Abstract
Transfer RNA (tRNA) is the most abundant cellular RNA family in terms of copy numbers. It not only folds into defined structures but also has complex cellular interaction networks involving aminoacyl-tRNA synthetases, translation factors, and ribosomes. The human tRNAome is comprised of chromosomal-encoded tRNAs with a large sequence diversity and mitochondrial-encoded tRNAs with A/U-rich sequences and noncanonical tertiary interactions. How tRNA folding and interactions in a eukaryotic cell respond to stress is poorly understood. Here, we develop DM-DMS-MaPseq, which utilizes in vivo dimethyl-sulfate (DMS) chemical probing and mutational profiling (MaP) coupled with demethylase (DM) treatment in transcriptome-wide tRNA sequencing to profile structures and the cellular interactions of human chromosomal and mitochondrial-encoded tRNAs. We found that tRNAs maintain stable structures in vivo, but the in vivo DMS profiles are vastly different from those in vitro, which can be explained by their interactions with cellular proteins and the ribosome. We also identify cytosolic and mitochondrial tRNA structure and interaction changes upon arsenite treatment, a type of oxidative stress that induces translational reprogramming, which is consistent with global translation repression in both compartments. Our results reveal variations of tRNA structurome and dynamic interactome that have functional consequences in translational regulation.
Similar content being viewed by others
Introduction
RNAs are both carriers of genetic information and adapters that mediate biomolecular interactions. In cells, RNAs fold into secondary and tertiary structures to detect metabolites1,2,3, recruit proteins4,5, and regulate phase separation6. Stable RNA structures also impact RNA–RNA interactions and affect RNA stabilities. In biological processes, including transcription, mRNA splicing, and translation, RNA structures play universal and profound regulatory roles. Although many in vitro RNA structures have been resolved through X-ray crystallography, NMR, and Cryo-EM, it was not until the development of in vivo chemical probing coupled with next-generation sequencing that mapping RNA structures in cellular contexts at the transcriptome level became possible7,8,9,10,11.
Chemical probing examines RNA nucleotide reactivity with certain chemicals to assess their structural accessibilities. Commonly applied chemical probing reagents include dimethyl sulfate (DMS)12, kethoxal13, selective hydroxyl acylation analyzed by primer extension (SHAPE) reagents14, and RNases15, which either introduce structure-dependent chemical adducts or strand scissions on RNAs. Among these reagents, DMS was the first to be applied for RNA in vivo structure mapping16, as its chemical properties endow excellent cell permeability, high reactivity with RNA bases in cells, and reaction times in just minutes13. DMS alkylates N1 of (A) and N3 of (C) on the Watson-Crick face of unpaired RNAs, which could be detected as stops or mutations by reverse transcriptases during cDNA synthesis9,14,15. In 2014, three parallel studies combining DMS chemical probing and deep sequencing reported the first cellular mRNA structuromes in A. thaliana, S. cerevisiae, and human cells8,9,10. Strikingly, mRNA structure mapping revealed that in vitro mRNA structures can be drastically different from their in vivo forms. Notably, many stable mRNA structural motifs in vitro are more unstructured in vivo. While the in vitro mRNA structures are largely driven by thermodynamic forces, in vivo mRNA structures are predominantly determined by energy-dependent unwinding processes and intricate interactions with metabolites, proteins, and other nucleic acids. Therefore, comprehending RNA functions and their regulatory roles requires a realistic view of RNA native structures within cells.
In vivo chemical probing can also be used to study RNA–protein interactions, as proteins can leave footprints by shielding RNA regions from chemical modifications10,11,17,18. It has been proposed that a large part of the differences between in vitro and in vivo chemical probing signals could be indicative of the protein-binding motifs on mRNAs11,17,18. However, the complexity of whole-cell RNA–protein interactions, the transient nature of many binding events, and the low coverage of individual mRNA structure elements limited these studies to the analyses of a few well-characterized mRNA–protein interactions and those between rRNAs and ribosomal proteins11,17,18.
RNA structures are also fundamental to the function of other types of RNAs, including long non-coding RNAs, microRNAs (miRNAs), small-nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and tRNAs. Luo et al. applied a SHAPE-based method to study the pre-miRNA substrates for Dicer and uncovered genetic elements crucial to Dicer binding and cleavage, providing an example of the small RNA structurome contributing to exciting biology19. tRNAs have highly conserved secondary and tertiary structures among the three kingdoms of life. They are often described as the classic cloverleaf secondary structures and “L-shaped” tertiary structures. Despite abundant studies on tRNA in vitro structures with X-ray crystallography and various chemical probing methods, the in vivo structures of tRNAs have been largely taken for granted to resemble their in vitro forms. However, tRNAs have many well-characterized protein interactions and modifications in vivo, which led us to hypothesize that their in vivo structural states could be more diverse and dynamic. Recently, Yamagami et al. applied tRNA structure-seq to examine in vivo structures of E. coli tRNAs and revealed dynamics of tRNA folding under heat stress20. tRNA structure-seq results significantly improved tRNA structure prediction under in vivo conditions and revealed tRNA structural dynamics under heat stress which also correlated with changes in tRNA abundance.
The reference human genome encodes a diverse set of over 600 putative tRNA gene21, among which 429 are considered tRNA genes with high confidence, i.e., they likely fold into the canonical tRNA structures based on their predicated base pairing and tertiary interaction properties. Nevertheless, whether chromosomal-encoded tRNA transcripts fold into a canonical tRNA structure in vivo has not been tested systematically. As tRNA function in translation requires the canonical structure, in vivo structural information would serve as a validation of tRNA functionality. Moreover, tRNAs represent an excellent model system for studying RNA–protein interactions by in vivo chemical probing since the structures of tRNAs and many of their interacting protein complexes are well characterized. Beyond cytosolic tRNAs, the human mitochondrial genome (mt) contains a distinct set of 22 mt-tRNAs. Mt-tRNAs have A/U-rich sequences, and most have noncanonical tertiary interactions. Thus, they are structurally weaker than cytosolic tRNAs22. Mapping the mt-tRNA structures in vivo would provide insights into the differences between cytosolic and mitochondrial translation systems. As demonstrated in a recent study using in organello DMS structure mapping, human mt-mRNA structures are actively regulated and contribute to mitochondrial translation kinetics23.
Here, we develop an in vivo structural mapping method for tRNA and other structured non-coding RNAs using DMS mapping with mutation profiling15 coupled with m1A/m3C demethylase (DM) treatment in tRNA-seq library construction to enhance the efficiency and accuracy of structural mapping for the highly structured tRNA. DMS reaction in cells with highly structured RNA generates lower reactivities than mRNA. As reverse transcriptases may also generate mutations at modified RNA nucleotides and structural motifs, an AlkB DM treatment was applied to selectively remove the m1A/m3C methyl groups before cDNA synthesis and thereby revealing DMS-specific mutations in sequencing reads. We term the method DM-DMS-MaPseq. We unveiled the in vivo tRNA structurome for cytosolic tRNAs at the isodecoder level and for mitochondrial tRNAs. We found that small sequence variations between isodecoders can have a visible impact on tRNA structural features. For mt-tRNAs with noncanonical base-pairing and tertiary interactions, the in vivo environment seemed to be crucial for stabilizing their structures. We derived tRNA-protein interaction insights by comparing in vivo and in vitro DMS signals. Furthermore, we revealed the dynamic cytosolic and mitochondrial tRNA folding and protein interaction during arsenite stress-induced translational reprogramming.
Results
DM-DMS-MaPseq development
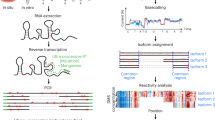
DM-DMS-MaPseq is a combination of in vivo DMS chemical probing and multiplexed small RNA sequencing (MSR-seq), which was recently developed to achieve efficient sequencing of highly structured and highly modified tRNA species24. Although tRNAs had been difficult to sequence due to their stable structure and numerous modifications, recent advances in tRNA-seq have overcome most of these challenges24,25,26,27,28. The major steps of DM-DMS-MaPseq are depicted in Fig. 1a. For in vivo structure profiling, the protocol started with DMS treatment on live human HEK293T cells. Total RNAs were extracted and used directly as the input for library preparation. For in vitro structure profiling, DMS treatment was performed on extracted, deproteinated total RNA. Following the MSR-seq protocol24, total RNAs, including deacylated tRNAs, were ligated to barcoded, biotinylated multiplexing adapters. Multiple barcoded samples were combined and immobilized on streptavidin beads. The combined samples were split into two, one mock and the other treated with two recombinant E. coli AlkB DM, which selectively demethylated m1A and m3C26,29. Library construction proceeded with reverse transcription using the thermostable Superscript IV reverse transcriptase and an optimized reaction condition, followed by on-bead second adapter ligation and PCR to generate DNA libraries for Illumina sequencing. The sequencing data were processed to obtain mutation signatures at each nucleotide position. We obtained 4.4–16.8 million reads mapped to human chromosomal and mitochondrial-encoded tRNA genes from these libraries, with mapping rates ranging from 55 to 71% (Supplementary Table S1). The read counts between biological replicates are highly correlated (Supplementary Fig. S1a). As expected, most mapped reads are tRNAs, with the remainder mostly mapped to 5S/5.8S rRNAs and other abundant noncoding RNAs, such as snRNAs and snoRNAs, that are of similar length to tRNAs (Supplementary Fig. S1b). Our data have very high positional coverage for many tRNAs, which enables in-depth analysis of the tRNA structure and interactions in vivo.
The parameters used here are: DMdiff = mutation rate (−DM) – mutation rate (+DM). DMS signal = DMdiff (+DMS) – DMdiff(–DMS). a Schematic diagram showing the demethylase, dimethylsulfate, mutation map sequencing (DM-DMS-MaPseq) experimental setup and data analysis workflow. b Line graphs of in vitro and in vivo positional DMdiff values of 5S rRNA with (blue) or without (red) 5% DMS treatment (n = 3 biological replicates for each). c Line graphs of in vitro and in vivo positional DMS signals of 5S rRNA with (blue) or without (red) 5% DMS treatment (n = 3 biological replicates for each). d Line graphs of in vivo positional DMS signals in the loop C region of 5S rRNA with 0% (red), 2% (light purple), or 5% DMS (blue) treatment (n = 3 biological replicates for each). e Cryo-EM structures of 5S rRNA in the human 80S ribosome (shown as 5S rRNA alone or in the ribosome, PDB: 4UG0) overlaid with in vitro and in vivo DMS signals of high (red), medium (orange), or low (yellow). f Line graphs of in vitro and in vivo positional DMS signals of tRNAiMet with (blue) or without (red) 5% DMS treatment (n = 3 biological replicates for each). g Secondary structure of tRNAiMet overlaid with in vitro and in vivo DMS signals (average of three biological replicates for each group). Each A or C nucleotide is colored based on DMS signal values: no color: <0.00125; light blue: 0.00125–0.0025; blue: 0.0025–0.005; yellow: 0.005–0.01; orange: 0.01–0.02; red: >0.02. h Line graphs of in vivo positional DMS signals in the anticodon region of tRNAiMet with 0% (red), 2% (light purple), or 5% DMS (blue) treatment (n = 3 biological replicates).
