na2GTP, 10 Na2-phosphocreatine, and 0.1 leupeptin, adjusted to pH 7.4 with KOH. The extracellular Tyrode’s solution contained (in mM) 138 NaCl, 3 KCl, 10 HEPES, 4 NaOH, 2 CaCl2, 1 MgCl2, and 11 glucose, adjusted to pH 7.4 with KOH. In all cortical neuron experiments, Tyrode’s solution contained 20 μM 6,7-dinitroquinoxaline-2,3-dione (DNQX, Tocris Bioscience, Ellisville, MO, USA), 25 μM d-(−)-2-amino-5-phosphonovaleric acid (D-AP5, Tocris), and 100 μM picrotoxin (Nacalai Tesque, Inc., Kyoto, Japan). The liquid junction potential was 16.3 mV and was compensated. Irradiations at 385, 423, 469 and 511 nm were performed by using a Colibri7 light source (Carl Zeiss, Oberkochen, Germany) controlled by computer software (SutterPatch, Sutter Instrument, Novato, CA, USA). Light power was directly measured through the objective lens of the microscope by a visible light-sensing thermopile (MIR-100Q, SSC Inc., Mie, Japan).
All data in the text and figures are expressed as mean ± SEM and were evaluated with the Mann–Whitney U test for statistical significance, unless otherwise noted. Data were judged as statistically insignificant when P > 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM density map and atomic coordinates for KnChR have been deposited in the Electron Microscopy Data Bank and the PDB, under accession codes EMD-61212 and 9J7W, respectively. A source Data file is available. Source data are provided with this paper.
References
Nagel, G. et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 100, 13940–13945 (2003).
Ernst, O. P. et al. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 (2014).
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005).
Volkov, O. et al. Structural insights into ion conduction by channelrhodopsin 2. Science 358, eaan8862 (2017).
Mattis, J. et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat. Methods 9, 159–172 (2012).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Bauer, J. et al. Limited functional convergence of eye-specific inputs in the retinogeniculate pathway of the mouse. Neuron 109, 2457–2468.e12 (2021).
Christoffel, D. J. et al. Input-specific modulation of murine nucleus accumbens differentially regulates hedonic feeding. Nat. Commun. 12, 2135 (2021).
Anisimova, M. et al. Spike-timing-dependent plasticity rewards synchrony rather than causality. Cereb. Cortex 33, 23–34 (2022).
Hooks, B. M., Lin, J. Y., Guo, C. & Svoboda, K. Dual-channel circuit mapping reveals sensorimotor convergence in the primary motor cortex. J. Neurosci. 35, 4418–4426 (2015).
Chiu, C. Q. et al. Input-specific NMDAR-dependent potentiation of dendritic GABAergic inhibition. Neuron 97, 368–377.e3 (2018).
Birdsong, W. T. et al. Synapse-specific opioid modulation of thalamo-cortico-striatal circuits. Elife 8, e45146 (2019).
Prasad, A. A. et al. Complementary roles for ventral pallidum cell types and their projections in relapse. J. Neurosci. 40, 880–893 (2020).
Xia, S.-H. et al. Cortical and thalamic interaction with amygdala-to-accumbens synapses. J. Neurosci. 40, 7119–7132 (2020).
Joffe, M. E. et al. Acute restraint stress redirects prefrontal cortex circuit function through mGlu5 receptor plasticity on somatostatin-expressing interneurons. Neuron 110, 1068–1083.e5 (2022).
Rindner, D. J., Proddutur, A. & Lur, G. Cell-type-specific integration of feedforward and feedback synaptic inputs in the posterior parietal cortex. Neuron 110, 3760–3773.e5 (2022).
Rindner, D. J. & Lur, G. Practical considerations in an era of multicolor optogenetics. Front. Cell Neurosci. 17, 1160245 (2023).
Marshel, J. H. et al. Cortical layer–specific critical dynamics triggering perception. Science 365, eaaw5202 (2019).
Kato, H. E. et al. Atomistic design of microbial opsin-based blue-shifted optogenetics tools. Nat. Commun. 6, 7177 (2015).
Fujimoto, K., Hayashi, S., Hasegawa, J. & Nakatsuji, H. Theoretical studies on the color-tuning mechanism in retinal proteins. J. Chem. Theory Comput. 3, 605–618 (2007).
Cembran, A. et al. Structure, spectroscopy, and spectral tuning of the gas-phase retinal chromophore: the β-Ionone “handle” and alkyl group effect. J. Phys. Chem. A 109, 6597–6605 (2005).