DMS predominantly methylates the Watson-Crick face of the unpaired A and C residues. The resulting m1A and m3C can be read as mutation signatures in our sequencing data, as thermostable RTs can read through them20,25,27,28. Given the presence of abundant and diverse tRNA modifications and the interference of tRNA structure to the RT reaction, it is critical to validate bona fide DMS signatures in our experiment. This is because compared to mRNA, DMS signals for tRNAs are substantially weaker, and many endogenous modifications present in tRNAs leave mutation signatures (Supplementary Fig. S1c). Additionally, some tRNA modifications contribute to the background mutation rates of nearby residues; therefore, enzyme treatment to remove these modifications is necessary. The E. coli wild-type AlkB DM selectively reverses the DMS products of m1A or m3C back to the unmodified A or C. Therefore, comparing sequencing libraries with and without DM treatment could more confidently assign the mutation signatures derived from m1A or m3C. In addition to the wild-type AlkB, we also included the E. coli AlkB D135S mutant during DM treatment, which demethylates m1G25, to further lower background mutation rates and reduce RT stops. Globally, the removal of natural modifications with DM is >95%, as shown by the largely reduced mutation rates of the native m1A modifications from a mean of ~70% to <1% with DM treatment (Supplementary Fig. S1d). When comparing the mutation rate without and with DM treatment (DMdiff = Mutation rate (–DM) – Mutation rate (+DM), Fig. 1a) at different nucleotides, only A and C residues show high and positive mutation rate differences in DMS-treated samples, validating the selectivity of demethylation (Supplementary Fig. 1e).
We present two abundant non-coding RNAs to illustrate the feasibility of DM-DMS-MaPseq. The 5S rRNA is part of the 60S ribosomal subunit in vivo and has a well-defined secondary structure and high-resolution structure in the ribosome. To showcase the effect of DM treatment, DMdiff under various treatment conditions in vitro and in vivo are shown for the 5S rRNA (Fig. 1b). Our demethylase treatment was highly effective in removing the methylation-induced mutation signatures for both in vitro and in vivo samples. To quantify the DMS reactivity at each nucleotide, we introduce “DMS signal,” which is equal to the difference between DMdiff with and without DMS reaction (DMS signal = DMdiff(+DMS) – DMdiff(–DMS), Fig. 1a). The in vivo DMS signal is drastically lower than the in vitro DMS signal (Fig. 1c), which could be due to the overall lower effective DMS concentration in vivo caused by inefficient diffusion and/or the fact that 5S rRNAs are protected as part of the ribosome in vivo. Zooming into the in vivo high reactivity region around A42, the three in vivo conditions show the expected results of zero DMS signal for untreated, high DMS signal for high DMS concentration, and intermediate DMS signal for low DMS concentration (Fig. 1d). The in vitro DMS signal pattern largely recapitulates the known 5S rRNA secondary structure (Fig. 1e). The residues with the highest DMS signals in vivo include A42, A90, A101, and A103, among which A42, A101, and A103 are located in the loop regions and on the surface of the ribosome, thus are expected to be more prone to DMS reaction (Fig. 1e). These results validate our experimental approach for the assessment of highly structured RNA in vitro and in vivo.
The DMS signals for the chromosomally encoded cytosolic initiator tRNAMet (tRNAiMet) are also much higher in vitro than in vivo (Fig. 1f). The known natural modifications m1A58 and m1G9 in this tRNA30 produce strong mutation rates, which are largely reversed upon demethylase treatment. DMS reaction generates additional mutation signatures primarily in the D, anticodon, variable, and anticodon loop regions in vitro, consistent with tRNAiMet folding into the canonical secondary structure (Fig. 1g). DMS signals in these regions are drastically reduced in vivo, consistent with tRNAiMet being protected in vivo, presumably through its interactions with the initiation factor eIF2B and the 40S ribosome31,32. Zooming into the anticodon loop region of the in vivo samples again shows the expected no DMS signal for untreated, high DMS signal for high DMS concentration, and intermediate DMS signal for low DMS concentration (Fig. 1h). The DMS signal at A37 is saturated at intermediate DMS treatment which may be related to tRNAiMet A37 being modified to N6-isopentenyladenosine (i6A). We therefore focus our downstream tRNA analysis on the in vivo untreated and high DMS concentration samples.
To further demonstrate that the in vivo DMS signals are not just a reduced reactivity version of the in vitro signals, we did three additional analyses. First, we compared the in vivo DMS signals for the cytosolic tRNA and mitochondrial tRNAs (Supplementary Fig. S1f). Because of the low structural stability, the mitochondrial tRNAs are expected to have higher DMS reactivity which is what we observe. However, in cells, mitochondrial tRNAs are enclosed in mitochondria with extra membrane protection against DMS diffusing in. Therefore, higher in vivo reactivity of mitochondrial tRNAs contradicts a model of systematic low reactivity of in vivo versus in vitro conditions. Second, we mapped the DMS reactivity in mRNA reads in our sequencing data. MSR-seq uses total RNA as input, so 0.5–1 million mRNA reads are still present together with the tRNA reads. We found that the mRNA median Adenosine mutation rates were ~10 times higher than those in tRNAs (Supplementary Fig. S1g), indicating that our in vivo DMS condition is within the range of reactivity in other mRNA structural mapping studies. Third, we made a Gini plot comparing the in vitro and in vivo DMS signal profiles of the same tRNAs (Supplementary Fig. 1h). The in vitro and in vivo profiles have drastically different shapes, consistent with different factors contributing to these profiles.
Chromosomal-encoded tRNA structurome in vivo
More than 600 tRNA genes are annotated in the human chromosome. Each tRNA gene is assigned a tRNAScan score in the genomic tRNA database, which correlates with its predicted secondary structure formation strength and conservation of residues that form tertiary interactions33. By convention, a tRNAScan score of >50 is considered a high confidence tRNA that likely forms a canonical tRNA structure, a cloverleaf secondary structure comprised of four stems and four loops with the nucleotides 14–21 (conventional tRNA nomenclature) in D, 32–39 in anticodon, 44–48 in variable, and 54–60 in T loops (Supplementary Fig. S2A). The tertiary structure is formed by a network of interactions among nucleotides in the D, T, and variable loops (Supplementary Fig. 2b). A total of 429 high-confidence tRNA genes are present in the hg38 genome, composed of 267 unique tRNA sequences (isodecoders)21. Given the diversity of tRNA sequences and the variation among their predicted structural stabilities, we systematically tested to what extent cytosolic tRNAs conform with canonical structures in vivo.
The in vitro and in vivo DMS signal profiles for the most abundant cytosolic tRNA isodecoders among each of the 47 tRNA families with different anticodons are shown in Fig. 2a. Both in vitro and in vivo DMS signals show clearly discernable patterns based on the tRNA secondary structure, where loop regions and the 3’CCA have significantly higher DMS signals than stem regions. The anticodon loop region shows the highest DMS signals which is consistent with this region forming the least extent of tertiary interactions among all loops: D, variable, and T loops form the tertiary structural core in the canonical tRNA tertiary structure. For these most abundant tRNAs in each anticodon family, the DMS signals are all consistent with the formation of the canonical secondary structure.
The parameters used here are: DMS signal = DMdiff (+DMS) – DMdiff(–DMS). a Heatmap showing the in vitro and in vivo positional DMS signals for the most abundant tRNA isodecoder in each anticodon family. Native modifications30 that are incompletely removed by demethylases are marked with “X”. tRNA positions are according to the standard tRNA nomenclature, and residues missing in specific tRNAs are marked in gray. b Box-whisker plots of DMS signal differences for each A and C residue in every tRNA using the formula (x – in vitro), where x = in vitro (1), or in vivo (2) in D, T, anticodon, variable loops, and the 3’ unpaired CCA regions. P-values are calculated with standard two-sided Wilcox rank-sum test; ****: p < 10-4. The exact P-values for each box-whisker plot are as follows: D-loop = 1.6 × 10–26, anticodon loop p = 1.3 × 10–31, variable loop p = 1.3 × 10–20, T loop = 1.9 × 0–21, CCA p = 4.9 × 10–05. In each box-whisker plot, the maxima represent the first quantiles, the middle lines represent the medians, the minima represent the 75th quantiles, the crosses in the center represent the means, and the whiskers represent the standard deviation. c Secondary structures of two tRNASer(AGA) isodecoders with different tRNAScan scores33: Ser-AGA-1, 90.4, and Ser-AGA-4, 78.8, overlaid with in vivo DMS signals. Sequence differences are indicated as circled nucleotides. d Tertiary structures of Ser-AGA-1 and Ser-AGA-4 overlaid with in vivo DMS signals (modified from the E site tRNA in the human 80S ribosome, PDB: 6Z6M)90. e Secondary structures of two tRNAVal(UAC) isodecoders with different tRNAScan scores33: Val-UAC-1, 82.9, and Val-UAC-4, 68.7, overlaid with in vivo DMS signals. Sequence differences are indicated as circled nucleotides. f Tertiary structures of Val-UAC-1 and Val-UAC-4 overlaid with in vivo DMS signals (modified from the tRNAVal in the human METTL1-WDR4-tRNA(Val) complex, PDB: 8CTI)91.
Putting aside the mutations derived from incomplete demethylation of the naturally occurring m1A, m3C, m1G, and m22G modifications (marked in Fig. 2a), the in vitro DMS signals are always higher than the in vivo DMS signals. This is also confirmed by examining the in vitro and in vivo averaged DMS signal distribution among all tRNAs in each loop region (Fig. 2b). Given that tRNAs form highly stable structures in vitro, lower DMS signal in vivo may be attributed to lower DMS reactivity in vivo, as DMS diffusion into cells may result in lower in cellulo DMS concentration. On the other hand, tRNAs interact extensively with proteins and ribosomes in cells, which can protect them from reacting with DMS. As an exemplar, the interaction model is more consistent with the much higher DMS signals seen in the 3’NCCA region in vitro than in vivo: for each tRNA, this region binds to the EF-1A protein or is buried in the ribosome in vivo, which can explain in part the dramatic difference.
To further explore how sequence variations between high-confidence isodecoders impact tRNA folding in vivo, we compared only the in vivo DMS signal profiles of two pairs of tRNA isodecoders with different tRNAScan scores. tRNASer(AGA)−1 and −4 have three nucleotide differences: C25 and U25 (standard tRNA nomenclature) that form a G10-C25 or G10-U25 pair at the end of D-stem, U39 and C39 that forms an A31-U39 pair in SerAGA-1, but an A31-C39 in SerAGA-4, and U50e and C50e in the loop of the variable stem-loop that is not involved in any base pairing in the tRNA. Despite just three sequence changes, tRNASer(AGA)−4 has significantly higher DMS signals, suggesting that it has a less stable structure in vivo compared to tRNASer(AGA)−1 (Fig. 2c). The conversion of A31-U39 in Ser(AGA)−1 to A31-C39 in Ser(AGA)−4 likely weakens the anticodon stem and leads to markedly increased DMS signals in the anticodon loop. The G10-U/C25 pair at the end of D stem is located ~15 Å away in the 3D structure to the end of the variable loop region. Thus, the conversion of G10-C25 to G10-U25 may be responsible for the DMS signal increase in the variable loop (Fig. 2d), perhaps transmitting long-range effects in structural change through the tertiary interaction network of tRNA. tRNAVal(UAC)−1 and tRNAVal(UAC)−4 differ at 15 out of 76 positions in acceptor, anticodon, and T stems and multiple loop regions. All stem sequence changes are matched with compensatory changes that maintained the Watson-Crick base pairs, whereas the loop changes can weaken noncanonical base pairs or tertiary interactions, such as U54 in Val(UAC)−1 to G54 in Val(UAC)−4, which can disrupt the G18-U54 tertiary interaction. These sequence changes result in higher DMS signals in Val(UAC)−4 than in Val(UAC)−1 (Fig. 2e). Again, the structural change permeates throughout the tRNA structure (Fig. 2f). In both tRNASer(AGA) and tRNAVal(UAC) cases, the respective high tRNAScan-scoring isodecoder indeed has lower DMS signals than the low-scoring isodecoder. On the other hand, the difference in structural accessibilities between these tRNA isodecoders does not translate to varied abilities to engage with the ribosome. Both tRNA isodecoders still fold like tRNA, as suggested by the participation of both isodecoders in translation at comparable ratios in the input and in the polysome (Supplementary Fig. S2c).
DM-DMS-MaPseq can characterize the in vivo tRNA structures for “rare” tRNA isodecoders as well, defined here as present at <10% levels of the most abundant isodecoder tRNA of the same anticodon family (Supplementary Fig. S2d). As an exemplar, we present the in vivo DMS profiles of seven isodecoders in the tRNAAla(AGC) family together with their tRNAScan scores to showcase the diverse structural accessibilities among them (Supplementary Fig. S2e).
In vivo-in vitro difference in DMS signals reflects cytosolic tRNA-protein/ribosome interactions
Previous structure mapping studies on mRNAs have concluded that mRNAs are prone to denature in vivo due to energy-driven cellular interactions, thereby showing higher DMS reactivities in vivo9,34,35. In our case, however, the in vitro DMS signal for chromosomal-encoded tRNA is substantially higher than in vivo. Cytosolic tRNA-protein interactions can reduce the in vivo DMS signals of the interacting tRNA motifs, resulting in a region-specific decrease in DMS signals36,37.
Dominant cytosolic tRNA binding partners include the highly abundant aminoacyl-tRNA synthetases (aaRSs), the elongation factor EF1A, and the ribosome. tRNAs are aminoacylated (a.k.a., charged) by 20 aaRSs, one for each amino acid. Cellular tRNA charging levels are high, but charged tRNAs turn over quickly during translation, so that tRNA-aaRS interaction occurs constantly38,39,40,41. Charged tRNAs are bound by EF1A, which delivers tRNAs to the ribosome. EF1A is among the most abundant cytosolic proteins in a human cell42 and the primary interacting protein for all cytosolic tRNAs. During translation, tRNAs occupy the A-, P-, and E-sites on the ribosome, and approximately one-third of all tRNAs can be bound by ribosomes in cells. As all three types of tRNA-protein interactions involve distinct combinations of tRNA structural elements, we set out to deduce these interaction modes by comparing the in vivo and in vitro DMS signals.
To assess the in vivo and in vitro differential DMS signals in individual tRNA regions and infer information on tRNA-protein interaction, we introduce the τ score, which is the normalized difference between in vitro and in vivo DMS signals at each nucleotide relative to the largest delta across the entire molecule for each tRNA isodecoder (Supplementary Fig. S3a). In part due to the inefficient diffusion of DMS into cells, which can lower the overall DMS reactivity in vivo, the normalization of τ score is critical to compensate for the global DMS reactivity differences between the in vivo and in vitro conditions. The τ score is between 0 and 1; a higher τ score implicates stronger protection of DMS reaction by tRNA interacting partners in vivo.
Our τ score normalization differs from other internal global normalization methods used in the literature43,44 which apply a single normalization factor for each experimental condition that is the mean of a certain percentile range (e.g., 90–98th or 90–95th). These methods are generally very effective when the data are more consistent internally and differ significantly under different conditions. In our tRNA study, however, such normalization methods can have two caveats: (i) they do not consider the heterogeneity between different tRNAs, which is far greater than mRNAs and other noncoding RNAs. (ii) They select a percentile range that is close to the maximum mutation rate in tRNAs, which are typically from the non-DMS-derived, heavily modified residues whose signals cannot be removed through demethylation, thus not relevant to “real” DMS signals.
The high tRNA variability in abundance, variety, and modification density necessitates choosing a balance between minimizing internal bias and cross-conditional bias in choosing in vivo versus in vitro normalization methods. In essence, the question comes down to whether (i) a specific tRNA is more comparable between in vitro and in vivo conditions or (ii) under either condition, different tRNAs are more comparable to each other. Given our interest in unveiling biological insights into the differences among tRNAs, we prioritized minimizing internal bias. The τ score first calculates the positional DMS signal differences between in vitro and in vivo conditions. Next, for each tRNA transcript, we normalized the positional DMS signal difference to the maximum difference. The advantage of our approach is that it compares tRNA by tRNA and position by position. For heavily modified tRNA positions, mutation signals tend to be very high both in vivo and in vitro, and the subtraction can partially reduce the gap between positions. The normalization to the maximum difference simply brings the scale of the τ score between 0 and 1.
The positional τ scores of the most abundant isodecoder from all tRNA anticodon families show that the loop regions tend to have higher τ scores, which are derived from their higher DMS reactivities in vivo and in vitro (Fig. 3a). Disregarding the incomplete demethylation of the natural tRNA modifications, we readily identify the D-loop, anticodon loop, T-loop, and the 3’CCA as hotspots of high τ scores. We subsequently used the τ scores from these regions to infer tRNA interacting with the aaRS, EF-1A, and the ribosome.
The parameters used here are: τ score = (DMS signalin vitro – DMS signalin vivo)/(DMS signalin vitro – DMS signalin vivo)highest value position on each isodecoder. a Heatmap showing the positional τ scores on the most abundant tRNA isodecoder in each anticodon family. Native modifications30 that are incompletely removed by demethylases are marked with “X”. b Box-whisker plot showing anticodon loop τ score medians of tRNAs with two distinct modes of aaRS interactions. Charging of tRNALeu, tRNASer, and tRNAAla (n = 13) does not require anticodon recognition, while others (n = 34) do. P-value is calculated with a standard two-sided t-test excluding the outlier tRNAArg(ACG) (p = 0.044 including tRNAArg(ACG)). In the box whisker plot, the maxima represent the first quantiles, the middle lines represent the medians, the minima represent the 75th quantiles, the crosses in the center represent the means, and the whiskers represent the standard deviation. c Scatter plot showing energetic contribution by tRNA binding or by amino acid binding to EF-Tu ΔG (kcal/mol) and τ scores at C75 averaged across all isoacceptors for each tRNA anticodon family. The x-axis scale is from Asahara et al.55,56. d Scatter plot showing D and T loop τ score medians for the most abundant isodecoders in each anticodon family. Pearson’s r is calculated excluding the outlier tRNAArg(ACG). e Box-whisker plot showing D loop τ score averages in the most abundant isodecoders grouped by the wobble nucleotide identity, either I/U at the wobble position (n = 23) or G/C (n = 24). P-value is calculated with a standard two-sided t-test excluding the outlier tRNAArg(ACG). In the box whisker plot, the maxima represent the first quantiles, the middle lines represent the medians, the minima represent the 75th quantiles, the crosses in the center represent the means, and the whiskers represent the standard deviation.
AaRSs recognize their cognate tRNA substrates through identity elements that involve many regions in tRNA in an idiosyncratic manner45,46. Seventeen aaRSs require anticodon recognition for charging. However, SerRS and LeuRS interact with the unique extra-long variable arm of their cognate tRNAs (Supplementary Fig. S3b), while AlaRS recognizes a G3:U70 base pair in the acceptor stem47,48,49,50,51,52,53. We find that the anticodon loop τ scores of tRNASer, tRNALeu, and tRNAAla as a group are significantly lower than τ scores of other tRNAs as a group, consistent with a lack of protein protection at the anticodon region for the tRNAs for these three amino acids (Fig. 3b). Unexpectedly, tRNAArg(ACG) shows a much higher τ score for the anticodon loop than all other tRNAArgs (Supplementary Fig. S3c). Human tRNAArg(ACG) has an unusual C13-A22 mismatch at the end of the D stem, whereas all other tRNAArgs have a conventional U13-A22 or C13-G22 base pair (Supplementary Fig. S3d). The C13-A22 mismatch may loosen the D loop structure that contains the U20 identity element for ArgRS recognition (Supplementary Fig. S3e), thereby increasing the τ score of tRNAArg(ACG).