Rajput, J. et al. Probing and modeling the absorption of retinal protein chromophores in vacuo. Angew. Chem. Int. Ed. 49, 1790–1793 (2010).
Tashiro, R. et al. Specific residues in the cytoplasmic domain modulate photocurrent kinetics of channelrhodopsin from Klebsormidium nitens. Commun. Biol. 4, 1–10 (2021).
Kato, H. E. et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature 482, 369–374 (2012).
Kato, H. E. et al. Structural mechanisms of selectivity and gating in anion channelrhodopsins. Nature 561, 349–354 (2018).
Tajima, S. et al. Structural basis for ion selectivity in potassium-selective channelrhodopsins. Cell 186, 4325–4344.e26 (2023).
Mohseni, M., Omar, Y., Engel, G. S. & Plenio, M. B. (eds) Quantum Effects in Biology (Cambridge University Press, 2014).
Sugiyama, Y. et al. Photocurrent attenuation by a single polar-to-nonpolar point mutation of channelrhodopsin-2. Photochem. Photobiol. Sci. 8, 328–336 (2009).
Man, D. et al. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 22, 1725–1731 (2003).
Akerboom, J. et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 6, 2 (2013).
Zhou, Q. et al. Common activation mechanism of class A GPCRs. eLife 8, e50279 (2019).
Borch, J. & Hamann, T. The nanodisc: a novel tool for membrane protein studies. Biol. Chem. 390, 805–814 (2009).
Kishi, K. E. et al. Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine. Cell 185, 672–689.e23 (2022).
Chazan, A. et al. Phototrophy by antenna-containing rhodopsin pumps in aquatic environments. Nature 615, 535–540 (2023).
Grinkova, Y. V., Denisov, I. G. & Sligar, S. G. Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Eng. Des. Sel. 23, 843 (2010).
Grote, M., Engelhard, M. & Hegemann, P. Of ion pumps, sensors and channels—perspectives on microbial rhodopsins between science and history. Biochim. Biophys. Acta 1837, 533–545 (2014).
Hirschi, S., Kalbermatter, D., Ucurum, Z., Lemmin, T. & Fotiadis, D. Cryo-EM structure and dynamics of the green-light absorbing proteorhodopsin. Nat. Commun. 12, 4107 (2021).
Tanaka, T. et al. The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4. Mol. Cell 84, 3530–3544.e6 (2024).
Kim, Y. S. et al. Crystal structure of the natural anion-conducting channelrhodopsin GtACR1. Nature 561, 343–348 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. 2021.10.04.463034 Preprint at https://doi.org/10.1101/2021.10.04.463034 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D. 66, 213–221 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Cryst. D. 74, 531–544 (2018).
Yamashita, K., Palmer, C. M., Burnley, T. & Murshudov, G. N. Cryo-EM single-particle structure refinement and map calculation using Servalcat. Acta Cryst. D. 77, 1282–1291 (2021).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Acknowledgements
We thank K. Ogomori, C. Harada, and Y. Kanazawa for technical assistance. This work was supported by grants from the Institute for Fermentation Osaka (W.S.), JSPS KAKENHI (grants 24K23232 to T.T., 23KJ0491 to F.K.S., 24KJ0906 to H.A.,19H05777 to W.S., 21H04969 to H.K., 18K06109 to S.P.T., 20K15900 to S.H. and 21H05037 to O.N.), a JST CREST grant (JPMJCR1753 to H.K. and JPMJCR20E2 to O.N.), a JST PRESTO grant (JPMJPR1688 to S.P.T.), Research Foundation for Opto-Science and Technology (W.S.), Brain Science Foundation (W.S.), and the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research) from the Japan Agency for Medical Research and Development (AMED) under grant number JP22ama121002 (support number 3272 to O.N.) and JP22ama121012 (support number 4893 to H.K.).
Author information
Authors and Affiliations
Contributions
Y.Z.W. performed the structural analysis with assistance from W.S., T.T. and H.A. F.K.S. assisted with the single particle analysis. K.N., S.H., R.T., S.P.T. and H.K. performed the electrophysiological characterization of the mutant proteins. W.S. made the expression vector for the structural analysis and designed the experiment. The manuscript was mainly prepared by Y.Z.W. and W.S. with input from all authors. W.S., H.K. and O.N. supervised the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Massimo Olivucci and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Y.Z., Natsume, K., Tanaka, T. et al. Cryo-EM structure of a blue-shifted channelrhodopsin from Klebsormidium nitens. Nat Commun 16, 5297 (2025). https://doi.org/10.1038/s41467-025-59299-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59299-9