EF1A delivers all aminoacylated tRNAs (aa-tRNAs) to the A-site of the elongating ribosome and forms extensive interactions with the T stem, the acceptor stem, and the 3’CCA of tRNAs54. Previous studies with prokaryotic tRNA-EF-Tu binding in vitro have established a near-uniform elongator protein binding affinities to all correctly aminoacylated aa-tRNAs which minimizes decoding bias and the loading of misacylated tRNAs on the ribosome55,56,57,58. tRNAs that bind to EF-Tu at high affinity alone have cognate amino acids with low binding affinity to EF-Tu and vice versa. This paradigm was established using prokaryotic systems in vitro55,56, however, it remains to be determined whether this model also works for human EF1A-aa-tRNA interactions in vivo. As the stem residues show very low DMS signals, we used the τ scores of the 3’ C74C75A as a proxy to study EF1A binding. The high-resolution structure of EF-1A-tRNA reveals that C75 and A76 interact directly with EF1A but not C74 (Supplementary Fig. S3f). However, A76 with the charged amino acid is completely masked by EF1A, so only the C75 τ score serves as a good proxy for EF1A-tRNA binding in vivo. Plotting the average C75 τ score across tRNAs for each amino acid with the estimated energy contribution by the tRNA body alone55 or by the amino acid alone56 (Fig. 3c) shows a positive correlation of tRNA (Pearson’s r = 0.64) and a negative correlation of the amino acid (Pearson’s r = −0.58) which is consistent with the prokaryotic tRNA-EF-Tu interaction paradigm.
Next, we examined tRNA engagement with the ribosome using the τ scores of the tRNA “elbow” region, where the D- and the T-loops form tertiary interactions; this region interacts mostly with the ribosome (Supplementary Fig. S3g)59. Ribosome interacts with the tRNA elbow region in all three of its tRNA binding sites to stabilize tRNA positioning, propel translation elongation, and enable proofreading59. Therefore, the tRNA D- and T-loop τ scores can be proxies to examine ribosome binding to aa-tRNAs. We find a strong positive correlation between the D- and T-loop τ scores of all most abundant tRNA isodecoders from each anticodon family (Pearson’s r = 0.95 excluding tRNAArg(ACG), Fig. 3d), which is consistent with stable D- and T-loop tertiary interactions in vivo. Recent findings suggest that the human ribosome decodes G/C-ending codons more efficiently than A/U-ending codons24,60. To see whether this decoding property also reflects tRNA-ribosome interactions, we group tRNAs based on their wobble anticodon nucleotide identities: A/U-ending codons are more readily decoded by I34/U34 tRNAs (A34 in tRNA gene is always modified to Inosine in tRNA transcript)61 and C/G-ending codons by G34/C34 tRNAs. We find tRNAs with G34/C34 with higher τ scores, consistent with stronger interaction with the ribosome (Fig. 3e). tRNAArg(ACG) is once again an outlier for both D-/T-loop τ score correlation and ribosome interaction, which may be derived from its C13-A22 mismatch in the D-stem altering its interaction with the ribosome.
Even though C74 shows a wide range of τ scores (Fig. 3a), we fail to identify a good correlation of it to anything specifically. One possibility is that because of its proximity to the interaction centers of tRNA, C74 is involved in interacting with the tRNA synthetase, EF-1A, and the ribosome in similar proportions.
Oxidative stress induces cytosolic tRNA structurome and interactome changes
In cells, global translation regulation is tightly linked to cellular metabolism and stress. As the critical components of the translation system, tRNAs undergo significant reprogramming in their expression, modification, charging, and fragmentation under stress24,62,63. To understand the dynamics of cellular tRNA structure and interaction in response to stress, we treated cells with sodium arsenite to induce oxidative stress, which is a common stress condition for the studies of translational stress response. Arsenite exposure induces strong phosphorylation of eIF2α, which leads to significant downregulation of translation initiation64. Indeed, under our experimental settings, global translation activities were reduced by ~12-fold (Supplementary Fig. S4a). Since arsenite stress inhibits translation initiation, we first examine the DMS signal changes of the initiator tRNAMet (tRNAiMet, Fig. 4a). We use the term “ΔDMS signal” to describe the DMS signal changes with and without stress (Supplementary Fig. 4b). Consistent with a high reduction of translation initiation, which corresponds to decreased utilization of tRNAiMet in the scanning ribosome65, the DMS signal strongly increases for the 5’ half of the anticodon loop of residues C31C32C33 in the presence of arsenite (anticodon nucleotides for tRNAiMet are C33AU). DMS signal in the D-loop also increases, which is consistent with decreased ribosome interaction with the tRNAiMet. Arsenite exposure leads to a strong reduction of the DMS signal at C70, consistent with an altered interaction between tRNAiMet with eIF2B (Supplementary Fig. S4c) and/or the scanning ribosome.
The parameters used here are: ΔDMS signal = DMS signalin vivo_AsO2 – DMS signalin vivo_control. a Heatmap of the positional ΔDMS signal on tRNAiMet, red: higher, blue: lower DMS signal with arsenite. Native modifications30 that are incompletely removed by demethylases are marked with “X”. The blue line and residue numbers indicate the residues that show a large DMS profile change under arsenite stress. The orange line indicates the anticodon nucleotides. b Heatmap of the positional ΔDMS signal of the most abundant isodecoder in each anticodon family, red: higher, blue: lower DMS signal with arsenite. Native modifications30 that are incompletely removed by demethylases are marked with “X”. tRNA positions are according to the standard tRNA nomenclature, residues missing in specific tRNAs are marked in gray. c Anticodon loop (excluding position 37) average ΔDMS signals in tRNA groups with different modification status at position 37. Only the most abundant isodecoders from each isoacceptor family are included. tRNAs are grouped as follows: A37 (n = 14): Asp, GluCTC, GluTTC, GlyCCC, GlyGCC, GlyTCC, GlnCTC, GlnTTC, ThrAGT, ThrCGT, ThrTGT, ValAAC, ValCAC, ValTAC; t6/i6A37 (n = 12): IleAAT, IleTAT, LysCTT, LysTTT, Met, iMet, Asn, SerAGA, SerCGA, SerGCT, SerTGA, ArgCCT, ArgTCT; m1G/m1I/W37 (n = 19): AlaAGC, AlaCGC, AlaTGC, ArgACG, ArgCCG, ArgTCG, Cys, His, LeuAAG, LeuCAG, LeuTAG, LeuCAA, LeuTAA, Phe, ProAGG, ProCGG, ProTGG, Trp, Tyr. P-values are calculated with two-sided Wilcoxon signed-rank test. In each box whisker plot, the maxima represent the first quantiles, the middle lines represent the medians, the minima represent the 75th quantiles, the crosses in the center represent the means, and the whiskers represent the standard deviation. d Box-whisker plot of DMS signal differences for each A and C residues in every tRNA in D, T, anticodon, variable loops, and the 3’ unpaired CCA regions. Depleted (=1) or enriched (=2): tRNAs that are significantly decreased or increased in the polysomes with arsenite, respectively. Average n = 42 for the depleted group and n = 14 for the enriched group for each region. P-values are calculated with a standard two-sided Wilcox rank-sum test; n.s.: not significant, **: p < 10-2. The exact P-values for each box-whisker plot are as follows: D loop p = 0.15, Anticodon loop p = 0.0046, Variable loop p = 0.67, T loop p = 0.31, CCA p = 0.018. In each box whisker plot, the maxima represent the first quantiles, the middle lines represent the medians, the minima represent the 75th quantiles, the crosses in the center represent the means, and the whiskers represent the standard deviation.
We examined the DMS signal changes for the most abundant isodecoders from all anticodon families (Fig. 4b). Arsenite stress induces widespread changes in cytosolic tRNA structures and/or protein/ribosome interactions. In general, the loop regions have the most DMS signal changes under stress, but changes in the acceptor stem are also detectable, and the direction of the changes is not consistent across tRNAs. To provide a better focus, we zoom into the anticodon loop region by grouping the tRNAs according to their modification status at position 37 (Fig. 4c), the immediate 3’ nucleotide to the anticodon nucleotides. Group 1 is A37 and unmodified, group 2 is A37 and modified to N6-threonylcarbamoyl adenosine (t6A) or N6-isopentenyl adenosine (i6A), and group 3 is G37 that are modified to m1G, m1I, or wybutosine (W)66. Intriguingly, group 1 tRNAs without A37 modifications generally have positive ΔDMS signals, i.e., higher DMS signals in the presence of arsenite, whereas group 2 tRNAs with A37 modifications have zero ΔDMS signals on average, and group 3 tRNAs with G37 modifications have negative ΔDMS signals on average. The purine at tRNA position 37 closely interacts with the 18S rRNA in the ribosome which stabilizes codon-anticodon interaction at the A site (Supplementary Fig. 4d)67,68. This result is consistent with tRNA modifications at position 37 positively contribute to ribosome usage of tRNA, and thus, tRNAs that lack modification at position 37 are the first to be affected by arsenite stress.
Cytosolic tRNA translational usage was profiled under the same arsenite stress condition in HEK293T cells with polysome-enriched tRNA sequencing in our previous study24. We re-analyzed the data and identified three significantly enriched and five significantly depleted tRNAs in polysome with or without arsenite treatment (Supplementary Fig. S4e). The three polysome-enriched tRNAs, tRNAIle(AAU), tRNAIle(UAU), and tRNALys(UUU), have been described in our previous study with Northern blot validation24. We analyzed the ΔDMS signal by comparing the group of significantly polysome-enriched tRNAs and the group of significantly polysome-depleted tRNAs by >1.5-fold (Fig. 4d). The ΔDMS signals at A and C residues in the anticodon loop and 3’ CCA are higher in the polysome-depleted tRNAs compared to the polysome-enriched tRNAs, consistent with the loss of tRNA-ribosome interactions due to the reduced translation usage for polysome-depleted tRNAs under arsenite stress.
Structural mapping of mitochondrial tRNAs in vivo
Human mitochondria have their own translation system that utilizes a separate set of 22 tRNA genes encoded by the mitochondrial DNA. Mitochondrial tRNAs (mt-tRNAs) have noncanonical tertiary interactions and lower melting temperatures in vitro due to higher numbers of A-U base pairs in the stems22. Our sequencing data contain both chromosomal and mitochondrial encoded tRNAs. To examine the folding and interaction properties of mt-tRNAs, we first plot the positional DMS signals for all 22 mt-tRNAs both in vitro and in vivo (Fig. 5a). Consistent with their weaker structures compared to cytosolic tRNAs, mt-tRNAs show high DMS signals throughout the tRNA bodies in vitro, including the stem regions. However, the in vivo DMS signals for mt-tRNAs are still much lower than those in vitro, reminiscent of the in vitro and in vivo differences seen for cytosolic tRNAs, consistent with mitochondrial protein and ribosome interactions making a major contribution to the stability of mt-tRNA structures in the cellular environment.
The parameters used here are: DMS signal = DMdiff (+DMS) – DMdiff(–DMS). a Heatmap showing the in vitro and in vivo positional DMS signals on the mitochondrial (mt) tRNAs. Native modifications30 that are incompletely removed by demethylases are marked with “X”. tRNA positions are according to the standard tRNA nomenclature, residues missing in specific tRNAs are marked in gray. b Secondary structure of mt-tRNALeu(UAA) overlaid with in vitro and in vivo DMS signals according to the colored scale bar. c Secondary structure of mt-tRNASer(GCU) overlaid with in vitro and in vivo DMS signals according to the colored scale bar. The acceptor stem, anticodon stem, and T stem are marked. d Secondary structure of mt-tRNALys(UUU) overlaid with in vitro and in vivo DMS signals according to the colored scale bar. The dashed lines connect the residues in this schematic for mt-tRNAs. The dashed lines connect the residues in this schematic for mt-tRNAs. Mt-tRNA positions are according to ref. 28. e Heatmap showing the in vitro and in vivo positional DMS signals of mt-tRNAVal. Native modifications30 that are incompletely removed by demethylases are marked with “X”. f The Cryo-EM structure of mt-tRNAVal in the human mt-ribosome (PDB: 6ZM692) overlaid with DMS signals of high (red), medium (orange), and low (yellow). g Scatter plot showing energetic contribution by mt-tRNA binding or by amino acid binding to EF-Tu ΔG (kcal/mol) and τ scores at C75 averaged across all isoacceptors for each tRNA anticodon family. The x-axis scale is from Asahara et al.55,56.
To examine the structure data in more detail, we overlayed the in vivo and in vitro DMS signals to the predicted secondary structures of all mt-tRNAs. The folding of mt-tRNAs is categorized into three types22,69. Type I secondary structure is represented by mt-tRNASer(UGA), which has only one nucleotide between the acceptor stem and the D stem, a small D loop, and an extended anticodon stem70. Most other mt-tRNAs fold into type II structures. Most of them lack the conserved interaction between G18G19 in the D-loop and U55C56 in the T loop, which maintains the stable “L-shape” tertiary structure of cytosolic tRNAs71,72. Type III structure is unique for mt-tRNASer(GCU), which lacks the entire D stem70,73. Mt-tRNASer(UGA) and most type II mt-tRNAs roughly resemble the reference structure but display various structural nuances in at least one of the stems (Supplementary Fig. 5a–c). Mt-tRNALeu(UAA) is considered to be one of the most stable mt-tRNAs, and the lack of in vivo DMS signals confirms its mostly stable stems. Despite that, we see DMS signals for the unpaired A12:C23 at the D stem and A31:C39 at the anticodon stem in vitro (Fig. 5b). Type III mt-tRNASer(GCU) is believed to be the least stable among all mt-tRNAs in vitro. However, its structure is largely stabilized in vivo, as shown by the reduced number of DMS reactive nucleotides in all three stems (Fig. 5c).
The mt-tRNALys(UUU) has been shown to misfold in the absence of the m1A9 modification in vitro74. In the presence of m1A9, mt-tRNALys folding is consistent with its predicted cloverleaf secondary structure in vitro and in vivo (Fig. 5d).
Mt-tRNAVal is unique among all mt-tRNAs because it is also a component of the mitochondrial ribosome in place of the 5S rRNA in the cytosolic ribosome75,76. The DMS signals for mt-tRNAVal have much higher values throughout this tRNA in vitro but were largely reduced and more restricted in the D and anticodon loops in vivo (Fig. 5e). The D and anticodon loop regions of mt-tRNAVal interact extensively with the components of the mitochondrial ribosome (Fig. 5f), suggesting that its ribosome interaction at least in part explains its reduced DMS signals in vivo.
We further examined mt-tRNA interactions by examining the positional τ scores, i.e., the normalized in vitro-in vivo differences for each mt-tRNA (Supplementary Fig. 5d). The mt-tRNA τ score interpretation is more complex than cytosolic tRNAs through its inclusion of both folding and interaction components. Focusing on the C75 τ scores, which are strongly associated with mt–tRNA interaction with mitochondrial EF-Tu protein (Supplementary Fig. 5e), we still find an inverse correlation between tRNA-EF-Tu and amino acid-EF-Tu interactions (Pearson’s r-values of −0.64 and 0.62, Fig. 5g). This result suggests that the principle of balancing tRNA and charged amino acid binding strength to EF-Tu still holds for human mitochondrial translation to maintain translation fidelity.
Arsenite stress induces mt-tRNA structural changes
Arsenite stress is known to significantly reduce mitochondrial activities, including translation77,78. Indeed, mt-tRNAMet, which works in both mitochondrial translational initiation and elongation, shows a large increase in DMS signal in the anticodon loop upon arsenite treatment, consistent with its decreased participation in mitochondrial translation (Fig. 6a). Analysis of previously published data under the same arsenite stress condition24 shows a reduction in mt-tRNAMet charging as well (Supplementary Fig. 6a), which may account for its loss of multiple types of protein interactions in vivo under arsenite stress. We also observe a reduction of the DMS signal for the equivalent of C71A70 residues in mt-tRNAMet, reminiscent of the C71 DMS signal reduction of cytosolic tRNAiMet (Fig. 4a). It remains to be seen whether this acceptor stem response to arsenite stress may also relate to mt-tRNAMet structural change and its contribution to the reduction of mitochondrial translation under stress.
The parameters used here are: ΔDMS signal = DMS signalin vivo_AsO2 – DMS signalin vivo_control. a Heatmap showing the positional ΔDMS signals on mt-tRNAMet, red: higher, blue: lower DMS signal with arsenite. Native modifications30 that are incompletely removed by demethylases are marked with “X”. tRNA positions are according to the standard tRNA nomenclature, residues missing in specific tRNAs are marked in gray. b Heatmap showing the positional ΔDMS signals on each mt-tRNA, red: higher, blue: lower DMS signal with arsenite. Native modifications30 that are incompletely removed by demethylases are marked with “X”. c Mt-tRNA abundance fold changes in polysome fractions under arsenite treatment. The dotted vertical lines indicate a 1.5-fold change, and the dotted horizontal line indicates p = 0.05. P value was determined by using a two-sided t-test with Bonferroni correction between mt-tRNA reads per million of Polysome-AsO2 over polysome-unstressed. Data from ref. 24 and NCBI GEO GSE198441. d Secondary structure of mt-tRNAAsp overlaid with in vivo DMS signals with or without arsenite treatment. e Box-whisker plots of DMS signal differences for each A and C residues in every tRNA in D, T, anticodon, variable loops, and the 3’ unpaired CCA regions. Depleted (=1) or enriched (=2): tRNAs that are significantly decreased or increased in the polysome fraction with arsenite, respectively. P-values are calculated with a standard two-sided Wilcox sum-rank test; n.s.: not significant, *: p < 0.05, **: p < 0.01; ****: p < 10-4. The exact P-values for each box-whisker plot are as follows: D loop p = 0.18, variable loop p = 2.5 × 0–6, T loop p = 2.2 × 0–2, CCA p = 0.54. In each box whisker plot, the maxima represent the first quantiles, the middle lines represent the medians, the minima represent the 75th quantiles, the crosses in the center represent the means, and the whiskers represent the standard deviation.
Arsenite stress also induces universal and notable alterations for all mt-tRNAs (Fig. 6b). Overall, mt-tRNAs show far more drastic changes throughout the tRNA bodies, whereas cytosolic tRNA structural changes are more concentrated in loop regions. This mitochondrial versus cytosolic tRNA change may be exacerbated by the severe loss of mitochondrial translation under arsenite stress79,80,81.
We also examined the polysome association of mitochondrial tRNAs with and without arsenite stress from our previous publication24. Strikingly, mt-tRNAAsp shows the highest enrichment in the polysome fraction, followed by several other mt-tRNAs (Fig. 6c). Asp accounts for only 1.85% of all codons in human mitochondrial proteins82; the mt-tRNAAsp enrichment in the polysome fraction is unlikely to be attributed to Asp codon usage in mitochondrial translation. Mt-tRNAAsp shows an increased DMS signal in the D-loop and anticodon loop and a reduced DMS signal in the acceptor stem under arsenite stress (Fig. 6d). Paradoxically, aside from mt-tRNAMet, mt-tRNAAsp is the only other mt-tRNA with reduced charging under arsenite stress (Supplementary Fig. 6a). Uncharged tRNA may accumulate in the E-site of the ribosome. One possible explanation would be a unique inability of mt-tRNAAsp to be released from the ribosome under arsenite stress, thereby contributing to a reduction of mitochondrial translation through its accumulation in the polysome.
Finally, we compared the ΔDMS signals of the polysome-enriched mt-tRNAs versus the polysome-depleted mt-tRNAs in different stem and loop regions (Fig. 6e). In contrast to cytosolic tRNAs in polysomes, the anticodon, variable, and T loops of the polysome-enriched mt-tRNAs show higher DMS signals under arsenite stress compared to no stress. This result suggests that polysome-associated mitochondrial tRNAs on the polysome respond to arsenite stress differently from the polysome-associated cytosolic tRNAs. It is possible that mitochondrial and cytosolic translation systems use different modes of tRNA interactions in arsenite stress response.
Discussion
The rapid development of high-throughput sequencing methods for highly structured RNAs made it possible to examine transcriptome-wide tRNA structures and interactions in living cells. With DM-DMS-MaPseq, we present comprehensive in vivo structure and interaction studies for human chromosomal and mitochondrial encoded tRNAs. Compared to DMS mapping of mRNAs, in vivo DMS signals for tRNA are much lower; therefore, additional considerations need to be implemented to ensure that the observed mutations are indeed from DMS reactions, not spurious reverse transcriptase events or other processes. This was accomplished here by treating the DMS-reacted RNA samples with a m1A/m3C demethylase before RT and considering only the mutation differences at nucleotides with and without demethylase treatment. This strategy ensures a more accurate interpretation of the DMS mapping results.
Our result is consistent with human chromosomal-encoded tRNAs folding into canonical tRNA structures in vivo, similar to the results in an E. coli tRNA study in vivo20. In contrast to E. coli, the human genome contains many tRNA genes that have the same anticodon but different body sequences termed isodecoders. This exceptional sequence diversity of human tRNAs offers an opportunity to compare the folding patterns of tRNA isodecoders that all have sufficiently high tRNAScan scores to predict to fold into canonical tRNA structures but possess distinct base pairing or tertiary interaction patterns. A brief inspection of all tRNAScan predicted “high confidence” tRNA isodecoders present at sufficiently high abundance in our sample did not identify a tRNA that may fold into a noncanonical structure in vivo. On the other hand, our results show that a mere three-nucleotide difference between two isodecoders can generate local structural variations in the proximity of the sequence changes. This result provides experimental evidence of the tRNAScan score as a valuable tool for predicting local structural variations.
The tRNA structural mapping differences between E. coli20 and our study on human cells can be attributed to both the different experimental conditions and the biological differences. HEK293T cells are adherent cells that grow as a thin layer attached to the bottom of the cell culture dish. E. coli cells were suspended when treated with DMS. Although the E. coli study used a gentler DMS treatment, human cells with more complex cell membranes may uptake DMS from the culture solution at different efficiencies than E. coli cells, leading to a lower intracellular DMS concentration in human cells. Furthermore, it is also possible that human tRNAs are better protected by proteins than E. coli due to the distinct tRNA processing and biological regulation of translation between eukaryotic and prokaryotic systems.
The most striking difference we observe is between the in vitro and in vivo DMS signatures, where tRNAs in vitro generally always have significantly higher DMS reactivities than in vivo, consistent with cellular protein/ribosome interactions with tRNA contributing greatly to the in vivo DMS footprints. The idea of using the DMS reactivity differences between in vitro and in vivo to study RNA–protein interactions has been implemented for well-characterized interactions between ribosomal proteins and rRNAs9,10, as well as RNA binding protein RBFOX2 with its mRNA binding targets17. Transcriptome-wide mRNA-protein interaction studies can be very challenging due to the low coverage of individual structural motifs in mRNAs and the transient nature of these interactions. Therefore, cytosolic tRNAs can be a promising model system to study RNA–protein interactions using DMS mapping. Indeed, we were able to approximate each of the three most predominant tRNA-protein/ribosome interactions to aaRS, EF1A, and the ribosome semi-quantitively by extracting DMS probing information from distinct sets of structural elements in tRNA, including using tRNA anticodon τ scores to relate with the known modes of aaRS substrate recognition and using tRNA D- and T-loop τ scores to support the model of C/G versus A/U-ending codon-tRNA interactions on the ribosome.
Our analysis of tRNA C75 τ scores supports the same EF1A-tRNA uniform binding paradigm originally identified from in vitro prokaryotic EF-Tu-tRNA studies. Given that the τ score reflects the average interaction strength of the tRNA with EF1A among a dynamic ensemble of in vivo tRNAs, our result suggests that the average interaction strength at the tRNA 3’ end also inversely correlates with the amino acid binding affinity and the tRNA body sequence binding affinity to EF1A. Since tRNAs are generally highly charged in vivo41 and C75 is located close to the charged amino acid, this result provides evidence for a similar EF1A-aa-tRNA interaction paradigm in human cells as previously described for E. coli EF-Tu-aa-tRNA.
To examine the tRNA structure and protein/ribosome-interaction dynamics in response to environmental changes, we applied arsenite treatment to induce oxidative stress in cells and compared the in vivo DM-DMS-MaPseq results with and without stress. Arsenite stress induces large-scale changes in the in vivo DMS footprints that can be interpreted in the context of severely reduced global translation activity and detailed translational reprogramming, such as the tRNAiMet DMS signal changes. Intriguingly, tRNAiMet exhibits a high in vivo signal of C71 (tRNA nomenclature; C70 in the tRNAiMet position, Fig. 1f) in the acceptor stem, which base pairs with G2 in its structure. This DMS signal is absent in vitro, suggesting that the C71-G2 base pair is weakened in vivo, potentially through its interaction with the eIF2B complex or the 48S scanning ribosome65. C71 forms the second base pair in the acceptor stem of tRNAiMet, so this result may implicate the melting of two base pairs in the eIF2B-tRNAiMet-40S or other higher-order complexes in vivo. tRNAiMet has an A1-U72 base pair instead of the most common G1-C72 base pair in elongator tRNAs, which may facilitate this melting at the end of the acceptor stem. An unexpected finding was that the modification status at position 37, the nucleotide immediately 3’ to the anticodon, determines the DMS sensitivity of tRNA towards stress. This result implicates a distinct modification-dependent translational response to arsenite stress for sub-groups of tRNA.
Human cells also contain 22 separate, mitochondrial-encoded tRNAs that are required for mitochondrial protein synthesis. Unlike chromosomal-encoded tRNAs, mt-tRNAs have A/U-rich sequences, and most do not have loop residues for forming canonical tertiary interactions. The human cytosolic tRNAs contain an average of 13 modifications, whereas the human mt-tRNAs contain an average of only 5–6 modifications83,84. This lower number of mt-tRNA modifications can also contribute to their lower stability. Our in vitro DMS mapping result shows that mitochondrial tRNAs do not fold into stable structures, but the in vivo DMS data show that they do fold into canonical structures in general. This result is consistent with mt-tRNA interacting very extensively with cellular proteins/mt-ribosomes in vivo. It is likely that the mt-tRNA only folds into a tRNA-like structure in vivo because of its intracellular interactions. We also find an inverse correlation between mt-tRNA and amino acid binding affinity to human EF-Tu, consistent with the binding paradigm originally identified from in vitro prokaryotic studies.
Arsenite stress induces large-scale changes in mt-tRNA DMS reaction profiles that include both stem and loop regions, consistent with mt-tRNA folding in vivo undergoing widespread change once mt-tRNA is no longer engaged in translation under arsenite stress, where they likely lose protein/mt-ribosome interaction. For the mt-tRNAs enriched in the polysome fraction under arsenite stress, their DMS reactivities are higher compared to the mt-tRNA depleted in the polysome fraction, pointing to an alternative mode of tRNA–ribosome interaction, in this case, perhaps in the possibly stalled mt-polysome. Human mt-tRNA abundance at the steady state is highly uneven. This abundance is mostly derived from differential degradation rates of individual mt-tRNAs, as mitochondrial transcription synthesizes equal amounts of 11 tRNAs in the plus strand and 8 mt-tRNAs in the minus strand22. How individual mt-tRNAs fold without protein/mt-ribosome interaction or upon altered mt-ribosome interaction likely contributes to their degradation rates.
In summary, we present the DM-DMS-MaPseq method to investigate the in vivo structure and interactions of tRNA. In addition to tRNAs, DM-DMS-MaPseq also captures reads for other abundant and highly structured noncoding RNAs, such as Y RNA and snRNAs in their cellular context. Similar analyses can be conducted to understand the structural dynamics of these RNAs and their associated RNPs. In the future, a simple add-on procedure would be to first fractionate different RNPs from DMS-treated cells before sequencing library construction. This would highly enrich the in vivo DMS footprints in specific cellular components such as the monosome, polysome, snRNPs, and larger snRNP complexes, which will facilitate more in-depth and accurate interpretation and reveal new biological insights about the cellular dynamics of these RNPs.
Methods
Cell culture and arsenite treatment
HEK 293T cells (ATCC, CRL-11268) were cultured with complete DMEM medium under standard conditions according to ATCC. Briefly, HEK 293T cells were grown in Hyclone DMEM medium (GE Healthcare Life Sciences, SH30022.01) with 10% FBS and 1% Pen–Strep (Penicillin–Streptomycin) to 80% confluency and passaged. Cells were collected, and total RNA was extracted using TRIzol (Thermo Fisher, 15596026) by following the manufacturer’s protocol when cells reached 80–90% confluency. Arsenite stress was induced by incubating cells in media containing 300 µM NaAsO2 at 37 °C for 2 h.
In vivo DMS treatment of HEK 293T cells
To a 10-cm dish of HEK 293T cells cultured in 15 ml media, 300 µL or 750 µL of DMS were added to reach a final concentration of 2% or 5%, respectively. After the addition of DMS, cells were incubated at 37 °C for 5 min. The media was then aspirated off, and the cells were washed with PBS twice and harvested for total RNA extraction with TRIzol (Thermo Fisher, 15596026) following the manufacturer’s protocol.
In vitro DMS treatment of HEK 293T total RNA
Total RNA was extracted from 80 to 90% confluent HEK 293T cells with TRIzol (Thermo Fisher, 15596026) following the manufacturer’s protocol. 4 µg of HEK 293T total RNA in 10 µL H2O was combined with 10 µL of the 2× refolding buffer (200 mM NaCl, 12 mM MgCl2, 20 mM Tris, pH 8.0) and incubated at 30 °C for 30 min. DMS was added to each sample to reach a final concentration of 5% at room temperature for 5 min. DMS treatment was stopped by adding an equal volume of 2× DMS quench buffer (60% β-mercaptoethanol, 0.6 M NaOAc, pH 5.5). DMS-treated total RNA samples were precipitated with ethanol.
Whole-cell translation assay by 35S pulse labeling
HEK 293T cells were cultured in a CO2 incubator at 37 °C with complete DMEM (Cytiva, SH30022.01) supplemented with 10% fetal bovine serum (Thermo, 16140071) and 1% Pen/Strep (Thermo, 15070063). For the whole-cell translation assay, 0.3 × 106 HEK 293T cells were seeded in each well of a six-well plate. Cells were incubated in a CO2 incubator at 37 °C for 2 days. Cells were treated with either sterile H2O or 300 µM NaAsO2 (Sigma, 1062771000) for 2 h in a CO2 incubator at 37 °C. Labeling medium without methionine was prepared by supplementing DMEM medium without methionine and cysteine (Thermo, 21013024) with 10% fetal bovine serum, 4 mM glutamine (25030081, Thermo), 0.063 g/L (0.2 mM) L-Cystine·2HCl (Sigma, C2526-100G). The labeling medium and normal complete medium were warmed in a cell incubator for >30 min. After NaAsO2 treatment, the culture medium was removed, and the cells were washed with warmed labeling media twice. 3 ml labeling medium was then added to each well, and cells were incubated in a CO2 incubator at 37 °C for 30 min. 62.5 µCi (1 mCi/16) 35S-labeled methionine (PerkinElmer, NEG709A001MC) was added into each well and incubated for 60 min. The labeling mixture was then removed from the plate and disposed of as radioactive waste. 3 ml of warmed normal medium was added to the plates and incubated for 10 min. The medium was then removed from the plate, and the cells were washed with ice-cold PBS three times. Cells were then harvested with cell lifter, and cell lysate was then extracted with CelLytic M lysis buffer (Sigma, C2978) supplemented with 1× protease inhibitor (Roche, 11873580001) and 25–250 U/ml Benzonase (Sigma, E1014) and rotate for 2 h or overnight at 4 °C. Equal amount of cell lysate from each sample was then loaded onto SDS-PAGE gel for protein separation. Gel was first stained with Coomassie blue to visualize the total protein loading amount. The gel was then dried using a gel dryer and exposed to a phosphorimaging plate for scanning and analysis.
tRNA sequencing with MSR-seq
Approximately 1 µg of in vivo or in vitro DMS treated-total RNA samples were used to build tRNA sequencing libraries, following a previously published MSR-seq protocol24. Briefly, RNAs were first deacylated by incubating at 37 °C in a 33 mM sodium tetraborate buffer for 30 min. Next, 5 µL of a PNK reaction stock (4 U/µL NEB T4 PNK, 40 mM MgCl2, 200 mM Tris-HCl, pH 6.8) was added to repair the 3’ end, and the sample was incubated at 37 °C for 20 min and then incubated at 65 °C for 10 min to inactivate the PNK enzyme. Following 3’ end repair, 30 µL of an RNA ligation reaction mix was added to the sample while still in the 3’ end repair mix (final concentration: 15% PEG 8000, 1× T4 RNA ligase I buffer, 50 µM ATP, 5% DMSO, 1 mM hexaamide cobalt (III) chloride, and 1 U/µL T4 RNA ligase I) and incubated overnight at 16 °C. This mix also included the barcoded RNA ligation linker/RT primer oligo at a 1.2:1 molar ratio to the input RNA. After ligating the RNA overnight, the sample was bound to streptavidin-coated MyOne C1 dynabeads (ThermoFisher) at room temperature on rotation for 15 min to facilitate the library construction process by minimizing sample loss and enabling rapid washes and buffer exchanges between reactions. “+DM” samples were treated according to the established demethylase protocol below. “+DM” and “−DM” samples were then dephosphorylated using a CIP reaction mix (0.2 U/µL CIP, 10 mM MgCl2, 0.5 mM ZnCl2, 20 mM HEPES, pH 7.5) for 30 min at 37 °C to liberate the 3’ end of the RT primer. Next, samples were resuspended in 25 µL of 1x SuperScript IV VILO mix and incubated at 55 °C for 10 min and then at 37 °C overnight. Next, samples were treated with an RNase H reaction mix (0.4 U/µL NEB RNaseH, 1× RNase H buffer) for 15 min at 37 °C. Subsequently, samples were treated with a solution of 50 mM sodium periodate in 15 mM sodium acetate, pH 5.0, for 30 min at room temperature to oxidize any non-extended RT primer. Following this, an overnight, room temperature cDNA ligation step was performed to enable PCR amplification of the cDNA (50 µL reaction; final concentrations: 2 U/µL T4 RNA ligase I, 25% PEG 8000, 7.5% DMSO, 50 µM ATP, 1 mM hexaamine cobalt (III) chloride, 2 mM barcoded cDNA ligation oligo, 1× T4 RNA ligase I buffer). Finally, the libraries were amplified by PCR with Illumina primers.
Sequencing was conducted using Illumina NovaX 6000, 100-bp paired-end.
AlkB demethylase treatment
Demethylase buffer conditions were modified from published protocols24,85. Three stock solutions were made fresh immediately before the reaction: l-ascorbic acid 200 mM, 2-ketoglutarate 3 mM, and ammonium iron sulfate 5 mM. The final reaction mixture contained 2 mM l-ascorbic acid, 1 mM 2-ketoglutarate, 0.3 mM ammonium iron sulfate, 100 mM KCl, 50 mM MES pH 6, 50 ng/µL BSA, 4 µM wild-type AlkB, and 4 µM AlkB-D135S (purified as described previously24). About 50 µL of the reaction mixture was added to 5–20 µL of decanted streptavidin bead slurry after ligation, immobilization, and washing. The reaction continued for 30 min at 37 °C. Following the reaction, beads were washed once with high salt wash buffer (1 M NaCl, 20 mM Tris-HCl pH 7.4) and once with low salt wash buffer (100 mM NaCl, 20 mM Tris-HCl pH 7.4).
Overexpression plasmids for recombinant E. coli AlkB and AlkB-D135S are available from Addgene (https://www.addgene.org/Tao_Pan/).
Data analysis
The data analysis followed the MSR-seq data processing pipeline. Libraries were sequenced from Illumina NovaSeq platforms as previously described. The resulting paired-end reads were demultiplexed by the identification of barcode sequences using Je demultiplex with the following parameters: BPOS = BOTH BM = READ_2 LEN = 6:4 FORCE = true C = false86. These options were optimized for samples where the barcode sequence is present on read 2. Barcode sequences were previously described24. Following demultiplexing, data was aligned using bowtie2 (version 2.3.3.1) with the following parameters: -q -p 10 --local --no-unal87. These reads were aligned to a curated human tRNA reference. This reference contained sequences of HG38 versions of 5S and 5.8S rRNA, snoRNA, Y tRNA, and tRNA genes that were curated for non-redundancy, with a tRNA-scan SE score >47, removing intron sequences, and 3’ “CCA” appended. The Bowtie2 output sam files were converted to bam files, which were then sorted by samtools’ sort function88. IGVtools count was used to collapse reads into 1nt windows using the following parameters: -z 5 -w 1 -e 250 —bases. The resulting IGV output wig files were reformatted using a custom Python script to obtain mutation rate and read coverage compatible with R for data visualization and analysis. To obtain canonical position information for all tRNA genes, the pair-end reads were processed by mim-tRNAseq28 with the following parameters: --species Hsap --cluster-id 0.95 --threads 4 --min-cov 0.0005 --max-mismatches 0.1 --control-condition --no-cca-analysis -n hek_mods_correl. The resulting mismatch table file contained deconvoluted tRNA gene sequences with canonical tRNA position information. For tRNA genes that could not be deconvoluted, canonical positioning was done by hand following tRNA nomenclature guidelines as described in Sprinzel et al. 89. For data visualization, a ≥ 600 read coverage filter was used to ensure only positions with sufficient coverage. Following this, the mutation rates were used for analysis as described below.
Formulas
1.
2.
3. In arsenite stress conditions:
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. The sequencing data generated for this study have been deposited to NCBI GEO accession number GSE262888. The sequencing data used for polysome unstressed can be retrieved from NCBI GEO accession number GSE198441.
Code availability
The MSR-seq analysis code, including mutation rate analysis with and without demethylase treatment, is available on GitHub (https://github.com/ckatanski/CHRIS-seq).
References
Winkler, W., Nahvi, A. & Breaker, R. R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002).
Nahvi, A. et al. Genetic control by a metabolite binding mRNA. Chem. Biol. 9, 1043–1049 (2002).
Mironov, A. S. et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756 (2002).
Jinek, M. et al. A programmable dual–RNA–guided DNA endonuclease in adaptive bacterial immunity. Science337, 816–821 (2012).
Pelletier, J. & Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325 (1988).
Langdon, E. M. et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science360, 922–927 (2018).
Kertesz, M. et al. Genome-wide measurement of RNA secondary structure in yeast. Nature 467, 103–107 (2010).
Ding, Y. et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700 (2014).
Rouskin, S., Zubradt, M., Washietl, S., Kellis, M. & Weissman, J. S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705 (2014).
Talkish, J., May, G., Lin, Y., Woolford, J. L. & McManus, C. J. Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA 20, 713–720 (2014).
Spitale, R. C. et al. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 9, 18–20 (2013).
Lempereur, L. et al. Conformation of yeast 18S rRNA. Direct chemical probing of the 5′ domain in ribosomal subunits and in deproteinized RNA by reverse transcriptase mapping of dimethyl sulfate-accessible sites. Nucleic Acids Res. 13, 8339–8357 (1985).
Swiatkowska, A. et al. Kinetic analysis of pre-ribosome structure in vivo. RNA 18, 2187–2200 (2012).
Kubota, M., Tran, C. & Spitale, R. C. Progress and challenges for chemical probing of RNA structure inside living cells. Nat. Chem. Biol. 11, 933–941 (2015).
Zubradt, M. et al. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 14, 75–82 (2016).
Climie, S. C. & Friesen, J. D. In vivo and in vitro structural analysis of the rplJ mRNA leader of Escherichia coli. protection by bound L10-L7/L12. J. Biol. Chem. 263, 15166–15175 (1988).
Spitale, R. C. et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490 (2015).
Smola, M. J., Calabrese, J. M. & Weeks, K. M. Detection of RNA–protein interactions in living cells with SHAPE. Biochemistry 54, 6867–6875 (2015).
Luo, Q. J. et al. RNA structure probing reveals the structural basis of Dicer binding and cleavage. Nat. Commun. 12, 3397 (2021).
Yamagami, R., Sieg, J. P., Assmann, S. M. & Bevilacqua, P. C. Genome-wide analysis of the in vivo tRNA structurome reveals RNA structural and modification dynamics under heat stress. Proc. Natl. Acad. Sci. USA 119, e2201237119 (2022).
Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016).
Suzuki, T., Nagao, A. & Suzuki, T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 45, 299–329 (2011).
Moran, J. C. et al. The human mitochondrial mRNA structurome reveals mechanisms of gene expression. Science 385, eadm9238 (2024).
Watkins, C. P., Zhang, W., Wylder, A. C., Katanski, C. D. & Pan, T. A multiplex platform for small RNA sequencing elucidates multifaceted tRNA stress response and translational regulation. Nat. Commun. 13, 2491 (2022).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Cozen, A. E. et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015).
Pinkard, O., McFarland, S., Sweet, T. & Coller, J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 11, 4104 (2020).
Behrens, A., Rodschinka, G. & Nedialkova, D. D. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol. Cell 81, 1802–1815.e7 (2021).
Fedeles, B. I., Singh, V., Delaney, J. C., Li, D. & Essigmann, J. M. The AlkB family of Fe(II)/α-Ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond *. J. Biol. Chem. 290, 20734–20742 (2015).
Clark, W. C., Evans, M. E., Dominissini, D., Zheng, G. & Pan, T. tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22, 1771–1784 (2016).
Weisser, M. et al. Structural and functional insights into human re-initiation complexes. Mol. Cell 67, 447–456.e7 (2017).
Adomavicius, T. et al. The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 10, 2136 (2019).
Chan, P. P., Lin, B. Y., Mak, A. J. & Lowe, T. M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49, 9077–9096 (2021).
Beaudoin, J.-D. et al. Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. Nat. Struct. Mol. Biol. 25, 677–686 (2018).
Mustoe, A. M. et al. Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell 173, 181–195.e18 (2018).
Tomezsko, P., Swaminathan, H. & Rouskin, S. Viral RNA structure analysis using DMS-MaPseq. Methods 183, 68–75 (2020).
Tijerina, P., Mohr, S. & Russell, R. DMS footprinting of structured RNAs and RNA–protein complexes. Nat. Protoc. 2, 2608–2623 (2007).
Dittmar, K. A., Sørensen, M. A., Elf, J., Ehrenberg, M. & Pan, T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 6, 151–157 (2005). 157.
Jakubowski, H. & Goldman, E. Quantities of individual aminoacyl-tRNA families and their turnover in Escherichia coli. J. Bacteriol. 158, 769–776 (1984).
Park, S. G., Schimmel, P. & Kim, S. Aminoacyl tRNA synthetases and their connections to disease. Proc. Natl. Acad. Sci. USA 105, 11043–11049 (2008).
Evans, M. E., Clark, W. C., Zheng, G. & Pan, T. Determination of tRNA aminoacylation levels by high-throughput sequencing. Nucleic Acids Res. 45, e133–e133 (2017).
Beck, M. et al. The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549 (2011).
Su, Z. et al. Genome-wide RNA structurome reprogramming by acute heat shock globally regulates mRNA abundance. Proc. Natl. Acad. Sci. USA 115, 12170–12175 (2018).
Mitchell, D. III, Cotter, J., Saleem, I. & Mustoe, A. M. Mutation signature filtering enables high-fidelity RNA structure probing at all four nucleobases with DMS. Nucleic Acids Res. 51, 8744–8757 (2023).
Giegé, R. & Eriani, G. The tRNA identity landscape for aminoacylation and beyond. Nucleic Acids Res. 51, 1528–1570 (2023).
Gomez, M. A. R. & Ibba, M. Aminoacyl-tRNA synthetases. RNA 26, 910–936 (2020).
Tamaki, S., Tomita, M., Suzuki, H. & Kanai, A. Systematic analysis of the binding surfaces between tRNAs and their respective aminoacyl tRNA synthetase based on structural and evolutionary data. Front. Genet. 8, 227 (2018).
Lenhard, B., Orellana, O., Ibba, M. & Weygand-Durasevic, I. tRNA recognition and evolution of determinants in seryl-tRNA synthesis. Nucleic Acids Res. 27, 721–729 (1999).
Shimada, N., Suzuki, T. & Watanabe, K. Dual mode recognition of two isoacceptor tRNAs by mammalian mitochondrial Seryl-tRNA synthetase *. J. Biol. Chem. 276, 46770–46778 (2001).
Chong, Y. E. et al. Distinct ways of G:U recognition by conserved tRNA binding motifs. Proc. Natl Acad. Sci. 115, 7527–7532 (2018).
Zeng, Q.-Y. et al. The G3-U70-independent tRNA recognition by human mitochondrial alanyl-tRNA synthetase. Nucleic Acids Res 47, 3072–3085 (2019).
Achsel, T. & Gross, H. J. Identity determinants of human tRNA(Ser): sequence elements necessary for serylation and maturation of a tRNA with a long extra arm. EMBO J. 12, 3333–3338 (1993).
Breitschopf, K., Achsel, T., Busch, K. & Gross, H. J. Identity elements of human tRNALeu: structural requirements for converting human tRNASer into a leucine acceptor in vitro. Nucleic Acids Res. 23, 3633–3637 (1995).
Holm, M. et al. mRNA decoding in human is kinetically and structurally distinct from bacteria. Nature 617, 200–207 (2023).
Asahara, H. & Uhlenbeck, O. C. The tRNA specificity of Thermus thermophilus EF-Tu. Proc. Natl. Acad. Sci. USA 99, 3499–3504 (2002).
Asahara, H. & Uhlenbeck, O. C. Predicting the binding affinities of misacylated tRNAs for Thermus thermophilus EF-Tu·GTP. Biochemistry 44, 11254–11261 (2005).
LaRiviere, F. J., Wolfson, A. D. & Uhlenbeck, O. C. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294, 165–168 (2001).
Schrader, J. M., Chapman, S. J. & Uhlenbeck, O. C. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc. Natl. Acad. Sci. USA 108, 5215–5220 (2011).
Zhang, J. & Ferré-D’amaré, A. R. The tRNA elbow in structure, recognition and evolution. Life 6, 3 (2016).
Benisty, H. et al. Genes enriched in A/T-ending codons are co-regulated and conserved across mammals. Cell Syst. 14, 312–323.e3 (2023).
Rafels-Ybern, À, Torres, A. G., Grau-Bove, X., Ruiz-Trillo, I. & Ribas de Pouplana, L. Codon adaptation to tRNAs with Inosine modification at position 34 is widespread among Eukaryotes and present in two Bacterial phyla. RNA Biol. 15, 500–507 (2018).
Endres, L., Dedon, P. C. & Begley, T. J. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 12, 603–614 (2015).
Liu, B. & Qian, S.-B. Translational reprogramming in cellular stress response. Wiley Interdiscip. Rev. RNA 5, 301–305 (2014).
McEwen, E. et al. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280, 16925–16933 (2005).
Wang, J. et al. Rapid 40S scanning and its regulation by mRNA structure during eukaryotic translation initiation. Cell 185, 4474–4487.e17 (2022).
Cappannini, A., et al. MODOMICS: a database of RNA modifications and related information. Nucleic Acids Res. 52, D239–D244 (2024).
Konevega, A. L. et al. Purine bases at position 37 of tRNA stabilize codon–anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 10, 90–101 (2004).
Khatter, H., Myasnikov, A. G., Natchiar, S. K. & Klaholz, B. P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015).
Krahn, N., Fischer, J. T. & Söll, D. Naturally Occurring tRNAs With Non-canonical Structures. Front. Microbiol. 11, 596914 (2020).
Yokogawa, T., Kumazawa, Y., Miura, K.-I. & Watanabe, K. Purification and characterization of two serine isoacceptor tRNAs from bovine mitochondria by using a hybridization assay method. Nucleic Acids Res. 17, 2623–38 (1989).
Messmer, M. et al. Tertiary network in mammalian mitochondrial tRNAAsp revealed by solution probing and phylogeny. Nucleic Acids Res. 37, 6881–6895 (2009).
Wakita, K. et al. Higher-order structure of bovine mitochondrial tRNA Phe lacking the ‘conserved’ GG and TΨCG sequences as inferred by enzymatic and chemical probing. Nucleic Acids Res. 22, 347–353 (1994).
Ueda, T., Ohta, T. & Watanabe, K. Large scale isolation and some properties of AGY-specific serine tRNA from bovine heart mitochondria1. J. Biochem. 98, 1275–1284 (1985).
Helm, M. & Attardi, G. Nuclear control of cloverleaf structure of human mitochondrial tRNALys. J. Mol. Biol. 337, 545–560 (2004).
Brown, A. et al. Structure of the large ribosomal subunit from human mitochondria. Science 346, 718–722 (2014).
Chrzanowska-Lightowlers, Z., Rorbach, J. & Minczuk, M. Human mitochondrial ribosomes can switch structural tRNAs – but when and why?. RNA Biol. 14, 1668–1671 (2017).
Haag, S. et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 35, 2104–2119 (2016)
Huber, S. M. et al. Arsenite toxicity is regulated by queuine availability and oxidation-induced reprogramming of the human tRNA epitranscriptome. Proc. Natl. Acad. Sci. USA 119, e2123529119 (2022).
Fritsch, R. M., Schneider, G., Saur, D., Scheibel, M. & Schmid, R. M. Translational repression of MCL-1 couples stress-induced eIF2α phosphorylation to mitochondrial apoptosis initiation. J. Biol. Chem. 282, 22551–22562 (2007).
Eiermann, N., Stoecklin, G. & Jovanovic, B. Mitochondrial inhibition by sodium azide induces assembly of eIF2α phosphorylation-independent stress granules in mammalian cells. Int. J. Mol. Sci. 23, 5600 (2022).
Rashad, S., Tominaga, T. & Niizuma, K. The cell and stress-specific canonical and noncanonical tRNA cleavage. J. Cell Physiol. 236, 3710–3724 (2021).
Jia, W. & Higgs, P. G. Codon usage in mitochondrial genomes: distinguishing context-dependent mutation from translational selection. Mol. Biol. Evol. 25, 339–351 (2008).
Pan, T. Modifications and functional genomics of human transfer RNA. Cell Res. 28, 395–404 (2018).
Suzuki, T. et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 11, 4269 (2020).
Li, M. et al. DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 25, 1047–1058 (2018).
Girardot, C., Scholtalbers, J., Sauer, S., Su, S.-Y. & Furlong, E. E. M. Je, a versatile suite to handle multiplexed NGS libraries with unique molecular identifiers. BMC Bioinform. 17, 419 (2016).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Sprinzl, M., Hartmann, T., Meissner, F., Moll, J. & Vorderwülbecke, T. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 15, r53–r188 (1987).
Wells, J. N. et al. Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes. PLoS Biol. 18, e3000780 (2020).
Li, J. et al. Structural basis of regulated m7G tRNA modification by METTL1–WDR4. Nature 613, 391–397 (2023).
Itoh, Y. et al. Mechanism of membrane-tethered mitochondrial protein synthesis. Science 371, 846–849 (2021).
Acknowledgments
This work was supported by grants from NIH RM1HG008935 and NCI R33CA272357 (to T.P.), and from 5T32AI007647 (to N.P.).
Author information
Authors and Affiliations
Contributions
N.P., C.P.W. and T.P. conceived the project. N.P., Y.H. and T.P. analyzed and interpreted data, with help from S.H. C.D.K. and C.P.W. performed the DMS experiments and built sequencing libraries, with help from W.Z. Y.H., N.P., C.P.W. and T.P. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peña, N., Hou, Y., Watkins, C.P. et al. In vivo structure profiling reveals human cytosolic and mitochondrial tRNA structurome and interactome in response to stress. Nat Commun 16, 5041 (2025). https://doi.org/10.1038/s41467-025-59435-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59435-5
This article is cited by
-
A model for propagation of RNA structural memory through biomolecular condensates
Nature Cell Biology (2025)